Abstract
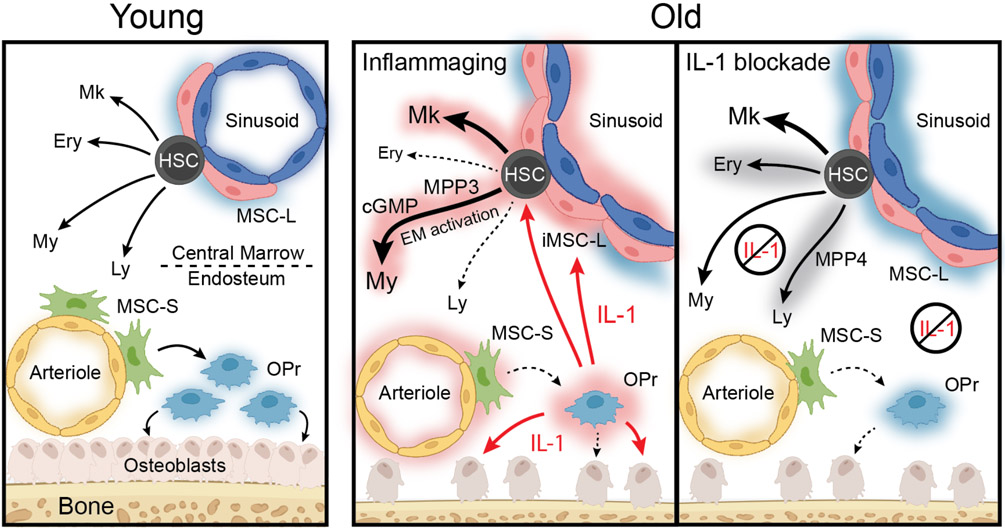
Hematopoietic aging is marked by a loss of regenerative capacity and skewed differentiation from hematopoietic stem cells (HSC) leading to impaired blood production. Signals from the bone marrow (BM) niche tailor blood production, but the contribution of the old niche to hematopoietic aging remains unclear. Here, we characterize the inflammatory milieu that drives both niche and hematopoietic remodeling. We find decreased numbers and functionality of osteoprogenitors (OPr) at the endosteum and expansion of central marrow LepR+ mesenchymal stromal cells (MSC-L) associated with deterioration of the sinusoidal vasculature, which together create a degraded and inflamed old BM niche. Niche inflammation, in turn, drives chronic activation of emergency myelopoiesis pathways in old HSCs and multipotent progenitors (MPP), which promotes myeloid differentiation and hinders hematopoietic regeneration. Moreover, we show how production of IL-1β by the damaged endosteum acts in trans to drive the proinflammatory nature of the central marrow with damaging consequences for the old blood system. Remarkably, niche deterioration, HSC dysfunction, and defective regeneration can all be ameliorated by blocking IL-1 signaling. Our results demonstrate that targeting IL-1 as a key mediator of niche inflammation is a tractable strategy to improve blood production during aging.
Introduction
Hematopoietic function declines with age, causing anemia, impaired adaptive immunity, and cancer1. Aging also leads to changes in bones and the bone marrow (BM) cavity, which provide specialized niches for hematopoietic stem and progenitor cells (HSPC)2-5. Blood production is tailored by differential production of multipotent progenitor (MPP) subsets with specific myeloid (MPP2, MPP3) or lymphoid (MPP4) lineage biases by hematopoietic stem cells (HSC), that give rise to all different lineage-restricted progenitors and mature cells6,7. Niche cells and secreted factors such as pro-inflammatory cytokines regulate HSC behavior, modulate stress hematopoiesis, and play context-dependent roles in leukemogenesis2,3,7,8. Circulating pro-inflammatory cytokines increase over time and contribute to declining tissue integrity with age9,10. Bones already display characteristic aging features by mid-life (~50 years in humans/~12 months in mice)11. In contrast, hematopoietic aging is not fully pronounced until later in life (~70 years in humans/~20-24 months in mice) and is marked by dysfunctional HSCs with reduced engraftment and blood production capabilities4. Many cell-intrinsic deregulations drive old HSC dysfunction4, which are not reverted by systemic rejuvenation interventions12. It is now well-appreciated that niche degradation contributes to hematopoietic aging4,5, but less is known about the crosstalk between old niche and old blood system in driving specific aging features.
The BM niche is comprised of many cell types including mesenchymal stromal cells (MSC) and their osteolineage cell (OLC) derivatives like osteoprogenitor cells (OPr), endothelial cells (EC), the sympathetic nervous system, adipocytes, megakaryocytes (Mk) and macrophages2,3. The marrow cavity is also partitioned between the endosteum and central marrow, with each compartment exhibiting distinct cellular compositions and functions in regulating hematopoiesis2-4. The BM niche has been extensively analyzed by imaging and single-cell molecular profiling13-16, with recent studies described age-related changes in the vasculature17,18, innervation19,20 and specific stromal populations21,22, as well as reporting heightened niche inflammation levels20,23,25. However, the molecular underpinnings of niche aging are still not fully understood. Here, we comprehensively dissect the aged BM niche in mice to determine its contribution to hematopoietic aging, and investigate stromal-derived interleukin-1 (IL-1) as a key targetable factor for anti-aging interventions.
Results
Aging remodels the BM niche
We first surveyed bones from young and old mice. Hematoxylin and eosin (H&E) staining showed accumulation of Mks with age that was accompanied by elevated TGF-β and TPO in BM fluids (Extended Data Fig. 1a,b), consistent with the known Mk bias of old HSCs26,27. In contrast, we only observed irregular accumulation of adipocytes, suggesting that aging in mice is not consistently associated with marrow adipogenesis as in humans. Micro-computed tomography (μCT) highlighted the decay of trabecular and cortical bones28, which corresponded to reduced bone-lining osteoblasts (Extended Data Fig. 1c,d), and confirmed the stereotypical changes associated with aging in mice.
We next employed a refined flow cytometric method29 to investigate in the same animal endosteal niche populations using bone chips, and central marrow niche populations using flushed BM plugs (Fig. 1a; Supplementary Fig. 1a). Regarding the vasculature, while the frequency, number and preferential endosteal location of CD31hi/Sca-1hi arteriolar ECs (AEC) was not affected with age, we observed a reduction in CD31lo/Sca-1lo sinusoidal EC (SEC) frequency and numbers both in the central marrow and at the endosteum in old mice (Fig. 1b). This contrasts with previous imaging studies17, and our own whole mount immunofluorescence analyses showing enhanced branching and dysmorphia of the old central marrow sinusoids (Fig. 1c), indicating poor recovery of old SECs for flow analyses and suggesting fragilization with age. Moreover, we confirmed unchanged vascular volume17,18 and found leakage of injected Dragon Green beads (DGB) into the BM cavity associated with decreased DGB endocytosis in old SECs (Extended Data Fig. 1e,f), which was consistent with vascular leakiness rather than increased transcytosis. Accordingly, age-related changes in vascular stiffness and BM hypoxia are known to contribute to degraded endothelial integrity30.
Figure 1 ∣. Remodeled BM microenvironment with age.
a, Experimental scheme to investigate endosteal (Endo) and central marrow (Marrow) stromal populations in young (~ 8 week of age) and old (~ 24 months of age) C57BL/6 wild type (WT) mice. Results are from 6 independently analyzed groups of 1 or 2 young or old mice. Cellularity data per mouse are for 1 BM plug for marrow populations and bone chips from 10 bones for endosteal populations. b, Changes in endosteal and marrow endothelial cell (EC) populations with age showing frequency (top) and total cell numbers (bottom). AEC, arteriolar endothelial cell; SEC, sinusoidal endothelial cell. c, Whole mount staining of the BM vasculature in young and old mice. Scale bar, 100 μm. d, Changes in endosteal and marrow mesenchymal populations with age showing frequency (top) and total cell numbers (bottom). MSC, Sca-1+ peri-arteriolar mesenchymal stroma cell; mOPr, PDGFRα+ multipotent osteoprogenitor; OPr, PDGFRα− osteoprogenitor; MSC-L, LepR+ peri-sinusoidal mesenchymal stroma cell; iMSC-L, Sca-1low inflammatory MSC-L. e, Representative image of MSC-L immunofluorescence staining in young and old BM. Scale bar, 38 μm. f, Representative pictures and quantification of fibroblast colony-forming units (CFU-F) from young and old MSC-S, osteoblastic lineage cell (OLC) and MSC-L. Results are from 5 independent cohorts. Data are means ± S.D.; P-values were obtained by two-tailed Welch’s t-test without adjustment for multiple comparisons.
Regarding the mesenchymal populations, old mice showed decreased endosteal periarteriolar Sca-1+ MSCs (MSC-S) and their OLC derivatives, both multipotent PDGFRα+ OPr (mOPr) and more committed PDGFRα− OPr (Fig. 1d). Conversely, in the central marrow, we found increased perisinusoidal LepR+ MSCs (MSC-L) and the emergence of an inflammatory Sca-1int MSC-L subset (iMSC-L) (Fig. 1d,e). Similar iMSC-L cells also arise following poly (I:C) treatment (Supplementary Fig. 1b), confirming that MSC-L can upregulate Sca-1 upon inflammatory exposure. Functional assessment revealed loss of fibroblastic colony-forming unit (CFU-F) potential and calcium deposition from old endosteal MSC-S and OLC populations, but no change in old central marrow MSC-L CFU-F potential (Fig. 1f; Extended Data Fig. 1g), in line with recent findings that age-related impairment of osteogenesis originates in endosteal MSCs31. We confirmed the cell-intrinsic nature of old MSC-S functional decline since both young and old BM cells could stimulate young MSC-S growth, but young BM cells could not rescue old MSC-S growth in co-culture experiments (Extended Data Fig. 1h). Finally, the changes in endosteal populations were already observed in 13-month-old middle-aged mice (Extended Data Fig. 1i), consistent with human aging where bone degradation precedes hematopoietic aging11. Collectively, these results reveal profound remodeling of the old niche, with loss and functional deterioration of endosteal mesenchymal populations, and expansion of inflammatory MSC-L alongside decaying sinusoids in the central marrow. They also indicate that the expanded MSC-L compartment does not compensate for age-related bone loss, likely due to their adipogenic bias32,33 and loss with age of an osteogenic subset22.
Abnormal inflammatory signaling in old BM niche cells
To understand the molecular changes driving niche remodeling, we performed plate-based single cell RNA sequencing (scRNAseq) on defined mesenchymal and endothelial populations from endosteum and central marrow of young and old mice. Uniform manifold approximation and projection (UMAP) representation confirmed preservation of the overall niche structure with age (Extended Data Fig. 2a). Further visualization of mesenchymal populations indicates that endosteal MSC-S/mOPr/OPr exist along a continuum of cell states distinct from marrow MSC-L, and revealed contamination of old endosteal populations by marrow MSC-L (Fig. 2a). Iterative clustering and guide-gene selection (ICGS) identified 16 cell clusters, with M1-2 associated with MSC-L identity, M3-5 with OPr identity, and M6-16 with MSC-S identity (Fig. 2b). While most clusters had similar contribution from young and old cells, two consisted overwhelmingly of old cells (M2 and M9) and 4 contained mostly young cells (M3, M4, M12 and M15). M3/M4/M12/M15 expressed many osteoblastic and chondrogenic genes (Supplementary Table 1), supporting a loss of osteogenic cells with age. M3/M4 also expressed the highest levels of collagens and extracellular matrix genes, identifying OPr-like cells as a more mature osteogenic population (Fig. 2b; Supplementary Table 1). M9 contained exclusively old cells with downregulated bone formation pathways, while M2 was dominated by old cells with reduced MSC-L identity gene and high adipogenic gene expression (Fig. 2b; Extended Data Fig. 2b; Supplementary Table 1). Many isolated old endosteal mOPr (45%) and OPr (58%) displayed M1 or M2 cluster identity gene expression, indicating they were MSC-L that were sorted as OLC due to reduced LepR surface expression with age (Extended Data Fig. 2c). Grouping clusters into MSC-L-like, OPr-like and MSC-S-like identity groups showed unaltered expression of HSC maintenance factors Kitl and Cxcl12 between young and old cells, with numerically expanded MSC-L likely contributing to elevated SCF production in old BM fluids (Extended Data Fig. 2d). Finally, gene set enrichment analysis (GSEA) of differentially expressed genes (DEG) between young and old cells within each identity group highlighted activation of inflammatory signaling pathways in all old mesenchymal populations (Extended Data Fig. 2e, Supplementary Table 2). These analyses demonstrate that the loss of endosteal populations and expansion of marrow MSC-L are more pronounced than quantified by flow cytometry due to surface marker infidelity in old MSC-L. They also uncover a loss of osteoblastic/chondrogenic commitment and chronic activation of inflammatory signaling that permeates the mesenchymal hierarchy, providing a molecular basis for the deterioration of the old niche.
Figure 2 ∣. Molecular characterization of the old BM niche.
a, UMAP visualization of plate-based scRNAseq analyses of mesenchymal (left) and endothelial (right) populations isolated from young (n = 3) and old (n = 4) mice. b, ICGS output of young and old mesenchymal populations with 16 clusters of cells (M1 to M16, horizontal) defined according to the expressing pattern of the 6 clusters of genes (A to F, vertical). Examples included in gene clusters A to F are shown. c, UMAP visualization of droplet-based scRNAseq analyses of endosteal and central marrow stroma fractions isolated from young (n = 2) and old (n = 1) mice. OLC, osteolineage cell; OPr(cart), cartilagenic osteoprogenitor; OPr(fibr), fibrogenic osteoprogenitor; Prc, pericyte.
UMAP visualization of endothelial populations separated marrow ECs (mostly SECs by index sorting data) from endosteal ECs (dominated by AECs) regardless of age (Fig. 2a). ICGS identified 7 clusters, with E3-4 associated with AEC-like identity and E7 with SEC-like identity (Extended Data Fig. 3a, Supplementary Table 1). Ingenuity Pathway Analysis (IPA) of DEGs between young and old populations revealed repression of extracellular matrix genes and activation of biosynthesis pathways in old AEC-like cells (Extended Data Fig. 3b; Supplementary Table 1). In contrast, old SEC-like cells exhibited broad downregulation of signaling pathways, decreased expression of endocytosis genes, and activation of cell death pathways. These molecular alterations likely underpin old SEC dysfunction and loss of vascular integrity with age.
Finally, we complemented these analyses with droplet-based scRNAseq of CD45−/Ter-119− endosteal and marrow stroma from young and old mice. Combined UMAP representation was indexed using a published dataset13 and annotated using projections of identity groups from our plate-based scRNAseq dataset (Supplementary Fig. 2-4). This approach confirmed the degradation of the endosteal niche with reduction in MSC-S and disappearance of many OLC and OPr subsets, the damaged vasculature with a loss of EC clusters, and the expansion of inflammatory MSC-L in old mice (Fig. 2c; Extended Data Fig. 3c). Collectively, these results provide a molecular atlas of the old BM niche and complement our flow cytometric and functional assays in demonstrating the loss of endosteal niche populations, vascular fragilization, and inflammatory remodeling of central marrow MSC-L.
Niche-mediated inflammation in the old BM milieu
To investigate the establishment of an inflamed BM milieu, we used a 200-plex array to screen cytokines in BM fluids from young and old mice (Fig. 3a). 81 analytes were detected at both ages, with 32 upregulated and 11 downregulated with age (Supplementary Table 3). The largest group represented inflammatory factors, some previously reported to increase with age such as RANTES23, Eotaxin (CCL11)33, and IL-124,25,34,35. The next group comprised endothelial cell ligands like ICAM-1 and E-selectin, whose increased levels indicate vascular activation and inflammation36. Others were involved in bone37 and fat38 production and function, likely reflecting loss of endosteal OPr and expanded adipogenic marrow MSC-L39. Cytokine bead-array measurements confirmed the pro-inflammatory state of the old BM milieu, with significantly elevated IL-1α/β, MIP1α, and TNFα levels (Fig. 3b). Although old BM fluid cytokines overlap with senescence-associated secretory pathway cytokines40, we did not detect senescence-related programs in old mesenchymal populations (Extended Data Fig. 4a; Supplementary Table 2), and old endosteal MSC-S and OLC were negative for senescence-associated β-galactosidase (SA-β-gal) staining (Extended Data Fig. 4b). These results reveal significant changes in secreted factors, complementing previous studies reporting inflammation in the aged niche20,23-25.
Figure 3 ∣. Inflammatory nature of the old BM niche.
a, Differentially secreted cytokines in young and old BM fluid measured by 200-plex array and clustered based on biological functions (n = 5). b, Changes in pro-inflammatory cytokines in young and old BM fluids. Results are from 3 independent cohorts. c, qRT-PCR-based analyses of Il1a, Il1b and Tnf expression in old BM cells and stromal fractions (n = 6). Results are represented as violin plots and are expressed as fold change compared to their respective young counterpart. Neu, Mac-1+/Ly-6G+/Ly-6Cmid neutrophil; Mac, Mac-1+/Ly-6G−/Ly-6C− macrophage; B cell, B220+/CD19+ B cell; T cell, CD4+/TCRβ+ T cell; Marrow, Ter119−/CD45− central marrow fraction; Endo, Ter119−/CD45− endosteal fraction. d, Violin plots representation of Il1a, Il1b, and Il1rn expression in the indicated young and old mesenchymal and endothelial like groups. Results are from the plate-based scRNAseq dataset; *padj ≤ 0.05, ** padj ≤ 0.01, *** padj ≤ 0.001. e, Caspase 1 activity (aCASP1) in the indicated young and old BM and stromal populations with representative FACS plots (top) and quantification (bottom). f, Experimental scheme for the indicated co-culture experiments showing the effects of young and old stroma on young HSC expansion as dependent on IL-1β. Data are means ± S.D.; P-values were obtained by two-tailed Welch’s t-test without adjustment for multiple comparisons (b) (c) (e), by two-tailed Student’s t-test adjusted for multiple comparisons using the Benjamini-Hochberg method (d), or by one-way Anova adjusted for multiple comparisons using Tukey’s method (f).
To determine which old niche populations contribute to marrow inflammation, we screened BM resident cells including neutrophils, macrophages, B cells and CD4+ T cells (Supplementary Fig. 5a) as well as CD45−/Ter-119− endosteal and central marrow stroma for Il1a, Il1b and Tnf expression by qRT-PCR. Strikingly, only endosteal stroma showed increased Il1b and Tnf expression with age, with plate-based scRNAseq confirming increased Il1b expression in endosteal OPr and AEC as well as marrow MSC-L (Fig. 3c,d; Extended Data Fig. 4c). While BM resident macrophages were previously identified as producing IL-1β in old mice24, caspase 1 (CASP1) activity, indicative of pro-IL-1β being processed into mature IL-1β, was only increased in old mOPr (Fig. 3e). To confirm that increased stromal IL-1β affects HSC behavior, we co-cultured young HSCs on confluent MSC-S/OLC with or without IL-1β blocking antibody. Consistent with the known impact of IL-1 on HSCs41, old stroma increased HSC expansion, an effect that was abrogated upon IL-1β blockade (Fig. 3f). These results identify endosteal mesenchymal cells as a relevant source of IL-1β with age.
Inflammatory remodeling of the old blood system
To investigate the consequences of niche inflammation, we first compared hematopoiesis in young and old mice (Fig. 4a; Extended Data Fig. 4d,e,f; Supplementary Fig. 5b). Old mice displayed increased white blood cells (WBC), unchanged red blood cells (RBC), and increased platelets (Pt) in the peripheral blood, mirroring the accumulation of Mks and Mk progenitors (MkP) in the BM24,27. BM resident myeloid cells and committed myeloid progenitors remained similar with age, while common lymphoid progenitors (CLP) and B cells showed strikingly diminished levels. Hematopoietic aging has long been characterized by an expanded Lin−/c-Kit+/Sca-1+ (LSK) compartment, which contains HSCs and all MPPs6. This reflects a massive expansion of CD34−/CD41+ Mk-biased HSCs that express additional Mk-lineage markers like P-selectin and von Willebrand Factor (vWF)26,42, as well as increased myeloid-biased MPP2 and MPP3, and decreased lymphoid-biased MPP443 (Fig. 4a and Extended Data Fig. 4f,g). To gain further insight into the HSPC compartment, we also performed droplet-based scRNAseq of LSK and Lin−/c-Kit+ (LK) BM from young and old mice. Combined UMAP representation was indexed with marker gene expression for cluster identification44,45 (Supplementary Fig. 6) and used to determine differentiation trajectories from HSCs, with MPP3 and MPP4 contributing to myeloid-primed MPP(my) prior to GMP commitment, and MPP2 sitting atop the bifurcation between MkP and erythroid progenitors (EryP) (Fig. 4b; Extended Data Fig. 4h). This confirmed HSC/MkP expansion and MPP4 contraction in old mice (Fig. 4c). Lastly, we used IPA Upstream Regulators to identify extracellular factors driving alterations in HSCs and MPPs with age, which identifed the strongest IL-1β signature in old HSCs (Extended Data Fig. 4i). These data confirm several established features of hematopoietic aging and demonstrate that inflammatory remodeling begins at the level of HSCs.
Figure 4 ∣. Activation of emergency myelopoiesis pathways in the old blood system.
a, Representative flow cytometry plots (top) and quantification (bottom) of HSCs and MPP populations in young and old mice. Results are from 4 independent cohorts with data expressed are means ± S.D; **p ≤ 0.01, ***p ≤ 0.001. b, UMAP visualization of droplet-based scRNAseq analyses of Lin−/c-Kit+ (LK) and Lin−/Sca-1+/c-Kit+ (LSK) BM fractions isolated from young (n = 2) and old (n = 1) mice. stHSC, short-term HSC; MPP(my), myeloid-primed MPP; GMP, granulocyte/macrophage progenitor; EryP, erythroid progenitor; MkP, megakaryocyte progenitor; CLP, common lymphoid progenitor; BcP, B cell progenitor. c, Quantification of the HSPCs and progenitors (Prog.) identified by droplet-based scRNAseq analyses. Results are expressed as percent LSK and LK, respectively. d, GSEA results for Reactome pathway analyses of isolated young and old MPP3 and MPP4 analyzed by microarray (FDR < 0.05; n = 3 per population). e, Pearson correlation of Fluidigm gene expression data comparing old and regenerative HSCs, MPP3 and MPP4 (n = 4 per population). Regenerating populations were isolated 2 and 3 weeks following young HSC transplantation (Tplx)6. f, GMP immunofluorescence staining in young and old BM. Representative of 2 independent experiments. Stars indicate self-renewing GMP patches. Scale bar, 60 μm. g, GSEA results for Gene Ontology analyses of affected pathways in old populations identified by droplet-based scRNAseq analyses. Data are means ± S.D.; P-values were obtained by two-tailed Welch’s t-test without adjustment for multiple comparisons (a) or Kolmogorov-Smirnov test (g); nd, not detected; ns, non-significant.
We next investigated the functional behavior of old MPP populations. Old MPP3 showed reduced clonogenic potential in methylcellulose and accelerated myeloid differentiation in liquid culture, whereas old MPP4 had elevated myeloid colony-forming activity reflecting of their myeloid bias (Extended Data Fig. 5a,b). All old HSPC populations exhibited delayed B cell differentiation in OP9/IL-7 co-cultures (Extended Data Fig. 5c), however, none of the old MPPs exhibited delayed replication46 or dampened apoptosis47 characteristic of old HSC dysfunction (Extended Data Fig. 5d,e). Furthermore, transplantation and short-term lineage-tracking indicated reduced reconstitution activity of old MPP3 and myeloid bias from every old HSPC population (Extended Data Fig. 5f). These results confirm altered differentiation potential with age, with enhanced myelopoiesis at the expense of lymphopoiesis reminiscent of engagement of emergency myelopoiesis programs in old MPPs7,48. They also indicate that old MPPs do not share key cell-intrinsic hallmarks of HSC aging4.
We next extended our published microarrays to include old HSPCs6. GSEA of old MPP4 showed upregulation of cell cycle and DNA repair genes, while old MPP3 overexpressed innate immunity and myeloid differentiation genes (Fig. 4d; Supplementary Table 4). Fluidigm analyses comparing aging and regeneration6 highlighted the strong correlation between old and 3-week post-transplantation MPP3 and MPP4, supporting constitutive activation of emergency myelopoiesis programs (Fig. 4e). Consistently, we observed self-renewing GMP patches in the old BM (Fig. 4f), which were never found in young mice absent a regenerative challenge48. Finally, GSEA of our scRNAseq confirmed enhancement of myeloid/megakaryocyte and reduction of erythroid/lymphoid commitment (Fig. 4g). They also highlighted differential metabolic/cell cycle activation and a global increase in ribosomal protein expression in old MPPs. These results illustrate profound alterations to hematopoiesis and constitutive emergency myelopoiesis engagement with age, consistent with chronic niche inflammation.
Impaired hematopoietic recovery in old mice
To understand how the old blood system responds to stress, we subjected young and old mice to myeloablative 5-fluorouracil (5FU) treatment. Old mice were exquisitely sensitive to 5FU with ~ 50% mortality likely due to massive thrombosis (Fig. 5a). Blood analyses revealed overproduction of WBCs and Pts, and persistent anemia, while BM analyses confirmed increased myelopoiesis with decreased B cell production (Fig. 5b,c,d). We also observed an amplified response to 5FU in old HSCs with increased and delayed MPP and myeloid progenitor activation, delayed GMP cluster differentiation, and impaired erythroid progenitor (EryP) production (Fig. 5e,f). Cytokine quantification indicated exacerbated niche inflammation in 5FU-treated old mice, with elevated production of IL-1α, IL-1β and MIP-1α (Fig. 5g). These results suggest that by constitutively activating emergency myelopoiesis in old HSCs, age-related niche inflammation compromises acute hematopoietic regeneration.
Figure 5 ∣. Impaired hematopoietic regeneration in old mice.
a-b, Summary of (a) survival and (b) blood regeneration in young and old mice following one 5FU injection. Results are from 3 independent cohorts started with 20 individual young and old mice injected with 5FU per group. c-d, Regeneration of (c) myeloid and (d) B cell populations post-5FU treatment of young and old mice with quantification of changes in the blood (left) and BM (right). e, Regeneration of the indicated BM progenitor populations post-5FU treatment of young and old mice. f, Representative image of GMP immunofluorescence staining at 12 days (D12) post-5FU treatment of young and old mice. Dotted lines indicate differentiating GMP clusters and stars self-renewing GMP patches. Representative of 3 independent experiments. Scale bar, 60 μm. g, Changes in IL-1α and IL-1β levels in BM fluids post-5FU treatment of young and old mice. Data are means ± S.D. except for CBC data shown in (b), (c) and (d) (± S.E.M.); P-values were obtained by log-rank test (a) or by two-tailed Student’s t-test without adjustment for multiple comparisons (b-g); *p ≤ 0.05.
Chronic IL-1β exposure promotes niche aging
Chronic inflammation produced by daily injections of IL-1β for 20 days in young mice is known to mimics features of blood aging including impaired HSC engraftment41 (Fig. 6a). We found that chronic IL-1β also phenocopies several effects of aging on niche cells, with expansion of a dysmorphic sinusoidal network in the central marrow, SEC loss upon isolation, and decreased endosteal MSC-S/OLC (Fig. 6b,c,d). In addition, chronic IL-1β induced changes in bone and reduction in OLC potency that mimic aging (Fig. 6e,f,g). These results indicate that chronically elevated IL-1 is sufficient to promote aging features of both niche and blood system.
Figure 6 ∣. Modulating IL-1 levels affects aging parameters.
a-g, Pro-aging effects of chronic IL-1β exposure in young mice with: (a) experimental scheme (left) and CBC values (right); (b) representative staining of the BM vasculature (scale bar, 100 μm); (c) changes in SEC frequency (left) and dragon green beads (DGB) retention (right); (d) changes in the indicated endosteal mesenchymal populations; (e) representative μCT images of trabecular bone; (f) quantification of μCT parameters; and (g) results of CFU-F assays. Results are from 4 independent cohorts of young and old mice injected daily with either PBS or IL-1β for 20 days. h-k, Pro-regenerative effects of acute IL-1 signaling blockade with Anakinra (Ana) in 5FU-treated old mice with: (h) experimental scheme; (i) representative H&E staining of sternum (scale bar, 100 μm); (j) quantification of the indicated BM HSPCs; and (k) quantification of the indicated BM mature populations and blood parameters. Results are from 3 independent cohorts started with 15 young and 11 old mice treated once with 5FU, injected daily with either PBS or Anakinra, and analyzed at day 12 post-5FU treatment. Data are means ± S.D.; P-values were obtained by two-tailed Student’s t-test without correction for multiple comparisons.
Improved regeneration upon acute IL-1 blockade
To determine whether reducing IL-1 signaling could revert some aging features, we administered human IL-1 receptor antagonist Anakinra (Ana) for 14 days in young and old mice (Extended Data Fig. 6a). We used this short-term exposure to limit immune rejection, and did not investigate an effect on BM niche cells given the slow turn-over of stromal populations29,49. We found negligible impact on steady-state hematopoiesis, with minor changes in HSPC composition but no effect on HSC function in transplantation experiments (Extended Data Fig. 6b,c; Supplementary Fig. 5c). We next investigated whether dampening IL-1 signaling could improve hematopoietic regeneration. Strikingly, daily treatment with Ana before and after 5FU injection dampened IL-1α/β levels and improved regeneration specifically in old mice (Fig. 6h,i ; Extended Data Fig. 6d), with increased B cell and RBC production accompanied by an expansion of MPP4 and Pre-MegE/CFU-E in 5FU-treated old mice (Fig. 6j,k). Ana treatment did not impair myeloid recovery from 5FU treatment, which contrast with genetic ablation of IL-1 signaling41, but also did not ameliorate age-related thrombocytosis (Fig. 6k; Extended Data Fig. 6e). Collectively, these results demonstrate that short-term IL-1 blockade improves hematopoietic regeneration in old mice.
Delayed aging upon long-term IL-1 inhibition
To test the necessity of IL-1 signaling in promoting aging we analyzed young and old Il1r1−/− mice together with age-matched Il1r1+/+ WT controls. Old Il1r1−/− mice displayed a more youthful BM niche, with limited loss of endosteal mOPr and expansion of marrow MSC-L occurring without iMSC-L induction (Fig. 7a). ScRNAseq analyses confirmed these observations and highlighted a complete rebalancing from the inflammatory MSC-L1 subset to a youthful MSC-L2 subset with decreased inflammatory signaling and normalized metabolism (Fig. 7b; Extended Data Fig. 6f). Old Il1r1−/− mice also showed reduced blood aging by immunophenotyping and scRNAseq analyses. No myeloid cell expansion, B cell loss, or anemia was observed in the blood of old Il1r1−/− mice, and despite persistence of an expanded HSC compartment, we found attenuated MPP3 expansion with diminished myeloid cell production and limited Pre-MegE loss in old Il1r1−/− BM (Fig. 7c). A youthful rebalancing of MkP and EryP and trend towards MPP4 maintenance was confirmed at the molecular level (Extended Data Fig. 6g,h). While this occurred with only modest reversion in lineage commitment pathways, old Il1r1−/− HSPCs showed recovered OXPHOS activity as well as youthful ribosomal protein expression (Fig. 7d; Extended Data Fig. 7a,b), and, most importantly, significantly improved regeneration potential in transplantation assays (Fig. 7e). To confirm the role of age-related stromal IL-1 signaling in driving enhanced myelopoiesis, we also transplanted young BM cells into young or old WT and Il1r1−/− recipient mice. Remarkably, the increased donor myelopoiesis observed in old WT recipients was completely rescued in old Il1r1−/− recipients (Fig. 7f; Extended Data Fig. 7c). Taken together, these results demonstrate that life-long blockade of IL-1 signaling delays niche aging and ameliorates specific aspects of blood aging. They also indicate that increased IL-1β production by endosteal stroma acts in trans on marrow MSC-L and affects blood production by old HSPCs, a conclusion supported by strong Il1r1 expression on stromal MSC-L and BM MPP4 (Extended Data Fig. 7d). The specificity of IL-1 in driving age-related niche inflammation was also substantiated by unaltered aging features in old Tnf−/− mice (Extended Data Fig. 7e,f,g,h,i). Collectively, these results offer a proof-of-concept for blocking specific niche-derived chronic inflammatory signals to improve blood production and hematopoietic regeneration with age.
Figure 7 ]. Blocking IL-1 signaling delays niche aging and improves old blood parameters.
a-b, Improved stromal aging features in old Il1r1−/− mice with: (a) color scheme (left) as well as endosteal (right) and central marrow (bottom) mesenchymal population frequencies; and (b) UMAP visualization of droplet-based scRNAseq analyses of endosteal and central marrow stromal fractions in young (n = 2) and old (n = 2) Il1r1+/+ wild type (WT) mice and old (n = 1) Il1r1−/− mice with quantification of percent of the different mesenchymal and endothelial populations (right). c-e, Delayed blood aging and improved HSC function in old Il1r1−/− mice with: (c) blood (top) and BM (bottom) parameters, (d) Rps29 expression in the droplet-based scRNAseq of old WT/Il1r1−/− LK/LSK dataset; and (e) engraftment over time (left) and lineage reconstitution (right) at 4 months (4mo) post-transplantation (Tplx) of the indicated HSC populations. Results are from 3 independent cohorts of young and old WT and age-matched Il1r1−/−, with HSCs isolated from the pooled BM of mice of the same genotype and transplanted into 3 to 5 recipients, each. f, Experimental scheme and donor myeloid cell output in young and old WT and Il1r1−/− recipients transplanted with young BM at 4 mo post-Tplx. Data are means ± S.D. except for engraftment results shown in (e) (± S.E.M.); P-values were obtained by two-tailed Student’s t-test without adjustment for multiple comparisons (a) and (c) or by one-way Anova followed by uncorrected Fisher’s LSD test (e) and (f).
Discussion
Chronic inflammation is a hallmark of aging but its consequences for tissue function remain unclear10. Here, we demonstrate that BM aging is defined by niche remodeling, increased IL-1β production by dysfunctional stromal cells, and activation of inflammatory response programs in both hematopoietic and niche cells (Extended Data Fig. 8). This chronic, low-grade inflammation directly contributes to the loss of endosteal mesenchymal populations, impaired osteogenesis, and vascular dysfunction. These changes, alongside expanded inflammatory MSC-L, drive lineage biases and regenerative defects from the old blood system. Niche inflammation also activates emergency myelopoiesis pathways in HSPCs, reinforcing myeloid cell production at the expense of lymphoid and erythroid commitment. This blunts regenerative responses relying on acute activation of these pathways, causing exacerbated phenotypes following stress in old mice.
A reduction of endosteal niches and expansion of neurovascular central marrow niches promoting megakaryopoiesis through increased production of pro-inflammatory cytokines, including IL-6 and IL-1β, has been previously described20. In addition, a role for IL-1β produced by aged macrophages has been implicated in promoting megakaryopoiesis24, and for IL-1β produced by myeloid cells in response to the aging microbiome in promoting myelopoiesis25. Here, we show that chronic IL-1β production by endosteal stromal cells acts in trans to induce inflammatory remodeling of marrow MSC-L and contributes to many aspects of altered blood production with age, in particular chronic engagement of emergency myelopoiesis. Conversely, blocking IL-1 signaling attenuates central marrow MSC-L niche inflammation and dampens HSC activation, recovering some differentiation biases and improving acute regenerative potential. We also found that blocking TNFα, another candidate mediator of these effects50 does not prevent niche or blood aging, highlighting the central role of IL-1.
Our findings add to the growing body of evidence for microenvironmental inflammation in driving blood aging, and the specific importance of IL-1β as a driver of this process4,5,20,24,25. They establish IL-1 as a central factor that damages the crosstalk between niche and blood system, with inflammatory remodeling of the central marrow likely having deleterious consequences for innervation19,20 and vascular function17,18. Consistently, IL-1β levels correlated with age-related mortality in human studies51. They indicate a potential application of IL-1 inhibitors to improve blood production in the elderly, especially in regenerative settings following chemotherapy or immunosuppression.
Methods
Mice.
All animal experiments were conducted at UCSF or CUIMC in accordance with IACUC protocols approved at each institution, and in compliance with all relevant ethical regulations. Young and old wild-type C57BL/6-CD45.2 (B6) and wild-type C57BL/6-CD45.1 (BoyJ) mice of both sexes were bred and aged in house. Some old B6 mice were also obtained from the National Institute on Aging (NIA) and from collaborators. β-actin–GFP C57BL/6-CD45.2 mice6, Il1r1−/− C57BL/6-CD45.2 mice41 and Tnf−/− C57BL/6-CD45.2 mice51 were previously described. At the time of analyses, young mice were 6-12 weeks old, middle-aged mice were 13 months old, and old mice were 18-31 months old. BoyJ recipient mice for transplantation assays were 8-12 weeks old at time of irradiation. No specific randomization or blinding protocol was used with respect to the identity of experimental animals, and both male and female animals were used indiscriminately in all experiments. Animal facilities were maintained at 71 ± 2°F and 50% ± 10% relative humidity on a 12/12 hour light/dark cycle. Mice were euthanized by CO2 asphyxiation followed by cervical dislocation.
In vivo assays.
For Dragon Green Bead (DGB; Bangs Laboratories, FSDG001) analyses, mice were injected retro-orbitally with 2.5 μl/g DGB solution under isoflurane anesthesia 10 min prior to euthanasia and immediately perfused with 20 ml PBS by cardiac puncture before bone harvest. For 5-fluorouracil (5FU; Sigma-Aldrich) treatment, mice were injected intraperitoneally with 150 mg/kg 5FU or vehicle (PBS). For chronic IL-1 treatment, mice were injected intraperitoneally with 0.5 μg IL-1β (Peprotech, 211-11B) in 100 μl PBS/0.2% BSA or vehicle (PBS/0.2% BSA) daily for 20 days. For Anakinra treatment, mice were injected intraperitoneally with 10 mg/kg Anakinra (Swedish Orphan Biovitrum AB, 666-58234-07) or PBS daily for 14 days with or without 5FU injection on the third day. For transplantation experiments, CD45.1 recipient mice were exposed to 9 Gy (sub-lethal) or 11 Gy (lethal) irradiation delivered in split doses 3 hours apart using either a 137Cs source (J. L. Shepherd, UCSF) or an X-ray irradiator (MultiRad225, Precision X-Ray Irradiation, CUIMC), and purified HSCs and MPPs were delivered via retro-orbital injection. For transplantation into sub-lethally irradiated recipients, mice were injected with 2,000-5,000 purified CD45.2 HSCs or MPPs. For transplantation in lethally irradiated CD45.1 recipients, mice were injected with 250 CD45.2 HSCs delivered together with 300,000 Sca-1-depleted CD45.1 helper BM cells. For reverse transplantations in lethally irradiated CD45.2 young and old WT and Il1r1−/− recipients, mice were injected with 2x106 young WT CD45.1 BM cells. Recipient mice were administered polymyxin/neomycin-containing water for 4 weeks following transplantation. Peripheral blood was collected under isoflurane anesthesia via retro-orbital bleeding and dispensed into EDTA-coated tubes (Becton Dickinson) for complete blood count (CBC) analyses using a Genesis (Oxford Science) hematology system. BM analyses were terminal analyses at the time of tissue harvest and BM cellularity was determined using a ViCell automated cell counter (Beckman-Coulter).
Flow cytometry of hematopoietic cells.
BM cells were obtained by crushing leg, arm, and pelvic bones (with sternum and spines for some experiments) in staining media composed of Hanks’ buffered saline solution (HBSS) containing 2% heat-inactivated FBS (Cellgro B003L52). Red blood cells were removed by lysis with ACK (150 mM NH4Cl/10 mM KHCO3) buffer, and single-cell suspensions of BM cells were purified on a Ficoll gradient (Histopaque 1119, Sigma-Aldrich). For HSC and progenitor isolation, BM cells were pre-enriched for c-Kit+ cells using c-Kit microbeads (Miltenyi Biotec, 130-091-224) and an AutoMACS cell separator (Miltenyi Biotec). Unfractionated or c-Kit-enriched BM cells were then incubated with purified rat anti-mouse lineage antibodies (CD3, BioLegend, 100202, 1:100; CD4, eBioscience, 16-0041-82, 1:800; CD5, BioLegend, 100602, 1:800; CD8, BioLegend, 1:800, 100702; CD11b, BioLegend, 101202, 1:800; B220, BioLegend, 103202, 1:800; Gr1, eBioscience, 14-5931-85, 1:800; Ter119, BioLegend, 116202, 1:400) followed by goat anti-rat-PE-Cy5 (Invitrogen, A10691, 1:400) and subsequently blocked with purified rat IgG (Sigma-Aldrich). Cells were then stained with c-Kit-APC-Cy7 (BioLegend, 105826, 1:800), Sca-1-PB (BioLegend, 108120, 1:800) or Sca-1-BV421 (BioLegend, 108128, 1:400), CD150-PE (BioLegend, 115904, 1:400) or CD150-BV650 (BioLegend, 115931, 1:200), CD48-A647 (BioLegend, 115904, 1:800) or CD48-A700 (BioLegend, 103426, 1:400), and Flk2-biotin (eBioscience, 13-1351-85, 1:100) or Flk2-PE (eBioscience, 12-1351-82, 1:100) followed by SA-PE-Cy7 (eBioscience, 25-4317-82, 1:400) for HSC/MPP staining, or together with CD34-FITC (eBioscience, 11-0341-85, 1:50) or CD34-biotin (BioLegend, 119304, 1:50) and FcγR-PerCP-Cy5.5 (eBioscience, 46-0161-82, 1:1600) or FcγR-PE-Cy7 (BioLegend, 101317, 1:800) followed by SA-BV605 (BioLegend, 405229, 1:400) for combined HSC/MPP/myeloid progenitor staining. For quantification of HSC surface marker expression, CD41-FITC (eBioscience, 11-0411-82, 1:100), CD62P-FITC (BD, 561923, 1:400) and vWF-FITC (EMFRET, P150-1, 1:400) were also used. For extended myeloerythroid progenitor staining, unfractionated BM cells were incubated with Lin/PE-Cy5 and then CD41-PE (BD, 558040, 1:100) or CD41-BV510 (BioLegend, 133923, 1:200), FcγR-PerCP-Cy5.5 or FcγR-PE-Cy7, CD150-APC (BioLegend, 115910, 1:400) or CD150-BV650, and CD105-A488 (BioLegend, 120406, 1:400) or CD105-BV605 (BD, 740425, 1:200). For CLP staining, unfractionated BM cells were incubated with Lin/PE-Cy5, then cKit-APC-Cy7, Sca-1-PB, Flk2-biotin, and IL7R-PE (eBioscience, 12-1271-82, 1:100) followed by SA-PE-Cy7. For mature cell analyses, depending on the experiments, BM cells were stained with Mac-1-PE-Cy7 (eBioscience, 25-0112-82, 1:1600), Gr-1-e450 (eBioscience, 57-5931-82, 1:400), B220-APC-Cy7 (eBioscience, 47-0452-82, 1:800), and CD3-FITC (BioLegend, 100306, 1:400). For myeloid cell isolation for qRT-PCR analyses, BM cells were stained with CD3-PE-Cy5 (eBioscience, 15-0031-63, 1:100), B220-PE-Cy5 (eBioscience, 15-0452-82, 1:800), NK1.1-PE-Cy5 (BioLegend, 108716, 1:200), CD11b-FITC (eBioscience, 11-0112-82, 1:800), Ly-6C-APC-Cy7 (BioLegend, 128025, 1:400) and Ly-6G-A700 (BioLegend, 127622, 1:400). For lymphoid cell isolation for qRT-PCR analyses, BM cells were stained with B220-APC-Cy7, CD19-PE (eBioscience, 12-0193-82, 1:800), TCRβ-PE-Cy7 (Invitrogen, 25-5961-80, 1:200), CD4-PE-Cy5 (eBioscience, 15-0041-82, 1:800), and CD8-A700 (Pharmigen, 557959, 1:200). For in vitro differentiation assays, cultured cells were stained with c-Kit-APC-Cy7, Sca-1-PB, Mac-1-PE-Cy7 and FcγR-PE (eBioscience, 12-0161-83, 1:800) for myeloid differentiation, or Mac-1-APC (eBioscience, 17-0112-82, 1:800) and CD19-PB (BioLegend, 115523, 1:400) for lymphoid differentiation on OP9 cells. For HSC chimerism analyses, HSCs were stained with Lin/PE-Cy5, c-Kit-APC-Cy7, Sca-1-BV421, CD150-BV650, CD48-A700, and Flk2-bio followed by SA-BV605, together with CD45.2-FITC (eBioscience, 11-0454-85, 1:400) and CD45.1-PE (eBioscience, 25-0453-83, 1:400). For peripheral blood chimerism analyses, cells were stained with Mac-1-PE-Cy7, Gr-1-e450, B220-APC-Cy7, CD3-APC (eBioscience, 17-0032-82, 1:200), and Ter-119-PE-Cy5 (eBioscience, 15-5921-83, 1:400) together with CD45.2-FITC and CD45.1-PE. Before isolation or analyses, stained cells were resuspended in staining media containing 1 μg/ml propidium iodide (PI) for dead cell exclusion. HSC and progenitor cell isolations were performed on a Becton Dickinson (BD) FACS Aria II (UCSF) or FACS Aria II SORP (CUIMC) using double sorting for purity. Mature cell isolations were performed on a FACS Aria II SORP using single sorting on purity mode. Cell analyses were performed either on the same FACS ARIA, or on a BD LSR II (UCSF), BD Celesta (CUIMC), Bio-Rad ZE5 (CUIMC) or Novacyte Quanteon (CUIMC) cell analyzer. Data collection was performed using FACSDiva (v9) or Everest (v1) and analysis was performed in FlowJo (v9/v10).
Flow cytometry of niche cells.
Endosteal stromal cells were isolated as previously described29. In brief, leg and pelvic bones were gently crushed and washed with HBSS until white. Stromal cells were released by treatment with 3 mg/ml Type I Collagenase (Worthington) in 2 ml HBSS for 1 hr at 37°C with shaking at 110 rpm. Stromal cells were washed with HBSS + 4% FBS and filtered through 70 μm mesh into a polypropylene tube. Isolation of central marrow stromal cells was adapted from a published protocol52 using individual femurs with the femoral head cut off and kneecap removed to expose the growth plate. Intact BM plugs were carefully flushed with HBSS into polypropylene tubes by inserting 1 ml syringes with 22G needles into the growth plate. HBSS was discarded and stromal cells were released by digestion with 3 mg/ml Type I Collagenase (Worthington) in 1 ml HBSS for 10 min twice at 37°C with shaking at 110 rpm. After the second incubation the plugs were resuspended by pipetting and filtered through 70 μm mesh into a new tube. Both endosteal and central marrow stromal cells were stained with Ter119-PE-Cy5, CD45-APC-Cy7 (BD, 557659, 1:400), CD31-PE (BD, 553373, 1:200), Sca-1-A700 (eBioscience, 56-5981-82, 1:800), CD105-BV786 (BD, 564746, 1:200), CD51-BV421 (BD, 740062, 1:100), PDGFRα-PE-Cy7 (eBioscience, 25-1401-82, 1:100) and LepR-biotin (R&D Systems, BAF497, 1:100) followed by SA-APC (eBioscience, 17-4317-82, 1:400), and resuspended in HBSS with 4% FBS and PI. Both cell isolations and analyses were performed on a Becton Dickinson (BD) FACS Aria II (UCSF) or FACS Aria II SORP (CUIMC) using single sorting on purity mode. Data collection was performed using FACSDiva (v9) and analysis was performed in FlowJo (v9/v10).
In vitro assays.
All cultures were performed at 37 °C in a 5% CO2 water jacket incubator (ThermoFisher). For methylcellulose colony assays, single cells were directly sorted into individual wells of a flat-bottom 96-well plate (Fisher Scientific, 353072) containing 100 μl methylcellulose (Stem Cell Technologies, M3231) supplemented with penicillin (50 U/ml)/streptomycin (50 μg/ml), 0.1 mM non-essential amino acids (Fisher Scientific, 11-140-050), 1 mM sodium pyruvate (Fisher Scientific, 11-360-070), 2 mM L-glutamine (Fisher Scientific 35-050-061), 50 μM 2-mercaptoethanol (Sigma, M7522) and the following cytokines (all from PeproTech): IL-3 (10 ng/ml), GM-CSF (10 ng/ml), SCF (25 ng/ml), IL-11 (25 ng/ml), Flt-3L (25 ng/ml), TPO (25 ng/ml) and EPO (4 U/ml). Colonies were visually scored after 7 days of culture. For myeloid differentiation assays, 1,000 cells were directly sorted into individual wells of a 96-well plate containing Iscove’s modified Dulbecco’s media (IMDM) medium (Invitrogen) supplemented with 5% FBS (StemCell Technology, 06200) and the same reagents and cytokines as methylcellulose assays (full cytokine medium). Cells were analyzed by flow cytometry after different culture periods. For cleaved caspase 3/7 (CC3/7) assays, 400-600 cells were directly sorted in triplicate into 40 μl full cytokine medium in a 384-well plate and incubated for 24 hr before adding 40 μl of Caspase-Glo 3/7 (Promega) to each well. Plates were then shaken for 30 s at 500 rpm, incubated for 45 min at RT and read on a luminometer (Synergy2, BioTek) to obtain relative units (RU). OP9 stromal cells (ATCC, CRL-2749) were maintained in Minimum Essential Medium Eagle alpha modification (αMEM) medium (Invitrogen) supplemented with 10% FBS (Cellgro, B003L52), L-glutamine (2 mM), and penicillin (50 U/ml)/streptomycin (50 μg/ml), and split at 1:4 every 3-4 days as needed to avoid over-confluence. For lymphoid differentiation assays, 500 cells were directly sorted into individual wells of a 24-well plate containing 10,000 OP9 stromal cells in OptiMEM medium (Invitrogen) supplemented with 5% FBS (StemCell Technology), L-glutamine (2 mM), penicillin (50 U/ml)/streptomycin (50 μg/ml), 2-mercaptoethanol (50 μM), SCF (10 ng/ml), Flt3L (10 ng/ml), and IL-7 (5 ng/ml). Following sequential withdrawal of Flt3L and SCF upon 2-day intervals, cultures were maintained in IL-7 and analyzed by flow cytometry at various intervals. For CFSE analyses, 1,000-1,500 cells were directly sorted into individual wells of a 96-well plate containing staining media, washed with PBS, resuspended in 100 μl PBS containing 5 μM CFSE (Molecular Probes, C-1157), incubated for 5 minutes at RT, quenched with FBS, incubated for 1 min at RT, washed twice with staining media, resuspended in 200 μl full cytokine medium, incubated for 72 hr and analyzed by flow cytometry. Stromal cells were grown in stroma media consisting of αMEM supplemented with 10% FBS (CellGro), penicillin (50 U/ml)/streptomycin (50 μg/ml), and 2-mercaptoethanol (50 μM). For fibroblastic colony-forming unit (CFU-F) assays, endosteal MSC-S (15-300 cells) or OLC (170-300 cells) were sorted directly in 6-well plates containing 1.5 ml medium/well and cultured for 11 days with medium exchange every 2-3 days, before staining with Giemsa-Wright to score colonies of 25 or more cells. In contrast, marrow MSC-L (100-300 cells) were sorted directly into 6-well plates containing 1.5 ml of Dulbecco’s Modified Eagle Medium (Gibco) supplemented with 20% FBS (CellGro), penicillin (50 U/ml)/streptomycin (50 μg/ml), 2-mercaptoethanol (50 μM), and 10 μM ROCK inhibitor (TOCRIS, Y-27632) and cultured in hypoxic conditions (5% O2) for 8 days with medium exchange every 2-3 days, before staining with Giemsa-Wright to score colonies of 25 or more cells. For Von Kossa staining, 1,000 cells were sorted into stroma media that was supplemented after 2 days of culture with β-glycerol phosphate (3mM, Sigma G9891) and ascorbic acid-2-phosphate (50 μg/ml, Sigma A8950). Cells were grown to confluence with media changes every 2-3 days, fixed in phosphate-buffered formalin (Fisher 23-245-684), stained with silver nitrate solution (1%, Electron Microscopy Sciences 26212-01) and rinsed with sodium thiosulfate (5%, Ricca Chemical R7866500), with PBS washes between steps, before imaging. For co-culture assays with β-actin-GFP MSC-S, 25 cells were directly sorted into 96-well plates containing 100 μl medium/well, then 1x105 unfractionated BM cells in 100 μl medium were added and co-cultures were incubated for 10 days without medium replacement. On day 10, medium was aspirated, and adherent cells were washed twice with PBS, liberated with 50 μl 0.25% trypsin/EDTA (Thermo, 25200056), and counted by flow cytometry using 25 μl Absolute Counting Beads (Life Technologies, C36950). For co-culture assays with endosteal CD45−/Ter119−/CD31−/CD51+ cells (MSC-S and OLC gates combined), 1,000 cells were sorted directly into 50% DMEM/50% αMEM supplemented with 20% FBS, penicillin (50 U/ml)/streptomycin (50 μg/ml), 2-mercaptoethanol (50 μM), and 10 μM ROCK inhibitor and grown to confluence for 7-10 days with medium changed every 2-3 days. Upon reaching confluence, half the medium was replaced with IMDM supplemented with 5% FBS (Stem Cell Technologies), penicillin (50 U/ml)/streptomycin (50 μg/ml), 0.1 mM non-essential amino acids, 1 mM sodium pyruvate, 2 mM L-glutamine, 50 μM 2-mercaptoethanol, 50 ng/ml SCF, 50 ng/ml TPO, and either 20 μg/ml anti-mouse/rat IL-1β (Bio Cell, BE0246) or 20 μg/ml isotype control (Armenian hamster IgG, Bio Cell, BE0091), and 100 β-actin-Gfp HSCs were seeded on top of this stromal layer. Co-cultures were grown for 7 days with media replacement every 2 days before cell counting and flow cytometry analysis. For SA-β-Gal (Cell Signaling, 9860S) staining, MSC-S and OLC were sorted directly onto poly-L-lysine coated slides (Sigma, P0425) and stained according to manufacturer’s protocol. For assessing Caspase 1 activity, analysis was performed using the FAM-FLICA Caspase-1 (YVAD) Assay Kit (ImmunoChemistry Technologies, 97) according to manufacturer’s instructions.
Bone analyses.
Bones for hematoxylin/eosin (H&E) staining were processed by the Mouse Pathology Core Facility at UCSF or the Molecular Pathology Core Facility at CUIMC. Tibia for MicroCT analyses were fixed in 10% neutral buffered formalin for 24 hours, then held in 70% ethanol at 4°C until further processing. CT scans were performed on a vivaCT40 MicroCT (Scanco; 55 kV x-ray energy, 10.0 μm voxels, 500 ms integration times) and bone densitometry analyses were performed as previously described29. Trabecular tissue volume (TV), mineralized bone volume (BV), and trabecular bone parameters were determined on 100 slices, starting 500 μm below the bottom edge of the growth plate using segmentation values of 0.5/2/350 which corresponds to 650 mg hydroxyapatite/cm3.
Immunofluorescence staining.
For whole mount imaging, mice were injected retro-orbitally under isoflurane anesthesia with 100 μl of CD31-A647 (BioLegend, 102416, 1:5) and VE-Cadherin-A647 (BioLegend, 138006, 1:5) antibody cocktail in PBS. Injected mice were euthanized 10 min later, perfused with 3 ml PBS and fixed by perfusion of 10 ml 4% paraformaldehyde (PFA) in PBS. Femurs were harvested, fixed in 4% PFA for 2 hr on ice, washed 3x with PBS for 10 min each, and then cleared by successive dehydration with sucrose (15% and then 30%) for either 1 hr or overnight at each step. Bones were then snap-frozen in a 100% ethanol/dry ice slurry and kept at −80 °C until sectioning. One side of the frozen femur was cryosectioned with a tungsten blade (Leica, CM3050 S) until the BM was exposed, then transferred to a 1.5mL Eppendorf tube, washed 3x with PBS at RT and blocked with 20% goat-serum in PBS/0.1% Tween-20 (PBST) overnight at 4°C. Bones were incubated with rabbit anti-mouse laminin (Sigma, L9393, 1:25) in 10% goat-serum in PBST for 3 days at 4°C, washed 3x with PBST, and incubated with goat anti-rabbit A594 (Thermo Fisher, A32740, 1:1600) for 2 days at 4°C. Finally, bones were washed 3x with PBST for 10 min each, mounted on a coverslip with silicone glue, always kept wet with PBS, and imaged on an SP8 inverted confocal microscope (Leica) with 20x objective using Leica Application Suite X (v3/v4) for data collection. For BM section imaging, mouse femurs were embedded in OCT, snap-frozen in a 100% ethanol/dry ice slurry and kept at −80 °C until sectioning. Thin 7 μm sections were obtained upon cryosection using the CryoJane tape transfer system (Leica, 39475205) and a tungsten blade. Sections were dried for 2-4 hr at room temperature and then frozen at −80 °C until stained. Before staining, sections were fixed with 100% acetone for 10 min at −20 °C, dried for 5 min at RT, blocked for 90 min with 10% goat-serum (Gibco) in PBS and washes 3x with PBS. For LepR staining, sections were first stained with LepR-biotin and rabbit anti-mouse laminin, and then with SA-A555 (Thermo Fisher, S21381, 1:400) and goat anti-rabbit A594 followed by blocking with rat IgG for 10 min at RT. Slides were then stained with A488-conjugated lineage markers Ter119 (BioLegend, 116215, 1:200), Mac-1 (BioLegend, 101217, 1:200), Gr-1 (BioLegend, 108417, 1:200), B220 (BioLegend, 103225, 1:200), CD41 (Thermo Fisher, 11-0411-82, 1:100) and CD3 (BioLegend, 100210, 1:100). For ALCAM staining, sections were stained with anti-ALCAM-Bio (R&D systems, BAF1172, 1:200) followed by SA-A555 blocked with rat IgG and then stained with A488-conjugated lineage markers. For laminin staining, sections were stained with rabbit anti-mouse laminin followed by goat anti-rabbit-A647 and DGB fluorescence was detected in the A488 channel. For GMP staining, sections were first stained with rat anti-mouse c-Kit (BioLegend, 135102, 1:25) followed with a goat anti-rat-Cy3 (Jackson ImmunoResearch, 112-165-167, 1:100), and then with A488-conjugated lineage markers, Sca-1-A488 (BioLegend, 108116, 1:50), CD150-A488 (BioLegend, 115916, 1:50) and FcγR-A647 (BioLegend, 101314, 1:50). After staining, all sections were counterstained with 1 μg/ml DAPI in PBS for 10 min at RT, mounted with Fluoromount G (Southern Biotech, 0100-01) and imaged on an SP5 upright confocal microscopes (Leica) with 20x objectives using Leica Application Suite X for data collection. For DGB staining of purified SECs, 2,000 cells were sorted directly onto poly-L-lysine coated slides (Sigma-Aldrich, P0425-72EA) and allowed to settle for 10 min before being fixed with 4% PFA for 10 min at RT, washed and counterstained with 1 μg/ml DAPI in PBS for 10 min at RT, mounted with VectaShield (Vector Laboratories, H-1200) and imaged on an A1 Ti Eclipse inverted microscope (Nikon) with 20x objectives using Nikon NIS Elements (v3) for data collection. Images were processed using Volocity (v6.2, Perkin Elmer) and analyzed with ImageJ (v1). The find objects function in Volocity was used to quantify vascular volume based on Laminin staining.
Cytokine profiling.
Per mouse, BM fluid was flushed out from femurs and tibiae using the same 200 μl of HBSS/2% FBS in a 1 ml syringe with 26G needle. BM cells were sedimented by centrifugation at 300 g for 5 min and supernatants were purified by an additional centrifugation at 15,300 g for 10 min before storage at −80°C until use. For 200-plex cytokine array, BM fluid samples were submitted to Quantibody Testing Service (Raybiotech) and diluted 4x prior to analyses. For bead array analysis, 50 μl of 2x diluted sample was analyzed using Mouse 20-Plex panel (Thermo Fischer Scientific) using either a Magpix (UCSF) or Luminex 200 (CUIMC) analyzer according to manufacturer’s protocol. For ELISA measurements, 4x-diluted (SCF, SDF1a, TPO; Raybiotech) and 90x-diluted (TGF-β; R&D Systems) samples were prepared according to the manufacturer’s instructions and analyzed on a Synergy 2 plate reader (Biotek).
Plate-based scRNAseq.
Samples were processed following modifications to the Smart-Seq2 protocol53 based in the mcSCRB-Seq protocol54. Single endosteal and central marrow stromal cells were directly sorted into individual wells of a 96-well PCR plate in 2.3 μl of lysis buffer containing 0.2% Triton X-100 (Sigma-Aldrich) and 1U of Superase-In RNase Inhibitor (Ambion). Of note, information regarding expression of surface markers was recorded for each cell when sorting. Cells were frozen immediately at −80C until further processing. After thawing on ice, 2 μl of an annealing mixture containing1 μM oligodT (IDT), 5 mM each dNTPs and a 1:6,000,000 dilution of ERCC RNA Spike-In Mix (Invitrogen) was added followed by incubation at 72 °C for 3 min. Then 5.7 μl of a Reverse Transcription mix containing 3.5 U/μl of Maxima H minus retrotranscriptase (ThermoFisher), 0.88 U/μl of Superase-In RNase Inhibitor, 1.75x Maxima RT Buffer, 3.5 μM TSO (Qiagen) and 13.15% PEG 8000 (Sigma-Aldrich) was added and the mixture was incubated at 42 °C for 90 min, followed by 70 °C for 15 min. cDNA was further amplified by adding 40 μl of a PCR mix containing 0.03 U/μl of Terra PCR direct polymerase (Takara Bio), 1.25x Terra PCR Direct Buffer, and 0.25 μM IS PCR primer (ID). PCR was as follows: 3 min at 98 °C for initial denaturation followed by 21 cycles of 15 s at 98 °C, 30 s at 65 °C, 4 min at 68 °C. Final elongation was performed for 10 min at 72 °C. Sequences of oligodT, TSO and IS PCR primers were as previously described53. Following preamplification, all samples were purified using Ampure XP beads (Beckman Coulter) at a ratio of 1:0.6 with a final elution in 25 μl of EB Buffer (Qiagen). The cDNA was then quantified using the Quant-iT PicoGreen dsDNA Assay Kit (Thermo Fisher). Size distributions were checked on high-sensitivity DNA chips (Agilent Bioanalyzer). Samples were used to construct Nextera XT libraries (Illumina) from 100 pg of preamplified cDNA. Libraries were purified and size selected (0.5x-0.7x) using Ampure XP beads. Then, libraries were quantified using KAPA qPCR quantification kit (KAPA Biosystems), pooled and sequenced in an Illumina HiSeq 4000. Reads were mapped to the Mus musculus genome (GRCm38.p4; https://www.ncbi.nlm.nih.gov/assembly/GCF_000001635.24/) and ERCC sequences using GSNAP (version 2015-09-29) with parameters: -A sam -B 5 -t 24 -n 1 -Q -N 1. HTseq-count55 was used to count reads mapped to each gene, with parameters: -s no. All cells with <100,000 reads mapping to endogenous RNA and >20% reads mapping to mitochondrial genes were considered low quality and removed from downstream analyses. Overall, 661 cells (67.38%) passed our quality controls distributed as follows: Young marrow EC (SEC), 70; Old marrow EC (SEC), 71; Young endosteal EC (AEC), 84; Old endosteal EC (AEC), 82; Young MSC-L, 53; Old MSC-L, 58; Young MSC-S, 59; Old MSC-S, 59; Young mOPr, 26; Old mOPr, 26; Young OPr, 44; Old OPr, 29. The mean of nuclear reads mapped per cell was 471,244. Data were normalized and highly variable genes were identified as previously described56, using a false discovery rate threshold equal to 0.1 for the chi-squared test. Only highly variable genes were considered to perform PCA analysis, using the prcomp function in R (v3.6.3). UMAP was calculated using umap function (v0.2.7) in R based on normalized expression value with 15 nearest neighbors. All genes were used for ICGS clustering (AltAnalyzer, v2.0)57 of mesenchymal and endothelial populations, using ‘stringent’ with cell cycle and all other parameters as default. For mesenchymal populations, 16 clusters of cells (denoted M1 to M16) and 6 clusters of genes (denoted A to F) were defined. For endothelial populations, 7 clusters of cells (denoted E1 to E7) and 6 clusters of genes (denoted a to g) were defined. For subsequent analysis, mesenchymal clusters of cells were pooled as follows: M1 and M2 for “MSC-L-like”; M3, M4 and M5 for “OPr-like”; M6 to M16 for “MSC-S-like”. Subsequent endothelial groups of cells were defined as follows: clusters E3 and E4 constituted “AEC-like”; cluster E7 “SEC-like”. For MSC-L gene identity comparison, the geometric mean of the expression of all genes contained in cluster A was calculated for each cell of clusters M1 and M2 and then clusters M1 and M2 were compared using the independent two-sided t test (p-value < 2.2e-16). Differential expression analysis of old and young cells within each group was performed using DESeq2 (v1.26.0)58. For violin plots representation, normalized expression values of Kitl, Cxcl12, Il1a, Il1b and Il1rn were plotted in various groups for young and old cells using ggplot2 (v3.3.0) in R with scale parameter set to ‘width’. Pathway analysis was conducted using Gene Set Enrichment Analysis (GSEA) software (v4.1.0). Gene symbols were mapped to MSigDB.v7.2.chip and overlaps with Hallmark (h.all.v7.2.symbols.gmt) gene sets were determined using the classic scoring scheme. For endothelial cell types, no FDR<0.05 significant enrichments were obtained using GSEA, and so Ingenuity Pathway Analyses software (v12-2020, Qiagen) was employed, where log 2 fold change of expression and p-adjusted values for all genes with nominal p values under 0.05 were imported into the software and IPA Canonical Pathways were determined.
Droplet-based scRNAseq for stroma analyses.
For both endosteal and central marrow populations, 30,000-50,000 Ter119−/CD45− stromal cells were sorted into 1.5 ml tubes containing 500 μl of filter-sterilized αMEM with 10% FBS and transferred to the Columbia Genome Center Single Cell Analysis Core for microfluidic cell processing, library preparation and sequencing. In brief, cells were re-counted, and viability was assessed using a Countess II FL Automated Cell Counter (Thermo), and samples were processed following manufacturer’s recommendations for Chromium Single Cell 3′ Library & Gel Bead Kit v2 (10X Genomics). 17,500 cells were loaded for each sample and 1 sample was loaded per condition. Samples were sequenced in an Illumina HiSeq4000 to obtain an average ~286 million reads per sample and alignment was performed using Cellranger (v2.1.1/v5.0.1). Top 2,300 barcodes were selected for samples corresponding to young and old central marrow and young endosteum. Top 2,858 barcodes were selected for the sample corresponding to old endosteum. The downstream analysis was done using Scanpy version (v1.6.0 and v1.7.0, Wolf et al., 2018) in Python (v3.7.1). For these samples, 757 doublets were estimated and removed using Scrublet (v0.2.1)59. Further quality control (QC) was performed based on 3 parameters: 1) cells with at least 200 and no more than 7,000 genes detected; 2) cells with less than 65,000 associated counts; 3) cells with less than 5.5% of UMI counts associated to mitochondrial genes. After QC, 8,735 cells were kept for subsequent analysis. In addition, only genes that have more than 1 UMI count in at least 5 cells were maintained in further analysis. Cells were then normalized to 10,000 UMIs per cell and logarithmically transformed. Highly variable genes (HVGs) were selected using “highly_variable_genes” method with “min_mean=0.0175, max_mean=3, min_disp=0.5”. Read depth, number of genes and number of mitochondrial counts were removed using the “regress_out” function. UMAP visualizations were obtained from 50 PCA components and 10 neighbors. Clusters were defined using Louvain clustering (v0.7.0) and subseted based on hematopoietic or stromal marker gene expression. Only clusters that contained cells that expressed stromal marker genes were kept for subsequent analysis. In total, 2378 cells were selected, distributed as follows: 188 cells came from the young central marrow sample, 726 cells from the old central marrow sample; 1320 cells came from the young endosteum sample and 144 cells from the old endosteum sample. Highly variable genes (HVGs) were then obtained for the selected cells and the effects of read depth, number of genes and number of mitochondrial counts were regressed, as indicated above. The UMAP visualization was obtained as before and again clusters were defined using Louvain clustering with “resolution 0.3”. Cell types were annotated using typical marker genes for the different populations.
Droplet-based scRNAseq for hematopoietic analyses.
For both LK and LSK populations, 30,000-50,000 cells were sorted into 1.5 ml tubes containing 500 μl of HBSS containing 2% FBS and transferred to the Columbia Genome Center Single Cell Analysis Core for microfluidic cell processing, library preparation and sequencing as described above for stromal analyses. Downstream analyses were done using Seurat (v4.0.3) in R (v4.1.0). Quality control (QC) was performed excluding cells with less than 200 genes detected or more than 7.5% of UMI counts associated to mitochondrial genes. In addition, only genes that have more than 1 UMI count in at least 3 cells were maintained in further analysis. In total, 28535 cells were selected, distributed as follows: 8489 cells came from the young WT sample, 9688 cells from the old WT sample; 10358 cells came from the old Il1r1−/− sample. Cells were then normalized to 10,000 UMIs per cell and logarithmically transformed. Highly variable genes (HVGs) were selected using the “FindVariableFeatures” method. Cell cycle variation was removed using the “CellCycleScoring” method followed by regressing out “S.Score” and “G2M.Score”. Young WT and Old WT samples were integrated using the “IntegrateData” method (Stuart et al., 2019). UMAP visualizations were obtained from 10 PCA components and clusters were defined at a resolution of 0.4 using K-nearest neighbor graph-based. Cell types were annotated using typical marker genes for the different hematopoietic populations. Differential gene expression was performed using “FindMarkers” method on non-cell cycle regressed expression data. Old Il1r1−/− data was analyzed by mapping to the Young WT/Old WT reference dataset using the “MapQuery” method60.
Quantitative RT-PCR analyses.
10,000-50,000 cells per population were sorted directly into RPE buffer (Qiagen) and stored at −80°C until purification with the RNeasy Plus Mini Kit (Qiagen) according to manufacturer’s protocol. Following column purification, RNA was immediately reverse transcribed using SuperScriptIII kit with random hexamers (Invitrogen). qPCR runs were performed on a QuantStudio 7 Flex Real-Time PCR System (Applied Biosystems) using SYBR Green reagents (Applied Biosystems), the cDNA equivalent of 200 cells per reaction template, and triplicate measurement per biological repeat. Sequences for qRT–PCR primers were: Il1a, forward GCACCTTACACCTACCAGAGT and reverse TGCAGGTCATTTAACCAAGTGG (NM_010554); Il1b, forward GCAACTGTTCCTGAACTCAACT and reverse ATCTTTTGGGGTCCGTCAACT (NM_008361); Tnf, forward AGGGATGAGAAGTTCCCAAAT and reverse GCTTGTCACTCGAATTTTGAG (NM_013693); and Gapdh, forward GACTTCAACAGCAACTCCCAC and reverse TCCACCACCCTGTTGCTGTA (NM_008084). Cycle threshold values were normalized to Gapdh.
Fluidigm analyses.
Gene expression analyses using the Fluidigm 96.96 Dynamic Array IFC were performed as previously described6. Briefly, HSC, MPP3 and MPP4 (100 cells/well) were directly sorted per well of a 96-well plate containing 5 μl CellsDirect lysis buffer (Invitrogen, 11753-100), reverse-transcribed and pre-amplified for 18 cycles using SuperScript III Platinum Taq Mix (Invitrogen, 12574-026) with a custom-made set of 96 proprietary target-specific primers (Fluidigm). The resulting cDNA was analyzed on a Biomark system (Fluidigm) using EvaGreen SYBR dye (Bio-Rad, 172-5211). Data were collected with Biomark Data Collection Software (Fluidigm) and analyzed using Biomark qPCR software with a quality threshold of 0.65 and linear baseline correction. Melt curves and melting temperature values for each assay reaction were checked individually, and reactions with melt curves showing multiple peaks or poor quality were discarded. Results for 2- and 3-weeks post-transplantation data were recalculated with Hprt as housekeeping gene and genes not analyzed in one of three conditions were removed, leaving 64-68 genes excluding housekeeping genes (Actb, Gapdh, Gusb and Hprt) for further analyses and calculation of Pearson’s correlation coefficients. For each dataset, values for each gene were averaged, and fold change values were calculated against young non-transplanted control and converted to Log2 value. Similarity matrices were visualized with Morpheus (version-less, Broad Institute).
Microarray analyses.
Microarray analyses of old HSCs, MPP3 and MPP4 were performed as previously described6. Three to five independent biological replicates were used for each population. Total RNA was isolated from 20,000 cells per population sorted directly into TRIzol-LS (Invitrogen) and purified using Arcturus PicoPure (Applied Biosystems) with RNase-free DNase (Qiagen). RNA was amplified, labeled, and fragmented using NuGEN Ovation Pico linear amplification kits (Nugen Technologies) and hybridized onto mouse Gene ST 1.0 arrays (Affymetrix). Gene expression microarray data were normalized using RMA followed by quantile normalization in R (v2.15.1) using a standard (lambda=1) exponential reference distribution. Significance Analysis of Microarrays (SAM) was then performed on young and old cells within a population to determine SAM delta scores61. Pathway analysis was performed using GSEA (v4.1.0) on the top 1000 up and downregulated genes in young versus old for each population. Gene symbols were mapped to MSigDB.v7.2.chip and overlaps with Reactome (c2.cp.reactome.v7.2.symbols.gmt) gene sets were determined using the classic scoring scheme.
Statistics and Reproducibility.
All experiments were repeated as indicated; n indicates the numbers of independent biological repeats. Data are expressed as mean ± standard deviation (S.D.) or standard error of the mean (S.E.M.) as indicated. Mice for treatment and transplantation were assigned to experimental groups based on age and genotype, randomized with respect to sex, samples were alternated whenever possible, and data collection and analysis were not performed blind to the conditions of the experiments. For experiments involving 5-FU treatment, animals that were found moribund at time of analysis were excluded from final presentation. For transplantations, recipients with <1% overall chimerism were excluded from lineage quantification. Approximate sample size was predetermined for most experiments based on our experience in murine HSC aging, where a standard deviation of less than 10% can be expected for most measurements. To show a difference of at least 10%, with a = 0.05 and 80% power, 6 to 11 independent biological replicates are required per group. Larger numbers of replicates were used in certain experiments with high variability such as 5-FU treatment and transplantation. Data were processed using Microsoft Excel (v16) and statistical significance was evaluated with GraphPad Prism (v9). Figures were made with GraphPad Prism. Data distribution was assumed to be normal but this was not formally tested.
Extended Data
Extended Data Figure 1 ∣. Gross analysis of the remodeled old BM cavity.
a, H&E staining of humeri and sterna of 2 individual young and old mice. Scale bar, 100 μm. b, TPO and TGF-β levels in young and old BM fluids. Results are from 2 independent cohorts. c, μCT analyses of young and old tibias with representative images of cortical and trabecular regions (left) and quantification of bone volume/total volume (BV/TV) and connectivity density (right). d, Representative image of bone lining ALCAM+ osteoblasts immunofluorescence staining in young and old mice. Scale bar, 100 μm. e, Representative images and quantification of immunofluorescence staining of vascular volume (laminin) and vascular leakage by dragon-green beads (DGB) diffusion assay in young and old BM. Scale bar, 50 μm. f, Representative images and quantification by flow cytometry of DGB endocytosis in young and old marrow SEC; scale bar, 5 μm. Results are from 3 independent cohorts. g, Representative images of Von Kossa staining in young and old endosteal MSC-S. h, Experimental scheme for the indicated co-culture experiments showing the effects of young or old BM cells on young MSC-S (top right), and young BM cells on young and old MSC-S (bottom). i, Frequency of endosteal and marrow mesenchymal populations in young and middle age (13-month-old) mice with changes in CFU-F from endosteal OLCs (bottom right). Data are means ± S.D; *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001.
Extended Data Figure 2 ∣. Molecular features of old mesenchymal populations.
a, UMAP visualization of the entire plate-based scRNAseq dataset of niche populations of mesenchymal and endothelial populations shown in Fig. 2a. b, Global changes in MSC-L gene identity in clusters M2 vs. M1 (***padj ≤ 0.001). c, Representative flow cytometry staining (top) and quantification of LepR and PDGF-Rα levels (bottom) in young and old MSC-L. Results are from 6 independently analyzed groups of 1 or 2 young or old mice. d, HSC niche factors with violin plots representation of Kitl and Cxcl12 expression in the indicated young and old mesenchymal populations (left) and SCF and SDF1α levels in young and old BM fluids (right). e, GSEA results for Hallmark biological processes significantly affected in old MSC-L-like, OPr-like and MSC-S-like groups (FDR < 0.05). Data are means ± S.D. except for (b) and (d); **p ≤ 0.01.
Extended Data Figure 3 ∣. Molecular features of old endothelial populations and further characteristics of the old niche.
a, ICGS output of young and old endothelial populations from the plate-based scRNAseq dataset shown in Fig. 2a with 7 clusters of cells (E1 to E7, horizontal) defined according to the expressing pattern of the 7 clusters of genes (a to g, vertical). Examples included in gene clusters a to g are shown. Star, contaminating mesenchymal/endothelial doublets; P, pericytes; A, arterioles; T, transition vessels. b, Ingenuity Pathway Analysis (IPA) canonical pathways enriched in old AEC-like and SEC-like groups (Z-score > 1; p < 0.01). c, Characteristic expression patterns for the indicated genes in the droplet-based scRNAseq dataset of young and old endosteal and central marrow stromal fractions shown in Fig. 2c. Cells in the UMAP were colored according to the expression levels of the indicated genes. Color scheme is based on ln scale of normalized counts from 0 (gray) to the indicated maximum value in the scale (red).
Extended Data Figure 4 ∣. Age-related changes in blood and BM populations and altered lineage bias in old MPPs.
a, Overlap between cytokines upregulated in old BM fluids and published SASP profile40. b, Representative SA-β-gal staining in control irradiated mouse embryonic fibroblasts (MEF) and isolated young and old MSC-S and OBC. Scale bar, 20 μm. c, Absolute expression of Il1b in different mature hematopoietic cell types and unfractionated (CD45−/Ter119−) endosteal and central marrow stroma from pooled young and old samples. Results are expressed as −log(dCt) relative to gapdh. d, Complete blood count (CBC) parameters in young and old mice. Results are from 7 independent cohorts. WBC, white blood cell; My, myeloid (neutrophil + basophil + eosinophil); Ly, lymphocyte; RBC, red blood cell; Pt, platelet. e, Cellularity and quantification of mature populations in young and old BM. Results are from 3 independent cohorts. Gr, Mac-1+/Gr-1+ granulocyte; B cell, B220+ B cell; T cell, CD3+ T cell. f, Quantification of progenitor populations in young and old BM. Results are from 3 independent cohorts. CMP, common myeloid progenitor; GMP, granulocyte-macrophage progenitor; MEP, megakaryocyte-erythrocyte progenitor; CLP, common lymphoid progenitor; CFU-E, erythroid colony-forming unit; Pre-GM, pre-granulocyte/macrophage; Pre-MegE, pre-megakaryocyte/erythrocyte; MkP, megakaryocyte progenitor. g, Representative staining and quantification of CD34, CD41, CD62P and vWF levels on young and old HSCs. Results are from 2 independent cohorts, with data represented as box and whiskers (min to max) and expressed as fold changes in mean fluorescence intensity (MFI) relative to young HSCs. h, Characteristic expression patterns of lineage determinant genes in the droplet-based scRNAseq young and old LK/LSK dataset shown in Fig. 4b. Cells in the UMAP were colored according to the expression levels of the indicated genes. Color scheme is based on ln scale of normalized counts from the indicated minimum (gray) to maximum (red) value in the scale. i, IPA Upstream Regulators analysis of young and old populations from the droplet-based scRNAseq young and old LK/LSK dataset shown in Fig. 4b filtered on cytokines and growth factors. Data are means ± S.D. except when indicated; *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001.
Extended Data Figure 5 ∣. Altered functionality of old HSCs and MPPs.
a, Colony formation in methylcellulose for young (Y) and old (O) HSCs, MPP3 and MPP4. Results are from 2 independent experiments. Mix: all lineages; GM: granulocyte/macrophage; G(or)M: granulocyte (or) macrophage; MegE: megakaryocyte/erythrocyte; CFU: colony-forming units. b, Myeloid differentiation in liquid culture for young and old MPP3 with quantification (right) of immature Sca-1+/c-Kit+ cells (left) and mature Mac-1+/FcgR+ macrophage (right). c, Representative flow cytometry staining of CD19+ lymphoid vs. Mac-1+ myeloid differentiation in OP9+IL7 culture conditions for young and old HSCs, MPP3 and MPP4. Results are representative of 3 independent experiments. d, Representative histograms of CFSE staining of cultured young and old HSCs, MPP3 and MPP4. Results are representative of 3 independent experiments. e, Cleaved caspase 3/7 (CC3/7) activity in cultured young and old HSCs, MPP3 and MPP4. f, Short-term lineage tracking following transplantations of young and old HSCs, MPP3 and MPP4 in sub-lethally irradiated recipients with experimental scheme (left) and quantification of overall blood donor chimerism (top graphs) and myeloid chimerism among donor cells (bottom graphs). Results are from 3 independent cohorts. Data are means ± S.D. except for (f) (± S.E.M.); *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001.
Extended Data Figure 6 ∣. Improved aging features with IL-1 signaling blockade.
a-c, Short-term blockade of IL-1 signaling upon Anakinra (Ana) treatment in young (Y) and old (O) mice with: (a) experimental scheme; (b) changes in HPSC frequency; and (c) engraftment over time (left) and lineage reconstitution (right) at 4 months (4 mo) post-transplantation (Tplx) of the indicated HSC populations. Results are from 3 independent cohorts of young and old mice injected with either PBS or Anakinra, with HSCs isolated from the pooled BM of mice from the same treatment group and transplanted into 3 to 5 recipients, each. d-e, Additional characterization of the effects of Anakinra blockade of IL-1 signaling during 5FU-mediated regeneration in young and old mice with: (d) changes in IL-1α, IL-1β and MIP1α levels in BM fluids; and (e) platelet (Plt) levels in peripheral blood. Results are from 3 independent cohorts started with 15 young and 11 old mice treated once with 5FU, injected daily with either PBS or Anakinra, and analyzed at day 12 post-5FU treatment. f, GSEA results for Hallmark biological processes significantly enriched in MSC-L2 vs. MSC-L1 groups (FDR < 0.05). Data are from the droplet-based scRNAseq analyses of endosteal and central marrow stromal fractions in young (n = 2) and old (n = 2) Il1r1+/+ wild type (WT) mice and old (n = 1) Il1r1−/− mice shown in Fig. 7b. g-h, Droplet-based scRNAseq analyses of Lin−/c-Kit+ (LK) and Lin−/Sca-1+/c-Kit+ (LSK) BM fractions isolated from young (n = 2) and old (n = 2) Il1r1+/+ wild type (WT) and old (n = 1) Il1r1−/− mice with (g) UMAP visualization and (h) quantification of percent of HSPCs and progenitors. Data are means ± S.D. except for engraftment results shown in (c) (± S.E.M.); *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001.
Extended Data Figure 7 ∣. Key role of IL-1 in the aging of both BM niche and blood system.
a-b, GSEA results in old Il1r1−/− HSPC population for the Gene Ontology pathways affected in either old WT HSPCs (a) or young WT HSPCs (b) identified by droplet-based scRNAseq analyses. n/a, non-available; ns, non-significant; *nominal p value ≤ 0.05, ** nominal p value ≤ 0.01, *** nominal p value ≤ 0.001. c, Peripheral blood CD45.1+ donor chimerism (left) and number of donor-derived GMPs (right) in young or old WT and Il1r1−/− CD45.2+ recipient mice at 4 months (mo) after lethal irradiation and transplantation (Tplx) with 2x106 young WT CD45.1+ donor BM cells. d, Il1r1 expression in the droplet-based scRNAseq of young and old WT stroma and LK/LSK datasets. Cells in the UMAP were colored according to the expression levels of the indicated genes. Color scheme is based on ln scale of normalized counts from the indicated minimum (gray) to maximum (red) value in the scale. e-i, Unchanged aging features in old Tnf−/− mice with: (e) color scheme; (f) blood parameters; (g) endosteal (left) and central marrow (right) mesenchymal population frequencies; (h) BM hematopoietic population frequencies; and (i) engraftment over time (left) and lineage reconstitution (right) at 4 mo post-Tplx of the indicated HSC populations. Results are from 3 independent cohorts of young and old WT and age-matched Tnf−/− mice, with HSCs isolated from the pooled BM of mice of the same genotype and transplanted into 3 to 5 recipients, each. Data are means ± S.D. except for engraftment results shown in (i) (± S.E.M.); *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001.
Extended Data Figure 8 ∣. Schematic of the crosstalk between the BM niche and hematopoietic system during physiological aging.
In youth, HSCs reside primarily in the central marrow where they are maintained by peri-sinusoidal MSC-L and produce a balanced output of all mature cell lineages (Mk, megakaryocytes; Ery, erythrocytes, My, myeloid cells; Ly, lymphoid cell). Abundant peri-arteriolar MSC-S at the endosteum efficiently produce OPr cells that support osteoblast development, ECM deposition and bone formation. With age, numerical loss and functional decline of MSC-S and OPr leads to bone thinning, with the remaining OPr constitutively producing IL-1. Chronic IL-1, in turn, reinforces niche degradation at the endosteum and contributes to dysfunction of the sinusoidal vasculature. Chronic IL-1 also acts in trans on central marrow MSC-L and HSPCs, driving the appearance of an inflammatory iMSC-L subset and steady-state engagement of emergency myelopoiesis (EM) programs with GMP cluster (cGMP) formation. Strikingly, acute IL-1 blockade with Anakinra enables more youthful blood production during 5FU-mediated regeneration, and life-long removal of IL-1 signaling in Il1r1−/− mice maintains MSC-L in a more youthful cell state associated with improved blood production and HSC function.
Supplementary Material
Supplementary Table 1 ∣ ICGS of young and old niche cells. Plate-based scRNAseq gene expression data of mesenchymal and endothelial cells were separated and subjected to unsupervised single-cell population identification using Iterative Clustering and Guide-gene Selection (ICGS). The relative expression of every guide gene in each cluster, as well as the relative expression of every guide gene in each pooled group of clusters corresponding to the indicated cell types, was calculated by averaging the relative expression of each gene across individual cells within a cluster or group.
Supplementary Table 2 ∣ DEGs and pathway analyses of young versus old niche cells. Plate-based scRNAseq gene expression data between young and old cells in pooled identity groups as defined by ICGS were analyzed for differentially expressed genes (DEG) using the DESeq2 package. Overlap with Hallmark gene sets was evaluated, and statistically significant (FDR<0.05) overlaps are indicated in bold for mesenchymal populations. Since no significant overlaps were observed for endothelial populations, Ingenuity Pathway Analysis software (Qiagen) was employed to determine IPA Canonical Pathways with absolute z-score ≥ 1 and −log10(p-value) ≥ 2 enriched in young and old AEC-like and SEC-like cells.
Supplementary Table 3 ∣ Quantibody array-based measurements of cytokine concentration in young and old BM fluids. BM fluids isolated from young (10 week of age) and old (27-29 months of age) mice (n=5) was analyzed using Quantibody Testing Service (Raybiotech). Samples were diluted 4-fold prior to analyses. Results are mean ± S.D. and are expressed as pg/ml concentration; *p≤0.05, **p≤0.05, ***p≤0.05.
Supplementary Table 4 ∣ DEGs and pathway analyses of young versus old HSCs. Significance Analysis of Microarrays (SAM) was performed on young and old cells within each population to determine SAM delta scores. The top 1000 most highly DEGs for each group were collected. SAM scores for these genes were used for GSEA and overlap with Reactome gene sets was evaluated for each population. The top 5 statistically significant (FDR<0.05) overlaps for each group are in bold.
Acknowledgments.
We thank S. Villeda (UCSF) for providing some old C57Bl/6 mice, D. Reynaud (UCSF) for initial analyses of old mice, A. Valencia (UCSF) for technical assistance with various experiments, S. Kinston (Cambridge) for scRNAseq library preparations, A. Li (Bone Imaging Research Core, UCSF) and both E. Guo and P.T. Shyu (CUIMC) for μCT analyses, M. Lee (UCSF) and M. Kissner (CUIMC) for management of our Flow Cytometry Core facilities, and all members of the Passegué laboratory for critical insights and suggestions. C.A.M was supported by NIH F31HL160207 and a NYSTEM training grant, E.V.V. by a Rubicon Grant from The Netherlands Organization for Scientific Research, a Stem Cell Grant from BD Biosciences, and a NYSTEM training grant, O.C.O by CRI/Margaret Dammann Eisner postdoctoral fellowship CRI3617, J.W.S. by long-term EMBO postdoctoral fellowship ALTF-2021-196, P.V.D by NIH F31HL151140, E.M.P. by NIH F32HL106989 and K01DK09831, S.T.B. by a CIRM postdoctoral fellowship, and T.T.H. by an AHA and Hillblom Center for the Biology of Aging predoctoral fellowships. F.J.C-N., X.W. and B.G. were supported by grants from the Wellcome (206328/Z/17/Z), CRUK (C1163/A21762) and core funding by the Wellcome to the Cambridge Stem Cell Institute. This work was funded by NIH R01CA184014, NIH R35HL135763, Glenn Foundation Research Award and LLS Scholar Award to E.P., and supported in part through the NIH/NCI Cancer Center Support Grant P30CA013696 to CUIMC. The funders of this work had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript. For the purpose of Open Access, the author has applied a CC BY public copyright license to any Author Accepted Manuscript version arising from this submission. Clipart in figures was created using BioRender.
Footnotes
Competing interests. The authors declare no competing financial interests.
Data Availability.
RNA-seq and microarray data that support the findings of this study have been deposited in the Gene Expression Omnibus under accession code GSE169162. Mus musculus genome GRCm38.p4 is available from the National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov/assembly/GCF_000001635.24/). Chip annotation MSigDB.v7.2.chip is available from the Broad Institute (https://software.broadinstitute.org/cancer/software/gsea/wiki/index.php/MSigDB_v7.2_Release_Notes). Gene sets used for GSEA h.all.v7.2.symbols.gmt and c2.cp.reactome.v7.2.symbols.gmt are available from the Broad Institute (https://data.broadinstitute.org/gsea-msigdb/msigdb/release/7.2/). Source data for all the figures are provided with the paper. All other data are available from the corresponding author upon reasonable request.
Code Availability.
All code and packages used to support the findings of this study are either publicly available or available from the corresponding author upon reasonable request.
References
- 1.Rossi DJ, Jamieson CHM & Weissman IL Stems cells and the pathways to aging and cancer. Cell 132, 681–696 (2008). [DOI] [PubMed] [Google Scholar]
- 2.Morrison SJ & Scadden DT The bone marrow niche for haematopoietic stem cells. Nature 505, 327–334 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pinho S & Frenette PS Haematopoietic stem cell activity and interactions with the niche. Nat. Rev. Mol. Cell Biol 20, 303–320 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Verovskaya EV, Dellorusso PV & Passegué E Losing sense of self and surroundings: hematopoietic stem cell aging and leukemic transformation. Trends Mol. Med 25, 494–515 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ho Y-H & Méndez-Ferrer S Microenvironmental contributions to hematopoietic stem cell aging. Haematologica 105, 38–46 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pietras EM et al. Functionally distinct subsets of lineage-biased multipotent progenitors control blood production in normal and regenerative conditions. Cell Stem Cell 17, 35–46 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Olson OC, Kang Y-A & Passegué E Normal hematopoiesis is a balancing act of self-renewal and regeneration. Cold Spring Harb. Perspect. Med 10, a035519 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schepers K, Campbell TB & Passegué E Normal and leukemic stem cell niches: insights and therapeutic opportunities. Cell Stem Cell 16, 254–267 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kovtonyuk LV, Fritsch K, Feng X, Manz MG & Takizawa H Inflamm-aging of hematopoiesis, hematopoietic stem cells, and the bone marrow microenvironment. Front. Immunol 7, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Soysal P, Arik F, Smith L, Jackson SE & Isik AT Inflammation, frailty and cardiovascular disease. in Frailty and Cardiovascular Diseases (ed. Veronese N) vol. 1216 55–64 (Springer International Publishing, 2020). [Google Scholar]
- 11.Almeida M & O’Brien CA Basic biology of skeletal aging: role of stress response pathways. J. Gerontol. A. Biol. Sci. Med. Sci 68, 1197–1208 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ho TT et al. Aged hematopoietic stem cells are refractory to bloodborne systemic rejuvenation interventions. J. Exp. Med 218, e20210223 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baryawno N et al. A Cellular taxonomy of the bone marrow stroma in homeostasis and leukemia. Cell 177, 1915–1932.e16 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tikhonova AN et al. The bone marrow microenvironment at single-cell resolution. Nature 569, 222–228 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baccin C et al. Combined single-cell and spatial transcriptomics reveal the molecular, cellular and spatial bone marrow niche organization. Nat. Cell Biol 22, 38–48 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kokkaliaris KD et al. Adult blood stem cell localization reflects the abundance of reported bone marrow niche cell types and their combinations. Blood 136, 2296–2307 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kusumbe AP et al. Age-dependent modulation of vascular niches for haematopoietic stem cells. Nature 532, 380–384 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Poulos MG et al. Endothelial transplantation rejuvenates aged hematopoietic stem cell function. J. Clin. Invest 127, 4163–4178 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maryanovich M et al. Adrenergic nerve degeneration in bone marrow drives aging of the hematopoietic stem cell niche. Nat. Med 24, 782–791 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ho Y-H et al. Remodeling of bone marrow hematopoietic stem cell niches promotes myeloid cell expansion during premature or physiological aging. Cell Stem Cell 25, 407–418.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guidi N et al. Osteopontin attenuates aging-associated phenotypes of hematopoietic stem cells. EMBO J. 36, 840–853 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shen B et al. A mechanosensitive peri-arteriolar niche for osteogenesis and lymphopoiesis. Nature 591, 438–444 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ergen AV, Boles NC & Goodell MA Rantes/Ccl5 influences hematopoietic stem cell subtypes and causes myeloid skewing. Blood 119, 2500–2509 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frisch BJ et al. Aged marrow macrophages expand platelet-biased hematopoietic stem cells via interleukin-1B. JCI Insight 4, e124213 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kovtonyuk LV et al. IL-1 mediates microbiome-induced inflammaging of hematopoietic stem cells in mice. Blood 139, 44–58 (2022). [DOI] [PubMed] [Google Scholar]
- 26.Grover A et al. Single-cell RNA sequencing reveals molecular and functional platelet bias of aged haematopoietic stem cells. Nat. Commun 7, 11075 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Poscablo DM, Worthington AK, Smith-Berdan S & Forsberg EC Megakaryocyte progenitor cell function is enhanced upon aging despite the functional decline of aged hematopoietic stem cells. Stem Cell Rep. 16, 1598–1613 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Glatt V, Canalis E, Stadmeyer L & Bouxsein ML Age-related changes in trabecular architecture differ in female and male C57BL/6J Mice. J. Bone Miner. Res 22, 1197–1207 (2007). [DOI] [PubMed] [Google Scholar]
- 29.Schepers K et al. Myeloproliferative neoplasia remodels the endosteal bone marrow niche into a self-reinforcing leukemic niche. Cell Stem Cell 13, 285–299 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu X et al. Age-related impairment of vascular structure and functions. Aging Dis. 8, 590 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ambrosi TH et al. Aged skeletal stem cells generate an inflammatory degenerative niche. Nature 597, 256–262 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nishikawa K et al. Maf promotes osteoblast differentiation in mice by mediating the age-related switch in mesenchymal cell differentiation. J. Clin. Invest 120, 3455–3465 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Singh L et al. Aging alters bone-fat reciprocity by shifting in vivo mesenchymal precursor cell fate towards an adipogenic lineage. Bone 85, 29–36 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Villeda SA et al. The ageing systemic milieu negatively regulates neurogenesis and cognitive function. Nature 477, 90–94 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mrak R Interleukin-1, neuroinflammation, and Alzheimer’s disease. Neurobiol. Aging 22, 903–908 (2001). [DOI] [PubMed] [Google Scholar]
- 36.Woollard KJ Soluble bio-markers in vascular disease: much more than gauges of disease? Clin. Exp. Pharmacol. Physiol 32, 233–240 (2005). [DOI] [PubMed] [Google Scholar]
- 37.Abdelmagid SM et al. Mutation in osteoactivin decreases bone formation in vivo and osteoblast differentiation in vitro. Am. J. Pathol 184, 697–713 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Villarroya F, Cereijo R, Villarroya J & Giralt M Brown adipose tissue as a secretory organ. Nat. Rev. Endocrinol 13, 26–35 (2017). [DOI] [PubMed] [Google Scholar]
- 39.Yue R, Zhou BO, Shimada IS, Zhao Z & Morrison SJ Leptin receptor promotes adipogenesis and reduces osteogenesis by regulating mesenchymal stromal cells in adult bone marrow. Cell Stem Cell 18, 782–796 (2016). [DOI] [PubMed] [Google Scholar]
- 40.Coppé J-P, Desprez P-Y, Krtolica A & Campisi J The senescence-associated secretory phenotype: the dark side of tumor suppression. Annu. Rev. Pathol. Mech. Dis 5, 99–118 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pietras EM et al. Chronic interleukin-1 exposure drives haematopoietic stem cells towards precocious myeloid differentiation at the expense of self-renewal. Nat. Cell Biol 18, 607–618 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gekas C & Graf T CD41 expression marks myeloid-biased adult hematopoietic stem cells and increases with age. Blood 121, 4463–4472 (2013). [DOI] [PubMed] [Google Scholar]
- 43.Young K et al. Progressive alterations in multipotent hematopoietic progenitors underlie lymphoid cell loss in aging. J. Exp. Med 213, 2259–2267 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rodriguez-Fraticelli AE et al. Clonal analysis of lineage fate in native haematopoiesis. Nature 553, 212–216 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dong F et al. Differentiation of transplanted haematopoietic stem cells tracked by single-cell transcriptomic analysis. Nat. Cell Biol 22, 630–639 (2020). [DOI] [PubMed] [Google Scholar]
- 46.Flach J et al. Replication stress is a potent driver of functional decline in ageing haematopoietic stem cells. Nature 512, 198–202 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gutierrez-Martinez P et al. Diminished apoptotic priming and ATM signalling confer a survival advantage onto aged haematopoietic stem cells in response to DNA damage. Nat. Cell Biol 20, 413–421 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hérault A et al. Myeloid progenitor cluster formation drives emergency and leukaemic myelopoiesis. Nature 544, 53–58 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Park D et al. Endogenous bone marrow MSCs are dynamic, fate-restricted participants in bone maintenance and regeneration. Cell Stem Cell 10, 259–272 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yamashita M & Passegué E TNF-α coordinates hematopoietic stem cell survival and myeloid regeneration. Cell Stem Cell 25, 357–372.e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Furman D et al. Expression of specific inflammasome gene modules stratifies older individuals into two extreme clinical and immunological states. Nat. Med 23, 174–184 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods-only References
- 52.Zhou BO, Yue R, Murphy MM, Peyer JG & Morrison SJ Leptin-receptor-expressing mesenchymal stromal cells represent the main source of bone formed by adult bone marrow. Cell Stem Cell 15, 154–168 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Picelli S et al. Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc 9, 171–181 (2014). [DOI] [PubMed] [Google Scholar]
- 54.Bagnoli JW et al. Sensitive and powerful single-cell RNA sequencing using mcSCRB-seq. Nat. Commun 9, 2937 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Anders S, Pyl PT & Huber W HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brennecke P et al. Accounting for technical noise in single-cell RNA-seq experiments. Nat. Methods 10, 1093–1095 (2013). [DOI] [PubMed] [Google Scholar]
- 57.Olsson A et al. Single-cell analysis of mixed-lineage states leading to a binary cell fate choice. Nature 537, 698–702 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Love MI, Huber W & Anders S Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wolock SL, Lopez R & Klein AM Scrublet: Computational identification of cell doublets in single-cell transcriptomic data. Cell Syst. 8, 281–291.e9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hao Y et al. Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587.e29 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tusher VG, Tibshirani R & Chu G Significance analysis of microarrays applied to the ionizing radiation response. PNAS. 98, 5116–5121 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1 ∣ ICGS of young and old niche cells. Plate-based scRNAseq gene expression data of mesenchymal and endothelial cells were separated and subjected to unsupervised single-cell population identification using Iterative Clustering and Guide-gene Selection (ICGS). The relative expression of every guide gene in each cluster, as well as the relative expression of every guide gene in each pooled group of clusters corresponding to the indicated cell types, was calculated by averaging the relative expression of each gene across individual cells within a cluster or group.
Supplementary Table 2 ∣ DEGs and pathway analyses of young versus old niche cells. Plate-based scRNAseq gene expression data between young and old cells in pooled identity groups as defined by ICGS were analyzed for differentially expressed genes (DEG) using the DESeq2 package. Overlap with Hallmark gene sets was evaluated, and statistically significant (FDR<0.05) overlaps are indicated in bold for mesenchymal populations. Since no significant overlaps were observed for endothelial populations, Ingenuity Pathway Analysis software (Qiagen) was employed to determine IPA Canonical Pathways with absolute z-score ≥ 1 and −log10(p-value) ≥ 2 enriched in young and old AEC-like and SEC-like cells.
Supplementary Table 3 ∣ Quantibody array-based measurements of cytokine concentration in young and old BM fluids. BM fluids isolated from young (10 week of age) and old (27-29 months of age) mice (n=5) was analyzed using Quantibody Testing Service (Raybiotech). Samples were diluted 4-fold prior to analyses. Results are mean ± S.D. and are expressed as pg/ml concentration; *p≤0.05, **p≤0.05, ***p≤0.05.
Supplementary Table 4 ∣ DEGs and pathway analyses of young versus old HSCs. Significance Analysis of Microarrays (SAM) was performed on young and old cells within each population to determine SAM delta scores. The top 1000 most highly DEGs for each group were collected. SAM scores for these genes were used for GSEA and overlap with Reactome gene sets was evaluated for each population. The top 5 statistically significant (FDR<0.05) overlaps for each group are in bold.
Data Availability Statement
RNA-seq and microarray data that support the findings of this study have been deposited in the Gene Expression Omnibus under accession code GSE169162. Mus musculus genome GRCm38.p4 is available from the National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov/assembly/GCF_000001635.24/). Chip annotation MSigDB.v7.2.chip is available from the Broad Institute (https://software.broadinstitute.org/cancer/software/gsea/wiki/index.php/MSigDB_v7.2_Release_Notes). Gene sets used for GSEA h.all.v7.2.symbols.gmt and c2.cp.reactome.v7.2.symbols.gmt are available from the Broad Institute (https://data.broadinstitute.org/gsea-msigdb/msigdb/release/7.2/). Source data for all the figures are provided with the paper. All other data are available from the corresponding author upon reasonable request.
All code and packages used to support the findings of this study are either publicly available or available from the corresponding author upon reasonable request.





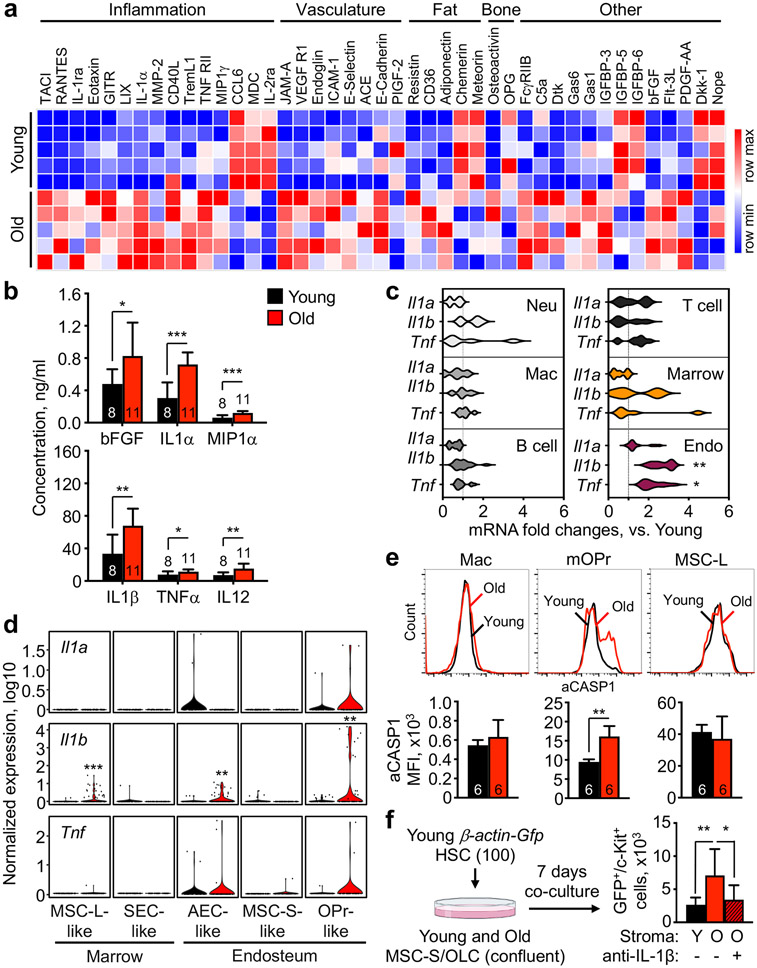



![Figure 7 ]](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/c1f4/10026258/5c3be38f31cb/nihms-1869586-f0007.jpg)