Abstract
Class II terpene cyclases, such as oxidosqualene and squalene-hopene cyclases, catalyze some of the most complex polycyclization reactions. They minimally exhibit a β,γ-didomain architecture that has been evolutionarily repurposed in a wide range of terpene-processing enzymes and likely resulted from a fusion of unidentified monodomain proteins. Although single domain class I terpene cyclases have already been identified, single domain class II terpene cyclases have not been previously reported. Here we report high-resolution X-ray structures of a monodomain class II cyclase, merosterolic acid synthase (MstE). With a minimalistic β-domain architecture, this cyanobacterial enzyme is able to construct four rings in cytotoxic meroterpenoids with a sterol-like topology. The structures with bound substrate, product, and inhibitor provide detailed snapshots of a cyclization mechanism largely governed by residues located in a noncanonical enzyme region. Our results complement the few known class II cyclase crystal structures, while also indicating that archaic monodomain cyclases might have already catalyzed complex reaction cascades.
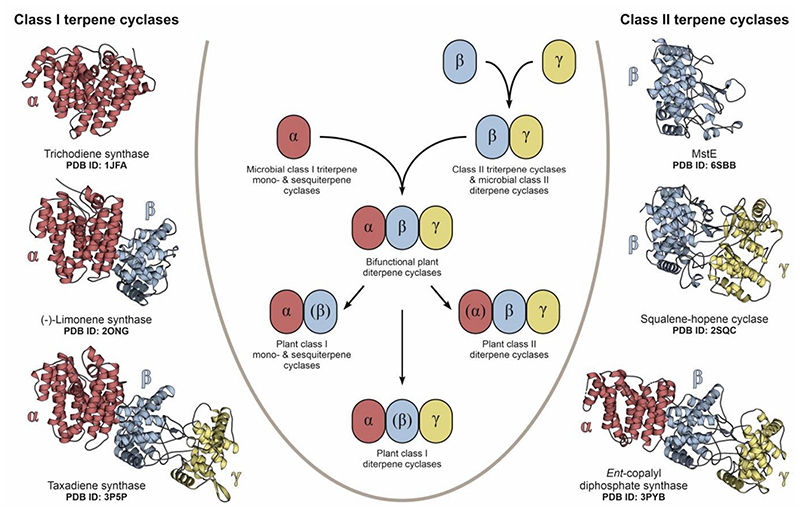
Terpene cyclases are key biosynthetic determinants for the vast diversity of isoprenoids, currently encompassing more than 76,000 compounds (Dictionary of Natural Products. http://dnp.chemnetbase.com). These enzymes convert acyclic substrates to cyclized products that can feature multiple new stereocenters and carbon-carbon bonds1,2. Among the two major classes of terpene cyclases3, class I enzymes dissociate pyrophosphorylated substrates to generate a carbocation for cyclization4, while class II enzymes protonate olefinic or epoxide moieties5. In canonical type II cyclases, an Asp residue in a conserved DXDD motif is responsible for Brønsted acid catalysis6. Available cyclase structures contain up to three domains, termed α, β, and γ (Fig. 1)7–9. Single-domain architectures are currently only known for the α-domain microbial class I terpene cyclases10–13. Other class I enzymes feature αβ or αβγ architectures, with the α-domain containing the active site14–16. For class II terpene cyclases, which generate some of the most complex terpene skeletons, only βγ or αβγ architectures are known17–20 It has been proposed that plant αβγ diterpene cyclases evolved from bacterial progenitors by fusion of the α and βγ enzymes and subsequently gave rise to αβ-didomain synthases by loss of the γ domain7. The class II βγ enzymes likely evolved from monodomain proteins, but as the catalytic acid residue is located in the β-region, and the γ-region provides most of the remaining residues needed for polycyclic product formation, it was unknown how a single domain might achieve catalysis.
Fig. 1. Architectural variants of class I and class II terpene cyclases.
The central scheme displays the proposed evolution of terpene cyclases from ancestral mono-domain enzymes3. Structurally related domains are colored and labeled α, β or γ accordingly, while parentheses indicate the absence of an active site. Cartoon representations to the left (class I) and right (class II) visualize the structural domain arrangement in selected examples with the respective PDB entry code noted below. So far, single domain architectures have only been described in microbial class I terpene cyclases, while MstE is a potential ancestor of βγ-assemblies in class II enzymes. Proposedly, bifunctional αβγ diterpene cyclases formed through fusion of α and βγ precursors and later diversified into a broad array of specialized enzymes.
Results and discussion
The structure of apo merosterolic acid synthase
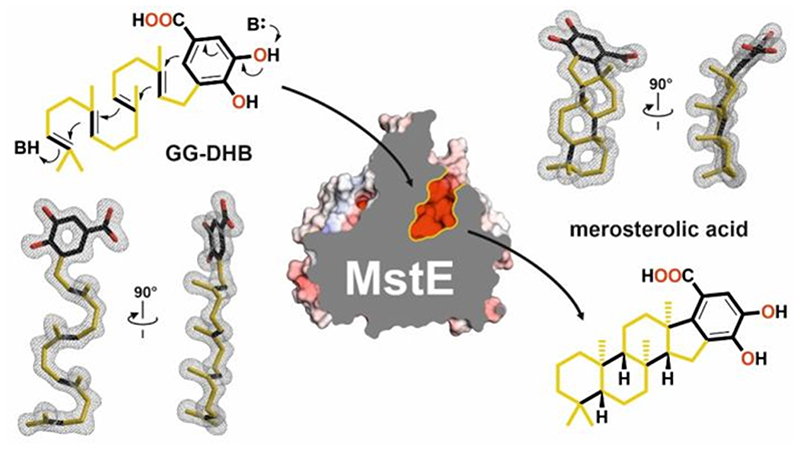
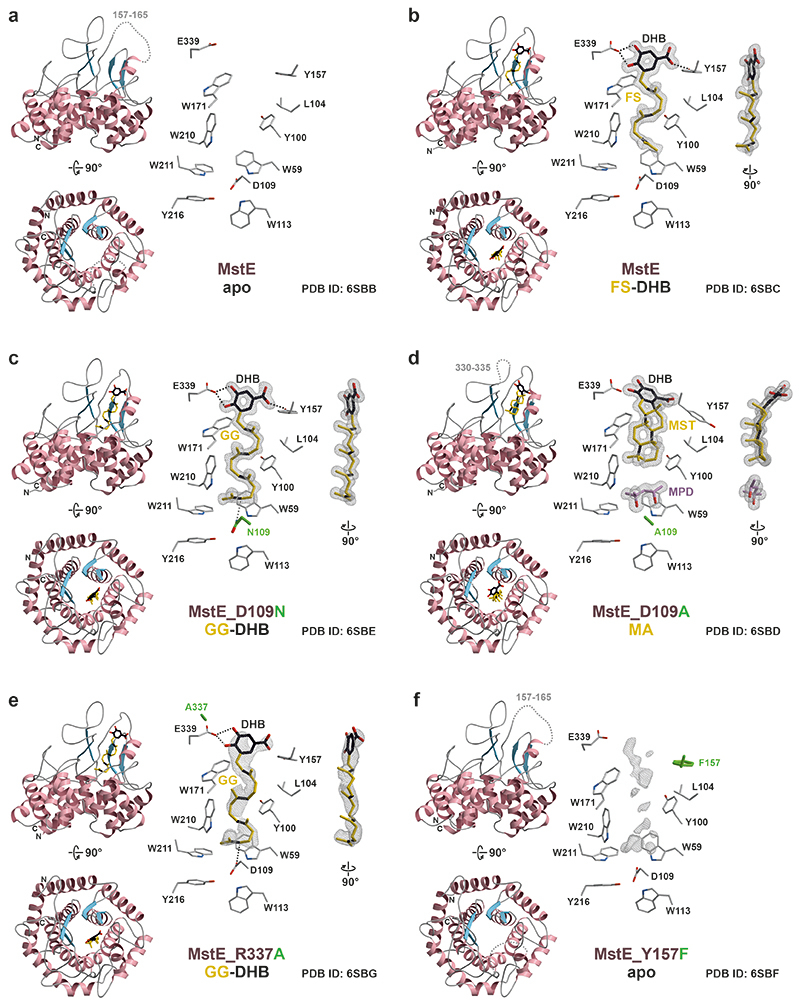
Here we report merosterolic acid synthase (MstE) as a founding member of the monodomain class II cyclases and present detailed structural and functional insights into the formation of its polycyclic steroid-like product. MstE constructs the core scaffold of the cyanobacterial cytotoxin merosterol A (Fig. 2A, 1), isolated from Scytonema sp. PCC 10023, by generating the pentacyclic intermediate merosterolic acid A (2)21. Although MstE produces a steroid-like skeleton, it does not act on a squalene-derived substrate but diverts the ubiquinone-type precursor 5-geranylgeranyl-3,4-dihydroxybenzoate (GG-DHB, 3) to specialized metabolism. X-ray structure analysis of MstE at resolutions up to 1.3 Å revealed atomic insights into this complex reaction (Supplementary Table 1). The monomeric state of MstE was confirmed by PISA calculations and size-exclusion chromatography22. It shows sequence homology to triterpene cyclases, such as squalene-hopene cyclases (SHCs, PDB-ID: 1H3B23, 2SQC24) and oxidosqualene synthases (OSCs, 1W6K18), classifying MstE as a class II cyclase (Extended Data Fig. 1–2). It is, however, significantly smaller than the homologs (Supplementary Fig. 1). MstE exhibits a conserved α6-α6 barrel fold with outer (o) and inner (i) helices forming a tilted circular arrangement (base subdomain, Fig. 3A).
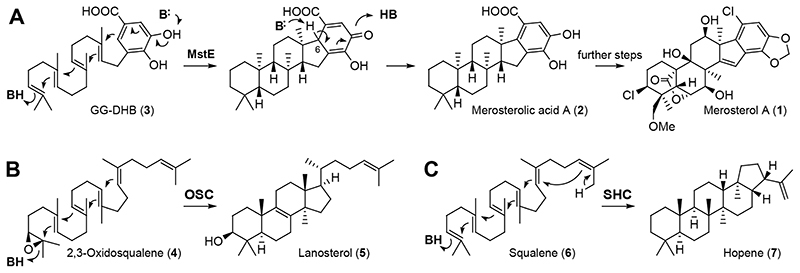
Fig. 2. Merosterol biosynthesis and comparison with the lanosterol and hopene cyclization pathways.
(A) MstE cyclizes GG-DHB (3) to generate the merosterol core skeleton for cytotoxin biosynthesis. Cyclization is initiated by direct protonation of the terminal double bond and the final carbocation is quenched by electrophilic aromatic substitution. Aromaticity of the carboxylic acid moiety is restored by deprotonation at C6. (B) Reaction catalyzed by the oxidosqualene cyclase (OSC) in steroid biosynthesis. In this case, a terminal epoxide is protonated for initiating the cyclization cascade. Additional methyl and hydrogen shifts are omitted. (C) Reaction catalyzed by squalene-hopene cyclase (SHC) in membrane hopanoid biosynthesis.
Fig. 3. The structure of MstE.
(A) Topological analysis of MstE in complex with GG-DHB (3, double bonds are highlighted,). Left: Top view with secondary structure elements shown as cartoon. The outer (oH1-6) and inner α6-ring (iH1-6) are colored in dark and light pink, respectively. The substrate is lined by an additional helix (H, gray) and β-sheets (S1-4, blue). N- and C-termini are labeled. Right: Topology diagram depicting polar (black) and aromatic (green) interactions between residues of MstE and 3 (close contacts indicated by green bars). Purple squares show the relative position of two distinct QW-motifs. (B) Side view of the terpene cyclase highlighting head and base subdomains. (C) Surface representation of MstE with colors indicating negative and positive electrostatic potentials. Left: MstE aligned with 3 from the D109N crystal structure. Right: MstE D109A in complex with merosterolic acid A (MA, 2). (D) Close-up view of the active site of MstE_D109N. The 2FO-FC electron density map (gray mesh, contoured to 1.0 σ) of 3 is shown together with residues engaged in ligand binding. (E) MstE_D109A retains the cyclic product (MST) carrying the dislocated DHB head-group (2) and 2-methyl-2,4-pentanediol (MPD, purple sticks). C-C bonds formed by cyclization are colored black.
Interestingly, helix oH1 of MstE corresponds closely to the N-terminal helix of the β-domain in canonical βγ-didomain triterpene cyclases (Extended Data Fig. 1). While the γ-domain of SHCs and OSCs mediates association and product incorporation into membranes, MstE solely uses the pronounced coils between each o and i helix (head subdomain, Fig. 3B) to form an extended central cavity about 17 Å deep and 6 Å wide (Fig. 3C). Residues 157 to 165, at the entrance of the head subdomain’s proposed terpene-binding site, are disordered in the apo structure (1.95 Å, PDB ID: 6SBB). Being lined with aliphatic and aromatic residues, the channel displays a negative electrostatic potential attracting hydrophobic molecules from the crystallization conditions (e.g. PEG) in absence of a natural ligand. Between the base and head subdomains, two Q-X5-W motifs located at the C-termini of oH4 and oH6 generate additional stability to endure the exergonic reaction. Found commonly in SHCs and OSCs17,25, they consist of tryptophan residues stacking with glutamine side-chains that form H-bonds towards the neighboring helix.
Closed-state structure of MstE with non-reactive substrate
First insights into the reaction cascade were gained by the co-crystal structure of MstE and non-reactive pseudo-substrate farnesyl dihydroxybenzoate (FS-DHB, 8) at 1.35 Å (PDB ID: 6SBC, Extended Data Fig. 3B, Supplementary Fig. 2, 3). The surrogate is fully defined in the 2FO-FC electron density map and located inside the binding channel with its C15-isoprenylic moiety oriented towards the protein core. Both hydroxy groups in DHB form strong H-bonds with Glu339. Upon ligand binding, residues 157 to 165 fold around the entrance in a substrate-induced fit, preventing access of solvent that could quench carbocations during the reaction. This is predominantly mediated by interactions between the DHB carboxylate and Tyr157 as well as the salt bridge between Asp162 and Arg337. Based on the structure of MstE:8, single-point mutants with impaired activity were constructed. Based on the protein alignment Asp109 was the likely protonating residue and was therefore chosen for mutagenesis (Extended Data Fig. 1). Tyr157 was selected for its interaction with the carboxylic acid of the DHB and Glu339 for its H-bonds with the DHB hydroxyl groups. Arg337 was chosen because of its interaction with Glu339.
Structures of mutant MstE with natural substrate and product
Crystallization trials in presence of the natural substrate GG-DHB (3) provided structures for Y157F (1.3 Å, PDB ID: 6SBF), R337A (2.3 Å, PDB ID: 6SBG), D109N (1.4 Å, PDB ID: 6SBE) and D109A (1.4 Å, PDB ID: 6SBD). MstE_Y157F resembles the apo state with diffuse electron density in the prenyl-binding cavity and supports the role of Tyr157 as a gatekeeper for substrate recognition (Extended Data Fig. 3F). 3 was identified in MstE_R337A (Extended Data Fig. 3E), but the lack of stabilization at the entrance resulted in low ligand occupancy. Intriguingly, MstE_D109N forms a complex with 3, where the ligand occupies the binding pocket entirely (Fig. 3D, Extended Data Fig. 3C, Supplementary Fig. 2). In accordance with the stereoisomeric configuration of the polycyclic product, all isoprenyl segments are forced into chair-like conformations. Together with the one-sided parallel alignment of all methyl groups, this compressed shape provides significant catalytic energy. In the superposition of MstE_D109N:3 and MstE:8 (RMSD-Cα: 0.12 Å) N109 takes the exact position of D109 and rests perpendicular to the terminal C=C bond of 3 at 3.2 Å. The residue is positioned in a DXD-polar motif conserved among class II cyclases and known to initiate olefin protonation8. Unexpectedly, MstE_D109A co-crystallized with 3 revealed cyclized product MA (2) in the binding pocket (Fig. 3E, Extended Data Fig. 3D, Supplementary Fig. 2). Presumably, the energy contribution through substrate coordination and product stabilization is sufficient for a minimal turnover within the time frame of crystallization experiments over several days (Supplementary Fig. 4). While the linear substrate observed in MstE_D109N:3 occupies the entire channel with a length of 17 Å, 2 is condensed to 12 Å and bent by methyl repulsions. Notably, 2-methyl-2,4-pentanediol (MPD) from crystallization buffers occupies the vacant space and seals the channel bottom. This might aid the selective binding of product even in excess of substrate.
Simultaneously, the DHB moiety of 2 loses H-bond contact to Glu339 at the meta-hydroxy group (Fig. 3E), and the carboxylate leaves the aromatic plane due to steric clashes with the generated C5-ring. As a result, the gatekeeper Tyr157 turns by 90° and Arg337 is displaced, forming a salt bridge with Glu339. The collective effect disorders the region between residues 330 and 335, supporting product release.
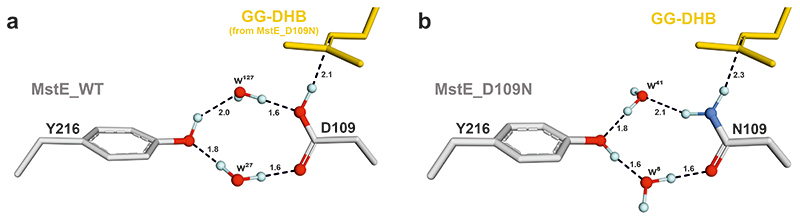
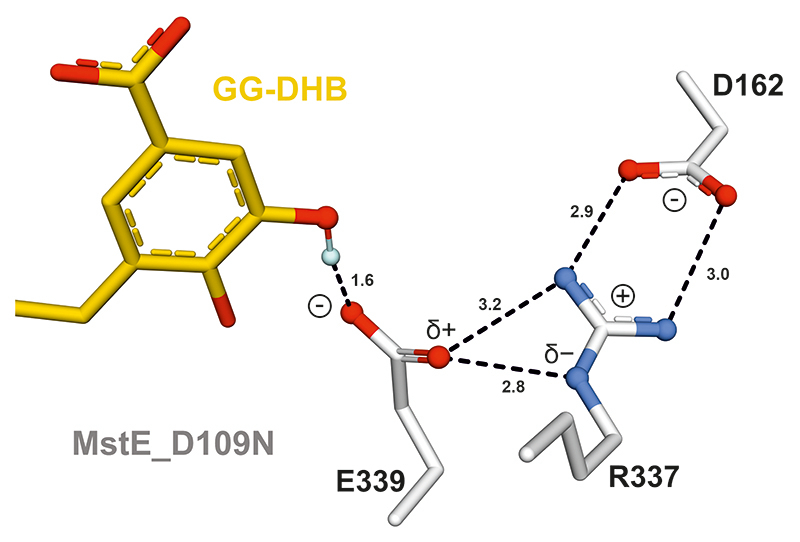
The structural snapshots at atomic resolution suggest that Asp109 initiates the reaction with its proton oriented in anti-configuration. This activated Brønsted acid forms a charge-relay system with Tyr216 and two water molecules (Extended Data Fig. 4, Supplementary Fig. 5). After protonation of the terminal C=C bond in 3 and concomitant formation of the first cycle26,27, the acid is likely regenerated by bulk water through solvent-accessible channels. During the subsequent cascade, precisely positioned aromatic residues of the binding pocket would stabilize intermediate carbocations until final cyclization onto the DHB π-system28. Mutation of aromatic residues likely fulfilling this role in MstE were mutated, resulting in enzymes no longer converting substrate to any product (Supplementary Fig. 6). Based on the MstE_R337A structure, the strong salt bridge between the Arg residue and Asp162 increases the basicity of Glu339, which deprotonates the DHB meta-hydroxy group, pushing π-electrons into the ring (Extended Data Fig. 5). Subsequently, spontaneous deprotonation at C6 and keto-enol tautomerism restores the aromatic system (Fig. 2A).
Mutant MstE with expanded product specificity
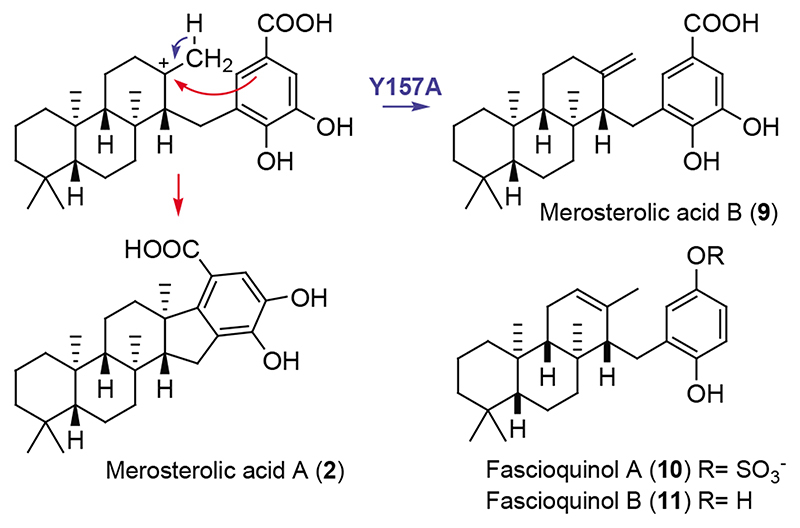
Members of the triterpene synthase family generate a plethora of structural scaffolds based on active-site geometries, with particularly impressive diversification found in plants29. This raises prospects that our structural insights might facilitate engineering of soluble cyclases yielding modified products. Therefore, we screened structure-guided mutants of MstE for alterations to the polycyclization reaction (Supplementary Fig. 6, 7). Among these, MstE_Y157A, predicted to exhibit weakened interactions with the DHB head group, formed a new product 9 after incubation with GG-DHB (Supplementary Table 2, Supplementary Fig. 8–16). Characterization of 9, termed merosterolic acid B, by NMR identified a tricyclic isoprenoid moiety containing an exo-methylene group. The structure is consistent with the proposed role of Y157 and suggests that dislocation of the DHB π-system prevents attack by the third-ring carbocation intermediate, resulting in deprotonation as an alternative reaction path (Fig. 4). Interestingly, the artificial compound 9 resembles fascioquinols (10-11), meroterpene antibiotics reported from a deep-water sponge30.
Fig. 4. Biosynthesis of new product by mutant MstE Y157A.
Without the stabilizing effect of Y157 the aromatic ring of the benzoic acid can rotate away from the carbocation, allowing for premature deprotonation at the methyl group and the production of merosterolic acid B (9) in addition to 2. The new product shows structural similarity to the antibiotic fascioquinols A and B (10, 11), for which only the relative configuration is known.
SHCs are ancient enzymes that already existed 2.7 billion years ago and have been regarded as the origin of the βγ-didomain fold31. While the β-domain is preserved in most terpene cyclases (Fig. 1), MstE is active despite lacking a γ-domain, present in all other characterized di- and triterpene cyclases, revealing that class II synthases can assemble complex polycyclic scaffolds with monodomain architectures. It remains unclear whether the βγ-fold evolved through insertion of a γ-domain between oH1 and iH2 of an ancestral protein, or whether MstE derived from a βγ predecessor by the deletion of helices 2-13. To date, triterpene synthase-ligand interactions have only been reported for SHC bound to an inhibitor and OSC complexed with its product17,18. This study on MstE visualizes crucial steps in terpenoid biosynthesis at atomic resolution. The availability of a soluble cyclase that accepts pyrophosphate-free substrates offers augmented perspectives for chemoenzymatic synthesis using engineered enzymes as shown for MstE.
Methods
Cloning of MstE for crystallization
The Scytonema sp. PCC10023 MstE gene (GenBank: ATN39899.1) was obtained from Eurofins Genomics (Ebersberg, Germany) after sequence optimization for recombinant expression in Escherichia coli. The gene was cloned into a pET-28b(+) vector (Invitrogen, Darmstadt, Germany) carrying an N-terminal H6-SUMO tag (Smt3p from Saccharomyces cerevisiae, residues 1-98). Single-point mutagenesis was carried out by QuikChange II site-directed mutagenesis (Agilent Technologies, Waldbronn, Germany). Sequence integrity was confirmed by Sanger sequencing at GATC Biotech (Konstanz, Germany).
Recombinant protein expression and purification for crystallization
Transformed E. coli BL21 (DE3) strains (Novagen/Merck, Darmstadt, Germany) was grown in 2 L lysogeny broth (50 mg/L kanamycin) at 130 rpm and 37 °C until an OD600 of 0.6 was reached. Expression was induced by addition of 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) and occurred over night at 20 °C. After harvest by centrifugation, cell pellets were washed in 0.9% (w/v) NaCl and stored at -20 °C. Cell pellets were resuspended in 60 mL buffer A (100 mM Tris/HCl, pH 7.5, 500 mM NaCl, 20 mM imidazole, 10 mM β-mercaptoethanol) with 1 mg DNAse I and 5 mg Pefabloc (Sigma-Aldrich, Taufkirchen, Germany) and disrupted by sonication with a Branson Digital Sonifier 250 (G. Heinemann, Schwäbisch Gmünd, Germany). After centrifugation (40.000 × g, 30 min, 4 °C) the supernatant was applied to a 5 mL HisTrap HP column equilibrated with buffer A using an ÄKTA prime plus system (GE Healthcare, Freiburg, Germany) at a flow-rate of 5 mL/min. The column was washed with 10 CV of buffer A prior to elution with a gradient of buffer B (0 - 100% in 50 mL). After addition of 0.5 mg SUMO protease (Ulp1 from S. cerevisiae), the eluate was dialyzed against 5 L buffer C (20 mM Tris/HCl, pH 7.5, 100 mM NaCl, 1 mM β- mercaptoethanol) overnight at 4 °C. Affinity chromatography was repeated, flow-through and wash fractions were collected and concentrated to 2 mL using Vivaspin ultrafiltration centrifugal concentrators (Sartorius, Göttingen, Germany). Protein concentrates were subjected to size-exclusion chromatography with a HiLoad Superdex 200 16/60 column equilibrated with buffer D (buffer C with 5 mM dithiothreitol instead of β-mercaptoethanol) on an ÄKTA purifier system (GE Healthcare, Freiburg, Germany) at 1.5 mL/min. Peak fractions were analyzed by SDS-PAGE, concentrated to at least 40 mg/mL using centrifugal filters and either applied directly in crystallography or flash frozen in liquid nitrogen and stored at -80 °C.
Protein crystallization and structure determination
Crystals of MstE were grown under conditions annotated in Supplementary Table 3 by the sitting drop vapor diffusion method. Co-crystallization was performed by adding 2 mM (final concentration) of the potential ligands. Crystals adequate for structure determination were soaked with cryo-protectant (50% reservoir solution with 50% glycerol) and frozen in liquid nitrogen. Diffraction datasets at 1.0 Å were obtained at the X-ray beamline X06SA (Swiss Light Source, Paul Scherrer Institute, Villigen, Switzerland). Diffraction intensities were evaluated by the XDS software package32. Molecular replacement was performed with Phaser by Patterson function calculations using coordinates of a squalene-hopene cyclase (PDB ID: 2SQC) truncated to a poly-alanine chain24,33. Restrained refinement of the structure with Refmac5 was alternated by model building with Coot34,35. ARP/wARP solvent was applied to add water molecules prior to finalization by TLS (Translation/Liberation/Screw) and restrained refinements until adequate values for Rwork and Rfree as well as all geometric parameters were reached (analyzed by the MolProbity webserver)36,37. Ramachandran compliance was additionally tested with Procheck38. The monomeric state of MstE was underlined by PISA webserver calculations 22. Structurally similar proteins were identified by a Dali search39. Figures were prepared using Molscript and PyMol40,41. The crystal structures were deposited at the RCSB Protein Data Bank under accession codes 6SBB, 6SBC, 6SBD, 6SBE, SBF and 6SBG, respectively.
Cloning, expression and purification of MstE for mutant screening
To generate point mutants of MstE, pET-29b MstE was used from the previous study21. Point mutations were introduced with the QuikChange mutagenesis method (Agilent Technologies) and confirmed by sequencing. Primers were designed using the PrimerX tool (http://www.bioinformatics.org/primerx/). The generated plasmids containing mutant MstE were transformed into E. coli BL21 tuner (DE3) cells (Novagen). For expressions, cells were grown in terrific broth supplemented with 50 μg/mL kanamycin to an OD600 of approximately 0.8 at 30 °C, induced with 0.1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) and incubated at 16 °C for another 20 h. Cells were harvested by centrifugation (20 min, 5000 × g, 4 °C), resuspended in buffer (50 mM phosphate buffer pH 7.8, 300 mM NaCl, 10 mM imidazole) and lysed by sonication (10 × 10 s pulses with 10 s breaks, on ice). After centrifugation at 15000 × g and 4 °C for 30 min, MstE was purified from the supernatant with Ni-NTA (Macherey-Nagel). The column was washed with buffer containing 20 mM imidazole and the protein was eluted stepwise with buffer containing 100 mM imidazole followed by 200 mM imidazole. Elution fractions were run on an SDS-PAGE and used for enzymatic assays directly.
Enzymatic assays
For quick assays, 1 mM of 3-geranylgeranyl-4,5-dihydroxybenzoic acid (3, 100 mM stock in DMSO, synthesized in previous study21) was dissolved in 50 μL buffer (50 mM phosphate pH 7.8, 300 mM NaCl). 50 μL of the protein elution fraction was added and the mixture was incubated at room temperature for 16 h.
The assays were quenched with 1 μL formic acid (FA) and extracted 3 times with 100 uL ethyl acetate. Organic phases were combined, dried in vacuo, resuspended in 100 uL methanol and analysed by liquid chromatography/high-resolution mass spectrometry (LC/HRMS) on a Thermo Scientific Q Exactive mass spectrometer coupled to a Dionex Ultimate 3000 UPLC system. We used a Phenomenex Kinetex C18 (4.6x150 mm) with a gradient from 5% acetonitrile + 0.1% FA in water + 0.1% FA to 100% acetonitrile + 0.1% FA in 10 minutes. To minimize substrate inhibition, protein concentrations were set to 200 μM and substrate concentrations to 0.5 mM.
Synthesis General Procedures
For the synthesis of 8 the procedure described by Lang M. and Steglich W. (2005) was followed42. For detailed procedures see supplementary information.
Purification of merosterolic acid B and NMR analysis
The ethyl acetate extracts of large-scale assays were separated by reversed-phase HPLC (Phenomenex Kinetex C18, 10 × 250 mm) with an isocratic run of 85% acetonitrile + 0.1% formic acid in water + 0.1% formic acid and a flow rate of 2 mL/min. The fraction containing the compound of interest was further purified by reversed-phase HPLC (Phenomenex phenyl-hexyl, 10 × 250 mm) with an isocratic run of 73% acetonitrile + 0.1% formic acid in water + 0.1% formic acid. Purifications from assays containing approximately 700 mg of MstE Y157A yielded 300 - 400 μg of product. NMR spectra were recorded on a Bruker Avance III spectrometer equipped with a cold probe at 600 MHz for 1H NMR and 150 MHz for 13C NMR. For details on the structure elucidation see supplementary information.
Protein alignments and phylogenetic analysis
Protein sequences were retrieved from the UniProt database, and β domains were identified based on alignment to available crystal structures for each domain architecture (MstE from this study, 1SQC, 1W6K, 2ONG, or 3PFP). Sequences corresponding to the β domain and up to 10 residues on either side were aligned using MUSCLE43. MEGA 7 was used to generate the tree (Extended Data Fig. 2) from the alignment using the Maximum Likelihood method based on the Whelan and Goldman model44,45. All positions with less than 20% site coverage were eliminated, and the resulting tree with the highest log likelihood is shown. Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using a JTT model, and then selecting the topology with superior log likelihood value. A discrete Gamma distribution was used to model evolutionary rate differences among sites (5 categories (+G, parameter = 3.5036)).
Extended Data
Extended Data Fig. 1. Protein alignment.
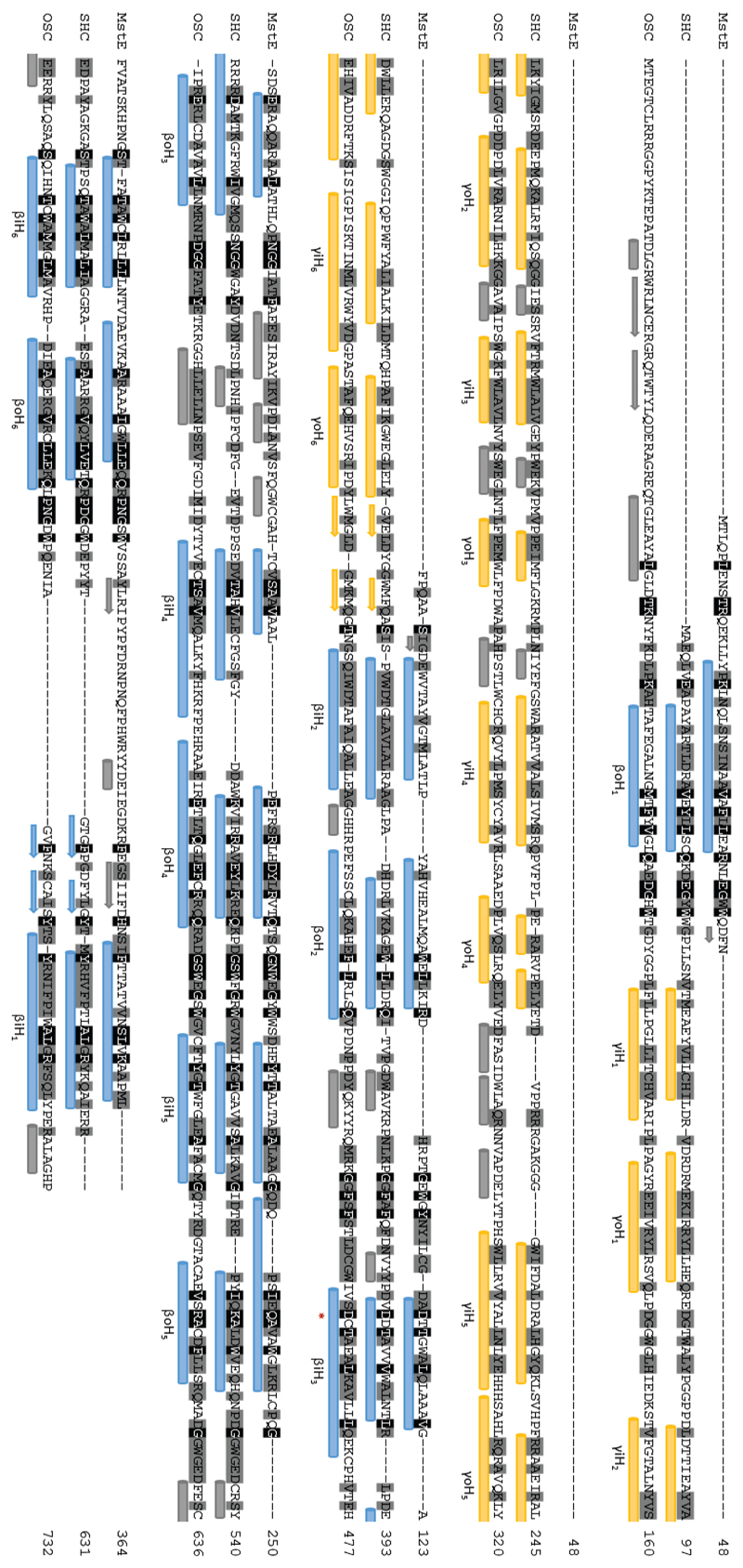
Structure-guided alignment of merosterolic acid synthase (MstE) with squalene-hopene cyclase (SHC; PDB 2SQC; 15.91 % identity*) and oxidosqualene cyclase (OSC; PDB 1W6K; 15.26 % identity*). Alpha helices are indicated with cylinders under the sequences, beta strands with arrows. These secondary structural elements are colored yellow if they are part of the core scaffold of the γ domain and blue if they are part of β domain. Non-conserved secondary structures are depicted in grey. They catalytic aspartate residue is indicated with a red asterisk. *Identity percentages were calculated without the γ-domains of SHC and OSC.
Extended Data Fig. 2. Phylogenetic tree of selected terpene cyclase β domains.
Maximum-likelihood tree of β domains from functionally characterized and unknown terpene cyclases, as described in the Materials and Methods above. Organisms are colored according to the phylogeny of their origin: Protists grey, animals gold, fungi red, plants green, and bacteria blue. Terpene enzyme class, domain architecture, and product are also indicated.
Extended Data Fig. 3. Structures of MstE in complex with different ligands.
Left: Side- and top-views (rotated by 90°) of MstE (A), MstE:FS-DHB (B), MstE_D109N:GG-DHB (C), MstE_D109A:MA (D), MstE_R337A:GG-DHB (E) and MstE_Y157F (F) as cartoons with helices and β-sheets colored in pink and blue, respectively. Dashed loops indicate disordered regions in the crystal structures and are labeled accordingly. Carbon atoms of ligands, if present, are shown as sticks in gold, with the DHB moieties in black. Double bonds and newly formed C-C bonds are highlighted in black. Right: Close-up views of the active sites. The 2Fo-FC electron density maps (gray mesh, contoured to 1.0 σ) of ligands, if present, are shown together with residues engaged in ligand binding. Side-views of all ligands (rotated by 90°) are depicted on the far right. Dashed lines represent important interactions. Mutated residues are colored green. Diffuse electron density in F indicates the presence of a ligand at low occupancy.
Extended Data Fig. 4. A proton relay conveys protonation of GG-DHB.
(A) The structure of MstE:FS-DHB superpositioned with MstE_D109N:GG-DHB (RMSD-Cα: 0.12 Å) reveals that Asp109 adopts anti-position for protonation of the terminal C=C in GG-DHB. Two water molecules align between Tyr216 and Asp109 to activate the Brønsted acid and replace the proton after reaction. Oxygen atoms are highlighted in red; the protons (light blue) were added with PyMol (37) and oriented according to potential interactions represented as dashed lines (distance given in Å). (B) The primary amine of Asn109 in MstE_D109N:GG-DHB displaces one of the water molecules by 0.6 Å, flipping the orientation of all protons in the cluster.
Extended Data Fig. 5. A charge relay supports deprotonation of GG-DHB.
The structure of MstE_D109N:GG-DHB reveals the formation of a triad between Glu339, Arg337 and Asp162. Asp162 forms a salt bridge with the guanidino group of Arg337. This pushes electrons towards the secondary amine which in turn polarizes the carboxylate of Glu339, strengthening the H-bond with the meta-hydroxy group of DHB and increasing basicity. After deprotonation, this cluster acts as a proton-relay, before it is promptly broken and reordered to accommodate the product. Heteroatoms are highlighted in red (O) and blue (N); the proton (light blue) was added with PyMol. Interactions are represented as dashed lines (distance given in Å).
Supplementary Material
Acknowledgments
We thank R. Ueoka for support with NMR and the staff of beamline X06SA at the Paul Scherrer Institute for assistance during data collection. We acknowledge funding by the Swiss National Science Foundation (205321_165695 and 205320_185077) and the Helmut Horten Foundation to J.P.
Footnotes
Author contributions: P.M., F.E., M.G., and J.P. designed, conducted, and analyzed the experiments, S.L.-M. and C.L.D. conducted organic syntheses, J.K.B.C. performed phylogenetic analyses, J.P., M.G, F.E., and P.M. wrote the paper. All authors edited the paper.
Competing interests: The authors declare no competing financial interests.
Data availability
Crystallographic data were deposited in the RCSB Protein Data Bank with the PDB IDs: 6SBB, 6SBC, 6SBD, 6SBE, 6SBF and 6SBG. All other data are available in the text or in the supplementary information.
References
- 1.Dickschat JS. Isoprenoids in three-dimensional space : the stereochemistry of terpene biosynthesis. Nat Prod Rep. 2011;28:1917–1936. doi: 10.1039/c1np00063b. [DOI] [PubMed] [Google Scholar]
- 2.Rudolf JD, Chang C-Y. Terpene synthases in disguise: enzymology, structure, and opportunities of non-canonical terpene synthases. Nat Prod Rep. 2019 doi: 10.1039/c9np00051h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wendt KU, Schulz GE. Isoprenoid biosynthesis: manifold chemistry catalyzed by similar enzymes. Structure. 1998;6:127–133. doi: 10.1016/s0969-2126(98)00015-x. [DOI] [PubMed] [Google Scholar]
- 4.Lesburg CA, Zhai G, Cane DE, Christianson DW. Crystal Structure of Pentalenene Synthase : Mechanistic Insights on Terpenoid Cyclization Reactions in Biology. Science. 1997;277:1820–1824. doi: 10.1126/science.277.5333.1820. [DOI] [PubMed] [Google Scholar]
- 5.Wendt KU, Schulz GE, Corey EJ, Liu DR. Enzyme Mechanisms for Polycyclic Triterpene Formation. Angew Chemie Int Ed. 2000;39:2812–2833. [PubMed] [Google Scholar]
- 6.Hammer SC, Marjanovic A, Dominicus JM, Nestl BM, Hauer B. Squalene hopene cyclases are protonases for stereoselective Brønsted acid catalysis. Nat Chem Biol. 2015;11:121–126. doi: 10.1038/nchembio.1719. [DOI] [PubMed] [Google Scholar]
- 7.Cao R, et al. Diterpene cyclases and the nature of the isoprene fold. Proteins Struct Funct Bioinforma. 2010;78:2417–2432. doi: 10.1002/prot.22751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Christianson DW. Structural and Chemical Biology of Terpenoid Cyclases. Chem Rev. 2017;117:11570–11648. doi: 10.1021/acs.chemrev.7b00287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oldfield E, Lin FY. Terpene biosynthesis: Modularity rules. Angew Chemie - Int Ed. 2012;51:1124–1137. doi: 10.1002/anie.201103110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Köksal M, Chou WKW, Cane DE, Christianson DW. Structure of 2-methylisoborneol synthase from Streptomyces coelicolor and implications for the cyclization of a noncanonical C -methylated monoterpenoid substrate. Biochemistry. 2012;51:3011–3020. doi: 10.1021/bi201827a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baer P, et al. Induced-fit mechanism in class I terpene cyclases. Angew Chemie Int Ed. 2014;53:7652–7656. doi: 10.1002/anie.201403648. [DOI] [PubMed] [Google Scholar]
- 12.Rynkiewicz MJ, Cane DE, Christianson DW. Structure of trichodiene synthase from Fusarium sporotrichioides provides mechanistic inferences on the terpene cyclization cascade. Proc Natl Acad Sci U S A. 2001;98:13543–13548. doi: 10.1073/pnas.231313098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Janke R, Görner C, Hirte M, Brück T, Loll B. The first structure of a bacterial diterpene cyclase: CotB2. Acta Cryst. 2014;D70:1528–1537. doi: 10.1107/S1399004714005513. [DOI] [PubMed] [Google Scholar]
- 14.Starks CM, Back K, Chappell J, Noel JP. Structural Basis for Cyclic Terpene Biosynthesis by Tobacco 5-epi-Aristolochene Synthase. Science. 1997;277:1815–1820. doi: 10.1126/science.277.5333.1815. [DOI] [PubMed] [Google Scholar]
- 15.Hyatt DC, et al. Structure of limonene synthase, a simple model for terpenoid cyclase catalysis. Proc Natl Acad Sci U S A. 2007;104:5360–5365. doi: 10.1073/pnas.0700915104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Köksal M, Jin Y, Coates RM, Croteau R, Christianson DW. Taxadiene synthase structure and evolution of modular architecture in terpene biosynthesis. Nature. 2011;469:116–120. doi: 10.1038/nature09628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wendt KU, Poralla K, Schulz GE. Structure and Function of a Squalene Cyclase. Science. 1997;277:1811–1815. doi: 10.1126/science.277.5333.1811. [DOI] [PubMed] [Google Scholar]
- 18.Thoma R, et al. Insight into steroid scaffold formation from the structure of human oxidosqualene cyclase. Nature. 2004;432:118–122. doi: 10.1038/nature02993. [DOI] [PubMed] [Google Scholar]
- 19.Rudolf JD, et al. Structure of the ent-Copalyl Diphosphate Synthase PtmT2 from Streptomyces platensis CB00739, a Bacterial Type II Diterpene Synthase. J Am Chem Soc. 2016;138:10905–10915. doi: 10.1021/jacs.6b04317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Köksal M, Hu H, Coates RM, Peters RJ, Christianson DW. Structure and mechanism of the diterpene cyclase ent-copalyl diphosphate synthase. Nat Chem Biol. 2011;7:431–433. doi: 10.1038/nchembio.578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moosmann P, et al. Cyanobacterial ent-Sterol-Like Natural Products from a Deviated Ubiquinone Pathway. Angew Chemie - Int Ed. 2017;56:4987–4990. doi: 10.1002/anie.201611617. [DOI] [PubMed] [Google Scholar]
- 22.Krissinel E, Henrick K. Inference of Macromolecular Assemblies from Crystalline State. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 23.Lenhart A, et al. Binding Structure and Potencies of Oxidosqualene Cyclase Inhibitors with the Homologous Squalene-Hopene Cyclase. J Med Chem. 2003;46:2083–2092. doi: 10.1021/jm0211218. [DOI] [PubMed] [Google Scholar]
- 24.Wendt KU, Lenhart A, Schulz GE. The structure of the membrane protein squalene-hopene cyclase at 2.0 Å resolution. J Mol Biol. 1999;286:175–187. doi: 10.1006/jmbi.1998.2470. [DOI] [PubMed] [Google Scholar]
- 25.Poralla K, et al. A specific amino acid repeat in squalene and oxidosqualene cyclases. Trends Biochem Sci. 1994;19:157–158. doi: 10.1016/0968-0004(94)90276-3. [DOI] [PubMed] [Google Scholar]
- 26.Corey EJ, et al. Methodology for the Preparation of Pure Recombinant S. cerevisiae Lanosterol Synthase Using a Baculovirus Expression System. Evidence That Oxirane Cleavage and A-Ring Formation Are Concerted in the Biosynthesis of Lanosterol from 2,3-Oxidosqualene. J Am Chem Soc. 1997;119:1277–1288. [Google Scholar]
- 27.Smentek L, Hess BAJ. Compelling computational evidence for the concerted cyclization of the ABC rings of hopene from protonated squalene. J Am Chem Soc. 2010;132:17111–17117. doi: 10.1021/ja1039133. [DOI] [PubMed] [Google Scholar]
- 28.Morikubo N, et al. Cation–π interaction in the polyolefin cyclization cascade uncovered by incorporating unnatural amino acids into the catalytic sites of squalene cyclase. J Am Chem Soc. 2006;128:13184–13194. doi: 10.1021/ja063358p. [DOI] [PubMed] [Google Scholar]
- 29.Thimmappa R, Geisler K, Louveau T, O’Maille P, Osbourn A. Triterpene Biosynthesis in Plants. Annu Rev Plant Biol. 2014;65:225–257. doi: 10.1146/annurev-arplant-050312-120229. [DOI] [PubMed] [Google Scholar]
- 30.Zhang H, Khalil ZG, Capon RJ. Fascioquinols A-F: Bioactive meroterpenes from a deep-water southern Australian marine sponge, Fasciospongia sp. Tetrahedron. 2011;67:2591–2595. [Google Scholar]
- 31.Brocks JJ, Buick R, Summons RE, Logan GA. A reconstruction of Archean biological diversity based on molecular fossils from the 2.78 to 2.45 billion-year-old Mount Bruce Supergroup, Hamersley Basin, Western Australia. Geochim Cosmochim Acta. 2003;67:4321–4335. [Google Scholar]
- 32.Kabsch W. XDS Acta Cryst. 2010;D66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murshudov GN, et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Cryst. 2010;D66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Langer G, Cohen SX, Lamzin VS, Perrakis A. Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat Protoc. 2008;3:1171–1179. doi: 10.1038/nprot.2008.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen VB, et al. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Cryst. 2010;D66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 39.Holm L, Laakso LM. Dali server update. Nucleic Acids Res. 2016;44:W351–W355. doi: 10.1093/nar/gkw357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kraulis PJ. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 41.The PyMOL Molecular Graphics System, Version 2.0. Schrödinger, LLC; [Google Scholar]
- 42.Lang M, Steglich W. An effective method for the synthesis of 13 C-labeled polyprenylhydroxy-benzoic acids. Synthesis (Stuttg) 2005;2005:1019–1027. [Google Scholar]
- 43.Madeira F, et al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019;47:W636–W641. doi: 10.1093/nar/gkz268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Whelan S, Goldman N. A General Empirical Model of Protein Evolution Derived from Multiple Protein Families Using a Maximum-Likelihood Approach. Mol Biol Evol. 2001;18:691–699. doi: 10.1093/oxfordjournals.molbev.a003851. [DOI] [PubMed] [Google Scholar]
- 45.Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol Biol Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Crystallographic data were deposited in the RCSB Protein Data Bank with the PDB IDs: 6SBB, 6SBC, 6SBD, 6SBE, 6SBF and 6SBG. All other data are available in the text or in the supplementary information.