Abstract
In prostate cancer, androgen receptor (AR)-targeting agents are very effective in various disease stages. However, therapy resistance inevitably occurs and little is known about how tumor cells adapt to bypass AR suppression. Here, we performed integrative multi-omics analyses on tissues isolated before and after 3 months of AR-targeting enzalutamide monotherapy from high-risk prostate cancer patients enrolled in a neoadjuvant clinical trial. Transcriptomic analyses demonstrated that AR inhibition drove tumors towards a neuroendocrine-like disease state. Additionally, epigenomic profiling revealed massive enzalutamide-induced reprogramming of pioneer factor FOXA1 – from inactive chromatin sites towards active cis-regulatory elements that dictate pro-survival signals. Notably, treatment-induced FOXA1 sites were enriched for circadian clock component ARNTL. Post-treatment ARNTL levels associated with poor outcome, and ARNTL knockout strongly decreased prostate cancer cell growth. Our data highlight a remarkable cistromic plasticity of FOXA1 following AR-targeted therapy, and revealed an acquired dependency on circadian regulator ARNTL, a novel candidate therapeutic target.
Keywords: Prostate cancer, neoadjuvant clinical trial, Androgen Receptor, enzalutamide, epigenetic plasticity, FOXA1, circadian rhythm, ARNTL
Introduction
Androgen ablation is the mainstay treatment for patients with metastatic prostate cancer (PCa), ever since the direct critical link between androgens and prostate tumor progression was first described (1). The androgen receptor (AR) is the key driver of PCa development and progression, and multiple therapeutic strategies have been developed over the years to effectively block the activity of this hormone-driven transcription factor. Upon androgen binding, AR associates with the chromatin at distal cis-regulatory enhancer elements, where it regulates the expression of genes through long-range chromatin interactions in three-dimensional genomic space (2,3). AR does not operate in isolation, but rather recruits a large spectrum of coregulators and other transcription factors to promote expression of genes that drive cancer cell proliferation (4). Critical AR interactors in the transcription complex are HOXB13 and FOXA1, which are both upregulated in primary PCa (4–6) and demarcate enhancers that drive not only primary tumorigenesis but also metastatic disease progression (7). Mechanistically, FOXA1 acts as a pioneer factor, rendering the chromatin accessible for AR to bind (8–11). FOXA1 is frequently mutated in PCa (12–16) which was shown to alter its pioneering capacities, perturb luminal epithelial differentiation programs, and promote tumor growth, further highlighting the critical role of FOXA1 in human prostate tumors (17,18).
Most patients are diagnosed with organ-confined PCa, which can potentially be cured through locoregional therapies, such as surgery (radical prostatectomy), radiotherapy and/or brachytherapy (19). However, approximately 30% of these patients experience a biochemical recurrence (BCR) – a rise in prostate-specific antigen (PSA) serum levels – indicating PCa relapse (20). At this stage of the disease, suppression of androgen production is a commonly applied therapeutic intervention that can delay further cancer progression for years (21,22). Nevertheless, the development of resistance to androgen deprivation is inevitable, resulting in castration-resistant prostate cancer (CRPC) for which there is no cure (23). Most CRPC tumors acquired molecular features that enable active AR signaling despite low circulating androgen levels, a finding that led to the development of several highly effective AR-targeted therapies. Enzalutamide (ENZ) is one of the most frequently used AR-targeting agents, which functions through a combined mechanism of blocked AR nuclear import, diminished AR chromatin binding and decreased transcription complex formation, effectively impairing AR-driven PCa growth (24). ENZ’s potent anti-tumor activity has been demonstrated in multiple clinical trials, which led to its FDA approval in various PCa disease stages – from metastatic CRPC (25,26), to metastatic hormone-sensitive (27), and even non-metastatic CRPC (28) – illustrating how AR-targeted therapies are being progressively introduced earlier in clinical practice. A clinical benefit of ENZ monotherapy as a neoadjuvant treatment prior to prostatectomy for patients with localized disease, has not been established. Although effective in the CRPC setting, resistance to AR pathway inhibition will ultimately develop, and the management of advanced PCa with this acquired resistance remains a major clinical challenge, especially since the underlying mechanisms are still not fully elucidated (29). Therefore, furthering our understanding of how ENZ affects PCa biology may lead to the identification of acquired cellular vulnerabilities that could be therapeutically exploited.
To study global drug-induced transcriptional and epigenetic plasticity in human prostate tumors and identify cellular adaptation mechanisms to evade drug treatment, we designed a phase 2 clinical trial to perform multi-omics studies in pre- and post-treatment samples from high-risk localized PCa patients, treated with neoadjuvant ENZ monotherapy. We identified transcriptional reprogramming after treatment, with deactivation of AR signaling and an activation of cell plasticity with neuroendocrine (NE)-like features upon 3 months of AR suppression. Post treatment, these tumors harbored a distinct set of 1,430 de novo occupied FOXA1-positive cis-regulatory elements, positive for – yet independent of – AR activity, which are dictated by circadian clock core regulator ARNTL to drive tumor cell proliferation instead. Using ARNTL knockout experiments we could further enhance ENZ sensitivity in cell line and xenograft models, revealing an unexpected biological interplay between hormonal resistance and circadian rhythm regulation, and identifying a novel highly promising candidate drug target in the clinical management of primary high-risk PCa.
Results
Neoadjuvant ENZ therapy for patients with high-risk localized PCa
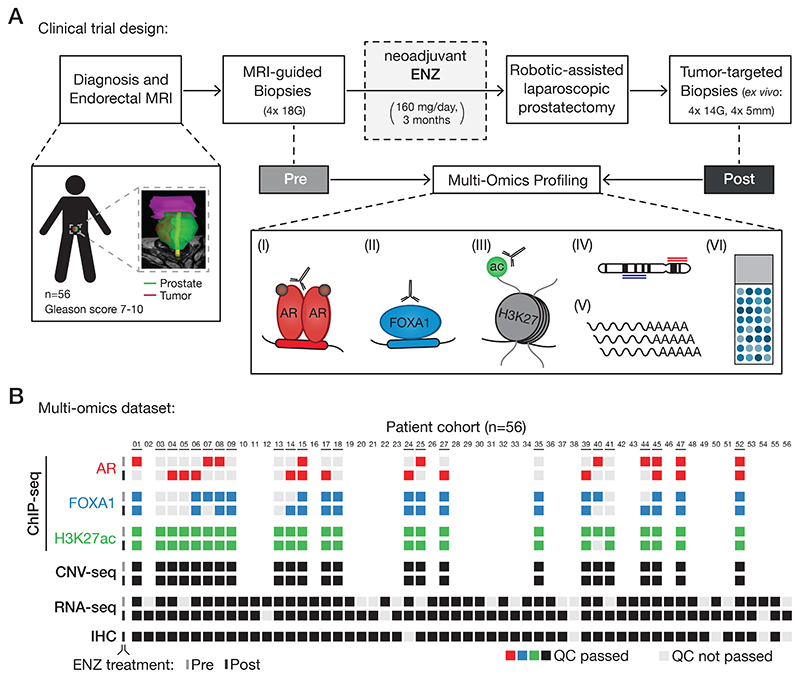
To study how early ENZ intervention affects prostate tumor biology in a non-castrate environment, we performed integrative multi-omics analyses as part of a single-arm, open-label phase 2 clinical trial: the DARANA study (Dynamics of Androgen Receptor Genomics and Transcriptomics After Neoadjuvant Androgen Ablation; ClinicalTrials.gov number, NCT03297385). In this trial, 56 men with primary high-risk (Gleason score ≥ 7) PCa were enrolled (Fig. 1A). Patient demographics and disease characteristics are summarized in Table 1, and clinical outcomes of this study are discussed in the Supplementary Data (Supplementary Fig. S1A–F; Supplementary Table S1). Prior to ENZ therapy, magnetic resonance imaging (MRI)-guided core needle tumor biopsies were taken – hereafter referred to as the pre-treatment setting. Subsequently, patients received neoadjuvant ENZ treatment (160 mg/day) without additional androgen deprivation therapy for three months, followed by robotic-assisted laparoscopic prostatectomy. Based on baseline MRI information and palpation, additional tumor-targeted core needle biopsies were taken ex vivo – representing the post-treatment setting. This pre- and post-treatment sampling allowed us to study the epigenetic, genomic, transcriptomic and proteomic effects of neoadjuvant ENZ therapy in individual patients (Fig. 1A). We generated chromatin immunoprecipitation (ChIP-seq) profiles of the prostate cancer drivers AR and FOXA1, as well as the histone modification H3K27ac before and after ENZ treatment, and integrated these cistromic findings with pre- and post-treatment gene expression (RNA-seq), copy number (CNV-seq) and immunohistochemistry (IHC) data from the same tumors. Stringent quality control (QC) analyses were performed on all data streams (Supplementary Fig. S2), and the following number of samples passed all QC measures (Fig. 1B): AR ChIP-seq (pre: n=10; post: n=12), FOXA1 ChIP-seq (pre: n=17; post: n=17), H3K27ac ChIP-seq (pre: n=24; post: n=23), CNV-seq (pre: n=24; post: n=24), RNA-seq (pre: n=42; post: n=52) and IHC (post: n=51).
Figure 1. Clinical trial design and omics data sample collection.
(A) Study design of the DARANA trial (NCT03297385). Multi-omics profiling, consisting of (I) Androgen Receptor (AR) ChIP-seq, (II) FOXA1 ChIP-seq, (III) H3K27ac ChIP-seq, (IV) DNA copy number sequencing (CNV-seq), (V) gene expression profiling (RNA-seq) and (VI) immunohistochemistry (IHC) analysis, was performed on MRI-guided biopsy samples prior to ENZ treatment (Pre) and tumor-target prostatectomy specimens after 3 months of neoadjuvant ENZ therapy (Post).
(B) Overview of data availability and quality control analyses for each sample. Individual data streams are indicated separately with ChIP-seq for AR (red), FOXA1 (blue), H3K27ac (green), CNV-seq, RNA-seq and IHC (all black). The ENZ treatment status indicates the pre-treatment (top) and post-treatment samples (bottom) per omics dataset. Samples not passing QC (light gray) were successfully applied for focused raw data analyses. Blank spots for ChIP-seq or CNV-seq samples indicate that the fresh-frozen material didn’t pass the tumor cell percentage cutoff of ≥ 50%.
Table 1. Characteristics of the DARANA cohort (n=56).
Table summarizing the patient baseline demographics, and pre- and post-treatment disease characteristics of the DARANA cohort. Shown are age (years), initial Prostate-specific antigen (PSA) serum levels (ng/mL) and International Society of Urological Pathology (ISUP) grade at diagnosis (with associated Gleason scores (GS)). In addition, T-stage (T) and Lymph node status (N) before (pre = at diagnosis) and after (post = at surgery) neoadjuvant ENZ therapy, as well as the surgical margin status of the prostatectomy specimens are shown. Pre-treatment measures are based on histological evaluation of biopsy material and radiographic evaluation (clinical grading; c), while post-treatment assessments are based on histological evaluations of prostatectomy specimens (pathological grading after neoadjuvant therapy; yp). Biochemical recurrence (BCR) was defined as a rise in PSA of ≥ 0.2 ng/mL at two consecutive time points, radiological recurrence (RR) was defined as detection of local or distant metastases by PSMA PET scanning, ADT salvage therapy was defined as the onset of Androgen deprivation therapy (ADT), and distant metastases (DM) were defined as detection of distant metastases by PSMA PET scanning (M1a-c). 5-year recurrence-free survival (% of patients and 95% CI) and time to last follow-up (months) are indicated. For continuous variables (age, baseline PSA, and time to last follow-up) the mean and 95% confidence interval (CI) are shown. For categorical variables (baseline ISUP, T-stage, N-status, surgical margins, BCR, RR, ADT, DM) the number of patients (no.) and percentages (%) are indicated.
| DARANA cohort (n= 56) | ||
|---|---|---|
| Age – years (95% CI) | 67 (65-68) | |
| Baseline PSA level – ng/mL (95% CI) | 12.8 (10.4-15.2) | |
| Baseline ISUP grade – no. (%) | ||
| ISUP 1 (GS 3+3) | 0 (0) | |
| ISUP 2 (GS 3+4) | 16 (28) | |
| ISUP 3 (GS 4+3) | 9 (16) | |
| ISUP 4 (GS 4+4, 3+5, 5+3) | 20 (36) | |
| ISUP 5 (GS 4+5, 5+4, 5+5) | 11 (20) | |
| T-stage (T) – no. (%) | Pre (cT) | Post (ypT) |
| T1 | 1 (2) | / |
| T2 | 25 (44) | 20 (36) |
| T3 | 29 (52) | 36 (64) |
| T4 | 1 (2) | 0 (0) |
| Lymph node status (N) – no. (%) | Pre (cN) | Post (ypN) |
| N0 | 53 (95) | 39 (70) |
| N1 | 3 (5) | 17 (30) |
| Surgical margins – no. (%) | ||
| Negative | 39 (70) | |
| Positive | 17 (30) | |
| Biochemical recurrence (BCR) – no. (%) | 23 (41) | |
| 5-year BCR-free survival – % (95% CI) | 38 (28-51) | |
| Radiological recurrence (RR) – no. (%) | 18 (32) | |
| 5-year RR-free survival – % (95% CI) | 64 (50-82) | |
| ADT salvage therapy (ADT) – no. (%) | 15 (27) | |
| 5-year ADT-free survival – % (95% CI) | 67 (53-85) | |
| Distant metastasis (DM) – no. (%) | 16 (28) | |
| 5-year DM-free survival – % (95% CI) | 74 (61-91) | |
| Mean time to last follow-up – months (95% CI) | 51 (47-55) | |
Collectively, we performed integrative multi-omics analyses as part of a clinical trial that enabled us to examine ENZ-induced oncogenomic changes to identify early epigenetic steps in treatment response, but also therapy-induced resistance.
Characterization of tissue ChIP-seq data
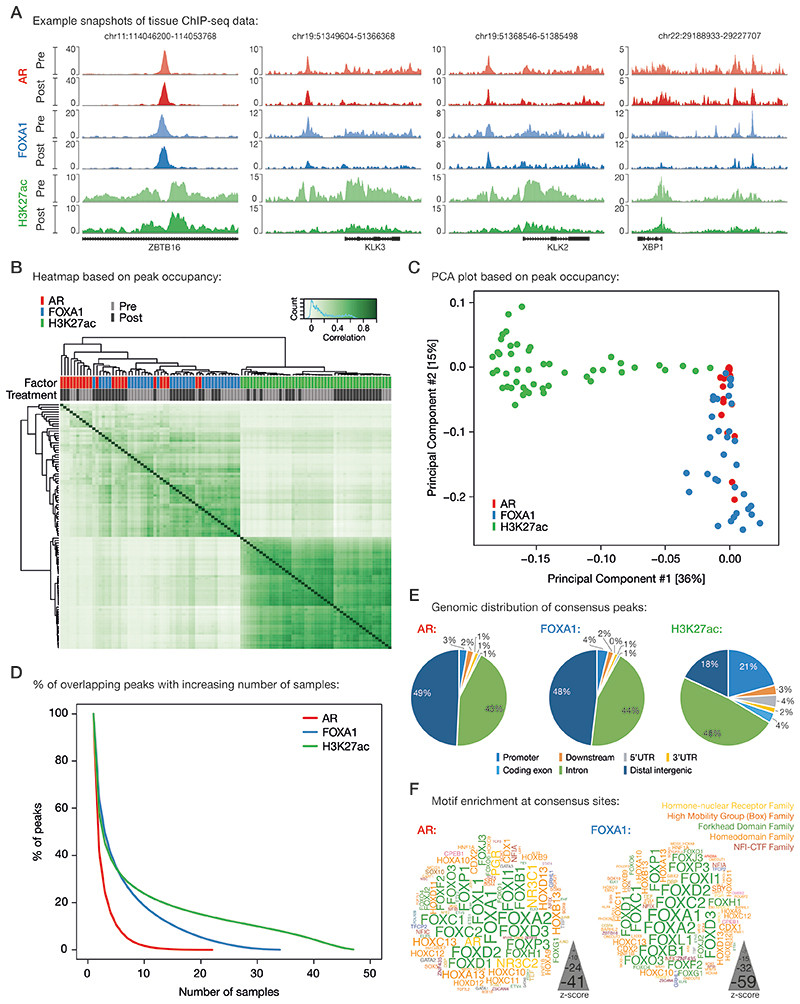
To assess how neoadjuvant ENZ treatment affects the cis-regulatory landscape in primary PCa, we generated human tumor ChIP-seq profiles for the transcription factors AR and FOXA1, along with the active enhancer/promoter histone mark H3K27ac before and after neoadjuvant intervention. ChIP-seq quality metrics are summarized in the Supplementary Data (Supplementary Fig. S3A–E and Supplementary Table S2). Visual inspection at known AR target genes showed high-quality data for all ChIP-factors in both clinical settings (Fig. 2A). On a genome-wide scale, the H3K27ac ChIP-seq profiles were highly distinct from the transcription factors (TFs) and divided the samples into two main clusters irrespective of their treatment status (Fig. 2B and 2C). Notably, AR and FOXA1 ChIP-seq datasets were intermingled in the clustering analysis, suggesting largely comparable binding profiles which is in line with FOXA1’s role as a canonical AR pioneer factor (Supplementary Fig. S4) (5,30). As described previously (31), highest Pearson correlation was found between H3K27ac samples, indicating comparable histone acetylation profiles among primary PCa samples (Fig. 2B; Supplementary Fig. S4). Much greater heterogeneity in chromatin binding was observed for the TFs AR and FOXA1, which is further supported by the steep decrease in the number of overlapping AR and FOXA1 peaks with increasing number of samples compared to H3K27ac (Fig. 2D; Supplementary Fig. S4). Heterogeneity was comparable when separately analyzing pre- versus post-treatment specimens, and in the same order of magnitude as compared to previous reports describing TF cistromics and epigenomics in clinical samples (31,32) (Supplementary Fig. S5A and S5B) with comparable overlap of peaks for AR and FOXA1 (Supplementary Fig. S5C and S5D). In order to maintain the high-confidence peaks that have been reproducibly identified in multiple patients without losing too much binding site heterogeneity between samples, we decided to generate consensus peaksets. To this end, we only considered binding sites that were present in at least 3 out of 22 AR samples, 7 out of 34 FOXA1 samples and 13 out of 47 H3K27ac samples, which corresponds to ~25% of all binding sites identified for each factor (Fig. 2D). Genomic distribution analyses of these consensus sites revealed distinct enrichments for annotated genomic regions: While AR and FOXA1 were almost exclusively found at intronic and distal intergenic regions, H3K27ac peaks were also enriched at promoters (Fig. 2E), which is in line with previously published genomic distributions of AR (5,31), FOXA1 (5,9), and H3K27ac (31,33). In addition, motif enrichment analyses at AR and FOXA1 consensus peaks identified, as expected, androgen and Forkhead response elements among the top-ranked motifs, respectively (Fig. 2F). Analyses on correlations between factors (Fig. 2B-D), genomic distributions (Fig. 2E) and motif enrichment (Fig. 2F) were repeated for the pre-treatment samples exclusively, supporting the same conclusions (Supplementary Fig. S6A–E).
Figure 2. Characterization of tissue ChIP-seq data streams.
(A) Representative example snapshots of AR (red), FOXA1 (blue) and H3K27ac (green) ChIP-seq data for four genomic loci in one patient. Pre- (light colors) and post-ENZ treatment (dark colors) is indicated. Y-axes indicate ChIP-seq signal in fragments per kilobase per million reads mapped (FPKM).
(B) Correlation heatmap based on peak occupancy. Clustering of the samples is based on all called peaks and represents Pearson correlations between individual ChIP-seq samples. The column color bars indicate the ChIP-seq factor (AR, FOXA1, H3K27ac) and treatment status (Pre, Post).
(C) Principal component analysis (PCA) plot based on peak occupancy. Each dot represents a ChIP-seq sample that is colored per factor.
(D) Elbow plot depicting the peak overlap between ChIP-seq samples per factor. Shown is the percentage of overlapping peaks with increasing number of samples. Consensus peaksets were designed by using a cutoff of peaks present in at least 3 AR, 7 FOXA1, or 13 H3K27ac samples.
(E) Pie charts showing the genomic distribution of AR (left), FOXA1 (middle) and H3K27ac (right) consensus peaks.
(F) Word clouds show motif enrichment at AR (left) and FOXA1 (right) consensus sites. The font size represents the z-score and colors correspond to transcription factor families.
Taken together, we generated multiple high-quality tissue ChIP-seq data streams that now allowed us to study ENZ-induced changes in primary PCa patients.
ENZ treatment enriches for newly acquired FOXA1-bound regulatory regions
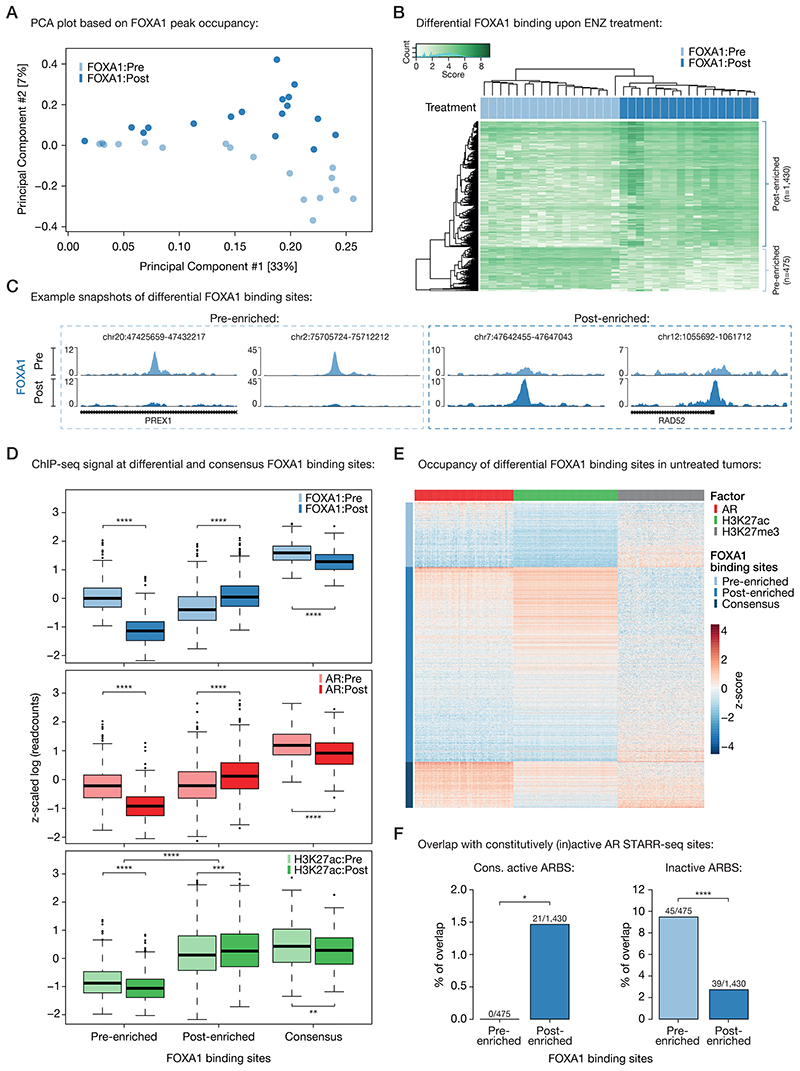
To identify ENZ-induced TF reprogramming and epigenetic changes, we performed differential binding analyses comparing the pre- and post-treatment tissue ChIP-seq samples. Therefore, we first ran occupancy-based unsupervised principal component analyses (PCA) to detect whether ENZ treatment led to differences in TF chromatin binding. While the sample size of the AR ChIP-seq data stream was not sufficient to observe significant differences in peak occupancy pre- versus post-treatment (Supplementary Fig. S7A), the FOXA1 data did show such differences, with a clear separation of pre- and post-treatment FOXA1 samples in the second principal component (Fig. 3A). Subsequent supervised analysis (pre vs. post) revealed a total of 1,905 genomic regions (475 pre-enriched, 1,430 post-enriched; Supplementary Table S3) that showed significant differential FOXA1 binding between both clinical settings (FDR < 0.05; Fig. 3B and 3C; Supplementary Fig. S7B–D). Further characterization of these differential FOXA1 regions showed that both sets of binding sites were still preferentially located in intronic and distal intergenic regions (with a slight enrichment for promoters at the post-enriched sites; Supplementary Fig. S7E). In addition, Forkhead domain family motifs were the top enriched motifs at both pre- and post-enriched sites, illustrating that treatment does not alter FOXA1 motif preference and still occupies canonical FOXA1 binding sites (Supplementary Fig. S7F).
Figure 3. Differential FOXA1 binding upon ENZ treatment.
(A) Principal component analysis (PCA) plot based on peak occupancy of FOXA1 ChIP-seq data. Color indicates pre-treatment (light blue) and post-treatment (dark blue) FOXA1 samples.
(B) Coverage heatmap depicting differential FOXA1 binding sites, selectively enriched in the pre-treatment (n=475) or post-treatment (n=1,430) setting.
(C) Representative example snapshots of FOXA1 ChIP-seq signal at two pre-enriched (left) and two post-enriched (right) FOXA1 sites in one patient (DAR45). Pre- (light blue) and post-ENZ treatment (dark blue) is indicated. Y-axes indicate ChIP-seq signal in FPKM.
(D) Boxplots indicating ChIP-seq signal (z-scaled readcounts) at pre-enriched (n=475), post-enriched (n=1,430) and consensus FOXA1 peaks (shared by ≥ 30 patients; n=338) for FOXA1 (blue), AR (red), and H3K27ac (green) ChIP-seq datasets before (Pre; light colors) and after (Post; dark colors) ENZ treatment. **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 (Mann-Whitney U-test adjusted for multiple testing using FDR).
(E) Coverage heatmap showing occupancy of differential (pre-/post-enriched) and consensus FOXA1 peaks in an external ChIP-seq dataset consisting of 100 untreated primary tumors (31). Heatmap color indicates region read counts (z-score) at pre-enriched, post-enriched and consensus FOXA1 sites (rows) in the AR (red), H3K27ac (green) and H3K27me3 (gray) ChIP-seq data streams (columns).
(F) Bar chart representing the overlap between differential FOXA1 sites (pre-enriched or post-enriched) and constitutively active (left) or inactive (right) AR binding sites (ARBS), based on STARR-seq. *, P < 0.05; ****, P < 0.0001 (Fisher’s exact test).
To examine whether structural variations are underlying these differential FOXA1 binding events, we performed CNV-seq on the same tumor specimens and then projected onto the differential FOXA1 cistromics the structural copy-number data. These analyses revealed a comparable level of CNV at pre- and post-treatment enriched FOXA1 sites before and after ENZ treatment, with an overall trend towards less CNV upon treatment (Supplementary Fig. S8A–C). However, in none of the matched sample pairs (pre and post CNV-seq, and FOXA1 ChIP-seq; n=15) a strong correlation between copy number difference and ChIP-seq signal difference was observed (R = 0.11; Supplementary Fig. S8D). In total, at only 44 out of 1,905 differential FOXA1 binding sites (< 2.5%), we observed copy number differences between post- and pre-treatment samples that could potentially explain binding site occupancy in 3 or more patients, indicating that the vast majority of these differential binding events is based on treatment-induced transcription factor reprogramming, rather than structural variation (Supplementary Fig. S8E).
As FOXA1 dictates AR chromatin binding capacity (5), epigenetic plasticity of FOXA1 induced by treatment may be associated with alterations in the AR cistrome. To assess this, and to explore the epigenetic landscape surrounding the differentially bound FOXA1 regions, we compared the ChIP-seq signal of all three factors (AR, FOXA1, H3K27ac) at differential (pre- / post-enriched) and consensus (shared by ≥ 30 patients; n=338) FOXA1 sites before and after ENZ therapy. While the FOXA1 ChIP-seq signal was highest at consensus binding sites, the pre- and post-treatment enriched regions followed the expected trend and showed significantly higher signal in the corresponding settings (Fig. 3D). Notably, we also observed less binding of FOXA1 to consensus sites when treated with ENZ, although the differences are much milder compared to the effects seen at pre-enriched FOXA1 sites (Padj = 3.62 × 10-22 at consensus vs. 3.76 × 10-130 at pre-enriched sites, Mann-Whitney U-test; Fig. 3D; Supplementary Fig. S9A). This could possibly be explained by decreased FOXA1 gene expression levels upon ENZ treatment (Supplementary Fig. S9B). The AR ChIP-seq signal followed the same patterns as observed for FOXA1, suggesting that relocated FOXA1 upon treatment functionally drives alterations in the AR cistrome (Fig. 3D). Unexpectedly, the pre-enriched FOXA1 sites were completely devoid of any H3K27ac signal in both pre- and post-treatment samples, while the post-enriched counterparts were positive for this active enhancer/promoter mark with a significant increase post-ENZ (Padj = 5.59 × 10-4, Mann-Whitney U-test; Fig. 3D; Supplementary Fig. S9C and S9D), suggesting that pre-ENZ FOXA1 sites are inactive. To validate these observations in an independent cohort, we analyzed previously published AR (n=87), H3K27ac (n=92) and H3K27me3 (n=76) ChIP-seq data from a cohort of 100 primary treatment-naïve PCa samples (31). Supporting our previous analyses, the vast majority of post-enriched FOXA1 sites were H3K27ac-positive and their histone acetylation status positively correlated with AR binding (R = 0.78) (Fig. 3E; Supplementary Fig. S9E). The pre-enriched FOXA1 sites, however, were again H3K27ac-negative, while the repressive histone modification H3K27me3 was present, which further points towards an inactive epigenetic state of these regulatory regions (Fig. 3E).
Recently, we reported that prostate cancers can reactivate developmental programs during metastatic progression (7). These sentinel enhancers appeared to be pre-marked by FOXA1 from prostate gland development, and albeit inactive in normal prostate and primary tumor specimens, the sites get reactivated by AR during metastatic outgrowth. Given the inactivity of the pre-enriched FOXA1 sites, we hypothesized that FOXA1 might be decommissioned at such developmental enhancers prior to hormonal intervention. To test this, we overlapped the differential FOXA1 binding sites with the metastasis-specific AR binding sites (met-ARBS; n=17,655), which revealed a strong enrichment for these developmental regulatory elements at pre-treatment FOXA1 sites (P = 2.13 × 10-16, Fisher’s exact test; Supplementary Fig. S9F). But are the inactive pre-enriched FOXA1 sites solely epigenetically suppressed, or are these regions intrinsically incapable of being active in this cellular context? To address this question and to further elucidate the role of AR at these differentially bound FOXA1 sites, we integrated our tissue ChIP-seq findings with previously identified tumor-specific AR binding sites (n=3,230) (5) that were functionally characterized using Self-Transcribing Active Regulatory Regions sequencing (STARR-seq), a massive parallel reporter assay to systematically annotate intrinsic enhancer activity (34). With this, three distinct classes of AR binding sites (ARBS) were identified (Supplementary Table S4): enhancers that were active regardless of AR stimulation (constitutively active; n=465), ARBS with no significant enhancer activity (inactive; n=2,479) and inducible AR enhancers that increase activity upon androgen treatment (inducible; n=286). Interestingly, we found that post-treatment FOXA1 sites were enriched for constitutively active ARBS, which further supports the high enhancer activity and H3K27ac positivity observed at these sites, but also illustrates that this activity is constitutive and AR-independent (Fig. 3F). Consistent with our postulated inactivity of the pre-treatment enriched FOXA1 sites, these regions overlapped highly significantly with inactive ARBS (P = 8.60 × 10-9, Fisher’s exact test), which implies that these DNA elements are intrinsically inactive and incapable to act as functional enhancers, and possibly explains why these AR-bound sites did not show active regulatory marks (Fig. 3E and 3F). As no enrichment of our differential FOXA1 sites was observed with inducible ARBS (pre-enriched: 4/475; post-enriched: 2/1,430), these data further support a conclusion that AR itself is not a driver at FOXA1 sites that are differentially occupied after ENZ exposure in patients.
Overall, these results suggest that prior to hormonal intervention, FOXA1 is decommissioned at inactive developmental enhancer elements, which based on their primary DNA sequence are intrinsically incapable of being active – at least in the tested hormone-sensitive disease setting. However, upon ENZ treatment, FOXA1 gets reprogrammed to highly active cis-regulatory regions, which act in an AR-independent manner.
Transcriptional rewiring upon neoadjuvant ENZ
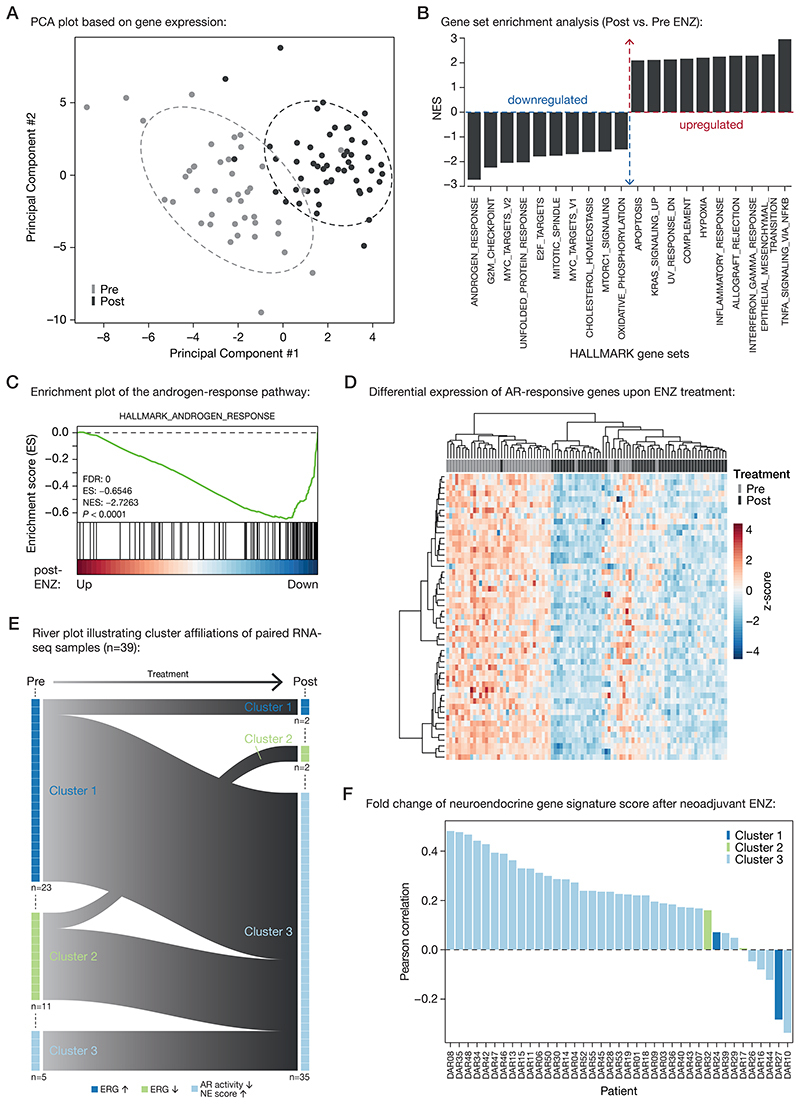
Having assessed the cistromic and epigenomic changes in response to neoadjuvant ENZ, we next determined how transcriptional programs were affected by this hormonal intervention. Principal component analysis (PCA) across both treatment states revealed that three months of ENZ therapy has a major effect on global gene expression profiles (Fig. 4A). Subsequently, we performed differential gene expression analysis, in which we compared pre- and post-treatment RNA-seq samples. Gene set enrichment analysis (GSEA) showed that AR signaling, along with mitosis and MYC signals, was strongly decreased upon treatment (Fig. 4B and C; Supplementary Fig. S10A). Since ENZ blocks the AR signaling axis, we analyzed the androgen-response pathway in more detail, which revealed a strong downregulation of AR target genes in almost every patient (Fig. 4D). In contrast to this, TNFα signaling, IFN-γ response and epithelial-mesenchymal transition (EMT) signals were most upregulated (Fig. 4B; Supplementary Fig. S10B).
Figure 4. Neoadjuvant ENZ deactivates AR signaling and induces neuroendocrine (NE)-like gene expression signatures.
(A) Principal component analysis (PCA) plot based on gene expression data. Color indicates pre-treatment (gray) and post-treatment (black) samples. Ellipses are based on the 80% confidence interval.
(B) Gene set enrichment analyses (GSEA) for Hallmark gene sets. Shown are the top differentially enriched pathways upon ENZ treatment. Y-axis indicates the normalized enrichment score (NES).
(C) Enrichment plot of the Hallmark Androgen Response pathway. Genes are ranked by differential expression upon ENZ treatment based on patient RNA-seq data (post vs. pre). Y-axis indicates enrichment score (ES). GSEA statistics (FDR, ES, NES, nominal P-value) are indicated.
(D) Unsupervised hierarchical clustering of pre- and post-treatment RNA-seq samples based on the expression of AR-responsive genes. Color scale indicates gene expression (z-score).
(E) River plot showing state transitions between Clusters 1 (dark blue), Cluster 2 (green) and Cluster 3 (light blue) for paired pre-treatment and post-treatment RNA-seq samples (n=39). Number of samples assigned to each cluster before and after treatment as well as the hallmarks per cluster are indicated.
(F) Waterfall plot depicting the Pearson correlation of neuroendocrine gene expression signature fold changes upon ENZ treatment per patient. Colors indicate the patients cluster affiliations after treatment.
Previously, we identified three distinct subtypes of primary treatment-naïve PCa (31), which we named Cluster 1-3 (Cl1-3). While Cl1 and Cl2 were mainly dominated by their ERG fusion status – with Cl1 expressing high ERG levels (ERG fusion-positive) and Cl2 expressing low ERG levels (ERG fusion-negative) – Cl3 was enriched for neuroendocrine (NE)-like features, including low AR activity and a high NE gene expression score. To assess the impact of neoadjuvant ENZ therapy on these PCa subtypes, we performed unsupervised hierarchical clustering in the pre- and post-treatment setting using the originally identified top 100 most differentially expressed genes per cluster. Prior to hormonal intervention, we could robustly assign the samples into all three clusters (Cl 1: n=23, Cl2: n=11, Cl3: n=8) with highly comparable distributions as we previously reported in another cohort of patients (31) (Supplementary Fig. S11A). Our pre- and post-treatment sampling now allowed us to investigate how individual tumors were affected by neoadjuvant therapy. This revealed that three months of ENZ therapy pushed almost all of the tumors towards our NE-like cluster 3 (Fig. 4E; Supplementary Fig. S11B). To assure that the observed effects are not solely driven by the treatment-induced reduction in AR activity (Fig. 4C and 4D), we used a well-established neuroendocrine PCa (NEPC) signature (35) to calculate gene expression fold changes pre- vs. post-ENZ, which confirmed an induction of NE-like signaling upon treatment (Fig. 4F). We further validated this transcriptional rewiring using gene sets that distinguish the three major lineages of prostate epithelial cells (luminal, basal, neuroendocrine) (36,37), which jointly illustrated reduced AR-driven luminal cell transcriptional activity accompanied by an enrichment of NE-like features along with a basal-type transcriptional program after treatment (Supplementary Fig. S12A). Along these lines, classical NEPC markers (38) and transcriptional disease drivers (39–41) were selectively upregulated upon treatment (CHGA, PEG10) with acquisition of promoter-enriched H3K27ac (Supplementary Fig. S12B–D), while others were not affected on expression level (SYP, N-MYC) or not even expressed in primary tumors – irrespective of neoadjuvant treatment status (BRN2, encoded by the POU3F2 gene). For classical NEPC IHC markers chromogranin A (CHGA) and synaptophysin (SYP), tissue microarrays (TMAs) were stained and analyzed, showing no change (SYP) or a modest non-significant increase (CHGA) upon neoadjuvant ENZ treatment (Supplementary Fig. S12E).
Recently, N-MYC ChIP-seq data was reported in models of NEPC (40), which however showed limited overlap with our post-treatment FOXA1 cistrome (Supplementary Fig. S12F). While a subset of NEPC markers was enriched on RNA-seq level, FOXA1 reprogramming did not seem to be a crucial driver in this phenomenon, based on limited overlap of our differential FOXA1 cistromes with a recently reported NEPC FOXA1 cistrome (42) (Supplementary Figure S13A), nor was FOXA1 ChIP-seq in our study enriched for classical NEPC signature genes (Supplementary Figure S13B). Jointly, these data suggest that altered FOXA1 cistromics after neoadjuvant ENZ treatment present a different biological state as compared to the fully developed NEPC-associated FOXA1 cistrome that presents in the advanced disease stage, and may represent an early intermediate state.
FOXA1 is frequently mutated in primary prostate cancer (14) and metastatic disease, where FOXA1 mutations were associated with loss of lineage-specific transcriptional programs and worse clinical outcome (17,18). Therefore, we determined the FOXA1 mutation status of our clinical samples using H3K27ac ChIP-seq and RNA-seq reads covering the FOXA1 gene (43), and tested for possible enrichment for poor clinical outcome and NE-like gene expression features specifically in the FOXA1 mutant cases. While we observed a significant enrichment of FOXA1 mutant tumors among ENZ non-responders (BCR ≤ 6 months after surgery), no such enrichment was observed at the transcriptomic level, likely affected by the almost-complete transition of all our tumor samples towards the NE-like cluster 3 – irrespective of FOXA1 mutation status (Supplementary Figure S13C-F).
Collectively, these results demonstrate that three months of neoadjuvant ENZ therapy not only uniformly diminish AR signaling, but also push practically all of our primary PCa samples to acquire some – but not all – features of NEPC, independently of their original subtype.
Post-treatment FOXA1 sites drive pro-survival gene programs, dictated by circadian clock component ARNTL
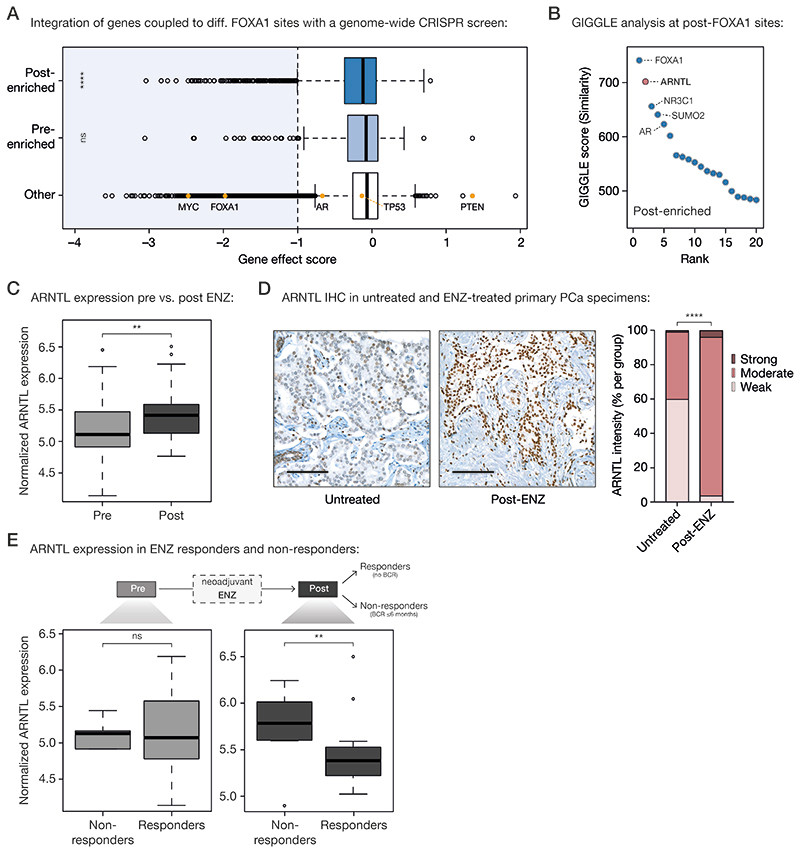
Having examined the global cistromic and transcriptomic changes upon ENZ therapy, we next sought to characterize the biological consequences of the observed FOXA1 reprogramming using integrative analyses. We hypothesized that the newly acquired FOXA1 sites would be driving expression of genes associated with tumor cell survival programs. Using H3K27ac HiChIP data generated in LNCaP cells (44), pre- and post-treatment FOXA1 sites were coupled to their corresponding gene promoters (Supplementary Table S5). Subsequently, genome-wide CRISPR knockout screen data from Project Achilles (DepMap 20Q1 Public; VCaP) were used to identify those genes essential for prostate cancer cell proliferation (45,46). While genes associated with pre-treatment FOXA1 sites were not enriched for essential genes (gene effect score < -1), genes under the control of post-treatment FOXA1 sites showed a significant enrichment (P = 8.66 × 10-8, Fisher’s exact test) for critical drivers of tumor cell proliferation (Fig. 5A), pointing towards a possible role of these sites in maintaining proliferative potential upon ENZ treatment. However, the factor regulating these genes to possibly drive proliferation remained elusive, especially since based on our STARR-seq and RNA-seq data, AR is likely not driving enhancer activity at post-treatment FOXA1 sites (Fig. 3F; Fig. 4C and 4D). Therefore, we sought to identify transcription factors involved in the activation of these regulatory regions that are selectively occupied by FOXA1 following treatment. To this end, we overlaid the genomic coordinates of the post-treatment enriched FOXA1 binding sites with those identified in publicly available ChIP-seq datasets (n= 13,976) as part of the Cistrome DB transcription factor ChIP-seq sample collection (47,48). Besides FOXA1 and AR, which were expected to bind at these regions (Fig. 3D), we also identified the glucocorticoid receptor (encoded by the NR3C1 gene), which has previously been described to be upregulated upon antiandrogen treatment and able to drive the expression of a subset of AR-responsive genes, conferring resistance to AR blockade (49–51). Unexpectedly, the second most enriched transcription factor after FOXA1 was circadian rhythm core component ARNTL (Aryl Hydrocarbon Receptor Nuclear Translocator Like; also known as BMAL1) which has not previously been implicated in PCa biology (Fig. 5B). Interestingly, ARNTL transcript levels were upregulated upon ENZ treatment (P = 6.4 × 10-3, Mann-Whitney U-test; Fig. 5C), which was accompanied by increased H3K27ac ChIP-seq signals at the ARNTL locus (Supplementary Fig. S14A). Consistent with this, TMA IHC analysis also revealed elevated ARNTL protein levels after treatment when comparing the prostatectomy specimens post-ENZ with those of matched untreated control patients (P = 6.89 × 10-19, Fisher’s exact test; Fig. 5D). To assess whether ARNTL levels are also associated with patient outcome, we compared the average ARNTL gene expression of patients that did not experience a BCR (responders, n=29) with those that experienced an early BCR within ≤ 6 months post-surgery (non-responders, n=8; Supplementary Table S1). While pre-treatment ARNTL levels were not significantly different between ENZ responders and non-responders, high ARNTL levels after treatment were associated with poor clinical outcome (P = 4.79 × 10-3, Mann-Whitney U-test; Fig. 5E). In agreement with this observation, ARNTL levels were exclusively found upregulated in non-responders (P = 3 × 10-4, paired Mann-Whitney U-test), while overall remaining unaffected upon neoadjuvant ENZ treatment in responders (P = 0.33; Supplementary Fig. S14B).
Figure 5. Acquired FOXA1 sites drive key-survival genes that are under control of circadian rhythm regulator ARNTL.
(A) Boxplot showing DepMap (20Q1) genome-wide loss-of-function CRISPR screen data for VCaP PCa cells, separately analyzing the gene effect score of genes associated with post-enriched FOXA1 sites (top), pre-enriched FOXA1 sites (middle) or all other tested genes (bottom). Differential FOXA1 binding sites were coupled to their respective target genes using H3K27ac HiChIP data. Indicated as controls are PCa-relevant driver genes: oncogenes MYC, FOXA1, AR, TP53 and tumor suppressor PTEN. The recommended stringent gene effect score cutoff of -1 is shown (dotted vertical line) and all genes passing the essentiality threshold are highlighted in light blue. ns, P > 0.05; ****, P < 0.0001 (Fisher’s exact test).
(B) Dot plot representing ranked GIGGLE similarity scores for transcriptional regulators identified at post-treatment FOXA1 sites. The top 20 identified factors are shown, and the 5 most enriched factors are labeled.
(C) Boxplot showing normalized ARNTL gene expression before and after 3 months of neoadjuvant ENZ treatment. **, P < 0.01 (Mann-Whitney U-test).
(D) Representative ARNTL immunohistochemistry (IHC) stainings (left) and quantification of ARNTL staining intensity (right) in tissue microarrays consisting of prostatectomy specimens from untreated patients (not receiving neoadjuvant ENZ; n=110) and DARANA patients post-ENZ (n=51). Scale bars, 100 μm. ****, P < 0.0001 (Fisher’s exact test).
(E) Boxplots depicting normalized ARNTL gene expression in ENZ non-responders (biochemical recurrence (BCR) ≤ 6 months; n=8) and responders (no BCR; n=29) in the pre- (left) and post- (right) treatment setting separately. ns, P > 0.05; **, P < 0.01 (Mann-Whitney U-test).
Interestingly, while the CLOCK and NPAS2 proteins, which form a heterodimer with ARNTL to activate transcription of core clock genes, didn‘t show differential gene expression upon ENZ treatment (Supplementary Fig. S14C), all downstream ARNTL targets were upregulated upon treatment – except for CRY1, which has recently been shown to be AR- and thus ENZ-responsive (52). In addition, the gene expression of these ARNTL dimerization partners was also not associated with clinical outcome (Supplementary Fig. S14D), hinting towards a treatment-induced role of ARNTL that is independent of its canonical function in the circadian machinery.
Notably, in two cohorts of metastatic castration-resistant PCa (mCRPC) (53,54), ARNTL expression was not associated with outcome (Supplementary Figure S15A-D), suggesting a context-dependent prognostic potential of this gene – being associated with outcome in high-risk primary PCa upon treatment with ENZ.
Taken together, these data suggest that the circadian clock regulator ARNTL may be functionally involved in ENZ resistance in high-risk primary PCa by driving tumor cell proliferation processes.
Acquired ARNTL dependency in ENZ-resistant PCa cells
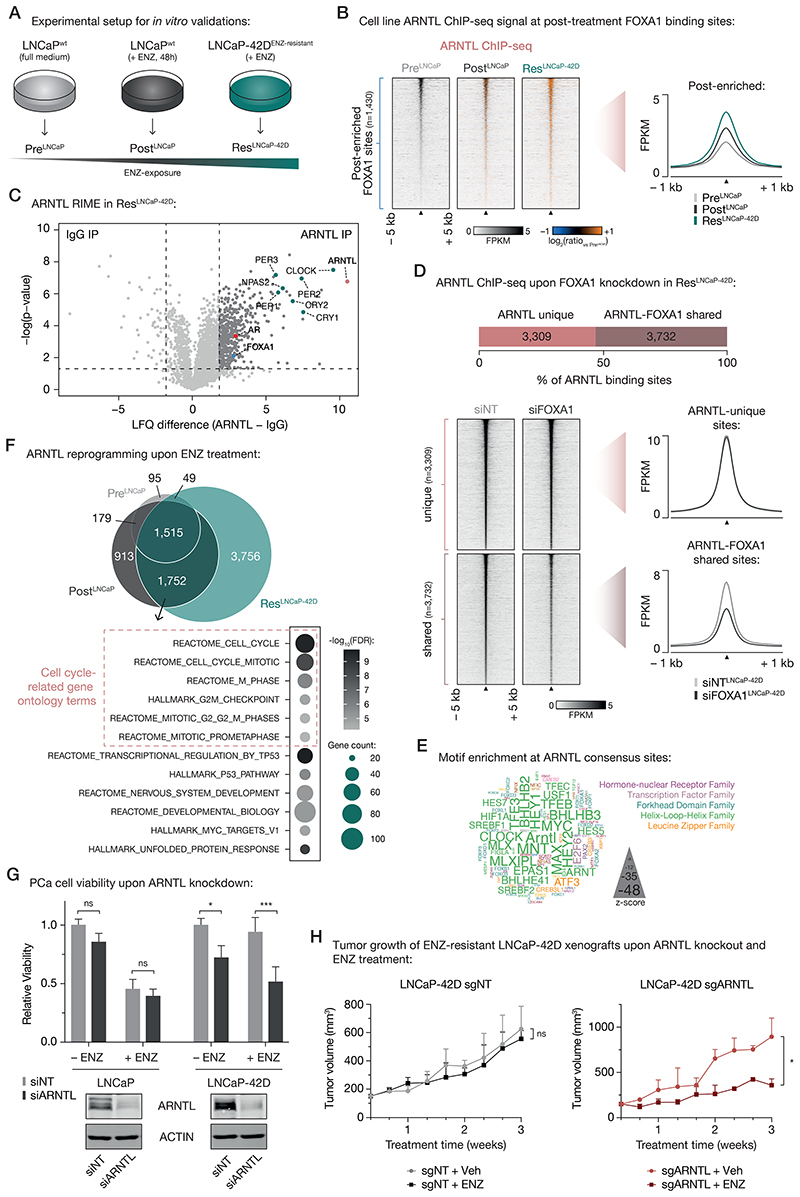
To further investigate the relevance of ARNTL as a transcriptional driver at post-treatment FOXA1 sites, we performed in vitro validation experiments. To this end, we used hormone-sensitive LNCaP PCa cells, which we either cultured in full medium alone (PreLNCaP) or with ENZ for 48 hours (PostLNCaP), mimicking our clinical trial setting (Fig. 6A). Based on the ENZ-induced acquisition of NE-like gene expression profiles in our patient cohort (Fig. 4E and 4F), we also included the ENZ-resistant LNCaP-42D model (41) that possesses NE-features (ResLNCaP-42D; Fig. 6A), allowing us to further validate our patient-derived findings in cell lines recapitulating the transcriptional features of post-treatment clinical specimens.
Figure 6. Treatment-induced dependency on ARNTL in ENZ-resistant PCa cells.
(A) Experimental setup for in vitro validation experiments.
(B) Tornado plots (left) and average density plot (right) visualizing ARNTL ChIP-seq signal (in FPKM) at post-enriched FOXA1 binding sites in untreated (PreLNCaP), short-term ENZ-treated (PostLNCaP), and ENZ-resistant NE-like LNCaP cells (ResLNCaP-42D). Data are centered at post-treatment FOXA1 peaks depicting a 5-kb (heatmaps) or 1-kb (density plots) window around the peak center. Heatmap color depicts the ChIP-seq signal compared to the untreated condition (PreLNCaP), with blue indicating lower peak intensity and orange indicating higher peak intensity. (n=2)
(C) Volcano plot depicting ARNTL interactors in ENZ-treated LNCaP-42D (ResLNCaP-42D) cells over IgG control. Significantly enriched interactors are highlighted and significance cutoffs are shown as dotted lines (label-free quantification (LFQ) difference ≥ 1.8; P ≤ 0.05; n = 4).
(D) Stacked bar chart (top) indicating the fraction of ARNTL binding sites in ENZ-treated LNCaP-42D (ResLNCaP-42D) cells that are ARNTL unique (n=3,309) or shared with FOXA1 (n=3,732). Tornado plots (lower left) and average density plot (lower right) visualize ARNTL ChIP-seq signal (in FPKM) at ARNTL unique or ARNTL-FOXA1 shared binding sites in LNCaP-42D cells upon transfection with non-targeting siRNA (siNT) or siFOXA1. Data are centered at ARNTL peaks depicting a 5-kb (heatmaps) or 1-kb (density plots) window around the peak center. (n=2)
(E) Word cloud shows motif enrichment at ARNTL consensus sites (n=1,515) shown in (E). The font size represents the z-score and colors correspond to transcription factor families. Since the human ARNTL motif is not part of the tested database, the homologous mouse motif (Arntl) was included.
(F) Venn diagram (top) indicating the overlap of ARNTL binding sites in all tested cell line conditions (PreLNCaP, PostLNCaP, ResLNCaP-42D). For each condition, only peaks present in both replicates were included. Gene ontology terms for ARNTL-bound gene sets uniquely shared between PostLNCaP and ResLNCaP-42D conditions are presented below. Overlapping ARNTL binding sites (n=1,752) were coupled to their respective target genes using H3K27ac HiChIP data. Color indicates the gene set enrichment (FDR q-value) and size depicts the number of genes that overlap with the indicated gene sets. Cell cycle-related gene ontology terms are highlighted.
(G) Bar chart (top) showing relative cell viability of LNCaP (left) and LNCaP-42D (right) cells upon transfection with non-targeting siRNA (siNT) or siARNTL, and exposure to ENZ. Treatment is indicated and data is shown relative to the untreated (– ENZ) siNT condition per cell line (n=3). Western blots (bottom) indicate ARNTL protein levels in LNCaP (left) and LNCaP-42D (right) cells following siRNA-mediated silencing of ARNTL for 48 h. Transfection with siNT and staining for ACTIN are included as controls for siRNA treatment and protein loading, respectively. Images are representative of three independent experiments. ns, P > 0.05; *, P < 0.05; ***, P < 0.001 (two-way ANOVA followed by Tukey’s multiple comparisons test).
(H) Growth curves depict tumor volume (measured 3 times per week using calipers) of non-targeting control (sgNT) or ARNTL knockout (sgARNTL) LNCaP-42D xenografts upon daily treatment with vehicle-alone (sgNT +Veh: n=4; sgARNTL +Veh: n=3) or ENZ (sgNT +ENZ: n=4; sgARNTL +ENZ: n=2). ns, P > 0.05; *, P < 0.05 (t-test).
We performed FOXA1 ChIP-seq experiments in all three cell line conditions (Supplementary Fig. S16A-D; Supplementary Table S6), which revealed highly similar FOXA1 chromatin binding dynamics as observed in our clinical samples: While the pre-enriched FOXA1 sites identified in vivo showed less binding upon treatment, we observed that merely 48 hours of ENZ exposure was sufficient to strongly induce binding at post-enriched sites, which was further increased in the long-term exposed, treatment-resistant LNCaP-42D cell line (Supplementary Fig. S16E and S16F). Similarly, genome-wide correlation analyses indicated that short-term ENZ treatment in cell lines induced FOXA1 reprogramming to regions that are FOXA1-bound in treatment-resistant but not in treatment-naïve cells (Supplementary Fig. S16G and S16H).
Having shown that differential FOXA1 chromatin binding in tumors could be recapitulated in vitro, we next sought to further assess the role of ARNTL in these pre-clinical models. Therefore, we first measured the intrinsic enhancer activity of our patient-derived and cell line-validated differential FOXA1 binding sites by STARR-seq for 1,209/1,905 differential regions in LNCaP cells. Notably, we identified a subset of regions (n=968) with sustained enhancer activity upon ENZ treatment (Supplementary Figure S17A), confirming our initial STARR-seq analysis (Fig. 3F). While GIGGLE analyses on the inactive regions showed enrichment for FOXA1 and AR, active enhancers – irrespective of treatment – were specifically enriched for ARNTL (Supplementary Figure S17B and S17C). These data are in full concordance with the tumor H3K27ac ChIP-seq (Fig. 3D) analyses, showing AR-independent activity at the post-treatment enriched FOXA1 sites, and uncovered once more ARNTL as a possible driver for transcriptional activity in case of AR suppression.
Next, we confirmed that treatment with ENZ increased ARNTL protein levels in PCa models (Supplementary Fig. S18A), recapitulating the clinical observations (Fig. 5C and 5D). Interestingly, this treatment-induced ARNTL upregulation appeared to be FOXA1-dependent, as FOXA1 knockdown abolished the ENZ-driven increase in ARNTL levels (Supplementary Fig. S18B).
Since cistromic ARNTL profiling has to date not been reported in PCa models, we generated ARNTL ChIP-seq data (Supplementary Fig. S19A–C) to validate its binding at post-treatment FOXA1 sites. Interestingly, while we already observed ARNTL binding to these regulatory regions in the pre-treatment setting, this was strongly enhanced upon ENZ exposure (Fig. 6B; Supplementary Fig. S19D–F).
Functional interactions between FOXA1 and ARNTL could be further validated using ARNTL RIME experiments in ENZ-treated LNCaP-42D and LNCaP cells, confirming interactions of ARNTL with AR and FOXA1, but also with other classical circadian rhythm components including CLOCK/NPAS2, CRYs and PERs (Fig. 6C; Supplementary Fig. S20A). As FOXA1 acts as a pioneer factor, enabling chromatin binding for other TFs including AR (9), we hypothesized that FOXA1 serves a comparable role for ARNTL. To test this hypothesis, we performed ARNTL ChIP-seq upon FOXA1 knockdown (Fig. 6D; Supplementary Fig. S20B–E), showing a significant decrease of ARNTL chromatin interactions exclusively for those regions co-occupied by FOXA1 – highlighting FOXA1’s critical role in determining ARNTL chromatin binding.
In agreement, at ARNTL consensus peaks, motifs were found to be enriched for CLOCK and MYC, but also FOXA1 and ARNTL itself (Fig. 6E). To identify functional differences in ARNTL cistromes induced upon treatment, we overlapped the ARNTL peaks identified in all tested cell line conditions, which revealed a massive cistromic reprogramming upon ENZ treatment (Fig. 6F; Supplementary Fig. S19E and S19F). Notably, ~70% of ENZ-gained ARNTL peaks (n=1,752) in LNCaP cells were captured by the ARNTL cistrome in treatment-resistant cells. Interestingly, upon ENZ treatment ARNTL binding was found to be enriched at promoter regions of key NEPC drivers, including BRN2 (POU3F2), FOXA2, EZH2, ASCL1 and SOX2 (Supplementary Fig. S21A), positioning ARNTL as a possible driver of the NE-like transcriptional program we identified. In addition, pathway over-representation analyses of genes coupled to PostLNCaP-ResLNCaP-42D-shared ARNTL binding sites revealed a treatment-induced enrichment for gene sets implicated in cell cycle progression and cell division, further supporting a possible functional involvement of ARNTL in sustaining tumor cell proliferation when AR is blocked by ENZ (Fig. 6F). To challenge this hypothesis, we assessed whether ARNTL-knockdown affects the viability of hormone-sensitive and in particular of long-term ENZ-exposed cell lines. While ARNTL-targeting had minimal effect on LNCaP cell proliferation (with or without ENZ), ARNTL knockdown significantly suppressed cell growth of ENZ-resistant LNCaP-42D cells in the absence (P = 0.031, two-way ANOVA) and even more so in the presence of ENZ (P = 7 × 10-4, two-way ANOVA), indicating that targeting ARNTL also partially restores ENZ-sensitivity in this treatment-resistant cell line model (Fig. 6G). While ARNTL was essential for sustaining cellular fitness upon ENZ treatment, exogenously introduced ARNTL did not suffice to further enhance cell proliferation when exposing LNCaP and LCNaP-42D cells to ENZ (Supplementary Fig. S21B), suggesting that ARNTL is required but not sufficient to drive the observed phenotype. Importantly, we could successfully validate the functional role of ARNTL in additional cell line models of ENZ resistance (LNCaP-ResV: originally referred to as LNCaP-ENZR (55), and LNCaP-ResA (56)) using ARNTL knockdown and CRISPR/Cas9-mediated ARNTL knockout (Supplementary Fig. S21C–E). In line with these in vitro validation experiments, ARNTL knockout also strongly inhibited the growth of LNCaP-derived ENZ-resistant xenografts (LNCaP-42D, LNCaP-ResA) in intact mice upon ENZ exposure (Fig. 6H; Supplementary Fig. 21F). Importantly, parental LNCaP cells were not affected in their proliferation potential by ARNTL knockout (Supplementary Fig. 21F), supporting the acquired dependency of ENZ-resistant cells on this circadian factor, instead of a general impact on cellular fitness. Jointly, these data further highlight the treatment-induced ARNTL dependency of high-risk PCa models, both in vitro and in vivo, and position ARNTL as a novel candidate therapeutic target.
Glucocorticoid Receptor (GR) was identified in the GIGGLE analysis as the third-most enriched factor at post-treatment FOXA1 sites, directly following ARNTL and FOXA1 itself (Fig. 5B). Given the known GR function in driving ENZ resistance in advanced CRPC (49,57,58), we next tested whether sustained tumor cell survival after short-term antiandrogen treatment was not only ARNTL, but also GR-dependent. Interestingly, GR (encoded by the NR3C1 gene) expression was upregulated upon neoadjuvant ENZ treatment in primary PCa patients (Supplementary Fig. S22A), but neither expression levels before nor after therapy were associated with clinical outcome (Supplementary Fig. S22B). Using publicly available GR ChIP-seq data from LNCaP-derived GR-positive LREX’ cells (49), we could identify GR occupancy at the majority of pre-treatment FOXA1 sites and at a subset of post-treatment sites (Supplementary Fig. S22C and S22D). However, GR knockdown did not affect cellular fitness after short-term ENZ treatment in the majority of cell line models we tested (Supplementary Fig. S22E), suggesting the observed ARNTL-driven early adaptation to ENZ exposure represents a different biological entity as compared to the known GR-driven treatment resistance described in CRPC.
Overall, these data confirm the ENZ-induced FOXA1 reprogramming as observed in PCa patients upon neoadjuvant antiandrogen therapy, and revealed an acquired dependency on circadian rhythm regulator ARNTL to drive tumor cell growth – positioning ARNTL as a highly promising new drug target in combination with ENZ for the treatment of high-risk PCa.
Discussion
In medicine, the evolutionary selection pressure as imposed by drug treatment has been a well-known clinical challenge, ever since the first antibiotics were discovered in the early 20th century. Also in oncology, clear escape mechanisms for both targeted therapeutics and systemic treatments are known for many years, involving ESR1 mutations in metastatic breast cancer (59), EGFR mutations in lung cancer (60), KRAS mutations in metastatic colorectal cancer (61), but also somatic amplification of the AR locus and/or an upstream AR enhancer in castration-resistant PCa (62,63). Apart from genetic alterations, also epigenetic rewiring (7,50) or transdifferentiation are reported as mechanisms of resistance, including treatment-emergent neuroendocrine (NE) prostate cancers that occur as an adaptive response under the pressure of prolonged AR-targeted therapy (64,65).
Our unique clinical trial design with paired pre- and post-treatment biopsies of high-risk primary PCa treated with ENZ monotherapy, allowed us to unravel global ENZ-induced alterations in gene regulation. We report that large-scale treatment-induced dedifferentiation in PCa may be a gradual process, of which the early signs are identified on transcriptomic level within the first months of treatment onset. While complete adenocarcinoma-to-neuroendocrine transdifferentiation was not observed in any of our samples, cellular plasticity characterized by the acquisition of cistromic, transcriptomic and proteomic features of NE disease may not only be present in primary tumors prior to treatment (31), but also become enriched upon short-term exposure to endocrine treatment, and thus represent an early intermediate disease state.
In PCa development (5,32) and progression (7), AR has been reported to expose substantial plasticity in its enhancer repertoire, and we now illustrate this is also the case in primary disease upon short-term treatment. Besides AR, FOXA1 is considered a master transcription factor and critical prostate lineage specific regulator acting in PCa, that upon overexpression during tumorigenesis gives rise to a tumor-specific AR cistrome. Also in NEPC, FOXA1 cistromes are reprogrammed (42), which indicates a direct AR-independent role of FOXA1 in PCa progression. Our study confirms these observations and shows that, while co-occupied by AR, the pre- and post-ENZ enriched FOXA1 sites appeared indifferent to AR signaling.
The functional implications of the pre-treatment FOXA1 sites remain unclear, as those regions were inactive, both in primary tissues as well as in reporter assays. A subset of these cis-regulatory elements demarcates developmental epigenomic programs, that we previously reported as being occupied by FOXA1 from prostate development to tumorigenesis and metastatic progression (7), whereas others may be relevant for different physiological processes.
The treatment-induced cistromic repositioning of pioneer factor FOXA1 initiated a thus far unknown transcriptional rewiring, in which ARNTL, a classical circadian rhythm regulator and dimerization partner of CLOCK, compensates for AR inhibition and becomes essential to rescue cellular proliferation signals. Recently, it has been reported that CRY1 – a transcriptional coregulator of ARNTL – is AR-regulated in PCa, and modulates DNA repair processes in a circadian manner (52). The current data illustrate that certain components of the circadian machinery may have a potential impact on drug response, as most clock components are not only temporally regulated at the transcriptional level, but are also dysregulated upon exposure to hormonal therapy. Our data now show that AR blockade forces tumor cells to adapt epigenetically, upon which these cells – over time – become dependent on ARNTL as a transcriptional regulator of proliferation processes. This acquired cellular vulnerability appears to be dependent on whether or not AR activity is inhibited and cells have had time to achieve full epigenetic reprogramming, explaining the limited effect of ARNTL knockdown in hormone-sensitive PCa cells, as compared to the long-term ENZ-exposed treatment-resistant models.
ARNTL expression did not correlate with outcome in mCRPC patients. Furthermore, post-treatment induced FOXA1 profiles showed limited overlap with NEPC-FOXA1 sites, and GR action – previously reported as a driver in CRPC – did not play a decisive role in our datasets to sustain cellular fitness following short-term enzalutamide exposure. Jointly, these data position the clinical state as induced by short-term neoadjuvant AR-targeted therapy in primary PCa as a separate biological entity, exposing already in this early clinical stage some – but not all – features of progressive therapy-resistant disease, that are invoked by drug-induced epigenetic plasticity.
With the identification of ARNTL as a rescue mechanism for tumor cells to evade AR blockade, the next question presents whether ARNTL could serve as a novel therapeutic target, which should be further pursued in future drug development and clinical research. Being critically relevant for circadian rhythm regulation, it would be imperative to balance ARNTL targeting in relation to any adverse side-effects. Additionally, we demonstrate that the surprisingly dynamic enhancer repertoire of FOXA1 is not only critical in prostate tumorigenesis (5) and neuroendocrine differentiation (42), but also appears crucial in evading AR therapy-induced growth inhibition, further supporting the rationale to intensify efforts in targeting this highly tissue-selective, yet critical transcriptional regulator, directly or indirectly (66).
Methods
Study design
Primary PCa tissues before and after enzalutamide (ENZ) treatment were acquired as part of the phase 2, prospective, single-arm DARANA study (ClinicalTrials.gov #NCT03297385) at the Netherlands Cancer Institute Antoni van Leeuwenhoek hospital. The primary clinical outcome measure of the trial was the positive margins rate after neoadjuvant ENZ treatment. To allow sample size calculation, we performed a survey into the surgical margins of 1492 in-house prostatectomy specimens (Gleason ≥ 7), not treated with antihormonal therapy prior to surgery, which revealed 34% not-radical resections. Earlier randomized studies on neoadjuvant androgen ablation showed reductions in positive surgical margin rate of at least 50% (67–69). To detect a reduction of positive surgical margins from 34% to 17% with a power of 80% and an alpha level set at 0.05, 55 patients needed to be included. Inclusion criteria were over 18 years of age, Gleason ≥ 7 PCa and planned for prostatectomy. Prior to treatment a multi-parametric MRI scan was made to identify tumors in the prostate (cT-stage) and pelvic lymph node metastasis (cN-stage). Patients were treated with ENZ, once daily 160 mg P.O. without androgen deprivation therapy, for three months prior to robotic-assisted laparoscopic prostatectomy (RALP) and a pelvic lymph node dissection. The resection specimen was assessed for tumor margins, prostate tumor stage (ypT-stage) and pelvic lymph node involvement (ypN-stage). Secondary endpoints included assessment of downstaging by comparison of pre-operative clinical cT and cN stage with post-treatment and post-operative ypT and ypN stage, and differences in pre- and post-treatment prostate cancer cleaved Caspase-3 and Ki-67 staining as markers of apoptosis and tumor cell proliferation, respectively. Moreover, various clinical time-to-event outcomes were included: time to biochemical recurrence, defined as time from trial inclusion to two consecutive rises of serum PSA with a minimal level of ≥ 0.2 ng/mL; ADT-free survival, defined as time from trial inclusion to the onset of ADT therapy; time to radiological recurrence, defined as time from trial inclusion until detection of local or distant metastases by PSMA PET scanning; and time to distant metastases, defined as time from trial inclusion until the detection of distant metastases by PSMA PET scanning. The trial was approved by the institutional review board of the Netherlands Cancer Institute, written informed consent was signed by all participants enrolled in the study, and all research was carried out in accordance with relevant guidelines and regulations.
Pre- and post-treatment sampling
Prior to ENZ intervention, 4 pre-operative MRI-guided 18-gauge core needle tumor biopsies were taken per patient. Directly after prostatectomy, 8 additional tumor-targeted core needle biopsies (4× 14-gauge, 4× 5-mm) were taken from prostatectomy specimens ex vivo, using previous MRI information and palpation. Biopsy and prostatectomy specimens were fresh frozen (FF) or formalin-fixed paraffin-embedded (FFPE) for ChIP-seq and CNV-seq, or RNA-seq and immunohistochemistry analyses, respectively. Prior to ChIP-seq experiments, FF material was cut in 30 μm sections, while FFPE material was cut in 10 μm sections prior to RNA extraction. Tissue sections were examined pathologically for tumor cell content and only samples with a tumor cell percentage of ≥ 50% were used for further downstream analyses.
ChIP-seq
Sample processing
Chromatin immunoprecipitations on PCa tissue specimens and cell line models were performed as previously described (70). In brief, cryosectioned tissue samples were double-crosslinked in solution A (50 mM HEPES-KOH, 100 mM NaCl, 1 mM EDTA, 0.5 mM EGTA), first supplemented with 2 mM disuccinimidyl glutarate (DSG; CovaChem) for 25 min at room temperature. Then, 1% formaldehyde (Merck) was added for 20 min and subsequently quenched with a surplus of 2.5 M glycine. Cell lines were crosslinked using single-agent fixation. Therefore, 1% formaldehyde was added to the cell culture medium and incubated at room temperature for 10 min, followed by glycine-quenching as described above. Tissue and cell line samples were lysed as described (71) and sonicated for at least 10 cycles (30 sec on; 30 sec off) using a PicoBioruptor (Diagenode). For each ChIP, 5 μg of antibody were conjugated to 50 μL magnetic protein A or G beads (10008D or 10009D, Thermo Fisher Scientific). The following antibodies were used: AR (06-680, Merck Millipore), FOXA1 (ab5089, Abcam), H3K27ac (39133, Active Motif), and ARNTL (ab93806, Abcam).
ChIP-seq
Immunoprecipitated DNA was processed for library preparation using a KAPA library preparation kit (KK8234, Roche) and generated libraries were sequenced on the Illumina HiSeq2500 platform using the single end protocol with a read length of 65-bp, and aligned to the human reference genome hg19 using Burrows-Wheeler Aligner (v0.5.10) (72). Reads were filtered based on mapping quality (MAPQ ≥ 20) and duplicate reads were removed.
Analysis of ChIP-seq
Peak calling over input controls (per tissue sample or cell line) was performed using MACS2 (v2.1.1) and Dfilter (v1.6) for tissues, and MACS2 (v2.1.2) for cell lines (73,74). For tissue samples, only the peaks shared by both peak callers were used for downstream analyses. DeepTools (v2.5.3) was used to calculated read counts in peaks (FRiP) (75). Read counts and the number of aligned reads, as well as normalized strand coefficient (NSC) and relative strand correlation (RSC), which were calculated using phantompeaktools (v1.10.1) (76), are shown in Supplementary Table S2 for tissue ChIP-seq data and Supplementary Table S6 for cell line ChIP-seq data. Tissue ChIP-seq samples that passed the following quality control measures were included in the final analyses; tumor cell percentage ≥ 50%, ChIP-qPCR enrichment, and more than 100 peaks called (Supplementary Fig. S2).
For visualization of cell line ChIP-seq data, an average enrichment signal was generated by merging mapped reads of replicate samples using SAMtools (v1.10-3) (77).
Genome browser snapshots, tornado and average density plots were generated using EaSeq (v1.101) (78). For snapshot overviews across multiple samples, bigWig files were generated from aligned bam files with the bamCoverage function from deepTools (v2.0), and snapshots were produced using pyGenomeTracks (v3.6) (79) with the added NCBI RefSeq genome track (80,81). Genomic distribution and motif enrichment analyses were performed using the CEAS and the SeqPos motif tools on Galaxy Cistrome (82), respectively. CistromeDB Toolkit was used to probe which TFs and chromatin regulators have a significant binding overlap with the differential FOXA1 peak sets (48). For this, genomic coordinates of high-confidence binding sites (FC ≥ 1.2) were converted between assemblies (from hg19 to hg38), using the UCSC genome browser liftOver tool (83). The DiffBind R package (v2.10) was used to generate correlation heatmaps and PCA plots based on occupancy, to perform differential binding analyses using a false discovery rate (FDR) < 0.05, and to generate consensus peaklists (84).
ChIP-seq signal of various datasets (FOXA1, AR and H3K27ac from this study; AR, H3K27ac and H3K27me3 from a previously reported study (31)) at differential and consensus FOXA1 sites was investigated by counting mapped reads in FOXA1 peak regions using bedtools multicov (v2.27.1) (85). Readcounts were subsequently z-transformed and visualized using the aheatmap function from the R package NMF (v0.21.0) (86) with a color scheme from RColorBrewer (v1.1-2; https://CRAN.R-project.org/package=RColorBrewer). To determine significance in binding site occupancy differences between pre- and post-treatment FOXA1 sites, median z-transformed readcounts were calculated per sample and compared using a Mann-Whitney U-test. These median readcounts per sample were also used to assess the correlation between ChIP-seq signals of AR, FOXA1 and H3K27ac at pre-enriched, post-enriched and consensus FOXA1 binding sites.
Bedtools intersect (v2.27.1) (85) was used to determine overlap of differential FOXA1 binding sites and inactive, constitutively active and inducible AR-binding sites.
To assign FOXA1 and ARNTL binding regions to potential target genes, we overlapped differential FOXA1 binding sites with H3K27ac HiChIP data (44) using bedtools intersect. To assess whether or not genes coupled to FOXA1 binding sites were considered to be essential for the VCAP prostate cancer cell line, we used the DepMap (Broad 2020) 20Q1 Public gene effect dataset (45) with a stringent gene effect score cutoff ≤ -1. Gene set overlaps between genes linked to ChIP-seq binding sites and the Molecular Signatures Database (v7.4) were computed using Gene Set Enrichment Analysis (GSEA) (87) with an FDR q-value cutoff ≤ 0.05.
RNA-seq
RNA isolation
Prior to RNA isolation, FFPE material was pathologically assessed. The expert pathologist scored tumor cell percentage and indicated most tumor-dense regions for isolation on a hematoxylin and eosin (H&E) slide. RNA and DNA from FFPE material were simultaneously isolated from 3-10 sections (depending on tumor size) of 10 μm using the AllPrep DNA/RNA FFPE isolation kit (80234, Qiagen) and the QIAcube according to the manufacturer’s instructions. cDNA was synthesized from 250 ng RNA using SuperScript III Reverse Transcriptase (Invitrogen) with random hexamer primers.
RNA-seq
Strand-specific libraries were generated with the TruSeq RNA Exome kit (Illumina) and sequenced on the Illumina HiSeq2500 platform using the single end protocol with a read length of 65-bp. Sequencing data was aligned to the human reference genome hg38 using TopHat (v2.1.0 using bowtie 1.1.0) (88) and the number of reads per gene was measured with HTSeq count (v0.5.3) (89).
For QC purposes, total readcounts per sample were determined and hierarchical clustering based on the Euclidean distance was applied. Samples with a readcount ≥ 2 standard deviations below the mean of all sample readcounts were removed, as well as samples that clustered in a separate branch.
Analysis of RNA-seq
Global gene expression differences between pre- and post-treatment samples passing QC were determined using DESeq2 (v1.22.2) (90). Significance of expression level differences between pre- and post-treatment samples was determined using a paired t-test.
Gene set enrichment was performed using pre-ranked GSEA (87) based on the Wald statistic provided by DESeq2. For visualization purposes, the data were Z-transformed per gene. Heatmaps of gene expression values were created using the aheatmap function from the R package NMF (v0.21.0) (86) with a color scheme from RColorBrewer (v1.1-2; https://CRAN.R-project.org/package=RColorBrewer).
To assign samples to previously described PCa subtypes (31), the z-transformed expression levels of the top ~100 most differentially expressed genes (n=285) in each of the three clusters were investigated. Using these values, samples were clustered based on their Pearson correlation. The resulting tree was divided into 3 clusters, corresponding to the previously published PCa subtypes. Potential transitioning of samples from one cluster to another after treatment was visualized using a riverplot (v0.6; https://CRAN.R-project.org/package=riverplot).
To calculate fold changes of neuroendocrine scores upon treatment, expression of 70 neuroendocrine signature genes were obtained from castration-resistant neuroendocrine and prostate adenocarcinoma samples as published previously (35). The expression of 5 of the 70 neuroendocrine signature genes were not included in the analysis (KIAA0408, SOGA3, LRRC16B, ST8SIA3, SVOP) because the genes are not expressed in these samples. Expression fold changes between paired pre- and post-treatment samples were calculated (n=39) and concordance in gene expression differences (fold change sign) were measured using Pearson correlation.
CNV-seq
CNV-seq
Low-coverage whole-genome samples (ChIP-seq inputs), sequenced single-end 65-bp on a HiSeq 2500 system were aligned to hg19 with Burrows-Wheeler Aligner backtrack algorithm (v0.5.10) (72). The mappability per 20-kb on the genome, for a samples’ reads, phred quality 37 and higher, was rated against a similarly obtained mappability for all known and tiled 65-bp subsections of hg19. Sample counts were corrected per bin for local GC effects using a non-linear loess fit of mappabilities over 0.8 on autosomes. Reference values were scaled according to the slope of a linear fit, forced to intercept at the origin, of reference mappabilities after GC correction. Ratios of corrected sample counts and reference values left out bins with mappability below 0.2 or overlapping ENCODE blacklisted regions (91).
Analysis of CNV-seq
Copy number log ratios were smoothed and segmented using the R package DNACopy (v1.50.1; https://bioconductor.org/packages/release/bioc/html/DNAcopy.html) with the parameters set to alpha=0.00000000001, undo.SD=2, and undo.splits=“sdundo”. Bedtools intersect (v2.27.1) (85) was used to determine overlap between copy number segments and differential FOXA1 binding sites. These data were subsequently visualized using the aheatmap function from the R package NMF (v0.21.0) (86) with a color scheme from RColorBrewer (v1.1-2; https://CRAN.R-project.org/package=RColorBrewer).
To correlate FOXA1 ChIP-seq signal with copy number status at differential FOXA1 sites, we employed the z-transformed FOXA1 ChIP-seq readcounts as described in the ChIP-seq section. The difference in transformed ChIP-seq readcounts and the difference in normalized segmented copy number data between matched post-treatment and pre-treatment samples was calculated for every patient. Subsequently, the Pearson correlation between these two sets of differences was calculated.
Immunohistochemistry
For immunohistochemistry (IHC) analysis, we matched our ENZ-treated patient cohort (n=51) in a 1:2 ratio to untreated control patients (not receiving ENZ prior to prostatectomy; n=110) based on clinicopathological parameters (initial PSA, Gleason score, TNM stage, age) using the R package MatchIt (v.4.1.0) (92).
Tissue microarrays (TMAs) were prepared containing 3 cores per FFPE tumor sample. Tumor-dense areas in FFPE megablocks were marked by an expert pathologist on a H&E slide. Cores were drilled in a receptor block using the TMA grandmaster (3D Histech/Sysmex). Next, cores were taken from the donor block and placed in the receptor block using the manual tissue arrayer (4508-DM, Beecher instruments). The filled receptor block was placed in a 70°C stove for 9 minutes and cooled overnight at RT.
IHC was applied to TMA slides using a BenchMark Ultra autostainer (Ventana Medical Systems). In brief, paraffin sections were cut at 3 μm, heated at 75°C for 28 minutes and deparaffinized in the instrument with EZ prep solution (Ventana Medical Systems). Heat-induced antigen retrieval was carried out using Cell Conditioning 1 (CC1, Ventana Medical Systems) for 24 minutes (cleaved-Caspase-3), 32 minutes (Chromogranin, Synaptophysin) or 64 minutes (ARNTL, Ki-67) at 95°C. The following antibodies and staining conditions were used: anti-ARNTL (ab230822, Abcam; 1:1000 dilution; 60 min at room temperature), anti-Ki-67 (M7240, Agilent; 1;100 dilution; 60 min at 37°C), anti-cleaved-Caspase-3 (9661, Cell Signaling; 1:100 dilution; 32 min at room temperature), anti-Chromogranin (760-2519, Ventana Medical Systems; undiluted; 32 min at 37°C), and anti-Synaptophysin (SYNAP-299-L-CE, Leica; 1:100; 32 min at 37°C). For Synaptophysin signal amplification was applied using the OptiView Amplification Kit (Ventana Medical Systems; 4 min). Bound antibody was detected using the OptiView DAB Detection Kit (Ventana Medical Systems). Slides were counterstained with hematoxylin and bluing reagent (Ventana Medical Systems).
Percentage of positive tumor cells or IHC staining intensity (weak, moderate, strong) in tumor cells were scored by an expert pathologist, and used for statistical analysis.
FOXA1 mutation status
FOXA1 mutation status was assessed from H3K27ac ChIP-seq and RNA-seq reads covering the gene. We focused our search to genomic coordinates with mutations previously reported in cBioPortal (https://www.cbioportal.org). cBioPortal was queried for all somatic mutations in FOXA1 (n=567 mutations) across all PCa samples (n=6,875 patients). Non-reference alleles were called from bam files with H3K27ac ChIP-seq or RNA-seq reads using the mpileup and call commands from bcftools (v1.9). The --prior variable for call was set to 0.05 to enhance sensitivity in the setting of low read coverage. The genomic coordinates of variants were listed in bed files and tested for overlap with FOXA1 mutations from cbioPortal.
Survival analysis in mCRPC cohorts
RNA-seq data from mCRPC was processed as previously described (53,54,93) and converted to transcripts per million (TPM) or fragments per kilobase per million reads mapped (FPKM). Patients were grouped by ARNTL expression levels as low (< median) or high (≥ median). Survival analysis was performed using the Kaplan-Meier method with endpoint overall survival from diagnosis of mCRPC and the Wald test was used to test for statistical significance.
Cell lines and cell culture
LNCaP human PCa cell line and HEK293T cells were purchased from the American Type Culture Collection (ATCC). Enzalutamide-resistant LNCaP-42D (41) and LNCaP-ResA (56) cells were described previously. LNCaP clones were maintained in RPMI-1640 medium (Gibco, Thermo Fisher Scientific) supplemented with 10% FBS (Sigma-Aldrich), with ENZ-resistant cell lines further supplemented with 10 μM ENZ (MedChemExpress). HEK293T cells were cultured in DMEM (Gibco, Thermo Fisher Scientific) supplemented with 10% FBS. Cell lines were subjected to regular mycoplasma testing and all cell lines underwent authentication by short tandem repeat profiling (Eurofins Genomics). For hormone stimulation with synthetic androgen, cells were treated with 10 nM R1881 (PerkinElmer) for 48 h. For in vitro AR blockade, cells were treated with 10 μM ENZ and harvested at the indicated time points.
STARR-seq
Generation of FOXA1-focused STARR-seq library
Pooled human male genomic DNA (Promega) was randomly sheared, end-repaired, and ligated with Illumina compatible xGen CS stubby adaptors (IDT) containing 3-bp unique molecular identifiers (UMI). The adaptor-ligated gDNA fragments (500-800 bp) were hybridized to a custom biotinylated oligonucleotide probe (Agilent) and captured by Dynabeads M-270 Streptavidin beads (NEB). The library was designed to capture regions from clinical ChIP-seq. Any overlapping reads were collapsed using BedTools’ ‘merge’ (v2.30.0) command to eliminate possible over-representation. Target regions were PCR-amplified with STARR_in-fusion_F (TAGAGCATGCACCGGACACTCTTTCCCTACACGACGCTCTTCCGATCT) and STARR_in-fusion_R (GGCCGAATTCGTCGAGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT) primers, and cloned into AgeI-HF (NEB) and SalI-HF (NEB) digested hSTARR-ORI plasmid (#99296, Addgene) by Gibson Assembly. STARR-seq capture library was transformed into MegaX DH10B T1R electrocompetent cells (Invitrogen). Plasmid DNA was extracted using the Qiagen Plasmid Maxi Kit.
STARR-seq
LNCaP cells (> 1.6 × 108 cells/replicate; 3 biological replicates for each cell line) were electroporated with the STARR-seq capture library (1 × 106 cells: 2 μg DNA; ~320 μg plasmid DNA/replicate) using the Neon Transfection System (Invitrogen). Electroporated LNCaP cells were immediately recovered in RPMI-1640 medium supplemented with 10% FBS and the culture medium was refreshed 24h after electroporation. LNCaP cells (~0.5 × 108 cells) were treated with dimethylsulfoxide (DMSO) or 10 μM ENZ for 48 h and then either EtOH or 10 nM DHT for 4 h. All electroporated LNCaP cells were harvested 72 h post electroporation. Cell samples were lysed with the Precellys CKMix Tissue Homegenizing Kit and Precellys 24 Tissue/Cell Ruptor (Berin Technologies). Total RNA was extracted using Qiagen Rneasy Maxi Kit (Qiagen) and poly-A mRNA was isolated using the Oligo (dT)25 Dynabeads (Thermo Fisher). FOXA1-focused STARR-seq cDNA was synthesized with the gene-specific primer (CTCATCAATGTATCTTATCATGTCTG) and amplified by junction PCR (15 cycles) with the RNA_jPCR_f (TCGTGAGGCACTGGGCAG*G*T*G*T*C) and jPCR-r (CTTATCATGTCTGCTCGA*A*G*C) primers. FOXA1-focused STARR-seq capture library plasmid DNA was extracted from 0.1 × 108 transfected but untreated cells using the QIAprep Spin Miniprep Kit (Qiagen). The extracted plasmid DNA and the input plasmid DNA were PCR amplified with the DNA-specific junction PCR primer (DNA_jPCR_f, CCTTTCTCTCCACAGGT*G*T*C) and jPCR-r primer. After purification with Ampure XP beads, Illumina compatible libraries were generated by PCR amplification with NEBNext universal and single indexing primers (NEB), and were sequenced on Illumina NovaSeq6000 (150-bp; paired-end).
Analysis of STARR-seq
STARR-seq sequencing data was analyzed using a custom Snakemake pipeline (https://github.com/birkiy/starr-pipe). Briefly, paired-end STARR-seq samples were aligned to hg19 genome using BWA (v0.7.17). Raw alignment files were converted into BEDPE format using BedTools (v2.30.0) ‘bamtobed -bedpe’ command. The start of the first paired read and the end of its mate defined the fragments from the BEDPE file. Any fragments overlapping with hg19 blacklisted regions (https://github.com/Boyle-Lab/Blacklist) or MAPQ scores < 30 were filtered from downstream analysis. Fragments containing unique genomic positions were counted using ‘uniq -c’ UNIX command. A count table of the unique fragment collection count was generated using a custom Julia script (v1.5.2) fragments.jl which uses library input samples to first generate the reference fragment population and then quantifies the frequencies of each fragment.
STARR-seq aligned files were downsampled using SAMtools (v1.10) to make files with equivalent readcounts across conditions. Next, count tables were generated from the downsampled files for all tested FOXA1 regions (n=1,209) using the deepTools (v2.0) multiBamSummary function. The most correlated replicates were chosen for further analysis using the cor function in R (v3.4.4). Regions with zero counts across samples were removed leaving 968 regions. These count tables were used as input for a differential expression analysis using DESeq2 (v1.22.2) in R. Regions with non-significant changes (FDR ≤ 0.05, logFC ≥ |2|) in readcounts upon ENZ treatment were identified and k-means clustering from the plotHeatmap function of deepTools was performed. To determine possible functional associations within these clusters the sets of regions were queried using the CistromeDB Toolkit to identify factors with significant overlap.
RIME
Sample processing
Following treatment of LNCaP and LNCaP-42D cells with ENZ (10 μM) for 48 h, cells were fixed, lysed and sonicated as previously described (94). The nuclear lysates were incubated with 50 μL magnetic protein A beads (10008D, Thermo Fisher Scientific) conjugated to 7.5 μg ARNTL antibody (ab93806, Abcam) or rabbit IgG control (12-370, Merck Millipore).
LC-MS/MS
Peptide mixtures were prepared and measured as previously described (4), with the following exceptions. Peptide mixtures (10% of total digest) were loaded directly onto the analytical column (ReproSil-Pur 120 C18-AQ, 2.4 μm, 75 μm × 500 mm, packed in-house) and analyzed by nanoLC-MS/MS on an Orbitrap Fusion Tribrid mass spectrometer equipped with a Proxeon nLC1200 system (Thermo Scientific). Solvent A was 0.1% formic acid/water and solvent B was 0.1% formic acid/80% acetonitrile. Peptides were eluted from the analytical column at a constant flow of 250 nL/min in a 120 min gradient, containing a 105 min step-wise increase from 7% to 34% solvent B, followed by a 15 min wash at 80% solvent B.
Analysis of RIME data
Raw data were analyzed by MaxQuant (v2.0.1.0) (95) using standard settings for label-free quantitation (LFQ). MS/MS data were searched against the Swissprot human database (20,397 entries, release 2021_01) complemented with a list of common contaminants and concatenated with the reversed version of all sequences. The maximum allowed mass tolerance was 4.5 ppm in the main search and 0.5 Da for fragment ion masses. False discovery rates for peptide and protein identification were set to 1%. Trypsin/P was chosen as cleavage specificity allowing two missed cleavages. Carbamidomethylation was set as a fixed modification, while oxidation and deamidation were used as variable modifications. LFQ intensities were log2-transformed in Perseus (v1.6.15.0) (96), after which proteins were filtered for at least 3 out of 4 valid values in at least one sample group. Missing values were replaced by imputation based on a normal distribution (width: 0.3 and downshift: 1.8). Differentially enriched proteins were determined using a Student’s t-test (threshold: P ≤ 0.05 and [x-y] ≥ 1.8 | [x-y] ≤ -1.8).
Transient cell line transfections
Transient transfections of cell lines were performed according to the manufacturer’s instructions using Lipofectamine 2000 (Invitrogen) or Lipofectamine RNAiMAX (Invitrogen) for overexpression or siRNA knockdown experiments, respectively. ARNTL containing expression plasmid was obtained from the CCSB-Broad Lentiviral Expression Library. siRNA oligos targeting ARNTL (M-010261-00-0005), FOXA1 (M-010319-01-0020), NR3C1 (M-003424-03-0005) and the non-targeting control (D-001206-14, D-001210-05-20) were purchased from Dharmacon. For ARNTL ChIP-seq upon FOXA1 knockdown, LNCaP and LNCaP-42D cells were reverse transfected with 50 nM siFOXA1 using Lipofectamine RNAiMAX. ENZ (10 μM) was added after 24 h, and cells were fixed and harvested for ChIP-seq analysis 72 h post-transfection.
CRISPR/Cas9-mediated knockout cell lines
Guide RNAs targeting human ARNTL (CTGGACATTGCGTTGCATGT) and a non-targeting control guide (AACTACAAGTAAAAGTATCG) were individually cloned into the lentiCRISPR v2 plasmid (97). CRISPR vectors were co-expressed with 3rd generation viral vectors in HEK293T cells using polyethyleneimine (PEI; Polysciences). After lentivirus production, the medium was harvested and transferred to the designated cell lines. Two days post infection, cells were put on puromycin (Sigma-Aldrich) selection for 3 weeks and knockout efficiency was tested using western blot analysis.
Western blotting
Total proteins were extracted from cells using Laemmli lysis buffer, supplemented with a complete protease inhibitor cocktail (Roche). Per sample, 40 μg of protein was resolved by SDS-PAGE (10%) and transferred on nitrocellulose membranes (Santa Cruz Biotechnology). The following antibodies were used for Western blot stainings: ARNTL (ab93806, Abcam), PSA (5365, Cell Signaling Technology), FOXA1 (ab5089, Abcam), GR (12041, Cell Signaling Technology) and ACTIN (MAB1501R, Merck Millipore). Blots were incubated overnight at 4°C with designated primary antibodies at 1:1000 (ARNTL, PSA) or 1:5000 (ACTIN) dilution, and visualized using the Odyssey system (Li-Cor Biosciences).
RNA isolation and mRNA expression
Total RNA from cell lines was isolated using TRIzol Reagent (Thermo Fisher Scientific), and cDNA was synthesized from 2 μg RNA using the SuperScript™ III Reverse Transcriptase system (Thermo Fisher Scientific) with random hexamer primers according to the manufacturer’s instructions. Quantitative PCR (qPCR) was performed using the SensiMix™ SYBR Kit (Bioline) according to the instructions provided by the manufacturer on a QuantStudio™ 6 Flex System (Thermo Fisher Scientific). Primer sequences for mRNA expression analyses are FOXA1 (forward: CGACTGGAACAGCTACTACG; reverse: TGGTGTTCATGGTCATGTAGGT) and ARNTL (forward: CTGGAGCACGACGTTCTTTCTT; reverse: GGATTGTGCAGAAGCTTTTTCG). mRNA levels are shown relative to the expression of housekeeping gene TBP (forward: GTTCTGGGAAAATGGTGTGC; reverse: GCTGGAAAACCCAACTTCTG).
Cell viability and proliferation assays
For cell viability assays, LNCaP, LNCaP-42D or LNCaP-ResA cells were seeded at 2 × 103 cells per well in 96-well plates (Greiner) ± 10 μM ENZ, and reverse transfected with 50-100 nM siRNA (Dharmacon) using Lipofectamine RNAiMAX (Invitrogen). Cell viability was assessed 7 days post-transfection using the CellTiter-Glo Luminescent Cell Viability Assay kit (Promega), as per the manufacturer’s instructions. Bar charts were plotted using GraphPad Prism 9 software.
Proliferation curves for stable ARNTL knockout clones were generated using a Lionheart FX automated microscope (BioTek). Cells (LNCaP, LNCaP-42D, LNCaP-ResA) were seeded at 2 × 103 cells per well in 96-well plates ± 10 μM ENZ. SiR-DNA (Spirochrome) live cell nuclear stain was added 2 h before imaging. Cell growth was recorded with a time resolution of 4 h for a total time span of 144 h. The microscope was maintained at 37 °C, 5% CO2 and live-cell imaging was performed using a 4x lens and a Sony CCD, 1,25-megapixel camera with two times binning (BioTek). Gen5 software (BioTek) was used to quantify cell numbers and growth curves were plotted using GraphPad Prism 9 software.
Xenograft studies
For in vivo tumor growth studies, 7.5 × 106 sgNT or sgARNTL (LNCaP, LNCaP-42D, LNCaP-ResA) cells in PBS with 50% BME (3536-005-02, Bio-Techne) were injected subcutaneously into one of the flanks of ~seven-week-old male NOD-SCID (NSG) mice. Once tumor size reached 150 mm3, mice were randomized and treated with either 10 mg/kg ENZ (MedChemExpress), or vehicle control (1% carboxymethylcellulose sodium salt, 0.1% Tween-80, 5% DMSO; Sigma-Aldrich) through oral gavage on a daily basis. Tumor volume was monitored by caliper measurements 3 times a week. Mice were kept under standard temperature and humidity conditions in individually ventilated cages, with food and water provided ad libitum. All animal experiments were approved by the Animal Welfare Committee of the Netherlands Cancer Institute and performed in accordance with institutional, national and European guidelines for animal research.
Statistical analysis
For differential binding and differential gene expression analyses (pre- vs. post-ENZ), an FDR cutoff < 0.05 (P < 0.01) and Padj < 0.01 was used, respectively. A Mann-Whitney U-test was used to determine differences in region readcounts (adjusted for multiple testing using FDR) and differences in gene expression levels before and after ENZ treatment. For peak set and gene set overlaps as well as to determine differences in IHC staining intensities, Fisher’s exact tests were applied. Differences in cell viability or cell/tumor growth were tested using a two-way or one-way ANOVA followed by Tukey’s multiple comparisons test, respectively (GraphPad Prism 9). Corresponding bar chart or growth curves show the mean with error bars representing the SD. All boxplots indicate the median (center line), upper-(75) and lower - (25) quartile range (box limits) and 1.5 × interquartile range (whiskers). Significance is indicated as follows: ns, P > 0.05; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. Further details of statistical tests are provided in the figure legends.
Supplementary Material
Significance.
Understanding how prostate cancers adapt to AR-targeted interventions is critical for identifying novel drug targets to improve clinical management of treatment-resistant disease. Our study revealed an enzalutamide-induced epigenomic plasticity towards pro-survival signaling, and uncovered circadian regulator ARNTL as an acquired vulnerability after AR-inhibition, presenting a novel lead for therapeutic development.
Acknowledgements
The authors would like to thank the NKI-AVL Core Facility Molecular Pathology & Biobanking (CFMPB), the NKI Proteomics/Mass Spectrometry Facility (supported by the Dutch NWO X-omics Initiative), and the NKI Preclinical Intervention Unit (MCCA) for experimental assistance; the NKI Genomics Core Facility (GCF) for next generation sequencing and bioinformatics support; and the NKI Research High Performance Computing facility (RHPC) for computational infrastructure. We also thank all Zwart/Bergman lab members for fruitful discussions and technical advice. Finally, we would like to thank all patients and clinical staff who were involved in the DARANA trial.
Grant Support
This work was supported by Movember (NKI01 to A.M. Bergman and W. Zwart), KWF Dutch Cancer Society (10084 ALPE to A.M. Bergman and W. Zwart), KWF Dutch Cancer Society/Alpe d’HuZes Bas Mulder Award (NKI 2014-6711 to W. Zwart), Netherlands Organization for Scientific Research (NWO-VIDI-016.156.401 to W. Zwart), and Astellas Pharma (to W. Zwart, A.M. Bergman, H. van der Poel).
Footnotes
Declaration of Potential Conflicts of Interest
W. Zwart, A.M. Bergman and H. van der Poel received research funding from Astellas Pharma B.V. (Leiden, the Netherlands). No potential conflicts of interest were disclosed by the other authors.
Data Availability
All tissue ChIP-seq and RNA-seq raw data generated in this study have been deposited in the European Genome-Phenome Archive (EGA) under the accession numbers EGAS00001006017 and EGAS00001006016, respectively. The cell line ChIP-seq, as well as all processed tissue ChIP-seq and RNA-seq data have been deposited in the Gene Expression Omnibus (GEO) database, under accession number GSE197781. The mass spectrometry proteomics (RIME) data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD032041. Public ChIP-seq datasets used in this study are available from GEO or the EGA under the following accession code: GSE120738 (AR, H3K27ac ChIP-seq), GSE51497 (GR ChIP-seq), GSE117306 (N-MYC ChIP-seq) and EGAS00001003928 (FOXA1 ChIP-seq).
References
- 1.Huggins C, Hodges CV. Studies on prostatic cancer. I. The effect of castration, of estrogen and androgen injection on serum phosphatases in metastatic carcinoma of the prostate. CA Cancer J Clin. 1972;22(4):232–40. doi: 10.3322/canjclin.22.4.232. [DOI] [PubMed] [Google Scholar]
- 2.Zhang Z, Chng KR, Lingadahalli S, Chen Z, Liu MH, Do HH, et al. An AR-ERG transcriptional signature defined by long-range chromatin interactomes in prostate cancer cells. Genome Res. 2019;29(2):223–35. doi: 10.1101/gr.230243.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stelloo S, Bergman AM, Zwart W. Androgen receptor enhancer usage and the chromatin regulatory landscape in human prostate cancers. Endocr Relat Cancer. 2019;26(5):R267–R85. doi: 10.1530/ERC-19-0032. [DOI] [PubMed] [Google Scholar]
- 4.Stelloo S, Nevedomskaya E, Kim Y, Hoekman L, Bleijerveld OB, Mirza T, et al. Endogenous androgen receptor proteomic profiling reveals genomic subcomplex involved in prostate tumorigenesis. Oncogene. 2018;37(3):313–22. doi: 10.1038/onc.2017.330. [DOI] [PubMed] [Google Scholar]
- 5.Pomerantz MM, Li F, Takeda DY, Lenci R, Chonkar A, Chabot M, et al. The androgen receptor cistrome is extensively reprogrammed in human prostate tumorigenesis. Nat Genet. 2015;47(11):1346–51. doi: 10.1038/ng.3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gerhardt J, Montani M, Wild P, Beer M, Huber F, Hermanns T, et al. FOXA1 promotes tumor progression in prostate cancer and represents a novel hallmark of castration-resistant prostate cancer. Am J Pathol. 2012;180(2):848–61. doi: 10.1016/j.ajpath.2011.10.021. [DOI] [PubMed] [Google Scholar]
- 7.Pomerantz MM, Qiu X, Zhu Y, Takeda DY, Pan W, Baca SC, et al. Prostate cancer reactivates developmental epigenomic programs during metastatic progression. Nat Genet. 2020;52(8):790–9. doi: 10.1038/s41588-020-0664-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cirillo LA, Lin FR, Cuesta I, Friedman D, Jarnik M, Zaret KS. Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol Cell. 2002;9(2):279–89. doi: 10.1016/s1097-2765(02)00459-8. [DOI] [PubMed] [Google Scholar]
- 9.Lupien M, Eeckhoute J, Meyer CA, Wang Q, Zhang Y, Li W, et al. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell. 2008;132(6):958–70. doi: 10.1016/j.cell.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sahu B, Laakso M, Ovaska K, Mirtti T, Lundin J, Rannikko A, et al. Dual role of FoxA1 in androgen receptor binding to chromatin, androgen signalling and prostate cancer. EMBO J. 2011;30(19):3962–76. doi: 10.1038/emboj.2011.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Teng M, Zhou S, Cai C, Lupien M, He HH. Pioneer of prostate cancer: past, present and the future of FOXA1. Protein Cell. 2021;12(1):29–38. doi: 10.1007/s13238-020-00786-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grasso CS, Wu YM, Robinson DR, Cao X, Dhanasekaran SM, Khan AP, et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012;487(7406):239–43. doi: 10.1038/nature11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barbieri CE, Baca SC, Lawrence MS, Demichelis F, Blattner M, Theurillat JP, et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat Genet. 2012;44(6):685–9. doi: 10.1038/ng.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cancer Genome Atlas Research N. The Molecular Taxonomy of Primary Prostate Cancer. Cell. 2015;163(4):1011–25. doi: 10.1016/j.cell.2015.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Robinson D, Van Allen EM, Wu YM, Schultz N, Lonigro RJ, Mosquera JM, et al. Integrative Clinical Genomics of Advanced Prostate Cancer. Cell. 2015;162(2):454. doi: 10.1016/j.cell.2015.06.053. [DOI] [PubMed] [Google Scholar]
- 16.Wedge DC, Gundem G, Mitchell T, Woodcock DJ, Martincorena I, Ghori M, et al. Sequencing of prostate cancers identifies new cancer genes, routes of progression and drug targets. Nat Genet. 2018;50(5):682–92. doi: 10.1038/s41588-018-0086-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Adams EJ, Karthaus WR, Hoover E, Liu D, Gruet A, Zhang Z, et al. FOXA1 mutations alter pioneering activity, differentiation and prostate cancer phenotypes. Nature. 2019;571(7765):408–12. doi: 10.1038/s41586-019-1318-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parolia A, Cieslik M, Chu SC, Xiao L, Ouchi T, Zhang Y, et al. Distinct structural classes of activating FOXA1 alterations in advanced prostate cancer. Nature. 2019;571(7765):413–8. doi: 10.1038/s41586-019-1347-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mottet N, van den Bergh RCN, Briers E, Van den Broeck T, Cumberbatch MG, De Santis M, et al. EAU-EANM-ESTRO-ESUR-SIOG Guidelines on Prostate Cancer-2020 Update. Part 1: Screening, Diagnosis, and Local Treatment with Curative Intent. Eur Urol. 2021;79(2):243–62. doi: 10.1016/j.eururo.2020.09.042. [DOI] [PubMed] [Google Scholar]
- 20.Roehl KA, Han M, Ramos CG, Antenor JA, Catalona WJ. Cancer progression and survival rates following anatomical radical retropubic prostatectomy in 3,478 consecutive patients: long-term results. J Urol. 2004;172(3):910–4. doi: 10.1097/01.ju.0000134888.22332.bb. [DOI] [PubMed] [Google Scholar]
- 21.Harris WP, Mostaghel EA, Nelson PS, Montgomery B. Androgen deprivation therapy: progress in understanding mechanisms of resistance and optimizing androgen depletion. Nat Clin Pract Urol. 2009;6(2):76–85. doi: 10.1038/ncpuro1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cornford P, van den Bergh RCN, Briers E, Van den Broeck T, Cumberbatch MG, De Santis M, et al. EAU-EANM-ESTRO-ESUR-SIOG Guidelines on Prostate Cancer. Part II-2020 Update: Treatment of Relapsing and Metastatic Prostate Cancer. Eur Urol. 2021;79(2):263–82. doi: 10.1016/j.eururo.2020.09.046. [DOI] [PubMed] [Google Scholar]
- 23.Zong Y, Goldstein AS. Adaptation or selection--mechanisms of castration-resistant prostate cancer. Nat Rev Urol. 2013;10(2):90–8. doi: 10.1038/nrurol.2012.237. [DOI] [PubMed] [Google Scholar]
- 24.Linder S, van der Poel HG, Bergman AM, Zwart W, Prekovic S. Enzalutamide therapy for advanced prostate cancer: efficacy, resistance and beyond. Endocr Relat Cancer. 2018;26(1):R31–R52. doi: 10.1530/ERC-18-0289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scher HI, Fizazi K, Saad F, Taplin ME, Sternberg CN, Miller K, et al. Increased survival with enzalutamide in prostate cancer after chemotherapy. N Engl J Med. 2012;367(13):1187–97. doi: 10.1056/NEJMoa1207506. [DOI] [PubMed] [Google Scholar]
- 26.Beer TM, Armstrong AJ, Rathkopf DE, Loriot Y, Sternberg CN, Higano CS, et al. Enzalutamide in metastatic prostate cancer before chemotherapy. N Engl J Med. 2014;371(5):424–33. doi: 10.1056/NEJMoa1405095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Armstrong AJ, Szmulewitz RZ, Petrylak DP, Holzbeierlein J, Villers A, Azad A, et al. ARCHES: A Randomized, Phase III Study of Androgen Deprivation Therapy With Enzalutamide or Placebo in Men With Metastatic Hormone-Sensitive Prostate Cancer. J Clin Oncol. 2019;37(32):2974–86. doi: 10.1200/JCO.19.00799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hussain M, Fizazi K, Saad F, Rathenborg P, Shore N, Ferreira U, et al. Enzalutamide in Men with Nonmetastatic, Castration-Resistant Prostate Cancer. N Engl J Med. 2018;378(26):2465–74. doi: 10.1056/NEJMoa1800536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Prekovic S, van den Broeck T, Linder S, van Royen ME, Houtsmuller AB, Handle F, et al. Molecular underpinnings of enzalutamide resistance. Endocr Relat Cancer. 2018;25(11):R545–R57. doi: 10.1530/ERC-17-0136. [DOI] [PubMed] [Google Scholar]
- 30.Gao N, Zhang J, Rao MA, Case TC, Mirosevich J, Wang Y, et al. The role of hepatocyte nuclear factor-3 alpha (Forkhead Box A1) and androgen receptor in transcriptional regulation of prostatic genes. Mol Endocrinol. 2003;17(8):1484–507. doi: 10.1210/me.2003-0020. [DOI] [PubMed] [Google Scholar]
- 31.Stelloo S, Nevedomskaya E, Kim Y, Schuurman K, Valle-Encinas E, Lobo J, et al. Integrative epigenetic taxonomy of primary prostate cancer. Nat Commun. 2018;9(1):4900. doi: 10.1038/s41467-018-07270-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mazrooei P, Kron KJ, Zhu Y, Zhou S, Grillo G, Mehdi T, et al. Cistrome Partitioning Reveals Convergence of Somatic Mutations and Risk Variants on Master Transcription Regulators in Primary Prostate Tumors. Cancer Cell. 2019;36(6):674–89.:e6. doi: 10.1016/j.ccell.2019.10.005. [DOI] [PubMed] [Google Scholar]
- 33.Kron KJ, Murison A, Zhou S, Huang V, Yamaguchi TN, Shiah YJ, et al. TMPRSS2-ERG fusion co-opts master transcription factors and activates NOTCH signaling in primary prostate cancer. Nat Genet. 2017;49(9):1336–45. doi: 10.1038/ng.3930. [DOI] [PubMed] [Google Scholar]
- 34.Huang CF, Lingadahalli S, Morova T, Ozturan D, Hu E, Yu IPL, et al. Functional mapping of androgen receptor enhancer activity. Genome Biol. 2021;22(1):149. doi: 10.1186/s13059-021-02339-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beltran H, Prandi D, Mosquera JM, Benelli M, Puca L, Cyrta J, et al. Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. Nat Med. 2016;22(3):298–305. doi: 10.1038/nm.4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Che M, Chaturvedi A, Munro SA, Pitzen SP, Ling A, Zhang W, et al. Opposing transcriptional programs of KLF5 and AR emerge during therapy for advanced prostate cancer. Nat Commun. 2021;12(1):6377. doi: 10.1038/s41467-021-26612-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Henry GH, Malewska A, Joseph DB, Malladi VS, Lee J, Torrealba J, et al. A Cellular Anatomy of the Normal Adult Human Prostate and Prostatic Urethra. Cell Rep. 2018;25(12):3530–42.:e5. doi: 10.1016/j.celrep.2018.11.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Epstein JI, Amin MB, Beltran H, Lotan TL, Mosquera JM, Reuter VE, et al. Proposed morphologic classification of prostate cancer with neuroendocrine differentiation. Am J Surg Pathol. 2014;38(6):756–67. doi: 10.1097/PAS.0000000000000208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim S, Thaper D, Bidnur S, Toren P, Akamatsu S, Bishop JL, et al. PEG10 is associated with treatment-induced neuroendocrine prostate cancer. J Mol Endocrinol. 2019;63(1):39–49. doi: 10.1530/JME-18-0226. [DOI] [PubMed] [Google Scholar]
- 40.Berger A, Brady NJ, Bareja R, Robinson B, Conteduca V, Augello MA, et al. N-Myc-mediated epigenetic reprogramming drives lineage plasticity in advanced prostate cancer. J Clin Invest. 2019;129(9):3924–40. doi: 10.1172/JCI127961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bishop JL, Thaper D, Vahid S, Davies A, Ketola K, Kuruma H, et al. The Master Neural Transcription Factor BRN2 Is an Androgen Receptor-Suppressed Driver of Neuroendocrine Differentiation in Prostate Cancer. Cancer Discov. 2017;7(1):54–71. doi: 10.1158/2159-8290.CD-15-1263. [DOI] [PubMed] [Google Scholar]
- 42.Baca SC, Takeda DY, Seo JH, Hwang J, Ku SY, Arafeh R, et al. Reprogramming of the FOXA1 cistrome in treatment-emergent neuroendocrine prostate cancer. Nat Commun. 2021;12(1):1979. doi: 10.1038/s41467-021-22139-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baca SC, Singler C, Zacharia S, Seo J-H, Morova T, Hach F, et al. Genetic determinants of chromatin reveal prostate cancer risk mediated by context-dependent gene regulation. bioRxiv. 2021:2021.05.10.443466. doi: 10.1101/2021.05.10.443466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Giambartolomei C, Seo JH, Schwarz T, Freund MK, Johnson RD, Spisak S, et al. H3K27ac HiChIP in prostate cell lines identifies risk genes for prostate cancer susceptibility. Am J Hum Genet. 2021;108(12):2284–300. doi: 10.1016/j.ajhg.2021.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Meyers RM, Bryan JG, McFarland JM, Weir BA, Sizemore AE, Xu H, et al. Computational correction of copy number effect improves specificity of CRISPR-Cas9 essentiality screens in cancer cells. Nat Genet. 2017;49(12):1779–84. doi: 10.1038/ng.3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dempster JM, Rossen J, Kazachkova M, Pan J, Kugener G, Root DE, et al. Extracting Biological Insights from the Project Achilles Genome-Scale CRISPR Screens in Cancer Cell Lines. bioRxiv. 2019:720243. doi: 10.1101/720243. [DOI] [Google Scholar]
- 47.Layer RM, Pedersen BS, DiSera T, Marth GT, Gertz J, Quinlan AR. GIGGLE: a search engine for large-scale integrated genome analysis. Nat Methods. 2018;15(2):123–6. doi: 10.1038/nmeth.4556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zheng R, Wan C, Mei S, Qin Q, Wu Q, Sun H, et al. Cistrome Data Browser: expanded datasets and new tools for gene regulatory analysis. Nucleic Acids Res. 2019;47(D1):D729–D35. doi: 10.1093/nar/gky1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Arora VK, Schenkein E, Murali R, Subudhi SK, Wongvipat J, Balbas MD, et al. Glucocorticoid receptor confers resistance to antiandrogens by bypassing androgen receptor blockade. Cell. 2013;155(6):1309–22. doi: 10.1016/j.cell.2013.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shah N, Wang P, Wongvipat J, Karthaus WR, Abida W, Armenia J, et al. Regulation of the glucocorticoid receptor via a BET-dependent enhancer drives antiandrogen resistance in prostate cancer. Elife. 2017;6 doi: 10.7554/eLife.27861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Palit SA, Vis D, Stelloo S, Lieftink C, Prekovic S, Bekers E, et al. TLE3 loss confers AR inhibitor resistance by facilitating GR-mediated human prostate cancer cell growth. Elife. 2019;8 doi: 10.7554/eLife.47430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shafi AA, McNair CM, McCann JJ, Alshalalfa M, Shostak A, Severson TM, et al. The circadian cryptochrome, CRY1, is a pro-tumorigenic factor that rhythmically modulates DNA repair. Nat Commun. 2021;12(1):401. doi: 10.1038/s41467-020-20513-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Quigley DA, Dang HX, Zhao SG, Lloyd P, Aggarwal R, Alumkal JJ, et al. Genomic Hallmarks and Structural Variation in Metastatic Prostate Cancer. Cell. 2018;175(3):889. doi: 10.1016/j.cell.2018.10.019. [DOI] [PubMed] [Google Scholar]
- 54.Abida W, Cyrta J, Heller G, Prandi D, Armenia J, Coleman I, et al. Genomic correlates of clinical outcome in advanced prostate cancer. Proc Natl Acad Sci U S A. 2019;116(23):11428–36. doi: 10.1073/pnas.1902651116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kregel S, Chen JL, Tom W, Krishnan V, Kach J, Brechka H, et al. Acquired resistance to the second-generation androgen receptor antagonist enzalutamide in castration-resistant prostate cancer. Oncotarget. 2016;7(18):26259–74. doi: 10.18632/oncotarget.8456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Handle F, Prekovic S, Helsen C, Van den Broeck T, Smeets E, Moris L, et al. Drivers of AR indifferent anti-androgen resistance in prostate cancer cells. Sci Rep. 2019;9(1):13786. doi: 10.1038/s41598-019-50220-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Isikbay M, Otto K, Kregel S, Kach J, Cai Y, Vander Griend DJ, et al. Glucocorticoid receptor activity contributes to resistance to androgen-targeted therapy in prostate cancer. Horm Cancer. 2014;5(2):72–89. doi: 10.1007/s12672-014-0173-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang Z, Zhou C, Li X, Barnes SD, Deng S, Hoover E, et al. Loss of CHD1 Promotes Heterogeneous Mechanisms of Resistance to AR-Targeted Therapy via Chromatin Dysregulation. Cancer Cell. 2020;37(4):584–98.:e11. doi: 10.1016/j.ccell.2020.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Robinson DR, Wu YM, Vats P, Su F, Lonigro RJ, Cao X, et al. Activating ESR1 mutations in hormone-resistant metastatic breast cancer. Nat Genet. 2013;45(12):1446–51. doi: 10.1038/ng.2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chabon JJ, Simmons AD, Lovejoy AF, Esfahani MS, Newman AM, Haringsma HJ, et al. Circulating tumour DNA profiling reveals heterogeneity of EGFR inhibitor resistance mechanisms in lung cancer patients. Nat Commun. 2016;7:11815. doi: 10.1038/ncomms11815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lievre A, Bachet JB, Le Corre D, Boige V, Landi B, Emile JF, et al. KRAS mutation status is predictive of response to cetuximab therapy in colorectal cancer. Cancer Res. 2006;66(8):3992–5. doi: 10.1158/0008-5472.CAN-06-0191. [DOI] [PubMed] [Google Scholar]
- 62.Takeda DY, Spisak S, Seo JH, Bell C, O’Connor E, Korthauer K, et al. A Somatically Acquired Enhancer of the Androgen Receptor Is a Noncoding Driver in Advanced Prostate Cancer. Cell. 2018;174(2):422–32.:e13. doi: 10.1016/j.cell.2018.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Viswanathan SR, Ha G, Hoff AM, Wala JA, Carrot-Zhang J, Whelan CW, et al. Structural Alterations Driving Castration-Resistant Prostate Cancer Revealed by Linked-Read Genome Sequencing. Cell. 2018;174(2):433–47.:e19. doi: 10.1016/j.cell.2018.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Beltran H, Rickman DS, Park K, Chae SS, Sboner A, MacDonald TY, et al. Molecular characterization of neuroendocrine prostate cancer and identification of new drug targets. Cancer Discov. 2011;1(6):487–95. doi: 10.1158/2159-8290.CD-11-0130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Davies AH, Beltran H, Zoubeidi A. Cellular plasticity and the neuroendocrine phenotype in prostate cancer. Nat Rev Urol. 2018;15(5):271–86. doi: 10.1038/nrurol.2018.22. [DOI] [PubMed] [Google Scholar]
- 66.Gao S, Chen S, Han D, Wang Z, Li M, Han W, et al. Chromatin binding of FOXA1 is promoted by LSD1-mediated demethylation in prostate cancer. Nat Genet. 2020;52(10):1011–7. doi: 10.1038/s41588-020-0681-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gleave ME, Goldenberg SL, Chin JL, Warner J, Saad F, Klotz LH, et al. Randomized comparative study of 3 versus 8-month neoadjuvant hormonal therapy before radical prostatectomy: biochemical and pathological effects. J Urol. 2001;166(2):500–6. discussion 6-7. [PubMed] [Google Scholar]
- 68.Klotz LH, Goldenberg SL, Jewett MA, Fradet Y, Nam R, Barkin J, et al. Long-term followup of a randomized trial of 0 versus 3 months of neoadjuvant androgen ablation before radical prostatectomy. J Urol. 2003;170(3):791–4. doi: 10.1097/01.ju.0000081404.98273.fd. [DOI] [PubMed] [Google Scholar]
- 69.Soloway MS, Pareek K, Sharifi R, Wajsman Z, McLeod D, Wood DP, Jr, et al. Neoadjuvant androgen ablation before radical prostatectomy in cT2bNxMo prostate cancer: 5-year results. J Urol. 2002;167(1):112–6. [PubMed] [Google Scholar]
- 70.Singh AA, Schuurman K, Nevedomskaya E, Stelloo S, Linder S, Droog M, et al. Optimized ChIP-seq method facilitates transcription factor profiling in human tumors. Life Sci Alliance. 2019;2(1):e201800115. doi: 10.26508/lsa.201800115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schmidt D, Wilson MD, Spyrou C, Brown GD, Hadfield J, Odom DT. ChIP-seq: using high-throughput sequencing to discover protein-DNA interactions. Methods. 2009;48(3):240–8. doi: 10.1016/j.ymeth.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–60. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kumar V, Muratani M, Rayan NA, Kraus P, Lufkin T, Ng HH, et al. Uniform, optimal signal processing of mapped deep-sequencing data. Nat Biotechnol. 2013;31(7):615–22. doi: 10.1038/nbt.2596. [DOI] [PubMed] [Google Scholar]
- 74.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9(9):R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ramirez F, Ryan DP, Gruning B, Bhardwaj V, Kilpert F, Richter AS, et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016;44(W1):W160–5. doi: 10.1093/nar/gkw257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kharchenko PV, Tolstorukov MY, Park PJ. Design and analysis of ChIP-seq experiments for DNA-binding proteins. Nat Biotechnol. 2008;26(12):1351–9. doi: 10.1038/nbt.1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–9. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lerdrup M, Johansen JV, Agrawal-Singh S, Hansen K. An interactive environment for agile analysis and visualization of ChIP-sequencing data. Nat Struct Mol Biol. 2016;23(4):349–57. doi: 10.1038/nsmb.3180. [DOI] [PubMed] [Google Scholar]
- 79.Lopez-Delisle L, Rabbani L, Wolff J, Bhardwaj V, Backofen R, Gruning B, et al. pyGenomeTracks: reproducible plots for multivariate genomic datasets. Bioinformatics. 2021;37(3):422–3. doi: 10.1093/bioinformatics/btaa692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Karolchik D, Hinrichs AS, Furey TS, Roskin KM, Sugnet CW, Haussler D, et al. The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 2004;32:D493–6. doi: 10.1093/nar/gkh103. Database issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pruitt KD, Brown GR, Hiatt SM, Thibaud-Nissen F, Astashyn A, Ermolaeva O, et al. RefSeq: an update on mammalian reference sequences. Nucleic Acids Res. 2014;42:D756–63. doi: 10.1093/nar/gkt1114. Database issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Liu T, Ortiz JA, Taing L, Meyer CA, Lee B, Zhang Y, et al. Cistrome: an integrative platform for transcriptional regulation studies. Genome Biol. 2011;12(8):R83. doi: 10.1186/gb-2011-12-8-r83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kuhn RM, Haussler D, Kent WJ. The UCSC genome browser and associated tools. Brief Bioinform. 2013;14(2):144–61. doi: 10.1093/bib/bbs038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ross-Innes CS, Stark R, Teschendorff AE, Holmes KA, Ali HR, Dunning MJ, et al. Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature. 2012;481(7381):389–93. doi: 10.1038/nature10730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Quinlan AR. BEDTools: The Swiss-Army Tool for Genome Feature Analysis. Curr Protoc Bioinformatics. 2014;47:11 2 1–34. doi: 10.1002/0471250953.bi1112s47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gaujoux R, Seoighe C. A flexible R package for nonnegative matrix factorization. BMC Bioinformatics. 2010;11:367. doi: 10.1186/1471-2105-11-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14(4):R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31(2):166–9. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ho D, Imai K, King G, Stuart EA. MatchIt: Nonparametric Preprocessing for Parametric Causal Inference. Journal of Statistical Software. 2011;42(8):1–28. doi: 10.18637/jss.v042.i08. [DOI] [Google Scholar]
- 93.Chen WS, Aggarwal R, Zhang L, Zhao SG, Thomas GV, Beer TM, et al. Genomic Drivers of Poor Prognosis and Enzalutamide Resistance in Metastatic Castration-resistant Prostate Cancer. Eur Urol. 2019;76(5):562–71. doi: 10.1016/j.eururo.2019.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mohammed H, Taylor C, Brown GD, Papachristou EK, Carroll JS, D’Santos CS. Rapid immunoprecipitation mass spectrometry of endogenous proteins (RIME) for analysis of chromatin complexes. Nat Protoc. 2016;11(2):316–26. doi: 10.1038/nprot.2016.020. [DOI] [PubMed] [Google Scholar]
- 95.Cox J, Hein MY, Luber CA, Paron I, Nagaraj N, Mann M. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol Cell Proteomics. 2014;13(9):2513–26. doi: 10.1074/mcp.M113.031591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tyanova S, Temu T, Sinitcyn P, Carlson A, Hein MY, Geiger T, et al. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat Methods. 2016;13(9):731–40. doi: 10.1038/nmeth.3901. [DOI] [PubMed] [Google Scholar]
- 97.Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods. 2014;11(8):783–4. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All tissue ChIP-seq and RNA-seq raw data generated in this study have been deposited in the European Genome-Phenome Archive (EGA) under the accession numbers EGAS00001006017 and EGAS00001006016, respectively. The cell line ChIP-seq, as well as all processed tissue ChIP-seq and RNA-seq data have been deposited in the Gene Expression Omnibus (GEO) database, under accession number GSE197781. The mass spectrometry proteomics (RIME) data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD032041. Public ChIP-seq datasets used in this study are available from GEO or the EGA under the following accession code: GSE120738 (AR, H3K27ac ChIP-seq), GSE51497 (GR ChIP-seq), GSE117306 (N-MYC ChIP-seq) and EGAS00001003928 (FOXA1 ChIP-seq).