Abstract
Replication and segregation of the genetic information is necessary for a cell to proliferate. In Bacillus subtilis, the Par system (ParA/Soj, ParB/Spo0J and parS) is required for segregation of the chromosome origin (oriC) region and for proper control of DNA replication initiation. ParB binds parS sites clustered near the origin of replication and assembles into sliding clamps that interact with ParA to drive origin segregation through a diffusion-ratchet mechanism. As part of this dynamic process, ParB stimulates ParA ATPase activity to trigger its switch from an ATP-bound dimer to an ADP-bound monomer. In addition to its conserved role in DNA segregation, ParA is also a regulator of the master DNA replication initiation protein DnaA. We hypothesized that in B. subtilis the location of the Par system proximal to oriC would be necessary for ParA to properly regulate DnaA. To test this model, we constructed a range of genetically modified strains with altered numbers and locations of parS sites, many of which perturbed chromosome origin segregation as expected. Contrary to our hypothesis, the results show that regulation of DNA replication initiation by ParA is maintained when a parS site is separated from oriC. Because a single parS site is sufficient for proper control of ParA, the results are consistent with a model where ParA is efficiently regulated by ParB sliding clamps following loading at parS.
Keywords: DNA replication initiation, chromosome segregation, ParB, ParA, parS, oriC, DnaA, Bacillus subtilis
Introduction
The ability for cells to coordinate DNA replication with chromosome segregation is critical for execution of the cell cycle. In most prokaryotes the processes of DNA replication and segregation overlap, with newly replicated DNA regions immediately separated from one another. Primary bacterial chromosomes generally harbour a single origin of replication (oriC). Binding of the master initiator protein DnaA to oriC promotes specific DNA unwinding and allows the loading of the replication machinery [1]. Following duplication of oriC, the organization and movement of this chromosomal region is critical for subsequent bulk chromosome segregation [2].
Bacterial DNA partitioning (par) genes were first identified as elements required for accurate segregation of low copy-number plasmids into daughter cells. The par locus consists of three elements, two trans-acting proteins (ParA and ParB) and a cis-acting binding sequence (parS). All three elements are essential for efficient inheritance of plasmid DNA into daughter cells [3–5]. Homologous partitioning genes are harboured within the majority of bacterial chromosomes and bioinformatic analysis revealed that parS sites generally cluster near oriC, suggesting that cellular Par systems are devoted to regulating processes involving the origin region of the bacterial genome [6].
ParB, known as Spo0J in B. subtilis because the spo0J mutation confers a classic sporulation defect, is a dimeric CTP-dependent DNA sliding clamp [7–14]. ParBCTP binds specifically to a parS site, which stimulates the ParB N-terminal nucleotide binding domain to dimerize, causing ParB to adopt an annular structure that can slide along the DNA [9, 15]. In B. subtilis, a parB null mutant both overinitiates DNA replication and produces anucleate cells, indicating that it plays roles in both DNA synthesis and chromosome segregation [8, 16–19].
ParA, known as Soj in B. subtilis (suppressor of spo0J), is a dynamic ATPase [8, 20] that changes oligomeric state and activity depending upon the bound adenine nucleotide [21]. It has been proposed that ParAADP is a monomer which can interact directly with DnaA to inhibit the initiation of DNA replication [16, 17, 22]. ParAATP forms a homodimer that binds DNA in a sequence non-specific manner and promotes DnaA-dependent DNA replication initiation [16, 17, 22]. ParB stimulates the ATPase activity of ParA to generate ParAADP monomers, thus triggering its switch from an activator to an inhibitor of DnaA [16, 17].
As in other Par systems, ParA plays a role in chromosome segregation, specifically facilitating bidirectional origin movement following DNA replication initiation [2]. Current evidence indicates that ParAATP binds to DNA in a non-specific manner, forming a gradient over the chromosome that attracts ParB: parS nucleoprotein complexes [23–26]. The interaction of ParB with dimeric ParA activates its ATPase activity, converting it to ParAADP monomers that cannot bind DNA and thus diffuse away from the ParB nucleoprotein complexes. The dispersion of ParAADP from the DNA creates a path of ParAATP dimers on the chromosome that can attract ParB, thereby working as a Brownian ratchet to couple the ATPase-dependent cycling of ParA to the directed movement of ParB:parS nucleoprotein complexes [23–26].
In addition to the Par system, the B. subtilis structural maintenance of chromosome (SMC) condensin complex is recruited to the oriC region by ParB:parS nucleoprotein complexes where it organizes the chromosome origin macrodomain and facilitates separation of newly replicated origins, particularly during rapid cell growth [27–32]. In nutrient rich media, null mutants in any of the genes encoding the SMC complex leads to growth inhibition, anucleate cell formation, and aberrant nucleoid structure [27–32].
It has been established that ParB:parS complexes must be located near oriC to promote origin region segregation through the ParA and condensin systems [27, 29, 30]. We hypothesized that origin proximal localization of ParB:parS nucleoprotein complexes might also be required for proper regulation of DNA replication initiation by ParA (via DnaA at oriC). In this model the Par system would act to coordinate the processes of DNA segregation and replication (Fig. 1A).
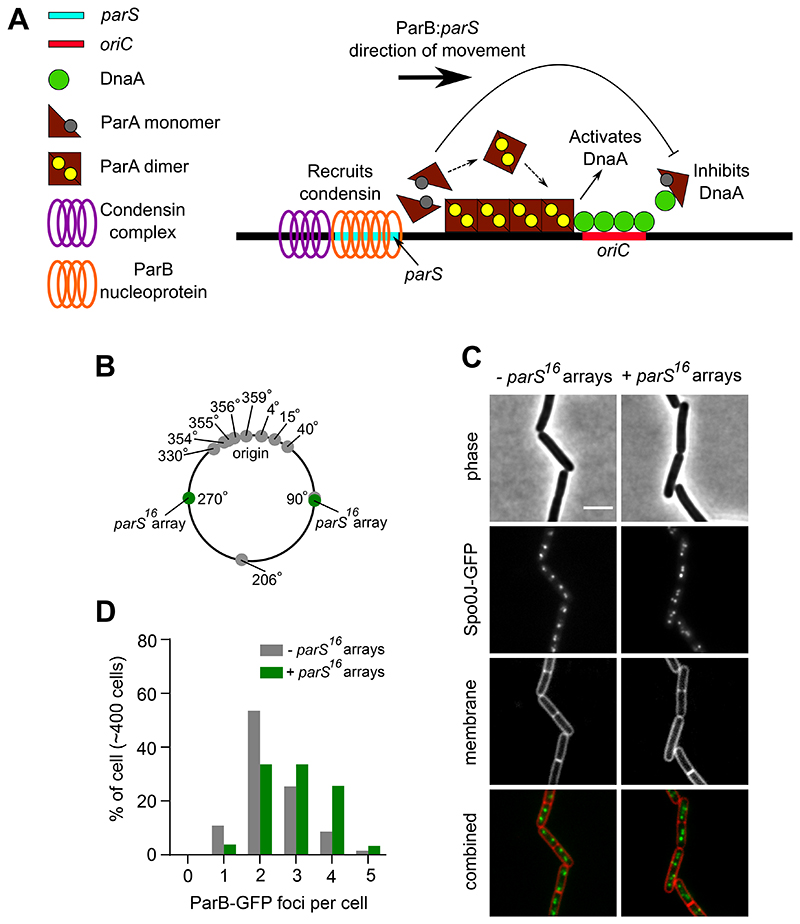
Figure 1. Chromosomal parS16 arrays redistribute ParB.
(A) Schematic of a two-dimensional ParB nucleoprotein complex along a parS with the left section showing ParB interacting with condensin for chromosome organization and segregation. The right section showing ParA spreading across the DNA in a gradient, with ParB regulating the activity cycle of ParA. Regulation of initiation of DNA replication depends on the oligomeric state of ParA. In addition, the diffusion ratchet manner of ParB-ParA interactions allow the segregation of replicated sister origins. For simplicity, ATP and ADP is shown as a yellow and grey circle respectively. (B) Schematic diagram showing the location of B. subtilis endogenous chromosomal parS (grey circles) and the parS16 arrays (green circles). (C) ParB-GFP localization in a wild-type strain and in a strain harbouring parS16 arrays. Phase contrast (top panel), ParB-GFP (middle panel) and outer membrane dye FM-595 (bottom panel). (D) The number of ParB-GFP foci per cell increased in the presence of parS16 arrays. Scale bar, 3 μm.
Results
Additional parS sites alter ParB localization and disrupt chromosome origin segregation
To investigate whether the localization of ParB:parS nucleoprotein complexes proximal to oriC is required for the proper regulation of DNA replication initiation, strains were constructed to redistribute ParB away from the endogenous parS sites located within the chromosome origin region. In one approach, two parS16 arrays (16 tandem repeats of the consensus parS sequence 5´-TGTTCCACGTGAAACA-3´ [19]) were integrated into the chromosome, one set in each chromosome arm (Fig. 1B). In a second approach, a single consensus parS site was engineered into a high copy-number plasmid (Fig. 2A). Note that in both strains the endogenous parS sites are unaltered.
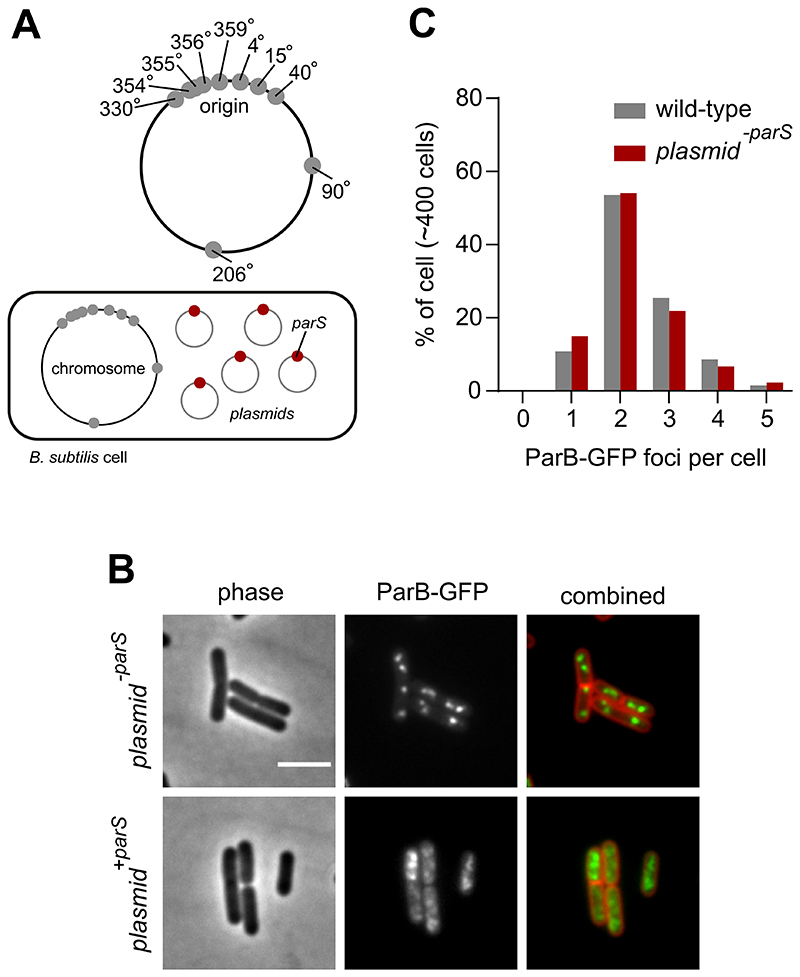
Figure 2. Extrachromosomal parS redistribute ParB.
(A) Schematic diagram showing the location of B. subtilis endogenous chromosomal parS (grey circles) and plasmid containing a single parS (red circles). (B) ParB-GFP localization in a wild-type strain harbouring plasmids that contain either a single or no parS. Phase contrast (left panel), ParB-GFP (middle panel) and membrane dye FM 5-95 combined with ParB-GFP (right panel). (C) The number of ParB-GFP foci per cell remains unchanged in the presence of the empty vector. Scale bar, 3 μm.
To determine the effect of additional parS sites on ParB activity, the endogenous parB gene was replaced by a functional parB-gfp fusion and epifluorescene microscopy was used to determine the localization of ParB-GFP. Under the slow growth conditions used in these experiments, the majority of wild-type cells contained a pair of ParB-GFP foci located towards the cell poles, which is characteristic of oriC localization in B. subtilis (Figs. 1C-D) [33, 34]. A control plasmid lacking parS did not alter this pattern (Figs. 2B-C). In contrast, both strains harbouring additional parS sites displayed marked differences. The chromosomal parS arrays resulted in an increase in the number of discreet ParB-GFP foci per cell (Figs. 1C-D), while the extrachromosomal parS site appeared to disturb ParB-GFP foci formation (Fig. 2B). Analysis of both ParB-GFP fluorescence in single cells and ParB levels using immunoblotting indicate that ParB expression is unchanged in strains with additional parS sites (Fig. S1). Together, the localization results suggest that additional parS sites recruit a population of ParB molecules away from oriC.
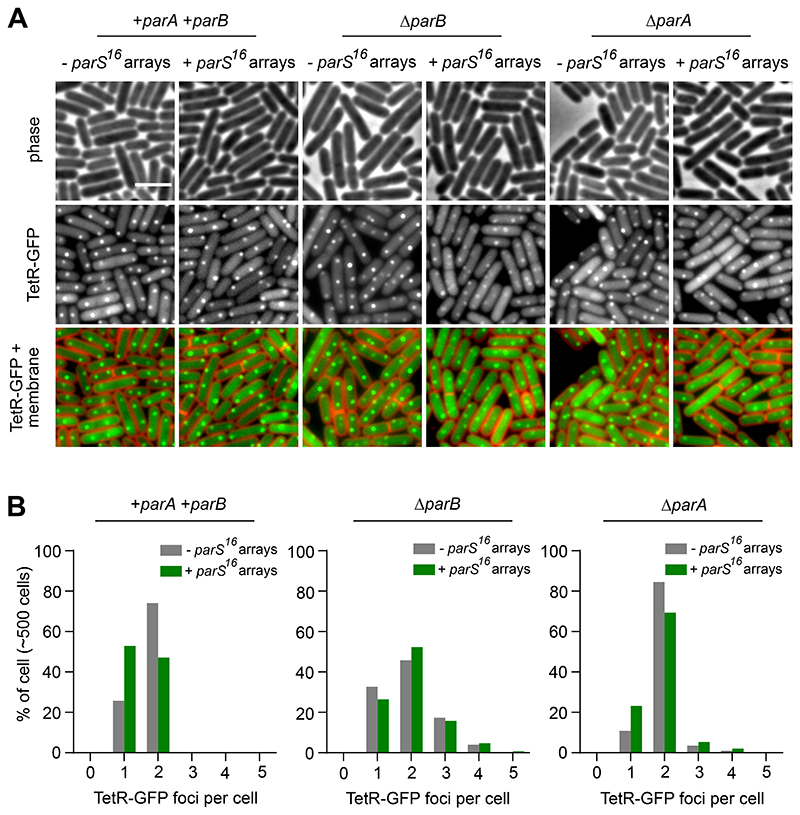
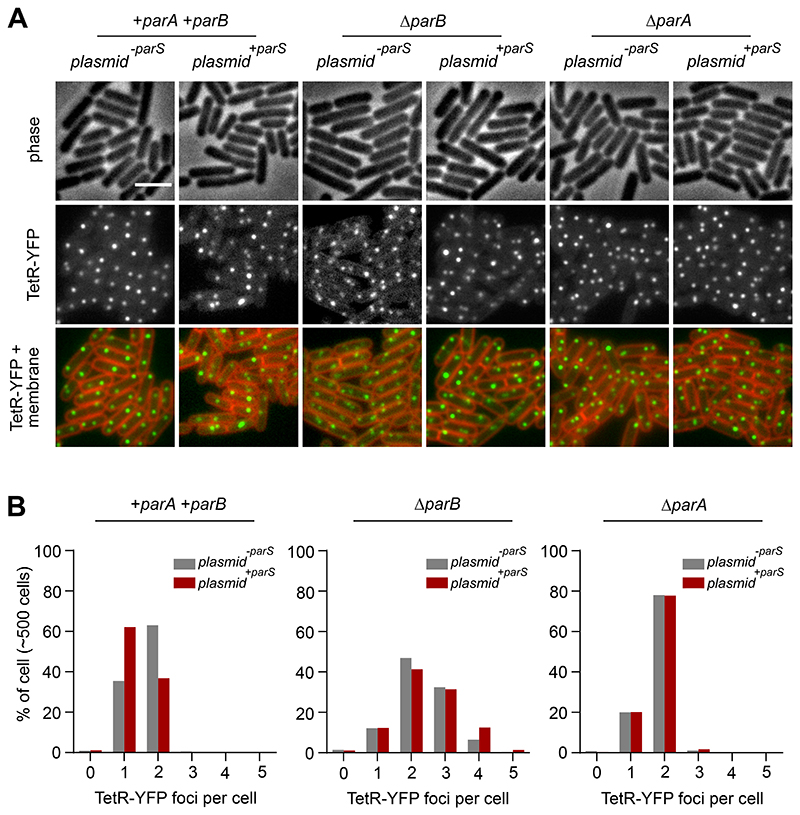
To investigate the consequence of ParB redistribution on oriC segregation, the origin region was visualised using a fluorescent repressor-operator system (tetO operators bound by TetR-GFP or TetR-YFP). As observed with ParB-GFP, wild-type cells contained two TetR-FP foci located near the cell poles (Figs. 3A-B, 4A-B). In contrast, when additional parS sites were present, there was an increase in the number of cells with a single fluorescent focus and a corresponding decrease in the number of cells with two foci, consistent with a defect in oriC separation (Figs. 3A-B, 4A-B). In a ΔparB mutant the number of origins per cell was increased independent of parS copy-number, indicating that the origin segregation defect is ParB-dependent and consistent with previous results showing that DNA replication initiation is stimulated in the absence of ParB (Figs. 3A-B, 4A-B) [16, 17, 22]. Interestingly, the origin separation defects caused by additional parS sites were diminished in a ΔparA mutant (Fig. 3A-B, 4A-B). This observation suggests that ParA exerts a negative effect on oriC segregation, possibly by perturbing the interaction between ParB and condensin near the chromosome origin [27, 28].
Figure 3. Chromosomal parS16 arrays redistribute ParB to disrupt chromosome origin segregation.
(A) Origin localization in a wild-type strain and in a strain harbouring parS16 arrays. The oriC region was labelled using an array of tetO operators bound by TetR-GFP. Phase contrast (top panel), TetR-GFP (middle panel) and membrane dye FM 5-95 combined with TetR-GFP (bottom panel). (B) ParB dependent decrease in the number of origin per cell in the presence of parS16 arrays. Scale bar, 3μm.
Figure 4. Extrachromosomal parS redistribute ParB to disrupt chromosome origin segregation.
(A) Origin localization in a wild-type strain and in a strain harbouring plasmid +parS. Origin region was labelled using an array of tetO operators bound by TetR-YFP. Phase contrast (top panel), TetR-YFP (middle panel) and membrane dye FM 5-95 combined with TetR-YFP (bottom panel). (B) ParB dependent decrease in the number of origin per cell in the presence of plasmid +parS. Scale bar, 3μm.
Control of DNA replication initiation by ParA is not affected by increasing the number of parS sites
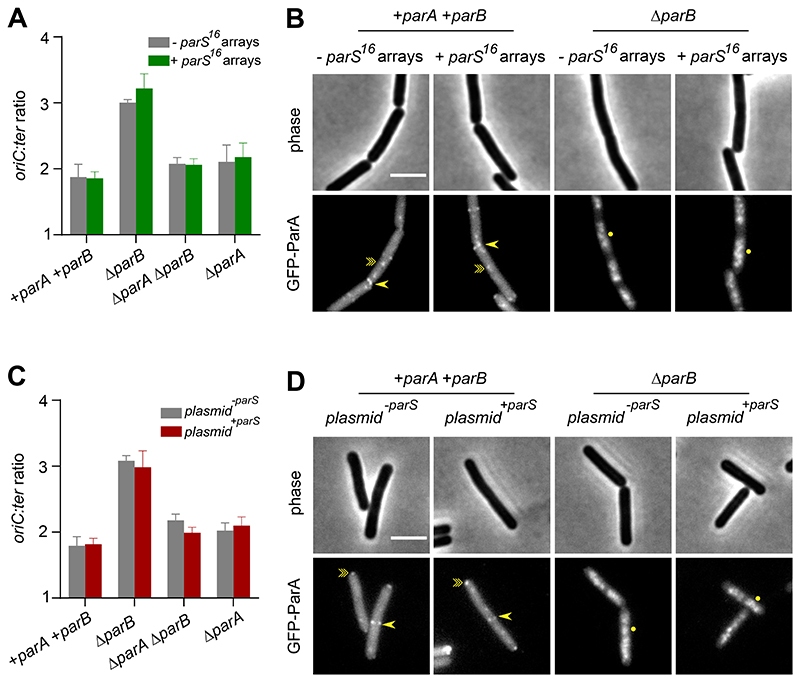
After establishing that additional parS sites redistribute ParB away from oriC and impair origin segregation, we next measured the rate of DNA replication initiation. Marker frequency analysis did not detect any significant change in the ori:ter ratio when additional parS sites were present, indicating that regulation of DnaA by ParA was retained (Figs. 5A, 5C). These results also suggest that the decrease in the number of TetR-FP foci per cell caused by additional parS sites was due to a defect in oriC segregation (Figs. 3A-B, 4A-B), rather than a decrease in the rate of DNA replication initiation. A significant ParA-dependent increase in the rate of replication initiation was observed when parB was deleted, showing that ParA remains competent to regulate DnaA in the presence of additional parS sites (Figs. 5A, 5C).
Figure 5. ParA activities are unperturbed in the presence of additional parS.
(A) The frequency of DNA initiation was not affected by chromosomal parS16 arrays. The oriC:ter ratio of each strain was determined using quantitative PCR. (B) GFP-ParA localization in strains containing chromosomal parS16 arrays. A single-point arrow (->) denotes localization at a septum, a double-point arrow (-≫) indicates localization as a focus and a circle (●) denotes nucleoid binding. (C) The frequency of DNA initiation was not affected by extrachromosomal parS. The oriC:ter ratio of each strain was determined using quantitative PCR. (D) GFP-ParA localization in strains containing plasmid +parS. A single-point arrow (->) denotes localization at a septum, a double-point arrow (-≫) indicates localization as a focus and a circle (●) denotes nucleoid binding. Scale bar, 3μm.
To confirm that ParA was being properly regulated in strains containing additional parS sites, the localization of GFP-ParA was determined. The parA gene was fused to gfp under the control of its native promoter and GFP-ParA was visualized using epifluorescence microscopy. Previous work has shown that in a wild-type strain GFP-ParA forms both DnaA-dependent foci at oriC and localizes to septa, while in a ΔparB mutant GFP-ParA colocalizes with the nucleoid (Figs. 5B, 5D) [16, 17, 21, 35, 36]. The pattern of GFP-ParA localization in strains containing additional parS sites was similar to the parent strains (Figs. 5B, 5D). Taken together with the marker frequency analysis, the results indicate that regulation of ParA by ParB, and thereby control of DNA replication initiation by ParA, is not affected by additional parS sites that titrate ParB away from oriC. Moreover, these experiments show that regulation of DnaA by ParA is retained under conditions when chromosome origin separation is disrupted, indicating that ParA does not mediate a causal relationship between DNA segregation and replication in B. subtilis.
A single parS site is necessary and sufficient to control ParA activity
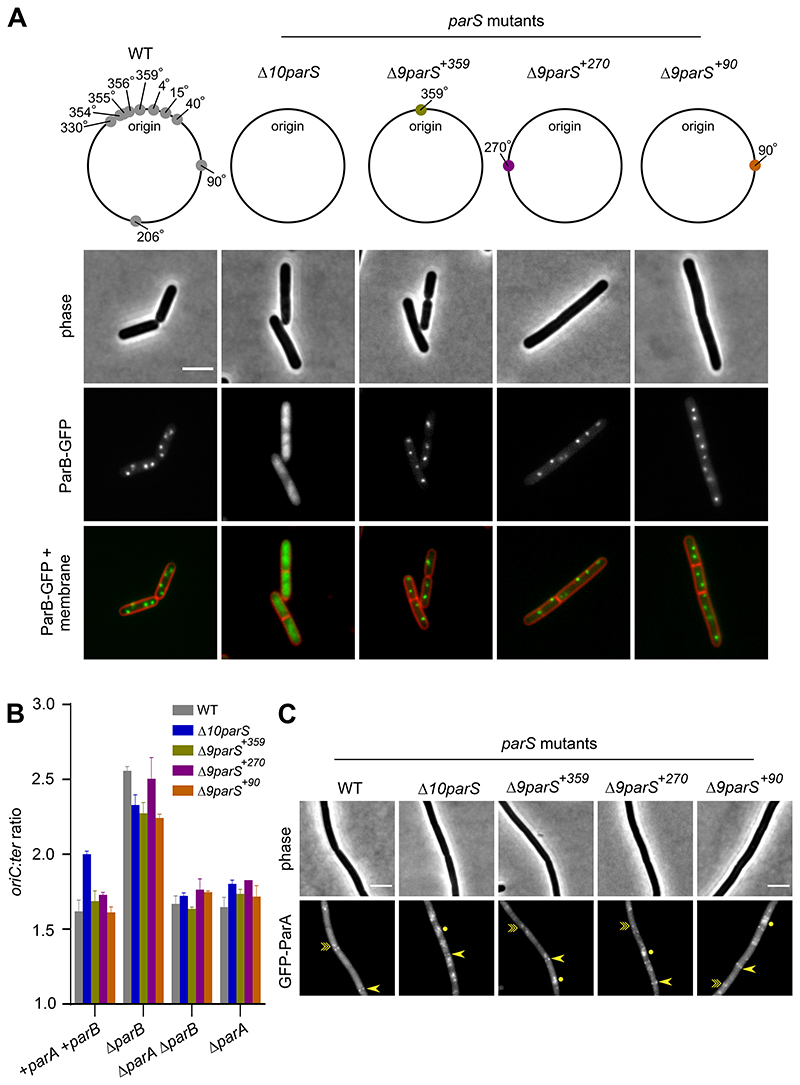
Although recruitment of ParB to additional parS sites appears sufficient to impair chromosome origin separation, in these strains the endogenous parS sites flanking oriC remained intact. Therefore, we wondered whether residual ParB binding to these oriC-proximal parS sites was responsible for regulating ParA. To address this question a strain was constructed with all ten parS sites deleted (Δ10parS). In the Δ10parS mutant, ParB-GFP foci formation was lost, confirming that no parS sites are present in the strain (Fig. 6A). Marker frequency analysis showed that the rate of DNA replication initiation was increased in the Δ10parS mutant, and deletion of parA from the Δ10parS strain confirmed that this effect was ParA-dependent (Fig. 6B). Consistent with the interpretation that ParA activity was altered in the absence of parS sites, the localization of GFP-ParA was shifted towards a nucleoid-bound state in the Δ10parS strain, a similar pattern to what is observed in a ΔparB mutant (Figs. 5B, 5D, 6C).
Figure 6. A single chromosomal parS is necessary and sufficient to regulate ParA activity.
(A) ParB-GFP localization in strains that differ in the location of parS. Phase contrast (top panel), ParB-GFP (middle panel) and membrane dye FM 5-95 combined with ParB-GFP (bottom panel). Schematic diagram showing the location of B. subtilis endogenous chromosomal parS (grey circles), parS at 359° (green circle), parS at 270° (purple circle) and parS at 90° (orange circle). (B) The frequency of DNA initiation was restored by ParB sliding clamps loading at a single parS. The oriC-ter ratio of each strain was determined using quantitative PCR. (C) GFP-ParA localization in strains that differ in the location of parS. A single-point arrow (->) denotes localization at a septum, a double-point arrow (-≫) indicates localization as a focus and a circle (●) denotes nucleoid binding. Scale bar, 3 μm.
To investigate whether a single parS site is sufficient to regulate ParA, the native parS359 site was re-introduced near oriC (Δ9parS+359). ParB-GFP foci were restored in the Δ9parS+359 strain, suggesting that ParB sliding clamps were being loaded at parS (Fig. 6A). Marker frequency analysis showed that the rate of DNA replication initiation in the Δ9parS+359 strain was comparable to wild-type (Fig. 6B). Deletion of parB in the Δ9parS+359 strain resulted in a ParA-dependent increase in the rate of DNA replication initiation (Fig. 6B), indicating that ParA is functional and being properly controlled by the single parS site. Consistent with the interpretation that ParA regulation remains intact in Δ9parS+359 strain, GFP-ParA localization was generally similar to its pattern in the wild-type strain (although there was an increase in the number of patchy fluorescent signals, likely representing interactions with the chromosome) (Fig. 6C). Together, these results indicate that a single oriC-proximal parS site is necessary and sufficient to regulate ParA.
To test whether the parS site needs to be located near oriC for ParA to regulate DNA replication initiation, a single parS site was reintroduced into the Δ10parS strain far from the origin region. We created two strains, with the parS site located in either the left or the right chromosome arm (Δ9parS+90 or Δ9parS+270). In both strains ParB-GFP formed foci within the cell, confirming that the reintroduced parS site was functional (Fig. 6A). As expected, locating a parS site away from oriC leads to cells displaying aberrant chromosome and cell morphologies (Fig. S2) [27–32]. In addition, examination of oriC number per cell in the Δ9parS+90 strain indicated a ParA-dependent origin separation defect (Figs. S3A-B). Critically however, marker frequency analysis showed that the rate of DNA replication initiation in both the Δ9parS+90 and Δ9parS+270 strains was similar to wild-type (Fig. 6B). Control of DNA replication initiation in the Δ9parS strains remained intact, as ParA-dependent over-initiation of DNA replication was observed when parB was deleted (Fig. 6B). In agreement with these observations, localization of GFP-ParA in either the Δ9parS+90 or Δ9parS+270 strain resembled the wild-type pattern, with the protein forming faint cytoplasmic foci and binding to septa (although like the Δ9parS+359 strain an increase in the number of patchy fluorescent signals was observed, likely representing chromosome binding) (Fig. 6C). Taken together, the results indicate that a parS site does not need to be located near oriC for it to dictate ParA activity at the chromosome origin.
Discussion
Par systems promote chromosome origin separation because parS sites are located near oriC. We hypothesized that parS positioning near oriC would also be important for the Par system to regulate the frequency of DNA replication initiation. Contrary to this proposed model, the data indicates that only a single parS site, independent of its location, is necessary and sufficient to maintain proper regulation of ParA. Moreover, control of DnaA activity by ParA was maintained even when the process of oriC separation was severely disrupted, indicating that ParA does not provide a functional link connecting origin segregation with DNA replication initiation in B. subtilis.
During the majority of the B. subtilis cell cycle, ParB acts to stimulate the ATPase activity of ParA and thereby promote monomer formation, which in turn inhibits DnaA activity [16, 17]. Because a parS site is required to properly regulate ParA, taken together with recent work describing the ParB:parS reaction cycle [9–14], the results suggest that ParB is most efficient at regulating ParA as a sliding clamp encircling DNA.
It remains unclear when or where ParA accumulates as a dimer to stimulate DnaA activity in B. subtilis. One possibility is that the rate of ParB clamp loading at parS could fluctuate, either during the cell cycle or between growth phases. We considered whether passage of the replisome through the origin region might displace ParB from the chromosome. Stimulation of ParA ATPase activity by ParB is 40-fold less efficient when the proteins are in solution without DNA [22]. However, in this scenario a new round of DNA replication would have just recently been initiated. Although this would appear to be an undesirable time to activate DnaA, it is important to note that DnaA is also a cell cycle regulated transcription factor [37]. For example, DnaA positively activates transcription of the developmental checkpoint gene sda to coordinate DNA synthesis with initiation of endosporulation, and dispersal of ParB from the chromosome might act as a signal to synchronize the DNA replication initiation with Sda expression [38]. Another possibility is that the ParB clamp could be modified to alter the interaction with ParA, either directly through interactions with proteins such as condensin or indirectly through changes in chromosome conformation [29, 30, 32].
Finally, current understanding of the opposing effects ParA has on DnaA is based mainly on genetic analysis of parA mutants [16, 17, 22]. It will be critical for future studies to analyze the dynamic behaviour of ParA, ParB, and DnaA within the context of a natural cell cycle to determine when and where they functionally interact.
Experimental Procedures
Strains and growth conditions
All strains used in this study are listed in Supplementary Table 1, and were routinely maintained on nutrient agar (NA) (Oxoid) with supplements. Selective media contained chloramphenicol (5 μg/ml), erythromycin (1 μg/ml), kanamycin (2 μg/ml), zeomycin (10 μg/ml), tetracycline (10 μg/ml), spectinomycin (50 μg/ml), phleomycin (1 μg/ml), ampicillin (100 μg/ml). For experiments in B. subtilis, overnight cells were grown in defined minimal medium base (Spizizen minimal salts supplemented with Fe-NH4-citrate (1 μg/ml), MgSO4 (6 mM), CaCl2 (100 μM), MnSO4 (130 μM), ZnCl2 (1 μM), thiamine (2 μM), casein hydrolysate (200 μg/ml), tryptophan (20 μg/ml) and succinate (2.0%) as carbon sources. Subsequent fresh media used for cell dilution were as mentioned above with either succinate (2.0%) or glucose (2.0%) as carbon source but without casein hydrolysate.
DNA manipulation and strain constructions
All B. subtilis transformations were carried out via a two-step starvation protocol as previously described (Anagnostopoulos and Spizizen 1961 and Hamoen 2002). DNA for transformation was provided as either genomic DNA from a B. subtilis strain or linearized plasmid DNA containing the desired genetic construct. Details of individual strain constructions and plasmids genotype are listed in Supplementary Table 1 and 2 respectively. Oligonucleotides used in plasmid construction are listed in Supplementary Table 3. All plasmids and strains were verified by sequencing.
Marker frequency analysis
For measurement of DNA replication, starter cultures were grown overnight at 37˚C in SMM based medium, before resuspension in fresh medium containing succinate (2.0%) as the carbon source the following day. Cultures were then allowed to undergo at least three doubling to early exponential phase at 37˚C. Sodium azide (0.5%; Sigma) was added to exponentially growing cells to prevent further metabolism. Chromosomal DNA was isolated using a DNeasy Blood and Tissue Kit (Qiagen). Either Rotor-Gene SYBR Green (Qiagen) or GoTaq (Promega) qPCR mix was used for PCR reactions. Q-PCR was performed in a Rotor-Gene Q Instrument (Qiagen). For quantification of the origin, the intergenic region between dnaA and dnaN was amplified using primers 5’-GATCAATCGGGGAAAGTGTG-3’ and 5’-GTAGGGCCTGTGGATTTGTG-3’. For quantification of the terminus, the region downstream of yocG was amplified using primers 5’-TCCATATCCTCGCTCCTACG-3’ and 5’-ATTCTGCTGATGTGCAATGG-3’. By use of crossing points (CT) and PCR efficiency a relative quantification analysis (∆∆CT) was performed using Rotor-Gene Software version 2.0.2 (Qiagen) to determine the ori/ter ratio of each sample. These results were normalized to the ori/ter ratio of a DNA sample from B. subtilis spores which only contain one chromosome and thus have an ori/ter ratio of 1. At least two biological repeats were performed for each experiments with error bars indicating the standard deviation of at least two technical replicates from one experimental set.
Epifluorescence microscopy
For fluorescent microscopy analysis, cells were grown at 37˚C in accordance with the marker frequency analysis using glucose (2.0%) as the carbon source for cell resuspension. Individual cell membrane were stained with 0.4 μg/ml FM5-95 (N-(3-Trimethylammoniumpropyl)-4-(6-(4(Diethylamino)phenyl)hexatrienyl) Pyridinium Dibromide) from ThermoFisher Scientific. Cells were immobilized onto 1.2% agarose in 0.25x SMM base medium spread thinly onto microscope slides. Microscopy was carried out with Nikon Eclipse Ti equipped with Nikon DM 100x/1.40 Oil Ph3 objective, Photometrics CoolSnap HQ2 cooled CCD camera and light source was Lambda LS (Sutter Instrument). Metamorph V.7.7 were used to acquire images. Cells were quantified using Fiji (Schindelin et al., 2012). At least three biological repeats were performed for each experiment.
Fluorescent intensity measurement
The average fluorescent intensity of ParB-GFP was measured as described [39]. Intensity was generated by averaging 11 pixels (0.72 μm) aligned perpendicular to the cell length axis, and by measuring along the length axis of the cell. Cells were quantified using Fiji [40].
Western blot analysis
For proteins visualization, cells were grown in accordance with general fluorescent microscopy analysis. Proteins were separated by electrophoresis using a NuPAGE 4-12% Bis-Tris gradient gel in 1 x MES buffer (Life Technologies) and transferred to a Hybond-P PVDF membrane (GE Healthcare, activated in 100% (v/v) methanol) using a semi dry apparatus (Hoefer Scientific Instruments). Membranes were blocked with 7.5% (w/v) skimmed milk powder in PBST (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4 [pH7.4], 0.1% (v/v) Tween 20), washed three times in PBST, and incubated with primary antibody in 1% bovine serum albumin in PBST for 2 hours at room temperature. Following three washes in PBST, incubation with secondary antibody (Anti-rabbit horseradish peroxidase-linked antibody) was carried out in milk-PBST for 1h at room temperature, followed by three final washes in PBST. Detection of proteins was by chemiluminescence using Pierce™ ECL Western Blotting Substrate (Thermo Fisher Scientific Inc.) and ImageQuant LAS 4000 mini digital imaging system (GE Healthcare).
Supplementary Material
Acknowledgements
We thank James Grimshaw for advice with microscopy image analysis.
Funding Information
Research support was provided to HM by a Wellcome Trust Senior Research Fellowship (204985/Z/16/Z). HS and AK were funded by a Biotechnology and Biological Sciences Research Council (BBSRC) grant BB/S00257X/1.
Footnotes
Author Contributions
AK carried out all laboratory work and data analysis; HM supervised all laboratory work and coordinated the research project; AK, HM and HS wrote the manuscript.
Conflicts of Interest
The authors declare that there are no conflicts of interest.
Suggested Reviewers:
Tung Le, John Innes Centre, Tung.Le@jic.ac.uk
Stephan Gruber, Universite de Lausanne, stephan.gruber@unil.ch
Dagmara Jakimowicz, University of Warsaw: Uniwersytet Warszawski, jolanta.zakrzewska@uwr.edu.pl
Alan Grossman, Massachusetts Institute of Technology, adg@mit.edu
Additional Information:
| Question | Response |
| Does this article report on work with humans or animals? | Not applicable (no human or animal experimentation is reported) |
| Does this article include details (names, initials, hospital numbers), images, or videos relating to an individual person? | No |
References
- 1.Messer W. The bacterial replication initiator DnaA. DnaA and oriC, the bacterial mode to initiate DNA replication. Fems Microbiology Reviews. 2002;26(4):355–374. doi: 10.1111/j.1574-6976.2002.tb00620.x. [DOI] [PubMed] [Google Scholar]
- 2.Wang X, Montero Llopis P, Rudner DZ. Bacillus subtilis chromosome organization oscillates between two distinct patterns. Proc Natl Acad Sci U S A. 2014 doi: 10.1073/pnas.1407461111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mierzejewska J, Jagura-Burdzy G. Prokaryotic ParA-ParB-parS system links bacterial chromosome segregation with the cell cycle. Plasmid. 2012;67(1):1–14. doi: 10.1016/j.plasmid.2011.08.003. [DOI] [PubMed] [Google Scholar]
- 4.Gerdes K, Howard M, Szardenings F. Pushing and pulling in prokaryotic DNA segregation. Cell. 2010;141(6):927–942. doi: 10.1016/j.cell.2010.05.033. [DOI] [PubMed] [Google Scholar]
- 5.Gerdes K, Moller-Jensen J, Bugge Jensen R. Plasmid and chromosome partitioning: surprises from phylogeny. Mol Microbiol. 2000;37(3):455–466. doi: 10.1046/j.1365-2958.2000.01975.x. [DOI] [PubMed] [Google Scholar]
- 6.Livny J, Yamaichi Y, Waldor MK. Distribution of centromere-like parS sites in bacteria: insights from comparative genomics. J Bacteriol. 2007;189(23):8693–8703. doi: 10.1128/JB.01239-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leonard TA, Butler PJ, Lowe J. Structural analysis of the chromosome segregation protein Spo0J from Thermus thermophilus. Mol Microbiol. 2004;53(2):419–432. doi: 10.1111/j.1365-2958.2004.04133.x. [DOI] [PubMed] [Google Scholar]
- 8.Ireton K, Gunther NWt, Grossman AD. spo0J is required for normal chromosome segregation as well as the initiation of sporulation in Bacillus subtilis. J Bacteriol. 1994;176(17):5320–5329. doi: 10.1128/jb.176.17.5320-5329.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Soh YM, Davidson IF, Zamuner S, Basquin J, Bock FP, et al. Self-organization of parS centromeres by the ParB CTP hydrolase. Science. 2019;366(6469):1129–1133. doi: 10.1126/science.aay3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Osorio-Valeriano M, Altegoer F, Das CK, Steinchen W, Panis G, et al. The CTPase activity of ParB determines the size and dynamics of prokaryotic DNA partition complexes. Mol Cell. 2021;81(19):3992–4007.:e3910. doi: 10.1016/j.molcel.2021.09.004. [DOI] [PubMed] [Google Scholar]
- 11.Osorio-Valeriano M, Altegoer F, Steinchen W, Urban S, Liu Y, et al. ParB-type DNA Segregation Proteins Are CTP-Dependent Molecular Switches. Cell. 2019;179(7):1512–1524.:e1515. doi: 10.1016/j.cell.2019.11.015. [DOI] [PubMed] [Google Scholar]
- 12.Jalal AS, Tran NT, Le TB. ParB spreading on DNA requires cytidine triphosphate in vitro. Elife. 2020;9 doi: 10.7554/eLife.53515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jalal AS, Tran NT, Stevenson CE, Chimthanawala A, Badrinarayanan A, et al. A CTP-dependent gating mechanism enables ParB spreading on DNA. Elife. 2021;10 doi: 10.7554/eLife.69676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Antar H, Soh YM, Zamuner S, Bock FP, Anchimiuk A, et al. Relief of ParB autoinhibition by parS DNA catalysis and recycling of ParB by CTP hydrolysis promote bacterial centromere assembly. Sci Adv. 2021;7(41):eabj2854. doi: 10.1126/sciadv.abj2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murray H, Ferreira H, Errington J. The bacterial chromosome segregation protein Spo0J spreads along DNA from parS nucleation sites. Molecular Microbiology. 2006;61(5):1352–1361. doi: 10.1111/j.1365-2958.2006.05316.x. [DOI] [PubMed] [Google Scholar]
- 16.Scholefield G, Whiting R, Errington J, Murray H. Spo0J regulates the oligomeric state of Soj to trigger its switch from an activator to an inhibitor of DNA replication initiation. Molecular Microbiology. 2011;79(4):1089–1100. doi: 10.1111/j.1365-2958.2010.07507.x. [DOI] [PubMed] [Google Scholar]
- 17.Murray H, Errington J. Dynamic Control of the DNA Replication Initiation Protein DnaA by Soj/ParA. Cell. 2008;135(1):74–84. doi: 10.1016/j.cell.2008.07.044. [DOI] [PubMed] [Google Scholar]
- 18.Ogura Y, Ogasawara N, Harry EJ, Moriya S. Increasing the ratio of Soj to Spo0J promotes replication initiation in Bacillus subtilis. J Bacteriol. 2003;185(21):6316–6324. doi: 10.1128/JB.185.21.6316-6324.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee PS, Lin DCH, Moriya S, Grossman AD. Effects of the chromosome partitioning protein SpoOJ (ParB) on oriC positioning and replication initiation in Bacillus subtilis. Journal of Bacteriology. 2003;185(4):1326–1337. doi: 10.1128/JB.185.4.1326-1337.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leonard TA, Butler PJ, Lowe J. Bacterial chromosome segregation: structure and DNA binding of the Soj dimer--a conserved biological switch. EMBO J. 2005;24(2):270–282. doi: 10.1038/sj.emboj.7600530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Quisel JD, Lin DCH, Grossman AD. Control of development by altered localization of a transcription factor in B-subtilis. Molecular Cell. 1999;4(5):665–672. doi: 10.1016/s1097-2765(00)80377-9. [DOI] [PubMed] [Google Scholar]
- 22.Scholefield G, Errington J, Murray H. Soj/ParA stalls DNA replication by inhibiting helix formation of the initiator protein DnaA. EMBO J. 2012;31(6):1542–1555. doi: 10.1038/emboj.2012.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vecchiarelli AG, Neuman KC, Mizuuchi K. A propagating ATPase gradient drives transport of surface-confined cellular cargo. Proc Natl Acad Sci U S A. 2014;111(13):4880–4885. doi: 10.1073/pnas.1401025111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lim HC, Surovtsev IV, Beltran BG, Huang F, Bewersdorf J, et al. Evidence for a DNA-relay mechanism in ParABS-mediated chromosome segregation. Elife. 2014;3:e02758. doi: 10.7554/eLife.02758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hwang LC, Vecchiarelli AG, Han YW, Mizuuchi M, Harada Y, et al. ParA-mediated plasmid partition driven by protein pattern self-organization. EMBO J. 2013;32(9):1238–1249. doi: 10.1038/emboj.2013.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vecchiarelli AG, Hwang LC, Mizuuchi K. Cell-free study of F plasmid partition provides evidence for cargo transport by a diffusion-ratchet mechanism. Proc Natl Acad Sci U S A. 2013;110(15):E1390–1397. doi: 10.1073/pnas.1302745110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang X, Tang OW, Riley EP, Rudner DZ. The SMC condensin complex is required for origin segregation in Bacillus subtilis. Current Biology. 2014;24(3):287–292. doi: 10.1016/j.cub.2013.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gruber S, Veening JW, Bach J, Blettinger M, Bramkamp M, et al. Interlinked sister chromosomes arise in the absence of condensin during fast replication in B. subtilis. Current Biology. 2014;24(3):293–298. doi: 10.1016/j.cub.2013.12.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sullivan NL, Marquis KA, Rudner DZ. Recruitment of SMC by ParB-parS Organizes the Origin Region and Promotes Efficient Chromosome Segregation. Cell. 2009;137(4):697–707. doi: 10.1016/j.cell.2009.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gruber S, Errington J. Recruitment of Condensin to Replication Origin Regions by ParB/SpoOJ Promotes Chromosome Segregation in B. subtilis. Cell. 2009;137(4):685–696. doi: 10.1016/j.cell.2009.02.035. [DOI] [PubMed] [Google Scholar]
- 31.Britton RA, Lin DC, Grossman AD. Characterization of a prokaryotic SMC protein involved in chromosome partitioning. Genes Dev. 1998;12(9):1254–1259. doi: 10.1101/gad.12.9.1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marbouty M, Le Gall A, Cattoni DI, Cournac A, Koh A, et al. Condensin- and Replication-Mediated Bacterial Chromosome Folding and Origin Condensation Revealed by Hi-C and Super-resolution Imaging. Mol Cell. 2015;59(4):588–602. doi: 10.1016/j.molcel.2015.07.020. [DOI] [PubMed] [Google Scholar]
- 33.Teleman AA, Graumann PL, Lin DCH, Grossman AD, Losick R. Chromosome arrangement within a bacterium. Current Biology. 1998;8(20):1102–1109. doi: 10.1016/s0960-9822(98)70464-6. [DOI] [PubMed] [Google Scholar]
- 34.Lin DCH, Levin PA, Grossman AD. Bipolar localization of a chromosome partition protein in Bacillus subtilis. Proceedings of the National Academy of Sciences of the United States of America. 1997;94(9):4721–4726. doi: 10.1073/pnas.94.9.4721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Autret S, Nair R, Errington J. Genetic analysis of the chromosome segregation protein Spo0J of Bacillus subtilis: evidence for separate domains involved in DNA binding and interactions with Soj protein. Molecular Microbiology. 2001;41(3):743–755. doi: 10.1046/j.1365-2958.2001.02551.x. [DOI] [PubMed] [Google Scholar]
- 36.Marston AL, Errington J. Dynamic movement of the ParA-like soj protein of B-subtilis and its dual role in nucleoid organization and developmental regulation. Molecular Cell. 1999;4(5):673–682. doi: 10.1016/s1097-2765(00)80378-0. [DOI] [PubMed] [Google Scholar]
- 37.Burkholder WF, Kurtser I, Grossman AD. Replication initiation proteins regulate a developmental checkpoint in Bacillus subtilis. Cell. 2001;104(2):269–279. doi: 10.1016/s0092-8674(01)00211-2. [DOI] [PubMed] [Google Scholar]
- 38.Veening JW, Murray H, Errington J. A mechanism for cell cycle regulation of sporulation initiation in Bacillus subtilis. Genes & Development. 2009;23(16):1959–1970. doi: 10.1101/gad.528209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Strahl H, Ronneau S, Gonzalez BS, Klutsch D, Schaffner-Barbero C, et al. Transmembrane protein sorting driven by membrane curvature. Nat Commun. 2015;6:8728. doi: 10.1038/ncomms9728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9(7):676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.