Abstract
The coexistence of discrete morphs that differ in multiple traits is common within natural populations of many taxa. Such morphs are often associated with chromosomal inversions, presumably because the recombination suppressing effects of inversions help maintain alternate adaptive combinations of alleles across the multiple loci affecting these traits. However, inversions can also harbor selected mutations at their breakpoints, leading to their rise in frequency in addition to (or independent from) their role in recombination suppression. In this review, we first describe the different ways that breakpoints can create mutations. We then critically examine the evidence for the breakpoint-mutation and recombination suppression hypotheses for explaining the existence of discrete morphs associated with chromosomal inversions. We find that the evidence that inversions are favored due to recombination suppression is often indirect. The evidence that breakpoints harbor mutations that are adaptive is also largely indirect, with the characterization of inversion breakpoints at the sequence level being incomplete in most systems. Direct tests of the role of suppressed recombination and breakpoint mutations in inversion evolution are thus needed. Finally, we emphasize how the two hypotheses of recombination suppression and breakpoint mutation can act in conjunction, with implications for understanding the emergence of supergenes and their evolutionary dynamics. We conclude by discussing how breakpoint characterization could improve our understanding of complex, discrete phenotypic forms in nature.
Keywords: chromosomal inversion, linkage, mutation, recombination, genome evolution
Supergenes and complex phenotypes
The origin and maintenance of discrete morphs within natural populations has long fascinated evolutionary biologists (Darwin, 1862; Fisher, 1930; Ford, 1971). Because of the discrete nature of the phenotypic differences observed, this variation was often assumed to have a genetic origin, making its study of primary interest for early geneticists (see Chapter 6 of Ford 1971 for an overview of genetic programs of the time). Moreover, the diverse nature of the traits that usually differ between these morphs (color, morphology, sexual-compatibility, etc.), led to the hypothesis that the genetic basis of this discrete variation is a functional unit containing multiple linked loci, termed a “Super-gene” (spelled supergene hereafter; see glossary) by Darlington and Mather in 1949 (Darlington & Mather, 1949; Fisher, 1930; Ford, 1965; Mather, 1955). Indeed, as early as 1930 Fisher had anticipated that genetic linkage between multiple selected loci could evolve under natural selection (Fisher, 1930) and Ford later suggested that linkage could be increased via translocation or chromosomal inversions (Ford, 1971). Chromosomal inversions can indeed strongly reduce recombination between the genomic segments they cover, a property leading to their discovery in 1921 by Sturtevant (Sturtevant, 1921). Specifically, single crossing-over events within the rearranged region in heterokaryotypic individuals produce non-viable or unfit gametes with unbalanced gene content (Hoffmann & Rieseberg, 2008; Kirkpatrick, 2010; Liehr et al., 2019). Recombination between chromosomal rearrangements is still possible when double crossing-over events occur, making recombination more likely to happen at the center of inversions than near breakpoints (Hoffmann & Rieseberg, 2008; Stump et al., 2007). Nonetheless, such double crossing-over events are relatively rare, estimated to occur with a frequency of the order of 10−4 to 10−3 per generation in Drosophila (Stevison, Hoehn, & Noor, 2011).
Examples of discrete morphs associated with a region of suppressed recombination (i.e., candidate supergenes; see glossary) are accumulating rapidly. In the majority of cases, suppressed recombination is due to an inversion (Schwander, Libbrecht, & Keller, 2014). However, in some instances, suppressed recombination is due to other mechanisms such as close physical proximity of highly diverged DNA sequences (Tetsuyuki Entani et al., 2003; Ushijima et al., 2003), proximity to a chromosomal centromere (T. Entani et al., 1999), or hemizygosity (Li et al., 2016). These findings have thus piqued interest in the evolution of supergenes and inversions, forming the central topic of the current review. Because there are already several excellent general reviews of supergenes and inversions (Faria, Johannesson, Butlin, & Westram, 2019; Schwander et al., 2014; Wellenreuther & Bernatchez, 2018), we focus here specifically on distinguishing two hypotheses for their evolution, a topic that has received less attention.
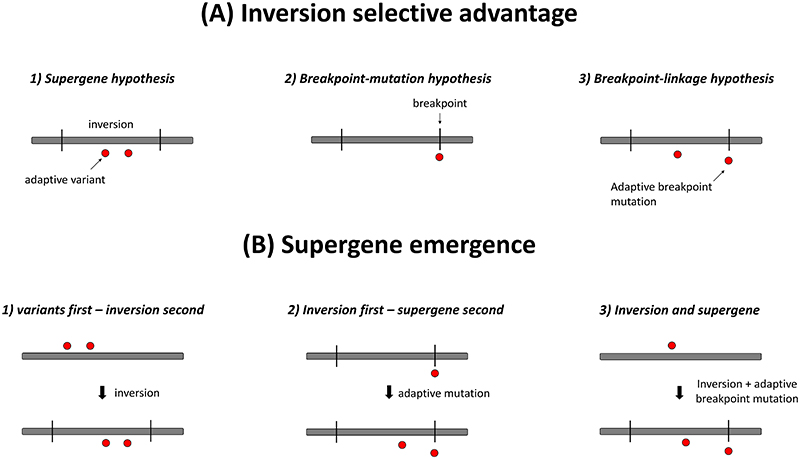
There are currently two core hypotheses for how and why supergenes and inversions evolve. The first and generally most accepted hypothesis is that inversions are selected due to their effect on suppressing recombination between sets of epistatic or locally adapted genes (i.e., supergene hypothesis; see glossary) (Charlesworth & Barton, 2018; Kirkpatrick, 2010; Kirkpatrick & Barton, 2006). The second hypothesis is that inversions create adaptive mutations at their breakpoints (i.e. breakpoint-mutation hypothesis; see glossary; Fig. 1A) (Dobzhansky, 1947, 1970; Kirkpatrick, 2010), leading to their rise in frequency via selection on breakpoint variants. As we discuss in more detail below, these two hypotheses are not mutually exclusive and might combine to drive inversion and supergene evolution. Specifically, inversions could be selected due to their effect on suppressing recombination between an adaptive breakpoint variant and an adaptive variant within the rearrangement (i.e. breakpoint-linkage hypothesis; see glossary; Fig. 1A). In a similar manner, breakpoint mutations could be favored because of their property to suppress recombination outside of the inversion (R. B. Corbett-Detig, 2016), locking together multiple variants favored in particular combinations, inside and/or outside the inversion.
Figure 1. Hypotheses underlying selective advantages to inversions and scenarios for supergene emergence.
A. Hypotheses explaining a selective advantage to inversions. B. Scenarios for supergene emergence. Grey squares represent a chromosome; red dots are adaptive variants affecting a selected trait; black bars are inversion breakpoints.
Distinguishing among these hypotheses (Fig. 1A) is important because it provides insight into the relative significance of novel mutation, standing genetic variation, and recombination in adaption, thus addressing a major issue in evolution concerning the relationship between adaptation and genome rearrangement (Fig. 1B). However, empirical testing of these hypotheses can be a challenging task as it requires the quantification and localization of adaptive variants in regions of suppressed recombination (here inversions). When multiple traits are associated with an inversion, a role for recombination suppression between multiple variants controlling different traits is usually assumed. However, a breakpoint mutation can be pleiotropic and affect multiple traits (Fisher, 1930), making the identification of adaptive variants within the region of suppressed recombination necessary.
While the supergene hypothesis and the breakpoint-linkage hypothesis seem similar, they imply different evolutionary histories for supergenes (Fig. 1B). Under the supergene hypothesis, divergently selected variants evolve first and subsequently become locked together in favorable combinations because of an inversion (Charlesworth & Barton, 2018; Kirkpatrick & Barton, 2006). Under the breakpoint-linkage hypothesis two main evolutionary scenarios for supergene emergence are possible. In the first scenario, inversions first establish because they generate an adaptive breakpoint mutation when they form. Only subsequently might inversions evolve into supergenes by the emergence of new variants within them (Navarro & Barton, 2003). In the second scenario, inversions establish because they couple an already existing adaptive variant with a new adaptive breakpoint mutation. This last scenario may facilitate the evolution of supergenes when many divergently selected variants exist by facilitating the initial rise in frequency of an inversion. Indeed, under the supergene hypothesis, theory implies that for new inversions to establish, they must capture most segregating variants at divergently selected loci in favorable combinations (Charlesworth & Barton, 2018; Kirkpatrick & Barton, 2006). However, an inversion with an adaptive breakpoint mutation may not need to lock all or a majority of the segregating variation in favorable combinations to initially rise in frequency due to the advantageous breakpoint mutation. If the inversion persists for a sufficient amount of time because of a selected breakpoint mutation, this may subsequently allow for the sorting out of any initially captured maladapted alleles (e.g., via double crossing-over events or noncrossover gene conversion) to create a supergene with a now higher selective value.
In this review, we critically examine the evidence for the supergene and breakpoint-mutation hypotheses, and their potential combination (i.e. the breakpoint-linkage hypothesis). We begin with a discussion of the mechanisms by which breakpoints can create adaptive mutations, as this is often a less appreciated facet of inversions than is recombination suppression. We then empirically review well-known cases of candidate supergenes (Box 2 – Fig. 1). Our collective findings lead us to propose that characterization of breakpoints at the sequence level could improve our understanding of supergenes. Specifically, a common molecular mechanism generating inversions also generates duplications at both ends of the inverted DNA segment (Ranz et al., 2007). This offers a unique opportunity to potentially date inversions independently from their content to test if they emerged before or after the divergently selected variants they lock together via suppressed recombination. Thus, breakpoint characterization could be important to understand the origin and maintenance of complex, discrete phenotypic forms in nature.
Molecular mechanisms by which inversion breakpoints can create mutations
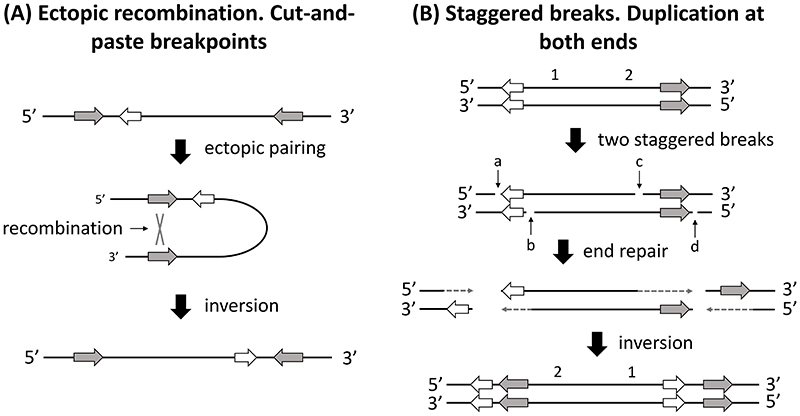
The idea that inversions might be selected because of an adaptive mutation at one of their breakpoints is not new (Dobzhansky, 1947). However, this hypothesis is yet to receive highly focused study in supergene evolution, despite known molecular mechanisms that are likely to create breakpoint mutations when inversions form. Two main processes can generate chromosomal inversions, ectopic recombination and staggered breaks. Ectopic recombination generates inversions via a recombination event between two homologous sequences oriented head-to-head along a chromosome (Box 1 for further details; Box 1 – Fig. 1A). Staggered breaks can generate inversions via the complete detachment of a DNA segment from a chromosome and its subsequent reattachment in the opposite orientation at the same position in the chromosome (Box 1; Box 1 – Fig. 1B). In the Drosophila melanogaster species subgroup, the group best characterized for inversion breakpoints at present (R. B. Corbett-Detig, 2016), the majority of inversions are generated by staggered breaks (Ranz et al., 2007). While ectopic recombination leaves little molecular trace, staggered breaks can generate duplications at both breakpoints of the newly formed inversion (Box 1; Box 1 – Fig. 1B). As explained in more detail below, these breakpoint duplications provide a possible means to accurately date inversions, independently from the divergently selected variants they contain and, thus, could potentially allow us to resolve among the competing scenarios for supergene emergence (Fig. 1B). Consequently, if staggered breaks are primarily responsible for generating inversions in groups in addition to D. melanogaster flies, this could be a helpful tool for characterizing the emergence of supergenes (for details see the section Future directions).
Box 1. Inversion breakpoints can create a diversity of types of genetic variation via mutation.
We here discuss how inversions are formed and the associated effects of induced mutations on genetic variation at breakpoints. Two known molecular mechanisms can generate inversions: (1) ectopic recombination and, (2) staggered breaks (Box 1. – Fig1) (Ling & Cordaux, 2010; Ranz et al., 2007).
The first mechanism occurs from recombination between homologous sequences, usually transposable elements (Ling & Cordaux, 2010) or paralogous genes, that are oriented head-to-head in the original sequence, subsequently resulting in inversion of the sequence located between these elements (Box 1 – Fig. 1A). This mechanism leaves little molecular trace and generates cut-and-paste type breakpoints. The second mechanism involves staggered breaks occurring in two positions of the original sequence (Ranz et al., 2007). Such breaks are usually staggered, meaning that they will result in fragments with stretches of single stranded DNA at their extremities (Box 1. – Fig. 1B). The DNA repair mechanism that synthesizes the reverse complement of these single stranded stretches then joins the DNA sequences back together in a non-homologous way, sometimes reinserting the DNA segment in the opposite orientation, creating an inversion. When the breaks are heavily staggered (i.e., with long single strand stretches), this results in inversions with duplicated sequences at both breakpoints in the derived sequence (Box 1. – Fig. 1B) (Ranz et al., 2007). In contrast, when the breaks are blunt or slightly staggered, cut-and-paste type breakpoints are created in the derived sequence, with no or small duplications, respectively.
Despite these clear mechanisms of inversion formation, we still know very little about genetic variation at breakpoints and the mutations that breakpoints generate (Table 1). This stems largely from the fact that inversion breakpoints often lie in very repetitive regions of the genome (Russell B. Corbett-Detig et al., 2019; da Silva et al., 2019) which are difficult to sequence and assemble and thus challenging to study (Fig. 3A). Nonetheless, some progress has been made, particularly in the Drosophila melanogaster species subgroup (R. B. Corbett-Detig, 2016; Russell B. Corbett-Detig & Hartl, 2012; McBroome, Liang, & Corbett-Detig, 2020; Ranz et al., 2007). In this system, staggered breaks appear to be the most common mechanism generating inversions, very often resulting in duplications at breakpoints (Ranz et al., 2007).
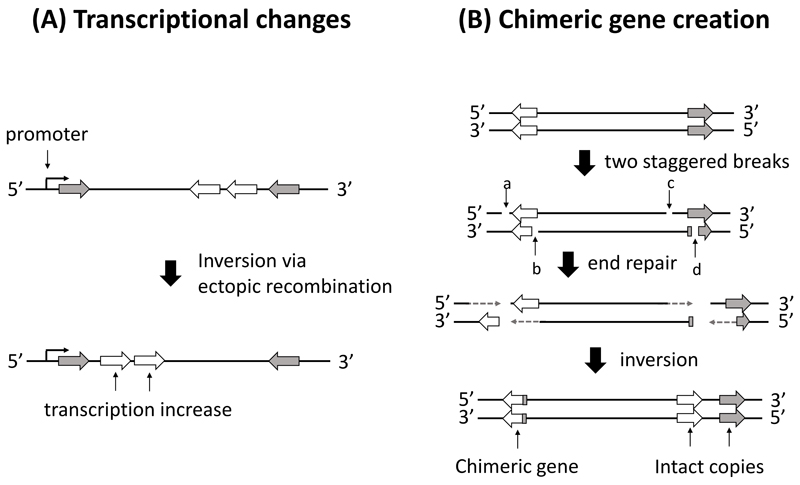
We note, however, that locating breakpoints is not sufficient to identify their phenotypic effects. Obtaining the derived DNA sequence at breakpoints is necessary to try and understand possible phenotypic effects. Understanding such effects from DNA sequence modification is however not always a straightforward task, as the disruption of a gene sequence by a breakpoint can sometimes lead to increases in expression of this gene, as seen in fire ants (Yan et al., 2020). Nonetheless, phenotypic effects may be expected, for example, because both ectopic recombination and staggered breaks can change the expression level of a set of genes (Fig. 2A). In addition, staggered breaks can have another potential effect, in which the duplication of sequences leads to the creation of a new chimeric gene. This occurs via merging the initial part of a gene with another coding sequence, such as another gene or pseudo-gene (Fig. 2B). This phenomenon can happen without the loss of the originally disrupted genes (via their duplication), mitigating fitness costs.
Thus, there are several possibilities by which breakpoints can create genetic variation with likely phenotypic effects. The observation that similar genomic regions are often re-used repeatedly as breakpoints for different inversions, including in different taxa, raises the possibility for a recurrent and even somewhat predictable role for breakpoints in evolution (Pevzner & Tesler, 2003).
Box 1 – Figure 1. Known molecular mechanisms generating inversions.
White and grey arrows represent genes, with their orientation indicated by the direction of the arrows. A. Ectopic recombination between similar elements (transposable elements or duplicated genes) in opposite orientation in a DNA segment leads to the inversion of the interspacing sequence. B. Fours single strand breaks (a, b, c, and d) lead to two staggered breaks. Through end repair and reattachment in the opposite orientation an inversion is generated with sequence duplication at both breakpoints (black and grey genes). The number above the DNA stretch are used here to illustrate the change in orientation of the sequence.
Both ectopic recombination and staggered breaks can generate important phenotypic effects, in principle, by changing the expression level of a gene (or set of genes) at breakpoints (Fig. 2A). However, while differential expression of genes at breakpoints is common, it is not always observed (Said et al., 2018). Furthermore, differential expression is usually not restricted to genes at breakpoints, but is also detected for many genes inside and outside inversions (Cheng, Tan, Hahn, & Besansky, 2018; Lavington & Kern, 2017; Said et al., 2018). This suggests that the differential expression of genes at breakpoints potentially works in conjunction with other genetic changes to affect phenotypic variation, or is sometimes a by-product of genome rearrangement rather than being the actual cause for observed phenotypic variation. Breakpoints can also create chimeric genes (see glossary; Fig. 2B), which might sometimes have a beneficial effect, as documented now in a diverse set of taxa (Carvunis et al., 2012; Guillén & Ruiz, 2012; Jones et al., 2012; Stewart & Rogers, 2019). For example, in Drosophila mojavensis a chimeric gene at an inversion breakpoint is associated with the detoxification of toxic compounds in cacti (Guillén & Ruiz, 2012). Another example is found in the threespine stickleback Gasterosteus aculeatus, where a gene located at an inversion breakpoint on chromosome XI harbors different exon sequences depending on the inversion orientation, and is associated with differences in muscular development between lake and oceanic phenotypes (Jones et al., 2012). While speculative, an inversion breakpoint could create a new pleiotropic chimeric gene that affects multiple traits. Moreover, staggered breaks often generates duplications at both breakpoints in the derived sequence (Box1 – Fig. 1), allowing for the creation of chimeric genes without loss of the donor genes (Fig. 2B). This property will also facilitate neo-functionalization of such duplicated genes.
Figure 2. Possible mechanisms creating mutations at inversion breakpoints.
A. An inversion generated through ectopic recombination affects the transcription level of two genes by changing their positions in the genome relative to a promoter. B. An inversion generated through staggered breaks generates a new chimeric gene, while avoiding the loss of the donor genes because of duplication. Grey and black arrows represent genes, with orientation indicated via arrow direction; a, b, c, d represent single strand breaks, resulting in two staggered breaks.
As we will see below, direct evidence that breakpoint mutations drive supergene evolution is lacking, but the mechanisms by which they might do so are established. With these considerations in mind, we now turn to evaluating the role of suppressed recombination and breakpoint mutation in the evolution of candidate supergenes associated with chromosomal inversions.
The supergene hypothesis: what is the evidence for recombination suppression among loci controlling selected trait variation?
A core prediction of the supergene hypothesis (i.e., that suppressed recombination is advantageous) is that multiple, linked genetic variants control trait variation that differs between inversion forms. In contrast, if variation is controlled by a single locus with pleiotropic effects on several traits, suppressed recombination is not necessarily favored or required to explain the existence of discrete morphs that differ in these traits.
One line of indirect evidence supporting the supergene hypothesis is that inversions of intermediate size usually reach higher frequencies than small inversions, suggesting that they are selectively favored more frequently than small inversions (Cáceres, Barbadilla, & Ruiz, 1997; R. B. Corbett-Detig, 2016; Wellenreuther & Bernatchez, 2018). This is expected if larger inversions are more likely to lock physically distant selected variants together via suppressed recombination. In contrast, if the selective nature of an inversion solely stems from a breakpoint mutation, shorter inversions should be favored because they have less chance of capturing recessive deleterious mutations when they form (Cáceres et al., 1997; R. B. Corbett-Detig, 2016; Kirkpatrick, 2010).
Despite these observations, direct evidence for the supergene hypothesis is still lacking in most systems. Indeed, our review of the literature revealed that we still know very little about the number, location and identity of selected variants within candidate supergenes associated with inversions. This is not surprising given that determining the number of adaptive loci within a region of suppressed recombination is difficult, because suppressed recombination frustrates attempts at fine-scale genetic mapping. The presence of two or more selected loci in a supergene was historically inferred from genetic crosses aimed at detecting rare recombinants (i.e., intermediate phenotypic forms) (Cain & Sheppard, 1954; Mather, 1950). For example, Mather described a genetic model where multiple selected loci locked within an inversion control heterostyly in Primula sinensis, a classic example of a putative supergene (Mather, 1950). However, recent work using modern genomic approaches suggests that Mather’s simple genetic model is erroneous and needs modification (Li et al., 2016). Specifically, a 178 kilo base-pair indel containing five predicted genes is associated with the different floral types (Li et al., 2016), with one floral type being hemizygous for this DNA segment and the other completely lacking it (Table 1) (Li et al., 2016).
Table 1. Examples of candidate supergene.
#: example number. Species: common and latin names. Multiple traits: Do morphs differ by multiple traits? Selection on phenotypes: Has selection on these traits been shown? Map to an inversion: Are these traits associated with an inversion? Multiple loci: Has the existence of multiple loci controlling the traits been demonstrated? Selection on recomb. sup.: Has the selective advantage of an inversion been demonstrated to come from its property of suppressing recombination between multiple selected loci? Adaptive breakpoint mutation: Is their evidence for selection acting on one or both of the breakpoints in the inversion? Breakpoint sequence: Have the breakpoints been characterized at the sequence level? Variants first?: Do adaptive variants in the putative supergene predate the emergence of the inversion? In each column, the term ‘likely’ represent cases were lines of evidence exist to support an item, but definitive evidence is currently lacking.
A revision of the classic genetic model also occurred for the iconic land snail Cepaea nemoralis (Table 1), where cryptic shell color-pattern was long thought to be associated with five loci locked within an inversion (Cain & Sheppard, 1954). Recent work showed an absence of recombination in the putative supergene, casting doubt on the number and linkage relationships of loci controlling shell color-pattern (Gonzalez, Aramendia, & Davison, 2019). These recent developments urge for more rigorous tests on the true number of selected variants within supergenes and their precise genomic location(s).
To our knowledge, the existence of multiple selected loci within a supergene associated with an inversion has been demonstrated in only six systems, three involving plants, two involving animals, and one involving a fungus. Of these six, three involve segregation distorters (Table 1; Box 2). In addition, indirect but fairly convincing evidence exist for two other animal systems. Details of all the examples mentioned above are presented in Box 2. Clearly, further work determining the number of loci within inversions and candidate supergenes that contribute to trait variation is warranted. In this regard, emerging techniques that allow functional genetic manipulation of structural variants (e.g., CRISPR-Cas9) hold promise for making such progress (Hopkins, Tyukmaeva, Gompert, Feder, & Nosil; Kraft et al., 2015).
Box 2. Determining the number of loci within inversions that contribute to selected trait variation.
Despite seventy years of research on supergenes, definitive evidence for the existence of multiple selected variants within a region of suppressed recombination containing an inversion exists only for a few systems, as described below.
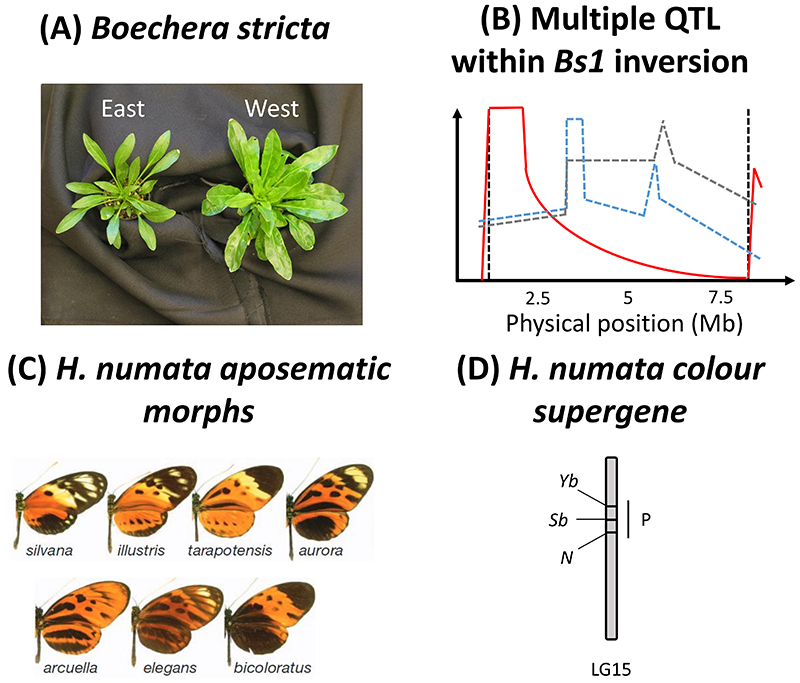
Evidence for multiple loci controlling trait variation within an inversion has been demonstrated in Boechera stricta (Box 2 – Fig. 1A), a perennial plant from the Rocky Mountains. Here, two subspecies adapted to different climates have come into secondary contact (Lee et al., 2017). The subspecies differ in multiple traits and through crosses between different genotypes lacking the inversion it was demonstrated that three different QTLs (i.e. Quantitative trait loci) are associated with these divergent traits and located within the inversion region (Box 2 – Fig. 2B). Interestingly, independent of the genotype at these three QTLs, the orientation of the DNA segment at the rearranged region explained a significant amount of variance for these traits, without any apparent effect of the inversion breakpoints. The molecular mechanism for this result is still unknown. The inversion is relatively young and originated after secondary contact, likely locking together pre-existing selected variants (Lee et al., 2017), but this interpretation is debated (Charlesworth & Barton, 2018).
In the Mimulus guttatus species complex, the existence of multiple selected loci within an inversion has also been confirmed by genetic crosses, where two life strategies (perennial vs annual) are maintained through divergent selection (Lowry & Willis, 2010). Specifically, through crosses between the Mimulus guttatus annual type and a closely related perennial species with a collinear gene order, Mimulus tilingii, Coughlan and Willis (Coughlan & Willis, 2018) identified two QTLs within the inversion associated with the traits experiencing divergent selection.
In the Brassicaceae self-incompatibility locus (S-locus), it was shown that two selected loci reside within a region of suppressed recombination containing inversions (Goubet et al., 2012; Takayama & Isogai, 2005). At the phenotypic level, self-incompatibility alleles are equivalent to mating types, where a plant stigma can recognize its own pollen via a specific interaction between a protein in the pollen coat and a receptor in the plant stigma, preventing fertilization (Durand et al., 2020; Takayama & Isogai, 2005). With this system, plants cannot self-fertilize and cannot be fertilized by pollen from plants expressing a similar mating type. Through knock-outs and gains of function experiments, the genes coding for the pollen protein (SCR) and the stigma receptor (SRK) have been identified (Schopfer, Nasrallah, & Nasrallah, 1999; Silva et al., 2001; Takasaki et al., 2000; Takayama & Isogai, 2005; Takayama et al., 1987; Takayama et al., 2000). These two genes are present in different positions and orientations in different S-locus haplotypes, suggesting that many inversions have occurred at the S-locus (Goubet et al., 2012). The S-locus region displays little sequence conservation (likely because of extensive degeneration), frustrating attempts at characterizing the exact breakpoints of these inversions or the ancestral orientation of the region (Goubet et al., 2012).
The existence of multiple selected loci within a region of suppressed recombination containing inversions has also been verified in a spore killer system (Neurospora intermedia spore killer element Sk-2) and two meiotic drive systems (Mus musculus t-haplotypes and Drosophila melanogaster SD locus). In these three systems, a combination of ‘killer’ loci (named driver loci hereafter) and ‘protective’ loci (named response loci hereafter) kills spores or gametes with specific genotypes, resulting in increased transmission (up to 99%) of the spore or gamete killing element. Suppressed recombination between driver and response loci is necessary for the maintenance of drive systems over many generations (Hammond, Rehard, Xiao, & Shiu, 2012), qualifying them as supergene examples.
The existence of two selected loci (one driver locus and one response locus) was demonstrated in Neurospora intermedia (element Sk-2) through gene knock-outs and complementation (Hammond, Rehard, Xiao, & Shiu, 2012; Rhoades et al., 2019). In this system, a succession of non-overlapping inversions interspaced with repeats tightly link the driver locus with the response locus (Svedberg et al., 2018). The existence of five selected loci (four driver loci and one response locus) (Herrmann & Bauer, 2012) was also demonstrated in Mus musculus t-haplotypes meiotic drive through the generation of rare recombinants via genetic crosses and mutagenesis (Herrmann & Bauer, 2012; Lyon, 1984; Lyon & Meredith, 1964). Multiple interspaced non-overlapping inversions are present in the t-haplotype locus but none contain both a driver and the response locus, suggesting that recombination suppression is achieved via additional factors in this system (Kelemen & Vicoso, 2018).
A similar technique to the one employed in the t-haplotype was used to decipher the genetic basis of SD meiotic drive in Drosophila melanogaster and confirmed the existence of five loci involved in this drive system (four driver loci and one response locus) (Hartl, 1974; Larracuente & Presgraves, 2012). Deletion of chromosomal segments via mutagenesis confirmed these results and allowed for fine mapping of the different loci (Ganetzky, 1977; Larracuente & Presgraves, 2012). As is the case for the t-haplotype locus, multiple interspaced non-overlapping inversions are present in the SD locus (different driver alleles are associated with different inversions), and inversions rarely contain a driver locus together with the response locus, suggesting that additional factors also prevent recombination in this system (Larracuente & Presgraves, 2012).
Less direct, but reasonably strong, evidence for multiple selected loci within an inversion has also been reported in two insect systems (Table 1). Here, the approach was to compare the genetic basis of traits in closely related taxa that exhibit versus lack inversion polymorphism. In the polymorphic butterfly Heliconius numata a region of suppressed recombination generated via multiple adjacent inversions controls variation in mimetic color morphs. The same genomic region is known to contain at least three recombining loci associated with color variation in other species of the genus (Box 2 – Fig. 1D) (Huber et al., 2015; Mathieu Joron et al., 2011; Van Belleghem et al., 2017). It is therefore likely that the adjacent inversions in H. numata harbor multiple variants at these three loci that recombine in other Heliconius species. A similar example stems from Timema, a genus of stick insect that is endemic to the western USA and Mexico and that shows within-population variation for cryptic coloration (i.e., green versus melanistic morphs). In several species, morphs are highly discrete and controlled by a region of suppressed recombination on linkage group eight (named ‘Mel-Stripe’) (Nosil et al., 2018; Villoutreix et al., 2020). However, a recent study in a species displaying more quantitative color variation (Timema chumash) and a lack of suppressed recombination revealed that multiple genetic variants, and likely multiple genes within Mel-Stripe, control color variation (Villoutreix et al., 2020).
Box 2 – Figure 1. Flagship examples of supergenes.
A. Boechera stricta subspecies types. The East subspecies has smaller leaves with more trichomes. Photo credit: Cheng-Ruei Lee. B. Multiple QTLs were detected in the Bs1 inversion. The red, blue dashed and grey-dashed lines correspond to different composite traits obtained through discriminant function analysis. See Lee et al., 2017 for more details. The two black-dashed lines correspond to the position of the inversion breakpoints. Breakpoints are of the cut-and-paste type for the Bs1 inversion. Figure redrawn and modified with the permission of the authors. C. Heliconius numata aposematic morphs, involved in Müllerian mimicry rings with other butterflies. D. Aposematic morphs in H. numata are associated with adjacent inversions (spanning the genomic region indicated by P) on linkage group 15 (LG15), encompassing at least three loci (Yb, Sb and N) associated with mimetic color morphs in H. melpomene.
The breakpoint-mutation hypothesis: what is the evidence that functional variants reside in or near inversion breakpoints?
A core assumption of the breakpoint-mutation hypothesis is that functional variants reside in or near inversion breakpoints. We found evidence consistent with this hypothesis in three systems (Papilio polytes, Solenopsis invicta and Timema cristinae), but expect this number to increase when breakpoint characterization is completed in more systems (Table 1).
In the mimetic butterfly Papilio polytes, an inversion breakpoint disrupts a transcriptional regulator and affects the expression in embryonic wings of three neighboring genes potentially involved in wing color-pattern development (UXT, U3X and prospero; Table 1) (Nishikawa et al., 2015). In the fire ant Solenopsis invicta, a region of suppressed recombination generated via three overlapping inversions (Yan et al., 2020) is associated with different social structures of colonies (Wang et al., 2013). A breakpoint of the second inversion disrupts a gene (SI2.2.0_02248) which is a strong candidate to explain differences in colony social structure, and the disrupted allele is associated with increased expression of this particular gene (Huang, Dang, Chang, & Wang, 2018; Yan et al., 2020). In the stick insect Timema cristinae a large deletion occurring at one breakpoint of a putative inversion deleted multiple loci known to affect multiple aspects of color in another species of the genus (Villoutreix et al., 2020).
Interestingly, with the exception of Timema cristinae, indications exist of putative selected variants within inversions at locations other than the breakpoints. Therefore, support for a strict version of the breakpoint-mutation hypothesis is scarce, with only one example potentially supporting it (T. cristinae). Even in the case of T. cristinae, genes controlling adaptive traits other than coloration, which combine with color alleles to determine fitness, could exist in locations in the inversion away from breakpoints. We discuss the implication of these observations in the following section.
Mixed hypotheses: The breakpoint-linkage hypothesis and its implications for supergene emergence
As mentioned previously, the supergene and breakpoint-mutation hypotheses are not mutually exclusive. The selective advantage of an inversion could come from its property of impeding recombination between variants within the inversion and a favorable breakpoint mutation. We refer to this mixed hypothesis as the breakpoint-linkage hypothesis (see glossary), which predicts that loci controlling trait variation will be localized both at one (or both) breakpoint(s) and other regions within the inversion.
In theory, the breakpoint-linkage hypothesis could involve different processes and dynamics than those predicted by the classic supergene hypothesis. A supergene composed of a selected locus within an inversion and an adaptive breakpoint mutation suggests two main evolutionary scenarios. In the first, an inversion could originally establish because of an adaptive breakpoint mutation (Guerrero, Rousset, & Kirkpatrick, 2012), and then subsequently evolve into a supergene by the emergence of additional genetic variants within it (Navarro & Barton, 2003) (Fig. 1B). This scenario could be initiated in allopatry, where an inversion with an adaptive breakpoint sweeps to high frequency or fixation and is selected for its recombination suppressing properties only later upon secondary contact (or introgression) and gene flow. In this case, allopatry allows for the evolution and capture of alternative sets of favorable alleles in alternate arrangements, bypassing the difficulties of supergene emergence when gene flow occurs and maladaptive alleles are segregating as standing variation within populations (Feder, Gejji, Powell, & Nosil, 2011).
In the second scenario, pre-existing genetic variants in the region where the inversion will occur could become associated with a new adaptive breakpoint mutation (Fig. 1B). Although formal modeling is required, it is possible that such a coupling could relax some of the restrictions of a model based strictly on suppressed recombination (Charlesworth & Barton, 2018; Kirkpatrick & Barton, 2006). For example, such coupling might mean that the initial inversion does not need to capture all or the majority of divergently selected variants in a favored combination in order to spread. Instead, variant sorting could occur after the establishment of the inversion, by double cross-over events (or noncrossover gene conversion (Korunes & Noor, 2017)) that disassociate adaptive and maladaptive alleles. In principle, the rise in frequency of the inversion and its equilibrium frequency could thus be dependent on the sum of adaptive values of the breakpoint and the variants it originally captures.
The breakpoint-linkage hypothesis predicts that loci controlling trait variation will be localized at one or both breakpoint(s) and within the inversion. Some empirical data are at least consistent with this prediction. For example, the butterfly (P. polytes) and fire ant (S. invicta) examples mentioned in the previous section as instances where functional loci reside in or near inversion breakpoints could involve the breakpoint-linkage hypothesis (Table 1). In P. polytes, the inversion associated with the breakpoint mutation, besides affecting the expression of the three genes UXT, U3X and prospero potentially involved in mimetic wing color-pattern (UXT, U3X and prospero), also contains a locus (away from any breakpoint) shown to affect mimetic wing color-pattern (doublesex) (Kunte et al., 2014; Nishikawa et al., 2015). In the fire ant S. invicta, in addition to the strong candidate gene for odor reception (SI2.2.0_02248) located at an inversion breakpoint, the region of suppressed recombination also contains (away from any breakpoints) a strong candidate gene for odor generation/reception (Gp-9), a mechanism at the root of social structure differences in fire ants (Wang et al., 2013).
Variations on the breakpoint-linkage hypothesis are possible. For example, another type of mixed model is one where the selective advantage of breakpoints stems from their property to suppress recombination outside of the inversion (R. B. Corbett-Detig, 2016). Recombination suppression outside of inversions could be achieved by a breakpoint disrupting a ‘sensitive site’ (a locus believed to be necessary to produce normal crossing-over frequencies in the surrounding genomic region) (R. B. Corbett-Detig, 2016). This could lock together multiple variants in particular combinations, inside and/or outside the inversion. This mechanism may be important in meiotic drive systems, where multiple selected loci are located within a region of suppressed recombination containing multiple, non-overlapping inversions. Mus musculus t-haplotypes and the Drosophila melanogaster SD locus could involve such a mixed model. In these two systems, individual inversions appear to very rarely contain more than one selected locus (Herrmann & Bauer, 2012; Larracuente & Presgraves, 2012) (See Box 2 for details).
While functional work is needed to better elucidate the role of different loci in controlling trait variation (P. polytes and S. invicta), or recombination suppression (M. musculus and D. melanogaster), these four systems provide some preliminary support for mixed hypotheses. Regardless of whether recombination suppression is most relevant for genes inside versus outside of an inversion or for both, these systems highlight the possibility for breakpoint mutations generated by a new chromosomal rearrangement to be integral members of the group of selected loci that form a supergene. The vast majority of candidate supergene systems still lack a complete characterization of inversion breakpoints (often one breakpoint is characterized at the DNA sequence level but not the other; Table 1). It is possible that more examples supporting the breakpoint-linkage hypothesis will emerge as breakpoints are better characterized in a greater number of systems.
Future directions
Future work could usefully focus in two directions. First, better information on the number, location, and identify of loci controlling trait variation within inversions is required. One way that this can be achieved is via genetic crosses to create recombinant forms, such as for the examples discussed in Box 2. Although the number of selected loci in an inversion determined through genetic crosses will tend to underestimate the actual number (because insufficient recombination may occur in these crosses to separate variants in close physical proximity), the results can still indicate whether or not multiple loci or variants are involved, thus confirming the existence of a supergene. As an alternative to controlled crosses, genome wide association mapping can be applied in species that are close relatives to those with discrete morphs and inversions, but that lack inversion polymorphism (Villoutreix et al., 2020). This will yield more precise estimates, but needs to be complemented with functional manipulation in the species with the inversion. Ultimately, functional manipulations such as those offered by CRISPR-Cas9 may be required to precisely and firmly establish the identities and numbers of variants functionally contributing to trait variation, possibly by ‘engineering’ recombinant alleles to validate the independent effects of individual mutations on a phenotype (Hopkins et al., 2020; Kraft et al., 2015). Such gene editing may be particularly required for identification of causal genetic variation at breakpoints, as many variants will be in complete association in these regions due to the lack of recombination (Hoffmann & Rieseberg, 2008; Matzkin, Merritt, Zhu, & Eanes, 2005).
Second, once the selected variants within a region of suppressed recombination are identified, further work can be implemented to understand how the supergene emerged (Fig. 1B). More specifically, when variants are located inside the inversion boundaries, one can test if the variants predate or postdate the inversion itself. The usual procedure to compare inversion age to the age of the selected variants within them is to compare their times to the most recent common ancestor (TMRCA) using coalescence theory (Lee et al., 2017). Inversion age is estimated by calculating TMRCA on the inverted haplotypes only, while ages for loci within the inversion are obtained comparing all haplotypes (inverted and non-inverted). While being the current standard, this method suffers from several caveats. First, it does not date the inversion totally independently from the variants it contains. Second, it can over-estimate inversion age relative to the selected variants within it and could thus result in the erroneous conclusion that the variants appeared after the inversion. The reason for such over-estimation is that double crossing-over at neutral loci within the rearranged region will reintroduce older alleles from non-inverted haplotypes into the inverted haplotypes, thus increasing the TMRCA for the inversion, and therefore the estimated age of the inversion. Duplications at breakpoints generated by staggered breaks, if neutral, could help circumvent these caveats. Indeed, breakpoint duplications would allow the inversion to be dated independently from its content, and the inversion age estimated using these duplications would not be as biased because double crossing-over events between non-inverted and inverted DNA sequences very rarely involve sequences at breakpoints (Hoffmann & Rieseberg, 2008). As a proof of concept, a breakpoint duplication was successfully used to date inversion age in H. numata butterflies (Jay et al., 2018).
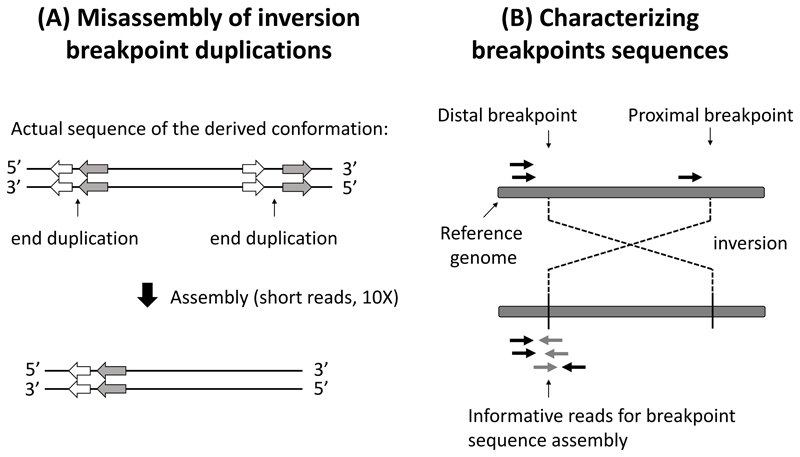
Nonetheless, even this method using breakpoint duplications is not without difficulties for at least two reasons. First, it requires sequence-level characterization of breakpoints, a difficult endeavor because breakpoints often reside in highly repetitive regions of the genomes (Russell B. Corbett-Detig et al., 2019; da Silva et al., 2019), which are difficult to assemble using short-read sequencing technologies (Fig. 3A). Second, obtaining accurate age estimates for loci experiencing selection is difficult, and breakpoint duplications may not be always neutral. Still, promise for progress exists in sequence-level characterization of breakpoints. Specifically, creative approaches such as those applied by Corbett-Detig and colleagues in Drosophila (Fig. 3B) (Russell B. Corbett-Detig, Cardeno, & Langley, 2012; Russell B. Corbett-Detig & Hartl, 2012) may help in characterizing breakpoints at the sequence level in other taxa. Long-read technologies such as Nanopore (Jain, Olsen, Paten, & Akeson, 2016) and PacBio (Eid et al., 2009) should also be of great help for such characterization.
Figure 3. Characterizing breakpoints at the DNA sequence level.
A. Using short-read sequencing technologies can lead to misassembly of inversion breakpoints, for example due to duplications generated through staggered breaks (Ranz et al., 2007). Grey and black arrows represent genes, with orientation indicated by the direction of arrows. B. A method to characterize breakpoint sequences using a single reference genome and short read data, as developed by Corbett-Detig and colleagues (Russell B. Corbett-Detig et al., 2012). Black arrows symbolize aligned reads on the reference genome, while grey arrows symbolize their unaligned paired reads. Breakpoint sequence characterization is possible by using the black and grey read pairs to generate de novo assembly of the breakpoint region. Figure redrawn and modified from (Russell B. Corbett-Detig et al., 2012) with the permission of the authors.
Concluding remarks
We outlined how inversions can facilitate supergene evolution by suppressed recombination, by inducing adaptive mutations at breakpoints or by a combination of these two mechanisms. Surprisingly, definitive evidence for the existence of multiple selected loci within candidate supergenes associated with an inversion exist in only six systems. Thus, direct evidence for the supergene hypothesis is still scarce, although indirect lines of evidence in several systems are consistent with the hypothesis. Little is known about the emergence of supergenes, specifically if divergently-selected variants predate inversions or if they accumulated within inversions after the inversion originated. Observations in multiple candidate supergene systems suggest a joint role for suppressed recombination and adaptive breakpoint mutation in supergene evolution. Thus, further tests of the breakpoint-linkage hypothesis are warranted. Distinguishing and confirming the role of breakpoints in supergene evolution will require characterization of inversion breakpoints at the sequence level, which has yet to be completed in most systems. We propose that further study of inversion breakpoints could improve our understanding of the origin and maintenance of complex, discrete phenotypic forms in nature, and the dynamics by which supergenes emerge. Although we focused here on morphs, similar processes could apply to the evolution of other discrete units of diversity such as ecotypes, species, and sexes.
Acknowledgements
This manuscript greatly benefited from discussions with Pr. Russel Corbett-Detig, Pr. Angus Davison, Pr. Haruhiko Fujiwara, Dr. Clemens Küpper, Pr. Cheng-Ruei Lee, Pr. Thomas Mitchell-Olds and Dr. John Wang on their respective study systems. We are grateful to Pr. Cheng-Ruei Lee for providing us photographs of Boechera stricta. We are grateful to Dr. Venera Tyukmaeva for insights on CRISPR-Cas9 and functional genomics. We are grateful to Dr. Paul Jay for a useful discussion on molecular dating methodologies. R.V. and P.N. were supported by a grant from the European Research Council (EE-Dynamics 770826, https://erc.europa.eu/). D.A. was supported by an ANR grant (ANR-18-CE35-0002-01 – WILDING). M.J. was supported by an ANR grant (ANR-18-CE02-0019-02 supergène). Z.G. was supported by a grant from the NSF (DEB 1638768). J.L.F. was supported by grants from the USDA NIFA program and NSF. All authors contributed to writing the manuscript. The authors declare having no competing interests.
Glossary
- Breakpoint-linkage hypothesis
the selective advantage of an inversion stems from both a favorable effect of a mutation at one of its breakpoints and the recombination suppression between this mutation and at least one other selected variant within the inversion.
- Breakpoint-mutation hypothesis
(also referred as ‘position effect hypothesis’ in the literature) the selective advantage of an inversion stems from a favorable effect associated with a mutation at one or both of its breakpoints.
- Candidate supergene
a case where multiple co-varying and discrete phenotypes are associated with a region of suppressed recombination (usually an inversion). The existence of multiple loci associated with these traits, within the region of suppressed recombination, must be demonstrated to advance the example from a candidate to a bonafide supergene.
- Chimeric gene
a new gene constituted of sequences not previously associated together, for example, as parts of two genes or parts of genes and non-coding DNA. Here, we consider cases where the different DNA segments are brought together by an inversion breakpoint (see Fig. 2B).
- Chromosomal inversion
a chromosomal rearrangement in which a segment of a chromosome is reversed end to end. This causes the inverted segment to have an opposite sequence order from the ancestral sequence and reduces recombination in individuals heterozygous for the inversion. Recombination is especially reduced near chromosomal inversion breakpoints.
- Chromosomal inversion breakpoint
(shortened to ‘inversion breakpoint’ or simply ‘breakpoint’ in the main text and hereafter) the position(s) on a chromosome where DNA sequence orientation shifts after the occurrence of a chromosomal inversion (i.e., the end of an inversion where the genome shifts from inverted back to collinear). An inversion generally has two breakpoints.
- (Chromosomal) breakpoint mutation
sequence modification(s) at breakpoints resulting from a chromosomal inversion. Mutations may also occur near breakpoints after an inversion has occurred, however, here, we refer to changes associated with the creation of the rearrangement itself.
- Discrete morphs
individuals of the same species that exhibit discrete phenotypic differences (e.g., in morphology, color, behavior, physiology, etc.). Here, we focus on cases where morphs are differentiated by multiple co-varying phenotypic traits such that multiple genes may be involved.
- Supergene
a group of genes affecting different phenotypes so seldom separated by crossing-over that in effect they operate as a single genetic entity (i.e., a non-recombining locus). This end may be achieved by close genetic proximity on a chromosome (i.e., tight physical linkage), by inclusion within an inversion, or by other molecular mechanisms impeding crossing-over. This definition is modified from that of E. B. Ford (Ford, 1965).
- Supergene hypothesis
supergenes evolve because suppressed recombination within them maintains favorable allelic combinations at multiple loci that control different traits, thereby favoring suppressed recombination.
Bibliography
- Albert AYK, Sawaya S, Vines TH, Knecht AK, Miller CT, Summers BR, Schluter D. The genetics of adaptative shape shift in stickleback: Pleiotropy and effect size. Evolution. 2007;62(1):76–85. doi: 10.1111/j.1558-5646.2007.00259.x. [DOI] [PubMed] [Google Scholar]
- Anderson JT, Lee CR, Mitchell-Olds T. Life-History Qtls and Natural Selection on Flowering Time in Boechera Stricta, a Perennial Relative of Arabidopsis. Evolution. 2011;65(3):771–787. doi: 10.1111/j.1558-5646.2010.01175.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JT, Lee CR, Rushworth CA, Colautti RI, Mitchell-Olds T. Genetic trade-offs and conditional neutrality contribute to local adaptation. Mol Ecol. 2013;22(3):699–708. doi: 10.1111/j.1365-294X.2012.05522.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayala D, Zhang S, Chateau M, Fouet C, Morlais I, Costantini C, et al. Besansky NJ. Association mapping desiccation resistance within chromosomal inversions in the African malaria vector Anopheles gambiae. Mol Ecol. 2019;28(6):1333–1342. doi: 10.1111/mec.14880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson WW. Natural-Selection for Mullerian Mimicry in Heliconius-Erato in Costa-Rica. Science. 1972;176(4037):936–939. doi: 10.1126/science.176.4037.936. [DOI] [PubMed] [Google Scholar]
- Berg PR, Star B, Pampoulie C, Sodeland M, Barth JMI, Knutsen H, et al. Jentoft S. Three chromosomal rearrangements promote genomic divergence between migratory and stationary ecotypes of Atlantic cod. Scientific Reports. 2016;6(23246) doi: 10.1038/srep23246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cáceres M, Barbadilla A, Ruiz A. INVERSION LENGTH AND BREAKPOINT DISTRIBUTION IN THE DROSOPHILA BUZZATII SPECIES COMPLEX: IS INVERSION LENGTH A SELECTED TRAIT? Evolution. 1997;51(4):1149–1155. doi: 10.1111/j.1558-5646.1997.tb03962.x. [DOI] [PubMed] [Google Scholar]
- Cain AJ, Sheppard PM. Selection in the polymorphic land snail Cepæa nemoralis. Heredity. 1950;4(3):275–294. doi: 10.1038/hdy.1950.22. [DOI] [PubMed] [Google Scholar]
- Cain AJ, Sheppard PM. The effects of natural selection on body colour in the land snail Cepaea nemoralis. Heredity. 1952;6(2):217–231. doi: 10.1038/hdy.1952.22. [DOI] [Google Scholar]
- Cain AJ, Sheppard PM. NATURAL SELECTION IN CEPAEA. Genetics. 1954;39(1):89–116. doi: 10.1093/genetics/39.1.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron R, Cook L. CORRELATED PHENOTYPIC RESPONSES TO HABITAT DIFFERENCE IN CEPAEA NEMORALIS (L.) FOLIA MALACOLOGICA. 2012;20(4):255–263. [Google Scholar]
- Carvunis A-R, Rolland T, Wapinski I, Calderwood MA, Yildirim MA, Simonis N, et al. Vidal M. Proto-genes and de novo gene birth. Nature. 2012;487(7407):370–374. doi: 10.1038/nature11184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B, Barton NH. The Spread of an Inversion with Migration and Selection. Genetics. 2018;208(1):377–382. doi: 10.1534/genetics.117.300426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng CD, Tan JC, Hahn MW, Besansky NJ. Systems genetic analysis of inversion polymorphisms in the malaria mosquito Anopheles gambiae. Proc Natl Acad Sci U S A. 2018;115(30):E7005–E7014. doi: 10.1073/pnas.1806760115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng CD, White BJ, Kamdem C, Mockaitis K, Costantini C, Hahn MW, Besansky NJ. Ecological Genomics of Anopheles gambiae Along a Latitudinal Cline: A Population-Resequencing Approach. Genetics. 2012;190(4):1417–1432. doi: 10.1534/genetics.111.137794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesley P, Dunn LC. THE INHERITANCE OF TAILLESSNESS (ANURY) IN THE HOUSE MOUSE. Genetics. 1936;21(5):525–536. doi: 10.1093/genetics/21.5.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chouteau M, Llaurens V, Piron-Prunier F, Joron M. Polymorphism at a mimicry supergene maintained by opposing frequency-dependent selection pressures. Proc Natl Acad Sci U S A. 2017;114(31):8325–8329. doi: 10.1073/pnas.1702482114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke CA, Sheppard FRS. The genetics of the mimetic butterfly Papilio polytes L. Philosophical Transactions of the Royal Society of London. B, Biological Sciences. 1972;263(855):431–458. doi: 10.1098/rstb.1972.0006. [DOI] [PubMed] [Google Scholar]
- Colosimo PF, Peichel CL, Nereng K, Blackman BK, Shapiro MD, Schluter D, Kingsley DM. The Genetic Architecture of Parallel Armor Plate Reduction in Threespine Sticklebacks. Plos Biology. 2004;2(5):e109. doi: 10.1371/journal.pbio.0020109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coluzzi M, Sabatini A, della Torre A, Di Deco MA, Petrarca V. A polytene chromosome analysis of the Anopheles gambiae species complex. Science. 2002;298(5597):1415–1418. doi: 10.1126/science.1077769. [DOI] [PubMed] [Google Scholar]
- Coluzzi M, Sabatini A, Petrarca V, Di Deco MA. Chromosomal differentiation and adaptation to humn environments in the Anopheles gambiae complex. Royal Society of Tropical Medicine and Hygiene. 1979;73(5):483–497. doi: 10.1016/0035-9203(79)90036-1. [DOI] [PubMed] [Google Scholar]
- Comeault Aaron A, Flaxman Samuel M, Riesch R, Curran E, Soria-Carrasco V, Gompert Z, et al. Nosil P. Selection on a Genetic Polymorphism Counteracts Ecological Speciation in a Stick Insect. Current Biology. 2015;25(15):1975–1981. doi: 10.1016/j.cub.2015.05.058. [DOI] [PubMed] [Google Scholar]
- Corbett-Detig RB, Said I, Calzetta M, Genetti M, McBroome J, Maurer NW, Petrarca V, della Torre A, Besansky NJ. Fine-mapping complex inversion break-points and investigating somatic pairing in the anopheles gambiae species complex using proximity-ligation sequencing. Genetics. 2019;213(4):1495–1511. doi: 10.1534/genetics.119.302385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett-Detig RB. Selection on Inversion Breakpoints Favors Proximity to Pairing Sensitive Sites in Drosophila melanogaster. Genetics. 2016;204(1):259–265. doi: 10.1534/genetics.116.190389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett-Detig RB, Cardeno C, Langley CH. Sequence-Based Detection and Breakpoint Assembly of Polymorphic Inversions. Genetics. 2012;192(1):131–137. doi: 10.1534/genetics.112.141622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett-Detig RB, Hartl DL. Population Genomics of Inversion Polymorphisms in Drosophila melanogaster. PLoS Genet. 2012;8(12):e1003056. doi: 10.1371/journal.pgen.1003056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coughlan JM, Willis JH. Dissecting the role of a large chromosomal inversion in life history divergence throughout the Mimulus guttatus species complex. Mol Ecol. 2018;28(6):1343–1357. doi: 10.1111/mec.14804. [DOI] [PubMed] [Google Scholar]
- Darlington CD, Mather K. The elements of Genetics. George Allen & Unwind LTD; London: 1949. [Google Scholar]
- Darwin C. On the Two Forms, or Dimorphic Condition, in the Species of Primula, and on their remarkable Sexual Relations. Journal of the Proceedings of the Linnean Society of London Botany. 1862;6(22):77–96. doi: 10.1111/j.1095-8312.1862.tb01218.x. [DOI] [Google Scholar]
- de Jong S, Chepelev I, Janson E, Strengman E, van den Berg LH, Veldink JH, Ophoff RA. Common inversion polymorphism at 17q21.31 affects expression of multiple genes in tissue-specific manner. BMC Genomics. 2012;13(458) doi: 10.1186/1471-2164-13-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Nettancourt D. Incompatibility and incongruity in wild and cultivated plants. Springer-Verlag; Berlin: 2001. [Google Scholar]
- Dobzhansky TG. Adaptive Changes Induced by Natural Selection in Wild Populations of Drosophila. Evolution. 1947;1(1/2):1–16. doi: 10.2307/2405399. [DOI] [Google Scholar]
- Dobzhansky TG. Genetics of the Evolutionary Process. University Press; Columbia: 1970. [Google Scholar]
- Durand E, Chantreau M, Le Veve A, Stetsenko R, Dubin M, Genete M, et al. Castric V. Evolution of self-incompatibility in the Brassicaceae: Lessons from a textbook example of natural selection. Evolutionary Applications. 2020;13(6):1279–1297. doi: 10.1111/eva.12933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eid J, Fehr A, Gray J, Luong K, Lyle J, Otto G, et al. Turner S. Real-Time DNA Sequencing from Single Polymerase Molecules. Science. 2009;323(5910):133–138. doi: 10.1126/science.1162986. [DOI] [PubMed] [Google Scholar]
- Entani T, Iwano M, Shiba H, Che F-S, Isogai A, Takayama S. Comparative analysis of the self-incompatibility (S-) locus region of Prunus mume: identification of a pollen-expressed F-box gene with allelic diversity. Genes to Cells. 2003;8(3):203–213. doi: 10.1046/j.1365-2443.2003.00626.x. [DOI] [PubMed] [Google Scholar]
- Entani T, Iwano M, Shiba H, Takayama S, Fukui K, Isogai A. Centromeric localization of an S-RNase gene in Petunia hybrida Vilm. Theoretical and Applied Genetics. 1999;99(3):391–397. doi: 10.1007/s001220051249. [DOI] [PubMed] [Google Scholar]
- Faria R, Johannesson K, Butlin RK, Westram AM. Evolving Inversions. Trends Ecol Evol. 2019;34(3):239–248. doi: 10.1016/j.tree.2018.12.005. [DOI] [PubMed] [Google Scholar]
- Feder JL, Gejji R, Powell THQ, Nosil P. ADAPTIVE CHROMOSOMAL DIVERGENCE DRIVEN BY MIXED GEOGRAPHIC MODE OF EVOLUTION. Evolution. 2011;65(8):2157–2170. doi: 10.1111/j.1558-5646.2011.01321.x. [DOI] [PubMed] [Google Scholar]
- Fisher RA. The Genetical Theory of Natural Selection. Oxford: 1930. [Google Scholar]
- Ford EB. Genetic Polymorphism. Studies Faber & Faber; London: 1965. [Google Scholar]
- Ford EB. Ecological Genomics. 3rd. Chapman and Hall LTD; London: 1971. [Google Scholar]
- Fouet C, Gray E, Besansky NJ, Costantini C. Adaptation to Aridity in the Malaria Mosquito Anopheles gambiae: Chromosomal Inversion Polymorphism and Body Size Influence Resistance to Desiccation. PLoS One. 2012;7(4):e34841. doi: 10.1371/journal.pone.0034841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganetzky B. On the components of segregation distortion in Drosophila melanogaster. Genetics. 1977;86(2 Pt. 1):321–355. [PMC free article] [PubMed] [Google Scholar]
- Gonzalez DR, Aramendia AC, Davison A. Recombination within the Cepaea nemoralis supergene is confounded by incomplete penetrance and epistasis. Heredity. 2019;123(2):153–161. doi: 10.1038/s41437-019-0190-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goubet PM, Bergès H, Bellec A, Prat E, Helmstetter N, Mangenot S, Castric V. Contrasted Patterns of Molecular Evolution in Dominant and Recessive Self-Incompatibility Haplotypes in Arabidopsis. PLoS Genet. 2012;8(3):e1002495. doi: 10.1371/journal.pgen.1002495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray EM, Rocca KAC, Costantini C, Besansky NJ. Inversion 2La is associated with enhanced desiccation resistance in Anopheles gambiae. Malaria Journal. 2009;8 doi: 10.1186/1475-2875-8-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero RF, Rousset F, Kirkpatrick M. Coalescent patterns for chromosomal inversions in divergent populations. Philosophical Transactions of the Royal Society B-Biological Sciences. 2012;367(1587):430–438. doi: 10.1098/rstb.2011.0246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillén Y, Ruiz A. Gene alterations at Drosophila inversion breakpoints provide prima facie evidence for natural selection as an explanation for rapid chromosomal evolution. BMC Genomics. 2012;13(1):53. doi: 10.1186/1471-2164-13-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond TM, Rehard DG, Xiao H, Shiu PKT. Molecular dissection of Neurospora Spore killer meiotic drive elements. Proceedings of the National Academy of Sciences. 2012;109(30):12093–12098. doi: 10.1073/pnas.1203267109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl DL. GENETIC DISSECTION OF SEGREGATION DISTORTION. I. SUICIDE COMBINATIONS OF SD GENES. Genetics. 1974;76(3):477–486. doi: 10.1093/genetics/76.3.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann BG, Bauer H. In: Evolution of the House Mouse. Piálek J, Macholán M, Munclinger P, Baird SJE, editors. Cambridge University Press; Cambridge: 2012. The mouse t-haplotype: a selfish chromosome – genetics, molecular mechanism, and evolution; pp. 297–314. [Google Scholar]
- Hiraizumi Y, Crow JF. HETEROZYGOUS EFFECTS ON VIABILITY, FERTILITY, RATE OF DEVELOPMENT, AND LONGEVITY OF DROSOPHILA CHROMOSOMES THAT ARE LETHAL WHEN HOMOZYGOUS. Genetics. 1960;45(8):1071–1083. doi: 10.1093/genetics/45.8.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann AA, Rieseberg LH. Revisiting the Impact of Inversions in Evolution: From Population Genetic Markers to Drivers of Adaptive Shifts and Speciation? Annual Review of Ecology, Evolution, and Systematics. 2008;39(1):21–42. doi: 10.1146/annurev.ecolsys.39.110707.173532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins DP, Tyukmaeva VI, Gompert Z, Feder JL, Nosil P. Functional genomics offers new tests of speciation hypotheses. Trends Ecol Evol. 2020;35(11):968–971. doi: 10.1016/j.tree.2020.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y-C, Dang VD, Chang N-C, Wang J. Multiple large inversions and breakpoint rewiring of gene expression in the evolution of the fire ant social supergene. Proceedings of the Royal Society B: Biological Sciences. 2018;285(1878):20180221. doi: 10.1098/rspb.2018.0221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber B, Whibley A, Poul YL, Navarro N, Martin A, Baxter S, et al. Joron M. Conservatism and novelty in the genetic architecture of adaptation in Heliconius butterflies. Heredity. 2015;114(5):515–524. doi: 10.1038/hdy.2015.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain M, Olsen HE, Paten B, Akeson M. The Oxford Nanopore MinION: delivery of nanopore sequencing to the genomics community. Genome Biology. 2016;17(1):239. doi: 10.1186/s13059-016-1103-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jay P, Whibley A, Frézal L, Rodríguez de Cara MÁ, Nowell RW, Mallet J, et al. Joron M. Supergene Evolution Triggered by the Introgression of a Chromosomal Inversion. Current Biology. 2018;28(11):1839–1845.:e1833. doi: 10.1016/j.cub.2018.04.072. [DOI] [PubMed] [Google Scholar]
- Jones FC, Grabherr MG, Chan YF, Russell P, Mauceli E, Johnson J, et al. Kingsley DM. The genomic basis of adaptive evolution in threespine sticklebacks. Nature. 2012;484:55–61. doi: 10.1038/nature10944. https://www.nature.com/articles/nature10944#supplementary-information . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joron M, Frezal L, Jones RT, Chamberlain NL, Lee SF, Haag CR, et al. ffrench-Constant RH. Chromosomal rearrangements maintain a polymorphic supergene controlling butterfly mimicry. Nature. 2011;477(7363):203–206. doi: 10.1038/nature10341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joron M, Wynne IR, Lamas G, Mallet J. Variable selection and the coexistence of multiple mimetic forms of the butterfly Heliconius numata. Evolutionary Ecology. 1999;13(7-8):721–754. doi: 10.1023/A:1010875213123. [DOI] [Google Scholar]
- Kapan DD. Three-butterfly system provides a field test of mullerian mimicry. Nature. 2001;409(6818):338–340. doi: 10.1038/35053066. [DOI] [PubMed] [Google Scholar]
- Kelemen RK, Vicoso B. Complex History and Differentiation Patterns of the t-Haplotype, a Mouse Meiotic Driver. Genetics. 2018;208(1):365–375. doi: 10.1534/genetics.117.300513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick M. How and Why Chromosome Inversions Evolve. Plos Biology. 2010;8(9):e1000501. doi: 10.1371/journal.pbio.1000501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick M, Barton N. Chromosome Inversions, Local Adaptation and Speciation. Genetics. 2006;173(1):419–434. doi: 10.1534/genetics.105.047985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirubakaran TG, Grove H, Kent MP, Sandve SR, Baranski M, Nome T, et al. Andersen O. Two adjacent inversions maintain genomic differentiation between migratory and stationary ecotypes of Atlantic cod. Mol Ecol. 2016;25(10):2130–2143. doi: 10.1111/mec.13592. [DOI] [PubMed] [Google Scholar]
- Korunes KL, Noor MAF. Gene conversion and linkage: effects on genome evolution and speciation. Mol Ecol. 2017;26(1):351–364. doi: 10.1111/mec.13736. [DOI] [PubMed] [Google Scholar]
- Kraft K, Geuer S, Will Anja J, Chan Wing L, Paliou C, Borschiwer M, et al. Andrey G. Deletions, Inversions, Duplications: Engineering of Structural Variants using CRISPR/Cas in Mice. Cell Reports. 2015;10(5):833–839. doi: 10.1016/j.celrep.2015.01.016. [DOI] [PubMed] [Google Scholar]
- Kunte K, Zhang W, Tenger-Trolander A, Palmer DH, Martin A, Reed RD, et al. Kronforst MR. doublesex is a mimicry supergene. Nature. 2014;507(7491):229–232. doi: 10.1038/nature13112. [DOI] [PubMed] [Google Scholar]
- Kupper C, Stocks M, Risse JE, dos Remedios N, Farrell LL, Mcrae SB, et al. Burke T. A supergene determines highly divergent male reproductive morphs in the ruff. Nature Genetics. 2016;48(1):79–83. doi: 10.1038/ng.3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamichhaney S, Fan GY, Widemo F, Gunnarsson U, Thalmann DS, Hoeppner MP, et al. Andersson L. Structural genomic changes underlie alternative reproductive strategies in the ruff (Philomachus pugnax) Nature Genetics. 2016;48(1):84–88. doi: 10.1038/ng.3430. [DOI] [PubMed] [Google Scholar]
- Larracuente AM, Presgraves DC. The Selfish Segregation Distorter Gene Complex of Drosophila melanogaster. Genetics. 2012;192(1):33–53. doi: 10.1534/genetics.112.141390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavington E, Kern AD. The Effect of Common Inversion Polymorphisms In(2L) and In(3R)Mo on Patterns of Transcriptional Variation in Drosophila melanogaster. G3: Genes|Genomes|Genetics. 2017;7(11):3659–3668. doi: 10.1534/g3.117.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CR, Wang BS, Mojica JP, Mandakova T, Prasad KYSK, Goicoechea JL, et al. Mitchell-Olds T. Young inversion with multiple linked QTLs under selection in a hybrid zone. Nature Ecology & Evolution. 2017;1(5) doi: 10.1038/s41559-017-0119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemopoulos A, Uusi-Heikkilä S, Huusko A, Vasemägi A, Vainikka A. Comparison of Migratory and Resident Populations of Brown Trout Reveals Candidate Genes for Migration Tendency. Genome Biology and Evolution. 2018;10(6):1493–1503. doi: 10.1093/gbe/evy102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Cocker JM, Wright J, Webster MA, McMullan M, Dyer S, et al. Gilmartin PM. Genetic architecture and evolution of the S locus supergene in Primula vulgaris. Nature Plants. 2016;2(12):16188. doi: 10.1038/nplants.2016.188. [DOI] [PubMed] [Google Scholar]
- Liehr T, Weise A, Mrasek K, Ziegler M, Padutsch N, Wilhelm K, Al-Rikabi A. Recombinant Chromosomes Resulting From Parental Pericentric Inversions—Two New Cases and a Review of the Literature. Frontiers in Genetics. 2019;10(1165) doi: 10.3389/fgene.2019.01165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling A, Cordaux R. Insertion Sequence Inversions Mediated by Ectopic Recombination between Terminal Inverted Repeats. PLoS One. 2010;5(12):e15654. doi: 10.1371/journal.pone.0015654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry DB, Willis JH. A Widespread Chromosomal Inversion Polymorphism Contributes to a Major Life-History Transition, Local Adaptation, and Reproductive Isolation. Plos Biology. 2010;8(9):e1000500. doi: 10.1371/journal.pbio.1000500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyon MF. Transmission ratio distortion in mouse t-haplotypes is due to multiple distorter genes acting on a responder locus. Cell. 1984;37(2):621–628. doi: 10.1016/0092-8674(84)90393-3. [DOI] [PubMed] [Google Scholar]
- Mallet J, Barton NH. Strong Natural-Selection in a Warning-Color Hybrid Zone. Evolution. 1989;43(2):421–431. doi: 10.2307/2409217. [DOI] [PubMed] [Google Scholar]
- Mather K. The Genetical Architecture of Heterostyly in Primula sinensis. Evolution. 1950;4(4):340–352. doi: 10.2307/2405601. [DOI] [Google Scholar]
- Mather K. Polymorphism as an Outcome of Disruptive Selection. Evolution. 1955;9(1):52–61. doi: 10.2307/2405357. [DOI] [Google Scholar]
- McBroome J, Liang D, Corbett-Detig R. Fine-scale position effects shape the distribution of inversion breakpoints in Drosophila melanogaster. Genome Biology and Evolution. 2020;12(8):1378–1391. doi: 10.1093/gbe/evaa103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrill RM, Wallbank RWR, Bull V, Salazar PCA, Mallet J, Stevens M, Jiggins CD. Disruptive ecological selection on a mating cue. Proceedings of the Royal Society B-Biological Sciences. 2012;279(1749):4907–4913. doi: 10.1098/rspb.2012.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro A, Barton NH. Chromosomal Speciation and Molecular Divergence--Accelerated Evolution in Rearranged Chromosomes. Science. 2003;300(5617):321–324. doi: 10.1126/science.1080600. [DOI] [PubMed] [Google Scholar]
- Nichols KM, Edo AF, Wheeler PA, Thorgaard GH. The genetic basis of smoltification-related traits in Oncorhynchus mykiss. Genetics. 2008;179(3):1559–1575. doi: 10.1534/genetics.107.084251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa H, Iijima T, Kajitani R, Yamaguchi J, Ando T, Suzuki Y, et al. Fujiwara H. A genetic mechanism for female-limited Batesian mimicry in Papilio butterfly. Nature Genetics. 2015;47(4):405–409. doi: 10.1038/ng.3241. [DOI] [PubMed] [Google Scholar]
- Nosil P, Villoutreix R, de Carvalho CF, Farkas TE, Soria-Carrasco V, Feder JL, et al. Gompert Z. Natural selection and the predictability of evolution in Timema stick insects. Science. 2018;359(6377):765–770. doi: 10.1126/science.aap9125. [DOI] [PubMed] [Google Scholar]
- O’Malley KG, Sakamoto T, Danzmann RG, Ferguson MM. Quantitative Trait Loci for Spawning Date and Body Weight in Rainbow Trout: Testing for Conserved Effects Across Ancestrally Duplicated Chromosomes. Journal of Heredity. 2003;94(4):273–284. doi: 10.1093/jhered/esg067. [DOI] [PubMed] [Google Scholar]
- Pearse DE, Miller MR, Abadia-Cardoso A, Garza JC. Rapid parallel evolution of standing variation in a single, complex, genomic region is associated with life history in steelhead/rainbow trout. Proceedings of the Royal Society B-Biological Sciences. 2014;281(1783) doi: 10.1098/rspb.2014.0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pevzner P, Tesler G. Human and mouse genomic sequences reveal extensive breakpoint reuse in mammalian evolution. Proc Natl Acad Sci U S A. 2003;100(13):7672. doi: 10.1073/pnas.1330369100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper J, Charlesworth B. The evolution of distyly in Primula vulgaris. Biological Journal of the Linnean Society. 2008;29(2):123–137. doi: 10.1111/j.1095-8312.1986.tb01827.x. [DOI] [Google Scholar]
- Puig M, Casillas S, Villatoro S, Caceres M. Human inversions and their functional consequences. Briefings in Functional Genomics. 2015;14(5):369–379. doi: 10.1093/bfgp/elv020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranz JM, Maurin D, Chan YS, von Grotthuss M, Hillier LW, Roote J, et al. Bergman CM. Principles of Genome Evolution in the Drosophila melanogaster Species Group. Plos Biology. 2007;5(6):e152. doi: 10.1371/journal.pbio.0050152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades NA, Harvey AM, Samarajeewa DA, Svedberg J, Yusifov A, Abusharekh A, et al. Hammond TM. Identification of rfk-1, a Meiotic Driver Undergoing RNA Editing in Neurospora. Genetics. 2019;212(1):93–110. doi: 10.1534/genetics.119.302122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robison BD, Wheeler PA, Sundin K, Sikka P, Thorgaard GH. Composite Interval Mapping Reveals a Major Locus Influencing Embryonic Development Rate in Rainbow Trout (Oncorhynchus mykiss) Journal of Heredity. 2001;92(1):16–22. doi: 10.1093/jhered/92.1.16. [DOI] [PubMed] [Google Scholar]
- Rocca KAC, Gray EM, Costantini C, Besansky NJ. 2La chromosomal inversion enhances thermal tolerance of Anopheles gambiae larvae. Malaria Journal. 2009;8 doi: 10.1186/1475-2875-8-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross KG, Keller L. Ecology and Evolution of Social-Organization - Insights from Fire Ants and Other Highly Eusocial Insects. Annual Review of Ecology and Systematics. 1995;26:631–656. doi: 10.1146/annurev.es.26.110195.003215. [DOI] [Google Scholar]
- Said I, Byrne A, Serrano V, Cardeno C, Vollmers C, Corbett-Detig R. Linked genetic variation and not genome structure causes widespread differential expression associated with chromosomal inversions. Proceedings of the National Academy of Sciences. 2018;115(21):5492–5497. doi: 10.1073/pnas.1721275115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandler L, Hiraizumi Y. MEIOTIC DRIVE IN NATURAL POPULATIONS OF DROSOPHILA MELANOGASTER. IV. INSTABILITY AT THE SEGREGATION-DISTORTER LOCUS. Genetics. 1960;45(9):1269–1287. doi: 10.1093/genetics/45.9.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandoval CP. Differential visual predation on morphs of Timema cristinae (Phasmatodeae:Timemidae) and its consequences for host range. Biological Journal of the Linnean Society. 1994;52(4):341–356. doi: 10.1111/j.1095-8312.1994.tb00996.x. [DOI] [Google Scholar]
- Sandoval CP, Nosil P. Counteracting selective regimes and host preference evolution in ecotypes of two species of walking-sticks. Evolution. 2005;59(11):2405–2413. [PubMed] [Google Scholar]
- Schopfer CR, Nasrallah ME, Nasrallah JB. The Male Determinant of Self-Incompatibility in Brassica. Science. 1999;286(5445):1697–1700. doi: 10.1126/science.286.5445.1697. [DOI] [PubMed] [Google Scholar]
- Schwander T, Libbrecht R, Keller L. Supergenes and Complex Phenotypes. Current Biology. 2014;24(7):R288–R294. doi: 10.1016/j.cub.2014.01.056. [DOI] [PubMed] [Google Scholar]
- Sharakhov IV, White BJ, Sharakhova MV, Kayondo J, Lobo NF, Santolamazza F, et al. Besansky NJ. Breakpoint structure reveals the unique origin of an interspecific chromosomal inversion 2La in the Anopheles gambiae complex. Proceedings of the National Academy of Sciences. 2006;103(16):6258–6262. doi: 10.1073/pnas.0509683103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva NF, Stone SL, Christie LN, Sulaman W, Nazarian KAP, Burnett LA, et al. Goring DR. Expression of the S receptor kinase in self-compatible Brassica napus cv. Westar leads to the allele-specific rejection of self-incompatible Brassica napus pollen. Molecular Genetics and Genomics. 2001;265(3):552–559. doi: 10.1007/s004380100446. [DOI] [PubMed] [Google Scholar]
- Sinclair-Waters M, Bradbury IR, Morris CJ, Lien S, Kent MP, Bentzen P. Ancient chromosomal rearrangement associated with local adaptation of a postglacially colonized population of Atlantic Cod in the northwest Atlantic. Mol Ecol. 2018;27(2):339–351. doi: 10.1111/mec.14442. [DOI] [PubMed] [Google Scholar]
- Stefansson H, Helgason A, Thorleifsson G, Steinthorsdottir V, Masson G, Barnard J, et al. Stefansson K. A common inversion under selection in Europeans. Nature Genetics. 2005;37(2):129–137. doi: 10.1038/ng1508. [DOI] [PubMed] [Google Scholar]
- Stevison LS, Hoehn KB, Noor MAF. Effects of Inversions on Within- and Between-Species Recombination and Divergence. Genome Biology and Evolution. 2011;3:830–841. doi: 10.1093/gbe/evr081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart NB, Rogers RL. Chromosomal rearrangements as a source of new gene formation in Drosophila yakuba. PLoS Genet. 2019;15(9):e1008314. doi: 10.1371/journal.pgen.1008314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stump AD, Pombi M, Goeddel L, Ribeiro JMC, Wilder JA, Torre AD, Besansky NJ. Genetic exchange in 2La inversion heterokaryotypes of Anopheles gambiae. Insect Molecular Biology. 2007;16(6):703–709. doi: 10.1111/j.1365-2583.2007.00764.x. [DOI] [PubMed] [Google Scholar]
- Sturtevant AH. A Case of Rearrangement of Genes in Drosophila. Proc Natl Acad Sci U S A. 1921;7(8):235–237. doi: 10.1073/pnas.7.8.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svedberg J, Hosseini S, Chen J, Vogan AA, Mozgova I, Hennig L, et al. Johannesson H. Convergent evolution of complex genomic rearrangements in two fungal meiotic drive elements. Nature Communications. 2018;9(1):4242. doi: 10.1038/s41467-018-06562-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takasaki T, Hatakeyama K, Suzuki G, Watanabe M, Isogai A, Hinata K. The S receptor kinase determines self-incompatibility in Brassica stigma. Nature. 2000;403(6772):913–916. doi: 10.1038/35002628. [DOI] [PubMed] [Google Scholar]
- Takayama S, Isogai A. SELF-INCOMPATIBILITY IN PLANTS. Annual Review of Plant Biology. 2005;56(1):467–489. doi: 10.1146/annurev.arplant.56.032604.144249. [DOI] [PubMed] [Google Scholar]
- Takayama S, Isogai A, Tsukamoto C, Ueda Y, Hinata K, Okazaki K, Suzuki A. Sequences of S-glycoproteins, products of the Brassica campestris self-incompatibility locus. Nature. 1987;326(6108):102–105. doi: 10.1038/326102a0. [DOI] [Google Scholar]
- Takayama S, Shiba H, Iwano M, Shimosato H, Che F-S, Kai N, et al. Isogai A. The pollen determinant of self-incompatibility in Brassica campestris. Proceedings of the National Academy of Sciences. 2000;97(4):1920–1925. doi: 10.1073/pnas.040556397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JW, Cáceres M, Lowman JJ, Morehouse CB, Short ME, Baldwin EL, et al. Martin CL. The Chromosomal Polymorphism Linked to Variation in Social Behavior in the White-Throated Sparrow (Zonotrichia albicollis) Is a Complex Rearrangement and Suppressor of Recombination. Genetics. 2008;179(3):1455–1468. doi: 10.1534/genetics.108.088229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorneycroft HB. Chromosomal Polymorphism in the White-Throated Sparrow, Zonotrichia albicollis (Gmelin) Science. 1966;154(3756):1571–1572. doi: 10.1126/science.154.3756.1571. [DOI] [PubMed] [Google Scholar]
- Throneycroft HB. A Cytogenetic Study of the White-Throated Sparrow, Zonotrichia albicollis (Gmelin) Evolution. 1975;29(4):611–621. doi: 10.2307/2407072. [DOI] [PubMed] [Google Scholar]
- Turner BC, Perkins DD. SPORE KILLER, A CHROMOSOMAL FACTOR IN NEUROSPORA THAT KILLS MEIOTIC PRODUCTS NOT CONTAINING IT. Genetics. 1979;93(3):587–606. doi: 10.1093/genetics/93.3.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuttle EM, Bergland AO, Korody ML, Brewer MS, Newhouse DJ, Minx P, et al. Balakrishnan CN. Divergence and Functional Degradation of a Sex Chromosome-like Supergene. Current Biology. 2016;26(3):344–350. doi: 10.1016/j.cub.2015.11.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ushijima K, Sassa H, Dandekar AM, Gradziel TM, Tao R, Hirano H. Structural and Transcriptional Analysis of the Self-Incompatibility Locus of Almond: Identification of a Pollen-Expressed F-Box Gene with Haplotype-Specific Polymorphism. The Plant Cell. 2003;15(3):771–781. doi: 10.1105/tpc.009290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Belleghem SM, Rastas P, Papanicolaou A, Martin SH, Arias CF, Supple MA, et al. Papa R. Complex modular architecture around a simple toolkit of wing pattern genes. Nature Ecology & Evolution. 2017;1:0052. doi: 10.1038/s41559-016-0052. http://www.nature.com/articles/s41559-016-0052#supplementary-information . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villoutreix R, de Carvalho CF, Soria-Carrasco V, Lindtke D, De-la-Mora M, Muschick M, et al. Nosil P. Large-scale mutation in the evolution of a gene complex for cryptic coloration. Science. 2020;369(6502):460–466. doi: 10.1126/science.aaz4351. [DOI] [PubMed] [Google Scholar]
- Wang J, Wurm Y, Nipitwattanaphon M, Riba-Grognuz O, Huang YC, Shoemaker D, Keller L. A Y-like social chromosome causes alternative colony organization in fire ants. Nature. 2013;493(7434):664–668. doi: 10.1038/nature11832. [DOI] [PubMed] [Google Scholar]
- Wellenreuther M, Bernatchez L. Eco-Evolutionary Genomics of Chromosomal Inversions. Trends Ecol Evol. 2018;33(6):427–440. doi: 10.1016/j.tree.2018.04.002. [DOI] [PubMed] [Google Scholar]
- Yan Z, Martin SH, Gotzek D, Arsenault SV, Duchen P, Helleu Q, et al. Keller L. Evolution of a supergene that regulates a trans-species social polymorphism. Nature Ecology & Evolution. 2020;4:240–249. doi: 10.1038/s41559-019-1081-1. [DOI] [PubMed] [Google Scholar]