Abstract
The drug discovery process involves designing compounds to selectively interact with their targets. The majority of therapeutic targets for low molecular weight (small molecule) drugs are proteins. The outstanding accuracy with which recent artificial intelligence methods compile the three dimensional structure of proteins has made protein targets more accessible to the drug design process. Here we present our perspective of the significance of accurate protein structure prediction on various stages of the small molecule drug discovery life cycle focusing on current capabilities and assessing how further evolution of such predictive procedures can have a more decisive impact in the discovery of new medicines.
Graphical Abstract.
Introduction
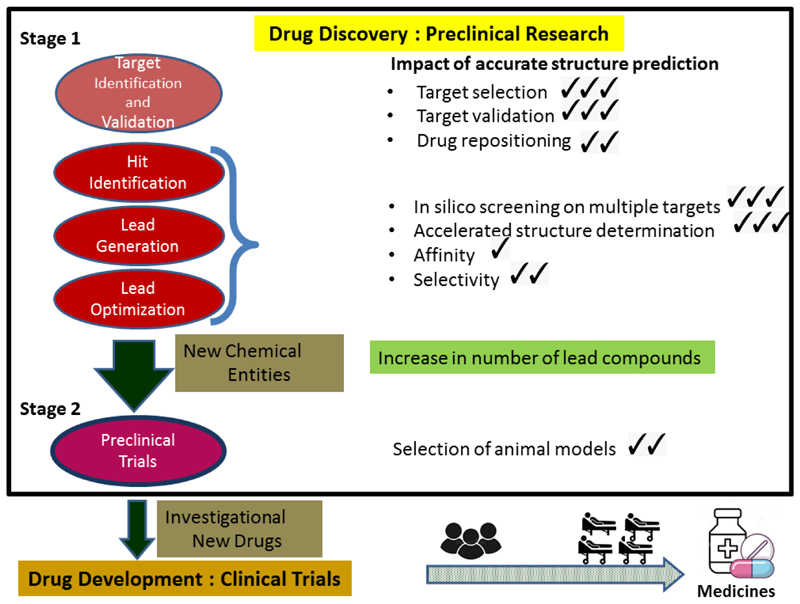
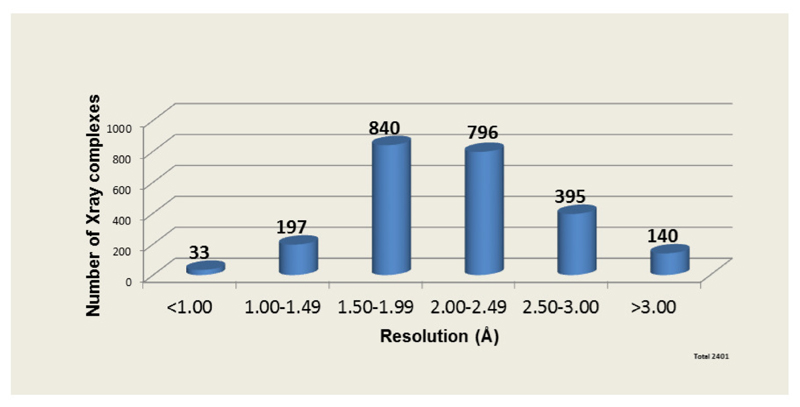
Drug discovery is a costly [1] process of research concerning the identification of new chemicals that have the potential to modulate disease [2]. The principal strategies utilised to identify potential new medicines in preclinical research are modification of natural substances, phenotype screening, biologic based methodologies and target -based selection [3]. Over the last few decades genomic, proteomic, and structural studies and access to their associated databases [4–6] have provided hundreds of new targets and opportunities for the drug discovery pipeline. Additionally, with the extensive use of combinatorial chemistry (CC), high-throughput screening (HTS) and virtual screening (VS) [7] together with the emergence of Artificial Intelligence (AI) and Machine Learning (ML) [8], the focus of drug discovery has shifted towards identifying specific molecular targets and structure based drug discovery (SBDD). An analysis of crystallographic data on drug-target complexes (See Figure 1) shows that 2401 X-ray complexes are available from 299 registered drugs (19% of total) bound to 501 unique proteins.The average resolution of these data is around 2.0 Å, suggesting that this resolution is sufficient for the purpose of SBDD. The recent advent of protein structure prediction with near experimental accuracy [9–11] has introduced a new paradigm for SBDD. The AlphaFold database, hosted at EMBL-EBI (https://alphafold.ebi.ac.uk/), provides free access for everyone to more than 200million protein structure predictions. In this review, we describe how such advances in protein structure prediction eg Alphafold2 [12], RoseTTA[13] are likely to impact research approaches in the various stages of the drug discovery process (See Graphical Abstract).
Figure 1. X-ray Crystallographic data on Drug-protein complexes.
An analysis X-ray data of marketed drugs in complex with their protein targets shows that the average resolution of these structures is around 2 Å.. A total of 2401 protein-ligand complexes contain 299 registered drug molecules and 501 unique proteins We note that the average resolution for drug targets has remained constant (between 2.0 and 2.15 Å) over time. Data from DrugPort[37] and DrugBank[38].
Target Identification and Validation
The first step of the drug discovery process is to identify a target and validate that it has an impact on the disease under study. Selection of targets for therapeutic intervention (protein, DNA, RNA) has traditionally been based on data from the scientific literature for insights into molecular pathways and biology of disease. Recently AI/ML, genomics and functional genomics [14–16] methods have employed to identify new molecular and cellular targets that have a role in pathogenicity or disease progression. In order to be viable, the target must have a confirmed role (via target validation) in the pathophysiology of a disease.
If the putative target has a known structure, then this can be used to assess whether the target is likely to be druggable, that is accessible to small molecules or biologicals which interact with it in order to modulate its activity. Given the accuracy of protein structure prediction by Alphafold2 [9], all protein targets identified on the basis of biology are now effectively complemented by three dimensional data for assessment of target tractability. We should note that almost all targets for approved drugs to date (especially in humans) have an experimental structure (either in native or in complex form) or a good model built by standard homology modelling methods. Therefore the new models, although sometimes more accurate than old predictions, will mainly find usage for completely new targets, especially in pathogens, which have not been widely studied to date and for which experimental data are not available.
To illustrate an application, the use of Alphafold2 (AF2) to model the replicase encoded by the 1708 amino acid polyprotein of the human-infecting Hepatitis E virus (HEV-3) has predicted five non-structural proteins [17]. The models have different levels of confidence/accuracy, provided by AF2, and this is critical to consider when using for SBDD (See Figure 2). These models provide a basis for ranking the suitability of the individual proteins as potential drug targets through ranking them according to
-
(a)
the confidence level (predicted local distance difference test or pLDDT score) of the structure predicted by Alphafold2 [9,12,18]
-
(b)
details of the size and accessibility of binding pockets [19]
-
(c)
access, through public [20–22] or private databases, to experimentally observed data on substrate- or ligand-binding sites on proteins with structures similar to that of the predicted target (Figure 2) or
-
(d)
if drug selectivity is the objective, the uniqueness of the predicted protein fold [18].
Figure 2. Alphafold structure prediction and comparison with experimental data.
(A) Alphafold2 prediction [18] of a segment of a viral polyprotein (Gen-Bank accession, HQ389543, amino acids 480-1253) shown in ribbon representation. Predictions are coloured from red (worst) to blue (best) according to pLDDT values. Three (mainly blue) clearly defined domains of secondary structure (Zinc binding domain, Macro domain and Helicase) are separated by linkers with low-confidence scores. The terminii are marked with their corresponding amino acid numbers.
(B) Middle domain of AF prediction for the viral polyprotein (amino acids 802-945) used as a query to find a similar structure in the PDB, giving the high scoring hit (green, PDB ID 3GPQ, TM-score 0.78 [21]), including the ligand__A-__A.
(C) An overlay of the structure of PDB ID 3GPQ with the Alphafold2 prediction (blue) of the second domain shown in Figure 2A. The similar three dimensional structures imply that ligand binding details available from experimental data can provide molecular modelling template(s) for ligand/drug design on the predicted protein. Foldseek [21] was for the database search of crystal structures. Maestro[53] and Pymol[54] were used for creating the figures.
These principles for prioritising protein targets using accurate models are also relevant for selecting targets for drug repurposing [23] or drug repositioning for diseases that share common genes or pathways [24].
As a rule-of-thumb, in order to be comparable to experimental data and useful for in silico modelling and VS purposes, AF2 predicted structures require values of pLDDT >80 (confident to very high, see Figure 2(A), Figure 3) [9,12]. The efficient use of AF2 generated models to decipher maps derived from both X-ray [25] and cryo-EM [26,27] data underscores the value of such reliable starting models in accelerating the delivery of the final refined structure. The technique of using AF2 models for obtaining phases and/or fitting electron density derived from experimental data will be the gold standard for providing observed structures of proteins and their ligand bound complexes for the Structure Based Drug Design stage of the drug discovery process. On the other hand, there are other uses where high accuracy is less important. Low pLDDT scoring linker regions with poor three-dimensional definition emphasise possible domain boundaries (See Figure 2(A)) which can provide vital information for expression constructs. These data can help produce stable and active proteins recombinantly, thus providing a platform for the initiation of structural (crystallisation) and functional (assay) studies.
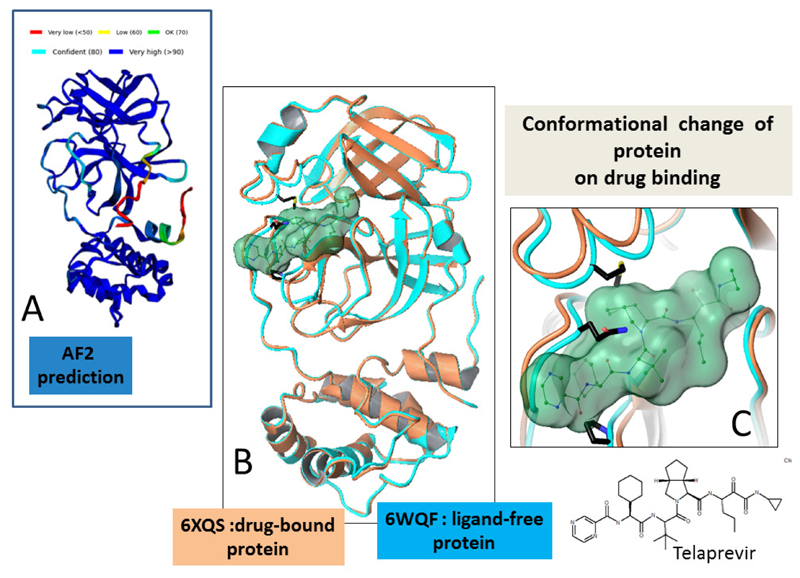
Figure 3. Structural plasticity at protein binding site.
(A) Alphafold2 predicted model (pLDDT score colours) of the structure of cysteine protease 3CL Mpro from SARS-CoV-2. The model is predicted with high pLDDT scores and is similar to the ligand-free form of the protein (PDB ID 6WQF, rmsd 2.0 Å2).
(B) A superposition, shown in ribbon representations, of the ligand-free of the protein 3CL Mpro (cyan, PDB ID 6WQF) with the structure of 3CL Mpro in complex with a drug (orange, PDB ID 6XQS). The bound compound Telaprevir (shown as light green surface) changes the protein conformation around the active site (rmsd 3.0 Å2 for all atoms, rmsd 8.8 Å2 for atoms within 8 Å of bound ligand) of the structure.
(C) Closeup of Panel B, illustrating ligand induced structural changes around the compound binding region of the protein.The ligand is shown in green ball and stick representation and light green surface. Protein residues of the ligand free structure (cyan ribbon) that present unfavourable interactions with the bound compound, influencing protein movement on ligand binding, are identified in black.
Hit Identification, Lead Generation and Lead Optimization
The next and crucial stage in preclinical drug discovery is to identify molecules (“hits”) that favourably interact with the target(s). The predictions of protein structures made by AF2 and RoseTTA do not include any ligands, so additional work is necessary for hit identification. In an ideal case, a drug, which could be a small molecule or an engineered macromolecule, would be selective and have its effect on disease by interacting solely with its selected target. Understanding the topography of the target (protein) is critical in the discovery of drugs, since chemical and shape complementarity between protein and ligand are the key drivers of drug affinity.AF2 based structure predictions can provide accurate templates for targets so that Structure Based Virtual screening (SBVS or VS) in silico explorations (using docking and molecular modelling [19,28]) of large libraries of low molecular weight compounds can be conducted for finding candidates that bind to the selected target. Access to the three-dimensional structure of the target enables the search for pockets [29] and functional relevant regions [30,31] on the protein. The success of these processes with experimental data holds promise for effective use of similar protocols on AF2 generated models.Computational methods [19,28] exploit these features of the target to define the path (hit-to-lead) for making carefully chosen chemical changes to the “hit” molecule, taking into account toxicological or selectivity liabilities, to a “lead” candidate with enhanced drug-like properties. The advances in energy perturbation (FEP) and AB-FEP (absolute binding free energy perturbation) methods [32,33] in predicting the energy of binding of molecules to target biomacromolecules can be used to provide a suitable filter for the judicious choice of candidate molecules. For a series of similar molecules, energy predictions of binding affinity to a given target are reasonable estimates and useful for developing the lead. However binding energies and models predicted for small molecules without exemplars are much less reliable.
Data on ligand(s) bound to proteins similar to the target can be used to kick-start hit discovery projects (See Figure 2(B)), without the use of computational screening protocols. Results from screening molecules using a panel of proteins with similar predicted structures [21] (implying an evolutionary relationship) can be used to identify potential off-target burdens. Conversely, characterising ligands that bind to proteins of different architecture (CATH annotations) [34,35] offers the promise of a drug repositioning programme [24].
Although most drugs are designed to modulate a specific target, in fact many bind to more than one protein. In most cases off-target binding lead to deleterious side effects which need to be resolved. However non-specific binding can be successfully exploited by optimising the compound scaffold to create a multitarget drug (MTD) that can simultaneously recognise more than one pertinent target in a specific disease pathway. Such MTD molecules are of particular relevance in treating multifactorial diseases such as Alzheimer’s disease, diabetes and cancer [36].
The easy access to accurate starting models of potential targets is likely to impact the output, efficiency and expenditure of the hit to lead process. Proficiency in designing molecules in vitro and cellular studies will lead to increased number of New Chemical Entities (NCEs) proposed for preclinical testing in animal models.
Preclinical trials
The rationale for advancing a NCE or potential drug into the clinic relies on it having demonstrated a safe and positive biological effect in animals. Preclinical development involves the testing of lead compounds for efficacy in animal models, to evaluate the therapeutic potential of the NCEs in terms of its pharmacological profile, biodistribution and safety/toxicology. Apart from the favoured murine [39] and canine species, porcine and primate preclinical animal models are also accessible [40]. Among other considerations, the choice of animal and model depends on the existing body of knowledge on the disease under consideration and features of the animal which have the best correlation to human traits. The availability of Alphafold 2 predicted structure of proteins known for all species [10] opens up the possibility of selecting preclinical models on the basis of the protein similarity of different species compared to humans Whether an AF2 predicted models would offer any advantage over standard homology modelling techniques which have, to date, been reliably used to predict the suitability of animal species for pharmacological testing remains to be established. The lack of public domain three dimensional data on membrane proteins could, as more data become available, necessitate modifying the current deep learning-based algorithms for more accurate AF2 predictions on members of the structurally less well represented families of the proteome.It may be possible to derive more predictive animals models by factoring in the comparison of target proteins and proteins that are associated with the metabolic pathway of the intended drug in the choice of species for preclinical studies.
Perspective
As explained in the previous sections, data from the Alphafold2 Database will be a valuable resource for structure biology and is likely to have the most impact on the hit and lead generation stage of preclinical stages of drug discovery. Discovery projects dealing with small molecule ligands are the most likely to benefit from these data.Although AF2 models have no information on solvent, ions or ligands, substrate- or ligand-binding sites on proteins with structures similar to that of the predicted target can be used to locate details about binding sites and protein ligand interactions [22] (see section above and Figure2). A major shortfall however is that AF2 cannot account for the structural plasticity of proteins and has been designed to predict only one conformational state of the protein. The predicted model may or may not be the form which binds its ligand (See Figures 3 and 4). A recent study of enzyme ligand interactions [41] provides evidence that almost two-thirds of enzymes show conformational changes on binding their ligands – which is sometimes local to the binding site or may involve large domain shifts.
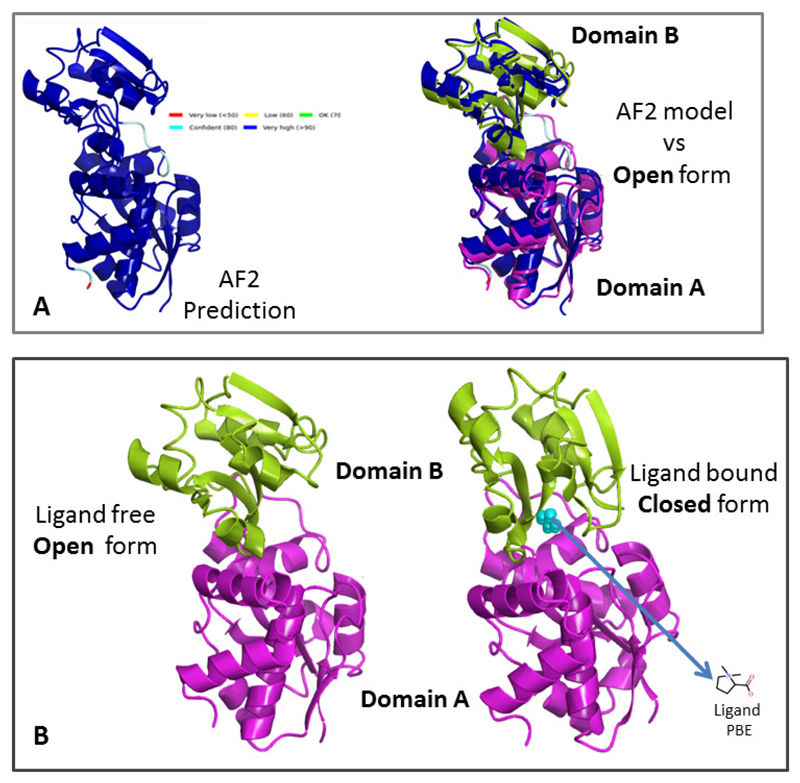
Figure 4. Ligand binding at domain interfaces.
(A) Alphafold2 predicted model (left, pLDDTscore colours) of the multidomain protein ProX from archeoglobus fulgidus. A superposition of this model (blue) with the crystal structure of the ligand free form of the protein (PDB ID 1SW5) shown in ribbon representation (two domains coloured magenta and green) is given on the right. The close match (rmsd 3.8 Å2) between the two structures indicates that the AF2 predicted model corresponds to the open ie ligand free form of the protein.
(B) Superposition of the ligand free open (PDB ID 1SW5, left) and ligand bound closed (PDB ID 1SW1, right) forms of ProX (rmsd 5.1 Å2). The two domains are identified in different colours.The ligand (PDB ID PBE) bound in the closed form of the protein is shown in cyan. Ligand binding causes a hinge-like rotation of Domain B towards Domain A, restricting access to the active site.
As shown in the example in Figure 3, although the AF2 prediction for the protein is ‘confidently predicted’ (as per pLDDT scores) and resembles its unliganded form, data on drug/ligand bound forms of the protein show that the protein alters conformation in order to accommodate ligands at its binding pocket. Therefore, in the absence of observed data, prediction of ligand-induced changes to the protein conformation need to be inferred using other tools such as molecular modelling or protein dynamics [42]. Similarly, the conformational diversity of proteins due to mutations and post translational changes [43] are not reflected in the current AF2 database, While efforts are being made to address this issue [44,45], the ability to identify conformationally variable residues at the ligand binding site or to predict the effect of mutations on the structure of the protein remains difficult.
The success of AF2 [26,46] in various structural biology applications has been mainly due to the local accuracy of the predicted models (pLTTD score). Individual domains are predicted accurately but the connection between these domains is often not determined precisely (Figure 2, Figure 4). This means that for targets which comprise more than one protein domain, AF2 models have to be used with caution. Multi domain proteins often have their ligand binding sites located at domain interfaces and global domain movement occurs on ligand binding (Figure 4). In such a scenario, the single conformer suggested by AF2 is clearly not fit for purpose. The problem is further highlighted when protein-protein interactions (PPIs) [47,48] are targeted, due to “cryptic” binding sites, which, when unoccupied by a ligand, are featureless and the protein pocket is only induced by ligand binding. Current algorithms [13,49,50] are much less accurate in modelling such protein-protein, protein-DNA or protein-RNA models.This poses problems for employing AF2 in antibody discovery, where the low success rate (11% success) of AF2 algorithms predicting correct antigen-antibody complexes precludes the ability of distinguishing across multiple antibodies to identify those that could bind a target in question,[51].
In conclusion, it is worth noting that 90% of clinical drug development fails due to lack of clinical efficacy (40%–50%), toxicity (30%), poor drug-like properties (10%–15%), and/or lack of commercial needs (10%) [52]. While it is true that AF2 predictions will straighten the road for part of the drug discovery process, the projected impact of accurate structure predictions needs to be augmented by the introduction of similar competencies in areas of drug delivery and clinical research in order for more disease modifying drugs to get to the market. Even so, with the further evolution of current machine learning processes in drug discovery and development, the potential of AI techniques to influence medical practices is enormous.
This research was funded in whole by European Molecular Biology Laboratory, which operates a fully open access policy. For the purpose of open access, the author has applied a CC BY public copyright licence to any Author Accepted Manuscript version arising from this submission.
Footnotes
Declaration of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Wouters OJ, McKee M, Luyten J. Estimated Research and Development Investment Needed to Bring a New Medicine to Market, 2009-2018. JAMA. 2020;323:844–853. doi: 10.1001/jama.2020.1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blass BE. In: Basic Principles of Drug Discovery and Development. Second Edition. Blass BE, editor. Academic Press; 2021. The drug discovery process: From ancient times to the present day; pp. 43–110. * Overview of the drug discovery process. [Google Scholar]
- 3.Vincent F, Nueda A, Lee J, Schenone M, Prunotto M, Mercola M. Phenotypic drug discovery: recent successes, lessons learned and new directions. Nat Rev Drug Discov. 2022;21:541. doi: 10.1038/s41573-022-00503-6. [DOI] [PubMed] [Google Scholar]
- 4.Sayers EW, Cavanaugh M, Clark K, Pruitt KD, Schoch CL, Shery ST, Karsch-MizrachiI GenBank. Nucleic Acids Research. 2022;50:D161–D164. doi: 10.1093/nar/gkab1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.The UniProt Consortium. UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Research. 2021;49:D480–D489. doi: 10.1093/nar/gkaa1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goodsell DS, Zardecki C, Di Costanzo L, Duarte JM, Hudson BP, Persikova I, Segura J, Shao C, Voigt M, Westbrook JD, Young JY, et al. RCSB Protein Data Bank: Enabling biomedical research and drug discovery. Protein Sci. 2020;29:52–65. doi: 10.1002/pro.3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ricci-Lopez J, Aguila SA, Gilson GK, Brizuela CA. Improving Structure-Based Virtual Screening with Ensemble Docking and Machine Learning. J Chem Inf and Mod. 2021;61:5362–5376. doi: 10.1021/acs.jcim.1c00511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Muller C, Rabal O, Gonzalez CD. In: Artificial Intelligence in Drug Design Methods in Molecular Biology. Heifetz AHumana., editor. Artificial Intelligence, Machine Learning, and Deep Learning in Real-Life Drug Design Cases. [DOI] [PubMed] [Google Scholar]
- 9.Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Žídek A, Potapenko A, Bridgland A. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–589. doi: 10.1038/s41586-021-03819-2. ** This study presents a deep learning method for protein structure prediction with near experimental accuracy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Varadi M, Anyango S, Deshpande M, Nair S, Natassia C, Yordanova G, Yuan D, Stroe O, Wood G, Laydon A, Žíidek A. AlphaFold Protein Structure Database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Research. 2022;50:D439–D444. doi: 10.1093/nar/gkab1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thornton JM, Laskowski RA, Borkakoti N. AlphaFold heralds a data-driven revolution in biology and medicine. Nat Med. 2021;27:1666–1669. doi: 10.1038/s41591-021-01533-0. [DOI] [PubMed] [Google Scholar]
- 12.Tunyasuvunakool K, Adler J, Wu Z, Green T, Zielinski M, Žídek A, Bridgland A, Cowie A, Meyer C, Laydon A, Velankar S, et al. Highly accurate protein structure prediction for the human proteome. Nature. 2021;596:590–596. doi: 10.1038/s41586-021-03828-1. ** AlphaFold2 protein structure prediction method ([9]**) applied to all proteins of the human proteome. Modelled structures are collated in a freely accessible database. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baek M, DiMaio F, Anishchenko I, Dauparas J, Ovchinnikov S, Lee GR, Wang J, Cong Q, Kinch LN, Schaeffer RD, et al. Accurate prediction of protein structures and interactions using a three-track neural network. Science. 2021;373:871–876. doi: 10.1126/science.abj8754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haley B, Roudnicky F. Functional Genomics for Cancer Drug Target Discovery. Cancer Cell. 2020;38:31–43. doi: 10.1016/j.ccell.2020.04.006. [DOI] [PubMed] [Google Scholar]
- 15.Kim H, Kim E, Lee I, Bae B, Park M, Nam H. Artificial Intelligence in Drug Discovery: A Comprehensive Review of Data-driven and Machine Learning Approaches. Biotechnol Bioprocess Eng. 2020;25:895–930. doi: 10.1007/s12257-020-0049-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Attwood MM, Jonsson J, Rask-Andersen M, Schiöth H. Soluble ligands as drug targets. Nat Rev Drug Discov. 2020;19:695–710. doi: 10.1038/s41573-020-0078-4. [DOI] [PubMed] [Google Scholar]
- 17.Goulet A, Cambillau C, Roussel A, Imbert I. Structure Prediction and Analysis of Hepatitis E Virus Non-Structural Proteins from the Replication and Transcription Machinery by AlphaFold2. Viruses. 2022;14:1537–1551. doi: 10.3390/v14071537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mirdita M, Schütze K, Moriwaki Y, Heo L, Ovchinnikov S, Steinegger M. ColabFold: Making protein folding accessible to all. Nature Methods. 2022;19:679–682. doi: 10.1038/s41592-022-01488-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goodsell DS, Sanner MF, Olson AJ, Forli S. The AutoDock suite at 30. Protein Sci. 2021;30:31–43. doi: 10.1002/pro.3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Armstrong DR, Berrisford JM, Conroy MJ, Gutmanas A, Anyango S, Choudhary P, Clark AR, Dana JM, Deshpande M, Dunlop R, et al. PDBe: improved findability of macromolecular structure data in the PDB. Nucleic Acids Res. 2020;48(D1):D335–D343. doi: 10.1093/nar/gkz990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Kempen M, Kim S, Tumescheit C, Mirdita M, Gilchrist CLM, Söding J, Steinegger M. Foldseek: fast and accurate protein structure search. bioRxiv. 2022 doi: 10.1038/s41587-023-01773-0. Foldseek: fast and accurate protein structure search . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hekkelman ML, de Vries I, Joosten RP, Perrakis A. AlphaFill: enriching the AlphaFold models with ligands and co-factors. bioRxiv. :2021.11.26.470110. doi: 10.1038/s41592-022-01685-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pushpakom S, Iorio F, Eyers PA, Escott KJ, Hopper S, Wells A, Doig A, Guilliams T, Latimer J, McNamee C, Norris A, et al. Drug repurposing: progress, challenges and recommendations. Nat Rev Drug Discov. 2019;18:41–58. doi: 10.1038/nrd.2018.168. [DOI] [PubMed] [Google Scholar]
- 24.Jourdan JP, Bureau R, Rochais C, Dallemagne P. Drug repositioning: a brief overview. J Pharm Pharmacol. 2020;72:1145–1151. doi: 10.1111/jphp.13273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu L, Salmen W, Sankaran B, Lasanajak Y, Smith DF, Crawford SE, Estes MK, Venkataram Prasad BV. Novel fold of rotavirus glycan-binding domain predicted by AlphaFold2 and determined by X-ray crystallography. Commun Biol. 2022;5:419–427. doi: 10.1038/s42003-022-03357-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mosalaganti M, Obarska-Kosinska A, Siggel M, Turonova B, Zimmerli C, Buczak K, Schmidt FH, Margiotta E, Mackmull M, Hagen W, Hummer G, et al. AI-based structure prediction empowers integrative structural analysis of human nuclear pores. Science. 2022;376:1178.:eabm9506. doi: 10.1126/science.abm9506. *Applicaton of accurate AI-based protein models to resolve a cryo-EM puzzle consisting of hundreds of proteins. [DOI] [PubMed] [Google Scholar]
- 27.Fontana P, Dong Y, Pi X, Tong AB, Hecksel CW, Wang L, Fu TM, Bustamante C, Wu H. Structure of cytoplasmic ring of nuclear pore complex by integrative cryo-EM and AlphaFold. Science. 2022;376:1176.:eabm9326. doi: 10.1126/science.abm9326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liebeschuetz JW, Cole JC, Korb O. Pose prediction and virtual screening performance of GOLD scoring functions in a standardized test. J Comput Aided Mol Des. 2012;26:737–748. doi: 10.1007/s10822-012-9551-4. [DOI] [PubMed] [Google Scholar]
- 29.Clark JJ, Orban ZJ, Carlson HA. Predicting binding sites from unbound versus bound protein structures. Sci Rep. 10:15856. doi: 10.1038/s41598-020-72906-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Akdel M, Pires DEV, Pardo EP, Jänes J, Zalevsky AO, Mészáros B, Bryant P, Good LL, Laskowski RA, et al. A structural biology community assessment of AlphaFold 2 applications. Nat Struct Mol Biol. 2022;29:1056–1067. doi: 10.1038/s41594-022-00849-w. * Evaluation of Alphafold 2 predictions in structural biology processes including the estimation of ligand binding sites. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Laskowski RA, Watson JD, Thornton JM. Protein function prediction using local 3D templates. J Mol Biol. 2005;351:614–626. doi: 10.1016/j.jmb.2005.05.067. [DOI] [PubMed] [Google Scholar]
- 32.Kuhn M, Firth-Clark S, Tosco P, Mey ASJS, Mackey M, Michel J. Assessment of Binding Affinity via Alchemical Free-Energy Calculations. J Chem Inf Model. 2020:3120–3130. doi: 10.1021/acs.jcim.0c00165. [DOI] [PubMed] [Google Scholar]
- 33.Tang H, Jensen K, Houang E, McRobb FM, Bhat S, Svensson M, Bochevarov A, Day T, Dahlgren MK, Bell JA, Frye L, et al. Discovery of a Novel Class of d-Amino Acid Oxidase Inhibitors Using the Schrödinger Computational Platform. Journal of Medicinal Chemistry. 2022;65(9):6775–6802. doi: 10.1021/acs.jmedchem.2c00118. [DOI] [PubMed] [Google Scholar]
- 34.Das S, Scholes HM, Sen N, Orengo C. CATH functional families predict functional sites in proteins. Bioinformatics. 2021;37:1099–1106. doi: 10.1093/bioinformatics/btaa937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sillitoe I, Bordin N, Dawson N, Waman VP, Ashford P, Scholes HM, Pang CSM, Woodridge L, Rauer C, Sen N, Abbasian M, et al. CATH: increased structural coverage of functional space. Nucleic Acids Res. 2021;49:D266–D273. doi: 10.1093/nar/gkaa1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zięba A, Stępnicki P, Matosiuk D, Kaczor AA. What are the challenges with multitargeted drug design for complex diseases? Expert Opinion on Drug Discovery. 2022;17:673–683. doi: 10.1080/17460441.2022.2072827. [DOI] [PubMed] [Google Scholar]
- 37.de Beer TAP, Berka K, Thornton JM, Laskowski RA. PDBsum additions. Nucleic Acids Res. 2014;42:D292–D296. doi: 10.1093/nar/gkt940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wishart DS, Feunang YD, Guo AC, Lo EJ, Marcu A, Grant JR, Sajed T, Johnson D, Li C, Sayeeda Z, Assempour N, et al. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 2018;46:D1074–82. doi: 10.1093/nar/gkx1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bryda EC. The Mighty Mouse: the impact of rodents on advances in biomedical research. Mo Med. 2013;110:207–11. [PMC free article] [PubMed] [Google Scholar]
- 40.Pehlivanovic B, Dina F, Emina A, Ziga Smajic N, Fahir B. Animal models in modern biomedical research. Eur J Pharm Med Res. 2019;6:35–38. [Google Scholar]
- 41.Riziotis IG, Ribeiro AM, Borkakoti N, Thornton JM. Conformational Variation in Enzyme Catalysis: A Structural Study on Catalytic Residues. Journal of Molecular Biology. 2022;434:167517. doi: 10.1016/j.jmb.2022.167517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miller MD, Phillips GN. Moving beyond static snapshots: Protein dynamics and the Protein Data Bank. JBiolChem. 2021;296:100749. doi: 10.1016/j.jbc.2021.100749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kumar A, Narayanan V, Sekhar A. Characterizing Post-Translational Modifications and Their Effects on Protein Conformation Using NMR Spectroscopy. Biochemistry. 2020;59:57–73. doi: 10.1021/acs.biochem.9b00827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.del Alamo D, Sala D, Mchaourab HS, Meiler J. Sampling the Conformational Landscapes of Transporters and Receptors with AlphaFold2. eLife. 2022;11:e75751. doi: 10.7554/eLife.75751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stein RA, Mchaourab HS. Modeling Alternate Conformations with Alphafold2 via Modification of the Multiple Sequence Alignment. bioRxiv. 2021 [Google Scholar]
- 46.Jones DT, Thornton JM. The impact of AlphaFold2 one year on. Nat Methods. 2022;19:15–20. doi: 10.1038/s41592-021-01365-3. [DOI] [PubMed] [Google Scholar]
- 47.Lu H, Zhou Q, He J, Jiang Z, Peng C, Tong R, Shi J. Recent Advances in the Development of Protein-Protein Interactions Modulators: Mechanisms and Clinical Trials. Signal Transduct Target Ther. 2020;5:213. doi: 10.1038/s41392-020-00315-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kuzmanic A, Bowman GR, Juarez-Jimenez J, Michel J, Gervasio FL. Investigating Cryptic Binding Sites by Molecular Dynamics Simulations. Acc Chem Res. 2020;53:654–661. doi: 10.1021/acs.accounts.9b00613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Evans R, O’Neill M, Pritzel A, Antropova N, Senior A, Green T, Žídek A, Bates R, Blackwell S, Yim J, et al. Protein complex prediction with AlphaFold-Multimer. bioRxiv. 2021 [Google Scholar]
- 50.Bryant P, Pozzati G, Elofsson A. Improved prediction of protein-protein interactions using AlphaFold2. Nat Commun. 2022;13:1265. doi: 10.1038/s41467-022-28865-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yin R, Feng BY, Varshney A, Pierce BG. Benchmarking AlphaFold for Protein Complex Modeling Reveals Accuracy Determinants. Protein Science. 2022;31:e4379. doi: 10.1002/pro.4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sun D, Gao W, Hu H, Zhou S. Why 90% of clinical drug development fails and how to improve it? Acta Pharmaceutica Sinica B. 2022;7:3049–3062. doi: 10.1016/j.apsb.2022.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schrödinger Release 2022-3. Maestro. Schrödinger, LLC; New York, NY: 2021. communMS Combi. [Google Scholar]
- 54.The PyMOL Molecular Graphics System, Version 12r3pre. Schrödinger, LLC; [Google Scholar]