Abstract
Background
P-element–induced wimpy testis (PIWI)–interacting RNAs (piRNAs) are short (21 to 35 nucleotides in length) and noncoding and are found almost exclusively in germ cells, where they regulate aberrant expression of transposable elements and postmeiotic gene expression. Critical to the processing of piRNAs is the protein poly(A)-specific RNase-like domain containing 1 (PNLDC1), which trims their 3’ ends and, when disrupted in mice, causes azoospermia and male infertility.
Methods
We performed exome sequencing on DNA samples from 924 men who had received a diagnosis of nonobstructive azoospermia. Testicular-biopsy samples were analyzed by means of histologic and immunohistochemical tests, in situ hybridization, reverse-transcriptase–quantitative-polymerase-chain-reaction assay, and small-RNA sequencing.
Results
Four unrelated men of Middle Eastern descent who had nonobstructive azoospermia were found to carry mutations in PNLDC1: the first patient had a biallelic stop–gain mutation, p.R452Ter (rs200629089; minor allele frequency, 0.00004); the second, a novel biallelic missense variant, p.P84S; the third, two compound heterozygous mutations consisting of p.M259T (rs141903829; minor allele frequency, 0.0007) and p.L35PfsTer3 (rs754159168; minor allele frequency, 0.00004); and the fourth, a novel biallelic canonical splice acceptor site variant, c.607-2A→T. Testicular histologic findings consistently showed error-prone meiosis and spermatogenic arrest with round spermatids of type Sa as the most advanced population of germ cells. Gene and protein expression of PNLDC1, as well as the piRNA-processing proteins PIWIL1, PIWIL4, MYBL1, and TDRKH, were greatly diminished in cells of the testes. Furthermore, the length distribution of piRNAs and the number of pachytene piRNAs was significantly altered in men carrying PNLDC1 mutations.
Conclusions
Our results suggest a direct mechanistic effect of faulty piRNA processing on meiosis and spermatogenesis in men, ultimately leading to male infertility.
Introduction
Worldwide, approximately one third of couples have fertility problems,1 with 50% arising from a male factor. Men with nonobstructive azoospermia have intact seminal ducts but impaired spermatogenesis, which results in no spermatozoa in the ejaculate. These men have very low chances of fathering a biologic child even after surgical testicular sperm extraction procedures. Known genetic causes of nonobstructive azoospermia include aberrations of sex chromosomes and an increasing number of rare deleterious variants in genes critical for spermatogenesis.2 P-element–induced wimpy testis (PIWI)–interacting RNAs (piRNAs) are a subgroup of small noncoding RNAs that are found abundantly in germ cells and are essential for spermatogenesis.3 The function of pachytene piRNAs, which make up approximately 95% of the piRNAs in the adult testis, is not well understood. It has been proposed that piRNAs promote the translation of repressed messenger RNAs (mRNAs) during differentiation of early postmeiotic spermatids4 and subsequently eliminate mRNAs in the later spermatid stages.5 The primary biogenesis of piRNAs is thought to take place in the intermitochondrial cement and chromatoid bodies, and the piRNA-processing protein TDRKH is thought to coordinate multiple steps in the process.6 Pachytene piRNAs are derived mainly from approximately 100 intergenic loci under the control of transcription factor MYBL17 and are produced as long single-stranded transcripts, which are then processed into pre-piRNAs (with lengths of approximately 30 to 40 bases)8 in a complex involving the protein PIWIL1.3,7 The 3’ ends of pre-piRNAs are subsequently trimmed by the protein poly(A)-specific RNase-like domain containing 1 (PNLDC1) in an exonucleolytic manner, resulting in mature piRNAs (with lengths of 21 to 35 bases). PNLDC1 is exclusively expressed in spermatocytes,9 and mice deficient in Pnldc1 have reduced testis size and are infertile.10-12 Spermatogenic arrest at meiotic and postmeiotic stages is observed in Pnldc1mt/mt mice, together with diminished expression of PIWIL1; these occurrences ultimately lead to longer (24 to 50 bases in length) and fewer piRNAs.10 We have identified four men carrying a defective PNLDC1 gene. Here we describe their clinical phenotype, the histologic features of their testicular tissue, and the expression pattern of piRNAs and key piRNA machinery proteins.
Methods
Study Design and Oversight
Ethical approval for the Genetics of Male Infertility Initiative (GEMINI) was granted in Denmark by the regional ethics committee, in Portugal by the ethics committee at Instituto Nacional de Saúde Dr. Ricardo Jorge, in the United States by the institutional review board at Weill Cornell Medical College, and in the Netherlands by Commissie Mensgebonden Onderzoek regio Arnhem–Nijmegen. The trial was designed by 10 of the authors, and the experimental procedures were performed by 11. The data were collected by all the authors. The manuscript was written by the first, second, and last authors, and all the authors participated in the review and correction of the manuscript and in the approval of the final version to be submitted for publication. The authors vouch for the accuracy and completeness of the data.
Patient Selection
The semen of the male partner of each infertile couple was analyzed, and his reproductive history was recorded. In the men with azoospermia, a complete history taking, physical examination, and hormonal evaluation were performed. Men with no clinical evidence of obstruction were characterized as having nonobstructive azoospermia. Standard genetic testing, including karyotype and Y chromosome microdeletion analysis, was performed in accordance with the guidelines of the American Urological Association and American Society for Reproductive Medicine (AUA–ASRM)13 and the guidelines of the European Association of Urology (EAU).14
Exome Sequencing
Three men with PNLDC1 mutations were identified in GEMINI (see the Supplementary Appendix). A total of 924 unrelated patients with nonobstructive azoospermia underwent exome sequencing at the McDonnell Genome Institute at Washington University in St. Louis with the use of an in-house exome targeting reagent at an average coverage of 80×. One additional man with a biallelic PNLDC1 mutation was identified in an independent cohort of trios from the Netherlands. The cohort included 99 subfertile or infertile men, of whom 55 had nonobstructive azoospermia (see the Supplementary Appendix). Each trio was composed of an affected man and his father and mother. Exome sequencing was performed with the Nextera DNA Exome Capture kit (Illumina) at an average coverage of 72× on the NovaSeq 6000 platform. Alignment of sequencing reads was followed by joint genotype calling of all samples within each cohort in accordance with the best practices of the Genome Analysis Toolkit.15
Identification of Pathogenic Variants
To prioritize the likely deleterious mutations among the sequenced DNA samples from the patients with nonobstructive azoospermia in GEMINI, a modified version of the population sampling probability (PSAP) software was applied. PSAP is a technique that evaluates the probability of sampling a genotype or a set of genotypes on the basis of pathogenicity scores and frequencies of variants observed in the unaffected population.16 Genotypes with a PSAP P value of less than 0.001 and minor allele frequency of less than 0.01 were selected through subsequent filtering across all populations in the Genome Aggregation Database (gnomAD), version 2.1.1 (https://gnomad.broadinstitute.org), whereas variants that occurred at a high frequency in the study cohort were excluded. Only genes with loss-of-function mutations or testis-enhanced expression (ProteinAtlas17) or genes known to cause infertility in humans18 or mice (http://www.informatics.jax.org) were considered. The probands of the Dutch cohort underwent screening for recessive compound heterozygous or homozygous pathogenic variants, which were present heterozygously in both parents, had a minor allele frequency of less than 0.01 in the gnomAD, had no more than one homozygote present in a control cohort (see the Supplementary Appendix), and were classified as being of uncertain significance, likely pathogenic, or pathogenic according to established guidelines.19 All genomic coordinates were based on the human genome assembly GRCh37 (hg19).
Bioinformatic Annotation
The ethnic origin of the patients in the GEMINI cohort was determined with the use of EthSeq software20 applied to variants with a minor allele frequency of greater than 0.2. Long runs of homozygosity (ROH) were detected independently for each ethnic origin with the use of the H3M2 tool.21 The longest autozygous stretches (class 5), which reflect recent inbreeding,22 were determined with the use of the mclust, version 5.4.3 (an R package for model-based clustering, classification, and density estimation; R Project for Statistical Computing), in a population-specific manner. Only the class 5 ROH regions (minimal length of 6.7 Mb on average across populations) were used to calculate the fraction of the autosome that was homozygous. In the Dutch patient, homozygosity calling was performed with the use of RareVariantVis (a tool used to analyze genome sequence data for rare variants),23 and genomic regions longer than 1 Mb with more than 85% homozygosity were classified as ROH. We used the R package masonmd24 to evaluate whether a protein-truncating mutation is predicted to trigger nonsense-mediated decay of the abnormal transcripts. We mapped identified mutations to structural features of PNLDC1 using VarMap.25 Protein sequence alignment was performed with Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/). Splice prediction was based on the SpliceSiteFinder-like, MaxEntScan, NNSplice, and GeneSplicer algorithms in Alamut Visual (version 2.13) (http://www.interactive-biosoftware.com).
Sanger Sequencing
The regions of interest were amplified with the use of primers and conditions as listed in Table S1 in the Supplementary Appendix. The polymerase-chain-reaction (PCR) products were then purified and sequenced.
Immunohistochemical Staining and In Situ Hybridization
Fixed testicular tissue samples were obtained from Patients 1, 2, and 4. Immunohistochemical staining was performed, as described previously,26 in all the samples from these patients at Rigshospitalet with the use of the conditions and antibodies that are listed in Table S2. In addition, biopsy samples in matching fixative from patients with complete spermatogenesis were used as positive controls. Negative controls were serial sections that were exposed to all treatments except the primary antibody. The mitotic index was obtained by inspecting the entire hematoxylin and eosin–stained sections in the samples from Patients 1 and 2 and counting the number of spermatogonia that were observed to be engaged in a mitotic division. The references of the mitotic index in control tissue samples were obtained from Almstrup et al.,27 and the differences were evaluated with the use of the Mann–Whitney test. In situ hybridization was performed on tissue samples from Patient 4 with the use of a custom-made PNLDC1 probe and the RNAscope 2.5 HD Assay-RED kit (Advanced Cell Diagnostics) according to the recommendations of the manufacturer.
RNA Isolation, RT-qPCR, and Small-RNA Sequencing
RNA was extracted (see the Methods section in the Supplementary Appendix and Table S3), and a reverse-transcriptase–quantitative-PCR (RT-qPCR) assay was performed with the use of the gene-specific primers that are listed in Table S1. Small RNAs were sequenced, and reads mapping to piRNAs were intersected with 205 piRNA genomic regions that had been detected previously in the adult human testis.28 The amount of detected piRNAs was inferred on the basis of the number of reads mapping to individual piRNA loci normalized to spike-in read count, and the differences between the respective patients and controls were assessed with the use of the Mann–Whitney test. To detect the percentage of piRNA reads arising from pachytene piRNA loci, piRNA reads were intersected with the genomic regions reported in Özata et al.3 Differences in the amount of pachytene piRNAs between the patients and matched controls were tested with the use of Fisher’s exact test.
Results
Characterization of Pathogenic Variants in PNLDC1
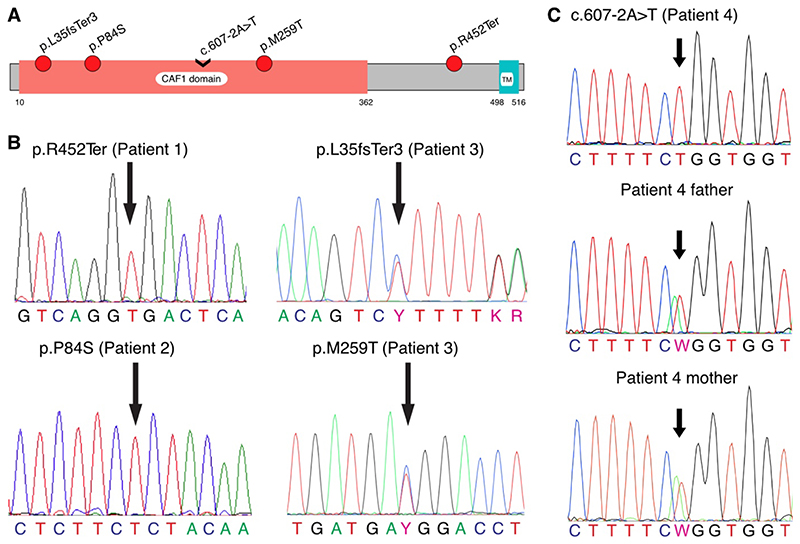
We performed exome sequencing on DNA samples from 924 patients with nonobstructive azoospermia in the GEMINI cohort and identified 3 patients from three centers who carried potentially pathogenic variants in PNLDC1. A fourth patient was identified in an independent Dutch cohort. Patient 1, a man from Denmark, carried a homozygous stop–gain mutation, p.R452Ter (chr6[GRCh37]:g.160240107C→T; NM_173516.2[PNLDC1]:c.1354C→T; rs200629089), which is rare (minor allele frequency, 0.00004) and represents the first observation of a homozygous carrier in human populations (according to gnomAD). This mutation is predicted to be highly pathogenic (Phred-scaled score [https://cadd.gs.washington.edu/info] of 38.0 on the Combined Annotation–Dependent Depletion [CADD] framework, version 1.3, which means that the variant belongs to the top 0.1% of the most pathogenic variants observed in the human genome) and to cause degradation of the abnormal transcripts through a nonsense-mediated decay mechanism, leading to a complete human knockout of PNLDC1. Patient 2, a man from the United States, carried a novel homozygous variant, p.P84S (chr6[GRCh37]:g.160225031C→T; NM_173516.2[PNLDC1]:c.250C→T; CADD score, 29.8), which has not been reported in gnomAD. This variant maps to a CAF1 domain common among mRNA deadenylases (Fig. 1A). Both mutations disrupt a site that is conserved across mammals (Fig. S1A and S1B). Patient 3, a man from Portugal, carried two compound heterozygous mutations mapping to the CAF1 domain. A likely pathogenic frameshift insertion, p.L35PfsTer3 (chr6[GRCh37]:g.160222146dup; NM_173516.2[PNLDC1]:c.103dup; rs754159168; minor allele frequency, 0.000036; CADD score, 34), may lead to loss of expression from one allele and affects a site highly conserved across mammals; on the other allele, a mildly pathogenic missense variation, p.M259T (chr6[GRCh37]:g.160231083T→C; NM_173516.2[PNLDC1]:c.776T→C; rs141903829; minor allele frequency, 0.00071; CADD score, 15.2), disrupts a partially conserved position (Fig. S1C). In addition, p.M259T is predicted to interact with the ligand guanosine diphosphateaccording to the VarMap annotation resources. All mutations were confirmed by Sanger sequencing (Fig. 1B). The ClinVar accession numbers for the mutations are SCV001760918 (p.R452Ter), SCV001760919 (p.P84S), and SCV001760920 (p.L35PfsTer3 and p.M259T). A fourth patient with nonobstructive azoospermia was identified in the Netherlands. Patient 4 carried a novel homozygous change (chr6[GRCh37]:g.160230045A→T; NM_173516.2[PNLDC1]:c.607-2A→T; CADD score, 6.8) in the canonical splice acceptor site of exon 9 (ClinVar accession number, SCV001760921) (Fig. S2). The variant is predicted to disrupt the splicing consensus sequence recognized by the splicing machinery. Both parents of the patient were heterozygous for the variant (Fig. 1C). None of the five PNLDC1 mutations were found in a Dutch control cohort of 5784 fertile men, except for p.M259T (in 13 heterozygous men), which indicates that these variants are rare. The homozygous state of the rare pathogenic PNLDC1 mutations p.R452Ter and p.P84S potentially indicates an increased rate of autozygosity in the carriers. In support of this possibility, both changes were found within long stretches of homozygosity (34.4 Mb in Patient 1 and 8.8 Mb in Patient 2). A cumulative burden of the long ROH regions per genome confirmed that both Patients 1 and 2 are descendants of third and fourth degree of consanguinity, with 4.9% and 2.9%, respectively, of autosomes being homozygous.29
Figure 1. Validation of the Pathogenic PNLDC1 Mutations by Sanger Sequencing.
Panel A shows a schematic diagram of PNLDC1 protein domains, depicted on the basis of the Pfam database (version 32.0), with the locations of the identified mutations. The CAF1 family RNase domain (PFAM:PF04857), common among messenger RNA deadenylases, is thought to contain the RNase activity of PNLDC1. TM denotes potential transmembrane domain. Panel B shows the identified mutations that were validated by means of Sanger sequencing. Homozygous genotypes were confirmed in the case of p.R452Ter (c.1354C→T) in Patient 1 and in the case of p.P84S (c.250C→T) in Patient 2; the compound variants p.L35fsTer3 (c.103dup) and p.M259T (c.776T→C) in Patient 3 were confirmed as being heterozygous. As shown in Panel C, Patient 4 was validated as a homozygous carrier of c.607-2A→T, whereas both of his parents were observed as being heterozygous for the change. In all the panels, the locations of the mutations are indicated by arrows.
Clinical Presentation
Patients 1, 2, and 4 descended from populations in the Greater Middle East and, although the ethnic origin of Patient 3 was unknown, his name is consistent with descent from a Middle Eastern population (Table 1). All patients were confirmed to have nonobstructive azoospermia on the basis of physical examination and histologic findings, according to the AUA–ASRM13 and EAU14 guidelines. The men had no anamnestic risk factors for obstruction, and they had a normal male phenotype (i.e., no gynecomastia, normal adult pubic hair distribution, no hypospadias, and testicles in the normal low-scrotal position). However, the sizes of the testes were slightly reduced (Table 1), and each patient had nondilated epididymides and palpable vasa deferentia on both sides, as expected in cases of nonobstructive azoospermia. All four men had a normal 46,XY karyotype, no Y chromosome microdeletions, and azoospermia with no sperm detected in repeated evaluations of the ejaculate after high-speed centrifugation. Reproductive hormone levels were within the normal range, except for low total and free testosterone levels in Patient 1 (Table 1). Fertility treatment was unsuccessful in all patients, and morphologically abnormal spermatozoa were observed only in the testicular sperm extraction sample from Patient 3 (Table 1). At consultation, all the patients appeared to be otherwise healthy, but Patient 1 had previously received a diagnosis of a mild psychotic phenotype (schizophrenia), which has been described before in a patient with heterozygous translocations involving PNLDC1.30 The coexisting psychotic condition is of unknown significance in the context of nonobstructive azoospermia.
Table 1. Clinical Presentation of Four Patients with Mutations in PNLDC1.*.
| Variable | Patient 1† | Patient 2 | Patient 3 | Patient 4 |
|---|---|---|---|---|
| Genetics | ||||
| PNLDC1 mutation | Homozygous nonsense | Homozygous missense | Compound heterozygous frameshift and missense | Homozygous splice acceptor variant |
| Karyotype | 46,XY | 46,XY | 46,XY | 46,XY |
| Y chromosome microdeletions | No | No | No | No |
| Patient history | ||||
| Age — yr | 32 | 42 | 39 | 36 |
| Ethnic origin‡ | Pakistani | Qatari | Unknown | Emirati |
| Congenital maldescended testicles | No | No | Unknown | No |
| Pubertal development | Normal | Normal | Unknown | Normal |
| Psychiatric diagnosis | Schizophrenia | Normal | Unknown | Normal |
| Phenotype | ||||
| Body-mass index§ | 30 | 26 | Unknown | 20 |
| Gynecomastia | No | No | Unknown | No |
| Pubic hair growth — Tanner stage¶ | 6 | 6 | Unknown | Unknown |
| Hypospadias | No | No | No | No |
| Left/right testis position in scrotum | Low/low | Low/low | Low/low | Low/low |
| Left/right testis size — ml | 15/12‖ | 10.9/13.0** | Unknown | 10/10‖ |
| Epididymis | Bilateral present; not enlarged | Bilateral present; not enlarged | Bilateral present; not enlarged | Bilateral present; not enlarged |
| Results of transrectal ultrasonography | Prostate normal; ejaculatory ducts normal | Unknown | Prostate normal; ejaculatory ducts normal | — |
| Semen characteristics | ||||
| Ejaculation abstinence — days | 3 | 3 | Unknown | 2 |
| Semen volume — ml | 1.3–1.5 | 3 | Unknown | 2.3 |
| Sperm concentration | Azoospermia | Azoospermia | Azoospermia | Azoospermia |
| Spermatozoa in ejaculate pellet | None | None | None | None |
| Semen pH | 8.5 | Alkaline | Unknown | 8.0 |
| Hormone levels | ||||
| Serum total testosterone — nmol/liter | 6–12 | 12 | 16 | 18 |
| Serum free testosterone — nmol/liter | 0.14–0.31 | 0.32 | Unknown | Unknown |
| Serum SHBG — nmol/liter | 21–23 | 19 | Unknown | Unknown |
| Serum FSH — IU/liter | 4.7–4.9 | 4.4 | 7.4 | 4.8 |
| Serum LH — IU/liter | 3.4–4.3 | 7.0 | 6.0 | Unknown |
| Serum inhibin B — pg/ml | 151–157 | 55 | Unknown | Unknown |
| Serum estradiol — pmol/liter | 39–49 | 73 | 115 | Unknown |
| Testicular-biopsy sample | ||||
| Fixative | Modified Stieve’s fixative | Bouin’s fixative | Unknown | Formalin |
| Leydig cell hyperplasia | No | No | Unknown | No |
| Spermatogenesis | Bilateral homogenous pattern with spermatogenic arrest at the late pachytene stage | Homogenous pattern with spermatogenic arrest at the late pachytene stage | No biopsy available (only tissue remnants after TESE were available) | Homogenous pattern with spermatogenic arrest at the late pachytene stage |
| RNA recovered | Yes | No | Yes, from remnants after TESE | Yes |
| Treatments | ||||
| TESE or microTESE | Unknown | No sperm identified after extensive TESE | TESE revealed morphologically abnormal spermatozoa | No sperm identified after extensive TESE |
| Treatment outcome | Unknown | No pregnancy | No pregnancy after ICSI with abnormal spermatozoa obtained from TESE | No pregnancy |
Patient 1 was from Denmark, Patient 2 from the United States, Patient 3 from Portugal, and Patient 4 from the Netherlands. To convert the values for estradiol to picograms per milliliter, divide by 3.671. To convert the values for testosterone to nanograms per milliliter, divide by 3.467. FSH denotes follicle-stimulating hormone, ICSI intracytoplasmic sperm injection, LH luteinizing hormone, SHBG sex hormone–binding globulin, and TESE testicular sperm extraction.
The reproductive hormones in Patient 1 were measured in several samples, and the range is indicated.
Information on ethnic origin was obtained from the medical records of the patients. The ethnic origin of Patient 3 was unknown, although he had an Arabic name.
The body-mass index is the weight in kilograms divided by the square of the height in meters.
Tanner stages of pubic hair range from 1 to 6, with higher stages indicating more advanced pubertal development.
Testis size was determined by palpation.
Testis size was determined by ultrasonograpy.
Testicular Pathological Characteristics
Testicular-biopsy samples from Patients 1, 2, and 4 showed that all seminiferous tubules had a very similar and homogeneous pattern of spermatogenic arrest at the late pachytene stage, although a few cells made it to diakinesis and metaphase I and through metaphase II to the spermatid stage (Fig. 2). Many pyknotic cells were observed in the lumen (Fig. 2C, 2G, and 2I). In some tubules, a few round spermatids of type Sa were seen (Fig. 2C and 2J), as well as occasional Sb spermatids, which appeared abnormally formed and pyknotic. We also observed a slight increase in the number of spermatogonia that appeared abnormal, but with no signs of malignant change, together with an increased number of mitotic divisions (Fig. 2D through 2F). In accordance with these observations, the mean mitotic index of spermatogonia tended to be slightly increased (Fig. S3) at 0.98%, as compared with the mitotic index of 0.25% (P = 0.06) that was reported previously in men with complete spermatogenesis.27 MAGE-A4 and TNP1 staining confirmed that the number of spermatogonia was similar to that in tissue with complete spermatogenesis, whereas the number of round spermatids was greatly reduced (Fig. S4).
Figure 2. Testicular Histologic Findings in Men with PNLDC1 Mutations.
Panel A shows a hematoxylin and eosin stain of a control testicular tissue sample in which seminiferous tubules had complete spermatogenesis. Asterisks indicate the seminiferous tubules with complete spermatogenesis. Panel B shows a hematoxylin and eosin stain of a testicular tissue sample from Patient 2. Hashtags indicate extensive arrest at the late pachytene stage in the tubules. Panels C and D show the sample from Patient 2 at higher magnifications. In Panel C, the arrow indicates round spermatids of stage Sa and the arrowheads pyknotic postmeiotic cells. In Panel D, the arrowhead indicates a spermatogonium that appears abnormal, and the arrow, arrest at diakinesis. The bars in Panels A through D represent 20 μm. Panels E through J also show the sample from Patient 2 at higher magnifications. The arrow in Panel E indicates an abnormal spermatogonium with enlarged nucleus but no signs of malignant change; in Panel F, spermatogonium in mitosis; in panel G, a meiotic spermatocyte that appears pyknotic; and in Panel H, a spermatocyte at diakinesis, the last stage of the meiotic prophase 1. The dashed outline in Panel I defines an area of pyknotic multinucleated cells; and in Panel J, an area of round spermatids of type Sa. The bars in Panels E through J represent 10 μm.
Expression of PNLDC1
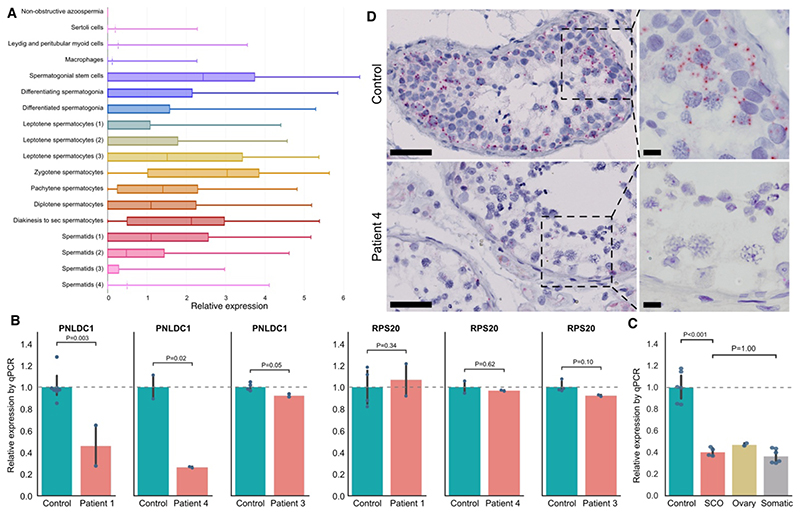
Interrogation of human single-cell RNA sequencing data31 indicated that PNLDC1 is absent in somatic testicular cells and is specifically transcribed in germ cells, with the highest levels occurring in spermatogonial stem cells and spermatocytes at the leptotene and zygotene stages (Fig. 3A). RT-qPCR analysis of PNLDC1 expression in testicular tissue showed significantly reduced transcript levels in Patients 1 and 4, as compared with matched controls (P = 0.003 and P = 0.02, respectively) (Fig. 3C), a finding that indicates the occurrence of nonsense-mediated decay. In Patient 3, who had compound heterozygous variants, a less pronounced decrease in PNLDC1 expression was observed. RNA was not available for Patient 2. No differences in expression of the housekeeping gene RPS20 were observed between the patients and controls, which indicates that RNA quality was not compromised.
Figure 3. Expression of PNLDC1.
Panel A shows the testicular cell types that express PNLDC1, as determined by interrogation of single-cell RNA sequencing on adult testis.31 In the horizontal box plots, the vertical solid lines are the medians and the left and right sides of the boxes are the interquartile ranges of the normalized read counts for each cell type. The I bars indicate the minimum and maximum counts, and when the third quartile is zero, the mean is indicated with a dotted line. Each cell type identified by the analysis31 is marked with a different color, and the number in brackets indicates sequential developmental stages of a given cell type. Panel B shows an in situ hybridization analysis performed with probes targeting PNLDC1 in a testicular-biopsy sample from a control with complete spermatogenesis (top) and a testicular-biopsy sample from Patient 4 (bottom). PNLDC1 shows high expression in pachytene spermatocytes (each red dot represents a single transcript) in the control but is nearly absent in Patient 4. The bars on the left images represent 50 μm, and on the right images 10 μm. Panel C shows a reverse-transcriptase–quantitative-polymerase-chain-reaction (RT-qPCR) analysis performed with primers targeting PNLDC1 on RNA isolated from fixed testis samples with complete spermatogenesis from controls (Table S3 in the Supplementary Appendix) and testicular RNA isolated from Patients 1, 3, and 4 (RNA was not available from Patient 2). The analysis was performed in duplicate. An RT-qPCR analysis of the housekeeping gene RPS20 showed nonsignificant differences in expression between the patients and controls, a finding that indicates that the RNA quality was not compromised. Panel D shows an RT-qPCR analysis performed with primers targeting PNLDC1 on three samples from controls with complete spermatogenesis, two samples of tissue devoid of germ cells (Sertoli cell–only pattern [SCO]), an ovary tissue sample, and different somatic tissue samples (liver, skin, and ductus deferens); the findings indicate that PNLDC1 expression is specific to germ cells. In Panels C and D, the data are presented as relative to the controls, as indicated by the dashed line. Each blue dot represents a measurement (performed in duplicate per sample), and the vertical bars indicate the 95% confidence intervals, which were calculated by means of bootstrapping. NS denotes not significant.
Immunohistochemical analysis of tissue samples with complete spermatogenesis showed major expression of PNLDC1 in pachytene spermatocytes, as expected (Fig. S5).9 However, an unexpected staining of interstitial cells (peritubular myoid cells, Leydig cells, and blood vessels) was observed even after extensive evaluation of experimental conditions and testing of two other commercial antibodies (Table S2). Negative controls showed no staining (Figs. S6 and S7). The intensity of PNLDC1 staining was, nevertheless, clearly reduced in pachytene spermatocytes of Patients 1 and 2 (Fig. S5), and we believe that the signals in interstitial cells are artifacts. To confirm that PNLDC1 transcription is specific to germ cells, we performed RT-qPCR assays on testicular tissue samples with and without germ cells. PNLDC1 transcripts were observed at high levels in tissue with complete spermatogenesis, but the levels were significantly reduced in tissue devoid of germ cells (Sertoli cell–only pattern) (P<0.001) and were similar to those in ovary tissue and somatic tissue with no piRNA production (Fig. 3D). Furthermore, in situ hybridization of the control testicular tissue confirmed PNLDC1 expression in the germ cells (primarily spermatogonia and spermatocytes) and showed loss of the expression in Patient 4, findings that are consistent with those from the RT-qPCR assays (Fig. 3B and Fig. S8).
Expression of Key piRNA-Processing Components
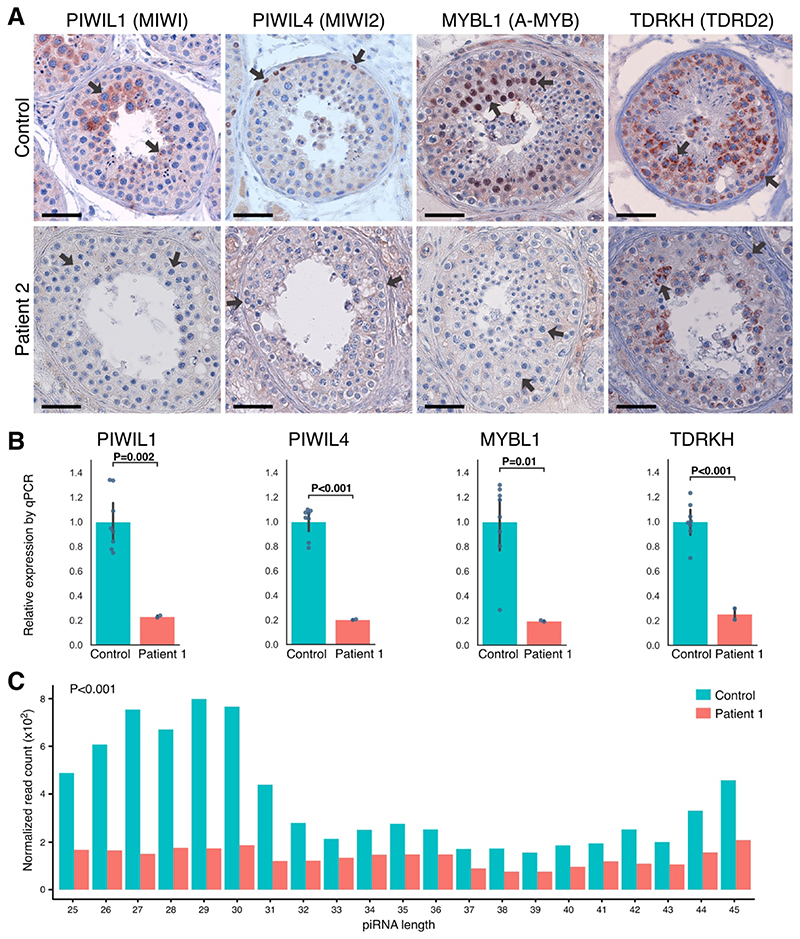
We investigated the effect of PNLDC1 mutations on the expression of several key piRNA-processing components. Although not all proteins were stained in all cases, the combined results indicated a greatly diminished expression of PIWIL1 in pachytene spermatocytes, PIWIL4 in spermatogonia, and transcription factor MYBL in spermatocytes (Fig. 4A and Fig. S9). TDRKH, which is found from the spermatogonia stage to late-stage pachytene spermatocytes, was detected in fewer cells and at lower levels. The observed changes in protein expression were in general mirrored at the transcript level. RT-qPCR assays of RNA isolated from testicular-biopsy samples from Patients 1 and 4 revealed greatly reduced levels of PIWIL1, PIWIL4, MYBL1, and TDRKH transcripts, whereas a less pronounced decrease was observed for Patient 3 (Fig. 4B and Fig. S10A and S10B).
Figure 4. Expression of Key piRNA-Processing Enzymes at Protein and Transcript Levels and Small-RNA Sequencing.
Panel A shows immunohistochemical staining for P-element–induced wimpy testis–interacting RNA (piRNA)–processing proteins PIWIL1, PIWIL4, MYBL1, and TDRKH in a control biopsy sample with complete spermatogenesis and in a sample from Patient 2, who carried the biallelic missense mutation p.P84S in PNLDC1. Arrows indicate the main sites of expression. In the control, we observed expression of PIWIL1 in pachytene spermatocytes, PIWIL4 in spermatogonia, and MYBL1 in spermatocytes, which in all cases were lost or greatly diminished in the patient. TDRKH was observed in the cytoplasm of spermatogonia and spermatocytes and appeared to be less expressed in Patient 2. The expression patterns were confirmed across five controls and also in Patient 1 for PIWIL1 and PIWIL4 and in Patient 4 for PIWIL1, MYBL1, and TDRKH (Fig. S9). A testicular tissue sample from Patient 3 was unavailable for staining. The bars represent 50 μm. Panel B shows that with the use of RNA isolated from a fixed testicular tissue sample from Patient 1, the transcript levels of piRNA- processing enzymes recapitulated the pattern that was observed at the protein level. A significant reduction in transcript levels was observed for PIWIL1, PIWIL4, MYBL1, and TDRKH. The RNA from four controls with complete spermatogenesis was also obtained from fixed testicular tissue and had a similar RNA quality as in Patient 1. The analysis was performed in duplicate. A similar pattern was observed in a sample from Patient 4, but a less pronounced pattern was observed in a sample from Patient 3 (Fig. S10A and S10B). Expression of the housekeeping transcript RPS20 was similar between the patients and their matched controls (Fig. 3C). As shown in Panel C, small-RNA sequencing of RNA isolated from fixed testicular tissue sample from Patient 1 revealed a significantly lower amount of piRNAs than that in the sample from a control with complete spermatogenesis (P<0.001), as well as a major loss of piRNAs with expected lengths of 26 to 31 bases, which were evident in the control samples. Read counts of piRNAs were normalized to the spike-in RNA. Control RNA was isolated from tissue preparations that were similar to those used for the patients. Data from Patient 4 that show a similar significant reduction and from Patient 3 that show a milder effect on piRNA biogenesis are provided in Fig. S10C and S10D.
Effects on piRNA Processing
We performed small-RNA sequencing on testicular RNA from Patients 1, 3, and 4. The results indicated a compromised biogenesis of piRNAs with significantly decreased amounts of piRNAs in all three patients, as compared with RNA in similarly treated tissue obtained from controls with complete spermatogenesis (Fig. 4C and Fig. S10C and S10D). A peak of expected piRNA lengths of 26 to 31 bases was observed in the controls, but this peak was absent in Patients 1 and 4, who had a significant loss of piRNAs (P<0.001) as well as a different piRNA length distribution. With respect to the compound heterozygous variants in Patient 3, a similar but milder effect on piRNA production was observed (P = 0.04) (Fig. S10D). In addition to impaired piRNA biogenesis, a significantly (P<0.001) reduced contribution of piRNAs originating from pachytene piRNA loci was detected in all sequenced RNA samples from the three patients as compared with their respective controls (Fig. S11).
Discussion
In our study, loss of the piRNA trimmer PNLDC1 was associated with severe impairment of spermatogenesis and nonobstructive azoospermia in men. Our results largely support the observations in mice with targeted ablation of Pnldc1.10 In addition, concomitant loss of PIWIL1 was observed in both men and mice with dysfunctional PNLDC1. We conclude that the three biallelic loss-of-function mutations described here led to a collapse of the piRNA machinery and that the heterozygous frameshift variant in combination with a milder missense variant (in Patient 3) caused a milder phenotype. In fetal germ cells, piRNAs are important for silencing transposable elements, and defective PNLDC1 could impair adult spermatogenesis because of testicular dysgenesis.32 However, we observed similar numbers of spermatogonia in the study patients and unaffected men. We therefore speculate that PNLDC1 may be more relevant for processing of piRNAs at the pachytene stage of adult spermatogenesis than during fetal development of the gonads. In accordance with this hypothesis, we observed loss of MYBL1, which is thought to be specifically involved in controlling the expression of more than half of piRNAs during pachytene,3,7,33 and diminished testicular transcription of MYBL1 and PIWIL1.3 Other studies have linked piRNA-processing enzymes to infertility in men. Deletions in the PIWIL4-interacting TDRD9 have been reported in men with cryptozoospermia originating from a consanguineous Bedouin family.34 Similarly, two patients from unrelated consanguineous families who had nonobstructive azoospermia were found to carry loss-of-function mutations in TDRD7.35 Furthermore, evidence from studies in mice suggests that mutations in other piRNA enzymes are likely causes of severe spermatogenic failure in men, because disruption of multiple piRNA-related proteins leads to meiotic arrest and infertility in male mice.36 Collectively, our findings indicate that mutations in PNLDC1 and other players in piRNA biogenesis are most likely an underlying cause of spermatogenic failure in a subgroup of men with azoospermia.
Supplementary Material
Support statement
Supported by a grant from Innovation Fund Denmark (14-2013-4, to Dr. Almstrup), ReproUnion, grants from the National Institutes of Health (R01HD078641, to Drs. Aston and Conrad, and P50HD096723, to Dr. Conrad), a starting grant from Fundação para a Ciência e a Tecnologia (IF/01262/2014, to Dr. Lopes), a grant from the Netherlands Organization for Scientific Research (918-15-667, to Dr. Veltman), and a grant from the Wellcome Trust (209451, to Dr. Veltman)
Contributor Information
Participating centres of the GEMINI consortium:
Donald F. Conrad, Liina Nagirnaja, Kenneth I. Aston, Douglas T. Carrell, James M. Hotaling, Timothy G. Jenkins, Moira K. O’Bryan, Rob McLachlan, Peter N. Schlegel, Michael L. Eisenberg, Jay I. Sandlow, Emily S. Jungheim, Kenan R. Omurtag, Alexandra M. Lopes, Susana Seixas, Filipa Carvalho, Susana Fernandes, Alberto Barros, João Gonçalves, Maris Laan, Margus Punab, Ewa Rajpert-De Meyts, Niels Jørgensen, Kristian Almstrup, Csilla G. Krausz, and Keith A. Jarvi
References
- 1.Barratt CLR, Björndahl L, De Jonge CJ, et al. The diagnosis of male infertility: an analysis of the evidence to support the development of global WHO guidance-challenges and future research opportunities. Hum Reprod Update. 2017;23:660–80. doi: 10.1093/humupd/dmx021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kasak L, Laan M. Monogenic causes of non-obstructive azoospermia: challenges, established knowledge, limitations and perspectives. Hum Genet. 2021;140:135–54. doi: 10.1007/s00439-020-02112-y. [DOI] [PubMed] [Google Scholar]
- 3.Özata DM, Yu T, Mou H, et al. Evolutionarily conserved pachytene piRNA loci are highly divergent among modern humans. Nat Ecol Evol. 2020;4:156–68. doi: 10.1038/s41559-019-1065-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dai P, Wang X, Gou L-T, et al. A translation-activating function of MIWI/piRNA during mouse spermiogenesis. Cell. 2019;179(7):1566–1581.:e16. doi: 10.1016/j.cell.2019.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gou L-T, Dai P, Yang J-H, et al. Pachytene piRNAs instruct massive mRNA elimination during late spermiogenesis. Cell Res. 2014;24:680–700. doi: 10.1038/cr.2014.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ding D, Liu J, Dong K, Melnick AF, Latham KE, Chen C. Mitochondrial membrane-based initial separation of MIWI and MILI functions during pachytene piRNA biogenesis. Nucleic Acids Res. 2019;47:2594–608. doi: 10.1093/nar/gky1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li XZ, Roy CK, Dong X, et al. An ancient transcription factor initiates the burst of piRNA production during early meiosis in mouse testes. Mol Cell. 2013;50:67–81. doi: 10.1016/j.molcel.2013.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saxe JP, Chen M, Zhao H, Lin H. Tdrkh is essential for spermatogenesis and participates in primary piRNA biogenesis in the germline. EMBO J. 2013;32:1869–85. doi: 10.1038/emboj.2013.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anastasakis D, Skeparnias I, Shaukat A-N, et al. Mammalian PNLDC1 is a novel poly(A) specific exonuclease with discrete expression during early development. Nucleic Acids Res. 2016;44:8908–20. doi: 10.1093/nar/gkw709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nishimura T, Nagamori I, Nakatani T, et al. PNLDC1, mouse pre-piRNA Trimmer, is required for meiotic and post-meiotic male germ cell development. EMBO Rep. 2018;19(3):e44957. doi: 10.15252/embr.201744957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ding D, Liu J, Dong K, et al. PNLDC1 is essential for piRNA 3’ end trimming and transposon silencing during spermatogenesis in mice. Nat Commun. 2017;8:819. doi: 10.1038/s41467-017-00854-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Y, Guo R, Cui Y, et al. An essential role for PNLDC1 in piRNA 3’ end trimming and male fertility in mice. Cell Res. 2017;27:1392–6. doi: 10.1038/cr.2017.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schlegel PN, Sigman M, Collura B, et al. Diagnosis and treatment of infertility in men: AUA/ASRM guideline part I. J Urol. 2021;205:36–43. doi: 10.1097/JU.0000000000001521. [DOI] [PubMed] [Google Scholar]
- 14.Jungwirth A, Giwercman A, Tournaye H, et al. European Association of Urology guidelines on male infertility: the 2012 update. Eur Urol. 2012;62:324–32. doi: 10.1016/j.eururo.2012.04.048. [DOI] [PubMed] [Google Scholar]
- 15.McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wilfert AB, Chao KR, Kaushal M, et al. Genome-wide significance testing of variation from single case exomes. Nat Genet. 2016;48:1455–61. doi: 10.1038/ng.3697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Uhlén M, Fagerberg L, Hallström BM, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 18.Oud MS, Volozonoka L, Smits RM, Vissers LELM, Ramos L, Veltman JA. A systematic review and standardized clinical validity assessment of male infertility genes. Hum Reprod. 2019;34:932–41. doi: 10.1093/humrep/dez022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–24. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Romanel A, Zhang T, Elemento O, Demichelis F. EthSEQ: ethnicity annotation from whole exome sequencing data. Bioinformatics. 2017;33:2402–4. doi: 10.1093/bioinformatics/btx165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Magi A, Tattini L, Palombo F, et al. H3M2: detection of runs of homozygosity from whole-exome sequencing data. Bioinformatics. 2014;30:2852–9. doi: 10.1093/bioinformatics/btu401. [DOI] [PubMed] [Google Scholar]
- 22.Pemberton TJ, Szpiech ZA. Relationship between deleterious variation, genomic autozygosity, and disease risk: insights from The 1000 Genomes Project. Am J Hum Genet. 2018;102:658–75. doi: 10.1016/j.ajhg.2018.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stokowy T, Garbulowski M, Fiskerstrand T, et al. RareVariantVis: new tool for visualization of causative variants in rare monogenic disorders using whole genome sequencing data. Bioinformatics. 2016;32:3018–20. doi: 10.1093/bioinformatics/btw359. [DOI] [PubMed] [Google Scholar]
- 24.Hu Z, Yau C, Ahmed AA. A pan-cancer genome-wide analysis reveals tumour dependencies by induction of nonsense-mediated decay. Nat Commun. 2017;8:15943. doi: 10.1038/ncomms15943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stephenson JD, Laskowski RA, Nightingale A, Hurles ME, Thornton JM. VarMap: a web tool for mapping genomic coordinates to protein sequence and structure and retrieving protein structural annotations. Bioinformatics. 2019;35:4854–6. doi: 10.1093/bioinformatics/btz482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nordkap L, Almstrup K, Nielsen JE, et al. Possible involvement of the glucocorticoid receptor (NR3C1) and selected NR3C1 gene variants in regulation of human testicular function. Andrology. 2017;5:1105–14. doi: 10.1111/andr.12418. [DOI] [PubMed] [Google Scholar]
- 27.Almstrup K, Nielsen JE, Mlynarska O, et al. Carcinoma in situ testis displays permissive chromatin modifications similar to immature foetal germ cells. Br J Cancer. 2010;103:1269–76. doi: 10.1038/sj.bjc.6605880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Girard A, Sachidanandam R, Hannon GJ, Carmell MA. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442:199–202. doi: 10.1038/nature04917. [DOI] [PubMed] [Google Scholar]
- 29.Sund KL, Zimmerman SL, Thomas C, et al. Regions of homozygosity identified by SNP microarray analysis aid in the diagnosis of autosomal recessive disease and incidentally detect parental blood relationships. Genet Med. 2013;15:70–8. doi: 10.1038/gim.2012.94. [DOI] [PubMed] [Google Scholar]
- 30.Bouwkamp CG, Kievit AJA, Olgiati S, et al. A balanced translocation disrupting BCL2L10 and PNLDC1 segregates with affective psychosis. Am J Med Genet B Neuropsychiatr Genet. 2017;174:214–9. doi: 10.1002/ajmg.b.32465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang M, Liu X, Chang G, et al. Single-cell RNA sequencing analysis reveals sequential cell fate transition during human spermatogenesis. Cell Stem Cell. 2018;23(4):599–614.:e4. doi: 10.1016/j.stem.2018.08.007. [DOI] [PubMed] [Google Scholar]
- 32.Skakkebaek NE, Rajpert-De Meyts E, Buck Louis GM, et al. Male reproductive disorders and fertility trends: influences of environment and genetic susceptibility. Physiol Rev. 2016;96:55–97. doi: 10.1152/physrev.00017.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu P-H, Fu Y, Cecchini K, et al. The evolutionarily conserved piRNA-producing locus pi6 is required for male mouse fertility. Nat Genet. 2020;52:728–39. doi: 10.1038/s41588-020-0657-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arafat M, Har-Vardi I, Harlev A, et al. Mutation in TDRD9 causes non-obstructive azoospermia in infertile men. J Med Genet. 2017;54:633–9. doi: 10.1136/jmedgenet-2017-104514. [DOI] [PubMed] [Google Scholar]
- 35.Tan Y-Q, Tu C, Meng L, et al. Loss-of-function mutations in TDRD7 lead to a rare novel syndrome combining congenital cataract and nonobstructive azoospermia in humans. Genet Med. 2019;21:1209–17. doi: 10.1038/gim.2017.130. [DOI] [PubMed] [Google Scholar]
- 36.Lehtiniemi T, Kotaja N. Germ granule-mediated RNA regulation in male germ cells. Reproduction. 2018;155(2):R77–R91. doi: 10.1530/REP-17-0356. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.