Abstract
Hepatitis C virus (HCV) infection remains a major global health problem, with 130-170 million chronically infected individuals at risk to develop severe liver disease, including hepatocellular carcinoma. Although the development of direct-acting antivirals offers cure in large majority of patients, there are still a number of clinical challenges. These include DAA failure in a significant subset of patients, difficult-to-treat genotypes and limited access to therapy due to high costs. Moreover, recent data indicate that the risk for liver cancer persists in patients with advanced fibrosis. These challenges highlight the need for continued efforts towards novel therapeutic strategies for HCV. Over the past two decades, advances in HCV model systems have enabled a detailed understanding of HCV entry and its clinical impact. Many of the virus-host interactions involved in HCV entry have now been identified and explored as antiviral targets. Furthermore, viral entry is recognized as an important factor for graft reinfection and establishment of persistent infection. HCV entry inhibitors, therefore, offer promising opportunities to address the limitations of DAAs. Here, we summarize recent advances in the field of HCV entry and discuss perspectives towards the prevention and cure of HCV infection and virus-induced liver disease.
Global impact of HCV infection
There are an estimated 130-170 million people worldwide who are chronically infected with hepatitis C virus (HCV) [1]. These individuals are at higher risk to develop severe liver disease, including cirrhosis and hepatocellular carcinoma (HCC) [1]. Although recent approval of direct-acting antivirals (DAAs) has improved the outlook for HCV patients, the risk for liver disease persists even after viral cure, once fibrosis has been established [2]. Furthermore, not all patients respond to therapy and the high costs of DAAs limit access to treatment even in high-resource countries [3–5]. A detailed, comprehensive knowledge of HCV entry will guide development of novel antiviral approaches [6, 7]. Here, we review recent insights into the HCV entry process and its clinical impact as an antiviral target. We also discuss perspectives to use our accumulating knowledge of HCV entry to develop strategies aimed at the prevention and cure of HCV infection and virus-induced liver disease.
Molecular virology and cell biology of HCV entry
HCV is a member of the flaviviridae family, classified in the hepacivirus genus. HCV, with a positive sense single-stranded RNA genome of 9.6 kilo-base pairs, is a cytoplasmic-replicating virus [8]. The HCV capsid is surrounded by a host-derived lipid envelope, in which the E1 and E2 glycoproteins are embedded, and is associated with serum lipoproteins such as apolipoprotein E (ApoE) [9].
The first step in HCV infection is low-affinity binding to heparan moieties in heparan sulfate proteoglycans on the surface of hepatocytes [10–12], an interaction at least partially mediated through virion-associated ApoE [13–15]. The cellular low-density lipoprotein receptor (LDL-R) also interacts with virion-associated apolipoproteins to facilitate further binding [16–19]. Furthermore, the scavenger receptor class B type I (SR-B1) binds to virion-associated lipoproteins [20] and the HCV E2 protein [21]. The lipid transfer activities of SR-BI may expose regions of E2 involved in interactions with other cellular factors, such as cluster of differentiation 81 (CD81) [22, 23]. CD81, the first receptor identified for HCV, binds directly to E2 [24] and also mediates critical post-binding events [25, 26], including activation of signaling pathways. Indeed, CD81 engagement was shown to activate signaling through the epidermal growth factor receptor (EGFR) [27] and Rho and Ras GTPases [28, 29].
Tight junction (TJ) proteins claudin-1 (CLDN1) and occludin (OCLN) have also been identified as HCV entry factors [30, 31]. CD81 interacts with CLDN1 to form a co-receptor complex [32], which along with the HCV particle is ultimately internalized into clathrin-containing endosomes [26]. Given that CD81-CLDN1 co-receptor complex formation could be detected at basolateral membranes but not in TJ-associated pools of CLDN1 [32], it is likely that the nonjunctional pool of CLDN1 predominantly contributes to HCV entry. OCLN is another TJ protein required for a post-binding step of HCV entry [31, 33], although its specific role in entry has not yet been elucidated.
Other cellular factors have been implicated in HCV entry, yet their functions remain enigmatic. For example, the Niemann-Pick C1-like 1 cholesterol absorption receptor interacts with virion-associated cholesterol to mediate binding or internalization steps [34]. Transferrin receptor 1 and cell death-inducing DFFA-like effector b are thought to be involved in late entry steps [35, 36]. Recently, the provirus integration site for Moloney murine leukemia virus (Pim1) kinase was identified as an additional HCV entry factor, perhaps by contributing to CD81-CLDN1 receptor complex formation via PI3K-Akt signaling [37]. The serum response factor binding protein 1 (SRFBP1) also interacts with CD81 to coordinate host cell penetration [38]. SRFBP1 was proposed to mediate actin-dependent translocation following HBV receptor binding [38], and may also contribute to CD81-CLDN1 co-receptor complex assembly.
Ultimately, these virus-host interactions lead to the internalization of HCV into Rab5-containing early endosomes, where low pH induces viral fusion [26, 39]. The HCV fusion protein is still unknown. Although initial predictions pointed to E2 as being the fusion protein, the recently solved crystal structure of the E2 core ectodomain suggests otherwise [40, 41]. Indeed, the E2 ectodomain is globular and did not undergo conformational changes when exposed to low pH [40, 41], suggesting that E2 acts through a novel mechanism or that E1 may be involved in inducing membrane fusion [42]. Further mechanistic studies, and likely a structure of the E1/E2 heterodimer, will be necessary to elucidate the fusion mechanism.
Another route of HCV entry relies on direct cell-to-cell spread [43]. In this context, CD81, CLDN1 and SR-BI likely play key roles [43, 44], as do the viral envelope glycoproteins [45]. However, CD81 may be dispensable for cell-to-cell spread, at least in hepatoma cell lines [45]. Signaling pathways activated by EGFR may also contribute to cell-to-cell spread [27, 29]. Furthermore, virion-associated host factors such as ApoE have been implicated in cell-to-cell transmission [46].
HCV entry and liver disease
Viral entry is thought to play a major role in the pathogenesis of HCV infection. In the context of liver transplantation – which is severely hampered by rapid reinfection of the graft – it has been shown that viral quasispecies are rapidly selected following transplantation [47] and the resulting selection of viral variants contributes to pathogenesis. Indeed, escape from antibody-mediated neutralization selects for viral variants with a highly efficient entry phenotype associated with altered receptor usage [48]. Mutations in E2 that modulate interactions with CD81 were implicated in mediating viral evasion at a post-binding step [49]. Altered usage of SR-B1 has also been observed [50], and increased levels of CLDN1 and OCLN modulate recurrence of HCV infection following liver transplantation [51]. These findings highlight viral entry as an important determinant for graft reinfection and the establishment of persistent infection. They also point to entry as an attractive therapeutic target, including preventing reinfection of the liver graft.
Entry as a therapeutic target to address current limitations of DAAs
HCV entry offers many advantages as an alternative antiviral target. Entry inhibitors block the virus life cycle at a step before persistent infection can be established. Indeed, in the absence of de novo infection, hepatocyte turnover likely results in the elimination of infected hepatocytes and leads to clearance of infection [52]. Furthermore, hosttargeting agents aimed at entry factors have a higher genetic barrier for resistance, as the targets are not encoded by highly mutable viral genomes. Entry inhibitors also act synergistically with DAAs [53, 54], which would allow their incorporation into combination regimens. Interestingly, adding an entry inhibitor to DAA therapy reduces breakthrough of DAA-resistant variants, and entry inhibitors have been shown to have strong antiviral activity against DAA-resistant variants [55, 56]. Furthermore, many of the entry inhibitors including natural compounds may be produced at low costs offering a perspective to improve access to therapy in particular in countries or patients with limited resources. The complex and multi-step HCV entry process offers many antiviral targets, and our accumulating knowledge of the virus-host interactions involved in HCV entry opens perspectives to develop antivirals targeting these steps.
Entry inhibitors in preclinical and clinical development
Several compounds have been shown to block HCV binding. These include negatively charged small molecules, such as heparin, heparin-like compounds and polyphenols, which non-specifically compete for binding to cell-surface HSPGs [10, 11, 57–63]. Other molecules target specific receptor binding. For example, the small molecule 281816 (a dibenzothiepin derivative) disrupts the interaction between the HCV E2 protein and CD81 [64]. Similarly, oleanane-type triterpenes and the terpenoid saikosaponin b2 bind to E2 and disrupt E2-CD81 interactions to inhibit HCV entry [65, 66]. Monoclonal antibodies against CD81 and SR-BI also interfere with HCV binding [67–69] and to protect human liver chimeric mice from HCV infection [67–69]. Antibodies targeting SR-BI also reduced viral spread in already infected mice [68, 69]. Conversely, neutralizing antibodies targeting highly conserved epitopes on the viral envelope are also able to inhibit viral binding to CD81 [70–72]. Polyclonal immunoglobulins against HCV have been shown to protect human liver chimeric mice from HCV infection [73,74] and are being evaluated in a clinical trial in the context of graft reinfection (NCT01804829). Antibodies targeting non-virally encoded virion-associated epitopes such as ApoE also interfere with virion binding [9], as do peptides derived from ApoE [75].
Following the binding step, CD81-CLDN1 co-receptor complex formation is a critical step in HCV entry and therefore a most promising antiviral target. Furthermore, the CD81-CLDN1 co-receptor complex has no known physiological role, thereby limiting off-target effects. Monoclonal antibodies targeting the extracellular loops of CLDN1 inhibit CD81-CLDN1 association at a post-binding step. These antibodies inhibit infection by all major genotypes of HCV as well as patient isolates [76–78]. Furthermore, anti-CLDN1 monoclonal antibodies prevent HCV infection in human liver chimeric mice [52, 79]. Notably, one such antibody cured chronically infected mice in monotherapy [52], providing proof-of-concept for the use of entry inhibitors to cure chronic viral infection. Similarly, peptides targeting CLDN1 inhibit HCV entry at a post-binding step [80]. Small molecules targeting EGFR and EphA2, kinases involved in CD81-CLDN1 association, block HCV infection in cell culture and in human liver chimeric mice [27]. An EGFR inhibitor, erlotinib, is in clinical trials for chronic HCV infection (NCT02126137).
Other post-binding steps in the HCV entry process are also targets. SR-BI receptor antagonists, including the arylketoamide ITX5061, inhibit HCV infection following binding [81]. ITX5061 is in a phase 1b clinical trial (NCT01560468). Arbidol (a synthetic indole) and silibinin (a flavonolignan from milk thistle) inhibit HCV clathrin-dependent endosomal trafficking by interfering with dynamin-2-mediated membrane scission [82, 83]. Ezetimibe, a small molecule in clinical use as a cholesterol-lowering agent, inhibits HCV infection by interfering with NPC1L1 internalization [34]. A clinical trial has been initiated to study its effects on chronically infected patients (NCT02126137). Recently, an antihistamine approved for allergy treatment, chlorcyclizine hydrochloride, was shown to inhibit infection by all genotypes of HCV, likely by targeting a late entry step linked to fusion [84].
Fusion is a critical step in the entry of enveloped viruses, including HCV. Indeed, the fusion inhibitor enfuvirtide – a peptide preventing conformational rearrangements of the human immunodeficiency virus (HIV) fusion protein – is approved to treat HIV infections. Our limited understanding of the HCV fusion mechanism currently prevents similar rational design approaches for HCV. However, peptides derived from E2 were shown to inhibit HCV infection at a post-binding step [85]. Furthermore, flunarizine (a piperazine derivative approved for the treatment of migraine headaches) inhibits HCV genotype 2 fusion by targeting E2 and a potential fusion peptide within E1 [42]. An antimalarial compound, ferroquine, inhibited HCV fusion, possibly through its interactions with E1 [86]. HCV infectivity inhibitor-1 (HCV-II1) is thought to lock the HCV envelope in a pre-fusion conformation, thus blocking HCV fusion [87]. An HCV-specific triazine inhibitor, EI-1, interacts with E2 to inhibit a post-binding pre-fusion entry step [88].
Given that lipids play a central role in membrane fusion, molecules that target lipids also modulate fusion of enveloped viruses [89]. Lipid-mimicking rigid amphipathic fusion inhibitors insert into the lipid core of virion envelopes, where they block curvature changes required for fusion of HCV [90, 91]. Membrane fluidity is another critical determinant of fusion. Indeed, modulators of membrane fluidity such as phenothiazine derivatives, benzhydrylpiperazines and curcumin inhibit HCV fusion [92–94]. Similarly, polyunsaturated endoplasmic reticulum-targeting liposomes deplete cellular cholesterol levels to inhibit HCV fusion [95]. Clinical cholesterol-lowering drugs such as statins and ezetimibe inhibit HCV infection [34, 96], perhaps at least partially by modulating membrane fluidity. Type II photosensitizers such as amphiphilic thiazolidine derivatives (e.g. LJ001) generate singlet oxygen species that oxidize phospholipids, leading to biophysical alterations in viral envelopes [97, 98]. These alterations were proposed to increase positive curvature and reduce membrane fluidity, both of which inhibit membrane fusion by increasing the energetics required.
Clearly, many compounds acting by distinct mechanisms show great promise in pre-clinical models; ultimately, clinical studies will determine the future role of HCV entry inhibitors. Several clinical trials are ongoing and will reveal the perspectives for entry inhibitors against HCV to prevent liver graft reinfection or to treat patients who fail DAA-based therapy.
Perspectives
Since the discovery of HCV approximately 25 years ago, major advances in HCV model systems have enabled a detailed understanding of HCV virology and virus-host interactions. These advances allowed the development of DAAs targeting virus replication steps, which have dramatically improved the standard of care for chronically infected patients. Recent advances in the understanding of HCV entry and its clinical impact have set the stage for further development of novel antiviral approaches, which could address the current limitations of DAAs including resistance/failure and access to therapy.
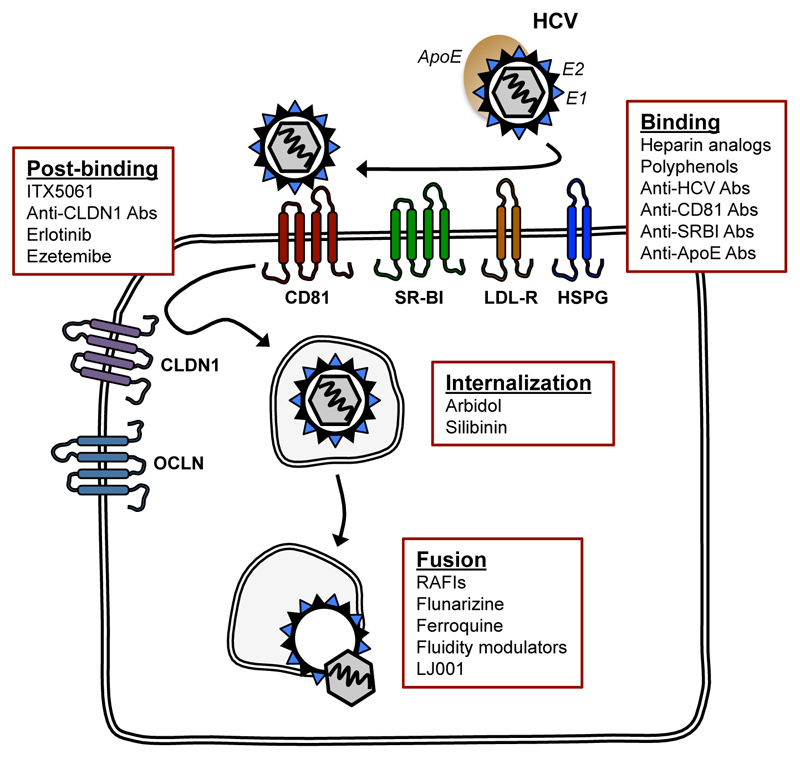
Figure 1.
A simplified scheme of the HCV entry pathway, showing major viral and cellular determinants of viral entry. Inhibitors targeting the main entry steps are also shown.
Table 1. HCV inhibitors targeting different steps of the HCV entry process. Inhibitors in bold are in clinical trials.
| Entry step | Compound | Target/Mechanism | References |
|---|---|---|---|
| Primary binding | Heparin-like molecules | HCV-HSPG interaction | [6, 7, 8, 53] |
| Epigallocatechin gallate | HCV-HSPG interaction | [54–56] | |
| Tannic acid | Docking of HCV at cell surface | [57] | |
| Gallic acid | Docking of HCV at cell surface | [58] | |
| Delphinidin | Docking of HCV at cell surface | [59] | |
| Specific binding | 281816 | HCV E2-CD81 interaction | [60] |
| Oleanane-type triterpenes | HCV E2-CD81 interaction | [61] | |
| Saikosaponin b2 | HCV E2-CD81 interaction | [62] | |
| Anti-CD81 antibody | HCV E2-CD81 interaction | [63] | |
| Anti-SRBI antibody | HCV-SRBI interaction | [64, 65] | |
| Anti-ApoE antibody | HCV-associated ApoE-HSPG interaction | [5] | |
| Neutralizing antibodies | HCV E1/E2 | [66–68] | |
| Polyclonal anti-HCV IgG | HCV E1/E2 | [69, 70] | |
| ApoE-derived peptide | HCV-associated ApoE-HSPG interaction | [71] | |
| Post-binding | Anti-CLDN1 antibodies | CD81-CLDN1 coreceptor complex | [48, 72–75] |
| CLDN1-derived peptide | CD81-CLDN1 coreceptor complex | [76] | |
| Erlotinib | CD81-CLDN1 coreceptor complex; signaling | [23] | |
| ITX5061 | SRBI lipid transfer activity | [77] | |
| Internalization | Arbidol | HCV endosomal trafficking | [78] |
| Silibinin | HCV endosomal trafficking | [79] | |
| Fusion | Flunarizine | HCV genotype 2 fusion (E1 and/or E2) | [38] |
| Ferroquine | HCV fusion (E1) | [81] | |
| RAFIs | HCV envelope curvature | [85, 86] | |
| Phenothiazines | HCV envelope fluidity | [87] | |
| Benzhydrylpiperazines | HCV envelope fluidity | [88] | |
| Curcumin | HCV envelope fluidity | [89] | |
| Polyunsaturated liposomes | Cellular membrane (cholesterol depletion) | [90] | |
| Statins | Cellular membrane (cholesterol depletion) | [91] | |
| Ezetimibe | Cellular membrane (cholesterol depletion) | [30] | |
| Photosensitizers (LJ001) | Lipid oxidation (viral envelope) | 92, 93] |
Acknowledgements
T.F.B. acknowledges support through funding by the European Union (ERC-2008-AdG-HEPCENT, ERC-2014-AdG-HEPCIR, FP7 HepaMAb, H2020 HEPCAR and Interreg IV FEDER-Hepato-Regio-Net 2012), the Agence Nationale de Recherches sur le SIDA (ANRS), the Direction Générale de l’Offre de Soins (A12027MS), Inserm and the University of Strasbourg Foundation. This work has been published under the framework of the LABEX ANR-10-LABX-0028_HEPSYS and benefits from funding from the state managed by the French National Research Agency as part of the Investments for the future program. C.C.C. is supported by a fellowship from the Canadian Institutes of Health Research (201411MFE-338606-245517).
Footnotes
Conflict of interest: the authors declare no conflict of interest. T.F.B. has served as an advisor on HCV antivirals for Biotest, Gilead and Vironexx. T.F.B is a co-inventor on a US patent on anti-Claudin-1 antibody for prevention and treatment of HCV infection filed by Inserm, University of Strasbourg and Genovac/Aldevron.
Author contributions: both authors contributed to the writing of the manuscript.
References
- 1.Thomas DL. Global control of hepatitis C: where challenge meets opportunity. Nat Med. 2013;19:850–858. doi: 10.1038/nm.3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van der Meer AJ, Veldt BJ, Feld JJ, Wedemeyer H, et al. Association between sustained virological response and all-cause mortality among patients with chronic hepatitis C and advanced hepatic fibrosis. JAMA. 2012;308:2584–2593. doi: 10.1001/jama.2012.144878. [DOI] [PubMed] [Google Scholar]
- 3.Ferenci P, Kozbial K, Mandorfer M, Hofer H. HCV targeting of patients with cirrhosis. J Hepatol. 2015;63:1015–1022. doi: 10.1016/j.jhep.2015.06.003. [DOI] [PubMed] [Google Scholar]
- 4.Roche B, Coilly A, Roque-Afonso AM, Samuel D. Interferon-Free Hepatitis C Treatment before and after Liver Transplantation: The Role of HCV Drug Resistance. Viruses. 2015;7:5155–5168. doi: 10.3390/v7092864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chung RT, Baumert TF. Curing chronic hepatitis C--the arc of a medical triumph. N Engl J Med. 2014;370:1576–1578. doi: 10.1056/NEJMp1400986. [DOI] [PubMed] [Google Scholar]
- 6.Colpitts CC, Verrier ER, Baumert TF. Targeting Viral Entry for Treatment of Hepatitis B and C Virus Infections. ACS Infectious Diseases. 2015;1:420–427. doi: 10.1021/acsinfecdis.5b00039. [DOI] [PubMed] [Google Scholar]
- 7.Fofana I, Jilg N, Chung RT, Baumert TF. Entry inhibitors and future treatment of hepatitis C. Antiviral Res. 2014;104:136–142. doi: 10.1016/j.antiviral.2014.02.001. [DOI] [PubMed] [Google Scholar]
- 8.Choo QL, Richman KH, Han JH, Berger K, et al. Genetic organization and diversity of the hepatitis C virus. Proc Natl Acad Sci U S A. 1991;88:2451–2455. doi: 10.1073/pnas.88.6.2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang KS, Jiang J, Cai Z, Luo G. Human apolipoprotein e is required for infectivity and production of hepatitis C virus in cell culture. J Virol. 2007;81:13783–13793. doi: 10.1128/JVI.01091-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barth H, Schafer C, Adah MI, Zhang F, et al. Cellular binding of hepatitis C virus envelope glycoprotein E2 requires cell surface heparan sulfate. J Biol Chem. 2003;278:41003–41012. doi: 10.1074/jbc.M302267200. [DOI] [PubMed] [Google Scholar]
- 11.Barth H, Schnober EK, Zhang F, Linhardt RJ, et al. Viral and cellular determinants of the hepatitis C virus envelope-heparan sulfate interaction. J Virol. 2006;80:10579–10590. doi: 10.1128/JVI.00941-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morikawa K, Zhao Z, Date T, Miyamoto M, et al. The roles of CD81 and glycosaminoglycans in the adsorption and uptake of infectious HCV particles. J Med Virol. 2007;79:714–723. doi: 10.1002/jmv.20842. [DOI] [PubMed] [Google Scholar]
- 13.Xu Y, Martinez P, Seron K, Luo G, et al. Characterization of hepatitis C virus interaction with heparan sulfate proteoglycans. J Virol. 2015;89:3846–3858. doi: 10.1128/JVI.03647-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang J, Cun W, Wu X, Shi Q, et al. Hepatitis C virus attachment mediated by apolipoprotein E binding to cell surface heparan sulfate. J Virol. 2012;86:7256–7267. doi: 10.1128/JVI.07222-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lefevre M, Felmlee DJ, Parnot M, Baumert TF, et al. Syndecan 4 is involved in mediating HCV entry through interaction with lipoviral particle-associated apolipoprotein E. PLoS One. 2014;9:e95550. doi: 10.1371/journal.pone.0095550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Monazahian M, Bohme I, Bonk S, Koch A, et al. Low density lipoprotein receptor as a candidate receptor for hepatitis C virus. J Med Virol. 1999;57:223–229. doi: 10.1002/(sici)1096-9071(199903)57:3<223::aid-jmv2>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 17.Agnello V, Abel G, Elfahal M, Knight GB, et al. Hepatitis C virus and other flaviviridae viruses enter cells via low density lipoprotein receptor. Proc Natl Acad Sci U S A. 1999;96:12766–12771. doi: 10.1073/pnas.96.22.12766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Molina S, Castet V, Fournier-Wirth C, Pichard-Garcia L, et al. The low-density lipoprotein receptor plays a role in the infection of primary human hepatocytes by hepatitis C virus. J Hepatol. 2007;46:411–419. doi: 10.1016/j.jhep.2006.09.024. [DOI] [PubMed] [Google Scholar]
- 19.Owen DM, Huang H, Ye J, Gale MJ. Apolipoprotein E on hepatitis C virion facilitates infection through interaction with low-density lipoprotein receptor. Virology. 2009;394:99–108. doi: 10.1016/j.virol.2009.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dao Thi VL, Dreux M, Cosset FL. Scavenger receptor class B type I and the hypervariable region-1 of hepatitis C virus in cell entry and neutralisation. Expert Rev Mol Med. 2011;13:e13. doi: 10.1017/S1462399411001785. [DOI] [PubMed] [Google Scholar]
- 21.Scarselli E, Ansuini H, Cerino R, Roccasecca RM, et al. The human scavenger receptor class B type I is a novel candidate receptor for the hepatitis C virus. EMBO J. 2002;21:5017–5025. doi: 10.1093/emboj/cdf529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zeisel MB, Koutsoudakis G, Schnober EK, Haberstroh A, et al. Scavenger receptor class B type I is a key host factor for hepatitis C virus infection required for an entry step closely linked to CD81. Hepatology. 2007;46:1722–1731. doi: 10.1002/hep.21994. [DOI] [PubMed] [Google Scholar]
- 23.Catanese MT, Ansuini H, Graziani R, Huby T, et al. Role of scavenger receptor class B type I in hepatitis C virus entry: kinetics and molecular determinants. J Virol. 2010;84:34–43. doi: 10.1128/JVI.02199-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pileri P, Uematsu Y, Campagnoli S, Galli G, et al. Binding of hepatitis C virus to CD81. Science. 1998;282:938–941. doi: 10.1126/science.282.5390.938. [DOI] [PubMed] [Google Scholar]
- 25.Bertaux C, Dragic T. Different domains of CD81 mediate distinct stages of hepatitis C virus pseudoparticle entry. J Virol. 2006;80:4940–4948. doi: 10.1128/JVI.80.10.4940-4948.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Farquhar MJ, Hu K, Harris HJ, Davis C, et al. Hepatitis C virus induces CD81 and claudin-1 endocytosis. J Virol. 2012;86:4305–4316. doi: 10.1128/JVI.06996-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lupberger J, Zeisel MB, Xiao F, Thumann C, et al. EGFR and EphA2 are host factors for hepatitis C virus entry and possible targets for antiviral therapy. Nat Med. 2011;17:589–595. doi: 10.1038/nm.2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brazzoli M, Bianchi A, Filippini S, Weiner A, et al. CD81 is a central regulator of cellular events required for hepatitis C virus infection of human hepatocytes. J Virol. 2008;82:8316–8329. doi: 10.1128/JVI.00665-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zona L, Lupberger J, Sidahmed-Adrar N, Thumann C, et al. HRas signal transduction promotes hepatitis C virus cell entry by triggering assembly of the host tetraspanin receptor complex. Cell Host Microbe. 2013;13:302–313. doi: 10.1016/j.chom.2013.02.006. [DOI] [PubMed] [Google Scholar]
- 30.Evans MJ, von Hahn T, Tscherne DM, Syder AJ, et al. Claudin-1 is a hepatitis C virus co-receptor required for a late step in entry. Nature. 2007;446:801–805. doi: 10.1038/nature05654. [DOI] [PubMed] [Google Scholar]
- 31.Ploss A, Evans MJ, Gaysinskaya VA, Panis M, et al. Human occludin is a hepatitis C virus entry factor required for infection of mouse cells. Nature. 2009;457:882–886. doi: 10.1038/nature07684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harris HJ, Davis C, Mullins JG, Hu K, et al. Claudin association with CD81 defines hepatitis C virus entry. J Biol Chem. 2010;285:21092–21102. doi: 10.1074/jbc.M110.104836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Benedicto I, Molina-Jimenez F, Bartosch B, Cosset FL, et al. The tight junction-associated protein occludin is required for a postbinding step in hepatitis C virus entry and infection. J Virol. 2009;83:8012–8020. doi: 10.1128/JVI.00038-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sainz BJ, Barretto N, Martin DN, Hiraga N, et al. Identification of the Niemann-Pick C1-like 1 cholesterol absorption receptor as a new hepatitis C virus entry factor. Nat Med. 2012;18:281–285. doi: 10.1038/nm.2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martin DN, Uprichard SL. Identification of transferrin receptor 1 as a hepatitis C virus entry factor. Proc Natl Acad Sci U S A. 2013;110:10777–10782. doi: 10.1073/pnas.1301764110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu X, Lee EM, Hammack C, Robotham JM, et al. Cell death-inducing DFFA-like effector b is required for hepatitis C virus entry into hepatocytes. J Virol. 2014;88:8433–8444. doi: 10.1128/JVI.00081-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Park C, Min S, Park EM, Lim YS, et al. Pim Kinase Interacts with Nonstructural 5A Protein and Regulates Hepatitis C Virus Entry. J Virol. 2015;89:10073–10086. doi: 10.1128/JVI.01707-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gerold G, Meissner F, Bruening J, Welsch K, et al. Quantitative Proteomics Identifies Serum Response Factor Binding Protein 1 as a Host Factor for Hepatitis C Virus Entry. Cell Rep. 2015;12:864–878. doi: 10.1016/j.celrep.2015.06.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lavillette D, Bartosch B, Nourrisson D, Verney G, et al. Hepatitis C virus glycoproteins mediate low pH-dependent membrane fusion with liposomes. J Biol Chem. 2006;281:3909–3917. doi: 10.1074/jbc.M509747200. [DOI] [PubMed] [Google Scholar]
- 40.Kong L, Giang E, Nieusma T, Kadam RU, et al. Hepatitis C virus E2 envelope glycoprotein core structure. Science. 2013;342:1090–1094. doi: 10.1126/science.1243876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Khan AG, Whidby J, Miller MT, Scarborough H, et al. Structure of the core ectodomain of the hepatitis C virus envelope glycoprotein 2. Nature. 2014;509:381–384. doi: 10.1038/nature13117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Perin PM, Haid S, Brown RJ, Doerrbecker J, et al. Flunarizine prevents hepatitis C virus membrane fusion in a genotype-dependent manner by targeting the potential fusion peptide within E1. Hepatology. 2015 doi: 10.1002/hep.28111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Timpe JM, Stamataki Z, Jennings A, Hu K, et al. Hepatitis C virus cell-cell transmission in hepatoma cells in the presence of neutralizing antibodies. Hepatology. 2008;47:17–24. doi: 10.1002/hep.21959. [DOI] [PubMed] [Google Scholar]
- 44.Catanese MT, Loureiro J, Jones CT, Dorner M, et al. Different requirements for scavenger receptor class B type I in hepatitis C virus cell-free versus cell-to-cell transmission. J Virol. 2013;87:8282–8293. doi: 10.1128/JVI.01102-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Witteveldt J, Evans MJ, Bitzegeio J, Koutsoudakis G, et al. CD81 is dispensable for hepatitis C virus cell-to-cell transmission in hepatoma cells. J Gen Virol. 2009;90:48–58. doi: 10.1099/vir.0.006700-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hueging K, Doepke M, Vieyres G, Bankwitz D, et al. Apolipoprotein E codetermines tissue tropism of hepatitis C virus and is crucial for viral cell-to-cell transmission by contributing to a postenvelopment step of assembly. J Virol. 2014;88:1433–1446. doi: 10.1128/JVI.01815-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brown RS. Hepatitis C and liver transplantation. Nature. 2005;436:973–978. doi: 10.1038/nature04083. [DOI] [PubMed] [Google Scholar]
- 48.Fafi-Kremer S, Fofana I, Soulier E, Carolla P, et al. Viral entry and escape from antibody-mediated neutralization influence hepatitis C virus reinfection in liver transplantation. J Exp Med. 2010;207:2019–2031. doi: 10.1084/jem.20090766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fofana I, Fafi-Kremer S, Carolla P, Fauvelle C, et al. Mutations that alter use of hepatitis C virus cell entry factors mediate escape from neutralizing antibodies. Gastroenterology. 2012;143:223–233.:e9. doi: 10.1053/j.gastro.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Grove J, Nielsen S, Zhong J, Bassendine MF, et al. Identification of a residue in hepatitis C virus E2 glycoprotein that determines scavenger receptor BI and CD81 receptor dependency and sensitivity to neutralizing antibodies. J Virol. 2008;82:12020–12029. doi: 10.1128/JVI.01569-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mensa L, Crespo G, Gastinger MJ, Kabat J, et al. Hepatitis C virus receptors claudin-1 and occludin after liver transplantation and influence on early viral kinetics. Hepatology. 2011;53:1436–1445. doi: 10.1002/hep.24110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mailly L, Xiao F, Lupberger J, Wilson GK, et al. Clearance of persistent hepatitis C virus infection in humanized mice using a claudin-1-targeting monoclonal antibody. Nat Biotechnol. 2015;33:549–554. doi: 10.1038/nbt.3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xiao F, Fofana I, Thumann C, Mailly L, et al. Synergy of entry inhibitors with direct-acting antivirals uncovers novel combinations for prevention and treatment of hepatitis C. Gut. 2015;64:483–494. doi: 10.1136/gutjnl-2013-306155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Padmanabhan P, Dixit NM. Modeling Suggests a Mechanism of Synergy Between Hepatitis C Virus Entry Inhibitors and Drugs of Other Classes. CPT Pharmacometrics Syst Pharmacol. 2015;4:445–453. doi: 10.1002/psp4.12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xiao F, Fofana I, Heydmann L, Barth H, et al. Hepatitis C virus cell-cell transmission and resistance to direct-acting antiviral agents. PLoS Pathog. 2014;10:e1004128. doi: 10.1371/journal.ppat.1004128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vercauteren K, Brown RJ, Mesalam AA, Doerrbecker J, et al. Targeting a host-cell entry factor barricades antiviral-resistant HCV variants from on-therapy breakthrough in human-liver mice. Gut. 2015 doi: 10.1136/gutjnl-2014-309045. [DOI] [PubMed] [Google Scholar]
- 57.Basu A, Kanda T, Beyene A, Saito K, et al. Sulfated homologues of heparin inhibit hepatitis C virus entry into mammalian cells. J Virol. 2007;81:3933–3941. doi: 10.1128/JVI.02622-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ciesek S, von Hahn T, Colpitts CC, Schang LM, et al. The green tea polyphenol, epigallocatechin-3-gallate, inhibits hepatitis C virus entry. Hepatology. 2011;54:1947–1955. doi: 10.1002/hep.24610. [DOI] [PubMed] [Google Scholar]
- 59.Calland N, Albecka A, Belouzard S, Wychowski C, et al. (-)-Epigallocatechin-3-gallate is a new inhibitor of hepatitis C virus entry. Hepatology. 2012;55:720–729. doi: 10.1002/hep.24803. [DOI] [PubMed] [Google Scholar]
- 60.Colpitts CC, Schang LM. A small molecule inhibits virion attachment to heparan sulfate-or sialic acid-containing glycans. J Virol. 2014;88:7806–7817. doi: 10.1128/JVI.00896-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu S, Chen R, Hagedorn CH. Tannic Acid Inhibits Hepatitis C Virus Entry into Huh7.5 Cells. PLoS One. 2015;10:e0131358. doi: 10.1371/journal.pone.0131358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hsu WC, Chang SP, Lin LC, Li CL, et al. Limonium sinense and gallic acid suppress hepatitis C virus infection by blocking early viral entry. Antiviral Res. 2015;118:139–147. doi: 10.1016/j.antiviral.2015.04.003. [DOI] [PubMed] [Google Scholar]
- 63.Calland N, Sahuc ME, Belouzard S, Pene V, et al. Polyphenols Inhibit Hepatitis C Virus Entry by a New Mechanism of Action. J Virol. 2015;89:10053–10063. doi: 10.1128/JVI.01473-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Al Olaby RR, Cocquerel L, Zemla A, Saas L, et al. Identification of a novel drug lead that inhibits HCV infection and cell-to-cell transmission by targeting the HCV E2 glycoprotein. PLoS One. 2014;9:e111333. doi: 10.1371/journal.pone.0111333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yu F, Wang Q, Zhang Z, Peng Y, et al. Development of oleanane-type triterpenes as a new class of HCV entry inhibitors. J Med Chem. 2013;56:4300–4319. doi: 10.1021/jm301910a. [DOI] [PubMed] [Google Scholar]
- 66.Lin LT, Chung CY, Hsu WC, Chang SP, et al. Saikosaponin b2 is a naturally occurring terpenoid that efficiently inhibits hepatitis C virus entry. J Hepatol. 2015;62:541–548. doi: 10.1016/j.jhep.2014.10.040. [DOI] [PubMed] [Google Scholar]
- 67.Meuleman P, Hesselgesser J, Paulson M, Vanwolleghem T, et al. Anti-CD81 antibodies can prevent a hepatitis C virus infection in vivo. Hepatology. 2008;48:1761–1768. doi: 10.1002/hep.22547. [DOI] [PubMed] [Google Scholar]
- 68.Meuleman P, Catanese MT, Verhoye L, Desombere I, et al. A human monoclonal antibody targeting scavenger receptor class B type I precludes hepatitis C virus infection and viral spread in vitro and in vivo. Hepatology. 2012;55:364–372. doi: 10.1002/hep.24692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lacek K, Vercauteren K, Grzyb K, Naddeo M, et al. Novel human SR-BI antibodies prevent infection and dissemination of HCV in vitro and in humanized mice. J Hepatol. 2012;57:17–23. doi: 10.1016/j.jhep.2012.02.018. [DOI] [PubMed] [Google Scholar]
- 70.Law M, Maruyama T, Lewis J, Giang E, et al. Broadly neutralizing antibodies protect against hepatitis C virus quasispecies challenge. Nat Med. 2008;14:25–27. doi: 10.1038/nm1698. [DOI] [PubMed] [Google Scholar]
- 71.Perotti M, Mancini N, Diotti RA, Tarr AW, et al. Identification of a broadly cross-reacting and neutralizing human monoclonal antibody directed against the hepatitis C virus E2 protein. J Virol. 2008;82:1047–1052. doi: 10.1128/JVI.01986-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Owsianka A, Tarr AW, Juttla VS, Lavillette D, et al. Monoclonal antibody AP33 defines a broadly neutralizing epitope on the hepatitis C virus E2 envelope glycoprotein. J Virol. 2005;79:11095–11104. doi: 10.1128/JVI.79.17.11095-11104.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Meuleman P, Bukh J, Verhoye L, Farhoudi A, et al. In vivo evaluation of the cross-genotype neutralizing activity of polyclonal antibodies against hepatitis C virus. Hepatology. 2011;53:755–762. doi: 10.1002/hep.24171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vanwolleghem T, Bukh J, Meuleman P, Desombere I, et al. Polyclonal immunoglobulins from a chronic hepatitis C virus patient protect human liver-chimeric mice from infection with a homologous hepatitis C virus strain. Hepatology. 2008;47:1846–1855. doi: 10.1002/hep.22244. [DOI] [PubMed] [Google Scholar]
- 75.Liu S, McCormick KD, Zhao W, Zhao T, et al. Human apolipoprotein E peptides inhibit hepatitis C virus entry by blocking virus binding. Hepatology. 2012;56:484–491. doi: 10.1002/hep.25665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fofana I, Krieger SE, Grunert F, Glauben S, et al. Monoclonal anti-claudin 1 antibodies prevent hepatitis C virus infection of primary human hepatocytes. Gastroenterology. 2010;139:953–64.:964.e1. doi: 10.1053/j.gastro.2010.05.073. [DOI] [PubMed] [Google Scholar]
- 77.Yamashita M, Iida M, Tada M, Shirasago Y, et al. Discovery of anti-claudin-1 antibodies as candidate therapeutics against hepatitis C virus. J Pharmacol Exp Ther. 2015;353:112–118. doi: 10.1124/jpet.114.217653. [DOI] [PubMed] [Google Scholar]
- 78.Paciello R, Urbanowicz RA, Riccio G, Sasso E, et al. Novel human anti-Claudin 1 monoclonal antibodies inhibit HCV infection and may synergize with anti-SRB1 mAb. J Gen Virol. 2015 doi: 10.1099/jgv.0.000330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fukasawa M, Nagase S, Shirasago Y, Iida M, et al. Monoclonal antibodies against extracellular domains of claudin-1 block hepatitis C virus infection in a mouse model. J Virol. 2015;89:4866–4879. doi: 10.1128/JVI.03676-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Si Y, Liu S, Liu X, Jacobs JL, et al. A human claudin-1-derived peptide inhibits hepatitis C virus entry. Hepatology. 2012;56:507–515. doi: 10.1002/hep.25685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Syder AJ, Lee H, Zeisel MB, Grove J, et al. Small molecule scavenger receptor BI antagonists are potent HCV entry inhibitors. J Hepatol. 2011;54:48–55. doi: 10.1016/j.jhep.2010.06.024. [DOI] [PubMed] [Google Scholar]
- 82.Blaising J, Levy PL, Polyak SJ, Stanifer M, et al. Arbidol inhibits viral entry by interfering with clathrin-dependent trafficking. Antiviral Res. 2013;100:215–219. doi: 10.1016/j.antiviral.2013.08.008. [DOI] [PubMed] [Google Scholar]
- 83.Blaising J, Levy PL, Gondeau C, Phelip C, et al. Silibinin inhibits hepatitis C virus entry into hepatocytes by hindering clathrin-dependent trafficking. Cell Microbiol. 2013;15:1866–1882. doi: 10.1111/cmi.12155. [DOI] [PubMed] [Google Scholar]
- 84.He S, Lin B, Chu V, Hu Z, et al. Repurposing of the antihistamine chlorcyclizine and related compounds for treatment of hepatitis C virus infection. Sci Transl Med. 2015;7:282ra49. doi: 10.1126/scitranslmed.3010286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Liu R, Tewari M, Kong R, Zhang R, et al. A peptide derived from hepatitis C virus E2 envelope protein inhibits a post-binding step in HCV entry. Antiviral Res. 2010;86:172–179. doi: 10.1016/j.antiviral.2010.02.316. [DOI] [PubMed] [Google Scholar]
- 86.Vausselin T, Calland N, Belouzard S, Descamps V, et al. The antimalarial ferroquine is an inhibitor of hepatitis C virus. Hepatology. 2013;58:86–97. doi: 10.1002/hep.26273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bush CO, Pokrovskii MV, Saito R, Morganelli P, et al. A small-molecule inhibitor of hepatitis C virus infectivity. Antimicrob Agents Chemother. 2014;58:386–396. doi: 10.1128/AAC.02083-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Baldick CJ, Wichroski MJ, Pendri A, Walsh AW, et al. A novel small molecule inhibitor of hepatitis C virus entry. PLoS Pathog. 2010;6:e1001086. doi: 10.1371/journal.ppat.1001086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Vigant F, Santos NC, Lee B. Broad-spectrum antivirals against viral fusion. Nat Rev Microbiol. 2015;13:426–437. doi: 10.1038/nrmicro3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.St Vincent MR, Colpitts CC, Ustinov AV, Muqadas M, et al. Rigid amphipathic fusion inhibitors, small molecule antiviral compounds against enveloped viruses. Proc Natl Acad Sci U S A. 2010;107:17339–17344. doi: 10.1073/pnas.1010026107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Colpitts CC, Ustinov AV, Epand RF, Epand RM, et al. 5-(Perylen-3-yl)ethynyl-arabino-uridine (aUY11), an arabino-based rigid amphipathic fusion inhibitor, targets virion envelope lipids to inhibit fusion of influenza virus, hepatitis C virus, and other enveloped viruses. J Virol. 2013;87:3640–3654. doi: 10.1128/JVI.02882-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chamoun-Emanuelli AM, Pecheur EI, Simeon RL, Huang D, et al. Phenothiazines inhibit hepatitis C virus entry, likely by increasing the fluidity of cholesterol-rich membranes. Antimicrob Agents Chemother. 2013;57:2571–2581. doi: 10.1128/AAC.02593-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chamoun-Emanuelli AM, Pecheur EI, Chen Z. Benzhydrylpiperazine compounds inhibit cholesterol-dependent cellular entry of hepatitis C virus. Antiviral Res. 2014;109:141–148. doi: 10.1016/j.antiviral.2014.06.014. [DOI] [PubMed] [Google Scholar]
- 94.Anggakusuma ColpittsCC, Schang LM, Rachmawati H, et al. Turmeric curcumin inhibits entry of all hepatitis C virus genotypes into human liver cells. Gut. 2014;63:1137–1149. doi: 10.1136/gutjnl-2012-304299. [DOI] [PubMed] [Google Scholar]
- 95.Pollock S, Nichita NB, Bohmer A, Radulescu C, et al. Polyunsaturated liposomes are antiviral against hepatitis B and C viruses and HIV by decreasing cholesterol levels in infected cells. Proc Natl Acad Sci U S A. 2010;107:17176–17181. doi: 10.1073/pnas.1009445107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ikeda M, Abe K, Yamada M, Dansako H, et al. Different anti-HCV profiles of statins and their potential for combination therapy with interferon. Hepatology. 2006;44:117–125. doi: 10.1002/hep.21232. [DOI] [PubMed] [Google Scholar]
- 97.Wolf MC, Freiberg AN, Zhang T, Akyol-Ataman Z, et al. A broad-spectrum antiviral targeting entry of enveloped viruses. Proc Natl Acad Sci U S A. 2010;107:3157–3162. doi: 10.1073/pnas.0909587107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Vigant F, Lee J, Hollmann A, Tanner LB, et al. A mechanistic paradigm for broad-spectrum antivirals that target virus-cell fusion. PLoS Pathog. 2013;9:e1003297. doi: 10.1371/journal.ppat.1003297. [DOI] [PMC free article] [PubMed] [Google Scholar]