Abstract
The endogenous circadian timekeeping system drives ~24 hour rhythm in the gene expression and rhythmically coordinates the physiology, metabolism and behavior in a wide range of organisms. Regulation at various levels is important for the accurate functioning of this circadian timing system. The core circadian oscillator comprises of an interlocked transcriptional-translational negative feedback loop (TTFL) that impose a substantial delay between the accumulation of clock gene mRNA and its protein to generate 24-hour oscillations. This TTFL mediated daily oscillation of clock proteins are further fine-tuned by post-translational modifications that regulate the clock protein stability, interaction with other proteins and subcellular localization. Emerging evidence from various studies indicates that besides TTFL and post-translational modifications, post-transcriptional regulation plays a key role in shaping the rhythmicity of mRNAs and to delay the accumulation of clock proteins in relation to their mRNA. In this review, we will summarize the current knowledge on the importance of post-transcriptional regulatory mechanisms such as splicing, polyadenylation, the role of RNA binding proteins, RNA methylation and microRNAs in the context of shaping the circadian rhythmicity in Drosophila and mammals. In particular, we will discuss microRNAs, an important player in post-transcriptional regulation of core-clock machinery, circadian neural circuit, clock input and output pathways. Furthermore we will provide an overview of the microRNAs that exhibit diurnal rhythm in expression and their role in mediating the rhythmic physiological processes.
Keywords: circadian, microRNA, post-transcriptional, input, output, mRNA
Introduction
Living organisms evolved by adapting to the daily cyclic fluctuations of environmental cues imposed by the rotation of the earth on its own axis. During the course of evolution, almost all organisms attained an endogenous timekeeping system to anticipate and adapt to these fluctuating environmental conditions (Johnson et al. 2003). This endogenous circadian clock perceives the environmental signals such as light, temperature and food through the input pathways. These input signals are transmitted to the central pacemaker that generates self-sustained oscillations of ~24 hours (h) and this timing information is further relayed to various downstream output pathways to generate overt rhythms in diverse behavioral, biochemical and physiological processes (Takahashi et al. 2001; Quintero et al. 2003; Lowrey and Takahashi 2004; Dubruille and Emery 2008; Allada and Chung 2010; Lowrey and Takahashi 2011; Mehta and Cheng 2013). The circadian pacemaker in Drosophila and mammals mainly comprises of conserved interlocked transcriptional/ translational feedback loops (TTFL) that drives self-sustained ~24-hour oscillation in the gene expression of the core clock components (Hardin et al. 1990; Allada et al. 2001; Yu and Hardin 2006; Lowrey and Takahashi 2011).
In Drosophila melanogaster, the TTFL consists of the positive limb with CLOCK (CLK) and CYCLE (CYC) heterodimeric transcription factor complex that activates the transcription of the repressor complex PERIOD (PER) and TIMELESS (TIM) (Figure 1). This repressor complex PER-TIM subsequently closes the loop by inhibiting the transcriptional activity of the positive limb components CLK-CYC (Konopka and Benzer 1971; Sehgal et al. 1994; Darlington et al. 1998; Allada et al. 1998; Rutila et al. 1998; Benito et al. 2007; Hardin 2011; Hurley et al. 2016). Additionally, the CLK-CYC complex activates the transcription of secondary feedback loop components vrille (vri), Pdp1ε and clockwork orange (cwo). VRI and PDP1ε rhythmically inhibit and activate the transcription of Clk respectively whereas CWO inhibits CLK-CYC transcriptional activity (Glossop et al. 2003; Cyran et al. 2003; Kadener et al. 2007). In addition to these primary and secondary feedback loops, CRYPTOCHROME (CRY) governs the light-mediated degradation of TIM and triggers the resetting of the circadian clock (Stanewsky et al. 1998; Dolezelova et al. 2007). In mammals, the transcriptional activators CLOCK and BMAL1 form the positive limb to activate the transcription of the PER (PER1, PER2, PER3) and CRY (CRY1, CRY2) repressor complex (King et al. 1997; Gekakis et al. 1998; Bunger et al. 2000; Zheng et al. 2001) (Figure 2). This repressor complex in turn inhibits the activity of CLOCK-BMAL1 (Kume et al. 1999; Griffin 1999; Sato et al. 2006). Rhythmic expression of BMAL1 is additionally regulated by a secondary feedback loop consisting of REV-ERBα and RORα proteins that inhibit or activate Bmal1 transcription respectively (Preitner et al. 2002; Guillaumond et al. 2005).
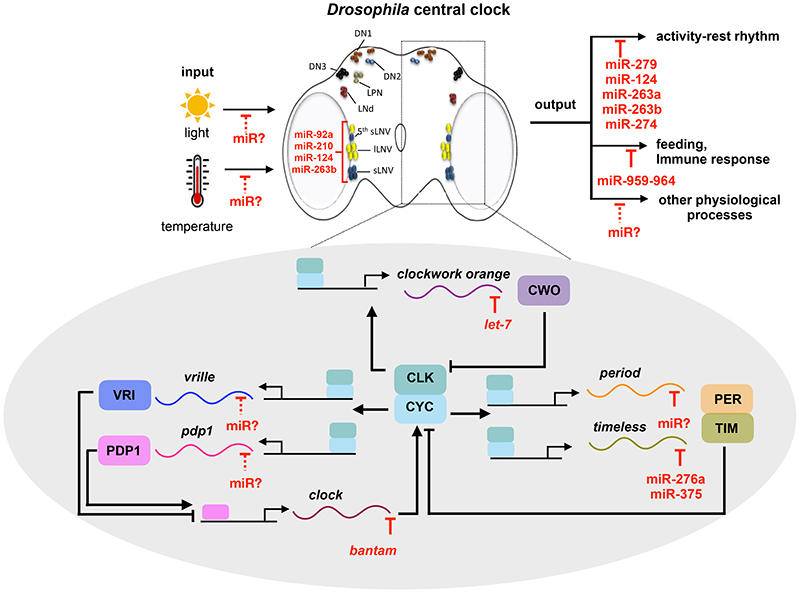
Figure 1. microRNA mediated regulation of circadian rhythm in Drosophila .
Circadian timing system is depicted with three primary components such as input pathways, the central oscillator and the output pathways. Environmental cues such as light and temperature entrain the central oscillator. Central oscillator shown in the schematic cross section of the fly brain comprises of ~150 clock neurons and it consists of large ventrolateral neurons (l-LNvs, yellow), Small ventrolateral neurons (s-LNvs, blue), lateral posterior neuron (LPN- green), dorsolateral neurons (LNds, red), and dorsal neurons (DN1- orange, DN2-blue open circle, DN3-black). The endogenous rhythmicity of the central oscillator is driven by interlocked primary and secondary transcriptional-translational feedback loop. The primary feedback loop consists of positive limb component CLK/CYC and a negative limb PER/TIM. VRI and PDP1 in the secondary feedback loop respectively inhibits and activates the transcription of Clk. In addition, the transcription repressor CWO inhibits the transcription of per and tim by competing with CLK-CYC for the E-box binding. Several microRNAs regulate the core clock components, clock neuron excitability, input and output pathways of Drosophila circadian timing system. Blunt end lines denote inhibitory effects. Solid and dashed lines denote known and putative pathways respectively.
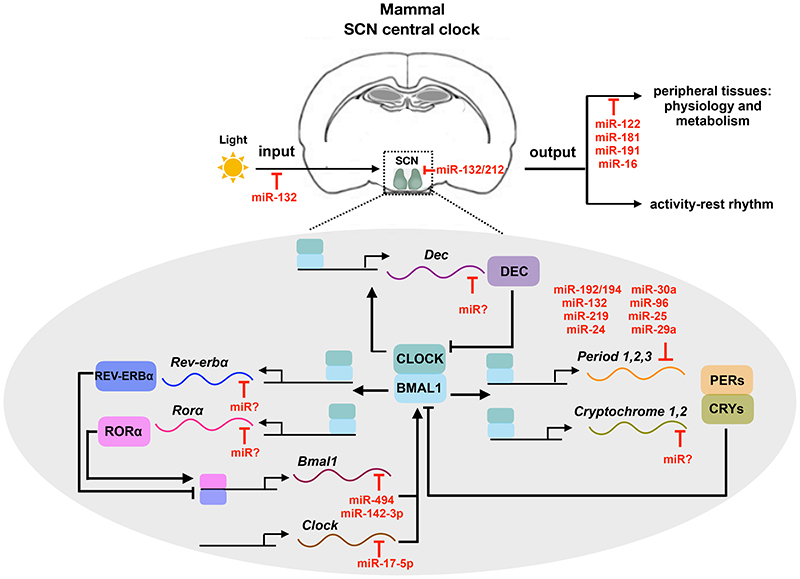
Figure 2. microRNA mediated regulation of circadian rhythm in mammals.
In mammals, light input is conveyed to the central oscillator suprachiasmatic nucleus (SCN). SCN is located at the base of the hypothalamus as shown in the schematic cross section of the adult mammalian brain. Primary and secondary feedback loops operating in the SCN neurons generates ~24 hour rhythmicity in behaviour and physiological outputs in the peripheral tissues. The primary TTFL consists of the negative arm PER/CRY and a positive arm CLOCK/BMAL1. The secondary feedback loop components REV-ERB and ROR repress and activate the Bmal1 transcription respectively. Furthermore Differentiated embryo chondrocyte (Dec) transcription factor interacts with Bmal1 and inhibits CLOCK/BMAL1 induced transcription. Various microRNAs regulate the core clock components, clock neuron morphology, input and output pathways of mammalian clock system. Blunt end lines denote inhibitory effects. Solid and dashed lines denote known and putative pathways respectively.
Apart from the TTFL mediated clock gene expression regulation, evidence from several studies indicates the importance of different post-translational modifications such as phosphorylation, acetylation, SUMOylation, ubiquitination, O-GlcNAcylation and poly (ADP-ribosyl)ation in mediating the stability, subcellular compartmentalisation and the rhythmic abundance of clock proteins for establishing the pace of the circadian clock with ~24h rhythmicity (Martinek et al. 2001; Lee et al. 2001; Miyazaki et al. 2004; Cyran et al. 2005; Hirayama et al. 2007; Asher et al. 2010; Reischl and Kramer 2011; Weber et al. 2011). For instance, kinase mediated phosphorylation governs the stability of clock proteins in both flies and mammals (Reischl and Kramer 2011). In Drosophila, DOUBLE TIME (DBT) and SHAGGY phosphorylate PER and TIM respectively to fine-tune circadian oscillations (Kloss et al. 1998; Martinek et al. 2001; Cyran et al. 2005). Another possible mode of circadian clock regulation is at the post-transcriptional level. In addition to understanding the importance of post-transcriptional regulation in the robustness of the circadian timing system, it is important to understand the role of different RNA specific regulatory mechanisms in governing the time lag between clock gene mRNA and protein oscillation. Recent studies recognised the relevance of a new class of post-transcriptional regulators, microRNAs (miRNAs) in regulating the diverse aspects of circadian rhythms (Kojima et al. 2011; Mehta and Cheng 2013; Mendoza-Viveros et al. 2017; Xue and Zhang 2018). In this review, we will provide an overview of the current knowledge on the post-transcriptional regulation of the circadian clock by emphasizing the findings on circadian relevant microRNAs in Drosophila and mammalian model systems. We will further discuss about the miRNAs that undergo circadian cycling and its impact on rhythmic physiological processes.
Post-transcriptional regulation of circadian rhythm
Multiple layers of post-transcriptional regulation ranging from splicing, polyadenylation at the 3’end and transport of mRNA from the nucleus have been reported to affect the phase, amplitude and free running periodicity of circadian rhythm (Green 2018). 3’ untranslated regions (3’ UTRs) of mRNAs have specific binding sites for RNA binding proteins (RBPs) or microRNAs (miRNAs) and these complicated series of post-transcriptional mechanisms collectively plays a key role in shaping the mRNA expression rhythmicity and in delaying the accumulation of clock proteins in relation to their respective mRNAs (Chen et al. 2013). This section summarizes various post-transcriptional regulatory processes and their relevance in the context of circadian rhythmicity.
Post-transcriptional regulation of circadian clock by alternative splicing
The functional importance of alternative splicing on the circadian clock has been recognized in Drosophila by studying the alternative splice variant isoforms of core clock genes. Studies showed that light and temperature modulate the alternative splicing of an intron (dmpi8 intron) located in the 3’ UTR of period mRNA and thus conferring the fly the ability to redistribute its activity in accordance with the seasonal changes in temperature and day length (Majercak et al. 1999; Majercak et al. 2004). Lower temperature enhances the splicing of dmpi8 intron. This enhanced splicing is associated with an advance in the steady-state phase of the PER protein that attributes to the earlier evening activity of flies under cold temperature (Majercak et al. 1999). Further studies showed that in addition to low temperature, short photoperiod also enhances dmpi8 intron splicing and no receptor potential A (norpA) gene that encodes phospoholipase C plays a non-photic role in regulating the dmpi8 splicing (Collins et al. 2004; Majercak et al. 2004). In addition, the serine/arginine family of protein splicing factor B52/SRp55 enhances the dmpi8 splicing efficiency and regulates mid-day sleep in Drosophila (Zhang et al. 2018).
Apart from affecting the per mRNA alternative splicing, temperature alters the splicing pattern of the core clock component timeless in Drosophila. Alternative splicing generates four major isoforms of tim (Martin Anduaga et al. 2019). The levels of the canonical transcript tim-long was reduced under lower temperature (18˚C) whereas the production of two tim cold-specific isoforms such as tim-cold and tim-short and cold were up-regulated under lower temperature. At higher temperature (29°C) tim-medium was abundantly present. The alternative splicing of tim intron sequences regulates the level of TIM at various temperatures and acts as the thermal sensor to adapt to the temperature changes by regulating the daily pattern of locomotor activity (Martin Anduaga et al. 2019). Further studies revealed that the splicing factor P-element somatic inhibitor (PSI) regulates the alternative splicing of tim and governs the pace of the circadian clock (Foley et al. 2019). Taken together, these studies collectively elucidated the importance of alternative splicing of per and tim mRNA on the environmental adaptation of the circadian timekeeping system.
In mice, the first evidence for light-induced rhythmic alternative splicing associated with the circadian clock was reported for the mRNA of U2-auxiliary-factor 26 (U2AF26). A circadian light-inducible splicing switch excludes exons 6 and 7 inducing a frameshift in U2af26 resulting in translation past into the 3’UTR and generates a C terminus with the TIM homology domain. This alternative U2AF26 isoform controls the stability of PER1 protein and clock resetting (Preußner et al. 2014). Additionally, temporal analysis of SCN by using RNA sequencing characterized a novel isoform of clock gene Cry1 expressed antiphasic to the canonical Cry1 oscillation that may be important in the SCN function (Pembroke et al. 2015). Studies in the human keratinocyte cell line provided the first evidence for the presence of the Per2S splice variant of the human Per2 gene (Avitabile et al. 2014). Although studies in mammalian system suggest that alternative splicing plays a key role in stabilizing the clock against the light changes (Preußner et al. 2014), the physiological importance of splicing event of various clock pre-mRNAs including Cry1 and Per2 remains elusive.
Relevance of poly(A) tail length in circadian rhythmicity
Among the post-transcriptional regulations, the poly(A) tail 3’ modification is not only important in mediating the mRNA stability and translation efficiency but also impacts the rhythmic gene expression in the context of circadian rhythmicity (Grima et al. 2019). Recent evidence suggests that mRNA with shorter poly(A) tails are translated more efficiently than the mRNA molecules with longer tails (Lima et al. 2017). In Drosophila, CCR4-NOT complex deadenylates mRNA and regulates the poly(A) length (Temme et al. 2014). Studies showed that the deadenylase component of CCR4-NOT complex encoded by Pop2 (homolog of Schizosaccharomyces pombe caf1) govern the mRNA oscillation and protein levels of the core clock gene timeless by regulating the polyadenylation of its mRNA. POP2 shortens the tim mRNA poly(A) tail and thus modulates the tim mRNA and protein level. Thus POP2 dependent deadenylation mechanism strongly contributes to the oscillation of core clock gene transcript and this deadenylation process is controlled by PERIOD (Grima et al. 2019).
In mouse, genome-wide poly(A) adenylome analysis by microarray studies showed that 2.5% of transcripts expressed in the liver undergo circadian oscillation in poly(A) tail length. This rhythmicity in the poly(A) length variation contributes to the rhythmic translation and protein abundance (Kojima et al. 2012). In addition, the mRNA of the deadenylase Nocturnin is rhythmically expressed in various circadian clock gene-expressing tissues such as the SCN, pineal gland, liver, kidney and testis indicating the importance of Nocturnin as a potential circadian regulatory component (Wang et al. 2001). It is likely that rhythmically expressing Nocturnin regulates the clock output pathway associated with metabolic regulation (Green et al. 2007). Although studies revealed the post-transcriptional contribution of circadian deadenylase Nocturnin in regulating the amplitude of metabolic pathways, it is worth to explore whether other deadenylases such as CNOT6/7/8 deadenylases of the mammalian CCR4–NOT complex play any role in posttranscriptional regulation of the core clock components (Stubblefield et al. 2018; Grima et al. 2019).
RNA binding proteins in circadian rhythm regulation
Accumulating evidence suggests that RNA binding proteins (RBP) and ribonucleoprotein (RNP) complexes regulate the expression of core clock genes by contributing to certain mRNA processing steps such as mRNA capping, splicing and intracellular transport of mRNA. A recent study reported the importance of the spliceosomal complex in the circadian regulation of Drosophila (Shakhmantsir et al. 2018). Spliceosome, RNA-protein complex is composed of five small nuclear ribonucleoprotein particles such as U1, U2, U4, U5, U6 snRNP and additional proteins associated with each snRNP complex (Wahl et al. 2009). RNA interference screen studies showed that pre-mRNA processing factor 4 (PRP4), a component of the U4/U5.U6 triple small nuclear ribonucleoprotein (tri-snRNP) complex and multiple tri-snRNP components regulate the molecular clock and free running periodicity. PRP4 contributes to the differential splicing of tim and thereby delays the accumulation of TIM in a 24h cycle (Shakhmantsir et al. 2018). This study shed light on a possible mechanism that delays the TIM protein accumulation in comparison to its mRNA oscillation. Another study reported the contribution of a novel gene twenty-four (tyf) in the lag between PER protein accumulation and mRNA in circadian pacemaker neurons. Mutants of tyf gene displayed weak behavioral rhythmicity associated with a reduction in the level of PER protein in Drosophila (Lim et al. 2011). TYF is associated with the 5′-cap binding complex, poly(A)-binding protein (PABP) and the clock gene per transcript, and it promotes the PER translation in pacemaker neurons in the fly brain to sustain the robust behavioral rhythmicity (Lim et al. 2011). Taken together, these studies decoded the importance of RBPs in post-transcriptional regulatory processes to delay the clock protein accumulation relative to that of its transcript oscillation.
A separate study deciphered the importance of RBP in clock output pathways. It was postulated that Drosophila RNA-binding protein LARK is required to transfer the temporal information from the molecular core clock to different output behavioral pathways through post-transcriptional repression of the distinct RNA targets (Newby and Jackson 1996; Schroeder et al. 2003; Huang et al. 2009). Biochemical and genetic analyses showed that LARK is associated with a large number of target RNA molecules and LARK selectively regulates the adult emergence rhythm by coordinating the temporal expression of Ecdysone induced protein-74 (E74) (Newby and Jackson 1996; Huang et al. 2007). While LARK regulates the output behavioral rhythm in Drosophila, it indeed acts as a post-transcriptional regulator of the mammalian circadian clock. In the mouse circadian pacemaker, LARK protein oscillates in phase with the PER1 protein. Further studies showed that LARK interacts with the Per1 3’UTR and post-transcriptionally regulates the expression of core clock component Per1 (Kojima et al. 2007).
Studies in cultured fibroblasts showed that temperature cycles govern the rhythmic expression of cold-inducible RNA-binding protein (CIRP). CIRP maintains the robustness of the circadian timing system by binding to the transcripts of various core clock genes such as Clock, Period 3, Rorα and clock associated gene Sirtuin 1 (Morf et al. 2012). Further studies identified another novel post-transcriptional mechanism in the thermal entrainment of the circadian clock mediated via two cold-induced RBPs, Cirbp and Rbm3 in mouse (Liu et al. 2013). This study showed that Cirbp and Rbm3 control the high-amplitude oscillation of temperature entrained clock gene expression by regulating the alternative polyadenylation of the target genes. Furthermore, low temperature lengthened the 3’UTR and significantly upregulated Cirbp and Rbm3. It was found that these two RBPs repress the usage of proximal polyadenylation site (PAS) by binding to the 3’UTR sites. This study implied the potential function of Cirbp and Rbm3 in temperature entrainment of the circadian clock by regulating the alternative polyadenylation of core clock components such as Bmal1, Clock, Per1 and Cry1 (Liu et al. 2013).
RNA methylation and circadian rhythm
RNA methylation is a lesser known process compared to DNA or protein methylation. However, the importance of RNA methylation as a key post-transcriptional regulatory mechanism is being increasingly studied in recent years. A landmark study conducted in mice embryonic fibroblasts showed that global down regulation of RNA methylation by reducing the methylation potential of the cells resulted in increased circadian period (Fustin et al. 2013). A similar increase in period of the locomotor activity rest rhythm was observed when RNA methylation was down regulated in the SCN. Transcriptome analysis by RNA-Sequencing in the same study revealed that the transcription of the RNA processing machinery is altered by RNA methylation and RNA immunoprecipitation with anti-m6A antibody (MeRIP) showed that a number of circadian relevant transcripts such as Per1, Per2, Per3, Clock, Arntl have methylation sites on their transcripts (Fustin et al. 2013). A subsequent study showed that the RNA methylation levels in the mice liver RNA exhibit circadian rhythm and the Cry 1/2- knockout mice had significantly lowered levels of m6A on their mRNAs and the circadian rhythm of m6A methylation was lost in them compared to the wild type mice liver (Wang et al. 2015). These studies suggest that the relationship between the m6A methylation and circadian clock is bidirectional with the RNA methylation status affecting the speed of the circadian clock and the circadian clock in turn regulating the levels of RNA methylation in the liver transcripts.
Another study shows an important role of RNA methylation on the circadian regulation of hepatic lipid metabolism. This study showed that the deletion of hepatic Bmal1 lead to altered expression and diurnal variations in the levels of methyl transferases, demethylases and methyl specific binding proteins and in turn lead to an increased mRNA methylation particularly of peroxisome proliferator-activator α (PPaRα), a major regulator of hepatic lipid metabolism (Zhong et al. 2018). The exact mechanism of how RNA methylation regulates the circadian clock or how it regulates the circadian control of metabolism is still unclear. However a recent study discusses some of the theoretically possible mechanisms by which RNA methylation can affect the period. This study suggests two possible mechanisms by which the decreased RNA methylation can result in period lengthening, either an increased stabilization of non oscillatory mRNAs such as casein kinase 1δ (CK1δ) that phosphorylates clock component and a subsequent distortion of the circadian waveform or an increased stabilization of oscillatory mRNAs such as Per and Cry (Gibo and Kurosawa 2020). These studies suggest that RNA methylation is critical for circadian function and for circadian regulation of downstream genes.
microRNA mediated regulation of circadian rhythm
miRNA mediated post-transcriptional regulation has emerged as another major player in circadian timekeeping. miRNAs bind with the complementary target sites within the 3’UTR of selected mRNAs and down-regulate the gene expression through mRNA degradation or translational repression (Bartel 2004). Studies indicate that miRNAs regulate the circadian gene expression and play an important role in generating the time delay in circadian feedback inhibition, thereby fine-tune 24-hr circadian rhythmicity (Kadener et al. 2009; Chen et al. 2013; Chen et al. 2014). In the following section, we summarize the current understanding of the miRNA-mediated regulation of core clock machinery, circadian neural circuit excitability, clock input, output pathways and peripheral clocks based on the studies carried out in flies (Figure 1, Table 1) and mammals (Figure 2, Table 1).
Table 1. miRNAs involved in circadian regulation.
List of miRNAs present in Drosophila and mammals that are involved in regulating different aspects of the circadian clock. The list includes the targets of miRNAs and the target validation techniques used.
| Organism | microRNA | Target | Target validation technique used | |
|---|---|---|---|---|
| Core clock machinery | Drosophila | bantam | Clk (Kadener et al. 2009) | Luciferase Reporter Assay |
| let-7 | cwo (Chen et al.2014) | Luciferase Reporter Assay | ||
| miR-276a | tim (Chen and Rosbash 2016) | Luciferase Reporter Assay | ||
| miR-375 | tim (Xia et al. 2020) | qRT-PCR and Western Blot | ||
| Mammals | miR-132 | (Cheng et al. 2007) | ||
| miR-219 | (Cheng et al. 2007) | |||
| miR-192/194 cluster | Per1,Per2,Per3(Nagel et al. 2009) | Luciferase Reporter Assay | ||
| miR-494 | Bmal1 (Shende et al. 2011) | Luciferase Reporter Assay | ||
| miR-142-3p | Bmal1 (Shende et al. 2011; Shende et al. 2013) | Luciferase Reporter Assay | ||
| miR-24 | Per1 (Chen et al. 2013), Per2(Chen et al. 2013;Yoo et al. 2017;Park et al. 2020) | Luciferase Reporter Assay | ||
| miR-30a | Per1 (Chen et al. 2013) | Luciferase Reporter Assay | ||
| miR-29a | Per1 (Chen et al. 2013) | Luciferase Reporter Assay | ||
| miR-17-5p | Clock (Gao et al. 2016) | Luciferase Reporter Assay | ||
| miR-25 | Per2 (Park et al. 2020) | Luciferase Reporter Assay | ||
| miR-96 | Per2 (Zhou et al. 2021) | Luciferase Reporter Assay | ||
| Clock neuronal circuitry | Drosophila | miR-92a | sirt2 (Chen and Rosbash 2017) | Luciferase Reporter Assay |
| miR-210 | (Cusumao et al. 2018) | |||
| miR-263b | Bx(Nian et al. 2019) | Luciferase Reporter Assay | ||
| Mammals | miR-132/212 | MeCP2 (Mendoza-Viveros et al. 2017) | Phenotypic Rescue Experiment | |
| Clock input pathway | Mammals | miR-132 | MeCP2, Jarid1a, Ep300,Btg2,Paip2a (Alvarez-Saavedra et al. 2011) | Luciferase Reporter Assay |
| Clock output pathway and peripheral tissues | Drosophila | miR-279 | Upd (Luo and Sehgal 2012) | Luciferase Reporter Assay |
| miR-959-964 Cluster | (Vodala et al. 2012) | |||
| miR-124 | (Zhang et al 2016) | |||
| miR-274 | MESK2, CG4328 (You et al. 2018) | Phenotype study using RNAi line | ||
| miR-263b | You et al.(2018) | |||
| miR-210 | Fas2(Niu et al. 2019) | Phenotype study in mutated Fas2 3’UTR flies | ||
| miR-263a | slo, homer(Nian et al 2020) | Luciferase Reporter Assay | ||
| Mammals | miR-122 | Pparβ/δ, Smarcd1/Baf60a (Gatfield et al. 2009), Nocturnin (Kojima et al. 2010) | Luciferase Reporter Assay | |
| miR-16 | (Balakrishnan et al. 2010) |
Regulation of core clock machinery by miRNAs
To assess the role of miRNA in Drosophila core clock machinery, studies were conducted by impairing the miRNA biogenesis pathway and it severely dampened the activity-rest rhythms indicating the importance of miRNAs for maintaining robust circadian rhythmicity (Kadener et al. 2009). Subsequent studies used tiling array to identify the potential miRNAs that were responsible for maintaining the robustness of circadian rhythm, and it was found that 27 miRNAs are expressed in the circadian tissue (Kadener et al. 2009). Out of these 27 miRNAs, further biochemical experiments focused on bantam showed that evolutionarily conserved bantam binding target sites were located in the core clock component Clk 3’UTR (Kadener et al. 2009). Overall, the results of this study indicated the importance of miRNAs in circadian rhythmicity, especially bantam regulating the Clk expression in the circadian pacemaker to sustain the pace of the circadian clock (Kadener et al. 2009). Further studies identified let-7 as a potential miRNA involved in the regulation of another core clock component in Drosophila. let-7 regulates locomotor activity circadian rhythm through repression of the circadian gene clockwork orange (cwo) expression in the central clock (Chen et al. 2014). cwo is rhythmically expressed and contributes to the robustness of core clock gene oscillation (Lim et al. 2007; Kadener et al. 2007; Matsumoto et al. 2007). Studies also reported a third microRNA, miR-276a that regulates robust circadian behavioral and molecular rhythms by binding to a single target site in the 3’UTR of the core clock gene tim and suppress the expression of TIM in Drosophila (Chen and Rosbash 2016). Furthermore, a recent study based on the CLEAR-CLIP analysis showed that miR-375 modulates circadian rhythm and sleep by targeting tim in Drosophila (Xia et al. 2020). This study also identified thousands of miRNA-mRNA interactions and disruption of Clk affected this miRNA-mRNA interactome. In particular, the miRNA-mRNA circadian relevant interactions involving tim, vri and pdp1 exhibited distinct changes in ClkJrk mutants. (Xia et al. 2020).
miRNAs that govern mammalian core clock timing mechanisms have also been identified. A forward genetic screen by using miRNA expression library in HeLA cells identified miR-192/194 gene cluster as a new mechanism for the post-transcriptional regulation of mammalian core clock machinery by negatively regulating the expression of the entire Per gene family, which consists of Per1, Per2 and Per3 (Nagel et al. 2009). Subsequent studies carried out in NIH3T3 cells also yielded similar results where overexpression of mR-192/194 altered the circadian periodicity by regulating the expression of Per genes (Nagel et al. 2009). Another study conducted in HEK293 cells showed that co-expression miR-132 and miR-219 augmented CLOCK/BMAL1 dependent Per1 transcription suggesting the role of these microRNAs in core clock timing mechanism by acting as the positive modulators of CLOCK/BMAL1 dependent Per1 transcription (Cheng et al. 2007). TargetScanMouse algorithm was used in a successive study to identify other miRNAs targeting the 3’-UTR of Per1 and Per2 mRNA. This study identified the three most promising miRNAs miR-24, 29a and 30a that interact with the 3’-UTR of Per1 and Per2 mRNA to affect the stability and translation efficiency of these core clock gene mRNAs (Chen et al. 2013). Furthermore, this study used Dicer mutant and showed that miRNAs delay the cytoplasmic accumulation of PER and thus miRNAs are critical to generate time delay for the feedback loop inhibition of the circadian oscillator (Chen et al. 2013).
Another study on Per2::LUCIFERASE (Per2::LUC) circadian reporter mouse lines assessed the role of Per2 3’-UTR in the post-transcriptional regulation of circadian clock. The Per2::LUC reporter mice retained the endogenous 3’-UTR region of the Per2 gene, whereas Per2 3′-UTR was replaced by an SV40 late poly(A) signal in Per2::LucSV mice. SCN and tissue explants from Per2::LucSV mice showed enhanced circadian bioluminescence level and oscillatory amplitude than the Per2::LUC reporter mice. Further molecular and genetic analysis showed that miR-24 binds to the Per2 3′-UTR to attenuate PER2 protein translation. This study provided important insights into the regulatory role of Per2 3′-UTR and miR-24 in core clock function (Yoo et al. 2017). A recent study used in silico algorithms and found that the novel miRNA-25 fine-tunes the circadian rhythmicity by repressing Per2 expression (Park et al. 2020). Luciferase reporter assays validated the predicted binding sites of miR-25 in the 3’ UTR of Per2 mRNA (Park et al. 2020). One more recent study used a genome-wide miRNA screen and identified miRNA cluster miR-183/96/182 that modulates circadian rhythms. Potential targets of this miRNA cluster were predicted by using computational methods miRanda and DIANA-microT-CDS. Both the methods predicted and luciferase reporter assay validated Per2 as the direct target of miR-96 (Zhou et al. 2021). In summary, these studies elucidated the role of miR-192/194, miR-132, miR-219, miR-24, miR-29a, miR-30a, miR-25 and miR-96 in post-transcriptional regulation of circadian rhythm by regulating the expression of the entire Per gene family.
miRNAs are also known to regulate various integral components of the mammalian core clock machinery other than the per. The pre-miRNA overexpression constructs of miR-494 and miR-142-3p in HEK293 cells and SCN cells repressed Bmal1 3′ UTR activity indicating the importance of these microRNAs as the posttranscriptional regulators of core clock element Bmal1 (Shende et al. 2011; Shende et al. 2013). Furthermore, studies showed that miR-17-5p is rhythmically expressed in SCN and inhibits the translation of Clock by binding the 3’UTR of its mRNA and shorten the circadian period. This study demonstrated CRY1 as an important output molecule for miR-17-5p mediated stabilization of the circadian period (Gao et al. 2016).
Regulation of clock neural circuit morphology and excitability
In Drosophila, approximately 150 neurons in the brain are important for orchestrating the rhythmicity in behavior. This circadian neuronal network consists of subsets of dorsal neurons (DNs), dorsal lateral neurons (LNd), small and large ventral lateral neurons (sLNv and lLNv), and lateral posterior neurons (LPN) (Helfrich-Förster 1997; Kaneko et al. 1997; Kaneko and Hall 2000; Klarsfeld 2004; Shafer et al. 2006). Four of the five sLNvs and all the lLNvs express circadian neuropeptide Pigment dispersing factor (PDF) (Shafer et al. 2008). PDF cells mediate the circadian period and the PDF cell termini undergo circadian morphological changes during the daily fasciculation and defasciculation remodelling cycles (Fernández et al. 2008; Guo et al. 2014). A growing body of evidence suggests that miRNAs regulate neuronal activity and synaptic plasticity in Drosophila and mammals (Tan et al. 2013; Verma et al. 2015). Extensive studies by using ArcLight, GCaMP6 imaging and In vivo calcium monitoring showed that miR-92a suppresses PDF neuronal excitability in flies. Adult stage-specific manipulation of miR-92a levels altered the fasciculation-defasciculation state of the PDF cell termini. Results obtained from translating ribosome affinity purification (TRAP) and miRNA target prediction tool Targetscan indicated that miR-92a suppresses neuronal excitability partly by targeting sirtuin 2 (sirt2) in PDF cells (Chen and Rosbash 2017).
A consecutive study investigated the effect of misexpressing miR-210 in circadian behavior and on circadian clock neurons. Overexpression of miR-210 altered the projections and morphology of the PDF expressing clock neurons. It also increased PER levels without affecting it’s cycling in the clock neurons and modulated the phase of activity-rest rhythm (Cusumano et al. 2018). miR-263b was also identified as an important regulator of circadian rhythm and structural plasticity of PDF positive sLNv axonal projections. In silico prediction algorithm was used to identify the potential targets of miR-263b and the results of subsequent genetic analysis showed that miR-263b regulates the arborisation rhythms of sLNv dorsal projections and activity-rest rhythm by suppressing the expression of LIM-only protein Beadex (Bx) (Nian et al. 2019).
In mammals, the suprachiasmatic nucleus (SCN) of the hypothalamus functions as the central circadian pacemaker. In addition to directing the daily behavioral rhythm, it senses changes in day length and seasonal adaptation relies predominantly on the structural plasticity of SCN neuronal network (Meijer et al. 2010). miR-132/212 modulates entrainment to various photoperiods and day length mediated regulation of PER2 expression in the SCN (Mendoza-Viveros et al. 2017). Proteomic analysis also showed that miR-132/212 play a key role in regulating the seasonal changes in the dendritic morphology of circadian pacemaker neurons. miR-132/212 mediate this dendritic morphology changes through the direct target, methyl CpG binding protein 2 (MeCP2) dependent mechanism and thus enables the SCN to encode photoperiod changes (Mendoza-Viveros et al. 2017).
Regulation of clock input pathways
In Drosophila, no miRNA has been implicated in the regulation of circadian input pathways yet. Whereas in mammals, miR-132 has been implicated in the photic input pathways of the circadian clock. Knockdown of miR-132 in the SCN enhanced the light-induced resetting effects of the clock and overexpression attenuated the clock resetting effects of light indicating that miR-132 acts as a negative regulator of photic entrainment (Cheng et al. 2007). A follow-up study used in silico analysis and in vitro 3’ UTR assays to reveal the potential targets of miR-132. miR-132 regulates the expression of chromatin remodelling related target genes such as Mecp2, Jarid1a and Ep300. This study collectively showed that miR-132 mediates the circadian clock entrainment through its downstream targets involved in chromatin function and protein translation within SCN (Alvarez-Saavedra et al. 2011). In the chicken retina, miR-26a exhibits diurnal oscillation regulated by CLOCK. miR-26a drive circadian rhythm in the production of the L-type voltage-gated calcium channel α1C subunit in chicken cone photoreceptors (Shi et al. 2009). Given that L-type calcium channels play a central role in rhythmic melatonin synthesis, miR-26a could play an important role in circadian retinal physiology and light input (Ivanova and Iuvone 2003)
Regulation of clock output and peripheral oscillators
In addition to regulating the core clock machinery and the clock neuron morphology, microRNAs are important regulators of various clock output pathways. In Drosophila, miR-279 governs the activity-rest rhythm behavioral output without affecting the central clock. Further studies by using computational algorithms such as PicTar, TargetScanFly5.1, MiRanda and RNAi knockdown studies on transgenic flies revealed that miR-279 target Unpaired (Upd), the ligand for JAK/STAT pathway regulates the activity-rest rhythm in Drosophila (Luo and Sehgal 2012). In addition, miR-124 has a striking impact on the phase of circadian locomotor activity rhythm. Flies lacking miR-124 advanced the phase of activity-rest rhythm under constant darkness without altering the phase of the circadian pacemaker in the brain that controls this behavioral rhythm. This indicates that miR-124 shapes the diurnal activity and determines the phase of the behavioral output without affecting the central clock (Zhang et al. 2016). A recent study identified miR-263a as an important regulator of activity-rest rhythm. Downregulation of miR-263a in the clock neurons reduced the morning peak of activity and the robustness of the circadian rhythm. Moreover, overexpression of miR-263a in the clock neurons lengthened the periodicity. Further in-vitro assays showed that miR-263a suppress the potential target slo that encodes the structural alpha subunit of a BK calcium-activated potassium channel and activates homer that acts as an adaptor binding to the group I metabotropic glutamates and other intracellular signaling proteins (Nian et al. 2020). Both slo and homer contribute to the circadian locomotor activity (Jepson et al. 2012; Naidoo et al. 2012).
While miR-279, miR-124 and miR-263a regulate activity-rest rhythm without affecting the central clock, let-7 regulate locomotor activity behavioral output through the circadian gene cwo in the central pacemaker (Chen et al. 2014). miR-92a modulates neuronal excitability of PDF cells and this regulation is not only restricted to PDF cells. miR-92a regulates sleep and phase shift response through the mRNA target sirt2 expressed in the dopaminergic neurons and PDF cells respectively (Chen and Rosbash 2017). Furthermore, miR-210 regulates the phase of activity rest rhythm through repression of the cell adhesion molecule Fasciclin 2 (Fas 2) (Niu et al. 2019). miR-210 modulates the morphology of clock neurons and the levels of PER without affecting the phase of its oscillation indicating that miR-210 plays a vital role in the circadian output pathways (Cusumano et al. 2018).
Accumulating evidence from various studies suggest that glial specific vesicle trafficking and release factors modulate the circadian rhythmic behavior (Ng et al. 2011; Ng and Jackson 2015; Ng et al. 2016). Glial secretion also modulates the PDF level within the s-LNv clock neuron projections indicating the importance of glial-neuronal communication in circadian rhythmicity (Ng et al. 2011). Studies addressed whether microRNAs expressed in astrocytes regulate circadian rhythmicity. The results of these studies showed that astrocyte-specific genetic inhibition of miR-263b and miR-274 in adult flies impedes the activity-rest rhythm without affecting the molecular clock and clock neuron viability (You et al. 2018). In silico procedures and genetic screens were used to identify the potential RNA targets of these microRNAs and genetic inhibition of miR-274 targets CG4328 (a predicted LIM-homeodomain transcription factor) and MESK2 (involved in Ras/ERK signaling and has homology with N-myc downstream-regulated gene 2) resulted in significantly reduced rhythmicity. This genetic screen identified the putative RNA targets of miR-274 in glia that might regulate circadian locomotor rhythm in Drosophila (You et al. 2018).
Apart from regulating the activity-rest rhythm, microRNAs also regulate other behavioral outputs. For example, a cluster of six microRNAs miR-959-964 undergo robust circadian cycling with a peak at ~ZT12 or close to lights off. Analysis of this cluster knock out and overexpression showed that these miRNAs impact circadian outputs such as innate immunity, metabolism, and feeding time. The circadian clock mediated timing of food intake in turn regulates the cluster miRNA cycling. These observations indicate that post-transcriptional feedback of cycling cluster miRNAs is involved in the timing of feeding (Vodala et al. 2012).
In mammals, studies were focused on the role of miRNA in peripheral clock tissues that drives the rhythm in physiological outputs such as lipid metabolism in the liver and enterocyte proliferation in the intestine. For instance, miR-122 was identified as a potential candidate that might regulate the circadian gene expression in the murine liver. miR-122 depletion resulted in upregulation of several circadian output genes specifically in the liver and had a striking effect on the circadian phase and amplitude of Smarcd1/Baf60a mRNA (Gatfield et al. 2009). SMARCD1/BAF60a is a component of the SWI/SNF chromatin-remodeling complex that regulates hepatic lipid metabolizing genes (Li et al. 2008). 3’ UTR assays confirmed peroxisome proliferator-activated receptor β/δ (Pparβ/δ) and the PPARα coactivator Smarcd1/Baf60a as the novel targets of miR-122. These results suggest the involvement of miR-122 in metabolic control via the metabolic regulators of the PPAR family (Gatfield et al. 2009). Additionally, a subsequent study showed that knockdown of miR-122 in the liver increased the expression and amplitude of Nocturnin rhythms (Kojima et al. 2010). Nocturnin is a circadian deadenylase enzyme that removes poly (A) tail from mRNAs and Nocturnin knockout mice have severe deficits in lipid metabolism (Green et al. 2007). These results suggest miR-122 as an important regulator of lipid metabolism by regulating the Nocturnin and PPAR family gene expression in the liver (Gatfield et al. 2009; Kojima et al. 2010). In addition to regulating the physiological output in the liver, miRNAs govern the circadian control of intestinal physiology. For example, circadian oscillators synchronize the enterocyte proliferation and miR-16 is expressed rhythmically in the intestinal crypts. miR-16 is anti-proliferative in the enterocytes and it may act as an important link in the circadian control of intestinal proliferation rhythm (Balakrishnan et al. 2010).
Extracellular vesicles are excellent candidates as transmitters of signals between the central and peripheral oscillators and among various peripheral clocks (Tao and Guo 2018). miRNA- 494, miR- 152 and miR-142-3p are reported to be present in the mice serum and among them, miR-494 and miR-152 exhibit diurnal oscillations with bimodal peak of expression (Shende et al. 2011). Studies examined the role of extracellular miRNAs miR-142-3p and miR-494 in mediating the rhythmicity among peripheral circadian oscillators. Overexpression of these microRNAs decreased the endogenous level of BMAL1 protein by modulating the 3’UTR activity of its mRNA in cultured fibroblasts (Shende et al. 2013). In fact, a careful look at the online database of miRNA composition of microvesicles (http://microvesicles.org/Archive/VESICLEPEDIA_MIRNA_DETAILS_4.1.txt) reveals that miRNAs such as miR-312, miR-142-3p and miR-494 that are implicated in circadian rhythm are enriched in the microvesicles. miR-142-3p communicate among cells by vesicular trafficking and inhibition of this vesicle trafficking modulated the ensemble clock gene rhythms. A recent study in human adipocytes treated with the exosomes containing plasma from night shift workers showed that circadian misalignment is conveyed to the peripheral tissues through circulating exosomal miRNAs and it plays an important role in metabolic dysfunction (Khalyfa et al., 2020). These results suggest that vesicle trafficking of miRNAs may act as extracellular regulatory signals of time for coordinating autonomous circadian oscillators in peripheral tissues (Shende et al. 2011; Shende et al. 2013; Shende et al., 2014).
II. Circadian regulation of microRNAs
While miRNA-mediated post-transcriptional regulation fine-tunes the circadian oscillations, the circadian timekeeping system, in turn, drives the diurnal rhythm in the expression of a number of miRNAs. For example, microarray analysis of miRNAs in wild type and cyc0 mutant fly heads showed that miR-263a and miR-263b exhibit robust diurnal changes in abundance with a peak during the night in wild type flies. This rhythm persisted even under constant darkness in wild type flies whereas it was abolished in cyc0 flies indicating the circadian regulation of these miRNAs (Yang et al. 2008). Both miR-263a and miR-263b are predicted to target Clk. Other clock relevant potential targets of miR-263a and miR-263b include doubletime (dbt) and cwo, respectively. The results of this study suggest that cycling miRNAs play a fundamental role in the circadian system by fine-tuning the daily changes in mRNA and protein levels. A subsequent study sequenced fly head RNA to identify miRNAs that exhibit robust circadian oscillations. A cluster of six miRNAs, miR-959-964 exhibited high amplitude circadian oscillation with a peak close to lights-off under the light-dark (LD) cycle and this rhythm was disrupted in ClkJrk and per0 clock mutants (Vodala et al. 2012). This study showed that the circadian clock regulates the timing of food intake, which in turn regulates the cluster miRNA cycling. These miRNA oscillation timings modulate the mRNA translation associated with the cycling of multiple physiological processes including the timing of feeding. Thus cycling cluster miRNAs contribute to a feedback circuit involved in the timing of feeding. (Vodala et al. 2012).
A successive study quantified the expression of 16 miRNAs in PDF- pacemaker neurons by using reverse transcriptase quantitative PCR (RT–qPCR) and among them, six microRNAs: miR-184, miR-276a, miR-999, miR-981, miR-210 and miR-92 exhibited cycling expression levels under LD cycles in wild type flies (Chen and Rosbash 2017). Among these six cyclers, only miR-92 oscillates with a peak during the night and circadian oscillations in mir-92a rhythmically regulate its functional target sirt2. SIRT2 levels expressed in a rhythmic manner govern the circadian fasciculation–defasciculation cycles in the PDF cell termini (Chen and Rosbash 2017). Circadian remodeling in the PDF cell termini propagates the time of the day information to the downstream targets underlying the activity-rest rhythm (Fernández et al., 2008). miR-210 exhibits rhythmic expression in the sLNvs and miR-210 regulates the timing of evening activity by inhibiting Fas 2 in the optic lobe (Chen and Rosbash 2017; Niu et al. 2019). miR-276a expression level oscillates under LD cycles and not under constant darkness (DD) and it appears that the expression is light-regulated (Chen and Rosbash 2016). miRNA let-7, a component of the let-7-complex was identified as the circadian rhythm regulator and the expression level of let-7 exhibited circadian oscillation with a peak during the day. Oscillation of let-7 is indirectly regulated by the CLK/CYC through a pathway involving circadian prothoracicotropic hormone (PTTH) and CLK mediated Ecdysone receptor (EcR) signaling (Chen et al. 2014). Rhythmically expressed let-7 regulates the amplitude of circadian oscillation by repressing the target gene cwo in the central oscillator (Chen et al. 2014).
In the murine brain, miR-219 exhibits robust circadian rhythm in expression with a peak during the subjective day and CLOCK/BMAL1 heterodimer regulates miR-219 expression. miR-219 plays a significant role in determining the circadian period and phase length (Cheng et al. 2007). PH domain and leucine rich repeat protein phosphatase (SCOP), a potential target of miR-219 is rhythmically expressed in the SCN with a peak during the subjective night and SCOP may be a physiologically relevant target of miR-219 within the SCN that mediate the effect of miR-219 on clock timing (Shimizu et al., 1999; Cheng et al. 2007). In addition, brain-specific miRNA miR-132 displays a modest significant rhythm with a peak during the subjective day and the expression of miR-132 is regulated by light and cAMP response element-binding protein (CREB). miR-132 acts as a negative regulator of the light-induced clock resetting (Cheng et al. 2007). The expression of putative miR-132 target regulatory factor X4 (RFX4), a member of the winged subfamily of helix-turn-helix transcription factors is light inducible and it is abundantly expressed in the SCN (Araki et al., 2004). Based on the in-vitro overexpression studies for target validation and by considering the light inducible nature of both miR-132 and RFX4, it is hypothesised that RFX4 may be the physiologically relevant target in the SCN that mediate the effect of miR-132 on clock entrainment (Cheng et al. 2007).
Furthermore, miR-17-5p is rhythmically expressed in mouse SCN implicating that its expression is under the circadian clock control (Gao et al. 2016). The expression rhythm of miR-17-5p and CLK was antiphasic with similar oscillatory periods and chromatin immunoprecipitation assay showed that CLOCK directly binds to the promoter of miR-17 and regulates its expression (Gao et al. 2016). miR-17-5p acts as a modulator of the circadian rhythm periodicity by interacting with Clock and potentially through CRY1 output molecule. In both flies and mammals, a small proportion of microRNAs exhibit clock regulated circadian rhythm in expression and it is likely that the rhythmic expression of miRNAs fine-tunes the level of specific proteins that generate robust circadian oscillations. These studies also suggest that circadian clock driven miRNA rhythmicity regulates the cyclic expression of their potential targets associated with diverse physiological functions and thus acts as critical mediators of rhythmic physiological processes.
Role of circadian relevant miRNAs in diseases
A number of disease conditions are known to have certain circadian misalignments and miRNA dysregulation as the underlying cause for disease onset and progression. Several studies have shown the involvement of circadian relevant miRNAs in cancer ontology. For example, downregulation of Per3 by miR-103 is observed in colorectal cancer cells and pancreatic clock disruption by the action of miR-135b on BMAL1 contributes to pancreatic cancer progression (Hong et al. 2014; Jiang et al. 2018). NPAS2, a core clock component is an oncogene in hepatocellular carcinogenesis and is up regulated in the Hepatocellular carcinoma cells due to the down regulation of miR-199b-5p (Yuan et al. 2017; Yuan et al. 2020). Another study shows the role of miR-206 targeting of Clock in glioma progression (Huang et al. 2019).
Altered expression of miRNAs is implicated in several sleep and circadian disorders. A study conducted in mice showed that the expression of nearly 50 miRNAs was affected following sleep loss (Davis et al. 2007). Another study showed that the REM sleep deprivation changes the hippocampal expression of miR-132, miR-182 and miR-124 in mice (Karabulut et al. 2019). In humans, the levels of circulating miR-125a, miR-126 and miR-146a were significantly lowered in habitual short sleep group compared to normal sleep group (Hijmans et al. 2019). Fatal familial insomnia shows significant association with a Single Nucleotide Polymorphism (SNP) in miR-146a and polymorphism in the pre-miR-182, a known circadian clock regulator, shows association with late insomnia in major depression patients (Saus et al. 2010; Gao et al. 2018). A study that was conducted in narcolepsy patients demonstrated that the levels of four miRNAs viz; miR-30c, let-7f, miR-26a and miR-130a were altered in the plasma compared to the controls (Holm et al. 2014).
Rhythmically expressed miRNAs are also associated with the disease progression of certain neurodegenerative diseases such as Alzheimer’s disease (AD) and Huntington’s disease (HD). For instance, miR-210 is rhythmically expressed in clock neurons and is known to regulate the phase circadian locomotor rhythms in Drosophila (Niu et al. 2019). miR-210 is evolutionarily conserved between Drosophila and mammals (Weigelt et al. 2019). It is also validated that miR-210 regulates various target genes involved in neurodegenerative pathways of Alzheimer’s, and Huntington’s diseases. The dysregulation of miR-210 in various brain tissues and cerebrospinal fluid has also been correlated with the pathology of Alzheimer’s disease (Watts et al. 2018). miR-146a shows rhythmic expression and is up regulated in various brain regions of AD patients and this miRNA has been suggested to play a role in the pathogenesis of AD by regulating the genes involved in cerebrovascular dysfunction and neuroinflammation (Wang et al. 2014; Swarbrick et al. 2019). miRNAs such as miR-132 and miR-124 are associated with Huntington’s disease. Photic entrainment cues induce the expression of miR-132 in mice and its expression is altered in the brains of HD patients (Cheng et al. 2007; Hoss et al. 2015). miR-124 is conserved across the animal kingdom and it is down-regulated in the brains of HD patients (Johnson et al. 2008; Xue et al. 2016). miR-124 is rhythmically expressed and regulates the circadian output in flies (Yang et al. 2008; Zhang et al. 2016). These results suggest that progression of various neurodegenerative diseases are linked to circadian rhythm and miRNA dysregulation.
Perspectives and conclusion
Accumulating evidence from various studies have shown that transcriptional-translational feedback loop regulation and post-translational regulation play a major role in circadian rhythmicity. Recent studies also showed that microRNA mediated post-transcriptional regulation is concomitantly important in shaping rhythmic gene expression. In this review, we summarized the importance of various post-transcriptional mechanisms such as splicing, polyadenylation, RNA binding proteins, RNA methylation and microRNAs in generating circadian rhythmicity of mRNAs and ultimately the rhythmic behavior and physiology of an organism. miR-132 and miR-26a are the only miRNAs that have been implicated in the photic input pathways of the circadian clock. The current understanding of miRNA mediated post-transcriptional regulation of clock input pathways is still rudimentary. It is possible that in addition to miR-132 and miR-26a, other microRNAs are involved in the clock input pathways and further characterization of miRNAs involved in the light, temperature and food entrainment will provide novel insights into the post-transcriptional control of the circadian clock input pathways.
miRNAs not only regulate the timekeeping function of the central clock, it also impacts the rhythmic clock gene expression in the peripheral tissues (Na et al. 2009; Shende et al. 2014). In addition, miRNAs are rhythmically expressed in both the central clock and in multiple peripheral tissues (Na et al. 2009; Balakrishnan et al. 2010). A previous study showed that miR-433 regulates glucocorticoid signaling and thus helps to maintain circadian rhythm in osteoblasts (Smith et al. 2016). Although studies indicate the importance of miRNAs in peripheral circadian rhythmicity, the broad impact of miRNAs in the coupling between the central and peripheral clocks present in various tissues remains to be explored.
In this review, we also discussed the specific role of miRNAs in regulating the core clock machinery, circadian neural circuit excitability, clock input and output pathways. Only a handful of miRNAs have been reported to be vital for circadian clock function. Apart from this, uncharacterized microRNAs may also play a key role in maintaining the circadian rhythm. Extensive future studies on novel circadian relevant miRNAs will provide insights into the post-transcriptional regulation of the circadian clock by non-coding RNAs. In addition to miRNA mediated post-transcriptional regulation, alternative splicing is currently recognized as an additional layer of regulation at the clock gene pre-mRNA level. However a comprehensive understanding of the physiological importance of splicing events of various clock pre-mRNAs including Cry1 and Per2 in mammals remains elusive. We summarized the current understanding on RBPs that regulate the core clock gene expression by modulating the mRNA processing steps. Apart from these evidence on RBPs regulating the circadian phenotypes, it is important to identify the remaining RBPs associated with circadian clocks and their functional relevance to provide valuable insights into the post-transcriptional regulation of the circadian timekeeping system.
Acknowledgements
The authors thank Swetha Gopal, Aishwarya S and Athira T M for their careful reading and inputs to improve the manuscript. This work was supported by the DBT/Wellcome Trust India Alliance Fellowship [IA/E/15/1/502329] awarded to NNK and intramural fund from Indian Institute of Science Education and Research, Thiruvananthapuram.
Footnotes
Disclosure statement
The authors report no conflicts of interest.
References
- Allada R, Chung BY. Circadian organization of behavior and physiology in Drosophila. Annu Rev Physiol. 2010;72:605–624. doi: 10.1146/annurev-physiol-021909-135815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allada R, Emery P, Takahashi JS, Rosbash M. Stopping Time: The Genetics of Fly and Mouse Circadian Clocks. Annu Rev Neurosci. 2001;24(1):1091–1119. doi: 10.1146/annurev.neuro.24.1.1091. [DOI] [PubMed] [Google Scholar]
- Allada R, White NE, So WV, Hall JC, Rosbash M. A Mutant Drosophila Homolog of Mammalian Clock Disrupts Circadian Rhythms and Transcription of period and timeless. Cell. 1998;93(5):791–804. doi: 10.1016/s0092-8674(00)81440-3. [DOI] [PubMed] [Google Scholar]
- Alvarez-Saavedra M, Antoun G, Yanagiya A, Oliva-Hernandez R, Cornejo-Palma D, Perez-Iratxeta C, Sonenberg N, Cheng H-YM. miRNA-132 orchestrates chromatin remodeling and translational control of the circadian clock. Hum Mol Genet. 2011;20(4):731–751. doi: 10.1093/hmg/ddq519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki R, Takahashi H, Fukumura R, Sun F, Umeda N, Sujino M, Inouye ST, Saito T, Abe M. Restricted expression and photic induction of a novel mouse regulatory factor X4 transcript in the suprachiasmatic nucleus. J Biol Chem. 2004;279:10237–10242. doi: 10.1074/jbc.M312761200. [DOI] [PubMed] [Google Scholar]
- Asher G, Reinke H, Altmeyer M, Gutierrez-Arcelus M, Hottiger MO, Schibler U. Poly(ADP-ribose) polymerase 1 participates in the phase entrainment of circadian clocks to feeding. Cell. 2010;142(6):943–953. doi: 10.1016/j.cell.2010.08.016. [DOI] [PubMed] [Google Scholar]
- Avitabile D, Genovese L, Ponti D, Ranieri D, Raffa S, Calogero A, Torrisi MR. Nucleolar localization and circadian regulation of Per2S, a novel splicing variant of the Period 2 gene. Cell Mol Life Sci. 2014;71(13):2547–2559. doi: 10.1007/s00018-013-1503-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balakrishnan A, Stearns AT, Park PJ, Dreyfuss JM, Ashley SW, Rhoads DB, Tavakkolizadeh A. MicroRNA mir-16 is anti-proliferative in enterocytes and exhibits diurnal rhythmicity in intestinal crypts. Exp Cell Res. 2010;316(20):3512–3521. doi: 10.1016/j.yexcr.2010.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Benito J, Zheng H, Ng FS, Hardin PE. Transcriptional Feedback Loop Regulation, Function, and Ontogeny in Drosophila . Cold Spring Harb Symp Quant Biol. 2007;72(1):437–444. doi: 10.1101/sqb.2007.72.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunger MK, Wilsbacher LD, Moran SM, Clendenin C, Radcliffe LA, Hogenesch JB, Simon MC, Takahashi JS, Bradfield CA. Mop3 Is an Essential Component of the Master Circadian Pacemaker in Mammals. Cell. 2000;103(7):1009–1017. doi: 10.1016/s0092-8674(00)00205-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, D’Alessandro M, Lee C. miRNAs Are Required for Generating a Time Delay Critical for the Circadian Oscillator. Curr Biol. 2013;23(20):1959–1968. doi: 10.1016/j.cub.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Liu Z, Li T, Zhang R, Xue Y, Zhong Y, Bai W, Zhou D, Zhao Z. Regulation of Drosophila circadian rhythms by miRNA let-7 is mediated by a regulatory cycle. Nat Commun. 2014;5(1):5549. doi: 10.1038/ncomms6549. [DOI] [PubMed] [Google Scholar]
- Chen X, Rosbash M. mir-276a strengthens Drosophila circadian rhythms by regulating timeless expression. Proc Natl Acad Sci U S A. 2016;113(21):E2965–2972. doi: 10.1073/pnas.1605837113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Rosbash M. MicroRNA-92a is a circadian modulator of neuronal excitability in Drosophila. Nat Commun. 2017;8:14707. doi: 10.1038/ncomms14707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H-YM, Papp JW, Varlamova O, Dziema H, Russell B, Curfman JP, Nakazawa T, Shimizu K, Okamura H, Impey S, Obrietan K. microRNA Modulation of Circadian-Clock Period and Entrainment. Neuron. 2007;54(5):813–829. doi: 10.1016/j.neuron.2007.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins BH, Rosato E, Kyriacou CP. Seasonal behavior in Drosophila melanogaster requires the photoreceptors, the circadian clock, and phospholipase C. Proc Natl Acad Sci U S A. 2004;101(7):1945–1950. doi: 10.1073/pnas.0308240100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cusumano P, Biscontin A, Sandrelli F, Mazzotta GM, Tregnago C, De Pittà C, Costa R. Modulation of miR-210 alters phasing of circadian locomotor activity and impairs projections of PDF clock neurons in Drosophila melanogaster. PLOS Genet. 2018;14(7):e1007500. doi: 10.1371/journal.pgen.1007500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyran SA, Buchsbaum AM, Reddy KL, Lin M-C, Glossop NRJ, Hardin PE, Young MW, Storti RV, Blau J. vrille, Pdp1, and dClock Form a Second Feedback Loop in the Drosophila Circadian Clock. Cell. 2003;112(3):329–341. doi: 10.1016/s0092-8674(03)00074-6. [DOI] [PubMed] [Google Scholar]
- Cyran SA, Yiannoulos G, Buchsbaum AM, Saez L, Young MW, Blau J. The double-time protein kinase regulates the subcellular localization of the Drosophila clock protein period. J Neurosci. 2005;25(22):5430–5437. doi: 10.1523/JNEUROSCI.0263-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darlington TK, Wager-Smith K, Ceriani MF, Staknis D, Gekakis N, Steeves TDL, Weitz CJ, Takahashi JS, Kay SA. Closing the Circadian Loop: CLOCK-Induced Transcription of Its Own Inhibitors per and tim. Science. 1998;280:6. doi: 10.1126/science.280.5369.1599. [DOI] [PubMed] [Google Scholar]
- Davis CJ, Bohnet SG, Meyerson JM, Krueger JM. Sleep loss changes microRNA levels in the brain: A possible mechanism for state-dependent translational regulation. Neurosci Lett. 2007;422(1):68–73. doi: 10.1016/j.neulet.2007.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolezelova E, Dolezel D, Hall JC. Rhythm Defects Caused by Newly Engineered Null Mutations in Drosophila’s cryptochrome Gene. Genetics. 2007;177(1):329–345. doi: 10.1534/genetics.107.076513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubruille R, Emery P. A Plastic Clock: How Circadian Rhythms Respond to Environmental Cues in Drosophila. Mol Neurobiol. 2008;38(2):129–145. doi: 10.1007/s12035-008-8035-y. [DOI] [PubMed] [Google Scholar]
- Fernández MP, Berni J, Ceriani MF. Circadian Remodeling of Neuronal Circuits Involved in Rhythmic Behavior. PLoS Biol. 2008;6(3):e69. doi: 10.1371/journal.pbio.0060069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley Lauren E, Ling J, Joshi R, Evantal N, Kadener S, Emery P. Drosophila PSI controls circadian period and the phase of circadian behavior under temperature cycle via tim splicing. Elife. 2019;8 doi: 10.7554/eLife.50063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fustin J-M, Doi M, Yamaguchi Y, Hida H, Nishimura S, Yoshida M, Isagawa T, Morioka MS, Kakeya H, Manabe I, Okamura H. RNA-methylation-dependent RNA processing controls the speed of the circadian clock. Cell. 2013;155(4):793–806. doi: 10.1016/j.cell.2013.10.026. [DOI] [PubMed] [Google Scholar]
- Gao C, Shi Qiang, Wei J, Zhou W, Xiao K, Wang J, Shi Qi, Dong X-P. The associations of two SNPs in miRNA-146a and one SNP in ZBTB38-RASA2 with the disease susceptibility and the clinical features of the Chinese patients of sCJD and FFI. Prion. 2018;12(1):34–41. doi: 10.1080/19336896.2017.1405885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Q, Zhou L, Yang S-Y, Cao J-M. A novel role of microRNA 17-5p in the modulation of circadian rhythm. Sci Rep. 2016;6(1):30070. doi: 10.1038/srep30070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatfield D, Le Martelot G, Vejnar CE, Gerlach D, Schaad O, Fleury-Olela F, Ruskeepaa A-L, Oresic M, Esau CC, Zdobnov EM, Schibler U. Integration of microRNA miR-122 in hepatic circadian gene expression. Genes Dev. 2009;23(11):1313–1326. doi: 10.1101/gad.1781009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gekakis N, Staknis D, Nguyen HB, Davis FC, Wilsbacher LD, King DP, Takahashi JS, Weitz CJ. Role of the CLOCK protein in the mammalian circadian mechanism. Science. 1998;280(5369):1564–1569. doi: 10.1126/science.280.5369.1564. [DOI] [PubMed] [Google Scholar]
- Gibo S, Kurosawa G. Theoretical study on the regulation of circadian rhythms by RNA methylation. J Theor Biol. 2020;490:110140. doi: 10.1016/j.jtbi.2019.110140. [DOI] [PubMed] [Google Scholar]
- Glossop NRJ, Houl JH, Zheng H, Ng FS, Dudek SM, Hardin PE. VRILLE Feeds Back to Control Circadian Transcription of Clock in the Drosophila Circadian Oscillator. Neuron. 2003;37(2):249–261. doi: 10.1016/s0896-6273(03)00002-3. [DOI] [PubMed] [Google Scholar]
- Green CB. Circadian Posttranscriptional Regulatory Mechanisms in Mammals. Cold Spring Harb Perspect Biol. 2018;10(6) doi: 10.1101/cshperspect.a030692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green CB, Douris N, Kojima S, Strayer CA, Fogerty J, Lourim D, Keller SR, Besharse JC. Loss of Nocturnin, a circadian deadenylase, confers resistance to hepatic steatosis and diet-induced obesity. Proc Natl Acad Sci U S A. 2007;104(23):9888–9893. doi: 10.1073/pnas.0702448104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin EA., Jr Light-Independent Role of CRY1 and CRY2 in the Mammalian Circadian Clock. Science. 1999;286(5440):768–771. doi: 10.1126/science.286.5440.768. [DOI] [PubMed] [Google Scholar]
- Grima B, Papin C, Martin B, Chélot E, Ponien P, Jacquet E, Rouyer F. PERIOD-controlled deadenylation of the timeless transcript in the Drosophila circadian clock. Proc Natl Acad Sci U S A. 2019;116(12):5721–5726. doi: 10.1073/pnas.1814418116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillaumond F, Dardente H, Giguère V, Cermakian N. Differential Control of Bmal1 Circadian Transcription by REV-ERB and ROR Nuclear Receptors. J Biol Rhythms. 2005;20(5):391–403. doi: 10.1177/0748730405277232. [DOI] [PubMed] [Google Scholar]
- Guo F, Cerullo I, Chen X, Rosbash M. PDF neuron firing phase-shifts key circadian activity neurons in Drosophila. Elife. 2014b;3 doi: 10.7554/eLife.02780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardin PE. Molecular genetic analysis of circadian timekeeping in Drosophila. Adv Genet. 2011;74:141–173. doi: 10.1016/B978-0-12-387690-4.00005-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardin PE, Hall JC, Rosbash M. Feedback of the Drosophila period gene product on circadian cycling of its messenger RNA levels. Nature. 1990;343(6258):536–540. doi: 10.1038/343536a0. [DOI] [PubMed] [Google Scholar]
- Helfrich-Förster C. Development of pigment-dispersing hormone-immunoreactive neurons in the nervous system of Drosophila melanogaster. J Comp Neurol. 1997;380(3):335–354. doi: 10.1002/(sici)1096-9861(19970414)380:3<335::aid-cne4>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- Hijmans JG, Levy M, Garcia V, Lincenberg GM, Diehl KJ, Greiner JJ, Stauffer BL, DeSouza CA. Insufficient sleep is associated with a pro-atherogenic circulating microRNA signature. Exp Physiol. 2019;104(6):975–982. doi: 10.1113/EP087469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirayama J, Sahar S, Grimaldi B, Tamaru T, Takamatsu K, Nakahata Y, Sassone-Corsi P. CLOCK-mediated acetylation of BMAL1 controls circadian function. Nature. 2007;450(7172):1086–1090. doi: 10.1038/nature06394. [DOI] [PubMed] [Google Scholar]
- Holm A, Bang-Berthelsen CH, Knudsen S, Kornum BR, Modvig S, Jennum P, Gammeltoft S. miRNA Profiles in Plasma from Patients with Sleep Disorders Reveal Dysregulation of miRNAs in Narcolepsy and Other Central Hypersomnias. Sleep. 2014;37(9):1525–1533. doi: 10.5665/sleep.4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Z, Feng Z, Sai Z, Tao S. PER3, a novel target of miR-103, plays a suppressive role in colorectal cancer in vitro. BMB Rep. 2014;47(9):500–505. doi: 10.5483/BMBRep.2014.47.9.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoss AG, Labadorf A, Latourelle JC, Kartha VK, Hadzi TC, Gusella JF, MacDonald ME, Chen J-F, Akbarian S, Weng Z, et al. miR-10b-5p expression in Huntington’s disease brain relates to age of onset and the extent of striatal involvement. BMC Med Genomics. 2015;8:10. doi: 10.1186/s12920-015-0083-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Genova G, Roberts M, Jackson FR. The LARK RNA-Binding Protein Selectively Regulates the Circadian Eclosion Rhythm by Controlling E74 Protein Expression. PLoS ONE. 2007;2(10):e1107. doi: 10.1371/journal.pone.0001107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Howlett E, Stern M, Jackson FR. Altered LARK expression perturbs development and physiology of the Drosophila PDF clock neurons. Mol Cell Neurosci. 2009;41(2):196–205. doi: 10.1016/j.mcn.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z, Zhao X, Wu X, Xiang L, Yuan Y, Zhou S, Yu W. LncRNA UCA1 facilitated cell growth and invasion through the miR-206/CLOCK axis in glioma. Cancer Cell Int. 2019b;19:316. doi: 10.1186/s12935-019-1023-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley JM, Loros JJ, Dunlap JC. Circadian Oscillators: Around the Transcription–Translation Feedback Loop and on to Output. Trends Biochem Sci. 2016;41(10):834–846. doi: 10.1016/j.tibs.2016.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova TN, Iuvone PM. Circadian rhythm and photic control of cAMP level in chick retinal cell cultures: a mechanism for coupling the circadian oscillator to the melatonin-synthesizing enzyme, arylalkylamine N-acetyltransferase, in photoreceptor cells. Brain Res. 2003;991(1-2):96–103. doi: 10.1016/j.brainres.2003.08.003. [DOI] [PubMed] [Google Scholar]
- Jepson JEC, Shahidullah M, Lamaze A, Peterson D, Pan H, Koh K. dyschronic, a Drosophila Homolog of a Deaf-Blindness Gene, Regulates Circadian Output and Slowpoke Channels. PLoS Genet. 2012;8(4):e1002671. doi: 10.1371/journal.pgen.1002671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Zhao S, Shen J, Guo L, Sun Y, Zhu Y, Ma Z, Zhang X, Hu Y, Xiao W, et al. The MiR-135b-BMAL1-YY1 loop disturbs pancreatic clockwork to promote tumourigenesis and chemoresistance. Cell Death Dis. 2018;9(2):149. doi: 10.1038/s41419-017-0233-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson CH, Elliott JA, Foster R. Entrainment of circadian programs. Chronobiol Int. 2003;20(5):741–774. doi: 10.1081/cbi-120024211. [DOI] [PubMed] [Google Scholar]
- Johnson R, Zuccato C, Belyaev ND, Guest DJ, Cattaneo E, Buckley NJ. A microRNA-based gene dysregulation pathway in Huntington’s disease. Neurobiol Dis. 2008;29(3):438–445. doi: 10.1016/j.nbd.2007.11.001. [DOI] [PubMed] [Google Scholar]
- Kadener S, Menet JS, Sugino K, Horwich MD, Weissbein U, Nawathean P, Vagin VV, Zamore PD, Nelson SB, Rosbash M. A role for microRNAs in the Drosophila circadian clock. Genes Dev. 2009;23(18):2179–2191. doi: 10.1101/gad.1819509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadener S, Stoleru D, McDonald M, Nawathean P, Rosbash M. Clockwork Orange is a transcriptional repressor and a new Drosophila circadian pacemaker component. Genes Dev. 2007;21(13):1675–1686. doi: 10.1101/gad.1552607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko M, Hall JC. Neuroanatomy of cells expressing clock genes in Drosophila: transgenic manipulation of the period and timeless genes to mark the perikarya of circadian pacemaker neurons and their projections. J Comp Neurol. 2000;422(1):66–94. doi: 10.1002/(sici)1096-9861(20000619)422:1<66::aid-cne5>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- Kaneko M, Helfrich-Förster C, Hall JC. Spatial and temporal expression of the period and timeless genes in the developing nervous system of Drosophila: newly identified pacemaker candidates and novel features of clock gene product cycling. J Neurosci. 1997;17(17):6745–6760. doi: 10.1523/JNEUROSCI.17-17-06745.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karabulut S, Korkmaz Bayramov K, Bayramov R, Ozdemir F, Topaloglu T, Ergen E, Yazgan K, Taskiran AS, Golgeli A. Effects of post-learning REM sleep deprivation on hippocampal plasticity-related genes and microRNA in mice. Behav Brain Res. 2019;361:7–13. doi: 10.1016/j.bbr.2018.12.045. [DOI] [PubMed] [Google Scholar]
- Khalyfa A, Gaddameedhi S, Crooks E, Zhang C, Li Y, Qiao Z, Trzepizur W, Kay SA, Andrade J, Satterfield BC, et al. Circulating Exosomal miRNAs Signal Circadian Misalignment to Peripheral Metabolic Tissues. Int J Mol Sci. 2020;21(17) doi: 10.3390/ijms21176396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King DP, Zhao Y, Sangoram AM, Wilsbacher LD, Tanaka M, Antoch MP, Steeves TD, Vitaterna MH, Kornhauser JM, Lowrey PL, et al. Positional cloning of the mouse circadian clock gene. Cell. 1997;89(4):641–653. doi: 10.1016/s0092-8674(00)80245-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klarsfeld A. Novel Features of Cryptochrome-Mediated Photoreception in the Brain Circadian Clock of Drosophila. J Neurosci. 2004;24(6):1468–1477. doi: 10.1523/JNEUROSCI.3661-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloss B, Price JL, Saez L, Blau J, Rothenfluh A, Wesley CS, Young MW. The Drosophila clock gene double-time encodes a protein closely related to human casein kinase Iepsilon. Cell. 1998;94(1):97–107. doi: 10.1016/s0092-8674(00)81225-8. [DOI] [PubMed] [Google Scholar]
- Kojima S, Gatfield D, Esau CC, Green CB. MicroRNA-122 Modulates the Rhythmic Expression Profile of the Circadian Deadenylase Nocturnin in Mouse Liver. PLoS ONE. 2010;5(6):e11264. doi: 10.1371/journal.pone.0011264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima S, Matsumoto K, Hirose M, Shimada M, Nagano M, Shigeyoshi Y, Hoshino S-i, Ui-Tei K, Saigo K, Green CB, et al. LARK activates posttranscriptional expression of an essential mammalian clock protein, PERIOD1. Proc Natl Acad Sci U S A. 2007;104(6):1859–1864. doi: 10.1073/pnas.0607567104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima S, Sher-Chen EL, Green CB. Circadian control of mRNA polyadenylation dynamics regulates rhythmic protein expression. Genes Dev. 2012;26(24):2724–2736. doi: 10.1101/gad.208306.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima S, Shingle DL, Green CB. Post-transcriptional control of circadian rhythms. J Cell Sci. 2011;124(3):311–320. doi: 10.1242/jcs.065771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopka RJ, Benzer S. Clock Mutants of Drosophila melanogaster. Proc Natl Acad Sci. 1971;68(9):2112–2116. doi: 10.1073/pnas.68.9.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kume K, Zylka MJ, Sriram S, Shearman LP, Weaver DR, Jin X, Maywood ES, Hastings MH, Reppert SM. mCRY1 and mCRY2 Are Essential Components of the Negative Limb of the Circadian Clock Feedback Loop. Cell. 1999;98(2):193–205. doi: 10.1016/s0092-8674(00)81014-4. [DOI] [PubMed] [Google Scholar]
- Lee C, Etchegaray J-P, Cagampang FRA, Loudon ASI, Reppert SM. Posttranslational Mechanisms Regulate the Mammalian Circadian Clock. Cell. 2001;107(7):855–867. doi: 10.1016/s0092-8674(01)00610-9. [DOI] [PubMed] [Google Scholar]
- Lim C, Chung BY, Pitman JL, McGill JJ, Pradhan S, Lee J, Keegan KP, Choe J, Allada R. clockwork orange Encodes a Transcriptional Repressor Important for Circadian-Clock Amplitude in Drosophila. Curr Biol. 2007;17(12):1082–1089. doi: 10.1016/j.cub.2007.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim C, Lee J, Choi C, Kilman VL, Kim J, Park SM, Jang SK, Allada R, Choe J. The novel gene twenty-four defines a critical translational step in the Drosophila clock. Nature. 2011;470(7334):399–403. doi: 10.1038/nature09728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima SA, Chipman LB, Nicholson AL, Chen Y-H, Yee BA, Yeo GW, Coller J, Pasquinelli AE. Short poly(A) tails are a conserved feature of highly expressed genes. Nat Struct Mol Biol. 2017;24(12):1057–1063. doi: 10.1038/nsmb.3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Hu W, Murakawa Y, Yin J, Wang G, Landthaler M, Yan J. Cold-induced RNA-binding proteins regulate circadian gene expression by controlling alternative polyadenylation. Sci Rep. 2013;3(1):2054. doi: 10.1038/srep02054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowrey PL, Takahashi JS. MAMMALIAN CIRCADIAN BIOLOGY: Elucidating Genome-Wide Levels of Temporal Organization. Annu Rev Genom Hum Genet. 2004;5(1):407–441. doi: 10.1146/annurev.genom.5.061903.175925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowrey PL, Takahashi JS. Genetics of circadian rhythms in Mammalian model organisms. Adv Genet. 2011;74:175–230. doi: 10.1016/B978-0-12-387690-4.00006-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W, Sehgal A. Regulation of Circadian Behavioral Output via a MicroRNA-JAK/STAT Circuit. Cell. 2012;148(4):765–779. doi: 10.1016/j.cell.2011.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majercak J, Chen W-F, Edery I. Splicing of the period gene 3’-terminal intron is regulated by light, circadian clock factors, and phospholipase C. Mol Cell Biol. 2004;24(8):3359–3372. doi: 10.1128/MCB.24.8.3359-3372.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majercak J, Sidote D, Hardin PE, Edery I. How a Circadian Clock Adapts to Seasonal Decreases in Temperature and Day Length. Neuron. 1999;24(1):219–230. doi: 10.1016/s0896-6273(00)80834-x. [DOI] [PubMed] [Google Scholar]
- Martin Anduaga A, Evantal N, Patop IL, Bartok O, Weiss R, Kadener S. Thermosensitive alternative splicing senses and mediates temperature adaptation in Drosophila. Elife. 2019;8 doi: 10.7554/eLife.44642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinek S, Inonog S, Manoukian AS, Young MW. A Role for the Segment Polarity Gene shaggy/GSK-3 in the Drosophila Circadian Clock. Cell. 2001;105(6):769–779. doi: 10.1016/s0092-8674(01)00383-x. [DOI] [PubMed] [Google Scholar]
- Matsumoto A, Ukai-Tadenuma M, Yamada RG, Houl J, Uno KD, Kasukawa T, Dauwalder B, Itoh TQ, Takahashi K, Ueda R, et al. A functional genomics strategy reveals clockwork orange as a transcriptional regulator in the Drosophila circadian clock. Genes Dev. 2007;21(13):1687–1700. doi: 10.1101/gad.1552207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta N, Cheng H-YM. Micro-Managing the Circadian Clock: The Role of microRNAs in Biological Timekeeping. J Mol Biol. 2013;425(19):3609–3624. doi: 10.1016/j.jmb.2012.10.022. [DOI] [PubMed] [Google Scholar]
- Meijer JH, Michel S, VanderLeest HT, Rohling JHT. Daily and seasonal adaptation of the circadian clock requires plasticity of the SCN neuronal network: Plasticity of the SCN neuronal network. Eur J Neurosci. 2010;32(12):2143–2151. doi: 10.1111/j.1460-9568.2010.07522.x. [DOI] [PubMed] [Google Scholar]
- Mendoza-Viveros L, Chiang C-K, Ong JLK, Hegazi S, Cheng AH, Bouchard-Cannon P, Fana M, Lowden C, Zhang P, Bothorel B, et al. miR-132/212 Modulates Seasonal Adaptation and Dendritic Morphology of the Central Circadian Clock. Cell Rep. 2017;19(3):505–520. doi: 10.1016/j.celrep.2017.03.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki K, Nagase T, Mesaki M, Narukawa J, Ohara O, Ishida N. Phosphorylation of clock protein PER1 regulates its circadian degradation in normal human fibroblasts. Biochem J. 2004;380(Pt 1):95–103. doi: 10.1042/BJ20031308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morf J, Rey G, Schneider K, Stratmann M, Fujita J, Naef F, Schibler U. Cold-Inducible RNA-Binding Protein Modulates Circadian Gene Expression Posttranscriptionally. Science. 2012;338(6105):379–383. doi: 10.1126/science.1217726. [DOI] [PubMed] [Google Scholar]
- Na YJ, Sung JH, Lee SC, Lee YJ, Choi YJ, Park WY, Shin HS, Kim JH. Comprehensive analysis of microRNA-mRNA co-expression in circadian rhythm. Exp Mol Med. 2009;41(9):638. doi: 10.3858/emm.2009.41.9.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagel R, Clijsters L, Agami R. The miRNA-192/194 cluster regulates the Period gene family and the circadian clock: Regulation of the circadian clock by miR-192/194. FEBS J. 2009;276(19):5447–5455. doi: 10.1111/j.1742-4658.2009.07229.x. [DOI] [PubMed] [Google Scholar]
- Naidoo N, Ferber M, Galante RJ, McShane B, Hu JH, Zimmerman J, Maislin G, Cater J, Wyner A, Worley P, Pack AI. Role of Homer Proteins in the Maintenance of Sleep-Wake States. PLoS ONE. 2012;7(4):e35174. doi: 10.1371/journal.pone.0035174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newby LM, Jackson FR. Regulation of a specific circadian clock output pathway by lark, a putative RNA-binding protein with repressor activity. J Neurobiol. 1996;31(1):117–128. doi: 10.1002/(SICI)1097-4695(199609)31:1<117::AID-NEU10>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Ng FS, Jackson FR. The ROP vesicle release factor is required in adult Drosophila glia for normal circadian behavior. Front Cell Neurosci. 2015;9:256. doi: 10.3389/fncel.2015.00256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng FS, Sengupta S, Huang Y, Yu AM, You S, Roberts MA, Iyer LK, Yang Y, Jackson FR. TRAP-seq Profiling and RNAi-Based Genetic Screens Identify Conserved Glial Genes Required for Adult Drosophila Behavior. Front Mol Neurosci. 2016;9:146. doi: 10.3389/fnmol.2016.00146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng FS, Tangredi MM, Jackson FR. Glial Cells Physiologically Modulate Clock Neurons and Circadian Behavior in a Calcium-Dependent Manner. Curr Biol. 2011;21(8):625–634. doi: 10.1016/j.cub.2011.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nian X, Chen W, Bai W, Zhao Z. Regulation of circadian locomotor rhythm by miR-263a. Biol Rhythm Res. 2020:1–11. [Google Scholar]
- Nian X, Chen W, Bai W, Zhao Z, Zhang Y. miR-263b Controls Circadian Behavior and the Structural Plasticity of Pacemaker Neurons by Regulating the LIM-Only Protein Beadex. Cells. 2019;8(8):923. doi: 10.3390/cells8080923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu Y, Liu Z, Nian X, Xu X, Zhang Y. Taghert PH, editor. miR-210 controls the evening phase of circadian locomotor rhythms through repression of Fasciclin 2. PLoS Genet. 2019;15(7):e1007655. doi: 10.1371/journal.pgen.1007655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park I, Kim D, Kim J, Jang S, Choi M, Choe HK, Choe Y, Kim K. microRNA-25 as a novel modulator of circadian Period2 gene oscillation. Exp Mol Med. 2020;52(9):1614–1626. doi: 10.1038/s12276-020-00496-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pembroke WG, Babbs A, Davies KE, Ponting CP, Oliver PL. Temporal transcriptomics suggest that twin-peaking genes reset the clock. eLife. 2015;4:e10518. doi: 10.7554/eLife.10518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preitner N, Damiola F, Lopez-Molina L, Zakany J, Duboule D, Albrecht U, Schibler U. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002;110(2):251–260. doi: 10.1016/s0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
- Preußner M, Wilhelmi I, Schultz A-S, Finkernagel F, Michel M, Möröy T, Heyd F. Rhythmic U2af26 Alternative Splicing Controls PERIOD1 Stability and the Circadian Clock in Mice. Mol Cell. 2014;54(4):651–662. doi: 10.1016/j.molcel.2014.04.015. [DOI] [PubMed] [Google Scholar]
- Quintero JE, Kuhlman SJ, McMahon DG. The Biological Clock Nucleus: A Multiphasic Oscillator Network Regulated by Light. J Neurosci. 2003;23(22):8070–8076. doi: 10.1523/JNEUROSCI.23-22-08070.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reischl S, Kramer A. Kinases and phosphatases in the mammalian circadian clock. FEBS Lett. 2011;585(10):1393–1399. doi: 10.1016/j.febslet.2011.02.038. [DOI] [PubMed] [Google Scholar]
- Rutila JE, Suri V, Le M, So WV, Rosbash M, Hall JC. CYCLE Is a Second bHLH-PAS Clock Protein Essential for Circadian Rhythmicity and Transcription of Drosophila period and timeless. Cell. 1998;93(5):805–814. doi: 10.1016/s0092-8674(00)81441-5. [DOI] [PubMed] [Google Scholar]
- Sato TK, Yamada RG, Ukai H, Baggs JE, Miraglia LJ, Kobayashi TJ, Welsh DK, Kay SA, Ueda HR, Hogenesch JB. Feedback repression is required for mammalian circadian clock function. Nat Genet. 2006;38(3):312–319. doi: 10.1038/ng1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saus E, Soria V, Escaramís G, Vivarelli F, Crespo JM, Kagerbauer B, Menchón JM, Urretavizcaya M, Gratacòs M, Estivill X. Genetic variants and abnormal processing of pre-miR-182, a circadian clock modulator, in major depression patients with late insomnia. Hum Mol Genet. 2010;19(20):4017–4025. doi: 10.1093/hmg/ddq316. [DOI] [PubMed] [Google Scholar]
- Schroeder AJ, Genova GK, Roberts MA, Kleyner Y, Suh J, Jackson FR. Cell-Specific Expression of the Lark Rna-Binding Protein in Drosophila Results in Morphological and Circadian Behavioral Phenotypes. J Neurogenet. 2003;17(2-3):139–169. [PubMed] [Google Scholar]
- Sehgal A, Price J, Man B, Young M. Loss of circadian behavioral rhythms and per RNA oscillations in the Drosophila mutant timeless. Science. 1994;263(5153):1603–1606. doi: 10.1126/science.8128246. [DOI] [PubMed] [Google Scholar]
- Shafer OT, Helfrich-Förster C, Renn SCP, Taghert PH. Reevaluation of Drosophila melanogaster’s neuronal circadian pacemakers reveals new neuronal classes. J Comp Neurol. 2006;498(2):180–193. doi: 10.1002/cne.21021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafer OT, Kim DJ, Dunbar-Yaffe R, Nikolaev VO, Lohse MJ, Taghert PH. Widespread Receptivity to Neuropeptide PDF throughout the Neuronal Circadian Clock Network of Drosophila Revealed by Real-Time Cyclic AMP Imaging. Neuron. 2008;58(2):223–237. doi: 10.1016/j.neuron.2008.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakhmantsir I, Nayak S, Grant GR, Sehgal A. Spliceosome factors target timeless (tim) mRNA to control clock protein accumulation and circadian behavior in Drosophila. eLife. 2018;7:e39821. doi: 10.7554/eLife.39821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shende VR, Goldrick MM, Ramani S, Earnest DJ. Expression and Rhythmic Modulation of Circulating MicroRNAs Targeting the Clock Gene Bmal1 in Mice. PLoS ONE. 2011;6(7):e22586. doi: 10.1371/journal.pone.0022586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shende VR, Kim S-M, Neuendorff N, Earnest DJ. MicroRNAs function as cis- and trans-acting modulators of peripheral circadian clocks. FEBS Lett. 2014;588(17):3015–3022. doi: 10.1016/j.febslet.2014.05.058. [DOI] [PubMed] [Google Scholar]
- Shende VR, Neuendorff N, Earnest DJ. Lyons LC, editor. Role of miR-142-3p in the Post-Transcriptional Regulation of the Clock Gene Bmal1 in the Mouse SCN. PLoS ONE. 2013;8(6):e65300. doi: 10.1371/journal.pone.0065300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L, Ko ML, Ko GY-P. Rhythmic Expression of MicroRNA-26a Regulates the L-type Voltage-gated Calcium Channel α1C Subunit in Chicken Cone Photoreceptors. J Biol Chem. 2009;284(38):25791–25803. doi: 10.1074/jbc.M109.033993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu K, Okada M, Takano A, Nagai K. SCOP, a novel gene product expressed in a circadian manner in rat suprachiasmatic nucleus. FEBS Lett. 1999;458:363–369. doi: 10.1016/s0014-5793(99)01190-4. [DOI] [PubMed] [Google Scholar]
- Smith SS, Dole NS, Franceschetti T, Hrdlicka HC, Delany AM. MicroRNA-433 Dampens Glucocorticoid Receptor Signaling, Impacting Circadian Rhythm and Osteoblastic Gene Expression. J Biol Chem. 2016;291(41):21717–21728. doi: 10.1074/jbc.M116.737890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanewsky R, Kaneko M, Emery P, Beretta B, Wager-Smith K, Kay SA, Rosbash M, Hall JC. The cryb Mutation Identifies Cryptochrome as a Circadian Photoreceptor in Drosophila. Cell. 1998;95(5):681–692. doi: 10.1016/s0092-8674(00)81638-4. [DOI] [PubMed] [Google Scholar]
- Stubblefield JJ, Gao P, Kilaru G, Mukadam B, Terrien J, Green CB. Temporal Control of Metabolic Amplitude by Nocturnin. Cell Reports. 2018;22(5):1225–1235. doi: 10.1016/j.celrep.2018.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarbrick S, Wragg N, Ghosh S, Stolzing A. Systematic Review of miRNA as Biomarkers in Alzheimer’s Disease. Mol Neurobiol. 2019;56(9):6156–6167. doi: 10.1007/s12035-019-1500-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi JS, Turek FW, Moore RY, editors. Handbook of Behavioral Neurobiology: Circadian Clocks. Vol. 12 Kluwer Acad./Plenum Publ; New York: 2001. [Google Scholar]
- Tan CL, Plotkin JL, Veno MT, von Schimmelmann M, Feinberg P, Mann S, Handler A, Kjems J, Surmeier DJ, O’Carroll D, et al. MicroRNA-128 Governs Neuronal Excitability and Motor Behavior in Mice. Science. 2013;342(6163):1254–1258. doi: 10.1126/science.1244193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao S-C, Guo S-C. Extracellular Vesicles: Potential Participants in Circadian Rhythm Synchronization. Int J Biol Sci. 2018;14(12):1610–1620. doi: 10.7150/ijbs.26518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temme C, Simonelig M, Wahle E. Deadenylation of mRNA by the CCR4-NOT complex in Drosophila: molecular and developmental aspects. Front Genet. 2014;5:143. doi: 10.3389/fgene.2014.00143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma P, Augustine GJ, Ammar M-R, Tashiro A, Cohen SM. A neuroprotective role for microRNA miR-1000 mediated by limiting glutamate excitotoxicity. Nat Neurosci. 2015;18(3):379–385. doi: 10.1038/nn.3935. [DOI] [PubMed] [Google Scholar]
- Vodala S, Pescatore S, Rodriguez J, Buescher M, Chen Y-W, Weng R, Cohen SM, Rosbash M. The Oscillating miRNA 959-964 Cluster Impacts Drosophila Feeding Time and Other Circadian Outputs. Cell Metab. 2012;16(5):601–612. doi: 10.1016/j.cmet.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl MC, Will CL, Lührmann R. The Spliceosome: Design Principles of a Dynamic RNP Machine. Cell. 2009;136(4):701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- Wang Y, Osterbur DL, Megaw PL, Tosini G, Fukuhara C, Green CB, Besharse JC. Rhythmic expression of Nocturnin mRNA in multiple tissues of the mouse. BMC Dev Biol. 2001:15. doi: 10.1186/1471-213X-1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C-Y, Yeh J-K, Shie S-S, Hsieh I-C, Wen M-S. Circadian rhythm of RNA N6-methyladenosine and the role of cryptochrome. Biochem Biophys Res Commun. 2015;465(1):88–94. doi: 10.1016/j.bbrc.2015.07.135. [DOI] [PubMed] [Google Scholar]
- Wang Q, Bozack SN, Yan Y, Boulton ME, Grant MB, Busik JV. Regulation of retinal inflammation by rhythmic expression of MiR-146a in diabetic retina. Invest Ophthalmol Vis Sci. 2014;55(6):3986–3994. doi: 10.1167/iovs.13-13076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts ME, Williams SM, Nithianantharajah J, Claudianos C. Hypoxia-Induced MicroRNA-210 Targets Neurodegenerative Pathways. Noncoding RNA. 2018;4(2) doi: 10.3390/ncrna4020010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber F, Zorn D, Rademacher C, Hung H-C. Post-translational timing mechanisms of the Drosophila circadian clock. FEBS Letters. 2011;585(10):1443–1449. doi: 10.1016/j.febslet.2011.04.008. [DOI] [PubMed] [Google Scholar]
- Weigelt CM, Hahn O, Arlt K, Gruhn M, Jahn AJ, Eßer J, Werner JA, Klein C, Büschges A, Grönke S, Partridge L. Loss of miR-210 leads to progressive retinal degeneration in Drosophila melanogaster . Life Science Alliance. 2019;2(1):e201800149. doi: 10.26508/lsa.201800149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia X, Fu X, Du J, Wu B, Zhao X, Zhu J, Zhao Z. Regulation of circadian rhythm and sleep by miR-375-timeless interaction in Drosophila. FASEB J. 2020;34(12):16536–16551. doi: 10.1096/fj.202001107R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Zhang Y. Emerging roles for microRNA in the regulation of Drosophila circadian clock. BMC Neurosci. 2018;19(1):1. doi: 10.1186/s12868-018-0401-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Lee JE, Padgett RW, Edery I. Circadian regulation of a limited set of conserved microRNAs in Drosophila. BMC Genomics. 2008;9(1):83. doi: 10.1186/1471-2164-9-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SH, Kojima S, Shimomura K, Koike N, Buhr ED, Furukawa T, Ko CH, Gloston G, Ayoub C, Nohara K, et al. Period2 3’-UTR and microRNA-24 regulate circadian rhythms by repressing PERIOD2 protein accumulation. Proc Natl Acad Sci U S A. 2017;114(42):E8855–E8864. doi: 10.1073/pnas.1706611114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You S, Fulga TA, Van Vactor D, Jackson FR. Regulation of Circadian Behavior by Astroglial MicroRNAs in Drosophila . Genetics. 2018;208(3):1195–1207. doi: 10.1534/genetics.117.300342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W, Hardin PE. Circadian oscillators of Drosophila and mammals. J Cell Sci. 2006;119(23):4793–4795. doi: 10.1242/jcs.03174. [DOI] [PubMed] [Google Scholar]
- Yuan P, Li J, Zhou F, Huang Q, Zhang J, Guo X, Lyu Z, Zhang H, Xing J. NPAS2 promotes cell survival of hepatocellular carcinoma by transactivating CDC25A. Cell Death Dis. 2017;8(3):e2704. doi: 10.1038/cddis.2017.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan P, Yang T, Mu J, Zhao J, Yang Y, Yan Z, Hou Y, Chen C, Xing J, Zhang H, Li J. Circadian clock gene NPAS2 promotes reprogramming of glucose metabolism in hepatocellular carcinoma cells. Cancer Lett. 2020;469:498–509. doi: 10.1016/j.canlet.2019.11.024. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Lamba P, Guo P, Emery P. miR-124 Regulates the Phase of Drosophila Circadian Locomotor Behavior. J Neurosci. 2016;36(6):2007–2013. doi: 10.1523/JNEUROSCI.3286-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Cao W, Edery I. The SR protein B52/SRp55 regulates splicing of the period thermosensitive intron and mid-day siesta in Drosophila. Sci Rep. 2018;8(1):1872. doi: 10.1038/s41598-017-18167-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng B, Albrecht U, Kaasik K, Sage M, Lu W, Vaishnav S, Li Q, Sun ZS, Eichele G, Bradley A, Lee CC. Nonredundant Roles of the mPer1 and mPer2 Genes in the Mammalian Circadian Clock. Cell. 2001;105(5):683–694. doi: 10.1016/s0092-8674(01)00380-4. [DOI] [PubMed] [Google Scholar]
- Zhong X, Yu J, Frazier K, Weng X, Li Y, Cham CM, Dolan K, Zhu X, Hubert N, Tao Y, et al. Circadian Clock Regulation of Hepatic Lipid Metabolism by Modulation of m6A mRNA Methylation. Cell Rep. 2018;25(7):1816–1828.:e4. doi: 10.1016/j.celrep.2018.10.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Miller C, Miraglia LJ, Romero A, Mure LS, Panda S, Kay SA. A genome-wide microRNA screen identifies the microRNA-183/96/182 cluster as a modulator of circadian rhythms. Proc Natl Acad Sci USA. 2021;118(1):e2020454118. doi: 10.1073/pnas.2020454118. [DOI] [PMC free article] [PubMed] [Google Scholar]