Abstract
Alveolar macrophages are the most abundant macrophages in the healthy lung where they play key roles in homeostasis and immune surveillance against air-borne pathogens. Tissue-specific differentiation and survival of alveolar macrophages relies on niche-derived factors, such as granulocyte-macrophage colony stimulating factor 2 (GM-CSF) and transforming growth factor beta (TGF-β). However, the nature of the downstream molecular pathways that regulate the identity and function of alveolar macrophages and their response to injury remains poorly understood. Here, we identify that the transcription factor EGR2 is an evolutionarily conserved feature of lung alveolar macrophages and show that cell-intrinsic EGR2 is indispensable for the tissue-specific identity of alveolar macrophages. Mechanistically, we show that EGR2 is driven by TGF-β and GM-CSF in a PPAR-γ-dependent manner to control alveolar macrophage differentiation. Functionally, EGR2 was dispensable for regulation of lipids in the airways, but crucial for the effective handling of the respiratory pathogen Streptococcus pneumoniae. Finally, we show that EGR2 is required for repopulation of the alveolar niche following sterile, bleomycin-induced lung injury and demonstrate that EGR2-dependent, monocyte-derived alveolar macrophages are vital for effective tissue repair following injury. Collectively, we demonstrate that EGR2 is an indispensable component of the transcriptional network controlling the identity and function of alveolar macrophages in health and disease.
Introduction
Alveolar macrophages provide a first line of defence against airborne pathogens, as well as maintaining lung homeostasis and orchestrating tissue repair following injury. However, in chronic lung pathologies such as allergic asthma, idiopathic pulmonary fibrosis (IPF) and chronic obstructive pulmonary disease (COPD), alveolar macrophages display aberrant activity and, in many cases, appear to perpetuate disease (1). Moreover, monocytes and macrophages appear to play a particular pathogenic role in the context of severe coronavirus disease 2019 (COVID-19) (2–4). Thus, understanding the environmental signals and downstream molecular pathways that control the tissue-specific imprinting of macrophages in different contexts may yield important insights into how lung-specific cues regulate homeostasis and susceptibility to disease.
Alveolar macrophages are derived from foetal progenitors that seed the lung during embryonic development (5–7). However, the characteristic phenotype and functional properties of alveolar macrophages do not develop until the first few days of postnatal life in parallel with alveolarisation of the lung and are controlled by GM-CSF (also known as CSF-2) (7, 8) and the immunoregulatory cytokine TGF-β (9). Together these cytokines induce expression of the transcription factor peroxisome proliferator-activated receptor gamma (PPAR-γ) to promote survival and tissue-specific specialisation, including upregulation of genes involved in lipid uptake and metabolism (8). Consequently, mice in which Csf2rb, Tgfbr2 or Pparg has been genetically ablated in myeloid cells develop spontaneous pulmonary alveolar proteinosis (8, 9). However, alveolar macrophages largely fail to develop in the absence of GM-CSF and TGF-β receptor signalling due to their key role in macrophage survival. Therefore, it remains unclear if or how these factors control the tissue-specific identity and function of alveolar macrophages. Moreover, while considered the ‘master transcription factor’ of alveolar macrophages, PPAR-γ has been implicated in the control of other tissue macrophages, including splenic red pulp macrophages (10, 11), and thus, the transcriptional network responsible for conferring specificity upon alveolar macrophage differentiation remains unclear. Finally, if and how additional transcriptional regulators are involved in regulating these processes in the context of inflammation and repair is largely unexplored.
Here, we have used single cell RNA sequencing (scRNA-seq) to identify the transcriptional regulators expressed by alveolar macrophages. We show that expression of the transcription factor EGR2 is a distinct feature of lung alveolar macrophages. Using cell-specific ablation of Egr2 and mixed bone marrow chimeric mice, we show that cell-intrinsic EGR2 is indispensable for the tissue-specific identity of alveolar macrophages and their ability to control infection with a major respiratory pathogen, Streptococcus pneumoniae. RNA sequencing (RNA-seq) shows that EGR2 controls a large proportion of the core transcriptional signature of alveolar macrophages, including expression of Siglec5, Epcam and Car4. Mechanistically, we show that EGR2 expression is induced by TGF-β and GM-CSF-dependent signalling, and acts to maintain expression of CCAAT-enhancer-binding protein beta (C/EBP-β) to control alveolar macrophage differentiation. Finally, using the bleomycin-induced model of lung injury and a combination of fate mapping approaches, we show that post-injury repopulation of the alveolar macrophage niche occurs via differentiation of bone marrow-derived cells in an EGR2-dependent manner and that these monocyte-derived macrophages are indispensable for effective tissue repair and resetting of tissue homeostasis.
Results
EGR2 expression is a selective property of alveolar macrophages
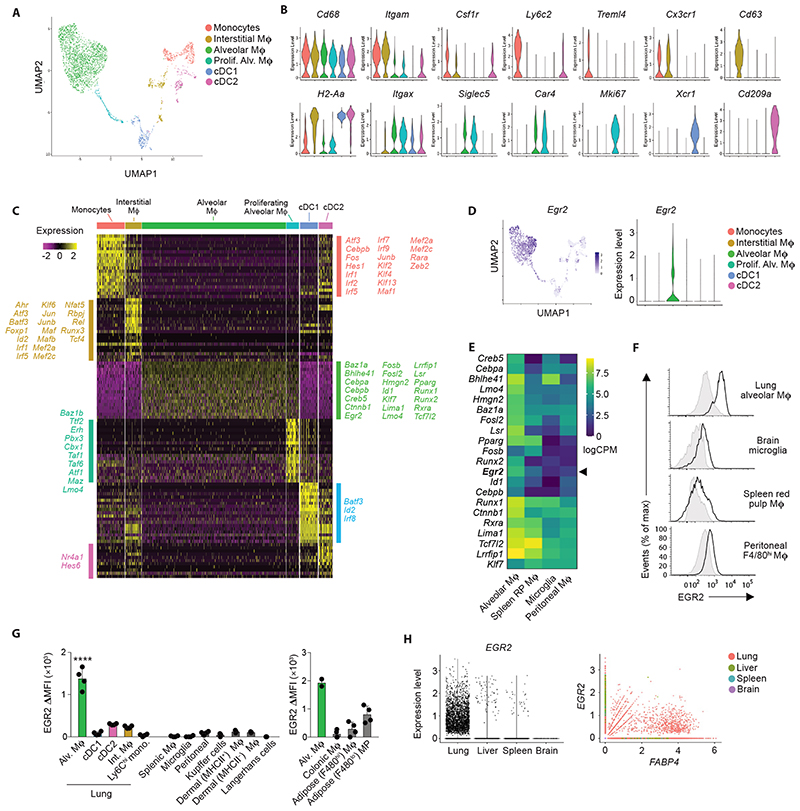
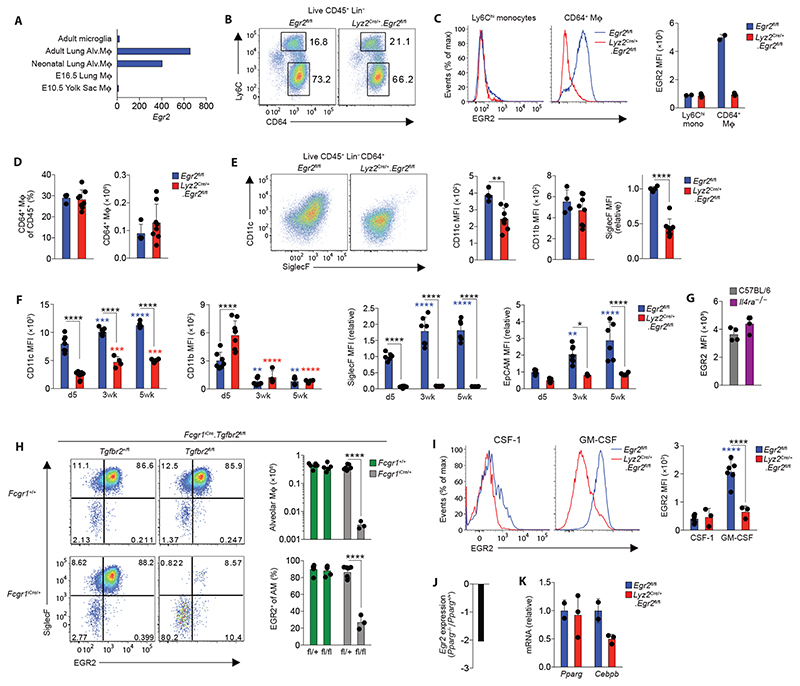
To begin to dissect the molecular pathways underlying the niche-specific imprinting of alveolar macrophages, we performed scRNA-seq of murine lung mononuclear phagocytes from lung digests to identify the transcriptional profile of alveolar macrophages. To this end, non-granulocytic CD45+ cells from lungs of Rag1 –/– mice were purified by FACS and sequenced using the 10x Chromium platform (Supplementary Figure 1A). Rag1 –/– mice were used to enrich for myeloid cells and reduce potential contamination by lymphocyte-macrophage doublets. 3936 cells passed quality control and were clustered using Uniform Manifold Approximation and Projection (UMAP) dimensionality reduction analysis within the Seurat R package. NK cells, identified by their expression of Ncr1, Nkg7 and Gzma, were excluded (Supplementary Figure 1A, B) and the remaining myeloid cells were re-clustered to leave six clusters of mononuclear phagocytes, and these were annotated using known landmark gene expression profiles (Figure 1A, B). Cluster 1 represented monocytes based on their expression of Itgam (encoding CD11b), Csf1r and Cd68, and could be divided into classical and non-classical monocytes based on expression of Ly6c2 and Treml4 respectively (Figure 1A, B). Cluster 2 represented interstitial macrophages based on their high expression of Cx3cr1, Cd68, Csf1r and H2-Aa and lack of the Xcr1 and Cd209a genes which defined cDC1 (cluster 5) and cDC2 (cluster 6) respectively. Alveolar macrophages (cluster 3) formed the largest population and could be defined by their expression of Itgax (encoding CD11c), Siglec5 (encoding SiglecF) and Car4, and lack of Cx3cr1 and Itgam. Cluster 4 was transcriptionally similar to cluster 3, but was defined by genes associated with cell cycle, including Mki67, Birc5 and Tubb5, suggesting these represent proliferating alveolar macrophages (Figure 1A, B).
Figure 1. EGR2 expression is a selective property of alveolar macrophages.
A. UMAP dimensionality reduction analysis of 3936 cells (non-granulocyte, myeloid cells) reveals six clusters of mononuclear phagocytes in murine lungs. Cells obtained from an individual Rag1 –/– mouse.
B. Feature plots displaying expression of individual genes by clusters identified in A.
C. Heatmap showing the top 20 most differentially expressed genes by each cluster defined in A. and annotated to show upregulated transcription factors/regulators within each cluster.
D. Overlay UMAP plot and feature plot showing expression of Egr2 by clusters identified in A.
E. Heatmap showing relative expression of selected transcription factors by lung alveolar macrophages, CD102+ peritoneal macrophages, brain microglia and red pulp splenic macrophages as derived from the ImmGen consortium.
F. Representative expression of EGR2 by lung alveolar macrophages, CD102+ peritoneal macrophages, brain microglia and red pulp splenic macrophages obtained from adult unmanipulated C57BL/6 mice. Shaded histograms represent isotype controls. Data are from one of three independent experiments.
G. Expression of EGR2 by the indicated macrophage and myeloid cell populations shown as relative MFI (MFI in Egr2 fl/fl – MFI in Lyz2 Cre/+.Egr2 fl/fl mice). Adipose & colonic macrophages shown on a separate graph due to measurements performed in an independent experiment with different flow cytometer settings. Repeat data for alveolar macrophages included as a reference. Data represent 3-4 mice (left graph) or 2-4 mice (right graph) per tissue. **** p<0.0001 (One-way ANOVA followed by Tukey’s multiple comparisons post-test).
H. In silico analysis of EGR2 and FABP4 expression by lung, liver, spleen and brain macrophages extracted on the basis of C1QA+ expression from (17–19).
Next, we compared gene expression profiles of these clusters, focussing on genes more highly expressed by alveolar macrophages relative to all other mononuclear phagocytes. 722 genes fitted these criteria, including Fapb1, Spp1 (encoding osteopontin) and Cidec which are known to be specifically and highly expressed by alveolar macrophages (Data File S1) (12, 13). Within this cassette of genes, we turned our attention to genes encoding transcription factors/regulators, as we hypothesised that these might control the tissue specific differentiation of alveolar macrophages. As expected, these included Pparg, Cebpb and Bhlhe41 which have been shown to control the development and self-renewal capacity of alveolar macrophages (8) (10, 14–16) (Figure 1C). However, this analysis also revealed transcription factors such as Id1, Klf7 and Egr2 which have not previously been implicated in the control of alveolar macrophage differentiation. We focussed on EGR2, which is part of a family of early growth response (EGR) transcription factors, comprising EGR1-4, as Egr2 appeared to be expressed in a particularly selective manner by alveolar macrophages (Figure 1D) when compared with other tissue macrophages at mRNA (Figure 1E) and protein level (Figure 1F, G & Supplementary Figure 2A). In contrast, while highly expressed by alveolar macrophages, Pparg was also expressed at a high level by splenic red pulp macrophages (Figure 1E), consistent with previous reports (10, 11). Of note, we did detect moderate EGR2 expression in F4/80lo mononuclear phagocytes in adipose tissue, whereas F4/80hi macrophage had low levels of EGR2 (Figure 1G). While our scRNA-seq analysis suggested that Egr2 was expressed at lower levels by proliferating alveolar macrophages, we could not confirm this at protein level, with Ki67+ alveolar expressing equivalent levels of EGR2 to their Ki67– counterparts (Supplementary Figure 2B). Next, we performed analogous analysis of EGR2 expression across a variety of human macrophage populations from scRNA-seq data sets within the Human Cell Atlas (17–19). Consistent with our analysis in the mouse, this showed that EGR2 expression was confined to lung macrophages, and in particular FABP4 + macrophages which correspond to airway macrophages (Figure 1H), and we confirmed this at protein level, showing that human CD163+HLA-DR+ bronchoalveolar lavage (BAL) macrophages uniformly express EGR2 (Supplementary Figure 2C). Thus, these data demonstrate that EGR2 expression is a constitutive, specific and evolutionarily conserved feature of alveolar macrophages.
EGR2 is required for the phenotypic identity of alveolar macrophages
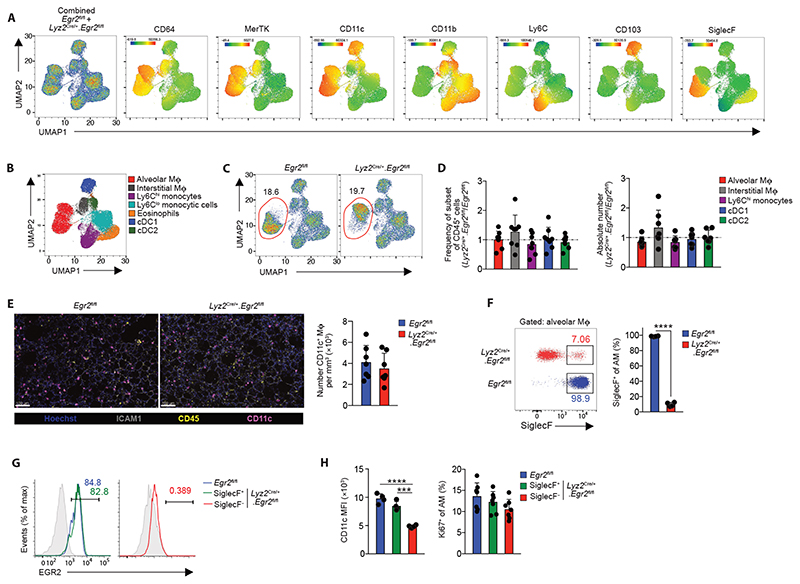
Previous work has suggested that EGR1 and EGR2 act in a redundant manner (20), while other studies have suggested EGR transcription factors are completely dispensable for macrophage differentiation (21). However, many of these studies were performed in vitro and the roles of EGRs in tissue-specific macrophage differentiation has not been assessed comprehensively in vivo, in part, due to the postnatal lethality of global Egr2–/– mice (22, 23). To determine the role of EGR2 in alveolar macrophage development and differentiation, we crossed Lyz2 Cre mice (24) with Egr2 fl/fl mice (25), to generate a strain in which myeloid cells, including monocytes, macrophages, dendritic cells and neutrophils, lack EGR2 in a constitutive manner. We performed unbiased UMAP flow cytometry analysis on lung leukocytes obtained from Lyz2 Cre.Egr2 fl/fl mice and Egr2 fl/fl littermate controls, focussing on ‘lineage’ negative (CD3–CD19– NK1.1–Ly6G–) CD11c+ and CD11b+ cells in lung digests (Figure 2A). Surface marker analysis of cells pooled from Egr2 fl/fl and Lyz2 Cre/+.Egr2 fl/fl mice confirmed the presence of alveolar and interstitial macrophages, eosinophils and subsets of dendritic cells and monocytes (Figure 2A) and this was validated by manual gating (Figure 2B & Supplementary Figure 3A). Due to their CD11chiCD11b– phenotype, alveolar macrophages clustered separately from the other CD11b+ myeloid cells (Figure 2A-C). All myeloid cells, including alveolar macrophages, were equally abundant in the lungs of Egr2 fl/fl and Lyz2 Cre/+.Egr2 fl/fl mice (Figure 2D, E). However, whereas alveolar macrophages from Egr2 fl/fl mice expressed high levels of SiglecF, the majority of alveolar macrophages obtained from Lyz2 Cre/+.Egr2 fl/fl mice lacked SiglecF expression (Figure 2F), explaining their distinct positioning within the alveolar macrophage cluster in the UMAP analysis. Indeed only ~5% of alveolar macrophages in Lyz2 Cre/+.Egr2 fl/fl mice expressed high levels of SiglecF, and further analysis showed that these expressed high levels of EGR2 (Figure 2G), suggesting that the SiglecF+ cells remaining in the Lyz2 Cre/+.Egr2 fl/fl mouse represent cells that have escaped Cre-mediated recombination. Consistent with this, SiglecF+ cells in the Lyz2 Cre/+.Egr2 fl/fl mouse expressed high levels of CD11c equivalent to alveolar macrophages from control mice, whereas SiglecF– alveolar macrophages expressed lower levels of CD11c (Figure 2H). We did not detect differences in the proliferative activity of Egr2-sufficient and -deficient alveolar macrophages (Figure 2H). Importantly and consistent with the lack of EGR2 expression by other tissue resident macrophages, we saw no effect on the cell number and expression of signature markers by resident macrophages in other tissues, including in the spleen where macrophages share a dependence on PPAR-γ (10, 11) and adipose tissue where we detected EGR2 expression (Supplementary Figure 3B-D). Thus, these data demonstrate that while EGR2 expression is dispensable for alveolar macrophages survival and self-maintenance, it is indispensable for imprinting key phenotypic features of the cells in the healthy lung.
Figure 2. EGR2 is required for the phenotypic identity of alveolar macrophages.
A. UMAP analysis of CD3–CD19–NK1.1–Ly6G–CD11b+/CD11c+ cells pooled from adult unmanipulated Egr2 fl/fl and Lyz2 Cre/+.Egr2 fl/fl mice (left panel). Heatmap plots showing the relative expression of the indicated markers by myeloid clusters.
B. Cluster identity confirmed by manual gating (see Fig. S3A).
C. Relative frequency of alveolar macrophages of all CD45+ leukocytes in unmanipulated adult Egr2 fl/fl and Lyz2 Cre/+.Egr2 fl/fl mice.
D. Relative frequency and absolute number of alveolar macrophages, cDC1, cDC2, Ly6Chi monocytes and CD64+MHCII+ interstitial macrophages in lung digests from adult unmanipulated Lyz2 Cre/+.Egr2 fl/fl mice compared with their abundance in Egr2 fl/fl littermates. Data are pooled from three independent experiments with 8 mice per group.
E. Confocal fluorescence imaging of Fixed Precision Cut Lung Slices from adult unmanipulated Egr2 fl/fl or Lyz2 Cre/+.Egr2 fl/fl mice stained with antibodies against ICAM1, CD45 and CD11c (left) and quantification of the number of CD11c+ macrophages per mm3 in each group (right). Data are pooled from three independent experiments with 7 mice per group.
F. Representative expression of SiglecF by CD11chiCD11blo alveolar macrophages (from F) obtained from lung digests from adult unmanipulated Egr2 fl/fl or Lyz2 Cre/+.Egr2 fl/fl mice (left), frequency of SiglecF+ macrophages in each strain (right). Data are from 4 mice per group from one of at least 5 independent experiments. SiglecF; ****p<0.0001 (unpaired Student’s t-test).
G. Representative expression of EGR2 by SiglecF-defined alveolar macrophages. Shaded histograms represent isotype controls. Data are from one of three independent experiments.
H. Mean fluorescence intensity (MFI) of CD11c expression (left) and frequency of Ki67+ SiglecF-defined CD11chiCD11blo alveolar macrophages (right) amongst lung digests from adult unmanipulated Egr2 fl/fl and Lyz2 Cre/+.Egr2 fl/fl mice. Data are from 4 mice per group from one of at least 5 independent experiments. *** p<0.001, **** p<0.0001 (One-way ANOVA followed by Tukey’s multiple comparisons post-test).
Symbols represent individual mice in all graphs and error bars represent the standard deviation.
EGR2 controls the tissue-specific transcriptional programme of alveolar macrophages
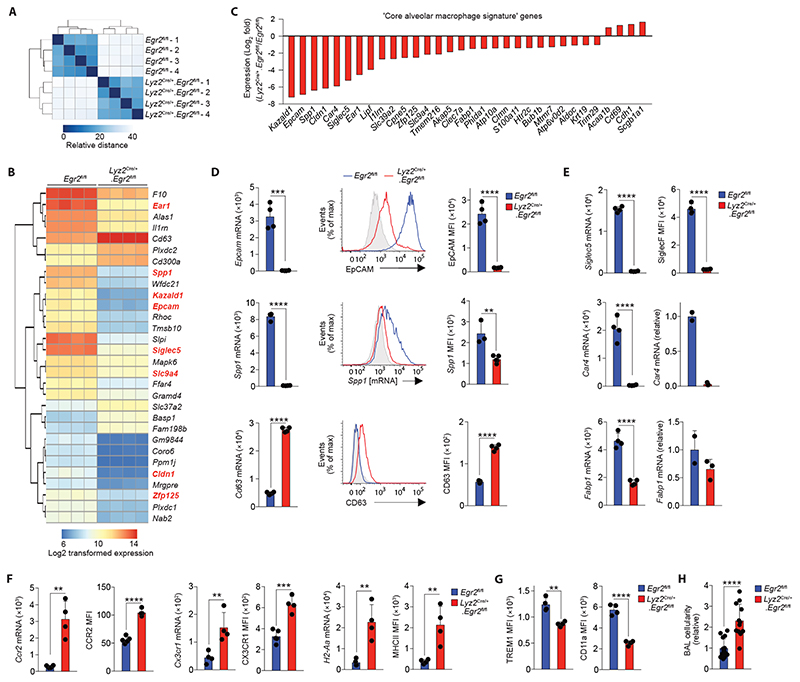
The failure of alveolar macrophages from Lyz2 Cre/+.Egr2 fl/fl mice to express SiglecF suggested that the tissue-specific differentiation programme of these cells may be altered by Egr2 deficiency. Hence, to ascertain the global effects of Egr2 deletion on alveolar macrophage differentiation, we next performed bulk RNA-seq of CD11chiCD11blo alveolar macrophages from lung digests of Egr2 fl/fl and Lyz2 Cre/+.Egr2 fl/fl mice (using only SiglecF– macrophages from Lyz2 Cre/+.Egr2 fl/fl mice to exclude confounding effects of EGR2-sufficient alveolar macrophages) (Supplementary Figure 4). Unbiased clustering confirmed the biological replicates from each group were highly similar (Figure 3A) and differential gene expression (DEG) analysis revealed that 858 genes were differentially expressed by at least 2-fold (417 and 440 genes downregulated and upregulated, respectively) (Data File S2). Consistent with our flow cytometry analysis, Siglec5, which encodes SiglecF, was one of the most downregulated genes in Egr2-deficient alveolar macrophages (Figure 3B). Many of the most differentially expressed genes formed part of the alveolar macrophage gene set identified in our scRNA-seq analysis. Moreover, approximately 30% of the core alveolar macrophage signature identified by the ImmGen consortium (12) was altered by Egr2 deficiency (32 genes) (Figure 3B, C), including the expression of Spp1, Epcam, Car4 and Fabp1, all of which were confirmed by flow cytometry or qPCR (Figure 3D, E). The vast majority of these ‘signature’ genes was downregulated in Egr2-deficient macrophages compared with their Egr2-sufficient counterparts. Gene Ontology (GO) analysis revealed that the top pathways affected by Egr2 deficiency were ‘Chemotaxis’, ‘Cell chemotaxis’ and ‘Immune system process’ (Supplementary Table 1). Consistent with this, the expression of chemokine receptors, such as Ccr2 and Cx3cr1, was elevated in alveolar macrophages from Lyz2 Cre/+.Egr2 fl/fl mice compared with their Egr2 fl/fl counterparts (Figure 3F). Genes encoding antigen presentation machinery, such as H2-Aa, H2-Eb1, Ciita and Cd74 were also upregulated in alveolar macrophages from Lyz2 Cre/+.Egr2 fl/fl mice. In parallel, there was significantly greater expression of MHCII at the protein level in Egr2-deficient alveolar macrophages (Figure 3F). Indeed, over 50 genes upregulated in Egr2-deficient alveolar macrophages were genes that defined interstitial macrophages in our scRNA-seq analysis, including Cd63, Mafb, Mmp12 and Msr1 (Figure 3D, Data File S2). Thus, EGR2 ablation renders alveolar macrophages transcriptionally more similar to their interstitial counterparts.
Figure 3. EGR2 controls tissue-specific transcriptional programme of alveolar macrophages.
A. Heatmap of RNA-seq data showing the euclidean distance between samples from adult Egr2 fl/fl and Lyz2 Cre/+.Egr2 fl/fl mice.
B. Heatmap showing log2 transformed expression of the 30 most differentially expressed genes by alveolar macrophages Egr2 fl/fl and Lyz2 Cre/+.Egr2 fl/fl mice. Each column represents a biological replicate with four mice per group. Genes highlighted in red appear in the ‘core signature’ of alveolar macrophages as defined by the ImmGen Consortium (12).
C. Log2-fold expression of differentially expressed genes that form part of the ‘core signature’ of alveolar macrophages as defined by the ImmGen Consortium (12).
D. Expression of Epcam, Spp1 and Cd63 from the RNA-seq dataset (left panels), representative flow cytometric validation of EpCAM, Spp1 (mRNA detected by PrimeFlow technology) and CD63 expression (middle panels) and replicate MFI expression data of each of these markers by alveolar macrophages from adult unmanipulated Egr2 fl/fl and Lyz2 Cre/+.Egr2 fl/fl mice. Data are from one of two independent experiments with 5 (Cre–) and 4 (Cre+) mice per group.
E. Expression of Siglec5, Car4 and Fabp1 from the RNA-seq dataset (left panels) and validation by flow cytometry (SiglecF) or qPCR (Car4, Fabp1). Data for SiglecF is from one of at least 10 independent experiments with 5 (Egr2 fl/fl) and 4 (Lyz2 Cre/+.Egr2 fl/fl) mice per group. Data for Car4 and Fabp1 represents 2 (Egr2 fl/fl) and 4 (Lyz2 Cre/+.Egr2 fl/fl) mice per group.
F. Expression of Ccr2, Cx3cr1 and H2-Aa from the RNA-seq dataset and replicate MFI expression data of CCR2, CX3CR1 and MHCII as determined by flow cytometry. Data are from one of two independent experiments with 5 (Egr2 fl/fl) and 4 (Lyz2 Cre/+.Egr2 fl/fl) mice per group.
G. Replicate MFI data of for CD11a and TREM1 expression as determined by flow cytometry. Data are from one of two independent experiments with 5 (Egr2 fl/fl) and 4 (Lyz2 Cre/+.Egr2 fl/fl) mice per group.
H. Absolute number of CD11chiCD11b– alveolar macrophages present in the BAL of adult unmanipulated Lyz2 Cre/+.Egr2 fl/fl mice relative to their abundance in Egr2 fl/fl littermates. Data are pooled from three independent experiments with 15 (Egr2 fl/fl) and 12 (Lyz2 Cre/+.Egr2 fl/fl) mice per group.
Symbols represent individual mice in all graphs and error bars represent the standard deviation. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001 (unpaired Student’s t test).
Further phenotypic analysis revealed reduced expression of ‘core signature’ alveolar macrophage markers TREM1 and CD11a at protein level in the context of Egr2 deficiency (Figure 3G). EpCAM and CD11a expression have been implicated in regulating adherence to and patrolling of the lung epithelium by alveolar macrophages (26), which suggested these behaviours may be altered by Egr2 deficiency. However, ex vivo analysis of live Precision-Cut Lung Slices (PCLS) showed that CD11c+ alveolar macrophages remained sessile in both strains, as compared with Ly6G+ CD11b+ neutrophils moving freely in sections (Data file S3 and Supplementary Figure 5A). Nevertheless, morphodynamics analysis of macrophages demonstrated increased changes in cell shape over time (as shown by the standard deviation of cell sphericity) indicating a more active behavior of Egr2-deficient macrophages (Data file S4 and Supplementary Figure 5B, C). In addition to this, while we found equivalent numbers of alveolar macrophages amongst tissue digests, we obtained consistently higher numbers of alveolar macrophages in the bronchoalveolar lavage (BAL) fluid of Lyz2 Cre/+.Egr2 fl/fl mice (Figure 3H). Taken together, these data suggest the EGR2-dependent differentiation programme may control the ability of alveolar macrophages to adhere to and interact with cells of their niche in the airways.
EGR2 controls distinct functional characteristics of alveolar macrophages
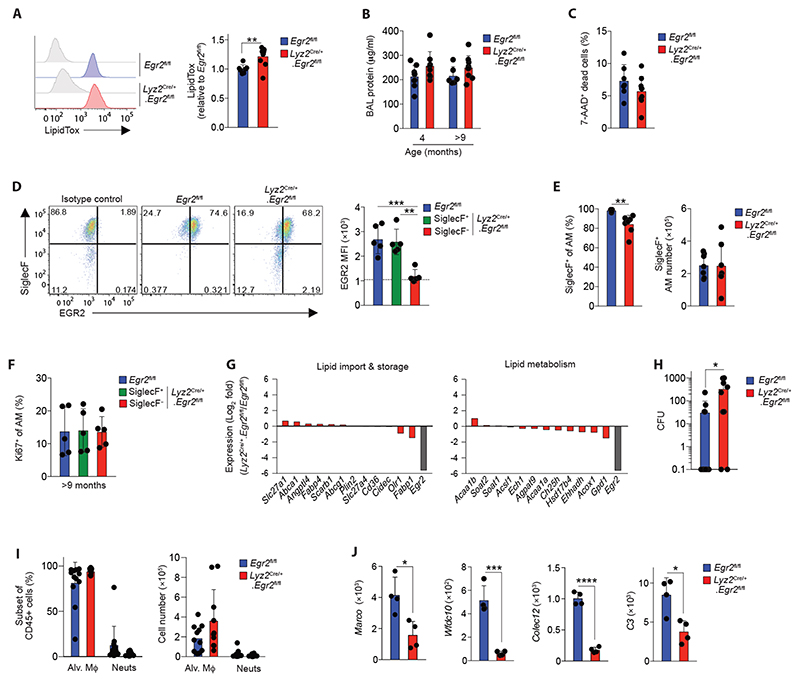
Individuals with mutations in EGR2 develop peripheral neuropathies due to the crucial role for EGR2 in Schwann cell function (26). However, many of these individuals also frequently encounter respiratory complications, including recurrent pneumonias and/or restrictive pulmonary disease, and in some cases respiratory failure (26). The cause of respiratory compromise in these individuals remains unexplained. To determine if alterations in alveolar macrophage behaviour may contribute to this, we next tested the function of Egr2-deficient alveolar macrophages. A major homeostatic function of alveolar macrophages is the regulation of pulmonary surfactant, and the absence of alveolar macrophages results in the development of pulmonary alveolar proteinosis (14, 28–32). We first examined neutral lipid context of alveolar macrophages using LipidTox. We found a small but significant increase in the neutral lipid context in alveolar macrophages from Lyz2 Cre/+.Egr2 fl/fl mice mice compared with Egr2 fl/fl littermates (Figure 4A). Despite this, Egr2 deficiency did not lead to spontaneous pulmonary alveolar proteinosis, as there were no differences in the levels of total protein in BAL fluid from Lyz2 Cre/+.Egr2 fl/fl and Egr2 fl/fl mice at either 4 or >9 months of age, a time at which proteinosis is detectable in Csf2rb –/– mice (32) (Figure 4B). Moreover, there was no detectable increase in the presence of dead cells in the BAL fluid, a common consequence of alveolar macrophage deficiency (Figure 4C). However, these results were confounded by the fact that the majority of alveolar macrophages in aged (>9 months) Lyz2 Cre/+.Egr2 fl/fl mice was now EGR2-sufficient, with most cells expressing high levels of SiglecF (Figure 4D). These findings suggested that the cells that had escaped Cre recombination may have a competitive advantage over their EGR2-deficient counterparts. Indeed, the absolute number of SiglecF+ alveolar macrophages no longer differed between aged Egr2 fl/fl and Lyz2 Cre/+.Egr2 fl/fl mice (Figure 4E). These data are consistent with other studies noting age-related repopulation of the alveolar niche with Cre ‘escapees’ in the Lyz2 Cre mouse (9). Notably, however, this preferential expansion of EGR2-sufficient ‘escapees’ did not relate to differences in the level of proliferation by EGR2-defined subsets, with identical frequencies of Ki67+ cells amongst EGR2-sufficient and -deficient macrophages in young adult and aged mice (Figure 2H & Figure 4F).
Figure 4. EGR2 controls distinct functional characteristics of alveolar macrophages.
A. Representative LipidTox staining of alveolar macrophages from the BAL fluid of unmanipulated adult Egr2 fl/fl or Lyz2 Cre/+.Egr2 fl/fl mice (left). Graph shows the mean fluorescence intensity (MFI) of LipidTox in alveolar macrophages from Lyz2 Cre/+.Egr2 fl/fl mice relative to those from Egr2 fl/fl mice. Data are from 7 (Egr2 fl/fl) and 10 (Lyz2 Cre/+.Egr2 fl/fl) mice per group pooled from three independent experiments. **p<0.01 (unpaired Student’s t-test).
B. Protein levels in the BAL fluid of Egr2 fl/fl or Lyz2 Cre/+.Egr2 fl/fl mice at 4 or 9-12 months of age. Data are from 6-9 mice per group pooled from two independent cohorts of aged mice.
C. Frequency of 7-AAD+ (dead) cells in the BAL fluid of Egr2 fl/fl or Lyz2 Cre/+.Egr2 fl/fl mice at 9-12 months of age. Data are from 6-9 mice per group pooled from two independent cohorts of aged mice.
D. Representative expression of SiglecF and EGR2 by CD11chiCD11blo macrophages and MFI of EGR2 by SiglecF-defined CD11chiCD11blo macrophages obtained from 11-12 month old Egr2 fl/fl or Lyz2 Cre/+.Egr2 fl/fl mice. Data are from 5 mice per group pooled from two independent cohorts of aged mice. ** p<0.01, *** p<0.001 (One-way ANOVA followed by Tukey’s multiple comparisons post-test.)
E. Frequency (left) and absolute number (right) of SiglecF+ cells amongst CD11chiCD11blo macrophages obtained from 11-12 month old Egr2 fl/fl or Lyz2 Cre/+.Egr2 fl/fl mice. Data are from 7 mice per group pooled from three independent cohorts of aged mice. **p<0.01 (unpaired Student’s t-test).
F. Frequency of Ki67+ cells amongst SiglecF-defined CD11chiCD11blo macrophages obtained from 11-12 month old Egr2 fl/fl or Lyz2 Cre/+.Egr2 fl/fl mice. Data are from 5 mice per group pooled from two independent experiments.
G. Log2-fold expression of genes that are implicated in lipid uptake or metabolism in alveolar macrophages as defined by (8). Expression of Egr2 is included as a reference.
H. Bacterial levels (colony forming units, CFU) in the BAL fluid of Egr2 fl/fl or Lyz2 Cre/+.Egr2 fl/fl mice 14hrs after infection. Data are from 10 (Lyz2 Cre/+.Egr2 fl/fl) or 12 (Egr2 fl/fl) mice per group pooled from three independent experiments. *p<0.05 (Mann Whitney test).
I. Frequency (left) and absolute number (right) of CD11chiCD11blo alveolar macrophages and Ly6G+ neutrophils in the BAL fluid of Egr2 fl/fl or Lyz2 Cre/+.Egr2 fl/fl mice 14hrs after infection. Data represent 10 (Lyz2 Cre/+.Egr2 fl/fl) or 11 (Egr2 fl/fl) mice per group pooled from three independent experiments.
J. Expression of Marco, Wfdc10, Colec12 and C3 from the RNA-seq dataset (left panels). *p<0.05, ***p<0.001, ****p<0.0001 (unpaired Student’s t-test).
Symbols represent individual mice in all graphs and error bars represent the standard deviation.
In an attempt to circumvent the confounding effects of these escapees, we generated a second strain to delete Egr2 from macrophages by crossing Egr2 fl/fl mice with mice expressing ‘improved’ Cre recombinase under control of the endogenous Fgcr1 promoter (Fcgr1 iCre)(13). By using Fcgr1 iCre.Rosa26 LSL-RFP reporter mice, we confirmed that this approach led to efficient Cre recombination in alveolar macrophages, as well as in other tissue macrophages, but not in other leukocytes (Supplementary Figure 6A). Importantly, alveolar macrophages from Fcgr1 iCre/+.Egr2 fl/fl mice phenocopied those from Lyz2 Cre/+.Egr2 fl/fl mice (Supplementary Figure 6B), but the frequency of Cre escapees was markedly lower in Fcgr1 iCre/+.Egr2 fl/fl mice compared with Lyz2 Cre/+.Egr2 fl/fl mice (Supplementary Figure 6C, D). Despite this, we did not detect the development of proteinosis or accumulation of dead cells in the BAL fluid of aged Fcgr1 iCre/+.Egr2 fl/fl mice compared to their littermate controls (Supplementary Figure 6E). Consistent with this, Egr2 deficiency had little if any effect on the expression of molecules associated with lipid uptake and metabolism that are characteristic of normal alveolar macrophages (8) (Figure 4G & Supplementary Figure 6F). Thus, while EGR2 is indispensable for the phenotypic identity of alveolar macrophages, it appears to be largely dispensable for lipid regulation.
We next sought to determine if EGR2-dependent differentiation controls protective immune functions of alveolar macrophages. To do so, we infected Egr2 fl/fl mice and Lyz2 Cre/+.Egr2 fl/fl mice with 1x104 colony forming units (CFU) Streptococcus pneumoniae, based on previous work showing that wild type alveolar macrophages efficiently clear infection at this dose (33, 34). This showed that the majority of Egr2 fl/fl mice (8 out of 12) had cleared infection at 14 hours post infection, whereas the majority of Lyz2 Cre/+.Egr2 fl/fl mice (8 out of 10) had detectable bacteria in the airways at this timepoint (Figure 4H). Importantly, the failure to handle bacteria did not reflect the loss of tissue resident macrophages that can occur during inflammation or infection, as alveolar macrophages continued to dominate the airways in both groups (Figure 4I, J). Similarly, increased bacteria in Lyz2 Cre/+.Egr2 fl/fl mice is also unlikely to reflect an effect in neutrophils, as neutrophil recruitment was negligible in both strains, and, although targeted in the Lyz2 Cre system, neutrophils failed to express EGR2 during health or in the context of S. pneumoniae infection (Supplementary Figure 7A). Instead, our RNA-seq analysis showed that expression of genes encoding molecules for the recognition, opsonisation and elimination of bacteria, including Colec12, Wfdc10, C3 and Marco, the latter of which has been shown to be indispensable for immunity to S. pneumoniae (35), were significantly reduced in Egr2-deficient alveolar macrophages (Figure 4K). Thus, EGR2-dependent differentiation is crucial for equipping alveolar macrophages with the machinery to capture and destroy pneumococci.
EGR2 expression by alveolar macrophages is dependent on TGFβ and GM-CSF
Alveolar macrophages derive from foetal monocytes that seed the developing lung in the late gestational period (7). To determine the point at which EGR2 is first expressed, we assessed EGR2 expression by E10.5 yolk sac macrophages, by macrophages in the embryonic lung (E16.5) and by CD11chiCD11blo alveolar macrophages in the neonatal and adult lung using the ImmGen database. This revealed that Egr2 was absent from yolk sac macrophages and macrophages in the embryonic lung at E16.5, but it was expressed by both neonatal and adult alveolar macrophages (Figure 5A), suggesting that it is induced during alveolarization in the neonatal period. Consistent with this, we found high expression of EGR2 at protein level by neonatal (d1) CD64+ lung macrophages (sometimes referred to as ‘pre-alveolar macrophages’) in Egr2 fl/fl mice; as expected, this expression was deleted efficiently in Lyz2 Cre/+.Egr2 fl/fl mice (Figure 5B, C). Importantly, Ly6Chi monocytes in the lung of d1 neonatal mice lacked any expression of EGR2 (Figure 5B, C), reinforcing the selectivity of EGR2 expression even at this highly dynamic stage of myeloid cell development in the lung. Consistent with our analysis of mature alveolar macrophages in adult mice, Egr2 deletion had no impact on the frequency and absolute number of pre-alveolar macrophages (Figure 5D). However, phenotypic differences were already apparent in macrophages from Lyz2 Cre/+.Egr2 fl/fl mice at this stage, with reduced CD11c and SiglecF expression which persisted into adulthood (Figure 5E, F). In parallel, EpCAM expression was absent from alveolar macrophages in the neonatal period and was progressively upregulated with age in an EGR2-dependent manner (Figure 5F). CD11b expression, which is downregulated in mature alveolar macrophages, was found on pre-alveolar macrophages in both Egr2 fl/fl and Lyz2 Cre/+.Egr2 fl/fl mice, and it was downregulated to the same extent with age in both strains.
Figure 5. TGFβ and CSF2 drive EGR2 expression.
A. Normalised expression (by DESeq2) of Egr2 by the indicated populations (data obtained from the ImmGen Consortium).
B. Representative expression of Ly6C and CD64 by live CD45+CD3–CD19–Ly6G– cells from the lungs of unmanipulated newborn Egr2 fl/fl or Lyz2 Cre/+.Egr2 fl/fl mice. Data are from one of two independent experiments performed.
C. Histograms show representative expression of EGR2 by CD64+ ‘pre-alveolar macrophages’ and Ly6Chi monocytes from the lungs of unmanipulated newborn Egr2 fl/fl or Lyz2 Cre/+.Egr2 fl/fl mice and bar chart shows the mean fluorescent intensity (MFI) of EGR2 expression by these cells. Data are from one of two independent experiments performed with 2 (Egr2 fl/fl) or 5 (Lyz2 Cre/+.Egr2 fl/fl) mice per group.
D. Frequency and absolute number of CD64+ ‘pre-alveolar macrophages’ from mice in B. Data are pooled from two independent experiments with 4 (Egr2 fl/fl) or 8 (Lyz2 Cre/+.Egr2 fl/fl) mice per group.
E. FACS plots show representative expression of CD11c and SiglecF by CD64+ ‘pre-alveolar macrophages’ from mice in B and bar chart shows the MFI of CD11c, SiglecF and CD11b expression by these cells. Data are pooled from two independent experiments with 4 (Egr2 fl/fl) or 8 (Lyz2 Cre/+.Egr2 fl/fl) mice per group.
F. MFI of CD11c and CD11b and relative MFI of SiglecF and EpCAM (relative to cells from d5 old Egr2 fl/fl mice) expression by CD11chiCD11blo alveolar macrophages obtained from unmanipulated Egr2 fl/fl or Lyz2 Cre/+.Egr2 fl/fl mice at the indicated ages. Data are pooled from two independent experiments with 4-9 mice per group. Coloured * denote significance between d5 and 3 and 5 weeks within the Egr2 fl/fl (blue) and Lyz2 Cre/+.Egr2 fl/fl (red) data. **p<0.01, ***p<0.001, ****p<0.0001 (Two-way ANOVA with Tukey’s multiple comparisons test).
G. Representative expression of EGR2 by alveolar macrophages from adult WT (C57BL/6) and Il4ra –/– adult mice. Data from one experiment with 4 mice per group.
H. Representative expression of EGR2 and SiglecF by CD11chiCD11blo alveolar macrophages obtained from lungs of neonatal (d8) Fcgr1 iCre/+.Tgfbr2 fl/fl and littermate controls. Bar charts show the absolute numbers of CD11chiCD11blo alveolar macrophages (upper) and the mean frequency of EGR2+ cells amongst CD11chiCD11blo alveolar macrophages (lower). Data are pooled from two independent experiments with 3-7 mice per group. **** p<0.0001 (One-way ANOVA followed by Tukey’s multiple comparisons post-test).
I. Representative expression of EGR2 (left) and MFI of EGR2 (right) by FACS-purified Ly6Chi monocytes cultured in vitro with recombinant CSF-1 (20ng/ml) or GM-CSF (20ng/ml) for five days. Symbols represent monocytes isolated from individual mice. Data are from 6 Egr2 fl/fl (Cre–) or 3 Lyz2 Cre.Egr2 fl/fl (Cre+) mice per group pooled from two independents experiment. **** p<0.0001 (Two-way ANOVA followed by Tukey’s multiple comparisons post-test). Coloured * denote significance between CSF-1 and GM-CSF within the Egr2 fl/fl (blue) and Lyz2 Cre.Egr2 fl/fl (red) data.
J. Relative expression of Egr2 by alveolar macrophages obtained from Pparg fl/fl or Itgax Cre.Pparg fl/fl mice from the ImmGen Consortium.
K. qPCR analysis of Pparg and Cebpb mRNA by BAL cells from unmanipulated adult Egr2 fl/fl or Lyz2 Cre.Egr2 fl/fl mice. Data represent 2 Egr2 fl/fl or 4 Lyz2 Cre.Egr2 fl/fl mice per group.
Symbols represent individual mice in all graphs and error bars represent the standard deviation.
We next set out to determine the environmental factors that drive EGR2 expression. Many studies employing in vitro culture systems have described EGR2 expression as a feature of ‘alternatively activated’ macrophages, dependent on IL-4R signalling (36, 37). Importantly, expression of EGR2 by alveolar macrophages was independent of IL-4R signalling (Figure 5G & Supplementary Figure 6G), as were key EGR2-dependent phenotypic traits, such as SiglecF and EpCAM expression (Supplementary Figure 6F). TGF-β has recently been shown to be crucial for the development of alveolar macrophages (9) and thus we next explored if the TGF-β-TGF-βR axis drives expression of EGR2. To do so, we generated a new mouse line by crossing Fcgr1 iCre mice to mice with LoxP sites flanking the Tgfbr2 allele (Tgfbr2 fl/fl). Consistent with the crucial role for TGF-βR in controlling alveolar macrophage development (9), there was a paucity of alveolar macrophages in the lungs of neonatal Fcgr1 iCre/+.Tgfbr2 fl/fl compared with Fcgr1 +/+.Tgfbr2 fl/fl and Fcgr1 iCre/+.Tgfbr2 fl/+ controls (Figure 5H). Strikingly, while CD11c+CD11blo alveolar macrophages expressed high levels of EGR2 in control groups, EGR2 expression was largely abolished in Fcgr1 iCre/+.Tgfbr2 fl/fl mice, demonstrating that TGF-βR signalling is vital for EGR2 induction in vivo. As Fcgr1 iCre/+.Tgfbr2 fl/fl developed fatal seizures between d14 and d21 of age, perhaps reflecting the indispensable role for TGF-βR in controlling microglia activity (38, 39), we were unable to carry out further analyses using this strain.
Given the central role for GM-CSF in alveolar macrophage development, we also assessed the role of GM-CSF in driving EGR2 expression using an in vitro culture system in which Ly6Chi monocytes from bone marrow were FACS-purified and cultured with recombinant CSF-1 or GM-CSF. This revealed that GM-CSF was also capable of driving EGR2 expression in this system (Figure 5I). Given that GM-CSF receptor and TGF-βR signalling is known to induce expression of PPAR-γ (8, 9, 14), we next determined if PPAR-γ is upstream of EGR2. Analysis of a publicly available dataset (ImmGen) comparing the transcriptional profile of Pparg-sufficient and -deficient alveolar macrophages revealed downregulation (2.1-fold change) of Egr2 in the context of Pparg deficiency (Figure 5j). In contrast, Pparg expression was unaffected in alveolar macrophages from Lyz2 Cre/+.Egr2 fl/fl mice (Figure 5K), suggesting EGR2 is downstream of PPAR-γ. Another transcription factor implicated in controlling alveolar macrophage differentiation is C/EBPβ (15) and EGR2 has been shown to modulate C/EBPβ in vitro (36). Egr2 deficiency led to reduced expression of C/β at mRNA (Figure 5K) and protein level (Supplementary Figure 6H). Taken together, these data support the premise that EGR2 expression by alveolar macrophages is induced by TGF-β and GM-CSF in a PPAR-γ-dependent manner in the neonatal period and this in turn induces expression of C/EBPβ to drive tissue-specific differentiation.
Egr2 deficiency confers a competitive disadvantage on alveolar macrophages
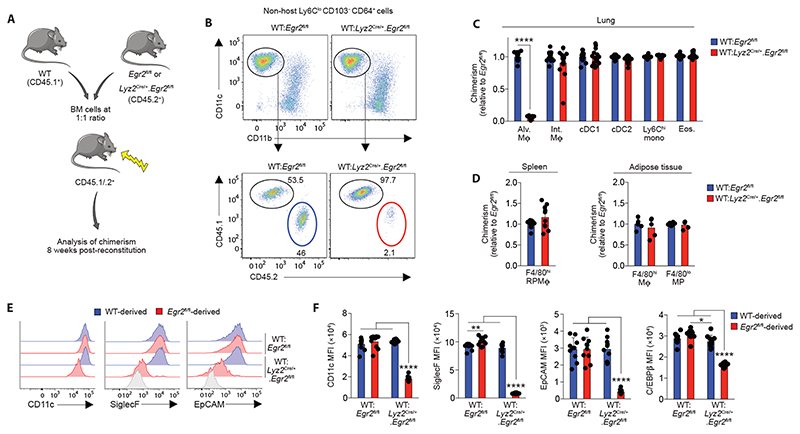
Given the observation that EGR2-sufficient alveolar macrophages come to dominate the airspace of Lyz2 Cre/+.Egr2 fl/fl mice, we next set out to determine if EGR2 deletion confers an intrinsic competitive disadvantage on alveolar macrophages. To this end, we generated mixed bone marrow chimeric mice by reconstituting lethally irradiated WT (CD45.1+/.2+) mice with a 1:1 ratio of WT (CD45.1+) and either Egr2 fl/fl or Lyz2 Cre/+.Egr2 fl/fl (CD45.2+) bone marrow cells (Figure 6A). 8 weeks after reconstitution, we found that Egr2-deficient and Egr2 sufficient bone marrow contributed equally to the pools of monocytes, interstitial macrophages and dendritic cell subsets in the lung (Figure 6B, C). In contrast, alveolar macrophages were derived almost exclusively from WT BM in WT:Lyz2 Cre/+.Egr2 fl/fl chimeric mice, whereas they were derived equally from both BM sources in WT:Egr2 fl/fl chimeric mice (Figure 6B, C). These effects were not a general feature of macrophages derived from Egr2-deficient bone marrow, as Egr2 deletion did not adversely affect the replenishment of splenic red pulp or adipose tissue macrophages (Figure 6D). The mixed BM chimeric model also confirmed that the phenotypic differences seen in alveolar macrophages from intact Lyz2 Cre/+.Egr2 fl/fl mice were due to cell intrinsic loss of EGR2, rather than effects of Egr2 deficiency on the lung environment (Figure 6E, F). We also used this system to confirm the reduced expression of C/EBPβ by alveolar macrophages deriving from Lyz2 Cre/+.Egr2 fl/fl bone marrow (Figure 6F).Taken together, these results demonstrate that cell intrinsic EGR2 is indispensable for the differentiation of alveolar macrophages and repopulation of the alveolar niche following radiation-induced depletion.
Figure 6. Egr2 deficiency confers a competitive disadvantage on alveolar macrophages.
A. Schematic of the generation of mixed bone marrow chimeric mice
B. Representative expression of CD11c and CD11b by Ly6CloCD64+ macrophages amongst live CD45+CD3–CD19–Ly6G–CD103– cells (upper panels) and representative expression of CD45.1 and CD45.2 by CD11chiCD11blo alveolar macrophages (lower panels) from WT:Egr2 fl/fl or WT:Lyz2 Cre/+.Egr2 fl/fl chimeric mice.
C. Contribution of Egr2 fl/fl BM to the indicated lung myeloid populations in WT:Lyz2 Cre/+.Egr2 fl/fl chimeric mice relative to WT:Egr2 fl/fl mice. Chimerism was normalised to Ly6Chi blood monocytes before normalisation of Lyz2 Cre/+.Egr2 fl/fl to Egr2 fl/fl. Data are from 15 (WT:Lyz2 Cre/+.Egr2 fl/fl) or 16 (WT:Egr2 fl/fl) mice per group pooled from three independent experiments. **** p<0.0001 (Student’s t-test with Holm-Sidak correction).
D. Contribution of Egr2 fl/fl BM to splenic red pulp F4/80hi macrophages and F4/80-defined mononuclear phagocytes in adipose tissue from chimeric in C. Spleen data are from 15 (WT:Lyz2 Cre/+.Egr2 fl/fl) or 16 (WT:Egr2 fl/fl) mice per group pooled from three independent experiments and adipose data from 5 (WT:Lyz2 Cre/+.Egr2 fl/fl) or 6 (WT:Egr2 fl/fl) mice per group pooled from two independent experiments.
E. Representative expression of CD11c, SiglecF and EpCAM by WT- and Egr2 fl/fl-derived alveolar macrophages in WT:Egr2 fl/fl or WT:Lyz2 Cre/+.Egr2 fl/fl chimeric mice. Shaded histograms represent FMO controls.
F. Mean fluorescent intensity (MFI) of CD11c, SiglecF, EpCAM and C/EBPβ expression by WT- and Egr2 fl/fl-derived alveolar macrophages in WT:Egr2 fl/fl or WT:Lyz2 Cre/+.Egr2 fl/fl chimeric mice. Data represent 10 mice per group from one experiment of three performed. **** p<0.0001 (One-way ANOVA followed by Tukey’s multiple comparisons post-test).
Symbols represent individual mice in all graphs and error bars represent the standard deviation.
Bone marrow-derived monocytes replenish the alveolar macrophage niche following lung injury
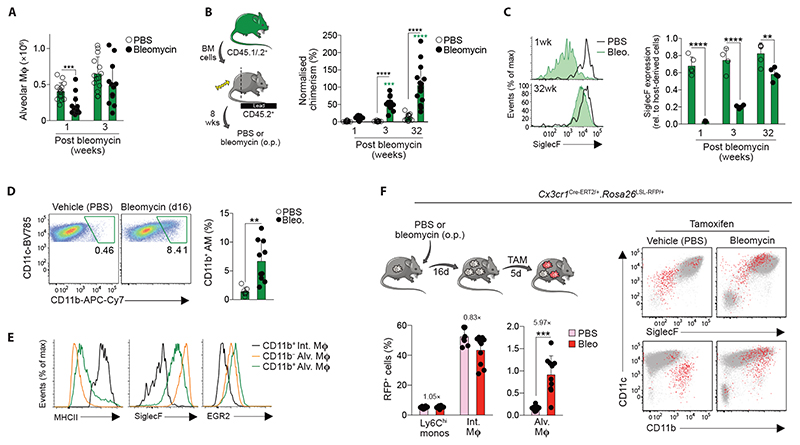
Loss of tissue resident macrophages is a frequent consequence of inflammation, including in the lung (40). Thus, given that Egr2-deficient macrophages failed to replenish the alveolar niche following radiation treatment, we next sought to determine if EGR2 plays a role in macrophage repopulation following lung injury. The chemotherapeutic agent bleomycin is a common model of chronic lung injury and self-resolving pulmonary fibrosis (41), which is characterised by initial loss of alveolar macrophages during the inflammatory phase (day 7), followed by repopulation during the fibrotic and resolution phases (from day 14 onwards) (Figure 7A). To determine if bone marrow-derived monocytes contribute to the alveolar macrophage compartment following bleomycin-induced injury, we used tissue protected bone marrow chimeric mice to assess replenishment kinetics without exposing the lung to the additional insult of ionising radiation (Figure 7B). Consistent with previous studies (42), we found that bleomycin instillation led to progressive replacement of resident alveolar macrophages by BM-derived cells, with the entire alveolar macrophage compartment being replaced at 32 weeks post injury (Figure 7B). Interestingly, recently arrived, monocyte-derived alveolar macrophages expressed low-intermediate levels of SiglecF, with acquisition of SiglecF requiring long-term residence in the airway (Figure 7C).
Figure 7. Monocyte-derived, CX3CR1+ parenchymal macrophages can replenish the alveolar macrophage niche following injury.
A. Absolute numbers of alveolar macrophages 1- and 3-weeks following bleomycin administration or PBS vehicle control. Data are pooled from two independent experiments at each time point with 13-15 mice per group. ***p<0.001 (unpaired Student’s t test with Holm-Sidak correction).
B. Non-host chimerism of alveolar macrophages in tissue protected bone marrow chimeric mice at 1-, 3- or 32-weeks following administration of bleomycin or PBS vehicle control. Chimerism is normalised to Ly6Chi blood monocytes. Data are pooled from two independent experiments at each time point with 13-15 mice per group. ***p<0.001, ****p<0.0001 (Two-way ANOVA with Tukey’s multiple comparisons test)
C. Expression of SiglecF by CD11chiCD11blo alveolar macrophages from the lung of mice in B. at 1 week and 32 weeks post bleomycin or PBS administration. Data are from one of two independent experiments at each time point with 4 mice per group. ***p<0.001, ****p<0.0001 (Two-way ANOVA with Tukey’s multiple comparisons test).
D. Representative expression of CD11c and CD11b by CD11chiCD64+ cells obtained by BAL from WT mice two weeks after instillation of bleomycin or vehicle control (left). Graph shows the mean frequency of CD11b+ alveolar macrophages (right). Data are pooled from two independent experiments with 7 (PBS) or 10 (bleomycin) mice per group. **p<0.01 (unpaired Student’s t test).
E. Representative expression of MHCII, SiglecF and EGR2 by CD11b+ interstitial macrophages and CD11b-defined CD11chi alveolar macrophages.
F. Experimental scheme for the induction of lung injury and tamoxifen administration in Cx3cr1 Cre-ERT2/+.Rosa26 LSL-RFP/+ fate mapping mice. Lower graphs show the levels of recombination in Ly6Chi monocytes, CD64+ interstitial macrophages and alveolar macrophages from Cx3cr1 Cre-ERT2/+.Rosa26 LSL-RFP/+ mice administered bleomycin or vehicle control. Representative expression of CD11c, SiglecF and CD11b by RFP+ (red) or RFP– (grey) cells present in the BAL fluid of Cx3cr1 Cre-ERT2/+.Rosa26 LSL-RFP/+ mice 3 weeks after bleomycin or vehicle instillation. Graphs show the mean fluorescent intensity (MFI) of CD11c and SiglecF expression by RFP+ cells.
Symbols represent individual mice in all graphs and error bars represent the standard deviation.
We next interrogated this process further to determine if monocyte-derived macrophages that accumulate in the lung parenchyma during injury can subsequently mature into alveolar macrophages during tissue repair (42, 43). Indeed, during the recovery phase of disease, we noted the presence of cells with features of both alveolar and interstitial macrophages (CD11chiCD11b+ MHCII+CD64hi) in the BAL fluid (Figure 7D), and these cells expressed intermediate levels of SiglecF (Figure 7E), indicative of recent monocyte origin. To examine the relationship of these ‘hybrid’ cells found in the airways to elicited monocyte-derived macrophages in the lung parenchyma more directly, we performed fate mapping studies using Cx3cr1 Cre-ERT2/+.Rosa26 LSL-RFP/+ reporter mice, in which administration of tamoxifen leads to irreversible expression of RFP by CX3CR1 expressing cells (44, 45) (Figure 7F). Administration of tamoxifen led to labelling of 40-50% of CD11b+ parenchymal macrophages in both healthy lung and at d21 post bleomycin administration (Figure 7F). No recombination was seen in Cx3cr1 Cre-ERT2/+.Rosa26 LSL-RFP/+ mice in the absence of tamoxifen (Supplementary Figure 8A). Although very low levels of recombination were detected in control alveolar macrophages, a clear population of RFP+ cells could be detected in the BAL of the recipients of bleomycin following tamoxifen treatment (Figure 7F). As monocytes are poorly labelled in this system and Cx3cr1 levels do not change in bona fide resident alveolar macrophages in response to bleomycin treatment (Supplementary Figure 8B), these RFP+ cells likely represent fate-mapped, monocyte-derived CX3CR1+ cells. In line with this, RFP+ cells had a ‘hybrid’ CD11chiCD11b+SiglecFint profile, supporting the idea that these represent transitional cells (Figure 7F). Thus, following bleomycin-induced injury, the alveolar macrophage compartment is restored, in part, by monocytes that transition through a CX3CR1hi state.
EGR2 is indispensable for alveolar macrophage repopulation and tissue repair following lung injury
Given that transitional CD11b+SiglecFint cells also expressed EGR2, contrasting with its restriction to SiglecFhi alveolar macrophages in health (Figure 7E), we examined whether EGR2 is necessary for the replenishment of the alveolar niche during recovery from bleomycin-induced injury. To do this, we administered bleomycin to Lyz2 Cre/+.Egr2 fl/fl mice and their Egr2 fl/fl littermates and assessed macrophage dynamics in total lung digests. The inflammatory phase of this disorder (day 7) was associated with accumulation of CD11b+ macrophages and this occurred to the same extent in both strains (Figure 8A, B). Consistent with recent reports (46), the CD11b+CD64+ interstitial macrophage population was heterogeneous during the fibrotic phase of disease (d14-d21), with MHCII+ and MHCIIloCD36+Lyve1+ subsets. This pattern was identical in between Egr2 fl/fl and Lyz2 Cre/+.Egr2 fl/fl groups, as were the numbers of Ly6Chi monocytes and neutrophils (Supplementary Figure 9A-C). We did however detect a significant reduction in eosinophils in the lung of Lyz2 Cre/+.Egr2 fl/fl mice compared with Egr2 fl/fl littermates, despite eosinophils lacking EGR2 expression and no differences in the level of eosinophil chemoattractant CCL11 (Supplementary Figure 9D-F).
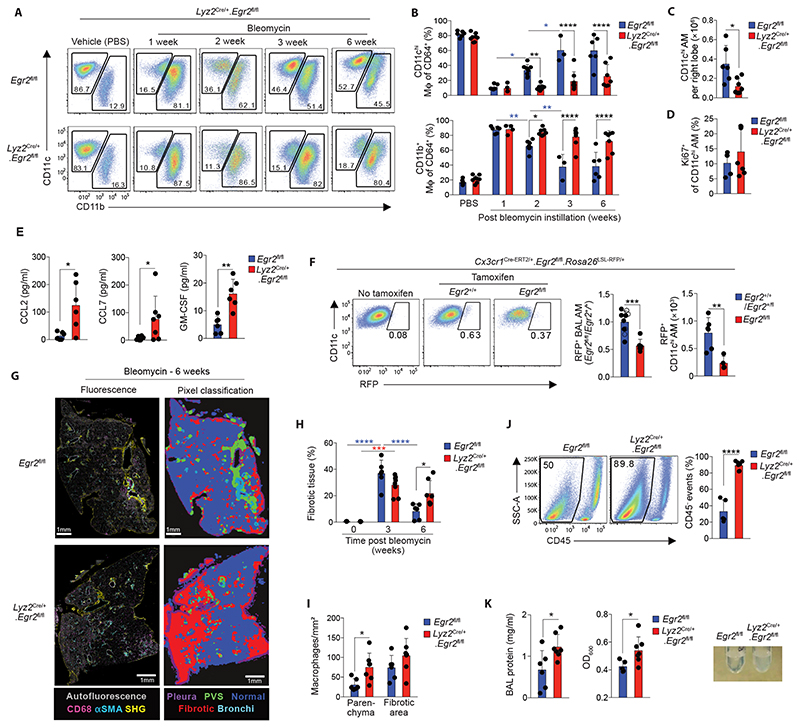
Figure 8. EGR2 is indispensable for the repopulation of the alveolar macrophage niche and tissue repair following lung injury.
A. Representative expression of CD11c and CD11b by live CD45+CD3–CD19–Ly6G–CD64+ cells from the lungs of Egr2 fl/fl and Lyz2 Cre/+.Egr2 fl/fl mice at 1, 2, 3 or 6 weeks post bleomycin or vehicle controls.
B. Frequency of CD11chiCD11blo alveolar macrophages and CD11cvarCD11b+ cells from mice in A. Data are pooled from at least two independent experiments at each time point with 3-7 mice per group. *p<0.05. **p<0.01, p<0.001, ****p<0.0001 (Two-way ANOVA with Tukey’s multiple comparisons test).
C. Absolute number of CD11chiCD11blo alveolar macrophages in lungs six weeks post bleomycin instillation. Data are pooled from two independent experiments with 6 (Egr2 fl/fl) or 7 (Lyz2 Cre/+.Egr2 fl/fl) mice per group. *p<0.05 (Mann Whitney test).
D. Frequency of Ki67+ CD11chiCD11blo alveolar macrophages in lungs six weeks post bleomycin instillation. Symbols represent individual mice. Data are pooled from two independent experiments with 6 (Egr2 fl/fl) or 7 (Lyz2 Cre/+.Egr2 fl/fl) mice per group.
E. CCL2, CCL7 and GM-CSF levels in BAL fluid obtained from Egr2 fl/fl and Lyz2 Cre/+.Egr2 fl/fl mice six weeks post bleomycin instillation. Data are pooled from two independent experiments with 6 mice per group. *p<0.05, Mann Whitney test (CCL2, CCL7), **p<0.01 (unpaired Student’s t test; GM-CSF).
F. Representative expression of RFP by CD11chiCD64+ alveolar macrophages present in the BAL fluid of Cx3cr1 Cre-ERT2/+.Rosa26 LSL-RFP/+.Egr2 fl/fl and their Cx3cr1 Cre-ERT2/+.Rosa26 LSL-RFP/+.Egr2 +/+ (open circles) or Cx3cr1 Cre-ERT2/+.Rosa26 LSL-RFP/+.Egr2 fl/+ (solid circles) controls 3 weeks following instillation of bleomycin or vehicle control. Graphs show the relative frequency (left) or absolute number (right) of RFP+ alveolar macrophages present in the BAL fluid. Data are from one experiment of two (number) with 6 (Egr2 +/+ [open symbols]/Egr2 fl/+ [filled symbols]) or 4 mice (Egr2 fl/fl) per group, or pooled from two independent experiments (frequency) at each time point with 10 (Egr2 +/+ [open symbols]/Egr2 fl/+ [filled symbols]) or 6 (Egr2 fl/fl) per group. ** p<0.01, ***p<0.001 (Unpaired Student’s t test).
G. 2-photon fluorescence imaging of lung tissue from adult Egr2 fl/fl and Lyz2 Cre/+.Egr2 fl/fl mice 6 weeks following bleomycin administration. Sections were stained with CD68, aSMA and DAPI. Autofluorescence is depicted in grey and collagen was detected by second harmonic generation (SHG). Pixel classification was used to segment lung regions of interest: (1) normal lung parenchyma/alveolar tissue, (2) pathologic/fibrotic tissue and (3) collagen rich areas (perivascular/bronchial spaces and pleura) were segmented to avoid false fibrotic region detection.
H. Quantification of fibrotic score of lung tissue from Egr2 fl/fl and Lyz2 Cre/+.Egr2 fl/fl 3 or 6 weeks following bleomycin administration or PBS controls (from 3 week time point). See Fig. S10. Data are pooled from two independent experiments with 6 (Egr2 fl/fl) or 7 (Lyz2 Cre/+.Egr2 fl/fl) mice per group. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001 (Two-way ANOVA followed by Tukey’s multiple comparisons test).
I. Quantification of macrophage density in the parenchyma and fibrotic areas of lung tissue from Egr2 fl/fl and Lyz2 Cre/+.Egr2 fl/fl 6 weeks following bleomycin administration. See Fig. S10. Data are pooled from two independent experiments with 6 (Egr2 fl/fl) or 7 (Lyz2 Cre/+.Egr2 fl/fl) mice per group. *p<0.05 (Student’s t test with Holm-Sidak correction for multiple tests).
J. SSC-A profile and expression of CD45 by BAL obtained from Egr2 fl/fl and Lyz2 Cre/+.Egr2 fl/fl 6 weeks following bleomycin administration. Graph shows the mean frequency of CD45+ cells amongst all live, single events. Symbols represent individual mice. Data are pooled from two independent experiments with 5 (Egr2 fl/fl) or 7 (Lyz2 Cre/+.Egr2 fl/fl) mice per group. ****p<0.0001 (unpaired Student’s t test).
K. Total protein concentration (left), turbidity (centre) and representative pictures (right) of BAL fluid from Egr2 fl/fl and Lyz2 Cre/+.Egr2 fl/fl 6 weeks following bleomycin administration. Symbols represent individual mice. Data are pooled from two independent experiments with 6 (Egr2 fl/fl) or 7 (Lyz2 Cre/+.Egr2 fl/fl) mice per group. *p<0.05 (Mann Whitney test).
Symbols represent individual mice in all graphs and error bars represent the standard deviation.
A reduction in alveolar macrophages was observed in both groups on day 7 after administration of bleomycin. Although this began to be restored by day 14 in Egr2 fl/fl control mice, this did not occur in Lyz2 Cre/+.Egr2 fl/fl mice and indeed, the alveolar macrophage compartment remained significantly reduced in Lyz2 Cre/+.Egr2 fl/fl mice compared with Egr2 fl/fl littermates even after 6 weeks (Figure 8A-C), suggesting EGR2 is indispensable for the repopulation of the alveolar macrophage niche following bleomycin-induced injury. The lack of repopulation in Lyz2 Cre/+.Egr2 fl/fl mice did not appear to reflect an inability of Egr2-deficient macrophages to proliferate, as the proportion of Ki67+ proliferating cells was equivalent across both strains (Figure 8D). Equally, this also did not reflect a lack of chemoattractants in the airways to recruit monocyte-derived cells, as both CCL2 and CCL7 were actually elevated in Lyz2 Cre/+.Egr2 fl/fl mice compared with control littermates (Figure 8E). Similarly, GM-CSF levels were elevated in the BAL fluid of Lyz2 Cre/+.Egr2 fl/fl mice, ruling out the possibility that lack of appropriate growth factors is responsible for defective alveolar macrophage differentiation in the absence of EGR2 (Figure 8E). Instead, these data suggested that Egr2 deficiency led to an intrinsic inability of bone marrow-derived cells to repopulate the macrophage niche. To test this directly, we crossed Cx3cr1 Cre-ERT2/+.Rosa26 LSL-RFP/+ mice with Egr2 fl/fl mice to allow for temporal RFP labelling of CX3CR1-expressing cells and Egr2 deficiency in the same animal. We administered tamoxifen during the period of alveolar macrophage reconstitution (d16 to d21) and assessed the presence of RFP-labelled cells amongst alveolar macrophages. Although labelling efficiencies were low, most likely reflecting the short period of tamoxifen induction and the dynamic nature of macrophage repopulation, compared with tamoxifen-treated controls (Cx3cr1 Cre-ERT2/+.Rosa26 LSL-RFP/+.Egr2 +/+ or Cx3cr1 Cre-ERT2/+.Rosa26 LSL-RFP/+.Egr2 fl/+ mice), we found a marked reduction in the frequency of RFP+ alveolar macrophages in the BAL of tamoxifen treated Cx3cr1 Cre-ERT2/+.Rosa26 LSL-RFP/+.Egr2 fl/fl mice during lung repair (Figure 8F), demonstrating that EGR2 controls the post-injury repopulation of the alveolar macrophage compartment by CX3CR1+ cells.
To determine the consequence of the failure of Egr2-deficient cells to reconstitute the alveolar niche, we assessed the fibrotic response and subsequent repair processes in Lyz2 Cre/+.Egr2 fl/fl mice. Notably, we did not detect differences in the degree of fibrosis or expression of key genes associated with fibrosis, including Col3a1 and Pdgfrb between Lyz2 Cre/+.Egr2 fl/fl mice and their Egr2 fl/fl littermate controls at day 21, a time considered ‘peak’ fibrosis (Figure 8H, Supplementary Figure 10A, B). However, analysis at 6 weeks post bleomycin showed that whereas the Egr2 fl/fl mice had largely repaired their lungs, Lyz2 Cre/+.Egr2 fl/fl mice had defective repair evidenced by persistent fibrosis and architectural damage (Figure 8G, H, Supplementary Figure 10A). This was paralleled by elevated numbers of macrophages in the lung parenchyma (Figure 8I, Supplementary Figure 10A) and parenchymal macrophage persistence correlated with the degree of fibrosis (Supplementary Figure 10C). Furthermore, homeostasis failed to be restored in the airways. Flow cytometric analysis of BAL fluid revealed that CD45+ leukocytes comprised only 10% of all events in Lyz2 Cre/+.Egr2 fl/fl mice compared with 60% in their Egr2 fl/fl littermates (Figure 8J). The vast majority of the CD45– fraction failed to express signature markers for cells of epithelial, endothelial or fibroblast origin, suggesting this may represent cellular debris, which could also be found amongst lung digests (Supplementary Figure 11A-C). This was paralleled by elevated BAL fluid protein levels and turbidity in the Lyz2 Cre/+.Egr2 fl/fl mice compared with controls, suggesting that the inability to replenish the alveolar macrophage niche following injury was associated with the development of alveolar proteinosis (Figure 8K). Thus, loss of EGR2-dependent, monocyte-derived alveolar macrophages leads to defective tissue repair, persistent cellular damage and failed restoration of lung homeostasis.
Discussion
Given the multifaceted role of macrophages in tissue homeostasis, inflammation and tissue repair, as well as many chronic pathologies, understanding the environmental signals and the downstream molecular pathways that govern macrophage differentiation is a key objective in the field of immunology. Here, we identify the transcription factor EGR2 as a selective and indispensable part of the tissue-specific differentiation of lung alveolar macrophages.
Our transcriptomic analysis identified EGR2 as a feature of murine lung alveolar macrophages, a finding consistent with previous studies using bulk transcriptomic analysis (5, 12) and a recent study using a similar scRNA-seq based approach (47). Our finding that EGR2 appears to represent an evolutionarily conserved transcriptional regulator is also consistent with previous studies (37, 48). While EGR2 has been implicated in controlling monocyte to macrophage differentiation in the past, these studies have often reached discrepant conclusions (20, 21). This could reflect the fact that most studies examining the role of EGR2 in monocyte-macrophage differentiation have employed in vitro culture systems due to the postnatal lethality of global Egr2-deficient mice (22, 23). By generating Lyz2 Cre.Egr2 fl/fl and Fcgr1 iCre.Egr2 fl/fl mice, we circumvented this lethality and demonstrated that EGR2 controls a large proportion of the alveolar macrophage ‘signature’. This is consistent with recent epigenetic analysis showing an overrepresentation of EGR motifs in the genes defining alveolar macrophages (49). Importantly, although previous work has suggested that there is redundancy between EGR family members, specifically EGR1 and EGR2, we found Egr1 expression was unaffected by EGR2 deficiency and was unable to rescue alveolar macrophage differentiation.
Notably, if assessed simply on the basis of their CD11chiCD11blo profile, the absolute number of alveolar macrophages was equivalent between adult Egr2 fl/fl and Lyz2 Cre.Egr2 fl/fl mice. This could explain why a recent study using an independent strain of Lyz2 Cre.Egr2 fl/fl mice concluded that EGR2 is dispensable for macrophage differentiation (37). Alternatively, this could reflect that the majority of their studies involved in vitro generated, CSF1-dependent macrophages. Indeed, we found that Egr2-deficient monocytes matured into macrophages equally well when cultured in vitro with CSF-1. However, in our hands, CSF-1 led to poor upregulation of EGR2 in maturing macrophages in vitro. Instead, we identified GM-CSF as a potent inducer of EGR2 expression, a finding consistent with the dependence of alveolar macrophages on alveolar epithelial cell-derived GM-CSF for their development and survival (7, 8). Importantly, deficiencies in Lyz2 Cre.Egr2 fl/fl mice did not reflect consistent differences in the expression of GM-CSF signalling molecules. EGR2 is often referred to as a feature of alternative macrophage activation on the basis that IL-4 can drive EGR2 upregulation in vitro in a STAT6-dependent manner (36, 37). However, we ruled out a role for IL-4 in EGR2 regulation in alveolar macrophages. Thus, the IL-4–IL-4R axis is sufficient, but not necessary, for inducing EGR2 expression in vivo. TGF-β also induced EGR2 and we confirmed that TGF-βR signalling is indispensable for the development of alveolar macrophages (9). If and how GM-CSF and TGF-β cooperate to promote alveolar macrophage differentiation is incompletely understood, however they both induce expression of PPAR-γ (8, 9) and Pparg-deficient alveolar macrophages expressed reduced EGR2 (8), suggesting EGR2 lies downstream of PPAR-γ. Whether an initial TGFβ signal is needed to induce EGR2 during alveolar macrophage development or if continual TGFβR signalling is needed to maintain EGR2 remains to be determined and will require new transgenic systems to allow inducible deletion of TGFβR. Notably, while genetic ablation of Pparg, Csf2rb or Tgfbr2 leads to defects in the development and self-maintenance of alveolar macrophages, this was not replicated by Egr2 deficiency. Thus, the EGR2-dependent programme appears to represent a discrete part of alveolar macrophage differentiation. Consistent with this, mice with myeloid or macrophage deletion of Egr2 did not develop spontaneous alveolar proteinosis, suggesting EGR2 is redundant for regulation of surfactant. However, Egr2-deficient mice displayed functional deficiencies in the ability to control low dose S. pneumoniae infection. Although we cannot rule out the possibility that this reflects differences in the killing capacity of Egr2-deficient alveolar macrophages, genes encoding e.g. reactive oxygen and nitrogen species were unaffected by Egr2 deficiency. Instead, genes encoding key pathogen recognition receptors and opsonins, were significantly downregulated in the absence of EGR2. These included MARCO and the complement component C3, both of which have been shown to be crucial for the effective elimination of S. pneumoniae (35, 50). Indeed, opsonisation is a critical factor in optimizing bacterial clearance by alveolar macrophages in health and disease (51). Thus, it is clear that EGR2-dependent differentiation equips alveolar macrophages with the machinery to recognise and engulf pneumococci, and this may explain the recurrent pneumonias in individuals with mutations in EGR2 (22). In future work, it will be important to determine if this extends to other respiratory pathogens. Moreover, given that MARCO appears to define a discrete subset of CXCL2-expressing alveolar macrophages with elevated pro-inflammatory features in the context of fungal infection (52), it will be of interest to determine if all alveolar macrophages are equal in their ability to eliminate S. pneumoniae or if an analogous CXCL2+ subset with superior anti-bacterial capacity exists in this context.
Loss of alveolar macrophages is a common feature of lung inflammation or injury. Consistent with previous work (42, 53), we found that the principal mechanism of macrophage replenishment was through recruitment of BM-derived cells which mature into bona fide alveolar macrophages with time. Using Cx3cr1-based genetic fate mapping, we also showed that CX3CR1+MHCII+ cells with a hybrid phenotype could be found in the airways during the fibrotic phase of injury, suggesting that monocyte-derived macrophages that accumulate in the lung parenchyma following injury may replenish the alveolar macrophage niche. Although we cannot rule out that monocytes, including Ly6Clow monocytes (54), enter the airways during this phase to give rise to alveolar macrophages directly, the phenotype of the RFP+ transitional cells was more aligned with the phenotype of elicited, monocyte-derived macrophages in the parenchyma, including high levels of MHCII. Importantly, repopulation of the alveolar macrophage compartment was dependent on EGR2, with constitutive deletion of EGR2 severely blunting the engraftment of monocyte-derived cells into the alveolar macrophage niche. This contrasts with initial population of the developing alveolar niche by foetal liver-derived monocytes, where Egr2 deficiency does not affect the development of alveolar macrophages. This could indicate differential dependence of developmentally distinct monocytes on EGR2, or the presence of compensatory pathways during development that are not present during repopulation and further work is required to fully understand this.
Interestingly, although previous work has suggested that monocyte-derived alveolar macrophages are key pro-fibrotic cells (42, 53), fibrosis appeared to develop normally in Egr2-deficient mice, despite the near absence of monocyte-derived alveolar macrophages. The reason for the discrepancy in our findings and those of Misharin et al. (42) is unclear, but it could reflect differences in the systems used. For instance, the Misharin study exploited the dependence of alveolar macrophages on Caspase-8 to impede monocyte differentiation into alveolar macrophages by using Lyz2 Cre.Casp8 fl/fl and Itgax Cre.Casp8 fl/fl mice. However, deletion of Caspase-8 also affects the ability of interstitial macrophages to repopulate following depletion, meaning that Casp8 deficiency may have wider effects on lung macrophage behaviour than disrupting the differentiation of monocyte-derived alveolar macrophages. In contrast, EGR2 expression is restricted to alveolar macrophages and deletion does not affect the reconstitution of the interstitial macrophage compartment. The location of interstitial macrophages in the parenchyma adjacent to fibroblasts and their production of the fibroblast mitogen PDGF-aa, suggests that interstitial macrophages are likely to be key to the fibrotic process (43). Indeed, depletion of interstitial macrophages using Cx3cr1 Cre-ERT2.Rosa26 LSL-DTA mice reduces lung fibrosis (43), although as we show here, this will also target CX3CR1+ cells destined to become monocyte-derived alveolar macrophages. Nevertheless, our data show a clear role for monocyte-derived macrophages in tissue repair processes, as Lyz2 Cre.Egr2 fl/fl mice failed to repair the lung after injury, a finding consistent with an older study using non-specific, clodronate-mediated depletion of lung macrophages (55) and a recent study implicating ApoE-producing, monocyte-derived alveolar macrophages in lung fibrosis resolution (56). These results may help explain the development of restrictive pulmonary disease in individuals with mutations in EGR2 (22).
In summary, our results demonstrate that EGR2 is an evolutionarily conserved transcriptional regulator of alveolar macrophage differentiation, loss of which leads to major phenotypic, transcriptional and functional deficiencies. By identifying EGR2 as a transcriptional regulator, we have begun to dissect how common factors such as GM-CSF and TGFβ confer specificity during macrophage differentiation. Our work reveals how distinct molecular modules appear to control the homeostatic versus immune protective functions of alveolar macrophages, which may be beneficial to the host by allowing these functions to be controlled independently. Importantly, given that recent studies using human systems have proposed that alveolar macrophage maintenance in humans requires monocyte input (57, 58), EGR2 may play a particularly important role in alveolar macrophage differentiation in man. Thus, further work is required to fully understand the molecular pathways downstream of EGR2 and whether this is conserved between mouse and humans, and if EGR2 plays distinct roles in different pathological settings.
Materials and Methods
Study Design
We performed phenotypic, transcriptomic and functional analysis of alveolar macrophages in the context of Egr2 deficiency to assess the features controlled by this transcription factor. Fate mapping techniques were used to assess the macrophage dynamics during bleomycin-induced injury and to test the cell intrinsic effects of Egr2 deficiency. Infection with Streptococcus pneumoniae was used to assess the immune protective features of alveolar macrophages. All imaging and associated analysis was blinded. Experimental replicate details are provided in figure legends.
Experimental Animals
Mice were bred and maintained in SPF facilities at the University of Edinburgh or University of Glasgow, UK. All experimental mice were age matched and both sexes were used throughout the study. The mice used in each experiment is documented in the appropriate figure legend. Experiments performed at UK establishments were permitted under license by the UK Home Office and were approved by the University of Edinburgh Animal Welfare and Ethical Review Body. Genotyping was performed by Transnetyx using real-time PCR. Mouse strains are detailed in Table S2.
Human cells
BAL fluid was obtained from patients attending the Edinburgh Lung Fibrosis Clinic. Ethical permission was granted from the NHS Lothian Research ethics board (LREC 07/S1102/20 06/S0703/53). BAL fluid cells were stained for flow cytometric analysis with antibodies listed in Table S3.
Tamoxifen-based fate mapping
For induction of Cre activity in Cx3cr1 Cre-ERT2/+ mice, tamoxifen was dissolved in sesame oil overnight at 50mg/ml in a glass vial and administered by oral gavage at 5mg per day for five consecutive days. In bleomycin experiments, tamoxifen was administered from d16 post bleomycin administration for 5 days. Fresh tamoxifen was prepared for each experiment.
Bleomycin lung injury
Bleomycin sulphate (Cayman chemicals) was prepared by first dissolving in sterile DMSO (Sigma) and further in sterile PBS at 0.66mg/ml. 8-12-week-old Lyz2 Cre.Egr2 fl/fl and Egr2 fl/fl littermate controls were anaesthetised with isofluorane and administered 50μl bleomycin (33μg) or vehicle control (DMSO/PBS) by oropharyngeal aspiration.
Streptococcus pneumoniae infection
Lyz2 Cre/+.Egr2 fl/fl mice and Egr2 fl/fl littermate control male mice (8–14-week-old) were anaesthetised ketamine/medetomidine and inoculated intratracheally with 50μl of PBS containing 104 CFU S. pneumoniae (capsular type 2 strain D39). 100μl of inoculum was plated on blood agar to determine exact dose. Mice were culled 14 h later and BAL fluid collected by lavage performed using sterile PBS. 100μl of lavage fluid was cultured for bacterial growth for 24 h. The remaining lavage fluid was centrifuged at 400g for 5 mins and the resulting cells counted and prepared for flow cytometric analysis.
BM chimeric mice
To generate WT:Lyz2 Cre.Egr2 fl/fl mixed chimeras, CD45.1+CD45.2+ WT mice were lethally irradiated with two doses of 5 Gy 1 hour apart before being reconstituted immediately WT (CD45.1+) and Lyz2 Cre/+.Egr2 fl/fl or Egr2 fl/fl (CD45.2+) bone marrow at a ratio of 1:1.Chimerism was assessed at 8 weeks after reconstitution.
Processing of tissues
Mice were sacrificed by overdose with sodium pentobarbitone followed by exsanguination. Mice were then gently perfused with PBS through the heart. In lung injury/fibrosis experiments, the right lobe was tied off, excised and stored in RPMI with 10% FCS on ice before being prepared for enzymatic digestion (see below). The left lung lobe was inflated with 600μl 4% PFA through an intra-tracheal canula. The trachea was tied off with thread and the lung and heart carefully excised and stored in 4% PFA overnight. Fixed lung tissue was moved to 70% ethanol before being processed for histological assessment. Right lung lobes were chopped finely and digested in pre-warmed RPMI1640 with ‘collagenase cocktail’ (0.625mg ml−1 collagenase D (Roche), 0.425mg ml−1 collagenase V (Sigma-Aldrich), 1mg ml−1 Dispase (ThermoFisher), and 30 U ml−1 DNase (Roche Diagnostics GmbH)) for 25 minutes in a shaking incubator at 37°C before being passed through a 100μm strainer followed by centrifugation at 300g for 5 mins. Erythrocytes were lysed using Red Blood Cell Lysing Buffer Hybri-Max (Sigma-Aldrich) for 2mins at room temperature, washed in FACS buffer (2% FCS/2mM EDTA/PBS) and resuspended in 5mls of FACS buffer, counted and kept on ice until staining for flow cytometry. In some experiments BAL fluid was obtained by lavaging the lungs with 0.8ml DPBS/2mM EDTA via an intra-tracheal catheter. This was repeated three times, with the first wash kept separate for analysis of BAL cytokines, turbidity and protein concentration. To obtain splenic leukocytes, spleens were chopped and digested in HBSS with 1mg/ml collagenase D for 45 mins in a shaking incubator at 37°C before being passed through a 100μm strainer followed by centrifugation at 400g for 5 mins. Erythrocytes were lysed as above. To obtain liver leukocytes, livers were perfused through the inferior vena cava with sterile PBS and liver tissue excised. Livers were then chopped finely and digested in pre-warmed collagenase ‘cocktail’ (5ml/liver) for 30 minutes in a shaking incubator at 37°C before being passed through an 100μm filter. Cells were washed twice in 50ml ice cold RPMI followed by centrifugation at 300g for 5 mins (59). Supernatants were discarded and erythrocytes were lysed. Epidermal and dermal leukocytes were isolated as described previously (60). Colonic and adipose tissue leukocytes were isolated as described previously (61–63). To obtain peritoneal leukocytes, the peritoneal cavity was lavaged with RPMI containing 2mM EDTA and 10mM HEPES (both ThermoFisher) as described previously (64). Cells were resuspended in FACS buffer, counted and kept on ice until staining for flow cytometry.
Flow cytometry
For analysis of unfixed cells, cells were first incubated with 0.025 μg anti-CD16/32 (2.4G2; Biolegend) for 10mins on ice to block Fc receptors and then stained with a combination of the antibodies detailed in Table S3. Where appropriate, cells were subsequently stained with streptavidin-conjugated BV650 (Biolegend). Dead cells were excluded using DAPI or 7-AAD (Biolegend) added 2mins before acquisition. When assessing intracellular markers, cells were first washed in PBS and then incubated with Zombie NIR fixable viability dye (Biolegend) for 10mins at room temperature protected from light before following the approach detailed above. Following the final wash step, cells were subsequently fixed and permeabilized using FoxP3/Transcription Factor Staining Buffer Set (eBioscience), and intracellular staining performed using antibodies detailed in Table S3. Samples were acquired using a FACS LSRFortessa or AriaII using FACSDiva software (BD) and analyzed with FlowJo software (version 9 or 10; Tree Star). Analysis was performed on single live cells determined using forward scatter height (FCS-H) versus area (FSC-A) and negativity for viability dyes. mRNA was detected by flow cytometry using PrimeFlow technology (ThermoFisher) using probes against Spp1 (AF647) according to the manufacturer’s guidelines. For staining controls in PrimeFlow analysis, the Target Probe Hybridization step was omitted with all other steps identical to samples.
BAL fluid analysis
The first BAL wash was centrifuged at 400g for 5mins and supernatant removed and stored at -80°C until analysis. Total protein concentrations in BAL fluid were measured by BCA Protein Assay according to the manufacturer’s instructions (ThermoFisher). Turbidity was determined following gentle mixing by diluting 25ul of sample with 75ul DPBS and measuring the optical density of 600nm and multiplying by the dilution factor. BAL cytokines were measured using 50ul undiluted sample and the Cytokine & Chemokine 26-Plex ProcartaPlex (Panel 1) assay according to manufacturer’s guidelines (ThermoFisher).
Lung histology
Formalin-inflated lungs were fixed overnight in 4% buffered formalin and stored in 70% ethanol. Paraffin-embedded sections of mouse lungs were stained with Masson’s trichome as per the manufacturer’s guidelines.
Statistics
Statistics were performed using Prism 7 (GraphPad Software). The statistical test used in each experiment is detailed in the relevant figure legend.
Details of transcriptional analysis and imaging can be found in the Supplementary Materials and Methods.
Supplementary Material
One Sentence Summary.
EGR2 controls alveolar macrophage function in health and disease.
Acknowledgements
We are grateful to Prof. Ping Wang and Dr. Su-Ling Li, QMUL, London for the kind gift of the Egr2-floxed strain and to Prof. Rick Maziels, University of Glasgow for the provision of Il4ra-deficient mice. Flow cytometry data were generated with support from the QMRI Flow Cytometry and Cell Sorting Facility, University of Edinburgh. mRNA sequencing was performed by Edinburgh Genomics, The University of Edinburgh. Edinburgh Genomics is partly supported through core grants from NERC (R8/H10/56), MRC (MR/K001744/1) and BBSRC (BB/J004243/1). Genewiz performed the bulk RNA-seq analysis. We would like to thank Beth Henderson for technical expertise in setting up 10X sequencing; Dr. Jordan Portman for initial processing of the scRNAseq raw data; the ShIELD (Sheffield, Edinburgh, Newcastle and Birmingham) consortium for access to bacterial stocks; and Dr. Brian McHugh and Dr Manuel Sánchez García for help and advice on bacterial studies. We would also like to thank the Core Services and Advanced Technologies at the Cancer Research UK Beatson Institute, with particular thanks to the Beatson Advanced Imaging Resource (BAIR). Finally, we would like to thank the Bioresearch and Veterinary Services at the University of Edinburgh for husbandry of our mice and other technical assistance. Servier Medical Art was used for the generation of some of the graphics.
Funding
This research was funded by a Sir Henry Dale Fellowship jointly funded by the Wellcome Trust and the Royal Society [Grant number 206234/Z/17/Z to C.C.B]. CGH is supported by a University of Edinburgh Chancellor’s Fellowship and CGH and RC are supported by Worldwide Cancer Research. The Lyz2Cre.Egr2fl/fl line was originally generated with funding from the Medical Research Council UK (MR/L008076/1 to S.J.J). GRJ is funded by a Wellcome Trust Clinical Career Development Fellowship (220725/Z/20/Z). SRW is funded by a Wellcome Trust Senior Clinical Fellowship (209220). PTKS received funding from the MRC (MR/N024524/1). FF and LMC are funded by Cancer Research UK core funding (A23983 & A31287), and Breast Cancer Now (2019DecPR1424). This research was funded in whole or in part by The Wellcome Trust [Grant number 206234/Z/17/Z], a cOAlition S organization. The author will make the Author Accepted Manuscript (AAM) version available under a CC BY public copyright license.
Footnotes
Author Contributions. J. McC. performed experiments, analysed the data and edited the manuscript. P.M.K. Performed scRNA-seq analysis. F.F. Designed and performed immunofluorescence analysis of lung tissue. W. T’J. Performed experiments, analysed data and edited the manuscript. L. M. H. & C.M.M performed experiments and analysed the data. R. C. and C. G. H provided bioinformatic analysis of ImmGen data. A.S.M. provided advice on and help with the execution of fibrosis experiments. A. H. performed analysis. D.H. advised and helped with the execution of infection experiments. G.R.J. performed histological analysis of lung sections. S.J.J. generated the Lyz2 Cre.Egr2 fl/fl mice. N. H. provided access to human bronchoalveolar samples. S.H. and B.M. generated and provided the Fcgr1 iCre mice. S.R.W. advised on the design of fibrosis experiments and provided reagents for infection experiments. D.D. advised on the design and execution of infection experiments. P.T.K.S. advised and provided infrastructure to perform scRNA-seq analysis. L.M.C. supervised F.F., advised and provided infrastructure and reagents to perform multi-parameter immunofluorescence analysis. C.C.B. conceived and performed experiments, analysed and interpreted the data, wrote the manuscript, obtained funding and supervised the project.
Competing Interests. The authors declare no competing interests.
Data and materials availability
All data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials, and RNA-seq data have been deposited in National Center for Biotechnology Information Gene Expression Omnibus public database (www.ncbi.nlm.nih.gov/geo/). Population-level RNA-seq (accession code: GSE182044) and scRNA-seq (accession code: GSE181894). Fcgr1iCre mice are available from Prof. Bernard Malissen under a material transfer agreement with the Centre d'lmmunologie de Marseille-Luminy, Aix Marseille Université. Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Calum Bain (calum.bain@ed.ac.uk).
References
- 1.Ogger PP, Byrne AJ. Macrophage metabolic reprogramming during chronic lung disease. Mucosal Immunol. 2021;14:282–295. doi: 10.1038/s41385-020-00356-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bost P, Giladi A, Liu Y, Bendjelal Y, Xu G, David E, Blecher-Gonen R, Cohen M, Medaglia C, Li H, Deczkowska A, et al. Host-Viral Infection Maps Reveal Signatures of Severe COVID-19 Patients. Cell. 2020;181:1475–1488.:e12. doi: 10.1016/j.cell.2020.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liao M, Liu Y, Yuan J, Wen Y, Xu G, Zhao J, Cheng L, Li J, Wang X, Wang F, Liu L, et al. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat Med. 2020;26:842–844. doi: 10.1038/s41591-020-0901-9. [DOI] [PubMed] [Google Scholar]
- 4.Merad M, Martin JC. Pathological inflammation in patients with COVID-19: a key role for monocytes and macrophages. Nature reviews. 2020;20:355–362. doi: 10.1038/s41577-020-0331-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mass E, Ballesteros I, Farlik M, Halbritter F, Günther P, Crozet L, Jacome-Galarza CE, Händler K, Klughammer J, Kobayashi Y, Gomez Perdiguero E, et al. Specification of tissue-resident macrophages during organogenesis. Science (New York, NY) 2016;353 doi: 10.1126/science.aaf4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gomez Perdiguero E, Klapproth K, Schulz C, Busch K, Azzoni E, Crozet L, Garner H, Trouillet C, de Bruijn MF, Geissmann F, Rodewald H-R. Tissue-resident macrophages originate from yolk-sac-derived erythro-myeloid progenitors. Nature. 2015;518:547–551. doi: 10.1038/nature13989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guilliams M, De Kleer I, Henri S, Post S, Vanhoutte L, DePrijck S, Deswarte K, Malissen B, Hammad H, Lambrecht BN. Alveolar macrophages develop from fetal monocytes that differentiate into long-lived cells in the first week of life via GM-CSF. The Journal of experimental medicine. 2013;210:1977–1992. doi: 10.1084/jem.20131199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schneider C, Nobs SP, Kurrer M, Rehrauer H, Thiele C, Kopf M. Induction of the nuclear receptor PPAR-γ by the cytokine GM-CSF is critical for the differentiation of fetal monocytes into alveolar macrophages. Nature immunology. 2014;15:1026–1037. doi: 10.1038/ni.3005. [DOI] [PubMed] [Google Scholar]
- 9.Yu X, Buttgereit A, Lelios I, Utz SG, Cansever D, Becher B, Greter M. The Cytokine TGF-β Promotes the Development and Homeostasis of Alveolar Macrophages. Immunity. 2017;47:903–912.:e4. doi: 10.1016/j.immuni.2017.10.007. [DOI] [PubMed] [Google Scholar]
- 10.Gautier EL, Chow A, Spanbroek R, Marcelin G, Greter M, Jakubzick C, Bogunovic M, Leboeuf M, Van Rooijen N, Habenicht AJ, Merad M, et al. Systemic analysis of PPARgamma in mouse macrophage populations reveals marked diversity in expression with critical roles in resolution of inflammation and airway immunity. J Immunol. 2012;189:2614–2624. doi: 10.4049/jimmunol.1200495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Okreglicka K, Iten I, Pohlmeier L, Onder L, Feng Q, Kurrer M, Ludewig B, Nielsen P, Schneider C, Kopf M. PPARγ is essential for the development of bone marrow erythroblastic island macrophages and splenic red pulp macrophages. The Journal of experimental medicine. 2021;218 doi: 10.1084/jem.20191314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gautier EL, Shay T, Miller J, Greter M, Jakubzick C, Ivanov S, Helft J, Chow A, Elpek KG, Gordonov S, Mazloom AR, et al. Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nature immunology. 2012;13:1118–1128. doi: 10.1038/ni.2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scott CL, T’Jonck W, Martens L, Todorov H, Sichien D, Soen B, Bonnardel J, De Prijck S, Vandamme N, Cannoodt R, Saelens W, et al. The Transcription Factor ZEB2 Is Required to Maintain the Tissue-Specific Identities of Macrophages. Immunity. 2018;49:312–325.:e5. doi: 10.1016/j.immuni.2018.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baker AD, Malur A, Barna BP, Ghosh S, Kavuru MS, Malur AG, Thomassen MJ. Targeted PPAR{gamma} deficiency in alveolar macrophages disrupts surfactant catabolism. J Lipid Res. 2010;51:1325–1331. doi: 10.1194/jlr.M001651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cain DW, O’Koren EG, Kan MJ, Womble M, Sempowski GD, Hopper K, Gunn MD, Kelsoe G. Identification of a tissue-specific, C/EBPβ-dependent pathway of differentiation for murine peritoneal macrophages. J Immunol. 2013;191:4665–4675. doi: 10.4049/jimmunol.1300581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rauschmeier R, Gustafsson C, Reinhardt A, A-Gonzalez N, Tortola L, Cansever D, Subramanian S, Taneja R, Rossner MJ, Sieweke MH, Greter M, et al. Bhlhe40 and Bhlhe41 transcription factors regulate alveolar macrophage self-renewal and identity. EMBO J. 2019;38:e101233. doi: 10.15252/embj.2018101233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Madissoon E, Wilbrey-Clark A, Miragaia RJ, Saeb-Parsy K, Mahbubani KT, Georgakopoulos N, Harding P, Polanski K, Huang N, Nowicki-Osuch K, Fitzgerald RC, et al. scRNA-seq assessment of the human lung, spleen, and esophagus tissue stability after cold preservation. Genome Biol. 2019;21(1) doi: 10.1186/s13059-019-1906-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.MacParland SA, Liu JC, Ma X-Z, Innes BT, Bartczak AM, Gage BK, Manuel J, Khuu N, Echeverri J, Linares I, Gupta R, et al. Single cell RNA sequencing of human liver reveals distinct intrahepatic macrophage populations. Nat Commun. 2018;9:4383. doi: 10.1038/s41467-018-06318-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Habib N, Avraham-Davidi I, Basu A, Burks T, Shekhar K, Hofree M, Choudhury SR, Aguet F, Gelfand E, Ardlie K, Weitz DA, et al. Massively parallel single-nucleus RNA-seq with DroNc-seq. Nat Methods. 2017;14:955–958. doi: 10.1038/nmeth.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laslo P, Spooner CJ, Warmflash A, Lancki DW, Lee H-J, Sciammas R, Gantner BN, Dinner AR, Singh H. Multilineage transcriptional priming and determination of alternate hematopoietic cell fates. Cell. 2006;126:755–766. doi: 10.1016/j.cell.2006.06.052. [DOI] [PubMed] [Google Scholar]
- 21.Carter JH, Tourtellotte WG. Early growth response transcriptional regulators are dispensable for macrophage differentiation. J Immunol. 2007;178:3038–3047. doi: 10.4049/jimmunol.178.5.3038. [DOI] [PubMed] [Google Scholar]
- 22.Swiatek PJ, Gridley T. Perinatal lethality and defects in hindbrain development in mice homozygous for a targeted mutation of the zinc finger gene Krox20. Genes Dev. 1993;7:2071–2084. doi: 10.1101/gad.7.11.2071. [DOI] [PubMed] [Google Scholar]
- 23.Topilko P, Levi G, Merlo G, Mantero S, Desmarquet C, Mancardi G, Charnay P. Differential regulation of the zinc finger genes Krox-20 and Krox-24 (Egr-1) suggests antagonistic roles in Schwann cells. J Neurosci Res. 1997;50:702–712. doi: 10.1002/(SICI)1097-4547(19971201)50:5<702::AID-JNR7>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 24.Clausen BE, Burkhardt C, Reith W, Renkawitz R, Förster I. Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Res. 1999;8(4):265–277. doi: 10.1023/a:1008942828960. [DOI] [PubMed] [Google Scholar]
- 25.Taillebourg E, Buart S, Charnay P. Conditional, floxed allele of the Krox20 gene. Genesis. 2002;32(2):112–113. doi: 10.1002/gene.10062. [DOI] [PubMed] [Google Scholar]
- 26.Neupane AS, Willson M, Chojnacki AK, Castanheira F Vargas E Silva, Morehouse C, Carestia A, Keller AE, Peiseler M, DiGiandomenico A, Kelly MM, Amrein M, et al. Patrolling Alveolar Macrophages Conceal Bacteria from the Immune System to Maintain Homeostasis. Cell. 2020;183:110–125.:e11. doi: 10.1016/j.cell.2020.08.020. [DOI] [PubMed] [Google Scholar]
- 27.Szigeti K, Wiszniewski W, Saifi GM, Sherman DL, Sule N, Adesina AM, Mancias P, Papasozomenos SC, Miller G, Keppen L, Daentl D, et al. Functional, histopathologic and natural history study of neuropathy associated with EGR2 mutations. Neurogenetics. 2007;8:257–262. doi: 10.1007/s10048-007-0094-0. [DOI] [PubMed] [Google Scholar]
- 28.Dranoff G, Crawford AD, Sadelain M, Ream B, Rashid A, Bronson RT, Dickersin GR, Bachurski CJ, Mark EL, Whitsett JA, et al. Involvement of granulocyte-macrophage colony-stimulating factor in pulmonary homeostasis. Science. 1994;264(5159):713–6. doi: 10.1126/science.8171324. [DOI] [PubMed] [Google Scholar]
- 29.Stanley E, Lieschke GJ, Grail D, Metcalf D, Hodgson G, Gall JA, Maher DW, Cebon J, Sinickas V, Dunn AR. Granulocyte/macrophage colony-stimulating factor-deficient mice show no major perturbation of hematopoiesis but develop a characteristic pulmonary pathology. Proc Natl Acad Sci USA. 1994;91(12):5592–6. doi: 10.1073/pnas.91.12.5592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martinez-Moczygemba M, Doan ML, Elidemir O, Fan LL, Cheung SW, Lei JT, Moore JP, Tavana G, Lewis LR, Zhu Y, Muzny DM, et al. Pulmonary alveolar proteinosis caused by deletion of the GM-CSFRalpha gene in the X chromosome pseudoautosomal region 1. The Journal of experimental medicine. 2008;205:2711–2716. doi: 10.1084/jem.20080759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Willinger T, Rongvaux A, Takizawa H, Yancopoulos GD, Valenzuela DM, Murphy AJ, Auerbach W, Eynon EE, Stevens S, Manz MG, Flavell RA. Human IL-3/GM-CSF knock-in mice support human alveolar macrophage development and human immune responses in the lung. Proc Natl Acad Sci USA. 2011;108:2390–2395. doi: 10.1073/pnas.1019682108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suzuki T, Arumugam P, Sakagami T, Lachmann N, Chalk C, Sallese A, Abe S, Trapnell C, Carey B, Moritz T, Malik P, et al. Pulmonary macrophage transplantation therapy. Nature. 2014;514:450–454. doi: 10.1038/nature13807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Preston JA, Bewley MA, Marriott HM, McGarry Houghton A, Mohasin M, Jubrail J, Morris L, Stephenson YL, Cross S, Greaves DR, Craig RW, et al. Alveolar Macrophage Apoptosis-associated Bacterial Killing Helps Prevent Murine Pneumonia. Am J Respir Crit Care Med. 2019;200:84–97. doi: 10.1164/rccm.201804-0646OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marriott HM, Daigneault M, Thompson AAR, Walmsley SR, Gill SK, Witcher DR, Wroblewski VJ, Hellewell PG, Whyte MKB, Dockrell DH. A decoy receptor 3 analogue reduces localised defects in phagocyte function in pneumococcal pneumonia. Thorax. 2012;67:985–992. doi: 10.1136/thoraxjnl-2012-201591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arredouani M, Yang Z, Ning Y, Qin G, Soininen R, Tryggvason K, Kobzik L. The scavenger receptor MARCO is required for lung defense against pneumococcal pneumonia and inhaled particles. The Journal of experimental medicine. 2004;200:267–272. doi: 10.1084/jem.20040731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Veremeyko T, Yung AWY, Anthony DC, Strekalova T, Ponomarev ED. Early Growth Response Gene-2 Is Essential for M1 and M2 Macrophage Activation and Plasticity by Modulation of the Transcription Factor CEBPβ. Front Immunol. 2018;9:2515. doi: 10.3389/fimmu.2018.02515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Daniel B, Czimmerer Z, Halasz L, Boto P, Kolostyak Z, Poliska S, Berger WK, Tzerpos P, Nagy G, Horvath A, Hajas G, et al. The transcription factor EGR2 is the molecular linchpin connecting STAT6 activation to the late, stable epigenomic program of alternative macrophage polarization. Genes Dev. 2020;34:1474–1492. doi: 10.1101/gad.343038.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Butovsky O, Jedrychowski MP, Moore CS, Cialic R, Lanser AJ, Gabriely G, Koeglsperger T, Dake B, Wu PM, Doykan CE, Fanek Z, et al. Identification of a unique TGF-β-dependent molecular and functional signature in microglia. Nat Neurosci. 2014;17:131–143. doi: 10.1038/nn.3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lund H, Pieber M, Parsa R, Grommisch D, Ewing E, Kular L, Han J, Zhu K, Nijssen J, Hedlund E, Needhamsen M, et al. Fatal demyelinating disease is induced by monocyte-derived macrophages in the absence of TGF-β signaling. Nature immunology. 2018;19:1–7. doi: 10.1038/s41590-018-0091-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guilliams M, Svedberg FR. Does tissue imprinting restrict macrophage plasticity? Nature immunology. 2021;496:445. doi: 10.1038/s41590-020-00849-2. [DOI] [PubMed] [Google Scholar]
- 41.Peng R, Sridhar S, Tyagi G, Phillips JE, Garrido R, Harris P, Burns L, Renteria L, Woods J, Chen L, Allard J, et al. Bleomycin induces molecular changes directly relevant to idiopathic pulmonary fibrosis: a model for “active” disease. PLoS ONE. 2013;8:e59348. doi: 10.1371/journal.pone.0059348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Misharin AV, Morales-Nebreda L, Reyfman PA, Cuda CM, Walter JM, McQuattie-Pimentel AC, Chen C-I, Anekalla KR, Joshi N, Williams KJN, Abdala-Valencia H, et al. Monocyte-derived alveolar macrophages drive lung fibrosis and persist in the lung over the life span. The Journal of experimental medicine. 2017;214:2387–2404. doi: 10.1084/jem.20162152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aran D, Looney AP, Liu L, Wu E, Fong V, Hsu A, Chak S, Naikawadi RP, Wolters PJ, Abate AR, Butte AJ, et al. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nature immunology. 2019;20:163–172. doi: 10.1038/s41590-018-0276-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yona S, Kim K-W, Wolf Y, Mildner A, Varol D, Breker M, Strauss-Ayali D, Viukov S, Guilliams M, Misharin A, Hume DA, et al. Fate mapping reveals origins and dynamics of monocytes and tissue macrophages under homeostasis. Immunity. 2013;38:79–91. doi: 10.1016/j.immuni.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Goldmann T, Wieghofer P, Müller PF, Wolf Y, Varol D, Yona S, Brendecke SM, Kierdorf K, Staszewski O, Datta M, Luedde T, et al. A new type of microglia gene targeting shows TAK1 to be pivotal in CNS autoimmune inflammation. Nat Neurosci. 2013;16:1618–1626. doi: 10.1038/nn.3531. [DOI] [PubMed] [Google Scholar]
- 46.Chakarov S, Lim HY, Tan L, Lim SY, See P, Lum J, Zhang X-M, Foo S, Nakamizo S, Duan K, Kong WT, et al. Two distinct interstitial macrophage populations coexist across tissues in specific subtissular niches. Science (New York, NY. 2019;363:eaau0964. doi: 10.1126/science.aau0964. [DOI] [PubMed] [Google Scholar]
- 47.Sajti E, Link VM, Ouyang Z, Spann NJ, Westin E, Romanoski CE, Fonseca GJ, Prince LS, Glass CK. Transcriptomic and epigenetic mechanisms underlying myeloid diversity in the lung. Nature immunology. 2020;21:221–231. doi: 10.1038/s41590-019-0582-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hirano S, Anuradha CD, Kanno S. Transcription of krox-20/egr-2 is upregulated after exposure to fibrous particles and adhesion in rat alveolar macrophages. Am J Respir Cell Mol Biol. 2000;23:313–319. doi: 10.1165/ajrcmb.23.3.4112. [DOI] [PubMed] [Google Scholar]
- 49.Lavin Y, Winter D, Blecher-Gonen R, David E, Keren-Shaul H, Merad M, Jung S, Amit I. Tissue-resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell. 2014;159:1312–1326. doi: 10.1016/j.cell.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brown JS, Hussell T, Gilliland SM, Holden DW, Paton JC, Ehrenstein MR, Walport MJ, Botto M. The classical pathway is the dominant complement pathway required for innate immunity to Streptococcus pneumoniae infection in mice. PNAS. 2002;99(26):16969–16974. doi: 10.1073/pnas.012669199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bewley MA, Budd RC, Ryan E, Cole J, Collini P, Marshall J, Kolsum U, Beech G, Emes RD, Tcherniaeva I, Berbers GAM, et al. COPDMAP, Opsonic Phagocytosis in Chronic Obstructive Pulmonary Disease Is Enhanced by Nrf2 Agonists. Am J Respir Crit Care Med. 2018;198:739–750. doi: 10.1164/rccm.201705-0903OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xu-Vanpala S, Deerhake ME, Wheaton JD, Parker ME, Juvvadi PR, MacIver N, Ciofani M, Shinohara ML. Functional heterogeneity of alveolar macrophage population based on expression of CXCL2. Sci Immunol. 2020;5 doi: 10.1126/sciimmunol.aba7350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Joshi N, Watanabe S, Verma R, Jablonski RP, Chen C-I, Cheresh P, Markov NS, Reyfman PA, McQuattie-Pimentel AC, Sichizya L, Lu Z, et al. A spatially restricted fibrotic niche in pulmonary fibrosis is sustained by M-CSF/M-CSFR signalling in monocyte-derived alveolar macrophages. Eur Respir J. 2020;55:1900646. doi: 10.1183/13993003.00646-2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Satoh T, Nakagawa K, Sugihara F, Kuwahara R, Ashihara M, Yamane F, Minowa Y, Fukushima K, Ebina I, Yoshioka Y, Kumanogoh A, et al. Identification of an atypical monocyte and committed progenitor involved in fibrosis. Nature. 2017;541:96–101. doi: 10.1038/nature20611. [DOI] [PubMed] [Google Scholar]
- 55.Gibbons MA, MacKinnon AC, Ramachandran P, Dhaliwal K, Duffin R, Phythian-Adams AT, van Rooijen N, Haslett C, Howie SE, Simpson AJ, Hirani N, et al. Ly6Chi monocytes direct alternatively activated profibrotic macrophage regulation of lung fibrosis. Am J Respir Crit Care Med. 2011;184:569–581. doi: 10.1164/rccm.201010-1719OC. [DOI] [PubMed] [Google Scholar]
- 56.Cui H, Jiang D, Banerjee S, Xie N, Kulkarni T, Liu R-M, Duncan SR, Liu G. Monocyte-derived alveolar macrophage apolipoprotein E participates in pulmonary fibrosis resolution. JCI Insight. 2020;5 doi: 10.1172/jci.insight.134539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Byrne AJ, Powell JE, O’Sullivan BJ, Ogger PP, Hoffland A, Cook J, Bonner KL, Hewitt RJ, Wolf S, Ghai P, Walker SA, et al. Dynamics of human monocytes and airway macrophages during healthy aging and after transplant. The Journal of experimental medicine. 2020;217 doi: 10.1084/jem.20191236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Evren E, Ringqvist E, Tripathi KP, Sleiers N, Rives IC, Alisjahbana A, Gao Y, Sarhan D, Halle T, Sorini C, Lepzien R, et al. Distinct developmental pathways from blood monocytes generate human lung macrophage diversity. Immunity. 2021;54:259–275.:e7. doi: 10.1016/j.immuni.2020.12.003. [DOI] [PubMed] [Google Scholar]
- 59.Lynch RW, Hawley CA, Pellicoro A, Bain CC, Iredale JP, Jenkins SJ. An efficient method to isolate Kupffer cells eliminating endothelial cell contamination and selective bias. Journal of leukocyte biology. 2018 doi: 10.1002/JLB.1TA0517-169R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tamoutounour S, Guilliams M, Montanana Sanchis F, Liu H, Terhorst D, Malosse C, Pollet E, Ardouin L, Luche H, Sanchez C, Dalod M, et al. Origins and functional specialization of macrophages and of conventional and monocyte-derived dendritic cells in mouse skin. Immunity. 2013;39:925–938. doi: 10.1016/j.immuni.2013.10.004. [DOI] [PubMed] [Google Scholar]
- 61.Scott CL, Bain CC, Mowat AM. Isolation and Identification of Intestinal Myeloid Cells. Methods Mol Biol. 2017;1559:223–239. doi: 10.1007/978-1-4939-6786-5_15. [DOI] [PubMed] [Google Scholar]
- 62.Bain CC, Mowat AM. CD200 receptor and macrophage function in the intestine. Immunobiology. 2011 doi: 10.1016/j.imbio.2011.11.004. [DOI] [PubMed] [Google Scholar]
- 63.Magalhaes MS, Smith P, Portman JR, Jackson-Jones LH, Bain CC, Ramachandran P, Michalidou Z, Stimson RH, Dweck MR, Denby L, Henderson NC, et al. Role of Tim4 in the regulation of ABCA1+ adipose tissue macrophages and post-prandial cholesterol levels. Nat Commun. 2021;12:4434–15. doi: 10.1038/s41467-021-24684-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bain CC, Jenkins SJ. Isolation and Identification of Murine Serous Cavity Macrophages. Methods Mol Biol. 2018;1784:51–67. doi: 10.1007/978-1-4939-7837-3_5. [DOI] [PubMed] [Google Scholar]
- 65.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 66.Dobie R, Wilson-Kanamori JR, Henderson BEP, Smith JR, Matchett KP, Portman JR, Wallenborg K, Picelli S, Zagórska A, Pendem SV, Hudson TE, et al. Single-Cell Transcriptomics Uncovers Zonation of Function in the Mesenchyme during Liver Fibrosis. Cell Rep. 2019;29:1832–1847.:e8. doi: 10.1016/j.celrep.2019.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Satija R, Farrell JA, Gennert D, Schier AF, Regev A. Spatial reconstruction of single-cell gene expression data. Nat Biotechnol. 2015;33:495–502. doi: 10.1038/nbt.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fercoq F, Remion E, Vallarino-Lhermitte N, Alonso J, Raveendran L, Nixon C, Le Quesne J, Carlin LM, Martin C. Microfilaria-dependent thoracic pathology associated with eosinophilic and fibrotic polyps in filaria-infected rodents. Parasit Vectors. 2020;13:551. doi: 10.1186/s13071-020-04428-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bankhead P, Loughrey MB, Fernández JA, Dombrowski Y, McArt DG, Dunne PD, McQuaid S, Gray RT, Murray LJ, Coleman HG, James JA, et al. QuPath: Open source software for digital pathology image analysis. Sci Rep. 2017;7:16878. doi: 10.1038/s41598-017-17204-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Luche H, Weber O, Nageswara Rao T, Blum C, Fehling HJ. Faithful activation of an extra-bright red fluorescent protein in “knock-in” Cre-reporter mice ideally suited for lineage tracing studies. Eur J Immunol. 2002;37(1):43–53. doi: 10.1002/eji.200636745. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials, and RNA-seq data have been deposited in National Center for Biotechnology Information Gene Expression Omnibus public database (www.ncbi.nlm.nih.gov/geo/). Population-level RNA-seq (accession code: GSE182044) and scRNA-seq (accession code: GSE181894). Fcgr1iCre mice are available from Prof. Bernard Malissen under a material transfer agreement with the Centre d'lmmunologie de Marseille-Luminy, Aix Marseille Université. Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Calum Bain (calum.bain@ed.ac.uk).