Summary
Centromeres exert vital cellular functions in mitosis and meiosis. A specialized histone and other chromatin-bound factors nucleate a dynamic protein assembly that is required for the proper segregation of sister chromatids. In several organisms including the fission yeast, Schizosaccharomyces pombe, the RNAi pathway contributes to the formation of silent chromatin in peri-centromeric regions. Little is known about how chromatin remodeling factors contribute to heterochromatic integrity and centromere function. Here we show that the histone chaperone and remodelling complex FACT is required for centromeric heterochromatin integrity and accurate chromosome segregation. We show that Spt16 and Pob3 are two subunits of the S. pombe FACT complex. Surprisingly, yeast strains deleted for pob3+ are viable and alleviate gene silencing at centromeric repeats and at the silent mating type locus. Importantly, like heterochromatin and RNAi pathway mutants, Pob3-null strains exhibit lagging chromosomes on anaphase spindles. Whereas processing of centromeric RNA transcripts into siRNAs is maintained in Pob3 mutants, Swi6-association with the centromere is reduced. Our studies provide the first experimental evidence for a role of the RNA polymerase II cofactor FACT in heterochromatin integrity and in centromere function.
Results and Discussion
FACT is an Evolutionary Conserved Nuclear Complex
Centromeres are composed of specialized chromatin in which the histone H3 variant CENP-A underpins the kinetochore and is flanked by heterochromatic regions. This heterochromatin is known to attract cohesin and contribute to centromere function by ensuring physical cohesion between sister chromatids [1]. Significant progress has been made in dissecting the connections between heterochromatin and centromere function. It is known that specific histone modifications [2] and RNAi-related processes [3] contribute to an ‘epigenetic’ mechanism that defines the heterochromatic nature of centromeric DNA.
We wish to further investigate how the correct nucleosomal structure is established and maintained over centromeric repeats. In fission yeast, Pol II is also required for centromere function as it transcribes complementary regions of the centromeric outer-repeats [4], but it is not known whether Pol II requires co-activating factors. Evidence already implicates the transcriptional cofactors / chromatin remodeling complexes RSC/PBAF in centromere-related functions [5, 6]. Pol II transcription is stimulated by the chromatin remodeling complex FACT [7]. In order to investigate the possible role of this factor in centromeric chromatin, we first identified the fission yeast FACT complex.
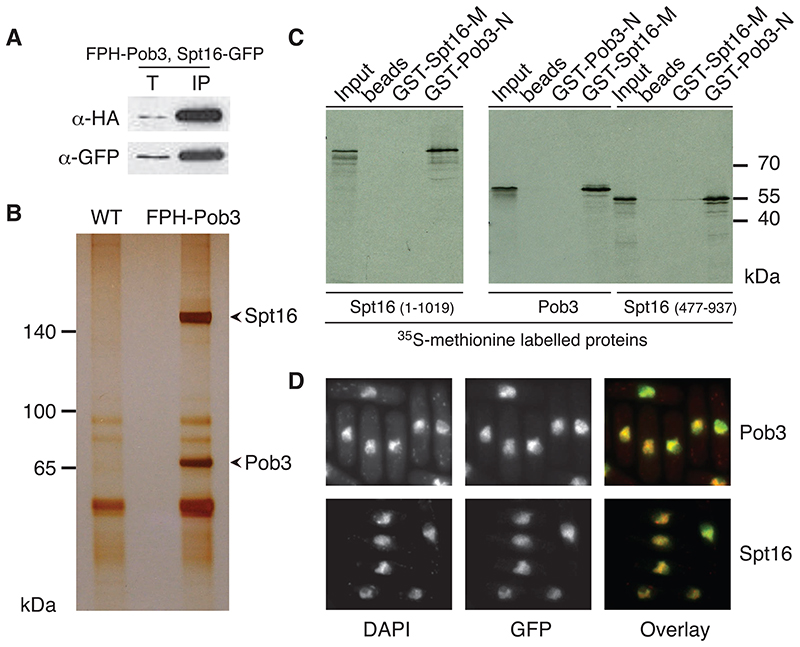
FACT from a variety of organisms contains two core proteins (Spt16 and Pob3/SSRP1). We identified a single set of closely related sequences for these subunits in the S. pombe genome (Figure S1). A strain was constructed to express S. pombe Pob3 fused to a FLAG-PreScission-HA epitope (FPH-Pob3). Western blots reveal expression of the functional FPH-Pob3 fusion protein. To check whether FPH-Pob3 forms a FACT-like complex with Spt16, we co-expressed GFP-tagged Spt16 (Figure S2). a-HA antibodies immunoprecipitate Spt16-GFP (Figure 1A). Thus Pob3 and Spt16 associate in vivo.
Figure 1. S. pombe FACT is a heterodimeric, nuclear protein complex.
(A) Co-immunoprecipitation between tagged S. pombe Pob3 and Spt16. Extracts prepared from FPH-Pob3 Spt16-GFP strains were incubated with HA antibody-coupled agarose. Fractions were analyzed by Western-blotting using either HA polyclonal or GFP monoclonal antibody. Lanes labeled ‘T’ show the equivalent of 10% extract used in the immunoprecipitation (IP).
(B) Biochemical purification of FPH-Pob3 and identification of co-purifying proteins. IPs from wild-type (WT) and FPH-pob3+ tagged strains were performed using FLAG-epitope affinity purification. Silver staining resolves two specific bands. Peptide sequencing by tandem MS identifies Pob3 and Spt16.
(C) GST-pulldowns map necessary interaction domains within SpFACT. Immobilized GST-fusion proteins were incubated with in vitro translated Pob3 and Spt16. GST-Pob3-N (1-448) and GST-Spt16-M (477-937). Input lane contains 10% of the 35Sproteins.
(D) Nuclear localization of Pob3-GFP and Spt16-GFP. The overlay shows the merge between DNA (DAPI) and SpFACT (GFP).
We next biochemically purified SpFACT from cellular extracts. SDS-PAGE analysis reveals two specific bands in the FPH-Pob3 extracts (Figure 1B). Mass spectrometry analysis identifies the two bands as Pob3 and Spt16. To verify whether the two FACT subunits interact, we performed GST-pulldowns. This demonstrates that Spt16 and Pob3 associate in vitro and that the interaction requires the Spt16-M domain (Figure 1C). Our biochemical data show that S. pombe contains a FACT complex similar to other eukaryotes.
To determine whether SpFACT localizes to the nucleus, we imaged functional Spt16- and Pob3-GFP fusions (Figure S2B). Pob3-GFP and Spt16-GFP are nuclear factors (Figure 1D). Together, our biochemical and localization data are consistent with nuclear functions of the SpFACT complex.
The Small FACT Subunit Pob3 is not Essential for Viability in S. pombe
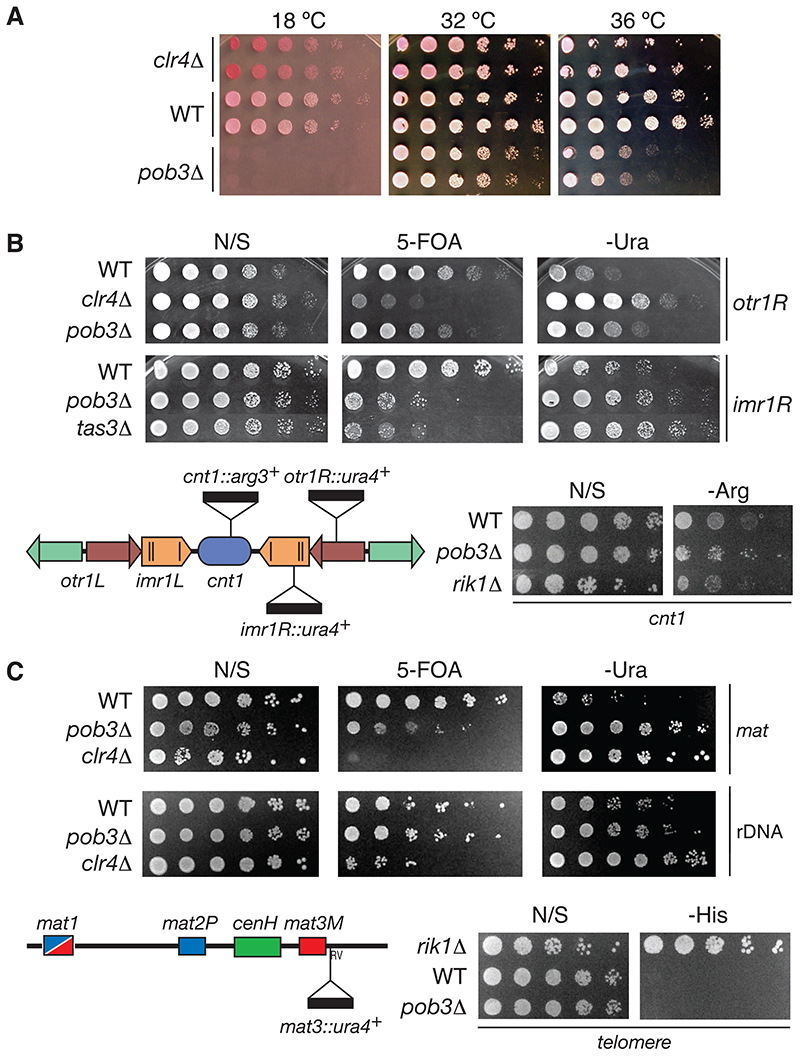
In S. cerevisae both subunits of the FACT complex are essential, and mutant alleles with phenotypes in genome stability have been described [8]. As expected, the S. pombe ortholog for the large FACT subunit, spt16+, is essential (Figure S3). Surprisingly, a strain bearing the deletion of pob3+ is viable (Figure 2A, S4 and S5), though it shows temperature sensitivity. No paralog that may account for genetic redundancy can be identified using genomic BLAST searches (data not shown). Since the pob3Δ strain is viable, we tested its role in distinct chromatin-based events. Cells lacking Pob3 are sensitive to HU, CPT, UV and (mildly) to 6-AU, suggesting DNA replication, DNA repair and transcription phenotypes (Figure S6). The sensitivity of pob3Δ to these stress agents indicates that SpFACT is involved in multiple chromatinbased cellular functions and participates in genome stability. Yet, its deletion in S. pombe is viable.
Figure 2. Viability and heterochromatic silencing phenotype of the S. pombe pob3+ deletion strain.
(A) Deletion of the small subunit of the chromatin remodeling complex FACT does not affect S. pombe viability. Wild-type (WT).
(B) pob3+ deletion alleviates heterochromatic silencing at imr1R and mildly at otr1R centromeric repeats and at the central core (cnt1). Mutants in the histone methyltransferase (clr4Δ), in rik1+ gene (rik1Δ) and in a RITS subunit (tas3Δ) serve as positive controls.
(C) Gene silencing phenotype of the pob3+ deletion in other heterochromatic regions such as the mating loci (mat), ribosomal DNA repeats (rDNA) and the telomere.
Deletion of pob3 Results in Transcriptional Silencing Defects
In vitro observations have shown that the FACT complex aids Pol II to overcome the nucleosome barrier to transcription [7]. In fission yeast it is known that Pol II is required to transcribe centromeric, non-coding outer repeats (otr) and form silent heterochromatin [4].
We therefore checked whether the loss of Pob3 affects transcriptional silencing within centromeres. Fission yeast strains with reporter genes inserted at distinct locations within the centromere (Figure 2) allow to assess the repressive state of chromatin at these locations [9]. We tested two mutant pob3Δ strains with ura4+ inserted either at the imrIR(NcoI)::ura4+ or at otrIR(SphI)::ura4+ repeats. Strains with an active ura4+ gene grow well in the absence of uracil (-Ura), but are unable to grow on counter-selective plates containing 5-Fluoro-orotic-acid (FOA) [9]. Plating assays show that pob3Δ strains grow slower than a WT strain on FOA medium (Figure 2B). Contrary to FACT’s known roles as a transcriptional elongation factor, our results reveal that the loss of Pob3 function allows higher levels of ura4+ gene expression relative to WT (Figure 2B), with a strong effect at imr1R and a weaker one at otr1R. This suggests that Pob3 has a novel, repressive role in centromeric transcription. In comparison, mutant strains clr4Δ and tas3Δ display complete alleviation of silencing at both loci. Further, ade6+ reporter assays show that pob3Δ loss of silencing is not a ura4+ gene-specific phenotype (Figure S2). Only a mild effect of pob3+ deletion is observed when arg3+ is inserted at the central core region cnt1::arg3+ (Figure 2B). The silencing assays show that the pob3Δ mutation distinctly affects the expression level of centromeric reporter genes depending on their location. Importantly, these data reveal an unexpected in vivo role for SpFACT in heterochromatin integrity.
We next determined whether pob3+ deletion affects the silencing of marker genes placed in other transcriptionally silent regions [10, 11]. The results show that pob3Δ causes de-repression of reporter silencing at the mating-type locus (Figure 2C). In contrast, the pob3Δ mutation does not affect the silencing of rDNA and telomeric reporters. Pob3 thus has a role in the formation or maintenance of heterochromatin at the mating locus and at centromeres.
FACT is a general transcription and remodeling factor. Recently, the human Pob3 orthologue, SSRP1, was shown to regulate the expression of a specific subset of genes [12]. The pob3Δ loss of silencing phenotype could therefore be indirect, altering the expression of specific heterochromatin factors. Thus, we performed gene expression profiling on pob3Δ cells. The results reveal that no such genes were up- or down-regulated significantly (Table S1). Importantly, the few genes whose expression is altered are similarly affected in mutants that play a role in heterochromatin integrity, such as clr1+, clr3+, clr6+ and rpb7+ (Table S2) [13]. For example, a significant fraction of pob3Δ up-regulated genes are also up-regulated in rpb7-G150D and in clr3Δ (Table S2). Since Clr3, Clr6 and Rpb7 are required for heterochromatin formation at centromeres, it is likely that FACT cooperates with these HDAC enzymes and Pol II in centromere function.
Our data show that the SpFACT complex has a new role in gene silencing at centromeric heterochromatin. Also, the transcriptional phenotype of pob3Δ significantly overlaps that of known heterochromatin mutants. Together, our data strongly suggest that Pob3 plays a specific and direct role in the establishment and/or maintenance of heterochromatin.
Pob3 is Required for Accurate Chromosome Segregation
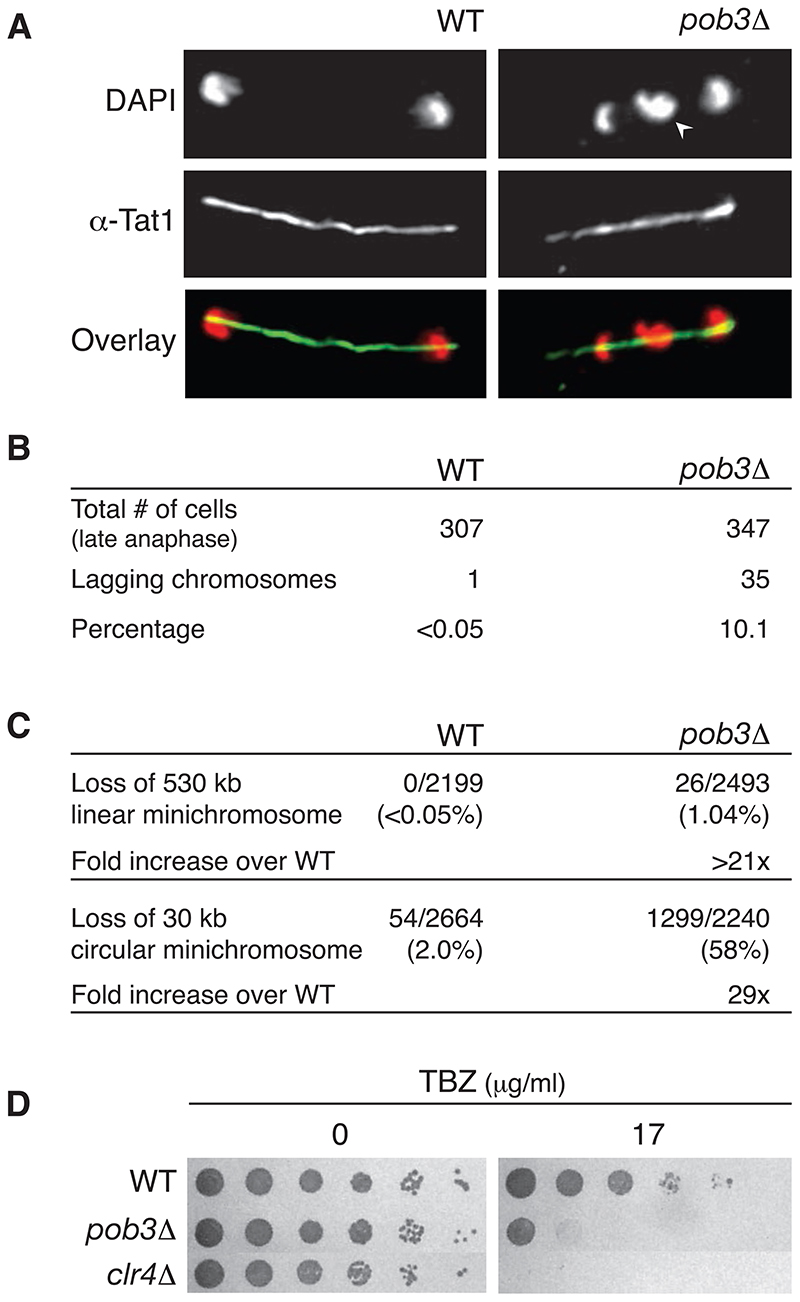
The observed centromeric silencing defects in pob3Δ cells suggest that centromeric heterochromatin is disrupted. It is well established that mutants affecting heterochromatin integrity at fission yeast centromeres also exhibit specific defects in mitotic segregation [14, 15]. We therefore conducted three types of test to identify mitotic defects. First, we checked for lagging chromosomes on anaphase spindles. Immunofluorescence staining shows that pob3Δ cells display a high incidence of lagging chromosomes (10%) in anaphase (Figure 3A). This represents a more than 200-fold increase over WT (Figure 3B).
Figure 3. S. pombe FACT subunit Pob3 is required for accurate chromosome segregation.
(A) pob3Δ mutant cells display lagging chromosomes in late anaphase. Cells grown at at 25°C were subjected to anti-tubulin (Tat1) immunodetection and DAPI staining. Arrowhead indicates a lagging chromosome in the midzone of the microtubule spindle in pob3Δ mutants. Wild-type (WT).
(B) pob3Δ deletion increases the frequency of abnormal anaphases. The percentage indicates the fraction of anaphase cells with lagging chromosomes.
(C) Enhanced minichromosome loss in pob3Δ mutant vs. WT strains.
(D) pob3+ deletion strains display a proncounced sensitivity to the tubulin depolymerizing drug TBZ. clr4Δ serves as positive control.
Second, we determined whether pob3+ is required for mini-chromosome maintenance over several cell divisions. We measured the fidelity of chromosome segregation using two distinct minichromosome-loss assays [14]. In WT cells, the 530-kb linear Ch16 minichromosome is mitotically stable [16]. Removal of Pob3 function increases the rate of minichromosome loss by >20-fold (Figure 3C). Since this phenotype could be due to defective telomere function in linear minichromosomes, we also tested the mitotic stability of the 30 kb circular minichromosome CM3112 [16]. The loss rate of CM3112 is increased by approximately 30-fold in pob3Δ compared to WT (Figure 3C). Thus, the mitotic segregation of both minochromosomes is severely affected in cells lacking pob3+.
pob3+ deletion may affect chromosome segregation by altering mitotic spindle function, as is seen in heterochromatin mutants [15]. We thus examined the growth and viability of pob3Δ in the presence of the microtubule-destabilizing drug thiabendazole (TBZ) [15]. The plating assays clearly reveal that, compared with WT, pob3Δ cells are TBZ-sensitive (Figure 3D), although to a lesser extent than clr4Δ cells.
Together, these assays demonstrate that pob3+ plays a new and important role in accurate chromosome segregation. Pob3’s role in centromeric silencing may account for its mitotic functions by contributing to heterochromatin integrity.
Loss of Pob 3 does not affect the RNAi pathway
To investigate the molecular mechanism underlying FACT’s novel repressive function, we analyzed the effect of pob3Δ on the RNAi-mediated heterochromatin formation at centromeres. In fission yeast, the RNAi pathway directs transcriptional gene silencing to the centromeric outer-repeats and is required to assemble intact centromeric heterochromatin [3]. In this pathway, RNA transcripts are generated from otr regions and are processed into siRNAs by Dcr1. These siRNAs are incorporated into the RITS effector complex, which is required to establish heterochromatin [3].
Mutants in the RNAi pathway, such as dcr1Δ and ago1Δ, are defective in processing non-coding centromeric outer repeat transcripts to homologous siRNA molecules. As a consequence, unprocessed otr transcripts accumulate [3]. In principle, a pob3Δ strain could either display altered levels of the primary transcript and/or show changes in siRNA accumulation. Either of these phenotypes could explain the centromere silencing and chromosome segregation defects.
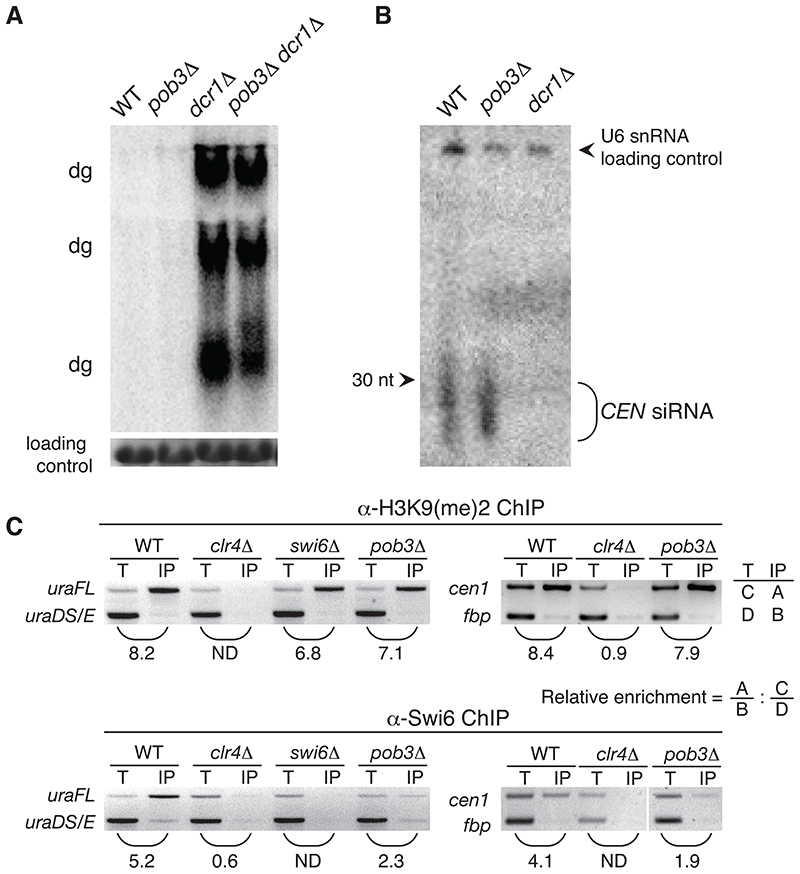
Northern blots show a clear accumulation of unprocessed otr transcripts in dcr1Δ cells, while they are not detected in pob3Δ or WT cells (Figure 4A). This could also be due to an important role of Pob3 in transcript generation. We therefore tested cells lacking both Dcr1 and Pob3. The results show that these transcripts appear to accumulate to the same extent as observed in dcr1Δ single mutants (Figure 4A). Outer-repeat transcripts thus accumulate in a dcr1Δ strain independently of Pob3. Consistently, centromere repeat-homologous siRNAs are detected at similar levels in WT and pob3Δ cells, but not in dcr1Δ cells (Figure 4B). Taken together, our results indicate that Pob3 function at the centromere does not appear to affect the production or accumulation of both unprocessed transcripts and siRNAs. FACT may thus affect centromeric silencing through changes in the integrity of silent chromatin itself and by acting either downstream of (or in parallel to) the RNAi machinery.
Figure 4. Pob3 likely acts downstream of Dicer and influences the deposition of the heterochromatic Swi6 protein on silenced chromosomal regions.
(A) Northern analysis of non-coding centromeric dg-dh transcripts detects no measurable changes in the accumulation of these transcripts in pob3Δ mutant strains, as opposed to dcr1+ deletion. pob3 Δ dcr1Δ double mutant does not alter the non-coding dg RNA levels. Centromeric transcripts were detected with a probe specific for the dg-dh repeat (top). RNA loading controls indicate total RNA added (ethidium bromide staining, bottom). Wild-type (WT).
(B) Centromeric siRNAs are unaffected by pob3+ deletion. Northern-blot of small RNAs extracted from WT, pob3Δ and dcr1Δ strains. The blot was probed with a centromeric (dg-dh) probe. U6 snRNA serves as a loading control.
(C) In pob3Δ mutants, Swi6 association is altered at centromeric outer repeats, while histone H3K9me2 levels are maintained. ChIP of H3K9me2 and Swi6 in WT, pob3Δ and heterochromatin mutants clr4Δ and swi6Δ. A significant loss of Swi6 is detected both at the uraFL transgene and at the endogenous cen1 locus (uraDS/E and fbp serve as euchromatic control regions, respectively). The figure shows a representative example of three independent biological experiments. The relative enrichment of IP/Input is calculated as shown.
The Swi6 Heterochromatin Mark is Altered in the pob3Δ Mutant
To further dissect FACT-mediated heterochromatinization, we analyzed chromatin structure at the centromeric repeats in the pob3Δ strain. Other fission yeast mutants, such as clr3Δ and sir2Δ, also affect RNAi-directed silent chromatin without affecting the production or abundance of centromeric repeat siRNAs (R. Allshire, pers. comm.). In such mutants, the H3K9me2 levels are reduced and consequently less Swi6 associates with centromeric repeats [17]. We therefore used chromatinimmunoprecipitation (ChIP) assays to determine the levels of H3K9me2 and Swi6 at centromeric outer repeats. The ChIP assays reveal normal levels of histone H3K9me2 on both the otrIR::ura4+ marker gene and directly on the outer repeats in cells lacking Pob3 (Figure 4C). Thus, Pob3 is not required to maintain normal levels of H3K9me2 methylation at centromeres. Since the H3K9me2 mark is unaffected, we expected the Swi6 protein levels to be maintained in pob3Δ. Surprisingly, we find that Swi6 association shows a moderate, but reproducible decrease over both the reporter gene and the endogenous centromeric region (Figure 4C). This shows that while centromeric Swi6 association still occurs to some level, its association is disturbed in pob3Δ strains. Consistent with this partial effect, we observe that Swi6-GFP remains localized to heterochromatic loci in pob3Δ cells, as is seen in several RNAi pathway mutants (Figure S7) [18]. Thus, although the key histone H3K9me2 mark is retained on centromeric repeats in cells lacking Pob3, Swi6 association is reduced. SpFACT may thus play a role in assembling or retaining Swi6 on centromeric heterochromatin.
Concluding Remarks
Here we have identified and characterized the SpFACT complex. Surprisingly, we show that deletion of the SpFACT subunit Pob3 is viable. This has allowed us to critically assess Pob3’s functions in vivo. Our experiments reveal a conserved biochemical protein assembly that functions in chromatin-based processes. Importantly, we provide the first biological evidence that Pob3 is required for accurate chromosome segregation. We find that this may be due to a novel role of the SpFACT complex in heterochromatin integrity at centromeres. pob3+ deletion does not affect H3K9me2 levels, but leads to decreases in Swi6 association at otr repeats. Together, our genetic and biochemical data implicate the chromatin remodeling complex FACT in forming functional centromeres.
FACT is known to facilitate transcription through chromatin. It has been proposed that FACT can disassemble H2A-H2B from nucleosomes in front of an advancing Pol II enzyme and reassemble H2A-H2B in its wake [19]. At centromeres, mutations in the SpFACT histone chaperone may affect H2A/H2B dimer incorporation and thus change the structural integrity of heterochromatin. Any alterations in the positioning, or composition of nucleosomes could interfere with Swi6 association/spreading. This could alleviate silencing without noticeably changing H3 K9 methylation, as we observe. Alternatively, FACT may recruit Swi6 directly to otr regions. Decreased binding of Swi6 to heterochromatic would be expected to impair sister chromatid cohesion, resulting in defective chromosome segregation [1].
In summary, our results show that the small subunit of the SpFACT complex is required to form normal silent chromatin on the centromeric repeats and for accurate chromosome segregation. Recent studies have shown that both subunits of the human FACT complex biochemically interact with centromeric CENP-A nucleosomes [20, 21]. While the biological role of this interaction is unclear, our in vivo data now suggest that FACT may use its histone chaperone activity to assemble and maintain the structural integrity of centromeric heterochromatin. Given the high degree of conservation in FACT subunit sequence and in biochemical functions, it is likely that our data point to an important and evolutionarily conserved role for FACT in maintaining centromere integrity.
Experimental Procedures
Strains, media, transformation and genetic techniques
Strains are listed in Table S3. Standard genetic techniques were used [22]. Cells were grown in yeast extracts supplemented with adenine (YES) or in synthetic minimal medium (PMG). When required, phloxin B, 6-azauracil (6AU), camptothecin (CPT), hydroxyurea (HU), thiabendazole (TBZ) was added. Damage assays [23], minichromosome loss rates [14], silencing assays, comparative plating and 5-fold serial dilution experiments [9] were performed as described.
Expression profiling and ChIP assays
Microarrays were carried out as described [13]. RNA was extracted using a standardized acid phenol protocol. cDNA was generated with S. pombe specific primers and random hexamers and labeled with Cy3/Cy5. Dye-swaps were done for all experiments. Hybridized slides were scanned (Biorad scanner), quantified (ImageQuant 4.2; Imagene) and analysed (Gene Spring; Silicon Genetics). Similar gene lists were identified using hyper-geometric distribution tests (Table S2). Swi6 and H3K9me2 ChIP performed as described [4]. Bands were quantified using Eastman Kodak EDAS 290 system and 1D image analysis software.
Immunofluorescence microscopy
Cell growth conditions, TAT1 immunofluorescence and staining protocol has been described [15]. Images were collected on a Carl Zeiss MicroImaging Axioplan 2 IE fluorescence microscope. Image acquisition was controlled using Metamorph (Universal Imaging Corp).
Northern blots
RNA was extracted from log phase cells by acid phenol protocol followed by PEG precipitation to separate high from low molecular weight RNA (HMW from LMW). 20 μg of HMW RNA and 40 μg of LMW RNA were resolved on 6% formaldehyde gels containing 1% agarose and on 8% urea-denaturating PAGE, respectively. Gels were blotted overnight to a Hybond-XL membrane (GE). DNA probes, complementary to centromeric dg-dh repeats and U6 snRNA were generated with High-Prime labeling (Roche) and T4 polynucleotide kinase (Promega), respectively.HMW and LMW RNA blots were hybridized overnight in a rotating oven at 65°C and 42°C, respectively. Phosphorscreens or films were exposed for 3h to 3 days.
Protein Methods
IPs with anti-HA agarose (Sigma) on whole-cell extracts performed as recommended (Sigma). For GST-pulldowns, 35S-Met proteins were expressed by TnT Quick-coupled in vitro transcription/translation (Promega). 20 μl of reaction and 160 μl buffer (1x HEMG, 0.15 M KCl, 1 mM DTT, 0.1% NP40) were added to 10-30 μg of immobilized GST fusions, incubated for 1h at 4°C and washed 5x. Gels were exposed on Kodak X-Omat AR. SpFACT was purified using yeast cell extracts [24]. Lysates were incubated with anti-FLAG M2 agarose (Sigma) for 6h at 4°C, washed in icecold PBS and elution performed with FLAG peptide (Sigma) followed by SDS-PAGE or MS analysis (Innova Proteomics).
Supplementary Material
Acknowledgements
We thank C. Bonilla for help with microarray experiments; S. Grewal for yeast strains; T. Urano for anti-histone H3K9me2 antibodies; A. Verdel for advice; D. Brunner for plasmids and advice; and members of the A.G.L., R.A. and D. Brunner labs for help and discussion. E.L. thanks M. Blondel for help. The EMBL, EU FP6 Marie Curie EST E-STAR (E.L.), Marie Curie RTN Chromatin Plasticity (A.G.L.) and NoE The Epigenome (A.G.L.) support our research. We apologize to all authors who we could not cite.
References
- 1.Bernard P, Maure JF, Partridge JF, Genier S, Javerzat JP, Allshire RC. Requirement of heterochromatin for cohesion at centromeres. Science. 2001;294:2539–2542. doi: 10.1126/science.1064027. [DOI] [PubMed] [Google Scholar]
- 2.Grewal SI, Rice JC. Regulation of heterochromatin by histone methylation and small RNAs. Curr Opin Cell Biol. 2004;16:230–238. doi: 10.1016/j.ceb.2004.04.002. [DOI] [PubMed] [Google Scholar]
- 3.Verdel A, Moazed D. RNAi-directed assembly of heterochromatin in fission yeast. FEBS Lett. 2005;579:5872–5879. doi: 10.1016/j.febslet.2005.08.083. [DOI] [PubMed] [Google Scholar]
- 4.Kato H, Goto DB, Martienssen RA, Urano T, Furukawa K, Murakami Y. RNA polymerase II is required for RNAi-dependent heterochromatin assembly. Science. 2005;309:467–469. doi: 10.1126/science.1114955. [DOI] [PubMed] [Google Scholar]
- 5.Xue Y, Canman JC, Lee CS, Nie Z, Yang D, Moreno GT, Young MK, Salmon ED, Wang W. The human SWI/SNF-B chromatin-remodeling complex is related to yeast RSC and localizes at kinetochores of mitotic chromosomes. Proc Natl Acad Sci U S A. 2000;97:13015–13020. doi: 10.1073/pnas.240208597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hsu JM, Huang J, Meluh PB, Laurent BC. The yeast RSC chromatin-remodeling complex is required for kinetochore function in chromosome segregation. Mol Cell Biol. 2003;23:3202–3215. doi: 10.1128/MCB.23.9.3202-3215.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Orphanides G, LeRoy G, Chang CH, Luse DS, Reinberg D. FACT, a factor that facilitates transcript elongation through nucleosomes. Cell. 1998;92:105–116. doi: 10.1016/s0092-8674(00)80903-4. [DOI] [PubMed] [Google Scholar]
- 8.Formosa T, Eriksson P, Wittmeyer J, Ginn J, Yu Y, Stillman DJ. Spt16-Pob3 and the HMG protein Nhp6 combine to form the nucleosome-binding factor SPN. Embo J. 2001;20:3506–3517. doi: 10.1093/emboj/20.13.3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Allshire RC, Javerzat JP, Redhead NJ, Cranston G. Position effect variegation at fission yeast centromeres. Cell. 1994;76:157–169. doi: 10.1016/0092-8674(94)90180-5. [DOI] [PubMed] [Google Scholar]
- 10.Ekwall K, Ruusala T. Mutations in rik1, clr2, clr3 and clr4 genes asymmetrically derepress the silent mating-type loci in fission yeast. Genetics. 1994;136:53–64. doi: 10.1093/genetics/136.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nimmo ER, Cranston G, Allshire RC. Telomere-associated chromosome breakage in fission yeast results in variegated expression of adjacent genes. Embo J. 1994;13:3801–3811. doi: 10.1002/j.1460-2075.1994.tb06691.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li Y, Zeng SX, Landais I, Lu H. Human SSRP1 has Spt16-dependent and independent roles in gene transcription. J Biol Chem. 2007;282:6936–6945. doi: 10.1074/jbc.M603822200. [DOI] [PubMed] [Google Scholar]
- 13.Wiren M, Silverstein RA, Sinha I, Walfridsson J, Lee HM, Laurenson P, Pillus L, Robyr D, Grunstein M, Ekwall K. Genomewide analysis of nucleosome density histone acetylation and HDAC function in fission yeast. Embo J. 2005;24:2906–2918. doi: 10.1038/sj.emboj.7600758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Allshire RC, Nimmo ER, Ekwall K, Javerzat JP, Cranston G. Mutations derepressing silent centromeric domains in fission yeast disrupt chromosome segregation. Genes Dev. 1995;9:218–233. doi: 10.1101/gad.9.2.218. [DOI] [PubMed] [Google Scholar]
- 15.Ekwall K, Nimmo ER, Javerzat JP, Borgstrom B, Egel R, Cranston G, Allshire R. Mutations in the fission yeast silencing factors clr4+ and rik1+ disrupt the localisation of the chromo domain protein Swi6p and impair centromere function. J Cell Sci. 1996;109:2637–2648. doi: 10.1242/jcs.109.11.2637. [DOI] [PubMed] [Google Scholar]
- 16.Niwa O, Matsumoto T, Chikashige Y, Yanagida M. Characterization of Schizosaccharomyces pombe minichromosome deletion derivatives and a functional allocation of their centromere. Embo J. 1989;8:3045–3052. doi: 10.1002/j.1460-2075.1989.tb08455.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SI. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science. 2001;292:110–113. doi: 10.1126/science.1060118. [DOI] [PubMed] [Google Scholar]
- 18.Hall IM, Noma K, Grewal SI. RNA interference machinery regulates chromosome dynamics during mitosis and meiosis in fission yeast. Proc Natl Acad Sci U S A. 2003;100:193–198. doi: 10.1073/pnas.232688099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Formosa T, Ruone S, Adams MD, Olsen AE, Eriksson P, Yu Y, Rhoades AR, Kaufman PD, Stillman DJ. Defects in SPT16 or POB3 (yFACT) in Saccharomyces cerevisiae cause dependence on the Hir/Hpc pathway: polymerase passage may degrade chromatin structure. Genetics. 2002;162:1557–1571. doi: 10.1093/genetics/162.4.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Foltz DR, Jansen LE, Black BE, Bailey AO, Yates JR, 3rd, Cleveland DW. The human CENP-A centromeric nucleosome-associated complex. Nat Cell Biol. 2006;8:458–469. doi: 10.1038/ncb1397. [DOI] [PubMed] [Google Scholar]
- 21.Obuse C, Yang H, Nozaki N, Goto S, Okazaki T, Yoda K. Proteomics analysis of the centromere complex from HeLa interphase cells: UV-damaged DNA binding protein 1 (DDB-1) is a component of the CEN-complex, while BMI-1 is transiently co-localized with the centromeric region in interphase. Genes Cells. 2004;9:105–120. doi: 10.1111/j.1365-2443.2004.00705.x. [DOI] [PubMed] [Google Scholar]
- 22.Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- 23.Edwards RJ, Carr AM. Analysis of radiation-sensitive mutants of fission yeast. Methods Enzymol. 1997;283:471–494. doi: 10.1016/s0076-6879(97)83038-8. [DOI] [PubMed] [Google Scholar]
- 24.Verdel A, Moazed D. Labeling and characterization of small RNAs associated with the RNA interference effector complex RITS. Methods Enzymol. 2005;392:297–307. doi: 10.1016/S0076-6879(04)92017-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.