Abstract
Background
The accumulation of somatic mutations contributes to ageing and cancer. Sunlight is the principal aetiological factor associated with skin cancer development. However, genetic and phenotypic factors also contribute to skin cancer risk. This study aimed at exploring the role of photoaging, as well as other well-known epidemiological risk factors, in the accumulation of somatic mutations in cancer-free human epidermis.
Material and Methods
We deeply sequenced 46 genes in normal skin biopsies from 127 healthy donors, from which phenotypic data (including age, pigmentation-related genotype and phenotype) and sun exposure habits were collected. We determined the somatic mutational burden, mutational signatures, clonal selection and frequency of driver mutations in all samples.
Results
Our results reveal an exponential accumulation of UV-related somatic mutations with age, matching skin cancer incidence. The increase of mutational burden is in turn modified by an individual’s skin phototype. Somatic mutations preferentially accumulated in cutaneous squamous cell carcinoma (cSCC) cancer genes and clonally expanded with age, with distinct mutational processes underpinning different age groups. Our results suggest loss of fidelity in transcription coupled repair later in life.
Conclusion
Our findings reveal that aging is not only associated with an exponential increase in the number of somatic mutations accumulated in normal epidermis, but also with selection and expansion of cancer-associated mutations. Aged, sun-exposed normal skin is thus an extended mosaic of multiple clones with driver mutations, poised for the acquisition of transforming events.
Keywords: Somatic mutation, Aging, Skin Phototype, Next-generation sequencing, UV exposure, normal epidermis, carcinogenesis, mutational spectrum
Introduction
Cancers arise as a result of somatic alterations occurring in the genomic DNA sequence of ‘normal’ cells. Somatic genomic alterations accumulate spontaneously in cells throughout a person's life as a result of errors occurring during cell replication and after exposure to mutagenic agents, such as certain chemicals in tobacco smoke or UV radiation from sunlight [1, 2].
The accumulation of clones of cells harbouring mutations across tissues may be expected to have functional consequences on the physiology of normal cells, contributing to ageing and promoting disease progression, as in the case of cancer. In this regard, recent sequencing studies have revealed that, in general, the somatic mutational burden and profile of physiological normal tissues seem to be surprisingly similar to those found in tumours from the same tissue [3–6]. These results suggest that the majority of somatic alterations may pre-date tumour formation, and only a small fraction of all somatic mutations in a cancer genome are, therefore, directly relevant in carcinogenesis, disease classification and treatment.
In the case of skin, a recent study analysing cancer-free epidermal samples from four individuals showed that the frequency of driver mutations in physiologically normal skin cells is remarkably high [7]. Sun-exposed epidermal cells carried a multitude of genetic alterations, and about 25-30% of these normal skin cells had already acquired at least one driver mutation, indicating that cancer-causing mutations are under strong positive selection even in epidermis maintaining normal physiological functions. The mutational profile found in eyelid samples, a chronically sun-exposed area, was similar to cutaneous squamous cell carcinomas (SCCs), but distinct from the profile characteristically observed in cutaneous melanoma [7]. This may be due to the low number of epidermal melanocytes (cells from which melanoma develops) in relation to keratinocytes [8], but also due to the fact that melanoma appears more frequently in sporadically, rather than chronically, sun-exposed areas of the body – those that are usually covered by clothing. This apparently paradoxical fact is attributable to injuries caused by an intermittent pattern of intense and acute sun exposure associated with recreational activities [9].
Skin cancer incidence worldwide reveals a clear relationship between pigmentation traits and sunlight damage, with individuals with fair skin and inability to tan showing greater cancer susceptibility. Cutaneous sensitivity to sunlight exposure (ability to tan versus tendency to burn) is defined by certain genetically determined pigmentation traits [10]. Thus, individuals carrying genetic variants associated with increased sun sensitivity should have higher somatic mutational rates, as they have reduced melanin levels, insufficient to protect the genome of epidermal cells from the mutagenic action of UV light [11, 12].
While previous studies have confirmed that seemingly normal cells harbour mutations, the key factors that determine which individuals are more prone to acquire and therefore accumulate somatic mutations remains unclear. In this study, we aimed at increasing our understanding of the accumulation of somatic mutations in the skin because of different patterns of sunlight exposure (Figure S1). Furthermore, we explored if there is an added risk to accumulate somatic mutations according to intrinsic characteristics of individuals (including age, and pigmentation-related genotype and phenotype) and to sun exposure habits.
Materials and Methods
We focused on sequencing healthy skin samples from a large cohort of 123 cancer-free individuals, aged 11 to 92 years (mean of 58.50 years) (Table S1). Skin samples were collected from different body areas, classified according to the pattern of sunlight exposure as chronically-photoexposed (n=44) and intermittently-photoexposed (n=79). Only one sample was collected from each donor due to ethical reasons.
Deep sequencing of 46 genes implicated in skin cancers was performed on each epidermal sample, obtaining an average on-target coverage across samples of 923.44x (range 377.96-1657.37x).
To identify somatic mutations in the skin biopsies, we developed a bespoke pipeline. In brief, we first applied the Mutect2 tumour-only mode, due to the absence of a matched control sample. Putative germline variants and technical artefacts were subsequently removed by applying several key filtering steps, which were applied with the aim of having the maximum specificity. To validate the specificity of our pipeline, we applied it to an external dataset with available germline data. We found a very low rate of false positives (<0.05%) (Figure S2A). The proportion of mutations likely missed after applying our stringent filtering procedure was estimated as likely between 10-20%.
Further details of sampling, sequencing, variant calling and filtering, and data analyses (including mutational burdens, mutational signatures, clonal selection, frequency of driver mutations, and copy number aberrations) are provided in Supplementary Methods.
Results
Variation of mutational burden across samples
A total of 5,214 somatic mutations were identified in our dataset, with an average of 42.39 mutations per sample (range 2-169) (Figure 1A), corresponding to an average rate of 132.47 mutations per megabase (range 6.25-528.13). Consistent with previous studies of normal tissue, most mutations were present only in a small fraction of cells, evidenced by the fact that nearly all mutations exhibited a variant allele frequency (VAF) lower than 5% (Figure S2B).
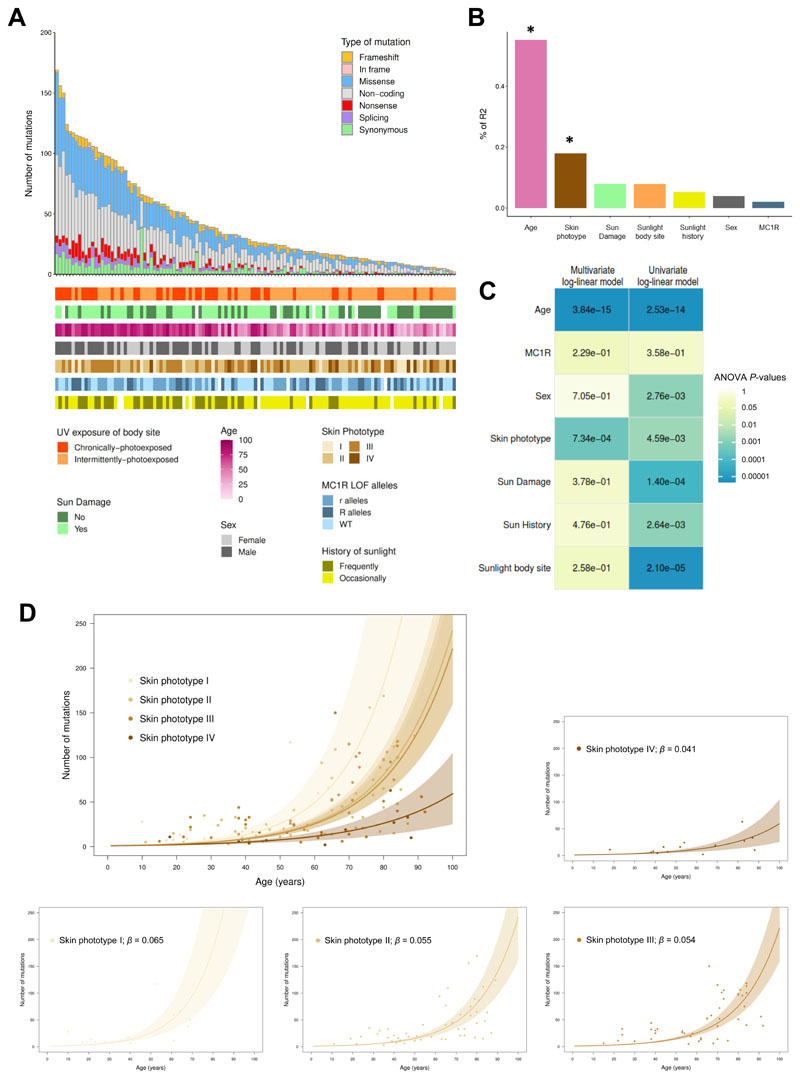
Figure 1. Risk factors contributing to the accumulation of somatic mutations in normal epidermis.
(A) Total number and type of somatic mutations detected across the 46 genes sequenced in each sample. Clinical and demographic characteristics are presented below. (B) Relative importance of predictors included in the log-linear multivariate regression model. The total variance explained by the model (adjusted-R2 = 49.88%) is decomposed in order to know the individual contribution (effect size) of each predictor. Asterisk denotes significant predictors. (C) Heatmap showing the P-values of univariate and multivariate log-linear model coefficients from the analyses-of-variance (ANOVA) tables. (D) A log-linear regression is used for analysing the age effect on the accumulation of somatic mutations for each skin phototype. Solid lines represent the bootstrapped mean of the slope, and shaded areas its bootstrapped 95% confidence intervals (CI95). Regression coefficients (β) for each skin phototype are also presented.
The size of our cohort offered us a unique opportunity to directly quantify the key factors associated with mutational burden in sun-exposed skin samples. Indeed, several phenotypic and behavioural risk factors were collected from the participants, including sex, age, Fitzpatrick’s skin phototype, history of sunlight exposure, body site pattern of sun exposure, signs of sun damage in the skin area biopsied and MC1R genotype.
To empirically assess the relative importance of each potential risk factor in contributing to the mutational burden of skin lesions, we first evaluated which type of model could best represent the data. A log-linear multivariate model was identified as providing the best fit, compared to linear, non-linear, quadratic and cubic models (Figure S3). When using this log-linear model, the total variance of mutational burden explained by all variables combined was high (adjusted-R2 = 49.88%). Age explained the largest proportion of the total variance (55.16%; Figure 1B). An individual’s skin phototype was, surprisingly, the second strongest predictor, explaining 17.92% of the mutational burden variance across samples. Our results suggested a significant decrease (β<0) in the number of somatic mutations accumulated in skin samples from individuals with high skin phototypes (skin types that normally tan after sunlight exposure), as compared with individuals with low skin phototypes (skin types that normally burn rather than tan after sunlight exposure) (Table S2). The lack of tanning capacity confers greater susceptibility to develop skin cancers, due in part to the inability to protect against UV-related DNA damage [10].
Contrary to previous studies [13], no association, in univariate or multivariate analysis, was found between the somatic mutational load and the genotype in MC1R, a key pigmentation-related gene determining the ability to respond to UV exposure [14]. Moreover, a lack of significant association between mutational burden and the pattern of body site photoexposure (chronic versus intermittent) was observed after including all risk factors in the model, explaining only 7.82% of the variance of the total number of mutations accumulated across samples. Our data suggested a comparable effect of intermittent exposure to sunlight, perhaps while on recreational activities, and continuous exposure, through spending a large amount of daylight time outdoors, with regards to the accumulation of somatic mutations in normal epidermis. None of the other risk factors remained significantly associated with mutational burden in the multivariate model (Figure 1C).
Given that age and phototype were the two most significant contributors to explain mutational burden, we quantitatively assessed age-related mutational burden according to skin phototype. For each skin phototype, we determined the mutation rate increased per year of life (β) in the panel of 46 genes (0.32 Mb) using a log-linear model (Figure 1D). Nonparametric bootstraps (1000 runs) were conducted to estimate the 95% confidence intervals (CI95) of the effect of age. Somatic accumulation rates per year of life increased across skin phototypes. Strikingly, by the age of 65 years old, we expect 68.63 mutations (CI95: 40.65-122.73) to have accumulated in the 46 selected genes for phototype I, whereas only 14.22 mutations (CI95: 8.16-20.54) are expected for type IV.
Taken together, these results suggest a profound influence of age on the accumulation of somatic mutations in normal skin. This age-associated rise of mutational burden is in turn modified by an individual’s skin phototype, reflecting the inability of UV-sensitive individuals to protect against UV-related DNA damage. We note that although sequencing coverage had an impact on the number of mutations called, our overall conclusions were not affected by discrepancies in sequencing coverage across samples (Supplementary Text and Figure S4).
Aging and the rise of UV-associated mutations
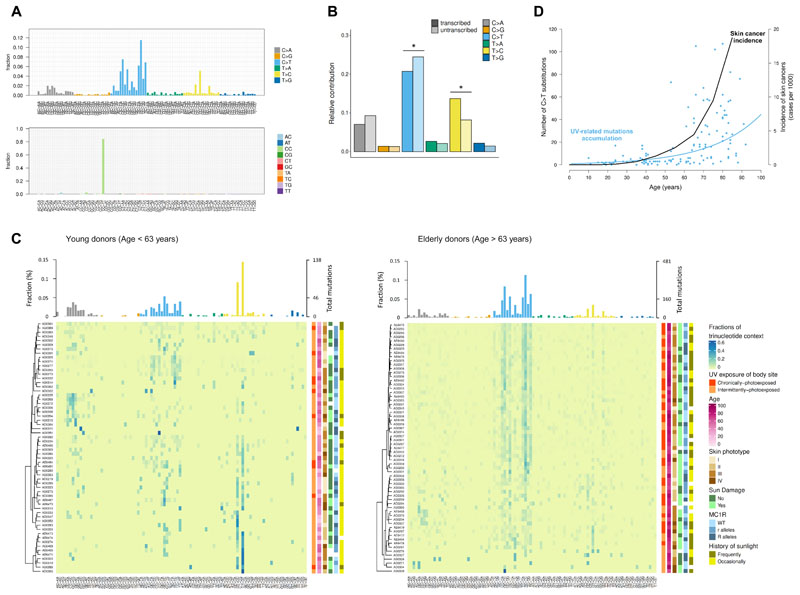
To explore the mutational processes underpinning the accumulation of mutations in sun-exposed samples, we considered the specific substitution types. In the cohort as a whole, we observed a predominance of C>T and CC>TT mutations at dipyrimidine sites (TpC or CpC), likely reflecting repair of 6,4-photoproducts and the production of cyclobutane pyrimidine dimers (CPDs) in response to UV-induced DNA damage [15](Figure 2A). These C>T substitutions were preferentially accumulated on the non-transcribed compared to the transcribed strand (P-value = 4.21×10-4), consistent with the activity of transcription-coupled nucleotide excision repair (Figure 2B). In addition, we also observed an enrichment of T>C/A>G mutations at CTT sites, potentially caused by indirect DNA damage after UV radiation – greater incorporation of G, rather than A, opposite thymidine and cytidine photodimers by translesion polymerases (Figure 2A). This is in line with the fact that these mutations tended to accumulate in the transcribed rather than in the non-transcribed strand (P-value = 5.43×10-5) (Figure 2B).
Figure 2. Spectrum of somatic mutations in normal epidermis.
(A) Bar plot showing the fraction of single (top) and doublet (bottom) base substitutions found in each of the possible 96 trinucleotide contexts (strand independent). (B) Relative number of each substitution type present on the transcribed (dark shading) and untranscribed strand (light shading). Asterisks indicate significant transcriptional strand asymmetries (Poisson test). (C) Mutational spectra in samples from young (left) and elderly donors (right). Heatmaps show the fraction of each trinucleotide change in each sample (middle). Bar plots represent the contribution mean of each 96-mutation type per age group (top). Clinical and demographic characteristics are presented next to each sample (right). (D) Age-dependent increase of both UV-mutation accumulation and skin cancer incidence. Data of skin cancer incidence in Spain was downloaded from the Global Cancer Observatory (http://gco.iarc.fr).
To quantify the presence of specific mutational signatures, which may reflect underlying mutational processes, we applied deconstructSigs [16] to the combined set of mutations within the cohort. The specific mutational spectrum observed in our samples mainly resembles the COSMIC base substitution signatures related to UV radiation (mutational signatures SBS7a-d) (Figure S5B).
The relative contribution of UV-related mutational pathogenesis varied significantly with age (Figure 2C). Mutational signatures directly related to UV contributed 68.14% of all mutations detected after the age of 63 (the cohort median age), but only 46.59% of the mutational burden in individuals younger than 63 (Figure S5B). This observation is in accordance with the fact that non-melanoma skin tumours typically occur at advanced age and are related to cumulative sun exposure [17]. In fact, we observed that the increase of UV-mutations in normal skin with age follows a similar exponential trend than skin cancer incidence in Spain (data downloaded from the Global Cancer Observatory, http://gco.iarc.fr)(Figure 2D). This exponential growth of UV-related mutations was not affected by the frequency of variant reads (Figure S4C). The main difference between the elderly and younger individuals in terms of mutational spectra, apart from the fraction of UV-related mutations, was related to the fraction of T>C/A>G mutations at CTT contexts, being proportionally higher in younger individuals. This mutational pattern closely matched the SBS17a signature, a COSMIC signature with unknown aetiology that has been shown to contribute to cutaneous melanoma [18]. To gain insight into the context dependency of this T>C/A>G substitutions, we explored the local sequence contexts (from -5 to +5 positions) and observed a specific pattern of contextual preference (CTTTT) in normal skin samples biopsied from younger individuals (Figure S7A). Additionally, the degree of transcriptional strand bias detected for T>C/A>G substitutions was substantially higher in younger individuals (Figure S7B). These differences in the mutational spectra between different age groups can be attributed to the exponential increase of C>T substitutions with age, coupled with a relatively constant, linear increase in T>C/A>G mutations with age (Figure S7D). These observations suggest that, after the age of 60, the mutagenic process related to sunlight is the predominant contributor to the accumulation of somatic mutations in skin.
To confirm that the differences shown in mutational spectra with age was not solely due to unequal representation of sunlight exposure patterns between subgroups (the proportion of chronically-photoexposed skin samples is higher in the elderly group than in the young group), we implemented deconstructSigs splitting samples of each age group according to the pattern of sunlight exposure of the skin tissue biopsied. These results confirmed that the sunlight exposure of the skin sample was not a confounding factor in the age-specific mutational pattern observed (Figure S7E).
Taken together, our results reveal that mutational processes may sculpt the genome in distinct ways as we age. While SBS17a signature seems to accumulate at a constant rate during life, UV-related mutational processes appear to become the dominant cause of mutational acquisition with age. This suggests a decline in the ability to repair UV induced mutagenic lesions later in life. Consistent with this, we observed a trend for decreased transcription coupled repair, as captured by transcription strand bias at C>T mutations, in older compared to younger individuals (P-value = 0.078; Figure S7C).
Positive selection of driver mutations in normal skin
We next considered whether protein-altering somatic mutations were subjected to positive selection and the interplay between age and selection. We evaluated the footprint of positive selection in two orthogonal ways, by quantifying the excess of non-synonymous mutations as well as by estimating clone sizes.
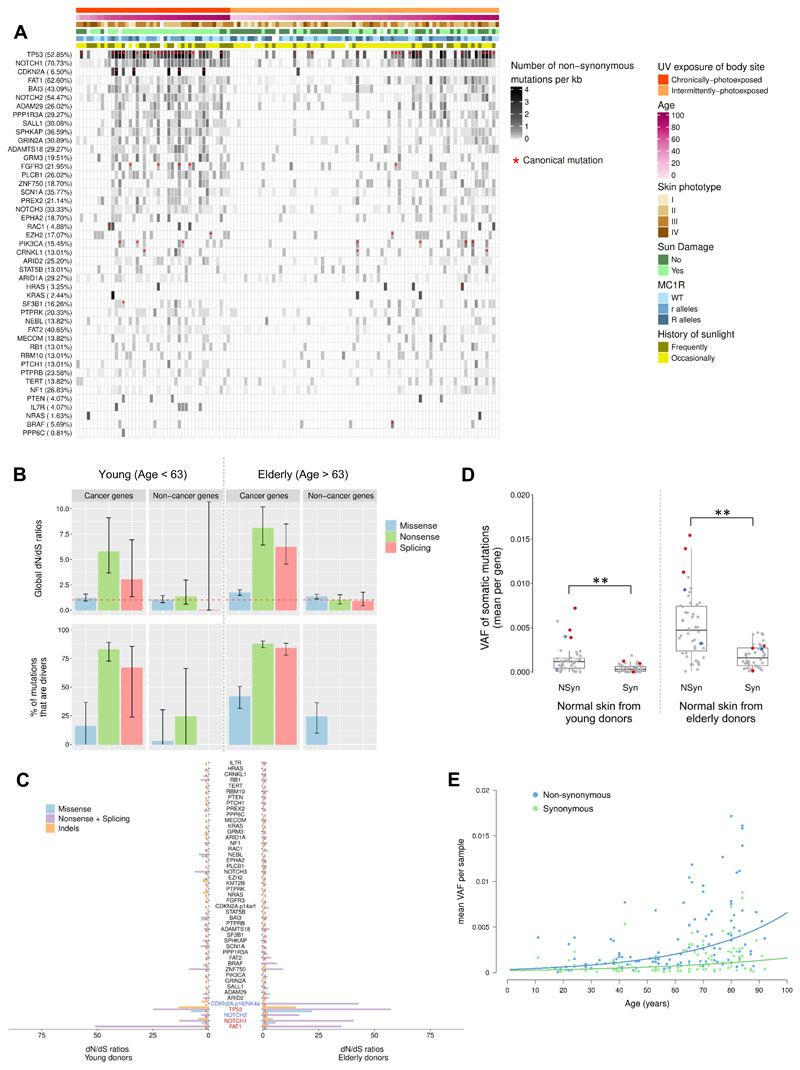
The majority of the normal sun-exposed skin samples harboured multiple protein-altering mutations, even though these epidermal samples were histologically benign (Figure 3A). The catalogue of recurrently mutated genes in normal skin was almost identical to that of cSCC, with TP53, NOTCH1, NOTCH2 and FAT1 being the most recurrently mutated genes. We also identified samples with canonical hotspot mutations with therapeutic relevance (according to the database of curated mutations) in several oncogenes such as BRAF (F595L, V600E), HRAS (G12D, G12V), PIK3CA (P471L, E542K, E726K, M1043I, H1047L), and FGFR3 (R248C, S249C, Y373C, A391E). In total, thirty-four (53.12%) skin samples from elderly individuals carried at least one of the 50 different relevant disease-causing mutations identified. Conversely, only eight (12.70%) young individuals harboured at least one of these canonical mutations (Figure 3A).
Figure 3. Occurrence, positive selection and expansion of driver mutations in normal skin with age.
(A) Heatmap showing the distribution of recurrent non-synonymous mutations per coding kilobase of sequence for each one of the 46 genes targeted across all normal skin samples. Genes are sorted from higher (top) to lower (bottom) number of non-synonymous mutations per kb. Percentage of normal skin samples carrying at least one non-synonymous mutation in each gene is shown in brackets. Clinical and demographic characteristics are presented above each sample. The KMT2B gene is not included in this plot since none non-synonymous mutation was found across samples. Red asterisks denote samples harbouring at least one canonical mutation in a specific gene. (B) Global dN/dS values (top) and frequency of driver mutations (bottom) calculated by taking together all cancer and non-cancer genes in normal skin biopsied from both young and elderly donors. Error bars represent 95% confidence intervals. Percentage of driver mutations was only calculated when dN/dS ratios denoted positive selection (dN/dS > 1). (C) dN/dS ratios for each of the 46 target genes. Genes under significant positive selection in both age groups are coloured in red, while genes positively selected only in the elderly group are coloured in blue (overall q-value < 0.05). Genes are sorted from higher (bottom) to lower (top) significant value in the elderly group. (D) Distribution of VAFs of somatic non-synonymous and synonymous mutations per gene. Red dots denote positively selected genes in both young and elderly groups (FAT1, NOTCH1, and TP53), and blue dots indicate genes under positive selection only in the elderly group (NOTCH2 and CDKN2A). Dots representing the other genes sequenced are coloured in grey. A Wilcoxon-Mann-Whitney test is used for testing differences among mutation types in each age group. (E) Expansion of non-synonymous and synonymous mutations with age. Each dot represents the mean VAF of somatic non-synonymous and synonymous mutations per sample. Fitted lines show that the variant allele frequencies of non-synonymous mutations (with a positive fitness effect) grew exponentially with age denoting selection, while synonymous mutations (with a neutral fitness effect) expanded linearly with age indicating fixation by drift.
In order to quantify the extent of positive selection driving minor clonal expansion in normal skin samples in both elderly and young donors, we considered the ratio of missense, nonsense and essential splicing mutations compared to synonymous mutations using the dNdScv package [19]. Our results provided evidence of significant positive selection when considering mutations in all known cancer genes as a whole, while a lack of selection pressure is observed in non-cancer genes (Figure 3B). Indeed, using the dN/dS ratios, we found that a considerable percentage of mutations (50.42%) accumulated in cancer genes may confer a growth advantage and thus may be positively selected in normal skin. However, non-cancer genes seemed to accumulate neutral mutations that may not be affected by natural selection and, hence, their growth/expansion can be attributed to genetic drift (Figure S9B).
At a single gene level, we had power to detect significant positive selection in three out of 46 sequenced genes for normal skin samples collected from both young and elderly individuals (TP53, NOTCH1, and FAT1) (Figure 3C). Notably, all three genes were also shown to have a significant excess of non-synonymous mutations in normal sun-exposed epidermis [7], and have been shown to be drivers of cSCC [20, 21]. Interestingly, NOTCH2 and CDKN2A, two tumour suppressor genes recurrently mutated in skin cancers [20–22], only had a significant excess of truncating mutations in samples from elderly individuals. NOTCH2 was also the gene most frequently subject to copy number aberrations (Supplementary Text and Figure S8).
The majority of sun-exposed samples (79.67%) carried at least one non-synonymous mutation in a cancer gene under positive selection (TP53, NOTCH1, and FAT1). Samples harbouring mutations in TP53, NOTCH1 and FAT1, as well as other canonical hotspot mutations, had a significant increase in the overall number of mutations. Although this increase seemed to be largely influenced by the participant’s age, the increase in mutation count associated with carrying protein-altering mutations in these positively selected genes was observed in both age groups (Figure S10A).
To confirm whether these protein-altering mutations are subject to positive selection, thereby resulting in clonal expansions, we scrutinized the VAF spectra of somatic mutations in each age group. The average VAF of non-synonymous mutations per gene was significantly higher than that of synonymous mutations in normal skin samples from both young (P-value = 9.96×10-6) and elderly donors (P-value = 6.21x10-8; Figure 3D). The highest average VAF was observed in cancer-associated genes under significant positive selection (NOTCH1, FAT1 and TP53), suggesting that these selectively advantageous mutations may appear early and expand with age in sun-exposed normal skin. However, the modest VAF of even these somatic mutations suggested that they are present in only a small subset of skin cells, therefore remaining in a minority of sun-exposed cells.
Overall, frequencies of both non-synonymous and synonymous mutations significantly increased in epidermal samples from elderly donors compared to those collected from young donors (P-value = 8.30×10-9 and P-value = 1.53×10-7, respectively; Figure 3D and S10B), suggesting tolerance and selection for larger clones with age. Intriguingly, while frequencies of synonymous mutations were found to exhibit a linear relationship with patient age, an age-dependent exponential growth was observed for non-synonymous mutations (Figure 3E). Conceivably, the linear expansion of synonymous mutations (which are generally expected to be neutral) with age reflects fixation by drift, while the exponential growth of clones carrying protein-affecting mutations (which are generally expected to increase cell fitness) with age indicates positive selection. Although there is a broad VAF spectrum in all samples, the association of clonal expansion with age and mutation effect was also observed when all mutations were analysed (Figure S9A).
Taken as a whole, our data suggest that sun-exposed normal skin already harbours protein-affecting mutations in cancer genes, with 5 genes subject to statistically significant positive selection. Furthermore, we find that skin from elderly individuals not only harbours more mutations, but a larger fraction of these reflect positive selection and expansion.
Discussion
Like other tissues, skin undergoes chronological aging that might lead to its functional decline, which is accelerated by chronic sun damage. This study aimed to explore the role of photoaging, as well as other well-known epidemiological risk factors, in the accumulation of somatic mutations in cancer-free human epidermis. Our experimental design was focused on analysing the mutational landscape in a large cohort of subjects with a wide range of ages and phenotypic characteristics.
A profound variability in terms of mutational burden and driver mutations was observed among individuals, which was largely explained by age. An exponential increase of mutation count with increasing age was observed in the panel of 46 genes. In addition, normal skin samples collected from donors with low skin phototypes (individuals with fair skin who tend to burn rather than tan after being exposed to sunlight) tended to accumulate a higher number of somatic mutations over time. Fair-skinned individuals are more severely affected by photoaging [24]. Therefore, the increased risk of developing skin cancer, at least for sporadic cancers, associated with these phenotypes may be due to the presence of a higher reservoir of mutant cells waiting to acquire more cancer-driving mutations, evade clonal growth control and initiate malignant transformation. By contrast, sun exposure habits were not significantly correlated to mutational burden variability across samples. Note that our samples were collected from Spanish individuals living in a region with relatively high UV index and pleasant weather throughout the year. Perhaps a higher effect of sun exposure pattern by body site would be found in populations from more northerly latitudes, exposed to sunlight mainly during summer vacations. Furthermore, the retrospective and subjective nature of some behavioural risk factors, particularly those related to sun exposure habits, raises the potential for recall bias. Therefore, validation in independent and larger cohorts will be needed to further analyse the association between behavioural risk factors and mutational burden. Additionally, future efforts should be done to explore other factors (i.e. repair mechanisms efficiency, antioxidant capability, etc.) that may impact on somatic mutation acquisition among individuals.
Analysing a large cohort with a wide age range has allowed us to investigate the timing of somatic mutation accumulation in normal skin. The number of T>C mutations accumulated in normal skin appeared to accumulate at a steady linear rate. In contrast, an exponential rise of UV-related mutations with increasing age was evident in normal epidermis. This exponential increase of UV-related mutations, together with the fact that the ratio of these mutations accumulated in the non-coding versus the coding strand tend to decrease with age, could be related to the decline of nucleotide excision repair (NER) capacity with age [25]. NER is a highly evolutionarily conserved mechanism for repairing bulky DNA lesions resulting, among others, from sunlight exposure. The importance of NER activity in the prevention of skin cancer is denoted by the extreme sensitivity to sunlight and severe predisposition to UV-induced skin cancers of patients with the inherited disorder xeroderma pigmentosum, in which genes encoding for the different components of the NER cascade are mutated [26].
The age-dependent exponential increase of skin cancer incidence in Spain followed a similar trend than the accumulation of UV-related mutations in normal skin. Apart from the decline of NER function, skin photoaging has also been linked to impaired skin homeostasis [27]. It is thought that strategies for tissue maintenance have a noteworthy impact on cancer incidence, consistent with the dramatic increase of cancer incidence with tissue and stem cell fitness decline [28]. Aged/damaged tissue microenvironment may therefore provide an opportunity for clones with a selective advantage to expand, and that may be the reason why mutational profile notably differs between normal skin samples from young and adult donors.
Our results revealed an enrichment of driver mutations in most normal sun-exposed skin samples, especially in those collected from elderly individuals. In addition, there was a marked overrepresentation of protein-altering mutations in several cSCC driver genes, especially in NOTCH1, TP53 and FAT1, likely reflecting positive selection. As previously seen in blood [29], the quantitative analysis of selection by measuring the excess of non-synonymous mutations (dN/dS ratios) were in line with clone size distributions. Analysing the VAF spectra of mutations accumulated in normal skin, we noted that clone size seems to be closely related to age (time during which it has been expanding) and to the variant impact on cell growth (rate at which it has been expanding).
However, because these positively-selected genes have been shown to be frequently mutated in normal skin [7, 23], it seems unlikely that these mutations alone confer a sufficient growth advantage to engender cancer development. Indeed, although clones carrying these selectively advantageous mutations have expanded in normal skin, it is notable that only 17 out of 123 donors (13.82%) carried more than one mutation with clinical relevance in their skin. These mutant clones also seem to be relatively small, suggesting even in these cases the somatic alterations are mostly in distinct clones. Indeed, using dN/dS ratio, the number of canonical mutations estimated per cell was only 0.21 (range of 0.07-0.72) in samples carrying at least two clinically relevant mutations. We speculate that most cells carrying driver mutations may not have yet acquired the right combination of mutated genes for expanding and culminating in the development of malignancy.
Cellular senescence, an irreversible proliferative arrest triggered by exogenous and endogenous stresses, may be a plausible explanation for the limited expansion of clones carrying cancer-causing mutations in normal tissues. Oncogenic-induced senescence is considered a crucial protective mechanism against cell transformation. Several reports from animal models support the idea that cell senescence may occur in tissues after acquiring a mitogenic mutation, preventing carcinogenesis at an initial step [30, 31]. The biology of naevi is a clear example of cellular senescence following an initial activating oncogenic mutation, normally in BRAF or NRAS [32, 33]. Melanocytic nevi are clonal proliferations of non-malignant melanocytic cells, which can remain non-growing for many years, but also can act as precursors of melanoma if cells overcome senescence. Interventions that favour oncogene-induced senescence may help restrict the growth of clones carrying cancer-causing mutations in normal tissues and thus tumour progression.
The question arising from our observations, together with those shown in different sequencing studies previously performed on healthy tissues [3–5, 7, 34, 35], is whether or not targeting these early mutations recurrently found in normal tissues will be relevant for preventing carcinogenesis. Further efforts should be done to delineate the succession of genetic alterations needed for malignant transformation of physiologically normal tissues to premalignant precursor lesions, and finally to tumours, with the aim of discriminating drivers of the disease from the non-pathogenic mutational landscape.
Supplementary Material
Highlights.
-
-
As skin cancer incidence, UV-related somatic mutations accumulate exponentially with increasing age in normal epidermis
-
-
UV-related mutations do not only accumulate with age, but also repair processes are less efficient in elderly individuals
-
-
Apart from age, an individual’s skin phototype is key in the build-up of somatic mutations in healthy skin
-
-
Distinct mutational processes may operate in different age groups
-
-
Aging is associated with positive selection and expansion of clones with cancer mutations in sun-exposed normal skin
Acknowledgments
We are extremely grateful to all the volunteers for giving their consent to take part in this study, as well as to all the medical specialists for supervising phenotype collection of all samples. This work is supported in part by the Jaume I University of Castellon (UJI-A2016-13). B.H. is funded by the Jaume I University of Castellon under a Postdoctoral Research contract (POSDOC-A/2018/07), and received additional funding for conducting a research exchange at the UCL Cancer Institute (E-2019-34). N.M is a Sir Henry Dale Fellow, jointly funded by the Wellcome Trust and the Royal Society (Grant Number 211179/Z/18/Z), and also receives funding from CRUK, Rosetrees, and the NIHR BRC at University College London Hospitals, and the CRUK University College London Experimental Cancer Medicine Centre.
Footnotes
Author Contributions
B.H. treated skin tissues, performed DNA extraction, prepared samples for sequencing, conducted bioinformatics analysis, and wrote the manuscript. M.D. helped with mutational signatures analysis. G.P. helped with variant calling and filtering, and copy number aberration analysis. M.G-B. helped with lab work. G.P, L.M. and F.V-C collected tissue samples and phenotypic data of donors. N.M. provided expertise in interpreting the results, supervised bioinformatics analyses, and wrote the manuscript. C.M-C conceived the project, supervised the study, and wrote the manuscript. All co-authors provided critical feedback and contributed to manuscript preparation.
Declaration of Interests
N.M. has received consultancy fees and has stock options in Achilles Therapeutics. All remaining authors have declared no conflicts of interest.
Data Availability
Sequencing data has been deposited at the European Genome–phenome Archive (EGA) under the accession code EGAS00001004279. The somatic mutations found in all samples are listed in the Supplementary Dataset S1. Clinical data of each donor can be found in the Supplementary Dataset S2. Any other relevant data can be obtained from the corresponding authors upon reasonable request.
References
- 1.Martincorena I, Campbell PJ. Somatic mutation in cancer and normal cells. Science. 2015;349(6255):1483–1489. doi: 10.1126/science.aab4082. [DOI] [PubMed] [Google Scholar]
- 2.Stratton MR, Campbell PJ, Futreal PA. The cancer genome. Nature. 2009;458(7239):719–724. doi: 10.1038/nature07943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brunner SF, Roberts ND, Wylie LA, et al. Somatic mutations and clonal dynamics in healthy and cirrhotic human liver. Nature. 2019;574(7779):538–542. doi: 10.1038/s41586-019-1670-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jaiswal S, Fontanillas P, Flannick J, et al. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med. 2014;371(26):2488–2498. doi: 10.1056/NEJMoa1408617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee-Six H, Olafsson S, Ellis P, et al. The landscape of somatic mutation in normal colorectal epithelial cells. Nature. 2019;574(7779):532–537. doi: 10.1038/s41586-019-1672-7. [DOI] [PubMed] [Google Scholar]
- 6.Martincorena I. Somatic mutation and clonal expansions in human tissues. Genome Med. 2019;11(1):35. doi: 10.1186/s13073-019-0648-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martincorena I, Roshan A, Gerstung M, et al. Tumor evolution. High burden and pervasive positive selection of somatic mutations in normal human skin. Science. 2015;348(6237):880–886. doi: 10.1126/science.aaa6806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoath SB, Leahy DG. The organization of human epidermis: functional epidermal units and phi proportionality. J Invest Dermatol. 2003;121(6):1440–1446. doi: 10.1046/j.1523-1747.2003.12606.x. [DOI] [PubMed] [Google Scholar]
- 9.Gandini S, Sera F, Cattaruzza MS, et al. Meta-analysis of risk factors for cutaneous melanoma: II. Sun exposure. Eur J Cancer Oxf Engl. 1990;41(1):45–60. doi: 10.1016/j.ejca.2004.10.016. 2005. [DOI] [PubMed] [Google Scholar]
- 10.Scherer D, Kumar R. Genetics of pigmentation in skin cancer--a review. Mutat Res. 2010;705(2):141–153. doi: 10.1016/j.mrrev.2010.06.002. [DOI] [PubMed] [Google Scholar]
- 11.Rijken F, Bruijnzeel PLB, van Weelden H, Kiekens RCM. Responses of black and white skin to solar-simulating radiation: differences in DNA photodamage, infiltrating neutrophils, proteolytic enzymes induced, keratinocyte activation, and IL-10 expression. J Invest Dermatol. 2004;122(6):1448–1455. doi: 10.1111/j.0022-202X.2004.22609.x. [DOI] [PubMed] [Google Scholar]
- 12.Brenner M, Hearing VJ. The Protective Role of Melanin Against UV Damage in Human Skin. Photochem Photobiol. 2008;84(3):539–549. doi: 10.1111/j.1751-1097.2007.00226.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Robles-Espinoza CD, Roberts ND, Chen S, et al. Germline MC1R status influences somatic mutation burden in melanoma. Nat Commun. 2016;7:12064. doi: 10.1038/ncomms12064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.García-Borrón JC, Abdel-Malek Z, Jiménez-Cervantes C. MC1R, the cAMP pathway, and the response to solar UV: extending the horizon beyond pigmentation. Pigment Cell Melanoma Res. 2014;27(5):699–720. doi: 10.1111/pcmr.12257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ravanat JL, Douki T, Cadet J. Direct and indirect effects of UV radiation on DNA and its components. J Photochem Photobiol B. 2001;63(1-3):88–102. doi: 10.1016/s1011-1344(01)00206-8. [DOI] [PubMed] [Google Scholar]
- 16.Rosenthal R, McGranahan N, Herrero J, et al. DeconstructSigs: delineating mutational processes in single tumors distinguishes DNA repair deficiencies and patterns of carcinoma evolution. Genome Biol. 2016;17:31. doi: 10.1186/s13059-016-0893-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leiter U, Eigentler T, Garbe C. Epidemiology of skin cancer. Adv Exp Med Biol. 2014;810:120–140. doi: 10.1007/978-1-4939-0437-2_7. [DOI] [PubMed] [Google Scholar]
- 18.Alexandrov LB, Kim J, Haradhvala NJ, et al. The repertoire of mutational signatures in human cancer. Nature. 2020;578(7793):94–101. doi: 10.1038/s41586-020-1943-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martincorena I, Raine KM, Gerstung M, et al. Universal Patterns of Selection in Cancer and Somatic Tissues. Cell. 2017;171(5):1029–1041.:e21. doi: 10.1016/j.cell.2017.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Inman GJ, Wang J, Nagano A, et al. The genomic landscape of cutaneous SCC reveals drivers and a novel azathioprine associated mutational signature. Nat Commun. 2018;9(1):3667. doi: 10.1038/s41467-018-06027-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pickering CR, Zhou JH, Lee JJ, et al. Mutational landscape of aggressive cutaneous squamous cell carcinoma. Clin Cancer Res Off J Am Assoc Cancer Res. 2014;20(24):6582–6592. doi: 10.1158/1078-0432.CCR-14-1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hayward NK, Wilmott JS, Waddell N, et al. Whole-genome landscapes of major melanoma subtypes. Nature. 2017;545(7653):175–180. doi: 10.1038/nature22071. [DOI] [PubMed] [Google Scholar]
- 23.Yizhak K, Aguet F, Kim J, et al. RNA sequence analysis reveals macroscopic somatic clonal expansion across normal tissues. Science. 2019 doi: 10.1126/science.aaw0726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fisher GJ, Kang S, Varani J, et al. Mechanisms of Photoaging and Chronological Skin Aging. Arch Dermatol. 2002;138(11):1462–1470. doi: 10.1001/archderm.138.11.1462. [DOI] [PubMed] [Google Scholar]
- 25.Gorbunova V, Seluanov A, Mao Z, Hine C. Changes in DNA repair during aging. Nucleic Acids Res. 2007;35(22):7466–7474. doi: 10.1093/nar/gkm756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lehmann J, Seebode C, Martens MC, Emmert S. Xeroderma Pigmentosum - Facts and Perspectives. Anticancer Res. 2018;38(2):1159–1164. doi: 10.21873/anticanres.12335. [DOI] [PubMed] [Google Scholar]
- 27.Panich U, Sittithumcharee G, Rathviboon N, Jirawatnotai S. Ultraviolet Radiation-Induced Skin Aging: The Role of DNA Damage and Oxidative Stress in Epidermal Stem Cell Damage Mediated Skin Aging. Stem Cells Int. 2016;2016:7370642. doi: 10.1155/2016/7370642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rozhok AI, DeGregori J. The evolution of lifespan and age-dependent cancer risk. Trends Cancer. 2016;2(10):552–560. doi: 10.1016/j.trecan.2016.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Watson CJ, Papula AL, Poon GYP, et al. The evolutionary dynamics and fitness landscape of clonal hematopoiesis. Science. 2020;367(6485):1449–1454. doi: 10.1126/science.aay9333. [DOI] [PubMed] [Google Scholar]
- 30.Collado M, Gil J, Efeyan A, et al. Tumour biology: senescence in premalignant tumours. Nature. 2005;436(7051):642. doi: 10.1038/436642a. [DOI] [PubMed] [Google Scholar]
- 31.Braig M, Lee S, Loddenkemper C, et al. Oncogene-induced senescence as an initial barrier in lymphoma development. Nature. 2005;436(7051):660–665. doi: 10.1038/nature03841. [DOI] [PubMed] [Google Scholar]
- 32.Bennett DC. Human melanocyte senescence and melanoma susceptibility genes. Oncogene. 2003;22(20):3063–3069. doi: 10.1038/sj.onc.1206446. [DOI] [PubMed] [Google Scholar]
- 33.Bennett DC, Medrano EE. Molecular regulation of melanocyte senescence. Pigment Cell Res. 2002;15(4):242–250. doi: 10.1034/j.1600-0749.2002.02036.x. [DOI] [PubMed] [Google Scholar]
- 34.Martincorena I, Fowler JC, Wabik A, et al. Somatic mutant clones colonize the human esophagus with age. Science. 2018;362(6417):911–917. doi: 10.1126/science.aau3879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suda K, Nakaoka H, Yoshihara K, et al. Clonal Expansion and Diversification of Cancer-Associated Mutations in Endometriosis and Normal Endometrium. Cell Rep. 2018;24(7):1777–1789. doi: 10.1016/j.celrep.2018.07.037. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data has been deposited at the European Genome–phenome Archive (EGA) under the accession code EGAS00001004279. The somatic mutations found in all samples are listed in the Supplementary Dataset S1. Clinical data of each donor can be found in the Supplementary Dataset S2. Any other relevant data can be obtained from the corresponding authors upon reasonable request.