Abstract
Life exists in three dimensions, but until the turn of the century most electron microscopy methods provided only 2D image data. Recently, electron microscopy techniques capable of delving deep into the structure of cells and tissues have emerged, collectively called volume electron microscopy (vEM). Developments in vEM have been dubbed a quiet revolution as the field evolved from established transmission and scanning electron microscopy techniques, so early publications largely focused on the bioscience applications rather than the underlying technological breakthroughs. However, with an explosion in the uptake of vEM across the biosciences and fast-paced advances in volume, resolution, throughput and ease of use, it is timely to introduce the field to new audiences. In this Primer, we introduce the different vEM imaging modalities, the specialized sample processing and image analysis pipelines that accompany each modality and the types of information revealed in the data. We showcase key applications in the biosciences where vEM has helped make breakthrough discoveries and consider limitations and future directions. We aim to show new users how vEM can support discovery science in their own research fields and inspire broader uptake of the technology, finally allowing its full adoption into mainstream biological imaging.
Introduction
Volume electron microscopy (vEM; also referred to as volume EM) describes a set of high-resolution imaging techniques used in biomedical research to reveal the 3D structure of cells, tissues and small model organisms at nanometre resolution. vEM techniques leverage the unique ability of electron microscopes to reveal the structure of cells and tissues with the contrast and resolution to distinguish cell membranes and membrane bilayers. In the context of this Primer, and to delineate boundaries with other fields, we consider vEM to include any method of generating serial images in an electron microscope from resin-embedded cells and tissues with a continuous depth greater than 1 μm. This includes techniques based on transmission electron microscopy (TEM), in which the electron beam passes through an ultra-thin slice of the sample to generate an image, and scanning electron microscopy (SEM), in which the electron beam is raster-scanned over the surface of a sample generating an image from secondary and/or backscattered electrons. TEM-based vEM techniques such as serial section TEM (ssTEM) and serial section electron tomography (ssET) and SEM-based vEM techniques such as array tomography, serial blockface SEM (SBF-SEM; also known as SBEM) and focused ion beam SEM (FIB-SEM) all generate a series of images that can be stacked to form a 3D digital representation of the original sample volume. Further, we cover emerging vEM technologies such as automated serial TEM (TEMCA and GridTape TEM), fast ion beam technologies (plasma and gas cluster ion beam systems) and massively parallel imaging in SEM (multibeam SEM (mSEM) and FAST-EM).
vEM techniques have emerged over the past 20 years, largely in response to the demands of the connectomics field 1. Connectomics has the dual challenge of imaging neurons in the brain with sufficient volume to capture their length and with sufficient resolution to image individual synapses, as structural indicators of functional connections between cells. The development of vEM was enabled by key technological advances in sample preparation, in automation of sample sectioning and TEM and SEM sources and detectors and, importantly, in scientific computing to facilitate acquisition and analysis of large image datasets. The biomedical applications of vEM quickly moved beyond connectomics, as the value of imaging complex biological processes in 3D in tissues and model organisms became clear. vEM is now widely used in studies of cell structure and non-brain tissue in fields as diverse as infection, immunity, cancer research, neurodegeneration, developmental biology, plant biology, synthetic biology, biomechanics, biomaterials and clinical research.
Scientists who develop and use vEM technology come from a wide variety of research disciplines. At first glance, it would seem to be difficult to form a coherent community from such a diverse group. However, over the past decade, grassroots initiatives have gathered technologists, microscopists, computational scientists, biologists and clinical scientists together to drive the development and application of vEM. The European Molecular Biology Organization (EMBO) and microscope manufacturers have been particularly active in supporting the organization of workshops and symposia 2, and more recently the community has self-organized around a set of working groups to collaborate globally on vEM research, development and application (see the volume EM community website).
In this Primer, we present the state of the art and future perspectives for new audiences who wish to leverage the power of vEM in their own research. We outline the principles and practical steps required to move from a living biological entity to an image volume representing the sample, covering sample preparation and imaging using a range of vEM modalities (Experimentation), consider how to handle and analyse the massive amounts of image data produced by vEM (Results) and illustrate how vEM is used in real-world research, revealing structure and function across scales from organelles to organisms (Applications). Limits to the throughput of vEM and the importance of sharing rich vEM datasets (Reproducibility and data deposition) and how bottlenecks might be overcome (Limitations and optimizations) are discussed. Finally, we consider the challenges and opportunities of vEM for mining new understanding from complex multiscale biological systems (Outlook). Although we do not include electron microscopy techniques that generate serial images from frozen hydrated samples, such as cryo electron tomography or serial imaging in the cryo FIB-SEM, we address the exciting and challenging crossover between the worlds of vEM and cryo electron microscopy (cryo-EM) in the final section of the primer.
Experimentation
A vEM experiment encompasses multiple steps (Fig.1), each of which must be carefully planned to maximize the chances of a successful outcome (Box 1). Steps to be decided on and optimized include the choice of biological sample and experimental conditions, considering that the number of replicates is limited by the complexity and throughput of the technique; the probes that will be used to mark the region of interest (ROI) within the sample and add molecular information to the ultrastructure; the sample preparation protocol(s) that will be used to preserve the sample as close to living state as possible whilst enhancing compatibility with electron imaging in a vacuum; the strategy for tracking and trimming the sample to the ROI for imaging; the choice of vEM imaging modality, considering the desired field of view and resolution of the image data; and the size and complexity of the data analysis task with respect to producing a useful biological interpretation of the results.
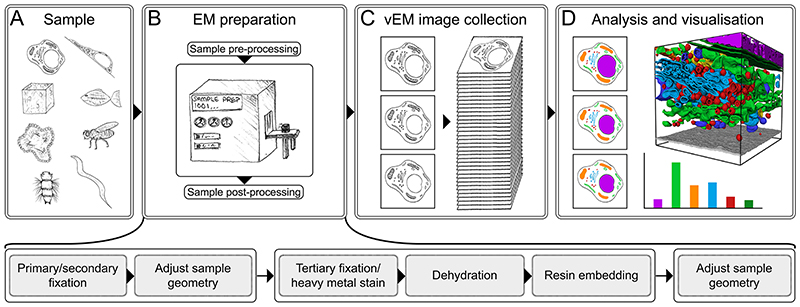
Figure 1. A typical vEM workflow.
Properties of the sample, the target structure and research question determine the overall design of a volume electron microscopy (vEM) workflow. A: A wide variety of sample types can be examined, including isolated cells and cell monolayers, individual model organisms (including Danio rerio, Drosophila melanogaster, Caenorhabditis elegans and Platynereis larvae) and model systems such as organoids and tissues. B: The sample undergoes numerous complex preparation steps to produce a specimen optimized for vEM imaging. The sample must first be preserved in as near a native state as possible using chemical or cryogenic fixation methods. In correlative vEM workflows, light microscopy is typically carried out before processing for vEM, either pre fixation or post fixation. The geometry of large samples must be modified to make them compatible with subsequent staining steps owing to limited penetration of reagents. The sample is then exposed to application-specific cocktails of heavy metals, dehydrated and embedded in resin to both introduce electron contrast to the features of interest, and stabilize the sample within the vacuum of the microscope. Once embedded, the samples then undergo a further process of application-specific geometry modification to meet constraints of the target imaging modality. C: All vEM modalities result in a stack of serial images. D: The image volume is then reconstructed, analysed and visualized using a varied mixture of manual, semi-automated and automated algorithms in opensource and commercial software. Imaged volume in panel D adapted from Heinrich et al. 158.
Box 1. Experimental design.
The choice of imaging modality for a research project should be carefully considered in view of the scientific question being asked. Key issues include the field of view required to image the entire structure of interest; the resolution required to observe the smallest feature of interest; conductivity of the sample after heavy metal staining and resin embedding, and hence tolerance of the sample to electron and/or ion beam exposure; variability of hardness of materials within the sample; and value of the sample, determining whether it can be imaged destructively (such as with tissue culture cells) or whether sample integrity should be maintained (in the case of human tissue).
Volume electron microscopy (vEM) approaches are slow and labour intensive, and so quality checks are crucial before the sample is subjected to extensive imaging and analysis. Thin section transmission electron microscopy (TEM) is appropriate to check quality of fixation and processing ahead of a vEM experiment. X-ray imaging of the resin-embedded specimens has also been used to assess overall quality for larger samples.
The low throughput of vEM limits the number of samples, and therefore the number of replicates, that can be analysed. This contributes to a difficulty in obtaining statistically robust data from biological systems with inherent variability in the population (for example, multiple cells). Quantification of objects (such as organelles and cells), lengths, surfaces, volumes, contacts between organelles and cell connectivity patterns inside a network would provide valuable information on the systems being studied, but the initial sampling strategy must mitigate the low throughput of vEM to reach significance. Often, vEM of defined regions must be used in parallel with more extensive quantitative analyses on multiple samples using 2D TEM and light microscopy.
A final issue relates to the significant challenges of processing, analysing and extracting features of interest from massive vEM image volumes. This requires specialist support, tools and software to extract meaning from the collected data. Preparations for data handling should start well before the first sample is imaged.
Sample preparation
In this primer, we refer to the initial biological material as the sample, and the prepared material that is ready for imaging as the specimen (Fig.1). Almost any biological sample can be imaged using vEM, from prokaryotic cells through to eukaryotic cells, tissues and model organisms, although the volume and resolution dictates how much of a sample can be imaged. vEM of biological samples is underpinned by extensive optimization of sample preparation 3. A successful experiment relies on protocols that ensure the sample is kept as close as possible to its native state but also in a form that can be imaged 4. The specimen must be able to withstand the vacuum inside the electron microscope, as well as the high energy and current of the electron and/or ion beam; to deliver sufficient image contrast; and to exhibit sufficient structural rigidity, especially for volume slicing 5. Protocols are often tweaked to account for the properties of each sample, target structure and imaging modality, and thus are highly variable within the community. We give a generalised overview of sample preparation below, covering optimization of the sample size, fixing the sample, staining with contrast dyes, dehydrating the sample and embedding it into a resin, and trimming the resin block.
Optimization of sample size
The maximum sample size is dictated by the depth of penetration of fixatives and stains. It is generally recommended that the sample is less than 1 mm thick in at least one dimension, although the use of customised microwave ovens to increase the permeability of samples and the diffusion of solutions is pushing this limit 6. Cell monolayers present no issues, with a thickness of around ten microns when grown on glass or plastic, and cell suspensions can be centrifuged to a pellet of suitable size 7,8. Model organisms in the millimetre range, like Caenorhabditis elegans 9,10, Drosophila melanogaster 11 and Danio rerio 12,13, may be dissected or imaged in their entirety. Tissue in the centimetre range is usually dissected into smaller pieces or cut into slices using a vibratome 14, although mouse brain has been successfully processed intact for connectomics 15.
Fixation
Two avenues can be taken for fixation, depending on the sensitivity of the biological structure under investigation and the size of the sample. Chemical fixation using combinations of methanol-free (para)formaldehyde (2–4%) and glutaraldehyde (1–4%) is straightforward; however, this may not capture fast subcellular events such as membrane tubulation 16. Alternatively, cryo-fixation using high pressure freezing (combined with freeze substitution to transition the sample to resin) captures fast biological events occurring at the scale of milliseconds and minimizes shrinkage artefacts, although this method requires complex equipment 17–19. The choice between chemical fixation and cryo-fixation is mostly determined by the initial size of the sample; high-pressure freezing is limited to samples with dimensions of approximately 200 μm, whereas millimetre-sized samples can be fixed through perfusion of chemical fixatives. As a result, cell monolayers, cell suspensions, C.elegans and D.melanogaster are particularly well suited to high-pressure freezing owing to their small size. Further detailed guidance on fixation choice is available elsewhere 3–4,14,17,19.
Contrasting
The addition of staining to enhance electron contrast of biological samples is a critical step in vEM to improve the visibility of membranes and make the sample conductive. The vEM imaging modality chosen influences the level of staining required: TEM-based techniques are compatible with lower contrast levels, partially owing to an improvement in detectors and because images are generated by scattering of primary electrons; ion beam techniques require medium levels of contrast because images are formed by backscattered electrons; and in situ microtome techniques require high levels of staining to reduce the beam dose required to generate a backscattered electron image to prevent charging artefacts and resin de-polymerization. Protocols used by the community are highly variable, but most involve immersion of the sample into solutions containing combinations of heavy metal salts to introduce contrast, including uranium, lanthanides 20, osmium (reduced or aqueous), thiocarbohydrazide, tannic acid 21 and pyrogallol 15,22. If an experiment can tolerate light staining, then uranium salts only may be used 23, but most samples require heavy staining with layers of reduced osmium, thiocarbohydrazide and osmium, followed by uranium salts and lead aspartate 24–27. These modern protocol variations are all based on historical work 28–30, with new variations optimized for specific sample types appearing regularly in the literature 6,15,22,31. Furthermore, the use of uranium is prohibited in several countries, leading to the development of replacement compounds 32,33.
Dehydration and resin embedding
To protect samples from the electron microscope vacuum and facilitate slicing of the sample into ultra-thin sections, water in the cells and tissues is removed by dehydration with a solvent that is compatible with subsequent liquid resin infiltration, which is then polymerised to form a hard block. Chemically fixed samples are dehydrated using a graded series of ethanol or acetone, whereas high pressure frozen samples are contrasted and dehydrated simultaneously during a process called freeze substitution 23,34,35. Following dehydration, a graded series of resins are used to infiltrate the sample before it is hardened using heat 36,37 or UV light 38,39. Epoxy resins such as Durcupan 40, Spurrs 41, Epon 812 and Hard Plus are most commonly used due to their good sample penetration properties and stability during imaging. Methacrylate resins such as Lowicryl HM20 and LR White may also be used, particularly when preserving the emission of fluorescent proteins in the sample.
Trimming
To prepare the sample for ultra-thin sectioning or milling, and to optimize its volume to the downstream imaging modality being used, the sample is manually trimmed using a razor blade or diamond knife in an ultramicrotome. Many samples require prior knowledge of the position of the target structure to guide trimming, which is usually achieved by integrating fluorescence microscopy and/or X-ray microscopy (Box 2) into a correlative workflow in order to locate and follow the target structure during trimming 23,35,42-46. Once trimmed, specimens may be mounted directly for vEM, or cut into ultra-thin sections using a diamond knife, depending on the downstream imaging modality. Removal of excess resin before polymerization 47–49, or use of a conductive coating 50 can minimize trimming and facilitate targeting of biological structures by revealing surface anatomy. Large resin-embedded tissue samples may also be subdivided using a hot knife for vEM of serial slabs 51,52.
Box 2. X-ray microscopy.
X-ray microscopy, and in particular computed tomography, provides 3D information about the structure and morphology of intact samples 283,284. In many ways, the data that computed tomography provides are similar to that of volume electron microscopy (vEM) but at lower resolution. For example, elements with a higher atomic number tend to have both higher X-ray attenuation and higher electron scattering properties. Therefore, many of the same biological structures are visible with both techniques - especially when contrasted with heavy metals - and images can have a very similar appearance 45,46,285–287. Tomography works by taking 2D images of a sample from many different angles and combining these via a reconstruction algorithm to give a 3D tomogram. X-ray tomography can be done in the laboratory using laboratory-based X-ray systems or at large synchrotron facilities. In general, laboratory based systems can reach sub-micron resolutions with scan times in the range of a few hours to a few days 44,283, whereas synchrotrons can achieve resolutions of tens to hundreds of nanometres 283,285,287 (depending on the set-up chosen and the technique employed) with scan times in the range of seconds or minutes 283,285, although the highest resolution scans still take several hours 287. However, the benefits of synchrotron X-rays come at the cost of reduced accessibility, and care must be taken to avoid damage to the sample from high doses of X-rays 288,289. X-ray microscopy has many applications when combined with vEM. For example, computed tomography can be used to quickly screen samples to assess structural preservation 173 and stain penetration 6 prior to vEM. With its large field of view, it is also very useful as a targeting technique for correlative light and volume electron microscopy (vCLEM), as in conjunction with fast scan times it can efficiently produce 3D maps of samples to guide electron microscopy acquisition of regions of interest 44–46,99,286. Finally, 3D X-ray imaging can be used in its own right as the primary technique for morphological studies of samples, as demonstrated in recent studies tracing neurons in Drosophila and mouse brain 285,287.
Probes
vEM techniques provide an opportunity to correlate morphological data with molecular characterization. The ideal labelling method for vEM should enable molecular localization throughout the depth of the sample, providing molecular resolution with the sensitivity to detect molecules at endogenous levels in their native non-perturbed environment. This is challenging but has been achieved through the development and application of a suite of genetically encoded tags.
Genetically encoded tags that produce electron-dense reaction products that are visible in vEM have been described, including metallothionein-based probes 53–56, reASH 57, miniSOG 58 and APEX/APEX2 59. Of these, APEX2 (an evolution of the original APEX probe that has higher activity in cells) is the most established tag. It is an ascorbate peroxidase that is active in both cytosolic and luminal/extracellular compartments, and is proving to be a robust method with a large number of applications including detection of directly tagged proteins of interest 60, nanobody-directed detection of GFP and RFP-tagged proteins 61 and labelling of specific protein complexes using a split GFP-based method 62 or split-APEX 63. APEX converts soluble diaminobenzidine (DAB) into an insoluble electron-dense reaction product 59. APEX can be used with chemically fixed specimens or in cryo-EM schemes (cryoAPEX) 64 and has widespread applicability in both cell and tissue/animal systems, even enabling multiplexed labelling of different cell populations in tissue 65. The APEX-directed DAB reaction product can also nucleate the formation of gold colloids 66,67, a method compatible with single-molecule detection as shown using genome-edited cells with tagged proteins at endogenous levels 67. Single molecule detection is also possible using an elegant technique based on a metallothionein tag that can be used to nucleate gold particles and, potentially, could have applications in different vEM modalities 68.
Fluorescent genetically encoded tags can also be used to locate structures in vEM, either indirectly or directly, using a range of correlative light and volume electron microscopy (vCLEM) workflows. In addition to genetically encoded tags, antibodies and neural tracers have been used to label organelles and cells for vEM. The recent development of a method for immunolabeling throughout larger volumes of tissue (sections up to 1 mm thick), without permeabilization, retains good ultrastructure and allows the collection of molecular information using vCLEM with SBF-SEM 69. Array tomography 70,71 allows labelling on sections, removing the limitation of depth penetration into hydrated tissue samples, but is dependent on the availability of electron microscopy-compatible antibodies and epitope preservation during sample preparation. Using a combination of neural tracers 72, expressed fluorescently-tagged proteins and on-section immunolabelling opens the possibility of multiplexed labelling in electron microscopy. Another strategy for multiplexed labelling with ssTEM is to pull out a few ultra-thin sections from the series, stain them using immunohistochemistry 73 or immunogold labelling 74 and, then, propagate the molecular information through the serial image stack.
TEM-based vEM
Volume imaging using TEM (Fig.2) combines the benefits of high resolution, non-destructive imaging with the speed of full-field (as opposed to scanning) acquisition using CCD or CMOS cameras. The geometry of the transmission electron microscope limits the size and thickness of the specimen that can be imaged so an ultramicrotome with a diamond knife is used to first cut the sample into serial ultra-thin sections, which are picked up on support grids 3 mm in diameter. For ssTEM, section thickness is generally between 50 and 100 nm. For ssET, slightly thicker sections of 200 to 300 nm are imaged, and resolution through the volume is increased by collecting a series of images at different tilt angles (usually ±70°), which are then reconstructed into a digital volume. To improve the reconstruction, dual tilts may be collected at 90° to each other. In both ssTEM and ssET, serial sections are imaged sequentially and the resulting images stacked to represent the sample volume.
Figure 2. vEM encompasses a collection of closely related imaging modalities.
Volume electron microscopy (vEM) is a collective term for numerous imaging modalities based on transmission electron microscopy (TEM) and scanning electron microscopy (SEM). Within each modality, overall sample position (dotted box, with a red spot indicating the location of electron beam interaction) and signal generation mechanisms remain broadly similar. For TEM, electrons that pass through the sample are collected by a downstream detector. For SEM, backscattered (BSE) and/or secondary electrons (SE) are generated by electron beam interaction with the sample and recorded using in-chamber or incolumn detectors (pale blue). However, the format of sample and mechanism used for generating serial images vary. For TEM, serial ultra-thin sections of 50-70 nm thickness are typically collected on substrates coated with an electron transparent support film such as formvar-coated copper slot grids (serial section TEM (ssTEM)). 3-D volumes can also be generated by imaging through serial sections of 200-300 nm thickness at multiple tilt angles, back projecting resultant images and reconstructing the volume (serial section electron tomography (ssET)). Alternatively, very extensive collections of serial ultra-thin sections can be imaged using a tape-based support system that feeds directly through the electron column (GridTape TEM). For SEM, options range from sequentially cutting and imaging the sample using a miniaturized in-chamber ultramicrotome (serial blockface SEM (SBF-SEM)) or directly milling from the imaged surface using a gallium ion beam (focused ion beam SEM (FIB-SEM)) or a variety of plasma moieties (pFIB-SEM). Enhancements such as sample bias, modified sample geometry and closed-circuit milling control are integral to some advanced workflows (enhanced FIB-SEM (eFIB-SEM)). Imaging of serial ultra-thin sections collected on a suitable substrate such as silicon wafer or indium tin oxide-coated glass (array tomography), or Kapton tape (array tomography with automated tape-collecting ultramicrotome (ATUM)) can also be achieved using SEM, with either a single electron beam, or parallel electron beamlets.
ssTEM delivered the first of the whole model organism electron microscopy volumes in the Mind of the Worm, a map of the neural connectivity of C.elegans 75. ssET has also delivered insights into the complexity of subcellular structures 76,77. However, these techniques are slow, low throughput, manual and technically challenging, severely limiting the volumes that can be imaged. Efforts to address these limitations in TEM-based vEM have focused on solving the dual challenges of the limited field of view imposed by commercial detectors and automation of section retrieval onto electron-transparent supports for imaging in transmission mode.
One of the first computational solutions for large-scale ssTEM imaging enabled the automated acquisition and assembly of thousands of individual images from each section into a large mosaic, to acquire a terabyte volume of rabbit retina 78. On the instrumentation side, a fruitful approach was the development of TEMCA 79, which used a commercial 120 keV TEM customised with an array of detectors combined into a single large sensor. The microscope was supported on a raised platform to increase the distance between the sample and the detector, thereby increasing the field of view. However, although detection was optimized, these approaches still relied on manual collection of thousands of sections onto classical TEM slot grids. This issue was partially addressed using an autoloader on the TEM, with which grid exchange was automated using a robot synchronized with the imaging process 80–82. Grid handling was further improved by the development of GridTape TEM 83,84 (Supplementary Movie 1), in which sections were cut using a modified ultramicrotome for collection on a tape with electron transparent film-covered holes. The tape was fed through a modified sample holder on a customised transmission electron microscope, and each section automatically imaged using a fast array of detectors, enabling the motor control circuits in adult D. melanogaster to be mapped 84. In parallel, a second system called piTEAM has been developed, consisting of a parallelized array of transmission electron microscopes optimized for GridTape imaging 85. The speed, automation and non-destructive nature of GridTape TEM has the potential to transform peta-scale vEM imaging while, simultaneously, creating new challenges in data handling and analysis.
SEM-based vEM
Volume imaging using SEM (Fig.2) combines the benefits of large fields of view and a microscope chamber compatible with large specimen holders and in situ specimen slicing mechanisms. SEM-based vEM has been widely adopted and includes some of the most user-friendly vEM technology, allowing routine acquisition of large-volume image data with ultrastructural resolution 5,86–89. Three complementary but distinct approaches have been developed in parallel: those using in situ microtomes such as SBF-SEM; those using ion beams such as FIB, plasma FIB (pFIB) 90 or enhanced FIB-SEM (eFIB-SEM) 52, and array tomography 5,70,87. These three approaches all require heavily contrasted specimens; a scanning electron microscope optimized for low voltage imaging (1-2 keV) with a field emission gun and backscattered or secondary electron detectors; and dedicated software and a stable system for autonomous imaging over days, weeks or even months.
In situ microtomes
There are currently three commercial SBF-SEM (Fig.2 and Supplementary Movie 2) products available, to our knowledge - 3View (Gatan Inc., now part of Ametek Inc.), VolumeScope (ThermoFisher Scientific Inc.) and Katana (ConnectomX Ltd) - as well as several home-made solutions 91. All rely on the same principle - a diamond knife held in a miniaturised ultramicrotome mounted in the SEM chamber moves back and forth across the resin block, with the block moving upwards in z in incremental steps that determine the section thickness 92,93. In all solutions, image tiling is possible by moving the entire microtome in the chamber. This technique has the advantage of imaging large volumes 50,91 but the disadvantage of suboptimal working distance, restricting resolution and choice of detector. Charging artefacts caused by poor conductivity of the sample blocks are a common problem 94. The first versions of in situ microtomes relied on variable pressure in SEM to mitigate charging 92, which is a particular problem for specimens that contain large areas of non-conductive empty resin such as cell pellets or lung tissue. Now, newer instruments use a focal charge compensation system 95 to inject a gas flow locally at the level of the blockface to mitigate charging, increasing the range of specimens that can be imaged.
Ion beams
Alongside the development of in situ microtomes, researchers and manufacturers explored the usage of a gallium FIB to remove thin sections from resin blocks inside a scanning electron microscope 96–98 (Fig.2 and Supplementary Movie 2) By replacing mechanical sectioning with electronically controlled ion milling, this technique has the advantage of shaving thinner slices than in situ microtomes (typically 50 nm for in situ microtomes versus 5 nm for FIB-SEM) and achieves higher resolution with a better signal-to-noise ratio 52. The possibility of obtaining isotropic voxels, where the slice thickness in z matches the pixel size in XY, makes FIB-SEM attractive for automatic segmentation 99. Uniquely, the direction of ion beam milling is perpendicular to that of other sectioning techniques, an important point to consider during experimental design, to ensure that the region to be imaged is accessible to the electron beam 100. However, the ion beam loses energy as it progressively mills down the blockface, limiting the depth that can be precisely cut 52. In order to improve speed and throughput, specimens have been imaged in parallel on multiple eFIB-SEM instruments that are more stable and resilient to errors over imaging runs of many months 52. As negative charges imparted to the imaged surface by the electron beam are mitigated by subsequent exposure to positive ions from the milling beam (and concomitant removal of this charged slice), FIB-SEM can be used to image poorly contrasted specimens in which the emission of fluorescent proteins has been preserved 23. Recently, pFIB instruments with other ion species have been used for milling biological specimens, including oxygen 90, xenon 101 and argon 102,103, which may offer finer or faster milling than gallium. Newer instruments use oxygen plasma 104, gas cluster ion beams 105 and multi-ion species to increase the milling speed and surface area.
Array tomography
Whereas in situ microtomes and ion beams both work by iterative imaging of the blockface, array tomography is based on SEM imaging of pre-cut ultra-thin serial sections arranged in two dimensions on a solid substrate (Fig.2 and Supplementary Movie 1). Serial sections are often obtained using a standard ultramicrotome and collected on either silicon wafers 106,107, or carbon-coated 108 or indium tin oxide-coated glass coverslips 107,109. Manual section pickup is challenging, requiring advanced expertise and manual dexterity, and is commonly used for smaller numbers of serial sections (< 500) 110. To overcome this limitation, various automated or semi-automated methods have been developed 71, including the Artos 3D (Leica Microsystems), the ASH-100 107 and ASH2 (RMC Boeckeler), fishing 111, MagC 112, a modified knife 106, AutoCUTS SEM 113 and the Arraybot 71. An alternative and reliable approach for producing large numbers of serial sections is the automated tape-collecting ultramicrotome (ATUM) 114. This system, as the name implies, collects the serial sections on tape, which is then mounted on wafers for imaging. Commercial solutions for array tomography are becoming more widely available, mainly facilitated by advances in software automation for serial imaging. For imaging large volumes, mSEM and FAST-EM offer fast imaging at nanometre resolution by splitting the electron beam into multiple beamlets that image the sample in parallel 115–117. Compared to in situ microtomes and ion beams, array tomography has a lower z-resolution (a minimum of 30 nm). However, because this approach preserves the sections, it has the advantage that the specimen can be re-imaged in different areas or at different magnifications. Furthermore, the sections can be imaged at the light level prior to electron microscopy, to collect molecular information from immunofluorescence, neural tracers or genetically encoded probes 70,71,107,108. Integrated light and scanning electron microscopes are a promising solution for efficient acquisition and registration of fluorescence microscopy and electron microscopy images from array tomography specimens 109,118,119.
Multi-vEM workflows
The research question, sample size and target feature may dictate a multiscale, multi-resolution workflow (Fig.3). In these circumstances, a multi-vEM approach is very powerful. For example, SBF-SEM can be used as a smart trimming tool to locate smaller regions for higher resolution imaging using FIB-SEM 12,120, TEM 12 or spatial compositional analysis using nanoSIMS 121. Other hybrid approaches have focused on the combination of thick serial sectioning and FIB-SEM to rapidly screen large volumes with acquisition of high-resolution isotropic information for targeted sub-volumes 122,123.
Figure 3. vEM as a multiscale multimodal imaging technique.
Volume electron microscopy (vEM) continually pushes the boundaries of high-resolution volume imaging. Transmission electron microscopy (TEM)-based imaging modalities (serial section electron tomography (ssET) and serial section TEM (ssTEM)) tend to deliver higher resolutions, whereas scanning electron microscopy (SEM)-based modalities (serial blockface SEM (SBF-SEM), array tomography and focused ion beam SEM (FIB-SEM)) tend to deliver larger volumes. Multimodal workflows, emerging vEM technologies such as GridTape TEM, multibeam SEM (mSEM), enhanced FIB-SEM (eFIB-SEM), plasma FIB-SEM (pFIB-SEM) and FAST-EM, and associated techniques including X-ray microscopy are further extending scales and resolutions over which volume imaging is possible.
vCLEM
Correlative microscopy techniques are an essential part of most vEM workflows, because the volume that can be imaged in electron microscopy is so much smaller than the size of most biological samples. vCLEM may be used to target and track a structure between two imaging modalities, or may be used to localise molecules to structures within the volume, providing clues to their function (Fig.4).
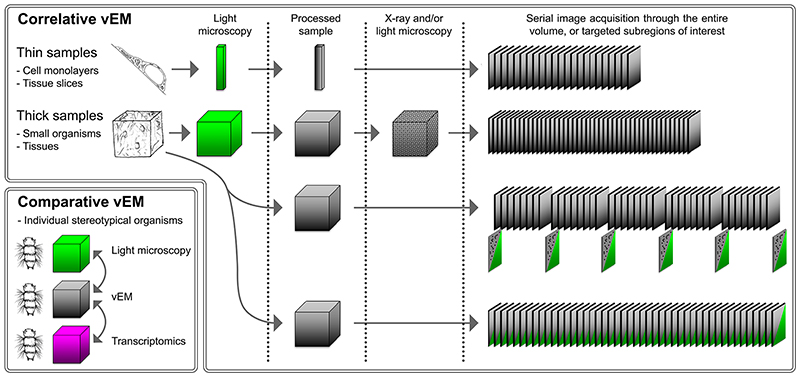
Figure 4. Correlative and comparative vEM for function-structure studies.
Volume electron microscopy (vEM) studies frequently use multimodal imaging pipelines. Light microscopy, in particular, can be used to obtain a wide variety of molecular, developmental or functional information from a sample prior to processing for electron microscopy. Light microscopy can also be used to locate a region of interest (ROI) for targeted imaging (correlation for relocation) or to localize molecules to structures (correlation for registration). In thin samples, such as cell monolayers or tissue slices, the final volume is relatively small and correlation between light and electron imaging modalities is more straightforward. For successful correlation in thick samples, intermediate imaging steps are often required such as X-ray and light microscopy, in order to provide a link between low and high-resolution datasets and help to relocate the target area of interest prior to serial image acquisition throughout the target volume. Alternatively, intercalated serial section transmission electron microscopy (ssTEM) data sets can be probed by computational molecular phenotyping 73, or multiplexed siGOLD immunolabelling (black dots) 74, thereby providing a wealth of functional information that can be translated across the whole dataset. To further enhance a multiplexed volumetric approach, all sections can be used for each imaging modality, combining immunofluorescence and electron imaging modalities to link functional identification to the underlying tissue ultrastructure 108. Finally, for small stereotypical organisms such as Platynereis larvae, comparative analysis can be used to draw powerful conclusions for populations by correlating multimodal data from individual organisms 50.
vCLEM for tracking at the single-cell level tends to rely on external coordinate systems such as gridded coverslips for coarse targeting 7,124 At the tissue scale, the problem becomes more complex because of the sheer volume of the sample compared to the size of the target structure, and workflows often rely on anatomical features such as blood vessels as landmarks for correlation 44,125–127. One option that has been employed to great effect in large specimens is the use of high-power lasers for branding marks within the specimen or on the resin block, laying a trail of breadcrumbs to individual cells and features of interest 23,44,109,128–131.
vCLEM for molecular localization uses high-resolution correlation to overlay fluorescent microscopy images of fluorescent proteins acquired post embedding with electron microscopy images for function-structure studies. Methods have been developed that allow retention of the fluorescence of proteins after resin embedding, delivering accurate correlation of fluorescence to ultrastructure 23,118,132–138. The discovery that fluorescent proteins showed blinking properties within the low-vacuum environment of SEM has allowed molecular localization at super-resolution scales 118. A recent study has carefully optimised the use of in-resin fluorescence, resulting in a complete and robust workflow for vCLEM 23. A special case of vCLEM is correlative array tomography 70–72,108,139, where molecular information from antibody immunolabeling, fluorescent proteins or neural tracers is acquired from the same ultra-thin sections as imaged using electron microscopy, thus facilitating registration of the different modalities and enabling multiplexed molecular labelling.
Results
Data acquisition
All commercial electron microscopes are bundled with proprietary acquisition control software that allows users to set parameters and manually acquire images. Parameters to be optimized include voltage, probe current, field of view, pixel size, slice thickness, dwell time and exposure (depending on the imaging modality used). For vEM, additional software is needed to manage the acquisition workflow and drive the stage, the ultramicrotome or the ion beam, for example, DigitalMicrograph (3View, Ametek Inc.), Maps (VolumeScope, ThermoFisher), Atlas 5 (Zeiss) and custom software with a tablet interface (Katana; ConnectomX). To extend the capabilities of commercial software and offer more flexibility, control, specialized features and ease of use, several open-source solutions have been developed. The earliest example is SerialEM, an acquisition control software developed to perform automated electron tomography with tilt series 140. WaferMapper is a MATLAB software package that automates image acquisition from sections collected on wafers for array tomography 141. SBEMimage is an open-source Python application designed to control complex image acquisition workflows with SBF-SEM 142 and array tomography 112. piTEAM 85 enables large-scale TEMCA image acquisitions, with core components controlling the microscope(s), a web-based graphical user interface, and a graphics processing unit-based image processing pipeline. Automated targeting of regions of interest for FIB-SEM can be achieved with CLEMSite 143.
Data processing
Data processing in vEM tends to be customized into application-specific pipelines, lifting modular components from different software platforms.
Volume reconstruction
After image acquisition, the series of individual images must be reconstructed into an image volume. This step may include alignment of serial images within a single volume, and subsequent stitching together of multiple image volumes, depending on the size of the original sample. Alignment of adjacent images should recover continuity across the image volume and retain the geometrical properties of the original sample, despite distortions that may occur during sample preparation and image acquisition. Cross correlation 92,144 and Scale Invariant Feature Transform (SIFT) 145 are commonly used techniques for the detection, characterization and matching of local features in image series. The SIFT concept was expanded to automatically register large image mosaics by globally minimizing the registration error, implemented as part of TrakEM2 in ImageJ/Fiji 146–149. Another method called Alignment to Median Smoothed Template (AMST) 150 was developed to align FIB-SEM datasets. Elaborating on sequential use of rigid and affine transformations, it has the advantage of correcting for local pixel size variations whilst preserving long-range alignment of structures. Following the general trend of exploiting deep learning for computer vision tasks, the largest currently available vEM volumes have been aligned through new learning-based procedures, combining defect detection, short-range and long-range alignment 151,152. In current practice, machine learning techniques are not used for alignment. However, there is ongoing research implementing this as a strategy 153.
Image enhancement
vEM datasets can be notoriously challenging to analyse due to their high dimensionality, texture, variations in intensity within and across volumes, noise and resolution loss due to sampling. Image enhancements to improve contrast or noise characteristics may be applied to the serial images. Traditional methods may be used to reduce noise within an image, for example contrast limited adaptive histogram equalization (CLAHE) 154, available as a standard image processing option in Fiji. Content-aware image restoration methods have also been applied to vEM data sets to improve the signal-to-noise ratio. In general, while supervised network training works well for SEM reconstruction, TEM data requires semi-supervised (Noise2Noise) or unsupervised (Noise2Void) training methods 155.
Multimodal registration
For vEM datasets collected as part of a correlative or multimodal workflow, an additional step is required to correlate the different datasets to each other. Here, we clarify correlation for relocation as the overlay of shared features or patterns between two or more imaging modalities, with the intention of targeting an ROI for imaging. On the other hand, correlation for registration is the accurate (and often computational) alignment of data sets from two or more modalities, with the aim of imaging cells or macromolecules that were preferentially labelled in a subset of the modalities. As an example, X-ray microscopy can reveal the position of a neuron or cell for downstream imaging by vEM, whereas overlay of fluorescence microscopy and vEM data sets acquired from the same volume can reveal the identity and ultrastructure of an organelle containing a fluorescently tagged molecule. Registration may also be used to align image volumes from different specimens from a population to create an average atlas of ultrastructure of a model organism 156,157.
To register multimodal data, a fiducial of some sort is required. Fiducials are often specific to - and constrained by - the sample type and biological question. With array tomography datasets, where different modalities come from the same ultra-thin sections, registration of fluorescence microscopy and vEM can be done with high precision in 2D, and thus in 3D, and cell nuclei are often used as fiducials 108. In vCLEM studies, mitochondria 35,158 and nuclei 50 are popular internal fiducials, as they are easily labelled with chemical dyes or fluorescent proteins and can generally be recognized in EM images by their morphology. Software packages such as 3D slicer, eC-CLEM 159, BigWarp 160 and elastix 161,162 offer varying degrees automation for multimodal image alignment.
Image segmentation
vEM image segmentation
Image segmentation - the detection and delineation of structures of interest - is required for extraction of quantitative information from rich vEM datasets. Non-discriminant contrast, diversity of appearance of structures, and large image volumes turn vEM segmentation into a highly non-trivial problem, where cutting-edge methods relying on state-of-the-art computer vision techniques are still far from reaching human parity in segmentation accuracy. Although significant progress has been made in recent years, largely due to the introduction of deep learning-based methods, there is not yet a single reliable and easy-to-use solution for fully automated segmentation of vEM images. Practitioners must choose between (or combine 163) manual, semi-automated and fully automated solutions based on the difficulty of the segmentation problem, the data size and the computational expertise and resources of their team or institution. Furthermore, almost all automated solutions rely on machine learning and may require large amounts of example segmentations to train a model, although in some cases models trained for the same task on similar datasets are available and can be applied directly.
Manual and semi-automated segmentation
For small datasets, for subsets of large datasets, and for datasets in which only a few structures are to be analysed, manual or semi-automated approaches are often preferable. Here, ready-made solutions that do not require computational expertise or example segmentations are available. Open-source solutions exist: the ilastik carving workflow 164 enables users to sequentially extract objects from a volume by labelling parts of the image that belong to the object and the background (everything else) and using a region growing algorithm for segmentation. Microscopy Image Browser (MIB) 165 also contains a set of tools for semi-automatic segmentation based on thresholding, morphological image operations and region-growing. The software suite IMOD 166 provides manual and semi-automatic drawing tools for tracing and refinement of organelles. Amira (ThermoFisher Scientific) and Imaris (Oxford Instruments) are commercial software packages that offer semi-automated segmentation combined with advanced visualization capabilities. The Fiji plugin TrakEM2 147, Reconstruct 167, Webknossos 168 and VAST 169 are also used. Note that these tools can often only load data up to a certain size. To overcome this limitation, one can downsample the data if the structures of interest are discernible at lower magnification, or crop regions of interest. Downsampling or cropping can be achieved with Fiji, for example using the BigDataProcesser plugin 170, or programmatically for even larger data using Dask or other libraries that provide easy access to task parallelization. These tools only support a certain set of image data formats. Bioformats 171, which is integrated within Fiji, can be used to convert between all common data formats.
Automated segmentation
If the data size exceeds the capabilities of semi-automatic segmentation, for example when structures of interest need to be fully and densely segmented in a large volume, fully automated methods must be employed. These methods are almost universally based on machine learning: they use example data in the form of ground-truth segmentations to estimate parameter values for the segmentation model, which can range from a simple classifier (rarely used in vEM due to the complexity of the task) to a very deep neural network with millions of parameters. Initially, fully automated segmentation was developed for connectomics. This application requires the complete segmentation of neurons and synapses in large volumes and is thus only feasible with a high degree of automation. Development has been significantly advanced through open challenges (such as ISBI2012 172) leading to methods that enable circuit reconstruction for the brains of small organisms 173 or brain regions 91. Recently, large-scale segmentations have also been achieved in other domains such as cells and selected organelles in a complete small organism 50 or many organelles in cells 99,158.
Machine learning methods can be trained to produce different flavours of segmentation, labelling the pixels either by semantics (for example, label all mitochondria pixels as 1 and the rest as 0) or by the objects they belong to (for example, label all pixels of the first mitochondrion as 1, of the second mitochondrion as 2, of the nth mitochondrion as n, with non-mitochondrion pixels as 0). Furthermore, a semantic segmentation can be transformed into an object segmentation by connected component labelling or a watershed transform. This approach is often used for separated objects such as mitochondria or nuclei and can be found, for example, in the Fiji plugin MorphoLibJ 174. Alternatively, semantic segmentation can be used to predict boundaries between objects. The object segmentation is then achieved by a watershed transform or graph-based agglomeration. This approach, often used to segment cells or densely packed vesicles, is also available in MorphoLibJ or in the ilastik boundary-based segmentation workflow 164. Finally, methods that directly predict an object segmentation have now been introduced for light microscopy 175,176 and, very recently and only in the form of research code, for vEM data 157.
Generally, machine learning-based segmentation models can be divided into two categories: feature-based learning and deep learning. Feature-based learning methods use a set of predefined features (usually linear and non-linear image filters) as input to a non-linear classifier such as a support vector machine or a random forest that outputs the (semantic) segmentation. They need few examples and are available via user-friendly tools. Methods using deep learning do not rely on pre-computed features but, instead, learn features and segmentation jointly. They can solve more difficult segmentation problems, but their superior accuracy requires much larger amounts of examples, and the training must be performed on graphics processing units. The U-Net 177 is a widely used deep learning model architecture for segmentation in vEM, employed as a backbone in most state-of-the-art segmentation methods (Fig.5).
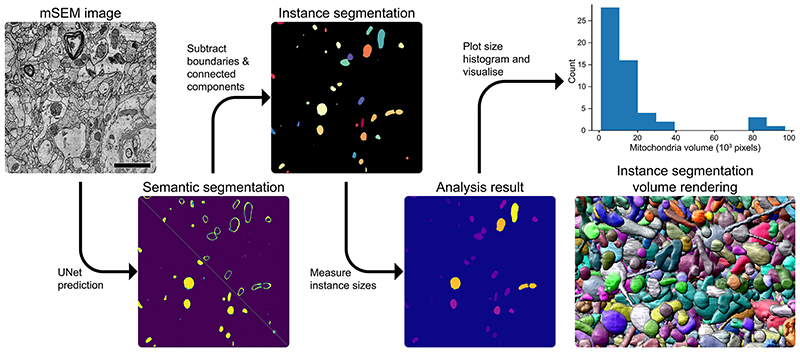
Figure 5. A typical vEM image analysis pipeline.
In this example, mitochondria have been segmented from an array tomography image stack acquired using multibeam SEM (mSEM) (MitoEM data challenge). The analysis pipeline contained the following steps: mitochondria pixel probabilities and boundaries were predicted with a pretrained UNet, boundary probabilities were subtracted from mitochondria probabilities and connected components applied to achieve an instance segmentation. Size per instance was measured and projected to the image and, finally, the extracted mitochondria volumes were plotted as a histogram. All analysis was performed in three dimensions. An example volume rendering of the instance segmentation is also shown, demonstrating number and complexity of mitochondria in a sub-volume of the data set. Scale bar in mSEM image = 2 μm. mSEM image courtesy of the Lichtman lab at Harvard University.
Feature-based segmentation methods can be trained interactively from brush-stroke annotations and are implemented in user-friendly tools such as the ilastik pixel classification and autocontext workflows 164 and the Fiji Weka plugin 178. Training deep learning models is more challenging than feature-based models, as these require significantly more training data, computational expertise and resources. Consequently, most user-friendly tools delegate the training step to method developers and implement prediction with pretrained models for end users. These tools include DeepImageJ 179, the ilastik neural network workflow 164, DeepMIB 180, CDeep3M 181, epanada 182 and APEER (Zeiss). For successful application, the model needs to be trained on data very similar to the data at hand, preferably using generalizability-enhancing tricks such as data augmentation, while unsupervised pretraining on large unlabelled and heterogeneous datasets may increase model performance overall 183. All tools listed above provide a set of pretrained models and the recent Bioimage Model Zoo community project hosts models in a cross-compatible format that is currently supported by DeepImageJ, ilastik and ZeroCostDL4Mic 184. While some of these tools (CDeep3M, APEER) also implement network training, they do not eliminate the need for many segmentation examples and a graphics processing unit. Given these requirements, for practitioners with some computational expertise it may be easier to train a new model directly using Python scripts, which provides more flexibility. An intermediate solution is the use of prepared Jupyter notebooks such as those provided by ZeroCostDL4Mic 184 that come with a running environment and use publicly available computational resources.
Efficient training and post-processing procedures for deep learning methods constitute an active area of research. Most of these methods are made available as Python code using one of the most popular deep learning frameworks (PyTorch or TensorFlow), and they can be retrained, extended and applied through programming in Python. Mala 185, MWS 186 and SegEM 187 were originally developed to segment neurons but can be used for other segmentation tasks as long as objects have discernible boundaries. The approach of flood filling networks 188 has been used for many large-scale connectomics segmentations 116,173, and synapse detection and synapse partner prediction tasks 189,190. Training deep learning methods for new data requires many segmentation examples; large and heterogenous training datasets may produce more generalist models 182. These examples can be generated using the manual/semi-automated tools introduced earlier or more general-purpose annotation tools such as napari 191 and Fiji plugins (like LabKit). Alternatively, proofreading tools can be used to correct an initial segmentation, for example FlyWire 192, Paintera 193 or Dojo 194. These tools enable faster generation of examples, but generally have a steeper learning curve and less documentation. They can also be used to correct errors in the final segmentation results.
Although the methods listed above can be wrapped in scripts and notebooks to be made accessible to practitioners of limited computational experience, application of these methods to large data sets, from tens of gigabytes to hundreds of terabytes, requires significant computational resources and experience with large-scale distributed computing. For such teams, open-source solutions for large-scale deployment exist and can be freely exploited 188,195–197.
Commercial solutions and crowdsourcing
If the segmentation problem at hand is not feasible with the tools introduced above, for example owing to missing pre-trained models or lack of computational expertise, the task can also be out-sourced to companies offering segmentation as a service, for example scalable minds or ariadne.ai. For large projects, crowdsourcing of the segmentation effort is also an option, which has successfully been pioneered for segmenting retinal neurons 198 as well as cells and organelles 199, and is currently underway for neurons in the fruit fly brain 202.
Examples of image segmentation workflows
Associated with this Primer, we provide two example data analysis workflows for neuron and mitochondria segmentation. The first example demonstrates how to use a pretrained network for neuron boundary segmentation in ilastik, followed by graph-based post-processing to obtain an instance segmentation. The second example demonstrates mitochondria segmentation for a large electron microscopy volume using a pretrained network and Python libraries. For both examples, alternative approaches are sketched.
Data exploration and visualization
Interactive visualization of large vEM datasets is challenging. Important features for visualization software are the ability to view the data from arbitrary (non-axis-aligned) angles, surface mesh rendering for displaying segmentation results, and support for viewing large datasets. Many of the software platforms already mentioned provide options for data visualization and exploration, including Fiji, napari, IMOD and MIB. However, exploration of very large datasets is not possible with many visualization tools, because they rely on loading all data into memory or only provide inefficient access to data out of memory. To overcome these limitations, support for chunk-based data access and a precomputed multi-scale image pyramid format is necessary and is offered by some microscopy vendors. In addition, support for on-demand access of data stored in the cloud is desirable. In Fiji, the BigDataViewer 200 and MoBIE 201, plugins can be used for exploring large data. For collaborative skeleton tracing and connectome analysis, Catmaid 202, Neuroglancer 203 and tools from the Knossos family (Knossos 204, pyKnossos 125 and webKnossos 168) provide similar functionality and extend support to collaborative annotation for volumetric segmentation.
Applications
Conventional 2D imaging with TEM requires the trained eye of an expert to interpret the intricate topology of organelles within the crowded environment of a cell. A particularly elegant, and accurate, interpretation of the sarcoplasmic reticulum was made by Porter and Palade in 1957 205. In this section, we have selected a few examples to illustrate the breadth of vEM applications following a scale logic, from single organelles through to cell-cell interactions, small organisms, and tissues. We do not include applications in the physical sciences, but it should be noted that FIB-SEM techniques, in particular, are commonly used in the semiconductor and materials sciences. This list of applications is far from being exhaustive, and we encourage readers to perform literature searches to find examples of vEM data representing their own organelles, cells and tissues in this fast-evolving field.
vEM within a single cell
The cytoskeleton
Most cytoskeletal elements and organelles span greater distances than the typical thickness of a TEM section (30 to 100 nm), and therefore require vEM to be seen in their entirety (Fig.6). Yet, they are usually challenging to detect by electron microscopy as their cross-sectional dimensions are often close to the minimum pixel size of the given acquisition technique. Microtubule spindles, for example, have been reconstructed using ssTEM tomography 206 at sufficient resolution to count microtubules, measure their length and visualize how they interact with the kinetochore (resolution of 2.3 nm). However, the acquisition and processing of serial tomograms, and, most importantly, the segmentation of individual microtubules, represent a major effort that very few laboratories have undertaken to date. More recently, state of the art FIB-SEM data at 4 nm isotropic resolution have reaffirmed the extent of single microtubules in cultured cells 158 and pancreatic β-cells 163, illustrating how microtubules connect with multiple organelles.
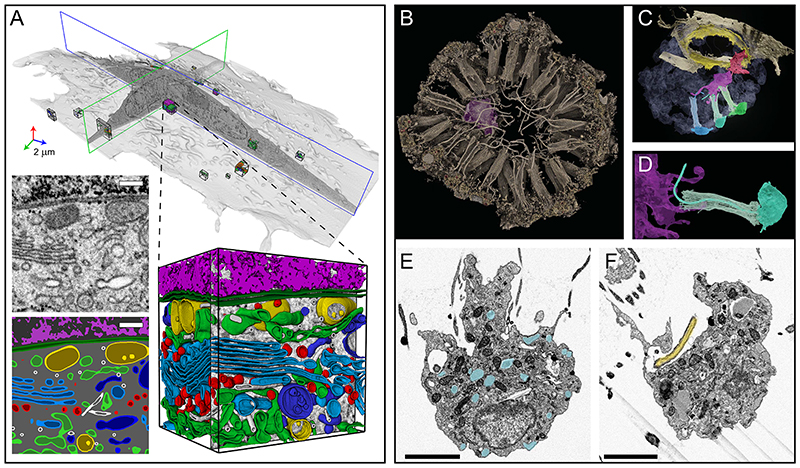
Figure 6. Examples of multiscale vEM on organelles and cells.
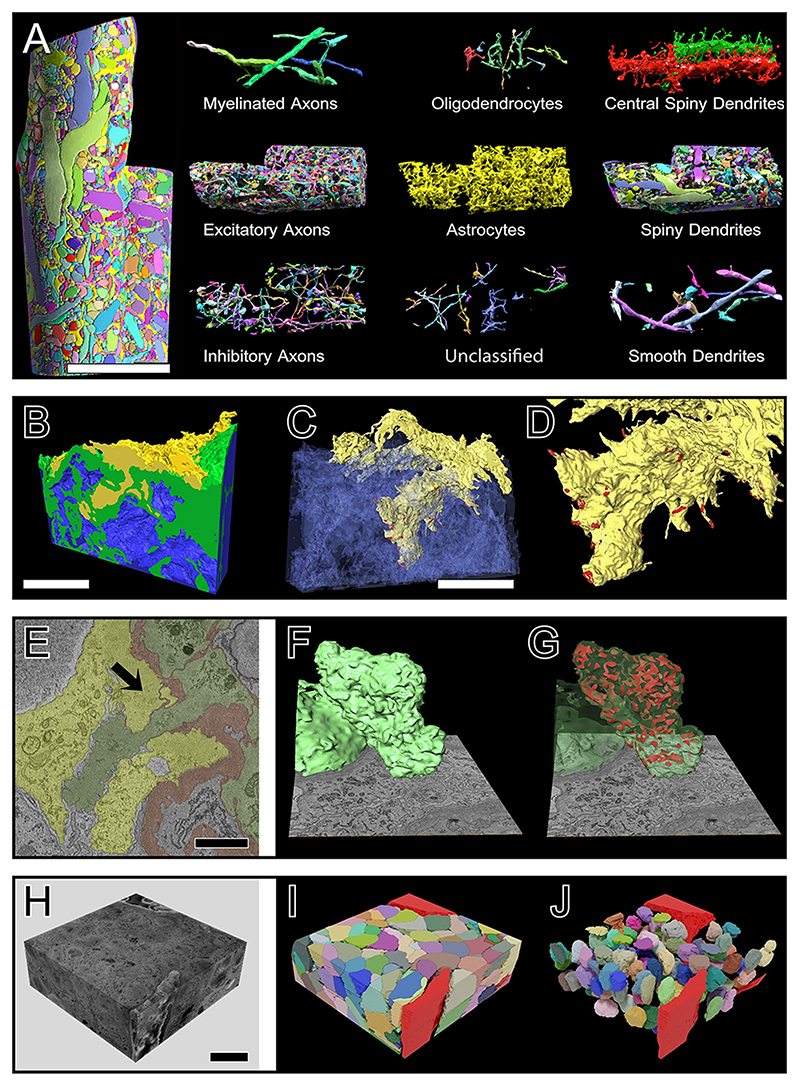
A:Segmentation and analysis of organelle volumes in a HeLa cell data set, acquired using enhanced focused ion beam scanning electron microscopy (eFIB-SEM) 150,158. Fifteen manually annotated training blocks were produced from a whole cell to train deep learning models to classify different organelles within the volume. Shown are a 3D rendering of one training block (right inset, 1.2 × 1.2 × 0.95 μm), with a single eFIB-SEM slice (top left inset), and annotation of every voxel within this slice (bottom left inset). Scale bars for insets = 200 nm. B-F: Analysis of cell diversity and nervous system evolution in sponges, data sets acquired using FIB-SEM to highlight neuroid-choanocyte interactions 46. 3D volume of entire choanocyte chamber segmented using machine learning with a neuroid cell, previously identified using light microscopy (violet), and interacting with multiple microvilli within the chamber (B); segmented volume showing the violet neuroid cell, and a second neuroid cell (red) closer to apopylar collar, contacting cilia and microvillar collars of three choanocytes (blue, turquoise and green) and apopylar cells (yellow) (C); higher magnification view of segmented neuroid cell (violet) with filopodia extending into microvillar collar (turquoise) (D); single slice from FIB-SEM data set showing a neuroid cell with secretory vesicles highlighted in cyan. F: Single slice from the same FIB-SEM dataset showing neuroid cell forming a pocket around tip of a choanocyte cilia (yellow) (F). Considered with other structural features, these findings are suggestive of a role for neuroid cells in bacteria and debris clearance and intercellular communication. Scale bars (E, F) = 2 μm. vEM, volume electron microscopy. Panel A adapted from Heinrich et al. 158. Panels B-F reproduced from Musser et al. 46.
Secretory organelles
Organelles of the secretory pathway have been studied in 3D with many vEM techniques. The complex architecture and connectivity of stacked endoplasmic reticulum sheets in rodent neuronal and secretory cells was unravelled by Terasaki et al. 207 using array tomography. Importantly, the section thickness (30-40 nm) was set to be lower than the ER lumen diameter and the inter-stack spacing. The changing morphology of the endoplasmic reticulum during cell division was studied by ssET and SBF-SEM, with or without electron-dense DAB labelling of the ER lumen 208,209. Further endoplasmic reticulum studies drove the introduction of image analysis tools to capture and model the complexity of the organelle 165,210, including a description of the curvature landscape that defines the various endoplasmic reticulum sub-compartments in cultured cells 158. The endoplasmic reticulum morphology of specialized cells such as neurons has also been investigated by SBF-SEM and FIB-SEM 211, and studies of endoplasmic reticulum exit sites using FIB-SEM unravelled the tubular (rather than vesicular) nature of this intermediate compartment in insect 212 and mammalian cells 213.
Further down the secretory pathway is the Golgi, the complexity of which justifies the need for 3D visualization. In the early 2000s, a detailed description of the Golgi complex visualised by ssET was published 77. ssET is still the method of choice to study this organelle as the distance between cisternae in a stack requires high-resolution imaging. More recent studies capitalized on the larger acquisition volumes offered by FIB-SEM to capture the full stretch of the Golgi ribbon 99,163. Further, the endo/lysosomal pathway has been extensively imaged by vEM, using vCLEM to assign molecular identity to otherwise ambiguously shaped structures 214,215.
Other subcellular components
vEM has made significant contributions to the study of inter-organelle interactions 216, identified by spatial proximity (< 20 to 30 nm) between the limiting membranes of two organelles. In cultured cells 158,163 and liver tissue, FIB-SEM has shown changes in organelle morphologies associated with changing physiological states 217. vEM is also the technique of choice to describe the complexity of pathogen-induced structures, such as virus replication organelles 8,218–220 and membrane perturbations associated with viral budding in HIV and SARS-CoV-2 infected cells 100,221–223.
vEM of cell-cell interactions
Cell-cell interaction in metazoans
At the cellular level of our scale logic, vEM workflows are ideally suited to studying the finer details of the complex interactions between individual cells and/or groups of cells. For instance, in Spongilla lacustris, a freshwater sponge, vCLEM was used with FIB-SEM to demonstrate that individual neuroid cells interact with multiple choanocytes, supporting a role in neuron-effector communication, and with their conserved genetic makeup, as putative neuronal ancestors 46. Using array tomography and SBF-SEM, fibre cells in the Placozoa Trichoplax adhaerens were shown to contact multiple adjacent cell types and display evidence of phagocytic activity, suggesting a macrophage-like role in innate immunity that appeared early in metazoan evolution224. In symbiotic microalgae (the algal symbiont Phaeocystis cordata, in its acantharean host), reconstructions of FIB-SEM image series showed significant alterations in morphometric measures, reflecting interaction between the symbiont and controlling host 225. Similar approaches have also been used to examine conserved morphometric features in various phytoplankton, suggesting that topology could be modulated by energy-management constraints 226.
Host-pathogen interactions
vEM techniques are also very powerful for studying host-pathogen interactions 227. vCLEM studies combining fluorescence microscopy and FIB-SEM have shown how Mycobacterium tuberculosis, first confined within a membrane-bound phagosome, could escape to the cytoplasm of their host macrophage 228,229. A multimodal vCLEM approach was used with ssTEM, SBF-SEM and FIB-SEM to validate zebrafish as a model system for studying Toxoplasma gondii infection, showing that macrophages have a role in parasite clearance in vivo 12. Likewise, FIB-SEM revealed the complex structure of T. gondii rosette formation and connectivity 230, and cell-cell interactions at the virological synapse in HIV infection 221.
Immune cell-cancer interactions
The whole cell volume of a mouse cytotoxic T lymphocyte forming an immunological synapse with an ovarian cancer cell was revealed using FIB-SEM 99; the topology of this system was previously difficult to study in detail. In more complex multicellular organisms, intercellular communication is crucial, and one such mediator of crosstalk and material transfer is thought to be tunnelling nanotubes. Using vEM, tunnelling nanotubes have been shown to play a role in the stroma-mediated protection of leukemic cells through cell-cell vesicle trafficking 231, and microtubule bridges connecting cells in the early mammalian embryo have been shown to facilitate membrane transport between cells 232.
Nerve cell interactions
vEM has been widely used to examine cell-cell interactions in neuroscience. In the mouse brain, ssTEM showed that ultrastructural features of the blood brain barrier, including increased pericyte-brain endothelial cell contact, are altered during aging 233, and single axons form clustered compound multi-synaptic contacts of distinct morphologies with individual neurons, thus maximizing their influence on the postsynaptic neuron 139. FIB-SEM has been used to demonstrate that some primary cilia form serotonergic axo-ciliary synaptic contacts in the hippocampus 234. SBF-SEM was used to reconstruct two individual cells from a large dataset of a leech ganglion, discovering new connections 235. Inferences were also made about previously unstudied pathways, validated by post hoc comparative electrophysiological measurements in matched living tissues. Tran et al. 236 demonstrated an elegant co-culture system, designed to examine neuron-neuron interactions in detail with FIB-SEM and ssTEM. Using array tomography, synaptic inputs and outputs of an inhibitory interneuron were mapped 237.
vEM at the organism and tissue scale
vEM reveals principles of tissue organization that are impossible to capture with 2D approaches or with lower resolution methods. For example, array tomography of mouse ovarian follicles uncovered previously unknown complex interactions between different cell types via cytoplasmic extensions and gap junctions 238. vEM has now been adopted for tissue scale investigations across species (Fig.7), including in plants 239,240, and a variety of insects 241,242 and other arthropods 243. vEM of the nervous system, in particular, has rapidly advanced in recent years, because mapping neuronal circuits at synaptic resolution is considered essential to understanding how the brain works and how specific behaviours are generated 244 (Fig.8). Thus, within the past 10 years an impressive number of large vEM datasets of brain tissue have been produced, including the full or partial connectomes of the mammalian retina 73,245, ascidian tadpole larva Ciona intestinalis 246, the larva and adult Drosophila brain 82,173,247, zebrafish brain 13,31, a cubic millimetre of mouse brain 248, a cubic millimetre of human brain 116 and the pathoconnectome of a degenerating human retina to study neurological diseases 249. This list continues to grow at pace, and it is now possible to image the connectomes of several individuals from the same species (C. elegans) to address questions of development and plasticity 250. Peripheral nervous system investigations have also benefited from vEM, for example by understanding the structure and connectivity of cell types within the mouse circumvallate taste bud 251. For smaller organisms, vEM of the entire body (Platynereis larvae 252,253) enables investigations of the structure and function of the nervous system in the context of the whole body and its interactions with the environment.
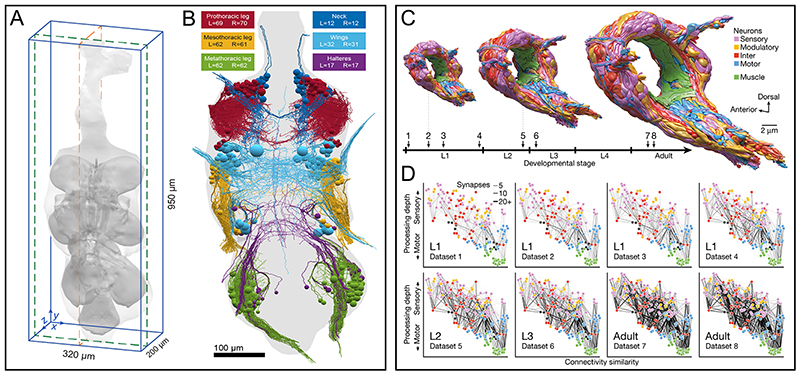
Figure 7. Examples of multiscale vEM on model organisms and connectomes.
A: Volumetric rendering of a Drosophila melanogaster ventral nerve cord (VNC) data set 84 showing the full extent of all imaged tissue (light grey) and the outline of the VNC neuropil (dark grey), with overall dimensions indicated. Data set acquired using GridTape transmission electron microscopy (TEM). B: All motor neurons in the thoracic segments of the VNC were reconstructed and analysed. Each motor neuron projects an axon to one peripheral nerve, leaving the bounds of the electron microscopy dataset, to innervate muscles. Cell bodies are represented as spheres, coloured along with their projections, in accordance with the target of innervation. C: Volumetric models of Caenorhabditis elegans brain 238,250, coloured by cell type, and shown at three stages in developmental timeline. The datasets were acquired using both serial section TEM (ssTEM), and single-beam array tomography with automated tape-collecting ultramicrotome (ATUM), and manually reconstructed using CATMAID. D: Wiring diagrams highlighting changes in complexity of connectivity during brain development for eight individuals indicated on the timeline in C. Each circle represents a cell, again coloured in accordance with cell type and target of innervation, and each line represents a connection between two cells with at least one chemical synapse. Vertical axis denotes signalling from sensory perception (top) to motor actuation (bottom). Horizontal axis denotes connectivity similarity; neurons that share partners are positioned more closely. Signal flow and connectivity similarity are based on accumulated connections from all datasets. Although brain geometry does not change substantially during development, with neuron number and position being fairly constant, complexity and relative strengths of synaptic connections evolve significantly during development. vEM, volume electron microscopy. Panels A and B adapted from Phelps et al. 84. Panels C and D reproduced from Witvliet et al. 250.
Figure 8. Examples of multiscale vEM in tissues and the clinical setting.
A: Analysis of a 1,500 μm3 volume of mouse neocortex, generated using array tomography with automated tape-collecting ultramicrotome (ATUM), imaged using single-beam scanning electron microscopy (SEM), and segmented automatically 254. The segmented volume was mined to study cellular composition and connectivity. The saturated reconstruction volume (left) had a varied composition which could be divided into individual subcategories, including cell types, axons, dendrites, and other unclassified processes. Scale bar = 7 μm. B-D: 3D ultrastructure of a kidney biopsy specimen from a patient with lupus nephritis studied using serial blockface SEM (SBF-SEM) 261. In this 3,700 μm3 data set, the glomerular basement membrane was found to be disrupted, with podocyte cytoplasmic processes (yellow) extending into the glomerular basement membrane and mesangial matrix (green) (B). Reconstructed in three dimensions, the podocyte cytoplasmic process (yellow) displayed highly complex topology with multiple spikes extending towards surrounding mesangial cells (blue), forming more than 100 contact sites (red) (C,D). Scale bars = 10 μm. E-G: SBF-SEM analysis of human biopsy samples from patients with immunoglobulin A nephropathy 262 highlighted the disruption of the glomerular basement membrane (red) and penetration of mesangial cellular processes (green) into the urinary space (highlighted by a black arrow with podocyte highlighted in yellow) (E); 3D reconstruction of the penetrating mesangial cell (F) revealed multiple contact sites with podocytes within the urinary space (red) (G). Scale bar = 1 μm. H-J: To study the 3D organization of patient- derived hepatoblastoma xenografts, SBF-SEM was used 267: one SBF-SEM data set (H) was used to develop a semi-automatic segmentation procedure, revealing 182 tumour cells (I) and 113 nuclei (J) within the volume, from which bioarchitectural parameters could be extracted to further study tumour tissue architecture. Scale bar = 20 μm. vEM, volume electron microscopy. Panel A reproduced from Kasthuri et al. 254. Panels B-D reproduced from Takaki et al. 261. Panels E-G reproduced from Nagai et al. 262. Panels H-J reproduced from de Senneville et al. 267.
In addition to addressing the research question that initially motivated their collection, such large vEM datasets have also proven to be a valuable resource for addressing unrelated questions at various scales. For example, a vEM dataset from mouse cortex was generated254 to investigate the rules of neocortical synaptic connectivity, and has also been used to define the 3D organization of the endoplasmic reticulum 255, the structure of mitochondria 256 and the distribution of myelin in axons of neocortical pyramidal neurons 257.
vEM in the clinical setting
Electron microscopy has long been used as a tool in the clinic, most commonly in diagnosis of ciliopathies and in renal pathology 258. Diagnostic electron microscopy generally uses TEM to deliver the resolution needed to measure defects in, for example, the central complex and microtubular arrangements in cilia in primary ciliary dyskinesia 259, and glomerular structures including the basement membrane in the kidney. A recent move has seen several teams investigate the potential of vEM techniques to reveal additional diagnostic features in volumes greater than that of a single ultra-thin section. Doubling the imaged volume using dual-axis electron tomography and sub-tomogram averaging demonstrated that a reduction in proximal outer dynein arm volume could be detected in archived specimens from patients with primary ciliary dyskinesia with a mutation in the DNAH11 gene, that could not be seen using standard ultrathin sections 260. Other work has demonstrated that SBF-SEM can reveal disruption of the glomerular basement membrane and associated changes in podocyte cell contacts in human biopsy samples from lupus nephritis 261 and patients with IgA nephropathy 264. In these cases, image volumes and segmentation were essential to trace and capture penetrations of cells through the basement membrane and rare cell-cell contact events. vEM techniques are now finding broader application in clinical research, to study synapse organization in post-mortem brain from patients with Alzheimer’s disease 263 and organelle changes in Parkinson’s disease 264, the tumour microenvironment in cancer biopsy samples 265–267, cell-cell contacts in the alveolar epithelium of the lung using archival material 268 and the osteocyte and lacuno-canalicular network in osteoarthritic and osteoporotic human bone 269.
Reproducibility and data deposition
Biological electron microscopy is still largely a manual discipline. Samples are typically prepared using customized protocols tailored to the research question and qualitative observations are often made on a small number of hand-selected images. This approach has led to seminal discoveries in biology yet limits throughput and complicates reproducibility. However, recent community efforts aim to produce data and tools that will support reproducibility in vEM.
Factors influencing reproducibility
The vast heterogeneity of biological samples and applications preclude a universal vEM sample preparation protocol, as evidenced by the daunting number of published protocols. Reproduction is confounded by historical differences in protocols between laboratories; the number of steps in sample preparation workflows, which deters users from optimizing adequately working protocols; variation in reagent batches; a lack of standard parameters and quality control metrics; and the absence of a repository of reference protocols with corresponding images showing examples of data of varying quality. To address these issues, reference protocols and consensus guidelines on optimization of workflows must be agreed within the expert vEM community. The production of benchmark specimens from different laboratories using these reference protocols will allow assessment of local variables. Public availability of these data will allow crowdsourcing or artificial intelligence approaches to derive quality control scores. Increased throughput of sample preparation using microwave processing and robotic devices will also support reproducibility.
The community currently lacks a standard set of quality control checks for the microscopes used to acquire vEM data, but inspiration can be taken from the introduction of guidelines and standards for quality control by the QUAREP-LiMi consortium in light microscopy 270. Empirical and heuristic choices made during image acquisition, plus (hidden) differences in the state of the instrument mean that imaging runs are rarely reproducible between laboratories. Benchmark specimens prepared from reference protocols would allow acquisition of benchmark datasets, which could be publicly deposited and used to better understand differences between microscopes and imaging parameters. Fixing parameters could result in reliably repeatable acquisition runs. Environmental stability during long imaging runs is critical for quality and reproducibility, and accurate monitoring of section thickness would improve the fidelity of downstream image volume reconstruction. Automation of imaging and internal quality control steps, such as on-the-fly focus and stigmation corrections, and feedback loops to maintain spatial fidelity are critical aspects of vEM 52. Lack of full reporting of image acquisition parameters has historically limited the reproducibility of vEM experiments, and therefore community consensus on parameters to be reported for each vEM imaging modality would provide valuable guidance and extend the recently published REMBI minimal metadata recommendations to support reproducible vEM data acquisition 271.
A lack of metrics for vEM data means that quality control and optimization are based on manual visual inspection. To improve this, the vEM community can take inspiration from the work of the cryo-EM community on gold standards 272. As with sample preparation, there is no silver bullet software package that can seamlessly process and analyse all vEM datasets. Most researchers use a mix and match/trial and error approach to visualize and mine their image data, slowing optimization and complicating reproducibility. Community guidelines on reporting methods for processing and analysis of vEM data are critical for reproducibility of published work 271,273. Image-to-image and multimodal registrations should be published and/or publicly deposited with accompanying landmark and transformation files for reproducing such registrations. Segmentation is still largely a manual process, despite progress in the development of machine learning algorithms, and known inter-expert variations on segmentation tasks raise questions around the fidelity of ground truth 274,275. Inter-expert variation is often due to inadequacies in the datasets; for example, sections may be too thick to resolve very thin axons, or to identify synapses that are sectioned parallel to the postsynaptic density, and missing sections and specimen contamination contributes to errors. Further development of machine learning approaches will succeed only with parallel improvement of data quality. Publication of usable models with accompanying weights within containerized workflows will allow reproduction of inferences.
Data handling and deposition
Community surveys suggest that many labs are collecting vEM datasets in the 10-100 GB range, with images saved to disk in proprietary or tiff file formats, transferred to a server and downloaded by users for downstream analysis using desktop software programs as cropped or binned image stacks. However, as vEM datasets reach the terabyte regime, this approach breaks down and data handling must be carefully planned with the support of scientific computing specialists. Institutions hosting vEM technology must consider related computational expenses and hire dedicated personnel to optimize and streamline data handling. Traditional file formats, such as mrc and tiff, fail at the terabyte scale and next generation file formats 276 are being developed that dynamically access chunks of data via cloud storage and compute, bypassing the need to download entire datasets. Public deposition of image data and associated metadata in image archives such as EMPIAR 277 and the BioImage Archive 278 are critical in driving full analysis and reanalysis of rich vEM datasets by a distributed user base, as well as aiding development of automated image analysis tools and supporting meta-studies into reproducibility.
Limitations and optimizations
As may be expected in a nascent field, there are currently a range of limitations affecting vEM, giving rise to an exciting and dynamic technology research, development and innovation environment. Stable, supported and accessible solutions will help overcome these obstacles and drive the adoption of vEM by the wider biological community.
Sample preparation
Ideally, cells and tissues would be cryofixed (to ensure that dynamic events are captured) and imaged at cold temperature (to avoid shrinkage and warping artefacts due to dehydration). However, the limitation in the depth of vitrification of biological samples is ~200 μm, far below the size of most samples destined for vEM. Serial imaging of small volumes (<10 μm) of vitrified samples is possible using cryo FIB-SEM 279,280 or cryo soft X-ray tomography 283. Reassuringly, images of 3D ultrastructure from cryo soft X-ray tomography show that most vitrified cell structures have a similar appearance to those seen in room-temperature vEM, suggesting that shrinkage artefacts will not invalidate interpretation for many biological questions. A further complication of working with human tissue samples for vEM is the incompatibility of clinical processing (wax embedding and snap freezing) with good ultrastructure, and the deterioration of ultrastructure of post-mortem samples prior to fixation, a complication that was critically evident in the early stages of the COVID-19 pandemic 282. A lack of protocols for introducing immunolabels deep into tissue without membrane-destabilizing permeabilization techniques (methanol, Triton and so on) 69 also hinders molecular localization in tissues, while high-contrast genetic probes that are still visible with multiple layers of heavy metal staining are only now being developed.
Imaging
vEM, similar to most imaging modalities, falls foul of the trade-off between speed of image acquisition, resolution of the images and the volume that can be imaged. Brute force speed-up of image acquisition results in lower signal and image quality, whereas targeted imaging may obscure important tissue context, and automated imaging of ROIs may lower the chance of serendipitous discovery by biasing acquisition. Long imaging runs and parallelization by installation of multiple instruments are possible but expensive and, currently, only implemented in a handful of dedicated facilities worldwide 52,85.
Data analysis
vEM data analysis is still a massive bottleneck, being largely manual, and therefore time and resource intensive. The volume that can be analysed and the number of features that can be extracted are limited. Furthermore, most vEM analysis workflows require some level of bespoke customization, depending on the research question, sample type, sample preparation, imaging modality and quality and format of the acquired data. A mishmash of constantly evolving open-source, in-house and commercial software packages add complexity for non-expert end users. The application of machine learning will alleviate some of these analysis burdens, but universal models are not yet a reality. The infancy of the field demands significant computational literacy in researchers to create or tweak algorithms and track and correct errors. vEM data analysis is a research area in flux, but a strong push toward open-source solutions will ideally prevent siloed computational solutions.
Outlook
The development of new tools and technologies has underpinned great advances in our understanding of how cells, tissues and organisms function in health and disease. Electron microscopy has been central to these advances, being the only technique that can reveal the structure of cells and tissues with the contrast and resolution to distinguish membranes and membrane bilayers. Whereas the challenge in genomics, proteomics and metabolomics has been to move from systems-level analysis to single cells, the challenge for electron microscopy has been to move from the single-cell level to a systems-level view of samples. vEM offers this opportunity.
The field is evolving rapidly, as more scientists realize the potential of vEM to contribute new insights into their research questions. This has attracted tool developers, in the form of microscopists, physicists, chemists, engineers and computational scientists, focused on improving the speed and throughput of vEM. Having originated in connectomics and neuroscience, the next grand challenge appears to be the mapping of the connectome of a single mouse brain. Rapid high-resolution image acquisition will also help to overcome the n-of-1 problem, allowing the analysis of multiple samples from a population to assess inter-organism variability. As we have already seen, technological advances focused on one challenge area (like the brain) quickly radiate out to drive discovery in other biological and clinical research areas.
A particular challenge lies in true multiscale imaging of tissues and organisms, to provide multidimensional views of samples with low-resolution and high-resolution information accessible from a single volume. The creation of multiscale digital twins will require parallel developments in light, X-ray and electron imaging, alongside improvements in sample preparation, multimodal probes and multiscale analysis and visualization. There are two crossover points in scales that currently block the creation of seamless image volumes from patient to protein. One is at the cryo-resin interface, at a sample thickness of between 1 μm and 200 μm, where 1 μm is the (somewhat generous) limit of continuous volume that can be imaged with cryo-ET to identify macromolecules in situ in cells, and 200 μm is the hard limit of vitrification, above which resin protocols effectively destroy or mask atomic structure. The second is at a sample thickness greater than 1 mm, above which diffusion limits the penetration of chemicals into the sample. Overcoming these blocks will require broad multidisciplinary collaboration between structural biologists, volume electron microscopists, light microscopists and pre-clinical and medical imaging scientists, with biologists, clinician scientists and the tool developers mentioned above.
The scale of this challenge requires investment into specialist facilities that gather a range of expertise and infrastructure. Deployment will then require roll out of new tools to specialist core facilities that will offer open access to a broad swathe of researchers, who will, in turn, depend on local facilities to perform fast effective sample preparation close to the end-user. Indeed, these local facilities are key in supporting the boom in vEM use, and are a source of valuable advice about experimental design and vEM modality selection for a specific biological question. However, the number of vEM experts is likely to become limiting as vEM technology is more widely adopted, a limitation already affecting the cryo-EM field, and it will be critical to develop and implement formal training programs, rather than the on-the-job training model currently used. Creation of a network of local, national and international open access vEM facilities will require a coordinated effort from the vEM community and funders, and will take time. In the meantime, vEM specialists and end-user researchers are encouraged to join the burgeoning community via the vEM community website or to contact their local electron microscopy facility for support and information.
If the effort to build a vEM infrastructure and community is successful, then we should see a move towards a new discipline of morphometrics, in which the number of samples imaged and analysed will provide statistically significant insights into the nanoscale spatial organization of organelles, cells, organs and systems. Crossing discipline boundaries, integration of spatial proteomics, transcriptomics and metabolomics data will add volume function to volume structure to drive a new era of functional morphometrics.
Supplementary Material
Supplementary information is available for this paper at https://doi.org/10.1038/s43586-022-00131-9
Acknowledgements
The authors thank E Duke (European Molecular Biology Laboratory (EMBL) Hamburg) for comments on the manuscript. The work of C.J.P. and L.M.C. was supported by the Francis Crick Institute, which receives its core funding from Cancer Research UK (FC001999), the UK Medical Research Council (FC001999), and the Wellcome Trust (FC001999. R.G.P is supported by the National Health and Medical Research Council of Australia (grant APP1150083 and fellowship APP1156489) and by the Australian Research Council (DP200102559). K.D.M. is supported by National Institutes of Health (grant NS094499). A.W. was supported by Howard Hughes Medical Institute, Janelia Research Campus. K.N. is funded in whole or in part with Federal funds from the National Cancer Institute, National Institutes of Health, under Contract No. 75N91019D00024. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Glossary
- Connectomics
An interdisciplinary field combining systems neuroscience, applied physics and computer science to map the physical and functional connections of the brain in a range of species.
- Vibratome
An instrument for sectioning tissue that utilizes a vibrating blade.
- Membrane tubulation
The process of membrane reorganization that produces tubular membrane invaginations or outward-facing tubular membrane projections.
- Ultra-thin sections
Sections thin enough to allow transmission of an electron beam (usually 100 nm or less).
- Diamond knife
A specialized tool used in electron microscopy where diamond is used to produce an extremely sharp and durable knife edge capable of cutting ultra-thin sections of tissue (down to about 30 nm).
- Ultramicrotome
An instrument for preparing ultra-thin sections for electron microscopy.
- Neural tracers
Fluorescent probes for imaging neuronal morphology.
- Charging
A process that occurs in poorly conducting specimens in the scanning electron microscope as electrons are trapped and their excess negative charge causes surface potentials that distort the signal amplitude, focus and image geometry.
- Isotropic voxels
3D pixels with the same size in x, y and z.
- Segmentation
The process of delineating objects within an image, usually with the aim of visualizing and/or analysing different object classes. Segmentation may be performed manually or using algorithms.
- Noise
Unwanted alterations of the pixel intensities in an image caused by different stages of the imaging process (electron gun, signal detection, amplification).
- Connectome
A map of the synaptic connections of the brain.
- Choanocytes
Specialised cells found in sponges, contributing to feeding function via water circulation and filtration.
- Tunnelling nanotubes
Extensions of the plasma membrane that form channels between cells whose proposed functions are to open communication channels between cells, for the exchange of organelles or other intracellular components.
- Vitrification
Transformation of a substance into a non-crystalline, amorphous, glass-like solid. In cryoelectron microscopy (cryo-EM) this involves cryopreservation of materials without hexagonal ice crystal formation, to maintain cellular components in a near-native state.
- Fiducial
An object added to the field of view of a microscope during imaging that can be used as a point of reference during sample preparation, imaging and image analysis.
- Watershed Transform
An image processing technique that can be used for image segmentation by modelling the intensity of greyscale images as topographical peaks.
- Classifier
An algorithm that categorises an observation based on quantifiable properties.
Declarations
Author contributions
Introduction (L.M.C. and P.V.); Experimentation (L.M.C., C.J.P., C.G., K.Me., K.D.M., R.G.P., N.L.S., Y.S., B.T. and P.V.); Results (L.M.C., C.G., K.Me., K.D.M., C.P., B.T. and A.W.); Applications (L.M.C., C.J.P., C.G., K.D.M., R.G.P., N.L.S., Y.S., P.V. and A.W); Reproducibility and data deposition (L.M.C., A.K., K.N., and Y.S.), Limitations and optimizations (L.M.C., K.N. and Y.S.) ; Outlook (L.M.C.); overview of the Primer (L.M.C.).
Competing Interests
K.D.M. has founder’s equity interests in Aratome, LLC (Menlo Park, CA), an enterprise that produces array tomography materials and services. All other authors declare no competing interests.
Related Links
3D slicer: www.slicer.org
APEER: https://www.apeer.com/home/
Ariadne: https://ariadne.ai/
BigDataProcessor: https://github.com/bigdataprocessor/bigdataprocessor2
BigWarp: https://imagej.net/plugins/bigwarp
BioFormats: https://www.openmicroscopy.org/bio-formats/
Bioimage Model Zoo: https://bioimage.io/#/
CDeep3M: https://github.com/CRBS/cdeep3m
CLEMsite: https://git.embl.de/lleti/clemsite
Dask: https://dask.org/
DeepImageJ: https://deepimagej.github.io/deepimagej/
DeepMIB: http://mib.helsinki.fi/
ec-CLEM: https://icy.bioimageanalysis.org/plugin/ec-clem/
Elastix: https://elastix.lumc.nl/
Example data analysis workflows: https://github.com/kreshuklab/vem-primer-examples
Fiji Weka plugin: https://imagej.net/plugins/tws/
Ilastik: https://www.ilastik.org/
Microscopy Image Browser: http://mib.helsinki.fi/
MitoEM Data Challenge: https://mitoem.grand-challenge.org/
MorphoLibJ: https://imagej.net/plugins/morpholibj
piTEAM: https://github.com/AllenInstitute/piTEAM
Reconstruct: https://synapseweb.clm.utexas.edu/software-0
SBEMImage: https://github.com/SBEMimage
Scalable minds: https://scalableminds.com/
SegEM: http://segem.brain.mpg.de/
SerialEM: https://bio3d.colorado.edu/SerialEM/
The Lichtman lab at Harvard University: https://lichtmanlab.fas.harvard.edu/
TrakEM2: https://imagej.net/plugins/trakem2/
VAST: https://lichtman.rc.fas.harvard.edu/vast/
vEM community website: https://www.volumeem.org/
WaferMapper: https://lichtman.rc.fas.harvard.edu/LGN/WaferMapper.html
Webknossos: https://webknossos.org/
ZeroCostDL4MIC: https://github.com/HenriquesLab/ZeroCostDL4Mic
Code availability
The two example data analysis workflows for neuron and mitochondria segmentation can be found at: https://github.com/kreshuklab/vem-primer-examples.Figure Legends
References
- 1.Abbott LF, et al. The Mind of a Mouse. Cell. 2020;182:1372–1376. doi: 10.1016/j.cell.2020.08.010. [DOI] [PubMed] [Google Scholar]
- 2.Guerin CJ, Lippens S. Correlative light and volume electron microscopy (vCLEM): How community participation can advance developing technologies. J Microsc. 2021;284:97–102. doi: 10.1111/jmi.13056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peddie CJ, Schieber NL. In: Correlative Imaging: Focusing on the Future. Collinson Lucy, Verkade Paul., editors. John Wiley & Sons Ltd; 2020. pp. 37–66. [Google Scholar]
- 4.Hayat MA. Principles and Techniques of electron microscopy: biological applications. Cambridge University Press; 2000. pp. 45–61. [Google Scholar]
- 5.Titze B, Genoud C. Volume scanning electron microscopy for imaging biological ultrastructure. Biology of the Cell. 2016;108:307–323. doi: 10.1111/boc.201600024. [DOI] [PubMed] [Google Scholar]
- 6.Ströh S, Hammerschmith EW, Tank DW, Seung HS, Wanner AA. In situ X-ray assisted electron microscopy staining for large biological samples. bioRxiv. 2021:2021.2006.2019.448808. doi: 10.1101/2021.06.19.448808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Russell MRG, et al. 3D correlative light and electron microscopy of cultured cells using serial blockface scanning electron microscopy. Journal of Cell Science. 2016;130:278–291. doi: 10.1242/jcs.188433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cortese M, et al. Integrative Imaging Reveals SARS-CoV-2-Induced Reshaping of Subcellular Morphologies. Cell host & microbe. 2020;28:853–866.:e855. doi: 10.1016/j.chom.2020.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Müller-Reichert T, Hohenberg H, O’Toole ET, McDonald K. Cryoimmobilization and three-dimensional visualization of C. elegans ultrastructure. Journal of Microscopy. 2003;212:71–80. doi: 10.1046/j.1365-2818.2003.01250.x. [DOI] [PubMed] [Google Scholar]
- 10.Mulcahy B, et al. A Pipeline for Volume Electron Microscopy of the Caenorhabditis elegans Nervous System. Frontiers in neural circuits. 2018;12:94. doi: 10.3389/fncir.2018.00094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Porrati F, Grewe D, Seybert A, Frangakis AS, Eltsov M. FIB-SEM imaging properties of Drosophila melanogaster tissues embedded in Lowicryl HM20. J Microsc. 2019;273:91–104. doi: 10.1111/jmi.12764. [DOI] [PubMed] [Google Scholar]
- 12.Yoshida N, et al. The zebrafish as a novel model for the in vivo study of Toxoplasma gondii replication and interaction with macrophages. Dis Model Mech. 2020;13 doi: 10.1242/dmm.043091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hildebrand DGC, et al. Whole-brain serial-section electron microscopy in larval zebrafish. Nature. 2017;545:345–349. doi: 10.1038/nature22356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Korogod N, Petersen CC, Knott GW, Häusser M. Ultrastructural analysis of adult mouse neocortex comparing aldehyde perfusion with cryo fixation. eLife. 2015;4:e05793. doi: 10.7554/elife.05793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mikula S, Denk W. High-resolution whole-brain staining for electron microscopic circuit reconstruction. Nature methods. 2015;12:541–546. doi: 10.1038/nmeth.3361. [DOI] [PubMed] [Google Scholar]
- 16.Polishchuk RS, Polishchuk EV, Luini A. Visualizing live dynamics and ultrastructure of intracellular organelles with preembedding correlative light-electron microscopy. Methods Cell Biol. 2012;111:21–35. doi: 10.1016/b978-0-12-416026-2.00002-9. [DOI] [PubMed] [Google Scholar]
- 17.McDonald K. Cryopreparation Methods for Electron Microscopy of Selected Model Systems. Methods Cell Biol. 2007;79:23–56. doi: 10.1016/s0091-679x(06)79002-1. [DOI] [PubMed] [Google Scholar]
- 18.McDonald KL, Morphew M, Verkade P, Muller-Reichert T. Recent advances in high-pressure freezing: equipment- and specimen-loading methods. Methods Mol Biol. 2007;369:143–173. doi: 10.1007/978-1-59745-294-6_8. [pii] 1-59745-294-7:143. [DOI] [PubMed] [Google Scholar]
- 19.McDonald KL. A review of high-pressure freezing preparation techniques for correlative light and electron microscopy of the same cells and tissues. J Microsc. 2009;235:273–281. doi: 10.1111/j.1365-2818.2009.03218.x. [DOI] [PubMed] [Google Scholar]
- 20.Odriozola A, et al. High contrast staining for serial block face scanning electron microscopy without uranyl acetate. bioRxiv. 2017:207472. doi: 10.1101/207472. [DOI] [Google Scholar]
- 21.Starborg T, et al. Using transmission electron microscopy and 3View to determine collagen fibril size and three-dimensional organization. Nature Protocols. 2013;8:1433–1448. doi: 10.1038/nprot.2013.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mikula S, Binding J, Denk W. Staining and embedding the whole mouse brain for electron microscopy. Nature methods. 2012;9:1198–1201. doi: 10.1038/nmeth.2213. [DOI] [PubMed] [Google Scholar]
- 23.Ronchi P, et al. High-precision targeting workflow for volume electron microscopy. The Journal of cell biology. 2021;220 doi: 10.1083/jcb.202104069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deerinck TJ, Bushong E, Thor A, Ellisman MH. NCMIR methods for 3D EM: A new protocol for preparation of biological specimens for serial block-face SEM. Microscopy. 2010:6–8. http://ncmir.ucsd.edu/sbfsem-protocol.pdf . [Google Scholar]
- 25.Tapia JC, et al. High-contrast en bloc staining of neuronal tissue for field emission scanning electron microscopy. Nature Protocols. 2012;7:193–206. doi: 10.1038/nprot.2011.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu Z, et al. En bloc preparation of Drosophila brains enables high-throughput FIB-SEM connectomics. bioRxiv. 2019:855130. doi: 10.1101/855130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hua Y, Laserstein P, Helmstaedter M. Large-volume en-bloc staining for electron microscopy-based connectomics. Nature communications. 2015;6:7923. doi: 10.1038/ncomms8923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seligman AM, Wasserkrug HL, Hanker JS. A new staining method (OTO) for enhancing contrast of lipid-containing membranes and droplets in osmium tetroxide-fixed tissue with osmiophilic thiocarbohydrazide (TCH) The Journal of cell biology. 1966;30:424–432. doi: 10.1083/jcb.30.2.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Malick LE, Wilson RB. Modified thiocarbohydrazide procedure for scanning electron microscopy: routine use for normal, pathological, or experimental tissues. Stain Technol. 1975;50:265–269. doi: 10.3109/10520297509117069. [DOI] [PubMed] [Google Scholar]
- 30.Willingham MC, Rutherford AV. The use of osmium-thiocarbohydrazide-osmium (OTO) and ferrocyanide-reduced osmium methods to enhance membrane contrast and preservation in cultured cells. J Histochem Cytochem. 1984;32:455–460. doi: 10.1177/32.4.6323574. [DOI] [PubMed] [Google Scholar]
- 31.Genoud C, Titze B, Graff-Meyer A, Friedrich RW. Fast Homogeneous En Bloc Staining of Large Tissue Samples for Volume Electron Microscopy. Frontiers in neuroanatomy. 2018;12:76. doi: 10.3389/fnana.2018.00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakakoshi M, Nishioka H, Katayama E. New versatile staining reagents for biological transmission electron microscopy that substitute for uranyl acetate. J Electron Microsc (Tokyo) 2011;60:401–407. doi: 10.1093/jmicro/dfr084. [DOI] [PubMed] [Google Scholar]
- 33.Kuipers J, Giepmans BNG. Neodymium as an alternative contrast for uranium in electron microscopy. Histochemistry and Cell Biology. 2020;153:271–277. doi: 10.1007/s00418-020-01846-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McDonald KL. Out with the old and in with the new: rapid specimen preparation procedures for electron microscopy of sectioned biological material. Protoplasma. 2013;251:429–448. doi: 10.1007/s00709-013-0575-y. [DOI] [PubMed] [Google Scholar]
- 35.Hoffman DP, et al. Correlative three-dimensional super-resolution and block-face electron microscopy of whole vitreously frozen cells. Science (New York, NY) 2020;367:eaaz5357. doi: 10.1126/science.aaz5357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luft JH. Improvements in epoxy resin embedding methods. J Biophys Biochem Cytol. 1961;9:409–414. doi: 10.1083/jcb.9.2.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Glauert AM, Rogers GE, Glauert RH. A New Embedding Medium for Electron Microscopy. Nature. 1956;178:803. doi: 10.1038/178803a0. [DOI] [PubMed] [Google Scholar]
- 38.Kellenberger E. Low denaturation embedding for electron microscopy of thin sections. Chemische Werke Lowi; 1980. [Google Scholar]
- 39.Armbruster B, et al. Specimen preparation for electron microscopy using low temperature embedding resins. Journal of microscopy. 1982;126:77–85. doi: 10.1111/j.1365-2818.1982.tb00358.x. [DOI] [PubMed] [Google Scholar]
- 40.Stäubli W. A new embedding technique for electron microscopy, combining a water-soluble epoxy resin (Durcupan) with water insoluble araldite. The Journal of cell biology. 1963;16:197–201. doi: 10.1083/jcb.16.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Spurr AR. A low-viscosity epoxy resin embedding medium for electron microscopy. Journal of Ultrastructure Research. 1969;26:31–43. doi: 10.1016/s0022-5320(69)90033-1. [DOI] [PubMed] [Google Scholar]
- 42.Maco B, Holtmaat A, Jorstad A, Fua P, Knott GW. Correlative In Vivo 2-Photon Imaging and Focused Ion Beam Scanning Electron Microscopy: 3D Analysis of Neuronal Ultrastructure. 2014;124 doi: 10.1016/B978-0-12-801075-4.00016-1. [DOI] [PubMed] [Google Scholar]
- 43.Maco B, et al. Semiautomated correlative 3D electron microscopy of in vivo-imaged axons and dendrites. Nature Protocols. 2014;9:1354–1366. doi: 10.1038/nprot.2014.101. [DOI] [PubMed] [Google Scholar]
- 44.Karreman MA, et al. Fast and precise targeting of single tumor cells in vivo by multimodal correlative microscopy. J Cell Sci. 2016;129:444–456. doi: 10.1242/jcs.181842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Karreman MA, et al. Find your way with X-Ray: using microCT to correlate in vivo imaging with 3D electron microscopy. BS:MCB; 2017. [DOI] [PubMed] [Google Scholar]
- 46.Musser JM, et al. Profiling cellular diversity in sponges informs animal cell type and nervous system evolution. Science (New York, NY) 2021;374:717–723. doi: 10.1126/science.abj2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schieber NL, et al. Minimal resin embedding of multicellular specimens for targeted FIB-SEM imaging. Methods Cell Biol. 2017;140:69–83. doi: 10.1016/bs.mcb.2017.03.005. [DOI] [PubMed] [Google Scholar]
- 48.Belu A, et al. Ultra-thin resin embedding method for scanning electron microscopy of individual cells on high and low aspect ratio 3D nanostructures. Journal of Microscopy. 2016;263:78–86. doi: 10.1111/jmi.12378. [DOI] [PubMed] [Google Scholar]
- 49.Van Donselaar EG, et al. Extremely thin layer plastification for focused-ion beam scanning electron microscopy: an improved method to study cell surfaces and organelles of cultured cells. J Microsc. 2018;270:359–373. doi: 10.1111/jmi.12694. [DOI] [PubMed] [Google Scholar]
- 50.Vergara HM, et al. Whole-body integration of gene expression and single-cell morphology. Cell. 2021;184:4819–4837.:e4822. doi: 10.1016/j.cell.2021.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hayworth KJ, et al. Ultrastructurally-smooth thick partitioning and volume stitching for larger-scale connectomics. Nature methods. 2015;12:319–322. doi: 10.1038/nmeth.3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xu CS, et al. Enhanced FIB-SEM systems for large-volume 3D imaging. eLife. 2017;6:e25916. doi: 10.7554/elife.25916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sano T, Glazer AN, Cantor CR. A streptavidin-metallothionein chimera that allows specific labeling of biological materials with many different heavy metal ions. Proceedings of the National Academy of Sciences. 1992;89:1534–1538. doi: 10.1073/pnas.89.5.1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mercogliano CP, DeRosier DJ. Concatenated metallothionein as a clonable gold label for electron microscopy. Journal of structural biology. 2007;160:70–82. doi: 10.1016/j.jsb.2007.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Diestra E, Fontana J, Guichard P, Marco S, Risco C. Visualization of proteins in intact cells with a clonable tag for electron microscopy. Journal of structural biology. 2009;165:157–168. doi: 10.1016/j.jsb.2008.11.009. doi:S1047-8477(08)00289-X [pii] [doi] [DOI] [PubMed] [Google Scholar]
- 56.Morphew MK, et al. Metallothionein as a clonable tag for protein localization by electron microscopy of cells. Journal of Microscopy. 2015;260:20–29. doi: 10.1111/jmi.12262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gaietta G, et al. Multicolor and Electron Microscopic Imaging of Connexin Trafficking. Science (New York, NY) 2002;296:503–507. doi: 10.1126/science.1068793. [DOI] [PubMed] [Google Scholar]
- 58.Shu X, et al. A Genetically Encoded Tag for Correlated Light and Electron Microscopy of Intact Cells, Tissues, and Organisms. PLoS Biology. 2011;9:e1001041-1001010. doi: 10.1371/journal.pbio.1001041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Martell JD, Deerinck TJ, Lam SS, Ellisman MH, Ting AY. Electron microscopy using the genetically encoded APEX2 tag in cultured mammalian cells. Nature Protocols. 2017;12:1792–1816. doi: 10.1038/nprot.2017.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lam SS, et al. Directed evolution of APEX2 for electron microscopy and proteomics. Nature methods. 2015;12:51–54. doi: 10.1038/nmeth.3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ariotti N, et al. Modular Detection of GFP-Labeled Proteins for Rapid Screening by Electron Microscopy in Cells and Organisms. Dev Cell. 2015;35:513–525. doi: 10.1016/j.devcel.2015.10.016. [DOI] [PubMed] [Google Scholar]
- 62.Ariotti N, et al. Ultrastructural localisation of protein interactions using conditionally stable nanobodies. PLoS Biol. 2018;16:e2005473. doi: 10.1371/journal.pbio.2005473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Han Y, et al. Directed Evolution of Split APEX2 Peroxidase. Acs Chem Biol. 2019;14:619–635. doi: 10.1021/acschembio.8b00919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sengupta R, Poderycki MJ, Mattoo S. CryoAPEX – an electron tomography tool for subcellular localization of membrane proteins. Journal of Cell Science. 2019;132:jcs222315. doi: 10.1242/jcs.222315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang Q, Lee W-CA, Paul DL, Ginty DD. Multiplexed peroxidase-based electron microscopy labeling enables simultaneous visualization of multiple cell types. Nature Neuroscience. 2019;22:828–839. doi: 10.1038/s41593-019-0358-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mavlyutov TA, et al. APEX2-enhanced electron microscopy distinguishes sigma-1 receptor localization in the nucleoplasmic reticulum. Oncotarget. 2017;8:51317–51330. doi: 10.18632/oncotarget.17906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rae J, et al. A robust method for particulate detection of a genetic tag for 3D electron microscopy. eLife. 2021;10:e64630. doi: 10.7554/elife.64630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jiang Z, et al. Genetically encoded tags for direct synthesis of EM-visible gold nanoparticles in cells. Nature methods. 2020;17:937–946. doi: 10.1038/s41592-020-0911-z. [DOI] [PubMed] [Google Scholar]
- 69.Fulton KA, Briggman KL. Permeabilization-free en bloc immunohistochemistry for correlative microscopy. Elife. 2021;10 doi: 10.7554/eLife.63392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Micheva KD, Smith SJ. Array tomography: a new tool for imaging the molecular architecture and ultrastructure of neural circuits. Neuron. 2007;55:25–36. doi: 10.1016/j.neuron.2007.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Smith SJ. Q&A: Array tomography. BMC biology. 2018;16:98. doi: 10.1186/s12915-018-0560-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Oberti D, Kirschmann MA, Hahnloser RH. Correlative microscopy of densely labeled projection neurons using neural tracers. Frontiers in neuroanatomy. 2010;4:24. doi: 10.3389/fnana.2010.00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Anderson JR, et al. Exploring the retinal connectome. Mol Vis. 2011;17:355–379. [PMC free article] [PubMed] [Google Scholar]
- 74.Shahidi R, et al. A serial multiplex immunogold labeling method for identifying peptidergic neurons in connectomes. Elife. 2015;4 doi: 10.7554/eLife.11147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.White JG, Southgate E, Thomson JN, Brenner S. The structure of the nervous system of the nematode Caenorhabditis elegans. Philosophical Transactions Royal Soc Lond B Biological Sci. 1986;314:1–340. doi: 10.1098/rstb.1986.0056. [DOI] [PubMed] [Google Scholar]
- 76.Marsh BJ, Volkmann N, McIntosh JR, Howell KE. Direct continuities between cisternae at different levels of the Golgi complex in glucose-stimulated mouse islet beta cells. Proc Natl Acad Sci U S A. 2004;101:5565–5570. doi: 10.1073/pnas.0401242101. [doi] 0401242101 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Marsh BJ. Lessons from tomographic studies of the mammalian Golgi. Biochim Biophys Acta. 2005;1744:273–292. doi: 10.1016/j.bbamcr.2005.04.002. doi:S0167-4889(05)00060-1 [pii] [doi] [DOI] [PubMed] [Google Scholar]
- 78.Anderson JR, et al. A Computational Framework for Ultrastructural Mapping of Neural Circuitry. PLOS Biology. 2009;7:e1000074. doi: 10.1371/journal.pbio.1000074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bock DD, et al. Network anatomy and in vivo physiology of visual cortical neurons. Nature. 2011;471:177–182. doi: 10.1038/nature09802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Potter CS, Pulokas J, Smith P, Suloway C, Carragher B. Robotic grid loading system for a transmission electron microscope. Journal of structural biology. 2004;146:431–440. doi: 10.1016/j.jsb.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 81.Lefman J, Morrison R, Subramaniam S. Automated 100-position specimen loader and image acquisition system for transmission electron microscopy. Journal of structural biology. 2007;158:318–326. doi: 10.1016/j.jsb.2006.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zheng Z, et al. A Complete Electron Microscopy Volume of the Brain of Adult <em>Drosophila melanogaster</em>. Cell. 2018;174:730–743.:e722. doi: 10.1016/j.cell.2018.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Graham BJ, et al. High-throughput transmission electron microscopy with automated serial sectioning. bioRxiv. 2019:657346. doi: 10.1101/657346. [DOI] [Google Scholar]
- 84.Phelps JS, et al. Reconstruction of motor control circuits in adult Drosophila using automated transmission electron microscopy. Cell. 2021;184:759–774.:e718. doi: 10.1016/j.cell.2020.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yin W, et al. A petascale automated imaging pipeline for mapping neuronal circuits with high-throughput transmission electron microscopy. Nature communications. 2020;11:4949. doi: 10.1038/s41467-020-18659-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Briggman KL, Denk W. Towards neural circuit reconstruction with volume electron microscopy techniques. Curr Opin Neurobiol. 2006;16:562–570. doi: 10.1016/j.conb.2006.08.010. doi:S0959-4388(06)00114-0 [pii] [doi] [DOI] [PubMed] [Google Scholar]
- 87.Peddie CJ, Collinson LM. Exploring the third dimension: volume electron microscopy comes of age. Micron. 2014;61:9–19. doi: 10.1016/j.micron.2014.01.009. [DOI] [PubMed] [Google Scholar]
- 88.Kubota Y, Sohn J, Kawaguchi Y. Large Volume Electron Microscopy and Neural Microcircuit Analysis. Frontiers in neural circuits. 2018;12:98. doi: 10.3389/fncir.2018.00098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Smith D, Starborg T. Serial block face scanning electron microscopy in cell biology: Applications and technology. Tissue Cell. 2019;57:111–122. doi: 10.1016/j.tice.2018.08.011. [DOI] [PubMed] [Google Scholar]
- 90.Sergey G, et al. Oxygen plasma focused ion beam scanning electron microscopy for biological samples. bioRxiv. 2018:457820. doi: 10.1101/457820. [DOI] [Google Scholar]
- 91.Motta A, et al. Dense connectomic reconstruction in layer 4 of the somatosensory cortex. Science (New York, NY) 2019;366:eaay3134. doi: 10.1126/science.aay3134. [DOI] [PubMed] [Google Scholar]
- 92.Denk W, Horstmann H. Serial block-face scanning electron microscopy to reconstruct three-dimensional tissue nanostructure. PLoS Biol. 2004;2:e329. doi: 10.1371/journal.pbio.0020329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lippens S, Kremer A, Borghgraef P, Guérin CJ. Serial block face-scanning electron microscopy for volume electron microscopy. Methods Cell Biol. 2019;152:69–85. doi: 10.1016/bs.mcb.2019.04.002. [DOI] [PubMed] [Google Scholar]
- 94.Titze B, Denk W. Automated in-chamber specimen coating for serial block-face electron microscopy. Journal of Microscopy. 2013;250:101–110. doi: 10.1111/jmi.12023. [DOI] [PubMed] [Google Scholar]
- 95.Deerinck TJ, et al. High-performance serial block-face SEM of nonconductive biological samples enabled by focal gas injection-based charge compensation. Journal of Microscopy. 2017 doi: 10.1111/jmi.12667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Knott G, Marchman H, Wall D, Lich B. Serial section scanning electron microscopy of adult brain tissue using focused ion beam milling. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2008;28:2959–2964. doi: 10.1523/JNEUROSCI.3189-07.2008. doi:28/12/2959 [pii] [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Knott G, Rosset S, Cantoni M. Focussed ion beam milling and scanning electron microscopy of brain tissue. J Vis Exp. 2011 doi: 10.3791/2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Narayan K, Subramaniam S. Focused ion beams in biology. Nature methods. 2015;12:1021–1031. doi: 10.1038/nmeth.3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Xu CS, et al. An open-access volume electron microscopy atlas of whole cells and tissues. Nature. 2021;599:147–151. doi: 10.1038/s41586-021-03992-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Baena V, et al. FIB-SEM as a Volume Electron Microscopy Approach to Study Cellular Architectures in SARS-CoV-2 and Other Viral Infections: A Practical Primer for a Virologist. Viruses. 2021;13 doi: 10.3390/v13040611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Burnett TL, et al. Large volume serial section tomography by Xe Plasma FIB dual beam microscopy. Ultramicroscopy. 2016;161:119–129. doi: 10.1016/j.ultramic.2015.11.001. [DOI] [PubMed] [Google Scholar]
- 102.Winiarski B, et al. Broad ion beam serial section tomography. Ultramicroscopy. 2017;172:52–64. doi: 10.1016/j.ultramic.2016.10.014. [DOI] [PubMed] [Google Scholar]
- 103.Gholinia A, et al. Coupled Broad Ion Beam-Scanning Electron Microscopy (BIB-SEM) for polishing and three dimensional (3D) serial section tomography (SST) Ultramicroscopy. 2020;214:112989. doi: 10.1016/j.ultramic.2020.112989. [DOI] [PubMed] [Google Scholar]
- 104.Wang J, et al. Reactive oxygen FIB spin milling enables correlative workflow for 3D super-resolution light microscopy and serial FIB/SEM of cultured cells. Scientific reports. 2021;11:13162. doi: 10.1038/s41598-021-92608-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hayworth KJ, et al. Gas cluster ion beam SEM for imaging of large tissue samples with 10 nm isotropic resolution. Nature methods. 2020;17:68–71. doi: 10.1038/s41592-019-0641-2. [DOI] [PubMed] [Google Scholar]
- 106.Burel A, et al. A targeted 3D EM and correlative microscopy method using SEM array tomography. Development. 2018;145:dev160879. doi: 10.1242/dev.160879. [DOI] [PubMed] [Google Scholar]
- 107.Wacker I, et al. Hierarchical imaging: a new concept for targeted imaging of large volumes from cells to tissues. BMC Cell Biol. 2016;17:38. doi: 10.1186/s12860-016-0122-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Coll man F, et al. Mapping synapses by conjugate light-electron array tomography. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2015;35:5792–5807. doi: 10.1523/jneurosci.4274-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Gabarre S, et al. A workflow for streamlined acquisition and correlation of serial regions of interest in array tomography. BMC biology. 2021;19:152. doi: 10.1186/s12915-021-01072-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Franke T, Kolotuev I. Array Tomography Workflow for the Targeted Acquisition of Volume Information using Scanning Electron Microscopy. J Vis Exp. 2021 doi: 10.3791/61847. [DOI] [PubMed] [Google Scholar]
- 111.Koike T, et al. A Device for Ribbon Collection for Array Tomography with Scanning Electron Microscopy. Acta Histochem Cytochem. 2017;50:135–140. doi: 10.1267/ahc.17013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Templier T. MagC, magnetic collection of ultrathin sections for volumetric correlative light and electron microscopy. Elife. 2019;8 doi: 10.7554/eLife.45696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Li X, et al. Large scale three-dimensional reconstruction of an entire Caenorhabditis elegans larva using AutoCUTS-SEM. Journal of structural biology. 2017;200:87–96. doi: 10.1016/j.jsb.2017.09.010. [DOI] [PubMed] [Google Scholar]
- 114.Schlek R, et al. ATUM-based SEM for High-Speed Large-Volume Biological Reconstructions. Microscopy and Microanalysis. 2012;18:572–573. doi: 10.1017/s1431927612004710. [DOI] [Google Scholar]
- 115.Eberle AL, et al. High-resolution, high-throughput imaging with a multibeam scanning electron microscope. Journal of Microscopy. 2015;259:114–120. doi: 10.1111/jmi.12224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Shapson-Coe A, et al. A connectomic study of a petascale fragment of human cerebral cortex. bioRxiv. 2021:2021.2005.2029.446289. doi: 10.1101/2021.05.29.446289. [DOI] [Google Scholar]
- 117.Kruit P, Zuidema W. A Dedicated Multi-Beam SEM for Transmission Imaging of Thin Samples. Microscopy and Microanalysis. 2019;25:1034–1035. doi: 10.1017/S1431927619005907. [DOI] [Google Scholar]
- 118.Peddie CJ, et al. Correlative super-resolution fluorescence and electron microscopy using conventional fluorescent proteins in vacuo. Journal of structural biology. 2017;199:120–131. doi: 10.1016/j.jsb.2017.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lane R, Wolters AHG, Giepmans BNG, Hoogenboom JP. Integrated Array Tomography for 3D Correlative Light and Electron Microscopy. Front Mol Biosci. 2021;8:822232. doi: 10.3389/fmolb.2021.822232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Guérin CJ, Kremer A, Borghgraef P, Shih AY, Lippens S. Combining serial block face and focused ion beam scanning electron microscopy for 3D studies of rare events. Methods Cell Biol. 2019;152:87–101. doi: 10.1016/bs.mcb.2019.03.014. [DOI] [PubMed] [Google Scholar]
- 121.Greenwood DJ, et al. Subcellular antibiotic visualization reveals a dynamic drug reservoir in infected macrophages. Science (New York, NY) 2019;364:1279–1282. doi: 10.1126/science.aat9689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kislinger G, et al. ATUM-FIB microscopy for targeting and multiscale imaging of rare events in mouse cortex. STAR Protoc. 2020;1:100232. doi: 10.1016/j.xpro.2020.100232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Hegermann J, et al. Volume-CLEM: a method for correlative light and electron microscopy in three dimensions. Am J Physiol Lung Cell Mol Physiol. 2019;317:L778–l784. doi: 10.1152/ajplung.00333.2019. [DOI] [PubMed] [Google Scholar]
- 124.Narayan K, et al. Multi-resolution correlative focused ion beam scanning electron microscopy: applications to cell biology. Journal of structural biology. 2014;185:278–284. doi: 10.1016/j.jsb.2013.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Wanner AA, Genoud C, Friedrich RW. 3-dimensional electron microscopic imaging of the zebrafish olfactory bulb and dense reconstruction of neurons. Sci Data. 2016;3:160100. doi: 10.1038/sdata.2016.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Maclachlan C, Sahlender DA, Hayashi S, Molnár Z, Knott G. Block Face Scanning Electron Microscopy of Fluorescently Labeled Axons Without Using Near Infra-Red Branding. Frontiers in neuroanatomy. 2018;12:88. doi: 10.3389/fnana.2018.00088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Luckner M, Wanner G. From Light Microscopy to Analytical Scanning Electron Microscopy (SEM) and Focused Ion Beam (FIB)/SEM in Biology: Fixed Coordinates, Flat Embedding, Absolute References. Microsc Microanal. 2018;24:526–544. doi: 10.1017/s1431927618015015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Lees RM, Peddie CJ, Collinson LM, Ashby MC, Verkade P. Correlative two-photon and serial block face scanning electron microscopy in neuronal tissue using 3D near-infrared branding maps. Methods Cell Biol. 2017;140:245–276. doi: 10.1016/bs.mcb.2017.03.007. [DOI] [PubMed] [Google Scholar]
- 129.Kremer A, et al. A workflow for 3D-CLEM investigating liver tissue. J Microsc. 2021;281:231–242. doi: 10.1111/jmi.12967. [DOI] [PubMed] [Google Scholar]
- 130.Thomas CI, et al. Targeting Functionally Characterized Synaptic Architecture Using Inherent Fiducials and 3D Correlative Microscopy. Microsc Microanal. 2021;27:156–169. doi: 10.1017/s1431927620024757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Dalecká M, et al. Invadopodia Structure in 3D Environment Resolved by Near-Infrared Branding Protocol Combining Correlative Confocal and FIB-SEM Microscopy. Int J Mol Sci. 2021;22 doi: 10.3390/ijms22157805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Agronskaia AV, et al. Integrated fluorescence and transmission electron microscopy. Journal of structural biology. 2008;164:183–189. doi: 10.1016/j.jsb.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 133.Kukulski W, et al. Correlated fluorescence and 3D electron microscopy with high sensitivity and spatial precision. The Journal of cell biology. 2011;192:111–119. doi: 10.1083/jcb.201009037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Liv N, et al. Simultaneous correlative scanning electron and high-NA fluorescence microscopy. PLoS One. 2013;8:e55707. doi: 10.1371/journal.pone.0055707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Zonnevylle AC, et al. Integration of a high-NA light microscope in a scanning electron microscope. Journal of Microscopy. 2013;252:58–70. doi: 10.1111/jmi.12071. [DOI] [PubMed] [Google Scholar]
- 136.Peddie CJ, et al. Correlative and integrated light and electron microscopy of in-resin GFP fluorescence, used to localise diacylglycerol in mammalian cells. Ultramicroscopy. 2014;143:3–14. doi: 10.1016/j.ultramic.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Luby-Phelps K, Ning G, Fogerty J, Besharse JC. Visualization of identified GFP-expressing cells by light and electron microscopy. J Histochem Cytochem. 2003;51:271–274. doi: 10.1177/002215540305100301. [DOI] [PubMed] [Google Scholar]
- 138.Nixon SJ, et al. A single method for cryofixation and correlative light, electron microscopy and tomography of zebrafish embryos. Traffic. 2009;10:131–136. doi: 10.1111/j.1600-0854.2008.00859.x. [DOI] [PubMed] [Google Scholar]
- 139.Bloss EB, et al. Single excitatory axons form clustered synapses onto CA1 pyramidal cell dendrites. Nat Neurosci. 2018;21:353–363. doi: 10.1038/s41593-018-0084-6. [DOI] [PubMed] [Google Scholar]
- 140.Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. Journal of structural biology. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 141.Hayworth KJ, et al. Imaging ATUM ultrathin section libraries with WaferMapper: a multi-scale approach to EM reconstruction of neural circuits. Frontiers in neural circuits. 2014;8:68. doi: 10.3389/fncir.2014.00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Titze B, Genoud C, Friedrich RW. SBEMimage: Versatile Acquisition Control Software for Serial Block-Face Electron Microscopy. Frontiers in neural circuits. 2018;12 doi: 10.3389/fncir.2018.00054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Serra Lleti JM, et al. CLEM<em>Site</em>, a software for automated phenotypic screens using light microscopy and FIB-SEM. bioRxiv. 2021:2021.2003.2019.436113. doi: 10.1101/2021.03.19.436113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Mastronarde DN, Held SR. Automated tilt series alignment and tomographic reconstruction in IMOD. Journal of structural biology. 2017;197:102–113. doi: 10.1016/j.jsb.2016.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Lowe DG. Distinctive Image Features from Scale-Invariant Keypoints. International Journal of Computer Vision. 2004;60:91–110. doi: 10.1023/B:VISI.0000029664.99615.94. [DOI] [Google Scholar]
- 146.Saalfeld S, Cardona A, Hartenstein V, Tomančák P. As-rigid-as-possible mosaicking and serial section registration of large ssTEM datasets. Bioinformatics. 2010;26:i57–i63. doi: 10.1093/bioinformatics/btq219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Cardona A, et al. TrakEM2 software for neural circuit reconstruction. PLoS One. 2012;7:e38011. doi: 10.1371/journal.pone.0038011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Rueden CT, et al. ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinformatics. 2017;18:529. doi: 10.1186/s12859-017-1934-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Schindelin J, Rueden CT, Hiner MC, Eliceiri KW. The ImageJ ecosystem: An open platform for biomedical image analysis. Molecular Reproduction and Development. 2015;82:518–529. doi: 10.1002/mrd.22489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Hennies J, et al. AMST: Alignment to Median Smoothed Template for Focused Ion Beam Scanning Electron Microscopy Image Stacks. Scientific reports. 2020;10:2004. doi: 10.1038/s41598-020-58736-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Mitchell E, Keselj S, Popovych S, Buniatyan D, Seung HS. Siamese Encoding and Alignment by Multiscale Learning with Self-Supervision. arXiv:1904.02643. 2019 https://ui.adsabs.harvard.edu/abs/2019arXiv190402643M . [Google Scholar]
- 152.Macrina T, et al. Petascale neural circuit reconstruction: automated methods. bioRxiv. 2021:2021.2008.2004.455162. doi: 10.1101/2021.08.04.455162. [DOI] [Google Scholar]
- 153.Haskins G, Kruger U, Yan P. Deep learning in medical image registration: a survey. Machine Vision and Applications. 2020;31:8. doi: 10.1007/s00138-020-01060-x. [DOI] [Google Scholar]
- 154.Pizer SM, et al. Adaptive histogram equalization and its variations. Computer Vision, Graphics, and Image Processing. 1987;39:355–368. doi: 10.1016/S0734-189X(87)80186-X. [DOI] [Google Scholar]
- 155.Buchholz T-O, et al. In: Methods Cell Biol. Müller-Reichert Thomas, Pigino Gaia., editors. Vol. 152. Academic Press; 2019. pp. 277–289. [DOI] [PubMed] [Google Scholar]
- 156.Bogovic JA, et al. An unbiased template of the Drosophila brain and ventral nerve cord. PLOS ONE. 2021;15:e0236495. doi: 10.1371/journal.pone.0236495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Mais L, et al. PatchPerPixMatch for Automated 3d Search of Neuronal Morphologies in Light Microscopy. bioRxiv. 2021:2021.2007.2023.453511. doi: 10.1101/2021.07.23.453511. [DOI] [Google Scholar]
- 158.Heinrich L, et al. Whole-cell organelle segmentation in volume electron microscopy. Nature. 2021:1–6. doi: 10.1038/s41586-021-03977-3. [DOI] [PubMed] [Google Scholar]
- 159.Paul-Gilloteaux P, et al. eC-CLEM: flexible multidimensional registration software for correlative microscopies. Nature methods. 2017;14:102–103. doi: 10.1038/nmeth.4170. [DOI] [PubMed] [Google Scholar]
- 160.Bogovic JA, Hanslovsky P, Wong A, Saalfeld S. 2016 IEEE 13th International Symposium on Biomedical Imaging (ISBI); pp. 1123–1126. [Google Scholar]
- 161.Klein S, Staring M, Murphy K, Viergever MA, Pluim JPW. <emphasis emphasistype=“mono”>elastix</emphasis>: A Toolbox for Intensity-Based Medical Image Registration. IEEE Transactions on Medical Imaging. 2010;29:196–205. doi: 10.1109/TMI.2009.2035616. [DOI] [PubMed] [Google Scholar]
- 162.Shamonin D, et al. Fast Parallel Image Registration on CPU and GPU for Diagnostic Classification of Alzheimer’s Disease. Frontiers in Neuroinformatics. 2014;7 doi: 10.3389/fninf.2013.00050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Müller A, et al. 3D FIB-SEM reconstruction of microtubule-organelle interaction in whole primary mouse β cells. Journal of Cell Biology. 2020;220:e202010039. doi: 10.1083/jcb.202010039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Berg S, et al. ilastik: interactive machine learning for (bio)image analysis. Nature methods. 2019;16:1226–1232. doi: 10.1038/s41592-019-0582-9. [DOI] [PubMed] [Google Scholar]
- 165.Belevich I, Joensuu M, Kumar D, Vihinen H, Jokitalo E. Microscopy Image Browser: A Platform for Segmentation and Analysis of Multidimensional Datasets. PLOS Biology. 2016;14:e1002340. doi: 10.1371/journal.pbio.1002340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. Journal of structural biology. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- 167.Fiala JC. Reconstruct: a free editor for serial section microscopy. J Microsc. 2005;218:52–61. doi: 10.1111/j.1365-2818.2005.01466.x. [DOI] [PubMed] [Google Scholar]
- 168.Boergens KM, et al. webKnossos: efficient online 3D data annotation for connectomics. Nature methods. 2017;14:691–694. doi: 10.1038/nmeth.4331. [DOI] [PubMed] [Google Scholar]
- 169.Berger DR, Seung HS, Lichtman JW. VAST (Volume Annotation and Segmentation Tool): Efficient Manual and Semi-Automatic Labeling of Large 3D Image Stacks. Frontiers in neural circuits. 2018;12:88. doi: 10.3389/fncir.2018.00088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Tischer C, et al. BigDataProcessor2: a free and open-source Fiji plugin for inspection and processing of TB sized image data. Bioinformatics. 2021;37:3079–3081. doi: 10.1093/bioinformatics/btab106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Linkert M, et al. Metadata matters: access to image data in the real world. The Journal of cell biology. 2010;189:777–782. doi: 10.1083/jcb.201004104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Arganda-Carreras I, et al. Crowdsourcing the creation of image segmentation algorithms for connectomics. Frontiers in neuroanatomy. 2015;9 doi: 10.3389/fnana.2015.00142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Scheffer LK, et al. A connectome and analysis of the adult Drosophila central brain. eLife. 2020;9:e57443. doi: 10.7554/elife.57443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Legland D, Arganda-Carreras I, Andrey P. MorphoLibJ: integrated library and plugins for mathematical morphology with ImageJ. Bioinformatics. 2016;32:3532–3534. doi: 10.1093/bioinformatics/btw413. [DOI] [PubMed] [Google Scholar]
- 175.Schmidt U, Weigert M, Broaddus C, Myers G. Springer International Publishing; pp. 265–273. [Google Scholar]
- 176.Stringer C, Wang T, Michaelos M, Pachitariu M. Cellpose: a generalist algorithm for cellular segmentation. Nature methods. 2021;18:100–106. doi: 10.1038/s41592-020-01018-x. [DOI] [PubMed] [Google Scholar]
- 177.Ronneberger O, Fischer P, Brox T. Springer International Publishing; pp. 234–241. [Google Scholar]
- 178.Arganda-Carreras I, et al. Trainable Weka Segmentation: a machine learning tool for microscopy pixel classification. Bioinformatics. 2017;33:2424–2426. doi: 10.1093/bioinformatics/btx180. [DOI] [PubMed] [Google Scholar]
- 179.Gómez-de-Mariscal E, et al. DeepImageJ: A user-friendly environment to run deep learning models in ImageJ. Nature methods. 2021;18:1192–1195. doi: 10.1038/s41592-021-01262-9. [DOI] [PubMed] [Google Scholar]
- 180.Belevich I, Jokitalo E. DeepMIB: User-friendly and open-source software for training of deep learning network for biological image segmentation. bioRxiv. 2020:2020.2007.2013.200105. doi: 10.1101/2020.07.13.200105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Haberl MG, et al. CDeep3M—Plug-and-Play cloud-based deep learning for image segmentation. Nature methods. 2018;15:677–680. doi: 10.1038/s41592-018-0106-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Conrad R, Narayan K. Instance segmentation of mitochondria in electron microscopy images with a generalist deep learning model. bioRxiv. 2022:2022.2003.2017.484806. doi: 10.1101/2022.03.17.484806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Conrad R, Narayan K. CEM500K, a large-scale heterogeneous unlabeled cellular electron microscopy image dataset for deep learning. eLife. 2021;10:e65894. doi: 10.7554/eLife.65894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.von Chamier L, et al. ZeroCostDL4Mic: an open platform to use Deep-Learning in Microscopy. bioRxiv. 2020:2020.2003.2020.000133. doi: 10.1101/2020.03.20.000133. [DOI] [Google Scholar]
- 185.Funke J, et al. Large Scale Image Segmentation with Structured Loss Based Deep Learning for Connectome Reconstruction. Ieee T Pattern Anal. 2019;41:1669–1680. doi: 10.1109/TPAMI.2018.2835450. [DOI] [PubMed] [Google Scholar]
- 186.Wolf S, et al. The Mutex Watershed and its Objective: Efficient, Parameter-Free Graph Partitioning. Ieee T Pattern Anal. 2021;43:3724–3738. doi: 10.1109/TPAMI.2020.2980827. [DOI] [PubMed] [Google Scholar]
- 187.Berning M, Boergens Kevin M, Helmstaedter M. SegEM: Efficient Image Analysis for High-Resolution Connectomics. Neuron. 2015;87:1193–1206. doi: 10.1016/j.neuron.2015.09.003. [DOI] [PubMed] [Google Scholar]
- 188.Januszewski M, et al. High-precision automated reconstruction of neurons with flood-filling networks. Nature methods. 2018;15:605–610. doi: 10.1038/s41592-018-0049-4. [DOI] [PubMed] [Google Scholar]
- 189.Heinrich L, Funke J, Pape C, Nunez-Iglesias J, Saalfeld S. Springer International Publishing; pp. 317–325. [Google Scholar]
- 190.Buhmann J, et al. Automatic detection of synaptic partners in a whole-brain Drosophila electron microscopy data set. Nature methods. 2021;18:771–774. doi: 10.1038/s41592-021-01183-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Sofroniew N, et al. napari. Zenodo. 2021 doi: 10.5281/zenodo.55878923. [DOI] [Google Scholar]
- 192.Dorkenwald S, et al. FlyWire: online community for whole-brain connectomics. Nature methods. 2022;19:119–128. doi: 10.1038/s41592-021-01330-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Leite V, et al. Paintera. Zenodo. 2021 doi: 10.5281/zenodo.5028021. [DOI] [Google Scholar]
- 194.Kaynig V, et al. Large-scale automatic reconstruction of neuronal processes from electron microscopy images. Medical Image Analysis. 2015;22:77–88. doi: 10.1016/j.media.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Pape C, et al. 2017 Ieee Int Conf Comput Vis Work Iccvw. :1–10. [Google Scholar]
- 196.Lu R, Zlateski A, Seung HS. Large-scale image segmentation based on distributed clustering algorithms. arXiv: 2106.10795. 2021 https://ui.adsabs.harvard.edu/abs/2021arXiv210610795L . [Google Scholar]
- 197.Madany M, Marcus K, Peltier S, Ellisman MH, Altintas I. NeuroKube: An Automated and Autoscaling Neuroimaging Reconstruction Framework using Cloud Native Computing and A.I. 2020 Ieee Int Conf Big Data Big Data. 2020:320–330. doi: 10.1109/bigdata50022.2020.9378053. 00. [DOI] [Google Scholar]
- 198.Kim JS, et al. Space-time wiring specificity supports direction selectivity in the retina. Nature. 2014;509:331–336. doi: 10.1038/nature13240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Spiers H, et al. Deep learning for automatic segmentation of the nuclear envelope in electron microscopy data, trained with volunteer segmentations. Traffic. 2021;22:240–253. doi: 10.1111/tra.12789. [DOI] [PubMed] [Google Scholar]
- 200.Pietzsch T, Saalfeld S, Preibisch S, Tomancak P. BigDataViewer: visualization and processing for large image data sets. Nature methods. 2015;12:481–483. doi: 10.1038/nmeth.3392. [DOI] [PubMed] [Google Scholar]
- 201.Tischer C, et al. MoBIE: A free and open-source platform for integration and cloud-based sharing of multi-modal correlative big image data. Microscopy and Microanalysis. 2021;27:2588–2589. doi: 10.1017/S1431927621009193. [DOI] [Google Scholar]
- 202.Saalfeld S, Cardona A, Hartenstein V, Tomančák P. CATMAID: collaborative annotation toolkit for massive amounts of image data. Bioinformatics. 2009;25:1984–1986. doi: 10.1093/bioinformatics/btp266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Maitin-Shepard J, et al. neuroglancer. Zenodo. 2021 zenodo.5573294. [Google Scholar]
- 204.Helmstaedter M, Briggman KL, Denk W. High-accuracy neurite reconstruction for high-throughput neuroanatomy. Nat Neurosci. 2011;14:1081–1088. doi: 10.1038/nn.2868. [DOI] [PubMed] [Google Scholar]
- 205.Porter KR, Palade GE. Studies on the Endoplasmic Reticulum : III. Its form and Distribution in Striated Muscle Cells. The Journal of Biophysical and Biochemical Cytology. 1957;3:269–300. doi: 10.1083/jcb.3.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Redemann S, et al. C. elegans chromosomes connect to centrosomes by anchoring into the spindle network. Nature communications. 2017;8:15288. doi: 10.1038/ncomms15288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Terasaki M, et al. Stacked Endoplasmic Reticulum Sheets Are Connected by Helicoidal Membrane Motifs. Cell. 2013;154:285–296. doi: 10.1016/j.cell.2013.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Puhka M, Vihinen H, Joensuu M, Jokitalo E. Endoplasmic reticulum remains continuous and undergoes sheet-to-tubule transformation during cell division in mammalian cells. The Journal of cell biology. 2007;179:895–909. doi: 10.1083/jcb.200705112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Puhka M, Joensuu M, Vihinen H, Belevich I, Jokitalo E. Progressive sheet-to-tubule transformation is a general mechanism for endoplasmic reticulum partitioning in dividing mammalian cells. Mol Biol Cell. 2012;23:2424–2432. doi: 10.1091/mbc.E10-12-0950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Pain C, Kriechbaumer V, Kittelmann M, Hawes C, Fricker M. Quantitative analysis of plant ER architecture and dynamics. Nature communications. 2019;10:984. doi: 10.1038/s41467-019-08893-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Wu Y, et al. Contacts between the endoplasmic reticulum and other membranes in neurons. Proc Natl Acad Sci U S A. 2017;114:E4859–e4867. doi: 10.1073/pnas.1701078114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Yang K, et al. ER exit sites in Drosophila display abundant ER-Golgi vesicles and pearled tubes but no megacarriers. Cell Rep. 2021;36:109707. doi: 10.1016/j.celrep.2021.109707. [DOI] [PubMed] [Google Scholar]
- 213.Weigel AV, et al. ER-to-Golgi protein delivery through an interwoven, tubular network extending from ER. Cell. 2021;184:2412–2429.:e2416. doi: 10.1016/j.cell.2021.03.035. [DOI] [PubMed] [Google Scholar]
- 214.Fermie J, et al. Single organelle dynamics linked to 3D structure by correlative live-cell imaging and 3D electron microscopy. Traffic. 2018;19:354–369. doi: 10.1111/tra.12557. [DOI] [PubMed] [Google Scholar]
- 215.Loginov S, et al. Correlative Organelle Microscopy: fluorescence guided volume electron microscopy of intracellular processes. bioRxiv. 2021:2021.2003.2029.437317. doi: 10.1101/2021.03.29.437317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Prinz WA, Toulmay A, Balla T. The functional universe of membrane contact sites. Nature reviews Molecular cell biology. 2020;21:7–24. doi: 10.1038/s41580-019-0180-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Parlakgül G, et al. High resolution 3D imaging of liver reveals a central role for subcellular architectural organization in metabolism. bioRxiv. 2020:2020.2011.2018.387803. doi: 10.1101/2020.11.18.387803. [DOI] [Google Scholar]
- 218.Cortese M, et al. Ultrastructural Characterization of Zika Virus Replication Factories. Cell Rep. 2017;18:2113–2123. doi: 10.1016/j.celrep.2017.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Hackstadt T, et al. Disruption of the Golgi Apparatus and Contribution of the Endoplasmic Reticulum to the SARS-CoV-2 Replication Complex. Viruses. 2021;13 doi: 10.3390/v13091798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Lee JH, Pasquarella JR, Kalejta RF. Cell Line Models for Human Cytomegalovirus Latency Faithfully Mimic Viral Entry by Macropinocytosis and Endocytosis. J Virol. 2019;93 doi: 10.1128/jvi.01021-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Felts RL, et al. 3D visualization of HIV transfer at the virological synapse between dendritic cells and T cells. Proc Natl Acad Sci U S A. 2010;107:13336–13341. doi: 10.1073/pnas.1003040107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Nkwe DO, Pelchen-Matthews A, Burden JJ, Collinson LM, Marsh M. The intracellular plasma membrane-connected compartment in the assembly of HIV-1 in human macrophages. BMC biology. 2016;14:50. doi: 10.1186/s12915-016-0272-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Aggarwal A, Stella AO, Henry CC, Narayan K, Turville SG. Embedding of HIV Egress within Cortical F-Actin. Pathogens. 2022;11:56. doi: 10.3390/pathogens11010056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Mayorova TD, et al. Placozoan fiber cells: mediators of innate immunity and participants in wound healing. Scientific reports. 2021;11:23343. doi: 10.1038/s41598-021-02735-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Decelle J, et al. Algal Remodeling in a Ubiquitous Planktonic Photosymbiosis. Current biology : CB. 2019;29:968–978.:e964. doi: 10.1016/j.cub.2019.01.073. [DOI] [PubMed] [Google Scholar]
- 226.Uwizeye C, et al. Cytoklepty in the plankton: A host strategy to optimize the bioenergetic machinery of endosymbiotic algae. Proc Natl Acad Sci USA. 2021;118 doi: 10.1073/pnas.2025252118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Weiner A, Enninga J. The Pathogen-Host Interface in Three Dimensions: Correlative FIB/SEM Applications. Trends Microbiol. 2019;27:426–439. doi: 10.1016/j.tim.2018.11.011. [DOI] [PubMed] [Google Scholar]
- 228.Beckwith MS, et al. Seeing a Mycobacterium-Infected Cell in Nanoscale 3D: Correlative Imaging by Light Microscopy and FIB/SEM Tomography. PLoS One. 2015;10:e0134644. doi: 10.1371/journal.pone.0134644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Bernard EM, et al. M. tuberculosis infection of human iPSC-derived macrophages reveals complex membrane dynamics during xenophagy evasion. J Cell Sci. 2020;134 doi: 10.1242/jcs.252973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Hunt A, et al. Differential requirements for cyclase-associated protein (CAP) in actin-dependent processes of Toxoplasma gondii. Elife. 2019;8 doi: 10.7554/eLife.50598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Kolba MD, et al. Tunneling nanotube-mediated intercellular vesicle and protein transfer in the stroma-provided imatinib resistance in chronic myeloid leukemia cells. Cell Death Dis. 2019;10:817. doi: 10.1038/s41419-019-2045-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Zenker J, et al. A microtubule-organizing center directing intracellular transport in the early mouse embryo. Science (New York, NY) 2017;357:925–928. doi: 10.1126/science.aam9335. [DOI] [PubMed] [Google Scholar]
- 233.Frías-Anaya E, et al. Age-related ultrastructural neurovascular changes in the female mouse cortex and hippocampus. Neurobiol Aging. 2021;101:273–284. doi: 10.1016/j.neurobiolaging.2020.12.008. [DOI] [PubMed] [Google Scholar]
- 234.Sheu S-H, et al. A serotonergic axon-cilium synapse drives nuclear signaling to maintain chromatin accessibility. bioRxiv. 2021:2021.2009.2027.461878. doi: 10.1101/2021.09.27.461878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Pipkin JE, Bushong EA, Ellisman MH, Kristan WB., Jr Verifying, Challenging, and Discovering New Synapses Among Fully EM-Reconstructed Neurons in the Leech Ganglion. Frontiers in neuroanatomy. 2018;12:95. doi: 10.3389/fnana.2018.00095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Tran HT, Lucas MS, Ishikawa T, Shahmoradian SH, Padeste C. A Compartmentalized Neuronal Cell-Culture Platform Compatible With Cryo-Fixation by High-Pressure Freezing for Ultrastructural Imaging. Frontiers in neuroscience. 2021;15:726763. doi: 10.3389/fnins.2021.726763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Morgan JL, Lichtman JW. An Individual Interneuron Participates in Many Kinds of Inhibition and Innervates Much of the Mouse Visual Thalamus. Neuron. 2020;106:468–481.:e462. doi: 10.1016/j.neuron.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Baena V, Terasaki M. Three-dimensional organization of transzonal projections and other cytoplasmic extensions in the mouse ovarian follicle. Scientific reports. 2019;9:1262. doi: 10.1038/s41598-018-37766-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Boulogne C, et al. Functional organisation of the endomembrane network in the digestive gland of the Venus flytrap: revisiting an old story with a new microscopy toolbox. Journal of Microscopy. 2020;280:86–103. doi: 10.1111/jmi.12957. [DOI] [PubMed] [Google Scholar]
- 240.Czymmek K, et al. Plant Endosomes, Methods and Protocols. Methods Mol Biology. 2020;2177:69–81. doi: 10.1007/978-1-0716-0767-1_7. [DOI] [PubMed] [Google Scholar]
- 241.Polilov AA, Makarova AA, Pang S, Xu CS, Hess H. Protocol for preparation of heterogeneous biological samples for 3D electron microscopy: a case study for insects. Scientific reports. 2021;11:4717. doi: 10.1038/s41598-021-83936-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Wang X-Q, et al. Three-dimensional reconstruction of a whole insect reveals its phloem sap-sucking mechanism at nano-resolution. eLife. 2021;10:e62875. doi: 10.7554/elife.62875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Kaji T, Kakui K, Miyazaki N, Murata K, Palmer AR. Mesoscale morphology at nanoscale resolution: serial block-face scanning electron microscopy reveals fine 3D detail of a novel silk spinneret system in a tube-building tanaid crustacean. Frontiers in Zoology. 2016;13:14. doi: 10.1186/s12983-016-0146-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Denk W, Briggman KL, Helmstaedter M. Structural neurobiology: missing link to a mechanistic understanding of neural computation. Nature Reviews Neuroscience. 2012;13:351–358. doi: 10.1038/nrn3169. [DOI] [PubMed] [Google Scholar]
- 245.Briggman KL, Helmstaedter M, Denk W. Wiring specificity in the direction-selectivity circuit of the retina. Nature. 2011;471:183–188. doi: 10.1038/nature09818. [DOI] [PubMed] [Google Scholar]
- 246.Ryan K, Lu Z, Meinertzhagen IA. The CNS connectome of a tadpole larva of Ciona intestinalis (L.) highlights sidedness in the brain of a chordate sibling. eLife. 2016;5:e16962. doi: 10.7554/elife.16962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Ohyama T, et al. A multilevel multimodal circuit enhances action selection in Drosophila. Nature. 2015;520:633–639. doi: 10.1038/nature14297. [DOI] [PubMed] [Google Scholar]
- 248.Consortium, M et al. Functional connectomics spanning multiple areas of mouse visual cortex. bioRxiv. 2021:2021.2007.2028.454025. doi: 10.1101/2021.07.28.454025. [DOI] [Google Scholar]
- 249.Pfeiffer RL, et al. A pathoconnectome of early neurodegeneration: Network changes in retinal degeneration. Experimental Eye Research. 2020;199:108196. doi: 10.1016/j.exer.2020.108196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Witvliet D, et al. Connectomes across development reveal principles of brain maturation. Nature. 2021;596:257–261. doi: 10.1038/s41586-021-03778-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Wilson CE, et al. Taste Bud Connectome: Implications for Taste Information Processing. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2021 doi: 10.1523/jneurosci.0838-21.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Randel N, et al. Inter-individual stereotypy of the Platynereislarval visual connectome. eLife. 2015;4:27. doi: 10.7554/elife.08069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Verasztó C, et al. Whole-animal connectome and cell-type complement of the three-segmented <em>Platynereis dumerilii</em> larva. bioRxiv. 2020:2020.2008.2021.260984. doi: 10.1101/2020.08.21.260984. [DOI] [Google Scholar]
- 254.Kasthuri N, et al. Saturated Reconstruction of a Volume of Neocortex. Cell. 2015;162:648–661. doi: 10.1016/j.cell.2015.06.054. [DOI] [PubMed] [Google Scholar]
- 255.Terasaki M. Axonal endoplasmic reticulum is very narrow. Journal of Cell Science. 2018;131:jcs210450. doi: 10.1242/jcs.210450. [DOI] [PubMed] [Google Scholar]
- 256.Yao PJ, et al. Mitochondrial Protrusions in Neuronal Cells. Iscience. 2020;23:101514. doi: 10.1016/j.isci.2020.101514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Tomassy GS, et al. Distinct Profiles of Myelin Distribution Along Single Axons of Pyramidal Neurons in the Neocortex. Science (New York, NY) 2014;344:319–324. doi: 10.1126/science.1249766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Erlandson RA. Role of Electron Microscopy in Modern Diagnostic Surgical Pathology. Mod Surg Pathology. 2009:71–84. doi: 10.1016/b978-1-4160-3966-2.00005-9. [DOI] [Google Scholar]
- 259.Shoemark A, et al. International consensus guideline for reporting transmission electron microscopy results in the diagnosis of Primary Ciliary Dyskinesia (BEAT PCD TEM Criteria) European Respiratory Journal. 2020:1900725. doi: 10.1183/13993003.00725-2019. [DOI] [PubMed] [Google Scholar]
- 260.Shoemark A, et al. Primary ciliary dyskinesia with normal ultrastructure: three-dimensional tomography detects absence of DNAH11. European Respiratory Journal. 2018;51:1701809. doi: 10.1183/13993003.01809-2017. [DOI] [PubMed] [Google Scholar]
- 261.Takaki T, Ohno N, Saitoh S, Nagai M, Joh K. Podocyte penetration of the glomerular basement membrane to contact on the mesangial cell at the lesion of mesangial interposition in lupus nephritis: a three-dimensional analysis by serial block-face scanning electron microscopy. Clin Exp Nephrol. 2019;23:773–781. doi: 10.1007/s10157-019-01701-0. [DOI] [PubMed] [Google Scholar]
- 262.Nagai M, et al. Glomerular Cellular Interactions Following Disruption of the Glomerular Basement Membrane in IgA Nephropathy: Ultrastructural Analyses by 3-Dimensional Serial Block-Face Scanning Electron Microscopy. Kidney Medicine. 2020;2:222–225. doi: 10.1016/j.xkme.2019.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Domínguez-Álvaro M, Montero-Crespo M, Blazquez-Llorca L, DeFelipe J, Alonso-Nanclares L. 3D Electron Microscopy Study of Synaptic Organization of the Normal Human Transentorhinal Cortex and Its Possible Alterations in Alzheimer’s Disease. Eneuro. 2019;6 doi: 10.1523/eneuro.0140-19.2019. ENEURO.0140-0119.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Shahmoradian SH, et al. Lewy pathology in Parkinson’s disease consists of crowded organelles and lipid membranes. Nat Neurosci. 2019;22:1099–1109. doi: 10.1038/s41593-019-0423-2. [DOI] [PubMed] [Google Scholar]
- 265.Riesterer JL, et al. A workflow for visualizing human cancer biopsies using large-format electron microscopy. Methods Cell Biol. 2020;158:163–181. doi: 10.1016/bs.mcb.2020.01.005. [DOI] [PubMed] [Google Scholar]
- 266.Johnson BE, et al. An Omic and Multidimensional Spatial Atlas from Serial Biopsies of an Evolving Metastatic Breast Cancer. bioRxiv. 2021:2020.2012.2003.408500. doi: 10.1101/2020.12.03.408500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.de Senneville BD, et al. Deciphering tumour tissue organization by 3D electron microscopy and machine learning. Communications Biology. 2021;4:1390. doi: 10.1038/s42003-021-02919-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Schneider JP, Hegermann J, Wrede C. Volume electron microscopy: analyzing the lung. Histochem Cell Biol. 2021;155:241–260. doi: 10.1007/s00418-020-01916-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Goggin P, et al. Development of protocols for the first serial block-face scanning electron microscopy (SBF SEM) studies of bone tissue. Bone. 2020;131:115107. doi: 10.1016/j.bone.2019.115107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Nelson G, et al. QUAREP-LiMi: A community-driven initiative to establish guidelines for quality assessment and reproducibility for instruments and images in light microscopy. Journal of Microscopy. 2021;284:56–73. doi: 10.1111/jmi.13041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Sarkans U, et al. REMBI: Recommended Metadata for Biological Images—enabling reuse of microscopy data in biology. Nature methods. 2021;18:1418–1422. doi: 10.1038/s41592-021-01166-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Scheres SH. RELION: implementation of a Bayesian approach to cryo-EM structure determination. Journal of structural biology. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Borrett S, Hughes L. Reporting methods for processing and analysis of data from serial block face scanning electron microscopy. Journal of Microscopy. 2016;263:3–9. doi: 10.1111/jmi.12377. [DOI] [PubMed] [Google Scholar]
- 274.Warfield SK, Zou KH, Wells WM. Simultaneous truth and performance level estimation (STAPLE): an algorithm for the validation of image segmentation. IEEE Transactions on Medical Imaging. 2004;23:903–921. doi: 10.1109/TMI.2004.828354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Destrempes F, et al. Assessment of Inter-Expert Variability and of an Automated Segmentation Method of 40 and 60 MHz IVUS Images of Coronary Arteries. PLOS ONE. 2017;12:e0168332. doi: 10.1371/journal.pone.0168332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Moore J, et al. OME-NGFF: a next-generation file format for expanding bioimaging data-access strategies. Nature methods. 2021;18:1496–1498. doi: 10.1038/s41592-021-01326-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Iudin A, Korir PK, Salavert-Torres J, Kleywegt GJ, Patwardhan A. EMPIAR: a public archive for raw electron microscopy image data. Nature methods. 2016;13:387–388. doi: 10.1038/nmeth.3806. [DOI] [PubMed] [Google Scholar]
- 278.Hartley M, et al. The BioImage Archive - Building a Home for Life-Sciences Microscopy Data. J Mol Biol. 2022:167505. doi: 10.1016/j.jmb.2022.167505. [DOI] [PubMed] [Google Scholar]
- 279.Vidavsky N, et al. Cryo-FIB-SEM serial milling and block face imaging: Large volume structural analysis of biological tissues preserved close to their native state. Journal of structural biology. 2016;196:487–495. doi: 10.1016/j.jsb.2016.09.016. [DOI] [PubMed] [Google Scholar]
- 280.Zhu Y, et al. Serial cryoFIB/SEM Reveals Cytoarchitectural Disruptions in Leigh Syndrome Patient Cells. Structure. 2021;29:82–87.:e83. doi: 10.1016/j.str.2020.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Groen J, Conesa JJ, Valcárcel R, Pereiro E. The cellular landscape by cryo soft X-ray tomography. Biophysical Reviews. 2019;11:611–619. doi: 10.1007/s12551-019-00567-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Dittmayer C, et al. Why misinterpretation of electron micrographs in SARS-CoV-2-infected tissue goes viral. Lancet. 2020;396:e64–e65. doi: 10.1016/s0140-6736(20)32079-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Withers PJ, et al. X-ray computed tomography. Nat Rev Methods Primers. 2021;1:18. doi: 10.1038/s43586-021-00015-4. [DOI] [Google Scholar]
- 284.Rawson SD, Maksimcuka J, Withers PJ, Cartmell SH. X-ray computed tomography in life sciences. BMC biology. 2020;18:21. doi: 10.1186/s12915-020-0753-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Bosch C, et al. Functional and multiscale 3D structural investigation of brain tissue through correlative <em>in vivo</em> physiology, synchrotron micro-tomography and volume electron microscopy. bioRxiv. 2021:2021.2001.2013.426503. doi: 10.1101/2021.01.13.426503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Bushong EA, et al. X-ray microscopy as an approach to increasing accuracy and efficiency of serial block-face imaging for correlated light and electron microscopy of biological specimens. Microsc Microanal. 2015;21:231–238. doi: 10.1017/s1431927614013579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Kuan AT, et al. Dense neuronal reconstruction through X-ray holographic nano-tomography. Nat Neurosci. 2020;23:1637–1643. doi: 10.1038/s41593-020-0704-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Immel A, et al. Effect of X-ray irradiation on ancient DNA in sub-fossil bones – Guidelines for safe X-ray imaging. Scientific reports. 2016;6:32969. doi: 10.1038/srep32969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Wallander H, Wallentin J. Simulated sample heating from a nanofocused X-ray beam. Journal of synchrotron radiation. 2017;24:925–933. doi: 10.1107/s1600577517008712. [DOI] [PMC free article] [PubMed] [Google Scholar]
References
- Ref 5: This comprehensive review of the main SEM-based vEM techniques is a must read for those who are starting in the field, as the authors clearly address the potential and limitations of FIB-SEM, SBF-SEM and array tomography, from sample preparation to image analysis.
- Ref 27: This important update in sample preparations for vEM imaging that addressed the uneven staining gradients of samples up to 1 mm in diameter.
- Ref 43: This work presents clever targeting of a ROI using an ultramicrotome to trim the sample and FIB-SEM to directly acquire the volume – an elegant example of vCLEM where in vivo imaging is registered to FIB-SEM.
- Ref 44: This work demonstrates the power of multimodal correlative microscopy for identifying, targeting, and acquiring a specific ROI within a large sample volume.
- Ref 52: This work is an example of how to optimize the use pattern of an instrument for a specific application by developing it into a fault-tolerant system, as opposed to re-engineering it to be fault free, in order to overcome some of the fundamental limitations of the hardware.
- Ref 59: This work is an example of the use, strengths and limitations of genetically encoded stains for vEM.
- Ref 71: This work is an accessible introduction to array tomography - light and electron microscopic applications.
- Ref 87: This work is the first articulation of the term vEM in the context that is now accepted and extended by the community.
- Ref 92: This work is one of the first examples of vEM imaging of resin-embedded cellular samples, effected by automated in situ serial sectioning and SEM imaging.
- Ref 96: This work is one of the first examples of vEM imaging of resin-embedded cellular samples, effected by automated in situ FIB milling and SEM imaging.
- Ref 116: This work is a tour de force on vEM and automated segmentation of a cubic millimetre of human cerebral cortex with excellent ultrastructural preservation that allows analysis of human cortical circuits at synaptic level, including identification of main neuron types, glia and blood vessels.
- Ref 138: This work is one of the first demonstrations of preservation of fluorescent protein emission after embedding in resin for electron microscopy, raising the possibility of high-accuracy postembedding vCLEM.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The two example data analysis workflows for neuron and mitochondria segmentation can be found at: https://github.com/kreshuklab/vem-primer-examples.Figure Legends