Abstract
An interplay of experimental and computational methods is required to achieve a comprehensive understanding of protein–RNA interactions. UV crosslinking and immunoprecipitation (CLIP) identifies endogenous interactions by sequencing RNA fragments that copurify with a selected RNA-binding protein under stringent conditions. Here we focus on approaches for the analysis of the resulting data and appraise the methods for peak calling, visualization, analysis, and computational modeling of protein–RNA binding sites. We advocate that the sensitivity and specificity of data be assessed in combination for computational quality control. Moreover, we demonstrate the value of analyzing sequence motif enrichment in peaks assigned from CLIP data and of visualizing RNA maps, which examine the positional distribution of peaks around regulated landmarks in transcripts. We use these to assess how variations in CLIP data quality and in different peak calling methods affect the insights into regulatory mechanisms. We conclude by discussing future opportunities for the computational analysis of protein–RNA interaction experiments.
Keywords: RNA-binding protein, ribonucleoprotein complexes, CLIP, peak calling, data quality, RNA map
Introduction
RNA-binding proteins (RBPs) are key orchestrators of posttranscriptional RNA regulation. They determine the fate of a transcript throughout its life cycle, directing regulatory stages including splicing, polyadenylation, localization, translation, stability, and degradation. Over a thousand human RBPs have been annotated and identified by mass spectroscopy studies (1, 2). RBPs specify their RNA binding sites by recognizing a combination of features, including RNA sequence motifs, RNA modifications, RNA structural motifs, and interactions with additional RBPs that bind at nearby loci (3). Each transcript interacts with many different RBPs to assemble into a ribonucleoprotein complex (RNP), which changes as the RNA passes through the various regulatory stages. RNP formation depends on the abundance of RNAs and RBPs in each cell type and on their posttranslational modifications and is sensitive to the competition between multiple factors for overlapping binding sites (4).
Due to the combinatorial and dynamic assembly of RNPs, it is crucial to identify the protein–RNA interactions that form within cells. Crosslinking between RNAs and proteins can be achieved by ultraviolet C (UVC) irradiation at 254 nm due to the photoreactivity of RNA bases (5). This has been exploited by the UV crosslinking and immunoprecipitation (CLIP) method that uses UV light to crosslink proteins covalently to RNAs in intact cells or tissues, followed by purification and sequencing of RNA fragments that were crosslinked to an RBP of interest (6). The development of CLIP follows a rich history of preceding methods, including RNA immunoprecipitation (RIP). Over the last 15 years, many variant protocols of CLIP have evolved, and in combination with high-throughput sequencing, they have led to a wealth of data, encapsulating transcriptome-wide binding profiles of hundreds of RBPs in multiple species, tissues, and cell lines (7). Proteomics approaches have also been developed to study protein–RNA interactions and have recently been reviewed (8); they provide a complementary approach to CLIP but are not discussed further here. The original CLIP and the derived variants all rely on sequencing; therefore, we use the term “CLIP” to refer generically to protocols that purify covalently crosslinked protein–RNA complexes and then sequence the bound RNA fragments. In contrast, we use the term “CLIP-seq” to refer to the protocol that was used by the first publication employing this term (9), which employed the original CLIP protocol (Table 1; see also Supplemental Table 1).
Table 1. The central features of CLIP methods from the perspective of data analysisa.
| Methods | Specificity | Resolution | Sensitivity |
|---|---|---|---|
| HTTS-CLIP, CLIP-seq, CRAC | ++ to
+++ Strong detergents and high-salt washes are used with further purification by SDS-PAGE and membrane transfer, which allows one to optimize RNase conditions and ensure that copurified RBPs and noncrosslinked RNAs are removed. Thus, only specific RNAs crosslinked to the immunoprecipitated RBP are normally isolated, but specificity depends on careful optimization and visualization of the purified complexes. |
Oligonucleotide corresponding to the size of readthrough cDNAs | ++ Limited by the loss of cDNAs truncated at crosslink sites |
| iCLIP, 4SU-iCLIP, FAST-iCLEP, BrdU-CLIP, irCLIP, Fr-iCLEP, sCLIP | ++ to +++, as in HTTS-CLIP | Nucleotide corresponding to the start of truncated cDNAs | ++ to +++ Increased due to amplification of truncated cDNAs, as well as other method-specific optimizations |
| eCLIP, FLASH | + to
+++ The purity of protein-RNA complexes is not validated by visualization. Blind cutting from the membrane is used in eCLEP, while SDS-PAGE separation is removed in sCLIP. Thus, copurification of nonspecific RBPs is not monitored, which could result in large variations in specificity. |
||
| iCLAP, uvCLAP | ++ to
+++ Expression of affinity-tagged proteins permits rigorous washing with denaturing conditions, which removes copurified RBPs. However, expression of tagged RBPs could in some cases affect RNA specificity or lead to artifacts associated with overexpression. |
||
| PAR-CLIP | ++ to +++, as in HTTS-CLIP | Nucleotide corresponding to the crosslink-induced mutations | + to
+++ Limited by the loss of cDNAs truncated at crosslink sites but offset to differing degrees by increased crosslinking efficiency for some RBPs |
| CIMS of HITS-CLIP | ++ to +++, as in HTTS-CLIP | + Limited by the loss of cDNAs truncated at crosslink sites and the low proportion of cDNAs with deletions |
|
| RIP-seq | + Preserves protein-protein interactions due to mild washing conditions or formaldehyde crosslinking, thus copurifying interacting RBPs |
Transcript-level resolution achieved by the original version of RIP-seq, since it doesn’t fragment the bound RNAs | ++ If no crosslinking is used, then transient weak interactions that take place in vivo can be lost during immunoprecipitation, and abundant RNAs tend to dominate the pulldown, leading to decreased coverage of introns or other low-abundant RNAs. |
| RIPiT-seq, DO-RIP-seq | ++ Use of RNase reduces copurification of RBPs that bind to the same transcripts, but such RBPs can still be purified due to the preserved protein-protein interactions. Sequential IP with two separate antibodies can specify RNPs composed of multiple RBPs in RIPiT-seq. |
Oligonucleotide corresponding to the size of cDNAs due to the use of RNase to fragment the bound RNAs | ++ Due to saturation with reads from abundant RNAs bound by copurified RBPs, the method has limited sensitivity for RBPs that primarily bind introns and other low-abundant RNAs. |
Abbreviations: +, moderate; ++, high; +++, best; 4SU, 4-thiouridine; BrdU, bromodeoxyuridine; cDNA, complementary DNA; CIMS, crosslink-induced mutation sites; CLAP, crosslinking and affinity purification; CLIP, UV crosslinking and immunoprecipitation; CRAC, crosslinking and cDNA analysis; DO, digestion-optimized; eCLIP, enhanced CLIP; FAST, fully automated and standardized; FLASH, FADD-like IL-1 (3-converting enzyme-associated huge protein; Fr, fractionation; HITS, high-throughput sequencing; iCLAP, individual-nucleotide resolution CLAP; iCLEP, individual-nucleotide resolution CLIP; IP, immunoprecipitation; irCLIP, infrared CLIP; PAR, photoactivatable ribonucleoside-enhanced; RBP, RNA-binding protein; RIP, RNA immunoprecipitation; RIPiT, RNA-protein IP in tandem; RNP, ribonucleoprotein complex; sCLIP, simphfied-platform CLIP; SDS-PAGE, sodium dodecyl sulfate polyacrylamide gel electrophoresis; seq, sequencing; uvCLAP, ultraviolet CLAP.
CLIP methods are grouped according to how the reads are used to identify binding sites. Associated technical features and limitations are summarized in terms of resolution, sensitivity, and specificity. Colors represent the quality of the parameter (red is poor, orange is adequate, and green is good).
Two orthogonal approaches to the analysis of CLIP data differ by their focus either on a specific RBP or on the interacting transcripts. In the RBP-centric approach, researchers aim to identify the RNA sequence, structure, and other features that are common to the binding sites of an RBP across the transcriptome in order to unravel the mechanisms underlying the specificity of these interactions and to identify functional relationships between the bound RNAs and their common regulatory principles. For example, the earliest CLIP studies of Nova proteins demonstrated that most RNA targets encode proteins with synaptic functions, identified the features of clustered YCAY motifs enriched at the endogenous binding sites, and defined the RNA map that demonstrated position-dependent activity of Nova at regulated exons and polyadenylation sites (6, 10, 11). The RNA-centric approach, on the other hand, examines the binding positions of a broad spectrum of RBPs on a specific transcript or set of transcripts. This approach requires integration of CLIP data sets for multiple RBPs, which can be achieved by comparing available CLIP data published by multiple research groups. The results of both approaches need to be integrated with other methods to unravel fully the functions and mechanisms of action of RNPs (see the sidebar titled RNA Maps: Integrating CLIP with Orthogonal Methods).
Here, we review computational and modeling methods and use visualization of enriched motifs and RNA maps to examine how the use of different methods impacts the biological insights gained from CLIP data. We start with a short evaluation of the experimental methods from a bioinformatic perspective to understand how the technical details of various CLIP protocols impact the specific requirements for the computational approaches. We then proceed through the primary stages of CLIP data analysis: (a) quality control, (b) peak calling, (c) binding site modeling, and (d) functional evaluation. At each stage, we explore the pertinent issues and potential pitfalls to elucidate how RBPs recognize and act on specific transcripts. In the penultimate section, we broach what will likely become an important avenue of study in the near future: the integration of CLIP data from different RBPs. This will lay the foundations for developing an understanding of the complex network of RBP-RNA interactions. In concluding, we propose a set of standards as a framework for CLIP data analysis.
RNA Maps: Integrating CLIP with Orthogonal Methods.
An RNA map is a conceptually simple yet powerful tool initially developed to explore the functional impact of Nova-binding motifs on splicing to predict Nova’s action genome-wide (10). It visualizes the positional distribution of assigned binding sites (commonly CLIP peaks or motifs) of the target RBP around regulated landmarks in transcripts (such as alternative exons for splicing regulators). Landmarks are defined by an orthogonal method, for example, by RNA-seq analysis of RBP knockout cells or tissues to identify the regulated exons. The distribution around each regulated landmark can be visualized as a heatmap, or summarized as a metaprofile (Figure 2d). To gain functional insight, researchers should also plot or use the distribution around control landmarks (such as unregulated exons) to determine binding enrichment, thus providing a sense of scale when comparing across experiments. The control variability can be examined using bootstrapping to determine the significance of enriched binding (97). To simplify implementation for general users, researchers have designed the rMAPS and expressRNA web servers to generate RNA maps using motifs or CLIP peaks around regulated exons and polyA sites (97, 98).
These maps are of great value not only in assessing RBP function, but also in validating CLIP experiments, since the enrichment of CLIP peaks around RNA features regulated by the same protein can serve as evidence of data specificity. The proportion of regulated RNAs with CLIP peaks at expected positions also provides insight into the sensitivity of data. In Figures 3 and 4, we use RNA maps to examine the sensitivity and specificity of CLIP peaks obtained by different CLIP methods and different peak calling tools or parameters.
Differences between CLIP Methods from the Perspective of Data Analysis
Despite the many variations of CLIP, its core principles mostly remain the same (7) (Figure 1). The covalent bond formed upon UV crosslinking allows the RNAs to be fragmented by a limited concentration of RNase after lysis, which is followed by purification of the RBP of interest under stringent conditions. Usually, an antibody is used to immunoprecipitate a specific RBP, which is separated via sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and visualized together with the crosslinked RNA fragments. The complex is then excised from the membrane and treated with proteinase K to remove the bulk of the RBP, leaving behind a short polypeptide at the crosslink site and releasing the RNA fragments. The fragments are then reverse-transcribed into cDNAs, and the resulting cDNA library is sequenced. Initially, CLIP relied on traditional Sanger sequencing to identify 340 RNA fragments that provided the first glimpse into the binding sites of the neuron-specific Nova proteins (6), but now, high-throughput sequencing enables us to gain a more comprehensive view across the transcriptome.
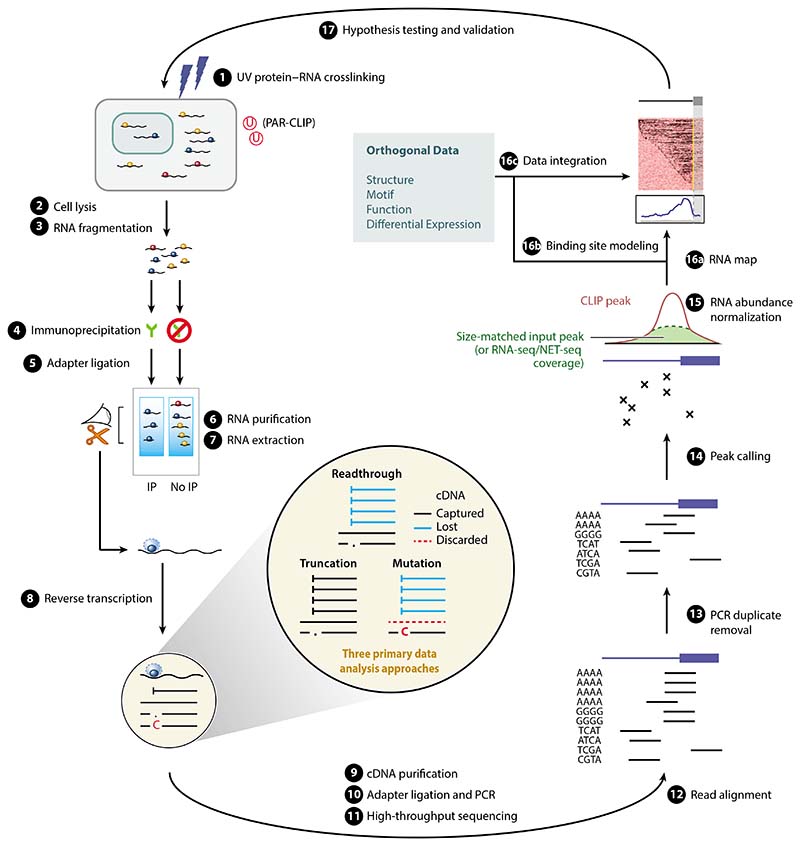
Figure 1.
A computational biologist’s overview of the CLIP method, and its key experimental (left) and computational (right) steps. The experimental steps, common across most methods, are numbered according to the scheme in Reference 7. Highlighted in the center are the three primary data analysis approaches that rely on cDNA readthrough, mutation, or truncation, depending on the type of CLIP protocol used to generate the data (related to Table 1). The cDNAs that are captured by representative protocols are in black, while those that are lost during reverse transcription are in blue, and those that are discarded during analysis are marked by dashed lines. Abbreviations: cDNA, complementary DNA; CLIP, UV crosslinking and immunoprecipitation; IP, immunoprecipitation; NET-seq, native elongating transcript sequencing; PAR-CLIP, photoactivatable ribonucleoside–enhanced CLIP; PCR, polymerase chain reaction; RNA-seq, RNA sequencing.
Resolution and Sensitivity
From the perspective of data analysis, CLIP methods can be divided into three principal approaches (Table 1, Figure 1). The division relates to the effect on reverse transcription of the polypeptide that remains at the crosslink site of fragmented RNAs. This can result in cDNAs that either (a) read through the peptide without any mutations, (b) read through the peptide but introduce a mutation at the crosslink site, or (c) truncate at the crosslink site.
The original CLIP method can only amplify cDNAs that fall into the first two categories because both adapters required for cDNA amplification are ligated to the RNA fragments, and therefore, the whole fragment needs to be reverse-transcribed along with its adapters. This method employs UVC light (254 nm) for crosslinking, which normally leads to only a minor proportion of cDNAs containing crosslink-induced mutations (12). Therefore, binding sites for CLIP and its derived methods such as HITS-CLIP are usually assigned on the basis of the whole-sequenced reads (6, 11). Nevertheless, mutations, and especially deletions in CLIP cDNAs, can help increase the method’s resolution (13).
In photoactivatable ribonucleoside–enhanced CLIP (PAR-CLIP), cells are preincubated with photoreactive ribonucleosides, usually 4-thiouridine (4SU), which enables the use of UVA light (365 nm) for crosslinking (14). Similar to CLIP, PAR-CLIP only amplifies cDNAs that fall into the first two categories, but it increases the proportion of cDNAs with mutations. About 50% of PAR-CLIP cDNAs normally contain thymidine-to-cytidine transitions at the crosslink site, which is the basis for binding site assignment used by most tools developed for PAR-CLIP analysis (14). However, a large proportion of PAR-CLIP cDNAs lacks transitions, and longer cDNAs may contain more than one transition, and thus, only a subset of cDNAs can be used for nucleotide-resolution studies (14).
Individual-nucleotide resolution CLIP (iCLIP) was developed to capture the third category of cDNAs that truncate at the crosslink site, in addition to the first two categories. This is achieved by ligating the second adapter to the cDNAs rather than the RNA fragments (15). In truncated cDNAs, the adapter is ligated exactly at the positions of their truncation. It has been estimated that approximately 90% of cDNAs in iCLIP truncate at the crosslink site (12, 16). Therefore, the nucleotide in the genome adjacent to the 5′ end of the aligned iCLIP cDNAs most often corresponds to the crosslink site. The same data analysis method applies to iCLIP and its more recent variants, including infrared CLIP (irCLIP) and enhanced CLIP (eCLIP) (17, 18), which also amplify truncated cDNAs.
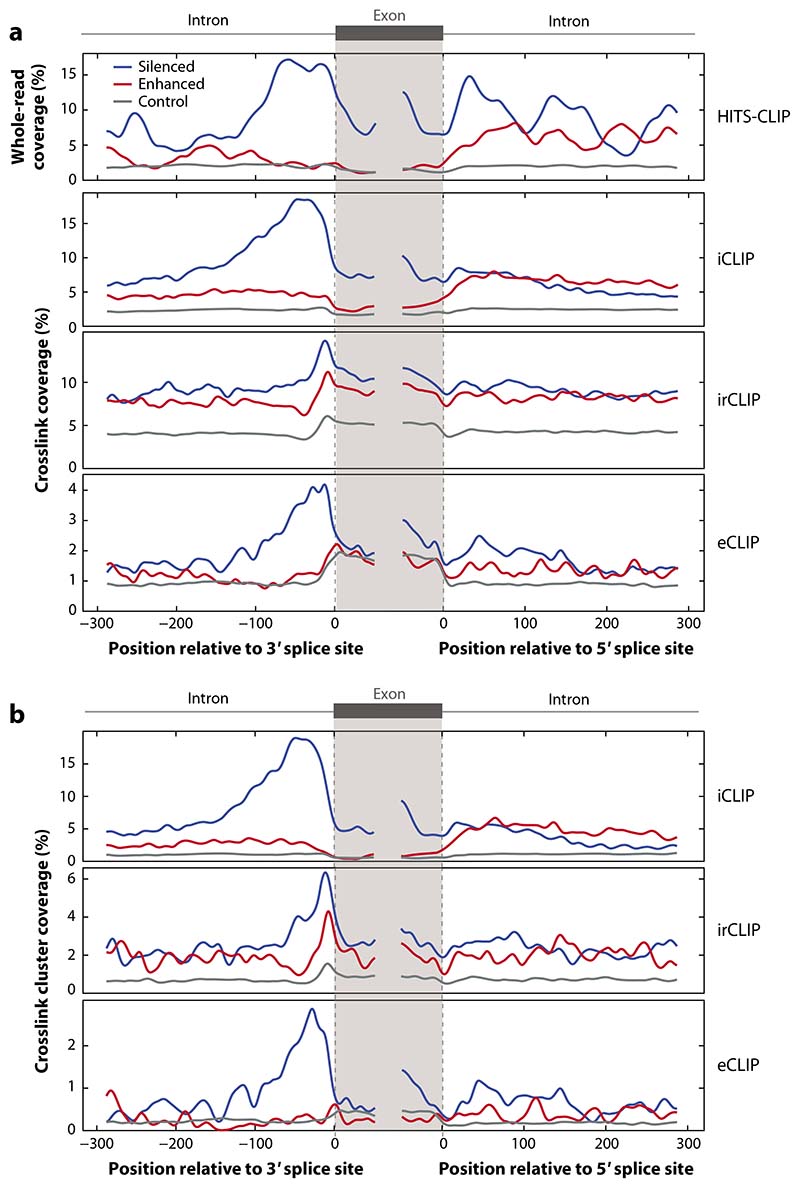
The sensitivity of all CLIP methods is driven to a large extent by this choice between the three principal approaches. The relative crosslinking efficiency with UVC or UVA differs between RBPs (19), and this affects the relative sensitivity of CLIP versus PAR-CLIP methods. Both CLIP and PAR-CLIP lead to the loss of cDNAs truncating at crosslink sites, which represent ∼90% of the total in most cases; therefore, it is expected that iCLIP increases the sensitivity of the method by amplifying the truncated cDNAs. If UVA crosslinking upon 4SU preincubation is beneficial, it can be combined with iCLIP in the variant termed 4SU-iCLIP (16, 20). Sensitivity can be considered at two levels: the first basic and the second functional. Crudely, the number of unique cDNAs in a CLIP experiment does give a basic indication of sensitivity. However, assessment with functional orthogonal data using RNA maps (Figure 2d) affords a more accurate delineation. Taking PTBP1 as an example, based on unique cDNA counts alone, eCLIP and iCLIP have similar sensitivities, while irCLIP’s is an order of magnitude greater (16). However, if we consider the distribution of raw crosslink sites, ∼18% of silenced exons contain an iCLIP crosslink site at the peak position at the 3∗ splice site, compared to ∼15% for irCLIP and ∼4% for eCLIP (Figure 3a). The choice of exons or cell lines does not account for the difference in eCLIP compared to iCLIP and irCLIP, as the sensitivity does not increase when using exons defined by RNA-seq analysis of knockout PTBP1 in the same cell lines used to produce the eCLIP data (see Supplemental Figure 1). Although only one example, this demonstrates that the number of unique cDNAs is only an approximate measure of sensitivity and that the crosslinking signal around regulated events should be used, whenever possible, as a more appropriate measure of data sensitivity. We particularly recommend this second functional approach when comparing different CLIP methods.
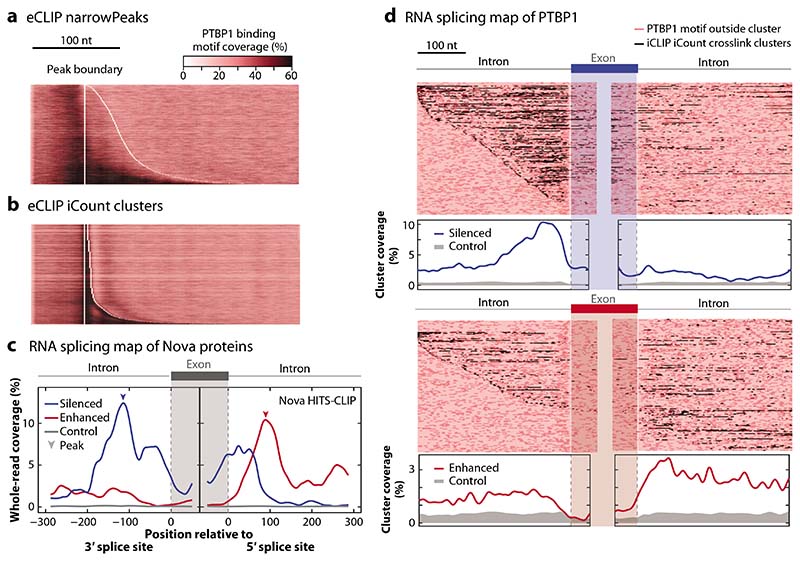
Figure 2.
Visualization of CLIP data: motif plots and RNA maps. (a) The distribution of PTBP1 motifs from Reference 16 are shown around eCLIP peaks that are defined as narrowPeaks and are available from the ENCODE website (https://www.encodeproject.org/). This algorithm relies on the use of whole reads, which leads to misalignment of motifs and peaks. (b) The iCount peak caller (15) uses the starts of aligned reads to define the crosslink positions and peaks, leading to good overlap with PTBP1 motifs. (c) Integrating CLIP and orthogonal data allows further exploration of data quality using an RNA splicing map, which examines the distribution of clusters of assigned binding sites around silenced (blue) and enhanced (red) exons. This approach was first used with HITS-CLIP reads for Nova in mouse brain (11). In this panel, we assign a binding site to all positions in transcripts that overlap with at least one raw read, based on the 168,632 reads obtained by the original HITS-CLIP publication (11); even though we do not use peak calling, this results in high position-dependent enrichment that agrees well with the computationally derived RNA map (10), thus highlighting the high specificity of raw CLIP data. (d) An RNA splicing map of PTBP1 iCLIP data from HeLa cells (16) is drawn in two ways, with peaks called using iCount with 3-nt clustering (15). Regulated exons are defined using microarray data upon knockdown of PTBP1/PTBP2 in HeLa cells (99). Each row of the heatmap is a regulated exon with its flanking region. The positions of peaks are shaded dark red; PTBP1 motifs inside or outside the clusters are shown as black and light red, respectively. The metaprofile of significant crosslink clusters is plotted below. The enrichment of peaks around regulated exons compared to control indicates the mechanisms of splicing regulation and the specificity of CLIP data. The code to reproduce this figure is available for download at https://github.com/jernejule/clip-data-science. Abbreviations: CLIP, UV crosslinking and immunoprecipitation; eCLIP, enhanced CLIP; HITS, high-throughput sequencing; iCLIP, individual-nucleotide resolution CLIP; nt, nucleotide.
Figure 3.
Using RNA maps to examine sensitivity and specificity of CLIP data. PTBP1 is an abundant RBP that crosslinks efficiently and follows a position-dependent regulatory mechanism, thus making it suitable for data analysis via an RNA map. The regulated exons were defined as those with an absolute dIrank of greater than 1 on analysis of splice junction microarray data with ASPIRE3 software upon knockdown of PTBP1/PTBP2 in HeLa cells (99). In panel a, we compare the raw data for different experimental methods, with whole reads from HITS-CLIP in HeLa cells (100), crosslink positions from irCLIP (18) and iCLIP (16) in HeLa cells, and eCLIP in HepG2 cells (25). This demonstrates that CLIP data can lead to strong enrichments even without peak calling, but this depends on the specificity of data. In panel b, we analyze the effects of peak calling on the crosslink positions from different experiments, with data from irCLIP (18) and iCLIP (16) in HeLa cells and eCLIP in HepG2 cells (25) analyzed using the iCount peak caller with 15 nucleotide clustering (15). The code to reproduce this figure is available for download at https://github.com/jernejule/clip-data-science. Abbreviations: CLIP, UV crosslinking and immunoprecipitation; eCLIP, enhanced CLIP; HITS, high-throughput sequencing; iCLIP, individual-nucleotide resolution CLIP; irCLIP, infrared CLIP; RBP, RNA-binding protein.
Specificity
The specificity of CLIP depends less on the choice of the three principal methods and more on the stringency and validation of the steps required for purification of the protein–RNA complex of interest. Many RBPs participate in stable RNPs that do not dissociate even under the relatively stringent immunoprecipitation conditions of the standard CLIP, especially in the presence of RNA fragments that could help to stabilize them. Copurified RBPs can have different RNA specificities and functions from the RBP of interest, and therefore ideally no additional RBPs should be copurified to ensure high specificity. A denaturing condition is used by some CLIP variants to disrupt interactions with copurified RBPs, but this is not possible when using antibodies that recognize the natively folded state of endogenous RBPs.
Separation of complexes by SDS-PAGE and membrane transfer followed by their visualization, along with the use of appropriate negative controls, is thus a crucial quality control step for methods that omit a denaturation step. Greater care needs to be taken when analyzing data from methods that neither denature nor visualize the complexes, such as eCLIP (17), since it cannot be assumed that the sequenced reads represent only RNAs in contact with the protein of interest. In these cases, careful computational quality control analyses, for example, orthogonal data and RNA maps, should be used to examine specificity on a protein-by-protein basis. As is evident by the analysis of PTBP1 RNA splicing maps (Figure 3b), the specificity of iCLIP is highest, since the silenced exons are specific in the enrichment at the 3′ splice site, while enhanced exons contain enrichment downstream of the exons. The specificity is also high for eCLIP despite its low sensitivity, with enrichment at silenced but not enhanced exons. However, specificity is low for irCLIP, where the enrichment at the 3′ splice site for silenced exons is only slightly larger than the enhanced exons.
The potential for nonspecific signal is higher for RBPs with low abundance or poor crosslinking efficiency. Crosslinking between RNAs and proteins requires close contacts between an amino acid and the nucleobase. Moreover, analyses of diverse CLIP data sets indicate that the crosslinking efficiency of uridines and uridine-rich motifs is highest (12, 16), and therefore RBPs that contain such motifs in their binding site are expected to crosslink best. Therefore, it is least challenging to produce highly specific CLIP data for RBPs that bind to uridine-rich sequences, especially if they are abundant, such as PTB1 and ELAV1. However, low-abundant or poorly crosslinking RBPs, which likely include many noncanonical or double-stranded RBPs, such as STAU1, may require denaturing conditions to ensure that the isolated RNA fragments are specific (21). Taken together, visualization of protein–RNA complexes with SDS-PAGE analysis can validate the specificity of purification across a broad range of RBPs and conditions, thus simplifying downstream computational analyses.
A Considered Clip Analysis Strategy
At its outset, CLIP data analysis follows a similar pipeline to most next-generation sequencing, but it diverges in its experimental quality assessment and the subsequent determination and functional integration of the identified binding sites (Figure 1). In this section, we first note the nuances of read alignment particular to CLIP. We then delve into the distinct analytical issues faced by the experimental choices, detailing the CLIP quality measures necessary to appraise any results. We then consider the many challenges encountered in elucidating binding sites from the aligned reads. Finally, we end by looking at ways to distill the properties of these sites and to relate them to biological functions.
Read Alignment
After a standard quality assessment of the sequencing run, the CLIP data analysis pipeline turns to read alignment. Comprehensive benchmarking of RNA-seq read aligners has recently been undertaken and is outside the scope of this review (22). However, there are three factors that should be considered when tailoring this step to a CLIP experiment.
The first decision is whether to align to the transcriptome or the genome. The main advantage of transcriptomic over genomic alignment is increased sensitivity, with the proviso that only annotated mature transcripts are considered. However, for the majority of cases, where there is usually sufficient experimental sensitivity, alignment to the genome is preferred. This ensures an appropriate assessment of the many RBPs that bind to precursor messenger RNA (pre-mRNA) transcripts, for example, in introns. Moreover, the use of a splice-aware aligner would accommodate those that bind to mature messenger RNA transcripts.
Second, the use of unique molecular identifiers (UMIs) in iCLIP and later methods accounts for the amplification biases introduced by polymerase chain reaction (PCR), but to be able to deconvolve UMIs for cDNAs that map to the same position, it is best to use only uniquely aligned reads. To maximize the fraction of reads that can be aligned uniquely, the originating cDNA needs to be sufficiently long. When cDNA lengths are greater than 35 nucleotides, high alignment rates can be achieved even with common RBP-bound repetitive elements, such as Alu elements (23, 24). However, if the RBP under study prefers other repetitive elements, such as microsatellite repeats or small nuclear RNA clusters, then a customized solution may be necessary. One option for these cases is to align reads to a consensus repetitive sequence (25). Another is to use expectation-maximization to assign multimapped reads (26), but with this approach, the mapping position cannot be used to identify PCR duplicates.
Third, more technically, it is important to fine-tune some of the alignment parameters to the CLIP method that has been used. The number of mismatches allowed must be chosen with care: Too lax a setting will align reads with multiple sequencing errors. These may subsequently be identified spuriously as originating from different cDNAs when collapsing duplicates. It will also affect the sensitivity and specificity of the mutation-based methods. Specifically for the truncation-based methods, it is important to disable soft-clipping to ensure that the crosslink site, reflected in the start of the read, is properly aligned.
Quality Control
Thorough quality assessment is imperative to understand the CLIP experiment, both for the appropriate assignment of binding sites and for integrating with other data sources. We propose that measures that evaluate sensitivity and specificity of data be explored in combination. The simplest measures of sensitivity and specificity are the number of unique cDNAs in the sequencing library and the number of significant peaks, respectively. In the future, it will be valuable to use a consistent computational approach to evaluate these basic quality measures in all publicly available data produced by the different variants of CLIP, thereby obtaining an estimate of the variation in existing data, and potentially create standards for future experiments.
Complementary DNA complexity
cDNA complexity provides an assessment of the sensitivity of the CLIP experiment. The total number of unique cDNAs helps one appreciate the dynamic range of RBP-RNA interactions that can be detected. Complexity reflects several biological and technical factors: the abundance and crosslinking efficiency of the RBP and the efficiency of immunoprecipitation, adapter ligation, and cDNA library preparation. PCR duplication, although necessary for the method, can create difficulties for monitoring library complexity. Amplification of cDNA fragments is not uniform but is affected by sequence content and length. In the original CLIP protocols, it is therefore necessary to remove the duplicate reads and consider only the unique reads to count the cDNAs reliably. The CLIP Tool Kit achieves this by collapsing the identical reads before alignment (27), but ideally, reads are collapsed after alignment based on identical genomic start positions, since this accounts for read variations that result from sequencing errors (13).
The current gold standard is a more sophisticated approach that experimentally labels each cDNA as it is reverse transcribed (15, 28). This is done by introducing a UMI, which is a randomized sequence of nucleotides (hence, it is also known as a random barcode or randomer), into the reverse transcription primer. After PCR amplification, the UMI remains as a hallmark of unique cDNAs. iCount and other tools developed for the analysis of iCLIP data use UMIs in combination with the read start position to count the unique cDNAs accurately and thus obtain reliable information about cDNA complexity and enable quantitative analysis of crosslinking at individual nucleotide positions. The use of UMIs is crucial to overcome the artifacts of PCR amplification and thus preserve the quantitative information present in the cDNA counts; this is particularly important in quantifying binding to high-affinity binding sites and in abundant RNAs.
Complementary DNA specificity
Establishing cDNA specificity is the most difficult evaluation, and yet the most important. It is mostly dictated by the purification of the RNA-RBP complex, hence the importance of optimizing this process. Ground truth is often not known, and the appropriate measurement may vary between RBPs due to differing sequence and structure specificities. In practice, often only circumspect or post hoc approaches can be used.
The percentage of crosslink sites that occur in peaks is a basic measure of the capacity of the cDNA library to identify binding sites, and it provides some indication of specificity. Enrichment of RBP-specific, binding-related k-mers within peaks, compared to a suitable background region, also provides some reassurance. Enrichment of motifs [ascertained using alternative methods, such as RNAcompete (29)] within clusters of peaks gives another independent, complementary assessment of specificity. However, these last two will only work for RBPs that bind particular sequences or motifs; for those that do not, there will be little enrichment regardless. Finally, the integration of CLIP results with orthogonal data provides the best measure of specificity but requires the availability of such data. RNA maps (detailed in the sidebar titled RNA Maps: Integrating CLIP with Orthogonal Methods) are an efficient approach for visualizing crosslinking around transcriptomic landmarks that are relevant for the function of the RBP: for example, exon–intron junctions of regulated exons for RBPs involved in splicing.
While overlapping cDNA starts are a measure of potentially high specificity of iCLIP data for crosslink clusters, they can also reflect the aforementioned sequence preferences of the UV crosslinking reaction, which needs to be taken into account. Moreover, overlapping cDNA ends in iCLIP (and both sides of cDNAs in HITS-CLIP and PAR-CLIP) reflect the preferences of the RNases used for fragmentation (30). The alignment of cDNA ends can lead to a generic misalignment of the starts of cDNAs of different lengths; while this was initially interpreted as possibly indicating the presence of readthrough cDNAs (31), the RNase fragmentation biases were found to be the more likely cause (16). The alignment of cDNA ends correlates with an enrichment of k-mers at cDNA ends, which is a useful tool to examine the biases introduced by RNA fragmentation. Optimized RNase fragmentation conditions produce a broad range of cDNA lengths and so avoid such biases, ensuring that the full binding sites can be defined (which is particularly important for long binding sites) (16). This also guarantees that the peaks identified by overlapping cDNA clusters are a true measure of data specificity, rather than an artifact of inappropriate RNase fragmentation.
Peak Calling
The main challenge of CLIP data analysis is related to the biological context of protein–RNA complexes. Binding cannot be classified into simple binary categories of specific and nonspecific; instead, RBPs bind RNAs with a range of affinities and kinetics (32). Some RBPs associate with RNA polymerase and transiently interact with many low-affinity sites on nascent transcripts before finding a high-affinity binding site, and others can spread over larger regions of RNA after finding a high-affinity sequence. Many assemble on RNAs combinatorially as part of larger complexes. While the probability that an RBP will crosslink repetitively to a clustered set of crosslink sites is generally increased at sites with high affinity and favorable binding kinetics, the exact threshold for defining functionally relevant types of crosslink clusters depends on many factors, such as the type of RBP under study, the type of bound RNA, the function under regulation, and the binding position relative to other regulatory complexes. Thus, there are no absolute thresholds that can be set to distinguish low-affinity, transient binding from high-affinity, functional binding. This challenge could become insurmountable if data contain many nonspecific sites that do not represent direct interactions of the specific RBP. It is thus of paramount importance to maximize the specificity of CLIP data experimentally, since this can ameliorate the computational analyses needed to identify the functionally relevant binding sites.
Peak calling is the first step toward identifying the RNA sites that are highly occupied by the RBP: those that are most likely of functional significance. The basic approach searches for the pileup of aligned reads at specific positions on transcripts. In methods such as ChIP-seq and RIP, which tend to purify large protein–protein complexes as well as free DNA or RNA, the purpose of this step is largely to isolate the signal from the inherent background noise of the techniques. CLIP employs many unique experimental steps to remove such noise, including covalent crosslinking, RNA fragmentation, stringent purification, and visualization of purified protein–RNA complexes, and thus, in a fully optimized experiment, the mapped reads should almost exclusively correspond to the sites of direct protein–RNA contacts. Therefore, noise from nonspecific backgrounds should not be a major concern for CLIP data analyses. As evidence of the high specificity of CLIP experiments, the raw whole reads from Nova (Figure 2c) and PTBP1 HITS-CLIP (Figure 3a) and the raw crosslink positions from PTBP1 iCLIP (Figure 3a) yield highly position-dependent enrichment on RNA splicing maps.
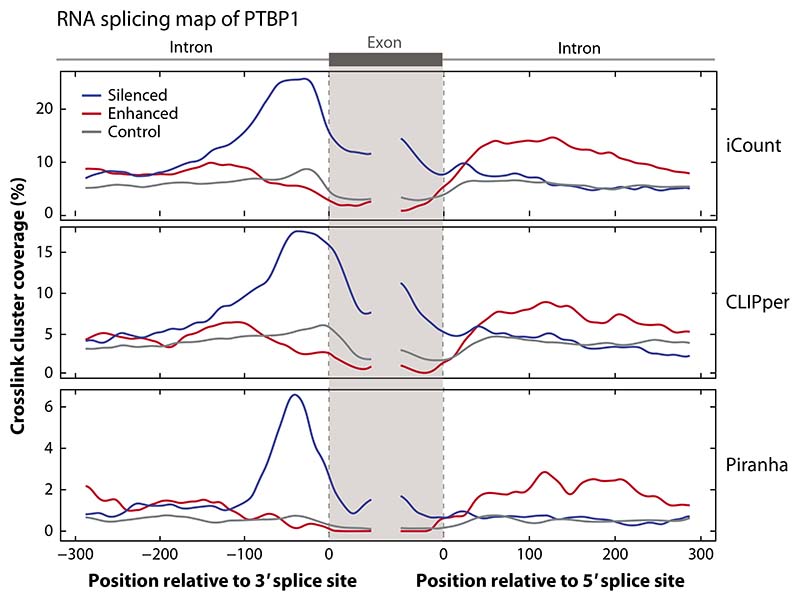
Many peak calling tools have been developed (33), some specific for particular CLIP protocols, others more generally applicable (detailed in Supplemental Table 2). The large number of peak calling tools, which often come with adjustable parameters, may present a bewildering set of possibilities. This is further complicated by the different strategies in identifying the crosslink sites by the various experimental protocols (Table 1; see also Supplemental Table 1). Benchmarking tools is challenging because of the differences in experimental protocols and our limited understanding of the ground truth regarding RNA binding sites in vivo (34, 35). Nevertheless, in this review, we attempt to demonstrate the impact of the different CLIP protocols and computational tools through use of the RNA maps, which combine CLIP with orthogonal functional data to derive an estimate of ground truth on the assumption that RNA landmarks regulated by an RBP should contain its nearby RNA binding sites (see the sidebar titled RNA Maps: Integrating CLIP with Orthogonal Methods). A comparison of peak calling by three tools in Figure 4 demonstrates that all have similar specificity when using iCLIP data as input, with iCount leading to highest sensitivity, since it detects significant crosslink clusters at the peak position at 3′ splice sites of 25% of the silenced exons.
Figure 4.
A comparison of different CLIP peak calling tools. RNA maps are used to demonstrate the differences in peak calling tools for the same iCLIP PTBP1 data set (16). To show that the RNA maps can be reproduced by exons defined using a different data source, we defined the regulated exons using RNA-seq data following PTBP1 CRISPR knockout in K562 cells from the ENCODE website. We identified the skipped exons detected using rMATS (101) using only junction counts and a p-value threshold of 0.05 and FDR threshold of 0.1. Silenced and enhanced exons were defined using an inclusion level difference threshold of 0.05; control exons were selected as those with a p-value greater than 0.1, an FDR value greater than 0.1, an inclusion level of less than 0.9, and an inclusion level difference less than 0.05. We compared the peaks called using iCount (15) (using a 15-nucleotide peak calling half-window and 30-nucleotide clustering window), Piranha (42) (using a 30-nucleotide bin size and 30-nucleotide merging window), and CLIPper (25, 44) (using default settings). For this data set, Piranha and iCount have runtimes of ∼2 minutes and ∼7 hours, respectively, using 1 processor; CLIPper has a runtime of ∼7 days using 20 processors. The code to reproduce this figure is available for download at https://github.com/jernejule/clip-data-science. Abbreviations: CLIP, UV crosslinking and immunoprecipitation; FDR, false discovery rate; iCLIP, individual-nucleotide resolution CLIP.
Challenge 1: What to use to call a peak?
The first consideration is how to use a read to define a peak. This differs for the mutation-based and the truncation-based CLIP methods. For mutation-based methods, it is important to distinguish a mutation from confounders, such as sequencing errors, single-nucleotide polymorphisms, or somatic mutations in cell lines. Early tools, such as PARalyzer, addressed this issue either by setting a minimum number of mutations at a site or by limiting the number of mismatches permitted during alignment (36). Although a simple and effective way of reducing false positives, this strategy has the disadvantage of also reducing the sensitivity of the experiment. PIPE-CLIP improves on this strategy by modeling each event with a binomial distribution, with a success rate calculated from the read coverage (37). As a further refinement, wavClusteR uses a nonparametric, two-component mixture model to distinguish crosslink-induced mutations from noise and integrates this using a Bayesian network representation (38, 39).
For truncation-based methods, peak calling seems more straightforward: The nucleotide upstream of the start of the read is the crosslink position (which we term the “cDNA start”) and can be used to call peaks. However, there is a caveat. For the clear majority of the cDNAs, reverse transcription stops at the crosslink site, but it does still read through at times (this provides the signal for the readthrough-based methods). In iCLIP experiments of most RBPs, however, over 90% of cDNAs terminate at the crosslink site (12). Therefore, as discussed above, provided that there are limited cDNA end constraints and that cDNA sizes cover a broad range of lengths, the use of the read starts assigns crosslink sites with no positional bias (16). Finally, 4SU-iCLIP uses 4SU for crosslinking, as is done in PAR-CLIP, but then employs iCLIP protocol to prepare the cDNA library, raising the question whether mutations (as in PAR-CLIP) or truncations (as in iCLIP) should be used. Analysis of PTBP1 binding motifs in 4SU-iCLIP cDNAs indicates that truncations report a more reliable estimate of crosslink sites than transitions (16). This still needs to be evaluated for additional RBPs.
It is important to use the appropriate marker to call peaks. The eCLIP narrowPeaks publicly available from the ENCODE consortium was defined using an algorithm that used whole reads. However, such use of whole reads leads to misalignment of binding sites and loss of resolution, as is evident from PTBP1 motif analysis (Figure 2a). This can be solved using the truncation-based approach of the iCount algorithm, which defines peaks based on the starts of mapped reads (Figure 2b), and a similar approach has also been implemented by the published eCLIP study (25). In summary, the use of whole reads is appropriate for the original variants of CLIP, and mutations can be used as alternative sources for peak calling, such as thymine-to-cytosine (T-to-C) mutations in PAR-CLIP, while the read starts should be used for iCLIP and other methods that are optimized for amplification of truncated cDNAs.
Challenge 2: What is a peak?
The next problem is defining what constitutes a peak: How high and how wide does the pileup of reads need to be? The height of a peak provides a guide as to the likelihood of a locus being a true binding site, while the width may indicate when one binding site should actually be considered as two adjacent ones. This is important because some RBPs have narrow, focused binding sites (e.g., PTBP1), whereas others bind more diffusely across a transcript (e.g., MATR3).
Peak height
The focus of most tools is calculating the probability that a given binding site does not belong to a background CLIP read distribution (34, 40). Generally, a probability distribution is fitted to the count data; differences in the tools arise from the generation of the background and the probability distribution function chosen to model the read counts.
The majority of available tools use variations on a negative binomial distribution. This is often used for count data because it can account for overdispersion (i.e., if the variance of data is greater than the mean). ASPeak uses this distribution unmodified (41). Piranha (42) and PIPE-CLIP (37) use a zero-truncated negative binomial distribution. It has been shown for a range of RBPs and CLIP methods that this zero-truncated negative binomial distribution fits the count data better than simple negative binomial or Poisson distributions (42). Piranha calculates the counts in user-defined bins across the genome; an appropriate size depends on the RBP. A zero-truncated negative binomial distribution is fitted to the data; bins where there is a higher read count than would be expected can then be selected as peaks using a p-value threshold.
The iCount tool, developed along with the iCLIP method, avoids fitting a specific distribution but uses permutation analysis (15; T. Curk, G. Rot, C. Gorup, J. Zmrzlikar, J. König, et al., manuscript in preparation, available at https://github.com/tomazc/iCount). The counts are randomly distributed a predefined number of times within a relevant region of interest (such as introns) on a gene-by-gene basis to generate a background. Then, the comparison of the observed distribution with the random one yields a false discovery rate. The primary disadvantage of this method is that, in order to generate meaningful random distributions, one needs an annotation to provide the regions of interest. A similar approach is used by the CLIP Tool Kit (27) and Pyicoclip (43).
CLIPper (44), the tool of choice of the ENCODE consortium (25), combines ideas from both these approaches. Similar to iCount, a false discovery rate is calculated in a first pass. However, by default, the reads are randomly distributed within the entire gene rather than a more localized region of interest (9, 44). (A user-defined window around a read can be used instead as a semiexperimental option.) In a second pass, similar to Piranha, peaks that have fewer reads than would be expected across the transcriptome are removed. However, a Poisson distribution is used rather than the zero-truncated negative binomial.
A different approach is used by PARalyzer for PAR-CLIP. Here, for a given position, a kernel density–based classifier estimates a Gaussian density profile for both T-to-C mutations (signal) and the absence of T-to-C mutations (background). Loci where the signal is greater than the background are called as binding sites.
Peak width
Demarcating the width of a peak is of important biological relevance. As already noted, different types of RBPs have differing binding preferences. Some tools, such as PIPE-CLIP, cluster adjacent overlapping reads to assign peak width, but this strategy lacks biological validity, as read length is more dependent on technical factors, such as RNase activity, than on RBP binding preferences.
The strategy to discern peak width from the crosslink positions usually needs to be adjusted to the RBP under study. As a result, several tools require the user to set this window or clustering size, e.g., PARalyzer, Piranha, iCount. However, prior knowledge of the RBP is needed to do so effectively. In cases where this is not available, it may be helpful to compare peak and motif distributions (Figures 2a,b) or RNA maps with different settings of clustering size (Figures 2d, 3b, and 4). Our current default conditions rely on three-nucleotide clustering windows for preliminary data exploration (Figure 2d), but crosslink sites from wider windows can be included to incorporate various types of RNA binding (Figure 3b). With this approach, it is evident that PTBP1 binding at 3∗ splice sites of silenced exons is highly clustered, and thus, the sensitivity at this position remains the same as for raw data, while sensitivity at control exons drops (compare Figures 3a and 3b). As further validation, this approach defines interaction sites with high sensitivity and specificity when using exons defined either by microarray (Figure 3b) or RNA-seq data (Figure 4).
Other methods utilize the read distribution to define the cluster boundaries on a statistical basis. wavClusteR uses a coverage-based algorithm called mini-rank norm to identify the boundaries by evaluating all putative clusters using a rank-based approach. The CLIP Tool Kit uses a valley-seeking algorithm, which uses user-defined thresholds based on the heights of local maxima within a cluster of peaks and the intervening valley read coverage to delineate adjacent peaks. Finally, CLIPper uses cubic spline fitting to fit a curve to the peak and defines the boundaries by excluding points on the curve that exceed the false discovery rate threshold. The precise margins for fitting the curve can be adjusted.
Taken together, the choice of the peak calling tool and settings for each tool can modify the sensitivity and specificity of data, thereby affecting the conclusions that are drawn (Figure 4). Thus, two principles can be used to determine the optimal approach for peak calling: settings should be tailored to the biology of the RBP under study, and when performing comparisons between data sets, the same tool and settings should be used.
Challenge 3: How to account for variable RNA abundance?
The read count is not a direct measure of RBP affinity or indeed even the importance of a binding site. It can be influenced by other factors, most notably RNA abundance. This varies from gene to gene, and so the count of CLIP cDNAs within a transcript, or within an intron, is a composite measure of both RBP binding affinity and the abundance of the transcript or the intron. This is confirmed by the correlation between CLIP read counts and RNA-seq read counts (42). A negative control lacking the specific antibody (usually replaced by nonspecific immunoglobulin G) is often performed as part of CLIP experiments, but due to the high stringency of the immunoprecipitation conditions in CLIP experiments, this negative control normally contains at least 100-fold fewer cDNAs than the specific experiments (15). Thus, if CLIP conditions are well optimized, the cDNA coverage from negative controls is too shallow to be used for correcting for RNA abundance.
To some extent, the CLIP data itself can be used to correct for the abundance of the different transcript regions. Most available data indicate that RBPs tend to crosslink quite broadly across their bound transcripts such that, in addition to the high-affinity binding sites that contain clustered crosslinking, many additional dispersed crosslink sites are present in the same transcripts, indicative of a low-affinity, scanning mode of binding. The density of such broadly dispersed crosslinking depends more on the abundance of transcript regions than on the presence of specific binding motifs. Thus, the randomization and permutation approach adopted by peak callers such as iCount, which uses the total number of CLIP cDNAs in each region to model the background distribution, implicitly models the variable RNA abundance between transcript regions.
In order to control for the impact of transcript abundance, one can obtain additional data in parallel with the CLIP experiment. RNA-seq data are the most commonly produced and have been used to normalize CLIP coverage within transcripts (42). Most peak calling algorithms cannot include RNA-seq or other independent count–based data for normalization, but Piranha and ASpeak are two exceptions. Piranha uses the data as a covariate in the zero-truncated negative binomial regression model for the counts, whereas ASpeak uses the data to calculate an expression-sensitive background.
There are limitations of using RNA-seq. Most commonly, polyadenylated or total RNA-seq data are used. However, many, if not most, RBPs strongly bind to pre-mRNA transcripts, especially to introns, which are not well covered by RNA-seq. In this case, it has been shown that normalizing the CLIP data using NET-seq (native elongating transcript sequencing), which captures nascent transcripts including pre-mRNA, improves recovery of binding motifs (45). An alternative approach is the generation of input libraries without immunoprecipitation (46). Here, the total lysate after treatment with RNase is loaded on the gel and transferred to the membrane. The RNAs that crosslink to all RBPs present in a selected section of the membrane are isolated and their cDNA libraries are prepared in the same way as for the specific immunoprecipitated RBP. A similar approach has been employed for the analysis of eCLIP data, where an enrichment score is calculated by dividing the cDNA count of a specific RBP at a given site by the size-matched input (SMI) read count (17).
Furthermore, it is not sufficient just to consider read counts per transcript for data normalization. The distribution of the reads along a transcript is also a factor. It has been observed with total RNA-seq that the abundance of reads along the long introns in the brain is variable, which results in a sawtooth pattern (47); interestingly, the long introns (especially introns longer than 100 kb) are strongly enriched in genes that are specifically expressed in the brain (48). Transcription of introns longer than 100 kb is expected to take over 30 minutes, which is much longer than the time needed for any nuclear RBPs to assemble on introns, regardless of whether this binding is cotranscriptional. It is this long delay that leads to increased RBP binding to 5′ regions compared to 3′ regions of introns and the resulting sawtooth pattern. Thus, it is expected that most nuclear RBPs should have the sawtooth binding pattern on long introns expressed in the brain—the possible exception being RBPs that bind introns only after splicing is completed, such as the branchpoint binding protein that binds to spliced intron lariats. Indeed, a study using iCLIP (49) reported that most nuclear RBPs that bind to long introns in the brain show the sawtooth pattern, including FUS, TDP-43, and U2AF2. However, a study using CLIP-seq (i.e., the original CLIP method) reported that only FUS, but not TDP-43, has such a pattern, which was the basis for the conclusion that FUS binds via cotranscriptional deposition (50).
The difference between conclusions reached by iCLIP (49) and CLIP-seq (50) might reflect the differences in the quantitative nature of the two methods. Overlapping cDNAs that map to the same position on transcripts are much more common for TDP-43 than FUS because the binding pattern of FUS is more broadly dispersed across introns. Due to its use of UMIs, iCLIP can quantify cDNAs that map to the same genomic locations, while the quantitative analysis of binding patterns across introns might be affected by PCR amplification artifacts in CLIP-seq. While the reasons for the observed differences remain to be further examined, it is clear that technical differences can affect the biological conclusions drawn from CLIP data, and thus data quality analyses are needed to aid their interpretation. Moreover, methods to normalize the data not only by the variable abundance of RNAs as a whole but also by variable abundance between exons and introns, between different introns, and across long introns are necessary to allow a more reliable interpretation of the binding profiles.
Finally, an important consideration for data analysis is that most RBPs are enriched in a specific cellular compartment, where the abundance of available RNAs is likely to be different from that seen in RNA-seq or SMI libraries. As our appreciation of RBP localization in subcellular compartments grows, with techniques that fractionate the cell before performing CLIP (51, 52), it will be valuable to produce SMI data for these compartments also, thus controlling for the compartmental variations in RNA abundance.
Challenge 4: How to account for crosslinking biases?
It is well established that there are inherent biases in the UV crosslinking reaction, with preferential crosslinking between certain peptides and certain nucleotides. UVC-induced crosslinking, as used in the truncation-based methods, occurs predominantly at uridines (12). Furthermore, analysis of the SMI controls from eCLIP experiments identified 10 generically enriched tetramers (16). These generic motifs were enriched at cDNA starts of eCLIP and iCLIP data of multiple RBPs, indicating that they might reflect increased efficiency of crosslinking rather than simply the presence of a few dominating RBPs in these different experiments. All the generic motifs have a high uridine content, which is consistent with the uridine enrichment seen in iCLIP when using UVC for crosslinking (12) but not with crosslinking induced by a mutant RNA methylase in methyl-5-cytosine methylation-iCLIP (53).
The SMI control can be used to account for these biases as well as to normalize for RNA abundance. However, it is not yet clear whether the normalization process is sufficient or whether peaks that overlap with those found in the SMI control should be subtracted. PureCLIP is one tool that uses a statistical framework to address this particular bias (54). It uses a hidden Markov model framework to incorporate experimental biases into the peak calling process. PureCLIP learns crosslink motifs from the SMI control data and incorporates this into the emission probability of the crosslink state. In this way, regions that correspond both to peaks and to generic motifs can be excluded to reduce the sequence artifacts that might arise from crosslinking preferences. However, this approach should be applied with care, since the binding preferences of many RBPs may include generic motifs; for example, proteins such as PTBP1 preferentially bind to uracil-cytosine-rich motifs, and therefore generic motifs are more strongly enriched at crosslink sites in PTBP1 iCLIP data (16).
Challenge 5: How reproducible are the data?
CLIP experiments should be replicated to ensure the robustness of the data and the resulting biological conclusions. The overall reproducibility can be assessed to some extent by correlating the number of crosslinks per peak between replicates. However, few tools explicitly leverage data across replicates in peak calling. One tool currently being developed, omniCLIP, aims to do so, in addition to modeling several confounding factors, including RNA abundance (55).
There are two ways to use replicates; the choice depends upon the quality of the experiment and the desired balance between sensitivity and specificity. If the sensitivity of the experiment is a concern, biological replicates can be merged before peak calling to boost it. This comes at a cost to the specificity of the results. An alternative that offsets this to some extent, but still increases sensitivity, calls peaks on each replicate separately, improving the signal-to-noise ratio. Then, taking the union of peaks from the replicates maximizes sensitivity. Of course, corroborative data would be needed to validate any resultant findings.
However, if specificity is of greater importance, then after peak calling on each replicate separately, the intersection of peaks can be used. Early studies took this route both to reduce the chance of peaks arising as an artifact of PCR duplication and to account for biological variation (11). The use of UMIs is now well established to deal with PCR duplicates. Nevertheless, replicate analysis does engender greatest confidence in the set of putative binding sites, particularly in experiments with greater expected biological variation, provided that the sensitivity of each replicate is sufficient to allow reliable peak calling within both the highly and the lowly abundant RNAs. Members of the ENCODE consortium have refined this approach by using the irreproducible discovery rate (56) originally implemented for ChIP-seq data to identify reproducible peaks across replicates using a statistical threshold (17). Given the great variation in RNA abundance levels, it remains to be tested if this approach introduces any bias for highly abundant RNAs.
Modeling Binding Sites and the False Negative Problem
Peak calling identifies putative binding sites, minimizing the false positive rate of the underlying experimental data. There are many tools for examining these results (Supplemental Table 3). Further simple analysis can reveal basic biological information about the RBP-RNA interaction: relationships with transcript regions or gene sets and ontologies. However, a more complex characterization is required for a fuller understanding. CLIP methods have an intrinsic biological and computational limitation: They can only generate data about binding sites on expressed transcripts and in regions that are mappable. Furthermore, these data are restricted by the sensitivity of the experiment, as already discussed. This is termed the false negative problem. A more complex characterization is required for one to generalize the findings beyond the cell, tissue, or biological state in which the experiment was performed or indeed beyond the limitations imposed by the quality of the experiment. This starts with basic motif finding but extends to computational modeling.
Sequence motif finding
The putative binding sites can be used to learn about the sequence preferences of the RBP under study. Motif-finding tools, such as DREME (discriminative regular expression motif elicitation) (57) and HOMER (hypergeometric optimization of motif enrichment) (58), generally work by comparing a positive (bound) and negative (background) set of sequences and assessing the enrichment of motifs statistically (Fisher’s exact test for DREME and a hypergeometric test for HOMER) to generate position weight matrices.
The motif recognition domain may not be the RNA binding domain in the protein; hence, on the transcript, the motif may not be at the binding site but adjacent to it. Thus, for the positive sequence, a predefined window around the putative binding site should be used. Care needs to be taken with the selection of the background sequences, as this has a large influence over the statistical assessment of the enrichment. An appropriate set of sequences should be chosen based on available knowledge of the RBP for maximizing both sensitivity and specificity. This could be designed in silico (59), but it is probably more straightforward to select relevant genomic sequences informed by the data set. For instance, if one is investigating an RBP involved in splicing, such as PTBP1, with the putative binding sites highlighting a preference for intronic binding just upstream of the intron–exon boundary, a suitable background would be the unbound deep intronic regions of the targeted genes. Easier options, such as shuffling the positive sequences (DREME) or generating a random sequence of nucleotides (HOMER), should be used only as a second option. Shuffling will reduce the sensitivity of the detection of short motifs. A random sequence will reduce the specificity, as spurious motifs may be called significant since the true distribution of nucleotides in the genome is not random. The majority of these tools were designed for transcription factors and ChIP-seq data. Often, however, RNA motifs are shorter and more degenerate than their DNA counterparts. Recently, in kpLogo, a more customized tool has been developed to look for shorter sequence motifs and consider positional information (60). This may prove to be more useful for CLIP data.
Sequence motifs generated from CLIP data can be used to predict possible binding sites in a genomic sequence of interest, using tools such as FIMO (find individual motif occurrences) (61). They can also be compared with those generated from in vitro experiments, such as RNAcompete (29) or RNA Bind-n-seq (62, 63) to corroborate the specificity of the CLIP experiment. Motifs are known for only ∼15% of RBPs (29), however, and poor experimental specificity should not be conflated with a lack of sequence specificity.
Although less well understood, it is known that structural context, in addition to sequence preference, plays a role in RBP binding preferences (64–67). This is likely one of the reasons for a lack of sequence specificity. Structural context should therefore be considered when predicting binding sites. However, it is difficult to incorporate adequately either the complexity of RNA structure or the interdependence between sequence and structure into motif discovery tools, despite attempts to do so in tools such as Zagros (67), MEMERIS (66) and RNAcontext (68). Recent programs have been more successful, at least in incorporating the interdependence, by using a hidden Markov model (ssHMM) (69), but computational modeling of binding sites is ideally placed to integrate multiple related features, as discussed next.
Computational binding site modeling
GraphProt was the first tool to use machine learning methods to incorporate sequence and structure into the analysis of CLIP data (70). The features are encoded using a graph kernel approach, and a support vector machine is used to build the model, which is essentially treated as a classification task. The utility of GraphProt in addressing the false negative problem has been demonstrated: Peaks not detected from the raw signal because they are in a poor mappability region were predicted using GraphProt, and furthermore, 90% have been experimentally validated (34). More advanced machine learning methods, such as deep boosting (DeBooster) (71), have helped to derive more accurate predictions using multiple binding site features.
Ideally, in vivo experimental data elucidating RNA structure would be used as inputs to these models. Despite the great advances that have recently been made by icSHAPE (in vivo click selective 2-hydroxyl acylation and profiling experiment; 72), DMS-seq (dimethyl sulfate sequencing; 73), DMS-MaPseq (dimethyl sulfate mutational profiling with sequencing; 74), and structure-seq (75, 76) in identifying paired or unpaired nucleotides and by hiCLIP (RNA hybrid and individual-nucleotide resolution CLIP; 21), PARIS (psoralen analysis of RNA interactions and structures; 77), LIGR-seq (ligation of interacting RNA with sequencing; 78), and SPLASH (sequencing of psoralen crosslinked, ligated, and selected hybrids; 79) in identifying RNA duplexes, these data are not yet comprehensive enough to use for modeling. Hence, computational predictions, often using thermodynamic free energy minimization, must instead be used, despite their fallibility (80, 81). Although SHAPE data can be incorporated into these predictions (82), their inherent limitations should be borne in mind when interpreting RBP-RNA interaction preferences.
RNA sequence and structure are not the only variables that drive RBP-RNA interactions. Other factors, such as cooperative binding, position in the gene relative to exons, and other features, also play a role (83). These parameters can be included in both unsupervised and supervised models. iONMF uses orthogonality-regularized non-negative matrix factorization to identify factors associated with RBP binding and to estimate the importance of their contributions (83). Alternative machine learning methods, such as iDeep and iDeepS, which use neural networks, have slightly improved these predictions (84, 85).
Integrative analysis of CLIP data across RNA-binding proteins
As increasing numbers of CLIP data sets are produced for an ever-widening range of RBPs, researchers naturally turn to exploring the RNA interactions of a given RBP in the context of all the others. Several studies have already exploited CLIP data to identify coregulatory interactions, such as the competition between hnRNP C and U2AF2 in controlling Alu exonization (23) and the interplay between PTBP1 and MATR3 in coregulating alternative splicing (86). Databases such as DoRiNA 2.0 (87) and POSTAR (88) have been set up to help. DoRiNA 2.0 uploads RBP binding sites as they are published. This places a severe limitation on the comparisons that can be meaningfully undertaken. As already demonstrated, the use of both different CLIP techniques and different CLIP peak callers has a significant impact on the number, location, and size of binding sites that are discovered. POSTAR reanalyzes all the raw data using a different peak caller for each kind of CLIP variant. Non-negative matrix factorization can then be used to group together RBPs that bind to the same sites to explore co-operativity (83, 89). This enables more reliable comparisons across experiments, but it is best to avoid comparing RBPs for which different peak calling tools or different CLIP methods were used, since this could result in differences that are of a technical nature (89). Ideally, if a comparison is being undertaken using publicly available data, the approach taken by POSTAR should be bolstered first by assessing whether the quality of the experiment is sufficient even to proceed with a comparison, and second, by using the same peak calling procedure for all the RBPs that are part of the same comparison. An alternative approach has been to use matrix factorization directly on the crosslink sites as input, thus combining binding site prediction with integration of data across RBPs (83).
Integrative analysis of CLIP with orthogonal functional data
RNA binding profiles need to be integrated with orthogonal data to gain functional insight into the role of a given RBP-RNA interaction. Throughout this review, we have used RNA maps to demonstrate analytical considerations. However, RNA maps are also a powerful tool for studying the functions of these interactions and understanding the position-dependent mechanisms behind these functions (see the sidebar titled RNA Maps: Integrating CLIP with Orthogonal Methods) (90). Integration with nonsequencing data, such as analysis of RNA specificity with RNA Bind-n-seq or RBP subcellular localization, can also provide new mechanistic hypotheses, such as the potential role of DHX30 in mitochondrial transcription termination (25).
iCLIP, (YO)U-CLIP, (w)eCLIP
We are in an era of integrative genomics. Fusing insights gleaned from CLIP data from multiple RBPs with orthogonal genomic and nongenomic approaches will be the cornerstone for further studies of RBP-RNA interaction networks. It is crucial to attend to the minutiae of each data set to avoid being misled in this unifying vision. In this review, we have considered the effects of the experimental choices on the sensitivity and specificity of CLIP data. Appreciating these limitations is necessary to adapt the computational analyses appropriately. We have discussed the need to examine sensitivity and specificity of data in combination in order to give credence to the biological conclusions drawn. A unique feature of CLIP (when compared with methods such as chromatin immunoprecipitation or RIP) is its capacity to assess specificity experimentally via visualization of the purified RBP-RNA complexes, which also can be used to check that RNase fragmentation conditions are appropriate. Moreover, computational quality controls can be performed by combining CLIP analysis with mechanistic (sequence motif) or functional data (RNA maps) concerning the RBP under investigation.
Several key steps can ensure a robust assignment of binding sites from CLIP data. First, peak calling is performed to distinguish high-occupancy sites from the more dispersed binding that is less likely to be of functional significance. Using cDNA starts in truncation-based methods to identify the crosslink position is crucial to maintain the single-nucleotide resolution in the peak calling step. Second, the peaks require normalizing for RNA abundance and assessing for crosslinking bias. Further studies are needed to better understand how the precise parameters of both these aspects should be defined with due consideration to the binding characteristics of the RBP under study. Third, using peaks as an input for computational predictive models of the binding sites can help generalize the findings and address the false negative problem. To achieve these goals, researchers should provide a well-annotated protocol for each published experiment so that computational biologists can examine the potential sources of technical variation in the data. Dozens of different CLIP protocols are already available, and further changes will likely continue to be introduced. To enable appropriate quality control analyses, we suggest that the submission of each CLIP data set to a public database is accompanied by a protocol file that describes how each of the core 11 experimental steps of the protocol were performed (7).
Due to the increasing amount of data across species, tissues, cell lines, and RBPs, the computational analysis of protein–RNA interactions is well positioned to address new questions. For example, it is still difficult to examine the different modes of RBP binding: Could one distinguish low-affinity, scanning modes of binding from high-affinity, anchored binding from CLIP data? Other methods have generated large data sets on protein–protein interactions (91), protein localization (92), in vitro binding preferences (29, 62), and RBP function (25). Integration of these diverse data sets is a present challenge but will yield significant advances in our understanding of the role of protein–RNA interactions.
Finally, RBPs have been implicated in a range of diseases, from cancers to neurological conditions (93, 94). Studies of RBPs have already led to major medical advances. Understanding the interactions between the RBPs hnRNPA1/A2 and the SMN2 pre-mRNA has led to a break-through, FDA-approved treatment for spinal muscular atrophy using the antisense oligonucleotide nusinersen (95, 96). Developing appropriate computational approaches hand-in-hand with further applications of CLIP to primary cells and tissues, pluripotent stem cell models, and disease model organisms will undoubtedly lead to further insights into protein–RNA interactions that could be targets for future therapies.
Summary Points.
Optimizing and visualizing purification of RBP-RNA complexes maximizes specificity.
Most current CLIP protocols can amplify truncated cDNAs, and analysis of cDNA starts forms the cornerstone of the analysis of data produced by these protocols.
UMIs identify PCR duplicates, reducing downstream biases in the peak calling stage.
Peak calling should ideally be performed by evaluating the crosslink clusters. The window size parameters for clustering need to be adapted to the RBP under study.
SMI libraries are valuable to normalize the peaks for variable RNA abundance.
Motif analysis, in addition to providing mechanistic insight, elucidates the quality and resolution of the data.
Computational modeling could help address the false negative problem and evaluate the contribution of RNA sequence, structure, and other features to endogenous RNA recognition.
It is best to use a consistent experimental and analytical approach when integrating multiple CLIP data sets.
Supplementary Material
- CLIP
the key experimental method for exploring protein–RNA interactions using covalent crosslinking and immunoprecipitation
- RNA map
a tool for visualizing the function of protein–RNA interactions by integrating orthogonal data sets
- Peak calling
the computational process of identifying statistically significant binding sites from the experimental sequencing data
- SDS-PAGE
a technique to isolate proteins by their molecular weight using sodium dodecyl sulfate to denature the protein and polyacrylamide gel electrophoresis to separate them
- UMI
a unique molecular identifier of random nucleotides that is introduced to the reverse transcription adapter to enable reads arising from polymerase chain reaction duplication to be collapsed
Acknowledgments
We would like to thank members of the Ule and Luscombe labs, in particular, Igor Ruiz de los Mozos, Flora Lee, and Federico Agostini for assistance with the tables and for valuable comments during the preparation of this manuscript. This work was supported by funding from the European Research Council (617837-Translate) to J.U., a Wellcome Trust Joint Investigator Award to J.U. and N.M.L. (103760/Z/14/Z), a Wellcome Trust PhD Training Fellowship for Clinicians Award to A.M.C. (110292/Z/15/Z), a University College London Grand Challenges Award to N.H., and the Francis Crick Institute, which receives its core funding from Cancer Research UK (FC001002), the UK Medical Research Council (FC001002), and the Wellcome Trust (FC001002).
Footnotes
Disclosure Statement
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
Literature Cited
- 1.Gerstberger S, Hafner M, Tuschl T. A census of human RNA-binding proteins. Nat Rev Genet. 2014;15(12):829–45. doi: 10.1038/nrg3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beckmann BM, Castello A, Medenbach J. The expanding universe of ribonucleoproteins: of novel RNA-binding proteins and unconventional interactions. Pflügers Arch. 2016;468(6):1029–40. doi: 10.1007/s00424-016-1819-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lewis CJT, Pan T, Kalsotra A. RNA modifications and structures cooperate to guide RNA–protein interactions. Nat Rev Mol Cell Biol. 2017;18(3):202–10. doi: 10.1038/nrm.2016.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jens M, Rajewsky N. Competition between target sites of regulators shapes post-transcriptional gene regulation. Nat Rev Genet. 2015;16(2):113–26. doi: 10.1038/nrg3853. [DOI] [PubMed] [Google Scholar]
- 5.Shetlar MD, Carbone J, Steady E, Hom K. Photochemical addition of amino acids and peptides to polyuridylic acid. Photochem Photobiol. 1984;39(2):141–44. doi: 10.1111/j.1751-1097.1984.tb03419.x. [DOI] [PubMed] [Google Scholar]
- 6.Ule J, Jensen KB, Ruggiu M, Mele A, Ule A, Darnell RB. CLIP identifies Nova-regulated RNA networks in the brain. Science. 2003;302(5648):1212–15. doi: 10.1126/science.1090095. [DOI] [PubMed] [Google Scholar]
- 7.Lee FCY, Ule J. Advances in CLIP technologies for studies or protein-RNA interactions. Mol Cell. 2018;69(3):354–69. doi: 10.1016/j.molcel.2018.01.005. [DOI] [PubMed] [Google Scholar]
- 8.Hentze MW, Castello A, Schwarzl T, Preiss T. A brave new world of RNA-binding proteins. Nat Rev Mol Cell Biol. 2018;19:327–41. doi: 10.1038/nrm.2017.130. [DOI] [PubMed] [Google Scholar]
- 9.Yeo GW, Coufal NG, Liang TY, Peng GE, Fu X-D, Gage FH. An RNA code for the FOX2 splicing regulator revealed by mapping RNA–protein interactions in stem cells. Nat Struct Mol Biol. 2009;16(2):130–37. doi: 10.1038/nsmb.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ule J, Stefani G, Mele A, Ruggiu M, Wang X, et al. An RNA map predicting Nova-dependent splicing regulation. Nature. 2006;444(7119):580–86. doi: 10.1038/nature05304. [DOI] [PubMed] [Google Scholar]
- 11.Licatalosi DD, Mele A, Fak JJ, Ule J, Kayikci M, et al. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456(7221):464–69. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sugimoto Y, König J, Hussain S, Zupan B, Curk T, et al. Analysis of CLIP and iCLIP methods for nucleotide-resolution studies of protein-RNA interactions. Genome Biol. 2012;13(8):R67. doi: 10.1186/gb-2012-13-8-r67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang C, Darnell RB. Mapping in vivo protein-RNA interactions at single-nucleotide resolution from HITS-CLIP data. Nat Biotechnol. 2011;29(7):607–14. doi: 10.1038/nbt.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141(1):129–41. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.König J, Zarnack K, Rot G, Curk T, Kayikci M, et al. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat Struct Mol Biol. 2010;17(7):909–15. doi: 10.1038/nsmb.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haberman N, Huppertz I, Attig J, König J, Wang Z, et al. Insights into the design and interpretation of iCLIP experiments. Genome Biol. 2017;18(1):7. doi: 10.1186/s13059-016-1130-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van Nostrand EL, Pratt GA, Shishkin AA, Gelboin-Burkhart C, Fang MY, et al. Robust transcriptome-wide discovery of RNA-binding protein binding sites with enhanced CLIP (eCLIP) Nat Methods. 2016;13(6):508–14. doi: 10.1038/nmeth.3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zarnegar BJ, Flynn RA, Shen Y, Do BT, Chang HY, Khavari PA. irCLIP platform for efficient characterization of protein–RNA interactions. Nat Methods. 2016;13(6):489–92. doi: 10.1038/nmeth.3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Castello A, Fischer B, Eichelbaum K, Horos R, Beckmann BM, et al. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell. 2012;149(6):1393–406. doi: 10.1016/j.cell.2012.04.031. [DOI] [PubMed] [Google Scholar]
- 20.Huppertz I, Attig J, D’Ambrogio A, Easton LE, Sibley CR, et al. iCLIP: protein–RNA interactions at nucleotide resolution. Methods. 2014;65(3):274–87. doi: 10.1016/j.ymeth.2013.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sugimoto Y, Vigilante A, Darbo E, Zirra A, Militti C, et al. hiCLIP reveals the in vivo atlas of mRNA secondary structures recognized by Staufen 1. Nature. 2015;519(7544):491–94. doi: 10.1038/nature14280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baruzzo G, Hayer KE, Kim EJ, Di Camillo B, FitzGerald GA, Grant GR. Simulation-based comprehensive benchmarking of RNA-seq aligners. Nat Methods. 2017;14(2):135–39. doi: 10.1038/nmeth.4106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zarnack K, König J, Tajnik M, Martincorena I, Eustermann S, et al. Direct competition between hnRNP C and U2AF65 protects the transcriptome from the exonization of Alu elements. Cell. 2013;152(3):453–66. doi: 10.1016/j.cell.2012.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Attig J, Ruiz de los Mozos I, Haberman N, Wang Z, Emmett W, et al. Splicing repression allows the gradual emergence of new Alu-exons in primate evolution. eLife. 2016;5:e19545. doi: 10.7554/eLife.19545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Van Nostrand EL, Freese P, Pratt GA, Wang X, Wei X, et al. A large-scale binding and functional map of human RNA binding proteins. bioRxiv. 2017:179648. doi: 10.1101/179648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Z, Xing Y. CLIP-seq analysis of multi-mapped reads discovers novel functional RNA regulatory sites in the human transcriptome. Nucleic Acids Res. 2017;45(16):9260–71. doi: 10.1093/nar/gkx646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shah A, Qian Y, Weyn-Vanhentenryck SM, Zhang C. CLIP Tool Kit (CTK): a flexible and robust pipeline to analyze CLIP sequencing data. Bioinformatics. 2017;33(4):566–67. doi: 10.1093/bioinformatics/btw653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.König J, Zarnack K, Luscombe NM, Ule J. Protein–RNA interactions: new genomic technologies and perspectives. Nat Rev Genet. 2012;13(2):77–83. doi: 10.1038/nrg3141. [DOI] [PubMed] [Google Scholar]
- 29.Ray D, Kazan H, Cook KB, Weirauch MT, Najafabadi HS, et al. A compendium of RNA-binding motifs for decoding gene regulation. Nature. 2013;499(7457):172–77. doi: 10.1038/nature12311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kishore S, Jaskiewicz L, Burger L, Hausser J, Khorshid M, Zavolan M. A quantitative analysis of CLIP methods for identifying binding sites of RNA-binding proteins. Nat Methods. 2011;8(7):559–64. doi: 10.1038/nmeth.1608. [DOI] [PubMed] [Google Scholar]
- 31.Hauer C, Curk T, Anders S, Schwarzl T, Alleaume A-M, et al. Improved binding site assignment by high-resolution mapping of RNA–protein interactions using iCLIP. Nat Commun. 2015;6:7921. doi: 10.1038/ncomms8921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jankowsky E, Harris ME. Specificity and nonspecificity in RNA–protein interactions. Nat Rev Mol Cell Biol. 2015;16(9):533–44. doi: 10.1038/nrm4032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.De S, Gorospe M. Bioinformatic tools for analysis of CLIP ribonucleoprotein data. Wiley Interdiscip Rev RNA. 2017;8(4):e1404. doi: 10.1002/wrna.1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Uhl M, Houwaart T, Corrado G, Wright PR, Backofen R. Computational analysis of CLIP-seq data. Methods. 2017;118–19:60–72. doi: 10.1016/j.ymeth.2017.02.006. [DOI] [PubMed] [Google Scholar]
- 35.Bottini S, Hamouda-Tekaya N, Tanasa B, Zaragosi L-E, Grandjean V, et al. From benchmarking HITS-CLIP peak detection programs to a new method for identification of miRNA-binding sites from Ago2-CLIP data. Nucleic Acids Res. 2017;45(9):e71. doi: 10.1093/nar/gkx007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Corcoran DL, Georgiev S, Mukherjee N, Gottwein E, Skalsky RL, et al. PARalyzer: definition of RNA binding sites from PAR-CLIP short-read sequence data. Genome Biol. 2011;12(8):R79. doi: 10.1186/gb-2011-12-8-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen B, Yun J, Kim MS, Mendell JT, Xie Y. PIPE-CLIP: a comprehensive online tool for CLIP-seq data analysis. Genome Biol. 2014;15(1):R18. doi: 10.1186/gb-2014-15-1-r18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sievers C, Schlumpf T, Sawarkar R, Comoglio F, Paro R. Mixture models and wavelet transforms reveal high confidence RNA-protein interaction sites in MOV10 PAR-CLIP data. Nucleic Acids Res. 2012;40(20):e160. doi: 10.1093/nar/gks697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Comoglio F, Sievers C, Paro R. Sensitive and highly resolved identification of RNA-protein interaction sites in PAR-CLIP data. BMC Bioinform. 2015;16:32. doi: 10.1186/s12859-015-0470-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang T, Xiao G, Chu Y, Zhang MQ, Corey DR, Xie Y. Design and bioinformatics analysis of genome-wide CLIP experiments. Nucleic Acids Res. 2015;43(11):5263–74. doi: 10.1093/nar/gkv439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kucukural A, Özadam H, Singh G, Moore MJ, Cenik C. ASPeak: an abundance sensitive peak detection algorithm for RIP-Seq. Bioinformatics. 2013;29(19):2485–86. doi: 10.1093/bioinformatics/btt428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Uren PJ, Bahrami-Samani E, Burns SC, Qiao M, Karginov FV, et al. Site identification in high-throughput RNA–protein interaction data. Bioinformatics. 2012;28(23):3013–20. doi: 10.1093/bioinformatics/bts569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Althammer S, González-Vallinas J, Ballaré C, Beato M, Eyras E. Pyicos: a versatile toolkit for the analysis of high-throughput sequencing data. Bioinformatics. 2011;27(24):3333–40. doi: 10.1093/bioinformatics/btr570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lovci MT, Ghanem D, Marr H, Arnold J, Gee S, et al. Rbfox proteins regulate alternative mRNA splicing through evolutionarily conserved RNA bridges. Nat Struct Mol Biol. 2013;20(12):1434–42. doi: 10.1038/nsmb.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Takeda J-I, Masuda A, Ohno K. Six GU-rich (6GUR) FUS-binding motifs detected by normalization of CLIP-seq by Nascent-seq. Gene. 2017;618:57–64. doi: 10.1016/j.gene.2017.04.008. [DOI] [PubMed] [Google Scholar]
- 46.Ule J, Jensen K, Mele A, Darnell RB. CLIP: a method for identifying protein–RNA interaction sites in living cells. Methods. 2005;37(4):376–86. doi: 10.1016/j.ymeth.2005.07.018. [DOI] [PubMed] [Google Scholar]
- 47.Ameur A, Zaghlool A, Halvardson J, Wetterbom A, Gyllensten U, et al. Total RNA sequencing reveals nascent transcription and widespread co-transcriptional splicing in the human brain. Nat Struct Mol Biol. 2011;18(12):1435–40. doi: 10.1038/nsmb.2143. [DOI] [PubMed] [Google Scholar]
- 48.Sibley CR, Emmett W, Blazquez L, Faro A, Haberman N, et al. Recursive splicing in long vertebrate genes. Nature. 2015;521(7552):371–75. doi: 10.1038/nature14466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rogelj B, Easton LE, Bogu GK, Stanton LW, Rot G, et al. Widespread binding of FUS along nascent RNA regulates alternative splicing in the brain. Sci Rep. 2012;2:603. doi: 10.1038/srep00603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lagier-Tourenne C, Polymenidou M, Hutt KR, Vu AQ, Baughn M, et al. Divergent roles of ALS-linked proteins FUS/TLS and TDP-43 intersect in processing long pre-mRNAs. Nat Neurosci. 2012;15(11):1488–97. doi: 10.1038/nn.3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brugiolo M, Botti V, Liu N, Müller-McNicoll M, Neugebauer KM. Fractionation iCLIP detects persistent SR protein binding to conserved, retained introns in chromatin, nucleoplasm and cytoplasm. Nucleic Acids Res. 2017;45(18):10452–65. doi: 10.1093/nar/gkx671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sanford JR, Coutinho P, Hackett JA, Wang X, Ranahan W, Caceres JF. Identification of nuclear and cytoplasmic mRNA targets for the shuttling protein SF2/ASF. PLOS ONE. 2008;3(10):e3369. doi: 10.1371/journal.pone.0003369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hussain S, Sajini AA, Blanco S, Dietmann S, Lombard P, et al. NSun2-mediated cytosine-5 methylation of vault noncoding RNA determines its processing into regulatory small RNAs. Cell Rep. 2013;4(2):255–61. doi: 10.1016/j.celrep.2013.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Krakau S, Richard H, Marsico A. PureCLIP: capturing target-specific protein–RNA interaction footprints from single-nucleotide CLIP-seq data. Genome Biol. 2017;18:240. doi: 10.1186/s13059-017-1364-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Drewe-Boss P, Wessels H-H, Ohler U. omniCLIP: Bayesian identification of protein-RNA interactions from CLIP-Seq data. bioRxiv. 2017:161877. doi: 10.1101/161877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li Q, Brown JB, Huang H, Bickel PJ. Measuring reproducibility of high-throughput experiments. Ann Appl Stat. 2011;5(3):1752–79. [Google Scholar]
- 57.Bailey TL. DREME: motif discovery in transcription factor ChIP-seq data. Bioinformatics. 2011;27(12):1653–59. doi: 10.1093/bioinformatics/btr261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38(4):576–89. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu SS, Hockenberry AJ, Lancichinetti A, Jewett MC, Amaral LAN. NullSeq: a tool for generating random coding sequences with desired amino acid and GC contents. PLOS Comput Biol. 2016;12(11):e1005184. doi: 10.1371/journal.pcbi.1005184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wu X, Bartel DP. kpLogo: Positional k-mer analysis reveals hidden specificity in biological sequences. Nucleic Acids Res. 2017;45:W534–38. doi: 10.1093/nar/gkx323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Grant CE, Bailey TL, Noble WS. FIMO: scanning for occurrences of a given motif. Bioinformatics. 2011;27(7):1017–18. doi: 10.1093/bioinformatics/btr064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dominguez D, Freese P, Alexis MS, Su A, Hochman M, et al. Sequence, structure and context preferences of human RNA binding proteins. bioRxiv. 2017:201996. doi: 10.1101/201996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lambert N, Robertson A, Jangi M, McGeary S, Sharp PA, Burge CB. RNA Bind-n-Seq: quantitative assessment of the sequence and structural binding specificity of RNA binding proteins. Mol Cell. 2014;54(5):887–900. doi: 10.1016/j.molcel.2014.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fukunaga T, Ozaki H, Terai G, Asai K, Iwasaki W, Kiryu H. CapR: revealing structural specificities of RNA-binding protein target recognition using CLIP-seq data. Genome Biol. 2014;15(1):R16. doi: 10.1186/gb-2014-15-1-r16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li X, Quon G, Lipshitz HD, Morris Q. Predicting in vivo binding sites of RNA-binding proteins using mRNA secondary structure. RNA. 2010;16(6):1096–107. doi: 10.1261/rna.2017210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hiller M, Pudimat R, Busch A, Backofen R. Using RNA secondary structures to guide sequence motif finding towards single-stranded regions. Nucleic Acids Res. 2006;34(17):e117. doi: 10.1093/nar/gkl544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bahrami-Samani E, Penalva LOF, Smith AD, Uren PJ. Leveraging cross-link modification events in CLIP-seq for motif discovery. Nucleic Acids Res. 2015;43(1):95–103. doi: 10.1093/nar/gku1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kazan H, Ray D, Chan ET, Hughes TR, Morris Q. RNAcontext: a new method for learning the sequence and structure binding preferences of RNA-binding proteins. PLOS Comput Biol. 2010;6:e1000832. doi: 10.1371/journal.pcbi.1000832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Heller D, Krestel R, Ohler U, Vingron M, Marsico A. ssHMM: extracting intuitive sequence-structure motifs from high-throughput RNA-binding protein data. Nucleic Acids Res. 2017;45(19):11004–18. doi: 10.1093/nar/gkx756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Maticzka D, Lange SJ, Costa F, Backofen R. GraphProt: modeling binding preferences of RNA-binding proteins. Genome Biol. 2014;15(1):R17. doi: 10.1186/gb-2014-15-1-r17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li S, Dong F, Wu Y, Zhang S, Zhang C, et al. A deep boosting based approach for capturing the sequence binding preferences of RNA-binding proteins from high-throughput CLIP-seq data. Nucleic Acids Res. 2017;45(14):e129. doi: 10.1093/nar/gkx492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Spitale RC, Flynn RA, Zhang QC, Crisalli P, Lee B, et al. Structural imprints in vivo decode RNA regulatory mechanisms. Nature. 2015;519(7544):486–90. doi: 10.1038/nature14263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rouskin S, Zubradt M, Washietl S, Kellis M, Weissman JS. Genome-wide probing of RNA structure reveals active unfolding of mRNA structures in vivo. Nature. 2014;505(7485):701–5. doi: 10.1038/nature12894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zubradt M, Gupta P, Persad S, Lambowitz AM, Weissman JS, Rouskin S. DMS-MaPseq for genome-wide or targeted RNA structure probing in vivo. Nat Methods. 2017;14(1):75–82. doi: 10.1038/nmeth.4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ding Y, Kwok CK, Tang Y, Bevilacqua PC, Assmann SM. Genome-wide profiling of in vivo RNA structure at single-nucleotide resolution using structure-seq. Nat Protoc. 2015;10(7):1050–66. doi: 10.1038/nprot.2015.064. [DOI] [PubMed] [Google Scholar]
- 76.Ritchey LE, Su Z, Tang Y, Tack DC, Assmann SM, Bevilacqua PC. Structure-seq2: sensitive and accurate genome-wide profiling of RNA structure in vivo. Nucleic Acids Res. 2017;45(14):e135. doi: 10.1093/nar/gkx533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lu Z, Zhang QC, Lee B, Flynn RA, Smith MA, et al. RNA duplex map in living cells reveals higher-order transcriptome structure. Cell. 2016;165(5):1267–79. doi: 10.1016/j.cell.2016.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sharma E, Sterne-Weiler T, O’Hanlon D, Blencowe BJ. Global mapping of human RNA-RNA interactions. Mol Cell. 2016;62(4):618–26. doi: 10.1016/j.molcel.2016.04.030. [DOI] [PubMed] [Google Scholar]
- 79.Aw JGA, Shen Y, Wilm A, Sun M, Lim XN, et al. In vivo mapping of eukaryotic RNA interactomes reveals principles of higher-order organization and regulation. Mol Cell. 2016;62(4):603–17. doi: 10.1016/j.molcel.2016.04.028. [DOI] [PubMed] [Google Scholar]
- 80.Babak T, Blencowe BJ, Hughes TR. Considerations in the identification of functional RNA structural elements in genomic alignments. BMC Bioinform. 2007;8:33. doi: 10.1186/1471-2105-8-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Eddy SR. Computational analysis of conserved RNA secondary structure in transcriptomes and genomes. Annu Rev Biophys. 2014;43:433–56. doi: 10.1146/annurev-biophys-051013-022950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lorenz R, Luntzer D, Hofacker IL, Stadler PF, Wolfinger MT. SHAPE directed RNA folding. Bioinformatics. 2016;32(1):145–47. doi: 10.1093/bioinformatics/btv523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Stražar M, Žitnik M, Zupan B, Ule J, Curk T. Orthogonal matrix factorization enables integrative analysis of multiple RNA binding proteins. Bioinformatics. 2016;32(10):1527–35. doi: 10.1093/bioinformatics/btw003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pan X, Shen H-B. RNA-protein binding motifs mining with a new hybrid deep learning based cross-domain knowledge integration approach. BMC Bioinform. 2017;18(1):136. doi: 10.1186/s12859-017-1561-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pan X, Rijnbeek P, Yan J, Shen H-B. Prediction of RNA-protein sequence and structure binding preferences using deep convolutional and recurrent neural networks. bioRxiv. 2017:146175. doi: 10.1101/146175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Coelho MB, Attig J, Bellora N, König J, Hallegger M, et al. Nuclear matrix protein Matrin3 regulates alternative splicing and forms overlapping regulatory networks with PTB. EMBO J. 2015;34(5):653–68. doi: 10.15252/embj.201489852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Blin K, Dieterich C, Wurmus R, Rajewsky N, Landthaler M, Akalin A. DoRiNA 2.0–upgrading the doRiNA database of RNA interactions in post-transcriptional regulation. Nucleic Acids Res. 2015;43:D160–67. doi: 10.1093/nar/gku1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hu B, Yang Y-CT, Huang Y, Zhu Y, Lu ZJ. POSTAR: a platform for exploring post-transcriptional regulation coordinated by RNA-binding proteins. Nucleic Acids Res. 2017;45(D1):D104–14. doi: 10.1093/nar/gkw888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Li YE, Xiao M, Shi B, Yang Y-CT, Wang D, et al. Identification of high-confidence RNA regulatory elements by combinatorial classification of RNA-protein binding sites. Genome Biol. 2017;18(1):169. doi: 10.1186/s13059-017-1298-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Witten JT, Ule J. Understanding splicing regulation through RNA splicing maps. Trends Genet. 2011;27(3):89–97. doi: 10.1016/j.tig.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Brannan KW, Jin W, Huelga SC, Banks CAS, Gilmore JM, et al. SONAR discovers RNA-binding proteins from analysis of large-scale protein-protein interactomes. Mol Cell. 2016;64(2):282–93. doi: 10.1016/j.molcel.2016.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Thul PJ, Åkesson L, Wiking M, Mahdessian D, Geladaki A, et al. A subcellular map of the human proteome. Science. 2017;356(6340):eaal3321. doi: 10.1126/science.aal3321. [DOI] [PubMed] [Google Scholar]
- 93.Nussbacher JK, Batra R, Lagier-Tourenne C, Yeo GW. RNA-binding proteins in neurodegeneration: Seq and you shall receive. Trends Neurosci. 2015;38(4):226–36. doi: 10.1016/j.tins.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Pereira B, Billaud M, Almeida R. RNA-binding proteins in cancer: old players and new actors. Trends Cancer Res. 2017;3(7):506–28. doi: 10.1016/j.trecan.2017.05.003. [DOI] [PubMed] [Google Scholar]
- 95.Cartegni L, Krainer AR. Disruption of an SF2/ASF-dependent exonic splicing enhancer in SMN2 causes spinal muscular atrophy in the absence of SMN1. Nat Genet. 2002;30(4):377–84. doi: 10.1038/ng854. [DOI] [PubMed] [Google Scholar]
- 96.Mercuri E, Finkel R, Kirschner J, Chiriboga CA, Kuntz N, et al. Interim analysis of the phase 3 CHERISH study evaluating nusinersen in patients with later-onset spinal muscular atrophy (SMA): primary and descriptive secondary endpoints. Eur J Paediatr Neurol. 2017;21(Suppl 1):e15. [Google Scholar]
- 97.Rot G, Wang Z, Huppertz I, Modic M, Lencč T, et al. High-resolution RNA maps suggest common principles of splicing and polyadenylation regulation by TDP-43. Cell Rep. 2017;19(5):1056–67. doi: 10.1016/j.celrep.2017.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Park JW, Jung S, Rouchka EC, Tseng Y-T, Xing Y. rMAPS: RNA map analysis and plotting server for alternative exon regulation. Nucleic Acids Res. 2016;44(W1):W333–38. doi: 10.1093/nar/gkw410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Cereda M, Pozzoli U, Rot G, Juvan P, Schweitzer A, et al. RNAmotifs: prediction of multivalent RNA motifs that control alternative splicing. Genome Biol. 2014;15(1):R20. doi: 10.1186/gb-2014-15-1-r20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Xue Y, Zhou Y, Wu T, Zhu T, Ji X, et al. Genome-wide analysis of PTB-RNA interactions reveals a strategy used by the general splicing repressor to modulate exon inclusion or skipping. Mol Cell. 2009;36(6):996–1006. doi: 10.1016/j.molcel.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Shen S, Park JW, Lu Z-X, Lin L, Henry MD, et al. rMATS: robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. PNAS. 2014;111(51):E5593–601. doi: 10.1073/pnas.1419161111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.