Summary
The gut microbiota contributes to diverse aspects of host physiology, ranging from immunomodulation to drug metabolism. Changes in the gut microbiota composition are associated with various diseases as well as with the response to medications. It is thus important to understand how different lifestyle and environmental factors shape gut microbiota composition. Beyond the commonly considered factor of diet, small-molecule drugs have recently been identified as major effectors of the microbiota composition. Other xenobiotics such as environmental or chemical pollutants can also impact gut bacterial communities. Here we review the mechanisms of interactions between gut bacteria and antibiotics, host-targeted drugs, natural food compounds, food additives and environmental pollutants. While xenobiotics can impact bacterial growth and metabolism, bacteria in turn can bioaccumulate or chemically modify these compounds. These reciprocal interactions can manifest in complex xenobiotic-host-microbiota relationships. Our review highlights the need to study mechanisms underlying interactions with pollutants and food additives towards deciphering the dynamics and evolution of the gut microbiota.
Introduction
The physiological importance of the gut microbiota − the largest microbial community in the human body − is apparent both in its form and function. A typical adult microbiota harbours an estimated 3.8 x 1013 bacteria, outnumbering the host somatic cells and contributing circa 200 g to the body weight1. At the genetic level, the gut microbiota contains approximately 30 times more genes compared to the human genome2, representing a vast metabolic capacity. Indeed, the gut microbiota contributes to various host functions, including food digestion, detoxification, endocrine function, neurological signalling, immune modulation and pathogen defence3–6. The essential role of gut bacteria in these functions is reflected in the associations between the gut microbiota composition and diseases such as inflammatory bowel disease7, cancer8, obesity, type 2 diabetes, cardio-metabolic diseases3 and neurological disorders9. The strong link to physiology and health argues for treating the gut microbiota as an organ of the human body (Box 1).
Box 1.
The gut microbiota as an organ/supra-organ/second liver
The human gut microbiota is often considered as an organ or supra-organ, as it is essential for host functions, such as detoxification, endocrine function, neurological signalling, immune modulation and pathogen defence3–6 and is involved in pathology and physiology43. The gut microbiota is often referred to as second liver, as it is involved in metabolic processes and can contribute to activation, deactivation or toxification of xenobiotics33–36.
Xenobiotics
Xenobiotics are compounds foreign to the human body. These include drugs, pollutants, toxins, and food additives and contaminants. It has been suggested that throughout a lifetime, an individual is exposed to around 10,000 to 100,000 different xenobiotics, at varying concentrations44.
Alpha diversity
Alpha diversity refers to the bacterial species diversity within an individual microbiota or within an individual sample.
Macronutrients
A macronutrient represent the nutritive/main component of our food that is used by the body for energy, such as protein, carbohydrates and fat.
Micronutrients
A micronutrient refers to chemical elements or substances only required in trace amounts for healthy growth and development, such as vitamins or minerals.
Bacteriophage (Phage)
Viruses that can infect bacteria and can thereby contribute to bacterial abundance and metabolism. Most phages in the gut replicate through incorporation of the phage genome into the bacterial genome, leading to formation of prophages. These prophages may be induced by DNA-damage, antibiotics or other xenobiotics, leading to expression of the phage genome45.
The interactions between this microbe-filled supra-organ (Box 1) with the rest of the body is largely determined by its microbial composition, predominantly through secreted metabolites. In recent years the impact of microbial metabolites on host physiology has been described by various reviews, showing how e.g. levels of short-chain fatty acids associate with diseases such as type 2 diabetes and neurological diseases10,11. Around 68 % of the gut metabolome can be explained by the microbiota composition12, with many of the bacteria-linked metabolites detected in the blood13. Given that, it is critical to understand how various intrinsic and extrinsic factors impact gut bacteria and their metabolism. While several such factors have been identified, the extent and the mechanistic basis of their impact is poorly understood. Known intrinsic factors include host genetic makeup, age, sex, and bowel movement14,15 (FIG. 1a).
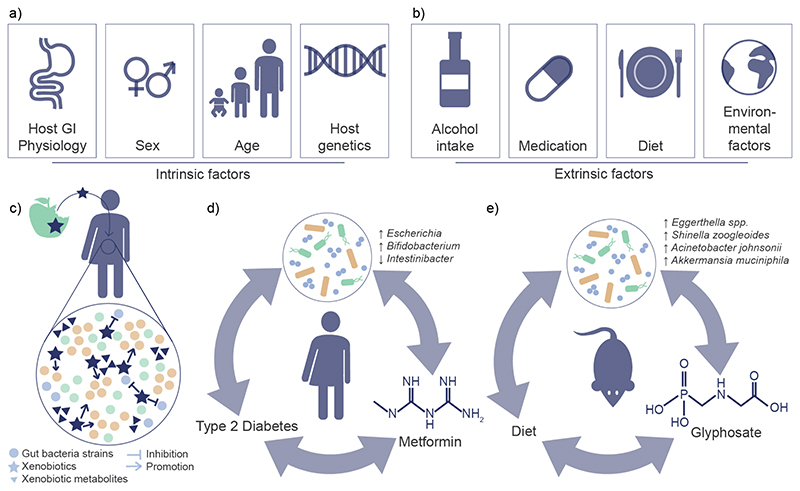
Figure 1. Reciprocal interactions between the gut microbiota and xenobiotics.
A) Intrinsic factors shown to influence gut microbiota composition include host-GI physiology, sex, age, and host genetics14,15. B) extrinsic factors shown to influence the gut microbiota composition include alcohol intake, medication, diet, and other environmental factors14,16,17. C) Ingested xenobiotics can affect the gut microbiota composition by inhibiting or promoting the growth of bacterial species. In turn, xenobiotics may metabolise xenobiotics and thereby alter their activity in the host. D) Metformin, a common treatment for type 2 diabetes has been shown to affect the gut microbiota composition. This altered composition has been shown to be at least in part responsible for the antidiabetic effect observed by metformin treatment21,54. E) The pesticide glyphosate, which may be ingested due to food contamination, has been shown to alter the gut microbiota composition78.
Extrinsic influences include diet, alcohol consumption, medications, and other environmental factors14,16,17 (FIG. 1b). Although environmental factors have been estimated to outweigh genetic differences in explaining gut microbiota variation18,19, the determination of effect sizes remains difficult due to numerous confounding factors18. Nevertheless, medications have been shown to contribute substantially to microbiota variation20. A good case in point is metformin, a widely used medication in type 2 diabetes treatment; the microbiota variation associated with the drug was found to be more pronounced than that associated with the disease21. Further, in contrast to most intrinsic factors, extrinsic factors such as diet and medications can be modified and adapted. Therefore, understanding the interactions between environment and gut microbiota will be critical for designing strategies to improve health and precision medicine.
The interactions between environmental factors such as diet and medications and the gut microbiota are reciprocal. Diet and drugs may modify microbial composition and in return, gut bacteria may chemically transform these compounds (FIG. 1c); examples include digoxin used for treating cardiovascular disease22,23 and levodopa for Parkinson’s24. The impact of environmental pollutants and other xenobiotics is also indicated by several studies25. These include pesticides26–28, heavy metals29,30 and nitrosamines31,32. The gut microbiota has thus been linked to the activation, inactivation and toxicity of xenobiotics33–36.
Xenobiotic exposure occurs on a day-to-day basis through skin contact, inhalation, or ingestion. We may ingest xenobiotics voluntarily, through supplementation or medication, or involuntarily through food and water contamination. Ingested compounds can be metabolized and modified by the gut microbiota before or after absorption37. Orally administered or ingested substances are either absorbed in the small intestine and metabolized in the liver, or pass through the small intestine into the colon, where they can be metabolized by the gut microbiota, which acts like a second liver (Box 1). In addition, previously absorbed substances or substances administered via a different route of exposure can circle back to the intestine via biliary secretion and then interact with the gut microbiota38,39. The metabolism of xenobiotics through gut bacteria may be performed by many different bacterial groups and species and therefore depend on the gut microbiota composition37,40,41. Given the vast variety and number of xenobiotics that are potentially encountered by the gut microbiota (>25,000; FIG. 2; Supplementary Table 1), understanding the extent and the nature of xenobiotic − microbiota interactions will be crucial in unravelling the mechanistic basis of microbiota variation, dynamics, and evolution across and within individuals.
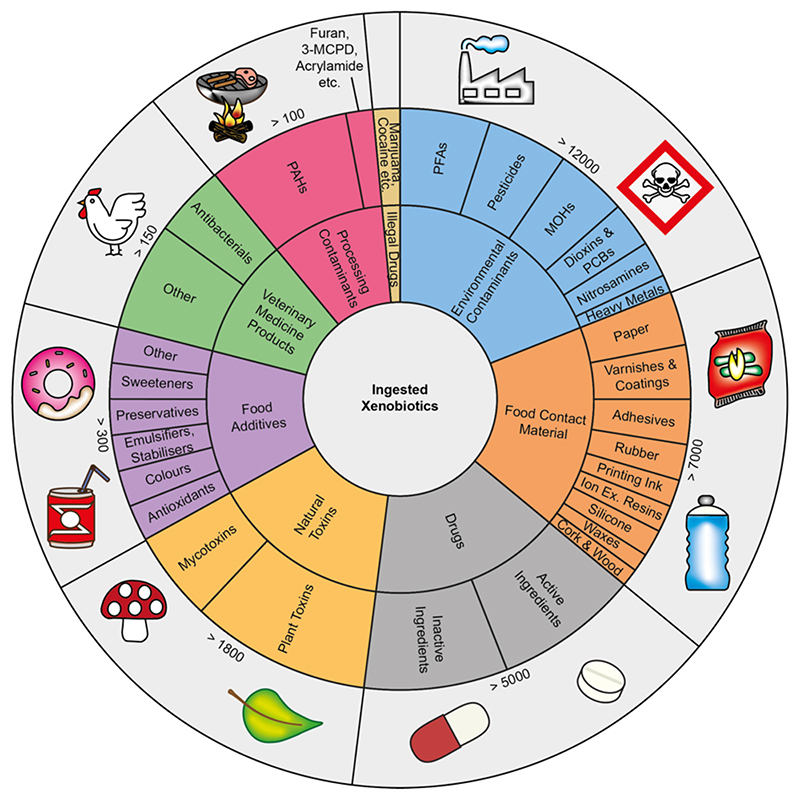
Figure 2. Overview of ingested xenobiotics.
Through our diet, we consume a vast number of natural compounds. The FooDB − the largest resource for food constituents, chemistry and biology − lists 797 different foods containing 15,750 detected natural compounds and a further 55,176 expected and predicted compounds60. Besides the natural compounds present in our food, we also consume many other xenobiotics on a day-to-day basis, many of which yet remain unknown. The shown chart is not comprehensive and is intended to provide an overview of the breadth of different xenobiotics that may be ingested regularly. The information shown is compiled from multiple different databases, which can be found in supplementary table 1. In addition, not everyone will be exposed to all of the here mentioned xenobiotics throughout their lifetime and exposures will vary depending on lifestyle, occupation and geographical location.
In this review, we discuss the influence of widely encountered xenobiotics on the gut microbiota composition and highlight known modes of xenobiotic − gut bacteria interactions. The compounds are grouped into four classes: (1) antibiotics, (2) host-targeted drugs, (3) natural food compounds and food additives, and (4) environmental pollutants and chemicals. Interactions revealed by multiple experimental approaches cover in vivo relevance as well as mechanistic insights. We focus on direct interactions and molecular mechanisms where possible, and span a much broader range of xenobiotic compounds than previously reviewed25,42.
Xenobiotics and gut microbiota composition
Collateral damage by antibiotics
Antibiotics have strong short-term effects on the gut microbiota and cause bacterial cell damage across numerous bacterial strains42,46. Both in vitro and human studies investigating the effects of broad-spectrum antibiotics rifaximin, ciprofloxacin, metronidazole and vancomycin showed their profound short-term effects in reducing total microbial biomass47 and alpha diversity and richness (Box1)48. In addition, these studies showed a large shift in gut microbiota composition with a decrease in Bacteroides-Firmicutes ratio, reduced abundance of several genera, including Bacteroides and Faecalibacterium, and a substantial reduction of butyrate-producing genera from 27 % to 0.3 % relative abundance47,48, which demonstrated the scale to which antibiotics might disrupt the gut microbiota.
Besides short-term effects, antibiotics have also been shown to be associated with long-term changes in gut microbiota composition. Ciprofloxacin and clindamycin have been shown to reduce species richness and alter microbial composition, including a decrease in Bacteroides and an increase in Faecalibacterium and Alistipes, lasting for up to 12 months49. The recovery of the gut microbiota to pre-intervention levels hereby depends on the antibiotic compound and host-specific effects50. The negative impact of antibiotics on commensals can open a niche for low abundant species, and sometimes even for pathogens51,52. A study in healthy men treated with a cocktail of meropenem, gentamycin and vancomycin observed blooms of enterobacteria and other pathobionts, and depletion of Bifidobacterium species and other butyrate producers, which are thought to be protective against pathogens53. These studies highlight that the damage antibiotic treatment has on the microbiota may further negatively impact the host through reduced production of beneficial metabolites or through the opening of a niche for pathogens.
Specific changes induced by human-targeted drugs
Compared to antibiotics, human-targeted drugs are a larger and more diverse group of medications. The exposure for some of the drugs, like antidiabetics and antidepressants, is often over a much longer period than to antibiotics. As the development of host-targeted drugs does not focus on bacteria, their effect on the gut microbiota composition is more unexpected and less studied. One of the few examples is that of metformin (FIG. 1d), which has been shown to influence microbial species abundance and metabolic pathways in the gut microbiota, possibly via metal homeostasis regulation54. Indeed, type 2 diabetes patients treated with metformin show a significantly different gut microbiota composition with an increase of Escherichia spp. and a decrease in Intestinibacter spp. compared to untreated patients, a difference that was previously erroneously linked to the disease state21. Further, this altered microbiota composition has been shown to be, at least in part, contributing to the antidiabetic effect of metformin. For example, faecal transplants from metformin treated patients to germ-free mice caused an improvement in glucose tolerance compared to mice receiving faecal transplants from type 2 diabetes patients before metformin treatment, but mechanisms remain unclear54. Besides this well-known example, a recent meta-analysis showed that 17 drug categories were associated with individual microbial taxa, with proton-pump inhibitors, antidiabetic drugs (metformin) and laxatives showing the strongest association with relative abundance of microbial taxa47,55. The effect of other host-targeted drugs on the gut microbiota have also been noted, including antipsychotics56, statins57, beta-blockers and ACE-inhibitors58. This growing evidence highlights that a wide range of host-targeted drugs can alter the gut microbiota composition in a drug-dependant manner and hence need to be considered for their role in microbiota dynamics and drug response.
Effect of natural food components and food additives
Not just macronutrients, but also micronutrients influence gut microbiota composition (Box 1)59. Beyond these, the number of compounds naturally occurring in foods is enormous. The FooDB − the largest resource for food constituents, chemistry and biology − lists >55,000 natural compounds60. Among the naturally occurring xenobiotics that have been shown to impact gut microbiota composition are phytochemicals. Herbal extracts of olive, camomile, cinnamon, and ginger were shown to influence gut microbiota composition. Common effects among these extracts were the inhibition of Escherichia and Shigella, which have been associated with gastrointestinal disorders, and the favoured growth of the butyrate producers Coprococcus and Butyricimonas61. In addition, single natural xenobiotic compounds, such as curcumin, have also been shown to alter the abundance of several gut bacterial species. Consumption of curcumin, a primary ingredient in turmeric, was associated with an increase in Clostridium and Bacteroides and a reduction in Blautia and Ruminococcus species62. Tea polyphenols increased Bifidobacterium species, which are associated with probiotic function, and reduced Bacteroides-Prevotella63, whereas berberine, an alkaloid found in several plants, reduced the abundance of Bacteroides uniformis47.
In addition to natural food ingredients, food additives may also interact with and shape gut microbiota composition. Artificial sweeteners and emulsifiers are some of the most widely used food additives. The commercially available sweeteners Splenda, saccharin and sucralose64–67 and the emulsifiers carbomethylcellusolse and polysorbate 8068–70 have been shown to promote changes in gut microbiota composition and dysbiosis in mice. These changes include a decrease in Firmicutes and an increase in Bifidobacterium by sucralose67, an increase in Proteobacteria and Escherichia coli overgrowth by Splenda65, and an increase in Bacteroidetes and decrease in Clostridiales by saccharin, which had been associated with diabetes mellitus66. In addition, both carbomethylcellusolse and polysorbate 80 promoted inflammation, reduced microbiota diversity and increased Bacteroidales while decreasing Clostridiales relative abundance68,69.
Impact of pesticides and other pollutants
Humans unintentionally consume a large number of environmental contaminants such as pesticides, environmental chemicals, processing contaminants, mycotoxins and heavy metals71 (FIG. 2). To date the effects of these contaminants on the human microbiota have not been thoroughly investigated. Nevertheless, the exposure to various environmental pollutants have been associated with changes in gut microbiota composition25.
Pesticides, including insecticides, fungicides, and herbicides, are widely used in food production and therefore represent a major group of food contaminants. In 2018, the Food and Drug Administration (FDA) and European Food Safety Authority (EFSA) detected 212 and 358 different pesticide residues in foods respectively72,73. In addition, in 2020 the UK Expert Committee on Pesticide Residues in Food detected 123 residues in 2460 food samples, 42 % of which contained at least one pesticide residue74. A recent study screening levels of 186 pesticides in a UK cohort (n=130) detected pyrethroid and/or organophosphorus insecticides in all urine samples. For example, the pyrethroids cypermethrin and permethrin were detected in >96 % of samples and the organophosphate diethylphosphate was detected in 75 % of samples. Glyphosate, the only herbicide detected, was present in 53 % of samples75, highlighting that the host, and thus likely the microbiota exposure to pollutants is widespread and should be systematically investigated. Various groups of pesticides have been shown to affect gut microbiota composition and microbiota-related metabolites, including short-chain fatty acids, bile acids and trimethylamine28. A study in mice reported that interaction with the gut microbiota may be contributing to the adverse effects observed for organophosphates, such as impaired glucose tolerance76.
Chlorpyrifos and glyphosate are among the most widely used organophosphates and have both been linked to changes in the gut microbiota composition in rodents26,27,77. Even at low concentrations relevant for dietary exposure (1 − 3.5 mg/kg/d), chlorpyrifos and glyphosate exposure led to alterations in gut microbiota composition26,27. Another recent study investigated the effect of glyphosate on the gut microbiota and metabolome in rats using a multi-omics approach (FIG. 1e). Glyphosate not only increased the abundance of Eggerthella spp., Shinella zoogleoides, Acinetobacter johnsonii, and Akkermansia muciniphila, but also caused changes in gut metabolite levels suggesting an inhibition of the shikimate pathway in gut bacteria, which is the primary mechanism through which glyphosate acts as a herbicide78. As the shikimate pathway is not present in human and animal cells, glyphosate is generally assessed as safe. However, the effect of the inhibition of this pathway in the human gut microbiota on host health remains unclear. Nevertheless, these findings suggest that biochemical pathways in the gut microbiota need to be considered when assessing the adverse health effects of pesticides and other environmental chemicals.
An important class of environmental pollutants are N-nitrosamines due to their carcinogenic and toxic potential. N-nitrosamines are a diverse class of chemicals including over 300 different compounds79. The total N-nitrosamine exposure has been estimated to be between 1900 and 25,000 ng/day depending on lifestyle choices, with tobacco-product consumption causing the highest exposure (~ 22,000 ng/day), followed by food (~ 1900 ng/day), alcoholic beverages (~ 1000 ng/day) and drinking water (~ 120 ng/day)79. In addition, in recent years nitrosamine levels in medications exceeding the acceptable daily intake of 26.5 ng/day, have led to recall of >1400 products by the FDA, e.g. metformin and the blood pressure medication valsartan80,81. While high concentrations in drinking water (mg/L) of various N-nitrosamines have been shown to be carcinogenic in rodent models82,83, the human exposure to single N-nitrosamines is usually orders of magnitudes lower (ng/L), and thus has not been extensively studied79. A study in male rats supplemented with an N-nitrosamine-mixture at environmentally relevant doses showed an appreciable shift in composition towards an obesogenic microbiota profile. This included an increased Firmicutes/Bacteroidetes-ratio and Alistipes and Ruminococcus enrichment32. In addition, supplementation with the nitrosamine dichloroacetonitrile altered microbiota composition and microbial metabolites, including metabolites involved in sphingolipid signalling pathway and fatty acid biosynthesis31. These results suggest that N-nitrosamines may affect the microbiota composition and host health at concentrations relevant for human exposure (ng/L).
Mycotoxins are toxic fungal secondary metabolites and are found as contaminants in food. More than 300 different mycotoxins have been identified, with some being regularly found in foods, such as aflatoxins, ochratoxins, fumonisins, patulin, zearalenone and trichothecenes84. A recent literature review and analysis of EFSA and other large global survey data estimated the prevalence of mycotoxins in foods to be >60 % and 20 % of the food samples to exceed the European Union regulatory limits85. A study in mice demonstrated that ochratoxin treatment (210μg/kg body weight) reduced microbiota diversity and increased Lactobacillus abundance, which has been shown to play a role in ochratoxin detoxification86. In addition a study exposing mice for 9 months to doses relevant for estimated human intake (10μg/kg body weight) also showed changed abundance in several bacterial phyla, families and genera, including a decrease in the genera Bifidobacterium and an increase in Odoribacter, which have been associated with gut inflammation87. These findings demonstrate that mycotoxins need to be considered regarding their interactions with the gut microbiota and in relation to host health.
Heavy metals are also common contaminants in food and drinking water. The human gut microbiota has been shown to contribute to the control and absorption of these toxic metals88. For example, the gut microbiota has been shown to protect mice from arsenic toxicity. Compared to antibiotic-treated or germ-free mice, mice with a gut microbiota accumulated less arsenic in organs and excreted more arsenic in their stool33, showing the possible importance of the gut microbiota in mediating heavy metal toxicity. In turn, heavy metal exposure has been shown to impact the gut microbiota diversity and composition88. Cadmium, arsenic, copper, lead and aluminium exposure have all been shown to alter the gut microbiota composition at both phylum and genus levels, and associate with reduced diversity29,30. In addition, zinc has also been shown to not only alter the gut microbiota composition in mice, but also increase susceptibility to C. difficile infection and severity of disease89, highlighting the importance of understanding the impact heavy metals have on our gut microbiota.
Residues of veterinary antibiotics and medications may also contaminate food, especially animal products. In 2018 the EFSA found that out of 354,517 samples 0.3 % were non-compliant and exceeding the permitted limits. These non-compliant samples contained veterinary medications such as antibacterials and substances with anabolic effects90. Although these residues are ingested in much lower concentrations than medications ingested for therapeutic use, they may also affect the gut microbiota composition91 and warrant further investigation.
Several other food contaminants, including food contact material, plant toxins or industrial chemicals can find their way into our food during food production, packaging, or preparation. As highlighted by the above examples, diverse classes of contaminants have the potential to alter the gut microbiota composition in vivo, stressing the need to consider microbiota-mediated impact on health in their safety assessment.
Xenobiotics − bacteria interactions mechanisms
The human cohort and animal studies have demonstrated numerous associations between the intake of xenobiotics and microbiota composition and function. However, understanding the underlying mechanisms is critical for leveraging these findings in a health context and requires in-depth investigation at species and strain level. Six major modes of xenobiotic − gut bacteria interactions have been described so far and are reviewed below (FIG. 3, FIG. 4, Table 1).
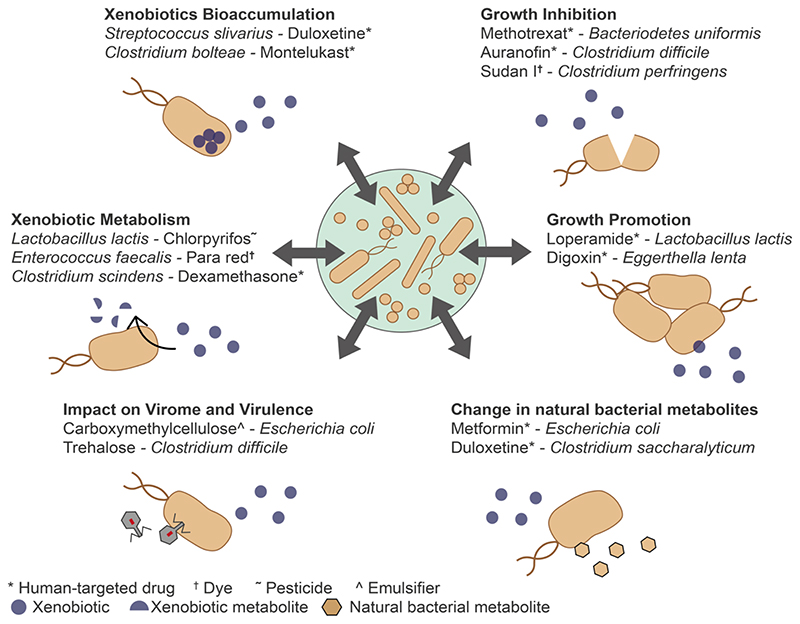
Figure 3. Xenobiotic − gut bacteria interactions.
There are six known modes of interaction between xenobiotics and the gut microbiota: (1) bacterial growth inhibition by xenobiotics92,96, (2) bacterial growth promotion by xenobiotics97,98, (3) change in natural bacterial metabolites by xenobiotics93,98, (4) impact of xenobiotics on the virome and virulence of bacteria106–108, (5) xenobiotic metabolism by bacteria35,36,40 and (6) xenobiotic bioaccumulation by bacteria97,98,. By inhibiting or promoting the growth of certain bacterial species xenobiotics contribute directly to shaping the gut microbiota composition, which in turn determines the function of the gut microbiota. In addition, gut bacteria can metabolize40,41 or bioaccumulate 97,98 and thereby change their activity or toxicity. This metabolism or bioaccumulation depends on the composition of the gut microbiota and the produced metabolites can in turn influence gut microbiota composition. All interactions may occur in various combinations and are dependent on gut microbiota composition. (For more xenobiotic-bacteria examples see Table 1)
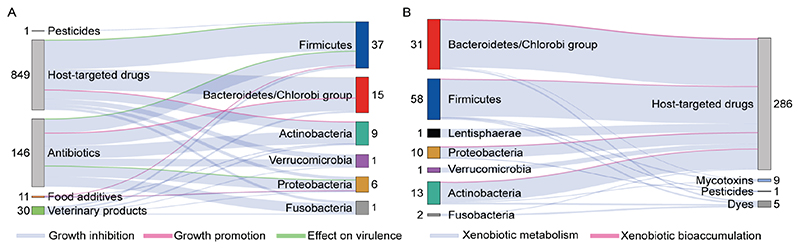
Figure 4. An overview of xenobiotic − gut bacteria interactions investigated at the species level.
A) Growth promoting or growth inhibiting effects and effects of xenobiotics on virulence of bacteria. B) Xenobiotic biotransformation or bioaccumulation by gut bacterial strains. Growth promotion and xenobiotic bioaccumulation, in contrast to growth inhibition and biotransformation, have only recently started being assessed, and, thus, are yet underrepresented. The research to date also focusses mainly on antibiotics and host-targeted drugs, leaving other xenobiotics such as environmental pollutants underrepresented. The information in this diagram was compiled from 8 systematic screens (Supplementary table 2). The numbers next to xenobiotic classes indicate the number of compounds in each class, whereas the numbers next to bacterial phyla indicate the number of bacterial species for which a mechanistic xenobiotic-species relationship has been demonstrated experimentally.
Table 1. Examples of xenobiotic-gut bacteria interactions.
| Interaction Type | Example | Mechanism | Ref. |
|---|---|---|---|
| Growth inhibition | B. vulgatus and B. uniformis & erythromycin (antibiotic) | Erythromycin causes cell shape defects (blebbing, cytoplasmic shrinkage, cell lysis) | 124 |
| Growth promotion | B. thetaiotaomicron & Alternaria toxins (mycotoxins) | Mycotoxins may be metabolised and used as carbon source by specific bacteria * | 97 |
| Change in native metabolites | Clostridium saccharolyticum & duloxetine (antidepressant) | Binding of duloxetine to metabolic enzymes. | 98 |
| Xenobiotic impact on virome and virulence | E.coli & carboxymethylcellulose and polysorbate-80 (emulsifiers) | Mediation of virulence factors flagella, type 1 pili and long polar fimbriae | 107 |
| Xenobiotic metabolism | B. thetaiotaomicron & diltiazem (oral calcium channel blocker) | Deacetylation of both diltiazem and diltiazem metabolites in the gut is bt4096-dependent. | 40 |
| Eggerthella lenta & digoxin (cardiac drug) | Reduction of digoxin to dihydrodigoxin through cgr operon. | 22 | |
| Xenobiotic bioaccumulation | Clostridium saccharolyticum & duloxetine (antidepressant) | After uptake duloxetine binds to proteins in bacteria. Differences in strain bioaccumulation may depend on different uptake and efflux systems. | 98 |
Proposed mechanisms
Growth inhibition by xenobiotics
The most frequently reported interaction mode is bacterial growth inhibition by xenobiotics. In a large-scale in vitro experiment, 78 % of 144 antibiotics showed anti-bacterial activity against at least one of the 40 tested gut bacterial strains, many of which are common members of the human gut, such as Bacteroides vulgatus and B. uniformis92. Most antibiotics affected several strains with no specificity pattern identified. The same study found that 24 % of 835 human targeted drugs were active against at least one of the 40 tested strains92. The human targeted drugs each affected a smaller number of strains compared to antibiotics, highlighting their specific effect as opposed to broad collateral damage of antibiotics. This suggests human-targeted drugs targeting more specific pathways within the bacterial cells, which may only be present in specific strains. Yet, human targeted drugs were found to affect more abundant species supporting their community-scale impact observed in cohort studies. As an example, metformin has been shown to inhibit E. coli growth by inhibiting folate metabolism93. In addition, the antipsychotic medication chlorpromazine and the antidepressant amitriptyline have been shown to inhibit ArcB-mediated efflux94.
Food-associated xenobiotics have also been shown to inhibit bacterial growth. The artificial sweeteners sucralose, saccharin and acesulfame potassium inhibited the growth of E. coli strains HB101 and K-1267. Further, an in vitro study looking at the effects of seven spice extracts (black pepper, cayenne pepper, cinnamon, ginger, oregano, rosemary, turmeric) on 88 different bacterial strains, found that the growth of all four tested Ruminococcus and some Fusobacterium and Clostridium species were inhibited by at least some of the tested spices95.
Other food contaminants with adverse effects on the growth of gut bacterial species include dyes and mycotoxins. The dyes Sudan I, II, III, IV and Para Red showed growth inhibition for at least one of the 11 tested gut bacterial strains96. A recent study demonstrated that a complex extract of Alternaria mycotoxins, toxic fungal metabolites, was able to inhibit the growth of diverse bacterial strains, including the abundant strain B. vulgatus97. These examples indicate that anti-gut bacterial activity is likely a common characteristic for a wide range of xenobiotics.
Growth promotion by xenobiotics
An in vitro study investigating the effects of seven spice extracts on 88 bacterial strains found that the growth of all 33 tested Bifidobacteria and Lactobacillus strains were promoted by at least one of the spice extracts95. In addition, another study found that some drugs may also promote the growth of certain bacterial species, such as B. uniformis by monekulast and Lactococcus lactis by loperamide98. The latter is an interesting observation as L. lactis is a probiotic and loperamide is used for diarrhea treatment. A complex Alternaria mycotoxin extract also promoted the growth of B. caccae and B. thetaiotaomicron at low concentrations (0.5 μg/mL) 97. For drugs and toxins growth inhibition is more frequently reported than growth promotion: Among 375 drug-bacteria interactions, 15 showed growth inhibition and only 5 promoted growth98, whereas among 56 Alternaria-bacteria interactions at different concentrations, 17 showed growth inhibition at high (50 μg/mL) or low (0.5 μg/mL) concentrations and only 2 showed growth promotion at low concentrations 97. There are also comprehensive reviews suggesting growth promoting effects of different natural xenobiotics 99 or food additives 100, however, most studies are performed on complex microbial communities, which makes it difficult to extrapolate single species growth effects. In addition, mechanisms underlying growth promotions are less well studied and understood. One possible explanation could be that xenobiotics serve as a nutrient source for bacteria101. For example, Klebsiella pneumoniae and Escherichia fergusonii have been shown to utilize the antibiotic chloramphenicol as a carbon source102. Taken together, growth promotion by xenobiotics is less frequently reported than inhibition (FIG. 4) yet may play a critical role in a community context when the concerned bacterium is central to the community ecology or function.
Modulation of bacterial native metabolism by xenobiotics
Beyond affecting growth, some xenobiotics have also been shown to alter the natural metabolism of gut bacteria. Despite the increasing evidence highlighting the importance of bacterial metabolism, there is still limited knowledge of how bacterial metabolism effects drug treatment and host health 103. Several studies reported a change in faecal metabolome after xenobiotic administration 31,78, yet only very few studies investigated this on a single strain level. Studies investigating the mechanisms behind metformin action reported that metformin not only altered bacterial folate and methionine metabolism 93, but also altered central pathways, including upregulation of the tricarboxylic acid cycle and downregulation of glycolysis and arginine degradation 104. In addition, a recent study performed metabolite profiling in six bacterial strains incubated with the antidepressant drug duloxetine. Of these, Clostridium saccharolyticum, Lactiplantibacillus plantarum, E. coli and L. lactis showed significant changes in extracellular metabolite abundances. Particularly C. saccharolyticum showed a large shift in its exo-metabolome, including changes in amino acids and nucleotide related pathways. This was also shown to be relevant for community composition through metabolic cross-feeding, as demonstrated by the indirect increase of Eubacterium rectale98. Beside drugs, the natural xenobiotic trigonelline, an alkaloid found in coffee, also showed potential in altering bacterial metabolism. Incubation of Citrobacter freundii with trigonelline in choline-enriched media significantly reduced trimethylamine production, which can be converted to the pro-atherosclerotic metabolite trimethylamine N-oxide in vivo 105. Overall, recent studies bring forward modulation of native bacterial metabolism as one of the fundamental modes of xenobiotic-bacteria interaction.
Xenobiotic impact on virome and virulence
The impact of xenobiotics on the gut virome and the virulence of gut bacteria is a mode of interaction that has been receiving increasing attention in recent years. The gut virome is dominated by bacteriophages (Box 1), which are viruses that can infect bacteria, thereby altering bacterial abundance and metabolism45. A recent study showed that several medications, including the antibiotic mitomycin, the chemotherapeutic fludarabine and the cardiac drug digoxin, lead to prophage induction in bacterial isolates from the human gut, demonstrating the potential of phage-mediated effects to alter the gut microbiota composition106. In addition, the emulsifiers carboxymethylcellulose and polysorbate-80 have been shown to promote chronic inflammation by directly inducing genes responsible for the mediation of virulence in adherent-invasive E.coli, such as the virulence factors flagella, type 1 pili and long polar fimbriae107. Dietary trehalose has also been shown to increase the disease severity and virulence of the C. difficile strains RT027 and RT078, possibly via increased toxin production108. Also, the artificial sweeteners saccharin, sucralose and aspartame have been shown to increase the ability of the model gut bacteria E. coli NCTC10418 and E. faecalis ATCC19433 to adhere to, invade and kill host epithelium in vitro, which is thought to be related to increased expression of virulence factors109. These examples demonstrate the importance of taking not only the microbiome but also the virome into account when investigating the relationship between xenobiotics, gut microbiota, and the host.
Xenobiotic metabolism by gut bacteria
Enzymatic modification of drugs by gut bacteria (biotransformation or metabolization) is a long known and widely recognised interaction mode 110. A well-known recent example is the cardiac drug digoxin, which demonstrates microbiota-dependent interpersonal effectiveness in drug response 22,23. In vitro and mouse studies have demonstrated that only specific strains of Eggerthella lenta, e.g. DSM2243, are responsible for the inactivation of digoxin in the gut 22, providing mechanistic insight into how the microbiota composition contributes to the varying effectiveness of digoxin treatment between patients. In addition, chemotherapeutic drugs, such as the fluoropyrimidine 5-fluorouracil or camptothecin, have been shown to have varying effects on C. elegans depending on their bacterial diets111,112. For example, worms fed E. coli HB101 showed a 80-times higher minimum inhibitory concentration on egg hatching compared to worms fed E. coli BL21G (8μM vs. 0.1μM)111. Other examples of microbial drug metabolism are the activation of the cancer prodrug irinotecan or the deactivation of the cancer drug gemcitabine. Irinotecan causes gastrointestinal toxicity, even if given intravenously, due to β-glucuronidase activation or re-activation in the gut113. Gemcitabine on the other hand has been shown to be deactivated by gammaproteobacteria, most of which belong to the common families of Enterobacteriaceae and Proteobacteriaceae114.
Recent studies indicate that transformation by gut bacteria is highly prevalent. An in vitro study screening 76 bacterial strains against 271 drugs found that around two thirds of these drugs were significantly reduced by at least one bacterial strain, and that each strain was able to metabolize between 11 and 95 drugs. In addition, they identified and validated 30 microbiota-encoded enzymes that converted 20 drugs to 59 metabolites, whose abundance in gut microbial communities partially explained the communities drug metabolising capacity 40. Another study using personalized gut microbiota cultures found that 57 out of 438 screened drugs spanning 28 pharmacological classes were metabolised, including 45 novel drug-microbiota interactions 41. These studies suggest that the gut microbiota is involved in the metabolism of a range of drugs and that by determining an individual’s drug metabolizing capacity one could predict the response to a specific drug or therapy. A recent study investigating the therapeutic efficacy of methotrexate, a common drug prescribed for rheumatoid arthritis, not only demonstrated a correlation between pre-treatment microbiota and response to treatment, but was also able to predict the response to methotrexate using machine learning approaches115. While some chemical similarities, such as amide-, nitro-, azo-or urea groups, have been identified between drugs metabolised by gut bacteria40, datasets to date are not large enough to enable generalisation.
Similar to therapeutic drugs, environmental contaminants may also be metabolized by the gut microbiota, which could affect their bioavailability or toxicity in the host 37. In vitro incubation of Lactococcus lactis, Lactobacillus fermentum and E. coli demonstrated the potential of these strains to degrade the pesticide chlorpyrifos within 15 days to 70, 61 and 16 % respectively. Interestingly, each of these strains produced different metabolites, indicating that multiple pathways may be involved in microbial chlorpyrifos degradation 35.
Mycotoxins, which are fungal secondary metabolites and found as contaminants in food, are also reported to be biotransformed by gut bacteria. A study investigating the metabolism of masked mycotoxins (i.e., mycotoxins conjugated to sugars, glutathione or sulphates) in human faecal samples demonstrated that these were efficiently hydrolysed in the faecal samples, which may lead to higher mycotoxin exposure, as unmasked mycotoxins are absorbed more efficiently 116. Another study investigated the metabolising capacity of 14 different bacterial strains to metabolise masked mycotoxins or to degrade free mycotoxins (i.e., deoxynivalenol, H2 and HT2 toxin). Several bacterial strains were shown to harbour these capacities, including B. adolescentis, an abundant strain in the human gut 117.
Different dyes, such as Sudan I, II, III, IV and Para Red, have also been shown to be metabolised by the gut microbiota. An in vitro study showed that 33 out of 35 tested bacterial species were able to degrade at least one of these dyes. This suggests that the ability to reduce these types of dyes are widely spread among human gut bacterial species 36. In addition, a recent study demonstrated that the azo dye Red 40, one of the most used food colorants worldwide, was shown to promote colitis in mice with dysregulated IL-23 expression in a microbiota dependent manner. The abundant bacterial strains E. faecalis and B. ovatus were shown to degrade Red 40 in vitro, and were also shown to contribute to Red 40 induced colitis in mice118. Another common group of food contaminants are polycyclic aromatic hydrocarbons (PAHs), which can be introduced during high-temperature cooking. PAHs have been shown to have toxic, carcinogenic and estrogenic effects in the human body. It has long been established that PAHs can be activated by cytochrome P450 enzymes in the human body to more reactive metabolites, which is thought to contribute to their carcinogenicity119. In addition to this, an in vitro study investigated the metabolising capacity of the gut microbiota for different PAHs in a simulated system of the human intestinal tract 120. Only microbial digested PAHs, and unmodified compounds, showed estrogenic activity indicating that the gut microbiota also contributes to the bioactivation of PAHs in the human body 120.
The examples in this section demonstrate the potential of the gut microbiota to metabolise a wide variety of compounds and thereby alter their bioavailability, efficacy, and toxicity in the host.
Xenobiotic bioaccumulation by gut bacteria
Bioaccumulation of xenobiotics by gut bacteria is only recently being appreciated. A study investigating the effect of 14 gut bacterial strains on a complex Alternaria mycotoxin extract showed different tendencies of these mycotoxins to associate with bacterial cell pellets. Alternariol, alternariol monomethylethyl ether and altersetin were not only found to be significantly depleted by many bacterial strains, but also to accumulate within the bacterial pellets. This accumulation was correlated with the lipophilicity of these compounds and was observed more often for gram-negative bacteria, such as B. vulgatus and B. thetaiotaomicron 97. Another study investigating drug-bacteria interactions found that 17 out of 29 interactions in which the drug was depleted, was not caused by biotransformation by bacteria, but by bioaccumulation. For example, duloxetine, a widely used antidepressant, was accumulated by several bacterial species, including Streptococcus salivarius, Clostridium saccharolyticum and E. coli98. This finding challenges the view that the main mode of bacteria-drug interaction is biotransformation. Also, some drugs showed potential to be either bioaccumulated or degraded depending on the bacterial strain, and most bacterial strains were capable of both interaction types depending on the drug98. These findings could explain results from other studies showing drug depletion without metabolite detection 41. Bioaccumulation thus needs to be considered in deciphering the effect of xenobiotics at species and community, and host level.
Xenobiotic mixtures
To date, most studies investigating microbiota-xenobiotic interactions focus on one compound at a time, a condition which does not occur in vivo. Medications are often taken in combination with food containing a large number of natural compounds and contaminants. As all ingested compounds will be co-present in the gut, it is important to determine not only single, but also combination effects of xenobiotics, as the effect of a single compound on a single strain or community may not be the same as the effect of a complex mixture on a single strain or community.
Several studies have shown that different dietary and environmental factors can influence drug action indicating combination effects 44. For example, bisphenol A, a chemical used in plastic food packaging, has been shown to decrease the efficacy of anti-cancer drugs, such as rapamycin 121,122. Also, the activation of the cancer drug fluoropyrimidine by gut bacteria has been shown to be regulated by dietary cues, such as pyrimidines or vitamins B6 or B9 in C. elegans111,112. In addition, an increasing number of studies show that the combination of xenobiotics has varying effects on the gut microbiota compared to single substances. The inactivation of digoxin by specific E. lenta strains has been shown to be inhibited by arginine, indicating that an increase in protein consumption may reduce in vivo digoxin reduction 22. The combination of chemicals in glyphosate formulations have also been shown to have a different effect on gut bacteria, including probiotic L. rhamnosus strains, than the pesticide glyphosate alone 78, suggesting a synergy between glyphosate and formulation compounds. Another study tested combinations of 79 compounds, including antibiotics, host-targeted drugs, and food additives on 6 bacterial strains, and found both synergistic and antagonistic effects, the latter being more frequent 123. Here antagonism was shown between drugs targeting different cellular processes, such as the food additive vanillin, which has been shown to increase the levels of aerobic respiration control protein ArcA, and the antibiotic chloramphenicol, an inhibitor of protein synthesis, in E. coli. The less frequently observed synergies only occurred between compounds targeting the same process, such as vanillin, effecting the multidrug transporter MdfA, and the antibiotic spectinomycin, a further inhibitor of protein synthesis, in E. col123. A search among the Prestwick drug library, containing FDA- and EMA-approved drugs, for antagonists of erythromycin, which is bactericidal to the common gut bacteria B. vulgatus by causing cell shape defects such as blebbing, cytoplasmic shrinkage and lysis, identified that dicoumarol and tolfenamic acid are able to rescue B. vulgatus without affecting the killing capability of erythromycin on pathogens 124. Further, an in vitro study found that 45 out of 186 bioactive compounds showed synergistic effects with antibiotics, such as triclosan, on E. coli MC1061 growth 125. These results bring forward the possibility of reducing adverse effects of antibiotics and other drugs using xenobiotic combinations 124, but also stress the need to systematically study xenobiotic combinations to better understand the extent of and mechanisms through which xenobiotics shape the microbiota and vice versa.
Conclusion and Future Perspectives
A wide range of xenobiotic compounds that can interact with the gut microbiota are consumed as part of diet or as medication. Several of these xenobiotics have been shown to influence bacterial physiology and thereby the gut microbiota composition, while the bacteria in turn modify xenobiotic activity and toxicity. Many of these interactions, e.g. digoxin22 or fluoropyrimidine111, have been shown to be variable across individuals depending on the gut microbiota composition. Further research of these inter-individual variations will advance the knowledge and enable development of personalised approaches for dietary or medical interventions.
In addition, most studies investigating environment-microbiota-host interactions have yet focussed on one environmental factor at a time, such as diet, drugs or environmental pollutants 40,41,95. To gain a complete understanding of environment-microbiota-host interactions, it will be important to simultaneously account for multiple contributing factors. Previous reviews have described and summarised system-level approaches that can be used to help elucidate drug-mircobiome-host interactions42. For example, it has been shown that in vitro and in vivo experiments in combination with computational strategies can be combined to disentangle host and microbial contributors for drug metabolism, an approach that could be adapted for other xenobiotic compounds38.
Another important aspect that needs to be considered when conducting or comparing studies is xenobiotic concentration. The effects of xenobiotics on single strains or the complete microbiota depend on the concentration at which they are tested. For example, a comprehensive drug screen showed that more species were inhibited at higher drug concentrations 92. In addition, growth inhibitory or promoting effects of spice extracts 95 or Alternaria mycotoxins 97 were dependant on the concentration. B. thetaiotaomicron, for example, exhibited growth promotion at low concentrations (0.5 μg/mL) and growth inhibition at high concentrations (50 μg/mL) 97, underscoring the critical importance of considering concentrations in assessing the xenobiotic effects. The observation that more bacterial species are inhibited at higher concentrations may be due to multiple factors, including changes in osmolarity or pH, or due to changes in compound uptake and binding to macromolecules. It is important to note that “high” or “low” concentrations are context and compound dependent. Besides this, the concentrations used in studies are also model dependent. For human or animal studies concentrations will usually be higher (mM range) than for in vitro cell studies (μM range).
While testing at concentrations relevant for the gastrointestinal tract would be ideal, this information is rarely available. The trend to include metabolomics analysis of faecal samples, urine and plasma in microbiota studies 12,78 would thus become more beneficial, as this would provide an idea of the concentration of specific compounds and also about the concentration range between individuals and time points. Additionally, screening of wastewater and wastewater-based epidemiology provide one possibility to monitor trends on drug consumption or environmental pollutants on a community wide basis126.
As more mechanistic data on xenobiotic-bacteria interactions become available, machine-learning approaches could help predict reciprocal xenobiotic-bacteria interactions given the information on the chemical structures and bacterial genomes. While some databases already cover >13,000 xenobiotic-gut bacteria interactions, >11,000 of these interactions concern pharmaceuticals 127,128. To enable the successful implementation of machine-learning approaches a large dataset covering diverse xenobiotics classes (FIG. 1) and diverse bacterial species is needed. Indeed, there are many more xenobiotic classes than could be reviewed here due to lack of studies. Future research should put a strong focus on substances that are commonly found in foods and medications and are regularly ingested, but also consider other substances and compounds that enter the body via inhalation, dermal absorption, and injection.
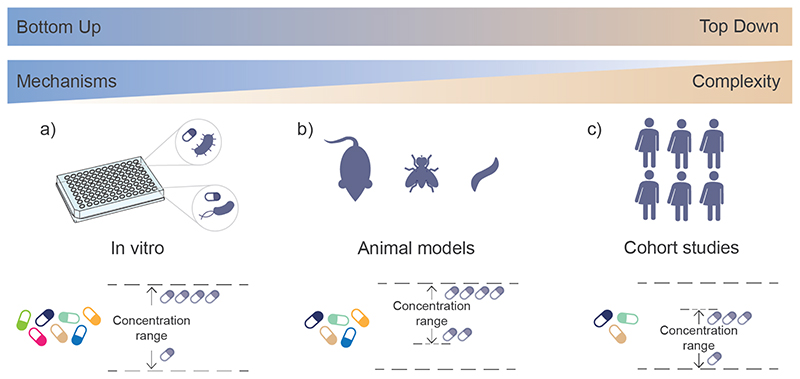
While many experimental approaches to determine microbiota-xenobiotic interactions exist, each comes with its own limitations, making translation of experimental findings into clinical or health advisory settings challenging (FIG. 5). Large scale human studies (top-down approach) are needed to establish associations between xenobiotics and the gut microbiota. These represent the physiological conditions and are directly relevant for both functional studies and applications. However, the complexity of the study design and relatively low control over the conditions mean that many confounding factors exist, such as lifestyle and reciprocal host-microbiota interactions. It is thus often difficult to establish causal links or pinpoint underlying mechanisms. In vitro studies (bottom-up approach) on the other hand with cultured gut bacterial strains can reveal molecular mechanisms, but it is challenging to extrapolate to in vivo conditions, as in vitro studies may not always be relevant in the in vivo contect. In addition, many gut bacterial strains are present in very low abundances and are difficult to cultivate, which hinders their identification and examination129. Animal models, such as mice, rats or C. elegans, allow an intermediate approach, where microbiota-host interactions can be investigated in vivo in controlled settings. However, translation of the findings from animals to humans may yet be difficult, as animal models of disease were not developed with the gut microbiota in mind130. In future, ingestible measurement and reporting devices, such as magnetic hydrogels containing living bacteria, or micro-bio-electronic devices, could provide information on in situ xenobiotic concentrations and thus shed light on xenobiotic-microbiota-human interactions under real-life conditions131,132. Understanding the possibilities and limitations of each model is crucial to enable linking of different study types to deepen our understanding of the gut microbiota. We envisage that emerging mechanistic insights would enable merging of top-down and bottom-up approaches towards model-guided prediction of xenobiotic-microbiota interactions.
Figure 5. Models used to investigate environment − microbiota − host interactions.
A) Bottom-up approaches based on in vitro studies using cultured gut bacterial strains can help reveal molecular mechanisms. On the other hand, in vitro studies may not always be relevant in the in vivo context. Additionally, the abundance of many strains is very low and cultivation can prove difficult, hindering strain identification and examination129. B) Animal models, including mice, flies and C. elegans, present an intermediate approach, allowing the investigation of microbiota interactions in vivo under controlled settings. Translation of findings from animal models to humans may yet be difficult, as animal models of disease were not developed with the gut microbiota in mind, leading to less informative results130. C) Investigation based on human cohorts, so called top-down approach, is directly relevant for both functional studies and applications. However, the complexity of the study design and relatively low control over the conditions means that many confounding factors exist, such as lifestyle and reciprocal host-microbiota interactions. It Is thus often difficult to establish causal links or pinpoint underlying mechanisms.
The xenobiotic concentrations used in studies often vary across the spectrum of bottom-up and topdown studies. In animal model studies, concentrations used are usually higher (mM range) than those used in the in vitro studies (μM range). While a wide range of concentrations can in principle be used in animal models, typically higher concentrations are chosen to ensure physiological readouts are possible and to restrict the number of animals to an ethically acceptable degree. Concentrations in human studies are constrained by the cohort access (in the case of observational studies) or by safety concerns (in the case of interventional studies). Further, while in human and animal models the compounds pass through host digestive processes before they reach the bacteria in the gut, in in vitro studies the bacteria are directly exposed to the compounds.
Supplementary Material
Acknowledgements
A.E.L. is supported by the Health Protection Research Unit in Chemical and Radiation Threats and Hazards, funded by the National Institute for Health Research (NIHR). K.R.P and A.E.L. acknowledge funding by UK Medical Research Council (project no. Mc_UU_00025/11). M.Z.-K. is supported by the postdoctoral fellowship from the AXA Research Fund. U. Hofer is acknowledged for helpful comments on the manuscript.
Footnotes
Competing interests
A.E.L. and M.Z.-K. declare no competing interests. K.R.P. is an inventor in two patent applications related to the findings and concepts discussed in this review (US patent application numbers 16966307 and 16966322).
References
- 1.Sender R, Fuchs S, Milo R. Revised Estimates for the Number of Human and Bacteria Cells in the Body. Plos Biol. 2016;14 doi: 10.1371/journal.pbio.1002533. ARTN e1002533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li JH, et al. An integrated catalog of reference genes in the human gut microbiome. Nature Biotechnology. 2014;32:834–841. doi: 10.1038/nbt.2942. [DOI] [PubMed] [Google Scholar]
- 3.Fan Y, Pedersen O. Gut microbiota in human metabolic health and disease. Nature Reviews Microbiology. 2021;19:55–71. doi: 10.1038/s41579-020-0433-9. [DOI] [PubMed] [Google Scholar]
- 4.Fung TC, Olson CA, Hsiao EY. Interactions between the microbiota, immune and nervous systems in health and disease. Nature Neuroscience. 2017;20:145–155. doi: 10.1038/nn.4476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hooper LV, Littman DR, Macpherson AJ. Interactions Between the Microbiota and the Immune System. Science. 2012;336:1268–1273. doi: 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kamada N, Chen GY, Inohara N, Núñez G. Control of pathogens and pathobionts by the gut microbiota. Nature Immunology. 2013;14:685–690. doi: 10.1038/ni.2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Franzosa EA, et al. Gut microbiome structure and metabolic activity in inflammatory bowel disease. Nat Microbiol. 2019;4:293–305. doi: 10.1038/s41564-018-0306-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rajagopala SV, et al. The Human Microbiome and Cancer. Cancer Prevention Research. 2017;10:226–234. doi: 10.1158/1940-6207.capr-16-0249. [DOI] [PubMed] [Google Scholar]
- 9.Cryan JF, O’Riordan KJ, Sandhu K, Peterson V, Dinan TG. The gut microbiome in neurological disorders. The Lancet Neurology. 2020;19:179–194. doi: 10.1016/s1474-4422(19)30356-4. [DOI] [PubMed] [Google Scholar]
- 10.Krautkramer KA, Fan J, Bäckhed F. Gut microbial metabolites as multi-kingdom intermediates. Nature Reviews Microbiology. 2021;19:77–94. doi: 10.1038/s41579-020-0438-4. [DOI] [PubMed] [Google Scholar]
- 11.Lee-Sarwar KA, Lasky-Su J, Kelly RS, Litonjua AA, Weiss ST. Metabolome–Microbiome Crosstalk and Human Disease. Metabolites. 2020;10:181. doi: 10.3390/metabo10050181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zierer J, et al. The fecal metabolome as a functional readout of the gut microbiome. Nature Genetics. 2018;50:790–795. doi: 10.1038/s41588-018-0135-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shuo Han WVT, Fischer Curt R, Merrill Bryan D, DeFelice Brian C, Sanchez Juan M, H SK, Guthrie Leah, Fall Lalla A, Dodd Dylan, Fischbach Michael A, Sonnenburg Justin L. A metabolomics pipeline for the mechanistic interrogation of the gut microbiome. Nature. 2021;595 doi: 10.1038/s41586-021-03707-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vujkovic-Cvijin I, et al. Host variables confound gut microbiota studies of human disease. Nature. 2020 doi: 10.1038/s41586-020-2881-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hughes DA, et al. Genome-wide associations of human gut microbiome variation and implications for causal inference analyses. Nat Microbiol. 2020;5:1079-+. doi: 10.1038/s41564-020-0743-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abdelsalam NA, Ramadan AT, ElRakaiby MT, Aziz RK. Toxicomicrobiomics: The Human Microbiome vs. Pharmaceutical, Dietary, and Environmental Xenobiotics. Front Pharmacol. 2020;11 doi: 10.3389/fphar.2020.00390. ARTN 390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kolodziejczyk AA, Zheng D, Elinav E. Diet–microbiota interactions and personalized nutrition. Nature Reviews Microbiology. 2019;17:742–753. doi: 10.1038/s41579-019-0256-8. [DOI] [PubMed] [Google Scholar]
- 18.Gacesa R, et al. The Dutch Microbiome Project defines factors that shape the healthy gut microbiome. Cold Spring Harbor Laboratory; 2020. [Google Scholar]
- 19.Rothschild D, et al. Environment dominates over host genetics in shaping human gut microbiota. Nature. 2018;555:210-+. doi: 10.1038/nature25973. [DOI] [PubMed] [Google Scholar]
- 20.Falony G, et al. Population-level analysis of gut microbiome variation. Science. 2016;352:560–564. doi: 10.1126/science.aad3503. [DOI] [PubMed] [Google Scholar]
- 21.Forslund K, et al. Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature. 2015;528:262–266. doi: 10.1038/nature15766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haiser HJ, et al. Predicting and Manipulating Cardiac Drug Inactivation by the Human Gut Bacterium Eggerthella lenta. Science. 2013;341:295–298. doi: 10.1126/science.1235872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lindenbaum J, Rund DG, Butler VP, Tse-Eng D, Saha JR. Inactivation of Digoxin by the Gut Flora: Reversal by Antibiotic Therapy. New England Journal of Medicine. 1981;305:789–794. doi: 10.1056/nejm198110013051403. [DOI] [PubMed] [Google Scholar]
- 24.Maini Rekdal V, Bess EN, Bisanz JE, Turnbaugh PJ, Balskus EP. Discovery and inhibition of an interspecies gut bacterial pathway for Levodopa metabolism. Science. 2019;364:eaau6323. doi: 10.1126/science.aau6323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chiu K, Warner G, Nowak RA, Flaws JA, Mei W. The Impact of Environmental Chemicals on the Gut Microbiome. Toxicological Sciences. 2020;176:253–284. doi: 10.1093/toxsci/kfaa065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mao QX, et al. The Ramazzini Institute 13-week pilot study on glyphosate and Roundup administered at human-equivalent dose to Sprague Dawley rats: effects on the microbiome. Environ Health-Glob. 2018;17 doi: 10.1186/s12940-018-0394-x. ARTN 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reygner J, et al. Inulin Supplementation Lowered the Metabolic Defects of Prolonged Exposure to Chlorpyrifos from Gestation to Young Adult Stage in Offspring Rats. Plos One. 2016;11 doi: 10.1371/journal.pone.0164614. ARTN e0164614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yuan XL, et al. Gut microbiota: An underestimated and unintended recipient for pesticide-induced toxicity. Chemosphere. 2019;227:425–434. doi: 10.1016/j.chemosphere.2019.04.088. [DOI] [PubMed] [Google Scholar]
- 29.Li X, et al. Heavy metal exposure causes changes in the metabolic health-associated gut microbiome and metabolites. Environment International. 2019;126:454–467. doi: 10.1016/j.envint.2019.02.048. [DOI] [PubMed] [Google Scholar]
- 30.Zhai QX, et al. Effects of subchronic oral toxic metal exposure on the intestinal microbiota of mice. Sci Bull. 2017;62:831–840. doi: 10.1016/j.scib.2017.01.031. [DOI] [PubMed] [Google Scholar]
- 31.Xue B, et al. Low-concentration of dichloroacetonitrile (DCAN) in drinking water perturbs the health-associated gut microbiome and metabolic profile in rats. Chemosphere. 2020;258 doi: 10.1016/j.chemosphere.2020.127067. ARTN 127067. [DOI] [PubMed] [Google Scholar]
- 32.Zhu JQ, et al. Consumption of drinking water N-Nitrosamines mixture alters gut microbiome and increases the obesity risk in young male rats. Environ Pollut. 2019;248:388–396. doi: 10.1016/j.envpol.2019.02.012. [DOI] [PubMed] [Google Scholar]
- 33.Coryell M, McAlpine M, Pinkham NV, McDermott TR, Walk ST. The gut microbiome is required for full protection against acute arsenic toxicity in mouse models. Nature Communications. 2018;9 doi: 10.1038/s41467-018-07803-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ilett KF, Tee LBG, Reeves PT, Minchin RF. Mebolism of drugs and other xenobiotics in the gut lumen and wall. Pharmacology & Therapeutics. 1990;46:67–93. doi: 10.1016/0163-7258(90)90036-2. [DOI] [PubMed] [Google Scholar]
- 35.Harishankar MK, Sasikala C, Ramya M. Efficiency of the intestinal bacteria in the degradation of the toxic pesticide, chlorpyrifos. 3 Biotech. 2013;3:137–142. doi: 10.1007/s13205-012-0078-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu H, Heinze TM, Paine DD, Cerniglia CE, Chen H. Sudan azo dyes and Para Red degradation by prevalent bacteria of the human gastrointestinal tract. Anaerobe. 2010;16:114–119. doi: 10.1016/j.anaerobe.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koppel N, Rekdal VM, Balskus EP. Chemical transformation of xenobiotics by the human gut microbiota. Science. 2017;356 doi: 10.1126/science.aag2770. ARTN eaag2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zimmermann M, Zimmermann-Kogadeeva M, Wegmann R, Goodman AL. Separating host and microbiome contributions to drug pharmacokinetics and toxicity. Science. 2019;363:eaat9931. doi: 10.1126/science.aat9931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roberts MS, Magnusson BM, Burczynski FJ, Weiss M. Enterohepatic Circulation. Clinical Pharmacokinetics. 2002;41:751–790. doi: 10.2165/00003088-200241100-00005. [DOI] [PubMed] [Google Scholar]
- 40.Zimmermann M, Zimmermann-Kogadeeva M, Wegmann R, Goodman AL. Mapping human microbiome drug metabolism by gut bacteria and their genes. Nature. 2019;570:462–467. doi: 10.1038/s41586-019-1291-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Javdan B, et al. Personalized Mapping of Drug Metabolism by the Human Gut Microbiome. Cell. 2020;181:1661–1679.:e1622. doi: 10.1016/j.cell.2020.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zimmermann M, Patil KR, Typas A, Maier L. Towards a mechanistic understanding of reciprocal drug–microbiome interactions. Molecular Systems Biology. 2021;17 doi: 10.15252/msb.202010116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baquero F, Nombela C. The microbiome as a human organ. Clinical Microbiology and Infection. 2012;18:2–4. doi: 10.1111/j.1469-0691.2012.03916.x. [DOI] [PubMed] [Google Scholar]
- 44.Pristner M, Warth B. Drug-Exposome Interactions: The Next Frontier in Precision Medicine. Trends Pharmacol Sci. 2020;41:994–1005. doi: 10.1016/j.tips.2020.09.012. [DOI] [PubMed] [Google Scholar]
- 45.Mirzaei MK, Maurice CF. Ménage à trois in the human gut: interactions between host, bacteria and phages. Nature Reviews Microbiology. 2017;15:397–408. doi: 10.1038/nrmicro.2017.30. [DOI] [PubMed] [Google Scholar]
- 46.Corinne Henry, Peter Xenobiotics Shape the Physiology and Gene Expression of the Active Human Gut Microbiome. Cell. 2013;152:39–50. doi: 10.1016/j.cell.2012.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li L, et al. RapidAIM: a culture- and metaproteomics-based Rapid Assay of Individual Microbiome responses to drugs. Microbiome. 2020;8 doi: 10.1186/s40168-020-00806-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Haak BW, et al. Long-term impact of oral vancomycin, ciprofloxacin and metronidazole on the gut microbiota in healthy humans. Journal of Antimicrobial Chemotherapy. 2019;74:782–786. doi: 10.1093/jac/dky471. [DOI] [PubMed] [Google Scholar]
- 49.Rashid M-U, et al. Determining the Long-term Effect of Antibiotic Administration on the Human Normal Intestinal Microbiota Using Culture and Pyrosequencing Methods. Clinical Infectious Diseases. 2015;60:S77–S84. doi: 10.1093/cid/civ137. [DOI] [PubMed] [Google Scholar]
- 50.Modi SR, Collins JJ, Relman DA. Antibiotics and the gut microbiota. Journal of Clinical Investigation. 2014;124:4212–4218. doi: 10.1172/jci72333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Theriot CM, et al. Antibiotic-induced shifts in the mouse gut microbiome and metabolome increase susceptibility to Clostridium difficile infection. Nature Communications. 2014;5 doi: 10.1038/ncomms4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kachrimanidou M, Tsintarakis E. Insights into the Role of Human Gut Microbiota in Clostridioides difficile Infection. Microorganisms. 2020;8:200. doi: 10.3390/microorganisms8020200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Palleja A, et al. Recovery of gut microbiota of healthy adults following antibiotic exposure. Nat Microbiol. 2018;3:1255–1265. doi: 10.1038/s41564-018-0257-9. [DOI] [PubMed] [Google Scholar]
- 54.Wu H, et al. Metformin alters the gut microbiome of individuals with treatment-naive type 2 diabetes, contributing to the therapeutic effects of the drug. Nat Med. 2017;23:850–858. doi: 10.1038/nm.4345. [DOI] [PubMed] [Google Scholar]
- 55.Vich Vila A, et al. Impact of commonly used drugs on the composition and metabolic function of the gut microbiota. Nature Communications. 2020;11 doi: 10.1038/s41467-019-14177-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Seeman MV. The gut microbiome and antipsychotic treatment response. Behavioural Brain Research. 2021;396:112886. doi: 10.1016/j.bbr.2020.112886. [DOI] [PubMed] [Google Scholar]
- 57.Vieira-Silva S, et al. Statin therapy is associated with lower prevalence of gut microbiota dysbiosis. Nature. 2020;581:310–315. doi: 10.1038/s41586-020-2269-x. [DOI] [PubMed] [Google Scholar]
- 58.Weersma RK, Zhernakova A, Fu J. Interaction between drugs and the gut microbiome. Gut. 2020;69:1510–1519. doi: 10.1136/gutjnl-2019-320204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.David LA, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.FooDB. FooDB. < www.foodb.ca>.
- 61.Pérez-Burillo S, Hinojosa-Nogueira D, Pastoriza S, Rufián-Henares JA. Plant extracts as natural modulators of gut microbiota community structure and functionality. Heliyon. 2020;6:e05474. doi: 10.1016/j.heliyon.2020.e05474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Peterson CT, et al. Effects of Turmeric and Curcumin Dietary Supplementation on Human Gut Microbiota: A Double-Blind, Randomized, Placebo-Controlled Pilot Study. Journal of Evidence-Based Integrative Medicine. 2018;23:2515690X1879072. doi: 10.1177/2515690x18790725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sun H, et al. The modulatory effect of polyphenols from green tea, oolong tea and black tea on human intestinal microbiota in vitro. Journal of Food Science and Technology. 2018;55:399–407. doi: 10.1007/s13197-017-2951-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bian X, et al. Saccharin induced liver inflammation in mice by altering the gut microbiota and its metabolic functions. Food and Chemical Toxicology. 2017;107:530–539. doi: 10.1016/j.fct.2017.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rodriguez-Palacios A, et al. The Artificial Sweetener Splenda Promotes Gut Proteobacteria, Dysbiosis, and Myeloperoxidase Reactivity in Crohn’s Disease–Like Ileitis. Inflammatory Bowel Diseases. 2018;24:1005–1020. doi: 10.1093/ibd/izy060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Suez J, et al. Artificial sweeteners induce glucose intolerance by altering the gut microbiota. Nature. 2014;514:181–186. doi: 10.1038/nature13793. [DOI] [PubMed] [Google Scholar]
- 67.Wang Q-P, Browman D, Herzog H, Neely GG. Non-nutritive sweeteners possess a bacteriostatic effect and alter gut microbiota in mice. PLOS ONE. 2018;13:e0199080. doi: 10.1371/journal.pone.0199080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chassaing B, et al. Dietary emulsifiers impact the mouse gut microbiota promoting colitis and metabolic syndrome. Nature. 2015;519:92–96. doi: 10.1038/nature14232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Viennois E, Merlin D, Gewirtz AT, Chassaing B. Dietary Emulsifier–Induced Low-Grade Inflammation Promotes Colon Carcinogenesis. Cancer Research. 2017;77:27–40. doi: 10.1158/0008-5472.can-16-1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chassaing B, Van De Wiele T, De Bodt J, Marzorati M, Gewirtz AT. Dietary emulsifiers directly alter human microbiota composition and gene expression ex vivo potentiating intestinal inflammation. Gut. 2017;66:1414–1427. doi: 10.1136/gutjnl-2016-313099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Commission E. Contaminants. < https://ec.europa.eu/food/safety/chemical_safety/contaminants_en>.
- 72.Medina-Pastor PT, Giuseppe The 2018 European Union report on pesticide residues in food. EFSA Journal. 2020;18 doi: 10.2903/j.efsa.2020.6057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.FDA. Pesticide Residue Monitoring Program Fiscal Year 2018 Pesticide Report 46. FDA - Food and Drug Administration; 2020. [Google Scholar]
- 74.Department for Environment, F. a. R. A. The Expert Committee on Pesticide Residues in Food (PRiF) Annual Report 2020. 2021 [Google Scholar]
- 75.Mesnage R, et al. Impacts of dietary exposure to pesticides on faecal microbiome metabolism in adult twins. Cold Spring Harbor Laboratory; 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Velmurugan G, et al. Gut microbial degradation of organophosphate insecticides-induces glucose intolerance via gluconeogenesis. Genome Biol. 2017;18 doi: 10.1186/s13059-016-1134-6. ARTN 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Aitbali Y, et al. Glyphosate based-herbicide exposure affects gut microbiota, anxiety and depression-like behaviors in mice. Neurotoxicol Teratol. 2018;67:44–49. doi: 10.1016/j.ntt.2018.04.002. [DOI] [PubMed] [Google Scholar]
- 78.Mesnage R, et al. Use of Shotgun Metagenomics and Metabolomics to Evaluate the Impact of Glyphosate or Roundup MON 52276 on the Gut Microbiota and Serum Metabolome of Sprague-Dawley Rats. Environmental Health Perspectives. 2021;129:017005. doi: 10.1289/ehp6990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gushgari AJ, Halden RU. Critical review of major sources of human exposure to N-nitrosamines. Chemosphere. 2018;210:1124–1136. doi: 10.1016/j.chemosphere.2018.07.098. [DOI] [PubMed] [Google Scholar]
- 80.Bharate SS. Critical Analysis of Drug Product Recalls due to Nitrosamine Impurities. Journal of Medicinal Chemistry. 2021;64:2923–2936. doi: 10.1021/acs.jmedchem.0c02120. [DOI] [PubMed] [Google Scholar]
- 81.FDA. Information about Nitrosamine Impurities in Medications. 2021. < https://www.fda.gov/drugs/drug-safety-and-availability/information-about-nitrosamine-impurities-medications#:~:text=Nitrosamine%20impurities%20may%20increase%20the,an%20increased%20risk%20of%20cancer.>.
- 82.Ha WS, Kim CK, Song SH, Kang CB. Study on mechanism of multistep hepatotumorigenesis in rat: development of hepatotumorigenesis. J Vet Sci. 2001;2:53–58. [PubMed] [Google Scholar]
- 83.Lijinsky W, Reuber MD. Dose-response study with N-nitrosodiethanolamine in F344 rats. Food and Chemical Toxicology. 1984;22:23–26. doi: 10.1016/0278-6915(84)90047-4. [DOI] [PubMed] [Google Scholar]
- 84.Alshannaq A, Yu JH. Occurrence, Toxicity, and Analysis of Major Mycotoxins in Food. Int J Env Res Pub He. 2017;14 doi: 10.3390/ijerph14060632. ARTN 632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Eskola M, et al. Worldwide contamination of food-crops with mycotoxins: Validity of the widely cited ‘FAO estimate’ of 25% Critical Reviews in Food Science and Nutrition. 2020;60:2773–2789. doi: 10.1080/10408398.2019.1658570. [DOI] [PubMed] [Google Scholar]
- 86.Guo M, et al. Combination of Metagenomics and Culture-Based Methods to Study the Interaction Between Ochratoxin A and Gut Microbiota. Toxicological Sciences. 2014;141:314–323. doi: 10.1093/toxsci/kfu128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Vignal C, et al. Chronic ingestion of deoxynivalenol at human dietary levels impairs intestinal homeostasis and gut microbiota in mice. Archives of Toxicology. 2018;92:2327–2338. doi: 10.1007/s00204-018-2228-6. [DOI] [PubMed] [Google Scholar]
- 88.Giambò F, et al. Influence of toxic metal exposure on the gut microbiota (Review) World Academy of Sciences Journal. 2021;3 doi: 10.3892/wasj.2021.90. [DOI] [Google Scholar]
- 89.Zackular JP, et al. Dietary zinc alters the microbiota and decreases resistance to Clostridium difficile infection. Nature Medicine. 2016;22:1330–1334. doi: 10.1038/nm.4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.EFSA. Report for 2018 on the results from the monitoring of veterinary medicinal product residues and other substances in live animals and animal products. EFSA Supporting Publications. 2020;17 doi: 10.2903/sp.efsa.2020.en-1775. [DOI] [Google Scholar]
- 91.Roca-Saavedra P, et al. Food additives, contaminants and other minor components: effects on human gut microbiota-a review. J Physiol Biochem. 2018;74:69–83. doi: 10.1007/s13105-017-0564-2. [DOI] [PubMed] [Google Scholar]
- 92.Maier L, et al. Extensive impact of non-antibiotic drugs on human gut bacteria. Nature. 2018;555:623–628. doi: 10.1038/nature25979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cabreiro F, et al. Metformin Retards Aging in C. elegans by Altering Microbial Folate and Methionine Metabolism. Cell. 2013;153:228–239. doi: 10.1016/j.cell.2013.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Grimsey EM, et al. Chlorpromazine and Amitriptyline Are Substrates and Inhibitors of the AcrB Multidrug Efflux Pump. mBio. 2020;11 doi: 10.1128/mbio.00465-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lu Q-Y, et al. Prebiotic Potential and Chemical Composition of Seven Culinary Spice Extracts. Journal of Food Science. 2017;82:1807–1813. doi: 10.1111/1750-3841.13792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Pan H, Feng J, He G-X, Cerniglia CE, Chen H. Evaluation of impact of exposure of Sudan azo dyes and their metabolites on human intestinal bacteria. Anaerobe. 2012;18:445–453. doi: 10.1016/j.anaerobe.2012.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Crudo F, et al. In vitro interactions of Alternaria mycotoxins, an emerging class of food contaminants, with the gut microbiota: a bidirectional relationship. Archives of Toxicology. 2021 doi: 10.1007/s00204-021-03043-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Klünemann M, et al. Bioaccumulation of therapeutic drugs by human gut bacteria. Nature. 2021 doi: 10.1038/s41586-021-03891-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Frame LA, Costa E, Jackson SA. Current explorations of nutrition and the gut microbiome: a comprehensive evaluation of the review literature. Nutrition Reviews. 2020;78:798–812. doi: 10.1093/nutrit/nuz106. [DOI] [PubMed] [Google Scholar]
- 100.Ruiz-Ojeda FJ, Plaza-Díaz J, Sáez-Lara MJ, Gil A. Effects of Sweeteners on the Gut Microbiota: A Review of Experimental Studies and Clinical Trials. Advances in Nutrition. 2019;10:S31–S48. doi: 10.1093/advances/nmy037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Teresita de J Bello González TZ, Bor Gerrit, Smidt Hauke, van Passel MarkWJ. Study of the Aminoglycoside Subsistence Phenotype of Bacteria Residing in the Gut of Humans and Zoo Animals. Frontiers in Microbiology. 2016 doi: 10.3389/fmicb.2015.01550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Xin Z, et al. Isolation, identification and characterization of human intestinal bacteria with the ability to utilize chloramphenicol as the sole source of carbon and energy. FEMS Microbiology Ecology. 2012;82:703–712. doi: 10.1111/j.1574-6941.2012.01440.x. [DOI] [PubMed] [Google Scholar]
- 103.Taguer M, Maurice C. The complex interplay of diet, xenobiotics, and microbial metabolism in the gut: Implications for clinical outcomes. Clinical Pharmacology & Therapeutics. 2016;99:588–599. doi: 10.1002/cpt.366. [DOI] [PubMed] [Google Scholar]
- 104.Pryor R, et al. Host-Microbe-Drug-Nutrient Screen Identifies Bacterial Effectors of Metformin Therapy. Cell. 2019;178:1299–1312.:e1229. doi: 10.1016/j.cell.2019.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Anwar S, et al. Trigonelline inhibits intestinal microbial metabolism of choline and its associated cardiovascular risk. Journal of Pharmaceutical and Biomedical Analysis. 2018;159:100–112. doi: 10.1016/j.jpba.2018.06.027. [DOI] [PubMed] [Google Scholar]
- 106.Sutcliffe SG, Shamash M, Hynes AP, Maurice CF. Common Oral Medications Lead to Prophage Induction in Bacterial Isolates from the Human Gut. Viruses. 2021;13:455. doi: 10.3390/v13030455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Viennois E, et al. Dietary Emulsifiers Directly Impact Adherent-Invasive E. coli Gene Expression to Drive Chronic Intestinal Inflammation. Cell Reports. 2020;33:108229. doi: 10.1016/j.celrep.2020.108229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Collins J, et al. Dietary trehalose enhances virulence of epidemic Clostridium difficile. Nature. 2018;553:291–294. doi: 10.1038/nature25178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Shil A, Chichger H. Artificial Sweeteners Negatively Regulate Pathogenic Characteristics of Two Model Gut Bacteria, E. coli and E. faecalis. International Journal of Molecular Sciences. 2021;22:5228. doi: 10.3390/ijms22105228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Chhabra RS. Intestinal absorption and metabolism of xenobiotics. Environ Health Perspect. 1979 doi: 10.1289/ehp.793361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Scott TA, et al. Host-Microbe Co-metabolism Dictates Cancer Drug Efficacy in C. elegans. Cell. 2017;169:442–456.:e418. doi: 10.1016/j.cell.2017.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.García-González AP. Bacterial Metabolism Affects the C. elegans Response to Cancer Chemotherapeutics. Cell. 2017;169:431–441.:e438. doi: 10.1016/j.cell.2017.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Md Masud Parvez AB, Jariwala Parth B, Gáborik Zsuzsanna, Kis Emese, Heyward Scott, Redinbo Matthew R, Prasad Bhagwat. Quantitative Investigation of Irinotecan Metabolism, Transport, and Gut Microbiome Activation. Drug Metabolism and Disposition. 2021 doi: 10.1124/dmd.121.000476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Geller LT, et al. Potential role of intratumor bacteria in mediating tumor resistance to the chemotherapeutic drug gemcitabine. Science. 2017;357:1156–1160. doi: 10.1126/science.aah5043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Artacho A, et al. The Pretreatment Gut Microbiome Is Associated With Lack of Response to Methotrexate in New-Onset Rheumatoid Arthritis. Arthritis & Rheumatology. 2021;73:931–942. doi: 10.1002/art.41622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Gratz SW, et al. Masked trichothecene and zearalenone mycotoxins withstand digestion and absorption in the upper GI tract but are efficiently hydrolyzed by human gut microbiota in vitro. Mol Nutr Food Res. 2017;61 doi: 10.1002/mnfr.201600680. ARTN 1600680. [DOI] [PubMed] [Google Scholar]
- 117.Daud N, et al. Prevalent Human Gut Bacteria Hydrolyse and Metabolise Important Food-Derived Mycotoxins and Masked Mycotoxins. Toxins. 2020;12 doi: 10.3390/toxins12100654. ARTN 654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.He Z, et al. Food colorants metabolized by commensal bacteria promote colitis in mice with dysregulated expression of interleukin-23. Cell Metabolism. 2021;33:1358–1371.:e1355. doi: 10.1016/j.cmet.2021.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Shimada T, Fujii-Kuriyama Y. Metabolic activation of polycyclic aromatic hydrocarbons to carcinogens by cytochromes P450 1A1 and1B1. Cancer Science. 2004;95:1–6. doi: 10.1111/j.1349-7006.2004.tb03162.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Van de Wiele T, et al. Human colon microbiota transform polycyclic aromatic hydrocarbons to estrogenic metabolites. Environmental Health Perspectives. 2005;113:6–10. doi: 10.1289/ehp.7259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Goodson WH, et al. Activation of the mTOR pathway by low levels of xenoestrogens in breast epithelial cells from high-risk women. Carcinogenesis. 2011;32:1724–1733. doi: 10.1093/carcin/bgr196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Sauer SJ, et al. Bisphenol A activates EGFR and ERK promoting proliferation, tumor spheroid formation and resistance to EGFR pathway inhibition in estrogen receptor-negative inflammatory breast cancer cells. Carcinogenesis. 2017;38 doi: 10.1093/carcin/bgx003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Brochado AR, et al. Species-specific activity of antibacterial drug combinations. Nature. 2018;559:259–263. doi: 10.1038/s41586-018-0278-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Maier L, et al. Dissecting the collateral damage of antibiotics on gut microbes. Cold Spring Harbor Laboratory; 2020. [Google Scholar]
- 125.Farha MA, Brown ED. Chemical Probes of Escherichia coli Uncovered through Chemical-Chemical Interaction Profiling with Compounds of Known Biological Activity. Chemistry & Biology. 2010;17:852–862. doi: 10.1016/j.chembiol.2010.06.008. [DOI] [PubMed] [Google Scholar]
- 126.Erickson TB, et al. “Waste Not, Want Not” — Leveraging Sewer Systems and Wastewater-Based Epidemiology for Drug Use Trends and Pharmaceutical Monitoring. Journal of Medical Toxicology. 2021;17:397–410. doi: 10.1007/s13181-021-00853-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Zeng X, et al. MASI: microbiota—active substance interactions database. Nucleic Acids Research. 2021;49:D776–D782. doi: 10.1093/nar/gkaa924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Aziz RK, Saad R, Rizkallah MR. PharmacoMicrobiomics or how bugs modulate drugs: an educational initiative to explore the effects of human microbiome on drugs. BMC Bioinformatics. 2011;12:A10. doi: 10.1186/1471-2105-12-s7-a10. [DOI] [Google Scholar]
- 129.Lynch SV, Ng SC, Shanahan F, Tilg H. Translating the gut microbiome: ready for the clinic? Nature Reviews Gastroenterology & Hepatology. 2019;16:656–661. doi: 10.1038/s41575-019-0204-0. [DOI] [PubMed] [Google Scholar]
- 130.Taroncher-Oldenburg G, et al. Translating microbiome futures. Nature Biotechnology. 2018;36:1037–1042. doi: 10.1038/nbt.4287. [DOI] [PubMed] [Google Scholar]
- 131.Liu X, et al. Magnetic Living Hydrogels for Intestinal Localization, Retention, and Diagnosis. Advanced Functional Materials. 2021;31:2010918. doi: 10.1002/adfm.202010918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Mark Mimee PN, Hayward Alison, Carim Sean, Flanagan Sarah, Jerger Logan, Collins Joy, McDonnell Shane, Swartwout Richard, Citorik RobertJ, Bulović Vladimir, Langer Robert, et al. An ingestible bacterial-electronic system to monitor gastrointestinal health. Science. 2018 doi: 10.1126/science.aas9315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.PA. PFAS Master List of PFAS Substances (Version 2) 2020. < https://comptox.epa.gov/dashboard/chemical_lists/PFASMASTER>.
- 134.EPA. PESTICIDES|EPA: List of Active Ingredients UPDATED 10/25/2019. 2019. < https://comptox.epa.gov/dashboard/chemical_lists/PESTACTIVES>.
- 135.EFSA. Scientific Opinion on Mineral Oil Hydrocarbons in Food. EFSA Journal. 2012;10 doi: 10.2903/j.efsa.2012.2704. [DOI] [Google Scholar]
- 136.EFSA. Results of the monitoring of dioxin levels in food and feed. EFSA Journal. 2010;8 doi: 10.2903/j.efsa.2010.1385. [DOI] [Google Scholar]
- 137.EFSA. Results of the monitoring of non dioxin-like PCBs in food and feed. EFSA Journal. 2010;8:1701. doi: 10.2903/j.efsa.2010.1701. [DOI] [Google Scholar]
- 138.EPA. LIST: Dioxins and dioxin-like compounds. < https://comptox.epa.gov/dashboard/chemical_lists/DIOXINS>.
- 139.EFSA. Metals as contaminants in food. < https://www.efsa.europa.eu/en/topics/topic/metals-contaminants-food>.
- 140.Groh KJ, Geueke B, Martin O, Maffini M, Muncke J. Overview of intentionally used food contact chemicals and their hazards. Environment International. 2021;150:106225. doi: 10.1016/j.envint.2020.106225. [DOI] [PubMed] [Google Scholar]
- 141.Groh KJ, et al. Overview of known plastic packaging-associated chemicals and their hazards. Science of The Total Environment. 2019;651:3253–3268. doi: 10.1016/j.scitotenv.2018.10.015. [DOI] [PubMed] [Google Scholar]
- 142.EPA. LIST: Azo Dyes. < https://comptox.epa.gov/dashboard/chemical_lists/AZODYES>.
- 143.EPA. NORMAN|LIST: Bisphenol Compounds. 2017. < https://comptox.epa.gov/dashboard/chemical_lists/BISPHENOLS>.
- 144.ELSIE. Extractables and Leachables Database. 2020. < https://www.elsiedata.org/elsie-database>.
- 145.FDA. Drugs@FDA. 2021. < https://www.fda.gov/drugs/drug-approvals-and-databases/drugsfda-data-files>.
- 146.FDA. Inactive Ingredient Search for Approved Drug Products. 2021. < https://www.fda.gov/drugs/drug-approvals-and-databases/inactive-ingredients-database-download>.
- 147.Agroscope. Toxic Plants − Phytotoxin (TPPT) Database. 2018. < https://www.agroscope.admin.ch/agroscope/en/home/publications/apps/tppt.html>.
- 148.EPA. LIST: Mycotoxins. < https://comptox.epa.gov/dashboard/chemical_lists/MYCOTOX2>.
- 149.FSA. Approved additives and E numbers. < https://www.food.gov.uk/business-guidance/approved-additives-and-e-numbers>.
- 150.EPA. PHARMACEUTICALS| WIKILIST: Veterinary Drugs. < https://comptox.epa.gov/dashboard/chemical_lists/VETDRUGS>.
- 151.EPA. CATEGORY: Polycyclic Aromatic Hydrocarbons (PAHs) < https://comptox.epa.gov/dashboard/chemical_lists/PAHLIST>.
- 152.NIH. Commonly Used Drugs Charts. 2020. < https://www.drugabuse.gov/drug-topics/commonly-used-drugs-charts>.
- 153.FAOSTAT. Fertilizers by Product. 2020. < http://www.fao.org/faostat/en/?#data/RFB>.
- 154.WHO. Exposome Explorer. 2021. < http://exposome-explorer.iarc.fr/>.
- 155.HMDB. Human Metabolome Database (HMDB) < https://hmdb.ca/metabolites?utf8=%E2%9C%93&filter=true&feces=1&food=1&plant=1&toxin=1&drug=1&drug_metabolite=1&filter=true> (
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.