Abstract
Objective
Automatic segmentation of vestibular schwannomas (VS) from magnetic resonance imaging (MRI) could significantly improve clinical workflow and assist patient management. Accurate tumour segmentation and volumetric measurements provide the best indicator to detect subtle VS growth but current techniques are labour-intensive and dedicated software is not readily available within the clinical setting. We aim to develop a novel artificial intelligence (AI) framework to be embedded in the clinical routine for automatic delineation and volumetry of VS.
Methods
Imaging data (contrast-enhanced T1-weighted (ceT1) and high-resolution T2-weighted (hrT2) MR images) from all patients meeting the study’s inclusion/exclusion criteria who had a single sporadic VS treated with Gamma Knife Stereotactic Radiosurgery were used to create our model. We developed a novel artificial intelligence (AI) framework based on a 2.5D convolutional neural network (CNN) to exploit the different in-plane and through-plane resolutions encountered in standard clinical imaging protocols. We used a computational attention module to enable the CNN to focus on the small VS target and propose a supervision on the attention map for more accurate segmentation. The manually-segmented target tumour volume (also tested for inter-observer variability) was used as the ground truth for training and evaluation of the CNN. We quantitatively measured the Dice score, average symmetric surface distance (ASSD) and relative volume error (RVE) of the automated segmentation results in comparison to manual segmentations to assess the model’s accuracy.
Results
Imaging data from all eligible patients (n=243) were randomly split into three non-overlapping groups for training (n=177), hyper-parameter tuning (n=20) and testing (n=46). Dice, ASSD and RVE scores were measured on the testing set for the respective input data types as follows: ceT1: 93.43%, 0.203mm, 6.96%; hrT2: 88.25%, 0.416mm, 9.77%; combined ceT1/hrT2: 93.68%, 0.199mm, 7.03%. Given a margin of 5% for the Dice score, the automated method was shown to achieve statistically equivalent performance in comparison to an annotator using ceT1 images alone (p=4e-13) and combined ceT1/hrT2 images (p=7e-18) as inputs.
Conclusions
We have a developed a robust AI framework for automatically delineating and calculating VS tumour volume achieving excellent results, equivalent to that achieved by an independent human annotator. This promising AI technology has the potential to improve the management of patients with VS and potentially other brain tumours.
Keywords: Vestibular schwannoma, artificial intelligence, convolutional neural network, segmentation, MRI, tumour
Introduction
Diagnosis of vestibular schwannoma (VS) has risen significantly in recent years and is now estimated to be between 14 and 20 cases per million per year 6,22,27. In the UK, this equates to approximately 1400 – 1500 new patients being diagnosed every year. The widespread availability of diagnostic MRI has notably resulted in a greater number of asymptomatic patients being diagnosed with small VS 27. For smaller tumours, expectant management with serial imaging is often advised 26 with treatment decisions based on the tumour’s maximal extra-meatal linear dimension 11,26.
However, linear measurements are not the most sensitive method of measuring a tumour’s size and several studies have demonstrated that a volumetric measurement is a more accurate method of calculating a vestibular schwannoma’s true size 18,24,29,33,34,35. Such methods are also superior at detecting subtle growth 33. The principal reason such volumetric methods have not been widely implemented is because the currently available tools makes segmenting (or contouring) the tumour and calculating volume a labour-intensive process with no dedicated software seamlessly implemented in the clinical scanners or reporting workstations and broadly available. MacKeith et al. recently described their experience of using a state-of-the-art commercially available semi-automated method for segmenting VS 18 highlighting the speed of the technique compared with older semi-automated 10,32 and manual segmentation methods 4,33. Nonetheless, it required the operator to identify the tumour and initiate the segmentation process 18 and thus potentially suffers from inter-operator variations.
An automated segmentation tool would also benefit the tumour contouring process that is key to the planning and treatment of vestibular schwannomas with Gamma Knife Stereotactic Radiosurgery (GK SRS). Current Gamma Knife planning software uses an in-plane semi-automated segmentation method enabling the user to manually segment each axial slice in turn. This is a relatively time-consuming task that could be improved by the availability of an automated segmentation tool.
In this study, we describe, to the best of our knowledge, the first fully automated method of segmenting VS from MRI, for which we developed a novel artificial intelligence (AI) deep machine learning framework. AI refers to computing technologies inspired by processes associated with human intelligence. Machine learning describes a system’s capability to acquire statistical knowledge by extracting patterns from training data and learning rules to make predictions for a given predefined task based on these patterns. Deep machine learning, or simply deep learning (DL), methods take this process a step further, enabling the computer to not only statistically reason on extracted patterns but also to build its own rich and complex visual representations out of simple mathematical operations cascaded into increasingly higher-level feature extractors 9. Convolutional Neural Networks (CNNs), often simply referred to as networks by machine learning practitioners, are the most commonly used deep learning models in medical image analysis and have achieved state-of-the-art performance for many segmentation tasks 2,15. These AI models for medical image segmentation applications are typically trained in a supervised manner with a set of annotated training images (e.g. manual segmentations) providing the network with expected input-output data pairs.
The choice of network structure is a key decision when designing CNN-based segmentation models. Most of the previously described CNN methods were designed to segment anatomical images with an isotropic resolution and are thus not very well suited to anisotropic routine clinical brain imaging protocols which present with high in-plane and low through-plane resolution 17. A two-dimensional (2D) CNN structure achieves a relatively low computational memory requirement by analysing data in a slice-by-slice manner but the network ignores three-dimensional (3D) information that ultimately limits its segmentation performance 25. 3D CNNs can better exploit 3D features but typically require a large amount of memory which may limit its representation power [15]. A 2.5D CNN, exploiting refined in-plane but coarse through-plane feature extraction, is a compromise between the 2D and 3D network; it has the advantage of being able to use inter-slice features absent in 2D networks but requires less memory than the more complex 3D networks 37.
Vestibular schwannomas are relatively small compared with the entire brain region. For VS segmentation, the ability of the CNN to segment small structures from large image contexts is thus highly desired. To address the segmentation of small structures, Yu et al. 42 used a coarse-to-fine approach with recurrent saliency transformation. Oktay et al.’s 23 method learned a computational attention map to locate the region of interest within the larger image context enabling the CNN model to then focus on target structures. However, the attention map used by Oktay et al. was not explicitly supervised during training so may not have been well-aligned with the target region, limiting segmentation accuracy. Complementary approaches to deal with small structures include the use of adapted loss functions such as Dice loss 21, generalized Dice loss 28 and Focal loss 16. These methods automatically address the imbalance between foreground and background voxels, but treat all the voxels equally during training. Considering the fact that some voxels are harder than the others to learn during training, we proposed a hardness-weighted Dice loss function to further improve the segmentation accuracy 40.
In this study, we describe a novel AI framework to automatically segment vestibular schwannomas using both contrast-enhanced T1-weighted (ceT1) and high-resolution T2-weighted images (hrT2). The method was trained and evaluated using MR images from patients with VS who had previously undergone Gamma Knife stereotactic radiosurgery.
Methods
Ethics statement
This study was approved by the NHS Health Research Authority and Research Ethics Committee (18/LO/0532). Because patients were selected retrospectively and the MR images were completely anonymised before analysis, no informed consent was required for the study.
Study population
Imaging data from consecutive patients with a single sporadic VS treated with GK SRS between October 2012 and January 2018 were screened for the study. All adult patients aged over 18 years with a single, unilateral VS treated with GK SRS were eligible for inclusion in the study, including patients who had previously undergone operative surgical treatment. Two hundred and forty-eight patients (M:F 97:151; median age 56 years, IQR 47 – 65 years) met this initial inclusion criteria. All patients had a MR performed on a 1.5T scanner (Avanto Siemens Healthineers, Erlangen, Germany) including a ceT1 MRI acquired with in-plane resolution of 0.4 x 0.4 mm, in-plane matrix of 512 x 512 and a slice thickness of 1.5mm (TR (Repetition time) = 1900 ms, TE (Echo time) = 2.97 ms, TI (Inversion time) = 1100 ms) and a hrT2 MRI with in-plane resolution of 0.5 x .0.5 mm, in-plane matrix of 384 x 384 and a slice thickness of 1.0 – 1.5 mm (TR = 9.4 ms, TE = 4.23 ms). Patients were only included in the study if their pre-treatment image acquisition dataset was complete; two patients were thus excluded because of incomplete datasets.
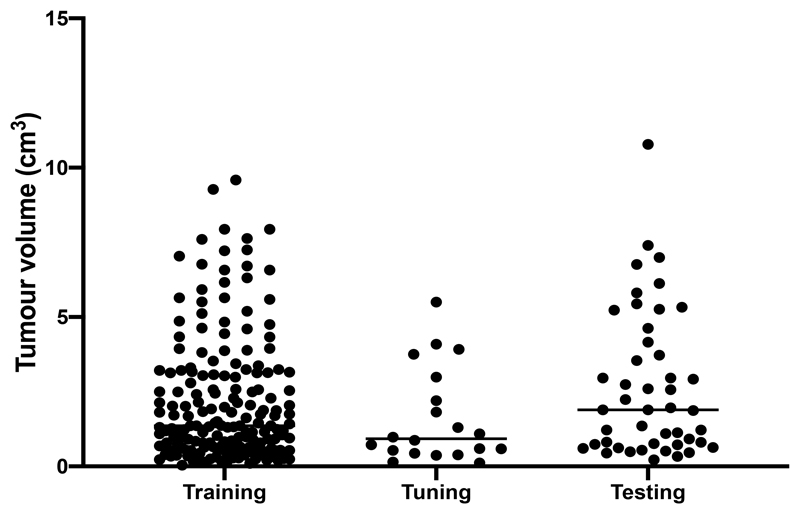
We randomly split the final 246 patients into three non-overlapping groups: 180 for training, 20 for hyper-parameter tuning and 46 for testing with median tumour volumes of 1.36 cm3 (range 0.04 – 9.59 cm3, IQR 0.63 – 3.15 cm3), 0.92 cm3 (range 0.12 – 5.50, IQR 0.51 – 2.40 cm3) and 1.89 cm3 (range 0.22 – 10.78, IQR 0.74 – 4.05 cm3), respectively (Figure 1). Thirty-five patients (19%) in the training dataset had undergone previous surgery compared to 2 patients (20%) in the hyper-parameter tuning set and 14 patients (30%) in the testing dataset.
Figure 1.
Vestibular schwannoma tumour volumes (cm3) of the training, hyper-parameter tuning and testing datasets used to develop the AI framework
Manual Segmentations
For each patient, the target tumour volume was manually segmented in consensus by the treating team including a neurosurgeon (RB/NK) and physicist (IP/AD) using both the ceT1 and hrT2 images. The target tumour volume was then considered as the ground truth for training and testing of our AI framework. Forty-six images were also manually segmented by a third neurosurgeon (JS), blinded to the original manual segmentation in order to test inter-observer variability. All manual segmentations were performed using Gamma Knife planning software (Leksell GammaPlan, Elekta, Sweden) that employs an in-plane semi-automated segmentation method. Using this software, each axial slice was manually segmented in turn.
Automated Segmentations using Artificial Intelligence
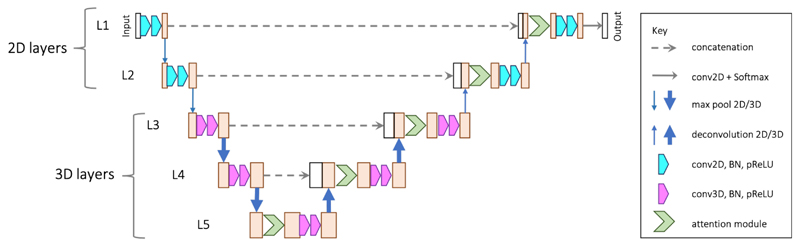
As described in more technical depth in our preliminary methodological study, we developed a novel attention-based 2.5D CNN combining 2D and 3D convolutions 40 to fully automate the process of segmenting VS. As shown in Figure 2, the main structure follows a typical encoder-decoder design as implemented in the widely used U-Net 25. The encoder contains five levels of convolutions. The first two levels (L1-L2) and the others use 2D and 3D convolutions/poolings, respectively. This is motivated by the fact that the in-plane resolution of our VS tumour images is 2 – 3 times that of the through-plane resolution. After the first two max-pooling layers that down-sample the feature maps only in 2D, the feature maps in L3 and followings have a near-isotropic 3D resolution. The output feature channel number of the convolutions at level l is denoted as Nl with Nl being set as 16l in our experiments.
Figure 2.
The proposed 2.5D U-Net with spatial attention for VS tumour segmentation from anisotropic MRI.
Note that our network is different from previous works that refer to fusing purely 2D networks in three orthogonal views as a 2.5D network 14,19. These indeed have a limited ability to exploit 3D features. Existing, more advanced, 2.5D CNNs 38 use inter-slice and intra-slice convolution to exploit 3D features but are limited to dealing with images with isotropic resolution. Our network goes beyond this limitation and is specifically designed to deal with anisotropic input volumes. It ensures a near-isotropic 3D physical receptive field (in terms of mm rather than voxels) along each axis.
To deal with the small target region, we added a spatial attention module to each level of the decoder. An attention module gives a score of relative importance for each spatial position. The module learns to give higher attention scores to voxels in the target region and lower attention scores to voxels in the background. Therefore, it enables the network to focus more on the small tumour target while suppressing irrelevant background. The proposed attention module consists of two convolutional layers followed by a sigmoid activation function to obtain the attention scores. As part of this attention module, we developed an attention loss to explicitly supervise the learning of spatial attention that broadly defined the tumour region of interest within the larger image. Previous works have shown that spatial attention can be automatically learned in CNNs to enable the network to focus on the target region in a large image context 23. We developed our network with explicit supervision on the attention map to further ensure more accurate results.
Thirdly, we adapted the usual Dice loss function frequently used to train CNNs 21. The Dice loss function has shown good performance in dealing with images that have an imbalanced foreground and background voxels. However, when segmenting small structures with low contrast, some voxels are harder than the others to learn. Treating all the voxels for a certain class equally may limit the performance of CNNs on hard voxels so our network defined a voxel-level difficulty weight that automatically gives higher weights to voxels mis-classified by the CNNs. Let pci represent the probability of voxel i belonging to class c predicted by the CNN and gci denote the corresponding probability value in the ground truth. The hardness-weighted Dice loss (HDL) is defined as:
where C is the class number that is 2 in our binary segmentation task. ϵ is a small number for numerical stability and set as 10−5 in our experiments. The weighting coefficient is defined as:
Network training
The networks were implemented in Tensorflow and NiftyNet 8 on a Ubuntu desktop with 32GB RAM and an NVIDIA GTX 1080 Ti GPU. For training, we used Adam optimizer with weight decay 10-7, batch size 2, and iteration number 30k where performance on the hyper-parameter tuning set stopped to increase. The learning rate was initialized to 10-4 and halved every 10k. We trained the networks respectively to segment vestibular schwannoma tumours from different modalities: 1) ceT1 images, 2) hrT2 images, and 3) a combined dataset including both imaging modalities.
Deep learning CNNs are nonlinear methods that learn via a stochastic training algorithm. This makes them sensitive to the specifics of the training dataset with the potential to generate results with high variance. One method to reduce this variance is to perform ensemble learning whereby the network is used to train multiple models and the results are combined 13. This may be achieved by varying the training dataset, the choice of models used in the ensemble or a combination thereof. For our network, we resampled the training dataset with replacement and trained the network five times using the final Baseline + SpvA + HDL model, taking their majority voting results as the final segmentations.
Testing and statistical analysis
For quantitative analysis, we measured the Dice, Average Symmetric Surface Distance (ASSD) and Relative Volume Error (RVE) scores. The Dice score is a proven statistical validation metric to evaluate the performance and spatial overlap between two sets of segmentations of the same anatomy 43. Dice is represented as a percentage with 100% being a perfect voxel-wise match between results. The ASSD is measured in millimetres and is determined by the average spatial distance between the border voxels of the automated segmentation results and the ground truth. Border voxels are defined as those voxels in the tumour that have at least one neighbour that does not belong to the tumour. A lower ASSD value indicates a better agreement, with ASSD = 0 representing a perfect agreement in the segmentation boundary. Finally, the RVE is an approximation error between an exact value and in this case, the network’s approximation to it. To calculate the RVE, the total volume of the segmentation is divided by the total volume of the ground truth and is then represented as a percentage. An RVE of 0 indicates a perfect segmentation. We calculated Dice, ASSD and RVE scores for the AI network using 1) ceT1 images, 2) hrT2 images, and 3) a combined dataset including both imaging modalities.
When describing our results, we refer to the basic network as the “Baseline” model with the additional supervised attention module (SpvA) and Hardness-weighted Dice Loss (HDL) function implemented and analysed sequentially. The model’s final Ensemble results are presented and computational times for each test are documented.
We also assessed if our AI model performed comparably to another independent neurosurgeon annotator (still considering the first manual segmentation in consensus as ground truth). Using bespoke software, we analysed this by testing the equivalence of the paired error means using a two one-sided test procedure for paired-samples (TOST-P) 30. For this experiment, we considered the mean error in Dice scores between the manual annotations and our algorithmic outputs (μalgo-gt) and that between the two clinical annotators (μinter-observer), with a margin δ of 5% deemed to be equivalent. Specifically, our null hypothesis stated that the difference between the two mean errors was expected to fall outside our selected equivalence interval (-δ,δ); H0: μinter-observer - μalgo-gt <-δ and μinter-observer - μalgo-gt >δ. Thus, if both of these one-sided tests are rejected, we may conclude that the paired means are equivalent.
Results
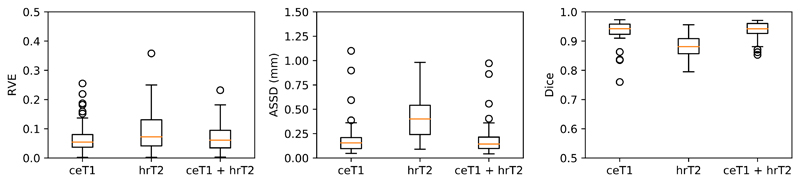
Compared to the ground truth annotations, the ensembled results for our AI framework generated a Dice score of 93.43% (SD 3.97%) for ceT1 images alone, 88.25% (SD 3.90%) for hrT2 images and 93.68 (SD 2.80%) for the combined dataset. Similarly, ASSD scores of 0.203 mm (SD 0.196), 0.416 mm (SD 0.209) and 0.199 mm (SD 0.181). The algorithm was more likely to over-estimate tumour volume and RVEs of 6.96% (SD 5.68%), 9.77% (SD 7.56%); and 7.03% (SD 5.04%) were obtained for ceT1, hrT2 and combined datasets, respectively (Table 1, Figure 3). The results also demonstrated an incremental improvement in segmentation accuracy with the use of a Supervised Attention module and a Hardness-weighted Dice Loss function compared with a baseline 2.5D U-Net (Table 1). Figures 4, 5 and 6 provide illustrative examples of the best, average and worst segmentation results using the AI framework across all patients and the five inferences used for ensemble learning. Mean computational times of 3.42 s (SD 0.37) to 3.87 s (SD 0.42) per image were observed in testing (Table 1). The same hardware was used for training and testing the AI framework.
Table 1.
Comparison of different AI methods for VS tumour segmentation from contrast-enhanced T1-weighted images (ceT1) alone, high resolution T2-weighted (hrT2) images alone and combined dataset (ceT1 and hrT2). ASSD: Average Symmetric Surface Distance, RVE: Relative Volume Error. Ground truth manually-calculated tumour volume: 2.66 ± 2.43 cm3
| Image sequence(s) | Method | Dice (%) | ASSD (mm) | RVE (%) | Volume (cm3) | Runtime (s) |
|---|---|---|---|---|---|---|
| ceT1 | Baseline | 92.21 ± 5.64 | 0.305 ± 0.507 | 8.90 ± 7.94 | 2.70 ± 2.51 | 3.46 ± 0.41 |
| Baseline + SpvA | 93.05 ± 4.61 | 0.226 ± 0.273 | 8.25 ± 6.36 | 2.70 ± 2.46 | 3.48 ± 0.41 | |
| Baseline + SpvA + HDL | 93.08 ± 4.85 | 0.218 ± 0.247 | 7.55 ± 8.33 | 2.67 ± 2.44 | 3.49 ± 0.40 | |
| Ensemble | 93.43 ± 3.97 | 0.203 ± 0.196 | 6.96 ± 5.68 | 2.68 ± 2.45 | 17.48 ± 2.02 | |
| hrT2 | Baseline | 85.71 ± 7.06 | 0.663 ± 0.451 | 15.98 ± 14.65 | 2.85 ± 2.55 | 3.42 ± 0.37 |
| Baseline + SpvA | 86.72 ± 4.98 | 0.525 ± 0.292 | 13.38 ± 9.33 | 2.85 ± 2.60 | 3.45 ± 0.41 | |
| Baseline + SpvA + HDL | 87.30 ± 4.89 | 0.433 ± 0.315 | 12.11 ± 8.92 | 2.67 ± 2.47 | 3.45 ± 0.42 | |
| Ensemble | 88.25 ± 3.90 | 0.416 ± 0.209 | 9.77 ± 7.56 | 2.67 ± 2.48 | 17.20 ± 2.08 | |
| Combined dataset (ceT1 + hrT2) | Baseline | 92.47 ± 5.39 | 0.492 ± 0.427 | 8.81 ± 7.02 | 2.80 ± 2.56 | 3.83 ± 0.45 |
| Baseline + SpvA | 92.91 ± 3.78 | 0.263 ± 0.385 | 8.37 ± 6.79 | 2.73 ± 2.49 | 3.87 ± 0.43 | |
| Baseline + SpvA + HDL | 93.19 ± 3.59 | 0.212 ± 0.219 | 7.57 ± 5.96 | 2.80 ± 2.53 | 3.87 ± 0.42 | |
| Ensemble | 93.68 ± 2.80 | 0.199 ± 0.181 | 7.03 ± 5.04 | 2.74 ± 2.50 | 19.26 ± 2.15 |
Figure 3.
Automated segmentation results for the testing dataset (n = 46). Dice, Average Symmetric Surface Distance (ASSD) and Relative Volume Error (RVE) scores for the testing dataset of 46 patients according to the input image. ceT1: contrast-enhanced T1-weighted image, hrT2: high-resolution T2-weighted image
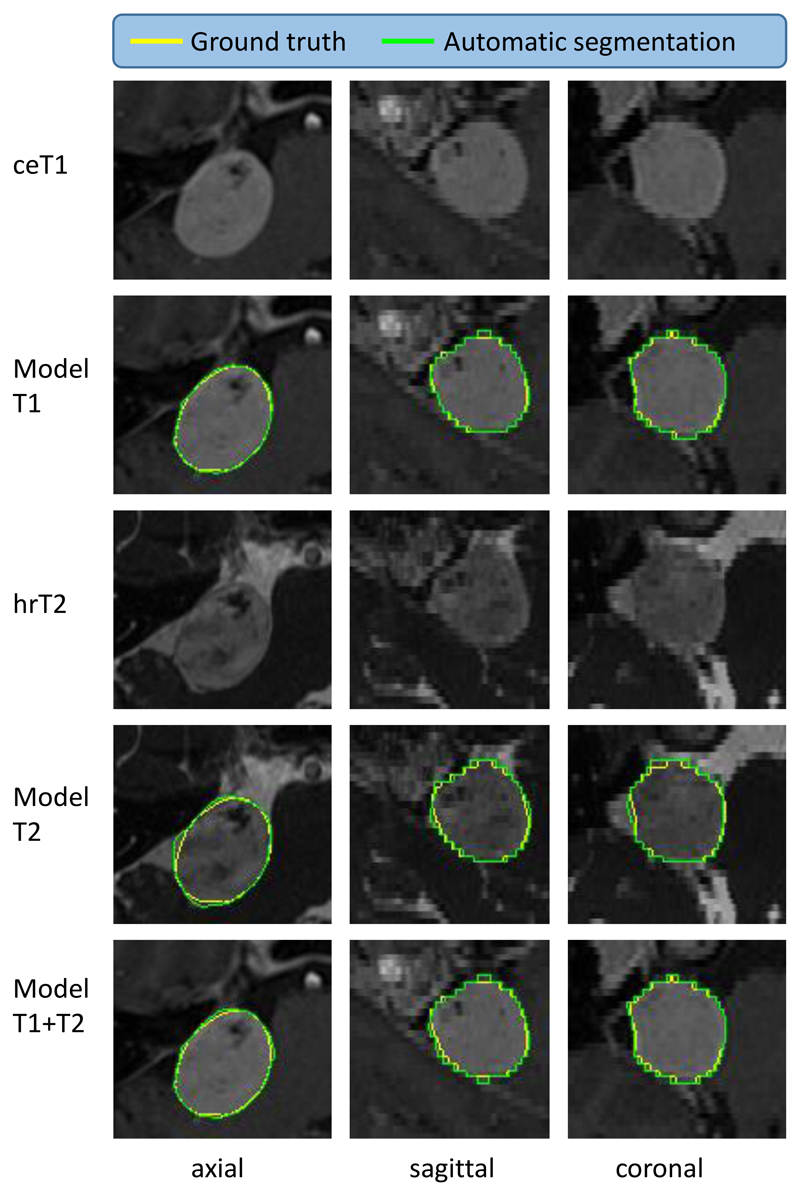
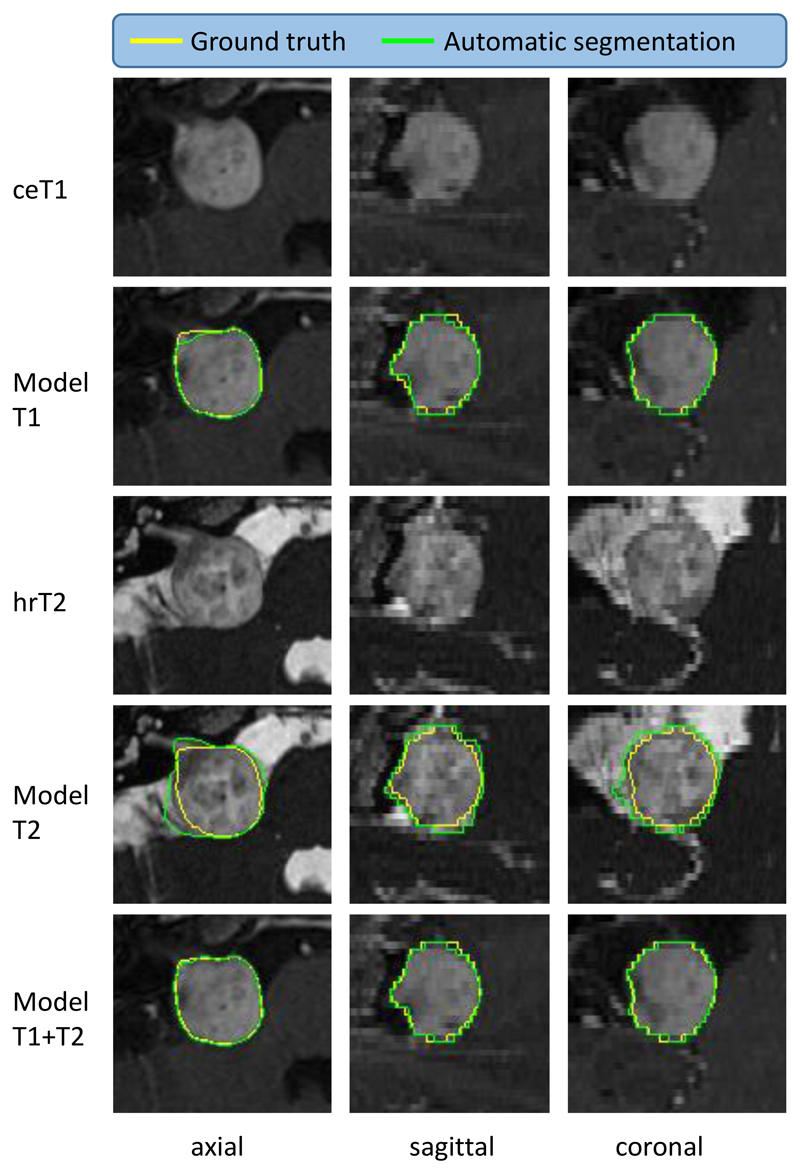
Figure 4.
Illustrative example of the best automated segmentation results (patient ID: 246). ceT1: contrast-enhanced T1-weighted image, hrT2: high-resolution T2-weighted image. Model results generated by AI model. Yellow: Manual ground truth, Green: Automated segmentation. Dice of Model T2 is 95.60%.
Figure 5.
Illustrative example of average automated segmentation results (patient ID: 206). ceT1: contrast-enhanced T1-weighted image, hrT2: high-resolution T2-weighted image. Model results generated by AI model. Yellow: Ground truth, Green: Segmentation. Dice of Model T2 is 84.80%.
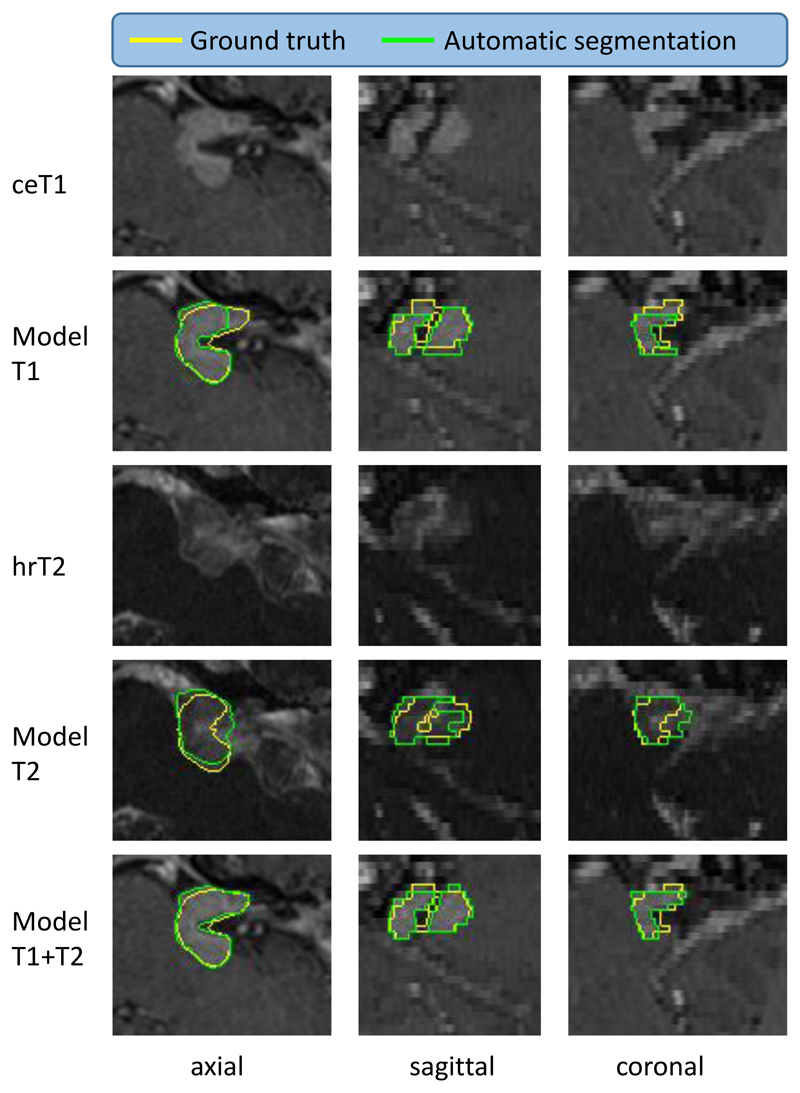
Figure 6.
Illustrative example of the worst automated segmentation results (patient ID: 238). ceT1: contrast-enhanced T1-weighted image, hrT2: high-resolution T2-weighted image. Model results generated by AI model. Yellow: Ground truth, Green: Segmentation. Dice of Model T2 is 79.50%. This patient underwent Gamma Knife treatment following a subtotal translabyrinthine resection of their VS tumour.
Finally, we determined if our AI model performed comparably to another independent neurosurgeon annotator. Inter-observer variability testing between clinical annotators recorded a Dice score of 93.82% (SD 3.08%), an ASSD score of 0.269mm (SD 0.095) and a RVE of 5.55% (SD 4.75%) between the two sets of manual annotations. Given a margin of 5% for the Dice score, our method is statistically equivalent to another annotator using ceT1 images alone as input (p=4e-13) and both the ceT1 and hrT2 images as inputs (p=7e-18).
Discussion
In this work, we describe the first fully automated method for segmenting vestibular schwannoma tumours using a deep learning AI model and show performance on par with inter-observer variability. We have developed a novel 2.5D CNN capable of generating automatic VS segmentations using standard clinical sequences requiring no user interaction. Our network is specifically designed for images with high in-plane resolution and low through-plane resolution and incorporates a multi-scale spatial attention mechanism and a novel hardness-weighted Dice loss function to deal with the small target tumour region. The choice of a 2.5D network was a trade-off between standard 2D and 3D CNNs however its lower memory demands will facilitate its implementation within current healthcare infrastructure. Applying 2D CNNs slice-by-slice will ignore inter-slice correlation and 3D context. To properly deal with anisotropic images, the application of standard 3D CNNs requires upsampling input images to isotropic 3D resolution. While this balances the physical receptive field along each axis, processing upsampled images requires more memory. This may limit the accuracy of the CNN by adding more stringent constraints on its depth and number of features. These limitations were highlighted in our preliminary study40 where we demonstrated that 2.5D networks outperform their 2D and 3D counterparts for VS tumour segmentation from anisotropic MRI. An attention module, designed with explicit end-to-end supervision was implemented to enable the CNN to focus on the target tumour region and we also introduced a hardness-weighted Dice loss function to boost the performance of the network.
The proposed AI model demonstrated excellent concordance between the automated results and the manually segmented ground truth. The network returned Dice scores of 93% for ceT1 and combined ceT1/hrT2 image datasets that were statistically equivalent to another clinical annotator. These results suggest that our network is sufficiently robust to perform tumour volumetry in clinical practice and a prospective evaluation of the network’s clinical utility is already planned.
This work has the potential to significantly change current clinical practice by altering the way VS are measured and managed. In 2003, it was agreed that a VS should be defined as either purely intrameatal (intracanalicular) or intrameatal with extrameatal extension11. By consensus, the size of a VS is currently defined by the tumour’s maximal extrameatal linear dimension 11,26 and an increase of at least 3mm in the largest extrameatal diameter is defines absolute growth 11. However, linear measurements are not the most sensitive method of measuring a tumour’s size and several studies have demonstrated that a volumetric measurement is a more reliable and accurate method of calculating a vestibular schwannoma’s true size 18,24,29,33,34,35. Such methods are also superior at detecting subtle growth 33. However, the principal reason such volumetric methods have not been widely implemented is because currently available tools make calculating tumour volume a labour-intensive process with no dedicated software readily available within the clinical setting. By providing a simple, fully automated tool to calculate vestibular schwannoma volume, this work has the ability to standardise a key part of clinical management of this disease, enabling accurate volumetric measurements to be performed easily in the clinic. An automated segmentation tool could also improve the process of contouring vestibular schwannomas for radiosurgery treatment. This could be used as the initialisation step of an interactive segmentation approach 39,41 and would speed up treatment planning.
This network was developed using a standardised dataset of images obtained on a routine clinical scanner. The next step would be to generalise the network to work with data from any type of MR scanner irrespective of the chosen sequence parameters in order to facilitate its widespread adoption within clinical practice. Future work will also focus on optimising the algorithm in post-surgical cases in order to improve the segmentation of residual tumour volumes.
Most patients in a VS surveillance programme have a ceT1 sequence performed as routine. However, there is increasing interest in using non-contrast imaging sequences in the surveillance of patients with VS because of the risks associated with gadolinium-containing contrast agents (GdCAs) including brain accumulation 5 and nephrogenic systemic fibrosis in patients with impaired renal function 20,31. In addition to improving patient safety, high-resolution T2 imaging is less expensive than ceT1 imaging so switching to non-contrast has the potential to deliver a 10-fold saving in scan costs 3.
Whilst the presence of a VS may be identified using hrT2 images alone, tumour-brain boundaries are sometimes difficult to determine on hrT2, particularly if the tumour is of a similar intensity to brain or when trying to segment cases following surgery (Figure 6). Consequently, the manual segmentation of tumours using hrT2 images alone is technically challenging. Our network demonstrated high accuracy using hrT2 images in isolation (Dice 88.25% (SD 3.90%), ASSD 0.416 mm (SD 0.209), 9.77% (SD 7.56%) but was notably less accurate than the segmentation results using ceT1 images alone or in combination with hrT2 data. Ongoing work is focused on optimising the segmentation of VS from hrT2 alone but this study has clearly demonstrated how emerging AI technology could be used to exploit non-contrast MR imaging data information.
It is postulated that AI technology has the potential to personalise medicine and significantly improve the management of patients of tumours including VS. To facilitate the delivery of patient-specific care, we now intend to introduce our AI model as part of the clinical decision making. Such a model would calculate the probability of an individual’s future tumour growth based on a number of past radiological characteristics (including higher-order features aka “radiomics”) and would then generate a suggested surveillance interval. A critical aspect of this work will be to establish a threshold for clinically significant volumetric growth. Some authors have suggested that volumetric growth of >20% should be considered clinically significant as this roughly equates to linear growth of >2mm year 18 whereas others concluded that a tumour’s volume doubling time (VDT) provided the best value to detect subtle growth 33.
Various groups are developing AI models to apply volumetric measurements in gliomas as evidenced by the myriad submissions to such technical challenges 1. However, there has been very little focus on optimising models for VS. To our knowledge, we have developed the first fully automated method of segmenting VS and our algorithm could be easily adapted to analyse other benign tumours in the cerebellopontine angle and skull base, such as meningiomas.
The main limitation of this study is common to most deep learning imaging studies; that it was developed using a uniform dataset and consequently may not immediately perform as well on images obtained with difference scan parameters. That said, ongoing research suggests that a clinically-robust, generalisable framework can be optimised. Secondly, this study was not designed to provide a measurement of uncertainty of the predictions. Such information would be helpful prior to its widespread clinical implementation and we plan to include analysis of epistemic uncertainty (from the model) and aleatoric uncertainty (due to corrupted data) 7,12,36 in future works.
Conclusion
We have developed a robust AI model to perform automated segmentations of vestibular schwannomas. The method was validated using ceT1 and hrT2 images, achieving excellent accuracy scores comparable to repeated measurements performed by clinicians. Validating our novel results on a larger-scale study in future and improving any model inaccuracy, this methodology has the potential to improve and personalise the surveillance management of patients with tumours in the skull base.
Acknowledgements
This work was supported by Wellcome Trust [203145Z/16/Z; 203148/Z/16/Z; WT106882], and EPSRC [NS/A000050/1; NS/A000049/1] funding. TV is supported by a Medtronic / Royal Academy of Engineering Research Chair [RCSRF1819\7\34].
Financial support:
This work was supported by Wellcome Trust [203145Z/16/Z; 203148/Z/16/Z; WT106882], and EPSRC [NS/A000050/1; NS/A000049/1] funding. TV is supported by a Medtronic / Royal Academy of Engineering Research Chair [RCSRF1819\7\34].
References
- 1.Bakas S, Reyes M, Jakab A, Bauer S, Rempfler M, Crimi A, et al. Identifying the Best Machine Learning Algorithms for Brain Tumor Segmentation, Progression Assessment, and Overall Survival Prediction in the BRATS Challenge. 2018 [Google Scholar]
- 2.Bello GA, Dawes TJW, Duan J, Biffi C, de Marvao A, Howard LSGE, et al. Deep learning cardiac motion analysis for human survival prediction. 2018 doi: 10.1038/s42256-019-0019-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coelho DH, Tang Y, Suddarth B, Mamdani M. MRI surveillance of vestibular schwannomas without contrast enhancement: Clinical and economic evaluation. Laryngoscope. 2018;128:202–209. doi: 10.1002/lary.26589. [DOI] [PubMed] [Google Scholar]
- 4.Cross JJ, Baguley DM, Antoun NM, Moffat DA, Prevost AT. Reproducibility of volume measurements of vestibular schwannomas - a preliminary study. Clin Otolaryngol. 2006;31:123–129. doi: 10.1111/j.1749-4486.2006.01161.x. [DOI] [PubMed] [Google Scholar]
- 5.Agency EM, editor. European Medicines Agency. Gadolinium-Containing Contrast Agents. EMA; 2017. [Google Scholar]
- 6.Evans DGR, Moran A, King A, Saeed S, Gurusinghe N, Ramsden R. Incidence of vestibular schwannoma and neurofibromatosis 2 in the North West of England over a 10-year period: higher incidence than previously thought. Otol Neurotol. 2005;26:93–7. doi: 10.1097/00129492-200501000-00016. [DOI] [PubMed] [Google Scholar]
- 7.Gal Y, Ghahramani Z. Dropout as a bayesian approximation: Representing model uncertainty in deep learning. International Conference on Machine Learning; 2016. pp. 1050–1059. [Google Scholar]
- 8.Gibson E, Li W, Sudre C, Fidon L, Shakir DI, Wang G, et al. NiftyNet: a deep-learning platform for medical imaging. 2017 doi: 10.1016/j.cmpb.2018.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goodfellow I, Bengio Y, Courville A, Bengio Y. Deep Learning. MIT press; Cambridge: 2016. [Google Scholar]
- 10.Harris GJ, Plotkin SR, MacCollin M, Bhat S, Urban T, Lev MH, et al. Three-dimensional volumetrics for tracking vestibular schwannoma growth in neurofibromatosis type II. Neurosurgery. 2008;62:1314–1320. doi: 10.1227/01.neu.0000333303.79931.83. [DOI] [PubMed] [Google Scholar]
- 11.Kanzaki J, Tos M, Sanna M, Moffat DA, Monsell EM, Berliner KI. New and modified reporting systems from the consensus meeting on systems for reporting results in vestibular schwannoma. Otol Neurotol. 2003;24:642–649. doi: 10.1097/00129492-200307000-00019. [DOI] [PubMed] [Google Scholar]
- 12.Kendall A, Gal Y. What uncertainties do we need in bayesian deep learning for computer vision? Advances in Neural Information Processing Systems. 2017:5574–5584. [Google Scholar]
- 13.Krizhevsky A, Sutskever I, Hinton GE. Imagenet classification with deep convolutional neural networks. Advances in Neural Information Processing Systems. 2012:1097–1105. [Google Scholar]
- 14.Li Y, Shen L. In: Deep Learning Based Multimodal Brain Tumor Diagnosis BT - Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries. Crimi A, Bakas S, Kuijf H, Menze B, Reyes M, editors. Cham: Springer International Publishing; 2018. pp. 149–158. [Google Scholar]
- 15.Lin L, Dou Q, Jin Y-M, Zhou G-Q, Tang Y-Q, Chen W-L, et al. Deep Learning for Automated Contouring of Primary Tumor Volumes by MRI for Nasopharyngeal Carcinoma. Radiology. 2019;291:677–686. doi: 10.1148/radiol.2019182012. [DOI] [PubMed] [Google Scholar]
- 16.Lin T-Y, Goyal P, Girshick R, He K, Dollar P. Focal loss for dense object detection. IEEE Trans Pattern Anal Mach Intell. 2018:1–1. doi: 10.1109/TPAMI.2018.2858826. [DOI] [PubMed] [Google Scholar]
- 17.Liu S, Xu D, Zhou SK, Pauly O, Grbic S, Mertelmeier T, et al. 3D Anisotropic Hybrid Network: Transferring Convolutional Features from 2D Images to 3D Anisotropic Volumes. International Conference on Medical Image Computing and Computer-Assisted Intervention; Cham: Springer; 2018. pp. 851–858. [Google Scholar]
- 18.MacKeith S, Das T, Graves M, Patterson A, Donnelly N, Mannion R, et al. A comparison of semi-automated volumetric vs linear measurement of small vestibular schwannomas. Eur Arch Otorhinolaryngol. 2018;275:867–874. doi: 10.1007/s00405-018-4865-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McKinley R, Wepfer R, Gundersen T, Wagner F, Chan A, Wiest R, et al. In: Nabla-net: A Deep Dag-Like Convolutional Architecture for Biomedical Image Segmentation BT - Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries. Crimi A, Menze B, Maier O, Reyes M, Winzeck S, Handels H, editors. Cham: Springer International Publishing; 2016. pp. 119–128. [Google Scholar]
- 20.MHRA, CHM. Gadolinium-Containing MRI Contrast Agents: Nephrogenic Systemic Fibrosis. 2007 [Google Scholar]
- 21.Milletari F, Navab N, Ahmadi S-A. V-Net: Fully Convolutional Neural Networks for Volumetric Medical Image Segmentation. 2016 [Google Scholar]
- 22.Moffat DA, Hardy DG, Irving RM, Viani L, Beynon GJ, Baguley DM. Referral patterns in vestibular schwannomas. Clin Otolaryngol Allied Sci. 1995;20:80–83. doi: 10.1111/j.1365-2273.1995.tb00018.x. [DOI] [PubMed] [Google Scholar]
- 23.Oktay O, Schlemper J, Le Folgoc L, Lee M, Heinrich M, Misawa K, et al. Attention U-Net: Learning Where to Look for the Pancreas. 1st Conference on Medical Imaging with Deep Learning: MIDL; 2018. [Google Scholar]
- 24.Roche PH, Robitail S, Régis J. Two- and three dimensional measures of vestibular schwannomas and posterior fossa – implications for the treatment. Acta Neurochir (Wien) 2007;149:267–273. doi: 10.1007/s00701-006-1093-x. [DOI] [PubMed] [Google Scholar]
- 25.Ronneberger O, Fischer P, Brox T. U-Net: Convolutional Networks for Biomedical Image Segmentation. Springer; Cham: 2015. pp. 234–241. [Google Scholar]
- 26.Shapey J, Barkas K, Connor S, Hitchings A, Cheetham H, Thomson S, et al. A standardised pathway for the surveillance of stable vestibular schwannoma. Ann R Coll Surg Engl. 2018;100:216–220. doi: 10.1308/rcsann.2017.0217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stangerup S-EE, Caye-Thomasen P. Epidemiology and natural history of vestibular schwannomas. Otolaryngol Clin North Am. 2012;45:257–68. doi: 10.1016/j.otc.2011.12.008. vii. [DOI] [PubMed] [Google Scholar]
- 28.Sudre CH, Li W, Vercauteren T, Ourselin S, Cardoso MJ. Generalised Dice overlap as a deep learning loss function for highly unbalanced segmentations. 2017 doi: 10.1007/978-3-319-67558-9_28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tang S, Griffin AS, Waksal JA, Phillips CD, Johnson CE, Comunale JP, et al. Surveillance After Resection of Vestibular Schwannoma. Otol Neurotol. 2014;35:1. doi: 10.1097/MAO.0000000000000459. [DOI] [PubMed] [Google Scholar]
- 30.Tango T. Equivalence test and confidence interval for the difference in proportions for the paired-sample design. Stat Med. 1998;17:891–908. doi: 10.1002/(sici)1097-0258(19980430)17:8<891::aid-sim780>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 31.Care H, S, editors. UK Government. Gadolinium-Containing Contrast Agents: Removal of Omniscan and Iv Magnevist, Restrictions to the Use of Other Linear Agents. 2017. www.gov.uk . [Google Scholar]
- 32.van de Langenberg R, de Bondt BJ, Nelemans PJ, Baumert BG, Stokroos RJ. Follow-up assessment of vestibular schwannomas: volume quantification versus two-dimensional measurements. Neuroradiology. 2009;51:517–24. doi: 10.1007/s00234-009-0529-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Varughese JK, Breivik CN, Wentzel-Larsen T, Lund-Johansen M. Growth of untreated vestibular schwannoma: a prospective study. J Neurosurg. 2012;116:706–712. doi: 10.3171/2011.12.JNS111662. [DOI] [PubMed] [Google Scholar]
- 34.Vokurka EA, Herwadkar A, Thacker NA, Ramsden RT, Jackson A. Using Bayesian tissue classification to improve the accuracy of vestibular schwannoma volume and growth measurement. AJNR Am J Neuroradiol. 2002;23:459–67. [PMC free article] [PubMed] [Google Scholar]
- 35.Walz PC, Bush ML, Robinett Z, Kirsch CFE, Welling DB. Three-Dimensional Segmented Volumetric Analysis of Sporadic Vestibular Schwannomas. Otolaryngol Neck Surg. 2012;147:737–743. doi: 10.1177/0194599812447766. [DOI] [PubMed] [Google Scholar]
- 36.Wang G, Li W, Aertsen M, Deprest J, Ourselin S, Vercauteren T. Aleatoric uncertainty estimation with test-time augmentation for medical image segmentation with convolutional neural networks. Neurocomputing. 2019;338:34–45. doi: 10.1016/j.neucom.2019.01.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang G, Li W, Ourselin SS, Vercauteren T. Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries. Springer; Cham: 2018. Automatic Brain Tumor Segmentation using Cascaded Anisotropic Convolutional Neural Networks; pp. 178–190. [Google Scholar]
- 38.Wang G, Li W, Ourselin S, Vercauteren T. Automatic Brain Tumor Segmentation Based on Cascaded Convolutional Neural Networks With Uncertainty Estimation. Front Comput Neurosci. 2019;13:56. doi: 10.3389/fncom.2019.00056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang G, Li W, Zuluaga MA, Pratt R, Patel PA, Aertsen M, et al. Interactive medical image segmentation using deep learning with image-specific fine tuning. IEEE Trans Med Imaging. 2018;37:1562–1573. doi: 10.1109/TMI.2018.2791721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang G, Shapey J, Li W, Dorent R, Demitriadis A, Bisdas S, et al. Automatic Segmentation of Vestibular Schwannoma from T2-Weighted MRI by Deep Spatial Attention with Hardness-Weighted Loss. 2019 [Google Scholar]
- 41.Wang G, Zuluaga MA, Li W, Pratt R, Patel PA, Aertsen M, et al. DeepIGeoS: a deep interactive geodesic framework for medical image segmentation. IEEE Trans Pattern Anal Mach Intell 1707. 2018 doi: 10.1109/TPAMI.2018.2840695. arXiv:1707.00652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu Q, Xie L, Wang Y, Zhou Y, Fishman EK, Yuille AL. Recurrent Saliency Transformation Network: Incorporating Multi-Stage Visual Cues for Small Organ Segmentation. 2018:8280–8289. [Google Scholar]
- 43.Zou KH, Warfield SK, Bharatha A, Tempany CMC, Kaus MR, Haker SJ, et al. Statistical validation of image segmentation quality based on a spatial overlap index. Acad Radiol. 2004;11:178–89. doi: 10.1016/S1076-6332(03)00671-8. [DOI] [PMC free article] [PubMed] [Google Scholar]