Abstract
DNA topoisomerases, capable of manipulating DNA topology, are ubiquitous and indispensable for cellular survival due to the numerous roles they play during DNA metabolism. As we review here, current structural approaches have revealed unprecedented insights into the complex DNA-topoisomerase interaction and strand passage mechanism, helping to advance our understanding of their activities in vivo. This has been complemented by single-molecule techniques, which have facilitated the detailed dissection of the various topoisomerase reactions. Recent work has also revealed the importance of topoisomerase interactions with accessory proteins and other DNA-associated proteins, supporting the idea that they often function as part of multienzyme assemblies in vivo. In addition, novel topoisomerases have been identified and explored, such as topo VIII and Mini-A. These new findings are advancing our understanding of DNA-related processes and the vital functions topos fulfil, demonstrating their indispensability in virtually every aspect of DNA metabolism.
Keywords: antibiotics, anti-cancer drugs, DNA gyrase, DNA supercoiling, DNA topoisomerase
Introduction
DNA structure and topology have profound consequences for metabolism
The DNA duplex is one of life’s fundamental molecules; therefore, maintaining its integrity is paramount. Potential topological issues associated with the double-helical structure were recognised soon after its structure was first elucidated in 1953 by James Watson, Francis Crick and Rosalind Franklin.[1,2] The consequences of topological perturbations in DNA are exemplified by DNA replication during which the strands of the duplex are separated. This separation leads to the formation of positive supercoils (DNA overwinding) ahead of the replication fork and intertwining of the daughter strands, forming precatenanes, behind (Figure 1A).[3,4] If the positive supercoils are not relaxed, progression of the replication fork is impeded, whereas failure to unlink the daughter strands prevents genome segregation, which is required for cell division.[5] Transcription also generates positive supercoiling ahead of, and negative supercoiling behind, the transcriptional complex, known as the twin-supercoiled domain model, first described in 1987 (Figure 1A).[6] These topological perturbations must be resolved for DNA metabolism to proceed, allowing the cell to efficiently replicate, transcribe and partition the genome to enable cellular division and vitality. However, in addition to the detrimental aspects of DNA topology that require resolution, beneficial aspects are harnessed by the cell to facilitate DNA melting and establish global genome architecture. For example, plasmid replication requires negative super-coiling of the origin, which facilitates local melting and exposes singlestranded DNA required for protein binding.[7] Furthermore, compaction of the E. coli genome is achieved in part by significant negative super-coiling.[8] The essential proteins responsible for performing these vital roles in controlling DNA topology are called the DNA topoisomerases (topos).
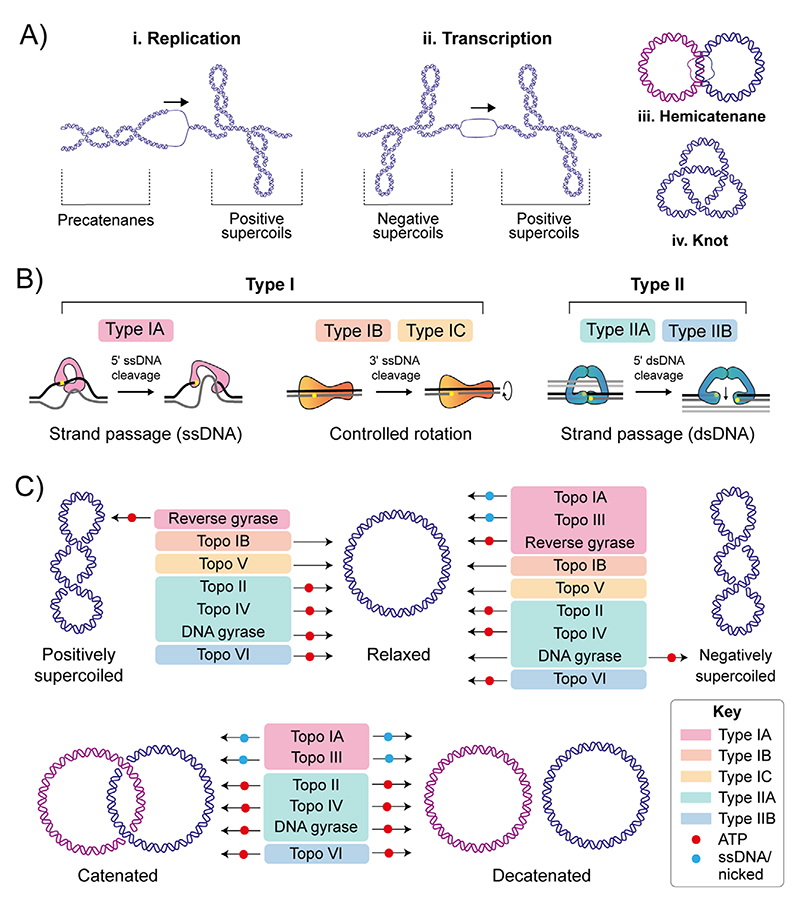
Figure 1. DNA topology and DNA topoisomerase mechanisms.
(A) Topological consequences of DNA metabolism. i) During DNA replication, strand separation leads to positive supercoiling ahead of the advancing protein machinery, and precatenane formation behind. Precatenanes form as the newly-synthesised duplexes wrap around one and other, and, if not removed prior to complete of replication, catenated DNA molecules are formed. ii) During transcription, strand separation leads to positive supercoiling ahead of the advancing protein machinery, and negative supercoil formation behind. iii) Hemicatenanes are a possible end result of replication, in which the parental strands of the replicated duplexes remain base-paired. iv: DNA knotting can also occur as a result of DNA replication in which a DNA molecule is intramolecularly linked. (B) Summary of topo categories and mechanism. The topos are categorised based on whether they catalyse single- (type I) or double-stranded (type II) DNA breaks. The type I topos are further subdivided to type IA, IB and IC. Type IA form a transient covalent bond to the 5′ DNA phosphate and function via a strand passage mechanism. Type IB and IC form a transient covalent bond to the 3′ DNA phosphate and function via a controlled-rotation mechanism. Type II topos are further subdivided into type IIA and IIB. Both form a transient covalent bond to the 5′ DNA phosphate of both strands of the duplex and function via a strand-passage mechanism. (C) Summary of the topological manipulations performed by DNA topoisomerases, namely relaxation of positive and negative supercoils and decatenation. Type IA topos are colour-coded pink, type IB are orange, type IC are yellow, type IIA are green, and type IIB are blue. Requirement of ATP or ssDNA for activity is denoted using a red or blue circle, respectively
Topos are structurally and mechanistically diverse
In general, all topos perform a similar task (i.e., interconverting the topological states of DNA), however, the precise ways in which this is achieved differs among enzyme classes (Figure 1B). A key feature linking all topos is the formation of a covalent DNA-topo intermediate in which the active site tyrosine of the topo forms a phosphotyrosyl linkage to the phosphate group in the DNA backbone via nucleophilic attack.[4] Topos are classified as type I or type II depending on whether they catalyse the formation and re-ligation of single-stranded (ss) or double-stranded (ds)DNA breaks, respectively.[4,9,10] The type I topos are further subcategorised as type IA, IB and IC. Type IA topos cleave the DNA backbone, generating a covalent linkage to the 5′-phosphate, in an Mg2+-dependent and ATP-independent manner (aside from reverse gyrase – see 2.1.3), and function via a strand passage mechanism.[4,9,10] The type IB and IC topos cleave the DNA backbone, generating a covalent linkage to the 3′-phosphate (albeit using distinct active sites), independently of both ATP and Mg2+, and function via a controlled rotation mechanism.[4,9,10] The type II topos are subcategorised as type IIA and IIB. Even though type IIA and IIB both catalyse dsDNA breaks through cleavage of the DNA backbone, generating a covalent linkage to the 5′-phosphate on both duplex strands, in an ATP/Mg2+ dependent manner, and function via a strand passage mech-anism (Figure 1B), they are structurally distinct.[4,9,10] In addition, type IIA topos cause dsDNA breaks with 4-base overhangs, while type IIB generate 2-base overhangs. The structural and mechanistic differences throughout the topo family impart certain activity preferences, for example: preferential decatenation rather than relaxation (Figure 1C).
These intriguing enzymes have been of keen scientific interest since the first was discovered in 1971.[11] As topos transiently disrupt the integrity of the DNA duplex in order to maintain it, they must function in a highly coordinated and precise manner to avoid generating permanent DNA breaks. Topos are important in human health and disease as this mechanism, along with their indispensability, makes them vulnerable to poisoning, which is exploited in the use and development of antimicrobial and anticancer therapeutics.[12,13] Explored below are the numerous ways in which topoisomerases employ their DNA cleavage/re-ligation mechanism in the preservation of genome integrity, with a focus on new results pertaining to structure, mechanism, and in vivo roles. Advancing our understanding of this crucial protein family has led to significant insights into numerous DNA processing pathways, with it becoming clear that the activity of topos pervades essentially every aspect of DNA metabolism.
Type Ia Dna Topoisomerases
Topo I relaxes transcription-induced negative supercoiling
Prokaryotic DNA topoisomerase I (topo IA), initially isolated from Escherichia coli, is a 97 kDa monomer that relaxes negative supercoils.[11] The main role in vivo for topo IA is thought to be preventing hyper-negative supercoil accumulation during transcription,[14] which can disrupt DNA metabolism and genome integrity by promoting stable R-loop formation (a DNA:RNA hybrid) due to the increased probability of forming ss-DNA regions.[15] Next-generation sequencing (NGS) techniques revealed that mycobacterial topo IA activity was highly correlated with RNA polymerase (RNAP) activity,[16,17] and E. coli topo IA has been demonstrated to physically interact with the β′ domain of RNAP via its CTD,[18] localising it to transcription sites. Recently, mycobacterial topo I has also been shown to alter RNA topology and modify ribosomal RNA precursors,[19] suggesting a potential RNA metabolism role in vivo. However, questions remain, including the detailed nature of the topo I and RNAP interaction in vivo, for example, does this only occur initially during topo I recruitment or do they remain bound throughout transcription, and what effect does this have on the processive removal of negative supercoils?
Structural characterisation of full-length E. coli topo IA complexed with ssDNA revealed 9 distinct domains in a toroidal arrangement (Figure 2A,B).[20] The N-terminal domains (NTDs) contribute to ssDNA binding and cleavage (domains 1, 3, 4), house the catalytic tyrosine (domain 3) and form a highly conserved hinge region with a flexible loop of charged residues, thought to play a role during strand passage (domain 2). The C-terminal domains (CTDs) include three 4-Cys zinc ribbon domains (domains 5–7) that interact with the DNA substrate, and two zinc ribbon-like domains (domains 8 and 9) that promote processivity by enhancing ssDNA binding.[21] Intriguingly, recent work on Helicobacter pylori topo IA, has suggested that the CTD alone can catalyse DNA relaxation. This raises the heretical possibility of catalysis without the canonical active-site tyrosine. Whether this occurs in other species and whether there is an independent topo IB-like activity in the CTD, remain to be determined. [22]
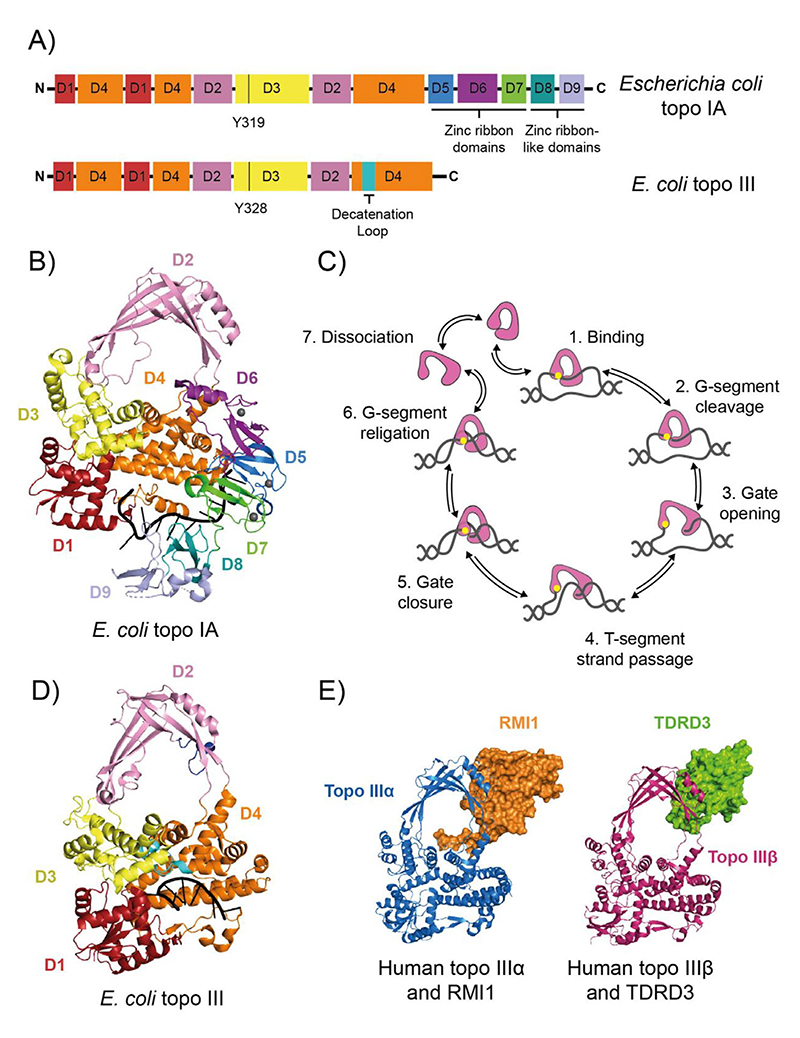
Figure 2. Type IA DNA topoisomerases.
(A) Protein domain organisation of Escherichia coli DNA topoisomerase IA (topo IA) and DNA topoisomerase III (topo III). Black vertical lines represent the active site tyrosines. (B) Crystal structure of E. coli topo I bound to ssDNA (PDB: 4RUL).[20] (C) Strand-passage mechanism for type IA topos. (1) topo binds G-segment ssDNA region, (2) the G-segment is cleaved. (3) The topo DNA-gate is opened, (4) which allows T-segment transfer through the cleaved G-strand. (5) The DNA gate is closed, (6) and the G-strand is re-ligated, changing the linking number by 1. (7) The topo can then go through another round of relaxation or dissociate from the DNA. Type IA topo (domains 1–4) is in pink, the active site tyrosine is yellow and the DNA is grey. (D) Crystal structure of E. coli topo III bound to ssDNA (PDB: 2O54).[26] (E) Crystal structures of human topo IIIα (blue) bound to RMI1(orange) (PDB: 4CGY),[39] and human topo IIIβ (magenta) bound to TDRD3 (green) (PDB: 5GVE).[60] For panels A, B and C, the topo I and III domains are colour coded as follows: D1 is red, D2 is pink, D3 is yellow, D4 is orange, D5 is marine blue, D6 is purple, D7 is green, D8 is teal, and D9 is light blue
Based on the topo IA structure, a mechanistic model was proposed.[20] A single DNA strand of the underwound duplex, known as the G (gate)-segment, is bound and cleaved at the 5' phosphate by the NTD, while the other strand, the T (transported)-segment, is bound by the CTD. The T-segment is then passed through the cleaved G-segment in a process termed strand passage, followed by G-segment re-ligation (Figure 2C). For strand passage to occur, topo IA must undergo a conformational change to open the DNA-gate and allow T-segment transfer. E. coli topo IA gate opening was observed directly using single-molecule magnetic tweezers, demonstrating that the DNA gate opens by 6.6 ± 1.0 nm and rapidly oscillates between open and closed conformations.[23] This is hypothesised to reflect its in vivo role in the efficient and processive removal of negative supercoils. Recent structural characterisation of Mycobacterium smegmatis topo I has shown that the CTD can bind ssDNA with higher affinity than the NTD, suggesting it may bind the T-segment before the G-segment.[24] This mechanism, coupled to the physical interaction with RNAP, makes topo I highly efficient in the relaxation of transcription-induced negative supercoiling, protecting genome integrity.
DNA topoisomerase III
Topo III resolves interlinked replication intermediates
DNA topoisomerase III (topo III) is highly conserved across prokaryotes and eukaryotes.[25] It closely resembles topo IA domains 1–4,[26] but with two additional loops, amino acids 502–519 and 241–255 (E. coli numbering), the former important for decatenation activity, possibly through interaction with duplex DNA (Figure 2D).[27] The mechanism is considered similar to topo IA (Figure 2C), but involves the intramolecular passage of a duplex T-segment, rather than a single DNA strand.[25] Topo III is hypothesised to function primarily in decatenation pathways in vivo, efficiently resolving precatenanes in vitro,[28] and recently shown to act at the E. coli replication fork in vivo, with topo III knock-outs markedly deficient in chromosome segregation.[29] This work also demonstrated that topo III interacts with the DnaX complex of the DNA III polymerase holoenzyme, and in vitro, topo III precatenane resolution was significantly stimulated by the DnaX complex. This interaction likely localises topo III to precatenanes in vivo, and is analogous to the topo I-RNAP interaction during transcription. E. coli topo III also maintained an open DNA-gate for longer than topo I, potentially reflecting the role of topo III in the intermolecular passage of a duplex during decatenation.[23] E. coli topo III is also known to cooperate with RecQ helicase and single-stranded DNA-binding protein in the resolution of stalled converging replication forks.[30] In addition, E. coli topo III can perform strand passage on RNA, suggesting potential roles in resolving RNA topology.[31,32]
Topo IIIα is crucial member of DNA-repair complexes
In metazoal and some fungal species, topo III exists as two isoforms, topo IIIα and IIIβ, which have been shown to play distinct roles in cellular development.[25] Murine topo IIIα knockouts are embryo-lethal, demonstrating a fundamental role in preserving cellular viability.[33] Human topo IIIα associates with BLM, a DNA repair-associated RecQ helicase,[34] and RMI1 (or BLAP75), an oligonu-cleotide/oligosaccharide binding (OB) protein, forming a complex known as the dissolvasome.[35] In human cells, RMI1 also interacts with RMI2 (or BLAP18), to form the RMI subcomplex, and RMI2 expression is interdependent on both RMI1 and topo IIIα expression.[36] The dis-solvasome is integral to the non-crossover resolution of double Holliday junctions (dHJ),[37] which are intermediates of the homologous recombination DNA-repair pathway.[38] The structural characterisation of the RMI1/topo IIIα interaction revealed a 23-residue loop from RMI1 inserted into the topo IIIα cavity (Figure 2E), stabilising topo IIIα DNA-gate opening and promoting dHJ dissolution.[39] The dissolva-some also interacts with FANCM (Fanconi anaemia group M protein), a DNA-repair protein that prevents the collapse of stalled replication forks.[40] This supresses the ALT (alternative lengthening of telomeres) pathway,[41] which is associated with DNA damage.[42,43] Human topo IIIα cellular levels inversely correlated with tumour growth rate, with topo IIIα demonstrated to physically interact with the tumour suppressor protein, p53, and stimulate expression by binding the p53 promoter.[44] In addition to modulating DNA-repair pathways, human topo IIIα was shown to interact with PICH DNA translocase (PIk1-interacting checkpoint helicase), generating positive supercoils via PICH-dependent loop extrusion of hypernegative supercoils that were relaxed by topo IIIα, potentially aiding in efficient centromere resolution.[45]
Topo IIIα is also localised to the mitochondria, and mutation of the mitochondrial import sequence in Drosophila melanogaster caused premature aging, mobility defects, and impaired fertility, caused by mitochondrial degeneration due to degradation of mitochondrial DNA (mtDNA).[46,47] A Met100Val mutation in human topo IIIα was identified in a patient with a mitochondrial disorder and was demonstrated to prevent resolution of mtDNA replication-specific hemicatenanes (Figure 1A).[48]
Topo IIIβ is involved in RNA metabolism
In contrast to topo IIIα, topo IIIβ knockout mice survive, albeit with a reduced life span, development of autoimmune reactions, aneuploidy and infertility caused by accumulating chromosomal mutations.[49–51] Recently in human cancer cells, the complete loss of topo IIIβ caused genome instability due to increased R-loop formation, functionally link-ing topo IIIβ to cancer suppression.[52] Topo IIIβ has also been shown to interact with RNA binding proteins (RBPs), TDRD3 (Tudor domain-containing 3) and FMRP (an RBP silenced in patients with Fragile X syndrome), localising topo IIIβ to polyribosome-bound mRNA.[53–56] Disruption of this interaction causes neurodevelopmental defects, suggesting that topo IIIβ plays a distinct role during translation of specific mRNAs. Deletion or mutation of the topo IIIβ gene is linked to schizophrenia and autism.[53,57,58] In Drosophila, neuronal synapse formation was disrupted when an autism patient-derived mutation in topo IIIβ was introduced.[54] In addition, a disease-linked mutation in FMRP (I304N) from a Fragile X syndrome patient was shown to disrupt the interaction between FMRP and TDRD3/topo IIIβ.[55,59] The structure of the topo IIIβ/TDRD3 complex reveals a largely hydrophobic inter-action between domain II of topo IIIβ and the OB-fold of TDRD3 (Figure 2E), reminiscent of the topo IIIα/RMI1 interaction.[39,60] However residues Arg96, Val109 and Phe139, along with the shorter TDRD3 insertion loop, were identified as crucial to the specific TDRD3/topo IIIβ interaction.[60] These recent results suggest that the main role of topo IIIβ may be as an RNA topoisomerase, particularly crucial during neurodevelopment, and this has become an active and exciting area of research.
Reverse gyrase positively supercoils DNA in thermophiles
Reverse gyrase, first discovered in Sulfolobus acidocaldarius, is a distinctive type IA topo that utilises ATP to introduce positive supercoils.[61,62] It is found in thermophilic and hyperthermophillic archaea and eubacteria, thought to be important in preventing ther-mal DNA denaturation and aiding DNA-repair processes.[63,64] Using magnetic tweezers, it was shown that Sulfolobus tokodaii reverse gyrase processively generates five positive supercoils s–1 on average,[65] with loose coupling to ATP hydrolysis (20 s–1), consistent with previous measurements.[66] Positive supercoiling by reverse gyrase is the combined activity of two distinct protein domains: the superfamily 2 helicase-like NTD and the topo IA-like CTD (Figure 3A,B).[67]
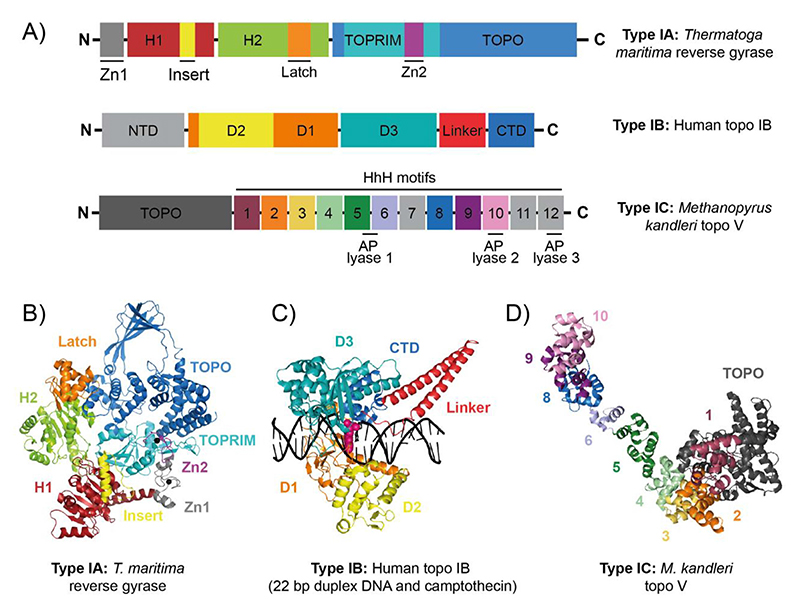
Figure 3. Reverse gyrase (type IA), topo IB (type IB) and topo V (type IC).
(A) Protein domain organisation of Thermatoga maritima reverse gyrase, human DNA topoisomerase IB (topo IB), and Methanopyrus kandleri DNA topoisomerase V (topo V). (B) Crystal structure of T. maritima reverse gyrase (PDB: 4DDU).[71] (C) Crystal structure of human topo IB in a cleavage complex with a 22 bp duplex DNA and camptothecin (PDB: 1T8I).[220] (D) Crystal structure of M. kandleri topo V (PDB: 5HM5).[89]
In the presence of ATP, the helicase domain of Thermatoga maritima reverse gyrase has high affinity for DNA and transiently destabilises the DNA duplex.[68,69] In the absence of the helicase-like domain, the topo I-like domain has nucleotide-independent DNA relaxation activity,[70] therefore the combination of DNA unwinding with strand passage permits positive supercoiling by reverse gyrase. The helicase and topo domains are coordinated via the latch domain, with bioinfor-matic analyses revealing significant latch sequence diversity amongst different reverse gyrases.[71] In the absence of the T. maritima latch domain, reverse gyrase is unable to positively supercoil due to preventing DNA unwinding,[69] although recently, a β-hairpin of the latch domain was demonstrated sufficient to maintain positive supercoiling activity.[72]
In addition to the reverse gyrase diversity among species, Sulfolobus solfataricus encodes two distinct copies of reverse gyrase, RG1 and RG2, which are alternatively regulated in vivo with separate biochemical activities.[73–75] RG1 expression is sensitive to thermal stress, relaxing negatively-supercoiled DNA independently of ATP hydrolysis and distributively generating moderately-overwound DNA; whereas RG2 expression is constitutive and it processively generates highly-overwound DNA with a strict dependence on ATP hydrolysis. Magnetic tweezers assays demonstrated that DNA unwinding by RG2 was nucleotide-independent, while ATP hydrolysis was strictly required for strand passage.[76] The physiological reasons for this variability in how reverse gyrases positively supercoil DNA remains to be determined.
Type Ib Dna Topoisomerases
Topo IB relieves torsional strain during transcription
The type IB topos, for example, eukaryotic DNA topoisomerase I (topo IB), were first discovered in 1972 and relax positive and negative supercoils via transient ssDNA cleavage of the DNA backbone, generating a covalent linkage to the 3’-phosphate.[77,78] An N-terminally truncated 70-kDa human topo IB crystallised in complex with a 22-bp DNA duplex (Figure 3C) revealed how topo IB binds DNA and led to the proposal that it functions via a ‘controlled-rotation’ mechanism, first described for Vaccinia topo I.[79,80] This involves topo IB creating a ssDNA nick, which permits DNA rotation of the free end around the intact strand, the speed controlled by friction within the enzyme cavity, before the nick is re-ligated.[81] In vivo, topo IB is thought to relieve torsional strain in DNA, particularly during transcription.[82] In line with this, the CTD of RNA polymerase II (RNAP II) is a potent activator of topo IB in vitro, and they have been shown to physically interact.[83,84] The binding of topo IB strongly correlates with RNAP II binding in vivo at transcription start sites, and topo IB catalytic activity is observed in gene bodies, positively correlated with the level of gene expression.[83] However, topo IB activity is also linked to transcription-associated mutations, characterised by 2–5 bp deletions in tandem repeats, particularly after topo IB cleavage at incorporated ribonucleotides.[85] In addition, topo IB activity has been associated with numerous human diseases, including several spinocerebellar ataxia disorders and the autoimmune condition scleroderma.[86] Inhibition of topo IB is used to suppress tumorigenesis and has also been shown to alleviate symptoms of Angelman syndrome (an autism spectrum disorder), potentially through preventing transcription of UBE3A-ATS, the RNA transcript of which causes pathogenesis.[86]
Type Ic Dna Topoisomerases
Topo V exhibits DNA relaxation and DNA repair activities
DNA topoisomerase V (topo V) was first isolated from the hyperther-mophillic methanogen, Methanopyrus kandleri and is the sole type IC member.[87–89] Like type IB topos, topo V relaxes positive and negative supercoils without ATP and Mg2+, forming a covalent intermediate with the 3'-phosphate of the DNA, and functioning via a controlled-rotation mechanism. However, topo V was classed type IC as it was demonstrated to contain unique protein folds and an atypical active site, indicating an alternative cleavage/re-ligation mechanism.[88] Topo V also exhibits DNA repair activity in vitro as an AP(apurinic or apyrimidinic)-lyase, potentially repairing abasic DNA damage.[90,91] This DNA-repair activity functions independently of the topo activity as mutation of the active site tyrosine did not affect DNA-repair.[90] Resolution of the 97-kDa topo V structure revealed a total of four active sites contained within a single polypeptide; one topo site and three AP lyase sites (Figure 3A).[89] Topo V has a globular N-terminal topo-like domain, followed by 12 helix-hairpin-helix, motif 2 ((HhH)2) domains, which harbour the AP-lyase sites (Figure 3D). While fascinating, topo V only appears in M. kandleri, living within hydrothermal vents of the deep ocean, and has therefore been postulated to have a viral origin as it is unlikely that it arose de novo in the ancestral lineage of M. kandleri.[92]
Type Iia Dna Topoisomerases
The type IIA topos include prokaryotic DNA gyrase (gyrase) and topoisomerase IV (topo IV), and eukaryotic topoisomerase II (topo II) (Figure 4A).[10] The general mechanism for the type II topos begins with the binding of one DNA duplex, termed the gate segment (G-segment), at the DNA gate. Another duplex, termed the transport segment (T-segment), is captured by the ATP-operated clamp (N-gate) and passed through a transient break in the G-segment before it is released through the C-gate and the G-segment is re-ligated. The N-gate then reopens resetting the enzyme for another round of strand passage or release from the DNA (Figure 4B). The specifics of this reaction vary amongst type II topos, each being both intrinsically (e.g., structure) and extrinsically (e.g., protein-protein interactions and temporal/spatial regulation) adapted to preferentially perform different DNA topology manipulations. The formation and resealing of the DSB is highly efficient to prevent extensive genotoxic damage,[93] but this also constitutes a juncture of vulnerability that is exploited by antimicrobial/anticancer drugs.[12,94]
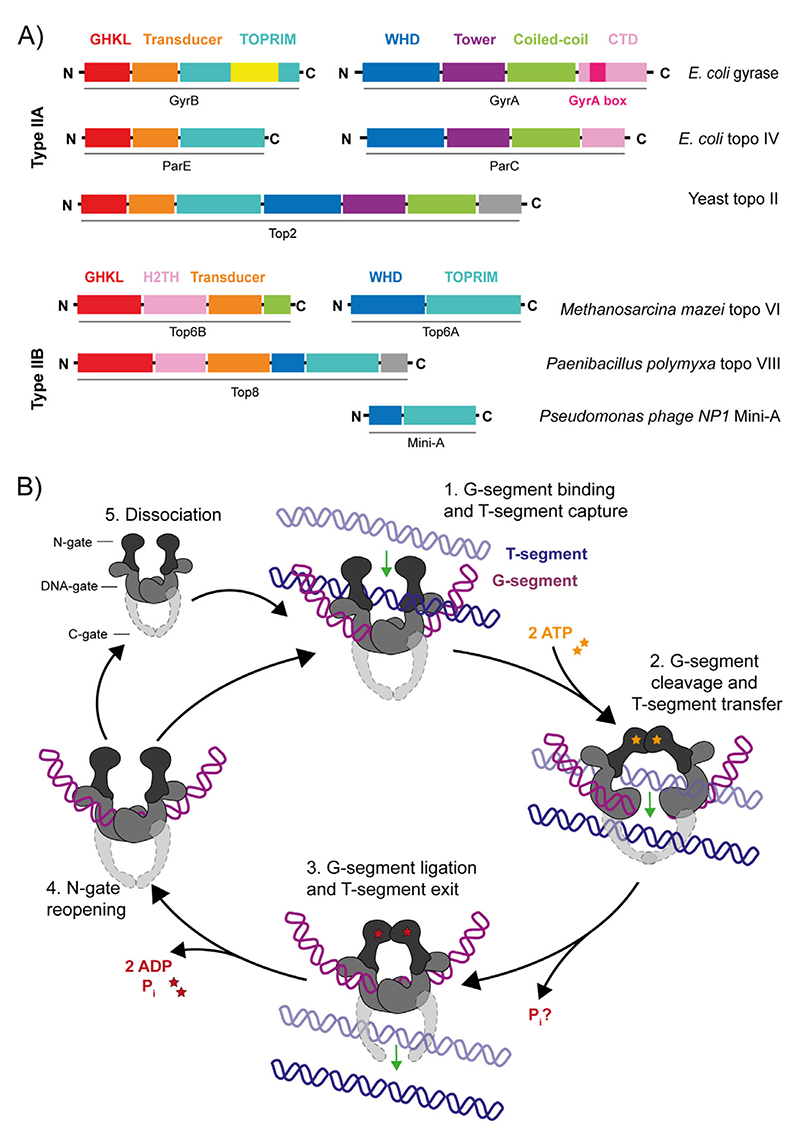
Figure 4. Type II DNA topoisomerases: domain organisation and mechanism.
(A) Protein domain organisation for the type IIA topos: E. coli DNA gyrase, E. coli DNA topoisomerase IV (topo IV), yeast DNA topoisomerase II (topo II), Methanosarcina mazei DNA topoisomerase VI (topo VI), Paenibacillus polymyxa DNA topoisomerase VIII (plasmid-borne), and Pseudomonas phage NP1 Mini-A. (B) type II topo strand passage mechanism. (1) G-segment is bound at the DNA-gate and the T-segment is captured. (2) ATP binding stimulates dimerisation of the N-gate, the G-segment is cleaved and the T-segment is passed through the break. (3) The G-segment is re-ligated and T-segment exits through the C-gate. For type IIB topos, there is no C-gate so once the T-segment passes through the G-segment, it is released from the enzyme. (4) Dissociation of ADP and Pi allows N-gate opening, a scenario where the enzyme either remains bound to the G-segment, ready to capture a consecutive T-segment, or (5) dissociates from the G-segment.
The key protein domains shared amongst the type IIA topos and present in pairs within the holoenzyme, are the WHD (winged-helix domain, or 5Y-CAP), the TOPRIM (topoisomerase/primase) domain and the GHKL (DNA Gyrase, Hsp90, bacterial CheA-family histidine kinases and MutL) ATPase domain.[95] The WHD contains a helix-turn-helix fold, commonly found in DNA-binding proteins, including the E. coli catabolite activator protein (CAP),[96] and houses both the catalytic tyrosine residue, which forms a reversible covalent bond with the 5'-scissile DNA phosphate,[97] and an isoleucine, which intercalates into the G-segment producing a ~150° bend,[98] promoting DNA cleavage.[99]
The TOPRIM domain chelates Mg2+ via the DxD motif, and contains a glutamate residue thought to act as a general acid during cleavage, donating a proton to the sugar hydroxyl, and a general base during religation, abstracting the proton from the 3′-OH.[100,101] Together, the TOPRIM DxD motif and the active site tyrosine of the WHD, form a bipartite active site capable of cleaving the DNA backbone.[95,102] The TOPRIM domain also contains conserved residues, namely the EGDS and PLRGK motifs, which interact with the G-segment and assist with DNA binding.[102,103] The type IIA-specific tower domain also interacts with the G-segment as it exits the WHD, anchoring the outer portion of the bent duplex and promoting both DNA binding and cleavage efficiency.[98]
Recently, the way in which type II topos utilise divalent metal ions during DNA cleavage has been questioned.[12] Early work supported the idea that each TOPRIM domain coordinated two Mg2+ ions in two pockets denoted sites “A” and “B,” with site A-bound Mg2+ participating in cleavage, and site B-bound Mg2+ anchoring the adjacent phosphate of DNA.[104,105] However, a moving metal ion mechanism is now gaining support; that is, following cleavage, the metal bound at site A moves to site B (associated with protein and DNA conformational changes) where it cannot participate in cleavage/re-ligation chemistry and protects the tyrosyl-phosphate linkage during strand passage.[12,106] However, confirmation of either model requires further structural and biochemical characterisation.
The GHKL ATPase domain binds ATP.[95,107] The precise role(s) of ATP in type II topo activity is still unclear; however, it is hypothesised that free energy of ATP enables the formation of a stable protein-protein interface, protecting against the formation of genotoxic DSBs when the DNA gate is opened during strand passage.[93] Explored below are the alternative ways in which the type IIA topos employ these protein domains to perform distinct roles in vivo.
DNA gyrase negatively supercoils DNA
Gyrase, discovered in 1976, is a unique type IIA topo found predominantly in bacteria, but also present in plants, apicomplexans and archaea.[108–112] Gyrase can introduce negative supercoils, relax positive supercoils and decatenate DNA in an Mg2+/ATP-dependent manner, and relax negative supercoils independently of nucleotide.[10] E. coli gyrase is a 374-kDa heterotetramer formed from two GyrA (97 kDa) and two GyrB (90 kDa) subunits.[113] As gyrase is essential for bacterial viability, and absent in humans, it has had significant and ongoing clinical success as an antibacterial target.[114,115]
It is thought that the fundamental role of gyrase in vivo is the introduction of negative supercoiling. Indeed, if gyrase is inhibited, the genome becomes relaxed, indicating that gyrase plays a role in the homeostatic maintenance of a negatively-supercoiled genome.[116–119] Negative supercoiling is important for the initiation of DNA replication and transcription as underwinding the DNA promotes melting of the origin and gene promoters.[120,121] In addition to this role, gyrase is also considered vital during the elongation phase of replication and transcription, relaxing positive supercoils ahead of the advancing protein machinery. This is supported by gyrase loss-of-function mutations causing a significant decline in replication and transcription.[122] An in vitro DNA replication system demon-strated that gyrase preferentially removed positive supercoils ahead of the fork.[123] Using NGS, the binding of mycobacterial gyrase was found to be enriched in areas of high transcriptional activity, directly correlated with the binding of RNA polymerase, and at the replication origin.[16] Another NGS-based study on E. coli gyrase also found increased activity downstream of highly transcribed operons.[124] Recent in vivo single-molecule imaging data suggest multiple gyrases (~12) cluster ahead of the DNA replication fork.[125] This is supported by magnetic tweezers data demonstrating that multiple gyrases were recruited to highly overwound DNA.[126]
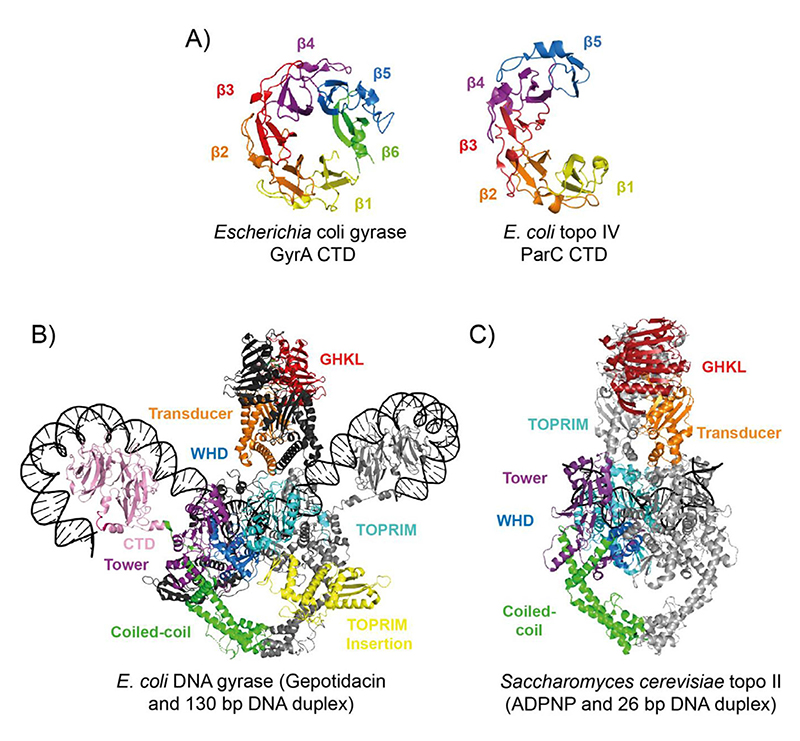
The unique negative supercoiling activity of gyrase arises from its capacity to wrap DNA via the CTDs of GyrA. The 35-kDa GyrA CTD has six β-strands in a β-pinwheel fold with a largely basic outer surface, indicating a role in DNA binding/bending, and the 7-residue GyrA-box (QRRGGKG), situated within a loop between β-strands 1 and 6, which is crucial for supercoiling (Figure 5A).[127–132] Supercoiling is thought to begin with G-segment binding to gyrase and chirally wrapped around one of the GyrA CTDs, before being presented over the G-segment at ~60° as the T-segment, forming a left-handed (positive) crossing.[133] Passage of the T-segment through the G-segment converts the crossing to a negative supercoil. Approximately ~130 bp of DNA is bound and wrapped by gyrase as measured by a variety of methods.[134] Recent work using Bacillus subtilis gyrase with a single catalytic tyrosine suggested supercoiling activity could instead function via a nicking-closing mechanism, however this remains to be substantiated by other methods.[135] Gyrase-DNA wrapping was recently demonstrated structurally using Cryogenic electron microscopy (cryo-EM), with a low-resolution (23 Å) structure of Thermus thermophilus gyrase in a cleavage complex with a 155 bp DNA duplex and ciprofloxacin, revealing asymmetric wrapping of DNA around the GyrA CTDs.[136] In 2019, the first full-length cryo-EM structure of E. coli gyrase in complex with DNA and gepotidacin was solved, with DNA-binding domain resolution approaching 3.0 Å (Figure 5B). [137] This landmark structure provided an in-depth view of the overall architecture of DNA gyrase, revealing the spatial organisation of the domains, the position of the GyrA-box, and insight into DNA-cleavage site conformational changes, particularly in regard to the position of the TOPRIM insertion domain.
Figure 5. Type IIA DNA topoisomerase structures.
(A) The E. coli gyrase GyrA CTD (PDB: 1ZI0)[129] and the E. coli topo IV ParC CTD (PDB: 1ZVT).[146] (B) CryoEM structure of full length E. coli gyrase complexed with a 130-bp DNA duplex and gepotidacin (PDB: 6RKW).[137] Colour coding for domains is as labelled in the figure with the second GyrA and GyrB coloured light grey and dark grey, respectively, and the DNA in black. (C) Crystal structure of Saccharomyces cerevisiae topo II with a 26 bp DNA duplex and ADPNP (PDB: 4GFH).[162] Colour coding of domains is as shown in the figure with second Top2 subunit coloured grey and the DNA in black
Topo IV is critical for chromosome segregation in bacteria
Topo IV, discovered in E. coli in 1990, is a ~308 kDa heterotetramer composed of two ParC (~84 kDa) and two ParE (~70 kDa) subunits, which can relax positive or negative supercoils and decatenate, in an ATP/Mg2+-dependent manner (Figure 4A).[138] The topo IV genes were discovered through DNA partitioning defects, which suggested that topo IV was involved in decatenation and chromosome segregation.[139] This was supported by an in vitro replication system that demonstrated topo IV was highly efficient at unlinking replicated daughter chromosomes.[140] Furthermore, using NGS, the binding/cleavage of topo IV was specifically enriched at the dif site, where E. coli chromosomes are unlinked.[141] This work also demonstrated a physical interaction between XerCD recombinases, modulated by MatP, indicating that topo VI is part of a multi-protein system required for efficient chromosome segregation.[141] MatP also regulates the physical interaction between topo IV and the E. coli SMC (structural maintenance of chromosomes) complex, MukBEF [142–145], at the origin of replication, enhancing topo IV decatenation.
In addition to protein-protein recruitment, the structure of topo IV also supports preferential decatenation activity. The topo IV ParC CTD has only five β-strands and no GyrA-box so does not permit supercoiling (Figure 5A). However, the outer surface is positively charged, suggesting a role in DNA binding and therefore potentially mediating topo IV substrate specificity.[146] Indeed, deletion of the ParC CTD results in significant reductions in relaxation and decatenation rates in vitro, which is far more profound for the relaxation of positive supercoils and decatenation, than negative supercoil relaxation.[146] Using single-molecule approaches, topo IV relaxed positive supercoils ~20-25-fold faster than negative, which has been attributed to the CTD of ParC stimulating high processivity during positive writhe relaxation, and suggests the topo IV CTDs recognise DNA geometries more common in positive supercoils and catenanes.[146–149] There is evidence that topo IV can complement the activity of type IA topos, and support replication and transcription fork progression in vivo in cells encoding temperature-sensitive mutants of gyrase.[150–153] However, topo IV seems to be a preferential decatenase in vivo through the combination of protein-protein recruitment, temporal regulation, and a structural preference for catenane geometries.[141–146,154]
Yeast topo II is involved in DNA segregation, replication and transcription
Topo II is the major and essential type IIA topo found in eukaryotes. Saccharomyces cerevisiae (yeast) topo II is a homodimer (Figure 5A) that relaxes positive and negative supercoils, decatenates and unknots DNA, in an ATP- and Mg2+ -dependent manner.[155] In vivo, the absence of yeast topo II prevented the completion of mitosis, suggesting a fundamental role resolving interlinked or knotted DNA, an activity which is promoted by the presence of condensin, an SMC complex.[156–158] Yeast topo II also plays a role in the relief of torsional strain during DNA replication, with a preference for the relaxation of positive supercoils ahead of the fork.[159,160] In addition, yeast topo II supports transcription of long genes (>3 kb), and its absence stalls fork progression, which cannot be rescued by topo I.[161]
The structure of yeast topo II (residues 408–1177) bound to a 30 bp G-segment, showed the DNA to be both A-form and bent to a ~150° angle, deformations thought to be important for DNA cleavage, correctly positioning the DNA backbone within the active site.[98] In addition, a minimally-truncated, fully-functional yeast topo II structure (residues 1–1177) bound to DNA and ADPNP (Figure 5C), revealed that a loop of the transducer domain, named the K-loop, interacted with the G-segment.[162] Mutagenesis within the K-loop didn’t affect DNA cleavage but caused a severe reduction in relaxation and decatenation activity for both yeast topo II and human topo IIα, implicating the K-loop in strand passage.[162]
Topo IIα is essential for DNA replication and segregation
In vertebrates, topo II exists as two isoforms, topo IIα and IIβ.[163] Topo IIα is critical for cellular viability, and has essential roles during DNA replication and mitosis, and a cell-cycle regulated expression pattern.[163–165] It is well-established that topo IIα is important for chromosome condensation,[165–169] however, it has recently been shown to also be important for maintenance of chromosome structure, despite previous results suggesting the contrary.[170,171] Chromatin compaction seems to arise, in part, from the interplay between topo IIα and the SMC complexes, such as condensin.[168] Topo IIα is also integral to chromosome segregation, removing catenanes along the chromosome arms prior to the onset of metaphase,[172,173] as well as at the centromere once cohesin has been removed by separase at the onset of anaphase.[174] As the chromatids are pulled apart, interlinked DNA at the centromere forms ultra-fine anaphase bridges (UFBs) that are bound by PICH, stimulating the decatenation activity of topo IIα.[175] Topo IIα is also involved in chromatid resolution at ribosomal DNA (rDNA) regions during anaphase alongside PICH, tankyrase and condensin II.[175,176] In addition to the complex protein-protein interaction profile for topo IIα, the CTD plays crucial roles in vivo, bearing a nuclear localisation signal,[177] the chromatin tether domain (crucial for activity during mitosis),[178] as well as sumoylation, acetylation, phosphorylation, and ubiquitination sites that regulate the activity of the enzyme in a cell-cycle-dependent manner.[179,180]
Topo IIβ has crucial roles in neurodevelopment
Topo IIα and IIβ have distinct roles in vivo, thought to be a consequence of the divergent CTDs imparting differential regulation and activity.[181] Whereas murine topo IIα knock-outs are embryonic lethal with expression restricted to proliferating cells, topo IIβ knock-outs die after birth due to respiratory failure and expression is detected in most adult tissues.[182–184] Numerous studies have since implicated topo IIβ activity in neuronal development and transcription.[181] Recently, activation of neuronal early-response gene expression, critical for external environment sensing, was linked to dsDNA break formation in the genes’ promoters, likely caused by topo IIβ.[185] In addition, two patients with autism spectrum disorder and profound neurodevelopmental delays were reported to have a de novo heterozygous His58Tyr topo IIβ mutation, strongly suggesting that impairment of topo IIβ activity has a severe effect on brain development.[186,187] In addition to its neurological role, topo IIβ has also been implicated in DNA repair, aging, HIV infection, and cancer,[188] and like topo IIα, the full extent of topo IIβ’s biological roles are beginning to be revealed.
In 2018, a 2.75-Å crystal structure of the open human topo IIβ DNA gate (residues 445–1201) revealed a fully opened G-segment, with no interactions between the separated DNA cleavage domains of the two subunits, and a channel large enough to allow passage of the T-segment. This has provided insight into the significant conformational changes type IIA topos undergo during strand passage.[189]
Type Iib Dna Topoisomerases
Topo VI is found in prokaryotes and eukaryotes
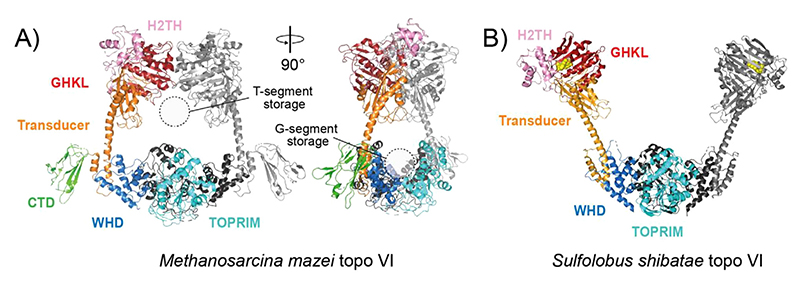
Topo VI, initially identified in the archaeal hyperthermophile Sulfolobus shibatae, has since been found throughout the archaea, a few bacterial species, and intriguingly, in eukaryotes such as plants and algae.[190,191] Topo VI is a heterotetramer formed from two Top6A (~45 kDa) and two Top6B (~60 kDa) subunits, that relaxes positive and negative supercoils, and decatenates DNA.[192] It is distinct from type IIA in terms of domain organisation, and having only two protein interfaces: the N-gate and the DNA-gate. Minimal homology to the type IIA topos is mainly found in the WHD, TOPRIM and GHKL domains (Figure 4A).[193,194] This simplicity in terms of structure has made topo VI of keen interest in the dissection of the type II mechanism, particularly the role of ATP in the opening/closing of the N-gate.[194]
All topo VI structural characterisation has been performed using the archaeal forms, and includes both Top6A and Top6B independently, as well as the full-length heterotetramer in a “closed” (Figure 6A) and “open” conformation (Figure 6B).[193–197] Together, these structures have revealed a clamp-like arrangement, with the Top6A dimer forming a positive electrostatic groove capable of accommodating the G-segment.[193] The Top6B dimer forms a cavity large enough for a DNA duplex, which has recently been shown biochemically to be crucial for T-segment sensing and tightly coupling Top6B ATPase activity to strand scission by Top6A.[198] Structural characterisation of archaeal Top6B in complex with ATP and hydrolysis-product analogues, allowed the ATP-mediated strand-passage mechanism to be modelled in more detail.[193,194,196] This revealed that the transducer domain alternates between a “restrained” and “relaxed” state mediated by the respective association and dissociation of a conserved lysine residue with the γ-phosphate of the bound nucleotide. This transducer domain movement is thought to be coupled to strand scission and DNA-gate opening. As Top6B and E. coli gyrase GyrB share highly conserved motifs, this mechanistic insight likely applies to type IIA topos.[194]
Figure 6. DNA topoisomerase VI (type IIB) structures.
(A) Crystal structure of Methanosarcina mazei topo VI (PDB: 2Q2E).[196] The domains are coloured as labelled in the figure on one TOP6A/Top6B heterodimer, with the second Top6A and Top6B coloured black and grey, respectively. (B) Crystal structure of Sulfolobus shibatae topo VI bound to radicicol (PDB: 2ZBK).[197] Colour coding is the same as in panel A except GHKL-bound radicicol is coloured yellow
Topo6A and Top6B homologues have been identified in plants such as Arabidopsis thaliana, named AtSPO11-3 and AtTOP6B, respectively.[199,200] Homozygous knockouts of AtSPO11-3 or AtTOP6B are associated with growth-stunted phenotypes, which were demonstrated to be caused by endoreduplication defects. Endoreduplication facilitates cellular enlargement through multiple rounds of genome replication in the absence of cellular division.[191,201,202] Why A. thaliana requires topo VI exclusively during endoreduplication remains unclear. Hypotheses include the resolution of endoreduplication-specific DNA structures, or endoreduplication-specific expression.[203]
Plant topo VI uniquely interacts with two accessory proteins named BIN4 (brassinosteroid-insensitive4) and RHL1 (roothairless-1), with mutants in either of these proteins also causing endoreduplication defects.[204,205] The basis of the interaction between BIN4, RHL1 and topo VI is still unknown. The in vivo roles for plant topo VI have recently been expanded, with the discovery of interactions with plant steroid hormone genes; a role in chromatin organisation and transcriptional silencing, via interaction with the MIDGET protein through RHL1; abscisic acid (ABA) resistance, high salt tolerance, dehydration resistance; and reactive oxygen-species response.[206–210] This demon-strates extensive roles for topo VI in mediating the plant’s response to endogenous and exogenous cues, integrating them through chromatin remodelling and transcriptional control.
Soon after the discovery of topo VI it was found that Top6A was highly homologous to the eukaryotic recombination factor, Spo11, which is responsible for the formation of dsDNA breaks during meiosis.[211,212] Recently, Top6B structural homologues have also been identified that interact with Spo11 in mouse and Arabidopsis thaliana, to form the Spo11 complex.[213,214] These important findings highlighted the evolutionary connection between topo VI and the meiotic machinery, and while Spo11 cannot reseal DNA cleavage, its structural similarity to topo VI suggests that the Spo11 complex may function in a similar manner.
There is uncertainty about whether a bone fide topo VI exists in plasmodia, as one study annotated Top6A and Top6B in the genome of Plasmodium falciparum [215], whereas, subsequent analysis concluded there was a Spo11, but no Top6A.[216] This was further confounded by the result that topo VI from P. falciparum complemented the function of topo II in S. cerevisiae when expressed transiently.[217] If topo VI is present in plasmodia, it is hypothesised to play a role in asexual reproduction and could have promise as a novel antimalarial drug target,[215] but more work is required.
Topo VIII is a novel type IIB topo
DNA topoisomerase VIII (topo VIII), discovered in 2014, is a novel member of the topo family. Identified through database screening using S. shibatae Top6B, topo VIII was classified as a highly divergent type IIB.[218] Currently, 77 topo VIII enzymes have been identified in nine bacterial phyla, four in archaea (euryarchaeota phylum), and one unclassified.[219] Topo VIII is distinct from topo VI as it is more common in bacteria than archaea, exhibits dramatic sequence divergence, is usually a homodimer, and encoding is dependent on plasmids and integrated elements.[219] In addition, distantly-related Mini-A proteins were also identified in archaeoviruses and bacteriophages as truncated homologues of Top6A (Figure 4A).[219]
Of the topo VIII enzymes currently characterised biochemically, two exhibited Mg2+-dependent relaxation of positive and negative supercoils (Microscilla marina and Paenibacillus polymyxa), however the M. marina topo VIII performed these reactions independent of ATP, a behaviour not typical of type IIB topos.[218] Topo VIII from Ammonifex degensii only demonstrated ATP-independent cleavage activity. In each case the activity was weak, moreover, many of the topo VIII enzymes appear to be in variable states of inactivation, so whether topo VIII plays considerable in vivo roles remains unclear.[219]
Conclusions and Future Directions
Since the remarkable discoveries of bacterial topo I and DNA gyrase, topos have taken centre-stage in a wide range of metabolic DNA processes beyond their crucial roles in replication and transcription.[11,108] It is becoming clear that topos play integral roles at multiple scales in the repair, segregation, and global organisation of nucleic acids. Continuing advances in cell biology and NGS approaches, that both provide spatial and temporal maps of protein activity, will undoubtedly expand and clarify our understanding of the central roles played by topos in many metabolic DNA processes. Complementing this, recent structural data has provided unprecedented insight into topo structure and function. These have included the full length DNA gyrase cryo-EM structure in complex with DNA, demonstrating the DNA wrapping behaviour; the open DNA gate of human topo II, informing further on the strand passage mechanism; and the M. smegmatis topo IA structure, indicating the T-strand may be bound prior to the G-strand.[24,136,137,189] The use of single-molecule technologies, such as magnetic tweezers, have revealed aspects of topo activity previously unseen, including the gate opening dynamics of E. coli topo I and III, and the detailed dissection of the S. solfataricus RG2 reaction mechanism.[23,76] It has also become clear that many topos form molecular complexes with other proteins that operate in a concerted way to maintain genome integrity, including the interactions of topo IIα and topo IIIα with PICH, and topo IIIβ with the RBPs, TDRD3 and FMRP.[45,53–56,175] Despite 50 years having passed since topos were first discovered, the true extent and intricacy of their activities are still being enthusiastically explored and expanded, further consolidating the significance of topos in cellular viability.
Acknowledgments
We thank Natassja Bush, Yeonee Seol and Rachel Kim for their editorial input. This work was funded by the Intramural Program of the National Heart, Lung, and Blood Institute of the National Institutes of Health (1ZIAHL001056-15; K.C.N. and S.J.M.), and through a Wellcome Trust and National Institutes of Health fellowship to S.J.M. AM’s lab is funded by an Investigator Award from the Wellcome Trust (110072/Z/15/Z), and bya BBSRC Institute Strategic Programme Grant (BB/P012523/1).
Funding information
Wellcome Trust, Grant/Award Number: 110072/Z/15/Z; Biotechnology and Biological Sciences Research Council, Grant/Award Number: BB/P012523/1
Footnotes
Conflict of Interest
None declared.
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
References
- 1.Watson JD, Crick FH. Genetical implications of the structure of deoxyribonucleic acid. Nature. 1953;171(4361):964–967. doi: 10.1038/171964b0. [DOI] [PubMed] [Google Scholar]
- 2.Watson JD, Crick FHC. A structure for deoxyribose nucleic acid. Nature. 1953;171:737–738. doi: 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]
- 3.Postow L, Crisona NJ, Peter BJ, Hardy CD, Cozzarelli NR. Topological challenges to DNA replication: Conformations at the fork. Proc Natl Acad Sci U S A. 2001;98(15):8219–8226. doi: 10.1073/pnas.11100699898/15/8219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bates AD, Maxwell A. DNA Topology. Oxford University Press; Oxford: 2005. [Google Scholar]
- 5.Racko D, Benedetti F, Goundaroulis D, Stasiak A. Chromatin loop extrusion and chromatin unknotting. Polymers (Basel) 2018;10(10) doi: 10.3390/polym10101126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu LF, Wang JC. Supercoiling of the DNA template during transcription. Proc Natl Acad Sci USA. 1987;84(20):7024–7027. doi: 10.1073/pnas.84.20.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schvartzman JB, Stasiak A. A topological view of the replicon. EMBO Rep. 2004;5(3):256–261. doi: 10.1038/sj.embor.7400101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Woldringh CL, Jensen PR, Westerhoff HV. Structure and partitioning of bacterial DNA: Determined by a balance of compaction and expansion forces? FEMS Microbiol Lett. 1995;131(3):235–242. doi: 10.1111/j.1574-6968.1995.tb07782.x. [DOI] [PubMed] [Google Scholar]
- 9.Champoux JJ. DNA topoisomerases: Structure, function, and mechanism. Annu Rev Biochem. 2001;70:369–413. doi: 10.1146/annurev.biochem.70.1.369. [DOI] [PubMed] [Google Scholar]
- 10.Bush NG, Evans-Roberts K, Maxwell A. DNA topoisomerases. EcoSal Plus. 2015;6(2) doi: 10.1128/ecosalplus.ESP-0010-2014. [DOI] [PubMed] [Google Scholar]
- 11.Wang JC. Interaction between DNA and an Escherichia coli protein w. J Mol Biol. 1971;55:523–533. doi: 10.1016/0022-2836(71)90334-2. [DOI] [PubMed] [Google Scholar]
- 12.Bax BD, Murshudov G, Maxwell A, Germe T. DNA topoisomerase inhibitors: Trapping a DNA-cleaving machine in motion. J Mol Biol. 2019;431(18):3427–3449. doi: 10.1016/j.jmb.2019.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pommier Y. Drugging topoisomerases: Lessons and challenges. ACS Chem Biol. 2013;8(1):82–95. doi: 10.1021/cb300648v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Masse E, Drolet M. Relaxation of transcription-induced negative supercoiling is an essential function of Escherichia coli DNA topoisomerase I. J Biol Chem. 1999;274(23):16654–16658. doi: 10.1074/jbc.274.23.16654. [DOI] [PubMed] [Google Scholar]
- 15.Drolet M, Broccoli S, Rallu F, Hraiky C, Fortin C, Masse E, Baaklini I. The problem of hypernegative supercoiling and R-loop formation in transcription. Front Biosci. 2003;8:d210–d221. doi: 10.2741/970. [DOI] [PubMed] [Google Scholar]
- 16.Ahmed W, Sala C, Hegde SR, Jha RK, Cole ST, Nagaraja V. Transcription facilitated genome-wide recruitment of topoisomerase I and DNA gyrase. PLoS Genet. 2017;13(5):e1006754. doi: 10.1371/journal.pgen.1006754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rani P, Nagaraja V. Genome-wide mapping of Topoisomerase I activity sites reveal its role in chromosome segregation. Nucleic Acids Res. 2019;47(3):1416–1427. doi: 10.1093/nar/gky1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheng B, Zhu CX, Ji C, Ahumada A, Tse-Dinh YC. Direct interaction between Escherichia coli RNA polymerase and the zinc ribbon domains of DNA topoisomerase I. J Biol Chem. 2003;278(33):30705–30710. doi: 10.1074/jbc.M303403200. [DOI] [PubMed] [Google Scholar]
- 19.Rani P, Kalladi S, Bansia H, Rao S, Jha RK, Jain P, et al. Nagaraja V. A Type IA DNA/RNA topoisomerase with RNA hydrolysis activity participates in ribosomal RNA processing. J Mol Biol. 2020 doi: 10.1016/j.jmb.2020.08.012. [DOI] [PubMed] [Google Scholar]
- 20.Tan K, Zhou Q, Cheng B, Zhang Z, Joachimiak A, Tse-Dinh YC. Structural basis for suppression of hypernegative DNA supercoiling by E. coli topoisomerase I. Nucleic Acids Res. 2015;43(22):11031–11046. doi: 10.1093/nar/gkv1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beran-Steed RK, Tse-Dinh Y-C. The carboxyl terminal domain of Escherichia coli DNA topoisomerase I confers higher affinity to DNA. Proteins: Struct Funct Genet. 1989;6:249–258. doi: 10.1002/prot.340060307. [DOI] [PubMed] [Google Scholar]
- 22.Kondekar SM, Gunjal GV, Radicella JP, Rao DN. Molecular dissection of Helicobacter pylori topoisomerase I reveals an additional active site in the carboxyl terminus of the enzyme. DNA Repair. 2020:91–92. doi: 10.1016/j.dnarep.2020.102853. [DOI] [PubMed] [Google Scholar]
- 23.Mills M, Tse-Dinh YC, Neuman KC. Direct observation of topoisomerase IA gate dynamics. Nat Struct Mol Biol. 2018;25(12):1111–1118. doi: 10.1038/s41594-018-0158-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cao N, Tan K, Zuo X, Annamalai T, Tse-Dinh YC. Mechanistic insights from structure of Mycobacterium smegmatis topoiso-merase I with ssDNA bound to both N- and C-terminal domains. Nucleic Acids Res. 2020;48(8):4448–4462. doi: 10.1093/nar/gkaa201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bizard AH, Hickson ID. The many lives of type IA topoisomerases. J Biol Chem. 2020;295(20):7138–7153. doi: 10.1074/jbc.REV120.008286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Changela A, DiGate RJ, Mondragon A. Structural studies of E. coli topoisomerase III-DNA complexes reveal a novel type IA topoisomerase-DNA conformational intermediate. J Mol Biol. 2007;368(1):105–118. doi: 10.1016/j.jmb.2007.01.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Z, Mondragon A, Hiasa H, Marians KJ, DiGate RJ. Identification of a unique domain essential for Escherichia coli DNA topoisomerase III-catalysed decatenation of replication intermediates. Mol Microbiol. 2000;35(4):888–895. doi: 10.1046/j.1365-2958.2000.01763.x. [DOI] [PubMed] [Google Scholar]
- 28.Nurse P, Levine C, Hassing H, Marians KJ. Topoisomerase III can serve as the cellular decatenase in Escherichia coli. J Biol Chem. 2003;278(10):8653–8660. doi: 10.1074/jbc.M211211200. [DOI] [PubMed] [Google Scholar]
- 29.Lee CM, Wang G, Pertsinidis A, Marians KJ. Topoisomerase III acts at the replication fork to remove precatenanes. J Bacteriol. 2019;201(7) doi: 10.1128/JB.00563-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suski C, Marians KJ. Resolution of converging replication forks by RecQ and topoisomerase III. Mol Cell. 2008;30(6):779–789. doi: 10.1016/j.molcel.2008.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.DiGate RJ, Marians KJ. Escherichia coli topoisomerase III-catalyzed cleavage of RNA. J Biol Chem. 1992;267:20532–20535. [PubMed] [Google Scholar]
- 32.Wang H, Di Gate RJ, Seeman NC. An RNA topoisomerase. Proc Natl Acad Sci USA. 1996;93(18):9477–9482. doi: 10.1073/pnas.93.18.9477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li W, Wang JC. Mammalian DNA topoisomerase IIIa is essential in early embryogenesis. Proc Natl Acad Sci USA. 1998;95:1010–1013. doi: 10.1073/pnas.95.3.1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu L, Hickson ID. The Bloom’s syndrome helicase suppresses crossing over during homologous recombination. Nature. 2003;426(6968):870–874. doi: 10.1038/nature02253. [DOI] [PubMed] [Google Scholar]
- 35.Yin J, Sobeck A, Xu C, Meetei AR, Hoatlin M, Li L, Wang W. BLAP75, an essential component of Bloom’s syndrome protein complexes that maintain genome integrity. EMBO J. 2005;24(7):1465–1476. doi: 10.1038/sj.emboj.7600622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Singh TR, Ali AM, Busygina V, Raynard S, Fan Q, Du CH, et al. Meetei AR. BLAP18/RMI2, a novel OB-fold-containing protein, is an essential component of the Bloom helicase-double Holliday junction dissolvasome. Genes Dev. 2008;22(20):2856–2868. doi: 10.1101/gad.1725108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Manthei KA, Keck JL. The BLM dissolvasome in DNA replication and repair. Cell Mol Life Sci. 2013;70(21):4067–4084. doi: 10.1007/s00018-013-1325-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Heyer WD. Recombination: Holliday junction resolution and crossover formation. Curr Biol. 2004;14(2):R56–R58. doi: 10.1016/j.cub.2003.12.043. [DOI] [PubMed] [Google Scholar]
- 39.Bocquet N, Bizard AH, Abdulrahman W, Larsen NB, Faty M, Cavadini S, Thoma NH, et al. Structural and mechanistic insight into Holliday-junction dissolution by topoisomerase IIIalpha and RMI1. Nat Struct Mol Biol. 2014;21(3):261–268. doi: 10.1038/nsmb.2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luke-Glaser S, Luke B, Grossi S, Constantinou A. FANCM regulates DNA chain elongation and is stabilized by S-phase checkpoint signalling. EMBO J. 2010;29(4):795–805. doi: 10.1038/emboj.2009.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lu R, O’Rourke JJ, Sobinoff AP, Allen JAM, Nelson CB, Tom-linson CG, et al. Pickett HA. The FANCM-BLM-TOP3A-RMI complex suppresses alternative lengthening of telomeres (ALT) Nat Commun. 2019;10(1):2252. doi: 10.1038/s41467-019-101806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bryan TM, Englezou A, Gupta J, Bacchetti S, Reddel RR. Telomere elongation in immortal human cells without detectable telomerase activity. EMBO J. 1995;14(17):4240–4248. doi: 10.1002/j.1460-2075.1995.tb00098.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wendorff TJ, Schmidt BH, Heslop P, Austin CA, Berger JM. The structure of DNA-bound human topoisomerase II alpha: Conformational mechanisms for coordinating inter-subunit interactions with DNA cleavage. J Mol Biol. 2012;424(3–4):109–124. doi: 10.1016/j.jmb.2012.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hsieh MY, Fan JR, Chang HW, Chen HC, Shen TL, Teng SC, et al. Li TK. DNA topoisomerase III alpha regulates p53- mediated tumor suppression. Clin Cancer Res. 2014;20(6):1489–1501. doi: 10.1158/1078-0432.CCR-13-1997. [DOI] [PubMed] [Google Scholar]
- 45.Bizard AH, Allemand JF, Hassenkam T, Paramasivam M, Sarlos K, Singh MI, Hickson ID. PICH and TOP3A cooperate to induce positive DNA supercoiling. Nat Struct Mol Biol. 2019;26(4):267–274. doi: 10.1038/s41594-019-0201-6. [DOI] [PubMed] [Google Scholar]
- 46.Tsai HZ, Lin RK, Hsieh TS. Drosophila mitochondrial topoisomerase III alpha affects the aging process via maintenance of mitochondrial function and genome integrity. J Biomed Sci. 2016;23:38. doi: 10.1186/s12929-016-0255-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu J, Feng L, Hsieh TS. Drosophila topo IIIalpha is required for the maintenance of mitochondrial genome and male germ-line stem cells. Proc Natl Acad Sci U S A. 2010;107(14):6228–6233. doi: 10.1073/pnas.1001855107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nicholls TJ, Nadalutti CA, Motori E, Sommerville EW, Gorman GS, Basu S, et al. Gustafsson CM. Topoisomerase 3alpha is required for decatenation and segregation of human mtDNA. Mol Cell. 2018;69(1):9–23.:e26. doi: 10.1016/j.molcel.2017.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kwan KY, Wang JC. Mice lacking DNA topoisomerase IIIbeta develop to maturity but show a reduced mean lifespan. Proc Natl Acad Sci U S A. 2001;98(10):5717–5721. doi: 10.1073/pnas.101132498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kwan KY, Greenwald RJ, Mohanty S, Sharpe AH, Shaw AC, Wang JC. Development of autoimmunity in mice lacking DNA topoisomerase 3beta. Proc Natl Acad Sci U SA. 2007;104(22):9242–9247. doi: 10.1073/pnas.0703587104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kwan KY, Moens PB, Wang JC. Infertility and aneuploidy in mice lacking a type IA DNA topoisomerase III beta. Proc Natl AcadSciUSA. 2003;100(5):2526–2531. doi: 10.1073/pnas.0437998100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang T, Wallis M, Petrovic V, Challis J, Kalitsis P, Hudson DF. Loss of TOP3B leads to increased R-loop formation and genome instability. Open Biol. 2019;9(12):190222. doi: 10.1098/rsob.190222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stoll G, Pietilainen OPH, Linder B, Suvisaari J, Brosi C, Hennah W, Palotie A, et al. Deletion of TOP3beta, a component of FMRP-containing mRNPs, contributes to neurodevelopmental disorders. Nat Neurosci. 2013;16(9):1228–1237. doi: 10.1038/nn.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ahmad M, Shen W, Li W, Xue Y, Zou S, Xu D, Wang W. Topoisomerase 3beta is the major topoisomerase for mRNAs and linked to neurodevelopment and mental dysfunction. Nucleic Acids Res. 2017;45(5):2704–2713. doi: 10.1093/nar/gkw1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xu D, Shen W, Guo R, Xue Y, Peng W, Sima J, et al. Wang W. Top3beta is an RNA topoisomerase that works with fragile X syn-drome protein to promote synapse formation. Nat Neurosci. 2013;16(9):1238–1247. doi: 10.1038/nn.3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang Y, McBride KM, Hensley S, Lu Y, Chedin F, Bedford MT. Arginine methylation facilitates the recruitment of TOP3B to chromatin to prevent R loop accumulation. Mol Cell. 2014;53(3):484–497. doi: 10.1016/j.molcel.2014.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Iossifov I, Ronemus M, Levy D, Wang Z, Hakker I, Rosenbaum J, et al. Wigler M. De novo gene disruptions in children on the autistic spectrum. Neuron. 2012;74(2):285–299. doi: 10.1016/j.neuron.2012.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xu B, Ionita-Laza I, Roos JL, Boone B, Woodrick S, Sun Y, et al. Karayiorgou M. De novo gene mutations highlight patterns of genetic and neural complexity in schizophrenia. Nat Genet. 2012;44(12):1365–1369. doi: 10.1038/ng.2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Linder B, Plottner O, Kroiss M, Hartmann E, Laggerbauer B, Meister G, Fischer U, et al. Tdrd3 is a novel stress granule-associated protein interacting with the fragile-X syndrome protein FMRP. Hum Mol Genet. 2008;17(20):3236–3246. doi: 10.1093/hmg/ddn219. [DOI] [PubMed] [Google Scholar]
- 60.Goto-Ito S, Yamagata A, Takahashi TS, Sato Y, Fukai S. Structural basis of the interaction between topoisomerase IIIbeta and the TDRD3 auxiliary factor. Sci Rep. 2017;7:42123. doi: 10.1038/srep42123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kikuchi A, Asai K. Reverse gyrase – a topoisomerase which introduces positive superhelical turns into DNA. Nature. 1984;309:677–681. doi: 10.1038/309677a0. [DOI] [PubMed] [Google Scholar]
- 62.Mirambeau G, Duguet M, Forterre P. ATP-dependent DNA topoisomerase from the Archaebacterium Sulfolobus acidocal-darius. J Mol Biol. 1984;179:559–563. doi: 10.1016/0022-2836(84)90080-9. [DOI] [PubMed] [Google Scholar]
- 63.Forterre P. A hot story from comparative genomics: Reverse gyrase is the only hyperthermophile-specific protein. Trends Genet. 2002;18(5):236–237. doi: 10.1016/s0168-9525(02)02650-1. [DOI] [PubMed] [Google Scholar]
- 64.Lulchev P, Klostermeier D. Reverse gyrase–recent advances and current mechanistic understanding of positive DNA supercoiling. Nucleic Acids Res. 2014;42(13):8200–8213. doi: 10.1093/nar/gku589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ogawa T, Yogo K, Furuike S, Sutoh K, Kikuchi A, Kinosita K., Jr Direct observation of DNA overwinding by reverse gyrase. Proc Natl Acad Sci U S A. 2015;112(24):7495–7500. doi: 10.1073/pnas.1422203112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rodriguez AC. Investigating the role of the latch in the positive supercoiling mechanism of reverse gyrase. Biochemistry. 2003;42(20):5993–6004. doi: 10.1021/bi034188l. [DOI] [PubMed] [Google Scholar]
- 67.Confalonieri F, Elie C, Nadal M, de La Tour C, Forterre P, Duguet M. Reverse gyrase: A helicase-like domain and a type I topoisomerase in the same polypeptide. Proc Natl Acad Sci U S A. 1993;90(10):4753–4757. doi: 10.1073/pnas.90.10.4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.del Toro Duany Y, Jungblut SP, Schmidt AS, Klostermeier D. The reverse gyrase helicase-like domain is a nucleotide-dependent switch that is attenuated by the topoisomerase domain. Nucleic Acids Res. 2008;36(18):5882–5895. doi: 10.1093/nar/gkn587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ganguly A, del Toro Duany Y, Klostermeier D. Reverse gyrase transiently unwinds double-stranded DNA in an ATP-dependent reaction. J Mol Biol. 2013;425(1):32–40. doi: 10.1016/j.jmb.2012.10.016. [DOI] [PubMed] [Google Scholar]
- 70.Declais AC, Marsault J, Confalonieri F, de La Tour CB, Duguet M. Reverse gyrase, the two domains intimately cooperate to promote positive supercoiling. J Biol Chem. 2000;275(26):19498–19504. doi: 10.1074/jbc.M910091199. [DOI] [PubMed] [Google Scholar]
- 71.Rudolph MG, del Toro Duany Y, Jungblut SP, Ganguly A, Klostermeier D. Crystal structures of Thermotoga maritima reverse gyrase: Inferences for the mechanism of positive DNA supercoiling. Nucleic Acids Res. 2013;41(2):1058–1070. doi: 10.1093/nar/gks1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Collin F, Weisslocker-Schaetzel M, Klostermeier D. A beta-hairpin is a minimal latch that supports positive supercoiling by reverse gyrase. J Mol Biol. 2020;432(16):4762–4771. doi: 10.1016/j.jmb.2020.06.018. [DOI] [PubMed] [Google Scholar]
- 73.Couturier M, Gadelle D, Forterre P, Nadal M, Garnier F. The reverse gyrase TopR1 is responsible for the homeostatic control of DNA supercoiling in the hyperthermophilic archaeon Sulfolobus solfataricus. Mol Microbiol. 2020;113(2):356–368. doi: 10.1111/mmi.14424. [DOI] [PubMed] [Google Scholar]
- 74.Shibata T, Nakasu S, Yasui K, Kikuchi A. Intrinsic DNA-dependent ATPase activity of reverse gyrase. J Biol Chem. 1987;262:10419–10421. [PubMed] [Google Scholar]
- 75.Bizard A, Garnier F, Nadal M. TopR2, the second reverse gyrase of Sulfolobus solfataricus exhibits unusual properties. J Mol Biol. 2011;408(5):839–849. doi: 10.1016/j.jmb.2011.03.030. [DOI] [PubMed] [Google Scholar]
- 76.Yang X, Garnier F, Debat H, Strick TR, Nadal M. Direct observation of helicase-topoisomerase coupling within reverse gyrase. Proc Natl Acad Sci U S A. 2020;117(20):10856–10864. doi: 10.1073/pnas.1921848117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Champoux JJ, Dulbecco R. An activity from mammalian cells that untwists superhelical DNA–a possible swivel for DNA replication (polyoma-ethidium bromide-mouse-embryo cells-dye binding assay) Proc Natl Acad Sci U S A. 1972;69(1):143–146. doi: 10.1073/pnas.69.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Soren BC, Dasari JB, Ottaviani A, Iacovelli F, Fiorani P. Topoisomerase IB: A relaxing enzyme for stressed DNA. Cancer Drug Resistance. 2020;3(1):18–25. doi: 10.20517/cdr.2019.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Stivers JT, Harris TK, Mildvan AS. Vaccinia DNA topoisomerase I: Evidence supporting a free rotation mechanism for DNA supercoil relaxation. Biochemistry. 1997;36:5212–5222. doi: 10.1021/bi962880t. [DOI] [PubMed] [Google Scholar]
- 80.Stewart L, Redinbo MR, Qiu X, Hol WGJ, Champoux JJ. A model for the mechanism of human topoisomerase I. Science. 1998;279:1534–1540. doi: 10.1126/science.279.5356.1534. [DOI] [PubMed] [Google Scholar]
- 81.Koster DA, Croquette V, Dekker C, Shuman S, Dekker NH. Friction and torque govern the relaxation of DNA supercoils by eukaryotic topoisomerase IB. Nature. 2005;434(7033):671–674. doi: 10.1038/nature03395. [DOI] [PubMed] [Google Scholar]
- 82.Cho J-E, Jinks-Robertson S. Topoisomerase I and genome stability: The good and the bad DNA Topoisomerases. Springer; 2018. pp. 21–45. [DOI] [PubMed] [Google Scholar]
- 83.Baranello L, Wojtowicz D, Cui K, Devaiah BN, Chung HJ, Chan-Salis KY, et al. Levens D. RNA polymerase II regulates topoi-somerase 1 activity to favor efficient transcription. Cell. 2016;165(2):357–371. doi: 10.1016/j.cell.2016.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wu J, Phatnani HP, Hsieh TS, Greenleaf AL. The phosphoCTD-interacting domain of topoisomerase I. Biochem Biophys Res Commun. 2010;397(1):117–119. doi: 10.1016/j.bbrc.2010.05.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cho JE, Jinks-Robertson S. Ribonucleotides and transcription-associated mutagenesis in yeast. J Mol Biol. 2017;429(21):3156–3167. doi: 10.1016/j.jmb.2016.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Li M, Liu Y. Topoisomerase I in human disease pathogenesis and treatments. Genomics Proteomics Bioinformatics. 2016;14(3):166–171. doi: 10.1016/j.gpb.2016.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Slesarev AI, Stetter KO, Lake JA, Gellert M, Krah R, Kozyavkin SA. DNA topoisomerase V is a relative of eukaryotic topoisomerase I from a hyperthermophilic prokaryote. Nature. 1993;364:735–737. doi: 10.1038/364735a0. [DOI] [PubMed] [Google Scholar]
- 88.Taneja B, Patel A, Slesarev A, Mondragon A. Structure of the N-terminal fragment of topoisomerase V reveals a new family of topoisomerases. EMBO J. 2006;25(2):398–408. doi: 10.1038/sj.emboj.7600922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Rajan R, Osterman A, Mondragón A. Methanopyrus kandleri topoisomerase V contains three distinct AP lyase active sites in addition to the topoisomerase active site. Nucleic Acids Res. 2016;44(7):3464–3474. doi: 10.1093/nar/gkw122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Belova GI, Prasad R, Kozyavkin SA, Lake JA, Wilson SH, Slesarev AI. A type IB topoisomerase with DNA repair activities. Proc Natl Acad Sci USA. 2001;98(11):6015–6020. doi: 10.1073/pnas.111040498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Rajan R, Prasad R, Taneja B, Wilson SH, Mondragón A. Identification of one of the apurinic/apyrimidinic lyase active sites of topoisomerase V by structural and functional studies. Nucleic Acids Res. 2012;41(1):657–666. doi: 10.1093/nar/gks1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Forterre P. DNA topoisomerase V: A new fold of mysterious origin. Trends Biotechnol. 2006;24(6):245–247. doi: 10.1016/j.tibtech.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 93.Bates AD, Berger JM, Maxwell A. The ancestral role of ATP hydrolysis in type II topoisomerases: Prevention of DNA doublestrand breaks. Nucleic Acids Res. 2011;39(15):6327–6339. doi: 10.1093/nar/gkr258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Pommier Y, Leo E, Zhang H, Marchand C. DNA topoisomerases and their poisoning by anticancer and antibacterial drugs. Chem Biol. 2010;17(5):421–433. doi: 10.1016/j.chembiol.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Corbett KD, Berger JM. Structure, molecular mechanisms, and evolutionary relationships in DNA topoisomerases. Annu Rev Biophys Biomol Struct. 2004;33:95–118. doi: 10.1146/annurev.biophys.33.110502.140357. [DOI] [PubMed] [Google Scholar]
- 96.Harrison SC, Aggarwal AK. DNA recognition by proteins with the helix-turn-helix motif. Annu Rev Biochem. 1990;59:933–969. doi: 10.1146/annurev.bi.59.070190.004441. [DOI] [PubMed] [Google Scholar]
- 97.Wang JC. DNA topoisomerases. Annu Rev Biochem. 1996;65:635–692. doi: 10.1146/annurev.bi.65.070196.003223. [DOI] [PubMed] [Google Scholar]
- 98.Dong KC, Berger JM. Structural basis for gate-DNA recognition and bending by type IIA topoisomerases. Nature. 2007;450(7173):1201–1205. doi: 10.1038/nature06396. [DOI] [PubMed] [Google Scholar]
- 99.Lee S, Jung SR, Heo K, Byl JA, Deweese JE, Osheroff N, Hohng S. DNA cleavage and opening reactions of human topoisomerase IIalpha are regulated via Mg2+-mediated dynamic bending of gate-DNA. Proc Natl Acad Sci USA. 2012;109(8):2925–2930. doi: 10.1073/pnas.1115704109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Aravind L, Leipe DD, Koonin EV. Toprim – a conserved catalytic domain in type IA and II topoisomerases, DnaG-type pri-mases, OLD family nucleases and RecR proteins. Nucleic Acids Res. 1998;26:4205–4213. doi: 10.1093/nar/26.18.4205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sissi C, Palumbo M. Effects of magnesium and related divalent metal ions in topoisomerase structure and function. Nucleic Acids Res. 2009;37(3):702–711. doi: 10.1093/nar/gkp024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Chang CC, Wang YR, Chen SF, Wu CC, Chan NL. New insights into DNA-binding by type IIA topoisomerases. Curr Opin Struct Biol. 2013;23(1):125–133. doi: 10.1016/j.sbi.2012.11.011. [DOI] [PubMed] [Google Scholar]
- 103.Laponogov I, Sohi MK, Veselkov DA, Pan X-S, Sawhney R, Thompson AW, et al. Sanderson MR. Structural insight into the quinolone-DNA cleavage complex of type IIA topoisomerases. Nat Struct Mol Biol. 2009;16(6):667–669. doi: 10.1038/nsmb.1604. [DOI] [PubMed] [Google Scholar]
- 104.Schmidt BH, Burgin AB, Deweese JE, Osheroff N, Berger JM. A novel and unified two-metal mechanism for DNA cleavage by type II and IA topoisomerases. Nature. 2010;465(7298):641–644. doi: 10.1038/nature08974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Noble CG, Maxwell A. The role of GyrB in the DNA cleavage-religation reaction of DNA gyrase: A proposed two metalion mechanism. J Mol Biol. 2002;318(2):361–371. doi: 10.1016/S0022-2836(02)00049-9. [DOI] [PubMed] [Google Scholar]
- 106.Laponogov I, Pan XS, Veselkov DA, McAuley KE, Fisher LM, Sanderson MR. Structural basis of gate-DNA breakage and resealing by type II topoisomerases. PLoS One. 2010;5(6):e11338. doi: 10.1371/journal.pone.0011338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Dutta R, Inouye M. GHKL, an emergent ATPase/kinase superfamily. Trends Biochem Sci. 2000;25(1):24–28. doi: 10.1016/s0968-0004(99)01503-0. [DOI] [PubMed] [Google Scholar]
- 108.Gellert M, Mizuuchi K, O’Dea MH, Nash HA. DNA gyrase: An enzyme that introduces superhelical turns into DNA. Proc NatlAcadSciUSA. 1976;73(11):3872–3876. doi: 10.1073/pnas.73.11.3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wall MK, Mitchenall LA, Maxwell A. Arabidopsis thaliana DNA gyrase is targeted to chloroplasts and mitochondria. Proc Natl Acad Sci U S A. 2004;101(20):7821–7826. doi: 10.1073/pnas.0400836101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Dar MA, Sharma A, Mondal N, Dhar SK. Molecular cloning of apicoplast-targeted Plasmodium falciparum DNA gyrase genes: Unique intrinsic ATPase activity and ATP-independent dimerization of PfGyrB subunit. Eukaryot Cell. 2007;6(3):398–412. doi: 10.1128/EC.00357-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Forterre P, Gadelle D. Phylogenomics of DNA topoisomerases: Their origin and putative roles in the emergence of mod-ern organisms. Nucleic Acids Res. 2009;37(3):679–692. doi: 10.1093/nar/gkp032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Yamashiro K, Yamagishi A. Characterization of the DNA gyrase from the thermoacidophilic archaeon Thermoplasma acidophilum. J Bacteriol. 2005;187(24):8531–8536. doi: 10.1128/JB.187.24.8531-8536.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Higgins NP, Peebles CL, Sugino A, Cozzarelli NR. Purification of subunits of Escherichia coli DNA gyrase and reconstitution of enzymatic activity. Proc Natl Acad Sci U S A. 1978;75(4):1773–1777. doi: 10.1073/pnas.75.4.1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Maxwell A. DNA gyrase as a drug target. Trends Microbiol. 1997;5(3):102–109. doi: 10.1016/S0966-842X(96)10085-8. [DOI] [PubMed] [Google Scholar]
- 115.Collin F, Karkare S, Maxwell A. Exploiting bacterial DNA gyrase as a drug target: Current state and perspectives. Appl Microbiol Biotechnol. 2011;92(3):479–497. doi: 10.1007/s00253-011-3557-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Drlica K, Snyder M. Superhelical Escherichia coli DNA: Relaxation by coumermycin. J Mol Biol. 1978;120:145–154. doi: 10.1016/0022-2836(78)90061-x. [DOI] [PubMed] [Google Scholar]
- 117.Steck T, Pruss G, Manes S, Burg L, Drlica K. DNA supercoiling in gyrase mutants. J Bacteriol. 1984;158:397–403. doi: 10.1128/jb.158.2.397-403.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lockshon D, Morris DR. Positively supercoiled plasmid DNA is produced by treatment of Escherichia coli with DNA gyrase inhibitors. Nucleic Acids Res. 1983;11(10):2999–3017. doi: 10.1093/nar/11.10.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Menzel R, Gellert M. Regulation of the genes for E. coli DNA gyrase: Homeostatic control of DNA supercoiling. Cell. 1983;34(1):105–113. doi: 10.1016/0092-8674(83)90140-x. [DOI] [PubMed] [Google Scholar]
- 120.Botchan P, Wang JC, Echols H. Effect of circularity and superhelicity on transcription from bacteriophagelambda DNA. Proc Natl Acad Sci U S A. 1973;70(11):3077–3081. doi: 10.1073/pnas.70.11.3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Funnell BE, Baker TA, Kornberg A. Complete enzymatic replication of plasmids containing the origin of the Escherichia coli chromosome. J Biol Chem. 1986;261(12):5616–5624. [PubMed] [Google Scholar]
- 122.Kreuzer KN, Cozzarelli NR. Escherichia coli mutants thermosensitive for deoxyribonucleic acid gyrase subunit A: Effects on deoxyribonucleic acid replication, transcription, and bacteriophage growth. J Bacteriol. 1979;140:424–435. doi: 10.1128/jb.140.2.424-435.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Hiasa H, Marians KJ. Two distinct modes of strand unlinking during theta-type DNA replication. J Biol Chem. 1996;271(35):21529–21535. doi: 10.1074/jbc.271.35.21529. [DOI] [PubMed] [Google Scholar]
- 124.Sutormin D, Rubanova N, Logacheva M, Ghilarov D, Severi-nov K. Single-nucleotide-resolution mapping of DNA gyrase cleavage sites across the Escherichia coli genome. Nucleic Acids Res. 2019;47(3):1373–1388. doi: 10.1093/nar/gky1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Stracy M, Wollman AJM, Kaja E, Gapinski J, Lee JE, Leek VA, Zawadzki P, et al. Single-molecule imaging of DNA gyrase activity in living Escherichia coli. Nucleic Acids Res. 2019;47(1):210–220. doi: 10.1093/nar/gky1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Agarwal R, Duderstadt KE. Multiplex flow magnetic tweezers reveal rare enzymatic events with single molecule precision. Nat Commun. 2020;11(1):4714. doi: 10.1038/s41467-020-18456-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Corbett KD, Shultzaberger RK, Berger JM. The C-terminal domain of DNA gyrase A adopts a DNA-bending {beta}- pinwheel fold. Proc Natl Acad Sci U S A. 2004;101(19):7293–7298. doi: 10.1073/pnas.0401595101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Hsieh TJ, Yen TJ, Lin TS, Chang HT, Huang SY, Hsu CH, et al. Chan NL. Twisting of the DNA-binding surface bya beta-strand-bearing proline modulates DNA gyrase activity. Nucleic Acids Res. 2010;38(12):4173–4181. doi: 10.1093/nar/gkq153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Ruthenburg AJ, Graybosch DM, Huetsch JC, Verdine GL. A superhelical spiral in the Escherichia coli DNA gyrase A C-terminal domain imparts unidirectional supercoiling bias. J Biol Chem. 2005;280(28):26177–26184. doi: 10.1074/jbc.M502838200. [DOI] [PubMed] [Google Scholar]
- 130.Tretter EM, Berger JM. Mechanisms for defining supercoiling set point of DNA gyrase orthologs: II The shape of the GyrA Subunit C-terminal domain (CTD) is not a sole determinant for controlling supercoiling efficiency. J Biol Chem. 2012;287(22):18645–18654. doi: 10.1074/jbc.M112.345736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kramlinger VM, Hiasa H. The “GyrA-box” is required for the ability of DNA gyrase to wrap DNA and catalyze the supercoiling reaction. J Biol Chem. 2006;281(6):3738–3742. doi: 10.1074/jbc.M511160200. [DOI] [PubMed] [Google Scholar]
- 132.Lanz MA, Klostermeier D. The GyrA-box determines the geometry of DNA bound to gyrase and couples DNA binding to the nucleotide cycle. Nucleic Acids Res. 2012;40(21):10893–10903. doi: 10.1093/nar/gks852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Nollmann M, Crisona NJ, Arimondo PB. Thirty years of Escherichia coli DNA gyrase: From in vivo function to single-molecule mechanism. Biochimie. 2007;89(4):490–499. doi: 10.1016/j.biochi.2007.02.012. [DOI] [PubMed] [Google Scholar]
- 134.Orphanides G, Maxwell A. Evidence for a conformational change in the DNA gyrase-DNA complex from hydroxyl radical footprinting. Nucleic Acids Res. 1994;22:1567–1575. doi: 10.1093/nar/22.9.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Gubaev A, Weidlich D, Klostermeier D. DNA gyrase with a single catalytic tyrosine can catalyze DNA supercoiling by a nicking-closing mechanism. Nucleic Acids Res. 2016;44(21):10354–10366. doi: 10.1093/nar/gkw740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Papillon J, Menetret JF, Batisse C, Helye R, Schultz P, Potier N, Lamour V. Structural insight into negative DNA supercoiling by DNA gyrase, a bacterial type 2A DNA topoisomerase. Nucleic Acids Res. 2013;41(16):7815–7827. doi: 10.1093/nar/gkt560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Vanden Broeck A, Lotz C, Ortiz J, Lamour V. Cryo-EM structure of the complete E. coli DNA gyrase nucleoprotein complex. Nat Commun. 2019;10(1):4935. doi: 10.1038/s41467-019-12914-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Kato J, Suzuki H, Ikeda H. Purification and characterization of DNA topoisomerase IV in Escherichia coli. J Biol Chem. 1992;267(36):25676–25684. [PubMed] [Google Scholar]
- 139.Kato J, Nishimura Y, Imamura R, Niki H, Hiraga S, Suzuki H. New topoisomerase essential for chromosome segregation in E. coli. Cell. 1990;63:393–404. doi: 10.1016/0092-8674(90)90172-b. [DOI] [PubMed] [Google Scholar]
- 140.Peng H, Marians KJ. Decatenation activity of topoisomerase IV during oriC and pBR322 DNA replication in vitro. Proc Natl Acad Sci U S A. 1993;90(18):8571–8575. doi: 10.1073/pnas.90.18.8571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.El Sayyed H, Le Chat L, Lebailly E, Vickridge E, Pages C, Cornet F, et al. Espeli O. Mapping topoisomerase IV binding and activity sites on the E. coli genome. PLoS Genet. 2016;12(5):e1006025. doi: 10.1371/journal.pgen.1006025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Nicolas E, Upton AL, Uphoff S, Henry O, Badrinarayanan A, Sherratt D. The SMC complex MukBEF recruits topoiso-merase IV to the origin of replication region in live Escherichia coli. MBio. 2014;5(1):e01001–01013. doi: 10.1128/mBio.01001-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Nolivos S, Upton AL, Badrinarayanan A, Müller J, Zawadzka K, Wiktor J, et al. Sherratt D. MatP regulates the coordinated action of topoisomerase IV and MukBEF in chromosome segregation. Nat Commun. 2016;7(1):10466. doi: 10.1038/ncomms10466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Hayama R, Marians KJ. Physical and functional interaction between the condensin MukB and the decatenase topoi-somerase IV in Escherichia coli. Proc Natl Acad Sci USA. 2010;107(44):18826–18831. doi: 10.1073/pnas.1008140107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Li Y, Stewart NK, Berger AJ, Vos S, Schoeffler AJ, Berger JM, et al. Oakley MG. Escherichia coli condensin MukB stimulates topoisomerase IV activity by a direct physical interaction. Proc Natl AcadSciUSA. 2010;107(44):18832–18837. doi: 10.1073/pnas.1008678107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Corbett KD, Schoeffler AJ, Thomsen ND, Berger JM. The structural basis for substrate specificity in DNA topoisomerase IV. J Mol Biol. 2005;351(3):545–561. doi: 10.1016/j.jmb.2005.06.029. [DOI] [PubMed] [Google Scholar]
- 147.Crisona NJ, Strick TR, Bensimon D, Croquette V, Cozzarelli NR. Preferential relaxation of positively supercoiled DNA by E. coli topoisomerase IV in single-molecule and ensemble measurements. Genes Dev. 2000;14(22):2881–2892. doi: 10.1101/gad.838900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Stone MD, Bryant Z, Crisona NJ, Smith SB, Vologodskii A, Bus-tamante C, Cozzarelli NR. Chirality sensing by Escherichia coli topoisomerase IV and the mechanism of type II topoisomerases. Proc Natl Acad Sci U S A. 2003;100(15):8654–8659. doi: 10.1073/pnas.1133178100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Neuman KC, Charvin G, Bensimon D, Croquette V. Mechanisms of chiral discrimination by topoisomerase IV. Proc Natl Acad Sci U S A. 2009;106(17):6986–6991. doi: 10.1073/pnas.0900574106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.McNairn E, Ní Bhriain N, Dorman CJ. Overexpression of the Shigella flexneri genes coding for DNA topoisomerase IV compensates for loss of DNA topoisomerase I: Effect on virulence gene expression. Molec Microbiol. 1995;15:507–517. doi: 10.1111/j.1365-2958.1995.tb02264.x. [DOI] [PubMed] [Google Scholar]
- 151.Raji A, Zabel D, Laufer C, Depew R. Genetic analysis of mutations that compensate for loss of Escherichia coli DNA topoisomerase I. J Bacteriol. 1985;162:1173–1179. doi: 10.1128/jb.162.3.1173-1179.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Reuss DR, Fasshauer P, Mroch PJ, Ul-Haq I, Koo BM, Pohlein A, et al. Stulke J. Topoisomerase IV can functionally replace all type 1A topoisomerases in Bacillus subtilis. Nucleic Acids Res. 2019;47(10):5231–5242. doi: 10.1093/nar/gkz260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Khodursky AB, Peter BJ, Schmid MB, DeRisi J, Botstein D, Brown PO, Cozzarelli NR. Analysis of topoisomerase function in bacterial replication fork movement: Use of DNA microarrays. Proc Natl Acad Sci USA. 2000;97:9419–9424. doi: 10.1073/pnas.97.17.9419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Espeli O, Levine C, Hassing H, Marians KJ. Temporal regulation of topoisomerase IV activity in E. coli. Mol Cell. 2003;11(1):189–201. doi: 10.1016/s1097-2765(03)00013-3. [DOI] [PubMed] [Google Scholar]
- 155.Goto T, Wang JC. Yeast DNA topoisomerase II. An ATP-dependent type II topoisomerase that catalyzes the catenation, decatenation, unknotting, and relaxation of double-stranded DNA rings. J Biol Chem. 1982;257(10):5866–5872. [PubMed] [Google Scholar]
- 156.Holm C, Goto T, Wang JC, Botstein D. DNA topoisomerase II is required at the time of mitosis in yeast. Cell. 1985;41(2):553–563. doi: 10.1016/s0092-8674(85)80028-3. [DOI] [PubMed] [Google Scholar]
- 157.Uemura T, Ohkura H, Adachi Y, Morino K, Shiozaki K, Yanagida M. DNA topoisomerase II is required for condensation and separation of mitotic chromosomes in S. pombe. Cell. 1987;50(6):917–925. doi: 10.1016/0092-8674(87)90518-6. [DOI] [PubMed] [Google Scholar]
- 158.Dyson S, Segura J, Martínez-García B, Valdés A, Roca J. Condensin minimizes topoisomerase II-mediated entanglements of DNA in vivo. EMBO J. 2020:e105393. doi: 10.15252/embj.2020105393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Baxter J, Diffley JFX. Topoisomerase II inactivation prevents the completion of DNA replication in budding yeast. Mol Cell. 2008;30(6):790–802. doi: 10.1016/j.molcel.2008.04.019. [DOI] [PubMed] [Google Scholar]
- 160.Le TT, Gao X, Park SH, Lee J, Inman JT, Lee JH, et al. Wang MD. Synergistic coordination of chromatin torsional mechanics and topoisomerase activity. Cell. 2019;179(3):619–631.:e615. doi: 10.1016/j.cell.2019.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Joshi RS, Pina B, Roca J. Topoisomerase II is required for the production of long Pol II gene transcripts in yeast. Nucleic Acids Res. 2012;40(16):7907–7915. doi: 10.1093/nar/gks626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Schmidt BH, Osheroff N, Berger JM. Structure of a topoisomerase II-DNA-nucleotide complex reveals a new control mechanism for ATPase activity. Nat Struct Mol Biol. 2012;19(11):1147–1154. doi: 10.1038/nsmb.2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Lee JH, Berger JM. cell cycle-dependent control and roles of DNA topoisomerase II. Genes. 2019;10(11):859. doi: 10.3390/genes10110859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Woessner RD, Mattern MR, Mirabelli CK, Johnson RK, Drake FH. Proliferation- and cell cycle-dependent differ-ences in expression of the 170 kilodalton and 180 kilodalton forms of topoisomerase II in NIH-3T3 cells. Cell Growth Differ. 1991;2(4):209–214. [PubMed] [Google Scholar]
- 165.Grue P, Grasser A, Sehested M, Jensen PB, Uhse A, Straub T, Boege F, et al. Essential mitotic functions of DNA topoisomerase IIalpha are not adopted by topoisomerase IIbeta in human H69 cells. J Biol Chem. 1998;273(50):33660–33666. doi: 10.1074/jbc.273.50.33660. [DOI] [PubMed] [Google Scholar]
- 166.Anderson H, Roberge M. Topoisomerase II inhibitors affect entry into mitosis and chromosome condensation in BHK cells. Cell Growth Differ. 1996;7(1):83–90. [PubMed] [Google Scholar]
- 167.Piskadlo E, Tavares A, Oliveira RA. Metaphase chromosome structure is dynamically maintained by condensin I-directed DNA (de)catenation. Elife. 2017;6:e26120. doi: 10.7554/eLife.26120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Nielsen CF, Zhang T, Barisic M, Kalitsis P, Hudson DF. Topoisomerase IIalpha is essential for maintenance of mitotic chromosome structure. Proc Natl Acad Sci U S A. 2020;117(22):12131–12142. doi: 10.1073/pnas.2001760117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Farr CJ, Antoniou-Kourounioti M, Mimmack ML, Volkov A, Porter AC. The alpha isoform of topoisomerase II is required for hypercompaction of mitotic chromosomes in human cells. Nucleic Acids Res. 2014;42(7):4414–4426. doi: 10.1093/nar/gku076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Spence JM, Phua HH, Mills W, Carpenter AJ, Porter AC, Farr CJ. Depletion of topoisomerase IIalpha leads to shortening of the metaphase interkinetochore distance and abnormal persistence of PICH-coated anaphase threads. J Cell Sci. 2007;120(Pt 22):3952–3964. doi: 10.1242/jcs.013730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Hirano T, Mitchison TJ. Topoisomerase II does not play a scaffolding role in the organization of mitotic chromosomes assembled in Xenopus egg extracts. J Cell Biol. 1993;120(3):601–612. doi: 10.1083/jcb.120.3.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Porter AC, Farr CJ. Topoisomerase II: Untangling its contribution at the centromere. Chromosome Res. 2004;12(6):569–583. doi: 10.1023/B:CHRO.0000036608.91085.d1. [DOI] [PubMed] [Google Scholar]
- 173.Nagasaka K, Hossain MJ, Roberti MJ, Ellenberg J, Hirota T. Sister chromatid resolution is an intrinsic part of chromosome organization in prophase. Nat Cell Biol. 2016;18(6):692–699. doi: 10.1038/ncb3353. [DOI] [PubMed] [Google Scholar]
- 174.Wang LH, Mayer B, Stemmann O, Nigg EA. Centromere DNA decatenation depends on cohesin removal and is required for mammalian cell division. J Cell Sci. 2010;123(Pt 5):806–813. doi: 10.1242/jcs.058255. [DOI] [PubMed] [Google Scholar]
- 175.Nielsen CF, Huttner D, Bizard AH, Hirano S, Li TN, Palmai-Pallag T, et al. Hickson ID. PICH promotes sister chromatid disjunction and co-operates with topoisomerase II in mitosis. Nat Commun. 2015;6:8962. doi: 10.1038/ncomms9962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Daniloski Z, Bisht KK, McStay B, Smith S. Resolution of human ribosomal DNA occurs in anaphase, dependent on tankyrase 1, condensin II, and topoisomerase IIalpha. Genes Dev. 2019;33(5–6):276–281. doi: 10.1101/gad.321836.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Mirski SE, Gerlach JH, Cummings HJ, Zirngibl R, Greer PA, Cole SP. Bipartite nuclear localization signals in the C terminus of human topoisomerase II alpha. Exp Cell Res. 1997;237(2):452–455. doi: 10.1006/excr.1997.3805. [DOI] [PubMed] [Google Scholar]
- 178.Lane AB, Gimenez-Abian JF, Clarke DJ. A novel chromatin tether domain controls topoisomerase IIalpha dynamics and mitotic chromosome formation. J Cell Biol. 2013;203(3):471–486. doi: 10.1083/jcb.201303045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Bedez C, Lotz C, Batisse C, Broeck AV, Stote RH, Howard E, Lamour V, et al. Post-translational modifications in DNA topoi-somerase 2alpha highlight the role of a eukaryote-specific residue in the ATPase domain. Sci Rep. 2018;8(1):9272. doi: 10.1038/s41598-018-27606-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Antoniou-Kourounioti M, Mimmack ML, Porter ACG, Farr CJ. The impact of the C-terminal region on the interaction of topoisomerase II alpha with mitotic chromatin. Int J Mol Sci. 2019;20(5):1238. doi: 10.3390/ijms20051238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Madabhushi R. The roles of DNA topoisomerase IIbeta in transcription. Int J Mol Sci. 2018;19(7) doi: 10.3390/ijms19071917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Yang X, Li W, Prescott ED, Burden SJ, Wang JC. DNA topoisomerase IIbeta and neural development. Science. 2000;287(5450):131–134. doi: 10.1126/science.287.5450.131. [DOI] [PubMed] [Google Scholar]
- 183.Akimitsu N, Kamura K, Tone S, Sakaguchi A, Kikuchi A, Hamamoto H, Sekimizu K. Induction of apoptosis by depletion of DNA topoisomerase IIalpha in mammalian cells. Biochem Biophys Res Commun. 2003;307(2):301–307. doi: 10.1016/s0006-291x(03)01169-0. [DOI] [PubMed] [Google Scholar]
- 184.Capranico G, Tinelli S, Austin CA, Fisher ML, Zunino F. Different patterns of gene expression of topoisomerase II isoforms in differentiated tissues during murine development. Biochim Biophys Acta. 1992;1132(1):43–48. doi: 10.1016/0167-4781(92)90050-a. [DOI] [PubMed] [Google Scholar]
- 185.Madabhushi R, Gao F, Pfenning AR, Pan L, Yamakawa S, Seo J, et al. Tsai LH. Activity-induced DNA breaks govern the expression of neuronal early-response genes. Cell. 2015;161(7):1592–1605. doi: 10.1016/j.cell.2015.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Lam CW, Yeung WL, Law CY. Global developmental delay and intellectual disability associated with a de novo TOP2B mutation. Clin Chim Acta. 2017;469:63–68. doi: 10.1016/j.cca.2017.03.022. [DOI] [PubMed] [Google Scholar]
- 187.Hiraide T, Watanabe S, Matsubayashi T, Yanagi K, Nakashima M, Ogata T, Saitsu H. A de novo TOP2B variant associ-ated with global developmental delay and autism spectrum disorder. Mol Genet Genomic Med. 2020;8(3):e1145. doi: 10.1002/mgg3.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Bollimpelli VS, Dholaniya PS, Kondapi AK. Topoisomerase IIbeta and its role in different biological contexts. Arch Biochem Biophys. 2017;633:78–84. doi: 10.1016/j.abb.2017.06.021. [DOI] [PubMed] [Google Scholar]
- 189.Chen SF, Huang NL, Lin JH, Wu CC, Wang YR, Yu YJ, Chan NL, et al. Structural insights into the gating of DNA passage by the topoisomerase II DNA-gate. Nat Commun. 2018;9(1):3085. doi: 10.1038/s41467-018-05406-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Gardner MJ, Hall N, Fung E, White O, Berriman M, Hyman RW, et al. Bowman S. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2002;419(6906):498. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Sugimoto-Shirasu K, Stacey NJ, Corsar J, Roberts K, McCann MC. DNA topoisomerase VI is essential for endoreduplication in Arabidopsis. Curr Biol. 2002;12(20):1782–1786. doi: 10.1016/s0960-9822(02)01198-3. [DOI] [PubMed] [Google Scholar]
- 192.Bergerat A, Gadelle D, Forterre P. Purification of a DNA topoisomerase II from the hyperthermophilic archaeon Sulfolobus shi- batae A thermostable enzyme with both bacterial and eucaryal features. J Biol Chem. 1994;269(44):27663–27669. [PubMed] [Google Scholar]
- 193.Nichols MD, DeAngelis K, Keck JL, Berger JM. Structure and function ofan archaeal topoisomerase VI subunit with homology to the meiotic recombination factor Spo 11. EMBO J. 1999;18:6177–6188. doi: 10.1093/emboj/18.21.6177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Corbett KD, Berger JM. Structure of the topoisomerase VI-B subunit: Implications for type II topoisomerase mechanism and evolution. EMBO J. 2003;22(1):151–163. doi: 10.1093/emboj/cdg008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Corbett KD, Berger JM. Structural dissection of ATP turnover in the prototypical GHL ATPase TopoVI. Structure. 2005;13(6):873–882. doi: 10.1016/j.str.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 196.Corbett KD, Benedetti P, Berger JM. Holoenzyme assembly and ATP-mediated conformational dynamics of topoiso-merase VI. Nat Struct Mol Biol. 2007;14(7):611–619. doi: 10.1038/nsmb1264. [DOI] [PubMed] [Google Scholar]
- 197.Graille M, Cladiere L, Durand D, Lecointe F, Gadelle D, Quevillon-Cheruel S, et al. van Tilbeurgh H. Crystal structure of an intact type II DNA topoisomerase: Insights into DNA transfer mechanisms. Structure. 2008;16(3):360–370. doi: 10.1016/j.str.2007.12.020. [DOI] [PubMed] [Google Scholar]
- 198.Wendorff TJ, Berger JM. Topoisomerase VI senses and exploits both DNA crossings and bends to facilitate strand passage. Elife. 2018;7:e31724. doi: 10.7554/eLife.31724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Hartung F, Puchta H. Molecular characterization of homologues of both subunits A (SPO 11) and B of the archaebacterial topoisomerase 6 in plants. Gene. 2001;271(1):81–86. doi: 10.1016/s0378-1119(01)00496-6. [DOI] [PubMed] [Google Scholar]
- 200.Hartung F, Puchta H. Molecular characterisation of two paralogous SPO11 homologues in Arabidopsis thaliana. Nucleic Acids Res. 2000;28(7):1548–1554. doi: 10.1093/nar/28.7.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Hartung F, Angelis KJ, Meister A, Schubert I, Melzer M, Puchta H. An archaebacterial topoisomerase homolog not present in other eukaryotes is indispensable for cell proliferation of plants. Curr Biol. 2002;12(20):1787–1791. doi: 10.1016/s0960-9822(02)01218-6. [DOI] [PubMed] [Google Scholar]
- 202.Sugimoto-Shirasu K, Roberts K. Big it up”: Endoreduplication and cell-size control in plants. Curr Opin Plant Biol. 2003;6(6):544–553. doi: 10.1016/j.pbi.2003.09.009. [DOI] [PubMed] [Google Scholar]
- 203.Corbett KD, Berger JM. Emerging roles for plant topoisomerase VI. Chem Biol. 2003;10(2):107–111. doi: 10.1016/s1074-5521(03)00027-9. [DOI] [PubMed] [Google Scholar]
- 204.Sugimoto-Shirasu K, Roberts GR, Stacey NJ, McCann MC, Maxwell A, Roberts K. RHL1 is an essential component of the plant DNA topoisomerase VI complex and is required for ploidy- dependent cell growth. Proc Natl Acad Sci U S A. 2005;102(51):18736–18741. doi: 10.1073/pnas.0505883102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Breuer C, Stacey NJ, West CE, Zhao Y, Chory J, Tsukaya H, Sugimoto-Shirasu K, et al. BIN4, a novel component of the plant DNA topoisomerase VI complex, is required for endoreduplication in Arabidopsis. Plant Cell. 2007;19(11):3655–3668. doi: 10.1105/tpc.107.054833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Yin Y, Cheong H, Friedrichsen D, Zhao Y, Hu J, Mora-Garcia S, Chory J. A crucial role for the putative Arabidopsis topoisomerase VI in plant growth and development. Proc Natl Acad Sci USA. 2002;99(15):10191–10196. doi: 10.1073/pnas.152337599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Kirik V, Schrader A, Uhrig JF, Hulskamp M. MIDGET unravels functions of the arabidopsis topoisomerase VI complex in DNA endoreduplication, chromatin condensation, and transcriptional silencing. Plant Cell. 2007 doi: 10.1105/tpc.107.054361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Schrader A, Welter B, Hulskamp M, Hoecker U, Uhrig JF. MIDGET connects COP1-dependent development with endoreduplication in Arabidopsis thaliana. Plant J. 2013;75(1):67–79. doi: 10.1111/tpj.12199. [DOI] [PubMed] [Google Scholar]
- 209.Jain M, Tyagi AK, Khurana JP. Overexpression of putative topoisomerase 6 genes from rice confers stress tolerance in transgenic Arabidopsis plants. FEBS J. 2006;273(23):5245–5260. doi: 10.1111/j.1742-4658.2006.05518.x. [DOI] [PubMed] [Google Scholar]
- 210.Simkova K, Moreau F, Pawlak P, Vriet C, Baruah A, Alexandre C, et al. Laloi C. Integration of stress-related and reactive oxygen species-mediated signals by topoisomerase VI in Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2012;109(40):16360–16365. doi: 10.1073/pnas.1202041109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Bergerat A, de Massy B, Gadelle D, Varoutas PC, Nicolas A, Forterre P. An atypical topoisomerase II from Archaea with implications for meiotic recombination. Nature. 1997;386(6623):414–417. doi: 10.1038/386414a0. [DOI] [PubMed] [Google Scholar]
- 212.Keeney S, Giroux CN, Kleckner N. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell. 1997;88(3):375–384. doi: 10.1016/s0092-8674(00)81876-0. [DOI] [PubMed] [Google Scholar]
- 213.Robert T, Nore A, Brun C, Maffre C, Crimi B, Bourbon HM, de Massy B. The TopoVIB-Like protein family is required for meiotic DNA double-strand break formation. Science. 2016;351(6276):943–949. doi: 10.1126/science.aad5309. [DOI] [PubMed] [Google Scholar]
- 214.Vrielynck N, Chambon A, Vezon D, Pereira L, Chelysheva L, De Muyt A, Grelon M, et al. A DNA topoisomerase VI-like complex initiates meiotic recombination. Science. 2016;351(6276):939–943. doi: 10.1126/science.aad5196. [DOI] [PubMed] [Google Scholar]
- 215.Aravind L, Iyer LM, Wellems TE, Miller LH. Plasmodium biology: Genomic gleanings. Cell. 2003;115(7):771–785. doi: 10.1016/s0092-8674(03)01023-7. [DOI] [PubMed] [Google Scholar]
- 216.Malik SB, Ramesh MA, Hulstrand AM, Logsdon JM., Jr Protist homologs of the meiotic Spo11 gene and topoiso-merase VI reveal an evolutionary history of gene duplication and lineage-specific loss. Mol Biol Evol. 2007;24(12):2827–2841. doi: 10.1093/molbev/msm217. [DOI] [PubMed] [Google Scholar]
- 217.Chalapareddy S, Chakrabarty S, Bhattacharyya MK, Bhattacharyya S. Radicicol-mediated inhibition of topoisomerase VIB-VIA activity of the human malaria parasite Plasmodium falciparum. mSphere. 2016;1(1) doi: 10.1128/mSphere.00025-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Gadelle D, Krupovic M, Raymann K, Mayer C, Forterre P. DNA topoisomerase VIII: A novel subfamily of type IIB topoisomerases encoded by free or integrated plasmids in Archaea and Bacteria. Nucleic Acids Res. 2014;42(13):8578–8591. doi: 10.1093/nar/gku568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Takahashi TS, Da Cunha V, Krupovic M, Mayer C, Forterre P, Gadelle D. Expanding the type IIB DNA topoisomerase family: Identification of new topoisomerase and topoisomerase-like proteins in mobile genetic elements. NAR Genomics and Bioinformatics. 2020;2(1):lqz021. doi: 10.1093/nargab/lqz021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Staker BL, Hjerrild K, Feese MD, Behnke CA, Burgin AB, Jr, Stewart L. The mechanism of topoisomerase I poisoning by a camptothecin analog. Proc Natl Acad Sci U S A. 2002;99(24):15387–15392. doi: 10.1073/pnas.242259599. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.