Abstract
CRISPR-Cas technologies have provided programmable gene editing tools that have revolutionized research. The leading CRISPR-Cas9 and Cas12a enzymes are ideal for programmed genetic manipulation, however, they are limited for genome-scale interventions. Here, we utilize a Cas3-based system featuring a processive nuclease for genome engineering. This minimal Cascade-Cas3 system (Type I-C), programmed with a single crRNA, was optimized to generate deletions with near-100% efficiency, and used to rapidly generate large deletions ranging from 7 - 424 kb in Pseudomonas aeruginosa. By comparison, Cas9 yielded small deletions and point mutations. Cas3-generated deletion boundaries were highly variable, but successfully specified by a homology-directed repair (HDR) template. HDR was much more efficient when lesions were generated by Cas3, compared to Cas9. The minimal Type I-C system is also portable; using an “all-in-one” vector, large deletions could be efficiently generated in Pseudomonas syringae, Escherichia coli, and Klebsiella pneumoniae. Notably, Cas3 generated bi-directional deletions originating from the programmed cut site, which was exploited to rapidly and iteratively reduce a P. aeruginosa genome by 837 kb (13.5 %) using 10 distinct crRNAs. We also enhance the utility of Cas3 systems by developing an “anti-anti-CRISPR” strategy to circumvent endogenous CRISPR-Cas inhibitor proteins. CRISPR-Cas3 could facilitate rapid strain manipulation for synthetic biology and metabolic engineering purposes, genome minimization, mobile genetic element removal, and the analysis of large regions of unknown function.
Introduction
CRISPR-Cas systems are a diverse group of RNA-guided nucleases1 that defend prokaryotes against viral invaders2,3. Gene-editing applications have focused on single subunit Class 2 CRISPR systems4 (i.e. Cas9 and Cas12a), but Class 1 systems hold great potential for editing technologies, despite consisting of multi-subunit complexes5–8. The signature gene in Class 1 Type I systems is Cas3, a 3’-5’ ssDNA helicase-nuclease enzyme that, unlike Cas9 or Cas12a, degrades target DNA processively5,6,9–14. This property of Cascade (CRISPR-associated complex for antiviral defense)-Cas3 systems has raised the possibility of its development as a tool for large genomic deletions, a task that current approaches using Class 2 systems are inefficient at.
Type I systems are the most prevalent CRISPR-Cas systems in nature15, which has enabled the use of endogenous CRISPR-Cas3 systems for genetic manipulation via self-targeting. This has been accomplished in Pectobacterium atrosepticum (Type I-F)16, Escherichia coli (Type I-E)17,18, Sulfolobus islandicus (Type I-A)19, various Clostridium species (Type I-B)20–22, Lactobacillus crispatus (Type I-E)23, Serratia sp. (Type I-F)24, Haloarcula hispanica (Type I-B)25, Streptococcus thermophilus (Type I-E)26, Pseudomonas aeruginosa (Type I-F)27, and Zymomonas mobilis (Type I-F)28, most frequently being used to generate small deletions. This multitude of different Type I systems have been shown to work to various degrees as editing tools in their native hosts, however, no Type I system has been optimized for efficient heterologous editing in bacteria, beyond the demonstration of the toxic effects of self-targeting29. Recent studies have repurposed Type I systems for use in human cells, via ribonucleoprotein (RNP) based delivery30 or plasmid-based expression31 of a Type I-E system, the fusion of FokI nuclease to Type I-E Cascade complex32, and Type I-F, I-E, and I-B systems have been used for transcriptional modulation33–35.
Here, we repurposed and optimized a Type I-C CRISPR system from Pseudomonas aeruginosa for both endogenous and heterologous genome engineering in four microbial species. Compared to other Type I systems, this presents a streamlined approach, requiring only four total proteins, which are easily transferable to different organisms. Importantly, by targeting the genome with a single crRNA and selecting only for survival after editing, this tool is a counter-selection-free approach to programmable large-scale genome engineering. We demonstrate this potential through iterative genome minimization of P. aeruginosa, achieving a 13.5 % total genome reduction on a ~30-day timescale. CRISPR-Cas3 is capable of efficient genome-scale deletions currently not achievable using other methodologies. It has the potential to serve as a powerful tool for basic research, discovery, and strain optimization.
Results
Implementation and optimization of genome editing with CRISPR-Cas3
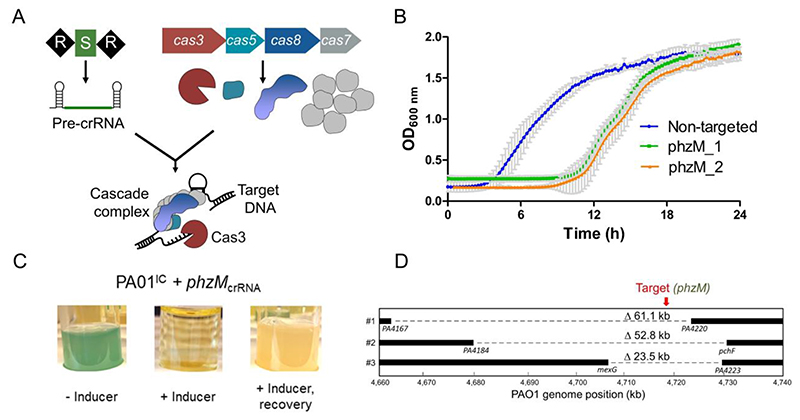
Type I-C CRISPR-Cas systems utilize just three cas genes (cas5, cas8, and cas7) to produce the crRNA-guided Cascade surveillance complex that can recruit Cas3 (Figure 1A), making it a compact system36,37 (see Supplementary Figure 1A for a comparison to other previously identified I-C systems). A previously constructed38 Pseudomonas aeruginosa PAO1 strain (PAO1IC) with inducible cas genes (cas5-8-7-3) and plasmid-expressed crRNAs targeting the genome was used to conduct genome manipulation. The initial transformation of the crRNA expression plasmids in uninduced conditions was not noticeably toxic, indicating tight regulation of the constructs (see Methods). Subsequent induced expression of crRNAs targeting the genome caused a transient growth delay (Figure 1B), but survivors were isolated after extended growth. By targeting phzM, a gene required for production of a blue-green pigment (pyocyanin), we observed yellow cultures (Figure 1C) for 10/18 and 6/18 surviving colonies, from two independent phzM targeting crRNAs. PCR of genomic DNA confirmed that the yellow cultures had lost this region, while blue-green survivors maintained it (Supplementary Figure 1B). Three of these deletion strains were sequenced, revealing deletions of 23.5 kb, 52.8 kb, and 60.1 kb, and each one was bi-directional relative to the crRNA target site (Figure 1D). This demonstrated the potential for Type I-C Cas3 systems to be used to induce large genomic deletions with random boundaries surrounding a programmed target site.
Figure 1.
A) A schematic of the Type I-C cas gene operon and CRISPR array. The surveillance complex is made up of Cas proteins (Cas51:Cas81:Cas77) and one crRNA, which recruits Cas3 upon target DNA recognition. Cas3 then degrades DNA through its dual helicase-nuclease activity. B) Growth curves of 2 PAO1IC strains expressing different crRNAs targeting phzM (green and orange) compared to a non-targeting strain (blue). Values are the mean of 8 biological replicates each, error bars indicate SD values. C) Cultures resulting from phzM targeting, in the absence of inducer (-ind), presence (+ind), and after recovery. D) Whole-genome sequencing of threePAO1ICself-targeted survivor strains. Bars indicate boundaries of deletions with ORF indicated below; red arrow indicates genomic position of targeted sequences.
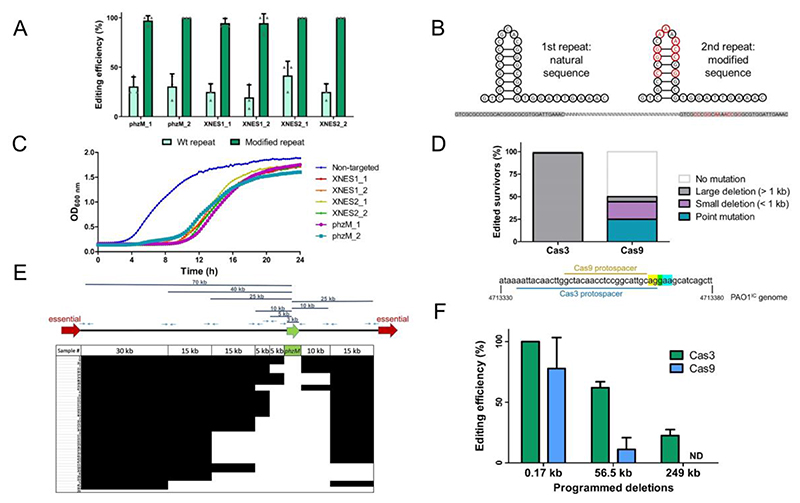
To determine the in vivo processivity of the Cas3 enzyme, we targeted 2 of the 16 extended non-essential (XNES) regions >100 kb in length (Supplementary Table 1) identified from a transposon sequencing (TnSeq) data set39. The frequency of deletions generated by crRNAs targeting XNES 1 and XNES 2 (along with additional targeting of phzM, which is found in XNES 15) was quantified revealing that 20-40 % of the surviving colonies had deletions (Figure 2A). To understand how cells lacking large deletions had survived self-targeting, three possibilities were considered: i) a cas gene mutation, ii) a PAM or protospacer mutation, or iii) a mutation to the plasmid expressing the crRNA. Three survivors lacking target deletions from each of the six self-targeting crRNAs were assayed. All had functional cas genes when the self-targeting crRNA was replaced with a phage-targeting crRNA (Supplementary Figure 2A), and target sequencing revealed no point mutations. PCR-amplification and sequencing of the crRNA-expressing plasmids isolated from the survivors revealed the primary escape mechanism: recombination between the direct repeats, leading to the loss of the spacer (Supplementary Figure 2B). An additional 17 survivors that lacked deletions were assayed via PCR and were also ~60 bp shorter (Supplementary Figure 2C-D), consistent with the loss of one repeat and spacer.
Figure 2.
A) Percentage of survivors with a genomic deletion at the location targeted. Six different crRNA constructs with either wild-type (Wt) repeat sequences (light green) or with the second repeat being modified (dark green). Values are means of 3 biological replicates each, where 12 individual surviving colonies were assayed per replicate, error bars show SD values. B) Sequence and structure of natural and modified repeat sequences. Specifically engineered modified nucleotides shown in red; repeat sequences highlighted in gray with an arbitrary intervening spacer sequence. C) Growth curves of PAO1IC strains expressing distinct self-targeting crRNAs flanked by modified repeats. Non-targeting crRNA expressing control is marked in blue. Values depicted are averages of 4 biological replicates each. D) Gene editing outcomes for distinct survivor cells targeted with either a Type II-A SpyCas9 system or a Type I-C Cas3 system (n=72). E) Deletion size distribution of 47 independently generated phzM deletion strains determined using a tiling PCR approach. Black segments indicate the presence of a PCR product for a given sample, while white segments indicate the absence, thus giving an estimate of the size for the generated deletions. F) Percentage of analyzed survivors with the specific deletion size present (0.17kb, 56.5 kb, or 249 kb) using homologous repair templates with the Cas3 system (green) or the SpyCas9 system (blue). Values are means of 3 biological replicates each, where 12 individual surviving colonies were assayed per replicate, and have been normalized to the total percentage of edited samples for each enzyme shown in Figure 2D (98.6 % for Cas3, 50 % for Cas9); error bars show SD values, ND: not detected.
Spacer excision was successfully prevented by engineering a modified repeat (MR), with six mutated nucleotides in the stem and three in the loop of the second repeat (Figure 2B), disrupting homology between the two direct repeats. A phage-targeting crRNA with this new design targeted phage as well or better than the same crRNA with unmodified repeats (Supplementary Figure 3A). Targeting of the same 6 genomic sites with MR-incorporating crRNAs resulted in encouraging growth delays in all cases (Figure 2C) and in a robust increase in editing efficiencies to 94-100 % (Figure 2A). Spacer excision was no longer detected. 211 of 216 (98 %) total survivor cells had large deletions based on PCR screening (i.e. > 1 kb), while the remaining 5 had inactive CRISPR-Cas systems when tested with the phage-targeting crRNA (Supplementary Figure 3B).
When targeting poorly characterized genomes, it is expected that essential genes may be unknowingly targeted leading to confounding editing outcomes. To assess the phenotype of such an event, we intentionally targeted an essential gene, rplQ (a 50S ribosomal subunit protein40). Two different MR crRNAs targeting rplQ led to a severely extended lag time compared to non-essential gene targeting. Only 8 out of 36 rplQ-targeting biological replicates grew after 24 hours, compared to the transient growth delay of ~12 hours when targeting non-essential genes (Supplementary Figure 4A). Subsequent analysis of these 8 survivor cultures with phage targeting assays revealed non-functional cas genes (Supplementary Figure 4B). Importantly, no spacer excision events were detected in this experiment, confirming the robustness of the crRNA engineering and of the deletion method, as the outcome of essential gene versus non-essential gene targeting is noticeably distinct.
Cas3 generates larger deletions than Cas9 and is recombinogenic
To determine whether large deletions are a direct consequence of the processive Cas3 enzyme, we compared Cas3-mediated self-targeting outcomes to those resulting from targeting using Streptococcus pyogenes Cas9, which lacks a helicase domain, expressed in an isogenic strain (PAO1IIA). It has previously been shown that large genomic island deletions can be naturally present within bacterial populations at small frequencies41. If the large deletions we observed in P. aeruginosa are preexisting, we would expect these to be selected for at comparable frequencies regardless of the system used to target the genome. To investigate this, we designed a Cas9 sgRNA that overlapped with one of the Cas3 crRNAs used to target phzM (Figure 2D, Supplementary Figure 5). PCR and sequencing analysis of these surviving cells revealed that deletions larger than 1 kb were a rare occurrence in the presence of Cas9 (5.6 % assayed survivor cells, n = 72) compared to 98.6 % with Cas3 (Figure 2D). Whole-genome sequencing (WGS) of two large deletion survivors selected for by Cas9 showed lesions of 5 kb and 23 kb around the target site, respectively. The more common modes of survival after Cas9 targeting were small deletions between 0.1 – 0.5 kb in length (25 % of all survivors), or 1-3 bp protospacer/PAM deletions/mutations (19.4 %), with the remaining 50 % of survivors unedited at their target sites. In sum, the apparent shift of deletions toward smaller size resulting from targeting with SpyCas9, compared to a nearly completely distinct set of outcomes when using Cas3, implicates Cas3’s enzymatic activity as the cause of large deletions.
To achieve a more granular measurement of the deletion sizes generated by Cas3-mediated editing, we examined 47 individual phzM-targeting events. Tiling PCR was used to determine the presence of flanking segments at various intervals spanning a total of 95 kb encompassing phzM (Figure 2E). Out of the 47 independent deletion outcomes, 44 had deletions of at least 5 kb, and 22 had deletions of at least 35 kb in size, with one strain having a deletion larger than 95 kb. The average deletion outcome was larger than 26.6 kb and smaller than 48.2 kb, based on the resolution of the tiling experiment. This comprehensive assessment confirms both the variability of Cas3-induced deletion outcomes and emphasizes how distinct they are from Cas9.
The processive ssDNA activity of nuclease-helicase Cas3 led us to hypothesize that it may promote recombination by exposing regions of ssDNA. To test this, we provided a repair template with 500 bp of the upstream and downstream regions flanking the desired deletion to enable homology directed repair (HDR). We chose 0.17 kb and 56.5 kb deletions around phzM, to model a gene and prophage deletion, respectively, and a large 249 kb deletion within XNES8 for the programmed deletions (Supplementary Figure 6). The recombination efficiencies were significantly higher with Cas3 than with Cas9 (Figure 2F). The 249 kb deletion was incorporated in 22 % of the Cas3-generated survivors, compared to 0% using Cas9 (χ2(1, N = 72) = 9, p = 2.7E-03). The 56.5 kb deletion had an efficiency of 61 % vs. 11 % (χ2(1, N = 72) = 25, p = 5.73E-07), and the 0.17 kb deletion had an efficiency of 100 % vs. 78 % when targeting with Cas3 or Cas9, respectively (χ2(1, N = 72) = 31.68, p = 1.82E-08). Of note, when Cas3 was used to generate 56.5 kb and 249 kb deletions, all of the isolates that did not produce a PCR fragment indicating successful HDR also lacked a wild-type PCR band. This indicates that non HDR-mediated deletions occurred as well. Most of the strains that survived Cas9 editing without incorporating the HDR template had no change at the target site (84.7 % and 80.6 % for the 56.5 kb and 249 kb deletions, respectively), similar to data reported above (Figure 2E). We presume that mutation or loss of Cas9 occurs more frequently than loss of the Cas3-based system under this experimental set-up. To account for this, we normalized the editing efficiency by ~2-fold (derived from the frequency of unedited clones in Figure 2D) to compare the ability of Cas3 and Cas9 to generate the desired outcome (Figure 2F). When assessing either the absolute percentage of colonies with the desired edits or the normalized value, Cas3 still outperforms Cas9, supporting the hypothesis that Cas3 has strong recombinogenic potential when generating larger lesions.
Rapid genome minimization of P. aeruginosa using CRISPR-Cas3 editing
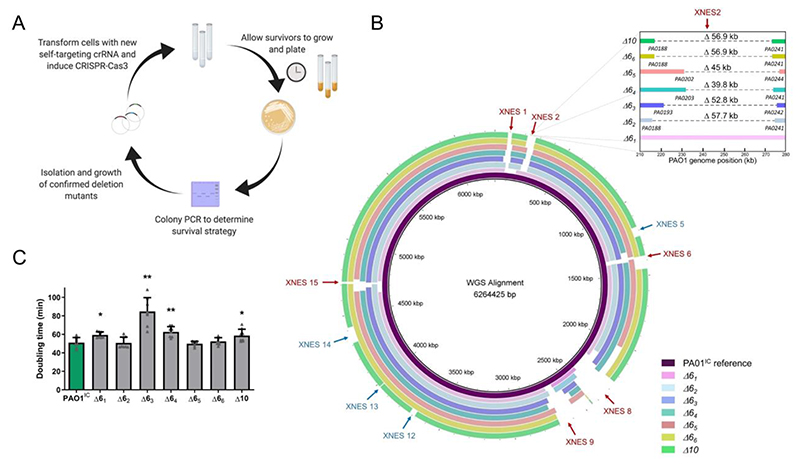
Large deletions with undefined boundaries provide an unbiased mechanism for genome streamlining, screening, and functional genomics. To demonstrate the potential for Cas3, we aimed to minimize the P. aeruginosa genome through a series of deletions of the XNES regions (Figure 3A). Six XNES regions (including XNES 15, carrying phzM) were iteratively targeted in six parallel lineages (Figure 3B), resulting in 35 independent deletions (WGS revealed no deletion at XNES 2 in one of the strains). Deletion efficiency remained high (> 80 %) throughout each round of self-targeting (Supplementary Figure 7A). WGS of these 6 multiple deletion strains (Δ61 -Δ66) revealed that no two deletions had the exact same coordinates, highlighting the stochastic nature of Cas3. The smallest isolated deletion was 7 kb and the largest 424 kb (mean: 92.9 kb, median: 58.2 kb). Of note, 4 genes (PA0123, PA1969, PA2024, and PA2156) previously identified as essential39 were deleted in at least one of the lineages. Most deletions appeared to be resolved by flanking microhomology regions ranging from 4-14 bp in length (Supplementary Figure 7B, Supplementary Table 2), implicating alternative-end joining42 as the dominant repair process.
Figure 3.
A) Schematic overview of the iterative deletion generating process. B) Whole-genome sequences of six PAO1IC strains that have been iteratively targeted at six distinct genomic positions and one (derived from strain Δ66) with ten total deletions (Δ10) aligned to the parental P. aeruginosa PAO1IC strain. The first six targeted sites are marked with red arrows, and the final four are marked with blue arrows. Inset shows deletion coordinates of XNES2 region of the various strains in finer detail. C) Calculated doubling times of the seven genome-reduced strains (strains Δ61 – 66 with six deletions, Δ10 with ten) compared to the parent PAO1ICstrain (green). Values are means of 8 biological replicates, error bars represent SD values, * p < 0.05, ** p < 0.01, paired T-test compared to PAO1IC.
To minimize the genome further, one of the already reduced strains was subjected to 4 additional rounds of deletions at XNES regions for a total of 10 genomic deletions (Δ10, Figure 3B). Whole-genome sequencing of the Δ10 strain showed a genome reduction of 849 kb (13.6 % of the genome). Generation of large deletions resulted in a growth defect in some cases, with significantly slower growth in 3 of the 6 deletion strains (Δ61, Δ63, and Δ64), with the other 3 growing normally (Figure 3C). Δ10 also displayed a slight decrease in fitness, showing a ~15 % increase in doubling time compared to the parent strain. Stronger growth defects were likely avoided by the selection of fast-growing colonies at each deletion round.
To again address whether these deletions may be pre-existing at low frequencies, we analyzed the presence or absence of a representative set of deletion-junction specific PCR products in wild-type cells. Primers designed to capture specific deletions at XNES1, 6, 8, and 9 revealed that these deletions were not detectable in wild-type cultures (Supplementary Figure 7C), with the exception of one non-specific PCR product. The lack of deletion-junction amplification products indicates the absence of naturally occurring deletion events, implicating the direct role of Cas3 in generating large deletions.
CRISPR-Cas3 editing in distinct bacteria
To enable expression of this system in other hosts, we constructed an all-in-one vector (pCas3cRh) carrying the I-C specific crRNA with a modified repeat sequence, cas3, cas5, cas8, and cas7, under rhamnose induction (Supplementary Figure 8A). As a pilot experiment, we transformed wild-type PAO1 with a non-targeting crRNA and crRNAs targeting phzM and XNES2. Expression of the crRNAs appeared to not be considerably leaky as transformation efficiency was similar comparing targeting and non-targeting constructs. Subsequent induction of the targeting crRNAs resulted in editing efficiencies between 95-100 % (Supplementary Figure 8B-D).
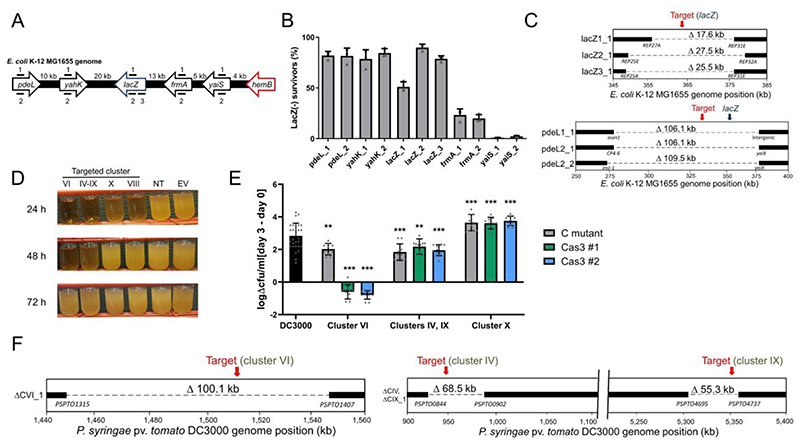
Having verified that pCas3cRh was functional, we tested this system in the model organism Escherichia coli K-12 MG1655. crRNAs were designed to target lacZ or its vicinity (Figure 4A), where it is flanked by non-essential DNA (124.5 kb upstream, 22.4 kb downstream). Transformations were plated directly on inducing media containing X-gal and scored using blue/white screening. Depending on the crRNA used, directly targeting lacZ or 30 kb upstream yielded 51-90 % or 82-85 % LacZ(-) survivors, respectively (Figure 4B). 95 of the 96 LacZ (-) survivors assayed by PCR showed an absence of the lacZ region. crRNAs downstream of lacZ, however, had reduced efficiency as they approached the essential gene, hemB. frmA targeting (13 kb downstream of lacZ) resulted in lower editing efficiencies (21-25 %) while targeting yaiS (18 kb downstream of lacZ) was even lower (2 %). This decrease in efficiency was independent of the strand being targeted (and therefore the predicted strand for Cas3 loading and 3’-5’ translocation), confirming the importance of Cas3 bi-directional deletions. WGS of selected ΔlacZ cells revealed bi-directional deletions ranging from 17.5-106 kb encompassing the targeted region (Figure 4C). Based on these findings, the nearby presence of an essential gene can significantly lower editing efficiency and must be considered when targeting a selected region.
Figure 4.
A) Schematic of the crRNA targeted sites in the E. coli MG1655 genome at the lacZ locus. B) lacZ deletion efficiencies using distinct crRNAs targeting the E. coil K-12 MG1655 chromosome. Efficiencies calculated based on LacZ activity. Values are averages of 3 biological replicates, error bars represent standard deviations. C) Whole-genome sequencing of E. coli deletion mutants targeted at lacZ and 30 kb upstream at pdeL. Bars indicate boundaries of deletions with ORF indicated below. D) Growth of P. syringae DC3000 strains expressing the I-C system and distinct crRNAs. Constructs VI, IV-IX, and VIII target P. syringae DC3000 non-essential chromosomal genes, non-targeting crRNA (NT), empty vector (EV). E) Bacterial growth of deletion mutants in Arabidopsis thaliana. Values are differences in colony forming units (cfu) / ml counted on day 0 of the experiment and day 3, shown on a logarithmic scale. The wild-type DC3000 strain is shown in black, while gray bars represent previously constructed polymutant control (C) strains of the different clusters (labeled at bottom), and green and blue bars show deletion mutants generated using Cas3 (two isolated strains for each targeted cluster, #1, #2). Values shown are means of 10 biological replicates each (30 for DC3000), error bars show SD values, ** p < 0.01, *** p < 0.005, ANOVA analysis (see methods). F) Whole-genome sequencing of P. syringae deletion mutants. Left panel shows virulence cluster VI targeting, while right panel shows virulence cluster IV and IX targeting with a single crRNA, as the clusters share sequence identity. Bars indicate boundaries of deletions with ORF indicated below.
Next, we tested Cas3-mediated editing in the plant pathogen Pseudomonas syringae pv. tomato DC3000, which does not naturally encode a CRISPR-Cas system43. P. syringae encodes many non-essential virulence effector genes whose activities are difficult to disentangle due to their redundancy44. We designed crRNAs targeting four chromosomal virulence effector clusters (IV, VI, VIII, and IX), or one plasmid cluster (pDC300045, cluster X) in P. syringae strain DC3000. Two clusters (IV and IX) shared identical sequences that could be targeted simultaneously using a single crRNA. Expression of targeting crRNAs led to a noticeable growth delay compared to non-targeted controls (Figure 4D). PCR analysis of surviving cells showed editing efficiencies of 67-92 % (Supplementary Figure 9A). In planta and in vitro growth assays of three deletion mutants effectively recapitulated the phenotypes of previously described cluster deletion polymutants45 (Figure 4E, Supplementary Figure 9B-G). Targeting cluster X cured the 73 kb plasmid (as observed by the absence of plasmid-specific reads in WGS) and simultaneous cluster IV and IX targeting led to dual deletions in 8 out of 12 survivors, with a sequenced representative having 68.5 kb and 55.3 kb deletions, respectively, at the target sites. The effector cluster VI Cas3-derived mutant (100.1 kb deletion) had a more severe growth defect in vitro and in planta than the control mutant, likely stemming from a fitness defect owing to the missing genetic material (Figure 4E, 4F, and Supplementary Figure 9B,9E). This outcome underscores the importance of considering multiple survivors using the appropriate assays to identify the desired mutant. In contrast to P. aeruginosa, we also note the presence of IS elements at the deletion junction sites, indicating the involvement of homologous recombination between IS-elements flanking the virulence gene clusters as a resolution to genomic targeting. In such instances, we have not ruled out that the loss of these large regions was not a natural occurrence in the population, as seen in Streptococcus thermophilus 41. In two out of three cases however, the deletions did entail significant fitness costs (Figure 4F), decreasing this likelihood. Using our portable streamlined system, we achieved three distinct applications in P. syringae: the single-step deletion of large virulence regions, multiplexed targeting, and plasmid curing.
Finally, we tested the feasibility of heterologous editing using the I-C system in the more distantly related and clinically relevant bacteria Klebsiella pneumoniae. Utilizing pCas3cRh, we targeted the genome of K. pneumoniae strain KPPR146 with 4 distinct crRNAs, with two each targeting the rfaH and sacX genes. These were selected as targets because the absence of the transcriptional elongation factor rfaH has been shown to lead to a smaller colony size compared to wild-type47, while sacX is located in an extended region of the genome with contiguous individually non-essential genes47. Analysis of induced targeted cultures of KPPR1 with all four crRNAs resulted in a significant growth compared to a non-targeted control, indicating functionality of the I-C system in K. pneumoniae (Supplementary Figure 10A). Individual colonies were isolated from the surviving targeted cultures that eventually grew and analyzed using PCR amplification of the genomic target sites (Supplementary Figure 10B). Editing efficiencies using the four different crRNAs ranged from 38-63 % showing the feasibility of Cas3 editing in K. pneumoniae as well (Supplementary Figure 10C). The phenotypic effect of rfaH deletion was confirmed, as mutants formed small colonies compared to wild-type (Supplementary Figure 10D). Overall, we have demonstrated portable Type I-C CRISPR-Cas3 editing to be a generally applicable tool capable of generating large genomic deletions in four distinct bacteria.
Repurposing endogenous CRISPR-Cas3 systems for gene editing
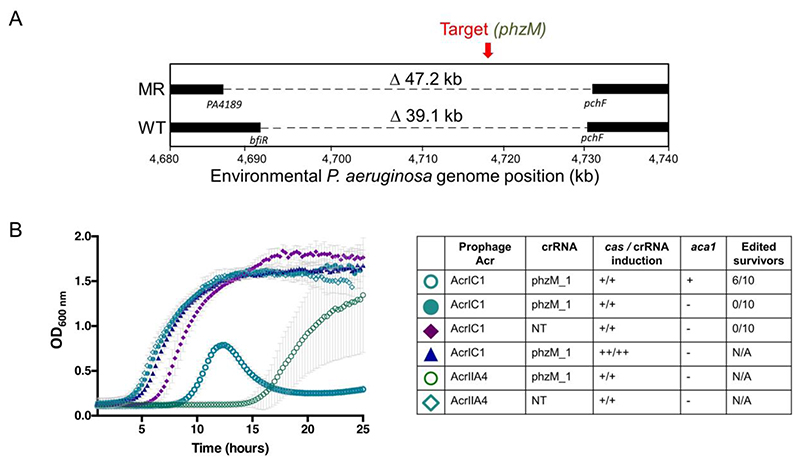
Type I CRISPR-Cas3 systems are the most common CRISPR-Cas systems in nature1. Therefore, many bacteria have a built-in genome editing tool to be harnessed. We first tested the environmental isolate (PaLML1) from which our Type I-C system was derived. Self-targeting phzM crRNAs led to the isolation of genomic deletions (Supplementary Figure 11A), with WGS revealing 33.7 (wild-type repeat) and 39 kb (MR) deletions of the target gene and surrounding regions (Figure 5A). Additionally, HDR-based editing with a single construct was again efficacious, with 7/10 survivors acquiring the specific 0.17 kb deletion (Supplementary Figure 11B).
Figure 5.
A) Schematic of whole genome sequencing of an environmental isolate of PAO1 with an endogenous Type I-C system. Two survivors were isolated post-targeting using either wild-type (WT) direct repeats flanking the spacer, or modified repeats (MR). Bars indicate boundaries of deletions with ORF indicated below. B) Growth curves of PAO1IC lysogenized by recombinant DMS3m phage expressing acrIIA4 or acrIC1 from the native acr locus. CRISPR-Cas3 activity is induced with either 0.5mM (+) or 5mM (++) IPTG and 0.1% (+) or 0.3% (++) arabinose. Edited survivors reflect number of isolated survivor colonies missing the targeted gene (phzM) and ‘N/A’ means that editing was not assessed as no growth defect was seen. ‘NT’ means a non-targeting crRNA was expressed. Each growth curve is the average of 10 biological replicates and error bars represent SD.
We next evaluated the feasibility of repurposing other Type I systems, using the naturally active Type I-F systems48 encoded by laboratory strain P. aeruginosa PA14, and the clinical strain P. aeruginosa z8. Plasmids with Type I-F specific crRNAs were expressed, targeting various genomic sites for deletion (Supplementary Table 3). HDR templates (600 bp arms on average) were included in the plasmids to generate deletions of defined coordinates ranging from 0.2 to 6.3 kb. Overall, at 5 different genomic target sites in strain z8 and 2 sites in PA14, we observed desired deletions in 29-100 % of analyzed survivor colonies (Supplementary Table 3). Together, these experiments demonstrate the capacity for different forms of high efficiency genome editing using a single plasmid and an endogenous CRISPR-Cas system.
Finally, one potential impediment to the implementation of any CRISPR-Cas bacterial genome editing tool is the presence of anti-CRISPR (acr) proteins that inactivate CRISPR-Cas activity49. In the presence of a prophage expressing AcrIC1 (a Type I-C anti-CRISPR protein38) from a native acr promoter, self-targeting was completely inhibited, but not by an isogenic prophage expressing a Cas9 inhibitor AcrIIA450 (Figure 5B). To attempt to overcome this impediment, we expressed aca1 (anti-CRISPR associated gene 1), a direct negative regulator of acr promoters51, from the same construct as the crRNA. Using this repression-based “anti-anti-CRISPR” strategy, CRISPR-Cas function was re-activated, allowing the isolation of edited cells despite the presence of acrIC1 (Figure 5B and Supplementary Figure 11C). In contrast, simply increasing cas gene and crRNA expression did not overcome AcrIC1-mediated inhibition as assessed by growth kinetics (Figure 5B). Therefore, using anti-CRISPR repressors presents a viable route towards enhanced efficiency of CRISPR-Cas editing and necessitates continued discovery and characterization of anti-CRISPR proteins and their cognate repressors.
Discussion
By repurposing a minimal Cascade-Cas3 system as either an endogenous and heterologous genome editing tool, we show that hurdles to generating large deletions can be overcome. We obtained high editing efficiencies after modifying a repeat sequence to prevent spacer loss. Using only a single crRNA, we isolated many deletions of variable sizes, one as large as 424 kb, without requiring the insertion of a selectable marker or HDR templates guiding the repair process. Notably, the mean (92.9 kb) and median (58.2 kb) deletion sizes are roughly in the range of the average size of Pseudomonas bacteriophage (35 – 100 kb for 92 % of sequenced genomes52), suggesting that the Cascade-Cas3 machinery efficiently degrades entire phage genomes. Additionally, the I-C system appears to produce bi-directional deletions, similar to what was previously observed with the I-F Cascade-Cas3 system53, but not with type I-E10,11,30. CRISPR-Cas3 presents a genome editing tool useful for the targeted removal of large, potentially undesirable elements (e.g. virulence clusters, plasmids54) and also for unbiased screening and genome streamlining. As a long-term goal of microbial gene editing has been genome minimization55,56, we used our optimized CRISPR-Cas3 system to generate ten iterative deletions, achieving >13 % genome reduction of the targeted strain. This spanned only 30 days while maintaining editing efficiency, a great improvement over previous genome reduction methods56. Some basic microbial applications of Cas3 include studying chromosome biology (e.g. replichore asymmetry57), pathogenesis58, the impact of the mobilome, and better understanding the essential building blocks for life.
An important outcome of this work is the high efficiency of recombination observed at cut sites when comparing Cas3 and Cas9 directly. The potential for Cas3 to be recombinogenic through the generation of exposed ssDNA may be advantageous for both programmed knock-outs and knock-ins. Although knock-ins were not systematically explored here, a preliminary attempt to affix a chromosomal mCherry tag to cas3, using the Type I-C system for counter-selection, was successful. The direct comparison presented here between Cas3 (large deletions) and Cas9 (small deletions), coupled with the high variability of deletions observed with Cas3 also confirms the causality of Cas3 in the deletion outcomes.
Our study has revealed some of the benefits and challenges of working with CRISPR-Cas3. While some of the iteratively edited strains demonstrated slight growth defects, the Cas3 editing workflow shows high potential for genome minimization efforts. Since many distinct deletion events are generated, screening various isolates for fitness benefits or defects is possible, and one can proceed with the strain that has the desired fitness property. Additionally, the nearby presence of an essential gene may greatly limit editing efficiencies, but may potentially be overcome by the addition of a repair template. Despite our success at transplanting the minimal Type I-C system, it remains to be seen whether the approach will be limited by differences in DNA repair mechanisms. Indeed, in E. coli and P. syringae, larger regions of homology, such as 34 bp long REP sequences were observed59, indicating the role of RecA-mediated homologous recombination60 in the repair process. Meanwhile in P. aeruginosa, the borders of the deletions showed either small (4-14 bp) micro-homology or no noticeable sequence homology. The former implies a role for alternative end-joining42, which has also been observed in Pectobacterium atrosepticum16, while the latter non-homologous end-joining61 in the repair process. Downstream studies are required to dissect the roles of each mechanism in the deletion generation process and variability of these between different organisms will also need to be considered for better predictable deletion outcomes.
CRISPR-Cas3 is an especially promising tool for use in eukaryotic cells as it would facilitate the interrogation of large segments of non-coding DNA, much of which has unknown function62. Additionally, it was recently shown that Cas9-generated “gene knockouts” (i.e. small indels causing out-of-frame mutations) frequently encode pseudo-mRNAs that may produce protein products, necessitating methods for full gene removal63,64. Encouragingly, Type I-E CRISPR-Cas systems were recently shown to generate large (up to 100 – 200 kb) deletions in human cells30–32, demonstrating the potential wide applicability of Cas3. Overall, the intrinsic properties of Cas3 make it a promising tool to fill a void in current gene editing capabilities. Employing Cas3 to make large genomic deletions will facilitate the manipulation of repetitive and non-coding regions, having a broad impact on genetics research by providing a tool to probe genomes en masse.
Methods
Bacterial strains, plasmids, DNA oligonucleotides, and media
A previously described38 environmental strain of Pseudomonas aeruginosa was used as a template to amplify the four cas genes of the Type I-C CRISPR-Cas system genes (cas3, cas5, cas7, and cas8). The genes were cloned into the pUC18-mini-Tn7T-LAC vector65 using the SacI-PstI restriction endonuclease cut sites in the order cas5, cas7, cas8, cas3 to generate the plasmid pJW31 (Addgene number:136423). This vector was introduced into Pseudomonas aeruginosa PAO166, inserting the cas genes into the chromosome, following previously described methods67. Following integration, the excess sequences, including the antibiotic resistance marker, were removed via Flp-mediated excision as described previously67. The resulting strain, dubbed PAO1IC, allowed for inducible expression of the I-C system through induction with isopropylβ-D-1 thiogalactopyranoside (IPTG). This same method was used to integrate the Cas3-Cas8 tether mutant in the order cas5, cas3, cas8, cas7. The linker amino acid sequence is RSTNRAKGLEAVS. An isogenic strain carrying Cas9 derived from Streptococcus pyogenes was constructed in the same fashion, resulting in the strain PAO1IIA. For experiments to test the system in Pseudomonas syringae, we employed the previously characterized strain DC300043. E. coli editing experiments were conducted with strain K-12 MG165568. Experiments conducted with K. pneumoniae were performed using strain KPPR146.
To construct the Cas3 helicase and nuclease mutant strains, the PAOIIC system was utilized to introduce point mutations. crRNAs were designed to target Cas3 along with a homology directed repair (HDR) template that included the desired mutation, and silent mutations to prevent CRISPR-Cas targeting of the final strain.
To achieve genomic self-targeting of the I-C CRISPR-Cas strains, crRNAs designed to target the genome were expressed from the pHERD20T and pHERD30T shuttle vectors69. So-called “entry vectors” pHERD20T-ICcr and pHERD30T-ICcr were first generated by cloning at the EcoRI and HindIII sites an annealed linear dsDNA template carrying the I-C CRISPR-Cas system repeat sequences flanking two BsaI Type IIS restriction endonuclease recognition sites. Additionally, a preexisting BsaI site in a non-coding site of the pHERD30T and pHERD20T plasmids was mutated using whole-plasmid amplification so it would not interfere with the cloning of the crRNAs38. Oligonucleotides with repeat-specific overhangs encoding the various spacer sequences were annealed and phosphorylated using T4 polynucleotide kinase (PNK) and cloned into the entry vectors using the BsaI sites. For experiments using Cas9, sgRNAs were expressed from the same pHERD30T vector, with the sgRNA construct cloned using the same restriction sites as with the I-C crRNAs.
The all-in-one vector pCas3cRh (Addgene number 133773) is a derivative of the pHERD30T-IC plasmid, with the 4 I-C system genes cloned downstream of the crRNA site. This was achieved by amplifying the genes cas3, cas5, cas8, and cas7 in two fragments with a junction within cas8 designed to eliminate an intrinsic BsaI site with a synonymous point mutation. The amplified fragments were cloned into pHERD30T-IC using the Gibson assembly protocol70. Finally, to guard against potential leaky toxic expression, we replaced the araC-ParaBAD promoter with the rhamnose-inducible rhaSR-PrhaBAD system71. The sequence for rhaSR-PrhaBAD was amplified from the pJM230 template71, provided by the lab of Joanna B. Goldberg (Emory University), and cloned into the pHERD30T-IC plasmid to replace araC-ParaBAD using Gibson Assembly (New England Biolabs). Without induction, transformation efficiencies of targeting constructs of assembled pCas3cRh were on average 5-10-fold lower when compared to non-targeting controls (Supplementary Figure 8C), indicating residual leakiness of the I-C system.
The aca1-containing vector pICcr-aca1 is a derivative of the pHERD30T-ICcr plasmid, with aca1cloned downstream of the crRNA site under the control of the pBAD promoter. The aca1 gene was cloned from P. aeruginosa phage DMS3m.
All oligonucleotides used in this study were obtained from Integrated DNA Technologies. For a complete list of all DNA oligonucleotides and a short description, see Supplementary Table 4.
P. aeruginosa, E. coli, and K. pneumoniae strains were grown in standard Lysogeny Broth (LB): 10 g tryptone, 5 g yeast extract, and 10 g NaCl per 1 L dH2O. Solid plates were supplemented with 1.5 % agar. P. syringae was grown in King’s medium B (KB): 20 g Bacto Proteose Peptone No. 3, 1.5 g K2HPO4, 1.5 g MgSO4•7H2O, 10 ml glycerol per 1 L dH2O, supplemented with 100 μg/ml rifampicin. The following antibiotic concentrations were used for selection: 50 μg/ml gentamicin for P. aeruginosa, P. syringae, and K. pneumoniae, 15 μg/ml for E. coli; 50 μg/ml carbenicillin for all organisms. Inducer concentrations were 0.5 mM IPTG, 0.1 % arabinose, and 0.1 % rhamnose. For transformation protocols, all bacteria were recovered in Super optimal broth with catabolite repression (SOC): 20 g tryptone, 5 g yeast extract, 10 mM NaCl, 2.5 mM KCl, 10 mM MgCl2, 10 mM, MgSO4, and 20 mM glucose in 1 L dH2O.
Bacterial transformations
Transformations of P. aeruginosa, E. coli, P. syringae, and K. pneumoniae strains were conducted using standard electroporation protocols. 10 ml of overnight cultures were centrifuged and washed twice in an equal volume of 300 mM sucrose (20 % glycerol for E. coli) and suspended in 1 ml 300 mM sucrose (20 % glycerol for E. coli). 100μl aliquots of the resulting competent cells were electroporated using a Gene Pulser Xcell Electroporation System (Bio-Rad) with 50 – 200 ng plasmid with the following settings: 200Ω, 25 μF, 1.8 kV, using 0.2 mm gap width electroporation cuvettes (Bio-Rad). Electroporated cells were incubated in antibiotic-free SOC media for 1 hour at 37 °C (28 °C for P. syringae), then plated onto LB agar (KB agar for P. syringae) with the selecting antibiotic, and grown overnight at 37 °C (28 °C for P. syringae). Cloning procedures were performed in commercial E. coli DH5α cells (New England Biolabs) or E. coli XL1-Blue (QB3 Macrolab Berkeley), according to the manufacturer’s protocols.
Construction of recombinant DMS3m acr phages
The isogenic DMS3macrIIA4 and acrlC1phages were constructed using previously described methods72. A recombination cassette, pJZ01, was constructed with homology to the DMS3m acr locus. Using Gibson Assembly (New England Biolabs), either acrIC1or acrIIA4were cloned upstream of aca1, and the resulting vectors were used to transform PAO1IC. The transformed strains were infected with WT DMS3m, and recombinant phages were screened for. Phages were stored in SM buffer at 4°C.
Isolation of PAO1IClysogens
PAO1IC was grown overnight at 37°C in LB media. 150 μl of overnight culture was added to 4 ml of 0.7 % LB top agar and spread on 1.5 % LB agar plates supplemented with 10 mM MgSO4.5 μl of phage, expressing either acrIC1 or acrIIA4were spotted on the solidified top agar and plates were incubated at 30°C overnight. Following incubation, bacterial growth within the plaque was isolated and spread on 1.5% LB agar plate. After an overnight incubation at 37°C, single colonies were assayed for the prophage. Confirmed lysogens were used for genomic targeting experiments.
Genomic targeting
Pseudomonas aeruginosa
Genomic self-targeting of P. aeruginosa PAO1IC was achieved by electroporating cells with pHERD30T (or pHERD20T) expressing the self-targeting spacer of choice. Cells were plated onto LB agar plates containing the selective antibiotic, without inducers, and grown overnight. Single colonies were then grown in liquid LB media containing the selective antibiotic, as well as IPTG to induce the genomic expression of the I-C system genes, and arabinose to induce the expression of the crRNA from the plasmid. The aca1-containing crRNA plasmids do not need additional inducers, as the pBAD promoter controls aca1. Cultures were grown at 37 °C in a shaking incubator overnight to saturation, then plated onto LB agar plates containing the selecting antibiotic, as well as the inducers, and incubated overnight again at 37 °C. The resulting colonies were then analyzed individually using colony PCR for any differences at the targeted genomic site compared to a wild-type cell. gDNA was isolated by resuspending 1 colony in 20 μl of H2O, followed by incubation at 95°C for 15 min. 1-2 μl of boiled sample was used for PCR. The primers used to assay the targeted sites were designed to amplify genomic regions 1.5 - 3 kb in size. In the event of a PCR product equal to or smaller than the wild-type fragment (as was often observed when analyzing Cas9-targeted cells), Sanger sequencing (Quintara Biosciences) was used to determine any modifications of the targeted sequences. In some cases, additional analysis of the crRNA-expressing plasmids of the surviving colonies was also performed, by isolating and reintroducing the plasmids into the original I-C CRISPR-Cas strain, where functional self-targeting could be determined based on a significant increase in the lag time of induced cultures, characteristic of self-targeting events.
In cases where a homology-directed repair template was used, homology arms ranging in size of 500-600 bp were cloned using a nested PCR-based approach where the two different arms were stitched together via 25 bp overlaps. These fragments were then cloned into the pHERD30T plasmid expressing self-targeting crRNAs at the NheI restriction sites. Genomic targeting was performed as described above. Surviving cells were analyzed using colony PCRs amplifying the desired deletion junction (verified with Sanger sequencing), as well as the wild-type target site. Editing efficiencies were counted as the number of colonies producing a desired deletion junction fragment from the total number of analyzed colonies.
Escherichia coli
Genomic self-targeting of E. coli was conducted in a similar fashion as P. aeruginosa, except using the pCas3cRh all-in-one vector. Electrocompetent E. coli cells were transformed with pCas3cRh expressing a crRNA targeting the genome. Individual transformants were selected and grown in liquid LB media containing the selecting antibiotic (gentamicin) overnight without any inducers added. The overnight cultures were then plated in the presence of inducer and X-gal to screen for functional lacZ (LB agar + 15 μg/ml gentamicin + 0.1 % rhamnose + 1 mM IPTG + 20 μg/ml X-gal) and blue/white colonies were counted the next day.
Pseudomonas syringae
Electrocompetent P. syringae cells were also transformed with pCas3cRh plasmids targeting selected genomic sequences. Initial transformants were plated onto KB agar + 100 μg/ml rifampicin + 50 μg/ml gentamicin plates, and incubated at 28 °C overnight. Single colony transformants were then selected and inoculated in KB liquid media supplemented with rifampicin, gentamicin, and 0.1 % rhamnose inducer, and grown to saturation in a shaking incubator at 28 °C. Cultures were finally plated onto KB agar plates with rifampicin, gentamicin, and rhamnose and incubated at 28 °C. Individual colonies were finally assayed with colony PCR to determine the presence of deletions at the targeted genomic sites.
Klebsiella pneumoniae
Electrocompetent K. pneumoniae cells were transformed with pCas3cRh plasmids targeting selected genomic sequences. Individual transformants were selected and grown in liquid LB media (containing 50 μg/ml gentamicin, as well as 0.1 % rhamnose inducer) overnight. Various dilutions from saturated cultures were then plated the next day onto LB agar plates containing the selective antibiotic (gentamicin 50 μg/ml) and 0.1 % rhamnose inducer. Individual colonies were then assayed for deletions using colony PCR.
Iterative genome minimization
Iterative targeting to generate multiple deletions in the P. aeruginosa PAO1IC strain was carried out by alternating the pHERD30T and pHERD20T plasmids each expressing different crRNAs targeting the genome. Each crRNA designed to target the genome was cloned into both the pHERD30T plasmid, which confers gentamicin resistance, as well as the pHERD20T plasmid, which confers carbenicillin resistance. After first transforming and targeting with a pHERD30T plasmid expressing a specific crRNA, deletion candidate isolates were transformed with a pHERD20T expressing a crRNA targeting a different genomic region. As the two plasmids are identical with the exception of the resistance marker, this eliminated the necessity for curing of the original plasmid to be able to target a different region. For the next targeting event, the pHERD30T plasmid could again be used, this time expressing another crRNA targeting a different genomic region. In this manner, pHERD30T and pHERD20T could be alternated to achieve multiple deletions in a rapid process. At each new transformation step, the cells were checked for any residual resistance to the given antibiotic from a previous cycle. Additionally, functionality of the CRISPR-Cas system of the edited cells could be determined through the introduction of a plasmid expressing crRNA targeting the D3 bacteriophage49, then performing a phage spotting assay to see if phage targeting was occurring or not.
Measurement of growth rates
Pseudomonas aeruginosa
Growth dynamics of various strains were measured using a Synergy 2 automated 96-well plate reader (Biotek Instruments) and the accompanying Gen5 software (Biotek Instruments). Individual colonies were picked and grown overnight in 300 μl volumes of LB in 96-well deep-well plates at 37 °C. The grown cultures were then diluted 100-fold into 100 μl of fresh LB in a 96-well clear microtitre plate (Costar) and sealed with Microplate sealing adhesive (Thermo Scientific). Small holes were punched in the sealing adhesive for each well for increased aeration. Doubling times were calculated as described previously73.
Pseudomonas syringae
To test bacterial growth in planta, we used the Arabidopsis thaliana ecotype Columbia (Col-0), which has previously been shown to be susceptible to infection by P. syringae DC3000. Plants were grown for 5-6 weeks in 9 h light/15 h darkness and 65 % humidity. For each inoculum, we measured bacterial growth in 10 individual Col-0 plants. Four leaves from each plant were infiltrated at OD600 = 0.0002, and cored with a #3 borer. The four cores from each plant were then ground, resuspended in 10 mM MgCl2 and plated in a dilution series on selective media for colony counts at both the time of infection and 3 days post-infection.
To test bacterial growth in vitro, we used both KB and plant apoplast mimicking minimal media (MM)74. Overnight cultures were prepared from single colonies of each strain, washed, and diluted to OD600 = 0.1 in 96-well plates using either KB or MM. Plates were incubated with shaking at 28 °C. OD600 was measured over the course of 24-25 hours using an Infinate 200 Pro automated plate reader (Tecan). Statistical analysis determined significantly different groups based on ANOVA analysis on the day 0 group of values and the day 3 group of values. Significant ANOVA results (p < 0.01) were further analyzed with a Tukey’s HSD post hoc test to generate adjusted p-values for each pairwise comparison. A significance threshold of 0.01 was used to determine which treatment groups were significantly different.
Bacteriophage plaque (spot) assays
Bacteriophage plaque assays were performed using 1.5 % LB agar plates supplemented with 10 mM MgSO4 and the appropriate antibiotic (gentamicin or carbenicillin, depending on the plasmid used to express the crRNA), and 0.7 % LB top agar supplemented with 0.5 mM IPTG and 0.1 % arabinose inducers added covering the whole plate. 150 μl of the appropriate overnight cultures was suspended in 4 ml molten top agar poured onto an LB agar plate leading to the growth of a bacterial lawn. After 10-15 minutes at room temperature, 3 μl of ten-fold serial dilutions of bacteriophage was spotted onto the solidified top agar. Plates were incubated overnight at 30 °C and imaged the following day using a Gel Doc EZ Gel Documentation System (BioRad) and Image Lab (BioRad) software. The following bacteriophage were used in this study: bacteriophage JBD3049, bacteriophage D375, and bacteriophage DMS3m76.
Whole-genome sequencing
Genomic DNA for whole-genome sequencing (WGS) analysis was isolated directly from bacterial colonies using the Nextera DNA Flex Microbial Colony Extraction kit (Illumina) according to the manufacturer’s protocol. Genomic DNA concentration of the samples was determined using a DS-11 Series Spectrophotometer/Fluorometer (DeNovix) and all fell into the range of 200-500 ng/μl. Library preparation for WGS analysis was done using the Nextera DNA Flex Library Prep kit (Illumina) according to the manufacturer’s protocol starting from the tagment genomic DNA step. Tagmented DNA was amplified using Nextera DNA CD Indexes (Illumina). Samples were placed overnight at 4 °C following the tagmented DNA amplification step, then continued the next day with the library clean up steps. Quality control of the pooled libraries was performed using a 2100 Bioanalyzer Instrument (Agilent Technologies) with a High Sensitivity DNA Kit (Agilent Technologies). The majority of samples were sequenced using a MiSeq Reagent Kit v2 (Illumina) for a 150 bp paired-end sequencing run using the MiSeq sequencer (Illumina). P. syringae and Cas9-generated P. aeruginosa deletion strains were sequenced using a NextSeq 500 Reagent Kit v2 (Illumina) for a 150 bp paired-end sequencing run using the NextSeq 500 sequencer (Illumina).
Genome sequence assembly was performed using Geneious Prime software version 2019.1.3. Paired read data sets were trimmed using the BBDuk (Decontamination Using Kmers) plugin using a minimum Q value of 20. The genome for the ancestral PAO1IC strain was de novo assembled using the default automated sensitivity settings offered by the software. The consensus sequence of PAO1IC assembled in this manner was then used as the reference sequence for mapping all of the PAO1IC strains with multiple deletions. As a control, the sequences were also mapped to the reference P. aeruginosa PAO1 sequence (NC_002516) to verify deletion border coordinates. Coverage of these sequenced strains ranged from 66 to 143-fold, with an average of 98.3-fold. The sequenced P. aeruginosa environmental strains were also mapped to the PAO1 (NC_002516) reference, while the sequenced E. coli strains were mapped to the E. coli K-12 MG1655 reference sequence (NC_000913). Finally, sequenced P. syringae strains were mapped to the P. syringae DC3000 (NC_004578) reference sequence, along with the pDC3000A endogenous 73.5 kb plasmid sequence (NC_004633). All of the remaining sequenced strains had > 100-fold coverage. All deletion junction sequences were manually verified by the presence of multiple reads spanning the deletions, containing sequences from both end boundaries.
WGS data was visualized using the BLAST Ring Image Generator (BRIG) tool77 employing BLAST+ version 2.9.0. In several cases, short sequences were aligned within previously determined large deletions at redundant sequences such as transposase genes. Such misrepresentations created by BRIG were manually removed to reflect the actual sequencing data.
Supplementary Material
Acknowledgements
We thank Joanna B. Goldberg (Emory University) for providing the plasmid pJM230, and Adair Borges (UCSF) for providing pAB04 to clone Type I-F crRNAs. We thank the Bondy-Denomy lab for productive conversations pertaining to this project.
Funding
B.C. is supported by the Eötvös National Scholarship of Hungary and a Marie Skłodowska-Curie Actions Individual Global Fellowship (‘GenDels’, no. 844093) of the Horizon 2020 Research Program of the European Commission. L.L. is supported by the HHMI Gilliam Fellowship for Advanced Study and the UCSF Discovery Fellowship. Research on plant immunity in the Lewis laboratory is supported by the USDA ARS 2030-21000-046-00D and 2030-21000-050-00D (J.D.L.), and the NSF Directorate for Biological Sciences IOS-1557661 (J.D.L.). I.J.C. is supported by a Grace Kase fellowship from UC Berkeley and the NSF Graduate Research Fellowship Program. A.V.R. is supported by funding from the Pew Charitable Trusts. E.D.C. is funded by the Chan Zuckerberg Biohub. CRISPR-Cas3 projects in the Bondy-Denomy Lab are supported by the UCSF Program for Breakthrough Biomedical Research funded in part by the Sandler Foundation, the Innovative Genomics Institute, and an NIH Director’s Early Independence Award DP5-OD021344.
Footnotes
Competing Interests:
J.B.-D. is a scientific advisory board member of SNIPR Biome and Excision Biotherapeutics and a scientific advisory board member and co-founder of Acrigen Biosciences.
Data availability
Raw whole-genome sequencing data associated with Figures 1D, 3B, 4A, 4C, 4F, and 5A) has been uploaded to GenBank (Accession numbers CP047061-CP047079) and is also available, along with bacterial strains, upon request from the corresponding author.
References
- 1.Makarova KS, et al. Evolutionary classification of CRISPR-Cas systems: a burst of class 2 and derived variants. Nat Rev Microbiol. 2020;18:67–83. doi: 10.1038/s41579-019-0299-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barrangou R, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 3.Garneau JE, et al. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010;468:67–71. doi: 10.1038/nature09523. [DOI] [PubMed] [Google Scholar]
- 4.Barrangou R, Doudna JA. Applications of CRISPR technologies in research and beyond. Nat Biotechnol. 2016:933–941. doi: 10.1038/nbt.3659. [DOI] [PubMed] [Google Scholar]
- 5.Wiedenheft B, et al. Structures of the RNA-guided surveillance complex from a bacterial immune system. Nature. 2011;477:486–489. doi: 10.1038/nature10402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Westra ER, et al. CRISPR immunity relies on the consecutive binding and degradation of negatively supercoiled invader DNA by Cascade and Cas3. Mol Cell. 2012;46:595–605. doi: 10.1016/j.molcel.2012.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brouns SJJ, et al. Small CRISPR RNAs Guide Antiviral Defense in Prokaryotes. Science. 2008;321:960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hidalgo-Cantabrana C, Barrangou R. Characterization and applications of Type I CRISPR-Cas systems. Biochem Soc Trans. doi: 10.1042/BST20190119. [DOI] [PubMed] [Google Scholar]
- 9.Sinkunas T, et al. Cas3 is a single-stranded DNA nuclease and ATP-dependent helicase in the CRISPR/Cas immune system. EMBO J. 2011;30:1335–1342. doi: 10.1038/emboj.2011.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sinkunas T, et al. In vitro reconstitution of Cascade-mediated CRISPR immunity in Streptococcus thermophilus. EMBO J. 2013;32:385–394. doi: 10.1038/emboj.2012.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mulepati S, Bailey S. In vitro reconstitution of an Escherichia coli RNA-guided immune system reveals unidirectional, ATP-dependent degradation of DNA target. J Biol Chem. 2013;288:22184–22192. doi: 10.1074/jbc.M113.472233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hochstrasser ML, et al. CasA mediates Cas3-catalyzed target degradation during CRISPR RNA-guided interference. Proc Natl Acad Sci U S A. 2014;111:6618–6623. doi: 10.1073/pnas.1405079111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Redding S, et al. Surveillance and Processing of Foreign DNA by the Escherichia coli CRISPR-Cas System. Cell. 2015;163:854–865. doi: 10.1016/j.cell.2015.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xiao Y, Luo M, Dolan AE, Liao M, Ke A. Structure basis for RNA-guided DNA degradation by Cascade and Cas3. Science. 2018;361:eaat0839. doi: 10.1126/science.aat0839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Makarova KS, et al. An updated evolutionary classification of CRISPR-Cas systems. Nat Rev Microbiol. 2015;13:722–736. doi: 10.1038/nrmicro3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vercoe RB, et al. Cytotoxic Chromosomal Targeting by CRISPR/Cas Systems Can Reshape Bacterial Genomes and Expel or Remodel Pathogenicity Islands. PLOS Genet. 2013;9:e1003454. doi: 10.1371/journal.pgen.1003454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gomaa AA, et al. Programmable removal of bacterial strains by use of genome-targeting CRISPR-Cas systems. mBio. 2014;5:e00928-00913. doi: 10.1128/mBio.00928-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kiro R, Shitrit D, Qimron U. Efficient engineering of a bacteriophage genome using the type I-E CRISPR-Cas system. RNA Biol. 2014;11:42–44. doi: 10.4161/rna.27766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Y, et al. Harnessing Type I and Type III CRISPR-Cas systems for genome editing. Nucleic Acids Res. 2016;44:e34. doi: 10.1093/nar/gkv1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pyne ME, Bruder MR, Moo-Young M, Chung DA, Chou CP. Harnessing heterologous and endogenous CRISPR-Cas machineries for efficient markerless genome editing in Clostridium. Sci Rep. 2016;6:25666. doi: 10.1038/srep25666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang J, Zong W, Hong W, Zhang Z-T, Wang Y. Exploiting endogenous CRISPR-Cas system for multiplex genome editing in Clostridium tyrobutyricum and engineer the strain for high-level butanol production. Metab Eng. doi: 10.1016/j.ymben.2018.03.007. [DOI] [PubMed] [Google Scholar]
- 22.Maikova A, Kreis V, Boutserin A, Severinov K, Soutourina O. Using endogenous CRISPR-Cas system for genome editing in the human pathogen Clostridium difficile. Appl Environ Microbiol. 2019:AEM.0141619. doi: 10.1128/AEM.01416-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hidalgo-Cantabrana C, Goh YJ, Pan M, Sanozky-Dawes R, Barrangou R. Genome editing using the endogenous type I CRISPR-Cas system in Lactobacillus crispatus. Proc Natl Acad Sci U S A. 2019;116:15774–15783. doi: 10.1073/pnas.1905421116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hampton HG, et al. CRISPR-Cas gene-editing reveals RsmA and RsmC act through FlhDC to repress the SdhE flavinylation factor and control motility and prodigiosin production in Serratia. Microbiology. 2016;162:1047–1058. doi: 10.1099/mic.0.000283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheng F, et al. Harnessing the native type I-B CRISPR-Cas for genome editing in a polyploid archaeon. J Genet Genomics Yi Chuan Xue Bao. 2017;44:541–548. doi: 10.1016/j.jgg.2017.09.010. [DOI] [PubMed] [Google Scholar]
- 26.Cañez C, Selle K, Goh YJ, Barrangou R. Outcomes and characterization of chromosomal self-targeting by native CRISPR-Cas systems in Streptococcus thermophilus. FEMS Microbiol Lett. 2019;366 doi: 10.1093/femsle/fnz105. [DOI] [PubMed] [Google Scholar]
- 27.Xu Z, et al. Native CRISPR-Cas-Mediated Genome Editing Enables Dissecting and Sensitizing Clinical Multidrug-Resistant P. aeruginosa. Cell Rep. 2019;29:1707–1717.:e3. doi: 10.1016/j.celrep.2019.10.006. [DOI] [PubMed] [Google Scholar]
- 28.Zheng Y, et al. Characterization and repurposing of the endogenous Type I-F CRISPR-Cas system of Zymomonas mobilis for genome engineering. bioRxiv. 2019:576355. doi: 10.1101/576355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Edgar R, Qimron U. The Escherichia coli CRISPR System Protects from λ Lysogenization, Lysogens, and Prophage Induction. J Bacteriol. 2010;192:6291–6294. doi: 10.1128/JB.00644-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dolan AE, et al. Introducing a Spectrum of Long-Range Genomic Deletions in Human Embryonic Stem Cells Using Type I CRISPR-Cas. Mol Cell. 2019;74:936–950.:e5. doi: 10.1016/j.molcel.2019.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morisaka H, et al. CRISPR-Cas3 induces broad and unidirectional genome editing in human cells. Nat Commun. 2019;10:1–13. doi: 10.1038/s41467-019-13226-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cameron P, et al. Harnessing type I CRISPR–Cas systems for genome engineering in human cells. Nat Biotechnol. 2019;37:1471–1477. doi: 10.1038/s41587-019-0310-0. [DOI] [PubMed] [Google Scholar]
- 33.Pickar-Oliver A, et al. Targeted transcriptional modulation with type I CRISPR–Cas systems in human cells. Nat Biotechnol. 2019:1–9. doi: 10.1038/s41587-019-0235-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen Y, et al. Repurposing type I-F CRISPR-Cas system as a transcriptional activation tool in human cells. Nat Commun. 2020;11:3136. doi: 10.1038/s41467-020-16880-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Young JK, et al. The repurposing of type I-E CRISPR-Cascade for gene activation in plants. Commun Biol. 2019;2:1–7. doi: 10.1038/s42003-019-0637-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nam KH, et al. Cas5d Protein Processes Pre-crRNA and Assembles into a Cascade-like Interference Complex in Subtype I-C/Dvulg CRISPR-Cas System. Structure. 2012;20:1574–1584. doi: 10.1016/j.str.2012.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hochstrasser ML, Taylor DW, Kornfeld JE, Nogales E, Doudna JA. DNA Targeting by a Minimal CRISPR RNA-Guided Cascade. Mol Cell. 2016;63:840–851. doi: 10.1016/j.molcel.2016.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marino ND, et al. Discovery of widespread type I and type V CRISPR-Cas inhibitors. Science. 2018;362:240–242. doi: 10.1126/science.aau5174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Turner KH, Wessel AK, Palmer GC, Murray JL, Whiteley M. Essential genome of Pseudomonas aeruginosa in cystic fibrosis sputum. Proc Natl Acad Sci. 2015;112:4110–4115. doi: 10.1073/pnas.1419677112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meek DW, Hayward RS. Nucleotide sequence of the rpoA-rplQ DNA of Escherichia coli: a second regulatory binding site for protein S4? Nucleic Acids Res. 1984;12:5813–5821. doi: 10.1093/nar/12.14.5813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Selle K, Klaenhammer TR, Barrangou R. CRISPR-based screening of genomic island excision events in bacteria. Proc Natl Acad Sci. 2015;112:8076–8081. doi: 10.1073/pnas.1508525112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chayot R, Montagne B, Mazel D, Ricchetti M. An end-joining repair mechanism in Escherichia coli. Proc Natl Acad Sci. 2010;107:2141–2146. doi: 10.1073/pnas.0906355107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Buell CR, et al. The complete genome sequence of the Arabidopsis and tomato pathogen Pseudomonas syringae pv. tomato DC3000. Proc Natl Acad Sci U S A. 2003;100:10181–10186. doi: 10.1073/pnas.1731982100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lindeberg M, Cunnac S, Collmer A. Pseudomonas syringae type III effector repertoires: last words in endless arguments. Trends Microbiol. 2012;20:199–208. doi: 10.1016/j.tim.2012.01.003. [DOI] [PubMed] [Google Scholar]
- 45.Kvitko BH, et al. Deletions in the repertoire of Pseudomonas syringae pv. tomato DC3000 type III secretion effector genes reveal functional overlap among effectors. PLoS Pathog. 2009;5:e1000388. doi: 10.1371/journal.ppat.1000388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Broberg CA, Wu W, Cavalcoli JD, Miller VL, Bachman MA. Complete Genome Sequence of Klebsiella pneumoniae Strain ATCC 43816 KPPR1, a Rifampin-Resistant Mutant Commonly Used in Animal, Genetic, and Molecular Biology Studies. Genome Announc. 2014;2 doi: 10.1128/genomeA.00924-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bachman MA, et al. Genome-Wide Identification of Klebsiella pneumoniae Fitness Genes during Lung Infection. mBio. 2015;6 doi: 10.1128/mBio.00775-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cady KC, Bondy-Denomy J, Heussler GE, Davidson AR, O’Toole GA. The CRISPR/Cas adaptive immune system of Pseudomonas aeruginosa mediates resistance to naturally occurring and engineered phages. J Bacteriol. 2012;194:5728–5738. doi: 10.1128/JB.01184-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bondy-Denomy J, Pawluk A, Maxwell KL, Davidson AR. Bacteriophage genes that inactivate the CRISPR/Cas bacterial immune system. Nature. 2013;493:429–432. doi: 10.1038/nature11723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rauch BJ, et al. Inhibition of CRISPR-Cas9 with Bacteriophage Proteins. Cell. 2017;168:150–158.:e10. doi: 10.1016/j.cell.2016.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stanley SY, et al. Anti-CRISPR-Associated Proteins Are Crucial Repressors of Anti-CRISPR Transcription. Cell. 2019;178:1452–1464.:e13. doi: 10.1016/j.cell.2019.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ha AD, Denver DR. Comparative Genomic Analysis of 130 Bacteriophages Infecting Bacteria in the Genus Pseudomonas. Front Microbiol. 2018;9:1456. doi: 10.3389/fmicb.2018.01456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rollins MF, et al. Cas1 and the Csy complex are opposing regulators of Cas2/3 nuclease activity. Proc Natl Acad Sci U S A. 2017;114:E5113–E5121. doi: 10.1073/pnas.1616395114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Caliando BJ, Voigt CA. Targeted DNA degradation using a CRISPR device stably carried in the host genome. Nat Commun. 2015;6:6989. doi: 10.1038/ncomms7989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pósfai G, et al. Emergent properties of reduced-genome Escherichia coli. Science. 2006;312:1044–1046. doi: 10.1126/science.1126439. [DOI] [PubMed] [Google Scholar]
- 56.Csörgő B, Nyerges Á, Pósfai G, Fehér T. System-level genome editing in microbes. Curr Opin Microbiol. 2016;33:113–122. doi: 10.1016/j.mib.2016.07.005. [DOI] [PubMed] [Google Scholar]
- 57.Képès F, et al. The layout of a bacterial genome. FEBS Lett. 2012;586:2043–2048. doi: 10.1016/j.febslet.2012.03.051. [DOI] [PubMed] [Google Scholar]
- 58.Ghosh S, O’Connor TJ. Beyond Paralogs: The Multiple Layers of Redundancy in Bacterial Pathogenesis. Front Cell Infect Microbiol. 2017;7 doi: 10.3389/fcimb.2017.00467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cui L, Bikard D. Consequences of Cas9 cleavage in the chromosome of Escherichia coli. Nucleic Acids Res. 2016 doi: 10.1093/nar/gkw223. gkw223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kowalczykowski SC, Eggleston AK. Homologous Pairing and Dna Strand-Exchange Proteins. Annu Rev Biochem. 1994;63:991–1043. doi: 10.1146/annurev.bi.63.070194.005015. [DOI] [PubMed] [Google Scholar]
- 61.Bowater R, Doherty AJ. Making Ends Meet: Repairing Breaks in Bacterial DNA by Non-Homologous End-Joining. PLOS Genet. 2006;2:e8. doi: 10.1371/journal.pgen.0020008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hnisz D, et al. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tuladhar R, et al. CRISPR-Cas9-based mutagenesis frequently provokes on-target mRNA misregulation. Nat Commun. 2019;10:1–10. doi: 10.1038/s41467-019-12028-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Smits AH, et al. Biological plasticity rescues target activity in CRISPR knock outs. Nat Methods. 2019:1–7. doi: 10.1038/s41592-019-0614-5. [DOI] [PubMed] [Google Scholar]
- 65.Choi K-H, et al. A Tn7-based broad-range bacterial cloning and expression system. Nat Methods. 2005;2:443–448. doi: 10.1038/nmeth765. [DOI] [PubMed] [Google Scholar]
- 66.Stover CK, et al. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature. 2000;406:959. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- 67.Choi K-H, Schweizer HP. mini-Tn7 insertion in bacteria with single attTn7 sites: example Pseudomonas aeruginosa. Nat Protoc. 2006;1:153–161. doi: 10.1038/nprot.2006.24. [DOI] [PubMed] [Google Scholar]
- 68.Blattner FR, et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 69.Qiu D, Damron FH, Mima T, Schweizer HP, Yu HD. PBAD-Based Shuttle Vectors for Functional Analysis of Toxic and Highly Regulated Genes in Pseudomonas and Burkholderia spp. and Other Bacteria. Appl Environ Microbiol. 2008;74:7422–7426. doi: 10.1128/AEM.01369-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gibson DG, et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 71.Meisner J, Goldberg JB. The Escherichia coli rhaSR-PrhaBAD Inducible Promoter System Allows Tightly Controlled Gene Expression over a Wide Range in Pseudomonas aeruginosa. Appl Environ Microbiol. 2016;82:6715–6727. doi: 10.1128/AEM.02041-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Borges AL, et al. Bacteriophage Cooperation Suppresses CRISPR-Cas3 and Cas9 Immunity. Cell. 2018;174:917–925.:e10. doi: 10.1016/j.cell.2018.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nyerges Á, et al. Directed evolution of multiple genomic loci allows the prediction of antibiotic resistance. Proc Natl Acad Sci. 2018;115:E5726–E5735. doi: 10.1073/pnas.1801646115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Huynh TV, Dahlbeck D, Staskawicz BJ. Bacterial blight of soybean: regulation of a pathogen gene determining host cultivar specificity. Science. 1989;245:1374–1377. doi: 10.1126/science.2781284. [DOI] [PubMed] [Google Scholar]
- 75.Kropinski AM. Sequence of the Genome of the Temperate, Serotype-Converting, Pseudomonas aeruginosa Bacteriophage D3. J Bacteriol. 2000;182:6066–6074. doi: 10.1128/jb.182.21.6066-6074.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Budzik JM, Rosche WA, Rietsch A, O’Toole GA. Isolation and Characterization of a Generalized Transducing Phage for Pseudomonas aeruginosa Strains PAO1 and PA14. J Bacteriol. 2004;186:3270–3273. doi: 10.1128/JB.186.10.3270-3273.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Alikhan N-F, Petty NK, Ben Zakour NL, Beatson SA. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics. 2011;12:402. doi: 10.1186/1471-2164-12-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw whole-genome sequencing data associated with Figures 1D, 3B, 4A, 4C, 4F, and 5A) has been uploaded to GenBank (Accession numbers CP047061-CP047079) and is also available, along with bacterial strains, upon request from the corresponding author.