Abstract
Nature regulates interference between cellular processes—allowing more complexity of life—by confining specific functions to organelles. Inspired by this concept, we designed an artificial organelle dedicated to protein engineering. We generated a membraneless organelle to translate only one type of messenger RNA—by recruiting an RNA-targeting system, stop codon–suppression machinery, and ribosomes—by means of phase separation and spatial targeting.This enables site-specific protein engineering with a tailored noncanonical function in response to one specific codon in the entire genome only in the protein of choice. Our results demonstrate a simple yet effective approach to the generation of artificial organelles that provides a route toward customized orthogonal translation and protein engineering in semisynthetic eukaryotic cells.
The ability to engineer orthogonal (non-cross-reactive) translation site-specifically into living cells enables the introduction of new functionalities into proteins. However this is a herculean task because translation is a complex multistep process in which at least 20 different aminoacylated tRNAs, their cognate tRNA synthetases (RSs), ribosomes, and diverse other factors work in concert to synthesize a polypeptide chain from the RNA transcript. An ideal orthogonal system would show no cross-reactivity with factors of the host machinery, minimizing its impact on the housekeeping translational activity and normal physiology of the cell.
Toward this goal, genetic code expansion (GCE) is a method that enables reprogramming of a specific codon. With GCE, an orthogonal suppressor RS can aminoacylate its cognate suppressor tRNA with noncanonical amino acids (ncAAs). These ncAAs are typically custom designed and harbor chemical functionalities that can, for example, enable protein function to be photocontrolled, encode posttranslational modifications, or allow the introduction of fluorescent labels for microscopy studies by using click chemistry. To introduce ncAAs site-specifically into a protein of interest (POI), the anticodon loop of the tRNA is chosen to decode and thus suppress one of the stop codons [(1–3), reviews]. To minimize the impact on the host cell machinery, the Amber stop codon (TAG) is often used, owing to its particularly low abundance in Escherichia coli, to terminate endogenous proteins (<10%). Nevertheless, in principle any Amber codon in the transcriptome can be suppressed, potentially leading to unwanted modification of nontargeted host proteins. If ncAA-modified proteins are recombinantly produced for in vitro applications, this background incorporation can be largely ignored as long as the yields of purified full-length protein are acceptable. However, the challenge is different if the host is considered more than just a bioreactor vessel that can be sacrificed for its protein. In order to study the function of a host-cell POI in situ, the physiological condition of that host cell is an important factor. In that context, minimization of background incorporation of the ncAA is required to ensure well-controlled experiments.
At least three elegant approaches have been developed to enable orthogonal translation in E. coli–that is, to decode a specific codon only for the RNA of the POI and not the entire genome. (i) Orthogonal ribosomes that recognize RNA with a specific Shine-Dalgarno sequence (4–6) have been developed to enhance codon specificity, to site-specifically encode an ncAA into a POI. (ii) Recently, genome engineering has advanced to the stage that E. coli strains can be depleted of selected native codons (7–10), providing a genetically clean (such as Amber codon-free) host background for selective decoding of specific codons only in the POI. (iii) Distinct noncanonical codons have been designed by using an artificial base pair encoded only in the coding sequence of the POI. This lowers the risk of non-specific decoding in other parts of the genome (11). However, because of genome complexity, it is not straightforward to transfer these orthogonal translation approaches to eukaryotes [(12), review], in which the Amber codon is highly abundant (~20% in human cells).
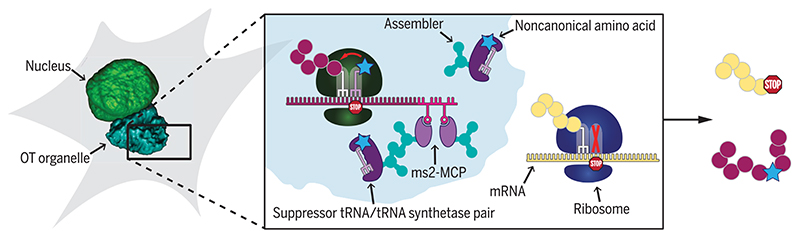
We hypothesized that it is possible to create an orthogonal translation system by spatially enriching specific components of the GCE machinery in an artificial orthogonally translating (OT) organelle/droplet/aggregate/condensate/dense phase. For an OT organelle to translate only the mRNA of the POI, it should be readily accessible to the translational machinery (>100 different biomolecules, such as canonical aminoacylated tRNAs, translation factors, and ribosomal subunits) and thus cannot be further easily membrane-encapsulated inside the cell. Another requirement is that the small cognate suppressor tRNA localizes efficiently to the OT organelle and is depleted from the rest of the cytoplasm.
The idea to create such an OT organelle was inspired by the concept of phase separation, which can generate high local concentrations of proteins and RNAs in cells (13, 14). Recently, phase separation has gained attention owing to the discovery of its prevalence in cell biology and its role in the formation of specialized organelles such as nucleoli, stress/RNA granules, and Balbiani bodies [(15), review]. Although our understanding of the design and functional principles of these organelles is emerging, it has been established that they are membraneless and thus are in direct contact and exchange with the surrounding cytoplasm and/or nucleoplasm. Despite lacking a membrane, these organelles can efficiently perform complex tasks, such as transcription in the nucleolus.
We aimed to create a new OT organelle in a living mammalian cell and envisioned use of a strategy in which we selectively target the RS and the mRNA of a POI to a spatially distinct site in the cytoplasm. We found that a combination of phase separation with spatial targeting by using motor proteins yields an organelle-like structure enriched in RS and mRNA, to which the cognate suppressor tRNA and ribosomes effectively copartition. This affords a set of spatially distinct ribosomes, forming an OT system that preferentially translates only our tagged mRNA, which enables site-specific recoding of a stop codon only in this mRNA. We show for a variety of proteins, including membrane proteins, that we can incorporate site-specific noncanonical functions only into a POI, whereas other mRNAs in the cytoplasm that contain the same stop codon are not translated efficiently.
Design of OT organelles
Our synthetic designer OT organelle (Fig. 1) is engineered with the following components.
-
(i)
An mRNA-targeting system in which two ms2 RNA stem loops are fused to the mRNA of choice, creating an mRNA::ms2 fusion coding for the POI. We denoted DNA in italics. The ms2 loops bind specifically to the phage-derived major capsid protein (MCP) (16), which will thus form a stable and specific mRNA::ms2–MCP complex in cells. The ms2 loops were always fused to the 3’ untranslated region (3’UTR) of the mRNA, which ensures translation to yield a scarless final POI.
-
(ii)
A tRNA/RS suppressor pair. We chose the orthogonal tRNA/RS pair from the Methanosarcina mazei pyrrolysyl system (tRNAPyl/PylRS) because it has enabled the encoding of more than 100 ncAAs with diverse functionalities into proteins by using GCE in a multitude of cell types and species, including E. coli, mammalian cells, and even living mice [(1–3), reviews].
-
(iii)
The assembler, the key component required to form an OT organelle. The purpose of the assembler is to create a dense phase or condensate, in which the mRNA::ms2-MCP complex is brought into close proximity of the tRNAPyl/PylRS pair.
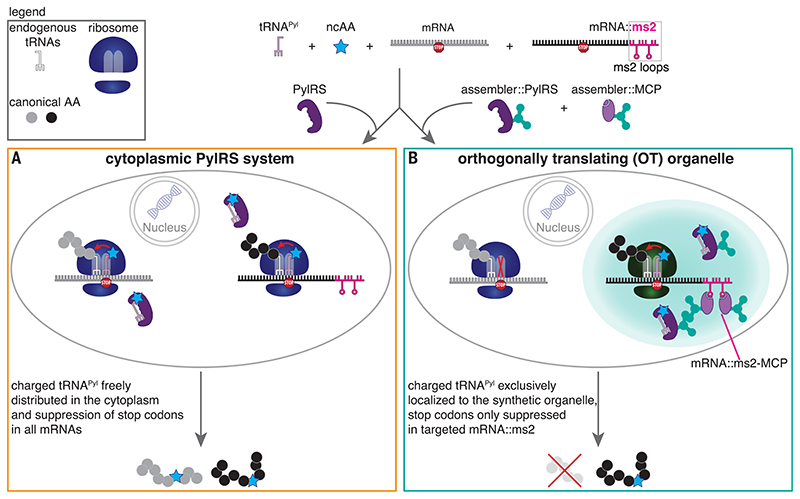
Fig. 1. Spatial separation of the necessary components to enable orthogonal translation to decode a specific stop codon in a specifically tagged mRNA.
(A) Expression of the synthetase PylRS leads to aminoacylation of its cognate stop codon suppressor tRNAPyl with a custom designed ncAA. This leads to site-specific ncAA incorporation whenever the respective stop codon occurs in the mRNA of the POI. Given that many endogenous mRNAs terminate on the same stop codon, using this approach in the cytoplasm potentially leads to misincorporation of the ncAA into unwanted proteins. (B) To avoid this, we propose to spatially enrich all components to an OT organelle, including the mRNA of the POI, the aminoacyl-tRNA synthetase, the tRNA, and ribosomes through the use of “assemblers.” Aminoacylated tRNAPyl should only be available in direct proximity of the OT organelle, so that only here stop codon suppression can occur. The corresponding stop codon in mRNAs that are not targeted to the OT organelle should not get translated. Whereas in (A) GCE is stop codon specific, in (B), it is stop codon– and mRNA-specific.
The simplest assembler strategy we tested is the bimolecular fusion of MCP::PylRS (termed B) (Fig. 2A). In addition, we tested strategies in which we expected to yield much larger assemblies. All of those assembly systems are composed of an assembler fusion to PylRS coexpressed with an assembler fusion to MCP. We expected assembler::PylRS•assembler::MCP to form large aggregates (we denote coexpression with a center dot, “•”). One tested assembly strategy was based on phase separation of proteins, and one was based on the assembly of kinesins, which we abbreviate here as P and K, respectively (Fig. 2A). Furthermore, for each P and K approach, we tested two different molecular designs: P1, P2 and K1, K2, respectively.
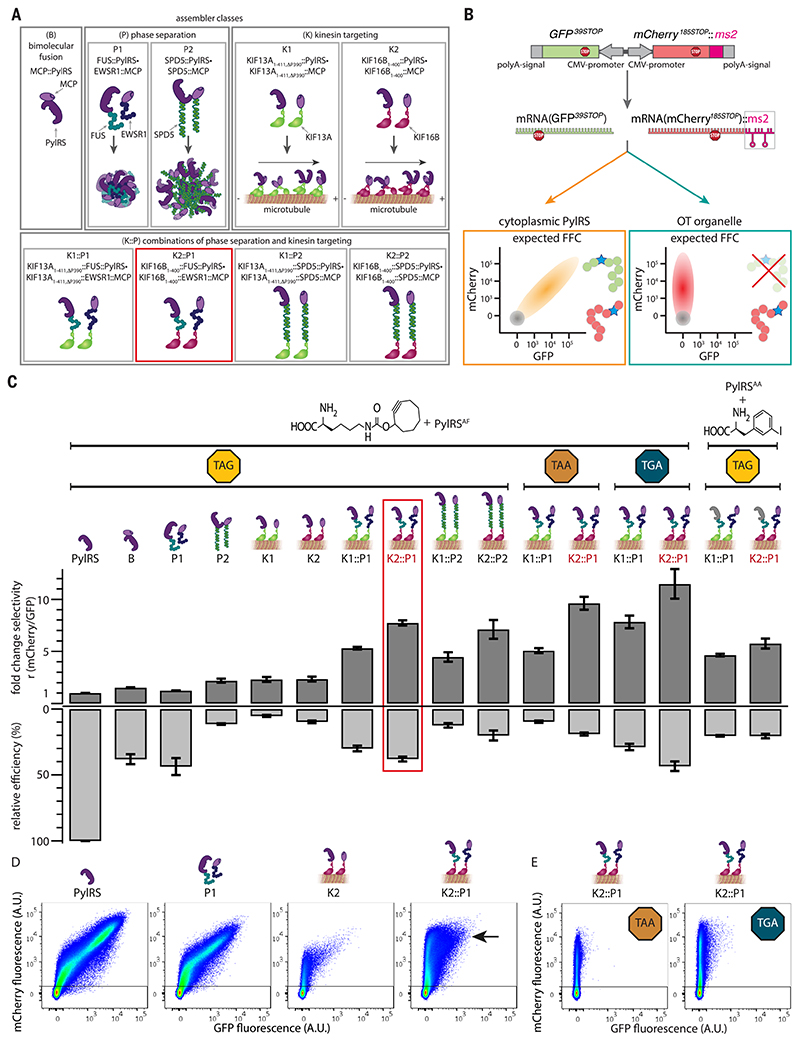
Fig. 2. Local enrichment by means of phase separation is a means to generate OT organelles.
(A) Schematic representation of different assembler classes. B, bimolecular MCP::PylRS fusion; P1, fusions to FUS and EWSR1; P2, SPD5; K1, truncation of kinesin KIF13A (KIF13A1-411,ΔP390); K2, truncation of kinesin KIF16B (KIF16B1-400) and combinations thereof (K1::P1, K1::P2, K2::P1, and K2::P2). (B) Schematic representation of the dual-color reporter. mRNAs encoding the fluorescent proteins GFP and mCherry, containing stop codons at permissive sites, are expressed from one plasmid, each with its own CMV promoter, ensuring a constant ratio of mRNA throughout each experiment. The mRNA of the mCherry reporter is tagged with two ms2 stem loops, mCherry:: ms2. In the presence of ncAA and tRNAPyl, in the case of cytoplasmic PylRS, both GFP39STOP and mCherry185STOP are produced, leading to a diagonal (schematically drawn in orange) in FFC analysis. However, under the same conditions, orthogonal translation in OTorganelles should enable selective stop codon suppression of mCherry::ms2 mRNA, resulting in an mCherry-positive and GFP-negative population (drawn schematically as a red vertical population). In both schemes, nontransfected HEK293T cells, which are also detected with FFC, are represented by a gray circle. (C) For all experiments, the indicated constructs were coexpressed with tRNAPyl (anticodon corresponding to the indicated codon) and the dual reporter (GFP39STOP, mCherry185STOP::ms2). GCE was performed in presence of the indicated ncAAs, and cells were analyzed by means of FFC. The dark gray bars (normalized to cytoplasmic PylRS) represent the fold change in the ratios r of the mean fluorescence intensities of mCherry versus GFP (derived from FFC) (Fig. 2, D and E, and fig. S1) for all the systems tested in this study. The light gray bars represent the relative efficiency as defined by the mean fluorescence intensity of mCherry for each condition divided by cytoplasmic PylRS control (derived from FFC) (Fig. 2, D and E, and fig. S1). Shown are the mean values of at least three independent experiments; error bars represent the SEM.The red box highlights the best performing OTorganelle (OTK2::P1). (D) FFC analysis of the dual-color reporter expressed with the four indicated systems in transfected HEK293Tcells and tRNAPyl in the presence of the ncAA SCO, a lysine derivative with a cyclooctyne side chain. Highly selective and efficient orthogonal translation was observed for the OT organelle (the black arrow indicates a bright, highly mCherry-positive population). Shown in the dot plots are the sums of at least three independent experiments. Axes indicate fluorescence intensity in arbitrary units (all FFC plots are summarized in fig. S1). (E) FFC plots for the OTorganelle selectively translating Opal and Ochre codons only of recruited mCherry185TGA::ms2 and mCherry185TAA::ms2 mRNA, respectively (corresponding cytoplasmic PylRS controls for those stop codons are provided in fig. S1).
P1
Previous studies have established the capacity of the proteins fused-in sarcoma (FUS) and Ewing sarcoma breakpoint region 1 (EWSR1) to form mixed dropletlike structures by means of phase separation. They both contain a prion-like disordered domain that facilitates phase separation into liquid, gel, and solid states (17, 18). In a phase-separated state, these proteins are locally highly concentrated (approximately orders of magnitude) compared with the remaining soluble fraction in the cell. FUS was fused to PylRS, and EWSR1 fused to MCP, and we speculated that this would lead to the formation of droplets in which MCP and PylRS are highly enriched. P1 is denoted FUS::PylRS•EWSR1::MCP.
P2
The Caenorhabditis elegans protein spindle-defective protein 5 (SPD5) has recently been shown to phase-separate into particularly large (several micrometer-sized) droplets (19). In a phase-separated state, SPD5 is locally highly concentrated compared with the remaining soluble fraction in the cytoplasm (by orders of magnitude). We speculate that a protein fused to SPD5 will condense into droplets. Similarly to FUS-EWSR1 droplets, we speculated that PylRS fused to SPD5 and MCP fused to SPD5 will be highly enriched. P2 is denoted SPD5::PylRS•SPD5::MCP.
K1
Certain kinesin truncations constitutively move toward microtubule-plus ends in living cells (20). One such truncated kinesin is KIF13A1-411,ΔP390, and we speculated that PylRS and MCP respectively fused to this kinesin truncation and co-expressed would be locally enriched, owing to spatial targeting to microtubule-plus ends. K1 is denoted KIF13A1-411,ΔP390::PylRS•KIF13A1-411,ΔP390::MCP. (Single-letter abbreviations for the amino acid residues are as follows: A, Ala; C, Cys; F, Phe; N, Asn; P, Pro; and Y, Tyr. In the mutants, other amino acids were substituted at certain locations; for example, Y306A indicates that tyrosine at position 306 was replaced by alanine).
K2
By analogy to K1, we also tested the truncated kinesin KIF16B1-400. K2 is denoted KIF16B1-400:: PylRS•KIF16B1-400::MCP.
We also tested whether combinations of these systems would lead to efficient OT organelles:
K1::P1 = KIF13A1-411,Δp390::FUS::PylRS•KIF13A1-411,ΔP390::EWSR1::MCP
K2::P1 = KIF16B1-400FUS:: PylRS•KIF16B1-400::EWSR1::MCP
K1::P2 = KIF13A1-411,ΔP390::SPD5::PylRS•KIF13A1-411,ΔP390::SPD5::MCP
K2::P2 = KIF16B1-400::SPD5::PylRS•KIF16B1-400::SPD5::MCP
In order to evaluate these assemblers for facilitating functional orthogonal translation of an ms2-tagged mRNA, we designed a dual-reporter construct, in which green fluorescent protein (GFP) and mCherry mutants are simultaneously expressed from two different expression cassettes from one plasmid, ensuring that the mRNA ratio between them is constant across all experiments. Stop codons were introduced at permissive sites into GFP at position 39 (GFP39STOP) and into mCherry at position 185 (mCherry185STOP) (Fig. 2B). Only if stop codon suppression is successful will the corresponding GFP or mCherry be produced. Transfected cells were analyzed by means of fluorescence flow cytometry (FFC); settings were adjusted so that an approximate diagonal results in the FFC plots if GFP and mCherry are expressed from this plasmid by using the conventional cytoplasmic PylRS system, which cannot differentiate mRNAs. A selective and functional OT organelle should selectively express mCherry only if the ms2 loops are fused to the 3’UTR of the mCherry mRNA, leading to appearance of a vertical line in the cytometry plot (Fig. 2B). Unless otherwise reported, all experiments were performed in the presence of tRNAPyl and the ncAA SCO, a widely used and well-characterized lysine derivative, the side chain of which carries a cyclooctyne that can be used in a variety of click-chemistry reactions to install diverse chemical groups onto the protein. As previously reported, this ncAA is efficiently encoded by a Y306A, Y384F double mutant of PylRS (for simplicity we refer to this PylRSAF mutant as PylRS unless otherwise specified) (21–23). Omission of the ncAA serves as a standard negative control and leads to no expression of GFP or mCherry (fig. S1).
We evaluated the performance of each OT system according to their selectivity and relative efficiency. We define selectivity as the ratio r of the mean mCherry FFC signal divided by the mean GFP signal. Final values are expressed as fold selectivity relative to that of cytoplasmic PylRS. We define relative efficiency as the mean mCherry signal of each system divided by the mean mCherry signal of the cytoplasmic PylRS system, which serves as the reference (here defined as 100%). All results on selectivity (Fig. 2C, dark gray positive bars) and efficiency (Fig. 2C, light gray negative bars) are summarized in the bar plot in Fig. 2C, and all corresponding FFC dot plots are summarized in fig. S1, where as selected FFC data are also shown in Fig. 2, D and E.
Combining two assembler strategies yields highly selective and efficient OT systems
The conceptually simplest assembly strategy B (MCP fused to PylRS) showed only a minor selectivity gain of about 1.5-fold, which is concomitant with a 60% decrease in efficiency (Fig. 2C and fig. S1). The OT system P1 (based on phase separation of FUS/EWSR1) performed similarly in terms of selectivity gain. In addition, a 50% decrease in efficiency was measured (Fig. 2, C and D). The P2 system (based on SPD5) showed an approximate twofold selectivity gain accompanied by an almost 90% decrease in efficiency (Fig. 2C and fig. S1). Analogously, we tested the kinesin-based assembly strategy and observed for K1 a twofold selectivity increase, with an efficiency decrease of ~90% (Fig. 2C and fig. S1). The K2 system behaved similarly (Fig. 2, C and D). In total, the selectivity gains were small but robustly detected, indicating that bringing the ncAA aminoacylation activity (the tRNAPyl/ PylRS in the presence of ncAA) in direct proximity of the target mRNA represents a pathway to more selective codon suppression.
Next, we tested the assembler combination strategies (K1::P1, K2::P1, K1::P2, and K2:: P2). For all combinations, we observed at least fivefold selectivity gain, indicating orthogonal translation [the observed selectivity effect is robust across a titration of Amber suppression efficiencies (fig. S2)]. The best performing system on the basis of the fusion of FUS/EWSR1 with KIF16B1-400, K2::P1, exhibited a selectivity of eightfold and 40% efficiency (Fig. 2C, red box). This was also directly obvious from the FFC data, in which the bright, mCherry-positive cell population was clearly retained, whereas GFP expression was minimal (Fig. 2D, black arrow).
To validate that the observed selectivity gain is specific to the ms2-MCP interaction, we further characterized OT organelles by expressing the RS assembler fusion of each OT system without MCP (figs. S1 and S3). As expected, no selective orthogonal translation of ms2-tagged mRNA was observed in those cases. Additionally, we performed a reporter inversion by moving the ms2 loops from the mCherry to the GFP cassette in the dual-color reporter, which as expected inverted selectivity of the system toward dominant GFP expression (fig. S3B). This establishes that the OT system acts selectively on the ms2-tagged RNA.
GCE can also be used to introduce multiple ncAAs into the same POI [(1–3), reviews]. However, only very few publications report on more than one—that is, two- or three-codon suppression in the same protein in eukaryotes—because yields typically suffer compared with single-codon suppression (24–26). Even dual- and triple-Amber proteins were still suppressed with the OT organelle (fig. S4).
To ensure that other ncAAs also can be incorporated by the OT system, we tested another structurally different ncAA (3-iodophenylalanine), which is a phenylalanine derivative instead of a lysine derivative and is encoded by a different PylRS mutant (N346A and C348A) (27). Consistent results were also observed for this system (Fig. 2C and fig. S1).
Because Opal (TGA) and Ochre (TAA) codons are highly abundant in eukaryotic genomes (~52% Opal, ~28% Ochre in the human genome), the Amber codon is by far the most used for GCE in eukaryotes. In addition, genomic approaches to orthogonal translation by removing these codons in the entire eukaryotic genome would be even more challenging than for the Amber codon and are currently beyond the state of the art. However, in the OT system, a simple mutation in the anticodon loop of the tRNAPyl, as well as in the respective codon in the mRNA::ms2, should allow orthogonal translation of these codons. FFC analysis revealed that the OT organelles provide freedom of choice with respect to the stop codon (Fig. 2, C and E, and fig. S1). In fact, Opal suppression showed an 11-fold selectivity increase at 50% efficiency (slightly outperforming Amber suppression). Ochre suppression still showed fivefold selectivity increase, with 20% efficiency.
The OTK2::P1 organelle enables orthogonal translation of proteins of various cellular compartments
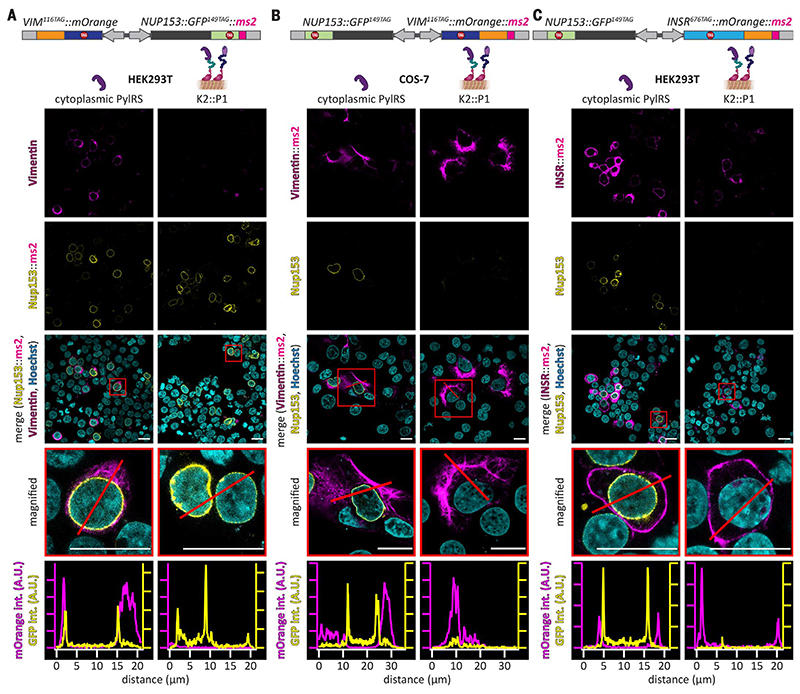
To visualize the power of the OTK2::P1 organelle (our best performing Amber suppression OT organelle in terms of selectivity and efficiency) beyond “simple” reporters, we next aimed to show differential expression of human nucleoporin 153 (Nup153) versus cytoskeletal vimentin. Nup153 locates to the nuclear pore complex and is more than 1500 amino acids long. Hence, its mRNA is approximately sixfold larger than those of the fluorescent protein reporters used above. We used a previously described C-terminal GFP fusion, with an Amber mutation (Nup153:: GFP149TAG) that gave rise to a characteristic nuclear envelope stain in confocal images only if Amber suppression was successful (28). Nup153:: GFP149TAG was then tagged with two ms2 loops (NUP153::GFP149TAG::ms2) and coexpressed from the same plasmid with vimentin containing an Amber codon at position 116 fused to mOrange (VIM116TAG::mOrange). Expression in human embryonic kidney (HEK) 293T cells resulted in production of both proteins in the presence of the cytoplasmic PylRS showing the characteristic nuclear envelope and cytoskeletal staining, respectively. In the presence of the OTK2::P1 organelle, only Nup153::GFP was visible (Fig. 3A, selective nuclear rim stain). Consistent results were also observed in Cercopithecus aethiops kidney (COS-7) cells (fig. S5). Swapping the ms2 loops to vimentin inverted the effect, so that only Vimentin116TAG::mOrange was visible (further experiments for COS-7 and for HEK293T cell experiments are shown in Fig. 3B and fig. S5). This showed that the OTK2::P1 worked for dramatically different mRNAs.
Fig. 3. A versatile OT organelle for selective and efficient orthogonal translation.
(A to C) Confocal images of cells transfected with constructs encoding PylRS (left column) or the OTK2::P1 organelle (right column) for different protein pairs. SCO and tRNAPyl were present in all cases. (A) HEK293T cells were transfected with NUP153::GFP149TAG::ms2 and VIM116TAG::mOrange. (B) VIM116TAG::mOrange::ms2 and NUP153::GFP149TAG transfected in COS-7 cells. More representative examples for (A) using COS-7 cells and for (B) using HEK293T cells are shown in fig. S5. Shown from top to bottom are Vimentin116TAG::mOrange (magenta, characteristic cytoskeletal stain), Nup153::GFP149TAG (yellow, characteristic nuclear envelope stain), overlay with Hoechst (cyan, nuclear stain), magnified images of representative cells (red boxes), and line profiles for the mOrange and GFP channel (red line, magenta and yellow curves, respectively), to highlight that only the ms2-tagged mRNA yields its respective expressed protein if the OTK2::P1 organelle is present. Scale bars, 20 mm. (C) HEK293T cells were transfected with INSR676TAG::mOrange::ms2 and NUP153::GFP149TAG. Shown from top to bottom are INSR676TAG::mOrange (magenta, characteristic plasma membrane stain), Nup153::GFP149TAG (yellow), overlay with Hoechst (cyan), magnified images of representative cells (red boxes), and line profiles for the mOrange and GFP channel (red line, magenta and yellow profiles, respectively), demonstrating selective translation of insr676TAG::mOrange::ms2 mRNA by the OTK2::P1 organelle. Scale bars, 20 μm.
Next, we asked whether it would be possible to selectively express transmembrane proteins by using our synthetic OTK2::P1 organelle. Membrane protein expression represents another layer of translational complexity because ribosomes need to bind the endoplasmic reticulum (ER) during translation, where the proteins are cotranslationally inserted into the membrane. To this end, we used a fusion of insulin receptor 1 with an Amber codon at position 676 with mOrange (INSR676TAG::mOrange), which locates to the plasma membrane and gives rise to a characteristic plasma membrane stain in HEK293T cells (21). This construct was tagged with ms2 loops in the 3’UTR and cloned with Nup153::GFP149TAG into one dual-cassette plasmid. We then expressed it in HEK293T cells either in the presence of the cytoplasmic PylRS system or in the presence of the OTK2::P1 organelle (Fig. 3C). In the presence of the synthetic OTK2::P1 organelle, we observed selective expression of the ms2-tagged protein and the expected plasma membrane localization of INSR676TAG::mOrange, indicating the potential of our organelle to participate in even more complex membrane-associated translational processes.
The OT organelle functions by recruiting ribosomes and tRNAPyl
The above experiments demonstrated the functionality of our synthetic OTK2::P1 organelle. Next, we aimed to study the spatial distribution of the different systems in the cell to understand the basic working mechanism of the OT organelle.
To assess the spatial distribution of PylRS in cells, we used immunofluorescence (IF). We also used fluorescence in situ hybridization (FISH) against tRNAPyl. In contrast to the dual-color reporter used in the FFC experiments above, in all IF/FISH experiments we used a single-color NLS::GFP39TAG reporter fused to ms2 loops to identify cells active in Amber suppression (this yields a green nucleus if Amber suppression is successful). IF and FISH stainings showed that in contrast to cytoplasmic PylRS, the P1 system formed small, intracellular assembler::PylRS droplets (Fig. 4A). This indicated the occurrence of phase separation. The tRNAPyl colocalized well with assembler::PylRS droplets (Fig. 4A), indicating that it could nicely partition into the assembler::PylRS phase. Additional stainings show further colocalization with assembler::MCP (fig. S6). Compared with P1, the P2 system (figs. S6 and S7) showed larger but still multiple dispersed dropletlike structures. Phase separation of proteins is based on exceeding the critical concentration up to which proteins are fully soluble in the cytoplasm. However, a soluble species coexists with the phase-separated species (29) that can contribute to stop codon suppression outside the droplet. The components of the K1 (fig. S7) and K2 (Fig. 4A and fig. S6) systems were mostly observed distributed across the cytoplasm, likely because of binding to the microtubule cytoskeleton, which appears rather distributed throughout the cytoplasm (fig. S8). Because small tRNAs can diffuse rapidly, we believe that a critical factor for the design of an OT organelle is how well the tRNAPyl is confined to few sites in the cell and thus spatially separated and sequestered from the rest of the cytoplasm. None of the P1, P2, K1, or K2 systems displaying high selectivity (Fig. 2C) is consistent with the observation that the systems showed a rather dispersed distribution in the cell.
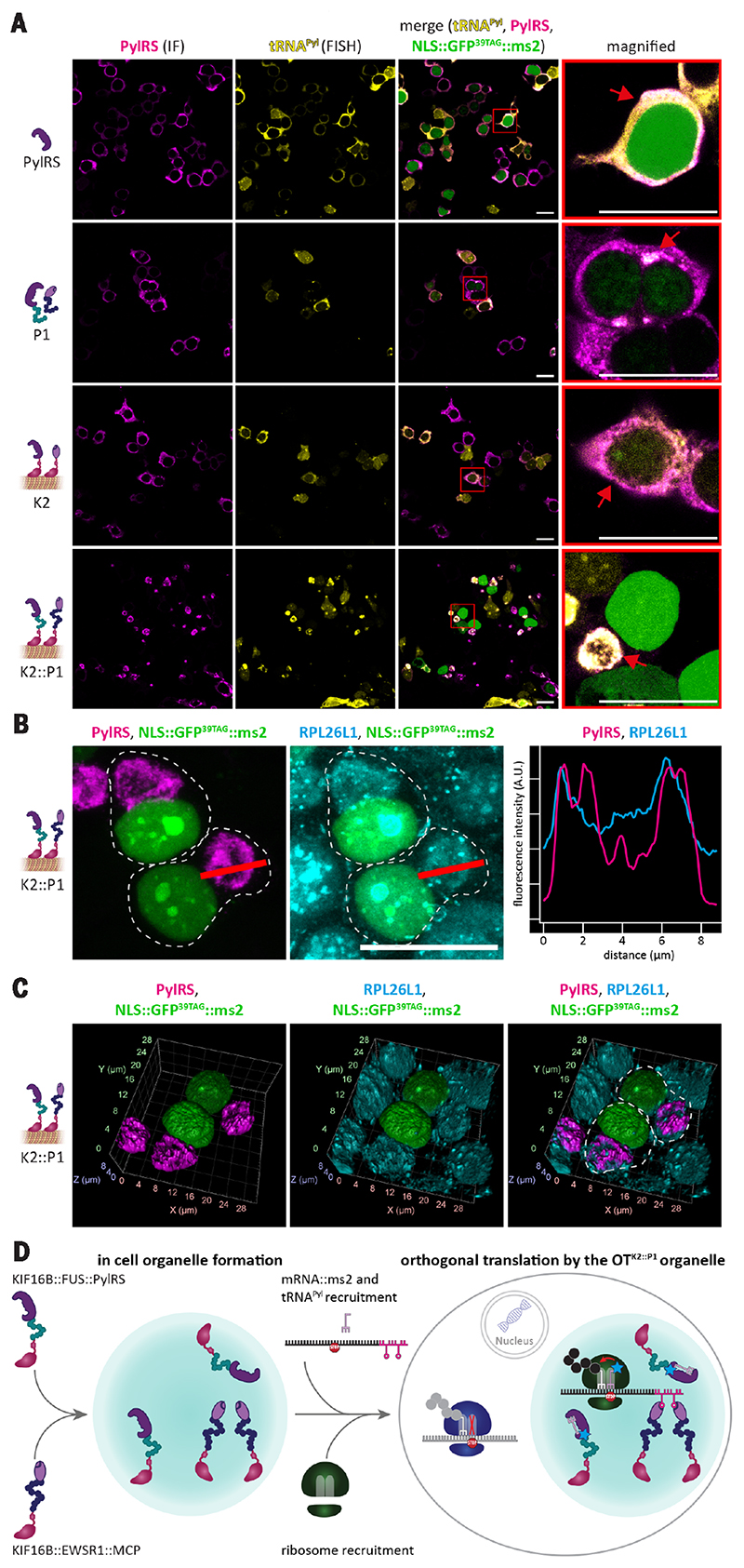
Fig. 4. OT organelles enrich tRNAPyl and ribosomes for orthogonal translation.
(A) IF and FISH imaging of HEK293T cells expressing tRNAPyl and the indicated system. For simplicity and to direct the eye, in all imaging experiments, a simple NLS::GFP39TAG::ms2 (nuclear staining) was used instead of the dual-color reporter. Green-colored nuclei report on faithful Amber suppression of the cells (shown only in overlay). Shown from left to right are IF against PylRS in magenta, FISH against tRNAPyl in yellow, overlay, and magnified images of representative cells (red boxes and red arrows highlight representative structures). Scale bars, 20 μm. (B) Maximum intensity Z-projection of IF image Z-stacks of HEK293T cells transfected with constructs encoding OTK2::P1 organelle and NLS::GFP39TAG::ms2 in the presence of SCO and tRNAPyl. Shown from left to right are IF against PylRS (magenta), IF against RPL26L1 (cyan), merge (NLS::GFP39TAG::ms2 is shown in green in all images), and line profiles for the PylRS and RPL26L1 channels (red line, magenta and cyan curves, respectively). Scale bar, 20 mm. (C) 3D reconstructions of IF images corresponding to those in (B) and movie S1. Shown from left to right are IF against PylRS (magenta), IF against RPL26L1 (cyan), and merge [NLS::GFP39TAG::ms2 in green; gray dashed lines highlight approximate cell boundaries in (B) and (C)]. RPL26L1 staining of the OT organelle demonstrates partial recruitment of ribosomes, which appear to be immersed into the organelle. (D) Working model of OTK2::P1-organelle-enabled orthogonal translation. KIF16B::FUS::PylRS and KIF16B::EWSR1::MCP form a spatially separated organelle inside a living cell. PylRS recruits tRNAPyl and largely depletes its availability in the cytoplasm, whereas MCP recruits ms2-tagged mRNA. Ribosomes and other translation factors are recruited to the organelle for orthogonal translation. Because the charged tRNAPyl is now in close proximity to only the recruited mRNA of the POI and the spatially distinct set of ribosomes, the selected stop codon can only be translated in the immediate vicinity of the synthetic OT organelle. Meanwhile, all other mRNAs that are not recruited to the OT organelle are subject to normal translational processing of the host machinery and available ribosomes in the remaining cytoplasm—that is, a stop codon will terminate translation. The assembly order of the OT organelle is unknown and is only shown here with arrows for illustrative purposes.
However, if we combined both assembler strategies (K1::P1, K2::P1, K1::P2, and K2::P2), we observed the formation of large micrometersized, organelle-like structures in the cytoplasm, which were in most cases localized to few or even a single position per cell. Association of the OT organelle with the microtubule cytoskeleton was also observed (fig. S8). As shown for K2::P1 in Fig. 4A and figs. S6 and S8, mRNA::ms2, tRNAPyl, assembler::PylRS, and assembler::MCP all colocalize to organelle-like structures (other combined assemblers are shown in figs. S6 and S7). The combination of the two assembler strategies—that is, phase separation paired with spatial targeting by kinesin truncations—yielded the best confinement as determined with FISH and IF and the highest selectivity increase.
This is consistent with our hypothesis that the higher spatial segregation and thus higher local concentration of aminoacylated tRNAPyl and mRNA correlates with higher selectivity. This effectively translates into a higher partition coefficient of tRNAPyl into the droplet and thus depletion of tRNAPyl from the cytoplasm, yielding a high concentration gradient between cytoplasm and OT organelle (fig. S7).
We next performed staining for ribosomes to see whether they colocalize to the OTK2::P1 organelle. IF staining of the ribosomal protein RPL26L1 showed its distribution throughout the cell but also an enrichment at the OTK2::P1 organelle [Fig. 4, B and C, two-dimensional (2D) projection and a 3D reconstruction; movie S1; and fig. S8), demonstrating partial ribosome recruitment, tentatively owing to binding to mRNA::ms2 during translation. We conclude that only ribosomes sufficiently immersed into the tRNAPyl gradient can perform codon suppression efficiently. This also visualizes the mobility of a set of ribosomes in the cell. High mobility can also explain why we were even able to express the membrane protein INSR (Fig. 3C) because for orthogonal translation of a membrane protein, two things must happen either sequentially or at the same time: The translating ribosome needs to interact with the ER and with the OT organelle. Together, this body of evidence strongly suggests that selective orthogonal translation happens within close proximity of the OT organelles, potentially even inside the organelle, by a set of recruited ribosomes that are near or fully immersed into a concentrated pool of tRNAPyl. tRNAPyl itself is recruited to the OTK2::P1 organelle because of its affinity for assembler::PylRS and can readily copartition into the droplet to be aminoacylated with its cognate ncAA, whereas assembler::MCP recruits ms2-tagged mRNA. This in turn attracts ribosomes to migrate to the dense phase formed by the dual assembler system (K2::P1 = KIF16B::FUS::PylRS and KIF16B::EWSR1::MCP), which maintains access to other translation factors for translation to function (Fig. 4D). Ribosomes elsewhere in the cytoplasm that are not exposed to tRNAPyl perform their canonical function to terminate translation whenever they encounter a stop codon.
Our route to enable orthogonal translation required only five extra components and represents an important step toward generating semisynthetic eukaryotic organisms that can potentially follow what has been dubbed the “orthogonal central dogma” (30). The OT organelle also represents a general strategy for tailoring complex functions in eukaryotes by mimicking the evolutionary concept to build distinct, but membraneless, organelles inside eukaryotic cells. Proteins such as FUS, EWSR1, and SPD5 have many vital functions in the cell, and we cannot exclude the possibility that the lower expression yield (half, in many cases) we observed is also due to over-expression of components of the OT organelle. The need to combine two different assembly strategies (phase separation-based assemblers with spatial targeting by using kinesin truncations) puts potentially an additional burden on the cell. However, our understanding of phase separation is developing dramatically, such as the identification of amino acid sequences that can be used to make layered droplets (31). We can thus expect future versions of this technology to afford even better selectivity, efficiency, and the ability to bestow additional functions on the OT organelle or for constructing other novel organelles with new functions.
Materials and methods
Cell culture
HEK293T cells (ATCC CRL-3216) and COS-7 cells (Sigma-Aldrich 87021302) were maintained in Dulbecco’s modified Eagle’s medium (DMEM, Gibco 41965-039) supplemented with 1% penicillin-streptomycin (Sigma-Aldrich P0781), 1% L-Glutamine (Sigma-Aldrich G7513), 1% sodium pyruvate (Life Technologies 11360), and 10% FBS (Sigma-Aldrich F7524). Cells were cultured at 37°C in a 5% CO2 atmosphere and passaged every 2-3 days up to 20 passages.
In all cases, cells were seeded 15-20 hours prior to transfection at a density resulting in 70-80% confluency at the time of transfection. Flow cytometry was performed using 24-well plates with plastic bottom (Nunclon Delta Surface ThermoScientific). IF labeling and FISH were performed on 24-well plates with glass bottom (Greiner Bio-One) or four-well chambered Lab-Tek #1.0 borosilicate coverglass (ThermoFisher).
Constructs, cloning, and mutagenesis
Dual-color reporters: The dual fluorescent protein reporters were cloned in a pBI-CMV1 vector (Clontech 631630), with ms2 tagged fluorescence protein (mRNA) version in one multiple cloning site and ms2 free version in the other. GFP39TAGI or mCherry185TAG were used as N-terminal fusions with nuclear localization sequences (NLS). Similar reporters for Ochre and Opal suppression were prepared (with GFP39TAA, mCherry185TAA and GFP39TGA, mCherry185TGA, respectively).
NLS::GFP39TAG::ms2 reporter: NLS::GFP39TAG was cloned with two copies of ms2 loops into the pBI-CMV1 vector as a reporter for successful Amber suppression for imaging experiments.
Double and triple Amber GFP: Position 149 and subsequently position 182 of GFP were mutated to TAG to obtain pBI-CMV constructs with multiple amber codons, these constructs did not have mCherry in the second multiple cloning site.
OT organelle constructs: Pyrrolysyl tRNA was cloned under the control of a human U6 (hU6) promoter, and all other constructs were under CMV (cytomegalovirus) promoters cloned in the pcDNA3.1 (Invitrogen V86020) vector. MCP was cloned from the Addgene plasmid #31230 and FUS from the Addgene plasmid #26374. In all FUS fusions, amino acids 1-478 were used, replacing the C-terminal NLS region by a Flag-tag. In all pyrrolysine synthetase fusions the previously reported efficient NES::PylRSAF (Y306A, Y384F) sequence was used (21, 28). NES::PylRSAA (N346A, C348A) was cloned via site-directed mutagenesis starting from NES::PylRSWT. The SPD5 gene was ordered from Genewiz and fused to MCP and PylRSAF via restriction cloning. KIF13A1-411 and KIF16B1-400 were cloned from human cDNA and inserted into pcDNA3.1 via restriction cloning. P390 of KIF13A1-411 was removed via site-directed mutagenesis. KIF13A1-411,ΔP390 and KIF16B1-400 fusions with MCP, PylRSAF, EWSR1::MCP, FUS::PylRSAF, FUS::PylRSAA, SPD5::MCP, and SPD5::PylRSAF were assembled via Gibson assembly (32).
Constructs for differential imaging experiments (Fig. 3): To selectively express Nup153:: GFP149TAG and Vimentin116TAG::mOrange, one gene was first inserted with ms2 loops into pBI-CMV1 (21). Subsequently, the other gene was inserted without ms2 loops. INSR676TAG::mOrange was fused to ms2 loops by replacing VIM116TAG::mOrange in the pBI vector bearing NUP153::GFP149TAG and VIM116TAG::mOrange::ms2 to yield a bicistronic vector with INSR676TAG::mOrange in one and NUP153::GFP149TAG in the other cassette.
Multicistronic Amber suppression vectors: For ease of experiments with the OTK2::P1 organelle we generated multicistronic vectors harboring all necessary components. To assemble multicistronic Amber suppression vectors, first one copy of tRNAPyl under the control of a hU6 promoter was inserted into the pBI-CMV1 vector via Gibson assembly. Subsequently, first KIF16B::FUS::PylRSAF and finally KIF16B:: EWSR1::MCP were inserted via Gibson assembly. Alternatively, a previously published pcDNA3.1 based construct (21) expressing NES::PylRSAF under a CMV promoter and tRNAPyl under a hU6 promoter was used. Alternatively, hU6-tRNAPyl and NES::PylRSAF or the components of OTK2::P1 were inserted into a pDonor vector (GeneCopoeia). These constructs were used for all experiments in COS-7 and for ribosome/tubulin imaging experiments.
OTK2::P1 construct tagging with iRFP (fig. S8): To exclude the possibility of staining artefacts we replaced the KIF16B::FUS::PylRSAF in the multicistronic pBI-CMV1 vector with a KIF16B:: iRFP::FUS::PylRSAF fusion via Gibson assembly. The final construct additionally encodes KIF16B:: EWSR1::MCP and tRNAPyl.
Transfections and used ncAAs
Transfections of HEK293T cells were performed with polyethylenimine (PEI, Sigma-Aldrich 408727) using 3 μg PEI per 1 μg DNA (1200 ng total DNA, diluted in DMEM without Phenol Red, Gibco11880-028). COS-7 cells were transfected using the JetPrime reagent (PeqLab) according to the manufacturer’s recommendations at a ratio of 1:2 (1000 ng total DNA).
For Amber suppression system tests, cells were transfected at a ratio of a 1:1:1:1 with POITAG vectors, tRNAPyl, PylRS assembler fusions and MCP assembler fusions or mock constructs. 4-6 hours after transfection, the medium was swapped to fresh one containing ncAA. HEK293T cells were analyzed one day after transfection, while COS-7 cells were processed after two days.
Stock and working solutions for all the used ncAAs were prepared as described in previous work (33). SCO (cyclooctyne lysine, SiChem SC-8000) was used at a final concentration of 250 μM, while 3-Iodophenylalanine (Chem-Impex International Inc.) was used at a final concentration of 1 mM. SCO is efficiently recognized by PylRSAF (Y306A, Y384F) (23) while 3-Iodophenylalanine is recognized by PylRSAA (C346A, N348A) (27).
Flow cytometry
HEK293T cells were harvested 1 day after transfection by removing the medium, resuspending the cells in 1xPBS (phosphate buffered saline) and passing them through 100 μm nylon mesh.
Data acquisition was performed on an LSRFortessa SORP Cell Analyzer (BD). Analysis was done using the FlowJo software (FlowJo). Cells were first gated by cell type (using FSC-A x SSC-A parameters) and then by single cell (SSC-A x SSC-W). The workflow of cell gating is shown in Fig. S9. Each shown FFC plot is the sum of three independent biological replicates from which the mean and SEM were calculated. At least 130000 single cells were analyzed per condition. Lastly, fluorescence was acquired in the 488-530/30 channel for GFP signal and in the 561-610/20 channel for mCherry signal.
IF labeling, FISH, and confocal imaging
IF: For immunolabeling experiments, cells were rinsed with PBS, fixed in 2% paraformaldehyde in 1xPBS at room temperature (RT) for 10 min.
Alternatively, if cells were stained for α-Tubulin, they were rinsed with DMEM without Phenol Red and subsequently fixed in a buffer to preserve microtubule structures (100 mM 1,4-Piperazinediethanesulfonic acid (PIPES), 1 mM MgCl2, 0.1 mM CaCl2, 0.1% Triton-x-100 and 2% PFA; pH = 7) for 10 min at RT.
Subsequently, cells were rinsed with PBS and permeabilized with 0.5% Triton-x-100 solution in 1xPBS for 15 min at RT and rinsed twice prior to blocking. Samples were blocked in 3% BSA in 1xPBS for 90 min at RT, after which incubation with the primary antibody was done overnight at 4°C in blocking solution (AbPylRS (1 μg/mL (21)), AbMCP (Merck ABE76-I, 1:333), Abα-Tubulin (Sigma-Aldrich T6199, 2 μg/mL) and/or AbRPL26L1 (Abcam ab137046, 1:200)). The next day, cells were rinsed with PBS and incubated with secondary antibody (ThermoFisher A-21471, A-31553 and/or A-21246, at 2 or 4 μg/mL in blocking solution) for 60 min at RT. Then, cells were rinsed with PBS and fresh PBS was added for imaging.
If only DNA was stained, cells were fixed and permeabilized the same way prior to staining with Hoechst 33342 (Sigma-Aldrich B2261) at 1 μg/mL in 1xPBS for 10 min at RT. Subsequently, cells were rinsed with PBS and fresh PBS was added for imaging.
FISH: FISH experiments were performed one day after transfection analogously to described previously (21). Briefly, the hybridization protocol was adapted for 24-well plates from Pierce et al. (34). In general, IF stainings appear crisper then FISH stainings.
For imaging of only tRNAPyl, the hybridization probe (5’-(Cy5)-CTAACCCGGCTGAACGGATTTAG-AGTCCATTCGATC-3’) was used at 0.25 μM (hybridization at 37°C, overnight). After four washes with saline sodium citrate buffer (SSC) and one wash with Tris-HCl•NaCl buffer (TN), cells were incubated for 1 hour at RT in 3% BSA prior to IF labeling. Cells were incubated with primary antibodies for 2 hours at RT, rinsed with PBS and incubated with secondary antibodies for 2 hours at RT (antibodies described above). Finally, cells were rinsed with PBS and fresh PBS was added for imaging.
For imaging of both tRNAPyl and mRNA::ms2, the hybridization probe for tRNAPyl (5’-(DIG)-CTAACCCGGCTGAACGGATTTAGAGTCCATTC-GATC-3’) was used at 0.16 μM, and the probe for ms2 (5’-(Alexa647)-CTGCAGACATGGGTGATCCTCA-TGTTTTCTA) was used at 0.75 μM. After the SSC washes, cells were incubated for 1 hour at RT in blocking buffer (0.1 M TrisHCl, 150 mM NaCl, 1x blocking reagent (Sigma-Aldrich 11096176001). Then, cells were incubated with anti-digoxigenin-fluorescein antibody (Sigma-Aldrich 11207741910) at a 1:200 dilution in blocking buffer overnight at 4°C. The next day, 3 washes of 5 min were done in Tween buffer (0.1 M TrisHCl, 150 mM NaCl, 0.5% Tween20), before cells were rinsed with PBS and fresh PBS was added for imaging.
Imaging: Confocal images were acquired on a Leica SP8 STED 3X microscope using the 405 nm (for Hoechst), 488 nm (for fluorescein, GFP), 548 nm (mOrange), 594 nm (for Alexa594) and 647 nm (for Alexa647, Cy5) laser lines for excitation. For HEK293T and COS-7 cells a 63x/1.40 oil immersion objective was used. IF images with ribosomes, microtubules, and/or iRFP were taken on an Olympus Fluoroview FV3000 microscope using 405 nm (Alexa405), 488 nm (GFP), 594 nm (for Alexa594), and 640 nm (for Alexa 647 and iRFP) lasers for excitation with a 60x/1.40 oil immersion objective for acquisition. Images were processed using FIJI software.
3D Reconstruction
3D reconstructions in Fig. 4C and corresponding movie S1 were made by using the arivis Vision4D software (arivis AG).
Supplementary Material
Research Article Summary.
Introduction
The ability to engineer translation of noncanonical (unnatural) amino acids (ncAAs) site-specifically into proteins in living cells greatly expands the chemical space that can be used to control, tailor, and study cellular function. However, translation is a complex multistep process in which at least 20 different aminoacylated tRNAs, their cognate tRNA synthetases, ribosomes, and other factors need to act in concert to synthesize a polypeptide chain encoded by an mRNA transcript. To minimize interference with the host machinery, we aimed to engineer fully orthogonal translation into eukaryotes: to encode a new functionality in response to a specific codon in only one targeted mRNA, leading to site-specific ncAA incorporation only into the selected protein of choice. Although codon specificity can be achieved with genetic code expansion (GCE), this technology relies on using an orthogonal tRNA/tRNA synthetase pair (one that does not cross-react with any of the endogenous pairs) to reprogram a stop codon. Most commonly, the Amber (UAG) stop codon is used (20% abundance in human cells), and in principle, stop codon suppression can happen for every cytoplasmic mRNA that terminates naturally on this codon. Here, we present a strategy to generate a distinctly expanded genetic code for only selected mRNAs.
Rationale
We hypothesized that it should be possible to create an orthogonal translation system by spatially enriching the key components of the GCE machinery in an orthogonally translating (OT) synthetic designer organelle and by targeting a specific mRNA to it. In order to perform protein translation, such an OT organelle would need to be readily accessible to the entire translational machinery of the host, thus precluding membrane encapsulation. Inspired by the concept of phase separation, which is used by cells to concentrate specific proteins and RNA locally, we hypothesized that it might be possible to use this principle to create such membraneless OT organelles. In our design, only a spatially distinct set of ribosomes associated with OT organelles can use the aminoacylated suppressor tRNA and thus will decode Amber codons only in the selected mRNA translated by the OT organelle, leading to a protein containing the ncAA.
Results
To bring the modified suppressor tRNA and the translated mRNA of choice in close proximity to each other, we used different strategies to generate highly concentrated assemblies and spatial separation inside cells:
-
(i)
proteins undergoing phase separation in cells [fused-in sarcoma (FUS), Ewing sarcoma breakpoint region 1 (EWSR1), and spindle-defective protein 5 (SPD5), which contain long intrinsically disordered domains] and
-
(ii)
kinesin motor proteins, which spatially enrich at microtubule plus ends (KIF13A and KIF16B). We fused each of these to the suppressor tRNA synthetase as well as an RNA-binding domain major capsid protein (MCP) that binds to a specific RNA motif (ms2 loops) engineered into the untranslated region of the mRNA of choice, forming an ms2-MCP complex. Each of these approaches yielded the desired local enrichment and preferential stop codon suppression of the mRNA tagged with ms2 loops. However, by far the best performing system was a combination of phase and spatial separation, which typically formed amicrometer-sized organelle-like structure per cell. Cells that contained this organelle efficiently and selectively performed Amber suppression of only the targeted mRNA. We were able to demonstrate the utility and robustness of these OT organelles by selectively decoding any of the three stop codons in a variety of proteins with different ncAA functionalities in two different mammalian cell lines.
Conclusion
Our results show how to combine phase and spatial separation inside cells to allow the concentration of a custom designed task into a distinct specially designed membraneless organelle. We successfully demonstrated that specific and selective protein translation could be achieved within these OT organelles, which allowed the introduction of noncanonical functionalities into proteins in a codon-specific and mRNA-selective manner. The system only requires engineering five components into the cell and can be reprogrammed to other stop codons in a single step. We expect this concept to be a scalable platform for further organelle engineering and to provide a route toward generation of semisynthetic eukaryotic cells and organisms.
Membraneless OT organelles enable mRNA-specific GCE in eukaryotes.
OT organelles are designed organelles enriched in a suppressor tRNA/tRNA synthetase pair and a specific mRNA binding domain (MCP) by means of using an assembler protein (such as FUS and/or KIFs). A spatially distinct set of ribosomes associated with the OT organelle preferentially translates recruited mRNAs tagged with ms2 loops to yield the selected protein with the targeted site-specific noncanonical functionality.
Acknowledgments
We thank I. Schneider as well as all members of the Lemke laboratory, in particular C. Koehler and J. Caria, for helpful discussions. We thank the European Molecular Biology Laboratory (EMBL) flow cytometry core facility and the advanced light microscopy facility for expert assistance.
Funding
The Lemke laboratory acknowledges generous support from European Research Council (ERC) SMPFv2.0, SPP1623, and SFB1129 (Projektnummer 240245660 funded by Deutsche Forschungsgemeinschaft) and the Gutenberg Research College (GRC).
Footnotes
Author contributions: E.A.L. conceived the project. C.D.R. and G.E.G. designed and performed all experiments. C.D.R., G.E.G., and E.A.L. analyzed all of the data and cowrote the manuscript.
Competing interests: C.D.R., G.E.G., and E.A.L. have filed a patent application on OT organelle technology (EP 19157257.7).
Data and materials availability
All data are available in the main text or the supplementary materials. All plasmids can be obtained upon reasonable request via an EMBL materials transfer agreement (free of charge for noncommercial purposes).
References and Notes
- 1.Liu CC, Schultz PG. Adding new chemistries to the genetic code. Annu Rev Biochem. 2010;79:413–444. doi: 10.1146/annurev.biochem.052308.105824. [DOI] [PubMed] [Google Scholar]
- 2.Lemke EA. The exploding genetic code. ChemBioChem. 2014;15:1691–1694. doi: 10.1002/cbic.201402362. [DOI] [PubMed] [Google Scholar]
- 3.Chin JW. Expanding and reprogramming the genetic code. Nature. 2017;550:53–60. doi: 10.1038/nature24031. [DOI] [PubMed] [Google Scholar]
- 4.Neumann H, Wang K, Davis L, Garcia-Alai M, Chin JW. Encoding multiple unnatural amino acids via evolution of a quadruplet-decoding ribosome. Nature. 2010;464:441–444. doi: 10.1038/nature08817. [DOI] [PubMed] [Google Scholar]
- 5.Orelle C, et al. Protein synthesis by ribosomes with tethered subunits. Nature. 2015;524:119–124. doi: 10.1038/nature14862. [DOI] [PubMed] [Google Scholar]
- 6.Fried SD, Schmied WH, Uttamapinant C, Chin JW. Ribosome Subunit Stapling for Orthogonal Translation in. E coli Angew Chem. 2015;127:12982–12985. doi: 10.1002/anie.201506311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Isaacs FJ, et al. Precise manipulation of chromosomes in vivo enables genome-wide codon replacement. Science. 2011;333:348–353. doi: 10.1126/science.1205822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lajoie MJ, et al. Genomically recoded organisms expand biological functions. Science. 2013;342:357–360. doi: 10.1126/science.1241459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ostrov N, et al. Design, synthesis, and testing toward a 57-codon genome. Science. 2016;353:819–822. doi: 10.1126/science.aaf3639. [DOI] [PubMed] [Google Scholar]
- 10.Wang K, et al. Defining synonymous codon compression schemes by genome recoding. Nature. 2016;539:59–64. doi: 10.1038/nature20124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Y, et al. A semi-synthetic organism that stores and retrieves increased genetic information. Nature. 2017;551:644–647. doi: 10.1038/nature24659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thompson DB, et al. The future of multiplexed eukaryotic genome engineering. ACS Chem Biol. 2018;13:313–325. doi: 10.1021/acschembio.7b00842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brangwynne CP, et al. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science. 2009;324:1729–1732. doi: 10.1126/science.1172046. [DOI] [PubMed] [Google Scholar]
- 14.Li P, et al. Phase transitions in the assembly of multivalent signalling proteins. Nature. 2012;483:336–340. doi: 10.1038/nature10879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hyman AA, Weber CA, Julicher F. Liquid-liquid phase separation in biology. Annu Rev Cell Dev Biol. 2014;30:39–58. doi: 10.1146/annurev-cellbio-100913-013325. [DOI] [PubMed] [Google Scholar]
- 16.Bertrand E, et al. Localization of ASH1 mRNA particles in living yeast. Mol Cell. 1998;2:437–445. doi: 10.1016/S1097-2765(00)80143-4. [DOI] [PubMed] [Google Scholar]
- 17.Altmeyer M, et al. Liquid demixing of intrinsically disordered proteins is seeded by poly(ADP-ribose) Nat Commun. 2015;6:8088. doi: 10.1038/ncomms9088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Patel A, et al. A liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell. 2015;162:1066–1077. doi: 10.1016/j.cell.2015.07.047. [DOI] [PubMed] [Google Scholar]
- 19.Woodruff JB, et al. The centrosome is a selective condensate that nucleates microtubules by concentrating tubulin. Cell. 2017;169:1066–1077.:e10. doi: 10.1016/j.cell.2017.05.028. [DOI] [PubMed] [Google Scholar]
- 20.Soppina V, et al. Dimerization of mammalian kinesin-3 motors results in superprocessive motion. Proc Natl Acad Sci USA. 2014;111:5562–5567. doi: 10.1073/pnas.1400759111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nikić I, et al. Minimal tags for rapid dual-color live-cell labeling and super-resolution microscopy. Angew Chem. 2014;53:2245–2249. doi: 10.1002/anie.201309847. [DOI] [PubMed] [Google Scholar]
- 22.Plass T, et al. Amino acids for Diels-Alder reactions in living cells. Angew Chem. 2012;51:4166–4170. doi: 10.1002/anie.201108231. [DOI] [PubMed] [Google Scholar]
- 23.Plass T, Milles S, Koehler C, Schultz C, Lemke EA. Genetically encoded copper-free click chemistry. Angew Chem. 2011;50:3878–3881. doi: 10.1002/anie.201008178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xiao H, et al. Genetic incorporation of multiple unnatural amino acids into proteins in mammalian cells. Angew Chem. 2013;52:14080–14083. doi: 10.1002/anie.201308137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schmied WH, Elsasser SJ, Uttamapinant C, Chin JW. Efficient multisite unnatural amino acid incorporation in mammalian cells via optimized pyrrolysyl tRNA synthetase/tRNA expression and engineered eRF1. J Am Chem Soc. 2014;136:15577–15583. doi: 10.1021/ja5069728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Z, et al. Construction of an inducible stable cell line for efficient incorporation of unnatural amino acids in mammalian cells. Biochem Biophys Res Commun. 2017;489:490–496. doi: 10.1016/j.bbrc.2017.05.178. [DOI] [PubMed] [Google Scholar]
- 27.Wang YS, et al. Genetic incorporation of twelve meta-substituted phenylalanine derivatives using a single pyrrolysyl-tRNA synthetase mutant. ACS Chem Biol. 2013;8:405–415. doi: 10.1021/cb300512r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nikic I, et al. Debugging eukaryotic genetic code expansion for site-specific click-PAINT super-resolution microscopy. Angew Chem. 2016;55:16172–16176. doi: 10.1002/anie.201608284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Banani SF, et al. Compositional control of phase-separated cellular bodies. Cell. 2016;166:651–663. doi: 10.1016/j.cell.2016.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu CC, Jewett MC, Chin JW, Voigt CA. Toward an orthogonal central dogma. Nat Chem Biol. 2018;14:103–106. doi: 10.1038/nchembio.2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Simon JR, Carroll NJ, Rubinstein M, Chilkoti A, López GP. Programming molecular self-assembly of intrinsically disordered proteins containing sequences of low complexity. Nat Chem. 2017;9:509–515. doi: 10.1038/nchem.2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gibson DG, et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 33.Nikic I, Kang JH, Girona GE, Aramburu IV, Lemke EA. Labeling proteins on live mammalian cells using click chemistry. Nat Protoc. 2015;10:780–791. doi: 10.1038/nprot.2015.045. [DOI] [PubMed] [Google Scholar]
- 34.Pierce JB, Chafe SC, Eswara MB, van der Merwe G, Mangroo D. Strategies for investigating nuclear-cytoplasmic tRNA dynamics in yeast and mammalian cells. Methods Cell Biol. 2014;122:415–436. doi: 10.1016/B978-0-12-417160-2.00019-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available in the main text or the supplementary materials. All plasmids can be obtained upon reasonable request via an EMBL materials transfer agreement (free of charge for noncommercial purposes).