Abstract
The genetic evolutionary features of solid tumour growth are becoming increasingly described but the spatial and physical nature of subclonal growth remains unclear. Here, we utilise 102 macroscopic whole tumour images from clear cell renal cell carcinoma patients, with matched genetic and phenotypic data from 756 biopsies. Utilising a digital image processing pipeline, a renal pathologist marked the boundaries between tumour and normal tissue and extracted positions of boundary line and biopsy regions to X- and Y-coordinates. We then integrated coordinates with genomic data to map exact spatial subclone locations, revealing how genetically distinct subclones grow and evolve spatially. We observed a phenotype of advanced and more aggressive subclonal growth in the tumour centre, characterised by an elevated burden of somatic copy number alterations, higher necrosis, proliferation rate and Fuhrman grade. Moreover, we found that metastasising subclones preferentially originate from the tumour centre. Collectively, these observations suggest a model of accelerated evolution in the tumour interior, with harsh hypoxic environmental conditions leading greater opportunity for driver SCNAs to arise and expand due to selective advantage. Tumour subclone growth is predominantly spatially contiguous in nature. We found only two cases of subclone dispersal, both associated with metastasis. The largest subclones spatially were dominated by driver somatic copy number alterations, suggesting a large selective advantage can be conferred to subclones upon acquisition of these alterations. In conclusion, spatial dynamics is strongly associated with genomic alterations and plays an important role in tumour evolution.
Keywords: tumour evolution, renal cell carcinoma, macroscopic imaging, spatial growth
Introduction
Clear cell renal cell carcinoma (ccRCC) has a well described evolutionary basis, with loss of chromosome 3p and inactivation of VHL representing pathognomonic features of the disease1–4. These founding clonal events are then classically followed by acquisition of widespread intratumour heterogeneity, which is characterised by the accrual of additional subclonal genetic alteration(s) including SETD2, PBRM1 and BAP1 mutation, and/or somatic copy number loss of chromosome arms including 14q and 9p5,6. This defined sequence of events, coupled with a general lack of extreme mutagenesis7 and strong evidence of selection8, renders ccRCC an excellent model for exploring the principles of tumour evolution. While genomic and phylogenetic reconstruction analyses have been extensively applied to study tumour evolution to date9, the spatial nature of tumour evolution at a macroscopic (whole tumour) level of resolution remain grossly underexplored. In particular, the modes of spatial growth (spatially contiguous versus dispersed), the origin of metastasising subclones (tumour interior versus tumour margin), and the relationship between genomic & spatial distance remain uncharacterised in large patient cohorts10. Moreover, in order to achieve a more complete understanding of cancer evolution within its natural ecological context11, spatially and genomic resolved analyses of clinical cohorts are urgently required.
To aid a greater understanding of the genomic and spatial basis of tumour evolution, we implemented a novel image processing pipeline within our multi-centre prospective longitudinal cohort study named TRAcking renal cell Cancer Evolution through Therapy (TRACERx Renal, https://clinicaltrials.gov/ct2/show/NCT03226886). As a feature of the study protocol, macroscopic images were taken at the time of surgery, which were then subject to quality control assessment and boundary annotation by a renal pathologist, digital image processing and integration with dense genomic/clinical data. In total, post quality control spatial imaging data, high-depth next generation sequencing results and Ki67/necrosis/pathological grade scores were captured from 756 tumour biopsies taken from 79 ccRCC primary tumour sections (mean 9.6 biopsies per tumour section). In addition, matched metastatic sequencing data was utilised from 79 biopsies, taken from 32 ccRCC metastatic tumours. Here we report findings from our integrated analysis, aiming to identify the spatial origin of metastasising subclones within the primary tumour, as well as determining the dominant mode of spatial growth (contiguous versus dispersed). In addition we assess the relationship between spatial and genetic distance, identify driver genotypes occupying maximal spatial area, and finally we utilise agent based modelling to explore the selective pressure of the local microenvironment.
Results
Macroscopic image workflow and data integration pipeline
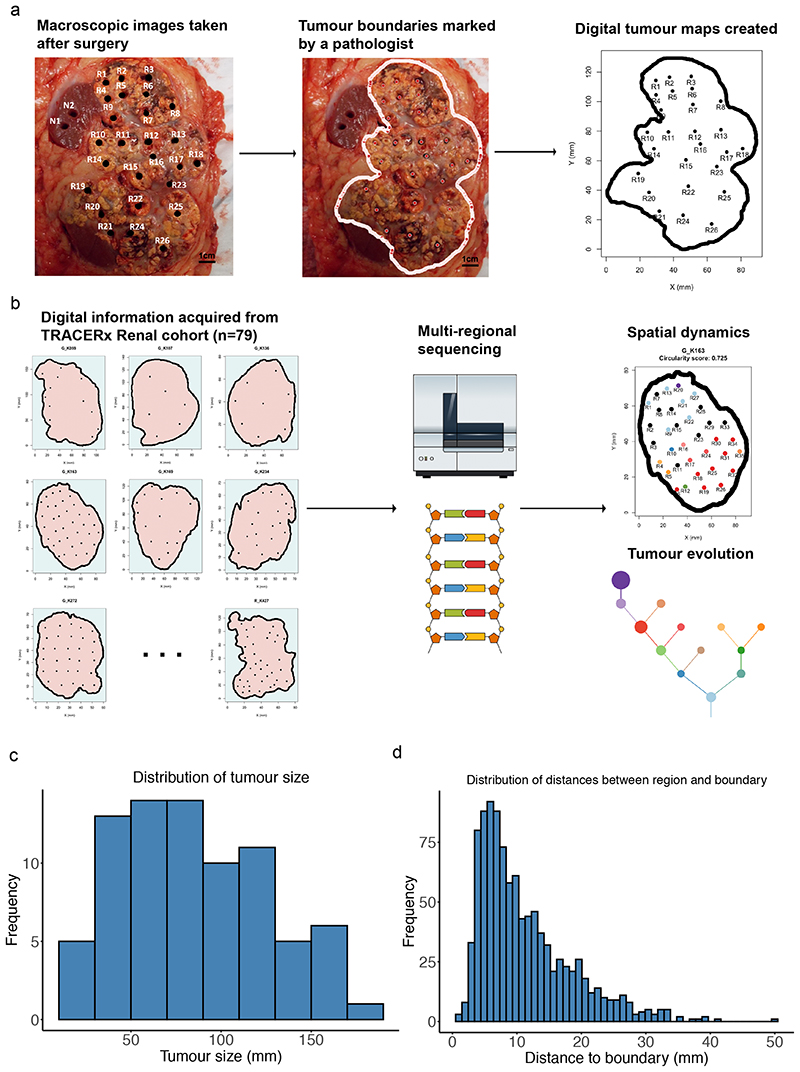
Macroscopic primary tumour photos (n = 102) were taken before and after multi-region biopsy sampling, for n=101 ccRCC patients from the renal TRACERx100 cohort5. Detailed image processing workflow is described in the methods section (Fig. 1a). This process yielded two-dimensional boundary shape and biopsy coordinates for n=79 tumour sections and n=1027 biopsy regions (Fig. 1b, Extended Data Fig. 1). Spatial coordinate data was then integrated with previously published5 high depth multi-region biopsy (n=756) sequencing data (median coverage 613X, driver gene panel, n=110 genes), together with biopsy level Ki67 immunohistochemistry (IHC) staining to measure proliferation, and pathologist (J.I.L.) scored biopsy level tumour necrosis and pathological Fuhrman grade data (Fig. 1b). Across the entire dataset the median tumour size (longest dimension) was 90mm (range 14mm to 180mm) (Fig. 1c), median distance from biopsy to boundary was 9mm (range 1mm to 50mm) (Fig. 1d) and median distance between biopsies was 34mm (range 2mm to 149mm).
Figure 1. Study overview.
A) Macroscopic images taken after surgery were reviewed by a pathologist to determine the physical boundary between tumour and normal tissues, and spatial information was subsequently gathered to generate digital tumour maps. Scale bar shows 1 cm in length on the photograph. B) Spatial data were combined with multi-regional sequencing data to study spatial dynamics and tumour evolution. C) Distribution of tumour size measured in millimetres. D) Distribution of distances between each biopsy region and its nearest tumour boundary on the tumour maps, measured in millimetres.
Somatic copy number alteration burden, necrosis and pathological grade are higher at the tumour centre
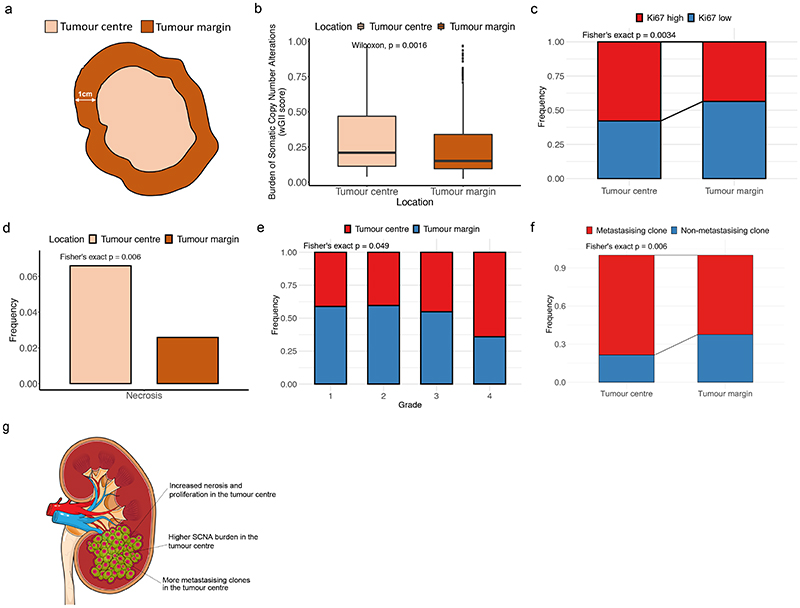
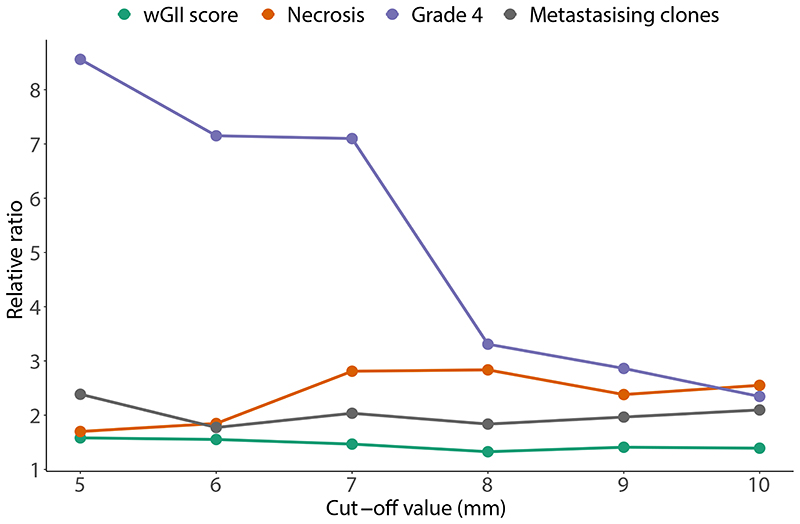
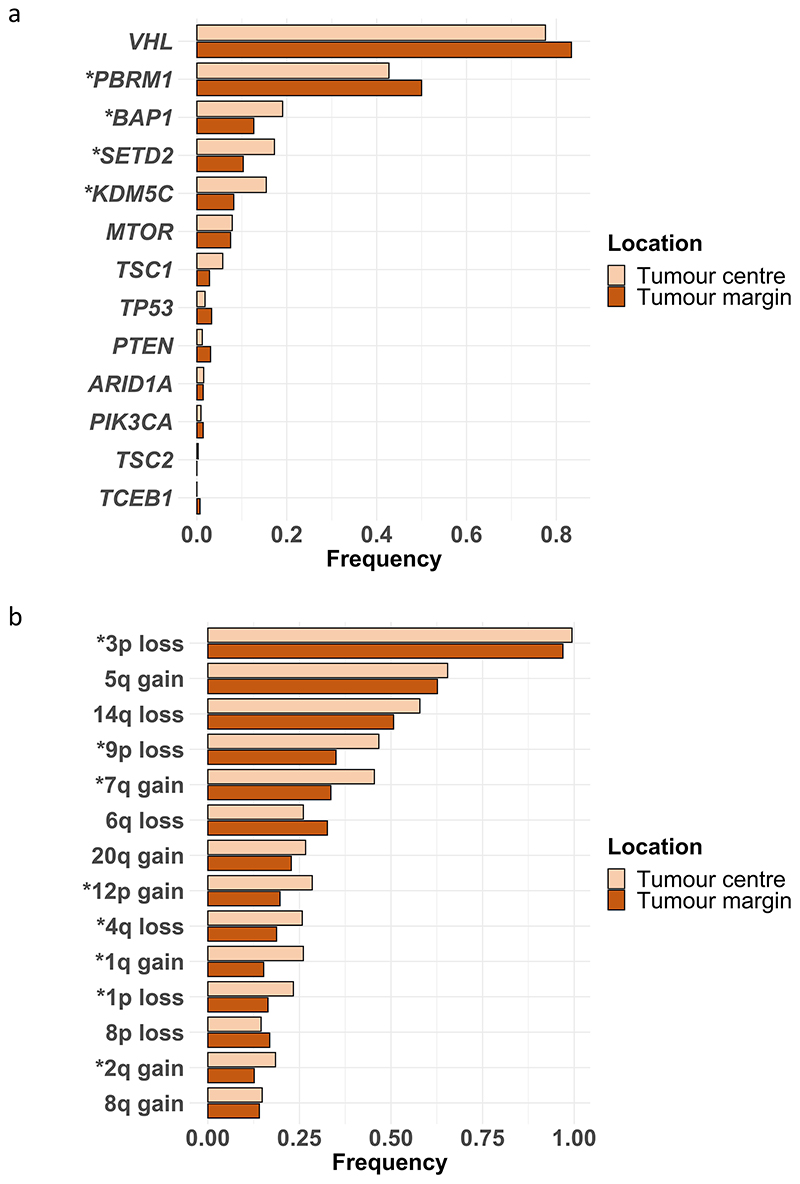
We began the analysis by assessing phenotypic differences between the tumour centre and tumour margin. Biopsies were classified as being at the tumour margin if they were 10mm or less from the nearest boundary (Fig. 2a), or otherwise classified as centrally located. A range of possible boundary threshold definitions produced consistent results, ruling out the possibility of bias due to the threshold used (Extended Data Fig. 2). First, we assessed any differences in the somatic copy number alteration (SCNA) burden measured by the Weighted Genome Integrity Index (wGII) score12 between tumour centre and margin since the prognostic significance of SCNAs has been demonstrated previously5. Surprisingly we found SCNA burden was significantly higher in the tumour centre (0.305 vs 0.251, p=0.0016, Fig. 2b). This difference remained significant after correcting for potential confounding factors including tumour size and purity (Table S2). Next we assessed for a difference in Ki67, as marker of cellular proliferation, and similarly this was found to be significantly higher in the tumour centre (odds ratio=1.77, p=0.0034, Fig. 2c). Necrosis, a feature associated with tumour aggressiveness13, harsh hypoxic environmental conditions13 and poor prognosis14, was additionally found to be significantly higher in the tumour centre (0.066 vs 0.026, p=0.006, Fig. 2d). Finally, the proportion of Furhman grade 4 regions, a clinical predictor of poor prognosis, was found to be higher in the tumour centre (grade 4 frequency 0.641 for tumour centre and 0.359 for tumour margin, Fig. 2e). Additionally, we assessed the frequency of each driver event between tumour centre and tumour margin, and found that the frequency of BAP1, SETD2 and KDM5C mutations, and multiple driver SCNA events were significantly higher in the tumour centre, while the frequency of PBRM1 mutations was significantly higher in the tumour margin (Extended Data Fig. 3).
Figure 2. Comparison between regions in the tumour centre and regions in the tumour margin.
A) Definition of tumour centre and tumour margin. Biopsies less than 1 centimetre to its boundary were defined as marginal regions, while biopsies more than 1 centimetre to its boundary were defined as central regions. B) Comparison of weighted genome integrity index (wGII) which is a measurement of burden of somatic copy number alterations between tumour centre (n=323) and tumour margin (n=425). C) Comparison of Ki67 between tumour centre (n=204) and tumour margin (n=250). D) Comparison of frequency of necrotic regions between tumour centre (n=379) and tumour margin (n=464). E) Comparison of frequencies of regions of different Fuhrman grades in clear cell renal cell carcinoma between tumour centre and tumour margin (n=495). F) Comparison of frequencies of metastasising clones and non-metastasising clones between tumour centre (n=131) and tumour margin (n=128) in a subset of metastatic tumours. G) A cartoon summarizing the differences observed between tumour centre and tumour margin, which might be explained by the differences of blood supply of different parts of the tumour. In all boxplots in this figure the centre line is the median, the bounds of the box represent the inter-quartile range, the lower whisker = max(min(x), Q_1 – 1.5 × IQR) and upper whisker = min(max(x), Q_3 + 1.5 × IQR).
ccRCC metastasising clones are preferentially located in the tumour centre
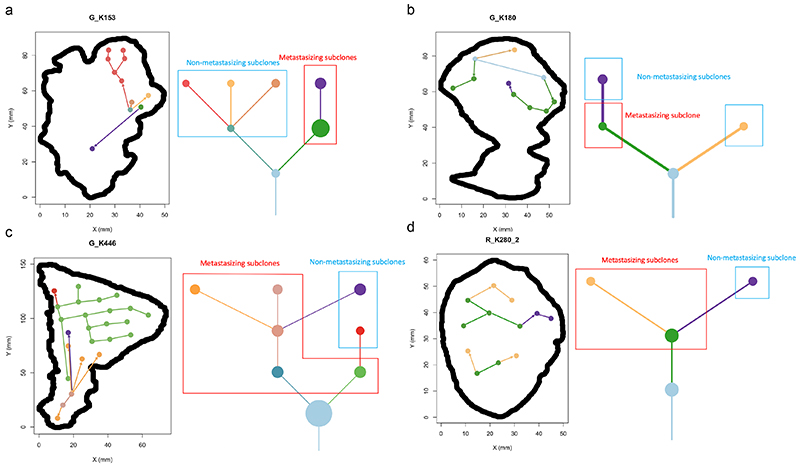
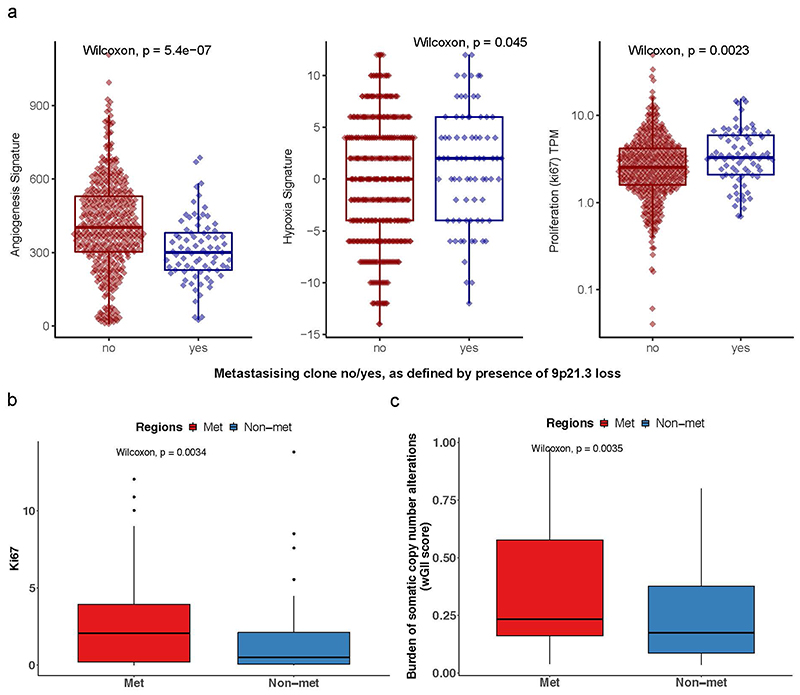
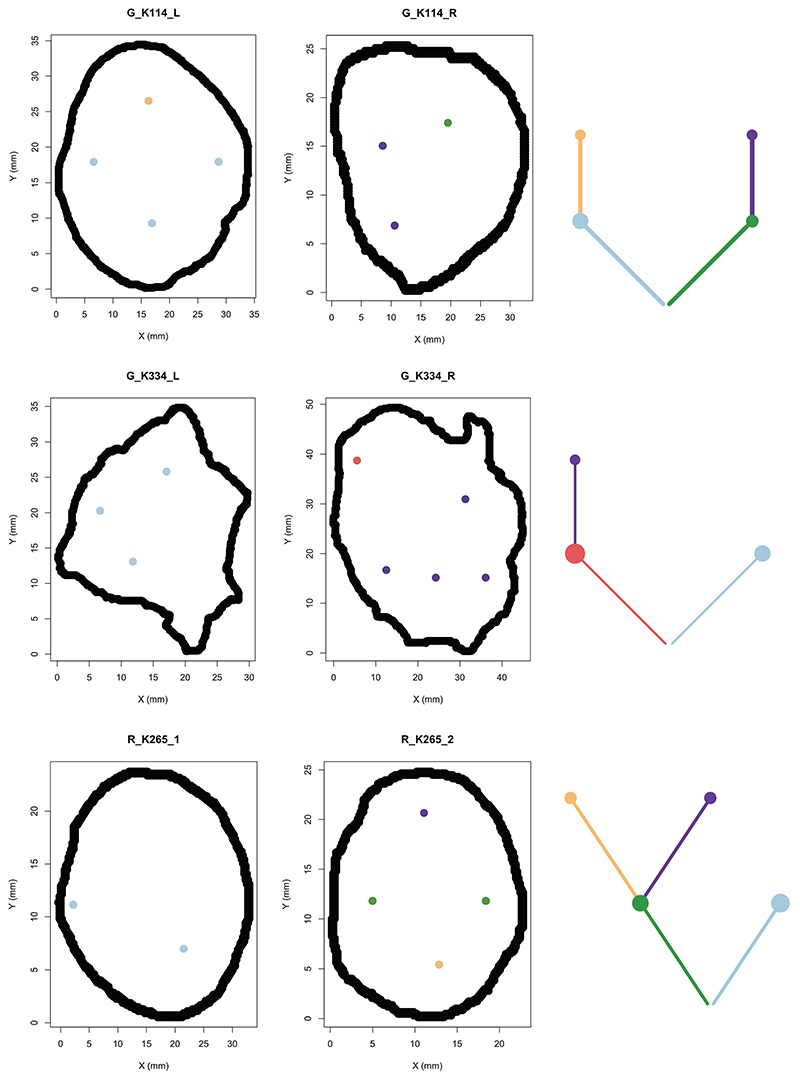
A key benefit of the renal TRACERx100 cohort is matched primary and metastatic biopsy sequencing data, and accordingly we obtained genetic data from an additional n=79 biopsies, taken from n=32 sites of metastatic disease. Using previously published phylogenetic data15, we linked genetically defined tumour subclones present in metastases to their spatial location within each primary tumour. Metastasising subclones were found to be significantly enriched in the tumour centre (odds ratio=2.20, p=0.006, Fig. 2f). We note this a controlled analysis only within metastatic cases, and the enrichment of metastasising subclones within the tumour centre is independent of subclone size as a confounding factor, as the analysis here is based on the proportion of metastasising versus non-metastasising subclones at each location (i.e. if the pattern is simply due to metastasising subclones being larger they would be expected to equally occupy a high proportion in both locations) (Fig. 2f). Collectively these results suggest a phenotype of advanced and accelerated subclonal growth present in the tumour centre, characterised by elevated wGII, Ki67, necrosis, Fuhrman grade and heightened metastatic potential (Fig. 2g). To assess the possibility that metastatic subclones simply had closer access to the vascular system, we measured angiogenesis signature scores using RNA-sequencing data from the cancer genome atlas (TCGA). As TCGA data does not have matched primary and metastatic tissue, we inferred metastasis/non-metastasis associated subclones based on the presence/absence of 9p21.3 loss, which is a genetic event strongly associated with metastasis in multiple studies (hazard ratio = 4.0-8.0)15,16. Interestingly, metastasis associated 9p21.3 loss samples had significantly lower rates of angiogenesis (p=5.4×10-7, median score metastasising/non- metastasising samples 300.0/402.7 respectively), suggesting that closer proximity to blood vessels is not the predominant explaining factor. Instead, metastasis associated 9p21.3 loss samples had significantly higher rates of hypoxia (p=0.045, median score metastasising/non-metastasising samples 2.0/0.0 respectively) and ki67 (p=0.0023, median score metastasising/non-metastasising samples 3.3/2.5 respectively) (Extended Data Fig. 4a). Returning to our TRACERx cohort, representative cases were next mapped in detail, to validate that patterns of metastasis from the tumour centre can clearly be observed. For example, in case K153 (Fig. 3a) the metastasising subclone shown in purple is centrally located, whereas subclones from the opposing branch are located close to the tumour margin but fail to metastasise. Similar patterns are observed in cases K180, K280 and K446 (Fig. 3b-d). Across the four representative cases (Fig. 3a-d) a dynamic range of growth patterns are observed, with later (terminal) phylogenetic clones often found in the tumour centre as well as tumour margin, and overall patterns appear to deviate from a simple model of linear growth from the tumour centre outwards. We note the pattern of metastasis from the tumour centre is an enrichment rather than an absolute rule, and metastasising clones are also capable of disseminating from the tumour periphery, albeit at a lower frequency than the tumour centre (Fig. 2f). Finally, Ki67 and wGII score were also found to be significantly higher in regions containing metastasising clones (2.65 vs 1.52 for Ki67, p=0.0034 and 0.35 vs 0.25 for wGII score, p=0.0035, Extended Data Fig. 4b-c).
Figure 3. Phylogenetic trees and tumour maps showing the possible metastasising routes of 4 example cases on a 2-dimention level.
A) In case K153, metastasising subclones and non-metastasising subclones originate from different branches of the phylogenetic tree. Regions containing metastasising subclones are more enriched in the tumour centre. B) In case K180, the 1st-level metastasising subclone and non-metastasising subclone originate from different branches of the phylogenetic tree, while the 1st-level metastasising subclone also gives rise to the 2nd-level non-metastasising subclone. Regions containing metastasising subclones are more enriched in the tumour centre. C) In case K446, the non-metastasising subclones are all originated from their parent metastasising subclones. Regions containing metastasising subclones are more enriched in the tumour centre and occupy more space. D) In case K280_2, the 1st-level metastasising subclone gives rise to a 2nd-level metastasising subclone and a 2nd-level non-metastasising subclone. Regions containing metastasising subclones are more enriched in the tumour centre and occupy more space.
Modelling associates necrosis with acquisition of SCNAs
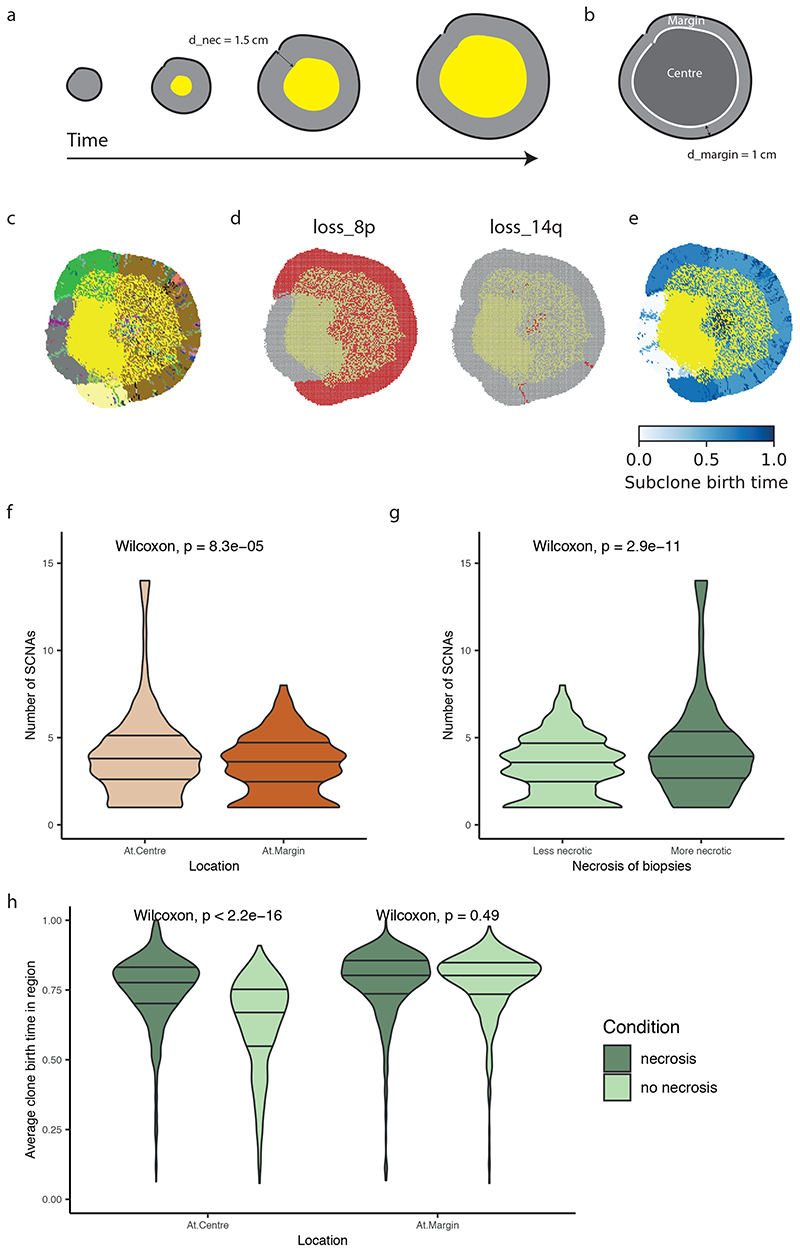
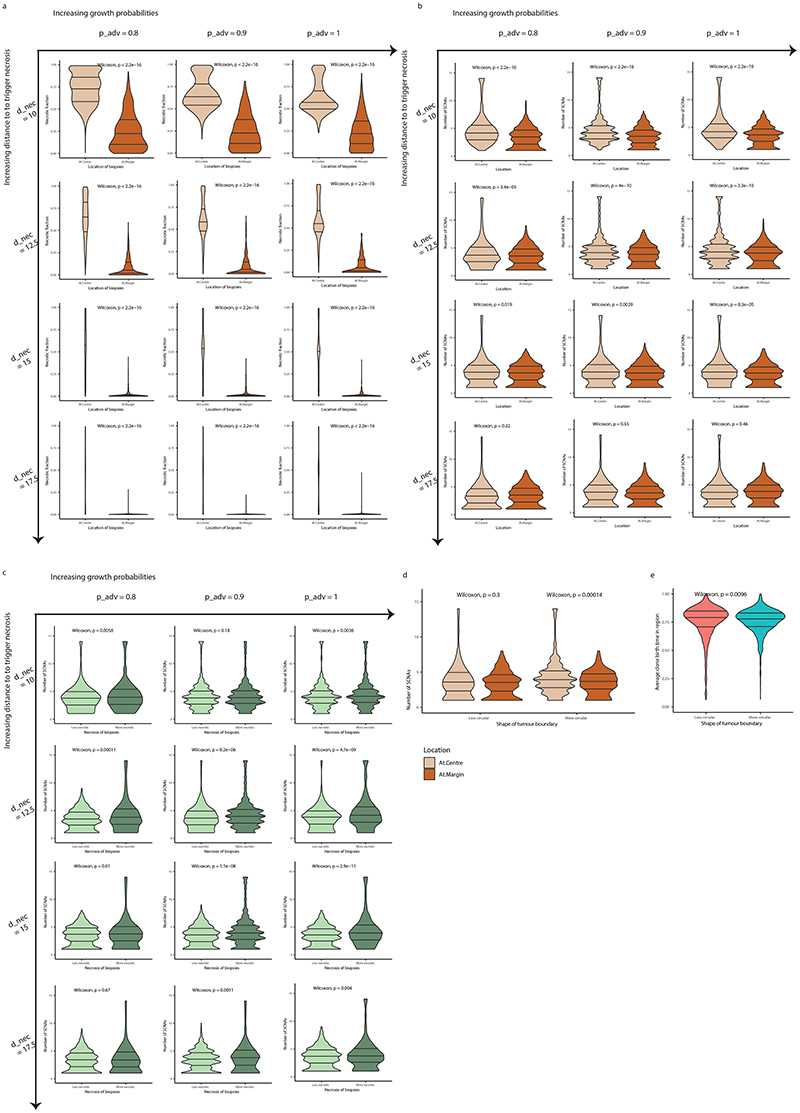
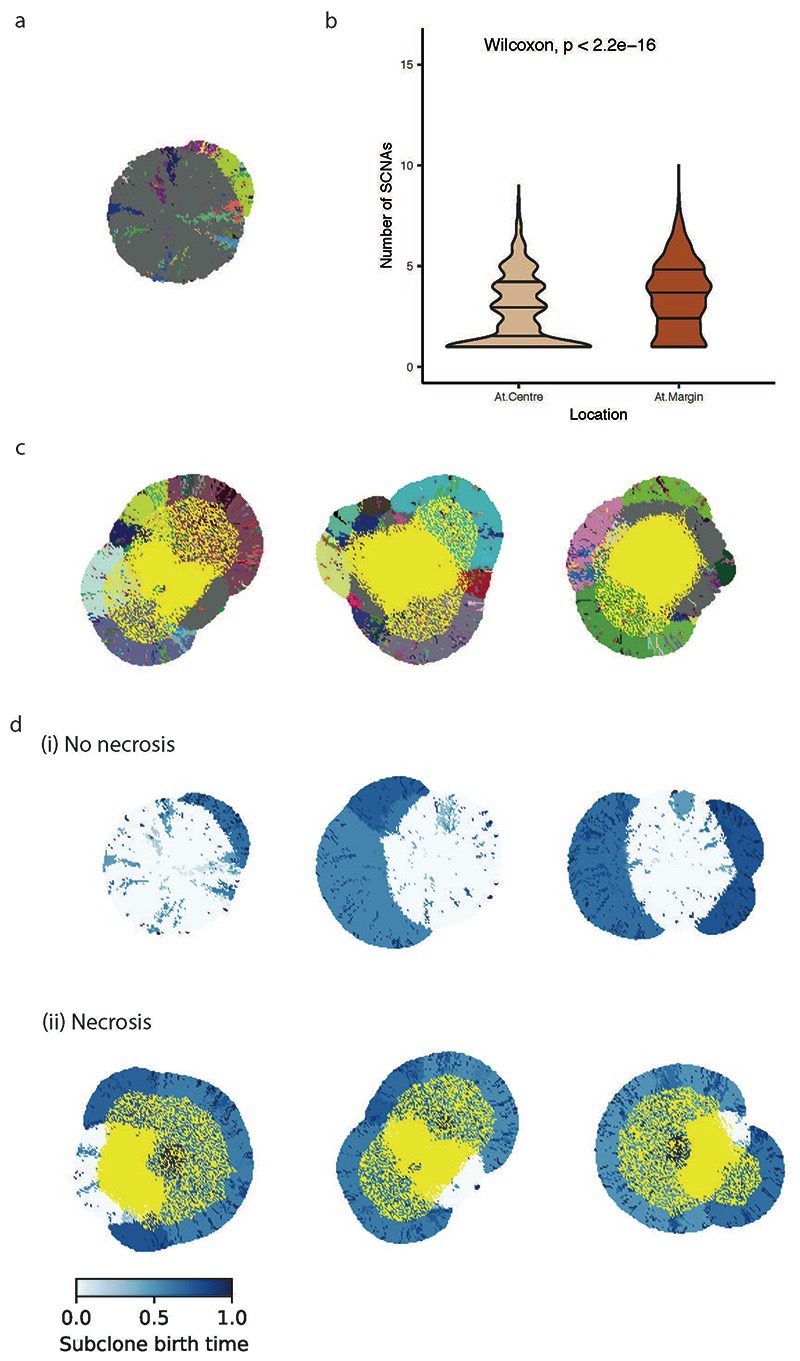
To further explore the relationship between necrosis and tumour location of SCNAs we utilised a coarse-grained cellular automata model to assess if necrosis could promote an elevated SCNA burden (methods). To assess the effect of higher necrosis we simulated the experimentally observed scenario by setting an elevated rate of necrosis at the tumour centre (probability of necrosis for each voxel = 0.5), triggered by a threshold distance from the tumour surface (d_nec=1.5 cm) (Fig. 4a), as compared to the tumour margin (necrosis probability = 0.0). Simulated tumours were then allowed to grow, and an in silico biopsy sampling protocol was implemented at the end to observe the results. Tumour centre and margin were defined using the same criterion as in the experimental analysis (Fig. 4b). Simulated tumours depict continuing clonal diversification and SCNA acquisition (Fig. 4c-e) at the necrotic tumour interior. Next, we assessed for any relationship between necrosis and central SCNA burden, and found that central biopsies had a higher SCNA burden consistent with experimental findings (4.03 for centre vs 3.36 for margin, p<0.0001, Fig. 4f) and that more necrotic biopsies indeed had a higher number of SCNAs (4.33 for more necrotic and 3.33 for less necrotic, p<0.0001, Fig. 4g). Note that the statistical significance holds for a range of model parameters evaluated (Extended Data Fig. 5a-c). To confirm necrosis was required for this result, we conducted additional simulations with necrosis removed and found the opposite result to the observed experimental data (i.e. higher SCNA burden at the margin, Extended Data Fig. 6). Finally, we assessed subclone birth time (Fig. 4e) as a measure of evolutionary age. This showed that presence of necrosis led to a later birth of new subclones at the tumour centre, indicating necrosis facilitates continuing clonal evolution (average clone birth time “necrosis” vs “no necrosis” at the centre: 0.75 vs 0.63, p<0.0001, Fig. 4h). This result was specific to the tumour centre, with no difference in birth time detected at the tumour margin (Fig. 4h). Overall, these observations, in a model-based framework, suggest that heightened necrosis at the tumour centre potentiates continuing clone evolution, and associates with greater opportunity for driver SCNAs to arise and expand due to selective advantage.
Figure 4. Computational modelling supports the preferential localisation of SCNA subclones in more necrotic regions of a tumour.
A) Schematic illustrating the emergence of necrosis in a growing tumour. Tumour areas in yellow experience a high death rate due to necrosis. B) Schematic illustrating the definition of central versus margin areas in a simulated tumour. C) Spatial pattern of subclones in a representative simulated tumour. Tumour areas in yellow are necrotic. Founder clone is in grey while other subclones are in randomly assigned colours. D) Spatial pattern of loss_8p and loss_14q in the representative simulated tumour shown in (C). E) Spatial maps of the birth time of subclones relative to the age of the simulated tumour. Darker blue reflects later birth. F) Number of SCNAs in central (“At.Centre”, n=1280) versus marginal (“At.Margin”, n=568) biopsies. G) Number of SCNAs in less versus more necrotic regional biopsies. “Less necrotic” biopsies (n=931) refer to the regional biopsies with a necrotic fraction less than or equal to median while “More necrotic” biopsies (n=917) refer to those with necrotic fraction greater than median. The 20% most necrotic biopsies are excluded from analysis. H) Average subclone birth time relative to the age of the tumour in central (“At.Centre”) versus marginal (“At.Margin”) biopsies, analysed in simulations without necrosis (“no necrosis”) and those with necrosis (“necrosis”). For the “At.Centre” group, N = 1527 biopsies for “necrosis” simulations and N = 1922 biopsies for “no necrosis” simulations. For the “At.Margin” group, N = 568 biopsies for “necrosis” simulations and N = 572 biopsies for “no necrosis” simulations.
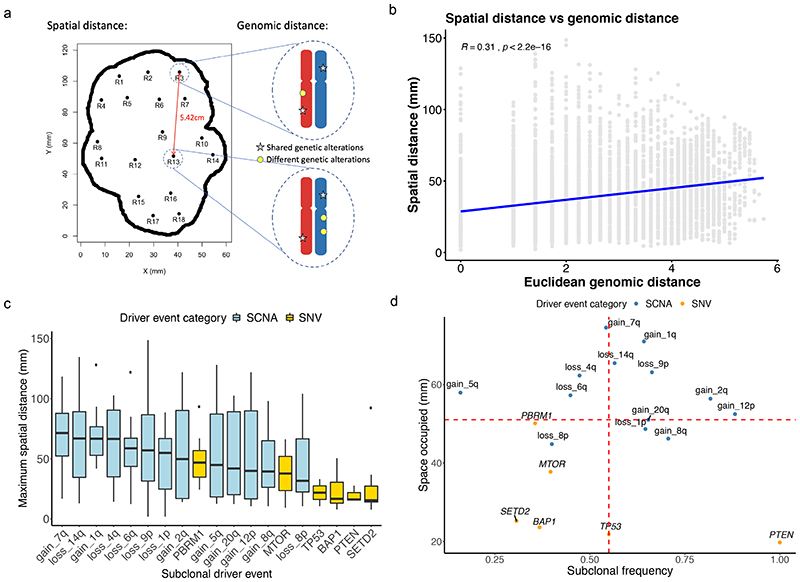
Spatial and genetic distance have a weak to moderate correlation
Next, returning to the experimental data, we compared the relationship between pairwise biopsy spatial distances (in mm) and biopsy genetic distances (Fig. 5a). Across the entire cohort, using all possible pairwise combinations, a weak-to-moderate but detectable correlation was observed (p<2.2×10-16, rho=0.31, Fig. 5b). An interesting question relates to how much spatial area a given subclonal driver genotype occupies, based on the assumption that a larger spatial expansion indicates positive selection due to subclonal expansion. We found driver SCNAs to dominate (Fig. 5c), with nine of the top ten driver events ranked by spatial distance being SCNAs, and these events included SCNAs previously shown to associate with reduced overall survival including 14q and 9p loss15 (Fig. 5c). By contrast the lower ranked events by spatial distance were enriched for mutational events (Fig. 5c), and intriguingly SETD2 mutations were lowest ranked. Spatial area occupied is also a function of event timing, for example a mutation occurring early will naturally propagate to larger size than those occurring late, assuming balanced fitness conditions. Accordingly, we plotted event timing (as measured by subclonal frequency across the cohort, where higher subclonal frequency indicates later occurrence of an event) versus spatial distance, and found driver SCNAs to be enriched in the top right quadrant (Fig. 5d). Thus, in spite of occurring later during tumour evolution, driver SCNA events including 9p and 14q loss still achieved dominant clonal expansion across the tumour surface area, supporting the notion of these events as potent drivers of malignant growth.
Figure 5. Integrated analysis of genomic and spatial distances.
A) A cartoon showing how spatial and genomic distances were measured. Spatial distances between each two biopsy regions were calculated using the spatial data previously retrieved, and genomic distances between each two biopsy regions were calculated based on the genomic alterations they harboured using Euclidean metrics. B) Correlation between spatial distance measured in millimetres and genomic distance across all samples is shown. C) Maximum spatial distance occupied by each subclonal driver event measured in millimetres is shown, ordered from the highest to the lowest. D) Maximum spatial distance occupied in millimetres versus subclonal frequency, showing the relationship between space occupied and timing of the event. Higher subclonal frequency indicates later occurrence of an event. In all boxplots in this figure the centre line is the median, the bounds of the box represent the inter-quartile range, the lower whisker = max(min(x), Q_1 – 1.5 × IQR) and upper whisker = min(max(x), Q_3 + 1.5 × IQR).
Spatially contiguous growth is a dominant feature of ccRCC evolution
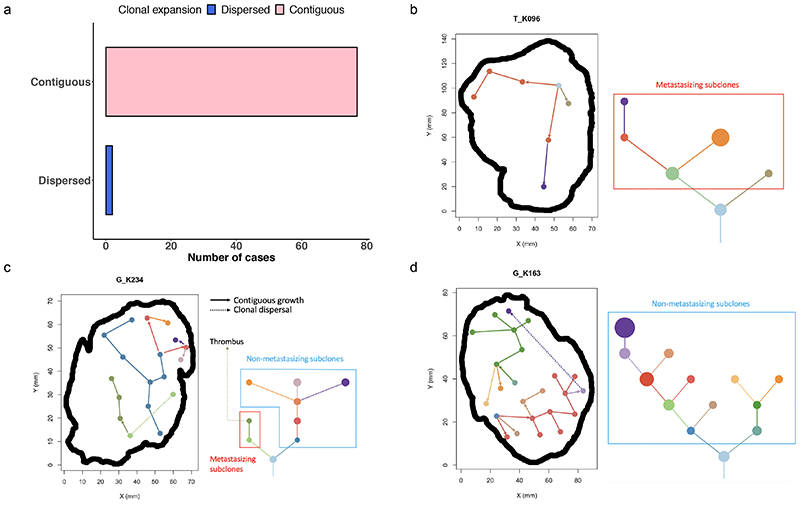
To understand the dynamic modes of spatial growth we examined for evidence of either spatially contiguous (all subclones are located together) or dispersal driven growth (subclones are dispersed and non-contiguous). Growth was found to be predominantly spatially contiguous, with n=77 tumour sections matching this pattern and a minority (n=2) showing evidence of dispersal (Fig. 6a-b). The two tumours showing evidence of clonal dispersal were assessed based on a strict criteria, whereby the dispersed subclone had to be surrounded by an uninterrupted block of genetically distinct subclone(s), mapping to a different branch in the phylogenetic tree. The subclonal differences had to be supported by distinct genetic mutation(s), rather than copy number alterations, which are at a greater risk of violating the infinite sites model17. Thus, we assume dispersal only when a subclone cannot visibly have reached a given spatial location without dispersing across a genetically different subclone. We acknowledge however our study is only powered to assess growth in the two-dimensional plane, and we cannot discount an abnormal growth pattern where dispersed clones are connected in three-dimensional space. Accepting this limitation, we mapped the two dispersing clones in detail (Fig. 6c) and inferred the directions of growth based on the parent->child relationships in the phylogenetic tree. Case K234 displayed dispersal from the left hand branch of the phylogenetic tree, crossing the genetically opposed right hand branch and achieving subsequent subclonal expansion in the tumour centre. Intriguingly, the dispersing subclone in this case was also the metastasising subclone, which seeded the tumour thrombus (Fig. 6c). In case K163, again dispersal from a subclone on the left hand branch was observed crossing a clone from the right hand side of the tree (Fig. 6d). The distance of dispersal in this case was large in nature, with the subclone crossing an area of 63mm in distance, which presumably would make unsampled spatially contiguous growth in the third dimension unlikely.
Figure 6. Phylogenetic trees and tumour maps showing the inferred clonal expansion pattern and tumour metastasising routes.
A) Most cases had a contiguous clonal expansion, while one case of clonal dispersal was seen in 2 out of 79 samples. B) A tumour map and a phylogenetic tree of case K096 showing a pattern of contiguous clonal expansion. C) Tumour maps and phylogenetic trees of cases G_K234 and D) K163 showing clonal dispersal (dotted line). In case K234, the dispersing clone formed the metastasising branch and gave rise to its descendant clone which was also found in a venous thrombus. In case K163, the dispersing clone wasn’t associated with metastasis.
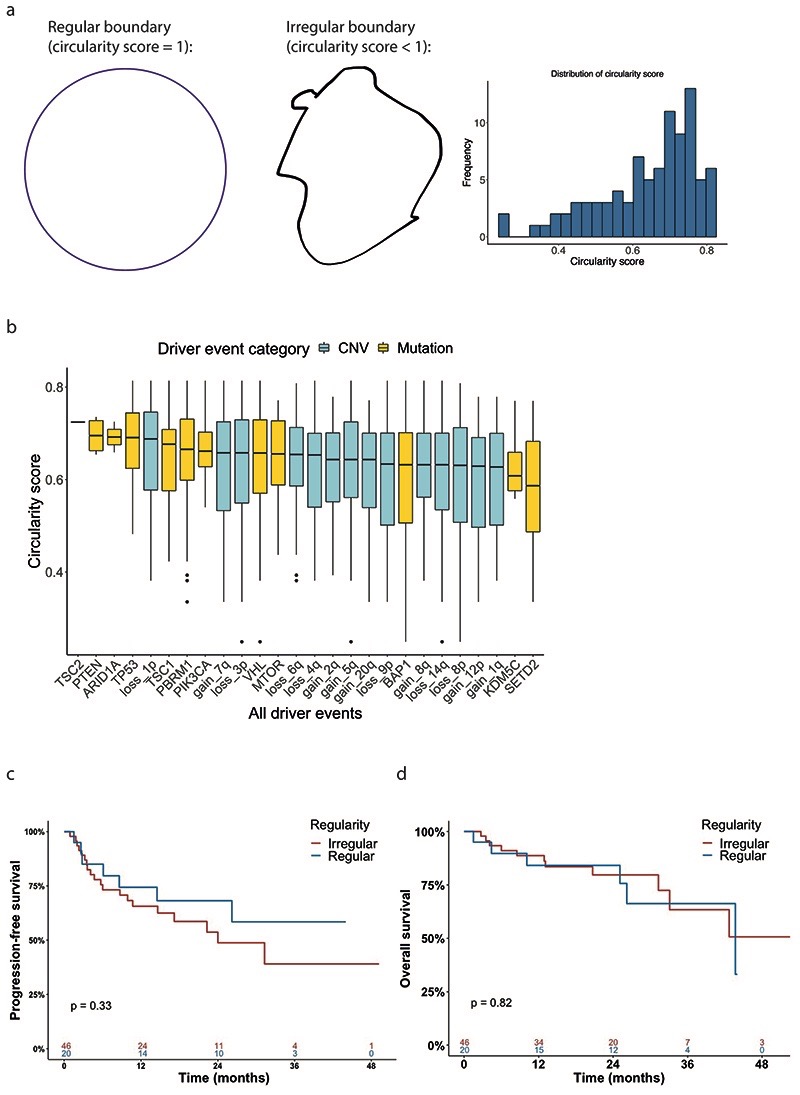
Irregularity of tumour boundary associates with SCNA burden
Lastly, we assessed the importance of boundary shape, based on the hypothesis that a highly irregular boundary shape may reflect sporadic growth driven by tumour intrinsic factors. For each tumour section the boundary circularity score was calculated (methods, Extended Data Fig. 7a) The median circularity score was 0.66 (range 0.25 to 0.81). We plotted driver genes and SCNAs in order of decreasing circularity and did not observe any notable patterns (Extended Data Fig. 7b). Similarly, circularity score was not prognostic for either overall or progression free survival (p=0.82 and p=0.33 respectively, Extended Data Fig. 7c-d).
Discussion
Here, we have conducted an integrated study of spatial evolution in ccRCC, analysing 756 biopsies across 79 primary tumour sections with spatial, genomic, IHC and clinical data. To our knowledge, this is the largest study of its kind to date, and it reveals insight into the spatial characteristics of solid tumour growth.
A key finding from this work is that metastatic ccRCC subclones preferentially originate from the interior of the tumour rather than the margin, with the tumour centre characterised by higher levels of proliferation, necrosis, Fuhrman grade and somatic copy number alteration (SCNA) burden. While not functionally validated, our computational simulation of these phenomena supports a model whereby heightened necrosis in the tumour interior facilitates increased SCNA acquisition thus creating a reservoir of subclones adapted to harsh microenvironmental conditions. Given these additional competencies, it is thus unsurprising that subclones from the tumour centre are better adapted to seed secondary metastatic sites. Necrotic primary tumour subclones have been shown to have greater propensity to seed metastases in pre-clinical models18, and clinically a recent systematic meta-analysis across 34 studies demonstrated tumour necrosis to be associated with reduced cancer-specific survival in RCC14. Biologically, tumour necrosis is classically associated with chronic hypoxia caused by rapid tumour growth and inadequate blood supply19. In addition hypoxia has a well-established role as a microenvironmental factor promoting metastasis20. Several lines of evidence link hypoxia as a cause of SCNAs and genome instability directly. Firstly, clinical data has demonstrated that patients with elevated hypoxia have a higher burden of focal copy number alterations, which can also be recapitulated in primary, nontransformed, and transformed human cells21, Secondly, there is evidence that hypoxia induces genome doubling in multiple cancer cell lines22. Thirdly, hypoxia has been associated with elevated rates of chromothripsis, allelic loss and shorter telomeres23. Lastly, hypoxia has been shown to be a potent fragile site inducer and a driver of double minutes24. Of separate note, ccRCC is additionally uniquely characterised by oxygen-independent so called “pseudohypoxia” as a universal feature of the disease due to VHL inactivation25. Moreover, as shown in our previous study, VHL evolutionary subtypes were found to be associated with increased tumour heterogeneity and high genomic instability5,26. Clinically, necrosis is associated with ischemia, either as a chronic process resulting in gradual tissue damage, or as an acute event such as infarction related to arterial or venous thrombosis. Tumour neovascularisation occurs through ingrowth of blood vessels from peripheral areas. These vessels are physiologically abnormal and thus susceptible to thrombosis rendering tumours or parts of them more prone to acute ischemic events. The chronic or acute nature of tumour necrosis may also in turn shape the nature of tumour evolution, either driving gradual subclonal adaption or a punctuated process of rapid cell death and repopulation by adapted subclones. While outside of the scope of this report, further functional study of necrotic events, hypoxia and processes associated with metastasis (such as epithelial-to-mesenchymal transition) in the tumour centre versus tumour margin would be of significant interest. In addition, comparison of the patterns observed here in ccRCC to other solid tumour types will be of future relevance, to understand if these findings are tumour-type specific (i.e. due to the large size of ccRCCs), or more broadly generalisable.
A unique benefit of this study is the integrated analysis of spatial and phylogenetic data, allowing visualisation of tumour subclones within their spatial location. Using this approach, we can show clear examples of metastasis from the tumour centre, as well as begin to understand the patterns and directions of spatial growth. While only descriptive in nature, the overall the patterns of growth do not appear to conform to a simplistic model of linear growth from the interior outwards towards the margins, instead more advanced evolutionary growth (i.e. later phylogenetic subclones) appear to be able to originate from central tumour regions as well as the margin (Fig. 3a-d, Fig. 6). The evolutionary basis of these patterns remains unclear, however it may be possible that the irregular/non-linear growth is as a result of environmental stimulus within the tumour, whereby subclonal genetic diversification and direction of growth is associated with environmental change (subclonal trophism).
Analysis of genetic versus spatial distance revealed a weak-to-moderate correlation, indicating that tumour subclones tend to be more genetically similar to their neighbouring subclones, as compared to ones further away. This result is congruent with the finding discussed below that spatial growth is predominantly spatially contiguous in nature. The weak-to-moderate strength of this correlation (rho=~0.3) suggests exceptions to this rule are frequent however, with clonal sweeps likely to be a key factor causing this correlation to breakdown and more rarely, clonal dispersal. It should be noted a limitation of this study is the panel based sequencing assay, which while well validated for driver mutation and genome-wide SCNA detection, lacks the dense number of genetic markers that would be available with whole exome or genome sequencing. In terms of individual driver events, we found the spatial area occupied was dominated by SCNAs rather than mutations, with loss of 14q, 9p and other known ccRCC SCNAs27 ranking highly. This adds a new dimension of evidence to support SCNAs as the primary driver of ccRCC growth, showing that they occupy large spatial areas of the primary tumour, despite occurring late within the evolutionary timeframe. Collectively these observations suggest a large selective advantage can be conferred to subclones upon acquisition of driver SCNAs, with growth disproportionally occurring after these events occur. This is consistent with previously reported survival analysis, where 9p loss is highly prognostic even in a setting where all cases have already metastasised15. It was reported that p16, encoded by CDKN2A on chromosome 9p, has been shown to modulate VEGF expression through interaction with HIF-1alpha, which was encoded by HIF1A on chromosome 14q28. Interestingly, the alternatively encoded CDKN2A protein p14ARF, as well as neighbouring gene CDKN2B, have both been associated with angiogenesis regulation29,30. Also, inhibition of AKT1 on the chromosome 14q was shown to promote invasion and metastasis in both non-small-cell lung cancer and breast cancer31,32. The driver event associated with minimal subclonal expansion was SETD2 mutation, with many subclones bearing this genetic event failing to achieve significant spatial expansion (i.e. event found only in one biopsy). By contrast however SETD2 is a frequently occurring subclonal event5, often found under strong parallel selection, indicating some beneficial growth advantage at a microscopic level which then reduces as clone size gets larger.
Lastly, we identified spatially contiguous growth as the dominant mode of ccRCC spatial growth, found in 77 of 79 tumour sections. This observation provides some insight into ccRCC tumour cell motility, suggesting a model of primary subclones growing predominantly within their localised microenvironment, with limited evidence of dynamic seeding between locations in the primary tumour. We note that n=32 of the tumours in our analysis had already achieved metastasis, so clearly subclones were present within the primary tumour with dispersal/motility competence. These observations would suggest that once motility competence is achieved those subclones are better able to seed and establish within secondary sites rather than other spatial locations in the primary. At a gene level, we note loss of CDKN2A on the chromosome 9p is strongly associated with enhanced motility33, and may act a key driver within this process. Previous evidence has indicated reseeding back from metastasis to the primary site in prostate cancer34, so we note this is another important model to consider. In addition, frequent rates of dispersal within the primary for glioblastoma have been reported35, so tumour specific patterns are likely to be present. In two cases dispersal was identified and was descriptively found to occur in the same subclone which achieved metastasis. In a second case dispersal was identified to have occurred across a large spatial distance, suggesting possible spread via the tumour vasculature. We note a limitation of this study is a lack of three-dimensional sampling. While feasible, three-dimensional mapping would likely require additional infrastructure requirements such as 3D imaging and improved analytical pipelines. In addition, we acknowledge the cause-and-effect relationship between necrosis, SCNA acquisition, proliferation and metastasis is supported by computational modelling, correlative analysis and prior literature, however alternative ordering of events is possible and further experimental investigation is required.
While the analysis presented here is exploratory in nature, we can put these results within a clinical context, and this study raises the point that there may be benefit of extra biopsy sampling (e.g. radiology guided) in the tumour centre, given higher tumour grade, proliferation and metastasis associated SNCAs were detected in central regions. However, a drawback of focusing biopsy sampling in the tumour centre is that central regions, if heavily necrotic, may have a poor diagnostic yield as they cannot be sequenced, and hence central biopsy sampling may only be appropriate as an addition to (where feasible) the current samples already being taken. This study seeks to highlight the importance of central regions as a source of metastatic subclones, and future technological improvements for sampling/profiling these necrotic areas will be of significant interest.
In summary, here we present the largest study to date mapping tumour subclones to their spatial locations within the primary tumour site. This study reveals unique insight into the metastatic process, indicating that ccRCC subclones preferentially originate from the tumour interior. Future studies resolved for spatial tumour location and subclonal structure are likely to continue to yield significant insight into the cancer evolutionary process.
Methods
Patient recruitment and sample collection
Patients were from the TRACERx Renal trial (NCT03226886), a multi-centre prospective longitudinal cohort study. The inclusion and exclusion criteria, along with full clinical, histological and follow-up data were described previously5,15. In brief, primary tumours were dissected along the longest axes and spatially separated regions sampled from the “tumour slice” using a 6mm punch biopsy needle. Primary tumour regions were labelled as R1, R2, R3... Rn with locations recorded. This study was ethically approved by the National Health Service Research Ethics Committee approval (11/LO/1996). Informed consent was obtained from all participants.
Distinct samples statement
Each biopsy region was measured only once and measurements were taken from distinct biopsy regions.
Macroscopic photo acquisition, quality control and pathological review
Photos were taken on the day of surgery from directly above the tumour, capturing the entire tumour specimen and any surrounding normal tissue (Fig. 1a). Quality control review of imaging data was conducted by a renal pathologist (J.I.L.), and images were rejected based on three criteria: i) image did not fully capture the entire tumour specimen (n=9), ii) the boundary between tumour and normal tissue could not be reliably determined (n=5), or iii) exact spatial position of any biopsy regions could not be reliably determined (n=9). Post quality control review n=79 images from 66 primary tumours were retained for further analysis. The remaining stages in the image processing workflow comprised: tracing of the tumour/normal boundary lines (by pathologist), automated digital capture of (X,Y) coordinates for biopsy and boundary locations, and finally integration of tumour width/length dimensions from the accompanying pathology reports (methods) (Fig. 1a).
Boundary drawing and digital tumour map processing
Boundaries between tumour and normal tissues were marked independently by a renal pathologist (JIL), after which the boundaries, along with the biopsy regions were extracted into X- and Y- coordinates using WebPlotDigitizer24. 2-dimensional size in millimetres of the tumours was set in accordance with the pathology reports after surgery. For those cases with only maximum length available, the length along the axis perpendicular to that of the maximum length was re-scaled based on the length on the photos. Digital tumour maps were then generated using R (version 3.6.2)25.
Microscopic pathology review, immunohistochemical staining and digital image analysis of Ki67
As previously described, histological sections, along with the immunohistochemical staining and digital image analysis of Ki67 of each region in each case were evaluated by the same pathologist (JIL)5,15. Briefly, tumour type, tumour architecture, tumour grading and the presence of necrosis and microvascular invasion were determined. Immunohistochemical staining of Ki67 was performed on 468 biopsy regions using a fully automated system and ready-to-use optimized reagents according to the manufacturer’s recommendations (Ventana Discovery Ultra, Ventana, Arizona, USA). Mouse intestine tissue was used as positive control, and regions containing tumour tissue were identified and marked by a pathologist and subsequently scanned into digital images, which were then subjected to automated image analysis for Ki67 quantification. Results were depicted as total percentage of Ki67-positive nuclei.
Targeted sequencing and genomic data processing
DNA extraction and multi-regional panel sequencing were as previously described5,15. Driver gene panels (Panel_v3, Panel_v5 and Panel_v6) were used in this study. Panel_v3, Panel_v5 and panel_v6 included 110, 119 and 130 putative driver genes, respectively. Driver genes were selected from genes that are frequently mutated in TCGA and other studies2–4. Mutation and somatic copy number alteration (SCNA) calling, clonality estimation of genomic alterations and phylogenetic tree reconstruction were done as previously described5,15. Subclonal frequency was defined as the proportion of an event that appeared to be subclonal across all samples. A matrix with presence and absence of nonsynonymous and synonymous point mutations, dinucleotide substitutions (DNVs), INDELs and arm level SCNAs was created for each tumour, and all the events were clustered as previously described5,15.
Definition of tumour centre and tumour margin
Tumour centre was defined as the area within the tumour where the distance between a biopsy region and the tumour boundary was more than or equal to 1cm, while tumour margin was defined as the area within the tumour where the distance between a biopsy region and the tumour boundary was less than 1cm (Fig. 2a).
Spatial and genomic distance calculation
Spatial distance between biopsy regions was measured using the Euclidean metric (Fig. 3a). Each biopsy region was represented as a point on the tumour map, with the X- and Y-coordinates referring to the actual position on the macroscopic photos. Distance between each biopsy region to the tumour boundary was calculated as the Euclidean distance between the biopsy region and its nearest boundary.
Genomic distance was calculated by taking the Euclidean distance of the detected genomic events (non-synonymous mutations and SCNAs). Mutations and SCNAs of all biopsy regions on a slice were represented as a matrix with 1 defined as an existing event and 0 as a non-existing event. Pairwise Euclidean distances were then calculated between all biopsy regions.
Calculation of space occupied by driver events
Driver event status of all biopsy regions was encoded in a matrix with 1 defined as an existing event and 0 as a non-existing event. The space occupied by a driver event was defined as the maximum spatial distance between any two regions containing the same driver event on a tumour slice, measured in millimetres.
Inference of metastasising routes on tumour maps
A matrix for all biopsy regions of all samples showing the relationship of clusters and biopsy regions was generated. Clusters containing nonsynonymous and synonymous point mutations, DNVs, INDELs and arm level SCNAs that appear in all the biopsy regions in a tumour were considered truncal clones, while clusters containing nonsynonymous and synonymous point mutations, DNVs, INDELs and arm level SCNAs that appear only in part of the biopsy regions in a tumour were considered subclones. The parent and child relationship of clusters were based on whether the genetic alterations each cluster contained were detected in other clusters. A secondary subclone was a cluster that contained not only point mutations, DNVs, INDELs and arm level SCNAs detected in the truncal clone, but also point mutations, DNVs, INDELs or arm level SCNAs that were not found in the truncal clone. A third subclone was a cluster that contained not only those genomic alterations detected in a secondary subclone, but also genomic alterations that were not found in the secondary subclone. The parent-and-child relationship went on and on until it reached a terminal clone which harboured genomic alterations that were not shared by any other clusters. Metastasising routes were inferred on tumour maps based on this parent-and-child relationship. Levels of the biopsy regions were set depending on the highest-level subclone it contained. Regions containing a parent clone as the highest-level subclone would point to regions containing a child clone as the highest-level subclone, while regions containing same levels of subclones were lined without arrows.
Definition of clonal expansion patterns
Types of clonal expansion patterns were defined based on the metastasising routes on the tumour maps. Contiguous growth was defined as the growth pattern where nearest subclones from the same branch of the phylogenetic tree were lined one by one, without any subclones from the other branch in between. Clonal dispersal was defined as the growth pattern where nearest subclones from the same branch of the phylogenetic tree crossed the border formed by the subclones from the other branch.
Circularity score calculation
Circularity score was calculated to assess the regularity of tumour boundaries:
where area and perimeter of the tumour slice were captured based on the tumour maps using ImageJ version 1.52q. A score of 1 indicates perfect circularity, and lower scores indicate increasing irregularity in shape.
Computational modelling
Simulation of tumour growth and evolution, as determined by the 26 ccRCC drivers, and in the context of necrosis, was achieved by the development of a coarse-grained cellular automata model (12 driver genes and 14 SCNAs). A model tumour comprises units called tumour voxels, each of which reflects a volume of 1 mm3. In the basic model, growth takes place in the form of probabilistic voxel duplication. A tumour voxel can only duplicate if surrounded by at least one empty site. Upon growth, a tumour voxel randomly acquires additional driver events.
The growth probability of a tumour voxel in the model further depends on the drivers harboured within. Two levels of relative driver advantage in growth are set to reflect the ranking of driver advantages by their association with regional Ki67 expression in the TRACERx Renal study. Four SCNAs (20q gain, 7q gain, 4q loss, and 8p loss) endow a tumour voxel with the strongest growth advantage; a tumour voxel harbouring any of these SCNAs grows with a probability of 1.0 (hereafter referred to as “p_adv”) every simulation step. Another seven SCNAs (2q gain, 5q gain, 8q gain, 12p gain, 1p loss, 9p loss, and 14q loss) and seven gene mutations (BAP1, MTOR, PBRM1, PIK3CA, SETD2, TP53, and TSC1) endow a tumour voxel with a lesser growth advantage; a tumour voxel harbouring any of these drivers, but not none of the above four strongest SCNAs, grows with a probability of 0.5 every simulation step. If a tumour voxel doesn’t harbour any of these eighteen drivers, it grows with a probability of 0.25 every simulation step.
The acquisition probability of driver gene mutations is 2×10-4 per simulation step. Mutation in either of the two driver genes, BAP1 and PBRM1, is assumed to promote the acquisition of SCNAs. In tumour voxels harbouring these two mutations, the acquisition probability of SCNAs is also 2×10-4 per simulation step. Otherwise, the acquisition of SCNAs is defined to take place with a probability of 2×10-7 per simulation step. The loss of 3p and VHL in the model are defined to be always truncal and already harboured in the first tumour voxel.
Extending the basic model, we implemented central necrosis to explore the impact of enhanced cell death at the tumour core on tumour growth and, of particular interest, the evolution of SCNA- harbouring subclones. Specifically, we defined that all tumour voxels located farther than 1.5 cm (hereafter referred to as “d_nec”) from the tumour surface become necrotic with a probability of 0.5 every simulation step. The chosen distance threshold of 1.5 cm reflects clinical observations; distances between any regional biopsies containing necrosis and the tumour contour, which lacks necrosis, was measured (mean: 12.83 mm; range: 3.71 – 27.11 mm). If a tumour voxel is chosen to undergo necrosis, the site occupied by that tumour voxel is labelled as necrotic; consequently, adjacent tumour voxels are allowed to grow into this necrotic site. Other than the default parameter set (p_adv, d_nec) = (1, 1.5 cm), and to assess the robustness of model observations, we also prepared additional sets of simulations with varying levels of p_adv (0.8, 0.9, 1) and d_nec (1, 1.25, 1.5, 1.75) cm.
Each three-dimensional (3D) simulation is permitted to run until reaching a size of at least 1 million tumour voxels after the last simulation step. Upon completion, a two-dimensional (2D) slice is collected from the middle of the 3D simulated tumour. in 3D. Within this 2D tumour slice, regional biopsies (each with a size reflecting a diameter of 1 cm) are collected uniformly, with a spacing of 1 cm. The necrotic status and driver composition within each regional biopsy, from these simulations, are recorded for further analysis.
To evaluate the association between the number of SCNAs within each region and their spatial location, regional biopsies are labelled with “At.Margin” versus “At.Centre” to indicate their marginal versus central localisation. Biopsies with a distance to the tumour contour greater than 1 cm (referred to as “d_margin”) are defined as central biopsies, while the rest marginal biopsies, consistent with the definition in the experimental analysis.
The necrotic fraction of a regional biopsy is calculated as the number of tumour voxels that are necrotic divided by the total number of tumour voxels in that biopsy. To evaluate the association between the number of SCNAs within each region and the necrotic status, all regional biopsies are ranked according to the necrotic fraction. Biopsies with a necrotic fraction greater than 0.8 are dropped from the analysis, due to their small amount of alive tumour voxels. The remaining biopsies are grouped into “less necrotic” (necrotic fraction between 0 and median) and “more necrotic” (necrotic fraction between median and 0.8) categories. The number of SCNAs in a regional biopsy is defined as the number of unique SCNAs found in any tumour voxels in that biopsy.
To evaluate the association between the evolutionary age of each regional biopsy and its spatial location, the average subclone birth time is calculated based on all subclones present within that regional biopsy.
Finally, to evaluate the association between tumour boundary shape and evolutionary age or SCNA burden, the circularity of each tumour boundary was calculated in the same way as experimentally determined. Tumours are then split into two equal-sized subsets according to their circularity value.
Statistical analyses
Non-parametric Wilcoxon signed-rank test was used to compare medians of groups of continuous variables. Fisher’s exact test was used to compare proportions of counts of categorical variables. Non-parametric Spearman’s rank correlation coefficient was used to assess the relationship between pairs of variables, as opposed to testing for a difference in medians between two continuous variables. Kaplan-Meier survival curves and log-rank tests were used to analyse patients’ survival. Progression-free survival was defined as the time to disease recurrence or relapse, or death without disease recurrence, whichever came first. Overall survival was defined as the time to cancer specific death. Significance level was set 0.05. All statistical analyses were conducted using R version 3.6.2.
Extended Data
Extended Data Fig. 1.
Extended Data Fig. 2.
Extended Data Fig. 3.
Extended Data Fig. 4.
Extended Data Fig. 5.
Extended Data Fig. 6.
Extended Data Fig. 7.
Extended Data Fig. 8.
Extended Data Fig. 9.
Extended Data Fig. 10.
Supplementary Material
Acknowledgements
Funding
This work was supported by the Francis Crick Institute that receives its core funding from Cancer Research UK (FC001003, FC001144, FC001169, FC001988), the UK Medical Research Council (FC001169), and the Wellcome Trust (FC001003, FC001144, FC001169, FC001988). This research was funded in whole, or in part, by the Wellcome Trust (FC001003, FC001144, FC001169, FC001988). For the purpose of Open Access, the author has applied a CC BY public copyright licence to any Author Accepted Manuscript version arising from this submission. K.L. is funded by the UK Medical Research Council (MR/P014712/1), the Rosetrees Trust and Cotswold Trust (A2437), the Royal Marsden Cancer Charity (thanks to the Ross Russell family and Macfarlanes donations), Melanoma Research Alliance, and Cancer Research UK (C69256/A30194). J.I.L is funded by MINECO, Spain grant: SAF2016-79847-R. X.F. and P.A.B. are funded by the Francis Crick Institute, which receives its core funding from Cancer Research UK (FC001003, FC001144), the UK Medical Research Council (FC001003, FC001144) and the Wellcome Trust (FC001003, FC001144). E.S. is funded by the Francis Crick Institute, which receives its core funding from Cancer Research UK (FC010144), the UK Medical Research Council (FC010144) and the Wellcome Trust (FC010144). S.T. is funded by Cancer Research UK (grant reference number C50947/A18176); the Francis Crick Institute, which receives its core funding from Cancer Research UK (FC10988), the UK Medical Research Council (FC10988), and the Wellcome Trust (FC10988); the National Institute for Health Research (NIHR) Biomedical Research Centre at the Royal Marsden Hospital and Institute of Cancer Research (grant reference number A109); the Royal Marsden Cancer Charity; The Rosetrees Trust (grant reference number A2204), Ventana Medical Systems Inc (grant reference numbers 10467 and 10530); the National Institute of Health (Bethesda) and Melanoma Research Alliance. C.S. is Royal Society Napier Research Professor (RP150154). C.S. is funded by Cancer Research UK (TRACERx, PEACE and CRUK Cancer Immunotherapy Catalyst Network), Cancer Research UK Lung Cancer Centre of Excellence, the Rosetrees Trust, Butterfield and Stoneygate Trusts, NovoNordisk Foundation (ID16584), Royal Society Professorship Enhancement Award (RP/EA/180007), the National Institute for Health Research (NIHR) Biomedical Research Centre at University College London Hospitals, the CRUK-UCL Centre, Experimental Cancer Medicine Centre, and the Breast Cancer Research Foundation (BCRF, USA). This research is supported by a Stand Up To Cancer-LUNGevity-American Lung Association Lung Cancer Interception Dream Team Translational Research Grant (Grant Number: SU2C-AACR-DT23-17). Stand Up To Cancer is a program of the Entertainment Industry Foundation. Research grants are administered by the American Association for Cancer Research, the Scientific Partner of SU2C. C.S. receives funding from the European Research Council (ERC) under the European Union’s Seventh Framework Programme (FP7/2007-2013) Consolidator Grant (FP7-THESEUS-617844), European Commission ITN (FP7-PloidyNet 607722), an ERC Advanced Grant (PROTEUS) from the European Research Council under the European Union’s Horizon 2020 research and innovation programme (grant agreement No. 835297), and Chromavision from the European Union’s Horizon 2020 research and innovation programme (grant agreement 665233). A.F. receives funding from the European Union’s Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie grant agreement No. 892360. L.A., S.T.C.S., L.P. and J.L. are supported by the National Institute for Health Research (NIHR) Biomedical Research Centre at the Royal Marsden Hospital and Institute of Cancer Research.
Footnotes
Author’s contributions
Study design: K.L., S.T., C.S., P.A.B.
Data analysis: Y.Z, X.F., J.I.L., M.A., S.H., H. X.
Data contribution and interpretation: All
Manuscript writing: K.L., Y.Z., X.F.
Competing Interests
S.T., and C.S. have a patent on indel burden and checkpoint inhibitor response pending, and a patent on targeting of frameshift neoantigens for personalised immunotherapy pending. K.L. has a patent on indel burden and CPI response pending and outside of the submitted work, speaker fees from Roche tissue diagnostics, research funding from CRUK TDL/Ono/LifeArc alliance, and a consulting role with Monopteros Therapeutics. S.T. has received speaking fees from Roche, Astra Zeneca, Novartis and Ipsen. S.T. has the following patents filed: Indel mutations as a therapeutic target and predictive biomarker PCTGB2018/051892 and PCTGB2018/051893 and Clear Cell Renal Cell Carcinoma Biomarkers P113326GB. C.S. acknowledges grant support from Pfizer, AstraZeneca, Bristol Myers Squibb, Roche-Ventana, Boehringer-Ingelheim, Archer Dx Inc (collaboration in minimal residual disease sequencing technologies) and Ono Pharmaceuticals. C.S is an AstraZeneca Advisory Board member and Chief Investigator for the MeRmaiD1 clinical trial, has consulted for Pfizer, Novartis, GlaxoSmithKline, MSD, Bristol Myers Squibb, Celgene, AstraZeneca, Illumina, Amgen, Genentech, Roche-Ventana, GRAIL, Medicxi, Bicycle Therapeutics, and the Sarah Cannon Research Institute, has stock options in Apogen Biotechnologies, Epic Bioscience, GRAIL, and has stock options and is co-founder of Achilles Therapeutics. C.S. holds patents relating to assay technology to detect tumour recurrence (PCT/GB2017/053289); to targeting neoantigens (PCT/EP2016/059401); identifying patent response to immune checkpoint blockade (PCT/EP2016/071471); determining whether HLA LOH is lost in a tumour (PCT/GB2018/052004); predicting survival rates of cancer patients (PCT/GB2020/050221); to treating cancer by targeting Insertion/deletion mutations (PCT/GB2018/051893); identifying insertion/deletion mutation targets (PCT/GB2018/051892); detecting tumor mutations (PCT/US2017/028013); and identifying responders to cancer treatment (PCT/GB2018/051912). E.S. receives research funding from Merck Sharp Dohme and Astrazeneca and is on the scientific advisory board of Phenomic.
We thank the TRACERx Renal trial team and the Skin and Renal Unit Research Team at The Royal Marsden NHS Foundation Trust, including Eleanor Carlyle, Lyra Del Rosario, Kim Edmonds, Karla Lingard, Mary Mangwende, Sarah Sarker, Charlotte Lewis, Fiona Williams, Hamid Ahmod, Tara Foley, Dilruba Kabir, Justine Korteweg, Aida Murra, Nahid Shaikh, Kema Peat, Sarah Vaughan and Lucy Holt. TRACERx Renal is funded by NIHR BRC at the Royal Marsden Hospital and Institute of Cancer Research (A109). The Francis Crick Institute, which receives its core funding from CRUK (FC010110), the UK Medical Research Council (FC010110), the Wellcome Trust (FC010110).
Contributor Information
The TRACERx Renal Consortium:
Lewis Au, Ben Challacombe, Ashish Chandra, Simon Chowdhury, William Drake, Archana Fernando, Nicos Fotiadis, Andrew Furness, Emine Hatipoglu, Karen Harrison-Phipps, Steve Hazell, Peter Hill, Catherine Horsfield, James Larkin, Jose Ignacio Lopez, Teresa Marafioti, David Nicol, Tim O’Brien, Jonathon Olsburgh, Lisa Pickering, Alexander Polson, Sergio Quezada, Sarah Rudman, Scott T.C. Shepherd, Charles Swanton, Samra Turajlic, Mary Varia, and Hema Verma
Code availability statement
Code used for analyses is available at both https://github.com/yuezhao97 and https://github.com/kevlitchfield1.
Source data availability statement
Sequencing data that supports this study have been deposited at the European Genome-phenome Archive (EGA), which is hosted by the European Bioinformatics Institute (EBI); accession number EGAS00001002793. Spatial X- and Y-coordinates and source data for all the figures are available at both https://github.com/yuezhao97 and https://github.com/kevlitchfield1.
References
- 1.Cancer Genome Atlas Research, N. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013;499:43–9. doi: 10.1038/nature12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dalgliesh GL, et al. Systematic sequencing of renal carcinoma reveals inactivation of histone modifying genes. Nature. 2010;463:360–3. doi: 10.1038/nature08672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scelo G, et al. Variation in genomic landscape of clear cell renal cell carcinoma across Europe. Nat Commun. 2014;5:5135. doi: 10.1038/ncomms6135. [DOI] [PubMed] [Google Scholar]
- 4.Sato Y, et al. Integrated molecular analysis of clear-cell renal cell carcinoma. Nat Genet. 2013;45:860–7. doi: 10.1038/ng.2699. [DOI] [PubMed] [Google Scholar]
- 5.Turajlic S, et al. Deterministic Evolutionary Trajectories Influence Primary Tumor Growth: TRACERx Renal. Cell. 2018;173:595–610.:e11. doi: 10.1016/j.cell.2018.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gerlinger M, et al. Genomic architecture and evolution of clear cell renal cell carcinomas defined by multiregion sequencing. Nat Genet. 2014;46:225–233. doi: 10.1038/ng.2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Priestley P, et al. Pan-cancer whole-genome analyses of metastatic solid tumours. Nature. 2019;575:210–216. doi: 10.1038/s41586-019-1689-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Williams MJ, Werner B, Barnes CP, Graham TA, Sottoriva A. Identification of neutral tumor evolution across cancer types. Nat Genet. 2016;48:238–244. doi: 10.1038/ng.3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gerstung M, et al. The evolutionary history of 2,658 cancers. Nature. 2020;578:122–128. doi: 10.1038/s41586-019-1907-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoefflin R, et al. Spatial niche formation but not malignant progression is a driving force for intratumoural heterogeneity. Nat Commun. 2016;7:ncomms11845. doi: 10.1038/ncomms11845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Axelrod R, Pienta KJ. Cancer as a Social Dysfunction-Why Cancer Research Needs New Thinking. Mol Cancer Res. 2018;16:1346–1347. doi: 10.1158/1541-7786.MCR-18-0013. [DOI] [PubMed] [Google Scholar]
- 12.Endesfelder D, et al. Chromosomal instability selects gene copy-number variants encoding core regulators of proliferation in ER+ breast cancer. Cancer Res. 2014;74:4853–4863. doi: 10.1158/0008-5472.CAN-13-2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bredholt G, et al. Tumor necrosis is an important hallmark of aggressive endometrial cancer and associates with hypoxia, angiogenesis and inflammation responses. Oncotarget. 2015;6:39676–91. doi: 10.18632/oncotarget.5344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang L, et al. Tumor necrosis as a prognostic variable for the clinical outcome in patients with renal cell carcinoma: a systematic review and meta-analysis. BMC Cancer. 2018;18:870. doi: 10.1186/s12885-018-4773-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Turajlic S, et al. Tracking Cancer Evolution Reveals Constrained Routes to Metastases: TRACERx Renal. Cell. 2018;173:581–594.:e12. doi: 10.1016/j.cell.2018.03.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.El-Mokadem I, et al. Significance of chromosome 9p status in renal cell carcinoma: a systematic review and quality of the reported studies. Biomed Res Int. 2014;2014:521380. doi: 10.1155/2014/521380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ma J, et al. The infinite sites model of genome evolution. Proc Natl Acad Sci U S A. 2008;105:14254–61. doi: 10.1073/pnas.0805217105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bonfil RD, Bustuoabad OD, Ruggiero RA, Meiss RP, Pasqualini CD. Tumor necrosis can facilitate the appearance of metastases. Clin Exp Metastasis. 1988;6:121–9. doi: 10.1007/BF01784843. [DOI] [PubMed] [Google Scholar]
- 19.Kumar V, Abbas AK, Aster JC, Robbins SL. Robbins Pathology. xii. Elsevier Saunders; Philadelphia, PA: 2013. Robbins basic pathology 1 online resource; p. 910. [Google Scholar]
- 20.Rankin EB, Giaccia AJ. Hypoxic control of metastasis. Science. 2016;352:175–80. doi: 10.1126/science.aaf4405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Black JC, et al. Hypoxia drives transient site-specific copy gain and drug-resistant gene expression. Genes Dev. 2015;29:1018–31. doi: 10.1101/gad.259796.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Young SD, Hill RP. Effects of reoxygenation on cells from hypoxic regions of solid tumors: anticancer drug sensitivity and metastatic potential. J Natl Cancer Inst. 1990;82:371–80. doi: 10.1093/jnci/82.5.371. [DOI] [PubMed] [Google Scholar]
- 23.Bhandari V, et al. Molecular landmarks of tumor hypoxia across cancer types. Nat Genet. 2019;51:308–318. doi: 10.1038/s41588-018-0318-2. [DOI] [PubMed] [Google Scholar]
- 24.Coquelle A, Toledo F, Stern S, Bieth A, Debatisse M. A new role for hypoxia in tumor progression: induction of fragile site triggering genomic rearrangements and formation of complex DMs and HSRs. Mol Cell. 1998;2:259–65. doi: 10.1016/s1097-2765(00)80137-9. [DOI] [PubMed] [Google Scholar]
- 25.Macklin PS, et al. Recent advances in the biology of tumour hypoxia with relevance to diagnostic practice and tissue-based research. J Pathol. 2020;250:593–611. doi: 10.1002/path.5402. [DOI] [PubMed] [Google Scholar]
- 26.Ricketts CJ, Linehan WM. Multi-regional Sequencing Elucidates the Evolution of Clear Cell Renal Cell Carcinoma. Cell. 2018;173:540–542. doi: 10.1016/j.cell.2018.03.077. [DOI] [PubMed] [Google Scholar]
- 27.Beroukhim R, et al. Patterns of gene expression and copy-number alterations in von-hippel lindau disease-associated and sporadic clear cell carcinoma of the kidney. Cancer Res. 2009;69:4674–81. doi: 10.1158/0008-5472.CAN-09-0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang J, Lu A, Li L, Yue J, Lu Y. p16 Modulates VEGF expression via its interaction with HIF-1alpha in breast cancer cells. Cancer Invest. 2010;28:588–97. doi: 10.3109/07357900903286941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zerrouqi A, Pyrzynska B, Febbraio M, Brat DJ, Van Meir EG. P14ARF inhibits human glioblastoma-induced angiogenesis by upregulating the expression of TIMP3. J Clin Invest. 2012;122:1283–95. doi: 10.1172/JCI38596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nanda V, et al. CDKN2B Regulates TGFbeta Signaling and Smooth Muscle Cell Investment of Hypoxic Neovessels. Circ Res. 2016;118:230–40. doi: 10.1161/CIRCRESAHA.115.307906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rao G, et al. Inhibition of AKT1 signaling promotes invasion and metastasis of non-small cell lung cancer cells with K-RAS or EGFR mutations. Sci Rep. 2017;7:7066. doi: 10.1038/s41598-017-06128-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li W, et al. Akt1 inhibition promotes breast cancer metastasis through EGFR-mediated beta-catenin nuclear accumulation. Cell Commun Signal. 2018;16:82. doi: 10.1186/s12964-018-0295-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zeng H, et al. Bi-allelic Loss of CDKN2A Initiates Melanoma Invasion via BRN2 Activation. Cancer Cell. 2018;34:56–68.:e9. doi: 10.1016/j.ccell.2018.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gundem G, et al. The evolutionary history of lethal metastatic prostate cancer. Nature. 2015;520:353–357. doi: 10.1038/nature14347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Foty RA. Tumor cohesion and glioblastoma cell dispersal. Future Oncol. 2013;9:1121–32. doi: 10.2217/fon.13.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Code used for analyses is available at both https://github.com/yuezhao97 and https://github.com/kevlitchfield1.
Sequencing data that supports this study have been deposited at the European Genome-phenome Archive (EGA), which is hosted by the European Bioinformatics Institute (EBI); accession number EGAS00001002793. Spatial X- and Y-coordinates and source data for all the figures are available at both https://github.com/yuezhao97 and https://github.com/kevlitchfield1.