Abstract
One of the fundamental processes that determine cellular fate is regulation of gene transcription. Understanding these regulatory processes is therefore essential for understanding cellular responses to changes in environmental conditions. At the core promoter, the regulatory region containing the transcription start site (TSS), all inputs regulating transcription are integrated. Here, we used Cap Analysis of Gene Expression (CAGE) to analyze the pattern of transcription start sites at four different environmental conditions (limited in ethanol, limited in nitrogen, limited in glucose and limited in glucose under anaerobic conditions) using the Saccharomyces cerevisiae strain CEN.PK113-7D. With this experimental setup we were able to show that the TSS landscape in yeast is stable at different metabolic states of the cell. We also show that the spatial distribution of transcription initiation events, described by the shape index, has a surprisingly strong negative correlation with measured gene expression levels, meaning that genes with higher expression levels tend to have a broader distribution of TSSs. Our analysis supplies a set of high quality TSS annotations useful for metabolic engineering and synthetic biology approaches in the industrially relevant laboratory strain CEN.PK113-7D, and provides novel insights into yeast TSS dynamics and gene regulation.
Introduction
Regulation of gene transcription is one of the fundamental processes that determine cellular fate. Transcription of protein encoding genes in eukaryotic cells is governed by RNA polymerase II in concert with the general transcription initiation factors (GTFs), namely TFIIA, TFIIB, TFIID, TFIIE, TFIIF and TFIIH (reviewed in (Smale and Kadonaga 2003)). These proteins assemble at the core promoter of a gene, which is commonly defined as the minimal region necessary to trigger transcription (Danino et al. 2015; Haberle and Lenhard 2016; Haberle and Stark 2018). This region encompasses the transcription start site (TSS), defined as the nucleotide position where transcription is initiated (Sandelin et al. 2007).
It was previously shown that transcription of a gene in eukaryotic cells is not always initiated from the same nucleotide, but that it can be initiated from a range of positions in the core promoter region, with an individual, sequence-influenced pattern for each gene (Suzuki et al. 2001; Sandelin et al. 2007; Haberle and Lenhard 2016; Haberle and Stark 2018). This important finding reshaped the view on transcription initiation showing that there is a higher complexity to this process than previously anticipated.
In addition to the TSS positions being a cornerstone of fundamental knowledge on genome organization, there are numerous applications where an exact mapping of TSS positions is important. One is in the field of synthetic biology, where synthetic promoters are created to obtain a variable range of expression levels. Synthetic promoters are designed by combining core promoters with different upstream regulatory sequences. In order to do this, accurate definition of the promoter regions are needed to place upstream regulatory sequences at the optimal distance to the core promoters. Another application is the modulation of gene expression by CRISPR interference (CRISPRi). An effective strategy for downregulation that has been documented to work for many genes is to target the catalytically inactive Cas9 protein directly to the TSS of the target gene (Qi et al. 2013).
The most accurate way to map transcription start sites is to selectively sequence intact capped 5’ ends of mRNA. In this study we choose the Cap Analysis of Gene Expression method (CAGE) (Shiraki et al. 2003), which was also shown to be the best performing method in a recent comparison of different 5’ end RNA sequencing methods by Adiconis et al., (Adiconis et al. 2018). This method gives a quantitative count of transcription start events with a single base pair resolution, allowing a more detailed interrogation of these events than with traditional RNA sequencing techniques. CAGE can also be used to determine the total expression of a given gene with results showing high correlations with traditional RNA sequencing techniques (Kawaji et al. 2014). With these high resolution data it is possible to accurately determine all TSSs of all expressed genes transcribed by RNA Polymerase II and to determine which TSS is the dominant one in a quantitative manner.
Previous work to annotate TSSs has been carried out in different yeast strains using techniques like SMORE-seq (Parky et al. 2014), or an earlier low-coverage protocol of CAGE (Wery et al. 2016). These studies used cells grown in shake flasks at only one environmental condition. Therefore, it was not possible to assess how the TSS landscape changes in response to environmental conditions.
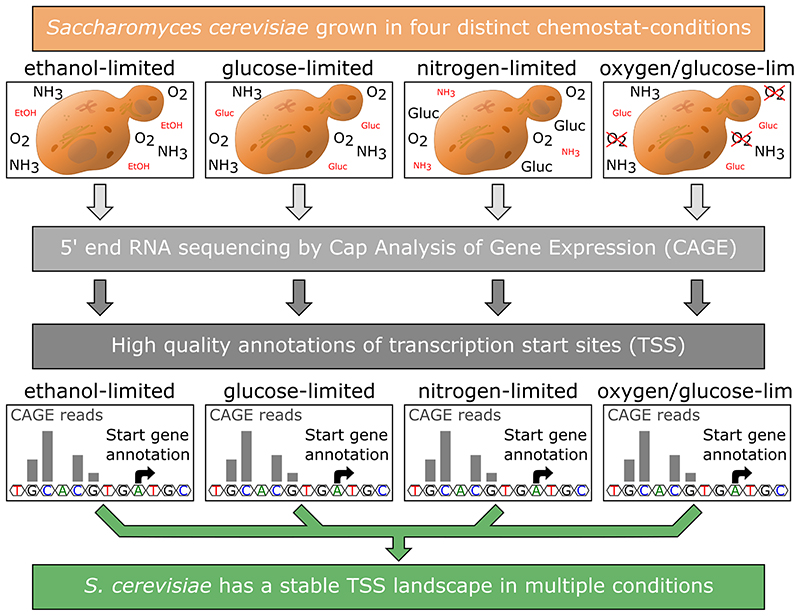
Here, we describe the first analysis of the content and dynamics of the yeast promoterome across four different metabolic states. For this, we used an updated CAGE protocol, called non-amplification non-tagging CAGE for Illumina sequencing (nAnT-iCAGE) (Murata et al. 2014), which is a more unbiased approach compared to the earlier protocol used by Wery et al. (Wery et al. 2016) as it omits the use of restriction enzymes to produce short tags and does not include a PCR amplification step of the cDNA. We performed CAGE on the industrially relevant S. cerevisiae laboratory strain CEN.PK113-7D (van Dijken et al. 2000), grown in four distinct chemostat conditions at a fixed dilution rate of 0.1/h. The four chemostat conditions were selected to cover a diverse range of metabolic states, namely: respiratory glucose metabolism using glucose limitation, gluconeogenic respiration using ethanol limitation, aerobic fermentation using nitrogen limitation and fermentative glucose metabolism using anaerobic conditions. With this setup, we were able to obtain highly reproducible condition-specific data and to assess changes in the TSS landscape in different environmental conditions of the cells, as well as providing a high quality set of TSS annotations for the research community.
Experimental Procedures
Gene annotations
To transfer the annotations from the reference genome of S288C (Engel et al. 2014) to the recently published genome of CEN.PK113-7D (Salazar et al. 2017), first the coding sequences for all verified and uncharacterized ORFs available in the Saccharomyces Genome Database (SGD, www.yeastgenome.org) (Cherry et al. 2012) were obtained using YeastMine, the data API of SGD. Then, using the NCBI software tool Blast+ (Camacho et al. 2009), every obtained sequence was blasted against the CEN.PK genome. Hits covering at least 95% of the sequence length showing at least a 95% sequence identity were retained and transferred if only a single hit existed for that sequence. In case of multiple strong hits, the hit that was found to be on the same chromosome and surrounded by the same neighboring genes as in the reference genome was transferred. In case of large genes where for multiple fragments a hit was found, a manual curation step was performed to check if these fragments could be reassembled into the full gene. Successfully reassembled genes were also transferred, all other hits were discarded. Using this approach, we were able to transfer 99% (5113 out of 5159) of the verified ORF annotations and 96% (727 out of 756) of all uncharacterized ORF annotations. The gene YCL018W, which was found to be duplicated in the originally published sequence, was also found to be duplicated using our approach (Salazar et al. 2017). In addition to that, the genes YHR055C and YHR054C were also found to be duplicated. The complete set of updated annotations can be found in Supplementary table S1.
Chemostat cultures and RNA extraction
The S. cerevisiae strain CEN.PK113-7D (van Dijken et al. 2000) was pre-cultured in a batch culture in 100 ml of minimal medium with 2% glucose (See Supplementary table S2 for media composition and recipe) at 30°C and 200 rpm in 250 ml shake flasks for 24 hours. The pre-culture was then transferred to the mini-bioreactors (40 ml volume) in triplicates to an initial OD600 above 3. For each of the four conditions, a single pre-culture was used. Four different media compositions with a limitation in a different nutrient were employed in the chemostat runs (See Supplementary table S2 for media composition and recipe). The medium volume in the chemostat runs was 40 ml and the temperature was set to 30°C. One hour after the transfer, the pumps where started with the dilution rate fixed to 0.1/h. Dissolved oxygen was kept above 30% of air saturation. For the anaerobic condition, the culture vessels were flushed with nitrogen gas. The cells were grown for 4 days to achieve stable cell numbers in the culture. An amount of cells corresponding to a 10 ml culture with an OD600 of 1 were collected, pelleted and snap frozen in liquid nitrogen for both RNAseq analysis and the CAGE experiment.
Cells were mechanically disrupted using a FastPrep®-24 from MPbio (Santa Ana, California, USA) in combination with the lysing matrix tubes type C from MPbio. The FastPrep was run 3 times for 20 seconds with 4.0 m/s settings and a 5 min break in between each run. RNA was subsequently extracted using the RNeasy® Mini Kit from QIAGEN (Hilden, Germany). RNA quality was assessed using a ThermoFischer NanoDrop (Waltham, Massachusetts, USA) and an Agilent2100 Bioanalyzer (Santa Clara, California, USA) to ensure high quality RNA.
RNA sequencing
All three biological replicates for each condition were sequenced using the NextSeq500 System from Illumina (San Diego, California, USA) at the Novo Nordisk Foundation Center for Biosustainability, Technical University of Denmark, with paired-end reads of 75 bp length. Library preparation was done using the Illumina TrueSeq stranded total RNA HT kit following the manufacturer’s instructions. Obtained reads were mapped to the CEN.PK113-7D genome using bowtie2 (Langmead and Salzberg 2012). Mapped reads were filtered using a quality threshold of 20 and converted to .bam files using samtools (Li et al. 2009). FeatureCounts was used to obtain expression values for each gene (Liao, Smyth and Shi 2014), which were subsequently converted into TPM values. The raw counts obtained by FeatureCounts were also used for differential gene expression analysis using DEseq2 (Love, Huber and Anders 2014), where the threshold for detecting differential expressed genes was set to an adjusted pValue of 0.001.
Cage
For the CAGE experiment the non-amplification non-tagging CAGE protocol for Illumina sequencing (nAnT-iCAGE) as previously published by Murata et al. (Murata et al. 2014), was used on two biological replicates of each condition, starting with 5 μg of extracted total RNA. The 8 barcoded samples were pooled together and sequenced using the Illumina HiSeq 2500 at Genomics Core Facility (MRC, London Institute of Medical Sciences). Between 2.6 to 22.1 million reads per sample were obtained, with an average of 9 million, showing a very high coverage of the yeast transcriptome. Sequencing reads were mapped to the CEN.PK113-7D genome using bowtie2 (Langmead and Salzberg 2012). Mapped reads were filtered using a quality threshold of 20 and converted to .bam files using samtools (Li et al. 2009). 29% of all reads mapped to a 7.2 kb region with ribosomal repeats on chromosome 12, which was excluded from further analysis, leaving an average mapped read count of 5.4 million reads per replicate. An overview of the sequencing read numbers is shown in Supplementary table S3.
CAGE data were analyzed using the R/Bioconductor package CAGEr (Haberle et al. 2015; Huber et al. 2015). The .bam files were imported into R and the biological replicates were merged together. Default CAGEr correction of the first G nucleotide was used. The data were then normalized for library size using the “powerLaw” method (Balwierz et al. 2009) with a fit range from 5 to 10000 and an alpha value of 1.10. The CAGE tags were clustered together using the “clusterCTSS” function of CAGEr with the “distclu” setting, a maximum distance of 20 and a TPM threshold of 1. These clusters were then aggregated across the conditions to obtain a set of consensus clusters using the “aggregateTagClusters” function with a TPM threshold of 3 and a maximum distance of 100. For each consensus cluster the expression level in every condition was calculated as TPM and the dominant TSS position was calculated based on the normalized tag count per base. Changes in TSS distribution patterns were calculated using the getShiftingPromoters function from CAGEr, which uses the Kolmogorov-Smirnov test to measure if the cumulative sum of TSS events is different between two conditions. As thresholds we used a minimal shifting score of 0.6 and an adjusted pValue from the Kolmogorov-Smirnov test of 0.01. Additionally, the shape index of each cluster was calculated by the formula described by Hoskins et al. (Hoskins et al. 2011): . A graphical example for this with two artificial promoters can be found in Supplementary Figure S1.
pi = proportion of counts at position i in the cluster
L = position with at least 1 tag
Annotation of CAGE clusters to genes
The obtained CAGE consensus clusters were assigned to the gene annotations using the following set of rules: The consensus cluster must be on the same strand as the gene annotation, the cluster is not more than 1 kb away from the start of the gene annotation and if the cluster is upstream of the gene annotation, RNAseq reads must be present covering the region between the cluster and the gene annotation.
Expression-based clustering of genes
For creating the four gene clusters based on the expression profiles, we normalized the RNAseq expression levels across the four conditions and a gene was assigned to one of the four clusters if it met the following requirements: Cluster “Always”: The expression level in each condition must account for 23 to 27% of the total observed expression (sum of TPM values from RNAseq of the four conditions). Cluster “Glu+Eth”: At least 83.3% of the observed total expression must come from the two respiratory conditions (respiratory glucose metabolism using glucose limitation and gluconeogenic respiration using ethanol limitation). Cluster “Nit”: At least 75% of the observed total expression must come from aerobic fermentation using nitrogen limitation. Cluster “Ana”: At least 75% of the observed total expression must come from fermentative glucose metabolism using anaerobic conditions.
Results and Discussion
CAGE data are highly reproducible and reveal promoters and TSS for 88% of all annotated genes
In order to gain insights into the promoter structure of the yeast strain CEN.PK113-7D and to obtain accurate positions of the transcription start sites (TSSs) we performed a cap analysis gene expression (CAGE) experiment on yeast grown in four different chemostat conditions. The conditions were: respiratory glucose metabolism using glucose limitation (Glu), gluconeogenic respiration using ethanol limitation (Eth), aerobic fermentation using nitrogen limitation (Nit) and fermentative glucose metabolism using anaerobic conditions (Ana).
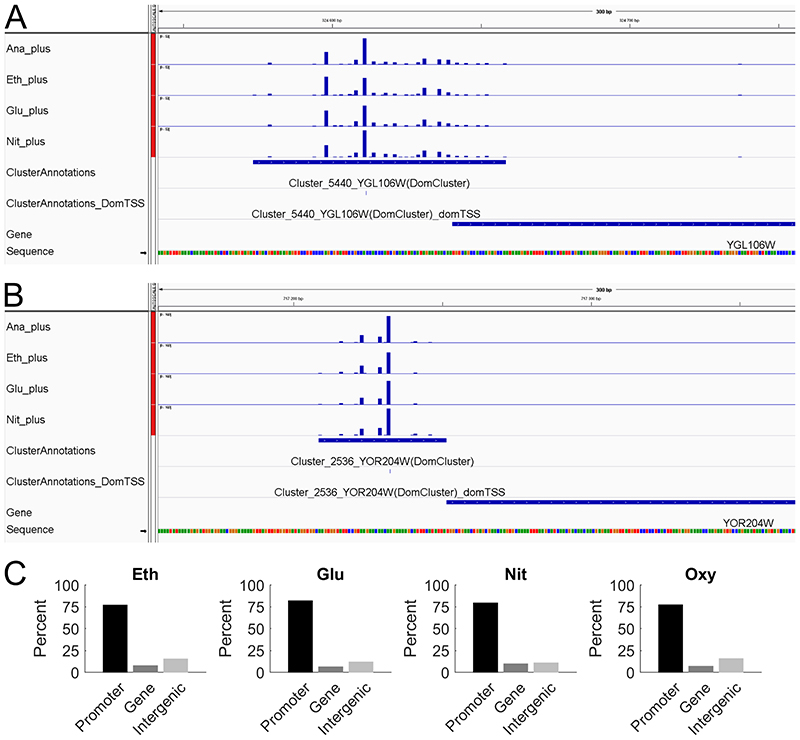
Using the R/Bioconductor package CAGEr (Haberle et al. 2015), for each of the four conditions we assembled the single position read tags into clusters and then merged overlapping clusters together to form a consensus cluster, combining information from all four conditions. This resulted in a total of 6565 consensus clusters which were then assigned to the gene annotations. For 5245 clusters a matching gene annotation could be found, of which 4975 genes were assigned a single cluster and 132 genes were assigned multiple clusters (ranging from 2 to 4 clusters per gene, a total of 270 clusters). This means that from a total of 5843 gene annotations in the genome, we could assign 5107 (88%) to at least one cluster. The complete set of results for each individual cluster can be found in Supplementary table S4. A representative display of the CAGE data, in the Integrative Genomics Viewer (IGV) (Robinson et al. 2011; Thorvaldsdottir, Robinson and Mesirov 2013), is shown in Figure 1A-B. The two genes were selected to showcase different distributions of CAGE reads for condition independent genes, with Figure 1A showing a broad TSS distribution, while Figure 1B displays a peaked TSS distribution. Screenshots for two condition dependent genes, showing also one broad and one peaked TSS distribution, can be found in Supplementary Figure S2, highlighting that a broad or peaked TSS distribution is not unique for condition independent or specific genes.
Figure 1. Overview of the obtained CAGE data.
A Screenshot from IGV showing the broad CAGE read distribution for the constitutively expressed gene YGL106W (MLC1). B Screenshot from IGV showing the peaked CAGE read distribution for the constitutively expressed gene YOR204W (DED1). C Intersection of mapped CAGE reads with gene annotations, the promoter region was defined as the 500 bp upstream of the start of the coding region and which was therefore not considered to be part of the intergenic region. (Eth = gluconeogenic respiration using ethanol limitation, Glu = respiratory glucose metabolism using glucose limitation, Nit = aerobic fermentation using nitrogen limitation, Ana = fermentative glucose metabolism using anaerobic conditions).
To assess the quality of our obtained CAGE data, we first analyzed the location of the sequencing reads in relation to the annotated genes (Figure 1E) and found that the majority of all reads (77% to 82%) mapped to the promoter region of annotated genes, which was defined as the 500 bp region upstream of the start of the coding sequence. As the TSS of a gene is expected to be upstream of the coding sequence, this also indicates that we obtained high quality CAGE data.
179 clusters were annotated as possible antisense transcription events to a total of 169 genes, as they were located at the 3’ end of the gene on the opposite strand. It has been shown before that anti-sense transcription occurs widely in yeast (Yassour et al. 2010). Yassour et al. reported for every gene what proportion of the coding sequence was covered by anti-sense transcription, and in their study 1523 genes had at least 10% of their sequence covered by anti-sense reads. Comparing these 1523 genes with our list of 169 genes with possible anti-sense initiation we find a high degree of overlap of 75% (122 out of 163 genes that are in both lists). As this finding is in line with the already known wide-spread anti-sense transcription, we focused on the sense transcription events. From the total of 6565 consensus CAGE clusters, 1115 clusters could not be assigned to any gene. These clusters could indicate missing gene annotations but it is more likely that they originate from non-annotated small RNA species and from cryptic unstable transcripts that have been shown to be transcribed widespread from the yeast genome (Berretta and Morillon 2009).
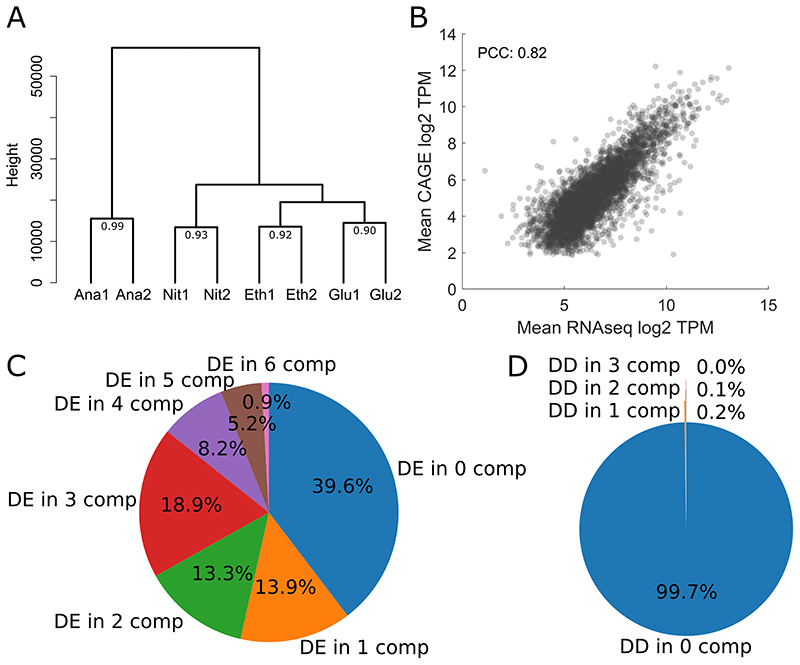
To further analyze the quality of the obtained CAGE data, the individual samples were clustered together using hierarchical clustering based on their genome-wide expression profile at each base pair. For each condition the replicates cluster together (Figure 2A), showing the high reproducibility of the data. This clustering also shows that the two respiratory conditions (respiratory glucose metabolism using glucose limitation, gluconeogenic respiration using ethanol limitation) are more similar to each other than to the two fermentative conditions, as one would expect. In addition, the correlations between the biological replicates were calculated and with a minimum Pearson correlation coefficient of 0.9 the replicates are in high agreement with each other (Figure 2A).
Figure 2. Quality control of CAGE expression levels and analysis of differential expression and CAGE tag distribution.
A Hierarchical clustering of the individual CAGE sequencing experiments based on normalized TSS tag values per base genome-wide. The number at the last branch points denotes the Pearson correlation between the replicates B Comparison between average expression values across the conditions obtained through RNAseq and CAGE, both in log2 TPM values, resulting in a PCC of 0.82. For genes with multiple CAGE clusters, all clusters where summed up to calculate the TPM value. C Results for differential gene expression analysis of RNAseq data, showing the proportions of genes that were detected as differentially expressed (using DEseq2, adj. pValue <0.001) in x pairwise comparisons of the four different conditions (4 conditions = 6 possible comparisons). D Results for detecting shifted promoters, showing the proportions of clusters associated to genes that were detected as differentially distributed (using getShiftingPromoters from CAGEr, shifting score > 0.6 and adj. pValue of Kolmogorov-Smirnov test < 0.01) in x pairwise comparisons of the different conditions.
Subsequently the expression values per gene promoter region obtained from CAGE was compared with gene expression values obtained from a control RNAseq experiment in the same chemostat conditions (Figure 2B). For genes with multiple assigned CAGE clusters, the expression values for all clusters were summed up prior to the comparison. The results show a strong correlation with a Pearson Correlation Coefficient of 0.82 between the RNAseq and the CAGE expression values, further demonstrating the high quality of our CAGE data.
The yeast TSS landscape shows stability across metabolic conditions in a variety of characteristics
To assess if and how much the TSS landscape changes between the four different conditions used in our study, a baseline of how much gene expression levels change between the conditions had to be calculated. For this we used the RNAseq dataset and determined differentially expressed genes using DEseq2 (Love, Huber and Anders 2014) for each pairwise condition comparison, resulting in 6 comparisons. The results for this analysis are shown in Figure 2C, where for each of the 5107 genes in our CAGE dataset, we counted in how many of the 6 pairwise comparison this gene was detected as differentially expressed based on an adjusted pValue threshold of 0.001. Only 0.9% of all genes were differentially expressed in all 6 comparison, meaning that those 46 genes showed completely different expression levels in all four condition. Furthermore, less than 40% of all genes were not changed between the four conditions. However, this means that more than 60% of the genes were differentially expressed in at least one comparison, and this demonstrates that the four condition chosen in our study led to very diverse gene expression patterns and were therefore suitable to assess changes in the TSS landscape.
For a first assessment of the stability of the TSS landscape we compared the distribution of TSS events in each consensus cluster assigned to a gene using the Kolmogorov-Smirnov test on the cumulative sum of TSS events. This was performed using the getShiftingPromoters function of CAGEr with a shifting score threshold of 0.6 and adj. pValue threshold of the Kolmogorov-Smirnov test of 0.01). As for the differential gene expression analysis we performed this analysis on all 6 pairwise comparisons and the results are shown in Figure 2D. Nearly all genes (99.7%) showed no differential distribution in any of the pairwise comparisons, showing that the TSS distribution was very stable in the four chosen conditions, even though the gene expression patterns were quite diverse.
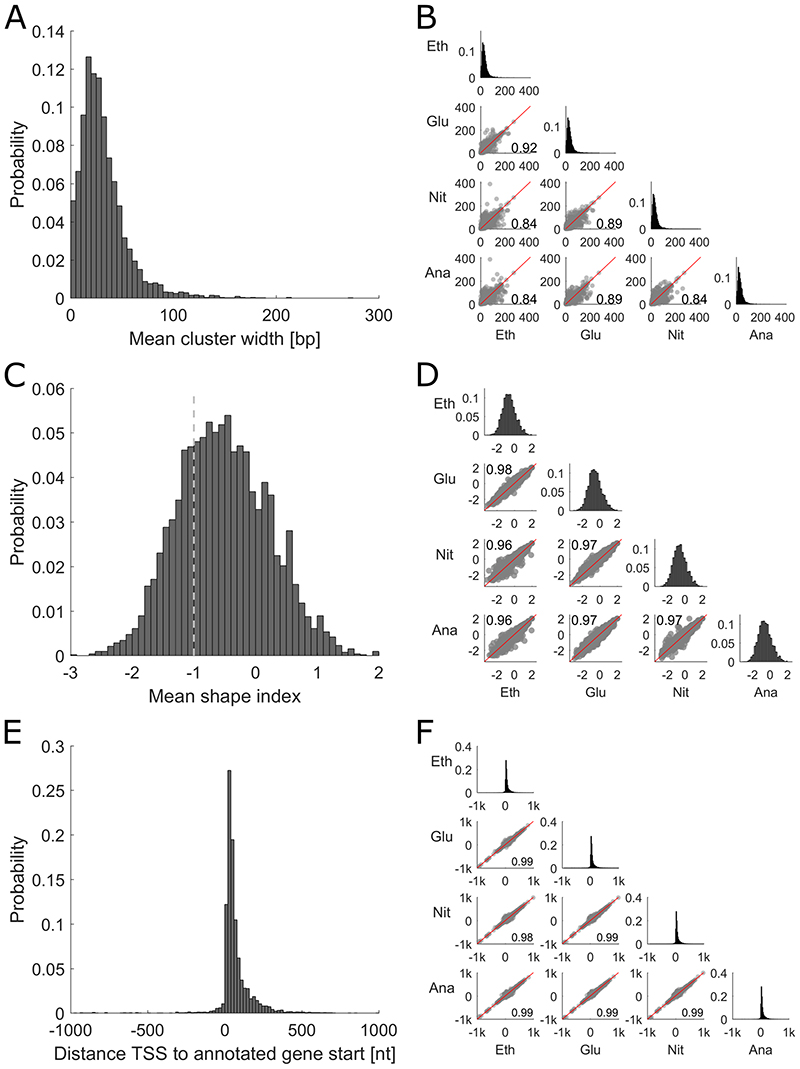
One rather cluster centered analysis of CAGE clusters is to look at the cluster width, which describes for each condition over how many bases the TSS are distributed in the consensus cluster, or to look at the interquantile cluster width for the quantiles 0.1 to 0.9, as established by Haberle et al. (Haberle et al. 2015). The interquantile cluster width was calculated based on the cumulative distribution of TSS events inside the consensus cluster and is the span of bases that contains every read from the 0.1 to 0.9 quantile. As this metric leaves out the extreme borders it is more robust to noise and therefore this approach was chosen for subsequent analysis. For each gene, an average cluster width across the four conditions was calculated. The distribution of the average widths (Figure 3A) shows a unimodal distribution with an average width of 31 bp. The interquantile cluster width is also stable across the four conditions, as shown in a pairwise comparison between conditions, with a minimum Pearson correlation coefficient of 0.84 (Figure 3B).
Figure 3. Overview of TSS cluster characteristics and their stability across conditions.
A Histogram showing distribution of mean interquantile cluster width across all conditions. B Comparison of the promoter width in different conditions. Middle axis showing the distribution in each condition and the lower half displaying the pairwise comparison of each condition together with the Pearson correlation coefficient. C Histogram showing the mean shape index of each cluster. The dashed line at -1 denotes the border, which separates clusters classified as peaked (shape index >-1) and clusters classified as broad (shape index ≤ -1). D Comparison of the shape index in different conditions. Middle axis showing the distribution in each condition and the lower half displaying the pairwise comparison. E Histogram showing the distribution of distances between the global TSS across conditions and the assigned genes. F Comparison of the distance between the condition-specific TSS and the assigned gene in different conditions. Middle axis showing the distribution in each condition and the lower half displaying the pairwise comparison.
It has been shown that the cluster width is not always sufficient to classify clusters as either peaked or broad. Clusters that are very wide but where the dominant positions contribute the majority of reads exist, as well as narrow clusters with multiple near equally strong positions. To overcome this issue, Hoskins et al. established the shape index as a more informative tool for this classification (Hoskins et al. 2011). The formula used to calculate the shape index can be found in the method section and an example classification of two artificial clusters is shown in Supplementary Figure S1. In short, for each gene the shape index is calculated based on the distribution of TSS in the consensus cluster for each individual condition. It results in a continuous variable with possible values from less than -5 up to 2 describing how peaked a TSS cluster is. As previously set by Hoskin et al., the threshold distinguishing genes with a peaked and broad TSS distribution is -1, therefore a gene with a shape index higher than -1 is a gene with a peaked TSS distribution, while a gene with a value lower than or equal to -1 has a broad TSS distribution. We classified the peaks detected by our analysis by the average shape index across the conditions and found that the majority of clusters classified as peaked, i.e. had a shape index higher than - 1 (Figure 3C). The shape index was also very stable between the conditions, with a minimal pairwise Pearson correlation coefficient of 0.96 (Figure 3D).
Calculating the 5’ UTR length (Figure 3E) showed that most clusters are quite close to the start of the coding sequence, with 70% of them being less than 75 bp away, which is in line with previously published average 5’ UTR lengths in yeast (Parky et al. 2014). This 5’ UTR length is again very stable across the different conditions with a minimal Pearson correlation coefficient of 0.99 between two individual conditions (Figure 3F). We further compared the published TSS dataset from Parky et. al. (Parky et al. 2014), obtained using the yeast strain BY4741 in YPD, with our TSS annotations for CEN.PK113-7D. For the 4872 genes that are present in both data sets we calculated the 5’ UTR lengths (using sacCer3 annotations for the dataset from Parky et al. and our CEN.PK113-7D annotations for our dataset) and compared them. The 5’ UTR lengths are in high agreement with each other, with an average difference of less than 9 bp. Both datasets have around 250 TSS annotations for genes not found in the other dataset, these differences are most likely due to different expression profiles caused by different media and growth conditions (YPD in shake flasks vs synthetic minimal media in chemostats) or strain differences (BY4741 vs CEN.PK113-7D). The high agreement between the two datasets highlights the quality of our TSS annotations for the industrially relevant strain CEN.PK113-7D.
Gene clustering by condition-specific expression shows no distinct promoter characteristics
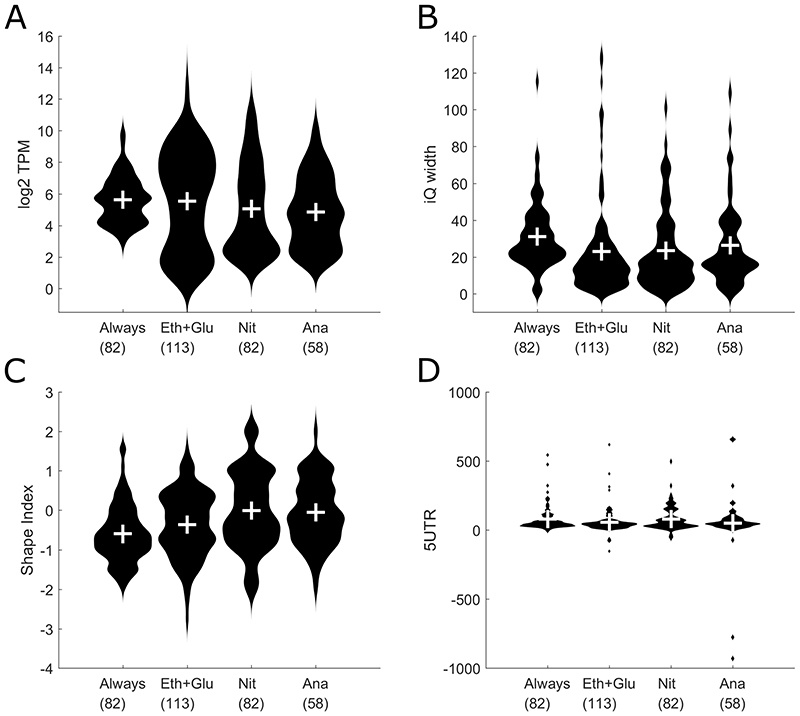
To further test the stability of the yeast transcriptional landscape in different conditions we clustered genes together based on their expression levels in different conditions. For this, we normalized the genes expression levels across the conditions and created the following four gene clusters: 1: Genes that are expressed under all four conditions (labeled as “always”); 2: Genes that are mostly active in the two respiratory conditions (respiratory glucose metabolism using glucose limitation and gluconeogenic respiration using ethanol limitation, labeled “Glu+Eth”); 3: Genes that are mainly active in aerobic fermentation using nitrogen limitation (labeled “Nit”) and 4: Genes that are mainly active in fermentative glucose metabolism using anaerobic conditions (labeled “Ana”). For each of these four groups, we analyzed the expression levels (Figure 4A), the interquantile widths (Figure 4B), the shape indices (Figure 4C) and the 5’ UTR length (Figure 4D). The overall picture shows that there are no clear differences in these characteristics between the four groups. The average gene expression levels are quite similar, with the genes expressed in all four conditions showing a slightly narrower distribution than the condition-specific genes, a trend that can also be seen in the distribution of shape indices. These differences however are not very strong.
Figure 4.
Comparison of condition-based gene clusters (Always = Genes that are expressed in all four conditions, Eth+Glu = Genes active in both respiratory conditions, Nit = Genes active under aerobic fermentation, Ana = Genes active in fermentative glucose metabolism). The number under the gene cluster denotes the number of genes in that cluster. A Violin plot showing distribution of gene expression levels for each gene cluster. B Violin plot showing distribution of the interquantile promoter width for each gene cluster. C Violin plot showing distribution of the shape index for each gene cluster. D Violin plot showing distribution of the 5’ UTR lengths.
Additionally we tested if there are differences in the promoter characteristics between TATA box-containing and TATA-less genes. For this classification we used the published list of TATA box-containing genes from Basehoar et al. (Basehoar, Zanton and Pugh 2004). Even though there are slight differences in gene expression level distributions, with TATA box-containing genes showing a higher maximal expression level, there are no differences in the distribution of shape indices, indicating that this promoter features is not affected by the presence or absence of a TATA box (see Supplementary Figure S3).
Cluster shape shows a high correlation to promoter expression levels
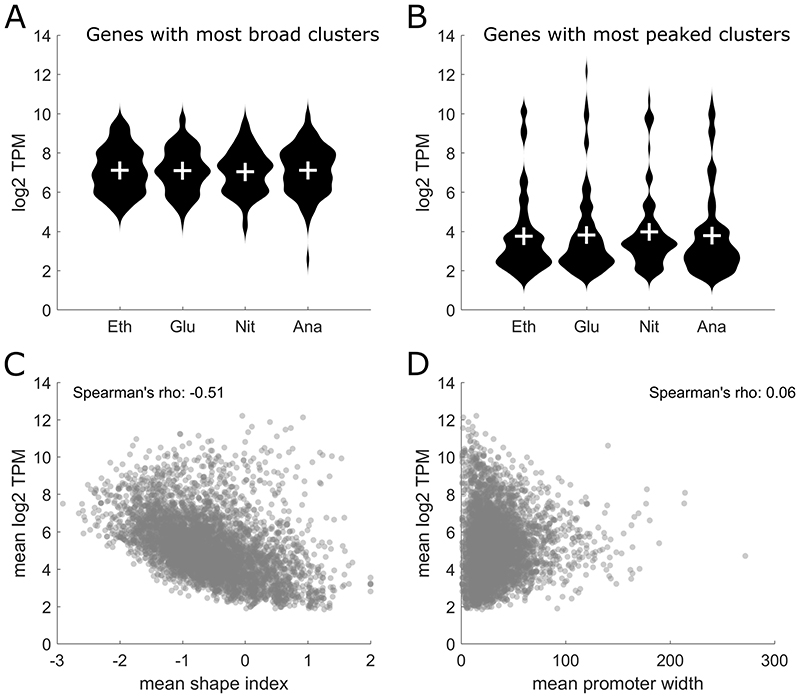
In higher organisms like Drosophila melanogaster, there is a remarkable connection between the shape index of a TSS cluster and the gene expression level during different developmental phases (Hoskins et al. 2011). Genes with a broad TSS cluster show a stable expression level throughout embryonic development, while genes with a peaked cluster show a transcription pattern that varies in time and space (Hoskins et al. 2011). To see if this relationship between shape index and gene expression variability also holds for yeast, we averaged the shape index of each cluster across the four conditions and then selected the 100 genes with the lowest shape index, i.e. the genes with the broadest clusters, and the 100 genes with the highest shape index, i.e. the genes with the most peaked clusters. For these selected genes, we then compared the expression values in each individual condition (Figure 5A-B). No significant differences in expression levels were observed when comparing the four different conditions. However, there was a marked difference in the overall expression levels between genes with a broad cluster and genes with a peaked cluster (comparing overall TPM levels in Figure 5A with 5B). Following this observation, the correlation between the mean shape index across the conditions and the mean expression levels was analyzed, as shown in Figure 5C. A striking anticorrelation with a Pearson correlation coefficient of -0.45 was observed, indicating that peaked clusters (clusters with a high shape index) in yeast are associated with lower expression levels. To check if that strong correlation was unique to the shape index, we also calculated the correlation between the mean interquantile promoter width with gene expression levels (Figure 5D) and we observe no correlation. This indicates that the strong correlation observed between the shape index and gene expression levels is a unique feature of the shape index.
Figure 5. Detailed analysis of the Shape Index.
A The 100 genes with the broadest clusters across all conditions were selected and their expression values in each condition are shown. B The 100 genes with the most peaked clusters across all conditions were selected and their expression values in each condition are shown. C Correlation of the mean shape index and the mean CAGE expression levels showing an anticorrelation with a Spearman’s Rho of -0.51 and a Pearson correlation coefficient of -0.45. D Correlation of the mean promoter width with mean CAGE expression levels showing no correlation.
Data availability and usage
To enable the easy usage of our data, we created custom data tracks and sessions for the Integrated Genomics Viewer (IGV, (Robinson et al. 2011; Thorvaldsdottir, Robinson and Mesirov 2013)), which can be found in the Supplementary information. After downloading the IGV from http://software.broadinstitute.org/software/igv/home, first the CEN.PK113-7D genome file (“CEN.PK113-7D.genome” part of the zipped Supplementary data file S1) has to be loaded via “Genomes” -> “Load Genome from File…” menu in IGV. After that, it is possible to load the session file for either the raw CAGE reads (“IGV_session_RawData.xml”, part of the zipped Supplementary data file S2) or the normalized CAGE reads (“IGV_session_NormData.xml”, part of the zipped Supplementary data file S3) using the “File” -> “Open Session…” menu.
After loading the session, a screen similar to the one shown in Figure 1A-D will be visible. For each condition, there are two tracks, one for reads on the plus strand and one for reads on the minus strand of the genome (labeled “_plus” and “_minus” respectively). In addition there are three different annotation tracks. The first one, labeled “ClusterAnnotations.bed”, will show each cluster with the full width, while the second one, labeled “ClusterAnnotationsDomTSS.bed”, will only show the position of the strongest TSS in each cluster. Both of these tracks include information about the cluster ID, and to which gene the cluster is annotated to (if any). For each gene, the strongest cluster is labeled as “(DomCluster)”. A third annotation track called “Gene” displays the blast based gene annotations for the CEN.PK113-7D genome.
Conclusion
In this study, we present a high quality CAGE dataset in four distinct chemostat conditions to accurately annotate the TSS of each gene. This resource will be valuable to the community as accurate TSS annotations, based on the dominant TSS position, are valuable for promoter engineering and implementation of CRISPRi approaches.
Analysis of the yeast promoterome in the different conditions shows a remarkable level of stability in terms of promoter characteristics like promoter width and shape index as well as individual TSS site usage. This is in contrast to higher organisms where strong changes can occur, especially during embryonal development stages (Haberle et al. 2014), and suggests that the basic regulatory events governing gene expression in yeast are quite distinct from other eukaryal cells.
Supplementary Material
Funding
This work was supported by the European Union’s Horizon 2020 research and innovation programme [Marie Skłodowska-Curie grant agreement No 722287], the Knut and Alice Wallenberg Foundation and the Novo Nordisk Foundation [grant number NNF10CC1016517].
Footnotes
Conflict of Interest
The authors declare that they have no conflicts of interest with the contents of this article.
Accession Numbers
The complete CAGE sequencing data and results can be found under the ArrayExpress accession code E-MTAB-6650.
The complete RNA sequencing data and results can be found under the ArrayExpress accession code E-MTAB-6722.
References
- Adiconis X, Haber AL, Simmons SK, et al. Comprehensive comparative analysis of 5′-end RNA-sequencing methods. Nat Methods. 2018;1 doi: 10.1038/s41592-018-0014-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balwierz PJ, Carninci P, Daub CO, et al. Methods for analyzing deep sequencing expression data: constructing the human and mouse promoterome with deepCAGE data. Genome Biol. 2009;10:R79. doi: 10.1186/gb-2009-10-7-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basehoar AD, Zanton SJ, Pugh BF. Identification and Distinct Regulation of Yeast TATA Box-Containing Genes. Cell. 2004;116:699–709. doi: 10.1016/s0092-8674(04)00205-3. [DOI] [PubMed] [Google Scholar]
- Berretta J, Morillon A. Pervasive transcription constitutes a new level of eukaryotic genome regulation. EMBO Rep. 2009;10:973–82. doi: 10.1038/embor.2009.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C, Coulouris G, Avagyan V, et al. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry JM, Hong EL, Amundsen C, et al. Saccharomyces Genome Database: the genomics resource of budding yeast. Nucleic Acids Res. 2012;40:D700–5. doi: 10.1093/nar/gkr1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danino YM, Even D, Ideses D, et al. The core promoter: At the heart of gene expression. Biochim Biophys Acta - Gene Regul Mech. 2015;1849:1116–31. doi: 10.1016/j.bbagrm.2015.04.003. [DOI] [PubMed] [Google Scholar]
- van Dijken J, Bauer J, Brambilla L, et al. An interlaboratory comparison of physiological and genetic properties of four Saccharomyces cerevisiae strains. Enzyme Microb Technol. 2000;26:706–14. doi: 10.1016/s0141-0229(00)00162-9. [DOI] [PubMed] [Google Scholar]
- Engel SR, Dietrich FS, Fisk DG, et al. The reference genome sequence of Saccharomyces cerevisiae: then and now. G3 (Bethesda) 2014;4:389–98. doi: 10.1534/g3.113.008995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberle V, Forrest ARR, Hayashizaki Y, et al. CAGEr: precise TSS data retrieval and high-resolution promoterome mining for integrative analyses. Nucleic Acids Res. 2015;43:e51. doi: 10.1093/nar/gkv054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberle V, Lenhard B. Promoter architectures and developmental gene regulation. Semin Cell Dev Biol. 2016;57:11–23. doi: 10.1016/j.semcdb.2016.01.014. [DOI] [PubMed] [Google Scholar]
- Haberle V, Li N, Hadzhiev Y, et al. Two independent transcription initiation codes overlap on vertebrate core promoters. Nature. 2014;507:381–5. doi: 10.1038/nature12974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberle V, Stark A. Eukaryotic core promoters and the functional basis of transcription initiation. Nat Rev Mol Cell Biol. 2018;19:621–37. doi: 10.1038/s41580-018-0028-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoskins RA, Landolin JM, Brown JB, et al. Genome-wide analysis of promoter architecture in Drosophila melanogaster . Genome Res. 2011;21:182–92. doi: 10.1101/gr.112466.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber W, Carey VJ, Gentleman R, et al. Orchestrating high-throughput genomic analysis with Bioconductor. Nat Methods. 2015;12:115–21. doi: 10.1038/nmeth.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaji H, Lizio M, Itoh M, et al. Comparison of CAGE and RNA-seq transcriptome profiling using clonally amplified and single-molecule next-generation sequencing. Genome Res. 2014;24:708–17. doi: 10.1101/gr.156232.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–9. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–9. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–30. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata M, Nishiyori-Sueki H, Kojima-Ishiyama M, et al. Detecting expressed genes using CAGE. Methods Mol Biol. 2014;1164:67–85. doi: 10.1007/978-1-4939-0805-9_7. [DOI] [PubMed] [Google Scholar]
- Parky D, Morrisy AR, Battenhouse A, et al. Simultaneous mapping of transcript ends at single-nucleotide resolution and identification of widespread promoter-associated non-coding RNA governed by TATA elements. Nucleic Acids Res. 2014;42:3736–49. doi: 10.1093/nar/gkt1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi LS, Larson MH, Gilbert LA, et al. Repurposing CRISPR as an RNA-Guided Platform for Sequence-Specific Control of Gene Expression. Cell. 2013;152:1173–83. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdóttir H, Winckler W, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–6. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salazar AN, de Vries ARG, van den Broek M, et al. Nanopore sequencing enables near-complete de novo assembly of Saccharomyces cerevisiae reference strain CEN.PK113-7D. FEMS Yeast Res. 2017;17 doi: 10.1093/femsyr/fox074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandelin A, Carninci P, Lenhard B, et al. Mammalian RNA polymerase II core promoters: insights from genome-wide studies. Nat Rev Genet. 2007;8:424–36. doi: 10.1038/nrg2026. [DOI] [PubMed] [Google Scholar]
- Shiraki T, Kondo S, Katayama S, et al. Cap analysis gene expression for high-throughput analysis of transcriptional starting point and identification of promoter usage. Proc Natl Acad Sci U S A. 2003;100:15776–81. doi: 10.1073/pnas.2136655100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smale ST, Kadonaga JT. The RNA Polymerase II Core Promoter. Annu Rev Biochem. 2003;72:449–79. doi: 10.1146/annurev.biochem.72.121801.161520. [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Taira H, Tsunoda T, et al. Diverse transcriptional initiation revealed by fine, large-scale mapping of mRNA start sites. EMBO Rep. 2001;2:388–93. doi: 10.1093/embo-reports/kve085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorvaldsdottir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14:178–92. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wery M, Descrimes M, Vogt N, et al. Nonsense-mediated decay restricts LncRNA Levels in Yeast Unless Blocked by Double-Stranded RNA Structure. Mol Cell. 2016;61:379–92. doi: 10.1016/j.molcel.2015.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yassour M, Pfiffner J, Levin JZ, et al. Strand-specific RNA sequencing reveals extensive regulated long antisense transcripts that are conserved across yeast species. Genome Biol. 2010;11:R87. doi: 10.1186/gb-2010-11-8-r87. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
To enable the easy usage of our data, we created custom data tracks and sessions for the Integrated Genomics Viewer (IGV, (Robinson et al. 2011; Thorvaldsdottir, Robinson and Mesirov 2013)), which can be found in the Supplementary information. After downloading the IGV from http://software.broadinstitute.org/software/igv/home, first the CEN.PK113-7D genome file (“CEN.PK113-7D.genome” part of the zipped Supplementary data file S1) has to be loaded via “Genomes” -> “Load Genome from File…” menu in IGV. After that, it is possible to load the session file for either the raw CAGE reads (“IGV_session_RawData.xml”, part of the zipped Supplementary data file S2) or the normalized CAGE reads (“IGV_session_NormData.xml”, part of the zipped Supplementary data file S3) using the “File” -> “Open Session…” menu.
After loading the session, a screen similar to the one shown in Figure 1A-D will be visible. For each condition, there are two tracks, one for reads on the plus strand and one for reads on the minus strand of the genome (labeled “_plus” and “_minus” respectively). In addition there are three different annotation tracks. The first one, labeled “ClusterAnnotations.bed”, will show each cluster with the full width, while the second one, labeled “ClusterAnnotationsDomTSS.bed”, will only show the position of the strongest TSS in each cluster. Both of these tracks include information about the cluster ID, and to which gene the cluster is annotated to (if any). For each gene, the strongest cluster is labeled as “(DomCluster)”. A third annotation track called “Gene” displays the blast based gene annotations for the CEN.PK113-7D genome.