Abstract
Genomic DNA residing in the nuclei of mammalian neurons can be as old as the organism itself. The life span of nuclear RNAs, which are critical for proper chromatin architecture and transcription regulation, has not been determined in adult tissues. Here, we identified and characterized nuclear RNAs that do not turn over for at least two years in a subset of postnatally-born cells in the mouse brain. These long-lived RNAs were stably retained in nuclei in a neural cell type-specific manner and were required for the maintenance of heterochromatin. Thus, the remarkable life span of neural cells may depend on both the molecular longevity of DNA for the storage of genetic information and also the extreme stability of RNA for the functional organization of chromatin.
After early development, most neurons survive for an organism’s entire life without ever being replaced. In the absence of mitotic nuclear disassembly and reassembly, the life span of some nuclear constituents such as genomic DNA, a subset of histones, and nuclear pore complex proteins extends to months and even years (1, 2). For example, the stability of genomic DNA has been used to determine the chronological age of neurons in humans over the course of decades (3, 4). The life-long stability of genomic DNA is necessary to safeguard the persistence of a cell’s genetic information. In recent years, non-coding RNAs have been identified as critical regulators of nuclear chromatin organization and transcription control (5, 6); however, little is known about the turnover of this class of nucleic acids in adult brain tissue.
Long-term retention of nuclear RNAs in the mouse brain
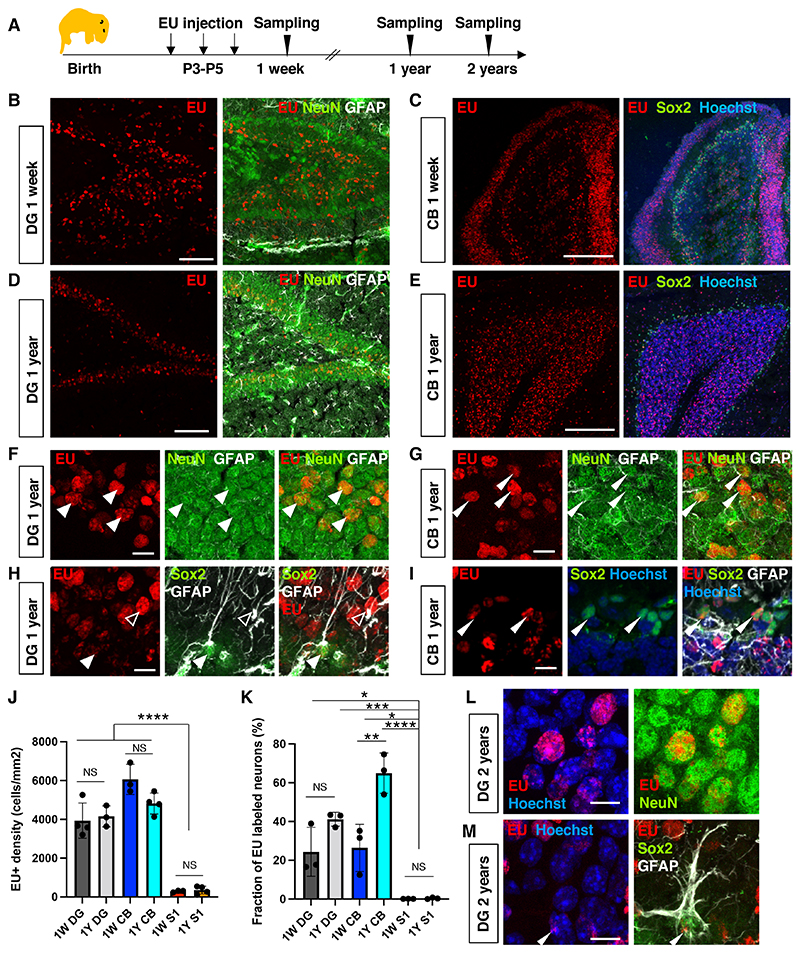
To explore RNA stability in the mouse brain, we performed in vivo pulse-chase labeling of transcripts with a modified uridine analog, 5-Ethynyl uridine (EU) (7). EU was injected into mice on postnatal days 3 to 5 (P3 to P5) to label de novo-synthesized RNAs during brain development (Fig. 1A). One day after the last injection of EU (defined as the 1-week sample), EU-labeled RNA was visualized by click chemistry using Alexa-555 fluorophores. EU signals were detectable in different brain regions such as the dentate gyrus (DG) of the hippocampus (Fig. 1B) and the cerebellum (CB) (Fig. 1C). The EU signal was sparse and not detectable in most cells in the primary somatosensory cortex (S1) (fig. S1). The density and fraction of EU+ cells varied among different brain tissues (Fig. 1J, 1K, ANOVA P < 0.0001). We confirmed that all cells in the brain were EU-labeled acutely after a single EU injection on P3 (30 min or 1 hour chase) (fig. S2-3) and detected no difference in EU incorporation into de novo-synthesized RNA between brain regions (fig. S3C), indicating that EU can be transported, metabolized, and used in transcription in all cells of the brain at this age. These data suggest that either the transcription or the retention of EU-incorporated transcripts varies between different cell types in the brain.
Fig. 1. Long-term retention of RNA in the mouse brain.
(A) Schematic diagram of experimental plan. RNA synthesis at P3-P5 was labeled using 5-Ethynyl-uridine (EU). (B) EU signals in the dentate gyrus (DG) of a 1-week-old animal. (C) EU signals in the cerebellum (CB) of a 1-week-old animal. (D) EU signals in the DG of a 1-year-old animal. (E) EU signals in the CB of a 1-year-old animal. (F-H) High magnification images with Sox2/GFAP immunostaining (arrowhead, RGL-ANSC; open arrowhead, astrocytes) or NeuN/GFAP staining (arrowheads, NeuN+ neurons). (J) Quantification of the density of EU+ cells. 1W (1-week-old) DG 3933 ± 911.9 cells/mm2; 1W CB 6060 ± 774.3 cells/mm2; 1W S1 291.8 ± 52.12 cells/mm2. 1Y (1-year-old) DG 4156 ± 542 cells/mm2; 1Y CB 4816 ± 537 cells/mm2; 1Y S1 359 ± 199 cells/mm2; NS not significant; ANOVA with post-hoc Tukey’s test. Data are presented as mean +/- standard deviation (SD). (K) Quantification of the percentage of EU+ neurons (NeuN+ cells). (L) EU signal in neurons of the DG in a 2-year-old animal. (M) EU signals in a neural stem cell (RGL-ANSC) in the DG of a 2-year-old animal. Data are presented as mean +/- SD. Scale bars, 100 μm (B, D), 10 μm (F-I, L, M), 200 μm (C, E). ANOVA followed by Tukey’s multiple comparison test; *P < 0.05, **P < 0.01, **P < 0.001, ****P < 0.0001.
The robust incorporation of EU in the DG and CB allowed us to determine whether the observed EU signals persisted for an extended period during adulthood. We analyzed brain samples one year after EU injection and detected strong EU signals in the DG and in the granular layer of CB (Fig. 1C-I). The density of EU+ cells was indistinguishable between 1-week-old and 1-year-old animals (Fig. 1J), indicating the existence of long-lived RNAs (LL-RNAs) in the brain. A comparison of EU intensities revealed that 68 ± 14.26% of the initial EU signals observed in the DG of 1-week-old animals persisted in 1-year-old animals (fig. S4). These data suggest that EU-labeled LL-RNAs are minimally turned over during adulthood. Notably, the EU signal was sensitive to RNase, although only to a limited extent even after harsh RNase treatment, suggesting either inaccessibility or remarkable stability in the tissue (fig. S5A-B). Consistent with EU being specifically incorporated into RNA, degradation of genomic DNA by DNase 1 treatment did not affect EU signal intensities (fig. S5C-H). Moreover, when EU was injected at the same time as bromodeoxyuridine (BrdU), which is incorporated into newly synthesized DNA, the spatial distribution of nuclear EU signals was distinct from BrdU signals (fig. S5I), providing further evidence for the specific incorporation of EU into RNA. We also studied animals 2 years after EU injections and were able to detect EU signals in DG and CB neurons (Fig. 1K, 1L, fig. S6A-B, D-F), radial glia-like adult neural stem cells (RGL-ANSCs)/cerebellar ANSCs (Fig. 1M, fig. S6H), and astrocytes (fig. S6C, E, G). This finding suggests that these RNAs persist throughout the life span of the animal.
Cell type-specificity of long-retained RNAs
To identify which cell types retained LL-RNA, we combined EU click chemistry with immunohistochemistry using cell type-specific markers. In 1-year-old mice, the majority of EU+ cells (83.7 ± 10.8%) in the DG were NeuN+ neurons (Fig. 1B, 1D, 1F). EU signals were also observed in RGL-ANSCs (Sox2+ with a GFAP+ radial fiber; 1.5 ± 0.9% of EU+ cells), adult neural progenitor cells (ANPCs) (Sox2+ without a GFAP+ radial fiber; 3.76 ± 1.93% of EU+ cells) and GFAP+ astrocytes (Fig. 1D, H; fig. S6C), suggesting that LL-RNAs can be maintained in neurons and somatic ANSCs in the DG. In 1-week-old mice (1 day after the last EU injection), only 25.6 ± 9.4% of EU+ cells were NeuN+ whereas 39.7 ± 3.8% were Sox2+ neural stem cell/progenitor cells (NSPCs) and 7.0 ± 1.5% were RGL-ANSCs (fig. S1I). In the first and second postnatal week, ANPCs in the DG give rise to neurons or become quiescent RGL-ANSCs (8, 9), suggesting that the retention/incorporation of EU+ transcripts could coincide with cell cycle exit during differentiation or transition into quiescence. Indeed, 4 h after a single EU injection, when most acutely-labeled RNA was already degraded (fig. S2-3), all high-intensity EU+ cells in the hippocampus were proliferating (Ki67+) NSPCs, whereas 24 h after the last injection, most EU+ cells had exited the cell cycle and 44.5% of EU+ cells expressed neuronal differentiation-associated marker NeuroD1 (fig. S7). Thus, EU-labeled RNA is retained long-term upon cell cycle exit in neurons and ANSCs in the DG.
Similar to the DG, in the CB of 1-week-old mice, 25.7 ± 15.4% of EU+ cells were NeuN+ neurons (fig. S1J) and 13.3 ± 4.4% of EU+ cells were Sox2+ adult NSPCs (10), indicating that EU+ transcripts are retained in neurons and stem cells of CB. 58% of EU+ cells in the CB did not express any tested markers at this age, but they were likely to be migrating immature granule cells based on their localization (Fig. 1F) (11). Indeed, in 1-year-old mice, 76.3 ± 8.5% of EU+ cells were NeuN+ neurons mostly located in the granular layer (Fig. 1G, fig. S1E, 1J). We also observed the retention of LL-RNA in Sox2+ ANSPCs in the CB of 1-year-old mice (Fig. 1H), suggesting that ANSPCs in both the DG and CB can retain LL-RNAs. In the S1, a total of 34.4 ± 14.6% and 43.0 ± 10.3% of EU+ cells were GFAP+ in 1-week-old and 1-year-old samples, respectively (fig. S1F-H, 1K), indicating that astrocytes in S1 retain EU-labeled transcripts. However, NeuN+ EU+ cells were rarely observed in S1 (fig. 1K). Thus, LL-RNAs are retained or transcribed in a cell type-specific manner.
To validate that our findings were independent of potential side effects caused by EU, we labeled nascent RNA using another uridine analogue, 5-Bromouridine (BrU). The spatial patterns of BrU-positive cells in 1-week-old and 1-months-old mice reflected those observed in EU-injected mice, with persistent BrU labeling in neurons and ANSPCs in the DG and CB but only very few BrU+ cells in S1 (fig. S8). Additionally, injection of triphosphorylated uridine (5-BrUTP), which is directly incorporated into RNA without metabolization, resulted in a similar distribution of label-retaining cells in the brain, providing further support that cell type-specific retention of RNA is not due to potential differences in cellular nucleotide metabolism.
Because most cortical neurons are generated during embryonic neurogenesis, we next asked whether cortical neurons retain EU signals when EU is injected during cortical neurogenesis. We injected EU into pregnant dams at E14.5-E16.5 and analyzed brains at E17.5 and at P6 (fig. S9A). We confirmed that EU injection labeled RNA synthesis in all embryonic brain cells at 1 h after injection (fig. S9B). At E17.5, EU signals were retained in Sox2- and Tbr2-positive NSPCs in the ventricular and subventricular zone of the neocortex (fig. S9B-E, S9G). In contrast, we found only very few and weakly labeled EU+ cells in the cortical plate - fewer cells than the expected number of new-born neurons based on the distribution of BrdU+ newborn cells at E17.5 (fig. S9E-F). Of those rare EU+ cells in the cortical plate, only 39.0 ± 3.2% and 5.1 ± 3.2% expressed neuronal markers NeuN and Ctip2, respectively (fig. S9E, S9H). Thus, cortical neurons have a limited capability to transcribe or retain LL-RNAs. Moreover, only sparse EU signals were detected in the cortex, hippocampus and CB at P6 when EU was injected embryonically (fig. S9). Thus, the long-term retention of RNA is cell type-specific, with cortical neurons showing much reduced capability to maintain RNAs compared to granule cells of the DG and CB.
We then addressed whether LL-RNAs can be transcribed in the brain at any age. EU injections at later postnatal ages (P7-9 and P13-15) resulted in patterns of EU+ cells in the hippocampus that were consistent with those obtained following EU injections at P3-5 (fig. S10). However, in the adult mouse brain, EU was not incorporated into nascent transcripts (fig. S11) and was, therefore, not suitable to label RNA synthesis. This is presumably due to the downregulation of enzymes of the pyrimidine salvage pathway in the adult brain (fig. S12), which are required for EU metabolism. No brain region-dependent or cell type-specific differences in the expression of the pyrimidine salvage pathway were detected in postnatal mice (fig. S12A-C), consistent with the ubiquitous EU signal observed acutely after EU injection in postnatal brains (fig. S2-3). This finding supports the notion that all cells in the postnatal brain can metabolize EU but that the retention of LL-RNAs is cell type-specific.
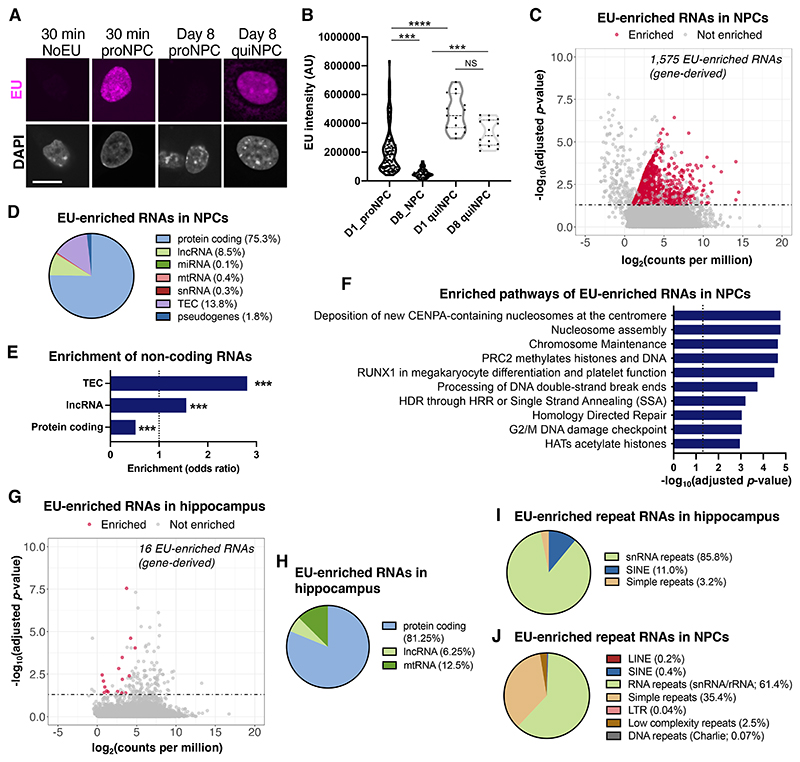
Having established the existence of nuclear LL-RNAs, we next aimed to determine their molecular identity and functions. To facilitate functional characterization, we utilized a tractable in vitro system in which we could recapitulate the long-term retention of transcripts and manipulate the identified RNAs. Because we observed EU signal retention in RGL-ANSCs in vivo, we tested whether quiescent neural progenitor cells (quiNPCs) retained EU-labeled transcripts in vitro. We used an established protocol to induce quiescence in mouse NPCs (12) and found that quiNPCs indeed retained EU+ transcripts for 8 days (Fig. 3A-B) and even 2 weeks (fig. S13), but not in proliferating conditions (Fig. 2A-B). EU signals in quiNPCs were depleted by RNase treatments (fig. S14A-B) and significantly reduced after inhibition of RNA polymerase II and III (fig. S14C), confirming that EU signals are derived from nascent RNA in quiNPCs. Inhibition of RNA polymerases after EU labeling did not reduce EU signals (fig. S14D-E), suggesting that EU signals are not derived from recycling of EU-labeled uridines. Moreover, DNase 1 treatment and ribonucleotide reductase inhibition did not reduce EU signals in quiNPCs (fig. S15), providing further evidence that EU signals are not derived from EU incorporation into DNA.
Fig. 2. Molecular identity of long-retained RNA.
(A) Retention of EU signals in quiescent NPCs (quiNPCs) in vitro. Proliferating NPC (proNPC). Scale bar, 10 μm. (B) Quantification of EU intensity in proNPCs and quiNPCs at day 1 (D1) or day 8 (D8) after EU labeling. Kruskal-Wallis test followed by Dunn’s multiple comparison test; ***P < 0.001, ****P < 0.0001, dots indicate individual cells. (C) MA plot showing EU-enriched (gene-derived) RNAs in quiNPCs at 8 days after EU labeling, as determined by EU-RNA-Sequencing. Significantly EU-enriched RNAs were defined as adjusted P < 0.05 and fold change > 2 compared to non-EU-labeled quiNPCs (n=3 experiments). (D) Gene class distribution of EU-enriched RNA in quiNPCs. (E) Protein-coding RNAs were underrepresented, whereas long non-coding RNAs (lncRNA) and uncharacterized transcripts (TEC) were over-represented in EU-enriched RNA. ***P < 0.001 (linear regression). (F) Top significantly enriched Reactome pathways among EU-enriched RNAs in quiNPCs (adjusted P < 0.05). (G) MA plot showing EU-enriched (gene-derived) RNAs in hippocampus tissue of 5-week-old mice that were injected with EU at postnatal days P3-P5 (n = 3 mice/group). (H) Gene class distribution of EU-enriched RNA in the hippocampus. (I-J) Distribution of EU-enriched RNAs that map to repeat RNAs in quiNPCs and hippocampus tissue.
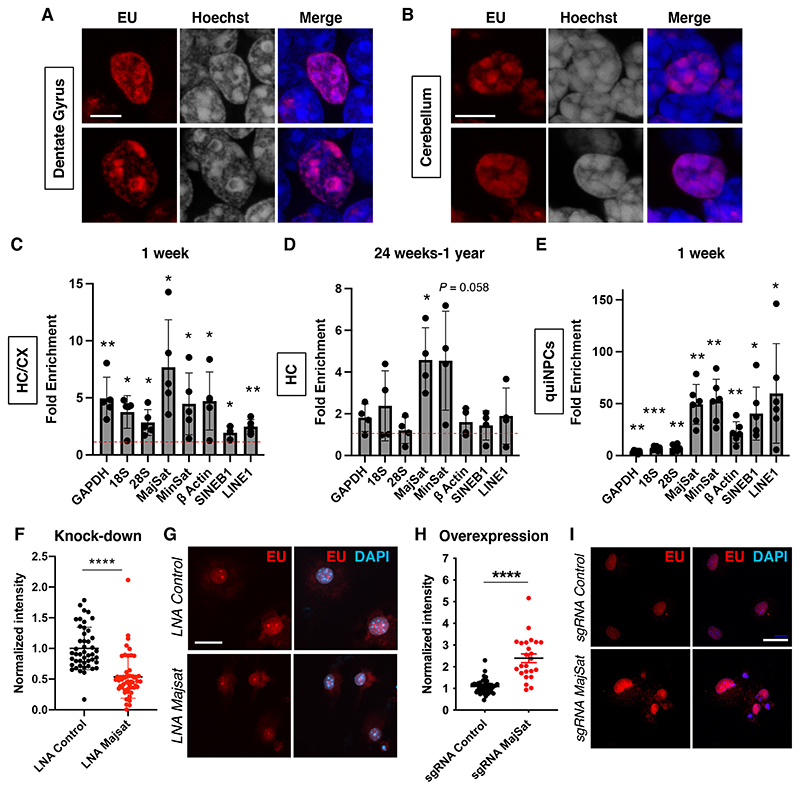
Fig. 3. Repeat RNAs are long retained in the mouse brain.
(A and B) Localization of EU signals in nuclei in the DG and CB of a 1-year-old animal. Scale bars, 5 μm. (C and D) Enrichment of EU-labeled transcripts in the hippocampus/cortex (HC/CX) of a 1-week-old animal (n = 5) and in the hippocampus (HC) of 24-week-old to 1-year-old animals (n = 4). One sample t-test, *P < 0.05, **P < 0.01. Fold enrichments compared to PBS-treated control: 1W Gapdh, 4.95 ± 1.85 ; 18S rRNA 3.76 ± 1.43; 28S rRNA 2.84 ± 1.13; major satellite 7.69 ± 4.16; minor satellite 4.48 ± 2.71; βactin 4.73 ± 2.54; SINEB1 1.95 ± 0.60; LINE-1 2.47 ± 0.62; n = 5 ; 24W-1 Y, Gapdh, 1.85 ± 0.66; 18S rRNA 2.38 ± 1.68; 28S rRNA 1.21 ± 0.63; major satellite 4.57 ± 1.55; minor satellite 4.54 ± 2.37; β actin 1.60 ± 0.68; SINEB1 1.44 ± 0.70; LINE-1 1.90 ± 1.34; n = 4. Data are presented as mean +/- SD. (E) Enrichment of EU-labeled transcripts in quiNPCs 8 days after EU treatment compared with PBS-treated cells. One sample t-test, *P < 0.05, **P < 0.01, ***P < 0.001. Fold enrichments compared to control: Gapdh, 3.53 ± 1.50; 18S rRNA 6.86 ± 2.06; 28S rRNA 7.38 ± 3.33; major satellite 49.3 ±19.1; minor satellite 52.57 ± 20.97; β actin 22.36 ± 10.2; SINEB1 40.45 ± 25.53; LINE-1 59.82 ± 47.95; n = 6. Data are presented as mean +/- SD. MajSat, major satRNA; MinSat, minor satRNA. (F) Quantification of EU signal intensity after LNA-GapmeR-mediated knock-down of major satRNAs in quiNPCs (LNA MajSat). ****P < 0.0001, t-test, dots indicate individual cells from 3 experiments. (G) Reduction of EU signals in quiNPCs after the application of LNA targeting major satRNAs. Scale bar, 20 μm. (H-I) Increased EU signals after CRISPRa-based overexpression of major satRNAs (sgRNA MajSat). ****P < 0.0001, t-test, dots indicate individual cells from 3 independent experiments. Scale bar, 20 μm.
Molecular identity of LL-RNAs
To determine the molecular identity of LL-RNAs, we performed EU-RNA-Sequencing of quiNPCs 1 week after EU labeling. We found 1,575 genes to be significantly enriched in the EU-labeled RNA fraction (Fig. 2C; Data S1; fig. S16A), identifying those as LL-RNAs in quiNPCs. The LL-RNAs comprised protein-coding RNAs and non-coding RNAs (Fig. 2D), with long non-coding RNAs (lncRNAs) and uncharacterized transcripts (TECs) being significantly over-represented in EU-enriched RNAs compared to all RNAs expressed in quiNPCs (Fig. 2E). LL-RNAs in quiNPCs were enriched in pathways related to nuclear architecture and epigenetic regulation, suggesting potential roles of LL-RNAs in these cellular functions (Fig. 2F). We also performed EU-RNA-Sequencing with hippocampal tissue from 1-month-old mice that were injected with EU at P3-P5, and we identified 16 gene-derived transcripts that were significantly enriched in the EU-labeled RNA fraction, including lncRNAs, mitochrondrial t-RNAs and protein-coding RNAs (Fig. 2G-H). Mapping of EU-RNA-Sequencing data to repetitive genomic regions revealed that LL-RNAs contained repetitive RNAs in both hippocampus and quiNPCs (Fig. 2I-J, fig. S16B-C), including small nuclear RNA repeats (snRNAs), SINEs and satellite RNAs (fig. S16D). Together, these results suggested that non-coding RNAs, including lncRNAs and repetitive RNAs, can be retained long-term in quiNPCs and in hippocampal granule cells. The difference in the detected numbers of LL-RNAs between quiNPCs and hippocampal tissue might result from differences in the length of the retention period or from cell type-specific molecular identities of LL-RNAs. Alternatively, the detection sensitivity of LL-RNAs may be reduced in hippocampal tissue since only a fraction of hippocampal cells retained EU-labeled transcripts whereas 100% of in vitro quiNPCs possessed EU-labeled RNAs. In the future, it would be important to compare LL-RNAs between different cell types with similar conditions.
Several studies suggest that repeat RNAs, including satellite RNAs (satRNAs), interact with heterochromatin (13–16). Consistent with this idea, high resolution imaging in DG and CB neurons from 1-year-old samples showed that EU signals were often associated with Hoechstdense heterochromatin areas (Fig. 3A-B), indicating that they might have functions in chromatin regulation. We confirmed the enrichment of repeat RNAs among EU-captured RNA using quantitative PCR (Fig. 3C) and found that a significant amount of satRNAs were retained for up to 1 year in the hippocampus (Fig. 3D). The PCR signals were diminished by RNase treatments (fig. S14F-H), supporting the notion that satRNAs are retained for years in the brain. We also found strong enrichments of major satRNAs and minor satRNAs in quiNPCs at 8 days after EU labeling compared with vehicle-treated quiNPCs (Fig. 3E). Other repeat sequences such as short interspersed nuclear element B1 (SINEB1) and long interspersed nuclear elements (LINE-1) were also enriched in quiNPCs. We also identified 18S and 28S rRNA to be moderately enriched, consistent with previous findings of ribosomal RNAs exhibiting longer half-lives (17). We validated that EU treatment did not alter repeat RNA levels (fig. S16E) and confirmed the long-term retention of repeat RNAs in quiNPCs using BrU labeling and BrU immunoprecipitation chase (BRIC)-qPCR (fig. S17). Altogether, our findings reveal transcript-specific long-term retention in quiNPCs.
Roles of LL-RNAs in heterochromatin maintenance
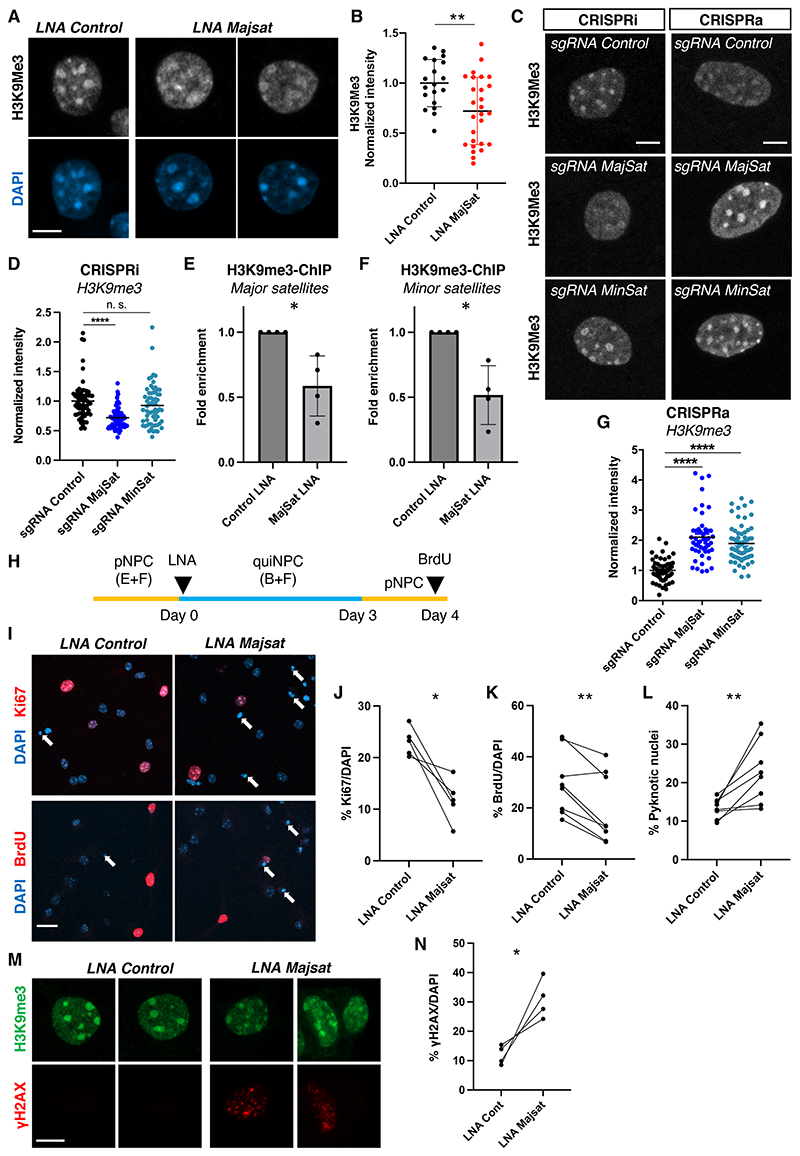
Finally, we addressed the biological function of LL-RNAs using quiNPCs as a model system and manipulated major satRNAs using previously published LNA-GapmeRs (18) and sgRNAs (CRISPRa and CRISPRi) (19). We observed a significant reduction in EU signals after major satRNA knock-down (Fig. 3F-G, fig. S18A), whereas overexpression of major satRNAs increased EU signals (Fig. 3H-I, fig. S18B), confirming the contribution of major satRNAs to LL-RNAs in quiNPCs. Because it has been shown that major satRNAs are associated with constitutive heterochromatin (14, 15), we investigated heterochromatin organization with the constitutive heterochromatin marker H3K9me3. After knock-down of major satRNAs, the signals of H3K9me3 around chromocenters became disorganized and reduced (Fig. 4A-D, fig. S18C, S19A-E), and we confirmed the reduction in H3K9me3 at satellite regions using chromatin immunoprecipitation (Fig. 4E-F). Conversely, overexpression of satRNAs led to an accumulation of H3K9me3 at chromocenters, suggesting that major satRNAs promoted heterochromatin formation (Fig. 4C, G). Knock-down of major satRNAs also reduced facultative heterochromatin levels (H3K27me3) but did not affect euchromatin mark H3K4me3 (fig. S19F-I). Because minor satRNAs were also enriched among LL-RNAs (Fig. 3C-E) and their overexpression increased long-retained EU signals in quiNPCs (fig. S18D-F), we analyzed their roles in heterochromatin retention. Although minor satRNA overexpression significantly increased H3K9me3 at chromocenters, their knock-down did not affect H3K9me3 levels (Fig. 4C-D, G, fig S18C). Altogether, our results suggest a critical role for major satRNAs in the maintenance of heterochromatin integrity in quiNPCs.
Fig. 4. Major satRNAs are essential for maintenance of NPC function.
(A) Distribution of H3K9me3 after LNA-mediated knock-down of major satRNAs in quiNPCs. Scale bar, 5 μm. (B) Quantification of H3K9me3 intensity after knock-down of major satRNAs. **P < 0.01, t-test, dots indicate individual cells from 3 experiments. (C) Distribution of H3K9me3 after CRISPR-mediated transcriptional inhibition (CRISPRi) or overexpression (CRISPRa) of major and minor satRNAs. Scale bar, 5 μm. (D) Transcriptional inhibition of major satRNAs using CRISPRi (dCas9-KRAB with sgRNAs) impaired heterochromatin retention in quiNPCs; minor satRNAs were dispensable. ***P < 0.001, n. s. not significant, t-test, dots represent individual nuclei from 3 independent experiments, group means are indicated. (E-F) LNA-mediated knock-down of major satRNAs reduced H3K9me3 abundance at major and minor satellite repeats. Shown are data from H3K9me3-ChIP-qPCR (*P < 0.05; one-sided t-test, n = 4). Data are presented as mean +/- SD. (G) Upregulation of satRNAs increased nuclear H3K9me3 in quiNPCs. ****P < 0.0001, t-test, dots indicate individual cells from 3 independent experiments, group means are indicated. (H) Experimental timeline for data shown in (I-N). (I) Cell cycle re-entry of quiNPCs after LNA-mediated knock-down of major satRNAs. Arrows indicate pyknotic cells. Scale bar, 25 μm. (J-K) Decreased percentage of Ki67+ and BrdU+ cells one day after re-activation of quiNPCs into proliferation. *P < 0.05, **P < 0.01, paired t-test, n = 5-8. Ki67, LNA control 23.1 ± 2.73%, LNA Majsat 11.8 ± 4.16%; BrdU, LNA control 29.6 ±12.35%, LNA Majsat 19.65 ± 13.64%. (L) Increased percentage of pyknotic cells after knock-down of major satRNAs. LNA control 13.4 ± 2.55%, LNA Majsat 22.76 ± 8.11%. (M-N) Increased percentage of γH2AX+ cells after knock-down of major satRNAs. *P < 0.05, ratio paired t-test, n = 4. LNA control 8.57 ± 3.25%, LNA Majsat 30.9 ± 6.66%. Scale bar in M, 5 μm.
Because satRNAs are generated from heterochromatic areas and bind heterochromatin-associated proteins (19, 20), we tested whether their long-term retention is controlled through association with heterochromatin. Perturbation of H3K9me3 by pharmacological inhibition of histone methyltransferases or by knock-down of heterochromatin-associated protein 1β (Hp1β) increased EU signal retention in quiNPCs (fig. S20), suggesting that heterochromatin is involved in LL-RNA regulation. Heterochromatin is essential to repress aberrant transcription of satRNAs, and increased satRNA expression is a hallmark of impaired epigenetic regulation in tumorigenesis (21, 22). Thus, the balanced level of LL-RNAs may be regulated by heterochromatin in cooperation with LL-RNAs themselves, yet its specific roles in LL-RNA transcription and/or retention require further investigation.
We next tested whether major satRNAs are important for maintaining quiNPCs. We knocked down major satRNAs using LNA-Majsat GapmeRs upon induction of quiescence and tested whether quiNPCs could be reactivated into proliferation (Fig. 4H). One day after reactivation of quiNPCs, the fraction of proliferating cells (Ki67+, BrdU + cells) was significantly reduced in LNA-Majsat-treated cells (Fig. 4I-K), suggesting attenuated reactivation of quiNPCs. Furthermore, significantly more pyknotic nuclei were observed in LNA-Majsat-treated cells (Fig. 4L). Consistent with this observation, the fraction of γH2AX+ cells was significantly higher in LNA-Majsat-treated cells (Fig. 4M, 4N). In contrast, knock-down of satRNAs in proliferating NPCs did not impair proliferative capacities or NPC maintenance (fig. S21). To further characterize the role of satRNAs in NPC maintenance, we assessed the consequences of their overexpression. Overexpression of both major and minor satRNAs was detrimental for NPCs, leading to reduced proliferation, increased apoptosis and DNA damage (fig. S22), indicating that proper control of satRNA levels is critical for NPC maintenance. Taken together, our results suggest that major satRNAs are retained long-term in quiNPCs and play critical roles in the maintenance of heterochromatin integrity and neural stem cell function. Since not all LL-RNAs are satRNAs, further investigations will be required to understand the biological roles of LL-RNAs.
Previously, several proteins had been discovered in the mammalian brain that can persist for years (1, 2), suggesting possible roles for long-lived cellular constituents in brain maintenance and aging. In this study, we found that a subset of RNAs could be retained for up to 2 years in certain brain cell types. Although, in contrast to DNA, RNA has not been considered a stable nuclear component (23), our findings are consistent with a previous study from dormant Xenopus oocytes showing that some RNAs can be retained for more than a year (24). The LL-RNAs we identified were enriched around heterochromatin and essential to maintain chromatin integrity in quiescent neural stem cells. These findings highlight a previously unrecognized temporal axis of RNA metabolism and possible functions for nuclear RNA in long-term epigenetic regulation and genome integrity. We propose that the lifelong maintenance of nuclear function in post-mitotic cells and somatic stem cells involves the dramatic extension of the life span of key molecular constituents, including LL-RNAs.
Supplementary Material
One sentence summary.
Long-lived RNAs in the mammalian brain may contribute to neuronal life span.
Acknowledgments
We thank Mary Lynn Gage for editorial comments, and current and previous members of the Toda and Hetzer labs for helpful discussions. SZ was supported by an Add-on fellowship from the Joachim Herz foundation. NGS data production and data analysis were carried out at the DRESDEN-concept Genome Center, supported by the DFG Research Infrastructure Program (Project 407482635) and part of the Next Generation Sequencing Competence Network NGS-CN (Project 423957469). We thank Maximilian Krause and Andreas Dahl from the DRESDEN-concept Genome Center for support with NGS data acquisition, Georg Girke for molecular cloning of sgRNAs, and Mareike Albert and Annika Kolodziejczyk for providing research materials.
Funding
Boehringer Ingelheim Foundation (TT)
European Research Council (ERC-2018-STG, 804468 EAGER; ERC-2023-COG, 101125034
NEUTIME) (TT)
DZNE (TT)
Deutsche Forschungsgemeinschaft (DFG, German Research Foundation (TO1347/4-1)) (TT)
Footnotes
Author contributions:
Conceptualization: MWH, AM, TT.
Methodology: AM, SZ, TT, RS, FHG, NR, UF, ML, AK.
Visualization: SZ, AM, RS, TT.
Funding acquisition: MWH, TT.
Investigation: SZ, AM, RS, TT.
Project administration: MWH, TT.
Resource: FHG, TT.
Supervision: MWH, TT.
Writing – original draft: TT, MHW.
Revision work: SZ, AK, TT.
Writing – revised manuscript: SZ, MHW, TT.
Writing – review & editing: TT, AM, RS, SZ, FHG, MHW.
Competing interest: The authors declare no competing or financial interests.
This is the author’s version of the work. It is posted here by permission of the AAAS for personal use, not for redistribution. The definitive version was published in Science on 2024, April 5th, DOI: [http://dx.doi.org/10.1126/science.adf3481]. https://www.science.org/doi/10.1126/science.adf3481
Data and materials availability
All data are available in the main text, figures or supplementary materials. EU-RNA-Sequencing data were submitted to GEO (GSE248101).
References
- 1.Savas JN, et al. Extremely long-lived nuclear pore proteins in the rat brain. Science. 2012;335:942. doi: 10.1126/science.1217421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Toyama BH, et al. Identification of long-lived proteins reveals exceptional stability of essential cellular structures. Cell. 2013;154:971–982. doi: 10.1016/j.cell.2013.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eriksson PS, et al. Neurogenesis in the adult human hippocampus. Nat Med. 1998;4:1313–1317. doi: 10.1038/3305. [DOI] [PubMed] [Google Scholar]
- 4.Spalding KL, et al. Dynamics of hippocampal neurogenesis in adult humans. Cell. 2013;153:1219–1227. doi: 10.1016/j.cell.2013.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bhat P, et al. Nuclear compartmentalization as a mechanism of quantitative control of gene expression. Nat Rev Mol Cell Biol. 2021;22:653–670. doi: 10.1038/s41580-021-00387-1. [DOI] [PubMed] [Google Scholar]
- 6.Loda A, et al. Gene regulation in time and space during X-chromosome inactivation. Nat Rev Mol Cell Biol. 2022;23:231–249. doi: 10.1038/s41580-021-00438-7. [DOI] [PubMed] [Google Scholar]
- 7.Jao CY, et al. Exploring RNA transcription and turnover in vivo by using click chemistry. Proc Natl Acad Sci USA. 2008;105:15779–15784. doi: 10.1073/pnas.0808480105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berg DA, et al. A Common Embryonic Origin of Stem Cells Drives Developmental and Adult Neurogenesis. Cell. 2019;177:654–668.:e615. doi: 10.1016/j.cell.2019.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nicola Z, et al. Development of the adult neurogenic niche in the hippocampus of mice. Front Neuroanat. 2015;9:53. doi: 10.3389/fnana.2015.00053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ahlfeld J, et al. Neurogenesis from Sox2 expressing cells in the adult cerebellar cortex. Sci Rep. 2017;7:6137. doi: 10.1038/s41598-017-06150-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yacubova E, et al. Cellular and molecular mechanisms of cerebellar granule cell migration. Cell Biochem Biophys. 2003;37:213–234. doi: 10.1385/cbb:37:3:213. [DOI] [PubMed] [Google Scholar]
- 12.Martynoga B, et al. Epigenomic enhancer annotation reveals a key role for NFIX in neural stem cell quiescence. Genes Dev. 2013;27:1769–1786. doi: 10.1101/gad.216804.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huo X, et al. The Nuclear Matrix Protein SAFB Cooperates with Major Satellite RNAs to Stabilize Heterochromatin Architecture Partially through Phase Separation. Mol Cell. 2020;77:368–383.:e367. doi: 10.1016/j.molcel.2019.10.001. [DOI] [PubMed] [Google Scholar]
- 14.Velazquez Camacho O, et al. Major satellite repeat RNA stabilize heterochromatin retention of Suv39h enzymes by RNA-nucleosome association and RNA:DNA hybrid formation. Elife. 2017;6 doi: 10.7554/eLife.25293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shirai A, et al. Impact of nucleic acid and methylated H3K9 binding activities of Suv39h1 on its heterochromatin assembly. Elife. 2017;6 doi: 10.7554/eLife.25317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Novo CL, et al. Satellite repeat transcripts modulate heterochromatin condensates and safeguard chromosome stability in mouse embryonic stem cells. Nat Commun. 2022;13:3525. doi: 10.1038/s41467-022-31198-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abelson HT, et al. Changes in RNA in relation to growth of the fibroblast: II. The lifetime of mRNA, rRNA, and tRNA in resting and growing cells. Cell. 1974;1:161–165. doi: 10.1016/0092-8674(75)90135-x. [DOI] [PubMed] [Google Scholar]
- 18.Probst AV, et al. A strand-specific burst in transcription of pericentric satellites is required for chromocenter formation and early mouse development. Dev Cell. 2010;19:625–638. doi: 10.1016/j.devcel.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 19.Zhu Q, et al. Heterochromatin-Encoded Satellite RNAs Induce Breast Cancer. Mol Cell. 2018;70:842–853.:e847. doi: 10.1016/j.molcel.2018.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maison C, et al. SUMOylation promotes de novo targeting of HP1alpha to pericentric heterochromatin. Nat Genet. 2011;43:220–227. doi: 10.1038/ng.765. [DOI] [PubMed] [Google Scholar]
- 21.Ting DT, et al. Aberrant overexpression of satellite repeats in pancreatic and other epithelial cancers. Science. 2011;331:593–596. doi: 10.1126/science.1200801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu Q, et al. BRCA1 tumour suppression occurs via heterochromatin-mediated silencing. Nature. 2011;477:179–184. doi: 10.1038/nature10371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rabani M, et al. High-resolution sequencing and modeling identifies distinct dynamic RNA regulatory strategies. Cell. 2014;159:1698–1710. doi: 10.1016/j.cell.2014.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ford PJ, et al. Very long-lived messenger RNA in ovaries of Xenopus laevis. Dev Biol. 1977;57:417–426. doi: 10.1016/0012-1606(77)90226-3. [DOI] [PubMed] [Google Scholar]
- 25.Toda T, et al. Nup153 Interacts with Sox2 to Enable Bimodal Gene Regulation and Maintenance of Neural Progenitor Cells. Cell Stem Cell. 2017;21:618–634. doi: 10.1016/j.stem.2017.08.012. [DOI] [PubMed] [Google Scholar]
- 26.Zocher S, Kempermann G. Generation of mouse hippocampal neural precursor cell lines with CRISPR/Cas9-mediated gene knockouts. STAR Protoc. 2021;2:100472. doi: 10.1016/j.xpro.2021.100472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Imamachi N, et al. BRIC-seq: A genome-wide approach for determining RNA stability in mammalian cells. Methods. 2014;67:55–63. doi: 10.1016/j.ymeth.2013.07.014. [DOI] [PubMed] [Google Scholar]
- 28.Palozola KC, et al. EU-RNA-seq for in vivo labeling and high throughput sequencing of nascent transcripts. STAR Protoc. 2021;2:100651. doi: 10.1016/j.xpro.2021.100651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liao Y, et al. FeatureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 30.Robinson MD, et al. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–40. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu G, He Q. Molecular BioSystems ReactomePA: an R/Bioconductor package for reactome pathway analysis and visualization. Mol Biosyst. 2016;12:477–479. doi: 10.1039/c5mb00663e. [DOI] [PubMed] [Google Scholar]
- 32.Smit A, et al. RepeatMasker Open-40. http://www.repeatmasker.org .
- 33.Pimentel H, et al. Differential analysis of RNA-seq incorporating quantification uncertainty. Nat Methods. 2017;14:687–690. doi: 10.1038/nmeth.4324. [DOI] [PubMed] [Google Scholar]
- 34.Rosenberg AB, et al. Single-cell profiling of the developing mouse brain and spinal cord with split-pool barcoding. Science. 2018;182:176–182. doi: 10.1126/science.aam8999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Berg DA, et al. A Common Embryonic Origin of Stem Cells Drives Developmental and Adult Neurogenesis. Cell. 2019;177:654–668.:e15. doi: 10.1016/j.cell.2019.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hochgerner H, et al. Conserved properties of dentate gyrus neurogenesis across postnatal development revealed by single-cell RNA sequencing. Nat Neurosci. 2018;21:290–299. doi: 10.1038/s41593-017-0056-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available in the main text, figures or supplementary materials. EU-RNA-Sequencing data were submitted to GEO (GSE248101).