Abstract
Background and Aims
To influence host and disease phenotype, compositional microbiome changes, which have been demonstrated in patients with primary sclerosing cholangitis (PSC), must be accompanied by functional changes. We therefore aimed to characterize the genetic potential of the gut microbiome in PSC compared to healthy controls (HCs) and inflammatory bowel disease (IBD).
Methods
Fecal DNA from two cohorts (one Norwegian and one German), in total comprising 136 patients with PSC (58% with IBD), 158 HCs and 93 IBD patients without PSC were subjected to metagenomic shotgun sequencing, generating 17 billion paired end sequences, which were processed using HUMAnN2 and MetaPhlAn2, and analyzed using generalized linear models and random effects meta-analyses.
Results
PSC patients had fewer microbial genes compared to HC (P<.0001). Compared to HC, PSC patients showed enrichment and increased prevalence of Clostridium species, and a depletion of e.g., Eubacterium spp. and Ruminococcus obeum. Patients with PSC showed marked differences in the abundance of genes related to vitamin B6 synthesis and branched chain amino acid (BCAA) synthesis (Qfdr<.05). Targeted metabolomics of plasma from an independent set of PSC patients and controls found reduced concentrations of vitamin B6 and BCAAs in PSC (P<.0001), which strongly associated with reduced liver transplantation-free survival (log-rank P<.001). No taxonomic or functional differences were detected between PSC patients with and without IBD.
Conclusion
The gut microbiome of PSC patients exhibits large functional differences compared to HC, including microbial metabolism of essential nutrients. Alterations in related circulating metabolites associated with disease course, suggesting that microbial functions may be relevant for the disease process in PSC.
Keywords: microbiome, primary sclerosing cholangitis, vitamin B, branched chain amino acids
Introduction
Primary sclerosing cholangitis (PSC) is a chronic progressive cholestatic liver disease of unknown etiology, characterized by multifocal inflammation and fibrosis of the biliary tree.1 Up to 80% of PSC patients are diagnosed with concomitant inflammatory bowel disease (IBD), thus PSC is a prototypical disease of the gut-liver axis.
Compositional changes in the fecal bacterial microbiome of PSC patients have been reported in several cross-sectional studies.2–5 Changes are also present in pediatric patients,6 but so far there are few established links between the gut microbiome and severe disease or late stage disease like cirrhosis.7 There are also disturbances in other important members of the microbiome, like fungi, and the interplay between these microbial communities.4 These data are all cross-sectional, limiting the possibility for conclusions on causality.
In mouse models, inducing a germ-free state may either improve or worsen the biliary disease, depending on disease mechanisms.8,9 In humans, trials with antibiotics improves liver biochemistry in PSC,10 while a pilot trial of fecal microbiota transplantation was too small to allow any firm conclusions.11 Furthermore, specific bacteria found in human PSC stool may modify experimental biliary disease in mice.12 Overall, both human and experimental evidence suggest that gut microbes may act as disease modifiers in PSC. The mechanisms underlying these effects are so far not known, but current data suggest that altered bile acid homeostasis, increased bacterial translocation or uncharacterized immune alterations could be relevant.7–10
A major limitation of the available data in PSC is the lack of microbe identification on the species level when using 16S rRNA gene sequencing. Furthermore, one important possibility is that bacterial functions and the by-products of their activity, e.g. metabolites, could be more important than the composition of the microbiome. The majority of compounds entering the body through the oral route may potentially be metabolized by gut microbes, resulting in a large number of microbial by-products entering the circulation of the host, potentially affecting human organs. One example for such a modified metabolite is the bacteria-dependent metabolite trimethylamine-N-oxide (TMAO), which has been associated with reduced liver transplantation-free survival.13 Both species level identification and quantification of the bacterial functional potential can be analyzed using full shotgun metagenomic sequencing. The present study therefore aimed to apply large-scale metagenomic sequencing in two independent cohorts of patients with PSC to define which species are associated with PSC, and whether specific microbial functions are altered, possibly pointing to pathways relevant for PSC pathogenesis and progression.
Materials And Methods
Participants
We collected fecal samples from non-transplanted PSC patients from two independent cohorts published previously14: The Norwegian cohort was included in the Norwegian PSC Research Center biobank at Oslo University Hospital, while PSC patients of the German cohort were recruited at the University Medical Center Hamburg-Eppendorf. The diagnosis of PSC was made in accordance with clinical guidelines and typical findings on cholangiography or liver biopsy, and all PSC patients had undergone screening for IBD.15,16 Plasma samples from 191 Norwegian PSC patients and 48 controls (collected 2008-2015) were included for analysis of metabolites and survival from sample date to liver transplantation or death. For an overview of the cohorts and the study design, see Supplementary Figure 1.
Norwegian healthy population controls were randomly selected from donors registered in the national Norwegian Bone Marrow Donor Registry (Oslo, Norway), while German healthy controls were recruited through the PopGen biobank17 and the FoCus cohort.18
Norwegian IBD patients without a medical history of liver disease, and in clinical remission, were recruited in an outpatient setting from Oslo University Hospital (Oslo, Norway). German patients with IBD included as part of the KINDRED program were recruited through the PopGen biobank. IBD diagnosis was based on colonoscopy and histology and accepted criteria.19,20 Routine biochemical parameters for all participants were retrieved through hospital databases and the respective biobank databases. Mayo risk scores were calculated using the algorithm for the revised Mayo risk score,21 while the Amsterdam-Oxford model for PSC was calculated according to the original formula.22 Transient elastography measurements were obtained with Fibroscan (Echosens, Paris, France), and ELF score calculated as previously described.23
Ethics
The study was performed in accordance with the declaration of Helsinki. Written informed consent was obtained from all study participants. Ethical approval was obtained from the respective Local Ethics Committees (Norway: Regional Committee for Medical and Health Research Ethics in South-Eastern Norway (reference number 2015/2140); Germany, Hamburg (reference number MC-111/15), Kiel (reference numbers A148/14, A117-13, A156-03)). All authors had access to the study data and reviewed and approved the final manuscript.
Fecal sample collection, DNA extraction and exclusion criteria
Fecal samples were collected and DNA extracted as described previously (see Supplementary Material for details).2,14 All participants with previous bowel resection, small-duct PSC, other chronic liver diseases, a gastrointestinal stoma, or exposed to antibiotics in preceding six weeks preceding sampling were excluded.
Metagenome library preparation, sequencing and quantitative real time (RT) PCR
Quality and quantity of DNA samples were determined using Qubit measurements and the Genomic DNA ScreenTape® (Agilent, Santa Clara, United States). Subsequently, metagenomic library preparation was performed as described previously by using Illumina Nextera DNA Library Preparation Kit (Illumina, San Diego, CA, USA).24 Sequencing was performed with 2x125 bp on a HiSeq 2500 platform (Illumina (German healthy controls and all Norwegian samples)) or with 2x150 bp on a HiSeq 4000 platform (Illumina (German PSC and IBD patients)). Primers for quantitative RT-PCR are given in Supplementary Table 1 and further details can be found in the Supplementary Methods.
Post-sequencing processing
Quality control and filtering of the raw reads was done with KneadData (v0.7.0), and Bowtie2 (v2.3.4.2). Taxonomic and functional profiling was performed using HUMAnN2 (v0.11.2),25 incorporating MetaPhlAn226 (v2.7.8) and DIAMOND (v0.8.38). Further details on bioinformatic handling can be found in the Supplementary Material.
Plasma metabolites
All analyses of metabolites in plasma were performed at BEVITAL (Bergen, Norway). Vitamers were analyzed by liquid chromatography-tandem mass spectroscopy (LC-MS/MS),27 whereas amino acids and related metabolites were analyzed by gas chromatography-tandem mass spectroscopy (GC-MS/MS).28
Statistical analysis
Comparison of categorical variables was performed using the Chi-square test, or Fisher’s exact test where appropriate, and the Cochran–Mantel–Haenszel (CMH) test across cohorts. Mann-Whitney U test was applied for continuous clinical variables, metabolomic data and analysis of gene-family-based alpha diversity measures. When adjusting for clinical co-variates in analyses of gene richness, linear regression was used. Kaplan-Meier plots with log-rank test and Cox proportional hazards regression analyses were used to investigate associations with endpoints (death or liver transplantation), all perfomed in SPSS (v25; IBM, New York, USA). Microbiome co-correlation networks were made using R (v3.6.1) using the SparCC algorithm with correlations cut-offs set at r>0.40 and unadjusted P<.05. Beta diversity analyses (non-constrained ordination of genus-level Bray-Curtis dissimilarities) were performed via permutational multivariate analysis of variance using the 'vegan' package in R with 10,000 permutations. For correlation analyses, Spearman’s rank correlation test was utilized. Analyses of differential abundance of single taxa and functional features (enzyme families (level-4 Enzyme Commission (EC) categories), KEGG ontology and MetaCyc pathways) were first performed within each country including age, BMI and gender as covariates in the model unless otherwise stated, followed by a meta-analysis. Samples with missing information for co-variates were excluded from the analyses. A generalized model with zero-inflated beta distribution was fitted using the R package 'GAMLSS' (see further details in Supplementray Methods). Lastly, a random effects meta-analyses was then performed using the R package 'metamicrobiomeR', allowing for variance in the effect sizes between the two cohorts, using the estimates and standard errors from the per-country analyses described above. An overview of the main health-state and drug usage comparisons analyzed (g1-g9), can be found in Supplementary Table 2. False-discovery rate (FDR) was calculated according to Benjamini-Hochberg, FDR-corrected P-values were denoted Q fdr. The machine-learning procedure random forest ('randomForest' package in R) was used to predict phenotypes based on the metagenome data. The cohorts were randomly split into training and prediction sets with a ratio of 8:2, or one country used for training and the other for prediction, and the procedure was repeated 100 times for each prediction task. Comparison of predicted outcomes was summarized with area under the receiver operating characteristic (ROC) curve (AUC), Matthew’s correlation coefficient (MCC) and F1 measure. For further details see the Supplemental Material.
Results
After sequencing and quality control of 17 billion paired end sequences, 5.5 terabytes of data were available for analysis from 387 individuals; in total 136 patients with PSC, 158 healthy controls, and 93 IBD controls (Table 1, Supplementary Table 3&4).
Table 1. Summary of PSC and HC characteristics in the study cohorts.
Demographics for IBD controls are given in Supplementary Table 4. None of the HCs used any of the stated medications. Continuous data given as median (min-max), count data as n (%).
| German cohort | Norwegian cohort | PSC G vs N |
HC G vs N |
|||||
|---|---|---|---|---|---|---|---|---|
| PSC (n = 67) |
HC (n = 120) |
P-value | PSC (n = 69) |
HC (n = 38) |
P-value | P-value | P-value | |
| Age, years | 47.0 (17–74) | 46.5 (21–72) | .353 | 48.0 (21–69) | 47.0 (35–61) | .852 | .880 | .269 |
| Sex, male | 43 (64.2) | 53 (44.2) | .010 | 45 (65.2) | 24 (63.2) | .836 | 1.000 | .064 |
| BMI, kg/m2* | 23.7 (15.7–32.9) | 23.6 (19.0–42.7) | .893 | 25.1 (17.7–36.9) | 26.0 (19.4–39.4) | .654 | .003 | .013 |
| Smoking, yes | 11 (16.4) | 48 (40.0) | <.001 | 1 (1.4) | 6 (15.8) | .008 | .002 | .011 |
| AB last 6 months, yes# | 16 (23.9) | NA | 8 (11.6) | 5 (13.2) | 1.000 | .098 | ||
| IBD, yes | 33 (49.3) | 0 | 46 (66.7) | 0 | .060 | |||
| Ulcerative colitis, yes | 32 (47.8) | 36 (52.2) | .730 | |||||
| Crohn’s disease, yes | 1 (1.5) | 10 (14.5) | .009 | Available for n | ||||
| Medication, yes | in G/N | |||||||
| PPI | 0 | 2 (2.9) | .500 | 67/69 | ||||
| Statins | NA | 0 | 67/69 | |||||
| Ursodeoxycholic acid | 63 (94.0) | 23 (33.3) | <.001 | 60/69 | ||||
| Prednisolone / cortisone | 13 (19.4) | 9 (13.0) | .420 | 60/69 | ||||
| 5-ASA | 29 (43.3) | 29 (42.0) | 1.000 | 61/69 | ||||
| Infliximab | 2 (3.0) | 1 (1.4) | .614 | 60/69 | ||||
| Azathioprine | 9 (13.4) | 9 (13.0) | 1.000 | 60/69 | ||||
| Budesonide | 4 (6.0) | 3 (4.3) | .709 | 57/69 | ||||
| PSC specific variables | ||||||||
| PSC duration, years | 7.0 (1.0–35.0) | 9.5 (1.6–31.7) | .057 | 61/69 | ||||
| IBD duration, years | 15.0 (1.0–40.0) | 14.2 (1.2–44.5) | .610 | 31/41 | ||||
| Other autoimmune disease, yes | NA | 18 (26.1) | 0/64 | |||||
| Mayo risk score | NA | -0.07 (-1.92–3.33) | 0/51 | |||||
| Cirrhosis, yes | 3 (4.5) | NA | 67/0 | |||||
| P-ANCA positive | NA | 26 (37.7) | 0/35 | |||||
| Creatinine, μmol/L | NA | 68.0 (42.0–100.0) | 0/63 | |||||
| Bilirubin, μmol/L | 10.3 (3.42–34.2) | 13.0 (5.0–114.0) | .007 | 43/62 | ||||
| Albumin, g/L | NA | 43.0 16.0–47.0 | 0/58 | |||||
| AST, U/L | NA | 42.0 (18.0–197.0) | 0/58 | |||||
| ALT, U/L | 37.5 (11.0–286.0) | 51.0 (14.0–331.0) | .021 | 44/63 | ||||
| ALP, U/L | 117.0 (44.0–590.0) | 152.5 (30.0–598.0) | .004 | 44/62 | ||||
| GGT, U/L | NA | 165.0 (12.0–1576.0) | 0/61 | |||||
BMI data missing for n = 10 HCs in the German cohort, other data complete unless otherwise stated.
None used AB <6 weeks before sample inclusion, data on AB use >six weeks <six months NA for German HCs. AB, antibiotics; ALP, alkaline phosphatase; ALT, alanine aminotransferase; AST, aspartate aminotransferase; BMI, body mass index; G, German cohort; GGT, gamma-glutamyltransferase; HC, healthy controls; IBD, inflammatory bowel disease; N, Norwegian cohort; NA, not available; PPI, proton pump inhibitors; PSC, primary sclerosing cholangitis; 5-ASA, 5-Aminosalicylic acid.
Overall microbial taxonomic and genetic diversity
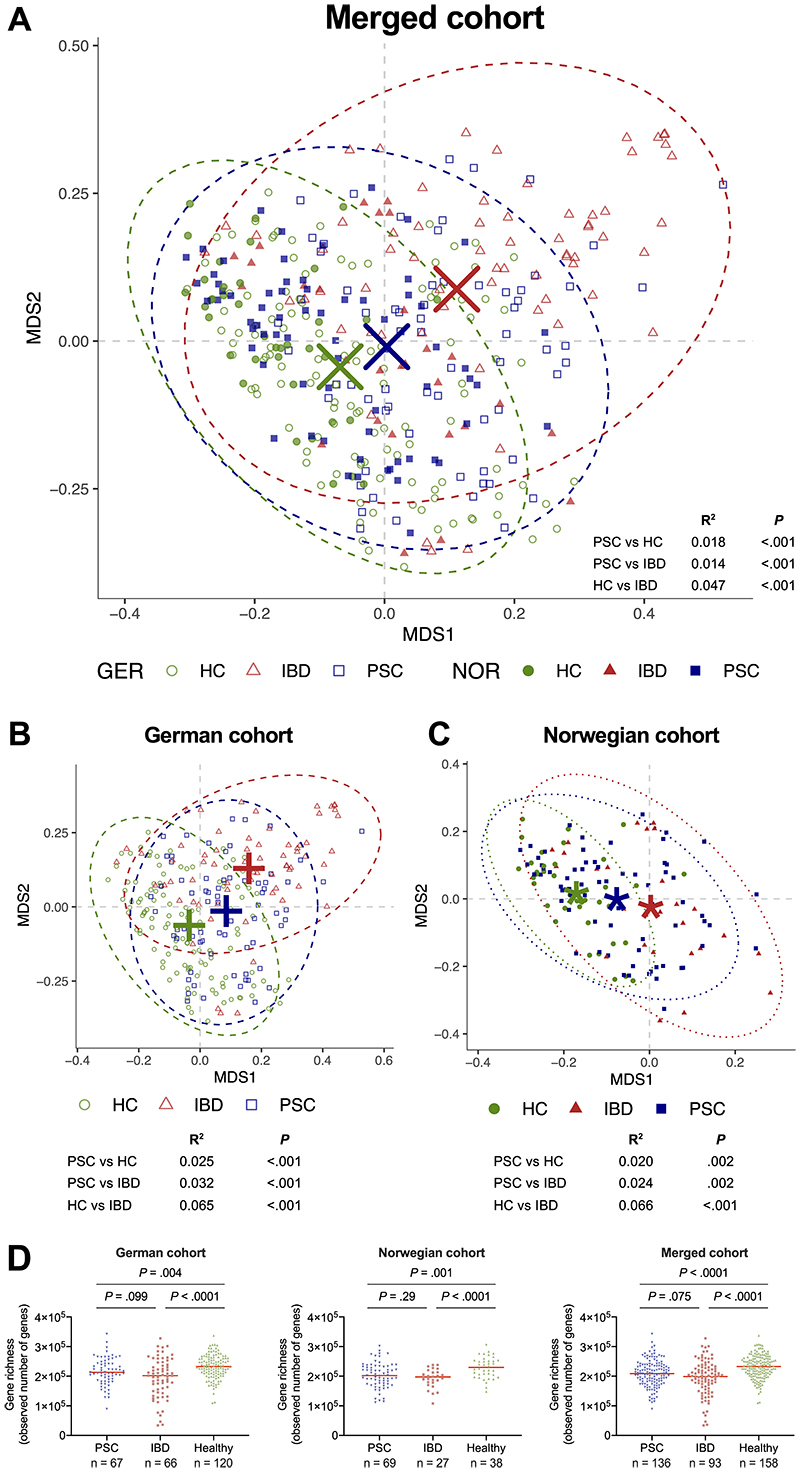
The overall microbiome composition showed significant shifts between phenotypes (PSC, healthy controls and IBD) in the merged cohort (Figure 1A) which was also apparent when analyzing each cohort separately (Figure 1B&C). Geography was also an important factor for the overall microbiome composition for all phenotypes (Figure 1A), including healthy controls (Supplementary Figure 2).
Figure 1. Overall bacterial taxonomic composition and gene richness in PSC patients and controls.
Non-constrained ordination of genus-level Bray-Curtis dissimilarities in the microbiome of (A) patients with PSC, as well as healthy and IBD controls, in the merged cohort, and in the (B) the German and (C) the Norwegian cohorts separately, showing shifts in the global microbiome composition. (D) Number of observed genes in PSC compared to controls. All group centroids in A-C represent the arithmetic mean of the groups' respective points in the ordination, ellipses represent 95% confidence level of the multivariate t-distributions. Red horizontal bar in D represents the median. GER, Germany; HC, healthy control; IBD, inflammatory bowel disease; NOR, Norway; MDS, multiple dimension scale; PSC, primary sclerosing cholangitis.
Patients with PSC had markedly reduced microbial gene richness (observed genes) compared to healthy controls (Figure 1D), also when adjusting for age, sex and BMI (linear regression, P<.016, P=.001, and P<.0001 in the German, Norwegian and merged cohort, respectively). Concomitant IBD had no effect on gene richness in patients with PSC (Supplementary Figure 3).
Individual species
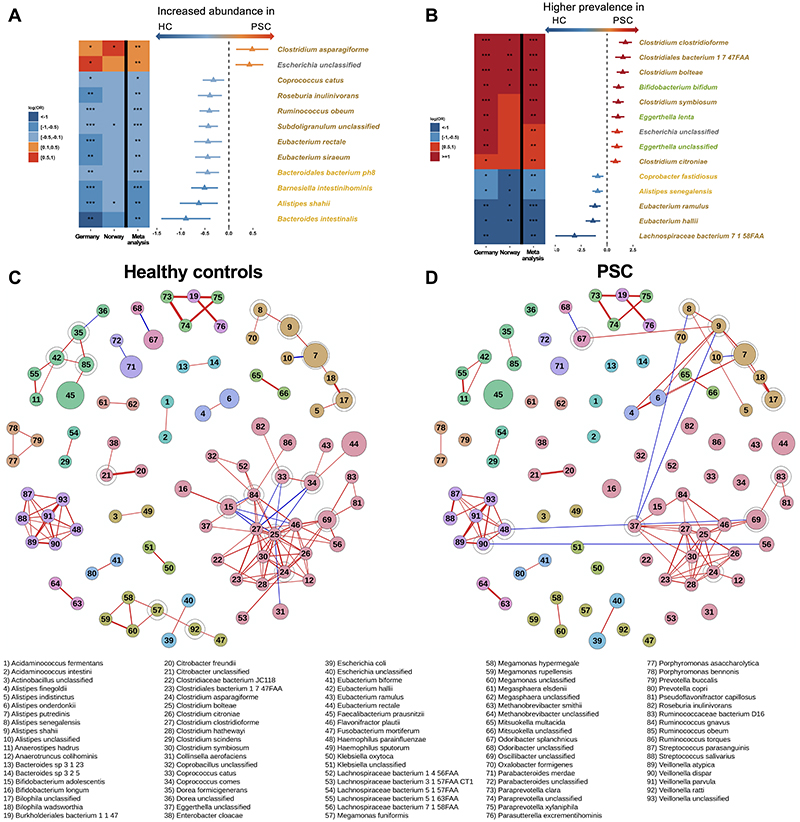
After quality control and filtering, a total of 413 taxa were identified in the full dataset (phylum to strain level). For each country, single taxa with less than 80% zeros across samples were analyzed both for a shift in distribution (mu) and a difference in zero abundance (nu). The analysis identified an enrichment of Clostridium asparagiforme and an unclassified Escherichia species in PSC compared to healthy controls, as well as a marked depletion of several species, e.g., Ruminococcus obeum, Eubacterium siraeum, and Eubacterium rectale (meta-analyses Q fdr<.05, Figure 2A). Furthermore, nine species showed an increased prevalence, while we identified five species with lower prevalence in patients with PSC compared to healthy controls (Figure 2B). While several species showed similar patterns in IBD, species like Ruminococcus obeum, Bacteroides intestinalis and several Clostridium species did not differ between IBD and healthy controls (Supplementary File 1).
Figure 2. Differences in species composition in PSC compared to healthy controls.
(A) Species that were differently abundant and (B) species with different prevalence in PSC and healthy controls. Species are colored by phyla affiliation. (C) Bacterial abundance co-correlation network in healthy controls and (D) PSC patients. In A&B the triangle/bar in the forest plot indicate log(OR)/SE. Red and blue lines in C&D indicate positive and negative correlations between species, respectively. Node-size reflects the square-root transformed median relative abundances in each phenotype, and encircled nodes mark articulations points. Only correlations with r>0.4 are shown, and increasing correlation coefficient are reflected by increasing line thickness. PSC, primary sclerosing cholangitis; HC, healthy controls. *Q fdr<.05, ** Q fdr<.005, *** Q fdr<.0005.
Previous works have found increased levels of the Veillonella genus in PSC.7 Both Veillonella atypica, Veillonella parvula and an unclassified Veillonella species showed trending increased prevalence in patients with PSC compared to healthy controls (meta-analyses P=.0062, .026 and .025, respectively, Q fdr=.058, .14 and 0.14, respectively, Supplementary Figure 4 and Supplementary File 1). Other PSC-related species include Klebsiella pneumoniae, in addition to Proteus mirabilis and Enterococcus gallinarum, previously linked to intestinal barrier function and immune responses in PSC.12 Klebsiella pneumoniae was left out from our initial analysis due to low abundance in the Norwegian cohort (see Methods). A targeted analysis detected Klebsiella pneumoniae in the microbiome of 25% of patients with PSC, but only in 4.4% of the healthy controls (CMH test, P<.001). Klebsiella pneumoniae was detected in the microbiome of 45.2% of IBD controls (CMH test, P=.0057 vs. PSC). Enterococcus gallinarum and Proteus mirabilis were detected in only a few patients (Supplementary Table 5). When applying a species-specific RT-PCR on a subset of the German samples (PSC n=36, healthy controls n=20, IBD n=17), Klebsiella pneumoniae showed similar prevalence in both PSC, IBD and healthy controls (77.8%, 94.1% and 75.0%, respectively, Supplementary Figure 5A). Enterococcus gallinarum and Proteus mirabilis were detected in a minority of samples (Supplementary Figure 5B&C).
To further delineate structural microbiome differences between the phenotypes, we generated bacterial species abundance co-correlation networks using SparCC.29 The network in healthy controls consisted of several large and small clusters (Figure 2C). When overlaying the co-correlations observed in PSC, there was a disruption of the network seen in the healthy controls in the PSC patients, with loss of several clusters and fewer co-correlations in others (Figure 2D). IBD controls without PSC also showed a disrupted network compared to healthy controls, but markedly different from what was observed in PSC (Supplementary Figure 6).
Extensive alterations in the genetic landscape of the microbiome in PSC, highlighting changes in biosynthesis of amino acids and B vitamins
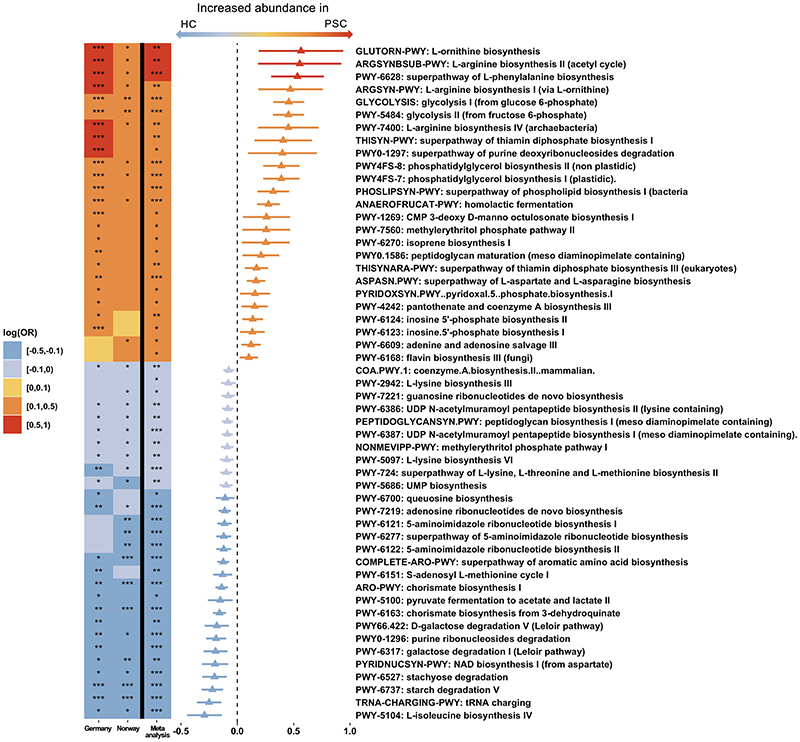
In total, after filtering we detected 121 metabolic pathways (MetaCyc pathways) and 424 enzyme families present in both cohorts. We identified 53 metabolic pathways differentially abundant in the microbiomes of patients with PSC compared to healthy controls, in addition to 106 enzyme families (all meta-analyses Q fdr<.05, Figure 3 and Supplementary File 1, respectively).
Figure 3. Alterations in metabolic pathways in the microbiome of PSC.
Heatmap and forest plot showing significant differences in microbiome metabolic pathways (Metacyc pathways, Q fdr<.05) between PSC and healthy controls from the individual cohorts (two left columns in the heatmap) and the meta-analysis (right column in the heatmap and the forest plot). Triangle/bar in forest plot indicate log(OR)/SE. HC, healthy controls; PSC, primary sclerosing cholangitis. *Q fdr<.05, **Q fdr<.005, ***Q fdr<.0005.
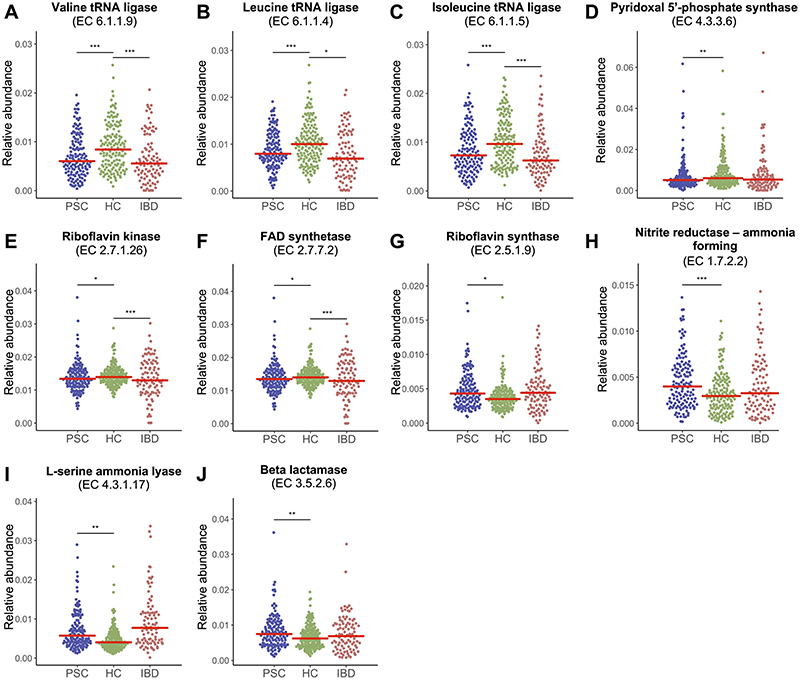
The microbiome of patients with PSC showed a marked depletion of the metabolic pathway related to the biosynthesis of the branched-chain amino acid (BCAA) isoleucine (Figure 3), as well as several enzyme families related to the BCAAs isoleucine, valine and leucine compared to healthy controls (Figure 4A-C and Supplementary File 1), while levels in patients with IBD were similar to levels in PSC. There were also differences in metabolic pathways related to biosynthesis of aromatic amino acids, but less consistent, with some pathways enriched in PSC (PWY6628 superpathway of L-phenylalanine biosynthesis) and others depleted, e.g. COMPLETE ARO-PWY superpathway of aromatic amino acid biosynthesis, as seen in Figure 3. Four of the seven most highly enriched metabolic pathways in PSC compared to healthy controls were related to biosynthesis of two other amino acids, i.e. arginine and ornithine (Figure 3). These four pathways had similar levels in PSC and IBD but in direct comparisons only two of these pathways were significantly enriched in IBD compared to healthy controls (Supplementary File 1).
Figure 4. Genetic potential of the gut microbiome in patients with PSC.
Dot-plots showing relative abundance of enzyme families (level-4 Enzyme Commission (EC) categories) related to microbial metabolism of branched chain amino acids (A) valine, (B) leucine and (C) isoleucine, (D) the active form of vitamin B6 (pyridoxal 5’-phosphate, PLP) and (E) the active forms of vitamin B2, flavin mononucleotide (FMN), and (F) flavin adenine dinucleotide (FAD), and (G) synthesis of riboflavin (vitamin B2) and related metabolites, (H-I) ammonia production and (J) antibiotic resistance. Red bar marks the median. EC, level-4 Enzyme Commission; FAD, flavin adenine dinucleotide; HC, healthy controls; IBD, inflammatory bowel disease; PLP, pyridoxal 5’-phosphate; PSC, primary sclerosing cholangitis. *Q fdr<.05, **Q fdr<.01, ***Q fdr<.001.
PSC patients showed higher levels of metabolic pathways related to biosynthesis of thiamine (vitamin B1) compared to healthy controls (e.g. THISYN-PWY Superpathway of thiamin diphosphate biosynthesis I, Figure 3). This pathway showed similar levels in PSC and IBD, while IBD patients showed higher levels compared to healthy controls (Supplementary File 1). On the other hand, three metabolic pathways for biosynthesis of 5'-Phosphoribosyl-5-aminoimidazole (PWY6122, PWY6277 and PWY6121), a key intermediate in the biosynthesis of thiamine (vitamin B1) were all depleted in PSC (Figure 3). Furthermore, patients with PSC showed marked differences compared to healthy controls of several enzyme families related to vitamin B6 metabolism, most prominent a marked depletion of EC4.3.3.6 pyridoxal-5-phosphate synthase (Figure 4D), an enzyme that alone constitutes the metabolic pathway pyridoxal 5'-phosphate biosynthesis II and the predominant biosynthetic route for the de novo production of the active form of vitamin B6, pyridoxal 5'-phosphate (PLP),30 and EC2.6.1.52 Phosphoserine transaminase (Supplementary File 1). The latter is one of seven enzyme families comprising the metabolic pathway PYRIDOXSYN-PWY: pyridoxal 5’-phosphate biosynthesis I, which was increased in PSC compared to healthy controls (Figure 3). As for the last six enzyme families in this metabolic pathway one (EC1.1.1.290) was enriched in PSC compared to healthy controls, three showed similar levels (EC2.6.99.2, 1.1.1.262 and 2.2.1.7), while the last two (EC1.2.1.72 and 1.4.3.5) did not pass our initial filtering (Supplementary File 1). A total of 84 species contributed to EC4.3.3.6 pyridoxal-5-phosphate synthase in the dataset (Supplementary File 2). Eubacterium rectale and Eubacterium siraeum, which were both depleted in PSC compared to healthy controls in the meta-analyses (see Figure 2A), were among the top five species contributing to the abundance of this enzyme. Twenty-eight species contributed to PYRIDOXSYN-PWY: pyridoxal 5’-phosphate biosynthesis I (Supplementary File 2), but none of the contributing species detected in the metanalysis showed a difference between PSC and healthy controls (all Bacteroides, Q fdr>.30, Supplementary File 2).
Regarding other B-vitamins, two enzymes, EC2.7.1.26 riboflavin kinase and EC2.7.7.2 FAD synthetase, responsible for synthesis of the active forms of vitamin B2 (flavin mononucleotide (FMN) and flavin adenine dinucleotide (FAD), respectively) were also reduced in PSC (Figure 4E&F), while riboflavin synthase (EC2.5.1.9), which is responsible for a reaction resulting in synthesis of riboflavin and 5-amino-6-d-ribitylaminouracil (5-A-RU), was enriched in PSC (Figure 4G). Furthermore, the microbiome of patients with PSC showed increased abundance of enzyme families related to ammonia production (Figure 4H&I). Enzyme families related to antibiotic resistance like EC3.5.2.6 beta-lactamase were also enriched in PSC compared to healthy controls (Figure 4J).
Metabolic pathways diminished in the microbiome of patients with PSC show low levels of related metabolites in plasma
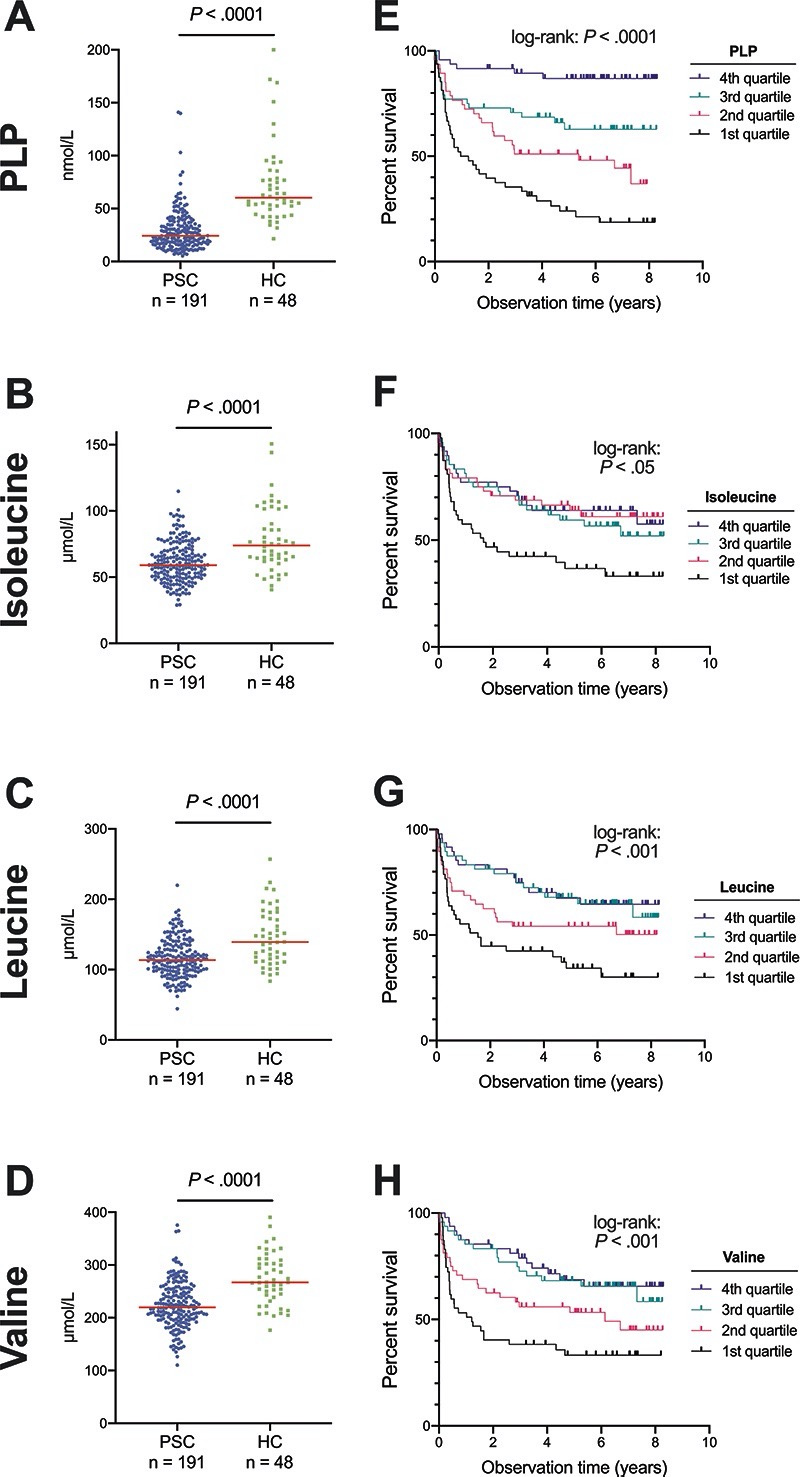
As the initial microbiome analyses identified potential alterations in bacterial metabolism of both branched-chain amino acids and B vitamins, and the gut microbiome could be an important source of these metabolites,31,32 we investigated selected vitamin B and branched-chain amino acid metabolites in plasma from another cohort of 191 PSC patients (27 were also recruited in the metagenome study cohort) and 48 healthy controls (see Supplementary Figure 1). The main active form of vitamin B6, PLP, and the BCAAs isoleucine, leucine and valine were clearly reduced in plasma of patients with PSC compared with healthy controls (Figure 5A-D). Other vitamin B6 metabolites (i.e. pyridoxal and 4-pyridoxic acid) were also lower in PSC compared to controls (Supplementary Figure 7). Furthermore, the vitamin B2 (riboflavin) metabolite FMN showed reduced levels in PSC corresponding to the reduction in EC2.7.1.26, while there was no difference for vitamin B2 itself (Supplementary Figure 8A&B). For vitamin B1, thiamine was similar in PSC patients compared with controls, while thiamine monophosphate was reduced in PSC (Supplementary Figure 8C&D). Plasma levels of these B-vitamins did not correlate with the relative abundance of associated metabolic pathways or enzyme families in the 27 patients with both microbiome and metabolomics data available (data not shown), possibly due to the long time interval between sampling of stool and plasma (median interval 2.9 years). The essential amino acid histidine was reduced in patients with PSC compared to healthy controls (P=.005, Supplementary Figure 9A). Among other measured amino acids (Supplementary Figure 9B-E), only ornithine was increased in PSC (P =.016).
Figure 5. Microbiome-related plasma metabolites and liver-transplantation free survival.
(A) The active vitamin B6 metabolite pyridoxal 5’-phosphate (PLP), (B) isoleucine, (C) leucine and (D) valine in plasma of patients with PSC compared to healthy controls. Kaplan-Meier plots showing that low levels (1st quartile) of (E) PLP, (F) isoleucine, (G) leucine and (H) valine were all associated with reduced liver-transplantation free survival in PSC. HC, healthy controls; PLP, pyridoxal 5’-phosphate; PSC, primary sclerosing cholangitis.
As circulating levels of BCAAs have been linked to the potential ability of the gut microbiome to transport these amino acids into bacteria,32 we specifically investigated five KEGG orthologue gene groups related to inward transportation of BCAAs into bacteria. These gene groups showed markedly higher abundance in PSC compared to healthy controls in the meta-analysis (K01995 Q fdr=.078; K01996, K01999, K01997 and K01998 Q fdr<.05), and similar levels in IBD compared to PSC (Supplementary Figure 10).
Microbial metabolites may modify the severity of diseases.32 We therefore investigated the relationship between metabolites and liver transplantation-free survival in PSC. The plasma concentrations of PLP and all BCAAs (i.e., isoleucine, leucine and valine) were lower in the patients reaching an endpoint than in those who did not (Supplementary Figure 11). When dividing the plasma concentrations into quartiles, low levels of these metabolites all strongly predicted reduced liver-transplantation free survival (Figure 5E-H). The effect seemed particularly strong for PLP (Figure 5A&E), which was associated with reduced risk of liver transplantation or death in a Cox regression model, independent of Mayo risk score and enhanced liver fibrosis (ELF) score (hazard ratio 0.66 per quartile increase in plasma PLP, 95% CI 0.51–0.87, P=.003, Supplementary Table 6). PLP was also associated with reduced risk of liver transplantation or death independent of other risk scores, i.e. Amsterdam-Oxford model for PSC (hazard ratio 0.67 per quartile increase in plasma PLP (95% CI 0.51–0.88), P=.003, Supplementary Table 7). PLP may be metabolized by alkaline phosphatase (ALP),33 and the two showed a moderate inverse correlation (rho -0.38, P<.001), but ALP was not independently associated with survival when added to the model (Supplementary Table 6). Isoleucine, leucine and valine were not independently associated with liver-transplantation free survival when corrected for Mayo risk score (data not shown).
Microbiome as predictor of disease phenotypes and clinical characteristics in PSC
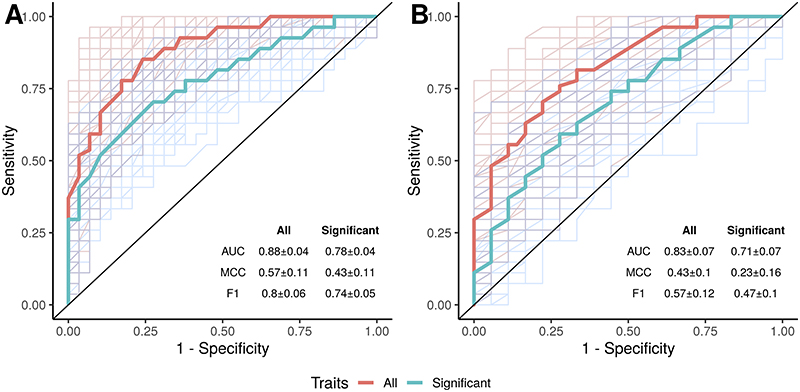
When applying a random forest model on the species data we were able to separate PSC from healthy controls with a high predictive power (AUC=0.88±0.04, Figure 6A), and achieved similar results when separating PSC from IBD (AUC=0.83±0.07, Figure 6B). Separating PSC from healthy controls and IBD at the same time gave similar predictive power (Supplementary Figure 12). Adding anthropometric information (i.e., age, gender and BMI) had minimal effect on the prediction of PSC vs. healthy controls (AUC=0.87±0.05, MCC=0.54±0.11, Supplementary Figure 12), and using species data generally performed better than enzyme families and metabolic pathways for separating the phenotypes (Supplementary Figure 12).
Figure 6. Separation of clinical phenotypes using random forest classification.
(A) PSC vs. healthy controls and (B) PSC vs. IBD. Results using all available species (red line) or only the significant species hits (blue line). AUC, area under the curve; F1, F1 measure; IBD, inflammatory bowel disease; MCC, Matthew’s correlation coefficient (range: -1– +1); PSC, primary sclerosing cholangitis.
Finally, we merged the two microbiome cohorts for analyses of differences between clinical entities in patients with PSC (n=136). Comparing PSC patients with and without IBD, we were unable to detect any differences in neither gene richness (P=.39, Supplementary Figure 3), species, metabolic pathways or enzyme families (all meta-analyses Q fdr> .1, Supplementary File 1). Neither was there a significant shift in overall bacterial composition between PSC patients with and without IBD (Supplementary Figure 13). There was no correlation between microbial gene richness and disease duration in PSC (rho=0.007, P=.94, n=135) or the Mayo risk score (rho=-0.07, P=.63, n=51). In a subset of the German PSC patients (n=41) with liver stiffness measurements available (measured by transient elastography), liver stiffness did not show significant correlation with gene richness (rho=-0.21, P=.19). When testing for associations of liver stiffness with enzyme families and metabolic pathways related to vitamin B6 or BCAA synthesis, there was a weak association between increased liver stiffness and reduced levels of EC6.1.1.4 leucine tRNA ligase (rho=-0.39, unadjusted P=.012, Supplementary Figure 14), but not for any other of the tested functional features.
Ursodeoxycholic acid (UDCA) usage in patients with PSC was associated with higher levels of Eubacterium rectale (estimate: 0.70, P=.004, Q fdr=.26, Supplementary File 1), which was depleted in PSC compared to healthy controls in the meta-analyses (Figure 2A). UDCA usage was also associated with reduced abundance of Veillonella dispar (estimate: −1.05, P =.01, Q fdr=.34, Supplementary File 1). Furthermore, there was a subtle shift in beta diversity (Bray-Curtis) between UDCA users and non-users (R2=0.030, P=.005, Supplementary Figure 13). We did not detect any significant effect by 5-Aminosalicylic acid (5-ASA) on any species (Q fdr>.50). Neither 5-ASA nor UDCA usage was associated with differences in enzyme families or metabolic pathways (all Q fdr>.19). Notably, UDCA usage in PSC was only analyzed in the Norwegian cohort, as 94% (n=63) of the German PSC patients were using UDCA at the time of sampling.
Discussion
In the present study we analyzed the stool microbiome in two cohorts of Norwegian and German subjects comprising PSC, IBD and healthy controls. Using high-resolution shotgun metagenomic sequencing and targeted metabolomics in a third independent cohort of Norwegian PSC patients and healthy controls we uncovered (i) a PSC-associated microbiome with a low gene count and multiple bacterial alterations unaffected by concurrent IBD, and which could now be defined at species level; (ii) marked alterations of gene functions related to e.g. vitamin B6 synthesis and branched chain amino acids synthesis in the microbiome of PSC; (iii) reduced plasma concentrations of vitamin B6 and branched-chain amino acids in PSC, and (iv) low concentrations of these metabolites strongly associated with reduced liver transplantation-free survival, suggesting that investigating altered microbial metabolism may help pinpoint biological changes relevant for the disease course in PSC.
By using shotgun metagenomic sequencing, we make a major leap forward in microbiome studies investigating PSC, providing detailed functional analysis of microbial genes encoding enzymes and metabolic pathways. Genes related to bacterial synthesis of several essential nutrients like BCAAs and vitamin B6 (e.g. PLP) showed clear changes in PSC patients compared to healthy controls. PLP is the active form of vitamin B6, an essential cofactor of numerous metabolic reactions, and contributes to intestinal immune regulation and lymphocyte trafficking into the intestines.34 PLP can be produced de novo via two independent pathways. The predominant route used by the majority of organisms capable of PLP production is via a single enzyme (EC4.3.3.6),30,31,35 which was markedly depleted in PSC. The less abundant, multi-enzyme pathway PYRIDOXSYN-PWY: pyridoxal 5'-phosphate biosynthesis was increased in PSC. Overall, the data suggest a reduced genetic potential to synthesize PLP in PSC, and calls for further evaluations in an experimental model system. The capacity of the gut bacteria to synthesize vitamin B6 has been estimated to meet 86% of the daily recommended intake,31 suggesting that alterations in the gut microbiome may dramatically influence dietary requirements. Targeted metabolite analyses in plasma showed reduced concentrations of both PLP and other B6 species (pyridoxal and 4-pyridoxic acid), indeed suggesting a true vitamin B6 deficiency in PSC.36 Notably, PLP is known to be reduced in end-stage liver disease of different etiologies,37 linked not only to deficiency per se, but also due to increased catabolism of PLP to pyridoxal, by e.g. ALP or other mechanisms in the context of chronic inflammation.38 Still, the predictive effect independent from Mayo risk score could point to a role of PLP in PSC beyond end-stage liver disease. Taken together, identification of altered bacterial functions pointed us to important biology that was altered also systemically. It should be emphasized that we do not show a direct link between the gut and blood, but further studies should investigate such a link and whether PLP represents a mechanism or only a marker of disease activity and severity in PSC.
Similar considerations regarding microbial synthesis and dietary requirements are relevant for BCAAs. Circulating BCAAs are known to be reduced in patients with cirrhosis, but also less advanced liver diseases, including PSC.39,40 In contrast, individuals with insulin resistance have been shown to have elevated circulating BCAAs levels, correlating with increased synthesis capacity in the microbiome as well as reduced capacity to import these BCAAs into bacteria.32 In PSC we found the opposite, with reduced bacterial synthesis and increased import capacity into bacteria. This calls for consideration as to whether the gut microbiome contributes to the observed reduced plasma BCAA concentrations in PSC. Low plasma BCAAs concentrations predicted reduced liver transplantation-free survival in our study, but not independently of the Mayo risk score, suggesting that alterations in BCAAs may relate to disease stage. Firm conclusions regarding the role of these metabolites in PSC in humans would require an interventional trial with e.g. supplementation,41 while the potential clinical value of vitamin B6 or BCAAs as predictive biomarkers in PSC would require independent validation studies. Notably, since deficiencies of vitamin B6 and BCAAs have been observed in liver diseases of several etiologies, similar gut microbiome-related modifier effects may be seen also in other liver diseases.37,42
For other measured metabolites, the relationship between the gut microbiome and plasma levels was less obvious. For thiamine and riboflavin, this could perhaps be explained by the low potential contribution from the gut microbes, estimated to meet only 2-3% of daily recommended intake.31 However, the alterations in bacterial genes related to riboflavin metabolism in PSC may be of interest, since these are the source of bacterial riboflavin metabolites that are essential in the activation of mucosal-associated invariant T (MAIT) cells, which are liver-abundant innate-like T cells relevant in PSC.43–45
Functional aspects of the gut microbiome are important, but the actions of specific bacterial species are also of interest. We observed an increase of the classical potential pathogen Escherichia, together with reductions in species typically classified as producers of the short-chain fatty acid butyrate. Previous work has highlighted an increased relative abundance of the Veillonella genus in PSC.7 Veillonella was less prevalent when investigated with the current methodology, but we observed an increased prevalence of several Veillonella species in PSC patients. In a study by Nakamoto et al., K. pneumonia strains isolated from PSC patients contributed to increased susceptibility to hepatobiliary damage in mice, and more bacterial translocation.12 In the shotgun sequencing data in the present study, K. pneumoniae showed higher prevalence in PSC patients compared to healthy controls, as reported by Nakamoto et al. in their study. Using a more sensitive method (qPCR) in a small subset of the German cohort, K. pneumoniae sequences were identified in a much higher proportion of the PSC patients compared to the shotgun sequencing data, but with similar prevalence in PSC and healthy controls. The clinical relevance of very low abundant taxa such as K. pneumoniae could be questioned, and the role of K. pneumoniae in human PSC is not well defined. As the experimental data from the paper by Nakamoto et al suggest,12 it is possible that only specific strains of the K. pneumoniae species are capable of inducing hepatobiliary damage, highlighting the need for increased resolution in further microbiome research. Interestingly, a similar concept was presented recently in patients with alcoholic hepatitis, who had higher frequency of fecal Enterococcus faecalis.46 The presence of E. faecalis strains with the ability to produce the exotoxin cytolysin was associated with more severe disease.46 Overall, it appears that both individual microbes and shifts in the collective metabolic capacity of the gut microbiome may modify human diseases, without necessarily being the triggering events. Such mechanisms may point towards new treatment opportunities.
One important observation was the striking similarity between the microbiome in PSC patients with and without IBD, both at species and functional level. In line with previous data,2,3 the global microbiome composition in IBD without liver disease was different from patients with PSC. However, few species and functions were significantly different between patients with PSC and IBD patients without PSC, suggesting that the differences separating PSC from IBD relate to smaller changes in multiple species or functions. Considering the microbiota literature, the differences between PSC and IBD are less consistent than between PSC and healthy controls.2,3,7 It is possible that the challenge of appropriately matching IBD patients with and without PSC for case-control studies make these comparisons difficult.
Differences depending on geography impose another challenge but may also be an important tool in case-control studies. In addition, sampling and DNA extractions were performed differently in the Norwegian and German cohorts. Geography does in some studies constitute the strongest predictor of microbiome composition,47 highlighting a need for identification and validation of core features in multi-cohort international studies to define true disease characteristics. However, in the present study it is not possible to assign with certainty differences between Norway and Germany to geography or methodology.
The major strengths of this study are the use of two independent case-control cohorts from different countries and the use of state-of-the art sequencing methods with high taxonomic and functional resolution. While this study, to our knowledge, is the largest to investigate the microbiome in PSC utilizing full metagenomic sequencing published so far, it is likely that by including additional patients and also liver disease controls, the robustness of the results could further increase. Notably, the link between our findings of microbiome changes and differences in metabolites and prognosis are, as of now, purely associative. Time- and subject-matched blood and stool-samples would be important in further studies, in addition to experimental studies, if these associations are to be further explored. Liver transplantation as an endpoint could vary in different countries, and could thus make it less useful as endpoint intended for clinical translation. Another limitation is that dietary data were only surveyed crudely through standard questionnaires, so it cannot be discounted that dietary factors could have influenced our results. In addition, due to the cross-sectional design we are unable to discern whether the changes we detect occur before or after disease onset, which limit the possibility to draw conclusions regarding cause or effect. As PSC is a rare disease it is difficult, if not impossible, to profile the microbiome over the time of disease onset. However, larger international cohorts with follow-up data should be established to help address these knowledge gaps.
In conclusion, the gut microbiome of PSC patients exhibits large functional differences compared to healthy controls, including microbial metabolism of essential nutrients, irrespective of the presence of IBD. Alterations in related circulating metabolites associated with liver transplantation-free survival, suggest that overall microbial functions may be relevant for the disease process in PSC.
Supplementary Material
Acknowledgements
Benedicte A. Lie and the Norwegian Bone Marrow Donor Registry are acknowledged for providing access to healthy controls, as are William Rosenberg at the University College London & Royal Free London and the NHS Foundation Trust for contributions to ELF score data. We are grateful to PSC Partners for funding metagenome sequencing. We would like to thank all technicians of the IKMB microbiome and NGS laboratory for excellent technical support.
Abbreviations
- 5-ASA
5-Aminosalicylic acid
- ALP
alkaline phosphatase
- ALT
alanine aminotransferase
- AST
aspartate aminotransferase
- AUC
area under the curve
- BCAA
branched-chain amino acid
- BMI
body mass index
- CD
Crohn's disease
- CMH
Cochran-Mantel-Haenszel
- EC
enzyme commission
- ELF
enhanced liver fibrosis
- FAD
flavin adenine dinucleotide
- FDR
false-discovery rate
- FMN
flavin mononucleotide
- HC
healthy controls
- GGT
gamma-glutamyltransferase
- IBD
inflammatory bowel disease
- LTx
liver transplantation
- MCC
Matthew’s correlation coefficient
- P-ANCA
perinuclear anti-neutrophil cytoplasmic antibodies
- PLP
pyridoxal 5’-phosphate
- PSC
primary sclerosing cholangitis
- PPI
proton pump inhibitors
- TMAO
trimethylamine-N-oxide
- UC
ulcerative colitis
- UDCA
ursodeoxycholic acid.
Footnotes
Author Contributions
MK, AF and JRH conceptualized the study. MK, MCR, TL, RZ, CSL, MLH, MV, ML, WL, THK, CB, CS, AF and JRH contributed to collection of samples and clinical data. ØM, AM and PMU were responsible for targeted metabolomics. KH performed the post-sequencing data processing. MK, LBT, MCR, SHH, LMS, AF and JRH planned the analyses. Data curation and formal analysis were performed by MK, LBT, MCR, KH, SHH, CSL and LMS, and CB. MK, LBT, MCR, SHH, LMS, CB, CS, AF and JRH interpreted the data. JRH and AF supervised the study and were responsible for project administration. MK and JRH wrote the original draft and LBT, MCR, KH, SHH, LMS, ØM, PMU, CB, CS and AF contributed to the writing of the manuscript. All authors read, critically revised for important intellectual content and approved the final manuscript. CS, AF and JRH acquired funding.
Disclosures
LBT is an employee and shareholder of BiomCare. PMU is a member of the steering board of the nonprofit foundation, which owns Bevital and R&D director of Bevital, and ØM and AM are employees of Bevital. MK, MCR, KH, SHH, LMS, TL, RZ, CSL, MLH, MV, MT, ML, WL, THK, CB, CS, AF, JRH: nothing to disclose.
Writing Assistance
none.
Author names in bold designate shared co-first authorship.
Grant support
MK was supported by a grant from Eastern Norway Regional Health Authority (grant number 2016067). JRH was funded by the Norwegian Research Council (project number 240787/F20) and the European Research Council (grant number 802544). The metagenome sequencing was funded in part by PSC Partners Seeking a Cure. CS is supported by the German Research Foundation (DFG, CRU 306) and by the Hannelore and Helmut Greve Foundation and the YAEL Foundation. LBT and PSC sample handling was supported by the Deutsche Forschungsgemeinschaft (DFG) Clinical Research Group 306 “Primary sclerosing cholangitis” (ID: DFG CRU306) and received infrastructure support from the DFG Cluster of Excellence “Precision Medicine in Chronic Inflammation” (PMI; ID: EXC2167) and the DFG Research Unit miTarget (ID: RU5042).
References
- 1.Karlsen TH, Folseraas T, Thorburn D, et al. Primary sclerosing cholangitis – a comprehensive review. J Hepatol. 2017;67:1298–1323. doi: 10.1016/j.jhep.2017.07.022. [DOI] [PubMed] [Google Scholar]
- 2.Kummen M, Holm K, Anmarkrud JA, et al. The gut microbial profile in patients with primary sclerosing cholangitis is distinct from patients with ulcerative colitis without biliary disease and healthy controls. Gut. 2017;66:611–619. doi: 10.1136/gutjnl-2015-310500. [DOI] [PubMed] [Google Scholar]
- 3.Sabino J, Vieira-Silva S, Machiels K, et al. Primary sclerosing cholangitis is characterised by intestinal dysbiosis independent from IBD. Gut. 2016;65:1681–1689. doi: 10.1136/gutjnl-2015-311004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lemoinne S, Kemgang A, Belkacem K Ben, et al. Fungi participate in the dysbiosis of gut microbiota in patients with primary sclerosing cholangitis. Gut. 2020;69:92–102. doi: 10.1136/gutjnl-2018-317791. [DOI] [PubMed] [Google Scholar]
- 5.Bajer L, Kverka M, Kostovcik M, et al. Distinct gut microbiota profiles in patients with primary sclerosing cholangitis and ulcerative colitis. World J Gastroenterol. 2017;23:4548–4558. doi: 10.3748/wjg.v23.i25.4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iwasawa K, Suda W, Tsunoda T, et al. Characterisation of the faecal microbiota in Japanese patients with paediatric-onset primary sclerosing cholangitis. Gut. 2017;66:1344–1346. doi: 10.1136/gutjnl-2016-312533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kummen M, Hov JR. The gut microbial influence on cholestatic liver disease. Liver Int. 2019;39:1186–1196. doi: 10.1111/liv.14153. [DOI] [PubMed] [Google Scholar]
- 8.Schrumpf E, Kummen M, Valestrand L, et al. The gut microbiota contributes to a mouse model of spontaneous bile duct inflammation. J Hepatol. 2016;66:382–389. doi: 10.1016/j.jhep.2016.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tabibian JH, O’Hara SP, Trussoni CE, et al. Absence of the intestinal microbiota exacerbates hepatobiliary disease in a murine model of primary sclerosing cholangitis. Hepatology. 2016;63:185–196. doi: 10.1002/hep.27927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shah A, Crawford D, Burger D, et al. Effects of Antibiotic Therapy in Primary Sclerosing Cholangitis with and without Inflammatory Bowel Disease: A Systematic Review and Meta-Analysis. Semin Liver Dis. 2019;1:432–441. doi: 10.1055/s-0039-1688501. [DOI] [PubMed] [Google Scholar]
- 11.Allegretti JR, Kassam Z, Carrellas M, et al. Fecal Microbiota Transplantation in Patients With Primary Sclerosing Cholangitis. Am J Gastroenterol. 2019;114:1071–1079. doi: 10.14309/ajg.0000000000000115. [DOI] [PubMed] [Google Scholar]
- 12.Nakamoto N, Sasaki N, Aoki R, et al. Gut pathobionts underlie intestinal barrier dysfunction and liver T helper 17 cell immune response in primary sclerosing cholangitis. Nat Microbiol. 2019;4:492–503. doi: 10.1038/s41564-018-0333-1. [DOI] [PubMed] [Google Scholar]
- 13.Kummen M, Vesterhus M, Trøseid M, et al. Elevated trimethylamine-N-oxide (TMAO) is associated with poor prognosis in primary sclerosing cholangitis patients with normal liver function. United Eur Gastroenterol J. 2017;5:532–541. doi: 10.1177/2050640616663453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rühlemann M, Liwinski T, Heinsen F-A, et al. Consistent alterations in faecal microbiomes of patients with primary sclerosing cholangitis independent of associated colitis. Aliment Pharmacol Ther. 2019;50:580–589. doi: 10.1111/apt.15375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.European Association for the Study of the Liver. EASL Clinical Practice Guidelines: management of cholestatic liver diseases. J Hepatol. 2009;51:237–267. doi: 10.1016/j.jhep.2009.04.009. [DOI] [PubMed] [Google Scholar]
- 16.Lindor KD, Kowdley KV, Harrison EM. ACG Clinical Guideline: Primary Sclerosing Cholangitis. Am J Gastroenterol. 2015;110:646–659. doi: 10.1038/ajg.2015.112. [DOI] [PubMed] [Google Scholar]
- 17.Krawczak M, Nikolaus S, von Eberstein H, et al. PopGen: population-based recruitment of patients and controls for the analysis of complex genotypephenotype relationships. Community Genet. 2006;9:55–61. doi: 10.1159/000090694. [DOI] [PubMed] [Google Scholar]
- 18.Müller N, Schulte DM, Türk K, et al. IL-6 blockade by monoclonal antibodies inhibits apolipoprotein (a) expression and lipoprotein (a) synthesis in humans. J Lipid Res. 2015;56:1034–1042. doi: 10.1194/jlr.P052209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lennard-Jones JE. Classification of inflammatory bowel disease. Scand J Gastroenterol. 1989;170(Suppl):2–6. doi: 10.3109/00365528909091339. [DOI] [PubMed] [Google Scholar]
- 20.Magro F, Gionchetti P, Eliakim R, et al. Third European Evidence-based Consensus on Diagnosis and Management of Ulcerative Colitis. Part 1: Definitions, Diagnosis, Extra-intestinal Manifestations Pregnancy, Cancer Surveillance, Surgery, and Ileo-anal Pouch Disorders. J Crohns Colitis. 2017;11:649–670. doi: 10.1093/ecco-jcc/jjx008. [DOI] [PubMed] [Google Scholar]
- 21.Kim WR, Therneau TM, Wiesner RH, et al. A revised natural history model for primary sclerosing cholangitis. Mayo Clin Proc. 2000;75:688–694. doi: 10.4065/75.7.688. [DOI] [PubMed] [Google Scholar]
- 22.Vries de EM, Wang J, Williamson KD, et al. A novel prognostic model for transplant-free survival in primary sclerosing cholangitis. Gut. 2018;67:1864–1869. doi: 10.1136/gutjnl-2016-313681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vesterhus M, Hov JR, Holm A, et al. Enhanced liver fibrosis score predicts transplant-free survival in primary sclerosing cholangitis. Hepatology. 2015;62:188–97. doi: 10.1002/hep.27825. [DOI] [PubMed] [Google Scholar]
- 24.Wang J, Thingholm LB, Skiecevičienė J, et al. Genome-wide association analysis identifies variation in vitamin D receptor and other host factors influencing the gut microbiota. Nat Genet. 2016;48:1396–1406. doi: 10.1038/ng.3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Franzosa EA, McIver LJ, Rahnavard G, et al. Species-level functional profiling of metagenomes and metatranscriptomes. Nat Methods. 2018;15:962–968. doi: 10.1038/s41592-018-0176-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Truong DT, Franzosa EA, Tickle TL, et al. MetaPhlAn2 for enhanced metagenomic taxonomic profiling. Nat Methods. 2015;12:902–903. doi: 10.1038/nmeth.3589. [DOI] [PubMed] [Google Scholar]
- 27.Midttun Ø, Hustad S, Ueland PM. Quantitative profiling of biomarkers related to B-vitamin status, tryptophan metabolism and inflammation in human plasma by liquid chromatography/tandem mass spectrometry. Rapid Commun Mass Spectrom. 2009;23:1371–1379. doi: 10.1002/rcm.4013. [DOI] [PubMed] [Google Scholar]
- 28.Midttun Ø, McCann A, Aarseth O, et al. Combined Measurement of 6 Fat-Soluble Vitamins and 26 Water-Soluble Functional Vitamin Markers and Amino Acids in 50 μL of Serum or Plasma by High-Throughput Mass Spectrometry. Anal Chem. 2016;88:10427–10436. doi: 10.1021/acs.analchem.6b02325. [DOI] [PubMed] [Google Scholar]
- 29.Ursell LK, Haiser HJ, Treuren W Van, et al. The intestinal metabolome: an intersection between microbiota and host. Gastroenterology. 2014;146:1470–1476. doi: 10.1053/j.gastro.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hanes JW, Keresztes I, Begley TP. 13C NMR snapshots of the complex reaction coordinate of pyridoxal phosphate synthase. Nat Chem Biol. 2008;4:425–430. doi: 10.1038/nchembio.93. [DOI] [PubMed] [Google Scholar]
- 31.Magnúsdóttir S, Ravcheev D, Crécy-Lagard de V, et al. Systematic genome assessment of B-vitamin biosynthesis suggests co-operation among gut microbes. Front Genet. 2015;6:148. doi: 10.3389/fgene.2015.00148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pedersen HK, Gudmundsdottir V, Nielsen HB, et al. Human gut microbes impact host serum metabolome and insulin sensitivity. Nature. 2016;535:376–381. doi: 10.1038/nature18646. [DOI] [PubMed] [Google Scholar]
- 33.Talwar D, Catchpole A, Wadsworth JM, et al. The relationship between plasma albumin, alkaline phosphatase and pyridoxal phosphate concentrations in plasma and red cells: Implications for assessing vitamin B6 status. Clin Nutr. 2020;39:2824–2831. doi: 10.1016/j.clnu.2019.12.012. [DOI] [PubMed] [Google Scholar]
- 34.Yoshii K, Hosomi K, Sawane K, et al. Metabolism of Dietary and Microbial Vitamin B Family in the Regulation of Host Immunity. Front Nutr. 2019;6:48. doi: 10.3389/fnut.2019.00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fitzpatrick TB, Amrhein N, Kappes B, et al. Two independent routes of de novo vitamin B6 biosynthesis: Not that different after all. Biochem J. 2007;407:1–13. doi: 10.1042/BJ20070765. [DOI] [PubMed] [Google Scholar]
- 36.Ueland PM, Ulvik A, Rios-Avila L, et al. Direct and Functional Biomarkers of Vitamin B6 Status. Annu Rev Nutr. 2015;35:33–70. doi: 10.1146/annurev-nutr-071714-034330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Henderson JM, Codner MA, Hollins B, et al. The fasting B6 vitamer profile and response to a pyridoxine load in normal and cirrhotic subjects. Hepatology. 1986;6:464–471. doi: 10.1002/hep.1840060324. [DOI] [PubMed] [Google Scholar]
- 38.Ulvik A, Midttun Ø, Pedersen ER, et al. Association of plasma B-6 vitamers with systemic markers of inflammation before and after pyridoxine treatment in patients with stable angina pectoris. Am J Clin Nutr. 2012;95:1072–1078. doi: 10.3945/ajcn.111.029751. [DOI] [PubMed] [Google Scholar]
- 39.Borg ter PCJ, Fekkes D, Vrolijk JM, et al. The relation between plasma tyrosine concentration and fatigue in primary biliary cirrhosis and primary sclerosing cholangitis. BMC Gastroenterol. 2005;5:11. doi: 10.1186/1471-230X-5-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morgan MY, Marshall AW, Milsom JP, et al. Plasma amino-acid patterns in liver disease. Gut. 1982;23:362–370. doi: 10.1136/gut.23.5.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang Z, Klipfell E, Bennett BJ, et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 2011;472:57–63. doi: 10.1038/nature09922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Holeček M. Branched-chain amino acid supplementation in treatment of liver cirrhosis: Updated views on how to attenuate their harmful effects on cataplerosis and ammonia formation. Nutrition. 2017;41:80–85. doi: 10.1016/j.nut.2017.04.003. [DOI] [PubMed] [Google Scholar]
- 43.Corbett AJ, Eckle SBG, Birkinshaw RW, et al. T-cell activation by transitory neo-antigens derived from distinct microbial pathways. Nature. 2014;509:361–365. doi: 10.1038/nature13160. [DOI] [PubMed] [Google Scholar]
- 44.Heymann F, Tacke F. Immunology in the liver - from homeostasis to disease. Nat Rev Gastroenterol Hepatol. 2016;13:88–110. doi: 10.1038/nrgastro.2015.200. [DOI] [PubMed] [Google Scholar]
- 45.von Seth E, Zimmer CL, Reuterwall-Hansson M, et al. Primary sclerosing cholangitis leads to dysfunction and loss of MAIT cells. Eur J Immunol. 2018;48:1997–2004. doi: 10.1002/eji.201847608. [DOI] [PubMed] [Google Scholar]
- 46.Duan Y, Llorente C, Lang S, et al. Bacteriophage targeting of gut bacterium attenuates alcoholic liver disease. Nature. 2019;575:505–511. doi: 10.1038/s41586-019-1742-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.He Y, Wu W, Zheng H-M, et al. Regional variation limits applications of healthy gut microbiome reference ranges and disease models. Nat Med. 2018;24:1532–1535. doi: 10.1038/s41591-018-0164-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.