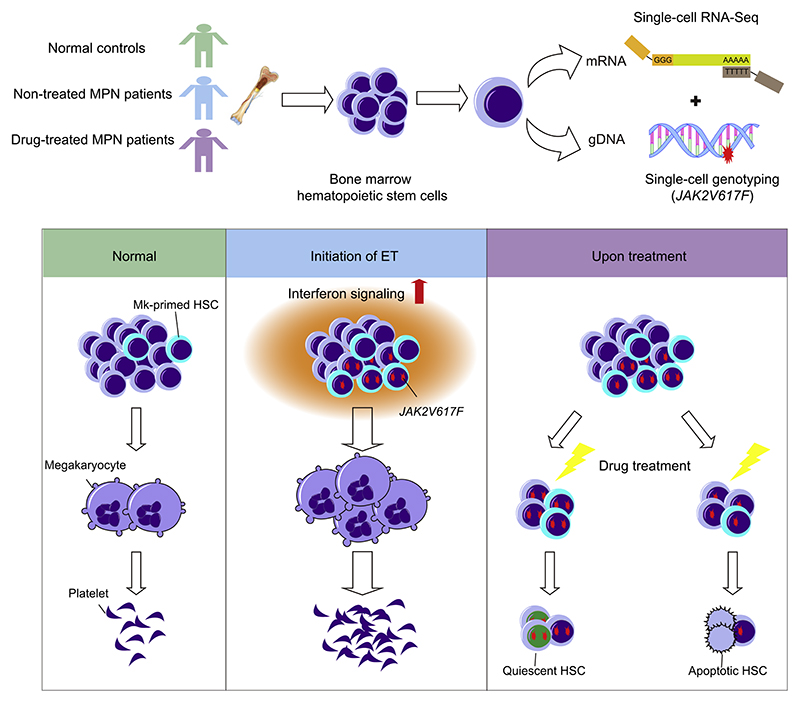
Summary
The implications of stem cell heterogeneity for disease pathogenesis and therapy are poorly defined. JAK2V617F+ myeloproliferative neoplasms (MPNs), harboring the same mutation in hematopoietic stem cells (HSCs), display diverse phenotypes, including polycythemia vera (PV), essential thrombocythemia (ET), and primary myelofibrosis (PMF). These chronic malignant disorders are ideal models to analyze the pathological consequences of stem cell heterogeneity. Single-cell gene expression profiling with parallel mutation detection demonstrated that the megakaryocyte (Mk)-primed HSC subpopulation expanded significantly with enhanced potential in untreated individuals with JAK2V617F+ ET, driven primarily by the JAK2 mutation and elevated interferon signaling. During treatment, mutant HSCs were targeted preferentially in the Mk-primed HSC subpopulation. Interestingly, homozygous mutant HSCs were forced to re-enter quiescence, whereas their heterozygous counterparts underwent apoptosis. This study provides important evidence for the association of stem cell heterogeneity with the pathogenesis and therapeutic response of a malignant disease.
Graphical Abstract.
Introduction
Stem cells in various tissues display considerable heterogeneity (Goodell et al., 2015), but the implications for pathogenesis (e.g., tumorigenesis) are largely unclear. Hematopoietic stem cells (HSCs) are a functionally heterogeneous population with intrinsic lineage biases (Jacobsen and Nerlov, 2019), and it would be instructive to investigate the pathogenic mechanisms involving HSC heterogeneity in hematological diseases.
Classic Philadelphia-negative (Ph−) myeloproliferative neoplasms (MPNs), which are often initiated by somatic mutations in HSCs, are classified into three main entities: polycythemia vera (PV), characterized by erythrocytosis and bone marrow panmyelosis; essential thrombocythemia (ET), characterized by thrombocytosis; and primary myelofibrosis (PMF), characterized by bone marrow myeloid proliferation (Nangalia and Green, 2017). The most recurrent mutation in Ph− MPNs is the Janus kinase 2 (JAK2) mutation JAK2V617F, occurring in more than 95% of individuals with PV and ~50%−60% of individuals with ET and PMF (Baxter et al., 2005; James et al., 2005; Kralovics et al., 2005; Levine et al., 2005). Studies have revealed that the JAK2V617F allele burden (Godfrey et al., 2012; Li et al., 2010, 2014; Scott et al., 2006; Tiedt et al., 2008), differential signal transducer and activator of transcription (STAT) activation (Chen et al., 2010; Grisouard et al., 2015; Walz et al., 2012), order of other somatic mutations (Ortmann et al., 2015), and inter-individual genetic variation (Grinfeld et al., 2018; Pardanani et al., 2008; Tapper et al., 2015) influence predisposition to specific MPN subtypes. Despite these tremendous advances, the mechanism by which identical mutations within stem cells instigate distinct MPN subtypes, especially the association between disease subtypes and HSC heterogeneity, is incompletely understood.
Interferon-alpha (IFNα) therapy can induce clinical, hematological, histopathological, and occasionally molecular remission in individuals with MPNs (Hasselbalch and Holmstrom, 2019; Kiladjian et al., 2016). In MPN mice, IFNα can directly eliminate malignant disease-initiating cells by inducing apoptosis or cell cycle progression (Hasan et al., 2013; Mullally et al., 2013). However, the therapeutic effects of IFNα on the stem cell compartment and heterogeneity in individuals with MPNs are elusive. Here we used single-cell RNA profiling and parallel mutation detection to determine the transcriptional heterogeneity within the HSC compartment of individuals with distinct JAK2V617F+ MPN subtypes with or without therapy.
Results
ET HSCs Exhibit Prominent Megakaryocyte (Mk) Lineage Priming and Elevated IFN Signaling
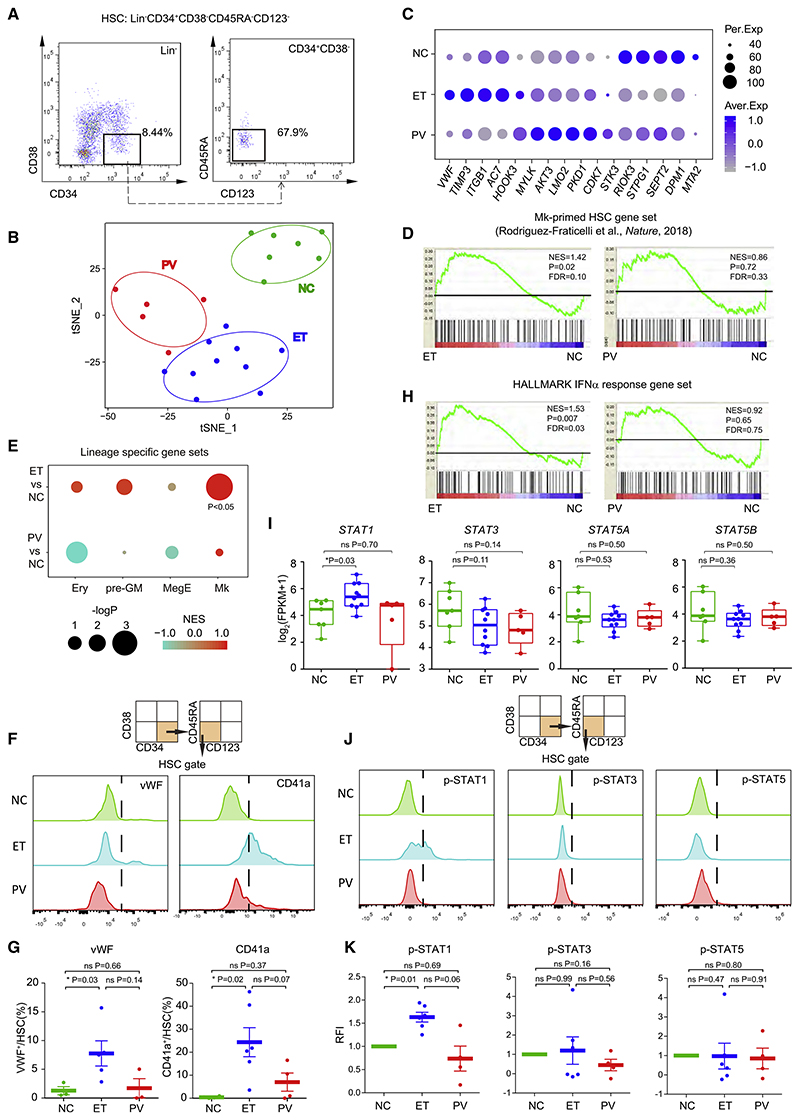
We performed low-input RNA sequencing (RNA-seq) of HSCs (Lin−CD34+CD38−CD45RA−CD123−) from individuals newly diagnosed with JAK2V617F+ ET (n = 10) and PV (n = 5) and normal controls (NCs) (n = 7) (Figure 1A; Figures S1A and S1B; Table S1). T-distributed stochastic neighbor embedding (t- SNE) analysis (Maaten and Hinton, 2008) indicated distinct transcriptomic profiles among MPN subtypes (Figure 1B). Self-organization maps (SOMs) (Wehrens and Buydens, 2007) showed clustering of the specifically expressed genes of distinct samples (Figure S1C) and revealed that von Willebrand factor (VWF), a hallmark of Mk-primed HSCs (Sanjuan-Pla et al., 2013), is elevated in ET HSCs (Figure 1C). Other genes differentially upregulated in vWF+ HSCs (Sanjuan-Pla et al., 2013; Table S2), including SELP and MEF2C, were also expressed at higher levels in ET HSCs than in PV HSCs (Figure S1 E). We thus hypothesized that ET HSCs are Mk lineage primed. Gene set enrichment analysis (GSEA) using gene sets associated with Mk progenitors (MkPs) (Pronk et al., 2007) and steady-state Mk-primed HSCs (Rodriguez-Fraticelli et al., 2018) showed that ET but not PV HSCs exhibited enriched Mk lineage gene expression (Figure 1D; Figures S1D and S1E; Table S2). In contrast, there was no enrichment of other hematopoietic lineages in PV or ET HSCs (e.g., pre-granulocytes and monocytes, Mk and erythroid precursors [Pronk et al., 2007], and erythroid precursors [Rodriguez-Fraticelli et al., 2018]) (Figure 1E).
Figure 1. ET HSCs Exhibit Prominent Mk Lineage Priming and Elevated Interferon (IFN) Signaling.
(A) Representative flow cytometry profiles for HSC isolation from bone marrow mononuclear cells (BM-MNCs) (Table S1).
(B) t-SNE analysis of transcriptomic profiles of HSCs from individuals with JAK2V617F+ ET and PV and normal controls (NCs).
(C) Selected feature genes from ET, PV, and NC HSCs. The percentage of expressing samples (Per.Exp) and average expression values (Aver.Exp) are shown.
(D) GSEA of Mk-primed HSC gene sets in HSCs of affected individuals and NCs (Table S2). Normalized enrichment score (NES), p value, and false discovery rate (FDR) are shown.
(E) GSEA of various lineage-restricted or primed gene sets in pre-granulocytes and monocytes (pre-GMs), Mk and erythroid precursors (MegEs), and erythroid precursors (Erys) (Table S2).
(F) Strategy and representative flow cytometry of vWF+ and CD41a+ HSC subpopulations.
(G) Proportions of vWF+ and CD41a+ HSC subpopulations shown in (F).
(H) GSEA of the HALLMARK IFNα response gene set in HSCs of affected individuals and NCs.
(I) STAT gene expression in ET, PV, and NC HSCs. Log2-transformed (fragments per kilobase million [FPKM]+1) are shown for individuals. Thick horizontal lines indicate median expression levels, and boxes represent the first and third quartiles.
(J) Strategy and representative flow cytometry for measurement of phosphorylated STAT1 (p-STAT1), p-STAT3, and p-STAT5 levels.
(K) Relative fluorescence intensity (RFI) of (J).
In (G) and (K), data represent mean ± standard error of the mean (SEM). The p values in (G), (I), and (K) were determined by Wilcoxon rank-sum test. *p < 0.05; ns, not significant.
To corroborate the uniquely enhanced Mk lineage priming of ET HSCs, we examined vWF+ cells in ET, PV, and NC HSCs by flow cytometry (Figure 1F). The mean proportion of vWF+ HSCs among ET HSCs (7.8%) was higher than among PV (1.7%) and NCs (1.3%) (ET versus NC, p = 0.03) (Figure 1G). Additionally, vWF+ HSCs could give rise to CD41a+ HSCs, resulting in greater restriction to Mk lineage differentiation in the hematopoietic hierarchy (Haas et al., 2015). CD41a+ HSCs also expanded in ET HSCs (25.0%) compared with that in PV (7.0%) and NCs (0.5%) (ET versus NC, p = 0.02) (Figures 1F and 1G).
To elucidate the mechanism of this Mk lineage bias, we evaluated the relevance of potentially critical regulatory pathways (Figure S1F). ET and PV HSCs exhibited signatures of active cell proliferation, with higher expression of cell cycle genes (e.g., cyclin-dependent kinase 1 [CDK1] and CDC45) (Figures S1 F and S1G). Altered expression of genes related to inflammatory signaling pathways and aberrant metabolic processes was also detected (Figure S1F). In particular, IFNα signaling was uniquely enriched in HSCs from individuals newly diagnosed with ET but not PV and NCs (Figure 1H; Figure S1F). Key IFN signaling components (e.g., IFI35, NMI, and STAT1), were highly expressed only in ET HSCs (Figure S1 E; Figure 1l), whereas there were no marked differences in STAT3 and STAT5 expression between PV and ET HSCs (Figure 1I).
Flow cytometry analysis revealed elevation of phosphorylated STAT1 (p-STAT1), but not p-STAT3 or p-STAT5, in ET HSCs compared with NC HSCs (Figures 1J and 1K) despite no apparent changes in the IFN receptors IFNAR1 and IFNGR1 (Figures S1H and S1I). Importantly, the percentage of p-STAT1 in CD41a+ Mk-primed HSCs was generally higher than in CD41a− HSCs of individuals with ET (Figure S1J), implying that the Mk-primed HSC subpopulation accounts for most of the enhanced p-STAT1 signal in ET HSCs. ET HSCs displayed a distinct signature that reflects prominent Mk lineage priming and hyperactivated IFN signaling.
JAK2V617F Expands the Mk-Primed Subpopulation of ET HSCs with Enhanced Potential
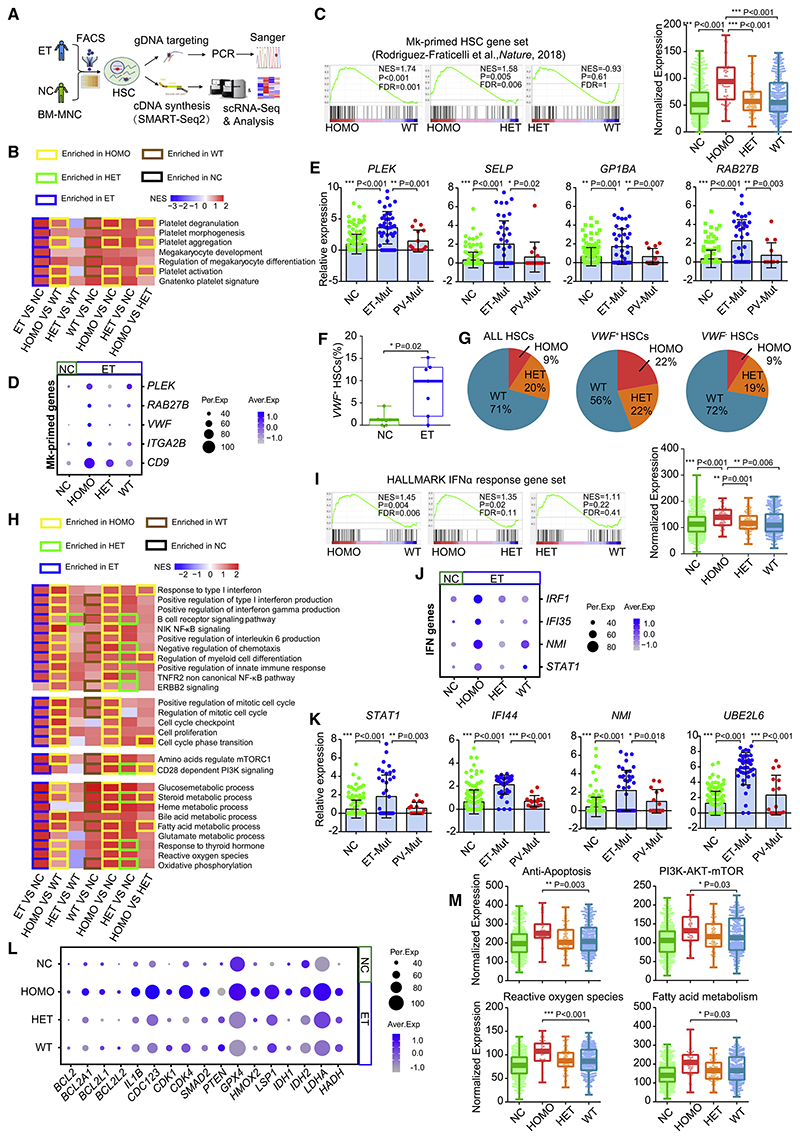
To assess whether mutant HSCs are responsible for elevating Mk priming and IFN response, we conducted modified 3′-TARGET-seq (Figure 2A; Psaila et al., 2020; Rodriguez-Meira et al., 2019), which allows precise identification of mutant cells from wild-type (WT) cells in a single individual. After quality control (Figure S2A), we selected 752 HSCs from seven individuals newly diagnosed with JAK2V617F+ ET and 485 HSCs from seven age-matched NCs (Table S1). In total, 308 HSCs, mainly from six individuals with ET, were categorized as homozygous mutant (HOMO), heterozygous mutant (HET), and non-mutant WT HSCs.
Figure 2. JAK2V617F Expands the Mk-Primed Subpopulation of ET HSCs with Enhanced Potential.
(A) Schematic of the modified 3’-TARGET-seq analysis of single HSCs from individuals with non-treated ET (NT-ET) and age-matched NCs (both n = 7).
(B) Enrichment of Mk- and platelet-related pathways among homozygous mutant (HOMO) ET HSCs, heterozygous mutant (HET) ET HSCs, non mutant WT ET HSCs (WT), and NC HSCs. The heatmap represents the NES for each pairwise comparison; border color indicates the group in which a specific pathway is enriched (p < 0.05).
(C) Enrichment of representative Mk-primed gene sets among distinct ET HSC subtypes by GSEA (left) and expression of the given Mk-primed gene set among distinct ET HSCs subtypes and NC HSCs (right). Dots represent the sum of the SCT transformed expression for the given gene set in single cells.
(D) Representative Mk-primed genes across distinct ET HSCs subtypes and and NC HSCs.
(E) Bee swarm plots of the expression (log2 transformed) of representative Mk-primed genes among JAK2V617F+ HSCs from individuals with ET and PV or NC HSCs, determined by single-cell quantitative real-time PCR; internal control: GAPDH.
(F) VWF Mk-primed HSC frequencies (count > 0) in individuals with ET and NCs.
(G) Proportion of mutant HSCs in the total HSC population and VWF+ and VWF HSC subpopulations in ET.
(H) GSEA of inflammatory pathways, metabolic processes, cell cycle processes, and key signaling pathways among HOMO, HET, and WT HSCs from ET and HSCs from NCs. The heatmap represents the NES for each pairwise comparison; border color indicates the group in which a specific pathway is enriched (p < 0.05).
(I) Enrichment of HALLMARK IFNα response genes among ETHSC subtypes (HOMO, HET, and WT), determined by GSEA (left) and IFNα gene expression among ET HSC subtypes and NC HSCs (right).
(J) Representative IFN gene expression among ET HSC subtypes and NC HSCs.
(K) Bee swarm plots of representative IFN response gene expression (log2 transformed) in JAK2V617F+ HSCs from individuals with ET and PV or NC HSCs, determined by single-cell quantitative real-time PCR; internal control: GAPDH.
(L) Expression of representative genes associated with cell survival (anti-apoptosis; BCL2, BCL2A1, BCL2L1, and BCL2L2), cell cycle progression (CDK4, CDK1, CDC123, and IL1B), and fatty acid metabolism and ROS production (HADH, LDHA, IDH1, IDH2, and LSP?) among distinct ET HSC subtypes and NC HSCs.
(M) Expression of the indicated gene sets among ET HSC subtypes and NC HSCs.
In (E) and (K), data represent mean ± SEM. The p values in (C), (E), (F), (I), (K), and (M) were determined by Wilcoxon rank-sum test. *p < 0.05, **p < 0.01, ***p < 0.001.
GSEA revealed that mutant ET HSCs, especially HOMO ones, were enriched in terms associated with Mk differentiation and platelet function (Figure 2B). The gene sets characterized previously as Mk-primed HSCs (Pronk et al., 2007; Rodriguez-Fraticelli et al., 2018; Sanjuan-Pla et al., 2013) were enriched significantly in HOMO HSCs compared with WT HSCs (Figure 2C; Figure S2B; Table S2), with higher expression of signature genes, including VWF, PLEK, and CD9 (Figure 2D). Single-cell quantitative real-time PCR with parallel mutation detection in ET, PV, and NC HSCs validated that mutant HSCs in ET highly and uniquely expressed Mk-primed genes (e.g., PLEK, SELP, and GP1BA) (Figures S2C and S2D; Figure 2E; Table S3), whereas the expression pattern in PV mutant cells resembled that in NC cells (Figure S2D). Mutant cells are thus the major cellular source and contributor to the elevated Mk lineage priming potential. Intriguingly, compared with age-matched NC HSCs, WT HSCs in individuals with ET also expressed higher levels of Mk-primed genes (Figure 2D). Therefore, the baseline of Mk differentiation potential in ET HSCs was elevated, implying that mutant HSCs in ET alter and instruct a permissive bone marrow microenvironment to favor Mk differentiation and platelet generation.
We also noted expansion of the VWF+ subpopulation of ET HSCs (median 9.9% versus 1.2%, p = 0.02), with a significantly higher proportion of HOMO cells (22%) than in the total HSC population (9%) and VWF− subpopulation (9%) (Figures 2F and 2G). To confirm the increased Mk-primed HSC subpopulation, we selected 50 genes constituting at least two of the previously used Mk-primed gene sets (Figure S2E; Table S2; Pronk et al., 2007; Rodriguez-Fraticelli et al., 2018; Sanjuan-Pla et al., 2013). A cell expressing at least 12 of these genes was deemed to be an Mk-primed HSC (Figure S2E). This analysis consistently revealed a higher proportion of Mk-primed HSCs in ET (median 24.2% versus 5.5%, p = 0.03) with an increased number of HOMO cells in this subpopulation (18% versus 9% of total HSCs) (Figure S2F; Figure 2G). Thus, the JAK2 mutation expanded the Mk-primed HSC subpopulation in ET.
To elucidate the molecular mechanisms underlying the enhanced Mk-priming potential of mutant HSCs and the enlarged Mk-primed HSC subpopulation in ET, we conducted GSEA of mutant cells (Figure 2H; Figure S2G). We discovered that mutant HSCs were the primary contributors to the enhanced inflammatory signaling (Figure 2H), especially IFNα signaling (e.g., IRF1 and IFI35) (Figures 2I and 2J), which was corroborated by single-cell quantitative real-time PCR (Figure 2K; Figure S2D). In addition, the JAK2V617F mutation enhanced pro-survival mechanisms (or anti-apoptosis) with higher expression of BCL2 family members (e.g., BCL2 and BCL2A1) (Figures 2L and 2M). This mutation also conferred a higher proliferative capacity by promoting cell cycle entry via induction of key cell cycle mediators (e.g., CDK4 and CDK1) (Figure 2H and 2L; Figure S2G). Various signaling pathways were altered in mutant cells as well, including phosphatidylinositol 3-kinase (PI3K)-serine/threonine protien kinase B (PKB; also known as AKT)-mechanistic target of rapamycin (mTOR), JAK-STAT, and nuclear factor κB (NF-κB) (Figure 2H; Figure S2G). It thus seems that active entry into the cell cycle can be partly ascribed to PI3K-AKT-mTOR pathway activation (Figures 2H and 2M; Figure S2G), which governs HSC cell cycle progression via the cyclin D-CDK complex (Hao et al., 2016; Pietraset al., 2011). Phosphatase and tensin homolog (PTEN), a dominant-negative regulator of the PI3K-AKT-mTOR pathway (Yilmaz et al., 2006; Zhang et al., 2006), was downregulated in mutant cells (Figure 2L). Thus, the PTEN-PI3K/AKT/mTOR-cyclin D/CDK regulatory axis is implicated in the higher proliferative capacity of mutant cells. Besides active proliferation, the inflammation (e.g., IFN)-induced increase in Mk priming potential could account for activation of the mTOR- and STAT1-dependent pathways (Figures 2H-2K; Figure S2G) implicated previously in megakaryopoiesis (Haas et al., 2015). The abundant expression of enzymes such as IDH1, IDH2, and LDHA (Figure 2L) provided additional evidence that the enhanced proliferation and differentiation of mutant HSCs is linked to active metabolic processes, such as oxidative phosphorylation, reactive oxygen species (ROS) production, and active fatty acid metabolism (Figures 2H, 2L, and 2M; Figure S2G).
Others have shown that intrinsic alterations (e.g., mutations) or extrinsic stimuli (e.g., inflammation) can induce quiescent HSCs to switch to a metabolically active status of oxidative phosphorylation, providing sufficient energy and the basic metabolites required to reshape the HSC transcriptional program essential for proliferation and differentiation (Verovskaya et al., 2019). Thus, it is likely that JAK2V617F and IFN signaling synergize to disrupt metabolism, promote cell cycle progression, and, ultimately, induce differentiation. These changes enhance the Mk priming potential of mutant HSCs and enlarge the Mk-primed HSC subpopulation in ET HSCs.
JAK2V617F+ HSCs in ET Are Hyper-Responsive to Transient, Low-Dose IFN Stimulation
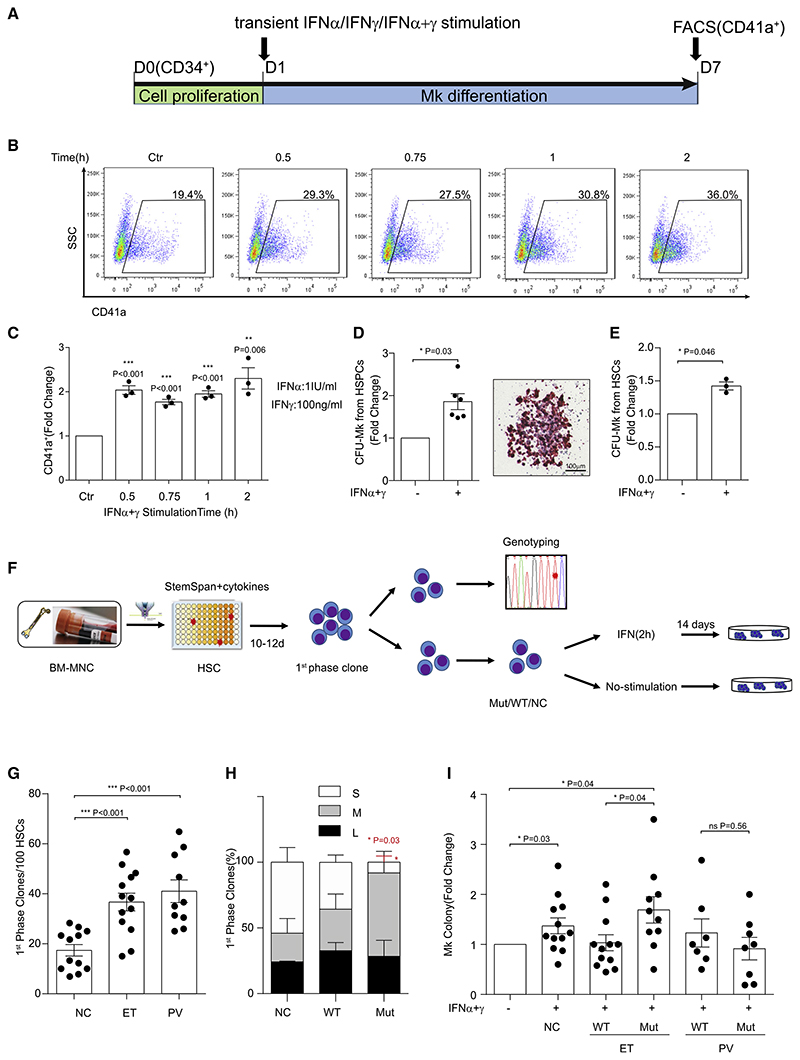
To validate the influence of IFNα and IFNγ signaling on ET, we analyzed the effect of IFN stimulation on Mk differentiation using cord blood CD34+ hematopoietic stem/progenitor cells (HSPCs) (Figure 3A; Figure S3A). Short-term (0.5−2 h) IFNα stimulation (1 IU/mL) promoted CD41a+ Mk differentiation, whereas prolonged stimulation at higher concentrations decreased Mk differentiation (Figure S3A). Although IFNγ alone had almost no effect (Figure S3B), combined IFNα+IFNγ stimulation (0.5−2 h) increased CD41a+ Mk differentiation synergistically (Figures 3B and 3C). Furthermore, greater numbers of Mk colonies were generated following IFNα+IFNγ stimulation for 2 h (p = 0.03) (Figure 3D). Importantly, we also observed this following identical stimulation in purified HSCs (Figure 3E), implying that transient, low-dose IFN stimulation directly enhances Mk lineage differentiation of HSCs. Transient IFNα+IFNγ stimulation also promoted Mk lineage differentiation in adult bone marrow CD34+ cells (Figures S3C−S3G).
Figure 3. JAK2V617F HSCs in ET Are Hyper-Responsive to Transient, Low-Dose IFN Stimulation.
(A) Schematic diagram showing the IFN stimulation strategy during Mk differentiation from human cord blood CD34+ hematopoietic stem and progenitor cells (HSPCs).
(B) Representative flow cytometry profiles of CD41a+ cells on day 7 with IFN (α+γ) stimulation. (C) Fold changes in CD41a+ Mks, quantified as shown in (B). Dots represent an independent experiment. The p values were determined by Student’s t test (n = 3 independent experiments).
(D) Fold changes in colony-forming unit (CFU) Mk numbers generated from cord blood CD34+ HSPCs with or without IFN (α+γ) stimulation for 2 h (left). Dots represents single replicates. The p values were determined by Student’s t test (n = 2 independent experiments). Right panel: a representative Mk colony. Scale bar, 100 μm.
(E) Fold changes in CFU-Mk numbers generated from sorted cord blood-derived HSCs with or without IFN (α+γ) stimulation. Dots represent independent experiments. The p values were determined by Student’s t test (n = 3 independent experiments).
(F) Schematic diagram of IFN stimulation and CFU-Mk assay using single HSCs from BM of individuals with JAK2V617F+ ET and PV and NCs.
(G) Average number of clones generated per 100 HSCs (WT and JAK2 mutant) during the first phase of proliferation. Dots represent single individuals. The p values were determined by Wilcoxon rank-sum test (n = 4 independent experiments).
(H) Percentage of different colony sizes generated from HSCs as shown in (G). L, large colonies (>100 cells); M, medium colonies (20-100 cells); S, small colonies (<20 cells). The p values were determined by Wilcoxon rank-sum test (n = 4 independent experiments).
(I) Influence of IFN stimulation on CFU-Mk generation from JAK2V617F+ and WT HSCs from individuals with ET and PV and NC HSCs. CFU-Mk colony numbers were normalized to the unstimulated control in the same group. Dots represent single individuals. The p values were determined by Wilcoxon rank-sum test (n = 4 independent experiments).
In (C)-(E) and (G)-(I), data represent mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001.
To clarify the influence of IFN on JAK2V617F+ and WT HSCs from individuals with ET, we conducted single-cell colony assays with parallel mutation detection (Figure 3F; Chen et al., 2010). During the first proliferation phase (Figure 3F), ET and PV HSCs exhibited greater proliferative capacity than NC HSCs (Figure 3G), with much larger clones generated from each mutant HSC (Figure 3H). This confirmed that the JAK2V617F mutation conferred a competitive advantage over WT HSCs regarding proliferative capacity. After inducing differentiation, mutant HSCs in ET generated the highest number of Mk colonies in response to IFN stimulation (Figure 3I), suggesting that JAK2V617F sensitized HSCs to Mk lineage differentiation upon IFN stimulation in ET. However, IFN had a negligible or even an adverse effect on mutant PV HSCs toward Mk differentiation (Figure 3I).
HET HSCs Undergo Apoptosis, whereas HOMO Ones Are Forced to Re-enter Quiescence during Treatment
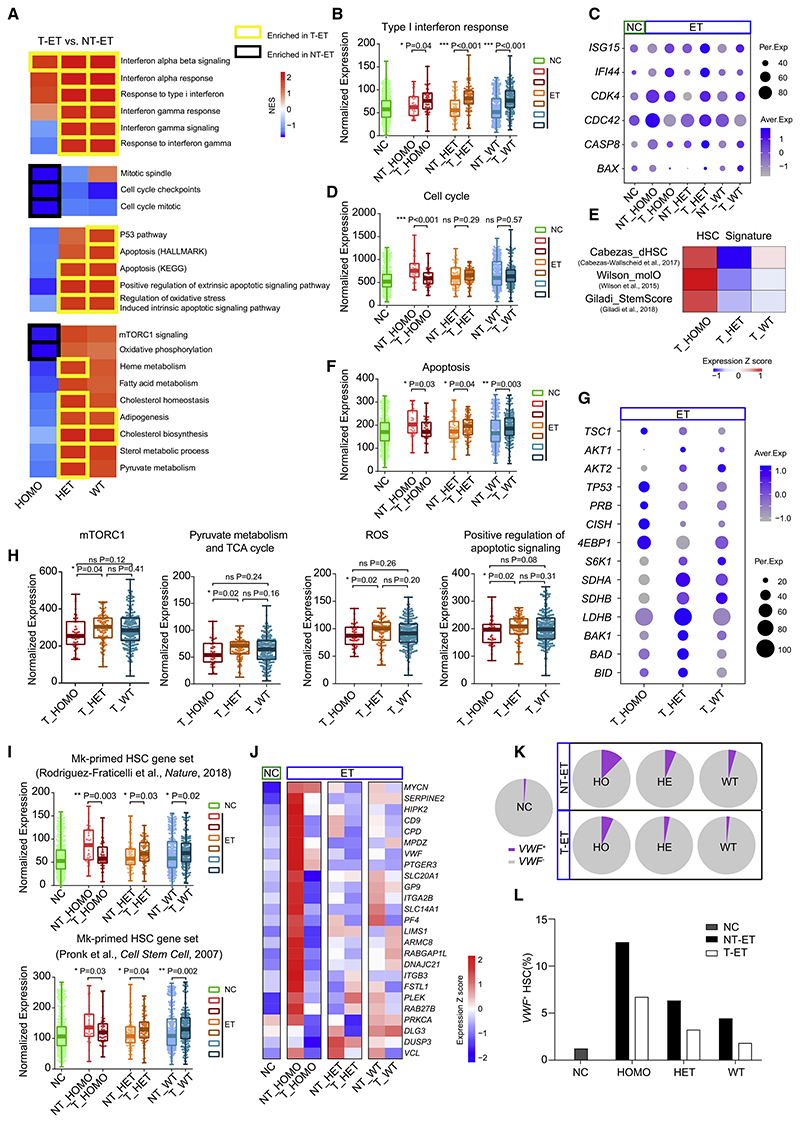
To clarify the mechanism of the therapeutic effects on HSCs in ET, especially JAK2 mutant Mk-primed HSCs, we conducted singlecell TARGET-seq analysis of 359 cells from six individuals with JAK2V617F+ ET treated with IFNα alone or in combination with hydroxyurea (HU) as well as seven age-matched controls (Table S1). Compared with individuals with non-treated ET (NT-ET), IFN signaling was upregulated significantly in all HSCs of individuals with treated ET (T-ET) (Figures 4A and 4B; Figure S4A), with corresponding upregulation of the key IFN effector genes ISG15 and IFI44 (Figure 4C). We also observed distinct behaviors of HOMO and HET HSCs in response to therapy. HOMO cells from treated individuals showed decreased expression of genes related to 96% (38% decreased significantly) of biological processes and signaling pathways compared with those from untreated individuals (Table S2). Considering that most cellular processes were inactive and the low expression of cell cycle-related genes (e.g., CDK4 and CDC42) in HOMO cells (Figures 4C and 4D), we hypothesized that HOMO HSCs re-enter the quiescent state during treatment. Indeed, treated HOMO cells showed the highest stemness score (Cabezas-Wallscheid et al., 2017; Giladi et al., 2018; Wilson et al., 2015; Figure 4E) and significantly decreased expression of the high-output HSC gene set (Figures S4B and S4C), characterized by lower stemness and higher differentiation potential (Rodriguez-Fraticelli et al., 2020). In contrast, HET HSCs after treatment were significantly enriched in terms associated with apoptosis (e.g., upregulation of BAX and CASP8) (Figures 4A, 4C, and 4F).
Figure 4. HET HSCs Undergo Apoptosis, whereas HOMO Ones Are Forced to Re-enter Quiescence during Treatment.
(A) GSEA of the indicated pathways in HOMO, HET, and WT HSCs in individuals with non-treated ET (NT-ET) and treated ET (T-ET) and heatmap of the NES for each pairwise comparison; border color indicates the group in which a specific pathway is enriched (p < 0.05).
(B) Gene Ontology (GO) type I IFN response gene expression among ET HSC subtypes with or without treatment and NC HSCs. Dots represent the sum of the SCT-transformed expression for the given gene set in single cells, which also applies to the following analogous figures.
(C) Expression of selected feature genes for IFNα signaling (ISG15 and IFI44), cell cycle process (CDK4 and CDC42), and apoptosis (CASP8 and BAX).
(D) REACTOME cell cycle gene expression among ET HSC subtypes with or without treatment and NC HSCs.
(E) Heatmap of the mean expression of the indicated gene sets, reflecting the HSC stemness signature among distinct HSC subtypes in ET with therapy (Table S2).
(F) HALLMARK apoptosis gene expression among ET HSC subtypes with or without treatment and NC HSCs.
(G) Dot plots of selected feature gene expression among ETHSC subtypes after treatment, including genes associated with HSC quiescence (TSC1, AKT1, AKT2, TP53, PRB, and CISH), the mTORC1 pathway (4EBP1 and S6K1), the TCA cycle and ROS generation (SDHA, SDHB, and LDHB), and positive regulation of apoptotic signaling (BAK1, BAD, and BID).
(H) Expression of genes associated with the HALLMARK mTORC1 pathway, REACTOME pyruvate metabolism and TCA cycle, HALLMARK ROS, and GO positive regulation of apoptotic signaling across distinct HSC subtypes in ET with therapy.
(I) Expression of different Mk-primed gene sets among ET HSC subtypes with or without treatment and NC HSCs.
(J) Heatmap of Mk-primed gene expression among ET HSC subtypes with or without treatment and NC HSCs.
(K and L) Proportion of VWF+ HSCs in NCs and HOMO, HET, and WT ET HSCs with or without therapy (K) and VWF+ HSC frequencies (L).
The p values in (B), (D), (F), (H), and (I) were determined by Wilcoxon rank-sum test. *p < 0.05, **p < 0.01, ***p < 0.001.
To delineate the molecular mechanisms underlying the induction of quiescence in treated HOMO cells, we analyzed the genes and pathways controlling HSC quiescence. Among the altered genes, upregulated tuberous sclerosis complex 1 (TSC1) (Figure 4G) would restrain the downstream mTOR signaling pathway (Figures 4A and 4H; Figure S4D); its upregulation might be attributed to reduced expression of its inhibitor AKTs (Figure 4G; Inoki et al., 2002). The TSC-mTOR pathway is needed to maintain HSC quiescence by repressing mitochondrial biogenesis and ROS production (Chen et al., 2008). In line with this, we observed low levels of oxidative phosphorylation and tricarboxylic acid (TCA) cycle activity as well as decreased ROS production in HOMO cells from treated individuals (Figures 4A and 4H). Additionally, TP53, a central regulator of HSC quiescence (Liu et al., 2009), was also upregulated in HOMO cells after treatment (Figure 4G). Thus, re-entry of HOMO mutant cells into quiescence could be associated with restoration of the TSC-mTOR pathway and TP53 activation.
Unlike HOMO cells from individuals with ET after treatment, HET cells seemed to be more active in metabolism-related processes (e.g., mTORC1, pyruvate metabolism, and TCA cycle) (Figures 4A and 4H) required for HSC division and differentiation (Ito et al., 2012), resulting in increased ROS production (Figure 4H). Given that the pro-apoptotic effect of IFNs is exerted mainly on active cells (Pietras et al., 2014), we propose that this treatment induced activation and subsequent death of HET cells by activating apoptotic signaling pathways (Figure 4G).
Terms associated with Mk differentiation and platelet function (e.g., coagulation) (Figure S4E) and expression of Mk-primed gene sets (Figures 4I and 4J) were also reduced in HOMO HSCs of individuals with ET after treatment. Conversely, expression of Mk-primed genes was increased in post-treatment HET cells (Figures 4I and 4J), reflecting their more active state. Furthermore, the VWF+ Mk-primed HSC population was also reduced after treatment (Figure S4F), with a marked decline in the HOMO cell subset (Figures 4K and 4L). These findings indicate that the cellular basis of therapeutic efficacy in ET is linked to the reduction of the Mk-primed HSC population.
Discussion
Many cancers, especially myeloid malignancies, are initiated at the stem cell level (Cheng, 2004). To investigate whether and how stem cell heterogeneity is linked to specific disease phenotypes, we found, using single-cell RNA profiling and parallel mutation detection in HSCs of individuals with JAK2V617F+ MPNs, that an enlarged, JAK2-mutant, Mk-primed HSC subpopulation with enhanced potential serves as the cellular basis of the pathogenesis of ET.
JAK2V617F occurs in approximately 0.18% of normal individuals (Genovese et al., 2014; Jaiswal et al., 2014; Xie et al., 2014), indicating that MPN pathogenesis requires additional triggers. Chronic inflammation involving multiple dysregulated inflammatory pathways has been identified in bone marrow of individuals with MPNs (Kleppe et al., 2018; Koschmieder et al., 2016; Sun et al., 2020). However, the specific pathways that synergize with JAK2V617F and their roles in MPN onset have yet to be explored. In this study, we found that IFNα signaling was uniquely upregulated in JAK2V617F+ HSCs of individuals with NT-ET. We also discovered that mutant, Mk-primed HSCs in NT-ET exhibited hyper-responsiveness to transient, low-dose IFN stimulation toward megakaryopoiesis.
Genetic mutations and/or inflammatory stimuli in the bone marrow provoke a metabolic switch in HSCs toward oxidative phosphorylation. The resulting increase in ATP and concomitant ROS generation promotes HSC proliferation and differentiation (Snoeck, 2017; Verovskaya et al., 2019). Among them, enhanced IFN signaling has been reported to promote differentiation of normal, Mk-primed HSCs (Haas et al., 2015) and even hematopoietic progenitors toward the Mk lineage via STAT1 activation (Chen et al., 2010). Our results further indicate that IFN-STAT1 has important effects on mutant, Mk-primed HSCs in individuals with NT-ET. Mechanistically, we found that JAK2 mutation and elevated IFN signaling induces a metabolic switch in Mk-primed HSCs toward more efficient oxidative phosphorylation. Consequently, various signaling pathways are activated that underpin the enhanced Mk priming potential and enlarge the Mk subpopulation during ET pathogenesis. For example, the activated PI3K-AKT-mTOR signaling pathway upregulates the cyclin D-CDK4/6 complex (Pietras et al., 2011) and could promote mutant cell proliferation. This finding might explain the competitive advantage of mutant HSCs over WT HSCs in Jak2V617F mice (Dunbar et al., 2017). Thus, mutant, Mk-primed HSCs and elevated IFN signaling are of importance in onset and progression of JAK2V617F+ ET. We propose a model in which JAK2V617F mutation occurs in Mk-primed HSCs, leading to expansion of this population and hypersensitivity to IFN signaling, which promotes Mk lineage differentiation. An alternative mechanism cannot be ruled out where the bone marrow microenvironment (e.g., enhanced IFN signaling) is altered to preferentially promote Mk differentiation with subsequent acquisition of JAK2V617F in Mk-primed HSCs, which further accelerates Mk production (Figure S4G).
The association between stem cell heterogeneity and therapeutic effects in ET was studied further here. We found that the JAK2V617F+ Mk-primed HSC compartment was reduced in individuals with ET after treatment. Prior studies in MPN mice (predominantly manifesting PV phenotypes) have demonstrated that IFNα can directly target Jak2V617F+ HSCs through pro-apoptosis or proliferation-associated exhaustion (Austin et al., 2020; Hasan et al., 2013; Mullally et al., 2013). We observed this pro-apoptotic effect in HET mutant HSCs in individuals with ET after treatment, whereas cell cycling was enhanced only moderately enhanced. Interestingly, HOMO cells seemed to re-enter quiescence through restoration of the TSC-mTOR signaling pathway or TP53 activation, implying that these mutant, quiescent cells are preserved and serve as residual disease-initiating stem cells. Molecular remission can be achieved by IFNα treatment (Hasselbalch and Holmstrom, 2019); however, rapid molecular relapse occurs in some individuals after IFNα discontinuation (Ishii et al., 2007). Our findings imply that relapsing cells might originate from quiescent mutant HSCs in individuals with T-ET. Thus, our results suggest that transient, low-dose IFN stimulation promotes proliferation and differentiation of JAK2V617F+ Mk-primed HSCs during disease onset, whereas, upon treatment (including a chronic, therapeutic dose of IFNα), the mutant Mk-primed HSC population was reduced by promoting apoptosis or quiescence of mutant cells.
Because HSC heterogeneity underlies the disparate phenotypes of MPNs harboring the same initiating mutation, malignant transformation of neoplasms might involve a specific subset of stem cells within a heterogeneous stem cell population. This concept might inform pathogenic mechanisms and potential therapeutic strategies for various cancer stem cell heterogeneities.
Limitations of Study
This study is limited by the small number of samples collected and the complexity of the therapies received by these individuals, which remains to be investigated further. Additionally, the cellular and molecular basis of the changes in HSCs of individuals with PV upon treatment remains to be established.
Star Methods
Key Resources Table.
| Reagent Or Resource | Source | Identifier |
|---|---|---|
| Antibodies | ||
| APC anti-CD34 | BD Biosciences | Cat# 555824 RRID:AB_398614 |
| PE-Cy7 anti-CD38 | BD Biosciences | Cat# 555483 RRID:AB_395875 |
| PerCP-Cy5.5 anti-CD123 | BD Biosciences | Cat# 560904 RRID:AB_10563250 |
| APC-H7 anti-CD45RA | BD Biosciences | Cat# 560674 RRID:AB_1727497 |
| APC anti-CD41 | BD Biosciences | Cat# 561852 RRID:AB_10895580 |
| PE-CF594 anti-phosphorylated STAT3 | BD Biosciences | Cat# 562673 RRID:AB_2737714 |
| Alexa Fluor 647 anti-phosphorylated STAT5 | BD Biosciences | Cat# 612599 RRID:AB_399882 |
| eFluor 450 anti-phosphorylated STAT1 | Thermo Fisher Scientific | Cat# 48-9008-42 RRID: AB_2574119 |
| BV421 anti-CD34 | BioLegend | Cat# 343610 RRID:AB_2561358 |
| BV785 anti-CD38 | BioLegend | Cat# 303530 RRID:AB_2565893 |
| BV605 anti-CD123 | BioLegend | Cat# 306026 RRID:AB_2563826 |
| BV510anti-CD41a | BioLegend | Cat# 303736 RRID:AB_2687213 |
| FITC anti-VWF | Abcam | Cat# ab8822 RRID:AB_306799 |
| PerCP anti-IFN-α/β R1 | R and D Systems | Cat# FAB245C RRID:AB_10972109 |
| Alexa Fluor 700 anti-IFN-γ R1/CD119 | R and D Systems | Cat# FAB6731N RRID:N/A |
| Biological Samples | ||
| Human bone marrow from MPN patients and normal controls | Biobank of the Institute of Hematology and Blood Diseases Hospital | N/A |
| Human umbilical cord blood | Institute of Hematology and Blood Diseases Hospital | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Histopaque-1077 | Sigma-Aldrich | Cat# 10771 |
| IMDM | Sigma-Aldrich | Cat# I3390 |
| StemSpan SFEM | STEMCELL Technologies | Cat# 09650 |
| StemSpan CC100 | STEMCELL Technologies | Cat# 02690 |
| Recombinant human IL-6 | STEMCELL Technologies | Cat# 78050.1 |
| Recombinant human IL-3 | Sigma-Aldrich | Cat# I1646 |
| Recombinant human SCF | Prospec Bio | Cat# CYT-255 |
| Recombinant human TPO | PeproTech | Cat# 300-18 |
| Recombinant human Flt3-Ligand | PeproTech | Cat# 300-19 |
| Recombinant human IFNα | Kaiyinyisheng | Cat# S20030030 |
| Recombinant human IFNγ | Thermo Fisher Scientific | Cat# PHC4031 |
| Phosphate Buffered Saline solution | Hyclone | Cat# SH30256.FS |
| Fetal Bovine Serum | GIBCO | Cat# 16000-044 |
| Penicillin Streptomycin | GIBCO | Cat# 15140-122 |
| UltraPure 0.5M EDTA | Invitrogen | Cat# 15575-038 |
| DAPI | Sigma-Aldrich | Cat# D9542 |
| RNase Inhibitor | Invitrogen | Cat# AM2684 |
| Triton X-100 | Sigma-Aldrich | Cat# T8787 |
| dNTP Mix | Thermo Fisher Scientific | Cat# R0193 |
| SuperScript II reverse transcriptase SuperScript II first-strand buffer DTT |
Invitrogen | Cat# 18064-071 |
| Betaine | Sigma-Aldrich | Cat# 61962 |
| Magnesium chloride | Sigma-Aldrich | Cat# M8266 |
| KAPA HiFi HotStart ReadyMix | Kapa Biosystems | Cat# KK2602 |
| AMPure XP | Beckman Coulter | Cat# A63881 |
| Dynabeads MyOne Streptavidin C1 | Invitrogen | Cat# 65001 |
| TransTaq-T PCR SuperMix | TransGen Biotech | Cat# AS122-01 |
| TaqMan SNP Genotyping Assay | Thermo Fisher Scientific | Cat# 4351374 |
| TaqMan Gene Expression Master Mix | Applied Biosystems | Cat# 4369016 |
| Critical Commercial Assays | ||
| Lineage Cell Depletion Kit, human | Miltenyi Biotec | Cat# 130-092-211 |
| CD34 MicroBead Kit, human | Miltenyi Biotec | Cat# 130-046-702 |
| MegaCult-C Complete Kit with Cytokines for human CFU-Mk assays | STEMCELL Technologies | Cat# 04971 |
| Transcription Factor Phospho Buffer Set | BD Biosciences | Cat# 563239 |
| Zombie NIR viability kit | BioLegend | Cat# 423105 |
| SMART-Seq Ultra Low Input RNA Kit | TaKaRa Bio | Cat# 634888 |
| Advantage 2 PCR kit | TaKaRa Bio | Cat# 639206 |
| NEBNext Ultra II DNA Library Prep Kit | New England Biolabs | Cat# E7645L |
| DNA Clean & Concentrator | Zymo Research | Cat# D4014 |
| Qubit dsDNA HS Assay Kit | Invitrogen | Cat# Q32854 |
| Bioanalyzer High Sensitivity DNA Kit | Agilent | Cat# 5067-4626 |
| KAPA Hyper Prep Kits with PCR Library Amplification Primer Mix | Kapa Biosystems | Cat# KK8504 |
| CellsDirect One-Step qRT-PCR Kit (Superscript III RT mix; 2 × Reaction mix) | Invitrogen | Cat# 11753-500 |
| Single-cell PCR Sample/Loading Kit for gene expression | Fluidigm | Cat# BMK-M10-48.48 |
| Deposited Data | ||
| Raw and analyzed RNA-seq data | This paper | GEO: GSE111410GSA: HRA000669 |
| Oligonucleotides | ||
| See Table S4 | N/A | |
| Software and Algorithms | ||
| FlowJo | FlowJo | https://www.flowjo.com |
| GraphPad Prism | GraphPad | https://www.graphpad.com/ |
| Seurat v3.0.2 | Butler et al., 2018 | https://satijalab.org/seurat/ |
| clusterProfiler v3.10.1 | Yu et al., 2012 | https://guangchuangyu.github.io/software/clusterProfiler/ |
| GSEA v4.0 | Subramanian et al., 2005 | https://www.gsea-msigdb.org/gsea/index.jsp |
| Trimmomatic v0.36 | Bolger et al., 2014 | http://www.usadellab.org/cms/?page=trimmomatic |
| Hisat2 v2.1.0 | Kim et al., 2015 | https://daehwankimlab.github.io/hisat2/ |
| Stringtie software v1.3.3b | Pertea et al., 2016 | http://ccb.jhu.edu/software/stringtie/ |
| R v3.6 | R-Project | https://www.r-project.org/ |
| Picard | N/A | https://broadinstitute.github.io/picard/ |
| Python v3.7 | N/A | https://www.python.org/ |
Resource Availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Lihong Shi (shilihongxys@ihcams.ac.cn).
Materials availability
This study did not generate new unique reagents.
Experimental Model And Subject Details
Human bone marrow (BM) from MPN patients and normal controls (NCs)
This study was approved by the Institutional Ethics Committee of the Institute of Hematology and Blood Diseases Hospital and written informed consent was obtained from all participants. BM cells were collected from 51 JAK2V617F+ ET patients, 36 JAK2V617F+ PV patients, and 35 NCs (Table S1). Among them, 45 ET and 35 PV patients were newly diagnosed and had not received prior therapy. The BM cell samples obtained were analyzed by bulk RNA-Seq (5 PV, 10 ETand 7 NC); TARGET-Seq (7 ET and 7 NC); single-cell quantitative realtime PCR (2 PV, 4 ET and 2 NC); Cytek flow cytometry (4 PV, 6 ET and 4 NC); Mk differentiation and colony assays (10 PV, 13 ET and 15 NC), and HSC population analysis (22 PV, 22 ET and 10 NC). TARGET-Seq analysis was conducted for six ET patients post-treatment with IFNα alone or in combination with HU. Combinational treatment was delivered to those patients showing either poor tolerance to the IFNα-induced side-effects or poor therapeutic outcomes following treatment with HU alone. All diagnoses were defined according to World Health Organization (WHO) criteria. The clinical and laboratory features of the patients at the time of sampling are shown in Table S1.
Human umbilical cord blood and cell culture
Human umbilical cord blood-derived mononuclear cells (MNCs) were isolated by Histopaque (Sigma-Aldrich) density gradient centrifugation. For enrichment of CD34+ cells, MNCs were incubated 30 min at 4°C with anti-CD34+ antibody coupled to magnetic beads and FcR blocking reagent (Miltenyi Biotec) followed by washing with PBS and selecting with a MACS MultiStand isolation system (Miltenyi Biotec). Cord blood-derived CD34+ hematopoietic stem and progenitor cells (HSPCs) or bone marrow-derived CD34+ HSPCs from NCs were cultured in StemSpan media (STEMCELL Technologies) supplemented with 1% penicillin & streptomycin and 1% CC100 cocktail (STEMCELL Technologies) containing recombinant human fms-like tyrosine kinase 3 ligand (Flt3L), recombinant human stem cell factor (SCF), recombinant human interleukin 3 (IL-3) and recombinant human interleukin 6 (IL-6) before inducing megakaryocytic differentiation or performing colony formation assay.
Method Details
Quantification of the JAK2V617Fallele burden in patient samples
Quantification of the JAK2V617Fallele burden in patient samples was performed as described previously (Fu et al., 2016). Screening for the JAK2V617F mutation was performed by quantitative real-time PCR using primers for the JAK2V617F mutation together with Taq-Man probes (Thermo Fisher Scientific; Table S4) specific for the WT or mutant JAK2 alleles. The quantitative real-time PCR analysis was performed using an ABI StepOne Detection System (Applied Biosystems). The sequences of the primers and probes are listed in Table S4.
Isolation of BM-MNCs and HSCs
Bone marrow-derived mononuclear cells (BM-MNCs) from PV and ET patients and NCs were isolated by Histopaque (Sigma-Aldrich) density gradient centrifugation. BM-MNCs were stained with a lineage-marker cocktail (biotinylated anti-CD2, -CD3, -CD11b, -CD14, -CD15, -CD16, -CD19, -CD56, -CD123, and -CD235a) and subsequently magnetically-labeled with anti-biotin MicroBeads (Miltenyi Biotec). Lineage-negative stem and progenitor cells (Lin- cells) were obtained by depleting magnetically-labeled cells using LS columns (Miltenyi Biotec). Lin- cells were stained with PE-Cy7-conjugated anti-CD38 (BD Biosciences), APC-conjugated anti- CD34 (BD Biosciences), PerCP-Cy5.5-conjugated anti-CD123 (BD Biosciences) and APC-H7-conjugated anti-CD45RA (BD Biosciences) antibodies. Cell washing and antibody incubations were performed as described previously (Zhang et al., 2017). Cells were stained with DAPI to exclude dead cells. HSCs (Lin-CD34+CD38-CD45RA-CD123-) were sorted using a FACS Aria III flow cytometer (BD Biosciences), and data were analyzed with FlowJo software.
Bulk RNA-seq
HSCs (Lin-CD34+CD38-CD45RA-CD123-) were sorted (10-100 cells) from each sample by FACS. RNA amplification, library production, and data pre-processing were performed by Novogene (Beijing, China). Briefly, the sorted HSCs were lysed in lysis buffer (No-vogene), and first-strand cDNA was synthesized directly from polyA-selected RNA with the SMART-Seq Ultra Low RNA Kit (TaKaRa Bio). The cDNA was then purified using AMPureXP beads followed by long-distance PCR to yield full-length first-strand cDNA. Double-stranded cDNA was synthesized and amplified with an Advantage 2 PCR kit (TaKaRa Bio). cDNA libraries were generated using the NEBNext Ultra II DNA Library Prep Kit (New England Biolabs), and then sequenced using an Illumina HiSeq 2000 sequencing system to generate 150-bp paired-end reads.
TARGET-seq
3′-TARGET-seq coupling single-cell RNA-seq and JAK2V617F mutation detection was performed using a previously described method with modifications (Rodriguez-Meira et al., 2019; Zhou et al., 2016). HSCs were sorted by FACS directly into individual wells of a 96-well plate, each containing 2.55 μL lysis buffer [2 U RNase inhibitor (Invitrogen), 0.095 μL 10% Triton X-100 (Sigma-Aldrich), 1 pmol barcode primer, 5 nmol dNTP Mix (Thermo Fisher Scientific) and 1.805 μL nuclease-free water]. For the RT-PCR, plates were vortexed thoroughly and incubated at 72°C for 3 min before adding 2.85 μL mixture media [50 U SuperScript II reverse transcriptase (Invitrogen), 1 μL 5 × Superscript II first-strand buffer (Invitrogen), 25 nmol DTT (Invitrogen), 5 U RNase Inhibitor (Invitrogen), 5 μmol betaine (Sigma-Aldrich), 0.03 μmol MgCl2 (Sigma-Aldrich), 5 pmol TSO and 0.145 μL nuclease-free water] to each well. Pre-amplification (22 cycles) of the cDNA samples was performed with 7.5 μL reaction mix [HiFi HotStart ReadyMix 6.25 μL (Kapa Biosystems), 6.25 pmol 3′ P2, 1.25 pmol IS PCR primers, 0.07 μL JAK2 gDNA primer mix, and 0.36 μL nuclease-free water] in each well. A sample (1 μl) of the amplification product was collected for JAK2V617F mutation detection and the remainder was used for construction of the SMART-Seq2 cDNA library (Zhou et al., 2016). For the construction of the SMART-Seq2 cDNA library, pooling 3 μL of amplified cDNA from each uniquely-barcoded well into an Ep-pendorf tube. Pooled cDNA libraries were purified using DNA Clean and Concentrator kit (Zymo Research) once and elute in 50 μL nuclease-free water, then purified with AMPure XP magnetic beads (Beckman Coulter) twice in a ratio of 0.8 to 1 with cDNA. The quality of cDNA libraries was assessed using High-Sensitivity Bioanalyzer (Agilent 2100). After purification, an additional of PCR (4 cycles) was performed to add index sequence with biotin modification to the 3′ ends of the amplified cDNAs. Then 300-400 ng cDNA was fragmented to approximately 300 bp by Covaris S2, and the 3′ terminal of the cDNA was captured by Dynabeads MyOne Streptavidin C1 beads (Invitrogen). The KAPA Hyper Prep Kits (Kapa Biosystems) were used to construct the libraries. The cDNA libraries were sequenced with an Illumina HiSeq 4000/NovaSeq 6000 sequencing system, and 150-bp paired-end reads were generated. For JAK2V617F mutation detection, we performed 35 cycles of regular PCR with a reaction mixture containing 1 μL amplicons, 10 μL 2 × TransTaq-T PCR SuperMix (TransGen Biotech), 1 μL JAK2 gDNA primer mix and 8 μL nuclease-free water before TaqMan qPCR or Sanger sequencing. The primers and oligos (listed in Table S4) used for single-cell RNA-seq were synthesized by BGI Technology.
After the quality control, 752 HSCs from seven newly diagnosed JAK2V617F+ ET patients, 359 from six post-treated JAK2V617F+ ET patients and 485 from seven age-matched NCs were analyzed further. HSCs from ET patients were further categorized into homogeneous (HOMO), heterogeneous (HET), and WT groups.
Pre-processing of bulk RNA-seq data
Clustering of the index-coded samples was performed using a cBot Cluster Generation System with the TruSeq PE Cluster Kit v3-cBot-HS (Illumina), according to the manufacturer’s instructions. After cluster generation, the library preparations were sequenced on an Illumina HiSeq 2000 platform, and 150-bp paired-end reads were generated.
Paired-end reads were trimmed for both PCR and sequencing adapters using the Trimmomatic-0.36 package (Bolger et al., 2014). We used the parameter “SLIDINGWINDOW:5:20, MINLEN:50” to ensure the average quality was more than 20, and reads of length greater than 50 bp were retained.
Alignment and quantification of expression for bulk RNA-seq
The genome sequence (GRCh38/hg38) and transcript annotation (Gencode V27) were obtained from NCBI. Trimmed reads were aligned using the Hisat2 package (Kim et al., 2015). We applied the parameter “-rna-strandness RF” to distinguish the first-strand specific cDNA library. Then, we used Picard (https://broadinstitute.github.io/picard/) to mark and remove the duplicate reads. The remaining reads were then used to quantify gene and transcript isoform expression with Stringtie software (Pertea et al., 2016).
After quality control analysis, HSCs from 10 ET patients, five PV patients, and seven NCs were analyzed. Genome sequences (GRCh38/hg38) and transcript annotations (Gencode V27) were obtained from NCBI. Gene expression was normalized to fragments per kilobase million (FPKM) and analyzed further by log2 (FPKM*1) transformation.
T-distributed stochastic neighbor embedding (t-SNE) analysis for bulk RNA-seq
Two-thousand highly variable genes were used in a t-distributed stochastic neighbor embedding analysis. These genes were generated using the “vst” method of the Seurat package in R, through an input of filtered genes (with cutoffs of fold change ≥ 2, p < 0.05, and FPKM ≥ 0.1). The first nine principal components and perplexity six were used.
Pre-processing and quality control of single-cell RNA-seq
FASTQ files for each cell were generated, corresponding to an 8-bp cell-specific barcode. Reads were aligned using Hisat2 (2.1.0). The genome sequence (GRCh38/hg38) and the transcript annotation (Gencode V27) were used as the index and reference, respectively. After conversion to.bam files using samtools (1.6), counts for each gene were obtained with Stringtie (1.3.3b). QC filtering was performed using the following parameters: library size > 50,000 reads; percentage of reads mapping to the mitochondrial chromosome < 25%; and number of detected genes per cell (count > 0) > 500. After applying these QC filters, we retained 1596 cells. Counts were normalized and scaled using the Seurat R package, with LogNormalize or SCTransform method.
Self-organizing maps (SOMs)
Self-organizing maps (SOMs) were generated using the R package Kohonen, which can support complex analysis through SOMs with multiple data layers and supervised learning (superSOMs). Gene counts were normalized and trained to map units and then clustered into classes. Each observation (gene expression) belonged to a map unit, and each map unit belonged to a class. In this study, divergent expression patterns of genes were clustered to a computation layer in SOM components. Then, the SOMs were visualized to RGB color data using RColorBrewer and ggplot2.
Gene set enrichment analysis (GSEA)
Gene sets derived from MSigDB and other articles in the literature (Pronk et al., 2007; Rodriguez-Fraticelli et al., 2018; Sanjuan-Pla et al., 2013) were used for GSEA (Mootha et al., 2003; Subramanian et al., 2005). For bulk analysis, we used the expression matrix generated from all individual samples of PV, ET, and NC, with genes of average FPKM ≥ 0.1, as the input. The permutation number was set to 1000 and the permutation type was set to gene set. For single-cell analysis, the gene expression value, normalized using the Seurat package in R, was used as the input. The permutation number was set to 1000 and the permutation type was set to phenotype. The threshold for significant enrichment was set to normal, p < 0.05.
Single-cell quantitative real-time PCR
Single-cell quantitative real-time PCR analysis was performed according to a previously described method with modifications (Wang et al., 2018). In total, 284, 119, and 161 single HSCs were collected by FACS from four ET patients, two PV patients, and two NCs, respectively. Cells were sorted into individual wells of a 96-well plate, in which each well contained 10 μL RT buffer (2.5 μL of 0.2 × primers, 5 μL2 × Reaction mix (Invitrogen), 0.5 μL Superscript III RT mix (Invitrogen), and 2 μLTris-EDTA buffer). Reverse transcription was performed at 50°C for 15 min. After incubating at 95°C for 2 min, samples were amplified in 22 cycles of 95°C for 15 s and 60°C for 4 min. Samples were diluted 1:5 with Tris-EDTA, and 2.7 μL of the diluted samples were mixed with 3 μL TaqMan Gene Expression master mix (Applied Biosystems), 0.3 μL sample loading buffer, 3 μL of each set of 20 × primers, and 3 μL assay loading reagent. The reaction mixture (10 μl) was loaded onto a single-cell PCR gene expression chip (format 48 × 48) (Fluidigm). The chip was placed into a Fluidigm Biomark system and single-cell quantitative real-time PCR performed at 95°C for 10 min followed by 40 cycles of 95°C for 15 s and 60°C for 60 s. The data were analyzed by Biomark real-time PCR analysis software (Fluidigm).
A specific primer pair designed to amplify the JAK2V617F mutant transcript was multiplexed during the reverse transcription and PCR amplification steps. The amplicons from each cell were analyzed by Sanger sequencing to identify mutant cells. In ET, 40 mutant cells were detected among 212 JAK2-expressing HSCs (19%), whereas in PV, 15 mutant cells were detected among 55 JAK2-expressing HSCs (27%) (Table S3). The primers and probes used for single-cell quantitative real-time PCR are listed in Table S4.
Ex vivo megakaryocyte differentiation
Cord blood-derived CD34+ hematopoietic stem and progenitor cells were cultured in StemSpan media (STEMCELL Technologies) supplemented with 1% CC100 cocktail (STEMCELL Technologies) containing recombinant human fms-like tyrosine kinase 3 ligand (Flt3L), recombinant human stem cell factor (SCF), recombinant human interleukin 3 (IL-3) and recombinant human interleukin 6 (IL-6) for 1 day before inducing megakaryocytic differentiation. During differentiation, 20 ng/mL recombinant human interleukin-3 (IL-3) (Sigma-Aldrich), 20 ng/mL recombinant human stem cell factor (SCF) (Prospec Bio), and 50 ng/mL recombinant human thrombopoietin (TPO) (PeproTech) were added to the culture medium. On day 1 of differentiation, the cells were stimulated with various concentrations of IFN [IFNα (Kaiyinyisheng) 1 IU/mL, 10 IU/mL, 150 IU/mLor 1000 IU/mL; IFNγ (Thermo Fisher Scientific) 1 ng/mL, 10 ng/mL or 100 ng/mL; IFNα+γ, 1 IU/mL and 100 ng/mL, respectively] for different periods (30 min, 1 h, 2 h, 12 h, 24 h or 72 h). Subsequently, the cells were washed to remove IFN before further differentiation. On day 7, cells were harvested and CD41a+ cells were analyzed by flow cytometry.
Bone marrow-derived CD34+ hematopoietic stem and progenitor cells from NCs were cultured using a similar protocol in StemSpan media supplemented with 1% CC100 cocktail for 1 day before inducing megakaryocytic differentiation. During differentiation, 20 ng/mL IL-3,20 ng/mL FLT-3 (PeproTech), 20 ng/mL SCF, 50 ng/mL of TPO, and 20 ng/mL recombinant human interleukin-6 (IL-6) (STEMCELL Technologies) were added to the culture medium. On day 1 of differentiation, the cells were stimulated with various concentrations of IFN [IFNα, 1 IU/mL; IFNγ 100 ng/mL; IFNα+γ, 1 IU/mL and 100 ng/mL, respectively] for 2 h. Subsequently, the cells were washed to remove IFN before further differentiation. Cells were maintained at a density between 5 × 104 and 5 × 105 cells/mL at 37°C under 5% CO2, with medium changed every 3 days. On day 11, cells were harvested and CD41a+ cells were analyzed by flow cytometry.
Colony formation assay
FACS-sorted HSCs (Lin-CD34+CD38-CD45RA-CD123-) or CD34+ HSPCs from cord blood were cultured in StemSpan containing 1% CC100 cocktail for 3 days. After stimulation with or without IFNα+γ (1 IU/mL+ 100 ng/mL) for 2 h, the cells were washed and resuspended in IMDM (GIBCO) and analyzed in a CFU-Mk colony assay (STEMCELL Technologies) according to manufacturer’s instructions. After 14 days of culture, CFU-Mk colonies were fixed and stained.
For the single-cell colony formation assays, FACS-sorted single HSCs (Lin-CD34+CD38-CD45RA-CD123-) from the bone marrow of 13 JAK2V617F ET patients, 10 JAK2V617F PV patients, and 12 NCs were cultured in StemSpan containing 1% CC100 cocktail for 10-12 days during the first phase of proliferation. At day 10-12, a subset of each first phase clone expanded from a single ET HSC was harvested to examine the JAK2V617F mutation by PCR, followed by TaqMan qPCR or Sanger sequencing. The JAK2V617F mutant ET HSCs, non mutant ET HSCs, JAK2V617F mutant PV HSCs, non mutant PV HSCs, and NC HSCs were stimulated with or without 1 IU/mL IFNα and 100 ng/mL IFNγ for 2 h. After washing away residual IFN from each group, CFU-Mk assays were conducted as described previously.
Cytek Aurora flow cytometry assay
BM-MNCs were first stained with a live/dead marker by Zombie NIR viability kit (BioLegend). After washing with staining buffer (including PBS, 2% FBS and 0.4% 0.5M EDTA), cells were stained with a cocktail of antibodies against a panel of surface markers (CD34, CD38, CD45RA, CD123, CD41A, IFNGR1, IFNαR1) for 30 min at 4°C. For each sample, half of the cells were analyzed immediately using a Cytek Aurora flow cytometer. The other half was washed with 1 × PBS, fixed with 1 × TFP Fix/Perm Buffer (BD Biosciences) at 4°C for 50 min and permeabilized using Perm Buffer III (BD Biosciences) for 20 min on ice before staining with antibodies (VWF, p-STAT1, p-STAT3, p-STAT5) for 40-50 min at 4°C according to the manufacturer’s instructions. Flow cytometry was carried on and analyzed by Cytek Aurora flow cytometer. RFI was used as an indicator of p-STAT1, p-STAT3 and p-STAT5 expression and was calculated by the following formula: RFI = (MFI of MPN cells - MFI of corresponding negative controls)/(MFI of normal control cells - MFI of corresponding negative controls), where MFI indicates the mean fluorescence intensity.
Immunocytochemical staining (ICC staining)
Cells were fixed in fixation buffer (50% methanol and 50% acetone) for 90 s at room temperature and air-dried. The cells were incubated with 30-50 μL biotinylated-CD41a primary antibody solution for 30-50 min at room temperature. After washing twice with PBS, the cells were incubated with 30-50 μL streptomycin-alkaline phosphatase buffer for another 20-40 min. Next, the cells were washed twice with PBS and stained with chromogenic substrate for 10-20 min. Hematoxylin was used for cytoplasm staining.
Quantification And Statistical Analysis
In this study, experimental data in cord blood or bone marrow from NCs, which can be assumed to be normally distributed, were analyzed using an unpaired two-tailed Student’s t test. For seq analysis or experimental data in patients, in which the distribution cannot be predicted, Wilcoxon rank-sum test was applied to determine statistical significance. p < 0.05 was considered to indicate statistical significance. For the GSEA, multiple comparisons were applied and p < 0.05 was considered to indicate statistical significance. Analyses were performed using GraphPad Prism or R software.
Supplementary Material
Highlights.
Single-cell analyses reveal links between HSC heterogeneity and thrombocythemia
JAK2-mutant HSCs in ET show strong megakaryocyte (Mk) lineage priming
JAK2-mutant HSCs in ET are more sensitive to IFN signaling to Mk differentiation
JAK2-mutant HSCs in ET exhibit distinct signatures upon treatment
Acknowledgements
This work was supported by the National Key Research and Development Program of China Stem Cell and Translational Research (2016YFA0102300, 2016YFA0100600, 2017YFA0103102, 2019YFA0801800, and 2019YFA0110802), the CAMS Initiative for Innovative Medicine (2016-I2M-1-018, 2016-I2M-3-002, 2016-I2M-3-015, 2016-I2M-1-017, 2019-I2M-1-006, and 2017-I2M-1-015), the CAMS Key Laboratory of Gene Therapy for Blood Diseases (2018PT31038), the National Natural Science Foundation (81870089, 81890990, 81470302, 81770104, 81600099, 81500084, 81770128, 82070109, 81730006, 62002153, 81700105, 81970121, and 82000136), the Tianjin Natural Science Foundation (19JCZDJC33000 and 18JCQNJC11900), the Beijing-Tianjin-Hebei Basic Research Project (18JCZDJC44600 and H2018206423), and Fundamental Research Funds for the Central Universities (3332020056). We thank Dr. H. Ema for insightful discussions and Dr. H. Shen (Cytek Biosciences) for technical support.
Footnotes
Author Contributions
J.T., T.S., S.M., Y.Z., M.J., Y.G., P.T., Ding Wang, Di Wang, and A.Z. performed the research and analyzed the data. P.Z., R.F., Z.X., J.Z., and R.Y. analyzed the data. Dong Wang, L.S., T.C., and L.Z. designed the experiments, analyzed the data, and wrote the paper. S.J.L., J.L., A.R.G., and E.H.B. designed the experiments and wrote the paper.
Declaration Of Interests
The authors declare no competing interests.
Data and code availability
The bulk RNA-seq data have been deposited in the NCBI Gene Expression Omnibus and are accessible through GEO Series accession number GEO: GSE111410. The single-cell RNA-seq data have been deposited in the Genome Sequence Archive in National Genomics Data Center, China National Center for Bioinformation / Beijing Institute of Genomics, Chinese Academy of Sciences GSA: HRA000669. All relevant data in this study are available from the corresponding authors upon reasonable request.
References
- Austin RJ, Straube J, Bruedigam C, Pali G, Jacquelin S, Vu T, Green J, Grasel J, Lansink L, Cooper L, et al. Distinct effects of ruxolitinib and interferon-alpha on murine JAK2V617F myeloproliferative neoplasm hematopoietic stem cell populations. Leukemia. 2020;34:1075–1089. doi: 10.1038/s41375-019-0638-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Project. Baxter EJ, Scott LM, Campbell PJ, East C, Fourouclas N, Swanton S, Vassiliou GS, Bench AJ, Boyd EM, Curtin N, et al. Acquired mutation of the tyrosine kinase JAK2 in human myeloproliferative disorders. Lancet. 2005;365:1054–1061. doi: 10.1016/S0140-6736(05)71142-9. [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 2018;36:411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabezas-Wallscheid N, Buettner F, Sommerkamp P, Klimmeck D, Ladel L, Thalheimer FB, Pastor-Flores D, Roma LP, Renders S, Zeisberger P, et al. Vitamin A-Retinoic Acid Signaling Regulates Hematopoietic Stem Cell Dormancy. Cell. 2017;169:807–823.:e19. doi: 10.1016/j.cell.2017.04.018. [DOI] [PubMed] [Google Scholar]
- Chen C, Liu Y, Liu R, Ikenoue T, Guan KL, Liu Y, Zheng P. TSC-mTOR maintains quiescence and function of hematopoietic stem cells by repressing mitochondrial biogenesis and reactive oxygen species. J Exp Med. 2008;205:2397–2408. doi: 10.1084/jem.20081297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen E, Beer PA, Godfrey AL, Ortmann CA, Li J, Costa-Pereira AP, Ingle CE, Dermitzakis ET, Campbell PJ, Green AR. Distinct clinical phenotypes associated with JAK2V617F reflect differential STAT1 signaling. Cancer Cell. 2010;18:524–535. doi: 10.1016/j.ccr.2010.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng T. Cell cycle inhibitors in normal and tumor stem cells. Oncogene. 2004;23:7256–7266. doi: 10.1038/sj.onc.1207945. [DOI] [PubMed] [Google Scholar]
- Dunbar A, Nazir A, Levine R. Overview of Transgenic Mouse Models of Myeloproliferative Neoplasms (MPNs) Curr Protoc Pharmacol. 2017;77:14.40.1–14.40.19. doi: 10.1002/cpph.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu R, Liu D, Cao Z, Zhu S, Li H, Su H, Zhang L, Xue F, Liu X, Zhang X, et al. Distinct molecular abnormalities underlie unique clinical features of essential thrombocythemia in children. Leukemia. 2016;30:746–749. doi: 10.1038/leu.2015.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genovese G, Kähler AK, Handsaker RE, Lindberg J, Rose SA, Bakhoum SF, Chambert K, Mick E, Neale BM, Fromer M, et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med. 2014;371:2477–2487. doi: 10.1056/NEJMoa1409405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giladi A, Paul F, Herzog Y, Lubling Y, Weiner A, Yofe I, Jaitin D, Cabezas-Wallscheid N, Dress R, Ginhoux F, et al. Single-cell characterization of haematopoietic progenitors and their trajectories in homeostasis and perturbed haematopoiesis. Nat Cell Biol. 2018;20:836–846. doi: 10.1038/s41556-018-0121-4. [DOI] [PubMed] [Google Scholar]
- Godfrey AL, Chen E, Pagano F, Ortmann CA, Silber Y, Bellosillo B, Guglielmelli P, Harrison CN, Reilly JT, Stegelmann F, et al. JAK2V617F homozygosity arises commonly and recurrently in PV and ET, but PV is characterized by expansion of a dominant homozygous subclone. Blood. 2012;120:2704–2707. doi: 10.1182/blood-2012-05-431791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodell MA, Nguyen H, Shroyer N. Somatic stem cell heterogeneity: diversity inthe blood, skin and intestinal stem cell compartments. Nat Rev Mol Cell Biol. 2015;16:299–309. doi: 10.1038/nrm3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grinfeld J, Nangalia J, Baxter EJ, Wedge DC, Angelopoulos N, Cantrill R, Godfrey AL, Papaemmanuil E, Gundem G, MacLean C, et al. Classification and Personalized Prognosis in Myeloproliferative Neoplasms. N Engl J Med. 2018;379:1416–1430. doi: 10.1056/NEJMoa1716614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grisouard J, Shimizu T, Duek A, Kubovcakova L, Hao-Shen H, Dirnhofer S, Skoda RC. Deletion of Stat3 in hematopoietic cells enhances thrombocytosis and shortens survival in a JAK2-V617F mouse model of MPN. Blood. 2015;125:2131–2140. doi: 10.1182/blood-2014-08-594572. [DOI] [PubMed] [Google Scholar]
- Haas S, Hansson J, Klimmeck D, Loeffler D, Velten L, Uckelmann H, Wurzer S, Prendergast AM, Schnell A, Hexel K, et al. Inflammation-Induced Emergency Megakaryopoiesis Driven by Hematopoietic Stem Cell-like Megakaryocyte Progenitors. Cell Stem Cell. 2015;17:422–434. doi: 10.1016/j.stem.2015.07.007. [DOI] [PubMed] [Google Scholar]
- Hao S, Chen C, Cheng T. Cell cycle regulation of hematopoietic stem or progenitor cells. Int J Hematol. 2016;103:487–497. doi: 10.1007/s12185-016-1984-4. [DOI] [PubMed] [Google Scholar]
- Hasan S, Lacout C, Marty C, Cuingnet M, Solary E, Vainchenker W, Villeval JL. JAK2V617F expression in mice amplifies early hematopoietic cells and gives them a competitive advantage that is hampered by IFNa. Blood. 2013;122:1464–1477. doi: 10.1182/blood-2013-04-498956. [DOI] [PubMed] [Google Scholar]
- Hasselbalch HC, Holmström MO. Perspectives on interferon alpha in the treatment of polycythemiavera and related myeloproliferative neoplasms: minimal residual disease and cure? Semin Immunopathol. 2019;41:5–19. doi: 10.1007/s00281-018-0700-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoki K, Li Y, Zhu T, Wu J, Guan KL. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat Cell Biol. 2002;4:648–657. doi: 10.1038/ncb839. [DOI] [PubMed] [Google Scholar]
- Ishii T, Xu M, Zhao Y, Hu WY, Ciurea S, Bruno E, Hoffman R. Recurrence of clonal hematopoiesis after discontinuing pegylated recombinant interferon-alpha 2a in a patient with polycythemia vera. Leukemia. 2007;21:373–374. doi: 10.1038/sj.leu.2404475. [DOI] [PubMed] [Google Scholar]
- Ito K, Carracedo A, Weiss D, Arai F, Ala U, Avigan DE, Schafer ZT, Evans RM, Suda T, Lee CH, Pandolfi PP. A PML-PPAR-δ pathway for fatty acid oxidation regulates hematopoietic stem cell maintenance. Nat Med. 2012;18:1350–1358. doi: 10.1038/nm.2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen SEW, Nerlov C. Haematopoiesis in the era of advanced single-cell technologies. Nat Cell Biol. 2019;21:2–8. doi: 10.1038/s41556-018-0227-8. [DOI] [PubMed] [Google Scholar]
- Jaiswal S, Fontanillas P, Flannick J, Manning A, Grauman PV, Mar BG, Lindsley RC, Mermel CH, Burtt N, Chavez A, et al. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med. 2014;371:2488–2498. doi: 10.1056/NEJMoa1408617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James C, Ugo V, Le Couédic JP, Staerk J, Delhommeau F, Lacout C, Garcon L, Raslova H, Berger R, Bennaceur-Griscelli A, et al. A unique clonal JAK2 mutation leading to constitutive signalling causes polycythaemia vera. Nature. 2005;434:1144–1148. doi: 10.1038/nature03546. [DOI] [PubMed] [Google Scholar]
- Kiladjian JJ, Giraudier S, Cassinat B. Interferon-alpha for the therapy of myeloproliferative neoplasms: targeting the malignant clone. Leukemia. 2016;30:776–781. doi: 10.1038/leu.2015.326. [DOI] [PubMed] [Google Scholar]
- Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleppe M, Koche R, Zou L, van Galen P, Hill CE, Dong L, De Groote S, Papalexi E, Hanasoge Somasundara AV, Cordner K, et al. Dual Targeting of Oncogenic Activation and Inflammatory Signaling Increases Therapeutic Efficacy in Myeloproliferative Neoplasms. Cancer Cell. 2018;33:29–43.:e7. doi: 10.1016/j.ccell.2017.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koschmieder S, Mughal TI, Hasselbalch HC, Barosi G, Valent P, Kiladjian JJ, Jeryczynski G, Gisslinger H, Jutzi JS, Pahl HL, et al. Myeloproliferative neoplasms and inflammation: whether to target the malignant clone or the inflammatory process or both. Leukemia. 2016;30:1018–1024. doi: 10.1038/leu.2016.12. [DOI] [PubMed] [Google Scholar]
- Kralovics R, Passamonti F, Buser AS, Teo SS, Tiedt R, Passweg JR, Tichelli A, Cazzola M, Skoda RC. A gain-of-function mutation of JAK2 in myeloproliferative disorders. N Engl J Med. 2005;352:1779–1790. doi: 10.1056/NEJMoa051113. [DOI] [PubMed] [Google Scholar]
- Levine RL, Wadleigh M, Cools J, Ebert BL, Wernig G, Huntly BJ, Boggon TJ, Wlodarska I, Clark JJ, Moore S, et al. Activating mutation in the tyrosine kinase JAK2 in polycythemia vera, essential thrombocythemia, and myeloid metaplasia with myelofibrosis. Cancer Cell. 2005;7:387–397. doi: 10.1016/j.ccr.2005.03.023. [DOI] [PubMed] [Google Scholar]
- Li J, Spensberger D, Ahn JS, Anand S, Beer PA, Ghevaert C, Chen E, Forrai A, Scott LM, Ferreira R, et al. JAK2 V617F impairs hematopoietic stem cell function in a conditional knock-in mouse model of JAK2 V617F-positive essential thrombocythemia. Blood. 2010;116:1528–1538. doi: 10.1182/blood-2009-12-259747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Kent DG, Godfrey AL, Manning H, Nangalia J, Aziz A, Chen E, Saeb-Parsy K, Fink J, Sneade R, et al. JAK2V617F homozygosity drives a phenotypic switch in myeloproliferative neoplasms, but is insufficient to sustain disease. Blood. 2014;123:3139–3151. doi: 10.1182/blood-2013-06-510222. [DOI] [PubMed] [Google Scholar]
- Liu Y, Elf SE, Miyata Y, Sashida G, Liu Y, Huang G, Di Giandomenico S, Lee JM, Deblasio A, Menendez S, et al. p53 regulates hematopoietic stem cell quiescence. Cell Stem Cell. 2009;4:37–48. doi: 10.1016/j.stem.2008.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maaten L, Hinton G. Visualizing Data using t-SNE. J Mach Learn Res. 2008;9:26. [Google Scholar]
- Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstråle M, Laurila E, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- Mullally A, Bruedigam C, Poveromo L, Heidel FH, Purdon A, Vu T, Austin R, Heckl D, Breyfogle LJ, Kuhn CP, et al. Depletion of Jak2V617F myeloproliferative neoplasm-propagating stem cells by interferon-a in a murine model of polycythemia vera. Blood. 2013;121:3692–3702. doi: 10.1182/blood-2012-05-432989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nangalia J, Green AR. Myeloproliferative neoplasms: from origins to outcomes. Hematology (Am Soc Hematol Educ Program) 2017;2017:470–479. doi: 10.1182/asheducation-2017.1.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortmann CA, Kent DG, Nangalia J, Silber Y, Wedge DC, Grinfeld J, Baxter EJ, Massie CE, Papaemmanuil E, Menon S, et al. Effect of mutation order on myeloproliferative neoplasms. N Engl J Med. 2015;372:601–612. doi: 10.1056/NEJMoa1412098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardanani A, Fridley BL, Lasho TL, Gilliland DG, Tefferi A. Host genetic variation contributes to phenotypic diversity in myeloproliferative disorders. Blood. 2008;111:2785–2789. doi: 10.1182/blood-2007-06-095703. [DOI] [PubMed] [Google Scholar]
- Pertea M, Kim D, Pertea GM, Leek JT, Salzberg SL. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat Protoc. 2016;11:1650–1667. doi: 10.1038/nprot.2016.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietras EM, Warr MR, Passegué E. Cell cycle regulation in hematopoietic stem cells. J Cell Biol. 2011;195:709–720. doi: 10.1083/jcb.201102131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietras EM, Lakshminarasimhan R, Techner JM, Fong S, Flach J, Binnewies M, Passegue E. Re-entry into quiescence protects hematopoietic stem cells from the killing effect of chronic exposure to type I interferons. J Exp Med. 2014;211:245–262. doi: 10.1084/jem.20131043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pronk CJ, Rossi DJ, Månsson R, Attema JL, Norddahl GL, Chan CK, Sigvardsson M, Weissman IL, Bryder D. Elucidation of the phenotypic, functional, and molecular topography of a myeloerythroid progenitor cell hierarchy. Cell Stem Cell. 2007;1:428–442. doi: 10.1016/j.stem.2007.07.005. [DOI] [PubMed] [Google Scholar]
- NIH Intramural Sequencing Center. Psaila B, Wang G, Rodriguez-Meira A, Li R, Heuston EF, Murphy L, Yee D, Hitchcock IS, Sousos N, O’Sullivan J, et al. Single-Cell Analyses Reveal Megakaryocyte-Biased Hematopoiesis in Myelofibrosis and Identify Mutant Clone-Specific Targets. Mol Cell. 2020;78:477–492.:e8. doi: 10.1016/j.molcel.2020.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Fraticelli AE, Wolock SL, Weinreb CS, Panero R, Patel SH, Jankovic M, Sun J, Calogero RA, Klein AM, Camargo FD. Clonal analysis of lineage fate in native haematopoiesis. Nature. 2018;553:212–216. doi: 10.1038/nature25168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Fraticelli AE, Weinreb C, Wang SW, Migueles RP, Jankovic M, Usart M, Klein AM, Lowell S, Camargo FD. Single-cell lineage tracing unveils a role for TCF15 in haematopoiesis. Nature. 2020;583:585–589. doi: 10.1038/s41586-020-2503-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Meira A, Buck G, Clark SA, Povinelli BJ, Alcolea V, Louka E, McGowan S, Hamblin A, Sousos N, Barkas N, et al. Unravelling Intratumoral Heterogeneity through High-Sensitivity Single-Cell Mutational Analysis and Parallel RNASequencing. Mol Cell. 2019;73:1292–1305.:e8. doi: 10.1016/j.molcel.2019.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjuan-Pla A, Macaulay IC, Jensen CT, Woll PS, Luis TC, Mead A, Moore S, Carella C, Matsuoka S, Bouriez Jones T, et al. Platelet-biased stem cells reside at the apex of the haematopoietic stem-cell hierarchy. Nature. 2013;502:232–236. doi: 10.1038/nature12495. [DOI] [PubMed] [Google Scholar]
- Scott LM, Scott MA, Campbell PJ, Green AR. Progenitors homozygous for the V617F mutation occur in most patients with polycythemia vera, but not essential thrombocythemia. Blood. 2006;108:2435–2437. doi: 10.1182/blood-2006-04-018259. [DOI] [PubMed] [Google Scholar]
- Snoeck HW. Mitochondrial regulation of hematopoietic stem cells. Curr Opin Cell Biol. 2017;49:91–98. doi: 10.1016/j.ceb.2017.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T, Ju M, Dai X, Dong H, Gu W, Gao Y, Fu R, Liu X, Huang Y, Liu W, et al. Multilevel defects in the hematopoietic niche in essential thrombocythemia. Haematologica. 2020;105:661–673. doi: 10.3324/haematol.2018.213686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapper W, Jones AV, Kralovics R, Harutyunyan AS, Zoi K, Leung W, Godfrey AL, Guglielmelli P, Callaway A, Ward D, et al. Genetic variation at MECOM, TERT, JAK2 and HBS1L-MYB predisposes to myeloproliferative neoplasms. Nat Commun. 2015;6:6691. doi: 10.1038/ncomms7691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiedt R, Hao-Shen H, Sobas MA, Looser R, Dirnhofer S, Schwaller J, Skoda RC. Ratio of mutant JAK2-V617F to wild-type Jak2 determines the MPD phenotypes in transgenic mice. Blood. 2008;111:3931–3940. doi: 10.1182/blood-2007-08-107748. [DOI] [PubMed] [Google Scholar]
- Verovskaya EV, Dellorusso PV, Passegué E. Losing Sense of Self and Surroundings: Hematopoietic Stem Cell Aging and Leukemic Transformation. Trends Mol Med. 2019;25:494–515. doi: 10.1016/j.molmed.2019.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walz C, Ahmed W, Lazarides K, Betancur M, Patel N, Hennighausen L, Zaleskas VM, Van Etten RA. Essential role for Stat5a/b in myeloproliferative neoplasms induced by BCR-ABL1 and JAK2(V617F) in mice. Blood. 2012;119:3550–3560. doi: 10.1182/blood-2011-12-397554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Dong F, Zhang S, Yang W, Yu W, Wang Z, Zhang S, Wang J, Ma S, Wu P, et al. TGF-β1 Negatively Regulates the Number and Function of Hematopoietic Stem Cells. Stem Cell Reports. 2018;11:274–287. doi: 10.1016/j.stemcr.2018.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehrens R, Buydens L. Self- and Super-organizing Maps in R: The Kohonen Package. J Stat Softw. 2007;21:19. [Google Scholar]
- Wilson NK, Kent DG, Buettner F, Shehata M, Macaulay IC, Calero-Nieto FJ, Sanchez Castillo M, Oedekoven CA, Diamanti E, Schulte R, et al. Combined Single-Cell Functional and Gene Expression Analysis Resolves Heterogeneity within Stem Cell Populations. Cell Stem Cell. 2015;16:712–724. doi: 10.1016/j.stem.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie M, Lu C, Wang J, McLellan MD, Johnson KJ, Wendl MC, McMichael JF, Schmidt HK, Yellapantula V, Miller CA, et al. Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat Med. 2014;20:1472–1478. doi: 10.1038/nm.3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz OH, Valdez R, Theisen BK, Guo W, Ferguson DO, Wu H, Morrison SJ. Pten dependence distinguishes haematopoietic stem cells from leukaemia-initiating cells. Nature. 2006;441:475–482. doi: 10.1038/nature04703. [DOI] [PubMed] [Google Scholar]
- Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Grindley JC, Yin T, Jayasinghe S, He XC, Ross JT, Haug JS, Rupp D, Porter-Westpfahl KS, Wiedemann LM, et al. PTEN maintains haematopoietic stem cells and acts in lineage choice and leukaemia prevention. Nature. 2006;441:518–522. doi: 10.1038/nature04747. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Zhang J, An W, Wan Y, Ma S, Yin J, Li X, Gao J, Yuan W, Guo Y, et al. Intron 1 GATA site enhances ALAS2 expression indispensably during erythroid differentiation. Nucleic Acids Res. 2017;45:657–671. doi: 10.1093/nar/gkw901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F, Li X, Wang W, Zhu P, Zhou J, He W, Ding M, Xiong F, Zheng X, Li Z, et al. Tracing haematopoietic stem cell formation at single-cell resolution. Nature. 2016;533:487–492. doi: 10.1038/nature17997. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The bulk RNA-seq data have been deposited in the NCBI Gene Expression Omnibus and are accessible through GEO Series accession number GEO: GSE111410. The single-cell RNA-seq data have been deposited in the Genome Sequence Archive in National Genomics Data Center, China National Center for Bioinformation / Beijing Institute of Genomics, Chinese Academy of Sciences GSA: HRA000669. All relevant data in this study are available from the corresponding authors upon reasonable request.