Summary
Viral infections in early pregnancy are a major cause of microcephaly. However, how distinct viruses impair human brain development remains poorly understood. Here we use human brain organoids to study the mechanisms underlying microcephaly caused by Zika Virus (ZIKV) and Herpes Simplex Virus (HSV-1). We find that both viruses efficiently replicate in brain organoids and attenuate their growth by causing cell death. However, transcriptional profiling reveals that ZIKV and HSV-1 elicit distinct cellular responses and HSV-1 uniquely impairs neuroepithelial identity. Furthermore, we demonstrate that while both viruses fail to potently induce the type I interferon system, the organoid defects caused by their infection can be rescued by distinct type I interferons. These phenotypes are not seen in 2D cultures, highlighting the superiority of brain organoids in modeling viral infections. Together, these results uncover virus-specific mechanisms and complex cellular immune defenses associated with virus-induced microcephaly.
Introduction
Viral infections during pregnancy are a major cause of fetal brain malformations. The transmission of infectious agents from mother to fetus can be devastating for fetal brain development (Bale and Murph, 1992; Brasil et al., 2016; Marquez et al., 2011). Infections account for up to 50% of cases of congenital microcephaly (Herber et al., 2019; Mlakar et al., 2016) and those occurring in the first trimester of pregnancy are typically associated with more severe outcomes (Coyne and Lazear, 2016). Despite these overlapping phenotypes, congenital infections are caused by distinct pathogens collectively referred to as TORCH and include Toxoplasma gondii, Other, Rubella, Human Cytomegalovirus (HCMV), Herpes Simplex Viruses 1 and 2 (HSV-1 and HSV-2), and Zika virus (ZIKV) (Schwartz, 2017). HCMV and HSV are the more common causes of newborn morbidity worldwide (Looker et al., 2017; Marsico and Kimberlin, 2017) and the newly emerged ZIKV remains a threat for pregnant women (“Zika: the continuing threat.,” 2019).
Yet, whether TORCH pathogens alter fetal brain development via similar mechanisms is unclear. Mechanistic understanding has been hampered by the lack of accurate models for individual TORCH infections. Although rodents can model in part microcephaly caused by ZIKV and CMV (C. Li et al., 2016; R. Y. Li and Tsutsui, 2000), they are not the natural hosts and do not faithfully recapitulate TORCH neuropathology (Cheeran et al., 2009; Ming et al., 2016). Human brain organoid systems derived from human pluripotent stem cell (hPSC) are emerging as human three-dimensional (3D) culture platforms for the study of viral infections and their impact on human neurodevelopment (J. Kim et al., 2019). Brain organoids recapitulate the cell composition and the 3D environment of the embryonic human brain (Di Lullo and Kriegstein, 2017; Lancaster et al., 2013), thus overcoming a major limitation of human two-dimensional (2D) culture systems (Baker and Chen, 2012; Duval et al., 2017). Organoid models are widely used to recapitulate the structural defects, cell depletion and molecular signatures associated with ZIKV-induced microcephaly (Cugola et al., 2016; Gabriel et al., 2017; Garcez et al., 2016; Qian et al., 2016; M. Watanabe et al., 2017) as well as to model part of the neuropathological defects caused by HCMV and HSV-1 (Brown et al., 2019; Qiao et al., 2020; Sison et al., 2019; Sun et al., 2020). However, a comparative analysis of organoid models of TORCH-induced microcephaly is lacking.
One potential common underlying mechanisms of TORCH-induced microcephaly is the activation of innate immune response and its detrimental effects on fetal brain development (Gottfried et al., 2015; Y. Watanabe et al., 2010). In particular, the activation of cytokines called type I interferons (IFN-I), which include several α species (IFNα) and one β species (IFNβ), upon infection is required to induce an array of antiviral effectors referred to as interferon-stimulated genes (ISGs) that can restrict viral spreading but also promote cell death (Schneider et al., 2014). Several studies using human 2D and 3D neural cultures have reported the upregulation of IFN-I, ISGs and immune signatures in response to ZIKV infection (Dang et al., 2016; Ferraris et al., 2019; Hanners et al., 2016; C. Li et al., 2017; Lima et al., 2019; Liu et al., 2019; Simonin et al., 2016; M. Watanabe et al., 2017; Zhang et al., 2016). However, the magnitude of this response varies substantially across studies. Furthermore, the function of this response in the pathogenesis of ZIKV infection remains debated, as evidence supporting both a neuroprotective function of IFN-I (C. Li et al., 2017; Lin et al., 2019) and a detrimental neuroinflammatory effect (Dang et al., 2016; Liu et al., 2019) exist. While these discrepancies are in part due to changes in cell differentiation stages and viral strains (Ferraris et al., 2019; Simonin et al., 2016), whether differences between 2D vs 3D culture systems and/or changes in cell type composition contribute to the magnitude and the function of the innate immune response remains unclear. Finally, little is known about the activation of the antiviral response in other human models of TORCH-induced microcephaly and thus a unifying view of the role of the innate immune response is missing.
Here we used human brain organoids to reproduce the microcephaly-like phenotype caused by multiple TORCH viruses, including ZIKV, HCMV and HSV-1, which differ in their viral genome structure, size and mode of replication (Table S1). We show that these organoid models exhibit major differences in their structure, transcriptional profiles, the engagement of the IFN-I system and sensitivity to IFN-I. These results argue for the existence of unique pathogenic mechanisms and a neuroprotective role of IFN-I responses underlying virus-induced microcephaly.
Results
ZIKV and HSV-1 infections impair the growth of early-stage brain organoids
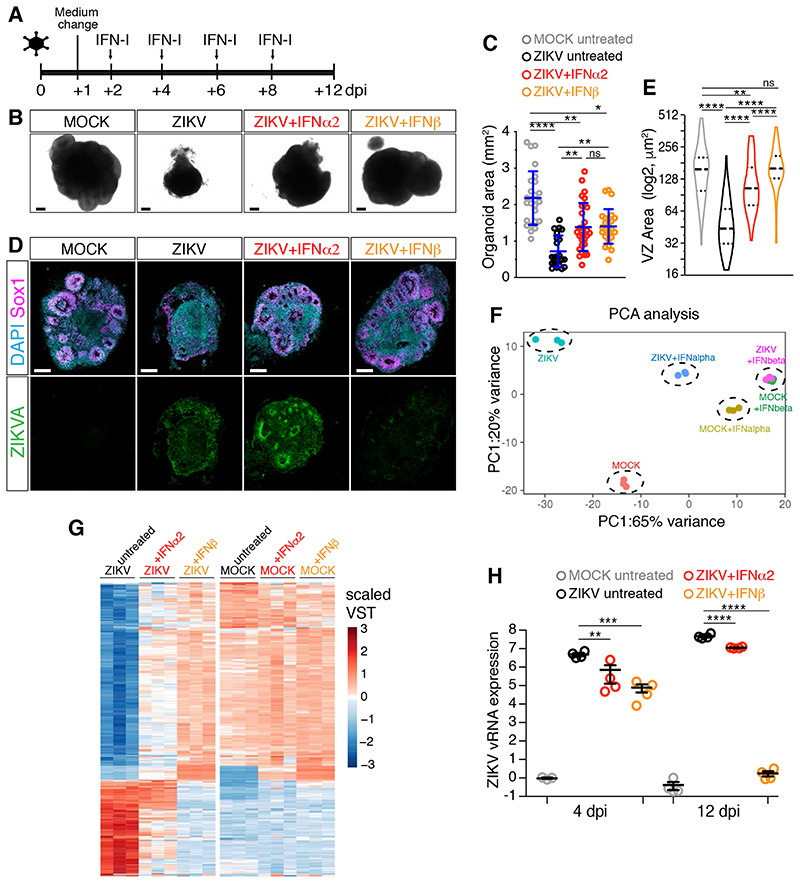
To compare the effects of distinct TORCH viruses on the early stages of human brain development, we generated brain organoids using our previously established protocol (Lancaster et al., 2013) and exposed them to a virus inoculum for 24 hours. We focused on day 10 organoids, an early organoid stage that expresses the neuroectodermal markers Nestin and Pax6 (Figure S1A–B) and is highly susceptible to viral infection (Dang et al., 2016; Gabriel et al., 2017).
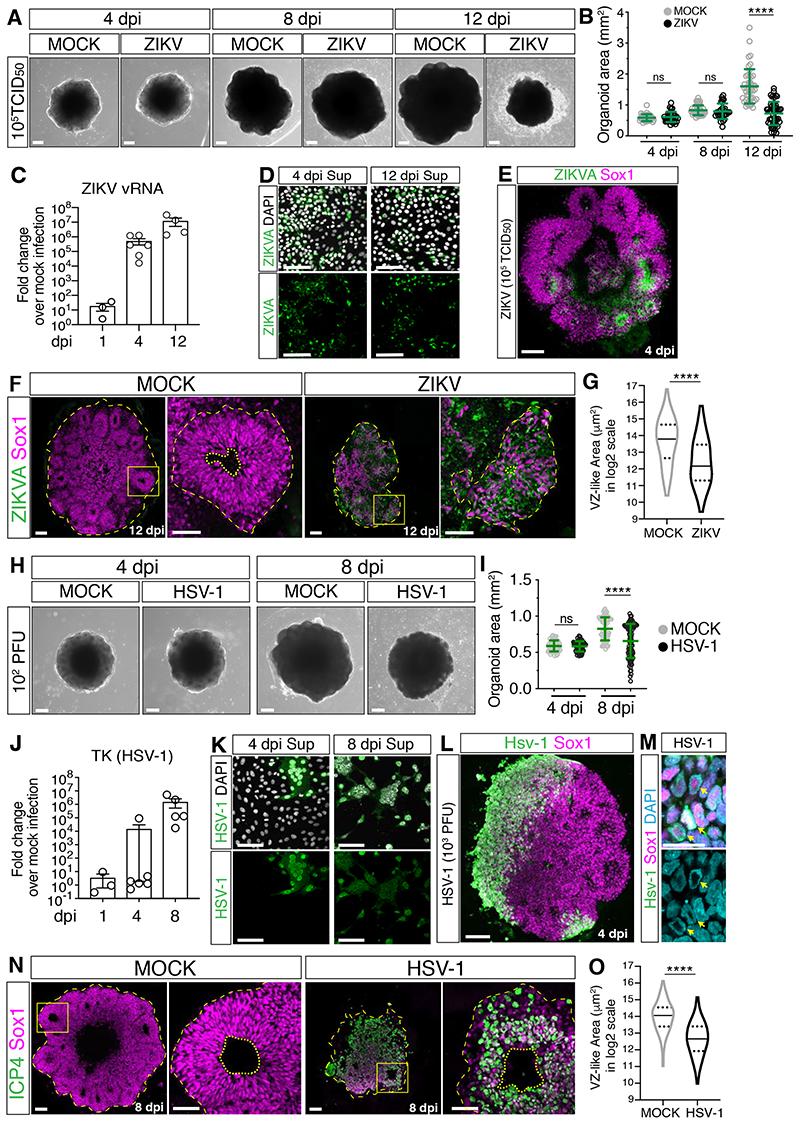
We first exposed brain organoids to ZIKV and after inoculum removal, organoids were examined at 4, 8 and 12 days post-infection (dpi). While MOCK-treated organoids grew over time, ZIKV-exposed organoids exhibited a time-dependent growth attenuation and were significantly smaller at 12 dpi (Figure 1A–B, Table S2). Concomitantly, we observed increasing expression of ZIKV viral RNA (vRNA) and the release of infectious ZIKV particles at 4 dpi and 12 dpi (Figure 1C–D), indicating a productive infection. Staining for the Zika virus antigen (ZIKVA) and the human neural progenitor cell (hNPC) marker Sox1 revealed foci of infection at 4 dpi (Figure 1E), confirming hNPCs as the main cellular targets of ZIKV infection. As a microcephaly phenotype can be described at the tissue level by a cell depletion phenotype, we analyzed hNPC abundance by measuring the area of ventricular zone (VZ)-like structures where hNPCs reside. At 12 dpi, while MOCK-treated organoids showed the typical organization of VZ-like structures, ZIKV- infected organoids contained fewer and smaller VZ-like regions (Figure 1F–G). Furthermore, analysis of apoptosis at 12 dpi by staining for the apoptotic marker cleaved caspase 3 (CC3) showed an increase in ZIKV-infected cultures, both in ZIKVA-positive and negative cells (Figure S1C–E). As hNPC depletion can result in lumen size changes, we also measured the lumen area and found it reduced in ZIKV-infected organoids (Figure S1F). These findings are in line with the ability of ZIKV to attenuate organoid growth and cause hNPC depletion (Cugola et al., 2016; Gabriel et al., 2017; Garcez et al., 2016; Qian et al., 2016; M. Watanabe et al., 2017) and thus validate our infection paradigm for the study of virus-induced microcephaly.
Figure 1. ZIKV and HSV-1 infections impair organoid growth.
A-B) Images (scale bars 200 μm) and area measurements of organoids exposed to ZIKV or MOCK-treated. Values are mean ± SD and represent individual organoids (p=0.7886 4 dpi; p=0.3190 8 dpi; **** is p<0.0001; Mann-Whitney test). TCID50, mean tissue culture infectious dose.
C) RT-qPCR analysis of ZIKV viral RNA (vRNA) in organoids exposed to ZIKV. Values are mean ± SEM (p=0.1 1 dpi; p=0.0022 4 dpi; p=0.0286 12 dpi; Mann-Whitney test over age-matched MOCK-treated).
D) Immunostaining of Vero cells (scale bars 100 μm) incubated with supernatants (sup) of ZIKV- infected organoids. ZIKVA, ZIKA Virus Antigen.
E-G) Immunostaining (scale bars are 100 μm) of organoids exposed to ZIKV or MOCK-treated. Dashed lines indicate organoid contour based on DAPI signal (not shown). Insets in F (scale bars 50 μm) show a magnified view of the ventricular zone (VZ)-like structures (dashed lines) and their lumens (dotted lines). Violin plots indicate median and quartiles (n=129 regions from 11 MOCK, n=109 from 13 ZIKV organoids from 3 experiments; **** is p<0.0001; Mann-Whitney test).
H-I) Images (scale bars 200 μm) and area measurements of organoids exposed to HSV-1 (102 PFU) or MOCK-treated. Values are mean ± SD and represent individual organoids (p=0.6625 4 dpi; **** is p<0.0001; Mann-Whitney test).
J) RT-qPCR analysis of HSV-1 gene Thymidine Kinase (TK) in organoids exposed to HSV-1 (102 PFU). Values are mean ± SEM (p>0.9999 1 dpi; p=0.4004 4 dpi; p=0.0079 8 dpi; Mann-Whitney test over age-matched MOCK-treated).
K) Immunostaining of Vero cells (scale bars 100 μm) incubated with supernatants (sup) from HSV- 1-infected organoids (102 PFU).
L-M) Immunostaining (scale bars 100 μm in L, 20 μm in M) of organoids exposed to HSV-1 (103 PFU) and analyzed at 4 dpi. Arrows indicate chromatin localization (marked by DAPI) at the nuclear periphery.
N-O) Immunostaining (scale bars 100 μm) of organoids exposed to HSV-1 (102 PFU) or MOCK- treated. Dashed lines indicate organoid contour based on DAPI signal (not shown). Insets (scale bars 50 μm) show a magnified view of ventricular zone (VZ)-like structures (dashed lines) around lumens (dotted lines). Inset from HSV-1 samples underwent a 180° rotation. Violin plots indicate median and quartiles (n=146 regions from 15 MOCK, n=69 from 16 HSV organoids from 3 experiments; **** is p<0.0001, Mann-Whitney test). ICP4, infected cell polypeptide 4 protein of HSV-1.
ns, non-significant; dpi, days post-infection. See also Figure S1, Table S1 and S2.
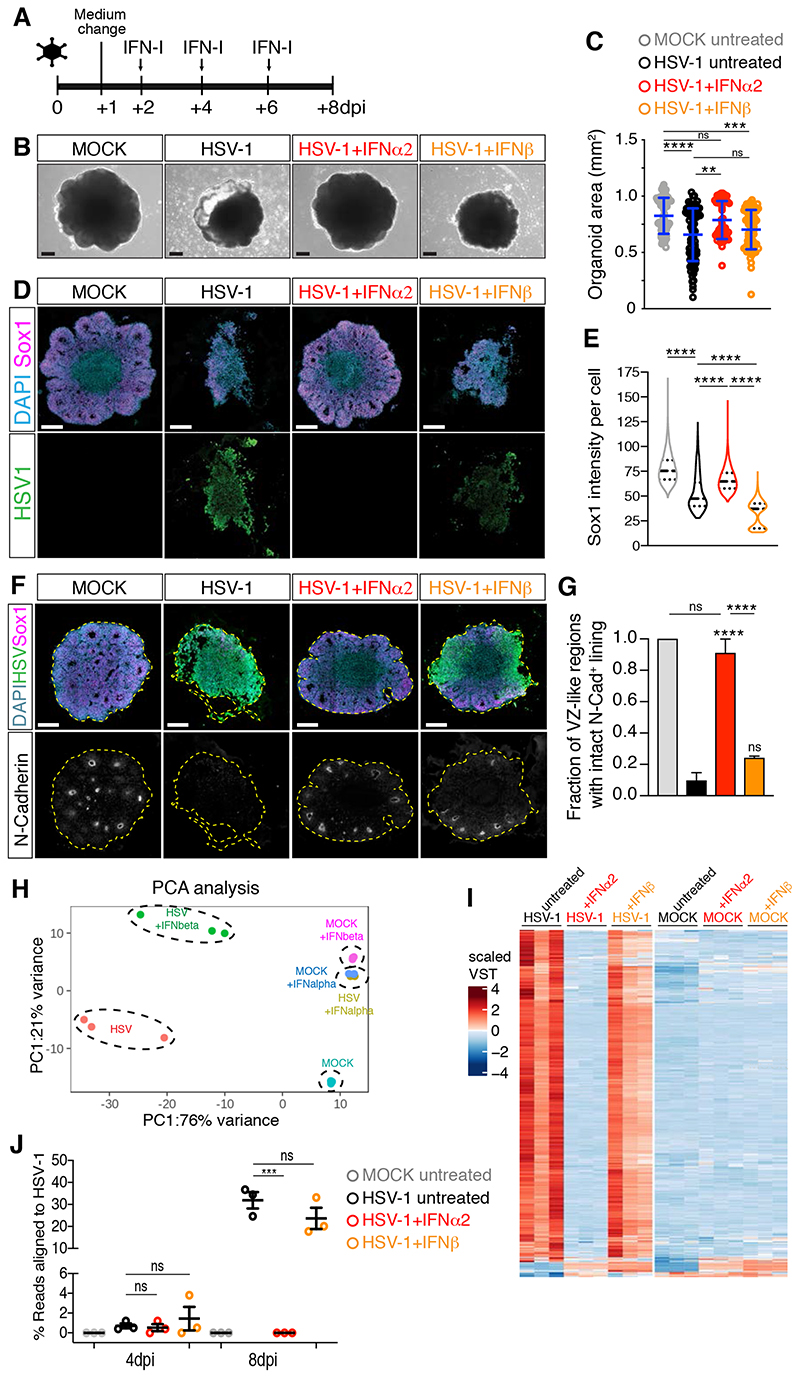
To study the effects of other TORCH viruses, we exposed organoids to HCMV virus expressing the mNeonGreen fluorescent reporter (HCMV-mNG, (Kasmapour et al., 2018) and to another Herpesvirus, HSV-1. We observed mNeonGreen fluorescence in HCMV-exposed organoids at 4 dpi and in a minority of Sox1+ cells after immunostaining (Figure S1G–I). However, HCMV infection remained limited to small clusters of cells at 12 dpi and did not attenuate organoid growth (Figure S1G–K). These observations indicate that HCMV does not replicate efficiently and does not cause a microcephaly-like phenotype in early-stage organoids. In contrast to this, organoids exposed to HSV-1 disintegrated at 8 dpi (Figure S1L–M), uncovering the destructive nature of HSV-1 infection. Organoids exposed to a lower dose of HSV-1 still showed impaired growth at 8 dpi (Figure 1H–I, Table S2) but preserved tissue integrity to a certain extent. We observed productive HSV-1 infection at this lower dose, as evidenced by time-dependent expression of the HSV-1 thymidine kinase (TK) gene and the production of infectious HSV-1 particles (Figure 1J–K). As TK expression at 4 dpi was low and variable, HSV-1 tropism at 4 dpi was analyzed in organoids exposed to the higher HSV-1 dose. This revealed infection of Sox1 + hNPCs and the typical chromatin marginalization to the nuclear periphery caused by HSV-1 replication (Aho et al., 2017) (Figure 1L–M). Immunostaining of tissues infected with low HSV-1 dose revealed a disrupted cytoarchitecture characterized by fewer and smaller VZ-like regions at 8 dpi (Figure 1N–O). Moreover, analysis of the fraction of apoptotic cells showed an increase in HSV-1-infected cultures at 8 dpi compared to MOCK-treated organoids and their accumulation in the lumen (Figure S1N–O). Unlike ZIKV, lumens of HSV-1-infected organoids appeared enlarged (Figure S1P), likely as a result of accumulation of apoptotic cells. Most apoptotic cells (92%) were negative for the expression of the immediate-early viral protein ICP4 (Figure S1Q–R), suggesting induction of apoptosis in bystander cells and/or abortively infected cells (Drayman et al., 2019). Together, these results indicate that ZIKV and HSV-1, but not HCMV, efficiently infect early-stage brain organoids and reduce their growth, thus mimicking ZIKV- and HSV-1-associated microcephaly.
ZIKV and HSV-1 infections elicit different transcriptional signatures
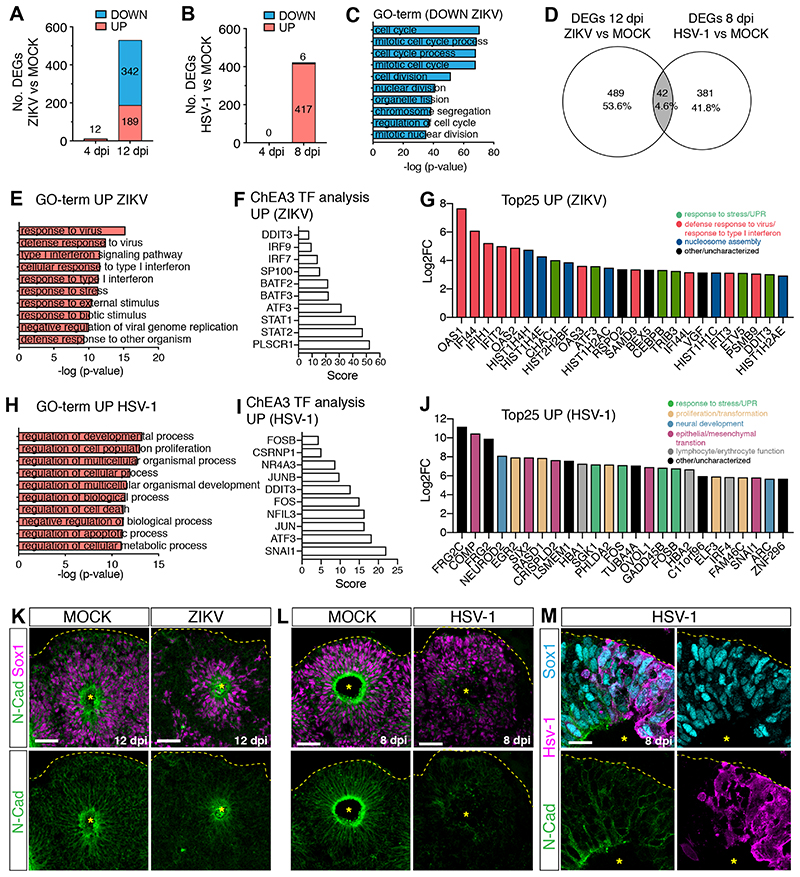
To investigate the molecular signatures underlying the different organoid growth defects, we analyzed the transcriptional profiles of ZIKV- and HSV-1-infected cultures by RNA-sequencing. We confirmed high expression of neural progenitor genes in these datasets (Figure S2A) and focused on the transcriptional changes at the late time points, as very few genes were deregulated at 4 dpi (Figure 2A–B), an early infection stage that is not associated with major structural defects (Figure 1). We identified 531 and 423 differentially expressed genes (DEGs) in ZIKV- and HSV-1-infected cultures in respect of their MOCK counterparts. Many DEGs in ZIKV- infected cultures were downregulated and involved in cell cycle and cell division (Figure 2C), while almost all DEGs found in HSV-1-infected cultures were upregulated (Figure 2B). Overlap between the two datasets was limited, since only 4.6% of DEGs were shared (Figure 2D).
Figure 2. ZIKV and HSV-1 infections elicit distinct transcriptional responses.
A-B) Graphs showing the number of differentially expressed genes (DEGs) in virus-infected organoids.
C) Top 10 Gene Ontology (GO)-terms of downregulated genes (log2FoldChange <1) in ZIKV vs. MOCK-exposed organoids (12 dpi).
D) Venn diagram showing limited overlap between the two datasets.
E and H) Top 10 GO-terms of upregulated genes (log2FoldChange >2) in virus-infected organoids (12 dpi in E, 8 dpi in H).
F and I) Top 10 results of ChEA3 transcription factor (TF) analysis performed on upregulated genes.
G and J) Expression of the top 25 upregulated genes in ZIKV-infected organoids (12 dpi, in G) or in HSV-1-infected organoids (8 dpi, in J). FC, fold change.
K-M) Immunostaining (scale bars 50 μm in K-L, 20 μm in M) of infected and MOCK-treated organoids. Dashed lines and asterisks indicate organoid surface based on DAPI signal (not shown) and lumen respectively.
GO-term analysis on genes upregulated in ZIKV-infected cultures revealed enrichment for antiviral defense pathways, response to interferon (IFN), stress or stimuli (Figure 2E). Furthermore, promoters of upregulated genes were enriched in binding sites for transcription factors involved in immune responses (IRF7/9 and STAT1/2) and in the unfolded protein response (UPR, DDIT3 and ATF3) (Figure 2F). Genes implicated in antiviral defense (Schneider et al., 2014) and UPR were the most upregulated at 12 dpi (Figure 2G, Table S3) and some were found upregulated already at 4 dpi (Figure S2B), confirming the activation of immune and stress-related signatures in ZIKV-infected cultures. In contrast, GO-term analysis on HSV-1-infected signatures revealed enrichment in the regulation of developmental and cellular processes, as well as cell death-related pathways (Figure 2H). Promoters of HSV-1-upregulated genes were enriched in binding sites of key regulators of proliferation, differentiation, apoptosis and transformation, including members of the transcription factor complex AP-1 (FOS, FOSB, JUN and JUNB) (Hess et al., 2004), NR4A3 and NFIL3 (Herring et al., 2019; Keniry et al., 2014), the neural crest regulator CSRNP1 (Simões-Costa and Bronner, 2015), the epithelial-mesenchymal transition (EMT) inducer SNAI1 (Carver et al., 2001) and stress response genes (DDIT3 and ATF3) (Figure 2I). Consistent with this, regulators of cell proliferation, apoptosis and stress response were upregulated in HSV-1-infected cultures (Figure 2J, Table S3). The most upregulated genes also included genes implicated in neural, neural crest and mesenchymal development and key regulators of lymphocyte and erythrocyte function (Figure 2J, Table S3). We confirmed increased expression of the EMT regulator Snai1 in HSV-1-infected cultures by immunostaining (Figure S2C). These results point towards the activation of multiple non-neural pathways in HSV-1- infected cultures.
We hypothesized that this could compromise the neural identity of brain organoids. Thus, we examined neuroepithelial polarity in infected organoids by immunostaining for N-cadherin, which is typically enriched at the apical side in cells surrounding the lumen in MOCK-treated organoids (Figure 2K–L) (Kadowaki et al., 2007). Strikingly, N-cadherin accumulation was lost in HSV-1-infected cultures but not in ZIKV-infected organoids (Figure 2K–L). Apical accumulation of N-cadherin was reduced in HSV-1-infected hNPCs (Figure 2M), suggesting a cell-autonomous effect. Loss of N-cadherin mainly reflected changes in its localization or protein expression, as its mRNA abundance was not severely reduced in infected organoids (Figure S2D). Furthermore, we observed decreased Sox1 protein intensity in HSV-1-infected organoids (Figure S2E), confirming impaired hNPC identity. We observed similar defects also in later-stage organoids infected with HSV-1 (Figure S2F–N), demonstrating the ability of HSV-1 to alter differentiation in more fate-restricted cells. Together, these results indicate that HSV-1 specifically activates alternative non-neural developmental programs and disrupts the neuroepithelial integrity of brain organoids, while ZIKV infection triggers antiviral and stress-related signatures but maintains cytoarchitecture.
ZIKV and HSV-1 infections differentially engage the IFN-I system
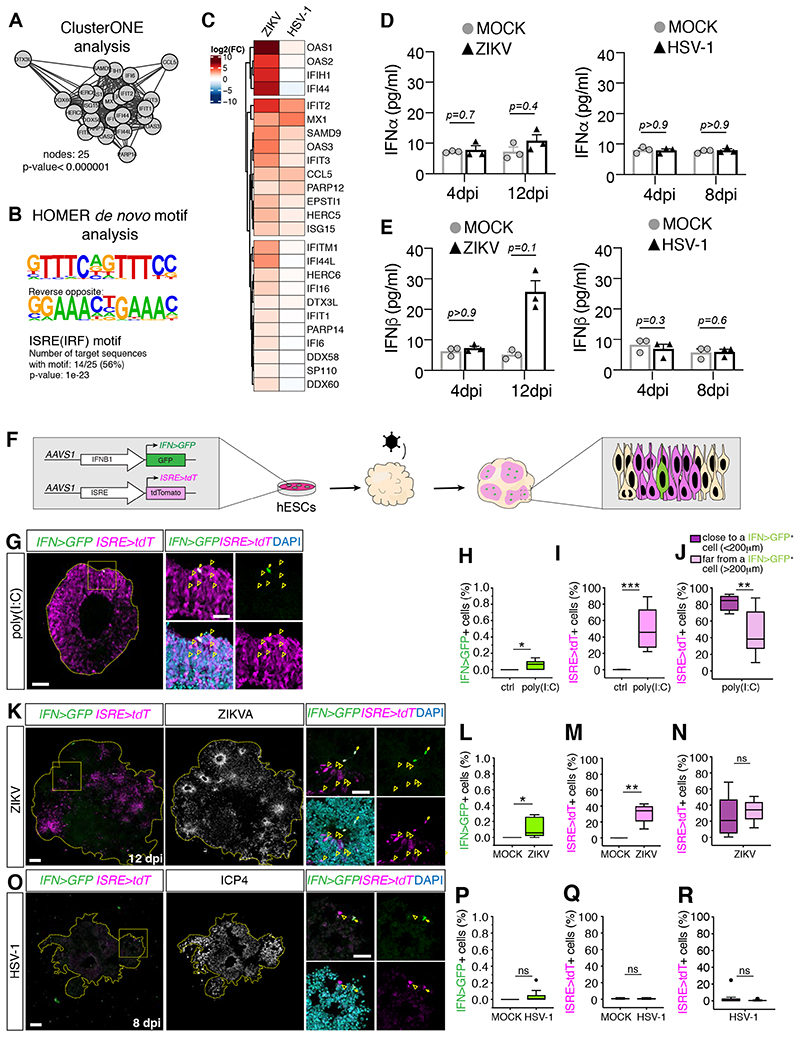
We next explored the mechanisms underlying the differential engagement of antiviral defense pathways by ZIKV and HSV-1 by examining the upregulated signatures from the RNA-sequencing data. Cluster analysis of genes upregulated in ZIKV-infected cultures revealed the presence of a highly connected cluster of genes enriched in the interferon-sensitive responsive element (ISRE) in their promoters (Figure 3A–B) and thus mapped to the ISG family (Schneider et al., 2014), in line with a previous study (Liu et al., 2019). Such a signature was not prominent in HSV-1-infected cultures (Figure 3C) and we confirmed the poor activation of ISGs by HSV-1 also at early time points by RT-qPCR analysis (Figure S3A–B). As ISGs are potently induced by IFN-I (Schneider et al., 2014), we measured production of all IFNα species and IFNβ in supernatants from ZIKV and HSV-1-infected organoids by ELISA assay. We detected a modest (but non-significant) increase of IFNβ specifically in the supernatants from ZIKV-infected cultures, while IFNα abundance was unchanged (Figure 3D–E). This analysis indicates the differential activation of ISGs and IFN-I in ZIKV and HSV-1-infected cultures.
Figure 3. ZIKV and HSV-1 infections differentially engage the IFN-I system.
A-B) Cluster of densely connected genes among genes upregulated in ZIKV-infected organoids at 12 dpi (in A) and their HOMER de novo motif analysis (in B).
C) Expression analysis of genes from A in infected organoids compared to their MOCK controls. D-E) Quantification of IFNα and IFNβ levels measured by ELISA assay. Values represent mean ± SEM (Mann-Whitney tests).
F) Schematic diagram of IFNB1>GFP (IFN>GFP) and ISRE>tdTomato (ISRE>tdT) reporters.
G-R) Immunostaining (scale bars 100 μm) of organoids generated from reporter cells and analyzed one day after stimulation with poly(I:C) in G, or after ZIKV or HSV-1 exposure (in K and O). Dashed lines indicate organoid contour based on DAPI (not shown). Insets (scale bars 50 μm) represent a magnified view of the area close to one single IFN>GFP+ cell (arrows). Arrowheads indicate ISRE>tdT+ cells. Graphs are Tukey plots (n≥3; p=0.0211 in H, p=0.0007 in I, p=0.0068 in J, p=0.0273 in L, p=0.0091 in M, p=0.3175 in N, p=0.0857 in P, p=0.6820 in Q, p=0.5994 in R, Mann-Whitney tests). Ctrl, control transfection. ns, non-significant.
dpi, days post-infection; See also Figure S3.
To simultaneously measure IFN-I and ISG expression at single-cell level and with spatial resolution in infected brain organoids, we built a dual fluorescent reporter system. We engineered hPSCs to carry a GFP cassette driven by the IFNB1 promoter (IFN>GFP) and a tdTomato cassette driven by the ISRE motif (ISRE>tdT) (Figure 3F). Immunostaining and flow cytometry analysis confirmed low basal expression of both reporters in control organoids and high ISRE>tdT expression in response to exogenous IFN-I (Figure S3C–D). To test the functionality of these reporters, we analyzed GFP and tdTomato expression by immunostaining in organoids stimulated with poly(I:C), a synthetic analog of double-stranded RNA (dsRNA) that acts as a potent IFN-I inducer in human NPCs (Lin et al., 2019). Consistent with induction, we detected high IFN>GFP signals and high ISRE>tdT expression (Figure 3G–J). However, this occurred only in 0.14% and 46% of the cells respectively, fractions that were smaller to those measured in human lung epithelial A549 cells engineered with the same reporter system (3.6% and 91% respectively; Figure S3E–G). These results validate the functionality the dual reporter systems and reveal a high degree of stochasticity of IFNB1 expression (Zhao et al., 2012) in brain organoids. We next analyzed reporter expression in response to viral infections by immunostaining. In tissues infected with ZIKV, we detected high IFN>GFP signals in very few cells (0.11%), indicating IFNB1 expression in a rare population of cells, and high ISRE>tdT expression in 30% of the cells (Figure 3K–N), consistent with ISG induction. Spatial analysis of reporter expression in ZIKV-infected organoids revealed that ISRE>tdT-expressing cells, which co-expressed the neural progenitor marker Sox1 (Figure S3H–I), appeared in clusters (Figure 3K and S3H) and located in areas far from high IFN>GFP-expressing cells (Figure 3N). These observations point towards ISG expression in the absence of high IFN-I production, in line with an IFN-independent ISG induction model (Liu et al., 2019; Schneider et al., 2014). Furthermore, the fraction of ISRE>tdT-expressing cells located far from or close to high IFN>GFP-expressing cells in ZIKV-infected organoids was similar, in contrast to the distribution observed in organoids stimulated with poly(I:C) (Figure 3N and J), suggesting viral inhibition of paracrine IFN-I signaling. Consistent with this, expression of the IFN-I signaling transducer Stat2, which is targeted for degradation by ZIKV (Grant et al., 2016; Kumar et al., 2016), was reduced in ZIKV-infected regions compared to non-infected areas (Figure S3J). These observations suggest an heterogenous and overall modest activation of the IFN-I response in ZIKV-infected organoids. In contrast to this, in HSV-1-infected organoids we found high IFN>GFP and ISRE>tdT signals only in a very small fraction of cells (0.03% and 0.7%, Figure 3O–R), indicating the lack of IFN-I and ISG induction in most cells. Altogether, these results reveal the differential induction of the IFN-I system in organoids infected by ZIKV and HSV-1.
IFN-I response in brain organoids is more attenuated than in 2D cultures
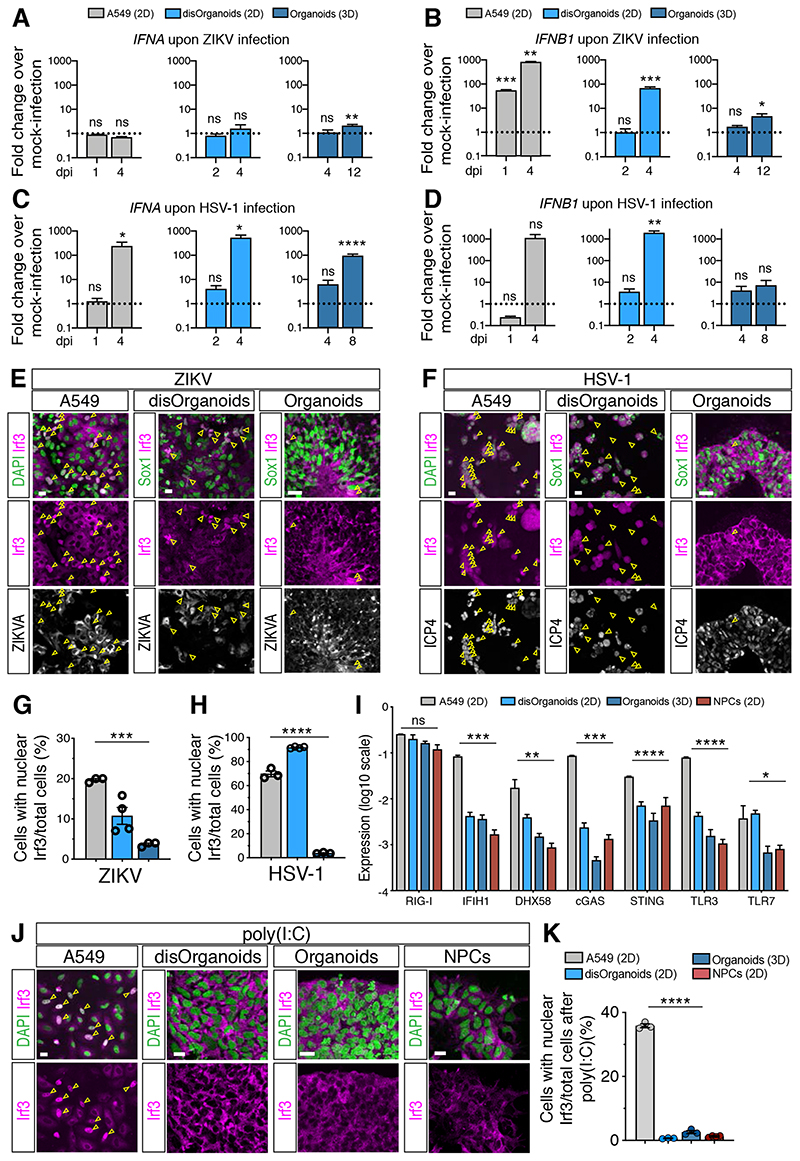
Our finding that IFN-I induction is modest in infected brain organoids was surprising given the strong IFN-I production measured in 2D hNPC cultures (Ferraris et al., 2019; Lima et al., 2019; Simonin et al., 2016; Zhang et al., 2016) and non-neural cultures (Hamel et al., 2015; C. Li et al., 2017). To test whether the magnitude of the IFN-I response is influenced by the culture system or by the cell type composition, we compared the organoid response to the one of 2D cultures obtained from the dissociation of brain organoids (called disOrganoids, Figure S4A) and 2D cultures of non-neural cells such as A549. ZIKV and HSV-1 efficiently replicated in both 2D cultures and these infections proceeded faster than in brain organoids (Figure S4B–E). RT-qPCR analysis at time points that showed similar ZIKV and HSV-1 infection levels across cultures revealed a more prominent induction of IFNA and IFNB1 in disOrganoids and A549 cells compared to the one measured in intact organoids (Figure 4A–D). The only exception was the induction of IFNA upon ZIKV infection, which was minimal in all culture systems tested (Figure 4A). Moreover, we analyzed the localization of Irf3, the major transcription factor that drives IFN-I transcription after nuclear translocation (Honda and Taniguchi, 2006) and found that the fraction of cells showing nuclear Irf3 accumulation was larger in disOrganoids and A549 cells compared to brain organoids (Figure 4E–H and S4F). These results confirm a more attenuated activation of the IFN-I response in brain organoids compared to 2D cultures.
Figure 4. The IFN-I response in brain organoids is more attenuated than in 2D cultures.
A-D) Quantification of IFNA and IFNB1 expression by RT-qPCR in cultures exposed to ZIKV (in A-B) or HSV-1 (in C-D). Dotted lines indicate the value of 1. Values are mean ± SEM (n=3 for A549, n=4 for disOrg, n=7 for ZIKV organoids, n=3 for HSV-1 organoids; ZIKV IFNA: p=0.8985 A549 1 dpi; p=0.0823 A549 4 dpi; p=0.6069 disOrg 2 dpi; p=0.9784 disOrg 4 dpi; p=0.7633 Org 4 dpi; p=0.0057 Org 12 dpi; ZIKV IFNB1: p=0.0005 A549 1 dpi; p=0.0015 A549 4 dpi; p=0.7036 disOrg 2 dpi; p=0.0008 disOrg 4 dpi; p=0.2135 Org 4 dpi; p=0.0333 Org 12 dpi; HSV-1 IFNA: p=0.5876 A549 1 dpi; p=0.0105 A549 4 dpi; p=0.1457 disOrg 2 dpi; p=0.0183 disOrg 4 dpi; p=0.2903 Org 4 dpi; p<0.0001 Org 8 dpi; HSV-1 IFNB1: p=0.0794 A549 1 dpi; p=0.1054 A549 4 dpi; p=0.3126 disOrg 2 dpi; p=0.0056 disOrg 4 dpi; p=0.4055 Org 4 dpi; p=0.2250 Org 8 dpi; Mann-Whitney test comparisons of infected samples vs age-matched MOCK samples).
E-H) Immunostaining (scale bars 100 μm) and quantification of Irf3 nuclear localization. A549 and disOrganoids were analyzed at 4 dpi, organoids at 12 dpi (in E) or 8 dpi (in F). ZIKVA, Zika virus Antigen; ICP4, infected cell polypeptide 4 protein of HSV-1. Color code as in A. Values are mean ± SEM (p=0.0007 in G; p<0.0001 in H; one-way ANOVA).
I) Expression of nucleic acid sensors measured by RT-qPCR. Values are mean ± SEM (n=3, p=0.3387 RIG-I; p=0.0063 DHX58; p=0.0432 TLR7; **** is p<0.0001; one-way ANOVA).
J-K) Immunostaining (scale bars 20 μm) and quantification of Irf3 nuclear accumulation after poly(I:C) treatment. Values are mean ± SEM (n=3; p<0.0001, one-way ANOVA).
ns, non-significant; dpi, days post-infection. See also Figure S4.
Efficient nuclear translocation of Irf3 relies on high expression of pattern recognition receptors (PRRs) mediating cytosolic viral sensing (Zhao et al., 2012). We found low expression of PRRs in early-stage organoids compared to later-stage organoids (Figure S4G), similar to the time-dependent expression timeline seen in vivo (Figure S4H). However, PRR expression in early-stage organoids was comparable to the one measured in disOrganoids as well as in 2D cultures of NPCs, and the levels were lower than those measured in A549 cells (Figure 4I). All neural cultures (organoids, disOrganoids and NPCs) also showed similar efficacy of Irf3 nuclear translocation upon poly(I:C) treatment and this was lower than that of A549 cells (Figure 4I–K and S4I). These results suggest cell type-specific modulation of viral sensing and argue against major changes in viral sensing between brain organoids and 2D neural cultures.
IFNβ treatment prevents ZIKV-induced organoid defects
The low levels of IFN-I measured in infected brain organoids could be responsible for their high susceptibility to ZIKV and HSV-1, despite some evidence suggesting that high IFN-I activity might exacerbate damage (Dang et al., 2016; Liu et al., 2019). Consistent with this, ISRE>tdT reporter expression correlated with lower levels of apoptosis in ZIKV-infected organoids (Figure S4J). To further demonstrate the neuroprotective effect of the IFN-I system in our infection models, we resorted to the administration of exogenous IFN-I, which inhibit ZIKV and HSV-1 replication in monolayer cultures (Contreras and Arumugaswami, 2016; Härle et al., 2002; Lafaille et al., 2012; Hamel et al., 2015; Lin et al., 2019; Gobillot et al., 2020). We confirmed the ability of two IFN-I subtypes, IFNα2 and IFNβ, to restrict ZIKV and HSV-1 infections in 2D disOrganoid cultures (Figure S5A–D), in line with their similar signaling downstream of the IFN-I receptor (Ivashkiv and Donlin, 2014). Given the major differences in viral kinetics and in the strength of innate immune signaling observed in brain organoids and in 2D cultures, we investigated whether IFN-I treatment could be efficacious also in brain organoid models.
We first applied IFN-I treatments to organoids that had been exposed to ZIKV and found that they both significantly ameliorated ZIKV-induced growth defects (Figure 5A–C). IFN-I treatments were well tolerated, since they did not compromise the growth of MOCK-treated organoids nor they induced large-scale apoptosis (Figure S5E–I). Strikingly, immunohistochemical analysis revealed that IFNβ treatment increased the area of VZ-like regions and reduced ZIKV infection, while IFNα2 showed much lower efficacy (Figure 5D–E). Poor efficacy of the IFNα2 treatment was unlikely due to a lower biological activity, since treatment with a higher dose of IFNα2 failed to further improve the rescue of ZIKV-infected cultures compared to the lower dose (Figure S5J). Also, neuroprotective activity was specific to IFN-I signaling, as treatment with type III IFNs did not ameliorate the growth phenotype nor it induced ISG expression (Figure S5J–K). We then performed RNA-sequencing on ZIKV-infected organoids treated with IFN-I. Principal component analysis (PCA) and differential gene expression data revealed that IFNβ treatment completely reverted the transcriptional changes caused by ZIKV, while IFNα2 treatment rescued gene expression only partially (Figure 5F–G and S5L), in line with our immunohistochemical data. Moreover, IFNβ treatment reduced ZIKV vRNA expression levels already at 4 dpi and even more dramatically at 12 dpi, while IFNα2 treatment failed to do so (Figure 5H). Together, these results reveal the neuroprotective function of the IFN-I system and the superiority of IFNβ over IFNα2 in ameliorating ZIKV-induced organoid defects.
Figure 5. IFNβ treatment prevents ZIKV-induced organoid defects.
A) Timeline of interferons (IFN-I) treatment. Organoids were analyzed at 12 dpi.
B-C) Images (scale bars 200 μm) of organoids treated as in A and area quantification. Values are mean ± SD and represent individual organoids (**** is p<0.0001, p=0.0063 ZIKV+IFNα2 vs ZIKV, p=0.0021 ZIKV+IFNβ vs ZIKV, p>0.9999 ZIKV+IFNα2 vs ZIKV+IFNβ, p=0.0015 ZIKV+IFNα2 vs MOCK, p=0.0117 ZIKV+IFNβ vs MOCK, Kruskal-Wallis test).
D-E) Immunostaining (scale bars 200 μm) of organoids and area quantification of ventricular zone (VZ)-like regions. Violin plots show median and quartiles (n=114 regions from 6 MOCK organoids, n=55 from 7 ZIKV organoids, n=73 from 7 ZIKV+IFNα2 organoids, n=106 from 7 ZIKV+IFNβ organoids; p=0.0026 ZIKV+IFNα2 vs MOCK; p>0.9999 ZIKV+IFNβ vs MOCK; **** is p<0.0001; Kruskal-Wallis test).
F-G) Principal Component Analysis (PCA) and expression (in scaled variance stabilizing transformation or VST) of differentially expressed genes.
H) Quantification of ZIKV viral RNA (vRNA) expression measured by RT-qPCR in organoids treated as in A. Values are mean ± SEM (4dpi: p=0.0017 ZIKV+IFNα2 vs ZIKV, p=0.0005 ZIKV+IFNβ vs ZIKV; **** is p<0.0001; one-way ANOVA Tukey’s multiple comparisons test). dpi, days post-infection; ns, non-significant. See also Figure S5 and Table S2.
IFNβ treatment fails to prevent HSV-1-induced organoid defects
We next tested the antiviral actions of IFNα2 and IFNβ against HSV-1 by comparing their ability to prevent growth and neuroepithelial defects in organoids that had been previously exposed to HSV-1 (Figure 6A). In contrast to the results obtained with ZIKV-infected organoids and HSV-1-infected 2D cultures, we found that only IFNα2 treatment ameliorated HSV-1-induced organoid growth defects (Figure 6B–C). Consistent with this, IFNα2 treatment improved organoid architecture, suppressed HSV-1 infection and rescued Sox1 expression as well as N-cadherin localization, while IFNβ treatment failed to do so (Figure 6D–G). Furthermore, transcriptional profiling of HSV-1-exposed cultures treated with IFN-I confirmed the inability of IFNβ treatment to rescue the transcriptional changes caused by HSV-1 (Figure 6H–I, S6A). Moreover, analysis of HSV-1 transcripts abundance measured by RNA sequencing confirmed that the IFNα2 treatment efficiently suppressed HSV-1 transcription at 8 dpi and that IFNβ exerted a minor effect (Figure 6J). Together, these results reveal the inability of IFNβ to perform its potent antiviral action against HSV-1, thus supporting differential functions of IFN-I subtypes in brain organoid models.
Figure 6. IFNβ treatment fails to prevent HSV-1-induced organoid defects.
A) Timeline of IFN-I administration. Organoids were analyzed at 8 dpi.
B-C) Images (scale bars 200 μm) and area quantification of organoids treated as in A. Values are mean ± SD (**** is p<0.0001; p=0.0043 HSV+IFNα2 vs HSV; p>0.9999 HSV+IFNβ vs HSV; p=0.0587 HSV+IFNα2 vs HSV+IFNβ; p>0.9999 HSV+IFNα2 vs MOCK; p=0.0007 HSV+IFNβ vs MOCK; Kruskal-Wallis test).
D and F) Immunostaining (scale bars 200 μm) of organoids. Dashed lines indicate organoid contour.
E) Quantification of Sox1 mean intensity per cell. Violin plots show median and quartiles (n>3000 cells from at least 3 organoids per condition, **** is p<0.0001, Kruskal-Wallis test).
G) Quantification of the ventricular zone (VZ)-like regions marked by N-Cadherin (N-Cad) apical accumulation. Data are mean ± SEM (n=3 experiments; **** is p<0.0001, ns is p>0.9999; Kruskal- Wallis test).
H-I) Principal Component Analysis (PCA) and expression (in scaled variance stabilizing transformation or VST) of genes differentially expressed in HSV-1-infected organoids.
J) Percentage of RNA-sequencing reads aligned to the HSV-1 genomic sequence. Values are mean ± SEM (for 4 dpi: p=0.8613 MOCK vs HSV, p=0.9975 HSV+IFNα2 vs HSV, p=0.8447 HSV+IFNβ vs HSV; for 8 dpi: p=0.003 MOCK vs HSV, p=0.0003 HSV+IFNα2 vs HSV, p=0.2943 HSV+IFNβ vs HSV, one-way ANOVA Tukey’s multiple comparisons test).
dpi, days post-infection; ns, non-significant. See also Figure S6 and Table S2.
HSV-1 selectively inhibits IFNβ activity
The poor antiviral activity of IFNβ against HSV-1 could be due to its inability to induce a subset of ISGs counteracting HSV-1 compared to IFNα2. To identify such a subset of ISGs, we analyzed the differential expression signatures of (uninfected) organoids treated with IFNα2 or IFNβ compared to untreated samples. These were enriched in genes implicated in defense response to virus and in response to IFN-I (Figure S6B–D). In contrast with our hypothesis, we found that all DEGs induced by IFNα2 were also found in the IFNβ dataset and that the pattern of differential expression was remarkably similar, with the exception that IFNβ showed higher potency (Figure S6E–F). Even at later time points, the DEGs induced by IFNα2 largely overlapped with those upregulated in the IFNβ dataset. Noticeably though, differences in ISG levels and a time-dependent ISG downregulation specific to IFNα2-treated cultures and reminiscent of the IFNα desensitization (Sandler et al., 2014) became prominent (Figure S6G–H). These data argue against the existence of a set of ISGs uniquely induced by IFNα2, thus ruling out our hypothesis.
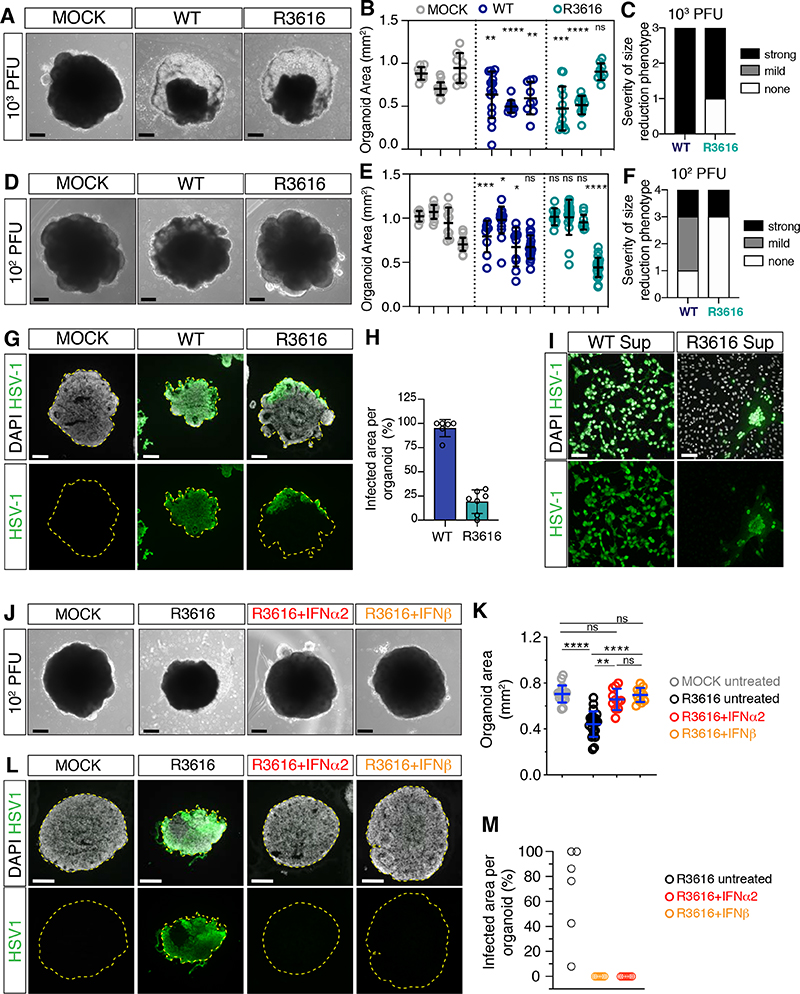
An alternative possibility is that HSV-1 might specifically block the activity of IFNβ through the action of one or multiple viral proteins (Danastas et al., 2020). Such an infection-dependent effect is supported by the observation that IFNβ showed potent antiviral activity against HSV-1 when administered to organoids before (but not after) viral exposure (Figure S7A–B), and by the diminished IFNβ-dependent ISG expression in HSV-1-infected cultures compared to MOCK- treated conditions (Figure S7C). We noticed that one of the affected ISGs was the protein kinase R (PKR), an antiviral effector that inhibits translation of viral mRNAs by phosphorylating eIF2a (Schneider et al., 2014) and whose activity is counteracted by the HSV-1 protein ICP34.5 (Chou et al., 1995). This observation led us to hypothesize that the HSV-1 protein ICP34.5 may counteract IFNβ activity via targeting the PKR pathway. This model predicts that loss of ICP34.5 would render HSV-1 sensitive to IFNβ. We thus performed infections with an HSV-1 mutant strain that carries the deletion of both copies of ICP34.5 genes and referred to as R3616 (Figure S7D) (Chou et al., 1990). ICP34.5-null mutants replicate well in cell culture but they are attenuated in the brains of mice and humans (Bolovan et al., 1994; Chou et al., 1990; Kaur et al., 2012; Leib et al., 1999). Consistent with in vivo neuroattenuation, the ICP34.5-null strain R3616 reduced organoid growth less efficiently than wild type (wt) HSV-1 especially at the low inoculum dose and exhibited limited viral spreading and viral particle release compared to wt HSV-1 (Figure 7A–I). Importantly, when R3616 showed prominent infection, its replication was efficiently blocked by IFNβ (Figure 7J–M), consistent with our prediction. R3616 infection did not change the expression levels of ISGs similar to wt HSV-1, ruling out a direct role of ICP34.5 in ISG induction (Figure S7E). Together, these results indicate that HSV-1 selectively inhibits IFNβ action in part via ICP34.5.
Figure 7. HSV-1 selectively counteracts IFNβ activity.
A-F) Images (scale bars 200 μm) and area quantifications of organoids exposed to HSV-1 wild type (WT), R3616 mutant or MOCK-treated and analyzed at 8 dpi. Values are mean ± SD and represent individual organoids (in B: for wt, p=0.0066 and p=0.0021; for R3616 p=0.0008 and p=0.7802; in E: for wt p=0.0002, p=0.02, p=0.0232 and p=0.1606; for R3616, p=0.8148, p=0.5701, p=0.7394; **** is p<0.0001; Mann-Whitney test comparisons to MOCK counterparts). Outcomes of infection experiments shown in C and F are based on statistical significance (strong if p<0.005, mild if 0.005<p<0.05 or none if p>0.05).
G-H) Immunostaining (scale bars 200 μm) and quantification of infected organoid area at 8 dpi. Dashed lines indicate organoid contour. Data are mean ± SD (n=6 organoids for WT, n=7 for R3616).
I) Immunostaining (scale bars 100 μm) of Vero cells incubated with supernatants (Sup) from HSV- 1 WT- or R3616-infected organoids at 8 dpi.
J-M) Images, immunostaining (scale bars 200 μm) and quantification of organoid infected area at 8 dpi. Refer to Figure 6 for comparison to wt HSV-1. Values in K are mean ± SD (n=6 untreated organoids, n=7 organoids for IFNα2 and IFNβ; **** is p<0.0001; ns is p p>0.9999; p=0.0020 R3616+IFNα2 vs R3616; Kruskal-Wallis test).
dpi, days post-infection; ns, non-significant. See also Figure S7, Table S1 and S2.
Discussion
Here we establish human brain organoid models for two distinct TORCH viruses, ZIKV and HSV-1. We show that these two models share features of microcephaly but exhibit major differences in the underlying structural defects and transcriptional profiles, as well as in the engagement of the antiviral system and sensitivity to IFN-I. These results argue for the existence of unique pathogenic mechanisms underlying virus-induced microcephaly and the use of tailored antiviral strategies against TORCH pathogens.
Both ZIKV and HSV-1 infect hNPCs leading to productive infection and causing reduced organoid size and cell depletion. These data implicate hNPC infection in TORCH neuropathogenesis and are consistent with other organoid models of ZIKV and HSV-1 infection (Cugola et al., 2016; Gabriel et al., 2017; Garcez et al., 2016; Qian et al., 2016; Qiao et al., 2020; M. Watanabe et al., 2017) as well as with the clinical findings associated with viral exposure in the first and second trimesters of pregnancy (Honein et al., 2017; Marquez et al., 2011). HSV-1 infection in organoids appears more destructive than that caused by ZIKV, in line with the high severity of in utero HSV infections (Marquez et al., 2011). In contrast to this, HCMV shows limited tropism for primitive hNPCs and poor replication in early-stage organoids. This phenotype is likely due to the lack of intermediate progenitors (Kanton et al., 2019), a population of hNPCs recently identified as the major cellular target of HCMV in a 45-days-old organoid model of microcephaly (Sun et al., 2020). These data suggest different neurotropism of TORCH viruses in the human fetal brain.
Mechanistically, we show that both ZIKV and HSV-1 infections trigger apoptosis, as previously reported (Cairns et al., 2020; Cugola et al., 2016; Hanners et al., 2016; Qian et al., 2016; Souza et al., 2016; M. Watanabe et al., 2017). Apoptosis likely represents a general cellular defense against viral spreading that could explain the neurotoxicity of congenital TORCH infections. Yet, our analysis of apoptosis and lumen size suggests that different mechanisms of tissue damage may be involved. Whereas ZIKV infection likely reduces the lumen size by basal delamination of apoptotic cells from the neuroepithelium, HSV-1 infection results in enlarged lumens possibly through apical extrusion of apoptotic cells. Furthermore, our data suggest that HSV-1 is unique in its ability to cause neuropathology by impairing neuroepithelial integrity, in line with the key roles of cell adhesion in controlling NPC proliferation and maintenance during brain development (Chenn and Walsh, 2002; Hatakeyama et al., 2014; Kadowaki et al., 2007; Rousso et al., 2012). HSV-1 may perturb neuroepithelial polarity either by promoting N-cadherin cleavage (Reiss et al., 2005) or by disrupting adherens junctions via binding to the cell adhesion molecule Nectin1 (Krummenacher et al., 2003; Richart et al., 2003; Simpson et al., 2005). Remarkably, these cytoarchitectural changes seen in organoids are distinct from the large multicellular structures observed in 2D neural cultures (this study and (Cairns et al., 2020)). These discrepancies are likely due to major changes in cell adhesion and polarity of NPCs grown in monolayer and 3D organoid cultures (Scuderi et al., 2021), confirming the superiority of brain organoids in mimicking the neuropathological features associated with HSV-1 infection.
The structural and cellular differences of ZIKV and HSV-1 organoid models are also reflected in their transcriptional profiles. While cell death is concomitant with the activation of cellular stress- and virus defense-related pathways in ZIKV-infected cultures in agreement with previous studies (Gladwyn-Ng et al., 2018; Liu et al., 2019; M. Watanabe et al., 2017), apoptosis is linked to the upregulation of ATF and Jun/Fos families of transcription factors and the activation of non-neural developmental pathways in HSV-1-infected cultures. In the case of HSV-1, it is possible that this transcriptional activation is required to ensure efficient replication, as HSV-1 induces similar profiles in other cells (Drayman et al., 2019; Hensel et al., 2019; Hu et al., 2016). Another option is that the deregulation of non-neural genes caused by HSV-1 could contribute to the loss of neuroepithelial identity. Indeed, the altered hNPC morphology, reduced apical N- cadherin localization and the disintegration of infected organoids are reminiscent of the morphological changes, lowered cadherin levels and weakened adhesion of mesenchymal cells (Thiery et al., 2009).
Our work highlights major differences in the cellular innate response against ZIKV and HSV-1 and their sensitivity to IFN-I. While these disparities are perhaps not surprising given the great diversity of the two viruses, these phenotypes are unique to brain organoids. We show that both ZIKV and HSV-1 infections of organoids attenuate IFN-I responses much strongly as compared to 2D cultures, helping resolve a discrepancy in the field on the magnitude of the antiviral response between 2D and 3D culture systems (C. Li et al., 2017; Lima et al., 2019; Liu et al., 2019; Simonin et al., 2016; Zhang et al., 2016). The attenuated IFN-I response of organoids is consistent with the low immune reactions described in the microcephalic brains of ZIKV-infected patients (Lima et al., 2019) and with the generally low IFN-I activity of the brain (Sorgeloos et al., 2013). Changes in the modulation of innate immune reactions between 3D and 2D culture systems may result from differences in infectivity levels and/or viral spreading, since we observed lower initial infection levels and much slower kinetics of viral replication in 3D compared to 2D culture. A slow kinetics often reflects a small fraction of infected cells and could thus explain the rare activation of IFNβ transcription measured by our reporter assay. These discrepancies could result from inefficient cell targeting due to limited cell surface accessibility and/or from reduced local spreading due to stronger cell-to-cell contacts in 3D as compared to 2D culture.
Moreover, our work reveals that ZIKV and HSV-1 replication in organoid cultures is differentially sensitive to the action of distinct IFN-I subtypes. These results lend support to the neuroprotective function of the IFN-I system against viral infections (Gorman et al., 2018; Leib et al., 1999; C. Li et al., 2017; Lin et al., 2019; Sorgeloos et al., 2013; J. P. Wang et al., 2012) and to the view that distinct IFN subtypes can mediate different biological outcomes (Ng et al., 2016). Nevertheless, it is surprising that distinct IFN-dependent viral phenotypes are detectable in 3D, while remaining indistinguishable in 2D cultures (this study and (Contreras and Arumugaswami, 2016; Härle et al., 2002; Lafaille et al., 2012)). A contributing factor could be the duration of the IFN-I treatment, since the pattern of ISG expression induced by IFNα/β starts to differ after repeated administrations. Viral phenotypes in 3D might also be strongly influenced by viral immune evasion mechanisms (Chou et al., 1990; Danastas et al., 2020; Gorman et al., 2018; Grant et al., 2016; Kumar et al., 2016). The reduced efficacy of IFN-I against ZIKV in 3D vs 2D, the neuroattenuation of the ICP34.5-null HSV-1 mutant and the poor IFNβ activity against HSV-1 in brain organoids are compatible with this possibility.
Finally, our work uncovers the existence of previously unappreciated differences in IFN-I activities. The superior antiviral activity of IFNβ over IFNα2 against ZIKV infection likely reflects the longer-lasting and more potent ISG induction by IFNβ over IFNα2, in line with the higher potency of IFNβ (Bolen et al., 2014; Gobillot et al., 2020). More intriguing is the poor antiviral activity of IFNβ against HSV-1 since this effect could be specific to the brain. In fact, IFNβ potently inhibits HSV-1 replication in other cells and in mice (Arao et al., 1997; Carr et al., 2003; Giraldo et al., 2020; Härle et al., 2001). Our pre and post-treatment IFNβ experiments with wt and ICP34.5-null HSV-1 viruses indicate that HSV-1 selectively neutralizes IFNβ action, in line with the well-described ability of HSV-1 to evade IFN-I signaling (Danastas et al., 2020). This mechanism may represent an efficient strategy to target the most potent IFN type in the brain when both IFNα species and IFNβ are produced (Sorgeloos et al., 2013). Our results suggest that such evasion mechanism is in part mediated by the viral protein ICP34.5, which possibly counteracts ISG induction and PKR activity downstream of IFN-I signaling (Chou et al., 1995; He et al., 1997). However, since PKR is induced and likely activated also in IFNα2-treated organoids, future work is needed to characterize the mechanisms determining the selectivity against IFNβ.
In conclusion, the organoid infection models described in this work have a great utility for evaluating therapeutics against ZIKV and HSV-1 and can serve as experimental platforms for better characterizing the activity of human IFNs. Although our results support the therapeutic use of specific IFN-I against ZIKV and HSV-1 infections, our understanding of IFN activities in humans is limited, as reflected in the toxicity and efficacy issues of IFN-based therapies (Fritz-French and Tyor, 2012; S.-F. Li et al., 2018). Mechanistic studies on ISG functions in these organoid infection systems will help guide the design of more efficacious IFN-I modulatory compounds. Furthermore, our infection models for HSV-1 could serve as platforms for further characterization of HSV-1 evasion mechanisms and for testing of HSV-1 mutants. This work will help improve the design of oncolytic HSV-1 vectors that could overcome the limited efficacy of current HSV-1 viruses in glioblastoma therapy (Kaur et al., 2012). More broadly, our work calls for a paradigm shift to human 3D systems for the study of viral mechanisms that will be instrumental for the development of more effective antiviral compounds for the treatment of neurological complications.
Limitation of the study
Our analysis of antiviral signaling is limited by the small number of antiviral genes analyzed and by the lack of time-resolved tissue dynamics of viral and host proteins expression. Such analyses in large 3D tissue-like brain organoids are technically challenging and the application of emerging techniques, such as spatial transcriptomics (Burgess, 2019) and live 3D tissue fluorescent imaging (Rios and Clevers, 2018) in an appropriate bio-safety setting, will allow greater insights into the tissue dynamics of viral spreading and antiviral signaling. Another limitation of the organoid models used in this study is their low cellular complexity, which does not recapitulate the full repertoire of neural cells seen in late-stage organoid cultures (Kanton et al., 2019), and their lack of the microglia, a limitation of current organoid models (Amin and Paşca, 2018). The development of co-culture systems of late-stage organoid cultures and microglia will be instrumental to better recapitulate fetal neuroimmune interactions that could be relevant for viral pathogenesis.
Star Methods
Experimental Model And Subject Details
Human embryonic stem cells and cell lines
Human embryonic stem cells (hESC) H9 were obtained from WiCell. Human lung epithelial carcinoma A549 cells and African green monkey kidney Vero cells were obtained from ATCC and maintained in regular Dulbecco’s minimal essential medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 2 mM L-Glutamine. All cells were authenticated using a short tandem repeat (STR) assay. All cells were maintained in a 5% CO2 incubator at 37°C and routinely tested for mycoplasma.
Viruses
The French Polynesian ZIKA virus strain (ZIKV, H/PF/2013) was propagated in Vero cells. Briefly, Vero cells were infected with ZIKV at multiplicity of infection (MOI) 0.1 and incubated at 37°C in a 5% CO2 incubator. At 3 days post-infection, cell supernatants from infected cells were harvested and purified by centrifugation at 1500 rpm for 10 min to remove cellular debris. The viral titre was determined by tissue culture infective dose (TCID) assay performed on Vero cells. Briefly, confluent Vero cells plated in 96-well plates were infected with serially diluted ZIKV stocks and incubated at 37°C in a 5% CO2 incubator. The assay was carried out in eight parallels wells for each dilution with the last column of 96-well plate as control cells without virus. At 5 days postinfection, the appearance of cytopathic effect (CPE) was examined by microscopy. The TCID50 was calculated from the CPE induced in the cell culture. The wild type Herpes Simplex virus 1 (wt HSV-1, strain F) and the R3616 mutant HSV-1 virus (HSV-1 R3616, strain F) were generated previously (Chou et al., 1990) and kindly provided by B. Roizman (University of Chicago, Chicago, IL). HSV-1 viruses were grown and subjected to titer determination by plaque assay on Vero cell monolayers as previously described (Calistri et al., 2003). Human Cytomegalovirus (HCMV, strain TB40/E) expressing mNeonGreen fluorescence protein under the control of the endogenous HCMV major immediate-early (MIE) promoter of UL122/123 genes (HCMV-UL122/123- mNeonGreen) was generated and kindly provided by L. Cicin-Sain (Helmholtz Centre of Infection Research, (Kasmapour et al., 2018). Note that this modified virus shows similar growth kinetics to the parental strain (Kasmapour et al., 2018). Viral stocks were aliquoted and stored at −80°C. Supernatants from uninfected Vero cells were prepared as performed during viral propagation and used as MOCK controls. Infection experiments were conducted under Biosafety Level 2 Plus containment.
Method Details
Maintenance of hESCs
H9 cells were cultured under feeder-free or feeder-dependent conditions. Feeder-free H9 cells were seeded onto 6-well plates coated with hES-qualified matrigel and maintained in mTeSR1 medium. Cells were fed daily and passaged every 3-4 days using 0.5 mM EDTA solution treatment and mechanical dissociation. Feeder-dependent H9 cells were cultured on CF-1-y- irradiated mouse embryonic fibroblasts seeded one day in advance onto gelatin-coated [0.1% (wt/vol) gelatin] 6-well plates. Cells were fed daily with stem cell medium containing DMEM-F12 supplemented with 20% KnockOut Serum Replacement, 3% FBS, 1X GlutaMAX, 1X minimum essential medium amino acids (MEM-NEAA), 0.1 mM beta-mercaptoethanol (BME), 20 ng/ml bFGF as previously described (Lancaster and Knoblich, 2014). Cells were passaged every 5-7 days following treatment with collagenase IV (0.1% wt/vol) for 15 minutes and mechanical dissociation.
Generation of interferon reporter lines
The reporter constructs were inserted into the AAVS1 safe-harbor locus of feeder-free H9 cells or A549 cells using TALEN technology as described before (Bagley et al., 2017). Donor plasmids were constructed to insert the following cassettes: (i) 2APuro-2xCHS4-IFNB1>eGFP- WPRE-SV40-2xCHS4 and (ii) 2ANeo-2xCHS4-ISRE>tdTomato-WPRE-SV40-2xCHS4. Fragments spanning -1425 base pairs (bp) downstream and 40 bp upstream of the transcription start site of the human IFNB1 gene and 353 bp downstream and 74 bp upstream of the transcription start site of the human ISG15 gene (Hummer et al., 2001) were used as promoters. All donor plasmids were verified by sequencing. For reporter integration in H9 cells, 106 single cells prepared using accutase were nucleofected with the Amaxa nucleofector (Lonza) and Human Stem Cell Nucleofector Kit 1 solutions containing 0.5 μg of each of the TALEN plasmids and 1 μg of each of the two donor plasmids following manufacturer’s guidelines. Nucleofected cells were grown for four days and then selected with 0.5 μg/ml puromycin and 100 μg/ml G418. For reporter integration in A549 cells, 1.2 × 106 cells were seeded and one day later transfected with Lipofectamine 3000 and 1 μg of each of the TALEN plasmids and 1.5 μg of each donor plasmid following manufacturer’s instructions. One day later, the medium was replaced and A549 cells were selected with 0.5 μg/ml puromycin and 750 μg/ml G418 starting from day 2. Surviving colonies of H9 and A549 were picked manually, transferred into 24-well plates and further expanded for genotyping and cryopreservation. For genotyping, DNA was extracted using the QuickExtract DNA Extraction Solution and a PCR assay was performed to identify correctly targeted AAVS1 insertions and loss of wild-type alleles in multiple clones. For ISRE>tdTomato expression analysis performed by flow cytometry (Figure S3C), organoids generated from reporter cells (12-days-old) were incubated with recombinant interferons or vehicle. One day later, organoids were manually dissociated into single cells after 5 minutes of incubation with an accutase:trypsin (2:1 v/v) mix at room temperature. Single cell suspensions were diluted into 300 μl of culture medium and tdTomato expression was analyzed by flow cytometry performed on a LSRFortessa cell analyzer (BD Biosciences) using BD FACSDIVA software. Data were analyzed using FlowJo.
Generation of cerebral organoids
Cerebral organoids were generated as previously described (Lancaster et al., 2013; Lancaster and Knoblich, 2014). Briefly, on day 0 hESCs were dissociated into single cells by accutase treatment (feeder-free cells) or by collagenase IV/dispase treatment followed by a short treatment with trypsin (feeder-dependent cells). Cells were transferred to an ultra-low binding 96-well plate (9000 cells/well) containing stem cell medium supplemented with 4 ng/ml bFGF and 50 μm Rho- associated protein kinase (ROCK) inhibitor. On day 3, the medium was replaced with fresh stem cell medium. From day 5 or 6, the medium was replaced daily with Neural Induction Medium containing DMEM-F12 supplemented with 1X N2 supplement, 1μg/ml heparin solution, 1X GlutaMAX and 1X MEM-NEAA. On Day 10 or 11, organoids with visible neuroepithelia were embedded into droplets of Matrigel and transferred into 6-cm dishes in Expansion Medium consisting of 50% DMEM-F12, 50% Neurobasal medium, 1X N2, 1X B27-Vitamin A, 2.5 μg/ml Insulin, 0.05 mM BME, 1X GlutaMAX, 1X MEM-NEAA and 1X Penicillin/Streptomycin. On day 15, media was replaced with Differentiation Medium consisting of 50% DMEM-F12, 50% Neurobasal medium, 1X N2, 1X B27, 2.5 μg/ml Insulin, 0.05mM BME, 1X GlutaMAX, 1X MEM-NEAA, 1X Penicillin/Streptomycin and organoids were cultures on an orbital shaker under 57 rpm rotating speed. The medium was changed every 2-3 days. From day 40 onwards, organoids were fed with Differentiation Medium supplemented with 1% (v/v) matrigel basement membrane. From day 65 onwards, organoids were fed with Differentiation Medium supplemented with BDNF (20 ng/ml), GDNF (20 ng/ml) and db-cAMP (1 mM). Organoids that passed the quality control criteria (Lancaster and Knoblich, 2014) were used. Criteria included visible surface brightening before embedding (as a proxy for an organized neuroepithelium) and formation of neural tube-like structures in matrigel.
Generation of 2D neural cultures
To generate dissociated 2D cultures of organoids (disOrganoids), 10-days-old organoids were plated onto matrigel-coated dishes in Neural Induction Medium. Three days later, plated organoids were incubated with accutase for 15 min at 37°C and manually dissociated into a single cell suspension. After a washing step, cells were resuspended in Neural Induction Medium supplemented with 10 μM ROCK inhibitor and plated as single cells onto matrigel-coated plates. One day later, the medium was replaced with Expansion Medium. Two days later (day 16), cells were dissociated again with accutase and plated onto matrigel-coated surfaces for further analysis. An outline of this protocol is presented in Figure S4A. To differentiate hESCs into hNPCs directly in 2D, we adapted a previously published protocol (Qi et al., 2017). Briefly, cells were dissociated with accutase and plated onto matrigel-coated plates at the density of 200,000 cells/cm2 in mTeSR1 supplemented with 10 μM ROCK inhibitor. Differentiation was started the next day using E6 medium. Inhibitors used in LSB+X induction in E6 included LDN193189 (100 nM), SB431542 (10 μM) and XAV939 (2 μM) for treatment of 3 days. Then starting from day 3, LDN193189 (50 nM), SB431542 (5 μM), XAV939 (1 μM), SU5402 (2 μM) and DAPT (5 μM) were added to E6. On day 6, cells were dissociated by incubation with accutase at 37°C for 15 minutes, resuspended in E6 medium supplemented with 10 μM ROCK inhibitor and plated onto poly-L- ornithine (50 μg/ml) and laminin (5 μg/ml)-coated surfaces for further analysis. An outline of this protocol is shown in Figure S4I.
Infection of cerebral organoids
Organoid infection experiments were performed by adapting previous ZIKV infection paradigms (Dang et al., 2016; Gabriel et al., 2017). On day 10 or 11, organoids were incubated with a virus inoculum diluted in 200 μl of Neural induction Medium in ultra-low binding 96-well plates. An equivalent volume of MOCK medium was used for uninfected controls. Plates were incubated at 37°C in a 5% CO2 incubator. One day later, organoids were embedded in Matrigel, transferred to 6-cm dishes in Expansion medium and cultured as described above. Details on the number of virus particles used in this study are provided in Table S1 and S2. For infection at later stages, 40-days-old organoids were exposed to 600 PFU of HSV-1 diluted in Differentiation medium. One day later, the medium was replaced with fresh Differentiation medium containing 1% (v/v) matrigel basement membrane and cultured as described above. Supernatants were collected at 4, 8 or 12 dpi and frozen at −80°C. 125 μl of each supernatant was used to infect Vero cells in 12-well plates. Cells were fixed 24 hours post-infection and the level of infection was assessed by viral antigen expression using immunostaining.
Infection of 2D cultures
disOrganoids (day 17) were seeded onto glass coverslips in 12-well plates at a seeding density of 150000 cells/well. One day later, cells were incubated with 150000 TCID50 units of ZIKV or with 187.5 PFU of HSV-1 for one day. For infection in A549 cells, 80000 cells were seeded onto a glass coverslip in 12-well plates. One day later, cells were incubated with 80000 TCID50 units of ZIKV or with 50 PFU of HSV-1 for one day to match the infection doses used in organoids. An equivalent volume of MOCK medium was used for control conditions.
Interferon and poly(I:C) treatments
Recombinant human interferons were reconstituted at 10 μg/ml according to the manufacturer’s instructions. Interferons were used at 10 ng/ml dose unless differently specified. Based on the biological activities provided by the manufacturer, 10 ng/ml dose of IFNα2 and IFNβ corresponds to activities of 1800 U/ml and 100 U/ml respectively. BSA solution (0.1%) was used as a vehicle for untreated conditions. For multiple-dose treatments, interferons or vehicle were freshly diluted into culture medium at 2, 4, 6 and 8 dpi. For poly(I:C) experiments, organoids (10-12 days-old) were transfected with 1 μg poly(I:C) diluted in 200 μl of Neural Induction Medium [corresponding to 5 μg/ml poly(I:C)] using Lipofectamine 3000 following manufacturer’s instructions and incubated at 37°C in a 5% CO2 humidified atmosphere for one day. For poly(I:C) treatment in 2D culture, cells were seeded onto glass coverslips in 12-well plates. One day later, cells were transfected with 25 ng/ml poly(I:C) using Lipofectamine 3000 Transfection Reagent. The medium was replaced after 6 hours and cells were further cultured at 37°C in a 5% CO2 incubator for additional 18 hours. poly(I:C) was replaced with water in control transfections.
Cryo-sectioning, immunostaining and imaging
Organoids were fixed in 4% paraformaldehyde (PFA) overnight at 4°C. After extensive washes with PBS, organoids were immersed in 30% sucrose solution overnight, embedded in 10% gelatin solution and frozen. Samples were sectioned at 20 μm thickness using a cryostat (Leica). Organoid cryo-sections were fixed with 4% PFA for 10 minutes. After extensive washing with PBS, cryo-sections were permeabilized and blocked with blocking solution [10% normal donkey serum (NDS) containing 0.5% Triton X-100] for 45 min at room temperature. Sections were then incubated with primary antibodies diluted in antibody solution [10% normal donkey serum containing 0.1% Triton X-100] overnight at 4°C. After three washes of 10 min with PBS, both sections were incubated with secondary antibodies diluted in antibody solution at room temperature for two hours and with DAPI solution (2 μg/ml) for 10 minutes. For immunostaining of 2D cultures, cells seeded on coverslips were fixed with 4% PFA for 30 minutes and washed extensively with PBS. Cells were permeabilized with PBS supplemented with 0.2% Triton X-100 for 5 min, washed with PBS and blocked with 10% NDS for one hour. Coverslips were then incubated with primary antibodies diluted in antibody solution for 2 hours at room temperature. Coverslips were washed with PBS and incubated with secondary antibodies for 45 minutes at room temperature, followed by incubation with DAPI solution for 3 minutes. Finally, both organoid sections and coverslips were washed three times with PBS containing 0.05% Triton X-100 and then mounted in fluorescent mounting medium. Primary antibodies used in this study and their dilutions are summarized in the Key Resource Table. Secondary antibodies AlexaFluor 488, 568 or 647 -conjugated donkey antibodies (Invitrogen) were used at 1:500 dilution. Immunostaining images were captured with Axio Imager confocal microscopes LSM700, 780 and 880 (Zeiss GmbH), using the “tile” and “stitching” functions in the Zen software when imaging large organoid areas. Immunostaining images of organoids in Figure 7 were acquired with an Axio Imager.Z2 microscope (Zeiss GmbH) equipped with an ORCA-Flash 4.0 V3 Digital CMOS Camera (Hamamatsu). Bright-field imaging of intact organoids was performed on a widefield microscope (AxioVert.A1, Zeiss GmbH) with a SONY Chameleon®3 CM3-U3-31S4M CMOS camera (Zeiss GmbH). Images were converted into maximal intensity projections, adjusted for brightness levels and cropped (if used for magnified insets) using Fiji. Image quantifications were performed in Fiji.
Quantification of cytokine levels by ELISA
Levels of all IFNα species and IFNβ in organoid supernatants were measured using ELISA Kits according to manufacturer’s instruction. Briefly, frozen supernatants from MOCK- and infected- organoids were thawed on ice and centrifuged at 13,000 rpm for 10 min to eliminate debris. Supernatants (3 ml for each sample) from three different experiments were concentrated using Centrifugal Filter Unit with Ultracel-3 membranes (UFC900308 Merck Millipore). For each sample (50 μl) absorbance at 450 nm and 540 nm were measured in duplicate using a microplate reader (Synergy H1 BioTek). Absorbance at 540 nm was subtracted from absorbance at 450 nm to correct for non-specific signals. Mean values of absorbance were multiplied for the concentration factor and used to estimate cytokine amounts (pg/ml) according to a standard curve of recombinant Human IFNα (0-125 μg/ml) or recombinant Human IFNβ (0-150 μg/ml).
RNA extraction and RT-qPCR analysis
For each condition and time point, 4-10 organoids were pooled into RNAse-free tubes and chilled on ice. Organoids were washed with PρBS and incubated with 1 ml of chilled Cell Recovery Solution for 1 hour on ice. Dissolved Matrigel was removed by rinsing twice in cold DEPC-treated PBS and samples were lysed in RLT buffer. RNA was extracted using the RNeasy Micro Kit and on-column DNAse digestion was performed using RNAse-Free DNAse I following the manufacturer’s protocol. cDNA synthesis was performed with SuperScript III and Random Hexamers using 500 ηg- 1 μg of total RNA according to the manufacturer’s instructions. qPCR reactions were performed using GoTaq qPCR master mix on 384-well (CXF384) or 96-well (CXF96) BioRad machines using the following reaction protocol: (i) 95 °C for 3 min, (ii) 95°C for 10 s, (iii) 62°C for 10 s, (iv) 72°C for 40 s, (v) go to 2, 40 cycles, (vi) 95°C for 1 min, and (vii) 50°C for 10 s. Quantification was performed in Excel by calculating the ΔCt value using TBP as a reference gene and the ΔΔCt value using age-matched untreated samples as calibrators. ACTIN was used as a reference gene in HSV-1 experiments. Data are presented as expression (2-ΔCt) or fold change (2-ΔΔCt). Primers used for RT-qPCR are provided in Table S4.
RNA-sequencing and analysis
For the ZIKV experiment, 3 replicates were used per condition and time point (4 and 12 dpi). For the HSV-1 experiment, 3 replicates were used per each condition and time point (4 and 8 days post-infection). Sample collection and RNA isolation were performed as described above. RNA concentration and quality were assessed with an RNA 6000 Nano Chip (Agilent Technologies). mRNA isolation was performed with NEBNext Poly(A) mRNA Magnetic Isolation Module. Barcoded samples were multiplexed and sequenced 50 bp single-end on a HighSeq 2500 (Illumina). mRNA sample isolation, library preparation, and sequencing were done at the VBCF NGS Unit (https://www.vbcf.ac.at). Reads were screened for ribosomal RNA by aligning with BWA (v0.7.12) (H. Li and Durbin, 2009) against known rRNA sequences (RefSeq). rRNA-subtracted reads were aligned with TopHat (D. Kim et al., 2013) against the Homo sapiens genome (hg38). Microexon-search was enabled. Additionally, a gene model was provided as GTF (UCSC, 2015_01, hg38). rRNA loci were masked on the genome for downstream analysis. Aligned reads were counted with HTSeq (v0.6.1; intersection-nonempty) (Anders et al., 2015). The samples were subjected to differential expression analysis with DESeq2 (v1.16.1) (Love et al., 2014). Furthermore, reads were subjected to TPM estimation with Kallisto (v0.43.0) (Bray et al., 2016). Unless differently specified, analysis was performed on the DESeq2 datasets filtered for TPM values >10, log2 Fold change absolute value of ≥1 and adjusted p-value < 0.05. Gene Ontology (GO) enrichment analysis was performed with Gene Ontology Consortium using the PANTHER classification system (Mi et al., 2013). Highly connected clusters were identified with ClusterONE plug-in for Cytoscape (Nepusz et al., 2012). Transcription factor enrichment analysis was performed with ChEA3 (Keenan et al., 2019). Motif analysis was performed with HOMER Software v4.10 on promoter sequences of input genes from 500 bp upstream and 100 bp downstream of the transcription start site (Heinz et al., 2010). For the analysis shown in Figure 6J, reads were aligned to the HSV-1 strain F complete genome (GeneBank GU734771) with Bowtie2 (v2.2.9) (Langmead and Salzberg, 2012) and further processed as described above. For heatmaps shown in Figure 5–6, only the late time points were included in the analysis and processed for DESeq2 analysis as described above. Principal component analysis on these samples was carried out with the top 1000 variable genes based on VST. Uncertainty of hierarchical clustering on the DESeq2 datasets was assessed with pvclust (Suzuki and Shimodaira, 2006). For data shown in Figure S6H, the comparison of ZIKV and HSV datasets (each a separate experiment = batch) was performed by DESeq2 with batch correction (~batch+condition). Variance-stabilizing transformations (VSTs) were batch corrected by limma with the design (~batch+condition) (Ritchie et al., 2015) and analysis was performed on genes with abs(log2FC)>1.
Quantification of apoptosis
To calculate the percentage of apoptotic cells, organoid cryosections immunostained with cleaved-caspase-3 (CC3) and DAPI were imaged under a confocal microscope. Random cortical structures on the surface of organoids were selected for quantification. The number of cells positive for CC3 in single z-plane images was counted manually using Fiji software. The total number of cells labeled by DAPI was estimated using the Fiji Plug-In GranFilter (using radius value set to 3 and step value set to 2) and watershed filter. The percentage of apoptosis is represented by the number of CC3+ cells divided by the total cell number labeled by DAPI.
Quantification of Sox1 levels
Organoid cryo-sections immunostained with Sox1 were imaged under a confocal microscope. Entire organoid sections were acquired using the “tile” and “stitching” functions in the Zen software (Zeiss). Single z-plane 8-bit images were used for quantification. Intensity (as mean gray value) of Sox1+ nuclei was measured using Fiji.
Quantification of IFN>GFP and ISRE>tdTomato positive cells
Organoid cryo-sections were immunostained with GFP and tdTomato antibodies and imaged under a confocal microscope using the “tile” and “stitching” functions in the Zen software (Zeiss). The total number of cells labeled by DAPI was estimated using the Fiji Plug-In GranFilter (using radius value set to 3 and step value set to 2) and watershed filter. Cells with high GFP signal were manually scored as IFN>GFP positive. Intensity (as mean gray value) of tdTomato signals was measured using Fiji and cells with tdTomato signal higher than the background intensity were scored as positive. Cells were scored to be within or outside the communication domain (200 μm in size) of a given IFN>GFP+ cell, a domain size compatible with paracrine signaling (Francis and Palsson, 1997).
PCR on viral nucleic acids
Viral nucleic acids were isolated from viral stocks using Purelink Viral RNA/DNA Mini Kit. PCR primer sequences are provided in the Key Resource Table. PCR amplification for TK was performed with GoTaq polymerase using the following program: 40 cycles of 30 sec at 95°C, 30 sec at 60°C, 1 min and 30 sec at 72°C, with an additional incubation of 5 min at 72°C. Due to the high GC content of the ICP34.5 target sequence and the failure of amplification by GoTaq polymerase-based approaches, PCR amplification of ICP34.5 was performed with KOD Xtreme Hot Start DNA polymerase. The program consisted of 2 min at 94°C, followed by 30 cycles of 30 sec at 98°C, 1 min at 60°C and 1 min at 68°C. PCR reactions were resolved on 1.25% agarose gels.
Quantification And Statistical Analysis
Statistical analysis was carried out with Prism software using unpaired t-test for comparison of two groups and ANOVA for comparisons of multiple groups. For non-normal distributions, non-parametric tests were used. The threshold for statistical significance was p<0.05. All details on sample size, the number of replicates, statistical tests and p values for each experiment are provided in the relevant figure, its legend or in Table S2. Unless differently specified in the figure legend, n refers to the number of replicates. Sample sizes of organoid experiments were estimated empirically based on previous experience. Organoids from the same batch were randomly allocated to different treatment groups during the first day of the experiment. Analyses were not performed blindly because of the substantial visual difference among groups.
Supplementary Material
Acknowledgements
We thank all members of the Knoblich laboratory for technical expertise and feedback. We thank the Organoid research unit, Lena Schwarz and Paul Möseneder for technical support, the IMBA/IMP BioOptics facility for flow cytometry and microscopy services, IMP/IMBA Bioinformatics for sequencing analysis, A. Kavirayani and the VBCF HistoPathology facility (www.viennabiocenter.org/facilities) for histological support and consultation, A. Sommer and VBCF Sequencing unit (www.viennabiocenter.org/facilities) for sequencing. We also thank the EVAg project, Anna Obenauf (IMP, Austria) and Luka Cicin-Sain (Helmholtz Centre for Infection Research, Germany) for providing reagents. V.K. received funding from an EMBO Fellowship (ALTF 1312-2015) and from the European Union’s Horizon 2020 research and innovation programme under the Marie Sklodowska-Curie grant agreement No. 703112. C.B. is a member of the VBC doctoral programme. Work in U.K's laboratory is supported by the Helmholtz Zukunftthema “Immunology & Inflammation” (ZT-0027). Work in A.M’s laboratory is supported by the Swedish Research Council (2018-05766). Work in J.A.K.’s laboratory is supported by the Austrian Federal Ministry of Education, Science and Research, the Austrian Academy of Sciences, the City of Vienna, the Austrian Science Fund (Special research program F78 Stem Cell, F 7803-B, and SFB-F78 P04) and a European Research Council Advanced Grant under the European Union’s Horizon 2020 program (No. 695642).
Footnotes
Author contributions
V.K. and J.A.K conceived the project and wrote the manuscript with input from all authors. V.K performed experiments, collected and analyzed the data with help from C.B. T.R.B. performed bioinformatic analysis. J.S. and U.K. contributed to the conceptualization of the project, provided reagents and technical expertise. A.C., C.S. and A.M. contributed reagents and technical expertise. R.R.C. and P.P.G. helped with experimental design and data interpretation. J.A.K. acquired funding.
Declaration of interests
J.A.K is inventor on a patent describing cerebral organoid technology and co-founder and scientific advisory board member of a:head bio AG.
Resource availability
Lead Contact
Information and requests regarding reagents and biological materials should be addressed to the Lead Contact, Dr. Jürgen Knoblich (juergen.knoblich@imba.oeaw.ac.at).
Materials availability
All unique reagents and biological materials generated in this study are available from the Lead Contact, Dr. Jürgen Knoblich (juergen.knoblich@imba.oeaw.ac.at), in compliance with Material Transfer Agreements (MTA).
Data and Code availability
RNA-seq data generated in this study have been deposited at NCBI Gene Expression Omnibus (GEO) under the accession numbers GSE123816 and GSE145496. The data that support the findings of this study are available from the Lead contact Dr. Jürgen Knoblich (juergen.knoblich@imba.oeaw.ac.at) upon reasonable request.
References
- Aho V, Myllys M, Ruokolainen V, Hakanen S, Mäntylä E, Virtanen J, Hukkanen V, Kühn T, Timonen J, Mattila K, Larabell CA, et al. Chromatin organization regulates viral egress dynamics. Sci Rep. 2017;7:3692. doi: 10.1038/s41598-017-03630-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amin ND, Paşca SP. Building Models of Brain Disorders with Three-Dimensional Organoids. Neuron. 2018;100:389–405. doi: 10.1016/j.neuron.2018.10.007. [DOI] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arao Y, Ando Y, Narita M, Kurata T. Unexpected correlation in the sensitivity of 19 herpes simplex virus strains to types I and II interferons. Journal of Interferon & Cytokine Research. 1997;17:537–541. doi: 10.1089/jir.1997.17.537. [DOI] [PubMed] [Google Scholar]
- Bagley JA, Reumann D, Bian S, Lévi-Strauss J, Knoblich JA. Fused cerebral organoids model interactions between brain regions. Nat Meth. 2017;14:743–751. doi: 10.1038/nmeth.4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker BM, Chen CS. Deconstructing the third dimension: how 3D culture microenvironments alter cellular cues. Journal of Cell Science. 2012;125:3015–3024. doi: 10.1242/jcs.079509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bale JF, Murph JR. Congenital Infections and the Nervous System. Pediatric Neurology. 1992;39:669–690. doi: 10.1016/s0031-3955(16)38370-5. [DOI] [PubMed] [Google Scholar]
- Bolen CR, Ding S, Robek MD, Kleinstein SH. Dynamic expression profiling of type I and type III interferon-stimulated hepatocytes reveals a stable hierarchy of gene expression. Hepatology. 2014;59:1262–1272. doi: 10.1002/hep.26657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolovan CA, Sawtell NM, Thompson RL. ICP34.5 mutants of herpes simplex virus type 1 strain 17syn+ are attenuated for neurovirulence in mice and for replication in confluent primary mouse embryo cell cultures. J Virol. 1994;68:48–55. doi: 10.1128/jvi.68.1.48-55.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bower JR, Mao H, Durishin C, Rozenbom E, Detwiler M, Rempinski D, Karban TL, Rosenthal KS. Intrastrain variants of herpes simplex virus type 1 isolated from a neonate with fatal disseminated infection differ in the ICP34.5 gene, glycoprotein processing, and neuroinvasiveness. J Virol. 1999;73:3843–3853. doi: 10.1128/jvi.73.5.3843-3853.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brasil P, Pereira JP, Raja Gabaglia C, Damasceno L, Wakimoto M, Ribeiro Nogueira RM, Carvalho de Sequeira P, Machado Siqueira A, Abreu de Carvalho LM, Cotrim da Cunha D, Calvet GA, et al. Zika Virus Infection in Pregnant Women in Rio de Janeiro -Preliminary Report. N Engl J Med. 2016 doi: 10.1056/NEJMoa1602412. NEJMoa1602412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray NL, Pimentel H, Melsted P, Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol. 2016;34:525–527. doi: 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
- Brown RM, Rana PSJB, Jaeger HK, O’Dowd JM, Balemba OB, Fortunato EA. Human Cytomegalovirus Compromises Development of Cerebral Organoids. J Virol. 2019 doi: 10.1128/JVI.00957-19. JVI.00957-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess DJ. Spatial transcriptomics coming of age. Nat Rev Genet. 2019;20:317–317. doi: 10.1038/s41576-019-0129-z. [DOI] [PubMed] [Google Scholar]
- Cairns DM, Rouleau N, Parker RN, Walsh KG, Gehrke L, Kaplan DL. A 3D human brain like tissue model of herpes-induced Alzheimer’s disease. Sci Adv. 2020;6:eaay8828. doi: 10.1126/sciadv.aay8828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calistri A, Parolin C, Palù G. Herpes simplex virus type 1 can either suppress or enhance human immunodeficiency virus type 1 replication in CD4-positive T lymphocytes. J Med Virol. 2003;70:163–170. doi: 10.1002/jmv.10350. [DOI] [PubMed] [Google Scholar]
- Carr DJJ, Al-khatib K, James CM, Silverman R. Interferon-beta suppresses herpes simplex virus type 1 replication in trigeminal ganglion cells through an RNase L-dependent pathway. J Neuroimmunol. 2003;141:40–46. doi: 10.1016/s0165-5728(03)00216-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carver EA, Jiang R, Lan Y, Oram KF, Gridley T. The mouse snail gene encodes a key regulator of the epithelial-mesenchymal transition. Mol Cell Biol. 2001;21:8184–8188. doi: 10.1128/MCB.21.23.8184-8188.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeran MC-J, Lokensgard JR, Schleiss MR. Neuropathogenesis of congenital cytomegalovirus infection: disease mechanisms and prospects for intervention. CliN Microbiol Rev. 2009;22:99–126. doi: 10.1128/CMR.00023-08. Table of Contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenn A, Walsh CA. Regulation of cerebral cortical size by control of cell cycle exit in neural precursors. Science. 2002;297:365–369. doi: 10.1126/science.1074192. [DOI] [PubMed] [Google Scholar]
- Chou J, Chen JJ, Gross M, Roizman B. Association of a M(r) 90,000 phosphoprotein with protein kinase PKR in cells exhibiting enhanced phosphorylation of translation initiation factor eIF-2 alpha and premature shutoff of protein synthesis after infection with gamma 134.5-mutants of herpes simplex virus 1. Proc Natl Acad Sci USA. 1995;92:10516–10520. doi: 10.1073/pnas.92.23.10516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou J, Kern ER, Whitley RJ, Roizman B. Mapping of herpes simplex virus-1 neurovirulence to gamma 134.5, a gene nonessential for growth in culture. Science. 1990;250:1262–1266. doi: 10.1126/science.2173860. [DOI] [PubMed] [Google Scholar]
- Chucair-Elliott AJ, Conrady C, Zheng M, Kroll CM, Lane TE, Carr DJJ. Microglia-induced IL-6 protects against neuronal loss following HSV-1 infection of neural progenitor cells. Glia. 2014;62:1418–1434. doi: 10.1002/glia.22689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contreras D, Arumugaswami V. Zika Virus Infectious Cell Culture System and the In Vitro Prophylactic Effect of Interferons. J Vis Exp. 2016 doi: 10.3791/54767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne CB, Lazear HM. Zika virus -reigniting the TORCH. Nat Rev Microbiol. 2016;14:707–715. doi: 10.1038/nrmicro.2016.125. [DOI] [PubMed] [Google Scholar]
- Cugola FR, Fernandes IR, Russo FB, Freitas BC, Dias JLM, Guimarães KP, Benazzato C, Almeida N, Pignatari GC, Romero S, Polonio CM, et al. The Brazilian Zika virus strain causes birth defects in experimental models. Nature. 2016:1–15. doi: 10.1038/nature18296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danastas K, Miranda-Saksena M, Cunningham AL. Herpes Simplex Virus Type 1 Interactions with the Interferon System. Int J Mol Sci. 2020;21:5150. doi: 10.3390/ijms21145150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang J, Tiwari SK, Lichinchi G, Qin Y, Patil VS, Eroshkin AM, Rana TM. Zika Virus Depletes Neural Progenitors in Human Cerebral Organoids through Activation of the Innate Immune Receptor TLR3. Cell Stem Cell. 2016 doi: 10.1016/j.stem.2016.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Lullo E, Kriegstein AR. The use of brain organoids to investigate neural development and disease. Nat Rev Neurosci. 2017;18:573–584. doi: 10.1038/nrn.2017.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drayman N, Patel P, Vistain L, Tay S. HSV-1 single-cell analysis reveals the activation of antiviral and developmental programs in distinct sub-populations. Elife. 2019;8:2503. doi: 10.7554/eLife.46339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duval K, Grover H, Han L-H, Mou Y, Pegoraro AF, Fredberg J, Chen Z. Modeling Physiological Events in 2D vs. 3D Cell Culture. Physiology (Bethesda) 2017;32:266–277. doi: 10.1152/physiol.00036.2016. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- Ferenczy MW, DeLuca NA. Epigenetic modulation of gene expression from quiescent herpes simplex virus genomes. J Virol. 2009;83:8514–8524. doi: 10.1128/JVI.00785-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferraris P, Cochet M, Hamel R, Gladwyn-Ng I, Alfano C, Diop F, Garcia D, Talignani L, Montero-Menei CN, Nougairède A, Yssel H, et al. Zika virus differentially infects human neural progenitor cells according to their state of differentiation and dysregulates neurogenesis through the Notch pathway. Emerg Microbes Infect. 2019;8:1003–1016. doi: 10.1080/22221751.2019.1637283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis K, Palsson BO. Effective intercellular communication distances are determined by the relative time constants for cyto/chemokine secretion and diffusion. Proc Natl Acad Sci USA. 1997;94:12258–12262. doi: 10.1073/pnas.94.23.12258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz-French C, Tyor W. Interferon-α (IFNα) neurotoxicity. Cytokine Growth Factor Rev. 2012;23:7–14. doi: 10.1016/j.cytogfr.2012.01.001. [DOI] [PubMed] [Google Scholar]
- Gabriel E, Ramani A, Karow U, Gottardo M, Natarajan K, Gooi LM, Goranci-Buzhala G, Krut O, Peters F, Nikolic M, Kuivanen S, et al. Recent Zika Virus Isolates Induce Premature Differentiation of Neural Progenitors in Human Brain Organoids. Cell Stem Cell. 2017 doi: 10.1016/j.stem.2016.12.005. [DOI] [PubMed] [Google Scholar]
- Garcez PP, Loiola EC, Madeiro da Costa R, Higa LM, Trindade P, Delvecchio R, Nascimento JM, Brindeiro R, Tanuri A, Rehen SK. Zika virus impairs growth in human neurospheres and brain organoids. Science. 2016;352:816–818. doi: 10.1126/science.aaf6116. [DOI] [PubMed] [Google Scholar]
- Giraldo D, Wilcox DR, Longnecker R. The Innate Immune Response to Herpes Simplex Virus 1 Infection Is Dampened in the Newborn Brain and Can Be Modulated by Exogenous Interferon Beta To Improve Survival. mBio. 2020;11 doi: 10.1128/mBio.00921-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladwyn-Ng I, Cordón-Barris L, Alfano C, Creppe C, Couderc T, Morelli G, Thelen N, America M, Bessières B, Encha-Razavi F, Bonnière M, et al. Stress-induced unfolded protein response contributes to Zika virus-associated microcephaly. Nat Neurosci. 2018;21:63–71. doi: 10.1038/s41593-017-0038-4. [DOI] [PubMed] [Google Scholar]
- Gobillot TA, Humes D, Sharma A, Kikawa C, Overbaugh J. The Robust Restriction of Zika Virus by Type-I Interferon in A549 Cells Varies by Viral Lineage and Is Not Determined by IFITM3. Viruses. 2020;12:503. doi: 10.3390/v12050503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman MJ, Caine EA, Zaitsev K, Begley MC, Weger-Lucarelli J, Uccellini MB, Tripathi S, Morrison J, Yount BL, Dinnon KH, Rückert C, et al. An Immunocompetent Mouse Model of Zika Virus Infection. Cell Host Microbe. 2018;23:672–685.:e6. doi: 10.1016/j.chom.2018.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottfried C, Bambini-Junior V, Francis F, Riesgo R, Savino W. The Impact of Neuroimmune Alterations in Autism Spectrum Disorder. Front Psychiatry. 2015;6:121. doi: 10.3389/fpsyt.2015.00121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant A, Ponia SS, Tripathi S, Balasubramaniam V, Miorin L, Sourisseau M, Schwarz MC, Sánchez-Seco MP, Evans MJ, Best SM, García-Sastre A. Zika Virus Targets Human STAT2 to Inhibit Type I Interferon Signaling. Cell Host Microbe. 2016;19:882–890. doi: 10.1016/j.chom.2016.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamel R, Dejarnac O, Wichit S, Ekchariyawat P, Neyret A, Luplertlop N, Perera-Lecoin M, Surasombatpattana P, Talignani L, Thomas F, Cao-Lormeau V-M, et al. Biology of Zika Virus Infection in Human Skin Cells. J Virol. 2015;89:8880–8896. doi: 10.1128/JVI.00354-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanners NW, Eitson JL, Usui N, Richardson RB, Wexler EM, Konopka G, Schoggins JW. Western Zika Virus in Human Fetal Neural Progenitors Persists Long Term with Partial Cytopathic and Limited Immunogenic Effects. CellReports. 2016;15:2315–2322. doi: 10.1016/j.celrep.2016.05.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatakeyama J, Wakamatsu Y, Nagafuchi A, Kageyama R, Shigemoto R, Shimamura K. Cadherin-based adhesions in the apical endfoot are required for active Notch signaling to control neurogenesis in vertebrates. Development. 2014;141:1671–1682. doi: 10.1242/dev.102988. [DOI] [PubMed] [Google Scholar]
- Härle P, Cull V, Guo L, Papin J, Lawson C, Carr DJJ. Transient transfection of mouse fibroblasts with type I interferon transgenes provides various degrees of protection against herpes simplex virus infection. Antiviral Res. 2002;56:39–49. doi: 10.1016/s0166-3542(02)00093-1. [DOI] [PubMed] [Google Scholar]
- Härle P, Lauret E, Pitha PM, De Maeyer E, Carr DJ. Expression of human and macaque type I IFN transgenes interferes with HSV-1 replication at the transcriptional and translational levels: IFN-beta is more potent than IFN-alpha 2. Virology. 2001;290:237–248. doi: 10.1006/viro.2001.1178. [DOI] [PubMed] [Google Scholar]
- He B, Gross M, Roizman B. The gamma(1)34.5 protein of herpes simplex virus 1 complexes with protein phosphatase 1 alpha to dephosphorylate the alpha subunit of the eukaryotic translation initiation factor 2 and preclude the shutoff of protein synthesis by double-stranded RNA-activated protein kinase. Proc Natl Acad Sci USA. 1997;94:843–848. doi: 10.1073/pnas.94.3.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Molecular Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hensel N, Raker V, Förthmann B, Detering NT, Kubinski S, Buch A, Katzilieris-Petras G, Spanier J, Gudi V, Wagenknecht S, Kopfnagel V, et al. HSV-1 triggers paracrine fibroblast growth factor response from cortical brain cells via immediate-early protein ICP0. Journal of Neuroinflammation. 2019;16:248–15. doi: 10.1186/s12974-019-1647-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herber S, Silva AA, Sanseverino MTV, Friedrich L, Ranieri TMS, Favreto C, Fraga LR, Terra AP, Schwartz IVD, Schuler-Faccini L. Prevalence and causes of congenital microcephaly in the absence of a Zika virus outbreak in southern Brazil. J Pediatr (Rio J) 2019;95:600–606. doi: 10.1016/j.jped.2018.05.013. [DOI] [PubMed] [Google Scholar]
- Herring JA, Elison WS, Tessem JS. Function of Nr4a Orphan Nuclear Receptors in Proliferation, Apoptosis and Fuel Utilization Across Tissues. Cells. 2019;8:1373. doi: 10.3390/cells8111373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess J, Angel P, Schorpp-Kistner M. AP-1 subunits: quarrel and harmony among siblings. Journal of Cell Science. 2004;117:5965–5973. doi: 10.1242/jcs.01589. [DOI] [PubMed] [Google Scholar]
- Honda K, Taniguchi T. IRFs: master regulators of signalling by Toll-like receptors and cytosolic pattern-recognition receptors. Nat Rev Immunol. 2006;6:644–658. doi: 10.1038/nri1900. [DOI] [PubMed] [Google Scholar]
- Honein MA, Dawson AL, Petersen EE, Jones AM, Lee EH, Yazdy MM, Ahmad N, Macdonald J, Evert N, Bingham A, Ellington SR, et al. US Zika Pregnancy Registry Collaboration Birth Defects Among Fetuses and Infants of US Women With Evidence of Possible Zika Virus Infection During Pregnancy. JAMA. 2017;317:59–68. doi: 10.1001/jama.2016.19006. [DOI] [PubMed] [Google Scholar]
- Hu B, Li X, Huo Y, Yu Y, Zhang Q, Chen G, Zhang Y, Fraser NW, Wu D, Zhou J. Cellular responses to HSV-1 infection are linked to specific types of alterations in the host transcriptome. Sci Rep. 2016;6:28075–14. doi: 10.1038/srep28075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hummer BT, Li XL, Hassel BA. Role for p53 in gene induction by double-stranded RNA. J Virol. 2001;75:7774–7777. doi: 10.1128/JVI.75.16.7774-7777.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivashkiv LB, Donlin LT. Regulation of type I interferon responses. Nat Rev Immunol. 2014;14:36–49. doi: 10.1038/nri3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadowaki M, Nakamura S, Machon O, Krauss S, Radice GL, Takeichi M. N-cadherin mediates cortical organization in the mouse brain. Dev Biol. 2007;304:22–33. doi: 10.1016/j.ydbio.2006.12.014. [DOI] [PubMed] [Google Scholar]
- Kanton S, Boyle MJ, He Z, Santel M, Weigert A, Sanchís-Calleja F, Guijarro P, Sidow L, Fleck JS, Han D, Qian Z, et al. Organoid single-cell genomic atlas uncovers human-specific features of brain development. Nature. 2019;574:418–422. doi: 10.1038/s41586-019-1654-9. [DOI] [PubMed] [Google Scholar]
- Kasmapour B, Kubsch T, Rand U, Eiz-Vesper B, Messerle M, Vondran FWR, Wiegmann B, Haverich A, Cicin-Sain L. Myeloid Dendritic Cells Repress Human Cytomegalovirus Gene Expression and Spread by Releasing Interferon-Unrelated Soluble Antiviral Factors. J Virol. 2018;92:795. doi: 10.1128/JVI.01138-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur B, Chiocca EA, Cripe TP. Oncolytic HSV-1 virotherapy: clinical experience and opportunities for progress. Curr Pharm Biotechnol. 2012;13:1842–1851. doi: 10.2174/138920112800958814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keenan AB, Torre D, Lachmann A, Leong AK, Wojciechowicz ML, Utti V, Jagodnik KM, Kropiwnicki E, Wang Z, Ma’ayan A. ChEA3: transcription factor enrichment analysis by orthogonal omics integration. Nucleic Acids Res. 2019;47:W212–W224. doi: 10.1093/nar/gkz446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keniry M, Dearth RK, Persans M, Parsons R. New Frontiers for the NFIL3 bZIP Transcription Factor in Cancer, Metabolism and Beyond. Discoveries (Craiova) 2014;2:e15. doi: 10.15190/d.2014.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Koo B-K, Yoon K-J. Modeling Host-Virus Interactions in Viral Infectious Diseases Using Stem-Cell-Derived Systems and CRISPR/Cas9 Technology. Viruses. 2019;11:124. doi: 10.3390/v11020124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krummenacher C, Baribaud I, Eisenberg RJ, Cohen GH. Cellular localization of nectin-1 and glycoprotein D during herpes simplex virus infection. J Virol. 2003;77:8985–8999. doi: 10.1128/JVI.77.16.8985-8999.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Hou S, Airo AM, Limonta D, Mancinelli V, Branton W, Power C, Hobman TC. Zika virus inhibits type-I interferon production and downstream signaling. EMBO Rep. 2016;17:1766–1775. doi: 10.15252/embr.201642627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafaille FG, Pessach IM, Zhang S-Y, Ciancanelli MJ, Herman M, Abhyankar A, Ying S-W, Keros S, Goldstein PA, Mostoslavsky G, Ordovas-Montanes J, et al. Impaired intrinsic immunity to HSV-1 in human iPSC-derived TLR3-deficient CNS cells. Nature. 2012;491:769–773. doi: 10.1038/nature11583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster MA, Knoblich JA. Generation of cerebral organoids from human pluripotent stem cells. Nat Protoc. 2014;9:2329–2340. doi: 10.1038/nprot.2014.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster MA, Renner M, Martin C-A, Wenzel D, Bicknell LS, Hurles ME, Homfray T, Penninger JM, Jackson AP, Knoblich JA. Cerebral organoids model human brain development and microcephaly. Nature. 2013;501:373–379. doi: 10.1038/nature12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanciotti RS, Kosoy OL, Laven JJ, Velez JO, Lambert AJ, Johnson AJ, Stanfield SM, Duffy MR. Genetic and serologic properties of Zika virus associated with an epidemic, Yap State, Micronesia, 2007 Emerging Infect. Dis. 2008;14:1232–1239. doi: 10.3201/eid1408.080287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Meth. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leib DA, Harrison TE, Laslo KM, Machalek MA, Moorman NJ, Virgin HW. Interferons regulate the phenotype of wild-type and mutant herpes simplex viruses in vivo. J Exp Med. 1999;189:663–672. doi: 10.1084/jem.189.4.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Deng Y-Q, Wang S, Ma F, Aliyari R, Huang X-Y, Zhang N-N, Watanabe M, Dong H-L, Liu P, Li X-F, et al. 25-Hydroxycholesterol Protects Host against Zika Virus Infection and Its Associated Microcephaly in a Mouse Model. Immunity. 2017;46:446–456. doi: 10.1016/j.immuni.2017.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Xu D, Ye Q, Hong S, Jiang Y, Liu X, Zhang N, Shi L, Qin C-F, Xu Z. Zika Virus Disrupts Neural Progenitor Development and Leads to Microcephaly in Mice. Cell Stem Cell. 2016;19:120–126. doi: 10.1016/j.stem.2016.04.017. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li RY, Tsutsui Y. Growth retardation and microcephaly induced in mice by placental infection with murine cytomegalovirus. Teratology. 2000;62:79–85. doi: 10.1002/1096-9926(200008)62:2<79::AID-TERA3>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- Li S-F, Gong M-J, Zhao F-R, Shao J-J, Xie Y-L, Zhang Y-G, Chang H-Y. Type I Interferons: Distinct Biological Activities and Current Applications for Viral Infection. Cell Physiol Biochem. 2018;51:2377–2396. doi: 10.1159/000495897. [DOI] [PubMed] [Google Scholar]
- Lima MC, de Mendonça LR, Rezende AM, Carrera RM, Aníbal-Silva CE, Demers M, D’Aiuto L, Wood J, Chowdari KV, Griffiths M, Lucena-Araujo AR, et al. The Transcriptional and Protein Profile From Human Infected Neuroprogenitor Cells Is Strongly Correlated to Zika Virus Microcephaly Cytokines Phenotype Evidencing a Persistent Inflammation in the CNS. Front Immunol. 2019;10:1928. doi: 10.3389/fimmu.2019.01928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J-Y, Kuo R-L, Huang H-I. Activation of type I interferon antiviral response in human neural stem cells. Stem Cell Res Ther. 2019;10:387–17. doi: 10.1186/s13287-019-1521-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Chen Z, Zhang X, Li S, Hui Y, Feng H, Du Y, Jin G, Zhou X, Zhang X. Protection of ZIKV infection-induced neuropathy by abrogation of acute antiviral response in human neural progenitors. Cell Death Differ. 2019;16:e119. doi: 10.1038/s41418-019-0324-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Looker KJ, Magaret AS, May MT, Turner KME, Vickerman P, Newman LM, Gottlieb SL. First estimates of the global and regional incidence of neonatal herpes infection. The Lancet Global Health. 2017;5:e300–e309. doi: 10.1016/S2214-109X(16)30362-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquez L, Levy ML, Munoz FM, Palazzi DL. A report of three cases and review of intrauterine herpes simplex virus infection. Pediatr Infect Dis J. 2011;30:153–157. doi: 10.1097/INF.0b013e3181f55a5c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsico C, Kimberlin DW. Congenital Cytomegalovirus infection: advances and challenges in diagnosis, prevention and treatment. Ital J Pediatr. 2017;43:38–8. doi: 10.1186/s13052-017-0358-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Muruganujan A, Casagrande JT, Thomas PD. Large-scale gene function analysis with the PANTHER classification system. Nat Protoc. 2013;8:1551–1566. doi: 10.1038/nprot.2013.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JA, Ding S-L, Sunkin SM, Smith KA, Ng L, Szafer A, Ebbert A, Riley ZL, Royall JJ, Aiona K, Arnold JM, et al. Transcriptional landscape of the prenatal human brain. Nature. 2014;508:199–206. doi: 10.1038/nature13185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ming G-L, Tang H, Song H. Advances in Zika Virus Research: Stem Cell Models, Challenges, and Opportunities. Cell Stem Cell. 2016;19:690–702. doi: 10.1016/j.stem.2016.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlakar J, Korva M, Tul N, Popović M, Poljšak-Prijatelj M, Mraz J, Kolenc M, Resman Rus K, Vesnaver Vipotnik T, Fabjan Vodušek V, Vizjak A, et al. Zika Virus Associated with Microcephaly. N Engl J Med. 2016:160210140106006. doi: 10.1056/NEJMoa1600651. [DOI] [PubMed] [Google Scholar]
- Nepusz T, Yu H, Paccanaro A. Detecting overlapping protein complexes in protein-protein interaction networks. Nat Meth. 2012;9:471–472. doi: 10.1038/nmeth.1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng CT, Mendoza JL, Garcia KC, Oldstone MBA. Alpha and Beta Type 1 Interferon Signaling: Passage for Diverse Biologic Outcomes. CELL. 2016;164:349–352. doi: 10.1016/j.cell.2015.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paijo J, Döring M, Spanier J, Grabski E, Nooruzzaman M, Schmidt T, Witte G, Messerle M, Hornung V, Kaever V, Kalinke U. cGAS Senses Human Cytomegalovirus and Induces Type I Interferon Responses in Human Monocyte-Derived Cells. PLoS Pathog. 2016;12:e1005546. doi: 10.1371/journal.ppat.1005546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y, Zhang X-J, Renier N, Wu Z, Atkin T, Sun Z, Ozair MZ, Tchieu J, Zimmer B, Fattahi F, Ganat Y, et al. Combined small-molecule inhibition accelerates the derivation of functional cortical neurons from human pluripotent stem cells. Nat Biotechnol. 2017;35:154–163. doi: 10.1038/nbt.3777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian X, Nguyen HN, Song MM, Hadiono C, Ogden SC, Hammack C, Yao B, Hamersky GR, Jacob F, Zhong C, Yoon K-J, et al. Brain-Region-Specific Organoids Using Mini-bioreactors for Modeling ZIKV Exposure. CELL. 2016:1–39. doi: 10.1016/j.cell.2016.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao H, Guo M, Shang J, Zhao W, Wang Z, Liu N, Li B, Zhou Y, Wu Y, Chen P. Herpes simplex virus type 1 infection leads to neurodevelopmental disorder-associated neuropathological changes. PLoS Pathog. 2020;16:e1008899. doi: 10.1371/journal.ppat.1008899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiss K, Maretzky T, Ludwig A, Tousseyn T, de Strooper B, Hartmann D, Saftig P. ADAM10 cleavage of N-cadherin and regulation of cell-cell adhesion and beta-catenin nuclear signalling. EMBO J. 2005;24:742–752. doi: 10.1038/sj.emboj.7600548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richart SM, Simpson SA, Krummenacher C, Whitbeck JC, Pizer LI, Cohen GH, Eisenberg RJ, Wilcox CL. Entry of herpes simplex virus type 1 into primary sensory neurons in vitro is mediated by Nectin-1/HveC. J Virol. 2003;77:3307–3311. doi: 10.1128/JVI.77.5.3307-3311.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rios AC, Clevers H. Imaging organoids: a bright future ahead. Nat Meth. 2018;15:24–26. doi: 10.1038/nmeth.4537. [DOI] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47-e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousso DL, Pearson CA, Gaber ZB, Miquelajauregui A, Li S, Portera-Cailliau C, Morrisey EE, Novitch BG. Foxp-mediated suppression of N-cadherin regulates neuroepithelial character and progenitor maintenance in the CNS. Neuron. 2012;74:314–330. doi: 10.1016/j.neuron.2012.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandler NG, Bosinger SE, Estes JD, Zhu RTR, Tharp GK, Boritz E, Levin D, Wijeyesinghe S, Makamdop KN, del Prete GQ, Hill BJ, et al. Type I interferon responses in rhesus macaques prevent SIV infection and slow disease progression. Nature. 2014;511:601–605. doi: 10.1038/nature13554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider WM, Chevillotte MD, Rice CM. Interferon-Stimulated Genes: A Complex Web of Host Defenses. Annu Rev Immunol. 2014;32:513–545. doi: 10.1146/annurev-immunol-032713-120231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz DA. The Origins and Emergence of Zika Virus, the Newest TORCH Infection: What’s Old Is New Again. Arch Pathol Lab Med. 2017;141:18–25. doi: 10.5858/arpa.2016-0429-ED. [DOI] [PubMed] [Google Scholar]
- Scuderi S, Altobelli GG, Cimini V, Coppola G, Vaccarino FM. Cell-to-Cell Adhesion and Neurogenesis in Human Cortical Development: A Study Comparing 2D Monolayers with 3D Organoid Cultures. Stem Cell Reports. 2021;13:265. doi: 10.1016/j.stemcr.2020.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonin Y, Loustalot F, Desmetz C, Foulongne V, Constant O, Fournier-Wirth C, Leon F, Molès J-P, Goubaud A, Lemaitre J-M, Maquart M, et al. Zika Virus Strains Potentially Display Different Infectious Profiles in Human Neural Cells. EBIOM. 2016;12:161–169. doi: 10.1016/j.ebiom.2016.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simões-Costa M, Bronner ME. Establishing neural crest identity: a gene regulatory recipe. Development. 2015;142:242–257. doi: 10.1242/dev.105445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson SA, Manchak MD, Hager EJ, Krummenacher C, Whitbeck JC, Levin MJ, Freed CR, Wilcox CL, Cohen GH, Eisenberg RJ, Pizer LI. Nectin-1/HveC Mediates herpes simplex virus type 1 entry into primary human sensory neurons and fibroblasts. J Neurovirol. 2005;11:208–218. doi: 10.1080/13550280590924214. [DOI] [PubMed] [Google Scholar]
- Sison SL, O’Brien BS, Johnson AJ, Seminary ER, Terhune SS, Ebert AD. Human Cytomegalovirus Disruption of Calcium Signaling in Neural Progenitor Cells and Organoids. J Virol. 2019:JVI.00954-19. doi: 10.1128/JVI.00954-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorgeloos F, Kreit M, Hermant P, Lardinois C, Michiels T. Antiviral type I and type III interferon responses in the central nervous system. Viruses. 2013;5:834–857. doi: 10.3390/v5030834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souza BSF, Sampaio GLA, Pereira CS, Campos GS, Sardi SI, Freitas LAR, Figueira CP, Paredes BD, Nonaka CKV, Azevedo CM, Rocha Vinicius PC, et al. Zika virus infection induces mitosis abnormalities and apoptotic cell death of human neural progenitor cells. 2016:1–13. doi: 10.1038/srep39775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun G, Chiuppesi F, Chen X, Wang C, Tian E, Nguyen J, Kha M, Trinh D, Zhang H, Marchetto MC, Song H, et al. Modeling Human Cytomegalovirus-Induced Microcephaly in Human iPSC-Derived Brain Organoids. Cell Reports Medicine. 2020:100002. doi: 10.1016/j.xcrm.2020.100002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki R, Shimodaira H. Pvclust: an R package for assessing the uncertainty in hierarchical clustering. Bioinformatics. 2006;22:1540–1542. doi: 10.1093/bioinformatics/btl117. [DOI] [PubMed] [Google Scholar]
- Thiery JP, Acloque H, Huang RYJ, Nieto MA. Epithelial-mesenchymal transitions in development and disease. CELL. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- Wang JP, Bowen GN, Zhou S, Cerny A, Zacharia A, Knipe DM, Finberg RW, Kurt-Jones EA. Role of Specific Innate Immune Responses in Herpes Simplex Virus Infection of the Central Nervous System. J Virol. 2012;86:2273–2281. doi: 10.1128/JVI.06010-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Mahalingam G, Hoover SE, Mont EK, Holland SM, Cohen JI, Straus SE. Diverse herpes simplex virus type 1 thymidine kinase mutants in individual human neurons and Ganglia. J Virol. 2007;81:6817–6826. doi: 10.1128/JVI.00166-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe M, Buth JE, Vishlaghi N, de la Torre-Ubieta L, Taxidis J, Khakh BS, Coppola G, Pearson CA, Yamauchi K, Gong D, Dai X, et al. Self-Organized Cerebral Organoids with Human-Specific Features Predict Effective Drugs to Combat Zika Virus Infection. CellReports. 2017;21:517–532. doi: 10.1016/j.celrep.2017.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y, Someya T, Nawa H. Cytokine hypothesis of schizophrenia pathogenesis: evidence from human studies and animal models. Psychiatry Clin Neurosci. 2010;64:217–230. doi: 10.1111/j.1440-1819.2010.02094.x. [DOI] [PubMed] [Google Scholar]
- Zhang F, Hammack C, Ogden SC, Cheng Y, Lee EM, Wen Z, Qian X, Nguyen HN, Li Y, Yao B, Xu M, et al. Molecular signatures associated with ZIKV exposure in human cortical neural progenitors. Nucleic Acids Res. 2016;44:8610–8620. doi: 10.1093/nar/gkw765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M, Zhang J, Phatnani H, Scheu S, Maniatis T. Stochastic expression of the interferon-β gene. PLoS Biol. 2012;10:e1001249. doi: 10.1371/journal.pbio.1001249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zika: the continuing threat. Zika: the continuing threat. Bull World Health Organ. 2019;97:6–7. doi: 10.2471/BLT.19.020119. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data generated in this study have been deposited at NCBI Gene Expression Omnibus (GEO) under the accession numbers GSE123816 and GSE145496. The data that support the findings of this study are available from the Lead contact Dr. Jürgen Knoblich (juergen.knoblich@imba.oeaw.ac.at) upon reasonable request.