Abstract
Steric exclusion is a key element of enzyme substrate specificity, including in polymerases. Such substrate specificity restricts the enzymatic synthesis of 2’-modified nucleic acids, which are of interest in nucleic acid-based drug development. Here we describe the discovery of a two-residue, nascent strand, steric control “gate” in an archaeal DNA polymerase. We show that engineering of the gate to reduce steric bulk in the context of a previously-described RNA polymerase activity unlocks the synthesis of 2’-modified RNA oligomers, specifically the efficient synthesis of both defined and random-sequence 2’-O-methyl-RNA (2’OMe-RNA) and 2’-O-(2-methoxyethyl)-RNA (MOE-RNA) oligomers up to 750 nt. This enabled the discovery of RNA endonuclease catalysts entirely composed of 2’OMe-RNA (“2’OMezymes”) for the allele-specific cleavage of oncogenic KRAS (G12D) and β-catenin CTNNB1 (S33Y) mRNAs, and the elaboration of mixed 2’OMe- / MOE-RNA aptamers with high affinity for Vascular Endothelial Growth Factor. Our results open up these 2’-modified RNAs—used in several approved nucleic acid therapeutics—for enzymatic synthesis and a wider exploration in directed evolution and nanotechnology.
Introduction
Chemical variations to the canonical (deoxy)ribonucleic acid have gained great interest in the overlapping fields of medicinal chemistry and nucleic acid-based therapeutics (including RNA vaccines), as well as in the synthetic biology of nucleic acids and chemical biology. These modifications encompass a wide range of isomer substitutions, sugar alterations, sugar substituent modifications, nucleobase modifications, including—but not limited to—alteration of the glycosidic linkage, unnatural base-pairing interactions, and modified backbone chemistries1, 2. Among these, modifications to the 2’-hydroxy group of ribose have been a specific focus.
Such 2’ modifications have been shown to preserve key physicochemical principles of nucleic acid function, such as helical structure and base pairing specificity, while enhancing the biophysical and pharmacological properties of the modified nucleic acids, which has driven their widespread incorporation into nucleic acid therapeutics. Among these, 2’-fluoro (2’F), 2’-O-methyl (2’OMe), 2’-O-(2-methoxyethyl) (MOE), and 2’,4’-locked, -bridged, or -constrained (e.g. tricyclo) nucleic acids have been extensively studied3.
2’OMe is a naturally-occurring RNA modification found in human rRNA, tRNAs, small nuclear RNA (snRNA)4 as well as both the Cap- and body of human mRNA5 and is therefore both inherently biocompatible and unlikely to trigger the innate immune system. Indeed, 2’OMe modifications of viral RNAs appear to exploited by some viruses as self-signal enabling evasion of interferon-mediated antiviral re-sponses6.
The 2’OMe and the related MOE modifications (Fig. 1a, 4a) display a range of favourable physicochemical, pharmacological and immunological properties and their clinical utility has been validated in recently approved nucleic acid drugs such as the silencing RNA (siRNA) drugs Patisiran and Givosiran (2’OMe) and the antisense oligonucleotide (ASO) drugs Nusinersen (Spinraza), Inotersen (Tegsedi) and Volanesorsen (Waylivra) (all MOE)7. Furthermore, 2’OMe-RNA modification at purine bases were found to be beneficial in the FDA-approved aptamer drug Peg-aptanib (Macugen) for the treatment of age-related macular degeneration8.
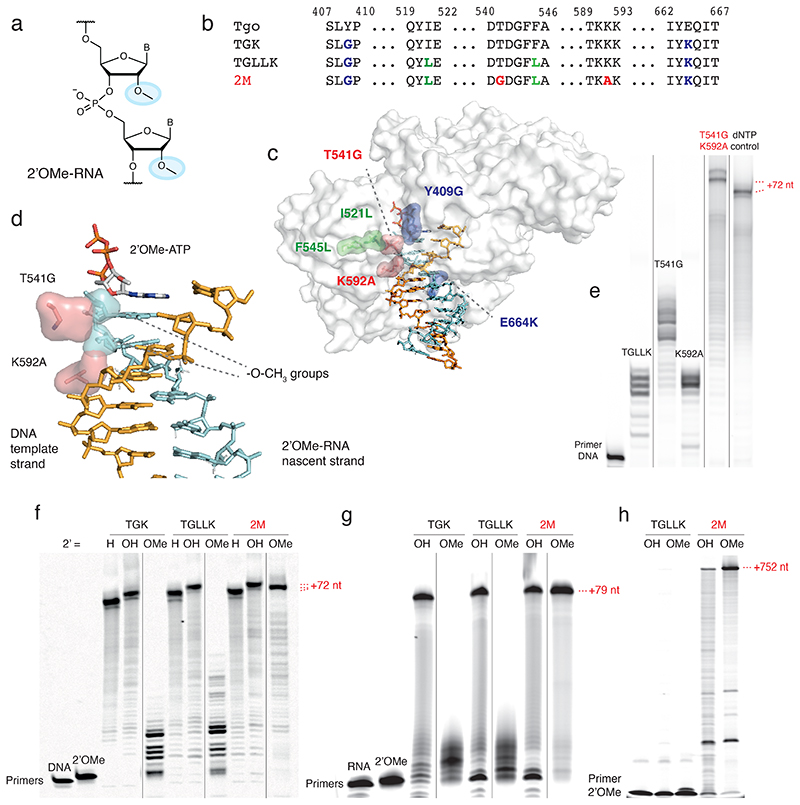
Fig. 1. The two-residue steric gate.
a) Chemical structure of 2’-O-methyl (2’OMe)-RNA. The 2-methoxy substituent is highlighted in cyan. b) Sequence alignment showing polymerases Tgo wildtype and engineered polymerases and respective key mutations in TGK (blue), TGLLK (green) and 2M (red). c) Space-filling model of the ternary structure of KOD DNA polymerase (PDB ID 5OMF) with respective mutations in TGK (blue), TGLLK (green) and 2M (red). d) Structural model of the active site of KOD DNA polymerase (PDB ID 5OMF) with DNA template strand (orange), active site 2’OMe-ATP (carbons shown in grey, nitrogens in blue, oxygens in red and phosphorus atoms in orange) and 2’OMe-RNA nascent strand (cyan) with 2’-methoxy groups of terminal 3’ and + 1 nucleotide shown as space-filling envelope. Key steric gate mutations (T541G, K592A) are displayed in pink (sticks) with wild-type side-chain residues shown as space-filling envelope highlighting the reduction in steric bulk. e) Denaturing PAGE of 2’OMe-RNA synthesis (DNA primer FD, template TempNpure, full length +72 nt) of steric gate single and double mutations. This experiment was performed once. Note the synergistic effect of T541G and K592A double mutation. Vertical black lines separate non-adjacent lanes from the same gel, see Source Data for full gel. f) Denaturing PAGE of DNA (H), RNA (OH) and 2’OMe-RNA (OMe) synthesis by TGK, TGLLK or 2M on defined-sequence template (DNA/2’OMe-RNA primer FD, template TempNpure, full length +72 nt). Similar, consistent results have been obtained twice as well as three times with 2M only. Vertical black lines separate non-adjacent lanes from the same gel, see Source Data for full gel. g) Denaturing PAGE of RNA (OH) and 2’OMe-RNA (OMe) synthesis by TGK, TGLLK or 2M on random N40 template (RNA/2’OMe-RNA primer A-Test2, template Tag3.3-N40-Test2, full length +79 nt), densitometry of N40 synthesis yield: TGLLK 2’OMe-RNA 0%, 2M 2’OMe-RNA 90% (SI Fig. 17). Similar, consistent results have been obtained twice. Vertical black lines separate non-adjacent lanes from the same gel, see Source Data for full gel. h) Denaturing PAGE of RNA (OH) and 2’OMe-RNA (OMe) long-range synthesis of a GFP transcript by TGLLK or 2M (2’OMe-RNA primer Synth-out1mm, template sfGFP, full length +752 nt). Similar, consistent results have been obtained three times (with 2M only).
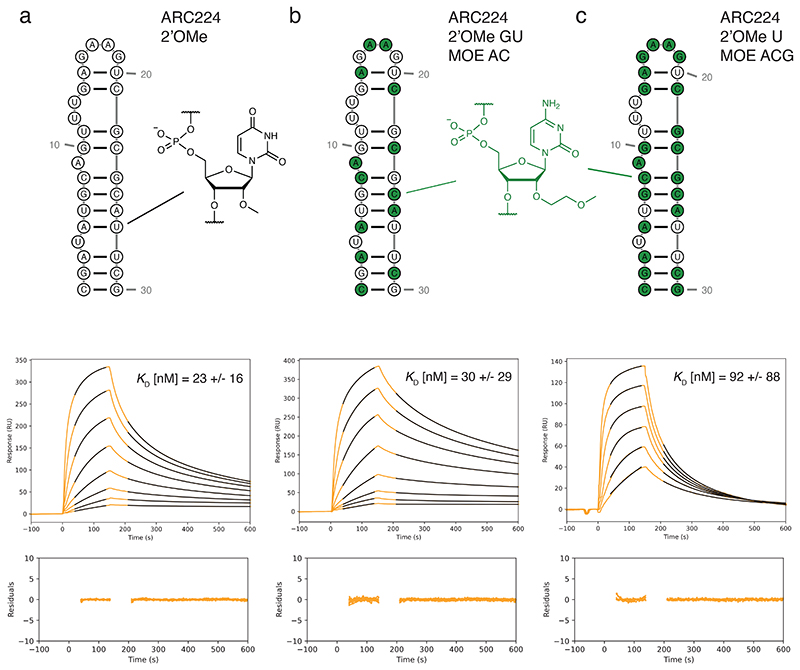
Fig. 4. 2’OMe/MOE-RNA aptamers and binding kinetics.
a-c) Sequence and secondary structure representation of anti-VEGF aptamer ARC224 12 (top panels) with respective SPR sensorgrams and average KD (middle) with residuals of the curves fit (bottom) for a) ARC224 2’OMe-GACU, b) ARC224 2’OMe-GU MOE-AC (MOE substitutions, green) and c) ARC224 2’OMe-U MOE-ACG (SPR binding kinetics: Supplementary Table 3). Two concentration series were measured for every aptamer. Note that ARC224 2’OMe-GACU and ARC224 2’OMe-GU MOE-AC showed virtually identical binding affinities to VEGF, whereas ARC224 2’OMe-U MOE-ACG bound VEGF with reduced affinity.
However, 2’OMe- and MOE-modified oligonucleotides are currently mainly synthesised via solid-phase phosphoramidite-based chemical synthesis, which is limited to short oligomers and a relatively small number of unique sequences and precludes their evolution. Thus, applicable sequences of 2’OMe- and MOE-modified oligonucleotides to be screened for a desired therapeutic effect have to be semi-rationally designed. This approach seems reasonable for ASO therapeutics designed to bind regulatory sequences on messenger RNA, but precludes the de novo discovery and development of aptamer and nucleic acid enzymes therapeutics in these important chemistries as well as hindering the development of nucleic acid nanotechnology objects and devices for both biotechnological and medical applications.
This has spurred the development of a range of engineered polymerases as tools for synthesis and reverse transcription of 2’OMe-RNA, including mutants of T7 RNA polymerase9, 10, 11, 12 or of the Stoffel fragment of Taq DNA polymerase13, which have enabled the discovery of partially as well as fully substituted 2’OMe-RNA aptamers12, 14. More recently, a mutant of KOD DNA polymerase has been described able to synthesize 1 kb 2’OMe-RNA fragments in the presence of Mn2+ ions and enabling the evolution of mixed LNAI2’OMe-RNA aptamers against Thrombin15.
Despite these advances, enzymatic synthesis of the bulkier MOE-RNA has not been described. Furthermore, due to the outstanding importance and potential of 2’OMe-RNA, tools for more efficient synthesis of longer or more complex 2’OMe-RNAs remain desirable. Here we describe progress towards these goals with the discovery of a two-residue steric gate in Tgo, the replicative DNA polymerase from the hyperthermophilic archaeon Thermococcus gorgonarius. Mutation of this steric gate in the context of an earlier engineered primer-dependent RNA polymerase activity in Tgo16, 17 enabled exceptionally efficient synthesis of 2’OMe-RNA as well as MOE-RNA, which had been as yet undescribed. This also allowed in vitro evolution of the first all-2’OMe-RNA catalysts (“2’OMezymes”) for mutation-specific cleavage of two oncogenic mRNA targets as well as the elaboration of mixed 2’OMe/OE-RNA aptamers with high affinity for Vascular Endothelial Growth Factor (VEGF).
Results
Elimination of steric hindrance improves 2’OMe-RNA synthesis
We had previously observed that engineered versions of Tgo, specifically TGK and TGLLK (Tgo: V93Q, D141A, E143A, Y409G, A485L, I521L, F545L, E664K)16, 17 (Fig. 1b) had a capacity for RNA, 2’F-DNA and to a lesser extent 2’OMe-RNA synthesis. However, 2’OMe-RNA synthesis by TGLLK was comparatively inefficient, especially on the more challenging N40 random-sequence templates often used in in vitro selection experiments. We sought to improve 2’OMe-RNA synthesis by quasi-rational design based on systematic elimination of unfavourable steric contacts between the bulky 2’-methoxy substituents of the 2’OMe-RNA nascent strand and the polymerase, using a simple static model of 2’OMe-RNA synthesis comprising the ternary structure of the homologous DNA polymerase from T. ko-dakarensis KOD1 (PDB ID 5OMF)18, and the structure of an RNA-DNA duplex19 augmented with 2’-O-methyl groups adjusted to C1’-C2’-O2’–CMethyl dihedral angles of 71° 20, 21 (gauche conformation).
This approach identified the sidechains of Tgo residues D540, T541, K592, D614, and E664 as proximal and potentially sterically clashing with 2’-methoxy groups in the 2’OMe-RNA nascent strand. These residues were targeted for site-saturation mutagenesis in the TGLLK framework and screened for 2’OMe-RNA synthesis activity (SI Fig. 1). Among these, T541 was of particular interest as it makes direct contact with the 3’-end nucleotide of the nascent (primer) strand, the positioning of which is crucial for catalysis, i.e., the nucleophilic attack of the nascent strand terminal 3’-OH on the α-phosphate of the incoming nucleoside triphosphate substrate. Indeed, the screen identified T541G as a mutation that increased 2’OMe-RNA synthesis activity, as well as mutations K592A and K664R, which led to slight increases in activity. Combining mutations revealed striking synergy of the T541G and K592A mutations for 2’OMe-RNA synthesis in the context of the previous TGLLK mutations (SI Fig. 1, Fig. 1e).
Polymerase TGLLK: T541G, K592A (henceforth named 2M) (Fig. 1) showed a striking increase in 2’OMe-RNA synthesis activity on a model DNA template containing all possible dinucleotide combinations (TempN)22 (Fig. 1f) as well as on a random-sequence N40 template (Fig. 1g). Furthermore, 2M enabled long-range 750 nt 2’OMe-RNA synthesis (Fig. 1h). This suggests that residues T541 and K592 together pose a strong block to 2’OMe-RNA synthesis, which is relieved by mutation to less bulky side-chains (T541G, K592A) (Fig. 1d). The 2M mutations also appear to reshape the polymerase primer-binding interface to the extent that 2’OMe-RNA synthesis is disfavoured from a DNA primer compared to a 2’OMe-RNA primer (SI Fig. 1). Nevertheless, these mutations do not seem to impede nu-cleobase discrimination as fidelity measurements suggest that the error rate of 2M synthesizing 2’OMe-RNA is in the same range as its parent polymerases TGLLK16 and TGK17 synthesizing 2’OMe-RNA and RNA, respectively (SI Table 4).
In vitro evolution of endonuclease 2’OMezymes
Poor efficiency of XNA synthesis and reverse transcription from random templates can cause synthetic biases and undersampling of the sequence space with concomitant loss of library diversity, which leads to suboptimal outcomes in repertoire selection experiments. We reasoned that the enhanced efficiency of 2’OMe-RNA synthesis by 2M (together with the recently described more efficient 2’OMe-RNA reverse transcriptase C823) might allow success in previously intractable in vitro evolution experiments. To this end, we pursued de novo selection of fully-2’OMe-RNA catalysts (henceforth called 2’OMezymes), which to our knowledge had not previously been described. Starting directly from random-sequence fully-2’OMe-RNA (N40) repertoires with RNA substrates covalently attached for cleavage in cis24, we sought to discover endonuclease 2’OMezymes targeted to the KRAS oncogene mRNA. After 15 rounds, the selection pool was deep sequenced, screened for RNA endonuclease activity, and the most abundant active sequence subjected to another five rounds of catalytic ‘maturation‘ selection from a doped sequence library (70% correct base, 10% each of the alternative bases). The most enriched 2’OMezyme sequence R15/5-KRAS (henceforth called R15/5-K) (Fig. 2a) was prepared by solid-phase synthesis for further characterization.
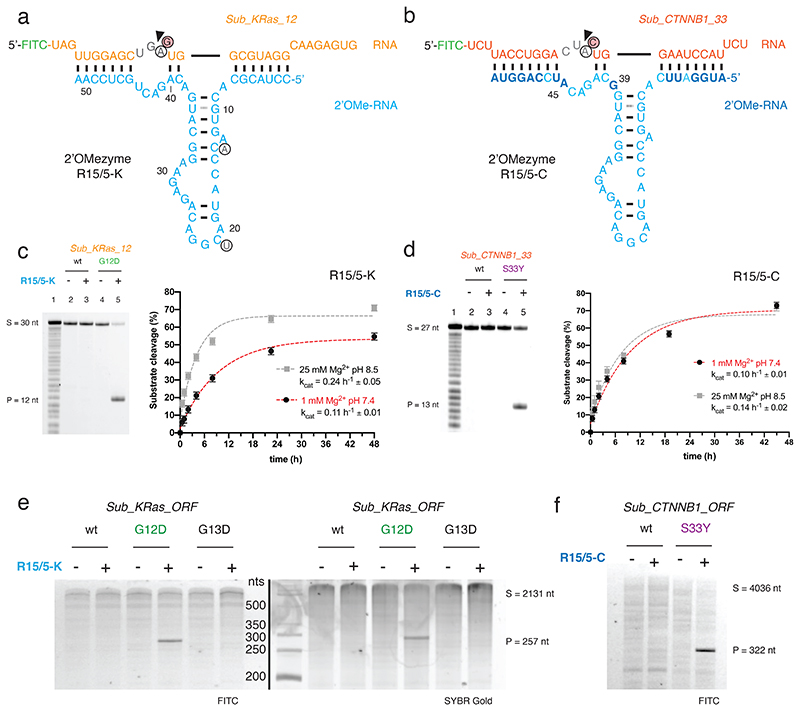
Fig. 2. Site-specific RNA endonuclease catalysts composed of 2’OMe-RNA.
a) Sequence and putative secondary structure of 2’OMezyme R15/5-K selected to target RNA “Sub_KRas_l2” [G12D] (residues 213-242 of the human KRAS mRNA bearing the c.35G>A (G12D) mutation) and b) variant 2’OMezyme R15/5-C re-targeted to an alternative RNA “Sub_CTNNB1_33” (residues 85-111 of the human CTNNB1 mRNA bearing the c.98C>A (S33Y) mutation). 2’OMe-RNA nucleotides are shown in cyan or blue (residues changes from R15/5-K to R15/5-C), RNA substrates in orange (KRAS) or red (CTNNB1). Black arrow denotes RNA cleavage site. Circled residues show bases in the “R15_1” parent 2’OMezyme changed during reselection. (below) c, d) (left panel) Urea-PAGE gels show 2’OMezymes (5 μM) performing allele-specific cleavage of substrate RNAs (1 μM) Sub_KRas_12 and Sub_CTNNB1 33 in a bimolecular reaction in trans under quasi-physiological conditions (37 °C, pH 7.4, 1 mM Mg2+, 17.5 h). Lane 1 shows partially hydrolyzed RNA substrate. (right panel) Graphs show pre-steady state single turnover reactions with substrate RNAs (1 μM), 2’OMezyme (5 μM) and reaction conditions indicated, at 37 °C. Error bars show standard error of the mean (s.e.m.) of three independent replicates. e, f) Reactions between (5 μM) 2’OMezyme and (0.5 μM) synthetic RNA transcripts of e) KRAS (“Sub_KRas_ORF”) and f) CTNNB1 (“Sub_CTNNB1_ORF”) bearing mutations as indicated, under quasi-physiological conditions (37 °C, pH 7.4, 1 mM Mg2+, 65 h). Similar, consistent results have been obtained once for e), whereas experiment f) was performed once.
R15/5-K is a highly sequence-specific RNA endonuclease that catalyzes cleavage of its cognate substrate, the KRAS G12D (cG35A) RNA, in a bimolecular reaction (kcat = 0.24 h-1 ± 0.05 in 25 mM Mg2+, pH 8.5, 37 °C) (Fig. 2c), and is capable of multiple-turnover catalysis (SI Fig. 2).
Cleavage is G12D (c.35G>A) mutation-specific with essentially no cleavage of ‘wild-type’ (wt) KRAS RNA, which differs by only one nucleotide (G35) (Fig. 2c). Furthermore, unlike comparable variants of the canonical 10-23 DNAzyme targeting the same KRAS sequence motif, R15/5-K was able to invade and cleave not just short model RNA substrates, but a long, structured 2.1 kb KRAS transcript, retaining its specificity for the G12D mutation (c.35G>A) (Fig. 2e), with virtually no cleavage of the wt KRAS transcript or a transcript with a similar nearby oncogenic mutation (G13D (c.38G>A)).
As observed previously in RNA endonuclease DNA- and XNAzymes (and some ribozymes), cleavage proceeds through transesterification and a 2’,3’-cyclic phosphate (>p) intermediate as shown by MALDI-ToF mass spectrometry and electro-phoretic mobility shift (EMSA) analysis of cleavage products (SI Fig. 3). However, while RNA endonuclease DNA- and XNAzymes are obligatory metalloenzymes, dependent on the presence of divalent cations (typically Mg2+) for both folding and catalysis, and therefore exhibit a substantial loss in catalytic activity under physiological conditions, the R15/5-K 2’OMezyme retained 70-80% activity under a qua-si-physiological low-Mg2+ regime (0.5–1 mM Mg2+) (Fig. 2c) over a broad pH range (SI Fig. 2). Indeed, even the single-turnover rate was only reduced by approximately 50% compared with optimal conditions (kcat = 0.11 h-1 ± 0.01 in 1 mM Mg2+, pH 7.4, 37 °C) (Fig. 2c). Furthermore, unlike the 10-23 DNAzyme, RNA cleavage activity of R15/5-K could even be observed in the absence of Mg2+, albeit at a very low rate (kcat = 0.001 h-1 ± 0.0002 in 5 mM EDTA, pH 7.4, 37 °C) (SI Fig. 2). Finally, as expected due to its all-2’OMe-RNA makeup, R15/5-K proved highly biostable with no significant degradation (or loss in activity) after incubation in human serum at 37 °C for 120 h (SI Fig. 4).
The potential for modularity, i.e. programmability of RNA target specificity through their binding arms, is an attractive feature of some nucleic acid catalysts like the 10-23 DNAzyme, but is not shared by all. We next explored whether the R15/5-K 2’OMezyme could be retargeted to an alternative mRNA substrate. Based on the putative secondary structure of R15/5-K (Fig. 2a) we reprogrammed nucleotides 1-7, 39-40, 45-51, flanking the central hairpin motif, to pair to the β-catenin (CTNNB1) proto-oncogene mRNA (c.85-111). The resulting 2’OMezyme R15/5-CTNNB1 was only weakly active, but an improved variant (R15/5-CTNNB1: A39G, U45A, henceforth called R15/5-C) (Fig. 2b) was readily discovered by screening mutations of residues flanking the recognition elements (position 9, 39, 42 & 45) (SI Fig. 5). The improved 2’OMezyme R15/5-C was highly specific and only able to cleave the oncogenic S33Y CTNNB1 (c.G99A) RNA substrate (Fig. 2d). It retained the capability for multi-turnover catalysis (SI Fig. 2) and invasion of long (4 kb), structured complete β-catenin transcript, while retaining its specificity (Fig. 2f). Although the R15/5-C turnover rate was ~40% lower compared to the parent R15/5-K under optimal conditions (kcat = 0.14 h-1 ± 0.02 in 25 mM Mg2+, pH 8.5, 37 °C), retargeting did not affect the rate under quasi-physiological low-Mg2+ conditions (kcat = 0.10 h-1 ± 0.01 in 1 mM Mg2+, pH 7.4, 37 °C) (Fig. 2d).
Enzymatic synthesis of MOE-RNA oligomers
Next we wondered if the 2M_polymerase would also be able to cope with more challenging 2’-modified RNA substrates. Among these, the 2’-O-(2-methoxyethyl) (MOE) modification (Fig. 3a) is of special interest because of the superior biophysical and pharmacological properties of the MOE-modified nucleic acid. In both 2’OMe- and MOE-RNA, the 2’-substituents favour a C3’-endo sugar conformation of the ribofuranose ring (akin to the ribose sugar puckering in RNA (A-form)) (Fig. 3b). The MOE ethylene glycol monomethyl ether modification is favoured in an extra gauche orientation along O2–C–C–O (Fig. 3c), extending the gauche effect from O4–C1–C2-O2 and thereby driving the rotational equilibrium to C3’-endo (Fig. 3b)25. This structural pre-organization (and rigidity of the MOE-RNA structure) enhances base-pairing and stacking interactions with target RNA and leads to a high antisense binding affinity of 2’OMe- and MOE-RNA to RNA. Indeed, every single MOE modification in a DNA oligo increases the Tm of the oligo bound to its complementary RNA by 0.9–1.2 °C25.
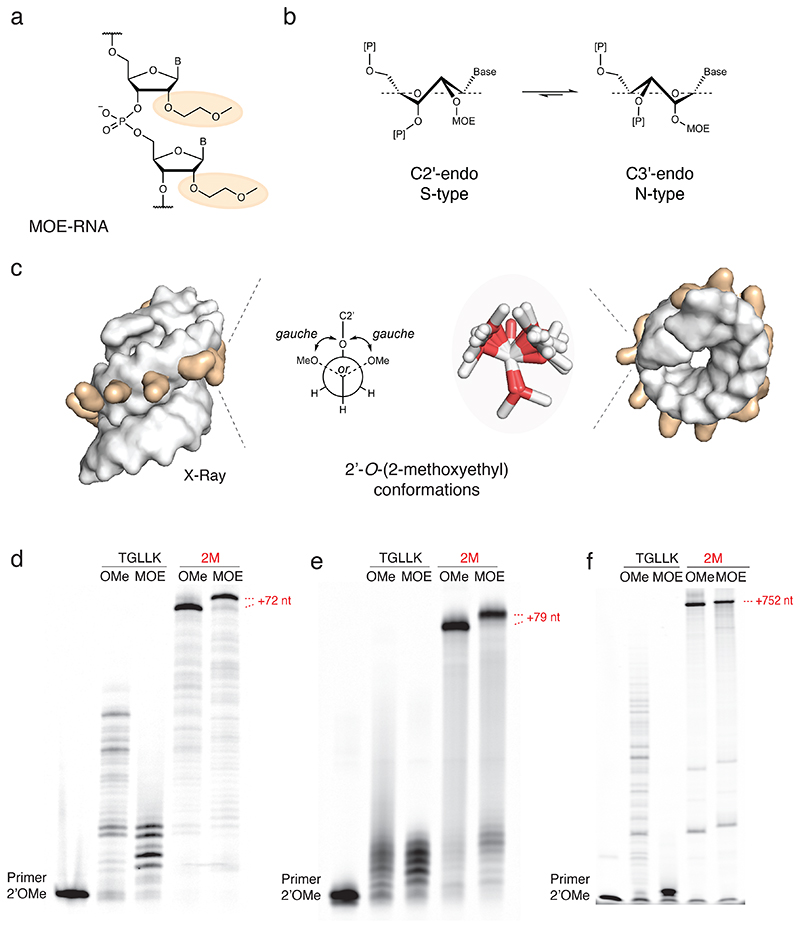
Fig. 3. MOE-RNA synthesis.
a) Chemical structure of 2’-O-(2-methoxyethyl)-RNA (MOE-RNA) with the 2’-O-(2-methoxyethyl) group highlighted (orange). b) Equilibrium of the ribose sugar puckering. The 2’-O-MOE modification shifts the equilibrium towards the C3’-endo (N-type) conformation, comparable to RNA. c) Space-filling representation of the X-ray structure of an MOE-RNA duplex (PDB ID 468D) viewed side (left) and top view (right) with 2’-O-(2-methoxyethyl) groups (orange) and overlay of observed 2’-O-(2-methoxyethyl) conformations (stick representation, middle) next to a Newman projection of the ethylene glycol monomethyl ether, which preferentially adopts a gauche conformation respective to the two oxygen atoms. d) Denaturing PAGE of 2’OMe-RNA (OMe) and MOE-RNA (MOE) synthesis by TGLLK or 2M on defined-sequence template (2’OMe-RNA primer FD, template TempNpure, full length +72 nt). Similar, consistent results have been obtained three times as well as five times with 2M only. e) Denaturing PAGE of 2’OMe-RNA (OMe) and MOE-RNA (MOE) synthesis by TGLLK or 2M on random N40 template (2’OMe-RNA primer A-Test2, template Tag3.3-N40-Test2, full length +79 nt), densitometry of N40 synthesis yield: TGLLK 2’OMe-RNA 1%, TGLLK MOE-RNA 0%, 2M 2’OMe-RNA 84%, 2M MOE-RNA 65% (SI Fig. 17). Similar, consistent results have been obtained five times as well as three times with 2M only. f) Denaturing PAGE of 2’OMe-RNA (OMe) and MOE-RNA (MOE) long-range synthesis of a GFP transcript by TGLLK or 2M (2’OMe-RNA primer Synth-outlmm, template sfGFP, full length +752 nt). Similar, consistent results have been obtained three times (with 2M only).
In addition, the gauche-oriented MOE moiety places an additional hydrogen bond acceptor in the minor groove, which favours the formation of a hydrogen bonding network. Thereby, the MOE modifications lead to stabilization of up to three water molecules trapped between the MOE moiety and the phosphodiester backbone26. This hydration “spine” together with steric hindrance introduced by the 2’-O-(2-methoxyethyl) group in the minor groove leads to shielding of the 5’-3’ phosphodiester linkage, resulting in exceptional biostability and in vivo half-life of MOE-RNA3, and the excessive hydration increases paracellular absorption and intestinal uptake rate of MOE-modified oligonucleotides compared to unmodified oligos27.
However, solution-state NMR28 and X-ray crystallography26 structures indicate a challenging steric envelope of the MOE-RNA helix for enzymatic synthesis with the bulky methoxyethyl groups, adopting the aforementioned gauche conformation and projecting away from the helical envelope (Fig. 3c). Nevertheless, we under-took chemical synthesis of MOE-NTPs to explore enzymatic MOE-RNA synthesis. Synthesis of the MOE-nucleosides29 and their phosphoramidites30 is established and commercial synthesis of MOE-oligonucleotides is available, but the 2’-O-(2-methoxyethyl)nucleoside triphosphates (MOE-NTPs) were neither commercially available nor was their synthesis established. We therefore first developed a synthetic route to the four MOE-NTPs starting from the commercially available 2’-O-(2-methoxyethyl)ribonucleosides by triphosphorylation based on the established Ludwig method31, 32 (SI Fig. 6, SI Materials & Methods).
Having synthesized all four MOE-NTPs (MOE-ATP, MOE-GTP, MOE-CTP, MOE-m5UTP), we proceeded to test the new engineered polymerase 2M for its ability to synthesize MOE-RNA oligomers. Unlike its predecessor TGLLK, 2M (SI Fig. 7)) was able to efficiently synthesize MOE-RNA on both a model DNA template (+72 nt) and a random N40 library template, and it was capable of long-range MOE-RNA synthesis of 750 nt oligomers (Fig. 3def, SI Fig. 7). The incorporation of the bulkier methoxyethyl substituents at full substitution resulted in an appreciable shift in electrophoretic mobility of MOE-oligomers compared to DNA or 2’OMe-RNA oligomers of the same length and sequence (SI Fig. 8).
Mixed 2’OMe- / MOE-RNA aptamers
MOE would be an attractive medicinal chemistry modification of RNA, 2’F-DNA or 2’OMe-RNA aptamers to modulate pharmacological properties and/or increase potency. Indeed, MOE-RNA and 2’OMe-RNA have similar conformational and helical preferences and similar base-pairing strength28, 33. On the other hand, 2’-O-(2-methoxyethyl) groups present a significantly larger steric envelope (Fig. 3c), which might lead to steric conflicts with other groups in tightly folded structures. Nevertheless, it seemed plausible that functional mixed 2’OMe/MOE-RNA aptamers could be elaborated from previously described all-2’OMe-RNA leads. To test this, we examined conversion of a well-characterized all-2’OMe-RNA aptamer against Vascular Endothelial Growth Factor (VEGF)12 to all-MOE-RNA or mixed 2’OMe/MOE-RNA aptamers and tested their respective binding activity by surface plasmon resonance (SPR). SPR revealed that while the aptamer in which two out of four 2’OMe-nucleotides were substituted with MOE-nucleotides showed virtually identical affinities to VEGF compared to the all-2’OMe-RNA aptamer, the aptamer in which three of the 2’OMe-nucleotides were replaced by MOE-nucleotides still bound VEGF, albeit with reduced affinity (Fig. 4, SI Table 3).
The all-MOE aptamer seemed to have lost virtually all of its binding activity (SI Fig. 9), perhaps in part due to the use of MOE-m5UTP, whereas the original VEGF aptamer had been evolved using 2’OMe-U. Indeed, when we replaced 2’OMe-U with 2’OMe-m5U in the original aptamer, its binding affinity was reduced (SI Fig. 9). The 2’OMe/MOE-RNA aptamers described here are mixed-chemistry aptamers, which have not previously been elaborated in such backbones. These findings suggest that MOE-modified nucleic acids are capable of folding into tight three-dimensional structures with high affinity for their protein target.
Discussion
Steric exclusion is a common determinant of enzyme and in particular polymerase specificity. This includes the “steric gate” residue found in the active site of most DNA polymerases thought to have evolved to exclude ribonucleoside triphosphates (present at much higher concentrations in the cell) from the polymerase active site in order to limit RNA incorporation into the genome. Kool and coworkers have shown that this may be a general mechanism of steric control of nucleobase pair dimension in the active site as an important component in replicative polymerase fidelity mechanisms34. Steric factors are also likely implicated in post-synthetic inhibition of nascent strand extension upon incorporation of mismatches35 or non-cognate nucleotides36 either through direct clashes with the nascent strand polymerase interface or by altering conformational equilibria of the nascent duplex. Finally, relaxation of steric control is a successful strategy for polymerase engineering, for example in the 9°N DNA polymerase variants engineered for incorporation of bulky 3’-substituents in Illumina next generation sequencing37 or in engineering DNA polymerases for RNA synthesis or reverse transcription17, 23
We had previously discovered key mutations in the polB family polymerase from T. gorgonarius that, in addition to the steric gate mutation (Y409G), enable efficient RNA synthesis (E664K)17 and incorporation of non-cognate 2’-5‘ linkages (I521L, F545L)16. The latter polymerase variant (named TGLLK) showed an increased, but still inefficient ability of 2’OMe-RNA synthesis, suggesting that aspects of the polymerase structure were still poorly adapted to 2’OMe-RNA synthesis. As RNA and 2’OMe-RNA share very similar conformational preferences, we suspected steric factors. Indeed, systematic evaluation of potential steric clashes of the polymerase with 2’-methoxy groups in the nascent strand identified a two-residue steric gate, mutation of which to less bulky side-chains (T541G, K592A) led to a dramatic increase in 2’OMe-RNA synthesis efficiency (Fig. 1) and enabled efficient MOE-RNA synthesis (Fig. 3) with full-length defined or random sequence (N40) products synthesized in <30 min (2’OMe-RNA, < 10 min) (SI Fig. 7) despite the considerably larger steric envelope of the 2’-O-(2-methoxyethyl) group of MOE-RNA. Incorporating T541G and K592A into TGLLK led to an increase in N40 synthesis yield as determined by densitometry from 1% to 90% (2’OMe-RNA) and from 0% to 65% (MOE-RNA, Figs. 1g and 3e, SI Fig. 17).
Both T541 and K592 are part of motifs (motif C38 and KxY39, respectively) that are very highly conserved both at the sequence and at the structural level (Fig. 5, SI Fig. 10) in polB polymerases of archaeal, eukaryotic, and even viral origin40. These motifs are thought to be part of a minor groove interaction motif that is involved in mismatch sensing41 and previous mutation to bulky, hydrophobic side-chains was shown to enhance mismatch discrimination42. Nevertheless, we find that fidelity of 2’OMe-RNA synthesis is essentially unaffected (SI Table 4) compared to parent polymerases TGK and TGLLK lacking these mutations16, 17. The fidelity of MOE synthesis is currently challenging to measure due to the poor efficiency of the available MOE-RNA RT23, but a dropout assay suggests specific processing of the correct MOE-NTPs (SI Fig. 11).
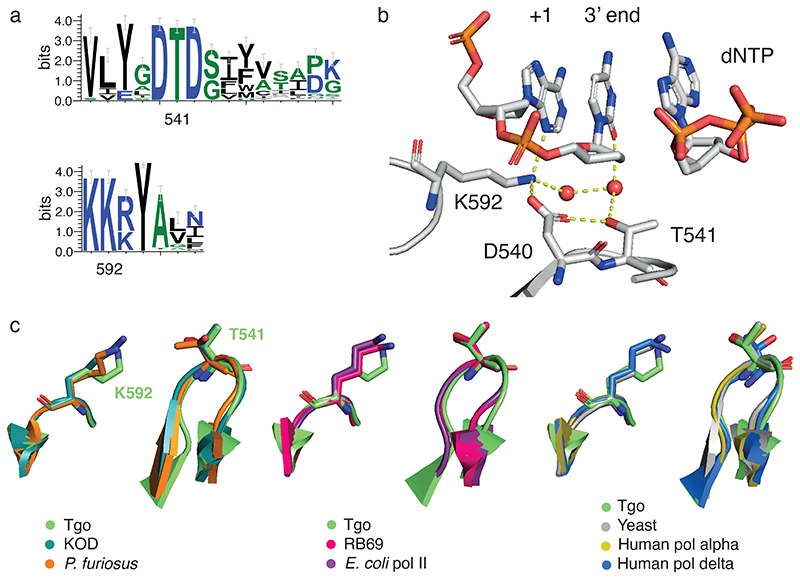
Fig. 5. Nascent strand steric gate and polymerase motifs.
a) Conserved sequence motifs in polB polymerase family showing sequence context and conservation of nascent strand steric gate in motif C (T541) and motif KxY (K592). b) Structural context with active site 2’OMe-ATP (KOD DNA polymerase (PDB ID 5OMF)) showing H-bonding network involving steric gate together with D540 as well as direct contact to +1 minor groove and indirect contact (via H2O) to 3’ end nucleotide (carbons shown in grey, nitrogens in blue, oxygens in red and phosphorus atoms in orange). c) Structural conservation of nascent strand steric gate across polB phylogeny from archaeal (left), bacterial (middle) to eukaryotic (right) polB polymerases.
According to the ternary complex structure of the closely related KOD polymerase18, both T541 and K592 are involved in H-bonding interactions with the nascent strand 3’ end (T541, via water) and +1 (K592) nucleobases, obstructing passage of 2’-modifications (Fig. 5b). Positive epistasis of the two mutations is in congruence with structural considerations. Relieving the steric block requires mutation of both, which yields a large free volume in this critical area proximal to the catalytic site and the nascent strand large enough to also accommodate the 2’-O-methyl groups of 2’OMe-RNA (Fig. 1) and the bulky 2’-O-(2-methoxyethyl) groups of MOE-RNA (Fig. 3).
A prediction of this structural model is that this two-residue steric gate of T541 and K592 mainly enhances the efficiency of the primer 3’-end extension rather than the nucleotide incorporation step of the polymerase catalytic cycle. Indeed, 2M single nucleotide incorporation steady-state kinetic parameters for ATP (from a 2’OMe-RNA primer) (SI Fig. 12) closely match that of the parent polymerase TGK (from an RNA primer)17. On the other hand, while the Vmax / kcat values for incorporation of ATP, 2’OMe-ATP and MOE-ATP are essentially identical, 2M has an approximately 5-fold improved KM value for both 2’OMe-ATP and MOE-ATP compared to ATP (SI Fig. 12) and compared to the parent polymerase TGK (KM = 13.3 μM for ATP)17. This may indicate that the steric gate improves the fit and positioning of 2’-modified nucleotide triphosphates into the polymerase active site, but does not accelerate the catalytic step.
While enzymatic MOE-RNA synthesis by a polymerase has not previously been described, a number of alternative engineering approaches to 2’OMe-RNA synthesis have been explored, including a variant of the closely related polB-family KOD polymerase (KOD: N210D / Y409G / A485L / D614N / E664K)15. While we find that 2’OMe-RNA synthesis by 2M is both more efficient (SI Fig. 14) and higher fidelity (SI Table 4) (not requiring forcing conditions such as Mn2+ ions), the DGLNK mutations represent an interesting alternative, non-steric strategy to enhance XNA-RNA synthesis. Starting from the same (or very similar) mutational background than 2M (including the Y409G active site steric gate, the E664K thumb subdomain mutation and the A485L “Therminator” mutation43, as well as a mutation (N210D) to inactivate the 3’-5’ exonuclease domain, DGLNK also comprises a critical D614N mutation in the thumb subdomain, which removes of a negative charge in proximity to the phosphodiester backbone of the nascent strand. This is highly reminiscent of the previously described Tgo: E664K mutation that was found to enable efficient RNA synthesis by expanding the positively charged polymerase interaction surface and enhancing affinity for the primertemplate duplex. While not demonstrated for DGLNK, it is plausible that the D614N mutation, which further reduces negative charge potential at the polymerase-nascent strand interface, also enhances affinity of the polymerase for the primer-template duplex. At the same time, our original model had identified D614 as a potential steric clash with the nascent strand methoxy groups, but our screen had not identified any strong positive effect on 2’OMe-RNA synthesis as an isolated mutation. Nevertheless, we re-examined the D614N mutation in the context of 2M (2MN; 2M: D614N) and found a small enhancement of 2’OMe-RNA and to a lesser extent MOE-RNA synthesis by 2MN (SI Fig. 14).
We also evaluated two other previously published polymerases, T7 RNA polymerase variant RGVG-M6 (T7: P266L, S430P, N433T, E593G, S633P, Y639V, V685A, H784G, F849I, F880Y)11 and Taq polymerase Stoffel fragment variant SFM4-6 (Taq SF: I614E, E615G, D655N, L657M, E681K, E742N, M747R)13, that had been reported to have 2’OMe-RNA synthesis activity. However, compared to 2M, the 2’OMe-RNA synthesis activity appeared to be modest in both cases and dependent on forcing conditions such as the presence of high concentrations of Mn2+ ions (SI Fig. 13).
Finally, as our initial screen also indicated that TGLLK: T541G, K664R (SI Fig. 1) also exhibited a (smaller) increase in 2’OMe-RNA synthesis efficiency compared to the single mutant T541G, we introduced K664R into the 2M polymerase, yielding TGLLK: T541G, K592A, K664R (henceforth named 3M). However, polymerases 2M and 3M exhibited virtually identical synthesis activity, full-length yield, and stalling pattern (SI Fig. 15).
Together with the discovery of a more efficient 2’OMe-RNA RT23, 2M has opened the door for more ambitious in vitro evolution experiments, including the discovery of the first 2’OMezymes. Unlike 2’OMe-RNA aptamers, no 2’OMezymes had previously been described, presumably due to the fact that catalysts generally appear to be more sparsely distributed in nucleic acid sequence space44, 45, 46. The RNA endonuclease 2’OMezymes R15/5-K and -C characterized herein differ in interesting ways from other RNA endonuclease DNA- and XNAzymes described. While highly specific, their maximal catalytic turnover is modest, possibly due to overly tight binding of the RNA substrate by 2’OMe-RNA, leading to product inhibition and/or a high proportion of 2’OMezymes trapped in non-catalytic conformations. However, unlike for example the canonical 10-23 DNAzyme, or some XNAzymes, 2’OMezymes retain much of their catalytic activity at low, physiologically relevant Mg2+ concentrations. This suggests that unlike the above, the 2’OMezymes are likely not obligate metalloenzymes, but may instead rely on acid-base catalysis akin to the classic hairpin ribozyme (Hpz). Intriguingly, the 2’OMezymes—despite lacking sequence homology—share some striking secondary structure and sequence segment similarities with the hairpin ribozyme47 (albeit with the hairpin and cleavage sites reversed) (SI Fig. 16). Like the Hpz, the 2’OMezymes also have the capacity to catalyze RNA ligation at low temperatures (SI Fig. 16) and exhibit activity in the absence of Mg2+ (SI Fig. 2). Consistent with this, mutations that increase the sequence identity with HPz are mostly benign (SI Fig. 16).
The 2M polymerase enables the templated enzymatic synthesis of MOE-RNA, which was previously elusive. MOE is a nucleic acid modification of great interest in nucleic acid therapeutics due to its unusual structural and pharmacological properties and extraordinary biostability, which have driven its application in FDA-approved ASO drugs7. This makes MOE a desirable medicinal chemistry modification of existing 2’OMe-RNA aptamers. In the case of an anti-VEGF 2’OMe-RNA aptamer12, chimeric versions in which two or three of the 2’OMe-nucleotides were replaced by MOE-nucleotides could be readily elaborated and showed identical or slightly reduced binding affinities for VEGF, respectively (Fig. 4), although full substitution of 2’OMe-with MOE-RNA abolished binding activity in this aptamer (SI Fig. 9).
In conclusion, our work underlines the importance of steric control in polymerase substrate specificity. Discovery of the two-residue nascent strand steric gate complements the classic active site steric gate in excluding 2’-modified nucleic acids from incorporation into the nascent strand and unlocks enzymatic synthesis of nucleic acid oligomers bearing bulky 2’-substituents. This has enabled the efficient synthesis and evolution of 2’OMezymes as well as MOE-RNA synthesis and elaboration of mixed 2’OMe- / MOE-RNA aptamers. We envisage a range of applications including the stereospecific synthesis of phosphorothioate (αPS)-MOE-RNA oligomers and the rapid iteration of variant aptamer and ASO sequences and chemistries towards enhanced potency.
Methods
Polymerase models and rational choice of mutagenesis sites
For construction of the mini-libraries introducing single mutants at specific polymerase residues, we used the ternary crystal structure of the closed form of Thermococcus kodakarensis KOD1 DNA polymerase in complex with a DNA primer–template duplex and an incoming dATP at the active site (PDB ID 5OMF)18, as this is a close B-family homologue of the Thermococcus gorgonarius polymerase mutants used in this study.
The crystal structure was loaded in Pymol and appropriate 2’-hydrogen atoms of primer nucleotides were manually replaced by oxygen atoms with Pymol’s “build” functionality. The hydrogen atoms on the newly introduced 2’-hydroxyl moieties were then replaced in the same manner by methyl groups. The added dihedral angles were adjusted manually to 71° (gauche conformation)20, 21. This model served as a structural guide to calculate distances from polymerase residues to the introduced primer 2’-O-methyl carbon atoms and identify sites of steric clashes. These were targeted for site-saturation mutagenesis to relieve the steric hindrance and increase polymerase processivity on 2’OMe-RNA.
Cloning of expression constructs and site-saturation mutagenesis
Inverse PCR (iPCR) was carried out using overlapping forward and reverse primers introducing a BsaI restriction site (see Supplementary Table 1) on pASK75 plasmid48 coding for Thermococcus gorgonarius (Tgo) polymerase mutant TGLLK (Tgo: V93Q, D141A, E143A, Y409G, A485L, I521L, F545L, E664K)16 as the parent plasmid. The cloning primers for site-saturation mutagenesis contained degenerate NNS codons (N for all bases, S for G and C) introducing mini-libraries of 32 codons coding for all 20 amino acids on a single residue (see Supplementary Table 1).
iPCR reactions were carried out with polymerase Q5 (New England Biolabs, NEB) with forward and reverse primers (0.5 μM each) and dNTPs (200 μM each) on 20 ng DNA template. The iPCR reactions were incubated in the thermocycler with the following programme: 98 °C, 30 s; 30 cycles of (98 °C, 10 s; 50-72 °C, 30 s; 72 °C, 3 min); 72 °C, 3 min. iPCR products were purified using the PCR Purification Kit (Qiagen). The products were restricted by BsaI and DpnI (NEB) and purified on an agarose gel if necessary. Products were ligated by T4 DNA ligase and purified by another clean-up kit (Bioline). The cloned constructs were transformed into chemically or electrocompetent E. coli 10-β cells (NEB) or E. coli BL21 CodonPlus-RIL cells (Agilent) and plated on TYE agar plates supplemented with the appropriate antibiotics.
Primer extension reactions
Analytical primer extension reactions were carried out in 1x Thermopol buffer (NEB) supplemented with MgSO4 (4 mM). Primer (100 nM) was extended on a template (200 nM) with appropriate nucleoside triphosphates (125–250 μM each) by purified polymerase (10–100 μg/mL) in a 10-μL reaction volume. Reactions were carried out at 65 °C. Primer extension products were analysed via urea-PAGE. All extensions with MOE-NTPs on defined-sequence template TempNpure required post-synthesis template capture with a ten-fold excess of antisense template, Turbo DNase (Invitrogen) treatment, subsequent Proteinase K (NEB) treatment, and loading on the urea-PAGE gel with a ten-fold excess of antisense template. Primer extensions with MOE-NTPs on template sfGFP required polymerase concentrations of 500 μg/mL.
Enzyme-linked oligonucleotide assay (ELONA) polymerase activity assay (PAA)
Site-saturation mutagenised polymerase mini-libraries were transformed in E. coli 10-β cells and plated on TYE agar plates supplemented with ampicillin. For every single mutant mini-library, 2x94 clones were manually picked from the agar plates and used to inoculate 2x94 liquid starting cultures of 1 mL 2xTY supplemented with ampicillin (100 μg/mL) in 96-deep well plates (Nunc) alongside two control wells per plate with parent polymerase TGLLK. The cultures were grown at 37 °C overnight. The next day, 100 μL of each culture was used to inoculate a new 1-mL culture on a new plate and the cultures were allowed to grow at 37 °C until they reached mid-log phase. Protein expression was then induced with anhydrotetracycline at 200 μg/L and carried out at 37 °C for 2 h. The cultures were stored at 4 °C overnight. The cells were harvested by centrifugation and then resuspended in 100 μL Thermopol buffer. The cells were transferred to a 200-μL 96-well plate and lysed at 75 °C for 30 min. Lysed cells were cooled in an ice-water bath and the lysates were cleared by centrifugation at 4 °C. The cleared lysates were transferred to a new 200-μL 96-well plate and stored at 4 °C.
Primer extension reactions were carried out in 1x Thermopol buffer (NEB) supplemented with MgSO4 (4 mM). Biotinylated primer FD (100 nM) was extended on template TempNpure (200 nM) with 2’-O-methylribonucleoside triphosphates (125 μM each) by polymerase mutants in whole-cell lysate in a 10-μL reaction volume. Reactions were carried out at 65 °C.
The biotinylated primer extension products were diluted in PBS supplemented with 0.1 % (v/v) Tween 20 (PBST) and bound on streptavidin-coated plates (Roche) for 1 h at room temperature. After every incubation step, the respective supernatant was discarded. Hybridised template was then removed by two 1-min denaturation steps with 0.1 M NaOH. After a neutralisation step with PBST, a digoxigenin-labelled oligonucleotide probe (DIGN25, 60 nM in PBST) was applied for 1 h, which hybridised to efficiently elongated primers only, exhibiting increasing affinity the longer the extension product was.
After three washing steps with PBST, an anti-digoxigenin antibody fragment bound to horseradish peroxidase (1:3,000 dilution in PBST, Roche) was bound on the plates for 1 h. After four PBST washes, the assay was developed by the addition of 3,3′,5,5′-tetramethylbenzidine (TMB, 1-Step Ultra TMB-ELISA, Thermo) and incubation until the blue colour formation was complete (judged by TGLLK control wells). The enzymatic reaction was stopped by the addition of 1 M H2SO4, which lead to a yellow colour switch. Absorbance was read on a plate reader at 450 nm.
Screen hits were mini-prepped and sequenced, and polymerase activity was verified with extension reactions of a fluorescently labelled primer FD as described above, where the amount of lysate added was adjusted by SDS-PAGE analysis and normalisation based on the polymerase band intensities. Primer extension products were analysed via urea-PAGE.
Expression and purification of polymerases
Polymerase expression and purification was essentially performed as described previously22. Briefly, a starting culture of E. coli BL21 CodonPlus-RIL cells (Agilent) was inoculated from a single colony and grown in 2xTY media supplemented with ampicillin (100 μg/mL) and chloramphenicol (25 μg/mL) at 37 °C overnight. This was used to inoculate 30 mL (small scale) or 1 L (large scale) of the same media the next day. The culture was grown until mid-log phase and expression was induced with anhydrotetracycline at 200 μg/L for 4 h at 37 °C. After storage at 4 °C overnight, harvested cells were lysed at 75 °C for 30 min and lysates were cleared by centrifugation. His-tagged polymerases were benchtop-purified via gravity flow on Ni-NTA agarose resin (Qiagen) while non-His-tagged polymerases were bench-top-purified via gravity flow on DEAE Sepharose fast flow anion exchange resin (GE Healthcare). Then eluted fractions were loaded onto a 16/10 Hi-Prep Heparin FF column (Cytiva Life Sciences) and eluted at 0.5–0.8 M NaCl. Appropriate fractions were filter-dialysed (Amicon Ultra Centrifugal Filters, Millipore) into 2x polymerase storage buffer (1M KCl, 2 mM EDTA, 20 mM Tris pH 7.4) and stored in 50 % glycerol at -20 °C.
Synthesis of long fluorophore-labelled RNA
Human cDNA clones for KRAS (transcript variant b, accession no. NM_004985) and CTNNB1 (transcript variant 1, accession no. NM_001904) in plasmids pCMV6-XL6 (SP6 promoter) (cat. no. SC109374) and pCMV6-XL5 (T7 promoter) (cat. no. SC107921), respectively, were obtained from OriGene, USA. Site-directed mutagenesis was performed using a QuikChange II kit (Agilent Technologies, USA), according to the manufacturer’s protocol; KRAS mutations G12D (c.35G>A) and G13D (c.38G>G), and CTNNB1 mutation S33Y (c.98C>A) were introduced using primer sets shown in Supplementary Table 2 (“Quik_KRAS_G12D_Fw/Rev”, “Quik_KRAS_G13D_Fw/Rev” or “Quik_CTNNB_G12D_Fw/Rev”) and resulting plasmids cloned and verified by Sanger sequencing (Source Biosciences, UK). Long RNA substrates equivalent to full KRAS and CTNNB1 mRNA transcripts bearing 5’ fluorescein (“Sub_KRas_ORF” and “Sub_CTNNB1_ORF”, respectively) were prepared using HiScribe T7 and SP6 RNA synthesis kits (NEB, USA), according to the manufacturer’s protocol, with a 4:1 ratio of 5’-Fluorescein-ApG dinucleotide (IBA Life Sciences, Germany) to GTP, using template plasmids linearised using XmaI (NEB, USA). Reactions were subsequently treated with TURBO DNase (Invitrogen / Thermo Fisher Scientific, USA) and RNA transcripts purified using RNeasy mini kits (Qiagen, Germany).
2’OMezyme selections
Broadly, chimeric RNA-2’OMe-RNA random-sequence libraries were prepared and selected using a similar strategy as previous XNAzymes24, 49. Initial library synthesis reactions were performed using 1 μM RNA primer “P1_KRas12[G12D]”, 2 μM DNA template “N40libtemp_KRas12”, 1.3 μM 2M polymerase and 0.125 mM (each) 2’OMe-ATP, 2’OMe-CTP, 2’OMe-GTP and 2’OMe-UTP, in Thermopol buffer (NEB, USA) for 1 h at 50 °C, 2 h at 65 °C. MyOne Streptavidin C1 Dynabeads (Invitrogen / Thermo Fisher Scientific, USA) were used to capture (5’ biotinylated) single-stranded chimeric RNA-2’OMe-RNA libraries, allowing (unbiotinyated) DNA template to be denatured using 0.1 N NaOH and removed, as described previously24; libraries were subsequently purified by Urea-PAGE. Selection reactions were performed by annealing libraries in nuclease-free water (Qiagen, Germany) for 60 s at 80 °C, 5 min RT then incubating at 37 °C in 2’OMezyme selection buffer (30 mM EPPS pH 7.4, 150 mM KCl, 1 mM MgCl2). Reaction times were varied as follows: rounds 1-11; overnight (≈16 h), rounds 11 & 12; 1 h, rounds 13-15; 30 min.
2’OMe-RNA reverse transcription was performed using 1 μM polymerase C823, with 0.2 μM 5’ biotinylated primer “RT_Ebo” in Thermopol buffer (NEB, USA) with an additional 2 mM MgCl2, 200 μM each dNTP, for 17 h at 65 °C. First-stand cDNA was isolated using streptavidin magnetic beads (C1 MyOne, Thermo Fisher Scientific, USA), eluted by incubation in nuclease-free water for 2 min at 80 °C, then amplified by a two-step nested PCR strategy using OneTaq Hot Start master mix (NEB, USA). The first ‘out-nested’ PCRs used 0.5 μM forward primer “dP2_KRas12” and 0.5 μM reverse primer “RT_Ebo_out”, cycling conditions were 94 °C for 1min, 20-35 x [94 °C for 30 s, 52 °C for 30 s, 72 °C for 30 s], 72 °C for 2 min. Following the first PCR, primers were digested using ExoSAP (Ambion/Life Technologies, USA), which was then heat inactivated, according to the manufacturer’s instructions. Second step (‘in-nest’) PCRs used 1 μl of unpurified out-nest PCR product as template in a 50 μl reaction with 0.5 μM forward primer “dP2_KRas12” and 0.5 μM reverse primer “RT_Ebo_in”, cycling conditions as above. Reactions were analysed by electrophoresis on 4% NGQT-1000 agarose (Thistle Scientific, UK) gels containing GelStar stain (Lonza, Switzerland). Bands of appropriate size were purified using a gel extraction kit (Qiagen, Germany) according to the manufacturer’s instructions. Purified DNA was used as the polyclonal template for either sequencing library PCR (see below) or preparative PCR (‘in-nest’ PCR scaled up to 500 μl) for generation of DNA templates for XNA synthesis. Single-stranded DNA templates were isolated using streptavidin beads and ethanol-precipitated before further use.
A ‘maturation’ selection was subsequently performed for five rounds (with 30 min reactions at 37 °C in 2’OMezyme reaction buffer) using the sequence of the most abundant clone at round 15 (comprising 84,674 of 3,942,063 deep sequencing reads; ≈2%) as the basis a spiked library, synthesised as described above, using DNA template “R15_1libtemp_KRasl2”. 2’OMezyme “R15/5-K” was the most abundant clone in round 5 of the maturation selection (comprising 1,291 of 5,507,023 deep sequencing reads; 0.02%).
Deep sequencing
Deep sequencing was performed using the MiSeq platform (Illumina, USA), as described previously24; 2’OMezyme selection pools were converted to sequencing libraries by PCR using primers “P5_P2_KRas12” and “P3_RT_Ebo_in” to append the necessary priming sites.
Synthesis of 2’OMezymes for characterisation
For initial screening of 2’OMezyme activity and evaluation of point mutations, 2’OMezymes were synthesised using polymerase 2M as described above, using RNA primer “P2_Ebo” and 3’ biotinylated DNA templates as shown in Supplementary Table 2, and isolated using MyOne Streptavidin C1 Dynabeads (Invitrogen / Thermo Fisher Scientific, USA), as described previously24. Following denaturation and removal of DNA template strands using 0.1 NaOH, 2’OMezymes were incubated in 0.8 N NaOH, 1 h at 65 °C, to fully hydrolyse primer RNA.
2’OMezymes for all other characterisation experiments were synthesised by solid phase phosphoramidite chemistry by Merck / MilliporeSigma (Germany).
2’OMezyme reactions
RNA cleavage assays were performed in trans using PAGE-purified 2’OMezymes and RNA substrates, annealed as described above and incubated at 37 °C in 2’OMezyme selection buffer (30 mM EPPS pH 7.4, 150 mM KCl, 1 mM MgCl2), or 30 mM EPPS pH 8.5, 150 mM KCl, 25 mM MgCl2, supplemented with RNasin ribonuclease inhibitor (Promega, USA). In Mg2+ titration experiments, 2’OMezyme selection buffer was supplemented with additional magnesium chloride (MgCl2); in pH titration experiments, 150 mM KCl, 1 mM MgCl2 plus 50mM buffer as follows was used: HEPES (pH 5.0 – 6.0), EPPS (pH 6.5–8.75), CHES (pH 9.0–12.0). For magnesium-free reactions, 30 mM EPPS pH 7.4, 150 mM KCl, 5 mM EDTA was used.
Pseudo first-order reaction rates (kobs) under single-turnover pre-steady-state (Km/kcat) conditions were determined from three independent reactions with (separately annealed) catalyst at 5 μM and substrate at 1 μM, as described previously49, fit using Prism 9 (GraphPad Software, USA). For multiple turnover reactions, 1 μM substrate was reacted with 10 nM 2’OMezyme at 37 °C in 2’OMezyme selection buffer.
For the reverse RNA ligation reactions, the products of a large-scale “Sub_KRas_12[G12D]” RNA cleavage reaction catalysed by 2’OMezyme “R15/5-K” were purified by Urea-PAGE and used as substrates. 5 μM 2’OMezyme “R15/5-K” and 1 μM (each) of the 5’ and 3’ RNA cleavage products were annealed in water as described above, then diluted into 2’OMezyme selection buffer with or without magnesium chloride, snap-frozen on dry ice then incubated reacted at -7 °C or 37 °C for 20 h. ‘Supercooled’ samples were incubated directly at -7 °C without prior freezing on dry ice.
Analysis of 2’OMezyme-catalysed RNA cleavage products
Substrate RNA “Sub_KRas_12 [G12D]” was reacted with 2’OMezyme “R15/5-K” under selection conditions and the 5’ RNA cleavage product was purified by Urea-PAGE. The cleavage product was analysed by MALDI-ToF mass spectrometry using an Ultraflex III TOF-TOF instrument (Bruker Daltonik, Bremen, Germany) in positive ion mode as described previously49.
Enzymatic removal of 3’ terminal phosphates was assayed by Urea-PAGE gel shift following incubation in Calf Intestinal Phosphatase (CIP)(NEB, USA) or T4 Polynucleotide Kinase (PNK)(NEB, USA) in manufacturer’s buffer for 30 min at 37 °C. Hydrolysis of cyclic phosphates was achieved by incubation in 10 mM glycine pH 2.5 for 30 min at room temperature.
Analysis of 2’OMezyme serum stability
PAGE-purified 2’OMezyme “R15/5-K” and DNAzyme “1023_KRasC” were annealed in water as described above, then incubated (at 5 μM) at 37 °C in 95% human serum (MilliporeSigma, Germany). Full-length catalyst remaining was quantified on Urea-PAGE gels stained with SYBR Gold (ThermoFisher Scientific, USA).
Analysis of aptamer binding by Surface Plasmon Resonance (SPR)
2’OMe/MOE-RNA aptamers were synthesized from RNA primer Prim1 and 3’-biotinylated DNA template Temp_ARC224 (Supplementary Table 1) as described in section “Synthesis of 2’OMezymes for characterization” using 2’OMe/MOE-NTPs.
2’OMe/MOE-RNA aptamers were annealed at 1–10 μM in nuclease-free water by heating to 95 °C for 5 min and equilibrating at RT for 10 min. They were then diluted and analysed in PBS + 0.1% (v/v) Tween20 (PBS-Tw). Surface Plasmon Resonance (SPR) measurements were made using a BIAcore 2000 instrument (GE Life Sciences, UK) at a flow rate of 20 μLmin-1 at 20 °C. CM4 sensor chip (GE Life Sciences, UK) surfaces were coated with Neutravidin (Pierce 31000, ThermoFisher Scientific, USA) surfaces (~8000 RU per flow cell) using an amine coupling kit (GE Life Sciences, UK) and flowing in 5 mM NaOAc (sodium acetate), pH 5.5. Chips were equilibrated in PBS-Tw and left to flow overnight until signal drift had settled. ~2000 RU biotinylated human VEGF165 (Bio-Techne, USA) was captured (except for the reference cell) before blocking with excess free biotin. 50 μL aptamer samples at a series of concentrations (500 nM, 250 nM, 125 nM, 62.5 nM, 31.3 nM, 15.6 nM, 7.8 nM, 3.9 nM) were injected for 150 s and dissociation was recorded for 600 s, in PBS-Tw. Single injections of aptamers outside of the concentration series were performed at 100 nM (50 μL) in PBS-Tw. After every injection, the sensor surface was regenerated using two 5 μL injections of 10 mM NaOH + saline (137 mM NaCl, 2.7 mM KCl).
To obtain optimal fits, SPR data had to be fit to a double-exponential heterogeneous dissociation/association model to determine kinetic parameters from two independent datasets per aptamer with on-line reference subtraction. For the ARC224 MOE-AGC aptamer, the lowest two concentration points were not included in the analysis and discarded as outliers due to insufficient binding signal. Deviation from homogeneous 1:1 binding models is established for nucleic acid-protein interactions, and a heterogeneous model describing two conformationally divergent populations of a DNA aptamer binding VEGF has been described50.
The rate constants of dissociation and association were obtained by fitting the observed response signal R using the two equations below.
Heterogeneous dissociation:
| (1) |
where R0 is the response at the start of dissociation (t0), R1 is the contribution to R0 from component 1 (floating parameter), and therefore, (R0 – R1) is the contribution to R0 from component 2. Kdi is the dissociation rate constant for component i (floating parameter).
Heterogeneous association:
| (2) |
where Reqi is the steady-state response level for component i (floating parameter), kai is the association rate constant for component i (floating parameter), kdi is the dissociation rate constant for component i, C is the molar concentration of analyte, and t0 is the start time for the association.
NGS for 2’OMe synthesis and RT fidelity analysis
For 2’OMe-RNA synthesis, ssDNA templates were generated by linearization of pASK_TGO plasmid using EcoR1 followed by by shrimp alkaline phosphatase treatment and restriction using BamHI. The 369 ntd dsDNA fragment is gel eluted and treated with lambda exonuclease (NEB) to generate single strand template for the RNA / 2’OMe-RNA synthesis. The 2’OMe-RNA synthesis is carried out in 20 μL reaction volumes, modFD-N25-TGO682F primer and the ssDNA template generated as mentioned above were annealed at 95 °C for two minutes followed by 55 °C for 5 minutes in 1x Thermopol buffer containing 200 μM rNTPs or 200 μM 2’OMe-NTPs. The RNA and 2’OMe-RNA syntheses were carried out using TGK polymerase (RNA) and TGLLK or 2M or 3M (2’OMe-RNA) synthesis, respectively.
The synthesised transcripts containing 5’ biotin modification were bound to Dyna-beads™ M-280 Streptavidin beads (Invitrogen) and purified by stripping off the template using 0.2 N NaOH. The magnetic beads immobilised with RNA or 2’OMe-RNA were used for reverse transcription using SSIII enzyme (ThermoFisherScientific). On bead RT reaction was performed using RT_primer TagR1-N25-TGO642R harbouring N25 internal barcode for PCR and sequencing error correction. RT reactions were carried out according to vendor’s guidelines for SSIII. The cDNA bound to the RNA or 2’OMe-RNA on the beads were washed twice using 1X BWBS, stripped using 0.2 N NaOH and neutralised using Tris buffer before using for sequencing library generation. RT was repeated three more times and the eluted cDNAs were used for library preparation for deep sequencing.
The cDNAs (25 μL) were added to 50 μL PCR reaction with primers HiSeq_ModFD, forward primer and HiSeq_TagR1xx, unique barcode identifier primer (Supplementary Table 5) to demultiplex samples and to introduce adaptors for Illumina sequencing using Q5 polymerase (NEB).
Barcoded fidelity libraries were pooled and sequenced on an Illumina MiSeq for PE read of 150 cycles. Fidelity analysis was performed using the Burrows-Wheeler Aligner (BWA)51, Samtools52 and custom scripts that do the following can be found at GitHub: https://github.com/holliger-lab/fidelity-analysis. Mean error rate (Supplementary Table 4) and base substitutions were calculated for RNA and 2’OMe-RNA per 106 bases sequenced (Supplementary Tables 6 & 7).23
Steady-State Kinetics
Steady-state kinetic parameters for NTP incorporation by 2M were determined by performing initial velocity measurements of single incorporations of either ATP, 2’OMe-ATP, or MOE-ATP. To generate the 2’OMe-RNA/DNA substrate, a 20-mer 2’OMe-RNA primer FD was 5’ 6-carboxyfluorescein end-labeled and annealed to the 52-mer DNA template BFL770 (Supplementary Table 1) at a 1:1.2 molar ratio. The reactions were performed at 50 °C in a mixture containing 1X Thermopol buffer, 6 mM Mg2+, 100 nM 2’OMe-RNA/DNA substrate, and at NTP concentrations ranging from 0.5–250 μM. Enzyme concentrations and reaction times were selected to main-tain initial velocity conditions. The 25 μL reactions were stopped by addition of a quenching solution containing 100 mM EDTA, 80% deionized formamide, 0.25 mg/ml bromophenol blue and 0.25 mg/ml xylene cyanol. Moreover, less than 20% of the primers were extended as required for steady-state conditions.
Product and substrate were separated on a 22% denaturing (8 M urea) polyacrylamide gel. The resulting bands were quantified using a Cytiva Typhoon RGB imager in fluorescence mode. Steady-state kinetic parameters (KM, kcat) were determined by fitting the data to the Michaelis–Menten equation. The data are the means and standard error from three independent experiments.
Transcription reactions with RGVG-M6
DNA template for transcription reactions was created by PCR-amplifying a 901-bp region on a plasmid encoding sfGFP under a T7 promoter. The PCR used 0.5 μM forward primer “5T7.for” and 0.5 μM reverse primer “pCUN_Do.rev”; cycling conditions were 95 °C for 30 s, 30 x [95 °C for 10 s, 69 °C for 30 s, 72 °C for 30 s], 72 °C for 2 min.
For very permissive conditions, reactions comprised 125 nM DNA template, 200 nM T7 RNAP WT or its variant RGVG-M611, 1.5 mM MnCl2, 7.5 mM each NTP or 1 mM each 2’OMe-NTP, 0.1 U yeast inorganic pyrophosphatase. In order to compare the yield of 2’OMe-RNA synthesis by 2M and RGVG-M6, reactions were run under equimolar nucleic acid input of 0.5 pmol primer (2M) and 0.5 pmol DNA template (50 nM, RGVG-M6), and 50 nM RGVG-M6 polymerase with a polymerase:template ratio of 1:1 as described in 11. Reactions were treated with Turbo DNase and Proteinase K followed by denaturing PAGE.
Supplementary Material
Acknowledgements
This work was supported by a Ph.D. fellowship from the Boehringer Ingelheim Fonds (N.F.), by the Medical Research Council (MRC) program grant program no. MC_U105178804 (A.I.T., S.-Y.P.-C., P. Holliger), a research collaboration between AstraZeneca UK Limited and the Medical Research Council-MRC-Astra Zeneca Blue Sky Grant (N.S., S.A.-F.), FWO (Flanders Research Foundation) Fund of Scientific Research and the Rega Institute, KU Leuven (M.A., P. Herdewijn), and by National Institute of Health grants R35-GM128562 (B.D.F.) and K99-ES031148 (A.M.W). The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Footnotes
Author contributions
N.F., S.A.-F., A.I.T. and P. Holliger conceived and designed experiments. N.F. performed polymerase studies. N.F. and S.A.-F. performed polymerase design and engineering. N.F., M.A. and P. Herdewijn synthesised MOE-nucleotides. N.F. and A.I.T. completed SPR measurements. A.I.T. performed 2’OMezyme selections and characterisation. N.S. performed polymerase fidelity measurements. S.- Y.P.-C. performed MS analysis; A.M.W. and B.D.F performed and analysed steady-state kinetic measurements. All authors analysed data, discussed results and co-wrote the manuscript.
Competing interests
UK Research and Innovation have filed a UK patent priority application on behalf of the inventors N.F., S.A.-F. and P. Holliger on the 2M/3M polymerase on 25 May 2022 (application number 2207699.6).
Data availability
All data generated or analysed during this study are included in this published article (and its supplementary information files), except raw sequencing reads, which are available in the NCBI SRA repository, BioProject ID PRJNA847930.
Code availability
Fidelity analysis of sequencing data for 2OMe-RNA synthesis by individual polymerases was performed using the Burrows-Wheeler Aligner (BWA-0.7.17), Samtools and custom scripts that can be found at GitHub: https://github.com/holliger-lab/fidelity-analysis.
References
- 1.Taylor AI, Houlihan G, Holliger P. Beyond DNA and RNA: The Expanding Toolbox of Synthetic Genetics. Cold Spring Harb Perspect Biol. 2019;11(6) doi: 10.1101/cshperspect.a032490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Freund N, Furst M, Holliger P. New chemistries and enzymes for synthetic genetics. Curr Opin Biotechnol. 2021;74:129–136. doi: 10.1016/j.copbio.2021.11.004. [DOI] [PubMed] [Google Scholar]
- 3.Wan WB, Seth PP. The Medicinal Chemistry of Therapeutic Oligonucleotides. J Med Chem. 2016;59(21):9645–9667. doi: 10.1021/acs.jmedchem.6b00551. [DOI] [PubMed] [Google Scholar]
- 4.Dimitrova DG, Teysset L, Carre C. RNA 2’-O-Methylation (Nm) Modification in Human Diseases. Genes (Basel) 2019;10(2) doi: 10.3390/genes10020117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dai Q, Moshitch-Moshkovitz S, Han D, Kol N, Amariglio N, Rechavi G, et al. Nm-seq maps 2’-O-methylation sites in human mRNA with base precision. Nat Methods. 2017;14(7):695–698. doi: 10.1038/nmeth.4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zust R, Cervantes-Barragan L, Habjan M, Maier R, Neuman BW, Ziebuhr J, et al. Ribose 2’-O-methylation provides a molecular signature for the distinction of self and non-self mRNA dependent on the RNA sensor Mda5. Nat Immunol. 2011;12(2):137–143. doi: 10.1038/ni.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aartsma-Rus A, Corey DR. The 10th Oligonucleotide Therapy Approved: Golodirsen for Duchenne Muscular Dystrophy. Nucleic Acid Ther. 2020;30(2):67–70. doi: 10.1089/nat.2020.0845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ruckman J, Green LS, Beeson J, Waugh S, Gillette WL, Henninger DD, et al. 2’-Fluoropyrimidine RNA-based aptamers to the 165-amino acid form of vascular endothelial growth factor (VEGF165). Inhibition of receptor binding and VEGF-induced vascular permeability through interactions requiring the exon 7-encoded domain. J Biol Chem. 1998;273(32):20556–20567. doi: 10.1074/jbc.273.32.20556. [DOI] [PubMed] [Google Scholar]
- 9.Chelliserrykattil J, Ellington AD. Evolution of a T7 RNA polymerase variant that transcribes 2’-O-methyl RNA. Nat Biotechnol. 2004;22(9):1155–1160. doi: 10.1038/nbt1001. [DOI] [PubMed] [Google Scholar]
- 10.Ibach J, Dietrich L, Koopmans KR, Nobel N, Skoupi M, Brakmann S. Identification of a T7 RNA polymerase variant that permits the enzymatic synthesis of fully 2’-O-methyl-modified RNA. J Biotechnol. 2013;167(3):287–295. doi: 10.1016/j.jbiotec.2013.07.005. [DOI] [PubMed] [Google Scholar]
- 11.Meyer AJ, Garry DJ, Hall B, Byrom MM, McDonald HG, Yang X, et al. Transcription yield of fully 2’-modified RNA can be increased by the addition of thermostabilizing mutations to T7 RNA polymerase mutants. Nucleic Acids Res. 2015;43(15):7480–7488. doi: 10.1093/nar/gkv734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burmeister PE, Lewis SD, Silva RF, Preiss JR, Horwitz LR, Pendergrast PS, et al. Direct in vitro selection of a 2’-O-methyl aptamer to VEGF. Chem Biol. 2005;12(1):25–33. doi: 10.1016/j.chembiol.2004.10.017. [DOI] [PubMed] [Google Scholar]
- 13.Chen T, Hongdilokkul N, Liu Z, Adhikary R, Tsuen SS, Romesberg FE. Evolution of thermophilic DNA polymerases for the recognition and amplification of C2’-modified DNA. Nat Chem. 2016;8(6):556–562. doi: 10.1038/nchem.2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu Z, Chen T, Romesberg FE. Evolved polymerases facilitate selection of fully 2’-OMe-modified aptamers. Chem Sci. 2017;8(12):8179–8182. doi: 10.1039/c7sc03747c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoshino H, Kasahara Y, Kuwahara M, Obika S. DNA Polymerase Variants with High Processivity and Accuracy for Encoding and Decoding Locked Nucleic Acid Sequences. J Am Chem Soc. 2020;142(51):21530–21537. doi: 10.1021/jacs.0c10902. [DOI] [PubMed] [Google Scholar]
- 16.Cozens C, Mutschler H, Nelson GM, Houlihan G, Taylor AI, Holliger P. Enzymatic Synthesis of Nucleic Acids with Defined Regioisomeric 2’-5’ Linkages. Angew Chem Int Ed Engl. 2015;54(51):15570–15573. doi: 10.1002/anie.201508678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cozens C, Pinheiro VB, Vaisman A, Woodgate R, Holliger P. A short adaptive path from DNA to RNA polymerases. Proc Natl Acad Sci U S A. 2012;109(21):8067–8072. doi: 10.1073/pnas.1120964109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kropp HM, Betz K, Wirth J, Diederichs K, Marx A. Crystal structures of ternary complexes of archaeal B-family DNA polymerases. PLoS One. 2017;12(12):e0188005. doi: 10.1371/journal.pone.0188005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perera RL, Torella R, Klinge S, Kilkenny ML, Maman JD, Pellegrini L. Mechanism for priming DNA synthesis by yeast DNA polymerase alpha. Elife. 2013;2:e00482. doi: 10.7554/eLife.00482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kawai G, Yamamoto Y, Kamimura T, Masegi T, Sekine M, Hata T, et al. Conformational rigidity of specific pyrimidine residues in tRNA arises from posttranscriptional modifications that enhance steric interaction between the base and the 2’-hydroxyl group. Biochemistry. 1992;31(4):1040–1046. doi: 10.1021/bi00119a012. [DOI] [PubMed] [Google Scholar]
- 21.Nishizaki T, Iwai S, Ohtsuka E, Nakamura H. Solution structure of an RNA.2’-O-methylated RNA hybrid duplex containing an RNA.DNA hybrid segment at the center. Biochemistry. 1997;36(9):2577–2585. doi: 10.1021/bi962297c. [DOI] [PubMed] [Google Scholar]
- 22.Pinheiro VB, Taylor AI, Cozens C, Abramov M, Renders M, Zhang S, et al. Synthetic genetic polymers capable of heredity and evolution. Science. 2012;336(6079):341–344. doi: 10.1126/science.1217622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Houlihan G, Arangundy-Franklin S, Porebski BT, Subramanian N, Taylor AI, Holliger P. Discovery and evolution of RNA and XNA reverse transcriptase function and fidelity. Nat Chem. 2020;12(8):683–690. doi: 10.1038/s41557-020-0502-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Taylor AI, Holliger P. Directed evolution of artificial enzymes (XNAzymes) from diverse repertoires of synthetic genetic polymers. Nat Protoc. 2015;10(10):1625–1642. doi: 10.1038/nprot.2015.104. [DOI] [PubMed] [Google Scholar]
- 25.Egli M, Minasov G, Tereshko V, Pallan PS, Teplova M, Inamati GB, et al. Probing the influence of stereoelectronic effects on the biophysical properties of oligonucleotides: comprehensive analysis of the RNA affinity, nuclease resistance, and crystal structure of ten 2’-O-ribonucleic acid modifications. Biochemistry. 2005;44(25):9045–9057. doi: 10.1021/bi050574m. [DOI] [PubMed] [Google Scholar]
- 26.Teplova M, Minasov G, Tereshko V, Inamati GB, Cook PD, Manoharan M, et al. Crystal structure and improved antisense properties of 2’-O-(2-methoxyethyl)-RNA. Nat Struct Biol. 1999;6(6):535–539. doi: 10.1038/9304. [DOI] [PubMed] [Google Scholar]
- 27.Khatsenko O, Morgan R, Truong L, York-Defalco C, Sasmor H, Conklin B, et al. Absorption of antisense oligonucleotides in rat intestine: effect of chemistry and length. Antisense Nucleic Acid Drug Dev. 2000;10(1):35–44. doi: 10.1089/oli.1.2000.10.35. [DOI] [PubMed] [Google Scholar]
- 28.Plevnik M, Cevec M, Plavec J. NMR structure of 2’-O-(2-methoxyethyl) modified and C5-methylated RNA dodecamer duplex. Biochimie. 2013;95(12):2385–2391. doi: 10.1016/j.biochi.2013.08.025. [DOI] [PubMed] [Google Scholar]
- 29.Martin P. Stereoselektive Synthese von 2’-O-(2-Methoxyethyl)ribonucleosiden: Nachbargruppenbeteiligung der Methoxyethoxy-Gruppe bei der Ribosylierung von Heterocyclen. Helvetica Chimica Acta. 1996;79(7):1930–1938. [Google Scholar]
- 30.Martin P. Ein neuer Zugang zu 2’-O-Alkylribonucleosiden und Eigenschaften deren Oligonucleotide. Helvetica Chimica Acta. 1995;78(2):486–504. [Google Scholar]
- 31.Gillerman I, Fischer B. An improved one-pot synthesis of nucleoside 5’-triphosphate analogues. Nucleosides Nucleotides Nucleic Acids. 2010;29(3):245–256. doi: 10.1080/15257771003709569. [DOI] [PubMed] [Google Scholar]
- 32.Ludwig J. A new route to nucleoside 5’-triphosphates. Acta Biochim Biophys Acad Sci Hung. 1981;16(3-4):131–133. [PubMed] [Google Scholar]
- 33.Freier SM, Altmann KH. The ups and downs of nucleic acid duplex stability: structure-stability studies on chemically-modified DNA:RNA duplexes. Nucleic Acids Res. 1997;25(22):4429–4443. doi: 10.1093/nar/25.22.4429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kool ET. Hydrogen bonding, base stacking, and steric effects in dna replication. Annu Rev Biophys Biomol Struct. 2001;30:1–22. doi: 10.1146/annurev.biophys.30.1.1. [DOI] [PubMed] [Google Scholar]
- 35.Wu EY, Beese LS. The structure of a high fidelity DNA polymerase bound to a mismatched nucleotide reveals an “ajar” intermediate conformation in the nucleotide selection mechanism. J Biol Chem. 2011;286(22):19758–19767. doi: 10.1074/jbc.M110.191130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang W, Wu EY, Hellinga HW, Beese LS. Structural factors that determine selectivity of a high fidelity DNA polymerase for deoxy-, dideoxy-, and ribonucleotides. J Biol Chem. 2012;287(34):28215–28226. doi: 10.1074/jbc.M112.366609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen CY. DNA polymerases drive DNA sequencing-by-synthesis technologies: both past and present. Front Microbiol. 2014;5:305. doi: 10.3389/fmicb.2014.00305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Redrejo-Rodriguez M, Ordonez CD, Berjon-Otero M, Moreno-Gonzalez J, Aparicio-Maldonado C, Forterre P, et al. Primer-Independent DNA Synthesis by a Family B DNA Polymerase from Self-Replicating Mobile Genetic Elements. Cell Rep. 2017;21(6):1574–1587. doi: 10.1016/j.celrep.2017.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Blasco MA, Mendez J, Lazaro JM, Blanco L, Salas M. Primer terminus stabilization at the phi 29 DNA polymerase active site. Mutational analysis of conserved motif KXY. J Biol Chem. 1995;270(6):2735–2740. doi: 10.1074/jbc.270.6.2735. [DOI] [PubMed] [Google Scholar]
- 40.Kazlauskas D, Krupovic M, Guglielmini J, Forterre P, Venclovas C. Diversity and evolution of B-family DNA polymerases. Nucleic Acids Res. 2020;48(18):10142–10156. doi: 10.1093/nar/gkaa760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Franklin MC, Wang J, Steitz TA. Structure of the Replicating Complex of a Pol α Family DNA Polymerase. Cell. 2001;105(5):657–667. doi: 10.1016/s0092-8674(01)00367-1. [DOI] [PubMed] [Google Scholar]
- 42.Rudinger NZ, Kranaster R, Marx A. Hydrophobic amino acid and single-atom substitutions increase DNA polymerase selectivity. Chem Biol. 2007;14(2):185–194. doi: 10.1016/j.chembiol.2006.11.016. [DOI] [PubMed] [Google Scholar]
- 43.Gardner AF, Jack WE. Determinants of nucleotide sugar recognition in an archaeon DNA polymerase. Nucleic Acids Res. 1999;27(12):2545–2553. doi: 10.1093/nar/27.12.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bartel DP, Szostak JW. Isolation of new ribozymes from a large pool of random sequences [see comment] Science. 1993;261(5127):1411–1418. doi: 10.1126/science.7690155. [DOI] [PubMed] [Google Scholar]
- 45.Wilson DS, Szostak JW. In vitro selection of functional nucleic acids. Annu Rev Biochem. 1999;68:611–647. doi: 10.1146/annurev.biochem.68.1.611. [DOI] [PubMed] [Google Scholar]
- 46.Mutschler H, Taylor AI, Porebski BT, Lightowlers A, Houlihan G, Abramov M, et al. Random-sequence genetic oligomer pools display an innate potential for ligation and recombination. Elife. 2018;7 doi: 10.7554/eLife.43022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fedor MJ. Structure and function of the hairpin ribozyme. J Mol Biol. 2000;297(2):269–291. doi: 10.1006/jmbi.2000.3560. [DOI] [PubMed] [Google Scholar]
- 48.Skerra A. Use of the tetracycline promoter for the tightly regulated production of a murine antibody fragment in Escherichia coli. Gene. 1994;151(1-2):131–135. doi: 10.1016/0378-1119(94)90643-2. [DOI] [PubMed] [Google Scholar]
- 49.Taylor AI, Pinheiro VB, Smola MJ, Morgunov AS, Peak-Chew S, Cozens C, et al. Catalysts from synthetic genetic polymers. Nature. 2015;518(7539):427–430. doi: 10.1038/nature13982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Potty AS, Kourentzi K, Fang H, Jackson GW, Zhang X, Legge GB, et al. Biophysical characterization of DNA aptamer interactions with vascular endothelial growth factor. Biopolymers. 2009;91(2):145–156. doi: 10.1002/bip.21097. [DOI] [PubMed] [Google Scholar]
- 51.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analysed during this study are included in this published article (and its supplementary information files), except raw sequencing reads, which are available in the NCBI SRA repository, BioProject ID PRJNA847930.
Fidelity analysis of sequencing data for 2OMe-RNA synthesis by individual polymerases was performed using the Burrows-Wheeler Aligner (BWA-0.7.17), Samtools and custom scripts that can be found at GitHub: https://github.com/holliger-lab/fidelity-analysis.