Abstract
Organ development is orchestrated by cell- and time-specific gene regulatory networks. In this study, we investigated the regulatory basis of mouse cerebellum development from early neurogenesis to adulthood. By acquiring snATAC-seq profiles for ~90,000 cells spanning eleven stages, we mapped cerebellar cell types and identified candidate cis-regulatory elements (CREs). We detected extensive spatiotemporal heterogeneity among progenitor cells and a gradual divergence in the regulatory programs of cerebellar neurons during differentiation. Comparisons to vertebrate genomes and snATAC-seq profiles for ~20,000 cerebellar cells from the marsupial opossum revealed a shared decrease in CRE conservation during development and differentiation, but also differences in constraint between cell types. Our work delineates the developmental and evolutionary dynamics of gene regulation in cerebellar cells and provides insights into mammalian organ development.
The cerebellum, known for its role in motor control, also contributes to other complex functions, such as language and memory (1–3). During development, two germinal zones – the ventricular zone and the rhombic lip – give rise to distinct neuronal and glial populations (2, 3). Cell type specification and differentiation processes are controlled by cis-regulatory elements (CREs), such as enhancers and promoters, and the transcription factors (TFs) that bind to them (4, 5). Although most CREs evolve rapidly (6–8), some, enriched near developmental genes, are conserved across vertebrates (9, 10). Studies of bulk CRE activity have explored gene regulation in the hindbrain and postnatal cerebellum (11, 12), as well as CRE evolutionary dynamics during organ development (13, 14). Single-cell approaches have been employed to study gene regulation in adult mouse organs (15), including the brain (16), and in a limited number of cells or stages for the developing mouse (17, 18) and human brain (19, 20), but a comprehensive characterization of the entire development of a mammalian organ has been lacking. In this study, we used single-nucleus Assay for Transposase Accessible Chromatin (snATAC-seq) (21) to profile the landscape of chromatin accessibility – a proxy for CRE activity – of ~90,000 cells across pre- and postnatal development of the mouse cerebellum and ~20,000 cerebellar cells from the marsupial opossum (https://apps.kaessmannlab.org/mouse_cereb_atac/). We leveraged this dataset to assess the developmental and evolutionary dynamics of gene regulation in the cerebellum.
Results
A snATAC-seq atlas of mouse cerebellum development
We generated snATAC-seq data for male and female mouse cerebella from eleven stages ranging from early neurogenesis (embryonic day 10 [E10]) to adulthood (postnatal day 63 [P63]; Fig. 1A, fig. S1, table S1). After applying quality controls (22), we acquired single-cell chromatin accessibility profiles for 91,922 cells with a median of 20,558 fragments per cell (Fig. 1B; table S2 and fig. S2A-D). We projected cells into a low dimensional embedding (excluding sex chromosomes) and identified clusters (22). Biological replicates show high similarity in this embedding (Fig. 1B), distribute evenly across clusters (fig. S2E), and have the highest correlations in the accessibility of autosomal CREs at the library level, followed by correlations between samples from adjacent developmental stages (fig. S2F).
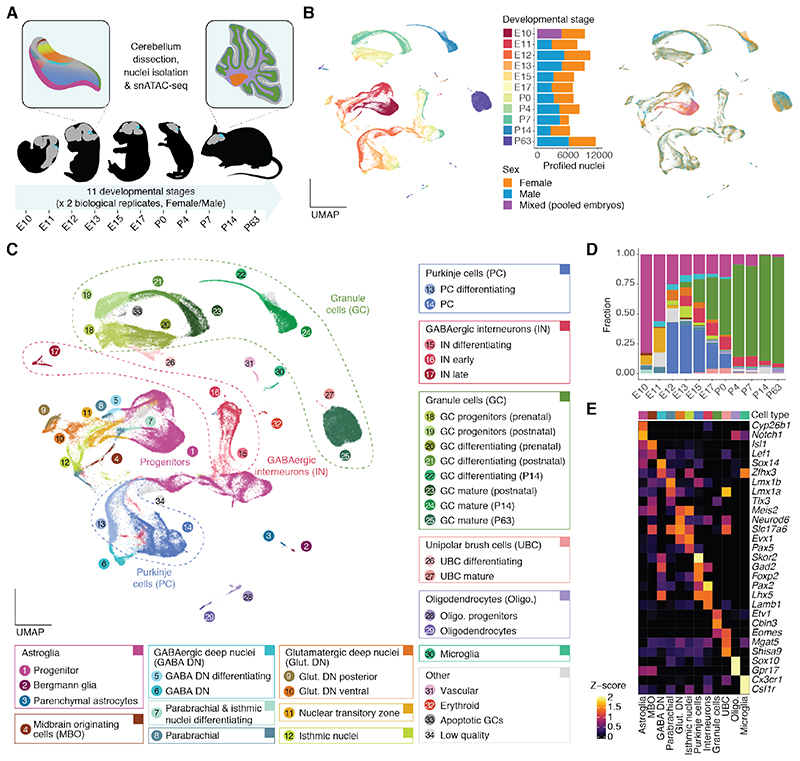
Fig. 1. A snATAC-seq atlas of mouse cerebellum development.
(A) Schematic overview of the dataset. Representative mouse silhouettes are shown for E11, E13, E17, P4 and P63 (brain in grey, cerebellum in cyan). The insets show the location of selected cell types in the cerebellum (colors are as in C). (B, C) UMAP projection of 91,922 cells colored by developmental stage (B, left) or sex (B, right), or cell type and state (C). Barplots in B show the number of profiled cells per stage and sex (each sex corresponding to one sample). In C, cell states or subtypes (numbered circles) are grouped into broad cell types (rectangles). Further progenitor and interneuron subtypes are shown in Fig. 3, fig. S11 and S13. (D) Proportions of broad cell types across developmental stages. (E) Activity scores of genes used for the annotation of broad cell types (Z-score, capped to 0-2). Broad cell type colors for D and E are as in C. Marker genes across subtypes and states in fig. S4.
Using iterative clustering (fig. S3) and approximating gene expression by the aggregated accessibility across a gene’s regulatory region (gene score) (22), we identified 12 broad cell types and 42 subtypes and cell states (Fig. 1C-E, fig. S4; table S2). To assess the use of gene scores as a proxy for gene expression and the quality of our cell type annotation, we reprocessed the data by developmental stage (fig. S5) and integrated with a single-cell RNA sequencing (scRNA-seq) atlas of the developing mouse hindbrain and cerebellum (23), transferring labels and imputing RNA expression for cells in our snATAC-seq atlas. Despite differences in sampling, we observed good correlations between gene scores and imputed RNA expression values (fig. S6A), and a concordance between cell type labels (fig. S6B-C). Discrepancies between the two annotations are explained by some scRNA-seq clusters containing mixtures of cell types (fig. S6D-E).
The earliest developmental stages in our dataset (E10-E11) are dominated by neural progenitors (key marker genes: Notch1, Cyp26b1; Fig. 1C-E). We also detected midbrain-originating cells (Isl1) and parabrachial neurons (Lmx1b), which later migrate out of the cerebellum (24, 25), as well as differentiating GABAergic deep nuclei neurons (Zfhx3, Sox14) and glutamatergic neurons (Meis2, Neurod6) of the nuclear transitory zone. From E12 onwards, the latter could be further resolved into posterior (Lmx1a) and ventral (Lmo3) populations of glutamatergic deep nuclei neurons, and into anteriorly located isthmic nuclei neurons (Pax5) (24). The same developmental window is marked by the generation of Purkinje cells (Skor2, Foxp2), which are the most abundant neuron type until E15. During the E10-E15 period the relative abundance of progenitor cells (excluding granule cell progenitors) decreases from 83% to 17% (Fig. 1D).
The next stages (E17-P4) are marked by the emergence of GABAergic interneurons (Pax2) and unipolar brush cells (UBC; Lmx1a, Eomes), and the rapid expansion of the proliferating (Atoh1, Gli2) and differentiating (Neurod1, Grin2b) granule cell populations (Fig. 1C-E). Small numbers of microglia (Ccr1, Hexb) and oligodendrocytes (Sox10), most of which are of extracerebellar origin (2), are detectable from E17 onwards. The last two stages (P14, P63) are dominated by mature granule cells (Etv1, Cbln3), with additional neuronal and glial populations being traceable, including mature astroglia such as Bergmann glia (Gdf10) and parenchymal astrocytes (Aqp4, Scl6a11). Throughout this study, we use the term astroglia to refer to cells transitioning along the lineage of neuroepithelial progenitors, radial glial progenitors and mature astrocytes (Fig. 1C-E), in agreement with their overlapping functions and molecular features (26). Overall, the developmental dynamics of cerebellar cell abundances observed in this study resemble those from scRNA-seq atlases (fig. S6F) (23, 27). However, some cell types, such as early-born neurons (GABAergic and glutamatergic deep nuclei, isthmic nuclei and parabrachial neurons) are better resolved in our snATAC-seq atlas.
The cis-regulatory landscape of cerebellar cell types
To characterize the regulatory profiles of cerebellar cell types we employed a cluster-specific and replicate-aware peak calling approach (22). We identified 261,643 high-confidence putative CREs (Fig. 2A and fig. S7A-B), most of which are intronic (51%) or intergenic (26%) (herein collectively referred to as distal). These putative CREs cover ~80% of enhancers active during hindbrain development (11), including 97% of experimentally validated elements in the E11.5 hindbrain (28), and 82% of enhancer RNAs (eRNAs) transcribed in the developing cerebellum (29), a significant enrichment compared to heterochromatin or enhancers active in non-neuronal tissues (fig. S7C-G). Comparisons with a single-cell ATAC-seq atlas of adult mouse organs (15) revealed the highest activity of the putative CREs in cells from the cerebellum (fig. S7H).
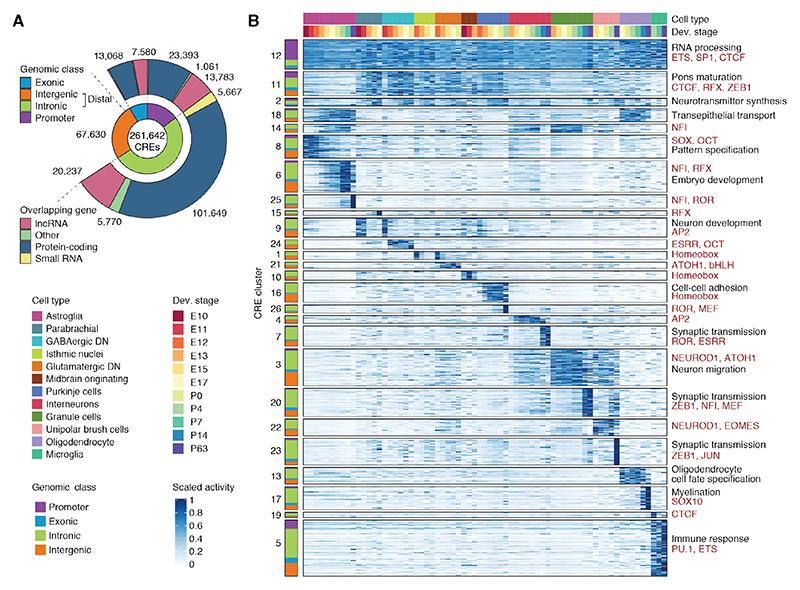
Fig. 2. CRE activity in cerebellum development.
(A) Genomic features of 261,642 putative CREs. Inner circle: genomic class; outer circle: biotype of the overlapping gene. (B) Clusters of CRE activity across cell types and developmental stages. CREs are grouped by activity cluster (k-means followed by hierarchical clustering) and genomic class (left). CRE clusters are arranged in decreasing order of pleiotropy (here: mean activity across rows) and then by cell type and developmental stage with maximum activity. Right: Representative enrichments (BH adjusted P < 0.05; hypergeometric test) for biological processes of adjacent genes (black) and motifs for TFs or TF families (red). 50,000 CREs confidently assigned to their cluster were chosen randomly for visualization.
We assigned 32,792 distal CREs to 5,766 putative target genes based on correlations with promoter accessibility and gene scores (fig. S8A, table S3). The CRE-gene pairs share more often the same topologically associating domain (TAD) in neural progenitors (30), are enriched for enhancer activity in the hindbrain (11, 28), and show higher correlations between promoter transcripts and eRNAs in the developing cerebellum (29) (fig. S8B-E). Although most genes were associated with a single distal CRE, 1,001 protein-coding genes were assigned to ten or more elements (fig. S8F). These genes are enriched for developmental processes, including cell fate commitment and neuron differentiation, as well as mature neuron functions like synapse organization and signaling (fig. S8G).
To describe patterns of CRE activity during cerebellum development, we used an iterative clustering procedure (fig. S9A-B). The majority of the 26 CRE clusters are cell type- and time-specific, whereas one shows constitutive activity (cluster 12; Fig. 2B). Most (67%) of these constitutive CREs correspond to promoters (fig. S9C) and among the remaining distal CREs, more than 50% contain a CTCF motif (fig. S9D), suggesting architectural roles, in accord with observations in humans (20). We further observed groups of CREs active in multiple early-born neuron types (clusters 2, 11), glial populations (cluster 18), and late-born cell types (cluster 14), affirming that a sizeable fraction of CREs have pleiotropic activity (31, 32).
CREs in cell type-specific clusters are close to genes associated with relevant gene ontology terms (e.g., myelination for oligodendrocyte-specific CREs) and are enriched for motifs of TFs known to be active in the respective cell types (e.g., ATOH1 for granule cells, SOX family TFs for progenitors, PU.1 for microglia; Fig. 2B and fig. S9E-F). We refined this analysis by identifying 50,815 differentially accessible CREs with significantly higher activity in a cell type and state (fig. S10A). Some of the distinct states we discovered distinguish cells from the same cell type based on the developmental stage (e.g., pre- and postnatal granule cell progenitors, P14 and P63 mature granule cells). The high similarity between biological replicates (fig. S2E-F) argues against technical confounders as a source of this separation. Indeed, we detected 3,988 differentially accessible CREs between pre- and postnatal granule cell progenitors (fig. S10B-C). CREs with increasing accessibility after birth are enriched for motifs of NFI, known to specify late-born neurons (33), and GLI2, consistent with the SHH-mediated expansion of the granule cell progenitor pool after birth (2). CREs with decreasing accessibility are enriched for motifs marking embryonic progenitors (SOX, MEIS) (2). Altogether, our analyses provide a comprehensive characterization of CRE activity during cerebellum development and highlight the cell- and time-specificity of gene regulation.
Progenitor heterogeneity reflects cell fate decisions
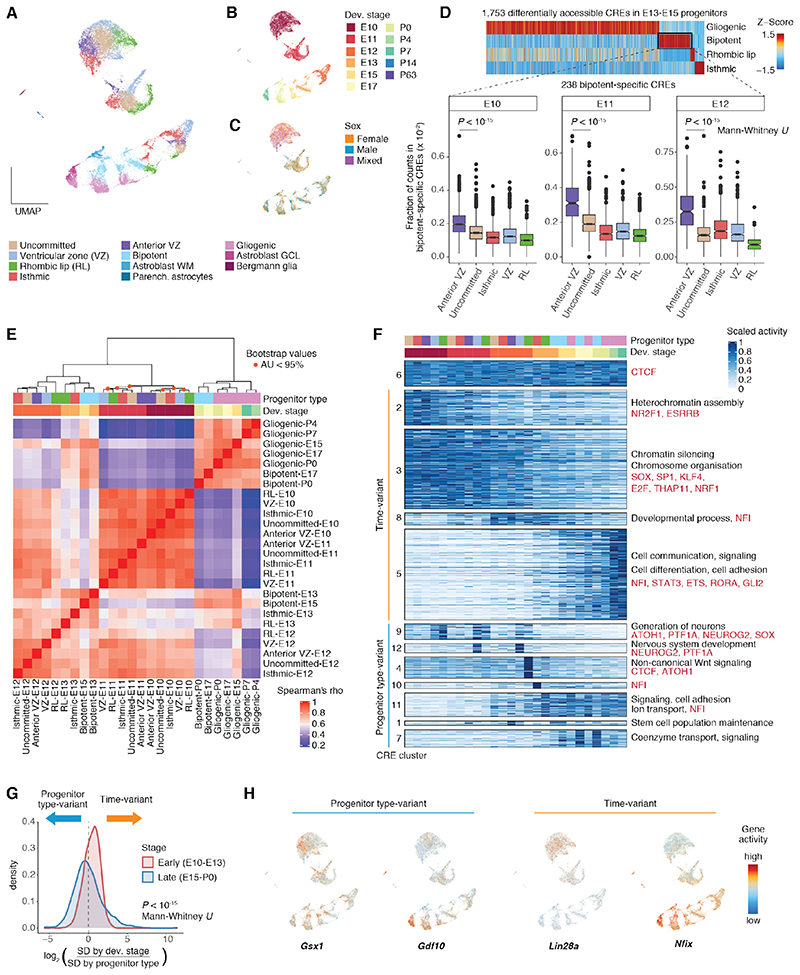
Cerebellum development relies on a spatially and temporally restricted pattern of cell type specification. GABAergic deep nuclei neurons, Purkinje cells, interneurons, and astrocytes are sequentially derived from progenitors in the ventricular zone, whereas the emergence of glutamatergic deep nuclei neurons from rhombic lip progenitors is followed by the generation of granule cells and UBCs (2). We asked whether this mode of cell fate specification was associated with heterogeneity amongst cerebellar progenitors. We subclustered cells from the astroglia lineage (progenitors and astrocytes) and identified progenitors from all germinal regions throughout cerebellum development, although without sharp boundaries within a given stage (Fig. 3A-C and fig. S11A-C). Early progenitors (E10-E12) include isthmic (En1, Pax5), ventricular zone (Dll1, Ptf1a from E11), and rhombic lip (Cdon, Atoh1) populations, as well as progenitors with no apparent commitment towards a cell fate (Fig. 3A-C and fig. S11A-C), in accord with previous reports (34). We additionally identified a Gsx1+ population that we traced to the anterior ventricular zone (fig. S11A-B) (35). E13-E15 are marked by the appearance of two late progenitor populations (fig. S11C) that broadly correspond to the previously described bipotent (Gsx1, Wnt8b) and gliogenic (Slc1a3, Grm3) progenitors (2, 36). Previous studies suggest that the bipotent progenitors generate interneurons and white matter astrocytes, whereas the gliogenic progenitors give rise to Bergmann glia and granule cell layer astrocytes (2, 36). Consistently, we detected two groups of differentiating parenchymal astrocytes at perinatal stages (E15-P7): white matter (Slc6a11, Olig2, Kcnd2) and granule cell layer (Aqp4, Tekt5) astroblasts, with higher similarity in CRE activity to bipotent and gliogenic progenitors, respectively (Fig. 3A-C and fig. S11A-D). We then investigated the relationship of the two Gsx1+ populations, early anterior ventricular zone progenitors and late bipotent progenitors. Besides additional shared marker genes (Wnt8b, Ndnf, Robo1), E10-E12 anterior ventricular zone progenitors show the highest similarity in CRE activity with E13-E15 bipotent progenitors across stage-matched progenitor types (Fig. 3D and fig. S11E-F). Previously, Gsx1+ progenitors were shown to arise from a temporal transition of Purkinje cell-producing ventricular zone progenitors (35). Our data support the existence of an additional, molecularly distinct, anterior population of early progenitors that is already primed to acquire bipotent progenitor identity.
Fig. 3. Spatiotemporal heterogeneity in cerebellar progenitor populations.
(A, B, C) UMAP projections of 21,830 astroglia cells (progenitors and astrocytes) colored by subtype (A), developmental stage (B) and sex (C). (D) Comparison of bipotent progenitors to earlier populations. Top: Activity profiles (Z-score) of progenitor type-specific CREs in E13-E15. Bottom: Fraction of fragments per cell in CREs specific to bipotent progenitors across progenitor types and developmental stages. (E) Hierarchical clustering based on Spearman’s correlation coefficients in CRE accessibility across progenitor types and developmental stages. Orange dots indicate nodes with approximately unbiased (AU) probability values < 95%. (F) Clusters of CRE activity across progenitor types and developmental stages. CREs are grouped by activity cluster (k-means followed by hierarchical clustering). Right: Representative enrichments (BH adjusted P < 0.05; hypergeometric test) for biological processes of adjacent genes (black) and motifs for TFs or TF families (red). 25,000 CREs confidently assigned to their cluster were chosen randomly for visualization. (G) Density distributions for the log2-ratio of gene score standard deviation (SD) across developmental stages and progenitor types for the 2,000 genes with the highest variance in early and late progenitor populations. (H) Gene scores (capped at 10th and 99th quantiles and log10 transformed) for genes with high variance across progenitor types (left) or developmental stages (right).
Despite our dense sampling and ability to identify the same germinal regions across consecutive stages, often based on the same marker genes (e.g., E10-E12 rhombic lip; Fig. S11A), progenitor cells primarily cluster by developmental stage and not by germinal zone (Fig. 3A-C, E), suggesting strong changes in their chromatin accessibility profiles during development. These temporal differences are strongest in early embryonic stages (E10-E12), a period coinciding with the sequential generation of distinct neuronal cell types (Fig. 3E and Fig. 1D). Unbiased clustering of CRE activity across progenitor groups and developmental stages revealed many temporally dynamic CREs that are shared between germinal zones (Fig. 3F), arguing for common factors in control of temporal transitions in progenitor competence (3, 33). These include a shift from CREs enriched for nuclear receptor and SOX motifs, and associated with chromatin silencing genes (E10-E12; clusters 2-3), towards CREs enriched for NFI motifs and adjacent to genes involved in developmental processes (E12-E13; cluster 8), signaling and cell adhesion (E13-P7; cluster 5). From E15 onwards, coinciding with the protracted generation of interneurons and astrocytes, temporal differences become smaller and progenitors from adjacent stages group by germinal region (gliogenic and bipotent; Fig. 3E).
To assess whether these temporal differences in chromatin accessibility also lead to changes in gene expression, we systematically identified genes with higher variance across developmental stages than between germinal zones (fig. S12A-B). Reassuringly, marker genes such as Gsx1 and Gdf10 show higher variance across germinal zones. However, most of the 2,000 highly variant genes in E10-E13 show higher variance across developmental stages, whereas variance by progenitor type is higher in E15-P0 (Fig. 3G), in agreement with our clustering analysis (Fig. 3E). Temporally-variant genes (310 in E10-E13 versus 61 in E15-P0; table S4) include the pluripotency factor Lin28a, which is expressed in E10 progenitors across all germinal zones, and Nfix (Fig. 3H), which increases in expression during development, in line with the enrichment of NFI motifs in late CRE clusters (Fig. 3F). We next clustered temporally-variant genes based on their activity profiles (decreasing, increasing; fig. S12C-D) and examined their expression in progenitor cells in a published scRNA-seq dataset (23). We observed significant temporal differences in the direction predicted by the snATAC-seq data (fig. S12E-F). Thus, although spatiotemporal heterogeneity among progenitors is prominent throughout cerebellum development, early stages are characterized by stronger temporal differences in CRE activity, which are often shared between germinal zones and lead to corresponding gene expression changes.
Regulatory activity during neuron differentiation
We next characterized the gene regulatory programs underlying the differentiation of the three most abundant neuron types in our dataset: granule cells, Purkinje cells, and interneurons. For each neuron type, we aligned cells from different developmental stages along their differentiation trajectories, which we modeled using diffusion pseudotime (fig. S13A-C). The observed differentiation dynamics ranged from stage-restricted (Purkinje cells, E12-E13) to protracted (granule cells, interneurons) (Fig. 4A-B and fig. S13D), consistent with the several week long generation of granule cells and interneurons (2). While granule cells form a homogeneous differentiating population (Fig. 4B), interneurons are stratified into distinct temporally-specified subtypes (fig. S13E-F): early-born interneurons (Zfhx4, Slit2) detected at E13-E15, mid-born Golgi cells (Chrm2), Purkinje layer interneurons (Nxph1, Klhl1) prevalent at E17-P7, and late-born molecular layer interneurons of type 1 (Sorcs3, Grm8) and 2 (MLI2; Nxph1, Pvalb), which are abundant at P14-P63 (2, 37).
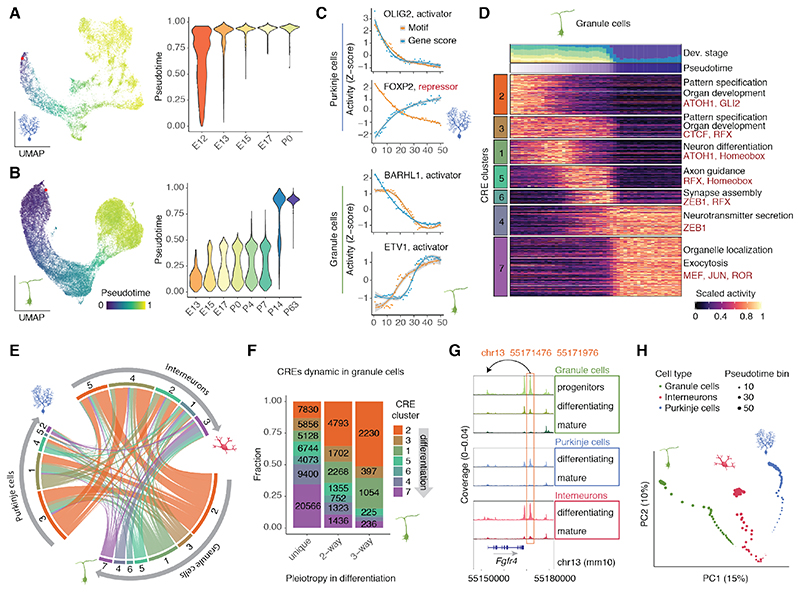
Fig. 4. Gene regulation in neuronal differentiation.
(A, B) Left : UMAP projections of 13,214 Purkinje cells (A) and 35,153 granule cells (B) aligned across developmental stages, colored by pseudotime value. Red points indicate the pseudotime root. Right : Distribution of pseudotime values across developmental stages. (C) Z-score scaled values for gene and motif activity across ranked pseudotime bins for examples of activator and repressor TFs in Purkinje cells (top) and granule cells (bottom). Curves drawn using LOESS regression, gray areas indicate 95% confidence intervals. (D) Z-score scaled activity of dynamic CREs during granule cell differentiation, averaged across 50 bins of increasing pseudotime ranks. Top: Contribution of developmental stages and mean pseudotime value for each bin. Right: Representative enrichments (BH adjusted P < 0.05; hypergeometric test) for biological processes of adjacent genes (black) and TF motifs (red). (E) Overlap between activity clusters for CREs dynamic in two or more neurons (pleiotropic). For each neuron type (outer sector) CRE clusters (as in D and fig. S14) are ordered from early (orange) to late (violet) activity during differentiation. Each node connects the activity clusters of two different neuron types for the same CRE. (F) Fraction of CRE clusters (as in D) across CREs dynamic in a single neuron type (unique) or shared across two or three cell types. (G) Example of a pleiotropic intergenic CRE, assigned to Fgfr4. Accessibility profiles for each cell type and state (from Fig. 1C) were aggregated across cells from all developmental stages and scaled by the total number of fragments in each group. (H) Principal component analysis of CRE accessibility during granule cell, Purkinje cell and interneuron differentiation. Percentage values show the proportion of variance explained by each component.
We used the modeled differentiation trajectories to characterize the gene regulatory dynamics in each neuron type. We identified TFs with dynamic activity during differentiation and correlated the accessibility of the respective binding motifs with their gene scores to classify TFs into putative activators and repressors (fig. S14A-C; table S5). These candidate TFs include known regulators, such as BARHL1 and ETV1 in granule cells, and OLIG2 and FOXP2 in Purkinje cells (Fig. 4C).
Across all three neuron types, we observed a gradual transition from CREs proximal to genes associated with embryonic development and cell fate commitment, towards genes with roles in neuron differentiation, migration and axon guidance, and eventually the formation of the synapse, neurotransmitter secretion and ion transport (Fig. 4D and fig. S14D-E). In support of this convergence in biological processes, 43% of protein-coding genes with dynamic activity across pseudotime are shared by at least two neuron types (fig. S14F), in accordance with observations in the developing neocortex (38). By contrast, only 20% of dynamic CREs are shared across neuron types (i.e., are pleiotropic), further highlighting the cell type-specificity of gene regulation compared to gene expression (fig. S14G). Pleiotropic CREs show similar activity profiles across neuron types (Fig. 4E; early-early: P < 10-15, late-late: P < 10-15, early-late: P > 0.99, hypergeometric test) and, with the exception of interneurons, are more common in early stages of neuron differentiation (Fig. 4F-G, S14H-I). The higher similarity of the early stages is supported by a principal component analysis in which the chromatin accessibility profiles of the three neuron types gradually diverge during differentiation (Fig. 4H).
Evolutionary dynamics of CREs in cerebellum development
Motivated by the temporal dynamics in CRE activity during development (Fig. 2), cell type specification (Fig. 3), and differentiation (Fig. 4), we next investigated whether such differences are also reflected in the evolutionary histories of CREs. A decrease in gene regulatory (13) and expression (39, 40) conservation as development progresses has been reported for whole organs. Given the profound changes of cell type abundances during development, it remains unclear whether whole organ patterns emerge from the parallel decline of conservation across cell types or instead reflect the higher conservation of cell types that are more abundant in earlier stages, such as progenitors and early-born cells. Hence, we sought to test for differences in the evolutionary dynamics of CREs both across cell types and throughout development.
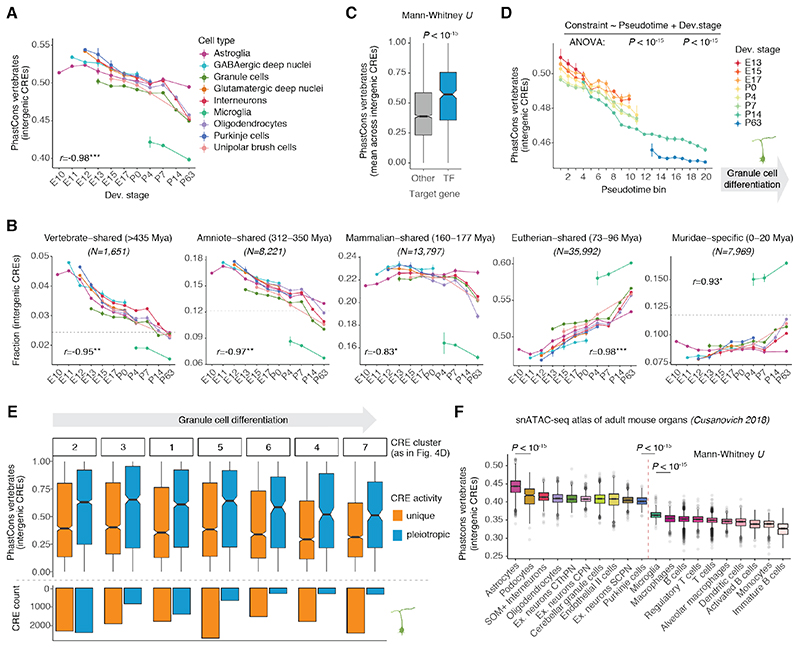
We assessed the selective (functional) constraints on CRE sequences using estimates of evolutionary conservation (phastCons scores) (41) and inferred their minimum age (i.e., when they first appeared) using syntenic sequence alignments between mouse and 16 other vertebrates at various phylogenetic distances (fig. S15; table S6). We assessed these conservation metrics for intergenic CREs at the single-cell level, enabling comparisons between cell types and developmental stages. Across all cell types, both sequence constraints and the predicted ages of intergenic CREs decreased significantly during development (Fig. 5A and fig. S16A). Thus, previous observations for whole organs (13) are mostly explained by decreasing conservation within cell types rather than changes in the relative abundance of cell types with pronounced differences in evolutionary constraints. Ancient CREs, shared across or even beyond mammals, show higher activity during embryonic development, when cell types begin their differentiation (Fig. 5B). As cell types mature and activate their terminal effector genes, elements specific to eutherians and murid rodents (Muridae) gradually increase their activity, potentially contributing to species/lineage-specific phenotypic innovations of ancestral cell types (Fig. 5B). Consistent with these observations, TF genes, which are central to cell type identity (42), are associated with older and more constrained CREs (Fig. 5C and fig. S16B).
Fig. 5. Evolutionary dynamics of CREs.
(A) Sequence constraint in intergenic CREs accessible per cell, averaged for each cell type and developmental stage. (B) Fraction of accessible intergenic CREs assigned to different age groups per cell, averaged for each cell type and developmental stage. Different y-ranges were used across age groups to facilitate comparisons between cell types and stages within each group, as the fraction depends on the number of CREs per group (indicated on top). The fraction of each age group across all intergenic CREs is marked by the dotted horizontal line. For A and B, Pearson’s r correlation coefficients between the estimates and development are shown (median across cell types; P<0.05*, P<0.01**, P<0.001***); vertical bars illustrate difference in average estimates between biological replicates. Only groups with at least 50 cells were considered. (C) Average sequence constraint of intergenic CREs per target gene for TFs and other genes. (D) Sequence constraint of intergenic CREs acessible per cell averaged for each developmental stage and pseudotime interval of differentiating granule cells (from Fig. 4B). Vertical bars illustrate the difference in average estimates between biological replicates. (E) Sequence constraint (top) and abundance (bottom) of cell type-specific and pleiotropic intergenic CREs active in different stages of granule cell differentiation (from Fig. 4D, ordered from early to late activity during differentiation). (F) Sequence constraint of intergenic CREs accessible per cell across cell types in the adult mouse (data from (15)). The ten most conserved (left) and all immune (right) cell types are shown.
Given the strong developmental signals in our dataset, we next assessed the contribution of cell type differentiation versus additional temporal differences, such as intrinsic patterning signals (33, 38) or interactions with morphogens and ligands (4), to the temporal decline in conservation. We focused on granule cells because these cells have a protracted differentiation trajectory (E13-P14) and, despite the different states observed in our data, are not known to form distinct temporally-specified subtypes (2). By stratifying granule cells based on their differentiation state (pseudotime) and developmental stage, we observed that both factors contribute to the decrease of intergenic CRE conservation during development, with cells at early differentiation states and developmental stages showing the most conserved regulatory program (Fig. 5D, and fig. S16C-D). We validated this observation in a pseudotime-independent framework by focusing on prenatal granule cell progenitors, which formed a single cluster without any alignment across developmental stages (fig. S16E). Despite the overall high similarity of these cells, we identified CREs with significant temporal activity changes and clustered them into two groups that decrease (7,527) or increase (11,972) in accessibility across prenatal stages (fig. S16E). CREs with decreasing accessibility are more conserved and are enriched for motifs recognized by pluripotency TFs (e.g., SOX2), whereas CREs with increasing accessibility are enriched for motifs of TFs active in granule cell progenitors, including ATOH1 and GLI2 (fig. S16F-G). Using published scRNA-seq data (23), we observed similar developmental patterns for genes adjacent to temporally dynamic CREs (fig. S16H), suggesting that these differences in CRE accessibility affect gene expression.
Both CRE pleiotropy (Fig. 4F-H) and sequence conservation (Fig. 5A) decrease during differentiation. Given that pleiotropy imposes constraints on the evolution of regulatory sequences (31, 43), we asked whether it could explain the decrease in CRE conservation. We found that pleiotropic CREs are significantly more conserved than those dynamic in a single neuron type, and that these differences are larger than those between early and late acting CREs with similar pleiotropy status (Fig. 5E). Consistently, CREs with decreasing accessibility across developmental stages in granule cell progenitors are more conserved (fig. S16F) and active in other types of progenitor and early differentiating cells, whereas CREs with increasing accessibility are specific to granule cell progenitors (fig. S16I). Thus, as CRE pleiotropy decreases during development and differentiation, constraint on CRE sequences relaxes, consistent with previous observations for gene expression (39, 40).
Differences in CRE constraint across cell types
We also observed differences in the conservation of intergenic CREs between cell types. CREs active in microglia show a faster evolutionary turnover compared to all other cerebellar cell types (Fig. 5A and fig. S16A), consistent with the rapid divergence in gene expression and morphology of microglia (44). In agreement with their younger age and reduced sequence constraint, CREs active in microglia are also enriched for genomic repeats, in particular for recently expanded transposable elements in rodents, such as short interspersed nuclear elements (SINEs) B1, B2 and B4, endogenous retrovirus sequences (ERVs), and L1 elements (fig. S17A-B). By contrast, astroglial cells (progenitors and astrocytes) show the most conserved intergenic CREs in the mature cerebellum (Fig. 5A and fig. S16A). They are enriched for CREs that originated 160-177 million years ago (Mya) in common mammalian and therian (i.e., eutherian-marsupial) ancestors (Fig. 5B) and have since been preserved by purifying selection (fig. S17C). We confirmed and extended these observations using a single-cell chromatin accessibility atlas of adult mouse organs (15). We found that astrocytes have the most conserved intergenic CREs, not only in the adult cerebellum but also across all adult organs (Fig. 5F). Moreover, eight of the ten most conserved cell types across all organs were neural (Fig. 5F), highlighting the overall stronger evolutionary constraints in the brain (39, 45). Consistently, despite having the fastest evolving regulatory landscape in the cerebellum, microglia constitute the most conserved immune cell type (Fig. 5F).
Differences among neurons are subtler than among glial cells (Fig. 5A and fig. S16A). Granule cells exhibit the lowest conservation levels compared with other stage-matched neuron types, especially during prenatal development (Fig. 5A and fig. S16A, E13: P < 10-15, Mann-Whitney U test). This difference might reflect the high turnover in developmental adaptations associated with granule cell amplification across vertebrates, including the emergence of the external granule cell layer, a secondary germinal zone specific to amniotes (3, 46). Collectively, our results reveal common temporal trends as well as cell type-specific differences in the evolutionary dynamics of CREs in the developing cerebellum.
CRE activity conservation across therian mammals
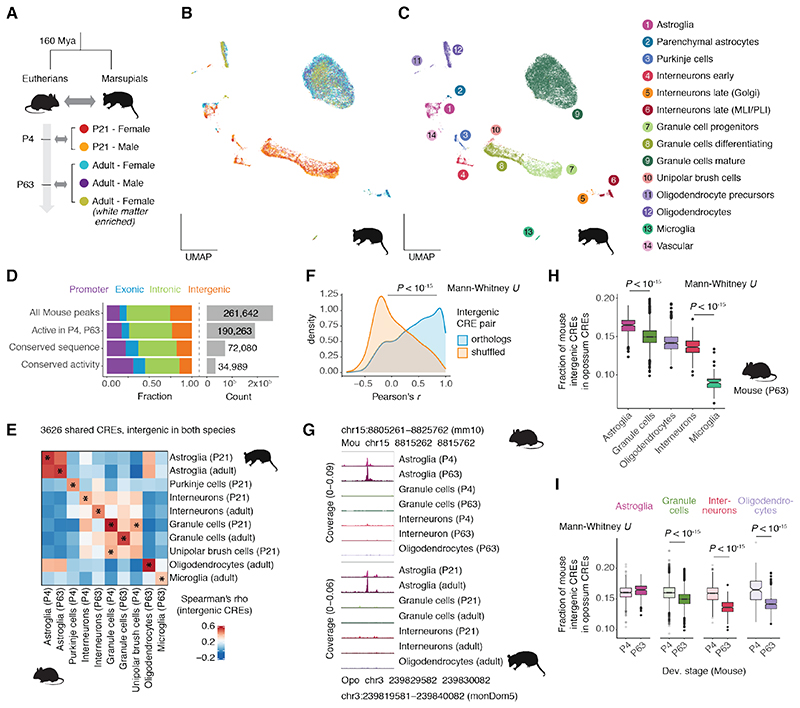
Although sequence constraint is a strong predictor of CRE activity conservation, comparative studies have shown that regulatory activity, especially for distal elements, evolves faster than DNA sequences (6–8, 43). To assess whether our observations on sequence constraints extend to CRE activity conservation, we examined the chromatin accessibility of cerebellar cells in the gray short-tailed opossum, a marsupial separated from mouse by ~160 million years of evolution. We profiled ~20,000 cells spanning two developmental stages: P21, which is transcriptionally similar to P4 in mouse (39), and adult (Fig. 6A). We detected the same cell types as in the mouse cerebellum at corresponding stages, with similar abundances, and with many shared marker genes (Fig. 6B-C and fig. S18A-B). We identified 167,340 putative CREs and compared them to mouse CREs based on reciprocal syntenic alignments. Nearly 50% of mouse CREs with conserved sequences also show conserved activity, with distal CREs evolving faster than promoters (Fig. 6D), as previously described (6, 7). Despite this turnover, all homologous cell types (except for UBCs) show the highest similarity in accessibility profiles of shared intergenic CREs (Fig. 6E), suggesting that most CREs that have been preserved during mammalian evolution maintain similar specificity patterns. Indeed, orthologous intergenic CREs show significantly higher correlations in their activity across matched cell types and developmental stages than shuffled homology pairs (Fig. 6F; table S7). For example, a mouse CRE assigned to the astroglia marker Slc1a3 and located 100 kb downstream of the gene has maintained the same astroglia-specific activity in both species (Fig. 6G). Overall, our data suggest that despite the high turnover of regulatory activity in terms of the emergence and loss of CREs (6, 7), radical repurposing of spatiotemporal CRE activity appears to be rare, at least between cell types in the same tissue.
Fig. 6. CRE activity conservation across therian mammals.
(A) Overview of opossum snATAC-seq dataset and correspondence to mouse developmental stages based on transcriptomic similarity (39). (B, C) UMAP projection of 19,204 opossum cells colored by sample (B) or cell type and state (C). (D) Distribution of genomic classes (left) and total number (right) for all mouse CREs, CREs active (≥ 5 CPM) in cell types and developmental stages corresponding to those sampled in opossum, and subsets showing conservation of sequence (CRE aligned to genome) and activity (CRE aligned to CRE) in opossum. (E) Spearman’s correlation of intergenic CRE activity between opossum and mouse corresponding cell types and stages. Stars mark the sample with the highest correlation for each row and column. (F) Pearson’s correlation coefficients of activity (in CPM) across corresponding cell types and stages for intergenic CRE pairs with true or shuffled orthology relationships. (G) Example of a shared intergenic CRE with conserved cell type-specificity between mouse and opossum. Accessibility profiles for each broad cell type and stage were aggregated across cells and scaled by the total number of fragments in each group. (H) Fraction of mouse intergenic CREs accessible per cell with an ortholog CRE in opossum, grouped by cell type in the adult mouse. (I) Fraction of mouse intergenic CREs accessible per cell with an ortholog CRE in opossum, grouped by cell type and developmental stage for cell types with at least 100 cells in both stages. MLI/PLI: molecular/Purkinje cell layer interneurons.
We next reexamined our evolutionary comparisons in the light of CRE activity conservation. In the adult mouse cerebellum, the fraction of intergenic CREs with a homolog active in the opossum cerebellum is the highest for astrocytes and the lowest for microglia (Fig. 6H), even when requiring the CRE to be active in the corresponding cell type and stage (fig. S18C). Except for astroglia, all cell types show higher conservation in P4 compared to the adult (Fig. 6I), confirming the temporal patterns observed based on sequence conservation (Fig. 5A). Finally, stratifying P4 granule cells according to their differentiation status revealed a significant, albeit small, decline in the fraction of mouse CREs shared with opossum (fig. S18D). Collectively, our analyses of regulatory activity conservation across ~160 million years of mammalian evolution reinforced our sequence-based conclusions regarding differences in CRE constraint across cell types and developmental stages.
Discussion
Our single-cell atlas of gene regulation in the mouse cerebellum spans development from the beginning of neurogenesis to adulthood, and is complemented with chromatin accessibility maps for the P21 and adult opossum cerebellum. We provide a comprehensive resource of the cis-regulatory profiles of cerebellar cell types, though not all cell types could be profiled at all stages due to low abundances. We detected extensive temporal differences in CRE activity between cells of the same cell type and even between matched differentiation states. Strong temporal differences in early progenitor cells are shared between germinal zones, suggesting that cell fates are induced through common temporal cues, which could be part of an intrinsic brain-wide temporal code (33) or involve common extrinsic signals, such as secreted factors (3).
Evolutionary comparisons revealed a decrease in sequence constraint and activity conservation of CREs during development across all cell types. This pattern is explained by both cell type differentiation and temporal differences between cells from matched differentiation states. The decrease in CRE evolutionary constraint parallels a decline in pleiotropy, as observed in gene expression studies (39, 40). Across cerebellar cell types, differences in regulatory conservation are most pronounced in the adult, where microglia – the immune cells of the brain – show the fastest evolutionary turnover. By contrast, mature astrocytes have the most conserved intergenic CREs, not only in the cerebellum but also across other adult mouse organs. This difference might be explained by astrocytes maintaining some of the properties of neural progenitors, including the ability to reactivate proliferation (47), and/or by pleiotropic constraints due to their bridging interactions with multiple cell types of the otherwise disconnected neuronal and vascular networks in the brain (48). Alternatively, higher sequence constraints could be associated with a more rigid motif grammar in the regulatory program of astrocytes (5). Collectively, our study illuminates how development and evolution shape the cis-regulatory profiles of cerebellar cells, and provides general insights into the dynamics of gene regulation during mammalian organ development.
Methods summary
Detailed information on materials and methods can be found in the supplement. In brief, snap-frozen cerebella samples from RjOrl:SWISS mice and grey short-tailed opossums were used for nuclei isolation and single cell library construction using the Chromium Single Cell ATAC Reagent kits (10x Genomics). Raw sequencing data were processed using cellranger-atac (21). ArchR (22) was used for quality control, dimensionality reduction, clustering and identification of open chromatin regions (CREs). Chromosome Y was excluded from all analyses. Chromosome X was excluded from dimensionality reduction but considered for subsequent analyses. Cell type identification was performed based on ArchR’s estimates of gene scores and comparison to known marker genes (2, 24) and in situ hybridization data from the Allen Brain Atlas resource (49).
Robust CREs were identified in a cluster-specific manner requiring reproducibility in at least two samples and accessibility in at least 5% of the cells of at least one cluster. Target gene assignment was performed based on promoter-peak co-accessibility and correlations with gene scores. CRE activity across groups (e.g., Fig. 2) was estimated by aggregating accessibility profiles across cells and scaling by sequencing depth (counts per million, CPM). For clustering of CRE activity, CPM values were standardized for each CRE as a fraction of its maximum CPM value across all groups and subjected to k-means clustering, followed by hierarchical clustering of the cluster means. For genes with temporal changes across progenitor types we estimated average gene scores in groups of cells from the same progenitor type, developmental stage and replicate. Time-variant genes were identified as those with a significantly higher standard deviation across stages compared to progenitor type and between replicates.
For modelling differentiation trajectories, we used Harmony (50) to align across developmental stages and diffusion pseudotime (51) to approximate differentiation. For CRE evolutionary dynamics, we estimated sequence constraint as the mean phastCons score (41) of the most conserved 100 bp region for each CRE. CRE minimum age was inferred based on the phylogenetic distance between mouse and the most distant vertebrate species for which a syntenic region could be detected. To estimate average values across cells we only used intergenic CREs to avoid biases from overlapping or adjacent protein-coding gene sequences. CREs conserved in mouse and opossum were identified based on syntenic alignments between the two species (minimum match of 10%) and requiring reciprocally best matches.
Supplementary Material
One Sentence Summary.
A single-cell chromatin accessibility atlas of the developing mouse cerebellum.
Acknowledgements
We thank Duncan Odom and Sebastian M. Waszak for critical reading of the manuscript, and Simon Anders, Daisuke Kawauchi, Francesco Lamanna, Florent Murat, Oliver Stegle, Judith Zaugg, and members of the Kaessmann lab for discussions. We thank Julia Schmidt, Céline Schneider, Annett Billepp and Petra Grimm for technical support.
Funding
This project has received funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (grant agreement No. 819894) and Seventh Framework Programme (FP7-2007-2013) (grant agreement No. 615253). M.C.M. was supported by the Francis Crick Institute, which receives its core funding from Cancer Research UK (FC011171), the UK Medical Research Council (FC011171), and the Wellcome Trust (FC011171). This research was funded in part by Wellcome Trust (FC011171), a cOAlition S organization. The author will make the Author Accepted Manuscript (AAM) version available under a CC BY public copyright license.
Footnotes
Author contributions: H.K., M.C.M., I.S. and M.S. originally designed the study; M.S. collected samples, P.G. maintained a breeding colony of opossums; M.S. and R.F. generated data; I.S. performed all analyses with contributions from M.S. and key input from R.F., K.L. and N.T; E.L., L.M.K., K.O. and P.J. provided important feedback and discussions; H.K. and S.M.P. provided funding; H.K., S.M.P. and M.C.M. supervised the study; I.S., M.S., M.C.M. and H.K. wrote the manuscript with input from all authors.
Competing interests: The authors report no conflict of interest.
Data and code availability
All data generated in this study are freely accessible in ArrayExpress with the accession codes E-MTAB-9765 (mouse) and E-MTAB-10533 (opossum). All other data are in the main paper or the Supplement. All code used to analyze the data is available online at https://github.com/ioansarr/mouse-cerebellum-atac/. Processed data can be interactively explored at https://apps.kaessmannlab.org/mouse_cereb_atac/. Active CREs across cell types and stages (Methods) can be inspected as custom UCSC genome browser tracks at: http://genome.ucsc.edu/s/ioansarr/Mouse%20cerebellum%20snATAC-seq%20cCREs.
References and notes
- 1.Sathyanesan A, Zhou J, Scafidi J, Heck DH, Sillitoe RV, Gallo V. Emerging connections between cerebellar development, behaviour and complex brain disorders. Nat Rev Neurosci. 2019;20:298–313. doi: 10.1038/s41583-019-0152-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leto K, Arancillo M, Becker EBE, Buffo A, Chiang C, Ding B, Dobyns WB, Dusart I, Haldipur P, Hatten ME, Hoshino M, et al. Consensus Paper: Cerebellar Development. Cerebellum. 2016;15:789–828. doi: 10.1007/s12311-015-0724-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Butts T, Green MJ, Wingate RJT. Development of the cerebellum: simple steps to make a ‘little brain.’. Development. 2014;141:4031–4041. doi: 10.1242/dev.106559. [DOI] [PubMed] [Google Scholar]
- 4.Nord AS, West AE. Neurobiological functions of transcriptional enhancers. Nat Neurosci. 2020;23:5–14. doi: 10.1038/s41593-019-0538-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Long HK, Prescott SL, Wysocka J. Ever-Changing Landscapes: Transcriptional Enhancers in Development and Evolution. Cell. 2016;167:1170–1187. doi: 10.1016/j.cell.2016.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Villar D, Berthelot C, Aldridge S, Rayner TF, Lukk M, Pignatelli M, Park TJ, Deaville R, Erichsen JT, Jasinska AJ, Turner JMA, et al. Enhancer evolution across 20 mammalian species. Cell. 2015;160:554–566. doi: 10.1016/j.cell.2015.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berthelot C, Villar D, Horvath JE, Odom DT, Flicek P. Complexity and conservation of regulatory landscapes underlie evolutionary resilience of mammalian gene expression. Nat Ecol Evol. 2018;2:152–163. doi: 10.1038/s41559-017-0377-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reilly SK, Yin J, Ayoub AE, Emera D, Leng J, Cotney J, Sarro R, Rakic P, Noonan JP. Evolutionary changes in promoter and enhancer activity during human corticogenesis. Science. 2015;347:1155–1159. doi: 10.1126/science.1260943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bejerano G, Pheasant M, Makunin I, Stephen S, Kent WJ, Mattick JS, Haussler D. Ultraconserved elements in the human genome. Science. 2004;304:1321–1325. doi: 10.1126/science.1098119. [DOI] [PubMed] [Google Scholar]
- 10.Dickel DE, Ypsilanti AR, Pla R, Zhu Y, Barozzi I, Mannion BJ, Khin YS, Fukuda-Yuzawa Y, Plajzer-Frick I, Pickle CS, Lee EA, et al. Ultraconserved Enhancers Are Required for Normal Development. Cell. 2018;172:491–499.:e15. doi: 10.1016/j.cell.2017.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gorkin DU, Barozzi I, Zhao Y, Zhang Y, Huang H, Lee AY, Li B, Chiou J, Wildberg A, Ding B, Zhang B, et al. An atlas of dynamic chromatin landscapes in mouse fetal development. Nature. 2020;583:744–751. doi: 10.1038/s41586-020-2093-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frank CL, Liu F, Wijayatunge R, Song L, Biegler MT, Yang MG, Vockley CM, Safi A, Gersbach CA, Crawford GE, West AE. Regulation of chromatin accessibility and Zic binding at enhancers in the developing cerebellum. Nat Neurosci. 2015;18:647–656. doi: 10.1038/nn.3995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nord AS, Blow MJ, Attanasio C, Akiyama JA, Holt A, Hosseini R, Phouanenavong S, Plajzer-Frick I, Shoukry M, Afzal V, Rubenstein JLR, et al. Rapid and pervasive changes in genome-wide enhancer usage during mammalian development. Cell. 2013;155:1521–1531. doi: 10.1016/j.cell.2013.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stergachis AB, Neph S, Reynolds A, Humbert R, Miller B, Paige SL, Vernot B, Cheng JB, Thurman RE, Sandstrom R, Haugen E, et al. Developmental fate and cellular maturity encoded in human regulatory DNA landscapes. Cell. 2013;154:888–903. doi: 10.1016/j.cell.2013.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cusanovich DA, Hill AJ, Aghamirzaie D, Daza RM, Pliner HA, Berletch JB, Filippova GN, Huang X, Christiansen L, De Witt WS, Lee C, et al. A Single-Cell Atlas of In Vivo Mammalian Chromatin Accessibility. Cell. 2018;174:1309–1324.:e18. doi: 10.1016/j.cell.2018.06.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li YE, Preissl S, Hou X, Zhang Z, Zhang K, Fang R, Qiu Y, Poirion O, Li B, Liu H, Wang X, et al. An Atlas of Gene Regulatory Elements in Adult Mouse Cerebrum. bioRxiv. 2020 doi: 10.1101/2020.05.10.087585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Preissl S, Fang R, Huang H, Zhao Y, Raviram R, Gorkin DU, Zhang Y, Sos BC, Afzal V, Dickel DE, Kuan S, et al. Single-nucleus analysis of accessible chromatin in developing mouse forebrain reveals cell-type-specific transcriptional regulation. Nat Neurosci. 2018;21:432–439. doi: 10.1038/s41593-018-0079-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Di Bella DJ, Habibi E, Stickels RR, Scalia G, Brown J, Yadollahpour P, Yang SM, Abbate C, Biancalani T, Macosko EZ, Chen F, et al. Molecular logic of cellular diversification in the mouse cerebral cortex. Nature. 2021 doi: 10.1038/s41586-021-03670-5. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ziffra RS, Kim CN, Wilfert A, Turner TN, Haeussler M, Casella AM, Przytycki PF, Kreimer A, Pollard KS, Ament SA, Eichler EE, et al. Single cell epigenomic atlas of the developing human brain and organoids. bioRxiv. 2019 doi: 10.1101/2019.12.30.891549. [DOI] [Google Scholar]
- 20.Domcke S, Hill AJ, Daza RM, Cao J, O’Day DR, Pliner HA, Aldinger KA, Pokholok D, Zhang F, Milbank JH, Zager MA, et al. A human cell atlas of fetal chromatin accessibility. Science. 2020;370:eaba7612. doi: 10.1126/science.aba7612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Satpathy AT, Granja JM, Yost KE, Qi Y, Meschi F, McDermott GP, Olsen BN, Mumbach MR, Pierce SE, Corces MR, Shah P, et al. Massively parallel single-cell chromatin landscapes of human immune cell development and intratumoral T cell exhaustion. Nat Biotechnol. 2019;37:925–936. doi: 10.1038/s41587-019-0206-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Granja JM, Corces MR, Pierce SE, Bagdatli ST, Choudhry H, Chang HY, Greenleaf WJ. ArchR is a scalable software package for integrative single-cell chromatin accessibility analysis. Nat Genet. 2021;53:403–411. doi: 10.1038/s41588-021-00790-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vladoiu MC, El-Hamamy I, Donovan LK, Farooq H, Holgado BL, Sundaravadanam Y, Ramaswamy V, Hendrikse LD, Kumar S, Mack SC, Lee JJYY, et al. Childhood cerebellar tumours mirror conserved fetal transcriptional programs. Nature. 2019;572:67–73. doi: 10.1038/s41586-019-1158-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wizeman JW, Guo Q, Wilion EM, Li JYH. Specification of diverse cell types during early neurogenesis of the mouse cerebellum. Elife. 2019;8:1–24. doi: 10.7554/eLife.42388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Millen KJ, Steshina EY, Iskusnykh IY, Chizhikov VV. Transformation of the cerebellum into more ventral brainstem fates causes cerebellar agenesis in the absence of Ptf1a function. Proc Natl Acad Sci. 2014;111:E1777–E1786. doi: 10.1073/pnas.1315024111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kriegstein A, Alvarez-Buylla A. The glial nature of embryonic and adult neural stem cells. Annu Rev Neurosci. 2009;32:149–184. doi: 10.1146/annurev.neuro.051508.135600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carter RA, Bihannic L, Rosencrance C, Hadley JL, Tong Y, Phoenix TN, Natarajan S, Easton J, Northcott PA, Gawad C. A Single-Cell Transcriptional Atlas of the Developing Murine Cerebellum. Curr Biol. 2018;28:2910–2920.:e2. doi: 10.1016/j.cub.2018.07.062. [DOI] [PubMed] [Google Scholar]
- 28.Visel A, Minovitsky S, Dubchak I, Pennacchio LA. VISTA Enhancer Browser - A database of tissue-specific human enhancers. Nucleic Acids Res. 2007;35:D88–D92. doi: 10.1093/nar/gkl822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dalby M, Rennie S, Andersson R. FANTOM5 transcribed enhancers in mm10 [Data set] 2018 doi: 10.5281/zenodo.1411211. [DOI] [Google Scholar]
- 30.Bonev B, Mendelson Cohen N, Szabo Q, Fritsch L, Papadopoulos GL, Lubling Y, Xu X, Lv X, Hugnot JP, Tanay A, Cavalli G. Multiscale 3D Genome Rewiring during Mouse Neural Development. Cell. 2017;171:557–572.:e24. doi: 10.1016/j.cell.2017.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sabarís G, Laiker I, Preger-Ben Noon E, Frankel N. Actors with Multiple Roles: Pleiotropic Enhancers and the Paradigm of Enhancer Modularity. Trends Genet. 2019;35:423–433. doi: 10.1016/j.tig.2019.03.006. [DOI] [PubMed] [Google Scholar]
- 32.Meuleman W, Muratov A, Rynes E, Halow J, Lee K, Bates D, Diegel M, Dunn D, Neri F, Teodosiadis A, Reynolds A, et al. Index and biological spectrum of human DNase I hypersensitive sites. Nature. 2020;584:244–251. doi: 10.1038/s41586-020-2559-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sagner A, Zhang I, Watson T, Lazaro J, Melchionda M, Briscoe J. Temporal patterning of the central nervous system by a shared transcription factor code. bioRxiv. 2020 doi: 10.1101/2020.11.10.376491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang T, Liu T, Mora N, Guegan J, Bertrand M, Contreras X, Hansen AH, Streicher C, Danda N, Tiberi L, Hippenmeyer S, et al. Generation of excitatory and inhibitory neurons from common progenitors via Notch signaling in the cerebellum. Cell Rep. 2021;35:109208. doi: 10.1016/j.celrep.2021.109208. [DOI] [PubMed] [Google Scholar]
- 35.Seto Y, Nakatani T, Masuyama N, Taya S, Kumai M, Minaki Y, Hamaguchi A, Inoue YU, Miyashita S, Fujiyama T, Yamada M, et al. Temporal identity transition from Purkinje cell progenitors to GABAergic interneuron progenitors in the cerebellum. Nat Commun. 2014;5:3337. doi: 10.1038/ncomms4337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cerrato V, Parmigiani E, Figueres-Oñate M, Betizeau M, Aprato J, Nanavaty I, Berchialla P, Luzzati F, De’Sperati C, López-Mascaraque L, Buffo A. Multiple origins and modularity in the spatiotemporal emergence of cerebellar astrocyte heterogeneity. PLoS Biol. 2018;16:1–38. doi: 10.1371/journal.pbio.2005513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kozareva V, Martin C, Osorno T, Rudolph S, Guo C, Vanderburg C, Nadaf N, Regev A, Regehr W, Macosko E. A transcriptomic atlas of the mouse cerebellum reveals regional specializations and novel cell types. bioRxiv. 2020 doi: 10.1101/2020.03.04.976407. [DOI] [Google Scholar]
- 38.Telley L, Agirman G, Prados J, Amberg N, Fièvre S, Oberst P, Bartolini G, Vitali I, Cadilhac C, Hippenmeyer S, Nguyen L, et al. Temporal patterning of apical progenitors and their daughter neurons in the developing neocortex. Science. 2019;364:eaav2522. doi: 10.1126/science.aav2522. [DOI] [PubMed] [Google Scholar]
- 39.Cardoso-Moreira M, Halbert J, Valloton D, Velten B, Chen C, Shao Y, Liechti A, Ascenção K, Rummel C, Ovchinnikova S, Mazin PV, et al. Gene expression across mammalian organ development. Nature. 2019;571:505–509. doi: 10.1038/s41586-019-1338-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sarropoulos I, Marin R, Cardoso-Moreira M, Kaessmann H. Developmental dynamics of lncRNAs across mammalian organs and species. Nature. 2019;571:510–514. doi: 10.1038/s41586-019-1341-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LDW, Richards S, Weinstock GM, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Arendt D, Musser JM, Baker CVH, Bergman A, Cepko C, Erwin DH, Pavlicev M, Schlosser G, Widder S, Laubichler MD, Wagner GP. The origin and evolution of cell types. Nat Rev Genet. 2016;17:744–757. doi: 10.1038/nrg.2016.127. [DOI] [PubMed] [Google Scholar]
- 43.Roller M, Stamper E, Villar D, Izuogu O, Martin F, Redmond AM, Ramachanderan R, Harewood L, Odom DT, Flicek P. LINE retrotransposons characterize mammalian tissue-specific and evolutionarily dynamic regulatory regions. Genome Biol. 2021;22:62. doi: 10.1186/s13059-021-02260-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Geirsdottir L, David E, Weiner A, Bohlen SC, Neuber J, Balic A, Giladi A, Sheban F, Dutertre CA, Pfeifle C, Peri F, et al. Cross-Species Single-Cell Analysis Reveals Divergence of the Primate Microglia Program. Cell. 2019;179:1609–1622.:e16. doi: 10.1016/j.cell.2019.11.010. [DOI] [PubMed] [Google Scholar]
- 45.Brawand D, Soumillon M, Necsulea A, Julien P, Harrigan P, Weier M, Liechti A, Kircher M, Albert FW, Zeller U, Khaitovich P, et al. The evolution of gene expression levels in mammalian organs. Nature. 2011;478:343–348. doi: 10.1038/nature10532. [DOI] [PubMed] [Google Scholar]
- 46.Iulianella A, Wingate RJ, Moens CB, Capaldo E. The generation of granule cells during the development and evolution of the cerebellum. Dev Dyn. 2019;248:506–513. doi: 10.1002/dvdy.64. [DOI] [PubMed] [Google Scholar]
- 47.Götz M, Sirko S, Beckers J, Irmler M. Reactive astrocytes as neural stem or progenitor cells: In vivo lineage, In vitro potential, and Genome-wide expression analysis. Glia. 2015;63:1452–1468. doi: 10.1002/glia.22850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Volterra A, Meldolesi J. Astrocytes, from brain glue to communication elements: The revolution continues. Nat Rev Neurosci. 2005;6:626–640. doi: 10.1038/nrn1722. [DOI] [PubMed] [Google Scholar]
- 49.Allen Institute for Brain Science. Allen Developing Mouse Brain Atlas. 2008 (available at http://developingmouse.brain-map.org/)
- 50.Korsunsky I, Millard N, Fan J, Slowikowski K, Zhang F, Wei K, Baglaenko Y, Brenner M, Raychaudhuri S. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat Methods. 2019;16:1289–1296. doi: 10.1038/s41592-019-0619-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Haghverdi L, Büttner M, Wolf FA, Buettner F, Theis FJ. Diffusion pseudotime robustly reconstructs lineage branching. Nat Methods. 2016;13:845–848. doi: 10.1038/nmeth.3971. [DOI] [PubMed] [Google Scholar]
- 52.Krishnaswami SR, Grindberg RV, Novotny M, Venepally P, Lacar B, Bhutani K, Linker SB, Pham S, Erwin JA, Miller JA, Hodge R, et al. Using single nuclei for RNA-seq to capture the transcriptome of postmortem neurons. Nat Protoc. 2016;11:499–524. doi: 10.1038/nprot.2016.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM, Hao Y, Stoeckius M, Smibert P, Satija R. Comprehensive Integration of Single-Cell Data. Cell. 2019;177:1888–1902.:e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nussbaum C, Myers RM, Brown M, Li W, Liu XS. Model-based Analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Haeussler M, Zweig AS, Tyner C, Speir ML, Rosenbloom KR, Raney BJ, Lee CM, Lee BT, Hinrichs AS, Gonzalez JN, Gibson D, et al. The UCSC Genome Browser database: 2019 update. Nucleic Acids Res. 2019;47:D853–D858. doi: 10.1093/nar/gky1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cunningham F, Achuthan P, Akanni W, Allen J, Amode MR, Armean IM, Bennett R, Bhai J, Billis K, Boddu S, Cummins C, et al. Ensembl 2019. Nucleic Acids Res. 2019;47:D745–D751. doi: 10.1093/nar/gky1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Quinlan AR, Hall IM. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schep AN, Wu B, Buenrostro JD, Greenleaf WJ. ChromVAR: Inferring transcription-factor-associated accessibility from single-cell epigenomic data. Nat Methods. 2017;14:975–978. doi: 10.1038/nmeth.4401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Weirauch MT, Yang A, Albu M, Cote AG, Montenegro-Montero A, Drewe P, Najafabadi HS, Lambert SA, Mann I, Cook K, Zheng H, et al. Determination and inference of eukaryotic transcription factor sequence specificity. Cell. 2014;158:1431–1443. doi: 10.1016/j.cell.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kent WJ, Zweig AS, Barber G, Hinrichs AS, Karolchik D. BigWig and BigBed: Enabling browsing of large distributed datasets. Bioinformatics. 2010;26:2204–2207. doi: 10.1093/bioinformatics/btq351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Karolchik D. The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 2004;32:493D–496. doi: 10.1093/nar/gkh103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Delignette-Muller ML, Dutang C. fitdistrplus: An R package for fitting distributions. J Stat Softw. 2015;64:1–34. [Google Scholar]
- 63.Robinson MD, McCarthy DJ, Smyth GK. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2009;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Trevino AE, Sinnott-Armstrong N, Andersen J, Yoon S, Huber N, Pritchard JK, Chang HY, Greenleaf WJ, Paşca SP. Chromatin accessibility dynamics in a model of human forebrain development. Science. 2020;367:eaay1645. doi: 10.1126/science.aay1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, Bejerano G. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple Combinations of Lineage-Determining Transcription Factors Prime cis-Regulatory Elements Required for Macrophage and B Cell Identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Supek F, Bošnjak M, Škunca N, Šmuc T. REVIGO Summarizes and Visualizes Long Lists of Gene Ontology Terms. PLoS One. 2011;6:e21800. doi: 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Suzuki R, Terada Y, Shimodaira H. pvclust: Hierarchical Clustering with P-Values via Multiscale Bootstrap Resampling. 2019 (available at https://cran.r-project.org/package=pvclust)
- 70.Futschik ME, Kumar L, Futschik ME. Introduction to Mfuzz package and its graphical user interface. Analysis. 2009:1–13. [Google Scholar]
- 71.Wolf FA, Angerer P, Theis FJ. SCANPY: Large-scale single-cell gene expression data analysis. Genome Biol. 2018;19:15. doi: 10.1186/s13059-017-1382-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pardy C. mpmi: Mixed-Pair Mutual Information Estimators. 2019 (available at https://cran.r-project.org/package=mpmi)
- 73.Scrucca L, Fop M, Murphy TB, Raftery AE. Mclust 5: Clustering, classification and density estimation using Gaussian finite mixture models. R J. 2016;8:289–317. [PMC free article] [PubMed] [Google Scholar]
- 74.Gu Z, Gu L, Eils R, Schlesner M, Brors B. Circlize implements and enhances circular visualization in R. Bioinformatics. 2014;30:2811–2812. doi: 10.1093/bioinformatics/btu393. [DOI] [PubMed] [Google Scholar]
- 75.Lê S, Josse J, Husson F. FactoMineR: An R package for multivariate analysis. J Stat Softw. 2008;25:1–18. [Google Scholar]
- 76.Berest I, Arnold C, Palla G, Rasmussen KD, Giles H, Bruch PM, Huber W, Dietrich S, Helin K, Zaugg JB. Quantification of Differential Transcription Factor Activity and Multiomics-Based Classification into Activators and Repressors: diffTF. Cell Rep. 2019;29:3147–3159.:e12. doi: 10.1016/j.celrep.2019.10.106. [DOI] [PubMed] [Google Scholar]
- 77.Hu H, Miao YR, Jia LH, Yu QY, Zhang Q, Guo AY. AnimalTFDB 3.0: A comprehensive resource for annotation and prediction of animal transcription factors. Nucleic Acids Res. 2019;47:D33–D38. doi: 10.1093/nar/gky822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Conesa A, Nueda MJ, Ferrer A, Talón M. maSigPro: A method to identify significantly differential expression profiles in time-course microarray experiments. Bioinformatics. 2006;22:1096–1102. doi: 10.1093/bioinformatics/btl056. [DOI] [PubMed] [Google Scholar]
- 79.R Core Team. R: A Language and Environment for Statistical Computing. 2020 (available at https://www.r-project.org/)
- 80.Wickham H, Averick M, Bryan J, Chang W, McGowan L, François R, Grolemund G, Hayes A, Henry L, Hester J, Kuhn M, et al. Welcome to the Tidyverse. J Open Source Softw. 2019;4:1686 [Google Scholar]
- 81.Dowle M, Srinivasan A. data.table: Extension of ‘data.frame’. 2017 [Google Scholar]
- 82.Bates D, Maechler M. Matrix: Sparse and Dense Matrix Classes and Methods. 2019 [Google Scholar]
- 83.Morgan M, Obenchain V, Hester J, Pages H. SummarizedExperiment: SummarizedExperiment container. 2019 [Google Scholar]
- 84.Baglama J, Reichel L, Lewis BW. irlba: Fast Truncated Singular Value Decomposition and Principal Components Analysis for Large Dense and Sparse Matrices. 2019 (available at https://cran.r-project.org/package=irlba)
- 85.Maechler M, Rousseeuw P, Struyf A, Hubert M, Hornik K. cluster: Cluster Analysis Basics and Extensions. 2019 [Google Scholar]
- 86.Gu Z, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32:2847–2849. doi: 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- 87.Kolde R. pheatmap: Pretty Heatmaps. 2019 (available at https://cran.r-project.org/package=pheatmap)
- 88.Chang W, Cheng J, Allaire J, Xie Y, McPherson J. shiny: Web Application Framework for R. 2020 [Google Scholar]
- 89.Attali D. shinyjs: Easily Improve the User Experience of Your Shiny Apps in Seconds. 2020 (available at https://cran.r-project.org/package=shinyjs)
- 90.Hahne F, Ivanek R. Visualizing Genomic Data Using Gviz and Bioconductor. Methods Mol Biol. 2016;1418:335–351. doi: 10.1007/978-1-4939-3578-9_16. [DOI] [PubMed] [Google Scholar]
- 91.Harmston N, Ing-Simmons E, Perry M, Baresic A, Lenhard B. GenomicInteractions: R package for handling genomic interaction data. 2020 doi: 10.1186/s12864-015-2140-x. (available at https://www.bioconductor.org/packages/release/bioc/html/GenomicInteractions.html) [DOI] [PMC free article] [PubMed]
- 92.Prekop HT, Kroiss A, Rook V, Zagoraiou L, Jessell TM, Fernandes C, Delogu A, Wingate RJT. Sox14 is required for a specific subset of cerebello-olivary projections. J Neurosci. 2018;38:9539–9550. doi: 10.1523/JNEUROSCI.1456-18.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chizhikov VV, Lindgren AG, Mishima Y, Roberts RW, Aldinger KA, Miesegaes GR, Spencer Currle D, Monuki ES, Millen KJ. Lmx1a regulates fates and location of cells originating from the cerebellar rhombic lip and telencephalic cortical hem. Proc Natl Acad Sci U S A. 2010;107:10725–10730. doi: 10.1073/pnas.0910786107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated in this study are freely accessible in ArrayExpress with the accession codes E-MTAB-9765 (mouse) and E-MTAB-10533 (opossum). All other data are in the main paper or the Supplement. All code used to analyze the data is available online at https://github.com/ioansarr/mouse-cerebellum-atac/. Processed data can be interactively explored at https://apps.kaessmannlab.org/mouse_cereb_atac/. Active CREs across cell types and stages (Methods) can be inspected as custom UCSC genome browser tracks at: http://genome.ucsc.edu/s/ioansarr/Mouse%20cerebellum%20snATAC-seq%20cCREs.