Abstract
The invasion of human erythrocytes by Plasmodium falciparum merozoites requires interaction between parasite ligands and host receptors. Interaction of PfRh5-CyRPA-Ripr protein complex with basigin, an erythrocyte surface receptor, via PfRh5 is essential for erythrocyte invasion. Antibodies raised against each antigen component of the complex have demonstrated erythrocyte invasion inhibition, making these proteins potential blood-stage vaccine candidates. Genetic polymorphisms present a significant challenge in developing efficacious vaccines, leading to variant-specific immune responses. This study investigated the genetic variations of the PfRh5 complex proteins in P. falciparum isolates from Lake Victoria islands, Western Kenya. Here, twenty-nine microscopically confirmed P. falciparum field samples collected from islands in Lake Victoria between July 2014 and July 2016 were genotyped by whole genome sequencing, and results compared to sequences mined from the GenBank database, from a study conducted in Kilifi, as well as other sequences from the MalariaGEN repository. We analyzed the frequency of polymorphisms in the PfRh5 protein complex proteins, PfRh5, PfCyRPA, PfRipr, and PfP113, and their location mapped on the 3D protein complex structure. We identified a total of 58 variants in the PfRh5 protein complex. PfRh5 protein was the most polymorphic with 30 SNPs, while PfCyRPA was relatively conserved with 3 SNPs. The minor allele frequency of the SNPs ranged between 1.9% and 21.2%. Ten high-frequency alleles (>5%) were observed in PfRh5 at codons 147, 148, 277, 410, and 429 and in PfRipr at codons 190, 255, 259, and 1003. A SNP was located in protein-protein interaction region C203Y and F292V of PfRh5 and PfCyRPA, respectively. Put together, this study revealed low polymorphisms in the PfRh5 invasion complex in the Lake Victoria parasite population. However, the two mutations identified on the protein interaction regions prompts for investigation on their impacts on parasite invasion process to support the consideration of PfRh5 components as potential malaria vaccine candidates.
Keywords: genetic variations, Rh5-CyRPA-Ripr invasion complex, malaria, vaccines, erythrocyte (RBC)
Background
The World Health Organization (WHO) estimates the latest global malaria health burden statistic at 627 000 deaths resulting from 241 million malaria infection cases (1). The scourge’s heaviest health and economic burden is borne by the developing countries in Sub-Saharan Africa, where an estimated 90% of all malaria deaths occur, with children under five accounting for 78% of all deaths (2). The emergence of multi-drug resistance Plasmodium falciparum (P. falciparum) resistant strains and insecticide resistance mosquitos remains a significant challenge in treating and eliminating malaria (3, 4). The lack of an effective vaccine remains one of the most critical gaps in the strategies developed to eliminate P. falciparum malaria (5).
The development of an effective P. falciparum vaccine focuses on targeting pre-erythrocytic or erythrocytic stages for parasite development and malaria pathogenesis in humans or the parasite development within the mosquito vector (6). Symptoms elicited by parasite infection originate from the erythrocytic stage of malaria infection. At this stage, the merozoite invades the erythrocytes, where at initial recognition of the human erythrocytes, the merozoite orients itself such that the apical region comes to direct contact with the host’s erythrocyte membrane, followed by irreversible attachment of merozoites to erythrocytes (7). Ring-like moving junction mediates complete parasite internalization to formation of an intracellular parasitophorous vacuole (8). The whole erythrocyte invasion process is mediated by multiple merozoite proteins mainly expressed on the surface, or in the apical organelles such as rhoptry and microneme (9). Since these proteins are essential for invasion and are exposed to host immune system, they are considered ideal targets for blood stage vaccines (BSV) (10–12). However, the exposure of candidate BSV antigens to human immune system during natural infections subjects them to selective pressure, that may result to high levels of polymorphisms (13). This presents a significant challenge for allele-specific immune responses as an ideal vaccine must be able to protect against multiple genetic variants of parasites (14, 15).
The P. falciparum reticulocyte binding homolog 5 complex (PfRh5) is a primary vaccine target for developing an effective malaria vaccine. The PfRh5 complex comprises four interacting proteins: Pf- reticulocyte binding homolog 5 (Rh5), Pf- interacting protein (PfRipr), Pf-Cysteine-rich protective antigen (CyRPA), and Pf-P113 protein (16). PfRh5 proteins bind to erythrocytes via the host receptor basigin, while the other three proteins interact within the complex to initiate erythrocyte invasion. PfCyRPA binds directly to PfRh5, while PfRipr interacts with PfCyRPA; thus, PfCyRPA forms the contact sites for PfRh5 and PfRipr. Studies have shown that PfP113 interacts with PfRh5 protein on the N-terminal, providing a releasable mechanism for anchoring PfRh5 to basigin (11, 17). The genes encoding for these proteins are highly maintained, as shown in gene knockout experiments suggesting they are vital for parasite survival (18, 19). Antibodies against PfRh5, PfRipr, and PfCyRPA have been shown to inhibit parasite erythrocyte invasion in non-human primates and mice, while antibodies against PfP113 protein have been associated with protection against clinical malaria in vivo (20, 21). These studies suggest that all proteins of the PfRh5 complex can be considered potential BSV targets.
Polymorphisms in all PfRh5 complex encoding genes could impede the development of an Rh5 malaria BSV. Like the PfRh5 complex, apical membrane antigen 1 (AMA1), once considered a potential malaria vaccine candidate is also essential for invasion. However, AMA1 is highly polymorphic, leading to allele-specific immune responses and limited efficacy in its Phase IIb trials (22). Investigation into polymorphisms on all members of the PfRh5 complex, their effects on the protein structure, and their association are significant considerations when designing a vaccine. Studies have demonstrated that P. falciparum parasites have a high within host genetic diversity in high transmission regions compared to low transmission settings (23, 24). This is due to the increased probability of recombination between genetically distinct variants in high transmission settings. This extensive genetic diversity is a major hindrance in malaria vaccine development as the host immune responses may fail to recognize all the variants of an antigen (25).
Here, to explore these questions, we analyzed the four genes of the PfRh5 complex by whole genome sequencing in a cross-sectional sample of parasites from two high malaria transmission regions in Kenya.
Methods
Sampling, DNA preparation, and whole genome sequencing
Parasite DNA was extracted from archived whole blood samples from patients recruited for a drug resistance surveillance study in local hospitals on four selected islands (Mfangano, Takawiri, Kibuogi, and Ngodhe) in Lake Victoria, a coastal mainland (Ungoye) between July 2014 and July 2016. The study’s approval was obtained from the Kenyatta National Hospital - University of Nairobi (KNH-UoN) ethical review committee (P609/10/2014) and the Mount Kenya University Ethics Review Committee (038/2014). Written consent was obtained from all the participants or guardians, and malaria cases were treated per the national malaria guidelines. The samples re-analyzed here were a subset of these studies which has been extensively described elsewhere (26, 27). Briefly, to increase the parasitemia, the field P. falciparum parasites were adapted for in vitro culture as previously described (28), and DNA was extracted from short-term cultures (1 month) at the schizont stage using QIAamp DNA mini kit (Qiagen, Valencia, CA). Paired-end sequencing libraries were prepared using Nextera XT DNA library preparation Kit according to the manufactures protocol. (Illumina, USA). Whole genome sequencing was performed on Illumina MiSeq technology (Illumina, USA) at 30X coverage generating reads of length 150 bps. These sequences are archived at the DDBJ BioProject, Accession number PRJDB12148. Quality control checks were performed using the FASTQC (Babraham Institute, UK) toolkit version 0.11.5.
Comparison of polymorphisms identified with other regions
For comparative analysis, we obtained previously reported whole genome sequences P. falciparum isolates collected from Kilifi, a malaria endemic region in coastal Kenya (29, 30). The mined sequences were generated from two drug trial studies that were conducted between 2005 and 2008, and the sequences deposited in the GenBank repository under accession numbers PfRipr: MW597717-MW597776, PfRh5: MW597550-MW597609, PfCyRPA : MW597610-MW597716, and PfP113: MW597459-MW597549.
We also accessed the catalogue of genetic variation in P. falciparum, of the global MalariaGEN database v6.0, for comparing and validating the SNPs identified from the Lake Victoria sample population. This dataset comprised of genomic variation records of 7,113P. falciparum samples from 28 malaria-endemic countries. The method used to retrieve the data was previously described by Amato et al., 2016. The dplyr v1.0.9 package (Wickham H, François R, Henry L, 2022) in R v4.2.1 was used to filter out the four genes of the PfRh5 complex using their PlasmoDB unique identifiers. SNPs identified were then filtered and analyzed.
Read mapping and coverage
Sequence reads were aligned against Plasmodium falciparum 3D7 reference genome (version 8.1) (https://plasmodb.org/common/downloads/release-46/Pfalciparum3D7/fasta/data/) using Burrows-Wheeler Alignment tool (BWA) (31) (http://bio-bwa.sourceforge.net) with default parameters. The resulting alignment was further processed with Samtools (32) and Picard v1.66 (33) to remove duplicates. SNPs were called using Genome Analysis Toolkit (GATK) HaplotypeCaller with the following parameters – genotyping mode DISCOVERY, –output mode EMIT_VARIANTS_ONLY, –stand_emit_conf 10, and – stand_call_conf 30. To improve the quality of variant calling, we further discarded genotyping calls with coverage of <5 reads. The resulting variant call format (VCF) files were then merged into one file using VCF tools (34).
Variant calling and analysis
The VCF file containing twenty-six samples that passed the quality test from read mapping analysis was used as the input file in VCF tools for variant analysis. High-quality SNPs in four target genes, PfRh5, PfCyRPA, PfRipr, and PfP113, were functionally annotated in the SNPEFF tool (35). Called variants were further analyzed in ARTEMIS software (36). MEGA 7 tool was used to perform multiple sequence alignment and translation of nucleotide sequences to amino acid sequences. Gene variants were identified by aligning the amino acid sequence reads to their corresponding 3D7 reference gene sequence. To test sensitivity of the above approach, we analyzed the sequences at different variant calling parameters to assess the impact of these settings on downstream analysis. Additionally, prior to variant calling, we performed base quality score recalibration to adjust the base quality scores of sequencing reads as well as local realignment around indels to reduce false-positive variant calls resulting from alignment artifacts.
Population genetics analysis
The population genetic tests for the neutral theory of evolution (37) and Tajima’s D and Fu & Li’s statistics and nucleotide diversity (Pi) were calculated using DnaSPv6.1 (38). Tajima’s D tested departure from neutrality based on allele frequency distribution in each gene. Fu and Li’s D test statistic calculated the variation between the observed number of singletons and the total number of mutations. Pi was used to test the genetic diversity of each gene of the PfRh5 complex within the parasite population from Lake Victoria region. The P.falciparum adenylosuccinate lyase gene, a house keeping gene and apical membrane antigen gene were used as control in this analysis. The sequences for these genes were obtained from Lake Victoria parasite population.
Protein structures
The structure of the Rh5-CyRPA-Ripr complex was retrieved from the Protein Data Bank (http://www.rcsb.org/) under the protein ID 6MPV. The datasets generated from this study were used to map the polymorphic sites of PfRh5 and PfCyRPA protein structures on the Rh5-CyRPA-Ripr complex 3D structure in Pymol (The PyMOL Molecular Graphics System, Version 2.2.0, Schrödinger, LLC) to determine the location of the SNPs in the 3D protein structure and whether the SNPs were localized in the protein-protein interaction regions of the complex.
Results
Genetic variation in the PfRh5 complex genes
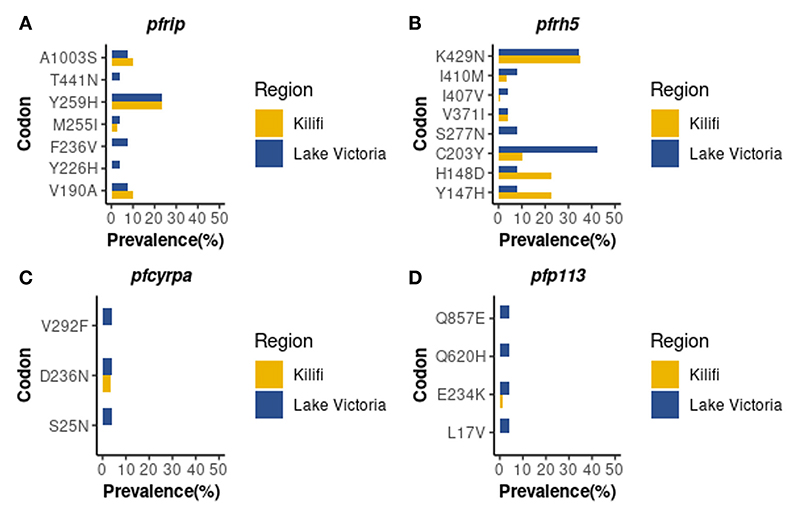
Whole genome sequence analysis data for the four genes of the PfRh5 complex were obtained from 26 samples from the Lake Victoria islands.A total of 45, 35, 25, and 3 Non-synonymous SNPs were identified within the PfP113, PfRipr, PfRh5, and in the PfCyRPA genes, respectively. The minor allele frequency in the four genes ranged from 0.7% to 24.06%. High-frequency alleles (>5%) were identified in codons Y147, H148, C203, S277, I410, and K429 of the PfRh5 gene and codons V190, M255, Y259, and A1003 of the PfRipr gene (Figure 1). Non-synonymous SNPs identified in PfCyRPA gene at codons S25, D236, and V292 and PfP113 gene at codons L17, E234, Q620, and Q857 occurred at low frequency (Figure 1).
Figure 1.
Prevalence of PfRh5 complex polymorphisms. Frequency in percentage of single nucleotide polymorphisms identified across four genes of the Rh5 invasion complex (A) PfRipr, (B) PfRh5, (C) PfCyRPA (D) PfP113 in Lake Victoria (n=26) and Kilifi (n=68) populations.
Comparison of polymorphisms identified with the Kilifi population
A total of nine non-synonymous previously not observed from the Kilifi population were identified across the four genes in Lake Victoria isolates (39). PfRipr gene at codon Y226, F236, and T441, PfRh5 at codon S277, PfP113 at codon L17, Q620, and Q857, and PfCyRPA gene at codon S25 and V292 (Figure 1).
Comparison of polymorphisms identified with global MalariaGEN
We further explored the MalariaGEN data to establish whether the variants identified were also present in the global database. We also screened for variants missed due to differences in analysis methodologies. This analysis confirmed that most of the variants observed from the Lake Victoria population had been previously observed elsewhere and deposited in the global MalariaGEN database giving confidence in our analysis methodology on the probability of missing variants (Table 1).
Table 1. List of high frequency SNPs identified in Lake Victoria and Kilifi parasite population.
| Gene | Position | Ref | Alt | S/NS | Lake Victoria (n=26) | Kilifi (n=68) |
MalariaGEN global dataset (n=7,113) |
|---|---|---|---|---|---|---|---|
| pfripr | pfripr_190 | V | A | NS | 7.7 | 10.3 | + |
| pfripr_226* | Y | H | NS | 3.8 | 0 | + | |
| pfripr_236* | F | V | NS | 3.8 | 0 | - | |
| pfripr_255 | M | I | NS | 7.7 | 2.8 | - | |
| pfripr_259 | Y | H | NS | 23.1 | 23.6 | + | |
| pfripr_441* | T | N | NS | 3.8 | 0 | - | |
| pfripr_1003 | A | S | NS | 7.7 | 10.3 | + | |
| pfrh5 | pfrh_147 | Y | H | NS | 7.7 | 22.6 | + |
| pfrh_148 | H | D | NS | 7.7 | 22.6 | + | |
| pfrh_203 | C | Y | NS | 42.3 | 10.3 | + | |
| pfrh_277 | S | N | NS | 7.7 | 0 | - | |
| pfrh_371* | V | I | NS | 3.8 | 4.1 | + | |
| pfrh_407* | I | V | NS | 3.8 | 0.7 | + | |
| pfrh_410 | I | M | NS | 7.7 | 3.4 | + | |
| pfrh_429 | K | N | NS | 34.6 | 34.9 | + | |
| pfcyrpa | pfcyrpa_25* | S | N | NS | 3.8 | 0 | - |
| pfcyrpa_236* | D | N | NS | 3.8 | 3.4 | - | |
| pfcyrpa_292* | V | F | NS | 3.8 | 0 | + | |
| pfp113 | pfp113_17* | L | V | NS | 3.8 | 0 | - |
| pfp113_234* | E | K | NS | 3.8 | 1.4 | + | |
| pfp113_620* | Q | H | S | 3.8 | 0 | - | |
| pfp113_857* | Q | E | NS | 3.8 | 0 | - |
Polymorphisms identified in both populations and the global MalariaGEN dataset are indicated with a plus +, and singleton SNPs are indicated by an asterisk*.
Ref refers to the 3D7 Plasmodium falciparum strain reference amino acid, while Alt refers to the amino acid variation. The SNPs are classified under S (synonymous or NS (non-synonymous) and frequency of SNPs is expressed as percentages.
Population genetics statistics
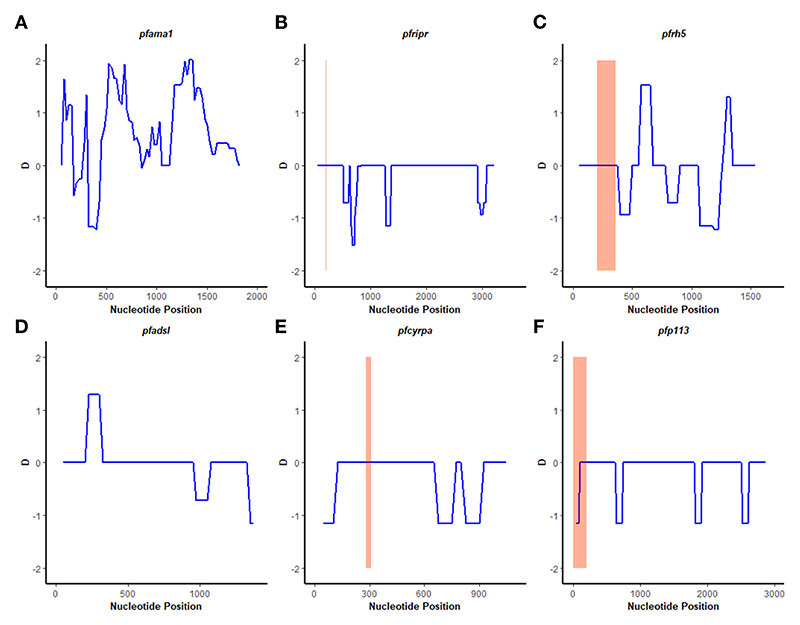
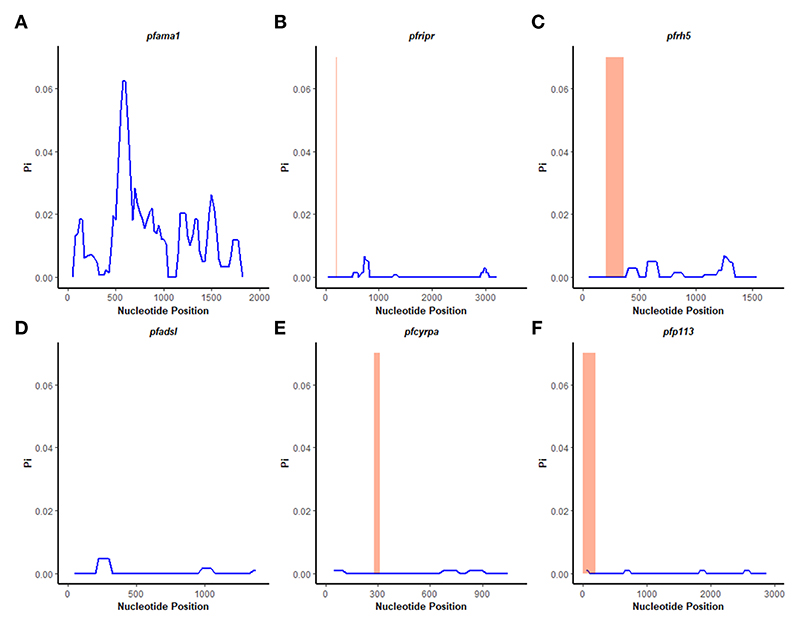
A sliding window approach was used to calculate the nucleotide diversity (Pi) and Tajima’s D statistics. All four genes had a negative neutrality summary statistic. Analysis revealed that the PfP113 gene was the most conserved relative to the other three genes of the PfRh5 complex and the positive control Pfama1 gene, with a Tajima’s D summary statistic of – 1.89 and a Pi of 0.00010 with 4 singleton mutations distributed along the 1578 bp nucleotide sequence. Relative to the negative control Pfadsl gene, the PfRh5 gene was the least conserved, with 25 non-synonymous polymorphisms distributed along the entire 2907 bp nucleotide sequence with a Tajima’s D summary value of -0.56 and a Pi of 0.00109 (Figures 2, 3). The Fu & Li’s statistics were not significant for PfRh5 and PfRipr genes. PfCyRPA and PfP113 gene had significant Fu & Li’s values, p < 0.05. (Table 2)
Figure 2.
Tajima’s D analysis. Sliding window analysis of Tajima’s D test for neutrality for the four genes of the Rh5 invasion complex of twenty-six samples obtained from Lake Victoria region. (A) Pfama1, (B) PfRipr, (C) pfrh5, and (D) pfadsl (E) pfcyrpa (F) pfp113 Tajimas’ D was calculated in DnaSP v6.1 software with a window length of 100 and a step size of 25 bases. D values are plotted against the mid-point of window length. The highlighted region indicates the basigin – Pfrh5 binding site and protein-protein interactions regions for Pfripr, Pfcyrpa, and Pfp113.
Figure 3.
Nucleotide diversity analysis. Sliding window analysis of nucleotide diversity (Pi) per site to compare genetic diversity in four genes of the Rh5 invasion complex of twenty-six samples obtained from Lake Victoria region. (A) pfama1, (B) pfripr, (C) pfrh5, and (D) pfadsl (E) pfcyrpa (F) pfp113. Pi is nucleotide diversity calculated using DnaSP ver. 6.1 with a window length of 100 bases and a step size of 25 bases, plotted against the window length’s midpoint. The highlighted region indicates the basigin – PfRh5 binding residues and protein-protein interactions regions for PfRipr, PfCyrpa, and PfP113.
Table 2. Fu & Li’s neutrality tests statistics based on Lake Victoria sample population.
| Gene | No. of Samples | S | Fu & Li’s D* | Fu & Li’s F* |
|---|---|---|---|---|
| pfcyrpa | 26 | 3 | -2.58495* | -2.7088* |
| pfp113 | 26 | 4 | -2.86849* | -2.99593* |
| pfrh5 | 26 | 8 | 0.10999 | -0.10381 |
| pfrirp | 26 | 7 | -1.11002 | -1.37673 |
| pf ama1 | 26 | 38 | 4.28469 | 3.98709 |
| pf adsl | 26 | 3 | -0.21602 | -0.25135 |
S refers to segregation sites. Asterisk * indicates significance, p > 0.05.
Polymorphisms on the PfRh5 protein complex
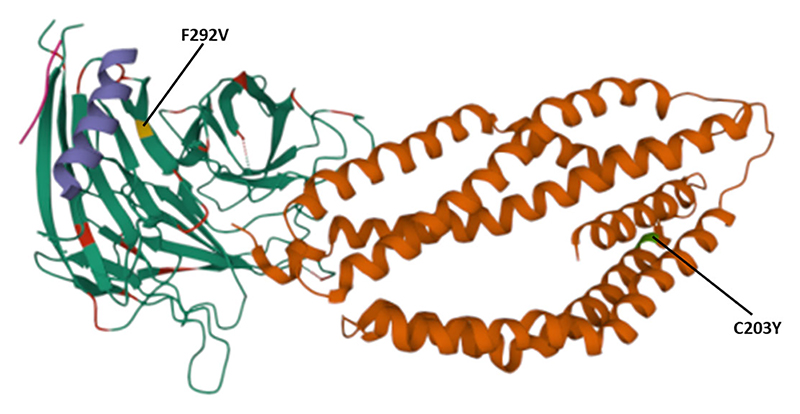
The polymorphisms established from our dataset were mapped on the PfRh5 protein complex to show whether they occurred within known protein-protein interacting regions and the PfRh5 – basigin interaction region. The previously published Rh5-CyRPA-Ripr invasion complex (16) was superimposed with the basigin structure to show the interaction of PfRh5 with basigin. PfRh5 binds to basigin via His- 102 linker, α -2, α -4, and a disulfide loop (Cys345–Cys351) (40). The F350 and W447 PfRh5 residues stabilize binding by packing into basigin hydrophobic bonds. Only one SNP corresponding to codon 203 within the PfRh5-basigin interacting region was identified. PfRipr binds to PfCyRPA blade 6 at amino acid residues 281 – 311. One mutation corresponding to this interaction region at codon 292 of PfCyRPA was identified. No mutation corresponding to PfCyRPA and PfRh5 binding regions was identified. Other polymorphisms were localized outside the protein interaction site (Figure 4).
Figure 4.
Location of polymorphisms on the 3D structure. Shows the SNPs identified that fall within the protein-protein and protein- basigin interaction regions of the Rh5 (brown), CyRPA (green) and Ripr (purple) protein complex. Polymorphic residues C203Y identified on Rh5 protein and F292V on CyRPA are highlighted in green and yellow, respectively. The Rh5 C203 mutation falls within the Basigin-Rh5 protein interaction region, while the CyRPA F292 mutation is located on blade 6, Ripr – CyRPA proteins interaction region.
Discussion
P. falciparum infects and replicates in human host erythrocytes leading to manifestation of clinical of malaria. The invasion process by invasive merozoites involves the interaction of PfRh5 protein and the basigin receptor localized on the erythrocyte membrane (41, 42). However, PfRh5 does not function alone. Upon secretion, it forms a heteromeric complex with two micronemal proteins, PfRipr and PfCyRPA. PfRipr and PfCyRPA proteins do not interact with basigin and have been shown to lack a membrane anchor (11). The rationale for developing a blood-stage malaria vaccine targeting the components of the PfRh5 complex has been supported by in-vitro and in-vivo studies in non-human primates. Antibodies raised against the PfRh5 proteins have been shown to block erythrocyte invasion by inhibiting its binding to basigin receptor (5, 40, 43–45). Genes coding for proteins of the PfRh5 complex are highly conserved in P. falciparum, suggesting their vital role in parasite survival (30, 46). Therefore, an PfRh5-complex-based vaccine would prove effective. In the present study, we identified the genetic variations of the proteins that make up the P. falciparum Rh 5 complex and determined the polymorphism’s locus on the protein complex in the parasite population from the Mfangano, Takawiri, Kibuogi, and Ngodhe Islands of Lake Victoria in Western Kenya and compared with Kilifi and global databases. All genes of the PfRh5 complex were relatively conserved, and the negative population genetics statistic suggests the parasite population has limited potential to retain these mutations.
The observed negative Tajima’s D statistics from the Lake Victoria population indicated an excess of rare variants and do not suggest balancing selection (30). Genes with a significant negative Tajima’s D value indicate that the parasites population has a limited potential to retain polymorphisms, especially PfP113 and PfRipr genes (47). These findings are consistent with previous studies of P. falciparum in the African population, which showed a majority of genes having a negative Tajima’s D value, suggesting a historical parasite population expansion event (48, 49).
In contrast to other merozoite antigens that are considered potential vaccine candidates such as Apical membrane antigen 1 (AMA1), merozoite surface protein 1 (MSP1), and merozoite surface protein 10 (MSP10) (50, 51), majority of polymorphisms of the PfRh5 complex components were rare variants and did not indicate balancing selection. Recent studies from Nigeria and Kenya reported one non-synonymous mutation on PfRh5 protein at codon C203Y (30, 52). We identified the C203Y mutation in the Lake Victoria population while mutations at codon Y147H, H148D, and K429N were reported in Kilifi samples and MalariaGEN global database as rare variants, which suggests a need for P. falciparum to maintain these mutations across various populations. Mutation at codon S277N observed from Lake Victoria isolates was not reported from the Kilifi population. Three singleton mutations at codons Y226H, F236V, and T441N of the PfRipr gene were identified in Lake Victoria. Among the three polymorphisms, only the mutation at codon Y226H was reported in MalariaGEN global dataset. The mutations were, however, absent from the Kilifi populations.
P. falciparum population from Uganda identified 16 SNPs in the PfRipr gene (53). Among the SNPs on PfRipr gene identified in our study, three were also observed in Uganda, where a negative population statistic on these variants was reported (53). Considering the geographical proximity between Uganda and Lake Victoria islands, the common variants across the two study sites should be investigated to determine if they affect the functionality of the PfRh5 complex. Mutations on PfCyRPA S25N, V292F, and PfP113 gene L17V, Q620H and Q857E were identified only in Lake Victoria isolates.
We identified two mutations at the basigin-PfRh5 interaction region and PfCyRPA-PfRipr proteins interaction regions. The mutation C203Y on PfRh5 protein was located on the Rh5- basigin interface, while mutation V292F located on blade 6 of PfCyRPA protein which is the region of interaction with PfRipr. Studies have demonstrated that recombinant PfRh5 with the C203Y mutation binds to basigin with the same affinity as the wild type (54).
The components of the PfRh5 complex are located in different subcellular locations; thus, the complex only forms during erythrocyte invasion when they are secreted from the rhoptries or micronemes (42). Field studies have demonstrated the PfRh5 complex components exhibit low immunogenicity suggesting the antigens are under limited immune pressure (55). This could explain the limited high-frequency and rare variants observed in this study, as the parasite has a limited need to acquire mutations to escape host immune responses.
Put together, developing an effective malaria vaccine remains a priority among strategies to eliminate and eradicate the disease. One major hindrance to achieving this is the emergence of polymorphisms within the various vaccine antigen targets leading to allele-specific immune responses. Among the PfRh5 complex, PfRh5 is the most advanced vaccine target. However, the presence of low-frequency mutations raises concerns about immune system evasion. This study recommends functional assay studies to investigate the immunological and biological relevance of the identified mutations.
Acknowledgments
We wish to acknowledge the Mount Kenya University research team for their insights in the improvement of this work.
Funding
This work was supported by the Royal Society of Tropical Medicine and Hygiene (RSTMH) small grants 2019 (HW). BK is an EDCTP Fellow under EDCTP2 programme supported by the European Union grant number TMA2020CDF-3203. JG received support from the African Academy of Sciences. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Ethics statement
The study’s approval was obtained from the Kenyatta National Hospital - University of Nairobi (KNH-UoN) ethical review committee (P609/10/2014) and the Mount Kenya University Ethics Review Committee (038/2014). Written informed consent to participate in this study was provided by the participants’ legal guardian/next of kin.
Author contributions
HW, BK, and JG conceived and designed the study; JG provided the analysed data sets; HW performed the bioinformatic and statistical analysis, under supervision of JOK and BK; HW and BK wrote the first draft of the manuscript, and the final version included edits from all authors. The final manuscript was read and approved by all authors.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ddbj.nig.ac.jp/, PRJDB12148.
References
- 1.WHO. World malaria report 2021. Vol. 2021. Geneva, Switzerland: World Health Organization, Geneva, Switzerland; 2021. p. 322. [Google Scholar]
- 2.Badmos AO, Alaran AJ, Adebisi YA, Bouaddi O, Onibon Z, Dada A, et al. What sub-Saharan African countries can learn from malaria elimination in China. Trop Med Health. 2021;49:86. doi: 10.1186/s41182-021-00379-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li J, Chen J, Xie D, Eyi UM, Matesa RA, Ondo Obono MM, et al. Limited artemisinin resistance-associated polymorphisms in plasmodium falciparum K13-propeller and PfATPase6 gene isolated from bioko island, equatorial Guinea. Int J Parasitol Drugs Drug Resist. 2016;6:54–9. doi: 10.1016/j.ijpddr.2015.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Frosch AEP, Laufer MK, Mathanga DP, Takala-Harrison S, Skarbinski J, Claassen CW, et al. Return of widespread chloroquine-sensitive plasmodium falciparum to Malawi. J Infect Dis. 2014;210:1110–4. doi: 10.1093/infdis/jiu216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bustamante LY, Bartholdson SJ, Crosnier C, Campos MG, Wanaguru M, Nguon C, et al. A full-length recombinant plasmodium falciparum PfRH5 protein induces inhibitory antibodies that are effective across common PfRH5 genetic variants. Vaccine. 2013;31:373–9. doi: 10.1016/j.vaccine.2012.10.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duffy PE, Patrick Gorres J. Malaria vaccines since 2000: Progress, priorities, products. NPJ Vaccines. 2020;5:1–9. doi: 10.1038/s41541-020-0196-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Volz JC, Yap A, Sisquella X, Thompson JK, Lim NTY, Whitehead LW, et al. Essential role of the PfRh5/PfRipr/CyRPA complex during plasmodium falciparum invasion of erythrocytes. Cell Host Microbe. 2016;20:60–71. doi: 10.1016/j.chom.2016.06.004. [DOI] [PubMed] [Google Scholar]
- 8.Bargieri DY, Andenmatten N, Lagal V, Thiberge S, Whitelaw JA, Tardieux I, et al. Apical membrane antigen 1 mediates apicomplexan parasite attachment but is dispensable for host cell invasion. Nat Commun. 2013;4:2552. doi: 10.1038/ncomms3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beeson JG, Drew DR, Boyle MJ, Feng G, Fowkes FJI, Richards JS. Merozoite surface proteins in red blood cell invasion, immunity and vaccines against malaria. FEMS Microbiol Rev. 2016;40:343–72. doi: 10.1093/femsre/fuw001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takala SL, Plowe CV. Genetic diversity and malaria vaccine design, testing and effficacy: Preventing and overcoming “vaccine resistant malaria”. Parasite Immunol. 2009;31:560–73. doi: 10.1111/j.1365-3024.2009.01138.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Galaway F, Drought LG, Fala M, Cross N, Kemp AC, Rayner JC, et al. P113 is a merozoite surface protein that binds the n terminus of plasmodium falciparum RH5. Nat Commun. 2017;8:1–11. doi: 10.1038/ncomms14333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miura K. Progress and prospects for blood-stage malaria vaccines. Expert Rev Vaccines. 2016;15:765–81. doi: 10.1586/14760584.2016.1141680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chan J-A, Fowkes FJI, Beeson JG. Surface antigens of plasmodium falciparum-infected erythrocytes as immune targets and malaria vaccine candidates. Cell Mol Life Sci. 2014;71:3633–57. doi: 10.1007/s00018-014-1614-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Opi DH, Kurtovic L, Chan J-A, Horton JL, Feng G, Beeson JG. Multi-functional antibody profiling for malaria vaccine development and evaluation. Expert Rev Vaccines. 2021;20:1257–72. doi: 10.1080/14760584.2021.1981864. [DOI] [PubMed] [Google Scholar]
- 15.Ouattara A, Takala-Harrison S, Thera MA, Coulibaly D, Niangaly A, Saye R, et al. Molecular basis of allele-specific efficacy of a blood-stage malaria vaccine: Vaccine development implications. J Infect Dis. 2013;207:511–9. doi: 10.1093/infdis/jis709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wong W, Huang R, Menant S, Hong C, Sandow JJ, Birkinshaw RW, et al. Structure of plasmodium falciparum Rh5–CyRPA–Ripr invasion complex. Nature. 2019;565:118–21. doi: 10.1038/s41586-018-0779-6. [DOI] [PubMed] [Google Scholar]
- 17.Crosnier C, Bustamante LY, Bartholdson SJ, Bei AK, Theron M, Uchikawa M, et al. Basigin is a receptor essential for erythrocyte invasion by plasmodium falciparum. Nature. 2011;480:534–7. doi: 10.1038/nature10606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen L, Lopaticki S, Riglar DT, Dekiwadia C, Uboldi AD, Tham W-H, et al. An EGF-like protein forms a complex with PfRh5 and is required for invasion of human erythrocytes by plasmodium falciparum. PloS Pathog. 2011;7:e1002199. doi: 10.1371/journal.ppat.1002199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reddy KS, Amlabu E, Pandey AK, Mitra P, Chauhan VS, Gaur D. Multiprotein complex between the GPI-anchored CyRPA with PfRH5 and PfRipr is crucial for plasmodium falciparum erythrocyte invasion. Proc Natl Acad Sci U S A. 2015;112(4):1179–84. doi: 10.1073/pnas.1415466112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Osier FH, Mackinnon MJ, Crosnier C, Kamuyu G, Fegan G, Wanaguru M, et al. New antigens for a multicomponent blood-stage malaria vaccine. Sci Transl Med. 2014;6:247ra102. doi: 10.1126/scitranslmed.3008705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ord RL, Rodriguez M, Lobo CA. Malaria invasion ligand RH5 and its prime candidacy in blood-stage malaria vaccine design. Hum Vaccin Immunother. 2015;11:1465–73. doi: 10.1080/21645515.2015.1026496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sagara I, Dicko A, Ellis RD, Fay MP, Diawara SI, Assadou MH, et al. A randomized controlled phase 2 trial of the blood stage AMA1-C1/Alhydrogel malaria vaccine in children in Mali. Vaccine. 2009;27:3090–8. doi: 10.1016/j.vaccine.2009.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mobegi VA, Loua KM, Ahouidi AD, Satoguina J, Nwakanma DC, Amambua-Ngwa A, et al. Population genetic structure of plasmodium falciparum across a region of diverse endemicity in West Africa. Malar J. 2012;11:223. doi: 10.1186/1475-2875-11-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Auburn S, Campino S, Miotto O, Djimde AA, Zongo I, Manske M, et al. Characterization of within-host plasmodium falciparum diversity using next-generation sequence data. PloS One. 2012;7:e32891. doi: 10.1371/journal.pone.0032891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ndila CM, Uyoga S, Macharia AW, Nyutu G, Peshu N, Ojal J, et al. Human candidate gene polymorphisms and risk of severe malaria in children in kilifi, Kenya: A case-control association study. Lancet Haematol. 2018;5:e333–45. doi: 10.1016/S2352-3026(18)30107-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Idris ZM, Chan CW, Kongere J, Gitaka J, Logedi J, Omar A, et al. High and heterogeneous prevalence of asymptomatic and Sub-microscopic malaria infections on islands in lake Victoria, Kenya. Sci Rep. 2016;6:36958. doi: 10.1038/srep36958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gitaka JN, Takeda M, Kimura M, Idris ZM, Chan CW, Kongere J, et al. Selections, frameshift mutations, and copy number variation detected on the surf4.1gene in the western Kenyan plasmodium falciparum population. Malar J. 2017;16:98. doi: 10.1186/s12936-017-1743-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schuster FL. Cultivation of plasmodium spp. Clin Microbiol Rev. 2002;15:355–64. doi: 10.1128/CMR.15.3.355-364.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Borrmann S, Sasi P, Mwai L, Bashraheil M, Abdallah A, Muriithi S, et al. Declining responsiveness of plasmodium falciparum infections to artemisinin-based combination treatments on the Kenyan coast. PloS One. 2011;6:e26005. doi: 10.1371/journal.pone.0026005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ndwiga L, Osoti V, Ochwedo KO, Wamae K, Bejon P, Rayner JC, et al. The plasmodium falciparum Rh5 invasion protein complex reveals an excess of rare variant mutations. Malar J. 2021;20:1–10. doi: 10.1186/s12936-021-03815-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li H, Durbin R. Fast and accurate short read alignment with burrows-wheeler transform. Bioinformatics. 2009;25:1754–60. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The sequence alignment / map format and SAMtools. Oxford Acad. 2009;25:2078–9. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, del Angel G, Levy-Moonshine A, et al. From fastQ data to high-confidence variant calls: The genome analysis toolkit best practices pipeline. Current Protocols in Bioinformatics. 2013:1–33. doi: 10.1002/0471250953.bi1110s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, et al. The variant call format and VCFtools. Bioinformatics. 2011;27:2156–8. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cingolani P, Platts A, Wang LL, Coon M, Nguyen T, Wang L, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carver T, Harris SR, Berriman M, Parkhill J, McQuillan JA. Artemis: An integrated platform for visualization and analysis of high-throughput sequence-based experimental data. Bioinformatics. 2012;28:464–9. doi: 10.1093/bioinformatics/btr703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–95. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Librado P, Rozas J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–2. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- 39.Njuguna P, Maitland K, Nyaguara A, Mwanga D, Mogeni P, Mturi N, et al. Observational study: 27 years of severe malaria surveillance in kilifi, Kenya. BMC Med. 2019;17:124. doi: 10.1186/s12916-019-1359-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wright KE, Hjerrild KA, Bartlett J, Douglas AD, Jin J, Brown RE, et al. Structure of malaria invasion protein RH5 with erythrocyte basigin and blocking antibodies. Nature. 2014;515:427–30. doi: 10.1038/nature13715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baum J, Chen L, Healer J, Lopaticki S, Boyle M, Triglia T, et al. Reticulocyte-binding protein homologue 5 - an essential adhesin involved in invasion of human erythrocytes by plasmodium falciparum. Int J Parasitol. 2009;39:371–80. doi: 10.1016/j.ijpara.2008.10.006. [DOI] [PubMed] [Google Scholar]
- 42.Wanaguru M, Liu W, Hahn BH, Rayner JC, Wright GJ. RH5-basigin interaction plays a major role in the host tropism of plasmodium falciparum. Proc Natl Acad Sci U S A. 2013;110:20735–40. doi: 10.1073/pnas.1320771110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nagaoka H, Kanoi BN, Ntege EH, Aoki M, Fukushima A, Tsuboi T, et al. Antibodies against a short region of PfRipr inhibit plasmodium falciparum merozoite invasion and PfRipr interaction with Rh5 and SEMA7A. Sci Rep. 2020;10:1–14. doi: 10.1038/s41598-020-63611-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Douglas AD, Williams AR, Knuepfer E, Illingworth JJ, Furze JM, Crosnier C, et al. Neutralization of plasmodium falciparum merozoites by antibodies against PfRH5. J Immunol. 2014;192:245–58. doi: 10.4049/jimmunol.1302045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Willcox AC, Huber AS, Diouf A, Barrett JR, Silk SE, Pulido D, et al. Antibodies from malaria-exposed malians generally interact additively or synergistically with human vaccine-induced RH5 antibodies. Cell Rep Med. 2021;2:100326. doi: 10.1016/j.xcrm.2021.100326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mian SY, Somanathan A, Chaddha K, Pandey AK, Singh H, Krishna S, et al. Plasmodium falciparum cysteine-rich protective antigen (CyRPA) elicits detectable levels of invasion-inhibitory antibodies during natural infection in humans. Infect Immun. 2022;90:e0037721. doi: 10.1128/IAI.00377-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Amambua-Ngwa A, Tetteh KKA, Manske M, Gomez-Escobar N, Stewart LB, Deerhake ME, et al. Population genomic scan for candidate signatures of balancing selection to guide antigen characterization in malaria parasites. PloS Genet. 2012;8:e1002992. doi: 10.1371/journal.pgen.1002992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ocholla H, Preston MD, Mipando M, Jensen ATR, Campino S, MacInnis B, et al. Whole-genome scans provide evidence of adaptive evolution in Malawian plasmodium falciparum isolates. J Infect Dis. 2014;210:1991–2000. doi: 10.1093/infdis/jiu349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mobegi VA, Duffy CW, Amambua-Ngwa A, Loua KM, Laman E, Nwakanma DC, et al. Genome-wide analysis of selection on the malaria parasite plasmodium falciparum in West African populations of differing infection endemicity. Mol Biol Evol. 2014;31:1490–9. doi: 10.1093/molbev/msu106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Polley SD, Tetteh KKA, Lloyd JM, Akpogheneta OJ, Greenwood BM, Bojang KA, et al. Plasmodium falciparum merozoite surface protein 3 is a target of allele-specific immunity and alleles are maintained by natural selection. J Infect Dis. 2007;195:279–87. doi: 10.1086/509806. [DOI] [PubMed] [Google Scholar]
- 51.Bendezu J, Villasis E, Morales Ruiz S, Garro K, Infante B, Gutierrez-Loli R, et al. Evaluation of plasmodium falciparum MSP10 and its development as a serological tool for the Peruvian Amazon region. Malar J. 2019;18:327. doi: 10.1186/s12936-019-2959-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Olusola A, Osuntoki A, Balogun E, Olukosi A, Iwalokun B, Oyebola K, et al. Genetic polymorphisms in malaria vaccine candidate plasmodium falciparum reticulocyte-binding protein homologue-5 among populations in Lagos, Nigeria. Malar J. 2020;19:6. doi: 10.1186/s12936-019-3096-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ntege EH, Arisue N, Ito D, Hasegawa T, Palacpac NMQ, Egwang TG, et al. Identification of plasmodium falciparum reticulocyte binding protein homologue 5-interacting protein, PfRipr, as a highly conserved blood-stage malaria vaccine candidate. Vaccine. 2016;34:5612–22. doi: 10.1016/j.vaccine.2016.09.028. [DOI] [PubMed] [Google Scholar]
- 54.Hjerrild KA, Jin J, Wright KE, Brown RE, Marshall JM, Labbeé GM, et al. Production of full-length soluble plasmodium falciparum RH5 protein vaccine using a drosophila melanogaster Schneider 2 stable cell line system. Sci Rep. 2016;6:30357. doi: 10.1038/srep30357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tran TM, Ongoiba A, Coursen J, Crosnier C, Diouf A, Huang C-Y, et al. Naturally acquired antibodies specific for plasmodium falciparum reticulocyte-binding protein homologue 5 inhibit parasite growth and predict protection from malaria. J Infect Dis. 2014;209:789–98. doi: 10.1093/infdis/jit553. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ddbj.nig.ac.jp/, PRJDB12148.