Abstract
Natural biomolecular systems have evolved to form a rich variety of supramolecular materials and machinery fundamental to cellular function. The assembly of these structures commonly involves interactions between specific molecular building blocks, a strategy that can also be replicated in an artificial setting to prepare functional materials. The self-assembly of synthetic biomimetic peptides thus allows the exploration of chemical and sequence space beyond that used routinely by biology. In this Review, we discuss recent conceptual and experimental advances in self-assembling artificial peptidic materials. In particular, we explore how naturally occurring structures and phenomena have inspired the development of functional biomimetic materials that we can harness for potential interactions with biological systems. As our fundamental understanding of peptide self-assembly evolves, increasingly sophisticated materials and applications emerge and lead to the development of a new set of building blocks and assembly principles relevant to materials science, molecular biology, nanotechnology and precision medicine.
Self-assembly in biological systems allows individual macromolecules to assemble into a wide set of supramolecular structures and architectures. In this manner, nature capitalizes on self-assembly to convert chemically simple building blocks into sophisticated materials and structures that function cooperatively in living systems1–3. The molecular interactions governing the formation of such systems are predominantly non-covalent, a key aspect that determines their microscale and macroscale properties4. In particular, the reversibility of the interactions confers dynamism on the molecular architectures, which can modulate their properties and confer an ability to respond to external stimuli5. In nature, a particularly diverse class of self-assembling materials are formed from proteins6. For instance, cellular motility and traction to surfaces are largely controlled through the self-assembly of cytoskeletal proteins7. These proteins reversibly self-assemble to enable highly regulated extension and contraction of cells, thus allowing their movement. Moreover, networks of such protein assemblies generate force for a wide range of active processes, including cell migration, movement of endocytic vesicles and other membrane-bound organelles within cells, along with intercellular transport of certain bacterial and viral pathogens1,8,9.
Much progress has been made in understanding the fundamental principles that govern native-protein self-assembly processes by studying naturally occurring building blocks10–15. A complementary approach is to use synthetic chemistry to explore the chemical space beyond that available to natural molecular building blocks. This has primarily been achieved by a bottom-up approach, whereby building blocks are designed to assemble into specific architectures with desired properties16. In natural systems, self-assembly benefits from the evolutionary processes that tune interactions to optimize the properties, morphology and functionality of the resulting biomaterials. Through evolution, nature exploits a narrow set of elementary motifs, including the α-helix and the β-sheet secondary structure of proteins and their complexes, to hierarchically assemble a remarkably complex set of structures17.
Even though nature commonly uses protein sequences of up to several hundred amino-acid residues as building blocks of functional materials, substantially shorter sequences can also exhibit highly sophisticated self-assembly behaviour18,19. In this context, biomimetic peptide-based motifs, including peptide amphiphiles, lipopeptides and conjugates with other organic and inorganic molecules, exemplify how design and chemistry can be successfully employed to generate multifunctional molecules that assemble predictably and interact with specific biological ligands. The recent emergence of the field of peptide biomimetics, which combines principles from disciplines including biology, chemistry and engineering, allows the preparation of synthetic materials with functions similar to or surpassing those of natural products20–22.
Bio-inspired peptides, especially short peptide building blocks, are minimal recognition modules used to mediate and facilitate processes of molecular recognition and self-assembly23,24. The synthesis and chemical modification of these building blocks are facile and they can assemble with remarkable efficiency into biocompatible and controllably biodegradable materials. Further, the extraordinary and often surprising chemical, physical and mechanical properties of these structures make them ideal for a wide range of applications, while opening new facets of molecular recognition, self-assembly and phase organization of these nanostructures.
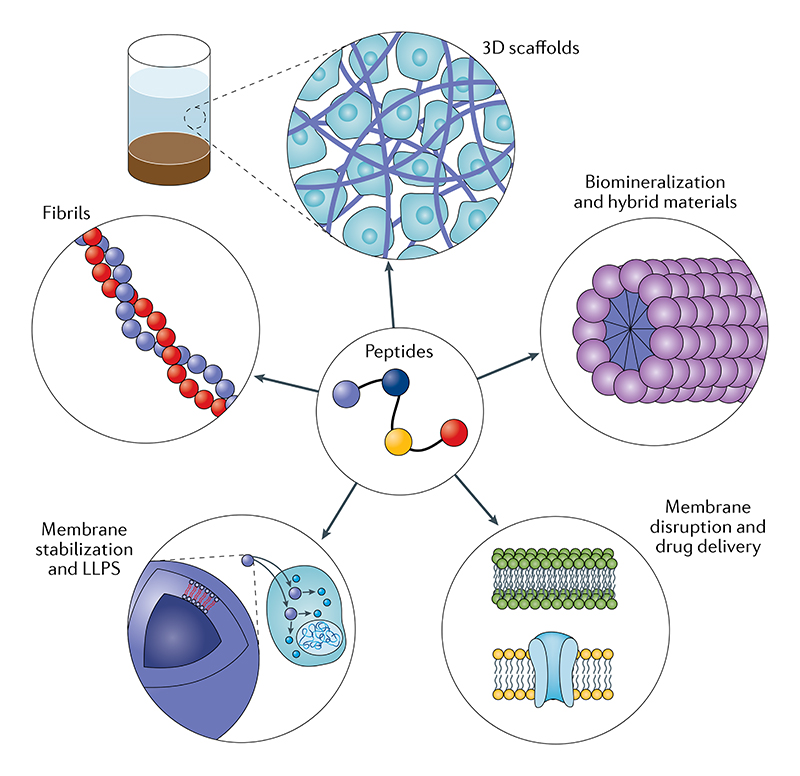
In this Review, we explore recent advances in biomimetic peptide self-assembly and discuss the properties and applications of the resulting materials, all the while comparing these to their more established protein-based counterparts25–27. We describe diverse applications of peptide-based systems and nanostructures, including surface modification, production of scaffolds for inorganic ultra-structures, generation of 3D hydrogel scaffolds for tissue engineering, drug delivery, production of new antimicrobial agents and active materials, and mimicking hierarchical self-assembly in protein-misfolding disorders (FIG. 1).
Fig. 1. Supramolecular chemical space accessible to biomimetic self-assembling peptides.
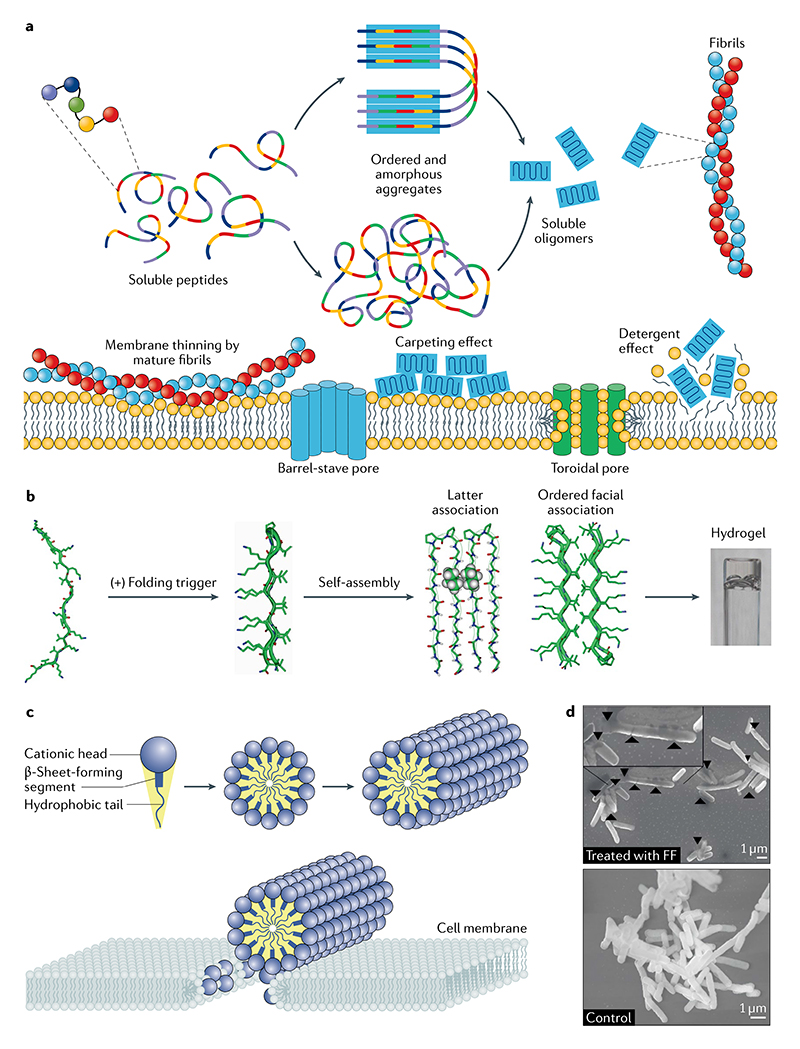
Chemicaiiy simple peptide sequences afford mechanistic understanding of molecular-level interactions in ordered supramolecular structures. Peptide building blocks have informed us about diverse phenomena, including the conversion of homogeneous solutions of peptide building blocks into discrete biomolecular condensates (liquid–liquid phase separation) and ordered fibrillar structures such as amyloid fibrils. A subset of peptides can assemble at interfaces to generate biomimetic membranes of artificial cells and organelles, while others disrupt the membranes of bacterial and cancer cells through pore formation, thus offering a wide range of therapeutic applications. The formation of ordered structures has given rise to the generation of biomimetic fibrils that can hierarchically assemble into complex structures, including 3D matrices used as scaffolds for cell growth and for forming organic–inorganic hybrid materials through incorporating peptide motifs known to be involved in biomineralization processes in nature. LLPS, liquid–liquid phase separation.
Peptide assembly into amyloid-like nanofibrils
Linear fibrils are common units from which supramolecular materials can be formed. Many of these β-sheet-rich units play functional or pathological roles in nature. This fibrillar self-assembly behaviour has been reproduced abiotically with diphenylalanine (FF), an archetypical model for elementary self-assembling units (BOX 1). Studying this di-homopeptide and its analogues (such as the tert-butoxycarbonyl (Boc) derivative) has generated key insights into the nucleation and oligomerization pathways, as well as the physical properties of the resulting amyloid and amyloid-like fibrils28,29. Furthermore, through their dynamic self-assembly behaviour, fibrils derived from FF have been used to generate forces of a similar order of magnitude to those of complex biological systems and synthetic polymers30.
Box 1. Amyloid-like peptide nanofibrils.
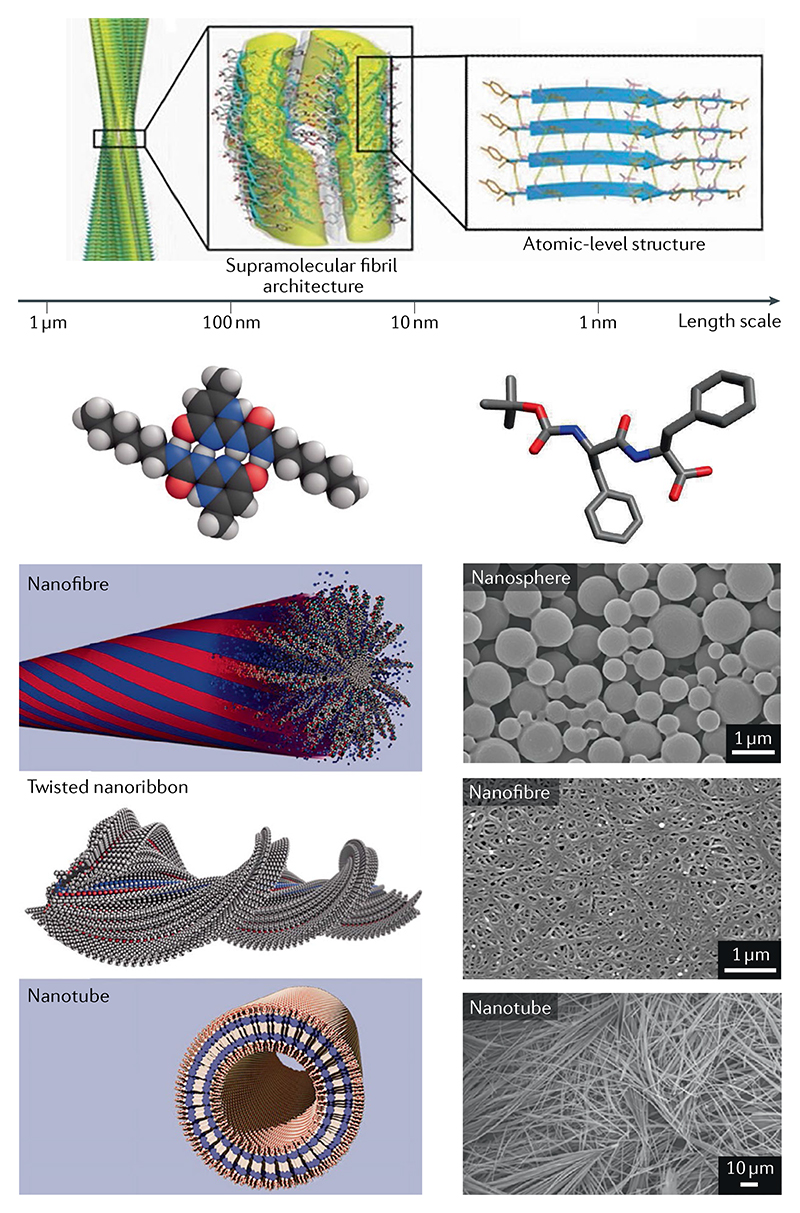
Linear assemblies of peptides and proteins serve as basic structural units for macroscopic materials in nature, such as collagen in skin and keratin in nails and hair. A particularly simple but common material forms when proteins or peptides assemble into β-sheets formed parallel to the fibril axis (see the figure, top) to give highly ordered H-bonded networks. The self-assembly of peptide systems into such ordered structures with supramolecular fibril architectures is commonly associated with the amyloid state of proteins linked to misfolding diseases in humans251. Artificial peptidic systems capable of such linear assembly can afford insights into the fundamental principles governing the formation of ordered structures through nucleation-dependent mechanisms. Self-assembly begins with primary nucleation252–254 and then growth of such structures by elongation and their replication by secondary nucleation on the surface of the initial fibrillar structures255. More recently, it has been found that, in addition to their pathological role in a range of human diseases, nature uses these structures as the basis for a diverse set of functional materials, including coatings and catalytic scaffolds27,256,257. The unique properties of fibrils composed of repeating sequences of short peptides are of considerable interest in nanotechnology and materials science, where they might serve as drug-delivery systems, tissue-engineering scaffolds, functionalized nanowires and bone-mimetic composites.
The propensity of short peptides to adopt amyloid-like structures can be enhanced by including features that promote aggregation in nature through hydrophobic and π–π interactions (see the figure, top27). Such interactions stabilize β-sheets involving one or more different components, as can be seen from the structures of specific aggregates, allowing them to form supramolecular systems structurally similar to amyloid fibrils (see the figure, bottom left5). Of the many peptides explored in this context, short peptides of 2–5 residues adopt stable fibrillar amyloid-like supramolecular structures. Thus, diphenylalanine fragments constituting the core of the Alzheimer disease β-amyloid polypeptides (Aβ) self-assemble into supramolecular systems and form nanotubes, nanospheres, nanofibrils and hydrogels (see scanning electron micrograph, bottom right28).
Top image adapted with permission from REF.27, Wiley. Bottom left image adapted with permission from REF.5, AAAS. Bottom right image adapted from REF.28, Springer Nature Limited.
Short peptides that assemble into amyloid-like supra-molecular structures can be used as drug-delivery systems either in the form of drugs that can themselves form fibrils31 or by conjugating the drug molecules to a sequence that forms β-sheets32. In this way, the monomeric drug units are slowly released as the ordered fibrils disassemble33. Another promising delivery strategy uses short peptides as gel matrices to encapsulate drug molecules and then enable their sustained release34,35. One such molecule is the dipeptide FΔF, which contains an α,β-dehydrophenylalanine (ΔF) residue and assembles into hydrogels consisting of a network of amyloid-like fibrils35 that traps and releases various structurally unrelated drug-like molecules. The potential of the FF motif in the formation of drug-releasing hydrogels has been further studied using the 9-fluorenylmethyloxycarbonyl (Fmoc)-protected monomer Fmoc-FF-konjac glucomannan (KGM)36. Here, self-assembly is driven by the Fmoc-FF motif, which forms peptide nanofibres interpenetrating and interwoven with KGM chains. This product has greater stability and mechanical strength than the hydrogel formed from Fmoc-FF alone.
Longer amino-acid sequences can exhibit higher complexity in their self-assembly behaviour. Thus, a peptide FFKLVFF, inspired by the amphiphilic KLVFF core section of β-amyloid (Aβ)(16–20) fibrils37, selfassembles in MeOH38. This process is likely to be influenced by interactions between the Ph sidechains of F residues, which are also responsible for its low solubility in H2O. This solubility can be increased by appending FFKLVFF with polyethylene glycol (PEG), whence cylindrical fibrils containing a peptide core and PEG corona can form in aqueous solution39. As longer PEG chains are used, hydration of these hydrophilic groups appears to influence self-assembly to a greater degree than the hydrophobic/aromatic stacking interactions of the F residues40,41.
Along with aromatic peptides, a class of tripeptides to hexapeptides with a characteristic sequential motif stimulate the process of fibre assembly and further condense to give amyloid fibrils42. These peptides consist of an aliphatic amino-acid tail capped by a polar head that self-assemble first into an α-helical intermediate before converting into cross-β amyloid fibrils. This class of aliphatic peptides have further been compared with natural amyloid core sequences, including Aβ, human amylin and calcitonin43. The designed aliphatic peptides self-assemble in a similar way to several natural sequences: through α-helical intermediates. Peptides containing the FF motif directly form β-sheet aggregates without going through α-helical intermediates.
In addition to the key role of the peptide sequence in driving self-assembly, the environment in which it takes place can also affect the final structure. Even small changes in humidity44 or O2 levels45 during assembly can have a distinct influence on the structures formed.
The self-assembly of FF dipeptides under various solution conditions has been particularly well explored. Although FF self-assembles into fibrillar structures in both H2O and MeOH, the crystal structures and properties of the products differ greatly46. The FF-NH2 peptide, which exists in its cationic ammonium form in solution, self-assembles into fibrillar structures that reversibly transition into spheres on dilution47. These spheres have been shown to facilitate nucleotide delivery48.
A particularly promising aspect of peptide self-assembly is our ability to control the formation of supramolecular structures by changing environmental conditions such as pH. For example, by adjusting the pH and concentration of aqueous peptide P11-4 (Ac-QQEFQWQFRQQ-NH2), one can manipulate an equilibrium between a nematic gel and an isotropic fluid phase49. This responsive behaviour is governed by the choice of amino-acid sidechains that enable hierarchical assembly of β-sheets through chemical and structural complementarity. Similarly, the role of electric charge in peptide self-assembly has been probed by designing and synthesizing the oppositely charged amyloid-inspired sequences Ac-EFFAAE-NH2 (AIP-1) and Ac-KFFAAK-NH2 (AIP-2), both of which self-assemble into amyloid-like nanofibrils at neutral pH50. Surfaces can also play a role in directing peptide self-assembly. Studies with QQEFQWQFRQQ (P11) conducted in the presence and absence of mica/highly oriented pyrolytic graphite substrates show differences in self-assembly kinetics and product morphologies50,51. The properties of the short peptides described in this section are summarized in TABLE 1.
Table 1. Examples of peptides that form amyloid-like fibrils and hydrogels.
| Peptide name | Number of residues | Associated protein/system | Self-assembled structure | Refs |
|---|---|---|---|---|
| Diphenylalanine (FF) | 2 | Aβ peptide | Peptide nanotubes | 28 |
| α,β-Dehydrophenylalanine (ΔF) | 2 | Aβ peptide | Hydrogel-forming fibrillar networks | 34,35 |
| Fmoc-FF-konjac glucomannan (KGM) | 2 | Aβ peptide | Fibrillar hydrogels | 36 |
| FF-NH2 | 2 | Aβ peptide | Reversible peptide nanotubes/spheres | 47,48 |
| Ac-EFFAAE-NH2 (AIP-1/2) | 6 | Aβ peptide | Amyloid fibrils | 50 |
| FFKLVFF | 7 | Aβ peptide | Amyloid fibrils | 37–41 |
| P11 (QQEFQWQFRQQ) | 11 | Aβ peptide | Amyloid antiparallel β-sheet tapes | 49,51 |
3D peptide matrices as cell-culture scaffolds
The biological extracellular matrix (ECM) serves as the main inspiration of engineered-tissue scaffolds that can support and sustain cells within a 3D matrix (BOX 2). Such biomimetic scaffolds enable cell binding and provide mechanical support by featuring a cell-adhesion peptide, a minimal amino-acid motif that promotes cell migration, differentiation and organization through the interactions of cells with the matrix52. Cell-adhesion peptides are key enablers of cell–matrix interactions, while the 3D nature of the material provides mechanical support for cell proliferation (FIG. 2a). Such peptide-based matrices must resist tensile forces acting on tissue and are, thus, required to mimic the properties of fibrillar assemblies of proteins such as collagens and glycosaminoglycans (GAGs). These supramolecular fibrillar networks must further be able to be deposited, remodelled and degraded as cells grow into ordered tissues, thus presenting additional challenges to their generation53.
Box 2. Natural and artificial extracellular matrices.
The extracellular matrix (ECM) is composed of proteins, carbohydrates and minerals, in combination with a wide variety of cell-adhesion molecules, including integrins, cadherins and transmembrane proteoglycans. The ECM provides external support to individual cells and facilitates interactions between cells, allowing their assembly and organization into functional tissue258. Depending on the nature of the tissue, differences in the composition and organization of the component proteins define its physical properties, such as elasticity, strength and influence on cell adhesion, all of which affects a cell’s ability to proliferate. Artificial cell-culture scaffolds and tissue-engineered constructs can enable improved cell viability and proliferation by mimicking the physicochemical conditions of the ECM. Self-assembly through non-covalent crosslinking can afford mouldable and injectable hydrogels as cell scaffolds. This approach has, however, so far, largely used polymers such as alginate259, poly(ethylene glycol)260 and poly(glycerol sebacate)261 in combination with a range of nanoparticles for controlled drug-release applications262. Common biological scaffolds include peptide-based and protein-based biopolymers, either in their natural forms, such as collagen, fibronectin and silk, or in related synthetic materials that can be used in various cell-culture technologies. Indeed, biomimetic materials are finding increasing appeal in biomedical applications owing to their ability to recapitulate the ECM both in architecture and in the capacity for cell signalling. In the case of self-assembled protein matrices, 3D fibrillar networks exhibit the potential to create scaffolds in tissue engineering. One notable commercially available macroscaffold is the Matrigel matrix263, which is produced from several proteins such as laminin, collagen IV and entactin, in combination with other growth factors and enzymes.
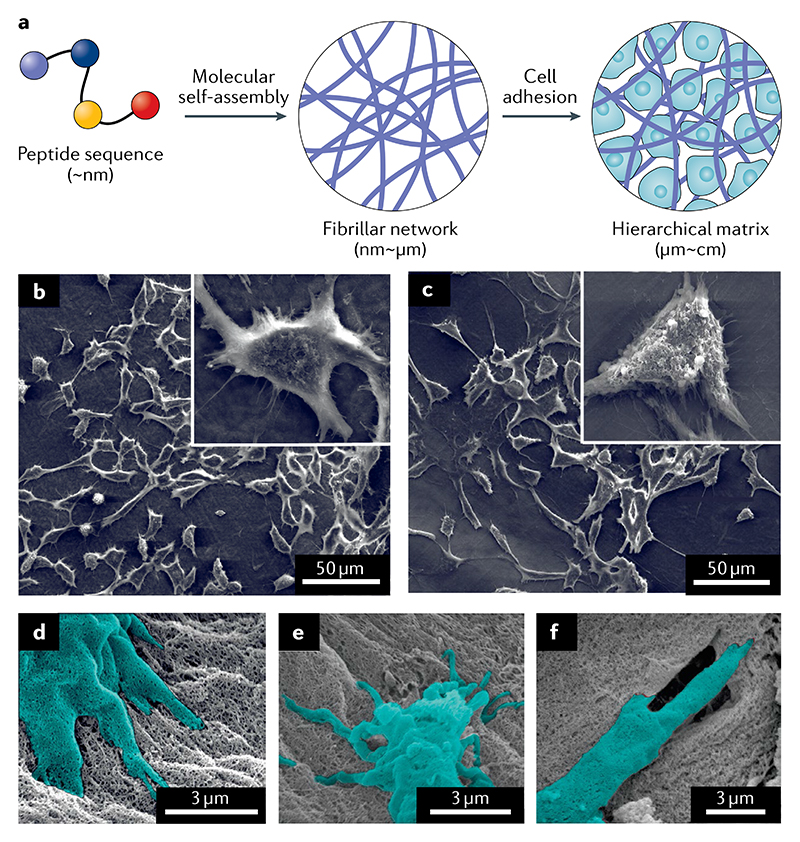
Fig. 2. Biomimetic supramolecular peptide scaffolds enable cell adhesion and proliferation.
a | Peptides can seif-assembie into biomimetic matrices that act as scaffolds to generate cell cultures. b,c | Scanning electron micrographs depict osteogenic cell viability and morphology when grown in glycosaminoglycan-mimetic peptide nanofibrils that promote biomineralization (scale bars represent 50 μm)56. b | Cells grown on sulfonated-peptide-amphiphile fibrils mimicking glycosaminoglycan sulfate. c | Cell proliferation is reduced when lauryl-VVAGE (E-PA) fibrils bearing carboxylate groups are used. This material mimics non-sulfated glycosaminoglycans. d | The cells, falsely coloured here in cyan, adhere to the self-assembled peptide nanofibrils61. e | The biocompatibility is evident from the cells extending into the peptide matrix. f | The cells can also remodel the matrix to best suit them. Parts b and c adapted with permission from REF.56, Elsevier. Parts d-f adapted with permission from REF.61, Elsevier.
The need for both biocompatibility and structural stability has motivated two decades of investigations into polypeptide matrices and gels that allow cell proliferation24,25. However, the self-assembly of short peptides can afford more diverse scaffolds that may offer optimal environments for different cell types. Control of composition, scaffold porosity and rigidity, along with the incorporation of growth factors, have now allowed further advances in cell-culture viability and improved tissue regeneration. Furthermore, self-assembled β-sheet matrices are stable across wide temperature and pH ranges and can resist high concentrations of denaturing agents, such as urea and guanidium hydrochloride19,24.
One route to robust hydrogels uses structurally well-defined peptides coupled to carbohydrate moieties54. These can be prepared through in vitro peptide glycosylation reactions, which enable systematic modifications to produce supramolecular hydrogels with diverse self-assembly behaviours. The glycopeptide-derived gels exhibit greater thermostability and biostability relative to the parent peptide gels55. In this way, the glycopeptidederived gels can have high H2O content and similar structural morphology and composition to the ECM in tissue, all the while exhibiting great potential as new biomimetic scaffolds for mammalian cell growth56 (FIG. 2b,c).
The major component of the ECM is collagen, whose multiscale hierarchical self-assembly we wish to replicate because of its potential biomedical applications in tissue engineering. Although many approaches to mimicking collagen self-assembly with synthetic peptides exist, until recently, none of these systems has simultaneously demonstrated all the different levels of structural assembly. This issue has been resolved using a peptide featuring collagen’s characteristic Pro-Hyp-Gly repeating unit, as well as salt bridges and H-bonds between Lys and Asp residues57, which can assemble into hierarchical nanofibres of several hundred nanometres in length with characteristic triple-helical packing58.
Amyloid-like peptide fibrils have recently been used to generate nanoscale biomaterials promoting cell adhesion and differentiation in vitro. The well-established cell-adhesion motif Arg-Gly-Asp (RGD) can be conjugated to an 11-residue peptide corresponding to residues 105–115 of the amyloidogenic protein transthyretin (TTR1) to promote specific cell–fibril interactions59. Similarly, the hen egg-white lysozyme peptide containing the tripeptide DGR, which is analogous to the integrin-binding RGD sequence, self-assembles into fibrillar networks that communicate force and signals between the ECM and cells60. More recently, other synthetic β-sheet-containing fibrous meshes have been shown to promote cell adhesion and proliferation61. In a similar manner, Fmoc-protected α-synuclein62 and β-amyloid-derived short peptides63 self-assemble into hydrogels composed of nanofibrils that promote stem cell adhesion and differentiation. These results strongly suggest that functionalized amyloid-derived fibrils have real potential as components in novel biomimetic materials or as tools to probe and exploit fundamental biological processes and cell behaviour.
As with the glycopeptides described above, incorporating peptide amphiphiles into hydrogel-forming networks can both promote cell viability and allow release of growth factors, making the networks useful for therapeutic applications (FIG. 2d–f). Nanofibrous matrices composed of two different self-assembling peptide amphiphiles have been designed as a coating for cardiovascular implants64. The nanofibrous matrix exhibits initial adhesion and proliferation of endothelial cells, while limiting the proliferation of smooth muscle cells and the adhesion of platelets. These characteristics are essential in promoting re-endothelialization, thus increasing the potential of this matrix for cardiovascular applications. Similarly, a nanofibrous network prepared from a heparin-mimetic peptide amphiphile (HM-PA) is a promising platform for pancreatic islet transplantation as a potential treatment for type 1 diabetes65.
In related work, a biomimetic peptide amphiphile derived from the extracellular glycoprotein tenascin C promotes neurite outgrowth66 by self-assembling into highly aligned supramolecular nanofibrils. Such peptide amphiphiles also increase the length and number of neurites extending from neurons differentiated from encapsulated cells. These bioactive gels could serve as artificial matrices that are delivered to regions of neuronal loss to guide neural stem cells and promote, through biochemical cues, neurite extension after differentiation. More recently, peptide-amphiphile–DNA conjugates have been shown to reversibly self-assemble into hydrogels67. By controlling this dynamic supramolecular system’s stiffness, changes in the architecture of the fibrous hydrogel networks can modulate important phenotypic transformations of neural cells in contact with these materials.
FF-containing short peptides have led to promising results in tissue engineering when incorporated with the RGD motif to facilitate cell growth and proliferation68. More complex cultures with multiple cell lines have further been studied with a scaffold assembled from the longer peptide Ac-ILVAGK-NH2 (REF.69). Incubated on this scaffold, human H1 embryonic stem cells proliferate into 3D spheroids while continuing to express various pluripotent nuclear transcription factors and surface biomarkers. Furthermore, multicellular constructs with human umbilical-vein endothelial cells, fibroblasts and keratinocytes can be used as a skin model.
The propensity of short Fmoc-protected peptides to produce rigid biocompatible gels has been studied in detail with varying degrees of success70–72. The two dipeptides Fmoc-3F-Phe-Arg and Fmoc-3F-Phe-Asp co-assemble into nanofibril hydrogels. The display of Arg and Asp residues at the nanofibril surface effectively mimics the integrin-binding RGD peptide of fibronectin without the need for covalent interactions, thereby supporting the viability and growth of fibroblasts73. This system forms a gel remarkably quickly and promotes adhesion of fibroblasts through specific RGD–integrin binding, thus providing a model 3D scaffold enabling culturing with anchor points for cell spreading and proliferation74.
Artificial scaffolds, even those based on biopolymers, can still sometimes offer only a suboptimal adhesion and proliferation environment for all cell types. The scaffold needs to exhibit the necessary physicochemical properties — porosity, rigidity and elasticity — at the composition required to promote the viability of specific cells. Peptide-based scaffolds can mimic microenvironments in the ECM to organize cells into different types of tissue. There is growing interest in minimal self-assembled peptides and amino acids because they can afford hydrogel networks for tissue engineering and surgical applications owing to their ability to undergo controlled sol–gel transitions, making them ideal injectable materials75. Indeed, we described above how the hierarchical self-assembly of basic peptide building blocks into final β-sheet-rich matrices affords 3D hydrogels with a fibrillar network that serves as a scaffold for cell growth. We now end our discussion on ECM models (TABLE 2) and describe the use of peptides to stabilize interfaces.
Table 2. 3D peptidic matrices can allow cell adherence, growth and proliferation.
| Peptide name | Number of residues | Associated protein/ system | Self-assembled structure | Refs |
|---|---|---|---|---|
| Fmoc-3F-Phe-Arg Fmoc-3F-Phe-Asp | 2 | Fibronectin | Nanofibrillar hydrogels | 70–72 |
| P1–P8 | 2–3 | β-Amyloid polypeptide | Nanofibre gels | 65 |
| A1–A7 | 5 | α-Synuclein | Nanofibre gels | 62 |
| Diphenylalanine-RGD | 5 | β-Amyloid polypeptide | Nanofibrillar matrix | 68 |
| Ac-ILVAGK-NH2 | 6 | Lys-containing peptide | Nanofibrillar hydrogels | 69 |
| HM-PA | 7 | Heparin | Nanofibre gels | 65 |
| TTR1-cycloRGDfK | 11 | Transthyretin | Nanofibrillar matrix | 59 |
| PA-YIGSR | 13 | Endothelial cell-adhesive ligand | Nanofibrillar matrix | 64 |
| EAK16 (Ala-Glu-Ala-Glu-Ala-Lys-Ala-Lys)2 | 16 | Zuotin | β-Sheet-containing membranes | 23,24 |
| (Pro-Lys-Gly)4(Pro-Hyp-Gly)4(Asp-Hyp-Gly)4 | 36 | Collagen | Triple-helix fibrils | 57,58 |
Peptides and their assemblies stabilize interfaces
A biological membrane composed of a lipid bilayer acts as a barrier to separate and protect a cell and its components from extracellular conditions and components, including ions, metabolites and pathogens. Along with the lipids forming the interface between the intracellular and extracellular environments, specific proteins are incorporated into the membrane, thus controlling permeability and interactions between the cell and its environment. These proteins manage a wide range of biological processes, such as active transport, signalling and energy dissipation, thereby allowing for controlled compartmentalization, which contributes to the proper function of their cellular machinery.
Over the past few years, new approaches to mimic cell surfaces have emerged, in part motivated by the prospect of biocompatible and bioactive drug-delivery systems, as well as for directed targeting (FIG. 3a,b). For example, self-assembling surfactant-like peptides are new alternatives to synthetic surfactants obtained from petrochemical sources76. Other applications of surfactant-like peptides stem from their antimicrobial activity based on micellar concentration and balanced amphiphilicity, consistent with their propensity for self-assembly and membrane lysis77 (FIG. 3c). Furthermore, these peptides can self-assemble at fluid interfaces to give cohesive films that stabilize foams and emulsions in applications where renewability, biocompatibility or added functionality may be desired. Sinapultide is the HOAc salt of KLLLLKLLLLKLLLLKLLLLK (KL4) and represents the first peptide-based replacement for the human lung surfactant protein B in pulmonary surfactant therapies approved for clinical use78. The penta-residue repeat of KL4 leads to adaptive peptide helicity and variation with partitioning depth, and its effectiveness suggests that structural plasticity may represent an important mechanism for differential lipid trafficking at air–H2O interfaces (FIG. 3d). More recently, a minimalistic approach to the design and synthesis of rigid helical peptides has afforded materials with the highest long-term stability among known peptide-based emulsifiers79. These peptide emulsifiers are composed of seven residues that mimic the rigid conformation of hydrophobins to afford stable oil–H2O emulsions80, the viscoelasticity of which can be high at relatively high peptide concentrations.
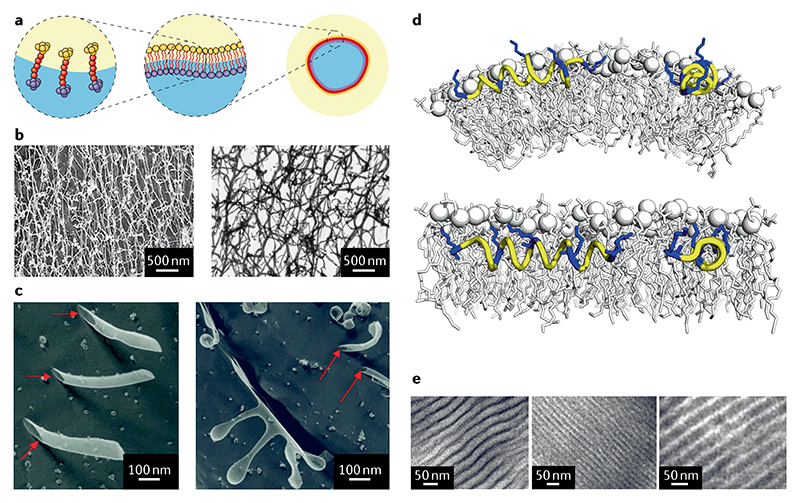
Fig. 3. Self-assembly of membrane and surfactant-like peptides at interfaces.
a | A peptide self-assembly can stabilize a liquid-liquid interface. b | Transmission electron micrographs of the surfactant peptides A6D (left) and V6D (right), which form a dense network several micrometres long77. c | On a smaller scale, these materials form open-ended tubes (left), micelles and spherical vesicles budding off the nanotubes in H2O (right). d | KL4 models built using backbone torsion angle restraints from solid-state NMR data. KL4 conformer from measurements with two different lipids, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) and 1,2-dipalmitoyl-sn-glycero-3-phosphocholine (DPPC) (top and bottom, correspondingly)78. e | Transmission electron micrographs of diblock copolypeptide-surfactant complexes, indicating a lamellar order of periodicity82. Parts b and c adapted with permission from REF.77, PNAS. Part d adapted with permission from REF.78, Elsevier. Part e adapted with permission from REF.81, American Chemical Society.
Related to our discussion on surfactants is the recent development of polymeric systems, not least amphiphilic block copolymers, that mimic biological membranes81 (FIG. 3e). Thus, copolymerization of natural and modified N-carboxy anhydrides alone or coupled with synthetic monomers enables the synthesis of an almost unlimited number of supramolecular structures82. In particular, separate studies considered how poly(Glu)83 and poly(Leu)84 diblock copolypeptides self-assemble in aqueous solution into vesicles known as peptosomes. In these systems, the hydrophilic block can form a well-defined α-helix whose hydrodynamic radius can be modified through varying the solution pH.
Bacterial lipopeptides are cyclic peptides containing a single fatty acyl chain. Such lipopeptides are secreted into growth media by a number of different microorganisms and are thought to play a role in bacterial swarming motility on semi-solid surfaces, as well as in the formation of structured biofilms on solid surfaces85. Lipopeptide amphiphiles are an important class of biomimetic surfactants readily synthesized from commercially available organics such as natural fatty and amino acids. In many cases, these amphiphiles can increase the rigidity of not only common organic solvents but also waxes, H2O and ionic liquids and can, thus, form hydrogels86.
Aside from the polymeric peptides described above, amphiphilic behaviour is also observed for short sequences in which a head group features charged residues and the tail group neutral ones. These surfactants are facially amphiphilic molecules that self-assemble at fluid interfaces to give cohesive films that stabilize foams and emulsions. Hydrophobic interactions between the amphiphilic peptides, along with interstrand H-bonds, are the main driving forces for self-assembly80. These interactions afford high-aspect-ratio structures such as ribbons, nanotubes, nanofibres and nanorods. Yet, a change in solution conditions can destabilize the interfacial film, leading to rapid foam or emulsion collapse80. Surfactant-like peptides composed of Ala residues as the tail group tend to form the most stable structures because it engages in very strong hydrophobic interactions87. The self-assembly of a cationic peptide A6R that consists of six consecutive hydrophobic Ala residues as a tail group with a cationic Arg head group affords ultra-thin sheets at low concentrations. At higher concentrations, the sheets first form helical ribbons that mature into nanotubes with an antiparallel arrangement of β-sheets that minimizes electrostatic repulsion between the Arg head groups88. By contrast, the oligopeptide A12R2 is twice as long and, instead, self-assembles into twisted fibres89. A similar system, A6K, forms lipid-like peptide nanovesicles, enabling drug delivery90. Furthermore, a simple amphiphilic decapeptide, with a phosphorylated Ser head located within a β-hairpin segment and linked to two hydrophobic tails, has recently been described91. This phospholipid-inspired peptide self-assembles into semi-elliptical nanosheets incorporating the FF motif, known to facilitate self-assembly and structure stability, as well as a β-hairpin for forming a hydrophilic phosphorylated head. The resulting bilayer crystal structure features interactions along all three axes: aromatic π–π interactions, H-bonding and β-sheet formation92. Thus, this demonstrates the capacity of peptides to mimic self-assembly in nature and gives us more information to help predict the intermolecular interactions in future oligopeptide designs.
Biomimetic peptides have yet to be widely used as membranes and surfactants thus far, but recent developments may facilitate the incorporation of these molecules into industrial and consumer products in the near future. This approach has recently allowed the conjugation of peptides onto stem cell membranes without affecting cell viability, proliferation or multipotency93. The systematic exploration of synthetic, genetically engineered peptides produced by conventional methodologies may afford a class of biomolecules that are superior to polymer-based materials.
Self-assembled peptide antimicrobial agents
In the previous section, we discussed several mechanisms by which peptide-based assemblies self-organize at surfaces and stabilize interfaces. However, the phenomenon of peptide self-assembly and the resulting structures can also destabilize interfaces, including those forming biological membranes. This has increasingly been explored in the context of the development of new antimicrobial agents to combat the rise of multidrug-resistant bacteria94. Antimicrobial peptides (AMPs), a growing class of natural and synthetic peptides active towards a large spectrum of microorganisms, provide a potential source of such agents95–97.
Endogenous AMPs represent the innate immune system’s first line of defence against pathogenic microorganisms. Produced by organisms found among all classes of life98,99, such peptides comprise a unique and diverse group of molecules formed by sequences generally shorter than 50 amino acids, sharing a net positive charge and containing a high fraction of hydrophobic residues100,101. This amino-acid sequence contributes to the amphipathicity and cationic nature of AMPs that allow them to partition into the anionic bacterial lipid bilayer membranes. This important feature of AMPs can enable membrane permeation, depolarization and destabilization100–102 (FIG. 4a). This characteristic mechanism of action, mediated through non-membrane-dependent mechanisms, enables AMPs to avoid the common resistance mechanisms observed for classical antibiotics103,104.
Fig. 4. Self-assembling biomimetic-peptide-based antimicrobial nanostructures.
a | Different peptide-membrane interactions are proposed to give rise to antibacterial functions. b | The MAX1 peptide undergoes environmentally triggered folding, selfassembly and non-covalent fibril-crosslinking processes to give a hydrogel114. c | The supramolecular nanofibres formed by self-assembling peptide amphiphiles present cationic peptide sequences that are essential to their proposed mode of action140. d | Scanning electron micrographs of Escherichia coli with and without diphenylalanine. This dipeptide forms nanostructures that have clear effects on bacterial morphology141. FF, diphenylalanine. Part b adapted from REF.113, Springer Nature Limited. Part c adapted with permission from REF.140, American Chemical Society. Part d adapted from REF.141, CC BY 4.0.
The development of natural AMPs into therapeutically relevant antibiotics has suffered from several problems, including their susceptibility to proteolysis, reduced efficacy, relatively high expense of manufacturing and limited tissue distribution and cell selectivity. Great strides have been made to overcome these limitations, by both rational and computer-aided design of enhanced functional biomimetics of the peptide sequences, which range from the optimization of natural amino acids to the development of synthetic mimics105–108. Because the interaction of AMPs with bacterial membranes depends primarily on the physicochemical properties of the peptides, and in particular the ordered structures formed on their self-assembly rather than their specific amino-acid sequences, many of these biomimetic sequences are much simpler than the innate AMPs evolved in nature105.
Biomimetic AMPs have been developed to harness self-organization to form hydrogels and nanostructures with intrinsic antimicrobial properties. The assembly process introduces relevant physicochemical features that are mostly absent from natural antibiotics. Indeed, one can readily modify the peptide sequence to tune the interactions between building blocks and the resulting supramolecular assemblies. Along with their antimicrobial functionalities, the resulting hydrogels and nanostructures can be highly dynamic and can demonstrate a wide range of structural properties, such as stimuli-responsiveness, improved stability and selectivity, injectability and sustained drug release109–111.
Antimicrobial hydrogels are formed by self-assembly of peptide building blocks on exposure to environmental stimuli such as changes in pH and the ionic composition of the surrounding solution. This induces interactions between the hydrophobic residues not commonly exposed to the environment, allowing for antimicrobial activity when it is most needed. One of the most prominent families of supramolecular macroscopic entities are the MAX peptides, which fold into an amphiphilic β-hairpin conformation to give hydrogels composed of fibril networks112–118 (FIG. 4b). These hydrogels can be used as coatings and/or injectable agents, assemble on specific external stimuli and exhibit antibacterial activity against multidrug-resistant Gram-positive and Gram-negative bacteria by disrupting inner and outer membranes. Additional self-assembling antimicrobial hydrogels include variants of the KLD-12 self-assembling peptide that enable rapid fracture healing and antimicrobial activity119. Further, naphthalene-based or Fmoc-based ultrashort peptide gelators display broad-spectrum antimicrobial activity due to the electrostatic interactions between the hydrogel and the anionic bacterial membrane120–122. The intrabacterial enzymatic triggering of self-assembly and subsequent hydrogelation of peptide amphiphiles also afford growth-inhibiting hydrogels in Escherichia coli123,124. A synergistic enhancement of the antibacterial activity of self-assembling hydrogels has also been achieved by incorporating classical antimicrobial agents125 and metals126,127 in these gels, with many additional strategies explored for the use of self-assembling antimicrobial-mimetic peptide-based hydrogels128,129.
Cyclic self-assembling AMPs are among the first examples of non-hydrogel-forming self-assembling antimicrobial functional structures. These antimicrobial agents were first introduced in the development of cyclic D,L-α-peptides exhibiting proteolytic stability and rapid nanotube formation in lipid membranes. These agents cause bacterial cell death and display potent activity against a wide range of bacteria and exhibit a near order-of-magnitude increase in antibacterial activity compared with their linear peptide counterparts130. Parameters such as the size and sequence of the peptides are important, with the octameric peptides generally displaying higher antimicrobial potency than their hexameric counterparts130. This strategy has been further expanded to antiviral cyclic D,L-α-peptides131,132 that are substantially less toxic to mammalian cells, yet maintain potent activities against multidrug-resistant bacteria. Cyclic lipodepsipeptides, as well as additional cyclic-peptide-based moieties, have been similarly developed and possess advanced antibacterial and antibiofilm activities133–135.
Although many different core–shell nanoparticles have been used as vehicles for drug delivery, those derived from the self-assembly of amphiphilic peptides further demonstrate strong antimicrobial properties against a broad spectrum of bacteria, yeast and fungi in vitro and in vivo136,137. These self-assembled nanoparticles are more potent than their free-peptide counterparts and have a high therapeutic effect in abolishing Staphylococcus aureus infections in mice while presenting reduced cytotoxicity. Furthermore, the peptide nanoparticles can cross the blood–brain barrier to suppress bacterial growth in S. aureus-infected brains of meningitis rabbits and suppress yeast growth136,137. Importantly, the nanostructures do not interfere with the balance of electrolytes in the blood or cause substantial damage to the liver and kidney functions.
Additional developments in self-assembling antimicrobial mimetics have been achieved in the design of antimicrobial lipopolypeptides and lipidomimetic peptides. Conjugating palmitic acid to the N terminus of very short cationic dipeptides and tripeptides composed of all L-amino acids and D,L-amino acids affords a diverse range of morphologically distinct potent antimicrobial agents in vitro and in vivo138. Success has also been had with amphiphilic self-assembling antimicrobial lipidomimetics based on peptides comprised of consecutive hydrophobic Ala residues linked to a hydrophilic charged Lys head group. There is a strong correlation between the propensity of the peptides to self-assemble, their membrane-penetration capabilities and their antimicrobial activity139.
Peptide-based nanofibres and nanorods have recently been developed as antimicrobial agents. Indeed, peptide amphiphiles featuring cationic peptide sequences can self-assemble into nanofibres to affect a broad spectrum of bacteria. These nanofibres have significantly higher antibacterial activities than those of soluble peptide molecules with identical sequences140 (FIG. 4c). Nanofibres and nanorods with substantial antibacterial activity can be generated from simple sequences, and, indeed, it is not complexity but, rather, the propensity to self-assemble that is most important. For example, FF forms nanostructures and has emerged as a minimal model for self-assembling, membrane-active AMPs141 (FIG. 4d). Similarly, truncated nanofibre-forming versions of natural self-assembling peptides also have impressive antibacterial capabilities142. The peptides described in our discussion are collated in TABLE 3.
Table 3. Peptides associated with antibacterial and anticancer activity through membrane disruption.
| Peptide | Number of residues | Associated protein/system | Self-assembled structure | Refs |
|---|---|---|---|---|
| FF | 2 | β-Amyloid polypeptide | β-Sheet-containing nanofibres | 141 |
| Cyclic D,L-α-peptides | 6–8 | Synthetic | Supramolecular peptide nanotubes | 131,132,135 |
| KLD | 12 | Synthetic | β-Sheet-containing nanofibres | 118 |
| PTP-7b | 13 | Synthetic | β-Sheet-containing nanofibres | 158 |
| (KLAKLAK)2 | 14 | Synthetic | α-Helix | 159–161 |
| MAX | 20 | Synthetic | β-Hairpin hydrogels | 112–118 |
Peptide assemblies in cancer diagnosis and therapy
The membranes of cancer cells can, in many cases, be enriched in anionic components in much the same way as bacterial outer membranes143–145. These anionic moieties include phosphatidylserines, GAGs and glycoproteins. Thus, the cationic and amphipathic features of peptides useful against bacteria sees them selectively bind cancerous cells through electrostatic interactions and effect cytotoxicity. Indeed, several AMP mimetics have been recognized as novel targeted cancer therapeutics because of their ability to disrupt cellular and organelle membranes143–145.
The majority of antitumour peptide therapeutics act in their monomeric form, yet their bioavailability and stability are often limited. Self-assembling peptide nanostructures show greater durability under physiological conditions. The ability of the monomeric peptides to adhere to and disrupt cancer cell membranes while undergoing controlled dissociation into monomeric subunits allows them to avoid unfavourable pharmacokinetic parameters that limit therapeutic efficacy and clinical translation146,147. Other strategies use self-assembled peptidic nanostructures to target cancer cells by binding receptors on cell surfaces148 and exposing specific epitopes related to cancer cells and angiogenesis inhibition149. An additional important application for such self-assembling peptide nanostructures is their use in drug delivery, in which they are able to penetrate cell membranes and deposit their cargo intracellularly. Thus, the release of Boc-FF spheres through an oil–H2O interface exemplifies how colloidal particles can encapsulate small hydrophobic and hydrophilic molecules, such as rhodamine and fluorescein, and transfer them through interfaces in a jet-like manner150. The above properties have seen peptide-based microcapsules and ordered structures recently find use in gene delivery for immunomodulation151,152 and chemotherapeutic agents153–158.
The examples we have described showcase peptide-based self-assembled nanostructures in a variety of therapeutic strategies. These developments have motivated many researchers to employ self-assembling nanostructures that themselves have specific membranedisruption properties, rather than having to find both a delivery agent and a bioactive species, or simply using soluble monomeric peptides. Recently, the peptide (KLAKLAK)2, known for its antitumour properties in its monomeric form, has been combined with elastin-like polypeptide (ELP) and the AP1 peptide to give polymer nanoparticles that target interleukin-4 receptors159. The polymer nanoparticles form at physiological temperatures while stabilizing their helical conformations, leading to membrane disruption of cancerous cells selectively. Similarly, combinations of hyaluronic acid and (KLAKLAK)2 peptide amphiphiles self-assemble into robust hybrid membranes to produce surface-bound cytotoxic agents or act as reservoirs for sustained release of such agents while avoiding their enzymatic degradation160. Furthermore, a different strategy has enabled (KLAKLAK)2 to assemble into nanoparticles that can be internalized and accumulate within cells. In this way, there is a 400-fold increase in the peptide’s antitumour activity, as the nanoparticles enable efficient disruption of mitochondrial membranes, causing excessive production of reactive oxygen species in cells156. Another strategy exploiting the specific properties of emerging self-assembled building blocks uses the peptide FLGALFKALSHLL (commonly denoted PTP-7b), which undergoes concentration-dependent self-assembly on cell surfaces157. Following self-assembly into exosome-like aggregates at specific locations on cell membranes, PTP-7b induces cell-tissue damage through cell lysis. This occurs because the assemblies can extract lipids from cell membranes and transport them into the cytoplasm.
We have described how self-assembled peptides can have anticancer effects on their own, but they can also show effects when triggered by external stimuli. Thus, short peptide sequences such as the FF motif can form ordered structures for photodynamic161 and photothermal162,163 therapies, either on their own or when conjugated to active chromophores such as porphyrins and metal ions. Moreover, such peptide–metal-ion assemblies allow the development of new cancer-cellimaging techniques. For example, the red shift observed in the yellow fluorescent protein, which results from π–π stacking, inspired the assembly of the Trp-Phe dipeptide into emissive nanoparticles164. These nanoparticles can be further functionalized with the MUC1 aptamer and doxorubicin payload, and the entire system can target cancer cells and image drug release in real time. These results exemplify the therapeutic possibilities emerging from peptide-based nanostructures. By harnessing the properties of these ordered self-assembling nanostructures, we can envisage novel anticancer and antibacterial mechanisms that allow for enhanced stability and cell selectivity of bioactive peptides for wide biomedical applications.
Peptides in liquid–liquid phase separation
Membrane-bound compartments provide spatial control over the localization of biomolecules in living cells. However, it has recently become apparent that many biomolecules can also spontaneously form spatially well-defined biological compartments as a result of liquid–liquid phase separation (LLPS, FIG. 5a), also referred to as coacervation or liquid-phase condensation165–167. This phase transition involves the demixing of proteins, RNA and other biomolecules from a homogeneous solution within the cytoplasm of a cell into dense, soft, colloidal liquid droplets that coexist as membraneless organelles or biomolecular condensates168 with the dilute phase in the cytoplasm. Liquid–liquid and liquid-solid phase transitions of such proteinaceous condensates are increasingly recognized to be at the heart of both biological function and malfunction169,170, motivating efforts towards understanding the physical principles that define these transitions in a biological context171,172,173 (FIG. 5b).
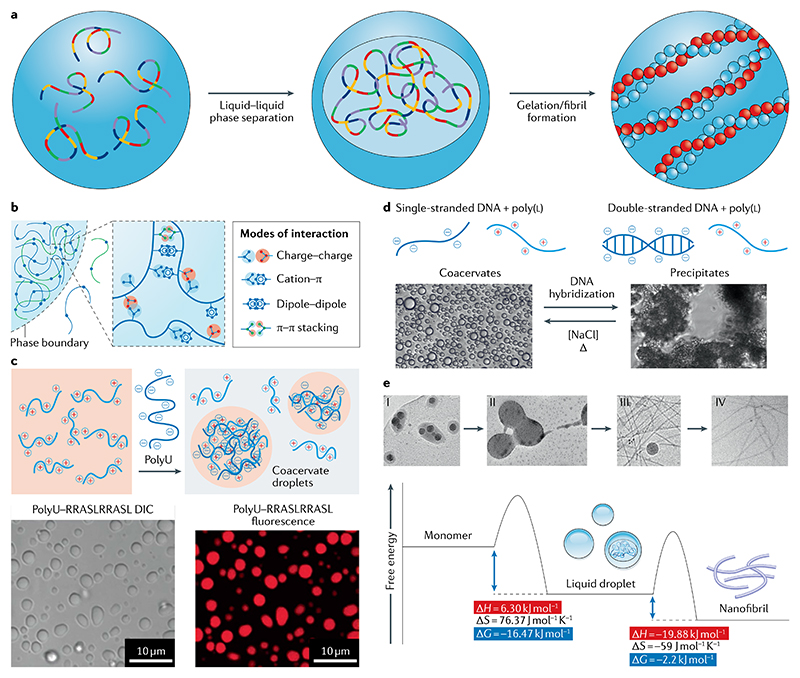
Fig. 5. Mechanisms of liquid–liquid phase separation and condensation.
a | A homogeneous peptide solution can undergo liquid–liquid phase separation (LLPS) to give metastable condensates. These, in turn, can undergo a phase transition to form thermodynamically favoured solid fibrils. b | LLPS involves several weak forces, including electrostatic, cation–π, dipole–dipole and π–π interactions171. c | Treating a solution of peptide RRASLRRASL with polyU RNA leads to complex coacervation on account of electrostatic forces, among other interactions197 (top). Bright-field (bottom left) and fluorescence (bottom right) images highlight aggregation into coacervate phase droplets. d | Schematics and bright-field-microscopy images presenting the effect of oligonucleotide hybridization, ion concentration and temperature on LLPS of poly(Lys) peptides199. e | Transmission electron micrographs of Fmoc-Ala undergoing LLPS and phase transition to form increasingly organized structures. The transition from the kinetically trapped nucleation precursors to the nanofibrils is accompanied by a decrease in Gibbs free energy201. DIC, differential interference contrast. Part b adapted with permission from REF.171, Elsevier. Part c adapted from REF.197, Springer Nature Limited. Part d adapted with permission from REF.199, American Chemical Society. Part e adapted with permission from REF.201, Wiley.
The majority of LLPS phenomena in cells have been attributed to the complex interactions between intrinsically disordered proteins themselves and other molecular species, such as RNA molecules174–176, which affords biomolecular condensates with liquid-like properties and membraneless organelles177. In a biophysical context, the structures formed through LLPS are of particular interest as they, despite not being enclosed by a membrane, have persistent sizes and shapes, even though the molecular building blocks exhibit dynamic exchange over timescales of minutes178. Moreover, the formation of such responsive condensate structures, either through precise control of protein mixing in bulk solution179 or using microfluidic approaches to generate condensates from Gly-rich RGG domain peptides180, has given rise to a wide range of materials science applications181,182. The formation of such synthetic organelles allows one to further generate confined membraneless organelles by combining proteins and mRNA to perform orthogonal translation of desired sequences to introduce new chemical functionalities into mammalian cells in a site-specific manner183. Yet, the study of the protein–RNA interactions leading to such phenomena remains challenging owing to the high diversity and sequence complexity of these biologically relevant building blocks. As such, using simpler short peptide building blocks as collated in TABLE 4 can help us to more easily explore the chemical and physical determinants leading to LLPS.
Table 4. Peptides template the formation of organic–inorganic hybrid materials.
| Peptide name | Number of residues | Associated protein/system | Self-assembled structure | Ref. |
|---|---|---|---|---|
| Fmoc-DOPA-DOPA | 2 | Synthetic | Nanofibrils | 221 |
| Fmoc-FFECG | 5 | Synthetic | Nanofibres | 212 |
| SO3-PA | 7 | Glycosaminoglycan | Nanofibrillar network | 56 |
| Surfactin | 7 | Synthetic | Surface coating | 216 |
| PA | 9 | Synthetic | β-Sheet-containing nanofibrils | 208 |
| A10H6 | 16 | Synthetic | β-Sheet-containing nanofibrils | 219 |
| R5 | 19 | Silaffin-1A1 | Micelle-like assemblies | 209 |
| Acidic/basic Leu zipper-like peptide | 36 | Synthetic | Left-handed coiled-coil structure | 220 |
| LRAP | 64 | Amelogenin | Nanofibrillar bundles | 206 |
Of key importance to LLPS is the presence of low-complexity (LC) protein domains, which have been shown to interact with RNA to form liquid droplets184–186. Such LC domains include repetitive polymers of Ser and Arg in many proteins involved in LLPS187,188. Based on this, model polypeptides containing Ser-Arg repeats have been recently used to monitor the formation of liquid droplets and hydrogels in vitro and in vivo. Specifically, a hexanucleotide repeat GGGGCC is the most common cause of amyotrophic lateral sclerosis and frontotemporal dementia. Thus, poly(Gly-Arg) (GR) and poly(Pro-Arg) (PR) peptide repeats have been found to interact with RNA-binding proteins and proteins with LC domains that often mediate the assembly of membraneless organelles189. LLPS phenomena play a crucial role in the formation of disease-relevant disorders that are challenging to study in vivo owing to the complexity of processes involved. Yet, chemistry comes to the fore because these complex systems can be modelled using short peptides to yield a mechanistic understanding of these interactions190,191.
Capitalizing on the above findings, the role of polypeptide repeats in LLPS further depends on the amino-acid sequence and repeat-length specificity. For example, repeats of the five dipeptides GA, GP, GR, PA and PR have been shown to undergo LLPS both in vitro and in vivo192,193, with as little as 50 or 20 repeat units. Such peptides, foremost PR repeats, promote cellular toxicity by binding polymeric forms of the LC domains at the N termini of intermediate filament proteins, thereby promoting direct interactions with RNA granules and further alter the properties of stress granules193,194. Indeed, RNA can cause the formation of intracellular droplets by complex coacervation, a type of phase separation that occurs owing to electrostatic attraction between oppositely charged macromolecules. For example, the polycationic peptide RRASLRRASL, inspired by LRRASLG (Kemptide, a model synthetic substrate for protein kinase), was used in combination with polyU as a model for the regulation of intracellular droplet formation by post-translational modifications195. Further, the polyU–RRASLRRASL system is extremely sensitive to peptide charge, and one can switch the ability to form droplets on or off by removing or adding a single phosphate196 (FIG. 5c).
Similarly, the effects of a variety of polymers and ion concentration on LLPS have recently been studied, exemplifying the role of coacervate interfacial tension and critical salt concentration in the formation of hierarchically organized multiphase droplets197. Similarly, oligonucleotide-peptide conjugates such as poly(Lys) peptides have been used to systematically explore nucleic acid hybridization during nucleic acid and cationic peptide complexation (FIG. 5d). The phase of the complexes formed is controlled by the hybridization of the nucleic acid — double-stranded nucleic acids form solid precipitates, while single-stranded oligonucleotides have lower charge density and, instead, give liquid coacervates198. This charge sensitivity can be crucial for cellular regulation of compartment formation in response to external stimuli. Similarly, I3V3A3G3K3, a surfactant-like peptide, can induce efficient DNA condensation into virus-mimicking structures in a two-step manner199. The peptide binds the DNA chain through electrostatic interactions and then self-assembles into β-sheets under hydrophobic interactions and H-bonding, thus mimicking the nature of the virus capsid in helping to package DNA.
More recently, the mechanism by which liquid condensates form has been explored using carboxybenzyl (Cbz)-protected FF and even Fmoc-protected single amino acids200. In the case of phase separation of Z-FF, one obtains low-enthalpy, solute-rich liquid droplets and high-entropy, solute-poor phases. The solute-rich liquid droplets act as nucleation sites, allowing the formation of thermodynamically favourable nanofibrils following Ostwald’s step rule, whereby metastable aggregates are converted into more ordered structures, thus, reducing the overall free energy of the system (FIG. 5e). This rule is exemplified here in that the nucleation barrier to self-assembled ordered structures is lowered when first transforming through a metastable liquid phase, as such droplets can serve as precursors in the formation of the thermodynamically more favourable supramolecular polymers.
Biomineralization and organic–inorganic hybrids
Evolutionary developments in biology have resulted in biomaterials with remarkable structural properties. Their assembly involves cooperative but relatively weak molecular interactions that contrast with the covalent interactions in synthetic polymers. Crucially, however, the biological materials produced by peptide and protein self-assembly can be structurally reinforced by subsequent biomineralization (FIG. 6). Thus, living organisms can build organic structures and then use ordered arrangements of inorganic materials to harden or stiffen existing tissues (FIG. 6a)201. Although Ca2+ is the main cation in biogenic minerals, biomineralization is widespread in various organisms and exploits a wide range of inorganic components such as CO32- and silicate anions. Bone, enamel and seashell nacre, for example, have proteins and inorganic platelets as common constituents202,203.
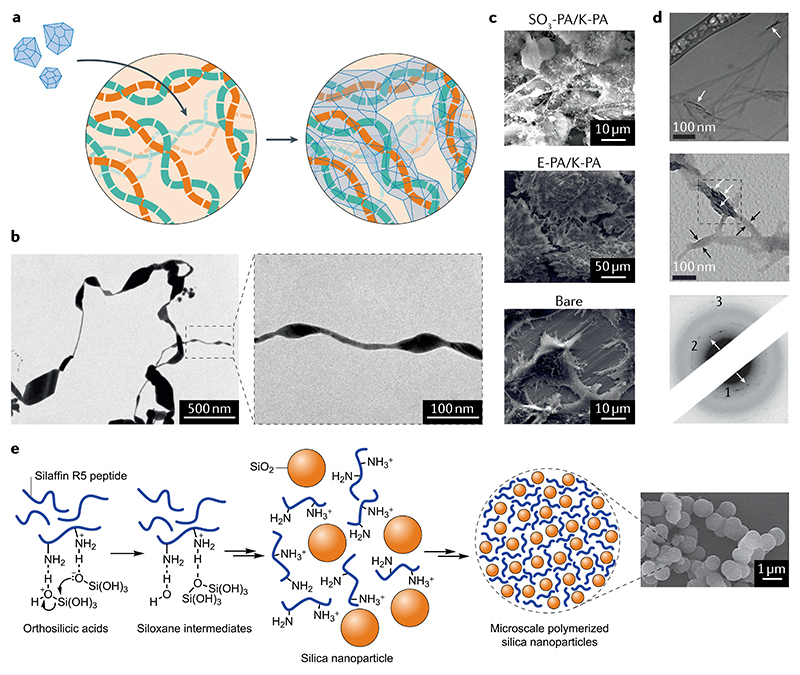
Fig. 6. Peptides as biomineralization scaffolds and organic–inorganic composite agents.
a | Self-assembled peptides can serve as templates for the deposition of inorganic materials. b | For example, Fmoc-protected 3,4-dihydroxy-L-phenylalanine dipeptide affords a hydrogel that reduces Ag+ ions over 3 days to give Ag crystals, as evidenced in transmission electron micrographs221. c | Scanning electron micrographs of mineralized bone-like nodules on nanofibres of a glycosaminoglycan-mimicking peptide57. d | Cryogenic transmission electron microscopy and selected-area electron diffraction (SAED) of hydroxyapatite mineralized at amphiphilic peptides. Black and white arrows indicate the location of the organic template and the position of inorganic crystals, respectively. SAED arrows indicate the oriented (002) reflection (1: (002), 2: {211}, 3: (004))208. e | Formation of SiO2 nanoparticles directed by self-assembled silaffin R5 peptide structures209. Part b adapted with permission from REF.221, American Chemical Society (https://pubs.acs.org/doi/10.1021/nn502240r). Part c adapted with permission from REF.56, Elsevier. Part d adapted with permission from REF.208, Wiley. Part e adapted with permission from REF.209, Wiley.
Natural biomineralization has stimulated much research in the field of biomimetic systems aimed at preparing complex materials with properties similar to those found in nature204,205. These materials are critically important to regenerative medicine and studies on tissue morphogenesis. In this regard, proteins and peptides are of interest in biomineralization processes owing to their high biocompatibility, structural stability, wide accessibility and sequence diversity. Moreover, their aptitude to template 0D to 3D structures and affinity for both hydrophobic and hydrophilic surfaces are very desirable203. Thus, the disadvantages of synthetic polymers in biological settings are well addressed by using peptides instead. In particular, complex peptide-based fibrillar networks that bind inorganic components have become attractive targets in materials design. One such system uses a supramolecular peptide nanofibre that can emulate both the nanofibrous architecture of collagenous ECM and the major chemical composition found on GAG for bone-tissue regeneration. GAGs constitute a significant portion of the ECM and have a substantial impact on regulating cellular behaviour, either directly or through encapsulation and presentation of growth factors to cells56 (FIG. 6c). This GAG and collagen-mimetic peptide assembles into nanofibres that interact with bone morphogenetic protein 2, which is a critical growth factor for osteogenic activity and mineralization by osteoblastic cells. The resulting structures sustain and direct the growth of bone cells and hydroxyapatite biominerals and, thus, can prove useful in the structural design of tissue-regenerating materials.
The biomineralization of enamel is regulated by amelogenin proteins such as Leu-rich amelogenin peptide (LRAP), which self-assembles and stabilizes amorphous Ca3(PO4)2 to promote enamel formation206. Further-more, phosphorylated LRAP not only stabilizes amorphous Ca3(PO4)2 but also prevents its transformation into Ca10(PO4)6(OH)2 (hydroxyapatite), aligned crystals of which form when non-phosphorylated LRAP is present. Furthermore, the non-phosphorylated N-terminal and C-terminal amelogenin domains are sufficient to template amorphous Ca3(PO4)2 transformation into ordered bundles of hydroxyapatite crystals, making LRAP an excellent candidate for biomimetic enamel regeneration207. Indeed, a peptidic amphiphile can self-assemble on a surface, thereby making it an amenable location for hydroxyapatite growth208. The highly aligned nanofibrillar bundles guide hydroxyapatite nucleation by varying the overall charge and propensity for β-sheet H-bonding. These cylindrical bundles allow mineralization in a specific orientation relative to the principal axis of the fibres, as found in mammalian bone structure (FIG. 6d). Thus, the controlled assembly of peptide amphiphiles and biomineralization at these sites can afford hierarchical structures that mimic bone. Similarly, the biomineralization of SiO2 plays a central role in the formation of structural exoskeletons in marine species such as diatoms and sponges209. This process takes place through specific deposition of SiO2 vesicles through interaction with a class of silaffin proteins at low pH. Inspired by this naturally evolved system, a variety of silaffin-mimicking peptides have recently been used to control the formation of SiO2 nanoparticles. The peptide SSKKSGSYSGSKGSKRRIL (R5) is of particular interest and promotes interactions with silicic acid through Lys residues, thereby leading to precipitation of SiO2 nanoparticles209. This self-assembling peptide has, thus, enabled the formation of ordered nanostructures onto which SiO2 shells can polymerize (FIG. 6e).
Another fascinating application of self-assembling systems is the ability to form organic–inorganic systems in which two types of building block are organized into intercalated layers to optimize mutual interactions and to combine their properties on greater length scales. Notable examples include protein-based self-assembling systems that incorporate nanoparticles into their structures210. In other studies, virus capsid-based peptides have been used as biotemplates to nucleate noble-metal nanoparticles and photoactive materials. This enabled controlled formation211,212, which is desirable in the case of thin films in energy-harvesting and energy-storage applications. Similarly, amyloid-related peptides template the formation of C and Au nanoparticles203,213,214. The resulting hybrid materials can adopt membrane, platelet and fibrillary gel morphologies and, thus, have a diverse set of properties, such as high toughness and strength, tunable fluorescence, conductivity and sensing. Combining self-assembled peptide nanostructures with Au and Ag also has fruitful outcomes215,216. For example, the cyclic antimicrobial lipopeptide surfactin self-assembles on photoluminescent Au nanodots to give a hybrid material, in which synergism between the two components efficiently inhibits the growth of various bacterial strains in vitro215. Furthermore, Fmoc-protected peptides self-assemble into nanofibres decorated with carboxylic acid and thiol groups — ideal coordination sites to act as scaffolds for the mineralization of Ag nanoparticles216. These composite materials exhibit highly effective and long-term antibacterial activity against both Gram-positive and Gram-negative bacteria and can maintain their structures.
The incorporation of metal ions during peptide self-assembly modulates the structures formed through coordination. Thus, adding Zn2+ ions to FF induces a structural transformation from β-sheet to a superhelix at a 1:1 Zn2+:FF ratio or a random coil at a 1:2 ratio, allowing specific control over the nature of the resulting metallohydrogel217. Similarly, short cationic peptides derived from FF spontaneously assemble into colloidal spheres in the presence of H3[PW12O40] (phosphotungstic acid)218. During the self-assembly of these spheres, they can host a variety of charged or uncharged guest molecules, along with hydrophobic and hydrophilic nanoparticles.
Au and Ni nanoparticles can bind short peptides such as an amyloid fibril model peptide containing a His6 tag219. This surfactant-like peptide undergoes a remarkable two-step self-assembly process at two distinct critical aggregation concentrations. When tagged with Au nanoparticles bearing Ni-NTA groups (where NTA is a tri(2-acetato)amine chelating derivative), one obtains functionalized amyloid fibrils as part of a pep-tide-nanoparticle hybrid. Peptides that mimic the native coiled-coil structure have similarly been used in Au nanoparticle functionalization220, as in the case of artificial Leu zipper-like peptides, which perform specific biomolecular recognition to assemble Au nanoparticles.
A different synthetic approach to organic–inorganic composites involves the reduction of metal ions in a controlled manner by self-assembled nanostructures. As demonstrated in the context of self-assembled short peptides, the non-coded aromatic amino acid 3,4-dihydroxy-L-phenylalanine (DOPA) can be introduced into peptides that self-assemble into a hydrogel with remarkable adhesive properties221. The potential utility of these structures was further explored in terms of spontaneously reducing metal cations into metal atoms. Thus, applying Ag+ to the hydrogel resulted in efficient reduction into Ag nanoparticles that formed a seamless metallic coating on the assemblies (FIG. 6b). Similarly, the T4P peptide from the metal-reducing Geobacter sulfurreducens bacterium has recently inspired the design of synthetic-peptide building blocks that self-assemble into T4P-like nanofibres222, bind metal oxide particles and reduce Au3+. The resulting peptide–AuNP nanocomposites exhibit enhanced thermal stability, electrical conductivity from the single-fibre level up and substrate-selective adhesion. Such nanoscale assemblies have unique properties and can serve as multifunctional platforms for biotechnological applications by combining the inherent structural properties of the peptides with those of the metal-based nanoparticles.
Peptide-derived assemblies can template other structures aside from inorganic species and can, for example, be combined with molecular metal complexes and organic species. Thus, the co-assembly of a guanine-rich nucleic acid with a His-rich peptide and haemin affords catalytic nanoparticles that mimic the active site and peroxidase activity of haem proteins223. The His-rich peptide provides the activating groups and haemin the active site, while the guanine-rich DNA acts as a scaffold for haemin coordination and stabilization. Peptide–porphyrin co-assemblies can similarly afford activity, including for photocatalytic H2 evolution. The peptides and porphyrins spontaneously self-organize into ordered hybrid fibres by molecular self-assembly and self-mineralization with the assistance of visible light224. Related peptide–porphyrin systems are catalysts for O2 evolution, thereby mimicking cyanobacteria225. Here, DOPA, in combination with a metalloporphyrin and Co3O4 nanoparticles, affords hybrid fibres that absorb light and oxidize H2O to O2, with quinones serving as the electron acceptors. A similar approach uses photooxidase-mimicking nanovesicles formed from amphiphilic amino acids such as Fmoc-His and phthalocyanines to give a catalytic material226. Overall, these model systems showcase the potential utility of simple building blocks — peptides and even single amino acids — to give complex reactivity that mimics that found in nature.
Hierarchical peptidic materials with nanoscale morphology
We have discussed how peptides self-assemble into 1D fibrils or 2D structures. The assembly is hierarchical in that these structures can further pack into multiscale functional materials. Nature often uses structural units beyond linear and planar geometries, such that a variety of nanoscale shapes have functional roles. This has inspired the exploration of artificial peptide-based materials that derive their functionality from their complex nanoscale shapes. For instance, the formation of hierarchical structures by peptide dimers and trimers adopting a coiled-coil motif allows the generation of globular protein mimics with well-defined molecular morphology and function227,228. Similarly, such hierarchically ordered structures have afforded peptide arrays used in bioelectronic, bioimaging and optical materials, as has recently been studied and reviewed229–236.
The formation of hierarchical peptide-based structures and materials has given rise to a field dubbed ‘peptide tectonics’237. Through introducing complementary units with selective association, peptide tectons allow for programmable self-assembly through selective interactions across domains, facilitating the development of new materials. Similarly, using abiological folded oligomers (foldamers) affords supramolecular architectures with diverse functions that extend beyond those found in nature238–240.
Among the more striking examples of biomimetic hierarchical peptide self-assembly is the formation of artificial viruses. The self-assembly of peptides into distinct 3D hierarchical structures mimics the distinct packing observed in viral capsids and their controlled disassembly. While initial research in this area focused on making peptide-based structures with dimensions similar to those of viruses, additional advances allowed the mimicry of linear viruses like the tobacco mosaic virus. Thus, the octapeptide lanreotide, synthesized as a growth hormone inhibitor, assembles into 20–30-nm-long nanotubes241. More recently, the β-annulus peptides from tomato bushy stunt virus have been observed to assemble into 30–50-nm viral-capsid-like nanocapsules242. These nanocapsules encapsulate various guest molecules and can be decorated with different molecules on their surface. In this way, one can prepare artificial viruses with human serum albumin or ribonuclease on their surfaces243,244.
A promising strategy in mimicking viral-capsid surfaces is using short peptides that assemble into filamentous nanoribbons to form an outer coat that encapsulates DNA or RNA245. Using this strategy, plasmid DNA has been combined with the peptide K3C6SPD to generate cocoon-like viral mimics through peptide self-assembly246. The nanococoon morphology, stability and ability to encapsulate DNA molecules can be further tuned by regulating the inter-nanofibril hydrophobic interactions to afford a cellular delivery system. Such nanococoons can also be made from the H4K5-HCBzl peptide, which assembles into subunit components of a low aspect ratio, thereby forming β-sheet nanodiscs247. A similar system has been demonstrated using TR4, a small molecule with four Arg residues and an N terminus functionalized with a tetraphenylethene and a lipophilic tail. The species self-assembles and hosts plasmid DNA248 in virus-mimicking nanoparticles that have low cytotoxicity, high stability and high transfection efficiency. The self-assembly process further induces bright fluorescence from tetraphenylethene groups packing together, allowing tracking of gene delivery. Further details regarding the use of such virus-mimicking assembly for therapeutic applications can be found in other recently published reviews249,250.
Conclusions and outlook
This Review has summarized new research into biomimicry that uses peptide self-assembly to afford ordered functional nanostructures with tunable physical, chemical and biological properties. Protein self-assembly is nature’s powerful tool to produce structures of varying length scales and functions, and with unique physical properties. The range of protein structures in natural systems is vast — ranging from oligomers and nanospheres to tubes and hierarchical assemblies that play key roles in biological functions such as cargo transport, microbial defence and structural support. These structures are held together by interactions that are predominantly non-covalent, thus conferring dynamism and flexibility on the structures. Great effort has been devoted to exploring self-assembly of natural and synthetic proteins, which allows the formation of materials that are functional but expensive and difficult to produce. New methods of studying these supramolecular structures, such as super-resolution microscopy and microfluidic platforms, have provided insights into the self-assembly process. But nature also uses short peptides composed of the minimal recognition modules, and these offer a unique platform for mimicking complex systems and phenomena with simple peptide-based model systems. These short peptide-based structures are more tractable and have shown great potential as materials for adhesives, cell scaffolds, drug-delivery systems, antimicrobial agents and surfaces, molecular machines and organic–inorganic matrices. The structural and functional diversity of such assemblies can be further expanded by incorporating inorganic molecules, such as inorganic materials and small molecules. Although simpler than protein derivatives, peptide-based biomimetic materials are still challenging to investigate and use. However, they present great promise for future research. We believe that exploring new modifications of short peptides will be the key to creating structures for new applications in even wider spread fields.
Acknowledgements
This project has received funding from the European Union’s Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie grant agreement no. 675007 (T.A.H., G.J.L.B. and T.P.J.K.), the European Research Council under the European Union’s Seventh Framework Programme (FP7/2007-2013) through the ERC grant PhysProt grant agreement 337969 (A.L. and T.P.J.K.) and the European Research Council under the European Union’s Horizon 2020 research and innovation programme BISON grant agreement 694426 (L.S. and E.G.). We also thank the Newman Foundation (T.P.J.K.), the Oppenheimer Early Career Fellowship (A.L.), the Israeli Ministry of Science, Technology and Space (L.S.), the BBSRC (T.P.J.K.), the Royal Society (URF\R\180019, G.J.L.B.), FCT Portugal (FCT Investigator IF/00624/2015 to G.J.L.B.), the Israeli National Nanotechnology Initiative and Helmsley Charitable Trust (E.G.), Elan Pharmaceuticals (T.P.J.K.) and the Centre for Misfolding Diseases (A.L. and T.P.J.K.) for financial support. We are grateful to our late colleague and friend Chris Dobson for input into this Review.
Footnotes
Author contributions
A.L., T.A.H and L.S. contributed equally to this work. A.L., G.J.L.B., E.G. and T.P.J.K. conceived the Review. All authors contributed to the discussion and writing of the Review.
Competing interests
The authors declare no competing interests.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Whitesides GM, Grzybowski B. Self-assembly at all scales. Science. 2002;295:2418–2421. doi: 10.1126/science.1070821. [DOI] [PubMed] [Google Scholar]
- 2.Philp D, Stoddart JF. Self-assembly in natural and unnatural systems. Angew Chem Int Ed Engl. 1996;35:1154–1196. [Google Scholar]
- 3.Schliwa M, Woehlke G. Molecular motors. Nature. 2003;422:759–765. doi: 10.1038/nature01601. [DOI] [PubMed] [Google Scholar]
- 4.Lehn JM. Perspectives in supramolecular chemistry — from molecular recognition towards molecular information processing and self-organization. Angew Chem Int Ed Engl. 1990;29:1304–1319. [Google Scholar]
- 5.Aida T, Meijer EW, Stupp SI. Functional supramolecular polymers. Science. 2012;335:813–817. doi: 10.1126/science.1205962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang S. Fabrication of novel biomaterials through molecular self-assembly. Nat Biotechnol. 2003;21:1171–1178. doi: 10.1038/nbt874. [DOI] [PubMed] [Google Scholar]
- 7.Granger E, McNee G, Allan V, Woodman P. The role of the cytoskeleton and molecular motors in endosomal dynamics. Semin Cell Dev Biol. 2014;31:20–29. doi: 10.1016/j.semcdb.2014.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ananthakrishnan R, Ehrlicher A. The forces behind cell movement. Int J Biol Sci. 2007;3:303–317. doi: 10.7150/ijbs.3.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krause M, Gautreau A. Steering cell migration: lamellipodium dynamics and the regulation of directional persistence. Nat Rev Mol Cell Biol. 2014;15:577–590. doi: 10.1038/nrm3861. [DOI] [PubMed] [Google Scholar]
- 10.Pegoraro AF, Janmey P, Weitz DA. Mechanical properties of the cytoskeleton and cells. Cold Spring Harb Perspect Biol. 2017;9:a022038. doi: 10.1101/cshperspect.a022038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Broedersz CP, et al. Cross-link-governed dynamics of biopolymer networks. Phys Rev Lett. 2010;105:238101. doi: 10.1103/PhysRevLett.105.238101. [DOI] [PubMed] [Google Scholar]
- 12.Knowles TPJ, Buehler MJ. Nanomechanics of functional and pathological amyloid materials. Nat Nanotechnol. 2011;6:469–479. doi: 10.1038/nnano.2011.102. [DOI] [PubMed] [Google Scholar]
- 13.Cohen SIA, Vendruscolo M, Dobson CM, Knowles TPJ. From macroscopic measurements to microscopic mechanisms of protein aggregation. J Mol Biol. 2012;421:160–171. doi: 10.1016/j.jmb.2012.02.031. [DOI] [PubMed] [Google Scholar]
- 14.Oosawa F, Kasai M. A theory of linear and helical aggregations of macromolecules. J Mol Biol. 1962;4:10–21. doi: 10.1016/s0022-2836(62)80112-0. [DOI] [PubMed] [Google Scholar]
- 15.Aguzzi A, Calella AM. Prions: protein aggregation and infectious diseases. Physiol Rev. 2009;89:1105–1152. doi: 10.1152/physrev.00006.2009. [DOI] [PubMed] [Google Scholar]
- 16.Reches M, Gazit E. Self-assembly of peptide nanotubes and amyloid-like structures by charged-termini-capped diphenylalanine peptide analogues. Isr J Chem. 2005;45:363–371. [Google Scholar]
- 17.Sunde M, Blake CCF. From the globular to the fibrous state: protein structure and structural conversion in amyloid formation. Q Rev Biophys. 1998;31:1–39. doi: 10.1017/s0033583598003400. [DOI] [PubMed] [Google Scholar]
- 18.Zhang S, Holmes T, Lockshin C, Rich A. Spontaneous assembly of a self-complementary oligopeptide to form a stable macroscopic membrane. Proc Natl Acad Sci USA. 1993;90:3334–3338. doi: 10.1073/pnas.90.8.3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang S, et al. Self-complementary oligopeptide matrices support mammalian cell attachment. Biomaterials. 1995;16:1385–1393. doi: 10.1016/0142-9612(95)96874-y. [DOI] [PubMed] [Google Scholar]
- 20.Mendes AC, Baran ET, Reis RL, Azevedo HS. Self-assembly in nature: using the principles of nature to create complex nanobiomaterials. Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2013;5:582–612. doi: 10.1002/wnan.1238. [DOI] [PubMed] [Google Scholar]
- 21.Bromley EHC, Channon K, Moutevelis E, Woolfson DN. Peptide and protein building blocks for synthetic biology: from programming biomolecules to self-organized biomolecular systems. ACS Chem Biol. 2008;3:38–50. doi: 10.1021/cb700249v. [DOI] [PubMed] [Google Scholar]
- 22.Wei G, et al. Self-assembling peptide and protein amyloids: from structure to tailored function in nanotechnology. Chem Soc Rev. 2017;46:4661–4708. doi: 10.1039/c6cs00542j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gazit E. A possible role for π-stacking in the self-assembly of amyloid fibrils. FASEB J. 2002;16:77–83. doi: 10.1096/fj.01-0442hyp. [DOI] [PubMed] [Google Scholar]
- 24.Reches M, Gazit E. Designed aromatic homo-dipeptides: formation of ordered nanostructures and potential nanotechnological applications. Phys Biol. 2006;3:S10. doi: 10.1088/1478-3975/3/1/S02. [DOI] [PubMed] [Google Scholar]
- 25.Gradišar H, Jerala R. Self-assembled bionanostructures: proteins following the lead of DNA nanostructures. J Nanobiotechnol. 2014;12:4. doi: 10.1186/1477-3155-12-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Desai MS, Lee S-W. Protein-based functional nanomaterial design for bioengineering applications. Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2015;7:69–97. doi: 10.1002/wnan.1303. [DOI] [PubMed] [Google Scholar]
- 27.Knowles TPJ, Mezzenga R. Amyloid fibrils as building blocks for natural and artificial functional materials. Adv Mater. 2016;28:6546–6561. doi: 10.1002/adma.201505961. [DOI] [PubMed] [Google Scholar]
- 28.Levin A, et al. Ostwald’s rule of stages governs structural transitions and morphology of dipeptide supramolecular polymers. Nat Commun. 2014;5:5219. doi: 10.1038/ncomms6219. [DOI] [PubMed] [Google Scholar]
- 29.Gazit E. Self-assembly of short aromatic peptides into amyloid fibrils and related nanostructures. Prion. 2007;1:32–35. doi: 10.4161/pri.1.1.4095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Levin A, et al. Elastic instability-mediated actuation by a supra-molecular polymer. Nat Phys. 2016;12:926–930. [Google Scholar]
- 31.Maji SK, et al. Amyloid as a depot for the formulation of long-acting drugs. PLoS Biol. 2008;6:e17. doi: 10.1371/journal.pbio.0060017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Standley SM, et al. Induction of cancer cell death by self-assembling nanostructures incorporating a cytotoxic peptide. Cancer Res. 2010;70:3020–3026. doi: 10.1158/0008-5472.CAN-09-3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aggeli A, et al. pH as a trigger of peptide β-sheet self-assembly and reversible switching between nematic and isotropic phases. J Am Chem Soc. 2003;125:9619–9628. doi: 10.1021/ja021047i. [DOI] [PubMed] [Google Scholar]
- 34.Friggeri A, Feringa BL, van Esch J. Entrapment and release of quinoline derivatives using a hydrogel of a low molecular weight gelator. J Control Release. 2004;97:241–248. doi: 10.1016/j.jconrel.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 35.Panda JJ, Mishra A, Basu A, Chauhan VS. Stimuli responsive self-assembled hydrogel of a low molecular weight free dipeptide with potential for tunable drug delivery. Biomacromolecules. 2008;9:2244–2250. doi: 10.1021/bm800404z. [DOI] [PubMed] [Google Scholar]
- 36.Huang R, Qi W, Feng L, Su R, He Z. Self-assembling peptide–polysaccharide hybrid hydrogel as a potential carrier for drug delivery. Soft Matter. 2011;7:6222–6230. [Google Scholar]
- 37.Tjernberg LO, et al. Arrest of β-amyloid fibril formation by a pentapeptide ligand. J Biol Chem. 1996;271:8545–8548. doi: 10.1074/jbc.271.15.8545. [DOI] [PubMed] [Google Scholar]
- 38.Krysmann MJ, Castelletto V, Hamley IW. Fibrillisation of hydrophobically modified amyloid peptide fragments in an organic solvent. Soft Matter. 2007;3:1401–1406. doi: 10.1039/b709889h. [DOI] [PubMed] [Google Scholar]
- 39.Hamley IW, et al. Nematic and columnar ordering of a PEG–peptide conjugate in aqueous solution. Chem Eur J. 2008;14:11369–11375. doi: 10.1002/chem.200800817. [DOI] [PubMed] [Google Scholar]
- 40.Castelletto V, Cheng G, Furzeland S, Atkins D, Hamley IW. Control of strand registry by attachment of PEG chains to amyloid peptides influences nanostructure. Soft Matter. 2012;8:5434–5438. [Google Scholar]
- 41.Castelletto V, Hamley I, Harris PJF. Self-assembly in aqueous solution of a modified amyloid beta peptide fragment. Biophys Chem. 2008;138:29–35. doi: 10.1016/j.bpc.2008.08.007. [DOI] [PubMed] [Google Scholar]
- 42.Hauser CAE, et al. Natural tri- to hexapeptides self-assemble in water to amyloid β-type fiber aggregates by unexpected α-helical intermediate structures. Proc Natl Acad Sci USA. 2011;108:1361–1366. doi: 10.1073/pnas.1014796108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lakshmanan A, et al. Aliphatic peptides show similar self-assembly to amyloid core sequences, challenging the importance of aromatic interactions in amyloidosis. Proc Natl Acad Sci USA. 2013;110:519–524. doi: 10.1073/pnas.1217742110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang J, et al. Trace water as prominent factor to induce peptide self-assembly: dynamic evolution and governing interactions in ionic liquids. Small. 2017;13:1702175. doi: 10.1002/smll.201702175. [DOI] [PubMed] [Google Scholar]
- 45.Ren X, et al. The dominant role of oxygen in modulating the chemical evolution pathways of tyrosine in peptides: dityrosine or melanin. Angew Chem Int Ed Engl. 2019;58:5872–5876. doi: 10.1002/anie.201814575. [DOI] [PubMed] [Google Scholar]
- 46.Mason TO, et al. Expanding the solvent chemical space for self-assembly of dipeptide nanostructures. ACS Nano. 2014;8:1243–1253. doi: 10.1021/nn404237f. [DOI] [PubMed] [Google Scholar]
- 47.Yan X, et al. Reversible transitions between peptide nanotubes and vesicle-like structures including theoretical modeling studies. Chem Eur J. 2008;14:5974–5980. doi: 10.1002/chem.200800012. [DOI] [PubMed] [Google Scholar]
- 48.Yan X, et al. Transition of cationic dipeptide nanotubes into vesicles and oligonucleotide delivery. Angew Chem Int Ed Engl. 2007;46:2431–2434. doi: 10.1002/anie.200603387. [DOI] [PubMed] [Google Scholar]
- 49.Riley JM, Aggeli A, Koopmans RJ, McPherson MJ. Bioproduction and characterization of a pH responsive self-assembling peptide. Biotechnol Bioeng. 2009;103:241–251. doi: 10.1002/bit.22274. [DOI] [PubMed] [Google Scholar]
- 50.Cinar G, et al. Amyloid inspired self-assembled peptide nanofibers. Biomacromolecules. 2012;13:3377–3387. doi: 10.1021/bm301141h. [DOI] [PubMed] [Google Scholar]
- 51.Du Q, et al. A comparative study on the self-assembly of an amyloid-like peptide at water–solid interfaces and in bulk solutions. Microsc Res Tech. 2015;78:375–381. doi: 10.1002/jemt.22483. [DOI] [PubMed] [Google Scholar]
- 52.Huettner N, Dargaville TR, Forget A. Discovering cell-adhesion peptides in tissue engineering: beyond RGD. Trends Biotechnol. 2018;36:372–383. doi: 10.1016/j.tibtech.2018.01.008. [DOI] [PubMed] [Google Scholar]
- 53.Collier JH, Rudraa JS, Gasiorowskia JZ, Jung JP. Multi-component extracellular matrices based on peptide self-assembly. Chem Soc Rev. 2010;39:3413–3424. doi: 10.1039/b914337h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu J, et al. Peptide glycosylation generates supramolecular assemblies from glycopeptides as biomimetic scaffolds for cell adhesion and proliferation. ACS Appl Mater Interfaces. 2016;8:6917–6924. doi: 10.1021/acsami.6b00850. [DOI] [PubMed] [Google Scholar]
- 55.Kisiday J, et al. Self-assembling peptide hydrogel fosters chondrocyte extracellular matrix production and cell division: implications for cartilage tissue repair. Proc Natl Acad Sci USA. 2002;99:9996–10001. doi: 10.1073/pnas.142309999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kocabey S, Ceylan H, Tekinay AB, Guler MO. Glycosaminoglycan mimetic peptide nanofibers promote mineralization by osteogenic cells. Acta Biomater. 2013;9:9075–9085. doi: 10.1016/j.actbio.2013.07.007. [DOI] [PubMed] [Google Scholar]
- 57.O’Leary LE, Fallas JA, Bakota EL, Kang MK, Hartgerink JD. Multi-hierarchical self-assembly of a collagen mimetic peptide from triple helix to nanofiber and hydrogel. Nat Chem. 2011;3:821–828. doi: 10.1038/nchem.1123. [DOI] [PubMed] [Google Scholar]
- 58.Kumar VA, et al. A nanostructured synthetic collagen mimic for hemostasis. Biomacromolecules. 2014;15:1484–1490. doi: 10.1021/bm500091e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bongiovanni MN, Scanlon DB, Gras SL. Functional fibrils derived from the peptide TTR1-cycloRGDfK that target cell adhesion and spreading. Biomaterials. 2011;32:6099–6110. doi: 10.1016/j.biomaterials.2011.05.021. [DOI] [PubMed] [Google Scholar]
- 60.Reynolds NP, Charnley M, Mezzenga R, Hartley PG. Engineered lysozyme amyloid fibril networks support cellular growth and spreading. Biomacromolecules. 2014;15:599–608. doi: 10.1021/bm401646x. [DOI] [PubMed] [Google Scholar]
- 61.Kumar VA, et al. Self-assembling multidomain peptides tailor biological responses through biphasic release. Biomaterials. 2015;52:71–78. doi: 10.1016/j.biomaterials.2015.01.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Das S, et al. Implantable amyloid hydrogels for promoting stem cell differentiation to neurons. NPG Asia Mater. 2016;8:e304. [Google Scholar]
- 63.Jacob RS, et al. Self healing hydrogels composed of amyloid nano fibrils for cell culture and stem cell differentiation. Biomaterials. 2015;54:97–105. doi: 10.1016/j.biomaterials.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 64.Kushwaha M, et al. A nitric oxide releasing, self assembled peptide amphiphile matrix that mimics native endothelium for coating implantable cardiovascular devices. Biomaterials. 2010;31:1502–1508. doi: 10.1016/j.biomaterials.2009.10.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Uzunalli G, et al. Improving pancreatic islet in vitro functionality and transplantation efficiency by using heparin mimetic peptide nanofiber gels. Acta Biomater. 2015;22:8–18. doi: 10.1016/j.actbio.2015.04.032. [DOI] [PubMed] [Google Scholar]
- 66.Berns EJ, et al. A tenascin-C mimetic peptide amphiphile nanofiber gel promotes neurite outgrowth and cell migration of neurosphere-derived cells. Acta Biomater. 2016;37:50–58. doi: 10.1016/j.actbio.2016.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Freeman R, et al. Reversible self-assembly of superstructured networks. Science. 2018;362:808–813. doi: 10.1126/science.aat6141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Panda JJ, Dua R, Mishra A, Mittra B, Chauhan VS. 3D cell growth and proliferation on a RGD functionalized nanofibrillar hydrogel based on a conformationally restricted residue containing dipeptide. ACS Appl Mater Interfaces. 2010;2:2839–2848. doi: 10.1021/am1005173. [DOI] [PubMed] [Google Scholar]
- 69.Azevedo HS, Pashkuleva I. Biomimetic supramolecular designs for the controlled release of growth factors in bone regeneration. Adv Drug Deliv Rev. 2015;94:63–76. doi: 10.1016/j.addr.2015.08.003. [DOI] [PubMed] [Google Scholar]
- 70.Loo Y, et al. Peptide bioink self-assembling nanofibrous scaffolds for three-dimensional organotypic cultures. Nano Lett. 2015;15:6919–6925. doi: 10.1021/acs.nanolett.5b02859. [DOI] [PubMed] [Google Scholar]
- 71.Jacob RS, et al. Cell adhesion on amyloid fibrils lacking integrin recognition motif. J Biol Chem. 2016;291:5278–5298. doi: 10.1074/jbc.M115.678177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vegas AJ, et al. Combinatorial hydrogel library enables identification of materials that mitigate the foreign body response in primates. Nat Biotechnol. 2016;34:345–352. doi: 10.1038/nbt.3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yesilyurt V, et al. Injectable self-healing glucoseresponsive hydrogels with pH-regulated mechanical properties. Adv Mater. 2016;28:86–91. doi: 10.1002/adma.201502902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Neal RA, et al. Three-dimensional elastomeric scaffolds designed with cardiac-mimetic structural and mechanical features. Tissue Eng Part A. 2013;19:793–807. doi: 10.1089/ten.tea.2012.0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Smith DJ, et al. A multiphase transitioning peptide hydrogel for suturing ultrasmall vessels. Nat Nanotechnol. 2016;11:95–102. doi: 10.1038/nnano.2015.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dexter AF, Middelberg APJ. Peptides as functional surfactants. Ind Eng Chem Res. 2008;47:6391–6398. [Google Scholar]
- 77.Vauthey S, Santoso S, Gong H, Watson N, Zhang S. Molecular self-assembly of surfactant-like peptides to form nanotubes and nanovesicles. Proc Natl Acad Sci USA. 2002;99:5355–5360. doi: 10.1073/pnas.072089599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Braide-Moncoeur O, Tran NT, Long JR. Peptide-based synthetic pulmonary surfactant for the treatment of respiratory distress disorders. Curr Opin Chem Biol. 2016;32:22–28. doi: 10.1016/j.cbpa.2016.02.012. [DOI] [PubMed] [Google Scholar]
- 79.Mondal S, et al. A minimal length rigid helical peptide motif allows rational design of modular surfactants. Nat Commun. 2017;8:14018. doi: 10.1038/ncomms14018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Macindoe I, et al. Self-assembly of functional, amphipathic amyloid monolayers by the fungal hydrophobin EAS. Proc Natl Acad Sci USA. 2012;109:804–811. doi: 10.1073/pnas.1114052109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hanski S, et al. Hierarchical ionic self-assembly of rod–comb block copolypeptide–surfactant complexes. Biomacromolecules. 2006;7:3379–3384. doi: 10.1021/bm0606770. [DOI] [PubMed] [Google Scholar]
- 82.Zheng X, et al. Expression, stabilization and purification of membrane proteins via diverse protein synthesis systems and detergents involving cell-free associated with self-assembly peptide surfactants. Biotechnol Adv. 2014;32:564–574. doi: 10.1016/j.biotechadv.2014.02.003. [DOI] [PubMed] [Google Scholar]
- 83.Chécot F, Lecommandoux S, Klok H-A, Gnanou Y. From supramolecular polymersomes to stimuli-responsive nano-capsules based on poly(diene-b-peptide) diblock copolymers. Eur Phys J E. 2003;10:25–35. doi: 10.1140/epje/e2003-00006-1. [DOI] [PubMed] [Google Scholar]
- 84.Bellomo EG, Wyrsta MD, Pakstis L, Pochan DJ, Deming TJ. Stimuli-responsive polypeptide vesicles by conformation-specific assembly. Nat Mater. 2004;3:244–248. doi: 10.1038/nmat1093. [DOI] [PubMed] [Google Scholar]
- 85.Chen C, et al. Molecular mechanisms of antibacterial and antitumor actions of designed surfactant-like peptides. Biomaterials. 2012;33:592–603. doi: 10.1016/j.biomaterials.2011.09.059. [DOI] [PubMed] [Google Scholar]
- 86.Delbecq F. Supramolecular gels from lipopeptide gelators: template improvement and strategies for the in-situ preparation of inorganic nanomaterials and for the dispersion of carbon nanomaterials. Adv Colloid Interface Sci. 2014;209:98–108. doi: 10.1016/j.cis.2014.02.018. [DOI] [PubMed] [Google Scholar]
- 87.Zhao X. Design of self-assembling surfactant-like peptides and their applications. Curr Opin Colloid Interface Sci. 2009;14:340–348. [Google Scholar]
- 88.Nagai A, Nagai Y, Qu HJ, Zhang SG. Dynamic behaviors of lipid-like self-assembling peptide A6D and A6K nanotubes. J Nanosci Nanotechnol. 2007;7:2246–2252. doi: 10.1166/jnn.2007.647. [DOI] [PubMed] [Google Scholar]
- 89.Dehsorkhi A, Castelletto V, Hamley IW. Self-assembling amphiphilic peptides. J Pept Sci. 2014;20:453–467. doi: 10.1002/psc.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Fatouros DG, et al. Lipid-like self-assembling peptide nanovesicles for drug delivery. ACS Appl Mater Interfaces. 2014;6:8184–8189. doi: 10.1021/am501673x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Pellach M, et al. Spontaneous structural transition in phospholipid-inspired aromatic phosphopeptide nanostructures. ACS Nano. 2015;9:4085–4095. doi: 10.1021/acsnano.5b00133. [DOI] [PubMed] [Google Scholar]
- 92.Pellach M, et al. A two-tailed phosphopeptide crystallises to form a lamellar structure. Angew Chem Int Ed Engl. 2017;56:3252–3255. doi: 10.1002/anie.201609877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cheng H, et al. Stem cell membrane engineering for cell rolling using peptide conjugation and tuning of cell-selectin interaction kinetics. Biomaterials. 2012;33:5004–5012. doi: 10.1016/j.biomaterials.2012.03.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lazzaro BP, Zasloff M, Rolff J. Antimicrobial peptides: application informed by evolution. Science. 2020;368:eaau5480. doi: 10.1126/science.aau5480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Jenssen H, Hamill P, Hancock REW. Peptide antimicrobial agents. Clin Microbiol Rev. 2006;19:491–511. doi: 10.1128/CMR.00056-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Giuliani A, Pirri G, Nicoletto SF. Antimicrobial peptides: an overview of a promising class of therapeutics. Cent Eur J Biol. 2007;2:1–33. [Google Scholar]
- 97.Seo M-D, Won H-S, Kim J-H, Mishig-Ochir T, Lee B-J. Antimicrobial peptides for therapeutic applications: a review. Molecules. 2012;17:12276–12286. doi: 10.3390/molecules171012276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Cederlund A, Gudmundsson GH, Agerberth B. Antimicrobial peptides important in innate immunity. FEBS J. 2011;278:3942–3951. doi: 10.1111/j.1742-4658.2011.08302.x. [DOI] [PubMed] [Google Scholar]
- 99.Zasloff M. Antimicrobial peptides of multicellular organisms. Nature. 2002;415:389–395. doi: 10.1038/415389a. [DOI] [PubMed] [Google Scholar]
- 100.Timofeeva L, Kleshcheva N. Antimicrobial polymers: mechanism of action, factors of activity, and applications. Appl Microbiol Biotechnol. 2011;89:475–492. doi: 10.1007/s00253-010-2920-9. [DOI] [PubMed] [Google Scholar]
- 101.Nguyen LT, Haney EF, Vogel HJ. The expanding scope of antimicrobial peptide structures and their modes of action. Trends Biotechnol. 2011;29:464–472. doi: 10.1016/j.tibtech.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 102.Bahar AA, Ren D. Antimicrobial peptides. Pharmaceuticals. 2013;6:1543–1575. doi: 10.3390/ph6121543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Cudic M, Otvos L. Intracellular targets of antibacterial peptides. Curr Drug Targets. 2002;3:101–106. doi: 10.2174/1389450024605445. [DOI] [PubMed] [Google Scholar]
- 104.Brogden KA. Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat Rev Microbiol. 2005;3:238–250. doi: 10.1038/nrmicro1098. [DOI] [PubMed] [Google Scholar]
- 105.Tew GN, Scott RW, Klein ML, DeGrado WF. De novo design of antimicrobial polymers, foldamers, and small molecules: from discovery to practical applications. Acc Chem Res. 2010;43:30–39. doi: 10.1021/ar900036b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Giuliani A, Rinaldi AC. Beyond natural antimicrobial peptides: multimeric peptides and other peptidomimetic approaches. Cell Mol Life Sci. 2011;68:2255–2266. doi: 10.1007/s00018-011-0717-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Godballe T, Nilsson LL, Petersen PD, Jenssen H. Antimicrobial β-peptides and α-peptoids. Chem Biol Drug Des. 2011;77:107–116. doi: 10.1111/j.1747-0285.2010.01067.x. [DOI] [PubMed] [Google Scholar]
- 108.Fjell CD, Hiss JA, Hancock REW, Schneider G. Designing antimicrobial peptides: form follows function. Nat Rev Drug Discov. 2012;11:37–51. doi: 10.1038/nrd3591. [DOI] [PubMed] [Google Scholar]
- 109.McCloskey A, Gilmore B, Laverty G. Evolution of antimicrobial peptides to self-assembled peptides for biomaterial applications. Pathogens. 2014;3:791–821. doi: 10.3390/pathogens3040791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Tian X, Sun F, Zhou XR, Luo SZ, Chen L. Role of peptide self-assembly in antimicrobial peptides. J Pept Sci. 2015;21:530–539. doi: 10.1002/psc.2788. [DOI] [PubMed] [Google Scholar]
- 111.Sun L, Zheng C, Webster TJ. Self-assembled peptide nanomaterials for biomedical applications: promises and pitfalls. Int J Nanomed. 2017;12:73–86. doi: 10.2147/IJN.S117501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Salick DA, Pochan DJ, Schneider JP. Design of an injectable β-hairpin peptide hydrogel that kills methicillin-resistant Staphylococcus aureus. Adv Mater. 2009;21:4120–4123. [Google Scholar]
- 113.Rajagopal K, Ozbas B, Pochan DJ, Schneider JP. Probing the importance of lateral hydrophobic association in self-assembling peptide hydrogelators. Eur Biophys J. 2006;35:162–169. doi: 10.1007/s00249-005-0017-7. [DOI] [PubMed] [Google Scholar]
- 114.Gupta K, et al. Mechanism of membrane permeation induced by synthetic β-hairpin peptides. Biophys J. 2013;105:2093–2103. doi: 10.1016/j.bpj.2013.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kretsinger JK, Haines LA, Ozbas B, Pochan DJ, Schneider JP. Cytocompatibility of self-assembled β-hairpin peptide hydrogel surfaces. Biomaterials. 2005;26:5177–5186. doi: 10.1016/j.biomaterials.2005.01.029. [DOI] [PubMed] [Google Scholar]
- 116.Haines LA, et al. Light-activated hydrogel formation via the triggered folding and self-assembly of a designed peptide. J Am Chem Soc. 2005;127:17025–17029. doi: 10.1021/ja054719o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Haines-Butterick L, et al. Controlling hydrogelation kinetics by peptide design for three-dimensional encapsulation and injectable delivery of cells. Proc Natl Acad Sci USA. 2007;104:7791–7796. doi: 10.1073/pnas.0701980104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Veiga AS, et al. Arginine-rich self-assembling peptides as potent antibacterial gels. Biomaterials. 2012;33:8907–8916. doi: 10.1016/j.biomaterials.2012.08.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Tripathi JK, et al. Variants of self-assembling peptide, KLD-12 that show both rapid fracture healing and antimicrobial properties. Biomaterials. 2015;56:92–103. doi: 10.1016/j.biomaterials.2015.03.046. [DOI] [PubMed] [Google Scholar]
- 120.Laverty G, et al. Ultrashort cationic naphthalene-derived self-assembled peptides as antimicrobial nanomaterials. Biomacromolecules. 2014;15:3429–3439. doi: 10.1021/bm500981y. [DOI] [PubMed] [Google Scholar]
- 121.McCloskey AP, Draper ER, Gilmore BF, Laverty G. Ultrashort self-assembling Fmoc-peptide gelators for anti-infective biomaterial applications. J Pept Sci. 2017;23:131–140. doi: 10.1002/psc.2951. [DOI] [PubMed] [Google Scholar]
- 122.Gahane AY, et al. Fmoc-phenylalanine displays antibacterial activity against Gram-positive bacteria in gel and solution phases. Soft Matter. 2018;14:2234–2244. doi: 10.1039/c7sm02317k. [DOI] [PubMed] [Google Scholar]
- 123.Hughes M, Debnath S, Knapp CW, Ulijn RV. Antimicrobial properties of enzymatically triggered self-assembling aromatic peptide amphiphiles. Biomater Sci. 2013;1:1138–1142. doi: 10.1039/c3bm60135h. [DOI] [PubMed] [Google Scholar]
- 124.Yang Z, Liang G, Guo Z, Guo Z, Xu B. Intracellular hydrogelation of small molecules inhibits bacterial growth. Angew Chem Int Ed Engl. 2007;46:8216–8219. doi: 10.1002/anie.200701697. [DOI] [PubMed] [Google Scholar]
- 125.Marchesan S, et al. Self-assembly of ciprofloxacin and a tripeptide into an antimicrobial nanostructured hydrogel. Biomaterials. 2013;34:3678–3687. doi: 10.1016/j.biomaterials.2013.01.096. [DOI] [PubMed] [Google Scholar]
- 126.Hu Y, et al. Self-assembled peptide nanofibers encapsulated with superfine silver nanoparticles via Ag+ coordination. Langmuir. 2015;31:8599–8605. doi: 10.1021/acs.langmuir.5b02036. [DOI] [PubMed] [Google Scholar]
- 127.Paladini F, et al. Silver-doped self-assembling di-phenylalanine hydrogels as wound dressing biomaterials. J Mater Sci Mater Med. 2013;24:2461–2472. doi: 10.1007/s10856-013-4986-2. [DOI] [PubMed] [Google Scholar]
- 128.Jiang L, Xu D, Sellati TJ, Dong H. Self-assembly of cationic multidomain peptide hydrogels: supramolecular nanostructure and rheological properties dictate antimicrobial activity. Nanoscale. 2015;7:19160–19169. doi: 10.1039/c5nr05233e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Liu Y, Yang Y, Wang C, Zhao X. Stimuli-responsive self-assembling peptides made from antibacterial peptides. Nanoscale. 2013;5:6413–6421. doi: 10.1039/c3nr00225j. [DOI] [PubMed] [Google Scholar]
- 130.Fernandez-Lopez S, et al. Antibacterial agents based on the cyclic d,l-α-peptide architecture. Nature. 2001;412:452–455. doi: 10.1038/35086601. [DOI] [PubMed] [Google Scholar]
- 131.Horne WS, et al. Antiviral cyclic d,l-α-peptides: targeting a general biochemical pathway in virus infections. Bioorg Med Chem. 2005;13:5145–5153. doi: 10.1016/j.bmc.2005.05.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Montero A, et al. Self-assembling peptide nanotubes with antiviral activity against hepatitis C virus. Chem Biol. 2011;18:1453–1462. doi: 10.1016/j.chembiol.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Bionda N, et al. Effects of cyclic lipodepsipeptide structural modulation on stability, antibacterial activity, and human cell toxicity. ChemMedChem. 2012;7:871–882. doi: 10.1002/cmdc.201200016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Fletcher JT, Finlay JA, Callow ME, Callow JA, Ghadiri MR. A combinatorial approach to the discovery of biocidal six-residue cyclic d,l-α-peptides against bacteria methicillin-resistant Staphylococcus aureus (MRSA) and E. coli and the biofouling algae Ulva linza and Navicula perminuta. Chemistry. 2007;13:4008–4013. doi: 10.1002/chem.200((......)). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Bionda N, et al. Identification of novel cyclic lipopeptides from a positional scanning combinatorial library with enhanced antibacterial and antibiofilm activities. Eur J Med Chem. 2016;108:354–363. doi: 10.1016/j.ejmech.2015.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Liu L, et al. Self-assembled cationic peptide nanoparticles as an efficient antimicrobial agent. Nat Nanotechnol. 2009;4:457–463. doi: 10.1038/nnano.2009.153. [DOI] [PubMed] [Google Scholar]
- 137.Xu K, et al. Efficacy of CG3R6TAT nanoparticles self-assembled from a novel antimicrobial peptide for the treatment of Candida albicans meningitis in rabbits. Chemotherapy. 2012;57:417–425. doi: 10.1159/000330855. [DOI] [PubMed] [Google Scholar]
- 138.Makovitzki A, Baram J, Shai Y. Antimicrobial lipopolypeptides composed of palmitoyl di- and tricationic peptides: In vitro and in vivo activities, self-assembly to nanostructures, and a plausible mode of action. Biochemistry. 2008;47:10630–10636. doi: 10.1021/bi8011675. [DOI] [PubMed] [Google Scholar]
- 139.Chen C, et al. Antibacterial activities of short designer peptides: A link between propensity for nanostructuring and capacity for membrane destabilization. Biomacromolecules. 2010;11:402–411. doi: 10.1021/bm901130u. [DOI] [PubMed] [Google Scholar]
- 140.Beter M, et al. Multivalent presentation of cationic peptides on supramolecular nanofibers for antimicrobial activity. Mol Pharm. 2017;4:3660–3668. doi: 10.1021/acs.molpharmaceut.7b00434. [DOI] [PubMed] [Google Scholar]
- 141.Schnaider L, et al. Self-assembling dipeptide antibacterial nanostructures with membrane disrupting activity. Nat Commun. 2017;8:1365. doi: 10.1038/s41467-017-01447-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Salinas N, Colletier JP, Moshe A, Landau M. Extreme amyloid polymorphism in Staphylococcus aureus virulent PSMα peptides. Nat Commun. 2018;9:3512. doi: 10.1038/s41467-018-05490-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Deslouches B, Di YP. Antimicrobial peptides with selective antitumor mechanisms: Prospect for anticancer applications. Oncotarget. 2017;8:46635–46651. doi: 10.18632/oncotarget.16743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Felício MR, Silva ON, Gonçalves S, Santos NC, Franco OL. Peptides with dual antimicrobial and anticancer activities. Front Chem. 2017;5:5. doi: 10.3389/fchem.2017.00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Gaspar D, Veiga AS, Castanho MARB. From antimicrobial to anticancer peptides. A review. Front Microbiol. 2013;4:294. doi: 10.3389/fmicb.2013.00294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Ding Y, et al. Improvement of stability and efficacy of C16Y therapeutic peptide via molecular self-assembly into tumor-responsive nanoformulation. Mol Cancer Ther. 2015;14:2390–2400. doi: 10.1158/1535-7163.MCT-15-0484. [DOI] [PubMed] [Google Scholar]
- 147.Ding Y, et al. Targeting vault nanoparticles to specific cell surface receptors. ACS Nano. 2009;3:27–36. doi: 10.1021/nn800638x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Tang A, Wang C, Stewart R, Kopecek J. Self-assembled peptides exposing epitopes recognizable by human lymphoma cells. Bioconjug Chem. 2000;11:363–371. doi: 10.1021/bc990133d. [DOI] [PubMed] [Google Scholar]
- 149.Zha RH, et al. Supramolecular assembly of multifunctional maspin-mimetic nanostructures as a potent peptide-based angiogenesis inhibitor. Acta Biomater. 2015;12:1–10. doi: 10.1016/j.actbio.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Levin A, et al. Self-assembly-mediated release of peptide nanoparticles through jets across microdroplet interfaces. ACS Appl Mater Interfaces. 2018;10:27578–27583. doi: 10.1021/acsami.8b09511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Ding Y, et al. Gene delivery and immunomodulatory effects of plasmid DNA associated with branched amphiphilic peptide capsules. J Control Release. 2016;241:15–24. doi: 10.1016/j.jconrel.2016.08.042. [DOI] [PubMed] [Google Scholar]
- 152.Mumcuoglu D, et al. Cellular internalization of therapeutic oligonucleotides by peptide amphiphile nanofibers and nanospheres. ACS Appl Mater Interfaces. 2016;8:11280–11287. doi: 10.1021/acsami.6b01526. [DOI] [PubMed] [Google Scholar]
- 153.Li Y, et al. Self-assembled peptide (CADY-1) improved the clinical application of doxorubicin. Int J Pharm. 2012;434:209–214. doi: 10.1016/j.ijpharm.2012.06.003. [DOI] [PubMed] [Google Scholar]
- 154.Sarangthem V, et al. Multivalent targeting based delivery of therapeutic peptide using AP1-ELP carrier for effective cancer therapy. Theranostics. 2016;6:2235–2249. doi: 10.7150/thno.16425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Zha RH, Sur S, Stupp SI. Self-assembly of cytotoxic peptide amphiphiles into supramolecular membranes for cancer therapy. Adv Healthc Mater. 2013;2:126–133. doi: 10.1002/adhm.201200118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Qiao ZY, et al. Self-assembled ROS-sensitive polymer-peptide therapeutics incorporating built-in reporters for evaluation of treatment efficacy. Biomacromolecules. 2016;17:1643–1652. doi: 10.1021/acs.biomac.6b00041. [DOI] [PubMed] [Google Scholar]
- 157.Chen L, Patrone N, Liang JF. Peptide self-assembly on cell membranes to induce cell lysis. Biomacromolecules. 2012;13:3327–3333. doi: 10.1021/bm301106p. [DOI] [PubMed] [Google Scholar]
- 158.Ding Y, et al. Smac therapeutic peptide nanoparticles inducing apoptosis of cancer cells for combination chemotherapy with doxorubicin. ACS Appl Mater Interfaces. 2015;7:8005–8012. doi: 10.1021/acsami.5b00329. [DOI] [PubMed] [Google Scholar]
- 159.Chen J, et al. Transmembrane delivery of anticancer drugs through self-assembly of cyclic peptide nanotubes. Nanoscale. 2016;8:7127–7136. doi: 10.1039/c5nr06804e. [DOI] [PubMed] [Google Scholar]
- 160.Soukasene S, et al. Antitumor activity of peptide amphiphile nanofiber-encapsulated camptothecin. ACS Nano. 2011;5:9113–9121. doi: 10.1021/nn203343z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Liu K, et al. Simple peptide-tuned self-assembly of photosensitizers towards anticancer photodynamic therapy. Angew Chem Int Ed Engl. 2016;55:3036–3039. doi: 10.1002/anie.201509810. [DOI] [PubMed] [Google Scholar]
- 162.Zou Q, et al. Biological photothermal nanodots based on self-assembly of peptide-porphyrin conjugates for antitumor therapy. J Am Chem Soc. 2017;139:1921–1927. doi: 10.1021/jacs.6b11382. [DOI] [PubMed] [Google Scholar]
- 163.Xing R, et al. Self-assembling endogenous biliverdin as a versatile near-infrared photothermal nanoagent for cancer theranostics. Adv Mater. 2019;31:1900822. doi: 10.1002/adma.201900822. [DOI] [PubMed] [Google Scholar]
- 164.Fan Z, Sun L, Huang Y, Wang Y, Zhang M. Bioinspired fluorescent dipeptide nanoparticles for targeted cancer cell imaging and real-time monitoring of drug release. Nat Nanotechnol. 2016;11:388–394. doi: 10.1038/nnano.2015.312. [DOI] [PubMed] [Google Scholar]
- 165.Hyman AA, Weber CA, Jülicher F. Liquid–liquid phase separation in biology. Annu Rev Cell Dev Biol. 2014;30:39–58. doi: 10.1146/annurev-cellbio-100913-013325. [DOI] [PubMed] [Google Scholar]
- 166.Banani SF, Lee HO, Hyman AA, Rosen MK. Biomolecular condensates: organizers of cellular biochemistry. Nat Rev Mol Cell Biol. 2017;18:285–298. doi: 10.1038/nrm.2017.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Shin Y, Brangwynne CP. Liquid phase condensation in cell physiology and disease. Science. 2017;357:eaaf4382. doi: 10.1126/science.aaf4382. [DOI] [PubMed] [Google Scholar]
- 168.Qamar S, et al. FUS phase separation is modulated by a molecular chaperone and methylation of arginine cation-π interactions. Cell. 2018;173:720–734. doi: 10.1016/j.cell.2018.03.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Alberti S, Dormann D. Liquid–liquid phase separation in disease. Annu Rev Genet. 2019;53:171–194. doi: 10.1146/annurev-genet-112618-043527. [DOI] [PubMed] [Google Scholar]
- 170.Boeynaems S, et al. Protein phase separation: a new phase in cell biology. Trends Cell Biol. 2018;28:420–435. doi: 10.1016/j.tcb.2018.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Lee K-H, et al. C9orf72 dipeptide repeats impair the assembly, dynamics, and function of membrane-less organelles. Cell. 2016;167:774–788. doi: 10.1016/j.cell.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Nakashima KK, Vibhute MA, Spruijt E. Biomolecular chemistry in liquid phase separated compartments. Front Mol Biosci. 2019;6:21. doi: 10.3389/fmolb.2019.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Bentley EP, Frey BB, Deniz AA. Physical chemistry of cellular liquid-phase separation. Chem Eur J. 2019;25:5600–5610. doi: 10.1002/chem.201805093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Wang J, et al. A molecular grammar governing the driving forces for phase separation of prion-like RNA binding proteins. Cell. 2018;174:688–699. doi: 10.1016/j.cell.2018.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Onuchic PL, Milin AN, Alshareedah I, Deniz AA, Banerjee PR. Divalent cations can control a switch-like behavior in heterotypic and homotypic RNA coacervates. Sci Rep. 2019;9:12161. doi: 10.1038/s41598-019-48457-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Sanders DW, et al. Competing protein–RNA interaction networks control multiphase intracellular organization. Cell. 2020;181:306–324. doi: 10.1016/j.cell.2020.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Alberti S, Gladfelter A, Mittag T. Considerations and challenges in studying liquid–liquid phase separation and biomolecular condensates. Cell. 2019;176:419–434. doi: 10.1016/j.cell.2018.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Shin Y, et al. Spatiotemporal control of intracellular phase transitions using light-activated optoDroplets. Cell. 2017;168:159–171. doi: 10.1016/j.cell.2016.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Schuster BS, et al. Controllable protein phase separation and modular recruitment to form responsive membraneless organelles. Nat Commun. 2018;9:2985. doi: 10.1038/s41467-018-05403-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Linsenmeier M, et al. Dynamics of synthetic membraneless organelles in microfluidic droplets. Angew Chem Int Ed Engl. 2019;58:14489–14494. doi: 10.1002/anie.201907278. [DOI] [PubMed] [Google Scholar]
- 181.Quiroz FG, Chilkoti A. Sequence heuristics to encode phase behaviour in intrinsically disordered protein polymers. Nat Mater. 2015;14:1164–1171. doi: 10.1038/nmat4418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Simon JR, Carroll NJ, Rubinstein M, Chilkoti A, López GP. Programming molecular self-assembly of intrinsically disordered proteins containing sequences of low complexity. Nat Chem. 2017;9:509–515. doi: 10.1038/nchem.2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Reinkemeier CD, Girona GE, Lemke EA. Designer membraneless organelles enable codon reassignment of selected mRNAs in eukaryotes. Science. 2019;363:eaaw2644. doi: 10.1126/science.aaw2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Uversky VN. Natively unfolded proteins: a point where biology waits for physics. Protein Sci. 2002;11:739–756. doi: 10.1110/ps.4210102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Martin EW, Mittag T. Relationship of sequence and phase separation in protein low-complexity regions. Biochemistry. 2018;57:2478–2487. doi: 10.1021/acs.biochem.8b00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Hughes MP, et al. Atomic structures of low-complexity protein segments reveal kinked β sheets that assemble networks. Science. 2018;359:698–701. doi: 10.1126/science.aan6398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Kato M, et al. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell. 2012;149:753–767. doi: 10.1016/j.cell.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Xiang S, et al. The LC domain of hnRNPA2 adopts similar conformations in hydrogel polymers, liquid-like droplets, and nuclei. Cell. 2015;5:829–839. doi: 10.1016/j.cell.2015.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Renton AE, et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72:257–268. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Wang Y, Lomakin A, Kanai S, Alex R, Benedek GB. Liquid–liquid phase separation in oligomeric peptide solutions. Langmuir. 2017;33:7715–7721. doi: 10.1021/acs.langmuir.7b01693. [DOI] [PubMed] [Google Scholar]
- 191.Ulijn RV, Lampel A. Order/disorder in protein and peptide-based biomaterials. Isr J Chem. 2019;59:1–13. [Google Scholar]
- 192.Lee KH, et al. C9orf72 dipeptide repeats impair the assembly, dynamics, and function of membrane-less organelles. Cell. 2016;167:774–788. doi: 10.1016/j.cell.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Lin Y, et al. Toxic PR poly-dipeptides encoded by the C9orf72 repeat expansion target LC domain polymers. Cell. 2016;167:789–802. doi: 10.1016/j.cell.2016.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Boeynaems S, et al. Phase separation of C9orf72 dipeptide repeats perturbs stress granule dynamics. Mol Cell. 2017;65:1044–1055. doi: 10.1016/j.molcel.2017.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Aumiller WM. Jr RNA-based coacervates as a model for membraneless organelles: formation, properties, and interfacial liposome assembly. Langmuir. 2016;32:10042–10053. doi: 10.1021/acs.langmuir.6b02499. [DOI] [PubMed] [Google Scholar]
- 196.Aumiller WM, Jr, Keating CD. Phosphorylation-mediated RNA/peptide complex coacervation as a model for intracellular liquid organelles. Nat Chem. 2016;8:129–137. doi: 10.1038/nchem.2414. [DOI] [PubMed] [Google Scholar]
- 197.Lu T, Spruijt E. Multiphase complex coacervate droplets. J Am Chem Soc. 2020;142:2905–2914. doi: 10.1021/jacs.9b11468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Vieregg JR, et al. Oligonucleotide–peptide complexes: phase control by hybridization. J Am Chem Soc. 2018;140:1632–1638. doi: 10.1021/jacs.7b03567. [DOI] [PubMed] [Google Scholar]
- 199.Cao M, et al. Peptide-induced DNA condensation into virus-mimicking nanostructures. ACS Appl Mater Interfaces. 2018;10:24349–24360. doi: 10.1021/acsami.8b00246. [DOI] [PubMed] [Google Scholar]
- 200.Yuan C, et al. Nucleation and growth of amino-acid and peptide supramolecular polymers through liquid–liquid phase separation. Angew Chem Int Ed Engl. 2019;58:18116–18123. doi: 10.1002/anie.201911782. [DOI] [PubMed] [Google Scholar]
- 201.Reznikov N, Steele JAM, Fratzl P, Stevens MM. A materials science vision of extracellular matrix mineralization. Nat Rev Mater. 2016;1:16041. [Google Scholar]
- 202.Espinosa AD, Rim JE, Barthelat F, Buehler BJ. Merger of structure and material in nacre and bone — perspectives on de novo biomimetic materials. Prog Mater Sci. 2009;54:1059–1100. [Google Scholar]
- 203.Chiti F, Dobson CM. Protein misfolding, amyloid formation, and human disease: A summary of progress over the last decade. Annu Rev Biochem. 2017;86:27–68. doi: 10.1146/annurev-biochem-061516-045115. [DOI] [PubMed] [Google Scholar]
- 204.Cao Y, Bolisetty S, Wolfisberg G, Adamcik J, Mezzenga R. Amyloid fibril-directed synthesis of silica core–shell nanofilaments, gels, and aerogels. Proc Natl Acad Sci USA. 2019;116:4012–4017. doi: 10.1073/pnas.1819640116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Li C, et al. Amyloid-hydroxyapatite bone biomimetic composites. Adv Mater. 2014;26:3207–3212. doi: 10.1002/adma.201306198. [DOI] [PubMed] [Google Scholar]
- 206.Le Norcy E, et al. Phosphorylated and non-phosphorylated leucine rich amelogenin peptide differentially affect ameloblast mineralization. Front Physiol. 2018;9:55. doi: 10.3389/fphys.2018.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Tavafoghi M, Cerruti M. The role of amino acids in hydroxyapatite mineralization. J R Soc Interface. 2016;13:20160462. doi: 10.1098/rsif.2016.0462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Newcomb CJ, Bitton R, Velichko YS, Snead ML, Stupp SI. The role of nanoscale architecture in supramolecular templating of biomimetic hydroxyapatite mineralization. Small. 2012;8:2195–2202. doi: 10.1002/smll.201102150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Lechner CC, Becker CFW. A sequence-function analysis of the silica precipitating silaffin R5 peptide. J Pept Sci. 2014;20:152–158. doi: 10.1002/psc.2577. [DOI] [PubMed] [Google Scholar]
- 210.Diaz D, Care A, Sunna A. Bioengineering strategies for protein-based nanoparticles. Genes. 2018;9:370. doi: 10.3390/genes9070370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Oh D, et al. M13 virus-directed synthesis of nanostructured metal oxides for lithium–oxygen batteries. Nano Lett. 2014;14:4837–4845. doi: 10.1021/nl502078m. [DOI] [PubMed] [Google Scholar]
- 212.Courchesne N-MD, et al. Constructing multifunctional virus-templated nanoporous composites for thin film solar cells: contributions of morphology and optics to photocurrent generation. J Phys Chem C. 2015;119:13987–14000. [Google Scholar]
- 213.Nyström G, Fernández-Ronco MP, Bolisetty S, Mazzotti M, Mezzenga R. Amyloid templated gold aerogels. Adv Mater. 2016;28:472–478. doi: 10.1002/adma.201503465. [DOI] [PubMed] [Google Scholar]
- 214.Bolisetty S, Mezzenga R. Amyloid–carbon hybrid membranes for universal water purification. Nat Nanotechnol. 2016;11:365–371. doi: 10.1038/nnano.2015.310. [DOI] [PubMed] [Google Scholar]
- 215.Ding Y, et al. Self-assembly of antimicrobial peptides on gold nanodots: against multidrug-resistant bacteria and wound-healing application. Adv Funct Mater. 2015;25:7189–7199. [Google Scholar]
- 216.Ding Y, et al. Silver mineralization on self-assembled peptide nanofibers for long term antimicrobial effect. J Mater Chem. 2012;22:2575–2581. [Google Scholar]
- 217.Ji W, et al. Metal-ion modulated structural transformation of amyloid-like dipeptide supramolecular self-assembly. ACS Nano. 2019;13:7300–7309. doi: 10.1021/acsnano.9b03444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Yan X, Zhu P, Fei J, Li J. Self-assembly of peptide-inorganic hybrid spheres for adaptive encapsulation of guests. Adv Mater. 2010;22:1283–1287. doi: 10.1002/adma.200901889. [DOI] [PubMed] [Google Scholar]
- 219.Hamely IA, Kirkham S, Dehsorkhi A, Castelletto V. Self-assembly of a model peptide incorporating a hexa-histidine sequence attached to an oligo-alanine sequence, and binding to gold NTA/nickel nanoparticles. Biomacromolecules. 2014;15:3412–3420. doi: 10.1021/bm500950c. [DOI] [PubMed] [Google Scholar]
- 220.Stevens MM, Flynn NT, Wang C, Tirrell DA, Langer R. Coiled-coil peptide-based assembly of gold nanoparticles. Adv Mater. 2004;16:915–918. [Google Scholar]
- 221.Fichman G, et al. Seamless metallic coating and surface adhesion of self-assembled bioinspired nanostructures based on di-(3,4-dihydroxy-l-phenylalanine) peptide motif. ACS Nano. 2014;8:7220–7228. doi: 10.1021/nn502240r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Guterman T, et al. Electrical conductivity, selective adhesion, and biocompatibility in bacteria-inspired peptide-metal self-supporting nanocomposites. Adv Mater. 2019;31:1807285. doi: 10.1002/adma.201807285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Liu Q, Wang H, Shi X, Wang Z-G, Ding B. Self-assembled DNA/peptide-based nanoparticle exhibiting synergistic enzymatic activity. ACS Nano. 2017;11:7251–7258. doi: 10.1021/acsnano.7b03195. [DOI] [PubMed] [Google Scholar]
- 224.Liu K, et al. Mimicking primitive photobacteria: sustainable hydrogen evolution based on peptide–porphyrin co-assemblies with a self-mineralized reaction center. Angew Chem Int Ed Engl. 2016;55:12503–12507. doi: 10.1002/anie.201606795. [DOI] [PubMed] [Google Scholar]
- 225.Liu K, Zhang H, Xing R, Zou Q, Yan X. Biomimetic oxygen-evolving photobacteria based on amino acid and porphyrin hierarchical self-organization. ACS Nano. 2017;11:12840–12848. doi: 10.1021/acsnano.7b08215. [DOI] [PubMed] [Google Scholar]
- 226.Han J, Liu K, Chang R, Zhao L, Yan X. Photooxidase-mimicking nanovesicles with superior photocatalytic activity and stability based on amphiphilic amino acid and phthalocyanine co-assembly. Angew Chem Int Ed Engl. 2019;58:2000–2004. doi: 10.1002/anie.201811478. [DOI] [PubMed] [Google Scholar]
- 227.Jiang L, Yang S, Lund R, Dong H. Shape-specific nanostructured protein mimics from de novo designed chimeric peptides. Biomater Sci. 2018;6:272–279. doi: 10.1039/c7bm00906b. [DOI] [PubMed] [Google Scholar]
- 228.Nambiar M, Wang L-S, Rotello V, Chmielewski J. Reversible hierarchical assembly of trimeric coiled-coil peptides into banded nano- and microstructures. J Am Chem Soc. 2018;140:13028–13033. doi: 10.1021/jacs.8b08163. [DOI] [PubMed] [Google Scholar]
- 229.Yuan C, et al. Hierarchically oriented organization in supramolecular peptide crystals. Nat Rev Chem. 2019;3:567–588. [Google Scholar]
- 230.Ardoña HAM, Tovar JD. Peptide π electron conjugates: organic electronics for biology? Bioconjugate Chem. 2015;26:2290–2302. doi: 10.1021/acs.bioconjchem.5b00497. [DOI] [PubMed] [Google Scholar]
- 231.Panda SS, Katz HE, Tovar JD. Solid-state electrical applications of protein and peptide based nanomaterials. Chem Soc Rev. 2018;47:3640–3658. doi: 10.1039/c7cs00817a. [DOI] [PubMed] [Google Scholar]
- 232.Tao K, Makam P, Aizen R, Gazit E. Self-assembling peptide semiconductors. Science. 2017;358:eaam9756. doi: 10.1126/science.aam9756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Sun B, et al. Photoactive properties of supramolecular assembled short peptides. Chem Soc Rev. 2019;48:4387–4400. doi: 10.1039/c9cs00085b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Gazit E. Aromatic dipeptides light up. Nat Nanotechnol. 2016;11:309–310. doi: 10.1038/nnano.2015.321. [DOI] [PubMed] [Google Scholar]
- 235.Ni N, et al. Self-assembling amyloid-like peptides as exogenous second harmonic probes for bioimaging applications. J Biophotonics. 2019;12:e201900065. doi: 10.1002/jbio.201900065. [DOI] [PubMed] [Google Scholar]
- 236.Chen Y, et al. High-efficiency fluorescence through bioinspired supramolecular self-assembly. ACS Nano. 2020;14:2798–2807. doi: 10.1021/acsnano.9b10024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Lou S, Wang X, Yu Z, Shi L. Peptide tectonics: encoded structural complementarity dictates programmable self-assembly. Adv Sci. 2019;6:1802043. doi: 10.1002/advs.201802043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.John EA, Massena CJ, Berryman OB. Helical anion foldamers in solution. Chem Rev. 2020;120:2759–2782. doi: 10.1021/acs.chemrev.9b00583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Yoo SH, Lee H-S. Foldectures: 3D molecular architectures from self-assembly of peptide foldamers. Acc Chem Res. 2017;50:832–841. doi: 10.1021/acs.accounts.6b00545. [DOI] [PubMed] [Google Scholar]
- 240.Martinek TA, Fülöp F. Peptidic foldamers: ramping up diversity. Chem Soc Rev. 2012;41:687–702. doi: 10.1039/c1cs15097a. [DOI] [PubMed] [Google Scholar]
- 241.Valery C, et al. Biomimetic organization: Octapeptide self-assembly into nanotubes of viral capsid-like dimension. Proc Natl Acad Sci USA. 2003;100:10258–10262. doi: 10.1073/pnas.1730609100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Matsuura K. Synthetic approaches to construct viral capsid-like spherical nanomaterials. Chem Commun. 2018;54:8944–8959. doi: 10.1039/c8cc03844a. [DOI] [PubMed] [Google Scholar]
- 243.Matsuura K, Honjo T. Artificial viral capsid dressed up with human serum albumin. Bioconjugate Chem. 2019;30:1636–1641. doi: 10.1021/acs.bioconjchem.9b00327. [DOI] [PubMed] [Google Scholar]
- 244.Matsuura K, Ota J, Fujita S, Shiomi Y, Inaba H. Construction of ribonuclease-decorated artificial virus-like capsid by peptide self-assembly. J Org Chem. 2020;85:1668–1673. doi: 10.1021/acs.joc.9b02295. [DOI] [PubMed] [Google Scholar]
- 245.Ni R, Chau Y. Structural mimics of viruses through peptide/DNA co-assembly. J Am Chem Soc. 2014;136:17902–17905. doi: 10.1021/ja507833x. [DOI] [PubMed] [Google Scholar]
- 246.Ni R, Chau Y. Tuning the inter-nanofibril interaction to regulate the morphology and function of peptide/ DNA co-assembled viral mimics. Angew Chem Int Ed Engl. 2017;56:9356–9360. doi: 10.1002/anie.201703596. [DOI] [PubMed] [Google Scholar]
- 247.Ni R, Chau Y. Nanoassembly of oligopeptides and DNA mimics the sequential disassembly of a spherical virus. Angew Chem Int Ed Engl. 2020;59:3578–3584. doi: 10.1002/anie.201913611. [DOI] [PubMed] [Google Scholar]
- 248.Zhang C, et al. Virus-inspired self-assembled nanofibers with aggregation-induced emission for highly efficient and visible gene delivery. ACS Appl Mater Interfaces. 2017;9:4425–4432. doi: 10.1021/acsami.6b11536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Parodi A, et al. Bio-inspired engineering of cell- and virus-like nanoparticles for drug delivery. Biomaterials. 2017;147:155–168. doi: 10.1016/j.biomaterials.2017.09.020. [DOI] [PubMed] [Google Scholar]
- 250.Cai Y, et al. Recent progress in supramolecular peptide assemblies as virus mimics for cancer immunotherapy. Biomater Sci. 2020;8:1045–1057. doi: 10.1039/c9bm01380f. [DOI] [PubMed] [Google Scholar]
- 251.Knowles TPJ, Vendruscolo M, Dobson CM. The amyloid state and its association with protein misfolding diseases. Nat Rev Mol Cell Biol. 2014;15:384–396. doi: 10.1038/nrm3810. [DOI] [PubMed] [Google Scholar]
- 252.Cohen SIA, Vendruscolo M, Dobson CM, Knowles TPJ. Nucleated polymerization with secondary pathways. III. Equilibrium behavior and oligomer populations. J Chem Phys. 2011;135:065107. doi: 10.1063/1.3608918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Knowles TPJ, et al. An analytical solution to the kinetics of breakable filament assembly. Science. 2009;326:1533–1537. doi: 10.1126/science.1178250. [DOI] [PubMed] [Google Scholar]
- 254.Dobson CM. Chemical space and biology. Nature. 2004;432:824–828. doi: 10.1038/nature03192. [DOI] [PubMed] [Google Scholar]
- 255.Cohen SIA, et al. Proliferation of amyloid-β42 aggregates occurs through a secondary nucleation mechanism. Proc Natl Acad Sci USA. 2013;110:9758–9763. doi: 10.1073/pnas.1218402110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Fowler DM, Koulov AT, Balch WE, Kelly JW. Functional amyloid — from bacteria to humans. Trends Biochem Sci. 2007;32:217–224. doi: 10.1016/j.tibs.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 257.Otzen D, Riek R. Functional amyloids. Cold Spring Harb Perspect Biol. 2019;11:a033860. doi: 10.1101/cshperspect.a033860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Appel EA, et al. Self-assembled hydrogels utilizing polymer–nanoparticle interactions. Nat Commun. 2015;6:6295. doi: 10.1038/ncomms7295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Benton G, Arnaoutova I, George J, Kleinman HK, Koblinski J. Matrigel: from discovery and ECM mimicry to assays and models for cancer research. Adv Drug Deliv Rev. 2014;79-80:3–18. doi: 10.1016/j.addr.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 260.Panda JJ, Chauhan VS. Short peptide based self-assembled nanostructures: implications in drug delivery and tissue engineering. Polym Chem. 2014;5:4418–4436. [Google Scholar]
- 261.Hamley IW, et al. Self-assembly of a model amphiphilic oligopeptide incorporating an arginine headgroup. Soft Matter. 2013;9:4794–4801. [Google Scholar]
- 262.Fleming S, Debnath S, Frederix PW, Hunt NT, Ulijn RV. Insights into the coassembly of hydrogelators and surfactants based on aromatic peptide amphiphiles. Biomacromolecules. 2014;15:1171–1184. doi: 10.1021/bm401720z. [DOI] [PubMed] [Google Scholar]
- 263.Liyanage W, Vats K, Rajbhandary A, Benoit DS, Nilsson BL. Multicomponent dipeptide hydrogels as extracellular matrix-mimetic scaffolds for cell culture applications. Chem Commun. 2015;51:11260–11263. doi: 10.1039/c5cc03162a. [DOI] [PMC free article] [PubMed] [Google Scholar]