Abstract
Genetic association results are often interpreted with the assumption that study participation does not affect downstream analyses. Understanding the genetic basis of participation bias is challenging as it requires genotypes of unseen individuals. Here we demonstrate that it is possible to estimate comparative biases by performing a genome-wide association study (GWAS) contrasting one subgroup versus another. For example, we show that sex exhibits artefactual autosomal heritability in the presence of sex-differential participation bias. By performing a GWAS of sex in ∼3.3 million males and females, we identify over 158 autosomal loci spuriously associated with sex and highlight complex traits underpinning differences in study participation between sexes. For example, the body mass index-increasing allele at FTO was observed at higher frequency in males compared to females (OR 1.02, P = 4.4 × 10-36). Finally, we demonstrate how these biases can potentially lead to incorrect inferences in downstream analyses and propose a conceptual framework for addressing such biases. Our findings highlight a new challenge that genetic studies may face as sample sizes continue to grow.
Individuals who enroll in research studies or who purchase direct-to-consumer genetic tests are often non-representative of the general population1,2,3. For example, the UK Biobank study invited ~9 million individuals and achieved an overall participation rate of 5.45%4. Enrolled individuals demonstrate an obvious ‘healthy volunteer bias’, with lower rates of obesity, smoking and self-reported health conditions than the population sampling frame4. Achieving good representation of the sampled population in any study is a difficult challenge. Some examples do exist, such as the iPSYCH study, which gathered a random population sample by extracting DNA from a nationwide routine collection of neonatal dried blood spots and linkage to national register data5. The benefits of good representation have been long debated6,7,8,9. Many researchers argue that non-representative studies can bias prevalence estimates but do not lead to substantial bias in exposure-disease associations10,11. Deliberately non-representative study designs can also be valuable, for example by enriching for cases who carry more disease-causing alleles in a case-control study to maximize the power to detect genetic effects12.
There is recent evidence that genetic factors are associated with degree of study engagement13,14,15. For example, within a study, individuals with high genetic risk for schizophrenia are less likely to complete health questionnaires, attend clinical assessments and continue to actively participate in follow-up than those with lower genetic risk13,16. It remains unclear to what extent genetic factors influence initial study enrollment, or what are the downstream consequences of such bias, although previous simulations have attempted to quantify this bias17. We hypothesised that study participation bias can be identified by performing a GWAS on a non-heritable trait. Given that there are no known biological mechanisms that can give rise to autosomal allele frequency differences between sexes at conception, any allele frequency difference between sexes highlights an impact of that locus on sex-differential survival or sex-differential study participation. Another way to describe this concept is, if any trait leads males and females to differentially participate in a study, we would observe artefactual associations between variants associated with that trait and sex (see Box 1 and Extended Data Fig. 1). Therefore, an autosomal GWAS of sex provides a unique negative control analysis for genetic association testing, and may provide novel insights into the factors that underlie non-representative study participation18.
Box 1.
Participation bias: Participation—also called “selection” or “sampling”—bias is observed when participation in a study is not random38,39 with respect to the reference population. Participation bias can impact prevalence estimates and results in biased association estimates. This latter phenomenon is caused because participation bias acts as a “collider”.
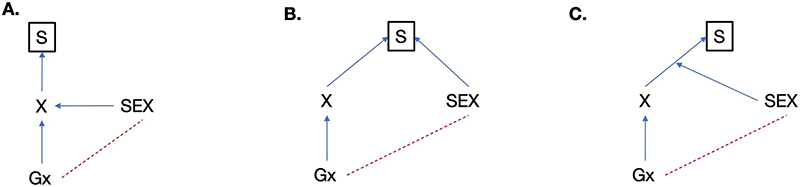
Collider bias: If two variables independently cause a third variable (the collider), then conditioning on the collider (i.e. conditioning on study participation) can cause a spurious association between the two variables40. In Extended Data Figure 1, we draw three path diagrams representing different types of participation bias.
Sex-differential participation bias: Sex-differential participation bias is a special case of participation bias where the determinants of study participation affect women and men to differing extents. While participation bias can be detected only if information on non-participating individuals is available, sex-differential participation bias can be detected by comparing genetic allele frequencies between males and females within a study.
Here we report the results from such a GWAS of sex, performed in ~3.3 million genotyped individuals. We identify more than 150 independent autosomal signals significantly associated with sex, highlighting several complex traits that contribute to sex-differential study participation. Furthermore, we demonstrate the potential impact of such bias on association testing and discuss a conceptual framework to address this issue.
Results
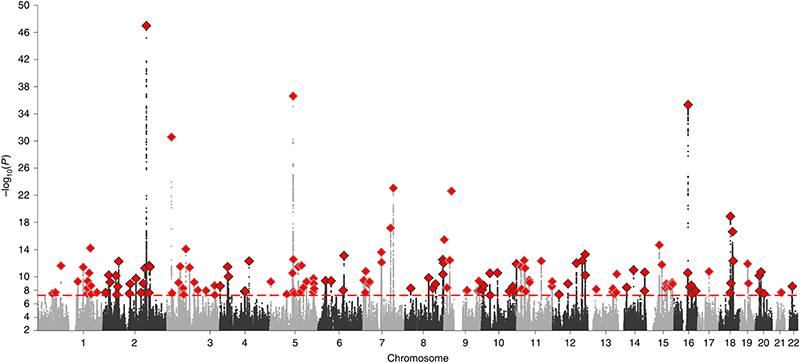
We performed a GWAS of sex (females coded as 1, males coded as 0) in 2,462,132 research participants from 23andMe using standard quality control procedures (Supplementary Note). We identified 158 independent genome-wide significant (P < 5 x 10-8) autosomal signals, indicating genetic variants that show significant allele frequency differences between sexes in this sample (Fig. 1 and Supplementary Table 1).
Fig. 1. Manhattan plot for a GWAS of sex in 2,462,132 participants from 23andMe.
The plot reports all identified loci, including those filtered by the extremely stringent quality control applied to directly genotyped SNPs.
Technical artefacts do not explain autosomal associations with sex
Additional conservative quality control procedures were performed to exclude any significant signals that might be caused by technical error (Supplementary Note). The most obvious reason for a false-positive association with sex is that the autosomal genotype array probe cross-hybridizes with a sex chromosome sequence. This issue has impacted similar previously published studies. For example, a GWAS in 8,842 South Korean males and females identified nine genetic variants strongly associated with sex19. The authors attributed their findings to biological mechanisms determining sex-selection; however, all of those nine associated variants are located within autosomal regions with significant homology to a sex chromosome sequence.
To evaluate the impact of this issue in our own data, we first identified directly genotyped variants that were both genome-wide significantly associated with sex and in LD (r 2 > 0.1) with one of our imputed top signals (n = 78; Supplementary Table 2). We then tested for sex chromosome homology with the genomic sequence (+/- 50bp) surrounding each genotyped variant, and found that one quarter (18/78) of our signals were potentially attributable to this technical issue. We further excluded additional loci due to low allele frequency (MAF < 5%), significant departure from Hardy-Weinberg equilibrium (P < 1 x 10-6) and/or low genotyping call rate (< 98%). Despite these very stringent filters, 49/78 directly genotyped genome-wide significant signals remained. These data suggest that the majority of signals that we identify represent true allele frequency differences between the sampled male and female participants in 23andMe, rather than genotyping errors.
Survival bias does not explain autosomal associations with sex
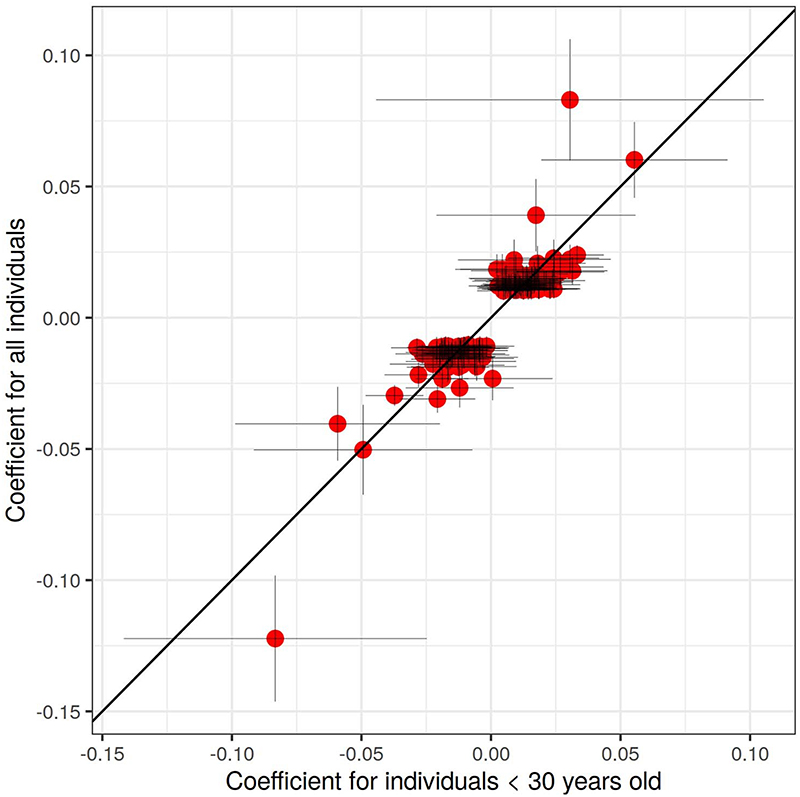
We next explored whether the observed signals for sex might arise due to sex-differential effects on mortality. To evaluate this, we repeated the GWAS of sex but restricted the sample to individuals aged 30 years or younger (n = 320,487), under the assumption that effects due to sex-differential mortality are less likely in younger than older age groups. While the substantially smaller sample size weakened the statistical significance of the signals, the magnitudes of effect across most signals remained consistent (Extended Data Fig. 2), with no significant difference in effect size observed for any of the 158 loci (Supplementary Table 3).
Participation bias results in autosomal associations with sex
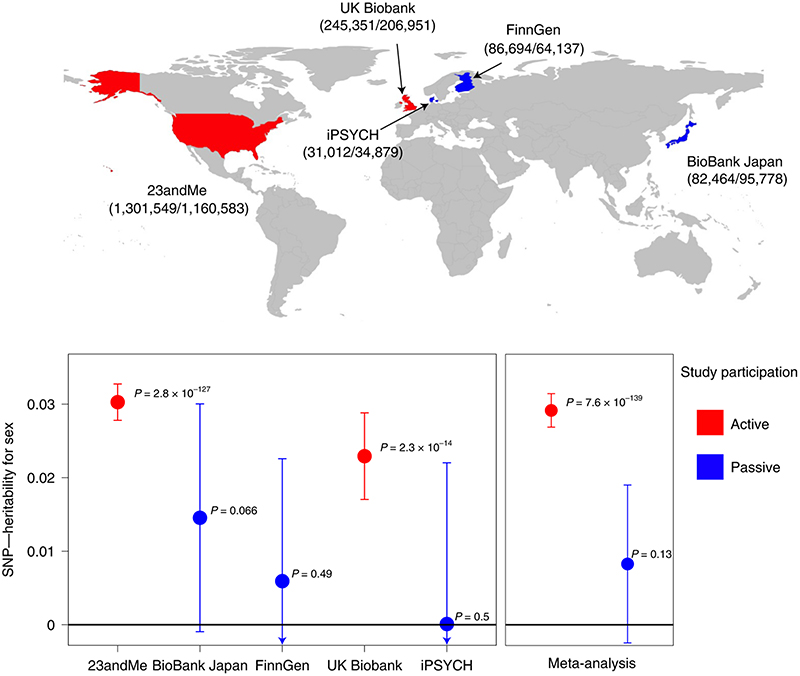
We next explored the hypothesis that many signals for sex that act by influencing sex-differential study participation rates may show markedly different associations with sex by study recruitment design (whereas effects due to sex-differential mortality would be consistent between studies). We therefore repeated the GWAS of sex in four additional studies (UK Biobank, FinnGen, Biobank Japan and iPSYCH; total n = 847,266) that varied by study recruitment design. As in 23andMe, UK Biobank required active participant engagement, albeit following a very different sampling and recruitment process. By contrast, FinnGen, Biobank Japan and iPSYCH required more passive participant involvement with no or little study engagement, as samples were collected from routine biospecimens or during clinical visits. We observed significant heritability of sex only in the studies that required more active participation (h 2 on liability scale = 3.0% (P = 3 x 10-127) and 2.3% (P = 2 x 10-14), in 23andMe and UK Biobank, respectively), while no significant heritability was detected in the three more passive studies (Fig. 2 and Supplementary Table 4).
Fig. 2. SNP heritability on the liability scale for sex across five studies.
Samples sizes were as follows: 23andMe, n = 2,462,132; BioBank Japan, n = 178,242; FinnGen, n = 150,831; UK Biobank, n = 452,302; iPSYCH, n = 65,891. The error bars represent the confidence interval for the SNP heritability estimate. For each study, we report in parentheses the number of females and males included in the analysis. Studies characterized by ‘active’ participation are shown in red, and studies with ‘passive’ participation are shown in blue. iPSYCH heritability is negative and therefore set to 0. Definitions of ‘active’ and ‘passive’ are ad hoc for this study and encompass heterogeneous enrollment strategies and consent modalities.
iPSYCH, in particular, showed the lowest heritability estimate, consistent with its study design that retrieved routinely collected neonatal dried blood spots from a random sample of individuals born between 1981 and 2005 who were alive and resident in Denmark on their first birthday, thus minimizing both participation and survival bias. In aggregate, these findings suggest that many autosomal signals for sex represent underlying mechanisms that influence sex-differential study participation rather than sex-differential pre-sampling mortality. We do not preclude the possibility that a small number of loci might influence sex-differential survival in utero, which should be explored in future studies.
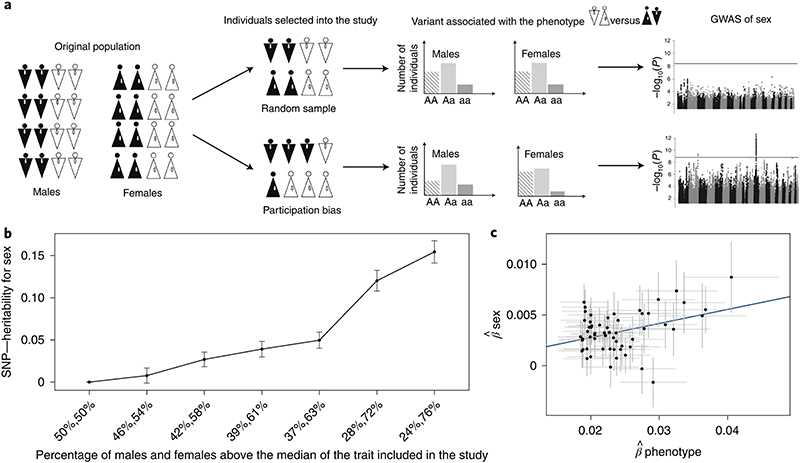
To demonstrate the statistical basis of our observed sex-differential participation bias, we simulated a phenotype that is uncorrelated with sex and has a heritability of 30% in 350,000 individuals, half males and half females (Fig. 3a). Under different sampling scenarios, we found that sex becomes significantly heritable on autosomes if study participation is dependent on the phenotype in a sex-differential manner (Fig. 3b). In the presence of this bias, autosomal variants associated with the phenotype are also associated with sex in a dose-response manner. As a consequence, Mendelian randomization (MR) analysis would wrongly identify a causal relationship between the phenotype and sex (Fig. 3c). An alternative explanation for our findings is that sex is a causal factor for the phenotype that influences study participation (Extended Data Fig. 1a) or that both sex and the phenotype drive participation independently (Extended Data Fig. 1b); however, we show using both real data and simulations that these models are less likely (see Supplementary Note).
Fig. 3. Illustration of the concept and consequences of sex-differential participation bias.
a, Schematic representation of sex-differential participation bias. Because males and females distribute differently for a certain trait in the selected study population, variants associated with the trait become associated with sex.
b, Heritability of sex increases as a function of sex-differential participation bias expressed as the percentage of males and females above the median of the phenotype included in the study (n = 350,000). If there is no bias, this value is 50% for both males and females. The dots represent the SNP effect size; the error bars represent the confidence intervals for the heritability estimate.
c, Variants associated with the phenotype are also associated with sex in a dose-dependent manner. MR would indicate a causal relationship between sex and phenotype. In this study, we considered only variants that were genome-wide significantly associated with the phenotype in the fourth scenario of b (39%, 61%). The dots represent the SNP effect size; the error bars represent the confidence intervals for the SNP effect size.
Genetic analyses reveal determinants of sex-differential participation bias
We then systematically tested complex traits for evidence of a shared genetic architecture with sex-differential participation bias in UK Biobank and 23andMe. By analyzing summary data from 4,155 publicly available GWASs20, we showed that sex-associated signals are enriched for pleiotropic associations (P < 2 x 10-16; Chi-square test comparing sex-associated SNPs vs. all SNPs); half of the genome-wide significant imputed signals for sex were associated with at least one complex trait, and one-fifth were associated with five or more traits (Supplementary Table 5). Genetically correlated traits spanned a diverse range of health outcomes, including blood pressure, type 2 diabetes, anthropometry, bone mineral density, autoimmune disease, personality traits and psychiatric diseases.
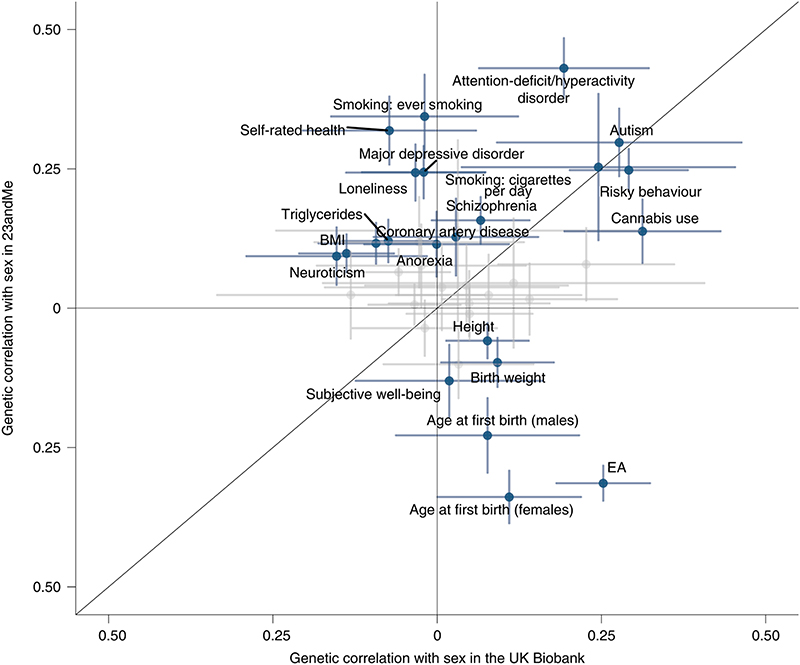
Genome-wide autosomal correlation analyses with 38 health and behavioral traits highlighted 22 significant associations with sex in 23andMe and 5 in UK Biobank (Fig. 4 and Supplementary Table 6). We noted that the genetic signals for sex overlapped only partially between 23andMe and UK Biobank (rg = 0.50, P = 4 x 10-34), which was reflected in several trait-specific study discordant associations. For example, higher educational attainment was associated with female sex in UK Biobank (rg = 0.25, P = 7 x 10-12), while the opposite direction of association was observed in 23andMe (rg = -0.31, P = 9 x 10-81). This finding demonstrates that the determinants of sex-differential participation bias may vary substantially between studies.
Fig. 4. Genetic correlation with being born female versus male and 38 traits in the UK Biobank and 23andMe.
Only correlations that were significant in at least one of the two studies are highlighted. The dots represent the genetic correlation estimates; the error bars represent the confidence intervals.
A notable autosomal signal for sex was at the obesity-associated FTO locus, where the body mass index (BMI)-increasing allele was observed in 23andMe at higher frequency in males compared to females (rs10468280, OR 1.02 (1.02-1.03), P = 4.4 x 10-36; Supplementary Table 1). The same direction and magnitude of effect at the FTO locus was also observed in UK Biobank (OR 1.02 (1.01-1.03), P = 3.6 x 10-5), and subsequent Mendelian randomization analyses supported a causal effect of BMI on sex in both 23andMe and UK Biobank (Supplementary Table 7). We note however that there was considerable heterogeneity in the dose-response relationship between genome-wide significant BMI variants and sex, and it remains unclear how genetically higher BMI leads to sex-differential study participation. Intriguingly, the genetic correlation between BMI and sex, which leverages the entirety of the genetic associations and not only genome-wide significant variants, was discordant between UK Biobank (rg = -0.13, P = 2 x 10-4) and 23andMe (rg = 0.10, P = 9 x 10-8), and this difference between studies appeared attributable to negative confounding by educational attainment (Supplementary Table 7). These results reinforce the need for caution when inferring causality from genetic correlations.
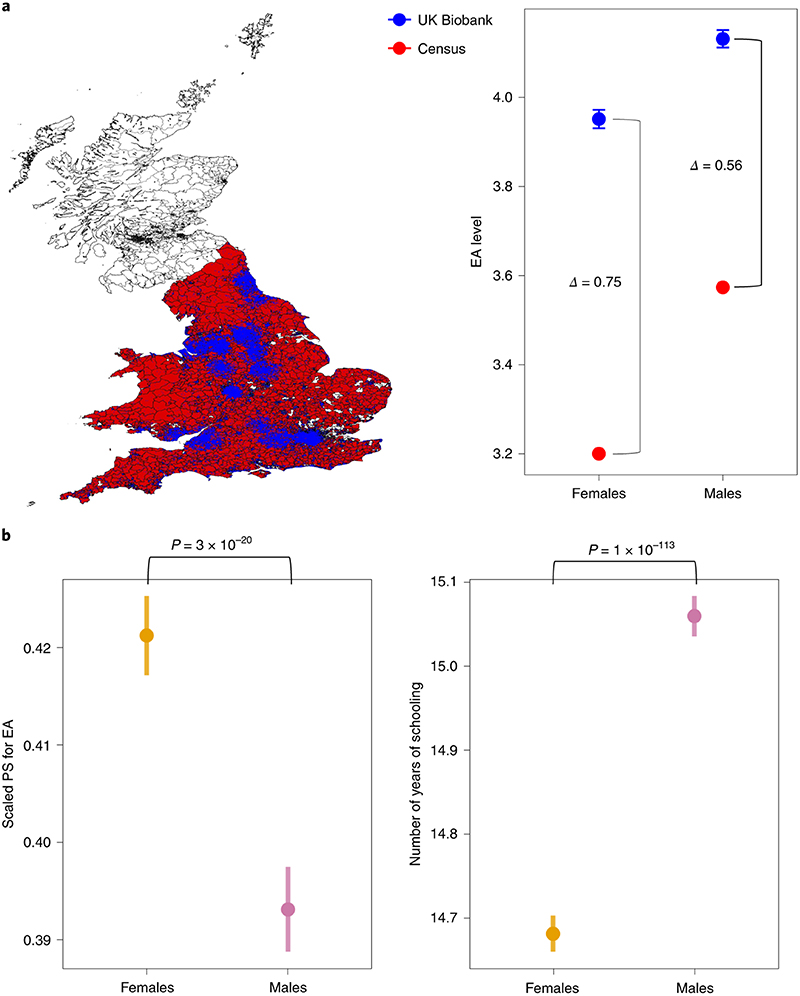
Traditional approaches to identify study participation bias compare the distribution of a phenotype in the study with that of a representative population. Using this approach, we confirmed our genetic inference that the difference in education level between UK Biobank participants and UK census data was larger in females than in males (Fig. 5a and Supplementary Table 8). Such greater differential participation by education among females can also be observed, without the need for census data, by comparing the distribution of polygenic scores for education between males vs. females. If we had a completely representative sample, we would not expect any differences in the distribution of the polygenic score for educational attainment between males and females (i.e. all the differences in measured educational attainment between the two sexes are expected to be due to environmental factors). Any difference in the polygenic score distribution needs therefore to be explained by selection acting on educational attainment that is either determined by sex or it has occurred differentially between men and women.
Fig. 5. PS distribution highlights sex-differential participation by educational level in the UK Biobank.
a, Comparing the highest educational level between the 2011 England and Wales census data (red, n = 29,492,209) with the UK Biobank (blue, n = 411,845). We only considered regional census districts with at least one UK Biobank participant. The difference (Δ) in the average educational level between males and females is higher in the general population than in participants in the UK Biobank. The dots represent the mean taking into account the sampling design; the error bars represent the confidence intervals. No confidence intervals were considered for the census data because the entire population was included.
b, PS for EA was significantly higher in females (n = 194,282) compared to males (n = 167,219) in the UK Biobank, whereas the number of years of schooling was higher in males (two-sided t-test). The dots represent the mean value; the error bars represent the s.d.
To test this hypothesis, we used data from the SSGAC consortium21, which did not include UK Biobank or 23andMe, and constructed a polygenic score for educational attainment. We first examined iPSYCH, where we do not expect participation bias, and indeed we saw no significant differences in the distribution of the polygenic score for educational attainment between males and females (P = 0.78). In UK Biobank, the mean polygenic score was higher in females than in males (P = 7 x 10-23; t-test), consistent with the census data comparison. We note that, opposite to the polygenic score, the reported education level in UK Biobank is significantly higher in males compared to females (P = 1 x 10-113; t-test) (Fig. 5b). Therefore, on its own, the distribution of the phenotype among study participants does not inform the direction and degree of sex-differential participation bias.
Educational attainment is one of few traits for which representative data are available, via the UK census. For other traits, where such information is not collected, genetic analysis in the form of polygenic scores provides a unique opportunity to identify novel sex-differential determinants of study participation.
Sex-differential participant bias can influence downstream genetic analyses
Next, we illustrated the potential effects of sex-differential participation bias on downstream genetic analyses using simulated and empirical data (Extended Data Figs. 3–4, Supplementary Figs. 2–5, Supplementary Note, and Supplementary Tables 9 and 10).
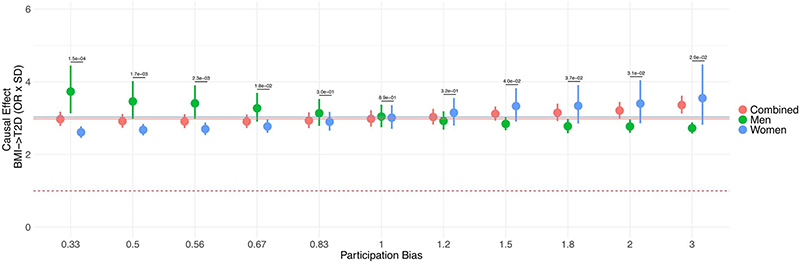
First, we performed simulation analyses to demonstrate that this bias can lead to spurious genetic correlations between two traits by exacerbating or attenuating the effects of overall participation bias (Extended Data Fig. 3). Furthermore, it can lead to an incorrect causal inference (in Mendelian randomization analyses) between two phenotypes in a sex-differential manner (Extended Data Fig. 4). For example, Censin and colleagues recently described sex differences in the causal effect of BMI on cardiometabolic outcomes in UK Biobank22. They concluded that the magnitude of increase in risk for type 2 diabetes (T2D) due to obesity differs between males and females. We attempted to confirm their results in light of our observations and found that their findings were likely biased due to reasons other than sex-differential participation bias (see Supplementary Note). However, we demonstrate through simulation analyses that sex-differential participation bias could indeed lead to incorrect inferences in such Mendelian randomization analyses (Supplementary Table 10). With only modest BMI-related sex-differential participation bias, we saw artificial sex differences in the association between a BMI genetic score and T2D and, in the most extreme sampling parameters, the direction of sex difference was swapped, with BMI genetic score-T2D effect estimates ranging from ORmale = 2.71 and ORfemale = 3.49 to ORmale = 3.86 and ORfemale = 2.61. These results highlight the challenges of performing and interpreting sex-specific analyses in studies where the exposure variable may be influenced by sex differences in participation bias.
Second, in a scenario where sex-differential participation exists, adjusting for sex as a covariate in a GWAS could bias effect estimates of any genetic analysis (Supplementary Fig. 3). To explore this possibility, we performed 565 GWASs of heritable traits in the UK Biobank and estimated the genetic correlation between each trait with and without inclusion of sex as a covariate. The results were highly consistent (Supplementary Fig. 4) between the two models, with sizable differences (indicated by lower genetic correlations) observed only for highly sex-differentiated traits (e.g. testosterone levels). Importantly, sex-differential participation bias did not impact the genetic correlation between males and females for each phenotype (Supplementary Fig. 5). We caution that, although inclusion of sex as covariate did not seem to impact most traits in these analyses, this issue might lead to significant differences between models as sample sizes continue to grow.
Discussion
Most large-scale biobank studies are not designed to achieve cohorts who are accurately representative of the general population23,24,25,26,27,28. Lack of representation is not per-se problematic if this is considered when interpreting study findings6. Here, we show an example of how sex-differential study participation bias could lead to spurious associations and ultimately incorrect biological inferences. In practice, the impact of differential participation bias on genetic results is hard to tease apart for most traits. We used sex, which provides a robust negative control as it has no autosomal determinants, to identify determinants of study participation bias that differentially impact males and females.
We demonstrate that sex-differential participation bias results in sex showing spurious heritability on the autosomes and being genetically correlated with the complex traits that underlie such bias. This is of importance for studies such as iPSYCH that focus on psychiatric disorders and traits strongly associated with gender such as, e.g., autism, ADHD, and depression, but the implications generalize to many other risk factors and phenotypes. For example, alleles genome-wide significantly associated with higher BMI are under-represented in females compared to males in both UK Biobank and 23andMe. This suggests that females with higher genetic susceptibility to obesity are less likely to participate in studies than their male equivalents (or that genetically lean males are more likely to), although the mechanism by which genetically determined BMI influences non-participation is unclear. These sex-differential biases may also have directionally opposite effects between studies—alleles associated with higher educational attainment were under-represented in 23andMe females but over-represented in UK Biobank females. While these results reflect differences in participation between men and women, we do not yet understand the mechanisms by which differences in BMI or education lead to differential participation between sexes. This may be due to clinical, social or cultural factors that lead to changes in the perception or expectations of individuals when deciding to engage in research studies. Our results are consistent with the larger effect—and larger bias—observed for the association between sex and cardiovascular mortality when UK Biobank is compared to a representative health survey29. We conclude that sex-differential participation can induce false sex-differential associations (or obscure true associations) and complicate the study of health disparities between males and females.
While study design and participant recruitment strategy are the most likely factors influencing participation bias, we showed that both novel and existing methods can be applied to reduce the impact of such bias. Inverse-probability-of-sampling-weighted (IPW) regression has been applied to achieve unbiased estimates from analyses of case-control data30,31. Dudbridge, Mahmoud and colleagues32,33 proposed a correction for selection that occurs when performing case-only analyses. However, the same technique can correct for selection that is conditioned on any trait as long as GWAS can be performed on it. We propose two additional conceptual frameworks and show how they can be implemented in genomicSEM34. First, we developed an application of Heckman correction for genetic data. Heckman correction35 is commonly used in econometrics to correct for the association between an exposure X and outcome Y when the outcome is observed only in study participants and thus is subject to participation bias. The intuition behind Heckman correction is that the predicted probability of study participation (S) can be used to adjust the association between Y and X.
Second, we propose a novel method that is based on the following intuition: the magnitude of participation bias introduced between X and Y under selection is proportional to their effects on the probability of study participation (S). By specifying a model where the bias and the effect that introduces the bias are forced through a single path, the correct genetic correlation between Y and X can be retrieved from the GWAS of Y and X in the selected samples and S. This method, unlike Heckman correction, does not require the predicted probability of study participation, but instead a GWAS of participating individuals vs. the population is sufficient. Details of both of these methods are provided in the Supplementary Note.
While we validated the two approaches via simulations (Supplementary Table 11), future work is needed to apply these methods to examples in real data. The biggest challenge to the implementation of both approaches to bias correction is that they require unbiased estimates of allele frequencies in the target population. The generation of such information, for example by establishing a “census of human genetic variation”, should be the primary focus of future activities in this area. Some extremely large genomic databases exist, such as gnomAD36. However, these are unlikely to be representative due to inclusion of data from studies with a wide range of designs and settings. Where legislation allows, designs such as used by iPSYCH could be implemented5. The iPSYCH study has already shown the value of generating accurate population-based estimates of rare copy number variants37. Future studies could valuably inform population allele frequencies using neonatal dried blood spots in a manner that protects anonymity, while significantly strengthening the inferences derived from other larger non-representative studies. Such an approach would be necessary to implement the bias correction frameworks proposed above.
In summary, we demonstrate that genetic analyses can uniquely profile the complex traits and behaviors that contribute to participation bias in epidemiological studies. We hope that future studies will build on these findings to create resources and tools that more systematically identify and correct for broader forms of participation bias and their effects on genetic association results.
Methods
Contributing cohorts GWAS
Genome wide association was conducted in five different cohorts (23andMe, UK Biobank, iPSYCH, FinnGen and Biobank Japan) for a total of 3,309,398 samples (1,747,070 female and 1,562,328 male). Detailed cohort description, recruitment and genotyping information can be found in the Supplementary Note. For all GWAS analyses, females were coded as 1 and males as 0.
Identification of independent loci and additional QC of results from 23andMe
To evaluate whether our sex-associated genome-wide significant signals were attributable to technical artefacts, we embarked in additional quality controls. First, we used the FUMA v1.3.5d pipeline42 to identify independent loci. In particular, we used pre-calculated LD (linkage disequilibrium) structure based on the European 1000 Genome panel to identify genome-wide significant SNPs independent from each other at r 2 < 0.6. If LD blocks of independent significant SNPs are located close to each other (< 250 kb based on the most right and left SNPs from each LD block), they are merged into one genomic locus. FUMA also identifies independent lead SNPs within a locus if they are independent of each other at r 2 < 0.1. Each genomic locus can thus contain multiple independent significant SNPs and lead SNPs. This approach resulted in 158 loci, which are reported in Supplementary Table 1.
For each locus, we identified one directly genotyped SNP with P < 5 x 10-8. This resulted in 78 SNPs since not all loci had a genome-wide significant directly genotyped SNP. We extracted 50 bp upstream and downstream of each SNP using h19 reference genome and the R function getSeq from the package BSgenome 1.58.0. We chose 50 bp as this is the probe length on the Illumina Global Screening array. We used BLAT v.407 (https://genome.ucsc.edu/cgi-bin/hgBlat?command=start) to search each extracted sequence vs. the human genome. We considered only matches on chromosome X and Y with 95% or greater similarity. We also considered stricter quality control metrics: Hardy-Weinberg disequilibrium P > 1 x 10-6, MAF > 5%, and call rate > 98%.
All downstream analyses looking at the aggregate effect of variants across the genome were done using all the variants that passed cohort-specific quality controls without considering the strict quality controls thresholds described above.
Pleiotropy analysis
To test the relevance of our sex-associated signals with other traits, we used the results from the analysis of Watanabe et al.20, which considered GWAS results from 4,155 publicly available GWASs. For each locus, we counted the number of associated traits and categorized as 0, 1, 2, 3, 4, or 5+. These results can be obtained by combining results from Supplementary Table 4 of Watanabe et al. together with all the SNPs tested for pleiotropy, which are available here: https://github.com/dsgelab/genobias. We then used a chi-square test to compare the count distribution for the number SNPs that were genome-wide significantly associated with sex vs. all SNPs considered by Watanabe et al.
Extracting results from the GWAS catalogue
We considered the most significant SNP for each of the 158 genome-wide significant loci and extracted all the SNPs in LD (r 2 > 0.2 and distance < ±500 Mb). To extract these SNPs, we used the R implementation of LDproxy (https://ldlink.nci.nih.gov/?tab=ldproxy) and used an LD reference panel from 1000 Genomes Europeans. To identify traits significantly associated with these proxy SNPs, we interrogated the GWAS catalog43 using the R package gwascat v. 2.22.0. The GWAS catalogue was extracted on 2 December 2019. We only considered reported association with P < 5 x 10-8 and extracted the EFO terms.
Comparison of full GWAS sample vs. individuals < 30 years old in 23andMe
To identify loci significantly associated with sex in individuals younger than 30 years old at recruitment, we used the same pipeline described above (“Identification of independent loci and additional QC of results from 23andMe”). To assess the difference in effect sizes between the two analyses, we used the following test:
where where Nall is the sample size and where N<30 is the full sample size for the people younger than 30. z-scores zall and z <30 are obtained from the corresponding GWAS results, and cti is the intercept from the LD-score genetic correlation between the two analyses. We can obtain z-scores for the difference between the two analyses reweighted by the corresponding sample size to allow for differences in sample sizes between the two analyses. The test is analogous to the test for a sum of z statistics forms dependent GWAS as presented in Baselmans et al.44 and Jansen et al.45, and similar to the method used by Nolte et al.46.
In order to test whether sample overlap would affect our results, we derived the expected z-scores for the GWAS run without the samples with age < 30. This was estimated as:
where z>30 is the expected z-score in people older than 30, and w>30 = . Differences tested between the >30 and <30 datasets showed no difference with the ones observed in the overall dataset.
Heritability estimation of sex
We used LD-score regression47 to estimate the proportion of variance in liability to sex at birth that could be explained by the aggregated effect of the SNPs. The method is based on the idea that an estimated SNP effect includes the effects of all SNPs in LD with that SNP. On average, a SNP that tags many other SNPs will have a higher probability of tagging a causal variant than a SNP that tags few other SNPs. Accordingly, for polygenic traits, SNPs with a higher LD-score have on average stronger effect sizes than SNPs with lower LD-scores. When regressing the effect size obtained from the GWAS against the LD-score for each SNP, the slope of the regression line gives an estimate of the proportion of variance accounted for by all analyzed SNPs. We included 1,217,312 SNPs (those available in the HapMap3 reference panel). We used stratified LD-score regression, including LD and frequency annotation, similar to what is used by Gazal et al.48 since this has been shown to reduce bias in heritability estimation49,50.
Since sex is a dichotomous trait whose frequency changes across studies, we have transformed the observed heritability into liability scale using the following formula51:
where K is the prevalence of sex in the population (50%), P is the proportion of females in the study and z is the height of the normal curve corresponding to the prevalence of sex in the population.
For estimation of heritability in Japan Biobank, we used a LD-score reference panel based on East Asian participants in 1000 Genomes.
Genetic correlations
We used cross-trait LD-score regression to estimate the genetic covariation between traits using GWAS summary statistics28. The genetic covariance is estimated using the slope from the regression of the product of z-scores from two GWAS studies on the LD-score. The estimate obtained from this method represents the genetic correlation between the two traits based on all polygenic effects captured by SNPs. Standard LD-scores were used as provided by Bulik-Sullivan et al.52 based on the 1000 Genomes reference set, restricted to European populations.
The decision of which summary statistics to include in the genetic correlation analysis was taken before analyzing the data by consensus across the authors of the paper.
Mendelian randomization analysis and genomicSEM regression for BMI and sex
We tested for possible causal effects of BMI on sex, induced by sex-differential participation bias, in both 23andMe and UK Biobank through Mendelian randomization. As instruments for the exposure, we used the 97 index SNPs associated with BMI reported by Locke and colleagues53. We tested different methods (MR Egger, Weighted median, Inverse variance weighted, Simple mode, Weighted mode) as implemented in the R package TwoSampleMR54.
We then further investigate whether the discordance in genetic correlations between BMI and sex in UK Biobank (rg = -0.13, P = 2 x 10-4) and 23andMe (rg = 0.10, P = 9 x 10-8) is due to a confounding effect of educational attainment. By using the respective GWAS summary statistics, we fitted the following multiple regression model in genomicSEM34 to estimate the genetic correlation between BMI and sex controlling for educational attainment:
Results for both analyses are reported in Supplementary Table 7.
Generation of genetic scores for educational attainment
We used summary statistics for a GWAS of years of education21, which did not include UK Biobank and 23andMe, to construct the polygenic score. This score was generated using PRSice v.2.0 55. Briefly, PRSice performs a pruning (distance = 250 kb and r 2 = 0.1) and thresholding approach. We then selected the P-value threshold that maximizes the r 2 between the score and educational attainment in UK Biobank (P=0.195, n SNPs = 39,014). The polygenic score was only constructed for a subset of the UK Biobank containing white-British unrelated individuals (n = 361,501) as described here: https://github.com/Nealelab/UK_Biobank_GWAS
We constructed the polygenic score on the dataset including both males and females and then we compared whether the average polygenic score differed between males and females using a t-test. Next, we compared the average years of education in the same dataset. We recorded the education level variable in UK Biobank (“6138”) into years of education following the approach used by the SSGAC consortium: 1 = 20 years; 2 = 15 years; 3 = 13 years; 4 = 12 years; 5 = 19 years; 6 = 17 years; -7 = 6 years; -3 = missing. We then tested for significant differences in education between males and females using a t-test.
Census data analysis
We obtained information about educational attainment from the UK Census from the year 2011. Data were extracted from the Office for National Statistics: https://www.nomisweb.co.uk/census/2011. We coded the qualification level collected in the census to match the corresponding levels in UK Biobank:
Census
No qualifications => 1
Level 1 qualifications => 2
Level 2 qualifications => 3
Apprenticeship => 4
Level 3 qualifications => 5
Level 4 qualifications and above: 6
Other qualifications => NA
UK Biobank
1: College or University degree => 6
2: A levels/AS levels or equivalent => 5
3: O levels/GCSEs or equivalent => 2.5
4: CSEs or equivalent => 2.5
5: NVQ or HND or HNC or equivalent => 6
6: Other professional qualifications eg: nursing, teaching => 6
-7: None of the above => 1
-3: Prefer not to answer => NA
Information from the 2011 census was grouped by three age bins (35-49, 50-64, 65+), sex and Middle Layer Super Output Area (MSOA) regions from England and Wales. In total, 6,050 MSOA regions with at least one UK Biobank participant were included. To map each individual to an MSOA region, we used the home location coordinates (variables 22702 and 22704) with the moving date that was closest to 2011. We then used the sp v.1.4-5 R package (over function) to map the coordinates to the MSOA region coordinates obtained from https://census.mimas.ac.uk/dataset/2011-census-geography-boundaries-middle-layer-super-output-areas-and-intermediate-zones-7. To estimate the average education level, separately in men and women in UK Biobank and in the census, we use the svydesign function from the survey v. 4.0 R package. This function implements different types of sampling designs and, in this analysis, we used a stratified sampling design with three strata: age, sex, and MSOA region.
Extended Data
Extended Data Fig. 1. Different participation bias scenarios that may lead to a correlation between sex and genetic variants.
S, selection (that is participation in the study); X, trait; Gx, genotype causing X. The assumed causal paths are shown in blue, and the induced correlations are shown in red. Three scenarios exist in which sex can become heritable due to selection. a, Sex causes X which in turn causes selection. b, X and sex influence the selection independently. c, The effect of X on selection is different between the two sexes. This is the scenario discussed in the paper. We have run simulations (Supplementary Fig. 3) and scenarios a and b are less likely to be observed because the effect of the trait on selection would need to be extremely large.
Extended Data Fig. 2. Effect size for association between SNPs and sex in 23andMe.
On the y-axis is the effect in the entire study population (n = 2,462,132), and on the x-axis is the effect only among those younger than 30 years (n = 320,366). Error bars represent the confidence intervals for the effect size estimates.
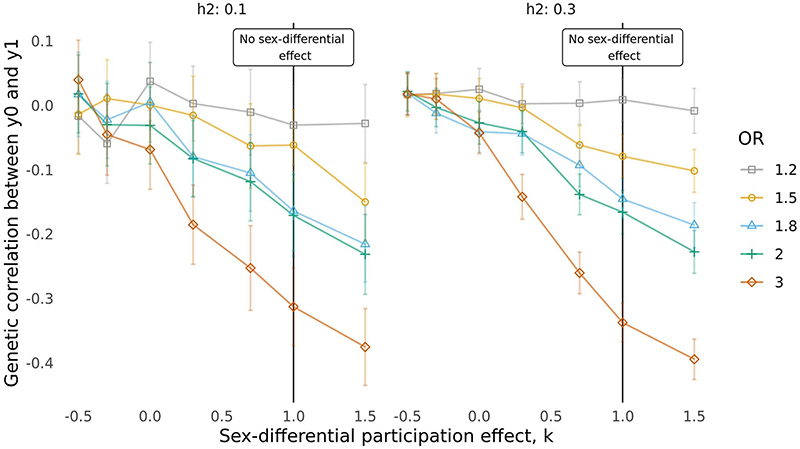
Extended Data Fig. 3. Effect of sex-differential participation bias on the genetic correlation between y0 and y1 when the phenotypes have h2 = 0.1 or h2 = 0.3.
Each line represents a different degree of participation bias, expressed as the odds ratio (OR) used for the sampling. The higher the OR, the higher the degree of participation bias. The x-axis represents different values for the parameter k that gives the sex-differential effect. The smaller k is, the higher is the degree of the sex-differential effect. Under no partecipation bias or sex-differential effect y0 and y1 have a genetic correlation equal to 0.
Extended Data Fig. 4. Effects of sex differential bias on the BMI→T2D relationship.
The forest plot shows the effect of sampling men and women differentially based on BMI. The x-axis represents different values of bias introduced. For higher values, heavier males and leaner women are randomly picked. The number on top of the segment represents the P-value of the difference in effect between the two sexes using the z-score method. The bias becomes large enough to be detected as ‘significant’ even at the lower values of bias applied. The straight lines represent the effect of BMI on T2D estimated without any sample selection.
Supplementary Material
Acknowledgements
We acknowledge George Davey Smith for insightful comments. This research was conducted by using the UK Biobank Resource under application 31063. A.G. was supported by the Academy of Finland Fellowship (323116). This work was supported by the Medical Research Council (Unit Programme number MC_UU_12015/2). M.G.N. is a fellow of the Jacobs Foundation and is supported by ZonMW grants 849200011 and 531003014 from The Netherlands Organisation for Health Research and Development and a VENI grant awarded by NWO (VI.Veni.191G.030). A. Abdellaoui is supported by the Foundation Volksbond Rotterdam and the ZonMw grant 849200011 from The Netherlands Organisation for Health Research and Development. The FinnGen project is funded by two grants from Business Finland (HUS 4685/31/2016 and UH 4386/31/2016) and 11 industry partners (AbbVie Inc, AstraZeneca UK Ltd, Biogen MA Inc, Celgene Corporation, Celgene International II Sàrl, Genentech Inc, Merck Sharp & Dohme Corp, Pfizer Inc., GlaxoSmithKline, Sanofi, Maze Therapeutics Inc., Janssen Biotech Inc). We acknowledge the following biobanks for collecting the FinnGen project samples: Auria Biobank (www.auria.fi/biopankki), THL Biobank (www.thl.fi/biobank), Helsinki Biobank (www.helsinginbiopankki.fi), Biobank Borealis of Northern Finland (https://www.ppshp.fi/Tutkimus-ja-opetus/Biopankki/Pages/Biobank-Borealis-briefly-in-English.aspx), Finnish Clinical Biobank Tampere (www.tays.fi/en-US/Research_and_development/Finnish_Clinical_Biobank_Tampere), Biobank of Eastern Finland (www.ita-suomenbiopankki.fi/en), Central Finland Biobank (www.ksshp.fi/fi-FI/Potilaalle/Biopankki), Finnish Red Cross Blood Service Biobank (www.veripalvelu.fi/verenluovutus/biopankkitoiminta) and Terveystalo Biobank (www.terveystalo.com/fi/Yritystietoa/Terveystalo-Biopankki/Biopankki/). All Finnish Biobanks are members of BBMRI.fi infrastructure (www.bbmri.fi). We thank the research participants and employees of 23andMe who contributed to this study.
Footnotes
Author contributions
Study design: N.P., M.C., P.N., C.C., M.D.V.d.Z., A. Abdellaoui, D.H., B.M.N., R.K.W., M.G.N., J.R.B.P., A.G. Data analysis: N.P., M.C., P.N., G.M., A. Abdellaoui, B.H., M.K., V.M.R., P.D.B.P., N.B, J.K., T.D.A., M.D.V.d.Z., R.B., A.D.B., A. Auton, D.H., M.G.N., J.R.B.P., A.G. Results interpretation: N.P., M.C., A. Abdellaoui, C.C., F.R.D., K.K.O., R.B., P.J., B.M.N., R.K.W., M.G.N., J.R.B.P., A.G. Provided data: P.N., A. Abdellaoui, V.M.R., T.D.A., T.M., E.d.G., Y.O., A.D.B., A. Auton, D.H., M.B.N., M.G.N., J.R.B.P., A.G. Wrote the manuscript: N.P., M.C., B.M.N., M.G.N., J.R.B.P., A.G.
Competing interests statement
P.N., A. Auton, and D.H. are employed at 23 and Me Inc. P.J. is a paid consultant to Global Gene Corp and Humanity Inc.
Contributor Information
FinnGen Study:
Mattia Cordioli, Pietro Della Briotta Parolo, Juha Karjalainen, and Andrea Ganna
23andMe Research Team:
Michelle Agee, Stella Aslibekyan, Robert K. Bell, Katarzyna Bryc, Sarah K. Clark, Sarah L. Elson, Kipper Fletez-Brant, Pierre Fontanillas, Nicholas A. Furlotte, Pooja M. Gandhi, Karl Heilbron, Barry Hicks, Karen E. Huber, Ethan M. Jewett, Yunxuan Jiang, Aaron Kleinman, Keng-Han Lin, Nadia K. Litterman, Marie K. Luff, Matthew H. McIntyre, Kimberly F. McManus, Joanna L. Mountain, Sahar V. Mozaffari, Elizabeth S. Noblin, Carrie A. M. Northover, Jared O’Connell, Aaron A. Petrakovitz, Steven J. Pitts, G. David Poznik, J. Fah Sathirapongsasuti, Janie F. Shelton, Suyash Shringarpure, Chao Tian, Joyce Y. Tung, Robert J. Tunney, Vladimir Vacic, Xin Wang, and Amir Zare
iPYSCH Consortium:
Preben Bo Mortensen, Ole Mors, Thomas Werge, Merete Nordentoft, David M. Hougaard, Jonas Bybjerg-Grauholm, and Marie Bækvad-Hansen
Data availability
GWAS results are available through GWAS catalogue accession numbers GCST90013473 (23andMe) and GCST90013474. Full summary statistics for 23andMe are available upon request from https://research.23andme.com/dataset-access/.
Code Availability
Scripts are available at: https://github.com/dsgelab/genobias
References
- 1.Prictor M, Teare HJA, Kaye J. Equitable participation in biobanks: the risks and benefits of a “dynamic consent” approach. Front Public Health. 2018;6:253. doi: 10.3389/fpubh.2018.00253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leitsalu L, et al. Cohort Profile: Estonian Biobank of the Estonian Genome Center, University of Tartu. Int J Epidemiol. 2015;44:1137–1147. doi: 10.1093/ije/dyt268. [DOI] [PubMed] [Google Scholar]
- 3.Klijs B, et al. Representativeness of the LifeLines Cohort Study. PLoS One. 2015;10:e0137203. doi: 10.1371/journal.pone.0137203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fry A, et al. Comparison of sociodemographic and health-related characteristics of UK Biobank participants with those of the general population. Am J Epidemiol. 2017;186:1026–1034. doi: 10.1093/aje/kwx246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pedersen CB, et al. The iPSYCH2012 case-cohort sample: new directions for unravelling genetic and environmental architectures of severe mental disorders. Mol Psychiatry. 2018;23:6–14. doi: 10.1038/mp.2017.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rothman KJ, Gallacher JEJ, Hatch EE. Why representativeness should be avoided. Int J Epidemiol. 2013;42:1012–1014. doi: 10.1093/ije/dys223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Keyes KM, Westreich D. UK Biobank, big data, and the consequences of non-representativeness. Lancet. 2019;393:1297. doi: 10.1016/S0140-6736(18)33067-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Swanson JM. The UK Biobank and selection bias. Lancet. 2012;380:110. doi: 10.1016/S0140-6736(12)61179-9. [DOI] [PubMed] [Google Scholar]
- 9.Elwood JM. Commentary: On representativeness. Int J Epidemiol. 2013;42:1014–1015. doi: 10.1093/ije/dyt101. [DOI] [PubMed] [Google Scholar]
- 10.Pizzi C, et al. Sample selection and validity of exposure-disease association estimates in cohort studies. J Epidemiol Community Health. 2011;65:407–411. doi: 10.1136/jech.2009.107185. [DOI] [PubMed] [Google Scholar]
- 11.Richiardi L, Pizzi C, Pearce N. Commentary: Representativeness is usually not necessary and often should be avoided. Int J Epidemiol. 2013;42:1018–1022. doi: 10.1093/ije/dyt103. [DOI] [PubMed] [Google Scholar]
- 12.Perry JRB, et al. Stratifying type 2 diabetes cases by BMI identifies genetic risk variants in LAMA1 and enrichment for risk variants in lean compared to obese cases. PLoS Genet. 2012;8:e1002741. doi: 10.1371/journal.pgen.1002741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martin J, et al. Association of genetic risk for schizophrenia with nonparticipation over time in a population-based cohort study. Am J Epidemiol. 2016;183:1149–1158. doi: 10.1093/aje/kww009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taylor AE, et al. Exploring the association of genetic factors with participation in the Avon Longitudinal Study of Parents and Children. Int J Epidemiol. 2018;47:1207–1216. doi: 10.1093/ije/dyy060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Adams MJ, et al. Factors associated with sharing e-mail information and mental health survey participation in large population cohorts. Int J Epidemiol. 2020;49:410–421. doi: 10.1093/ije/dyz134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tyrrell J, et al. Genetic predictors of participation in optional components of UK Biobank. Nat Commun. 2021;12:886. doi: 10.1038/s41467-021-21073-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Munafò MR, Tilling K, Taylor AE, Evans DM, Davey Smith G. Collider scope: when selection bias can substantially influence observed associations. Int J Epidemiol. 2018;47:226–235. doi: 10.1093/ije/dyx206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boraska V, et al. Genome-wide meta-analysis of common variant differences between men and women. Hum Mol Genet. 2012;21:4805–4815. doi: 10.1093/hmg/dds304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ryu D, Ryu J, Lee C. Genome-wide association study reveals sex-specific selection signals against autosomal nucleotide variants. J Hum Genet. 2016;61:423–426. doi: 10.1038/jhg.2015.169. [DOI] [PubMed] [Google Scholar]
- 20.Watanabe K, et al. A global overview of pleiotropy and genetic architecture in complex traits. Nat Genet. 2019;51:1339–1348. doi: 10.1038/s41588-019-0481-0. [DOI] [PubMed] [Google Scholar]
- 21.Lee JJ, et al. Gene discovery and polygenic prediction from a genome-wide association study of educational attainment in 1.1 million individuals. Nat Genet. 2018;50:1112–1121. doi: 10.1038/s41588-018-0147-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Censin JC, et al. Causal relationships between obesity and the leading causes of death in women and men. PLoS Genet. 2019;15:e1008405. doi: 10.1371/journal.pgen.1008405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sudlow C, et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015;12:e1001779. doi: 10.1371/journal.pmed.1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gaziano JM, et al. Million Veteran Program: A mega-biobank to study genetic influences on health and disease. J Clin Epidemiol. 2016;70:214–223. doi: 10.1016/j.jclinepi.2015.09.016. [DOI] [PubMed] [Google Scholar]
- 25.Chen Z, et al. China Kadoorie Biobank of 0.5 million people: survey methods, baseline characteristics and long-term follow-up. Int J Epidemiol. 2011;40:1652–1666. doi: 10.1093/ije/dyr120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dewey FE, et al. Distribution and clinical impact of functional variants in 50,726 whole-exome sequences from the DiscovEHR study. Science. 2016;354:aaf6814. doi: 10.1126/science.aaf6814. [DOI] [PubMed] [Google Scholar]
- 27.Gottesman O, et al. The Electronic Medical Records and Genomics (eMERGE) Network: past, present, and future. Genet Med. 2013;15:761–771. doi: 10.1038/gim.2013.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.All of Us Research Program Investigators et al. The “All of Us” Research Program. N Engl J Med. 2019;381:668–676. doi: 10.1056/NEJMsr1809937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Batty GD, Gale CR, Kivimäki M, Deary IJ, Bell S. Comparison of risk factor associations in UK Biobank against representative, general population based studies with conventional response rates: prospective cohort study and individual participant meta-analysis. BMJ. 2020;368:m131. doi: 10.1136/bmj.m131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Richardson DB, Rzehak P, Klenk J, Weiland SK. Analyses of case-control data for additional outcomes. Epidemiology. 2007;18:441–445. doi: 10.1097/EDE.0b013e318060d25c. [DOI] [PubMed] [Google Scholar]
- 31.Monsees GM, Tamimi RM, Kraft P. Genome-wide association scans for secondary traits using case-control samples. Genet Epidemiol. 2009;33:717–728. doi: 10.1002/gepi.20424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dudbridge F, et al. Adjustment for index event bias in genome-wide association studies of subsequent events. Nat Commun. 2019;10:1561. doi: 10.1038/s41467-019-09381-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mahmoud O, Dudbridge F, Davey Smith G, Munafo M, Tilling K. Slope-Hunter: A robust method for index-event bias correction in genome-wide association studies of subsequent traits. bioRxiv. 2020:2020.01.31.928077. doi: 10.1101/2020.01.31.928077. [DOI] [Google Scholar]
- 34.Grotzinger AD, et al. Genomic structural equation modelling provides insights into the multivariate genetic architecture of complex traits. Nat Hum Behav. 2019;3:513–525. doi: 10.1038/s41562-019-0566-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heckman JJ. Sample selection bias as a specification error. Econometrica. 1979;47:153. [Google Scholar]
- 36.Karczewski KJ, et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–443. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Olsen L, et al. Prevalence of rearrangements in the 22q11.2 region and population-based risk of neuropsychiatric and developmental disorders in a Danish population: a case-cohort study. Lancet Psychiatry. 2018;5:573–580. doi: 10.1016/S2215-0366(18)30168-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Henn BM, et al. Cryptic distant relatives are common in both isolated and cosmopolitan genetic samples. PLoS One. 2012;7:e34267. doi: 10.1371/journal.pone.0034267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Loh P-R, et al. Efficient Bayesian mixed-model analysis increases association power in large cohorts. Nat Genet. 2015;47:284–290. doi: 10.1038/ng.3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zheng X, et al. A high-performance computing toolset for relatedness and principal component analysis of SNP data. Bioinformatics. 2012;28:3326–3328. doi: 10.1093/bioinformatics/bts606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Manichaikul A, et al. Robust relationship inference in genome-wide association studies. Bioinformatics. 2010;26:2867–2873. doi: 10.1093/bioinformatics/btq559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Watanabe K, Taskesen E, van Bochoven A, Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat Commun. 2017;8:1826. doi: 10.1038/s41467-017-01261-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Buniello A, et al. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 2019;47:D1005–D1012. doi: 10.1093/nar/gky1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baselmans BML, et al. Multivariate genome-wide analyses of the well-being spectrum. Nat Genet. 2019;51:445–451. doi: 10.1038/s41588-018-0320-8. [DOI] [PubMed] [Google Scholar]
- 45.Jansen IE, et al. Genome-wide meta-analysis identifies new loci and functional pathways influencing Alzheimer’s disease risk. Nat Genet. 2019;51:404–413. doi: 10.1038/s41588-018-0311-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nolte IM, et al. Missing heritability: is the gap closing? An analysis of 32 complex traits in the Lifelines Cohort Study. Eur J Hum Genet. 2017;25:877–885. doi: 10.1038/ejhg.2017.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bulik-Sullivan BK, et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet. 2015;47:291–295. doi: 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gazal S, et al. Linkage disequilibrium-dependent architecture of human complex traits shows action of negative selection. Nat Genet. 2017;49:1421–1427. doi: 10.1038/ng.3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gazal S, Marquez-Luna C, Finucane HK, Price AL. Reconciling S-LDSC and LDAK functional enrichment estimates. Nat Genet. 2019;51:1202–1204. doi: 10.1038/s41588-019-0464-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Evans LM, et al. Comparison of methods that use whole genome data to estimate the heritability and genetic architecture of complex traits. Nat Genet. 2018;50:737–745. doi: 10.1038/s41588-018-0108-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee SH, Wray NR, Goddard ME, Visscher PM. Estimating missing heritability for disease from genome-wide association studies. Am J Hum Genet. 2011;88:294–305. doi: 10.1016/j.ajhg.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bulik-Sullivan B, et al. An atlas of genetic correlations across human diseases and traits. Nat Genet. 2015;47:1236–1241. doi: 10.1038/ng.3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Locke AE, et al. Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518:197–206. doi: 10.1038/nature14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hemani G, et al. The MR-Base platform supports systematic causal inference across the human phenome. Elife. 2018;7 doi: 10.7554/eLife.34408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Choi SW, O’Reilly PF. PRSice-2: Polygenic Risk Score software for biobank-scale data. Gigascience. 2019;8 doi: 10.1093/gigascience/giz082. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
GWAS results are available through GWAS catalogue accession numbers GCST90013473 (23andMe) and GCST90013474. Full summary statistics for 23andMe are available upon request from https://research.23andme.com/dataset-access/.
Scripts are available at: https://github.com/dsgelab/genobias