Abstract
Metabolic changes contribute to cancer initiation and progression through effects on cancer cells, the tumour microenvironment and whole-body metabolism. Alterations in serine metabolism and the control of one-carbon cycles have emerged as critical for the development of many tumour types. In this Review, we focus on the mitochondrial folate cycle. We discuss recent evidence that in addition to supporting nucleotide synthesis, mitochondrial folate metabolism also contributes to metastasis through support of antioxidant defence, mitochondrial protein synthesis and the overflow of excess formate. These observations offer potential therapeutic opportunities, including the modulation of formate metabolism through dietary interventions and the use of circulating folate cycle metabolites as biomarkers for cancer detection.
Introduction
The observation that cancer cells show increased nutrient uptake and alterations in metabolism was noted by Otto Warburg almost a century ago1. Since then, alterations in many metabolic pathways have been shown to underpin the acquisition of malignant behaviour, highlighting numerous potential options for therapeutic intervention2. The first – and arguably still the most clinically impactful – metabolic therapy was pioneered in the 1950’s by Sidney Farber, who used the folate analogue, aminopterin3, to induce remission of childhood acute lymphocytic leukaemia (ALL). The original rationale for this approach was based on Farber’s empirical observation that folic acid stimulated the progression of ALL. Since then, the anti-folates have been shown to function as competitive inhibitors of dihydrofolate reductase (DHFR), the enzyme that produces tetrahydrofolate (THF) from folate. THF is an essential cofactor in nucleotide synthesis4 and the inhibition of DHFR results in depletion of intracellular pools of folate species5, leading to reduced synthesis of purines and pyrimidines6. Today, inhibitors of folate metabolism such as methotrexate are used extensively as first line chemotherapy for many types of cancer7. However, antifolate drugs are accompanied by many adverse effects due to the requirement of folate in normal, non-malignant cells – leading to a search for strategies beyond direct inhibition of DHFR that retain the efficacy of anti-folates without the limiting toxicities.
Folate metabolism is part of the broader network of one-carbon (1C) metabolism, which describes the pathways that transfer 1C units for numerous physiological processes, such as nucleotide synthesis, methylation reactions, and redox defence. The 1C units that sustain the intersecting folate and methionine cycles are derived from several sources, including serine and glycine, tryptophan, choline and sarcosine metabolism (Figure 1). The methionine cycle is critically important for the production of S-adenosyl methionine (SAM), used for methylation reactions that modify and regulate nucleic acids, proteins and lipids, as well as the production of phospholipids, polyamines and cysteine (Figure 1). As such, the methionine cycle impacts all aspects of normal and diseased states, including cancer8,9. However, in this Review we will focus on the closely linked folate cycle, and recent advances in our understanding of its function in tumorigenesis.
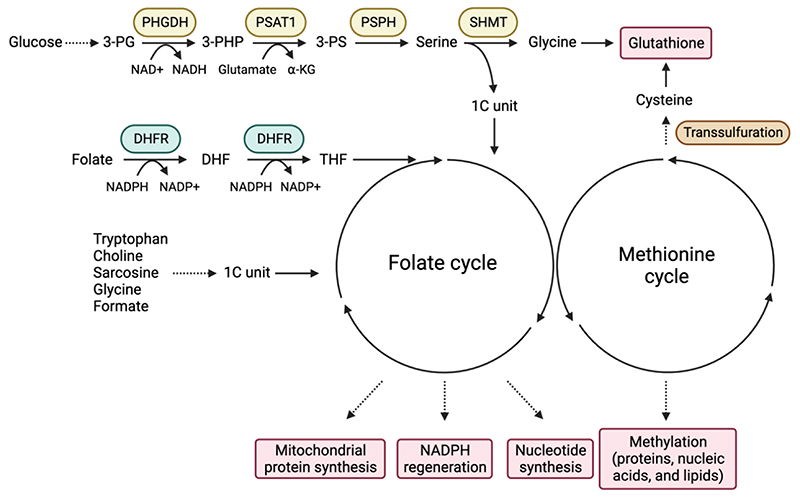
Figure 1. The serine synthesis pathway, folate cycle and methionine cycle.
The serine synthesis pathway (SSP) uses glycolytic intermediate 3-phosphoglycerate (3-PG) in three sequential reactions to generate serine. First, phosphoglycerate dehydrogenase (PHGDH) catalyses the NAD+-dependent oxidation of 3-PG to 3-phosphohydroxypyruvate (3-PHP). Second, phosphoserine aminotransferase 1 (PSAT1) converts 3-PHP into 3-phosphoserine (3-PS) in a glutamate-dependent transamination reaction. Third, phosphoserine phosphatase (PSPH) generates serine through hydrolysis of 3-PS. Serine hydroxymethyltransferase (SHMT) converts serine into glycine by donating a one-carbon (1C) unit to tetrahydrofolate (THF). Glycine functions as a precursor for glutathione. Folate is reduced to dihydrofolate (DHF) and THF through dihydrofolate reductase (DHFR)-mediated NADPH-dependent reactions. THF serves as a carrier of 1C unit through the folate cycle. The folate cycle contributes to mitochondrial protein synthesis, NADPH regeneration and nucleotide synthesis. The folate cycle is coupled with the methionine cycle that generates the universal methyl-donor SAM for the methylation of proteins, nucleic acids and lipids and provides cysteine, a precursor of glutathione. Tryptophan, choline, sarcosine, glycine and formate can all generate 1C units.
While targeting folate metabolism to impair nucleotide synthesis is a well-established therapeutic paradigm, the folate cycle contributes to a wide range of metabolic reactions beyond nucleotide synthesis. As we will discuss, recent studies have revealed how the folate cycle can influence cancer progression and the tumour microenvironment through regulation of antioxidant defence, mitochondrial translation and formate overflow, revealing contributions of folate cycle intermediates on tumour growth, metastatic dissemination and the microenvironment.
Serine and glycine metabolism
Serine and glycine are freely interconvertible, non-essential amino acid (NEAA) that support several metabolic pathways, including the production of phospholipids such as sphingolipids and phosphatidylserine, glutathione, porphyrins, creatine, purines and pyrimidines (Figure 2). However, serine is also a major donor of 1C units to the folate cycle. Serine can be taken up from the environment using several different transporters10 or generated through the de novo serine synthesis pathway (SSP). Briefly, the glycolytic intermediate 3-phosphoglycerate (3-PG) is converted to serine by three consecutive enzymatic reactions that are catalysed by phosphoglycerate dehydrogenase (PHGDH), phosphoserine aminotransferase 1 (PSAT1), and phosphoserine phosphatase (PSPH)11 (Figure 1).
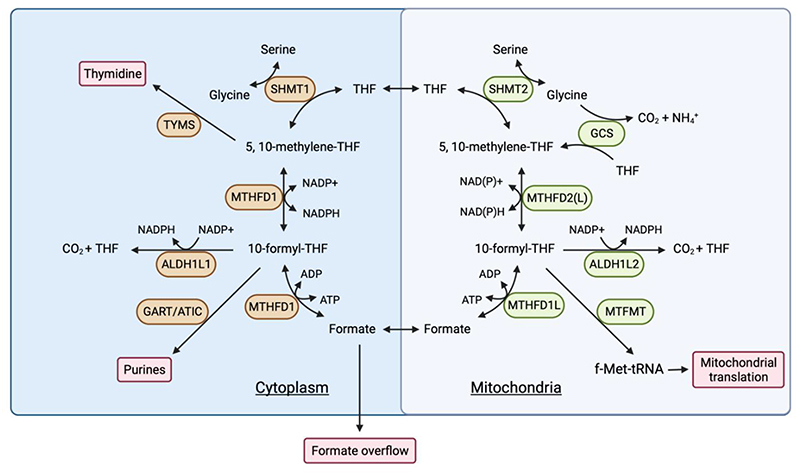
Figure 2. Folate metabolism in cytoplasm and mitochondria.
Folate metabolism occurs in both the cytoplasm and mitochondria. Serine hydroxymethyltransferase 1 (SHMT1) in the cytoplasm or SHMT2 in the mitochondria generates glycine and donates one-carbon (1C) unit to THF, producing 5,10-methylene-THF. In the mitochondria, methylenetetrahydrofolate dehydrogenase 2 (MTHFD2) or MTHFD2-like (MTHFD2L) oxidize 5,10-methylene-THF to 5,10-methenyl-THF and 10-formyl-THF, regenerating NADPH or NADH. Mitochondrial 10-formyl-THF functions as the substrate for the generation of formate by MTHFD1-like (MTHFD1L), formylation of the methionine loaded mitochondrial initiator tRNA by mitochondrial methionyl-tRNA formyltransferase (MTFMT) for mitochondrial translation18,19 and the release of the 1C unit as CO2 by aldehyde dehydrogenase 1 family member L2 (ALDH1L2) in a reaction that regenerates NADPH. Mitochondrial formate is transported to the cytoplasm and used to regenerate 10-formyl-THF and 5,10-methylene-THF by MTHFD1. Surplus formate is excreted from the cell in a process termed formate overflow. The glycine cleavage system (GCS) can donate 1C units to the folate cycle. In the cytoplasm, cytosolic 5,10-methylene-THF serves as the substrate for thymidylate synthesis by thymidylate synthetase (TYMS). Cytosolic 10-formyl-THF functions as the substrate for the synthesis of purine nucleotides by phosphoribosylglycinamide formyltransferase (GART) and 5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase (ATIC).
Folate metabolism takes place in both the cytosol and mitochondria, using distinct but similar enzymes localised to each subcellular compartment (Figure 2). Mammalian cells are unable to produce folate (also called vitamin B9), which is present in standard culture media in vitro or provided in the diet or by the microbiota in vivo. Folate is reduced to tetrahydrofolate (THF – the biologically active form of folate) and dihydrofolate through an NADPH-dependent reaction catalysed by DHFR (the target of methotrexate). The conversion of serine to glycine by serine hydroxymethyltransferase 1 (SHMT1) in the cytosol or SHMT2 in the mitochondria produces glycine and donates a 1C unit to THF, producing 5,10-methylene-THF.
Interestingly, two recent studies have shown that the major source of folate in physiological circulation is 5-methyl THF, with unmetabolized folic acid contributing less than 10% of total folate species12,13. Dietary folates, mainly found in vegetables, exist mostly under the polyglutamyl form, which is cleaved into the corresponding monoglutamyl forms by glutamate carboxypeptidase II (GCPII) present at the brush-border of enterocytes. The monoglutamyl forms - predominantly 5-methylTHF - are then absorbed by the enterocytes and delivered into the circulation14,15. Cells taking up this form of folate utilise the vitamin B12 (cobalamin)-dependent enzyme methionine synthase (MTR) to transfer the methyl group from 5-methylTHF to homocysteine - producing methionine and supplying THF to the folate cycle16,17. This pathway for the acquisition of folate renders tumour cells less dependent on DHFR - and therefore resistant to methotrexate - but more sensitive to loss of MTR, which leads to folate trapping and reduction of purine and pyrimidine synthesis16,17.
In the mitochondria, 5,10-methylene-THF is oxidized by methylenetetrahydrofolate dehydrogenase 2 (MTHFD2) or MTHFD2-like (MTHFD2L) in a two-step reaction, the first producing 5,10-methenyl-THF – a reaction that also generates NADPH (or NADH) - and the second to produce 10-formyl-THF. Mitochondrial 10-formyl-THF serves as the substrate for three further reactions; the generation of formate by MTHFD1-like (MTHFD1L), formylation of the mitochondrial initiator tRNA by mitochondrial methionyl-tRNA formyltransferase (MTFMT)18,19, and the release of the 1C unit as CO2 by aldehyde dehydrogenase 1 family member L2 (ALDH1L2) – a second step in mitochondrial 1C metabolism to generate NADPH. A similar series of cytosolic enzymes also have the potential to generate 10-formyl THF. However, in most cells, the directionality of 1C metabolism is from the mitochondria to cytoplasm20,21. As 1C loaded THF conjugates cannot be transported across the mitochondrial membrane, mitochondrial formate is transported to the cytosol, where it serves to regenerate formyl-THF pools for purine synthesis and methylene-THF pools for thymidine synthesis, or is excreted from the cell in a process termed formate overflow22,23.
Interestingly, despite the predominance of the mitochondrial serine metabolism in supplying 1C unit for nucleotide synthesis, the cytosolic 1C flux from serine can compensate to sustain tumour growth when the mitochondrial pathway is inhibited20. However, ablating mitochondrial 1C metabolism can result in folate degradation in the cytosol, an effect that is also seen in response to methotrexate21. Overall, it has been suggested that the two connected cycles in the mitochondria and cytosol allow for the maintenance of both glycine and 10-formyl/ 5,10-methylene-THF synthesis20.
Alternative sources of 1C units
While serine is the major donor of 1C units to fuel the folate cycle, other potential sources of 1C units can be found12. Both the production of glycine through choline metabolism and catabolism of glycine can result in the transfer of 1C units onto THF to produce 5,10-methylene-THF24. Glycine donates a 1C unit to THF to produce 5,10-methylene-THF in a reaction catalysed by a mitochondrial enzymic complex called the glycine cleavage system (GCS)25. However, the principal role of the GCS appears to be to limit glycine accumulation, with germline mutations in GLDC (a major component of the GCS) resulting in glycine encephalopathy26. Most recently, the 1C units produced by GCS have been suggested to drive reverse flux of SHMT2, producing serine and clearing excess mitochondrial glycine in the liver27. Choline contributes 1C units to support both the methionine cycle and – through the production of sarcosine – the mitochondrial folate cycle. Histidine catabolism can also produce 5,10-methylene-THF through the intermediate of 5-formimino-THF28,29 although the contribution of histidine degradation to the pool of 10-formyl-THF remains to be fully quantified.
Several other pathways sustain the 1C pool in cells through the production of formate. The catabolism of tryptophan produces N-formyl-kynurenine, which subsequently releases formate to form kynurenine. Formate produced through this pathway was shown to contribute to de novo purine synthesis and formate overflow30. Formate can also be directly produced by demethylation reactions, notably during cholesterol biosynthesis where lanosterol is demethylated to cholestatrienol by lanosterol 14α-demethylase31. The demethylation of endogenous metabolites or xenobiotics, and methanol metabolism also produce formaldehyde, a toxin that is either directly oxidized to formate by the mitochondrial NAD+ dependent aldehyde dehydrogenase (ALDH) 2 or indirectly through a detoxification pathway involving reduced glutathione and ALDH532,33. Interestingly, formate generation from formaldehyde is sufficient to sustain the growth of cells that cannot utilize serine33. Finally, formate can also be produced and utilized by numerous bacterial species found in the colonic microbiome34–37.
Alterations of serine & 1C metabolisms in cancer
Upon amino acid deprivation, eukaryotic cells activate an adaptive response called the integrated stress response to maintain cellular homeostasis38. This leads to the increased expression of the transcription factor activating transcription factor 4 (ATF4), which induces expression of serine and 1C metabolism-related genes, such as PHGDH, PSAT1, PSPH and SHMT239–42. Serine starvation therefore upregulates expression of SSP genes to compensate for the loss of extracellular serine40,43. Activation of mammalian target of rapamycin complex1 (mTORC1), a major metabolic regulator, or induction of nuclear factor erythroid 2-related factor 2 (NRF2), a major antioxidant transcription factor, both also promote expression of SSP enzymes through ATF439,41,42,44. Serine starvation activated ATF4 also induces the transcription factor ATF3, which in turn drives expression of the SSP enzymes45.
Serine itself can also modulate central carbon metabolism, by binding to and activating the M2 isoform of pyruvate kinase (PKM2) a glycolytic enzyme that controls flux of pyruvate to lactate. Decreased serine levels lower PKM2 activity, allowing increased channelling of glucose-derived carbon to de novo SSP40,46.
Genetic alteration in cancer
The importance of serine as a precursor of diverse pathways that control biosynthesis, epigenetic regulation, redox homeostasis, ATP generation, and TCA cycle regulation explains why cancer cells – and many normal cells – have a high demand for either endogenous or exogenous serine47–49. Consequently, genetic alterations of the SSP and 1C metabolism genes are frequently observed in cancers..
Copy number gain or overexpression of PHGDH and PSPH are found in several cancer types, including ER-negative breast cancers, melanoma, lung adenocarcinoma, T-cell ALL and osteosarcoma48,50. Chromosomal regions 1p12 and 7p11 - containing PHGDH and PSPH respectively - are amplified in cancers, with 7p11 amplifications potentially also encompassing the closely located oncogene, epidermal growth factor receptor (EGFR)48,51–53. In many cases, increased expression of the SSP enzymes correlates with poor prognosis54–57. In contrast to amplification, 5-methylthioadenosine phosphorylase (MTAP) - an enzyme that contributes to methionine salvage from S-adenosylmethionine (SAM) - is frequently deleted in cancer patients due to its proximity to the tumour suppressor gene, cyclin dependent kinase inhibitor 2A (CDKN2A), on chromosome 9p2158,59. Loss of MTAP leads to the accumulation of MTA and renders these cancer cells vulnerable to inhibition of protein arginine methyltransferase 5 (PRMT5).
Most cells can take up exogenous serine and in normal tissue loss of PHGDH (leading to loss of de novo serine synthesis) is generally well tolerated, although some changes in liver ceramide and lipid profiles are detected60. Interestingly, cancer cells can become dependent on PHGDH activity, even when supplied with exogenous serine, suggesting that serine uptake cannot always compensate from endogenously produced serine or that PHGDH has functions beyond serine synthesis (see below). By contrast, cells can become dependent on exogenous serine when decreased NAD+/NADH ratios impede the use of the SSP47. This was recently confirmed in mitochondrial malate/aspartate shuttle (MAS) deficient cells in which lower cytosolic NAD+/NADH ratios were accompanied by lower de novo serine synthesis. Supplementation of pyruvate restored the availability of NAD+ in MAS deficient cells, resulting in increased PHGDH activity61.
Downstream of serine, enzymes of the folate cycle are also found to be upregulated in multiple cancers. For example, RNA profiling of metabolic enzymes across 19 cancer types found MTHFD2 is frequently overexpressed 62. Enhanced expression of mitochondrial folate cycle enzymes such as SHMT2, MTHFD2, and MTHFD1L was also detected in cancer cell lines using metabolic profiling, and this was associated with higher mortality in breast cancer patients63. In glioma, increased SHMT2 expression was found to be critical for cancer cell survival and adaptation to the ischaemic tumour microenvironment (TME)64, a response that made cancer cells dependent on glycine decarboxylase (GLDC) to prevent the accumulation of excess glycine. The SHMT2 gene is amplified in B cell lymphomas due to copy number gain, contributing to lymphoma development by epigenetic modulation of tumour suppressor gene expression9. Progression to metastatic disease has also been linked to increased expression of mitochondrial serine and 1C metabolism genes65.
Enhanced expression of the GCS enzyme GLDC is detected in various cancers63,66, although this reflects a requirement of cancer cells to remove glycine rather than a need for 1C units. Interestingly, in vitro studies showed that in several cancer cell lines, glycine could not substitute for serine in supporting cell growth – rather increased glycine in the absence of serine inhibited proliferation67. In hepatocellular carcinoma (HCC), the GCS glycine cleavage system also plays a role in maintaining protein lipoylation and mitochondrial activity, rather than supplying 1C units68. Most recently, the 1C units produced by GCS have been suggested to drive reverse flux of SHMT2 to clear excess mitochondrial glycine in the healthy liver. This regulation of homeostatic glycine levels could be an important factor when dietary depletion of serine is considered for cancer therapy27.
Oncogene and tumour suppressor mediated control of serine metabolism
Reflecting the importance of serine metabolism in cancer development, several oncogenes and tumour suppressor have been shown to regulate serine and formate metabolism. MYC, a transcription factor frequently deregulated in cancers, increases the expression of SSP and mitochondrial 1C metabolism related enzymes, alone or in collaboration with other transcription factors such as activating transcription factor 4 (ATF4) or hypoxia-inducible factor1α (HIF1α)69–72. Activation of the internal tandem duplication of FMS-like tyrosine kinase 3 (FLT3-ITD) - an alteration frequently observed in acute myeloid leukaemia patients - also upregulates the SSP in a mechanistic target of rapamycin complex 1 (mTORC1)-ATF4-dependent manner, making these tumours sensitive to serine biosynthesis inhibition73. Similarly, inhibitor of nuclear factor kappa B (IkB) kinase ε, a regulator of cytokine secretion, can induce the SSP enzymes through ATF4 activation74. Mitochondrial respiratory chain defects – a common feature of malignancies75 – result in decreased flux through the mitochondrial 1C cycle but also lead to an ATF4 -dependent upregulation of SSP enzymes76. Activated KRAS increases the dependence of cancer cells on folate metabolism and concurrently upregulates expression of SSP enzymes through nuclear factor erythroid 2-related factor 2 (NRF2)39,49,77,78. NRF2 responds to oxidative stress and its ability to enhance serine metabolism has been linked to an increased production of NADPH for antioxidant defense39. In a pancreas cancer model, loss of liver kinase B1 (LKB1) with KRAS activation induced serine and 1C metabolism, altering the epigenetic landscape by upregulating SAM levels and DNA methylation44.
p53 is a tumour suppressor protein that is lost or mutated in most cancers79. p53 responds to oncogenic stress and - depending on the context - either helps cells to survive and adapt to mild or transient stress or eliminates cells exposed to persistent or severe stress80. Consistently, p53 can both support cell survival under conditions of serine starvation81 and sensitize cells to serine starvation by downregulating the expression of PHGDH82. The ubiquitin ligase, mouse double minute 2 (MDM2), is a transcriptional target of p53 that functions as an oncogene - in part by restraining p53 activity. MDM2 inhibits DHFR by catalysing its monoubiquitination83, but can also be recruited to chromatin to enhance serine and 1C metabolism independently of p5384.
Another key regulator of metabolic homeostasis that can function to suppress or support tumorigenesis is AMP-activated protein kinase (AMPK). AMPK indirectly downregulates expression of the 1C metabolism enzymes, MTHFD1, MTHFD1L and MTHFD2 in breast cancer, raising the possibility that AMPK activators may sensitize cancers to anti-folate therapy85. Atypical isoforms of protein kinase C (PKC) also regulate serine and 1C metabolism. PKCζ represses the expression of SSP enzymes, PHGDH and PSAT1 and inhibits PHGDH enzymatic activity by phosphorylation. PKCζ loss therefore promotes the ability of cancer cells to synthesize serine86. Additionally, loss of PKCλι induces serine metabolism through the mTORC1/ATF4-mediated mechanism, which contributes to neuroendocrine prostate cancer progression via increased DNA methylation87.
How does serine and 1C metabolism support cancer development and progression?
The observation that many cancers depend on enhanced serine and 1C metabolism prompts the question of how they support tumorigenesis. Uncontrolled proliferation of cancer cells fuels a demand for nucleotide synthesis that can be met – in part – by enhanced 1C metabolism67,88, the importance of which has been well described and reviewed89,90. For example, increased mitochondrial 1C metabolism induced by the growth regulating kinase mTORC1 – which is frequently activated in cancer - is required for purine production and cell growth42. Upregulation of serine and 1C metabolism confers therapeutic resistance to 5-fluorouracil (5-FU), a commonly used chemotherapy that inhibits DNA synthesis91. As a result, 5-FU-resistant colorectal cancer cells become dependent on serine metabolism to support purine biosynthesis92, highlighting a potentially targetable induced vulnerability.
Despite the clear importance of nucleotide production, it is becoming increasingly clear that other aspects of serine, glycine and folate metabolism may contribute to cancer development, progression and metastasis.
Redox defence
Serine and 1C metabolism can contribute to redox balance by either providing intermediates such as glycine and cysteine or by contributing to reactions that generate NADPH. Serine-derived glycine (from the SHMT reaction) and cysteine (from the transsulfuration pathway (Figure 1) are components of glutathione (GSH) - a major antioxidant molecule. Under limiting serine conditions, cancer cells prioritize the synthesis of GSH to maintain antioxidant defense81. 1C metabolism also generates NADPH, which is used to reduce the oxidized forms of GSH and thioredoxin – another antioxidant. As described above, a number of reactions in the 1C cycle use or generate NADPH, depending on the directionality of cycle12. Under most conditions, flow through the mitochondrial 1C pathway generates mitochondrial NADPH through the activity of MTHFD2 and ALDH1L2 (Figure 2). Perturbation of mitochondrial 1C metabolism decreases the NADPH/NADP+ and GSH/GSSG ratios93,94 and numerous studies have underpinned the role of serine metabolism in antioxidant defence in different contexts72,94–98. This activity becomes important during tumorigenesis, where oncogene activation, loss of normal stromal support and hypoxia lead to increased levels of reactive oxygen species (ROS) that result in a requirement for enhanced antioxidant activity to maintain viability (Figure 3). This requirement can be met by increased 1C metabolism, for example, hypoxia due to insufficient blood supply leads to the upregulation of SHMT2, leading to the maintenance of NADPH production and redox balance in multiple cancer cell lines72,99. In melanoma, inhibition of folate metabolism with methotrexate or ALDH1L2 knockdown decreases NADPH production and increases ROS100. Similarly, ALDH1L2 depletion has been shown to increase mitochondrial ROS levels in breast cancer cells20,94,97,100,101.
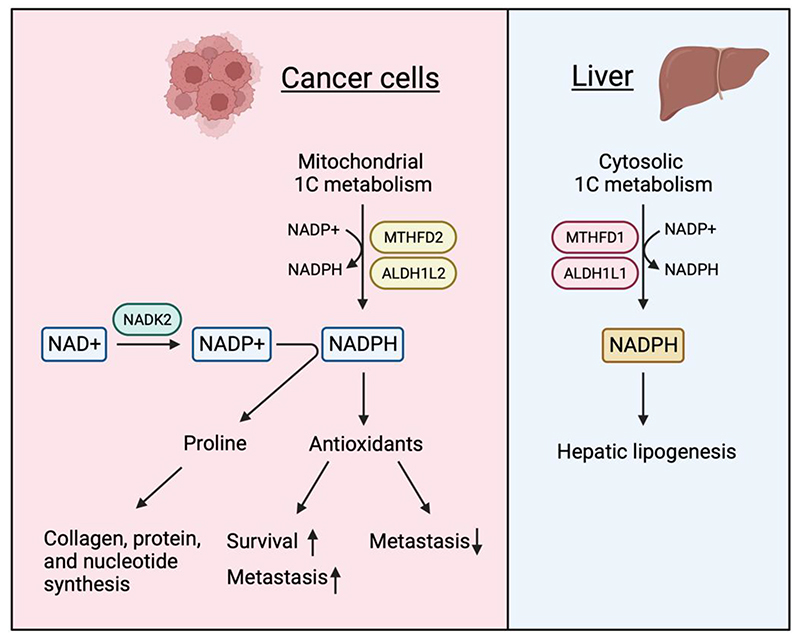
Figure 3. Mitochondrial and cytosolic NADPH use in cancer cells and hepatocytes.
Cancer cells generate mitochondrial NADPH through methylenetetrahydrofolate dehydrogenase 2 (MTHFD2) and aldehyde dehydrogenase 1 family member L2 (ALDH1L2)-mediate mitochondrial one-carbon (1C) metabolism. Mitochondrial NAD kinase 2 (NADK2) produces NADP+ from NAD+, leading to an increase in the pool of NADPH. Increased NADPH pools in cancer cells are required for proline synthesis that supports the synthesis of collagen, protein and nucleotide and for antioxidant defence. Limitation of reactive oxygen species (ROS) can promote or impede metastasis in a context-dependent manner. In the liver, cytosolic NADPH generated through MTHFD1 and ALDH1L2-mediated 1C metabolism contributes to hepatic lipogenesis.
The upregulation of 1C metabolism-mediated antioxidant defence may also contribute to therapy resistance102. However, there are other pathways for mitochondrial NADPH regeneration (such as malic enzyme 2/3 and isocitrate dehydrogenase 2) or production (such as NAD kinase). Furthermore, NADPH itself is important for other functions, such as proline or lipid production95,98,103,104. How cells respond to the modulation of NADPH production through 1C metabolism is therefore highly dependent on the system and context.
Mitochondria translation
Serine-derived sphingolipids such as ceramides are required for mitochondrial function and cell death induced by serine starvation may be partly due to structural defects of the mitochondria mediated by a reduction in ceramide. Serine and mitochondrial 1C metabolism also make important contributions to mitochondrial protein production105. MTFMT uses 10-formyl-THF to formylate methionine loaded tRNA (fMet-tRNA), which initiates mitochondrial protein synthesis18. Deletion of SHMT2 in mice reduces the availability of fMet-tRNA, leading to a decrease in the production of mitochondrial proteins and defects in mitochondrial respiration106 that result in a failure of cancer cells to adapt under glucose starvation19. Interestingly, the conversion of 10-formyl-THF to THF and CO2 by ALDH1L2 not only produces NADPH, but also limits other 10-formyl-THF fuelled reactions, including the production of formyl-methionine (fMet) and formate101 (Figure 2). As a result, while deletion of ALDH1L2 in breast cancer cell lines led to more ROS, it also increased mitochondrial protein synthesis101.
Formate overflow
Formate is a key product of mitochondrial 1C metabolism, illustrated by the observation that the embryonic lethality resulting from deletion of MTHFD1L or the impaired T-cell function in aging mice that is associated with reduced mitochondrial metabolism can be at least partially rescued by formate supplementation107,108. Intriguingly, the rate of folate metabolism in normal cells and tissue exceeds the demand of 1C units for nucleotide synthesis, leading to an excess production of formate, which is excreted from the cell. Cancer cells show an even higher rate of formate overflow23, contributing to an increase in circulating formate in tumour bearing mice that is dependent on serine23,109 (Figure 4). However, circulating formate levels can be either increased or decreased in cancer patients, depending on the tumour type110. While serine starvation and inhibition of 1C metabolism enzymes such as MTHFD1L and MTHFD2 decrease formate production, depletion of ALDH1L2 allows for an increased use of 10-formyl-THF for formate production101. Several studies show an important role for formate overflow in supporting malignancies, with multiple explanations for the benefit of this apparently wasteful metabolic activity. Firstly, formate overflow may simply be a by-product of a high rate of flux through the mitochondrial 1C cycle to generate ATP and contribute NADH to support mitochondrial respiration. There is evidence that inhibiting MTHFD1L (the step producing formate from 10-formyl-THF) leads to a switch in energy production to glycolysis23 and, consistently, formate overflow was shown to be a hallmark of oxidative cancers, with formate levels directly correlating to NADH/NAD+ ratios in a large panel of cancer cells types109. Additionally, there is accumulating evidence that formate itself provides several advantages to cancer cells. Formate may induce a metabolic switch to favour nucleotide and energy production111, with formate availability leading to increased purine production, while formate deficiency results in a reduction of AICAR, an intermediate of de novo purine synthesis and activator of AMPK111.
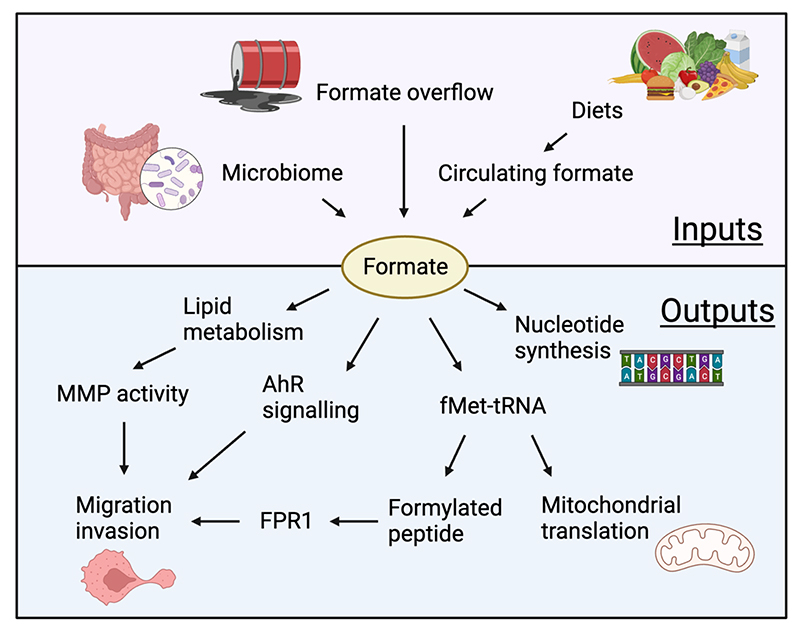
Figure 4. Inputs and outputs of formate metabolism.
Formate can be derived from the microbiome, diet and formate overflow and can contribute to the behaviour of cancer cells through several mechanisms. Formate supports proliferation by functioning as substrate of nucleotide synthesis. Formate-derived formylated methionine-loaded tRNA (fMet-tRNA) initiates mitochondrial protein translation and migration and formylated peptides drive the invasion of cancer cells through formyl-peptide receptor (FPR) signalling. In addition, formate can increase migration and invasion of cancer cells through aryl hydrocarbon receptor (AhR) signalling and lipid metabolism/ matrix metalloproteinase (MMP) activity.
In the context of cancer, the contribution of the formate dependent metabolic switch to cancer development could explain, to some extent, the dependency of these cancers on serine and 1C enzymes. Furthermore, recent studies have highlighted growth-independent consequences of formate overflow and have uncovered an ability of formate to increase the migrative and invasive potential of cancer cells, contributing to metastasis as discussed further below 101,112.
A role for 1C metabolism intermediates in metastasis
Cancer mortality predominantly results from metastatic spread of the primary cancer to other organs, a point at which therapeutic interventions show limited efficacy. Disseminating cancer cells face numerous obstacles as they move out of the tumour mass, circulate in the blood or lymphatic system and enter distant organs to re-establish growth. One key requirement for metastasis is the ability to move into and out of the circulation (intravasation and extravasation), partially reflected in model systems by the ability to migrate and invade. A second requirement is the ability to survive increased oxidative stress that accompanies the changing environment.
Several studies have shown a link between availability of exogenous and endogenously produced serine and migration. For example, production of SSP derived α-ketoglutarate (α-KG) was shown to be responsible for enhanced breast cancer metastasis113. Interestingly, the pro-tumorigenic roles of PHGDH are balanced by the observation that loss of PHGDH also enhanced metastasis, in this case by promoting aberrant protein glycosylation114. Consequently, heterogeneity of PHGDH expression is linked to enhanced aggressiveness in primary cancers. Numerous other studies have implicated mitochondrial 1C metabolism activity in the metastatic process. Inhibition of purine synthesis was shown to accelerate cell migration by increasing serine synthesis and 1C metabolism, despite reducing proliferation115. Furthermore, hypoxia induced activation of PHGDH and 1C cycle enzymes supported the maintenance of metastasis-inducing breast cancer stem cells (BCSCs)116. Aggressive breast cancer cell lines also show upregulation of mitochondrial serine and 1C metabolism and in patients, expression of SHMT2 is correlated with poor survival65. Indeed, a common theme emerging from these studies is the observation that enhanced SSP and folate cycle activity can promote metastasis without impacting cancer cell proliferation or the growth of the primary tumour101,112,113.
These observations raise the question of how folate metabolism supports metastasis. Evidence is now emerging that control of ROS, formate production and mitochondrial protein synthesis may all play a role. The production of mitochondrial NADPH and consequent protection from excessive mitochondrial ROS have been shown to be critical in allowing metastasis of some tumour types (Figure 3). For example, the PHGDH-dependent protection of BCSCs mentioned above is linked to a switch to 1C cycle supported mitochondrial antioxidant defence116. Loss of the NADPH-producing enzyme ALDH1L2 leads to increased ROS and a decrease in tumour sphere formation of glioblastoma cells117 and metastatic capacity of melanoma cells100. However, the consequences of loss of antioxidant capacity can be context dependent, with increased ROS also promoting enhanced metastasis in some cancers118.
Formate is emerging as a potentially important regulator of metastasis, correlating with the high levels of formate overflow seen in many cancers (Figure 4). Exogenous formate can directly enhance migration and invasion of different cancer cell lines101,109 and a reduction of formate overflow in response to MTHFD1L silencing reduces migration in vitro and the incidence of lung metastasis in breast cancer models109,112. As mentioned above, ALDH1L2 expression can shift the balance between NADPH, formate and fMet production101, with decreased ALDH1L2 expression leading to lower NADPH production (and more ROS) but enhanced formate and fMet synthesis. In some models, both increased ROS and decreased formate/ fMet production contributed to increased cell migration and enhanced metastasis. Increased formate or decreased ALDH1L2 activity both resulted in an increase in the production of formylated peptides – produced from proteins initiated with fMet. A better-known source of formylated peptides are bacterial proteins (also initiated by fMet), which bind to the formyl-peptide receptors expressed on immune cells to promote immune cell migration and anti-bacterial defense119–121. Interestingly, cancer cells also express formyl-peptide receptors and the increased migration of these cells in response to formate or ALDH1L2 depletion reflects – at least in part – the production of formylated peptides and activation of formyl-peptide receptor signalling101. Expression of ALDH1L2 in tumours is variable, and while increased ALDH1L2 can promote metastasis by limiting mitochondrial ROS, in other cancers, metastases showed lower ALDH1L2 expression compared to the primary tumour, suggesting that in some cases there is a selection for enhanced formate production during dissemination. However, additional mechanisms through which formate functions have been described in other cancer types. For example, microbiome-derived formate can activate AHR to support colorectal cancer stemness and migration34. Formate has also been shown to favour migration of glioblastoma cells lines through upregulation of matrix metalloproteinase 2 (MMP2). In this case, depletion of MTHFD1L – which decreases formate levels - led to reduced MMP2 activity. Additionally, formate supplementation can increase the expression of genes implicated in lipid metabolism, resulting in reduced invasion of glioblastoma cells and decreased levels of MMP2 in response to fatty acid synthesis inhibitors. Targeting lipid metabolism may therefore provide a mechanism to limit the effect of formate overflow on migration122.
Taken together these studies highlight the role of formate in promoting invasion but also show that formate availability can be sustained by many different mechanisms. More work is needed to disentangle the role of each pathway that contributes to formate overflow and the different consequences of this phenomenon.
Interplay between cancer cells and the TME
It is well established that the normal stromal cells in the TME play a key role in supporting the genesis and progression of tumours, including supporting the nutritional demands of tumour cells123. For example, while increased activity of the SSP can support tumour growth in conditions of low serine124, other cells in the TME – such as neurons - can also provide serine to the tumour125. Cancer cells themselves can induce or enhance the supportive role of most cell types in their periphery126 to support their survival and progression126. Tumour mediated stress signals trigger the induction of PHGDH in endothelial cells that drives a shift in central metabolism, triggering proliferation and vessel sprouting of the tumour associated endothelial cells that promote tumour growth127. Tumours also affect their microenvironment by excreting metabolites into the tumour interstitial fluid (TIF). TIF contains nutrient levels distinct from those found in the circulation128, with evidence for depletion or increase in specific nutrients depending on tumour type and location.
Serine and folate metabolism-derived compounds that are excreted by the cancer cells can have an impact on all the cells of the TME (Figure 5). Metabolites such as glycine and formate have been shown to accumulate in the TIF, with levels of formate potentially reaching up to 500μM in mouse models of breast cancer, compared to normal circulating formate levels of 30μM101,129. The induction of indoleamine 2,3-dioxygenase 1 (IDO1), a tryptophan degrading enzyme, in pancreatic ductal adenocarcinoma leads to increased formate overflow, which was shown to be utilized by surrounding pancreatic stellate cells for purine synthesis30. However, possibly the most important effect of cancer cell excreted 1C metabolites is the modulation of immune cells. Immune cells play a complex role in tumorigenesis, with different branches of the immune system showing pro- and anti-tumour activity130. Critical mediators of anti-tumour immunity are T-cells and while mitochondrial 1C metabolism is required for optimal T-cell activation, the field is just beginning to explore the effect of tumour derived metabolites such as formate or glycine on the immune population. A recent study showed that fusobacterium derived formate can contribute not only to colorectal cancer promotion but also Th17 T-cell expansion34. Furthermore, a role for formate in supporting T-cell activity and improving the responses to immune checkpoint inhibitors, such as anti-programmed cell death 1 (PD1), has also been reported131,132. As described above, silencing of ALDH1L2 in tumour cells leads to increased production of both formate and fMet, accompanied by enhanced signalling through the formyl-peptide receptor expressed by cancer cells. It is possible, therefore, that these tumour cell derived formylated peptides will also promote the migration of various immune cell populations and play a critical role in shaping the landscape of immune infiltration in tumours. Supporting this idea, overexpression of ALDH1L2 in cancer cells, decreasing the production of fMet, led to reduced numbers of neutrophils in a murine breast cancer models101. Taken together, these studies are beginning to reveal the complexity of the potential responses to tumour-derived formate, which may both promote tumour cell migration and enhance anti-tumour immunity. Overall, the effect of formate and fMet production by tumours on broader immune cell function – or even other cells of the TME - remains to be explored (Figure 5).
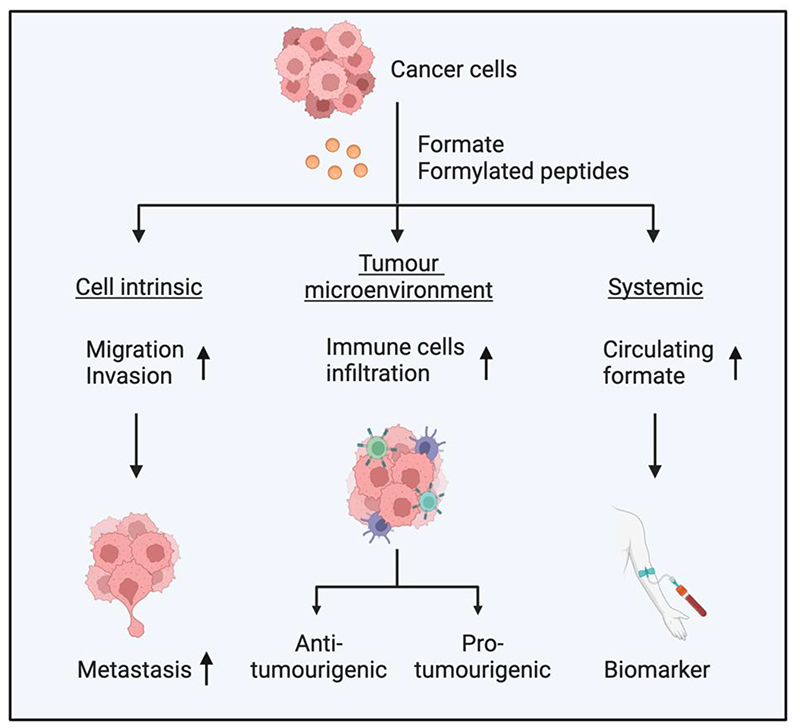
Figure 5. Potential consequences of formate metabolism in cancer.
Cancer cells produce formate and formylated peptides, which can affect cancer cell-intrinsic properties, the tumour microenvironment (TME) and systemic metabolism. Formate increases migration and invasion of cancer cells, which leads to increased metastasis. Increased formate levels in the TME can promote infiltration of various immune cells, which can lead to pro- or anti-tumorigenic effects depending on the type of immune cells affected. Cancer cells-excreted formate can increase circulating formate levels, which have the potential to be used as a biomarker.
Therapeutic approaches
The success of antifolates has driven attempts to target folate metabolism through different routes, with the aim of retaining therapeutic efficacy while reducing dose limiting toxicities. While the effectiveness of serine and folate cycle limitation can be explained by the restriction of nucleotide, NADPH and fMet production, other consequences of reduced serine availability that may contribute to the therapeutic effect have been described. These include an accumulation of toxic metabolites such as deoxysphingolipids and mitochondrial fragmentation/ceramide accumulation105,133.
Cancer associated changes that support malignant progression can also impose targetable vulnerabilities. For example, PHGDH overexpressing breast cancer cell lines require elevated NAD+ salvage pathway activity to meet the high demand for NAD+-dependent PHGDH activity. ER-negative, basal-like breast cancers show increased expression of both PHGDH and nicotinamide phosphoribosyltransferase (NAMPT), the rate-limiting enzyme of NAD+ salvage, and are sensitive to NAMPT inhibition134. While most cells use the mitochondrial folate cycle to capture 1C units, some cancer cells become dependent on cytosolic folate metabolism – a phenotype that has been linked to intracellular folate levels. These tumours become selectively sensitive to SHMT1 inhibition135. Moreover, PHGDH expression is required in breast cancer metastases – but not primary tumours – to allow α-KG-dependent mTORC1 signalling, making these cells sensitive to rapamycin, an mTORC1 inhibitor113. Increased serine metabolism leads to glycine accumulation and a dependence of cells on glycine removal systems such as the GCS – inhibition of which results in the production of toxic products such as aminoacetone and methylglyoxal64.
Dietary intervention
In mice, dietary limitation of serine has been shown to reduce circulating serine levels and have a therapeutic effect in multiple tumour models49,124,136,137. However, the response to this approach depends on various cancer cell intrinsic and extrinsic factors, including the tumour environment. Cancer cells with upregulated SSP activity, such as those driven by KRAS activation or those adapted to growth in serine depleted environments, are less likely to be affected by limitations of exogenous serine49,124. Dietary intervention may also be inadequate to effectively limit a high serine containing TME, or in situations where serine is provided by stromal cells. On the other hand, cancer cells with limited SSP activity could be highly responsive to dietary serine limitation. For example, this approach may be effective in luminal breast tumours, which tend to express low level of PSAT1 due to epigenetic silencing137, or the subset of pancreas cancer that show low PHGDH expression125. A subgroup of platinum resistant ovarian cancer showed decreased PHGDH expression at relapse – increasing the sensitivity of these tumours to serine free diets138 and suggesting this approach may become effective at later stages of tumorigenesis. p53-null cancers also show increased sensitivity to serine starvation and serine limiting diets81. Cancers harbouring Kelch-like ECH-associated protein 1 (KEAP1) mutations show enhanced NRF2-dependent antioxidant defence but become dependent on the uptake of several NEAA, including serine. In mouse models, KEAP1 mutations render tumours highly sensitive to a serine depleted diet136. Importantly, serine free diets can decrease metastases, even when primary tumour growth is only slightly affected101. Building on the successful impact of dietary serine depletion in mice, bespoke diets lacking a number of NEAA are now undergoing clinical trials to test for tolerability and efficacy in combination with standard of care chemotherapy in metastatic pancreatic cancer patients139.
Small molecule inhibitors
Inhibitors of several different steps of serine and folate metabolism have been developed and shown to be effective in preclinical models (Table 1). While several of these drugs were developed to inhibit different targets and were only subsequently shown to inhibit enzymes of the folate cycle (which may therefore be considered off-target effects), others were specifically designed to inhibit serine synthesis and folate metabolism. These include a number of PHGDH inhibitors, which are effective in limiting tumour growth in various preclinical models43,140–142. An SHMT/1/2 inhibitor, SHIN1, also showed efficacy in B-cell lymphoma and T-cell ALL models143, with evidence for a synergistic effect between SHIN2 and methotrexate144. Several MTHFD1 and MTHFD2 inhibitors have been developed and shown to limit tumour development145,146. One of these has a unique function in trapping folate and promoting the toxic accumulation of 10-formyl-THF145. Additionally, a study using artificial intelligence to predict collateral lethal metabolic pathways, revealed the targeting of MTHFD2 as a vulnerability for ovarian cancers lacking ubiquinol-cytochrome c reductase, complex III subunit XI (UQCR11)147. As noted for dietary approaches, use of these inhibitors will be limited by the metabolic landscape of the cancer. Tumours growing in serine limited environments – such as the mammary gland – are dependent on de novo serine production124 and tumours that metastasise to the brain – another serine limited environment – are highly responsive to treatment with a PHGDH inhibitor148. Another interesting approach is to inhibit GCPII, thereby reducing the folate pools available to cancer cells. Prostate cancers frequently show upregulation of GCPII, leading to an exploration of the efficacy of GCPII inhibition as a therapeutic option in these tumours149.
Table 1. Drugs targeting serine and one-carbon metabolism in cancers.
| Agent | Target (s) | Status | Reference |
|---|---|---|---|
| Azacoccone E | PHGDH | Preclinical | 150 |
| BI-4916/4924 | PHGDH | Preclinical | 151 |
| CBR-5884 | PHGDH | Preclinical | 152 |
| Disulfiram | PHGDH | FDA-approved to treat alcohol dependence | 153 |
| Ixocarpalactone A | PHGDH | Preclinical | 154 |
| NCT-502/503 | PHGDH | Preclinical | 140 |
| Oridonin | PHGDH | Preclinical | 155 |
| PH-739/755 | PHGDH | Preclinical | 43,148 |
| PKUMDL-WQ-2101 | PHGDH | Preclinical | 141 |
| Compound C25 | PHGDH | Preclinical | 156 |
| Compound D8 | PHGDH | Preclinical | 157 |
| Thimerosal | PHGDH | Preclinical | 158 |
| Withangulatin A | PHGDH | Preclinical | 159 |
| Chlopromazine | PSPH | FDA-approved to treat schizophrenia, bipolar disorder and acute psychosis | 160 |
| Clofazimine | PSPH | FDA-approved to treat leprosy and tuberculosis | 161 |
| CMPSA | PSPH | Preclinical | 162 |
| Glycerophosphorylcholine | PSPH | Clinical trial for dementia and physical and psychomotor performance | 162 |
| Jung11 | PSPH | Preclinical | 161 |
| L-AP3 D-AP3 |
PSPH | Preclinical | 163 |
| AGF291 AGF320 AGF347 |
SHMT1/2 | Preclinical | 164 |
| AM-807/42004511 AM-807/40675298 AM-807/42004633 |
SHMT2 | Preclinical | 165 |
| Compound 2.12 | SHMT1 | Preclinical | 166 |
| Lometrexol, pemetrexed | SHMT2 | Lometrexol (clinical trial), pemetrexed (FDA-approved as chemotherapy) | 167 |
| Metformin | SHMT2 | FDA-approved to treat type 2 diabetes mellitus | 168 |
| RZ-2994 | SHMT1/2 | Preclinical | 169 |
| Sertraline | SHMT1/2 | FDA-approved as antidepressant | 158 |
| SHIN1 | SHMT1/2 | Preclinical | 143 |
| SHIN2 | SHMT1/2 | Preclinical | 144 |
| 3-bromopyruvate | SHMT1/2 | Preclinical | 170 |
| Carolacton | MTHFD1/2 | Preclinical | 171 |
| Compound 1, 2, 3, 4R, 4S, 5 | MTHFD2 | Preclinical | 172 |
| C18 | MTHFD2 | Preclinical | 173 |
| DS18561882 | MTHFD2 | Preclinical | 146 |
| DS44960156 | MTHFD2 | Preclinical | 174 |
| LY345899 | MTHFD1/2 | Preclinical | 175 |
| TH7299 TH9028 TH9619 |
MTHFD1/2/2L | Preclinical | 176,177 |
| Xanthine derivative 15 | MTHFD2 | Preclinical | 178 |
| Standardized Nonessential Amino Acid Restriction | Nonessential Amino Acid | Clinical trial | 139 |
Currently, we are not aware of ongoing clinical trials of drugs that have been designed to target serine or folate metabolism. It is possible that their use will be limited by on-target toxicities resulting from the systemic inhibition of these pathways or by difficulty in achieving sufficient impact on highly expressed metabolic enzymes. Some of the drugs listed in Table 1 that were designed to hit other targets have been clinically tested, but their pleiotropic functions make it difficult to assess how much of their impact is due to the inhibition of the folate cycle enzymes.
Effect on stromal cells
Dietary serine starvation or treatment with folate cycle inhibitors will have systemic impacts on normal, as well as tumour cells - especially cells with high proliferation rates such as blood, gut or activated immune cells. While mice were shown to tolerate serine starvation in the short-term, long-term maintenance on a serine free diet led to peripheral neuropathy, which was accelerated by high fat intake179. T-cells, which are critical mediators of anti-tumour immunity are highly dependent on serine and likely to be compromised by serine-limiting therapies. Indeed, dietary restriction of serine can impair T-cell expansion upon pathogen infection180 while serine synthesis and mitochondrial 1C metabolism is necessary for T-cell survival and antigen specific T-cell abundance in vivo181,182. Perturbation of folate metabolism by depletion of MTHFD2 promotes the differentiation of regulatory T-cells (Tregs), which can have tumour supportive function183. Consistently, increased serine metabolism in Tregs can restrict their function, a response that was rescued by limiting serine184. Defective 1C metabolism is observed in aged mice, contributing to a compromised T-cell response and consistent with the age associated increase in cancer burden107. Serine synthesis is also important in macrophages185, where high levels of PHGDH expression drives the polarization of these cells towards an immunosuppressive and cancer supporting phenotype186. The SSP is also required for germinal centre formation and antibody production, with overexpression of PHGDH necessary to allow the survival of large B-cell lymphoma142. Similarly, while formate production may be an attractive therapeutic target to limit metastatic potential of cancer cells, the role of formate and formylated peptides in provoking an immune response raises the possibility that such approaches could also blunt anti-cancer immunity.
Taken together, it will be critical to assess whether systemic serine metabolism modulating therapies dampen an effective anti-tumour immune response.
Combinational therapy
Dietary modulation will only reduce – but not eliminate – the availability of NEAA to fuel folate metabolism and so is unlikely to be effective as a monotherapy. However, approaches that combine specific diets with matched small molecule inhibitors have shown promise in preclinical trials187,188. Serine free diets combined with a PHGDH inhibitor showed enhanced anti-tumour activity in mice, although this combination therapy also led to dose limiting weight loss, most likely reflecting a systemic drop in serine that was not apparent with either treatment alone43. As mentioned earlier, serine starvation may also help to overcome 5-FU resistance92. Intriguingly, although histidine can be a source of 1C units, histidine catabolism in cancer cells led to the production of 5-formyl-THF and the depletion of the intracellular THF pool – resulting in enhanced response to methotrexate in a leukaemia xenograft model189.
Lack of serine leads to increased intracellular ROS, due to a reduction in components of antioxidant defence mechanisms and has been shown to sensitize tumours to ROS-inducing radiotherapy190. Similarly, serine depletion synergises with biguanides such as phenformin or metformin to limit cancer growth in mice49,191. While this response may reflect the activity of biguanides in limiting mitochondrial activity and increasing ROS, the ability of metformin to directly inhibit SHMT2 suggests another mechanism by which this drug can enhance the effectiveness of serine limitation168.
Cancer therapy has been transformed over the past ten years by the availability of immune checkpoint inhibitors that target PD1 or PD1 ligand 1 (PDL1). These drugs release the brake on T-cell activation imposed by cancer cells and restore an effective anti-tumour immune response192. As discussed above, serine and folate metabolism are important for the activation of effective T-cell responses, so it is surprising and encouraging that pemetrexed – an antifolate that is standard of care for non-small cell lung cancer patients – was shown to synergise with anti-PD1193. PSPH was shown to be overexpressed in HCC, promoting the SSP and modulating the immune response to these tumours by attracting tumour supporting monocytes/macrophages while limiting T-cell recruitment. Inhibition of PSPH reversed these immune-limiting effects, a response that was enhanced by anti-PD1 treatment194. Intriguingly, metformin was shown to inhibit PSPH in this study, suggesting another potential mechanism for biguanide function. A beneficial effect of formate supplementation in combination with checkpoint inhibitors on CD8+ T cell antitumor activity suggests that in some contexts, enhanced folate cycle activity in immune or tumour cells may be beneficial for cancer therapy131,132.
Checkpoint inhibition has curative potential, but many patients fail to respond to these therapies. While the efficacy of combinations with folate pathway inhibitors remains to be fully validated in clinical trials, the data support a broader view that understanding the metabolic requirements and vulnerabilities of cancer cells will allow for the development of targeted therapeutic options to exploit these cancer specific changes. The concept of specific dietary modulation matched to tumour type, oncogenic changes and location (precision nutrition) is especially attractive to cancer patients who frequently search for diets that will help improve their survival187.
Future Directions
Early success in modulating folate metabolism in cancer therapy has suggested that additional therapeutic targets may be identified in these pathways. Preclinical studies are revealing numerous potential points of intervention and effective combinatorial approaches, although most of these await clinical validation. Several considerations may be pertinent to future success in this area. Firstly, the complexities of how different cell types in different environments show differences in their requirements and use of various metabolites will need careful assessment. As an example, the production of mitochondrial NADPH has been shown to play a key role in limiting ROS (as described here), but in some cell systems – where mitochondrial NADPH can be obtained from alternative sources – 1C cycle derived NADPH is more important in supporting lipogenesis or proline synthesis20,93,95,103,104. The plasticity of metabolism may also represent a barrier to the development of effective therapeutics. However, metabolic plasticity may be more evident in nutrient rich tissue culture conditions than in vivo, where nutrient limitation may restrict the available options for metabolic rewiring. Indeed, much of the work in this area has been based on the response of cancer cells grown in tissue culture and while these studies have been successful, the use of more physiologically accurate conditions – such as culture media that contain physiological levels of various nutrients - may more effectively point to targetable vulnerabilities129,195. In support of this suggestion, the dependence of many cancer cells on exogenous serine was to some degree mitigated in physiological media due to the presence of hypoxanthine to support nucleotide synthesis196. Future use of media reflecting nutrient composition of TIFs or organ sites of various cancers and metastases, coupled with analysis of organoids – which are more reflective of the architecture of cancers in vivo and can accommodate several cell types to mimic at least some aspects of tumour/stromal interactions - may help to identify new therapeutic options. Interestingly, up to 500μM of formate has been measured in TIF of 4T1 breast cancers compared to 10-50μM in plasma, and circulating formate levels were shown to be increased in different genetically engineered mouse models of cancer109. It is therefore possible that increased formate could be used as a circulating biomarker for early cancer detection or monitoring of therapeutic response, although detection of both increased and decreased circulating formate levels in cancer patients110 shows that further work is needed to establish such an approach. One limitation at present is the requirement for robust systems of metabolite detection. The detection of formate – for example – is not usually part of a routine metabolomics analysis and the failure to measure certain metabolites will lead to an underestimation of the role they play. However, great strides have been made and while the therapeutic success of metabolic interventions has to date been limited, we are optimistic that an increased understanding of metabolic demands, vulnerabilities and plasticity of cancer cells will soon lead to improved therapy.
Acknowledgements
The work was funded by Cancer Research UK grant C596/A26855, ERC-2020-ADG PancObese 10102064 and supported by the Francis Crick Institute which receives its core funding from Cancer Research UK (CC2073), the UK Medical Research Council (CC2073) and the Wellcome Trust (CC2073). Figures were created with Biorender.com.
Footnotes
Competing Interests
KHV is on the board of directors and shareholder of Bristol Myers Squibb and on the science advisory board (with stock options) of PMV Pharma, RAZE Therapeutics, Volastra Pharmaceuticals and Kovina Therapeutics. She is on the SAB of Ludwig Cancer and a co-founder and consultant of Faeth Therapeutics. She has been in receipt of research funding from Astex Pharmaceuticals and AstraZeneca and contributed to CRUK Cancer Research Technology filing of patent application WO/2017/144877. The other authors declare no conflicts of interest.
References
- 1.Warburg O, Wind F, Negelein E. The Metabolism of Tumors in the Body. J Gen Physiol. 1927;8:519–530. doi: 10.1085/jgp.8.6.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 3.Farber S, Diamond LK. Temporary remissions in acute leukemia in children produced by folic acid antagonist, 4-aminopteroyl-glutamic acid. N Engl J Med. 1948;238:787–793. doi: 10.1056/NEJM194806032382301. [DOI] [PubMed] [Google Scholar]
- 4.Osborn MJ, Freeman M, Huennekens FM. Inhibition of dihydrofolic reductase by aminopterin and amethopterin. Proc Soc Exp Biol Med. 1958;97:429–431. doi: 10.3181/00379727-97-23764. [DOI] [PubMed] [Google Scholar]
- 5.Schalinske KL, Steele RD. Methotrexate alters carbon flow through the hepatic folate-dependent one-carbon pool in rats. Carcinogenesis. 1996;17:1695–1700. doi: 10.1093/carcin/17.8.1695. [DOI] [PubMed] [Google Scholar]
- 6.Huennekens FM, Duffy TH, Vitols KS. Folic acid metabolism and its disruption by pharmacologic agents. NCI Monogr. 1987:1–8. [PubMed] [Google Scholar]
- 7.Chabner BA, Roberts TG., Jr Timeline: Chemotherapy and the war on cancer. Nat Rev Cancer. 2005;5:65–72. doi: 10.1038/nrc1529. [DOI] [PubMed] [Google Scholar]
- 8.Sedillo JC, Cryns VL. Targeting the methionine addiction of cancer. Am J Cancer Res. 2022;12:2249–2276. [PMC free article] [PubMed] [Google Scholar]
- 9.Parsa S, et al. The serine hydroxymethyltransferase-2 (SHMT2) initiates lymphoma development through epigenetic tumor suppressor silencing. Nat Cancer. 2020;1:653–664. doi: 10.1038/s43018-020-0080-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Geeraerts SL, Heylen E, De Keersmaecker K, Kampen KR. The ins and outs of serine and glycine metabolism in cancer. Nat Metab. 2021;3:131–141. doi: 10.1038/s42255-020-00329-9. [DOI] [PubMed] [Google Scholar]
- 11.Yang M, Vousden KH. Serine and one-carbon metabolism in cancer. Nat Rev Cancer. 2016;16:650–662. doi: 10.1038/nrc.2016.81. [DOI] [PubMed] [Google Scholar]
- 12.Ducker GS, Rabinowitz JD. One-Carbon Metabolism in Health and Disease. Cell Metab. 2017;25:27–42. doi: 10.1016/j.cmet.2016.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pfeiffer CM, et al. Folate status and concentrations of serum folate forms in the US population: National Health and Nutrition Examination Survey 2011-2. Br J Nutr. 2015;113:1965–1977. doi: 10.1017/S0007114515001142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chandler CJ, Wang TT, Halsted CH. Pteroylpolyglutamate hydrolase from human jejunal brush borders. Purification and characterization. J Biol Chem. 1986;261:928–933. [PubMed] [Google Scholar]
- 15.Wright AJ, Dainty JR, Finglas PM. Folic acid metabolism in human subjects revisited: potential implications for proposed mandatory folic acid fortification in the UK. Br J Nutr. 2007;98:667–675. doi: 10.1017/S0007114507777140. [DOI] [PubMed] [Google Scholar]
- 16.Sullivan MR, et al. Methionine synthase is essential for cancer cell proliferation in physiological folate environments. Nat Metab. 2021;3:1500–1511. doi: 10.1038/s42255-021-00486-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ghergurovich JM, et al. Methionine synthase supports tumour tetrahydrofolate pools. Nat Metab. 2021;3:1512–1520. doi: 10.1038/s42255-021-00465-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tucker EJ, et al. Mutations in MTFMT underlie a human disorder of formylation causing impaired mitochondrial translation. Cell Metab. 2011;14:428–434. doi: 10.1016/j.cmet.2011.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Minton DR, et al. Serine Catabolism by SHMT2 Is Required for Proper Mitochondrial Translation Initiation and Maintenance of Formylmethionyl-tRNAs. Mol Cell. 2018;69:610–621.:e615. doi: 10.1016/j.molcel.2018.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ducker GS, et al. Reversal of Cytosolic One-Carbon Flux Compensates for Loss of the Mitochondrial Folate Pathway. Cell Metab. 2016;23:1140–1153. doi: 10.1016/j.cmet.2016.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zheng Y, et al. Mitochondrial One-Carbon Pathway Supports Cytosolic Folate Integrity in Cancer Cells. Cell. 2018;175:1546–1560.:e1517. doi: 10.1016/j.cell.2018.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brosnan ME, Brosnan JT. Formate: The Neglected Member of One-Carbon Metabolism. Annu Rev Nutr. 2016;36:369–388. doi: 10.1146/annurev-nutr-071715-050738. [DOI] [PubMed] [Google Scholar]
- 23.Meiser J, et al. Serine one-carbon catabolism with formate overflow. Sci Adv. 2016;2:e1601273. doi: 10.1126/sciadv.1601273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barlowe CK, Appling DR. In vitro evidence for the involvement of mitochondrial folate metabolism in the supply of cytoplasmic one-carbon units. Biofactors. 1988;1:171–176. [PubMed] [Google Scholar]
- 25.Kikuchi G, Motokawa Y, Yoshida T, Hiraga K. Glycine cleavage system: reaction mechanism, physiological significance, and hyperglycinemia. Proc Jpn Acad Ser B Phys Biol Sci. 2008;84:246–263. doi: 10.2183/pjab.84.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nowak M, Chuchra P, Paprocka J. Nonketotic Hyperglycinemia: Insight into Current Therapies. J Clin Med. 2022;11 doi: 10.3390/jcm11113027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McBride MJ, Hunter CJ, Rabinowitz JD. Glycine homeostasis requires reverse SHMT flux. bioRxiv. 2023 doi: 10.1101/2023.01.11.523668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Solans A, Estivill X, de la Luna S. Cloning and characterization of human FTCD on 21q22.3, a candidate gene for glutamate formiminotransferase deficiency. Cytogenet Cell Genet. 2000;88:43–49. doi: 10.1159/000015483. [DOI] [PubMed] [Google Scholar]
- 29.Mao Y, et al. Structure of the bifunctional and Golgi-associated formiminotransferase cyclodeaminase octamer. EMBO J. 2004;23:2963–2971. doi: 10.1038/sj.emboj.7600327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Newman AC, et al. Immune-regulated IDO1-dependent tryptophan metabolism is source of one-carbon units for pancreatic cancer and stellate cells. Mol Cell. 2021;81:2290–2302.:e2297. doi: 10.1016/j.molcel.2021.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sen K, Hackett JC. Peroxo-iron mediated deformylation in sterol 14alpha-demethylase catalysis. J Am Chem Soc. 2010;132:10293–10305. doi: 10.1021/ja906192b. [DOI] [PubMed] [Google Scholar]
- 32.Dorokhov YL, Shindyapina AV, Sheshukova EV, Komarova TV. Metabolic methanol: molecular pathways and physiological roles. Physiol Rev. 2015;95:603–644. doi: 10.1152/physrev.00034.2014. [DOI] [PubMed] [Google Scholar]
- 33.Burgos-Barragan G, et al. Mammals divert endogenous genotoxic formaldehyde into one-carbon metabolism. Nature. 2017;548:549–554. doi: 10.1038/nature23481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ternes D, et al. The gut microbial metabolite formate exacerbates colorectal cancer progression. Nat Metab. 2022;4:458–475. doi: 10.1038/s42255-022-00558-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ergal I, et al. Formate Utilization by the Crenarchaeon Desulfurococcus amylolyticus. Microorganisms. 2020;8 doi: 10.3390/microorganisms8030454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Laverde Gomez JA, et al. Formate cross-feeding and cooperative metabolic interactions revealed by transcriptomics in co-cultures of acetogenic and amylolytic human colonic bacteria. Environ Microbiol. 2019;21:259–271. doi: 10.1111/1462-2920.14454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hughes ER, et al. Microbial Respiration and Formate Oxidation as Metabolic Signatures of Inflammation-Associated Dysbiosis. Cell Host Microbe. 2017;21:208–219. doi: 10.1016/j.chom.2017.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pakos-Zebrucka K, et al. The integrated stress response. EMBO Rep. 2016;17:1374–1395. doi: 10.15252/embr.201642195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.DeNicola GM, et al. NRF2 regulates serine biosynthesis in non-small cell lung cancer. Nat Genet. 2015;47:1475–1481. doi: 10.1038/ng.3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ye J, et al. Pyruvate kinase M2 promotes de novo serine synthesis to sustain mTORC1 activity and cell proliferation. Proc Natl Acad Sci U S A. 2012;109:6904–6909. doi: 10.1073/pnas.1204176109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Torrence ME, et al. The mTORC1-mediated activation of ATF4 promotes protein and glutathione synthesis downstream of growth signals. Elife. 2021;10 doi: 10.7554/eLife.63326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ben-Sahra I, Hoxhaj G, Ricoult SJH, Asara JM, Manning BD. mTORC1 induces purine synthesis through control of the mitochondrial tetrahydrofolate cycle. Science. 2016;351:728–733. doi: 10.1126/science.aad0489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tajan M, et al. Serine synthesis pathway inhibition cooperates with dietary serine and glycine limitation for cancer therapy. Nat Commun. 2021;12:366. doi: 10.1038/s41467-020-20223-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kottakis F, et al. LKB1 loss links serine metabolism to DNA methylation and tumorigenesis. Nature. 2016;539:390–395. doi: 10.1038/nature20132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li X, et al. ATF3 promotes the serine synthesis pathway and tumor growth under dietary serine restriction. Cell Rep. 2021;36:109706. doi: 10.1016/j.celrep.2021.109706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chaneton B, et al. Serine is a natural ligand and allosteric activator of pyruvate kinase M2. Nature. 2012;491:458–462. doi: 10.1038/nature11540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Diehl FF, Lewis CA, Fiske BP, Vander Heiden MG. Cellular redox state constrains serine synthesis and nucleotide production to impact cell proliferation. Nat Metab. 2019;1:861–867. doi: 10.1038/s42255-019-0108-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Possemato R, et al. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature. 2011;476:346–350. doi: 10.1038/nature10350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maddocks ODK, et al. Modulating the therapeutic response of tumours to dietary serine and glycine starvation. Nature. 2017;544:372–376. doi: 10.1038/nature22056. [DOI] [PubMed] [Google Scholar]
- 50.Locasale JW, et al. Phosphoglycerate dehydrogenase diverts glycolytic flux and contributes to oncogenesis. Nat Genet. 2011;43:869–874. doi: 10.1038/ng.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Beroukhim R, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sato K, et al. Phosphoserine Phosphatase Is a Novel Prognostic Biomarker on Chromosome 7 in Colorectal Cancer. Anticancer Res. 2017;37:2365–2371. doi: 10.21873/anticanres.11574. [DOI] [PubMed] [Google Scholar]
- 53.Frattini V, et al. The integrated landscape of driver genomic alterations in glioblastoma. Nat Genet. 2013;45:1141–1149. doi: 10.1038/ng.2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liao L, et al. Upregulation of phosphoserine phosphatase contributes to tumor progression and predicts poor prognosis in non-small cell lung cancer patients. Thorac Cancer. 2019;10:1203–1212. doi: 10.1111/1759-7714.13064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhu J, et al. High Expression of PHGDH Predicts Poor Prognosis in Non-Small Cell Lung Cancer. Transl Oncol. 2016;9:592–599. doi: 10.1016/j.tranon.2016.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang B, et al. PHGDH Defines a Metabolic Subtype in Lung Adenocarcinomas with Poor Prognosis. Cell Rep. 2017;19:2289–2303. doi: 10.1016/j.celrep.2017.05.067. [DOI] [PubMed] [Google Scholar]
- 57.Rathore R, et al. Metabolic compensation activates pro-survival mTORC1 signaling upon 3-phosphoglycerate dehydrogenase inhibition in osteosarcoma. Cell Rep. 2021;34:108678. doi: 10.1016/j.celrep.2020.108678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Christopher SA, Diegelman P, Porter CW, Kruger WD. Methylthioadenosine phosphorylase, a gene frequently codeleted with p16(cdkN2a/ARF), acts as a tumor suppressor in a breast cancer cell line. Cancer Res. 2002;62:6639–6644. [PubMed] [Google Scholar]
- 59.Kryukov GV, et al. MTAP deletion confers enhanced dependency on the PRMT5 arginine methyltransferase in cancer cells. Science. 2016;351:1214–1218. doi: 10.1126/science.aad5214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kang YP, et al. PHGDH supports liver ceramide synthesis and sustains lipid homeostasis. Cancer Metab. 2020;8:6. doi: 10.1186/s40170-020-00212-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Broeks MH, et al. The malate-aspartate shuttle is important for de novo serine biosynthesis. Cell Rep. 2023;42:113043. doi: 10.1016/j.celrep.2023.113043. [DOI] [PubMed] [Google Scholar]
- 62.Nilsson R, et al. Metabolic enzyme expression highlights a key role for MTHFD2 and the mitochondrial folate pathway in cancer. Nat Commun. 2014;5:3128. doi: 10.1038/ncomms4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jain M, et al. Metabolite profiling identifies a key role for glycine in rapid cancer cell proliferation. Science. 2012;336:1040–1044. doi: 10.1126/science.1218595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kim D, et al. SHMT2 drives glioma cell survival in ischaemia but imposes a dependence on glycine clearance. Nature. 2015;520:363–367. doi: 10.1038/nature14363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li AM, et al. Metabolic Profiling Reveals a Dependency of Human Metastatic Breast Cancer on Mitochondrial Serine and One-Carbon Unit Metabolism. Mol Cancer Res. 2020;18:599–611. doi: 10.1158/1541-7786.MCR-19-0606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang WC, et al. Glycine decarboxylase activity drives non-small cell lung cancer tumor-initiating cells and tumorigenesis. Cell. 2012;148:259–272. doi: 10.1016/j.cell.2011.11.050. [DOI] [PubMed] [Google Scholar]
- 67.Labuschagne CF, van den Broek NJ, Mackay GM, Vousden KH, Maddocks OD. Serine, but not glycine, supports one-carbon metabolism and proliferation of cancer cells. Cell Rep. 2014;7:1248–1258. doi: 10.1016/j.celrep.2014.04.045. [DOI] [PubMed] [Google Scholar]
- 68.Mukha D, et al. Glycine decarboxylase maintains mitochondrial protein lipoylation to support tumor growth. Cell Metab. 2022;34:775–782.:e779. doi: 10.1016/j.cmet.2022.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sun L, et al. cMyc-mediated activation of serine biosynthesis pathway is critical for cancer progression under nutrient deprivation conditions. Cell Res. 2015;25:429–444. doi: 10.1038/cr.2015.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xia Y, et al. Metabolic Reprogramming by MYCN Confers Dependence on the Serine-Glycine-One-Carbon Biosynthetic Pathway. Cancer Res. 2019;79:3837–3850. doi: 10.1158/0008-5472.CAN-18-3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vazquez A, Tedeschi PM, Bertino JR. Overexpression of the mitochondrial folate and glycine-serine pathway: a new determinant of methotrexate selectivity in tumors. Cancer Res. 2013;73:478–482. doi: 10.1158/0008-5472.CAN-12-3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ye J, et al. Serine catabolism regulates mitochondrial redox control during hypoxia. Cancer Discov. 2014;4:1406–1417. doi: 10.1158/2159-8290.CD-14-0250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bjelosevic S, et al. Serine Biosynthesis Is a Metabolic Vulnerability in FLT3-ITD-Driven Acute Myeloid Leukemia. Cancer Discov. 2021;11:1582–1599. doi: 10.1158/2159-8290.CD-20-0738. [DOI] [PubMed] [Google Scholar]
- 74.Xu R, et al. The breast cancer oncogene IKKepsilon coordinates mitochondrial function and serine metabolism. EMBO Rep. 2020;21:e48260. doi: 10.15252/embr.201948260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Smith ALM, Whitehall JC, Greaves LC. Mitochondrial DNA mutations in ageing and cancer. Mol Oncol. 2022;16:3276–3294. doi: 10.1002/1878-0261.13291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bao XR, et al. Mitochondrial dysfunction remodels one-carbon metabolism in human cells. Elife. 2016;5 doi: 10.7554/eLife.10575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.DeNicola GM, et al. Oncogene-induced Nrf2 transcription promotes ROS detoxification and tumorigenesis. Nature. 2011;475:106–109. doi: 10.1038/nature10189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Moran DM, et al. KRAS mutation status is associated with enhanced dependency on folate metabolism pathways in non-small cell lung cancer cells. Mol Cancer Ther. 2014;13:1611–1624. doi: 10.1158/1535-7163.MCT-13-0649. [DOI] [PubMed] [Google Scholar]
- 79.Kandoth C, et al. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502:333–339. doi: 10.1038/nature12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kruiswijk F, Labuschagne CF, Vousden KH. p53 in survival, death and metabolic health: a lifeguard with a licence to kill. Nat Rev Mol Cell Biol. 2015;16:393–405. doi: 10.1038/nrm4007. [DOI] [PubMed] [Google Scholar]
- 81.Maddocks OD, et al. Serine starvation induces stress and p53-dependent metabolic remodelling in cancer cells. Nature. 2013;493:542–546. doi: 10.1038/nature11743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ou Y, Wang SJ, Jiang L, Zheng B, Gu W. p53 Protein-mediated regulation of phosphoglycerate dehydrogenase (PHGDH) is crucial for the apoptotic response upon serine starvation. J Biol Chem. 2015;290:457–466. doi: 10.1074/jbc.M114.616359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Maguire M, et al. MDM2 regulates dihydrofolate reductase activity through monoubiquitination. Cancer Res. 2008;68:3232–3242. doi: 10.1158/0008-5472.CAN-07-5271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Riscal R, et al. Chromatin-Bound MDM2 Regulates Serine Metabolism and Redox Homeostasis Independently of p53. Mol Cell. 2016;62:890–902. doi: 10.1016/j.molcel.2016.04.033. [DOI] [PubMed] [Google Scholar]
- 85.Audet-Walsh E, et al. The PGC-1alpha/ERRalpha Axis Represses One-Carbon Metabolism and Promotes Sensitivity to Anti-folate Therapy in Breast Cancer. Cell Rep. 2016;14:920–931. doi: 10.1016/j.celrep.2015.12.086. [DOI] [PubMed] [Google Scholar]
- 86.Ma L, et al. Control of nutrient stress-induced metabolic reprogramming by PKCzeta in tumorigenesis. Cell. 2013;152:599–611. doi: 10.1016/j.cell.2012.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Reina-Campos M, et al. Increased Serine and One-Carbon Pathway Metabolism by PKClambda/iota Deficiency Promotes Neuroendocrine Prostate Cancer. Cancer Cell. 2019;35:385–400.:e389. doi: 10.1016/j.ccell.2019.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mehrmohamadi M, Liu X, Shestov AA, Locasale JW. Characterization of the usage of the serine metabolic network in human cancer. Cell Rep. 2014;9:1507–1519. doi: 10.1016/j.celrep.2014.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.De Vitto H, Arachchige DB, Richardson BC, French JB. The Intersection of Purine and Mitochondrial Metabolism in Cancer. Cells. 2021;10 doi: 10.3390/cells10102603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Shi DD, Savani MR, Abdullah KG, McBrayer SK. Emerging roles of nucleotide metabolism in cancer. Trends Cancer. 2023;9:624–635. doi: 10.1016/j.trecan.2023.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Montrose DC, et al. Exogenous and Endogenous Sources of Serine Contribute to Colon Cancer Metabolism, Growth, and Resistance to 5-Fluorouracil. Cancer Res. 2021;81:2275–2288. doi: 10.1158/0008-5472.CAN-20-1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Pranzini E, et al. SHMT2-mediated mitochondrial serine metabolism drives 5-FU resistance by fueling nucleotide biosynthesis. Cell Rep. 2022;40:111233. doi: 10.1016/j.celrep.2022.111233. [DOI] [PubMed] [Google Scholar]
- 93.Lewis CA, et al. Tracing compartmentalized NADPH metabolism in the cytosol and mitochondria of mammalian cells. Mol Cell. 2014;55:253–263. doi: 10.1016/j.molcel.2014.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Fan J, et al. Quantitative flux analysis reveals folate-dependent NADPH production. Nature. 2014;510:298–302. doi: 10.1038/nature13236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhang Z, et al. Serine catabolism generates liver NADPH and supports hepatic lipogenesis. Nat Metab. 2021;3:1608–1620. doi: 10.1038/s42255-021-00487-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.He L, et al. Serine is required for the maintenance of redox balance and proliferation in the intestine under oxidative stress. FASEB J. 2020;34:4702–4717. doi: 10.1096/fj.201902690R. [DOI] [PubMed] [Google Scholar]
- 97.Balsa E, et al. Defective NADPH production in mitochondrial disease complex I causes inflammation and cell death. Nat Commun. 2020;11:2714. doi: 10.1038/s41467-020-16423-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhang Z, TeSlaa T, Rabinowitz JD. Reply to: revisiting the role of serine metabolism in hepatic lipogenesis. Nat Metab. 2023;5:762–764. doi: 10.1038/s42255-023-00793-z. [DOI] [PubMed] [Google Scholar]
- 99.Engel AL, et al. Serine-dependent redox homeostasis regulates glioblastoma cell survival. Br J Cancer. 2020;122:1391–1398. doi: 10.1038/s41416-020-0794-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Piskounova E, et al. Oxidative stress inhibits distant metastasis by human melanoma cells. Nature. 2015;527:186–191. doi: 10.1038/nature15726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hennequart M, et al. ALDH1L2 regulation of formate, formyl-methionine, and ROS controls cancer cell migration and metastasis. Cell Rep. 2023;42:112562. doi: 10.1016/j.celrep.2023.112562. [DOI] [PubMed] [Google Scholar]
- 102.Wei L, et al. Genome-wide CRISPR/Cas9 library screening identified PHGDH as a critical driver for Sorafenib resistance in HCC. Nat Commun. 2019;10:4681. doi: 10.1038/s41467-019-12606-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Tran DH, et al. Mitochondrial NADP(+) is essential for proline biosynthesis during cell growth. Nat Metab. 2021;3:571–585. doi: 10.1038/s42255-021-00374-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zhu J, et al. Mitochondrial NADP(H) generation is essential for proline biosynthesis. Science. 2021;372:968–972. doi: 10.1126/science.abd5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Gao X, et al. Serine Availability Influences Mitochondrial Dynamics and Function through Lipid Metabolism. Cell Rep. 2018;22:3507–3520. doi: 10.1016/j.celrep.2018.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Tani H, et al. Mice deficient in the Shmt2 gene have mitochondrial respiration defects and are embryonic lethal. Sci Rep. 2018;8:425. doi: 10.1038/s41598-017-18828-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ron-Harel N, et al. Defective respiration and one-carbon metabolism contribute to impaired naive T cell activation in aged mice. Proc Natl Acad Sci U S A. 2018;115:13347–13352. doi: 10.1073/pnas.1804149115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Momb J, et al. Deletion of Mthfd1l causes embryonic lethality and neural tube and craniofacial defects in mice. Proc Natl Acad Sci U S A. 2013;110:549–554. doi: 10.1073/pnas.1211199110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Meiser J, et al. Increased formate overflow is a hallmark of oxidative cancer. Nat Commun. 2018;9:1368. doi: 10.1038/s41467-018-03777-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Pietzke M, et al. Stratification of cancer and diabetes based on circulating levels of formate and glucose. Cancer Metab. 2019;7:3. doi: 10.1186/s40170-019-0195-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Oizel K, et al. Formate induces a metabolic switch in nucleotide and energy metabolism. Cell Death Dis. 2020;11:310. doi: 10.1038/s41419-020-2523-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kiweler N, et al. Mitochondria preserve an autarkic one-carbon cycle to confer growth-independent cancer cell migration and metastasis. Nat Commun. 2022;13:2699. doi: 10.1038/s41467-022-30363-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Rinaldi G, et al. In Vivo Evidence for Serine Biosynthesis-Defined Sensitivity of Lung Metastasis, but Not of Primary Breast Tumors, to mTORC1 Inhibition. Mol Cell. 2021;81:386–397.:e387. doi: 10.1016/j.molcel.2020.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Rossi M, et al. PHGDH heterogeneity potentiates cancer cell dissemination and metastasis. Nature. 2022;605:747–753. doi: 10.1038/s41586-022-04758-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Soflaee MH, et al. Purine nucleotide depletion prompts cell migration by stimulating the serine synthesis pathway. Nat Commun. 2022;13:2698. doi: 10.1038/s41467-022-30362-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Samanta D, et al. PHGDH Expression Is Required for Mitochondrial Redox Homeostasis, Breast Cancer Stem Cell Maintenance, and Lung Metastasis. Cancer Res. 2016;76:4430–4442. doi: 10.1158/0008-5472.CAN-16-0530. [DOI] [PubMed] [Google Scholar]
- 117.Quere M, et al. ALDH1L2 Knockout in U251 Glioblastoma Cells Reduces Tumor Sphere Formation by Increasing Oxidative Stress and Suppressing Methionine Dependency. Nutrients. 2022;14 doi: 10.3390/nu14091887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Cheung EC, Vousden KH. The role of ROS in tumour development and progression. Nat Rev Cancer. 2022;22:280–297. doi: 10.1038/s41568-021-00435-0. [DOI] [PubMed] [Google Scholar]
- 119.Itagaki K, et al. Formyl Peptide Receptor-1 Blockade Prevents Receptor Regulation by Mitochondrial Danger-Associated Molecular Patterns and Preserves Neutrophil Function After Trauma. Crit Care Med. 2020;48:e123–e132. doi: 10.1097/CCM.0000000000004094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Shao G, et al. Formyl peptide receptor ligands promote wound closure in lung epithelial cells. Am J Respir Cell Mol Biol. 2011;44:264–269. doi: 10.1165/rcmb.2010-0246RC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wenceslau CF, et al. Mitochondrial N-formyl peptides cause airway contraction and lung neutrophil infiltration via formyl peptide receptor activation. Pulm Pharmacol Ther. 2016;37:49–56. doi: 10.1016/j.pupt.2016.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Delbrouck C, et al. Formate promotes invasion and metastasis in reliance on lipid metabolism. Cell Rep. 2023;42:113034. doi: 10.1016/j.celrep.2023.113034. [DOI] [PubMed] [Google Scholar]
- 123.Lyssiotis CA, Kimmelman AC. Metabolic Interactions in the Tumor Microenvironment. Trends Cell Biol. 2017;27:863–875. doi: 10.1016/j.tcb.2017.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Sullivan MR, et al. Increased Serine Synthesis Provides an Advantage for Tumors Arising in Tissues Where Serine Levels Are Limiting. Cell Metab. 2019;29:1410–1421.:e1414. doi: 10.1016/j.cmet.2019.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Banh RS, et al. Neurons Release Serine to Support mRNA Translation in Pancreatic Cancer. Cell. 2020;183:1202–1218.:e1225. doi: 10.1016/j.cell.2020.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Anderson NM, Simon MC. The tumor microenvironment. Curr Biol. 2020;30:R921–R925. doi: 10.1016/j.cub.2020.06.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Zhang D, et al. PHGDH-mediated endothelial metabolism drives glioblastoma resistance to chimeric antigen receptor T cell immunotherapy. Cell Metab. 2023;35:517–534.:e518. doi: 10.1016/j.cmet.2023.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Sullivan MR, et al. Quantification of microenvironmental metabolites in murine cancers reveals determinants of tumor nutrient availability. Elife. 2019;8 doi: 10.7554/eLife.44235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Vande Voorde MG, et al. Improving the metabolic fidelity of cancer models with a physiological cell culture medium. Sci Adv. 2019;5:eaau7314. doi: 10.1126/sciadv.aau7314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.de Visser KE, Eichten A, Coussens LM. Paradoxical roles of the immune system during cancer development. Nat Rev Cancer. 2006;6:24–37. doi: 10.1038/nrc1782. [DOI] [PubMed] [Google Scholar]
- 131.Rowe JH, et al. Formate supplementation enhances anti-tumor CD8+ T cell fitness and efficacy of PD-1 blockade. Cancer Discov. 2023 doi: 10.1158/2159-8290.CD-22-1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Xu X, et al. One-carbon unit supplementation fuels tumor-infiltrating T cells and augments checkpoint blockade. bioRxiv. 2023 doi: 10.1101/2023.11.01.565193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Muthusamy T, et al. Serine restriction alters sphingolipid diversity to constrain tumour growth. Nature. 2020;586:790–795. doi: 10.1038/s41586-020-2609-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Murphy JP, et al. The NAD(+) Salvage Pathway Supports PHGDH-Driven Serine Biosynthesis. Cell Rep. 2018;24:2381–2391.:e2385. doi: 10.1016/j.celrep.2018.07.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Lee WD, et al. Tumor Reliance on Cytosolic versus Mitochondrial One-Carbon Flux Depends on Folate Availability. Cell Metab. 2021;33:190–198.:e196. doi: 10.1016/j.cmet.2020.12.002. [DOI] [PubMed] [Google Scholar]
- 136.LeBoeuf SE, et al. Activation of Oxidative Stress Response in Cancer Generates a Druggable Dependency on Exogenous Non-essential Amino Acids. Cell Metab. 2020;31:339–350.:e334. doi: 10.1016/j.cmet.2019.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Choi BH, et al. Lineage-specific silencing of PSAT1 induces serine auxotrophy and sensitivity to dietary serine starvation in luminal breast tumors. Cell Rep. 2022;38:110278. doi: 10.1016/j.celrep.2021.110278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Van Nyen T, et al. Serine metabolism remodeling after platinum-based chemotherapy identifies vulnerabilities in a subgroup of resistant ovarian cancers. Nat Commun. 2022;13:4578. doi: 10.1038/s41467-022-32272-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. ( https://classic.clinicaltrials.gov/show/NCT05078775)
- 140.Pacold ME, et al. A PHGDH inhibitor reveals coordination of serine synthesis and one-carbon unit fate. Nat Chem Biol. 2016;12:452–458. doi: 10.1038/nchembio.2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Wang Q, et al. Rational Design of Selective Allosteric Inhibitors of PHGDH and Serine Synthesis with Anti-tumor Activity. Cell Chem Biol. 2017;24:55–65. doi: 10.1016/j.chembiol.2016.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.D’Avola A, et al. PHGDH is required for germinal center formation and is a therapeutic target in MYC-driven lymphoma. J Clin Invest. 2022;132 doi: 10.1172/JCI153436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Ducker GS, et al. Human SHMT inhibitors reveal defective glycine import as a targetable metabolic vulnerability of diffuse large B-cell lymphoma. Proc Natl Acad Sci U S A. 2017;114:11404–11409. doi: 10.1073/pnas.1706617114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Garcia-Canaveras JC, et al. SHMT inhibition is effective and synergizes with methotrexate in T-cell acute lymphoblastic leukemia. Leukemia. 2021;35:377–388. doi: 10.1038/s41375-020-0845-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Green AC, et al. Formate overflow drives toxic folate trapping in MTHFD1 inhibited cancer cells. Nat Metab. 2023;5:642–659. doi: 10.1038/s42255-023-00771-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Kawai J, et al. Discovery of a Potent, Selective, and Orally Available MTHFD2 Inhibitor (DS18561882) with in Vivo Antitumor Activity. J Med Chem. 2019;62:10204–10220. doi: 10.1021/acs.jmedchem.9b01113. [DOI] [PubMed] [Google Scholar]
- 147.Achreja A, et al. Metabolic collateral lethal target identification reveals MTHFD2 paralogue dependency in ovarian cancer. Nat Metab. 2022;4:1119–1137. doi: 10.1038/s42255-022-00636-3. [DOI] [PubMed] [Google Scholar]
- 148.Ngo B, et al. Limited Environmental Serine and Glycine Confer Brain Metastasis Sensitivity to PHGDH Inhibition. Cancer Discov. 2020;10:1352–1373. doi: 10.1158/2159-8290.CD-19-1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Barinka C, Rojas C, Slusher B, Pomper M. Glutamate carboxypeptidase II in diagnosis and treatment of neurologic disorders and prostate cancer. Curr Med Chem. 2012;19:856–870. doi: 10.2174/092986712799034888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Guo J, et al. Azacoccone E inhibits cancer cell growth by targeting 3-phosphoglycerate dehydrogenase. Bioorg Chem. 2019;87:16–22. doi: 10.1016/j.bioorg.2019.02.037. [DOI] [PubMed] [Google Scholar]
- 151.Weinstabl H, et al. Intracellular Trapping of the Selective Phosphoglycerate Dehydrogenase (PHGDH) Inhibitor BI-4924 Disrupts Serine Biosynthesis. J Med Chem. 2019;62:7976–7997. doi: 10.1021/acs.jmedchem.9b00718. [DOI] [PubMed] [Google Scholar]
- 152.Mullarky E, et al. Identification of a small molecule inhibitor of 3-phosphoglycerate dehydrogenase to target serine biosynthesis in cancers. Proc Natl Acad Sci U S A. 2016;113:1778–1783. doi: 10.1073/pnas.1521548113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Spillier Q, et al. Anti-alcohol abuse drug disulfiram inhibits human PHGDH via disruption of its active tetrameric form through a specific cysteine oxidation. Sci Rep. 2019;9:4737. doi: 10.1038/s41598-019-41187-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Zheng M, et al. Ixocarpalactone A from dietary tomatillo inhibits pancreatic cancer growth by targeting PHGDH. Food Funct. 2019;10:3386–3395. doi: 10.1039/c9fo00394k. [DOI] [PubMed] [Google Scholar]
- 155.Tan Y, et al. Biophysical and biochemical properties of PHGDH revealed by studies on PHGDH inhibitors. Cell Mol Life Sci. 2021;79:27. doi: 10.1007/s00018-021-04022-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Zhang FM, et al. Discovery of PHGDH inhibitors by virtual screening and preliminary structure-activity relationship study. Bioorg Chem. 2022;121:105705. doi: 10.1016/j.bioorg.2022.105705. [DOI] [PubMed] [Google Scholar]
- 157.Gao D, et al. Discovery of Novel Drug-like PHGDH Inhibitors to Disrupt Serine Biosynthesis for Cancer Therapy. J Med Chem. 2023;66:285–305. doi: 10.1021/acs.jmedchem.2c01202. [DOI] [PubMed] [Google Scholar]
- 158.Geeraerts SL, et al. Repurposing the Antidepressant Sertraline as SHMT Inhibitor to Suppress Serine/Glycine Synthesis-Addicted Breast Tumor Growth. Mol Cancer Ther. 2021;20:50–63. doi: 10.1158/1535-7163.MCT-20-0480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Chen C, et al. Identification of a novel PHGDH covalent inhibitor by chemical proteomics and phenotypic profiling. Acta Pharm Sin B. 2022;12:246–261. doi: 10.1016/j.apsb.2021.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Yadav GP, et al. Characterization of M. tuberculosis SerB2, an essential HAD-family phosphatase, reveals novel properties. PLoS One. 2014;9:e115409. doi: 10.1371/journal.pone.0115409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Haufroid M, Wouters J. Targeting the Serine Pathway: A Promising Approach against Tuberculosis? Pharmaceuticals (Basel) 2019;12 doi: 10.3390/ph12020066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Hawkinson JE, Acosta-Burruel M, Ta ND, Wood PL. Novel phosphoserine phosphatase inhibitors. Eur J Pharmacol. 1997;337:315–324. doi: 10.1016/s0014-2999(97)01304-6. [DOI] [PubMed] [Google Scholar]
- 163.Hawkinson JE, Acosta-Burruel M, Wood PL. The metabotropic glutamate receptor antagonist L-2-amino-3-phosphonopropionic acid inhibits phosphoserine phosphatase. Eur J Pharmacol. 1996;307:219–225. doi: 10.1016/0014-2999(96)00253-1. [DOI] [PubMed] [Google Scholar]
- 164.Dekhne AS, et al. Novel Pyrrolo[3,2-d]pyrimidine Compounds Target Mitochondrial and Cytosolic One-carbon Metabolism with Broad-spectrum Antitumor Efficacy. Mol Cancer Ther. 2019;18:1787–1799. doi: 10.1158/1535-7163.MCT-19-0037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Han Y, et al. Identification of three new compounds that directly target human serine hydroxymethyltransferase 2. Chem Biol Drug Des. 2021;97:221–230. doi: 10.1111/cbdd.13774. [DOI] [PubMed] [Google Scholar]
- 166.Marani M, et al. A pyrazolopyran derivative preferentially inhibits the activity of human cytosolic serine hydroxymethyltransferase and induces cell death in lung cancer cells. Oncotarget. 2016;7:4570–4583. doi: 10.18632/oncotarget.6726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Scaletti E, Jemth AS, Helleday T, Stenmark P. Structural basis of inhibition of the human serine hydroxymethyltransferase SHMT2 by antifolate drugs. FEBS Lett. 2019;593:1863–1873. doi: 10.1002/1873-3468.13455. [DOI] [PubMed] [Google Scholar]
- 168.Tramonti A, et al. Metformin Is a Pyridoxal-5’-phosphate (PLP)-Competitive Inhibitor of SHMT2. Cancers (Basel) 2021;13 doi: 10.3390/cancers13164009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Pikman Y, et al. Targeting serine hydroxymethyltransferases 1 and 2 for T-cell acute lymphoblastic leukemia therapy. Leukemia. 2022;36:348–360. doi: 10.1038/s41375-021-01361-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Paiardini A, et al. Differential 3-bromopyruvate inhibition of cytosolic and mitochondrial human serine hydroxymethyltransferase isoforms, key enzymes in cancer metabolic reprogramming. Biochim Biophys Acta. 2016;1864:1506–1517. doi: 10.1016/j.bbapap.2016.08.010. [DOI] [PubMed] [Google Scholar]
- 171.Fu C, et al. The natural product carolacton inhibits folate-dependent C1 metabolism by targeting FolD/MTHFD. Nat Commun. 2017;8:1529. doi: 10.1038/s41467-017-01671-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Jha V, Eriksson LA. Binding Modes of Xanthine-Derived Selective Allosteric Site Inhibitors of MTHFD2. ChemistryOpen. 2023;12:e202300052. doi: 10.1002/open.202300052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Zhou F, et al. Pharmacological targeting of MTHFD2 suppresses NSCLC via the regulation of ILK signaling pathway. Biomed Pharmacother. 2023;161:114412. doi: 10.1016/j.biopha.2023.114412. [DOI] [PubMed] [Google Scholar]
- 174.Kawai J, et al. Structure-Based Design and Synthesis of an Isozyme-Selective MTHFD2 Inhibitor with a Tricyclic Coumarin Scaffold. ACS Med Chem Lett. 2019;10:893–898. doi: 10.1021/acsmedchemlett.9b00069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Gustafsson R, et al. Crystal Structure of the Emerging Cancer Target MTHFD2 in Complex with a Substrate-Based Inhibitor. Cancer Res. 2017;77:937–948. doi: 10.1158/0008-5472.CAN-16-1476. [DOI] [PubMed] [Google Scholar]
- 176.Bonagas N, et al. Pharmacological targeting of MTHFD2 suppresses acute myeloid leukemia by inducing thymidine depletion and replication stress. Nat Cancer. 2022;3:156–172. doi: 10.1038/s43018-022-00331-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Scaletti ER, et al. The First Structure of Human MTHFD2L and Its Implications for the Development of Isoform-Selective Inhibitors. ChemMedChem. 2022;17:e202200274. doi: 10.1002/cmdc.202200274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Lee LC, et al. Xanthine Derivatives Reveal an Allosteric Binding Site in Methylenetetrahydrofolate Dehydrogenase 2 (MTHFD2) J Med Chem. 2021;64:11288–11301. doi: 10.1021/acs.jmedchem.1c00663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Handzlik MK, et al. Insulin-regulated serine and lipid metabolism drive peripheral neuropathy. Nature. 2023;614:118–124. doi: 10.1038/s41586-022-05637-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Ma EH, et al. Serine Is an Essential Metabolite for Effector T Cell Expansion. Cell Metab. 2017;25:345–357. doi: 10.1016/j.cmet.2016.12.011. [DOI] [PubMed] [Google Scholar]
- 181.Ron-Harel N, et al. Mitochondrial Biogenesis and Proteome Remodeling Promote One-Carbon Metabolism for T Cell Activation. Cell Metab. 2016;24:104–117. doi: 10.1016/j.cmet.2016.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Ma EH, et al. Metabolic Profiling Using Stable Isotope Tracing Reveals Distinct Patterns of Glucose Utilization by Physiologically Activated CD8(+) T Cells. Immunity. 2019;51:856–870.:e855. doi: 10.1016/j.immuni.2019.09.003. [DOI] [PubMed] [Google Scholar]
- 183.Sugiura A, et al. MTHFD2 is a metabolic checkpoint controlling effector and regulatory T cell fate and function. Immunity. 2022;55:65–81.:e69. doi: 10.1016/j.immuni.2021.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Kurniawan H, et al. Glutathione Restricts Serine Metabolism to Preserve Regulatory T Cell Function. Cell Metab. 2020;31:920–936.:e927. doi: 10.1016/j.cmet.2020.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Rodriguez AE, et al. Serine Metabolism Supports Macrophage IL-1beta Production. Cell Metab. 2019;29:1003–1011.:e1004. doi: 10.1016/j.cmet.2019.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Wilson JL, et al. Inverse Data-Driven Modeling and Multiomics Analysis Reveals Phgdh as a Metabolic Checkpoint of Macrophage Polarization and Proliferation. Cell Rep. 2020;30:1542–1552.:e1547. doi: 10.1016/j.celrep.2020.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Tajan M, Vousden KH. Dietary Approaches to Cancer Therapy. Cancer Cell. 2020;37:767–785. doi: 10.1016/j.ccell.2020.04.005. [DOI] [PubMed] [Google Scholar]
- 188.Mendez-Lucas A, et al. Identifying strategies to target the metabolic flexibility of tumours. Nat Metab. 2020;2:335–350. doi: 10.1038/s42255-020-0195-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Kanarek N, et al. Histidine catabolism is a major determinant of methotrexate sensitivity. Nature. 2018;559:632–636. doi: 10.1038/s41586-018-0316-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Falcone M, et al. Sensitisation of cancer cells to radiotherapy by serine and glycine starvation. Br J Cancer. 2022;127:1773–1786. doi: 10.1038/s41416-022-01965-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Gravel SP, et al. Serine deprivation enhances antineoplastic activity of biguanides. Cancer Res. 2014;74:7521–7533. doi: 10.1158/0008-5472.CAN-14-2643-T. [DOI] [PubMed] [Google Scholar]
- 192.Kalbasi A, Ribas A. Tumour-intrinsic resistance to immune checkpoint blockade. Nat Rev Immunol. 2020;20:25–39. doi: 10.1038/s41577-019-0218-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Schaer DA, et al. The Folate Pathway Inhibitor Pemetrexed Pleiotropically Enhances Effects of Cancer Immunotherapy. Clin Cancer Res. 2019;25:7175–7188. doi: 10.1158/1078-0432.CCR-19-0433. [DOI] [PubMed] [Google Scholar]
- 194.Peng ZP, et al. Downregulation of phosphoserine phosphatase potentiates tumor immune environments to enhance immune checkpoint blockade therapy. J Immunother Cancer. 2023;11 doi: 10.1136/jitc-2022-005986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Cantor JR, et al. Physiologic Medium Rewires Cellular Metabolism and Reveals Uric Acid as an Endogenous Inhibitor of UMP Synthase. Cell. 2017;169:258–272.:e217. doi: 10.1016/j.cell.2017.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Hennequart M, et al. The impact of physiological metabolite levels on serine uptake, synthesis and utilization in cancer cells. Nat Commun. 2021;12:6176. doi: 10.1038/s41467-021-26395-5. [DOI] [PMC free article] [PubMed] [Google Scholar]