Abstract
Purpose
Mongolia has the world’s highest incidence of hepatocellular carcinoma (HCC), with ˜100 cases/105 inhabitants, although the reasons for this feature have not been thoroughly delineated.
Experimental Design
We performed a molecular characterization of Mongolian (n=192) compared to Western HCCs (n=187) by RNA-seq and WES to unveil distinct genomic and transcriptomic features associated with environmental factors in this population.
Results
Mongolian patients were younger, with higher female prevalence, and with predominantly HBV-HDV co-infection etiology. Mongolian HCCs presented significantly higher rates of protein-coding mutations (121 vs 70 mutations per tumor in Western), and in specific driver HCC genes (i.e. APOB, TSC2). Four mutational signatures characterized Mongolian samples, one of which was novel (SBS Mongolia) and present in 25% of Mongolian HCC cases. This signature showed a distinct substitution profile with a high proportion of T>G substitutions and was significantly associated with exposure to the environmental agent dimethyl sulfate (DMS, 71%), a 2A carcinogenic associated with coal combustion. Transcriptomic-based analysis delineated three molecular clusters, two not present in Western HCC; one with a highly inflamed profile and the other significantly associated with younger female patients.
Conclusions
Mongolian HCC has unique molecular traits with a high mutational burden and a novel mutational signature associated with genotoxic environmental factors present in this country.
Keywords: Liver Cancer; Mongolia; Molecular characterization; Mutational Signatures, Environmental Carcinogens
Introduction
Liver cancer is the second leading cause of cancer-related mortality, and its global burden has increased in recent years1,2. Hepatocellular carcinoma (HCC) accounts for 90% of liver cancer cases and arises almost unfailingly in the setting of chronic liver diseases. The worldwide incidence of HCC cases is heterogeneous, reflecting the distribution of known liver disease risk factors such as hepatitis B virus (HBV), hepatitis C virus (HCV), alcohol consumption, and non-alcoholic steatohepatitis2. Mongolia, a landlocked East Asian country between Russia and China, shows the world’s highest incidence of HCC, with a burden of 86 cases per 100,000 inhabitants1. This incidence far exceeds that of the surrounding countries (4x and >20x compared to China and Russia, respectively) or any other country worldwide1, and has been attributed to a historical high prevalence of HBV (10.6%) and HCV (6.4%) viruses, as well as alcohol consumption3–5. Indeed, 90% of Mongolian HCC cases are positive for HBV, HCV, or both,6 and co-infection with hepatitis delta virus (HDV), a defective virus that needs HBV for its replication cycle and has been associated with liver fibrosis and HCC development, occurs in 50-80% of HBV-infected individuals7. Despite the implementation of universal infant HBV vaccination in 19913 and the improved control of HCV blood-based transmission, the burden of HCC in Mongolia is increasing year after year, and its incidence is now ˜10x compared to the 1960s1. Overall, the reasons for this extreme incidence have never been thoroughly understood.
During the last 10 years, large-scale next-generation sequencing studies have been key in deciphering the transcriptomic-based HCC subtypes and the molecular alterations occurring in HCC8,9. The presence of mutational signatures consisting of unique nucleotide substitution patterns has allowed to track the exposure of endogenous and exogenous factors in cancer10. For instance, HCC risk factors such as aristolochic acid, aflatoxin, alcohol, or tobacco smoking can be related to specific signatures8,11. In this context, assessing the genetic alterations, mutational frequencies and transcriptomic profile of Mongolian HCC could provide relevant information that may help unveil the genetic factors and environmental exposures underlying the high incidence in this population. Previous studies have provided valuable data about the molecular landscape in Mongolian HCC12. However, further analyses comparing Mongolian tumors with a Western cohort are needed to understand the relevance of the molecular traits in Mongolian HCC. Additionally, viral characteristics in Mongolian HCC remain underexplored.
To identify the unique molecular features of Mongolian HCC, we performed whole-exome (WES) and RNA sequencing (RNA-seq) in 379 HCC tumors from Mongolian and Western origins. Herein, we provide a comprehensive characterization of the molecular profile in Mongolian HCC and reveal unique features consisting in an increased number of mutations, as well as specific mutational and transcriptomic patterns. These findings could pave the way for the identification of environmental or genetic factors associated with the increased incidence in this country.
Materials and Methods
Study design
A total of 219 paired HCC/non-tumoral liver samples from distinct patients undergoing HCC resection were collected at the National Cancer Center, Ulaanbaatar, Mongolia (Fig. 1a). Samples were collected from October 2015 to October 2017, in accordance with Mongolian regulations, the National Cancer Center, and the Ministry of Health of Mongolia. Written informed consent was obtained from all participants.
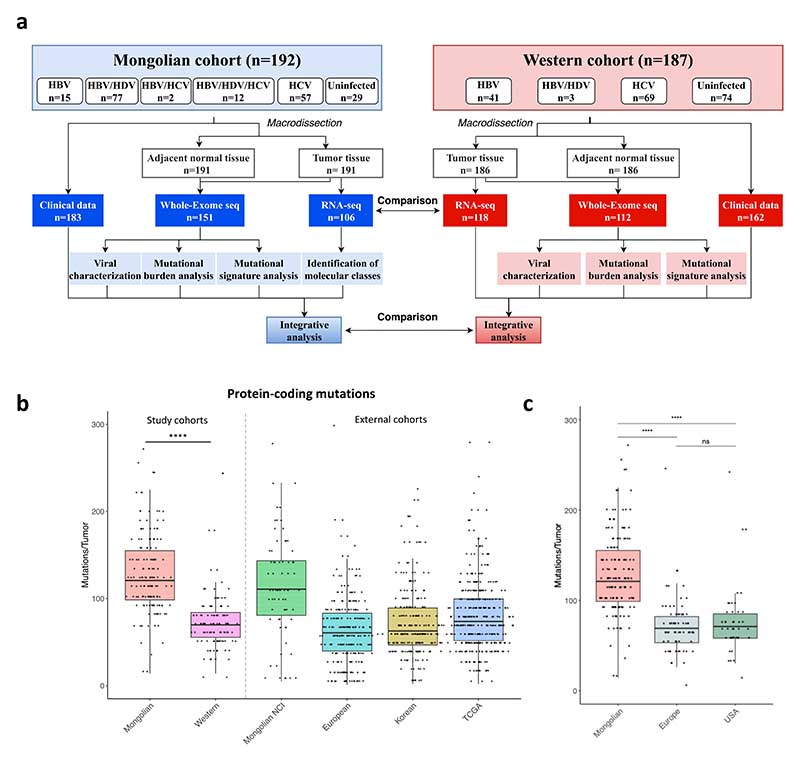
Figure 1. Flow chart of the study and mutational profile in Mongolian HCC.
a A total of 192 HCC samples from Mongolia were used in this study. A Western cohort including 187 HCCs was used as internal control. b Mutations per tumor in the Mongolian (n=151) and Western (n=112) cohorts. Mongolian NCI (n=71), European (n=241), Korean (n=231) and TCGA (n=363) external cohorts are shown as reference. c Mutations per tumor in Mongolian HCC (n=151) compared to Western HCC from Europe (n=69) and USA (n=43). Y axis was cut at 300 mutations/tumor to facilitate data interpretation. P-value corresponds to Kruskal-Wallis test. Box plots indicate median (middle line), 25th, 75th percentile (box) and 5th and 95th percentile (whiskers).
A Western cohort was used as internal control, including tumor and matched non-tumoral liver samples (n=187) from patients undergoing resection (Fig. 1a). Samples were collected from two institutions of the HCC Genomic Consortium: IRCCS Istituto Nazionale dei Tumori (Milan, Italy; n=110) and Icahn School of Medicine at Mount Sinai (New York, USA; n=77), with written informed consent upon Institutional Review Board approval.
This study complies with the Declaration of Helsinki and was performed according to ethics committee approval.
Whole exome sequencing
WES analysis was run in NovaSeq 6000 (Illumina, San Diego, CA) in the New York Genome Center facilities. WES data was used for mutation calling, mutational signature analysis, and tumor mutational burden (TMB) evaluation. TMB was calculated based on protein-coding mutations assuming an average exome size of 30 Mb, in accordance with previously published studies10. Mutations were called by comparing the tumor with its paired non-tumoral counterpart. Molecular variant calling was performed by Sema4 (Stamford, CT, USA), using the Tigris pipeline (v2.0.1), which carries out modified GATK4 (4.0.11.0) best practices (https://software.broadinstitute.org/gatk/).
In addition, WES data from a European (n=241)8, Korean (n=231)13, TCGA (n=363)14 and Mongolian NCI (n=71)12 HCC cohorts were used for mutation calling and TMB evaluation.
Mutational signature analysis
Somatic SNVs in exome region (defined by coding exons in Ensembl GRCh37 built) at allelic frequency cutoff of 0.05 and with gnomAD population frequency or ethnic-specific frequency ≤ 0.5% were selected. Tumor samples with total SNV count ≥ 50 (after the aforementioned filtering) were used in downstream mutational signature analyses, resulting in a total of 254 samples (148 from the Mongolian cohort plus 106 from the Western cohort). The remaining samples (n=9) were considered negative for the signatures. All signature fitting and de novo signature extraction analyses were performed using exome region SNVs and trinucleotide frequencies normalized via `exome2genome` approach in the deconstructSigs R package15.
RNA sequencing
RNA-seq data was processed by the RAPiD pipeline at the Mount Sinai Genomics Core Facility. Briefly, Fastq files were aligned using STAR (v 2.7.0f)16 to hg19 with gencode annotation v19. QoRTs (v1.3.6)17 was used for QC and obtaining raw counts. Batch correction was performed using RUVSeq18. Factor analysis of read counts (RUVg procedure with 10,000 low expressed genes) and normalization for subsequent analysis was performed using VST method from DESeq219.
Unsupervised clustering analysis of the whole cohort (n=224) was performed using the Non-negative matrix factorization (NMFc) module from GenePattern and Euclidean distance and Ward’s agglomerative procedure. Clustering of the Mongolian (n=118) and NCI Mongolian cohort12 (n=70) was also performed using NMFc. Gene expression characterization was performed using Nearest Template Prediction (NTP), Gene Set Enrichment Analysis (GSEA), and single sample GSEA (ssGSEA) modules from GenePattern. To this end, Molecular Signature Database (MSigDB, www.broadinstitute.org/msigdb) and previously reported gene sets were used (Supplementary Table 1). Class comparison between molecular clusters was performed using subclass mapping analysis and NTP in the in-house and external cohorts. Finally, the stromal infiltration and relative tumor purity were assessed using the ESTIMATE R package20.
Statistical analysis
Statistical analyses were performed using either SPSS software package (version 24.0; SPSS Inc, Chicago, IL, USA) or scipy (v 1.2.1) and matplotlib (v3.0.3) modules from Python (v3.7.3). Differences between qualitative variables were assessed with the Fisher exact test and corrected for multiple comparisons using false discovery rate. Differences between quantitative variables were analyzed with a two-sided non-parametric test (Mann-Whitney or Kruskal-Wallis, were appropriate), and adjustments for multiple comparison analysis were performed using Dunn’s test.
Results
Clinico-pathological characteristics of the cohorts
HCC patients from the Mongolian cohort were younger (61 vs 66 years old, p<0.001), with a higher rate of HBV/HDV co-infection (84% of HBV infected vs 7% in the Western cohort, p<0.001), and lower rate of non-infected cases (15% vs 40%, p<0.001) (Table 1, Supplementary Table 2). In line with previous data21. 54% of the HCC patients in Mongolia were male compared to 80% in the Western cohort (p<0.001). The rate of advanced hepatic fibrosis (F3-4) and cirrhosis (F4) was significantly lower in Mongolian compared to Western cases (38% vs 79% and 16% vs 60%, respectively), independently of etiology (Supplementary Fig. 1a). Tumor characteristics were similar in both cohorts, with most tumors within Barcelona Clinic Liver Cancer (BCLC) stages 0-A, and with alpha fetoprotein (AFP) <400 (IU/mL). However, tumors in the Mongolian cohort showed a lower differentiation grade (Table 1). No significant survival differences were observed between cohorts.
Table 1. Baseline characteristics of the Mongolian and Western cohorts.
| Mongolian Cohort (n=192) | Western Cohort (n=187) | p value | |
|---|---|---|---|
| < 50 years (n, %) | 20 (10.9) | 7 (4.1) | 0.017 |
| Gender (male, %) | 98 (53.6) | 137 (79.7) | <0.001 |
| Non-infected (n, %) | 29 (15) | 74 (39.6) | |
| Bilirubin (mg/dL) | 0.6 (0.1-3.7) | 0.9 (0.3-3.8) | <0.001 |
| Albumin (g/L) | 41 (29-49) | 40 (22-54) | ns |
| < 150x109/L (n, %) | 50 (28.6) | 81 (47.4) | <0.001 |
| > 400 IU/mL (n, %) | 32 (20.9) | 26 (16.4) | ns |
| > 5 cm (n, %) | 93 (52.8) | 67 (41.4) | 0.039 |
| BCLC stage (0-A, %) | 132 (78.1) | 129 (79.6) | ns |
| Multinodular disease (n, %) | 26 (15.4) | 42 (25.8) | 0.021 |
| Cirrhosis (F4, %) | 27 (16.1) | 81 (60) | <0.001 |
| Microvascular invasion (yes, %) | 72 (47.7) | 80 (46.5) | ns |
| Tumor grade (G3-4, %) | 11 (10.6) | 41 (28.7) | <0.001 |
HBV, hepatitis B virus; HCV, hepatitis C virus; HDV, hepatitis delta virus; AFP, alfa-fetoprotein; BCLC, Barcelona Clinic Liver Cancer The following variables have missing values for the Mongolian and Western cohorts, respectively: Age: 8 and 15 patients. Gender: 9 and 15 patients. Bilirubin, albumin, and platelets: 19 and 17 patients. AFP: 39 and 28 patients. Tumor size: 16 and 25 patients. BCLC stage: 23 and 25 patients. Tumor number: 23 and 24 patients. Liver fibrosis in 24 and 52 patients. Microvascular invasion in 41 and 15 patients. Tumor grade in 88 and 44 patients.
Viral characterization of Mongolian and Western individuals
To understand whether the particular clinicopathological features of the Mongolian HCCs were due to unique viral characteristics, we analyzed the phenotypes of HBV and HDV in infected patients. HBV genomes can be classified into 9 genotypes (A to I), according to differences in nucleotide sequences. HBV genotypes have a characteristic geographical and ethnic distribution, and HBV genotype D is known to be almost universal in Mongolia22. In the Western cohort, patients were infected mostly by genotypes C and D (27% and 43%, respectively, p<0.001) whereas in the Mongolian cohort, all HBV-infected individuals were genotype D, including 2 patients with recombinant forms of genotype C and D (Supplementary Table 3). Interestingly, genotype D has been previously associated with reduced HCC development as compared to genotype C23. The most frequent HBV sub-genotypes in Mongolian and Western cases were D1 (77%) and D3 (39%), respectively (Supplementary Fig. 1b). Among Western cohort samples, the most prevalent genotypes in European patients was genotype D3 (71%), whereas in patients from USA it was genotype C (60%) (Supplementary Fig. 1c). Regarding HDV infection, all Mongolian patients were genotype 1 (Supplementary Table 3). We then evaluated the presence of basal core promoter (BCP) and precore HBV mutations, which have been associated with liver disease progression and HCC development24. It has been previously suggested that these mutations are more frequent in HBV-infected patients with genotype D, with only 11% of patients with such genotype being wild-type25. We detected BCP A1762T, BCP G1764A and precore G1896A mutations in 14%, 21% and 29% of HBV-infected Mongolian patients, respectively (Supplementary Fig. 1d, Supplementary Table 3). Interestingly, the prevalence of these mutations was significantly higher in the Western cohort (60%, 66%, and 46%, respectively), (p<0.001, p<0.001, and p=0.089, respectively, Supplementary Fig. 1d). The percentages observed in our Western patients were similar to those previously reported26. In line with previously reported data27, precore G1896A mutations were less frequent in HBV/HDV co-infected versus HBV mono-infected in Mongolia (20% vs 64%, p=0.002); however, we found no differences for both BCP A1762T and G1764A mutations (14% vs 14%, and 19% vs 29%, respectively, p=ns). Overall, these data suggest that the rate of BCP and precore HBV pro-oncogenic mutations in Mongolia is particularly low, despite the predominance of genotype D in this population.
The interaction between HBV and HDV viruses is complex and not fully understood. It has been proposed that HDV can suppress HBV28,29, but HBV and HDV levels can fluctuate over time30. To further investigate this, we determined HBV-DNA and HDV-RNA levels in tumor-adjacent liver tissue in Mongolian and Western samples. Of note, high HBV-DNA levels in blood have been associated with more aggressive liver disease, including HCC development31. First, we analyzed the differences between HDV positive and negative samples. Intrahepatic HBV-DNA load was significantly higher in samples with HBV/HDV co-infection than those with HBV mono-infection (5.0 vs 3.8 log copies/μg total DNA, p=0.001, Supplementary Fig. 2a). Furthermore, high HBV-DNA levels were associated with advanced liver fibrosis, advanced tumor stage, and worse survival in Mongolian individuals (Supplementary Table 4, Supplementary Fig. 2b). Conversely, patients with high HDV-RNA levels showed significantly higher alanine aminotransferase levels, suggesting greater inflammation (Supplementary Table 5). Finally, no significant differences in HBV-DNA load were found between Mongolian and Western samples (4.85 vs. 4.97 log copies/μg total DNA, respectively p=0.23, Supplementary Fig. 2c, Supplementary Table 6). Taken together, our analysis suggests that HBV and HDV viral characteristics in Mongolia are highly homogeneous and with low oncogenic potential.
Analysis of the genomic landscape in Mongolian HCC
To gain further insights into the molecular landscape of Mongolian HCC, we performed mutation and copy number variation (CNV) analysis. The pattern of broad gains and losses in our Western cohort was consistent with previous reports in HCC8, with 1q and 8q gains and 8p losses being the most common alterations (Supplementary Fig. 3a-b, Supplementary Table 7). When we compared the broad chromosomal variation profiles between Mongolian and Western HCC patients, no difference was observed in terms of overall CNV burden (Supplementary Fig. 3c-e). Nonetheless, Mongolian HCCs showed a significantly higher occurrence of 1p gains, 9q gains, as well as fewer 9q losses, 1q losses, and 8p losses (Supplementary Table 7).
The average mutations per tumor was significantly higher in Mongolian patients compared to Western, with a median of 121 and 70 mutations/tumor, respectively (p<0.001) (Fig. 1b). Accordingly, the median tumor mutational burden (TMB) in the Mongolian and Western cohorts was 4.0 and 2.3 mutations/Mb, respectively (p<0.001). No significant differences were observed depending on the origin of Western samples (0.645, Fig. 1c). To rule out the possibility that the observed difference was due to random selection bias, we then compared the median protein-coding mutations with previously published cohorts in Western and Asian countries, applying the same filtering criteria8,12–14. The median number of mutations per tumor was 111 in the Mongolian NCI cohort12, 76 in TCGA14, 61 in the European8 cohort, and 63 in the Korean cohort32, corresponding to a TMB of 3.7, 2.5, 2.0, and 2.1, respectively (Fig. 1b, Supplementary Table 8) confirming that the number of mutations in both Mongolian HCC cohorts was significantly higher than in other countries (all <0.001). There was a positive association between the number of mutations and tumor grade in Mongolian cases, with a median of 100, 120, and 171 mutations/tumor in samples with good, moderate, or poor differentiation grades (p=0.004). We did not observe any significant difference in the median of mutations according to etiology in either cohort (Supplementary Fig. 4a-c).
Previous studies in HCC have suggested that highly mutated tumors (TMB ≥4 mutations/Mb) are enriched with mutations in DNA damage response (DDR) genes33. In our Mongolian cohort, 84 (56%) of samples showed ≥4 mutations/Mb, compared to 7 (6.3%) in the Western cohort (p<0.001). However, no association was observed with the presence of mutations in DDR genes (Supplementary Table 9).
Mutational profile of Mongolian HCC
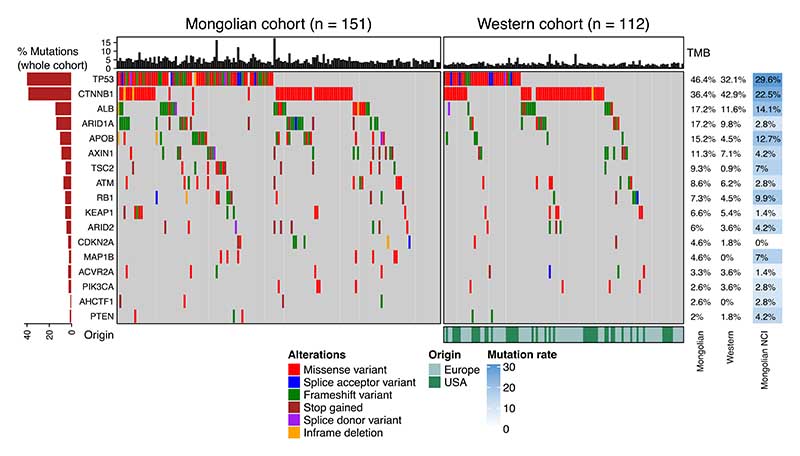
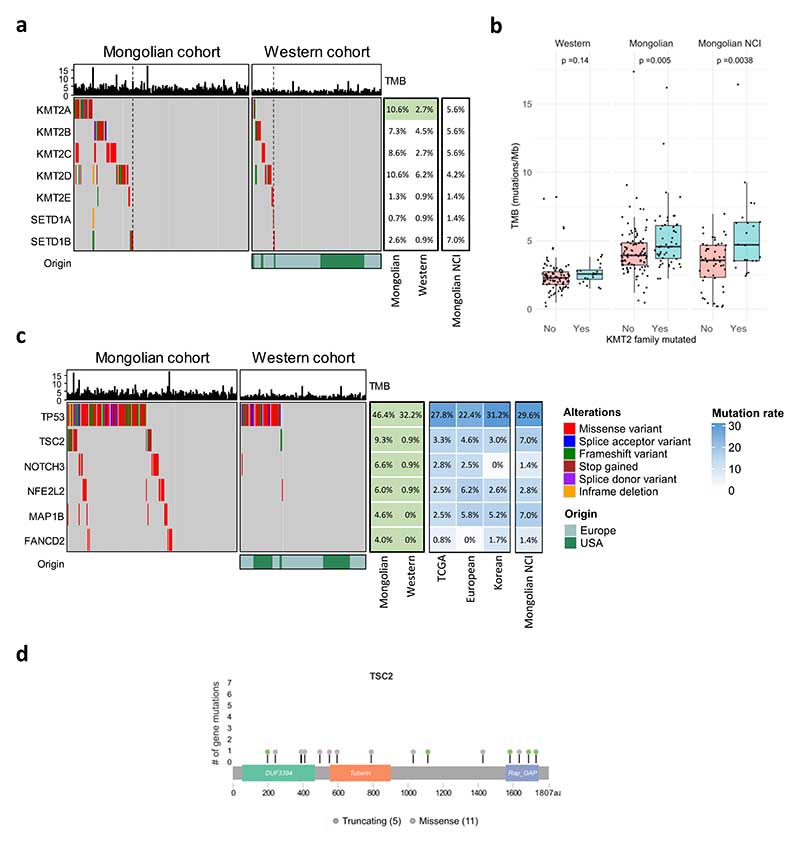
We then explored whether the higher frequency of mutations in the Mongolian cohort was due to enrichment in specific genes. Among the 100 genes with significant differences between cohorts, 89 were more mutated in Mongolian vs Western HCCs, suggesting that the higher number of mutations was broadly occurring across the whole genome and not concentrated in specific loci (Supplementary Table 10). Overall, we detected a significant mutational increase in multiple known HCC driver genes in Mongolia compared to the Western cohort, including TP53 (46% vs 32%), APOB (15% vs 5%), TSC2 (9% vs 1%), and NFE2L2 (6% vs 1%), (p<0.05, Fig. 2, Supplementary Fig. 5, Supplementary Tables 11-12). Furthermore, genes belonging to the KMT2 histone lysine methyltransferase family were significantly more mutated in Mongolian HCC (34% vs 18%, p=0.005, Fig. 3a). Mongolian HCC with mutations in the KMT2 gene family displayed higher TMB than patients without these mutations in both Mongolian cohorts (median TMB 4.6 and 4.7 vs 3.9, p=0.005 and p<0.0001, Fig. 3b). The mutation rate of HCC drivers was also assessed in external HCC cohorts for comparison, confirming a similar frequency of mutations in the Mongolian NCI cohort for most HCC drivers including APOB, TSC2, NFE2L2, and KMT2 family (all n.s. vs in-house Mongolian cohort) (Fig. 2, Supplementary Table 12).
Figure 2. Mutational landscape of Mongolian and Western HCC.
Mutations present in HCC samples from the Mongolian (n=151) and Western (n=112) cohorts. The frequency of mutations in the Western and Mongolian NCI cohort are indicated for comparison (left). Top panel shows tumor mutational burden (TMB, mutations/Mb) per sample. Middle panel indicates the presence of mutations per sample (right) and overall percentage (left) in the most frequently mutated genes.
Figure 3. Potential driver and KMT2 family mutations in Mongolian and Western cohorts.
a Mutations in the KMT2 gene family in the Mongolian (n=151) and Western cohorts (n=112). b TMB in samples with KMT2 family mutations and wild type (wt). Box plots indicate median (middle line), 25th, 75th percentile (box) and 5th and 95th percentile (whiskers). P-value corresponds to Kruskal-Wallis test. c Mutations in potential driver genes in the Mongolian and Western cohorts according to enrichment in damaging alterations. Percentage of mutations in external cohorts is indicated in the right panel. Top panel shows tumor mutational burden (TMB, mutations/Mb) per sample. Middle panel indicates the presence of mutations per sample (left) and overall percentage (right) in the Mongolian and Western cohorts. Significant differences between the Mongolian and Western cohorts are indicated in green (Fisher p < 0.05). d Mutation distribution in the TSC2 gene.
Next, potential drivers in Mongolian HCC were further assessed by OncodriveCLUSTL and dN/dScv algorithms34,35. Among the genes significantly more mutated in the Mongolian cohort, 6 were enriched for damaging alterations, suggesting that they could exert a driver role in Mongolian HCC (q<0.05; Fig. 3c-d), including TSC2 (9%). Similar TSC2 mutation rates were confirmed in the Mongolian NCI cohort (7%) (Fig. 3c).
Finally, differences in the mutation profile depending on etiology were detected in the Mongolian cohort (Supplementary Fig. 4d-e). CDKN2A mutations were enriched in patients with HBV (7.4% vs 0%) and HDV (8.0% vs 0%) infection, while ARID2 mutations were more common in HCV-infected patients (11.9% vs 1.2%) (p<0.05, Supplementary Fig. 4d). No significant differences between Western samples from Europe and USA were observed (Supplementary Fig. 6, Supplementary Table 11).
Overall, our results indicate that Mongolian HCC shows a significantly higher tumor mutational burden and, although the mutational and chromosomal spectrum highly resembled that of Western HCC, significant differences were observed for key driver genes including APOB, KMT2 family, and TSC2.
Mutational signature analysis
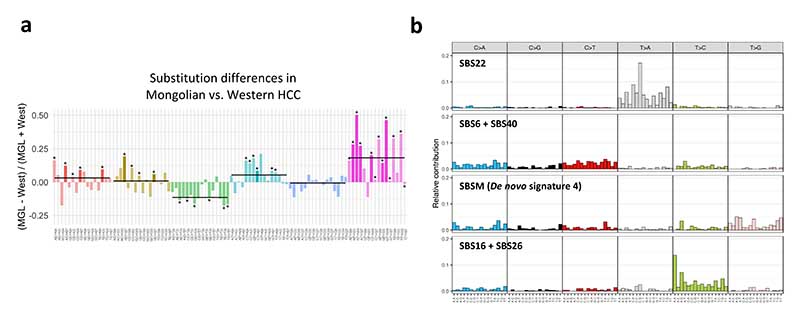
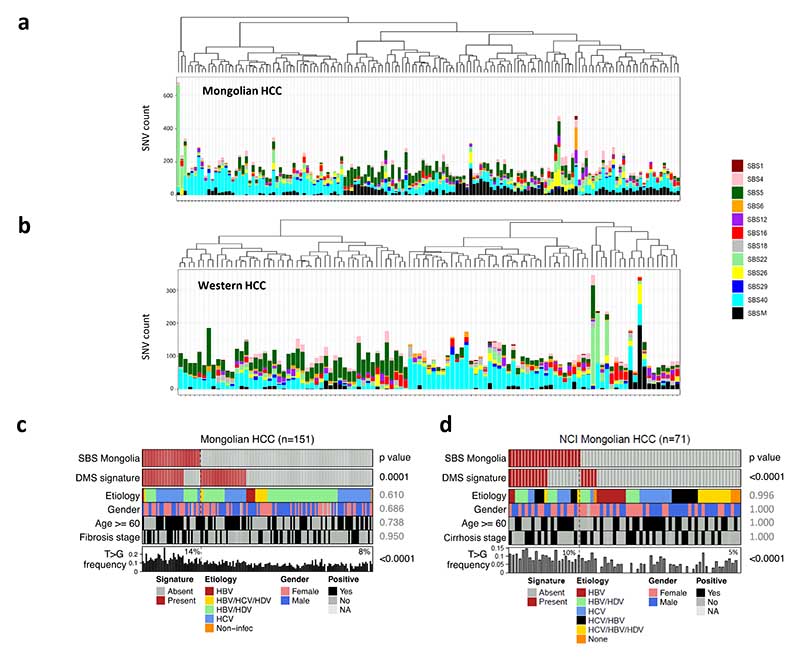
We then analyzed the pattern of single base substitutions in the Mongolian and Western cohorts. Notably, Mongolian HCC was characterized by a higher proportion of T>G substitutions compared to Western tumors (Fig. 4a,Supplementary Fig. 7a-b, Supplementary Table 13). This was also observed in the Mongolian NCI cohort (Supplementary Fig. 7c).
Figure 4. Single-base substitution profile in Mongolian HCC.
a Differences in trinucleotide substitution frequency between the in-house Mongolian and Western cohorts. Bars indicate the median values for each substitution group. Significant substitutions differences in both in-house and NCI Mongolian cohorts are highlighted in brighter colors. * p < 0.05 (Kruskal-Wallis test). b De novo single-base substitution (SBS) signatures found in Mongolian and Western HCC samples. For each signature, the 96-substitution classification is displayed, including the substitution type and sequence context.
De novo mutational signature extraction from the Mongolian and Western cohorts revealed four signatures (Fig. 4b), three of which were mapped to COSMICv3 signatures previously found in liver cancer: SBS22, a combination of SBS6-SBS40 and of SBS16-SBS26 (cosine similarity >0.90 in all cases)36. The fourth signature did not present strong similarities with any of the COSMIC signatures and was therefore considered novel (Supplementary Tables 14-15). We then performed signature fitting using the de novo signature 4 and HCC-specific COSMICv3 signatures by applying a bootstrap approach. Interestingly, the Mongolian cohort was enriched in the de novo signature 4, henceforth renamed SBS Mongolia (SBSM) (25.2% [38/151 vs 4.5% [5/112] in Mongolian vs Western cohorts; p<0.0001) (Fig. 5a-b). Notably, Mongolian HCC presenting the SBSM signature showed a distinct substitution profile consisting of a high proportion of T>G substitutions (14% vs 8% in SBSM positive and negative samples, respectively [p<0.001], and 6% in Western HCC, Fig. 5c). Other COSMICv3 signatures previously reported in HCC36 presented similar prevalence between cohorts (Fig. 5a-b, Supplementary Fig. 7d-e, Supplementary Table 16). High presence of the SBSM signature in Mongolian HCC was confirmed in the external NCI Mongolian cohort (31% of positive cases, 22/71), but not in another Asian (Korean) cohort (4.3% of positive cases, 10/231, Supplementary Table 17). No differences in the frequency of SBSM were detected in Western HCC depending on the sample origin (Supplementary Table 16). However, SBS22, associated with exposure to aristolochic acid, was more frequent in samples from the USA (6/43 vs 0/69 in Europe, p=0.02), likely due to the presence of Asian-American patients, which reportedly present higher exposure to this compound37.
Figure 5. Mutational signatures in Mongolian and Western HCC.
a,b Signature fitting results in Mongolian (a, n=151) and Western HCC (b, n=112) using HCC-specific COSMIC signatures and SBS Mongolia (SBSM). c,d Distribution of samples harboring SBSM in the Mongolian cohort (c) and in the NCI Mongolian cohort (d). Presence of dimethyl sulfate (DMS) mutational signature, main clinical variables and T>G substitution frequency are shown for each sample. FDR-adjusted p-values comparing SBSM positive and negative samples are indicated. Percentages in the lower panel refer to median T>G frequency.
To further characterize SBSM-positive samples, we assessed the presence of mutational signatures linked to the effects of known or suspected environmental mutagens from the Compendium of Mutational Signatures of Environmental Agents38. Samples presenting SBSM were significantly enriched for the mutational signature associated with exposure to dimethyl sulfate (DMS) (71.1% [27/38] vs 26.5% [30/113], p<0.0001, Fig. 5c), as further confirmed in the Mongolian NCI cohort (Fig. 5d). In line with this, DMS was the only environmental-related signature significantly enriched in Mongolian HCC compared to Western (37.7% [57/151] vs 18.8% [21/112], Supplementary Table 18), a feature confirmed in external HCC cohorts (Supplementary Table 19), with no significant differences depending on the origin of Western samples (Supplementary Table 18). Patients presenting the DMS signature were older (64.3 vs 59.5 years) and predominantly HCV-positive; Supplementary Fig. 8). No association between SBSM and TMB, etiology, fibrosis or other clinical and molecular variables were found (Fig. 5c).
Finally, we investigated the mutational profile in adjacent matched liver tissue of Mongolia and Western HCCs. Due to the small number of SNVs observed in the adjacent tissues, mutational signature fitting was performed on pooled variants from the Mongolian and Western cohorts using HCC-specific COSMICv3 signatures plus SBSM (Supplementary Fig. 9). SBSM was the only dominant signature in adjacent tissue from the Mongolian cohort (Supplementary Fig. 9c-d), suggesting that non-tumoral liver tissue in Mongolia presents the signature before HCC arises. SBS5, associated with age-related clock-like mutations36, was the main signature in Western non-tumoral tissue. The DMS signature was the most dominant environmental signature in Mongolian non-tumoral tissues, while it was not detected in Western non-tumoral samples (Supplementary Figure 9e-f).
Overall, Mongolian HCC presents a unique substitution profile characterized by a novel mutational signature, SBSM, which is associated with the DMS-related signature. Considering this, DMS exposure warrants further investigation as a potential environmental factor for HCC in Mongolia.
Identification of unique gene expression patterns in Mongolian HCC
We then investigated the transcriptome profiling of HCC samples using RNA-seq data to define the molecular patterns in Mongolian HCC tumors. Unsupervised clustering analysis of Mongolian and Western samples using non-negative matrix factorization (NMFc) identified two robust clusters (Supplementary Fig. 10a-c). Notably, 80% of Western tumors were included in one cluster, while the second cluster showed a strong enrichment of Mongolian HCC samples, suggesting that Mongolian HCC may present a distinct molecular profile. No clustering differences were observed between Western samples from Europe and USA (Supplementary Fig, 9c). Furthermore, NMFc analysis of the Mongolian HCC samples alone revealed three main clusters -MGL1, MGL2 and MGL3- (Supplementary Fig. 10b-c), which overlapped with the classification of the whole cohort. Unsupervised clustering showed similar results, thus confirming robustness of our findings (Supplementary Fig. 10d).
To elucidate the transcriptomic differences between Mongolian and Western HCC, we performed ssGSEA and NTP analyses (Supplementary Fig. 11a). The Mongolian cohort presented an enrichment in Hoshida S1 and Proliferation classes (39% vs 20% and 36% vs 14%, p<0.01). In addition, Mongolian tumors showed enhanced inflammatory signaling (i.e., IFN and HCC Immune class), response to viral infection, and growth factor-related pathways (all p<0.05). Comparatively, the Western cohort was enriched in the Hoshida S3 class (26% vs 47%, p<0.01) and liver-related metabolic activation. Proliferative and inflammatory profiles similar to the in-house cohorts were confirmed in the Mongolian NCI cohort compared to non-Mongolian external HCC cohorts (Supplementary Figure 12a). Overall, Western HCC samples from Europe and USA showed a similar molecular profile (Supplementary Fig. 11a)
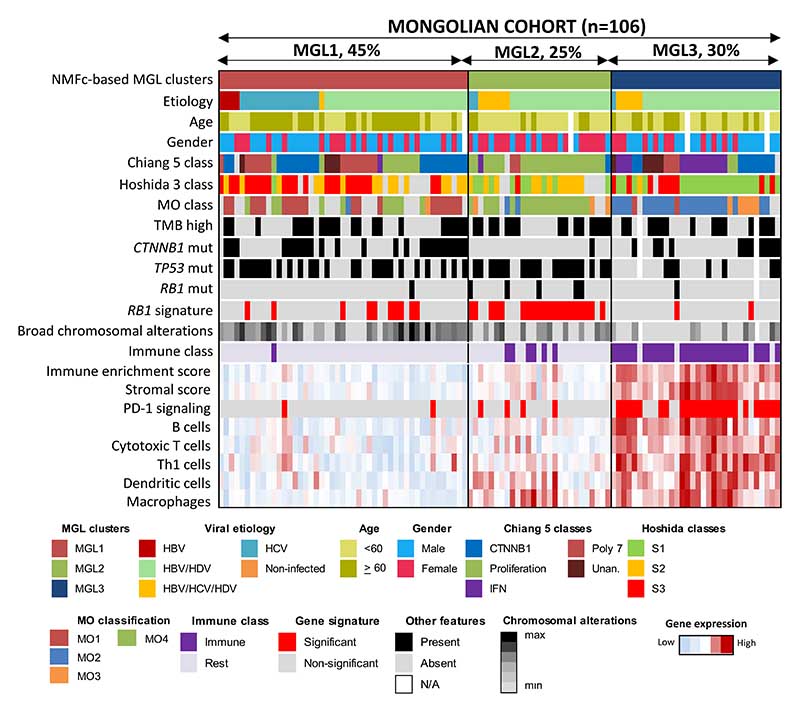
We then characterized each one of the MGL clusters. Patients belonging to MGL1 class (44% of the cohort) were more frequently HCV-infected, older and mostly males (female:male ratio of 1:2), (Supplementary Table 20) and with a molecular profile closer to Western HCCs than the rest of Mongolian HCC (Fig. 6). On the other hand, patients of the MGL2 (26%) and MGL3 clusters (30%) were significantly younger than MGL1 and enriched in HBV/HDV infection and triple infections (HBV/HDV/HCV). Interestingly, while patients of the MGL3 class showed a female:male ratio similar to MGL1 (1:2), the MGL2 class showed a female:male ratio of 2:1 and higher AFP levels. None of the MGL clusters was associated with specific outcomes (Fig. 6, Supplementary Fig. 13, Supplementary Table 20).
Figure 6. Molecular classification of Mongolian HCC.
Consensus-clustered classification and inflammatory profile of Mongolian HCC samples. In the heatmap, clinico-pathological characteristics, nearest template prediction and gene set enrichment in each sample are shown.
In terms of molecular features (Supplementary Tables 21-23), MGL1 cluster was characterized by enrichment in CTNNB1 class and activation of metabolic and liver-specific pathways (all p<0.05). MGL2 HCC cases displayed a proliferative HCC phenotype (i.e enrichment of Proliferation, G3, and Cluster A gene signatures), higher rates of RB1 mutations, and lower rates of CTNNB1 mutations (Fig. 6). Finally, MGL3 tumors were characterized by lower rates of TP53 mutations, fewer broad chromosomal alterations, and were particularly enriched in immune-related features (i.e immune class, interferon and inflammatory pathways and PD1 signaling).
To validate these results, we performed NMFc analysis using RNA-seq data from the Mongolian NCI cohort12. The clinico-pathological and molecular features of each MGL cluster aligned with what was described in the in-house Mongolian cohort (Supplementary Fig. 12, Supplementary Table 24). Subclass mapping and NTP analyses in the in-house Mongolian cohort indicated a good overlap of the MGL clusters with the previously published MO classification of Mongolian HCC12 (Supplementary Fig. 14). Specifically, MGL1 aligned with the MO1 class, associated with good prognosis and metabolic processes (FDR < 0.018). MGL2 overlapped with the MO4 class (FDR = 0.012), characterized by poor prognosis and activation of proliferative pathways. Finally, MGL3 encompassed both MO2, a good prognosis class with inflammatory signaling, and MO3, associated with poor prognosis and metabolic pathway enrichment12 (both FDR = 0.012; Supplementary Fig. 14). Overall, our results suggest that three distinct gene expression patterns characterize Mongolian HCC of whom two present unique clinicopathological features not observed in HCC samples from Western countries.
Immune characterization of the molecular classes of Mongolian HCC
We then further explored the inflammatory profile of Mongolian HCC using ESTIMATE20 and ssGSEA in our in-house Mongolian cohort and the NCI cohort12. In both Mongolian cohorts, MGL3 showed significantly higher immune enrichment score, presence of both an innate and adaptive immune response, and signatures predicting response to immunotherapy (Fig. 6, Supplementary Fig. 12c). A certain degree of inflammation was also observed in MGL2, even if significantly lower than MGL3. Our previously reported HCC immune class was detected in most patients belonging to MGL3 (88% in the Mongolian cohort, 100% in the NCI Mongolian cohort12, Fig. 6, Supplementary Fig. 12c). Compared to Western HCC, the immune class in Mongolian tumors was larger (42% versus 29%, p=0.05) with an inverted ratio of Exhausted/Active subtypes (65/35 vs 30/70, Supplementary Fig. 11b), indicating a more prominent immunosuppressive phenotype. Overall, we observed a higher presence of immune signaling in the Mongolia clusters MGL2 and MGL3. Considering that most patients belonging to these clusters were HBV/HDV infected and younger, this suggests that HDV infection could accelerate disease progression through inflammatory mechanisms.
Discussion
Our study entails a comprehensive characterization of the molecular profile of HCC in Mongolia, the country with the highest global incidence. Mongolia has many particularities that might play a role in HCC burden, including specific risk factors, socioeconomic particularities, and genetic profile12,39. Despite a strikingly high prevalence of HBV (10.6%), HCV (6.4%), and HDV (70% of HBV-positive individuals) infections and alcohol consumption3–5, it is unclear whether HCC incidence is completely explained by the unique combination of risk factors, or eventually other unknown factors might be responsible. In addition, a direct comparison with a comprehensive Western cohort is necessary to understand the relevance of newly identified molecular traits. By analyzing WES and RNA-seq data from 379 new Mongolian and Western HCC samples, we identified unique genomic and transcriptomic footprints in Mongolian tumors that suggest a role of specific genetic and environmental factors in the country. The study provides novel information in three major areas: a) Clinical characteristics of Mongolian cases, b) Unique tumor mutational burden and mutational profile associated with environmental agents, and c) Unique transcriptomic-based molecular classes.
Regarding the clinicopathological particularities of Mongolian HCC patients, we confirmed the high prevalence in females (up to 46% of the cohort), consistent with the reported male/female 1.5/1 ratio21, as opposed to that observed globally2 and in the surrounding countries (2.6/1 in Russia, 3.4/1 in China, and 3/1 in East Asia)21. Mongolian HCC also occurred in younger patients with milder underlying liver fibrosis (F1-2 stages in >60% of cases) and with a dominant viral-related etiology (85% of either HBV, HBV-HDV or HCV-positive).
In our study, HBV characteristics such as genotype D1 and precore mutations in <30% of cases revealed traits associated with low oncogenic potential of the virus in Mongolia23,25, whereas HBV load was similar to Western samples. Thus, other factors not associated with HBV infection might be responsible for the high HCC incidence rates in Mongolia23,25. In this sense, 84% of the HBV-infected Mongolian patients showed co-infection with HDV, which contrasts with the Western data (less than 7% of coinfection)40. This particular co-infection profile was associated with two molecular subclasses only identified in Mongolian patients (MGL2 and MGL3), thus pointing towards a potential role of HDV in the oncogenic process, despite it is not currently considered a carcinogenic agent7.
At the genomic standpoint, Mongolian HCCs had a high rate of protein-coding mutations, which almost doubled that in the Western in-house cohort (121 vs 70 mutations per tumor) and publicly available datasets8,12–14. This suggests the presence of intrinsic and/or extrinsic factors promoting mutagenesis in Mongolian HCC. Notably, while several HCC driver genes presented similar mutation rates in the Mongolian cohort compared to the Western tumors and previous studies8,13,14 (e.g.CTNNB1 and ARID1A), others such as APOB, and the KMT2 gene family were significantly more mutated in Mongolian HCC (e.g. APOB in 15.2% vs 4.5% in Western). For instance, KMT2 family mutations (34% vs 17% in Western) were associated with higher TMB in Mongolian HCC, suggesting that they could be partially responsible for the increased mutational burden. In line with this, loss of function in KMT2 methyltransferases has been proposed to induce DNA damage due to aberrant chromatin remodeling41. TSC2 (9%) mutations were identified as potential drivers in Mongolian tumors. TSC2, a known cancer-related gene participating in the mTOR oncogenic pathway 8,42, has been proposed as an actionable alteration with level 2B evidence, as it could be a predictor of response to the FDA-approved drug everolimus42.
To understand whether Mongolian HCCs present differential genomic and genotoxic footprints, we assessed the presence of distinct mutational signatures10,38. We identified a new mutational signature (SBS Mongolia) with no similarities with previously reported COSMIC signatures. SBS Mongolia was significantly enriched in Mongolian HCC (25% vs 4.5%), indicating unique substitution patterns characterized by increased T>G substitutions. Overall, this mutational landscape points towards specific exposure to environmental factors in Mongolian patients. In this regard, Mongolian HCC samples presenting the novel SBS Mongolia were significantly enriched for the mutational signature associated with exposure to DMS (71.1% vs 26.5%). DMS has been classified as a probable carcinogenic hazard to humans by the International Agency for Research on Cancer43 (category 2A carcinogen) and its production has been associated with coal combustion. Most of Mongolian population is currently exposed to coal combustion, as coal is used to fight against the intense cold weather both in urban and rural areas. Half of the 3-million population of Mongolia lives in Ulaanbaatar, an overpopulated capital with dismal environmental conditions44, whereas the rest is still predominantly nomad and lives in traditional tents or gers, where coal is used both for cooking and heating. This fact has been recognized by international organizations as a major health threat in this country44. Considering this, we hypothesize that long-term exposure to DMS from coal combustion could be a risk factor for HCC development in Mongolia. Future functional and epidemiological studies will be required to confirm the association between the DMS-related signature and exposure to coal combustion in Mongolia. Specifically, DMS could act as a cofactor and exacerbate the risk of HCC in patients exposed to viral infection. A similar phenomenon is observed with the fungal compound aflatoxin B1, which acts synergistically with HBV to induce HCC in Southeast Asia and Sub-Saharan Africa37.
The transcriptomic profile of Mongolian tumors was consistent with known HCC features2. Nonetheless, we observed two striking differences a) Mongolian HCC was characterized by an enhanced proliferative and immunological signaling, with a proportion of tumors belonging to proliferative/progenitor HCC classes (39%) doubling the one in Western HCC (20%) and previously published studies45; and b) Two out of the three identified molecular classes presented distinct molecular features compared to Western HCC, and thus are deemed unique for Mongolian tumors. Effectively, MGL2 (26%) and MGL3 classes (30%) were specific for Mongolian tumors but not for Western HCC and were enriched in HBV/HDV infection. Interestingly, MGL2 class, was associated with both clinical and molecular features of aggressiveness and showed a female:male ratio of 2:1. The presence of this molecular class could be due to the increased HCC incidence in females in this population21. Finally, MGL3 presented an inflamed profile, potentially linked to the response to HBV/HDV infection7. Inflamed HCC tumors from the MGL3 class could be ideal candidates to receive immune checkpoint inhibitors and combination therapy approaches46.
In conclusion, we provided an exhaustive comparison of the genomic and transcriptomic characteristics of Mongolian HCC with an in-house Western cohort. Mongolian HCC is characterized by high mutational rates, a distinct mutational signature profile, and the presence of two unique molecular subclasses. Finally, environmental factors such as DMS need to be further explored as a potential risk factor in this population.
Supplementary Material
Statement of Translational Relevance.
Mongolia has the world’s highest incidence of hepatocellular carcinoma (HCC), with ˜100 cases per 100,000 inhabitants, far exceeding that of the rest of the world and surrounding countries. However, it is unclear whether this HCC burden is completely explained by the combination of risk factors (e.g. HBV, HCV, and HDV infections), or eventually other unknown agents might play a role. By performing a thorough molecular characterization of Mongolian HCC, we identified distinct genomic and transcriptomic footprints in Mongolian HCC, including higher number of mutations per tumor, a novel mutational signature, and two molecular clusters not observed in Western HCC. The presence of mutational fingerprints associated with exposure to the carcinogenic agent dimethyl sulfate − a derivate from coal combustion − suggests a role of environmental factors in this country. Thus, our findings could pave the way for the identification of environmental or genetic factors partially responsible for the increased HCC incidence in Mongolia.
Acknowledgmenents
We thank Clara Rossi, Fellow at Mount Sinai, and Luca Niebla, an MD student at UB, for their help.
Financial Support
This study was partially supported by Bristol-Myers Squibb. MP received a scholarship grant from Asociación Española para el Estudio del Hígado (AEEH). PKH is the recipient of a grant from the German Research Foundation (DFG, HA 8754/1-1). MGL is supported by the i-PFIS program (fellowship IFI18/00006) of the Instituto de Salud Carlos III (ISCIII), co-funded by the European Social Fund (ESF). REF is supported by a doctoral training grant (BES-2017-081286) from MCIN/AEI/10.13039/501100011033 and the European Social Fund (ESF). AV is supported by the US Department of Defense grant (CA150272P3) and the Tisch Cancer Institute (Cancer Center grant P30 CA196521). SPP is supported by the ISCIII through the Plan Estatal de Investigación Científica y Técnica y de Innovación 2013-2016 and 2017-2020 co-funded by the European Regional Development Fund (ERDF) (PI16/00111 and PI19/00036). JZR’s team is supported by Inserm, Labex OncoImmunology Investissement d’Avenir and is “Equipe labellisée par la Ligue Nationale Contre le Cancer». XF is supported by the ISCIII through the Plan Estatal de Investigación Científica y Técnica y de Innovación 2013-2016 and 2017-2020 co-funded by the ERDF (PI18/00079), by the Secretaria d’Universitats i Recerca del Departament d’Economia i Coneixement (grant 2017_SGR_1753) and by CERCA Programme/Generalitat de Catalunya. DS is supported by the Gilead Sciences Research Scholar Program in Liver Disease. JML is supported by grants from the Samuel Waxman Cancer Research Foundation, the Spanish National Health Institute (MICINN, PID2019-105378RB-I00), NIH (R01 DK128289-01), HUNTER (Ref. C9380/A26813) through a partnership between Cancer Research UK, Fondazione AIRC and Fundación Científica de la Asociacion Española Contra el Cáncer and by the Generalitat de Catalunya (AGAUR, SGR-1358).
Footnotes
Author Contributions
M.P. and L.T. contributed equally to this work. M.P., D.S., and J.M.L. developed the study concept and design. M.P., L.T., M.T.M., H.W., M.M.., T.L., M.G.L., W.Q.L., C.M., S.T., A.R., P.T., C.E., E.T., A.Y., G.C., S.P.P., S.T., and D.S. acquired the experimental data. M.P., L.T., M.T.M., H.W., M.M., P.K.H., T.L., M.G.L., R.E.F., W.Q.L., C.M., S.T., A.R.V., P.T., G.C., S.P.P., A.V., E.L., J.Z.R., A.U., J.N., X.F., S.R., D.S., and J.M.L. conducted the analysis and interpretation of the data. M.P., L.T., D.S., M.T.M., H.W., and A.U. performed statistical analyses. M.P., L.T., M.T.M., P.K.H., D.S., and J.M.L. drafted the manuscript. M.P., L.T., M.T.M., P.K.H., R.E.F., S.P.P., A.V., S.T., J.C., E.L., J.Z.R., A.U., J.N., X.F., S.R., D.S., and J.M.L. provided critical revision of the manucript. D.S. and J.M.L. supervised the study.
Conflict of Interest
HW, ARV and PT are employed by Sema4. AV has received consulting fees from Guidepoint, Fujifilm, Boehringer Ingelheim, FirstWord, and MHLife Sciences; advisory board fees from Exact Sciences, Nucleix, Natera, Gilead and NGM Pharmaceuticals; and research support from Eisai. AU reports employment and stock ownership from Sema4. JN is employed by Bristol-Myers Squibb. JML is receiving research support from Bayer HealthCare Pharmaceuticals, Eisai Inc, Bristol-Myers Squibb, Boehringer-Ingelheim and Ipsen, and consulting fees from Eli Lilly, Bayer HealthCare Pharmaceuticals, Bristol-Myers Squibb, Eisai Inc, Celsion Corporation, Exelixis, Merck, Ipsen, Genentech, Roche, Glycotest, Nucleix, Sirtex, Mina Alpha Ltd and AstraZeneca. The remaining authors declare no competing interests.
Data Sharing
Total RNA-seq data and the original WES data from the Mongolia and Western cohorts have been deposited at the European Genome-Phenome Archive (EGA), which is hosted by the European Bioinformatics Institute (EBI) and the Centre for Genomic Regulation (CRG), under the study accession code EGAS00001005364. HBV and HDV genotypes are deposited in Genebank (accession IDs MZ984226 - MZ984323 and OM981174 - OM981226, respectively). The remaining data are available in the Article, Supplementary Information, or available from the authors upon reasonable request”.
Additional detailed information is provided in the Supplementary Materials and Methods.
References
- 1.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: A Cancer Journal for Clinicians. 2021;71:209–49. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Llovet JM, Kelley RK, Villanueva A, Singal AG, Pikarsky E, Roayaie S, et al. Hepatocellular carcinoma. [cited 2021 May 21];Nature Reviews Disease Primers. 2021 7:7. doi: 10.1038/s41572-020-00240-3. [Internet] [DOI] [PubMed] [Google Scholar]
- 3.Polaris Observatory Collaborators. Global prevalence, treatment, and prevention of hepatitis B virus infection in 2016: a modelling study. The Lancet Gastroenterology and Hepatology. 2018;3:383–403. doi: 10.1016/S2468-1253(18)30056-6. [DOI] [PubMed] [Google Scholar]
- 4.Blach S, Zeuzem S, Manns M, Altraif I, Duberg AS, Muljono DH, et al. Global prevalence and genotype distribution of hepatitis C virus infection in 2015: a modelling study. The Lancet Gastroenterology and Hepatology. 2017;2:161–76. doi: 10.1016/S2468-1253(16)30181-9. [DOI] [PubMed] [Google Scholar]
- 5.Demaio AR, Dugee O, de Courten M, Bygbjerg IC, Enkhtuya P, Meyrowitsch DW. Exploring knowledge, attitudes, and practices related to alcohol in Mongolia: a national population-based survey. BMC Public Health. 2013;13:178. doi: 10.1186/1471-2458-13-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dondog B, Lise M, Dondov O, Baldandorj B, Franceschi S. Hepatitis B and C virus infections in hepatocellular carcinoma and cirrhosis in Mongolia. Eur J Cancer Prev. 2011;20:33–9. doi: 10.1097/cej.0b013e32833f0c8e. [DOI] [PubMed] [Google Scholar]
- 7.Puigvehí M, Moctezuma-Velázquez C, Villanueva A, Llovet JMJ, Moctezuma-Velazquez C, Villanueva A, et al. The oncogenic role of hepatitis delta virus in hepatocellular carcinoma. [cited 2020 Jul 1];JHEP Rep. 2019 1:120–30. doi: 10.1016/j.jhepr.2019.05.001. [Internet] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schulze K, Imbeaud S, Letouzé E, Alexandrov LBL, Calderaro J, Rebouissou S, et al. Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and potential therapeutic targets. Nat Genet. 2015;47:505–11. doi: 10.1038/ng.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zucman-Rossi J, Villanueva A, Nault JC, Llovet JM. Genetic Landscape and Biomarkers of Hepatocellular Carcinoma. Gastroenterology. 2015;149:1226–39. doi: 10.1053/j.gastro.2015.05.061. [DOI] [PubMed] [Google Scholar]
- 10.Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SAJR, Behjati S, Biankin Av, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–21. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Letouzé E, Shinde J, Renault V, Couchy G, Blanc JF, Tubacher E, et al. Mutational signatures reveal the dynamic interplay of risk factors and cellular processes during liver tumorigenesis. [cited 2020 Jul 1];Nature Communications. 2017 8:1315. doi: 10.1038/s41467-017-01358-x. [Internet] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Candia J, Bayarsaikhan E, Tandon M, Budhu A, Forgues M, Tovuu LO, et al. The genomic landscape of Mongolian hepatocellular carcinoma. [cited 2020 Oct 29];Nature Communications. 2020 11:4383. doi: 10.1038/s41467-020-18186-1. [Internet] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ahn S-M, Jang SJ, Shim JH, Kim D, Hong S-M, Sung CO, et al. Genomic portrait of resectable hepatocellular carcinomas: implications of RB1 and FGF19 aberrations for patient stratification. Hepatology. 2014;60:1972–82. doi: 10.1002/hep.27198. [DOI] [PubMed] [Google Scholar]
- 14.Ally A, Balasundaram M, Carlsen R, Chuah E, Clarke A, Dhalla N, et al. Comprehensive and Integrative Genomic Characterization of Hepatocellular Carcinoma. [cited 2017 Jul 17];Cell. 2017 169:1327–1341.:e23. doi: 10.1016/j.cell.2017.05.046. [Internet], Available from: /pmc/articles/PMC5680778/?report=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rosenthal R, McGranahan N, Herrero J, Taylor BS, Swanton C. deconstructSigs: Delineating mutational processes in single tumors distinguishes DNA repair deficiencies and patterns of carcinoma evolution. Genome Biology. 2016;17 doi: 10.1186/s13059-016-0893-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hartley SW, Mullikin JC. QoRTs: a comprehensive toolset for quality control and data processing of RNA-Seq experiments. BMC Bioinformatics. 2015;16:224. doi: 10.1186/s12859-015-0670-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Risso D, Ngai J, Speed TP, Dudoit S. Normalization of RNA-seq data using factor analysis of control genes or samples. Nat Biotechnol. 2014;32:896–902. doi: 10.1038/nbt.2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yoshihara K, Shahmoradgoli M, Martínez E, Vegesna R, Kim H, Torres-Garcia W, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nature Communications. 2013;4 doi: 10.1038/ncomms3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Petrick JL, Braunlin M, Laversanne M, Valery PC, Bray F, McGlynn KA. International trends in liver cancer incidence, overall and by histologic subtype, 1978-2007. Int J Cancer. 2016;139:1534–45. doi: 10.1002/ijc.30211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baatarkhuu O, Gerelchimeg T, Munkh-Orshikh D, Batsukh B, Sarangua G, Amarsanaa J. Epidemiology, Genotype Distribution, Prognosis, Control, and Management of Viral Hepatitis B, C, D, and Hepatocellular Carcinoma in Mongolia. Euroasian Journal of Hepato-Gastroenterology. 2018;8:57–62. doi: 10.5005/jp-journals-10018-1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wong GL-H, Chan HL-Y, Yiu KK-L, Lai JW-Y, Chan VK-K, Cheung KK-C, et al. Meta-analysis: The association of hepatitis B virus genotypes and hepatocellular carcinoma. Aliment PharmacolTher. 2013;37:517–26. doi: 10.1111/apt.12207. [DOI] [PubMed] [Google Scholar]
- 24.Wei F, Zheng Q, Li M, Wu M. The association between hepatitis B mutants and hepatocellular carcinoma: A meta-analysis. Medicine. 2017;96:e6835. doi: 10.1097/MD.0000000000006835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sonneveld MJ, Rijckborst V, Zeuzem S, Heathcote EJ, Simon K, Senturk H, et al. Presence of precore and core promoter mutants limits the probability of response to peginterferon in hepatitis B e antigen-positive chronic hepatitis B. Hepatology. 2012;56:67–75. doi: 10.1002/hep.25636. [DOI] [PubMed] [Google Scholar]
- 26.Chen B-F, Liu C-J, Jow G-M, Chen P-J, Kao J-H, Chen D-S. High prevalence and mapping of pre-S deletion in hepatitis B virus carriers with progressive liver diseases. Gastroenterology. 2006;130:1153–68. doi: 10.1053/j.gastro.2006.01.011. [DOI] [PubMed] [Google Scholar]
- 27.Jardi R, Rodriguez F, Buti M, Costa X, Cotrina M, Galimany R, et al. Role of hepatitis B, C, and D viruses in dual and triple infection: Influence of viral genotypes and hepatitis B precore and basal core promoter mutations on viral replicative interference. Hepatology. 2001;34:404–10. doi: 10.1053/jhep.2001.26511. [DOI] [PubMed] [Google Scholar]
- 28.Giersch K, Homs M, Volz T, Helbig M, Allweiss L, Lohse AW, et al. Both interferon alpha and lambda can reduce all intrahepatic HDV infection markers in HBV/HDV infected humanized mice. Scientific Reports. 2017;7 doi: 10.1038/s41598-017-03946-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schaper M, Rodriguez-Frias F, Jardi R, Tabernero D, Homs M, Ruiz G, et al. Quantitative longitudinal evaluations of hepatitis delta virus RNA and hepatitis B virus DNA shows a dynamic, complex replicative profile in chronic hepatitis B and D. Journal of Hepatology. 2010;52:658–64. doi: 10.1016/j.jhep.2009.10.036. [DOI] [PubMed] [Google Scholar]
- 30.Schaper M, Rodriguez-Frias F, Jardi R, Tabernero D, Homs M, Ruiz G, et al. Quantitative longitudinal evaluations of hepatitis delta virus RNA and hepatitis B virus DNA shows a dynamic, complex replicative profile in chronic hepatitis B and D. Journal of Hepatology. 2010;52:658–64. doi: 10.1016/j.jhep.2009.10.036. [DOI] [PubMed] [Google Scholar]
- 31.Chen CJ, Yang HI, Su J, Jen CL, You SL, Lu SN, et al. Risk of hepatocellular carcinoma across a biological gradient of serum hepatitis B virus DNA Level. J Am Med Assoc. 2006;295:65–73. doi: 10.1001/jama.295.1.65. [DOI] [PubMed] [Google Scholar]
- 32.Ahn S-M, Jang SJ, Shim JH, Kim D, Hong S-M, Sung CO, et al. Genomic portrait of resectable hepatocellular carcinomas: implications of RB1 and FGF19 aberrations for patient stratification. Hepatology. 2014;60:1972–82. doi: 10.1002/hep.27198. [Internet] [DOI] [PubMed] [Google Scholar]
- 33.Totoki Y, Tatsuno K, Covington KR, Ueda H, Creighton CJ, Kato M, et al. Trans-ancestry mutational landscape of hepatocellular carcinoma genomes. Nature Genetics. 2014;46:1267–73. doi: 10.1038/ng.3126. [DOI] [PubMed] [Google Scholar]
- 34.Martincorena I, Raine KM, Gerstung M, Dawson KJ, Haase K, Van Loo P, et al. Universal Patterns of Selection in Cancer and Somatic Tissues. Cell. 2017;171:1029–1041.:e21. doi: 10.1016/j.cell.2017.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arnedo-Pac C, Mularoni L, Muiños F, Gonzalez-Perez A, Lopez-Bigas N, Schwartz R. OncodriveCLUSTL: A sequence-based clustering method to identify cancer drivers. Bioinformatics. 2019;35:4788–90. doi: 10.1093/bioinformatics/btz501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alexandrov LB, Kim J, Haradhvala NJ, Huang MN, Tian Ng AW, Wu Y, et al. The repertoire of mutational signatures in human cancer. Nature. 2020;578:94–101. doi: 10.1038/s41586-020-1943-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.European Association For The Study Of The Liver, European Organisation For Research And Treatment Of Cancer. EASL−EORTC Clinical Practice Guidelines: Management of hepatocellular carcinoma. Journal of Hepatology. 2012;56:908–43. doi: 10.1016/j.jhep.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 38.Kucab JE, Zou X, Morganella S, Joel M, Nanda AS, Nagy E, et al. A Compendium of Mutational Signatures of Environmental Agents. [cited 2020 Nov 3];Cell. 2019 177:821–836.:e16. doi: 10.1016/j.cell.2019.03.001. [Internet] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bai H, Guo X, Zhang D, Narisu N, Bu J, Jirimutu J, et al. The Genome of a Mongolian Individual Reveals the Genetic Imprints of Mongolians on Modern Human Populations. Genome Biology and Evolution. 2014;6:3122–36. doi: 10.1093/gbe/evu242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tsatsralt-Od B, Takahashi M, Nishizawa T, Endo K, Inoue J, Okamoto H. High prevalence of dual or triple infection of hepatitis B, C, and delta viruses among patients with chronic liver disease in Mongolia. Journal of Medical Virology. 2005;77:491–9. doi: 10.1002/jmv.20482. [DOI] [PubMed] [Google Scholar]
- 41.Rao RC, Dou Y. Nature Reviews Cancer. Vol. 15. Nature Publishing Group; 2015. Hijacked in cancer: The KMT2 (MLL) family of methyltransferases; pp. 334–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chakravarty D, Gao J, Phillips S, Kundra R, Zhang H, Wang J, et al. OncoKB: A Precision Oncology Knowledge Base. JCO Precision Oncology. 2017:PO.17.00011. doi: 10.1200/PO.17.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.International Agency for Research on Cancer. Agents Classified by the IARC Monographs, Volumes 1−127 − IARC Monographs on the Identification of Carcinogenic Hazards to Humans. 2018. [cited 2020 Nov 5]. [Internet], Available from: https://monographs.iarc.fr/agents-classified-by-the-iarc.
- 44.WHO. Air pollution in Mongolia. Bull World Health Organ. 2019;97:79–80. doi: 10.2471/BLT.19.020219. [Internet], Available from: https://www.who.int/bulletin/volumes/97/2/19-020219/en/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chiang DY, Villanueva A, Hoshida Y, Peix J, Newell P, Minguez B, et al. Focal gains of VEGFA and molecular classification of hepatocellular carcinoma. Cancer Research. 2008;68:6779–88. doi: 10.1158/0008-5472.CAN-08-0742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Montironi C, Castet F, Haber PK, Pinyol R, Torres-Martin M, Torrens L, et al. Inflamed and non-inflamed classes of HCC: a revised immunogenomic classification. Gut. 2022:gutjnl-2021-325918. doi: 10.1136/gutjnl-2021-325918. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Total RNA-seq data and the original WES data from the Mongolia and Western cohorts have been deposited at the European Genome-Phenome Archive (EGA), which is hosted by the European Bioinformatics Institute (EBI) and the Centre for Genomic Regulation (CRG), under the study accession code EGAS00001005364. HBV and HDV genotypes are deposited in Genebank (accession IDs MZ984226 - MZ984323 and OM981174 - OM981226, respectively). The remaining data are available in the Article, Supplementary Information, or available from the authors upon reasonable request”.
Additional detailed information is provided in the Supplementary Materials and Methods.