Abstract
Streptococcus pneumoniae is a significant cause of morbidity and mortality worldwide, causing life-threatening diseases such as pneumonia, bacteraemia, and meningitis, with an annual death burden of over one million. Discovered over a century ago, pneumococcal serotype 1 (S1) is a significant cause of these life-threatening diseases. Our understanding of the epidemiology and biology of pneumococcal S1 has significantly improved over the past two decades, informing the development of preventative and surveillance strategies. However, many questions remain unanswered. Here, we review the current state of knowledge of pneumococcal S1, with a special emphasis on clinical epidemiology, genomics, and disease mechanisms.
Keywords: Pneumococcus, serotype 1, genomics, outbreaks, invasiveness, virulence
Introduction
Streptococcus pneumoniae, also known as the pneumococcus, is an α-haemolytic Grampositive, opportunistic pathogen which colonises the human nasopharynx [1]. Colonisation with the pneumococcus is typically asymptomatic and more common in infants compared to adults. Rates of colonisation tend to vary by age, for example, on average 7% and 11% prevalence in adults in the United Kingdom and Malawi, respectively, and approximately 42% in children [2,3] reaching rates as high as 100% during the first year of life [4]. Colonisation is also believed to be a pre-requisite for the onset of pneumococcal disease and may confer, albeit weak, natural immunity against pneumococcal infection [5,6]. The pneumococcus can spread from the nasopharynx to other sites of the body such as the lung, blood, and meninges, to cause both non-invasive and invasive pneumococcal diseases (IPD) [1]. Recent global estimates report that S. pneumoniae is the largest cause of death due to lower respiratory tract infections, resulting in over one million casualties annually [7,8], of which ca. 320,000 are children under the age of 5, affecting particularly low- and middle-income countries (LMICs) [8].
Thus far, 100 pneumococcal capsular serotypes have been identified [9,10]. Distinct capsular serotypes may differ in their ability to colonise the nasopharynx [11] or cause IPD [12]. Pneumococcal S1 was among the first serotypes discovered in the early 20th century [13] and ranks among the most common serotypes associated with IPD globally - particularly in Sub-Saharan Africa (SSA), South America and Asia [14] - where it is widely associated with outbreaks (Table 1). Although bacteraemia, pneumonia, and meningitis are the main IPD caused by pneumococcal S1, it is also widely recovered in patients with empyema [15].
Table 1. Summary of known outbreaks and epidemics associated with pneumococcal serotype 1.
| Year of outbreaks | Place of outbreak | Continent | Specific location | Outbreak setting | Clinical manifestation | Age group | Conjugate vaccination period | Dominant clone | Reference |
|---|---|---|---|---|---|---|---|---|---|
| 1917 | USA | North America | Rochester | Boys’ asylum and hospital | Pneumonia | All ages | Pre-PCV | Unknown | [38] |
| February to June 1931 | Norway | Europe | Bærum county | Orphanage | Croupous pneumonia | Boys (5 to 20 years) | Pre-PCV | Unknown | [43] |
| January to March 1937 | USA | North America | Worcester, Massachusetts | Hospital setting | Lobar pneumonia | Adults | Pre-PCV | Unknown | [44] |
| Early 1978 | USA | North America | Boston city | Homeless men’s shelter | Bacteraemia | Adults | Pre-PCV | Unknown | [45] |
| April 1988 to March 1989 | France | Europe | Paris | Homeless men’s shelters | Acute pneumonia | Adults | Pre-PCV | Unknown | [34] |
| Winter of 1991 | Australia | Australia | Alice Springs region | Mainly Aboriginal men | Bacteraemia pneumonia | Children and adults | Pre-PCV | Unknown | [39] |
| March 1997 | Israel | Asia | City of Dimona, Southern Israel | Closed community | Lobar pneumonia and bacteraemia | Children and young adults | Pre-PCV | Unknown | [46] |
| 1998 to 2003 | Ghana | Africa | Kassena-Nankana District, northern Ghana | Community | Meningococcal- like meningitis | All ages | Pre-PCV | ST217 | [47] |
| August 2000 to 2002 | Canada | North America | Nunavick, northern Canada | Community | Severe pneumonia | Adults (20 to 64 years) | Pre-PCV | Unknown | [48][49] |
| 2000 to 2007 | South Pacific | Oceania | New Caledonia, Wallis and Futuna, and French Polynesia | Community | Bacteraemia and pneumonia | All ages | Pre- PCV | ST306 | [50] |
| August 2001 to April 2002 | Tunisia | Africa | Tunis | Jail and community | Acute lower respiratory tract infection | Adults | Pre-PCV13 | Unknown | [51] |
| May 2002 to February 2005 | Burkina Faso | Africa | District 15, 22, and Houndé | Community | Meningitis | Children and adults | Pre-PCV13 | ST618 | [52] |
| Between 2002 and 2004 | Australia | Australia | Tiwi islands | Aboriginal population | Unknown | Children and adults | Pre-PCV13 | Unknown | [53] |
| 2004 to 2006 | The Gambia | Africa | Nation-wide | Community | Several IPD | All ages | Pre-PCV | ST217 | [54] |
| 10 to 13 October 2006 | United Kingdom | Europe | North Tyneside | Primary school | Lobar pneumonia | Children | Pre-PCV13 | Unknown | [36] |
| 1 June 2008 to 31 May 2010 | Netherlands | Europe | Nation-wide | Community | Several IPD | Young female adults | Pre-PCV13 | Unknown | [55] |
| 2010 to 2013 | Australia | Australia | Northern and central Australia | Mostly indigenous population | Several IPD | All ages (median age 15 years) | Pre- and post-CV13 | ST306 | [56] |
| October 2010 to 2012 | Australia | Australia | Central Australia | Community | Several IPD | Older children | PCV10 | ST306 | [35] |
| 2000-2004, and 2010 | Kenya | Africa | Kilifi | Community | Bacteraemia and meningitis | Children and young adults | Pre-PCV | Unknown | [57] |
| 2006-2007, 2010, and 2015 | Malawi | Africa | Blantyre | Community | Bacteraemia and meningitis | All ages | Pre-PCV, and post-PCV13 | ST217 | [58] |
| December 2015 to April 2016 | Ghana | Africa | Brong-Ahafo region | Community | Meningitis | Older children and adults | Post-PCV13 | ST217 | [40] |
| 2016 to 2017 | Ghana | Africa | Northern and upper west region | Community | Meningitis | All ages | Post-PCV13 | Unknown | [59] |
| October 2016 to April 2017 | Central African Republic | Africa | North-western region | Community | Meningitis | All ages | Post-PCV13 | Unknown | [41] |
Pneumococcal serotype 1 is an atypical commensal and an adept invader
The duration of pneumococcal carriage varies significantly across epidemiological settings - with age being one of the strongest determinants [11,16,17]. Unlike most pneumococcal serotypes, which are proficient asymptomatic colonisers presenting modest invasive potential, S1 typically exhibits high invasive disease potential i.e. invasiveness [18]. Invasiveness is classically defined as the odds ratio of IPD relative to asymptomatic nasopharyngeal carriage events, whereby a value >1 indicate a higher propensity of a given strain to be isolated in IPD patients vs. asymptomatic carriers. Several studies have consistently reported high invasive potential for pneumococcal S1 [18], temporally as well as geographically [19]. Studies have shown that S1 is more invasive compared to other serotypes, although the odds ratios vary globally. For example, - 9.6 in the United Kingdom [12], 15.6 in Belgium [20], 46.68 in Sweden [21], and 22.3 in Mozambique [22]. Such variable invasiveness estimates may reflect differences in sample sizes, host factors, circulating clones and distribution of serotypes within a given setting [14]. Despite high invasiveness - which translates to a high IPD burden - pneumococcal S1 may not necessarily be the most lethal strain e.g., compared to serotype 3 [23,24]. Current evidence on the lethality of pneumococcal serotypes primarily originates from high income settings where the incidence of S1 is low, hence further studies are warranted.
By virtue of its ability to establish nasopharyngeal colonisation over a very short duration (up to 1-2 weeks [11,16,25] compared to other serotypes, pneumococcal S1 does not appear to behave like a typical commensal [26,27]. Previous studies showed that the duration of carriage and invasive potential of pneumococci were positively correlated with the degree of encapsulation [28]. Indeed, the polysaccharide capsule is known to protect against immune-mediated clearance, e.g., by blocking the deposition and function of opsonins, evading neutrophil extracellular traps (NETs), or reducing mucus-mediated clearance. The zwitterionic structure of the pneumococcal S1 capsule is particularly intriguing in that surface charge-switching (i.e. zwitterionic) carriers are regarded as promising delivery systems to traverse the mucus layer and reach the underlying epithelium [29]. The S1 capsule was found to be significantly more resistant to opsonisation and complement deposition [30] with an enhanced capacity to translocate across the nasopharynx to reach the olfactory tissues and, ultimately, the brain compartments [31]. A rapid nose-to-brain translocation was reported to occur via an inward flow of fluid transporting S. pneunoniae through the cribriform plate and to the dura within minutes [32]. The hypervirulence of pneumococcal S1 was also found to be associated with the rapid release of pneumolysin and consequent enhanced dissemination [31,33]. Other serotypes, such as 5 and 7F, were also documented for their short duration of carriage and small capsule size [28]. Further investigations on the properties shared by all these serotypes may bring further insight into the peculiarity of S1.
Pneumococcal serotype 1 outbreaks and epidemics
Although rare, outbreaks of pneumococcal disease do occur and are commonly associated with vulnerable groups, such as the homeless or alcoholic individuals [34], those living in closed communities or endemic regions such as the African meningitis belt. Although non-S1 serotypes have shown potential to cause similar outbreaks, a large majority of reported outbreaks were associated with S1 [35,36] and serotype 5 [37]. Interestingly, the two earliest recorded outbreaks of S1 were associated with lobar pneumonia and occurred at a boys asylum and at the Rochester State Hospital both in 1917 and in New York, USA [38] and resulted in fatality rates of 50% and 66%, respectively. Table 1 provides a detailed summary of known reported outbreaks of S1 globally. Despite the apparent increase in the number of outbreaks reported over the past 50 years - likely due to the increased availability of typing techniques and the expansion of global surveillance programmes, the widespread use of antibiotics and the rollout of pneumococcal vaccines have led to an overall decrease in the occurrence of outbreaks [39]. Based on current data, most of the reported pneumococcal S1 outbreaks seem to be associated with only a select number of sequence types (ST) (or clones) as defined by pneumococcal multilocus sequence typing or MLST (Table 1). Indeed, S1 outbreaks in Europe, North America, and South America are typically associated with ST306 clones, while in Africa, especially SSA, pneumococcal ST217 and its single locus variants - namely ST303 and ST612 - were documented as the main culprits. Worryingly, S1 outbreaks continue to occur in some parts of the world even after the introduction of PCVs, for example, in Ghana [40] and Central African Republic [41] in the West African meningitis belt. These outbreaks will likely become less common due to the expansion of routine immunisation programmes. Although the ST217 clone is predominantly found in both West and Southern Africa, it forms distinct clades, which suggest that this clone may present limited transmission and local adaptation [42] (Figure 1a). In-depth investigations of different outbreak-causing S1 strains using whole-genome sequencing (WGS) datasets have the potential to reveal genetic patterns associated with virulence.
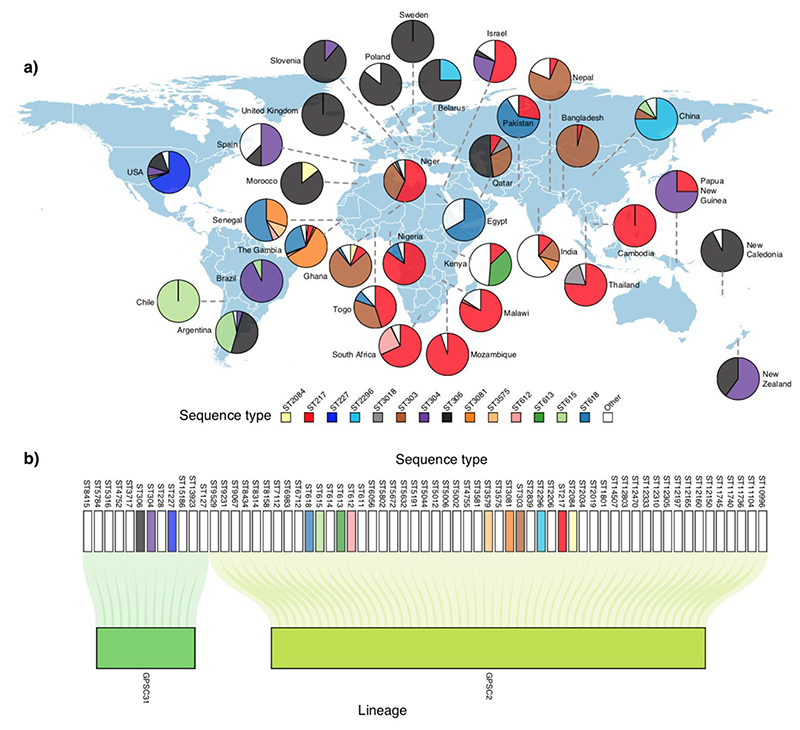
Figure 1. Serotype 1 sequence types (ST) and lineages globally.
a) Geographical distribution of STs defined using MLST. The S1 isolates shown were obtained from both the Pneumococcal African Genomics (PAGe) [70] and Global Pneumococcal Sequencing (GPS) consortium projects [69]. The pie charts show the proportion of the STs in each country. The number of serotype 1 isolates used to estimate the proportion were small (less than 10) for some countries such as Slovenia, Spain, Poland, Morocco, Egypt, Peru, New Zealand, United Kingdom, Papua New Guinea, and Belarus. b) Association between serotype 1 STs and lineages defined using the GPSC nomenclature [69].
Global distribution of pneumococcal serotype 1 clones
To date, several clones associated with pneumococcal S1 have been characterised, and their distribution varies globally. These clones were primarily defined using molecular techniques such as MLST [60], enabling consistent definition of clones suitable for surveillance internationally. Whole genome sequencing (WGS) is rapidly becoming a more prevalent approach as it provides higher resolution and more in-depth genome-wide information [61]. Using the MLST approach, Brueggemann et al. showed that S1 isolates belong to geographically distinct sequence types (STs) which form three genetic clusters designated as lineage A, B, and C [62]; lineage A was exclusively found in North America and Europe, while lineage B was mainly identified in Africa and Israel, and lineage C in South America. Since then, further S1 lineage diversity was defined (Figure 1): ST306 lineage A expands across Europe, Australia, South Pacific, and North Africa; ST217 lineage Bis is found in SSA, Middle East, and Asia while ST303 lineage B in West Africa and Asia; ST304 lineage B in Oceania, Europe, and South America; and ST615 lineage C is mostly isolated in South America (Figure 1a). Other clones tend to be geographically restricted: for example, the ST613 clone is associated with Eastern Africa, while ST227 and ST2296 are commonly found in the U.S.A and China, respectively. Although the temporal distribution of the S1 clones is remarkably stable, replacement of clones has been noted in some countries, including in The Gambia where ST3081 superseded ST618 [63], in the United Kingdom where ST306 succeeded ST227 [64], and in Brazil where ST304 became the dominant clone (no major clone identified prior to that) [65]. With the increasing availability of WGS data, there has been a shift towards defining pneumococcal S1 clones and lineages using genome-wide data. However, there have been challenges in assigning consistent and epidemiologically meaningful nomenclature for the lineages, making it difficult to compare findings over different studies. To address this, a consistent international nomenclature for the pneumococcal lineages known as Global Pneumococcal Sequence Clusters (GPSC), was proposed by Gladstone et al. [66] using the PopPUNK framework [67]. Using this GPSC nomenclature, the most common lineages of S1 were found to be GPSC2 and GPSC31 (Figure 1b).
Population genomics of pneumococcal serotype 1
Genomic studies have revealed a phylogeographically structured population of S1 with infrequent inter-mixing of isolates from different countries, suggesting a rare spread of clones between settings [68,69]. Whether such limited dissemination of clones between countries is a consequence of the rare and short duration of carriage remains to be investigated. As a naturally competent bacterium, the pneumococcus reshuffles its genomic DNA through a process known as recombination [70] . For example, recombination of the capsule biosynthesis genes may alter the antigenicity of the expressed capsule, i.e. capsule switching [71]. An important consequence of capsule switching is vaccine escape, a phenomenon which occurs when strains presenting capsules targeted by PCVs switch to an antigenically distinct capsule not targeted by existing PCV formulations [72]. Recombination in S1 is considered to be rare compared to other serotypes which are capable of establishing longer durations of nasopharyngeal carriage [27,28] . A stable colonisation may indeed prolong exposure to co-colonising pneumococcal strains or related species, hence increasing the likelihood of genetic exchange (Box 1) [28]. Experimental studies have reported challenges in transforming S1 strains using a suicide plasmid instead of linear DNA [73,74], possibly supporting the notion that recombination rates are low in these strains.
Phylogenetically, S1 isolates belong to genetically related lineages sharing a common ancestor. To date, no isolates bearing the S1 capsule have been found in other distinct lineages [66,69], further highlighting both the purported low recombination levels of pneumococcal S1, and the possibility that capsule biosynthesis loci from S1 strains are less likely to be taken up by other lineages to generate capsule-switched S1 lineages. One recent study by Lessa et al. has, nevertheless, found homologs of S1 specific capsule biosynthesis genes in Streptococcus mitis, a closely related commensal of the pneumococcus sharing overlapping niches [74]. This raises important questions regarding the contribution of commensal streptococci to natural immunity against S1 and other pneumococcal serotypes.
Acquired antimicrobial resistance (AMR) has been widely regarded as rare in S1 isolates and this was attributed to low recombination rates. Low AMR rates were indeed reported in S1 isolates originating from high-income countries (HICs); however, in other parts of the world, such as in SSA, higher AMR rates were documented. Multidrug resistance rates (MDR) among S1 isolates in Malawi was nearly 82%, the highest recorded for any serotype [75], in contrast with an MDR of 4% in South Africa [61]. Similarly, high AMR rates were reported among S1 for cotrimoxazole and tetracycline but not chloramphenicol, penicillin, and cefotaxime, which are widely used to treat pneumococcal diseases in The Gambia [54]. Further studies are required to understand the factors driving the differences in the AMR rates of S1.
Genome-wide association studies (GWAS) have paved the way for exploratory investigations to identify genomic variation likely to affect bacterial phenotypes, including disease susceptibility [76,77] and antimicrobial resistance [78]. For example, a recent study comparing S1 isolates collected from the cerebrospinal fluid vs. non-CNS tissues of IPD patients revealed statistically significant allelic variants within the gene encoding the surface-exposed choline-binding protein A (CbpA or PspC) associated with neurotropism [79]. Cornick et al. also used whole-genome analysis to investigate the species-wide distribution of vaccine candidate genes in a global collection of S1 isolates to inform vaccine design [77]. Other studies have combined genomic analysis and in vivo modelling to investigate phenotypic differences leading to the clonal replacement of ST618 with ST3081 S1 isolates in The Gambia [80].
Dynamics of pneumococcal S1 colonisation, shedding and transmission
While acquisition rate and carriage duration are known to be serotype-dependent, an additional layer of complexity resides in the observation that the human nasopharynx can harbor multiple pneumococcal serotypes simultaneously. While multiple pneumococcal serotypes can either simultaneously or sequentially colonise the human nasopharynx [81], previous studies showed that the current colonising serotype usually prevails [82,83]. Population-based studies conducted on samples collected in Gambian infants reported that co-colonisation with multiple pneumococcal serotypes was observed in over 40% of infants at any given sampling time point [16]. S1 was found at a prevalence of 0.93% (compared to 11.42% for type 19A at the highest end) and was also frequently associated with several serotypes.
Using murine models, the propensity of S1 to colonise the nasopharynx was shown to be reduced in the presence of a prior colonizer such as serotype 19F or 6B [33]. In high transmission settings, the persistence of invasive disease and ongoing outbreaks caused by pneumococci S1 has raised questions over the need to introduce a booster dose [52,84]. It is generally accepted that pneumococcal transmission occurs primarily through indirect contact via inhalation of airborne droplets, mainly prevailing in high-density living settings, e.g., daycare centres, prisons, and nursing homes [85], and in the presence of concomitant viral respiratory tract infections [90]. The type and amount of capsular polysaccharide were shown to play a critical role in the dynamics of pneumococcal shedding and transmission [86]; however, other factors such as density and duration of colonisation, as well as ex vivo survival [87,88] are also contributing influences. Murine models have been developed to aid the understanding of pneumococcal transmission and disease susceptibility [82,89]. These animal models were primarily developed using lab-adapted strains such as serotype 2 (D39) and 4 (TIGR4), or clinical isolates such as serotypes 6A, 19F, 23F, 7F, and 14. An adult murine model of pneumococcal transmission has been developed [32] which could further the understanding of factors promoting transmission of S1 and other serotypes to aid in development of better control measures.
High attack rate, pneumolysin and haemolytic activity
As early as 1937, Heffon found that S1 was responsible for 22% of pneumococcal pneumonia cases in children and 33% in adults, which is higher than the median attack rates of 7% seen in outbreaks of other pneumococcal serotypes [92]. The high attack rates of S1 could also be related to circumstances that may contribute to the development of IPD, such as high population density [93], viral co-infections [94] and environmental factors such as pollutants [95], cigarette vapour [96], airborne dust and high temperatures, as shown in Figure 2 [42]. In areas such as the African meningitis belt, the incidence of meningitis is over ten times higher than in Western Europe and the United States, with S1 accounting for 76% of all isolates causing meningitis [47]. Exposure of S1-colonised mice to high temperatures, representative of those in SSA, resulted in greater levels of bacterial dissemination and increased invasiveness [42]. Evidence suggests that the role of pneumolysin in S1 pathogenesis is complex, and mechanistic studies have been hampered by the inability to genetically modify S1. Recently, Terra et al. have successfully transformed S1 (ST5316, European lineage A) to deplete the pneumolysin gene [73]. Murine models of infection using this pneumolysin-deficient S1 suggest that S1 can still cause pneumonia in mice. Another isolate from lineage A (ST228), despite being cultured from the blood of a patient with pneumonia, was found not to express pneumolysin but could still disseminate from lungs into the blood, suggesting that other virulence factors play a more prominent role in the pathogenesis of these S1 sequence types. This contrasts with the depletion of pneumolysin in an ST615 clone which renders the isolate completely avirulent in murine models of pneumonia [97]. The pore-forming ability of pneumolysin was described as a critical virulence factor for pneumococci [98]. S1 strains expressing a fully haemolytic pneumolysin appear to induce higher secretion levels of type 1-interferon in mouse lungs, which drives dissemination into the blood, thus aiding pneumococcal S1 virulence [99]. In contrast, the absence of haemolytic activity was associated with reduced inflammation; for example, poor activation of the NLRP3 inflammasome, which drives IL-1β production and reduction in pro-inflammatory cytokines such as KC and IL-6, led to a decrease in neutrophil recruitment in the lungs [100]. Interestingly, however, S1 clones such as ST306 cause high rates of pneumococcal disease while expressing a non-cytolytic pneumolysin variant [64] and was commonly associated with a high incidence of non-lethal empyema, particularly in recurrent paediatric infections [101]. While the ST306 clone was first described almost two decades ago, the evolutionary significance of non-cytolytic pneumolysin variants remain elusive.
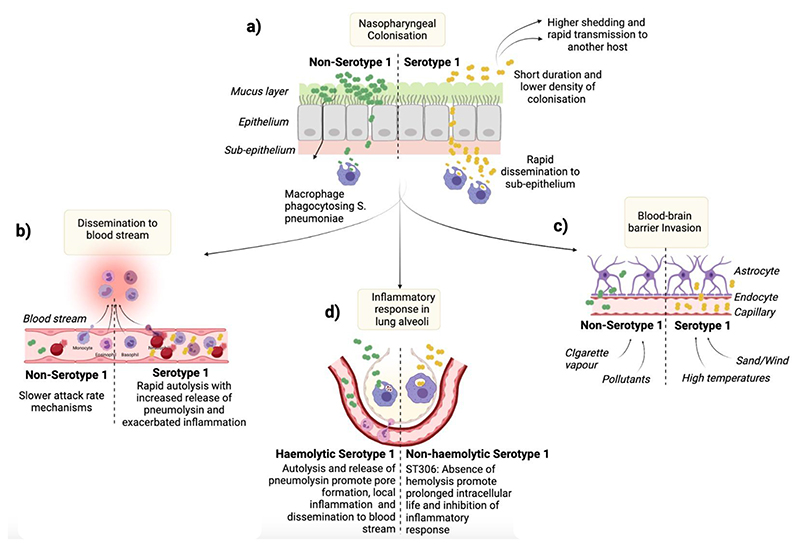
Figure 2. Hypothetical virulence mechanisms and host immunity to pneumococcal serotype 1.
S1 is an atypical commensal known for its short duration (1-2 weeks) and lower density of colonisation in the human nasopharynx, potentially accompanied by higher shedding and host-to-host transmission (a). Its zwitterionic capsule promotes its ability to traverse the mucus layer and migrate deeper into the sub-epithelium to either reach the bloodstream - where it induces acute haemolysis due to its rapid autolysis (b), or cross the olfactory system and the blood-brain barrier to invade the central nervous system – especially in the presence of environmental factors such as sand, dust, and high temperature (c). The immune recognition of S1 and release of pneumolysin leads to the activation of inflammatory response that promotes the invasion process (d).
Badgujar et al. suggested that the loss of haemolytic activity enables ST306 to adopt an atypical intracellular lifestyle due to improved cellular invasion and attenuation of inflammatory responses, including autophagy evasion thus promoting long term survival in the lower airways [102]. Further work is warranted to clarify the contributory role of pneumolysin in S1 pathogenicity.
Concluding remarks and future perspectives
Remarkable advances have been made regarding understanding the epidemiology and biology of pneumococcal S1, including the critical role of the antigenic outer cell wall polysaccharide capsule and virulence factors, such as pneumolysin and autolysin, on colonisation and disease. Nevertheless, challenges remain to fully understand the atypical hyper-invasiveness of S1 pneumococci compared to other serotypes. Recent advances in transforming S1 isolates will pave the way for further studies to examine the role of the capsule and genetic background in the invasiveness of pneumococcal strains. And although the IPD burden due to S1 has significantly decreased globally owing to the introduction of higher-valency PCVs (PCV10 and PCV13), it remains as important as ever to understand the determinants and mechanisms of virulence and pathogenicity of highly invasive pneumococcal serotypes such as S1. Such knowledge will inform the implementation of improved public health interventions, and the development of broader-acting vaccines to prevent IPD caused by serotypes other than those included in the existing PCV formulation, including those poorly controlled by PCVs e.g. serotype 3 [108] and niche replacement serotypes e.g. serotype 12F [109].
The increasing availability of omics (genomic, transcriptomics, proteomics, metabolomics) datasets of pneumococcal isolates, from small [68,110,111] and large-scale projects such as the PAGe [70] and GPS consortiums [66], will provide unprecedented opportunities to understand the complex epidemiology and biology of S1. Additionally, the development of in vitro and in vivo translational models closely mimicking human physiology, combined with the ability to genetically manipulate the pneumococcal S1 genome, will promote further understanding of the hyper-invasiveness of pneumococcal S1. Outstanding questions pertaining to pathogen, host and environmental factors will be addressed through an integrated approach encompassing multi- and interdisciplinary approaches combining epidemiological, bioinformatical, experimental and clinical studies. Ultimately, addressing these pertinent questions will inform prevention and control strategies for pneumococcal diseases.
Highlights.
Pneumococcal serotype 1 (S1) is a major cause of invasive pneumococcal disease (IPD) globally, especially in Africa, South America, and Asia
Pneumococcal S1 strains are frequently found in the tissues and fluids of IPD patients while they are rarely isolated from the nasopharynx of asymptomatic carriers, including in high pneumococcal carriage and disease settings
A select number of S1 clones are frequently associated with outbreaks and epidemics, especially in closed communities and highly vulnerable groups
Whole genome sequencing (WGS) has provided valuable insights on the clones, lineages, evolutionary dynamics, and phylogeographic structure of pneumococcal S1 globally
Box 1. Hypothetical scenarios for the rarity of serotype 1 in the nasopharynx.
The paradox of how pneumococcal S1 persists in the human population despite its rare nasopharyngeal carriage rate, including in IPD endemic settings, remains unresolved [63]. Several hypothetical scenarios have been put forward. Firstly, S1 isolates may inactivate capsule expression enabling transient colonisation of the nasopharynx as a non-encapsulated non-typeable pneumococcus [90]. Such inactivation of the capsule would in turn reduce detection of S1 by widely used tests such as latex agglutination and Quellung reaction. However, in this scenario, due to the capsule biosynthesis locus remaining in the genome, pneumococcal S1 would still be detectable using molecular tests such as polymerase chain reaction (PCR). Secondly, it has also been speculated that S1 strains may immediately acquire the capsule of another serotype upon colonisation. This would result in infrequent detection of S1 during carriage as the strains may be disguised as a non-S1 serotype when typing is done using microbiological or PCR tests. However, this scenario is unlikely as capsule-switching is a rare event, and to date, there has been no evidence of capsule-switching between pneumococcal strains expressing an S1 capsule. A third scenario is that pneumococcal S1 can colonise in an alternative compartment of the human body, where it can seed to the nasopharynx to facilitate person-to-person transmission. There is increasing evidence, using murine models, to suggest that pneumococcal S1 colonises niches such as olfactory tissues. If this phenomenon also occurs in humans, then oral-nasopharyngeal swabs would likely be unable to detect pneumococci in these niches and account for reports of low S1 carriage prevalence [31,91].
Box 2. Impact of PCV on the incidence of pneumococcal serotype 1 IPD.
Pneumococcal S1 is one of the serotypes included in the existing PCV10 and PCV13 vaccine formulations. Since the introduction of PCV vaccines into infant immunisation programmes in 2010, there has been a substantial global reduction in S1 outbreaks and IPD in both immunised children and unvaccinated older children and adults. Consistent with earlier data on PCV effectiveness in low disease burden settings [103], recent data describe a 95% reduction in disease incidence in settings presenting high S1 disease burden, six years following the introduction of PCV10 or PCV13 [104–106] . S1 epidemics persist in some settings despite high vaccine uptake, for example, an increase of S1 IPD was documented five years after the introduction of PCV13 in Malawi [58]. In the African Meningitis belt, whilst the overall incidence of pneumococcal meningitis has decreased in children under the age of five, S1 persisted as the main causative agent [40,59,107]. There have been speculations that the persistence of S1 post-vaccination may be attributed to the “3+0” immunisation schedule (no booster), which may be less effective compared to high income settings where a booster dose is given in the second year of life [84]. Another contributing factor may be that unvaccinated older siblings may be the reservoir for transmission to younger infants. The cyclical nature of S1 disease outbreaks has made it challenging to assess the true impact of PCV13 and will require a longer monitoring period with accurate documentation on vaccine uptake. Further studies are required to assess the impact of different PCV vaccine coverage and schedules on the incidence of S1 disease.
Outstanding questions.
Can genome-wide association studies comparing isolates sampled from asymptomatic healthy individuals vs. IPD patients reveal further clues about the hyper-invasiveness and pathogenicity of pneumococcal S1?
In the longer term, how will the introduction of PCVs impact pneumococcal S1 disease incidence in outbreak settings, and will it drive the emergence of vaccine-escape S1 variants?
Given its seemingly low prevalence and short duration in nasopharyngeal colonisation, how does pneumococcal S1 persist in the human population as one of the prevailing causes of IPD?
If the transformation rate of pneumococcal S1 differs from that of other serotypes, what are the underlying determinants of these differences?
Does inter-serotype competition play a major role in the disease and carriage dynamics of pneumococcal S1 strains?
How do variations in lineages and sequence types determine the differences observed in the mortality rate or disease severity observed among different pneumococcal S1 strains?
What are the host and environmental factors that influence the transmission and global distribution of pneumococcal S1 lineages?
Glossary.
Attack rate: Refers to the proportion of susceptible individuals who get infected during a specified period, for example, during an outbreak
Capsule biosynthesis locus: A cluster of genes delineated by the dexB and aliA genes in the pneumococcal genome, which are involved in the synthesis of the serotype-defining antigenic polysaccharide surrounding pneumococcal cells
Global pneumococcal sequence clusters (GPSC): Consistent international nomenclature for pneumococcal lineages defined by the Global Pneumococcal Sequencing (GPS) project
Multilocus sequence typing (MLST): A portable method for identifying bacterial clones based on the unique combination of alleles of the internal fragments of specific house-keeping genes (usually seven) for each species
Pneumococcal conjugate vaccine (PCV): A pneumococcal vaccine containing purified capsular polysaccharides of a subset of serotypes conjugated to a carrier protein to induce a robust immune response
Pneumolysin: A potent cholesterol-dependent cytolysin and key pneumococcal virulence factor, which creates pores in cell membranes and depending on its concentration, can result in the lysis of host cells.
Sequence type (ST): An ST refers to a unique combination of alleles for the internal fragments of housekeeping genes used to define the MLST scheme of a bacterial species
Acknowledgments
The authors would like to acknowledge funding from the Joint Initiative for Antimicrobial Resistance (Grant no. MR/R003076/1), the Bill and Melinda Gates Foundation (OPP1034556), and the UK Medical Research Council Programme Grant (grant no. MR/P011284/1), Wellcome Trust (2016-2021 core award grant no. 206194), and Meningitis Now.
Footnotes
Declaration of interests
The authors declare no competing interests.
References
- 1.Bogaert D, et al. Streptococcus pneumoniae colonisation: the key to pneumococcal disease. Lancet Infect Dis. 2004;4:144–154. doi: 10.1016/S1473-3099(04)00938-7. [DOI] [PubMed] [Google Scholar]
- 2.Gordon SB, et al. Poor potential coverage for 7-valent pneumococcal conjugate vaccine, Malawi. Emerg Infect Dis. 2003;9:747–749. doi: 10.3201/eid0906.030020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adler H, et al. Pneumococcal Colonization in Healthy Adult Research Participants in the Conjugate Vaccine Era, United Kingdom, 2010-2017. J Infect Dis. 2019;219:1989–1993. doi: 10.1093/infdis/jiz034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hill PC, et al. Nasopharyngeal carriage of Streptococcus pneumoniae in Gambian villagers. Clin Infect Dis. 2006;43:673–679. doi: 10.1086/506941. [DOI] [PubMed] [Google Scholar]
- 5.Ramos-Sevillano E, et al. Mechanisms of Naturally Acquired Immunity to Streptococcus pneumoniae. Front Immunol. 2019;10:358. doi: 10.3389/fimmu.2019.00358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ferreira DM, et al. Controlled human infection and rechallenge with Streptococcus pneumoniae reveals the protective efficacy of carriage in healthy adults. Am J Respir Crit Care Med. 2013;187:855–864. doi: 10.1164/rccm.201212-2277OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collaborators, G.B.D.L.R.I. Estimates of the global, regional, and national morbidity, mortality, and aetiologies of lower respiratory infections in 195 countries, 1990-2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet Infect Dis. 2018;18:1191–1210. doi: 10.1016/S1473-3099(18)30310-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wahl B, et al. Burden of Streptococcus pneumoniae and Haemophilus influenzae type b disease in children in the era of conjugate vaccines: global, regional, and national estimates for 2000-15. Lancet Glob Health. 2018;6:e744–e757. doi: 10.1016/S2214-109X(18)30247-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bentley SD, et al. Genetic analysis of the capsular biosynthetic locus from all 90 pneumococcal serotypes. PLoS Genet. 2006;2:e31. doi: 10.1371/journal.pgen.0020031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ganaie F, et al. A New Pneumococcal Capsule Type, 10D, is the 100th Serotype and Has a Large cps Fragment from an Oral Streptococcus. mBio. 2020;11 doi: 10.1128/mBio.00937-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abdullahi O, et al. Rates of acquisition and clearance of pneumococcal serotypes in the nasopharynges of children in Kilifi District, Kenya. J Infect Dis. 2012;206:1020–1029. doi: 10.1093/infdis/jis447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brueggemann AB, et al. Clonal relationships between invasive and carriage Streptococcus pneumoniae and serotype- and clone-specific differences in invasive disease potential. J Infect Dis. 2003;187:1424–1432. doi: 10.1086/374624. [DOI] [PubMed] [Google Scholar]
- 13.Inverarity DJ, Diggle MA. Observations regarding historical accounts of pneumococcal diseases due to serotypes 1 and 3. J R Coll Physicians Edinb. 2010;40:354–361. doi: 10.4997/JRCPE.2010.408. [DOI] [PubMed] [Google Scholar]
- 14.Johnson HL, et al. Systematic evaluation of serotypes causing invasive pneumococcal disease among children under five: the pneumococcal global serotype project. PLoS Med. 2010;7 doi: 10.1371/journal.pmed.1000348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burgos J, et al. The spectrum of pneumococcal empyema in adults in the early 21st century. Clin Infect Dis. 2011;53:254–261. doi: 10.1093/cid/cir354. [DOI] [PubMed] [Google Scholar]
- 16.Chaguza C, et al. Carriage Dynamics of Pneumococcal Serotypes in Naturally Colonized Infants in a Rural African Setting During the First Year of Life. Front Pediatr. 2020;8:587730. doi: 10.3389/fped.2020.587730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Almeida ST, et al. Dynamics of Pneumococcal Carriage in Adults: A New Look at an Old Paradigm. J Infect Dis. 2021;223:1590–1600. doi: 10.1093/infdis/jiaa558. [DOI] [PubMed] [Google Scholar]
- 18.Balsells E, et al. The relative invasive disease potential of Streptococcus pneumoniae among children after PCV introduction: A systematic review and meta-analysis. J Infect. 2018;77:368–378. doi: 10.1016/j.jinf.2018.06.004. [DOI] [PubMed] [Google Scholar]
- 19.Brueggemann AB, et al. Temporal and geographic stability of the serogroup-specific invasive disease potential of Streptococcus pneumoniae in children. J Infect Dis. 2004;190:1203–1211. doi: 10.1086/423820. [DOI] [PubMed] [Google Scholar]
- 20.Desmet S, et al. In-depth analysis of pneumococcal serotypes in Belgian children (2015-2018): Diversity, invasive disease potential, and antimicrobial susceptibility in carriage and disease. Vaccine. 2021;39:372–379. doi: 10.1016/j.vaccine.2020.11.044. [DOI] [PubMed] [Google Scholar]
- 21.Browall S, et al. Clinical manifestations of invasive pneumococcal disease by vaccine and non-vaccine types. Eur Respir J. 2014;44:1646–1657. doi: 10.1183/09031936.00080814. [DOI] [PubMed] [Google Scholar]
- 22.Massora S, et al. Invasive disease potential of Streptococcus pneumoniae serotypes before and after 10-valent pneumococcal conjugate vaccine introduction in a rural area, southern Mozambique. Vaccine. 2019;37:7470–7477. doi: 10.1016/j.vaccine.2019.09.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weinberger DM, et al. Association of serotype with risk of death due to pneumococcal pneumonia: a meta-analysis. Clin Infect Dis. 2010;51:692–699. doi: 10.1086/655828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pletz MW, et al. The paradox in pneumococcal serotypes: highly invasive does not mean highly lethal. Eur Respir J. 2010;36:712–713. doi: 10.1183/09031936.00041210. [DOI] [PubMed] [Google Scholar]
- 25.Dube FS, et al. Longitudinal characterization of nasopharyngeal colonization with Streptococcus pneumoniae in a South African birth cohort post 13-valent pneumococcal conjugate vaccine implementation. Sci Rep. 2018;8:12497. doi: 10.1038/s41598-018-30345-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weiser JN. The pneumococcus: why a commensal misbehaves. J Mol Med (Berl) 2010;88:97–102. doi: 10.1007/s00109-009-0557-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ritchie ND, et al. What is different about serotype 1 pneumococci? Future Microbiol. 2012;7:33–46. doi: 10.2217/fmb.11.146. [DOI] [PubMed] [Google Scholar]
- 28.Chaguza C, et al. Recombination in Streptococcus pneumoniae Lineages Increase with Carriage Duration and Size of the Polysaccharide Capsule. mBio. 2016;7 doi: 10.1128/mBio.01053-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leal J, et al. Physicochemical properties of mucus and their impact on transmucosal drug delivery. Int J Pharm. 2017;532:555–572. doi: 10.1016/j.ijpharm.2017.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Melin M, et al. Serotype-related variation in susceptibility to complement deposition and opsonophagocytosis among clinical isolates of Streptococcus pneumoniae. Infect Immun. 2010;78:5252–5261. doi: 10.1128/IAI.00739-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jacques LC, et al. Increased pathogenicity of pneumococcal serotype 1 is driven by rapid autolysis and release of pneumolysin. Nat Commun. 2020;11:1892. doi: 10.1038/s41467-020-15751-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Audshasai T, et al. Streptococcus pneumoniae rapidly translocates from the nasopharynx through the cribriform plate to invade and inflame the dura. bioRxiv. 2021:2021.2009.2028.462246. doi: 10.1101/2021.09.28.462246. [DOI] [Google Scholar]
- 33.Bricio-Moreno L, et al. Lower Density and Shorter Duration of Nasopharyngeal Carriage by Pneumococcal Serotype 1 (ST217) May Explain Its Increased Invasiveness over Other Serotypes. mBio. 2020;11 doi: 10.1128/mBio.00814-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mercat A, et al. An outbreak of pneumococcal pneumonia in two men’s shelters. Chest. 1991;99:147–151. doi: 10.1378/chest.99.1.147. [DOI] [PubMed] [Google Scholar]
- 35.Lai JY, et al. Surveillance of pneumococcal serotype 1 carriage during an outbreak of serotype 1 invasive pneumococcal disease in central Australia 2010-2012. BMC Infect Dis. 2013;13:409. doi: 10.1186/1471-2334-13-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gupta A, et al. Outbreak of Streptococcus pneumoniae serotype 1 pneumonia in a United Kingdom school. BMJ. 2008;337:a2964. doi: 10.1136/bmj.a2964. [DOI] [PubMed] [Google Scholar]
- 37.Miller RR, et al. Genomic Analysis of a Serotype 5 Streptococcus pneumoniae Outbreak in British Columbia, Canada, 2005-2009. Can J Infect Dis Med Microbiol. 2016;2016:5381871. doi: 10.1155/2016/5381871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stillman EG. Further Studies on the Epidemiology of Lobar Pneumonia. J Exp Med. 1917;26:513–535. doi: 10.1084/jem.26.4.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gratten M, et al. An outbreak of serotype 1 Streptococcus pneumoniae infection in central Australia. Med J Aust. 1993;158:340–342. doi: 10.5694/j.1326-5377.1993.tb121794.x. [DOI] [PubMed] [Google Scholar]
- 40.Kwambana-Adams BA, et al. An outbreak of pneumococcal meningitis among older children (>/=5 years) and adults after the implementation of an infant vaccination programme with the 13-valent pneumococcal conjugate vaccine in Ghana. BMC Infect Dis. 2016;16:575. doi: 10.1186/s12879-016-1914-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Coldiron ME, et al. Outbreak of Pneumococcal Meningitis, Paoua Subprefecture, Central African Republic, 2016-2017. Emerg Infect Dis. 2018;24:1720–1722. doi: 10.3201/eid2409.171058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jusot JF, et al. Airborne dust and high temperatures are risk factors for invasive bacterial disease. J Allergy Clin Immunol. 2017;139:977–986.:e972. doi: 10.1016/j.jaci.2016.04.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Strom A. An Epidemic of Croupous Pneumonia Caused by Pneumococcus Type I. The Journal of Infectious Diseases. 1932;50:430–436. [Google Scholar]
- 44.Smillie WG, et al. A Study of a Type I Pneumococcus Epidemic at the State Hospital at Worcester, Mass. Am J Public Health Nations Health. 1938;28:293–302. doi: 10.2105/ajph.28.3.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.DeMaria A, Jr, et al. An outbreak of type 1 pneumococcal pneumonia in a men’s shelter. JAMA. 1980;244:1446–1449. [PubMed] [Google Scholar]
- 46.Dagan R, et al. An outbreak of Streptococcus pneumoniae serotype 1 in a closed community in southern Israel. Clin Infect Dis. 2000;30:319–321. doi: 10.1086/313645. [DOI] [PubMed] [Google Scholar]
- 47.Leimkugel J, et al. An outbreak of serotype 1 Streptococcus pneumoniae meningitis in northern Ghana with features that are characteristic of Neisseria meningitidis meningitis epidemics. J Infect Dis. 2005;192:192–199. doi: 10.1086/431151. [DOI] [PubMed] [Google Scholar]
- 48.Ndiaye AA, et al. Impact of a mass immunization campaign to control an outbreak of severe respiratory infections in Nunavik, northern Canada. Int J Circumpolar Health. 2006;65:297–304. doi: 10.3402/ijch.v65i4.18120. [DOI] [PubMed] [Google Scholar]
- 49.Proulx JF, et al. Pneumonia epidemic caused by a virulent strain of Streptococcus pneumoniae serotype 1 in Nunavik, Quebec. Can Commun Dis Rep. 2002;28:129–131. [PubMed] [Google Scholar]
- 50.Le Hello S, et al. Invasive serotype 1 Streptococcus pneumoniae outbreaks in the South Pacific from 2000 to 2007. J Clin Microbiol. 2010;48:2968–2971. doi: 10.1128/JCM.01615-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mehiri-Zghal E, et al. Molecular epidemiology of a Streptococcus pneumoniae serotype 1 outbreak in a Tunisian jail. Diagn Microbiol Infect Dis. 2010;66:225–227. doi: 10.1016/j.diagmicrobio.2009.05.008. [DOI] [PubMed] [Google Scholar]
- 52.Yaro S, et al. Epidemiological and molecular characteristics of a highly lethal pneumococcal meningitis epidemic in Burkina Faso. Clin Infect Dis. 2006;43:693–700. doi: 10.1086/506940. [DOI] [PubMed] [Google Scholar]
- 53.Mackenzie GA, et al. Epidemiology of nasopharyngeal carriage of respiratory bacterial pathogens in children and adults: cross-sectional surveys in a population with high rates of pneumococcal disease. BMC Infect Dis. 2010;10:304. doi: 10.1186/1471-2334-10-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Antonio M, et al. Seasonality and outbreak of a predominant Streptococcus pneumoniae serotype 1 clone from The Gambia: expansion of ST217 hypervirulent clonal complex in West Africa. BMC Microbiol. 2008;8:198. doi: 10.1186/1471-2180-8-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Van Mens SP, et al. Increased incidence of serotype-1 invasive pneumococcal disease in young female adults in The Netherlands. Epidemiol Infect. 2014;142:1996–1999. doi: 10.1017/S0950268813002860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cook HM, et al. An outbreak of serotype-1 sequence type 306 invasive pneumococcal disease in an Australian Indigenous population. Commun Dis Intell. 2020;44 doi: 10.33321/cdi.2020.44.66. 2018. [DOI] [PubMed] [Google Scholar]
- 57.Hammitt LL, et al. Effect of ten-valent pneumococcal conjugate vaccine on invasive pneumococcal disease and nasopharyngeal carriage in Kenya: a longitudinal surveillance study. Lancet. 2019;393:2146–2154. doi: 10.1016/S0140-6736(18)33005-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bar-Zeev N, et al. Impact and effectiveness of 13-valent pneumococcal conjugate vaccine on population incidence of vaccine and non-vaccine serotype invasive pneumococcal disease in Blantyre, Malawi, 2006-18: prospective observational time-series and case-control studies. Lancet Glob Health. 2021;9:e989–e998. doi: 10.1016/S2214-109X(21)00165-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bozio CH, et al. Continued occurrence of serotype 1 pneumococcal meningitis in two regions located in the meningitis belt in Ghana five years after introduction of 13-valent pneumococcal conjugate vaccine. PLoS One. 2018;13:e0203205. doi: 10.1371/journal.pone.0203205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Enright MC, Spratt BG. A multilocus sequence typing scheme for Streptococcus pneumoniae: identification of clones associated with serious invasive disease. Microbiology (Reading) 1998;144(Pt 11):3049–3060. doi: 10.1099/00221287-144-11-3049. [DOI] [PubMed] [Google Scholar]
- 61.du Plessis M, et al. Phylogenetic Analysis of Invasive Serotype 1 Pneumococcus in South Africa, 1989 to 2013. J Clin Microbiol. 2016;54:1326–1334. doi: 10.1128/JCM.00055-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brueggemann AB, Spratt BG. Geographic distribution and clonal diversity of Streptococcus pneumoniae serotype 1 isolates. J Clin Microbiol. 2003;41:4966–4970. doi: 10.1128/JCM.41.11.4966-4970.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ebruke C, et al. Temporal changes in nasopharyngeal carriage of Streptococcus pneumoniae serotype 1 genotypes in healthy Gambians before and after the 7-valent pneumococcal conjugate vaccine. PeerJ. 2015;3:e903. doi: 10.7717/peerj.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kirkham LA, et al. Identification of invasive serotype 1 pneumococcal isolates that express nonhemolytic pneumolysin. J Clin Microbiol. 2006;44:151–159. doi: 10.1128/JCM.44.1.151-159.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chiou AC, et al. Molecular assessment of invasive Streptococcus pneumoniae serotype 1 in Brazil: evidence of clonal replacement. J Med Microbiol. 2008;57:839–844. doi: 10.1099/jmm.0.47612-0. [DOI] [PubMed] [Google Scholar]
- 66.Gladstone RA, et al. International genomic definition of pneumococcal lineages, to contextualise disease, antibiotic resistance and vaccine impact. EBioMedicine. 2019;43:338–346. doi: 10.1016/j.ebiom.2019.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lees JA, et al. Fast and flexible bacterial genomic epidemiology with PopPUNK. Genome Res. 2019;29:304–316. doi: 10.1101/gr.241455.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cornick JE, et al. Region-specific diversification of the highly virulent serotype 1 Streptococcus pneumoniae. Microb Genom. 2015;1:e000027. doi: 10.1099/mgen.0.000027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chaguza C, et al. Understanding pneumococcal serotype 1 biology through population genomic analysis. BMC Infect Dis. 2016;16:649. doi: 10.1186/s12879-016-1987-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Croucher NJ, et al. Rapid pneumococcal evolution in response to clinical interventions. Science. 2011;331:430–434. doi: 10.1126/science.1198545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Croucher NJ, et al. Selective and genetic constraints on pneumococcal serotype switching. PLoS Genet. 2015;11:e1005095. doi: 10.1371/journal.pgen.1005095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Golubchik T, et al. Pneumococcal genome sequencing tracks a vaccine escape variant formed through a multi-fragment recombination event. Nat Genet. 2012;44:352–355. doi: 10.1038/ng.1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Terra VS, et al. Constructing Mutants in Serotype 1 Streptococcus pneumoniae strain 519/43. J Vis Exp. 2020 doi: 10.3791/61594. [DOI] [PubMed] [Google Scholar]
- 74.Lessa FC, et al. Streptococcus mitis Expressing Pneumococcal Serotype 1 Capsule. Sci Rep. 2018;8:17959. doi: 10.1038/s41598-018-35921-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chaguza C, et al. Population genetic structure, antibiotic resistance, capsule switching and evolution of invasive pneumococci before conjugate vaccination in Malawi. Vaccine. 2017;35:4594–4602. doi: 10.1016/j.vaccine.2017.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Young BC, et al. Panton-Valentine leucocidin is the key determinant of Staphylococcus aureus pyomyositis in a bacterial GWAS. Elife. 2019;8 doi: 10.7554/eLife.42486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cornick JE, et al. The global distribution and diversity of protein vaccine candidate antigens in the highly virulent Streptococcus pnuemoniae serotype 1. Vaccine. 2017;35:972–980. doi: 10.1016/j.vaccine.2016.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chewapreecha C, et al. Comprehensive identification of single nucleotide polymorphisms associated with beta-lactam resistance within pneumococcal mosaic genes. PLoS Genet. 2014;10:e1004547. doi: 10.1371/journal.pgen.1004547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chaguza C, et al. Bacterial genome-wide association study of hyper-virulent pneumococcal serotype 1 identifies genetic variation associated with neurotropism. Commun Biol. 2020;3:559. doi: 10.1038/s42003-020-01290-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bricio-Moreno L, et al. Comparative Genomic Analysis and In Vivo Modeling of Streptococcus pneumoniae ST3081 and ST618 Isolates Reveal Key Genetic and Phenotypic Differences Contributing to Clonal Replacement of Serotype 1 in The Gambia. J Infect Dis. 2017;216:1318–1327. doi: 10.1093/infdis/jix472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Valente C, et al. The blp Locus of Streptococcus pneumoniae Plays a Limited Role in the Selection of Strains That Can Cocolonize the Human Nasopharynx. Appl Environ Microbiol. 2016;82:5206–5215. doi: 10.1128/AEM.01048-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shen P, et al. Pneumococcal quorum sensing drives an asymmetric owner-intruder competitive strategy during carriage via the competence regulon. Nat Microbiol. 2019;4:198–208. doi: 10.1038/s41564-018-0314-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cobey S, Lipsitch M. Niche and neutral effects of acquired immunity permit coexistence of pneumococcal serotypes. Science. 2012;335:1376–1380. doi: 10.1126/science.1215947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Swarthout TD, et al. High residual carriage of vaccine-serotype Streptococcus pneumoniae after introduction of pneumococcal conjugate vaccine in Malawi. Nat Commun. 2020;11:2222. doi: 10.1038/s41467-020-15786-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Connor V, et al. Hands are vehicles for transmission of Streptococcus pneumoniae in novel controlled human infection study. Eur Respir J. 2018;52 doi: 10.1183/13993003.00599-2018. [DOI] [PubMed] [Google Scholar]
- 86.Wolter N, et al. High nasopharyngeal pneumococcal density, increased by viral coinfection, is associated with invasive pneumococcal pneumonia. J Infect Dis. 2014;210:1649–1657. doi: 10.1093/infdis/jiu326. [DOI] [PubMed] [Google Scholar]
- 87.Zafar MA, et al. Host-to-Host Transmission of Streptococcus pneumoniae Is Driven by Its Inflammatory Toxin, Pneumolysin. Cell Host Microbe. 2017;21:73–83. doi: 10.1016/j.chom.2016.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Verhagen LM, et al. Genome-wide identification of genes essential for the survival of Streptococcus pneumoniae in human saliva. PLoS One. 2014;9:e89541. doi: 10.1371/journal.pone.0089541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Weiser JN, et al. Streptococcus pneumoniae: transmission, colonization and invasion. Nat Rev Microbiol. 2018;16:355–367. doi: 10.1038/s41579-018-0001-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Andam CP, Hanage WP. Mechanisms of genome evolution of Streptococcus. Infect Genet Evol. 2015;33:334–342. doi: 10.1016/j.meegid.2014.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.McNeela EA, et al. Pneumolysin activates the NLRP3 inflammasome and promotes proinflammatory cytokines independently of TLR4. PLoS Pathog. 2010;6:e1001191. doi: 10.1371/journal.ppat.1001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zivich PN, et al. Streptococcus pneumoniae outbreaks and implications for transmission and control: a systematic review. Pneumonia (Nathan) 2018;10:11. doi: 10.1186/s41479-018-0055-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Feemster KA, et al. Risk of invasive pneumococcal disease varies by neighbourhood characteristics: implications for prevention policies. Epidemiol Infect. 2013;141:1679–1689. doi: 10.1017/S095026881200235X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Berry I, et al. Association of Influenza Activity and Environmental Conditions With the Risk of Invasive Pneumococcal Disease. JAMA Netw Open. 2020;3:e2010167. doi: 10.1001/jamanetworkopen.2020.10167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Shears RK, et al. Exposure to diesel exhaust particles increases susceptibility to invasive pneumococcal disease. J Allergy Clin Immunol. 2020;145:1272–1284.:e1276. doi: 10.1016/j.jaci.2019.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Miyashita L, et al. E-cigarette vapour enhances pneumococcal adherence to airway epithelial cells. Eur Respir J. 2018;51 doi: 10.1183/13993003.01592-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Panagiotou S, et al. Hypervirulent pneumococcal serotype 1 harbours two pneumolysin variants with differential haemolytic activity. Sci Rep. 2020;10:17313. doi: 10.1038/s41598-020-73454-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kadioglu A, et al. The role of Streptococcus pneumoniae virulence factors in host respiratory colonization and disease. Nat Rev Microbiol. 2008;6:288–301. doi: 10.1038/nrmicro1871. [DOI] [PubMed] [Google Scholar]
- 99.Hughes CE, et al. Development of primary invasive pneumococcal disease caused by serotype 1 pneumococci is driven by early increased type I interferon response in the lung. Infect Immun. 2014;82:3919–3926. doi: 10.1128/IAI.02067-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Fatykhova D, et al. Serotype 1 and 8 Pneumococci Evade Sensing by Inflammasomes in Human Lung Tissue. PLoS One. 2015;10:e0137108. doi: 10.1371/journal.pone.0137108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Harvey RM, et al. The impact of pneumolysin on the macrophage response to Streptococcus pneumoniae is strain-dependent. PLoS One. 2014;9:e103625. doi: 10.1371/journal.pone.0103625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Badgujar DC, et al. Structural insights into loss of function of a pore forming toxin and its role in pneumococcal adaptation to an intracellular lifestyle. PLoS Pathog. 2020;16:e1009016. doi: 10.1371/journal.ppat.1009016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Waight PA, et al. Effect of the 13-valent pneumococcal conjugate vaccine on invasive pneumococcal disease in England and Wales 4 years after its introduction: an observational cohort study. Lancet Infect Dis. 2015;15:629. doi: 10.1016/S1473-3099(15)00028-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bennett JC, et al. Changes in Invasive Pneumococcal Disease Caused by Streptococcus pneumoniae Serotype 1 Following Introduction of PCV10 and PCV13: Findings from the PSERENADE Project. Microorganisms. 2021;9 doi: 10.3390/microorganisms9040696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Gambia Pneumococcal Surveillance, G et al. Impact of the introduction of pneumococcal conjugate vaccination on invasive pneumococcal disease and pneumonia in The Gambia: 10 years of population-based surveillance. Lancet Infect Dis. 2021;21:1293–1302. doi: 10.1016/S1473-3099(20)30880-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.von Gottberg A, et al. Effects of vaccination on invasive pneumococcal disease in South Africa. N Engl J Med. 2014;371:1889–1899. doi: 10.1056/NEJMoa1401914. [DOI] [PubMed] [Google Scholar]
- 107.Kambire D, et al. Early impact of 13-valent pneumococcal conjugate vaccine on pneumococcal meningitis-Burkina Faso, 2014-2015. J Infect. 2018;76:270–279. doi: 10.1016/j.jinf.2017.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Horacio AN, et al. Serotype 3 Remains the Leading Cause of Invasive Pneumococcal Disease in Adults in Portugal (2012-2014) Despite Continued Reductions in Other 13-Valent Conjugate Vaccine Serotypes. Front Microbiol. 2016;7:1616. doi: 10.3389/fmicb.2016.01616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Nakano S, et al. Streptococcus pneumoniae Serotype 12F-CC4846 and Invasive Pneumococcal Disease after Introduction of 13-Valent Pneumococcal Conjugate Vaccine, Japan, 2015-2017. Emerg Infect Dis. 2020;26:2660–2668. doi: 10.3201/eid2611.200087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Williams TM, et al. Genome analysis of a highly virulent serotype 1 strain of Streptococcus pneumoniae from West Africa. PLoS One. 2012;7:e26742. doi: 10.1371/journal.pone.0026742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Betts M, et al. Complete Genome Sequence of Streptococcus pneumoniae Strain BVJ1JL, a Serotype 1 Carriage Isolate from Malawi. Microbiol Resour Announc. 2021;10:e0071521. doi: 10.1128/MRA.00715-21. [DOI] [PMC free article] [PubMed] [Google Scholar]