Summary
DNA replication initiates at genomic locations known as origins of replication, which, in S. cerevisiae, share a common DNA consensus motif. Despite being virtually nucleosome-free, origins of replication are greatly influenced by the surrounding chromatin state. Here, we show that histone H3 lysine-37 mono-methylation (H3K37me1) is catalyzed by Set1p and Set2p, and that it regulates replication origin licencing. H3K37me1 is uniformly distributed throughout most of the genome but it is scarce at replication origins, where it increases according to the timing of their firing. We find that H3K37me1 hinders Mcm2 interaction with chromatin maintaining low levels of MCM outside of conventional replication origins. Lack of H3K37me1 results in defective DNA replication from canonical origins whilst promoting replication events at inefficient and non-canonical sites. Collectively, our results indicate that H3K37me1 ensures correct execution of the DNA replication programme by protecting the genome from inappropriate origin licencing and spurious DNA replication.
Introduction
The eukaryotic genome is compacted into chromatin, whose fundamental structural unit is the nucleosome. Nucleosomes consist of approximately 150 base pairs of DNA wrapped around a core histone octamer, which is composed of two copies each of histones H2A, H2B, H3 and H4 (Luger et al., 1997). Dynamic remodelling of nucleosomes, predominantly by ATP-driven machineries, affects chromatin organisation, controlling the association of protein-complexes to the genome. In addition, histones are decorated by a plethora of covalent post-translational modifications that can directly affect chromatin structure or regulate the association of effector proteins to chromatin. Thus, the organization of the genome into chromatin provides an instructive regulatory platform for all DNA processes, including replication.
In S. cerevisiae, DNA replication initiates at particular sites in the genome known as Autonomous Replication Sequences (ARSs), which are bound by the Origin Recognition Complex (ORC) throughout most of the cell cycle. Initiation of DNA replication can be divided into two distinct steps. First, during the G1 phase, ORC coordinates the recruitment of Cdc6, followed by the Cdt1-MCM replicative DNA helicase (Mcm2–7) onto ARS to form an inactive pre-replicative complex (pre-RC). Second, as cells enter S-phase, DNA replication is initiated by the action of S-phase kinases. Firing of each replication origin requires the loading of two MCM complexes as a head-to-head double hexamer around the DNA. Phosphorylation of these MCM dimers by the S-phase kinases leads to structural changes and recruitment of the activating factors Sld3/Cdc45, GINS and Mcm10, among others, to assemble a complete replisome and initiate DNA synthesis (for review, Bell and Labib, 2016).
In S. cerevisiae, origins of replication share a 17bp AT-rich motif, called ARS Consensus Sequence (ACS), that is necessary but not sufficient for origin activity. The ACS is followed by a T-rich element B1, and two A-rich elements, B2 and B3, which are important for proper origin function (for review, Bell and Labib, 2016). Although the ACS occurs more than 10,000 in the yeast genome (Breier et al., 2004), the current estimation is that only 400-600 ARSs are commonly used (Siow et al., 2012). On average, at the population level, replication origins fire according to a very coordinated and reproducible temporal-program, although in individual cells they actually fire in a probabilistic manner. During S-phase, some origins fire early, some fire later, whilst others are passively replicated from neighbouring ones. As a general trend, early/efficient ARSs are located near transcriptionally active open reading frames, whereas late/inefficient ARSs are close to the telomeric chromosomal ends. Regardless of their firing time, most ACSs are located in intergenic regions which minimizes collisions between RNA-polymerases and components of the replication machinery (Eaton et al., 2010; Mori and Shirahige, 2007).
Numerous mathematical models and much experimental work has sought to explain the stochastic nature of ARS firing. The likelihood that an origin will fire is directly proportional to the number of MCM dimers recruited to it, which in turn is influenced by the interaction between the MCM helicase and ACS adjacent nucleosomes (Das et al., 2015; Das and Rhind, 2016). Accordingly, repositioning nucleosomes adjacent to an ARS results in firing defects due to impaired assembly of the pre-replication complex (Lipford and Bell, 2001). Reciprocally, binding of ORC to the ACS defines nucleosome positioning around ARS (Eaton et al., 2010). In addition to recruitment, helicase activation also depends on the interaction between DNA-loaded MCM and adjacent chromatin (Belsky et al., 2015).
Over the past few years, the importance of proximal histone modifications in defining origin use and regulating replication initiation has become increasingly apparent. For instance, acetylation of several lysines within H3 and H4 at origin-proximal nucleosomes promotes origin firing by recruiting Cdc45 (Vogelauer et al., 2002, Unnikrishnan et al., 2010). Moreover, mono-methylation, but not di- or tri-methylation of H3K36 at ARS flanking nucleosomes also contributes towards MCM helicase activation by Cdc45 (Pryde et al., 2009). Here, we show that H3K37me1 is important for the regulation of origin licencing. Specifically, H3K37me1 hinders Mcm2 interaction with chromatin, maintaining low levels of MCM outside of replication origins and preventing DNA replication events at non-canonical sites.
Results
Set2p and Set1p (COMPASS) are required for H3K37me1 in vivo
Various mass spectrometry studies have identified H3K37me1, both in yeast and human cells (Li et al., 2017; Ren et al., 2007; Unnikrishnan et al., 2010). However, its function remains unknown. To investigate possible roles of H3K37me1, we raised a highly specific antibody against H3K37me1. Dot-blot analyses confirmed that the antibody recognizes H3K37me1 peptides, but not unmodified ones, and that it does not cross-react with H3K36me1 (Fig.S1A). Importantly though, it does recognize H3K37me1 in combination with H3K36me1, the actual context in which H3K37me1 was reported to occur in yeast (Unnikrishnan et al., 2010) (Fig.S1A). In addition, ELISA revealed that the antibody specificity towards H3K37me1 modified peptides was several orders of magnitude higher than that towards unmodified peptides (Fig.S1B). Immunoblot studies further showed that the antibody reacts with H3 purified from wild-type yeast, but not from an isogenic strain in which lysine 37 is replaced by an alanine (H3K37A) (Fig.S1C). Similarly, chromatin immunoprecipitation (ChIP) experiments revealed that the antibody detects H3 in chromatin isolated from wild-type (H3WT) cells, but not from H3K37A mutant cells (Fig.S1D). Furthermore, immunoblot studies demonstrated that the antibody strongly detects H3 in yeast total protein extracts, but it shows only very weak reactivity towards unmodified recombinant H3 produced in bacteria (Fig.S1E and S1F). Moreover, the antibody also detects H3K37me1 in a wide variety of mammalian cell lines (Fig.S1G). Taken together, these results confirm that the antibody specifically recognizes H3K37me1, and that this modification is conserved from yeast to human.
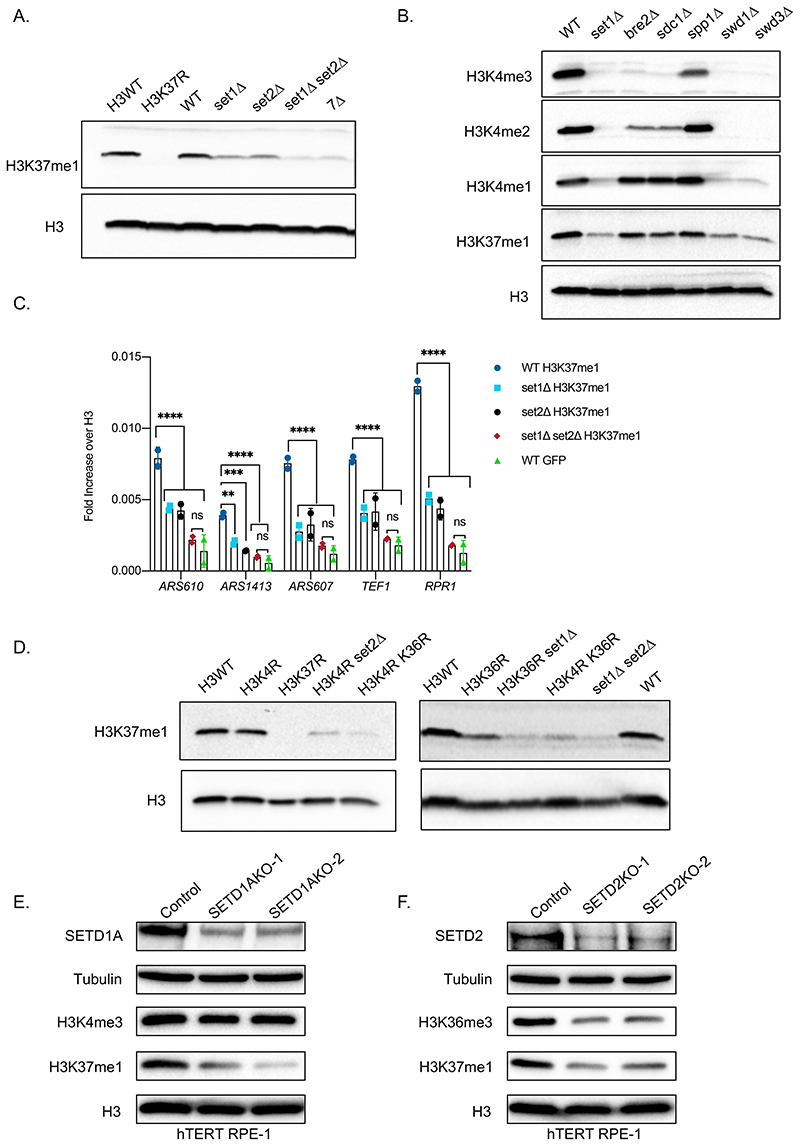
To identify the enzyme(s) responsible for H3K37me1 deposition, we immunoblotted extracts prepared from individual yeast knock-out strains of all non-essential proteins harbouring an S-adenosyl-methionine (SAM) binding domain (Fig.S2A). Since SAM is the only known methyl-group donor in yeasts, the list includes all bona fide and putative methyltransferases (Petrossian and Clarke, 2009). No single deletion completely abolished H3K37me1, but deletion of either SET1 or SET2 lysine methyltransferases resulted in a decrease of H3K37me1 levels (Fig.S2A). To confirm this finding, we deleted de novo SET1, SET2, both together and the entire SET family of lysine methyltransferases (7Δ: deletion of all 7 family members, including SET1 and SET2). We found that individual deletion of SET1 or SET2 decreased H3K37me1, but the signal was further reduced when both were deleted together (Fig.1A). Indeed, the deletion of both SET1 and SET2 reduced the level of H3K37me1 as effectively as deleting the entire SET family (7Δ) (Fig.1A). Of note, the residual signal in the double set1Δ set2Δ mutant was comparable to that of unmodified recombinant H3 (Fig.S2B). These results strongly suggested that Set1p and Set2p are redundant enzymes in the catalysis of H3K37me1. Furthermore, we found that deletion of SWD1 and SWD3, two members of the Set1 protein complex (COMPASS) required for its activity, phenocopied SET1 deletion (Fig.1B), further supporting the notion that Set1p is important for H3K37me1 in vivo. Consistent results were observed in chromatin immunoprecipitation experiments using wild-type, set1Δ, set2Δ and set1Δ set2Δ double-mutant cells. Both single mutants showed reduced levels of H3K37me1, with respect to H3, at all tested locations (Fig.1C). Importantly, the modification was decreased to background levels in the set1Δ set2Δ double-mutant (Fig.1C, compare H3K37me1 to GFP). Thus, simultaneous deletion of SET1 and SET2 eliminates H3K37me1.
Figure 1. Set2 and Set1 (COMPASS) are necessary for H3K37me1 in vivo.
(A and B) Immunoblot analysis of total protein extracts from different yeast isogenic mutants as specified. Equal amounts of proteins were separated by SDS-PAGE in 16% acrylamide gels. Blots were probed with anti-H3K37me1 or anti-H3K4me1/me2/me3 antibodies and then re-probed with anti-H3 antibody. (C) ChIP qPCR experiments showing H3K37me1 levels at different genomic locations. Chromatin from isogenic strains was immunoprecipitated using anti-H3K37me1, anti-H3 and anti-GFP (negative control) antibodies. Statistical analysis was performed using Two-way ANOVA multiple comparisons and Tukey’s multiple comparison test (Alpha: 0.05); * - P ≤ 0.05, ** - P ≤ 0.01, *** - P ≤ 0.001, **** - P ≤ 0.0001. Error bars represent the mean ± SD of 2 independent experiments. (D) Immunoblot analysis of total protein extracts from different yeast isogenic strains as specified. Equal amounts of proteins were separated by SDS-PAGE in 16% acrylamide gels. Blots were probed with anti-H3K37me1 antibody and then re-probed with anti-H3 antibody as indicated. (E and F) Immunoblot analysis of total protein extracts from (E) SETD1A or (F) SETD2 CRISPR knockout cell pools. Human RPE1 cells stably expressing Cas9 were transfected with two guide RNAs targeting different exons of each gene or a non-targeting control. Total protein extracts were prepared 7 days after transfection, and proteins were separated by SDS-PAGE in 12% acrylamide gels or 3-8% Tris-Acetate gels.
Notwithstanding the above, it was possible that the lack of H3K37me1 in the set1Δ set2Δ strain was actually due to a transcriptional defect of an essential methyltransferase. S. cerevisiae encodes 9 SAM binding proteins that are essential for cell viability (Petrossian and Clarke, 2009). However, we found no significant differences in the relative amounts of mRNAs derived from these genes in wild-type and the set1Δ set2Δ double knocked-out strains, except for a mild increase of NOP2 and DIM1 (Fig.S2C). Altogether, the above results indicate that both Set2p and Set1p (COMPASS) are responsible for H3K37me1 in vivo.
It is well established that Set1p (COMPASS) catalyzes mono-, di- and tri -methylation of H3K4 (Schneider et al., 2005) and Set2p catalyzes mono-, di- and tri-methylation of H3K36 (Strahl et al., 2002). However, none of these modifications are essential for H3K37me1, as mutation of either K4 or K36 to arginine (K4R and K36R, respectively) does not eliminate H3K37me1 (Fig.1D). We suspected that the residual H3K37me1 observed in each of the “R mutants” was actually catalysed by the enzyme whose known target-site was not mutated. To test this hypothesis, we deleted SET2 in the H3K4R strain and SET1 in the H3K36R strain. H3K37me1 was further reduced in both cases (Fig.1D). We conclude that, although K4 and K36 methylation are dispensable, Set1p needs a K at position 4, and Set2p a K at position 36, to facilitate H3K37 methylation.
Given that H3K37me1 is conserved throughout evolution, we explored the identity of the enzymes responsible for this modification in higher eukaryotes. SETD1A and SETD1B, which are required for H3K4 methylation, are the closest homologues of yeast Set1p (Shilatifard, 2012), while SETD2, which is responsible for H3K36me3 in vivo, is the closest homologue of yeast Set2p (McDaniel and Strahl, 2017). We used CRISPR/Cas9 technology to knock out SETD1A and SETD2 in hTERT RPE-1 cells targeting two different exons per gene. Consistent with previous reports, knocking out SETD1A did not affect H3K4me3 (Tajima et al., 2015, 2019). However, it clearly decreased the levels of H3K37me1 (Fig.1E). As expected, knocking out SETD2 reduced H3K36me3 levels, and, importantly, also H3K37me1 (Fig.1F). Together, these data support the notion that the mammalian SETD1A and SETD2 enzymes are orthologues of Set1p and Set2p regarding H3K37 methylation.
Set2p and Set1p (COMPASS) methylate H3K37 in vitro
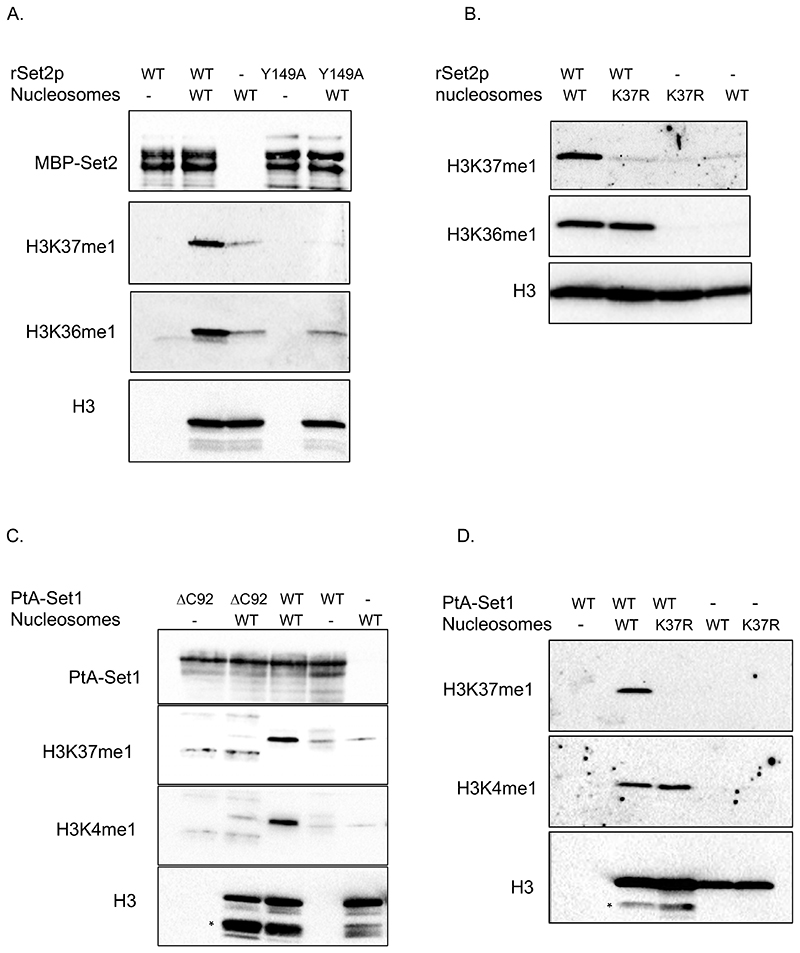
Thereafter, we addressed whether Set1p (COMPASS) and Set2p display H3K37 methyltransferase activity in vitro. Recombinant Set2p is active as a methyltransferase (Strahl et al., 2002), so we produced in E.coli full length wild-type Set2p, as well as two different mutant Set2p protein, each containing a mutation of a conserved residue within the catalytic domain (Set2 Y149A and Set2 N198Q). Wild-type Set2p, but neither of the mutants, methylated H3 in a radioactive methyltransferase reaction using 3H-SAM as a methyl donor (Fig.S3A). However, such an approach did not identify the site of methylation within H3. To address this, we performed reactions on nucleosomes reconstituted with either recombinant wild-type (H3WT) or H3K37R mutant proteins and analyzed them by immunoblot using H3K36me1 and H3K37me1 specific antibodies. Firstly, wild-type Set2p, but not Set2 Y149Ap, methylated H3K37 and H3K36 in nucleosomes harbouring H3WT (Fig.2A), indicating that methylation of both of these K residues depends on the Set2p catalytic domain. In a second experiment, we assessed Set2p activity on wild-type versus H3K37R mutant nucleosomes. Set2p methylated H3K36 in both nucleosome preparations (Fig.2B). However, it could only methylate H3K37 in H3WT containing nucleosomes and not in H3K37R mutant nucleosomes (Fig.2B). Thus, Set2p mono-methylates H3K37 in vitro.
Figure 2. Set2 and Set1 (COMPASS) methylate H3K37 in vitro.
(A) In vitro methyltransferase assay. Recombinant wild-type Set2p and Set2Y149Ap mutant proteins were incubated with wild-type (WT) H3.1 nucleosomes in the presence of SAM. Reactions were separated by SDS-PAGE in 16% acrylamide gels. Blots were probed with anti-H3K37me1 antibody and then re-probed sequentially with anti-H3K36me1 and anti-H3 antibodies. Finally, the membrane was blotted with anti-MBP antibody to monitor the amount of enzyme in each reaction. (B) In vitro methyltransferase assay. Equal amount of recombinant wild-type Set2p was incubated with H3.1WT or H3.1K37R nucleosomes in the presence of SAM. Reactions were separated by SDS-PAGE in 16% acrylamide gels. Blots were probed with anti-H3K37me1 antibody and then re-probed sequentially with anti-H3K36me1 and anti-H3 antibodies. (C) In vitro methyltransferase assay. Wild-type PtA-Set1p and PtA-set1ΔC92p yeast purified complexes were incubated with wild-type H3.1 nucleosomes in the presence of SAM. Reactions were separated by SDS-PAGE in 16% acrylamide. Blots were probed with anti-H3K37me1 antibody and sequentially with anti-H3K4me1 and anti-H3 antibodies in this order. * indicates Δ tail–H3. PtA-Set1p and PtA-set1ΔC92p in each reaction were detected using anti-PAP antibody. (D) In vitro methyltransferase assay. Equal amount of wild-type PtA-Set1p complex was incubated with H3.1WT or H3.1K37R nucleosomes in the presence of SAM. Reactions were separated by SDS-PAGE in 16% acrylamide gels. Blots were probed with anti-H3K37me1 antibody and sequentially with anti-H3K4me1 and anti-H3 antibodies in this order. * indicates Δ tail–H3.
As recombinantly produced Set1p is not active in vitro (Takahashi et al., 2011), we purified the Set1 complex (COMPASS) from a yeast strain expressing either a protein A-tagged Set1p wild-type allele (PtA-Set1) or a mutant lacking the C-terminal 92 amino acids (PtA-Set1ΔC92), which corresponds to the post-SET domain (Fig.S3B). As previously shown (Santos-Rosa et al., 2002), in vitro methyltransferase reactions using H3 and 3H-SAM as donor revealed that PtA-Set1ΔC92 is not active, in contrast to wild-type Set1p (Fig.S3C). We then performed in vitro reactions using PtA-Set1 and PtA-Set1ΔC92 on wild-type nucleosomes (Fig.2C, top panel) and analyzed them by immunoblotting with H3K37me1 and H3K4me1 specific antibodies. We detected H3K37me1 and H3K4me1 only in the reactions catalyzed by the wild-type enzyme (Fig.2C, middle panels). Furthermore, H3K37me1 methylation by wild-type PtA-Set1p was lost when H3K37R-containing nucleosomes were assayed (Fig.2D, top panel). These results show that Set1p (COMPASS) mono-methylates H3K37 in vitro. Altogether, the above results indicate that both Set2p and Set1p (COMPASS) are indeed H3K37 mono-methyltransferases.
H3K37me1 correlates with replication origin firing
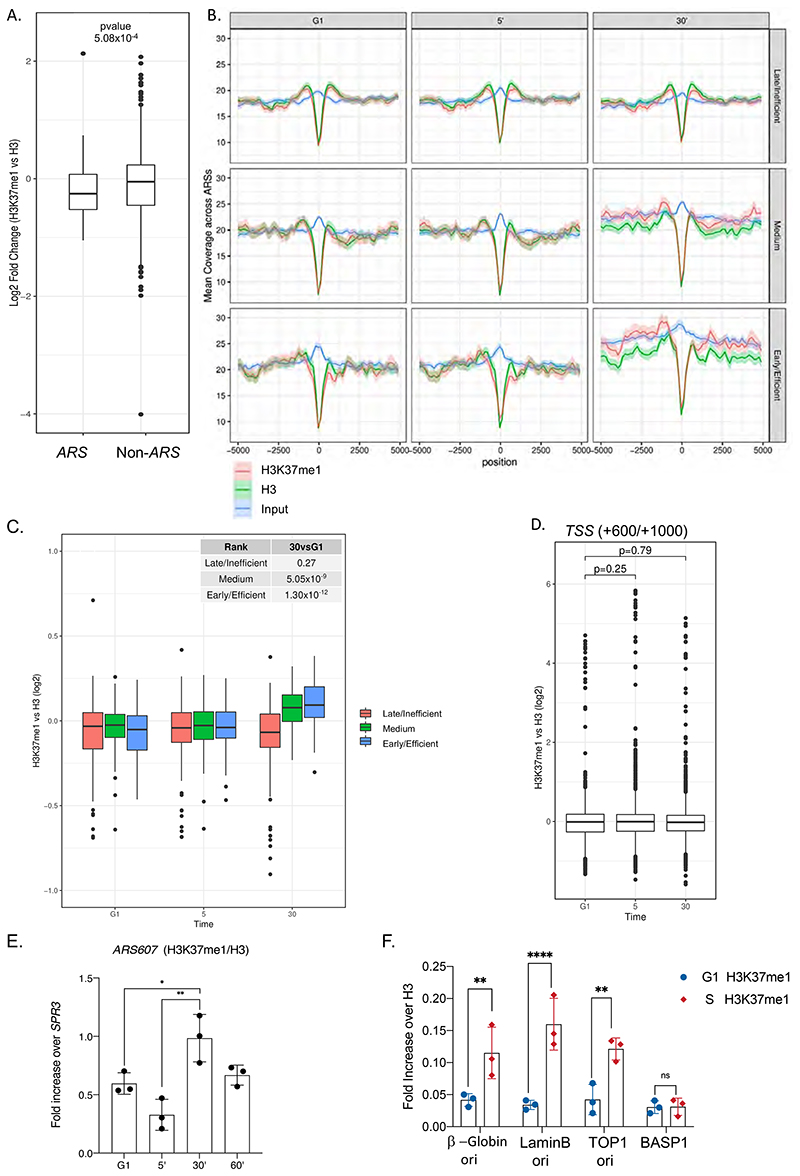
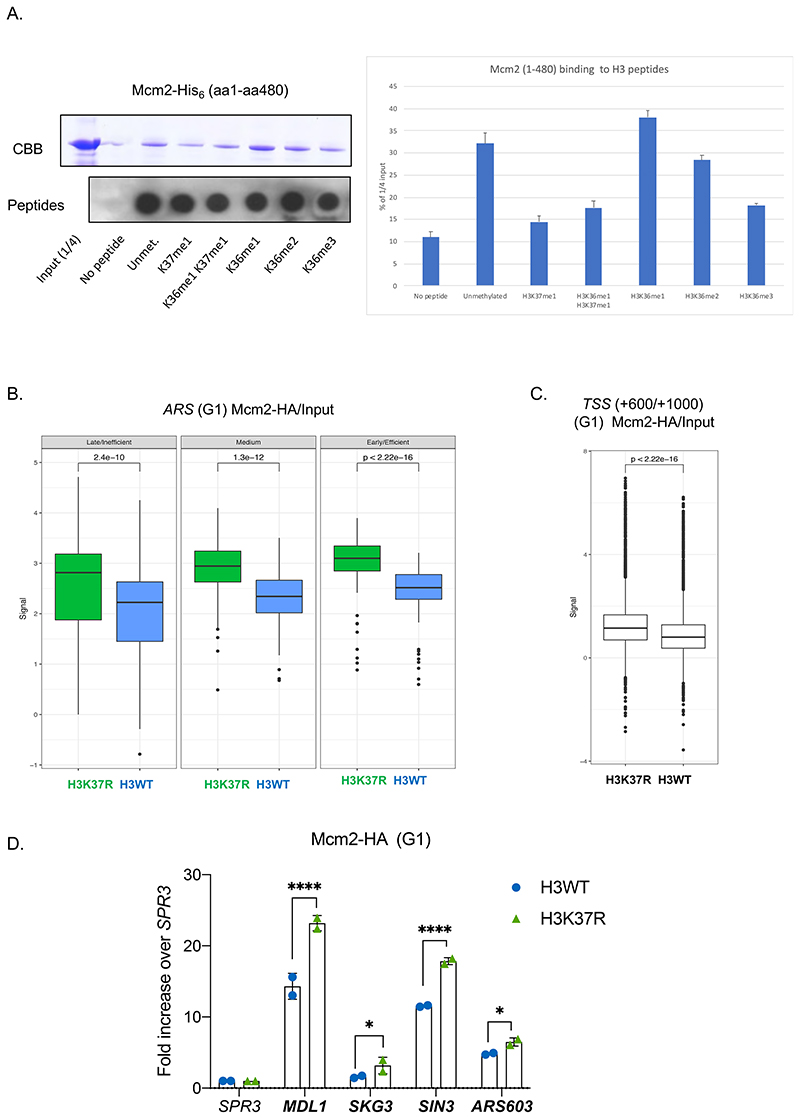
To investigate the function of H3K37me1, we first analyzed its genome-wide distribution by ChIP followed by high-throughput DNA sequencing (ChIP-seq). H3K37me1 was broadly distributed throughout the genome of asynchronous yeast cells, with no apparent enrichment at specific locations. However, previous mass spectrometry studies had identified H3K37me1 at nucleosomes flanking the plasmid-borne replication origin ARS1 (Unnikrishnan et al., 2010), thus we focused our attention towards origins of replication. Surprisingly, we found that ARSs were significantly H3K37 hypo-methylated compared to the rest of the genome (Fig.3A), suggesting that H3K37me1 is specifically restrained from these locations. To explore whether H3K37me1 deposition at ARSs is cell cycle regulated, we arrested wild-type cells in G1 using α factor and then released them into the cycle. We prepared ChIP-seq libraries from G1, G1/S and S phase synchronized cells, which correspond to times 0, 5 and 30 minutes after release, respectively (Fig.S4A). Origins were classified as early/efficient, medium and late/inefficient, based upon their firing efficiency according to our DNA replication profiling dataset (see Fig.S5C). As expected, early origins showed a clear increase in DNA copy number during S phase, whereas medium origins showed a moderate increase, and late origins no change at all (Fig.3B, 30’ Input). Notably, we observed a pronounced increase of H3K37me1, relative to H3, during S phase specifically at early origins, whilst medium and late origins showed a milder increase or no change, respectively (Fig.3B and 3C, 30’). We chose “non-ARS control region” as the window + 600 to 1000bp from Transcription Start Sites, based on the ORF average size in S.cerevisiae (1.2Kb) and the premise that most ARS are located outside ORFs. In contrast to the origins of replication, non-ARS showed a constant H3K37me1/H3 ratio throughout the experiment (Fig.3D and Fig.S4B). We independently confirmed these results at different locations by qPCR (Fig.3E and Fig.S4C). From these experiments, we conclude that H3K37me1 is under-represented at origins of replication in G1, but its presence at these regions is cell cycle-regulated and correlates with the timing of their firing.
Figure 3. Histone H3K37me1 correlates with Replication Origin firing.
(A) Box-plot representing H3K37me1 enrichment over H3 (log2) of genomic windows overlapping replication origins (ARS +/- 2kb) versus non-ARS (mean of the region from Transcription Start Sites+600 to TSS+1000). (B) Coverage plot showing the mean normalised ChIP-signal (+/- s.e.m.) for H3, H3K37me1 and Input at ARS +/-5kb (total number of ARSs: 408). ARSs were grouped into 3 categories according to firing efficiency/time. (C and D) Box-plot showing the mean H3K37me1 enrichment over H3 (log2) at ARS +/-2kb (left panel) and at non-ARS control regions (TSS +600 to TSS+1000, right panel). p-values were calculated with the Mann-Whitney-Wilcoxon test. (E) ChIP qPCR experiments showing H3K37me1 levels at ARS607. Chromatin from the time points shown in Figure S4A was immunoprecipitated using anti-H3K37me1 and anti-H3 antibodies. H3K37me1/H3 levels were normalized to a non-ARS region (SPR3). Statistical analysis was performed using Two-way ANOVA multiple comparisons and Tukey’s multiple comparison test (Alpha: 0.05); * - P ≤ 0.05, ** - P ≤ 0.01, *** - P ≤ 0.001. Error bars represent the mean ± SD of 3 independent experiments. (F) ChIP qPCR experiments showing H3K37me1 enrichment at different origins of replication in G1/S and S-phase synchronized human RPE1 cells as shown in Figure S4D. Statistical analysis was performed using Two-way ANOVA corrected for the comparisons using the Holm-Sidak method (Alpha: 0.05); * - P ≤ 0.05, ** - P ≤ 0.01, *** - P ≤ 0.001. Error bars represent the mean ± SD of 3 independent experiments.
To address whether H3K37me1 is also cell cycle regulated at human origins of replication, we synchronized non-transformed but immortalized retinal pigment epithelial cells (hTERT RPE-1) at the G1/S boundary and released them into the cell cycle (Fig.S4D). We then performed ChIP experiments using G1/S and S-phase synchronized cells and analyzed H3K37me1 at different well-characterized replication origins. We observed that H3K37me1 levels significantly increased with respect to H3 at origins of replication, but not at an unrelated region, during S phase (Fig.3F). This result suggests that the distribution and cell cycle regulation of this modification is conserved in mammalian cells.
H3K37me1 regulates DNA replication origin firing
Set1p and Set2p are involved in many cellular processes due to their K4 and K36 methyltransferase activities, respectively (for review, Freitag, 2017). This prevented us from studying the function of H3K37 methylation in vivo by mutating the responsible enzymes. Instead, we probed the function of this modification by mutating the modified residue. Mutation of H3K37 to arginine retains the residue’s positive charge but prevents its methylation by Set1p and Set2p. Thus, we compared the DNA replication profiles of H3WT and H3K37R mutant strains by performing genome-wide immunoprecipitation of nascent BrdU-labelled DNA (Lengronne et al., 2001). Once transformed with the thymidine kinase (TK) and nucleoside transporter (hENT1), both H3WT and H3K37R yeast strains incorporated BrdU with similar efficiencies (Fig.S5A). Also, we specifically immunoprecipitated DNA only from cells incubated with BrdU (Fig.S5B), and the signal was higher at replication origins compared to other regions of the genome (Fig.S5B), testifying to the specificity of our IPs.
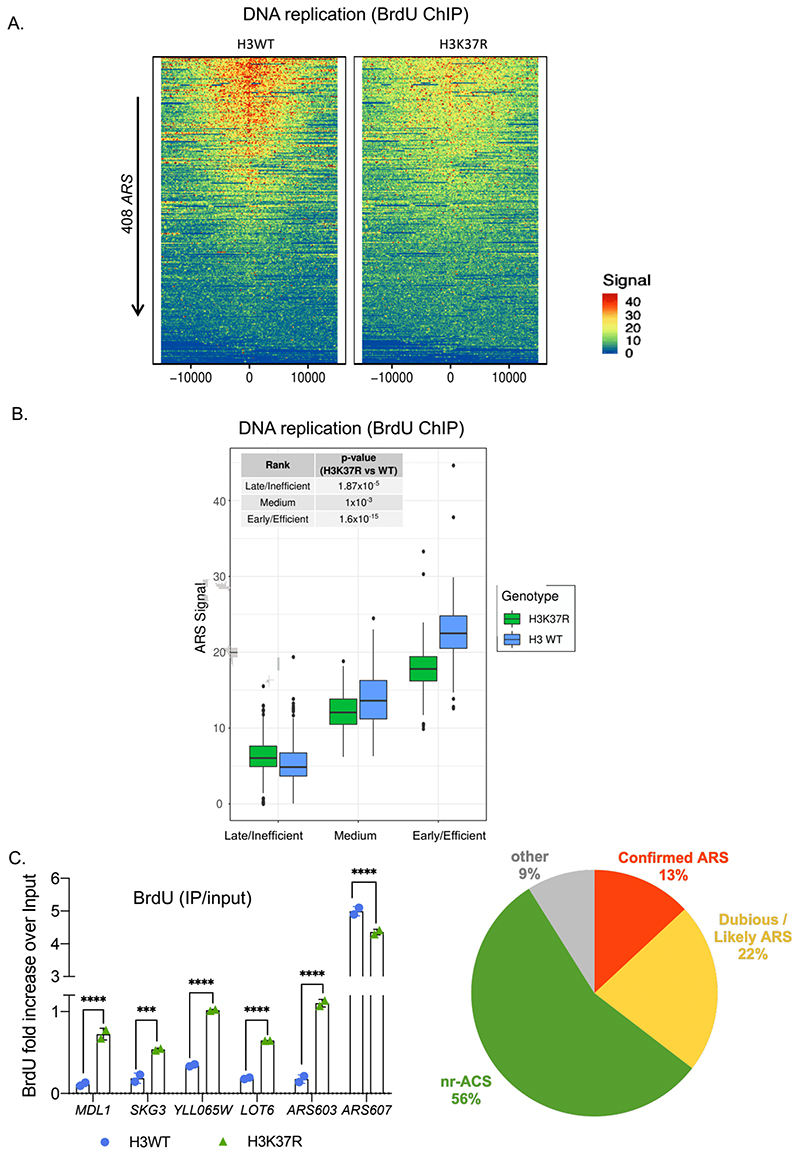
To investigate the genome-wide effects of H3K37R, we analyzed BrdU incorporation at replication origins. ARSs were ranked top to bottom according to their firing efficiencies in the H3WT strain. Remarkably, we observed an evident general decrease in the replication efficiency of the H3K37R mutant strain (Fig.4A and Fig.S6A). Next, we sorted all replication origins into early/efficient, medium and late/inefficient ARSs as before (Fig.S5C). We found that early/efficient and medium replication origins were less active in the H3K37R mutant strain than in the wild-type strain (Fig.4B). Interestingly, the late/inefficient group, which includes the ‘dormant replication origins’, showed an increased BrdU signal in the H3K37R mutant strain in comparison to the H3WT strain (Fig.4B). These results indicate that H3K37me1 is required for the correct execution of the DNA replication programme.
Figure 4. H3K37me1 regulates DNA replication.
(A) Heatmap showing the distribution of BrdU incorporation at replication origins in wild-type H3 and H3K37R mutant cells. Origins were aligned from highest to lowest BrdU signal in the wild-type strain and centred at ARS ACS (Ars Consensus Sequence). For visualisation purposes the heatmaps’ colour scale was saturated at the 99th percentile of the distribution of signal intensities. The plot represents the average of 4 independent experiments. (B) Box-plot showing the distribution of mean BrdU signal incorporated at the replication origins (ARS +/- 1kb) shown in (A). The p-values were calculated with the Mann-Whitney-Wilcoxon test. ARSs were classified into early/efficient, medium and late/inefficient following the BrdU distribution shown in Figure S5C. (C) ChIP qPCR experiments showing incorporation of BrdU in H3WT and H3K37R mutant cells. IPs were analysed by qPCRs at the early/efficient ARS607, late/inefficient ARS603 and “H3K37R-unique” firing locations, using specific primers. Statistical analysis was performed using Two-way ANOVA corrected for the comparisons using the Holm-Sidak method (Alpha: 0.05); * - P ≤ 0.05, ** - P ≤ 0.01, *** - P ≤ 0.001, **** - P ≤ 0.0001. Error bars represent the mean ± SD of 2 independent experiments. (D) Piechart showing proportion of H3K37R unique replication events occurring at different genomic locations.
Surprisingly, comparison of the H3WT and H3K37R DNA replication profiles outside of canonical ARS revealed numerous genomic locations displaying stronger or exclusive BrdU labeling in the mutant strain (Fig.S6B). These replication events occur several kilobases away from the nearest firing ARS, supporting the notion that they result from de novo firing rather than from interference with nearby replicating ARS and/or differences in the speed of the replication fork (Fig.S6C). Importantly, BrdU-IP followed by qPCR using specific primers confirmed that DNA replication initiates substantially better at these distinct locations in the H3K37R mutant (Fig.4D and Fig.S6D), suggesting that H3K37me1 may serve to restrict the initiation of DNA replication to bona fide origins of replication.
To further investigate the nature of the H3K37R unique events we first calculated, as a proxy, the average width of the replication peaks of the middle ARS group (2.0957 kb) We then assumed that all H3K37R unique peaks occurring within a 2kb size window initiated from a single event. Using these stringent criteria, we identified 326 replication events, 91% of which occurred at previously predicted “ACS-matches” (Breier et al., 2004). Specifically, H3K37R unique peaks included (a) confirmed late/inefficient ARS which were not firing in the WT strain under our experimental conditions (13%), (b) dubious/likely ARS (22%) and (c) non replicative ACS (nr-ACS) which, under our experimental conditions, were able to drive significant replication in the H3K37R mutant (56%) (Fig.4D). All together, these results are consistent with a model in which H3K37me1 restricts origin firing potential to conventionally used origins of replication, preventing non-canonical ones from firing.
H3K37me1 modulates MCM association to chromatin
It has been reported that Mcm2, a subunit of the MCM replicative helicase, physically interacts with nucleosomes through association of its N-terminus to histone H3 (Ishimi et al., 1998). Thus, we considered the possibility that methylation of H3K37 might affect this interaction. As a preliminary analysis, we performed in vitro pull-down experiments using H3 biotinylated peptides (unmodified and methylated at K37, K36 or both) and recombinant Mcm2aa1-aa480-His6, which has been shown to mediate the Mcm2 to H3 interaction (Ishimi et al., 1998). We found that Mcm2 has a slight preference to bind to H3 when K36 is mono-methylated (Fig.5A; Fig.S7A & S7B). This is in agreement with in vivo data showing that K36me1, but not K36me2 or K36me3, is present at ARSs upon firing (Pryde et al., 2009). Notably, the presence of one methyl group at H3K37 compromises the interaction between the H3 and Mcm2 (Fig.5A; Fig.S7A & S7B), suggesting that H3K37me1 creates an unfavorable environment for the interaction between MCM and chromatin.
Figure 5. H3K37me1 regulates origin establishment.
(A) Left: Mcm2-His6 in vitro binding to biotinylated H3 peptides, modified as indicated. Input and peptide-bound Mcm2 were resolved by SDS-PAGE in 8% acrylamide and detected by Coomasie-Brilliant Blue (CBB) staining. 1/5 of each bead slurry was spotted onto PVDF membrane and biotin-peptides detected with anti-biotin antibody. Right: Average ImageJ quantification of assays shown in left and in Fig.S7A. Binding is represented as % of the signal corresponding to ¼ of the input. (B) Box plot of Mcm2-HA occupancy at early/efficient, medium and late/inefficient ARS +/- 1Kb. Blue: H3WT, green: H3K37R. (C) Box plot of Mcm2 occupancy at non-ARS (mean of the region from TSS+600 to TSS+1000) in H3WT and H3K37R mutant cells. The plots in (B) and (C) represent the average of the 3 independent cultures. (D) ChIP qPCR experiments showing Mcm2-HA levels at “H3K37R unique” replication sites. H3WT and H3K37R yeast cells expressing Mcm2-HA were arrested in G1, crosslinked and chromatin was immunoprecipitated (IP) with anti-HA antibody. The IP material was analyzed by qPCR with primers specific for each location. Data were normalized to the non-ARS region (SPR3). Statistical analysis was performed using Two-way ANOVA corrected for the comparisons using the Holm-Sidak method (Alpha: 0.05); * - P ≤ 0.05, ** - P ≤ 0.01, *** - P ≤ 0.001, **** - P ≤ 0.0001. Error bars represent the mean ± SD of 2 independent experiments.
Next, we investigated the in vivo recruitment of the MCM complex to chromatin in the absence of H3K37me1. To do so, we HA-tagged Mcm2 as a proxy for the binding of the MCM complex in H3WT and H3K37R cells. Both strains expressed similar levels of Mcm2-HA (Fig.S7C). In addition, we could ChIP Mcm2-HA at ARSs in the HA-tagged strain, but not in its isogenic untagged strain, attesting to the specificity of our ChIPs (Fig.S7D). Then, we performed genome-wide Mcm2-HA ChIP-seq in G1 arrested H3WT and H3K37R cells. We “spiked” our yeast cultures with an identical number of S. pombe cells expressing Mcm3-HA tagged, for use as an ‘external normalizer’ (Orlando et al., 2014). Importantly, we found that lack of H3K37me1 resulted in a significant increase in the association of MCM to all ARSs groups (Fig.5B; Fig.S7E) and, notably, all over the genome (Fig.5C; Fig.S7F). ChIP-qPCR confirmed the increase in MCM recruitment, which was particularly pronounced at those specific genomic locations that triggered DNA replication in the H3K37R mutant strain, but not in the WT strain (Fig.5D). Importantly, however, we found that histone occupancy was not affected by mutating H3K37 (Fig.S7G).
These results, in agreement with the in vitro data, support the notion that H3K37me1 negatively regulates the association of MCM to chromatin.
Concurring with the above, ChIP experiments during a replication time course showed that changes in H3K37me1 and MCM anticorrelate at the efficient origin ARS607 but do not at the inactive ARS1333 (Fig.S8A). In fact, we found a global anti-correlation between H3K37me1 and MCM at active ARSs in vivo (Fig.S8B, early/efficient origins). We then investigated whether H3K37me1 may also impact MCM dynamics upon G1 release. We found that H3K37R mutation led to retention and/or better detection of MCM at origins of replication, particularly at late/inefficient ones (Fig.S8C). This result is intriguing, as it is well-established that MCM recruitment to ARS is restricted to late M/ G1 phase (Diffley, 2011). We did not find significant difference between H3WT and H3K37R chromatin states upon G1 release (as measure by H3 occupancy, Fig.S8D). Yet, we can’t exclude that MCM structural changes occurring during G1->S transition in the H3K37R mutant may increase epitope accessibility and detection of the already G1-recruited helicase. Alternatively, lack of H3K37me1 may result in longer retention of the inactive MCM complexes, particularly at late/inefficient ARS, increasing the probability of those to encounter a helicase travelling with the replication forks from adjacent origins.
Altogether, these results are consistent with a model in which H3K37me1 modulates the association and dynamics of the replicative helicase to chromatin.
Depletion of H3K37me1 results in competition between conventional and non-replicative ARS for rate limiting MCM activators
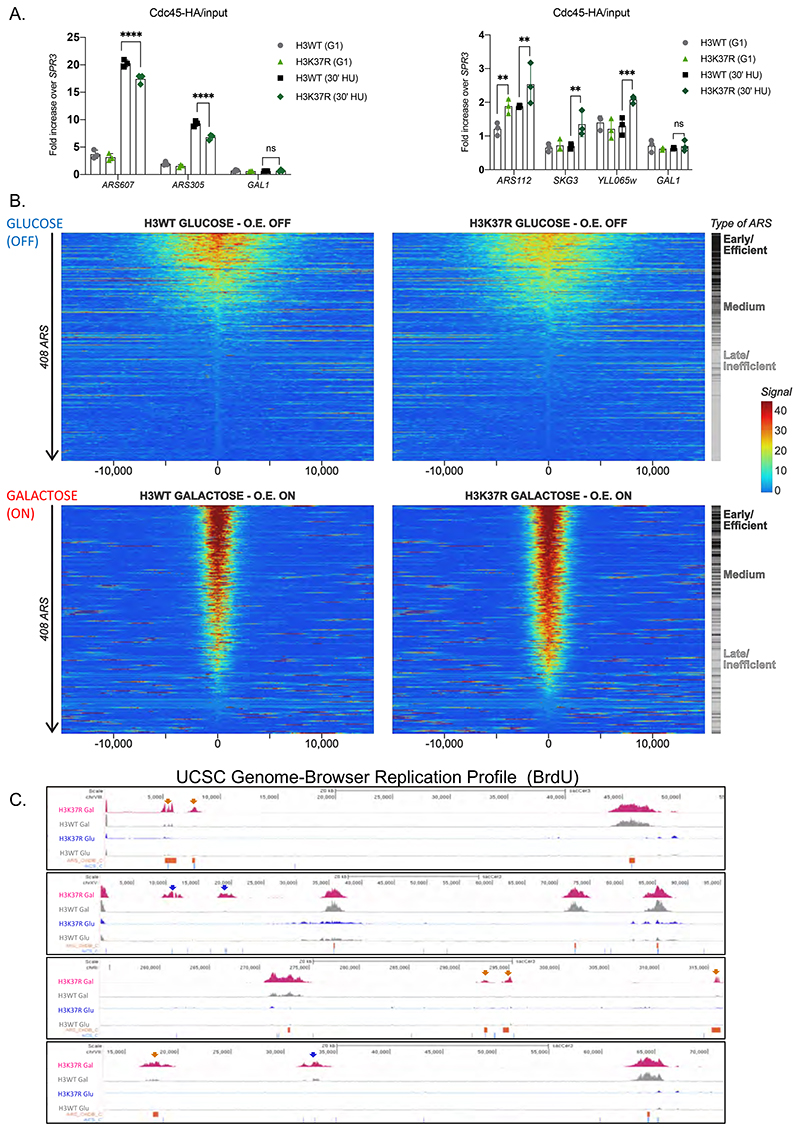
Our results show that lack of H3K37me1 increases MCM chromatin binding across the genome, but reduces DNA replication from most commonly used ARS. It has been reported that recruitment of limiting helicase activators (Sld2 Sld3, Sld7, Dpb11, Dbf4 and Cdc45) define origin efficiency and firing time (Mantiero et al., 2011; Tanaka et al., 2011). Thus, we hypothesized that competition between canonical ARSs and novel replication sites for these rate limiting factors might underlie the observed H3K37R phenotypes. To test this, we first performed ChIP analysis of Cdc45 in cells arrested in G1 arrested and released into S-phase in the presence of hydroxyurea. We found lower levels of Cdc45 at efficient ARS, such as ARS607 or ARS305 (Fig.6A, left panel), in the H3K37R mutant strain compared to the WT strain, but increased Cdc45 recruitment to the late/inefficient ARS112, and also to genomic locations that supported DNA replication exclusively in the H3K37R mutant (Fig.6A, right panel). These results suggest that in the absence of H3K37me1, limiting helicase activators are diverted from canonical ARS to unconventional and non-replicative ACS.
Figure 6. Over-expression of MCM activators suppresses H3K37R replication defects.
(A) ChIP qPCR experiments showing Cdc45-HA levels at efficient ARS (left panel) and “H3K37R unique” replication sites (right panel). H3WT and H3K37R yeast cells expressing Cdc45-HA were arrested in G1 and released into HU containing medium for 30’. Samples were crosslinked and chromatin was immunoprecipitated (IP) with anti-HA antibody. The IP material was analyzed by qPCR with primers specific for each location. Data were normalized to a non-ARS region (SPR3). Statistical analysis was performed using Two-way ANOVA corrected for the comparisons using the Holm-Sidak method (Alpha: 0.05); * - P ≤ 0.05, ** - P ≤ 0.01, *** - P ≤ 0.001, **** - P ≤ 0.0001. Error bars represent the mean ± SD of 2 independent experiments. Note the difference in the scale between left and right panels. (B) Heatmap showing the distribution of BrdU incorporation in wild-type H3 and H3K37R mutant cells under non-overexpression (GLUCOSE, OFF) and over-expression (GALACTOSE, ON) of MCM activators. Replication origins were aligned from highest to lowest BrdU signal in the wild-type strain and centred at ACS. ARSs were classified into early/efficient, medium and late/inefficient following the BrdU distribution shown in Figure S5C. For visualisation purposes the heatmaps’ colour scale was saturated at the 99th percentile of the distribution of signal intensities. The plot represents the average of 3 independent experiments. (C) Representative genome browser snapshots of BrdU incorporation in H3WT and isogenic H3K37R mutant cells at different genomic locations. ARS (OriDB) are represented as orange boxes; non-replicative ACS matches are represented as blue lines. H3K37R exclusive firing event are indicated by the corresponding color-coded arrows. The plots represent the average of 3 independent cultures per strain and condition.
Next, we engineered H3WT and H3K37R strains to express an additional copy of Sld2, Sld3, Sld7, Dpb11, Dbf4 and Cdc45 under the control of the strong inducible GAL1-10 promoter, as previously performed (Mantiero et al., 2011; Tanaka et al., 2011; Material and Methods). Importantly, immunoblot analysis revealed that both strains induced the expression of all factors to the same extent (Fig.S9A). We performed BrdU immunoprecipitation under non-inducing conditions (glucose, OFF) and over-expressing conditions (galactose, ON) and analyzed the immunoprecipitated DNA with primers specific to efficient (ARS607) and inefficient (ARS603) origins. In agreement with previous reports (Mantiero et al., 2011; Tanaka et al., 2011), overexpression of MCM activators (+galactose) resulted in stronger replication from inefficient and, to certain extent, also from efficient ARS in the wild-type strain (Fig.S9B, compare H3WT Glu with H3WT Gal).
Thus, we analyzed DNA replication genome-wide. As in previous experiments, ARSs were ranked top to bottom according to their firing efficiencies in the H3WT strain. We found that overexpression of the limiting helicase activators suppressed the overall firing deficiency otherwise occurring in the H3K37R mutant (Fig.6B), suggesting that competition for limiting MCM activators underlies the strain’s DNA replication defects. We sorted all replication origins into early/efficient, medium and late/inefficient ARSs, as before. Notably, over-expression of MCM activators largely rescued DNA replication from early/efficient origins (Fig.6B and Fig.S9C). However, under the same conditions, late/inefficient origins still fired more in the H3K37R mutant than in the wild-type strain (Fig.6B and Fig.S9C). Similar results were obtained using ChIP-qPCR analysis of ARS607 (early/efficient) and ARS603 (late/inefficient), respectively (Fig.S9B). These results argue that the increased activity of inefficient ARS in the H3K37R mutant is not merely a response to palliate the deficient firing from conventional ones. In fact, “ACS sites” that remained inactive in wild-type cells, even under excess helicase activators, fired in the absence of H3K37me1 (Fig.6C).
Altogether, the above experiments are consistent with a model in which the absence of H3K37me1 causes a redistribution of origin firing towards unconventional and non-replicative ACSs, diverting limiting helicase activators from canonical ARS.
Discussion
In late mitosis/early G1 of each cell cycle, ARSs are bound by ORC and the replicative helicase (MCM) in a process known as origin licensing, necessary but not sufficient for an origin to fire. Commitment to fire occurs upon MCM activation by recruitment of rate limiting factors such as Sld2, Sld3, Sld7, Dpb11, Dbf4 and Cdc45 at the G1/S transition (for review, Bell and Labib, 2016; Riera et al., 2017). ORC binding to the ACS (ARS Consensus Sequence) and recruitment of the replicative helicase depend on the organization of adjacent nucleosomes, whose epigenetic state can influence origin specificity, licensing and helicase activation (Azmi et al., 2017; Belsky et al., 2015; Soriano et al., 2014; Soudet and Stutz, 2019). In particular, histone acetylation and H3K36 mono-methylation promote ARS firing and recruitment of the helicase activating protein Cdc45 (Knott et al., 2009; Pryde et al., 2009; Unnikrishnan et al., 2010; Vogelauer et al., 2002). Also, H4K20 mono-methylation plays an important role, at the exit of mitosis, in establishing the necessary chromatin compaction to limit DNA replication licensing (Shoaib et al., 2018).
Mcm2, a component of the replicative helicase, directly binds to a region of H3 spanning amino acids 26 to 67 (Ishimi et al., 1998). Our results suggest that regulation of this interaction by H3K37me1 is important for origin licensing. We showed that H3K37me1 is uniformly distributed throughout the vast majority of the genome during G1, but is particularly sparse at origins of replication, where its absence might improve ORC binding and/or stabilize MCM-ORC interactions on chromatin, thus facilitating MCM loading. In agreement with this possibility, depletion of H3K37me1 results in increased MCM chromatin binding across the entire genome, particularly at ACS locations where cooperativity between ORC and nucleosomes may occur. Perhaps surprisingly, we observed that this increase in MCM chromatin association is accompanied by a general decrease in firing from canonical replication origins. Although the S. cerevisiae genome harbors around 10,000 ACS motifs (Breier et al., 2004), only around 400 are effectively used during standard growth conditions (Eaton et al., 2010; Xu et al., 2006). Notably, we found that mutation of H3K37 causes a redistribution of origin firing potential from ARS efficiently used towards inefficient and non-canonical ACS, suggesting that H3K37me1-dependent regulation of MCM chromatin loading (and perhaps activation) is important for the proper execution of the origin firing programme. In line with this, it has been reported that recruitment of limiting helicase activators (Sld2 Sld3, Sld7, Dpb11, Dbf4 and Cdc45) define origin efficiency and firing time (Mantiero et al., 2011; Tanaka et al., 2011). Our results suggest that competition for these rate limiting factors, between canonical and non-canonical ARSs, underlies the DNA replication defects observed in the absence of H3K37me1.
Altogether, our results support a model in which the absence of H3K37me1 makes chromatin broadly more permissive to interactions with the replicative helicase, facilitating MCM loading and origin licensing at cryptic genomic ACS, thereby rendering such normally non-replicative locations competent to fire. Hence, we propose a role for H3K37me1 in safeguarding a proper origin firing programme by protecting the genome from unscheduled association of MCM to chromatin, thereby preventing spurious initiation of DNA replication from cryptic origin locations.
The identification of Set1p and Set2p as H3K37 methyltransferases was quite unexpected, as these enzymes were first discovered due to their ability to methylate H3K4 and H3K36, respectively. However, Set1p also methylates the kinetochore protein Dam1, whilst SETD2, the Set2p human homologue, methylates tubulin during mitosis (Park et al., 2016; Zhang et al., 2005). Thus, the panel of Set1p and Set2p substrates is broader than first anticipated. Importantly, several reports implicate the two enzymes in cell cycle regulation and DNA replication. Firstly, the enzymatic activities of Set1p and Set2p are required for cell cycle progression via transcriptional, as well as non-transcriptional pathways (Biswas et al., 2008; Park et al., 2016; Rizzardi et al., 2012; Zhang et al., 2005). Secondly, H3K36me1 and H3K4me2 are specifically enriched at replication origins where they contribute to firing, although the precise mechanisms are not fully understood (Biswas et al., 2008; Pryde et al., 2009; Rizzardi et al., 2012). Moreover, although Set1p and Set2p are recruited to chromatin by their interaction with RNA polymerase II, yeast COMPASS physically interacts with Orc2p (Kan et al., 2008) and a Set2 mutant lacking its phospho-CTD interaction domain still binds to chromatin (Youdell et al., 2008), arguing that both enzymes can be recruited to chromatin in a transcription-independent manner. Hence, while Set1p and Set2p are involved in a plethora of cellular functions, a more direct role in controlling DNA replication is increasingly supported.
Although methylation of H3K4 and/or H3K36 are not a prerequisite for H3K37me1, the lysines appear important for Set1p and Set2p activity, respectively. Whilst the positioning of H3K36 directly adjacent to H3K37 might explain the relevance of both lysines to Set2p’s substrate recognition, it is intriguing that mutation of H3K4 affects Set1p activity towards K37. There is no apparent reason to indicate that Set1p uses distinct mechanisms to catalyze H3K37 and H3K4 mono-methylation, as the requirement of COMPASS members for methylation is the same for both sites. It was recently shown that COMPASS operates as a dimer in vivo (Choudhury et al., 2019), so one possibility is that the enzyme is recruited by binding of one COMPASS to H3K4 while the second module catalyzes methylation of H3K37.
During the course of this work, it was reported that S. pombe Set7 is a H3K37 methyltransferase and that this modification affects gametogenesis in fission yeast (Shen et al., 2019). Pombe Set7 shares a similar degree of homology with all S. cerevisiae SET family members, as it does with human counterparts, so there is no clear orthologue. Nevertheless, we tested and found that H3K37me1 is also upregulated at the onset of premeiotic DNA replication in S. cerevisiae (Fig.S10). In this regard, it is notable that deletion of SET1 leads to sporulation defects due to impaired initiation of pre-meiotic DNA replication, rather than to transcriptional defects (Sollier et al., 2004). Interestingly, this Set1p function requires its catalytic domain but not H3K4 methylation (Sollier et al., 2004). Moreover, set1Δ sporulation defect is exacerbated by simultaneous deletion of SET2 (Morohashi et al., 2005). These data support a model in which H3K37me1 deposition by Set1 and Set2 regulates DNA replication not only during mitosis, but also during meiosis.
Our results point to the fact that H3K37me1 and its cell cycle regulation are evolutionary conserved, raising the possibility that H3K37me1 may participate in the selection of replication origins in higher eukaryotes, (for review, Cayrou et al., 2011; Ding and MacAlpine, 2011). This might be particularly relevant in human cancer, where deregulated intragenic origin firing generates DNA replication stress that contributes critically to genomic instability (Macheret and Halazonetis, 2018). Given that SETD1A and SETD2 are implicated in human malignancies, the unravelling of a function for these enzymes in coordinating origin firing may be of therapeutic importance.
Limitations of study
Histone-modifying enzymes are involved in the regulation of diverse DNA-templated processes, often modifying more than one histone residue or even, additional non-histone substrates. Hence, phenotypes resulting from enzyme mutation can’t always be owed to the lack of a particular histone mark. Here, we mutated histone H3K37 to arginine in order to investigate the role of H3K37me1 in DNA replication. While retaining the residue’s positive charge, H3K37R mutation would prevent not only methylation but also any other possible post-translational modification at lysine 37, which may also contribute to the observed phenotypes. We note, however, that no other post-translation modification has been identified so far at this particular site.
Star Methods
Contact for reagent and resource sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by Helena Santos-Rosa (Hsrs2@gurdon.cam.ac.uk).
Experimental model and subject details
All Saccharomyces cerevisiae strains used in this study are derivatives of W303 background except for those used in the screen shown in Fig.S3A, which are BY4743 & BY4741. Integrations and deletions were performed using one-step PCR-based methods (Janke et al., 2004). Histone point mutants were shuffled by counter-selection on 5-FOA.
Yeast strains overexpressing the helicase activators where constructed by genomic integration of the following plasmids (Mantiero et al., 2011): bpZ46 (pRS305 Gal1-10 Cdc45/Sld7, KasI digested); bpZ109 (pRS304 Gal1-10 Sld2/Dpb11, HindIII digested); bpZ287 (pRS306 Sld3/Dbf4, NdeI digested).
Genotypes are listed in Table S1. All strains were routinely grown in YPAD at 30°C.
Method details
Yeast histone purification
Yeast cells were grown in 1l of YPD for 3-4 generations to OD600 = 1. Cells were then collected, washed with water and frozen in liquid nitrogen. The cell pellet was resuspended in SP buffer (1M sorbitol; 50mM potassium phosphate pH 6.8, 14mM β-mercaptoethanol) and spheroplasted by zymolyase digestion. Nuclei were then prepared by Douncing in Lysis buffer (18% Ficoll-400 [w/v]; 20mM potassium phosphate pH 6.8; 1mM MgCl2; 0.5mM EDTA) supplemented with both protease and phosphatase inhibitors (Roche). After spinning down in a benchtop centrifuge, supernatant was recovered and nuclei were pelleted by spinning at 50,000g in a Beckman SW-41 Ti rotor. Nuclei were then resuspended in NP buffer (0.34M sucrose; 20mM Tris-HCl pH 7.4; 50mM KCl; 5mM MgCl2) supplemented with both protease and phosphatase inhibitors (Roche) and pelleted by spinning at 30,000g in a Beckman SW-41 Ti rotor. Nuclei were washed three times with Buffer A (10mM Tris-HCl pH 8.0; 0.5% NP-40 [v/v]; 75mM NaCl) supplemented with both protease and phosphatase inhibitors (Roche). Histones were then extracted in Buffer B (10mM Tris-HCl pH 8; 400mM NaCl; 0.2M H2SO4) and TCA precipitated.
Western blot and dot-blot
Total protein extracts were prepared as previously described (Rossmann and Stillman, 2013). Protein extracts were separated by sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) in the specified acrylamide % gels. Biotinylated peptides were separated in 17% Tris-Tricine gels and transferred to PVDF membranes as described (Kirmizis et al., 2007). Dot-blot analysis of peptides was performed by spotting serial dilutions onto a PVDF membrane, air drying them and processed following manufacturer’s instructions. Western blots were developed within the linear range of exposure using a ChemiDoc MP imaging system (Bio-Rad).
Flow cytometry analysis
Cell pellets were resuspended in ice-cold 70% [v/v] ethanol and incubated at 4°C overnight. Cells were then washed with 50mM Tris-HCl pH 8.0 and incubated with 0.4mg/ml RNAse A at 37°C overnight. After treatment with pepsin for 30 min at 37°C, cells were resuspended in 1μM Sytox Green solution and analyzed using BD FACSCalibur flow cytometer.
Recombinant protein purification
Plasmids used for recombinant expression are described in Table S2. Expression of MBP-SET2, MBP-SET2N198Q and MBP-SET2Y149A fusions in BL21 (DE3) cells was induced with 0.3mM IPTG over night at 18°C. Cells were then lysed by sonication in TBS + 1% Triton-X100 supplemented with protease inhibitors, and clarified lysate was incubated with Amylose Resin for 2h at 4°C. Immobilized fusion proteins were washed extensively with TBS + 1% Triton-X100 and then with TBS. Immobilized recombinant proteins were then used for methyltransferase reactions.
Expression of MCM2 (aa1-aa480)-His6 fusions in BL21 (DE3) cells was induced with 1mM IPTG over night at 18°C. Cells were then lysed by sonication in TBS + 1% Triton-X100 +10mM imidazole supplemented with protease inhibitors, and clarified lysate was incubated with Ni-Sepharose beads for 2h at 4°C. Immobilized fusion proteins were washed extensively with TBS + 1% Triton-X100, then with TBS+20mM imidazole and eluted with TBS+400mM imidazole. Full length Mcm2 was dialyzed O/N at 4C against TBS+10% glycerol. Mcm2 (aa1-aa480) was further purified by HPLC. Fractions containing Mcm2(1-480) were pooled together and its concentration measured by Bradford assay.
PtA-Set1p Complex (COMPASS) Purification
PtA-Set1p and PtA-set1ΔC92 under the control of the PNOP promoter, were expressed in C13 yeasts. Purification of the fusion proteins and complex interacting factors, onto IgG-Sepharose resin, were performed as previously described (Santos-Rosa et al., 2002). Beads bound purified complexes were then used for methyltransferase reactions.
Reconstitution of recombinant nucleosomes
Recombinant human core histone proteins H2A, H2B, H3.1, and H4 were expressed in E. coli BL21(DE3)/RIL cells from pET21b(+) (Novagen) vectors and purified by denaturing gel filtration and ion exchange chromatography essentially as described (Dyer et al., 2003). The H3.1 K36R mutant was generated from the wild type pET21-hH3.1 expression plasmid by site directed mutagenesis. Histone octamers (WT and H3 K37R mutant) were refolded from the purified histones, purified by size exclusion chromatography on a Superdex 200 gel filtration column (GE Healthcare), and then assembled into nucleosomes with biotinylated 601 DNA via salt deposition dialysis as previously described (Bartke et al., 2010; Dyer et al., 2003). Biotinylated nucleosomal DNA containing the 601 nucleosome positioning sequence was prepared from a pUC19 vector containing 16 tandem repeats of the 601 sequence as described (Bartke et al., 2010; Lowary and Widom, 1998).
In vitro methyltransferase assays
Methylation reactions were performed for 1h at 30°C in JB buffer (50 mM Tris-HCl pH 8.5; 20mM KCl; 10mM MgCl2; 0.5mM DTT; 250mM sucrose; 0.2% β-maltoside) with either 32μM SAM (NEB) or 0.1MBq 3H-SAM, using 35 ng of MBP-Set2 (full length WT) or MBP-Set2 Y149A (full length YA) enzyme and 250ng nucleosomes (equivalent to 62.5 ng human H3.1 WT or human H3.1 K37R). Reactions on histones were separated by SDS-PAGE in 16% polyacrylamide gels, whereas reactions on peptides were resolved in 17% polyacrylamide Tricine gels. For radioactive assays, gels were stained with Coomassie brilliant blue to visualize proteins and peptides, dried and exposed to film. For non-radioactive assays, gels were transferred to nitrocellulose membranes and analyzed by western blot as described above. Set1 catalyzed methyltransferase reactions were performed as above but replacing recombinant MBP-Set2 by PtA-Set1p and PtA-set1ΔC92 purified from 1gr spheroplasts /reaction.
In vitro peptide pull-downs
15μg of each biotinylated H3 peptide were diluted in 0.5ml of TBS+1% [v/v] NP-40 and incubated with 150μl of Dynabeads MyOne Streptavidin T1 for 60 minutes at room temperature. Beads were washed once with TBS/500mM NaCl+1% [v/v] NP-40, once with TBS/250mM NaCl+1% [v/v] NP-40, twice with TBS+1% [v/v] NP-40 and resuspended in 150μl TBS+1% [v/v] NP-40. 5μl beads slurry was spotted onto PVDF membrane and stained with Coomassie Blue to monitor binding for each peptide. For the pull downs, 50 μl Mcm2(aa1-aa480)-His6, corresponding to 5μg of protein, were incubated with 20μl beads in a total volume of 300μl TBS/200mM NaCl+1% [v/v] NP-40 during 1 hour at room temperature on a bench wheel. Beads were washed 4 times with binding buffer and bound material eluted by boiling the beads in 25 μl SDS Laemmli buffer, resolved by SDS-PAGE and visualized by Coomasie staining.
Yeast chromatin immunoprecipitation
Yeast strains were grown in YPAD medium at 30°C and synchronized to G1 by addition of α-factor (10μg/ml final concentration). 100ml of yeast cultures per ChIP experiment were collected at the indicated times. Cells were cross-linked with 1% formaldehyde for 15 minutes at room temperature, and the reaction was quenched with 125mM glycine. Cells were resuspended in ChIP SDS buffer (1% SDS, 10mM EDTA, 50mM Tris-HCl pH 8.0) supplemented with protease inhibitors (Roche) and disrupted with glass beads by using a FastPrep instrument (MP Biomedicals). Chromatin was sonicated (Bioruptor Pico, Diagenode; 9 cycles, 30 s on/off) to yield an average DNA fragment of 300-500 base pairs, and diluted 10 times in ChIP IP buffer (0.01% SDS, 1.1%Triton X-100, 167mM NaCl, 1.2mM EDTA, 16.7mM Tris-HCl pH 8.0) supplemented with protease inhibitors (Roche) prior to overnight immunoprecipitation on rotation at 4°C. Next day, 50μl of protein A Dynabeads were added, and samples were incubated again on rotation at 4°C for 2 hours. Dynabeads were washed twice with the following buffers: TSE 150 (1% Triton X-100, 0.1%SDS, 150mM NaCl, 2mM EDTA, 20mM Tris-HCl pH 8.0), TSE 500 (1% Triton X-100, 0.1%SDS, 500mM NaCl, 2mM EDTA, 20mM Tris-HCl pH 8.0) and LiCl buffer (0.25M LiCl, 1% NP-40, 1% deoxycholate, 1mM EDTA, 10mM Tris-HCl pH 8.0). DNA was eluted for 30 min at 37°C in elution buffer (100mM NaHCO3, 1% SDS), and cross-linking was reverted by overnight incubation at 65°C. Samples were treated with 0.5mg/ml of RNase A at 37°C for 2 h. DNA was purified using the ChIP DNA Clean & Concentrator kit (Zymo Research). Relative DNA amounts were determined by qPCR using Fast SYBR Green Master Mix (Applied Biosystems). Primer pairs used for amplification are available upon request. For each strain and/or condition, three independent colonies were grown and processed. The mean values +SEM derived from three biological replicates were plotted using Prism (GraphPad Software, Inc.).
S. pombe spike
S.cerevisiae Mcm2-HA ChIPs were spiked with S.pombe expressing a Mcm3-HA allele as independent internal normalizer. Crosslink treatment (1% folmaldehyde, 15 minutes) and sonication conditions (9 cycles 30 sec ON/30 sec OFF) yielded similar fragment size for both yeasts strains as monitored by tape station (256-261 bp, below). Thus, a volume of S.pombe, corresponding to 5% of each S.cerevisiae culture, was added to each WT and H3K37R culture prior to formaldehyde treatment. All samples (containing S.pombe and S.cerevisiae cells) were processed for ChIP in parallel, following our standard procedure.
BrdU-DNA labelling and immunoprecipitation
Immunoprecipitation of BrdU nascent DNA was adapted from Lengronne et al., 2001. Per each strain, 500ml of culture was synchronized in G1 by addition of α-factor (10μg/ml final concentration) for 1h and 30’. Half culture (250ml) remained untreated (-BrdU) whereas 5ml BrdU (25mg/ml stock) was added to the other half (+BrdU). Both flasks were incubated for a further 30min at 30°C, then cells were collected by centrifugation and resuspended into 30°C warmed YPAD medium containing 25ml HU (2M stock) and incubated for 1.25 hours. BrdU (+) and (-) cultures were immediately transferred to ice/water bath and 2.5ml NaN3 (10% stock) was added to each one. After 10min, cells were collected by centrifugation, washed twice with TBS and stored at - 20°C. Incorporation of BrdU into DNA was monitored by dot blot with an anti-BrdU antibody as described (Lengronne et al., 2001).
For BrdU Immunoprecipitation, each of the above cell pellets was shaken five times for 2 min in NIB buffer (17% [v/v] glycerol, 50mM MOPS buffer, 150mM potassium acetate, 2mM MgCl2, 500μM spermidine, 150μM spermine at pH 7.2) with zirconium beads on a Vibrax (VXR basic, Ika) at 4°C. Genomic DNA extraction, from the above cultures was performed using Qiagen Genomic DNA Isolation Kit and Qiagen genomic-tip 100/G (Cat. No. 19060 and Cat. No.10243). DNA was fragmented using sonication (∼200- to 500-base-pair [bp] size range), boiled at 95°C for 10 minutes, placed on ice for 10 minutes and precipitated O/N at 4°C with 5μg mouse anti-BrdU antibody coupled to Dynabeads M-280 Sheep anti-mouse IgG. Immunoprecipitated material was washed and eluted as described for ChIP.
Specificity of the IPs was tested comparing samples +BrdU and -BrdU at replication origins (ARS) and comparing samples + BrdU at ARS versus non replication origins. For the immunoprecipitation of BrdU nascent DNA under overexpression of the helicase activators, 3 independent colonies of HSR727 and HRS729 were grown in YPA-Raffinose to OD600nm=0.4 and then synchronized in G1 by addition of α-factor (10μg/ml final concentration) for 1.5 hours. To half of a culture (100ml), glucose (2% final concentration) and 1.3ml BrdU (50mg/ml stock) was added. To the other half, galactose (2% final concentration) and 1.3ml BrdU (50mg/ml stock) was added. Both flasks were incubated for a further 30min at 30°C, then cells were collected by centrifugation and resuspended into 100ml of 30°C pre-warmed YPA-2%glucose or YPA-2%galactose medium containing 25ml HU (2M stock) plus 1.2ml BrdU (50mg/ml stock) and incubated for 70 minutes. YPA-Glucose and YPA-Galactose cultures were immediately transferred to ice/water bath and 2.5ml NaN3 (10% stock) added to each one. After 10min, cells were collected by centrifugation, washed twice with TBS and store at -20°C. Incorporation of BrdU onto DNA was monitored by dot blot analysis with an anti-BrdU antibody as described (Lengronne et al., 2001).
Quantitative PCR was performed with primers specific to each genomic location. All sequences are available upon request.
Library preparation
Library preparations followed the NEXTflex Rapid DNA-Seq protocol (2015; Bioo Scientific) with a few adjustments. Illumina adaptors were substituted with Bioo NEXTflex adaptors. Agilent Technologies High Sensitivity D1000 was used for determining the fragment size on the Agilent 4200 TapeStation. Libraries were quantified using Invitrogen Qubit dsDNA HS Assay. Equal molarity of each barcoded library was pulled together to multiplex. Single-end 50 bp Next-generation sequencing was performed on a HiSeq 4000 (Illumina).
Analysis of BrdU IP data
Raw reads were processed with Trimmomatic (v0.39) to remove adapters (Bolger et al., 2014), retaining reads longer than 20nt. Reads were then mapped to the reference genome (sacCer3) with BWA (-n 3 -k 2 -R 300) (Li and Durbin, 2009) and filtered to remove multimappers based on the alignments quality (samtools view -q 10) (Li et al., 2009). Finally, PCR duplicates were removed with Picard MarkDuplicates (Broad Institute; GitHub repository, 2019).
In order to estimate the library fragment size we used the correlateReads function of the csaw package (Lun and Smyth, 2015) to estimate the cross-correlation coefficients. We then selected as a fragment size the distance that maximised the loess-smoothed cross-correlation.
Coverage plots were produced by computing the read coverage of annotated ARS (see below) with the featureScores function of the Repitools package (Statham et al., 2010), using a window size of 150 and a smoothing size equal to the fragment length.
To identify enriched binding regions of BrdU-IP-seq outside canonical ARS, we followed the method in Yoshida et al. (Yoshida et al., 2014) but improved the bin size to a single base pair resolution. We then normalized the binned tag densities of IP sample by the densities of the matched input sample. MACS2 with default setting was used for peak calling for the histone mutation samples versus wild-type samples, and p-value <= 0.05 was used to identify confident peaks.
Co-localization of H3K37R unique peaks on ARS or ACS
To determine how many H3K37R unique peaks are occurring at ARS, we downloaded ARS table from ORiDB (Siow et al., 2012) and used the first “status (ARS)” column to generate (1) “Confirmed ARS” and (2) “Dubious/Likely ARS” in Figure 6F. We used nr-ACS list from Eaton et al. (Eaton et al., 2010) for (3) “nr-ACS”. We downloaded ACS genomic location data from OriScan database (Breier et al., 2004) and lift-over the reference genome to SacCer3 for (4) “ACS like”. The (5) “Other” indicate any peaks that falls outside of any regions of 1-4.
ARS annotation and efficiency estimates
To define ARS efficiency, we averaged the coverage of experimental replicates and then further summarized the coverage of each ARS by averaging all the coverage of all windows within 1kb of the ARS center. We then fitted a Gaussian Mixture Model to find the parameters of two normal distributions, representing inactive and active origins respectively. We defined as “Late/Inefficient” all ARS having a coverage less than 2 standard deviations above the mean of the first normal distribution. The remaining ARS were then equally split into the “Medium” and “Early/Efficient” categories based on their coverage (see Figure S5C).
The annotation of ARS was obtained from OriDB. All ARS with “Confirmed” status in OriDB were consider, with the exception of ARS1216.5 which was excluded due to its overlap with repetitive rDNA and its extreme coverage in the Input samples.
Analysis of H3K37me1 ChIP-seq
Raw reads were pre-processed and mapped as outlined for the BrdU libraries. To calculate the K37me1 around ARS, BAM files were converted to GRanges (Lawrence et al., 2013) objects and replicates were average using the mergeReplicates function of Repitools (Statham et al., 2010). We then used the featureScores function of Repitools package to calculate the coverage of 150nt windows around ARS, using as a smoothing distance the library fragmented size estimated as explained before.
To test for significant difference at 5min and 30min vs G1, we averaged the coverage of all windows within 2kb of the ARD mid-point, and then applied the Wilcoxon rank sum test on the base 2 logarithm of the coverage ratio between H3K37me1 and H3.
To calculate the coverage at TSSs, we repeated the same procedure as above, but calculating coverage of regions within 2.5kb of SGD annotated TSSs, using a window size of 50 and the same smoothing distance as before.
Analysis of Mcm2 ChIP-seq
Reads were adapter-trimmed as outlined before and then mapped with bwa to a combined reference database consisting of both S. cerevisiae and S. pombe genomes. Uniquely mapping reads were filtered as outlined before and the resulting BAM files were then split by reference chromosomes with the split utility of bamtools. After splitting, BAM files were combined again into two new BAM files containing alignments belonging to S. cerevisiae and S. pombe, respectively. To calculate Mcm2 coverage around ARS, we first converted the BAM files containing S. cerevisiae alignments to GRanges objects, and then used the featureScores function of the Repitools package to calculate the coverage of 50nt windows around ARS, using as a smoothing distance the library fragmented size estimated as explained before. The resulting window-coverage counts were automatically normalised by library size by the featureScores function; therefore, to normalise by the coverage of the S. Pombe spike-in chromatin the window counts were divided by the total number of reads mapped to S. Pombe and multiplied by the total number of reads mapped to S. cerevisiae (this latter step is required in order to revert the automatic normalisation performed by featureScores). Downstream analysis and statistical testing were then done as outlined for the H3K37me1 ChIP-seq experiment.
Cell culture
hTERT immortalized RPE-1 cells were cultured in Dulbecco’s Modified Eagle’s Medium/Nutrient Mixture F-12 Ham (D8437; Sigma-Aldrich) supplemented with 10% (vol/vol) fetal bovine serum (Sigma-Aldrich), 100U/ml penicillin, 100μg/ml streptomycin (Sigma-Aldrich) and 2mM L-glutamine.
Human cell chromatin immunoprecipitation
Exponentially growing cells were plated in 150mm dishes at 30–40% confluence and incubated at 37°C overnight. Next day, 2mM thymidine was added and cells were incubated for 18h at 37°C. Thymidine was removed by washing with PBS and cells were incubated with fresh medium for 9h at 37°C before a second round of 2mM thymidine block. Cells were then washed with PBS and released into the cycle. Cells were cross-linked with 1% formaldehyde for 10 minutes at 37°C, and the reaction was quenched with 125mM glycine for 5 minutes at room temperature. Cells were washed twice with cold PBS and scraped in cold PBS supplemented with protease inhibitors (Roche). Cells were lysed in in ChIP lysis buffer I (5mM PIPES KOK pH 8; 85mM KCl; 0.5% NP-40) for 10 minutes on ice, spun down and resuspended in ChIP lysis buffer II (1% SDS, 10mM EDTA, 50mM Tris-HCl pH 8.0) supplemented with protease inhibitors (Roche). Chromatin was sonicated (Bioruptor, Diagenode; 20 cycles, 30s on/off) to yield an average DNA fragment of 300-500 base pairs, and diluted 10 times in ChIP IP buffer (0.01% SDS, 1.1%Triton X-100, 167mM NaCl, 1.2mM EDTA, 16.7mM Tris-HCl pH 8.0) supplemented with protease inhibitors (Roche) prior to overnight immunoprecipitation with rotation at 4°C. Next day, 50μl of protein A Dynabeads were added, and samples were incubated again with rotation at 4°C for 2 hours. Dynabeads were washed twice with the following buffers: TSE 150 (1% Triton X-100, 0.1%SDS, 150mM NaCl 2mM EDTA, 20mM Tris-HCl pH 8.0), TSE 500 (1% Triton X-100, 0.1%SDS, 500mM NaCl, 2mM EDTA, 20mM Tris-HCl pH 8.0) and LiCl buffer (0.25 M LiCl, 1% NP-40, 1% deoxycholate, 1mM EDTA, 10mM Tris-HCl pH 8.0). DNA was eluted for 30min at 37°C in elution buffer (100mM NaHCO3, 1% SDS), and cross-linking was reverted by overnight incubation at 65°C. Samples were treated with 0.5mg/ml of RNase A at 37°C for 2 h. DNA was purified using the Qiagen PCR purification kit. Relative DNA amounts were determined by qPCR using SYBR Green Master Mix (Applied Biosystems). Primer pairs used for amplification are available upon request.
Direct enzyme-linked immunosorbent assay (ELISA)
Serial dilutions of biotinylated unmodified and K37me1 peptides were immobilized on streptavidin-coated 96-well plates and the ELISA was performed using a 1:5000 dilution of the anti-H3K37me1 antibody. The ELISA was developed using horseradish peroxidase (HRP)-coupled secondary anti-rabbit antibody and SuperSignal ELISA Pico and quantified in using a ClarioStar plate reader.
Key resources table.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-H3 K37me1 | This study/Abcam | Ab215728 |
| Anti-H3 | Abcam | RRID:AB_302613 |
| Anti-H3 K36me1 | Abcam | RRID:AB_306964 |
| Anti-H3 K36me2 | Abcam | RRID:AB_1280939 |
| Anti-H3 K36me3 | Abcam | RRID:AB_306966 |
| Anti-H3 K4me1 | Abcam | RRID:AB_306847 |
| Anti-H3 K4me2 | Abcam | RRID:AB_2560996 |
| Anti-H3 K4me3 | Abcam | RRID:AB_306649 |
| Anti-SETD2 | Abcam | RRID:AB_2185782 |
| Anti-SETD1A | Abcam | RRID:AB_1951955 |
| Anti-TUBULIN | SIGMA | RRID:AB_477593 |
| Anti-HA | Abcam | RRID:AB_307019 |
| Anti-GFP | Abcam | RRID:AB_303395 |
| Anti-BrdU | BD Bioscience | RRID:AB_395993 |
| Anti-ORC6 | Stillman B. | - |
| Anti-Biotin | SIGMA | RRID:AB_258625 |
| Anti-PAP | SIGMA | RRID:AB_1079562 |
| Bacterial Strains | ||
| BL21 (DE3) | NEB | Cat.No. C2571 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Biotinylated H3 K37me1 peptide (aa28-aa48) | GeneCust | This study |
| Biotinylated H3 K36me1peptide (aa28-aa48) | GeneCust | This study |
| Biotinylated H3 K36me1 K37me1 peptide (aa28-aa48) | GeneCust | This study |
| Biotinylated H3 unmodified peptide (aa28-aa48) | GeneCust | This study |
| Biotinylated H3 K36me2 peptide (aa28-aa48) | GeneCust | This study |
| Biotinylated H3 K36me3 peptide (aa28-aa48) | GeneCust | This study |
| Sulfolink Coupling Resin | ThermoFisher | Cat.No. 20402 |
| Protease inhibitors | Roche | Cat.No. 11697498001 |
| SAM | NEB | B9003S |
| 3H-SAM | PerkinElmer | NET155V250UC |
| 5-Bromo-2’-deoxyuridine | Sigma | Cat.No. 10280879001 |
| α-factor | GenScript | Cat.No.RP01002 |
| Sytox Green | Invitrogen | Cat.No. S7020 |
| Amylose Resin High Flow | NEB | E8022L |
| IgG Sepharose 6 Fast Flow | GE Healthcare | Cat.No. 17-0969-01 |
| Ni Sepharose 6 Fast Flow | GE Healthcare | Cat.No. 17-5318-01 |
| Dynabeads M-280 | DYNAL | Cat.No. 112.02 |
| Calf histones | Roche | Cat.No. 10223565001 |
Supplementary Material
Acknowledgments
Our gratitude to F. Puddu, S. Siniossoglou and P. Zegerman for critically reading the manuscript and for technical help. We are very grateful to Hiroyuki Araki for the generous gift of the antibodies to detect the MCM activators. We are most indebted to A.J. Bannister for valuable discussions and for the critical reading/editing of this manuscript.
Funding
The Kouzarides laboratory is supported by Cancer Research UK (grant references RG72100/ RG96894) and benefits from core support from the Wellcome Trust (Core Grant reference WT203144) and Cancer Research UK (grant reference C6946/A24843). For the purpose of open access, the author has applied a CC BY public copyright licence to any Author Accepted Manuscript version arising from this submission. G. Millan-Zambrano was funded by an EMBO long-term fellowship (ALTF907-2014). K.Tzelepis was supported by a Sir Henry Wellcome Fellowship (grant reference RG94424). T. Bartke was supported by the European Research Council (ERC StG 309952) and the Helmholtz Association.
Footnotes
Authors Contributions
H.S.-R.: Conceptualized idea, designed Methodology, performed Experiments, Wrote and Edited the paper; G.M.-Z.: Designed Methodology, performed Experiments, Wrote and Edited the paper; N.H: Software and Bioinformatic Analysis of Mcm2, BrdU replication profiling and contributed to Editing ; T.L.: Software and Bioinformatic Analysis of H3K37me1, Mcm2 and replication profiling; L.P.: Supported software, ARS annotation and efficiency estimates; M.K.: Performed Experiments and contributed to Editing; S.N and T.B: Provided in vitro reconstituted recombinant nucleosomes and contributed to Writing the manuscript; K.Z: Provided MOLM13/IOC2 Cre cell extracts; T.K: Provided Funding and General Supervision.
Declaration of Interests
T.K. is a co-founder and shareholder of Abcam Plc and co-founder of Storm Therapeutics Ltd., Cambridge, UK. T.L. is a consultant for Storm Therapeutics Ltd., Cambridge, UK.
References
- Azmi IF, Watanabe S, Maloney MF, Kang S, Belsky JA, MacAlpine DM, Peterson CL, Bell SP. Nucleosomes influence multiple steps during replication initiation. Elife. 2017;6:e22512. doi: 10.7554/eLife.22512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartke T, Vermeulen M, Xhemalce B, Robson SC, Mann M, Kouzarides T. Nucleosome-interacting proteins regulated by DNA and histone methylation. Cell. 2010;143:470–484. doi: 10.1016/j.cell.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell SP, Labib K. Chromosome duplication in Saccharomyces cerevisiae. Genetics. 2016;203:1027–1067. doi: 10.1534/genetics.115.186452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belsky JA, Macalpine HK, Lubelsky Y, Hartemink AJ, Macalpine DM. Genome-wide chromatin footprinting reveals changes in replication origin architecture induced by pre-RC assembly. Genes Dev. 2015;29:212–224. doi: 10.1101/gad.247924.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas D, Takahata S, Xin H, Dutta-Biswas R, Yu Y, Formosa T, Stillman DJ. A role for Chd1 and Set2 in negatively regulating DNA replication in Saccharomyces cerevisiae. Genetics. 2008;178:649–659. doi: 10.1534/genetics.107.084202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breier AM, Chatterji S, Cozzarelli NR. Prediction of Saccharomyces cerevisiae replication origins. Genome Biol. 2004;5:R22. doi: 10.1186/gb-2004-5-4-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broad Institute; GitHub repository. Picard Toolkit. 2019 [Google Scholar]
- Cayrou C, Coulombe P, Vigneron A, Stanojcic S, Ganier O, Peiffer I, Rivals E, Puy A, Laurent-Chabalier S, Desprat R, et al. Genome-scale analysis of metazoan replication origins reveals their organization in specific but flexible sites defined by conserved features. Genome Res. 2011;21:1438–1449. doi: 10.1101/gr.121830.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhury R, Singh S, Arumugam S, Roguev A, Francis Stewart A. The set1 complex is dimeric and acts with jhd2 demethylation to convey symmetrical h3k4 trimethylation. Genes Dev. 2019;33:550–564. doi: 10.1101/gad.322222.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das SP, Rhind N. How and why multiple MCMs are loaded at origins of DNA replication. BioEssays. 2016;38:613–617. doi: 10.1002/bies.201600012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das SP, Borrman T, Liu VWT, Yang SCH, Bechhoefer J, Rhind N. Replication timing is regulated by the number of MCMs loaded at origins. Genome Res. 2015;25:1886–1892. doi: 10.1101/gr.195305.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diffley JFX. Quality control in the initiation of eukaryotic DNA replication. Philos Trans R Soc B Biol Sci. 2011;366:3545–3553. doi: 10.1098/rstb.2011.0073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Q, MacAlpine DM. Defining the replication program through the chromatin landscape. Crit Rev Biochem Mol Biol. 2011;46:165–179. doi: 10.3109/10409238.2011.560139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer PN, Edayathumangalam RS, White CL, Bao Y, Chakravarthy S, Muthurajan UM, Luger K. Reconstitution of Nucleosome Core Particles from Recombinant Histones and DNA. Methods Enzymol. 2003;375:23–44. doi: 10.1016/s0076-6879(03)75002-2. [DOI] [PubMed] [Google Scholar]
- Eaton ML, Galani K, Kang S, Bell SP, MacAlpine DM. Conserved nucleosome positioning defines replication origins. Genes Dev. 2010;24:748–753. doi: 10.1101/gad.1913210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitag M. Histone Methylation by SET Domain Proteins in Fungi. Annu Rev Microbiol. 2017;71:413–439. doi: 10.1146/annurev-micro-102215-095757. [DOI] [PubMed] [Google Scholar]
- Ishimi Y, Komamura Y, You Z, Kimura H. Biochemical function of mouse minichromosome maintenance 2 protein. J Biol Chem. 1998;273:8369–8375. doi: 10.1074/jbc.273.14.8369. [DOI] [PubMed] [Google Scholar]
- Janke C, Magiera MM, Rathfelder N, Taxis C, Reber S, Maekawa H, Moreno-Borchart A, Doenges G, Schwob E, Schiebel E, et al. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 2004;21:947–962. doi: 10.1002/yea.1142. [DOI] [PubMed] [Google Scholar]
- Kan J, Zou L, Zhang J, Wu R, Wang Z, Liang C. Origin recognition complex (ORC) mediates histone 3 lysine 4 methylation through cooperation with Spp1 in Saccharomyces cerevisiae. J Biol Chem. 2008;283:33803–33807. doi: 10.1074/jbc.C800182200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane SM, Roth R. Carbohydrate metabolism during ascospore development in yeast. J Bacteriol. 1974;118:8–14. doi: 10.1128/jb.118.1.8-14.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirmizis A, Santos-Rosa H, Penkett CJ, Singer MA, Vermeulen M, Mann M, Bähler J, Green RD, Kouzarides T. Arginine methylation at histone H3R2 controls deposition of H3K4 trimethylation. Nature. 2007;449:928–932. doi: 10.1038/nature06160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knott SRV, Viggiani CJ, Tavaré S, Aparicio OM. Genome-wide replication profiles indicate an expansive role for Rpd3L in regulating replication initiation timing or efficiency, and reveal genomic loci of Rpd3 function in Saccharomyces cerevisiae. Genes Dev. 2009;23:1077–1090. doi: 10.1101/gad.1784309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence M, Huber W, Pagès H, Aboyoun P, Carlson M, Gentleman R, Morgan MT, Carey VJ. Software for Computing and Annotating Genomic Ranges. PLoS Comput Biol. 2013;9:e1003118. doi: 10.1371/journal.pcbi.1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengronne A, Pasero P, Bensimon A, Schwob E. Monitoring S phase progression globally and locally using BrdU incorporation in TK + yeast strains. Nucleic Acids Res. 2001;29:1433–1442. doi: 10.1093/nar/29.7.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li QQ, Hao JJ, Zhang Z, Krane LS, Hammerich KH, Sanford T, Trepel JB, Neckers L, Agarwal PK. Proteomic analysis of proteome and histone post-translational modifications in heat shock protein 90 inhibition-mediated bladder cancer therapeutics. Sci Rep. 2017;7:201. doi: 10.1038/s41598-017-00143-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipford JR, Bell SP. Nucleosomes Positioned by ORC Facilitate the Initiation of DNA Replication text can significantly alter origin function. For example, many potential origins of replication function well in the context of a plasmid, but poorly or not at all in their. Mol Cell. 2001;7:21–30. doi: 10.1016/s1097-2765(01)00151-4. [DOI] [PubMed] [Google Scholar]
- Lowary PT, Widom J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J Mol Biol. 1998;276:19–42. doi: 10.1006/jmbi.1997.1494. [DOI] [PubMed] [Google Scholar]
- Luger K, Mader A, Richmond R, Sargent D, Richmond T. Crystal structure of the nucleosome core particle at 2.8 angstrom resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- Lun ATL, Smyth GK. Csaw: A Bioconductor package for differential binding analysis of ChIP-seq data using sliding windows. Nucleic Acids Res. 2015;44:e45. doi: 10.1093/nar/gkv1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macheret M, Halazonetis TD. Intragenic origins due to short G1 phases underlie oncogene-induced DNA replication stress. Nature. 2018;555:112–116. doi: 10.1038/nature25507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantiero D, MacKenzie A, Donaldson A, Zegerman P. Limiting replication initiation factors execute the temporal programme of origin firing in budding yeast. EMBO J. 2011;30:4805–4814. doi: 10.1038/emboj.2011.404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDaniel SL, Strahl BD. Shaping the cellular landscape with Set2/SETD2 methylation. Cell Mol Life Sci. 2017;74:3317–3334. doi: 10.1007/s00018-017-2517-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori S, Shirahige K. Perturbation of the activity of replication origin by meiosis-specific transcription. J Biol Chem. 2007;282:4447–4452. doi: 10.1074/jbc.M609671200. [DOI] [PubMed] [Google Scholar]
- Morohashi N, Mitchell AP, Shimizu M. Effect of histone methyltransferase gene mutations on sporulation in S. cerevisiae. Nucleic Acids Symp Ser (Oxf) 2005:325–326. doi: 10.1093/nass/49.1.325. [DOI] [PubMed] [Google Scholar]
- Orlando DA, Chen MW, Brown VE, Solanki S, Choi YJ, Olson ER, Fritz CC, Bradner JE, Guenther MG. Quantitative ChIP-Seq normalization reveals global modulation of the epigenome. Cell Rep. 2014;9:1163–1170. doi: 10.1016/j.celrep.2014.10.018. [DOI] [PubMed] [Google Scholar]
- Park IY, Powell RT, Tripathi DN, Dere R, Ho TH, Blasius TL, Chiang YC, Davis IJ, Fahey CC, Hacker KE, et al. Dual Chromatin and Cytoskeletal Remodeling by SETD2. Cell. 2016;166:950–962. doi: 10.1016/j.cell.2016.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrossian TC, Clarke SG. Multiple Motif Scanning to Identify Methyltransferases from the Yeast Proteome. Mol Cell Proteomics. 2009;8:1516–1526. doi: 10.1074/mcp.M900025-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryde F, Jain D, Kerr A, Curley R, Mariotti FR, Vogelauer M. H3 K36 methylation helps determine the timing of Cdc45 association with replication origins. PLoS One. 2009;4:e5882. doi: 10.1371/journal.pone.0005882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puddu F, Herzog M, Selivanova A, Wang S, Zhu J, Klein-Lavi S, Gordon M, Meirman R, Millan-Zambrano G, Ayestaran I, et al. Genome architecture and stability in the Saccharomyces cerevisiae knockout collection. Nature. 2019;573:416–420. doi: 10.1038/s41586-019-1549-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren C, Liu S, Ghoshal K, Hsu P-H, Jacob ST, Marcucci G, Freitas MA. Simultaneous metabolic labeling of cells with multiple amino acids: Localization and dynamics of histone acetylation and methylation. PROTEOMICS – Clin Appl. 2007;1:130–142. doi: 10.1002/prca.200600631. [DOI] [PubMed] [Google Scholar]
- Riera A, Barbon M, Noguchi Y, Reuter LM, Schneider S, Speck C. From structure to mechanism— understanding initiation of DNA replication. Genes Dev. 2017;31:1073–1088. doi: 10.1101/gad.298232.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzardi LF, Dorn ES, Strahl BD, Cook JG. DNA replication origin function is Promoted by H3K4 di-methylation in Saccharomyces cerevisiae. Genetics. 2012;192:371–384. doi: 10.1534/genetics.112.142349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossmann MP, Stillman B. Immunoblotting Histones from Yeast Whole-Cell Protein Extracts. Cold Spring Harb Protoc. 2013;8:625–630. doi: 10.1101/pdb.prot067116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, Emre NCT, Schreiber SL, Mellor J, Kouzarides T. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419:407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- Schneider J, Wood A, Lee JS, Schuster R, Dueker J, Maguire C, Swanson SK, Florens L, Washburn MP, Shilatifard A. Molecular regulation of histone H3 trimethylation by COMPASS and the regulation of gene expression. Mol Cell. 2005;19:849–856. doi: 10.1016/j.molcel.2005.07.024. [DOI] [PubMed] [Google Scholar]
- Shen Y, Mevius DEHF, Caliandro R, Carrozzini B, Roh Y, Kim J, Kim S, Ha SC, Morishita M, di Luccio E. Set7 Is a H3K37 Methyltransferase in Schizosaccharomyces pombe and Is Required for Proper Gametogenesis. Structure. 2019;27:631–638.:e8. doi: 10.1016/j.str.2019.01.011. [DOI] [PubMed] [Google Scholar]
- Shilatifard A. The COMPASS Family of Histone H3K4 Methylases: Mechanisms of Regulation in Development and Disease Pathogenesis. Annu Rev Biochem. 2012;81:65–95. doi: 10.1146/annurev-biochem-051710-134100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoaib M, Walter D, Gillespie PJ, Izard F, Fahrenkrog B, Lleres D, Lerdrup M, Johansen JV, Hansen K, Julien E, et al. Histone H4K20 methylation mediated chromatin compaction threshold ensures genome integrity by limiting DNA replication licensing. Nat Commun. 2018;9:3704. doi: 10.1038/s41467-018-06066-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siow CC, Nieduszynska SR, Müller CA, Nieduszynski CA. OriDB, the DNA replication origin database updated and extended. Nucleic Acids Res. 2012;40:D682–D686. doi: 10.1093/nar/gkr1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollier J, Lin W, Soustelle C, Suhre K, Nicolas A, Géli V, De La Roche Saint-André C. Set1 is required for meiotic S-phase onset, double-strand break formation and middle gene expression. EMBO J. 2004;23:1957–1967. doi: 10.1038/sj.emboj.7600204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano I, Morafraile EC, Vázquez E, Antequera F, Segurado M. Different nucleosomal architectures at early and late replicating origins in Saccharomyces cerevisiae. BMC Genomics. 2014;15:791. doi: 10.1186/1471-2164-15-791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soudet J, Stutz F. Regulation of Gene Expression and Replication Initiation by Non-Coding Transcription: A Model Based on Reshaping Nucleosome-Depleted Regions: Influence of Pervasive Transcription on Chromatin Structure. BioEssays. 2019;41:e1900043. doi: 10.1002/bies.201900043. [DOI] [PubMed] [Google Scholar]
- Statham AL, Strbenac D, Coolen MW, Stirzaker C, Clark SJ, Robinson MD. Repitools: An R package for the analysis of enrichment-based epigenomic data. Bioinformatics. 2010;26:1662–1663. doi: 10.1093/bioinformatics/btq247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl BD, Grant PA, Briggs SD, Sun Z-W, Bone JR, Caldwell JA, Mollah S, Cook RG, Shabanowitz J, Hunt DF, et al. Set2 Is a Nucleosomal Histone H3-Selective Methyltransferase That Mediates Transcriptional Repression. Mol Cell Biol. 2002;22:1298–1306. doi: 10.1128/mcb.22.5.1298-1306.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima K, Yae T, Javaid S, Tam O, Comaills V, Morris R, Wittner BS, Liu M, Engstrom A, Takahashi F, et al. SETD1A modulates cell cycle progression through a miRNA network that regulates p53 target genes. Nat Commun. 2015;6:8257. doi: 10.1038/ncomms9257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima K, Matsuda S, Yae T, Drapkin BJ, Morris R, Boukhali M, Niederhoffer K, Comaills V, Dubash T, Nieman L, et al. SETD1A protects from senescence through regulation of the mitotic gene expression program. Nat Commun. 2019;10:2854. doi: 10.1038/s41467-019-10786-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi YH, Westfield GH, Oleskie AN, Trievel RC, Shilatifard A, Skiniotis G. Structural analysis of the core COMPASS family of histone H3K4 methylases from yeast to human. Proc Natl Acad Sci U S A. 2011;108:20526–20531. doi: 10.1073/pnas.1109360108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S, Nakato R, Katou Y, Shirahige K, Araki H. Origin association of Sld3, Sld7, and Cdc45 proteins is a key step for determination of origin-firing timing. Curr Biol. 2011 doi: 10.1016/j.cub.2011.11.038. [DOI] [PubMed] [Google Scholar]
- Tessarz P, Kouzarides T. Histone core modifications regulating nucleosome structure and dynamics. Nat Rev Mol Cell Biol. 2014;15:703–708. doi: 10.1038/nrm3890. [DOI] [PubMed] [Google Scholar]
- Unnikrishnan A, Gafken PR, Tsukiyama T. Dynamic changes in histone acetylation regulate origins of DNA replication. Nat Struct Mol Biol. 2010;17:430–437. doi: 10.1038/nsmb.1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viggiani CJ, Aparicio OM. New vectors for simplified construction of BrdU-Incorporating strains of Saccharomyces cerevisiae. Yeast. 2006;23:1045–1051. doi: 10.1002/yea.1406. [DOI] [PubMed] [Google Scholar]
- Vogelauer M, Rubbi L, Lucas I, Brewer BJ, Grunstein M. Histone acetylation regulates the time of replication origin firing. Mol Cell. 2002;10:1223–1233. doi: 10.1016/s1097-2765(02)00702-5. [DOI] [PubMed] [Google Scholar]
- Xu W, Aparicio JG, Aparicio OM, Tavaré S. Genome-wide mapping of ORC and Mcm2p binding sites on tiling arrays and identification of essential ARS consensus sequences in S. cerevisiae. BMC Genomics. 2006;7:276. doi: 10.1186/1471-2164-7-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K, Bacal J, Desmarais D, Padioleau I, Tsaponina O, Chabes A, Pantesco V, Dubois E, Parrinello H, Skrzypczak M, et al. The Histone Deacetylases Sir2 and Rpd3 Act on Ribosomal DNA to Control the Replication Program in Budding Yeast. Mol Cell. 2014;54:691–697. doi: 10.1016/j.molcel.2014.04.032. [DOI] [PubMed] [Google Scholar]
- Youdell ML, Kizer KO, Kisseleva-Romanova E, Fuchs SM, Duro E, Strahl BD, Mellor J. Roles for Ctk1 and Spt6 in Regulating the Different Methylation States of Histone H3 Lysine 36. Mol Cell Biol. 2008;28:4915–4926. doi: 10.1128/MCB.00001-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Lin W, Latham JA, Riefler GM, Schumacher JM, Chan C, Tatchell K, Hawke DH, Kobayashi R, Dent SYR. The Set1 methyltransferase opposes Ipl1 Aurora kinase functions in chromosome segregation. Cell. 2005;122:723–734. doi: 10.1016/j.cell.2005.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.