Abstract
Protein recruitment to DNA break sites is an integral part of the DNA damage response (DDR). Elucidation of the hierarchy and temporal order with which DNA damage sensors as well as repair and signaling factors assemble around chromosome breaks has painted a complex picture of tightly regulated macromolecular interactions that build specialized compartments to facilitate repair and maintenance of genome integrity. While many of the underlying interactions, e.g. between repair factors and damage-induced histone marks, can be explained by lock-and-key or induced fit binding models assuming fixed stoichiometries, structurally less well defined interactions, such as the highly dynamic multivalent interactions implicated in phase separation, also participate in the formation of multi-protein assemblies in response to genotoxic stress. Although much remains to be learned about these types of cooperative and highly dynamic interactions and their functional roles, the rapidly growing interest in material properties of biomolecular condensates and in concepts from polymer chemistry and soft matter physics to understand biological processes at different scales holds great promises. Here, we discuss nuclear condensates in the context of genome integrity maintenance, highlighting the cooperative potential between clustered stoichiometric binding and phase separation. Rather than viewing them as opposing scenarios, their combined effects can balance structural specificity with favorable physicochemical properties relevant for the regulation and function of multilayered nuclear condensates.
Keywords: Biomolecular condensates, DNA damage response (DDR), DNA repair, Genome stability, Liquid-liquid phase separation (LLPS), Intrinsically disordered regions (IDR), Low complexity domains (LCD), Multivalent interactions, Higher-order assemblies
1. Introduction
According to Richard Dawkins’ ‘The Blind Watchmaker: Why the Evidence of Evolution Reveals a Universe without Design’, “biology is the study of complicated things that give the appearance of having been designed for a purpose”, whereas “physics is the study of simple things that do not tempt us to invoke design” [1]. Oil droplets in water, from this perspective, could be considered simple things, with no sign of design. Living cells, on the other hand, with their intricate architecture and organizational complexity, are complicated things. As tiny high-precision machines, cells use energy to exert spatio-temporal control over the numerous biochemical reactions that take place every millisecond inside the intracellular space. Compartmentalization is key to such spatio-temporal control, and recent years have seen resurging interest in the biological, chemical and physical principles that govern cellular compartmentalization [2–5].
The cell nucleus is the biggest cellular compartment, a membrane-enclosed organelle and home of the chromosomes and the embedded genetic code. It comprises various layers of organizational complexity to maintain genome structure and function, including chromatin loops and higher-order chromatin architecture, but also a large number of chromatin-associated and non-associated nuclear proteins and protein complexes, which can form biomolecular condensates and thereby subdivide the nuclear space. Biomolecular condensates can be generally defined as intracellular compartments that lack surrounding membranes but function to concentrate biological molecules, and the term was chosen in part because it provides a link to concepts from condensed matter physics [5]. Such assemblies, despite the absence of a lipid membrane, can be considered physical entities, in which certain molecules are enriched, while others are excluded. Biomolecular condensates can form by different means and have different material properties, ranging from highly dynamic liquid droplets to dense and sometimes irreversible aggregates [6–8]. They can form through stoichiometric binding of molecules to one another, e.g. with the chromatin scaffold and histone modifications serving as binding site clusters for cooperative or non-cooperative protein binding, or, at the other end of the spectrum, by thermodynamically-driven liquid-liquid phase separation (LLPS). Lava lamps and vinaigrettes (‘simple things’) have served as popular examples for liquid demixing and to illustrate behavioral features of coexisting liquid phases. But of course, biomolecular condensates in living cells are in many aspects far more complex than oil droplets in water or than the ‘lava’ in lava lamps. They are composed of hundreds of molecules of different kind that make inter- and intra-molecular contacts, often in a highly dynamic manner and employing bonds of varying interaction strength. Their compositional control is energy dependent, and enzymatic reactions, including post-translational protein modifications, are involved in regulating their formation and functions [9]. Structured domains provide high affinity interaction surfaces for binding partners, and these interactions can be critical for the formation or maintenance of a compartment. Energy dependence does not refute a role for local thermodynamics, however, and lock-and-key type interactions involved in clustered binding can cooperate with dynamic multivalent weak interactions to form functional condensates [10]. Stable protein complexes with fixed stoichiometries represent a minority in protein interaction networks, which are instead dominated by weak, nonstoichiometric interactions [11]. On the other extreme, intrinsically disordered proteins that form liquid droplets in vitro typically show more complicated behavior in their natural habitat inside cells, where homotypic interactions are dominated by various heterotypic interactions that involve multiple interaction partners [12]. A lot of biology (‘complicated things’) likely happens between the extremes of stable complexes with fixed stoichiometries and purely entropy-driven phase separation in idealized few component systems (Fig. 1).
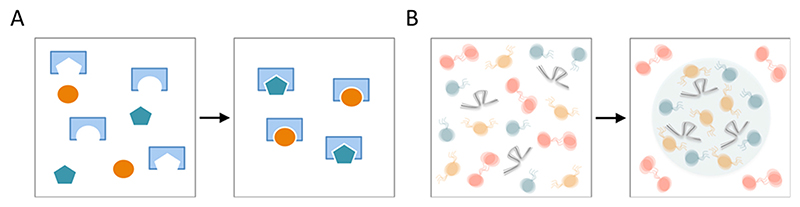
Fig. 1. Binding modes.
(A) Stoichiometric, in this case monovalent one-to-one binding, e.g. by lock-and-key or induced fit association. (B) Highly dynamic multivalent interactions involved in phase separation. Note that in both cases dynamic association and dissociation can occur and, importantly, high binding affinities can be reached.
Maintenance of genome integrity depends on local multi-protein assemblies, e.g. around sites of DNA damage, to mark genomic lesions and coordinate DNA repair reactions with other vital functions such as transcription and cell cycle progression. Denominating DNA repair foci as biomolecular condensates does not serve a purpose on its own, but focusing on material properties of these compartments and on the role of disordered protein sequences and their highly dynamic multivalent interactions complements classical biochemical and structural approaches and can open new areas of research. Here, we focus on and discuss connections between cellular mechanisms important for the maintenance of genome integrity and phase separation in the nucleus of eukaryotic cells, highlighting emerging examples of how protein properties involved in phase separation contribute to DNA damage sensing and compartmentalization of lesions, to cell cycle checkpoint activation, to the clustering and movement of DNA breaks, to replication initiation and replication stress signaling, and to telomere maintenance in cancer. We draw parallels to phase separated compartments outside the nucleus and discuss opportunities and future perspectives related to a better understanding, based on new conceptual frameworks and advancing technologies, of the specific interactions that form condensates and how such knowledge may be used to modulate condensate functions and to exploit condensate biology therapeutically.
2. Condensate formation by phase separation
Phase separation as principle implicated in the formation of biomolecular condensates has been discussed for a wide range of organisms [13] including bacteria [14], yeast [15], plants [16], animals [17], and also for viruses [18]. Example condensates include nucleoli [19–24], Cajal bodies [25–27], histone locus bodies [28], nuclear speckles and paraspeckles [29–34], PML nuclear bodies and APBs [35–38], SAM68 nuclear bodies [39], chromatin domains [40–51], transcription condensates [52–64], centromeres [65–67], DNA replication and repair condensates [68–79], stress granules and P-granules [80–82], and virus particles [83–92]. Several of these reside in the cell nucleus [93] and have genome-related functions (Table 1). Although attributing functional roles to phase separation has been difficult in some cases and requires further attention and dedicated research efforts, a rapidly growing list of cellular processes has been linked to phase separation properties and includes ribosome biogenesis and RNA metabolism, protein quality control and degradation, signal transduction, response to stress, cell division, endocytosis, autophagy, cytoskeleton regulation and cargo transport [94–111]. In the context of these processes, phase separation has been implicated in the enhancement or suppression of biochemical reactions and regulation of enzyme activities, signal amplification, storage, sequestration and molecular buffering, sensing of changes in the environment, control of viscoelasticity and viscoadaptation, and exertion of mechanical forces [112–119].
Table 1. Biomolecular condensates in the nucleus and their main function(s).
| Nuclear condensates | Main function(s) | References |
|---|---|---|
| Nucleoli | rRNA transcription and ribosome biogenesis | [19,20,21,22,23,24] |
| Cajal bodies | Biogenesis and maturation of snRNPs | [25,26,27] |
| Histone locus bodies | Processing of histone pre-mRNAs | [28] |
| Nuclear speckles | Regulation of pre-mRNA splicing | [29,30] |
| Paraspeckles | Protein and (lnc)RNA sequestration, gene regulation | [31,32,33,34] |
| PML nuclear bodies | Genome organization, transcriptional regulation, viral defense | [35,36] |
| APBs in ALT cells | Telomere clustering and telomere maintenance by ALT | [37,38] |
| SAM68 nuclear bodies | RNA metabolism | [39] |
| Chromatin domains | Genome organization, gene regulation, X-inactivation | [40,41,42,43,44,45,46,47,48,49,50,51] |
| Transcriptional condensates | Transcription initiation and elongation | [52,53,54,55,56,57,58,59,60,61,62,63,64] |
| Centromeres | Chromosome segregation during cell division | [65,66,67] |
| DNA repair compartments | DNA damage signaling and repair | [68,69,70,71,72,73,74,75,76] |
| Replication condensates | Replication initiation, coordination of origin firing | [77,78,79] |
Listed are membraneless nuclear compartments, which have been linked to protein and nucleic acid phase separation, together with their main cellular function(s). Note that condensate formation does not necessarily rely on phase separation alone, and that for some condensates the causal relationship between phase separation properties of the involved molecules and cellular condensate function(s) is still debated.
In the context of equilibrium thermodynamics, a phase can be defined as form of matter, which within its boundaries is homogeneous in relevant properties such as chemical composition and physical state. As living cells are not at equilibrium, the definition of a phase broadens, yet without losing its general meaning or applicability. Accordingly, a polymer core is not per se incompatible with multi-layered condensate organization through percolation (an example for a cellular compartment with multiple co-existing phases being the nucleolus [20]), and local thermodynamic equilibria exist at mesoscopic scales despite living cells being out of equilibrium [120–122]. Phase separation and the regulation of intracellular condensates by active processes are therefore not mutually exclusive mechanisms, and a number of energy-consuming processes, including different post-translational modifications (PTMs), have been shown to participate in the spatio-temporal regulation of phase separation [9,117]. The relative abundance of the participating molecules determines composition and morphology of condensates [12,60,123], and PTMs not only change chemical properties of modified proteins, but are also central to the regulation of the abundance and local concentration of proteins and RNA. Although the exact nature, or molecular grammar, of interactions that drive phase separation and their regulation by PTMs is only starting to emerge, it is important to appreciate that, while they are often highly dynamic and may be considered ‘fuzzy’, they are not ‘unspecific’. The multivalent interactions that participate in phase separation are sequence-encoded and often conserved [124,125], yet they differ from stoichiometric lock-and-key type interactions, for which structure-guided exchange of a single amino acid may be sufficient to abrogate binding (Fig. 1). This poses inherent difficulties to classic biochemical and cell biological approaches, such as the generation of separation-of-function mutants by single amino acid exchange, or structure-based design of small molecule inhibitors. However, these challenges should not only be seen as a disadvantage, but also as a chance for novel approaches in biochemistry and drug development. As the field moves towards a better understanding of the dynamic interplay between different types of interactions that contribute to condensate formation in different cellular contexts (Table 2), the easier it will become to modulate condensate behavior and tune condensate functions in a targeted manner.
Table 2. Interactions implicated in (liquid-liquid) phase separation.
| Interaction | Residues involved | References |
|---|---|---|
| π-π | Phe-Phe; Tyr-Tyr; Phe-Tyr | [126,127,128,129,130,131] |
| Cation-π | Tyr-Lys; Phe-Lys; Tyr-Arg; Phe-Arg | [131,132] |
| Charge-charge | Asp/Glu-Arg/Lys; Phospho-Arg/Lys | [133,134,135,136,137] |
| Hydrophobic | Ala/Ile/Leu/M/Phe/Trp/Tyr/Val- or Pro-rich | [138,139,140,141,142,143] |
| Dipole-Dipole | Gly/Gln/Asn/Ser-Gly/Gln/Asn/Ser | [143,144] |
Listed are interactions implicated in phase separation, together with residues involved. For example, attractive π-stacking between aromatic rings (aromatic stacking, π-π interactions), electrostatic (charge-charge) interactions, or dipoledipole associations including hydrogen bonding can promote protein assembly by phase separation. Note that multiple types of interactions can cooperate for phase separation, and that besides proteins and their indicated residues and sequence motifs also nucleic acids participate in phase separation and provide multivalent binding surfaces.
A useful general framework for phase transitions driven by multivalent protein and nucleic acid molecules is provided by the ‘stickers and spacers’ model, adapted from the field of associative polymers [145,146]. In simplified terms, stickers provide intra- or intermolecular attraction between discrete sequence elements and are interspersed by spacers, which do not contribute significantly to the attractive interactions that drive condensate formation. Of note, stickers can be folded binding domains (e.g. SH2, SH3, SUMO, Chromo, Bromo, RRMs), labile structures (e.g. LARKS, low-complexity aromatic-rich kinked segments; RACs, reversible amyloid cores), or short linear motifs and even single residues that form attractive condensate-driving interactions [145–153]. Likewise, disordered sequence stretches can serve as neutral spacers between motifs that drive phase transitions. Intrinsically disordered sequences are thus neither necessary nor sufficient for phase separation [154]. Rather than sequence disorder per se, valence and patterning, e.g. of aromatic residues in prion-like domains, are important features associated with phase separation [129,140, 155]. In multi-component condensates, nodal scaffold molecules that are critical for condensate formation can be discriminated from low valency clients, although the distinction, rather than being binary, might be more gradual and also vary depending on the cellular context [156]. Nevertheless, identifying driver molecules and driver interactions that form biomolecular condensates and elucidating their context-dependent regulation is among the key challenges in this area.
The three intrinsically disordered FET proteins, Fused in sarcoma (FUS), Ewing sarcoma breakpoint region 1 (EWSR1/EWS), and TATA box binding protein associated factor 15 (TAF15), with their aminoterminal prion-like domains and their carboxy-terminal RNA-binding motifs and RG/RGG-repeats, play a prominent role as drivers of phase separation. The FET proteins are abundant nucleic acid binding proteins, which bind to thousands of transcripts and affect multiple steps in mRNA biogenesis [157]. They can be seen as prototype intrinsically disordered proteins (IDPs), and their low complexity domains serve as translocation partners for DNA-binding domains of various transcription factors in several different cancers [2]. Point mutations, on the other hand, cause neurodegenerative diseases such as amyotrophic lateral sclerosis (ALS) and frontotemporal lobar dementia (FTLD) [2]. As prototype IDPs with high medical relevance, their self-assembly features have attracted significant attention in recent years. Dissecting the molecular grammar of FET protein phase separation revealed that multivalent interactions among tyrosine residues from prion-like domains and arginine residues from RNA-binding domains and RG/RGG-repeats are critical [125]. Interestingly, when minimalist polymers of the positively charged amino acids arginine or lysine were investigated together with mono- or polynucleotides, poly-arginine condensates exhibited 100-fold higher viscosity as compared to poly-lysine droplets, and arginine outcompeted lysine for anionic binding, resulting in condensate immiscibility and inversion [135]. Consistently, lysine cannot substitute for arginine in the context of nuclear speckle condensation by intrinsically disordered mixed-charge domains, underpinning the specificity of interactions involved in condensate properties [30]. These results suggest that arginine motifs with their guanidium groups that enable simultaneous formation of charge-charge, π-π, and cation-π contacts play particularly important roles as stickers that drive condensate formation and govern their viscoelastic properties. On the other hand, such themes cannot be overly generalized, as sequence determinants of phase separation, both at the level of participating proteins and at the level of participating nucleic acids, are condensate type and context dependent. RNAs, for instance, not only act as molecular seeds that trigger protein phase separation, but based on their properties, such as abundance, sequence, length, secondary structures, and covalent modifications, also directly tune biophysical features of phase separated condensates, including size and shape, viscosity and surface tension, and molecular composition [31,60,81,158–161]. Such features can change over time and can result in what is typically referred to as ‘aging’ of phase-separated compartments towards increased condensate viscosity, enabling dynamic and sometimes adaptable transitions from liquid to gel and to solid [21,81,158,162–164]. While condensate solidification and protein aggregation may be more difficult (although not impossible) to reverse and are often associated with disease [165,166], a rather broad spectrum of (mixed) material property states is used in biological systems under physiological conditions and in response to cell stress. Within this broad spectrum, low complexity domains can even form labile, reversible cross-β polymer cores inside liquid droplets and reversible hydrogels [167,168]. The formation of these labile amyloid-like fibers is regulated by PTMs and may thereby respond to changing cellular conditions, including cell stress.
A particularly dangerous type of stress occurs in the nucleus when cells experience replication stress or DNA damage, and emerging evidence supports that replication stress and DNA damage responses not only involve stoichiometric high affinity interactions at sites of DNA damage and among signaling molecules, but also make use of phase separation properties of proteins, nucleic acids, and of the DNA damage-induced polymer poly(ADP-ribose) (PAR) for spatio-temporal and physico-chemical control of genome integrity maintenance.
3. Phase separation and maintenance of genome integrity
3.1. DNA damage sensing and PAR formation
Among the first cellular responses to DNA damage is the synthesis of PAR by enzymes of the PARP family [169,170]. PAR formation occurs locally at sites of DNA damage and is a transient response due to the antagonizing activity of PARG. Due to two phosphate groups per ADP-ribose unit, PAR is a highly negatively charged polymer, which during the peak of PAR production recruits a plethora of PAR-binding proteins by multivalent electrostatic interactions, including FUS, EWS and TAF15 [171]. While the prion-like N-termini of the FET proteins undergo concentration-dependent phase separation and form liquid droplets in cells, their positively charged RG/RGG-rich C-termini confer affinity to anionic PAR [68,172,173]. Interestingly, polyelectrolyte interactions can lead to ultrahigh affinity complexes in which the binding partners retain their structural disorder and highly dynamic character [174,175], implying that high affinity binding and structural disorder within complexes are not mutually exclusive. Hence, cooperative weak interactions can lead to very high binding affinities despite a high degree of conformational flexibility. PAR can reach chain lengths of several dozens to hundreds of units and can thereby generate a highly anionic environment at sites of PAR synthesis. FUS, on the other hand, with its multiple RG/RGG motifs is among the top 5% of proteins in terms of cellular abundance and has an estimated nuclear concentration between 4 and 8 μM [69]. The local induction of PAR chains nucleates the assembly of FUS both in vitro and in vivo [68,69]. Under physiologic conditions, intracellular FUS condensates stay liquid, but liquid to solid phase transition and aggregation into fibrous structures occurs for disease-associated mutations of FUS [68,69]. FUS-deficient or mutated cells have DSB repair defects and accumulate DNA damage, and they are sensitive to ionizing radiation and display chromosomal instability [176–180]. As FUS is involved in gene expression and transcription-associated processes, some of these effects might be due to altered transcription programs and transcription-associated DNA damage. Importantly, however, the PARP1-PAR-FUS system was recently reconstituted in vitro and analyzed at the single molecule level by atomic force microscopy [70]. This confirmed FUS recruitment to DNA damage through PAR-binding, and interestingly also revealed PAR- and FUS-dependent compartmentalization of damaged DNA [70]. Although direct in vivo evidence for PAR-dependent tethering of DNA ends by alteration of viscoelastic properties is currently missing, PARylation was shown to promote DNA repair by end-joining mechanisms that rely on close proximity of broken DNA ends [181,182]. Moreover, phase separation of FUS at DNA damage sites was recently reported to be involved in the spatial organization and clustering of DNA damage-induced γH2AX nanofoci detected by 3D-SIM (structured illumination microscopy), further strengthening the link between DNA damage-induced PAR formation, protein condensation and damage clustering [183]. Due to the short-lived nature of PAR and the transient recruitment of PAR-binding proteins, the proposed tethering of broken DNA ends might primarily assist early events in the DDR (e.g. comparatively fast end-joining reactions), whereas more long-lived modifications and proteins recruitments have the potential to stabilize the damaged chromatin domain in a more sustained manner (see below).
The FUS-related proteins EWS and TAF15 show very similar transient recruitment behavior and co-assemble into PAR-seeded compartments [68,172,173], and several additional RNA- and PAR-binding proteins, including hnRNPs and hnRNP-like proteins, also get transiently enriched in PAR-seeded condensates [171,184]. While according to the scaffold and client model some of these RNA- and PAR-binding proteins likely play more important roles than others for the local demixing that occurs around DNA break sites, it seems reasonable to assume that collectively they shape the local environment and its viscoelastic properties. One example is the nuclear matrix protein SAFB, which through an R/G-rich region at its C-terminus is recruited to sites of DNA damage in a PAR-dependent manner and promotes γH2AX spreading and DNA damage signaling [185]. More recently, SAFB was shown to phase separate in vitro and its interaction with heterochromatin-associated repeat transcripts via its R/G-rich region is required for maintaining interchromosomal interactions adjacent to pericentromeric heterochromatin and heterochromatin condensation [186]. In response to DNA damage, PAR may thus compete with repeat transcripts such as major satellite RNAs for SAFB binding, and it will be interesting to address whether recruitment of SAFB (and potentially of other non-coding RNA-binding proteins) to sites of DNA damage is associated with compromised heterochromatin architecture.
The transient nature of PAR-seeded condensates may balance such risks, and the FET proteins, SAFB and other RNA- and PAR-binding proteins are later released and excluded from sites of DNA damage [68,185,187–191]. DNA damage-induced phosphorylation regulates this exclusion, and phosphorylation was shown to disrupt FUS phase separation by reducing self-interaction [167,192]. The bimodal assembly and exclusion dynamics that are observed for FET and FET-like proteins can help to remove nascent RNA and RNA-DNA hybrids from DNA break sites to facilitate repair [188,193]. In addition to exclusion from the damaged area, phosphorylation by the DDR kinase DNA-PK also promotes nuclear export and cytoplasmic accumulation of the FET proteins [194]. Interestingly, in ataxia telangiectasia (A-T) cells, which show deficiency in the DDR kinase ATM, neurotoxic protein aggregation occurs due to increased levels of reactive oxygen species (ROS) and hyperactivation of PARylation [195]. Elevated levels of PAR have also been reported for Parkinson’s disease (PD), Alzheimer’s disease (AD), Huntington’s disease (HD) and for amyotrophic lateral sclerosis (ALS), and inhibition of PARylation was shown to be beneficial in PD, AD, HD, and ALS disease models [196–201]. Of note, while PAR formation is most strongly induced by DNA damage in the nucleus, it is also involved in cytoplasmic phase separation and protein condensation into stress granules, with important implications for health and disease [202–206]. As PARP inhibitors impact PAR-triggered phase separation and can prevent toxic protein aggregation, their therapeutic value may go beyond modulating DNA repair. In the context of DNA damage, the dynamic nature and timely dissolution of PAR-seeded condensates facilitates the handover to downstream compartmentalization of the damaged area.
3.2. Chromatin topology and the chromatin response to DNA damage
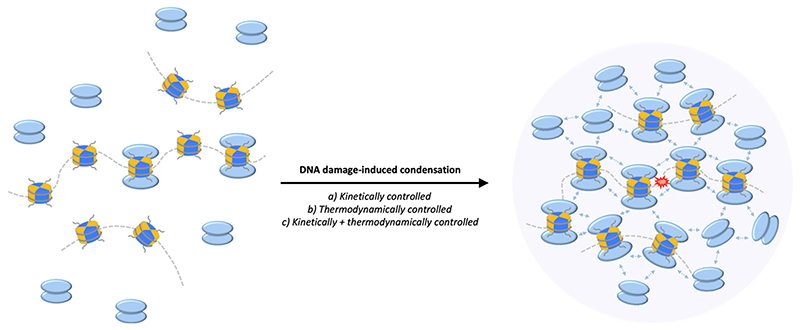
The chromatin response to DNA double-strand breaks (DSBs) has been investigated in great detail [207–210]. Around the break sites, multiple DNA damage-induced chromatin modifications work together to remodel the chromatin and recruit repair factors. Many of the proteins that get recruited to damaged chromatin contain structurally defined chromatin reader domains with affinity for specific DNA damage-induced histone marks. An apical mark that is induced by DSBs and spreads at megabase scale is the phosphorylation of H2AX by ATM to form γH2AX compartments. The formation of these large domains depends on cohesin-mediated loop extrusion at DSBs, which results in bidirectional spreading of γH2AX within the boundaries of topologically associating domains (TADs) marked by CTCF [211]. In yeast, DNA-cohesin clustering resembles bridging-induced (polymer-polymer) phase separation, which may stabilize cohesin-CTCF complexes [212], and thereby potentially also stabilize the γH2AX domain. Downstream of γH2AX and its reader protein MDC1 the large scaffolding protein 53BP1 gets recruited. 53BP1 is a multivalent chromatin reader and its recruitment depends on γH2AX, on the abundant histone mark H4K20me2 (a mark that discriminates unreplicated from replicated chromatin and guides DSB repair pathway choice accordingly), and on DNA damage-induced de novo deposition of H2AK15ub [213–216]. 53BP1 binds modified chromatin as a dimer, but preformed 53BP1 dimers quickly self-assemble into dynamic higher order oligomers upon cooperative H4K20me2 and H2AK15ub binding [217,218]. Consistently, apart from H4K20me2 and H2AK15ub binding, also the oligomerization domain of 53BP1 is required for 53BP1 assembly into DNA damage-induced foci [219]. Stoichiometric one-to-one binding models can therefore explain the initial recruitment of 53BP1 dimers to modified nucleosomes, but simplified linear ‘beads on a string’ illustrations fall short of capturing the ensuing assembly and maturation of 53BP1 compartments. Of note, there is no dichotomy between initial, kinetically controlled, stoichiometric binding, i. e. one 53BP1 dimer per nucleosome, and the ensuing, thermodynamically driven nonstoichiometric assembly to generate the 53BP1 compartment (Fig. 2). Rather than viewing them as mutually exclusive, these mechanisms likely synergize to stabilize the domain and endow it with favorable properties.
Fig. 2. Model of 53BP1 condensation at sites of DNA damage.
At the spatial and temporal scales relevant for the DNA damage response, 53BP1 condensation may be favored both kinetically, through enzymatically-controlled deposition of histone marks, and thermodynamically, through self-assembly of 53BP1 into mature oligomers to form a denser 53BP1 compartment.
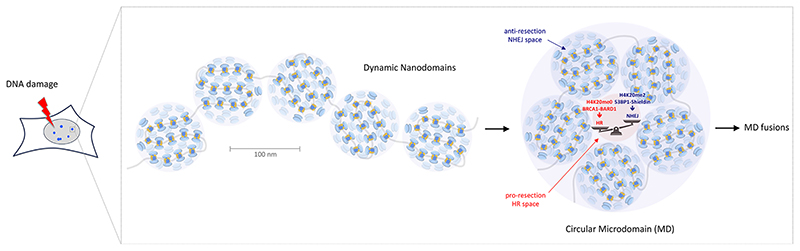
Indeed, the 53BP1 compartment stabilizes chromatin topology at multiple levels: In the sub-micrometer range, 53BP1 forms dynamic nanodomains, which overlap with TADs, similar to γH2AX [220–222]. These nanodomains then cluster together to form circular microdomains, which provide an environment that favors DSB repair by non-homologous end-joining (NHEJ) and confines limited DNA end resection and homologous recombination (HR) factors to its center. 53BP1 compartments, which can also undergo fusions to form even larger repair condensates, are thus multi-layered, and multiple types of interactions, weak and strong, cooperate in their formation and functionality (Fig. 3).
Fig. 3. Model of 53BP1 nano- and microdomains.
The propensity of 53BP1 to self-interact may play a role for the formation of 53BP1 nanodomains, for the assembly of multiple nanodomains into bigger microdomains, and for the fusion of microdomains into large DNA repair centers. The multi-layered organization of these domains enables tuning of DNA end resection and repair pathway choice according to replication and chromatin status, cell cycle position, and lesion complexity.
This multi-layered organization is disrupted easily, in fact more easily than the upstream γH2AX and MDC1 foci, by changes in osmotic pressure, temperature, salt concentration, or inhibition of hydrophobic interactions [71]. This is important to keep in mind when detergents or methanol are used before or simultaneously with cell fixation, as such treatments may disrupt critical interactions. The self-assembly features of 53BP1, which are triggered by DNA damage, can be recapitulated in DNA damage-independent optoDroplet experiments using the Arabi-dopsis thaliana photoreceptor cryptochrome 2 (Cry2) as controllable and reversible optogenetic seed module [71]. On its own, this module does not result in light-induced protein clustering, but when fused to protein sequences that can phase separate by multivalent interactions, rapid formation of liquid droplets can be observed in living cells [71,223]. The C-terminus of 53BP1, which contains the oligomerization domain and is particularly rich in amino acids that have been implicated in phase separation, is sufficient to promote optoDroplet formation in this system, and the formed condensates are highly dynamic, frequently fuse, and once formed remain for several minutes in the absence of blue light [71]. When combined with a localized, chromatin-tethered seeding event such as endonuclease-mediated DSBs, blue light induction triggers rapid assembly of additional 53BP1 molecules specifically at these sites. 53BP1 also phase separates in vitro [71,72], and 53BP1 foci in irradiated cells display behavior consistent with nucleation, growth and coarsening, both when expressed ectopically and also in cells in which the endogenous gene locus had been edited to express fluorescently labeled 53BP1 [71,72]. Interestingly, DNA damage induced 53BP1 condensates also ‘age’ towards increased viscosity, and their formation and properties are modulated by additional macromolecules, including DNA damage-induced long non-coding RNAs (dilncRNA) synthesized by RNA Pol II from DSBs [72], and by the large 53BP1-interacting scaffolding protein AHNAK [73]. As more functions for RNA and RNA-binding proteins as regulators of genome stability are emerging [224–226], it will be interesting to elucidate exactly how different RNA species, their modifications and their abundance and dynamics around DNA break sites affect DDR condensate properties and repair outcomes.
53BP1 condensates also selectively attract clients, including the tumor suppressor p53 [71], which itself has phase separation properties and forms nuclear condensates to amplify target gene expression [29,227]. Consistently, p53 target gene expression and p21 induction are dependent on 53BP1 and can be tuned by 53BP1 regulators, which in turn determines cell fate decisions [71,73,228–231]. While structural and biochemical studies suggest that dimethylated p53 binds stoichiometrically to the 53BP1 tudor domain [228,232], cellular experiments have implicated the oligomerization domain and the BRCT domain of 53BP1 in p53 activation [71,73,229], raising the possibility that structural diversity and interaction multiplicity within a larger conformational space may contribute to full p53 activation.
DNA damage compartments not only form larger repair centers by fusion, but also relocalize within the nuclear space [233]. Nuclear actin polymerization, the actin-nucleating ARP2/3 complex, and nuclear myosins have been implicated in the directed motion of a subset of DSBs into discrete clusters to promote homology-directed repair [234,235]. Cell membrane-associated actin assembly by the ARP2/3 complex depends on phase separation and stoichiometry-dependent dwell times [236]. Potentially, relocalization of the subset of DNA breaks, which are difficult to repair, e.g. due to the chemical nature of the break site itself, due to heterochromatic environment, or due to iterative cutting by an activated endonuclease, also depends on dwell times of the actin polymerizing machinery at the damaged domain. Thus, how long a DNA break remains unrepaired may work as a timer to eventually trigger mobilization of the domain and initiate a different type of repair, e.g. to switch from unsuccessful NHEJ attempts to relocalization and repair by HR.
In yeast, the meiotic DNA break machinery assembles by a DNA-driven condensation process that shares several features with phase separation [74]. Furthermore, the recombination protein Rad52, the functional homolog of the HR factor BRCA2 in mammalian cells, accumulates at induced DSBs to form liquid droplets with features reminiscent of phase separation [75,76]. Rad52 exhibits a slower diffusion coefficient and confined motion inside the condensate, and the diffusion coefficient changes sharply when molecules cross the phase boundary [76]. Furthermore, foci fusions result in condensates with twice the volume of individual foci, and Rad52 molecules inside fused domains explore the entire compartment. Together, these results support that rather than clustered binding, a nucleation core and phase separation cooperate to form the repair compartment [76].
Of note, Rad52 condensates nucleate DNA damage-inducible intranuclear microtubule filaments (DIMs), which are important for condensate fusions and, upon enrichment of tubulin in fused domains, also mediate the mobilization of damaged DNA to the nuclear envelope for repair [75]. Functional cooperation between membraneless liquid droplets, microtubule filaments, and lipid membranes is an exciting new concept, and the mechanical connections between membraneless and membrane-bound compartments, e.g. as can be observed for instance for phase separated RNA granules that ‘hitchhike’ on lysosomes for long-distance RNA transport in neurons [237], is largely unexplored in the context of mammalian DNA repair and maintenance of genome integrity.
3.3. DNA replication and replication stress signaling
DNA replication and DNA replication stress are a major source of DNA damage and genome instability in cancer [238], and fine-tuned mechanisms have evolved to coordinate the firing of replication origins, replication fork speed, fork stability and repair with cell cycle progression and checkpoint signaling [239]. In mammalian cells, replication initiation involves licensing of replication origins and formation of the pre-replication complex (pre-RC), followed by origin firing in S-phase. While it is understood that replication initiation and origin firing are tightly regulated and well coordinated in space and time, the dynamic control of interactions involved in forming initiation clusters and replication factories in eukaryotic cells remains nebulous. Recent work on replication initiation in the fruit fly Drosophila melanogaster revealed that the origin recognition complex (ORC) and the initiation factors Cdc6 and Cdt1 contain intrinsically disordered regions that drive DNA-nucleated phase separation, which selectively promotes assembly and loading of the Mcm2-7 complex of the replicative helicase [77]. Cyclin-CDK-mediated phosphorylation disrupts phase separation of these condensates, likely by abolishing electrostatic interactions between charged disordered protein sequences and DNA [77]. In mammalian cells, a very similar mechanism seems to be in place, because short linear motifs (SLiMs) within intrinsically disordered regions of ORC1 and CDC6 drive phosphorylation state-dependent interactions and liquid-liquid phase separation on DNA [78]. Also here, the process is neither untargeted nor independent of energy, but rather part of a multi-step initiator condensation process.
Downstream of origin licensing and formation of the pre-RC, origin firing during S-phase progression is controlled by the cell cycle checkpoint kinase ATR [240]. ATR activation in response to replication stress depends on the scaffolding protein TopBP1 and its intrinsically disordered ATR activation domain (AAD). Similar to the DSB repair factor 53BP1, TopBP1 was recently shown to form light-inducible optoDroplets with properties resembling phase separation [79]. Intriguingly, TopBP1 optoDroplets not only enriched the ATR kinase, but could also be used to reversibly switch ATR signaling on and off, resulting in blue light-controlled activation of the major downstream checkpoint kinase CHK1 and a slowdown of replication forks [79]. All these effects were mediated by the disordered AAD of TopBP1 and were abrogated by mutation of an essential aromatic tryptophan residue. TopBP1 is phosphorylated in response to replication stress, and mutation of a phosphorylation site in close proximity of the critical tryptophan to alanine reduced TopBP1 condensation and ATR signaling, suggesting a positive feedback loop between TopBP1 and ATR in TopBP1 condensates for signal amplification [79]. Consistent with the multi-step processes and multi-layered condensate organization discussed above, the authors propose the formation of a nucleation complex (based on stoichiometric interactions, e.g. between RPA and ssDNA and ATR-ATRIP, TopBP1 and the 9-1-1 complex), which stabilizes TopBP1 at stalled or damaged replication forks, and an ensuing condensation into dynamic higher-order assemblies by multivalent cooperative interactions to achieve robust ATR activation and signal amplification [79].
3.4. Alternative lengthening of telomeres (ALT)
Certain regions in the genome are particularly fragile, e.g. due to DNA secondary structures, repetitive sequence elements, heterochromatic environment, late replication timing, low density of replication origins, or a combination of these features [230]. Telomeres at chromosome ends fall into this category, and replication stress at telomeres is frequently seen in cancer cells. A subset of cancer cells uses alternative lengthening of telomeres (ALT) as telomerase-independent mechanism of telomere maintenance and show particularly high levels of telomere-associated replication stress [241]. ALT is characterized by telomere clustering in ALT-associated PML bodies (APBs) and by recombination-based telomere elongation involving break-induced replication (BIR), a noncanonical form of DNA synthesis [242]. APB formation is dependent on SUMOylation and on SUMO recognition by SUMO interaction motifs (SIMs), and recent work has implicated phase separation by SUMO-SIM binding modules in the formation of APBs [37, 38]. Poly(SUMO) and poly(SIM) scaffolds, or chemically induced tethering of SIMs at telomeres, drives liquid condensation and telomere clustering by coalescence, and induces ALT-like phenotypes such as mitotic DNA synthesis (MiDAS) at telomeres, BLM-dependent C-circle formation, and heterogeneous telomere length [37,38].
Interestingly, ALT-associated homology-directed repair also depends on carefully controlled PAR formation at broken telomeres [243]. PARylation at ALT telomeres assembles various PAR-binding proteins, including the repair factors XRCC1 and LIG3 and the chromatin remodelers ALC1 and CHD7, but also the disordered FET proteins FUS, EWS and TAF15, several hnRNPs, SAFB1, SATB1, other RNA- and PAR-binding proteins such as RBMX and NONO, and the F-actin nucleation complex ARP2/3 [243]. Consistent with a role in PAR-triggered phase separation at ALT telomeres, loss of these proteins impairs APB formation and telomere clustering [243].
Finally, recent findings have suggested that telomeric BIR in APBs is a self-perpetuating process, due to BLM helicase-dependent BLM enrichment and BIR-dependent TRF2 SUMOylation to sustain SUMOSIM interactions and DNA repair factor recruitment [244]. These findings are consistent with the phase separation model of APB assembly, and self-perpetuation of biochemical processes might be a more general feature of biomolecular condensates during their respective lifetimes.
4. Opportunities and future perspectives
Investigating spatio-temporal aspects of genome integrity maintenance has been a long-standing endeavor, not least due to the clinical relevance of cellular replication stress and DNA damage responses. Resurgent interest in phase separation and its different flavors (e.g. liquid-liquid, liquid-gel, liquid-solid, polymer-polymer) has started to broaden the focus from stoichiometric binding between structurally defined binding partners towards studying multivalent interactions and structurally less well defined complexes. This has also spurred new interest in material properties of membraneless compartments and how these relate to function. Rather than being seen as mutually exclusive, stoichiometric binding between structured domains and multivalent interactions that can drive phase separation likely cooperate for condensate formation in the complex environment of the cell nucleus, and several intriguing examples related to different aspects of maintenance of genome integrity have recently emerged (Fig. 4).
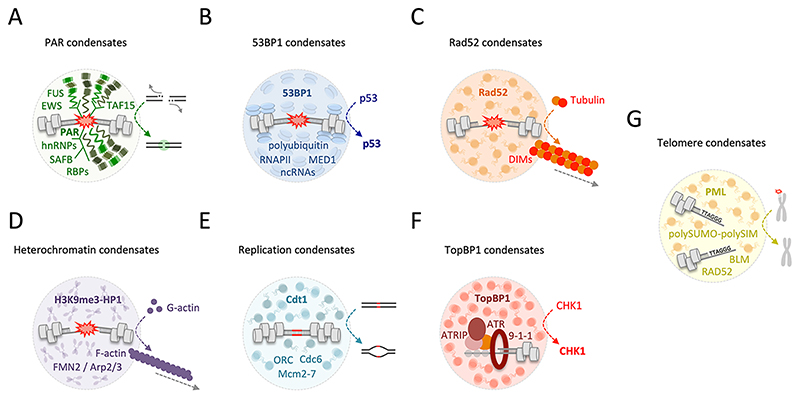
Fig. 4. Emerging examples of nuclear condensates with phase separation properties and roles in maintenance of genome integrity.
(A) Poly(ADP-ribose) (PAR) triggers assembly of multiple intrinsically disordered and RNA-binding proteins (RBPs) including FUS/TLS, EWS/EWSR1, TAF15, hnRNPs, SAFB1/2 and SLTM and initiates their local demixing at sites of DNA damage, which may promote tethering and religation of broken chromosome ends. (B) 53BP1 condensates form around DSBs in a manner that requires multivalent chromatin binding and oligomerization and is promoted by DNA damage-induced transcription of long noncoding RNAs (ncRNAs). 53BP1 condensation is linked to activation of p53 signaling, and p53 dynamically co-assembles in 53BP1 condensates. (C) In yeast, the DNA repair protein Rad52 forms phase-separated condensates, which concentrate tubulin and project DNA damage-inducible intranuclear microtubule filaments (DIMs). DIMs mobilize the liquid Rad52 compartment to the nuclear periphery for repair. (D) In mouse and human cells, heterochromatin domains get mobilized when they experience DNA damage through FMN2/ARP2/3-mediated actin polymerization. Nuclear F-actin and myosin re-localize heterochromatic DSBs to the nuclear periphery for repair. (E) For initiation of DNA replication, DNA-dependent phase separation of the origin recognition complex (ORC), Cdt1 and Cdc6 facilitates Mcm2-7 loading. (F) TopBP1 condensation at DNA replication impediments switches on ATR/CHK1 kinase signaling to initiate a replication stress response. (G) Through multivalent polySUMO-polySIM interactions at chromosome ends in cells that use alternative lengthening of telomeres (ALT), ALT-associated PML bodies (APBs) form by phase separation for telomere clustering and telomere recombination.
Unlike phase separation in few component systems, the formation of biological condensates involved in the maintenance of genome integrity is more complex and typically follows a multi-step process, but in this process makes use of phase separation properties of some of the key components involved [245]. Studying these properties in vitro, using purified proteins, can be revealing (e.g. to identify relevant interactions and important sequence features, or to study mixing behavior in well controlled settings), and just like the biochemical characterization of an enzymatic activity in a test tube calls for an in vivo counterpart to provide context, cellular assays ideally performed at endogenous protein concentrations as was done in several cases, provide an important counterpart to phase separation experiments with purified components.
Existing technologies are being further developed and new ones are rapidly emerging, which can complement and extend classical biochemistry to inform about structure-function relationships in the context of biomolecular condensates. These include structure elucidation techniques such as solution and solid-state nuclear magnetic resonance spectroscopy (NMR, ssNMR), small-angle x-ray scattering (SAXS), cryo-electron microscopy (cryo-EM) and atomic force microscopy (AFM), as well as single molecule fluorescence microscopy (SMFM) techniques including fluorescence fluctuation spectroscopy (FFS) and single molecule Förster resonance energy transfer (smFRET). Moreover, in vitro turbidity assays, microfluidics and optical tweezers, as well as light-inducible phase separation (optoDroplets) to study phase behavior under controlled conditions in living cells have emerged as informative techniques. Computational tools for sequence analysis and prediction of phase separation potential, for structural modeling, molecular dynamics simulations, and to calculate phase diagrams for multicomponent systems have become increasingly important, and intravital microscopy (IVM) may be used to examine condensates in their native environment [246–255]. As technologies advance and become more quantitative, a clearer picture of the relative contribution and functional relevance of phase separation properties in different biological contexts, also with regard to alternative mechanisms and interaction modes [256–259], is going to emerge. As with any single technology, limitations exist and have to be taken into account. For instance, as mentioned above, the choice of cell permeabilisation, pre-extraction and fixation has an impact on cellular architecture, and weak interactions involved in condensate formation may get easily lost. In live cell imaging experiments of endogenously labeled proteins (which, if the tag does not interfere with protein functions, is preferable over ectopic expression of fluorescently labeled proteins), signal intensity depends on target protein abundance, with limiting consequences for exposure times and phototoxicity. The development of particularly bright monomeric fluorescent proteins and advanced image segmentation tools using deep learning may attenuate some of these limitations in the future. While single cell and single molecule techniques provide insight into phenotypic and structural heterogeneity, this information is lost when population averages are analyzed. Moreover, microscopy-based approaches such as fluorescence in situ hybridization (FISH) and genomics-based approaches such as chromatin conformation capture (3C) techniques do not always yield congruent results, and it was suggested that 3C measurements may not always reflect spatial proximity [260]. Such limitations need to be taken into account when functions of biomolecular condensates are studied in the context of genome stability and organization.
Over the last couple of years the concept of phase separation has revived significant interest in and spurred new research on multivalent interactions, disordered protein sequences, dynamic protein and nucleic acid complexes and material properties of intracellular condensates. In multiple cellular contexts, including the processes involved in DNA repair and maintenance of genome integrity, there is mounting experimental support for intracellular phase boundaries, coarsening, and differential viscoelastic properties of condensates versus the surrounding milieu. At the same time there is experimental support for the notion that in nuclear multi-component compartments not all components are distributed uniformly and that their assembly resembles a tightly controlled and energy-dependent multi-step process, which often starts from a nucleation core. This process, however, once initiated, uses self-assembly properties of key components to foster percolation.
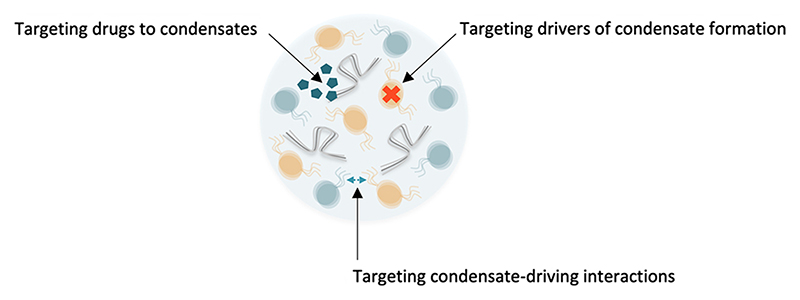
Working towards identifying and characterizing the specific interactions that are critical for the dynamics and for the material properties of intracellular biomolecular condensates may offer new opportunities to modulate their functions (Fig. 5). First examples of condensate-specific enrichment of cancer therapeutics have been reported [261,262], and the better we understand the physico-chemical, viscoelastic properties of intracellular condensates and the molecular interactions that define them, the more specific the targeting of drugs to their relevant sites of action may work. This might be particularly relevant as derailed phase separation could well be a unifying theme for the development of a broad spectrum of conditions ranging from neurodegeneration to cancer and aging [263–266]. Finally, the development of bio-inspired materials such as designer organelles and nanocarriers can benefit from a deeper understanding of biomolecular condensates and their varied properties [267,268]. Although applying concepts from polymer chemistry and soft matter physics to cell biology and, vice versa, using knowledge from biological systems to engineer functional biomaterials comes with inherent challenges, the chances and new opportunities prevail.
Fig. 5. Emerging opportunities for condensate therapeutics.
Condensate properties and functions may be modulated by targeting the molecular drivers of condensate formation, or by interfering with the intra- and intermolecular interactions that are important for condensate formation and maturation. Conversely, the physico-chemical properties of condensates may be exploited to facilitate partitioning of drugs into target compartments and enhance their efficacy.
Acknowledgements
We acknowledge research funding from the Swiss National Science Foundation (PP00P3_179057, 310030_197003) and the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program (ERC-2016-STG 714326). We apologize to all authors whose work could not be cited due to space limitations and sincerely thank Dr. Priya R. Banerjee and Dr. Evi Soutoglou for insightful comments on the manuscript and members of the Altmeyer lab for fruitful discussions.
Footnotes
Declaration of Competing Interest
The authors declare no conflict of interest.
References
- [1].Dawkins R. The Blind Watchmaker: Why the Evidence of Evolution Reveals a Universe Without Design. 1988 [Google Scholar]
- [2].Aguzzi A, Altmeyer M. Phase separation: linking cellular compartmentalization to disease. Trends Cell Biol. 2016;26:547–558. doi: 10.1016/j.tcb.2016.03.004. [DOI] [PubMed] [Google Scholar]
- [3].Alberti S. Phase separation in biology, Curr. Biol. 2017;27:R1097–R1102. doi: 10.1016/j.cub.2017.08.069. [DOI] [PubMed] [Google Scholar]
- [4].Boeynaems S, Alberti S, Fawzi NL, Mittag T, Polymenidou M, Rousseau F, Schymkowitz J, Shorter J, Wolozin B, Van Den Bosch L, Tompa P, et al. Protein phase separation: a new phase in cell biology. Trends Cell Biol. 2018;28:420–435. doi: 10.1016/j.tcb.2018.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Banani SF, Lee HO, Hyman AA, Rosen MK. Biomolecular condensates: organizers of cellular biochemistry, Nat. Rev Mol Cell Biol. 2017;18:285–298. doi: 10.1038/nrm.2017.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Shin Y, Brangwynne CP. Liquid phase condensation in cell physiology and disease. Science. 2017;357 doi: 10.1126/science.aaf4382. [DOI] [PubMed] [Google Scholar]
- [7].Lyon AS, Peeples WB, Rosen MK. A framework for understanding the functions of biomolecular condensates across scales. Nat Rev Mol Cell Biol. 2020 doi: 10.1038/s41580-020-00303-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Alberti S, Hyman AA. Biomolecular condensates at the nexus of cellular stress, protein aggregation disease and ageing. Nat Rev Mol Cell Biol. 2021;22:196–213. doi: 10.1038/s41580-020-00326-6. [DOI] [PubMed] [Google Scholar]
- [9].Snead WT, Gladfelter AS. The control centers of biomolecular phase separation: how membrane surfaces, PTMs, and active processes regulate condensation. Mol Cell. 2019;76:295–305. doi: 10.1016/j.molcel.2019.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Schmit JD, Feric M, Dundr M. How hierarchical interactions make membraneless organelles tick like clockwork. Trends Biochem Sci. 2021;46(7):525–534. doi: 10.1016/j.tibs.2020.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Hein MY, Hubner NC, Poser I, Cox J, Nagaraj N, Toyoda Y, Gak IA, Weisswange I, Mansfeld J, Buchholz F, Hyman AA, et al. A human interactome in three quantitative dimensions organized by stoichiometries and abundances. Cell. 2015;163:712–723. doi: 10.1016/j.cell.2015.09.053. [DOI] [PubMed] [Google Scholar]
- [12].Riback JA, Zhu L, Ferrolino MC, Tolbert M, Mitrea DM, Sanders DW, Wei MT, Kriwacki RW, Brangwynne CP. Composition-dependent thermodynamics of intracellular phase separation. Nature. 2020;581:209–214. doi: 10.1038/s41586-020-2256-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Feric M, Misteli T. Phase separation in genome organization across evolution. Trends Cell Biol. 2021 doi: 10.1016/j.tcb.2021.03.001. Online ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Cohan MC, Pappu RV. Making the case for disordered proteins and biomolecular condensates in bacteria. Trends BiochemSci. 2020;45:668–680. doi: 10.1016/j.tibs.2020.04.011. [DOI] [PubMed] [Google Scholar]
- [15].Franzmann TM, Jahnel M, Pozniakovsky A, Mahamid J, Holehouse AS, Nuske E, Richter D, Baumeister W, Grill SW, Pappu RV, Hyman AA, et al. Phase separation of a yeast prion protein promotes cellular fitness. Science. 2018;359 doi: 10.1126/science.aao5654. [DOI] [PubMed] [Google Scholar]
- [16].Emenecker RJ, Holehouse AS, Strader LC. Emerging roles for phase separation in plants. DevCell. 2020;55:69–83. doi: 10.1016/j.devcel.2020.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Hyman AA, Weber CA, Julicher F. Liquid-liquid phase separation in biology. AnnuRev Cell Dev Biol. 2014;30:39–58. doi: 10.1146/annurev-cellbio-100913-013325. [DOI] [PubMed] [Google Scholar]
- [18].Brocca S, Grandori R, Longhi S, Uversky V. Liquid-Liquid phase separation by intrinsically disordered protein regions of viruses: roles in viral life cycle and control of virus-host interactions. Int J Mol Sci. 2020;21 doi: 10.3390/ijms21239045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Brangwynne CP, Mitchison TJ, Hyman AA. Active liquid-like behavior of nucleoli determines their size and shape in Xenopus laevis oocytes. ProcNatl Acad Sci U S A. 2011;108:4334–4339. doi: 10.1073/pnas.1017150108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Feric M, Vaidya N, Harmon TS, Mitrea DM, Zhu L, Richardson TM, Kriwacki RW, Pappu RV, Brangwynne CP. Coexisting liquid phases underlie nucleolar subcompartments. Cell. 2016;165:1686–1697. doi: 10.1016/j.cell.2016.04.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Frottin F, Schueder F, Tiwary S, Gupta R, Korner R, Schlichthaerle T, Cox J, Jungmann R, Hartl FU, Hipp MS. The nucleolus functions as a phase-separated protein quality control compartment. Science. 2019;365(6451):342–347. doi: 10.1126/science.aaw9157. [DOI] [PubMed] [Google Scholar]
- [22].Ide S, Imai R, Ochi H, Maeshima K. Transcriptional suppression of ribosomal DNA with phase separation. Sci Adv. 2020;6 doi: 10.1126/sciadv.abb5953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Lafontaine DLJ, Riback JA, Bascetin R, Brangwynne CP. The nucleolus as a multiphase liquid condensate. NatRev Mol Cell Biol. 2021;22:165–182. doi: 10.1038/s41580-020-0272-6. [DOI] [PubMed] [Google Scholar]
- [24].Abraham KJ, Khosraviani N, Chan JNY, Gorthi A, Samman A, Zhao DY, Wang M, Bokros M, Vidya E, Ostrowski LA, Oshidari R, et al. Nucleolar RNA polymerase II drives ribosome biogenesis. Nature. 2020;585:298–302. doi: 10.1038/s41586-020-2497-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Neugebauer KM. Special focus on the cajal body. RNA Biol. 2017;14:669–670. doi: 10.1080/15476286.2017.1316928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Razin SV, Gavrilov AA. The role of liquid-Liquid phase separation in the compartmentalization of cell nucleus and spatial genome organization. Biochemistry Mosc. 2020;85:643–650. doi: 10.1134/S0006297920060012. [DOI] [PubMed] [Google Scholar]
- [27].Courchaine EM, Lu A, Neugebauer KM. Droplet organelles? EMBO J. 2016;35:1603–1612. doi: 10.15252/embj.201593517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Hur W, Kemp JP, Jr, Tarzia M, Deneke VE, Marzluff WF, Duronio RJ, Di Talia S. CDK-regulated phase separation seeded by histone genes ensures precise growth and function of histone locus bodies. Dev Cell. 2020;54:379–394.:e376. doi: 10.1016/j.devcel.2020.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Alexander KA, Cote A, Nguyen SC, Zhang L, Gholamalamdari O, Agudelo-Garcia P, Lin-Shiao E, Tanim KMA, Lim J, Biddle N, Dunagin MC, et al. p53 mediates target gene association with nuclear speckles for amplified RNA expression. MolCell. 2021;81:1666–1681.:e1666. doi: 10.1016/j.molcel.2021.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Greig JA, Nguyen TA, Lee M, Holehouse AS, Posey AE, Pappu RV, Jedd G. Arginine-enriched mixed-charge domains provide cohesion for nuclear speckle condensation. Mol Cell. 2020;77:1237–1250.:e1234. doi: 10.1016/j.molcel.2020.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Fox AH, Nakagawa S, Hirose T, Bond CS. Paraspeckles: where long noncoding RNA meets phase separation. Trends BiochemSci. 2018;43:124–135. doi: 10.1016/j.tibs.2017.12.001. [DOI] [PubMed] [Google Scholar]
- [32].Hennig S, Kong G, Mannen T, Sadowska A, Kobelke S, Blythe A, Knott GJ, Iyer KS, Ho D, Newcombe EA, Hosoki K, et al. Prion-like domains in RNA binding proteins are essential for building subnuclear paraspeckles. J Cell Biol. 2015;210:529–539. doi: 10.1083/jcb.201504117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Yamazaki T, Souquere S, Chujo T, Kobelke S, Chong YS, Fox AH, Bond CS, Nakagawa S, Pierron G, Hirose T. Functional domains of NEAT1 architectural lncRNA induce paraspeckle assembly through phase separation. MolCell. 2018;70:1038–1053.:e1037. doi: 10.1016/j.molcel.2018.05.019. [DOI] [PubMed] [Google Scholar]
- [34].Wang Y, Chen LL. Organization and function of paraspeckles. Essays Biochem. 2020;64:875–882. doi: 10.1042/EBC20200010. [DOI] [PubMed] [Google Scholar]
- [35].Corpet A, Kleijwegt C, Roubille S, Juillard F, Jacquet K, Texier P, Lomonte P. PML nuclear bodies and chromatin dynamics: catch me if you can! Nucleic Acids Res. 2020;48(21):11890–11912. doi: 10.1093/nar/gkaa828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Banani SF, Rice AM, Peeples WB, Lin Y, Jain S, Parker R, Rosen MK. Compositional control of phase-separated cellular bodies. Cell. 2016;166:651–663. doi: 10.1016/j.cell.2016.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Min J, Wright WE, Shay JW. Clustered telomeres in phase-separated nuclear condensates engage mitotic DNA synthesis through BLM and RAD52. Gene Dev. 2019;33:814–827. doi: 10.1101/gad.324905.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Zhang HY, Zhao RW, Tones J, Liu MC, Dilley RL, Chenoweth DM, Greenberg RA, Lampson MA. Nuclear body phase separation drives telomere clustering in ALT cancer cells. MolBiol Cell. 2020;31:2048–2056. doi: 10.1091/mbc.E19-10-0589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Pankivskyi S, Pastre D, Steiner E, Joshi V, Rynditch A, Hamon L. ITSN1 regulates SAM68 solubility through SH3 domain interactions with SAM68 proline-rich motifs. CellMol Life Sci. 2021;78:1745–1763. doi: 10.1007/s00018-020-03610-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Keenen MM, Brown D, Brennan LD, Renger R, Khoo H, Carlson CR, Huang B, Grill SW, Narlikar GJ, Redding S. HPI proteins compact DNA into mechanically and positionally stable phase separated domains. Elife. 2021 doi: 10.7554/eLife.64563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Larson AG, Elnatan D, Keenen MM, Trnka MJ, Johnston JB, Burlingame AL, Agard DA, Redding S, Narlikar GJ. Liquid droplet formation by HP1alpha suggests a role for phase separation in heterochromatin. Nature. 2017;547:236–240. doi: 10.1038/nature22822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Gibson BA, Doolittle LK, Schneider MWG, Jensen LE, Gamarra N, Henry L, Gerlich DW, Redding S, Rosen MK. Organization of chromatin by intrinsic and regulated phase separation. Cell. 2019;179:470–484.:e421. doi: 10.1016/j.cell.2019.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Li CH, Coffey EL, Dall’Agnese A, Hannett NM, Tang X, Henninger JE, Platt JM, Oksuz O, Zamudio AV, Afeyan LK, Schuijers J, et al. MeCP2 links heterochromatin condensates and neurodevelopmental disease. Nature. 2020;586:440–444. doi: 10.1038/s41586-020-2574-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Strickfaden H, Tolsma TO, Sharma A, Underhill DA, Hansen JC, Hendzel MJ. Condensed chromatin behaves like a solid on the mesoscale in vitro and in living cells. Cell. 2020;183:1772–1784.:e1713. doi: 10.1016/j.cell.2020.11.027. [DOI] [PubMed] [Google Scholar]
- [45].Strom AR, Emelyanov AV, Mir M, Fyodorov DV, Darzacq X, Karpen GH. Phase separation drives heterochromatin domain formation. Nature. 2017;547:241–245. doi: 10.1038/nature22989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Wang L, Gao Y, Zheng X, Liu C, Dong S, Li R, Zhang G, Wei Y, Qu H, Li Y, Allis CD, et al. Histone modifications regulate chromatin compartmentalization by contributing to a phase separation mechanism. Mol Cell. 2019;76:646–659.:e646. doi: 10.1016/j.molcel.2019.08.019. [DOI] [PubMed] [Google Scholar]
- [47].Shakya A, Park S, Rana N, King JT. Liquid-liquid phase separation of histone proteins in cells: role in chromatin organization. Biophys J. 2020;118:753–764. doi: 10.1016/j.bpj.2019.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Williams JF, Surovtsev IV, Schreiner SM, Nguyen H, Hu Y, Mochrie SGJ, King MC. Phase separation enables heterochromatin domains to do mechanical work. bioRxiv. 2020 doi: 10.1101/2020.07.02.184127. [DOI] [Google Scholar]
- [49].Sanulli S, Trnka MJ, Dharmarajan V, Tibble RW, Pascal BD, Burlingame AL, Griffin PR, Gross JD, Narlikar GJ. HP1 reshapes nucleosome core to promote phase separation of heterochromatin. Nature. 2019;575:390–394. doi: 10.1038/s41586-019-1669-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Tatavosian R, Kent S, Brown K, Yao T, Duc HN, Huynh TN, Zhen CY, Ma B, Wang H, Ren X. Nuclear condensates of the Polycomb protein chromobox 2 (CBX2) assemble through phase separation. J Biol Chem. 2019;294:1451–1463. doi: 10.1074/jbc.RA118.006620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Pandya-Jones A, Markaki Y, Serizay J, Chitiashvili T, Leon WRM, Damianov A, Chronis C, Papp B, Chen CK, McKee R, Wang XJ, et al. A protein assembly mediates Xist localization and gene silencing. Nature. 2020;587:145–151. doi: 10.1038/s41586-020-2703-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Boehning M, Dugast-Darzacq C, Rankovic M, Hansen AS, Yu T, Marie-Nelly H, McSwiggen DT, Kokic G, Dailey GM, Cramer P, Darzacq X, et al. RNA polymerase II clustering through carboxy-terminal domain phase separation. Nat Struct Mol Biol. 2018;25:833–840. doi: 10.1038/s41594-018-0112-y. [DOI] [PubMed] [Google Scholar]
- [53].Boija A, Klein IA, Sabari BR, Dall’Agnese A, Coffey EL, Zamudio AV, Li CH, Shrinivas K, Manteiga JC, Hannett NM, Abraham BJ, et al. Transcription factors activate genes through the phase-separation capacity of their activation domains. Cell. 2018;175:1842–1855.:e1816. doi: 10.1016/j.cell.2018.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Cai DF, Feliciano D, Dong P, Flores E, Gruebele M, Porat-Shliom N, Sukenik S, Liu Z, Lippincott-Schwartz J. Phase separation of YAP reorganizes genome topology for long-term YAP target gene expression. Nat Cell Biol. 2019;21:1578–1589. doi: 10.1038/s41556-019-0433-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Cho WK, Spille JH, Hecht M, Lee C, Li C, Grube V, Cisse II. Mediator and RNA polymerase II clusters associate in transcription-dependent condensates. Science. 2018;361:412–415. doi: 10.1126/science.aar4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Hnisz D, Shrinivas K, Young RA, Chakraborty AK, Sharp PA. A phase separation model for transcriptional control. Cell. 2017;169:13–23. doi: 10.1016/j.cell.2017.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Sabari BR, Dall’Agnese A, Boija A, Klein IA, Coffey EL, Shrinivas K, Abraham BJ, Hannett NM, Zamudio AV, Manteiga JC, Li CH, et al. Coactivator condensation at superenhancers links phase separation and gene control. Science. 2018;361 doi: 10.1126/science.aar3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Cramer P. Organization and regulation of gene transcription. Nature. 2019;573:45–54. doi: 10.1038/s41586-019-1517-4. [DOI] [PubMed] [Google Scholar]
- [59].Fasciani A, D’Annunzio S, Poli V, Fagnocchi L, Beyes S, Michelatti D, Corazza F, Antonelli L, Gregoretti F, Oliva G, Belli R, et al. MLL4-associated condensates counterbalance Polycomb-mediated nuclear mechanical stress in Kabuki syndrome. Nat Genet. 2020;52:1397–1411. doi: 10.1038/s41588-020-00724-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Henninger JE, Oksuz O, Shrinivas K, Sagi I, LeRoy G, Zheng MM, Andrews JO, Zamudio AV, Lazaris C, Hannett NM, Lee TI, et al. RNA-mediated feedback control of transcriptional condensates. Cell. 2021;184:207–225.:e224. doi: 10.1016/j.cell.2020.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Kent S, Brown K, Yang CH, Alsaihati N, Tian C, Wang HB, Ren XJ. Phase-separated transcriptional condensates accelerate target-search process revealed by live-cell single-molecule imaging. Cell Rep. 2020;33 doi: 10.1016/j.celrep.2020.108248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Lu Y, Wu T, Gutman O, Lu H, Zhou Q, Henis YI, Luo K. Phase separation of TAZ compartmentalizes the transcription machinery to promote gene expression. NatCell Biol. 2020;22:453–464. doi: 10.1038/s41556-020-0485-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Wei MT, Chang YC, Shimobayashi SF, Shin Y, Strom AR, Brangwynne CP. Nucleated transcriptional condensates amplify gene expression. NatCell Biol. 2020;22:1187–1196. doi: 10.1038/s41556-020-00578-6. [DOI] [PubMed] [Google Scholar]
- [64].Schneider N, Wieland FG, Kong D, Fischer AAM, Horner M, Timmer J, Ye H, Weber W. Liquid-liquid phase separation of light-inducible transcription factors increases transcription activation in mammalian cells and mice. Sci Adv. 2021;7 doi: 10.1126/sciadv.abd3568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Liu X, Liu X, Wang H, Dou Z, Ruan K, Hill DL, Li L, Shi Y, Yao X. Phase separation drives decision making in cell division. J Biol Chem. 2020;295:13419–13431. doi: 10.1074/jbc.REV120.011746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Trivedi P, Palomba F, Niedzialkowska E, Digman MA, Gratton E, Stukenberg PT. The inner centromere is a biomolecular condensate scaffolded by the chromosomal passenger complex. NatCell Biol. 2019;21:1127–1137. doi: 10.1038/s41556-019-0376-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Trivedi P, Stukenberg PT. A condensed view of the chromosome passenger complex. Trends Cell Biol. 2020;30:676–687. doi: 10.1016/j.tcb.2020.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Altmeyer M, Neelsen KJ, Teloni F, Pozdnyakova I, Pellegrino S, Grofte M, Rask MBD, Streicher W, Jungmichel S, Nielsen ML, Lukas J. Liquid demixing of intrinsically disordered proteins is seeded by poly(ADP-ribose) Nat Commun. 2015;6 doi: 10.1038/ncomms9088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Patel A, Lee HO, Jawerth L, Maharana S, Jahnel M, Hein MY, Stoynov S, Mahamid J, Saha S, Franzmann TM, Pozniakovski A, et al. A liquid-to-Solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell. 2015;162:1066–1077. doi: 10.1016/j.cell.2015.07.047. [DOI] [PubMed] [Google Scholar]
- [70].Singatulina AS, Hamon L, Sukhanova MV, Desforges B, Joshi V, Bouhss A, Lavrik OI, Paste D. PARP-1 activation directs FUS to DNA damage sites to form PARG-reversible compartments enriched in damaged DNA. Cell Rep. 2019;27:1809–1821. doi: 10.1016/j.celrep.2019.04.031. [DOI] [PubMed] [Google Scholar]
- [71].Kilic S, Lezaja A, Gatti M, Bianco E, Michelena J, Imhof R, Altmeyer M. Phase separation of 53BP1 determines liquid-like behavior of DNA repair compartments. EMBO J. 2019;38(16):e101379. doi: 10.15252/embj.2018101379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Pessina F, Giavazzi F, Yin Y, Gioia U, Vitelli V, Galbiati A, Barozzi S, Garre M, Oldani A, Flaus A, Cerbino R, et al. Functional transcription promoters at DNA double-strand breaks mediate RNA-driven phase separation of damage-response factors. Nat Cell Biol. 2019;21:1286–1299. doi: 10.1038/s41556-019-0392-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Ghodke I, Remisova M, Furst A, Kilic S, Reina-San-Martin B, Poetsch AR, Altmeyer M, Soutoglou E. AHNAK controls 53BP1-mediated p53 response by restraining 53BP1 oligomerization and phase separation. MolCell. 2021;81(12):2596–2610.:e7. doi: 10.1016/j.molcel.2021.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Claeys Bouuaert C, Pu S, Wang J, Oger C, Daccache D, Xie W, Patel DJ, Keeney S. DNA-driven condensation assembles the meiotic DNA break machinery. Nature. 2021;592:144–149. doi: 10.1038/s41586-021-03374-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Oshidari R, Huang R, Medghalchi M, Tse EYW, Ashgriz N, Lee HO, Wyatt H, Mekhail K. DNA repair by Rad52 liquid droplets. Nat Commun. 2020;11:695. doi: 10.1038/s41467-020-14546-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Mine-Hattab J, Heltberg M, Villemeur M, Guedj C, Mora T, Walczak AM, Dahan M, Taddei A. Single molecule microscopy reveals key physical features of repair foci in living cells. Elife. 2021;10 doi: 10.7554/eLife.60577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Parker MW, Bell M, Mir M, Kao JA, Darzacq X, Botchan MR, Berger JM. A new class of disordered elements controls DNA replication through initiator self-assembly. Elife. 2019;8 doi: 10.7554/eLife.48562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Hossain M, Bhalla K, Stillman B. Multiple, short protein binding motifs in ORC1 and CDC6 control the initiation of DNA replication. Mol Cell. 2021;81:1951–1969.:e1956. doi: 10.1016/j.molcel.2021.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Frattini C, Promonet A, Alghoul E, Vidal-Eychenie S, Lamarque M, Blanchard MP, Urbach S, Basbous J, Constantinou A. TopBP1 assembles nuclear condensates to switch on ATR signaling. MolCell. 2021;81(6):1231–1245.:e8. doi: 10.1016/j.molcel.2020.12.049. [DOI] [PubMed] [Google Scholar]
- [80].Yang P, Mathieu C, Kolaitis RM, Zhang P, Messing J, Yurtsever U, Yang Z, Wu J, Li Y, Pan Q, Yu J, et al. G3BP1 is a tunable switch that triggers phase separation to assemble stress granules. Cell. 2020;181:325–345.:e328. doi: 10.1016/j.cell.2020.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Guillen-Boixet J, Kopach A, Holehouse AS, Wittmann S, Jahnel M, Schlussler R, Kim K, Trussina I, Wang J, Mateju D, Poser I, et al. RNA-induced conformational switching and clustering of G3BP drive stress granule assembly by condensation. Cell. 2020;181:346–361.:e317. doi: 10.1016/j.cell.2020.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Brangwynne CP, Eckmann CR, Courson DS, Rybarska A, Hoege C, Gharakhani J, Julicher F, Hyman AA. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science. 2009;324:1729–1732. doi: 10.1126/science.1172046. [DOI] [PubMed] [Google Scholar]
- [83].Carlson CR, Asfaha JB, Ghent CM, Howard CJ, Hartooni N, Safari M, Frankel AD, Morgan DO. Phosphoregulation of phase separation by the SARS-CoV-2 N protein suggests a biophysical basis for its dual functions. MolCell. 2020;80:1092–1103.:e1094. doi: 10.1016/j.molcel.2020.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Cubuk J, Alston JJ, Incicco JJ, Singh S, Stuchell-Brereton MD, Ward MD, Zimmerman MI, Vithani N, Griffith D, Wagoner JA, Bowman GR, et al. The SARS-CoV-2 nucleocapsid protein is dynamic, disordered, and phase separates with RNA. Nat Commun. 2021;12:1936. doi: 10.1038/s41467-021-21953-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Iserman C, Roden CA, Boerneke MA, Sealfon RSG, McLaughlin GA, Jungreis I, Fritch EJ, Hou YJ, Ekena J, Weidmann CA, Theesfeld CL, et al. Genomic RNA elements drive phase separation of the SARS-CoV-2 nucleocapsid. Mol Cell. 2020;80:1078–1091.:e1076. doi: 10.1016/j.molcel.2020.11.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Jack A, Ferro LS, Trnka MJ, Wehri E, Nadgir A, Costa K, Schaletzky J, Yildiz A. SARS CoV-2 nucleocapsid protein forms condensates with viral genomic RNA. bioRxiv. 2021 doi: 10.1101/2020.09.14.295824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Lu S, Ye Q, Singh D, Cao Y, Diedrich JK, Yates JR, 3rd, Villa E, Cleveland DW, Corbett KD. The SARS-CoV-2 nucleocapsid phosphoprotein forms mutually exclusive condensates with RNA and the membrane-associated M protein. Nat Commun. 2021;12:502. doi: 10.1038/s41467-020-20768-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Perdikari TM, Murthy AC, Ryan VH, Watters S, Naik MT, Fawzi NL. SARS-CoV-2 nucleocapsid protein phase-separates with RNA and with human hnRNPs. EMBO J. 2020;39 doi: 10.15252/embj.2020106478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Savastano A, de Opakua AI, Rankovic M, Zweckstetter M. Nucleocapsid protein of SARS-CoV-2 phase separates into RNA-rich polymerase-containing condensates. Nat Commun. 2020;11 doi: 10.1038/s41467-020-19843-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Seim I, Roden CA, Gladfelter AS. Role of spatial patterning of N-protein interactions in SARS-CoV-2 genome packaging. Biophys J. 2021;S0006-3495(21) doi: 10.1016/j.bpj.2021.06.018,00505-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Guseva S, Milles S, Jensen MR, Salvi N, Kleman JP, Maurin D, Ruigrok RWH, Blackledge M. Measles virus nucleo- and phosphoproteins form liquid-like phase-separated compartments that promote nucleocapsid assembly. Sci Adv. 2020;6 doi: 10.1126/sciadv.aaz7095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Seyffert M, Georgi F, Tobler K, Bourqui L, Anfossi M, Michaelsen K, Vogt B, Greber UF, Fraefel C. The HSV-1 transcription factor ICP4 confers liquid-like properties to viral replication compartments. Int J Mol Sci. 2021;22 doi: 10.3390/ijms22094447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Sabari BR, Dall’Agnese A, Young RA. Biomolecular condensates in the nucleus. Trends Biochem Sci. 2020;45(11):961–977. doi: 10.1016/j.tibs.2020.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Zhao YG, Zhang H. Phase separation in membrane biology: the interplay between membrane-bound organelles and membraneless condensates. DevCell. 2020;55:30–44. doi: 10.1016/j.devcel.2020.06.033. [DOI] [PubMed] [Google Scholar]
- [95].Wilfling F, Lee CW, Erdmann PS, Zheng Y, Sherpa D, Jentsch S, Pfander B, Schulman BA, Baumeister W. A selective autophagy pathway for phase-separated endocytic protein deposits. Mol Cell. 2020;80:764–778.:e767. doi: 10.1016/j.molcel.2020.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Bergeron-Sandoval L, Kumar S, Heris HK, Chang C, Cornell CE, Keller SL, François P, Hendricks AG, Ehrlicher AJ, Pappu RV, Michnick SW. Proteins with prion-like domains can form viscoelastic condensates that enable membrane remodeling and endocytosis. bioRxiv. 2021 doi: 10.1101/145664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Day KJ, Kago G, Wang L, Richter JB, Hayden CC, Lafer EM, Stachowiak JC. Liquid-like protein interactions catalyse assembly of endocytic vesicles. NatCell Biol. 2021;23:366–376. doi: 10.1038/s41556-021-00646-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Nandana V, Schrader JM. Roles of liquid-liquid phase separation in bacterial RNA metabolism. CurrOpin Microbiol. 2021;61:91–98. doi: 10.1016/j.mib.2021.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Dao TP, Castaneda CA. Ubiquitin-modulated phase separation of shuttle proteins: does condensate formation promote protein degradation? Bioessays. 2020;42:e2000036. doi: 10.1002/bies.202000036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Yasuda S, Tsuchiya H, Kaiho A, Guo Q, Ikeuchi K, Endo A, Arai N, Ohtake F, Murata S, Inada T, Baumeister W, et al. Stress- and ubiquitylation-dependent phase separation of the proteasome. Nature. 2020;578:296–300. doi: 10.1038/s41586-020-1982-9. [DOI] [PubMed] [Google Scholar]
- [101].Lin C, Suen KM, Jeffrey P, Wieteska L, Stainthorp A, Seiler C, Koss H, Molina-París C, Miska E, Ahmed Z, Ladbury JE. Receptor tyrosine kinases regulate signal transduction through a liquid-liquid phase separated state. bioRxiv. 2019 doi: 10.1101/783720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Li P, Banjade S, Cheng HC, Kim S, Chen B, Guo L, Llaguno M, Hollingsworth JV, King DS, Banani SF, Russo PS, et al. Phase transitions in the assembly of multivalent signalling proteins. Nature. 2012;483:336–340. doi: 10.1038/nature10879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Su X, Ditlev JA, Hui E, Xing W, Banjade S, Okrut J, King DS, Taunton J, Rosen MK, Vale RD. Phase separation of signaling molecules promotes T cell receptor signal transduction. Science. 2016;352:595–599. doi: 10.1126/science.aad9964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Huang WYC, Alvarez S, Kondo Y, Lee YK, Chung JK, Lam HYM, Biswas KH, Kuriyan J, Groves JT. A molecular assembly phase transition and kinetic proofreading modulate Ras activation by SOS. Science. 2019;363:1098–1103. doi: 10.1126/science.aau5721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Riback JA, Katanski CD, Kear-Scott JL, Pilipenko EV, Rojek AE, Sosnick TR, Drummond DA. Stress-triggered phase separation is an adaptive, evolutionarily tuned response. Cell. 2017;168:1028–1040.:e1019. doi: 10.1016/j.cell.2017.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Peran I, Sabri N, Mittag T. Managing hyperosmotic stress through phase separation. Trends BiochemSci. 2020;45:721–723. doi: 10.1016/j.tibs.2020.05.004. [DOI] [PubMed] [Google Scholar]
- [107].Franzmann TM, Alberti S. Protein phase separation as a stress survival strategy. Csh Perspect Med. 2019;9 doi: 10.1101/cshperspect.a034058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Ong JY, Torres JZ. Phase separation in cell division. MolCell. 2020;80:9–20. doi: 10.1016/j.molcel.2020.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Wen W. Phase separation in asymmetric cell division. Biochemistry. 2020;59:47–56. doi: 10.1021/acs.biochem.9b00813. [DOI] [PubMed] [Google Scholar]
- [110].Case LB, Ditlev JA, Rosen MK. Regulation of transmembrane signaling by phase separation. AnnuRev Biophys. 2019;48:465–494. doi: 10.1146/annurev-biophys-052118-115534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Agudo-Canalejo J, Schultz SW, Chino H, Migliano SM, Saito C, Koyama-Honda I, Stenmark H, Brech A, May AI, Mizushima N, Knorr RL. Wetting regulates autophagy of phase-separated compartments and the cytosol. Nature. 2021;591:142–146. doi: 10.1038/s41586-020-2992-3. [DOI] [PubMed] [Google Scholar]
- [112].Klosin A, Oltsch F, Harmon T, Honigmann A, Julicher F, Hyman AA, Zechner C. Phase separation provides a mechanism to reduce noise in cells. Science. 2020;367:464–468. doi: 10.1126/science.aav6691. [DOI] [PubMed] [Google Scholar]
- [113].Zhang Y, Narlikar GJ, Kutateladze TG. Enzymatic reactions inside biological condensates. J Mol Biol. 2021;433(12):166624. doi: 10.1016/j.jmb.2020.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Yoo H, Triandafillou C, Drummond DA. Cellular sensing by phase separation: using the process, not just the products. J Biol Chem. 2019;294:7151–7159. doi: 10.1074/jbc.TM118.001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Persson LB, Ambati VS, Brandman O. Cellular control of viscosity counters changes in temperature and energy availability. Cell. 2020;183:1572–1585. doi: 10.1016/j.cell.2020.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].O’Flynn BG, Mittag T. The role of liquid-liquid phase separation in regulating enzyme activity. Curr Opin Cell Biol. 2021;69:70–79. doi: 10.1016/j.ceb.2020.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Hondele M, Heinrich S, De Los Rios P, Weis K. Membraneless organelles: phasing out of equilibrium. Emerg Top Life Sci. 2020;4:331–342. doi: 10.1042/ETLS20190190. [DOI] [PubMed] [Google Scholar]
- [118].Jalihal AP, Pitchiaya S, Xiao LB, Bawa P, Jiang X, Bedi K, Parolia A, Cieslik M, Ljungman M, Chinnaiyan AM, Walter NG. Multivalent proteins rapidly and reversibly phase-separate upon osmotic cell volume change. Mol Cell. 2020;79:978–990.:e975. doi: 10.1016/j.molcel.2020.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Peeples W, Rosen MK. Mechanistic dissection of increased enzymatic rate in a phase-separated compartment. NatChem Biol. 2021;17:693–702. doi: 10.1038/s41589-021-00801-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Fritsch AW, Diaz-Delgadillo AF, Adame-Arana O, Hoege C, Mittasch M, Kreysing M, Leaver M, Hyman AA, Jülicher F, Weber CA. Local thermodynamics governs the formation and dissolution of protein condensates in living cells. bioRxiv. 2021 doi: 10.1101/2021.02.11.430794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Falahati H, Haji-Akbari A. Thermodynamically driven assemblies and liquid-liquid phase separations in biology. Soft Matter. 2019;15:1135–1154. doi: 10.1039/c8sm02285b. [DOI] [PubMed] [Google Scholar]
- [122].Pappu RV. Phase separation-a physical mechanism for organizing information and biochemical reactions. DevCell. 2020;55:1–3. doi: 10.1016/j.devcel.2020.09.023. [DOI] [PubMed] [Google Scholar]
- [123].Kaur T, Raju M, Alshareedah I, Davis RB, Potoyan DA, Banerjee PR. Sequence-encoded and composition-dependent protein-RNA interactions control multiphasic condensate morphologies. NatCommun. 2021;12:872. doi: 10.1038/s41467-021-21089-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Martin EW, Holehouse AS. Intrinsically disordered protein regions and phase separation: sequence determinants of assembly or lack therev of. EmergTop Life Sci. 2020;4:307–329. doi: 10.1042/ETLS20190164. [DOI] [PubMed] [Google Scholar]
- [125].Wang J, Choi JM, Holehouse AS, Lee HO, Zhang XJ, Jahnel M, Maharana S, Lemaitre R, Pozniakovsky A, Drechsel D, Poser I, et al. A molecular grammar governing the driving forces for phase separation of prion-like RNA binding proteins. Cell. 2018;174:688–699.:e616. doi: 10.1016/j.cell.2018.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Li HR, Chiang WC, Chou PC, Wang WJ, Huang JR. TAR DNA-binding protein 43 (TDP-43) liquid-liquid phase separation is mediated by just a few aromatic residues. J Biol Chem. 2018;293:6090–6098. doi: 10.1074/jbc.AC117.001037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Vernon RM, Chong PA, Tsang B, Kim TH, Bah A, Farber P, Lin H, Forman-Kay JD. Pi-Pi contacts are an overlooked protein feature relevant to phase separation. Elife. 2018;7 doi: 10.7554/eLife.31486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Gabryelczyk B, Cai H, Shi X, Sun Y, Swinkels PJM, Salentinig S, Pervushin K, Miserez A. Hydrogen bond guidance and aromatic stacking drive liquid-liquid phase separation of intrinsically disordered histidine-rich peptides. Nat Commun. 2019;10:5465. doi: 10.1038/s41467-019-13469-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Martin EW, Holehouse AS, Peran I, Farag M, Incicco JJ, Bremer A, Grace CR, Soranno A, Pappu RV, Mittag T. Valence and patterning of aromatic residues determine the phase behavior of prion-like domains. Science. 2020;367:694–699. doi: 10.1126/science.aaw8653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Lin Y, Currie SL, Rosen MK. Intrinsically disordered sequences enable modulation of protein phase separation through distributed tyrosine motifs. J Biol Chem. 2017;292:19110–19120. doi: 10.1074/jbc.M117.800466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Wang J, Choi JM, Holehouse AS, Lee HO, Zhang X, Jahnel M, Maharana S, Lemaitre R, Pozniakovsky A, Drechsel D, Poser I, et al. A molecular grammar governing the driving forces for phase separation of prion-like RNA binding proteins. Cell. 2018;174:688–699.:e616. doi: 10.1016/j.cell.2018.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Qamar S, Wang G, Randle SJ, Ruggeri FS, Varela JA, Lin JQ, Phillips EC, Miyashita A, Williams D, Strohl F, Meadows W, et al. FUS phase separation is modulated by a molecular chaperone and methylation of arginine cation-pi interactions. Cell. 2018;173:720–734.:e715. doi: 10.1016/j.cell.2018.03.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Nott TJ, Petsalaki E, Farber P, Jervis D, Fussner E, Plochowietz A, Craggs TD, Bazett-Jones DP, Pawson T, Forman-Kay JD, Baldwin AJ. Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Mol Cell. 2015;57:936–947. doi: 10.1016/j.molcel.2015.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Lin YH, Chan HS. Phase separation and single-chain compactness of charged disordered proteins are strongly correlated. Biophys J. 2017;112:2043–2046. doi: 10.1016/j.bpj.2017.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Fisher RS, Elbaum-Garfinkle S. Tunable multiphase dynamics of arginine and lysine liquid condensates. Nat Commun. 2020;11:4628. doi: 10.1038/s41467-020-18224-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Pak CW, Kosno M, Holehouse AS, Padrick SB, Mittal A, Ali R, Yunus AA, Liu DR, Pappu RV, Rosen MK. Sequence determinants of intracellular phase separation by complex coacervation of a disordered protein. Mol Cell. 2016;63:72–85. doi: 10.1016/j.molcel.2016.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Brady JP, Farber PJ, Sekhar A, Lin YH, Huang R, Bah A, Nott TJ, Chan HS, Baldwin AJ, Forman-Kay JD, Kay LE. Structural and hydrodynamic properties of an intrinsically disordered region of a germ cell-specific protein on phase separation. Proc Natl Acad Sci U S A. 2017;114:E8194–E8203. doi: 10.1073/pnas.1706197114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Reichheld SE, Muiznieks LD, Keeley FW, Sharpe S. Direct observation of structure and dynamics during phase separation of an elastomeric protein. Proc Natl Acad Sci U S A. 2017;114:E4408–E4415. doi: 10.1073/pnas.1701877114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Lin Y, Fichou Y, Longhini AP, Llanes LC, Yin P, Bazan GC, Kosik KS, Han S. Liquid-Liquid phase separation of tau driven by hydrophobic interaction facilitates fibrillization of tau. J Mol Biol. 2021;433:166731. doi: 10.1016/j.jmb.2020.166731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Dignon GL, Best RB, Mittal J. Biomolecular phase separation: from molecular driving forces to macroscopic properties. Annu Rev Phys Chem. 2020;71:53–75. doi: 10.1146/annurev-physchem-071819-113553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Krainer G, Welsh TJ, Joseph JA, Espinosa JR, Wittmann S, de Csillery E, Sridhar A, Toprakcioglu Z, Gudiskyte G, Czekalska MA, Arter WE, et al. Reentrant liquid condensate phase of proteins is stabilized by hydrophobic and non-ionic interactions. Nat Commun. 2021;12:1085. doi: 10.1038/s41467-021-21181-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Rauscher S, Pomes R. The liquid structure of elastin. Elife. 2017;6 doi: 10.7554/eLife.26526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Murthy AC, Dignon GL, Kan Y, Zerze GH, Parekh SH, Mittal J, Fawzi NL. Molecular interactions underlying liquid-liquid phase separation of the FUS low-complexity domain. NatStruct Mol Biol. 2019;26:637–648. doi: 10.1038/s41594-019-0250-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Brangwynne CP, Tompa P, Pappu R. Polymer physics of intracellular phase transitions. NatPhys. 2015;11:899–904. [Google Scholar]
- [145].Choi JM, Holehouse AS, Pappu RV. Physical principles underlying the complex biology of intracellular phase transitions. AnnuRev Biophys. 2020;49:107–133. doi: 10.1146/annurev-biophys-121219-081629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Peran I, Mittag T. Molecular structure in biomolecular condensates. CurrOpin Struct Biol. 2020;60:17–26. doi: 10.1016/j.sbi.2019.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Hughes MP, Sawaya MR, Boyer DR, Goldschmidt L, Rodriguez JA, Cascio D, Chong L, Gonen T, Eisenberg DS. Atomic structures of low-complexity protein segments reveal kinked beta sheets that assemble networks. Science. 2018;359:698–701. doi: 10.1126/science.aan6398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Guenther EL, Ge P, Trinh H, Sawaya MR, Cascio D, Boyer DR, Gonen T, Zhou ZH, Eisenberg DS. Atomic-level evidence for packing and positional amyloid polymorphism by segment from TDP-43 RRM2. NatStruct Mol Biol. 2018;25:311–319. doi: 10.1038/s41594-018-0045-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Gui X, Luo F, Li Y, Zhou H, Qin Z, Liu Z, Gu J, Xie M, Zhao K, Dai B, Shin WS, et al. Structural basis for reversible amyloids of hnRNPA1 elucidates their role in stress granule assembly. Nat Commun. 2019;10:2006. doi: 10.1038/s41467-019-09902-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Tinti M, Kiemer L, Costa S, Miller ML, Sacco F, Olsen JV, Carducci M, Paoluzi S, Langone F, Workman CT, Blom N, et al. The SH2 domain interaction landscape. Cell Rep. 2013;3:1293–1305. doi: 10.1016/j.celrep.2013.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Zhong S, Muller S, Ronchetti S, Freemont PS, Dejean A, Pandolfi PP. Role of SUMO-1-modified PML in nuclear body formation. Blood. 2000;95:2748–2752. [PubMed] [Google Scholar]
- [152].Martin EW, Thomasen FE, Milkovic NM, Cuneo MJ, Grace CR, Nourse A, Lindorff-Larsen K, Mittag T. Interplay of folded domains and the disordered low-complexity domain in mediating hnRNPA1 phase separation. Nucleic Acids Res. 2021;49:2931–2945. doi: 10.1093/nar/gkab063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Chong PA, Vernon RM, Forman-Kay JD. RGG/RG motif regions in RNA binding and phase separation. J Mol Biol. 2018;430:4650–4665. doi: 10.1016/j.jmb.2018.06.014. [DOI] [PubMed] [Google Scholar]
- [154].Borcherds W, Bremer A, Borgia MB, Mittag T. How do intrinsically disordered protein regions encode a driving force for liquid-liquid phase separation? Curr Opin Struct Biol. 2021;67:41–50. doi: 10.1016/j.sbi.2020.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Sanders DW, Kedersha N, Lee DSW, Strom AR, Drake V, Riback JA, Bracha D, Eeftens JM, Iwanicki A, Wang A, Wei MT, et al. Competing protein-RNA interaction networks control multiphase intracellular organization. Cell. 2020;181:306–324.:e328. doi: 10.1016/j.cell.2020.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Ditlev JA, Case LB, Rosen MK. Who’s in and who’s out-compositional control of biomolecular condensates. J Mol Biol. 2018;430:4666–4684. doi: 10.1016/j.jmb.2018.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Schwartz JC, Cech TR, Parker RR. Biochemical properties and biological functions of FET proteins. Annu Rev Biochem. 2015;84:355–379. doi: 10.1146/annurev-biochem-060614-034325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Frank L, Rippe K. Repetitive RNAs as regulators of chromatin-associated subcompartment formation by phase separation. J Mol Biol. 2020;432:4270–4286. doi: 10.1016/j.jmb.2020.04.015. [DOI] [PubMed] [Google Scholar]
- [159].Langdon EM, Qiu YP, Niaki AG, McLaughlin GA, Weidmann CA, Gerbich TM, Smith JA, Crutchley JM, Termini CM, Weeks KM, Myong S, et al. mRNA structure determines specificity of a polyQ-driven phase separation. Science. 2018;360:922–927. doi: 10.1126/science.aar7432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Roden C, Gladfelter AS. RNA contributions to the form and function of biomolecular condensates. Nat Rev Mol Cell Biol. 2021;22:183–195. doi: 10.1038/s41580-020-0264-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].Maharana S, Wang J, Papadopoulos DK, Richter D, Pozniakovsky A, Poser I, Bickle M, Rizk S, Guillen-Boixet J, Franzmann TM, Jahnel M, et al. RNA buffers the phase separation behavior of prion-like RNA binding proteins. Science. 2018;360:918–921. doi: 10.1126/science.aar7366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162].Jawerth L, Fischer-Friedrich E, Saha S, Wang J, Franzmann T, Zhang X, Sachweh J, Ruer M, Ijavi M, Saha S, Mahamid J, et al. Protein condensates as aging Maxwell fluids. Science. 2020;370:1317–1323. doi: 10.1126/science.aaw4951. [DOI] [PubMed] [Google Scholar]
- [163].Nair SJ, Yang L, Meluzzi D, Oh S, Yang F, Friedman MJ, Wang S, Suter T, Alshareedah I, Gamliel A, Ma Q, et al. Phase separation of ligand-activated enhancers licenses cooperative chromosomal enhancer assembly. Nat Struct Mol Biol. 2019;26:193–203. doi: 10.1038/s41594-019-0190-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Audas TE, Audas DE, Jacob MD, Ho JJ, Khacho M, Wang M, Perera JK, Gardiner C, Bennett CA, Head T, Kryvenko ON, et al. Adaptation to stressors by systemic protein amyloidogenesis. Dev Cell. 2016;39:155–168. doi: 10.1016/j.devcel.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [165].Alberti S, Dormann D. Liquid-Liquid phase separation in disease. AnnuRev Genet. 2019;53:171–194. doi: 10.1146/annurev-genet-112618-043527. [DOI] [PubMed] [Google Scholar]
- [166].Zbinden A, Perez-Berlanga M, De Rossi P, Polymenidou M. Phase separation and neurodegenerative diseases: a disturbance in the force. DevCell. 2020;55:45–68. doi: 10.1016/j.devcel.2020.09.014. [DOI] [PubMed] [Google Scholar]
- [167].Murray DT, Kato M, Lin Y, Thurber KR, Hung I, McKnight SL, Tycko R. Structure of FUS protein fibrils and its relevance to self-assembly and phase separation of low-complexity domains. Cell. 2017;171:615–627.:e616. doi: 10.1016/j.cell.2017.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [168].Kato M, McKnight SL. Cross-beta polymerization of low complexity sequence domains. Cold Spring Harb Perspect Biol. 2017;9 doi: 10.1101/cshperspect.a023598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [169].Pellegrino S, Altmeyer M. Interplay between ubiquitin, SUMO, and poly(ADP-Ribose) in the cellular response to genotoxic stress. Front Genet. 2016;7:63. doi: 10.3389/fgene.2016.00063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [170].Ray Chaudhuri A, Nussenzweig A. The multifaceted roles of PARP1 in DNA repair and chromatin remodelling. Nat Rev Mol Cell Biol. 2017;18:610–621. doi: 10.1038/nrm.2017.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].Teloni F, Altmeyer M. Readers of poly(ADP-ribose): designed to be fit for purpose. Nucleic Acids Res. 2016;44:993–1006. doi: 10.1093/nar/gkv1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [172].Lee SG, Kim N, Kim SM, Park IB, Kim H, Kim S, Kim BG, Hwang JM, Baek IJ, Gartner A, Park JH, et al. Ewing sarcoma protein promotes dissociation of poly(ADP-ribose) polymerase 1 from chromatin. EMBO Rep. 2020;21:e48676. doi: 10.15252/embr.201948676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [173].Mastrocola AS, Kim SH, Trinh AT, Rodenkirch LA, Tibbetts RS. The RNA-binding protein fused in sarcoma (FUS) functions downstream of poly(ADP-ribose) polymerase (PARP) in response to DNA damage. J Biol Chem. 2013;288:24731–24741. doi: 10.1074/jbc.M113.497974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [174].Sottini A, Borgia A, Borgia MB, Bugge K, Nettels D, Chowdhury A, Heidarsson PO, Zosel F, Best RB, Kragelund BB, Schuler B. Polyelectrolyte interactions enable rapid association and dissociation in high-affinity disordered protein complexes. NatCommun. 2020;11:5736. doi: 10.1038/s41467-020-18859-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [175].Borgia A, Borgia MB, Bugge K, Kissling VM, Heidarsson PO, Fernandes CB, Sottini A, Soranno A, Buholzer KJ, Nettels D, Kragelund BB, et al. Extreme disorder in an ultrahigh-affinity protein complex. Nature. 2018;555:61–66. doi: 10.1038/nature25762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [176].Hicks GG, Singh N, Nashabi A, Mai S, Bozek G, Klewes L, Arapovic D, White EK, Koury MJ, Oltz EM, Van Kaer L, et al. Fus deficiency in mice results in defective B-lymphocyte development and activation, high levels of chromosomal instability and perinatal death. NatGenet. 2000;24:175–179. doi: 10.1038/72842. [DOI] [PubMed] [Google Scholar]
- [177].Kuroda M, Sok J, Webb L, Baechtold H, Urano F, Yin Y, Chung P, de Rooij DG, Akhmedov A, Ashley T, Ron D. Male sterility and enhanced radiation sensitivity in TLS-/-mice. EMBO J. 2000;19:453–462. doi: 10.1093/emboj/19.3.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [178].Wang WY, Pan L, Su SC, Quinn EJ, Sasaki M, Jimenez JC, Mackenzie IR, Huang EJ, Tsai LH. Interaction of FUS and HDAC1 regulates DNA damage response and repair in neurons. NatNeurosci. 2013;16:1383–1391. doi: 10.1038/nn.3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [179].Hill SJ, Mordes DA, Cameron LA, Neuberg DS, Landini S, Eggan K, Livingston DM. Two familial ALS proteins function in prevention/repair of transcription-associated DNA damage. Proc Natl Acad Sci U S A. 2016;113:E7701–E7709. doi: 10.1073/pnas.1611673113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [180].Wang HB, Guo WT, Mitra J, Hegde PM, Vandoorne T, Eckelmann BJ, Mitra S, Tomkinson AE, Van den Bosch L, Hegde ML. Mutant FUS causes DNA ligation defects to inhibit oxidative damage repair in Amyotrophic Lateral Sclerosis. Nat Commun. 2018;9 doi: 10.1038/s41467-018-06111-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [181].Rother MB, Pellegrino S, Smith R, Gatti M, Meisenberg C, Wiegant WW, Luijsterburg MS, Imhof R, Downs JA, Vertegaal ACO, Huet S, et al. CHD7 and 53BP1 regulate distinct pathways for the re-ligation of DNA double-strand breaks. Nat Commun. 2020;11:5775. doi: 10.1038/s41467-020-19502-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [182].Luijsterburg MS, de Krijger I, Wiegant WW, Shah RG, Smeenk G, de Groot AJL, Pines A, Vertegaal ACO, Jacobs JJL, Shah GM, van Attikum H. PARP1 links CHD2-mediated chromatin expansion and H3.3 deposition to DNA repair by non-homologous end-joining. Mol Cell. 2016;61:547–562. doi: 10.1016/j.molcel.2016.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [183].Levone BR, Lenzken SC, Antonaci M, Maiser A, Rapp A, Conte F, Reber S, Mechtersheimer J, Ronchi AE, Muhlemann O, Leonhardt H, et al. FUS-dependent liquid-liquid phase separation is important for DNA repair initiation. J Cell Biol. 2021;220 doi: 10.1083/jcb.202008030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [184].Klaric JA, Wüst S, Panier S. New faces of old friends: emerging new roles of RNA-Binding proteins in the DNA double-strand break response. Front Mol Biosci. 2021;8 doi: 10.3389/fmolb.2021.668821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [185].Altmeyer M, Toledo L, Gudjonsson T, Grofte M, Rask MB, Lukas C, Akimov V, Blagoev B, Bartek J, Lukas J. The chromatin scaffold protein SAFB1 renders chromatin permissive for DNA damage signaling. Mol Cell. 2013;52:206–220. doi: 10.1016/j.molcel.2013.08.025. [DOI] [PubMed] [Google Scholar]
- [186].Huo XR, Ji LZ, Zhang YW, Lv P, Cao X, Wang QF, Yan ZX, Dong SS, Du D, Zhang F, Wei G, et al. The nuclear matrix protein SAFB cooperates with major satellite RNAs to stabilize heterochromatin architecture partially through phase separation. Mol Cell. 2020;77:368–383.:e367. doi: 10.1016/j.molcel.2019.10.001. [DOI] [PubMed] [Google Scholar]
- [187].Beli P, Lukashchuk N, Wagner SA, Weinert BT, Olsen JV, Baskcomb L, Mann M, Jackson SP, Choudhary C. Proteomic investigations reveal a role for RNA processing factor THRAP3 in the DNA damage response. Mol Cell. 2012;46:212–225. doi: 10.1016/j.molcel.2012.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [188].Britton S, Dernoncourt E, Delteil C, Froment C, Schiltz O, Salles B, Frit P, Calsou P. DNA damage triggers SAF-A and RNA biogenesis factors exclusion from chromatin coupled to R-loops removal. Nucleic Acids Res. 2014;42:9047–9062. doi: 10.1093/nar/gku601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [189].Adamson B, Smogorzewska A, Sigoillot FD, King RW, Elledge SJ. A genomewide homologous recombination screen identifies the RNA-binding protein RBMX as a component of the DNA-damage response. NatCell Biol. 2012;14:318–328. doi: 10.1038/ncb2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [190].Polo SE, Blackford AN, Chapman JR, Baskcomb L, Gravel S, Rusch A, Thomas A, Blundred R, Smith P, Kzhyshkowska J, Dobner T, et al. Regulation of DNA-end resection by hnRNPU-like proteins promotes DNA double-strand break signaling and repair. Mol Cell. 2012;45:505–516. doi: 10.1016/j.molcel.2011.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [191].Izhar L, Adamson B, Ciccia A, Lewis J, Pontano-Vaites L, Leng Y, Liang AC, Westbrook TF, Harper JW, Elledge SJ. A systematic analysis of factors localized to damaged chromatin reveals PARP-Dependent recruitment of transcription factors. Cell Rep. 2015;11:1486–1500. doi: 10.1016/j.celrep.2015.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [192].Monahan Z, Ryan VH, Janke AM, Burke KA, Rhoads SN, Zerze GH, O’Meally R, Dignon GL, Conicella AE, Zheng WW, Best RB, et al. Phosphorylation of the FUS low-complexity domain disrupts phase separation, aggregation, and toxicity. EMBO J. 2017;36:2951–2967. doi: 10.15252/embj.201696394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [193].Chou DM, Adamson B, Dephoure NE, Tan X, Nottke AC, Hurov KE, Gygi SP, Colaiacovo MP, Elledge SJ. A chromatin localization screen reveals poly (ADP ribose)-regulated recruitment of the repressive polycomb and NuRD complexes to sites of DNA damage. ProcNatl Acad Sci U S A. 2010;107:18475–18480. doi: 10.1073/pnas.1012946107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [194].Deng QD, Holler CJ, Taylor G, Hudson KF, Watkins W, Gearing M, Ito D, Murray ME, Dickson DW, Seyfried NT, Kukar T. FUS is phosphorylated by DNA-PK and accumulates in the cytoplasm after DNA damage. J Neurosci. 2014;34:7802–7813. doi: 10.1523/JNEUROSCI.0172-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [195].Lee JH, Ryu SW, Ender NA, Paull TT. Poly-ADP-ribosylation drives loss of protein homeostasis in ATM and Mre11 deficiency. Mol Cell. 2021;81:1515–1533.:e1515. doi: 10.1016/j.molcel.2021.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [196].Duan Y, Du A, Gu J, Duan G, Wang C, Gui X, Ma Z, Qian B, Deng X, Zhang K, Sun L, et al. PARylation regulates stress granule dynamics, phase separation, and neurotoxicity of disease-related RNA-binding proteins. Cell Res. 2019;29:233–247. doi: 10.1038/s41422-019-0141-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [197].McGurk L, Mojsilovic-Petrovic J, Van Deerlin VM, Shorter J, Kalb RG, Lee VM, Trojanowski JQ, Lee EB, Bonini NM. Nuclear poly(ADP-ribose) activity is a therapeutic target in amyotrophic latera sclerosis. Acta Neuropathol Com. 2018;6 doi: 10.1186/s40478-018-0586-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [198].Cardinale A, Paldino E, Giampa C, Bernardi G, Fusco FR. PARP-1 inhibition is neuroprotective in the R6/2 mouse model of Huntington’s disease. PLoS One. 2015;10 doi: 10.1371/journal.pone.0134482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [199].McGurk L, Rifai OM, Bonini NM. Poly(ADP-Ribosylation) in age-related neurological disease. Trends Genet. 2019;35:601–613. doi: 10.1016/j.tig.2019.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [200].Liu C, Fang Y. New insights of poly(ADP-ribosylation) in neurodegenerative diseases: a focus on protein phase separation and pathologic aggregation. BiochemPharmacol. 2019;167:58–63. doi: 10.1016/j.bcp.2019.04.028. [DOI] [PubMed] [Google Scholar]
- [201].Kam TI, Mao X, Park H, Chou SC, Karuppagounder SS, Umanah GE, Yun SP, Brahmachari S, Panicker N, Chen R, Andrabi SA, et al. Poly(ADP-ribose) drives pathologic alpha-synuclein neurodegeneration in Parkinson’s disease. Science. 2018;362 doi: 10.1126/science.aat8407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [202].Leung AKL. Poly(ADP-ribose): a dynamic trigger for biomolecular condensate formation. Trends Cell Biol. 2020;30:370–383. doi: 10.1016/j.tcb.2020.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [203].Leung AK, Vyas S, Rood JE, Bhutkar A, Sharp PA, Chang P. Poly(ADP-ribose) regulates stress responses and microRNA activity in the cytoplasm. Mol Cell. 2011;42:489–499. doi: 10.1016/j.molcel.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [204].Leung AK. Poly(ADP-ribose): an organizer of cellular architecture. J Cell Biol. 2014;205:613–619. doi: 10.1083/jcb.201402114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [205].McGurk L, Gomes E, Guo L, Mojsilovic-Petrovic J, Tran V, Kalb RG, Shorter J, Bonini NM. Poly(ADP-Ribose) prevents pathological phase separation of TDP-43 by promoting liquid demixing and stress granule localization. Mol Cell. 2018;71:703–717.:e709. doi: 10.1016/j.molcel.2018.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [206].Hayashi Y, Ford LK, Fioriti L, McGurk L, Zhang M. Liquid-Liquid phase separation in physiology and pathophysiology of the nervous system. J Neurosci. 2021 doi: 10.1523/JNEUROSCI.1656-20.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [207].Wilson MD, Durocher D. Reading chromatin signatures after DNA double-strand breaks. Philos Trans R Soc Lond, B, Biol Sci. 2017;372 doi: 10.1098/rstb.2016.0280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [208].Schwertman P, Bekker-Jensen S, Mailand N. Regulation of DNA double-strand break repair by ubiquitin and ubiquitin-like modifiers. NatRev Mol Cell Biol. 2016;17:379–394. doi: 10.1038/nrm.2016.58. [DOI] [PubMed] [Google Scholar]
- [209].Clouaire T, Legube G. A snapshot on the Cis chromatin response to DNA doublestrand breaks. Trends Genet. 2019;35:330–345. doi: 10.1016/j.tig.2019.02.003. [DOI] [PubMed] [Google Scholar]
- [210].Hauer MH, Gasser SM. Chromatin and nucleosome dynamics in DNA damage and repair. Gene Dev. 2017;31:2204–2221. doi: 10.1101/gad.307702.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [211].Arnould C, Rocher V, Finoux AL, Clouaire T, Li K, Zhou FL, Caron P, Mangeot PE, Ricci EP, Mourad R, Haber JE, et al. Loop extrusion as a mechanism for formation of DNA damage repair foci. Nature. 2021;590 doi: 10.1038/s41586-021-03193-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [212].Ryu JK, Bouchoux C, Liu HW, Kim E, Minamino M, de Groot R, Katan AJ, Bonato A, Marenduzzo D, Michieletto D, Uhlmann F, et al. Bridging-induced phase separation induced by cohesin SMC protein complexes. Sci Adv. 2021;7 doi: 10.1126/sciadv.abe5905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [213].Pellegrino S, Michelena J, Teloni F, Imhof R, Altmeyer M. Replication-coupled dilution of H4K20me2 guides 53BP1 to pre-replicative chromatin. Cell Rep. 2017;19:1819–1831. doi: 10.1016/j.celrep.2017.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [214].Michelena J, Pellegrino S, Spegg V, Altmeyer M. Replicated chromatin curtails 53BP1 recruitment in BRCA1-proficient and BRCA1-deficient cells. Life Sci Alliance. 2021;4 doi: 10.26508/lsa.202101023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [215].Nakamura K, Saredi G, Becker JR, Foster BM, Nguyen NV, Beyer TE, Cesa LC, Faull PA, Lukauskas S, Frimurer T, Chapman JR, et al. H4K20me0 recognition by BRCA1-BARD1 directs homologous recombination to sister chromatids. NatCell Biol. 2019;21:311–318. doi: 10.1038/s41556-019-0282-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [216].Saredi G, Huang H, Hammond CM, Alabert C, Bekker-Jensen S, Forne I, Reveron-Gomez N, Foster BM, Mlejnkova L, Bartke T, Cejka P, et al. H4K20me0 marks post-replicative chromatin and recruits the TONSL-MMS22L DNA repair complex. Nature. 2016;534:714–718. doi: 10.1038/nature18312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [217].Wilson MD, Benlekbir S, Fradet-Turcotte A, Sherker A, Julien JP, McEwan A, Noordermeer SM, Sicheri F, Rubinstein JL, Durocher D. The structural basis of modified nucleosome recognition by 53BP1. Nature. 2016;536:100–103. doi: 10.1038/nature18951. [DOI] [PubMed] [Google Scholar]
- [218].Lou J, Priest DG, Solano A, Kerjouan A, Hinde E. Spatiotemporal dynamics of 53BP1 dimer recruitment to a DNA double strand break. NatCommun. 2020;11:5776. doi: 10.1038/s41467-020-19504-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [219].Zgheib O, Pataky K, Brugger J, Halazonetis TD. An oligomerized 53BP1 tudor domain suffices for recognition of DNA double-strand breaks. MolCell Biol. 2009;29:1050–1058. doi: 10.1128/MCB.01011-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [220].Ochs F, Karemore G, Miron E, Brown J, Sedlackova H, Rask MB, Lampe M, Buckle V, Schermelleh L, Lukas J, Lukas C. Stabilization of chromatin topology safeguards genome integrity. Nature. 2019;574:571–574. doi: 10.1038/s41586-019-1659-4. [DOI] [PubMed] [Google Scholar]
- [221].Ghodke I, Soutoglou E. 53BP1-RIF1: sculpting the DNA repair focus in 3D. NatStruct Mol Biol. 2019;26:1087–1088. doi: 10.1038/s41594-019-0348-1. [DOI] [PubMed] [Google Scholar]
- [222].Caron P, Polo SE. Reshaping chromatin architecture around DNA breaks. Trends BiochemSci. 2020;45:177–179. doi: 10.1016/j.tibs.2019.12.001. [DOI] [PubMed] [Google Scholar]
- [223].Shin Y, Berry J, Pannucci N, Haataja MP, Toettcher JE, Brangwynne CP. Spatiotemporal control of intracellular phase transitions using light-activated optoDroplets. Cell. 2017;168:159–171.:e114. doi: 10.1016/j.cell.2016.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [224].D’Alessandro G, d’Adda di Fagagna F. Transcription and DNA damage: holding hands or crossing swords? JMol Biol. 2017;429:3215–3229. doi: 10.1016/j.jmb.2016.11.002. [DOI] [PubMed] [Google Scholar]
- [225].Lesage E, Clouaire T, Legube G. Repair of DNA double-strand breaks in RNAPI- and RNAPII-transcribed loci. DNA Repair (Amst) 2021;104:103–139. doi: 10.1016/j.dnarep.2021.103139. [DOI] [PubMed] [Google Scholar]
- [226].Zong D, Oberdoerffer P, Batista PJ, Nussenzweig A. RNA: a double-edged sword in genome maintenance. NatRev Genet. 2020;21:651–670. doi: 10.1038/s41576-020-0263-7. [DOI] [PubMed] [Google Scholar]
- [227].Kamagata K, Kanbayashi S, Honda M, Itoh Y, Takahashi H, Kameda T, Nagatsugi F, Takahashi S. Liquid-like droplet formation by tumor suppressor p53 induced by multivalent electrostatic interactions between two disordered domains. Sci Rep. 2020;10:580. doi: 10.1038/s41598-020-57521-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [228].Parnandi N, Rendo V, Cui G, Botuyan MV, Remisova M, Nguyen H, Drane P, Beroukhim R, Altmeyer M, Mer G, Chowdhury D. TIRR inhibits the 53BP1-p53 complex to alter cell-fate programs. MolCell. 2021;81(12):2583–2595.:e6. doi: 10.1016/j.molcel.2021.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [229].Cuella-Martin R, Oliveira C, Lockstone HE, Snellenberg S, Grolmusova N, Chapman JR. 53BP1 integrates DNA repair and p53-Dependent cell fate decisions via distinct mechanisms. Mol Cell. 2016;64:51–64. doi: 10.1016/j.molcel.2016.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [230].Lezaja A, Altmeyer M. Dealing with DNA lesions: when one cell cycle is not enough. Curr Opin Cell Biol. 2020;70:27–36. doi: 10.1016/j.ceb.2020.11.001. [DOI] [PubMed] [Google Scholar]
- [231].Lezaja A, Altmeyer M. Inherited DNA lesions determine G1 duration in the next cell cycle. Cell Cycle. 2018;17:24–32. doi: 10.1080/15384101.2017.1383578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [232].Roy S, Musselman CA, Kachirskaia I, Hayashi R, Glass KC, Nix JC, Gozani O, Appella E, Kutateladze TG. Structural insight into p53 recognition by the 53BP1 tandem tudor domain. J Mol Biol. 2010;398:489–496. doi: 10.1016/j.jmb.2010.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [233].Oshidari R, Mekhail K, Seeber A. Mobility and repair of damaged DNA: random or directed? Trends Cell Biol. 2020;30:144–156. doi: 10.1016/j.tcb.2019.11.003. [DOI] [PubMed] [Google Scholar]
- [234].Schrank BR, Aparicio T, Li Y, Chang W, Chait BT, Gundersen GG, Gottesman ME, Gautier J. Nuclear ARP2/3 drives DNA break clustering for homology-directed repair. Nature. 2018;559:61–66. doi: 10.1038/s41586-018-0237-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [235].Caridi CP, D’Agostino C, Ryu T, Zapotoczny G, Delabaere L, Li X, Khodaverdian VY, Amaral N, Lin E, Rau AR, Chiolo I. Nuclear F-actin and myosins drive relocalization of heterochromatic breaks. Nature. 2018;559:54–60. doi: 10.1038/s41586-018-0242-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [236].Case LB, Zhang X, Ditlev JA, Rosen MK. Stoichiometry controls activity of phase-separated clusters of actin signaling proteins. Science. 2019;363:1093–1097. doi: 10.1126/science.aau6313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [237].Liao YC, Fernandopulle MS, Wang GZ, Choi H, Hao L, Drerup CM, Patel R, Qamar S, Nixon-Abell J, Shen Y, Meadows W, et al. RNA granules hitchhike on lysosomes for long-distance transport, using annexin A11 as a molecular tether. Cell. 2019;179:147–164.:e120. doi: 10.1016/j.cell.2019.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [238].Macheret M, Halazonetis TD. DNA replication stress as a hallmark of Cancer. Annu Rev Pathol-Mech. 2015;10:425–448. doi: 10.1146/annurev-pathol-012414-040424. [DOI] [PubMed] [Google Scholar]
- [239].Panagopoulos A, Altmeyer M. The hammer and the dance of cell cycle control. Trends Biochem Sci. 2021;46(4):301–314. doi: 10.1016/j.tibs.2020.11.002. [DOI] [PubMed] [Google Scholar]
- [240].Saldivar JC, Cortez D, Cimprich KA. The essential kinase ATR: ensuring faithful duplication of a challenging genome. NatRev Mol Cell Biol. 2017;18:622–636. doi: 10.1038/nrm.2017.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [241].Hoang SM, O’Sullivan RJ. Alternative lengthening of telomeres: building bridges to connect chromosome ends. Trends Cancer. 2020;6:247–260. doi: 10.1016/j.trecan.2019.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [242].Zhang JM, Zou L. Alternative lengthening of telomeres: from molecular mechanisms to therapeutic outlooks. Cell Biosci. 2020;10 doi: 10.1186/s13578-020-00391-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [243].Hoang SM, Kaminski N, Bhargava R, Barroso-Gonzalez J, Lynskey ML, Garcia-Exposito L, Roncaioli JL, Wondisford AR, Wallace CT, Watkins SC, James DI, et al. Regulation of ALT-associated homology-directed repair by polyADP-ribosylation. Nat Struct Mol Biol. 2020;27:1152–1164. doi: 10.1038/s41594-020-0512-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [244].Zhang JM, Genois MM, Ouyang J, Lan L, Zou L. Alternative lengthening of telomeres is a self-perpetuating process in ALT-associated PML bodies. MolCell. 2021;81:1027–1042. doi: 10.1016/j.molcel.2020.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [245].Laflamme G, Mekhail K. Biomolecular condensates as arbiters of biochemical reactions inside the nucleus. Commun Biol. 2020;3 doi: 10.1038/s42003-020-01517-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [246].Choi JM, Dar F, Pappu RV. LASSI: a lattice model for simulating phase transitions of multivalent proteins. PLoS ComputBiol. 2019;15:e1007028. doi: 10.1371/journal.pcbi.1007028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [247].Ginell GM, Holehouse AS. Analyzing the sequences of intrinsically disordered regions with CIDER and localCIDER. Methods MolBiol. 2020;2141:103–126. doi: 10.1007/978-1-0716-0524-0_5. [DOI] [PubMed] [Google Scholar]
- [248].Hardenberg M, Horvath A, Ambrus V, Fuxreiter M, Vendruscolo M. Widespread occurrence of the droplet state of proteins in the human proteome. Proc Natl Acad Sci U S A. 2020;117:33254–33262. doi: 10.1073/pnas.2007670117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [249].Alberti S, Gladfelter A, Mittag T. Considerations and challenges in studying liquid-Liquid phase separation and biomolecular condensates. Cell. 2019;176:419–434. doi: 10.1016/j.cell.2018.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [250].Alshareedah I, Kaur T, Banerjee PR. Methods for characterizing the material properties of biomolecular condensates. Methods Enzymol. 2021;646:143–183. doi: 10.1016/bs.mie.2020.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [251].Babinchak WM, Surewicz WK. Studying protein aggregation in the context of liquid-liquid phase separation using fluorescence and atomic force microscopy, fluorescence and turbidity assays, and FRAP. Bio Protoc. 2020;10 doi: 10.21769/BioProtoc.3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [252].Bracha D, Walls MT, Wei MT, Zhu L, Kurian M, Avalos JL, Toettcher JE, Brangwynne CP. Mapping local and global liquid phase behavior in living cells using photo-oligomerizable seeds. Cell. 2018;175:1467–1480.:e1413. doi: 10.1016/j.cell.2018.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [253].Bremer A, Mittag T, Heymann M. Microfluidic characterization of macromolecular liquid-liquid phase separation. Lab Chip. 2020;22:4225–4234. doi: 10.1039/d0lc00613k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [254].Burke KA, Janke AM, Rhine CL, Fawzi NL. Residue-by-Residue view of in vitro FUS granules that bind the C-Terminal domain of RNA polymerase II. MolCell. 2015;60:231–241. doi: 10.1016/j.molcel.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [255].Dannenhoffer-Lafage T, Best RB. A data-driven hydrophobicity scale for predicting liquid-Liquid phase separation of proteins. J Phys Chem B. 2021;125:4046–4056. doi: 10.1021/acs.jpcb.0c11479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [256].Chong SS, Dugast-Darzacq C, Liu Z, Dong P, Dailey GM, Cattoglio C, Heckert A, Banala S, Lavis L, Darzacq X, Tjian R. Imaging dynamic and selective low-complexity domain interactions that control gene transcription. Science. 2018;361 doi: 10.1126/science.aar2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [257].Erdel F, Rademacher A, Vlijm R, Tunnermann J, Frank L, Weinmann R, Schweigert E, Yserentant K, Hummert J, Bauer C, Schumacher S, et al. Mouse heterochromatin adopts digital compaction states without showing hallmarks of HP1-Driven liquid-Liquid phase separation. Mol Cell. 2020;78:236–249.:e237. doi: 10.1016/j.molcel.2020.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [258].McSwiggen DT, Hansen AS, Teves SS, Marie-Nelly H, Hao Y, Heckert AB, Umemoto KK, Dugast-Darzacq C, Tjian R, Darzacq X. Evidence for DNA-mediated nuclear compartmentalization distinct from phase separation. Elife. 2019;8 doi: 10.7554/eLife.47098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [259].McSwiggen DT, Mir M, Darzacq X, Tjian R. Evaluating phase separation in live cells: diagnosis, caveats, and functional consequences. Gene Dev. 2019;33:1619–1634. doi: 10.1101/gad.331520.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [260].Williamson I, Berlivet S, Eskeland R, Boyle S, Illingworth RS, Paquette D, Dostie J, Bickmore WA. Spatial genome organization: contrasting views from chromosome conformation capture and fluorescence in situ hybridization. Genes Dev. 2014;28:2778–2791. doi: 10.1101/gad.251694.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [261].Klein IA, Boija A, Afeyan LK, Hawken SW, Fan MY, Dall’Agnese A, Oksuz O, Henninger JE, Shrinivas K, Sabari BR, Sagi I, et al. Partitioning of cancer therapeutics in nuclear condensates. Science. 2020;368:1386–1392. doi: 10.1126/science.aaz4427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [262].Howard TP, Roberts CWM. Partitioning of chemotherapeutics into nuclear condensates-opening the door to new approaches for drug development. Mol Cell. 2020;79:544–545. doi: 10.1016/j.molcel.2020.07.029. [DOI] [PubMed] [Google Scholar]
- [263].Mathieu C, Pappu RV, Taylor JP. Beyond aggregation: pathological phase transitions in neurodegenerative disease. Science. 2020;370:56–60. doi: 10.1126/science.abb8032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [264].Boija A, Klein IA, Young RA. Biomolecular condensates and cancer. Cancer Cell. 2021;39(2):174–192. doi: 10.1016/j.ccell.2020.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [265].Pessina F, Gioia U, Brandi O, Farina S, Ceccon M, Francia S, d’Adda di Fagagna F. DNA damage triggers a new phase in neurodegeneration. Trends Genet. 2021;37(4):337–354. doi: 10.1016/j.tig.2020.09.006. [DOI] [PubMed] [Google Scholar]
- [266].Cai D, Liu Z, Lippincott-Schwartz J. Biomolecular condensates and their links to Cancer progression. Trends Biochem Sci. 2021;46(7):535–549. doi: 10.1016/j.tibs.2021.01.002. [DOI] [PubMed] [Google Scholar]
- [267].Reinkemeier CD, Girona GE, Lemke EA. Designer membraneless organelles enable codon reassignment of selected mRNAs in eukaryotes. Science. 2019;363 doi: 10.1126/science.aaw2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [268].Levin A, Hakala TA, Schnaider L, Bernardes GJL, Gazit E, Knowles TPJ. Biomimetic peptide self-assembly for functional materials. IntRev Chem Eng. 2020;4:615–634. [Google Scholar]