Summary
KRAS-mutant pancreatic ductal adenocarcinoma (PDAC) is highly immunosuppressive and resistant to targeted and immunotherapies. Among the different PDAC subtypes, basal-like mesenchymal PDAC, which is driven by allelic imbalance, increased gene-dosage and subsequent high expression levels of oncogenic KRAS, shows the most aggressive phenotype and strongest therapy resistance. Here, we perform a systematic high-throughput combinatorial drug screen and identify a synergistic interaction between the MEK inhibitor trametinib and the multi-kinase inhibitor nintedanib, which targets KRAS-directed oncogenic signaling in mesenchymal PDAC. This combinatorial treatment induces cell cycle arrest and cell death, and initiates a context-dependent remodeling of the immunosuppressive cancer cell secretome. Using a combination of single cell RNA sequencing, CRISPR screens and immunophenotyping, we show that this combination therapy promotes intra-tumor infiltration of cytotoxic and effector T cells, which sensitizes mesenchymal PDAC to PD-L1 immune checkpoint inhibition. Overall, our results open new avenues to target this aggressive and therapy-refractory mesenchymal PDAC subtype.
Keywords: Pancreatic cancer, combination therapy, immune checkpoint inhibition, scRNA-seq
Introduction
With a ten-year survival rate of approximately 1%, PDAC is almost universally fatal 1 . Recently, it surpassed breast cancer becoming the third leading cause of cancer related deaths in the western world 2 . PDAC is highly refractory to all available treatment options, including chemo- and immunotherapies. Meaningful treatment responses are limited almost exclusively to the classical glandular subtype of the disease, characterized by an epithelial morphology and gene expression program 3–5 . Undifferentiated non-glandular PDACs display a mesenchymal morphology and a basal-like transcriptional program 3–8 . These tumors are characterized by a particularly poor prognosis, often not responding to standard of care chemotherapy, therefore representing an unmet clinical need 3–5,7 .
Novel therapeutic approaches, such as immune checkpoint blockade (ICB), have demonstrated no effect in trials of PDAC patients 9 . This lack of response likely results from the relatively low mutational burden, leading to a low amount of immunogenic neo-antigens. Combined with multiple immunosuppressive features of the PDAC tumor microenvironment (TME), this leads to scarcity of tumor infiltrating lymphocytes (TILs) 9 . Recent studies reported rare cases of PDAC patients presenting tumors with high T cell infiltration, a feature associated with prolonged overall survival 10–12 . These reports highlighted the potential to treat PDAC more effectively by targeting the immunosuppressive TME and recruiting TILs via rational combination therapies.
KRAS is mutationally activated in over 90% of PDAC patients. However, to date no clinically applicable strategy has been developed to effectively treat KRAS-mutant PDAC. Downstream of oncogenic KRAS, the RAF-MEK-ERK pathway plays a central role in tumor initiation 13 . Although MEK inhibitors yielded therapeutic value in RAS-mutant melanoma and lung cancer 14,15 , unstratified trials failed in PDAC patients. Recently, we and others showed that an increased gene-dosage (iGD) and expression of mutant KRAS (KRAS-mut) drives the disease, with the mesenchymal non-glandular basal-like subtype displaying the highest KRAS-mut gene-expression levels 5,16,17 . Based on the strong effects of KRAS-mut expression on PDAC phenotypes, we set out to develop a combination therapy for mesenchymal tumors that targets KRAS-driven tumor cell intrinsic signaling and in parallel reprograms the TME. By high-throughput drug screening combining the MEK inhibitor (MEKi) trametinib with 418 drugs, we identified a synergistic combination with the multikinase inhibitor nintedanib, inducing cell death and widespread reprogramming of the immunosuppressive microenvironment. The combination therapy activates an antitumor immune response, resulting in the recruitment of cytotoxic T cells, sensitizing mesenchymal tumors to ICB.
Results
Mesenchymal PDAC is resistant to MEK pathway inhibition
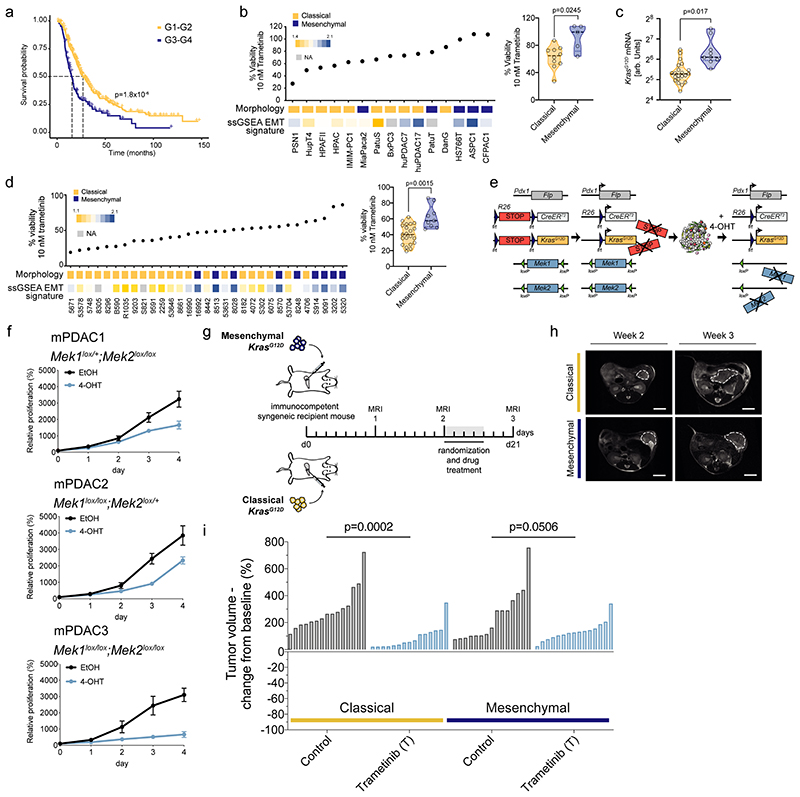
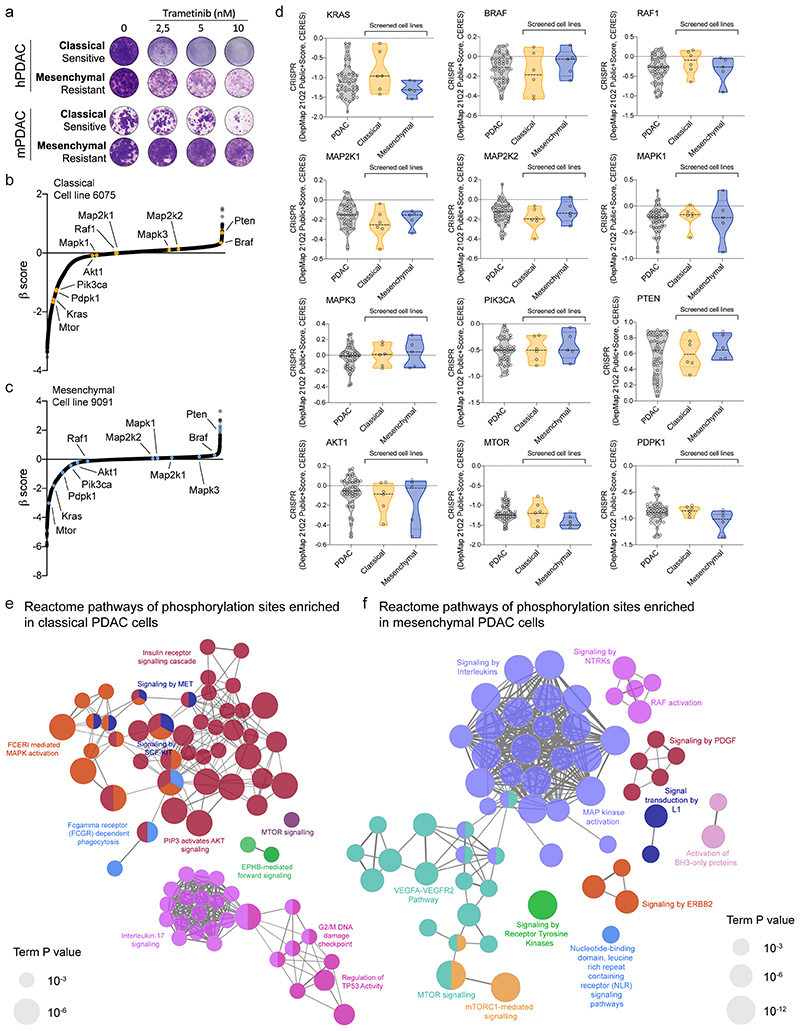
Recent data show that KRAS-mut expression levels have a strong impact on PDAC differentiation and phenotype, including the response to standard of care polychemotherapy 5,16,17 . In addition, the mesenchymal basal-like phenotype is linked to poor prognosis (figure 1a and 3,5–8 ). Based on the hypothesis that oncogenic KRAS overexpression provides a unique vulnerability in mesenchymal tumors, we systematically explored canonical RAF-MEK-ERK pathway inhibition. To this end, a panel of primary patient-derived cell cultures and conventional human PDAC (hPDAC) cell lines was screened with the MEKi trametinib. Unexpectedly, mainly cell lines with a classical epithelial morphology and gene expression signature 18,19 showed high sensitivity towards MEKi (figure 1b, extended data fig. 1a). Human PDAC cells of complete mesenchymal morphology, representing the most undifferentiated and aggressive subtype, are underrepresented and commonly not observed in surgically resectable cancers. Therefore, we extended our screen to mouse primary cell cultures (mPDAC) isolated from tumors of mice expressing KrasG12D in the pancreas 16 . Mesenchymal mPDAC cells expressed KrasG12D to the highest levels (figure 1c and 16 ). In line with hPDAC, mainly classical mPDAC cells were sensitive to MEKi, whereas almost all mesenchymal cells showed remarkable resistance (figure 1d, extended data fig.1a). These differences were not due to different dependencies on the driving oncogene KRAS, nor direct KRAS-downstream targets, as indicated by similar dependency scores of CRISPR/Cas9 based negative selection screens (extended data fig. 1b-d). In contrast, we observed fundamental differences in signaling output. Site-specific protein-phosphorylation profiling revealed enrichment of PDGF, ERBB2, mTORC1, RTK, NTRK signaling and the VEGFA/VEGFR pathway in mesenchymal tumor cells, whereas classical showed e.g. higher levels of the insulin receptor and AKT signaling cascade (extended data fig. 1e,f and supplementary table 1).
Figure 1. Resistance to MEK inhibition in vitro and in vivo .
a, Kaplan–Meier analysis comparing survival of surgically resected patients having either G1/G2 or G3/G4 tumor grading. We combined data from 6–8 .
b, Left, Percentage of cell viability at 10 nM trametinib in hPDAC cell lines. Cell morphology and single sample gene set enrichment analysis (ssGSEA) EMT signature are integrated below. Cells showing a classical phenotype are marked in yellow, those presenting a mesenchymal phenotype in blue. huPDAC17 and huPDAC7 are primary human patient derived PDAC cell cultures generated from PDX models in our laboratory 58 . Right, Violin plots comparing the percentage of cell viability at 10 nM trametinib between classical and mesenchymal hPDAC cell cultures.
c, Allele-specific KrasG12D mRNA expression in classical (n=21 mice) and mesenchymal (n=9 mice) tumors by combining amplicon-based RNA-seq and qRT-PCR.
d, Left, Percentage of cell viability at 10 nM trametinib in mPDAC cell cultures. Cell morphology and ssGSEA EMT signature are integrated below. Right, Comparison of the percentage of cell viability at 10 nM trametinib between classical and mesenchymal mPDAC cell cultures.
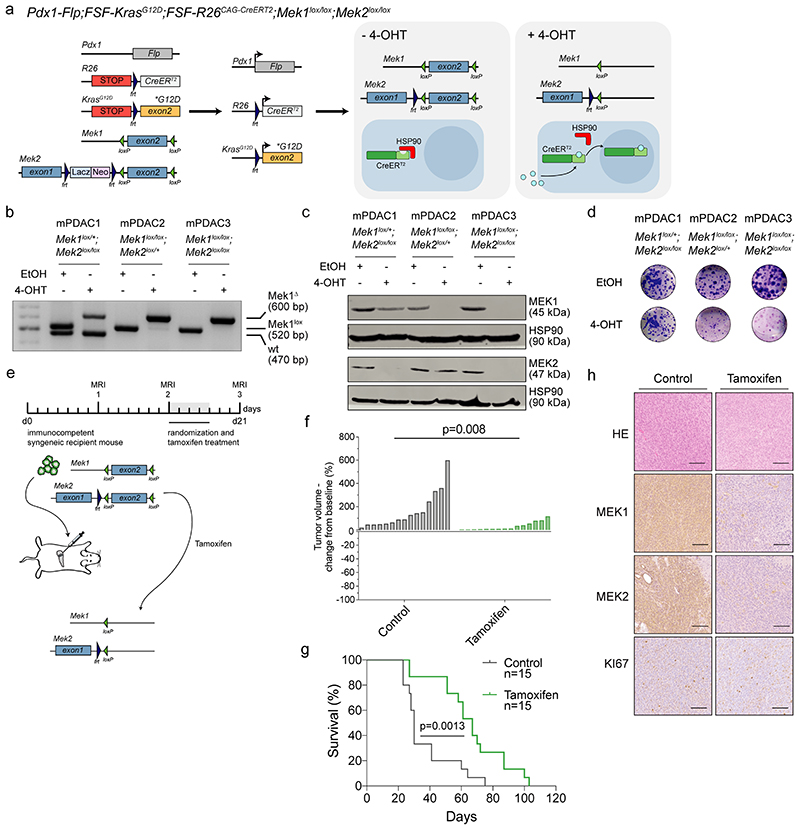
e, Schematic representation of the dual-recombinase system to inducible delete floxed Mek1 and Mek2 in established tumors using a tamoxifen activatable CreERT2 allele.
f, Viability assay of hydroxytamoxifen (4-OHT)-treated Mek1lox/+;Mek2lox/lox (mPDAC1), Mek1lox/lox;Mek2loxl+ (mPDAC2), Mek1lox/lox;Mek2lox/lox (mPDAC3) cell cultures compared to vehicle (ethanol, EtOH). Data are shown as mean ±SD; n=3 independent experiments.
g, Schematic representation of the experimental set up to test treatment efficacy of trametinib in vivo using subtype-specific models. Mesenchymal (9091) and classical (8661) mPDAC cells were used for the orthotopic transplantation experiments.
h, Representative magnetic resonance imaging (MRI) of trametinib treated mice before (week 2) and after one week of treatment (week 3). Scale bar, 5mm.
i, Waterfall plot of the fold change in tumor volume compared to baseline (determined by MRI-based volumetric measurements) of orthotopically transplanted classical and mesenchymal tumors after one week of trametinib treatment.
P values in (b, right), (c), (d, right) and (i) were calculated by two-tailed unpaired t test. P value in (a) was calculated with log-rank (Mantel-Cox) test.
To exclude insufficient MEK1/2 inhibition and/or feedback/cross-activation of the pathway in primary resistant cells, we developed a dual-recombinase based PDAC mouse model 20 allowing the inducible permanent genetic inactivation of the MEK pathway in established tumors (figure 1e and extended data fig. 2a): we crossed Pdx1-Flp;FSF-KrasG12D/+;FSF-R26CAG-CreERT2/+ mice with animals harboring loxP-flanked Mek1 and Mek2 alleles and subsequently ablated MEK1/2 in PDAC cells by tamoxifen administration in vitro and in vivo after orthotopic transplantation (figure 1e,f and extended data fig. 2a-h). Loss of MEK1/2 reduced cell proliferation in vitro, without triggering cell death. In line, ablation of MEK1/2 in vivo did not induce tumor regression or a complete growth arrest as assessed by KI67 staining, but delayed tumor progression of the respective animals (figure 1f, extended data fig. 2b-h). In parallel, we found that pharmacological MEKi delayed disease progression in classical epithelial PDAC significantly (p=0.0002 vs controls), but only moderately in the mesenchymal subtype (p=0.0506 vs controls). Of note, trametinib did not induce tumor regression as observed in other cancer types, such as non-small cell lung cancer (figure 1g-i and 14,21 ). Therefore, neither MEKi nor complete sustained genetic disruption of canonical KRAS-downstream signaling is sufficient to induce PDAC regression, demonstrating the need to develop combinatorial treatment strategies.
Drug screening identifies novel therapies for mesenchymal PDAC
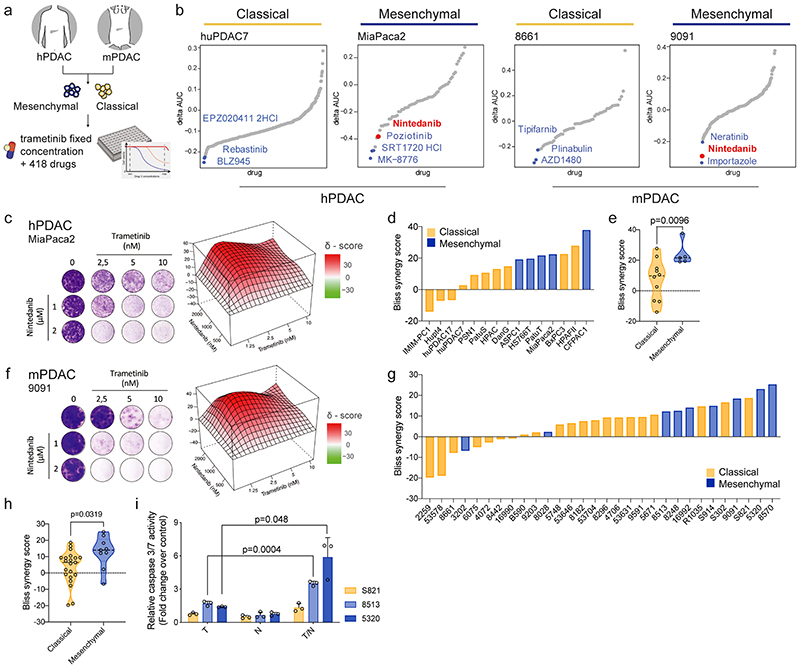
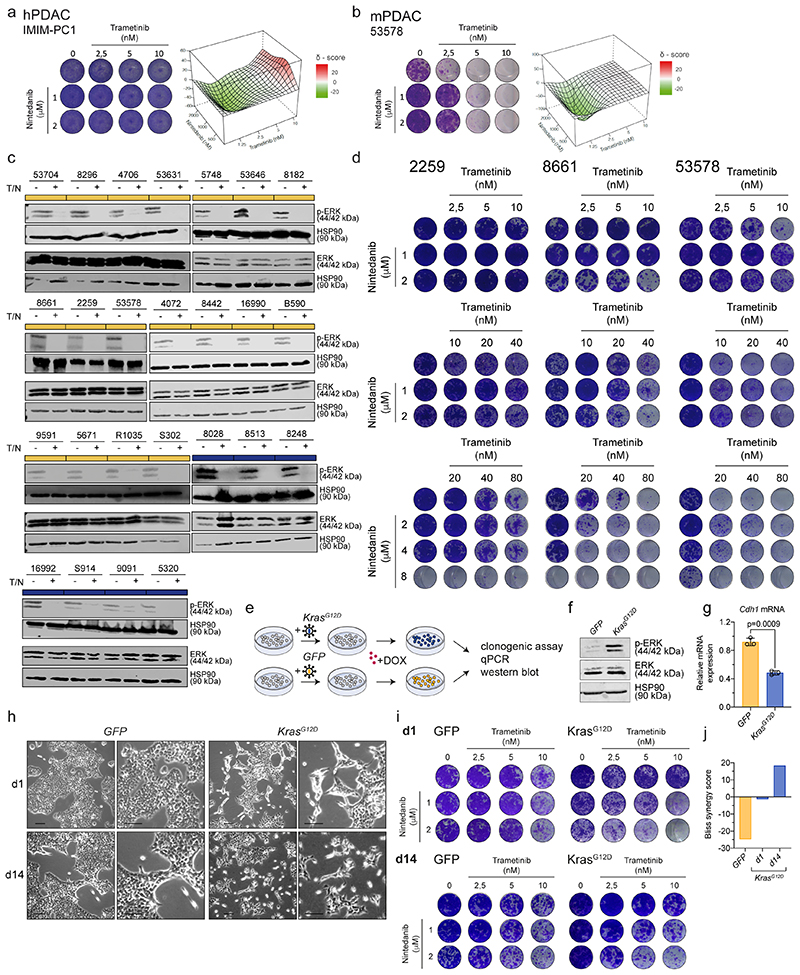
We performed a systematic high-throughput combinatorial compound screen to identify drugs synergizing with MEKi. We screened two hPDAC and two mPDAC cultures, representing both classical and mesenchymal KRAS-subtypes, with trametinib in combination with 418 drugs in preclinical and clinical investigation (figure 2a and supplementary table 2). One of the top-hits in mesenchymal PDAC was the clinically approved RTK-inhibitor nintedanib (figure 2b). We validated our findings in long-term clonogenic assays in a larger cohort and observed synergistic interactions of trametinib and nintedanib in 11/15 hPDAC cultures, with the strongest effect in the five mesenchymal cultures (figure 2c-e; p=0.009). We extended the combinatorial screen to 30 additional mPDAC cultures 16 . A high synergism was achieved in most of the models, with the mesenchymal Kras-mut overexpressing cells predominantly benefitting from the treatment (figure 2f-h). Antagonism was observed in 3/15 human and 6/30 mouse cell cultures, respectively, which were characterized almost exclusively by a classical epithelial morphology. In line with the heterogeneity of responses observed in the clinical setting even within molecularly stratified cohorts, we detected synergism in some classical PDAC cultures, demonstrating heterogeneity within subtypes (figure 2c-h and extended data fig. 3a,b). Trametinib and nintedanib (T/N) treated epithelial and mesenchymal PDAC cells revealed similar changes in ERK phosphorylation, indicating that primary sensitivity and resistance to T/N is not due to differences in canonical pathway blockade (extended data fig. 3c). Increasing the dosage of both inhibitors did not alter the antagonistic action of the drug combination substantially, arguing for fundamental biological differences between the tumors (extended data fig. 3d). Strikingly, we observed cell death upon T/N treatment, with the strongest effects in mesenchymal PDAC (figure 2i).
Figure 2. Systematic combinatorial drug screens identify novel therapies for non-glandular mesenchymal PDAC.
a, Experimental set up of the high-throughput drug screen.
b, Combinatorial drug screen on two mPDAC (9091, 8661) and two hPDAC cell cultures (MiaPaca2, huPDAC7).
c, f Clonogenic assay and synergy map of representative hPDAC (c) and mPDAC (f) cultures treated with the indicated concentrations of trametinib and nintedanib.
d, g, Bliss synergy scores integrated with cell morphology for hPDAC (d) and mPDAC (g) cell cultures (classical subtype in yellow, mesenchymal in blue).
e, h, Comparison of the Bliss synergy scores, from panels (d) and (g), between classical and mesenchymal hPDAC (e) and mPDAC (h) cells.
i, Induction of caspase 3/7 activity upon treatment with trametinib (10 nM), nintedanib (2 μM) or the combination of both for 24 hours relative to the vehicle treated control. Data are shown as mean ±SD; n=3 independent experiments.
P values in (e), (h) and (i) were calculated by two-tailed unpaired t test.
T: trametinib, N: nintedanib, T/N: trametinib+nintedanib.
We assessed the consequences of oncogenic KRAS-mut expression on cell morphology and treatment response using classical mPDAC cells transduced with a doxycycline-inducible KRASG12D vector or GFP as control. KRASG12D overexpression led to increased ERK1/2 phosphorylation, decreased E-cadherin expression (Cdh1), and morphological changes of the epithelial PDAC cells towards a mesenchymal spindle shaped morphology, which increased from day 1 to day 14 (extended data fig. 3e-h). T/N treatment revealed a remarkable change in drug sensitivity. While the controls showed antagonism, this was reverted to synergism in the KRASG12D overexpression system (extended data fig. 3i,j), supporting the notion that KRAS levels impact on cellular differentiation states 16 , are main drivers of mesenchymal PDAC and provide therapeutic vulnerabilities.
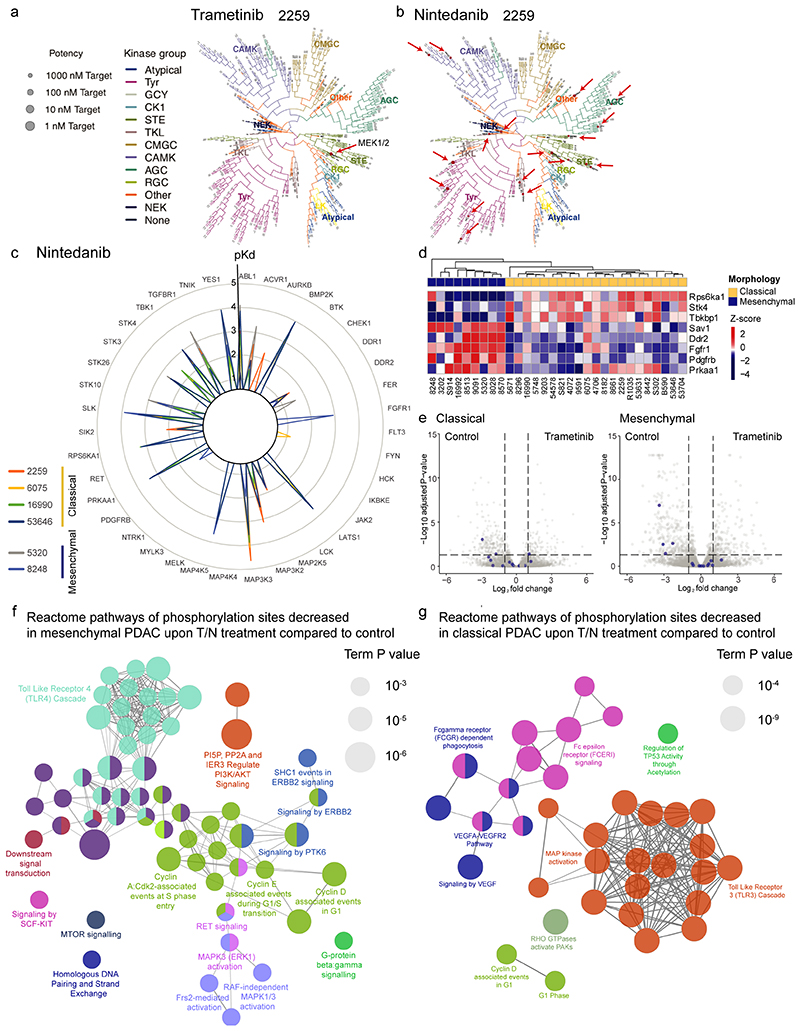
To uncover the direct targets of trametinib and nintedanib and to elucidate the drivers of treatment response and resistance, we performed kinobead pulldown assays on six mPDAC cultures of both subtypes. While trametinib showed selectivity for binding MEK1/2, nintedanib displayed a broad range of targets, enriched in RTKs and cell surface receptors (extended data fig. 4a-c). Importantly, expression of the nintedanib targets was subtype-dependent. Four were selective for mesenchymal PDAC, including PDGFRB, FGFR1 and DDR2, and 24 were shared by both subtypes. Further, expression of these targets remained largely unchanged after drug perturbation (extended data fig. 4d,e). Therefore, differences in the basal gene expression program underlie synergistic drug-action in PDAC subtypes.
To identify the functional relevant pathways mediating response to T/N, we analyzed changes in the phosphoproteome. In mesenchymal PDAC, we observed a decreased activity of a broad range of important cancer-relevant pathways, such as the cell cycle regulators CDK2, Cyclin D and Cyclin E; PP2A and IER3 that regulate PI3K/AKT signaling; ERBB2, mTOR and KIT downstream signaling, as well as RAF-dependent and independent ERK1/2 activation (extended data fig. 4f and supplementary table 3). These findings support the view that mesenchymal PDAC cells depend on broad RTK-driven signaling input. In contrast, we found a more limited spectrum of altered pathways in classical tumors, including distinct downregulation of VEGFA/VEGFR pathway and RHO GTPases, as well as decreased MAPK activation (extended data fig. 4g and supplementary table 3). This confirmed and extended our initial observation of fundamental differences in signaling between classical and mesenchymal tumors under basal conditions (see extended data fig. 1), reinforcing the notion that multiple kinases need to be targeted to achieve meaningful responses in the mesenchymal subtype.
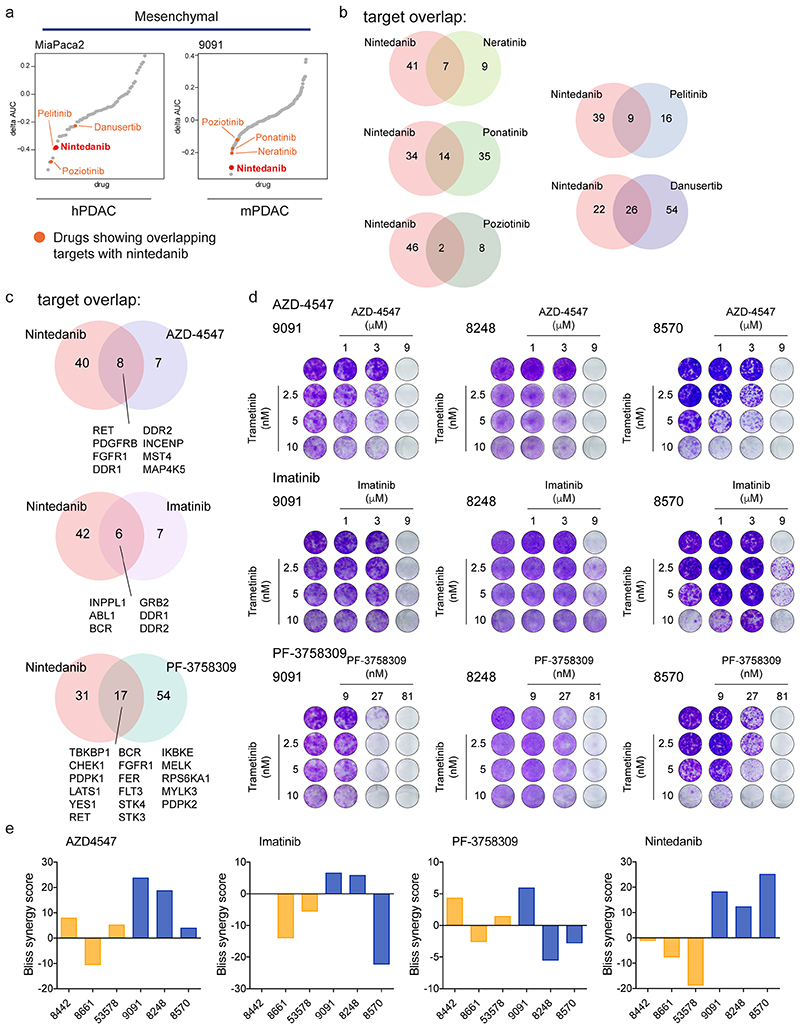
Next, we assessed other drugs sharing targets with nintedanib. Strikingly, in mesenchymal PDAC several top hits of our combinatorial drug screen are multikinase inhibitors that display an overlapping target spectrum with nintedanib (extended data fig. 5a,b and supplementary table 2 and 4). Additionally, we tested novel compounds with overlapping targets (extended data fig. 5c, https://www.proteomicsdb.org). AZD-4547, which shares with nintedanib amongst others the targets PDGFRB, FGFR1, DDR1 and DDR2, mimics the synergistic responses observed with T/N and could potentially be used in combination with trametinib. In contrast, other drugs that share targets with nintedanib, including imatinib, display highly heterogeneous responses, showing cell type and contextspecific synergisms (extended data fig. 5c-e).
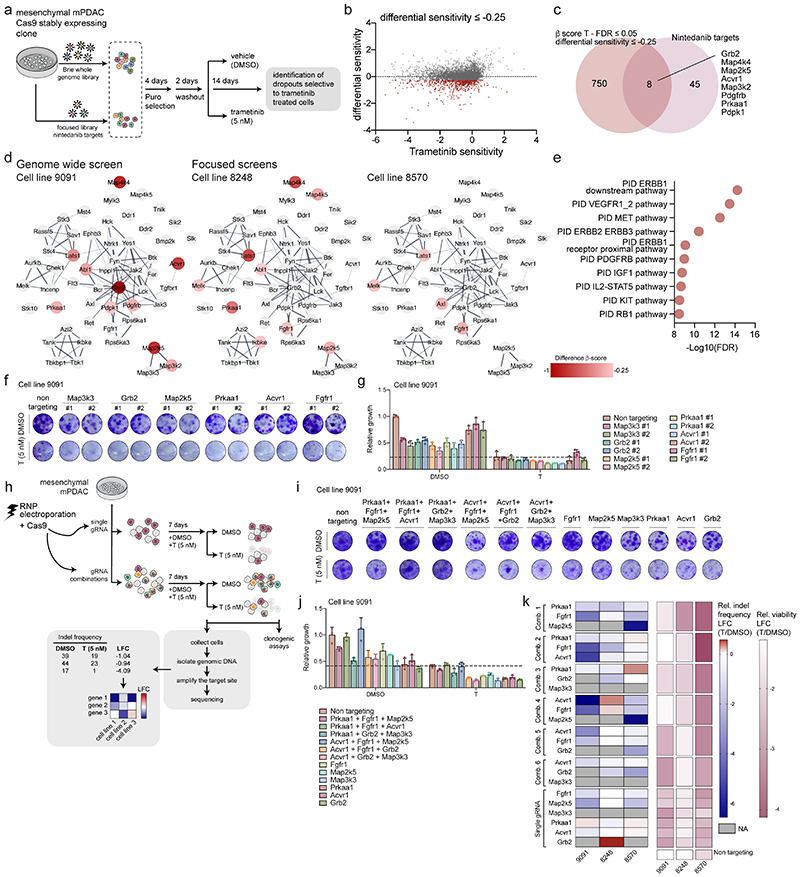
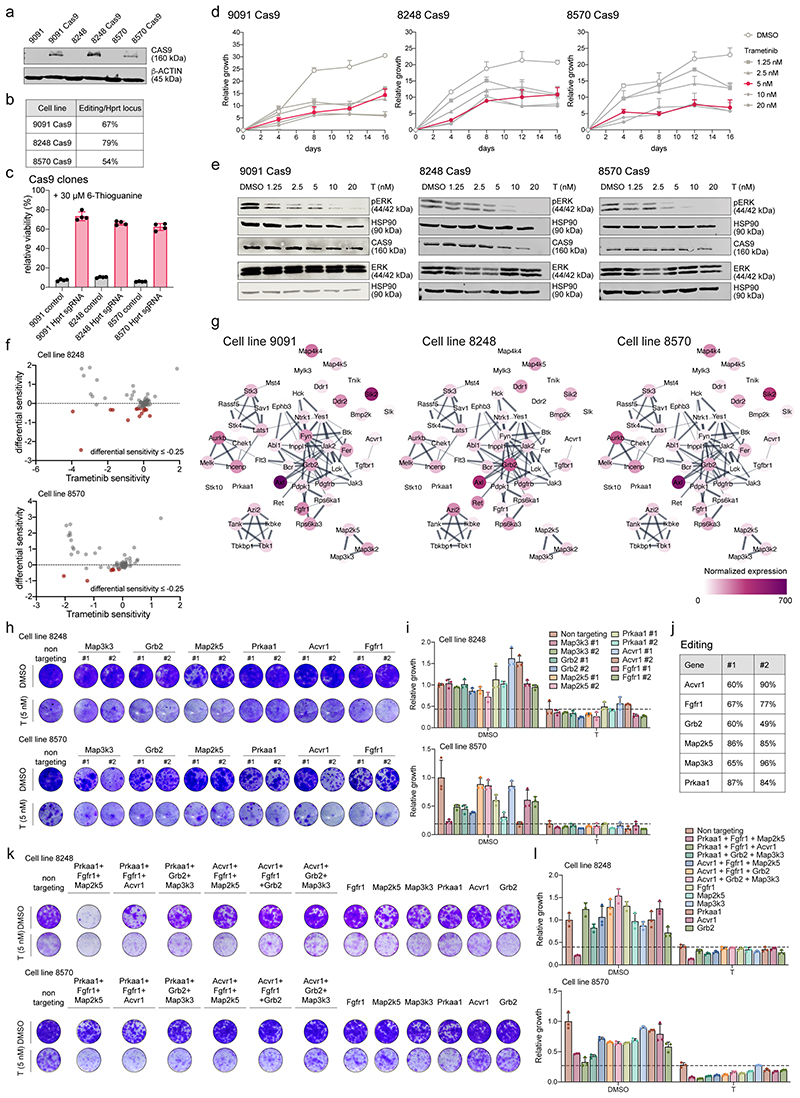
To functionalize the targets of nintedanib and decipher key gens synergizing with MEKi, we employed pooled genome-wide as well as focused CRISPR/Cas9-based negative selection (viability) screens in three mesenchymal mPDAC cell cultures. We performed a genome-scale screen in 9091 cells and an in-house developed nintedanib-target-focused screen in 8248 and 8570 cells, with and without trametinib (figure 3a-e, extended data fig. 6a-f and supplementary table 5 and 6). To identify genes altering trametinib sensitivity, we calculated differential sensitivity scores, as the difference in β-score between the trametinib- and DMSO-treated arms. We focused on genes displaying a negative differential sensitivity score, indicating enhanced depletion in presence of trametinib (figure 3b and extended data fig. 6f).
Figure 3. Genetic-screens uncover nintedanib targets that sensitize mesenchymal PDAC towards trametinib.
a, Schematic representation of genome-scale and nintedanib-target focused CRISPR/Cas9 screens.
b, Genome-scale screen in mesenchymal mPDAC 9091 cells. Trametinib sensitivity (x-axis) represents β-scores calculated as sgRNA representation difference between trametinib-treated cells and their initial representation. Differential sensitivity (y-axis) indicates β-score differences between trametinib- and DMSO-treated arms. In red, genes presenting differential sensitivity ≤-0.25.
c, Venn-diagram of overlap of genome-wide screening hits (b) (differential sensitivity ≤-0.25 and FDR ≤0.05) and the nintedanib targets.
d, Network of nintedanib targets of CRISPR/Cas9 screens in 9091 (genome-wide), 8248 and 8570 (focused) cells built on the string database and visualized using Cytoscape. Nintedanib targets are color-coded according to the differential sensitivity between trametinib- and DMSO-treated arms.
e, Pathway enrichment within the MSigDB canonical-pathways database of genomewide screening hits of (b) showing a differential sensitivity ≤-0.25.
f, Lentiviral CRISPR/Cas9-mediated deletion of selected top-scoring nintedanib targets in 9091 cells. Knock-out cells were treated with trametinib (5 nM) or DMSO and viability was assessed through clonogenic assays.
g, Quantification of panel (f). Data are normalized to DMSO-treated non-targeting controls (mean ±SD; n=3 biological replicates). The dashed line represents the mean of trametinib-treated non-targeting controls.
h, Combinatorial deletion of nintedanib targets via ribonucleoprotein (RNP) electroporation. Mesenchymal mPDAC cells 9091, 8248 and 8570 were electroporated to deliver the Cas9-sgRNA complex. The resulting cells were treated for 7-9 days with DMSO or trametinib (5 nM). Cell viability was assessed via clonogenic assays (panel i,j and extended data fig. 6) and indel frequencies via sequencing (panel k). The indels were used to determine the log2-fold-change (LFC) of the indel frequency in panel (k).
i, Clonogenic assays of 9091 cells electroporated with RNPs targeting the indicated nintedanib targets. Knock-out cells were treated with trametinib (5 nM) or DMSO.
j, Quantification of panel (i). Data are normalized to DMSO-treated non-targeting controls (mean ±SD; n=3 biological replicates). Dashed line represents the mean of trametinib-treated non-targeting controls.
k, Left, Heatmap of the indel frequencies LFC (trametinib/DMSO) as described in (h). Right, Heatmap of relative viability (trametinib/DMSO) of the clonogenic experiments described in (i,j).
In the genome-scale CRISPR screen we identified 8 nintedanib targets out of a total of 758 hits with statistically significant β-scores, whose inactivation resulted in the specific depletion of trametinib treated cells (figure 3b-d). In the focused screens in 8248 and 8570 cells, nine and four nintedanib targets cooperated with trametinib, respectively (figure 3d). Altogether, of 53 nintedanib targets identified in kinobead pulldown experiments, 15 showed functional relevance in the presence of trametinib in mesenchymal PDAC cells (figure 3d). Importantly, the three different mesenchymal cell cultures showed some degree of heterogeneity across relevant nintedanib targets, indicating potential differences in underlying genetic, epigenetic and phenotypic characteristics of this subtype. Further analysis of the targets and their interaction using the STRING database (http://string-db.org) revealed a high degree of interconnectivity and convergence on FGFR, MEK/ERK family members and PDGFR regulated networks with different members of these pathways observed in the three screens (figure 3d). Unexpectedly, gene expression profiling revealed no clear correlation between mRNA abundance of nintedanib targets and trametinib sensitization upon depletion (extended data fig. 6g). Taken together, these data indicate that no single kinase but rather a specific spectrum of targets, such as PDGFR, FGFR and MEK/ERK family members, act in concert to mediate therapeutic efficacy in a context-specific manner.
Analysis of the 758 hits of the genome-scale CRISPR screen allowed us to identify pathways in mesenchymal PDAC, which globally cooperated with MEKi, such as ERBB, PDGFRB, KIT, and RB1 (figure 3e). These pathways correlated to a high degree with the phospho-proteomic analysis of the T/N-treated mesenchymal PDAC cells, thereby cross-validating these results functionally (extended data fig. 4).
To extend and validate the top-scoring nintedanib targets of our negative selection screens, we exploited single and combinatorial CRISPR/Cas9 sgRNA-based approaches. First, we individually depleted Acvr1, Grb2, Map2k5, Map3k3, Prkaa1 and Fgfr1, and assessed cell growth using clonogenic assays with and without trametinib. In-line with our negative selection screens, we observed heterogeneity in the cooperation of these nintedanib targets with MEKi across the different models (figure 3f,g and extended data fig. 6h-j). To probe the cooperation among the identified targets in mediating trametinib-sensitization, we next depleted Acvr1, Grb2, Map2k5, Map3k3, Prkaa1 and Fgfr1 in triple combinations with and without trametinib, using a transfection-based Cas9-sgRNA ribonucleoprotein (RNP) approach (figure 3h-k and extended data fig. 6k,l). Additionally, we correlated the relative indel frequency of each combination with the loss of viability in presence of trametinib (figure 3h,k). Consistently, we observed a heterogeneous response across the three cellular models, with the combinatorial depletion of Prkaa1, Fgfr1 and Map2k5 being most efficient in two out of three mesenchymal cell cultures in presence of trametinib. This confirmed our hypothesis that the depletion, not of one, but of a combination of multiple targets is important to sensitize mesenchymal PDAC towards MEKi. Thus, broad targeting is needed to efficiently and comprehensively treat mesenchymal PDAC.
T/N triggers a T cell-dependent anti-tumor immune response
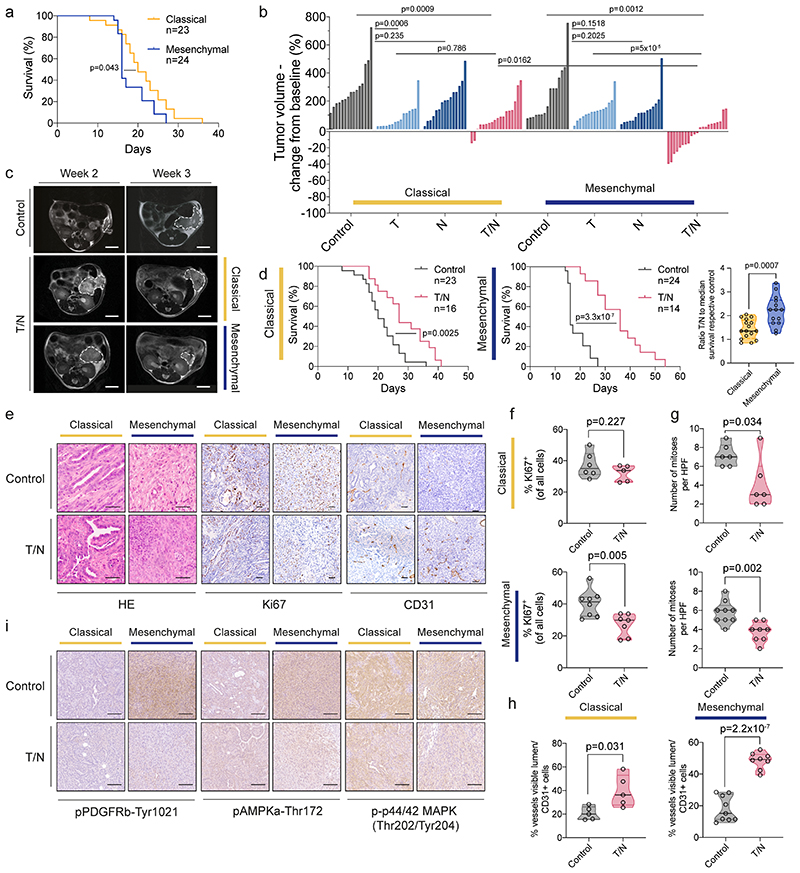
Our in vitro findings prompted us to explore the combination treatment in vivo in syngeneic orthotopic transplantation models of classical and mesenchymal PDAC (see figure 1g). These models resemble the clinical course of PDAC patients, with the mesenchymal subtype model being more aggressive, displaying rapid and uniform tumor progression to death (figure 4a).
Figure 4. The combination treatment prolongs survival and reprograms the tumor microenvironment in vivo .
a, Kaplan-Meier curve comparing the survival of classical and mesenchymal orthotopic PDAC models. The number of mice per treatment condition is indicated.
b, Quantification of tumor volume changes of the classical and mesenchymal subtype after one week of indicated treatment assessed by MRI. Each column represents one mouse, in comparison with baseline MRI measurement before treatment. Control, T, N and T/N cohorts are shown.
c, Representative MRI of vehicle (Control) and T/N treated mice before (week 2) and after 1 week of treatment (week 3). Scale bar, 5 mm.
d, Left and middle panel, Kaplan-Meier survival curve of classical and mesenchymal orthotopic models. The number of mice per treatment condition is indicated. Right, Comparison of the ratio between the survival of mice treated with T/N and the median survival of their respective vehicle treated controls.
e, Representative H&E and immunohistochemical staining for KI67 and CD31 of tumor sections from orthotopically transplanted classical and mesenchymal PDAC subtype models treated with vehicle (Control) or T/N for 1 week. Scale bars, 50 μm.
f, Quantification of KI67+ cells (classical: control n=6, T/N n=5; mesenchymal: control n=8, T/N n=7).
g, Quantification of the number of mitoses per high power field (classical: control n=6, T/N n=6; mesenchymal: control n=9, T/N n=8).
h, Quantification of vessels CD31+ (classical: control n=5, T/N n=5; mesenchymal: control n=9, T/N n=8).
i, Representative immunohistochemical staining for pPDGRFb-Tyr1021, pAMPKa-Thr172 and p-p44/42 (Thr202/Tyr204) of tumor sections from orthotopically transplanted classical and mesenchymal PDAC subtype models treated with vehicle (Control) or T/N for 1 week. Scale bars, 100 μm.
T: trametinib, N: nintedanib, T/N: trametinib+nintedanib.
P values in (b), (d, right), (f), (g) and (h) were calculated by two-tailed unpaired t test. P values in (a) and (d, left and middle) were calculated with log-rank (Mantel-Cox) test.
Note: The classical and mesenchymal cohorts in panel (a) are the same shown in panel (d). Vehicle and trametinib-treated cohorts shown in panel (b) are the same as shown in figure 1, panel (i).
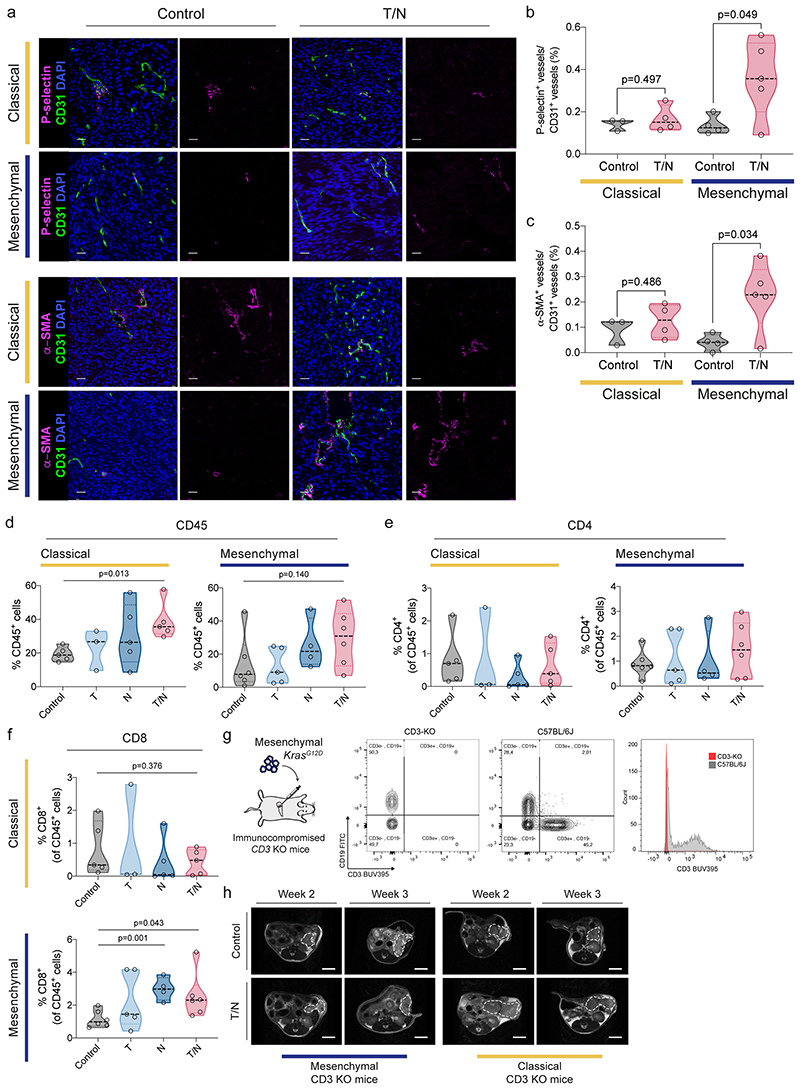
We randomized tumor-bearing mice and observed that the T/N combination led to a remarkable response of mesenchymal PDAC with a significant reduction in tumor volume of up to ~40% and doubled survival (figure 4b-d). Moreover, we observed not only a decrease in tumor cell proliferation, but also vascular remodeling as evidenced by an increased amount of CD31+ vessels (figure 4e-h), as well as markers of angiogenesis and endothelial cell activation (extended data fig. 7a-c).
Unexpectedly, also the classical subtype responded to the combination therapy; however, this effect was mainly mediated by trametinib as evidenced by similar tumor volumes of trametinib and T/N treated cancers (p=0.786; figure 4b). In addition, mesenchymal tumors showed a superior overall response with two partial remissions and stable disease in 12/21 mice treated with T/N, according to the Response Evaluation Criteria in Solid Tumors (RECIST) (figure 4b,c). In contrast, classical tumors showed no partial remission and only 3/18 mice displayed stable disease. Accordingly, the difference in T/N-induced tumor regression between both subtypes is statistically significant (p=0.0162, figure 4b). This translated into improved overall survival, which was doubled in the mesenchymal subtype (36 days for T/N vs 16 days for controls), whereas classical tumors displayed only a 50% increase (27 days for T/N vs 20 days for controls; figure 4d). This difference in survival benefit is statistically significant (p=0.0007; figure 4d, right panel). Thus, the novel combination is to our knowledge the first preclinical therapy that induces tumor regression and an increase in overall survival in Kras-mut iGD-driven mesenchymal PDAC. In line with the differential response, we observed a subtype-specific decrease in PDGFRB-Tyr1021 and AMPKα-Thr172 (PRKAA1) phosphorylation of T/N treated mesenchymal tumors in vivo (figure 4i). Both kinases are important nintedanib targets identified in the proteomic kinobead-based pulldown approach (extended data fig. 4).
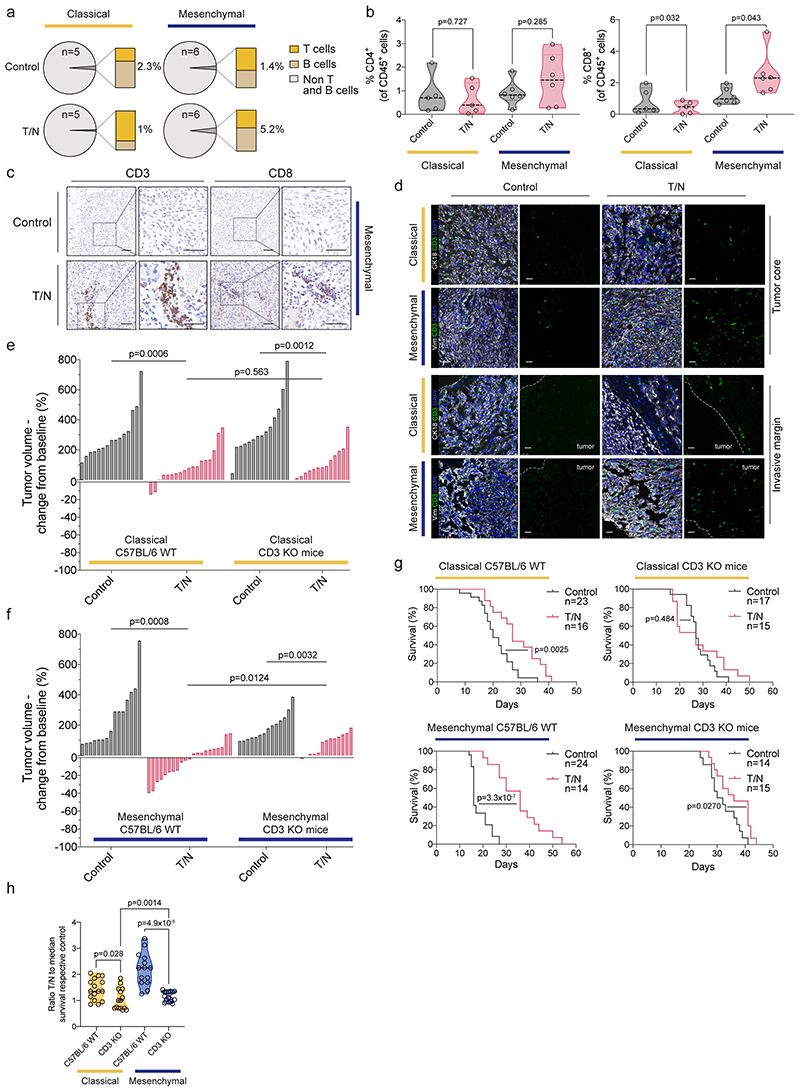
PDAC is considered immunologically “cold”: its microenvironment is characterized by a lack of cytotoxic T cells and infiltration of immunosuppressive immune cells 9,21 . T/N treatment substantially increased T cell infiltration into mesenchymal tumors. In contrast, classical tumors displayed features of immune exclusion – showing only a moderate enrichment of T cells at the tumor margins (figure 5a-d and extended data fig. 7d-f). Immunophenotyping of mesenchymal tumors revealed increased infiltration of CD8+ T cells, localized preferentially around vessels, arguing that vascular remodeling upon drug perturbation contributes to cytotoxic T cell infiltration (extended data fig. 7a-c).
Figure 5. The combination treatment enhances tumor immune infiltration specifically in the mesenchymal subtype.
a, Fraction of adaptive immune cell populations in tumors from vehicle (Control) and T/N treated mice. Classical and mesenchymal orthotopic transplanted tumors, originating from 8661 and 9091 mPDAC cells, respectively, were analyzed by flow cytometry. The number of tumors/condition analyzed is depicted in the corresponding panel.
b, FACS staining for CD4+ and CD8+ T cells of tumors treated with vehicle or T/N combination for 1 week. Individual tumors are shown as single points in the graph (classical: control n=5, T/N n=5; mesenchymal: control n=6, T/N n=6).
c, Representative images of IHC staining for CD3+ and CD8+ T cells of tumor sections from orthotopically transplanted mesenchymal models treated with vehicle or T/N for 1 week. Scale bars, 50 μm.
d, Representative images of tissue sections stained for CD3+ cells (green). Keratin 18 detects epithelial PDAC cells in classical tumors while vimentin detects undifferentiated tumor cells in mesenchymal tumors (white). DAPI was used for nuclear staining (blue). Tumor borders are marked by a white dotted line and tumor localization is indicated. Scale bars, 25 μm.
e, f, Waterfall plot of the response of classical (e) and mesenchymal (f) tumors orthotopically transplanted in immunocompetent C57BL/6 wild-type (C57BL/6 WT) and T cell deficient CD3ε-knockout (CD3ε-KO) mice after 1 week of T/N treatment (fold-change compared to baseline before treatment based on MRI-volumetric measurements, y axis).
g, Kaplan-Meier survival curve of CD3ε-KO and C57BL/6 WT mice orthotopically transplanted with classical (upper panel) or mesenchymal (lower panel) tumor cells. The number of mice per treatment condition is shown.
T: trametinib, N: nintedanib, T/N: trametinib+nintedanib.
h, Comparison of the ratio between the survival of mice treated with T/N and the median survival of their respective vehicle-treated controls.
P values in (b), (e), (f) and (h) were calculated by two-tailed unpaired t test. P values in (g) were calculated with log-rank (Mantel-Cox) test.
Note: The immunocompetent cohorts shown in panel (e, left), (f, left), (g, left) and (h) are the same shown in figure 3, panels (a), (b) and (d, left and middle panel).
To investigate the role of T cells in therapy response, we employed CD3ε knockout mice on a C57BL/6 background, lacking all T cells 22 (figure 5e-h and extended data fig. 7g,h). T cell deficiency blunted T/N response and diminished survival benefits of mesenchymal tumors (figure 5f-h and extended data fig. 7h). The tumor volume decreased significantly in wild-type vs. CD3ε knockout mice (p=0.0124; figure 5f). In addition, T/N treatment prolonged survival in comparison to controls by 20 days in wild-type mice, but only marginally by 5 days in mice lacking T cells (p=4.9x10-5, figure 5g,h). These data support the notion that T cells contribute significantly to the in vivo efficacy of the T/N combination in the mesenchymal subtype. However, treatment response is not mediated by T cells alone, but depends on TME reprogramming and tumor cell intrinsic drug action.
In contrast to mesenchymal PDAC, the classical subtype showed a mixed response in T cell deficient animals. There was no difference in tumor volume (p=0.563, wild-type vs CD3ε knockout mice; figure 5e), but a slight effect of T cell deficiency on survival, which was reduced by 5 days in T/N treated CD3ε knockout mice (p=0.028, figure 5g,h), indicating immune surveillance also in this model upon therapy, but to a significantly smaller extent (p=0.0014, figure 5h).
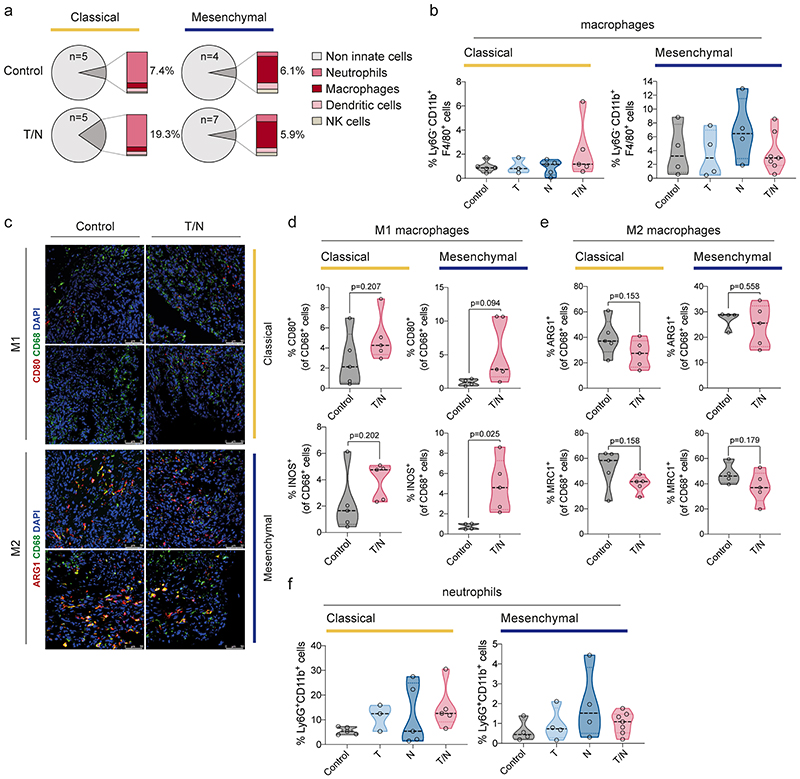
Cancer therapies can affect macrophage functions and alter their recruitment or polarization states 23 . The combination treatment did not change the overall number of macrophages substantially (extended data fig. 8a,b). However, their polarization changed from the pro-tumorigenic M2- to an anti-tumorigenic M1-like state (extended data fig. 8c-e), suggesting that they might contribute to anti-tumor responses. However, this effect was present in both subtypes. Therefore, these changes do not explain the observed subtype-specific differences.
Distinctively, epithelial tumors displayed an increase of neutrophils upon treatment (extended data fig. 8a,f). The role of tumor associated neutrophils is controversial; some studies have shown their ability to block anti-tumor immune responses and mediate CD8+ T cell suppression, others have highlighted their various anti-tumor properties, including direct cytotoxicity and inhibition of metastasis 24–26 . Therefore, distinct immune cell types and mechanisms seem to contribute to the anti-tumor effects of the drug combination in both subtypes, with T cells being the key contributor to the strong effects observed in mesenchymal PDAC.
The T/N combination sensitizes mesenchymal PDAC towards ICB
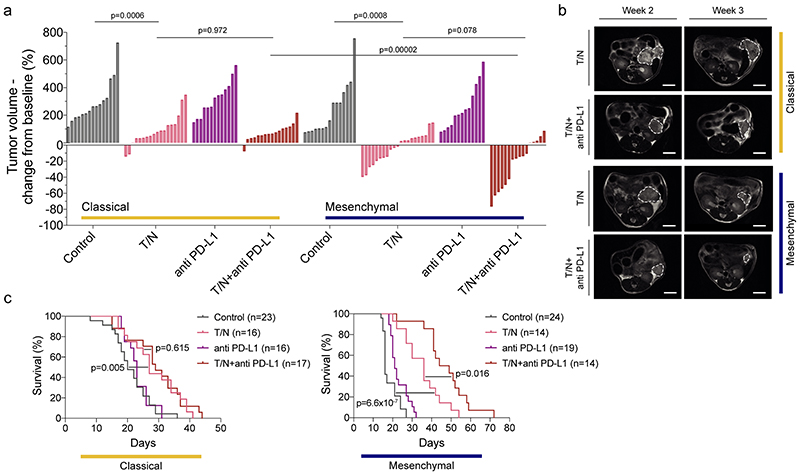
Our in vivo findings prompted us to investigate whether the combination therapy could sensitize the highly aggressive mesenchymal subtype towards ICB. Compared with controls, anti-PD-L1, and T/N treated groups, T/N+anti PD-L1 therapy induced tumor regression up to ~80% and increased survival selectively in mesenchymal PDAC (p=0.016 T/N+anti PD-L1 vs T/N; figure 6a-c). The median survival benefit by adding anti PD-L1 to the combination was 10.5 days compared to T/N alone and 30.5 days compared to vehicle, representing an almost 3-fold increase in survival (figure 6c). 6/16 mice showed objective tumor regression with a partial response according to RECIST. Of the remaining 10 mice, 8 displayed stable and only 2 progressive disease (p=0.078 T/N+anti PD-L1 vs T/N; figure 6a). In contrast, no tumor regression, survival benefit or increased response rates were observed in classical PDAC, and both subtypes did not respond to ICB with PD-L1 alone (figure 6a,c).
Figure 6. The trametinib/nintedanib combination sensitizes mesenchymal PDAC towards anti PD-L1 immune checkpoint blockade.
a, Waterfall plot showing tumor response of classical and mesenchymal PDAC to T/N+anti PD-L1 vs vehicle control, T/N, and baseline anti PD-L1 therapy after one week of treatment (values represent fold-change compared to baseline before treatment based on MRI-volumetric measurements, y axis). P values calculated with two-tailed unpaired t test.
b, Representative MRI of vehicle and T/N+anti PD-L1 treated mice before (week 2) and after 1 week treatment (week 3). Scale bar, 5 mm.
c, Kaplan-Meier survival curves of classical and mesenchymal orthotopically transplanted models of the indicated treatment arms. The number of mice is indicated in the corresponding panels. P value was calculated with log-rank (Mantel-Cox) test.
T: trametinib, N: nintedanib, T/N: trametinib+nintedanib, T/N+anti PD-L1: trametinib+nintedanib+anti PD-L1 antibody.
Note: The classical and mesenchymal cohorts, control and T/N, in panel (a) and (c) are the same shown in figure 3, panels (a), (b) and (d, left and middle panel).
In summary, the triple treatment with ICB markedly improves anti-tumor responses offering a clear survival benefit selectively in mesenchymal PDAC.
Single cell RNA sequencing reveals context-specific T/N driven changes
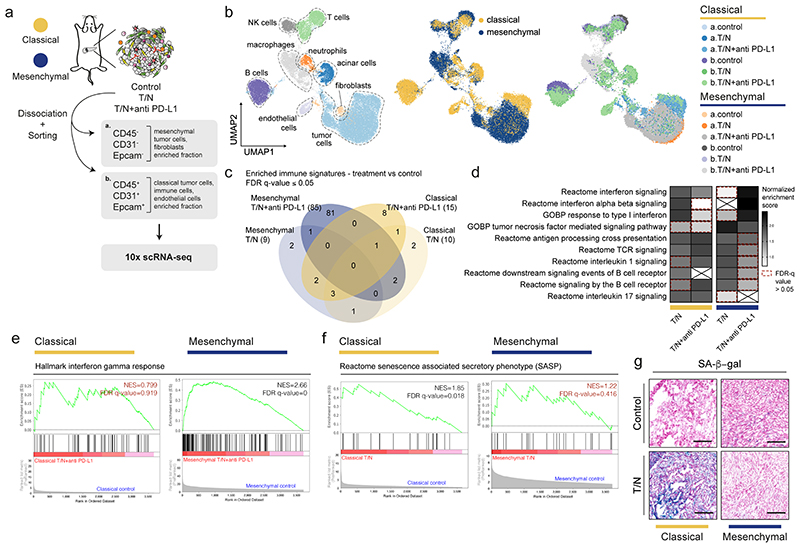
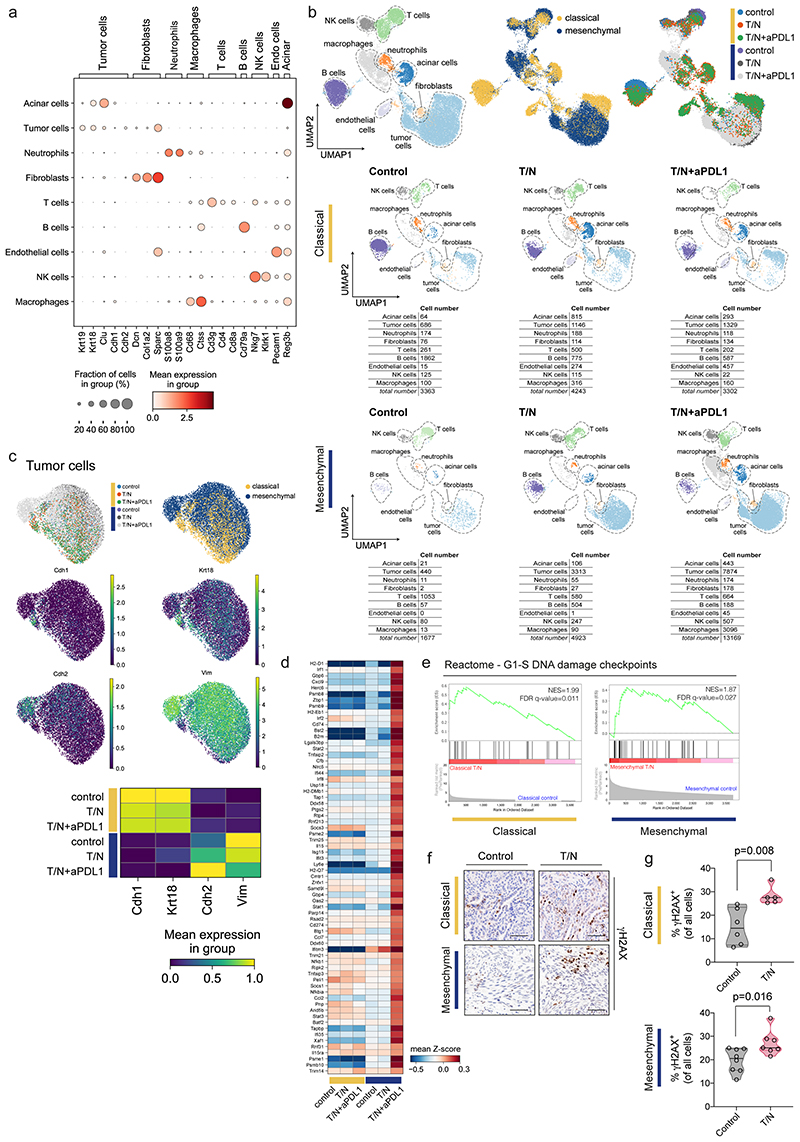
To investigate therapy-induced TME changes in a global and unbiased fashion, and to mechanistically decipher the drug action on epithelial and mesenchymal tumor cells and their environment in vivo, we performed single cell RNA sequencing (scRNA-seq) of whole tumors. 1-2 PDAC per model and treatment condition were dissociated, sorted into mesenchymal/fibroblast and epithelial/immune enriched cell fractions and sequenced (figure 7a). To define cell populations, we combined the data from both subtypes and all treatment groups, representing a total of 30677 cells (1677–13169 cells per model and treatment condition, figure 7b). In both subtypes we identified tumor cells, acinar cells, T cells, natural killer cells, B cells, myeloid populations such as macrophages and neutrophils, and fibroblasts (extended data fig. 9a). In the classical tumors we additionally identified a cluster of endothelial cells (figure 7b and extended data fig. 9a,b).
Figure 7. Single cell RNA-seq analysis reveals context-specific responses of tumor cells and their microenvironment upon combinatorial drug treatment.
a, Scheme of the experimental strategy of the scRNA-seq experiment. 1-2 tumors per model and treatment condition were dissociated and sorted into mesenchymal/fibroblast and epithelial/immune enriched fractions and subjected to scRNA-seq analysis (10x Chromium).
b, Left, UMAP plot showing all identified cell populations within the scRNA-seq experiment. Middle, UMAP plot showing classical (yellow) and mesenchymal (blue) tumors from all groups. Right, UMAP plot showing the treatment induced changes in cell type composition among the identified cell populations across subtypes.
c, Venn-diagram showing the overlap of immune-related signatures obtained from gene set enrichment analysis (GSEA) of the tumor cell cluster across treatment conditions for both subtypes. Only those presenting a false discovery rate (FDR) q value ≤0.05 are shown.
d, GSEA signatures presenting at least one overlap in panel (c). The normalized enrichment scores (NES) are plotted in the heatmap. The red dotted line marks the signatures showing an FDR q value >0.05.
e, GSEA of the differentially expressed genes induced by T/N+anti PD-L1 in classical and mesenchymal tumor cells in vivo. For the mesenchymal subtype, the top immune-related, positively regulated “hallmark” signature is “Interferon gamma response” and is shown for both classical and mesenchymal tumors. NES and FDR-q values are indicated.
f, GSEA of the differentially expressed genes induced by T/N in classical and mesenchymal tumor cells in vivo. The “reactome” signature “Senescence Associated Secretory Phenotype (SASP)” is shown. NES and FDR-q values are indicated.
g, Representative images of three independent experiments of senescence associated (SA)-β-gal stainings of tissue sections of classical and mesenchymal control and T/N treated tumors. Scale bar, 70 μm.
T: trametinib, N: nintedanib, T/N: trametinib+nintedanib, T/N+anti PD-L1: trametinib+nintedanib+anti PD-L1 antibody.
Treatment-induced cancer cell transcriptional changes
We assessed the impact of the combination therapies on gene expression of cancer cells and observed no evidence of epithelial to mesenchymal transition (EMT) or the reverse process (MET) in T/N treated tumors with and without ICB (extended data fig. 9c). Gene set enrichment analysis (GSEA) uncovered widespread and in part context-specific treatment-induced alterations of immune related pathways in tumor cells (figure 7c-e). For both subtypes, we observed an induction of antigen processing and cross-presentation (figure 7d). Additionally, mesenchymal PDAC showed an exclusive and striking enrichment for interferon signaling signatures, including an interferon gamma response as well as interferon regulatory factor 2 (IRF2) induction in the context of the triple therapy (figure 7d,e and extended data fig. 9d).
A growing body of evidence connects immune responses to errors in DNA replication and genomic instability 27,28 . GSEA revealed treatment-induced DNA-damage in both subtypes. We validated this observation by γH2AX immunohistochemistry, a marker for DNA-damage. Interestingly, induction of DNA-damage was stronger in the classical subtype, indicating that the remarkable treatment response of mesenchymal PDAC is not mediated by DNA-damage alone (extended data fig. 9e-g). Several studies link DNA-damage to the senescence associated secretory phenotype (SASP) as well anti-tumor immune responses 29 . GSEA showed that SASP was strongly enriched exclusively in classical PDAC and this was maintained across treatments (figure 7f). Senescence-associated β-galactosidase (SA-β-gal) staining confirmed this phenotype (figure 7g). This indicates that the therapeutic effects observed in the classical subtype are mediated by a complex combination of mechanisms, which include induction of DNA damage, SASP, and reduction in cell proliferation.
Drug treatment-induced immune responses
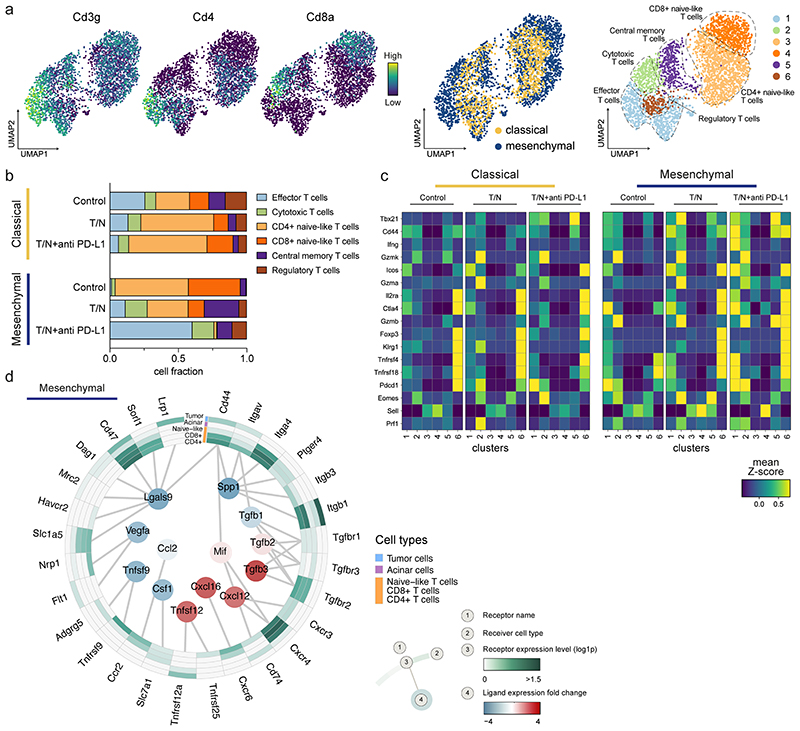
Our in vivo immune profiling shows a robust anti-tumor immune program in mesenchymal PDAC centered on T cells upon T/N treatment. Analysis of the T cells (3260 cells) of our scRNA-seq dataset (figure 8) revealed six subpopulations for both PDAC subtypes (cluster 1 to 6) (figure 8a and methods).
Figure 8. The combinatorial therapy induces a T cell mediated anti-tumor immune response in mesenchymal PDAC.
a, Left, UMAP plots displaying Cd3g, Cd4 and Cd8a marker gene expression across the whole population of T cells identified by scRNA-seq in classical and mesenchymal tumors. Center, UMAP plots of classical (yellow) and mesenchymal (blue) T cells from all treatment and vehicle groups are highlighted. Right, UMAP plots showing the six T cell subpopulations identified by scRNA-seq.
b, Proportion of cells divided by treatment condition and PDAC subtype as identified by scRNA-seq analysis of the T cell clusters annotated in (a).
c, Heatmap displaying expression of selected genes across the identified T cell clusters (1-6) for both classical and mesenchymal tumors. The different treatment conditions are shown separately.
d, Circos plot showing the key communication signals from tumor cells to T cell subpopulations, tumor cells and acinar cells in mesenchymal mPDAC. The ligand protein expression fold-change, identified from secretome experiments, between T/N and control is shown in the middle. Normalized receptor expression levels obtained from scRNA-seq data are shown in the outer concentric circles.
T/N: trametinib+nintedanib, T/N+anti PD-L1: trametinib+nintedanib+anti PD-L1 antibody.
T/N treated mesenchymal tumors showed a substantial decrease of CD4+ and CD8+ T cells with a gene expression signature resembling naïve T cells, and a dramatic increase in T cells with functional cytotoxic, effector and memory gene expression signatures (figure 8b,c). Addition of anti PD-L1 to T/N resulted in a further increase of cytotoxic and effector T cells up to almost 75% of all T cells (figure 8b).
In contrast to recent studies indicating that SASP induces vascular remodeling leading to T cell infiltration and anti-tumor immunity 21 , the T/N combination reprogrammed the TME of classical PDAC towards a decrease of regulatory and effector T cells and an increase in the naïve-like CD4+ compartment (figure 8b). Furthermore, the functional gene expression signature was much weaker in the cytotoxic and effector clusters in classical compared to mesenchymal tumors (figure 8c).
Together, our findings show that in mesenchymal tumors the T/N combination alone leads to a considerable increase of effector-like, activated, and cytotoxic T cells, indicating an effective antitumor immune response, which was further enhanced by anti PD-L1 treatment. This contrasts the classical subtype, which showed a reduction of regulatory and effector T cells upon treatment despite displaying a SASP phenotype.
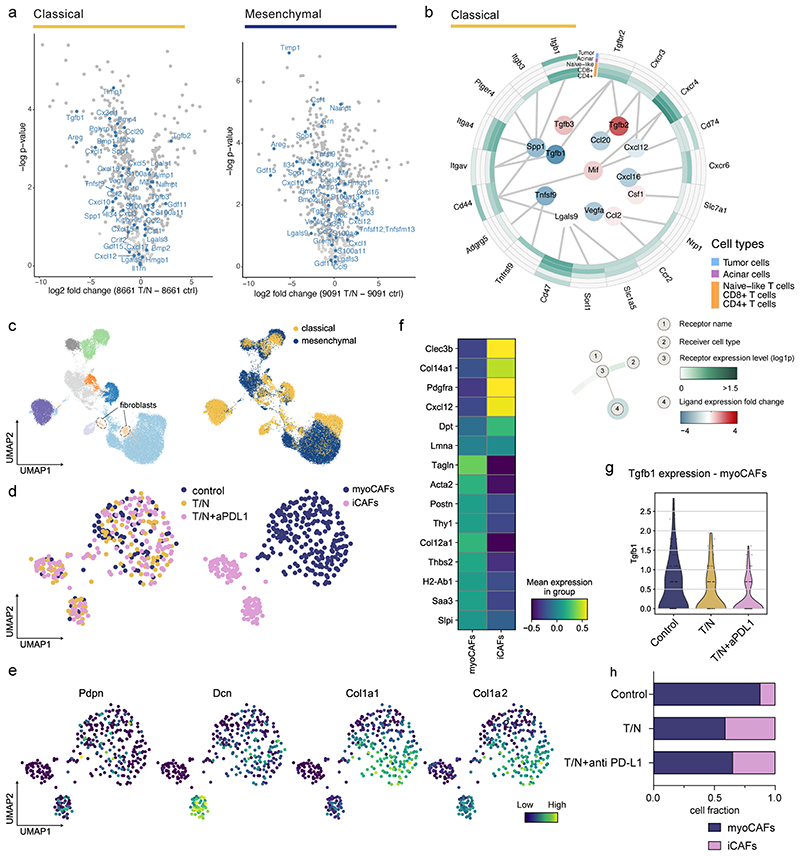
To gain insights into changes that could mediate T cell infiltration upon therapy, we analyzed the secretomes of T/N treated epithelial and mesenchymal tumor cells with mass spectrometry-based proteomics 30 . We observed profound T/N-induced changes in the secretion of immunomodulatory chemokines and cytokines between both subtypes (extended data fig. 10a). To get a comprehensive overview of the resulting intercellular communication networks, we integrated tumor cell secreted proteins across treatment conditions with cell populations expressing the corresponding receptors as identified by scRNA-seq. This revealed key differences in the secretion of factors previously shown to modulate immune cell recruitment, differentiation and expansion, as well as tumor immune responses (figure 8d and extended data fig. 10b, inner circle) 31–36 . In mesenchymal PDAC, the T/N combination specifically induced CXCL12, CXCL16 and TNFSF12 secretion, whereas CCL2, CSF1 and LGALS9 were downregulated (figure 8d). This contrasts the classical subtype, which showed reduced levels of secreted CXCL12, CXCL16 and CXCL20 as well as an increase in CCL2 and CSF1 upon T/N treatment (extended data fig. 10a,b). CXCL16, which is upregulated in mesenchymal PDAC upon T/N treatment, is a key chemoattractant for TILs and high expression levels correlate with a favorable prognosis and increases TILs e.g. in colorectal and breast cancer 31,32,37 . Accordingly, PDAC tumors with high CXCL16 mRNA expression levels display an increased overall survival (n=176 samples, log rank test, p=0.042; https://www.proteinatlas.org/ENSG00000161921-CXCL16/pathology/pancreatic+cancer 38 ). CXCL12 increases cytotoxic T cell infiltration in osteosarcoma 33 and CCL2 and CSF1 mediate immunosuppression in various cancer types 34,35 . Therefore, T/N treatment induces a tumor-cell derived favorable reprogramming of the immunosuppressive TME thereby priming mesenchymal PDAC for ICB.
Cancer associated fibroblasts reprogramming
PDAC subtypes differ in cancer-associated fibroblasts (CAFs), desmoplastic stroma and extracellular matrix content, most abundant in classical while almost absent in mesenchymal tumors 39 . CAFs are highly heterogeneous, displaying diverse effects on tumor progression and therapeutic response 40,41 . Recently, at least three different CAF subpopulations, myofibroblast-like cells (termed myoCAFs), inflammatory (iCAFs) and antigen presenting CAFs (apCAFs), were characterized in PDAC 40,42 .
To investigate the dynamics of CAF subtypes in the PDAC TME, we analyzed their number and polarization making use of our scRNA-seq dataset (extended data fig. 10c-h). Fibroblasts comprised only 9% of sorted classical tumors, in line with recent human PDAC datasets, which contain only <2% CAFs out of all cells in scRNA-seq experiments 42 . Mesenchymal tumors consist of densely packed tumor cells that lack the classical desmoplastic stroma 43,44 . Accordingly, only very few CAFs were detected in vivo preventing the analysis of this population.
In classical PDAC, T/N treated tumors displayed a substantially reduced amount of myoCAFs and a remarkable increase in iCAFs. In addition, myoCAFs presented reduced Tgfb1 expression upon dual- and triple treatment (extended data fig. 10c-h). In line with its well-known role in blocking immune responses within the TME 45 , TGFβ1 downregulation was paralleled by decreased regulatory T cells (figure 8b).
Collectively, our data highlight distinct treatment- and context-specific reprogramming of the TME between subtypes. Exploiting these changes for additional rational therapeutic intervention could lead to improved treatment efficacy also in classical PDAC.
Discussion
PDAC is a complex disease with diverse molecular and morphological subtypes showing TME heterogeneity. Classical PDAC is characterized by a dense desmoplastic stroma, which fosters an immunosuppressive hypovascularized TME, believed to play a major role in primary therapy resistance 21,46,47 . Conversely, the mesenchymal KRAS-mut iGD PDAC subtype shows high tumor cellularity, sparse stroma, is commonly more aggressive and unresponsive to all to date attempted polychemotherapies, targeted therapies and ICB 3–5,7,17,48 .
Here, we identified a novel targeted therapy for mesenchymal PDAC by high-throughput combinatorial drug screening. We show how this therapy remodels the immune landscape in vivo and how it creates new vulnerabilities towards ICB in mesenchymal tumors. The T/N combination reprogrammed the immunosuppressive mesenchymal cancer cell secretome and downregulated cytokines and chemokines, including CCL2 and CSF1, capable of attracting and inducing expansion of macrophages and myeloid derived suppressor cells. In parallel, it induced secretion of T cell modulators, such as CXCL16 and CXCL12, important for TIL recruitment 32–35,37 and provoked upregulation of antigen processing and presentation pathways in mesenchymal PDAC cells in vivo. Finally, the combination strongly impacts on the TME by increasing blood vessel density, inducing endothelial cell activation, vascular remodeling and facilitating cytotoxic and effector T-cell infiltration. Thus, we show for the first time that reprogramming of the immunologically “cold” to a “hot” tumor stroma of the highly aggressive mesenchymal subtype is possible and can be exploited therapeutically by adding anti PD-L1 ICB to the T/N combination.
The context-specific impact of the T/N combination on the immune system was not expected. While a link between the DNA damage response, induction of antigen processing and presentation, and the activation of strong immune responses is well established 49–52 , the lack of these effects in classical PDAC was surprising. Although the combination induces DNA damage, it also induces immunosuppression by increasing the secretion of CCL2 and CSF1. Differences in oncogenic KRAS signaling outputs might drive immune signaling divergence between subtypes. High levels of KRAS expression, as observed in mesenchymal tumors, were shown to repress interferon gamma signaling in vivo as evidenced by reduction in IRF2 53 . Blockade of KRAS signaling by MEKi in combination with broad RTK and PD-L1 inhibition might release this important pathway and - in combination with therapy-induced DNA damage - boost neoantigen presentation while inducing anti-tumor immunity in mesenchymal tumors 53 . In line, we observed selective upregulation of IRF2 in PDAC cells upon T/N+anti PD-L1 therapy in this subtype in vivo.
Although being highly antagonistic in vitro, the combination showed substantial antitumor effects also in classical PDAC in vivo, indicating an impact on the TME 54 . The failure of the in vitro drug screen to predict in vivo responses of this subtype is remarkable. Therefore, treatment-mediated TME remodeling should be considered for the transition of in vitro screening hits to the (pre)clinical setting.
We observed an antiproliferative effect of the combination in classical PDAC, as well as the selective induction of SASP. SASP is characterized by the secretion of chemokines, cytokines, matrix metalloproteinases and other paracrine factors 21,29,55 . In classical KRAS-driven PDAC, it has been shown that the combination of MEKi and CDK4/6 inhibitors induces a strong SASP response with the release of pro-angiogenic factors that promote tumor vascularization, endothelial cell activation and VCAM-1 expression. This in turn promotes T cell extravasation into tumors and sensitizes this subtype to ICB 21 . In contrast, we found no evidence for SASP-induced vascular remodeling in classical PDAC upon T/N therapy. In line, classical T/N-treated tumors did not show T cell infiltration into the tumor core. This might be due to insufficient levels of SASP-induction or other treatment-induced context-specific effects on the cancer cells and their environment counteracting vascular remodeling, T cell extravasation and reactivity. Of note, our study supports the notion that SASP-induction is context-specific 29 as we were not able to detect therapy-induced senescence in vivo in mesenchymal KRAS-mut driven PDAC. This demonstrates that the biology, stromal composition and treatment response of PDAC is highly contextdependent and differs substantially between PDAC subtypes.
Trametinib and nintedanib are both clinically approved by the FDA/EMA. Trametinib is used for treatment of patients with unresectable or metastatic BRAF-mutant melanoma, NSCLC and anaplastic thyroid cancer. Nintedanib in combination with docetaxel has been approved for the second-line treatment of advanced NSCLC and first-line therapy for idiopathic pulmonary fibrosis (IPF) (www.accessdata.fda.gov/scripts/cder/daf/index.cfm?event=BasicSearch.process). IPF is characterized by the activation of fibroblasts towards a myofibroblast differentiation 56,57 . Accordingly, blocking myofibroblast activation by nintedanib, or pirfenidone, which targets the TGFβ pathway, slows down pulmonary disease progression 56 . Classical PDAC harbor a dense stroma, which is composed in large parts by activated myoCAFs and TGFβ expression (extended data fig. 10) 42,43 We therefore hypothesized that adding nintedanib to trametinib might reprogram myoCAFs, thereby contributing to stromal normalization in classical PDAC. Using scRNA-seq we observed that the amount of myoCAFs decreased upon T/N therapy, and TGFβ1 expression, which can contribute to immunosuppression via regulatory T cells, is downregulated. This demonstrates that reprogramming of the fibrotic microenvironment of the classical subtype is feasible and could be exploited for further improved therapeutic responses.
To identify the therapeutically relevant targets of the multikinase inhibitor nintedanib, we performed multiscale analyses ranging from kinobead-based proteomic identification of the nintedanib-bound kinases, to phosphoproteomic analyzes of drug action, and genome-scale and focused CRISPR/Cas9-based negative selection screens. This revealed a key set of nintedanib targets, including FGFR, kinases belonging to the MEK/ERK family and PDGFR regulated networks that cooperate with trametinib in mesenchymal PDAC. However, it also revealed heterogeneity of the functionally relevant targets within this subtype. This is remarkable and indicates that indeed broad multikinase inhibition is needed to target this highly aggressive and therapy-resistant subtype efficiently across the whole spectrum of its phenotypes. In addition, it supports the notion that no single target gene is responsible for the synergistic effect of the T/N combination and challenges the one biomarker per drug paradigm in oncology. Indeed, mesenchymal cancer cell morphology and the underlying genetic program is the strongest predictor of therapeutic response towards T/N.
In summary, our work sets the basis for the combination of T/N with immunotherapy in the treatment of mesenchymal PDAC and provides a first step towards molecularly stratified combinatorial therapies in the clinic. Considering the widespread occurrence of RAS-driven tumor entities with an increased RAS gene dosage and a mesenchymal differentiation state, our data suggest that combining T/N with ICB or other forms of immunotherapy might trigger anti-tumor immunity and improve therapeutic outcomes across entities.
Methods
Our study complies with all relevant ethical regulations. Animal experiments were approved by the Institutional Animal Care and Use Committees (IACUC) of the local authorities of Technische Universität München and the Regierung von Oberbayern.
Primary PDAC cell culture, clonogenics and inhibitors
Primary mPDAC cell cultures were isolated from autochthonous PDAC and cultured as described previously 59 . All cells were cultivated for less than 30 passages, authenticated by genotyping and tested for mycoplasma contamination by PCR. Conventional human PDAC cell lines and primary patient derived low passaged PDAC cell cultures were established and cultured as previously reported 58 .
For long-term clonogenic cell proliferation assays, cells were seeded into 24-well plates (density of 1-2×103 cells/well, depending on growth rate. The following day, plates were treated with different concentrations of drugs as indicated. Every 7 days, media and drug were refreshed. Cells were fixed and stained with 0.2% crystal violet in an ethanol/water solution 7 to 13 days after the start of treatment, according to the confluence reached by the untreated control. Crystal violet was solubilized with 10% acetic acid and absorbance was quantified at 595 nm. The resulting values were used to calculate the Bliss synergy score with the online software Synergy Finder (v1.0) 60 . All assays were performed independently at least three times. Trametinib, nintedanib, AZD-4547, imatinib and PF-3758309 were obtained from Selleckchem, 4-OHT from Sigma, murine anti PD-L1 mAb (Anti PD-L1-mIgG1e3 InvivoFit™) was purchased from InvivoGen, and tamoxifen for in vivo treatment from Sigma.
Caspase 3/7 assay
To assess apoptosis, 1000 cell/well were seeded in 96-well plates and treated after 24 hours with trametinib (10 nM) or nintedanib (2 μM) alone or the combination of both as indicated. After 24 hours from the start of treatment, caspase 3/7 activity was determined measuring luminescence using Caspase 3/7 assay (Promega), according to the kit instruction. All assays were performed independently at least three times.
Kinobead Pull Downs
Kinobead pulldown assays were performed as previously described 61 . In-gel digestion was performed according to standard procedures 62 . Peptide/protein identification and quantification were performed with MaxQuant (v.1.5.7.4) by utilizing the Swissprot database (murine, 16,996 entries, downloaded on 23.11.2018) using the search engine Andromeda. Data analysis was performed as previously described 63 .
Automated combinatorial drug screen
The screened compounds were purchased from SelleckChem either in DMSO or water. 1000-2000 cells were seeded in 96-well plates using a Multidrop Combi (Thermo Fisher) dispenser. The optimal cell number for each cell culture was determined to ensure that each cell line has undergone at least two cell doublings at the end of the assay 96h after seeding (~85% confluency). After overnight incubation, cells were treated with a 7 point dilution series (7 concentrations of each compound, 3-fold dilutions, highest concentration 10μM) and DMSO as control, using liquid handling robotics (CyBio Felix), and assayed 72h later. Each cell culture was treated with a library of 418 compounds either in monotherapy or in combination with a fixed concentration of trametinib (final concentration: 5nM). Viability was measured using CellTiter-Glo® Luminescent Assay (Promega). CellTiter-Glo reagent was added using the Multidrop Combi dispenser, cells were shaken and incubated for 10 minutes in the dark. Luminescence was measured using an Infinite Pro 2000 Lumi (Tecan) Luminometer.
Dose-response curves were generated for both monotherapy and combination using the R package GRmetrics (v3.14) 64,65 to derive both growth-rate adjusted and traditional measures of drug sensitivity (half-maximal inhibitory concentration (IC50), area under the curve (AUC), efficacy (Emax), half-maximal effective concentration (EC50)). Only drugs for which a sigmoid curve could be fitted (coefficient of determination, r 2 >0.9) were considered for further analysis. For each drug we calculated an expected effect of the combination with trametinib using the Bliss independence model. We used the delta of the AUC between the expected and the measured response to the combination as a proxy for synergy. Drug sensitivity parameters for each cell culture are summarized in supplementary table 2.
Phosphoprotein array sample preparation and analysis
mPDAC cells 9091 and 8661 were plated in 10 cm dishes. The next day, they were treated for 6 hours with DMSO (vehicle) or T/N (10 nM trametinib and 2 μM nintedanib) and analyzed using the Phospho Explorer antibody microarray, which contains 1,318 antibodies (Full Moon Biosystems), according to the protocol of the manufacturer.
Pathway enrichment analysis was based on the Reactome gene-set and performed through Cytoscape (v3.8.2) with ClueGO (v2.5.8) 66 , a Cytoscape plug-in to decipher functionally grouped pathway annotation networks. The functionally grouped networks used for visualization present terms as nodes and are linked based on their kappa score level (≥0.4). The node size represents the term-enrichment significance and functionally related groups are depicted by similar colors.
Cloning of focused Cas9 sgRNA library
For the custom sgRNA Cas9 library, 4 sgRNAs (from the Brie library (Addgene ID #73633)) per target, non-targeting controls, common essential genes and trametinib sensitizing/resistance genes were included to a total of 350 sgRNA sequences which were embedded into an oligo sequence with flanking PCR handles and BsmBI restriction sites 67 . The oligo pool (Twist Bioscience) was then amplified at 5 nM input with NEBNext Ultra II polymerase and primers binding the PCR handles, followed by purification with AmpureXP beads (Beckman Coulter). For Golden Gate cloning, BsmBI-v2 (NEB)-digested pLenti-guide puro (Addgene ID #52963) was mixed with T4 DNA ligase (NEB), 10x T4 buffer, BsmBI-v2, linearized backbone and amplified insert (1:3 molar ratio) (30 cycles Golden Gate). The assembled product was purified with AmpureXP beads and electroporated into Endura Competent cells (Lucigen) using a BioRad MicroPulser (1.8 kV in 0.1 cm gap cuvettes (Sigma-Aldrich)). Bacteria were grown at 33°C overnight (LB-medium (Sigma) with 100 μg/ml Ampicillin). DNA was extracted using the NucleoBond Xtra Midi EF Kit (Macherey-Nagel). Libraries for NGS were constructed according to the protocol given below to determine sgRNA abundance. Sequences of oligonucleotides for all described methods from this study are provided in supplementary table 7.
Lentivirus production and titration
For virus production of sgRNA libraries, HEK293FT cells were seeded into 15cm dishes to reach confluency of 60% the following day. Cells were transfected with 14.3 μg library plasmid, 10.9 μg psPAX2 (Addgene ID #12260) and 7.1 μg pMD2.G (Addgene ID #12259) per plate using 119 μl TransIT-LT1 (Mirus Bioscience) in 850 μl OptiMEM (Gibco). 48h and 72h post transfection, supernatant was collected and filtered (0.45 μm). For other constructs, HEK293FT were seeded in 10 cm plates and transfected with 2 μg viral plasmid, 1.25 μg psPAX2 and 0.75 μg pMD2.G per plate using 18 μl TransIT-LT1 in 270 μl OptiMEM (Gibco) the next day.
Lentiviral sgRNA libraries were functionally titrated by spinfection (2h, 1000g, 33°C) of target cells with varying amounts of lentiviral supernatant in 12-well plates with 3x10^6 cells per well as described in syngeneic 68 . Amount of lentivirus needed per 12-well for a target MOI of 0.3 was calculated as survival of 0.25 relative to unselected control.
Whole-genome and focused CRISPR/Cas9 screens
CRISPR/Cas9 screens were performed in clonal Cas9-expressing cells, with the focused Cas9 library at 1000x coverage and with the genome-wide Brie library (pLenti-guide puro) at 500x coverage. Parental cell lines were infected with pLenti Cas9-2A-BSD (Addgene ID #52962) and selected with BlasticidinS (Invivogen) (10 μg/ml). Single clones were obtained by limited-dilution in 96-well plates, tested for Cas9 expression by immunoblot and validated by resistance to 6-Thioguanine (Sigma) treatment in cells edited at the Hprt-locus (mediated by a sgRNA cloned into pLenti-guide puro).
The final dose of trametinib for the screens was determined by culturing the cells with different concentrations of trametinib (1.25 nM, 2.5 nM, 5 nM, 10 nM and 20 nM) and assessing cell proliferation and ERK1/2 phosphorylation. 500000 cells were seeded in 10 cm dishes and trametinib at the indicated concentrations was added. Cells were passaged every 3-4 days and counted at each passage. Protein samples were harvested at each count and phospho-ERK levels were assessed by immunoblot analysis.
The CRISPR/Cas9 screens were performed in side-by-side duplicates (genomewide) or triplicates (focused Cas9). Cas9 expressing cells transduced with the lentiviral libraries and selected in medium containing Puromycin (Sigma; final concentration 9091 and 8248: 4μg/ml; 8570: 12μg/ml). 4 days after infection, Puromycin was washed out and cells were allowed to recover for 2 days. Then, cells were assigned to control (vehicle, DMSO) and experimental arms (5 nM trametinib). Cells were passaged every 3-4 days for 2 weeks, thereby refreshing the drug treatment and maintaining representation. At the final timepoint, cells were harvested and genomic DNA (gDNA) was extracted using the DNeasy Blood & Tissue kit (focused libraries) or the Blood & Cell Culture DNA Maxi Kit (both Qiagen) according to the manufacturer’s instructions.
sgRNA library construction, Next-generation Sequencing and MaGECK analysis
Approx. 230 μg (genome-wide screen) and 9 μg (focused screen) of gDNA was used for library preparation. One PCR reaction (50 μl) contained 6 μg gDNA, 25 μl 2x KAPA HiFi HotStart ReadyMix (Roche) and 2 μl each of forward/reverse primer (10 μM) with unique sequencing-barcode indices. PCRs were purified using NEB Monarch PCR-cleanup kit, pooled and quantified using the KAPA library quantification kit for Illumina. Pooled libraries were sequenced on Illumina NextSeq 500 (custom read and indexing primers spiked in). Read depth was intended to maintain library coverage (35 Mio reads for genome-wide, 350000 reads for custom library).
Downstream processing was conducted with MAGeCK v0.5.9.4 69 . Reads were aligned to the reference of sgRNA sequences and counted. Beta scores were calculated using the maximum likelihood estimation (mle) method utilizing the information from non-targeting control guides, resulting in a single score for each gene (beta score). The final score represents enrichment (+) or depletion (-) of the sgRNAs with respect to their initial abundance.
To investigate significantly depleted nintedanib targets in the presence of trametinib, the score difference between both arms of the experiment was calculated (DMSO and trametinib). Values falling in the left and right tails of this distribution (< 5th and > 95th percentile) were considered as conferring resistance or synergizing with drug treatment.
Lentiviral CRISPR/Cas9-induced deletion of individual genes
sgRNAs were designed using CRISPick 70 (https://portals.broadinstitute.org/gppx/crispick/public) and synthesized as complementary forward and reverse oligos (Eurofins Genomics GmbH; see supplementary table 7). Oligos were annealed in T4 DNA-ligase buffer (NEB), cloned into CRISPR expression vectors (either pLenti CRISPR V2, Addgene ID # 52961, or pLenti-guide puro) using BsmBI-v2 and T4-DNA ligase (both NEB), transformed into chemically competent bacteria (Stbl3 strain) using 5x KCM buffer (500 mM KCl, 150 mM CaCl2, 250 mM MgCl2) and grown on Ampicillin-containing (100 μg/ml) LB-plates overnight. Plasmid DNA was isolated using the Monarch Plasmid MiniPrep Kit (NEB) and used for lentivirus production.
For lentiviral transduction of target cell lines, 1 x 10^5 cells were seeded into 6-well plates. The following day, medium was replaced with 1 ml lentiviral supernatant, 1 ml fresh medium and polybrene (final concentration of 8 μg/ml). 48h post transduction, selection with puromycin was initiated. Indel analysis was performed by genomic DNA extraction using the DNeasy Blood & Tissue kit (Qiagen). PCR-amplification was done with primers flanking the genomic sgRNA binding site using 2x KAPA HiFi HotStart ReadyMix (Roche). PCR product was purified (Monarch PCR cleanup kit, NEB), submitted to Sanger sequencing (Eurofins) and analyzed for CRISPR edits using the web-based application ICE (Synthego, v2.0, https://ice.synthego.com/). Edited cells were seeded and clonogenic assays performed in 24-well plates (1000 cells/well) in presence and absence of trametinib as described above.
Electroporation of CRISPR/Cas9 Ribonucleoproteins (RNPs) and Indel depletion assay
For multiplexed validation of targets conferring sensitivity to trametinib, sgRNAs were synthesized as crRNAs (Integrated DNA Technologies, IDT; see supplementary table 7) and complexed with a tracrRNA (IDT) according to manufacturer’s instructions. The resulting crRNA::tracrRNA duplex was complexed at 22 pmol with Alt-R Cas9 enzyme (Alt-R S.p. Cas9 Nuclease V3, IDT) at 18 pmol at room temperature for 20 min, thus forming ribonucleoproteins (RNPs).
Cells were detached trypsin-free by using PBS-EDTA (0.046%) and cell number was adjusted to obtain 400000 cells per transfection (10 μl volume). Complexed RNPs were added to the cell solution and electroporated using the Neon Transfection system (Thermo Fisher) according to the manufacturer’s instructions. Next, cells were seeded for clonogenic growth as described above. To determine indel shift due to drug exposure, 20000 cells were seeded in 6-well plates and subjected to trametinib (5 nM) or vehicle (DMSO) treatment the next day. Cells were passaged if necessary and drug was renewed after 4 days. After 7-9 days, gDNA was extracted and PCR-amplified as described above. Indel frequency was determined by Sanger Sequencing and ICE analysis (Synthego, v2.0) for DMSO and trametinib treated samples.
Mouse strains, tumor models and in vivo treatment
LSL-KrasG12D/+ 71 , Pdx1-Cre 72 , Ptf1aCre/+ 73 , Pdx1-Flp, FSF-R26CAG-CreERT2/+ and FSF-KrasG12D/+ 20 and Map2k1lox/lox 74 mice have been described previously. Map2k2lox/lox (Map2k2tm1e(EUCOMM)Wtsi) mice were obtained from EUCOMM. The strains were on a mixed C57Bl/6;129S6/SvEv genetic background and interbred to obtain compound mutant mice that develop autochthonous tumors in the pancreas.
For transplantation experiments, cancer cells (2500 to 10000) were orthotopically grafted into the pancreas of syngeneic immunocompetent C57Bl/6J wild-type, or T cell deficient C57BL/6 CD3ε-knockout mice 22 . When tumors grew to size of ~100 mm 3 mice were randomized into the different treatment arms. The following drugs were used: trametinib (3 mg/kg, 5 days a week, oral gavage), nintedanib (50 mg/kg, 5 days a week, oral gavage), anti PD-L1 antibody (200 μg/mouse, every third day, intraperitoneal injections) and tamoxifen (4 mg/mouse, every third day, intraperitoneal injections). One week after implantation, mice were scanned by magnetic resonance imaging (MRI) for the presence of tumors. Animals were sacrificed when individual mice reached the human endpoint or at the completion of treatment.
All animal studies were conducted in compliance with the ARRIVE and the European guidelines for the care and use of laboratory animals and were approved by the Institutional Animal Care and Use Committees (IACUC) of the local authorities of Technische Universität München and the Regierung von Oberbayern. A maximal tumor size of 1,5 cm and a specific burden score (determined by a cumulative burden score) permitted by the IACUC and Regierung von Oberbayern was not exceeded in this study.
Animals were kept in a dedicated facility, with a light-dark cycle or 12:12 hours, a housing temperature between 20 and 24°C and a relative air humidity of 55%.
Magnet Resonance Imaging (MRI) and Quantification
Animals were anesthetized with isoflurane to perform MRI of the pancreas using Bruker Biospec 7T MRI scanner to scan 35 consecutive sections. Tumor volume was quantified using the Horos medical image viewer software to reconstruct MRI volumetric measurements (v3.3.6). Horos is an open source code software (FOSS) program that is distributed free of charge under the LGPL license at Horosproject.org and sponsored by Nimble Co LLC d/b/a Purview in Annapolis, MD USA. Acquisition of the MRI signal was adapted according to cardiac and respiratory cycles to minimize motion effects during imaging.
Histology and immunohistochemistry
Mouse pancreatic tumors were fixed in 4% PFA (Carl Roth), embedded in paraffin and cut into 1 μm sections. Hematoxylin and eosin (H&E) staining of tissue sections was performed according to standard protocols. The following primary antibodies were used for immunohistochemistry (IHC): Rat anti-CK19 (DHSB, 1:250), Rabbit anti-KI67 (Thermo Scientific, 1:50), Rat anti-CD31 (Optistain, 1:50), Rabbit anti-CD3 (Zytomed Systems, 1:100), Rat anti-CD8 (Dianova, 1:100), Rabbit anti-Phospho-Histone H2A.X (Ser139) (Cell Signaling Technology, 1:500), Rabbit anti-MEK1 (30C8) (Cell Signaling Technology, 1:50), Rabbit anti-MEK2 (13E3) (Cell Signaling Technology, 1:50), Rabbit anti-pERK (p-p44/42 MAPK Thr202/Tyr204) (Cell Signaling Technology, 1:50), Rabbit anti-pPDGF Receptorβ (Tyr1021) (Cell Signaling Technology, 1:50) and Rabbit anti-pAMPKα (Thrl72) (Cell Signaling Technology, 1:50). Antibody detection was performed with the Bond Polymer Refine Detection Kit (Leica) or Rabbit anti-rat IgG (Vector Laboratories, 1:200) secondary antibody or followed by a secondary antibody conjugated to biotin (Vector Laboratories). Detailed protocols of individual stainings are available upon request. Images were acquired using Leica AT2 Scanner (Leica) and processed by Aperio Image Scope (Leica, v12.3.3) and FIJI (NIH, v2.1.0). For quantification of KI67, CD31 and γH2AX stainings five fields of view of individual tumors per treatment condition were analyzed in a blinded fashion. In at least 6 individual tumors per treatment condition, mitoses were counted per HPF in areas showing increased mitotic activity on scanning magnification. Moritz Jesinghaus a board certified pathologist performed all quantifications.
Senescence β-Galactosidase (SA-β-gal) staining
PFA-fixed OCT-embedded tumor tissues were cut into 5 μm sections and mounted on slides. SA-β-gal staining was performed using a Senescence β-Galactosidase Staining Kit (Cell Signaling Technology) at pH 6.0. Images were acquired using Aperio Versa Scanner (Leica) and were processed by FIJI (NIH, v2.1.0).
Immunofluorescence staining and imaging
PFA-fixed OCT-embedded tumor tissues were cut into 5 or 10 μm sections and mounted on slides. The slides were incubated with Aceton (Sigma) for 6 min at 4°C. After rehydration with PBS for 10 min, the tissues were blocked for 1h at RT with 10% goat serum and 10% donkey serum in PBS. The following primary antibodies were used for immunofluorescence staining of T cells, endothelial cells, epithelial and mesenchymal tumor cells: Rat anti-CD3 (Biolegend, 1:50), Armenian hamster anti-CD31 (Abcam, 1:400), Rabbit anti-aSMA (Abcam, 1:100), Rabbit anti-P-selectin (LSbio, 1:200), Rabbit anti-CK18 (Sigma, 1:800) and Chicken anti-Vimentin (Invitrogen, 1:100). Primary antibodies were incubated each for 3h at RT and diluted in 3% BSA (Sigma) in PBS. Goat anti-rat AF680 (Invitrogen, 1:200), Donkey anti-rat AF488 (Invitrogen, 1:200), Goat anti-Armenian Hamster IgG Cy™3 (Jackson Immuno, 1:200), Goat anti-rabbit AF488 (1:200) and Goat anti-chicken AF680 (1:200) were used as secondary antibodies (staining for 1h at RT diluted in in 3% BSA in PBS). Nuclear staining was performed for 10 min at RT with DAPI (Biotium, 1:500) in 3% BSA in PBS.
The following primary antibodies were used for IF staining of macrophage subpopulations: Rat anti-CD68 (Bio-Rad, 1:150), Rabbit anti-ARG1 (ThermoFisher, 1:300) and Rabbit anti-CD80 (Abcam, 1:300). Donkey anti-rat AF594 (Invitrogen, 1:200) and Goat anti-rabbit AF488 (Invitrogen, 1:200) were used as secondary antibodies (staining for 1h at RT diluted in in 3% BSA + 6% TritonX in PBS). Nuclear staining was performed with DAPI (Biotium, 1:1000) in 0,25% BSA in PBS.
After three washes in PBS, slides were mounted with Vectashield Mounting Medium (Vector Laboratories). Images were acquired using a TCS SP8 Confocal Laser Scanning Microscope (Leica) and were processed by FIJI (NIH, v2.1.0). For Quantification of T Cells, ten fields of view of 4 individual tumors per treatment condition were analyzed. For Quantification of Macrophage subpopulations five fields of view of 5 individual tumors per treatment condition were analyzed.
Immunophenotyping by flow cytometry
Fresh tumor samples were minced and enzymatically digested with the tumor dissociation kit (Miltenyi #130-096-730) for 40 min at 37°C with agitation. The cell suspension was strained through a 100 μm strainer, spun down and resuspended in 2% FCS/PBS. Cells were blocked for 10 min on ice with anti-mouse CD16/CD32 FC block (Biolegend, 1:100) and stained with Zombie Aqua Fixable Viability Kit (Biolegend, 1:500) and the following antibody cocktails: CD4 BUV805 (BD, 1:100), CD3εBUV395 (BD, 1:20), CD8a BV785 (Biolegend, 1:100), CD45 PerCP Cy5.5 (Biolegend, 1:100), CD19 FITC (Biolegend, 1:100), EpCAM APC/AF647 (Biolegend, 1:200) for acquisition of adaptive immune cells; CD11c BUV737 (BD, 1:30), NK1.1 BUV395 (BD, 1:25), Ly6C BV785 (Biolegend, 1:200), CD11b BV650 (Biolegend, 1:100), F4/80 BV421/PB (Biolegend, 1:30), CD45 PerCP Cy5.5 (Biolegend, 1:100), Ly6G PE (Biolegend, 1:200), CD68 APC-CY7 (Biolegend, 1:20), EpCAM APC/AF647 (Biolegend, 1:200) for acquisition of innate immune cells. Per panel 1,000,000 events were acquired on the BD LSRFortessa. Flow cytometry data was analyzed using FlowJo software (v10.6.2).
Whole cell lysates and western blot
Protein extraction from cells and tissues, western blots and subsequent detection of proteins were performed as described 58,59 . The following primary antibodies were used: HSP90 (1:1000, Cat# sc-13119, Santa Cruz Biotechnology), MEK1 rabbit mAb (1:1000, Cat # 9146, Cell Signaling Technology), MEK2 rabbit mAb (1:1000, Cat # 9147, Cell Signaling Technology), p-ERK (1:1000, Cat # 4377, Cell Signaling Technology), ERK (1:1000, Cat #610123, BD), CAS9 (1:1000, Cat #14697, Cell Signaling Technology), β-ACTIN (1:1000, Cat #4970, Cell Signaling Technology).
KRASG12D induction after lentiviral transduction of mPDAC cells
The pINDUCER20 vector system comprising a puromycin resistance gene was used for doxycycline-inducible KRASG12D overexpression as previously described 16 .
Gene expression profiling and amplicon-based deep sequencing at the Kras locus or of Kras mRNA expression
Drug perturbed cells were seeded in a 10 cm dish and treated the following day with 10 nM trametinib or vehicle. RNA was isolated with the RNeasy kit (Qiagen) from 80% confluent primary cells and immediately transferred into RLT buffer (Qiagen) containing β-mercaptoethanol.
RNA-seq library preparation, amplicon-based deep sequencing at the Kras locus or of Kras mRNA was performed as previously described 16 .
Analyses were carried out using R version 3.6.2 (R Core Team, 201) and Bioconductor v3.1. Differential gene expression analysis was performed using DESeq2 version 1.26.0. A gene was considered to be differentially expressed with a Benjamini-Hochberg adjusted p-value of 0.05 and an absolute fold change >1.
We used the single sample Gene Set Enrichment Analysis (ssGSEA) function from the gene set variation analysis (GSVA, v3.14) 18 and the Hallmark gene set EMT 19 to estimate the classical or mesenchymal gene expression programs in our collection of human and mouse PDAC cell cultures.
Sample preparation for single cell RNA sequencing (scRNA-seq)
Tumor specimens were dissociated and enzymatically digested with the tumor dissociation kit as described above. The cell suspension was strained through a 100 μm strainer, spun down and resuspended in 2% FCS/PBS including RNase inhibitor (NEB, #M0314L,1:100). Debris removal solution (Miltenyi #130-109-398) was used to remove cell debris from the dissociated tissue. Then the dead cell removal kit (Miltenyi #130-090-101) was used to enrich for live cells. The cell suspension was spun down and then resuspended in PBS and blocked for non-antigen-specific binding for 10 min on ice with anti-mouse CD16/CD32 FC block (Biolegend, 1:100). Cells were stained with the following antibodies for FACS sorting: TER-119 BV421 (Biolegend, 1:100), CD45-AF647 (Biolegend, 1:20), CD31-AF647 (Biolegend, 1:20) and EPCAM-AF647 (Biolegend, 1:20) for 30 min on ice. Cell sorting was performed using the BD FACS Aria Fusion. The sorted cells from the TER-119-negative/CD45-/CD31-/EPCAM-positive fraction (for enrichment of immune, endothelial and epithelial tumor cells and exclusion of erythrocytes) as well as TER-119-/CD45-/CD31-/EPCAM-negative fraction (for enrichment of fibroblasts/mesenchymal tumor cells and exclusion of erythrocytes). Sorted cells were collected in 2% FCS/PBS.
scRNA-seq library preparation and sequencing
The sorted cells were counted, diluted in 2% FCS/PBS and up to 20,000 cells were loaded per lane on a 10x Chromium chip to generate gel beads in emulsion (GEMs). Single cell GEM generation, barcoding and library construction was performed by using 10x Chromium Single Cell 3’ v3 chemistry according to the manufacturer’s instructions. cDNA and generated libraries were checked for sample size and quality on an Agilent Bioanalyzer 2100 using HS DNA Kit (Agilent). Libraries were sequenced on Illumina NovaSeq 6000 S2 (PE, 28+94 bp).
Data preprocessing and quality control
Alignment of the scRNA-seq data to the mouse reference genome (mm10, release 108.20200622), filtering, barcode and unique molecular identifier (UMI) counting was performed using the 10x Genomics Cell Ranger software (v3.1.0). Python (v3.8.3) and the Python software package SCANPY was employed for all further analyses (v1.6.0) 75 . Cells that expressed less than 200 genes or had more than 10% mitochondrial gene counts were excluded from the analysis. We also filtered out genes with less than 20 counts. Counts were per-cell normalized and (log+1)-transformed. Highly variable genes were computed using the first N=4000 most variable genes for the analyses across cell types, tumor cells, T cells, fibroblasts, macrophages and treatment conditions. Batch-effect correction was performed using BBKNN (batch balanced k nearest neighbors, v1.5.1).
Dimensionality reduction and clustering
The Leiden algorithm (v0.8.1) was used for cell clustering and Uniform Manifold Approximation and Projection (UMAP, v0.4.6) for dimensionality reduction. The clusters were further annotated by assessment of known cell-type specific markers. Principal component analysis was performed with default parameters. Neighborhood graphs were computed based on n=10 principal components and k=30 neighbors. The calculation of all UMAP projections was based on default parameters. The number of Leiden clusters was adjusted according to the sample of consideration.
Gene Set Enrichment Analysis (GSEA)
Differential gene expression analysis was performed using the tool rank_genes_groups, which is part of the SCANPY package (v1.6.0, https://github.com/theislab/scanpy). The Benjamini-Hochberg method was used to correct for multiple testing. Subsequent GSEA was performed using GSEA v4.1.0 jar package and MSigDB v7.1 gene sets provided by Broad Institute, Massachusetts Institute of Technology and Harvard University. GSEA was conducted on a pre-ranked gene list output of the differential gene expression analysis, genes were ranked based on “t test” metric. Parameters were set as follows: number of permutations was set to 1000 and enrichment statistic for scoring was set as “weighted”; other parameters were set as default. The cut-off for a significant FDR q-value was set at 0.05 and NOM p-value at 0.05.
Cell type-specific analysis
CD4+ and CD8+ T-cell identification
In classical and mesenchymal tumors, scRNA-seq uncovered six different T cell clusters. The identified CD4 naive-like populations expressed Cd4 and the naive T cell marker Sell, additionally they lacked the expression of Cd44 and T cell activation genes, such as Icos, Lag3, Havcr2 (Tim-3), Pdcd1, Tnfrsf4 and Ctla4. The identified activated/effector T cells showed the highest levels of activated markers such as Icos, Tim-3, Ctla4 and Pdcd1, and intermediate levels of Sell and Cd44. Regulatory T cells expressed high levels of Cd4 and Foxp3, and intermediate levels of T cell activation genes Icos, Ctla4 and Pdcd1. Central memory T cells were positive for Cd4, Cd27, Cd28, Cd44, Il7r and displayed a unique central memory marker signature including Sell and Ccr7. CD8 naive-like T cells expressed high levels of Cd8a and Sell. Cytotoxic T cells showed the highest levels of T cell activation genes Lag3, Tim-3, Pdcd1, Tnfrsf18 and Ifng, and cytotoxic markers Gzma, Gzmb and Prf1.
CAFs identification
scRNA-seq uncovered a sufficient number of CAFs for downstream analysis only in classical tumors. We defined CAFs based on the expression of the following genes: Vim, S100a4, Acta2, Col6a1, Col1a2, Fap, Pdgfra, Cspg4. MyoCAFs expressed Tagln, Acta2, Postn, Thy1, Col12a1, Thbs2, apCAFs H2-Ab1, Cd74, Saa3, Slpi and iCAFs Clec3b, Col14a1, Il6, Pdgfra, Cxcl12, Dpt, Lmna, Cxcl1, Cxcl2, Ccl2.
Conditioned media collection
mPDAC cells 9091 and 8661 were plated in 10 cm dishes, and subsequently treated after 24 hours for 3 days in the presence of DMSO (vehicle) or T/N (10 nM trametinib and 2 μM nintedanib). At day three of drug treatment, the medium was removed, cells were washed once with PBS, twice with serum/phenol-red free medium, and incubated for 6 hours in 5 ml serum/phenol-red free medium with DMSO or T/N. The resulting conditioned medium was then collected, filtered with 0.2 μm pores and used for downstream secretome analysis.
MS-based secretomics
Cell supernatants were concentrated to ~250 μl and washed with 50 mM Tris, pH 8 using Amicon Ultra 3 kDa cutoff filter units (Merck) at 4°C, 4000 x g. 50 μl of concentrated supernatants were supplemented with 10 mM TCEP and 40 mM 2-chloroacetamide and then heated in a thermoshaker for 10 minutes at 95°C, 1000 rpm. Afterwards, samples were digested with 1.5 μg trypsin/ LysC mix for 16 h at 37°C, 1000 rpm. Samples were acidified by adding 100 μl isopropanol, 1% TFA and desalted using in-house made SDB-RPS StageTips.
Desalted peptide mixtures were reconstituted in buffer A (0.1% formic acid) and analyzed with an EASY-nLC 1200 ultrahigh-pressure system (Thermo Fisher Scientific) coupled to an Orbitrap Exploris 480 instrument (Thermo Fisher Scientific). 300 ng peptide were loaded onto a 50-cm in-house made column with 75 μm inner diameter, packed with C18 1.9 μm ReproSil beads (Dr. Maisch GmbH). Peptides were eluted with a linear gradient from 5% to 30% buffer B (0.1% formic acid, 80% acetonitrile) in 95 minutes at flow rate of 300 nl min-1. An in-house made column oven maintained the temperature at 60°C.
Data were acquired with a data-dependent MS/MS method. Full scans (300 to 1650 m/z, R = 60,000 at 200 m/z) at a normalized AGC target of 300% were followed by 15 MS/MS scans with higher energy collisional dissociation (normalized AGC target 100%, maximum injection time 28 ms, isolation window 1.4 m/z, HCD collision energy 30%, R = 15,000 at 200 m/z). Dynamic exclusion of 30 s was enabled.
Data analysis
MS raw files were processed with the Andromeda search engine built into MaxQuant3 (v1.6.2.10) and MS/MS spectra were matched against the mouse UniProt FASTA database (June 2019) with an FDR of 0.01 at the protein and peptide level and a minimum peptide length of seven amino acids. Match between runs was enabled and the minimal ratio count for label-free quantification was set to one. Proteins were filtered for extracellular annotation (GOCC terms “extracellular space” and “extracellular matrix”, UniProt keywords “secreted”). Missing values were replaced from a Gaussian distribution (30 % width and downshift by 1.8 standard deviations of measured values) and t-tests were performed with a permutation-based FDR of 0.05.
Intercellular communication analysis
Cell-to-cell interactions were inferred based on the expression of known ligandreceptor pairs in different cell types. Specifically, we used the ligand-receptor interactions from CellPhoneDB (v2.0) 76 complemented by proteins with secretomics-derived experimental evidences 77 . Communication networks were devised by integrating quantitative MS secretome data of “sending” tumor cells with scRNA-seq data of “receiving” cell populations.
Statistics and reproducibility
No statistical method was used to predetermine sample size. No data were excluded from the analyses. If possible, the experiments were randomized and the Investigators blinded to allocation during experiments and outcome assessment. Graphical depiction, data correlation and statistical analysis were performed with GraphPad Prism (v8). Unless otherwise indicated, all data were determined from at least 3 independent experiments. For comparisons between data sets, log-rank test or two-tailed t-test with Welch's correction was employed and resulting p-values are indicated in the respective figures. The significance level was set to 0.05. If more than one statistical test was performed simultaneously on a single data set, a Bonferroni-adjusted significance level was calculated to account for the increased possibility of false-positive results. Survival analysis was carried out by the log-rank test.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Extended Data
Extended Data Fig. 1. Assessment of differential pharmacologic and genetic dependencies and signaling pathway activities in PDAC subtypes.
a, Clonogenic assay of two hPDAC cell lines (top) and two mPDAC cell cultures (bottom) treated with the MEK inhibitor trametinib. The shown cell lines represent the drug-response of the epithelial and mesenchymal subtypes to trametinib treatment.
b, c, β-score distribution of CRISPR/Cas9 genome wide negative-selection (viability) screens performed in one classical (6075, panel (b)) and one mesenchymal (9091, panel (c)) mPDAC cell line. Highlighted in yellow, for the classical line, and blue, for the mesenchymal line, are the β-scores of KRAS and the core genes involved in direct KRAS downstream signaling.
d, CRISPR/Cas9 dependency scores of KRAS and core genes involved in direct KRAS downstream signaling. The dependency scores of all hPDAC cell lines were obtained from the DepMap database and are shown in grey. Dependency scores corresponding to classical and mesenchymal cell lines included in the T/N drug screen are represented in the yellow and blue violin plots. Data were obtained from the CRISPR dataset and analyzed through the DepMap release DepMap 21Q2 Public (https://depmap.org/portal/download/).
e, f, Mesenchymal (9091) and classical (8661) PDAC cell cultures were used to generate site-specific phospho-array datasets (Phospho Explorer antibody microarray, Full Moon Biosystems). Phospho-array data (supplementary table 1) were used to test for the enrichment of differentially phosphorylated sites between classical and mesenchymal mPDAC cell lines. Functionally grouped networks with reactome terms as nodes, showing pathways overrepresented in classical (e) and mesenchymal (f) cells are represented with the ClueGO plugin of Cytoscape. Only the pathways with an adjusted p value (calculated by two-sided hypergeometric test, Bonferroni corrected) ≤ 0.05 are depicted. The node size represents the term enrichment significance.
Extended Data Fig. 2. Genetic depletion of Mek1/2 in established PDAC.
a, Genetic strategy to delete Mek1 and Mek2 by 4-hydroxitamoxifen (4OHT)-mediated CreERT2 activation. Pdx1-Flp;FSF-KrasG12D/+;FSF-R26CAG-CreERT2/+ mice were crossed with mice harboring loxP-flanked Mek1 and Mek2 alleles. This allowed MEK1/2 deletion in established PDAC by tamoxifen administration in vitro and in vivo after orthotopic transplantation.
b, Genotyping PCR of PDAC cells with indicated genotypes to analyze recombination of the floxed Mek1 allele. Non-recombined mutant, recombined mutant and wild-type PCR products are indicated on the right side. Representative gel of three independent experiments.
c, Western blot analysis of MEK1 and MEK2 expression in primary PDAC cell cultures with indicated genotypes after 4 days of tamoxifen (4-OHT) and vehicle (ethanol, EtOH) treatment. HSP90 served as loading control. Representative gel of three independent experiments.
d, Clonogenic assays of mPDAC cells with indicated genotypes. Control cells treated with vehicle (ethanol; EtOH) are shown in the upper row, 4-OHT treated cells in the lower row.
e, Schematic representation of the experimental set-up to test the effect of Mek1/2 knockout in vivo by tamoxifen administration using syngeneic immunocompetent PDAC models. mPDAC cells with conditional Mek1lox/lox;Mek2lox/lox alleles were used for the orthotopic transplantation experiments.
f, Waterfall plot showing tumor response of vehicle and tamoxifen treated animals after one week of treatment (fold-change compared to baseline before treatment based on MRI-volumetric measurements, y-axis). Each bar represents one mouse. P values calculated with two-tailed unpaired t test.
g, Kaplan-Meier survival curve of vehicle and tamoxifen treated PDAC models. Number of mice is indicated in the corresponding panels. P value was calculated with log-rank (Mantel-Cox) test.
h, Representative images of HE and IHC for MEK1, MEK2 and KI67 of tissue sections of tumors from orthotopically transplanted Mek1lox/lox;Mek2lox/lox models treated with vehicle or tamoxifen. Representative pictures of three independent experiments. Scale bars, 100 µm.
Extended Data Fig. 3. Pharmacologic and genetic modulation of drug sensitivity in classical and mesenchymal PDAC cell cultures.
a, b, Clonogenic assays of a representative human (left) and mouse (right) PDAC cell culture showing antagonism to the trametinib/nintedanib (T/N) combination. Cell cultures were treated with indicated concentrations of T/N.
c, Western blot of phospho-ERK and ERK in T/N (10 nM trametinib + 2 µM nintedanib) and vehicle treated classical and mesenchymal primary mPDAC cell lines. HSP90 served as loading control. Classical cell lines are marked in yellow, mesenchymal in blue. Representative gels of three independent experiments.
d, Clonogenic assays using increased drug concentrations of the T/N combination of three of the most antagonistic cell lines, as depicted in figure 2, panel (g).
e, Doxycycline-induced overexpression of KRASG12D in mouse PDAC cells. 2259 mPDAC cells representative of the classical subtype was transduced with lentivirus carrying doxycycline-inducible KRASG12D or GFP-control expression constructs. KRASG12D or GFP expression were induced by doxycycline (100 ng/ml) for one or 14 days.
f, Western blot of phospho-ERK and total ERK in cells overexpressing KRASG12D or GFP for one day. HSP90 served as loading control.
g, Expression of the marker gene Cdh1 for epithelial cell differentiation was evaluated by qRT-PCR (normalized to Cyclophilin B). Data are shown as mean ±SD; n=3 biological replicates. P value was calculated with two-tailed unpaired t test.
h, Representative picture of three independent experiments of the morphological changes of PDAC cells upon KRASG12D induction for one or 14 days of doxycycline treatment. Scale bars, 200 µm.
i, Representative clonogenic assays of mPDAC cells treated with the indicated concentrations of trametinib and nintedanib upon KRASG12D (right panel) or GFP (left panel) overexpression.
j, Bliss synergy scores for the mPDAC cell line treated with the combination of trametinib and nintedanib upon KRASG12D or GFP overexpression.
Extended Data Fig. 4. Kinobead-based proteomic identification of the trametinib and nintedanib targets and treatment-induced changes in the phosphoproteome of classical and mesenchymal PDAC.
a, b, Representative pictures of the target space of trametinib (a) and nintedanib (b) for 2259 PDAC cells. A phylogenetic tree of all kinases for the 2259 primary mouse PDAC cell culture is shown. The indicated circle sizes indicate potency (apparent dissociation constants (Kdapp)), the color code specifies protein-drug interaction with the designated or other targets. Arrows highlight the identified targets.
c, Radar plot showing the overlay of the pKd (−log10Kd) for the targets of nintedanib in the 6 PDAC cell cultures tested. PDAC cells of the classical (n=4) and mesenchymal (n=2) subtype are indicated with the color code.
d, Heatmap showing the differentially expressed genes between epithelial and mesenchymal cell cultures identified as targets of nintedanib. The color code indicates the Z score.
e, Volcano plots representing the change in gene expression of the nintedanib targets (in blue) upon trametinib treatment. The x-axis log2 fold change (treated/control), the y-axis shows the per test adjusted p values, which were calculated by differential expression test (using the DESeq2 package). A gene was considered to be differentially expressed with a Benjamini-Hochberg adjusted p-value of 0.05 and an absolute fold change >1.
f, g, Mesenchymal (9091) and classical (8661) PDAC cell cultures were used to generate site-specific phospho-array datasets (Phospho Explorer antibody microarray, Full Moon Biosystems). The cell lines were analyzed at basal condition and in presence of T/N (trametinib 10 nM + nintedanib 2 μM). Phospho-array data (supplementary table 3) were used to test for the decrease of differentially phosphorylated sites between T/N and vehicle (DMSO) treated classical and mesenchymal mPDAC cells. Functionally grouped maps, obtained with the ClueGO plugin of Cytoscape, representing pathways overrepresented in mesenchymal (f) and classical (g) mPDAC upon T/N treatment are shown. Only the pathways with an adjusted p value (calculated by two-sided hypergeometric test, Bonferroni corrected) ≤ 0.05 are represented. The node size represents the term enrichment significance.
Extended Data Fig. 5. Pharmacologic assessment of nintedanib targets.
a, Combinatorial drug screen on mesenchymal hPDAC cell line MiaPaca2 and mPDAC cell line 9091, as shown in Figure 2, panel (b). The MEK inhibitor trametinib was used in fixed concentration and combined with 418 additional drugs under preclinical and clinical investigation. Highlighted in orange are the drugs in the high-throughput drug screen showing overlapping targets with nintedanib.
b, Venn diagrams showing the target overlap between the drugs identified in (a) and nintedanib (see supplementary table 4) as reported from the ProteomicsDB database (https://www.proteomicsdb.org).
c, Venn diagrams showing the target overlap between nintedanib and additional drugs with an overlapping target profile chosen for further target assessment. The overlapping targets are listed below each figure. The target information was downloaded from the ProteomicsDB database (https://www.proteomicsdb.org).
d, Representative clonogenic assays of mesenchymal mPDAC cell cultures treated with trametinib in combination with the drugs shown in (c) as compared to nintedanib. The cell lines were treated with the indicated concentrations of trametinib and the indicated experimental drug.
e, Bliss synergy scores of clonogenic assays shown in (d) integrated with cell morphology for the treated mPDAC cell cultures (classical subtype depicted in yellow and mesenchymal in blue).
Extended Data Fig. 6. Genetic screens to identify relevant nintedanib targets sensitizing mesenchymal PDAC towards trametinib.
a, Western blot of Cas9 expression in the clonal cell lines used for CRISPR/Cas9 screens. β-Actin served as loading control.
b, Editing efficiency at the Hprt locus.
c, Relative viability upon 6-Thioguanine treatment to validate Cas9 function in Hprt proficient and deficient Cas9 cells (mean ±SD; n=3 biological replicates).
d, Relative cell growth (y-axis), assessed by cell counting, in the presence of different concentrations of trametinib (mean ±SD; n=3 biological replicates). The pink line indicates the trametinib concentration used for the CRISPR/Cas9 screens.
e, Phospho-ERK, ERK and Cas9 Western blots of clones used for CRISPR/Cas9 screens. Cells were treated with DMSO or trametinib (1.25 nM, 2.5 nM, 5 nM, 10 nM and 20 nM) for 4 days. HSP90 served as loading control.
f, Focused CRISPR/Cas9-based genetic screening in mesenchymal mPDAC cells 8248 and 8570. Trametinib sensitivity (x-axis) represents β-scores calculated as sgRNA representation difference between trametinib-treated cells and their initial representation. Differential sensitivity (y-axis) indicates β-score differences between trametinib- and DMSO-treated arms. In red, genes presenting differential sensitivity ≤-0.25.
g, Network visualization of normalized gene expression (assessed by RNA-seq) of nintedanib targets shown in figure 3 (d).
h, Lentiviral CRISPR/Cas9-mediated deletion of selected top-scoring nintedanib targets in 8248 and 8570 cells. Knock-out cells were treated with trametinib (5 nM) or DMSO and viability was assessed through clonogenic assays.
i, Quantification of panel (h). Data are normalized to DMSO-treated non-targeting controls (mean ±SD; n=3 biological replicates). The dashed line represents the mean of trametinib-treated non-targeting controls.
j, Editing efficiency of each sgRNA used in figure 2 (f and g) and in panels (h) and (i) of this figure.
k, Combinatorial deletion of nintedanib targets via ribonucleoprotein (RNP) electroporation. Mesenchymal 8248 and 8570 knock-out cells were treated with trametinib (5 nM) or DMSO and viability was assessed through clonogenic assays.
l, Quantification of panel (k). Data are normalized to DMSO-treated non-targeting controls (mean ±SD; n=3 biological replicates). Dashed line represents the mean of trametinib-treated non-targeting controls.
The shown gels are representative of three independent experiments.
Extended Data Fig. 7. Characterization of context-specific changes of the tumor vasculature and the adaptive immune system in classical and mesenchymal PDAC subtypes upon therapy.
a, Orthotopically transplanted tumors of the indicated subtypes were treated with vehicle (control) and the T/N combination. Representative images of immunofluorescence stainings of tissue sections for P-selectin (upper panel) and α-SMA (lower panel) (magenta). CD31 was used to detect endothelial cells (green). DAPI was used for nuclear staining (blue). Scale bars, 25 µm.
b, c, Quantification of the P-selectin+ vessels (b) and α-SMA+ vessels (c) of the immunofluorescence stainings depicted in (a). Individual tumors are shown as single dots in the graph (classical: control n=3, T/N n=4; mesenchymal: control n=4, T/N n=5).
d, Orthotopically transplanted tumors of the indicated subtypes were treated with vehicle (control) and the indicated drugs and drug combinations, explanted, single cell suspended and analyzed by flow cytometry. Panel (d) shows the staining for CD45+ cells. Individual tumors are shown as single points in the graph.
e, f, Graphs representing the percentage of CD4+ (e) and CD8+ (f) cells in the PDAC control cohort and in the different treatment conditions as analyzed by flow cytometry. Single points represent individual tumors.
g, Left, scheme of the in vivo experimental strategy using orthotopic PDAC cell transplantations into T cell deficient CD3ε knockout (KO) mice. Right, representative FACS plot of immunodeficient CD3ε-KO and wild-type C57BL/6 mice, highlighting the lack of T cells in the CD3ε-KO animals.
h, Representative MRI picture of vehicle (control) and T/N treated PDAC bearing CD3ε-KO mice before (week 2) and after 1 week treatment (week 3).
P values in (b), (c), (d) and (f) were calculated with two-tailed unpaired t test.
T: trametinib, N: nintedanib, T/N: trametinib+nintedanib.
Extended Data Fig. 8. Characterization of context-specific changes of the innate immune system in classical and mesenchymal PDAC subtypes upon therapy.
a, Pie charts representative of the fraction of innate immune cell populations in PDAC from vehicle control and mice that received the combination (T/N) for both classical and mesenchymal orthotopically transplanted tumors as analyzed by flow cytometry. The number of tumors per condition analyzed is depicted in the corresponding panel.
b, Graphs representing the percentage of Ly6G- CD11b+ F4/80+ macrophages in PDAC of the control cohort and in the treatment conditions as analyzed by flow cytometry. Single points represent individual tumors.
c, Representative immunofluorescence staining for CD80/CD68 and ARG1/CD68 cells in both classical and mesenchymal tumors treated with the T/N combination therapy or vehicle as control. Scale bars, 50 µm.
d, Quantification of the M1-like polarization macrophage markers CD80/CD68+ and INOS/CD68+ from the immunofluorescence stainings depicted in (c). Individual tumors are shown as single points in the graph (classical: control n=5, T/N n=5; mesenchymal: control n=4, T/N n=5).
e, Quantification of the M2-like polarization macrophage markers ARG1/CD68+ and MRC1/CD68+ from the staining depicted in (c). Individual tumors are shown as single points in the graph (classical: control n=5, T/N n=5; mesenchymal: control n=4, T/N n=5).
f, Graphs representing the percentage of Ly6G+ CD11b+ neutrophils in the control cohort and in the treatment conditions as analyzed by flow cytometry. Single points represent individual tumors.
P values in (d) and (e) were calculated with two-tailed unpaired t test.
T: trametinib, N: nintedanib, T/N: trametinib+nintedanib.
Extended Data Fig. 9. scRNA-seq reveals treatment-induced changes in TME cell subpopulations and activation of the DNA damage pathway in cancer cells.
a, Dotplot displaying marker gene expression across each identified cluster of cancer cells and corresponding tumor microenvironment for both classical and mesenchymal tumors. The clusters are indicated on the y axis and the main markers for each identified population are indicated on the x axis.
b, Left, UMAP plot showing all identified cell populations within the scRNA-seq experiment. Middle, UMAP plot showing classical (yellow) and mesenchymal (blue) tumors from all treatment and vehicle groups. Right, UMAP plot showing the treatment induced changes in cell type composition among the identified cell populations across subtypes. Lower part, UMAP density plots showing distribution of annotated clusters upon treatment, cell numbers for each condition are integrated below.
c, UMAP plot showing the identified tumor cell clusters. The expression of Cdh1 and Krt18, epithelial markers, and of Cdh2 and Vim, mesenchymal markers, across treatment conditions are shown below.
d, Heatmap of the most differentially expressed genes from the gene expression signature in figure 7 across subtypes and treatment conditions.
e, Gene set enrichment analysis (GSEA) of scRNA-seq data of cancer cells reveals enrichment of DNA damage in both classical and mesenchymal tumors upon treatment with the T/N combination. NES and FDR-q values are indicated.
f, Representative images of immunohistochemical staining for γH2AX of control and T/N treated tumor sections for both classical and mesenchymal subtypes. Scale bar, 70 µm.
g, Quantification of γH2AX positive cells in (f). Individual tumors are shown as single points in the graph (classical: control n=6, T/N n=5; mesenchymal: control n=8, T/N n=7). P values were calculated with two-tailed unpaired t test.
Endo cells: endothelial cells. T/N: trametinib+nintedanib. T/N+aPD-L1: trametinib+nintedanib+anti PD-L1 antibody.
Extended Data Fig. 10. Context-dependent reprogramming of the cancer cell derived secretome and cancer associated fibroblasts (CAFs) by the T/N combination therapy.
a, Volcano plots highlighting the changes in secreted factors upon T/N treatment in classical (left) and mesenchymal (right) PDAC cells. The x-axis shows log2 fold change (treated/control), the y-axis the per test adjusted p values, which were calculated by differential expression test (two-sided t test).
b, Circos plot showing the key communication signals from tumor cells to T cell subtypes, tumor cells and acinar cells in classical mPDAC. The ligand protein expression fold change, identified from secretome experiments, between T/N and control is shown in the middle. Normalized receptor expression levels obtained from scRNA-seq data are shown in the outer concentric circles.
c, UMAP plot highlighting the whole population of CAF cells identified in classical and mesenchymal tumors.
d, Left, UMAP plot showing the CAF population across different treatment conditions in classical tumors. Right, UMAP plots displaying the identified CAF clusters and resulting subpopulations for classical tumors.
e, UMAP plots of the CAF cluster displaying selected marker gene expression.
f, Heatmap displaying expression of selected genes in CAFs across the identified clusters. The y axis shows the selected marker genes, the x axis represents each of the identified clusters in (d).
g, Violin plot showing Tgfb1 expression by myoCAFs across the different treatment conditions.
h, Proportion of CAF subtypes in the indicated different treatment conditions. CAF subpopulations were identified in the fibroblast cell clusters and annotated with the markers described in (f).
T/N: trametinib+nintedanib, T/N+aPD-L1: trametinib+nintedanib+anti PD-L1 antibody.
Supplementary Material
Acknowledgments
We would like to thank Drs. Hassan Nakhei, Tyler Jacks, David Tuveson, Manuela Baccarini, Roland Schmid and Allan Bradley for providing transgenic animals, and the TUM animal facility and the imaging core facility of the Department of Nuclear Medicine, Klinikum rechts der Isar, for excellent technical support. This study was supported by the German Cancer Consortium (DKTK), Deutsche Forschungsgemeinschaft (DFG SA 1374/4-2, SFB 824 C09 to G.S. and D.S., SFB 1321 Project-ID 329628492 P06 to D.S. and M.S.S, P11 to M.S.R. and D.S., and S01 to D.S., M.R., R.R., and G.S., and SFB 1371 Project-ID 395357507 P12 to DS), the Wilhelm Sander-Stiftung (2020.174.1), and the European Research Council (ERC CoG No. 648521, to D.S.).
Footnotes
Author contributions
C.F., S.B., S.A.W., C.S., J.J.M., A.T., J.M., T.K., J.H., J.J.S., B.T., O.R., C.S., K.G., A.C., C.V., M.Z., A.A.V., W.H.P., R.M., R.Ö., T.A., and J.R. performed research; M.S.R., B.K., K.S., F.M., M.R., M.F., R.R., M.S.S. and D.S. contributed new reagents and analytic tools; C.F., S.B., S.A.W., C.S., J.M., J.J.S., J.C., A.C., F.B., M.J., K.S., O.B., J.R, F.M., M.F., R.R., M.S.S., G.S. and D.S. analyzed data; C.F. and D.S. wrote the paper.
Competing Interests
The authors declare that they have no competing interests. Correspondence and requests for materials should be addressed to D.S.
Data availability
The RNA-sequencing dataset has been deposited to the EBIArrayExpress repository with the dataset identifier E-MTAB-11187. The mass spectrometry kinobead pulldown and the mass spectrometry secretomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository with the dataset identifier PXD023267 and PXD027877, respectively. The scRNA-seq data have been deposited to the EBIArrayExpress repository with the dataset identifier E-MTAB-9954. The human pancreatic cancer data were derived from previous studies and are available in the supplementary information of the respective publications 6–8 . All other data have been provided as supplementary tables or source data files. Mice and cell lines are available from the corresponding author on reasonable request. Key resources are listed in supplementary table 8.
Code availability
Analyses were performed using open-source software, and in-house scripts in R v3.6.2 and Python v3.8.3, which are available from the corresponding author on reasonable request. Codes are provided via the GitHub repository at the following link: https://github.com/stefanie-baerthel/combinatorial_treatment_analysis.
References
- 1.Quaresma M, Coleman MP, Rachet B. 40-year trends in an index of survival for all cancers combined and survival adjusted for age and sex for each cancer in England and Wales, 1971-2011: a population-based study. Lancet (London, England) 2015;385:1206–1218. doi: 10.1016/s0140-6736(14)61396-9. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70:7–30. doi: 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- 3.Aung KL, et al. Genomics-Driven Precision Medicine for Advanced Pancreatic Cancer: Early Results from the COMPASS Trial. Clinical cancer research : an official journal of the American Association for Cancer Research. 2018;24:1344–1354. doi: 10.1158/1078-0432.Ccr-17-2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kalimuthu SN, et al. Morphological classification of pancreatic ductal adenocarcinoma that predicts molecular subtypes and correlates with clinical outcome. Gut. 2020;69:317–328. doi: 10.1136/gutjnl-2019-318217. [DOI] [PubMed] [Google Scholar]
- 5.Chan-Seng-Yue M, et al. Transcription phenotypes of pancreatic cancer are driven by genomic events during tumor evolution. Nature genetics. 2020;52:231–240. doi: 10.1038/s41588-019-0566-9. [DOI] [PubMed] [Google Scholar]
- 6.Bailey P, et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature. 2016;531:47–52. doi: 10.1038/nature16965. [DOI] [PubMed] [Google Scholar]
- 7.Dijk F, et al. Unsupervised class discovery in pancreatic ductal adenocarcinoma reveals cell-intrinsic mesenchymal features and high concordance between existing classification systems. Scientific reports. 2020;10:337. doi: 10.1038/s41598-019-56826-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Puleo F, et al. Stratification of Pancreatic Ductal Adenocarcinomas Based on Tumor and Microenvironment Features. Gastroenterology. 2018;155:1999–2013.:e1993. doi: 10.1053/j.gastro.2018.08.033. [DOI] [PubMed] [Google Scholar]
- 9.Morrison AH, Byrne KT, Vonderheide RH. Immunotherapy and Prevention of Pancreatic Cancer. Trends in cancer. 2018;4:418–428. doi: 10.1016/j.trecan.2018.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Balachandran VP, et al. Identification of unique neoantigen qualities in long-term survivors of pancreatic cancer. Nature. 2017;551:512–516. doi: 10.1038/nature24462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen DS, Mellman I. Elements of cancer immunity and the cancer-immune set point. Nature. 2017;541:321–330. doi: 10.1038/nature21349. [DOI] [PubMed] [Google Scholar]
- 12.Ino Y, et al. Immune cell infiltration as an indicator of the immune microenvironment of pancreatic cancer. British journal of cancer. 2013;108:914–923. doi: 10.1038/bjc.2013.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Collisson EA, et al. A central role for RAF-->MEK-->ERK signaling in the genesis of pancreatic ductal adenocarcinoma. Cancer Discov. 2012;2:685–693. doi: 10.1158/2159-8290.CD-11-0347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blumenschein GR, Jr, et al. A randomized phase II study of the MEK1/MEK2 inhibitor trametinib (GSK1120212) compared with docetaxel in KRAS-mutant advanced non-small-cell lung cancer (NSCLC) Annals of Oncology. 2015;26:894–901. doi: 10.1093/annonc/mdv072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Caunt CJ, Sale MJ, Smith PD, Cook SJ. MEK1 and MEK2 inhibitors and cancer therapy: the long and winding road. Nat Rev Cancer. 2015;15:577–592. doi: 10.1038/nrc4000. [DOI] [PubMed] [Google Scholar]
- 16.Mueller S, et al. Evolutionary routes and KRAS dosage define pancreatic cancer phenotypes. Nature. 2018;554:62–68. doi: 10.1038/nature25459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miyabayashi K, et al. Intraductal Transplantation Models of Human Pancreatic Ductal Adenocarcinoma Reveal Progressive Transition of Molecular Subtypes. Cancer Discovery. 2020;10:1566–1589. doi: 10.1158/2159-8290.Cd-20-0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-Seq data. BMC Bioinformatics. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liberzon A, et al. The Molecular Signatures Database Hallmark Gene Set Collection. Cell Systems. 2015;1:417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schönhuber N, et al. A next-generation dual-recombinase system for time- and hostspecific targeting of pancreatic cancer. Nature medicine. 2014;20:1340–1347. doi: 10.1038/nm.3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ruscetti M, et al. Senescence-Induced Vascular Remodeling Creates Therapeutic Vulnerabilities in Pancreas Cancer. Cell. 2020;181:424–441.:e421. doi: 10.1016/j.cell.2020.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.De Jarnette JB, et al. Specific requirement for CD3epsilon in T cell development. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:14909–14914. doi: 10.1073/pnas.95.25.14909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pathria P, Louis TL, Varner JA. Targeting Tumor-Associated Macrophages in Cancer. Trends Immunol. 2019;40:310–327. doi: 10.1016/j.it.2019.02.003. [DOI] [PubMed] [Google Scholar]
- 24.Nywening TM, et al. Targeting both tumour-associated CXCR2(+) neutrophils and CCR2(+) macrophages disrupts myeloid recruitment and improves chemotherapeutic responses in pancreatic ductal adenocarcinoma. Gut. 2018;67:1112–1123. doi: 10.1136/gutjnl-2017-313738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Steele CW, et al. CXCR2 Inhibition Profoundly Suppresses Metastases and Augments Immunotherapy in Pancreatic Ductal Adenocarcinoma. Cancer cell. 2016;29:832–845. doi: 10.1016/j.ccell.2016.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gerrard TL, Cohen DJ, Kaplan AM. Human neutrophil-mediated cytotoxicity to tumor cells. Journal of the National Cancer Institute. 1981;66:483–488. [PubMed] [Google Scholar]
- 27.Bakhoum SF, Cantley LC. The Multifaceted Role of Chromosomal Instability in Cancer and Its Microenvironment. Cell. 2018;174:1347–1360. doi: 10.1016/j.cell.2018.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mackenzie KJ, et al. cGAS surveillance of micronuclei links genome instability to innate immunity. Nature. 2017;548:461–465. doi: 10.1038/nature23449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Faget DV, Ren Q, Stewart SA. Unmasking senescence: context-dependent effects of SASP in cancer. Nature Reviews Cancer. 2019;19:439–453. doi: 10.1038/s41568-019-0156-2. [DOI] [PubMed] [Google Scholar]
- 30.Meissner F, Scheltema RA, Mollenkopf H-J, Mann M. Direct Proteomic Quantification of the Secretome of Activated Immune Cells. Science. 2013;340:475–478. doi: 10.1126/science.1232578. [DOI] [PubMed] [Google Scholar]
- 31.Matsumura S, et al. Radiation-induced CXCL16 release by breast cancer cells attracts effector T cells. Journal of immunology (Baltimore, Md : 1950) 2008;181:3099–3107. doi: 10.4049/jimmunol.181.5.3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hojo S, et al. High-Level Expression of Chemokine CXCL16 by Tumor Cells Correlates with a Good Prognosis and Increased Tumor-Infiltrating Lymphocytes in Colorectal Cancer. Cancer research. 2007;67:4725–4731. doi: 10.1158/0008-5472.Can-06-3424. [DOI] [PubMed] [Google Scholar]
- 33.Li B, et al. Epigenetic Regulation of CXCL12 Plays a Critical Role in Mediating Tumor Progression and the Immune Response In Osteosarcoma. Cancer research. 2018;78:3938–3953. doi: 10.1158/0008-5472.Can-17-3801. [DOI] [PubMed] [Google Scholar]
- 34.Nagarsheth N, Wicha MS, Zou W. Chemokines in the cancer microenvironment and their relevance in cancer immunotherapy. Nature Reviews Immunology. 2017;17:559–572. doi: 10.1038/nri.2017.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mehta AK, et al. Targeting immunosuppressive macrophages overcomes PARP inhibitor resistance in BRCA1-associated triple-negative breast cancer. Nature Cancer. 2021;2:66–82. doi: 10.1038/s43018-020-00148-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peng DH, et al. Th17 cells contribute to combination MEK inhibitor and anti-PD-L1 therapy resistance in KRAS/p53 mutant lung cancers. Nature communications. 2021;12:2606. doi: 10.1038/s41467-021-22875-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Di Pilato M, et al. CXCR6 positions cytotoxic T cells to receive critical survival signals in the tumor microenvironment. Cell. 2021;184:4512–4530.:e4522. doi: 10.1016/j.cell.2021.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Uhlén M, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 39.Steins A, et al. High-grade mesenchymal pancreatic ductal adenocarcinoma drives stromal deactivation through CSF-1. EMBO reports. 2020;21:e48780. doi: 10.15252/embr.201948780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sahai E, et al. A framework for advancing our understanding of cancer-associated fibroblasts. Nat Rev Cancer. 2020;20:174–186. doi: 10.1038/s41568-019-0238-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schneider G, Schmidt-Supprian M, Rad R, Saur D. Tissue-specific tumorigenesis: context matters. Nat Rev Cancer. 2017;17:239–253. doi: 10.1038/nrc.2017.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Elyada E, et al. Cross-Species Single-Cell Analysis of Pancreatic Ductal Adenocarcinoma Reveals Antigen-Presenting Cancer-Associated Fibroblasts. Cancer Discov. 2019;9:1102–1123. doi: 10.1158/2159-8290.Cd-19-0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hosein AN, Brekken RA, Maitra A. Pancreatic cancer stroma: an update on therapeutic targeting strategies. Nature reviews Gastroenterology & hepatology. 2020;17:487–505. doi: 10.1038/s41575-020-0300-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ligorio M, et al. Stromal Microenvironment Shapes the Intratumoral Architecture of Pancreatic Cancer. Cell. 2019;178:160–175.:e127. doi: 10.1016/j.cell.2019.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Batlle E, Massagué J. Transforming Growth Factor-β Signaling in Immunity and Cancer. Immunity. 2019;50:924–940. doi: 10.1016/j.immuni.2019.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Olive KP, et al. Inhibition of Hedgehog signaling enhances delivery of chemotherapy in a mouse model of pancreatic cancer. Science. 2009;324:1457–1461. doi: 10.1126/science.1171362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sherman MH, et al. Vitamin D receptor-mediated stromal reprogramming suppresses pancreatitis and enhances pancreatic cancer therapy. Cell. 2014;159:80–93. doi: 10.1016/j.cell.2014.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hayashi A, et al. A unifying paradigm for transcriptional heterogeneity and squamous features in pancreatic ductal adenocarcinoma. Nature Cancer. 2020;1:59–74. doi: 10.1038/s43018-019-0010-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brzostek-Racine S, Gordon C, Van Scoy S, Reich NC. The DNA damage response induces IFN. Journal of immunology (Baltimore, Md : 1950) 2011;187:5336–5345. doi: 10.4049/jimmunol.1100040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou F. Molecular Mechanisms of IFN-γ to Up-Regulate MHC Class I Antigen Processing and Presentation. International Reviews of Immunology. 2009;28:239–260. doi: 10.1080/08830180902978120. [DOI] [PubMed] [Google Scholar]
- 51.Respa A, et al. Association of IFN-γ Signal Transduction Defects with Impaired HLA Class I Antigen Processing in Melanoma Cell Lines. Clinical Cancer Research. 2011;17:2668–2678. doi: 10.1158/1078-0432.Ccr-10-2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kearney CJ, et al. Tumor immune evasion arises through loss of TNF sensitivity. Science Immunology. 2018;3:eaar3451. doi: 10.1126/sciimmunol.aar3451. [DOI] [PubMed] [Google Scholar]
- 53.Liao W, et al. KRAS-IRF2 Axis Drives Immune Suppression and Immune Therapy Resistance in Colorectal Cancer. Cancer cell. 2019;35:559–572.:e557. doi: 10.1016/j.ccell.2019.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tape CJ, et al. Oncogenic KRAS Regulates Tumor Cell Signaling via Stromal Reciprocation. Cell. 2016;165:910–920. doi: 10.1016/j.cell.2016.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kuilman T, Peeper DS. Senescence-messaging secretome: SMS-ing cellular stress. Nature Reviews Cancer. 2009;9:81–94. doi: 10.1038/nrc2560. [DOI] [PubMed] [Google Scholar]
- 56.Lederer DJ, Martinez FJ. Idiopathic Pulmonary Fibrosis. N Engl J Med. 2018;378:1811–1823. doi: 10.1056/NEJMra1705751. [DOI] [PubMed] [Google Scholar]
- 57.Richeldi L, et al. Efficacy and safety of nintedanib in idiopathic pulmonary fibrosis. N Engl J Med. 2014;370:2071–2082. doi: 10.1056/NEJMoa1402584. [DOI] [PubMed] [Google Scholar]
- 58.Eser S, et al. Selective requirement of PI3K/PDK1 signaling for Kras oncogene-driven pancreatic cell plasticity and cancer. Cancer cell. 2013;23:406–420. doi: 10.1016/j.ccr.2013.01.023. [DOI] [PubMed] [Google Scholar]
- 59.von Burstin J, et al. E-cadherin regulates metastasis of pancreatic cancer in vivo and is suppressed by a SNAIL/HDAC1/HDAC2 repressor complex. Gastroenterology. 2009;137:361–371.:371 e361-365. doi: 10.1053/j.gastro.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 60.Ianevski A, Giri AK, Aittokallio T. SynergyFinder 2.0: visual analytics of multidrug combination synergies. Nucleic acids research. 2020;48:W488–W493. doi: 10.1093/nar/gkaa216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Klaeger S, et al. The target landscape of clinical kinase drugs. Science. 2017;358 doi: 10.1126/science.aan4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shevchenko A, Tomas H, Havli J, Olsen JV, Mann M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nature protocols. 2006;1:2856–2860. doi: 10.1038/nprot.2006.468. [DOI] [PubMed] [Google Scholar]
- 63.Vizcaíno JA, et al. The PRoteomics IDEntifications (PRIDE) database and associated tools: status in 2013. Nucleic acids research. 2013;41:D1063–1069. doi: 10.1093/nar/gks1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hafner M, Niepel M, Chung M, Sorger PK. Growth rate inhibition metrics correct for confounders in measuring sensitivity to cancer drugs. Nature methods. 2016;13:521–527. doi: 10.1038/nmeth.3853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Clark NA, et al. GRcalculator: an online tool for calculating and mining doseresponse data. BMC cancer. 2017;17:698. doi: 10.1186/s12885-017-3689-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bindea G, et al. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25:1091–1093. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.De Weirdt PC, et al. Optimization of AsCas12a for combinatorial genetic screens in human cells. Nature biotechnology. 2021;39:94–104. doi: 10.1038/s41587-020-0600-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Joung J, et al. Genome-scale CRISPR-Cas9 knockout and transcriptional activation screening. Nature protocols. 2017;12:828–863. doi: 10.1038/nprot.2017.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li W, et al. MAGeCK enables robust identification of essential genes from genomescale CRISPR/Cas9 knockout screens. Genome Biol. 2014;15:554. doi: 10.1186/s13059-014-0554-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Doench JG, et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nature biotechnology. 2016;34:184–191. doi: 10.1038/nbt.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jackson EL, et al. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes & development. 2001;15:3243–3248. doi: 10.1101/gad.943001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hingorani SR, et al. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer cell. 2003;4:437–450. doi: 10.1016/s1535-6108(03)00309-x. [DOI] [PubMed] [Google Scholar]
- 73.Nakhai H, et al. Ptf1a is essential for the differentiation of GABAergic and glycinergic amacrine cells and horizontal cells in the mouse retina. Development (Cambridge, England) 2007;134:1151–1160. doi: 10.1242/dev.02781. [DOI] [PubMed] [Google Scholar]
- 74.Catalanotti F, et al. A Mek1-Mek2 heterodimer determines the strength and duration of the Erk signal. Nature structural & molecular biology. 2009;16:294–303. doi: 10.1038/nsmb.1564. [DOI] [PubMed] [Google Scholar]
- 75.Wolf FA, Angerer P, Theis FJ. SCANPY: large-scale single-cell gene expression data analysis. Genome Biology. 2018;19:15. doi: 10.1186/s13059-017-1382-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Efremova M, Vento-Tormo M, Teichmann SA, Vento-Tormo R. CellPhoneDB: inferring cell–cell communication from combined expression of multi-subunit ligand– receptor complexes. Nature protocols. 2020;15:1484–1506. doi: 10.1038/s41596-020-0292-x. [DOI] [PubMed] [Google Scholar]
- 77.Phulphagar K, et al. Proteomics reveals distinct mechanisms regulating the release of cytokines and alarmins during pyroptosis. Cell reports. 2021;34:108826. doi: 10.1016/j.celrep.2021.108826. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-sequencing dataset has been deposited to the EBIArrayExpress repository with the dataset identifier E-MTAB-11187. The mass spectrometry kinobead pulldown and the mass spectrometry secretomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository with the dataset identifier PXD023267 and PXD027877, respectively. The scRNA-seq data have been deposited to the EBIArrayExpress repository with the dataset identifier E-MTAB-9954. The human pancreatic cancer data were derived from previous studies and are available in the supplementary information of the respective publications 6–8 . All other data have been provided as supplementary tables or source data files. Mice and cell lines are available from the corresponding author on reasonable request. Key resources are listed in supplementary table 8.
Analyses were performed using open-source software, and in-house scripts in R v3.6.2 and Python v3.8.3, which are available from the corresponding author on reasonable request. Codes are provided via the GitHub repository at the following link: https://github.com/stefanie-baerthel/combinatorial_treatment_analysis.