Abstract
Gastrulation controls the emergence of cellular diversity and axis patterning in the early embryo. In mammals, this transformation is orchestrated by dynamic signalling centres at the interface of embryonic and extraembryonic tissues1–3. Elucidating the molecular framework of axis formation in vivo is fundamental for our understanding of human development4–6 and to advance stem-cell-based regenerative approaches7. Here, we illuminate early gastrulation of marmoset embryos in utero by spatial transcriptomics and stem cell-based embryo models. Gaussian process regression-based 3D-transcriptomes delineate the emergence of the anterior visceral endoderm, which is hallmarked by conserved (HHEX, LEFTY2, LHX1) and primate-specific (POSTN, SDC4, FZD5) factors. WNT signalling spatially coordinates primitive streak formation in the embryonic disc and is counteracted by SFRP1/2 to sustain pluripotency in the anterior domain. Amnion specification occurs at the boundaries of the embryonic disc through ID1/2/3 in response to BMP-signalling, providing a developmental rationale for amnion differentiation of primate pluripotent stem cells (PSCs). Spatial identity mapping demonstrates that primed marmoset PSCs exhibit highest similarity to the anterior embryonic disc, while naïve PSCs resemble the preimplantation epiblast. Our 3D-transcriptome models reveal the molecular code of lineage specification in the primate embryo and provide an in vivo reference to decipher human development.
Introduction
In mammalian embryogenesis, gastrulation establishes the body plan and navigates the coordinated transition of the pluripotent embryonic disc (EmDisc) into three germ layers. Most of our knowledge on mammalian gastrulation has been derived from mouse1–3, but primate embryogenesis differs in anatomical architecture, developmental timing, molecular configuration and sequence of extraembryonic lineage segregations4,5,8. Recent transcriptomic characterisation of a late human gastrulation stage embryo9 provided insights into cell populations present during gastrulation, however the molecular framework coordinating axis patterning at the onset of primate gastrulation remains elusive. While blastocyst culture to postimplantation stages10–14 and stem cell-based embryo models15–19 have opened promising avenues, these approaches are currently limited by the absence of a reliable in vivo reference. Molecular comparisons to in vivo developed tissues are crucial to ensure that lineage specification, morphology and kinetics of in vitro models faithfully recapitulate embryonic development. To address these issues, we performed spatial transcriptome profiling of in utero developed and implanted marmoset (Callithrix jacchus) embryos. Marmosets present an emerging genetic model20–22 whose embryonic development is conserved with rhesus and human5. We reveal the signalling landscape surrounding the EmDisc in early gastrulation, track amnion (Am) and primitive streak formation and map the spatiotemporal identity of embryo-derived stem cells and stem cell-based embryo models.
Spatial embryo profiling in utero
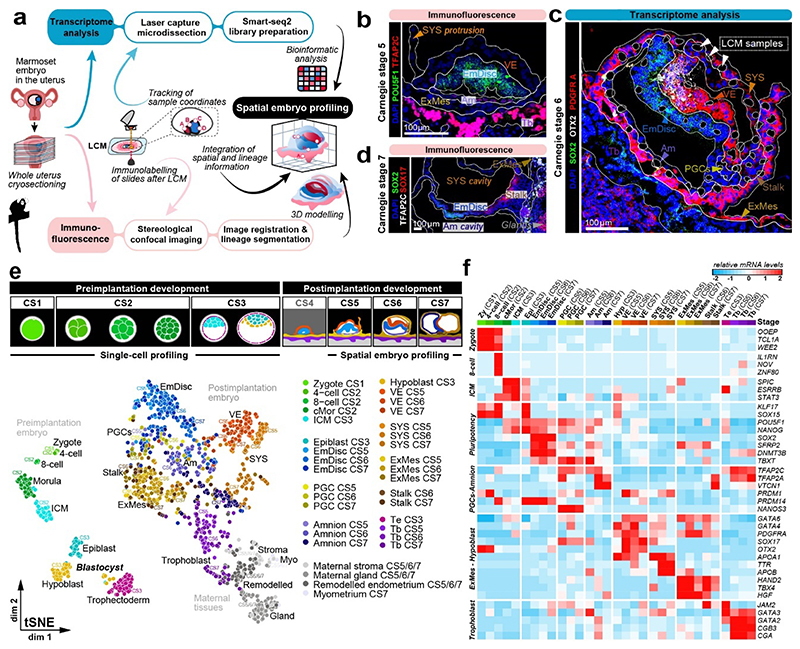
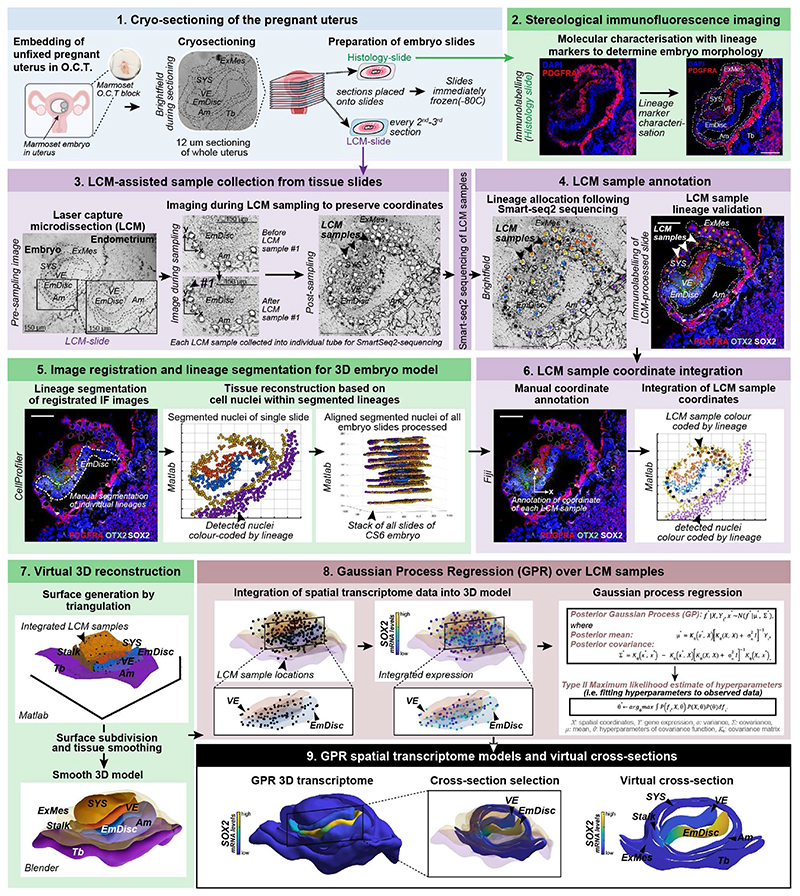
We established SpaTial Embryo Profiling (STEP) to delineate early marmoset postimplantation development (Fig. 1a, Extended Data Fig.1). STEP combines laser capture microdissection (LCM)-assisted sample collection for Smart-Seq2 profiling with virtual reconstruction of implanted marmoset embryos from stereological immunofluorescence stainings. Individual marmoset uteri were extracted after natural matings, snap frozen and cryosectioned (Supplementary Fig. 1a, Methods). At the implantation site, tissue samples of one to three cells were captured by LCM and processed individually by Smart-seq223 for full-length transcriptome profiling (Fig. 1b-d). We generated 1912 (1564 after quality control) transcriptomes from seven implanted marmoset embryos at Carnegie stages(CS) 5-7 and maternal tissues (Supplementary Table 1). STEP-transcriptomes were sequenced to high saturation, detecting on average >8,000 genes per sample and showed homogenous read-depths throughout lineages (Supplementary Fig. 1b-h). Tracking the position of each LCM-sample allowed us to assign lineages according to transcriptome location within the embryo (Fig. 1c).
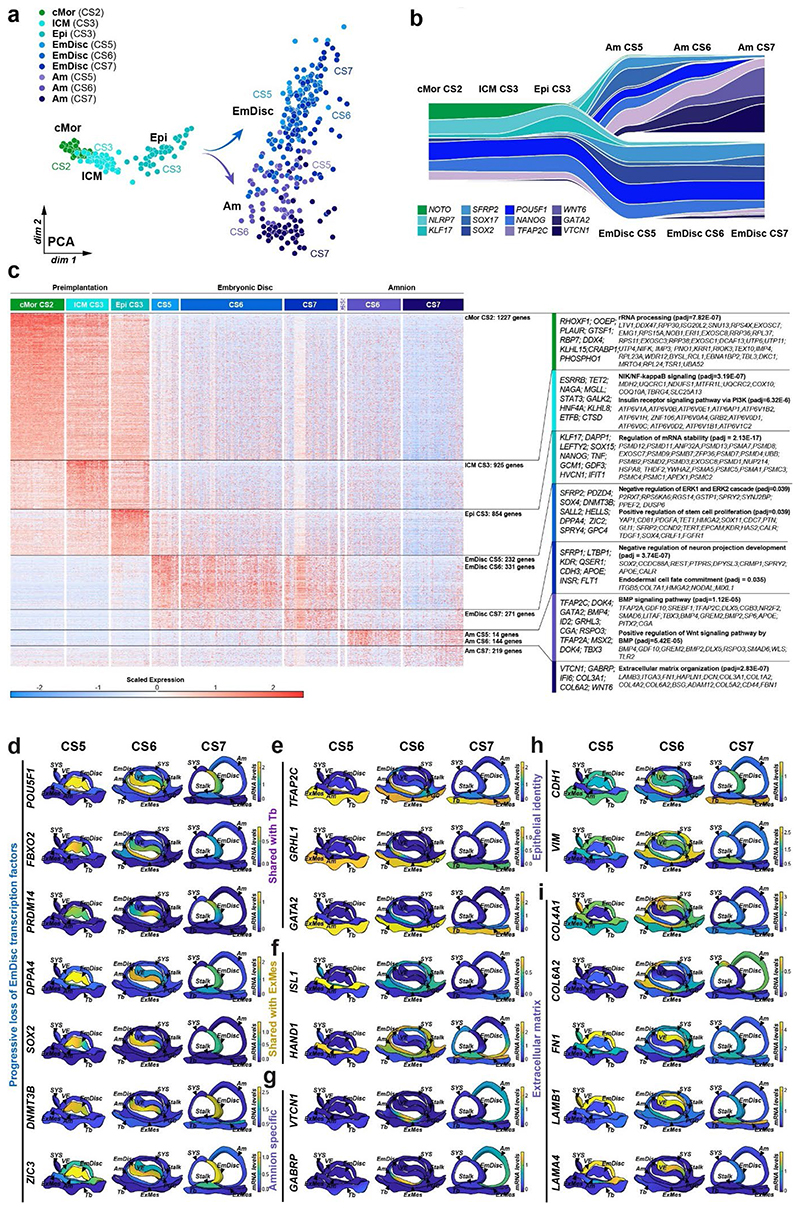
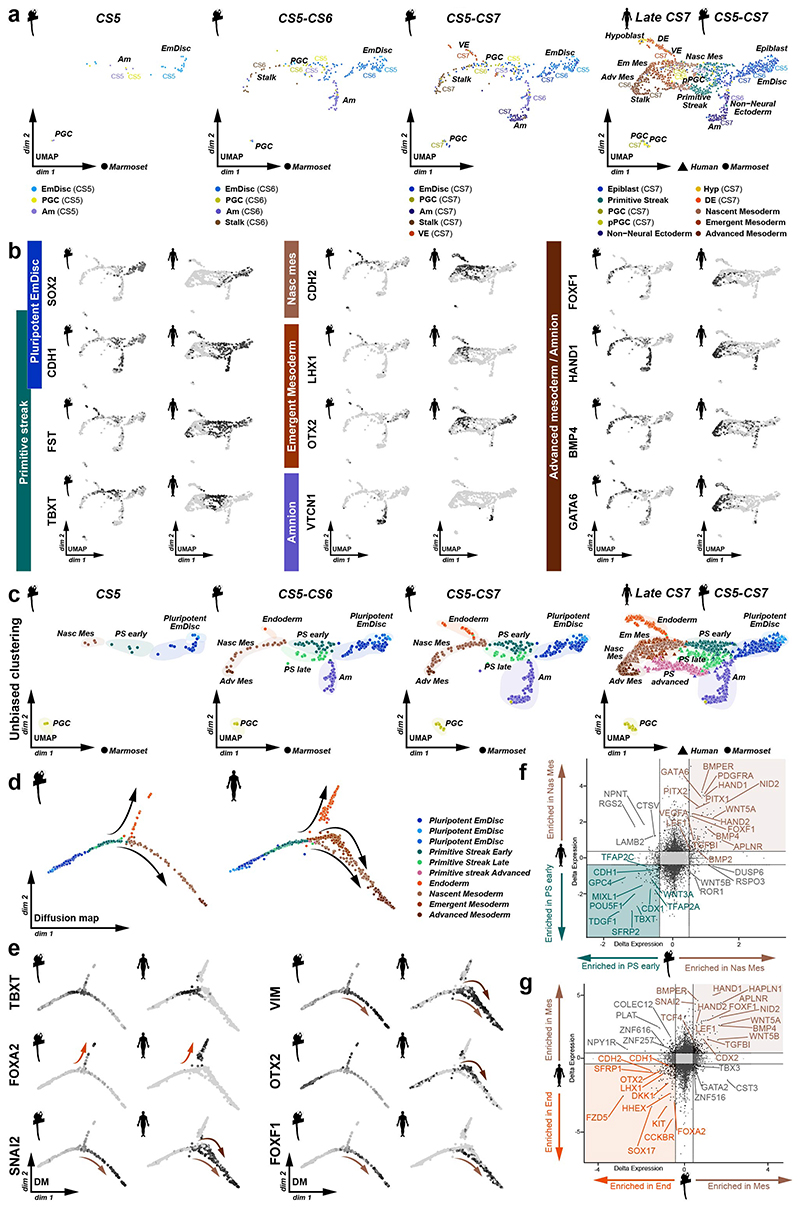
Figure 1. Spatial profiling of marmoset embryogenesis.
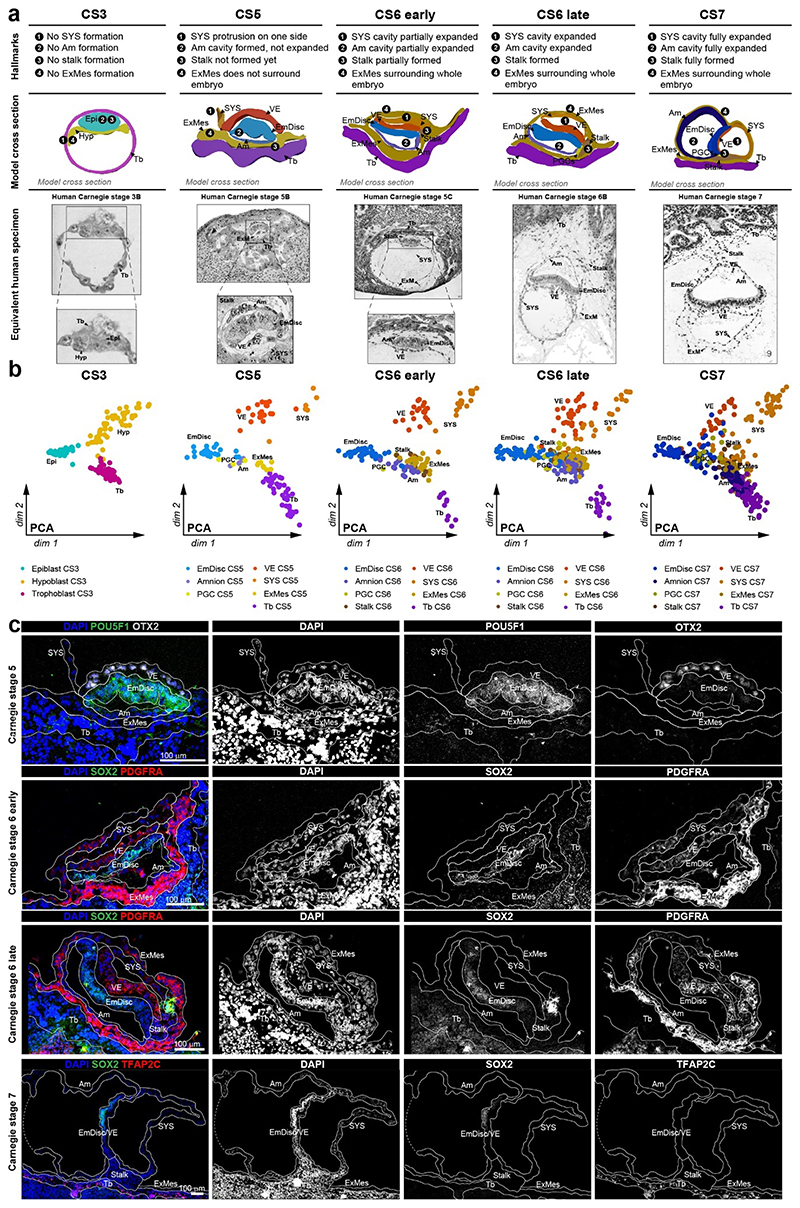
a, SpaTial Embryo Profiling (STEP) to delineate early marmoset postimplantation development. Transcriptome samples are collected by laser capture microdissection (LCM) and processed by Smart-Seq2. Sample coordinates are determined from consecutive cryosections and embedded in virtually reconstructed embryo models based on stereological confocal immunofluorescence images. b-d, Confocal immunofluorescence stainings of marmoset implantation stages at Carnegie stages (CS) 5,6,7. b, Pluripotency factor POU5F1 and Tb / early Am marker TFAP2C at CS5. c, Lineage marker analysis after LCM-processing using pluripotency marker SOX2, VE marker OTX2 and hypoblast/VE/mesoderm marker PDGFRA of a CS6 embryo cryosection. The locations of harvested LCM-samples are indicated with white dashed circles. d, Immunofluorescence of CS7 for SOX2, SOX17 and TFAP2C. e, Schematic overview of marmoset embryonic stages (top panel) from zygote (CS1) to gastrula (CS7); CS4 was not included in transcriptome analysis. The tSNE plot shows the combined Smart-seq2 embryo atlas consisting of 279 preimplantation single-cell samples as well as 866 postimplantation embryo and 193 maternal tissue STEP samples. Lineage colour code was used for all following figures (preimplantation stages=green, embryonic lineage and derivatives=blue, amnion=lavender, hypoblast-derived lineages=yellow, trophoblast-derived lineages=purple, maternal tissue=grey). f, Heatmap of embryonic and extraembryonic lineage markers. Relative mRNA levels were centred and scaled across samples. Zy, Zygote; cMor, Compacted Morula; ICM, Inner cell mass; Epi, Epiblast; Hyp, Hypoblast; Te, Trophectoderm; Am, Amnion; EmDisc, Embryonic disc; VE, Visceral endoderm; ExMes, Extraembryonic Mesoderm; SYS, Secondary Yolk Sac; Tb, Trophoblast; PGCs, Primordial Germ Cells; Myo, Myometrium.
We expanded a previous single-cell marmoset preimplantation dataset24 to include trophectoderm and cleavage stage samples. Combining pre- and postimplantation samples generated a uniform, Smart-seq2-processed atlas of marmoset development from zygote to gastrulation (Fig. 1e, Supplementary Video 1 and Table 2). Global dimensionality reduction separated preimplantation, postimplantation and maternal tissues (Fig. 1e). Preimplantation development concluded in the formation of three lineages by the late blastocyst stage (CS3): the pluripotent epiblast, which gives rise to the embryo proper, and extraembryonic hypoblast and trophectoderm. In the implanted embryo (CS5), the blastocyst lineages underwent substantial diversification and the epiblast-derived EmDisc segregated from extraembryonic samples (Fig. 1b-e). The trophoblast secreted pregnancy hormones CGA and CGB3 (Fig. 1f), which induced widespread remodelling of KRT7- maternal endometrial glands into KRT7+ branching cell clusters to promote vasculogenesis (Supplementary Fig. 2a-f). Consistent with previous reports13,14,25, we detected transcriptional overlap of early postimplantation lineages, in particular for Am, extraembryonic mesoderm (ExMes) and gastrulating EmDisc. In contrast, STEP identified postimplantation lineages according to the anatomy of the primate embryo in vivo (Fig. 1e, f). The resulting molecular signatures showed continuous lineage progression and diversification according to Carnegie staging (Extended Data Fig. 2a-c).
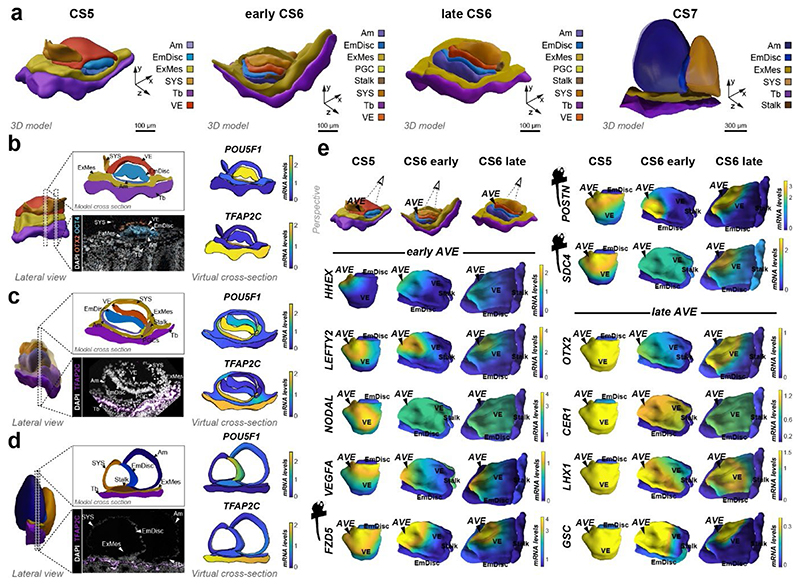
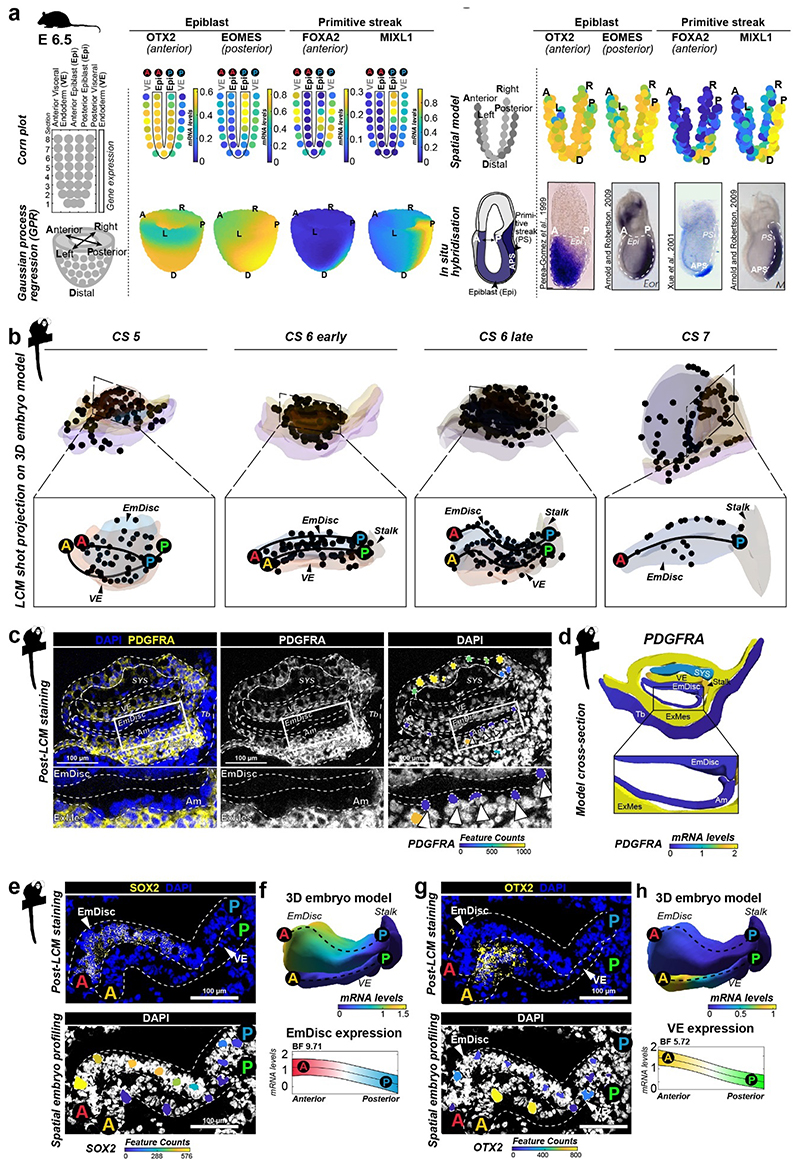
To generate virtual models of CS5, 6 and 7, we performed stereological confocal microscopy, image registration and lineage segmentation of the implanted marmoset embryos (Fig. 2a, Extended Data Fig. 1, Supplementary Fig. 3-5, Supplementary Videos 2–4). Embryo models increased in size according to developmental time and stage (EmDisc lengths CS5: 370μm, CS6: 430μm, CS7: 1240μm). We integrated spatial transcriptomes into the virtual embryos and redeployed Gaussian process regression (GPR), a nonparametric Bayesian approach widely used in machine learning26, geostatistics27 and most recently 2D spatial transcriptomics28, to model gene expression between discrete samples. We validated GPR as a tool to build 3D-transcriptomes in the gastrulating mouse egg-cylinder29,30 (Extended Data Fig. 3a). GPR-based 3D-transcriptomes accurately rendered well-characterised marker patterns from individual data points, including Otx2, Eomes, Foxa2, and Mixl1. We then applied GPR to marmoset LCM-samples with defined locations within the embryo models (Extended Data Fig. 3b). Marmoset 3D-transcriptomes faithfully recapitulated lineage marker expression (Fig. 2b-d) and specific expression patterns observed by immunofluorescence (Extended Data Fig. 3c-h, Supplementary Fig. 6a,c,e). Embryonic architecture and lineage marker profiles were consistent with in situ hybridisations of stage-matched cynomolgus embryos (Supplementary Fig. 6b,d,f). We conclude that GPR-based 3D-transcriptomes reliably emulate in situ marker expression, presenting a unique opportunity to determine genome-wide expression gradients.
Figure 2. Virtual reconstruction of gastrulating marmoset embryos.
a,Virtual 3D-reconstructions of postimplantation implantation stages at CS5, early CS6, late CS6, and CS7 based on stereological confocal imaging and lineage segmentation. ExMes of CS6 early and late embryos is vertically sectioned to expose underlying structures. ExMes of CS7 that overlays amnion and SYS are not displayed, and both amnion and yolk sac are partially transparent. b-d, GPR-based 3D-transcriptome expression of pluripotency marker POU5F1 and Tb/Am marker TFAP2C in CS5 (b), late CS6 (c), and CS7 (d) embryos. Cross-sections are indicated in lateral views of virtual 3D-reconstructions with immunofluorescence staining of corresponding embryo sections. (e) Anterior visceral endoderm (AVE) genes depicted in EmDisc/VE for the stages indicated. Marmoset symbols indicate primate-specificity. GPR, Gaussian process regression; CS, Carnegie stage; Am, Amnion; EmDisc, Embryonic disc; ExMes, Extraembryonic Mesoderm; SYS, Secondary Yolk Sac; Tb, Trophoblast; VE, Visceral endoderm; PGC, Primordial Germ Cells.
Emergence of the AVE and yolk sac
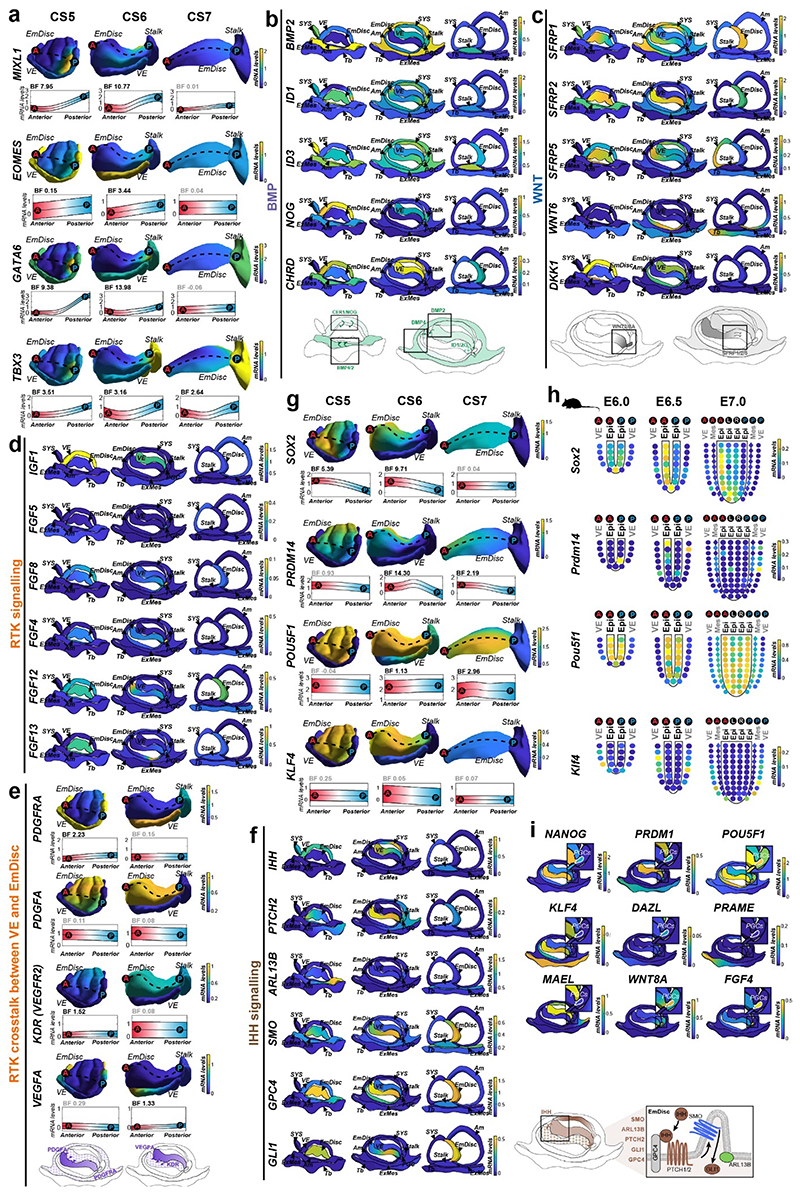
In mouse, signals from the anterior visceral endoderm (AVE) restrict gastrulation towards the posterior egg-cylinder2,3,31,32. However, the dynamics and molecular attributes of the primate AVE, as well as secondary yolk sac (SYS) formation in vivo have remained elusive4,5,8,33. Early (CS5) visceral endoderm (VE) robustly expressed conserved hypoblast markers (GATA6, SOX17, APOA1, PDGFRA) (Fig. 1f, Supplementary Fig. 7a) 24. To delineate primate AVE formation, we extracted anteriorly expressed genes in the VE at CS5/6 (Supplementary Table 3). The top 50 most significantly enriched genes in the AVE included conserved AVE markers HHEX, OTX2, GSC, LEFTY2, VEGFA and LHX1 (Lim1) (Fig. 2e). We identified additional primate-specific AVE-associated genes SDC4, POSTN, and FZD5. NODAL was anterior at CS5, but expressed throughout the VE by early CS6. In contrast, OTX2, LHX1 and GSC were transcribed throughout the early VE and then became progressively restricted towards the AVE. Prior to the emergence of the SYS-cavity at CS6, we observed protrusions from the CS5 VE (Fig. 2a, Extended Data Fig. 2c, Supplementary Fig. 3). SYS-protrusions expressed VE markers (SOX17, APOA1), but also showed specific expression of later SYS marker BMP6, RBP4, TTR and GC (Supplementary Fig. 7b), which may suggest SYS formation from edges of the early VE. At late CS6, most of the SYS and Am was surrounded by ExMes. PDGFRA was expressed across SYS, VE and ExMes, in contrast to OTX2, which was specific to VE (Fig. 1c, f, Extended Data Fig. 2c). We determined the transcriptional signatures for VE (CER1, CXCR4, SOX17) and SYS (FABP1, VCAN, GSTA1) (Supplementary Fig. 7c-d). ExMes formation was hallmarked by epithelial to mesenchymal transition (SNAI2, HGF), extracellular matrix and, WNT and BMP-ligand expression (Supplementary Fig. 7c-e).
The molecular framework of gastrulation
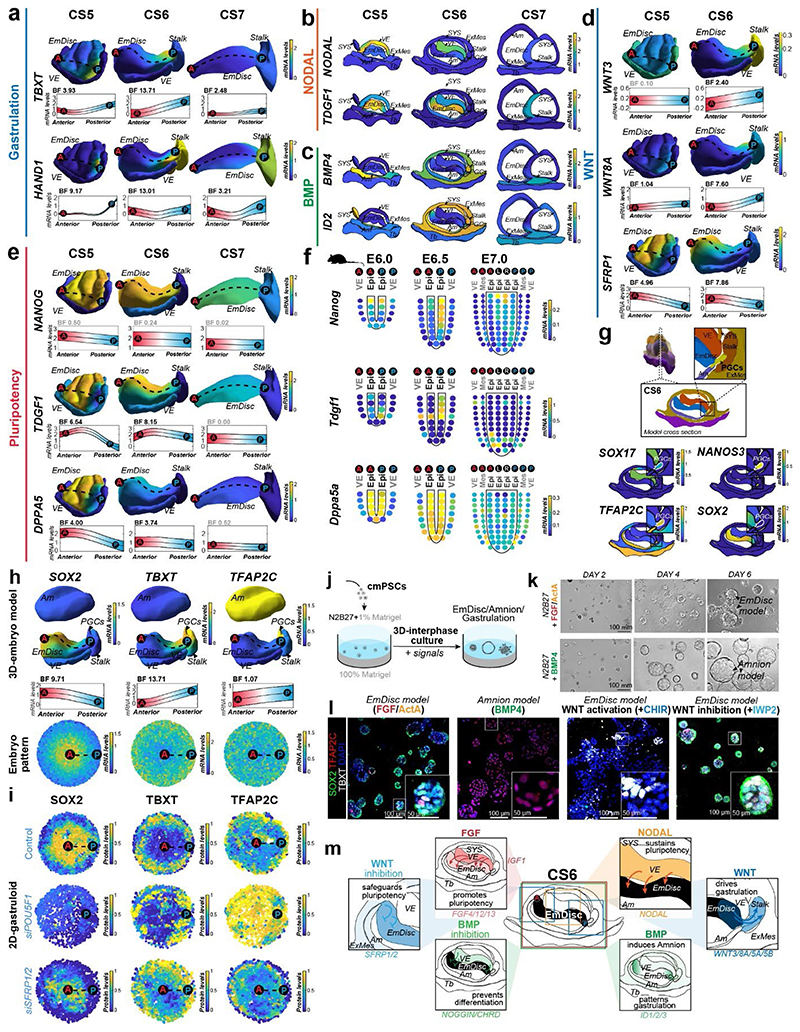
Gastrulation establishes the body axis by patterning prospective anterior (head) and posterior (tail) regions1–3. To elucidate the acquisition of regionalised identity, we extracted anterior and posterior genes in the EmDisc (Supplementary Table 3). We readily identified primitive streak marker TBXT, MIXL1, EOMES and HAND1 (Fig. 3a, Extended Data Fig. 4a) in the posterior EmDisc as early as CS5. NODAL was strongly expressed in the VE, while its co-receptor TDGF1 (CRIPTO) was confined to the EmDisc (Fig. 3b). ExMes and Am expressed BMPs, while the VE secreted BMP-inhibitors NOG (Noggin), CER1 and CHRD (Chordin) (Fig. 2e, 3c, Extended Data Fig. 4b). There was a surge of WNT ligands (WNT3, WNT8A) in the posterior EmDisc, in contrast to WNT inhibitors SFRP1/2/5, which localised anteriorly (Fig. 3d, Extended Data Fig. 4c). Interestingly, the primate EmDisc upregulated non-secreted FGF12 and FGF13, and VE expressed high levels of IGF1 (Extended Data Fig. 4d), which may stimulate MAPK-signalling in the adjacent EmDisc34. We observed reciprocal expression of PDGFA and PDGFRA in EmDisc and VE, respectively, and detected core Hedgehog-signalling pathway components across the entire EmDisc (Extended Data Fig. 4e,f).
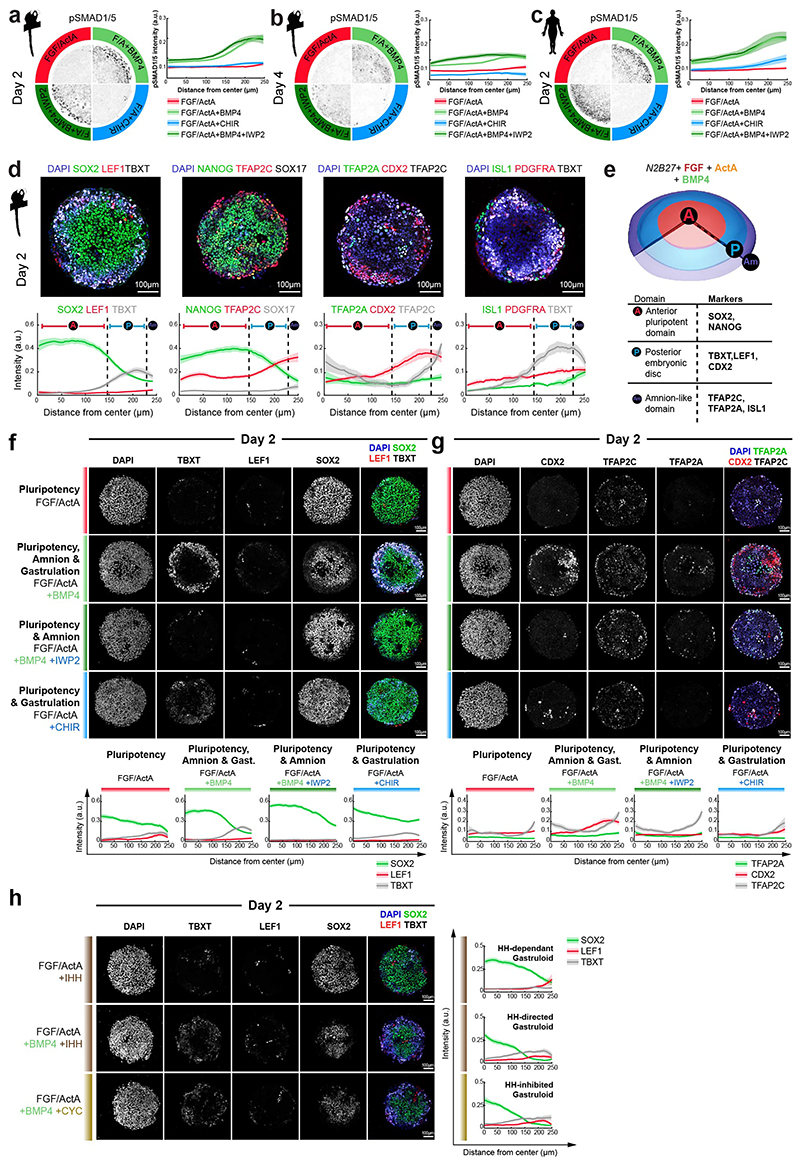
Figure 3. 3D-transcriptomes and stem cell-based embryo models delineate body axis formation.
a, GPR-based 3D-transcriptomes in EmDisc/VE showing gastrulation marker expression in the posterior EmDisc. Upper panels: Relative mRNA levels in EmDisc, VE, and stalk. Lower panels: mRNA expression change along anterior-posterior axis (dashed line; anterior (red, A) to posterior (blue, P)) in EmDisc, quantified by Bayesian factor (BF). b-c, Relative mRNA levels of (b) NODAL and (c) BMP in virtual embryo cross sections. d, WNT signalling pathway components shown in EmDisc/VE model. e, GPR-models for EmDisc/VE displaying regionalised pluripotency factor transcription in the anterior EmDisc. f, Spatial expression of pluripotency factors in gastrulating mouse embryos at E6.0, E6.5, and E7.0 according to Geo-seq30. g, Virtual cross-sections of the PGCs at CS6. GPR, Gaussian process regression; CS, Carnegie stage; EmDisc, Embryonic disc; VE, Visceral endoderm; ExMes, Extraembryonic Mesoderm. Am, Amnion; SYS, Secondary Yolk Sac; Tb, Trophoblast; PGC, Primordial Germ Cells. h, GPR-models for CS6 embryo of anterior marker (SOX2), posterior marker (TBXT) and amnion/PGC maker (TFAP2C). Upper panels: Relative mRNA levels for gene expression in EmDisc, VE, PGCs, Stalk, and Amnion. Am is displayed separately for visualisation. Middle panels: mRNA expression change along anterior-posterior axis (dashed line, anterior (red, A) to posterior (blue, P)) in EmDisc, quantified by Bayesian factor (BF). Lower panel: Virtual embryo pattern of EmDisc expression patterns generated by axis expression in middle panel. i, Expression patterns of in vitro 2D gastruloids segmented nuclei stained for anterior marker (SOX2), posterior marker (TBXT) and amnion/PGC maker (TFAP2C) after differentiation in BMP4 (50 ng/mL), FGF (10ng/mL), and Activin A (20ng/mL) under control conditions (top panel) and following siRNA transfection (bottom panels). Each intensity profile is normalized log expression levels are standardized so that they vary within [0,1]. j, Schematic representation of 3D-interphase culture system. cmPSCs are seeded on a bed of 100% Matrigel and overlaid with 1% Matrigel supplemented N2B27-based culture medium with and without signalling molecules. k, Brightfield images of EmDisc-like structures (N2B27 + 100 ng/mL FGF + 20 ng/mL Activin A) or Amnion-like structures (N2B27 + 50 ng/mL BMP4). l, Immunofluorescence images of structures generated after 4 days in EmDisc- or Amnion-promoting conditions or in EmDisc conditions with WNT modulation through 3 μM CHIR99021 (WNT activator) or 3 μM IWP-2 (WNT production inhibitor). m, Schematic summary diagram of BMP, WNT, FGF and NODAL signalling pathway activities in the marmoset embryo at CS6. PSCs, Pluripotent Stem Cells.
We and others25 have noticed the co-existence of pluripotent (NANOG+/POU5F1+/SOX2+) cells and gastrulating (TBXT+/MIXL1+) cells in early primate implantation stages. 3D-trancriptome analysis revealed co-localisation of core pluripotency factors NANOG, POU5F1, SOX2 and several other essential pluripotency factors35,36 (TGDF1, DPPA5, PRDM14, KLF4) in the anterior EmDisc (Fig. 3e, Extended Data Fig. 4g). This is in stark contrast to mouse development, where Nanog and Tdgf1 are expressed posteriorly (Fig. 3f, Extended Data Fig. 4h). Marmoset PGCs exhibited primate-specific marker expression (TFAP2C+/SOX17+/NANOG+/POU5F1+)37–39 and localised within Am at CS5 and within the posterior Am and EmDisc boundary at CS6 (Fig. 3g), similar to cynomolgus39. Early PGCs expressed ETS transcription factors (ETS2, ETV3L, ETV5), but were negative for SOX2 and germ cell markers of later developmental stages (Fig. 3g, Extended Data Fig. 4i).
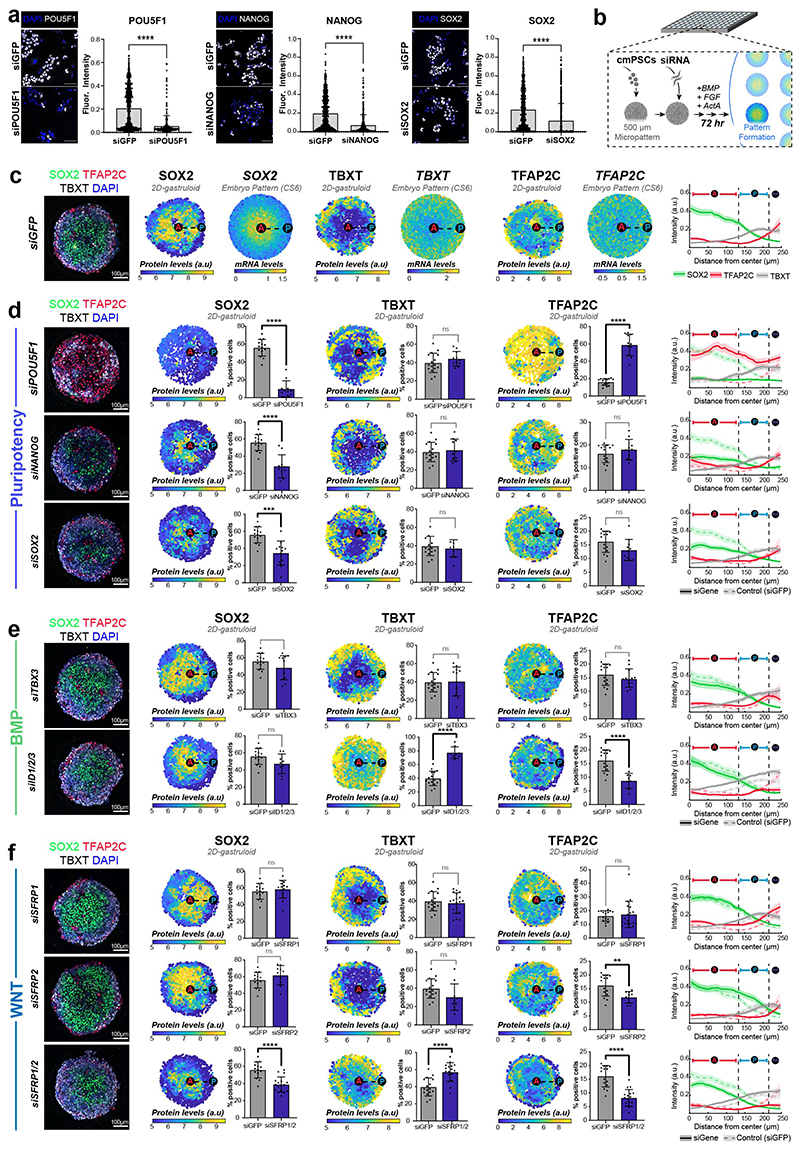
To functionally interrogate the role of the signalling environment in the primate embryo, we established two stem cell-based embryo models with common marmoset (cm)PSCs (Fig. 3h-l). Micropatterned6,18,40 cmPSCs (2D-gastruloids) emulated key aspects of EmDisc patterning (Fig. 3h,i, Extended Data Fig. 6a-e, 7c), whilst 3D-interphase-culture modelled pluripotency, gastrulation and amniogenesis (Fig.3j-l, Extended Data Fig. 8, Supplementary Fig. 8). Marmoset 2D-gastruloids acquired anterior-posterior patterning from the centre to the edge of the colony, consisting of an anterior (SOX2+/NANOG+) pluripotent domain in the centre, posterior (TBXT+) regions in the middle and an Am-like (TFAP2C+/TFAP2A+/ISL1+) domain at the edge, in accordance with the CS6 embryo (Fig. 3h,i). WNT signalling induced – and was required for – primitive streak formation in both models (Extended Data Fig. 6f,g, 8i-n). In the absence of BMP4 or WNT, the pluripotent compartment extended over the entire 2D-gastruloid (Extended Data Fig. 6f). We also tested the functional relevance of IHH expression from VE but did not detect significant differences in Hedgehog signalling perturbation experiments (Extended Data Fig. 6h). siRNA knockdown of core pluripotency factors reduced the pluripotent compartment, but only loss of POU5F1 resulted in strong upregulation of amnion marker TFAP2C (Extended Data Fig. 7a-d). Double-knockdowns of the anteriorly located WNT-inhibitors SFRP1 and SFRP2 showed an expansion of the (TBXT+) posterior domain at the expense of the (SOX2+) anterior pluripotent compartment. These results suggest that endogenous WNT inhibition through SFRP1/2 promotes pluripotent epiblast identity in the anterior EmDisc, in contrast to the posterior domain, where WNT induces primitive streak formation (Fig. 3m).
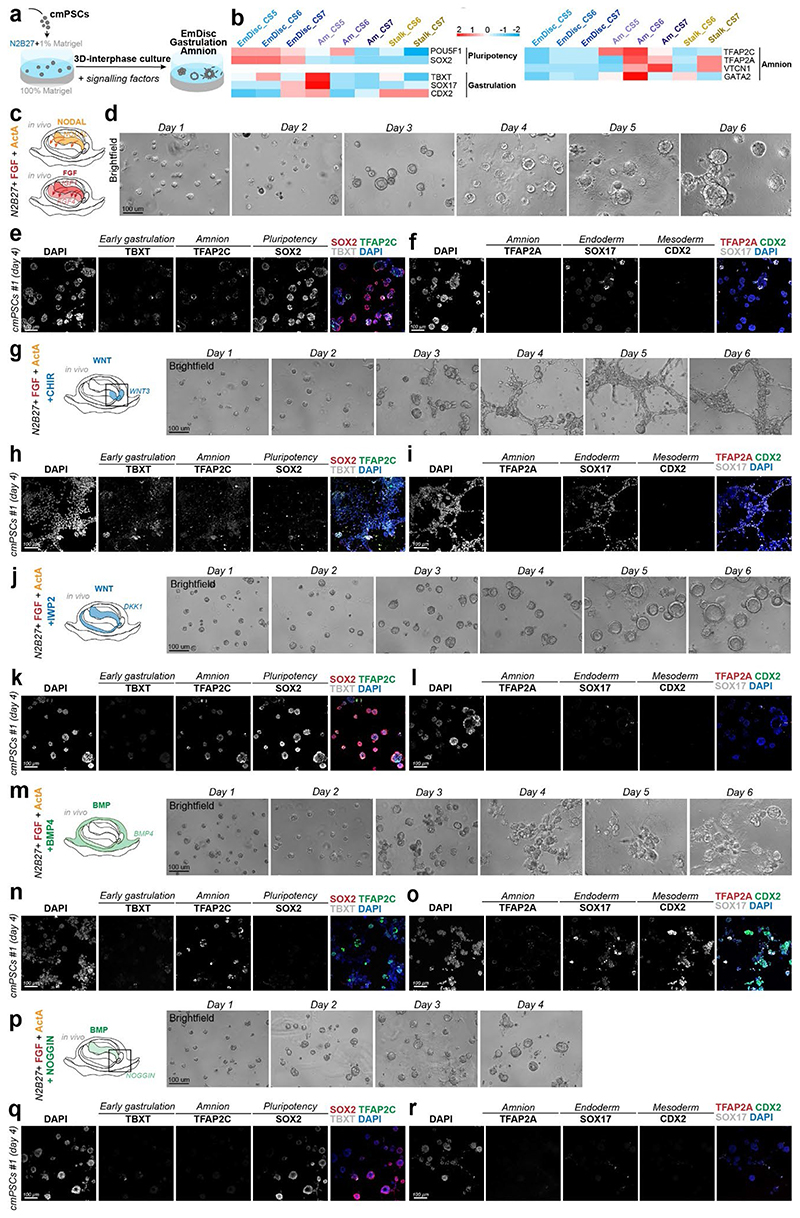
Amniogenesis in vivo
Primate amniogenesis has been exclusively studied in vitro12–17,41, therefore we leveraged STEP to track Am formation in vivo. At CS5, the amniotic cavity was small and both EmDisc and early Am expressed pluripotency factors POU5F1, NANOG and PRDM14 (Fig. 1f, Extended Data Fig. 2c, Supplementary Fig. 6a). Nascent Am rapidly lost pluripotency marker SOX2 and upregulated TFAP2C, TFAP2A, GATA2, ID2, ID3 and signalling pathway components BMP4, GDF10 and WNT6 (Fig. 1b,f, Extended Data Fig. 5a-c). Enriched GO-terms for early Am (CS5/6) included “BMP-signalling”, more mature Am (CS7) showed “ECM organization”. Virtual cross-sections demonstrated progressive loss of pluripotency regulators during Am formation and revealed Am markers shared with Tb (GATA2, GRHL1, TFAP2C) and ExMes (ISL1, HAND1). Importantly, our spatial profiling approach enabled us to univocally identify Am-specific markers (VTCN1, GABRP, ARSI, ENSCJAG00000003246), not expressed in other lineages (Extended Data Fig. 5d-g). Am featured epithelial identity and ECM-associated genes, including COL6A2 and COL4A1 (Extended Data Fig. 5h,i), in accordance with its role as an elastic extraembryonic membrane. The signalling environment during Am formation was dominated by dorsal BMP signals from the surrounding ExMes (Fig. 3c), while the EmDisc was exposed to secreted BMP inhibitors (NOG, CER1, CHRD) from the ventrally located VE (Extended Data Fig. 4b). To gauge BMP signalling activity, we examined BMP downstream targets ID1, ID2 and ID342 and found them to be present in SYS, ExMes and Am (Fig. 3c, Extended Data Fig. 4b). Virtual cross-sections showed regionalised expression of ID1/2/3 at the boundary between EmDisc and Am at CS6, suggesting a regulatory role of ID-genes in marmoset amnion specification.
In both marmoset stem cell-based embryo models, BMP4 signalling induced Am formation, in contrast to NODAL and FGF signals (Fig. 4i-l, Extended Data Fig. 6f,g, 8, Supplementary Fig. 8). Am induction was independent of WNT signalling (Extended Data Fig. 6f,g, Supplementary Fig. 8j-l). POU5F1, but neither SOX2 or NANOG knockdown, promoted Am formation, despite the early loss of SOX2 in nascent Am (Extended Data Fig. 7d). In contrast, triple-knockdown of ID1/2/3 reduced Am formation and increased TBXT expression (Extended Data Fig. 7e). Collectively, these results suggest a functional role for POU5F1 in EmDisc versus Am formation and that BMP-signalling through ID1/2/3 promotes Am.
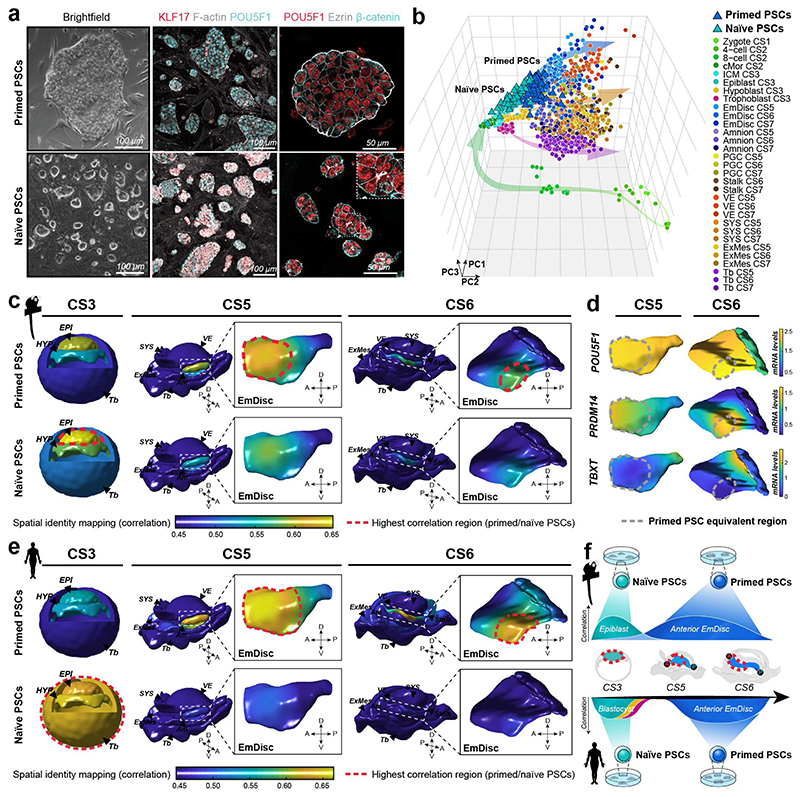
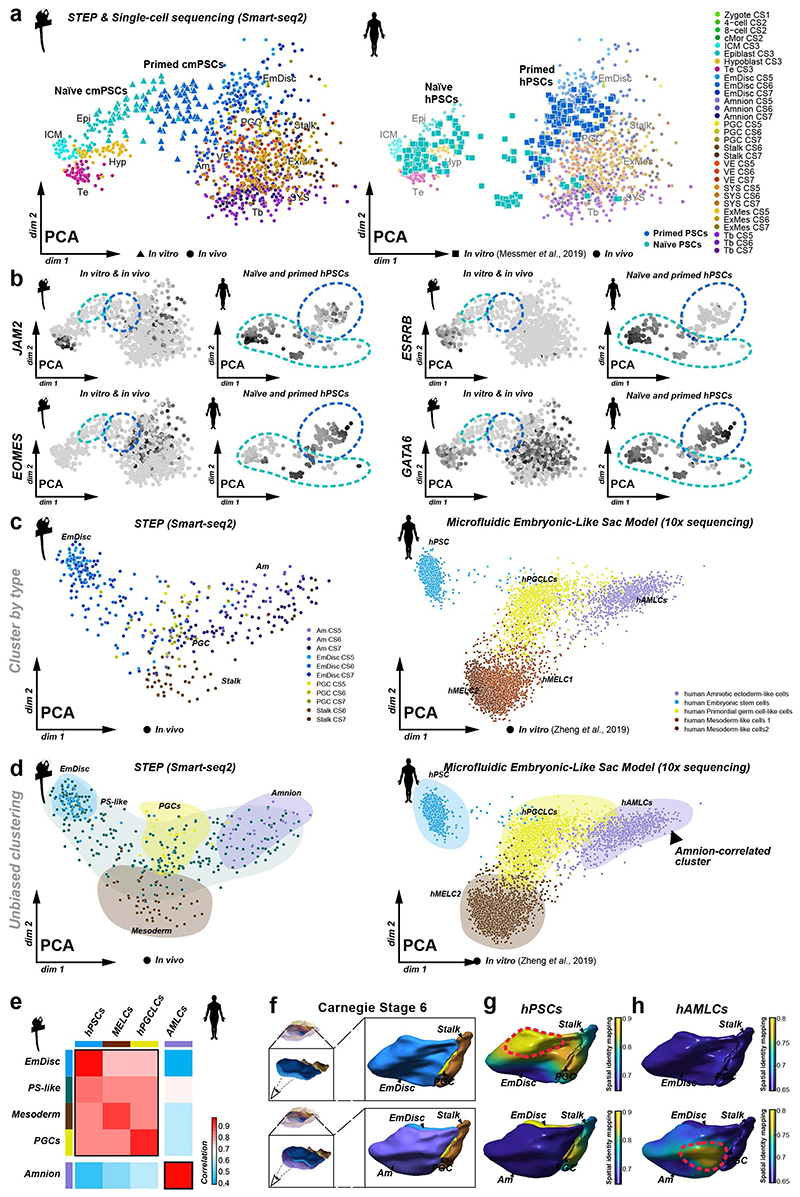
Figure 4. Spatial identity mapping of in vitro cultured cells.
a, Brightfield and immunofluorescence images of primed and naïve marmoset pluripotent stem cells (PSCs) cultured in PLAXA medium. b, Principal component analysis (PCA) of embryonic and extraembryonic cells in vivo and naïve and primed PSCs in vitro. PCA based on the top 2000 most variable genes, PC1=22.0%, PC2 =12.4%, PC3=7.5%. c-e, Spatial identity mapping of marmoset (c) and human (e) naïve and primed PSCs. Colour scale represents projection of correlation values onto embryo model surfaces followed by Gaussian process regression mapping. Blastocyst model is a schematic representation with bulk correlation plotted for each lineage. Gene expression in regions of highest correlations for primed PSCs in the pluripotent anterior indicated by dotted line (d). f, Summary of PSC mapping of marmoset and human PSCs to the marmoset in vivo atlas. A, anterior; P, posterior; D, dorsal; V, ventral; EPI, Epiblast; HYP, Hypoblast; Tb, Trophoblast; EmDisc, Embryonic disc; Am, Amnion; SYS, Secondary Yolk Sac; VE, Visceral Endoderm; ExMes, Extraembryonic mesoderm; PGCs, Primordial Germ Cells.
Conserved features of gastrulation
We sought to investigate divergent and overarching features of primate embryogenesis. Integrated analysis of marmoset, cynomolgus25 and human embryo datasets9,12 showed that pre- and postimplantation samples grouped according to embryonic stage with developmental trajectories for epiblast, hypoblast and trophoblast along principal component one (Supplementary Fig. 9). A subset of human postimplantation CS5 epiblast samples grouped closer to CS3/4 preimplantation stages, which may either suggest slower lineage maturation in human or could be a result of postimplantation embryo culture in vitro. We extracted conserved preimplantation-specific markers for epiblast (ANPEP, KLF17, DAPP1), hypoblast (HNF1B, GSG1, GATM) and trophectoderm (NOTO, FXYD3 and SLC30A2) and identified the earliest population of hemogenic endothelial progenitors in the marmoset yolk sac at CS7 (Supplementary Fig. 9,10).
We next examined the path to primate gastrulation (Extended Data Fig. 9a). Integrated analysis showed that marmoset EmDisc and EmDisc-derived lineages robustly aligned to the germ layer populations of the late CS7 human gastrula9. ‘Primitive streak’, ‘Nascent mesoderm’, ‘Endoderm’ and ‘Am populations’ were well conserved between marmoset and human. We demonstrate the emergence of a TBXT+/CDX2+ ‘Primitive streak early’ and a SFRP1+/SHISA2+ ‘Primitive streak late’ population, in addition to ‘Nascent mesoderm’ (PDGFRA+), ‘Advanced Mesoderm’ (HAND1+), ‘Endoderm’ (FOXA2+/SOX17+), ‘Am’ (VTCN1+/GABRP+) and ‘PGC’ (TFAP2C+/SOX17+/SOX2-) populations (Extended Data Fig. 9b,c, Supplementary Fig. 11c-f). ‘Primitive streak early’ cells appeared from CS5 onwards, while the ‘Primitive streak late’ population emerged at CS6/7 (Extended Data Fig. 9c). In contrast to human9, we did not detect OTX2+ ‘Emergent mesoderm’ in the marmoset (Extended Data Fig. 9c-g, Supplementary Fig. 11c-f). Diffusion maps showed that TBXT is transiently expressed prior to and during mesoderm-endoderm segregation, FOXA2 labels endoderm and VIM/SNAI2 highlight mesoderm populations in both species. Marmoset mesoderm differentiated via a FOXF1-positive population, as opposed to the human gastrula, where mesoderm formed through an OTX2-positive population (Extended Data Fig. 9d-g). Comparing ‘PGC’ and ‘Primitive streak early’ populations between human and marmoset showed that human PGCs secreted more GDF3 and NODAL (Supplementary Fig. 11a,b). Nevertheless, we observed only few species-specific differences between ‘Primitive streak early’, ‘Primitive streak late’, and ‘Am’ populations (Supplementary Fig. 11c-f), indicating the conserved molecular framework in primate embryogenesis.
Spatial identity mapping
Human and non-human primate PSCs resemble the postimplantation epiblast9,25, but their regional identity has remained obscure. We therefore investigated the spatiotemporal identity of marmoset and human (h)PSCs. In conventional medium (KSR/bFGF), cmPSCs exhibited characteristic features of primed pluripotency, including co-expression of core pluripotency and specification markers, epithelial colony morphology and absence of naïve pluripotency factors (Fig. 4a). We established a protocol to reset cmPSCs to naïve pluripotency in PLAX (PD03 (MEK-inhibitor), LIF, Ascorbic Acid, XAV939 (WNT inhibitor)) (Supplementary Fig. 12a). Interestingly, long-term propagation required addition of Activin A (PLAXA), consistent with primate-specific NODAL expression in the preimplantation epiblast24 (Supplementary Fig. 12a-b). Naïve cmPSCs lost epithelial polarity, upregulated preimplantation epiblast-specific genes and downregulated EmDisc-associated transcripts (Fig. 4a, Supplementary Fig. 12c-h).
To assess developmental states at the transcriptome level, we performed single-cell Smart-seq2 of naïve cmPSCs, primed cmPSCs and somatic cells from the neonate forebrain. PCA aligned samples into a developmental continuum from zygote to late gastrulation (Fig. 4b), consistent with cross-species analysis (Supplementary Fig. 9). Somatic forebrain cells clustered far away from both embryonic and extraembryonic tissues (Supplementary Fig. 12i). In contrast, primed cmPSCs grouped with EmDisc samples at CS5 and CS6 (Fig. 4b). Naïve cmPSCs were most similar to the CS3 preimplantation epiblast, with some cells clustering in between embryo samples (Fig. 4b). This demonstrates EmDisc identity of primed cmPSCs and genome-wide resetting of the transcriptional machinery towards the preimplantation epiblast in naïve conditions.
We used spatial identity mapping to determine spatial correlations between the embryo and in vitro cultured cells. The highest correlation of primed cmPSCs was observed in the anterior domain of the EmDisc at CS5 and, to a lesser degree, the anterior domain at CS6 (Fig. 4c,d). Naïve cmPSCs exhibited highest correlation scores in the preimplantation epiblast. Primed hPSCs43 also mapped to the anterior EmDisc, but stronger at CS6, suggesting a slightly later developmental stage. Naïve hPSCs43 showed high similarity to all preimplantation lineages (Fig. 4e), in line with their capacity to give rise to trophoblast44,45 and extraembryonic endoderm46 stem cells. Moreover, integrated PCA demonstrated that naïve hPSCs contained several subpopulations expressing extraembryonic differentiation markers (Extended Data Fig. 10a,b). As a proof of principle for spatial identity mapping of extraembryonic in vitro cultures, we analysed human Am-like cells (hAMLCs) from microfluidic embryonic-like sac models16. hAMLCs formed coherent clusters with marmoset postimplantation lineages in integrated PCAs (Extended Data Fig. 10c-e). Spatial identity mapping showed that hAMLCs closely correlated with CS6 Am, but not EmDisc (Extended Data Fig. 10f-h). These results identify the anterior EmDisc as the closest counterpart of primate PSCs and highlight the power of STEP datasets to determine the regional identity of in vitro cultured cells and embryo models.
Discussion
Analysis of marmoset 3D-transcriptomes identified a pluripotent compartment in the anterior EmDisc, which is sustained by NODAL expression from the AVE/VE and shielded from WNT-induced gastrulation by endogenous SFRP1/2. Co-expression of all core pluripotency factors in the anterior compartment of the marmoset EmDisc contrasts from mouse, where Sox2 is expressed anteriorly and Nanog posteriorly29,30. It is tempting to speculate that the early physical separation of core pluripotency factor expression may present a rodent-specific adaptation to accelerate lineage acquisition, but further studies are required to delineate the kinetics of primate versus rodent development. STEP elucidated the dynamics of regionalised expression in the primate AVE, although its functional attributes remain to be validated. We reveal the expression of BMP downstream targets at the boundary between EmDisc and Am and demonstrate their requirement for Am specification in 2D-gastruloids. These results indicate the continuous formation of Am at the periphery of the EmDisc, presenting a developmental rationale for Am specification from human PSCs15–17. Collectively, this work provides the missing link to illuminate early primate gastrulation and to benchmark the developmental authenticity of in vitro cultured cells and embryo models16,43. 3D-transcriptomes will be of critical importance to expand the repertoire of embryo-derived stem cell lines corresponding to distinct developmental stages and regional identities for the functional interrogation of human embryogenesis.
Methods
Animal experiments
Animal husbandry
Marmoset preimplantation embryos were obtained at Central Institute for Experimental Animals (CIEA). The animal experiments were approved by the Institutional Animal Care and Use Committee of the CIEA (approved numbers: 17029A for year of 2017, 18031A for year of 2018, 19033A for year of 2019) and were performed in accordance with CIEA standard guidelines which are in accordance with the guidelines for the ”Proper Conduct of Animal Experiments” by the Science Council of Japan.
Postimplantation embryos were collected at the German Primate Centre. All animal studies were performed according to the German Animal Protection Law and approved by German Primate Center (Deutsches Primatenzentrum – Leibniz Institute for Primate Research) ethics committee. Animals were obtained from the self-sustaining marmoset monkey (Callithrix jacchus) breeding colony of the German Primate Center and housed according to the standard German Primate Center practice for common marmoset monkeys.
The females used in this study (n = 8, age range 8-11 years) had a history as successful breeders and were housed together with fertile males to allow natural mating. All animal work was performed by experienced veterinarians and trained staff in agreement with the requirements of the German Animal Protection Law (Deutsches Tierschutzgesetz). Animal procedures to obtain the marmoset embryos used in this study were approved by the Niedersächsisches Landesamt für Verbraucherschutz und Lebensmittelsicherheit, LAVES, under licence number 33.19-42502-04-16/2130 “Gewinnung früher Implantationsembryonen des Weißbüschelaffen zur molekularen Charakterisierung frühembryonaler Differenzierungsschritte bei Primaten”, which included a positive ethics statement.
Pregnancy timing and hysterectomy
Early postimplantation embryos used in this study were from embryonic day (E)15 and E25, roughly equivalent to the embryonic period in human development between Carnegie stages 5 and 7, respectively48. Timed pregnancies were obtained from animals in which the stage of gestation was established from the post-ovulatory rise in progesterone50, which was determined after blood collection from the Vena femoralis from female marmoset monkeys twice weekly. Pregnancies were confirmed and monitored by progesterone determination and ultrasonography. Those with unclear staging were terminated by prostaglandin F2α injection and only animals with clearly gravid uteri were used for hysterectomy.
For hysterectomy, which delivers embryos in an optimal histological condition in their natural in situ position, animals were deeply anesthetized with an i.m. injection of 0.5 ml/kg bodyweight Göttinger Mischung II [50 mg/ml ketamine (WDT, Garbsen, Germany), 10 mg/mL Xylazin (Bayer, Leverkusen, Germany), 10 mg/mL atropin (Eifelfango, Bad Neuenahr-Ahrweiler, Germany)] and 0.05 mL/animal diazepam (Ratiopharm, Ulm, Germany). The gravid uterus and the ovaries were delivered through a ventral midline incision in the abdominal wall under sterile conditions. To avoid possible perioperative pain, an analgesic (meloxicam 0.5 mg/kg body weight) was administered approximately 30 minutes before the anesthetic. An incision of about 2-4 cm, depending on the size of the uterus, was made in the linea alba, and the uterus, the oviducts, and the ovaries were removed from the abdominal cavity. The uterus was removed lege artis after ligation of the blood vessels. The whole uterus including the implanted embryos was removed from the live animal in order to preserve the embryos’ transcriptomes optimally. After removal of the uterus, the animals were injected intracardially in deep and irreversible anesthesia with 1.0 to 2.0 mL Narcoren® (pentobarbital 160mg/kg body weight) for painless euthanasia. The females’ organs were collected and stored for additional follow-up studies in accordance with approved animal study plans.
All surgical procedures on the animals including anesthesia and analgesia were performed by specialized and experienced veterinarians.
Whole uterus embedding
Uteri containing implanted embryos were immediately cryopreserved in toto by shock-freezing in liquid nitrogen. Transfer of the tissue samples was performed on dry ice under temperature monitoring. Uteri were embedded unfixed into optimum cutting temperature (O.C.T) compound (4583, TissueTek) and snap-frozen to preserve RNA integrity.
Spatial Embryo Profiling (STEP)
Cryosectioning
Each O.C.T. block containing uteri with implanted embryos was sectioned fully at a thickness of 12 μm using a Leica cryostat microtome (CM3050) to obtain consecutive slices of the whole organ. During the cutting process, embryo tissue within the uterine cavity was confirmed using light microscopy of haematoxylin and eosin stainings prepared during sectioning. Embryo-tissue containing sections were collected either on Naphthalate (PEN) membrane slides (Zeiss, 1.0PEN) or histological slides (Superfrost Plus, Thermo Scientific) for laser capture microdissection and immunostaining, respectively, and immediately transferred to dry ice.
Laser capture microdissection
Membrane slides were processed using a Zeiss PALM microbeam laser capture station coupled to a Zeiss Axiovert 200 microscope. Unfixed sections were taken from dry ice storage and briefly (around 5 seconds) thawed at room temperature. Identification of tissue types was based on morphology and topology using the PALM brightfield set-up in 5x and 20x magnification.
Clusters of 1-3 cells (around 20-30 μm2) were cut at 20x magnification and pulsed directly into the lid of an 0.5 mL Eppendorf tube containing 15 μl RLTplus lysis buffer (1053393, Qiagen). Laser cutting speed, pulse intensity and focus was adjusted for each sample according to tissue type. Samples were incubated in lysis buffer for 2 minutes, spun down and immediately transferred to dry ice to prevent RNA degradation.
Brightfield microscope images at 20x magnification were taken prior to and subsequent to each sample’s collection (before and after microdissection of each cell) to preserve its spatial identity, allowing lineage allocation of each sample after RNA-sequencing.
Transcriptome library preparation
Sequencing libraries were prepared according to an adapted version of the G&T protocol previously described (adapted from 51, based on 52). LCM-processed samples were transferred to a 96-well plate and prepared for both transcriptome and methylome sequencing, by physical separation of genomic DNA (gDNA) from messenger RNA (mRNA). Genomic DNA was frozen and stored for processing in a separate study.
For each sample, RNA separation was performed using biotinylated oligo-dT30VN-tailed oligonucleotides (IDT) conjugated to Dynabeads Streptavidin C1 (65001, Invitrogen) in an RNAse-inhibitor (RNAsin; N2615, Promega) supplemented buffer solution.
For transcriptome libraries, complementary DNA (cDNA) was synthesized by reverse transcription using Superscript II (Invitrogen, 200 U/μL) and template-switching oligos (TSO; Exiqon, 100 μM) in 5x Superscript II first strand buffer (Invitrogen) containing RNAse-inhibitor (Promega, 1U/ μL), MgCl2 (Invitrogen, 1M), Betaine (Sigma, 5M), DTT (Invitrogen, 100 mM) and dNTPs (Roche, 10mM). Subsequently, material was amplified by PCR using the KAPA HiFi HotStart Readymix (KK2601, Kapa) and IS PCR primers (IDT, 10μM).
Sample clean-up was performed with AMPure XP beads (A63881, Beckman Coulter) at RT, using 80% ethanol, and cDNA samples were eluted in 20 μL elution buffer (Qiagen). For quality control, the DNA concentration of eleven randomly chosen samples per plate was measured using the Agilent Bioanalyser high sensitivity chip system (5067-4626, Agilent Technologies) according to the manufacturer’s protocol. Following successful quality control indicated by cDNA between 0.5 and 3 kb, tagmentation reaction was performed using the Nextera XT DNA kit (FC-131-2001, Illumina).
Samples were indexed using the Nextera XT 96-index kit (Illumina) and adapter-ligated fragments were amplified using the Nextera PCR master mix according to the manufacturer’s instructions. According to their quality, measured by the Bioanalyzer, samples volumes equivalent to a concentration in the range of 200-500pg were collected and pooled. Following a two-step library purification of the pooled samples with Ampure XP beads and 80% ethanol solution at RT (1:0.5 ratio and 1:0.2 ratio of beads to original volume), cDNA was eluted in 22 μl elution buffer and quality control was performed using the Bioanalyzer. Pooled libraries were sequenced on an Illumina HiSeq4000 platform with a read length of PE 150 bp.
Mapping of reads
Reads were trimmed of adapter sequences using TrimGalore!53 and mapped to the common marmoset genome (Callithrix jacchus 3.2.1) using STAR54 aligner v2.5.4. Only samples with >100,000 mapped reads and mapping efficiency >40% were used for downstream analysis. Gene counts were quantified using FeatureCounts55 v1.6.0 using a modified Ensembl gene annotation file (release 91).
Modification of marmoset genome annotation
Gene annotation files for common marmoset were downloaded from Ensembl (release 91) and gene models extended similarly to the approach of 56. Two modifications were considered, extending the transcription end site (TES) by 1kb or 5kb respectively. Transcripts <300bp apart that would overlap following extension were only extended by 500bp/2500bp.
Haematoxylin and Eosin staining of marmoset embryos
Slides were thawed briefly at RT and fixed for 7 min in fixation solution (4% PFA/PBS) as described previously. Fixation solution was removed by two gentle washing steps using PBS. Importantly, all following washing and staining steps were carried out using a P200 pipette due to the fragility of the tissue. For nuclei staining, filtered haematoxylin (10052574, Fisher Scientific) was applied for precisely 18 sec and then thoroughly washed with tap water several times. Samples were incubated in a drop of tap water on the slide for 1 min, followed by three wash steps using Millipore MQ water. Counterstaining of extracellular matrices and cytoplasm with eosin (Fisher Scientific) was performed for 23 sec. Slides were washed with MQ water and samples were dehydrated sequentially in 25%, 50%, 70%, 90% and 100% ethanol (three washes each). Slides were dried at RT and mounted using distyrene/plasticiser/xylene mounting medium (DPX; 06522, Sigma-Aldrich), and coverslips. After hardening of the mounting medium, slides were stored at RT until imaging.
Immunofluorescence staining of marmoset embryos
Slides were thawed at room temperature (RT) and fixed for 8 min in 4% paraformaldehyde (PFA)/phosphate-buffered saline (PBS) solution (15714S, Electron microscopy sciences/Thermo Fisher), followed washing steps using PBS (3x). Permeabilisation was performed in 0.25% Triton X100 (13444259, Thermo Scientific) in 0.3% polyvinyl pyrrolidone/PBS (Fisher Scientific) for 30 min at RT. Slides were rinsed (3x, PBS), and incubated for 30 min in blocking buffer (2% donkey serum (116-4101Fisher Scientific)), 0.1% bovine serum albumin (BSA; A9418, Sigma), 0.01% Tween20 (BP337-100Fisher Scientific) in PBS) at RT. Primary antibodies in blocking buffer at given concentration were incubated in a humidified chamber overnight at 4C.
Secondary antibodies supplemented with nuclear staining DAPI (4′,6-diamidino-2-phenylindole, Sigma) in blocking buffer were applied after washing steps (3x with PBS) and incubated for 60 min at RT. Slides were rinsed and mounted using Vectashield mounting medium (H-1200, Vector laboratories) and coverslips (12343138, Fisher Scientific). All washing steps were carried out by manual pipetting using a P200 pipette due to the fragility of the tissue.
Primary antibodies
SOX2 (1:100; R&D systems, MAB2018), TFAP2C (1:500, Abcam, ab218107 or 1:500, Santa Cruz, Sc12762 or 1:100, R&D systems, AF5059), OCT4 (1:200; Santa Cruz, sc5279 or 1:300, Santa Cruz, sc8628), TBXT (1:500; Abcam, ab209665), GATA2 (1:500; Abcam, ab173817), NANOG (1:400, Cell Signalling, 4893), KRT7 (1:100, Thermo Fisher, MA5-11986), GATA6 (1:100, R&D systems, AF1700), SOX17 (1:100, R&D systems, AF1924), OTX2 (1:300, R&D systems, AF1979).
Secondary antibodies
Alexa Fluor 488 Donkey anti-mouse (1:500; ThermoFisher, A21202), Alexa Fluor 555 Donkey anti-rabbit (1:500; ThermoFisher, A32727), Alexa Fluor 647 Donkey anti-goat (1:500, ThermoFisher, A21447).
Lineage annotation of STEP-transcriptomes
LCM-sample lineage identity was assigned based on the position within the embryo. Sample annotation was performed manually, side-by-side with phase contrast images acquired during sample collection and the confocal image with lineage markers (e.g. PDGFRA, OTX2, SOX2) of the same section. In addition, annotations were guided by the density and orientation of DAPI-labelled nuclei, which allowed us to discriminate between neighbouring tissues. We refined annotations by integration of lineage marker expression from immunofluorescence stainings and STEP-transcriptome data. Samples with more than one lineage signature were annotated as mixed and removed from downstream analysis. For LCM-sections without immunolabelling, we extrapolated lineage identity from immunofluorescence stainings of adjacent sections.
EmDisc
Nuclei density, pseudostratified arrangement and immunolabelling allowed us to distinguish EmDisc from neighbouring VE (EmDisc is PDGFRA-negative, VE is PDGFRA-positive). Core pluripotency markers POU5F1, SOX2, NANOG25 and primed pluripotency markers SFRP243, FZD757, DNMT3B24 validated EmDisc identity in the STEP-transcriptomes. Anterior SOX225 and posterior TBXT12, MIXL113 and EOMES13 expression confirmed anterior-posterior patterning in the EmDisc.
Am
At CS5 and CS6 Am was characterised by its squamous epithelial morphology and inner position towards the amniotic cavity. Am was readily distinguished from underlying ExMes by the absence of hypoblast-lineage marker PDGFRA24. At CS7, Am and ExMes were analysed jointly.
VE
VE appeared as cuboidal epithelium overlying the EmDisc and exhibited a substantially lower density of nuclei. At CS5 and CS6, IF markers for VE included SOX1725and OTX224. At CS7, VE is replaced by definitive endoderm and was analysed jointly with EmDisc.
SYS
At CS5, SYS samples were identified as protrusions from the VE pointing towards the central cavity and consistently observed on subsequent sections. At CS6, SYS markers SOX1725 and APOA124 were used to discriminate between ExMes and SYS. At CS7, SYS and ExMes were analysed together.
ExMes
For CS5 and CS6, ExMes was readily identifiable as cells of mesenchymal character (PDGFRA+) with random nuclear orientation, predominantly located between Am and Tb. At CS6, ExMes further expanded around the SYS and stalk. At CS7, ExMes samples were taken from the stalk region. At CS7, we were not reliable able to discriminate between Am and its surrounding ExMes as well as SYS and surrounding ExMes, which were extremely thin, often consisting of single-cell layers. As we could not reliably exclude ExMes in CS7-Am and CS7-SYS samples we analysed them jointly.
Tb
Tb predominately appeared as multinucleated syncytium, showing clusters of densely packed nuclei, in contrast to the adjacent less dense ExMes and maternal stroma. Tb was demarcated by TFAP2C58 and GATA214 immunofluorescence stainings.
PGCs
PGC samples were identified based on transcriptomic expression of TFAP2C, TBXT, SOX17, NANOG, NANOS3 and absence of SOX237,39.
Myometrium (Myo)
Myo samples were annotated based on the characteristic appearance of smooth muscles and the location in the outer region of the uterine wall
Endometrial Glands (Gland)
Glands appeared as columnar epithelium in the endometrium. Glands were SOX17+by immunofluorescence staining (Supplementary Fig. 1b, f).
Remodelled glands (ReGlands)
ReGlands lost their epithelial character and downregulated SOX17, while upregulating KRT7 (Supplementary Fig. 2).
Data reporting
No statistical methods were used to predetermine sample size. Experiments were not randomised and investigators were not blinded to allocation during experiments and outcome assessment.
Virtual marmoset embryo reconstruction
Image acquisition
Stained immunofluorescence slides of CS5 and CS6 embryo were imaged with an Inverted Leica SP8 confocal microscope with 2μm z-intervals with a x0.75 air objective at 20x magnification using acquisition software LASX (Leica microsystems). Tile scanning was performed to image the uterine cavity with embryo in its entirety, tile-scanned images were automatically merged by the acquisition software. Images obtained by confocal imaging were processed using ImageJ/Fiji software. Histogram of images were appropriately adjusted (values of grey with low number of pixels excluded) and ‘remove outliers’ option was applied where necessary to remove background speckles for presentation purposes. For z-projections, where indicated, ‘maximum intensity projection’ was applied.
Immunofluorescence-stained slides of CS7 embryos were imaged with a Zeiss Apotome 2 (widefield microscope with motorised stage) using an 0.8 M27 air plan apochromat objective to obtain whole uterine cavity images. Several images were acquired to cover the whole endometrial area and merged automatically by the Apotome software. Images were processed similarly to CS5 and 6 using ImageJ software.
For each embryo, all stained serial transverse sections with visible, intact embryo structures were used for embryo reconstruction. Broken sections were removed.
Image registration/alignment
Images were registered using the Fiji package MultiStackReg whereby each image was registered to the DAPI channel of the previous image using an affine transformation and applied to all other channels. Registered images were cropped to an identical region of interest. For sections processed by LCM, LCM-sample locations were manually annotated and tracked in Fiji.
Lineage segmentation
Lineage segmentation was performed by sequential nuclear segmentation and lineage annotation using CellProfiler 2.0 (open-source and available from the Broad Institute at www.cellprofiler.org)59. Nuclei were first segmented with the “IdentifyPrimaryObjects” module on DAPI images using intensity to identify objects and propagation methods to declump. The “MeasureObjectIntensity” module was used on all stained channels to store fluorescence intensity, and composite images were manually traced with the “MaskObjects” module to segment lineages based on morphology and marker expression. Lineage segmentation was documented by pre-drawing traces on confocal images (Supplementary Fig. 3-5) and comparison to output images of traced nuclei from “MaskObjects”.
Lineage-annotated nuclei coordinates were compiled into matrices in MATLAB and integrated with spatial sample information. Segmentation was further evaluated by plotting antibody fluorescence intensity stored from “MeasureObjectIntensity” to ensure marker expression corresponded to the expected lineage (i.e. TFAP2C showed high expression in trophoblast nuclei and mid-level expression in amnion nuclei). Spatial LCM sample coordinates were integrated into the nuclear segmentation matrix. Nuclei and spatial sample coordinates were aligned according to tissue center of mass on adjacent slides using custom scripts and scaled to appropriate dimensions between sections.
3D surface generation and modelling
Surfaces were generated from segmented nuclei coordinates using MATLAB built-in functions (Delauney triangulation) and custom scripts. Tissues were extended by scaling the last processed cross section using an inverse function with custom MATLAB scripts.
Generated surfaces were imported into Blender 2.81 (open-source 3D modelling and animation software, https://www.blender.org/) as object files (.obj) for smoothing into representative models. Sculpting was guided by imported confocal images placed at equivalent coordinates in the model. Subdivision-surface (Catmull Clark) was first applied to reduce poly-count and produce smooth surfaces between sections. A variety of digital sculpting tools and Blender add-ons were employed to generate non-overlapping tissues, with Boolean modifiers employed to generate tight seams between lineages. Final surfaces were made manifold using Mesh Clean-up tools and vertex editing. Surfaces were converted to quad-mesh with the “Remesh” tool to generate even topology with defined voxel size and exported as object files. Spatial LCM samples were re-projected onto final surfaces using custom scripts in MATLAB (https://github.com/Boroviak-Lab/SpatialModelling).
2D cross section generation
Completed 3D models were used for virtual sectioning through applying Boolean modifiers to each tissue at defined coordinates and exported as object files (.obj).
Animation
Animations of models were produced using standard Blender tools in the animation workspace. The “3D Print Toolbox” and built-in measurement tools were used to extract morphometric measurements of complete models.
Generation of 3D-transcriptomes in mouse and marmoset
Gaussian progress regression
Gaussian process regression (GPR)26,60 represents a Bayesian nonparametric approach to regression that is ideally suited to the analysis of spatial or temporal data, due to its flexibility and explicit treatment of uncertainty. Here we outlined the use GPR to reconstruct spatial expression patterns in developing embryos from potentially sparse or non-uniformly sampled spatial transcriptomics data. Initially we consider an embryo with only a single tissue. Individual cells (or sections) were assumed to have been sampled from known positions within the embryo, where the spatial domain of the embryo is denoted D ∈ R3. For any given gene, i, we have a vector of observations representing appropriately normalised gene expression measurements with a corresponding vector of spatial coordinates X = (x(1),…,x(N)) where x(j) ∈ D ∀j. Gene expression observations correspond to noisy instances of an unknown potentially nonlinear function:
| (1) |
where ϵ represents Gaussian additive noise. The functional form of fi(·) is unknown and may vary between genes. The aim is to infer it from the noisy observations in a nonparametric way. We first assigned the unknown function a Gaussian process prior, denoted fi(x)∼GP(μ(x), Cθ(x, x′)), where μ(x) represents the mean function, Cθ(x, x′) represents the covariance function, and θ represents any hyperparameters of the covariance function. If we assume gene expression varies smoothly over space we can use a squared-exponential covariance function: cθ(x, x′) = σ2 exp (–(x – x′)2/2l2), where θ = {σ, 1} represent the process-variance and length-scale hyperparameters, which respectively represent how varied the (normalised) gene expression levels are, and how rapidly gene expression can change over increments in space. Given our observations the posterior distribution at an arbitrary position within the embryo x* ∈ D can be inferred using Bayes rule and has the following Gaussian form:
| (2) |
where
where Kθ(.,.) denotes the covariance matrix, I the identify matrix, and σn the noise hyperparameter. Since prediction is dependent on the choice of hyperparameters θ, these are tuned in a data-driven way by maximising the marginal likelihood of the model:
| (3) |
This provides a comprehensive, scalable and data-driven approach for inferring nonlinear gene expression patterns within an embryo.
3D-transcriptomes of mouse E7.0 embryos
GPR for embryo reconstruction was implemented using the gpml package61 and initially evaluated on existing spatial transcriptomic data taken from E7.0 gastrulating mice29 (GSE65924). For each gene of interest, the expression levels for each section could be mapped to a position in 3D space corresponding to the surface of a concentric half-sphere. This geometry was chosen to recapitulate the cup-shaped epiblast of the E7.0 mouse embryo, with non-dimensionalised height h = 3, and inner and outer radii r1 = 0.75, and r2 = 1.2 respectively. Prior distributions over hyperparameters were set as: In(l)∼N(0,1), In (σf)~N(0,1), In(σn)∼N(ln(1/3),1). Hyperparameters were optimised via gradient descent, and GPR used to predict the posterior mean expression of specific genes at 20,000 randomly sampled position within the embryo. Inferred expression patterns for key genes were compared to existing in situ hybridisations.
3D-transcriptomes in marmoset CS5, CS6, and CS7 embryos
Following benchmarking in mouse datasets, GPR was used to spatially reconstruct gene expression patterns in marmoset embryos. Here we note that, unlike the mouse dataset, the marmoset CS5, CS6 and CS7 embryos contained multiple embryonic and extraembryonic tissues. Since expression patterns may be discontinuous across tissue types, we inferred an independent Gaussian process model for each tissue at each stage. The spatial domain for each tissue was predefined using blender denoted: {DEmDisc_CS5, DErnDisc_cs6,…DTb_cs7}. The position vector of each individual cell was mapped to the surface of the appropriate tissue domain e.g., for the CS5 EmDisc we have XEmDisc_cs5 = (xEmDisc_cs5(1),⋯,xEmDisc_cs5(N)) where xEmDisc_cs5(j) ∈ S(DEmDisc_cs5) ∀j, with a corresponding vector of normalised expression values . Prior distributions over hyperparameters were set as In(l)∼N(0,2), In(σf)∼N(0,2), In (σn)∼N(In(0.5),2), and a posterior Gaussian process could be evaluated using Equation (2). In some cases where it was expected different tissues formed a continuum of change and joint GP model could be considered by taking the union of observations. This was the case for EmDisc_CS7 and the stalk of ExMes_CS7.
For visualisation of results we evaluated the posterior mean of the GP at one of four sets of locations:
For overall visualisation of the embryos we constructed a high-resolution surface for each of the tissues in blender, denoted {S(DEmDisc_CS5), S(DEmDisc_CS6),…, S(DTb_cS7)},and evaluated the posterior mean of the GP at each point on the surface for every gene of interest. This provides a full 3D representation of the expression pattern for each tissue which could be visualised using the inbuilt MATALAB patch or mesh functions.
A 3D surface of points was also constructed corresponding to a section bisecting the embryo along a specific plane of interest. The posterior mean of the GP at these points was evaluated and visualised using the patch or mesh functions, corresponding to a virtual immunofluorescent staining.
For quantification and visualisation of anterior-posterior gradients in the EmDisc, we defined a curve s(x) using splines or polynomials that ran from the presumptive posterior end of the embryo (TBXT high) to the anterior end (SOX2 high) with 100 uniformly sampled points. The posterior mean and variance of the GP were evaluated along these increments and visualised as a line plots with uncertainty.
Similarly, for the visualisation of anterior-posterior gradients in VE we defined a curve that ran approximately parallel to the anterior-posterior curve in the EmDisc but following the natural shape of the VE. The posterior mean and variance of the GP was calculated at 100 uniformly sampled points along this curve and visualised as a line plot.
The inferred 1D patterns along a specified curve e.g., an AP axis, was projected onto a 2D circle to generate a virtual 2D-gastruloid pattern.
Crucially, due to the nature of GPs, we note that the underlying probabilistic model for all visualisations were consistent i.e., for each gene they represent the posterior mean (and variance) of the same GP evaluated at a different sets of test locations.
Statistical identification of spatially varying genes in the EmDisc and VE
Using GPR we could identify genes that significantly varied over their spatial domain as a basis for identifying anterior or posterior expressed genes, similarly to the approach outlined for time series data in62. Within a Bayesian setting the marginal likelihood, sometimes referred to as evidence, is often used to pick between two or more competing models. As described above, the marginal likelihood of a GP model, M1, with hyperparameter θ1, is given as:
| (4) |
This model corresponds to a GP model with a squared exponential covariance function as previously outlined. An alternative, simpler model could be considered where there exists no spatial variation in the expression levels of a given gene: any variance observed between points and between replicates are entirely down to biological variance in a set of flat functions and due to measurement noise. For this model we fit an alternative GP model, M2, in which the length-scale of the covariance function is set arbitrarily high compared to the length-scale of the system itself i.e., l → ∞. We can optimise the process variance and noise hypermeters using Equation (3), as previously outlined, to give θ1.
Within a Bayesian setting, the Bayes factor (BF) is used to determine the evidence in favour of one model over another and defined as:
| (5) |
where P (Mj) denotes the prior probability of model j which, since there is no strong a priori evidence in favour of one model or another, may be set to 0.5 for both models. When this ratio is equal to (or close to) one there exists no strong evidence in favour of a spatially-varying model over a simpler (flat) model, and Occam’s razor therefore favours simpler model. Likewise when this value BF ≫ 1 there is some evidence for a spatially-varying model over the simpler one, with the magnitude determining the strength of that evidence as outlined by Jeffreys63,64. Here we choose a stringent cut-off In In(BF) > 1 and overall variance var(Yi) > 0.5 to identify spatially varying genes. Finally, to determine which of the spatially varying genes showed anterior or posterior bias, we used the different in the predicted posterior mean at the anterior vs posterior ends of the EmDisc and VE, Δ, with Δ> 0.5 representing posterior genes Δ< -0.5 representing anterior genes.
Cell culture
Marmoset pluripotent stem cells (PSCs)
Embryo-derived conventional marmoset PSC lines no. 40, New2, and New4 (established at the CIEA) were maintained in KSR/bFGF media, which comprises Dulbecco's Modified Eagle Medium (DMEM)/F12 (21331, Gibco) supplemented with 20% Knockout Serum Replacement (KSR) (10828028, ThermoFisher Scientific), 1% GlutaMAX (35050061, ThermoFisher Scientific), 1% MEM non-essential amino acids (11140035, ThermoFisher Scientific), 100μM β-mercaptoethanol (21985023, ThermoFisher Scientific), and 10ng/mL bFGF (Cambridge Stem Cell Institute). Cells were routinely cultured on mitomycin C (M4287, Sigma) inactivated mouse embryonic fibroblast (MEF) feeder cells (Cambridge Stem Cell Institute) under 10% O2 and 5% CO2 at 37°C. Medium was changed daily, and cells were passaged every 2-4 days by dissociation with Accutase® (00-4555-56, ThermoFisher) for 5 minutes.
Marmoset naïve PSCs
For conversion to naïve pluripotency, conventional marmoset PSCs were seeded as clumps of 2-5 cells one day prior to resetting at 50,000 cells/12W (1.3 x 104 cells/cm2) on MEFs. After 24 hours, media was changed to PLAXA, which comprised N2B27 media (NDiff®, Y40002 Takara Bio) supplemented with 1μM PD0325901 (Cambridge Stem Cell Institute), 10 ng/mL recombinant human LIF (Cambridge Stem Cell Institute), 50 μg/mL L-ascorbic acid (Sigma), 2μM XAV939 (SM38-200, Cell Guidance Systems), and 20 ng/mL Activin A (Cambridge Stem Cell Institute). Throughout conversion, cells were passaged with Accutase® (00-4555-56, ThermoFisher) 1:1.5 every 3-4 days. Dome-shaped colonies first emerge at day 4-5 and naïve conversion is complete by day 9.
Marmoset neonate forebrain-derived cells
Neural cells were derived from the frontal lobe of a freshly culled marmoset neonate. Marmosets were bred at the Innes Marmoset Colony (Behavioral and Clinical Neuroscience Institute) in Cambridge, UK. The animals were housed in pairs under temperature (24 °C) and humidity (55%) controlled conditions. Marmosets were provided with a balanced diet and water ad libitum and cared for by experienced animal technicians. Research at this facility is regulated under the Animals (Scientific Procedures) Act 1986 Amendment Regulations 2012 following ethical review by the University of Cambridge Animal Welfare and Ethical Review Body.
The frontal lobe was surgically extracted, washed in N2B27 (NDiff®, Y40002 Takara Bio) and cut into small pieces using a scalpel. Dissociation was performed in a 1:1 mixture of 0.025% trypsin plus EDTA (25200056, Invitrogen) and 0.025% trypsin (Invitrogen) plus 1% chick serum (C5405, Sigma) by repetitive pipetting. Frontal lobe cells were seeded at low density on Fibronectin coated plates in N2B27 supplemented with bFGF (20ng/mL). Fibronectin coating was performed at 37 °C for at least 2 h using freshly prepared 20 ug/mL fibronectin (FC010, Millipore). Neonate forebrain-derived were collected for single-cell RNA-seq at passage 3.
Conventional human PSCs
Conventional SHEF665 were cultured on vitronectin-coated dishes (10 μg/mL; A14700, Thermo Fisher Scientific) in E8 medium (A1517001, Thermo Fisher Scientific) under hypoxic conditions (37°C, 5% CO2, 5% O2). Cells were routinely passaged in clumps using 0.5 mM EDTA. hPSC experiments were approved by the UK Stem Cell Bank Steering Committee and comply with the regulations of the UK Code of Practice for the Use of Human Stem Cell Lines.
Cell seeding and pattern induction in 2D-gastruloids
PEG-micropatterned plates were prepared as previously described66. Briefly, 500-μm diameter circular patterns were transferred to the surface of the PEG-coated side of the coverslip by photo-oxidizing select regions of the substrate using Deep UV exposure. Patterned slides were glued to bottomless 96-well plates to produce microtiter plates with patterned cell-culture surfaces. Prior to seeding cells onto the plates, the wells were activated with N-(3-Dimethylaminopropyl)-N'-ethylcarbodiimide hydrochloride (03450, Sigma) and N-hydroxysuccinimide (130672, Sigma) for 20 minutes. The plates were thoroughly washed 5 times with Phosphate Buffered Saline (PBS) and incubated with growth factor reduced Matrigel (Corning, CLS356230, diluted 1:100) overnight at 4°C. After coating, the plate was washed with PBS at least 2 times to get rid of any passively adsorbed extracellular matrix (ECM) before cell seeding.
Conventional marmoset PSCs were feeder depleted by 3 days of feed passaging cells 1:2 to feeder free culture. For transfer to feeder-free culture, 80-90% confluent PSCs were passaged by incubation with Accutase for 7 min at 37°C, at which point MEF lift from the tissue culture plate and are in suspension in Accutase but PSCs remain attached. Accutase was discarded, and attached PSCs were resuspended in MEF-conditioned KSR/bFGF (described above) and seeded on dishes coated with 5 μg/mL iMatrix-511 (Takara, T303). Conditioned media was prepared daily and supplemented with 10ng/mL bFGF (Cambridge Stem Cell Institute) and 2μM XAV939 (SM38-200, Cell Guidance Systems).
To seed marmoset PSCs on micropatterned plates, cells were dissociated by incubation with Accutase for 7 min at 37°C. Accutase was discarded and cells were resuspended in KSR/bFGF medium, then filtered through a 40μm cell strainer to achieve a single-cell suspension. PSCs were centrifuged and resuspended at a concentration of 1 × 106 cells/mL in feeder conditioned medium supplemented with 10 ng/mL bFGF (Cambridge Stem Cell Institute) and 10 μM ROCK inhibitor Y-27632 (#1254, Tocris). Wells were seeded at a density of 60,000 cells/well and incubated for 2 to 3 hours at 37°C. After 2 to 3 h, the medium was changed to conditioned media without ROCK inhibition. When confluent colonies were observed (12–18 h after seeding), wells were washed once with N2B27 (NDiff®, Y40002 Takara Bio) media to remove residual unattached cells and experimental conditions were applied. To seed feeder-free hPSCs on micropatterns, hPSCs were dissociated by incubation with Accutase for 5 min at 37°C, spun down at 200xg for 3 minutes, and resuspended in KSR/bFGF medium before proceeding to follow an identical protocol to cmPSCs, as previously described67,68. Cells were fixed at 2, 3, or 4 days after experimental conditions were applied for an alysis. Small molecules and growth factors used in cluded bFGF (10 ng/mL, Cambridge Stem Cell Institute), Activin A (20ng/mL, Cambridge Stem Cell Institute), BMP4 (50 ng/mL BMP4, R&D, #314-BP), IWP-2 (3 μM, Sigma Aldrich, 10536-5MG), CHIR99021 (3 μM, Cambridge Stem Cell Institute), Indian Hedgehog (IHH) (200 ng/mL, Abcam ab205517), and Cyclopamine (5 μM, Santa Cruz, CAS 4449-51-8) as specified in figure legends.
siRNA transfection
siRNA transfection was performed with PSCs seeded on either micropatterned plates as described above, or in μ-Slide 8 well dishes (80826, Ibidi) for control stainings. Pools of four siRNAs were used against each target gene (Dharmacon, Zoonome, SiPOU5F1: 5’-AAGCGAACCAGTATCGAGA-3’, 5’-GCAGCTTGGGCTCGAGAAG-3’, 5’-AGAATTTGTTCCTGCAGTG-3’, 5’-AACCCACACTTCAGCAGAT-3’; siNANOG: 5’-CTAAACTACTCTACGAACA-3’, 5’-CTGAAAGGAGGGTGGGGTA-3’, 5’-GAGTATGGATCCAGTTTGT-3’, 5’-GCATCCGACTGTGGAGAAT-3’; siSOX2: 5’-CGGAAAACCAAGACGCTCA-3’, 5’-ACATGAACGGCTGGAGCAA-3’, 5’-GCTCGCAGACCTACATGAA-3’, 5’-CGGAGATCAGCAAGCGCCT-3’; siTBX3: 5’-AGTGAGATGTTCTGGGCTA-3’, 5’-CATCGAACCTCAAAGATTT-3’, 5’-GGACAAACATGGATTTACT-3’, 5’-TGATGACTGTCGTTATAAA-3’; siID1: 5’-GACTTTAGAGGGTGGGATT-3’, 5’-GTTTGGTGCTTCTCAGATT-3’, 5’-CAAGAGGAATTAAGTTGCT-3’, 5’-CAGTGGTAGTTGCGCGCTG-3’; silD2: 5’-TTTCAAAGGTGGAGCGTGA-3’, 5’-CGTTAAAATCACAAGGAAT-3’, 5’-GGACCAGTGCTTTGATTTT-3’, 5’-GTATAGTGGCAGAGATGTC-3’; silD3: 5’-GTTCTGATGCCCTGATTTA-3’, 5’-GGAAGGTGACTTTCTGTAA-3’, 5’-GTATATAGCTTTTGTACCT-3’, 5’-AGCTTAGCCAGGTGGAAAT-3’, siSFRPI: 5’- CGGCCAGCGAGTACGACTA-3’, 5’-GCTTAAGTGTGACAAGTTC-3’, 5’- TCATGCAGTTCTTCGGCTT-3’, 5’-TCTCTGCGCCAGCGAGTTT-3’; siSFRP2: 5’- CGAGGAAGCTCCAAAGGTA-3’, 5’-ACTGAGACTCGGTGTGTAA-3’, 5’- GTGAGGAGATGAACGACAT-3’, 5’-GGAGATAACCTACATCAAT-3’; siGFP: 5’- GCAAGCTGACCCTGAAGTTC-3’). 25nM siRNA pool was complexed with 1.5 μL lipofectamine 2000 (Invitrogen, #11668030) diluted in 20 μL in Opti-MEM Reduced Serum Medium ThermoFisher, #31985062) for 15 minutes. For micropattern assays, 6x104 cells/well were seeded as described above in conditioned medium suppleme nted with Y-27. After 2-3 hours, micropatterns were washed once with KSR/bFGF and replaced with 80 μL conditioned media. In Ibidi 8-well dishes, similarly 6x104 cells were seeded per well, and media was changed to 80 μL conditioned media after 3 hours. Diluted siRNA/lipofectami ne was added to cells and incubated for 14 hours. Then, medium was changed to N2B27 (NDiff®, Y40002 Takara Bio) +10ng/mL bFGF (Cambridge Stem Cell Institute) +Activin A (Cambridge Stem Cell Institute) +50 ng/mL BMP4 (R&D, #314-BP) and analysed after 3 days or changed to N2B27 +bFGF +Activin A and analysed after 24 hours for micropattern or Ibidi control experiments, respectively.
Interphase set-up
Cells were feeder-depleted for interphase experiments as described above. μ-Slide 8-well ibiTreat dishes (80826, Ibidi) wells were coated with 40-45 μl of 100% growth factor reduced (GFR) Matrigel (Corning, CLS356230-1EA) and kept at 37°C to solidify until cell seeding. Cells cultured in 6-well plates were dissociated by incubation with 500 μL Accutase (00-4555-56, ThermoFisher) for 8 mins at 37°C, centrifuged at 200xg for 5 min and resuspended in 9.5 mL KSR/bFGF per well. Cells were coun ted to obtain 50,000 cells per experimental well, spun down again and seeded onto the GFR Matrigel beds resuspended in 100 μl N2B27 (NDiff ¢, Y40002 Takara Bio) +10 μM ROCK inhibitor Y-27632 (#1254, Tocris) and kept at 37°C for 30 minutes to attach. Next, medium replaced with 80-90 μL N2B27 supplemented with 1% GFR-Matrigel and signalling factors of respective experimental condition (a small amount of medium stayed in well during medium change). All experimental conditions were based in N2B27 and supplemented with small molecules and growth factors including bFGF (10 ng/mL, Cambridge Stem Cell Institute), Activin A (20ng/mL, Cambridge Stem Cell Institute), BMP4 (50 ng/mL BMP4, R&D, #314-BP), Noggin (200 ng/mL, Qkine, Qk034), IWP- 2 (3 μM, Sigma Aldrich, 10536-5MG), and CHIR99021 (3 μM, Cambridge Stem Cell Institute) as specified in figure legends. Medium was changed daily and medium with signalling factors was prepared freshly every second day.
Immunofluorescence of cultured cells
Cells were cultured in μ-Slide 8 well dishes (80826, Ibidi) or micropatterned plates (described above) and subjected to standard immunofluorescence staining protocols. Briefly, cells were fixed with 4% paraformaldehyde in PBS for 10 minutes, or for phospho-SMAD1/5 staining cells were fixed with ice cold methanol for 3 minutes. Next, samples were rinsed, and permeabilized in PBS/PVP containing 0.25% Triton™ X-100 (X100-100ML, Sigma) for 30 minutes, then blocked in blocking buffer (2% donkey serum (116-4101, Fisher Scientific)), 0.1% bovine serum albumin (BSA; A9418, Sigma), 0.01% Tween 20 (BP337-100, Fisher Scientific) in PBS) at RT. Primary antibodies were diluted in blocking buffer and stained overnight at 4°C. Secondary antibodies and DAPI were incubated in blocking buffer and stained for 60-90 min at room temperature.
Primary antibodies
KLF17 (1:100; Atlas Antibodies, HPA024629), SOX2 (1:100; R&D systems, MAB2018), TFAP2C (1:500, Abcam, ab218107), OCT4 (1:200; Santa Cruz, sc5279), TBXT (1:500; Abcam, ab209665), GATA2 (1:500; Abcam, ab173817), KLF4 (1:200; Santa Cruz, sc20691), NANOG (1:400, Cell Signalling, 4893), PAR6 (1:200, Santa Cruz, sc67393), aPKC (1:200, Santa Cruz, sc216), β-catenin (1:300, Cell signalling, cs8480), ZO-1 (1:300, Thermo Fisher, ZO1-1A12), OTX2 (1:300, R&D systems, AF1979), CDX2 (1:100, Cell Signalling, 3977), Phospho-SMAD1/5 (1:100, Cell Signalling, 9516), LEF1 (1:100, Cell Signalling, 2230), ISL1 (1:50, Developmental Studies Hybridoma, 39.4DS), TFAP2A (1:100, Invitrogen, MA1-872), VTCN1 (1:250, Abcam, ab209242).
Secondary antibodies
Alexa Fluor 488 Donkey anti-mouse (1:500; ThermoFisher, A21202), Alexa Fluor 555 Donkey anti-rabbit (1:500; ThermoFisher, A32727), Alexa Fluor 647 Donkey anti-goat (1:500, ThermoFisher, A21447)
F-actin staining: Acti-Stain 670 (1:100; Cytoskeleton, Inc., PHDN1-A)
Immunofluorescence images were acquired on an inverted SP8 confocal microscope (Leica Microsystems) with the 20x/0.75 IMM CS2 (HC PL APO) objective with a z-stack of 2 μm. Image preparation was conducted in Fiji software69. Laser power and detector gain were maintained constant within a single experiment.
Brightfield images were acquired on an EVOS XL Core Imaging System (AMEX1100, ThermoFisher).
Image analysis for 2D-gastruloids
For image analysis and quantification of micropatterned 2D gastruloids, each 500 μm diameter colony was captured with a single 20x image acquired on an inverted SP8 as described above in 5-6 z-slices. Maximum projection images of individual micropatterned 2D gastruloids were prepared for each channel and exported as TIFFs in Fiji69. Nuclei segmentation and antibody fluorescence measurements were conducted in CellProfiler59, which preserves the X,Y coordinate of each nucleus measured. Data were imported into MATLAB and each colony was centred to its centroid with nuclei assigned to hexagonal bins for colony overlay. Protein expression was normalized to maximum expression from the experiment, and protein expression was calculated as the number of cells that expressed the protein of interest as a percentage of the total number of cells within the colony. The frequencies of fluorescence intensity values were plotted for each experiment to determine gates for calculation of the percentage of cells expressing the protein of interest. Spatial expression trends within colonies were visualized as line plots, in which cells are grouped by the Euclidean distance between a cell and the centroid of the colony. The plot profile extracted was then run through a Savitzky–Golay smoothing filter in MATLAB and represented as a function of distance from the colony centre. Pairwise two-tailed Mann Whitney test was used to compare pairwise to control in GraphPad Prism.
Single-cell RNA-seq
Single-cell RNA sequencing of embryo-derived cell lines
Cells were dissociated in Accutase (00-4555-56, ThermoFisher) for 5 minutes and pipetted vigorously to obtain a single cell suspension in normal culture media. Single cells were picked by mouth pipette using a blunted microcapillary with internal diameter 15 μm and under a dissection microscope. Cells were collected into 0.2 mL tubes with 4 μL lysis buffer and immediately snap frozen on dry ice. Smart-seq2 library preparation was carried out in 96W format as previously described23. Library quality was assessed using the High Sensitivity DNA Analysis Kit (5067-4626, Agilent) on the 2100 Bioanalyzer system (Agilent).
Single-cell RNA sequencing of preimplantation samples
Marmoset preimplantation trophoblast and 4-cell stage embryos newly profiled in this study were obtained from self-sustaining colonies at the CIEA. Marmoset embryos were obtained by in vitro fertilization for 4-cell stage or non-invasive uterine flushing for blastocysts, as previously described20,70. Marmoset embryos were staged based on embryonic day, diameter and blastocoel formation as previously described 71. Zona pellucidae were removed using acidic Tyrode’s solution (T1788, Sigma). 4-cell embryos were dissociated to single cells in 1:1 0.025% trypsin plus EDTA and 0.025% trypsin plus 1% chick serum while early blastocysts were dissociated in 0.025% trypsin plus EDTA (Invitrogen). Early blastocysts and 4-cell embryos were incubated for 5–20min at 37°C, until cell boundaries appeared rounded and distinct. Embryos were subsequently washed in N2B27 supplemented with 10μM HEPES (15630056, Gibco) and 1.5mg/mL BSA (A9418, Sigma) and dissociated in a small drop of medium using blunted microcapillaries pulled to an inner diameter large enough to accommodate approximately 2–3 cells. Single cells were transferred immediately into 4 μl single cell lysis buffer and processed identically to embryo-derived cells.
scRNA-seq data from preimplantation marmoset data was mapped along with existing preimplantation marmoset data 24 (ArrayExpress accession E-MTAB-7078) using an identical pipelines to the CS5, CS6 and CS7 embryos. Briefly, reads were trimmed of adapter sequences using Trim Galore! and mapped to Callithrix jacchus 3.2.1 using STAR. Gene counts for cells passing QC were quantified using FeatureCounts using the modified Ensembl gene annotation file.
Bioinformatics
Data visualisation and analysis
Marmoset samples that passed QC were analysed using Seurat54,55 v3.1.2. Feature counts were normalised and standardised using the NormalizeData and ScaleData function using either the top 2,000, 5000 or 20,000 most varied genes for downstream analysis. Principle component analysis was run using the inbuilt RunPCA function, with nonlinear dimensionality reduction techniques generated using RunUMAP or RunTSNE (perplexity=40). Dimensionality reduction was visualised in 2D or 3D using ggplot or scatterplot3D. Marker genes were identified using FindMarkers for cell or cluster specific comparisons or FindAllMarkers using a Wilcoxon Rank Sum test (only positive, minimum percent 50%, log FC threshold 0.25). Lineage specific expression profiles for key marker genes were calculated using the AverageExpression function and visualised using pheatmap 1.0.12 or using the DoHeatmap function using all DEGs with adjusted p-value <0.05.
GO terms for individual lineages were generated with all DEGs with p val adj <0.05 as input to GO_Biological_Process_2017b 72,73 in enrichR (https://cran.r-project.org/web/packages/enrichR/index.html).
Stream plots were plotted as proportional stacked area using ggplot274 and ggalluvial75 in R. Track widths represent proportional gene expression normalized to the mean expression across developmental stages displayed.
Mouse spatial transcriptomics
Data of existing mouse spatial transcriptomics29,30 (GSE65924, GSE120963) were reprocessed: reads were trimmed using Trim Galore! and aligned to mouse genome mm10 using STAR aligner. Gene counts were quantified using FeatureCounts and normalised in Seurat. Data was visualised in MATLAB following 3D GP reconstruction or in 2D using corn plots30.
Cross-species data integration and analysis
Data from human peri-implantation in vitro cultures and from cynomolgus pre- and post-implantation embryos were processed and analysed alongside our marmoset data. Data from the human peri-implantation in vitro cultures12 (GSE136447) were trimmed and aligned to human genome hg19 using Trim Galore! and STAR respectively. Reads for cynomolgus pre- and post-implantation embryos56(GSE74767) were trimmed and aligned to Macaca fascicularis genome 5.0 using Bowtie3 v1.2.2. Gene counts for human and cynomolgus datasets were quantified using FeatureCounts. Normalised expression data from the human CS7 gastrula was downloaded from http://www.human-gastrula.net.
Genes for marmoset and cynomolgus were first annotated according to their closest human orthologues using the Ensembl BioMart database (https://m.ensembl.org/info/data/biomart/index.html). Count data from marmoset, Cynomolgus, and human in vitro cultures were jointly analysed using Seurat based on Canonical Correlation Analysis (CCA) and mutual nearest neighbour (MNN) approaches. Specifically, FindIntegrationAnchors was run using 4000 features and IntegrateData (with 20 dimensions) was used to calculate corrected gene expression matrices for the three datasets. Datasets were visualised using PCA on the corrected gene expression matrix. Conserved markers for lineages were identified using the FindMarkers function on the 4000 integration genes. Marker expression of key markers was visualised using a scatter plot on the first two principle components. Where applicable, diffusion maps were generated using corrected expression matrices with using destiny 2.12.0, and the reduced dimensional representation loaded into a custom slot in Seurat for visualisation.
Identification of conserved and species-specific genes was achieved by first sub-clustering cells following integrative analysis, and then comparing a subcluster of interest with a reference cluster for each species and visualising as a 2-species scatter plot. Specifically, the x-axis shows log fold change of a cluster of interest in marmoset cells versus a reference cluster in marmoset cells, with the y-axis showing log fold change of the cluster of interest in human CS7 cells versus the reference cluster in human CS7 cells. In most cases, the reference cluster was chosen to be the joint cluster expressing pluripotency markers. Such plots can split into four quadrants, with the upper right hand quadrant representing genes up-regulated in both species (compared to their reference cell type), the lower left hand quadrant representing genes down regulated in both species (compared to their reference), and the upper left and lower right hand quadrants representing species-specific behaviour.
Spatial mapping of cell lineages to embryo stages and spatial domains
In vitro cultured cells were mapped to appropriate stage and spatial domains using a two-stage approach. Primed and naïve ESCs were mapped separately to CS3, CS5, and CS6 embryos. Specifically, for conventional ESCs, the Pearson correlation coefficient between each of the m in vitro cultured PS cells and each individual cell from the embryo was first calculated using R’s inbuilt cor function, to yield a vector of correlations YConv = (RES_cell(1),EmDisc_CS5(l), RES_cell(l),EmDisc_CS5(2),,⋯, RES_cell(n),Tb_CS6(m)), for these observations a corresponding spatial position exists, corresponding the position of each the embryonic cell, XConv = (xEmDisc_CS5(l), xEmDisc_CS5(2),,⋯,xTb_CS6(m)). To visualise the overall correlation pattern over all cells and at intermediate locations, an independent GP model could be constructed for each tissue in each embryo. A similar analysis could be used to infer identity patterns of marmoset naïve ESCs.
For the spatial mapping of microfluidic derived embryonic sac data16, 10X data was aligned to our marmoset dataset using Seurat and subclustered. Average expression of the corrected expression matrix for the PSC and amnion subclusters were then calculated and used to construct heatmaps or to project to the marmoset CS6 embryo as described above.
Extended Data
Extended Data Figure 1. Overview of the STEP method.
1. Cryosectioning of the pregnant uterus: Pregnant marmoset uteri were extracted, embedded in O.C.T. and snap-frozen. To provide the best possible RNA quality, the tissue was processed unfixed for laser capture microdissection (LCM)-mediated Smart-Seq2 sequencing. 2. Stereological immunofluorescence imaging: Immunostainings for established lineage markers were performed for every other to every third section containing the implanted embryo. Tile-scan images were generated tile-scan with a confocal or apotome microscope. 12μm-thick cryosections were catalogued in ascending order, which was used to determine the Z-coordinate of LCM-transcriptomes. 3. LCM assisted sample collection: Every other to every third section was processed for LCM sample collection and all LCM-processed sections were subjected to immunofluorescence (IF) with lineage markers afterwards. For transcriptome sample acquisition, a photo was taken of the section. Then, one to three cells in a region of interest were selected using the LCM software Zeiss PALM and cut out by the laser. In a second step, a pulse laser catapulted the sample into a collection tube with lysis buffer. Then, a second picture was taken of the section at the matching position with the sample removed and matched to the collection tube and image file. Each STEP-transcriptome was assigned an individual ID and lineage identity based on the original location within the embryo cryosection, i.e. the cut-out location of the LCM-sample. 4. Sample annotation: Collected LCM-samples were subjected to the Smart-Seq2 protocol and sequenced to an average depth of > 2 million 150bp paired reads. LCM-sample lineage identity was assigned based on the position within the embryo. Sample annotation was performed manually, side-by-side with phase contrast images acquired during sample collection and the confocal image with lineage markers (e.g. PDGFRA, OTX2, SOX2) of the same section. In addition, annotations were guided by the density and orientation of DAPI-labelled nuclei, which allowed us to discriminate between neighbouring tissues. We refined annotations by integration of lineage marker expression from immunofluorescence stainings or STEP-transcriptome data. Samples with more than one lineage signature were annotated as mixed and removed from downstream analysis. 5. Image registration and lineage segmentation: Images were aligned by image registration in Fiji, whereby each image was registered to the DAPI channel of the previous image. Next, nuclei were segmented into individual objects using Cell Profiler. For lineage segmentation, the segmented nuclei were assigned lineages based on lineage marker immunostaining (e.g. POU5F1 to demarcate the EmDisc and Amnion, PDGFRA for ExMes, VE and SYS, TFAP2C for trophoblast, Amnion and PGCs), and taking into account known anatomical features of the embryo (e.g. EmDisc resides in-between VE and Amnion). 6. Transcriptome coordinate integration: The X and Y coordinates of the annotated transcriptomes were compiled into MATLAB matrices. 7.Virtual 3D embryo reconstruction: We generated primary surfaces in MATLAB by triangulation (see Methods). In a second step, embryonic and extraembryonic surfaces were smoothened in Blender, an open-source 3D modelling and animation software. 8. Gaussian Process Regression (GPR) over LCM samples: LCM spatial transcriptome sample coordinates were integrated into the 3D embryo models and continuous expression patterns between discrete LCM samples were inferred using Gaussian Process Regression (see Methods). Since expression patterns may be discontinuous across tissue types, we inferred an independent GPR model for each tissue at each stage. 9. GPR spatial transcriptome models and virtual cross sections: Final smooth GPR gene expression patterns could be displayed in 3D on embryo models. Defined coordinates were used to extract expression patterns in each lineage for visualisation of virtual cross sections. Scale bars represent 100 μm. cs, Carnegie stage; O.C.T, Optimal cutting temperature compound (used to mount uteri); EmDisc, Embryonic disc; Am, Amnion; SYS, Secondary Yolk Sac; VE, Visceral Endoderm; Tb, Trophoblast; ExMes, Extraembryonic mesoderm; PGCs, Primordial Germ Cells.
Extended Data Figure 2. Staging of marmoset postimplantation embryos.
a, Staging of marmoset embryos based on listed hallmarks allowed us to stage blastocysts (Carnegie Stage (CS) 3), CS5, CS6 early and late, and CS7 embryos. Middle panel: illustrative cross section of embryo models for each stage with stage-specific differences in (1) secondary yolk sac, (2) Am, (3) stalk, and (4) ExMes formation indicated. Bottom panel: Representative images from human embryos at corresponding Carnegie Stages. CS3 reprinted from47, CS5, CS6 late, and CS7 from48, and CS6 early from49.
b, Principal component analysis of marmoset development of CS5-7 integrated by stage based on whole transcriptome (>20,000 genes).
c, Immunofluorescence stainings of marmoset embryo sections. Scale bars represent 100 μm. EmDisc, Embryonic disc; Am, Amnion; SYS, Secondary Yolk Sac; VE, Visceral Endoderm; Tb, trophoblast; ExMes, Extraembryonic mesoderm; PGCs, Primordial Germ Cells.
Extended Data Figure 3. Gaussian Process Regression models of spatial transcriptomics in mouse and marmoset embryos.
a, Spatial modelling of mouse postimplantation embryos based on published mouse embryo expression data shown for representative genes markers of anterior/posterior (A/P) epiblast (Epi), primitive streak (PS) and anterior primitive streak (APS). Each kernel on corn plots represents a micro-dissected and sequenced section of the mouse embryo from ref4,5. Corn plots were transformed into spatial models to match anterior-posterior and left-right axis. Gaussian process regression allowed visualisation of gene expression gradients, which were compared to in-situ hybridisation of postimplantation mouse embryos for validation of gaussian process regression approach. Source publication indicated next to individual images.
b, Projection of shots on 3D virtual reconstructions. LCM-samples projected as black dots on 3D virtual models of each developmental stage used for downstream gene expression analysis. EmDisc, Embryonic Disc; VE, Visceral Endoderm; ExMes, Extraembryonic Mesoderm; A, Anterior; P, Posterior.
c-d, Spatial embryo profiling LCM-sample lineage assignment examples. LCM-samples that were spatially close were assigned to amnion or ExMes based on PDGFRA expression. PDGFRA immunostaining of the CS6 early embryo demonstrates that amnion (nuclei adjacent to the amniotic cavity) is PDGFRA-negative (highlighted in inset below). Raw PDGFRA feature counts of individual LCM-samples overlaid on DAPI recapitulate immunostaining pattern, showing high expression in ExMes samples and no expression in amnion samples. LCM-samples from other lineages, that showed mixed lineage identity, or did not pass QC are not displayed. Arrowheads indicate PDGFRA-negative amnion. Cross-section of 3D-model (right) represents gaussian process regression-based modelling of all lineages in CS6 early embryo, recapitulating specific PDGFRA expression pattern observed by immunofluorescence and raw transcriptome data.
e-h, Spatial embryo profiling processing example. Representative example of a CS6 section processed by spatial embryo profiling and immunofluorescence staining for SOX2 (e) and OTX2 (g). Gaussian process regression-based modelling of EmDisc and VE for SOX2 recapitulates the anterior EmDisc expression pattern observed by immunofluorescence and raw transcriptome data (f), and recapitulated anterior expression of OTX2 recapitulates the anterior VE expression pattern observed by immunofluorescence and raw transcriptome data (h). Upper panel: Relative mRNA levels for gene expression across the model. Lower panel: mRNA expression changes along anterior-posterior axis (dashed line, anterior EmDisc (red, A) to posterior EmDisc (blue, P), anterior VE (yellow, A) to posterior VE (green, P)) change along A-P axis quantified by Bayesian factor (BF).
Extended Data Figure 4. Expression gradients and signalling environment in marmoset gastrulation.
a, Posterior markers depicted in Gaussian process regression-based 3D models of CS5-7 EmDisc and VE. Upper panels: Relative mRNA levels across the model. Lower panels: mRNA expression change along EmDisc anterior-posterior axis (indicated by dashed line; anterior (red, A) to posterior (blue, P)), quantified by Bayes Factor (BF) (relates to Figure 3a).
b, BMP signalling-related gene expression depicted in CS5-7 model cross sections. Schematic (bottom) summarises BMP signalling pathways in the context of amnion differentiation from EmDisc boundaries in CS5 and 6.
c, WNT signalling genes shown in CS5-7 in model cross sections. Schematic summarises WNT signalling patterning in the CS6 EmDisc during gastrulation.
d, RTK-related gene expression depicted in CS5-7 model cross sections displays VE is the primary source of IGF1, low expression of FGFs involved in mouse gastrulation (FGF8, FGF5), and presence of FGF4 and intracellular FGFs (FGF12, FGF13).
e, RTK-related gene expression depicted in EmDisc and VE in CS5 and 6 3D models. Schematic summarizes PDGFA and VEGFA in the CS6 embryo.
f, IHH signalling-related gene expression shown in CS5-7 in model cross sections. Schematic summarises proposed paracrine IHH signalling pathways.
g, Representative anterior pluripotency genes depicted in CS5-7 EmDisc and VE 3D models (relates to Fig 3e)
h, Corn plots of matched pluripotency genes in the gastrulating mouse embryo at E6.0, E6.5, and E7.05. Each kernel represents the average transcriptome of micro-dissected, spatially-defined sections of mouse embryos (relates to Figure 3c).
i, Early (NANOG, PRDM1, POU5F1, KLF4) and late (DAZL, MAEL, PRAME) PGC marker and enriched signalling components (FGF4, WNT8A) expression in PGCs, depicted in CS6 model cross sections.
EmDisc, Embryonic Disc; SYS, Secondary Yolk Sac; VE, Visceral Endoderm; ExMes, Extraembryonic Mesoderm; Am, Amnion; Tb, Trophoblast.
Extended Data Figure 5. Amnion segregation from the Embryonic disc.
a, PCA of EmDisc and Am based on the top 5000 most variable genes, PC1=20.7%, PC2=13.6%.
b, Marker expression delineates the divergence of EmDisc and Am. Genes enriched in EmDisc and Am are marked in blue and purple, respectively; preimplantation genes are depicted in green. Stream plot track width is scaled to relative expression normalized to the mean across all stages displayed.
c, Heatmap of expression of differentially expressed genes (DEG) in embryonic and extraembryonic lineages displayed in (a, b). Representative genes (left panel) and key gene ontology (GO) enrichment analysis (right panel) are shown. Genes shown in heatmap from Seurat function FindAllMarkers (minimum percent 50%, minimum log fold change 0.25) and filtered by adjusted p-value <0.05.
d-i, Virtual cross-sections of 3D-transcriptomes at CS5, 6 and 7 depicting mRNA levels of representative genes for (d) pluripotency factor expression in the nascent Am, (e) Am-Tb shared genes, (f) Am-ExMes shared genes, (g) Am-specific genes, (h) epithelial genes, (i) ECM-related genes.
Categories indicated on the left of each panel. EmDisc, embryonic disc; Am, amnion.
Extended Data Figure 6. Endogenous WNT required for posterior patterning in marmoset 2D gastruloids models.
a-c, pSMAD1/5 (phosphorylated SMAD 1/5) activity in marmoset 2D-gastruloids detected by immunostaining at day 2 (a) or day 4 (b) compared to human 2D-gastruloids under conventional conditions at day 2 (c). Micropatterned colonies were treated with self-renewal conditions (10 ng/mL FGF + 20 ng/mL Activin A), conventional gastruloid conditions (10 ng/mL FGF + 20 ng/mL Activin A + 50 ng/mL BMP4) or WNT modulatory conditions (10 ng/mL FGF + 20 ng/mL Activin A + 3 μM IWP-2 or 10 ng/mL FGF + 20 ng/mL Activin A + 3 μM CHIR99021). Representative maximum projection of immunofluorescence images (left). Quantification plots (right) mean +/- SEM across a minimum of 10 gastruloids across 2 wells. F/A = FGF/Activin A. pSMAD1/5 gradient indicates that in the marmoset system, similar to the human, FGF/ActivinA/BMP4 induce a graded response to BMP signalling at day 2, with the highest signalling in the outermost ring of the colony.
d, Molecular characterisation of 2D-gastruloids. Representative immunofluorescence images of gastruloids differentiated in conventional gastruloid conditions (10 ng/mL FGF + 20 ng/mL Activin A + 50 ng/mL BMP4) for 2 days. Quantification plots (bottom) display mean +/- SEM across a minimum of 10 gastruloids across 2 experiments. Anterior domain (A, SOX2+, TBXT-, TFAP2C-), posterior domain (P, TBXT high, CDX2 heterogeneous, SOX17 sparse, LEF1 sparse), and amnion domain (Am, TFAP2C high, SOX2-) demarcated. ISL1 and TFAP2A are observed heterogeneously predominantly in the amnion region. PDGFRA expression is low, indicating lack of mature mesoderm.
e, Schematic of lineage identities present in 2D-gastruloids and marker patterns that define each region.
f, WNT-associated anterior/posterior patterning phenotypes of 2D-gastruloids. Representative immunofluorescence images of gastruloids differentiated in conditions listed at left for 2 days. Quantification plots (bottom) mean +/- SEM across a minimum of 10 gastruloids across 2 wells.
g, WNT-associated amniogenesis phenotypes of 2D-gastruloids. Representative immunofluorescence images of gastruloids differentiated in conditions listed at left for 2 days. CDX2 expression is lost upon 3 μM IWP2 treatment, but TFAP2C and TFAP2A remain expressed. 3 μM CHIR treatment (WNT agonist) leads to low CDX2 expression but does not support upregulation of TFAP2C or TFAP2A. Quantification plots (bottom) mean +/- SEM across a minimum of 10 gastruloids across 2 wells.
h, No evident change in expression profiles associated with Hedgehog signalling manipulation in 2D-gastruloids. Representative immunofluorescence images of gastruloids differentiated in conditions listed at left for 2 days. Exogenous Indian Hedgehog (IHH, 200 ng/mL) did not lead to loss of pluripotency under FGF/Activin self-renewal conditions, or change expression patterns under conventional gastruloids conditions. Inhibition of hedgehog signalling with Cyclopamine (5 μM) also did not lead to evident changes in expression patterns. Quantification plots (bottom) mean +/- SEM across a minimum of 10 gastruloids across 2 wells.
Extended Data Figure 7. siRNA in 2D marmoset gastruloids.
a, siRNA knockdown efficiency. Representative immunofluorescence images (left) and quantification of mean fluorescence intensity (right) 24 hours following transfection with siRNA against POU5F1, SOX2, or NANOG. Comparisons to siGFP (green fluorescent protein) control conducted with two-tailed Mann-Whitney test (****, p<0.0001). siRNA: small interfering RNA
b, Schematic of siRNA screening approach. cmPSCs (common marmoset pluripotent stem cells) were seeded in micropatterned 96-well plates on day -1 and transfected overnight with siRNA. On day 0, media was changed to gastruloid induction media (10 ng/mL FGF + 20 ng/mL Activin A + 50 ng/mL BMP4). 2D-gastruloids were fixed after 72 hours and stained to assess pattern formation.
c, Comparison of siGFP phenotype to in vivo EmDisc patterns. Representative maximum projection immunofluorescence image of siGFP-treated gastruloids differentiated in conventional gastruloid conditions (10 ng/mL FGF + 20 ng/mL Activin A + 50 ng/mL BMP4) for 3 days (left). 2D-gastruloid log expression patterns normalized to maximum intensity plotted for individual channels (SOX2, TBXT, TFAP2C) side by side with virtual embryo pattern of CS6 EmDisc expression patterns generated by Gaussian process regression of anterior-posterior axis expression. Quantification plot (right) shows mean +/- SEM across a minimum of 10 gastruloids with anterior domain (A, SOX2+, TBXT-, TFAP2C-), posterior domain (P, TBXT high), and amnion domain (Am, TFAP2C high, SOX2-) demarcated.
d-f. siRNA knockdown phenotypes of pluripotency factors (siPOU5F1, siNANOG, siSOX2), BMP-related genes (siID1/2/3, siTBX3) and WNT-related genes (siSFRP1, siSFRP2, siSFRP1/2). For each siRNA, a representative maximum projection immunofluorescence is shown (left). Representative expression patterns are plotted for individual channels (SOX2, TBXT, TFAP2C) adjacent to quantification of the percent of nuclei positive for each marker per gastruloid (center). Comparison conducted with two-tailed Mann-Whitney test (ns: not significant; p<0.0332 (*); p <0.0021 (**); p<0.0002,(***); p<0.1; p < 0.0001 (****)).
Quantification plot (right) shows mean +/- SEM across a minimum of 10 gastruloids across 2 wells. SOX2 = green, TFAP2C = red, TBXT = grey. siGFP patterns plotted for comparison in reduced opacity and dashed line with control anterior domain (A, SOX2+, TBXT-, TFAP2C-), posterior domain (P, TBXT high), and amnion domain (Am, TFAP2C high, SOX2-) demarcated.
Extended Data Figure 8. 3D in vitro modelling of the marmoset EmDisc.
a, Schematic overview of interphase culture. To model self-organisation of conventional cmPSCs (common marmoset pluripotent stem cells) in 3D culture, cmPSCs were seeded on a 100% Matrigel base overlaid with 1% Matrigel dissolved in N2B27 media supplemented with signalling factors. Interphase culture was amenable to probing signalling requirements that promote anterior embryonic disc-like pluripotency or differentiation into the germ layers of the gastrulating embryo. All experiments were performed in two different cell lines (cell line #1 (New4f), N=2 and cell line #2 (New2f), N=2) showing consistent results.
b, Heatmap of marker genes used for molecular characterisation of interphase culture structures. Relative mRNA levels were centred and scaled across all marmoset in vivo samples.
c, Summary schematic of NODAL and FGF signalling in the marmoset embryo. The visceral endoderm is the primary source of NODAL and IGF1 in the marmoset embryo, while the EmDisc expresses low levels of FGF4. Relative mRNA expression gradients summarised in CS6 cross section.
d, Time series brightfield images of interphase culture with FGF (100ng/ml) and Activin A (20ng/ml). FGF/Activin A culture provides a signalling environment that mimics high NODAL and IGF1 from the VE. Structures formed columnar epithelial cysts, reminiscent of the embryonic disc. Structures first open a lumen at day 3 and expand up to day 6.
e-f, Molecular characterisation of EmDisc-like structures at day 4. Representative maximum projection images from immunostaining at day 4 for pluripotency (SOX2), early gastrulation (TBXT), amnion (TFAP2C, TFAP2A), endoderm (SOX17) or mesoderm (CDX2) markers. EmDisc-like structures homogenously expressed SOX2, with heterogeneous low expression of TBXT, SOX17 and TFAP2C indicative of priming toward gastrulation and rare emergence of endoderm. Pluripotent EmDisc-like structures support a role for FGF and Activin/NODAL signalling in promoting pluripotency in the EmDisc.
g, Summary schematic of canonical WNT signalling in the marmoset embryo. The posterior EmDisc, stalk and PGCs express WNT3. mRNA expression gradients summarised in CS6 cross section (left). Time series brightfield images of interphase culture with FGF and Activin A + CHIR (CHIR99021, WNT agonist) (right). The emergence of differentiated populations was evident at day 4.
h-i, Molecular characterisation of WNT-treated EmDisc-like structures at day 4. Representative maximum projection images from immunostaining at day 4 from staining for pluripotency (SOX2), early gastrulation (TBXT), amnion (TFAP2C, TFAP2A), endoderm (SOX17) or mesoderm (CDX2) markers. Structures exhibited loss of SOX2 expression and upregulation of TBXT and SOX17 in comparison to FGF/Activin A culture, consistent with differentiation into amnion, endoderm, and mesoderm populations.
j, Summary schematic of WNT inhibition in the marmoset embryo. The VE and Amnion express canonical WNT inhibitor DKK1. mRNA expression gradients summarised in CS6 cross section (left). Time series brightfield images of interphase culture with FGF and Activin A + IWP-2 (right). Similar to FGF/Activin A culture, structures first open a lumen at day 3 and expand up to day 6.
k-l, Molecular characterisation of WNT-inhibited EmDisc-like structures at day 4. Representative maximum projection images from immunostaining at day 4 from staining for pluripotency (SOX2), early gastrulation (TBXT), amnion (TFAP2C, TFAP2A), endoderm (SOX17) or mesoderm (CDX2) markers. EmDisc-like structures homogenously expressed SOX2 and downregulated TBXT and SOX17 in comparison to FGF/Activin A culture, consistent with a role for WNT inhibition in preserving pluripotency in the EmDisc.
m, Summary schematic of BMP signalling in the marmoset embryo.The ExMes, amnion, and PGCs are sources of BMP4 in the embryo. mRNA expression gradients summarised in CS6 cross section (left). Time series brightfield images of interphase culture with FGF and Activin A + BMP4 (right).The emergence of disorganized, differentiated populations was evident at day 4.
n-o, Molecular characterisation of BMP-treated EmDisc-like structures at day 4.Representative maximum projection images from immunostaining at day 4 from staining for pluripotency (SOX2), early gastrulation (TBXT), amnion (TFAP2C, TFAP2A), endoderm (SOX17) or mesoderm (CDX2) markers. Structures exhibited loss of SOX2 expression and upregulation of TFAP2C, TFAP2A, CDX2 and SOX17 in comparison to FGF/Activin A culture, consistent with a mixed amnion and posteriorized primitive streak-like fate.
p, Summary schematic of BMP inhibition in the marmoset embryo. The VE expresses BMP inhibitor NOGGIN in the embryo. mRNA expression gradients summarised in CS6 cross section (left). Time series brightfield images of interphase culture with FGF and Activin A + BMP4 (right).Similar to FGF/Activin A culture, structures first open a lumen at day 3 and form homogenous spheroids.
q-r, Molecular characterisation of BMP-inhibited EmDisc-like structures at day 4.Representative maximum projection images from immunostaining at day 4 from staining for pluripotency (SOX2), early gastrulation (TBXT), amnion (TFAP2C, TFAP2A), endoderm (SOX17) or mesoderm (CDX2) markers. Scale bars represent 100 μm. EmDisc-like structures homogenously expressed SOX2 and downregulated TBXT, SOX17 and TFAP2C in comparison to FGF/Activin A culture. This is consistent with a role for BMP inhibition in preserving pluripotency in the EmDisc and inhibiting amnion differentiation.
Extended Data Figure 9. Cross-species analysis of primate mesoderm differentiation in vivo.
a, Alignment of EmDisc-derived postimplantation lineages in marmoset and human. Visualization based on alignment of embryo in vivo and in vitro datasets of pre- to postimplantation cynomolgus monkey25, in vitro-cultured human12, in vivo human CS79, preimplantation marmoset (ref24 and this study), and postimplantation marmoset embryo data (this study) were aligned. UMAP: Uniform Manifold Approximation and Projection for Dimension Reduction.
b, UMAP plots of marmoset CS5-7 or human late CS7 lineages showing normalized log expression of marker genes from 9. Pluripotent EmDisc (Embryonic Disc): SOX2, CDH1; Primitive Streak: CDH1, FST, TBXT; Nasc Mes (Nascent mesoderm): TBXT, CDH2, Emergent Mesoderm: OTX2, LHX1; Advanced Mesoderm: FOXF1, HAND1, BMP4, GATA6; Amnion: VTCN1, HAND1, BMP4; Endoderm subcluster also indicated by co-expression of CDH2, OTX2, GATA6.
c, Unbiased clustering of gastrulation stage lineages represented in UMAP in (a) resolves 9 clusters by shared nearest neighbour clustering: Pluripotent EmDisc (Embryonic Disc), PS early (Primitive Streak early), PS late (Primitive Streak late), PS advanced (Primitive Streak advanced), Endoderm, Nasc Mes (Nascent Mesoderm), Em Mes (Emergent Mesoderm), Adv Mes (Advanced Mesoderm), PGCs (Primordial Germ Cells).
d., Bifurcation of endoderm and mesoderm from the primitive streak represented in a diffusion map. Human CS7 shows an additional route from primitive streak to advanced mesoderm through emergent mesoderm. Alignment includes data plotted in (c) with amnion and PGC clusters excluded where cluster identity is given by colour. The first 2 diffusion components are shown (dim 1, dim 2).
e, Diffusion maps of marmoset CS5-7 or human late CS7 lineages showing normalized log expression of marker genes for primitive streak (TBXT), endoderm (FOXA2), pan-mesoderm (SNAI2, CDH2), emergent mesoderm (OTX2) or nascent mesoderm (FOXF1).
f-g, Human vs. marmoset scatterplots of primitive streak vs. nascent mesoderm (f) or nascent mesoderm vs. endoderm (g). Highlighted quadrants show human-marmoset conserved markers for each lineage, whereas white quadrants show species-specific expression patterns. Gene names for transcription factors, ligands, and extracellular matrix molecules are labelled.
Extended Data Figure 10. Spatial identity mapping of human in vitro models.
a, Alignment of naïve and primed human and marmoset PSCs with in vivo marmoset data. Marmoset in vivo datasets of pre- to postimplantation development and naïve/primed PSCs and human naïve/primed PSCs43 were aligned. Visualisation of aligned datasets by principal component analysis shows separation of preimplantation (on the left) and postimplantation (on the right) samples, with marmoset PSCs sitting between pre- and post-implantation. Human PSCs are plotted overlayed with transparent marmoset embryo data. Naïve human cells mapped earlier on PC1 than naïve marmoset cells and showed greater heterogeneity in mapping to ICM, Hypoblast, and Tb. Primed human cells mapped closely to the marmoset EmDisc. PCA, Principal component analysis; PSCs, pluripotent stem cells. ICM, inner cell mass; Hyp, hypoblast; SYS, Secondary Yolk Sac; VE, Visceral Endoderm; ExMes, Extraembryonic Mesoderm; EmDisc, Embryonic Disc; Am, Amnion; Tb, Trophoblast.
b, Expression of extraembryonic markers in marmoset in vivo data (left) and human naïve and primed PSCs (right) represented in aligned PCAs from (a) showing integrated log expression of trophoblast markers (JAM2), endoderm markers (EOMES, GATA6) and ICM/preimplantation blastocyst marker (ESRRB). Turquoise lines indicate location of naïve cells, blue lines indicate primed cells on the PCA.
c, Alignment of human microfluidic embryonic-like sac model to marmoset STEP data. Smart-seq2 profiling of marmoset in vivo lineages (EmDisc, PGC, Amnion, and Stalk populations) was aligned to 10x sequencing profiling of microfluidic embryonic-like sac model16. Visualisation of aligned datasets by principal component analysis shows separation hPSC, mesoderm, PGCLCs, and AMLCs. PCA, Principal component analysis; PSC, human pluripotent stem cell; hPGCLC, human primordial germ cell-like cell, hAMLC; human amnion-like cell; hMELC1/2, human mesoderm-like cell population 1 or 2.
d. Unbiased Clustering represented in UMAP in (c) resolves 5 clusters by shared nearest neighbour clustering in marmoset data. Four clusters aligned to subpopulation of the human microfluidic embryonic-like sac model and were annotated EmDisc, PGC, Amnion, and Mesoderm. A fifth subcluster in marmoset data contained cells from the gastrulating EmDisc, PGCs, and amnion and was therefore annotated PS-like (primitive streak-like).
e, Pearson’s correlation of marmoset and human microfluidic embryonic-like sac model of clusters identified in (d).
f-h. Spatial identity mapping of human microfluidic embryonic-like sac model. Subpopulations of the human microfluidic embryonic-like sac model were mapped to the marmoset EmDisc, PGC, Stalk, and Amnion. Spatial identity displayed in the orientations described in (f) for hPSCs (g), which mapped to the anterior EmDisc and hAMLCs (h) which mapped most strongly to the posterior amnion.
Supplementary Material
Acknowledgements
We thank Prof. Angela Roberts and the Cambridge marmoset research facility for providing marmoset neonates. We also thank the Endocrinology Laboratory of the German Primate Center for hormone analyses and the members of the Platform Degenerative Diseases for excellent assistance. Songyang Li, Clara Munger, Magdalena Schindler, Ioakeim Ampartzidis, Geraldine Jowett, Christos Kyprianou and Prof. Graham Burton have contributed with fruitful discussions and helpful comments on the manuscript. Peter Humphreys and Darran Clements supported imaging. Prof. Azim Surani and Prof. John Gurdon have kindly provided access to computational facilities at the Gurdon Institute. We would like to thank Charles Bradshaw for help with High Performance Computing and Dr Xiaohui Zhao and Dr. Russell Hamilton for valuable advice and bioinformatics support. This research is generously supported by the Wellcome Trust (WT RG89228), the Centre for Trophoblast Research, the Isaac Newton Trust and JSPS KAKENHI 15H02360, 19H05759.
Footnotes
Author contributions
T.E.Boroviak, S.Bergmann, E.Slatery, C.A.Penfold and D.Siriwardena designed and conducted experiments and wrote the manuscript, C.Drummer set up timed marmoset matings and extracted uteri, T.N.Kohler performed staining and imaging of PSCs, C.A.Penfold, E. Slatery and D.Siriwardena performed bioinformatics, S.Bergmann, D.Siriwardena, S.E.Strawbridge and E.Slatery established virtual reconstructions, C.A.Penfold pioneered Gaussian process regression to model 3D-transcriptomes, K.Kishimoto and E.Sasaki provided marmoset preimplantation embryos. R.Behr provided marmoset postimplantation embryos, A. Vickers and M. Tewary provided expertise and produced micropattern plates, F. Hollfelder, W.Reik and S.Clarke provided expertise and facilities for imaging and high-throughput RNA-seq library preparation, T.E.Boroviak conceived, coordinated and supervised the project.
Competing interests
The authors declare no competing interests.
Code
Code is available from the GitHub repository: https://github.com/Boroviak-Lab/SpatialModelling
Data
STEP- and single-cell RNA-seq data are available in the ArrayExpress repository under accession numbers:
E-MTAB-9367 Spatial Embryo Profiling of primate gastrulation
E-MTAB-9349 Primed and naïve marmoset pluripotent stem cells as a model for primate development
References
- 1.Tam PP, Loebel DA. Gene function in mouse embryogenesis: get set for gastrulation. Nat Rev Genet. 2007;8:368–381. doi: 10.1038/nrg2084. [DOI] [PubMed] [Google Scholar]
- 2.Rossant J, Tam PPLL. Blastocyst lineage formation, early embryonic asymmetries and axis patterning in the mouse. Development. 2009;136:701–713. doi: 10.1242/dev.017178. [DOI] [PubMed] [Google Scholar]
- 3.Arnold SJ, Robertson EJ. Making a commitment: cell lineage allocation and axis patterning in the early mouse embryo. Nat Rev Mol Cell Biol. 2009;10:91–103. doi: 10.1038/nrm2618. [DOI] [PubMed] [Google Scholar]
- 4.Rossant J, Tam PPL. Exploring early human embryo development. Science (80-.) 2018;360:1075–1076. doi: 10.1126/science.aas9302. [DOI] [PubMed] [Google Scholar]
- 5.Boroviak T, Nichols J. Primate embryogenesis predicts the hallmarks of human naïve pluripotency. Development. 2017;144:175–186. doi: 10.1242/dev.145177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Simunovic M, Brivanlou AH. Embryoids, organoids and gastruloids: New approaches to understanding embryogenesis. Development (Cambridge) 2017 doi: 10.1242/dev.143529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tabar V, Studer L. Pluripotent stem cells in regenerative medicine: Challenges and recent progress. Nature Reviews Genetics. 2014 doi: 10.1038/nrg3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ross C, Boroviak TE. Origin and function of the yolk sac in primate embryogenesis. Nat Commun. 2020 doi: 10.1038/s41467-020-17575-w. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tyser RCV, et al. Single-cell transcriptomic characterization of a gastrulating human embryo. Nature. 2021;600 doi: 10.1038/s41586-021-04158-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deglincerti A, et al. Self-organization of the in vitro attached human embryo. Nature. 2016;533:251–254. doi: 10.1038/nature17948. [DOI] [PubMed] [Google Scholar]
- 11.Shahbazi MN, et al. Self-organization of the human embryo in the absence of maternal tissues. Nat Cell Biol. 2016;18:700–708. doi: 10.1038/ncb3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xiang L, et al. A developmental landscape of 3D-cultured human pre-gastrulation embryos. 577:537–542. doi: 10.1038/s41586-019-1875-y. [DOI] [PubMed] [Google Scholar]
- 13.Ma H, et al. In vitro culture of cynomolgus monkey embryos beyond early gastrulation. Science (80-.) 2019:eaax7890. doi: 10.1126/science.aax7890. [DOI] [PubMed] [Google Scholar]
- 14.Niu Y, et al. Dissecting primate early post-implantation development using long-term in vitro embryo culture. Science (80-.) 2019:eaaw5754. doi: 10.1126/science.aaw5754. [DOI] [PubMed] [Google Scholar]
- 15.Shao Y, et al. Self-organized amniogenesis by human pluripotent stem cells in a biomimetic implantation-like niche. Nature Materials. 2017 doi: 10.1038/NMAT4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zheng Y, et al. Controlled modelling of human epiblast and amnion development using stem cells. Nature. 2019 doi: 10.1038/s41586-019-1535-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shao Y, et al. A pluripotent stem cell-based model for post-implantation human amniotic sac development. Nat Commun. 2017 doi: 10.1038/s41467-017-00236-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Warmflash A, Sorre B, Etoc F, Siggia ED, Brivanlou AH. A method to recapitulate early embryonic spatial patterning in human embryonic stem cells. Nat Methods. 2014 doi: 10.1038/nMeth.3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martyn I, Siggia ED, Brivanlou AH. Mapping cell migrations and fates in a gastruloid model to the human primitive streak. Development. 2019 doi: 10.1242/dev.179564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sato K, et al. Generation of a Nonhuman Primate Model of Severe Combined Immunodeficiency Using Highly Efficient Genome Editing. Cell Stem Cell. 2016;19:127–138. doi: 10.1016/j.stem.2016.06.003. [DOI] [PubMed] [Google Scholar]
- 21.Okano H, Hikishima K, Iriki A, Sasaki E. The common marmoset as a novel animal model system for biomedical and neuroscience research applications. Semin Fetal Neonatal Med. 2012;17:336–340. doi: 10.1016/j.siny.2012.07.002. [DOI] [PubMed] [Google Scholar]
- 22.Sasaki E, et al. Generation of transgenic non-human primates with germline transmission. Nature. 2009;459:523–527. doi: 10.1038/nature08090. [DOI] [PubMed] [Google Scholar]
- 23.Picelli S, et al. Full-length RNA-seq from single cells using Smart-seq2. Nat Protoc. 2014;9:171–181. doi: 10.1038/nprot.2014.006. [DOI] [PubMed] [Google Scholar]
- 24.Boroviak T, et al. Single cell transcriptome analysis of human, marmoset and mouse embryos reveals common and divergent features of preimplantation development. Dev. 2018 doi: 10.1242/dev.167833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakamura T, et al. A developmental coordinate of pluripotency among mice, monkeys and humans. Nature. 2016;537:57–62. doi: 10.1038/nature19096. [DOI] [PubMed] [Google Scholar]
- 26.Rasmussen CE, Williams CKI. Chapter 2 - Regression. Gaussian Processes for Machine Learning. 2006 doi: 10.1093/bioinformatics/btq657. [DOI] [Google Scholar]
- 27.G M. Krigeage d’un panneau rectangulaire par sa périphérie [Kriging of a rectangular panel via its periphery] 1960 [Google Scholar]
- 28.V S, SA T, O S. SpatialDE: identification of spatially variable genes. Nat Methods. 2018;15:343–346. doi: 10.1038/nmeth.4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peng G, et al. Spatial Transcriptome for the Molecular Annotation of Lineage Fates and Cell Identity in Mid-gastrula Mouse Embryo. 2016;36:681–697. doi: 10.1016/j.devcel.2016.02.020. [DOI] [PubMed] [Google Scholar]
- 30.Peng G, et al. Molecular architecture of lineage allocation and tissue organization in early mouse embryo. Nature. 2019;572 doi: 10.1038/s41586-019-1469-8. [DOI] [PubMed] [Google Scholar]
- 31.Rivera-Pérez JA, Magnuson T. Primitive streak formation in mice is preceded by localized activation of Brachyury and Wnt3. Dev Biol. 2005 doi: 10.1016/j.ydbio.2005.09.012. [DOI] [PubMed] [Google Scholar]
- 32.Perea-Gomez A, et al. Nodal antaginists in the anterior visceral endoderm prevent the formation of multiple primitive streaks. Dev Cell. 2002;3:745–756. doi: 10.1016/s1534-5807(02)00321-0. [DOI] [PubMed] [Google Scholar]
- 33.Molè MA, et al. A single cell characterisation of human embryogenesis identifies pluripotency transitions and putative anterior hypoblast centre. Nat Commun. 2021;12:1–12. doi: 10.1038/s41467-021-23758-w. 2021 121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wamaitha SE, et al. IGF1-mediated human embryonic stem cell self-renewal recapitulates the embryonic niche. Nat Commun. 2020;11 doi: 10.1038/s41467-020-14629-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zorzan I, et al. The transcriptional regulator ZNF398 mediates pluripotency and epithelial character downstream of TGF-beta in human PSCs. doi: 10.1038/s41467-020-16205-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakaki F, Saitou M. PRDM14: a unique regulator for pluripotency and epigenetic reprogramming. Trends Biochem Sci. 2014;39:289–298. doi: 10.1016/j.tibs.2014.04.003. [DOI] [PubMed] [Google Scholar]
- 37.Irie N, et al. SOX17 is a critical specifier of human primordial germ cell fate. Cell. 2015;160:253–268. doi: 10.1016/j.cell.2014.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kobayashi T, Surani MA. On the origin of the human germline. Development. 2018;145:dev150433. doi: 10.1242/dev.150433. [DOI] [PubMed] [Google Scholar]
- 39.Sasaki K, et al. The Germ Cell Fate of Cynomolgus Monkeys Is Specified in the Nascent Amnion. Dev Cell. 2016 doi: 10.1016/j.devcel.2016.09.007. [DOI] [PubMed] [Google Scholar]
- 40.Tewary M, et al. A stepwise model of reaction-diffusion and positional information governs self-organized human peri-gastrulation-like patterning. 2017 doi: 10.1242/dev.149658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang R, et al. Amnion signals are essential for mesoderm formation in primates. Nat Commun. 2021;12:1–14. doi: 10.1038/s41467-021-25186-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hollnagel A, Oehlmann V, Heymer J, Rüther U, Nordheim A. Id genes are direct targets of bone morphogenetic protein induction in embryonic stem cells. J Biol Chem. 1999 doi: 10.1074/jbc.274.28.19838. [DOI] [PubMed] [Google Scholar]
- 43.Messmer T, et al. Transcriptional Heterogeneity in Naive and Primed Human Pluripotent Stem Cells at Single-Cell Resolution. 2019;26:815–824.:e4. doi: 10.1016/j.celrep.2018.12.099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guo G, et al. Human naive epiblast cells possess unrestricted lineage potential. Cell Stem Cell. 2021:1–17. doi: 10.1016/j.stem.2021.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dong C, et al. Derivation of trophoblast stem cells from naïve human pluripotent stem cells. Elife. 2020 doi: 10.7554/eLife.52504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Linneberg-Agerholm M, et al. Naïve human pluripotent stem cells respond to Wnt, Nodal and LIF signalling to produce expandable naïve extra-embryonic endoderm. Dev. 2019 doi: 10.1242/dev.180620. [DOI] [PubMed] [Google Scholar]
- 47.Hertig A. On the preimplantation stages of the human ovum : a description of four normal and four abnormal specimens ranging from the second to the fifth day of development. Contrib to Embryol. 1954 [Google Scholar]
- 48.R O, et al. Developmental Stages in Human Embryos: Revised and New Measurements. Cells Tissues Organs. 2010;192:73–84. doi: 10.1159/000289817. [DOI] [PubMed] [Google Scholar]
- 49.Hertig AT, Rock J. Two human ova of the pre-villous stage, having an ovulation age of about eleven and twelve days respectively. Contrib Embryol Carnegie Inst Washingt. 1941;29:127–156. [Google Scholar]
- 50.Harlow CR, Hearn JP, Hodges JK. Ovulation in the marmoset monkey: endocrinology, prediction and detection. J Endocrinol. 1984;103:17–24. doi: 10.1677/joe.0.1030017. [DOI] [PubMed] [Google Scholar]
- 51.Clark SJ, et al. scNMT-seq enables joint profiling of chromatin accessibility DNA methylation and transcription in single cells. Nat Commun. 2018 doi: 10.1038/s41467-018-03149-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Macaulay IC, Ponting CP, Voet T. Single-Cell Multiomics: Multiple Measurements from Single Cells. Trends in Genetics. 2017 doi: 10.1016/j.tig.2016.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Krueger F. Trim Galore. Babraham Bioinformatics. 2016 http://www.bioinformatics.babraham.ac.uk/projects/trimgalore/ [Google Scholar]
- 54.Dobin A, et al. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liao Y, Smyth GK, Shi W. FeatureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 56.Nakamura T, et al. Single-cell transcriptome of early embryos and cultured embryonic stem cells of cynomolgus monkeys. Sci Data. 2017 doi: 10.1038/sdata.2017.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fernandez A, et al. The WNT receptor FZD7 is required for maintenance of the pluripotent state in human embryonic stem cells. Proc Natl Acad Sci U S A. 2014 doi: 10.1073/pnas.1323697111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Okae H, et al. Derivation of Human Trophoblast Stem Cells. Cell Stem Cell. 2018;22:50–63.:e6. doi: 10.1016/j.stem.2017.11.004. [DOI] [PubMed] [Google Scholar]
- 59.McQuin C, et al. CellProfiler 3.0: Next-generation image processing for biology. PLoS Biol. 2018;16 doi: 10.1371/journal.pbio.2005970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.O’Hagan A. Curve Fitting and Optimal Design for Prediction. J R Stat Soc Ser B. 1978 doi: 10.1111/j.2517-6161.1978.tb01643.x. [DOI] [Google Scholar]
- 61.Rasmussen CE, Nickisch H. Gaussian processes for machine learning (GPML) toolbox. J Mach Learn Res. 2010 [Google Scholar]
- 62.Kalaitzis AA, Lawrence ND. A simple approach to ranking differentially expressed gene expression time courses through Gaussian process regression. BMC Bioinformatics. 2011 doi: 10.1186/1471-2105-12-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jeffreys H. Some Tests of Significance, Treated by the Theory of Probability. Math Proc Cambridge Philos Soc. 1935 doi: 10.1017/S030500410001330X. [DOI] [Google Scholar]
- 64.Kass RE, Raftery AE. Bayes factors. J Am Stat Assoc. 1995 doi: 10.1080/01621459.1995.10476572. [DOI] [Google Scholar]
- 65.Adewumi O, et al. Characterization of human embryonic stem cell lines by the International Stem Cell Initiative. Nat Biotechnol. 2007;257(25):803–816. doi: 10.1038/nbt1318. 2007. [DOI] [PubMed] [Google Scholar]
- 66.Tewary M, et al. High-throughput micropatterning platform reveals Nodal-dependent bisection of peri-gastrulation–associated versus preneurulation-associated fate patterning. 2019 doi: 10.1371/journal.pbio.3000081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tewary M, et al. High-throughput micropatterning platform reveals Nodal-dependent bisection of peri-gastrulation-associated versus preneurulation-associated fate patterning. PLOS Biol. 2019;17:e3000081. doi: 10.1371/journal.pbio.3000081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vickers A, et al. Plating human iPSC lines on micropatterned substrates reveals role for ITGB1 nsSNV in endoderm formation. Stem Cell Reports. 2021;16:2628–2641. doi: 10.1016/j.stemcr.2021.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nature Methods. 2012 doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Thomson JA, Kalishman J, Hearn JP. Nonsurgical uterine stage preimplantation embryo collection from the common marmoset. J Med Primatol. 1994;23:333–336. doi: 10.1111/j.1600-0684.1994.tb00295.x. [DOI] [PubMed] [Google Scholar]
- 71.Boroviak T, et al. Lineage-Specific Profiling Delineates the Emergence and Progression of Naive Pluripotency in Mammalian Embryogenesis. Dev Cell. 2015;35:366–382. doi: 10.1016/j.devcel.2015.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ashburner M, et al. Gene ontology: Tool for the unification of biology. Nature Genetics. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Carbon S, et al. The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res. 2019;47:D330–D338. doi: 10.1093/nar/gky1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wickham H. ggplot2: Elegant Graphics for Data Analysis. Springer-Verlag Media; New York: 2009. [DOI] [Google Scholar]
- 75.Brunson J. ggalluvial: Layered Grammar for Alluvial Plots. J Open Source Softw. 2020 doi: 10.21105/joss.02017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Code is available from the GitHub repository: https://github.com/Boroviak-Lab/SpatialModelling
STEP- and single-cell RNA-seq data are available in the ArrayExpress repository under accession numbers:
E-MTAB-9367 Spatial Embryo Profiling of primate gastrulation
E-MTAB-9349 Primed and naïve marmoset pluripotent stem cells as a model for primate development