Abstract
Mechanical signals are pivotal ingredients in how cells perceive and respond to their microenvironments, and to synthetic biomaterials that mimic them. In spite of increasing interest in mechanobiology, probing the effects of physical cues on cell behavior remains challenging for a cell biology laboratory without experience in fabrication of biocompatible materials. Hydrogels are ideal biomaterials recapitulating the physical cues that natural Extracellular Matrices (ECM) delivers to cells. Here we streamlined protocols for the synthesis and functionalization of cell adhesive Polyacrylamide-based (PAA-OH) and fully-defined Polyethyleneglycol-based (PEG-RGD) hydrogels tuned at various rigidities for mechanobiology experiments, from 0.3 to >10kPa. We investigated the mechanosignaling properties of these hydrogels in distinct cell types by monitoring the activation state of YAP/TAZ. By independently modulating substrate stiffness and adhesiveness, we found that although ECM stiffness represents an overarching mechanical signal, the density of adhesive sites does impact on cellular mechanosignaling at least at intermediate rigidity values, corresponding to normal and pathological states of living tissues. Using these tools, we found that YAP/TAZ nuclear accumulation occurs when the projected area of the nucleus surpasses a critical threshold of approximatively 150 μm2. This work suggests the existence of distinct checkpoints for cellular mechanosensing.
Keywords: Biomaterials, Hydrogels, Mechanobiology, YAP/TAZ, cell culture
Introduction
Recent years have witnessed a renaissance of mechanobiology,[1] leading to the view that potentially every aspect of cell behavior is under control of the physical, architectural cues of the cell’s microenvironment, and of the cell’s own shape. On a more applicative side, mechanobiology is key for tissue engineering and regenerative medicine.[2][3][4] Yet, a major limitation is represented by the fact that probing the effects of forces on living cells requires a multidisciplinary effort, and the implementation of a dedicated suite of tools and bioassays.[5][6][7][8][9][10] This is too often a deterrent for a biological laboratory willing to investigate the effects of cell mechanics on a given biological process but lacking materials science expertise. For example, 2D hydrogel substrates recapitulating biophysical and adhesive cues of natural ECM are ideal tools for mechanobiology investigations.[11][12][13] In particular, the stiffness of hydrogel is a critical parameter in dictating cell behavior, fate and gene expression. That said, other structural features may also impact on cellular decisions, and in particularly the density and distribution of cell adhesive sites.[4][14] That said, relatively few studies have attempted to independently investigate stiffness from adhesiveness; for example, it would be interesting to appreciate whether, and to what extent, these correspond to molecularly distinct signaling cascade, or, rather, converge on the same mechanosensing pathway.[15] Moreover, there is not consensus between authors on what substrate formulations should be adopted, or straightforward interpretation of the results obtained with different substrates.[4][5][8][16][17][18][19][20][21] We posit that these apparent inconsistencies stem from two interconnected problems: the technical complexity of substrate fabrication, and the fact that there is not a univocal consensus on immediate readers and predictors of the mechanical attributes that a fabricated niche imparts to cells. In turn the latter is essential for substrate testing and tailoring before any specific biological application.
Here we have addressed these limitations by using YAP/TAZ, universal mechanical rheostats of mammalian cells, to monitor the mechanosignaling attributes of various hydrogel formulations.[22][23][24] YAP/TAZ is known to translocate to the nucleus in response to a host of mechanical stimulations.[1][3] Using YAP/TAZ activity as beacon, here we have optimized the synthesis of hydrogels substrates, and controlled their biological effects, as such offering protocols that could be implemented by any cell biology laboratory. This includes fabrication of fully synthetic PEG-RGD hydrogels over a gradient of rigidities and adhesiveness. Improved control over these features revealed that ECM stiffness and adhesiveness play distinct and mutually modifying roles on YAP/TAZ regulation. We also discovered that the degree of YAP/TAZ nuclear localization on distinct substrates invariably correlates with the nuclear shape of individual cells, indicating the existence of a “nuclear ruler” for YAP/TAZ mechanosensing.
Results
With this background in mind, we started this investigation aiming to define simple and scalable protocols for the production of PAA- and PEG-based hydrogel systems that do not require prior expertise in material science or specialized equipment. We considered that an ideal hydrogel should fulfill a number of key criteria: i) be composed of readily available and relatively inexpensive reagents; ii) require an easily reproducible synthesis procedure; iii) provide a versatile mean to control cell adhesiveness; iv) allow hassle-free scalability for high throughput studies. For this, we first optimized chemical formulations for a Polyacrylamide (PAA)-based hydrogel system. The use of PAA coated with natural ECM proteins, pioneered by Pelham and Wang and popularized by the protocols of Tse and Engler colleagues,[11][12][13] has indeed been seminal for a broad number of discoveries in mechanobiology.[1][3] However, even the preparation of this classic substrate remains artisanal, limited by difficulties in obtaining a homogeneous adhesive coating, batch-to-batch reproducibility, limited scalability, and shelf-life. These limitations stem either from the use of unstable compounds (such as succinimide containing monomers), or from the adoption of specialized protein coating strategies (such as sulfo-SANPAH), in turn requiring a multistep procedure and dedicated equipment for UV activation, or direct protein printing.[11][20][21][25] Altogether, these drawbacks also negatively impact on feasibility of experiments on a large scale.
To overcome these limitations, we adopted an OH-functionalized PAA hydrogel (PAA-OH, see Figure 1a) and whose composition is reported in Table 1 (and see Methods). This fabrication employs readily available and inexpensive reagents; PAA-OH hydrogels are stable for days and have mechanical properties very close to those of classic PAA (Table S1 and Figure S1). Compared to the latter, however, PAA-OH chemistry holds the advantage that it does not require any functionalization step for attachment of ECM proteins, as these spontaneously form stable hydrogen-bonds with the OH-functionalized hydrogel surface (see Figure 1c for a scheme of the process). We tested coating of PAA-OH hydrogels with both Fibronectin (FN) and Laminin, suggesting that different ECM proteins may be used as adhesive substrates (Figure S2).
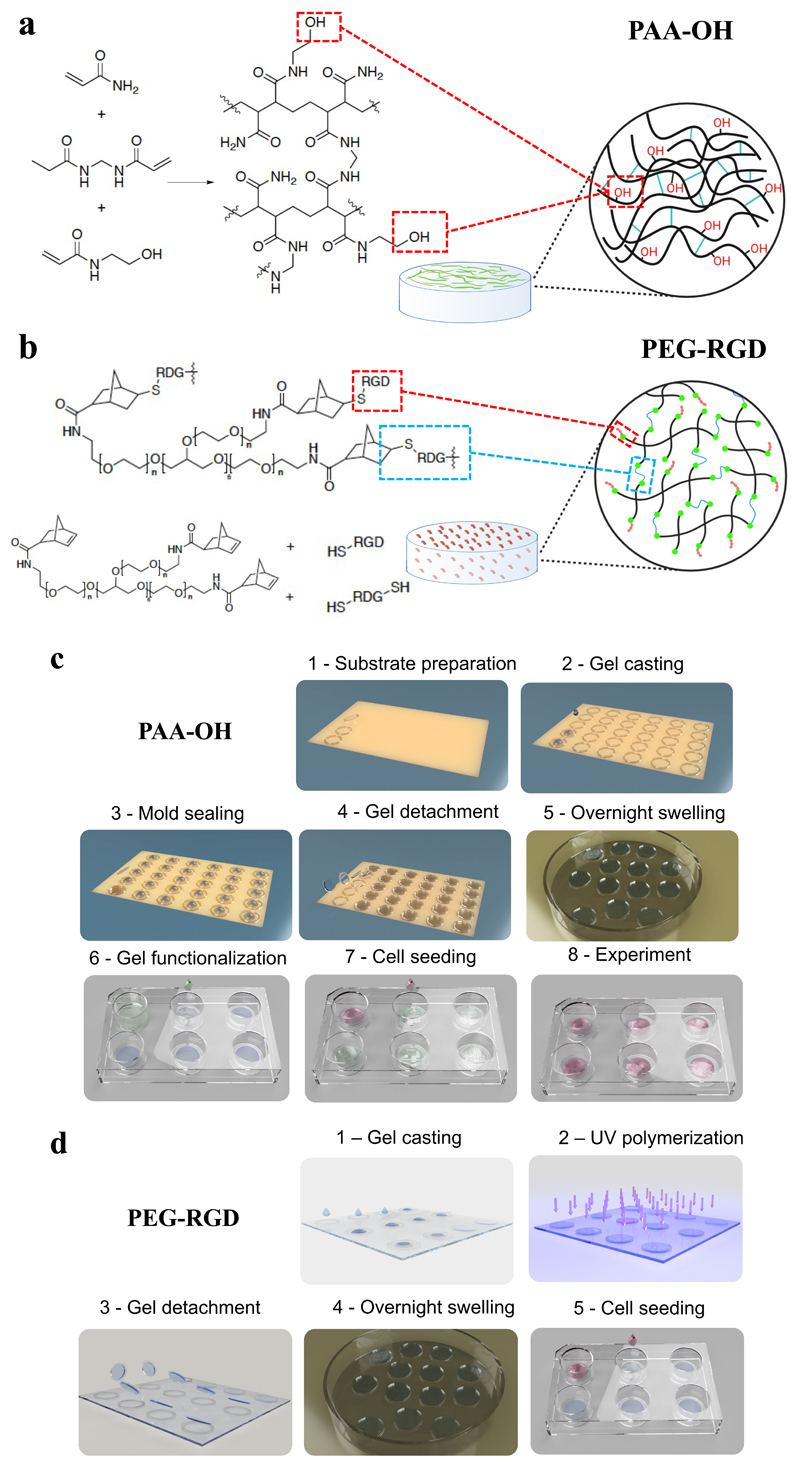
Figure 1.
Scheme of the a) PAA-OH and b) PEG-RGD hydrogels. c) Scheme of PAA-OH hydrogel fabrication process, from the synthesis to bio-functionalization and cell seeding. d) Scheme of PEG-RGD hydrogel synthesis, UV-curing and cell seeding.
Table 1. PAA-OH and PAA-RGD Hydrogel formulations and elastic modulus measurements.
| 1AA wt% | 2BA wt% | PAA-OH Hydrogel | 3Elastic modulus PAA-OH | PAA-RGD Hydrogel | 3Elastic modulus PAA-RGD |
|---|---|---|---|---|---|
| 3.5 | 0.03 | PAA-OH1 | 0.32 ± 0.03 | PAA-RGD1 | 2.79 ± 0.38 |
| 3 | 0.15 | PAA-OH2 | 3.15 ± 0.27 | PAA-RGD2 | 4.86 ± 1.02 |
| 5 | 0.15 | PAA-OH3 | 6.09 ± 1.35 | PAA-RGD3 | 13.81 ± 1.52 |
| 5 | 0.225 | PAA-OH4 | 13.39 ± 1.73 | PAA-RGD4 | 21.07 ± 3.41 |
| 8 | 0.48 | PAA-OH5 | 50.01 ± 3.07 | PAA-RGD5 | 57.48 ±4.00 |
AA represent the sum of the monofunctional acrylates Acrylamide and N-hydroxyethyl acrylamide (HEA) for PAA-OH gels, and Acrylamide alone for PAA-RGD gels. The amount of HEA is fixed at 0.1M. A fixed amount of ACR-PEG-MAL-RGD of 3mM has been added to the composition.
BA wt% is the weight percent of bisacrylamide
Stiffness measurements by micropipette aspiration.[40] The data indicate that the effect of the maleimide-modified acrylate addition is to increase gel stiffnesses.
By monitoring YAP/TAZ nuclear vs. cytoplasmic localization of FN-coated gel preparations, we empirically found that a critical parameter to obtain an even, homogeneous ECM coating is the control of the substrate drying timing before cell seeding (Figure S3). We found that an ideal drying time is 5 min; when we extended it beyond 15 min, we found that cells abnormally turned-on YAP/TAZ even when the substrate bulk modulus was soft, in principle unable to support YAP/TAZ function.[22][23] Investigating the reason of this odd result, we found that excessive drying causes formation of fibrous structures, adhesive bundles or nets (Figure 2b, 2e). Although these are intrinsic to natural FN molecules,[26] the formation of these structures disproportionally bias cell attachment to such ECM fibers, leading to aberrant YAP/TAZ nuclear localization (Figure 2h, 2l). In other words, surface fibrosity, a hardly controllable parameter, can confound the effects of substrate rigidity. We conclude that the PAA-OH hydrogel system is easily implementable, and scalable (Table S2), without special equipment or prior expertise, thus overcoming some typical drawbacks associated to hydrogel production. In terms of reproducibility, we repeated these fabrications and YAP/TAZ validations at least 10 times, with completely comparable results between distinct experiments.
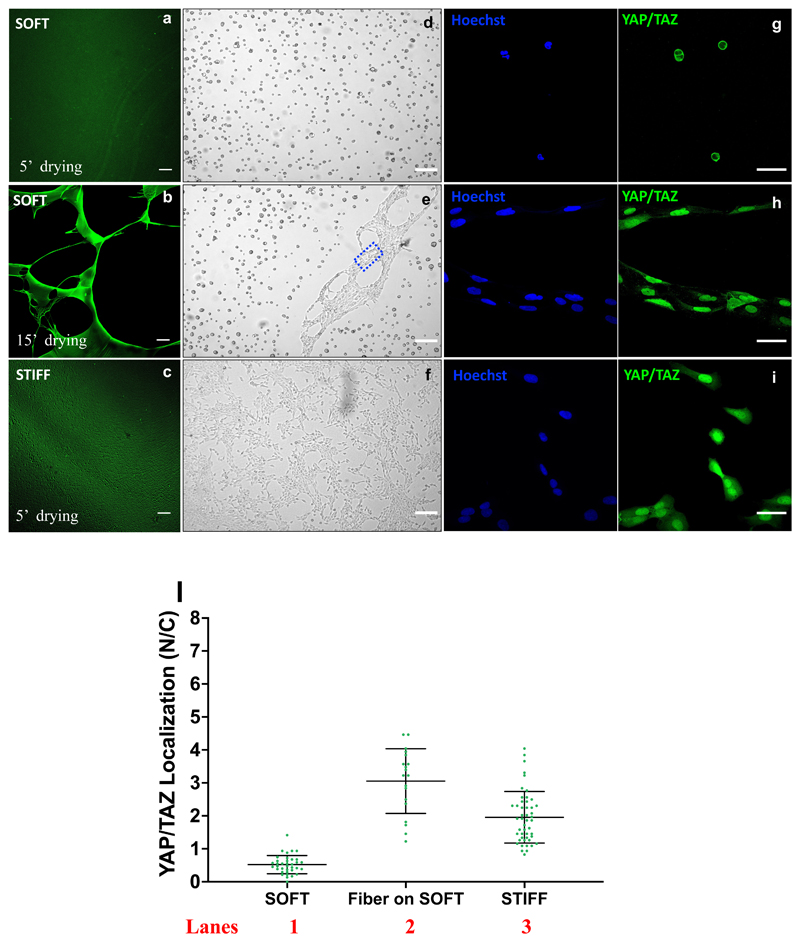
Figure 2.
Fluorescence images of the PAA-OH substrates coated with a solution of fibronectin (25μg/mL) and Alexa488 conjugated Fibrinogen (2μg/mL), for two different stiffnesses (a,b: soft 0.32kPa, c: stiff, 50kPa) and two drying timing (for the soft hydrogel) after FN incubation. For each picture it is shown an example of cell (MCF10A) seeded on that substrate (bright field images d, e, f). FN fibers are visible in the rectangle of figure e. From the comparison between a,d and b,e pictures it is evident that softer hydrogels are more sensitive to the drying time and require shorter (5’) drying times to avoid formation of adhesive bundles or nets instead of a homogeneous protein coating on a relatively large area hydrogel.
Immunofluorescence images of YAP/TAZ subcellular localization of U2OS cells seeded on: soft PAA-OH (g), FN fibers conjugated to soft PAA-OH (h) and stiff PAA-OH (i). From the staining are visible: nuclei (in blue) and YAP/TAZ (in green).
1) quantifications of the nuclear/cytoplasmic (N/C) ratio of YAP/TAZ subcellular localization in U2OS seeded on soft PAA-OH (0.32 kPa), FN fibers conjugated to soft PAA-OH (0.32 kPa) and stiff PAA-OH (50 kPa). Number of cells for each lane in Figure 2l is: lane 1:36; lane 2:19; lane 3:48. Scale bar: 100 μm (a,b and c), 200 μm (d,e and f) and 50 μm (g,h and i).
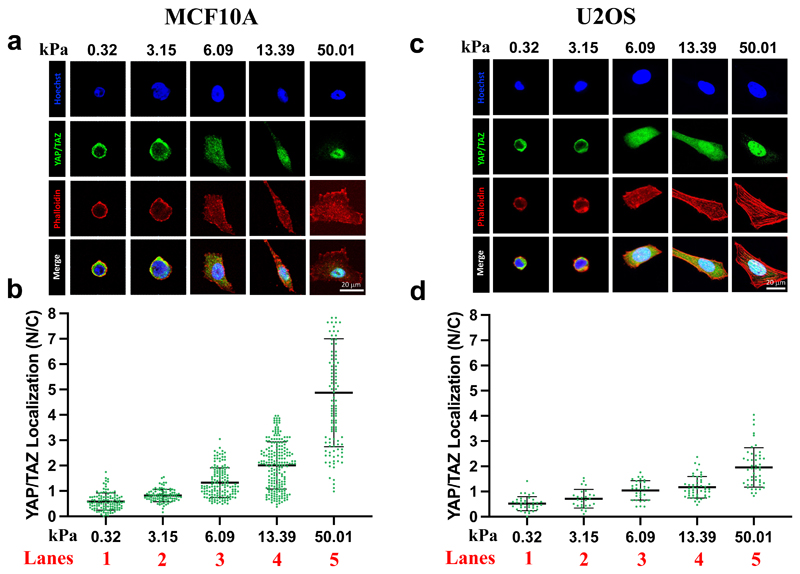
Next, we used the localization of the YAP/TAZ mechanotransducer as immediate, quantitative proxy of biomaterial-cell interaction, monitoring the effects of stiffness of PAA-OH hydrogels in different cell types. For this, we selected two different cell lines - that is, immortalized mammary epithelial cells (MCF10A), and transformed U2OS osteosarcoma cells – as such reflecting distinct cell states and tissues of origin. Each of these cell types was seeded on PAA-OH hydrogels over a gradient of different rigidities. We quantified YAP/TAZ activity by measuring its subcellular localization through immunofluorescence and software-assisted quantification of nuclear and cytoplasmic signal. We found that, as expected, YAP/TAZ activity nicely matched the rigidity gradient of the hydrogels, as such validating our fabrication protocol (Figure 3).
Figure 3.
Representative immunofluorescence images (a, c) and quantifications (b, d) of the nuclear/cytoplasmic (N/C) ratio of YAP/TAZ subcellular localization in MCF10A (a, b) or U2OS (c, d) cells, after seeding on PAA-OH substrates, coated with FN of five different stiffness. From the staining are visible: nuclei (in blue), F-actin (in red), YAP/TAZ (in green). F-actin was stained with fluorescently labeled phalloidin to serve as cell shape reference. Each dot in b) and d) corresponds to quantification of the N/C ratios of YAP/TAZ subcellular localization in individual cells, as obtained with Software-assisted Imaging processing of confocal images (see methods). Number of cells for each lanes are: b) lane 1: 116; lane 2: 93; lane 3: 146; lane 4: 210; lane 5: 120. d) lane 1: 36; lane 2: 27; lane 3: 31; lane 4: 48; lane 5: 48.
A critical and only partially addressed question in mechanobiology relates to whether cells read physical stiffness per se, or, rather, a combination of substrate rigidity and density, or distribution, of adhesive features.[14][16][17] Addressing this type of questions required the development of dedicated implementations of hydrogel fabrication. For this, we switched to fully synthetic hydrogels in which adhesiveness is controlled by RGD containing peptides, as such bypassing any risk of confounding effects from fibrosity or spatial arrangement of adhesive spots of natural ECM molecules. For this, we first attempted to copolymerize acrylamide (AA) and bisacrylamide (BA) in presence of a maleimide-modified acrylate monomer conjugated with RGD (Mal-PEG-RGD) to obtain PAA-RGD, as previously reported in literature. [16][19] However, using U2OS as paradigm, we found that the levels of YAP/TAZ activity are, in absolute values, reduced when compared to FN-coated PAA at the same stiffness (compare PAA-RGD 2, 3 and 5 with PAA-OH 2, 4 and 5 in Figure S4). This suggests that availability of RGD adhesive sites in FN greatly boosts cellular mechanotransduction if compared with PAA-RGD gels. In the latter, accessibility and/or presentation of the RGD ligand is ostensibly hindered, likely due, as previously noticed, to the presence of too long PAA brushes at the gel surface, preventing cell attachment.[27][28]
These suboptimal results prompted us to switch to PEG macromer-based hydrogels aiming to generate a fully synthetic adhesive hydrogel of different rigidities, PEG-RGD. For this, we used a Norbornene-terminated 8-Arms-Polyethyleneglycol (NB-8ArmPEG, 40 kDa), in which RGD moieties are precisely attached at the extremity of the arms by a thiol-ene click reaction between NB functionalities and cysteine groups in the peptide. Simultaneously, the gel is crosslinked through the same reaction mechanism with di-cysteine terminated synthetic peptides (see Figure 1b and Figure 1d for a scheme of the process). A potential caveat of PEG-RGD formulation is the fact that RGD containing peptides and crosslinking peptides compete with each other; this has so far limited the range of elastic moduli obtainable with these hydrogels to values that are below few kPa (see Figure S5a), that are typically insufficient for most mechanobiology tests. However, here we empirically optimized PEG macromer precursor concentration and crosslinking peptide concentrations to greatly expand the range of rigidities obtainable with PEG-RGD hydrogels, even in presence of high RGD molar content (see composition recipes in Table 2). Using YAP/TAZ localization in U2OS cells, we found that this went hand-in-hand with substrate stiffness: YAP/TAZ activation on PEG-RGD is comparable to PAA-OH and greatly enhanced when compared to PAA-RGD at comparable stiffness (Figure 4a and Figure S4).
Table 2. PEG-RGD hydrogel formulations and elastic modulus measurements.
In the Table is reported the comparison of stiffness of PEG-RGD Hydrogel formulations achieved with the synthesis procedure used in the present work (column 4) and as reported in literature[35] (column 5), with the highest RGD molar concentration (3mM). The latter is achieved by pre-reacting the RGD containing peptide with NB functionalities and then by UV-crosslinking. In our procedure all peptides (the one containing RGD and the crosslinking peptide) are reacted simultaneously, allowing a more homogeneous distribution of RGD containing peptide among 8ArmPEG macromers (i.e. avoiding possible “saturation” of the NB terminated arms) and therefore achieving a more efficient crosslinking degree.
| PEG-RGD Hydrogel | Molar ratio 1Cys/NB | 2NB-8ArmPEG wt% | 3Elastic modulus PEG-RGD | 3*Elastic modulus PEG-RGD |
|---|---|---|---|---|
| PEG-RGD1 | 0.4/1 | 4.7 | 0.30 ± 0.13 | 0.02 ± 0.08 |
| PEG-RGD2 | 0.5/1 | 5.0 | 0.87 ± 0.13 | 0.11 ± 0.01 |
| PEG-RGD3 | 0.6/1 | 5.2 | 1.17 ± 0.31 | 0.20 ± 0.03 |
| PEG-RGD4 | 0.7/1 | 5.5 | 2.63 ± 0.38 | 0.71 ± 0.08 |
| PEG-RGD5 | 0.8/1 | 9 | 7.71 ± 0.38 | 1.62 ± 0.03 |
| PEG-RGD6 | 0.8/1 | 12.5 | 13.7 ± 0.48 | 4.47 ± 0.65 |
Cys=Cysteine terminal group of the crosslinking peptide. RGD concentration is fixed to 3mM
NB-8ArmPEG wt% is the weight% of macromer used to synthesize PEG-RGD hydrogels.
Stiffness measurements by micropipette aspiration.[40]
Figure 4.
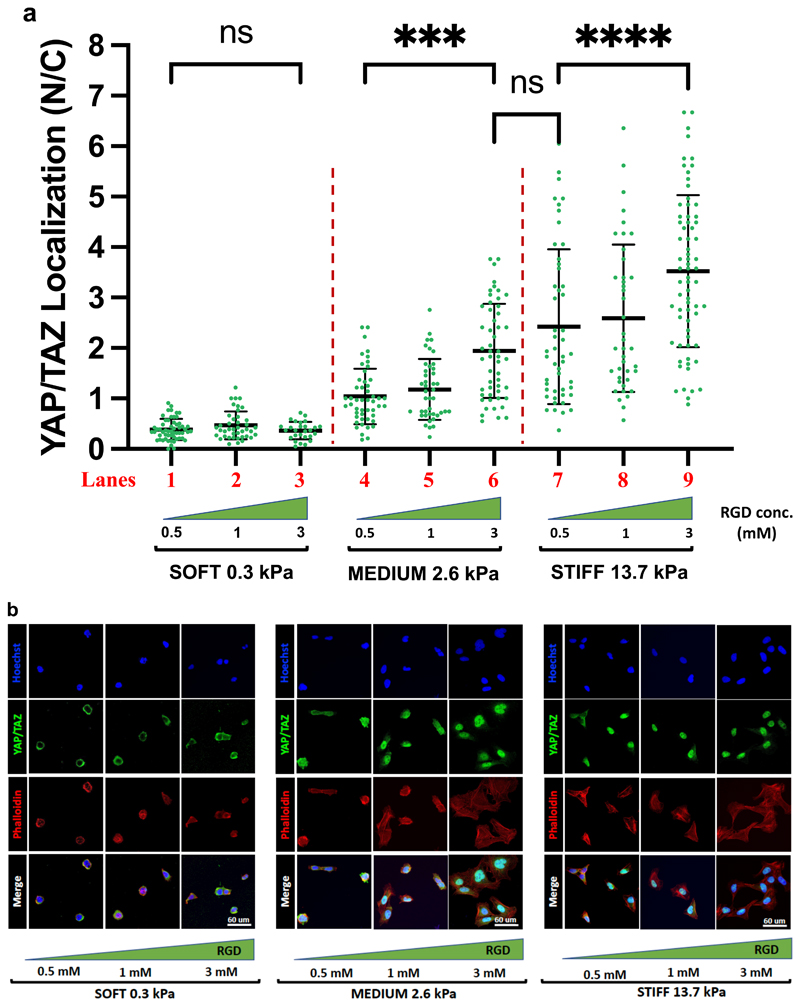
a) Quantifications of the ratio nuclear/cytoplasmic YAP/TAZ subcellular localization in U2OS plated on PEG-RGD hydrogels at three different values of stiffness and RGD concentration. Statistically significative differences were evaluated comparing two groups performing a one-way ANOVA with Tukey’s multiple comparison test using GraphPad Prism 9 and considering a confidence interval of 95% (p-values obtained comparing: lanes 1 vs. 3, p=0.99; lane 4 vs. 6, p<0.001; lane 6 vs. 7, p=0.3; lane 7 vs. 9 p<0.0001). b) Immunofluorescence of U2OS plated on PEG-RGD hydrogels at three different values of stiffness and RGD concentration. From the staining are visible: nuclei (in blue), F-actin (in red), YAP/TAZ (in green). F-actin was stained with fluorescently-labeled phalloidin to serve as cell shape reference. Single cell immunofluorescence is reported in Figure S6. Number of cells for each lanes are: lane 1: 56; lane 2: 41; lane 3: 27; lane 4: 49; lane 5: 42; lane 6: 52; lane 7: 49; lane 8: 41; lane 9: 69.
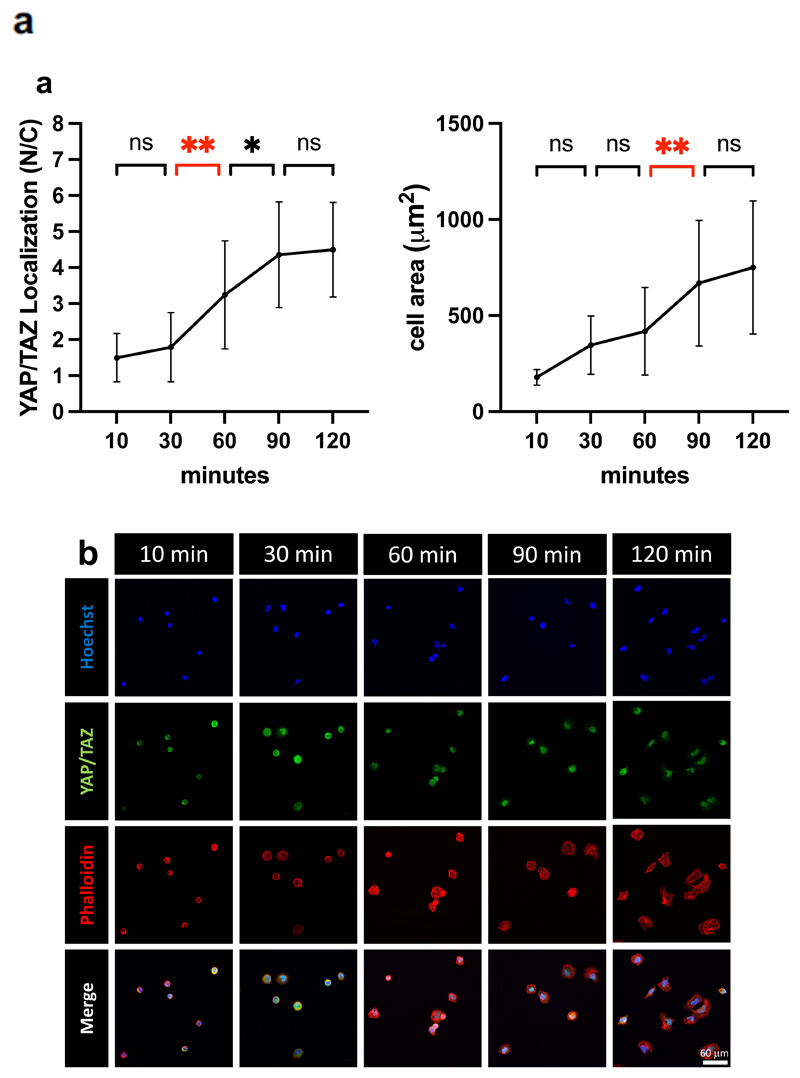
PEG-RGD substrates represent ideal tools to dissect the effects of rigidity and adhesiveness on mechanosignaling. To address this, we generated hydrogels at low (0.3 kPa, soft), medium (2.6 kPa, intermediate) and high (13.7, stiff) modulus, each carrying a gradient of adhesiveness (as obtained by stoichiometrically controlling the concentration of RGD ligand attached to PEG polymers at 0.5, 1, and 3 mM). By quantifying the ratio of YAP/TAZ localization in individual cells experiencing these distinct stimulations (dots in Figure 4a), we made several interesting observations. On the one hand, we found that, as previously reported,[16] matrix modulus is an overarching signal in the control of YAP/TAZ activity (compare YAP/TAZ levels at the three rigidities at constant RGD: lanes 3, 6, 9 with 2, 5 and 8, or 1, 4 and 7 of Figure 4a). However, on the other hand, we also found that raising adhesiveness can also enhances mechanosensing (compare lanes 4 and 6, and lanes 7 and 9 in Figure 4a), and may even compensate for reduced ECM modulus (compare lanes 6 and 7, blurring the border between intermediate and high stiffness in Figure 4a), although, differently from previous observations,[17][21] a minimal threshold of stiffness is required to impart the necessary pulling feedback to cells (no effects of RGD dosage at low E, lanes 1-3 in Figure 4a). In these experiments, changes in YAP/TAZ activity are supported by variations in cell spreading, as gauged from the F-actin staining (Figure 4b and S6). Intriguingly, we found that the degree of YAP/TAZ nuclear localization could be appreciated immediately after cells attain a firm attachment to the material surface (Figure 5a left), but before any cell spreading per se (see cell area in Figure 5a right and the phalloidin staining in Figure 5b), suggesting that mechanosensing occurs irrespectively of whole cell shape, likely reflecting a hierarchy of events from cell attachment to nuclear spreading and then cell spreading. Indeed, cells attain plateau of YAP/TAZ mechanosensing between 90 and 120 min of exposure to a biomaterial.
Figure 5.
a) Quantifications of nuclear to cytoplasmic ratio of YAP/TAZ (N/C) of U2OS cells, as a function of the post-seeding time, on stiff PEG-RGD (13.7 kPa) after 10, 30, 60, 90 and 120 minutes. Statistically significative differences were evaluated by one-way ANOVA with Tuckey multiple comparison. P-values for YAP/TAZ localization: 10 vs 30 min p=0.95; 30 vs 60 min p=0.003; 60 vs 90 p=0.047; 90 vs 120 p=0.99. p-values for cell dimension: 10 vs 30 min p=0.068; 30 vs 60 min p=0.77; 60 vs 90 min p=0.003; 90 vs 120 min p=0.755. Number of cells for each times are: 10m: 21; 30m: 20; 60m: 24; 90m: 20; 120m: 25.
b) Representative immunofluorescence images of the corresponding YAP/TAZ subcellular localization in U2OS cells. Staining shows nuclei (in blue), YAP/TAZ (in green) and F-actin (in red).
In terms of reproducibility, we repeated these experiments at least 12 times, all with comparable results. Mechanobiology assays frequently require preparation of multiple hydrogels, prompting us to compare scalability; for this, the reader may refer to Table S2, where we provide back-to-back comparison of the time required to prepare from 10 to 50 gels at once using PAA-OH, PEG-RGD, also compared to traditional PAA gels. PEG-RGD scalability is lower than PAA-OH, but still comparable with PAA gels.
We also controlled for mesh size (Table S3 and Figure S7) of PEG-RGD and PAA-OH hydrogels and their swelling behavior. By comparing these measurements, it appears that the crosslinking degree decreases mesh size although we found no effect on swelling. Notably, PEG-RGD hydrogels do not show relevant differences in swelling at various rigidity values (Figure S8), suggesting that stoichiometric concentrations of RGD adhesive points, measured as molar ratio in the liquid gel, are well maintained during gelification. This has relevance, because, on the one hand, it indicates that the variation of different RGD densities is preserved within the same value of stiffness (e.g., from 0.5 to 3mM in the experiments shown in Figure 4); and, on the other hand, it also ascertains that the same value of RGD density is comparable at different stiffnesses. All in all, this further validates the notion that PEG-RGD are valid tools to independently tune stiffness and adhesiveness.
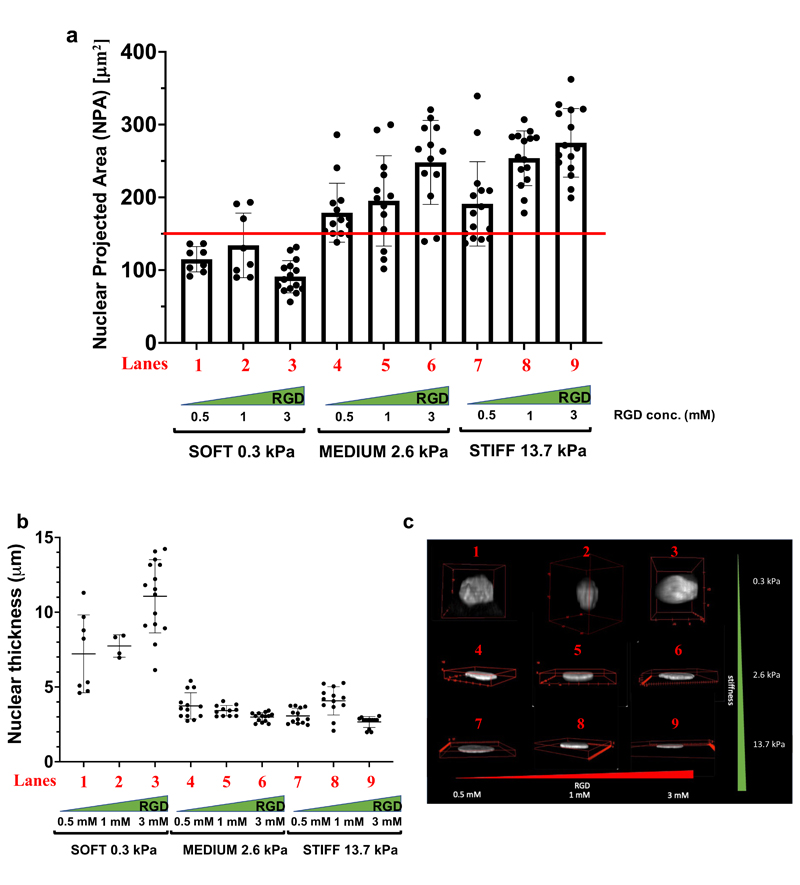
Next, we took advantage of PEG-based tunable hydrogels to probe the contribution of biomaterial rigidity and its modulation by adhesiveness on the shape of subcellular structures. In particular, the nucleus has recently emerged as a key element in cellular mechanotransduction and YAP/TAZ activity.[29] A continuum of force transmission from ECM rigidity to the nucleus mediated by the actomyosin-contractility and the LINC complex profoundly affects nuclear shape.[29][30] Notably, this is integral to changes in YAP/TAZ localization, to the extent that experimentally compressing the nucleus has been shown to be sufficient to increase YAP/TAZ nuclear localization.[31] Complementarily, the nucleus has been recently shown to play also an active and upstream role in cellular mechanotransduction: dendritic cells use their nucleus as “ruler”,[32] a term intended by those authors as a functional entity, by which external mechanical strains that deform nuclei beyond a certain threshold (the ruler) to foster actomyosin-contractility. Cell stretching is known to induce nuclear spreading, in turn correlating with YAP/TAZ nuclear entry; this was originally shown by Rocha-Cusachs and colleagues[31], when they measured YAP nuclear accrual in cells experiencing substrates of 5, 29 and 150kPa and correlating it with the ratio of nuclear length and thickness (height). Yet, at the starting point of such experimental conditions (i.e., 5kPa substrate stiffness), YAP was already substantially nuclear, having a Nuclear/Cytoplasmic (N/C) ratio of 1.[31] That said, it remains still unclear whether critical thresholds exist for YAP/TAZ nuclear entry, and, as such, of their activation from a prevalently cytoplasmic (OFF) to nuclear (ON) states. To explore this issue, we used confocal microscopy to measure nuclear shape parameters in U2OS cells plated on PEG-RGD hydrogels at various stiffnesses and degrees of adhesiveness. As shown in Figure 6a, we found that the nuclear projected area (NPA) represents a sensitive parameter by which cells read biomaterial mechanics: at low rigidity (0.3 kPa), NPA remains around 100 μm2, and this value is unaffected by changes in adhesiveness. On stiffer substrates (13.7 kPa), and at the highest RGD density, NPA raises close to a mean value of 270 μm2. Intriguingly, at intermediate rigidity values, NPA ranges from about 180 to 240 μm2 depending on RGD concentration (from 0.5 to 3 mM).
Figure 6.
a) Nuclear projected area (NPA) of U2OS plated on PEG-RGD hydrogels at three different values of stiffness and RGD concentration. b) Nuclear Thickness of U2OS plated on PEG-RGD hydrogels at three different values of stiffness and RGD concentration. c) Representative images of nuclear shapes of U2OS seeded on PEG-RGD hydrogels. Movies showing a complete view of the nuclear shape are present in SI. Number of cells for each lanes are: a) lane 1: 8; lane 2: 8; lane 3: 15; lane 4: 14; lane 5: 13; lane 6: 13; lane 7: 15; lane 8: 15; lane 9: 15 b) lane 1: 8; lane 2: 4; lane 3: 15; lane 4: 13; lane 5: 11; lane 6: 14; lane 7: 13; lane 8: 13; lane 9: 13.
The data above suggest the existence of a critical threshold for YAP/TAZ activation set at a threshold of NPA of 150 μm2 (red line in Figure 6a). The results are consistent with the view that ECM mechanosensing to the nucleus is key for nuclear shape and concomitant YAP/TAZ regulation, as previously reported [30,31]; at the same time, these data specify that the NPA is a critical permissive checkpoint for efficient YAP/TAZ nuclear entry, and thus to activate YAP/TAZ mechanosignaling from an otherwise inhibited cell state in which YAP/TAZ are cytoplasmic (N/C << 1) (Figure 4a). Interestingly, however, another nuclear shape parameter, nuclear thickness (or height), as obtained by measuring with confocal z-stacking the space between the basal and dorsal nuclear surfaces, abruptly decreases as soon as cells experience sufficient traction from their substrate, and notably in a manner that is ostensibly independent from adhesiveness (Figures 6b); as shown in Figure 6c and Movies (see SI), cells on soft substrates display dome-like nuclei that dramatically flatten with a sharp curvature at their outer border at 2.6 kPa or stiffer substrates.
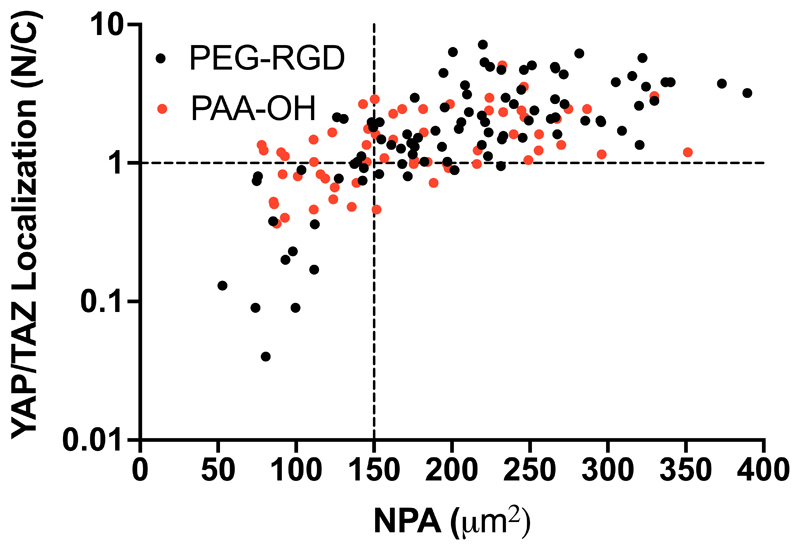
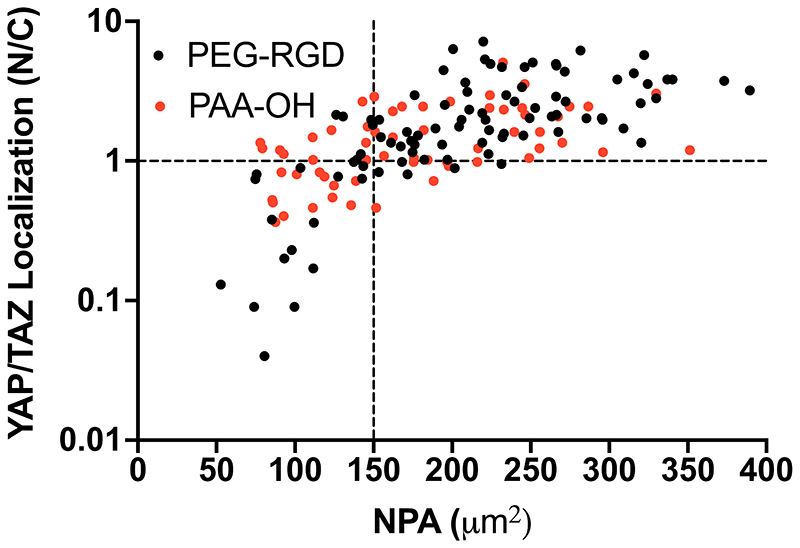
The above findings predict that changing nuclear shape to a NPA more than 150 μm2 needs to be achieved to activate efficient mechanosensing, as visualized by YAP/TAZ nuclear level. We tested this prediction by measuring NPA of individual U2OS cells and scored it as superior vs. inferior to 150 μm2, and then asked whether this associated to a prevalently nuclear vs. even/cytoplasmic YAP/TAZ localization in the same cell (i.e., Nuclear to Cytoplasmic ratio higher or lower than about 1). As shown in Figure 7, we found a very strong positive correlation between YAP/TAZ activity and NPA [e.g. only 7 cells (7%) in the right bottom quadrant of Figure 7 display N/C<1 in spite having suprathreshold NPA, whereas >90 cells (>93%) surpassing the 150 μm2 threshold have N/C = or >1], and in a manner independent from biomaterial chemistry (as it occurred in both PEG-RGD and PAA-OH hydrogels). We conclude from these results that cells read biomaterial mechanics by modulating NPA and nuclear thickness, although only the former is influenced by the density of adhesive sites. We also conclude that NPA is a sensitive read-out of cellular mechanosensing, as such directly correlating with the cellular YAP/TAZ status. Thus, a critical threshold of NPA regulated by the surrounding ECM needs to be surpassed for YAP/TAZ nuclear accrual.
Figure 7.
YAP/TAZ Nuclear to Cytoplasmic ratio measured for individual U2OS cells (plated on PEG-RGD and PAA-OH hydrogels with different values of stiffness and RGD concentration) as a function of their individual NPA. Black dashed line at N/C=1 give an indication of cells with even YAP/TAZ localization and at NPA=150 μm2 indicates a threshold for the efficient activation of YAP/TAZ. Statistical significance for monotonic correlation between nuclear to cytoplasmic YAP/TAZ ratio (N/C) and nuclear projected area (NPA) was evaluated by Spearman’s correlation analysis using 95% as confidence interval (rs=0.7277, p<0.0001 and rs=0.5337, p<0.0001 for PEG-RGD and PAA-OH respectively). Number of cells analyzed is 153.
Discussion and conclusions
In the present work, we optimized fabrication of PAA based hydrogel substrates, and present a protocol to synthesize fully defined PEG-RGD substrate of varying rigidities and adhesiveness. As for PAA, we adopted a variation of the traditional procedure, this time using PAA-OH. The latter eliminates the need of sulfo-SANPAH and UV treatment for substrate functionalization with the ECM. [21][11] To this end, we used OH-functionalized PAA that can be readily coated with adhesive proteins without additional chemistry or need of special equipment (Figure 1a, c and Table 1). Although PAA-OH based hydrogels represent easily applicable tools for mechanobiology experiments, they provide effective adhesiveness to cells only after coating them with natural ECM proteins. A limitation with ECM proteins is variability in the homogeneity of the adhesive surface; a second limitation with natural fibrous proteins is the variability of adhesive spots that is an intrinsic attribute of each ECM protein and its structure.
Fully synthetic PEG-RGD hydrogels overcome the above limitations, particularly in experimental settings requiring to independently control adhesiveness and substrate bulk modulus. Using YAP/TAZ activity as immediate readout of the effects of substrate mechanics, we found that, in PEG-RGD hydrogels stiffness is the dominant cue controlling cellular mechanosensing, and that a high modulus can compensate for lower density of adhesion sites. That said, ligand density is relevant to tune such response at intermediate levels (few kPa) of substrate rigidities (Figure 4). These data potentially connect to prior work on the role of adhesiveness as obtained by modulation of natural ECM fibers concentration[21] (such as of Matrigel or collagen), or ECM tethering;[17] for example, a “tethering effect” (i.e., a cell pulling on a short vs. long ECM fiber segments, as defined by the distance intervening between the two neighboring anchoring points of that fiber) has been proposed to induce cell spreading even on soft substrates, even overriding the effect of stiffness on embryonic or epithelial stem cell differentiation.[17] Other work has instead implied that the long-term biological effects (e.g., stem cell differentiation, requiring several days in culture) of the physicality of the microenvironment occurs according to stiffness and not ECM tethering.[16] None of these prior studies controlled for YAP/TAZ patterning, and thus a direct comparison with the present work remains speculative. Yet, we note that our PEG-RGD system does not allow for tethering and can still pattern YAP/TAZ activity over a gradient of rigidity, as such remarking the cardinal role of stiffness. At the same time, finding that the density of adhesion sites can modulate YAP/TAZ mechanosignaling also suggests that the distance between anchoring sites on fibrous proteins may still be, particularly in vivo, of paramount relevance to sustain YAP/TAZ function. Thus, our data suggest that adhesiveness and tethering would impact on cell behavior not as cues independent from stiffness, but only to the extent to which they contribute to YAP/TAZ mechanosignaling. In this line, our observation that ligand density may contribute to overall YAP/TAZ function may be consistent with a recently proposed model of mechanosignaling based on force loading among individual RGD-bound integrins:[14][15] maximal integrin force load and YAP/TAZ activation, comparable to that one obtained by stiffer substrates,[22] may be achieved by a more compliant substrate deformation and appropriate spacing of integrin-bound RGD (Figure 4).
Finally, mechanical signaling affects the whole cell and its organelles. In particular, the nucleus is the stiffest organelle and the nuclear envelope provides anchoring sites for cytoskeletal filaments and microtubules in a manner conceptually not dissimilar from focal adhesions.[33] Not surprisingly, nuclear mechanics is increasingly appreciated as a key element in a continuum of forces that link the ECM and integrins to the nuclear envelope, and potentially chromatin.[29] Indeed, the loading of YAP/TAZ in the nucleus is intimately associated to nuclear deformation and nuclear flattening.[31] Here, by using a gradient of PEG-RGD hydrogels, we further extended these conclusions to show how a critical threshold in the nuclear projected area (NPA) parameter need to be surpassed to allow YAP/TAZ entry in the nucleus (Figure 6a). These results are also consistent with recent work suggesting that specialized integrin adhesive sites occur in the plasma membrane just below the nucleus, and that these sites are in fact those required for YAP/TAZ activity and mechanotransduction.[33] These adhesions organize a specialized F-actin substructure, called the nuclear actin cap, that distends the nucleus into a disk-like morphology (Figure 6b and c, Movies in SI).[34] Here we found that the nucleus flattens on the z-axis abruptly at 2.6 kPa, but in a manner ostensibly independent from ligand density (Figure 6b). Thus, NPA and nuclear thickness are distinct, although partially overlapping, elements of mechanosensing, and their relative value may depend on the nature of the cellular microenvironment. NPA is dependent on organization and expression of nuclear lamins and the continuum between the nuclear envelope and the endoplasmic reticulum.[29] An overarching limitation of our results is their correlative nature, that, although sufficient to validate the usefulness of new biomaterials, by no mean implies specific mechanisms; for example, we have not addressed here the causal connections between NPA and YAP/TAZ nuclear entry. It remains an open question whether our findings involve opening of nuclear pores, as elegantly shown by Elosegui-Artola et al., under conditions of nuclear deformations and in cells experiencing moderate level of substrate stiffness (e.g. >5kPa, associated to partially nuclear YAP) [31]; or, rather, whether we are here merely monitoring the consequences of a restructured cytoskeleton of cells under tension, in which nuclear entry of YAP/TAZ reflects other or additional mechanisms. In this respect, it is worth noticing that the stiffness ranges along a wide spectrum allowed by the biomaterials here described would allow to address this type of outstanding questions in future studies.
As final note, here we iteratively used YAP/TAZ as proxy for mechanosensing throughout the optimization of the hydrogel preparations here presented. This approach differs from most applications of biomaterials, whereby the material is first designed and then used to investigate its long-term effects on cell behavior, such as stem cell differentiation, migration and other effects. However, we note that biomaterial fabrication without appropriate testing of its effective mechanosignaling properties may be risky, as the latter may be profoundly affected by a host of additional features beyond stiffness, including fibrosity and adhesiveness, some manifest but other potentially hidden in the idiosyncrasies of each fabrication protocols. The consequence is what appears today a complex literature, with myriads of variegations in procedures that are not straightforward to replicate from lab to lab, all in all complicating the interpretation, and ultimately limiting the use of hydrogels for mechanobiology studies. Here we propose a relatively simple solution to this hurdle, that is the incorporation of YAP/TAZ activity as guidance during the biomaterials' design and synthesis.
Methods
PAA-OH and PAA-RGD hydrogel synthesis
Acrylamide (AA) solution (40 wt% in water), bisacrylamide (BA) solution (2 wt% in water) and water are mixed in the proper ratio to obtain the prepolymer solution. For PAA-OH part of the AA was substituted with N-hydroxyethyl acrylamide (HEA) to a final concentration of 0.1M, as reported in literature.[25],[36] For the synthesis of PAA-RGD, the solution of AA and BA is copolymerized with an Acrylate-PEG-Maleimide monomer (Lysan Bio, MW 3400) previously conjugated with a mono-cysteine terminated synthetic peptide containing the adhesive sequence RGD, GRGDSPC. The conjugation reaction is carried out in water at room temperature: the RGD peptide and Acrylate-PEG-Maleimide are dissolved to a final molar ratio of 1.1 and a final concentration of 10 wt%.
Both solutions are degassed for 15 minutes at 0.1 bar, to remove the oxygen dissolved in the solution which would inhibit the polymerization. Ammonium persulfate (APS) dissolved in water (10 wt% or 20 wt% for gels 1, 2, and 3 or 4 and 5 respectively). Once completely degassed, the monomer solutions are mixed with 1% V/V of the APS solution and 0.1% V/V of Tetramethylethylenediamine (TEMED). The final solution is then mixed and poured inside PDMS gaskets on Kapton substrates, with an internal diameter of 22mm and height of 250um. Functionalized coverslips are used to seal the molds. Once polymerized (about 10-15 min), the gels are detached from the molds and placed in a petri dish submerged with milliQ water. Gels are left overnight to reach the swelling equilibrium before protein functionalization (for PAA-OH) and cell seeding.
To increase the number of PAA gels obtainable simultaneously, the precursor solutions are polymerized in PDMS rings between a Kapton film (non-PAA adhesive) and a functionalized (PAA adhesive) coverslip, as such minimizing time of exposure to atmospheric O2 (that inhibits PAA polymerization). After preparation, coverslips-bound hydrogels are recovered and stored in H2O for at least 24 hours to allow homogeneous swelling (see Figure 1c for a scheme of the process, used for both PAA-OH and PAA-RGD gels. For PAA-RGD synthesis step 6 in Figure 1c is not required). This procedure warrants scalability in terms of number and size of hydrogels, as up to 50 hydrogels can be prepared simultaneously in about 30 min, in a straightforward and easily reproducible manner, so expanding the versatility and range of downstream applications (see also Table S2).
PEG-RGD hydrogel synthesis
A hydrogel system based on formulations reported in literature was used,[36] modifying the synthesis procedure in order to achieve a broad range of stiffnesses able to generate large differences in YAP/TAZ localization. Stock solutions of 8-Arm Norbornene terminated macromers (40kDa, Creative PEGWorks, 250mg/mL), bi-cysteine terminated synthetic peptides CRDGQPGYSGQDRC (crosslinking peptide, 40mg/mL), mono-cysteine terminated adhesive and non-adhesive peptides GRGDSPC and GRDGSPC (37.5mg/mL) and a photoinitiator (Lithium phenyl-2,4,6- trimethylbenzoylphosphinate, LAP, 31.7mg/mL) are mixed together to the final PEG concentration and molar ratio between NB functionalities and the cysteine terminal groups of the crosslinking peptide reported in Table 2. The mono-cysteine terminated adhesive peptide GRGDSPC has a concentration in the final gel volume of 0.5, 1 or 3mM. The non-adhesive peptide GRDGSPC is added to have a total concentration of mono-cysteine terminated peptides of 3mM in each gel. LAP photoinitiator has a concentration of 1% w/w respect to PEG. All peptides are supplied by CRIBI Biotechnology Center, University of Padova. Gasket of PDMS (250μm thick, diameter 22mm) are arranged on the non-adhesive glass substrates and 40 μL drop of the final solution is poured in the gasket. An adhesive functionalized glass coverslip is then placed on top of the solution to homogeneously spread the drop and handle the gels.
The gel solutions are then exposed under a UV led light (Delolux 20) with the emission peak centered at 400 nm. A light dose that depends on the gel composition is used: 10 min exposure at 65 mW/cm2 or 2 min exposure at 26 mW/cm2 for the softer (PEG-RGD 1- 4) and stiffer (PEG-RGD 5 and 6) gel respectively.
Non-adhesive PEG-based hydrogels, called NA-PEG in the Figure S5, are synthesized starting from a more concentrated stock solutions of 8-Arm Norbornene terminated macromers (40kDa, Creative PEGWorks, 300mg/mL) and bi- cysteine peptide crosslinker (100 mg/mL), without adding RGD peptide.
After the polymerization, the gels are peeled from the mold, placed on a petri dish, submerged with 1X PBS and left at room temperature overnight to reach the swelling equilibrium.
We optimized the set-up to increase the number of gels that can be UV-polymerized simultaneously. PDMS rings are used between a non-adhesive and a gel-adhesive glass coverslip which allow gel handling (see Figure 1d for a scheme of the process). Polymerized gels are stored in 1X PBS for at least 24 hours to allow a homogeneous swelling.
Mechanical characterization
Micropipette aspiration is used to measure hydrogel stiffness. In brief, hydrogel samples are prepared attached to an adhesive glass coverslip and are left in 1X PBS overnight to reach the swelling equilibrium. For the measurement, a glass capillary connected to a syringe pump and a pressure sensor, is mounted on top of an inverted microscope with the sample holder placed perpendicular to it. The gel is placed on the holder and the capillary is moved toward the surface of the gel to achieve a complete contact. A negative pressure is then applied to the sample surface through the capillary and is detected and registered using the pressure sensor. Simultaneously, an image of the gel aspirated length inside the glass pipette is taken with the inverted microscope. The analysis of the elastic modulus is done using a model that correlates the aspirated length and the pressure exerted with the Young modulus of the hydrogel. Details on the micropipette aspiration technique can be found in ref.[40]
Kapton polymerization platform
An array of Polydimethylsiloxane (PDMS) round gaskets with an internal diameter of 22 mm and an external diameter of 24 mm is placed on flat Kapton sheet (Advent research material Ltd), see Figure 1c. The gaskets are prepared from a 250 μm height PDMS sheet (Specialty manufacturing inc.) using hollow punches. The PDMS circles are washed extensively with pure ethanol and disposed on the surface of the Kapton sheet while still wet. The following evaporation of the ethanol promotes the adhesion between Kapton and PDMS. A 140 μL drop of the desired PAA or PAA-OH solution is poured inside every PDMS gasket and a silanized coverslip is put on top to seal the gaskets.
Adhesive silanization
Glass coverslips with a diameter of 24 mm are washed with isopropanol and acetone and air dried to remove any residual grease or powder on the surface. After activation with a plasma cleaner (Harricks Expanded Plasma Cleaner 230V) they are silanized 15 min with a solution of 3-(trimethoxysilyl)propyl methacrylate (TMSPM) composed by pure ethanol (950 μL), acetic acid glacial (50 μL), TMSPM (20 μL). The coverslips are then washed three times with pure acetone and air dried.
Non-adhesive glass substrates
Glass substrates are washed with isopropanol and acetone and air dried to remove any residual grease or powder on the surface. Then they are cleaned using a plasma cleaner and left 2h in a dry box.
PAA-OH protein functionalization and imaging
Gels are placed in a multiwell dish inside the sterile hood and UV sterilized for 15 minutes (Figure 1c). Once the sterilization is completed, 1.5mL of a fibronectin (human plasma, Corning) solution in 1X PBS (25 μg/mL) is pipetted inside each well. The gels are put at 37°C inside an incubator overnight. The following day, gels are extensively washed with sterile 1X PBS to remove any excess of fibronectin and residual monomers. Laminin (EHS murine sarcoma, Sigma-Aldrich) coating is performed dispensing a drop of a 25 μg/mL laminin solution on a sterile Parafilm sheet and placing the gels upside down on top to ensure the complete coverage of the surface. Fluorescence imaging of the fibronectin coating on PAA-OH gels is obtained by adding fluorescent fibrinogen to the coating solution (2 μg/mL AF488 labelled fibrinogen, Thermo Scientific). Laminin coating is visualized by indirect immunofluorescence staining: using a primary anti-laminin antibody (Invitrogen, PA1-16730), diluted 1:50 and a AF488-conjugated secondary antibody (Molecular Probes) diluted 1:200. Images are collected right after the drying step for Fibronectin and after the immunofluorescence for Laminin using a Leica Stellaris confocal microscope equipped with a 10x air objective.
Cell seeding
After protein functionalization (PAA-OH) and UV sterilization for 15 minutes, the PBS solution is aspirated with a vacuum pump equipped with a thin nozzle without touching the gel surface. Gels are then left drying in air under sterile conditions without the lid for 5 mins (PAA-OH1 and PAA-OH2), 10mins (PAA-OH3 and PAA-OH4) or 15 mins (PAA-OH5).
Cell lines are then seeded on the gels. For PAA-OH and PAA-RGD gels, immortalized mammary gland cells (MCF10A) and transformed osteosarcoma cells (U2OS) are seeded pipetting 200 μl of cell suspension in culturing medium (concentration respectively of 200000 cell/mL and 40000 cells/mL) on the surface of each hydrogel. When the adhesion of the cell to the substrate is completed, the wells can be flushed with culture medium submerging the whole hydrogel. PEG-RGD hydrogels were placed in 12 wells multiwells and seeded with 1.5 mL U2OS cells in culturing medium (concentration 5300 cells/mL).
Immunofluorescence
Immunofluorescence (IF) is performed on PFA-fixed cells. Primary antibody is YAP/TAZ (Santa Cruz Biotechnology no. sc-101199). F-actin is stained with Alexa Fluor 488 Phalloidin (Thermo Fisher Scientific). Secondary antibodies (1:200) are from Molecular Probes. Samples are counterstained with Hoechst 33342 dye (Thermo scientific no. 62249) to label cell nuclei. Confocal images were obtained with a Leica Stellaris with a CCD camera and analyzed using LASX (Leica). The YAP/TAZ nuclear to cytosolic ratio is calculated creating a pipeline sequence in CellProfiler software. The CellProfiler sequence is prepared to obtain a mask of the NPA analyzing the nuclear signal, a mask of the cell based on the phalloidin channel, and a mask of the cytosolic area obtained subtracting the NPA to the cell projected area. The nuclear to cytosolic ratio is then calculated by the software on the YAP/TAZ channel as the ratio between the mean signal intensity on the nuclear mask and the mean signal intensity on the cytosolic area.
Mesh size measurement
The gels are placed in a glass bottom petri dish, submerged with 1X PBS and left at RT overnight to reach the swelling equilibrium.
Solutions of fluorescent dextrans or fluorescent polystyrene nanoparticles with different dimensions are prepared in 1X PBS. The dextrans employed (Sigma-aldrich) have different MW to fill a range of hydrodynamic diameter from 9.33 nm up to 61.83 nm (see the Table M1 and Figure S7).
Table M1. Dimension (molecular weight, Mw) of dextrans employed to measure mesh sizes and parameters (intrinsic viscosity, η, and hydrodynamic radius, Rh) calculated with the equations 1.1 and 1.2.
| Mw (g/mol) | [η] (mL/g) | Rh (cm) | Rh (nm) | d (nm) |
|---|---|---|---|---|
| 40k | 1.60E+01 | 4.67E-07 | 4.67 | 9.33 |
| 70k | 2.06E+01 | 6.12E-07 | 6.12 | 12.23 |
| 250k | 3.66E+01 | 1.13E-06 | 11.32 | 22.63 |
| 500k | 4.99E+01 | 1.58E-06 | 15.82 | 31.64 |
| 2000k | 9.32E+01 | 3.09E-06 | 30.92 | 61.83 |
The equations use to calculate the intrinsic viscosity [η] and the hydrodynamic radius Rh are the following:
| (1.1) |
| (1.2) |
where [η] is expressed in (mL/g) and is related to molecular weight by an empirical relation, the Mark-Houwink equation, and Rh is the hydrodynamic radius obtained by the equation 1.2. [37][38] The K and a parameter (K = 0.1361 mL/g and a = 0.45) are polymer-specific and depend on the temperature and the solvent in which the polymer is dissolved and the values are reported in the references.[37][38] A diluted solution of monodisperse fluorescent nanoparticles (purchase from Polysciences) is, instead, used to analyze the 100 nm cut-off.
After the swelling, the buffer is replaced by one of the fluorescent solutions and a confocal image of the gel edge is acquired. The incubation is carried out for 24 hours at RT to permit the dextrans or nanoparticles diffusion inside the gels. Finally, the fluorescent solution is replaced by new 1X PBS and another image is acquired to evaluate the diffusion and to define a mesh size cut-off.
Swelling characterization
Hydrogel swelling is measured using a Netsch lab+ rheometer with a plate-plate configuration and an upper geometry with a diameter of 20 mm. The hydrogel samples, with height of approximately 300 μm, are casted on an adhesive glass coverslip matching the diameter of the upper plate. The initial height of the gel is measured immediately after synthesis. During the custom sequence exploited for the measurement, the upper plate of the rheometer approaches the gel surface with a speed of 10 μm/s until a normal force of 0.1N is detected. The same measurement is performed after an overnight swelling of the gel in 1X PBS. The vertical swelling is evaluated as the percentual ratio between the final and initial heights after deducting the glass coverslip thickness. The lateral swelling of the gel is considered as negligible due to the adhesion to the underlying glass substrate.
Nuclear parameters, nuclear to cytosolic YAP/TAZ ratio and cell area
For the assessment of the nuclear parameters (nuclear thickness, projected area (NPA) and nuclear to cytosolic ratio (N/C)), z-stacks comprising the full thickness of the cells is acquired with a Leica Stellaris confocal microscope on fixed cells stained for YAP/TAZ and F-actin and counterstained with the Hoechst nuclear dye.
The nuclear thickness has been manually measured analyzing the Hoechst signal and using the orthogonal views function of Fiji.[39] For each cell, the thickness is assessed on the cross-section exhibiting the highest value.
For the NPA and cell area, the z-stack of the nuclear signal and of phalloidin signal respectively is projected on a single plane using the Z-project function in Fiji. The projection is then converted in a binary image and analyzed using the analyze particle plugin to define the nuclear and cell outlines and areas.
The data obtained are correlated with the YAP/TAZ nuclear to cytosolic ratio calculated creating a pipeline sequence in CellProfiler software, as described in “immunofluorescence” methods section.
Statistical analysis
Nuclear to cytosolic YAP/TAZ ratio: the number of analyzed cells for each analysis considered for the quantification reported in Figures 3, 4, S4, 5 and 7 is indicated in the relative Figures legends. The values reported (excluded those on Figure 7) are the means and standard deviations obtained after the signal background subtraction.
Statistically significative differences in Figures 4 and 5 are evaluated with a one-way ANOVA with Tukey’s multiple comparison test using GraphPad Prism 9 and considering a confidence interval of 95%. P-values are indicated in each Figure legends.
The Statistical significance in Figure 7 for monotonic correlation between nuclear to cytoplasmic YAP/TAZ ratio (N/C) and nuclear projected area (NPA) is evaluated by Spearman’s correlation analysis using GraphPad Prism 9 and considering a confidence interval of 95%.
Supplementary Material
Acknowledgements
GB acknowledges the Fondazione Cariparo for financial support through the project “Sensing Cell Mechanics” Bando “Ricerca Scientifica di Eccellenza 2018” and University of Padova for financial support through the project SID 2020 “New biomaterials and microtechnologies to reverse engineer the microenvironment of tissue-specific stem cells”.
The work by SP is funded by the European Research Council (ERC, DENOVOSTEM grant agreement No. 670126) and Fondazione AIRC under the 5 per Mille 2019 programme (ID No. 22759).
References
- [1].Panciera T, Azzolin L, Cordenonsi M, Piccolo S. Nat Rev Mol Cell Biol. 2017;18:758. doi: 10.1038/nrm.2017.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Kim S, Uroz M, Bays JL, Chen CS. Developmental Cell. 2021;56:180. doi: 10.1016/j.devcel.2020.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Brusatin G, Panciera T, Gandin A, Citron A, Piccolo S. Nature Mater. 2018;17:1063. doi: 10.1038/s41563-018-0180-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Vining KH, Mooney DJ. Mechanical Forces Direct Stem Cell Behaviour in Development and Regeneration. 2017 doi: 10.1038/nrm.2017.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Caliari SR, Burdick JA. Nat Methods. 2016;13:405. doi: 10.1038/nmeth.3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Gjorevski N, Sachs N, Manfrin A, Giger S, Bragina ME, Ordóñez-Morán P, Clevers H, Lutolf MP. Nature. 2016 doi: 10.1038/nature20168. [DOI] [PubMed] [Google Scholar]
- [7].Enemchukwu NO, Cruz-Acuña R, Bongiorno T, Johnson CT, García JR, Sulchek T, García AJ. Journal of Cell Biology. 2016;212:113. doi: 10.1083/jcb.201506055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Rosales AM, Anseth KS. Nat Rev Mater. 2016;1:15012. doi: 10.1038/natrevmats.2015.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Polacheck WJ, Chen CS. Nat Methods. 2016;13:415. doi: 10.1038/nmeth.3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Caliari SR, Vega SL, Kwon M, Soulas EM, Burdick JA. Biomaterials. 2016;103:314. doi: 10.1016/j.biomaterials.2016.06.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Tse JR, Engler AJ. Current Protocols in Cell Biology. 2010;47 doi: 10.1002/0471143030.cb1016s47. [DOI] [PubMed] [Google Scholar]
- [12].Engler AJ, Sen S, Sweeney HL, Discher DE. Cell. 2006;126:677. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- [13].Pelham RJ, Wang YL. Proc Natl Acad Sci. 1997;94:13661. doi: 10.1073/pnas.94.25.13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Oria R, Wiegand T, Escribano J, Elosegui-Artola A, Uriarte JJ, Moreno-Pulido C, Platzman I, Delcanale P, Albertazzi L, Navajas D, et al. Nature. 2017;552:219. doi: 10.1038/nature24662. [DOI] [PubMed] [Google Scholar]
- [15].Kechagia JZ, Ivaska J, Roca-Cusachs P. Nat Rev Mol Cell Biol. 2019;20:457. doi: 10.1038/s41580-019-0134-2. [DOI] [PubMed] [Google Scholar]
- [16].Wen JH, Vincent LG, Fuhrmann A, Choi YS, Hribar KC, Taylor-Weiner H, Chen S, Engler AJ. Nature Mater. 2014;13:979. doi: 10.1038/nmat4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Trappmann B, Gautrot JE, Connelly JT, Strange DGT, Li Y, Oyen ML, Stuart MA Cohen, Boehm H, Li B, Vogel V, et al. Nature Mater. 2012;11:642. doi: 10.1038/nmat3339. [DOI] [PubMed] [Google Scholar]
- [18].Chaudhuri O, Gu L, Klumpers D, Darnell M, Bencherif SA, Weaver JC, Huebsch N, Lee H, Lippens E, Duda GN, et al. Nature Mater. 2016;15:326. doi: 10.1038/nmat4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Stanton AE, Tong X, Lee S, Yang F. ACS Appl Mater Interfaces. 2019;11:8849. doi: 10.1021/acsami.8b21270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Stanton AE, Tong X, Yang F. Acta Biomaterialia. 2019;96:310. doi: 10.1016/j.actbio.2019.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Lee S, Stanton AE, Tong X, Yang F. Biomaterials. 2019;202:26. doi: 10.1016/j.biomaterials.2019.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Dupont S, Morsut L, Aragona M, Enzo E, Giulitti S, Cordenonsi M, Zanconato F, Le Digabel J, Forcato M, Bicciato S, et al. Nature. 2011;474:179. doi: 10.1038/nature10137. [DOI] [PubMed] [Google Scholar]
- [23].Aragona M, Panciera T, Manfrin A, Giulitti S, Michielin F, Elvassore N, Dupont S, Piccolo S. Cell. 2013;154:1047. doi: 10.1016/j.cell.2013.07.042. [DOI] [PubMed] [Google Scholar]
- [24].Panciera T, Citron A, Di Biagio D, Battilana G, Gandin A, Giulitti S, Forcato M, Bicciato S, Panzetta V, Fusco S, et al. Nat Mater. 2020;19:797. doi: 10.1038/s41563-020-0615-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Grevesse T, Versaevel M, Circelli G, Desprez S, Gabriele S. Lab Chip. 2013;13:777. doi: 10.1039/c2lc41168g. [DOI] [PubMed] [Google Scholar]
- [26].Mitsi M, Handschin S, Gerber I, Schwartländer R, Klotzsch E, Wepf R, Vogel V. Biomaterials. 2015;36:66. doi: 10.1016/j.biomaterials.2014.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Wörz A, Berchtold B, Moosmann K, Prucker O, Rühe J. J Mater Chem. 2012;22:19547 [Google Scholar]
- [28].Wilson MJ, Liliensiek SJ, Murphy CJ, Murphy WL, Nealey PF. Soft Matter. 2012;8:390. doi: 10.1039/C1SM06589K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Kirby TJ, Lammerding J. Nat Cell Biol. 2018;20:373. doi: 10.1038/s41556-018-0038-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Aureille J, Buffière-Ribot V, Harvey BE, Boyault C, Pernet L, Pernet T, Pernet G, Balland M, Fraboulet S, Van Landeghem L, Guilluy C. EMBO Reports. 2019;20:e48084. doi: 10.15252/embr.201948084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Elosegui-Artola A, Andreu I, Beedle AEM, Lezamiz A, Uroz M, Kosmalska AJ, Oria R, Kechagia JZ, Rico-Lastres P, Le Roux A-L, et al. Cell. 2017;171:1397. doi: 10.1016/j.cell.2017.10.008. [DOI] [PubMed] [Google Scholar]
- [32].Lomakin AJ, Cattin CJ, Cuvelier D, Alraies Z, Molina M, Nader GPF, Srivastava N, Sáez PJ, Garcia-Arcos JM, Zhitnyak IY, et al. Science. 2020;370:eaba2894. doi: 10.1126/science.aba2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Shiu J-Y, Aires L, Lin Z, Vogel V. Nat Cell Biol. 2018;20:262. doi: 10.1038/s41556-017-0030-y. [DOI] [PubMed] [Google Scholar]
- [34].Khatau SB, Hale CM, Stewart-Hutchinson PJ, Patel MS, Stewart CL, Searson PC, Hodzic D, Wirtz D. PNAS. 2009;106:19017. doi: 10.1073/pnas.0908686106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Singh SP, Schwartz MP, Lee JY, Fairbanks BD, Anseth KS. Biomaterials Science. 2014;2:1024. doi: 10.1039/C4BM00022F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Grevesse T, Versaevel M, Gabriele S. JoVE. 2014:51010. doi: 10.3791/51010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Armstrong JK, Wenby RB, Meiselman HJ, Fisher TC. Biophysical Journal. 2004;87:4259. doi: 10.1529/biophysj.104.047746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Masuelli MA. Journal of Polymer and Biopolymer Physics Chemistry. 2013;1:13. [Google Scholar]
- [39].Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. Nat Methods. 2012;9:676. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Gandin A, Murugesan Y, Torresan V, Ulliana L, Citron A, Contessotto P, Battilana G, Panciera T, Ventre M, Netti AP, Nicola L, et al. Scientific Report. doi: 10.1038/s41598-021-01036-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.