Abstract
All multicellular organisms rely on differential gene transcription regulated by genomic enhancers, which function through the transcription-factor-mediated recruitment of cofactors1,2. Emerging evidence suggests that not all cofactors are required at all enhancers3–5, yet if these observations reflect more general principles or distinct types of enhancers has remained unknown. Here, we categorize human enhancers by their cofactor dependencies and show that these categories provide a framework to understand the sequence and chromatin diversity of enhancers and their roles in different gene-regulatory programmes. We quantify enhancer activities along the entire human genome using STARR-seq6 in HCT116 cells, following the rapid degradation of eight cofactors. This identifies different types of enhancers with distinct cofactor requirements, sequence and chromatin properties, including enhancers that are insensitive to the depletion of the core Mediator subunit MED14 or the bromodomain protein BRD4, respectively, and appear to regulate distinct transcriptional programmes. In particular, canonical Mediator7 seems dispensable for P53-responsive enhancers and MED14-depleted cells are able to induce endogenous P53 target genes. Similarly, BRD4 is not required for the transcription of CCAAT- and TATA-box-bearing genes, including histone genes and LTR12 retrotransposons, and for the induction of heat-shock genes. This first categorization of enhancers via cofactor dependencies reveals distinct enhancer types that are able to bypass broadly utilized cofactors, illustrating how alternative ways to activate transcription separate gene expression programmes and provide a conceptual framework to understand enhancer function and regulatory specificity.
Introduction
Multicellular organisms depend on differential gene transcription mediated by enhancers, which bind transcription factors (TFs) and recruit cofactors (COFs) to activate transcription1. Both COFs and the DNA-binding TFs are critical for enhancer function2 and transcription activation at the initiation, pause-release or elongation step7,8. Prominent COFs include the acetyltransferase P300 and the Mediator complex, which mediate histone modifications, RNA polymerase II (Pol II) recruitment and transcription initiation7,9, and the bromodomaincontaining protein 4 (BRD4) and cyclin-dependent kinase 9 (CDK9), which mediate transcriptional pause-release and elongation8,10.
Although COFs generally localize to active enhancers and promoters11,12 and have long been thought to be universially required, emerging evidence suggests that different regulatory elements and genes might require different COFs13,14. For example, pharmacological inhibition of COFs shows gene-specific rather than global effects (e.g. for BRD415,16, CDK75 and CDK817), and cells can for example acquire resistance to BRD4 inhibition by deploying a BRD4-independent enhancer3. Similarly, several Mediator subunits are not necessary for the transcription of all genes4,18. These findings suggest that even essential COFs that localize to most or all active genes are not globally required for transcription and that individual enhancers can bypass some of the COFs. However, whether such examples reflect more general gene-regulatory principles, such as different enhancer types with distinct properties and regulatory roles, has remained unknown and systematic analyses of COF requirements for enhancer-mediated transcription activation are still lacking.
To systematically discern the dependency of enhancers on various COFs, we measured genome-wide enhancer activities in human HCT116 cells in the presence and absence of specific COFs. Since many COFs are essential and their prolonged depletion impacts cell viability15,18, we used the auxin-inducible-degron (AID) system19 to rapidly and inducibly deplete the COF proteins. We coupled this to the quantitative assessment of enhancer activities for millions of fragments across the entire human genome using the plasmid-based massively parallel reporter assay STARR-seq6 (Fig. 1a).
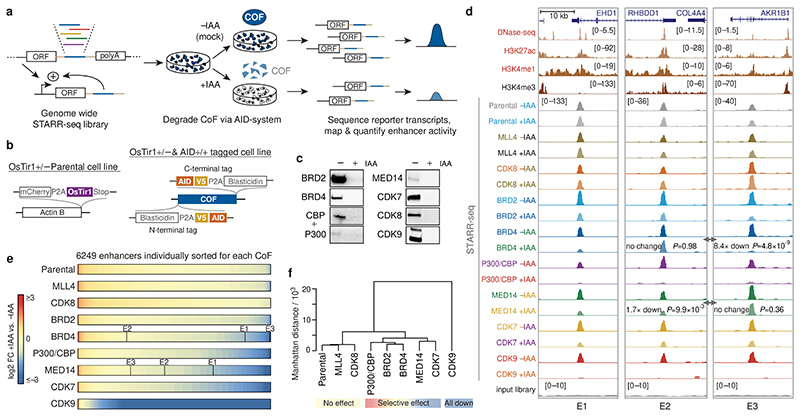
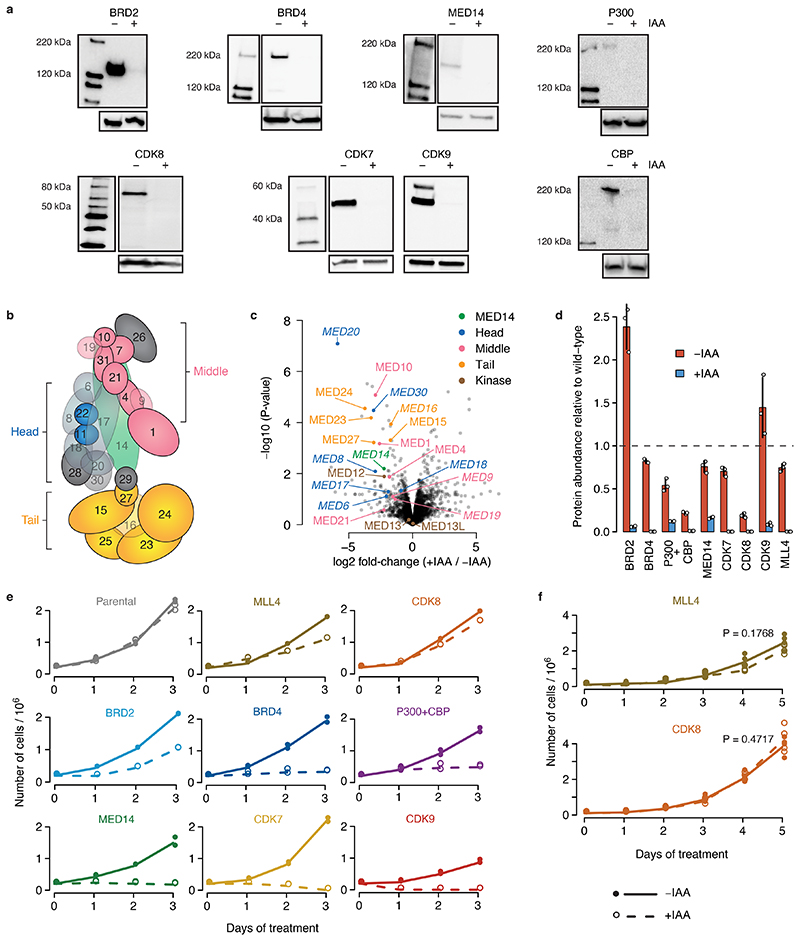
Figure 1. Rapid cofactor degradation coupled to STARR-seq reveals cofactor-specific effects on enhancer activity.
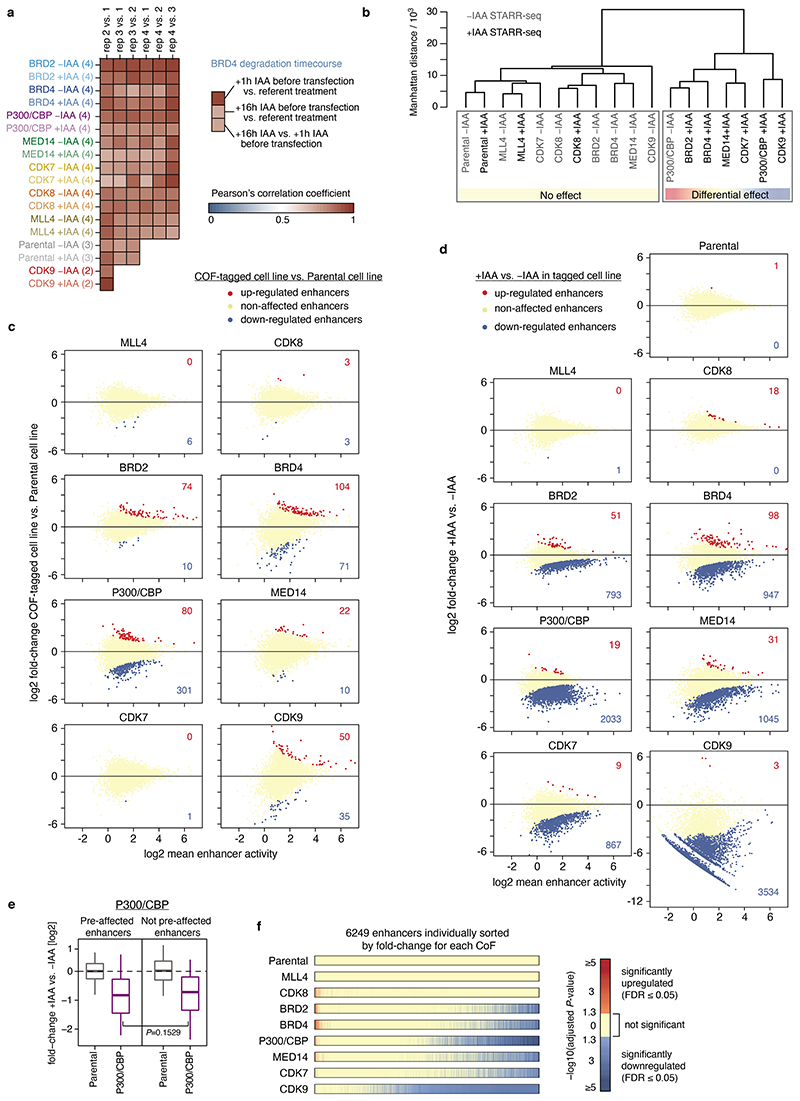
a, HCT116 cells with a cofactor (COF) of interest tagged by an auxin inducible degron (AID) are transfected with a genome-wide STARR-seq library and treated with either auxin (IAA) to degrade the COF or with a mock control. Enhancer activity across the entire human genome is quantified in the two conditions by sequencing and mapping reporter transcripts. b, COF tagging strategy. Parental HCT116 cell line carries heterozygous insertion of OsTir1 ligase downstream of Actin B gene (left). AID-tagged cell line was created for each COF by homozygous insertion of a cassette containing an AID to either N- or C-terminus of the respective COF gene in the Parental cell line (right). c, Western blots of denoted COFs in the cell line where the respective COF is tagged by AID, without and with IAA treatment for 1h. done once, validated by mass spectrometry; gel source data: Extended Data Fig. 1a and Supplementary Figure 1. d, Activity of three enhancers (E1-E3) measured by STARR-seq in different COF-AID cells with and without IAA treatment (normalized STARR-seq signal for merged replicates; adjusted P-values of the edgeR negative binomial model). Endogenous chromatin accessibility and histone modifications in wild-type HCT116 cells are shown on top. e, Log2 fold-change for a reference set of 6249 enhancers is shown, sorted individually for each COF-AID cell line from the least affected (or most upregulated) enhancers on the left to the most downregulated enhancers on the right. Three enhancers shown in d are marked for BRD4 and MED14 cell lines. f, Hierarchical clustering of Parental and COF-AID cell lines based on log2 fold-change of enhancer activity in IAA treated vs. untreated cells shown in e.
Results
COF-AID cells allow rapid COF depletion
To generate COF-AID-tagged cell lines, we first created a Parental cell line uniformly expressing the OsTir1 ligase (Fig. 1b; left), and subsequently knocked-in the AID-tag homozygously at individual COF genes19 (Fig. 1b; right). We created eight cell lines to deplete various COFs that regulate critical steps of transcription: the bromodomain-containing BRD2 and BRD4, the structural core Mediator subunit 14 (MED14), the acetyltransferases P300 and CBP (both tagged in a single cell line), the cyclin-dependent kinase CDK7 (a core TFIIH subunit), the Mediator kinase CDK8, the pTEFb kinase CDK9 and the methyltransferase MLL4 (as HCT116 cells lack the MLL4 paralog MLL320, MLL4 depletion should deplete MLL3/4 functionality).
Treatment with auxin (3-indoleacetic acid) strongly depleted all tagged COFs after 1h (Fig. 1c, Extended Data Fig. 1a). Shot-gun mass-spectrometry of auxin-treated MED14-AID cells revealed a >2-fold depletion of all detectable Mediator subunits, suggesting that Mediator is disintegrated as expected (Extended Data Fig. 1b,c and refs 4,21). A targeted mass spectrometry approach for all COFs after 3h of auxin treatment revealed no (BRD4, CBP, CDK7, CDK8 and MLL4) or low (<15%; BRD2, P300, MED14, and CDK9) residual levels (Extended Data Fig. 1d). After two days, COF degradation strongly affected proliferation for all COFs except CDK8 and MLL4, for which proliferation was not affected even after five days (Extended Data Fig. 1e,f), consistent with reports that CDK8 and MLL4 are not essential in HCT116 cells22,23.
Enhancers have distinct COF dependencies
To assess enhancer-activity changes upon loss of each COF, we performed STARR-seq in the Parental and the eight COF-AID-tagged cell lines upon mock or auxin treatment (Fig. 1a). Briefly, we transfected the cells with a genome-wide STARR-seq library comprising >50 million genomic fragments of 1.2kb6 (~22X genome coverage), treated half of the cells with water (mock) or auxin, and harvested the cellular RNAs after 6 hours (see Extended Data Fig. 2a for different timepoints of BRD4 depletion). We added spike-in RNAs to total cellular RNA for normalization, isolated, amplified and quantified the poly-adenylated reporter transcripts by deep sequencing.
We performed three replicates per condition for the Parental cell line, two for CDK9-AID cells and four for all other COF-AID cells (replicates had pairwise Pearson’s correlation coefficients ≥0.7; Extended Data Fig. 2a). We first defined a set of enhancers that were strongly active in at least one condition using all replicates and stringent thresholds (see Methods), which detected between 141 and 1979 enhancers per condition (fewer in COF depleted conditions) and 6249 enhancers total.
Without auxin, STARR-seq in COF-AID-tagged cells was similar to the Parental controls (Extended Data Fig. 2b), suggesting that COF and enhancer functions were maintained. The only exception were the double-tagged P300/CBP cells that showed reduced enhancer activity in the absence of auxin (Extended Data Fig. 2c), potentially due to significant pre-degradation of both COFs (Extended Data Fig. 1d). However, the loss of enhancer activity was marginal compared to the effects after auxin-induced COF degradation (≤15% of enhancers, compare Extended Data Fig. 2c and d), and auxin treatment downregulated pre-affected and non-pre-affected enhancers to similar extents (Extended Data Fig. 2e), suggesting that P300/CBP-dependent enhancers can be studied.
Overall, COF depletion revealed different effects for different COFs: degradation of CDK8 and MLL4 showed no effect on enhancer activity (Fig. 1d-f; Extended Data Fig. 2b,d,f), consistent with unaltered proliferation and reports that CDK8 and MLL4 are dispensable in HCT116 cells (Extended Data Fig. 1e,f and refs 22,23). In contrast, CDK9 depletion led to global inactivation of enhancers (Fig. 1d,e, Extended Data Fig. 2d,f), consistent with the role of CDK9 during pause-release and elongation8,10.
Degradation of the remaining COFs had more selective effects that differed between individual COFs, with some COFs having more similar effects, such as BRD2 and BRD4, than others (Fig. 1f): some enhancers were down-regulated, whereas others were unaffected or even up-regulated (Fig. 1d,e, Extended Data Fig. 2d,f). For instance, BRD4 loss had no effect on an enhancer in the RHBDD1 gene, but strongly impaired an enhancer in AKR1B1, while the opposite was true for MED14. Taken together, rapid COF degradation coupled to STARR-seq revealed differential COF dependencies for individual enhancers.
COF dependencies define 4 enhancer types
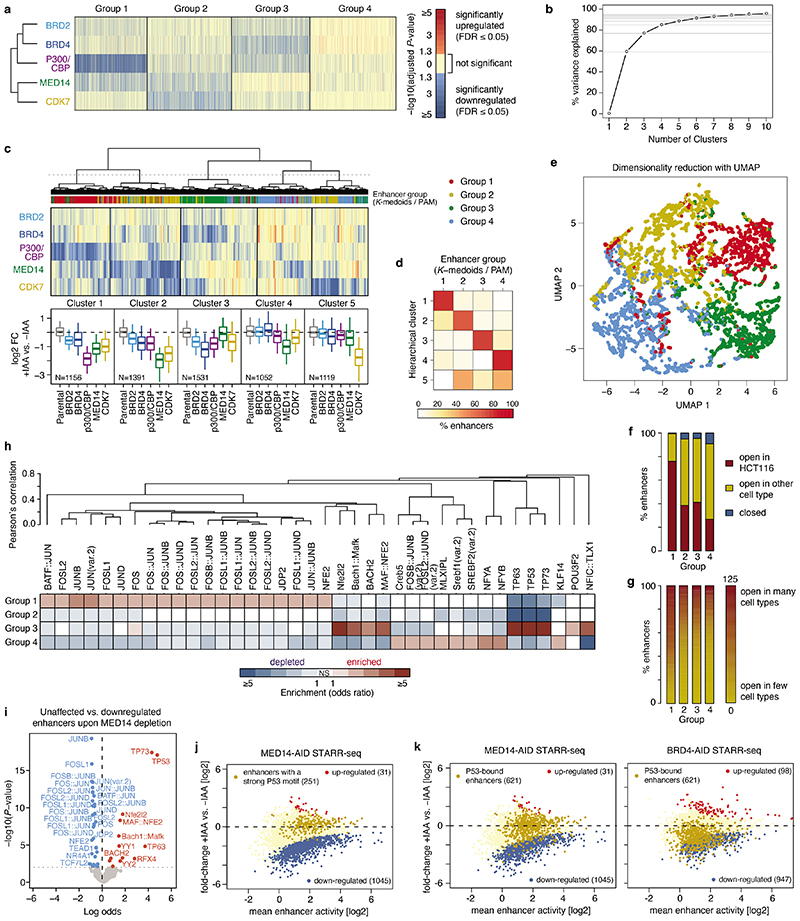
The result that not all enhancers depended similarly on all COFs suggests the existence of enhancer groups with specific COF requirements. To reveal such groups, we clustered the 6249 enhancers based on enhancer-activity change upon degradation of each of the five COFs with selective effects (BRD2, BRD4, P300/CBP, MED14, CDK7). Using partitioning-around-medoids (PAM, K-medoids), we defined four distinct groups of enhancers (Fig. 2a, Extended Data Fig. 3a) that accounted for ≥85% of the variance in the data (Extended Data Fig. 3b) and were reproducible with alternative clustering approaches (Extended Data Fig. 3c-e). The first two groups required all five COFs for full activity, with Group 1 being more strongly dependent on P300/CBP and Group 2 on CDK7 (Fig. 2a,b). Interestingly, the enhancers of Groups 3 and 4 were not impaired by the degradation of MED14 or BRD4, respectively, thereby defining enhancer types that can function with limiting levels, or potentially entirely independently, of these two COFs (Fig. 2a,b).
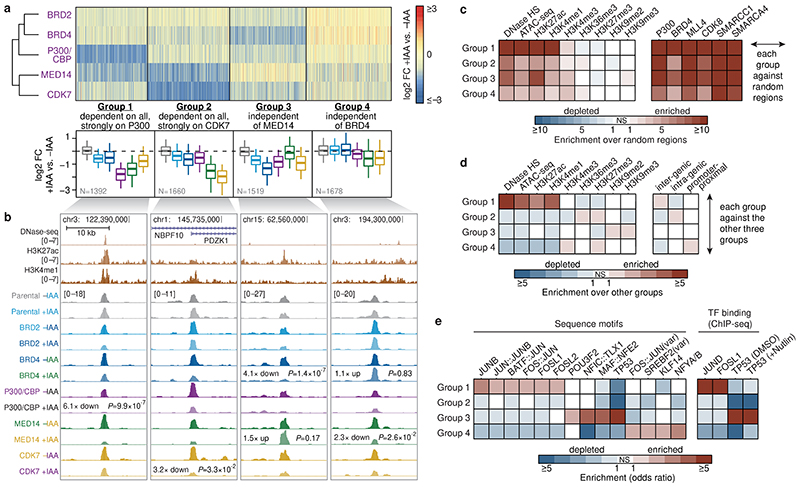
Figure 2. Differential cofactor requirements define distinct enhancer types with distinguishing sequence and chromatin features.
a, Log2 fold-change of enhancer activity upon individual cofactor degradation for four groups of enhancers defined by partitioning-around-medoids clustering. Boxplots summarize the values per COF for each group. N = 1392, 1660, 1519, 1678 for Groups 1 to 4, respectively. Boxes: median and interquartile range; whiskers: 5th and 95th percentiles. b, Examples of enhancers from each of the four groups showing activity in different COF-AID cell lines with and without auxin (IAA) treatment (normalized STARR-seq signal for merged replicates; adjusted P-values of the edgeR negative binomial model). c, Enrichment of chromatin accessibility and histone modification ChIP-seq peaks (left) and various cofactor ChIP-seq peaks (right) from HCT116 cells for the four groups of enhancers against random control regions. d, e, Mutual enrichment of chromatin accessibility and histone modification ChIP-seq peaks (d, left), genomic localization (d, right), transcription factor (TF) motifs (e, left) and TF ChIP-seq peaks (e, right) for the four groups of enhancers. The enrichment for each group is calculated against the remaining three groups. Statistically significant (twosided Fisher’s exact test; P-value ≤0.05) enrichments and depletions are colored in shades of red and blue, respectively. Non-significant (NS) fields are shown in white.
Endogenous enhancer chromatin features in HCT116 cells were enriched in all four groups of enhancers compared to random control regions, including DNA accessibility, H3K27ac, H3K4me1 and COF binding (Fig. 2c; published data sources see Methods). However, the groups differed in relative levels of chromatin marks and in genomic localization (Fig. 2d). Group 1 contained the highest proportion of endogenously accessible enhancers (open across many cell types; Extended Data Fig. 3f,g) and were most highly enriched for H3K27ac and H3Kme1 (Fig. 2c,d). In contrast, Group 2 enhancers were subtly enriched for H3K36me3, a gene-body mark, and intra-genic localization (Fig. 2c,d). Groups 3 and 4 contained enhancers accessible in HCT116 cells and enhancers accessible only in other cell types (Extended Data Fig. 3f), indicative of chromatin-mediated silencing in HCT116 cells6. Indeed, both groups displayed a relative enrichment of repressive H3K27me3 (Group 4) and H3K9me2/3 marks (Group 3) (Fig. 2d).
The four groups most notably differed in their sequences and contained specific TF motifs. Group 1 enhancers were highly enriched for the AP-1 family (FOS & JUN) motifs and their combinations (Fig. 2e, Extended Data Fig. 3h), while Group 3 enhancers were most strongly enriched for P53 motifs, and Group 4 enhancers for NFY (CCAAT-box) motifs. Published ChIP-seq datasets confirmed preferential binding of these TFs to endogenous enhancers of the different groups (Fig. 2e), suggesting that trans-activation by different TFs requires different sets of COFs.
Mediator-independence of P53-targets
The finding that enhancers characterized by P53 motifs and endogenous P53 binding are insensitive to MED14 depletion (Fig. 2a,b,e) suggests that P53-mediated activation might be Mediator-independent, consistent with reports that some active or stress-inducible promoters do not associate with Mediator in yeast24. However, it is also unexpected, as P53 directly interacts with Mediator7,25,26 and most activators of stress-responsive genes do recruit Mediator24.
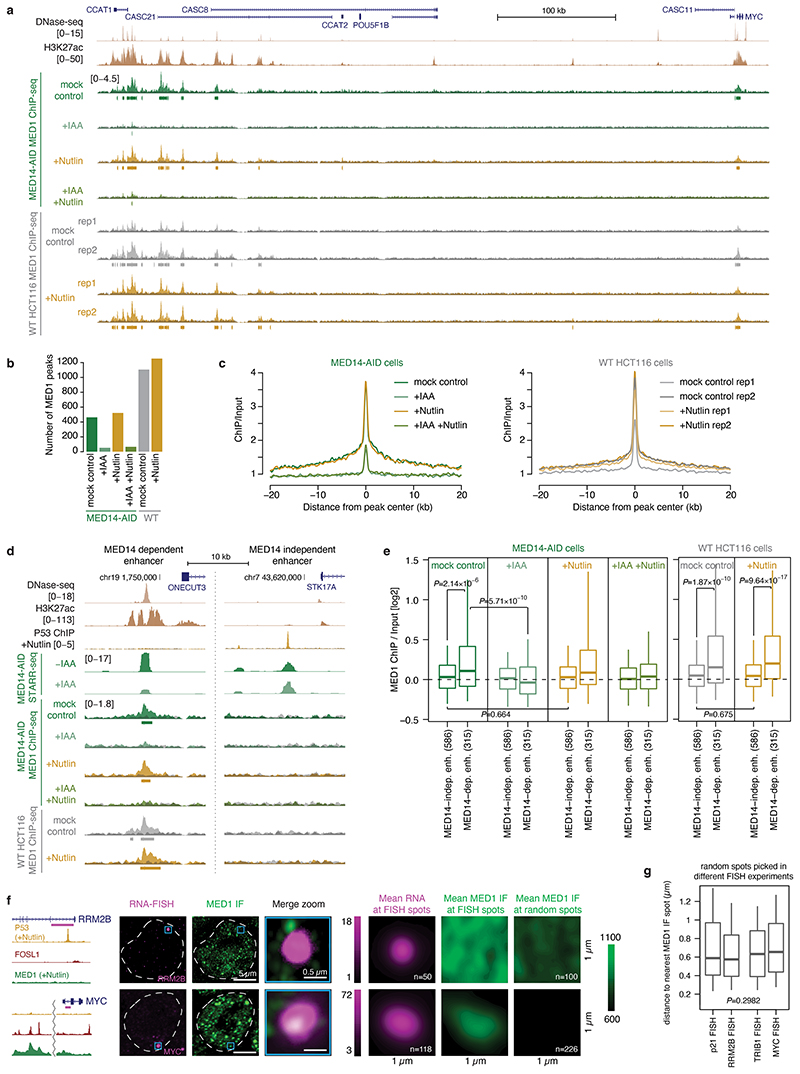
We first confirmed that P53 motifs and P53 binding27 are most strongly enriched in enhancers that show the least dependence on MED14 (Fig. 3a), whereas motifs for FOS and JUN for example were enriched in MED14-dependent enhancers (Extended Data Fig. 3i,j). Consistently, MED14 depletion did not affect P53-bound enhancers, whereas the activity of enhancers not bound by P53 decreased ~2-fold on average (Fig. 3b, Extended Data Fig. 3k). This difference is specific to MED14 depletion, while for example BRD4 depletion reduced enhancer activity irrespective of P53 binding (Fig. 3b; Extended Data Fig. 3k) as exemplified by an enhancer in the first intron of the P53-target gene RRM2B, which was strongly affected by depletion of BRD4 but not MED14 (Fig. 3c).
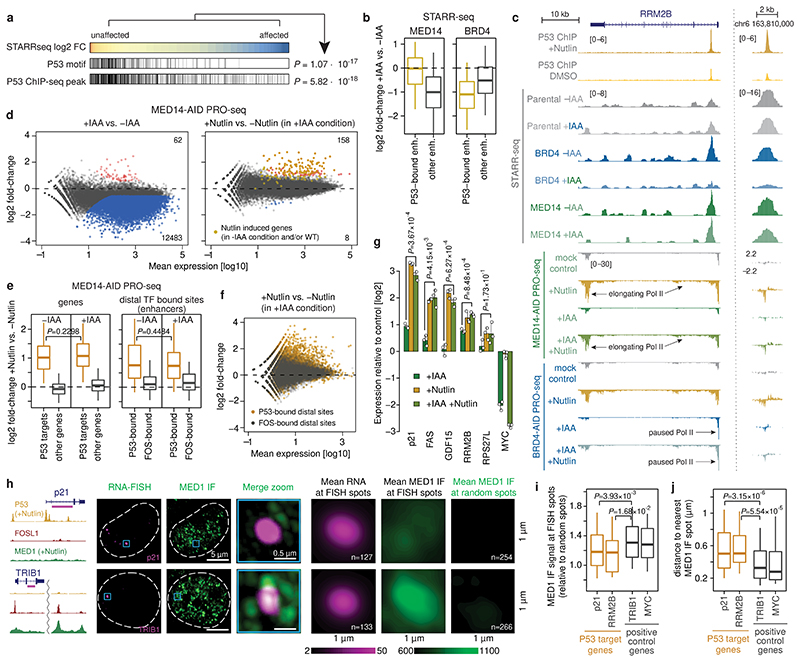
Figure 3. P53-bound enhancers and target genes are insensitive to MED14 depletion.
a, P53 motifs and ChIP-seq peaks in STARR-seq enhancers sorted from least to most affected upon MED14 depletion. P-values: one-sided Fisher’s exact test (top against bottom 20%). b, Activity change for P53-bound (N=621) vs. other (N=5628) enhancers in MED14- (left) and BRD4-AID (right) cells. c, Enhancer activity (merged normalized STARR-seq replicates) and nascent transcription (merged normalized PRO-seq replicates) in RRM2B locus upon P53 induction with Nutlin-3a in MED14- and BRD4-AID cells with and without auxin (IAA). d, Differential gene PRO-seq in MED14-AID cells (left: +/- auxin; right: auxin+Nutlin-3a vs. auxin-only; FDR≤0.05; fold-change≥2; N=2 independent replicates; yellow: 151 Nutlin-3a-induced genes [Extended Data Fig. 4b]). PRO-seq fold-change for P53 targets (left; N=243 [Extended Data Fig. 4c]) and distal P53-bound sites around targets (right) in MED14-AID cells upon Nutlin-3a with (+IAA) or without auxin (-IAA). N = 243, 20964, 233, 346 for P53 targets, other genes, P53- and FOS-bound enhancers, respectively. f, Differential PRO-seq analysis for distal P53- or FOS-bound enhancers upon Nutlin-3a in auxin-treated MED14-AID cells. g, Expression (qPCR) of P53 targets in auxin- or/and Nutlin-3a-treated MED14-AID cells. N=3 independent replicates; mean +/- SD; P-values: two-sided Student’s t-test. h, MED1 IF with concurrent RNA-FISH against P53 target p21 (top) and control TRIB1 gene (bottom) in Nutlin-3a-treated HCT116 cells. Left: gene loci with P53, FOSL1 and MED1 ChIP-seq signal and intronic FISH target sequence (magenta). Dashed line: nuclear periphery. Right: mean RNA-FISH and MED1-IF signals centred on FISH spots, or random spots (n=number of spots). i, MED1 IF signal at FISH spots, normalized to mean MED1 IF signal at random regions. j, Distance between FISH spot and nearest MED1 IF spot. In i and j, N = 127, 50, 133, 118 FISH spots for p21, RRM2B, TRIB1 and MYC, respectively. In b, e, i and j, boxes: median and interquartile range; whiskers: 5th and 95th percentiles; P-values: two-sided Wilcoxon rank-sum test.
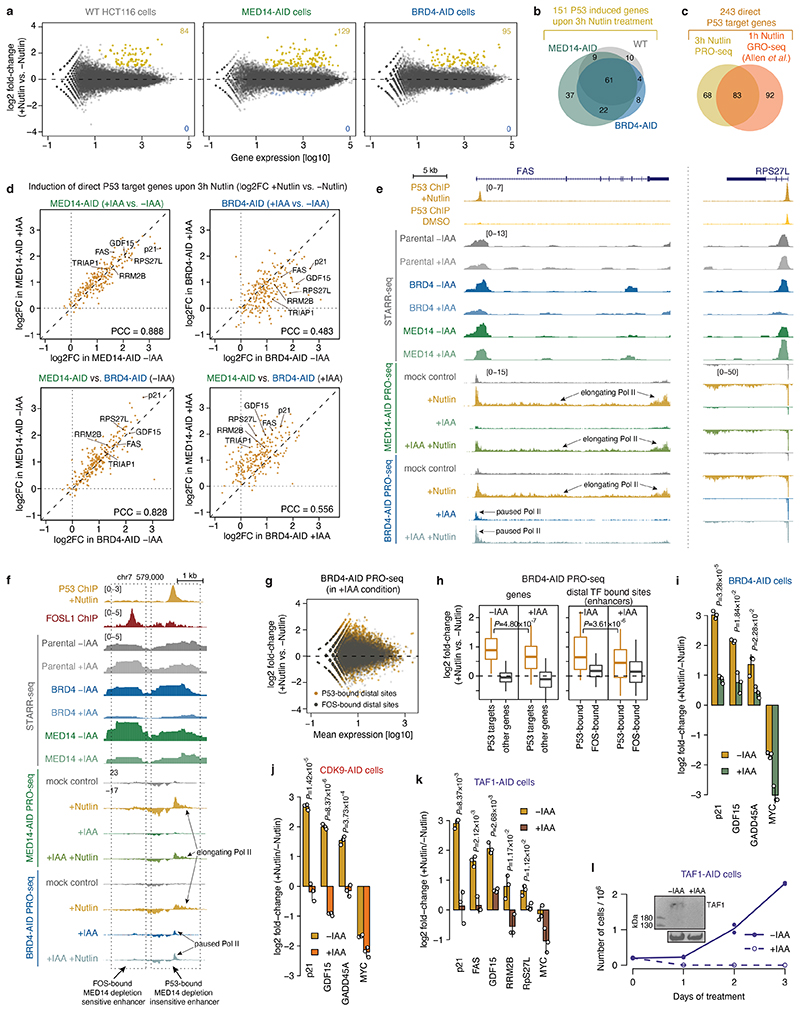
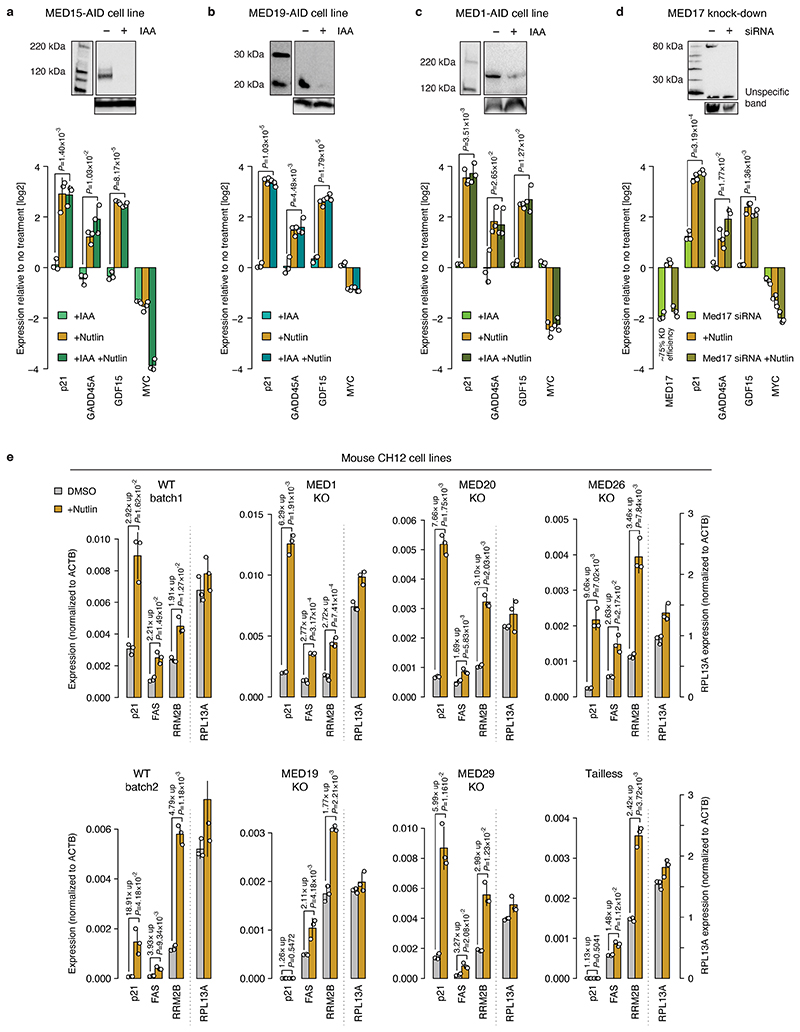
We next assayed the transcriptional response of endogenous P53 target genes using PRO-seq after depleting MED14. Auxin treatment for 3h led to a global transcriptional downregulation of almost all genes (Fig. 3d, left), which is consistent with Mediator-dependence of most HCT116 enhancers (Fig. 2a) and confirms effective depletion of Mediator. However, when we treated MED14-depleted cells with Nutlin-3a, which activates P53 signaling27, the transcriptional response was essentially identical as in MED14-non-depleted cells and in WT HCT116 cells (Fig. 3d, right; Extended Data Fig. 4a,b). Indeed, direct P53-target genes activated by Nutlin-3a treatment in WT HCT116 cells (Extended Data Fig. 4c), were upregulated to the same extent in both MED14-depleted and control cells, including the well known P53-targets FAS, RPS27L and RRM2B (Fig. 3c,e left, Extended Data Fig. 4d,e). Consistent with the induction of P53-target genes, we also observed the specific upregulation of nascent bidirectional transcription from P53-bound enhancers in the vicinity of those genes (Fig. 3c, Extended Data Fig. 4f) to the same extent in both MED14-depleted and control cells (Fig. 3e right), confirming that the endogenous enhancers are activated despite MED14 depletion (Fig. 3f). Additionally, we confirmed the induction at the mature mRNA level for several well-known P53 targets, including p21, via qPCR (Fig. 3g). After MED14 depletion, Nutlin-3a lead to a robust induction of all assayed P53 targets to similar final levels as without depletion, while the transcription of Mediator-dependent control genes, including MYC, was failing.
In contrast to MED14 depletion, BRD4 depletion significantly impaired the induction of both P53 target genes and P53-bound enhancers as measured by PRO-seq and qPCR (Fig. 3c, Extended Data Fig. 4a,d-i), demonstrating that, unlike MED14, BRD4 is required for a robust P53 response. Furthermore, degradation of either TAF1 or CDK9 completely abolished the induction of P53 target genes (Extended Data Fig. 4j-l), indicating that P53-mediated activation depends on functioning initiation and pause-release steps, both of which seem to occur in MED14-depleted cells.
Taken together, these results show that P53-mediated activation is insensitive to limiting levels of MED14, consistent with models that P53-target enhancers are either highly efficient in recruiting residual MED14 (Extended Data Fig. 1d) or function independently of MED14 through non-canonical Mediator sub-complexes, presumably containing MED1 or MED17 that can directly interact with P5325,26,28. To discern between these possibilities, we performed MED1 ChIP-seq in MED14-AID and in WT HCT116 cells upon auxin and/or Nutlin-3a treatment. In unperturbed cells, MED1 bound to many endogenously active enhancers, including a previously described enhancer cluster at the MYC locus (Extended Data Fig. 5a-c). MED1-ChIP signals were elevated at endogenous MED14-dependent enhancers compared to MED14-independent enhancers, and the vast majority, including those in the MYC locus and at MED14-dependent enhancers, were lost upon MED14 depletion (Extended Data Fig. 5d,e). Thus, Mediator-dependent enhancers bind detectable levels of Mediator, which is effectively depleted by MED14 degradation. In contrast, we did not detect MED1 ChIP-seq signals at P53-target enhancers in any condition, suggesting that these enhancers do not recruit high levels of MED1, at least not like MED14-dependent enhancers (e.g. MYC enhancers; Extended Data Fig. 5e).
To assess Mediator binding to P53-target genes (p21 and RRM2B) and Mediator-dependent control genes (TRIB1 and MYC) by an independent approach, we combined MED1 immunofluorescence (IF) with RNA FISH against nascent transcripts in WT HCT116 cells treated with Nutlin-3a for 3h. In this condition, the gene loci of both groups of genes were robustly detected by FISH, allowing the quantification of MED1 IF signals at 127 p21 and 133 TRIB1 gene loci, respectively (Fig. 3h; see Extended Data Fig. 5f for RRM2B and MYC). Consistent with the ChIP-seq data, the MED1 signal at individual gene loci was significantly lower for P53-target genes than controls (Fig. 3h,i) and MED1 spots were significantly farther from P53-target genes than from controls (Fig. 3j), which is not due to overall differences in the number of MED1 spots (Extended Data Fig. 5g). This demonstrates that P53-target genes do not recruit substantial amounts of MED1 and suggests that P53-mediated activation does not require the full/canonical MED14- and MED1-containing Mediator complex7.
To assess if the P53 response is independent of additional Mediator subunits, we measured the induction of known P53-target genes by qPCR in cells depleted of different Mediator subunits from the head, tail and middle modules, including the two subunits previously reported to interact with P53, MED1 and MED1725,26,28. Depletion of all targeted subunits by AID or siRNAs had no effect on P53-target gene induction, which was the same as in unperturbed cells (Extended Data Fig. 6a-d). To extend our findings to another cell type and organism and to cells that are permantently devoid of non-essential Mediator subunits, we chose knock-out (KO) mouse lymphoma CH12 cells, lacking the MED1, MED19, MED20, MED26 or MED29 Mediator subunit, respectively, or the entire Mediator tail (MED15, MED16, MED23, MED24 and MED25)18. The known P53-target genes p21, FAS and RRM2B were induced in all KO cells, including cells lacking the P53-interacting subunit MED1 (MED17 is essential and could not be tested; Extended Data Fig. 6e). Only the MED19-KO and tailless cells had undetectable levels of p21 in all conditions, potentially a result of clonal selection, but both strongly induced FAS and RRM2B.
Overall, the results on enhancer activities and nascent transcription after MED14 depletion, the lack of detectable MED1 binding, and the dispensability of various Mediator subunits for P53-targets in human and mouse cells suggest that P53-mediated transcription activation is independent of full/canonical Mediator7 (see Discussion).
TATA-boxes confer BRD4 independence
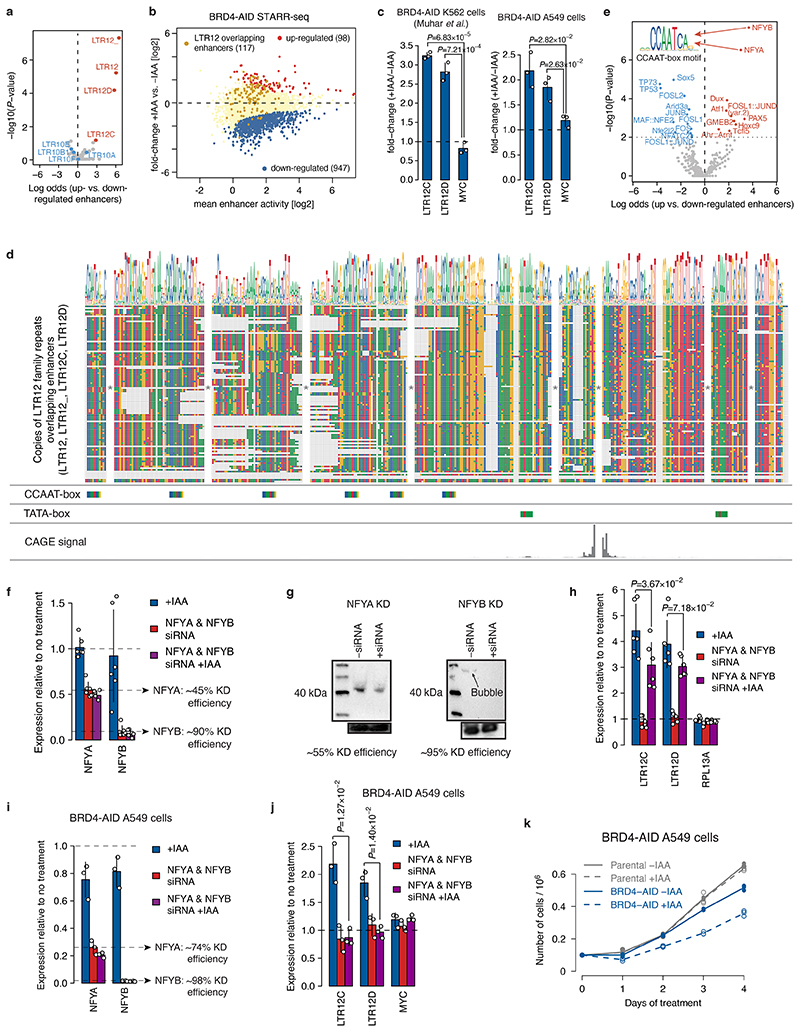
Group 4 enhancers remained active or even increased in activity in the absence of BRD4 (Fig. 2a) and were often associated with closed chromatin, repressive histone marks (Fig. 2d), and individual repeat elements (Fig. 4a). In particular, the long terminal repeat families LTR12/C/D were enriched in up-regulated enhancers (Extended Data Fig. 7a) and LTR12 elements detected in STARR-seq displayed strongly increased enhancer activity upon BRD4 depletion, unlike the related LTR10 elements and most enhancers that generally lost activity (Fig. 4b, Extended Data Fig. 7b). Furthermore, endogenous LTR12C/D were strongly upregulated in qPCR after prolonged BRD4 degradation, consistent with effects of inhibiting histone-deacetylases29,30, but not after MED14 depletion (Fig. 4c); and the upregulation upon BRD4 depletion also occurred in K562 and A549 cells (Extended Data Fig. 7c).
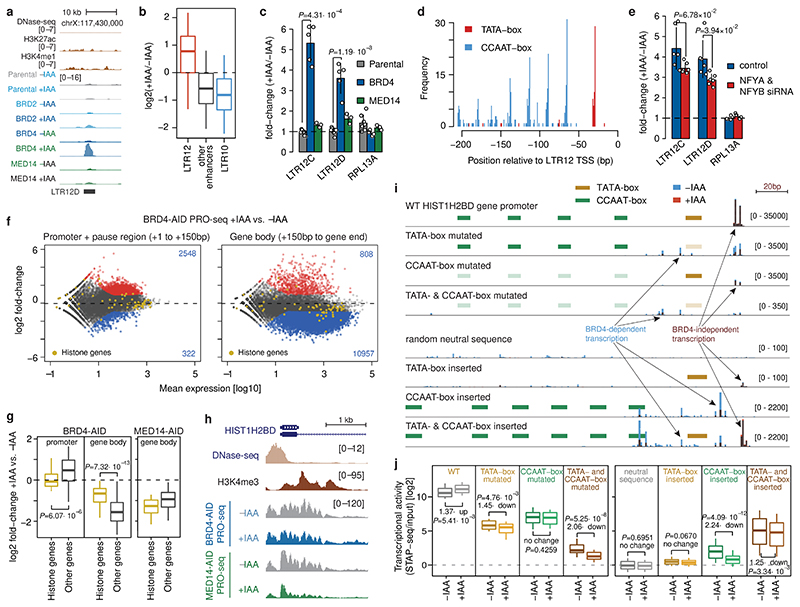
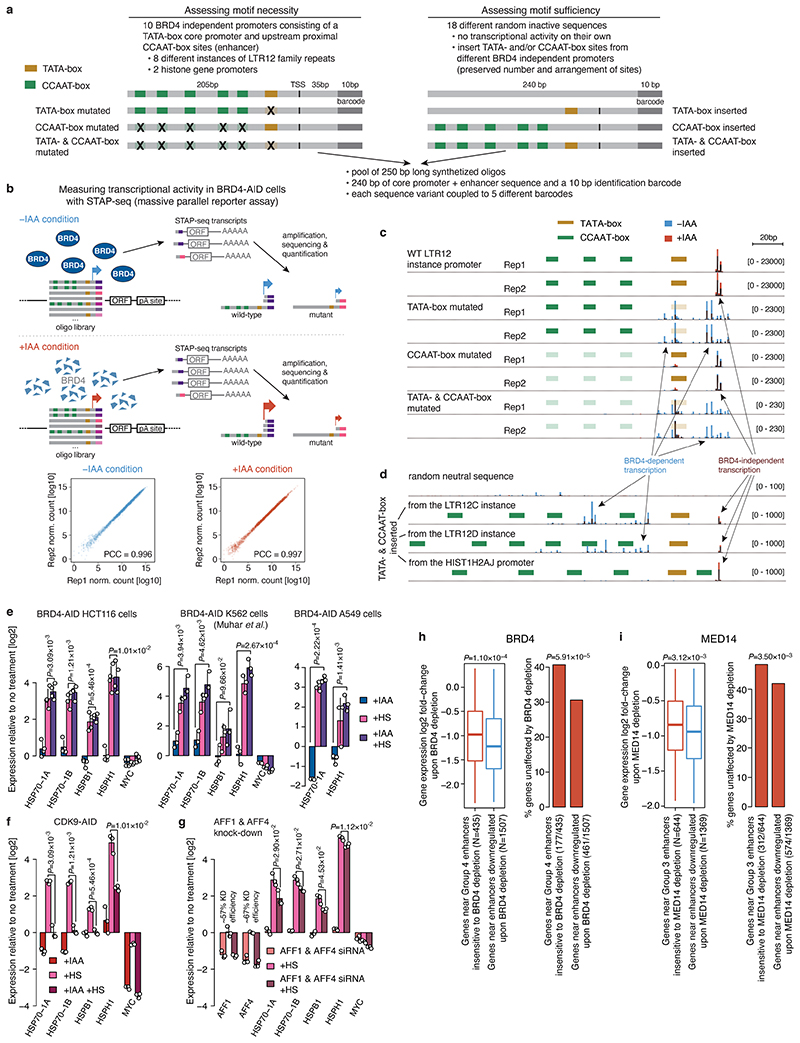
Figure 4. Combination of TATA- and CCAAT-boxes renders transcription of LTR12 retrotransposons and histone genes independent of BRD4.
a, LTR12D element with increased enhancer activity upon BRD4 degradation. b, Enhancer-activity change upon BRD4 depletion for LTR12- (N=117), LTR10-overlapping (N=198) and all other (N=5935) enhancers. c, Change in endogenous LTR12 expression (qPCR) upon auxin treatment of BRD4- or MED14-AID cells. N = 7, 5, 3 independent replicates for Parental, BRD4- and MED14-AID cells, respectively. d, Occurrence of TATA- and CCAAT-boxes in LTR12 repeats with STARR-seq activity, relative to their endogenous TSSs. e, Change in endogenous LTR12 expression (qPCR) upon BRD4 depletion before and after NFYA & NFYB knock-down. N=6 independent replicates. f, Differential analysis (+/-auxin) of PRO-seq in promoter+pause region (left) and gene body (right) for BRD4-AID cells (FDR≤0.05; fold-change≥2; yellow: histone genes; N=2 independent replicates). g, Change of PRO-seq signal in promoter+pause region and gene body in BRD4-AID cells (left) and gene body in MED14-AID cells (right) for histone genes (N=50) vs. all other expressed genes (N=11869). h, PRO-seq signal at HIST1H2BD in BRD4- and MED14-AID cells +/-auxin (normalized signal for merged replicates). i, Transcription (base-pair resolution; Extended Data Fig. 9b) from WT and mutant HIST1H2BD promoters (top) and from neutral sequences with inserted LTR12-derived TATA- and/or CCAAT-boxes (bottom). Mean normalized STAP-seq signal across barcodes and replicates (N=2 independent replicates, 5 barcodes per sequence) in +auxin (red) vs. -auxin (blue) BRD4-AID cells is overlaid. j, STAP-seq signal for WT and mutated versions of histone and LTR12 promoters (left; N=50), and for random neutral sequences with inserted TATA- and/or CCAAT-boxes (right; N = 90, 120, 900 for WT, single insertions and double insertions, respectively). In b, g, j, boxes: median and interquartile range; whiskers: 5th and 95th percentiles; P-values: two-sided Wilcoxon rank-sum test. In c, e, mean +/- SD; P-values: two-sided Student’s t-test.
LTR12 elements contain a TATA-box promoter and multiple CCAAT-boxes29,30 (Fig. 4d, Extended Data Fig. 7d), which were also the most highly enriched motifs in BRD4-independent enhancers (Fig. 2e) and in enhancers up-regulated upon BRD4 depletion (Extended Data Fig. 7e). As CCAAT-boxes in LTR12 bind the NFY TFs30, which maintain nucleosomal depleted regions31, we tested whether NFY is required for LTR12 expression by depleting the NFY subunits A and B via RNAi in BRD4-depleted HCT116 (Extended Data Fig. 7f-h) and A549 cells (Extended Data Fig. 7i-k). Indeed, NFYA/B depletion significantly reduced the up-regulation of LTR12C/D after BRD4 depletion in both cell types (Fig. 4e, Extended Data Fig. 7h,j). Thus, NFY contributes to the upregulation of LTR12C/D upon BRD4 loss and is potentially involved in the mechanism that confers BRD4-independence.
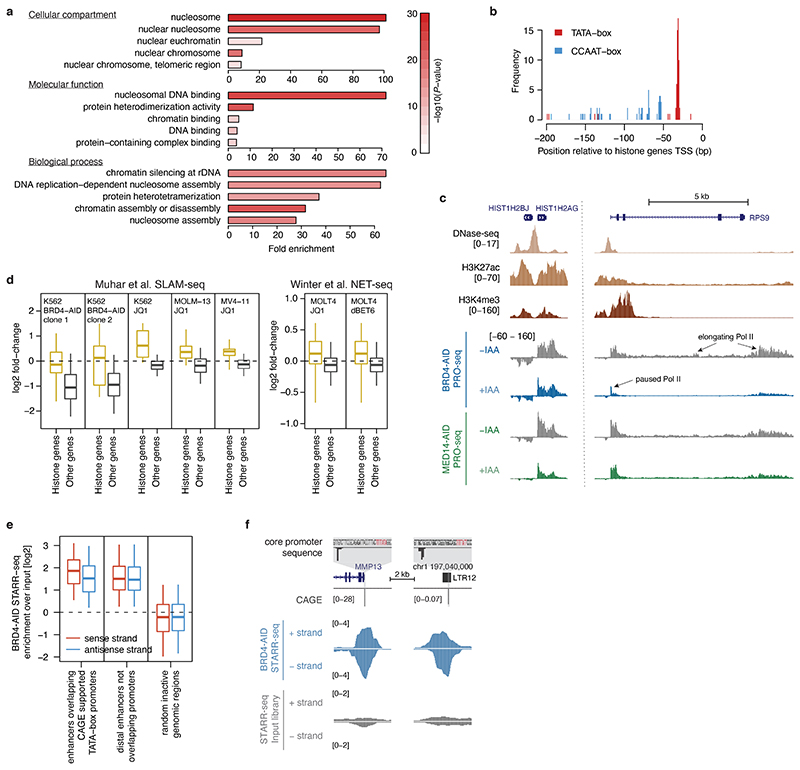
Gene-ontology analysis for genes with a CCAAT- and TATA-box promoter structure revealed terms related to nucleosome-assembly and DNA packaging (Extended Data Fig. 8a), identifying histone genes as top hits. Indeed, promoters of histone genes have a precisely positioned TATA-box and proximal upstream CCAAT-boxes (Extended Data Fig. 8b). To test if histone genes are transcribed in absence of BRD4, we performed PRO-seq upon BRD4 depletion. Consistent with the function of BRD4 in pause-release and in line with previous reports32,33, BRD4 depletion led to a global pause-release defect characterized by loss of Pol II signal in gene bodies and gain in the promoter-proximal pause region (Fig. 4f). However, histone genes were much less affected compared to other genes after BRD4 depletion and to histone genes after MED14 depletion (Fig. 4g,h, Extended Data Fig. 8c), suggesting that histone gene transcription is independent of BRD4, but dependent on MED14. Indeed, re-analysis of published datasets using nascent transcription after BRD4 inhibition or degradation32,33 confirmed that transcription of histone genes appears BRD4 independent (Extended Data Fig. 8d).
The results above suggest that LTR12 elements and histone gene promoters contain TATA-box-compatible enhancers that can activate the heterologous TATA-box promoter in STARR-seq and their cognate TATA-box promoters in vivo in a BRD4 independent manner (the elements are also orientation independent in STARR-seq as expected for bona fide enhancers; Extended Data Fig. 8e,f). To dissect a functional link between the TATA- and CCAAT-boxes and BRD4-independent transcription, we made use of the fact that these elements function as autonomous promoters and assessed the transcriptional activity of hundreds of wild-type and mutant sequences in BRD4-AID cells with or without auxin (Extended Data Fig. 9a,b), employing a massively parallel reporter assay with single base-pair resolution34 with a synthetic oligo library comprising 240bp-long fragments, each with five unique barcodes. To test motif necessity, we selected ten BRD4-independent promoters, including LTR12 elements and histone gene promoters, and generated wild-type sequences and variants mutant for either TATA- or CCAAT-boxes or both (Extended Data Fig. 9a). To test motif sufficiency, we inserted the TATA- and/or CCAAT-boxes into 18 different transcriptionally inactive random sequences, preserving the arrangement of these motifs in BRD4-independent promoters.
This resulted in highly reproducible transcriptional activities and initiation patterns (Extended Data Fig. 9b,c) that confirmed BRD4-independent transcription of histone gene promoters and LTR12 elements (Fig. 4i,j, Extended Data Fig. 9c). Mutating TATA-boxes impaired transcription from the cognate TSS and BRD4-independence, as seen by a further reduction in transcription upon auxin treatment. In contrast, mutating CCAAT-boxes resulted in a strong loss of transcription, but the remaining transcription was still BRD4-independent. Mutating both motifs reduced the transcriptional activity even further and any remaining transcription was strongly BRD4-dependent (Fig. 4i,j, Extended Data Fig. 9c).
Consistently, inserting a TATA-box into inactive sequences resulted in very low levels of BRD4-independent transcription from a single TSS (Fig. 4i,j), in line with observations that TATA-boxes on their own support only low levels of transcription34. Inserting only CCAAT-boxes increased transcription from dispersed ectopic initiation sites, and this transcription was strongly dependent on BRD4. Inserting both motifs together resulted in robust transcription from a single TSS that was less dependent on BRD4 and to varying levels of BRD4-dependent transcription from ectopic sites (Fig. 4i,j, Extended Data Fig. 9d).
Taken together, these results demonstrate that a TATA-box promoter is necessary and sufficient to confer BRD4 independence, while CCAAT-boxes act as enhancers to boost BRD4-independent transcription but cannot themselves confer BRD4 independence. Since STARR-seq uses a promoter with mixed features and multiple TSSs6, we speculate that BRD4 independent enhancers activate TATA-box-associated TSSs, while BRD4 dependent enhancers are presumably not compatible with the TATA-box and activate other TSSs within the same promoter.
To further investigate the role of TATA-boxes in conferring BRD4 independence, we analyzed heat-shock genes, which are well-studied models of TATA-box promoters and proximally bound activators35. Briefly, we induced heat-shock for 1h at 43°C in BRD4-AID cells pre-treated with water (mock) or auxin and analyzed the expression of four heat-shock genes via qPCR. In three different cell lines, all tested genes were strongly induced after heat-shock irrespective of BRD4 depletion (Extended Data Fig. 9e, ref. 36), while CDK9 depletion abolished gene induction as expected (Extended Data Fig. 9f). This dependence on CDK9 but not on BRD4 suggests that the CDK9-containing complex pTEFb is recruited by other means, presumably by the super elongation complex (SEC) that functions at stress-related genes37. Indeed, the simultaneous depletion of the two SEC subunits AFF1 and AFF4 led to a mild but significant reduction in heat-shock gene induction (Extended Data Fig. 9g), arguing that SEC might aid in recruiting CDK9 to support full inducibility of heat-shock genes independently of BRD436.
Taken together, our data show that transcription from TATA-box promoters is insensitive to BRD4 depletion and allows BRD4 independent transcription of different types of genes via different TATA-box-compatible enhancers. Thus, specific classes of genes and their associated enhancers have distinct COF requirements and can function independently of broadly deployed COFs, possibly via alternative mechanisms, to regulate specific steps in transcription.
Discussion
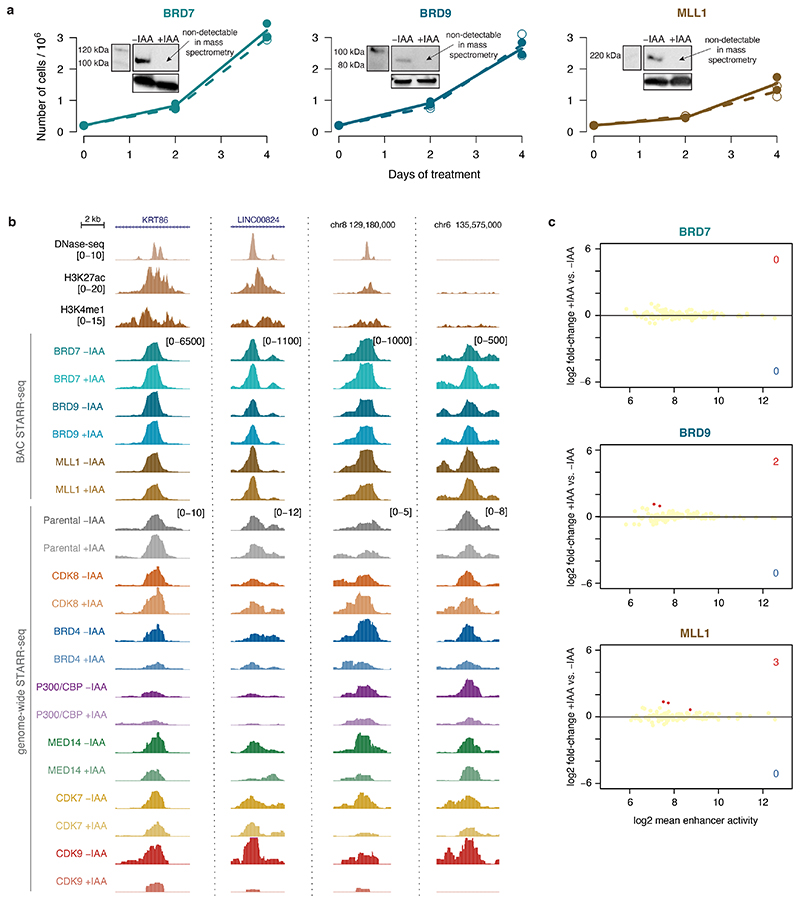
Here, we report distinct enhancer types with different COF dependencies that further differ in TF binding, chromatin modifications, genomic localization and the transcriptional response of nearby genes to COF depletion (Extended Data Fig. 9h,i). We anticipate that enhancer classifications will be refined when additional COFs are considered. However, when we AID-tagged and depleted three additional COFs (BRD7, BRD9 and MLL1; Extended Data Fig. 10a), STARR-seq with a focused library covering ~0.4% of the human genome (11.7 Mb) did not reveal any enhancer-activity changes (Extended Data Fig. 10b,c). In steady-state HCT116 cells, these factors might act redundantly with others or could only be required upon stimuli38 or during cellular transitions39.
The results for MED14 suggest that P53-mediated transcription might be independent of the Mediator complex, a finding that is difficult or impossible to formally prove given the essentiality of Mediator: residual MED14 or partial Mediator complexes may allow activation of P53-target genes in MED14-depleted cells. While selective rescue of P53 targets by residual MED14 seems less likely given that Mediator does not preferentially localize to these genes in any condition (Fig. 3h; Extended Data Fig. 5d-f), diverse Mediator sub-complexes indeed exist in yeast40 and human21,41 and could be recruited, e.g. via the interaction of MED17 with P5326. While the depletion of individual Mediator subunits by AID (four subunits), RNAi (MED17), or genetic depletion in stable knock-out cells18 (five subunits) and the combined depletion of five Mediator tail subunits in stable knock-out cells did not impair P53-target-gene transcription (Extended Data Fig. 6a-e), it is possible that these subunits function partially redundantly or in subcomplexes of variable composition. Redundancy between Mediator subunits has indeed been observed in yeast42–44 and stable partial human Mediator complexes could be reconstituted21,41, including a Mediator head and middle module that included MED17 but not MED1421. Alternatively, P53 targets might require only minimal levels of Mediator below the detection limits of this study, or other factors and conditions such as high local Pol II concentrations45, bypass via BRD4 and/or CDK9 (which are both required), or compensation by mobilized CDK932 might partially substitute for Mediator function at these genes. Finally, Pol II may initiate at these promoters via different mechanisms with distinct rate-limiting steps, potentially involving PICs with differential protein composition46.
The finding that TATA-boxes can confer BRD4-independence to LTR12 repeats, histone genes, and heat-shock genes, a classical model of TATA-box promoter genes regulated mainly at the pause-release step, suggests the existance of alternative mechanisms to recruit CDK9, e.g. via the SEC complex47,48 or TFs49. Interestingly, many enhancers required either MED14 or BRD4 (Fig. 3b, Fig. 2a - compare Groups 3 and 4). As these COFs function mainly in initiation or pause-release, respectively, Groups 2 and 3 enhancers might regulate distinct steps of transcription. The fact that both Mediator and BRD4 independent enhancers relate to genes activated upon stress suggests that rapidly inducible genes might have exploited this concept by circumventing certain regulatory steps (regulatory shortcutting) or by overcoming particular steps prior to actual induction (regulatory priming). Priming and regulation at pause-release step is for instance well known for heat-shock inducible genes50.
Together with the recent finding that promoters show distinct compatibilities towards different enhancers and specific COFs51, the finding that enhancers differ consistently in their COF dependencies and that gene-regulatory programmes differentially utilize these enhancer types is an important step towards understanding gene-regulatory specificities and determining innovative targets for the precise modulation of gene expression.
Materials and Methods
Cell culture
HCT116 cells were purchased from ATCC (#CCL-247) and cultured in DMEM with 10% heat inactivated fetal calf serum (FCS) (SigmaAldrich #F7524) and 1% L-Glutamine (LifeTech Austria/Invitrogen #25030024). HCT116 cells are near-diploid, chromosomally stable (P53 wild-type) and do not elicit interferon response upon reporter plasmid transfection6. For proliferation assays, cells were seeded into 6-well plates with 2x105 cells/well starting seeding density with or without the addition of Indole-3-acetic acid sodium salt (IAA/auxin, SigmaAldrich #I5148-2G) 500 μM final concentration. For up to 5 consecutive days cells were counted (Countess II Thermo Fisher #AMQAX1000) in 24 h intervals. K562 BRD4-AID cells were obtained from ref. 33 and cultured in RPMI-1640 with 10% FCS (SigmaAldrich #F7524). CH12 mouse lymphoma cell lines (wild-type and knock-out for different Mediator subunits) were obtained from ref. 18 and were cultured in RPMI-1640 with 10% FCS (SigmaAldrich #F7524), 1% Penicillin-Streptomycin and 50 μM of β-mercaptoethanol (Thermo Fisher Scientific). All cell lines tested negative for mycoplasma.
Cloning and characterization of genome editing events
SpCas9 knock-in homology dependent recombination (HDR) strategy and cloning of vectors are based on ref. 33. Parental cell line was generated via the insertion of the knock-in cassette “500 bp 5’HA -mCherry-P2A-OsTir1-3xmyc-500 bp 3’HA” downstream of the ActinB gene. 500 bp homology arms (HA) flanking the regions up and downstream of the ActinB stop codon were obtained via PCR on human genomic DNA (Promega # G304A). A total of 20 μg of the knock-in cassette (cloned into a MCS of a pbluescript vector) and the lentiCRISPR v2 vector comprising SpCas9 and gRNA (Addgene plasmid #52961) against ActinB stopcodon were equimolar electroporated into 5x106 HCT116 cells via the Maxcyte STX electroporation device (Cat No. GOC1). After 25 min recovery phase, medium was added, and cells were grown for 3 days. Afterwards cells were single cell sorted based on mCherry signal (~0,5-1% of total population). After 14 days, outgrowing clones were lysed (Biozym #101094) genotyped and potential knock-in candidates further validated via western blot against 3xmyc tag (Merck #05-724). Within an established Oryza sativa Tir1 (OsTir1) heterozygote tagged parental clone Ostir+/-, tagging of individual cofactors with the auxin inducible degradation system (AID) was performed. Auxin inducible destabilization domain constructs were cloned into lentiviral vector (Addgene plasmid #14748; ref. 33) for either N-terminal cofactor tagging :“5’HA-Blasticidin-P2A-V5-AID-spacer-3’HA” or C-terminal cofactor tagging “5’HA-spacer-AID -V5-P2A-Blasticidin-3’HA”. N or C-terminal tagging constructs were electroporated with the lentiCRISPR v2 containing gRNA against individual cofactors via Maxcyte STX. After 25 min recovery at 37 °C, medium (DMEM with 10% FCS and 1% L-Glutamine) was added and cells grown for 3 days. Furthermore, cells were trypsinized, transferred (1x106) into 6-well plates and selected for 10 days on Blasticidin (10 μg/ml) (eubio #ant-bl-10p). Outgrowing colonies were harvested, and single cell sorted for mCherry and against GFP. (As described in ref. 33 the Addgene plasmid #14748 construct expresses a constitutive active GFP which allowed negative FACS selection against potential vector backbone integrations.) After 14 days, grown out colonies were individually harvested, lysed with DNA extraction solution (Biozym #101094) and genotyped via sanger sequencing. Potential candidates were investigated via western blot against integrated V5-tag (Thermo Fisher #R960-25) or antibodies against endogenous proteins (Supplementary Table 1 - Antibodies).
PITCh-knock-in HCT116 cells
Cloning of PITCh vectors is based on ref. 52. “pX330S-2-PITCh” (Addgene, plasmid no. 63670) containing PITCh gRNA was cloned via Golden Gate assembly into the “pX330A-1x2” vector (Addgene, plasmid no. 58766) expressing Cas9 and the gRNA against a target locus. Knock-in cassette flanked by 40 bp micro-homology arms were cloned into the “pCRIS-PITChv2-FBL” vector (Addgene, Plasmid no. 63672). 20 μg total (13 μg pX330A-1x2 : 7 μg pCRIS-PITChv2-FBL) were electroporated into 5x106 cells via the Maxcyte STX. Follow up steps were similarly performed as described in the previous section “Cloning and characterization of genome editing events”.
Western blot
1x106 cells were harvested, centrifuged with 300 g for 5 min, washed with 1x PBS and lysed in 75 μl RIPA buffer containing protease inhibitor (Roche #11836170001). For complete lyses, cells were incubate on ice for 30 min, sonicated 4 x 30 s with a sonicator (Diagenode Bioruptor) and treated with 1 μl Benzonase endonuclease (SigmaAldrich #E1014-5KU) for 30 min to solubilize the chromatin-bound proteins. Afterwards, samples were centrifugated 10 min with 12,000 rcf at 4 °C and 40 μl 2x Laemmli buffer (BioRad #1610737) was added. Samples were vortexed, boiled for 5 min at 95 °C and centrifuged for 2 min at 12,000 rcf. Next, samples and marker (Invitrogen #LC5602) were loaded on protein gel (BioRad #4561083) using 1x SDS running buffer with 120V for 1:20h. Separated proteins were transferred via wet-transfer (BioRad #1703930) onto methanol activated membrane (Millipore, PVDF, 0.45 μm, IPFL00010); Transfer time: 1 h at 100V. After transfer, membrane was incubated for 10 min with TBST and blocked for 30 min in TBST + 5% milk (BioRad #1706404) on a rotating platform at room temperature. Next, the membrane was incubated in TBST + 5% milk comprising the primary antibody (Supplementary Table 1 - Antibodies) overnight (O.N.) 4 °C. After O.N. incubation, membrane was washed 3x with TBST for 15 min and incubated with secondary antibody (Supplementary Table 1 - Antibodies) for 2 h on rotating platform at RT. Last, membrane was washed 3x for 15 min in TBST before protein visualization via ECL detection (ChemiDOC Imager-Bio-Rad #170-5060).
Mass spectrometry analysis of COF depleted cell nuclei
1x106 cells were treated with water (mock) or 500 μM IAA for 1 or 3 h. Afterwards, cells were harvest with 1x trypsin, washed 1x PBS and centrifugation 3 min, at RT, 500g. Supernatant was removed and cell pellet was resuspended in ~100 μl of cytoplasmic extraction buffer (CE) (1X solution: 10 mM HEPES, 60 mM KCl, 1 mM EDTA, 0.075% (v/v) NP40, 1 mM DTT and 1 mM PMSF). Cells were incubate on ice for 3 min and centrifuged for 5 min, 4 °C with max speed. Cytoplasmic extract was removed from nuclei pellet and washed 3x with 100 μl CE without detergent NP40. Next, pellets were frozen in liquid nitrogen and stored at −80 °C for the following processing step.
Sample preparation for mass spectrometry
Samples for mass spectrometry analysis were prepared by using of iST kit (PreOmics GmbH #P.O.00027), according to the manufacturer’s instructions. Frozen pellets from nuclear extraction were 10 min incubated with 50 μl of lysis buffer at 95 °C. To share long DNA fragments RT cold lysate was sonicated with ultrasonication probe for 20s (amplitude 50%, cycle 0.5s; UP100H; Hielscher). Total protein concentration was determined by measurement of tryptophan fluorescence. The protein lysate was transferred into the cartridge, mixed with 50 μl lysate buffer and digested overnight at 37 °C. Digestion was quenched with 100 μl of Stop solution. Peptides were bind to sorbent in the cartridge by centrifugation at RT with 3800 × g for 3 min. Then a wash with 200 μl of Wash1 and then with of Wash2 solution was performed. The flow through was discarded and cleaned peptides were eluted from the cartridge in two steps by adding 100 μl of Elute buffer and centrifugation at RT with 3800 × g for 3 min. Peptides solution was placed into the SpeedVac machine until completely dry. Then resuspended in 50 μl of 0.1% TFA and sonicated in ultrasonication bath for 5 min to facilitate peptide solubilization. Peptides solution was stored in −80 °C prior further use.
Peptides separation
The nano HPLC system used was an UltiMate 3000 RSLC nano system coupled to a Q Exactive HF-X mass spectrometer, equipped with an EASY-spray ion source (Thermo Fisher Scientific) and JailBreak 1.0 adaptor insert for a spray emitter (Phoenix S&T, Inc., USA). Peptides were loaded onto a trap column (Thermo Fisher Scientific, PepMap C18, 5 mm × 300 μm ID, 5 μm particles, 100 Å pore size) at a flow rate of 25 μL/min using 0.1% TFA as mobile phase. After 10 min, the trap column was switched in line with the analytical column (Thermo Fisher Scientific, PepMap C18, 500 mm × 75 μm ID, 2 μm, 100 Å). For shotgun mass spectrometry analysis peptides were eluted using a flow rate of 230 nl/min, and a binary 3 h gradient, respectively 220 min. The gradient starts with the mobile phases: 98% A (water/formic acid, 99.9/0.1, v/v) and 2% B (water/acetonitrile/formic acid, 19.92/80/0.08, v/v/v), increases to 35%B over the next 180 min, followed by a gradient in 5 min to 90%B, stays there for 5 min and decreases in 2 min back to the gradient 98%A and 2%B for equilibration at 30 °C. For parallel reaction monitoring peptides were eluted using a flow rate of 230 nl/min, and a binary 1 h gradient, respectively 105 min. The gradient starts with the mobile phases: 98% A (water/formic acid, 99.9/0.1, v/v) and 2% B (water/acetonitrile/formic acid, 19.92/80/0.08, v/v/v) and hold for 10 min, increases to 35% B over the next 60 min, followed by a gradient in 5 min to 95% B, stays there for 5 min and decreases in 2 min back to the gradient 98% A and 2% B for equilibration at 30 °C.
Shotgun mass spectrometry analysis
The Q Exactive HF-X mass spectrometer was operated in data-dependent mode, using a full scan (m/z range 380-1500, nominal resolution of 60,000, target value 1E6) followed by MS/MS scans of the 10 most abundant ions. MS/MS spectra were acquired using normalized collision energy of 28, isolation width of 1.0 m/z, resolution of 30.000 and the target value was set to 1E5. Precursor ions selected for fragmentation (exclude charge state 1, 7, 8, >8) were placed on a dynamic exclusion list for 60 s. Additionally, the minimum AGC target was set to 5E3 and intensity threshold was calculated to be 4.8E4. The peptide match feature was set to preferred and the exclude isotopes feature was enabled. For peptide identification, the RAW-files were loaded into Proteome Discoverer (version 2.3.0.522, Thermo Scientific). All hereby created MS/MS spectra were searched using MSAmanda v2.0.0.9849 (ref. 53). For the 1st step search the RAW-files were searched against the SwissProt-human database (2019-02-23; 20,333 sequences; 11,357,489 residues), using following search parameters: The peptide mass tolerance was set to ±5 ppm and the fragment mass tolerance to 15ppm. The maximal number of missed cleavages was set to 2. The result was filtered to 1 % FDR on protein level using Percolator algorithm integrated in Thermo Proteome Discoverer. A sub-database was generated for further processing. For the 2nd step the RAW-files were searched against the created sub-database called Neumayr_20190223_QExHFX4_med14_human_step1.fasta. The following search parameters were used: beta-methylthiolation on cysteine was set as a fixed modification, oxidation on methionine, deamidation on N, Q, acetylation on lysine, phosphorylation on S, T, Y, methylation on K, R, di-methylation on K, R, tri-methylation on lysine, ubiquitinylation residue on lysine, biotinylation on lysine were set as variable modifications. Monoisotopic masses were searched within unrestricted protein masses for tryptic enzymatic specificity. The peptide mass tolerance was set to ±5 ppm and the fragment mass tolerance to ±15 ppm. The maximal number of missed cleavages was set to 2. The result was filtered to 1% FDR on peptide level using Percolator algorithm integrated in Thermo Proteome Discoverer. Peptide areas have been quantified using IMP-apQuant54. Statistical significance of differentially abundant peptide/proteins between different conditions was determined using a paired LIMMA test55.
Parallel reaction monitoring (PRM)
The Q Exactive HF-X mass spectrometer was operated by a mixed MS method which consisted of one full scan (m/z range 380-1,500; 15,000 resolution; target value 1e6) followed by the PRM of targeted peptides from an inclusion list (isolation window 0.7 m/z; normalized collision energy (NCE) 30; 30,000 resolution, AGC target 2e5). The maximum injection time variably changed based on the number of targets in the inclusion list to use up the total cycle time of 3s. The scheduling window were set to 4 min for each precursor. List of peptides including basic mass spectrometry information used for PRM analysis of proteins of interest and 7 normalization proteins are displayed in the Supplementary Table 1 - Mass Spec peptide sequences. Data processing and manual evaluation of results were performed in Skyline-daily56 (64-bit, v19.0.9.190). For the data processing peptides which had at least 3 specific peptide fragments were used. Proteins of interest were quantified based on integrated ion intensities over retention time of peptides from inclusion list. To account for different amounts between samples, these values were normalized based on a set of seven abundant/house-keeping proteins (Supplementary Table 1 - Mass Spec peptide sequences).
STARR-seq
Cells were grown in square plates (Thermo Scientific #166508) with a seeding density ~20 mill. cells 2 days/square plate before transfection. For genome-wide screens 4x108 and for BAC screens 4x107 cells were used. Genome wide (Addgene #99296) or BAC STARR-seq library utilizing the ORI as a core promoter6 was electroporated via Maxcyte STX into 85% confluent OsTir1+/- - COF-AID+/+ tagged cells. After 30 min recovery phase cells were split in 2 conditions, receiving medium containing water or IAA (500 μM final conc.; 2x108 cells). After 6 h cells were harvested, and total RNA was isolated using RNeasy Maxi kit (Qiagen #75162) containing β-mercaptoethanol supplemented RLT buffer. Spike-in control was added in a 1:1000 ratio to the isolated total RNA. Following steps were carried out as described in refs 6,57. Briefly, mRNA was isolated via Oligo-dT25 beads (Invitrogen #61005) followed by 1 h 37 °C TurboDNase I treatment (Invitrogen #AM2238). Subsequent, mRNA was cleaned via AMPure XP beads (Beckman Coulter #A63882) 1 : 1.8 ratio (RNA : beads) followed by reverse transcription via SuperScript III (Invitrogen #18080093) using a gene specific primer (GSP): 50 °C for 1h, 70 °C for 15 min, 4 °C for 10 min. Afterwards cDNA was treated with RNaseA (Thermo Fisher #EN0531) for 1 h at 37 °C followed by cleanup via AMPure XP beads 1:1.8 ratio. Next, “junction PCR”, which allows enrichment of reporter transcripts, was performed using KAPA 2x HiFi (KapaBiosystems #KK2601) utilizing the thermocycler program: 98 °C- 45 s, 98 °C- 15 s, 65 °C- 30 s 16 cycles, 72 °C- 70 s, 72 °C- 120 s followed by purification with AMPure XP 1 : 0.8 ratio (DNA: beads). Afterwards, “sequencing ready PCR” which amplifies STARR-seq transcripts was performed on the junction PCR products using Illumina primers with the thermocycler program: 98 °C- 45 s, 98 °C- 15 s, 65 °C- 30 s 5 cycles, 72 °C- 45 s, 72 °C- 120 s. Illumina adapter-containing STARR-seq library fragments were cleaned using SPRIselect beads (Beckman Coulter #B23318) with a stringent ratio of 1:0.5 (DNA: beads) and deep sequenced paired-end on an Illumina HiSeq2500 or NextSeq550 platform following manufacturer’s protocol recovering 15-20 mill. (genome-wide) or 1.5-2 mill. (BAC) reads per sample. Deep sequencing base-calling was performed with CASAVA 1.9.1.
STARR-seq spike-in controls
To accurately quantify changes in enhancer activity upon COF degradation and allow detection of potential global loss, we used spike-in controls for normalization of STARR-seq signal. In total 13 neutral/enhancer sequences (Supplementary Table 2 – STARR-seq spike-in sequences) from either the human or mouse genome were cloned into the STARR-seq vector6 (Addgene #99296) downstream of the ORI into the 3’UTR. Five human spike-in sequences were flanked by a 25 bp unique D. melanogaster sequence to distinguish spike-in reads from genome-wide STARR-seq reads and cloned in one orientation. Four promoter-proximal mouse enhancers were cloned in both orientations. All individually cloned vectors were pooled equimolar and electroporated into HCT116 cells. Total RNA was harvested after 6 h and stored at −80 °C. Spike-in was added to each genome wide STARR-seq screen in a ratio of 1:1000 at the total RNA isolation step.
PRO-seq
PRO-seq protocol was adapted from ref. 58 as follows. 1x107 COF-AID-tagged or WT HCT116 cells per replicate were harvested and nuclei were isolated after following treatments: (1) 3 h DMSO (mock), (2) 3 h 500 μM IAA (MED14- & BRD4-AID), (3) 3 h 10 μM Nutlin-3a (Sigma #SML0580) or (4) 3 h 500 μM IAA and subsequent 3 h 10 μM Nutlin-3a (MED14- & BRD4-AID). Spike-in control (S2 cells; 1% of total human cells) were added at the level of nuclei permeabilization step. Subsequent nuclear-run-on was performed for 3 min at 37 °C with biotin labeled CTPs (Perkin Elmer #NEL542001EA) followed by RNA extraction and base hydrolysis. Biotin nuclear-run-on RNA was enriched via M280 streptavidin beads (Invitrogen #112.06D) and precipitated via Phenol-Chloroform treatment. Next, 3’RNA adapters were ligated and second biotin RNA enrichment followed by RNA 5’ cap modification via TAP (Biozym #187005) treatment was performed. Furthermore, 5’ hydroxyl repair via PNK (NEB #M0201S) and subsequent 5’ adapter ligation was carried out. Afterwards, cDNA was generated from enriched RNA via reverse transcription (Super Script III Reverse Transcriptase, Invitrogen #18080-044). 10 μl of the cDNA library was amplified via KAPA Amplification reaction (Roche #7959028001) on a qPCR machine (Biorad CFX Connect RealTime System). KAPA reaction: 10 μl cDNA, 1 μl forward primer 35 μM (RP1-RP20), 1 μl of reverse primer 35 μM (RP1: 5’- AATGATACGGCGACCACCGAGATCTACAGTTCAGAGTTCTACAGTCCGA-3’), 25 μl 2x KAPA SYBER master mix, 13 μl water. PCR program: 98 °C 45 s, 98 °C 15 s, 60 °C 30 s, 72 °C 30 s, 72 °C 10 s. Samples were removed from the qPCR machine after 12-15 cycles and cleaned with Ampure beads (Beckman #A63881) in a 1 (sample) to 1.4 (beads) ratio. DNA bound to the beads was eluted in 11 μl water and deep sequenced single-end on an Illumina HiSeq2500 platform following manufacturer’s protocol. Deep sequencing base-calling was performed with CASAVA 1.9.1.
P53 induction for qPCR
HCT116 COF-AID-tagged cells (5x105 per replicate) were treated for 3 h (MED14-, BRD4-, CDK9- and TAF1-AID cells) or 12 h (MED15-, MED19- and MED1-AID cells) with 500 μM IAA (SigmaAldrich #I5148-2G) or water (mock) at 37 °C. This was followed by 6 h treatment with 10 μM Nutlin-3a (Sigma #SML0580) or DMSO (mock). Mouse CH12 knock-out cells were treated for 6 h with 30 μM Nutlin-3a (Sigma #SML0580) or DMSO (mock).
Oxidative stress induction
HCT116 MED14-AID-tagged cells (5x105 cells per replicate) were treated for 3 h with 500 μM IAA (SigmaAldrich #I5148-2G) or water (mock) at 37 °C. This was followed by 4 h treatment with 100 μM H2O2 or water (mock).
Heat shock induction
HCT116 (Parental, BRD4-, CDK9- and MED14-AID), K562 (BRD4-AID) and A549 (BRD4-AID) cells (5x105 cells per replicate) were treated for 3 h with 500 μM IAA (SigmaAldrich #I5148-2G) or water (mock) at 37 °C. This was followed by heat shock for 1 h at 43 °C.
Induction of LTR12 transcription
BRD4-AID-tagged cells (HCT116, K562 and A549) were treated for 18 h with 500 μM IAA (SigmaAldrich #I5148-2G) or water (mock) at 37 °C, to observe robust induction of LTR12 transcription upon BRD4 depletion.
siRNA-mediated knockdown
For gene knock-down by siRNA 3x105 cells were plated into single 6 well plates 5 h before transfection. 5 μl Lipofectamine 2000 (Thermo Fisher #11668027) was added to 250 μl OptiMEM (Invitrogen #31985062) and incubated for 5 min. Meanwhile siRNAs against target genes (10 nM final conc., IDT) were mixed with 250 μl OptiMEM, the mixes were combined, incubated for 20 min and dropwise added to the cells. For NFYA and NFYB knock-down BRD4-AID-tagged cells (HCT116 or A549) were used. 6 h after addition of NFYA and NFYB siRNAs, IAA (500 μM final conc.) or water (mock) was added for 18 h for a total of 24 h knockdown. For AFF1 and AFF4 knock-down Parental HCT116 cells (containing OsTir1) were used. After 24 h knockdown cells were heat shocked for 1 h at 43 °C. For MED17 knockdown Parental HCT116 cells were used. 18 h after addition of MED17 siRNA, Nutlin-3a (10 μM final conc.) or DMSO (mock) was added for 6 h for a total of 24 h knockdown.
qPCR
Following the different treatments cells were washed with 1x PBS, trypsinized for 3 min at 37 °C with 500 μl Trypsin and harvested after the addition of 500 μl medium. Cells were centrifuged at 500 g and washed with 1x PBS. PBS was removed and cells were lysed using Qiashredder columns (Qiagen #79654) followed by total RNA extraction via the RNeasy mini prep kit (Qiagen #74104), with β-mercaptoethanol supplemented RLT buffer. 2 μg of isolated total RNA was treated with 2 μl TurboDNase and 2 μl TurboDNase buffer (Invitrogen #AM2238) for 30 min at 37 °C in a thermocycler. Afterwards, 2 μl DNase inactivation reagent (Ambion #AM1906) was added, samples were vortexed for 2 min with 20 s breaks within and centrifuged for 5 min at 10,000 g. 10 μl of RNA was used for reverse transcription: 1 μl d(T)18 primer (NEB #S1316S) for mRNA or random hexamers (Bioline #38028) for LTRs, 1 μl dNTPs (NEB #4475), 1 μl RNase Inhibitor (Thermo Fisher #EN0531), 1 μl SuperScript III (Invitrogen #18080093), 1 μl DTT (Invitrogen #18080093; within SSIII kit), 4 μl forward strand buffer (Invitrogen #18080093, within SSIII kit), 1 μl water. Reaction was mixed and heated to: 25 °C for 5 min, 50 °C for 50 min, 70 °C for 15 min, 4 °C for 10 min in a thermocycler. Afterwards samples were diluted to total of 100 μl and 2 μl were used for qPCR. Reaction setup/sample: 10 μl SybrGreen (Promega #A6002), 1 μl forward (10 μM final conc.), 1 μl reverse primer (10 μM final conc.), 7 μl water and 2 μl DNA. qPCR setup/whole plate: 95 °C 2 s, 95 °C 3 min, 60 °C 30 s, read plate, go back to step 2 for 39 times (40 cycles in total).
MED1 ChIP-seq
MED14-AID-tagged HCT116 cells were cultured as described above. Media was removed and 1% formaldehyde in PBS for 15 min was used to fix cells. 0.5 ml 2.5M Glycine was added to each plate and let sit for 5 min. Media was dumped and plates were washed with PBS. 10 ml PBS was added to plate and scraped. Cell pellet was spun down and flash frozen in liquid nitrogen and stored at −80 °C with ~140 mill. cells in each tube. All buffers contained freshly prepared cOmplete protease inhibitors (Roche #11873580001). Frozen crosslinked cells were thawn on ice and then resuspended in lysis buffer I (50 mM HEPES-KOH, pH 7.5, 140 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% NP-40, 0.25% Triton X-100, protease inhibitors) and rotated for 10 min at 4 °C, then spun at 1350 rcf. for 5 min at 4 °C. The pellet was resuspended in lysis buffer II (10 mM Tris-HCl, pH 8.0, 200 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, protease inhibitors) and rotated for 10 min at 4 °C and spun at 1350 rcf. for 5 min at 4 °C. The pellet was resuspended in sonication buffer (20 mM Hepes pH 7.5, 140 mM NaCl, 1 mM EDTA 1 mM EGTA, 1% Triton X-100, 0.1% Na-deoxycholate, 0.1% SDS, protease inhibitors) and then sonicated on a Misonix 3000 sonicator for 10 cycles at 30 s each on ice (18-21 W) with 60 s on ice between cycles. Sonicated lysates were cleared once by centrifugation at 16,000 rcf. for 10 min at 4 °C. Input material was reserved and the remainder was incubated overnight at 4 °C with magnetic beads bound with MED1 antibody (Bethyl #A300-793A) to enrich for DNA fragments bound by MED1. Beads were washed with each of the following buffers: washed twice with sonication buffer (20 mM Hepes pH 7.5, 140 mM NaCl, 1 mM EDTA 1 mM EGTA, 1% Triton X-100, 0.1% Na-deoxycholate, 0.1% SDS), once with sonication buffer with high salt (20 mM Hepes pH 7.5, 500 mM NaCl, 1 mM EDTA 1 mM EGTA, 1% Triton X-100, 0.1% Na-deoxycholate, 0.1% SDS), once with LiCl wash buffer (20 mM Tris pH 8.0, 1 mM EDTA, 250 mM LiCl, 0.5% NP-40, 0.5% Na-deoxycholate), and once with TE buffer. DNA was eluted off the beads by incubation with agitation at 65 °C for 15 min in elution buffer (50 mM Tris-HCl pH 8.0, 10 mM EDTA, 1% SDS). Cross-links were reversed for 12 h at 65 °C. To purify eluted DNA, 200 ml TE was added and then RNA was degraded by the addition of 2.5 ml of 33 mg/ml RNase A (Sigma, R4642) and incubation at 37 °C for 2h. Protein was degraded by the addition of 4 μl of 20 mg/ml proteinase K (Invitrogen #25530049) and incubated at 55 °C for 30 min. DNA was purified using Qiagen PCR purification kit, eluted in Buffer EB, and deep sequenced single-end on an Illumina HiSeq2500 platform following manufacturer’s protocol.
MED1 immunofluorescence with RNA FISH
Immunofluorescence (IF) with concurrent RNA FISH was performed as described previously59,60. Briefly, coverslips were coated at 37 °C with 5 μg/ml poly-l-ornithine (Sigma-Aldrich, P4957) for 30 min and 5 μg/ml of laminin (Corning, 354232) for 2 h. HCT116 cells were plated on the pre-coated cover slips and grown for 24 h. For the last 3 h the cells were treated with 10 μM Nutlin-3a (Sigma #SML0580) or DMSO (mock) followed by fixation using 4% paraformaldehyde (PFA) (VWR, BT140770) in PBS for 10 min. After washing cells 3 times in PBS, the coverslips were put into a humidifying chamber or stored at 4 °C in PBS. Permeabilization of cells was performed using 0.5% Triton X100 (Sigma Aldrich, X100) in PBS for 10 min, followed by 3 PBS washes. Cells were blocked with 4% IgG-free bovine serum albumin (VWR, 102643-516) for 30 min and MED1 antibody (Bethyl #A300-793A) was added at a concentration of 1:500 in PBS for 4-16 hours. Cells were washed with PBS 3 times, followed by incubation with secondary antibody at a concentration of 1:5000 in PBS for 1 h. After washing twice with PBS, cells were fixed using 4% PFA (VWR, BT140770) in PBS for 10 min. After two washes of PBS, wash buffer A (20% Stellaris RNA FISH wash buffer A (Biosearch Technologies SMF-WA1-60), 10% deionized formamide (EMD Millipore S4117)) in RNase-free water (Life Technologies, AM9932) was added to cells and incubated for 5 min. 12.5 μM RNA probe (Biosearch Technologies, Stellaris RNA FISH Probe) in hybridization buffer (90% Stellaris RNA FISH hybridization buffer (Biosearch Technologies, SMF-HB1-10) and 10% deionized formamide) was added to cells and incubated overnight at 37 °C. After washing with wash buffer A for 30 min at 37 °C, the nuclei were stained in 20 mg/ml Hoechst 33258 (Life Technologies, H3569) for 5 min, followed by a 5-min wash in wash buffer B (Biosearch Technologies, SMF-WB1-20). Cells were washed once in water, followed by mounting the coverslip onto glass slides with Vectashield (VWR, 101098-042), and finally by sealing the cover slip with nail polish (Electron Microscopy Science Nm, 72180). Images were acquired on the RPI Spinning Disk confocal microscope with 100× objective using MetaMorph acquisition software and a Hammamatsu ORCA-ER CCD camera (W. M. Keck Microscopy Facility, MIT). Images were post-processed using Fiji Is Just ImageJ (FIJI). RNA FISH probes were custom-designed and generated by Biosearch Technologies (Stellaris RNA FISH) to target p21, RRM2B, TRIB1 and MYC intronic regions to visualize nascent RNA (Supplementary Table 1 – RNA FISH probes).
TATA- and CCAAT-box motif mutations oligo library
Eight instances of LTR12 elements overlapping a STARR-seq peak and promoters of two histone genes insensitive to BRD4 depletion were used as representative BRD4 independent promoters. For each candidate, the extended promoter sequence consisting of 205 nt upstream and 35 nt downstream of the CAGE-defined TSS was selected and scored against the TATA-box (TBP binding motif) and CCAAT-box (NFYA/B binding motif) PWM from the JASPAR database61 with R package seqPattern v.1.14.0. All motif instances with match above 90% were replaced by a fixed, low scoring sequence with similar nucleotide content as follows: CCAATCAS → AACTGACC for CCAAT-box motifs and STATAWAWRS → TGCAAGTCTT for the TATA-box motif, creating mutants for either TATA-box, CCAAT-box or both motifs together. For gain of function approach, 18 transcriptionally inert 240 bp long genomic regions were randomly selected. TATA- and/or CCAAT-box motif instances from the 10 BRD4 independent promoters were inserted into these neutral backgrounds by preserving the original number and arrangement of the motifs. Double motif insertions were designed for all 18 random sequences and motifs from all 10 BRD4 independent promoters promoters, and single motif insertions for 6 random sequences and motifs from 4 promoters. Each 240 nt long candidate sequence is present in the library 5 times and is barcoded with a unique 10 nt random barcode at the 3’ end. Barcode sequences were designed to match the GC content of the human 5’ UTRs62 and to differ from each other by at least 3 nucleotides. Designed 250 nt long candidate sequences are provided in Supplementary Table 7 – Oligos info and input counts. Sequences were flanked by the Illumina i5 (25 bp; 5’-TCCCTACACGACGCTCTTCCGATCT) and i7 (25 bp; 5’- GTTCAGACGTGTGCTCTTCCGATCT) adaptor sequences upstream and downstream, respectively, serving as constant linkers for amplification and cloning. The pool of 2,000 synthesized 300-mer oligonucleotides was obtained from Twist Biosciences Inc.
STAP-seq
STAP-seq input library was generated by cloning the amplified synthetic oligo pool into a human STAP-seq screening vector (Addgene ID 125150) as described previously34,51. 80 μg of input library was transfected into 4x107 BRD4-AID-tagged HCT116 cells using MaxCyte STX. Two independent transfections (biological replicates) were performed. After 30 min recovery phase cells were split in 2 conditions, receiving medium containing water or IAA (500 μM final conc.). Total RNA was isolated 6 h post electroporation followed by polyA+ RNA purification and turbo DNase treatment (Ambion; AM2238). Spike-in control was added in a 1:100 ratio to the isolated total RNA. STAP-seq RNA processing and cDNA amplification was performed as described previously51. Samples were sequenced paired-end on an Illumina NextSeq 550 platform following manufacturer’s protocol and base-calling was performed with CASAVA 1.9.1.
STAP-seq spike in controls
To accurately quantify changes in transcriptional activity upon BRD4 degradation, we used spike-in controls for normalization of STAP-seq signal. Previously described spike-in mix consisting of 9 mouse extended promoters cloned into a human STAP-seq spike-in vector (Addgene ID 125152) was used51. WT HCT116 cells were electroporated with the spike-in plasmid mix and total RNA was isolated after 6 h as described above and stored at −80 °C. Spike-in RNA was added to each STAP-seq screen in a ratio of 1:100 at the total RNA isolation step.
STARR-seq data processing
Paired-end 50 bp long STARR-seq reads were mapped using Bowtie63 v.1.2.2, first to the reference hg19 genome allowing up to 3 mismatches and then to the reference consisting of 5 human (flanked by D. melanogaster) and 4 mouse spike-in sequences allowing 1 mismatch. Only read pairs mapping uniquely were kept. Mapped reads were sorted and indexed with samtools v.0.1.19 and combined into paired-end fragments with R/Bioconductor64 package GenomicAlignments v.1.18.1. Summary of reads mapping to the reference genome and spike-in sequences for each sample is provided in Supplementary Table 2 – Reads statistics per sample.
STARR-seq normalization by spike-in
For each spike-in sequence, number of paired-end fragments mapping exactly to sequence ends and spanning the entire cloned spike-in sequence in the correct orientation was counted. For mouse spike-in sequences that were cloned in both orientations, mappings in the two orientations were considered separately. For each individual STARR-seq sample, relative abundance (proportion) of each of the 13 cloned spike-in sequences was calculated and scaled by dividing with the mean across the 13 sequences. These relative abundances were used to normalize the STARR-seq signal between auxin (+IAA) treated and control condition for each AID-tagged COF as follows. For each individual sample (replicate) the median of scaled relative abundances across 13 spike-in sequences was taken and used to calculate the ratio between paired treated and control samples (these samples stem from the same STARR-seq library transfection and differ only in the treatment). The control sample was then set to 1 and the scaling factor for the treated sample was expressed relative to the control using the calculated ratio. Finally, for each AID-tagged COF, mean scaling factor across the replicates was taken to make the normalization more robust and less sensitive to variability between replicates. For P300+CBP-AID we did not use spike-in for normalization because it is not reliable in this case. P300/CBP regulates transcription of rRNAs by Pol I65 and thus depletion of P300/CBP leads to drastic changes in total cellular RNA abundance. Our normalization approach relies on adding spike-in RNA in a fixed ratio to total RNA and assumes that the bulk of total cellular RNA is not changing, so it cannot be used in case of P300/CBP depletion. All spike-in counts, relative abundances and calculations of scaling factors are provided in Supplementary Table 2 – Spikein counts & norm. factors. Final scaling factor for each AID-tagged COF was used to normalize the STARR-seq coverage in auxin treatment relative to control and was supplied as custom scaling factor in differential analysis.
Detection and quantification of enhancer activity
For each AID-tagged COF and condition, unique STARR-seq fragments (after removing duplicates) from all replicates were combined and used for peak calling with MACS2 v.2.1.2.1. Genome-wide STARR-seq library input was sequenced previously6 and used here as background for peak calling. Only peaks at 1% FDR with enrichment over input ≥3 on both strands and at least 3 tags per million (corresponding to ~25 fragments) were kept and combined into a reference set of 6249 STARR-seq enhancers. Number of unique fragments for peak calling and peaks called per COF/condition is provided in Supplementary Table 2 – Called peaks per CoF&condition. Note that due to COF depletion, the number of peaks called per condition varies, yet all enhancer-acitvity changes are re-evaluated independently of these initial peak calling for each of the 6249 enhancers in the reference set. To quantify enhancer activity, the number of STARR-seq fragments overlapping each enhancer in the reference set was counted in each individual STARR-seq sample (replicate). Raw count table is provided in Supplementary Table 3 - STARR-seq raw counts, and was used for subsequent differential analysis.
Differential analysis of COF-AID STARR-seq
Differential analysis between auxin treated and control condition was performed per COF-AID cell line with R/Bioconductor package edgeR66 v.3.24.3, always using the same reference set of 6249 STARR-seq enhancers. Scaling factor calculated from spike-in was supplied as custom scaling factor for normalization to allow accurate assessment of changes in enhancer activity and possible detection of global effects. Significant changes in enhancer activity were called at 5% FDR (Extended Data Fig. 2d). Corrected log2 fold-change values and multiple-testing adjusted P-values from edgeR for all enhancers in the reference set were used for downstream analyses and are provided in Supplementary Table 3. To assess the effect of COF tagging on enhancer activity (in the absence of auxin), we also performed differential analysis between control condition of each COF and the Parental cell line with edgeR, calling significant changes at 5% FDR (Extended Data Fig. 2c).
Clustering of COF-AID STARR-seq screens
To group the different COF-AID-tagged cell lines based on enhancer activity, we used normalized COF STARR-seq signals from merged replicates per COF and condition (auxin treatment and control). Hierarchical clustering was performed using Manhattan distance between normalized STARR-seq signals (Extended Data Fig. 2b). To group the COF-AID-tagged cell lines based on changes in enhancer activity upon auxin treatment, we performed hierarchical clustering using Manhattan distance between log2 fold-change values (Fig. 1f).
Clustering of STARR-seq enhancers
We clustered enhancers based on change in their activity upon depletion of 5 individual COFs (BRD2, BRD4, P300+CBP, MED14 and CDK7) with K-medoids (Fig. 2a). Partitioning around medoids (PAM, K-medoids) was performed on log2 fold-change values using PAM algorithm implemented in the R package cluster v.2.0.7-1. To determine the optimal number of clusters, PAM was initially run with varying number of clusters from 1 to 10, and for each run the proportion of variance explained by clustering was calculated as ratio of within-cluster variance and between-cluster variance. Clustering into 4 clusters explained more than 85% of the variance and further increasing the number of clusters led to less than 5% gain (Extended Data Fig. 3b), so we selected 4 as the optimal number of clusters. To make the clustering robust, we run PAM with k=4 clusters independently 1000 times, each time using different randomly chosen data points as initial centroids. For each enhancer we then calculated the number of times it was assigned to each of the 4 clusters and assigned it to the most frequent cluster. The clustering was robust with majority of enhancers (>86%) being assigned to the same cluster >50% of the time. To further confirm the robustness of the defined enhancer groups (size of groups and enhancer group membership), we used two alternative clustering approaches. We performed hierarchical clustering using Euclidean distance metric, and defined 5 clusters by cutting the dendrogram. For each hierarchical cluster we calculated the percentage of enhancers that are assigned to each of the four originally defined PAM enhancer groups. This revealed an almost 1-to-1 correspondence between hierarchical clusters and originally defined PAM clusters, with more than 80% of enhancers in each hierarchical cluster belonging to a single originally defined enhancer group (Extended Data Fig. 3c,d). We also employed Uniform Manifold Approximation and Projection (UMAP) algorithm to reduce the dimensionality and visualize the data. This revealed a clear separation of originally defined enhancer groups in two-dimensional UMAP representation (Extended Data Fig. 3e).
Annotation of enhancers with TF motifs and transposable elements
All TF motifs from the JASPAR 2020 vertebrate core collection61 of 579 non-redundant motifs were considered and the occurrence of these motifs at different score thresholds in the hg19 genome assembly was downloaded directly from the JASPAR database (https://jaspar2020.genereg.net/download/data/2020/CORE/JASPAR2020_CORE_non-redundant_pfms_jaspar.zip). Only the most highly scoring motif occurrences, with a score in the top 1 percentile of the scores for the respective motif, were kept. These motif occurrences were overlapped with STARR-seq enhancers, and a binary matrix denoting which motifs are present in each enhancer was constructed. For annotation of enhancers with transposable elements, the annotation of repeats from RepatMasker for hg19 genome assembly was downloaded from the UCSC Table Browser67.
Annotation of enhancers with TF/COF binding and histone modifications
Various published datasets for HCT116 cell line were downloaded from the GEO repository and ENCODE database, including chromatin accessibility68,69, ChIP-seq for different histone modifications68,70, TFs27,68 and COFs23,38,70,71. All accession numbers of used published datasets are listed in Supplementary Table 4. Raw sequencing data was downloaded from GEO/SRA and reads were mapped with Bowtie v.1.2.2 to hg19 genome assembly allowing only unique mapping. Peaks were called with MACS2 v.2.1.2.1. against matching input (if available) using only unique reads and default MACS2 parameters, keeping peaks at 5% FDR. For datasets from ENCODE, the peaks files were downloaded and used directly in downstream analyses.
ChIP-seq peaks from individual datasets were overlapped with STARR-seq enhancers, and a binary matrix denoting which TF, COF or histone modification peaks are present in each enhancer was constructed.
Motif, TF/COF binding and histone modification enrichment analysis
For enrichment analysis a binary matrix denoting which enhancers overlap which motifs, repeat elements, TF/COF binding sites or histone modifications was used. To create a random background for assessing enrichment, STARR-seq peaks were shifted by 10kb and the resulting shifted regions were annotated with motifs, TF/COF binding sites and histone modifications as described above. Two-sided Fisher’s exact test was used to assess the enrichment/depletion of a particular feature in a specific group of enhancers, either against random regions or against enhancers in other groups. Enrichment/depletion values (odds ratios) of different features across different groups of enhancers were visualized in form of a heatmap, showing only significant enrichments (P-value ≤ 0.05; Fig. 2c-e, Extended Data Fig. 3h).
Multiple alignment of LTR12 elements
Sequences of LTR12 family retrotransposons overlapping STARR-seq enhancers were multiple aligned using ClustalW algorithm implemented in the R package msa v.1.14.0. Multiple alignment was visualized with ggmsa v.0.0.2 package (Extended Data Fig. 7d).
Gene and TSS annotation
To obtain a non-redundant set of genes and their precise associated TSSs for accurate quantification of PRO-seq signal in different gene regions, we pre-processed and refined gene annotation as follows. We took all coding and long non-coding transcripts from Ensembl version 82 for hg19 genome assembly and removed transcripts shorter than 300 bp. For each group of transcripts that have the same annotated TSS we kept only the longest one. We annotated these non-redundant transcripts with CAGE TSS clusters from FANTOM572 as follows. For each transcript (unique annotated TSS) we identified the strongest CAGE TSS within a window encompassing 500 bp upstream and 500 bp downstream of the annotated TSS, excluding the CDS. Then, for each selected CAGE TSS (that was possibly associated with multiple annotated transcripts) we kept the closest transcript and corrected its annotated TSS to the CAGE TSS. The resulting non-redundant transcript/gene annotation with precise CAGE-corrected TSSs was used in all downstream analyses.
Gene ontology analysis
We assessed whether genes with CCAAT- and TATA-box-containing promoters are enriched for a particular gene ontology (GO) term by calculating hypergeometric P values for every GO term with R/Bioconductor package GOstats73 v.2.48.0, using CCAAT- and TATA-box-containing genes as a foreground and all other annotated genes as a background. Only terms with P-value ≤10-4 were considered significant and sorted by the enrichment. Top 5 enriched terms for each of the 3 GO categories (biological process, molecular function and cellular compartment) were shown (Extended Data Fig. 8a).
PRO-seq data processing
Single-end 50 bp long PRO-seq reads contain a 8 bp long UMI at the 5’ end, which was removed before mapping and kept track of. From the remaining 42 bp the Illumina adapter was trimmed of with cutadapt v.1.18. Reads longer than 15 bp after adapter trimming were mapped using Bowtie63 v.1.2.2 to a reference consisting of hg19 and dm3 (spike-in) genome allowing up to 2 mismatches. Multimapping was allowed to up to 1000 positions and all multimapping reads were randomly assigned to one mapping position. For reads that mapped to the same genomic position, we collapsed those that have identical UMIs as well as those for which the UMIs differed by 1 nucleotide to ensure the counting of unique nascent RNA molecules. To generate the coverage of PRO-seq signal, i.e. exact positions of Pol II molecules associated with 3’ end of nascent transcripts, only the first nucleotide of each read was considered, and the strand was swapped to match the direction of transcription. Summary of reads mapping to the reference genome and spike-in genome, and counts of reads with unique UMIs for all PRO-seq samples is provided in Supplementary Table 5 – PRO-seq (MED14-, BRD4-AID & WT).
Differential analysis of PRO-seq
Differential analysis was performed using a non-redundant set of genes with CAGE corrected TSSs. For each gene the region from the TSS up to 150 bp downstream (+1 to +150) was defined as “promoter + pause region”, and the rest of the annotated gene was defined as “gene body”. For BRD4 depletion in BRD4-AID cells (Fig. 4f,g), the number of unique (UMI collapsed) PRO-seq read 5’ ends falling into these two regions was counted for each gene. Differential analysis was performed with DESeq2 v.1.22.2 (ref. 74) for “promoter + pause” and “gene body” region separately to capture the pause-release defect. For MED14 depletion in MED14-AID cells and induction of P53 target genes by Nutlin-3a in WT, MED14- and BRD4-AID cells (Fig. 3d,e, Extended Data Fig. 4a,d), number of unique (UMI collapsed) PRO-seq read 5’ ends falling into the whole gene region was counted and differential analysis was performed on the entire gene. Raw PRO-seq counts used for differential analysis are provided in Supplementary Table 6. To allow accurate assessment of changes in enhancer activity upon different treatments and possible detection of global effects, we used spike-in based normalization. Scaling factor for normalization between conditions was calculated from relative abundance of reads mapping to spike-in genome (dm3) in combined replicates for each condition. Spike-in normalization factors were supplied as custom scaling factors to DESeq2, with all replicates of the same condition receiving the same scaling factor. These scaling factors were also used to normalize PRO-seq coverage of combined replicates per condition for visualization in the genome browser. Spike-in read counts, relative abundances, and calculations of scaling factors are provided in Supplementary Table 5 – PRO-seq (MED14-, BRD4-AID & WT).
qPCR data analysis
All treatments for qPCR were done in at least 3 independent biological replicates and each sample was measured at least 2 times (technical replicates). Raw CT values of technical replicates were averaged and then normalized to a reference gene: GAPDH for all human wild-type and AID-tagged cell lines and Actb for mouse CH12 wild-type and knock-out cell lines. When calculating a ratio to a control (no treatment) condition, the normalized value for each individual replicate of the treated condition was divided by the normalized value for the corresponding replicate of the control condition. Obtained ratios thus account for the variance in both treated and control conditions and were used to calculate the standard deviation shown in all qPCR barplots and to perform two-sided Student’s t-test (Fig. 3g, Fig. 4c,e, Extended Data Fig. 4i,j,k, Extended Data Fig. 6a-d, Extended Data Fig. 7c,f,h-j, Extended Data Fig. 9e-g).
MED1 ChIP-seq data processing and analysis
Single-end 50 bp long reads were mapped using Bowtie v.1.2.2 to the reference hg19 genome allowing up to 3 mismatches and only uniquely mapping reads were retained. Summary of reads mapping to the reference genome for each sample is provided in Supplementary Table 5 – MED1 ChIP-seq (MED14-AID & WT). To generate genome-wide coverage mapped reads were extended to 500 bp with GenomicRanges v1.34.0. and the coverage was normalized to reads per million. Unique reads were used to call peaks with MACS2 v2.1.2.1 for each condition/treatment against the respective input, using default MACS2 settings (adjusted P-value ≤ 0.05). For WT HCT116 cells unique reads from two independent biological replicates were combined prior to peak calling to obtain a common set of peaks per condition. Peaks from different conditions were sequentially combined to obtain a non-redundant set of reference peaks and ChIP-seq signal (ChIP-seq coverage over input) from different datasets centered at the reference peak summits was visualized (Extended Data Fig. 5c). MED1 ChIP-seq signal was quantified and compared at two different types of STARR-seq enhancers: (1) P53-bound (overlapping a P53 ChIP-seq peak in HCT116 cells upon Nutlin-3a treatment27) enhancers insensitive to MED14 depletion according to differential analysis of MED14 STARR-seq, and (2) accessible (according to DHS-seq) and H3K27ac-marked enhancers significantly downregulated upon MED14 depletion according to differential analysis of MED14 STARR-seq (Extended Data Fig. 5e).
Analysis of MED1 IF with RNA FISH
3D image data gathered in RNA FISH and IF channels for ~120 cells per FISH probe (gene) was processed with custom Python and MATLAB scripts as described previously59,60. Briefly, FISH foci were manually identified in individual z stacks through intensity thresholds, centered along a box of size l = 1 μm, and stitched together in 3-D across z stacks. Only cells with 1 or 2 FISH foci were considered for downstream analyses. For every RNA FISH focus identified, signal from the corresponding location in the IF channel is gathered in the l x l square centered at the RNA FISH focus at every corresponding z-slice. The IF signal centered at FISH foci for each FISH and IF pair are then combined and an average intensity projection is calculated, providing averaged data for IF signal intensity within a l x l square centered at FISH foci. The same process was carried out for the FISH signal intensity centered on its own coordinates, providing averaged data for FISH signal intensity within a l x l square centered at FISH foci. As a control, this same process was carried out for IF signal centered at randomly selected nuclear positions within the nucleare volume determined from DAPI staining through the z stack image as described in detail in ref. 59. Average MED1 IF intensity projections centered at FISH foci were visualized using the same intensity-color range for all genes, ranging from minimal to maximal observed IF intensity within the 1 μm x 1 μm area (Fig. 3h, Extended Data Fig. 5f). For quantitative comparison of MED1 IF signal between different genes (Fig. 3i), MED1 IF signal at each FISH focus was normalized to the average signal at random spots from the same dataset to account for the difference in overall MED1 IF intensity between different datasets.
STAP-seq reads processing
STAP-seq sequencing reads were processed as described previously51. Briefly, paired-end STAP-seq reads were mapped to a reference containing 250 bp long sequences of 2,000 barcoded wild-type and mutant promoter oligos and to the 9 mouse spike-in promoter sequences using Bowtie63 v.1.2.2 allowing only 1 mismatch. Prior to mapping the 10 nt long UMI was removed from the 5’ end of the forward read and kept track of for later counting. Only uniquely mapping read pairs where the reverse read maps exactly to the oligo end were kept, ensuring they correspond to reporter transcripts transcribed from that particular cloned barcoded promoter candidate. For read pairs that mapped to the same positions, we collapsed those that have identical UMIs as well as those for which the UMIs differed by 1 nucleotide to ensure the counting of unique reporter transcripts. Tag counts at each position represent the sum of the 5’-most position of UMI collapsed fragments. Total read counts mapping to promoter oligo library and spike-in promoters are summarized in Supplementary Table 5 – STAP-seq (BRD4-AID).
STAP-seq data analysis
Tag counts at each position in each screened promoter candidate were quantified in different conditions/datasets as described above and represent the number of unique RNA molecules initiated at that position (Supplementary Table 7). Raw counts were normalized by the spike-in as described previously51. Briefly, number of unique RNA molecules originating from each of the 9 spike-in mouse promoters was quantified as described above and the counts were used to calculate the scaling factor from each individual spike-in promoter. Final normalization factor was calculated as median of factors derived from individual spike-in promoters and is provided in Supplementary Table 5 – STAP-seq (BRD4-AID). For comparison of transcriptional output between wild-type promoters or neutral sequences and their mutated variants, the sum of normalized counts in the 5 bp window centered at the cognate/expected TSS (position 206 in the 250 bp long promoter candidate) was considered and was corrected for the abundance of each promoter sequence in the input STAP-seq library (Fig. 4j). For visualization of transcriptional output per position in a specific promoter variant (wild-type or mutant), the signal from 5 instances of that promoter variant present in the library (each barcoded with a different unique barcode) was combined (Fig. 4i, Extended Data Fig. 9c,d).
Statistics and data visualization
All statistical calculations and graphical displays have been performed in R statistical computing environment75 v.3.5.1. In all box plots, the central line denotes the median, the box encompasses 25th to 75th percentile (interquartile range) and the whiskers extend from 5th to 95th percentile of the data. In all bar plots, bar height denotes the mean and error bars denote standard deviation. Heatmaps were created with R package gplots version 3.0.1. Coverage data tracks have been visualized in the UCSC Genome Browser67 and used to create displays of representative genomic loci.
Extended Data
Extended Data Fig. 1. Validation of cofactor degradation and effect on cell growth.
a, Western blots of denoted cofactors (COF) in the cell line where the respective COF is tagged by AID, without and with auxin (IAA) treatment for 1h. done once; validated by mass spectrometry; gel source data: Supplementary Figure 1. b, Schematic of the Mediator complex structure with head, middle and tail domains shown in different colors. Core structural subunit MED14 targeted in this study is shown in green. Subunits that cannot be detected anymore in mass-spectrometry upon MED14 depletion are semi-transparent. c, Protein abundance change as measured by shot-gun mass-spectrometry upon MED14 depletion by auxin treatment. All detected Mediator subunits are marked and colored according to different Mediator modules/domains shown in b. Subunits marked in italic were not detected anymore (i.e. were below detection limit) in all replicates of auxin treatment. N=3 independent replicates. d, Protein abundance of denoted COFs as measured by targeted mass-spectrometry approach in the cell line where the respective COF is tagged by AID, without and with auxin treatment for 3h. N=3 independent replicates; mean +/- SD shown. e, Growth curves over a course of 3 days comparing untreated (solid line) and auxin treated (dashed line) cells, for each COF-AID cell line. N=2 independent replicates. f, Growth curves over a course of 5 days comparing untreated (solid line) and auxin treated (dashed line) cells for MLL4- and CDK8-AID cell line. N=3 independent replicates. P-value of two-sided Student’s t-test at final day 5 timepoint is shown.
Extended Data Fig. 2. Effect of cofactor tagging and targeted cofactor degradation on enhancer activity.
a, Pearson’s correlations for pair-wise comparisons of replicates for each cofactor (COF) with and without auxin (IAA) treatment calculated across a reference set of 6249 enhancers. For majority of COFs there are 4 independent replicates in each condition, except for our positive and negative controls, CDK9 and the Parental cell line, that have 2 and 3 replicates per condition, respectively. Inset on the right shows correlations between BRD4 samples pre-treated with auxin before STARR-seq library transfection, i.e. with an extended period of protein degradation. b, Hierarchical clustering of untreated and auxin treated Parental and different COF-AID cell lines based on enhancer activity for a reference set of 6249 enhancers. All untreated cell lines (except p300/CBP which shows high level of COF pre-degradation in absence of auxin) cluster together with the Parental cell line, as well as auxin treated MLL4- and CDK8-AID cell lines. c, Differential analysis of STARR-seq enhancer activity between each individual COF-AID cell line and Parental cell line without any treatment to assess the effect of COF-tagging on enhancer activity. Number of significantly up- or down-regulated enhancers is denoted (FDR≤0.05). d, Differential analysis of STARR-seq enhancer activity for each COF-AID cell line with and without auxin treatment to assess the effect of COF degradation on enhancer activity. Number of significantly up- or down-regulated enhancers is denoted (FDR≤0.05). e, Log2 fold-change in enhancer activity for enhancers pre-affected by P300 and CBP tagging (left; N=301) and the rest of non-affected enhancers (right; N=5948) in Parental and P300/CBP-AID cells upon auxin treatment. Boxes: median and interquartile range; whiskers: 5th and 95th percentiles. P-values: two-sided Wilcoxon rank-sum test. f, Significance of change in enhancer activity (P-values from differential analysis corrected for multiple testing/FDR) for a reference set of 6249 enhancers sorted individually by fold-change in each COF-AID cell line, from unaffected (or upregulated) enhancers on the left to most downregulated enhancers on the right.
Extended Data Fig. 3. Features of the four different groups of enhancers.
a, Significance of change in enhancer activity (P-values from differential analysis corrected for multiple testing/FDR) upon individual cofactor degradation for four groups of enhancers defined by PAM (partitioning-around-medoids) clustering. Significant P-values (FDR≤0.05) for down- and up-regulated enhancers are shown in shades of blue and red, respectively. Non-significant P-values are shown in yellow. N = 1392, 1660, 1519, 1678 for Groups 1-4, respectively. b, Percent of variance explained by clustering of 6249 enhancers with partitioning around medoids (PAM) algorithm into different number of clusters. Four clusters explain ~85% of the variance. c, Hierarchical clustering of enhancers based on change in enhancer activity upon individual cofactor degradation. Boxplots summarize the log2 fold-change values per COF for each of the 5 clusters defined by cutting the dendrogram as denoted with a dashed line. Enhancer group assignment (from PAM clustering shown in Fig. 2a) is denoted by the coloured stripe below the dendrogram. N = 1156, 1391, 1531, 1052, 1119 for Groups 1-5, respectively. Boxes: median and interquartile range; whiskers: 5th and 95th percentiles. d, Agreement between clusters defined by hierarchical clustering and enhancer groups defined by PAM. For each hierarchical cluster (row) percent of enhancers falling into each PAM enhancer group is shown. e, Two-dimensional visualization of the data after dimensionality reduction with UMAP algorithm. Points represent individual enhancers coloured by their group membership (from PAM clustering). f, Percent of enhancers accessible/open according to DNase-seq in HCT116 cells or in other cell types in the four groups of enhancers defined in Fig. 2a. g, Percent of enhancers accessible/open according to DNase-seq in different number of cell lines ranging from enhancers closed in all cell lines (0 - yellow) to enhancers open in many/all (125 - red) cell lines assayed by DNase-seq in ENCODE. h, Mutual enrichment of transcription factor motifs for the four groups of enhancers. For each motif from the JASPAR vertebrate core collection of 579 non-redundant TF motifs (https://jaspar2020.genereg.net/download/data/2020/CORE/JASPAR2020_CORE_non-redundant_pfms_jaspar.zip) the enrichment/depletion in each group is assessed against the remaining three groups using two-sided Fisher’s exact test and only motifs with P-value≤0.001 and odds-ratio≥2 are shown. The motifs are hierarchically clustered based on pair-wise Pearson’s correlation between motif position-weight matrices (PWMs) to group together similar motifs. A selection of representative motifs from these groups of similar motifs is shown in Fig. 2e. i, Enrichment analysis of 579 non-redundant TF motifs from the JASPAR vertebrate core collection (https://jaspar2020.genereg.net/download/data/2020/CORE/JASPAR2020_CORE_non-redundant_pfms_jaspar.zip) between unaffected and down-regulated enhancers upon MED14 depletion. Significantly enriched and depleted motifs (two-sided Fisher’s exact test; P-value ≤0.01) are shown in red and blue, respectively. j, Differential analysis of enhancer activity upon MED14 depletion with enhancers containing a P53 motif marked in yellow. k, Differential analysis of enhancer activity upon MED14 (left) or BRD4 (right) depletion with enhancers overlapping a P53 ChIP-seq peak marked in yellow.
Extended Data Fig. 4. Induction of P53 target genes and enhancers is insensitive to MED14 depletion, but sensitive to BRD4 depletion.
a, Differential analysis of PRO-seq signal at genes between Nutlin-3a treated and untreated WT HCT116 (left), MED14- (middle) or BRD4-AID (right) cells. Number of significantly upregulated genes in each cell line is denoted in yellow (FDR≤0.05 and fold-change≥2). N=2 independent replicates for each condition. b, Venn diagram showing overlap of significantly upregulated genes in the 3 cell lines shown in panel a, defining in total 151 P53 target genes induced after 3h of Nutlin-3a treatment. c, Venn diagram showing overlap of 151 P53 target genes induced after 3h of Nutlin-3a (this study) and 175 P53 target genes defined previously after 1h of Nutlin-3a treatment76, defining a set of 243 direct P53 target genes used in panels d and h, and in Fig. 3e. d, Comparison of induction of direct P53 target genes (defined in panel c) in different cell lines and conditions. Top row compares induction in MED14- (left) or BRD4-AID (right) cells when the respective factor is present (-IAA) or depleted (+IAA). P53 targets are induced to the same extent upon MED14 depletion, but their induction is impeded upon BRD4 depletion. Bottom row compares induction between the two cell lines in the condition without auxin (left) or with auxin (right). Without auxin both MED14- and BRD4-AID cells induce P53 target genes to the same extent, however with auxin the induction in the BRD4-AID cells is impeded compared to the MED14-AID cells. e, Loci of the P53 target genes FAS (left) and RPS27L (right) with intronic P53-bound enhancers. Enhancer activity in different COF-AID cell lines with and without auxin (IAA) treatment is shown (normalized STARR-seq signal for merged replicates), together with nascent transcription (normalized PRO-seq signal for merged replicates) upon induction of P53 signalling with Nutlin-3a in MED14- and BRD4-AID cells with and without auxin treatment. Transcription of both genes is induced upon Nutlin treatment in both conditions with MED14 present (-IAA) or degraded (+IAA), but is strongly reduced with BRD4 degraded due to a pause-release defect that persists upon Nutlin treatment. Activity of their associated P53-bound enhancers is unchanged upon MED14 depletion but is abolished upon BRD4 depletion. f, Locus with a FOS-bound MED14-depletion sensitive (left) and a P53-bound MED14-depletion insensitive (right) enhancer. Activity in different COF-AID cell lines with and without auxin (IAA) treatment is shown (normalized STARR-seq signal for merged replicates), together with nascent transcription (normalized PRO-seq signal for merged replicates) upon induction of P53 signalling with Nutlin-3a in MED14- and BRD4-AID cells with and without auxin treatment. Activity of the FOS-bound enhancer is strongly reduced by both MED14 and BRD4 depletion, whereas the activity of the P53-bound enhancer is unchanged upon MED14 depletion but is abolished upon BRD4 depletion. Endogenous bidirectional transcription of the P53-bound enhancer is induced upon Nutlin treatment in both conditions with MED14 present (-IAA) or degraded (+IAA), but is reduced with BRD4 degraded due to a pause-release defect that persists upon Nutlin treatment. g, Differential analysis of PRO-seq signal at distal P53 or FOS bound sites (enhancers) upon Nutlin-3a treatment in auxin treated BRD4-AID cell line. h, Log2 fold-change of PRO-seq signal for direct P53 target genes (left; genes defined in panel c) and distal P53 bound sites around direct P53 target genes (enhancers; right) in BRD4-AID cell line upon Nutlin-3a induction in background with BRD4 present (-IAA) or depleted (+IAA). N = 151, 20964, 244, 359 for P53 targets, other genes, P53- and FOS-bound enhancers, respectively. Boxes: median and interquartile range; whiskers: 5th and 95th percentiles; P-values: two-sided Wilcoxon rank-sum test. i-k, Endogenous induction of known P53 target genes with Nutlin-3a as measured by qPCR in BRD4- (i), CDK9- (j) or TAF1-AID (k) cells without or with auxin treatment, i.e. with the respective factor present or degraded. N=3 independent replicates; fold-change for each replicate calculated independently by dividing the treatment value with the corresponding control value; mean +/- SD shown; P-values: two-sided Student’s t-test. l, Growth curves over a course of 3 days comparing untreated (solid line) and auxin treated (dashed line) TAF1-AID cells. N=2 independent replicates. Inset shows Western blot for TAF1 in cells without and with auxin (IAA) treatment for 1h. gel source data: Supplementary Figure 1.
Extended Data Fig. 5. P53 target genes and enhancers are not bound by MED1.
a, Locus of the MYC gene with an upstream cluster of endogenously active MED1-bound enhancers. ChIP-seq signal and called MED1 peaks in MED14-AID cells treated with auxin or/and Nutlin-3a and in WT HCT116 cells treated with Nutlin-3a are shown. b, Number of MED1 peaks called in each condition in MED14-AID and WT cells (MACS2, FDR≤0.05). c, Average plot of MED1 ChIP-seq enrichment over input for a common set of MED1 peaks called in MED14-AID (638 peaks; left) and in WT HCT116 cells (1545 peaks; right). d, Example of an endogenously active MED14-dependent enhancer bound by MED1 (left) and a P53-bound MED14-independent enhancer not bound by MED1 (right). MED14-dependent enhancer is bound by MED1 in both WT and MED14-AID cells and this binding is abolished upon auxin treatment, i.e. upon MED14 depletion. P53-bound enhancer shows no MED1 binding in any condition, not even upon P53 induction with Nutlin-3a in either WT or MED14-AID cells. e, MED1 ChIP-seq enrichment over input for 2 groups of STARR-seq enhancers: 1) MED14-independent, P53-bound enhancers (N=586) and 2) endogenously open and H3K27ac-marked MED14-dependent enhancers (N=315), upon Nutlin-3a treatment in control and MED14-depleted MED14-AID cells (left) or in WT cells (right). While MED14-dependent enhancers show some MED1 binding in both WT and MED14-AID cells, which is abolished upon MED14 depletion (i.e. auxin treatment), P53-bound enhancers show no binding in any condition, including after Nutlin-3a treatment when these enhancers are activated. Boxes: median and interquartile range; whiskers: 5th and 95th percentiles. P-values: two-sided Wilcoxon rank-sum test. f, MED1 IF with concurrent RNA FISH against P53 target gene RRM2B (top row) and Mediator-regulated positive control gene MYC (bottom row) in Nutlin-3a treated WT HCT116 cells. Examples of individual cells with merged view of the FISH and MED1 IF signal at the FISH spot are shown on the left. Hoechst staining was used to determine the nuclear periphery, highlighted with a dashed white line. Mean RNA FISH and mean MED1 IF signal in 1x1μm window centred at FISH spots, or at random spots is shown on the right. Number of spots analysed is indicated in the lower right corner (n). g, Distribution of distance between each random spot and the nearest MED1 IF spot for random spots picked in different FISH experiments. Boxes: median and interquartile range; whiskers: 5th and 95th percentiles. P-value: Kruskal-Wallis rank sum test.
Extended Data Fig. 6. P53 target gene induction is independent of multiple Mediator subunits in human and mouse cells.
a-c, Endogenous expression of known P53 target genes as measured by qPCR in auxin or/and Nutlin-3a treated MED15- (a, tail module), MED19- (b, middle module) or MED1-AID (c, middle module) cells. Western blot of the denoted Mediator subunit in the respective COF-AID cell line, without and with auxin (IAA) treatment for 3h is shown on top. gel source data: Supplementary Figure 1. d, Endogenous expression of known P53 target genes as measured by qPCR upon Nutlin-3a treatment before and after MED17 (head module) knock-down via RNAi in WT HCT116 cells. e, Endogenous expression of P53 target genes as measured by qPCR in DMSO or Nutlin treated mouse CH12 cells, either wild-type (WT) or knock-out (KO) cell lines for different Mediator subunits (cell lines from ref. 18). Experiment was performed in two batches (shown in two rows), each time using a re-thawed WT cell line as a control. Tailless = quintuple knock-out for MED15, MED16, MED23, MED24 and MED25 subunits. In all panels, N=3 independent replicates; fold-change for each replicate calculated independently by dividing the treatment value with the corresponding control value; mean +/- SD shown; P-values: two-sided Student’s t-test.
Extended Data Fig. 7. LTR12 family repeats act as BRD4 independent enhancers/promoters that contain a combination of TATA-box and multiple CCAAT-box motifs.
a, Enrichment of retrotransposons in enhancers up- vs. down-regulated upon BRD4 depletion. b, Differential analysis of enhancer activity upon BRD4 depletion with LTR12 family repeat-overlapping enhancers marked in yellow. c, Fold-change of endogenous LTR12 expression as measured by qPCR in auxin treated vs. untreated BRD4-AID K562 (left) and A549 (right) cells. In both cell lines BRD4 depletion leads to upregulation of LTR12C and D. d, Multiple alignment of LTR12 family repeats with detected enhancer activity in STARR-seq. Occurrences of CCAAT-box and TATA-box motifs, and the endogenous transcription initiation previously mapped by CAGE are marked below the alignment. e, Enrichment analysis of 579 non-redundant TF motifs from the JASPAR vertebrate core collection (https://jaspar2020.genereg.net/download/data/2020/CORE/JASPAR2020_CORE_non-redundant_pfms_jaspar.zip) between upregulated and down-regulated enhancers upon BRD4 depletion in HCT116 cells. Significantly enriched and depleted motifs (two-sided Fisher’s exact test; P-value ≤0.05) are shown in red and blue, respectively. Logo of the most highly enriched CCAAT-box motif bound by NFYA/B is shown. f, Endogenous expression of NFYA and NFYB as measured by qPCR without or with NFYA & NFYB siRNA treatment in BRD4-AID HCT116 cells. g, Western blots of NFYA (left) and NFYB (right) with and without treatment with the respective siRNA. gel source data: Supplementary Figure 1. h, Endogenous expression of LTR12C and D as measured by qPCR in auxin or/and NFYA & NFYB siRNA treated BRD4-AID HCT116 cells. i, Endogenous expression of NFYA and NFYB as measured by qPCR without or with NFYA & NFYB siRNA treatment in BRD4-AID A549 cells. j, Endogenous expression of LTR12C and D as measured by qPCR in auxin or/and NFYA & NFYB siRNA treated BRD4-AID A549 cells. k, Growth curves over a course of 4 days comparing untreated (solid line) and auxin treated (dashed line) BRD4-AID and Parental A549 cells. N=3 independent replicates. In c, f, h, i and j, mean +/- SD shown; P-values: two-sided Student’s t-test. N=3 (c, i and j) or N=6 (f and h) independent replicates; fold-change for each replicate calculated independently by dividing the treatment value with the corresponding control value.
Extended Data Fig. 8. Histone genes have a promoter with TATA-box and CCAAT-box motifs and do not require BRD4 for productive transcription.
a, Gene ontology term enrichment for genes with promoters containing both TATA-box and CCAAT-box motifs. Top 5 terms for cellular compartment (top), molecular function (middle) and biological process (bottom) categories are shown. Bars show fold-enrichment and are colored according to the P-value of the one-sided hypergeometric test. b, Occurrence of TATA- and CCAAT-boxes in histone genes promoters relative to TSSs. c, Loci of the histone genes HIST1H2BJ and HIST1H2AG (left) and ribosomal protein gene RPS9 (right) with nascent transcription (normalized PRO-seq signal for merged replicates) in BRD4- and MED14-AID cells with and without auxin treatment. While RPS9 shows typical pause release defect with loss of RNA polymerase II signal throughout the gene body and increase at the promoter, the two histone genes do not lose signal in the gene body and still have high levels of actively elongating RNA polymerase II. d, Log2 fold-change of endogenous nascent transcription for histone genes from previously published datasets. Left: SLAM-seq in different cell lines upon rapid BRD4 degradation via AID system or BRD4 inhibition by JQ1 (from ref. 33); Right: NET-seq in MOLT4 cell line upon BRD4 inhibition by JQ1 or dBET6 (from ref. 32). e, STARR-seq signal enrichment over input in BRD4-AID cell line separated by strand for enhancers overlapping TATA-box promoters (N=190), distal enhancers not overlapping promoters (N=4917) and random inactive regions (negative control; N=5151). Sense strand corresponds to orientation of the gene for enhancers overlapping promoters and is randomly assigned for distal enhancers and random regions. In d and e, boxes: median and interquartile range; whiskers: 5th and 95th percentiles. f, Examples of STARR-seq enhancers overlapping TATA-box promoters with evidence of endogenous initiation (CAGE): promoter of the MMP13 gene (left) and an instance of LTR12 repeat element (right). STARR-seq signal in BRD4-AID cell line and input library coverage is shown for + and − strands separately. Fragments from both strands are enriched over input, i.e. these promoter-overlapping fragments work as enhancers in both orientations.
Extended Data Fig. 9. Combination of a TATA-box core promoter and CCAAT-box-containing proximal enhancer is required and sufficient to drive high levels of BRD4 independent transcription.
a, Design of a sequence library to assess the requirement and sufficiency of the TATA-box and CCAAT-box motifs in the core and proximal promoter region, respectively, for the BRD4-independent transcriptional activity with massive parallel reporter assay. For the loss of function approach (left) 10 different BRD4-independent promoters (from LTR12 repeats and histone genes) were selected and variants with either TATA- and/or CCAAT-box motifs mutated were designed. For the gain of function approach (right) the TATA- and/or CCAAT-box motifs from the 10 selected promoters were inserted into 18 randomly picked neutral sequences. Each sequence variant is present in the library 5 times, coupled to a different 10bp long barcode at the 3’ end. b, Schematic of the massive parallel reporter assay (STAP-seq) to measure transcriptional activity at a single base-pair resolution in BRD4-AID cells without or with auxin (IAA) treatment. 5’ ends of transcripts arising from each sequence present in the library are captured, amplified and sequenced, and the sequenced tags are uniquely mapped to the sequence variant of origin via the 10bp identification barcode. Correlation between transcriptional activity across all sequences in the library measured in two independent replicates for auxin treated (right) and untreated (left) cells is shown at the bottom. c, Transcriptional activity at single base-pair resolution measured by STAP-seq for wild-type (WT) and different mutant versions of the LTR12 promoter instance. Transcription from each sequence variant was assessed 5 times in the library (coupled to 5 different barcodes) and the mean normalized STAP-seq signal across different barcodes is shown for the 2 independent replicates. STAP-seq signal in auxin treated (red) vs. untreated (blue) BRD4-AID cells is shown as semi-transparent overlay. d, Transcriptional activity at single base-pair resolution measured by STAP-seq for a random neutral sequence upon insertion of TATA- and CCAAT-box motifs from an LTR12C, an LTR12D instance or from the HIST1H2AJ promoter. e-f, Endogenous expression of known heat-shock responsive genes as measured by qPCR in auxin or/and heat-shock treated BRD4-AID HCT116 (left), K549 (middle) and A549 (right) cells (e), and CDK9-AID HCT116 cells (f). In all three BRD4-AID cell lines heat-shock genes are equally strongly induced with BRD4 present or depleted but fail to get induced with CDK9 depleted. g, Endogenous expression of AFF1, AFF4 and known heat-shock responsive genes as measured by qPCR without or with AFF1 & AFF4 siRNA treatment in HCT116 cells. The induction of heat-shock genes is decreased after AFF1 & AFF4 knock-down. In e-g, N=3 independent replicates; fold-change for each replicate calculated independently by dividing the treatment value with the corresponding control value; mean +/- SD shown; P-values: two-sided Student’s t-test. h, i, Changes in gene expression (log2 fold-change in PRO-seq signal) upon BRD4 (h) or MED14 (i) depletion for two groups of genes: (1) genes that have an enhancer insensitive to respective COF depletion (Group 4 enhancer for BRD4 or Group 3 enhancer for MED14) and (2) genes that have an enhancer downregulated upon respective COF depletion within 50 kb of their TSS. Number of genes in each group (N) is denoted in parentheses. Boxes: median and interquartile range; whiskers: 5th and 95th percentiles; P-values: one-sided Wilcoxon rank-sum test. Barplots show percentage of genes in each group that are unaffected (not significantly downregulated) by COF depletion in PRO-seq. P-values: one-sided Fisher’s exact test.
Extended Data Fig. 10. STARR-seq for additional AID-tagged cofactors shows no effect on enhancer activity.
a, Growth curves over a course of four days comparing untreated (solid line) and auxin treated (dashed line) cells, for BRD7- (left), BRD9- (middle) and MLL1-AID (right) cell line. N=2 independent replicates. Insets show Western blot for the respective cofactor in cells without and with auxin (IAA) treatment for 3h. Upon auxin treatment none of the cofactors were detectable either in Western blot or in mass spectrometry. b, Examples of four enhancers detected by STARR-seq in the BAC library. For each enhancer the activity in BRD7-, BRD9- and MLL1-AID cell lines in the BAC-STARR-seq screen with and without IAA treatment is shown (normalized STARR-seq signal for merged replicates), alongside with endogenous chromatin accessibility and histone modifications in wild-type HCT116 cells. For comparison, enhancer activity in different COF-AID cell lines from the genome-wide STARR-seq screen is shown. None of the enhancers are affected by the loss of neither BRD7, BRD9 nor MLL1, while they are sensitive to depletion of other COFs (e.g. BRD4, MED14 or CDK9). c, Differential analysis of STARR-seq enhancer activity for 114 enhancers detected in the BAC library in each COF-AID cell line with and without auxin treatment to assess the effect of COF degradation on enhancer activity. Number of significantly up- or down-regulated enhancers is denoted (FDR≤0.05). Depletion of none of the three COFs has an effect on enhancer activity, suggesting that they are not required for enhancer activity in the unperturbed HCT116 cells.
Supplementary Material
a-n, Source images of Western blots shown in Fig. 1c (a-g), ED Fig. 1a (a-g), ED Fig. 5l (h), ED Fig. 7a-d (i-l) and ED Fig. 11a (m-n), detecting BRD2 (a), BRD4 (b), P300 (c), CBP (d), MED14 (e), CDK7 (f), CDK9 (f), CDK8 (g), TAF1 (h), MED15 (i), MED19 (j), MED1 (k), MED17 (l), BRD7 (m), MLL1 (m) or BRD9 (n) in the cell line where the respective cofactor is tagged by an AID tag, comparing control (-IAA) and auxin (+IAA) treatment for 1h (a-k,m-n), or in Parental HCT116 cell line comparing control and MED17 siRNA treatment for 24h (l). o-p, Source images of Western blots shown in ED Fig. 8f detecting NFYA (o) or NFYB (p) in BRD4-AID tagged cells comparing control and combined NFYA and NFYB siRNA treatment for 24h. In each panel, top image always shows immunoblot with antibody against the V5 Tag or the endogenous protein (denoted in the top right corner). Bottom image always shows Tubulin, which was blotted from same gel and serves as a loading control. Regions cropped for presentation in final figures are boxed in red.
List of sequences of gRNAs to establish HCT116 parental cell line; List of sequences of gRNAs to target individual COFs within the parental cell line; Table of mass spectrometry peptide sequences utilized to measure abundance of individual COFs; List of primary and secondary antibodies used to measure COF-AID degradation, to assess siRNA knockdown efficiency and to perform ChIP and IF experiments; List of utilized qPCR primers; List of custom designed intronic RNA FISH probes.
Summary of total sequenced reads, mapped reads and spike-in reads for genome-wide and BAC STARR-seq screens; Individual spike-in counts and calculated normalization factor used to scale each COF-AID STARR-seq screen; List of selected STARR-seq spike-in sequences (mouse enhancers and human enhancers with D. melanogaster flanking sequence) used for normalizing STARR-seq counts Number of called peaks with MACS2 for each COF/condition from merged replicates.
Raw counts for a referent set of 6,249 enhancers in all STARR-seq experiments; Table of log2FC values between treatment and control in each COF-AID STARR-seq experiment for a referent set of enhancers; Table of adjusted P-values (FDR) from the differential analysis between treatment and control in each COF-AID STARR-seq experiment for a referent set of enhancers.
Table of used, previously published STARR-seq input libraries (genome-wide and BAC); List of all previously published datasets analyzed in this study, with respective references and GEO or ENCODE database accessions.
Summary of total sequenced and mapped reads for PRO-seq experiments in MED14-AID, BRD4-AID and WT cells; MED1 ChIP-seq in MED14-AID and WT cells; and STAP-seq in BRD4-AID cells. Where applicable, statistics of spike-in reads and derived normalization factors are provided.
Raw PRO-seq counts in promoter and gene body regions of 21,116 analyzed genes for MED14-AID, BRD4-AID and WT HCT116 cell lines with different treatments (auxin and/or Nutlin-3a).
Raw STAP-seq counts per position for 2,000 promoter candidate sequences each 250bp long, including wild-type CCAAT- and TATA-box containing promoters, their mutated versions and insertions of these motifs into random neutral sequences.
Reporting summary.
Further information on research design is available in the Nature Research Reporting Summary linked to this paper.
Acknowledgements
We thank Felix Muerdter, Matthias Muhar, Johannes Zuber and Ursula Schoeberl (all IMP) for advice and help establishing the AID system and PRO-seq, respectively, Richard Imre (IMP/IMBA) for help with analyzing MS data, Denes Hnisz (MPI Berlin) for help with Mediator ChIP-seq, Rafael C. Casellas, Jens Kalchschmidt and Seol Kyoung Jung (NIH NIAMS) for sharing the mouse Mediator KO cell lines and data, Dylan Taatjes (U. of Colorado), Carrie Bernecky (IST) and Richard Young (MIT) for discussions, Ben Sabari (UTSW) for feedback and help, and Christa Buecker and Henry Thomas (MPL), Clemens Plaschka (IMP) and A. Andersen (Life Science Editors) for comments on the manuscript. We thank the IMP/IMBA in-house FACS facility, in particular Gerald Schmauss, and Molecular Biology Service. Deep sequencing was performed at the Vienna Biocenter Core Facilities GmbH. V.H. is supported by the Human Frontier Science Program (grant no. LT000324/2016-L) and A.B. by the Swedish Research Council Postdoctoral Fellowship (VR 2017-00372). Research in the Stark group is supported by the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (grant agreement no. 647320) and by the Austrian Science Fund (FWF, F4303-B09). Basic research at the IMP is supported by Boehringer Ingelheim GmbH and the Austrian Research Promotion Agency (FFG). For the purpose of Open Access, the author has applied a CC BY public copyright licence to any Author Accepted Manuscript (AAM) version arising from this submission.
Footnotes
Author contributions
C.N. and A.S. conceived the project. C.N, K.B., O.H and L.S. generated CRISPR/Cas9 edited cell lines. M.P., M.R. and K.B. cultured cells and performed transfections. C.N., K.K. and C.D.A performed STARR-seq, C.N., C.D.A, L.S., O.H. and M.P. performed PRO-seq and C.D.A. performed STAP-seq. A.B. and C.H.L. performed MED1 ChIP-seq and A.B and J.E.H. performed and analyzed MED1 IF with RNA FISH. K.S. performed Mass spectrometry experiments under supervision of K.M.. C.N., L.S., C.D.A., M.P. and O.H. performed qPCR experiments. V.H. and G.L. performed the computational analyses. C.N., V.H. and A.S. interpreted the data and wrote the manuscript. A.S. supervised the project.
Competing interests
The authors declare no competing interests.
Additional information
Reprints and permissions information is available at www.nature.com/reprints.
Data availability
All raw deep sequencing data (STARR-seq, PRO-seq, ChIP-seq and STAP-seq) and associated processed data generated in this study have been deposited in the NCBI Gene Expression Omnibus (GEO) database under accession number GSE156741.
Previously published datasets re-analyzed in this study are available in the GEO repository under the following accession numbers: GSE100432 (genome-wide STARR-seq input library), GSE97889 (ATAC-seq), GSE71510 (H3K4me1, H3K4me3, H3K27ac, SMARCC1 and SMARCA4 ChIP-seq), GSE51176 (P300 and MLL4 ChIP-seq), GSE57628 (BRD4 ChIP-seq), GSE38258 (CDK8 ChIP-seq) and GSE86164 (P53 ChIP-seq). Peak files for the following ChIP-seq datasets are available from ENCODE (https://www.encodeproject.org/): DNase-seq (ENCFF001SQU, ENCFF001WIJ, ENCFF001WIK, ENCFF175RBN, ENCFF228YKV, ENCFF851NWR, ENCFF927AHJ, ENCFF945KJN, ENCFF360XGA), H3K36me3 (ENCFF467KXG, ENCFF742ZBG, ENCFF922EIA), H3K27me3 (ENCFF237TTT, ENCFF991HKN, ENCFF029ZPV), H3K9me2 (ENCFF586SOS, ENCFF808XMV, ENCFF346SOF), H3K9me3 (ENCFF751VFZ, ENCFF577FKU, ENCFF909UTX), JUND (ENCFF001UDY, ENCFF001UDZ, ENCFF950JTT, ENCFF088WYS) and FOSL1 (ENCFF001UDW, ENCFF001UDX).
Vertebrate transcription factor motifs collection is available from the JASPAR database (https://jaspar2020.genereg.net/download/data/2020/CORE/JASPAR2020_CORE_non-redundant_pfms_jaspar.zip). SwissProt-human database is available at: https://www.uniprot.org/proteomes/UP000005640. No restrictions on data availability apply.
Code availability
All custom code used for data processing and computational analyses is available from the authors upon request.
References
- 1.Reiter F, Wienerroither S, Stark A. Combinatorial function of transcription factors and cofactors. Current Opinion in Genetics & Development. 2017;43:73–81. doi: 10.1016/j.gde.2016.12.007. [DOI] [PubMed] [Google Scholar]
- 2.Nakagawa T, Yoneda M, Higashi M, Ohkuma Y, Ito T. Enhancer function regulated by combinations of transcription factors and cofactors. Genes Cells. 2018;23:808–821. doi: 10.1111/gtc.12634. [DOI] [PubMed] [Google Scholar]
- 3.Rathert P, et al. Transcriptional plasticity promotes primary and acquired resistance to BET inhibition. Nature. 2015;525:543–547. doi: 10.1038/nature14898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jaeger MG, et al. Selective Mediator dependence of cell-type-specifying transcription. Nat Genet. 2020;52:719–727. doi: 10.1038/s41588-020-0635-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chipumuro E, et al. CDK7 Inhibition Suppresses Super-Enhancer-Linked Oncogenic Transcription in MYCN-Driven Cancer. Cell. 2014;159:1126–1139. doi: 10.1016/j.cell.2014.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Muerdter F, et al. Resolving systematic errors in widely used enhancer activity assays in human cells. Nat Methods. 2018;15:141–149. doi: 10.1038/nmeth.4534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Allen BL, Taatjes DJ. The Mediator complex: a central integrator of transcription. Nat Rev Mol Cell Biol. 2015;16:155–166. doi: 10.1038/nrm3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Adelman K, Lis JT. Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nat Rev Genet. 2012;13:720–731. doi: 10.1038/nrg3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vo N, Goodman RH. CREB-binding protein and p300 in transcriptional regulation. J Biol Chem. 2001;276:13505–13508. doi: 10.1074/jbc.R000025200. [DOI] [PubMed] [Google Scholar]
- 10.Gressel S, et al. CDK9-dependent RNA polymerase II pausing controls transcription initiation. eLife. 2017;6:R106. doi: 10.7554/eLife.29736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Visel A, et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;457:854–858. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hnisz D, et al. Super-Enhancers in the Control of Cell Identity and Disease. Cell. 2013;155:934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heintzman ND, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 14.Krebs AR, Karmodiya K, Lindahl-Allen M, Struhl K, Tora L. SAGA and ATAC Histone Acetyl Transferase Complexes Regulate Distinct Sets of Genes and ATAC Defines a Class of p300-Independent Enhancers. Mol Cell. 2011;44:410–423. doi: 10.1016/j.molcel.2011.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zuber J, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478:524–528. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Filippakopoulos P, et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pelish HE, et al. Mediator kinase inhibition further activates super-enhancer-associated genes in AML. Nature. 2015;526:273–276. doi: 10.1038/nature14904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.El Khattabi L, et al. A Pliable Mediator Acts as a Functional Rather Than an Architectural Bridge between Promoters and Enhancers. Cell. 2019;178:1145–1158.:e20. doi: 10.1016/j.cell.2019.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nishimura K, Fukagawa T, Takisawa H, Kakimoto T, Kanemaki M. An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat Methods. 2009;6:917–922. doi: 10.1038/nmeth.1401. [DOI] [PubMed] [Google Scholar]
- 20.Watanabe Y, et al. Frequent Alteration of MLL3 Frameshift Mutations in Microsatellite Deficient Colorectal Cancer. PLoS ONE. 2011;6:e23320. doi: 10.1371/journal.pone.0023320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cevher MA, et al. Reconstitution of active human core Mediator complex reveals a critical role of the MED14 subunit. Nat Struct Mol Biol. 2014;21:1028–1034. doi: 10.1038/nsmb.2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liang J, et al. CDK8 Selectively Promotes the Growth of Colon Cancer Metastases in the Liver by Regulating Gene Expression of TIMP3 and Matrix Metalloproteinases. Cancer Research. 2018;78:6594–6606. doi: 10.1158/0008-5472.CAN-18-1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu D, et al. The MLL3/MLL4 Branches of the COMPASS Family Function as Major Histone H3K4 Monomethylases at Enhancers. Mol Cell Biol. 2013;33:4745–4754. doi: 10.1128/MCB.01181-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fan X, Chou DM, Struhl K. Activator-specific recruitment of Mediator in vivo. Nat Struct Mol Biol. 2006;13:117–120. doi: 10.1038/nsmb1049. [DOI] [PubMed] [Google Scholar]
- 25.Meyer KD, Lin S-C, Bernecky C, Gao Y, Taatjes DJ. p53 activates transcription by directing structural shifts in Mediator. Nat Struct Mol Biol. 2010;17:753–760. doi: 10.1038/nsmb.1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ito M, et al. Identity between TRAP and SMCC Complexes Indicates Novel Pathways for the Function of Nuclear Receptors and Diverse Mammalian Activators. Mol Cell. 1999;3:361–370. doi: 10.1016/s1097-2765(00)80463-3. [DOI] [PubMed] [Google Scholar]
- 27.Andrysik Z, et al. Identification of a core TP53 transcriptional program with highly distributed tumor suppressive activity. Genome Res. 2017;27:1645–1657. doi: 10.1101/gr.220533.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Drané P, Barel M, Balbo M, Frade R. Identification of RB18A, a 205 kDa new p53 regulatory protein which shares antigenic and functional properties with p53. Oncogene. 1997;15:3013–3024. doi: 10.1038/sj.onc.1201492. [DOI] [PubMed] [Google Scholar]
- 29.Brocks D, et al. DNMT and HDAC inhibitors induce cryptic transcription start sites encoded in long terminal repeats. Nat Genet. 2017;49:1052–1060. doi: 10.1038/ng.3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krönung SK, et al. LTR12 promoter activation in a broad range of human tumor cells by HDAC inhibition. Oncotarget. 2016;7:33484–33497. doi: 10.18632/oncotarget.9255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oldfield AJ, et al. NF-Y controls fidelity of transcription initiation at gene promoters through maintenance of the nucleosome-depleted region. Nat Commun. 2019;10:411–12. doi: 10.1038/s41467-019-10905-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Winter GE, et al. BET Bromodomain Proteins Function as Master Transcription Elongation Factors Independent of CDK9 Recruitment. Mol Cell. 2017;67:5–18.:e19. doi: 10.1016/j.molcel.2017.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Muhar M, et al. SLAM-seq defines direct gene-regulatory functions of the BRD4-MYC axis. Science. 2018;360:800–805. doi: 10.1126/science.aao2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arnold CD, et al. Genome-wide assessment of sequence-intrinsic enhancer responsiveness at single-base-pair resolution. Nat Biotechnol. 2017;35:136–144. doi: 10.1038/nbt.3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lis J. Promoter-associated Pausing in Promoter Architecture and Postinitiation Transcriptional Regulation. Cold Spring Harb Symp Quant Biol. 1998;63:347–356. doi: 10.1101/sqb.1998.63.347. [DOI] [PubMed] [Google Scholar]
- 36.Zheng B, et al. Acute perturbation strategies in interrogating RNA polymerase II elongation factor function in gene expression. Genes Dev. 2021;35:273–285. doi: 10.1101/gad.346106.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen FX, Smith ER, Shilatifard A. Born to run: control of transcription elongation by RNA polymerase II. Nat Rev Mol Cell Biol. 2018;19:464–478. doi: 10.1038/s41580-018-0010-5. [DOI] [PubMed] [Google Scholar]
- 38.Galbraith MD, et al. HIF1A Employs CDK8-Mediator to Stimulate RNAPII Elongation in Response to Hypoxia. Cell. 2013;153:1327–1339. doi: 10.1016/j.cell.2013.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kubo N, Hu R, Ye Z, Ren B. MLL3/MLL4 Histone Methyltranferase Activity Dependent Chromatin Organization at Enhancers during Embryonic Stem Cell Differentiation. bioRxiv. 2021:2021.03.17.435905-43. doi: 10.1101/2021.03.17.435905. [DOI] [Google Scholar]
- 40.Kang JS, et al. The Structural and Functional Organization of the Yeast Mediator Complex*. J Biol Chem. 2001;276:42003–42010. doi: 10.1074/jbc.M105961200. [DOI] [PubMed] [Google Scholar]
- 41.Rengachari S, Schilbach S, Aibara S, Dienemann C, Cramer P. Structure of the human Mediator–RNA polymerase II pre-initiation complex. Nature. 2021;594:129–133. doi: 10.1038/s41586-021-03555-7. [DOI] [PubMed] [Google Scholar]
- 42.Lee D, Kim S, Lis JT. Different upstream transcriptional activators have distinct coactivator requirements. Gene Dev. 1999;13:2934–2939. doi: 10.1101/gad.13.22.2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Petrenko N, Jin Y, Wong KH, Struhl K. Evidence that Mediator is essential for Pol II transcription, but is not a required component of the preinitiation complex in vivo. eLife. 2017;6:155. doi: 10.7554/eLife.28447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Anandhakumar J, Moustafa YW, Chowdhary S, Kainth AS, Gross DS. Evidence for Multiple Mediator Complexes in Yeast Independently Recruited by Activated Heat Shock Factor. Mol Cell Biol. 2016;36:1943–1960. doi: 10.1128/MCB.00005-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cho W-K, et al. RNA Polymerase II cluster dynamics predict mRNA output in living cells. eLife. 2016;5:1123. doi: 10.7554/eLife.13617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hochheimer A, Zhou S, Zheng S, Holmes MC, Tjian R. TRF2 associates with DREF and directs promoter-selective gene expression in Drosophila. Nature. 2002;420:439–445. doi: 10.1038/nature01167. [DOI] [PubMed] [Google Scholar]
- 47.Lin C, et al. AFF4, a Component of the ELL/P-TEFb Elongation Complex and a Shared Subunit of MLL Chimeras, Can Link Transcription Elongation to Leukemia. Mol Cell. 2010;37:429–437. doi: 10.1016/j.molcel.2010.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lin C, et al. Dynamic transcriptional events in embryonic stem cells mediated by the super elongation complex (SEC) Genes Dev. 2011;25:1486–1498. doi: 10.1101/gad.2059211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bugai A, et al. P-TEFb Activation by RBM7 Shapes a Pro-survival Transcriptional Response to Genotoxic Stress. Mol Cell. 2019;74:254–267.:e10. doi: 10.1016/j.molcel.2019.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lis JT, Mason P, Peng J, Price DH, Werner J. P-TEFb kinase recruitment and function at heat shock loci. Genes Dev. 2000;14:792–803. [PMC free article] [PubMed] [Google Scholar]
- 51.Haberle V, et al. Transcriptional cofactors display specificity for distinct types of core promoters. Nature. 2019;570:122–126. doi: 10.1038/s41586-019-1210-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sakuma T, Nakade S, Sakane Y, Suzuki K-IT, Yamamoto T. MMEJ-assisted gene knock-in using TALENs and CRISPR-Cas9 with the PITCh systems. Nature Protocols. 2016;11:118–133. doi: 10.1038/nprot.2015.140. [DOI] [PubMed] [Google Scholar]
- 53.Dorfer V, et al. MS Amanda, a Universal Identification Algorithm Optimized for High Accuracy Tandem Mass Spectra. J Proteome Res. 2014;13:3679–3684. doi: 10.1021/pr500202e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Doblmann J, et al. apQuant: Accurate Label-Free Quantification by Quality Filtering. J Proteome Res. 2019;18:535–541. doi: 10.1021/acs.jproteome.8b00113. [DOI] [PubMed] [Google Scholar]
- 55.Smyth GK. Linear Models and Empirical Bayes Methods for Assessing Differential Expression in Microarray Experiments. Statistical Applications in Genetics and Molecular Biology. 2004;3:1–25. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 56.MacLean B, et al. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010;26:966–968. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Neumayr C, Pagani M, Stark A, Arnold CD. STARR-seq and UMI-STARR-seq: Assessing Enhancer Activities for Genome-Wide-, High-, and Low-Complexity Candidate Libraries. Curr Protoc Mol Biol. 2019;128:e105. doi: 10.1002/cpmb.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mahat DB, et al. Base-pair-resolution genome-wide mapping of active RNA polymerases using precision nuclear run-on (PRO-seq) Nature Protocols. 2016;11:1455–1476. doi: 10.1038/nprot.2016.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Boija A, et al. Transcription Factors Activate Genes through the Phase-Separation Capacity of Their Activation Domains. Cell. 2018 doi: 10.1016/j.cell.2018.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Guo YE, et al. Pol II phosphorylation regulates a switch between transcriptional and splicing condensates. Nature. 2019;13:720–6. doi: 10.1038/s41586-019-1464-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fornes O, et al. JASPAR 2020: update of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2020;48:D87–D92. doi: 10.1093/nar/gkz1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang L, Kasif S, Cantor CR, Broude NE. GC/AT-content spikes as genomic punctuation marks. Proceedings of the National Academy of Sciences. 2004;101:16855–16860. doi: 10.1073/pnas.0407821101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gentleman RC, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pelletier G, et al. Competitive recruitment of CBP and Rb-HDAC regulates UBF acetylation and ribosomal transcription. Mol Cell. 2000;6:1059–1066. doi: 10.1016/s1097-2765(00)00104-0. [DOI] [PubMed] [Google Scholar]
- 66.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2009;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kent WJ, et al. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ponnaluri VKC, et al. NicE-seq: high resolution open chromatin profiling. Genome Biol. 2017;18:122–15. doi: 10.1186/s13059-017-1247-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mathur R, et al. ARID1A loss impairs enhancer-mediated gene regulation and drives colon cancer in mice. Nat Genet. 2017;49:296–302. doi: 10.1038/ng.3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Baranello L, et al. RNA Polymerase II Regulates Topoisomerase 1 Activity to Favor Efficient Transcription. Cell. 2016;165:357–371. doi: 10.1016/j.cell.2016.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.The FANTOM Consortium and the RIKEN PMI and CLST (DGT) A promoter-level mammalian expression atlas. Nature. 2014;507:462–470. doi: 10.1038/nature13182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Falcon S, Gentleman R. Using GOstats to test gene lists for GO term association. Bioinformatics. 2007;23:257–258. doi: 10.1093/bioinformatics/btl567. [DOI] [PubMed] [Google Scholar]
- 74.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.The R Development Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; 2013. pp. 1–3079. [Google Scholar]
- 76.Allen MA, et al. Global analysis of p53-regulated transcription identifies its direct targets and unexpected regulatory mechanisms. eLife. 2014;3:R106. doi: 10.7554/eLife.02200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
a-n, Source images of Western blots shown in Fig. 1c (a-g), ED Fig. 1a (a-g), ED Fig. 5l (h), ED Fig. 7a-d (i-l) and ED Fig. 11a (m-n), detecting BRD2 (a), BRD4 (b), P300 (c), CBP (d), MED14 (e), CDK7 (f), CDK9 (f), CDK8 (g), TAF1 (h), MED15 (i), MED19 (j), MED1 (k), MED17 (l), BRD7 (m), MLL1 (m) or BRD9 (n) in the cell line where the respective cofactor is tagged by an AID tag, comparing control (-IAA) and auxin (+IAA) treatment for 1h (a-k,m-n), or in Parental HCT116 cell line comparing control and MED17 siRNA treatment for 24h (l). o-p, Source images of Western blots shown in ED Fig. 8f detecting NFYA (o) or NFYB (p) in BRD4-AID tagged cells comparing control and combined NFYA and NFYB siRNA treatment for 24h. In each panel, top image always shows immunoblot with antibody against the V5 Tag or the endogenous protein (denoted in the top right corner). Bottom image always shows Tubulin, which was blotted from same gel and serves as a loading control. Regions cropped for presentation in final figures are boxed in red.
List of sequences of gRNAs to establish HCT116 parental cell line; List of sequences of gRNAs to target individual COFs within the parental cell line; Table of mass spectrometry peptide sequences utilized to measure abundance of individual COFs; List of primary and secondary antibodies used to measure COF-AID degradation, to assess siRNA knockdown efficiency and to perform ChIP and IF experiments; List of utilized qPCR primers; List of custom designed intronic RNA FISH probes.
Summary of total sequenced reads, mapped reads and spike-in reads for genome-wide and BAC STARR-seq screens; Individual spike-in counts and calculated normalization factor used to scale each COF-AID STARR-seq screen; List of selected STARR-seq spike-in sequences (mouse enhancers and human enhancers with D. melanogaster flanking sequence) used for normalizing STARR-seq counts Number of called peaks with MACS2 for each COF/condition from merged replicates.
Raw counts for a referent set of 6,249 enhancers in all STARR-seq experiments; Table of log2FC values between treatment and control in each COF-AID STARR-seq experiment for a referent set of enhancers; Table of adjusted P-values (FDR) from the differential analysis between treatment and control in each COF-AID STARR-seq experiment for a referent set of enhancers.
Table of used, previously published STARR-seq input libraries (genome-wide and BAC); List of all previously published datasets analyzed in this study, with respective references and GEO or ENCODE database accessions.
Summary of total sequenced and mapped reads for PRO-seq experiments in MED14-AID, BRD4-AID and WT cells; MED1 ChIP-seq in MED14-AID and WT cells; and STAP-seq in BRD4-AID cells. Where applicable, statistics of spike-in reads and derived normalization factors are provided.
Raw PRO-seq counts in promoter and gene body regions of 21,116 analyzed genes for MED14-AID, BRD4-AID and WT HCT116 cell lines with different treatments (auxin and/or Nutlin-3a).
Raw STAP-seq counts per position for 2,000 promoter candidate sequences each 250bp long, including wild-type CCAAT- and TATA-box containing promoters, their mutated versions and insertions of these motifs into random neutral sequences.
Data Availability Statement
All raw deep sequencing data (STARR-seq, PRO-seq, ChIP-seq and STAP-seq) and associated processed data generated in this study have been deposited in the NCBI Gene Expression Omnibus (GEO) database under accession number GSE156741.
Previously published datasets re-analyzed in this study are available in the GEO repository under the following accession numbers: GSE100432 (genome-wide STARR-seq input library), GSE97889 (ATAC-seq), GSE71510 (H3K4me1, H3K4me3, H3K27ac, SMARCC1 and SMARCA4 ChIP-seq), GSE51176 (P300 and MLL4 ChIP-seq), GSE57628 (BRD4 ChIP-seq), GSE38258 (CDK8 ChIP-seq) and GSE86164 (P53 ChIP-seq). Peak files for the following ChIP-seq datasets are available from ENCODE (https://www.encodeproject.org/): DNase-seq (ENCFF001SQU, ENCFF001WIJ, ENCFF001WIK, ENCFF175RBN, ENCFF228YKV, ENCFF851NWR, ENCFF927AHJ, ENCFF945KJN, ENCFF360XGA), H3K36me3 (ENCFF467KXG, ENCFF742ZBG, ENCFF922EIA), H3K27me3 (ENCFF237TTT, ENCFF991HKN, ENCFF029ZPV), H3K9me2 (ENCFF586SOS, ENCFF808XMV, ENCFF346SOF), H3K9me3 (ENCFF751VFZ, ENCFF577FKU, ENCFF909UTX), JUND (ENCFF001UDY, ENCFF001UDZ, ENCFF950JTT, ENCFF088WYS) and FOSL1 (ENCFF001UDW, ENCFF001UDX).
Vertebrate transcription factor motifs collection is available from the JASPAR database (https://jaspar2020.genereg.net/download/data/2020/CORE/JASPAR2020_CORE_non-redundant_pfms_jaspar.zip). SwissProt-human database is available at: https://www.uniprot.org/proteomes/UP000005640. No restrictions on data availability apply.
All custom code used for data processing and computational analyses is available from the authors upon request.