Abstract
With the recent rapid progress in the aging field, there is increasing evidence that many features commonly attributed to mechanisms/drivers of aging in fact represent adaptations. Here, we examine several such features, including cellular senescence, epigenetic aging and stem cell alterations. We draw a distinction between causes and consequences of aging and define short-term consequences as “responses” and long-term ones as “adaptations”. We also discuss “damaging adaptations”, which despite having beneficial effects short-term, lead to the exacerbation of the initial insult and acceleration of aging. Features commonly recognized as “basic mechanisms of the aging process” are critically examined here for the possibility of their adaptation-driven emergence from the processes like cell competition and wound-like appearance of the aging body. Finally, we speculate on the meaning of these interactions for the aging process and their relevance for the development of anti-aging interventions.
Introduction
Aging is a highly complex biological process associated with a plethora of changes. However, not all changes observed in old tissues are necessarily drivers of the aging process or are even important for it. The distinction between causes and consequences of aging is especially relevant for designing interventions aimed at extending healthy lifespan. The reason is that targeting features that change with age but are not drivers of aging might be ineffective or even harmful. However, drawing the distinction between causes and consequences of aging is not easy. This is because many of the cellular and molecular phenotypes found in aged tissues represent adaptations of cells to their internal changes or the everchanging environment (Fig. 1). These adaptations, while downstream of the degenerative events that truly represent aging, themselves play a crucial role in the process. A comprehensive description of aging-related adaptations requires comprehensive understanding, which we dive into in this paper.
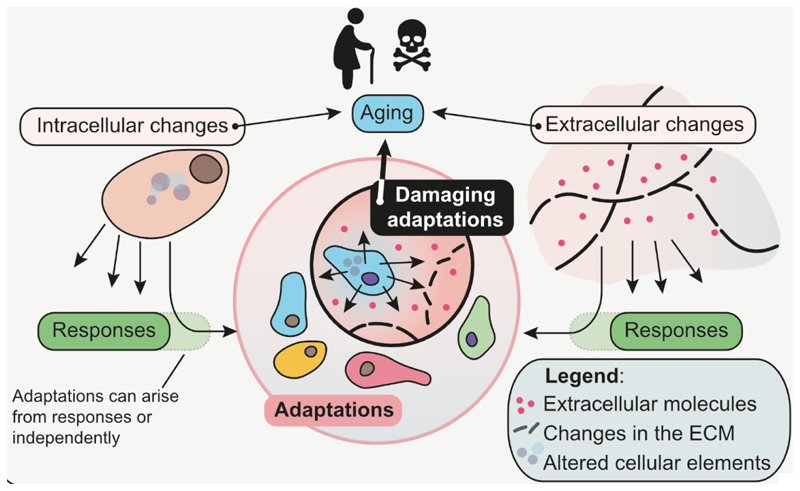
Figure 1. Relationships between causes and consequences of the aging process.
Aging leads to changes at all levels of organismal complexity including damage accumulation, alterations of the extracellular matrix (ECM), inflammation etc. These and other changes lead to short-term responses and long-term adaptations of cells, allowing them to thrive and survive. A subset of these adaptations is “damaging”, i.e. with the negative consequences outweighing the benefits and leading in the long-term to the exacerbation of the aging process.
Since the beginning of cellular life, cells have been maximizing their chances of survival and propagation. The transition to multi-cellular life imposed spatiotemporal restrictions on the division rate, motility and function of individual cells to favor survival and propagation of cell communities/organisms. Thus, for cells hosted within a multi-cellular organism, survival is not only about nutrient availability and cell integrity but also about the proper response to microenvironmental cues. Failure to properly respond to signals from the microenvironment leads to the elimination of irresponsive cells. For example, neurons that do not manage to connect to their tissues of destination during development suffer deprivation of the nerve growth factor (synthesized by target tissues) and undergo apoptosis1, the fate of ~1/3 of all neurons within two weeks after birth2. In contrast, if a cell manages to become “independent” and resistant to signals from the microenvironment, the outcome is cancer, collapse of the whole multi-cellular system, and death. Thus, effective responsiveness to the local microenvironment is the basis for multi-cellular existence, for example with beta cells producing insulin in response to glucose, muscle cells contracting upon stimulation by nerves, immune cells becoming pro-inflammatory in response to foreign antigens, etc. When environmental cues are aberrant or missing, properly responding cells will not be able to function effectively. For example, for the effective uptake of glucose, cells need to be co-stimulated with insulin, and in diabetes the reduction in the release of insulin can lead to starvation of cells in an otherwise nutrient-full environment. However, cellular responses can also occur due to cell-internal changes. Skin cells exposed to excessive sunlight/UV show profound DNA damage and respond with apoptosis3. Other types of cells can respond to DNA damage with cell cycle arrest, usually a temporal one4. In this context, we define “response” as a short-term change of cellular phenotype to match signals from the microenvironment or from the inside of cells.
A long-term or a permanent change of cellular phenotype upon an internal or microenvironmental stimulation is defined here as “adaptation”. The microenvironmental drivers of adaptations include changes of the extracellular matrix (ECM), presence or absence of secretory factors (e.g. cytokines or hormones), electrical or mechanical stimuli, etc. For example, repeated/continuous stimulation of neurons leads to structural and biochemical changes in their synapses and the long-lasting increase in signal transmission5. Upon wounding, fibroblasts sense alterations in the local microenvironment, becoming pro-inflammatory and pro-migratory for a prolonged period of time6. The cell-internal drivers of adaptation include a wide range of structural changes of molecules, changes in their quantity, localization or properties, changes that are usually defined as “damage” such as mutations, epimutations, aggregates, certain post-translational modifications, macromolecular breaks, etc. (for more details see7). Damage and other cell-internal changes including fragmented mitochondria or a leaky nuclear envelope increase the risk of failed cell division and the overall risk of death7–9. In terms of coping with damage, a cell can be induced to undergo senescence (see later) or commit to differentiation10, in both cases preventing cell cycle progression. Thus, similar to adaptations stimulated by the microenvironment, adaptations originating from cell-internal changes are set to maximize survival of cells. As for the origin of such adaptations, they could be a direct consequence of the responses or be response-independent. For the prior, responses transition to adaptations when the stimuli are repetitive (an “aggregate” of responses) or of a prolonged duration, such as adaptations to cold, starvation etc. The latter could be due to stimuli which are below the threshold to trigger a response or do not have a dedicated machinery needed for an effective response and mild damage forms would fulfil these criteria7,11. Following this line of thought, one can also find a “grey zone” of processes, which are challenging to be assigned to binary categories of responses and adaptations. Especially, with the prolonged or an “aggregate” of responses it is currently unclear what the threshold for a response to develop into an adaptation is, and how such a transition looks like. In this perspective piece we will focus on the end points of these transitions, thus discuss adaptations that arise during aging.
While adaptations are generally set to maximize cell survival, some adaptations can have negative long-term consequences for their host. Based on that, we define “damaging adaptations” as a subset of adaptations that emerge during aging and further accelerate and/or exacerbate the aging process. In the following sections we will dissect three areas of extensive research in the field of aging and discuss what types of adaptations they represent and whether they are of internal origin or driven by the microenvironment.
Cellular senescence
Cellular senescence is a cell cycle arrest that brings a variety of phenotypic changes to cells including a pro-inflammatory phenotype12. Senescence is commonly associated with aging, but its appearance has also been frequently attributed to healing, regeneration and development10. The first thing worth clarifying about senescence is whether this process is just a response or an adaptation, i.e. whether senescence in vivo represents a short-term or a long-term change. Most studies on senescent cells examine a snapshot of an organ’s physiology (e.g. using frozen or formalin-fixed piece of tissue) making it impossible to tell how long the cells currently positive for senescence markers have been in this state. Nonetheless, much evidence collected from in vitro studies as well as numerous cross-sectional in vivo studies have reported an elevated level of senescence markers persisting for a period of time, suggesting that senescence is long-term, and as such an adaptation13. In addition, while responses underlying senescence such as DNA damage response or others12 have their dedicated cellular machinery, cellular senescence as such is not definable by any specific protein and rather develops gradually as an indirect consequence to prolonged and/or multiple responses, once again matching the criteria of an adaptation. Although in vitro studies showed that senescent cells do not recover from a high load of damage, overexpression of oncogenes or prolonged cell culture, there is currently no evidence for the notion that senescence in vivo pertains a permanent cell cycle arrest.
It is a matter of ongoing debate whether cellular senescence observed in older organisms is caused by cell-intrinsic changes or the microenvironment. On the one hand, it is well-established that a variety of damage forms cause cellular senescence in vitro and in vivo11, suggesting that senescence is a cell-internal adaptation. On the other hand, appearance of senescent cells, for example during development and in wound healing, is tightly regulated in a spatiotemporal manner10,14–16, suggesting that senescence in vivo can be an adaptation caused by the microenvironment. There has been a number of hypotheses on the origin of cellular senescence in aging, including protection against cancer17, accumulation of damage incompatible with cell division11, high activity of cell expansion pathways18, a combination of both damage and expansion stimuli19, dysfunction of the immune system20, and a wound-like signature of aging organs10. The matter is, however, more complex as senescence can arise not only from cell-internal or non-cellular environmental stimuli, but can also be induced by other senescent cells in a paracrine manner21,22. Additionally, there is a “grey zone” for senescence induction as senescence can be triggered and maintained by reactive oxygen species (ROS)23, which can originate from cell-internal, environmental or paracrine triggers24.
With the evidence supporting both cell-intrinsic and microenvironmental-driven senescence induction, it is worth noting that they are not mutually exclusive and that there is also a positive feedback loop between cellular and extracellular factors that drive senescence. For example, inflammation and ECM damage contribute to upregulation of metabolic pathways and damage accumulation inside the cells25,26 and vice versa, damaged cells are more proteolytic and pro-inflammatory, leading to disruption of their microenvironment10,12. Thus, as in any other aspects of aging, both the dysfunctional microenvironment and cell-internal changes can drive adaptations of cells, including the induction of cellular senescence.
Another key question is whether this adaptation (senescence during aging) can be considered a “damaging adaptation”, and thus a promising target for anti-aging interventions. It might take many years of research and clinical trials before a reliable answer can be established, but the results reported so far are promising. It was reported that disabling the machinery required for cellular senescence induction can alleviate certain age-related dysfunctions; for example, mice with dysfunctional telomeres show improved tissue maintenance and survival following p21 depletion27. A number of studies in mice showed that the elimination of senescent cells is sufficient to alleviate a number of age-related diseases and to increase average lifespan12,28, albeit without a prominent effect on maximum lifespan11,29. However, there is increasing concern that the elimination of senescent cells could bring with it detrimental long-term consequences: recent research has revealed beneficial functions of senescence10,14–16, and elimination of p16-expressing senescent cells has been shown to lead to deterioration of health30. This pleiotropic role of senescence could be because the cell cycle arrest that underlies senescence is a response that has a context-specific function, with consequences varying between cell types and conditions (Fig. 2). For example, in health, cell cycle inhibitors are expressed when cells need to become more specialized, with arrested beta cells producing more insulin31, and arrested macrophages becoming more fit to fight off infections32. Inhibition of cell cycle is usually also coupled with the inhibition of cell death pathways, a feature prominent for both senescence and differentiation10. When it comes to aging, there are reports suggesting that cells acquiring features of senescence such as cell cycle arrest, for example neurons, do so in order to avoid death10,33. Interventions used to target senescent cells are not cell-type-specific (and often not senescence-specific) and the number of studies that investigated in detail what type of cells are and are not killed by senescence-targeting treatments is limited. Thus, more research is needed to assess the impact of senescence in aging and beyond to develop more selective anti-senescence interventions. Overall, senescence appears to be a type of damaging adaptation of aging cells resulting in an exacerbation of the aging phenotype.
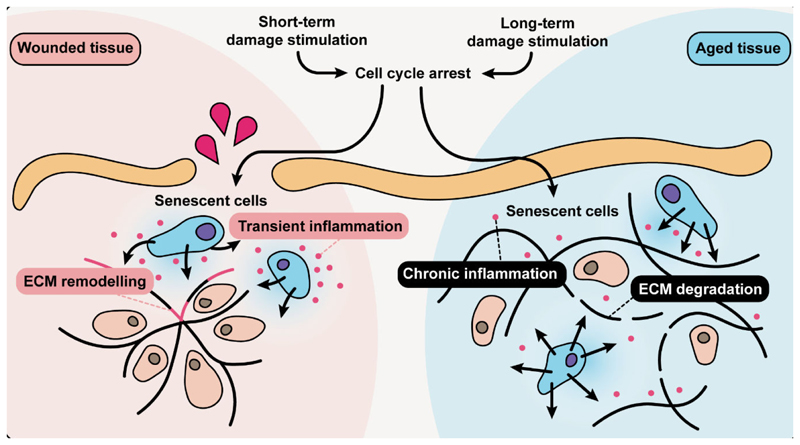
Figure 2. Cellular senescence as an adaptation to damage in tissues.
A burst of damage that is a consequence of wounding results in an adaptive response, cell cycle arrest and induction of cellular senescence needed to cause transient inflammation and to remodel the extracellular matrix (ECM). Damage is also present during aging, but its irreparability leads to the persistence of cellular senescence, chronic inflammation and ECM degradation.
Epigenetic changes
DNA methylation at CG dinucleotides (CpG sites) is a type of epigenetic modification that has a number of functions, most notably the regulation of accessibility of transcription factors to the DNA34,35. A quantitative approach that relates age-associated changes in the DNA methylome with chronological age has been coined the “epigenetic aging clock” with the approximation of age it measures being called the “epigenetic age”36. Epigenetic clocks are now viewed as currently the best available tools for the estimation of biological age, however, the origin of epigenetic changes that underlies the clocks remains mysterious. One of the reasons why it is difficult to decipher the meaning of epigenetic clocks originates in the methodology behind their design. Essentially, epigenetic clocks are made by attributing a value to each relevant methylation site, such that multi-variate machine learning models enable a prediction of an individual’s age as accurately as possible. There are now multiple types of clocks designed to predict the age of tissues such as the blood37, liver38 or any tissue (multi-tissue)39,40 of mice and humans. There are also clocks that work with bulk samples and at a single cell level41, as well as clocks that are trained for phenotypic or functional changes, future mortality and the rate of aging42–44. The equations that these clocks are based on rely on different CpG sites and attribute different weights to them. In other words, clocks are not made to trace methylation sites of any specific genes, cell types, or biological processes, but rather are made so that the algorithm gives the most accurate age prediction, regardless of the origin or meaning of the variables it uses.
The DNA methylome consists of modifications that are often different between cell types and further change when cells execute specific functions34. Furthermore, some methylation patterns can be rapidly altered, for example during responses to metabolites, pathogens or other microenvironmental cues45–48, while other methylation patterns change slowly or persist, for example those responsible for cell identity or for supporting constitutive heterochromatin34,35. From this perspective, some methylome modifications would be “responses”, and others would be “adaptations”. When detected at the level of organs (e.g. in tissue lysates), the parameters used for the measurement of the epigenetic age of tissues might thus represent a mixture of reversible responses, long-term adaptations, alterations of gene expression, modifications of the accessibility to the non-coding regions of the DNA or changes in the proportions between populations of cells inhabiting a given tissue. One solution to distinguish cell-from population-level methylation changes would be with single-cell methylome sequencing41. However, even then, the diversity of information used to design the clocks is overwhelming, making it highly challenging to understand the role of clocks in aging and clarify whether epigenetic age could be considered a damaging adaptation.
Instead of investigating the clocks’ origin, let’s examine closely the biological phenomena for which the reliability of clocks has been tested. The clocks are designed to measure age-related changes, not only associated with “healthy aging”, but also with conditions known to accelerate aging such as obesity, smoking, trauma and certain genetic disorders36. Would epigenetic changes underpinning the clock models be driven by cell-internal changes or microenvironmental stimulations? In support of the latter, it was recently shown that some forms of damage such that DNA breaks do not increase the clock readout and that conditions leading to a rapid induction of senescence such as X-ray irradiation and overexpression of oncogenes do not increase epigenetic age49.
While a rapid induction of senescence does not elevate the predicted epigenetic age, a gradual senescence induction as in the case of replicative senescence does increase it49. Importantly, an increase in epigenetic age during replicative aging was not strictly related to the induction of cell senescence (i.e. conditions of late passages), but instead showed a gradual increase over time throughout the duration of cell culture49. Together with the observation that even immortal cells, e.g. telomerase reverse transcriptase (TERT)-expressing fibroblasts, show an increase in epigenetic age in cell culture over time, it can be assumed that epigenetic aging is caused, in part, by the progressive adaptations to the cell culture conditions49. Similar to unicellular organisms, primary as well as immortal cells maximize their survival and propagation chances while competing for resources and space in a cell culture dish. Such adaptations require changes in the DNA methylome and thus must be contributing to epigenetic aging.
Another causal factor behind progression of epigenetic clocks is differentiation. Not only less differentiated cells, such as those in the muscle and intestine50,51, exhibit younger epigenetic age than their more differentiated counterparts, but mixed populations of differentiated and non-differentiated cells show the epigenetic age matching the proportion between populations49. Similarly, differentiating retinal cells exhibit epigenetic aging52. Even more importantly, while undifferentiated stem cells do not show epigenetic aging, the process of epigenetic aging starts as soon as stem cells start differentiating49. These observations match the recent findings that the epigenetic clock in mice starts ticking around ~7.5E of development41, which roughly corresponds to the formation of three germ layers and the initiation of the differentiation processes, such as the transition of the inner cell mass to epiblast34 and epithelial-to-mesenchymal transitions53. Thus, the epigenetic age could be to a certain degree driven by the aging-induced progressive differentiation of cells10 and depletion of stem cells54. This, however, cannot be the sole source of aging-driven alterations of the methylome as epigenetic clocks also work very well for tissues that do not have many known stem cells, such as liver and blood. These changes in the methylome could be driven by systemic or microenvironmental adaptations as epigenetic aging has been shown to be influenced by the alterations and damage of the ECM, changes in nutrient availability and sensing, cellular metabolism and mitochondria biogenesis40,47,49,55.
While there is a lot of evidence that epigenetic clocks measure aging-induced microenvironmental adaptations, it remains unclear whether these adaptations, or more generally, age-related changes in the methylome, are damaging. In this respect, it was recently shown that a model of dysfunctional epigenetics shows accelerated aging and higher readouts of the epigenetic clocks56. However, as the model is not specific to epigenetic clock-related methylations, it is unclear whether clock-specific methylations could be considered “damaging”.
A recent study leveraged large-scale genetic data in combination with epigenome-wide Mendelian Randomization in an attempt to identify CpG sites causal to age-related traits, such as lifespan, healthspan and longevity57. It also developed a framework for integrating causal knowledge into epigenetic clock models that measure age-related damaging and adaptive changes. Interestingly, the former model was associated with various adverse conditions (e.g., mortality risk), whereas the latter was related to beneficial adaptations. Both clock models could be used to predict epigenetic age, but they performed differently in response to interventions57. This study further reinforces the idea of adaptive changes in the epigenome during aging.
As the clocks are integrative biomarkers, it is likely that they include signals of both damaging and non-damaging adaptations (Fig. 3). This is consistent with the deconstruction of clocks resulting in differentially behaving modules58. However, further research is needed to deconvolute epigenetic clocks and identify their subsets representing diverse adaptations. While total epigenetic changes may maximize chances for survival of cells in the face of systemic and microenvironmental changes that underly aging, some of them may be the consequences of the other. Although being amazing tools to characterize and quantitively measure the process of aging, the diversity of epigenetic clocks and their complex composition make them unlikely targets for anti-aging interventions.
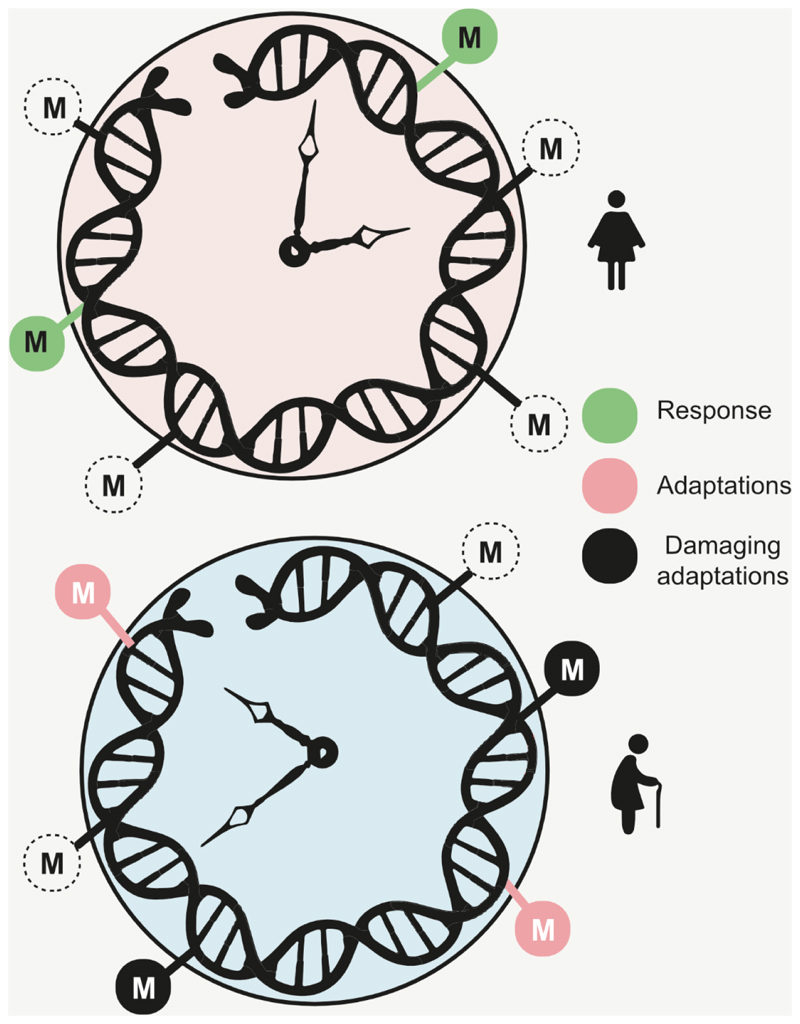
Figure 3. Age-related changes in DNA methylation are associated with diverse processes.
Changes in DNA methylation status of CpG sites between young (upper) and old (lower) individuals may reflect short-term responses and long-term adaptations, including damaging adaptations. M is a methyl group. Open circles show CpGs that previously were methylated, and filled circles show currently methylated sites.
Stem cell alterations
Numerous changes and dysfunctions of the stem cell compartment occur during aging, including skewing of the differentiation lineage of stem cells, a reduction in proliferation, and an overall decline in stem cell number54. Remarkably, these age-related deficiencies come as a surprise, as stem cells themselves seem to be more resistant to the aging process than other cell types. They have a high repair capacity59,60, an ability to regulate the length of their telomeres61 and robust mechanisms to remove an already accumulated damage via asymmetric distribution62–64. Empirically, at least for some types of stem cells the cell-internal consequences of the aging process, such as cell cycle arrest and DNA damage, are infrequent or even absent65–67. This disparity between the functionality and the state of stem cells in aged tissues can be explained by the dependency of stem cells on their microenvironment. Like no other cell type, stem cells strictly rely on their surroundings, called the “niche”. The microenvironment of stem cells does not only embed them within their tissue of origin, but also regulates their functionality including quiescence, proliferation rate, commitment towards the differentiation lineage and the responsiveness to systemic stimuli54. There is abundant experimental evidence supporting aging-induced degeneration of the niches, including an increase in their stiffness68, degeneration of the surrounding ECM69–71, a reduction in blood supply72, an increase in fat deposition73, an infiltration of immune cells and the consequent chronic inflammation74. We hypothesize that the majority of age-related changes in the function of stem cells are due to their adaptations to the conditions of aging niches. As the cellular machinery underlying adaptations originates from the cellular changes to the physiological stimuli, it would be safe to assume that there are certain conditions aside from aging where such adaptations are beneficial.
In addition to their role during tissue homeostasis, stem cells are extremely important during tissue injury, even to the extent that depletion of some types of stem cells has no effect on homeostasis, but impedes regeneration75. Skin stem cells, residing in hair follicles and in the basal layer of the epidermis, are mobilized out of their niches during wound healing in order to support re-epithelialization, among others76,77. It has been observed that in the aged skin, stem cells residing within hair follicles migrate to the epidermis, which has been causally linked to age-related hair loss70. As age-related changes of skin include an increase in inflammation and degradation of the ECM78, conditions that are highly resemblant of skin wounding6, it is possible that aged skin bears phenotypic resemblance to wounded skin10. Thus, the age-related changes in skin stem cells could be potentially attributed to their adaptations to the wound-like conditions of the aged skin.
Another example is the bone marrow providing a niche for hematopoietic stem cells (HSCs). These stem cells are responsible for the generation of new blood cells, both the red blood cells like erythrocytes originating in the process called “myelopoiesis” and the white blood cells like lymphocytes originating in the process of “lymphopoiesis”54,79. While the processes of myelopoiesis and lymphopoieses are balanced during homeostasis, certain physiological conditions can shift the balance towards the preference of one process over the other that skews the differentiation profile of HSCs. Such conditions include blood loss/hemorrhage when HSCs are stimulated to skew the differentiation profile with a concomitant increase in the generation of erythrocytes over lymphocytes80. In a surprising similarity, the aging process of the HSC compartment is well-known to result in the skewing of the differentiation process to generate fewer lymphoid and more myeloid progeny79. Knowing that the process of aging is associated with a general decline in the fitness and oxygen carrying capacity of erythrocytes81–83, it is possible that the commitment of HSCs towards myeloid differentiation is an adaptation compensating for the decrease in the functionality of erythrocytes in the aging body.
Finally, muscle stem cells (MuSCs) are known to be responsible for muscle regeneration in conditions of injury and minor damage originating from exercise84,85. While small injuries result in MuSCs differentiating into muscle cells, deep injuries cause muscle fibrosis86. Similarly, the aging process results in MuSCs being “primed” towards differentiation87,88, with a fraction of progeny becoming pro-fibrotic89. As the aging process of the muscle is accompanied by the degradation of the ECM and an increase in inflammation71,90, the aging-induced changes in MuSCs could originate from an injury-like state of the muscle tissue. In summary, aging-associated changes in stem cell function could derive from the degradation and inflammation of their niches resembling the conditions of the wounded tissues and leading to adaptations aimed at aiding their regeneration.
Another question is whether the aging-associated changes in stem cell function are a long-term adaptation or just a short-term response. Treatments such as parabiosis have proven that at least some of the age-induced changes in the stem cell compartment depend on the presence and absence of systemic factors and are reversible over a short period of treatment time91,92. This suggests that at least some aging-associated features of stem cells are direct responses to the current state of the body. However, many types of stem cell seem to be resistant to the presence of rejuvenating factors and/or reduction in pro-aging factors in the circulation93. Similarly, experiments involving transplantation of stem cells show that even when they are integrated into youthful niches, stem cells from aged animals retain certain aging-related features68,94,95, suggesting that the aging process has at least a long-term if not permanent impact on them. Overall, stem cells react to the aging process in a complex manner, with the short-term and reversible responses alongside the long-term adaptations (Fig. 4).
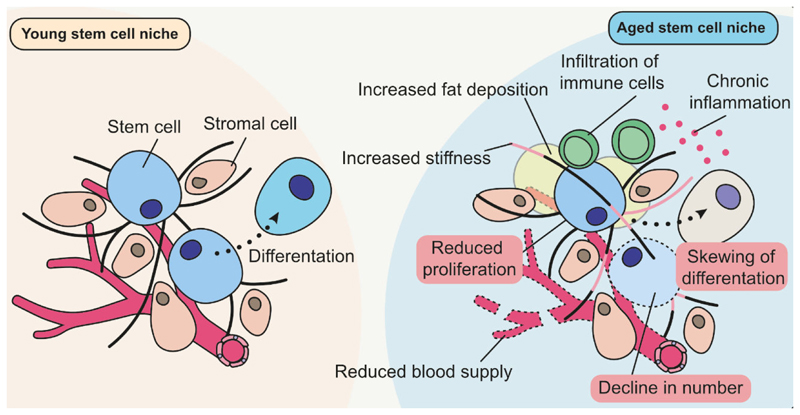
Figure 4. The age-associated alterations of stem cells are driven to a large extent by adaptations to a degrading niche.
Adaptations of stem cells found in aged organisms are highlighted in red.
Finally, it is challenging to estimate how “damaging” these stem cell adaptations are to the aging process. Certainly, these adaptations have negative consequences, such as a decline in the propensity of HSCs to differentiate into lymphocytes, thereby increasing the risk of infection96, and the excessive migration of epidermal stem cells out of hair follicles, thereby causing hair loss70. However, it is not clear whether the alternatives are better. If stem cells were to be prevented from changing their phenotypes in relation to the wound-like conditions of the aged tissues would that bear beneficial effects or accelerate tissue degradation and aging even further? To illustrate, it has been hypothesized that the selection of mutant HSC clones makes them more pro-inflammatory97, which could potentially be beneficial for tissue regeneration or to counteract infections, but on the other hand these clones seem to exacerbate age-related conditions such as cardiac infarction97.
In summary, the aging process of the stem cell compartment appears to be tailored to the needs of the aging body. With a progressive degradation of the ECM and systemic cueing of tissue damage, stem cells in aged tissues are geared towards regeneration rather than homeostasis. Thus, it is difficult to assess whether treating aging-associated changes in stem cells would provide a robust target for anti-aging interventions.
Linking it back together: adapt to survive at all costs
Each and every aspect of our lives, whether in health or disease, bears witness that our cells constantly strive to thrive and survive. It is thus rather unlikely that aging conditions would be an exception, where a malicious program sets in motion systems and means to drive disease and death. Instead, aging manifests as a progressive accumulation of intra- and extracellular damage and a consequential change in function in many types of cells that attempt to adapt and survive by any means necessary. Some of these means might work short-term, but in the long run they become damaging, and further exacerbate and accelerate the aging process.
The examples of adaptations to the aging process discussed above include cellular senescence, methylome alterations used to measure the epigenetic age, and changes in the stem cell compartment, but it is clear that such adaptations are widespread in other systems too. The strategies behind these adaptations are versatile, with the justification for cellular senescence being the most obvious, as the cell cycle arrest it imposes provides an alternative fate to cell death. When it comes to stem cells, there is evidence that their adaptations provide an advantage during aging-induced clonal selection in tissues such as the skin98, bone marrow99 and muscle100. Interestingly, it was shown that some of the genes positively selected include epigenetic regulators such as DNA methyltransferase 3A101. Despite some data suggesting that senescence does not accelerate epigenetic aging in vitro49, overall, senescence induction leads to prominent changes in the DNA methylome102, therefore it is possible that both these adaptations could contribute to epigenetic aging as measured by the epigenetic clocks. Another element positively selected for the survival of stem cells during aging is their attachment to the matrix. While during aging there is a decline in the ECM supporting the niche, skin stem cells that show a compensatory increase in expression of ECM proteins103 are better attached to the ECM and thus are selected for survival69. Similarly, genes of the MuSCs positively selected during aging are responsible for integrin signaling71 and other regulators of stem cell-niche interactions50. Senescent cells are not only strongly attached to the surface104, but also tend to inhibit the detachment and death of the surrounding cells105. While preserving lives of cells, this paracrine effect of senescent cells can be tumorigenic105, thus negative long-term. Overall, the experimental evidence suggests that cells of aging organisms employ any means necessary to avoid death including niche-driven alterations of stem cells and senescence induction of differentiated and progenitor cells.
In this piece, we use the term “adaptations” in relation to events happening within a single lifespan of an organism, in contrast to “evolutionary adaptations” that drive changes across generations of organisms enabling them to thrive in a given environment. Nonetheless, the processes described here may be complementary to the ideas on evolution of aging, e.g. postulates of the Medawar’s and Williams’s evolutionary theories106,107. Briefly, Medawar proposed that organisms evolved features that are neutral early in life while selection is actively taking place, but are detrimental later, where they become unfixable by evolution106,107. Going one step further, Williams hypothesized that evolution selects for features providing benefits early in life even if these features have negative consequences late in life. Likewise, we argue that some adaptations while increasing survival over the course of lifespan contribute to aging in the long-term. Also, integral components of the three types of adaptations we described above can be found throughout the early life of an organism: senescent cells are present during development and healing, epigenetic clocks tick during these processes, and stem cells show phenotypic similarities during the processes of regeneration and aging. Even when beneficial in early life, these phenotypes can become damaging later on, in line with the Williams’s concept of antagonistic pleiotropy.
The matter of causality and targeted interventions in aging
While the identification of causal factors is straightforward for processes such as wound healing and fighting off infection, aging is multi-factorial and presents a landscape of intra- and extracellular causes in the context of responses and adaptations of cells. With the dense network of interactions between aging cells and their host tissues it is challenging to distinguish what comes first. In this respect, we offer the perspective that features of aging assigned as adaptations cannot be the primary drivers of the aging process, but some of them may nonetheless exacerbate the aging process. This perspective further assumes that the adaptations found in aging are not there to drive the aging process, but are an attempt to alleviate age-related changes regardless of whether this is possible or not (i.e. pro-healing activation of cells due to the wound-like appearance of an aged tissue is unlikely to actually heal it). Finally, the category of “damaging adaptations” describes adaptations which are predominantly detrimental to the aging process, making them a tempting target for anti-aging interventions. However, as the processes underlying “damaging adaptations” are not specific to aging, targeting them could have side effects for conditions where such adaptations are beneficial (such as tissue regeneration). Moreover, targeting these adaptations is unlikely to have a major effect on the aging process itself, as the underlying processes are not going to be affected.
Ideally, we would want to target the very basal and specific causal factors behind the aging process, as there could be no side effects coming from their selective targeting. While the current progress of the science of biogerontology seems promising when it comes to defining a variety of adaptations to the aging process such as senescence, stem cell alterations and methylome changes, much less is known about the processes causal to adaptations, and the science behind these processes is still in its infancy. The variety of mild damage forms accumulating in cells and in the ECM and likely other molecular processes are mostly beyond the detection limit, without any possibility of specific targeting. In this respect, we encourage scientists to expand the technical, conceptual and experimental scope of their research to pioneer the science of the drivers of aging.
Conclusions
The three exemplary adaptations described here were intentionally selected to be highly diverse and seemingly unrelated. The conceptual framework we created here is aimed at showing that essentially any of the prominent targets of aging research and anti-aging interventions can be dissociated into sets of phenotypes representing responses and adaptations to the cell-internal and external changes occurring during aging. However, even if secondary to the basal processes of aging and set for survival of individual cells, these adaptations might cause more damage that exacerbate the initial insult. While each one of such damaging adaptations appears to be a promising target for anti-aging treatments, at this point it is not obvious which of these are “sufficiently” damaging that their alteration or elimination would be considered beneficial long-term. With the advent of single-cell omics, advanced genetic tools and high-resolution, multi-target spatial histology, it becomes more realistic to spatiotemporally map the plethora of changes occurring during the aging process and establish their long-term consequences, thus shedding light on the causal factors in aging providing the most optimal therapeutic targets.
Acknowledgements
We would like to thank Hanna Salmonowicz who developed illustrations used in the paper and Nadja Ring for proofreading of the manuscript. The Research Group Senescence and Healing of Wounds (SHoW) is a collaboration between the Ludwig Boltzmann Gesellschaft GmbH and the Austrian Workers’ Compensation Board (AUVA) with support of the Austrian Nationalstiftung. Supported by NIA grants and the Impetus grant program. M.O. is supported by Der Wissenschaftsfonds grant P 35382 and FEBS Excellence Award.
Footnotes
Author contributions:
M.O. wrote the first draft.
M.O. and V.N.G. extended, revised, and finalized the manuscript.
References
- 1.Kristiansen M, Ham J. Programmed cell death during neuronal development: the sympathetic neuron model. Cell Death Differ. 2014;21:1025–1035. doi: 10.1038/cdd.2014.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wright LL, Cunningham TJ, Smolen AJ. Developmental neuron death in the rat superior cervical sympathetic ganglion: cell counts and ultrastructure. J Neurocytol. 1983;12:727–738. doi: 10.1007/BF01258147. [DOI] [PubMed] [Google Scholar]
- 3.Mohania D, et al. Ultraviolet Radiations: Skin Defense-Damage Mechanism. Adv Exp Med Biol. 2017;996:71–87. doi: 10.1007/978-3-319-56017-5_7. [DOI] [PubMed] [Google Scholar]
- 4.Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lisman J, Yasuda R, Raghavachari S. Mechanisms of CaMKII action in long-term potentiation. Nat Rev Neurosci. 2012;13:169–182. doi: 10.1038/nrn3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rodrigues M, Kosaric N, Bonham CA, Gurtner GC. Wound Healing: A Cellular Perspective. Physiol Rev. 2019;99:665–706. doi: 10.1152/physrev.00067.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gladyshev VN, et al. Molecular damage in aging. Nature Aging. 2021;1:1096–1106. doi: 10.1038/s43587-021-00150-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lindenboim L, Zohar H, Worman HJ, Stein R. The nuclear envelope: target and mediator of the apoptotic process. Cell Death Discov. 2020;6:29. doi: 10.1038/s41420-020-0256-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karbowski M. Mitochondria on guard: role of mitochondrial fusion and fission in the regulation of apoptosis. Adv Exp Med Biol. 2010;687:131–142. doi: 10.1007/978-1-4419-6706-0_8. [DOI] [PubMed] [Google Scholar]
- 10.Ring NAR, Valdivieso K, Grillari J, Redl H, Ogrodnik M. The role of senescence in cellular plasticity: Lessons from regeneration and development and implications for age-related diseases. Dev Cell. 2022;57:1083–1101. doi: 10.1016/j.devcel.2022.04.005. [DOI] [PubMed] [Google Scholar]
- 11.Ogrodnik M, Salmonowicz H, Gladyshev VN. Integrating cellular senescence with the concept of damage accumulation in aging: Relevance for clearance of senescent cells. Aging Cell. 2019;18:e12841. doi: 10.1111/acel.12841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Deursen JM. The role of senescent cells in ageing. Nature. 2014;509:439–446. doi: 10.1038/nature13193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ogrodnik M. Cellular aging beyond cellular senescence: Markers of senescence prior to cell cycle arrest in vitro and in vivo. Aging Cell. 2021;20:e13338. doi: 10.1111/acel.13338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Demaria M, et al. An essential role for senescent cells in optimal wound healing through secretion of PDGF-AA. Dev Cell. 2014;31:722–733. doi: 10.1016/j.devcel.2014.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Munoz-Espin D, et al. Programmed cell senescence during mammalian embryonic development. Cell. 2013;155:1104–1118. doi: 10.1016/j.cell.2013.10.019. [DOI] [PubMed] [Google Scholar]
- 16.Storer M, et al. Senescence is a developmental mechanism that contributes to embryonic growth and patterning. Cell. 2013;155:1119–1130. doi: 10.1016/j.cell.2013.10.041. [DOI] [PubMed] [Google Scholar]
- 17.Rodier F, Campisi J. Four faces of cellular senescence. J Cell Biol. 2011;192:547–556. doi: 10.1083/jcb.201009094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blagosklonny MV. Cell senescence, rapamycin and hyperfunction theory of aging. Cell Cycle. 2022;21:1456–1467. doi: 10.1080/15384101.2022.2054636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ogrodnik M, Salmonowicz H, Jurk D, Passos JF. Expansion and Cell-Cycle Arrest: Common Denominators of Cellular Senescence. Trends Biochem Sci. 2019;44:996–1008. doi: 10.1016/j.tibs.2019.06.011. [DOI] [PubMed] [Google Scholar]
- 20.Ovadya Y, et al. Impaired immune surveillance accelerates accumulation of senescent cells and aging. Nat Commun. 2018;9:5435. doi: 10.1038/s41467-018-07825-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nelson G, et al. A senescent cell bystander effect: senescence-induced senescence. Aging Cell. 2012;11:345–349. doi: 10.1111/j.1474-9726.2012.00795.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Acosta JC, et al. A complex secretory program orchestrated by the inflammasome controls paracrine senescence. Nat Cell Biol. 2013;15:978–990. doi: 10.1038/ncb2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Passos JF, et al. Feedback between p21 and reactive oxygen production is necessary for cell senescence. Mol Syst Biol. 2010;6:347. doi: 10.1038/msb.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nelson G, Kucheryavenko O, Wordsworth J, von Zglinicki T. The senescent bystander effect is caused by ROS-activated NF-kappaB signalling. Mech Ageing Dev. 2018;170:30–36. doi: 10.1016/j.mad.2017.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martins SG, Zilhao R, Thorsteinsdottir S, Carlos AR. Linking Oxidative Stress and DNA Damage to Changes in the Expression of Extracellular Matrix Components. Front Genet. 2021;12:673002. doi: 10.3389/fgene.2021.673002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fisher GJ, et al. Collagen fragmentation promotes oxidative stress and elevates matrix metalloproteinase-1 in fibroblasts in aged human skin. Am J Pathol. 2009;174:101–114. doi: 10.2353/ajpath.2009.080599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Choudhury AR, et al. Cdkn1a deletion improves stem cell function and lifespan of mice with dysfunctional telomeres without accelerating cancer formation. Nat Genet. 2007;39:99–105. doi: 10.1038/ng1937. [DOI] [PubMed] [Google Scholar]
- 28.Baker DJ, et al. Naturally occurring p16(Ink4a)-positive cells shorten healthy lifespan. Nature. 2016;530:184–189. doi: 10.1038/nature16932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kowald A, Kirkwood TBL. Senolytics and the compression of late-life mortality. Exp Gerontol. 2021;155:111588. doi: 10.1016/j.exger.2021.111588. [DOI] [PubMed] [Google Scholar]
- 30.Grosse L, et al. Defined p16(High) Senescent Cell Types Are Indispensable for Mouse Healthspan. Cell Metab. 2020;32:87–99.:e86. doi: 10.1016/j.cmet.2020.05.002. [DOI] [PubMed] [Google Scholar]
- 31.Helman A, et al. p16(Ink4a)-induced senescence of pancreatic beta cells enhances insulin secretion. Nat Med. 2016;22:412–420. doi: 10.1038/nm.4054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Behmoaras J, Gil J. Similarities and interplay between senescent cells and macrophages. J Cell Biol. 2021;220 doi: 10.1083/jcb.202010162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fielder E, von Zglinicki T, Jurk D. The DNA Damage Response in Neurons: Die by Apoptosis or Survive in a Senescence-Like State? J Alzheimers Dis. 2017;60:S107–S131. doi: 10.3233/JAD-161221. [DOI] [PubMed] [Google Scholar]
- 34.Brevini TAL, Manzoni EFM, Gandolfi F. Methylation mechanisms and biomechanical effectors controlling cell fate. Reprod Fertil Dev. 2017;30:64–72. doi: 10.1071/RD17348. [DOI] [PubMed] [Google Scholar]
- 35.Moore LD, Le T, Fan G. DNA methylation and its basic function. Neuropsychopharmacology. 2013;38:23–38. doi: 10.1038/npp.2012.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Horvath S, Raj K. DNA methylation-based biomarkers and the epigenetic clock theory of ageing. Nat Rev Genet. 2018;19:371–384. doi: 10.1038/s41576-018-0004-3. [DOI] [PubMed] [Google Scholar]
- 37.Hannum G, et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol Cell. 2013;49:359–367. doi: 10.1016/j.molcel.2012.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Horvath S, et al. Obesity accelerates epigenetic aging of human liver. Proc Natl Acad Sci U S A. 2014;111:15538–15543. doi: 10.1073/pnas.1412759111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meer MV, Podolskiy DI, Tyshkovskiy A, Gladyshev VN. A whole lifespan mouse multi-tissue DNA methylation clock. Elife. 2018;7 doi: 10.7554/eLife.40675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stubbs TM, et al. Multi-tissue DNA methylation age predictor in mouse. Genome Biol. 2017;18:68. doi: 10.1186/s13059-017-1203-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Trapp A, Kerepesi C, Gladyshev VN. Profiling epigenetic age in single cells. Nature Aging. 2021;1:1189–1201. doi: 10.1038/s43587-021-00134-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Levine ME, et al. An epigenetic biomarker of aging for lifespan and healthspan. Aging (Albany NY) 2018;10:573–591. doi: 10.18632/aging.101414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Belsky DW, et al. DunedinPACE, a DNA methylation biomarker of the pace of aging. Elife. 2022;11 doi: 10.7554/eLife.73420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lu AT, et al. DNA methylation GrimAge strongly predicts lifespan and healthspan. Aging (Albany NY) 2019;11:303–327. doi: 10.18632/aging.101684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Verdikt R, Allard P. Metabolo-epigenetics: the interplay of metabolism and epigenetics during early germ cells development. Biol Reprod. 2021;105:616–624. doi: 10.1093/biolre/ioab118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bierne H, Hamon M, Cossart P. Epigenetics and bacterial infections. Cold Spring Harb Perspect Med. 2012;2:a010272. doi: 10.1101/cshperspect.a010272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lelievre SA. Contributions of extracellular matrix signaling and tissue architecture to nuclear mechanisms and spatial organization of gene expression control. Biochim Biophys Acta. 2009;1790:925–935. doi: 10.1016/j.bbagen.2009.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Qin W, Scicluna BP, van der Poll T. The Role of Host Cell DNA Methylation in the Immune Response to Bacterial Infection. Front Immunol. 2021;12:696280. doi: 10.3389/fimmu.2021.696280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kabacik S, et al. The relationship between epigenetic age and the hallmarks of aging in human cells. Nature Aging. 2022;2:484–493. doi: 10.1038/s43587-022-00220-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hernando-Herraez I, et al. Ageing affects DNA methylation drift and transcriptional cell-to-cell variability in mouse muscle stem cells. Nat Commun. 2019;10:4361. doi: 10.1038/s41467-019-12293-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lewis SK, et al. DNA Methylation Analysis Validates Organoids as a Viable Model for Studying Human Intestinal Aging. Cell Mol Gastroenterol Hepatol. 2020;9:527–541. doi: 10.1016/j.jcmgh.2019.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hoshino A, Horvath S, Sridhar A, Chitsazan A, Reh TA. Synchrony and asynchrony between an epigenetic clock and developmental timing. Sci Rep. 2019;9:3770. doi: 10.1038/s41598-019-39919-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hernandez-Martinez R, Ramkumar N, Anderson KV. p120-catenin regulates WNT signaling and EMT in the mouse embryo. Proc Natl Acad Sci U S A. 2019;116:16872–16881. doi: 10.1073/pnas.1902843116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Brunet A, Goodell MA, Rando TA. Ageing and rejuvenation of tissue stem cells and their niches. Nat Rev Mol Cell Biol. 2022 doi: 10.1038/s41580-022-00510-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thompson MJ, et al. A multi-tissue full lifespan epigenetic clock for mice. Aging (Albany NY) 2018;10:2832–2854. doi: 10.18632/aging.101590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang JH, et al. Loss of epigenetic information as a cause of mammalian aging. Cell. 2023;186:305–326.:e327. doi: 10.1016/j.cell.2022.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ying K, et al. Causal Epigenetic Age Uncouples Damage and Adaptation. bioRxiv. 2022:2022.2010.2007.511382. doi: 10.1101/2022.10.07.511382. [DOI] [Google Scholar]
- 58.Levine ME, Higgins-Chen A, Thrush K, Minteer C, Niimi P. Clock Work: Deconstructing the Epigenetic Clock Signals in Aging, Disease, and Reprogramming. bioRxiv. 2022:2022.2002.2013.480245. doi: 10.1101/2022.02.13.480245. [DOI] [Google Scholar]
- 59.Weeden CE, Asselin-Labat ML. Mechanisms of DNA damage repair in adult stem cells and implications for cancer formation. Biochim Biophys Acta Mol Basis Dis. 2018;1864:89–101. doi: 10.1016/j.bbadis.2017.10.015. [DOI] [PubMed] [Google Scholar]
- 60.Vitale I, Manic G, De Maria R, Kroemer G, Galluzzi L. DNA Damage in Stem Cells. Mol Cell. 2017;66:306–319. doi: 10.1016/j.molcel.2017.04.006. [DOI] [PubMed] [Google Scholar]
- 61.Li JSZ, Denchi EL. How stem cells keep telomeres in check. Differentiation. 2018;100:21–25. doi: 10.1016/j.diff.2018.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pattabiraman S, et al. Vimentin protects differentiating stem cells from stress. Sci Rep. 2020;10:19525. doi: 10.1038/s41598-020-76076-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Morrow CS, et al. Vimentin Coordinates Protein Turnover at the Aggresome during Neural Stem Cell Quiescence Exit. Cell Stem Cell. 2020;26:558–568.:e559. doi: 10.1016/j.stem.2020.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rujano MA, et al. Polarised asymmetric inheritance of accumulated protein damage in higher eukaryotes. PLoS Biol. 2006;4:e417. doi: 10.1371/journal.pbio.0040417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sun D, et al. Epigenomic profiling of young and aged HSCs reveals concerted changes during aging that reinforce self-renewal. Cell Stem Cell. 2014;14:673–688. doi: 10.1016/j.stem.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sacma M, et al. Haematopoietic stem cells in perisinusoidal niches are protected from ageing. Nat Cell Biol. 2019;21:1309–1320. doi: 10.1038/s41556-019-0418-y. [DOI] [PubMed] [Google Scholar]
- 67.Kalamakis G, et al. Quiescence Modulates Stem Cell Maintenance and Regenerative Capacity in the Aging Brain. Cell. 2019;176:1407–1419.:e1414. doi: 10.1016/j.cell.2019.01.040. [DOI] [PubMed] [Google Scholar]
- 68.Segel M, et al. Niche stiffness underlies the ageing of central nervous system progenitor cells. Nature. 2019;573:130–134. doi: 10.1038/s41586-019-1484-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liu N, et al. Stem cell competition orchestrates skin homeostasis and ageing. Nature. 2019;568:344–350. doi: 10.1038/s41586-019-1085-7. [DOI] [PubMed] [Google Scholar]
- 70.Matsumura H, et al. Hair follicle aging is driven by transepidermal elimination of stem cells via COL17A1 proteolysis. Science. 2016;351:aad4395. doi: 10.1126/science.aad4395. [DOI] [PubMed] [Google Scholar]
- 71.Schuler SC, et al. Extensive remodeling of the extracellular matrix during aging contributes to age-dependent impairments of muscle stem cell functionality. Cell Rep. 2021;35:109223. doi: 10.1016/j.celrep.2021.109223. [DOI] [PubMed] [Google Scholar]
- 72.Ichijo R, et al. Vasculature atrophy causes a stiffened microenvironment that augments epidermal stem cell differentiation in aged skin. Nature Aging. 2022;2:592–600. doi: 10.1038/s43587-022-00244-6. [DOI] [PubMed] [Google Scholar]
- 73.Shimabukuro MK, et al. Lipid-laden cells differentially distributed in the aging brain are functionally active and correspond to distinct phenotypes. Sci Rep. 2016;6:23795. doi: 10.1038/srep23795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dulken BW, et al. Single-cell analysis reveals T cell infiltration in old neurogenic niches. Nature. 2019;571:205–210. doi: 10.1038/s41586-019-1362-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fry CS, et al. Inducible depletion of satellite cells in adult, sedentary mice impairs muscle regenerative capacity without affecting sarcopenia. Nat Med. 2015;21:76–80. doi: 10.1038/nm.3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ito M, et al. Stem cells in the hair follicle bulge contribute to wound repair but not to homeostasis of the epidermis. Nat Med. 2005;11:1351–1354. doi: 10.1038/nm1328. [DOI] [PubMed] [Google Scholar]
- 77.Plikus MV, et al. Epithelial stem cells and implications for wound repair. Semin Cell Dev Biol. 2012;23:946–953. doi: 10.1016/j.semcdb.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pils V, et al. Promises and challenges of senolytics in skin regeneration, pathology and ageing. Mech Ageing Dev. 2021;200:111588. doi: 10.1016/j.mad.2021.111588. [DOI] [PubMed] [Google Scholar]
- 79.Yamamoto R, et al. Large-Scale Clonal Analysis Resolves Aging of the Mouse Hematopoietic Stem Cell Compartment. Cell Stem Cell. 2018;22:600–607.:e604. doi: 10.1016/j.stem.2018.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kelly LS, Darden DB, Fenner BP, Efron PA, Mohr AM. The Hematopoietic Stem/Progenitor Cell Response to Hemorrhage, Injury, and Sepsis: A Review of Pathophysiology. Shock. 2021;56:30–41. doi: 10.1097/SHK.0000000000001699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kumar D, Rizvi SI. Markers of oxidative stress in senescent erythrocytes obtained from young and old age rats. Rejuvenation Res. 2014;17:446–452. doi: 10.1089/rej.2014.1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kosower NS. Altered properties of erythrocytes in the aged. Am J Hematol. 1993;42:241–247. doi: 10.1002/ajh.2830420302. [DOI] [PubMed] [Google Scholar]
- 83.Samaja M, Rovida E, Motterlini R, Tarantola M. In: Red Blood Cell Aging. Magnani Mauro, De Flora Antonio., editors. Springer US; 1991. pp. 115–123. [DOI] [PubMed] [Google Scholar]
- 84.Bazgir B, Fathi R, Rezazadeh Valojerdi M, Mozdziak P, Asgari A. Satellite Cells Contribution to Exercise Mediated Muscle Hypertrophy and Repair. Cell J. 2017;18:473–484. doi: 10.22074/cellj.2016.4714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Karalaki M, Fili S, Philippou A, Koutsilieris M. Muscle regeneration: cellular and molecular events. Vivo. 2009;23:779–796. [PubMed] [Google Scholar]
- 86.Mahdy MAA. Skeletal muscle fibrosis: an overview. Cell Tissue Res. 2019;375:575–588. doi: 10.1007/s00441-018-2955-2. [DOI] [PubMed] [Google Scholar]
- 87.Garcia-Prat L, et al. FoxO maintains a genuine muscle stem-cell quiescent state until geriatric age. Nat Cell Biol. 2020;22:1307–1318. doi: 10.1038/s41556-020-00593-7. [DOI] [PubMed] [Google Scholar]
- 88.Chakkalakal JV, Jones KM, Basson MA, Brack AS. The aged niche disrupts muscle stem cell quiescence. Nature. 2012;490:355–360. doi: 10.1038/nature11438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Brack AS, et al. Increased Wnt signaling during aging alters muscle stem cell fate and increases fibrosis. Science. 2007;317:807–810. doi: 10.1126/science.1144090. [DOI] [PubMed] [Google Scholar]
- 90.Lightfoot AP, McCormick R, Nye GA, McArdle A. Mechanisms of skeletal muscle ageing; avenues for therapeutic intervention. Curr Opin Pharmacol. 2014;16:116–121. doi: 10.1016/j.coph.2014.05.005. [DOI] [PubMed] [Google Scholar]
- 91.Conboy IM, et al. Rejuvenation of aged progenitor cells by exposure to a young systemic environment. Nature. 2005;433:760–764. doi: 10.1038/nature03260. [DOI] [PubMed] [Google Scholar]
- 92.Katsimpardi L, et al. Vascular and neurogenic rejuvenation of the aging mouse brain by young systemic factors. Science. 2014;344:630–634. doi: 10.1126/science.1251141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Rebo J, et al. A single heterochronic blood exchange reveals rapid inhibition of multiple tissues by old blood. Nat Commun. 2016;7:13363. doi: 10.1038/ncomms13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kuribayashi W, et al. Limited rejuvenation of aged hematopoietic stem cells in young bone marrow niche. J Exp Med. 2021;218 doi: 10.1084/jem.20192283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sousa-Victor P, et al. Geriatric muscle stem cells switch reversible quiescence into senescence. Nature. 2014;506:316–321. doi: 10.1038/nature13013. [DOI] [PubMed] [Google Scholar]
- 96.Linton PJ, Dorshkind K. Age-related changes in lymphocyte development and function. Nat Immunol. 2004;5:133–139. doi: 10.1038/ni1033. [DOI] [PubMed] [Google Scholar]
- 97.Jaiswal S, Libby P. Clonal haematopoiesis: connecting ageing and inflammation in cardiovascular disease. Nat Rev Cardiol. 2020;17:137–144. doi: 10.1038/s41569-019-0247-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Martincorena I, et al. Tumor evolution. High burden and pervasive positive selection of somatic mutations in normal human skin. Science. 2015;348:880–886. doi: 10.1126/science.aaa6806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Jaiswal S, Ebert BL. Clonal hematopoiesis in human aging and disease. Science. 2019;366 doi: 10.1126/science.aan4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Tierney MT, Stec MJ, Rulands S, Simons BD, Sacco A. Muscle Stem Cells Exhibit Distinct Clonal Dynamics in Response to Tissue Repair and Homeostatic Aging. Cell Stem Cell. 2018;22:119–127.:e113. doi: 10.1016/j.stem.2017.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Tovy A, et al. Tissue-Biased Expansion of DNMT3A-Mutant Clones in a Mosaic Individual Is Associated with Conserved Epigenetic Erosion. Cell Stem Cell. 2020;27:326–335.:e324. doi: 10.1016/j.stem.2020.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Yang N, Sen P. The senescent cell epigenome. Aging (Albany NY) 2018;10:3590–3609. doi: 10.18632/aging.101617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ge Y, et al. The aging skin microenvironment dictates stem cell behavior. Proc Natl Acad Sci U S A. 2020;117:5339–5350. doi: 10.1073/pnas.1901720117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Cristofalo VJ. Cellular biomarkers of aging. Exp Gerontol. 1988;23:297–307. doi: 10.1016/0531-5565(88)90032-0. [DOI] [PubMed] [Google Scholar]
- 105.Igarashi N, et al. Hepatocyte growth factor derived from senescent cells attenuates cell competition-induced apical elimination of oncogenic cells. Nat Commun. 2022;13:4157. doi: 10.1038/s41467-022-31642-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Medawar PB. Old age and natural death. Mod Q. 1946;1:30–56. [Google Scholar]
- 107.Williams GC. Pleiotropy, Natural Selection, and the Evolution of Senescence. Science of Aging Knowledge Environment. 2001;2001:cp13. doi: 10.1126/sageke.2001.1.cp13. [DOI] [Google Scholar]