Abstract
RNA-catalysed RNA methylation was recently shown to be part of the catalytic repertoire of ribozymes. The methyltransferase ribozyme MTR1 catalyses the site-specific synthesis of 1-methyladenosine (m1A) in RNA, using O6-methylguanine (m6G) as methyl group donor. Here we report the crystal structure of MTR1 at a resolution of 2.8 Å, which reveals a guanine binding site reminiscent of natural guanine riboswitches. The structure represents the postcatalytic state of a split ribozyme in complex with the m1A-containing RNA product and the demethylated cofactor guanine. The structural data suggest the mechanistic involvement of a protonated cytidine in the methyl transfer reaction. A synergistic effect of two 2'-O-methylated ribose residues in the active site results in accelerated methyl group transfer. Supported by these results, it seems plausible that modified nucleotides may have enhanced early RNA catalysis and that metabolite-binding riboswitches may resemble inactivated ribozymes that have lost their catalytic activity during evolution.
Natural ribozymes are known to catalyse a narrow range of chemical reactions, while a large diversity of natural riboswitches are known to bind nucleotide metabolites, including S-adenosylmethionine (SAM), cobalamin, nicotinamide adenine dinucleotide (NAD+), flavine mononucleotide (FMN), and purines, such as guanine and pre-queuosine (preQ1).1,2 Several of these metabolites are essential coenzymes for methyl transferase enzymes in contemporary biology. These coenzymes are thought to have evolved in the RNA world, and primordial RNA catalysts may have utilized coenzymes to achieve greater chemical diversity.3-7 However, only one natural cofactor-utilizing ribozyme has yet been found, that is the glmS riboswitch-ribozyme, which uses glucosamine-6-phosphate to assist the site-specific cleavage of an RNA phosphodiester bond.8 In contrast, ribozymes generated in the laboratory by in vitro selection from random nucleic acid libraries have been shown to catalyse a wider variety of reactions,2,9 and some synthetic ribozymes use natural cofactors.10-13
While natural RNA-cleaving ribozymes are well characterized, only a few structures are known of artificial ribozymes that catalyse other reactions,14-17 limiting our general structural and mechanistic view of the scope of RNA catalysis. Recently, we identified the synthetic methyl transferase ribozyme MTR1 that catalyses the site-specific methylation of RNA to generate 1-methyladenosine (m1A) using O6-methylguanine (m6G) as methyl group donor.18 Subsequently, a natural preQ1 riboswitch was shown to enable a similar methyl transfer reaction, using the non-natural O6-methyl-preQ1 ligand as cofactor.19 Micura and coworkers demonstrated the formation of 3-methylcytidine (m3C) in the preQ1 riboswitch RNA, with the methyl group being transferred from the ligand to a precisely positioned cytidine in the ligand binding site.19 In analogy, one may anticipate that the active site of MTR1 could mimic the ligand binding site of guanine riboswitches,20 which was earlier shown to accommodate m6G with only slight structural perturbations,21 but a transfer of the methyl group to the RNA had not been observed.
To address the question of how the laboratory-evolved ribozyme MTR1 binds and activates m6G for synthesis of m1A in the target RNA, we solved the crystal structure of the MTR1 ribozyme and examined the chemical mechanism of the methyl transfer reaction. We report the co-crystal structures of MTR1 bound to the methylated RNA product and to the remaining guanine at a resolution of 2.8 Å. The sample for crystallization was prepared with unmethylated RNA and m6G, however, the methyl group was stoichiometrically transferred to the RNA during crystallization. Therefore, the structure represents a post-catalytic state. The catalytic core harbours m1A and guanine in the centre of a 3-helix junction structure, and the H-bonding interactions of the core nucleotides with the bound guanine resemble those found in natural guanine riboswitch aptamers. Additional post-catalytic structures were obtained from experiments with m6G derivatives that mimic the substrate used for in vitro selection. We found the same guanine binding site architecture with the alkyl groups from O6-benzylguanine (bn6G) and O6-(p-aminomethyl)benzylguanine (ab6G) being transferred to adenine. These structural insights suggested the chemical mechanism of MTR1-catalyzed RNA methylation, which was supported by structure-guided mutations of the ribozyme core and analysis of the methylation rates. We found evidence for general acid catalysis and discovered a methylated ribozyme variant (MTR1m2) with strongly enhanced catalytic activity. Our findings uncovered a surprising similarity in the architecture of natural guanine riboswitches20 and the in vitro selected ribozyme, and provide experimental support for the hypothesis that the presence of modified nucleotides may have provided a catalytic advantage for early RNA catalysis.22 Moreover, these results nourish the thoughts that more diverse ribozymes may have existed in an RNA world.
Results
Design and activity of the crystallization construct
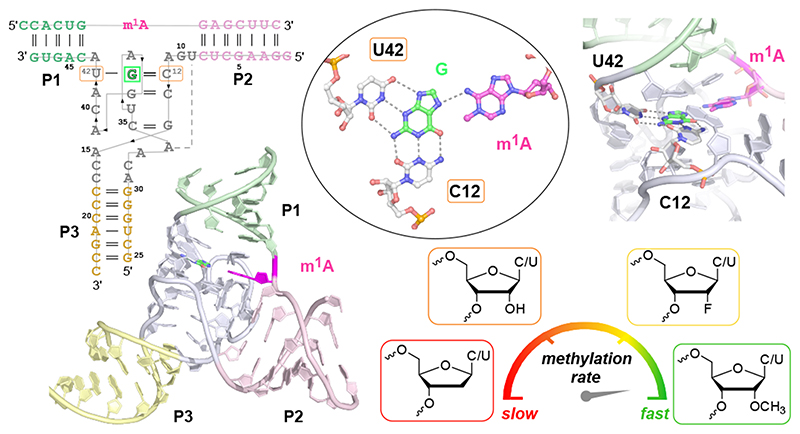
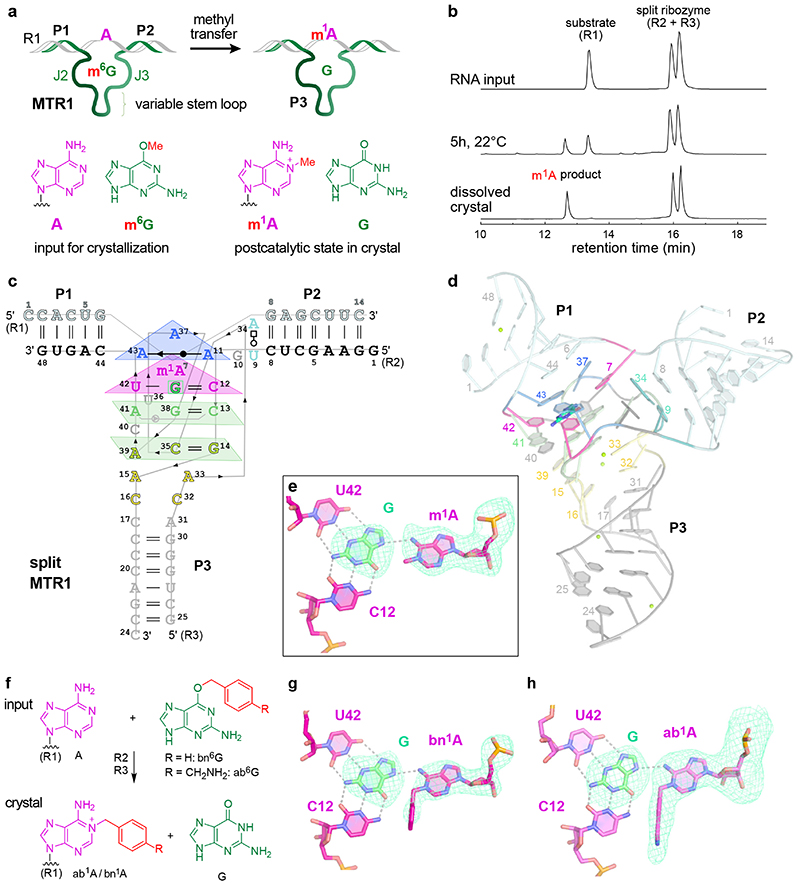
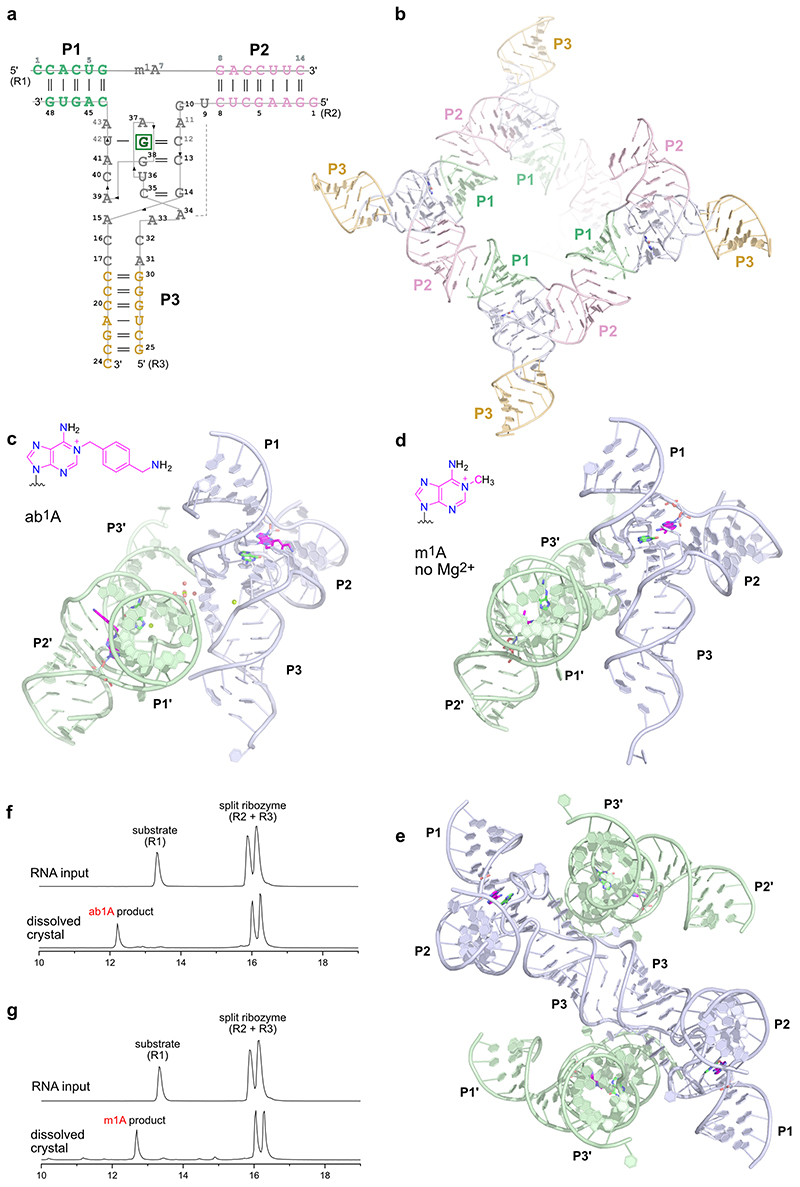
The MTR1 ribozyme contains two binding arms (P1 and P2) that hybridize to the RNA substrate (R1) with the methylation site being located at the single adenosine in the junction between P1 and P2 (Fig 1a). An internal stem-loop in MTR1, earlier confirmed by structure probing and covariation,18 is connected to the binding arms via the 9 and 13 nt long junctions J2 and J3 that form the catalytic core. The stem P3 is variable and the loop can be removed, resulting in a split ribozyme that assembles into an active ribozyme from two 24-nt long RNAs (R2 and R3).
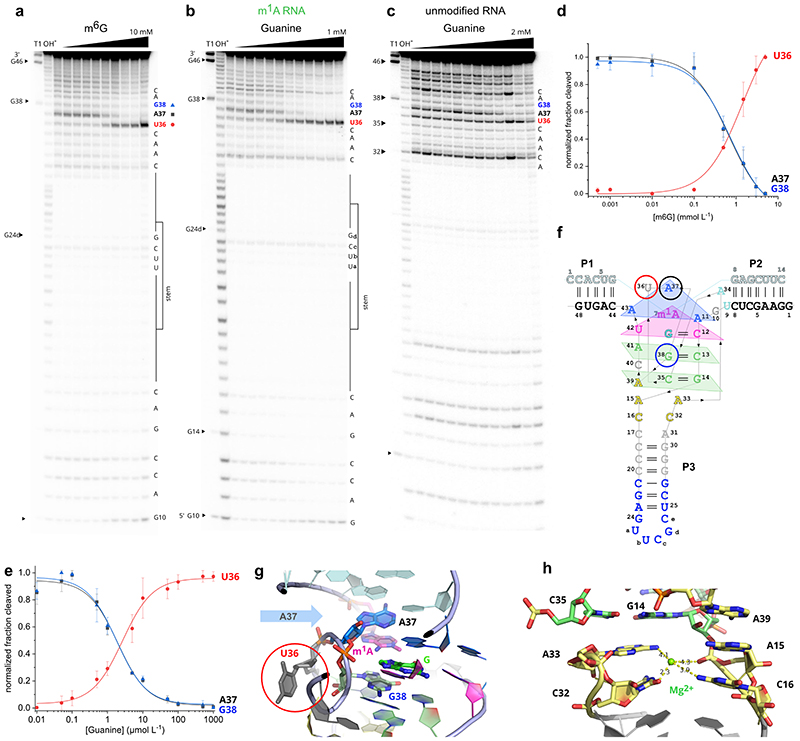
Fig.1. MTR1 ribozyme-catalysed methyl transfer reaction and overall structure.
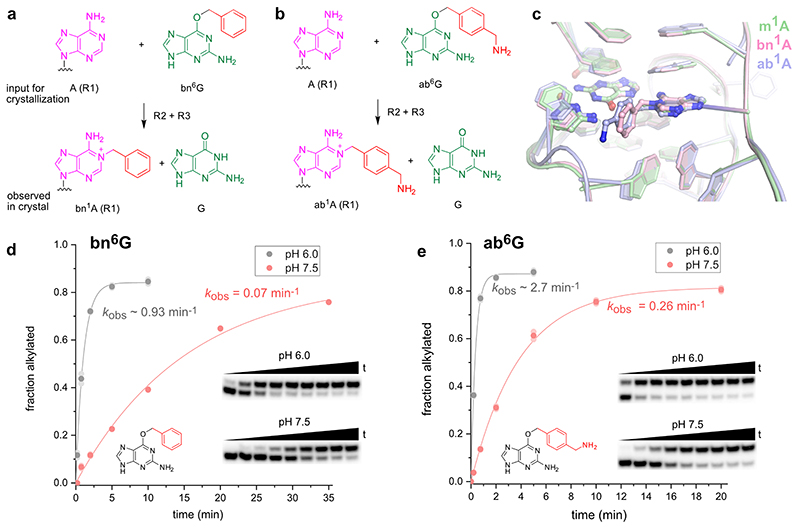
Schematic depiction of unmodified RNA (R1) hybridized to MTR1, which binds m6G and catalyses methyl transfer to N1 of adenosine. Guanine and m1A are present in the post-catalytic state captured in the crystal. (b) Anion-exchange HPLC analysis of a dissolved crystal confirms methyl transfer and shows the presence of three RNA strands. (c) Sequence and scheme of split MTR1 bound to m1A-RNA and G (boxed). (d) Ribbon diagram of the 3D structure, color-coded and numbered as in c). (e) Zoom into the Guanine binding site. (f) MTR1 also acts as alkyltransferase ribozyme with O6-benzylguanine cofactors bn6G and ab6G. (g-h) MTR1 active sites show guanine and alkylated adenosines. The green mesh represents Fo-Fc omit maps for guanine and m1A (e), bn1A (g), and ab1A (h) contoured at 3σ.
We screened crystallization conditions for several bi- and trimolecular constructs with variation in binding arm length, composition and overhangs, and found highly reproducible crystallization for the split MTR1 ribozyme complex (R2 & R3) bound to a 14-nt long substrate RNA R1. Diffraction quality crystals were formed when the complex was supplemented with O6-methylguanine (m6G), O6-benyzlguanine (bn6G) or O6-(p-aminomethyl)benzylguanine (ab6G), but not with guanine (G). Anion exchange HPLC analysis of a dissolved crystal grown with m6G confirmed the presence of all three RNA strands and revealed that product formation of m1A RNA had occurred (Fig 1b). Initial phases were obtained by iterative molecular replacement with A-form double helix fragments,23 followed by model building and refinement. Additional crystals were grown with heavy atom derivatives, including a 2'-selenomethyluridine modified RNA substrate,24 and co-crystallization with thallium acetate. Anomalous diffraction data were used for structure solution by MR-SAD and for checking the final structures for consistency (Extended Data Fig. 1, Supplementary Table 1). The structure of the m1A containing MTR1 complex was refined at a resolution of 2.8 Å and revealed the bound guanine in the active site (Fig. 1d). Likewise, the structures of MTR1 with bound guanine and the RNA products containing bn1A and ab1A were refined at resolutions of 2.9 and 3.3 Å, respectively (Supplementary Table 1). The electron density maps clearly indicated the presence of guanine and the alkyl groups always attached to N1 of adenine (Fig. 1e, g, h). The structures with m1A and bn1A contained one copy in the asymmetric unit (ASU), while the ab1A-modified complex had two copies in the ASU, and their arrangement was found to be similar to the m1A containing structure of a crystal grown in the absence of Mg2+ (Extended Data Fig. 2).
Overall structure of the MTR1 ribozyme
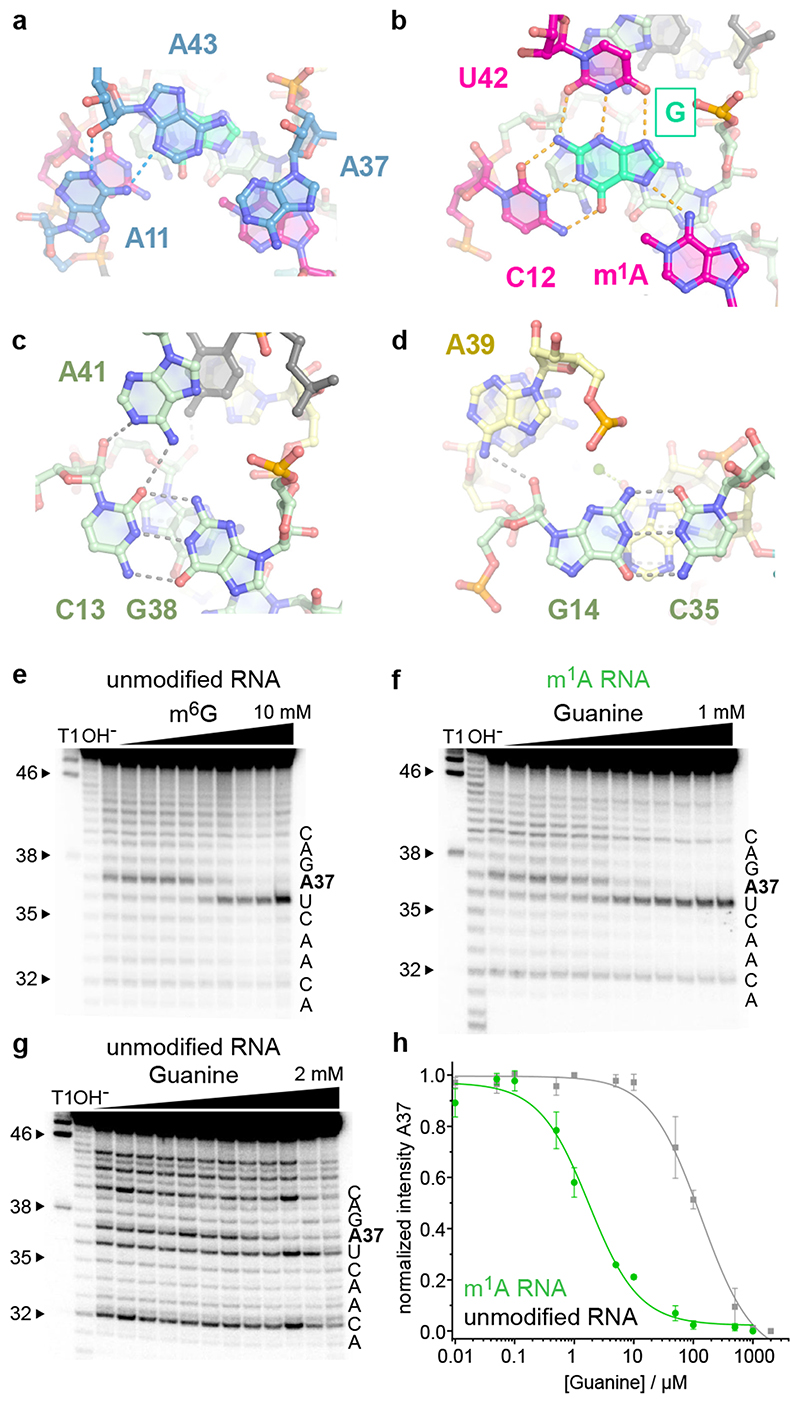
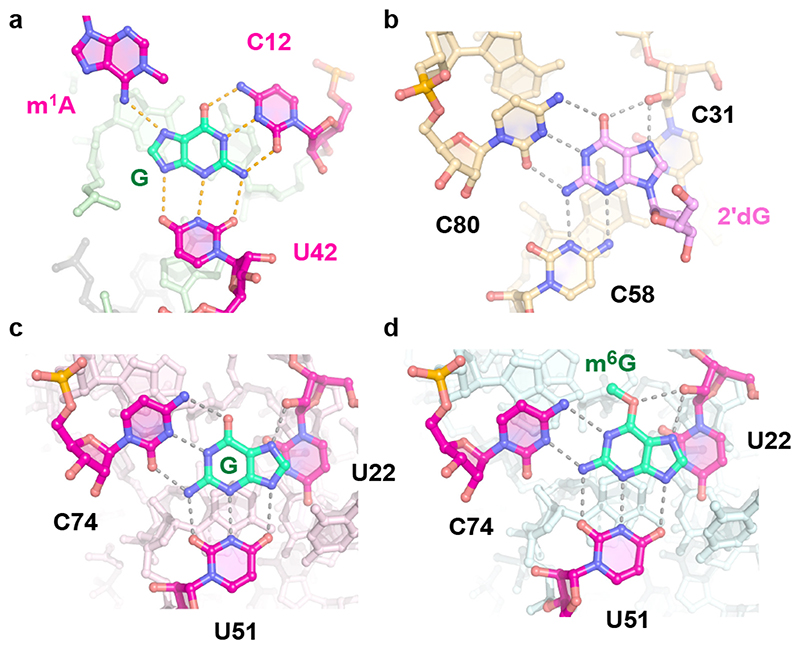
The overall structures of the MTR1 ribozyme reflects a 3-helix junction with the three A-form helices P1, P2 and P3 radiating from the central catalytic core, which positions the adenosine substrate (A7 of R1) and the m6G cofactor. In the crystal structure, the reaction products m1A and guanine are held in close proximity and form extensive tertiary contacts with the ribozyme. A schematic of the tertiary fold of MTR1 is depicted in Fig. 1c, and its 3D structure is shown in a ribbon representation in Fig. 1d. The complementary 5’ overhanging nucleotides in P1 and P2 form a semi-continuous duplex in the crystal lattice (Extended Data Fig. 2). The catalytic domain contains four layers of stacked base pairs and base triples that are connected to P1 and P3 via continuous π stacking interactions, while P2 is oriented almost perpendicularly to the core domain, and is connected via stacking with the reverse Hoogsteen U9:A34 base pair, followed by G10. The top layer of the core domain is formed by a cis Watson-Crick sugar edge base-pair A11:A43 and A37 (Fig. 2a). The second layer contains the products of the RNA catalysed reaction, i.e. m1A7 of the 14-mer RNA strand R1 and the demethylated guanine ligand, which is immobilized by extensive H-bonding with C12 and U42 (Fig. 2b). In the third layer, A41 forms an A-minor interaction with the Watson-Crick base pair C13:G38 (Fig. 2c), and the fourth layer contains the Watson-Crick base pair G14:C35, which is contacted in the minor groove by the Hoogsteen edge and phosphate of A39 (Fig. 2d). In this way, A39 mediates a continuous staple of C42, A41, C40, to A15 and C16, which transitions into P3 via the C17:A31 base interaction (Fig. 1d). A parallel staple is formed on top of A31, involving C32 and A33. In this arrangement, A33 stacks with the G14:C35 base pair and is placed in the same layer as A15, with their Hoogsteen edges oriented toward each other, but too distant for H-bonding. Similarly, C16 and C32 are oriented toward each other, but are too distant for direct interactions. Instead, this loose junction between the core domain and P3 is stabilized by coordinating metal ions. The structures suggest a magnesium ion interacting with C32 and A15, and additional metal ion binding sites were confirmed by anomalous signals from bound Tl+ ions (Extended Data Fig. 1). However, magnesium is not essential for structure formation and activity of the ribozyme, since the reaction products m1A and guanine were also found in the crystal grown in a buffer that contained only monovalent ions Na+, K+ and Li+ (Extended Data Fig. 2).
Fig.2. The catalytic core domain of MTR1.
(a) – (d) Stick representations of the four layers of the core domain from top to bottom: (a) adenosine triple A11, A37, A43. (b) active site with m1A and G interacting by base pairing with C12 and U42. (c) C13:G38 base pair with A-minor interaction of A41. (d) G14:C35 base pair interacting with A39. (e-g) Representative excerpts of in-line probing gels of MTR1 (pH 8.0, 20 °C, 36 h): (e) hybridized to unmodified R1 in the absence and presence of m6G (0.5 μM–10 mM). (f) hybridized to methylated m1A-RNA, or (g) unmodified R1 in the absence and presence of guanine (0.01–1000 μM in (f), 2000 μM in (g). (h) Normalized in-line probing band intensities for A37 in the m1A-containing RNA complex (f, green) compared to unmodified RNA (g, grey). The lines represent a fit to a one-site binding model. Error bars denote ± s.d. of the mean for n = 3 (grey) or 4 (green) independent replicates. See Extended Data Fig 3 for full gel images and additional analyses for U36 and G38.
The overall fold seen in the crystal structure is consistent with results from in-line probing experiments in solution, which monitor changes in backbone flexibility upon RNA folding and ligand binding events.25 When 5'-32P-labeled MTR1 was hybridized to R1 and incubated with varying concentrations of m6G at pH 8.0, increasing backbone cleavage was observed at U36, which is not involved in any tertiary interactions in the MTR1 structure (Fig. 2e). On the other hand, the cleavage bands at A37 and G38 significantly decreased, consistent with their stacked positions in the catalytic core. Comparable cleavage patterns were obtained with MTR1 hybridized to m1A-RNA and incubation with increasing concentrations of guanine, resulting in an apparent Kd of 2 μM (Fig. 2f, Extended Data Fig. 3). Surprisingly, in the absence of m1A (i.e. when MTR1 was hybridized to unmodified RNA substrate), the affinity for guanine was weaker by almost two orders of magnitude (Fig 2g,h), as observed by the inline probing pattern of A37 which stacks directly on m1A in the crystal structure (Fig. 2a).
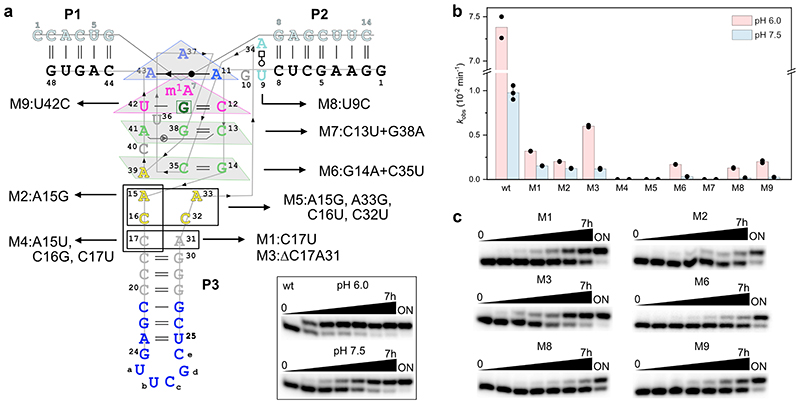
The functional significance of individual base pairs observed in the crystal structure was supported by activity analyses of several ribozyme mutants (Extended Data Fig. 4). Individual base changes in the bubble connecting the core domain and P3 (such as C17U (M1) or A15G (M2)) as well as deletion of the loose C17:A31 base pair (M3) were tolerated, but resulted in 5-10-fold reduced methyl transfer rates at pH 7.5. Attempts to close the stem by base pairing (M4), as well as transition mutations of the AC stacks (M5) impaired the ribozyme activity. The compensatory mutation of the base pair G14:C35 (M6) was tolerated, while changing the base pair C13:G38 (M7) was detrimental.
Mechanism of RNA-catalysed RNA methylation
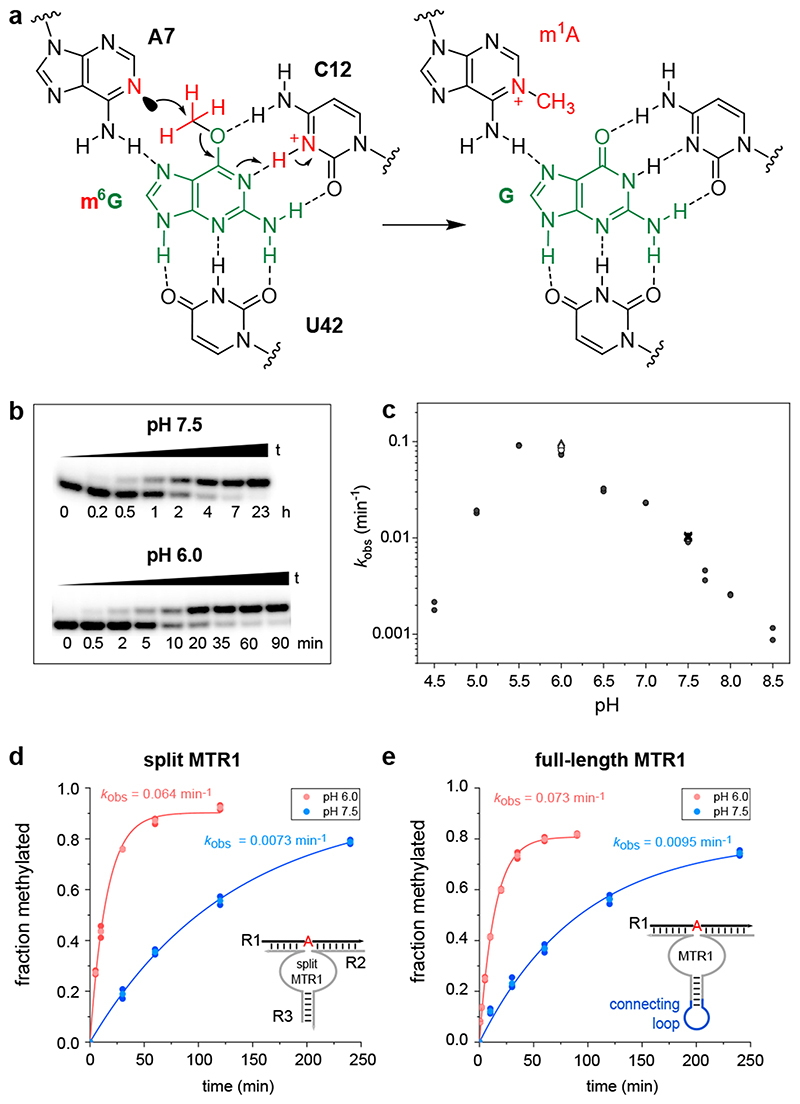
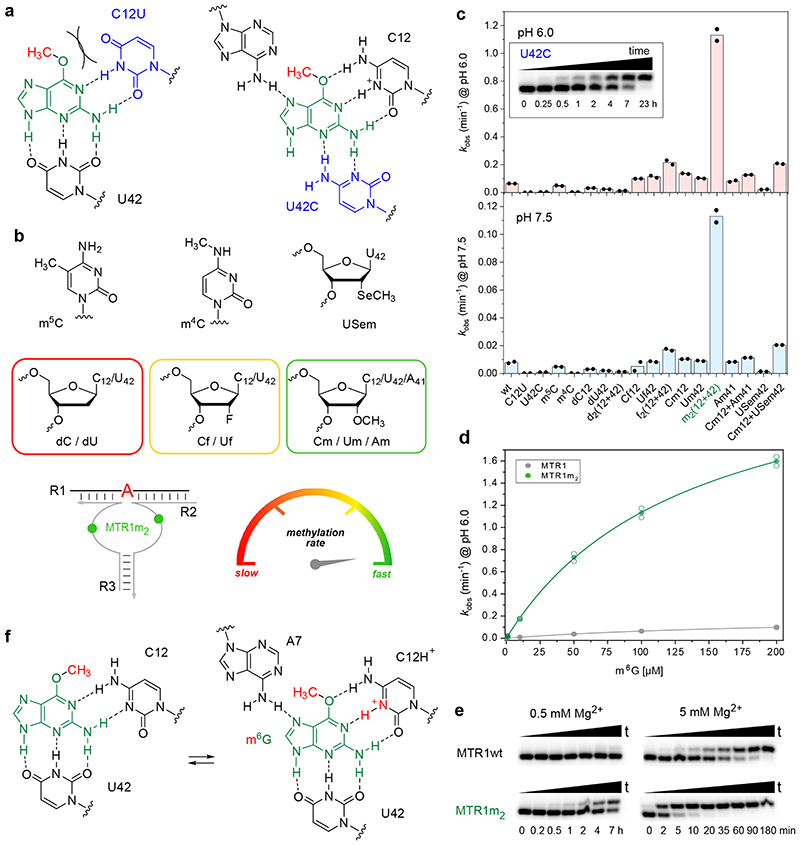
The structure represents the post-catalytic state with the methyl group transferred to N1 of A7, which is sandwiched between A37 and G38 of MTR1. The key observation that the bound guanine in the centre forms extensive hydrogen bonds with C12 and U42 of MTR1 and m1A7 of the methylated RNA product suggests a plausible mechanism for the methyl transfer reaction. The Watson-Crick interaction of guanine with C12 in the product state provides a favourable arrangement of the reactive groups, with the nucleophilic N1 of A7 in line with the electrophilic CH3 of m6G in the pre-catalytic state. This scenario likely requires a protonated C12 as H-bonding partner for m6G to maintain a Watson-Crick like orientation, thus suggesting a protonated cytidine as general acid catalyst in the methyl transfer reaction (Fig. 3a). Consistent with this hypothesis, the MTR1-catalysed methyl transfer was accelerated by lowering the pH of the reaction medium (Fig. 3b). The pH dependence of the reaction was analysed with the full-length MTR1 as well as with the split ribozyme construct used for crystallization, and in both cases, the reaction rate was ca 8-fold enhanced at pH 6.0 compared to pH 7.5 (Fig. 3d,e). The pH rate profile showed an optimal pH range near pH 5.5 (Fig. 3c). The analogous alkyl transfer reactions with bn6G and ab6G were also accelerated at pH 6.0 (Extended Data Fig. 5). These results likely reflect a modest increase in pKa for C12 in MTR1 compared to free cytidine, similar to the apparent pKa for protonation of a m6G:C base pair in a DNA duplex.26
Fig. 3. The methyltransferase ribozyme employs general acid catalysis.
(a) A protonated cytidine is suggested in the active site for RNA-catalysed methyl group transfer. The mechanism involves the nucleophilic attack of the nitrogen lone pair (N1 of A7) on the methyl carbon (at O6 of m6G). The guanine leaving group accepts a proton from cytidine C12+, resulting in a positive charge on m1A and a standard G:C hydrogen bonding pattern in the product state. (b) Representative PAGE images of the kinetic assays from 3 independent experiments with split MTR1 at pH 6.0 and pH 7.5 under single-turnover conditions. (c) pH rate profile for split MTR1 (n = 2 or 3, individual data points for each buffer (different symbols for same pH indicate different buffers used; for details see methods section and source data). (d, e) Comparison of methyl transfer rates for split (d) and full-length (e) MTR1, reaction with m6G at pH 6.0 (red) and pH 7.5 (blue). Conditions in b-e) 5'-32P-labeled R1, 10 μM MTR1, 100 μM m6G, 40 mM MgCl2, 25°C.
Rate acceleration by ribose methylation
Consistent with the functional importance of the base triple, both the C12U mutant and the U42C mutant showed strongly reduced reaction rates (Fig. 4a). At pH 6.0, the U42C mutant still generated 65% methylated product after 7 h, but less than 15% product was formed by the C12U mutant after 24 h. Replacing C12 by m5C lowered the rate by less than a factor of two, while a variant containing m4C was also significantly impaired (30-fold slower rate than wt), likely due to steric crowding in the active site (Fig 4b,c). Replacing C12 and U42 individually or simultaneously with the corresponding 2'-deoxynucleotides was tolerated, although the reaction rates were slightly reduced, suggesting that nucleotides with a 3’-endo conformation are preferred at positions 12 and 42. This was further confirmed by installation of the analogous 2'-fluoro-2'-deoxy nucleotides, which restored the activity, when inserted individually, but enabled 2-3-fold faster methyl transfer when incorporated simultaneously. An even more surprising rate acceleration was observed with 2'-OMe nucleotides at C12 and U42. Individual substitutions were slightly favourable (less than 2-fold higher kobs), but replacing both nucleotides at the same time and allowing them to act in concert enhanced the kobs by a factor of 15. In other words, the Cm12Um42-MTR1 variant (from here on called MTR1m2, Fig. 4b) produced 90% m1A RNA within 5 min under single turnover conditions at pH 6.0. The K1/2 (apparent Km) for m6G is on the order of 130 μM (obtained from kobs at variable m6G concentrations, Fig. 4d). Importantly, the synergistic effect of the two ribose methylations was specific for C12 and U42, both of which are directly involved in ligand/cofactor binding and activation. The 2'-OMe substitution was also tolerated at A41, but its combination with C12 was only additive at best. Moreover, we found that MTR1m2 retained significant activity at a strongly reduced Mg2+ concentration down to 0.5 mM Mg2+, where MTR1wt barely produced any methylated product (Fig. 4e). These observations strongly confirm the specific roles of C12 and U42 for the activity of the MTR1 ribozyme in recruiting, aligning and activating the cofactor m6G (Fig. 4f).
Fig. 4. Acceleration of methyl transfer by synergistic effects of Cm12 and Um42.
(a) Schematic of C12U and U42C active site mutants with strongly reduced activity. (b) Structures of modifications used at positions 12 and 42, including schematic of simultaneous 2'-O-methylation of C12 and U42 (green symbols in MTR1m2) and color code (red to green) for rate accelerations. (c) Comparison of kobs values for modified split MTR1 ribozymes, at pH 6.0 (top) and at pH 7.5 (bottom); individual data points shown for two independent replicates. Inset shows representative gel image (of two replicates) for slow but complete reaction of U42C mutant. (d) MTR1m2 kinetics (green) at various m6G concentrations at pH 6.0 in comparison to unmodified MTR1 (grey). The lines represent the curve fits kobs = kmax[m6G]/(K1/2+[m6G]) to the mean (filled symbol) of two individual replicates shown as empty data points (see source data). (e) Representative PAGE images (from two independent experiments) of kinetic assays with MTR1 wt and MTR1m2 at 0.5 mM Mg2+ (left) and 5 mM Mg2+ (right) with 5'-32P-labeled R1, 10 μM split MTR1 wt or split MTR1m2, 100 μM m6G, pH 6.0, 25°C. (f) Possible conformational equilibrium in the active site involving syn/anti orientation of m6G, and C12 in unprotonated or protonated state.
Comparison of MTR1 guanine binding site with purine riboswitches
The bound guanine cofactor is located in the same position in all three structures, immobilized by H-bonds to the Watson-Crick edges of C12 and U42 and the Hoogsteen side of the N1-alkylated adenine. This active site architecture has striking similarities to guanine binding sites found in natural riboswitches (Fig. 5). In the purine riboswitch family, guanine recognition is mediated by two universally conserved pyrimidine residues,20,27 such as U51 and C74 in the B. subtilis xpt-pbuX riboswitch (pdb 1Y27). Mutation of these two residues changed the specificity of the guanine riboswitch. While C74U allowed binding of adenine instead of guanine, changing U51 to cytidine shifted the base pairing pattern and created room for binding of N9-substituted guanine, similar to 2'-deoxyguanosine (2’dG) riboswitches.28 In the 2’dG riboswitch from M. florum, the ligand forms a Watson-Crick base pair with C80, and C58 contacts N3 and N2 at the sugar edge of the purine nucleoside (pdb 3SKI, Fig. 5b). In MTR1, the C12U mutant strongly reduced the methyl transferase activity, while the U42C mutation was better tolerated, suggesting a similarly shifted H-bonding arrangement as seen in the 2’dG riboswitch.
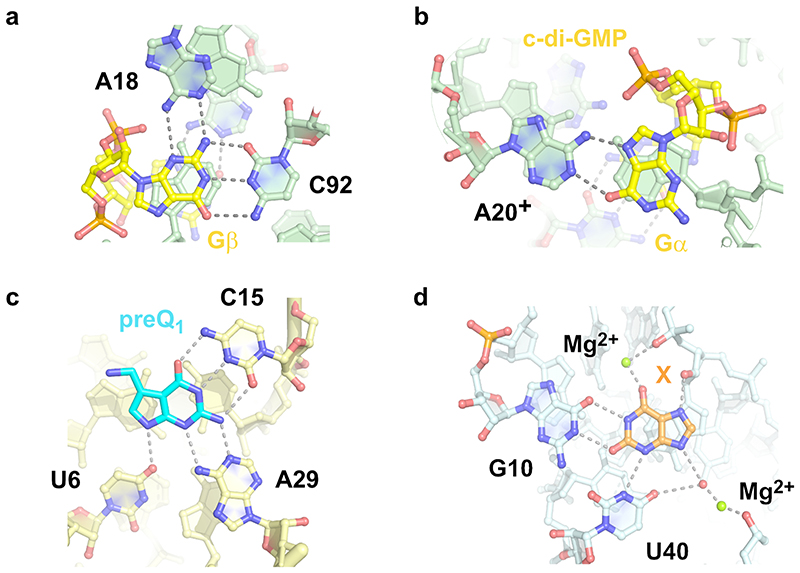
Fig. 5. Comaprison of MTR1 guanine binding site to purine riboswitches.
(a) MTR1 active site with m1A and guanine (this work, pdb 7q7x). (b) M. florum riboswitch with 2'-deoxyguanosine (pdb 3ski). (c) B. subtilis xpt-pbuX riboswitch with guanine (pdb 1y27). (d) B. subtilis xpt-pbuX with m6G (pdb 3fo6).
Interestingly, the guanine riboswitch was earlier shown to bind m6G, however, with significantly decreased binding affinity relative to guanine21, while bn6G was not bound, and no alkyl transfer was reported from any of the guanine derivatives. The crystal structure with m6G revealed that C74 shifted downwards to form new H-bonds with N1 and N2 of m6G and make room for the methyl group, in the otherwise tight binding pocket, in which the Hoogsteen edge of the ligand is contacted by the 2'-OH of U22 (Fig. 5d). In principle, a similarly shifted H-bonding pattern for C12 with m6G is conceivable in the MTR1 pre- catalytic state (Fig. 4f). However, the relative arrangement of m1A and G in the post-catalytic state suggest that the nucleophilic attack is likely to happen to the methyl group in the anti orientation (Fig. 4f, right). Moreover, the protonated C12+:m6G base pair is more consistent with the observed pH activity profile.
Interestingly, also riboswitches make use of protonated nucleobases, e.g. for binding of cyclic-diGMP, which involves a stably protonated adenine (Extended Data Fig. 6a,b).29 Protonated nucleobases with highly shifted pKa values were also found as key structural elements in synthetic RNA aptamers, including the GTP-binding RNA aptamer.30 Alternatively, the pKa of the functional groups in the ligand can be changed upon binding to RNA, as for example described for aminoglycosides, including neomycin-B.31
A conceptually similar scenario is likely operating in the preQ1 riboswitch, which was recently shown to mediate a methyl group transfer from an O6-methylated cofactor analog to N3 of C15, which is involved in a Watson-Crick base pair with the non-methylated cofactor preQ1 (Extended Data Fig. 6c). With m6preQ1, the methyl transfer reaction was accelerated by shifting the pH of the reaction medium from pH 7.5 to pH 6.0, consistent with protonation of the ligand and stabilization of the leaving group. The methyl transfer rate of the preQ1 riboswitch is, however, significantly slower than observed for MTR1. PreQ1 yields ca 50% methylated RNA after 48 h (at pH 6.0 with 100 μM m6preQ1). For comparison, MTR1 provides 90% methylated RNA in 50 min under comparable conditions (pH 6.0, 100 μM m6G). This difference is not too surprising, given that the preQ1 riboswitch evolved in Nature for tight binding to the un-methylated ligand preQ1, which has also been shown to be a strong competitive inhibitor for the preQ1-mediated RNA methylation.19 In contrast, MTR1 was evolved in vitro by enrichment based on catalytic activity, and not based on tight binding or high affinity. This is also reflected in the Kd for guanine, which was found by in-line probing to be in the range of 2 μM for the methylated ribozyme-product complex and >100 μM for the unmethylated ribozyme-substrate complex, in contrast to a Kd of 5 nM for guanine binding to the natural riboswitch.32,33 The weak affinity for guanine also supports the potential of MTR1 to perform multiple turnover catalysis of RNA methylation, especially in the tri-molecular format, with the RNA substrate transiently hybridized to the split ribozyme.
Discussion
The crystal structure of the MTR1 ribozyme indicates that the RNA forms an intricate cofactor binding site that enabled highly efficient methyl transfer from m6G to the RNA (before or) during crystallization. The structure revealed the post-catalytic state of the RNA catalyst, with the m1A-containing methylated RNA product and unmodified guanine bound in the catalytic core. Analogous results were obtained with two additional O6-alkylated guanine co-substrates that transferred a benzyl group to N1 of A7 in R1. The benzyl/(p-aminomethyl)benzyl groups appeared in a perpendicular orientation to the adenine nucleobase and pointed outward from the core domain, which is consistent with their likely orientation during in vitro selection, when biotin was conjugated to the p-aminomethyl group.
We found that the methyl group transfer is significantly accelerated at pH 6.0 compared to pH 7.5, which hints at the involvement of a protonated nucleobase in the catalytic mechanism. Indeed, the organization of the active site is highly consistent with a protonated cytidine in the pre-catalytic state that facilitates binding through H-bonding of m6G and at the same time servers as donor for protonation of the leaving group, resulting directly in the stable G:C base pair found in the product state. The pH rate profile suggests that the pKa of C12 is moderately enhanced compared to the pKa of cytidine in single-stranded RNA. Shifted pKa values toward neutrality are commonly observed in ribozyme catalysis of RNA cleavage,34 and the pKa of nucleobases are considered key determinants of chemical evolution.35
Moreover, the methyl transfer rate of MTR1 was strongly enhanced upon simultaneous presence of two 2'-O-methylated nucleosides C12 and U42 in the catalytic core. In combination with the lower pH, the methylated ribozyme achieved at least a 120-fold faster methylation compared to the rates observed under in vitro selection conditions. Analogous replacement with 2'-fluoro-2'-deoxynucleosides was also beneficial, but the accelerating effect was much less pronounced. Ribose methylation is known to affect the preferred ribose pucker and the flexibility and dynamics of conformational states,36 and in addition, 2'-ribose modifications have a small effect on the pKa of the nucleobase.37 A combination of these forces likely causes the unexpected synergistic effect of Cm12 and Um42. To our knowledge, a comparable accelerating effect by two natural methylated nucleotides acting in concert to enhance cofactor binding and catalytic activity has not been previously observed for any other ribozyme. In previous work, 2'-modified nucleotides were used to explore the importance and the involvement of the 2'-OH groups in the catalytic mechanisms of ribozymes. 38,39 While placement of 2'-OMe or LNA modifications in the ribozymes binding arms usually increased activity of RNA-cleaving ribozymes and deoxyribozymes by enhancing the hybridization strengths, introduction into the catalytic core was often detrimental, or in the best case tolerated. Only recently a synergistic effect of two 2'-fluoroarabino nucleosides introduced into an engineered version of the RNA-cleaving 10-23 deoxyriboyzme was shown to be beneficial for multiple turnover under cellular conditions.40
Our data indicate that MTR1 does not require divalent metal ions for catalysis. The post- catalytic state of MTR1 also occurred in a crystal that was grown in a buffer without added magnesium ions. On first glance, this was surprising, since previous kinetic experiments revealed a significant drop in the methyl transfer rate below 5 mM Mg2+.18 Likely, the higher concentrations and the longer reaction time during crystallization rescued the slower rate in the absence of Mg2+. In principle, Mg2+ ions could have assisted in cofactor binding, as recently shown in the structure of a xanthine riboswitch.41 However, none of the six structures analyzed in this work revealed any Mg2+ ions in contact with the purine cofactor. Our data rather support the view that Mg2+ assists folding and/or affects RNA dynamics in the ribozyme core. The observation that the methylated ribozyme MTR1m2 retained high activity with less than 1 mM Mg2+ is consistent with the interpretation that the 2'-O-methyl nucleotides impart some degree of preorganization to the catalytic core.
In summary, we reported the structure and the chemical mechanism of the methyltransferase ribozyme MTR1. The crystal structure captured the post-catalytic state with the methylated RNA product and the released guanine bound in close proximity. We found that the guanine binding site shares common structural features with natural purine riboswitches. The ligand RNA interactions were confirmed by mutagenesis. The synergistic effect of the two methylated pyrimidine nucleosides directly in contact with the cofactor suggests that small changes of a few key nucleotides can have profound beneficial effects for catalysis. Based on these results, we anticipate that a wide range of activities for cofactor-utilizing ribozymes can be identified in the laboratory. Our results also support the hypothesis that methylated nucleotides in RNA may be evolutionary leftovers from a time when RNA catalysts were engaged in a wider scope of chemical reactivity.
Materials and Methods
RNA synthesis
The MTR1 ribozyme and point mutants were prepared by in vitro transcription with T7 RNA polymerase from synthetic DNA templates (purchased from Microsynth), as described previously.42 Typical reaction conditions included 1 μM DNA template, 4 mM NTPs, 30 mM MgCl2, 2 mM spermidine and 10 mM DTT. RNA oligonucleotides for the split MTR1 complexes were prepared by solid-phase synthesis using 2'-O-TOM-protected ribonucleotide phosphoramidites. Modified phosphoramidites were purchased (2'-OMe-U, 2'-OMe-C, 2'-F-U, 2'-F-C, m5C) or prepared in-house following known procedures (2'-SeMe-U,24 m4C43). Deprotection was performed with a 1:1 (v/v) mixture of 28% aqueous ammonia and 40% aqueous methylamine (known as AMA) at 37°C for 4-6 h, followed by 1 M tetrabutyl-ammonium fluoride in THF at 25°C for 12-16 h. 2'-SeMe-U containing RNA was treated with 2 mM DTT in water for 5 min at 37°C to reduce partially oxidized 2'-methylselenoxide groups back to the desired 2'-selenomethyl moiety.44 RNAs were purified by denaturing PAGE, followed by extraction and ethanol precipitation. Purity was confirmed by anion-exchange HPLC (Dionex DNAPac PA200, 2 × 250 mm, at 60 °C; solvent A: 25 mM Tris-HCl (pH 8.0) and 6 M urea; solvent B: 25 mM Tris-HCl (pH 8.0), 6 M urea and 0.5 M NaClO4; linear gradient, 0-40% solvent B, with a slope of 4% solvent B per column volume), and the identity of the RNAs was confirmed by HR-ESI-MS (negative ion mode).
Complex formation and crystallization
Crystallization complexes were formed by mixing RNA strands in a 1:1:1 molar ratio in 50 mM KCl and 10 mM HEPES (pH7.5). The sample was heated to 95 °C for 3 min and cooled at 20 °C for 15 min. 5 mM MgCl2 and 240 μM substrate (m6G, bn6G or ab6G (also known as BG-NH2)) were added to a final complex concentration of 200 μM. MgCl2 was omitted for crystallization in the absence of magnesium ions.
Crystals were grown at 20 °C using the hanging drop vapor-diffusion method by mixing the RNA complex in a 1:1 ratio with solutions containing 100 mM NaCl, 100 mM LiCl, 10 mM MgCl2, 50 mM MES (pH 6.4-6.7) and 36-42% MPD.
Drops with volumes of 1 μL produced crystals after 3-4 days (5-6 days when no MgCl2 was present) and were harvested after 14 days. Crystals were flash frozen in liquid nitrogen without any additional cryoprotection.
Incorporation of thallium ions was achieved by co-crystallization. The crystallization complex was prepared as described before, but with 50 mM KOAc instead of KCl to prevent precipitation of TlCl. The reservoir buffer contained 100 mM NaOAc, 100 mM LiOAc, 50 mM MES (pH 6.4-6.7) and 36-42% MPD. Crystals appeared after 5-6 days and were harvested after 14 days, and flash frozen in liquid nitrogen without any additional cryoprotection.
Data collection and structure determination
X-ray diffraction data were collected at 100K on PILATUS 6M and EIGER 2X 16 M detectors at the ID23-01 (European Synchrotron Radiation Facility) or P11 (DESY) beamlines. Diffraction data were indexed, integrated, scaled with XDS,45 and reduced with POINTLESS, AIMLESS, and CTRUNCATE within the CCP4 package.46 Initial phases for the Crystal I data (wavelength 0.9198 Å) were determined by molecular replacement (MR) using Phaser in Phenix.47 Search models were double-stranded RNA A-form helix fragments as described previously.23 Here we used the 7-bp bottom stem from the Chili RNA aptamer48 (PDB: 7OAX, chain A nt 2-8/45-51), searched for three copies in the ensemble and obtained a MR solution with a Z-score (TFZ) of 14.4 (TFZ >8 indicates a definite solution). The initial electron density map obtained from Phaser was used for automated model building with the AutoBuild module in Phenix.49 The model from AutoBuild was completed by iterative rebuilding in COOT50 and refinement with Phenix.refine.51 The final structures of Crystal II (bn6A), Crystal III (ab6A), and Crystal IV (no Mg) were solved by MR using the model built with the data of Crystal I (m1A). The N1-modified adenosine residue restraints were prepared using JLigand in the CCP4 suite52 or eLBOW in Phenix.53
The final structures of Crystal V (with SeMeU) and Crystal VI (with thallium) were determined by MR-single anomalous dispersion (SAD) using the anomalous data collected at the Selenium K-edge (0.9786 Å) and Thallium L-III edge (0.9751 Å). Data collection and refinement statistics are summarized in ED Table 1. Figures displaying molecular models were prepared using PyMol (Schrödinger).
HPLC analysis of the RNA methylation reaction and the crystal content
For analysis of the methylation status and the composition of the crystals, a crystal was harvested from the drop, briefly washed in buffer, then dissolved in 15 μL H2O, and analyzed by anion-exchange HPLC on Dionex DNAPac PA200 column, 2 × 250 mm at 60 °C with UV detection at 260 nm. Solvent A was 25 mM Tris-HCl (pH 8.0), 6 M urea and solvent B was 25 mM Tris-HCl (pH 8.0), 6 M urea and 0.5 M NaClO4 with a linear gradient 0–48% solvent B in 12 column volumes.
Kinetic assays of RNA-catalyzed methyltransfer reaction
Single-turnover kinetic assays were carried out as described previously.18 In a standard reaction, 100 pmol ribozyme were mixed with 10 pmol of trace-labeled 5'-32P-RNA R1 in 10 μl of reaction buffer (120 mM KCl, 5 mM NaCl and 50 mM Bis-Tris (pH 6.0) or HEPES (pH 7.5)) and annealed (3 min at 95 °C and 10 min at 25 °C), followed by addition of 100 μM m6G and 40 mM MgCl2. The reaction mixture was incubated at 25 °C and aliquots (1 μl) were taken at different time points, quenched immediately with loading dye (4 μl) and half of each time-point sample was resolved on 20% denaturing PAGE. Methylation yields were determined by measuring band intensities using phosphorimaging. The data was fit with the pseudo first-order kinetic equation Y = Ymax(1 – e-kobs*t) to obtain the observed reaction rate kobs and the final yield Ymax using Origin (2021). For slow ribozymes with ≤ 20% methylated RNA product after overnight reaction, kobs values were approximated linearly using the 7h methylation yield. For measuring the pH-dependence of the MTR1 catalyzed methylation reaction between pH 4.5-8.5, the following buffers were used (50 mM NaOAc pH 4.5, 5.0, 5.5; 50 mM Bis-Tris pH 6.0, 6.5; 50 mM MES pH 6.0; 50 mM sodium cacodylate pH 6.0; 50 mM HEPES pH 7.0, 7.5, 8.0; 50 mM MOPS pH 7.5, 50 mM PIPES pH 7.5; 50 mM Tris-HCl pH 7.7, 8.0; pH determined at 25°C). Additional kinetic experiments were performed at pH 6.0 (50 mM Bis-Tris) using only 0.5, 1, or 5 mM MgCl2. All kinetic assays were carried out with at least two independent replicates.
Preparative scale methylation of RNA
The methylated RNA m1A-R1 was prepared in a preparative scale ribozyme-mediated methylation reaction. 1 nmol target RNA was mixed with 1.1 nmol MTR1 in 20 μl of reaction buffer (120 mM KCl, 5 mM NaCl and 50 mM Bis-Tris, pH 6.0) including 100 μM m6G as small-molecule substrate and 40 mM MgCl2. Before addition of m6G and MgCl2, an annealing step (3 min at 95 °C and 10 min at 25 °C) was performed. After overnight incubation at 25 °C, the methylated RNA was purified by denaturing PAGE yielding ca. 75% isolated product.
In-line probing
In-line probing was performed using the 5'-32P-labeled full-length MTR1 ribozyme in complex with the substrate RNA R1 (unmodified or methylated). In general, 125 IPS ribozyme were annealed to 25 pmol R1 or m1A-R1 (3 min at 95 °C and 10 min at 25 °C). 100 mM KCl and 100 mM Tris-HCl (pH 8.0) were added along with 20 mM MgCl2 and m6G or G in various concentrations to yield a final volume of 5 μL. After incubation at 20°C for 36 h, the reactions were quenched by adding loading dye (5 μL), resolved on 20% denaturing PAGE, and visualized by autoradiography. As a reference, an RNase T1 digestion (150 IPS of 5'-32P-RNA in 10 μl 50 mM Tris (pH 7.5) digested with 1U RNase T1 at 37 °C for 45 s) and an alkaline hydrolysis lane (250 IPS of 5′-32P-RNA in 10 μl 20 mM NaOH incubated for 4 min at 95 °C) were used. Band intensities of cleaved RNA at three selected positions (U36, A37, G38) were quantified and normalized, plotted against ligand concentration and fitted with a one-site-binding model using Origin (2019 & 2021).
Extended Data
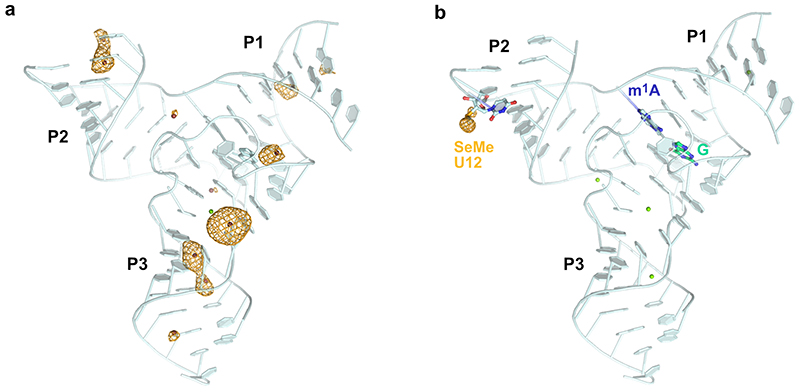
Extended Data Fig. 1. Heavy atom derivatives of MTR1 crystal structure.
(a) Overall structure of MTR1 co-crystallized with Tl+. (b) MTR1 containing 2'-Selenomethyl-uridine modified residue in the RNA substrate. Yellow mesh indicates anomalous difference Fourier map contoured at (a) 3σ and (b) 5σ. The difference maps were computed from data collected at the thallium L-III edge and selenium K-edge, respectively.
Extended Data Fig. 2. Overall structures of MTR1 and crystal contacts.
(a) Secondary structure scheme of trimolecular MTR1. (b) Packing of overhangs of P1 and P2 to form a semi-continuous double helix. Arrangement of two copies in the asymmetric unit for crystals grown with(c) ab6G (ab1A in the product) and (d) with m6G (m1A in the crystal) but no added Mg2+. (e) Crystal contact via stacking of P3 in the structure shown in (d). (f, g) Anion exchange HPLC analyses of dissolved crystals corresponding to (c) and (d).
Extended Data Fig. 3. In-line probing of MTR1.
(a-c) Full gel images for the excerpts shown in Fig 2. (a) In-line probing of MTR1 hybridized to unmethylated RNA R1, with increasing concentrations of m6G. (b) In-line probing of guanine binding to the MTR1 product complex containing m1A. (c) In-line probing of guanine binding to the MTR1 starting complex hybridized to unmethylated RNA R1. (a, b, c) Incubation at pH 8.0, 20°C, 36 h. (d) Normalized band intensities seen in (a), shown for U36 (red), A37 (black), G38 (blue). The [m6G]1/2 value is ca 800 μM, however, this value cannot be interpreted as a Kd or Km because of multiple overlapping equilibria (with m6G and G since partial/slow methylation occurred during incubation). (e) Normalized in-line probing band intensities seen in (b) for U36 (red), A37 (black), G38 (blue). Kd,app = 2.0 ± 0.3 μM. Data in d) and e) are fitted to a one-site binding model. Error bars denote ± s.d. of the mean for n = 3 (d) or 4 (e) independent replicates. (f) Secondary structure scheme of MTR1 used in in-line probing experiments (with connecting loop at P3; numbers of nucleotides correspond to the split version). (g) Excerpt of the catalytic core showing solvent exposed location of U36 and stacking of A37 on m1A. (h) Excerpt of the metal ion binding site in the transition of the catalytic core to P3.
Extended Data Fig. 4. MTR1 mutagenesis of the core nucleotides and pH dependence.
(a) Scheme of MTR1 with mutated nucleotides and base-pairs indicated. (b) Summary of rate constants determined at pH 6.0 and 7.5. Individual data points of two independent replicates are shown as black dots. Reaction of MTR1-wt at pH 7.5 war repeated three times. (c) Representative gel images of kinetic experiments of two independent replicates. Reaction conditions: 5'-32P-labeled R1, 10 μM MTR1 or mutant, 100 μM m6G, 40 mM MgCl2, 25°C. Timepoints: 0, 0.2, 0.5, 1, 2, 4, 7, 23 h (ON).
Extended Data Fig. 5. MTR1 as alkyltransferase ribozyme and pH dependence.
(a, b) Chemical structures of substrates (adenosine and benzyl guanine cofactor) before the RNA-catalyzed reaction, and after transfer to the target adenosine in R1 and release of guanine. (c) Overlay of active sites containing m1A, bn1A or ab1A in stick representation. (d) Kinetics of benzyl group transfer at pH 6.0 and pH 7.5, with exemplary gel images as inset. (e) Same as (d), but reaction with ab6G. The fastest reaction was observed with ab6G at pH 6.0, yielding ca 90% ab1A-RNA within 2 min. All reactions were performed in duplicate, individual data points and representative gel images are shown (timepoints shown in gels up to 60 min).
Extended Data Fig. 6. Comparison to binding sites of purine riboswitches also involving protonated nucleobases or metal ion binding.
(a, b) G20A mutant of Vc2 riboswitch with c-di-GMP (pdb 3mum), showing (a) Gß and (b) G(/ bound via stably protonated A20. (c) T. tengcongensis preQ1 riboswitch with bound preQ1 (pdb 3q50). (d) NMT1 riboswitch in complex with xanthine (pdb 7elr).
Supplementary Material
Acknowledgements
This work was supported by the European Research Council (ERC / No 682586), the Deutsche Forschungsgemeinschaft (DFG, HO4436/3-1) and the University of Würzburg. We thank Celine Pfeuffer and Ann-Kathrin Lenz for technical assistance, and the beamline staff at DESY (PETRA III, P11) and ESRF (ID23) for assistance with data collection.
Footnotes
Author Contributions
CPMS and CH designed the study, HS and CPMS collected diffraction data, MM solved the structure, CPMS performed biochemical experiments, CH wrote the paper with input from all authors. CPMS and MM contributed equally.
Competing Interest Statement
The authors declare no competing interest.
Data availability
Structural data obtained by X-ray crystallography were deposited in the Protein Data Bank (PDB) and are available with the following accession codes: 7Q7X, 7Q7Y, 7Q7Z, 7Q80, 7Q81, 7Q82. All relevant data are provided in the Figures, Extended Data Fig 1-6, Supplementary Tables 1-2, Supplementary Fig 1, and Source Data files. Publicly available datasets from rcsb.org used in this study: 7OAX, 3SKI, 1Y27, 3FO6, 3MUM, 3Q50, 7ELR.
References
- 1.Panchapakesan SSS, Breaker RR. The case of the missing allosteric ribozymes. Nat Chem Biol. 2021;17:375–382. doi: 10.1038/s41589-020-00713-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Micura R, Höbartner C. Fundamental studies of functional nucleic acids: aptamers, riboswitches, ribozymes and DNAzymes. Chem Soc Rev. 2020;49:7331–7353. doi: 10.1039/d0cs00617c. [DOI] [PubMed] [Google Scholar]
- 3.Kirschning A. Coenzymes and their role in the evolution of life. Angew Chem Int Ed. 2021;60:6242–6269. doi: 10.1002/anie.201914786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wilson TJ, Lilley DMJ. The potential versatility of RNA catalysis. Wiley Interdiscip Rev RNA. 2021;12:e1651. doi: 10.1002/wrna.1651. [DOI] [PubMed] [Google Scholar]
- 5.Chen X, Li N, Ellington AD. Ribozyme catalysis of metabolism in the RNA world. Chem Biodivers. 2007;4:633–655. doi: 10.1002/cbdv.200790055. [DOI] [PubMed] [Google Scholar]
- 6.Breaker RR. Imaginary Ribozymes. ACS Chem Biol. 2020;15:2020–2030. doi: 10.1021/acschembio.0c00214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jadhav VR, Yarus M. Coenzymes as coribozymes. Biochimie. 2002;84:877–888. doi: 10.1016/s0300-9084(02)01404-9. [DOI] [PubMed] [Google Scholar]
- 8.Winkler WC, Nahvi A, Roth A, Collins JA, Breaker RR. Control of gene expression by a natural metabolite-responsive ribozyme. Nature. 2004;428:281–286. doi: 10.1038/nature02362. [DOI] [PubMed] [Google Scholar]
- 9.Silverman SK. Catalytic DNA: Scope, Applications, and Biochemistry of Deoxyribozymes. Trends Biochem Sci. 2016;41:595–609. doi: 10.1016/j.tibs.2016.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cernak P, Sen D. A thiamin-utilizing ribozyme decarboxylates a pyruvate-like substrate. Nat Chem. 2013;5:971–977. doi: 10.1038/nchem.1777. [DOI] [PubMed] [Google Scholar]
- 11.Tsukiji S, Pattnaik SB, Suga H. An alcohol dehydrogenase ribozyme. Nat Struct Mol Biol. 2003;10:713–717. doi: 10.1038/nsb964. [DOI] [PubMed] [Google Scholar]
- 12.Ishida S, Terasaka N, Katoh T, Suga H. An aminoacylation ribozyme evolved from a natural tRNA-sensing T-box riboswitch. Nat Chem Biol. 2020;16:702–709. doi: 10.1038/s41589-020-0500-6. [DOI] [PubMed] [Google Scholar]
- 13.Jadhav VR, Yarus M. Acyl-CoAs from coenzyme ribozymes. Biochemistry. 2002;41:723–729. doi: 10.1021/bi011803h. [DOI] [PubMed] [Google Scholar]
- 14.Serganov A, et al. Structural basis for Diels-Alder ribozyme-catalyzed carbon-carbon bond formation. Nat Struct Mol Biol. 2005;12:218–224. doi: 10.1038/nsmb906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Robertson MP, Scott WG. The structural basis of ribozyme-catalyzed RNA assembly. Science. 2007;315:1549–1553. doi: 10.1126/science.1136231. [DOI] [PubMed] [Google Scholar]
- 16.Shechner DM, et al. Crystal structure of the catalytic core of an RNA-polymerase ribozyme. Science. 2009;326:1271–1275. doi: 10.1126/science.1174676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ponce-Salvatierra A, Wawrzyniak-Turek K, Steuerwald U, Höbartner C, Pena V. Crystal structure of a DNA catalyst. Nature. 2016;529:231–234. doi: 10.1038/nature16471. [DOI] [PubMed] [Google Scholar]
- 18.Scheitl CPM, Ghaem Maghami M, Lenz AK, Höbartner C. Site-specific RNA methylation by a methyltransferase ribozyme. Nature. 2020;587:663–667. doi: 10.1038/s41586-020-2854-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flemmich L, Heel S, Moreno S, Breuker K, Micura R. A natural riboswitch scaffold with self-methylation activity. Nat Commun. 2021;12:3877. doi: 10.1038/s41467-021-24193-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Serganov A, et al. Structural basis for discriminative regulation of gene expression by adenine- and guanine-sensing mRNAs. Chem Biol. 2004;11:1729–1741. doi: 10.1016/j.chembiol.2004.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gilbert SD, Reyes FE, Edwards AL, Batey RT. Adaptive ligand binding by the purine riboswitch in the recognition of guanine and adenine analogs. Structure. 2009;17:857–868. doi: 10.1016/j.str.2009.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wolk SK, et al. Modified nucleotides may have enhanced early RNA catalysis. Proc Natl Acad Sci USA. 2020;117:8236–8242. doi: 10.1073/pnas.1809041117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Robertson MP, Scott WG. A general method for phasing novel complex RNA crystal structures without heavy-atom derivatives. Acta Cryst D. 2008;D64:738–744. doi: 10.1107/S0907444908011578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Höbartner C, Micura R. Chemical synthesis of selenium-modified Oligoribonucleotides and their enzymatic ligation leading to an U6SnRNA stem-loop segment. J Am Chem Soc. 2004;126:1141–1149. doi: 10.1021/ja038481k. [DOI] [PubMed] [Google Scholar]
- 25.Regulski EE, Breaker RR. In-line probing analysis of riboswitches. Meth Mol Biol. 2008;419:53–67. doi: 10.1007/978-1-59745-033-1_4. [DOI] [PubMed] [Google Scholar]
- 26.Gaffney BL, Goswami B, Jones RA. Nitrogen-15-labeled oligodeoxynucleotides. 7. Use of nitrogen-15 NMR to probe hydrogen bonding in an O6MeG:C base pair. J Am Chem Soc. 2002;115:12607–12608. [Google Scholar]
- 27.Batey RT, Gilbert SD, Montange RK. Structure of a natural guanine-responsive riboswitch complexed with the metabolite hypoxanthine. Nature. 2004;432:411–415. doi: 10.1038/nature03037. [DOI] [PubMed] [Google Scholar]
- 28.Pikovskaya O, Polonskaia A, Patel DJ, Serganov A. Structural principles of nucleoside selectivity in a 2'-deoxyguanosine riboswitch. Nat Chem Biol. 2011;7:748–755. doi: 10.1038/nchembio.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Keller H, Weickhmann AK, Bock T, Wohnert J. Adenine protonation enables cyclic-di-GMP binding to cyclic-GAMP sensing riboswitches. RNA. 2018;24:1390–1402. doi: 10.1261/rna.067470.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wolter AC, et al. A Stably protonated adenine nucleotide with a highly shifted pKa value stabilizes the tertiary structure of a GTP-binding RNA aptamer. Angew Chem Int Ed. 2017;56:401–404. doi: 10.1002/anie.201609184. [DOI] [PubMed] [Google Scholar]
- 31.Freire F, et al. A simple NMR analysis of the protonation equilibrium that accompanies aminoglycoside recognition: dramatic alterations in the neomycin-B protonation state upon binding to a 23-mer RNA aptamer. Chem Commun. 2007:174–176. doi: 10.1039/b611597g. [DOI] [PubMed] [Google Scholar]
- 32.Mandal M, Boese B, Barrick JE, Winkler WC, Breaker RR. Riboswitches control fundamental biochemical pathways in Bacillus subtilis and other bacteria. Cell. 2003;113:577–586. doi: 10.1016/s0092-8674(03)00391-x. [DOI] [PubMed] [Google Scholar]
- 33.Noeske J, et al. An intermolecular base triple as the basis of ligand specificity and affinity in the guanine- and adenine-sensing riboswitch RNAs. Proc Natl Acad Sci USA. 2005;102:1372–1377. doi: 10.1073/pnas.0406347102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wilcox JL, Ahluwalia AK, Bevilacqua PC. Charged nucleobases and their potential for RNA catalysis. Acc Chem Res. 2011;44:1270–1279. doi: 10.1021/ar2000452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krishnamurthy R. Role of pK(a) of nucleobases in the origins of chemical evolution. Acc Chem Res. 2012;45:2035–2044. doi: 10.1021/ar200262x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abou Assi H, et al. 2'-O-Methylation can increase the abundance and lifetime of alternative RNA conformational states. Nucl Acids Res. 2020;48:12365–12379. doi: 10.1093/nar/gkaa928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chatterjee S, et al. The chemical nature of the 2'-substituent in the pentose-sugar dictates the pseudoaromatic character of the nucleobase (pKa) in DNA/RNA. Org Biomol Chem. 2006;4:1675–1686. doi: 10.1039/b601460g. [DOI] [PubMed] [Google Scholar]
- 38.Olsen DB, Benseler F, Aurup H, Pieken WA, Eckstein F. Study of a hammerhead ribozyme containing 2'-modified adenosine residues. Biochemistry. 1991;30:9735–9741. doi: 10.1021/bi00104a024. [DOI] [PubMed] [Google Scholar]
- 39.Schubert S, et al. RNA cleaving ‘10-23’ DNAzymes with enhanced stability and activity. Nucl Acids Res. 2003;31:5982–5992. doi: 10.1093/nar/gkg791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang Y, Nguyen K, Spitale RC, Chaput JC. A biologically stable DNAzyme that efficiently silences gene expression in cells. Nat Chem. 2021;13:319–326. doi: 10.1038/s41557-021-00645-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu X, et al. Insights into xanthine riboswitch structure and metal ion-mediated ligand recognition. Nucl Acids Ress. 2021;49:7139–7153. doi: 10.1093/nar/gkab486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ghaem Maghami M, Scheitl CPM, Höbartner C. Direct in Vitro Selection of Trans-Acting Ribozymes for Posttranscriptional, Site-Specific, and Covalent Fluorescent Labeling of RNA. J Am Chem Soc. 2019;141:19546–19549. doi: 10.1021/jacs.9b10531. [DOI] [PubMed] [Google Scholar]
- 43.Höbartner C, et al. The synthesis of 2'-O-[(triisopropylsilyl)oxy] methyl (TOM) phosphoramidites of methylated ribonucleosides (m1G, m2G, m22G, m1I, m3U, m4C, m6A, m62A) for use in automated RNA solid-phase synthesis. Monatsh Chem (Chemical Monthly) 2003;134:851–873. [Google Scholar]
- 44.Moroder H, Kreutz C, Lang K, Serganov A, Micura R. Synthesis, oxidation behavior, crystallization and structure of 2'-methylseleno guanosine containing RNAs. J Am Chem Soc. 2006;128:9909–9918. doi: 10.1021/ja0621400. [DOI] [PubMed] [Google Scholar]
- 45.Kabsch W., Xds Acta Crystallogr D. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Evans PR. An introduction to data reduction: space-group determination, scaling and intensity statistics. Acta Crystallogr D. 2011;67:282–292. doi: 10.1107/S090744491003982X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mieczkowski M, et al. Large Stokes shift fluorescence activation in an RNA aptamer by intermolecular proton transfer to guanine. Nat Commun. 2021;12:3549. doi: 10.1038/s41467-021-23932-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Terwilliger TC, et al. Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr D. 2008;64:61–69. doi: 10.1107/S090744490705024X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 51.Afonine PV, et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D. 2012;68:352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lebedev AA, et al. JLigand: a graphical tool for the CCP4 template-restraint library. Acta Crystallogr D. 2012;68:431–440. doi: 10.1107/S090744491200251X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Moriarty NW, Grosse-Kunstleve RW, Adams PD. electronic Ligand Builder and Optimization Workbench (eLBOW): a tool for ligand coordinate and restraint generation. Acta Crystallogr D. 2009;65:1074–1080. doi: 10.1107/S0907444909029436. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Structural data obtained by X-ray crystallography were deposited in the Protein Data Bank (PDB) and are available with the following accession codes: 7Q7X, 7Q7Y, 7Q7Z, 7Q80, 7Q81, 7Q82. All relevant data are provided in the Figures, Extended Data Fig 1-6, Supplementary Tables 1-2, Supplementary Fig 1, and Source Data files. Publicly available datasets from rcsb.org used in this study: 7OAX, 3SKI, 1Y27, 3FO6, 3MUM, 3Q50, 7ELR.