Abstract
The Type VI secretion system (T6SS) is a bacterial nanomachine which delivers toxic effectors to kill competitors or subvert some of their key functions. Here we use transposon directed insertion-site sequencing (TraDIS) to identify T6SS toxins associated with the H1-T6SS, one of the three T6SS machines found in Pseudomonas aeruginosa. This approach identified several putative toxin-immunity pairs, including Tse8-Tsi8. Full characterization of this protein pair demonstrated that Tse8 is delivered by the VgrG1a spike complex into prey cells where it targets the transamidosome, a multiprotein complex involved in protein synthesis in bacteria that lack either one, or both, of the asparagine and glutamine tRNA synthases. Biochemical characterization of the interactions between Tse8 and the transamidosome components GatA, GatB and GatC suggests that the presence of Tse8 alters the fine-tuned stoichiometry of the transamidosome complex, and in vivo assays demonstrate that Tse8 limits the ability of prey cells to synthesize proteins. These data expand the range of cellular components targeted by the T6SS by identifying a T6SS toxin affecting protein synthesis proteins and validate the use of a TraDIS-based global genomics approach to expand the repertoire of T6SS toxins in T6SS-encoding bacteria.
Bacteria rarely exist in a single-species planktonic state and instead form complex polymicrobial structures, called biofilms1,2. Within this context bacteria often compete with other microorganisms to secure space and nutrients. The Type VI secretion system (T6SS) is a Gram-negative bacterial nanomachine that delivers toxins into neighbouring competitors to either kill or subvert their key functions in order to attain dominance within a given niche3–5. The T6SS is composed of 13 core components, several of which are structurally related to proteins from the T4 bacteriophage tail6. The Hcp tube-like structure is capped by a VgrG-PAAR tip complex, or spike, and encapsulated within a TssBC (also known as VipAB) contractile sheath 7–9. Upon extension of the sheath within the cytoplasm and subsequent contraction, the spike is thought to facilitate the puncturing of the cell membranes of both the producing and target cells, allowing delivery of the attached toxins8,10. T6SS toxins have been shown to be secreted in association with the VgrG tip complex, the Hcp tube, or as extension domains of the VgrG, PAAR or Hcp proteins11–14. Importantly, neighbouring bacterial sister cells are protected from the effects of the toxins by production of cognate immunity proteins, which are usually encoded adjacent to the toxin gene in the genome15. The major identified targets of T6SS toxins to date are components of the cell wall, as well as the cell membrane and nucleic acids16. These T6SS toxins have mainly been identified by searching in the genomic proximity of known T6SS components, or by detection of toxins in the secretome11,14,17.
Pseudomonas aeruginosa is a highly antibiotic-resistant Gram-negative pathogen and ranked second by the World Health Organization in the list of bacteria that require immediate attention. It is also equipped with three independent T6SS systems (H1- to H3-T6SS)18. In the current study we used a global genomics-based approach called TraDIS (Transposon directed insertion-site sequencing) to identify toxins associated with the P. aeruginosa H1-T6SS 19. A previous study has used Tn-Seq, a similar global transposon mutagenesis approach, and confirmed the presence of three T6SS toxin-immunity genes which are located in the vicinity of vgrG genes in V. cholerae 41. Our TraDIS approach identified several remote and previously unidentified putative T6SS toxin-immunity pairs. We found that one of the identified toxins, Tse8 (Type six exported 8), targets the bacterial transamidosome complex, which is required for protein synthesis in bacteria that lack the asparagine and/or glutamine tRNA synthases20. This is a previously unidentified target for a T6SS toxin, demonstrating that T6SS toxins can impair bacterial protein synthesis.
TraDIS identifies known and previously unidentified H1-T6SS toxin-immunity pairs
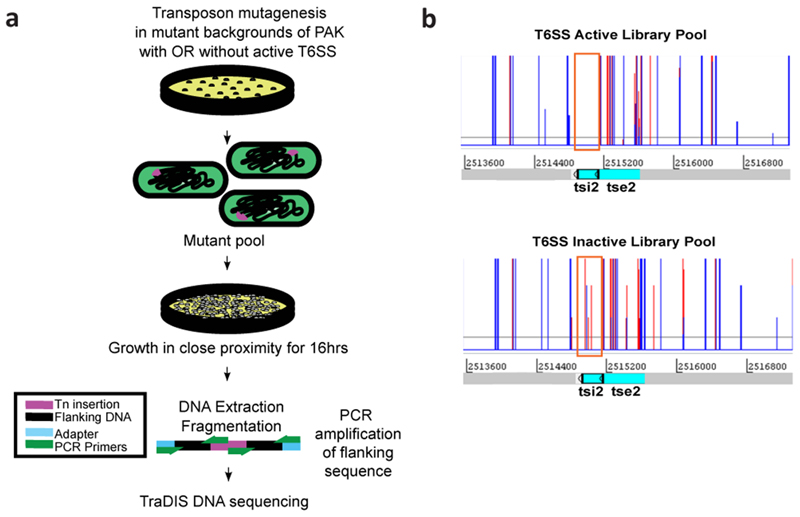
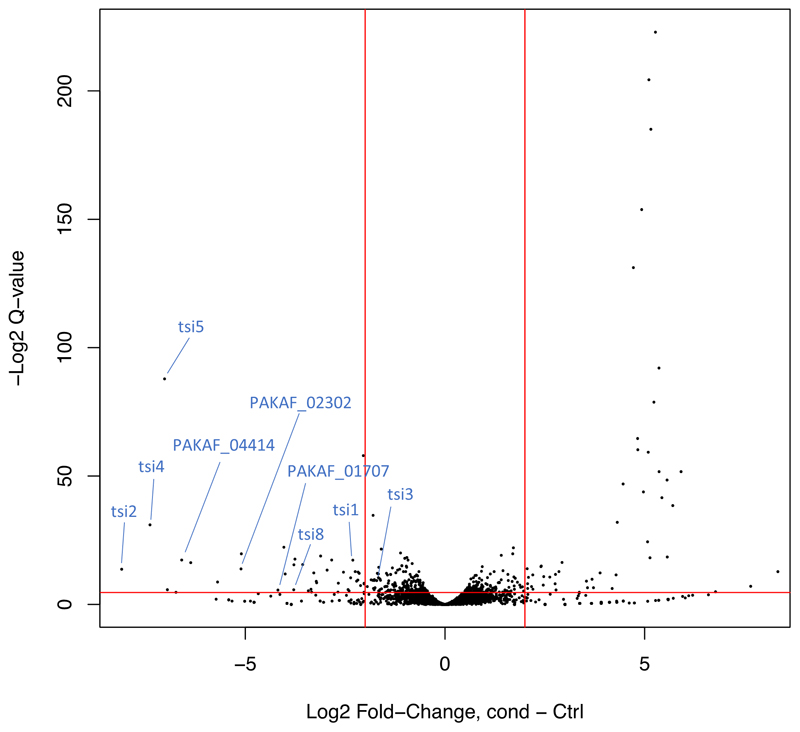
To systematically identify P. aeruginosa PAK H1-T6SS associated immunity genes we generated duplicate high-density insertion transposon mutant libraries consisting of ~2 million mutants in a H1-T6SS active (PAKΔretS) and a H1-T6SS inactive (PAKΔretSΔH1) background. We reasoned that transposon insertions in immunity genes would only be tolerated in the H1-T6SS inactive library, while in the H1-T6SS active library, cells lacking an immunity protein would be killed upon injection of the cognate toxin from neighbouring sister cells or due to self-intoxication. Each duplicate library was plated separately at high-contact density on agar plates and passaged in an overnight incubation step to promote T6SS-mediated killing of mutants with transposon insertions in immunity genes (Extended data Fig. 1). The genomic DNA of mutants which were not killed in both the H1-T6SS active and inactive libraries were then separately sequenced using a mass-parallel approach as described previously21,22 (Extended Data Fig. 1). The relative frequencies of transposon insertion in genes in the H1-T6SS active and inactive libraries revealed a large number of genes which had changes in relative numbers of transposon insertions. Forty-five genes which had a significantly greater number of normalized transposon insertions in the H1-T6SS inactive library background, compared to the H1-T6SS active library background, were identified (Supplementary Table 1), and considered as potential H1-T6SS immunity proteins. Our approach is validated by our ability to identify five (tsi1-tsi5) out of the seven known H1-T6SS immunity genes, whose gene products protect against cognate toxins acting in both the cytoplasm and periplasm (Table 1). Our screen was unable to identify tsi6 as this gene is deleted in our PAKΔretSΔH1 strain, thus there is no possibility to assess the relative frequency of transposon insertions in this gene between the two library backgrounds. In the case of tsi7 we did not see any difference in the levels of insertions between the two libraries (Supplementary Table 1). It is not clear why this was the case, but we cannot exclude the possibility that one of the uncharacterized proteins encoded by the vgrG1b cluster23 containing the tse7-tsi7 pair, or a gene elsewhere in the genome, can also confer protection against the Tse7 toxin in the absence of Tsi7.
Table 1. TraDIS allows identification of known and putative previously unidentified H1-T6SS immunity genes.
| Immunity gene PAK/PA number | Immunity | Toxin | Log fold change* | Toxin activity/target |
|---|---|---|---|---|
| PAKAF_RS16410/PA1845 | tsi1 | tse1 | -2.30 | Amidase/peptidoglycan |
| PAKAF_RS11975/PA2703 | tsi2 | tse2 | -7.30 | Unknown cytoplasmic target |
| PAKAF_RS07460/PA3485 | tsi3 | tse3 | -1.28 | Muramidase/peptidoglycan |
| PAKAF_RS11540/PA2775 | tsi4 | tse4 | -7.30 | Unknown periplasmic target |
| PAKAF_RS12070/PA2683.1 | tsi5 | tse5 | -7.02 | Unknown periplasmic target |
| PAKAF_RS22000/PA0802 | PA0802 | PA0801 | -6.60 | Putative M4 peptidase regulator |
| PAKAF_RS11515/PA2779 | PA2279 | PA2778 | -5.50 | Putative C39 peptidase |
| PAKAF_RS08570/PA3274 | PA3274 | PA3272 | -4.70 | Putative nucleoside triphosphate hydrolase |
| PAKAF_RS03995/PA4164 | tsi8 | tse8 (PA4163) | -3.30 | Putative amidase |
Log fold change compared to normalized levels of insertions in T6SS inactive and T6SS active libraries
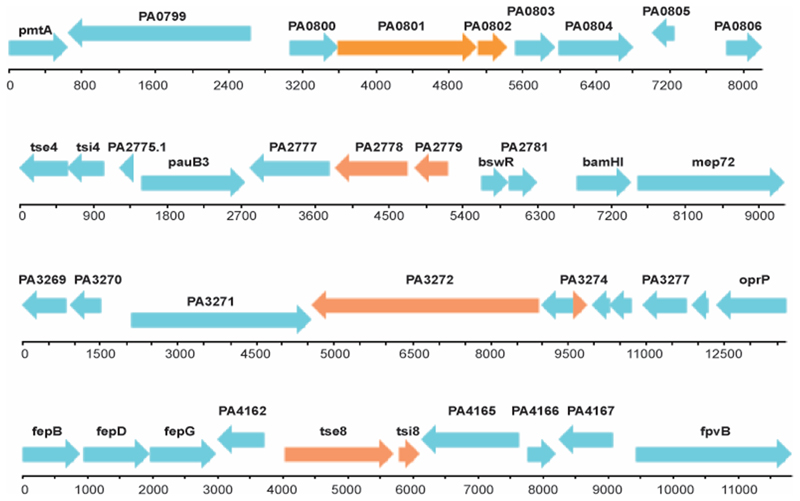
In addition to known H1-T6SS associated immunity genes, our TraDIS approach identified multiple uncharacterised small coding sequences that displayed a decrease in transposon insertions in the H1-T6SS active, compared to the inactive, background (represented by a negative log fold change), suggesting a role for these genes in protecting against H1-T6SS mediated killing (Supplementary Table 1). Upstream of several of these loci were genes encoding proteins with putative enzymatic activity which could be T6SS toxins: PAKAF_04415 (PA0801) encodes a putative M4 peptidase regulator; PAKAF_02303 (PA2778) encodes a putative C39 peptidase domain-containing protein; PAKAF_01709 (PA3272) encodes a putative nucleoside triphosphate hydrolase; and PAKAF_00798 (PA4163) encodes a putative amidase (Table 1 and Extended Data Fig. 2). In the present study, we selected the putative toxin/immunity pair PAKAF_00798/PAKAF_00797 (PA4163/PA4164) for further characterization, and we refer to it as tse8-tsi8 (type six exported 8-type six immunity 8) in all subsequent sections.
Tse8-Tsi8 is a toxin-immunity pair
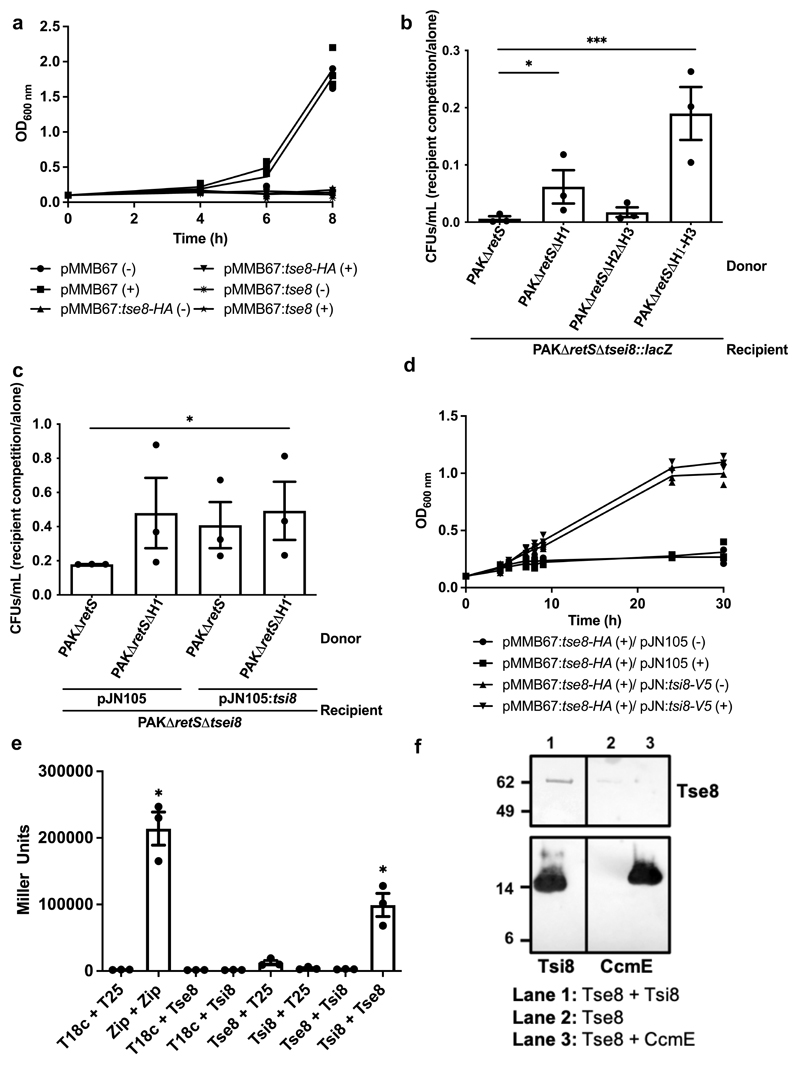
To assess the toxic role of Tse8, a strain lacking both tse8 and the downstream putative immunity gene (tsi8) was generated in a PAKΔretS background, yielding PAKΔretSΔtsei8. In this mutant, expression of tse8 from pMMB67HE with and without a C-terminal HA tag affected growth (Fig. 1a). Furthermore, in a competition assay this mutant strain carrying a lacZ reporter gene (recipient PAKΔretSΔtsei8::lacZ) was outcompeted only by donor strains with an active H1-T6SS, i.e. PAKΔretS or PAKΔretSΔH2ΔH3 (Fig. 1b). The observed killing of the receiver strain was further demonstrated to be Tse8-dependent in competition assays with a donor lacking Tse8 (Extended Data Fig. 3a). The PAKΔretS strain lacking either tsei8 or tse8 could be complemented in a competition assay by expression of tsei8 from pBBR-MCS5 or tse8 from pBBR-MCS4 (Extended Data Fig. 3b, c).
Figure 1. Tse8-Tsi8 is a H1-T6SS toxin-immunity pair.
a-b, Expression of Tse8 (either HA tagged or untagged) in PAKΔretSΔtsei8 is toxic when expressed in trans from pMMB67HE ((-) no induction; (+) with induction) (a) or when delivered by the H1-T6SS into a recipient strain lacking tsi8 (b). c-d, Tsi8 can rescue Tse8 toxicity in competition assays with donors PAKΔretS or PAKΔretSΔH1 and recipient PAKΔretSΔtsei8 expressing either pJN105 or pJN:tsi8 (c) and in growth assays with PAKΔretSΔtsei8 expressing pMMB:tse8 or pJN:tsi8 (d). e, Bacterial-Two-Hybrid (BTH) assays were used to quantify the level of interaction between Tse8 and Tsi8 with β-galactosidase activity assays performed on the cell lysates of each interaction pair. f, Tse8-HA-Strep interacts directly and specifically with Tsi8-His. Proteins were added to His-Tag Dynabeads as indicated. Lane 1: Tsi8-His (as bait) interacts with Tse8-HA-Strep. Lane 2: Tse8-HA-Strep alone does not interact with the Dynabeads. Lane 3: Tse8-HA-Strep does not interact with a different His-tagged bait protein, CcmE. Molecular weight markers positions are indicated on the left in kDa. Black vertical lines indicate where a lane was removed. Statistical analyses: (a) mean OD600 ± SEM is plotted over time from three independent replicates; (b) Mean CFUs/mL ± SEM of recipient cells in competition/alone are represented from three independent replicates performed in triplicate (n=3). Two-tailed student’s t-test, *** P<0.001; * P<0.05; ns between PAKΔretS and PAKΔretSΔH2ΔH3 (P=0.436); (c) Mean CFUs/mL ± SEM of recipient cells in competition/alone are represented from three independent replicates performed in triplicate (n=3). Two-tailed student’s t-test, * P<0.05 for each sample to PAKΔretS and ns between PAKΔretSΔH1 [pJN105] and PAKΔretS [pJN:tsi8] (P=0.598); (d) Mean OD600 ± SEM is plotted over time from three independent replicates; (e) Mean ± SEM of three biological replicates performed in triplicate (n=3). One-way Anova with Tukey’s multiple comparison post-test, * P<0.05 compared to the Miller units for T18c + T25 for Zip + Zip, or compared to Tsi8 + T25 and T18c + Tse8 for Tsi8 + Tse8; (f) Representative blot from one independent replicate (n=1).
The toxicity associated with the H1-T6SS-dependent delivery of Tse8 into a sensitive receiver strain could be rescued by expressing the tsi8 immunity gene from pJN105 in both a competition assay (Fig. 1c) and a growth assay (Fig. 1d), further confirming the protective role of Tsi8. In several cases, T6SS immunity proteins have been shown to directly interact with their cognate toxins17,24,25. Here, bacterial-two-hybrid (BTH) assays demonstrate that indeed Tse8 interacts strongly with Tsi8 (Fig. 1e). In addition, pull-down experiments using Tsi8-His as a bait, show direct interaction of the two proteins (Fig. 1f); this interaction is specific to Tsi8 as almost no Tse8-HA-Strep elutes from the pull-down beads in the absence of Tsi8 or in the presence of the non-specific binding control, CcmE-His (Fig. 1f).
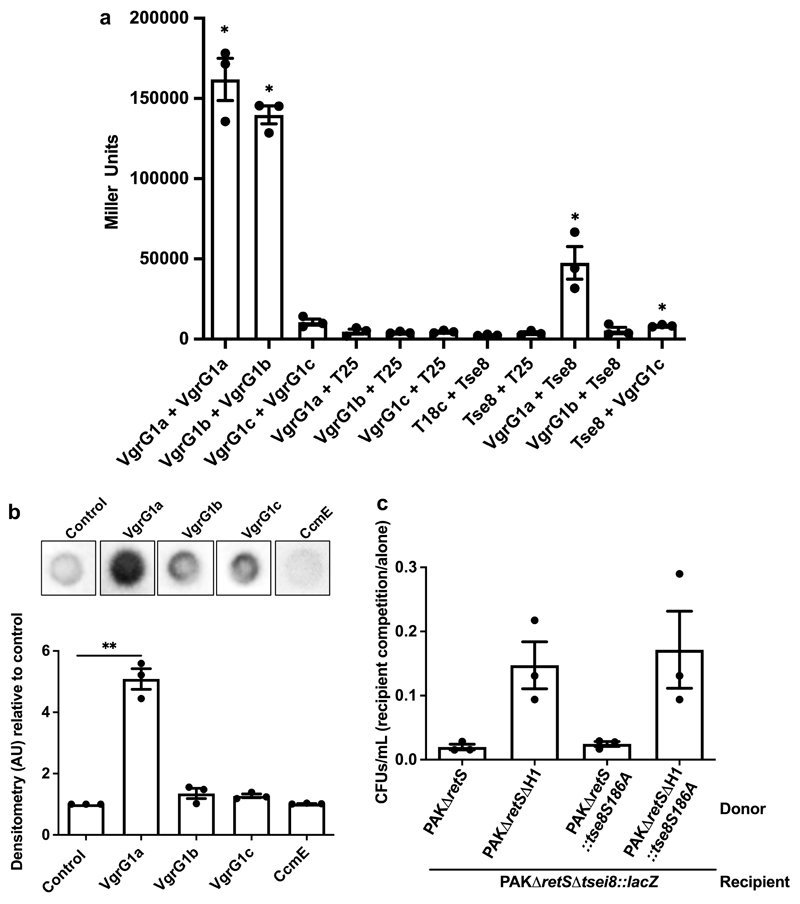
T6SS toxin delivery frequently relies on a direct interaction between the toxin and components of the T6SS spike11,14. BTH assays (Fig. 2a), as well as dot blot assays, revealed that Tse8 interacts strongly with VgrG1a (Fig. 2b). While the interaction of Tse8 with VgrG1c was significant in the BTH assay (Fig. 2a), no interaction above the non-specific binding control (CcmE-His) was observed in the dot blot assay (Fig. 2b). Finally, no interaction between Tse8 and VgrG1b was observed in BTH (Fig. 2a) or dot blot assays (Fig. 2b).
Figure 2. Tse8 interacts with VgrG1a and does not require putative catalytic residue for toxicity.
a, BTH assays were used to quantify the level of interaction between Tse8 and VgrGs with β-galactosidase activity assays performed on the cell lysates of each interaction pair. b, Tse8 interacts with VgrG1a in dot blot assays (top panel). Densitometry quantifications of Tse8 interactions with respective partners (bottom panel). CcmE-His is used as a non-specific binding control. c, Tse8 toxicity is not dependent on the conserved putative catalytic residue S186. Competition assays were performed with donors PAKΔretS, PAKΔretSΔH1, PAKΔretS::tse8S186A or PAKΔretSΔH1::tse8S186A and recipient PAKΔretSΔtsei8::lacZ. Statistical analyses: (a) Mean ± SEM of three biological replicates performed in triplicate (n=3). One-way Anova with Tukey’s multiple comparison post-test, * P<0.05 compared to the Miller units for each of VgrG1a, VgrG1b, VgrG1c and Tse8 with the respective T18c or T25 partner. (b) Densitometry measurements normalized to the control and represented as the Mean ± SEM from three independent replicates (n=3). Two-tailed student’s t-test, ** P<0.005 compared to control; ns between control and VgrG1b (P=0.169), VgrG1c (P=0.067) and CcmE (P=0.159). (c) Mean CFUs/mL ± SEM of recovered recipient are represented from three independent replicates performed in triplicate (n=3). Two-tailed student’s t-test, * P<0.05 for PAKΔretS compared to PAKΔretSΔH1 and PAKΔretSΔH1::tse8S186A; ns between PAKΔretS and PAKΔretS::tse8S186A (P=0.226).
Overall, the above results demonstrate that Tse8-Tsi8 is an antibacterial toxin-immunity pair associated with the H1-T6SS, and that Tse8 interacts with the VgrG1a tip to facilitate delivery into target cells.
Tse8 is a predicted amidase family enzyme
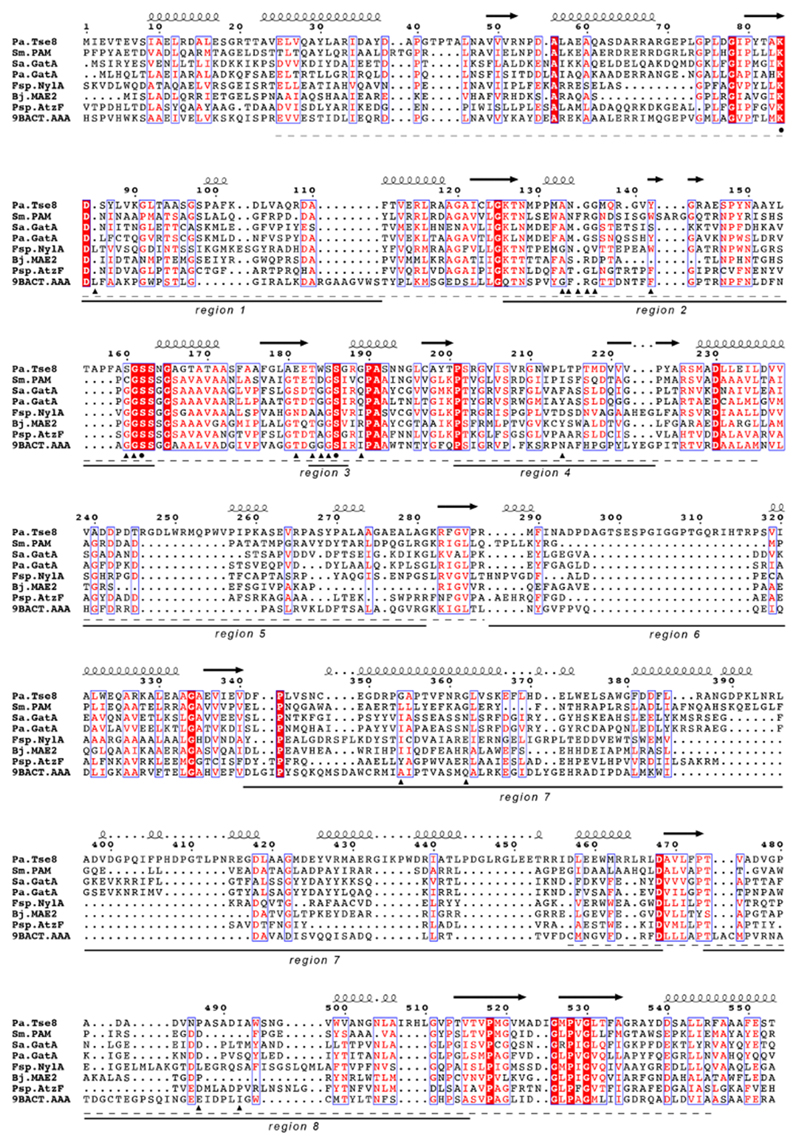
Using Phyre226 we found that the closest 3D homologs of Tse8 are the Stenotrophomonas maltophilia Peptide amidase (Pam)27 (sequence identity 29%), the Staphylococcus aureus Gln-tRNA(Gln) transamidosome subunit A (GatA)28 (sequence identity 20%), the P. aeruginosa Asn-tRNA(Asn) transamidosome subunit A (GatA)29 (sequence identity 25%), the Flavobacterium sp. 6-aminohexanoate cyclic dimer hydrolase (NylA)30 (sequence identity 24%), the Bradyrhizobium japonicum malonamidase E2 (MAE2)31 (sequence identity 25%), the Pseudomonas sp. allophanate hydrolase (AtzF)32 (sequence identity 30%), and the Bacterium csbl00001 Aryl Acylamidase (AAA)33 (sequence identity 22%). Amino acid sequence analysis indicates that Tse8 contains an Amidase Signature (AS) domain (Pfam PF01425) (Extended Data Fig. 4). AS sequences are characterized by a stretch rich in glycine and serine residues, as well as a highly conserved Ser-cisSer-Lys catalytic triad27,28,34–37. The catalytic Lys is located in the C-terminal end of a conserved β-strand (region 1) (Extended Data Fig. 4), while the cisSer is located at the C-terminus of region 2 (Extended Data Fig. 4). Finally, the nucleophilic Ser residue is located in a highly conserved short loop of region 3. All these AS signature sequence characteristics (underlined by a dashed line in Extended Data Fig. 4) are present in Tse8 and its closest 3D homologues.
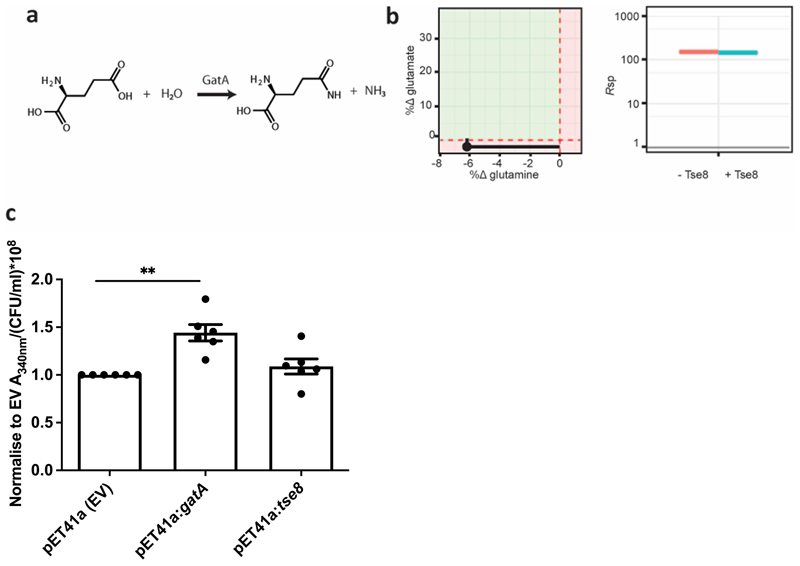
Given that Tse8 possesses the conserved catalytic features of amidase family enzymes (Extended Data Fig. 4), we tested whether it has amidase activity. Tse8 was purified and confirmed to be intact (Extended Data Fig. 5). Subsequently, its capacity to hydrolyse carbon-nitrogen bonds was tested on two molecules, epinecidin-1 and glutamine, which are substrates for Pam from S. maltophilia and GatA of the transamidosome, respectively. The amidase activities of Pam and Tse8 were analysed by Mass Spectrometry (MS) by monitoring the modifications of epinecidin-1 in the presence and absence of the tested proteins and of the small nucleophile hydroxylamine (Extended Data Fig. 6). While the C-terminus of epinecidin-1 was deaminidated in the presence of Pam (Extended Data Fig. 6b), it remained amidated in the presence of Tse8, suggesting that Tse8 has no amidase activity on this substrate (Extended Data Fig. 6 and Extended Data Fig. 7). The amidase activity of Tse8 was also tested on the GatA substrate glutamine (Extended Data Fig. 8) and no modification was detected by MS (Extended Data Fig. 8b). In addition, whole-cell glutaminase assays were performed and the amidase activity of E. coli whole cell lysates expressing GatA or Tse8 on L-glutamine was determined by monitoring the accumulation of NADPH. These experiments demonstrated that while GatA expressed from plasmid pET41a had a significant amidase activity, whole cells expressing Tse8 from the same vector produced a level of NADPH which was not significantly different to the empty vector-carrying control strain (Extended Data Fig. 8c). Overall, these data demonstrate that the substrates for Pam and GatA are not substrates for Tse8, suggesting that Tse8 is highly specific or unlikely to utilize amidase activity to elicit toxicity.
To assess whether Tse8 toxicity is mediated through amidase activity in vivo, we replaced the tse8 gene on the chromosome by an allele encoding a putative catalytic site mutant of Tse8 with a Ser186Ala (S186A) substitution. This conserved Ser186 residue (Extended Data Fig. 4) acts as the catalytic nucleophile in homologous amidases, and is necessary for enzymatic function38. PAKΔretS and PAKΔretSΔH1 donor strains encoding either wild-type Tse8 or Tse8S186A were competed against the recipient strain PAKΔretSΔtsei8::lacZ. This showed that there was no difference in the recovered CFUs/mL of the recipient when the attacking strain delivered either wild-type Tse8 or Tse8S186A (Fig. 2c), further suggesting that Tse8 does not utilize amidase activity to elicit toxicity in vivo.
Tse8 elicits toxicity by interacting with the bacterial amidotransferase complex
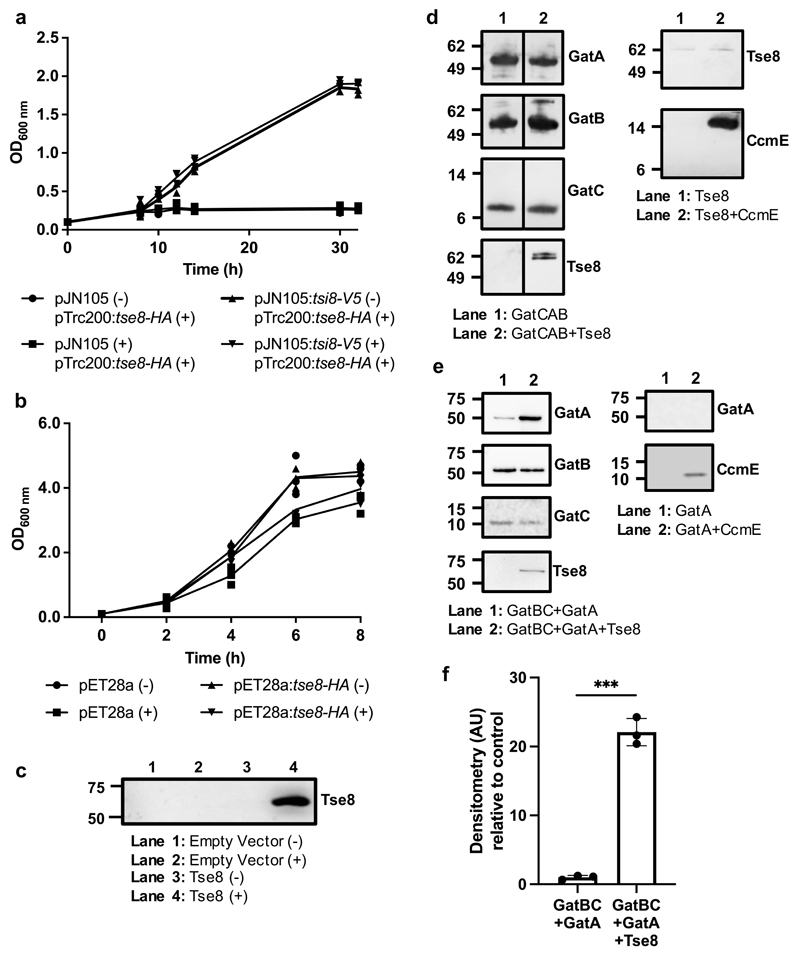
Since Tse8 toxicity does not appear to depend on it having amidase activity (Fig. 2c), we hypothesized that Tse8 could instead be eliciting toxicity by competing with a functional amidase either within the cell, or within a complex in the cell. Two 3D homologues of Tse8 are the A subunit of the S. aureus Gln-tRNA(Gln) transamidosome and the P. aeruginosa Asn-tRNA(Asn) transamidosome. Both of these proteins are the A subunit of transamidosome complexes, which are used by bacteria that lack the cognate tRNA synthases for asparagine (Asn) and/or glutamine (Gln)20. These bacteria utilize a two-step pathway instead, whereby a non-discriminating tRNA synthase generates a misacetylated aspartate- or glutamate-loaded tRNA which is then transaminated by the heterotrimeric amidotransferase enzyme GatCAB, within the transamidosome complex, to leave asparagine or glutamine correctly loaded onto their cognate tRNA. Given that not all bacteria rely on the transamidosome for protein synthesis, we reasoned that if Tse8 toxicity is directed at this enzymatic complex, then expression of Tse8 should only be toxic in bacteria which use the transamidosome. P. aeruginosa relies on the transamidosome for Asn-tRNA synthesis39 and we see a growth defect when Tse8 is expressed from a plasmid or delivered into a strain lacking Tsi8 (Fig. 1a-d). Agrobacterium tumefaciens lacks both Asn-tRNA and Gln-tRNA synthases and generates these cognate tRNAs through the transamidosome (Supplementary Table 4), while E. coli possesses both the Asn- and Gln-tRNA synthases and does not have a transamidosome complex (Supplementary Table 4). The effect of Tse8 expression was examined for both A. tumefaciens and E. coli. A growth defect was observed for A. tumefaciens, which could be rescued by co-expression of Tsi8 (Fig. 3a), but no growth defect was observed for E. coli (Fig. 3b) despite Tse8 expression at high levels from pET28a (Fig. 3c). Taken together these data suggest that Tse8 toxicity depends on the presence of the transamidosome.
Figure 3. Tse8 targets the transamidosome.
a-c, Tse8 is only toxic in bacteria which rely on the transamidosome for protein synthesis. Expression of Tse8 in A. tumefaciens is toxic but can be rescued by coexpression of Tsi8 ((-) no induction; (+) with induction) (a). Expression of Tse8 in E. coli is not toxic ((-) no induction; (+) with induction) (b), despite Tse8 being expressed (c). d-e, Proteins were added to His-Tag Dynabeads as indicated. d, Left panel, lane 1: His-GatB (as bait) interacts with GatC-HA and GatA-V5. Left panel, lane 2: Tse8 HA-Strep interacts with the GatCAB complex, but does not displace GatA-V5, even at a 15-fold molar excess. Right panel, lane 1: Tse8-HA-Strep alone does not interact with the Dynabeads. Right panel, lane 2: Tse8-HA-Strep does not interact with a different His-tagged bait protein, CcmE. e, Left panel, lane 1: His-GatB (as bait) interacts with GatC-HA and GatA-V5. Left panel, lane 2: The presence of Tse8-HA-Strep leads to drastic increase of the amount of GatA-V5 interacting with His-GatB and GatC-HA. Right panel, lane 1: GatA-V5 alone does not interact with the Dynabeads. Right panel, lane 2: GatA-V5 does not interact with a different His-tagged bait protein, CcmE. f, Quantification of the amount of GatA-V5 bound to His-GatB and GatC-HA in the presence or absence of Tse8-HA-Strep by densitometry. For panels (c-e) molecular weight markers positions are indicated on the left in kDa. Statistical analyses: (a-b) Mean OD600 ± SD is plotted over time from three independent replicates; (c) Representative blot from three independent replicates (n=3); (d) Representative blots from one independent replicate (n=1); (e) Representative blots from three independent replicates (n=3) for the left panel and one independent replicate (n=1) for the right panel; (f) Mean densitometry ± SEM from three independent replicates (n=3). Two-tailed student’s t-test, *** P<0.001 for GatBC+GatA compared to GatBC+GatA+Tse8.
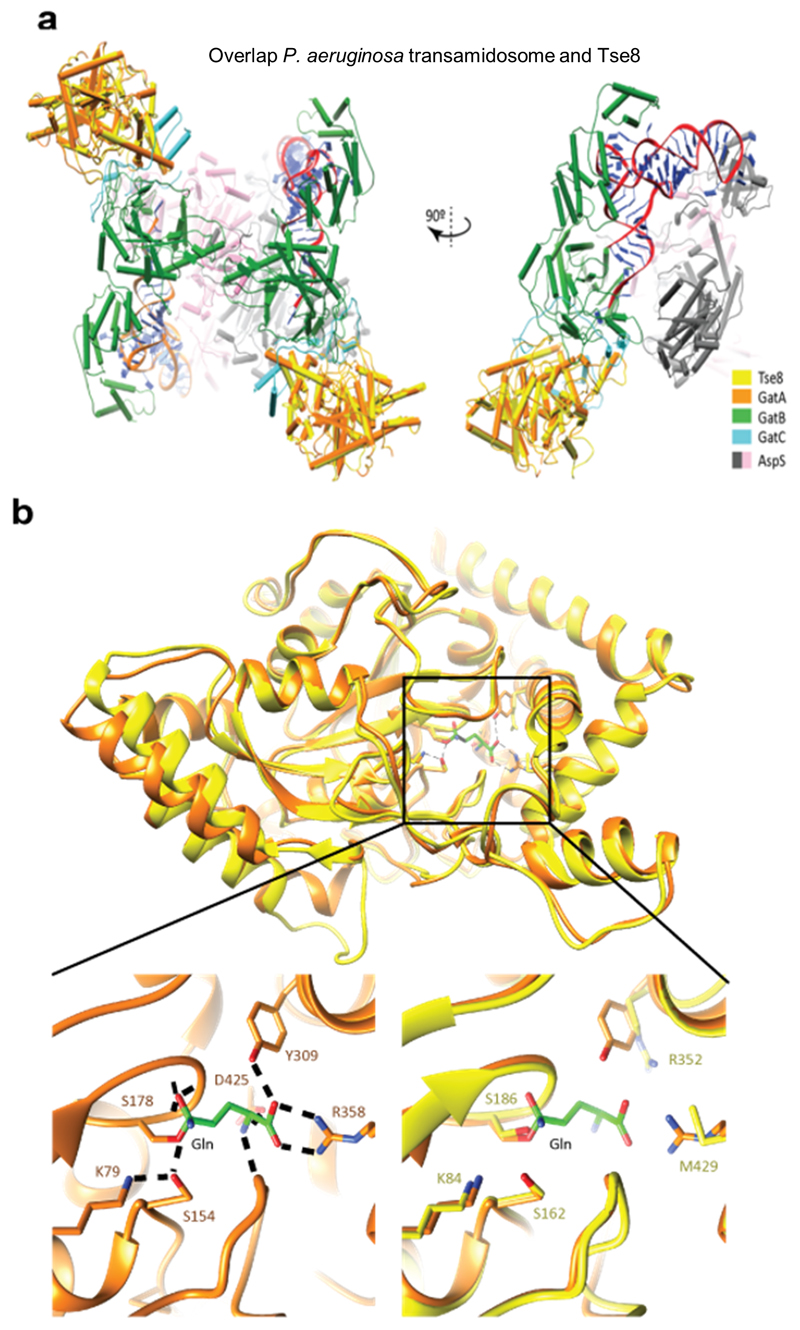
We generated a structural homology model of Tse8 based on the solved S. aureus GatA 3D structure (PDB: 2F2A). By overlaying this model with the A subunit of the P. aeruginosa transamidosome structure (PDB: 4WJ3) (Extended Data Fig. 9a), we found that Tse8 shares a high level of structural similarity to the A subunit of the complex. Further, comparison of the homologous residues within the substrate binding pockets of SaGatA and PaTse8 revealed that while the catalytic triad residues are conserved, the substrate binding residues (Tyr309, Arg358 and Asp425 in SaGatA)24 are not (Extended Data Fig. 9b), supporting our data and hypothesis that Tse8 does not have the same substrate as GatA (Extended Data Fig. 8). While this manuscript was in preparation a structure for Tse8 was published (RDB: 6TE4)40 that agrees with the overall conclusions from our homology modelling data.
Given the high level of predicted structural similarity between GatA and Tse8 we hypothesized that Tse8 may be able to interact with the transamidosome and could be eliciting toxicity by altering the functionality of this complex. The most likely scenario was that Tse8 replaces GatA, thus rendering the GatCAB complex inactive. To investigate this, we performed a pull-down experiment using purified proteins. GatCAB was purified as a complex using a Ni-affinity column through histidine-tagged GatB (His-GatB); GatA and GatC also had tags which were appropriate for their detection by western blot (GatA-V5 and GatC-HA). Tse8 was purified separately through a StrepII tag (Tse8-HA-Strep). GatCAB was pulled down in the presence and absence of a 15-fold molar excess of Tse8 via His-GatB on His-Tag Dynabeads. Tse8 was found to co-purify with GatCAB (lane 2, Fig. 3d). This interaction is specific to GatCAB, as minimal amounts of Tse8 elute from the pull-down beads in the absence of the transamidosome or in the presence of the non-specific binding control (CcmE-His) (Fig. 3d). However, even though a large molar excess of Tse8 was used in our pull-down experiment, the amount of GatA detected in the GatCAB complex remained largely unaffected (lane 2, Fig. 3d) excluding the possibility that Tse8 displaces GatA.
Another possibility was that Tse8 interacts with transamidosome components as the GatCAB complex assembles and that this interaction disrupts transamidosome function. To test this hypothesis, we purified GatBC as a complex using a Ni-affinity column through histidine-tagged GatB (His-GatB) and used this complex in pull-down experiments with cell lysates containing GatA and Tse8; GatA, GatC and Tse8 also had tags which were appropriate for their detection by western blot (GatA-V5, GatC-HA and Tse8-HA-Strep). We found that that the presence of Tse8, rather than inhibiting the binding of GatA to GatBC as we initially hypothesized, promotes it (lane 2, Fig. 3e), leading to a drastic accumulation of GatA on the GatBC complex (Fig. 3f). This GatA accumulation is specific to the presence of Tse8 and GatBC, as no GatA elutes from the pull-down beads in the absence of these proteins or in the presence of the non-specific binding control (CcmE-His) (Fig. 3e). The fact that we did not observe GatA accumulation upon Tse8 exposure in our pull down using intact GatCAB (Fig. 3d), suggests that Tse8 is more effective when it is acts on transamidosome components during the assembly of this complex. The structure of the P. aeruginosa GatCAB transamidosome reveals it to be a symmetric complex comprising an aspartyl-tRNA synthase (ND-AspRS), GatCAB, and tRNAAsn in a defined 2:2:2 stoichiometry29 (as represented in Extended Data Fig. 9aa). The function of this complex relies on large conformational changes between the ND-AspRS and the GatCAB components that are fine-tuned to accommodate the movement of the tRNAAsn between the domains of the transamidosome super-complex29. As such, additional Tse8 and GatA domains attached to the optimal transamidosome complex structure would likely inhibit transamidosome function by obstructing the communication between the ND-AspRS, GatCAB and the tRNAAsn. This in turn would result in a decrease in the production of Asn-tRNAAsn, ultimately impairing protein synthesis.
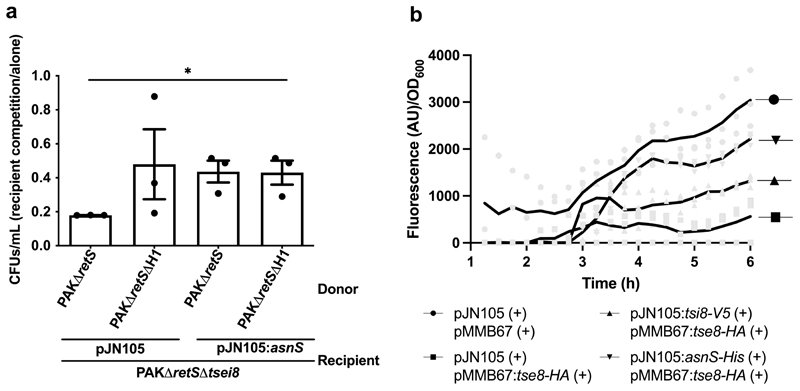
To further support our data suggesting that Tse8 exerts its toxicity by impairing protein synthesis through inhibition of the transamidosome, we hypothesized that if we were able to override the need for transamidosome function by providing the bacterium with the tRNA synthase it lacked, we would be able to rescue the observed growth defect when Tse8 is either expressed from a plasmid (Fig. 1a,d) or delivered by an attacker (Fig. 1b,c). P. aeruginosa only lacks the asparagine tRNA synthase39 (Supplementary Table 4), thus in this case Tse8 toxicity should be rescued by simply providing the cell with this tRNA synthase. To investigate this possibility, the Asn-tRNA synthase (asnS) from E. coli was expressed in PAKΔretSΔtsei8 from pJN105, and the strain competed against PAKΔretS and PAKΔretSΔH1. Expression of AsnS was able to rescue Tse8 toxicity (Fig. 4a) to the same extent as expression of the cognate immunity protein, Tsi8 (Fig. 1c). Furthermore, to directly test the effect of Tse8 expression on protein synthesis in vivo we expressed superfolder Gfp (sfGfp) from the Tn7 site of the P. aeruginosa chromosome in a Tse8-sensitive strain (PAKΔretSΔtsei8) that also expressed Tse8 or harboured the empty pMMB67HE vector. We found that the strain expressing Tse8 produces less sfGfp compared to the empty vector control (sfGfp signal was normalised to OD600; Fig. 4b), while this effect is specific to Tse8, since the decrease in sfGfp/total cells level in the presence of Tse8 could be rescued by co-expression of Tsi8 (Fig. 4b). Finally, co-expression of Tse8 with E. coli AsnS, also rescues the production of sfGfp/total cells (Fig. 4b), demonstrating that the decrease in fluorescent signal observed in the presence of Tse8 alone is originating from the specific interaction of Tse8 with its target, the transamidosome complex. Together these data demonstrate that strains containing Tse8 are less able to produce sfGfp, which, in turn, indicates that protein synthesis is inhibited by this T6SS toxin.
Figure 4. Tse8 impacts on protein synthesis in vivo.
a, Asn tRNA synthase (asnS) can rescue Tse8 toxicity. Competition assays were performed with donors PAKΔretS or PAKΔretSΔH1 and recipient PAKΔretSΔtsei8 expressing either pJN105 or pJN:asnS. b, Cells expressing Tse8 produce less sfGfp/total cells compared to an empty vector control. This effect can be rescued by expression of Tsi8 or AsnS. sfGfp levels normalised to total cells (measured by OD600) were monitored over time in PAKΔretSΔtsei8 with sfGFP expressed from the vacant Tn7 chromosomal site in cells containing the indicated plasmids ((+) with induction). Statistical analyses: (a) Mean CFUs/mL ± SEM of recipient cells in competition/alone are represented from represented from three independent replicates performed in triplicate (n=3). Two-tailed student’s t-test, * P<0.05; ns for PAKΔretSΔH1 [pJN105] vs PAKΔretS [pJN:asnS] (P=0.687) or vs PAKΔretSΔH1 [pJN:asnS] (P=0.631). (b) Mean fluorescent AU/OD600 ± SEM is plotted over time from three independent replicates performed in 8 technical replicates (n=3).
Discussion
In the current study we demonstrate that our global genomic approach can be used to identify T6SS toxin-immunity pairs associated with the H1-T6SS of P. aeruginosa. Our approach not only confirmed previously characterized P. aeruginosa T6SS toxin-immunity pairs, but also revealed several previously unidentified putative toxin-immunity pairs, including Tse8-Tsi8, which would probably not have been found using targeted approaches or bioinformatics. Characterization of the Tse8-Tsi8 pair, revealed that Tsi8 is the cognate immunity protein for the Tse8 toxin, and that Tse8 interacts with VgrG1a, hence it is likely delivered into target cells via the VgrG1a-tip complex.
Tse8 was also found to interact with GatCAB of the bacterial transamidosome complex, which is required for protein synthesis in certain bacteria that lack one or both of the asparagine or glutamine tRNA synthases20. Our pull-down data (Fig. 3e,f) demonstrate that Tse8 interaction with transamidosome components leads to accumulation of GatA onto GatBC, resulting in an amidotransferase complex with altered stoichiometry. Transamidosome function depends on a series of interactions between its ND-AspRS, GatCAB and the tRNAAsn components. These interactions are, in turn, reliant on the optimal architecture of the transamidosome that allows for extensive conformational changes to take place in order for the tRNAAsn to efficiently move between the domains of the super-complex29. It would be expected that Tse8-mediated precipitation of several additional GatA molecules on this complex will impact upon its fine-tuned architecture, resulting in functional deficits. According to our pull-down data, very little Tse8 is pulled with GatBC (Fig. 3e; all the blots in this figure have been exposed for the same amount of time using comparable commercial antibodies). This small amount of toxin is sufficient to nucleate the accumulation of GatA in significant amounts (Fig. 3f) and impair transamidosome function. Overall, this is in agreement with the logistics of Tse8 being delivered through the VgrG1a-tip complex, since only a maximum of three molecules of toxin can be delivered per T6SS firing event through VgrG. Based on this data, we propose that in bacteria where the transamidosome is essential (i.e. in bacteria lacking one or both of the Asn- or Gln-tRNA synthases), activity of Tse8 results in reduced fitness due to decreased levels of protein synthesis. In agreement with this, Tse8 toxicity can be rescued if the transamidosome function is bypassed upon provision of the transamidosome-independent tRNA-synthase lacked by the bacterium (i.e. AsnS for P. aeruginosa (Fig. 4a and 4b)).
Future work, focusing on further characterization of the specifics of the Tse8-GatCAB interaction, could point to ways of inhibiting the transamidosome and may provide a basis for the development of antibacterial agents against this target. Such agents might be useful in inhibiting the growth of important pathogens that rely on the transamidosome, without affecting the viability of many commensal bacteria which produce their proteins without depending on this pathway. Moreover, investigation of the other putative toxins detected in this study could also open new therapeutic avenues; elucidation of the substrates of these putative toxins could offer insights into pathways that are naturally validated antibacterial targets against P. aeruginosa. Looking beyond the T6SS of P. aeruginosa, there are many Gram-negative bacteria that infect human and animal hosts, or are plant pathogens or plant-associated organisms and possess at least one, if not multiple T6SSs clusters41–44. Furthermore, in several cases it has been demonstrated that distinct T6SS machines deliver a specific subset of toxins into target cells, often under certain conditions9,12,16, suggesting that toxins are not only bacterial specific, but potentially even niche specific. Given this diversity, we predict that our TraDIS approach could be useful for drastically expanding the repertoire of known T6SS toxins across a range of bacteria and ecologically or clinically relevant growth environments.
Methods
Bacterial strains, plasmids and growth conditions
Bacterial strains and plasmids used in this study are reported in Supplementary Table 2. P. aeruginosa PAK was used for TraDIS library generation and subsequent assays using mutant strains generated by allelic exchange mutagenesis as described previously45,46. P. aeruginosa strains were grown in tryptone soy broth (TSB), Lysogeny Broth (LB) or M9 or MOPS minimal media (with indicated supplements), supplemented with antibiotics as appropriate (streptomycin 2000 μg/mL, carbenicillin 100 μg/mL, gentamicin 50 μg/mL) at 37 °C with agitation. E. coli strains DH5α, SM10, CC118λpir and BL21(DE3) were used for cloning, conjugation and protein expression steps. E. coli cells were grown in TSB, LB, Terrific Broth or M9 minimal media (with indicated supplements), supplemented with antibiotics as appropriate (streptomycin 50 μg/mL, ampicillin 100 μg/mL, kanamycin 50 μg/mL) at 37 °C with agitation. A. tumefaciens C58 was grown in LB or M9 minimal media (with indicated supplements), supplemented with antibiotics as appropriate (gentamicin 50 μg/mL, spectinomycin 100 μg/mL) at 30 °C with agitation.
DNA manipulation
DNA isolation was performed using the PureLink Genomic DNA mini kit (Life Technologies) except for TraDIS library genomic DNA isolation (see below). Isolation of plasmid DNA was carried out using the QIAprep spin miniprep kit (Qiagen). Primers (Sigma) used are shown in Supplementary Table 3. DNA fragments were amplified with either KOD Hot Start DNA Polymerase (Novagen) or standard Taq polymerase (NEB) as described by the manufacturer, with the inclusion of Betaine (Sigma) or DMSO (Sigma). Restriction endonucleases (Roche) were used according to the manufacturer’s specifications. DNA sequencing was performed by GATC Biotech.
TraDIS library generation
A highly saturated transposon mutant library was generated in P. aeruginosa PAKΔretS or PAKΔretSΔH1 strains by large scale conjugation with an E. coli SM10 [pBT20] donor which allowed for random insertion of a mariner transposon throughout the genome and conferred gentamicin resistance in the recipient PAK strain. The E. coli donor strain was grown in LB supplemented with gentamicin (15 μg/mL) overnight at 37 °C and the recipient PAK strain was grown overnight at 37 °C in LB. Equivalent amounts of both strains were spread uniformly on separate LB agar plates and incubated overnight at 37 °C for E. coli and at 43 °C under humid conditions for the PAK recipient. The next day one E. coli donor plate was harvested and combined by extensive physical mixing on a fresh LB agar plate with one plate of harvested recipient PAK strain. Conjugation between the two strains was achieved by incubation of the high-density mixture of both strains at 37 °C for 2 hrs under humid conditions. The conjugation mix was then harvested, pelleted by centrifugation (10,000 g, 10 mins, 4 °C), and resuspended in LB. The resuspended cells were recovered onto large square (225 mm) Vogel-Bonner Media (VBM) (MgSO4.7H2O (8 mM), citric acid (anhydrous) (9.6 mM), K2HPO4 (1.7 mM), NaNH5PO4.4H20 (22.7 mM), pH 7) agar plates supplemented with gentamicin (60 μg/mL) and incubated for 16 hrs at 37 °C. The numbers of mutants obtained were estimated by counting a representative number of colonies across multiple plates. Mutants for each library background on plates were recovered as two separate pools (T6SS active and T6SS inactive), resuspended in LB, then pelleted by centrifugation (10,000 g, 10 mins, 4 °C), and then finally resuspended in LB plus glycerol (15% (v/v)) and stored at -80 °C. The protocol was repeated on a large scale until ~2 million mutants were obtained in each library background. For the TraDIS assay glycerol stocks of harvested PAKΔretS or PAKΔretSΔH1 TraDIS libraries were combined at normalized cell density for each separate replicate (i.e two final pools in total) and spread onto large square (225 mm) VBM agar plates supplemented with gentamicin (60 μg/mL) and incubated for 16 hrs at 37 °C to facilitate T6SS delivery of toxins and subsequent killing/self-intoxication of mutants lacking immunity genes for the cognate toxin. Cells were then harvested into 5 mL LB and pelleted by centrifugation (10,000 g, 15 mins, 4 °C). Cell pellets were resuspended in 1.4 mL LB and 1 mL was retained for subsequent genomic DNA extraction (see ‘TraDIS library genomic DNA extractions’ section below).
TraDIS library assay
Glycerol stocks of harvested PAKΔretS or PAKΔretSΔH1 TraDIS libraries were combined at normalized cell density for each separate replicate and spread onto large square (225 mm) VBM agar plates supplemented with gentamicin (60 μg/mL) and incubated for 16 h at 37 °C. Cells were then harvested into 5 mL LB and pelleted by centrifugation (10,000 g, 15 min, 4 °C). Cell pellets were resuspended in 1.4 mL LB and 1 mL was taken for subsequent genomic DNA extraction (see below).
TraDIS library genomic DNA extractions
Genomic DNA from the harvested pooled library pellets either before or after undergoing the ‘TraDIS library assay’ (above) were resuspended in 1.2 mL lysis solution (10 mM Tris-HCl, 400 mM NaCl and 2 mM Na2EDTA, supplemented with Proteinase K in storage buffer (50 mM Tris-HCl, 50% (v/v) glycerol, 100 mM NaCl, 0.1 mM EDTA, 10mM CaCl2, 0.1% (v/v) Triton X-100 and 1 mM DTT) to a concentration of 166 μg/mL. Cell lysis was achieved by incubation at 65 °C for 1 h, with occasional vortexing. The samples were then cooled to room temperature and RNA removed by addition of RNase A (5 μg/mL) and incubation at 37 °C for 80 min. Samples were then placed on ice for 5 min. Each lysate was then split into 2 eppendorf tubes at ~600 μL per tube, and 500 μL NaCl (5 M) were added to each tube. Cell debris were removed by centrifugation (10,000 g, 10 min, 4 °C) and 500 μL from each tube was added to 2 volumes of isopropanol to precipitate the DNA. DNA was then collected by centrifugation (10,000 g, 10 min, 4 °C), and DNA pellets were washed twice in 70% (v/v) ethanol. The fully dried DNA pellet was finally resuspended in Tris-EDTA buffer.
PAK reference genome
The PAK genome under the NCBI number accession number LR657304, also listed in the European Nucleotide Archive (ENA) under accession number ERS195106, was used. See details in Cain et. al. (2019)47. PAK loci in Table 1, Extended Data Fig. 9 and throughout the text are the corresponding loci names from this genome.
Generation of TraDIS sequencing libraries, sequencing and downstream analysis
TraDIS sequencing was performed using the method described previously22, with some minor modifications for this study, as described below. Also see Extended Data Fig. 10, and Supplementary Table 1.
PCR primers were designed for library construction and used for both the PAK libraries (5′: AATGATACGGCGACCACCGAGATCTACACACAGGAAACAGGACTCTAGAGG ATCACC and 3′: AATGATACGGCGACCACCGAGATCTACACCTTCTGTATGGAACG GGATGCG) and the sequencing TraDIS primers (5′: CAGCTTTCTTGTACACTAGA GACCGGGGACTTATCAG, and 3′: AAGCCTGCTTTCTAGAGACCGGGGACTTAT CAG). During library production, a post-ligation double digest with restriction enzymes AgeI and SgrAI was performed according to the manufacturer’s instructions (New England Biolabs) to prevent amplification of plasmid background. The T6SS TraDIS sequencing was performed on a HiSeq2500 Illumina platform on the RAPID 50bp SE read setting. Reads were mapped onto the PAK genome (accession number: ERS195106), and comparisons were performed using the TraDIS Toolkit informatics package22. 10% of the 3′ end of each gene was discounted, and a 10 read minimum cut-off was used to be included in analysis. On average there was a unique transposon insertion site every 53 bp over the whole genome for each of the T6SS active and T6SS inactive backgrounds and, thus the genome was highly saturated in each library. The distribution of transposon insertions across the genome based upon the normalized transposon insertions in a H1-T6SS inactive library background, compared to the H1-T6SS active library background is shown in Extended Fig. 10. The resulting sequences of the T6SS TraDIS assays are available from the European Nucleotide archive (ENA) under study accession number PRJEB1597.
To pinpoint genes involved in protection of T6SS-mediated killing, EdgeR48 was used to identify significant differences in read counts of genes in strains with (PAKΔretS) and without (PAKΔretSΔH1) an active H1-T6SS. Then the trimmed mean of M values (TMM) normalization was used to account for differences between then libraries, and tagwise dispersion was estimated. Only genes exhibiting greater than 5 reads in both replicates of the conditions or control sets were examined for differences in the prevalence of mutants. Genes with zero read counts in the other condition were offset using the prior count function in EdgeR48 so that fold changes could be estimated. P values were corrected for multiple testing using the Benjamini-Hochberg method, and genes with a corrected P value (Q value) of <0.05 (5% false discovery rate) and an absolute log2 fold change (log2FC) of >2 were considered significant (see Tab 2 in Supplementary Table 1). A list of 49 genes resulted having statistically significant decreased insertions in the T6SS active library PAKΔretS compared to normalized values in the PAKΔretSΔHI library. These genes were then interrogated as potential immunities, based firstly on gene size (the known H1-T6SS associated immunity genes (tsi1-6) at the time of analysis are all less than 600bp, thus this was used as a guide to shorten the list to 29 genes) (see Tab 3 in Supplementary Table 1) and also on whether a protein upstream these genes appeared to have a predicted enzymatic or putative toxin function.
Bacterial growth assays
Growth assays were performed as follows. For Fig. 1a, overnight cultures of PAKΔretSΔtsei8 were diluted down to OD600 = 0.1 in M9 minimal media (supplemented with MgSO4 (2 mM), CaCl2 (0.1 mM), glucose (0.4% (w/v)) and FeSO4.7H2O (0.01 mM)) and grown shaking at 37 °C. Expression of Tse8 was induced with IPTG (1 mM) at 4 h. For Fig 1d, PAKΔretSΔtsei8 cells carrying both pJN105 and pMMB67HE plasmids (+/- Tsi8/Tse8) were grown in MOPS minimal media (MOPS (40mM, pH 7.5), Tricine (4 mM, pH 7.5), NH4CL (9.52 mM), CaCL2 (0.5 uM), MgCl2.7H20 (0.52 mM), NaCl (50 mM), FeSO4.7H2O 20 mM (0.01 mM), K2HPO4 (1.32 mM) supplemented with 1x micronutrient mix (100x: Ammonium molybdate tetrahydrate (3 uM), Boric acid (400 uM), Cobalt chloride (30 uM), Cupric sulphate (10 uM), Manganese chloride (80 uM), Zinc sulphate (10 uM) and Nickel chloride hexahydrate (0.1% (w/v/)) and glucose (0.4% (w/v)) and L-Glutamine (0.05% (w/v)) shaking at 37 °C (without antibiotics). Expression of Tse8 was induced with IPTG (1 mM) and Tsi8 with arabinose (0.2% (w/v)) at 5 h. For Fig. 3a, overnight cultures of A. tumefaciens with pTrc200/pJN105 plasmids (+/- Tse8/Tsi8) were diluted down to OD600 = 0.1 in MOPS media without antibiotics as above and grown shaking at 30 °C. Expression of Tse8 was induced with IPTG (1 mM) and Tsi8 with arabinose (0.2% (w/v)) at 8 h. For Fig. 3b, overnight cultures of E. coli were diluted down to OD600 = 0.1 in M9 minimal media (supplemented with MgSO4 (2 mM), CaCl2 (0.1 mM), FeSO4.7H2O (0.01 mM) and glucose (0.4% (w/v)) and grown shaking at 37 °C. Tse8 expression was induced with IPTG (1 mM) after 2 h. For Fig. 4b, overnight cultures of the indicated P. aeruginosa strain were diluted down to OD600 = 0.1 in LB (without antibiotics) and grown shaking at 37 °C. Expression of Tse8 was induced with IPTG (0.25 mM) and Tsi8 or AsnS with arabinose (0.2% (w/v)) at 0 h.
T6SS competition assays
T6SS competition assays were performed as described previously49 with modifications as indicated. Briefly, overnight cultures of donor and recipient bacteria alone or in a 1:1 ratio were combined and spot plated on LB agar plates for 5 h at 37 °C and recovered in serial dilution on LB agar plates supplemented with Xgal (5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside) (100 μg/mL) to differentiate recipient (PAKΔretSΔtsei8::lacZ seen as blue) from donor (white). For recovery of competition assays between donor and recipient PAKΔretSΔtsei8 [pBBR1-MCS5] and [pBBR1:tsei8], the competition assay was plated onto LB agar plates with gentamicin (50 μg/mL) to differentiate donor from recipient (GmR). For recovery of competition assays between donor and recipient PAKΔretSΔtsei8 [pBBR1-MCS4] and [pBBR4:tse8], the competition assay was plated onto LB agar plates with carbenicillin (50 μg/mL) to differentiate donor from recipient (CarbR). In other cases, expression of Tsi8 or AsnS in the recipient strains was induced in the overnight cultures by addition of arabinose (0.2% (w/v)). These overnight cultures of donor and induced recipient alone or in a 1:1 ratio, were combined and spot plated onto LB agar supplemented with arabinose (1% (w/v)) for induction of Tsi8-V5 or AsnS-His for 5 h, with the competition assay finally being recovered on LB agar plates supplemented with gentamycin (50 μg/mL) and arabinose (1% (w/v)).
Bacterial Two Hydrid (BTH) and β-Galactosidase assays
Protein-protein interactions were analysed using the BTH system as described previously50. Briefly, the DNA region encoding the protein of interest were amplified by PCR and were then cloned into plasmids pKT25 and pUT18C, which each encode for complementary fragments of the adenylate cyclase enzyme, as previously described50 resulting in N-terminal fusions of T25/T18 from the adenylate cyclase to the protein of interest. Recombinant pKT25 and pUT18c plasmids were simultaneously used to transform the E. coli DHM1 strain, which lacks adenylate cyclase, and transformants were spotted onto Xgal (40 μ/mL) LB agar plates supplemented with IPTG (1 mM), Km (50 μg/mL) and Amp (100 μg/mL). Positive interactants were identified after incubation at 30 °C for 48 h. The positive controls used in the study were pUT18C or pKT25 derivatives encoding the leucine zipper from GCN4, which forms a dimer under the assay conditions. The strength of the interactions in the BTH assays was quantified from the β-galactosidase activity of co-transformants scraped from Xgal plates and measured as described previously; activity was calculated in Miller units50.
Western Blot analysis
SDS-PAGE and western blotting were performed as described previously11. Proteins were resolved in 8%, 10%, 12% or 15% gels using the Mini-PROTEAN system (Bio-Rad) and transferred to nitrocellulose membrane (GE Healthcare) by electrophoresis. Membranes were blocked in 5% (w/v) milk (Sigma) before incubation with primary antibodies (anti-His at 1:1000 dilution and anti-V5 or anti-HA at 1:5000 dilution). Membranes were washed with TBST (0.14 M NaCl, 0.03 M KCl and 0.01 M phosphate buffer plus Tween 20 (0.05% v/v)) before incubation with HRP-conjugated secondary antibodies (Sigma; anti-mouse at 1:5000 dilution). The resolved proteins on the membrane blots were detected using the Novex ECL HRP Chemiluminescent substrate (Invitrogen) or the Luminata Forte Western HRP substrate (Millipore) using a Las3000 Fuji Imager. For Fig. 3c, samples were taken after 8 h of growth and expression of Tse8 was assessed by Western blot as above; detection of Tse8 was performed using anti-HA antibody (1:5000 dilution).
Dot blotting
For Tse8 interactions with VgrG1a, VgrG1b and VgrG1c purified untagged Tse8 was spotted on nitrocellulose membrane (3 mg/ml) and dried at room temperature. Membranes were blocked with TBST with 5% (w/v) milk or 2.5% (w/v) bovine serum albumin for 7 h at room temperature. E. coli overexpressing VgrG1a-V5, VgrG1b-V5, VgrG1c-V5 or CcmE-His (equivalent 150 OD600 units) were pelleted and then resuspended in 10 mL 100 mM NaCl, 20 mM Tris, 10% (w/v) glycerol, 2% (w/v) milk powder and 0.1% (v/v) Tween-20 (Tween-20 was added after sonication) (pH 7.6) and sonicated. 10 mL of the crude lysates were applied directly to the membranes and incubated overnight at room temperature. The membranes were immunoblotted with anti-V5 (1:5000 Invitrogen) or anti-His (1:1000 Sigma) overnight at 4 °C and anti-mouse secondary (1:5000). Quantification of dot blots was performed using the Gel Analyzer plugin in ImageJ51. Levels were normalised to the control signal based on 3 independent experiments.
Pull-down experiments
E. coli BL21(DE3) strains expressing simultaneously GatA-V5, GatB-His and GatC-HA or GatB-His and GatC-HA were grown in LB at 37°C to an OD600 of 0.8 and expression was subsequently induced using 1 mM IPTG (Sigma) for 16 h at 18 °C. E. coli BL21(DE3) cells expressing Tse8-HA-Strep were grown in Terrific Broth at 37°C to an OD600 of 0.8 and expression was subsequently induced using 1 mM IPTG (Sigma) for 16 h at 30 °C. The same expression strategy used for Tse8-HA-Strep was also used for E. coli BL21(DE3) strains expressing Tsi8-His or CcmE-His except that TSB medium was used. Cell pellets resulting during expression of GatCAB, GatBC, Tsi8 or CcmE were resuspended in buffer A (50 mM Tris-HCl, 150 mM NaCl, 20 mM imidazole (pH 7.5)) and lysed by sonication after the addition of protease inhibitors (Roche). Cell debris were eliminated by centrifugation (48,000 g, 30 min, 4 °C). Proteins were purified by immobilized metal affinity chromatography using nickel-Sepharose resin (GE Healthcare) equilibrated in buffer A. Proteins were then eluted off the resin with buffer A containing 200 mM instead of 20 mM imidazole. Cell pellets resulting during expression of Tse8 were resuspended in 50 mM Tris-HCl, 150 mM NaCl (pH 7.5) and lysed by sonication after the addition of protease inhibitors (Roche). Tse8-HA-Strep was purified using Strep-Tactin Sepharose (IBA), according to the manufacturer’s specifications.
For pull-down experiments using pure-proteins, the above purified protein solutions and His-Tag Isolation & Pull Down Dynabeads (ThermoFischer Scientific) were used. Briefly, the appropriate protein mixtures were generated by mixing 40 μM of the bait protein with equimolar amounts of Tse8-HA-Strep (Tsi8 bait) or 15-fold molar excess of Tse8-HA-Strep (GatCAB bait); a condition containing solely the same amount of Tse8-HA-Strep was also tested as a negative binding control. Mixtures were added to a 25 μL bed of Dynabeads and incubated at 25 °C with agitation for 1 h, before the beads were washed 8x with 800 μL of wash buffer (50 mM Tris pH 7.5, 150 mM NaCl, 0.01% Tween 20) and resuspended in elution buffer (50 mM Tris pH 7.5, 150 mM NaCl, 0.01% Tween 20, 200 mM imidazole).
For pull-down experiments using purified GatCB and cell lysates containing GatA-V5 and Tse8-HA-StrepII, 150 OD600 units of cells expressing GatA-V5 and Tse8-HA-StrepII were resuspended in binding buffer (20 mM Tris pH 7.5, 100 mM NaCl, 10% (v/v) glycerol and 3% (w/v) bovine serum albumin) and lysed by sonication. Pull-downs were performed by adding a total volume of 6 mL of cell lysate (3 mL GatA-V5 lysate mixed with 3 mL binding buffer or 3 mL GatA-V5 lysate mixed with 3 mL Tse8-HA-StrepII lysate) to a 25 μL bed of His-Tag Isolation & Pull Down Dynabeads (ThermoFischer Scientific) loaded with 40 μg of purified GatCB. Mixtures were incubated at 25 °C with agitation for 1 h before the beads were washed 8x with 800 μL of wash buffer (50 mM Tris pH 7.5, 150 mM NaCl, 0.01% Tween 20) and resuspended in elution buffer (50 mM Tris pH 7.5, 150 mM NaCl, 0.01% Tween 20, 200 mM imidazole).
For all experiments, eluted samples were denatured in 4 x Laemmli buffer and subjected to western blotting as described above. Anti-V5 (1:5000 Invitrogen), anti-HA (1:5000 Biolegend) or anti-His (1:1000 Sigma) primary antibodies were used along with an anti-mouse secondary (1:5000 Sigma). For detection of StrepII tags a Strep-Tactin HRP conjugate was used (1:3000 IBA Lifesciences). Quantification of Western blot bands was performed using the Gel Analyzer plugin in ImageJ51.
Whole-cell glutaminase assays
The whole-cell glutaminase activity was measured as described previously52 with some modifications as follows. E. coli B834 cells containing empty vector, gatA or tse8 in pET41a were grown to OD600 ~ 0.6 when expression was induced by addition of IPTG (0.5 mM) and grown at 18 °C for 16 h. Cells pellets equivalent to 45 OD600 units were washed in sodium acetate solution (sodium acetate (100 mM, pH 6), L-glutamine (20 mM)) and resuspended in a final volume of 600 μL sodium acetate solution, and incubated at 37 °C for 30 min. 20 μL of cells were retained and serially diluted to quantify the CFUs present. The remaining cell volume was then lysed by heating at 99 °C for 3 min. Once cooled to room temperature 100 μL of cell lysate was added to 2 mL of glutamate dehydrogenase solution (sodium acetate (10 mm), NAD+ (4 mM), hydroxylamine HCl (400 mM), 30 U of glutamate dehydrogenase (GDH) enzyme (Sigma) in potassium phosphate buffer (100 mM, pH 7.2)) and incubated at 60 °C for 60 min. 150 μL of the reaction was added to a 96 well clear plate and the relative accumulation of NADPH was calculated using the measured absorbance at 340 nm.
Expression and purification of Tse8 used for activity measurements
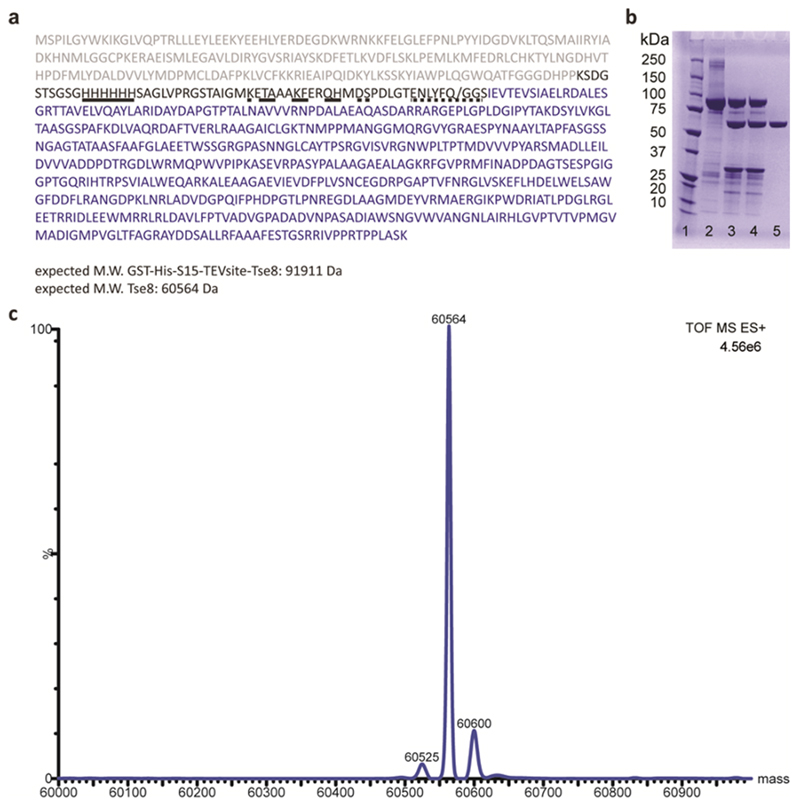
The pET41a::GST-TEV-Tse8 vector coding for P. aeruginosa Tse8 was obtained by FastCloning53 using pET41a:GST-Tse8 (see Supplementary Table 2) as template. This construct was subcloned using the forward primer 5′-AACCTGTATTTTCAGGGCGGATCC ATCGAGGTCACCGAGGTTTCCATCG-3′ and reverse primer 5′-CCTGAAAATACAGG TTTTCGGTACCCAGATCTGGGCTGTCCATGTGCTGG-3′ in order to exchange the Human Rhinovirus (HRV) 3C cleavage site (LEVLFQ/GP) with a TEV protease cleavage site (ENLYFQ/G). The resulting construct includes (i) a 651-nucleotide sequence encoding a N-terminal GST tag, (ii) an 18-nucleotide sequence encoding a 6x histidine tag, (iii) a 45-nucleotide sequence encoding a S15 tag and a 21-nucleotide sequence encoding the optimal tobacco etch virus (TEV) protease cleavage site Glu-Asn-Leu-Tyr-Phe-Gln-Gly (Extended Fig. 5). For protein expression, E. coli BL21(DE3) cells were transformed with the pET41a::GST-TEV-Tse8 plasmid and grown in 2xYT (Yeast Extract Tryptone) medium (supplemented with 50 μg/ml kanamycin) at 37 °C. When the culture reached an OD600 value of 0.7, Tse8 expression was induced by adding 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) and the temperature was dropped to 18°C. After 18 hr, cells were harvested and frozen for later use.
For protein purification, each 1 L pellet was resuspended in 50 ml of 50 mM Tris-HCl pH 8, 500 mM NaCl, 20 mM imidazole, 0.5 mM EDTA and 2 μL of benzonase endonuclease (without addition of protease inhibitors). Cells were then disrupted by sonication and the suspension was centrifuged for 40 mins at 56,000 g. The supernatant was filtered with a 0.2 μm syringe filter and subjected to immobilized metal affinity chromatography using a 1 ml HisTrap HP column (GE Healthcare), on a fast protein liquid chromatography system (ÄKTA FPLC; GE Healthcare) equilibrated with 5 ml of 50 mM Tris-HCl pH 8, 500 mM NaCl and 20 mM imidazole (buffer A). The column was washed with buffer A at 1 ml/min until no absorbance at 280 nm was detected. Elution was performed with a linear gradient between 0-50% of 50 mM Tris-HCl pH 8, 500 mM NaCl and 500 mM imidazole in 30 mL and at 1 ml/min. Fractions containing GST-TEV-Tse8 fusion protein were pooled and protein concentration was measured. The cleavage of the GST-His-S15 tag was performed with TEV protease (1 mg per 10 milligrams of protein) overnight at 18 °C in buffer 50 mM Tris–HCl pH 7.5, 2 mM DTT, at a protein concentration between 0.3-0.5 mg/mL. The cleaved Tse8, non-cleaved Tse8 and TEV protease were collected, filtered and applied onto a HisTrap HP column (5 ml; GE Healthcare) equilibrated with 25 ml of 50 mM Tris-HCl pH 7.5. The cleaved Tse8 was eluted in the flow-through and applied onto a Mono Q column of 5 mL (GE Healthcare) equilibrated with 25 ml of 50 mM Tris-HCl pH 7.5. The protein was eluted in a single step using 500 mM NaCl in 50 mM Tris-HCl pH 7.5. The Tse8 protein was dialyzed with 20 mM sodium phosphate buffer pH 7.6 and concentrated using Centricon centrifugal filter units of 30 kDa molecular mass cut-off (Millipore) to a final concentration of 5 mg/mL for enzymatic assays. The purity of the protein was verified by SDS-PAGE (Extended Data Fig. 5) and protein integrity was evaluated following desalting with stage-tip C4 microcolumns (Zip-tip, Millipore) by electrospray ionization mass spectrometry (ESI-MS). The sampling cone energy was set at 35 V. The m/z data were then deconvoluted into MS-data using the MaxEnt software (MaxEnt Solutions Ltd, Cambridge, UK) with a resolution of the output mass of 0.5 Da/channel and Uniform Gaussian Damage Model at the half height of 0.5 Da. The analysis indicates that 90% of the protein sample corresponds to the expected Tse8 molecular weight (60,564 Da; Extended Data Fig. 5).
Tse8 substrate activity assays
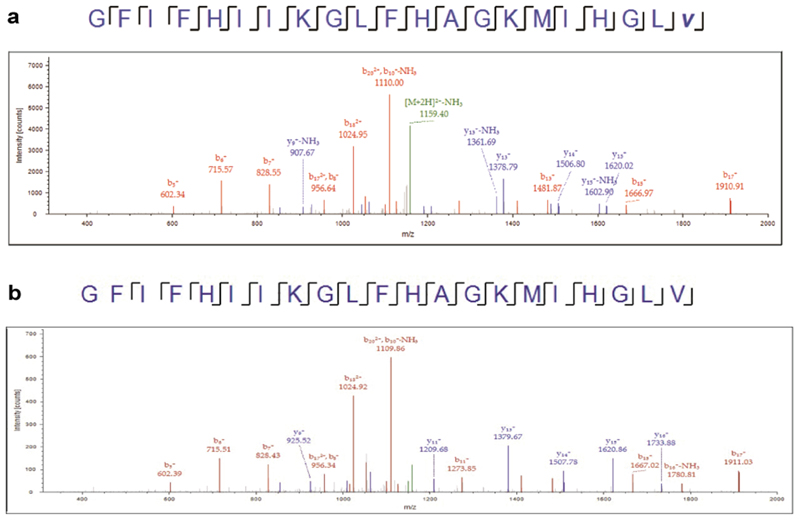
Putative Tse8 substrates were selected based on the predicted GatA and PAM homology. Thus, the capacity of Tse8 to hydrolyse carbon-nitrogen bonds was analysed by mass spectrometry (MS) using as putative substrates the free amino acid glutamine and the C-terminally amidated peptide epinecidin-1 (sequence: GFIFHIIKGLFHAGKMIHGLV-NH2) (Bachem AG). Glutamine (10 mM) was incubated with 2 μM of freshly purified Tse8. Reactions were carried out in two different buffers to test the possible effect of pH; one set of reactions was carried out in 10 mM sodium phosphate buffer (pH 7.6) and another set of reactions was carried out in 20 mM Tris-HCl buffer (pH 8.3). For epinecidin-1, 5 μM of freshly-purified Tse8 or the positive control protein Pam (purified as described previously54), were incubated with 50 μM of putative substrate in 10 mM sodium phosphate buffer (pH 7.2); control reactions, lacking Tse8 or Pam, were also tested. Reactions were incubated overnight at 30 °C, followed by MS analysis. For full details on the MS analysis see the relevant section below for use of epinecidin-1 or glutamine as a substrate.
Mass spectrometry analysis of Tse8/Pam enzymatic assays using epinecidin-1 as a substrate
Samples were desalted and peptides were isolated using stage-tip C18 microcolumns (Zip-tip, Millipore) and further resuspended in 0.1% formic acid prior to MS analysis. Peptide separation was performed on a nanoACQUITY UPLC System (Waters) on-line connected to an LTQ Orbitrap XL mass spectrometer (Thermo Electron). An aliquot of each sample was loaded onto a Symmetry 300 C18 UPLC Trap column (180 μm x 20 mm, 5 μm (Waters)). The precolumn was connected to a BEH130 C18 column (75 μm x 200 mm, 1.7 μm (Waters), and equilibrated in 3% acetonitrile and 0.1% FA. Peptides were eluted directly into an LTQ Orbitrap XL mass spectrometer (Thermo Finnigan) through a nanoelectrospray capillary source (Proxeon Biosystems), at 300 nl/min and using a 120 mins linear gradient of 3-50% acetonitrile. The mass spectrometer automatically switched between MS and MS/MS acquisition in DDA mode. Full MS scan survey spectra (m/z 400-2000) were acquired in the orbitrap with mass resolution of 30,000 at m/z 400. After each survey scan, the six most intense ions above 1,000 counts were sequentially subjected to collision-induced dissociation (CID) in the linear ion trap. Precursors with charge states of 2 and 3 were specifically selected for CID. Peptides were excluded from further analysis during 60 s using the dynamic exclusion feature. RAW files were searched with the Mascot search engine (www.matrixscience.com) through Proteome Discoverer v1.4 (Thermo) against a FASTA database containing the protein and peptide sequences of interest, together with a Pichia pastoris database from Uniprot/Swissprot as a background. Search parameters were: 10 ppm peptide mass tolerance, 0.5 Da fragment mass tolerance, carbamydomethylation of cysteines as fixed modification, and oxidation of methionine, amidation and deamidation of protein C-terminus as variable modifications. Only highly reliable hits (p<0.01) were considered.
Mass spectrometry analysis of Tse8 enzymatic assay using glutamine as substrate
Overnight incubations were quenched by addition of 150 μL 20% acetonitrile (MeCN). Controls for the experiment were prepared by first adding MeCN to the reaction blank and subsequently adding enzyme. In order to determine LC-MS performance, 100 μM stock solutions of glutamine substrate in 2:3 water/MeCN were injected before the experimental samples. Quenched incubations and controls were shaken in the tubes for 30 mins at 4°C and 1,000 g. Next, samples were centrifuged for 30 mins at 4°C and 25,000 g. The resulting solutions were immediately injected in the LC-MS. Samples were measured with a UPLC system (Acquity, Waters Inc., Manchester, UK) coupled to a Time of Flight mass spectrometer (ToF MS, SYNAPT G2, Waters Inc.). A 2.1 x 100 mm, 1.7 μm BEH amide column (Waters Inc.), thermostated at 40 °C, was used to separate the analytes before entering the MS. Mobile phase solvent A (aqueous phase) consisted of 99.5% water, 0.5% formic acid and 20 mM ammonium formate while solvent B (organic phase) consisted of 29.5% water, 70% MeCN, 0.5% formic acid and 1 mM ammonium formate. In order to obtain a good separation of the analytes the following gradient was used: from 5% A to 50% A in 2.4 mins in curved gradient (#8, as defined by Waters), from 50% A to 99.9% A in 0.2 mins constant at 99.9% A for 1.2 mins, back to 5% A in 0.2 mins. The flow rate was 0.250 mL/min and the injection volume was 2 μL. The MS was operated in positive (ESI+) and negative (ESI-) electrospray ionization in full scan mode. The cone voltage was 25 V and capillary voltage was 250 V for ESI+ and 500 V for ESI-. Source temperature was set to 120 °C and capillary temperature to 450 °C. The flow of the cone and desolvation gas (both nitrogen) were set to 5 L/h and 600 L/h, respectively. A 2 ng/mL leucine-enkephalin solution in water/acetonitrile/formic acid (49.9/50/0.1% (v/v/v)) was infused at 10 μL/min and used for a lock mass which was measured each 36 seconds for 0.5 seconds. Spectral peaks were automatically corrected for deviations in the lock mass.
Bioinformatics analysis of prokaryotic organisms encoding AsnS and GlnS
Escherichia coli AsnS and GlnS protein sequences were interrogated against the National Center for Biotechnology Information (NCBI) collection of non-redundant protein sequences of bacteria and archaea (non-redundant Microbial proteins, update: 2017/11/29) using the pBLAST search engine. The search was further restricted for non-redundant RefSeq proteins, with a 20,000-hit limit, the BLOSUM62 matrix scoring function and an Expect threshold value (E-value) of 1e-5. Hits were selected if sequence identity was above 50% with respect to the query sequences and those associated with bacterial species Agrobacterium tumefaciens, Escherichia coli and Pseudomonas aeruginosa were extracted (Supplementary Table 4).
Bioinformatics analysis of prokaryotic organisms predicted to encode the amidotransferase GatCAB complex
A none-exhaustive search for organisms encoding GatCAB was carried out using the National Center for Biotechnology Information (NCBI) database. Pseudomonas aeruginosa GatA and GatB protein sequences were interrogated against the NCBI collection of non-redundant protein sequences of bacteria and archaea (non-redundant Microbial proteins, update: 2017/11/29) using the pBLAST search engine. The search was further restricted for non-redundant RefSeq proteins, with a 20,000-hit limit, BLOSUM62 matrix scoring function and an Expect threshold value (E-value) of 1e-5. Hits were selected if annotated as Asp-tRNA(Asn)/Glu-tRNA(Gln) amidotransferase subunits, and the results for Agrobacterium tumefaciens, Escherichia coli and Pseudomonas aeruginosa were extracted (Supplementary Table 4).
Statistical analyses
Statistical analyses were performed using GraphPad Prism version 9 and are detailed in the figure legends.
Extended Data
Extended data Fig. 1.
Extended data Fig. 2.
Extended data Fig. 3.
Extended data Fig. 4.
Extended data Fig. 5.
Extended data Fig. 6.
Extended data Fig. 7.
Extended data Fig. 8.
Extended data Fig. 9.
Extended data Fig. 10.
Supplementary Material
Acknowledgements
The Authors state that they have no conflict of interest. L.M.N. was supported by MRC Grant MR/N023250/1 and a Marie Curie Fellowship (PIIF-GA-2013-625318). A.F. was supported by Medical Research Council (MRC) Grants MR/K001930/1 and MR/N023250/1 and Biotechnology and Biological Sciences Research Council (BBSRC) Grant BB/N002539/1. R.C.D.F and D.A.I.M. were supported by the MRC Career Development Award MR/M009505/1. D.A.J acknowledges support by the MINECO Contract CTQ2016-76941-R. M.A.S.P. was supported by the MINECO under the “Juan de la Cierva Postdoctoral program” (position FJCI-2015-25725). Technical support from the CIC bioGUNE Metabolomics and Proteomics platforms are gratefully acknowledged.
Footnotes
Author contributions
L.M.N designed the overall experimental plan for the manuscript, performed the majority of the experiments presented and wrote the manuscript; A.K.C performed all TraDIS sequencing and associated bioinformatic analyses; T.C and R.C.D.F performed pull down experiments and bacterial growth assays; E.M. assisted with protein pull-down assays; M.A.S.P and D.A.J performed protein purification and MS enzymatic assays; D.A.J performed homology modelling and bioinformatic analyses; G.D and J.P contributed to project management and supported TraDIS sequencing and associated bioinformatic analyses; D.A.I.M. contributed to project management, designed experiments, performed protein purification and pull-down experiments, and contributed to the writing of the manuscript; A.F contributed to project management, designed the overall experimental plan for the manuscript, and contributed to writing the manuscript.
Competing interests
The authors state that there are no competing interests to declare.
Data availability statement
PAK genome NCBI number is LR657304 and in the ENA (European Nucleotide Archive) is ERS195106 (https://www.ebi.ac.uk/ena/browser/view/ERS195106). The resulting sequences of the T6SS TraDIS assays are available from the ENA under study accession number ERS577921 (https://www.ebi.ac.uk/ena/browser/view/ERS577921).
References
- 1.Sibley CD, et al. A polymicrobial perspective of pulmonary infections exposes an enigmatic pathogen in cystic fibrosis patients. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:15070–15075. doi: 10.1073/pnas.0804326105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peters BM, Jabra-Rizk MA, O’May GA, Costerton JW, Shirtliff ME. Polymicrobial interactions: impact on pathogenesis and human disease. Clinical microbiology reviews. 2012;25:193–213. doi: 10.1128/CMR.00013-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bingle LE, Bailey CM, Pallen MJ. Type VI secretion: a beginner’s guide. Current opinion in microbiology. 2008;11:3–8. doi: 10.1016/j.mib.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 4.Pukatzki S, McAuley SB, Miyata ST. The type VI secretion system: translocation of effectors and effector-domains. Current opinion in microbiology. 2009;12:11–17. doi: 10.1016/j.mib.2008.11.010. [DOI] [PubMed] [Google Scholar]
- 5.Cianfanelli FR, Monlezun L, Coulthurst SJ. Aim, load, fire: the type VI secretion system, a bacterial nanoweapon. Trends in microbiology. 2016;24:51–62. doi: 10.1016/j.tim.2015.10.005. [DOI] [PubMed] [Google Scholar]
- 6.Filloux A. Microbiology: a weapon for bacterial warfare. Nature. 2013;500:284–285. doi: 10.1038/nature12545. [DOI] [PubMed] [Google Scholar]
- 7.Nazarov S, et al. Cryo-EM reconstruction of type VI secretion system baseplate and sheath distal end. The EMBO journal. 2018;37 doi: 10.15252/embj.201797103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shneider MM, et al. PAAR-repeat proteins sharpen and diversify the type VI secretion system spike. Nature. 2013;500:350–353. doi: 10.1038/nature12453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang J, et al. Cryo-EM structure of the extended type VI secretion system sheath-tube complex. Nat Microbiol. 2017;2:1507–1512. doi: 10.1038/s41564-017-0020-7. [DOI] [PubMed] [Google Scholar]
- 10.Kudryashev M, et al. Structure of the type VI secretion system contractile sheath. Cell. 2015;160:952–962. doi: 10.1016/j.cell.2015.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hachani A, Allsopp LP, Oduko Y, Filloux A. The VgrG proteins are “a la carte” delivery systems for bacterial type VI effectors. The Journal of biological chemistry. 2014;289:17872–17884. doi: 10.1074/jbc.M114.563429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ma J, et al. The Hcp proteins fused with diverse extended-toxin domains represent a novel pattern of antibacterial effectors in type VI secretion systems. Virulence. 2017:1–14. doi: 10.1080/21505594.2017.1279374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Silverman JM, et al. Haemolysin coregulated protein is an exported receptor and chaperone of type VI secretion substrates. Molecular cell. 2013;51:584–593. doi: 10.1016/j.molcel.2013.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Whitney JC, et al. Genetically distinct pathways guide effector export through the type VI secretion system. Molecular microbiology. 2014;92:529–542. doi: 10.1111/mmi.12571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Russell AB, et al. A widespread bacterial type VI secretion effector superfamily identified using a heuristic approach. Cell host & microbe. 2012;11:538–549. doi: 10.1016/j.chom.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Russell AB, Peterson SB, Mougous JD. Type VI secretion system effectors: poisons with a purpose. Nature reviews. Microbiology. 2014;12:137–148. doi: 10.1038/nrmicro3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hood RD, et al. A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell host & microbe. 2010;7:25–37. doi: 10.1016/j.chom.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Allsopp LP, et al. RsmA and AmrZ orchestrate the assembly of all three type VI secretion systems in Pseudomonas aeruginosa . Proceedings of the National Academy of Sciences of the United States of America. 2017;114:7707–7712. doi: 10.1073/pnas.1700286114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cain AK, et al. A decade of advances in transposon-insertion sequencing. Nature reviews Genetics. 2020;21:526–540. doi: 10.1038/s41576-020-0244-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ibba M, Becker HD, Stathopoulos C, Tumbula DL, Soll D. The adaptor hypothesis revisited. Trends in biochemical sciences. 2000;25:311–316. doi: 10.1016/s0968-0004(00)01600-5. [DOI] [PubMed] [Google Scholar]
- 21.Langridge GC, et al. Simultaneous assay of every Salmonella typhi gene using one million transposon mutants. Genome research. 2009;19:2308–2316. doi: 10.1101/gr.097097.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barquist L, et al. The TraDIS toolkit: sequencing and analysis for dense transposon mutant libraries. Bioinformatics (Oxford, England) 2016;32:1109–1111. doi: 10.1093/bioinformatics/btw022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pissaridou P, et al. The Pseudomonas aeruginosa T6SS-VgrG1b spike is topped by a PAAR protein eliciting DNA damage to bacterial competitors. Proceedings of the National Academy of Sciences of the United States of America. 2018;115:12519–12524. doi: 10.1073/pnas1814181115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang F, Waterfield NR, Yang J, Yang G, Jin Q. A Pseudomonas aeruginosa Type VI secretion phospholipase D effector targets both prokaryotic and eukaryotic cells. Cell host & microbe. 2014;15:600–610. doi: 10.1016/j.chom.2014.04.010. [DOI] [PubMed] [Google Scholar]
- 25.Whitney JC, et al. An interbacterial NAD(P) glycohydrolase toxin requires elongation factor Tu for delivery to target cells. Cell. 2015 doi: 10.1016/j.cell.2015.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. The Phyre2 web portal for protein modeling, prediction and analysis. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Labahn J, Neumann S, Buldt G, Kula MR, Granzin J. An alternative mechanism for amidase signature enzymes. Journal of molecular biology. 2002;322:1053–1064. doi: 10.1016/s0022-2836(02)00886-0. [DOI] [PubMed] [Google Scholar]
- 28.Nakamura A, Yao M, Chimnaronk S, Sakai N, Tanaka I. Ammonia channel couples glutaminase with transamidase reactions in GatCAB. Science (NewYork NY) 2006;312:1954–1958. doi: 10.1126/science.1127156. [DOI] [PubMed] [Google Scholar]
- 29.Suzuki T, et al. Structure of the Pseudomonas aeruginosa transamidosome reveals unique aspects of bacterial tRNA-dependent asparagine biosynthesis. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:382–387. doi: 10.1073/pnas.1423314112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yasuhira K, et al. X-ray crystallographic analysis of the 6-aminohexanoate cyclic dimer hydrolase: catalytic mechanism and evolution of an enzyme responsible for nylon-6 byproduct degradation. The Journal of biological chemistry. 2010;285:1239–1248. doi: 10.1074/jbc.M109.041285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shin S, et al. Characterization of a novel Ser-cisSer-Lys catalytic triad in comparison with the classical Ser-His-Asp triad. The Journal of biological chemistry. 2003;278:24937–24943. doi: 10.1074/jbc.M302156200. [DOI] [PubMed] [Google Scholar]
- 32.Balotra S, et al. X-ray structure of the amidase domain of AtzF, the allophanate hydrolase from the cyanuric acid-mineralizing multienzyme complex. Applied and environmental microbiology. 2015;81:470–480. doi: 10.1128/AEM.02783-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee S, et al. Crystal structure analysis of a bacterial aryl acylamidase belonging to the amidase signature enzyme family. Biochemical and biophysical research communications. 2015;467:268–274. doi: 10.1016/j.bbrc.2015.09.177. [DOI] [PubMed] [Google Scholar]
- 34.Ruan LT, Zheng RC, Zheng YG. Mining and characterization of two amidase signature family amidases from Brevibacterium epidermidis ZJB-07021 by an efficient genome mining approach. Protein expression and purification. 2016;126:16–25. doi: 10.1016/j.pep.2016.05.006. [DOI] [PubMed] [Google Scholar]
- 35.Valina AL, Mazumder-Shivakumar D, Bruice TC. Probing the Ser-Ser-Lys catalytic triad mechanism of peptide amidase: computational studies of the ground state, transition state, and intermediate. Biochemistry. 2004;43:15657–15672. doi: 10.1021/bi049025r. [DOI] [PubMed] [Google Scholar]
- 36.Wei BQ, Mikkelsen TS, McKinney MK, Lander ES, Cravatt BF. A second fatty acid amide hydrolase with variable distribution among placental mammals. The Journal of biological chemistry. 2006;281:36569–36578. doi: 10.1074/jbc.M606646200. [DOI] [PubMed] [Google Scholar]
- 37.Shin S, et al. Structure of malonamidase E2 reveals a novel Ser-cisSer-Lys catalytic triad in a new serine hydrolase fold that is prevalent in nature. The EMBO journal. 2002;21:2509–2516. doi: 10.1093/emboj/21.11.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Patricelli MP, Cravatt BF. Clarifying the catalytic roles of conserved residues in the amidase signature family. The Journal of biological chemistry. 2000;275:19177–19184. doi: 10.1074/jbc.M001607200. [DOI] [PubMed] [Google Scholar]
- 39.Akochy PM, Bernard D, Roy PH, Lapointe J. Direct glutaminyl-tRNA biosynthesis and indirect asparaginyl-tRNA biosynthesis in Pseudomonas aeruginosa PAO1. Journal of bacteriology. 2004;186:767–776. doi: 10.1128/JB.186.3.767-776.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gonzalez-Magana A, et al. Structural insights into Pseudomonas aeruginosa type six secretion system exported effector 8. Journal of structural biology. 2020;212:107651. doi: 10.1016/j.jsb.2020.107651. [DOI] [PubMed] [Google Scholar]
- 41.Bernal P, Allsopp LP, Filloux A, Llamas MA. The Pseudomonas putida T6SS is a plant warden against phytopathogens. The ISME journal. 2017;11:972–987. doi: 10.1038/ismej.2016.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Filloux A, Hachani A, Bleves S. The bacterial Type VI secretion machine: yet another player for protein transport across membranes. Microbiology. 2008;154:1570–1583. doi: 10.1099/mic.0.2008/016840-0. [DOI] [PubMed] [Google Scholar]
- 43.Shalom G, Shaw JG, Thomas MS. In vivo expression technology identifies a type VI secretion system locus in Burkholderia pseudomallei that is induced upon invasion of macrophages. Microbiology. 2007;153:2689–2699. doi: 10.1099/mic.0.2007/006585-0. [DOI] [PubMed] [Google Scholar]
- 44.Zhang W, et al. Modulation of a thermoregulated type VI secretion system by AHL-dependent quorum sensing in Yersinia pseudotuberculosis . Archives of microbiology. 2011;193:351–363. doi: 10.1007/s00203-011-0680-2. [DOI] [PubMed] [Google Scholar]
- 45.Kaniga K, Delor I, Cornelis GR. A wide-host-range suicide vector for improving reverse genetics in gram-negative bacteria: inactivation of the blaA gene of Yersinia enterocolitica . Gene. 1991;109:137–141. doi: 10.1016/0378-1119(91)90599-7. [DOI] [PubMed] [Google Scholar]
- 46.Hachani A, et al. Type VI secretion system in Pseudomonas aeruginosa: secretion and multimerization of VgrG proteins. The Journal of biological chemistry. 2011;286:12317–12327. doi: 10.1074/jbc.M110.193045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cain AK, et al. Complete genome sequence of Pseudomonas aeruginosa reference strain PAK. Microbiology resource announcements. 2019;8 doi: 10.1128/MRA.00865-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics (Oxford, England) 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hachani A, Lossi NS, Filloux A. A visual assay to monitor T6SS-mediated bacterial competition. Journal of visualized experiments : JoVE. 2013:e50103. doi: 10.3791/50103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Karimova G, Pidoux J, Ullmann A, Ladant D. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:5752–5756. doi: 10.1073/pnas.95.10.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schindelin J, et al. Fiji: an open-source platform for biological-image analysis. Nature methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sato I, et al. Cryptococcus nodaensis sp nov, a yeast isolated from soil in Japan that produces a salt-tolerant and thermostable glutaminase. Journal of Industrial Microbiology and Biotechnology. 1999;22:127–132. doi: 10.1038/sj.jim.2900623. [DOI] [Google Scholar]
- 53.Li C, et al. FastCloning: a highly simplified, purification-free, sequence- and ligation-independent PCR cloning method. BMC biotechnology. 2011;11:92. doi: 10.1186/1472-6750-11-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu B, et al. Versatile peptide C-terminal functionalization via a computationally engineered peptide amidase. ACS Catalysis. 2016;6:5405–5414. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
PAK genome NCBI number is LR657304 and in the ENA (European Nucleotide Archive) is ERS195106 (https://www.ebi.ac.uk/ena/browser/view/ERS195106). The resulting sequences of the T6SS TraDIS assays are available from the ENA under study accession number ERS577921 (https://www.ebi.ac.uk/ena/browser/view/ERS577921).