Abstract
Background
Previously identified transcriptomic signatures have been based on primary and metastatic melanomas with relatively few AJCC stage I tumors given difficulties in sampling small tumors. The advent of adjuvant therapies has highlighted the need for better prognostic and predictive biomarkers especially for AJCC stage I and II disease.
Patients and Methods
687 primary melanoma transcriptomes were generated from the Leeds Melanoma Cohort (LMC). The prognostic value of existing signatures across all the AJCC stages was tested. Unsupervised clustering was performed and the prognostic value of the resultant signature was compared with that of sentinel node biopsy (SNB) and tested as a biomarker in three published immunotherapy datasets.
Results
Previous Lund and TCGA signatures predicted outcome in the LMC dataset (P=10-8 to 10-4) but showed a significant interaction with AJCC stage (P=0.04) and did not predict outcome in stage I tumors (P=0.3 to 0.7). Consensus-based classification of the LMC dataset identified six classes which predicted outcome, notably in stage I disease. LMC class was a similar indicator of prognosis when compared to SNB and it added prognostic value to the genes reported by Gerami et al. One particular LMC class consistently predicted poor outcome in patients receiving immunotherapy in two of three tested datasets. Biological characterisation of this class revealed high JUN and AXL expression and evidence of epithelial to mesenchymal transition.
Conclusion
A transcriptomic signature of primary melanoma was identified with prognostic value, including in stage I melanoma and in patients undergoing immunotherapy.
Keywords: Consensus clustering, Sentinel Node Biopsy, EMT, JUN, AXL
Introduction
Cutaneous melanoma continues to increase in incidence worldwide. Although earlier diagnosis has been documented with correspondingly better outcomes, the rising incidence of thinner tumors means that, counterintuitively, one fifth of deaths now occur in patients presenting initially with early disease (1). In the UK, 91% of melanomas are diagnosed at AJCC stage I to II (2). Therefore, better prognostic biomarkers are needed to identify early stage disease requiring adjuvant therapies, as well as predictive biomarkers of response to checkpoint blockade.
Previous transcriptomic analyses of cutaneous melanoma have generated gene signatures with a prognostic value independent of AJCC stage (3–7). The prognostic signature developed by Jonsson et al. (3) classifies metastatic melanomas into four classes (Lund 4-classes), later simplified into two classes (Lund 2-grades, (4)), and the signature developed by the TCGA (The Cancer Genome Atlas) consortium classified melanomas into three classes (TCGA 3-classes) (8). The prognostic significance of the Lund 4-class and TCGA 3-class signatures have been replicated in relatively small datasets, notably with few AJCC stage I patients (5, 9). Another transcriptomic signature based on 27 genes was developed by Gerami et al. (6) to classify primary melanoma patients into tumors which were high or low-risk for metastasis.
In this study, the first aim was to test the prognostic value of the Lund and TCGA signatures, as well as the gene list of Gerami et al’s signature (6) in a large population-based cohort of primary melanomas with a good proportion of stage I patients and extensive phenotypic annotations (Leeds Melanoma Cohort, LMC). Since the dataset was well powered for discovery of novel tumor subtypes, unsupervised clustering of the tumor transcriptomes of the LMC was performed and the prognostic value of the resultant signature was compared with that of SNB in analyses stratified by AJCC stage. Finally, the association between the Leeds signature and outcome was tested in published data from patients receiving immunotherapy (10–12).
Materials and Methods
Leeds Melanoma Cohort
As described previously (13), 2184 primary melanoma patients were recruited to the Leeds Melanoma Cohort (LMC) in the period of 2000-2012. This was a population-ascertained cohort which therefore recruited patients treated at multiple clinical centres (recruitment rate 67%). During this period SNB was neither offered nor accepted universally. The study was ethically approved (ethical approval MREC 1/3/57, PIAG 3-09(d)/2003) and in accordance with the Declaration of Helsinki. Participants were consented to sampling of their FFPE (formalin fixed paraffin embedded) tumor blocks which were stored in the NHS (UK National Health Service) histopathology departments of the respective hospitals. Haemotoxylin and eosin (H&E)-stained slides were generated and examined to facilitate subsequent sampling of the blocks using a 0.6mm diameter tissue microarray needle as previously reported (5, 13). Prior to sampling, all the tumor blocks were reviewed, and if there was only a small amount of tumor left in the block then the block was not sampled, lest a clinically important block be destroyed. Up to two cores were sampled from each block, and, to increase the comparability between tumors, the samples were consistently extracted from the least inflamed, least stromal regions of the invasive front of the tumor. The tumor infiltrating lymphocytes were scored using Clark et al.’s classification system (14). As previously reported (13), 703 tumor transcriptomes were profiled and in the current study 16 samples were removed in quality control leaving a cohort of 687 patients, henceforth referred to as the whole LMC dataset. The dataset contained 251 patients who had a SNB test (Supplementary table S10), and only 16 patients are known so far to have been treated with checkpoint blockade. The LMC patients were assigned an AJCC stage based on the AJCC staging 8th edition (15). Where patients did not have a SNB, the AJCC staging used was derived from clinical staging and pathological examination of the wide local excision sample.
mRNA extraction and expression data generation
Both mRNA and DNA were extracted from the tumor samples derived from cores following a previously described protocol (5, 13). The whole genome mRNA expression profiling was carried out using Illumina’s DASL-HT12-v4 array. As described previously, for quality control, the mRNA was extracted from up to 2 cores for a number of patients (117 duplicates in total); gene expression data from only one extraction per patient was used in subsequent analyses (13). The raw transcriptomic data were extracted from the image files using GenomeStudio (Illumina Inc., San Diego) and were pre-processed as previously reported (13). Briefly, after background correction and quantile normalisation (R package LUMI (16)), singular value decomposition (SVD) was used to remove the batch effect (R package SWAMP (17)) (13).
Quality control in LMC
The array included 29,262 probes corresponding to 20,715 unique genes. For genes with multiple probes, the probe detected in the largest number of tumors was retained, and two additional filters were applied: genes had to be detected with P<0.05 in at least 40% of tumors and had to have a standard deviation (SD)>0.40. This SD threshold was chosen based on the overall distribution across the 20,715 genes on the log2 scale. The median SD was 0.68. The data were standardized to give each gene a mean of 0 and SD of 1.
Procedures
The LMC tumors were classified into the Lund 4-classes, Lund 2-grades and TCGA 3-classes using the supervised nearest centroid classification (NCC) as previously described (5). All the 27 genes of the Gerami et al. gene signature (6) were present in LMC dataset and were analysed using a univariable survival analysis in the whole LMC dataset and stage I tumors. Unsupervised clustering was performed using the consensus Partitioning Around Medoids clustering method in the R-package ConsensusClusterPlus (18, 19) with Euclidean distance as the dissimilarity measure and a resampling fraction of 0.8 for both genes and samples in 1000 iterations (Supplementary methods).
Statistical analysis
Cox proportional hazard models and Kaplan-Meier curves were used to test the association with survival (R-package Survival) (20). The survival time was calculated from time of diagnosis to time of last follow-up or time of death from melanoma, whichever occurred first, referred to as melanoma-specific survival (MSS). Patients with deaths caused by factors other than melanoma were censored at the time of death. Receiver Operating Characteristic (ROC) analysis was performed using AJCC stage pre-SNB and AJCC stage post-SNB for patients who had SNB. Clinical staging prior to SNB is described as AJCC pre-SNB. The AJCC stage post-SNB includes additional information on regional lymph node metastasis. The analysis used AJCC staging 8th edition, and MSS up to 3 years was chosen as cut-off based upon the inclusion of the majority of the deaths without loss of data as a result of censoring (Supplementary table S11). Patients who were censored before 3 years were not included in the analysis. The analysis was performed using R-packages pROC, plotROC, and ggplot2 (21–23).
Pathway enrichment analysis
The differentially expressed genes (DEG) were identified using the Significance Analysis of Microarrays (R-package SAMR) by comparing each class versus all others (24). Pathway enrichment and biological network analysis of DEGs with a q-value equal to 0 were performed using ReactomeFiviz in Cytoscape (25). The central nodes of the biological network were identified using a centrality measure (betweenness) in Gephi (26) (Supplementary methods).
Copy Number Alterations (CNA)
The CNA data were generated in a subset of LMC tumors using Illumina’s next-generation sequencing platform as reported in Filia et al. (in revision) (Supplementary methods). Among the 687 transcriptome-profiled patients of LMC, CNA data were available for 272 patients. The CNA were assessed in the regions spanning the genes identified as hubs in network enrichment analysis. The ratio between mean of the window read counts in the region mapping to a gene and the average read count of the 10 flanking regions around that gene was used to estimate the copy number changes. The windows (5k) corresponding to a gene locus were identified using the R packages biomaRt (27, 28). The cut-off for calling a region amplified was chosen as a value greater than 0.4 while a value less than -0.4 was used to identify a deletion. The 272 samples in the CNA dataset were at AJCC stages I (n=80), II (n=147), and III (n=45) (similar distribution to the whole LMC dataset).
Lund validation dataset
For replication, a primary melanoma transcriptomic dataset of 223 tumors from a Lund cohort (Sweden) was used (Harbst et al. (4)). The samples were classified using the newly generated signature by the supervised NCC approach (5). Out of those 223 patients, 200 had recorded information on melanoma relapse in the follow-up time post-diagnosis and were used to test the association between patient subgroups and relapse-free survival (using Cox proportional hazard models, Kaplan-Meier curves and log-rank test).
Immunotherapy datasets
Three publicly available transcriptome datasets (Hugo Cohort: GSE78220, Ulloa-Monotoya cohort: GSE35640, Riaz Cohort: https://github.com/riazn/bms038_analysis) were downloaded (10–12), samples were quantile normalised and classified using the NCC method (Pearson’s correlation coefficient). The Riaz cohort was a mixture of samples from various melanoma types (cutaneous melanoma, mucosal melanoma, acral melanoma, uveal/ocular melanoma, others). In this study the samples labelled as cutaneous melanoma were analysed. In all the three cohorts, the association with response to immunotherapy was tested using Fisher’s exact test.
Results
Existing signatures showed no association with survival in stage I melanoma
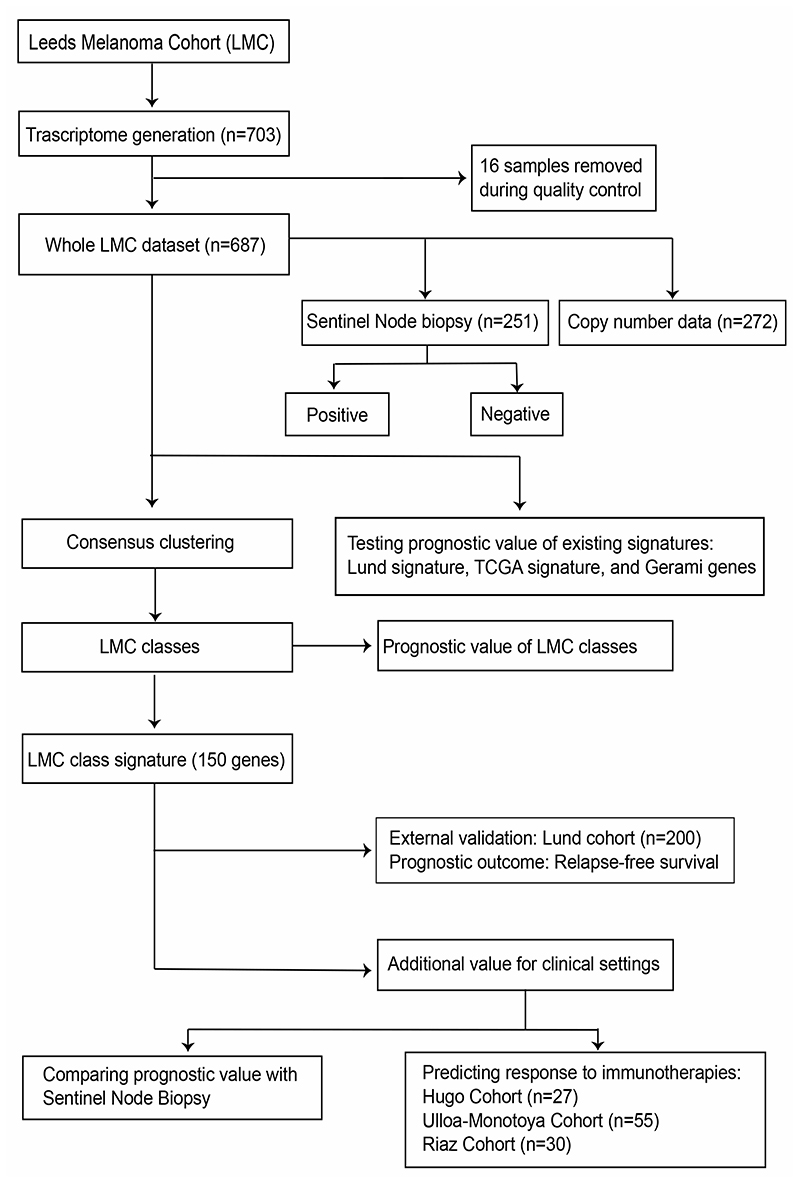
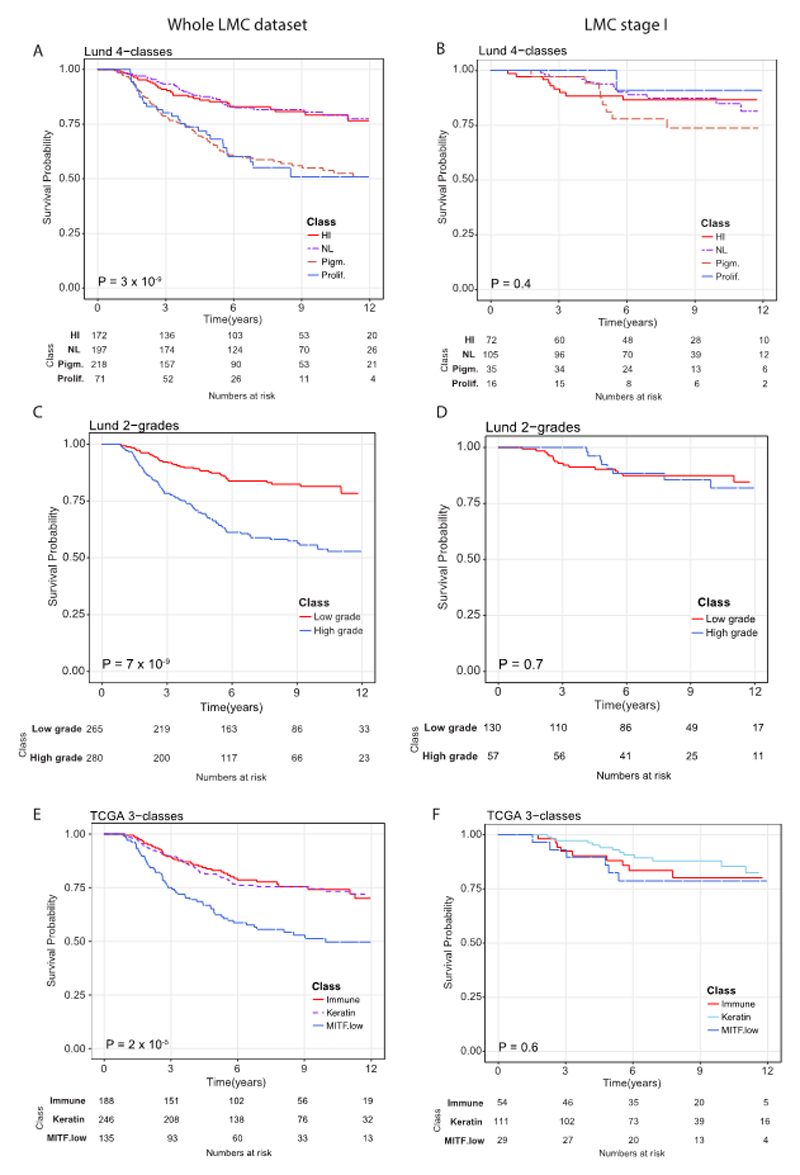
The structure of datasets used in this study are depicted in Figure 1. When applied to the whole LMC dataset (n=687), the three formerly published signatures (Lund 4-class, Lund 2-grade, TCGA 3-class) replicated previously observed associations with MSS (Figure 2A, 2C, and 2E). However, upon stratifying LMC patients on the basis of AJCC stage, the Lund and TCGA signatures showed no association with prognosis for LMC stage I patients (Figure 2B, 2D, and 2F). The Lund 2-grade signature had the highest statistical power (since based on only two groups) and showed a statistically significant interaction with AJCC stage (P=0.02, Supplementary table S1), suggesting that the lack of association in stage I was not solely due to low sample size. Because the full details of Gerami et al’s (6) commercial signature were not published, we were limited in the scope of its replication in the LMC dataset. However, analysing the 27 Gerami genes identified 23 genes as predictors of prognosis in the whole LMC dataset (Supplementary table S2). However, in keeping with the Lund and TCGA signatures, none of these genes showed a significant association with prognosis in stage I tumors (Supplementary table S2).
Figure 1. Analysis workflow of the study.
Figure 2.
Replicating Lund and TCGA signatures using LMC dataset. Kaplan-Meier plots showing the Melanoma-specific survival (MSS) for (A) Lund 4-classes (HI- high-immune, NL- normal-like, Pigm.- pigmentation, Prolif.- proliferative), (B) Lund 2-grades (low grade and high grade) and (C) TCGA 3-classes (immune, keratin, MITF low) across the whole LMC dataset. In LMC stage I subset, Kaplan-Meier plots showing the MSS for (D) Lund 4-classes, (E) Lund 2-grades, and (F) TCGA 3-classes. Pvalues are from log-rank test. Samples which could not be classified into any of the classes were not used in survival analysis.
Generating novel LMC classes and their clinical characteristics
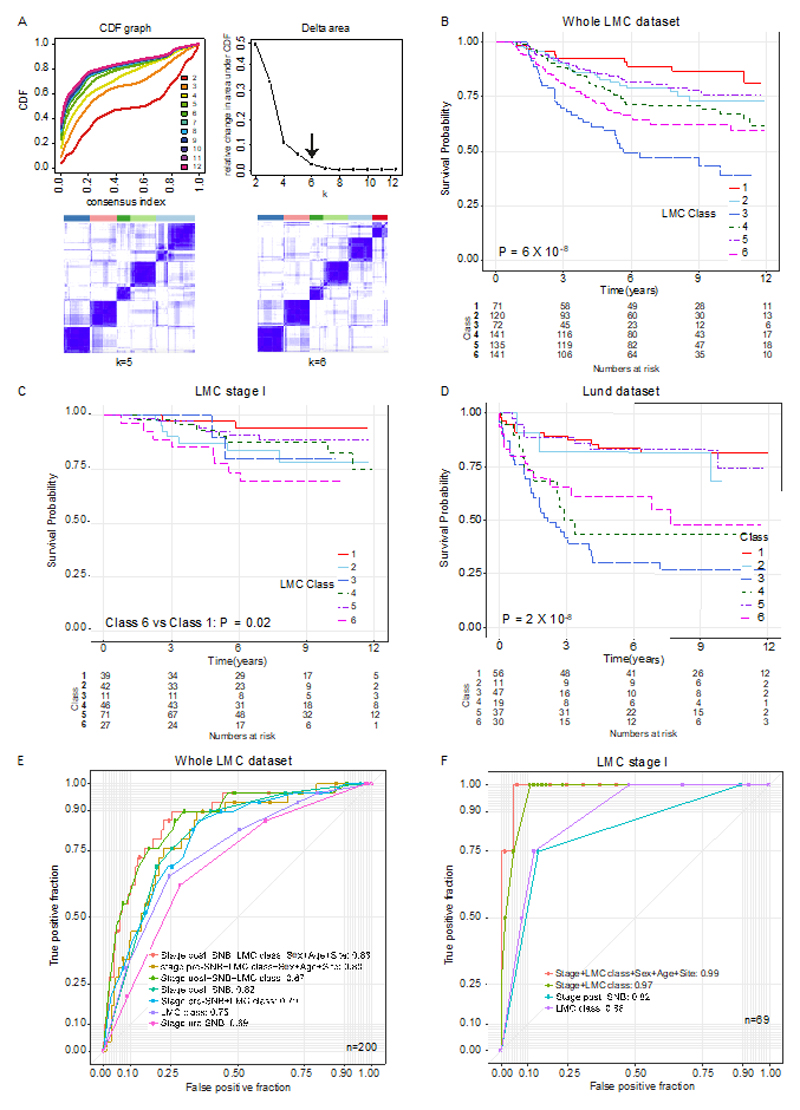
Consensus clustering of the LMC dataset was performed, and following additional quality control measures (Supplementary table S3), six distinct, novel molecular classes were identified (Figure 3A). These classes were associated with clinico-pathological variables known to have prognostic value, including tumor site (P=0.03), age at diagnosis (P=0.03), mitotic rate (P=0.002), ulceration (P=0.01), AJCC stage (P=6x10-10), tumor infiltrating lymphocytes (TILs) (P=6x10-4), and Breslow thickness (P=9x10-14) (Table 1). The LMC classes 1 and 5 tumors tended to be thin and non-ulcerated, whilst classes 2 and 4 tumors were thicker. Class 3 and 6 tumors were the thickest and most frequently ulcerated. The six classes showed strong association with BRAF (P=6x10-5) and NRAS mutation status (P=3x10-4): classes 1, 5, and 6 tumors were frequently BRAF mutated, while classes 2, 3, and 4 tumors were frequently NRAS mutated (Table 1).
Figure 3. Defining LMC signature and its prognostic value.
(A) The area under the CDF and its relative change with increasing k. The delta area graph shows little variation at k=6. Heatmap of consensus matrices at k=5 and 6. The blue color indicates high consensus score and the white color indicates low consensus (B) Kaplan-Meier plot showing the MSS for the six classes in (B) the whole LMC dataset, (C) the LMC stage I, and (D) relapse-free survival in the Lund cohort (Pvalue from log-rank test, or Wald test for two-groups comparison). Seven mucosal tumors were excluded from analysis. (E) ROC curves comparing the prognostic value of the LMC signature to that of Sentinel Node Biopsy (SNB) in the whole dataset. The AUCs for LMC class+ stage pre-SNB and stage post-SNB were not significantly different (DeLong’s test P=0.7). (F) The ROC curve comparing prognostic value of LMC signature with SNB in the stage I pre-SNB group. All but one patient were stage IB pre-SNB, therefore AUC for LMC signature alone was compared to stage post-SNB and the difference was not significant (DeLong’s test P=0.7). The difference in AUCs between stage post-SNB alone and LMC class +stage post-SNB was also not significant (DeLong’s test P=0.1).
Table 1. The LMC classes association with clinico-histopathological variables.
| Histopathological variables | Whole dataset n=687 (%) |
LMC classes | ||||||
|---|---|---|---|---|---|---|---|---|
| Class 1 (n=71) |
Class 2 (n=122) |
Class 3 (n= 73) |
Class 4 (n=143) |
Class 5 (n=136) |
Class 6 (n=142) |
Pa | ||
| Sex : male n (%) | 310 (45) | 39 (55) | 51 (42) | 34 (47) | 56 (39) | 55 (40) | 75 (52) | 0.07 |
| Tumor site: limbs n (%) | 289 (42) | 37 (52) | 58 (48) | 26 (36) | 58 (41) | 64 (47) | 46 (32) | 0.03 |
| Age at diagnosis (years) m(r) | 58 (18, 81) | 59 (21,76) | 59 (22,79) | 60 (20,77) | 58 (18,81) | 53 (25,76) | 59 (22,81) | 0.03 |
| Breslow thickness (mm) m(r) | 2.3 (0.3, 20) | 1.7 (0.7, 5.5) | 2.1 (0.8, 8.9) | 3.2 (0.8, 20) | 2.3 (0.3, 15) | 1.8 (0.7, 12) | 3.0 (0.8, 18) | 9 × 10-14 |
| AJCC stage (%) b I | 236 (35) | 39 (55) | 42 (35) | 11 (15) | 46 (64) | 71 (53) | 21 (19) | 6 × 10-10 |
| Ulceration (present) n (%) | 228 (33) | 16 (23) | 32 (26) | 30 (41) | 53 (37) | 38 (28) | 59 (42) | 0.01 |
| Mitotic rate (/mm2) | 1 (0,25) | 0 (0,11) | 1 (0,17) | 2 (0,25) | 1 (0,13) | 1 (0,12) | 1 (0,18) | 0.002 |
| TILs (%) Absent | 76 (15) | 2 (4) | 13 (14) | 17 (32) | 14 (16) | 15 (16) | 15 (13) | 6 × 10-4 |
| BRAF mutant yes (%) | 266 (47) | 26 (43) | 38 (30) | 23 (40) | 44 (36) | 63 (59) | 72 (61) | 6 × 10-5 |
| NRAS mutant yes (%) | 138 (25) | 8 (14) | 35 (34) | 17 (30) | 41 (34) | 20 (19) | 17 (15) | 3 × 10-4 |
The associations were tested using Pearson’s chi-squared test for categorical variables and the Kruskal-Wallis test for continuous variables. Symbol n is the number of samples, m is the median and r is the range.
7 patients had mucosal melanoma and, although they were classified, they were not included in survival analyses. Their AJCC stage was not reported. Each of LMC class 2 and 4 contained 2 of these, while class 3, 5 and 6 had 1 each.
LMC classes predicted prognosis in primary melanoma and in stage I subset
The LMC classes predicted MSS in the whole LMC dataset and notably, across AJCC stages I, II and III subsets (Figure 3B-3C, Supplementary figure S1). In the unadjusted analysis of the whole dataset (Figure 3B, Supplementary table S4), class 1 (baseline) had the best prognosis, class 2 (HR=1.7, 95% confidence interval (CI) 0.8-3.5) and class 5 (HR=1.5, 95% CI 0.7-3.1) showed intermediate prognosis, while class 3 (HR=5.0, 95% CI 2.5-10.1), class 4 (HR=2.4, 95% CI 1.2-4.7), and class 6 (HR=3.1, 95% CI 1.6-6.1) had the worst prognosis. In multivariable analysis, classes 3, 4, and 6 remained significant predictors of poor prognosis after including AJCC stage, sex, age at diagnosis, mitotic rate (Table S4) and when the AJCC stage was replaced by ulceration and Breslow thickness in the model (Table S6). In the LMC stage I subset, class 6 (HR=6.6, 95% CI 1.4-31.2) significantly predicted poor prognosis in unadjusted analysis (Figure 3C and Table S5) and it remained significant when sex, age at diagnosis, mitotic rate, ulceration and Breslow thickness were adjusted (HR=9.8, 95% CI 1.1-86.2, Table S6). Since Gerami signature was not available to us in full, we ran unsupervised clustering of the LMC dataset using the 27 Gerami genes to generate the 2 tumor groups analysed by Gerami et al. (6), referred to as the Gerami clusters. This analysis showed that the LMC classes and Gerami clusters had independent prognostic effects in the whole LMC dataset (Supplementary table S7); however, the Gerami clusters showed no prognostic value in stage I tumors while LMC class 6 remained a significant predictor in the multivariable model (Supplementary table S8).
To validate the prognostic value of the LMC classes in an independent dataset, a 150-gene based signature (LMC signature), generated after refining ~13,000 genes (Supplementary figure S2), was applied to the Lund dataset (4). In keeping with the observations made in the LMC dataset, class 3, class 4, and class 6 predicted worse prognosis in the Lund dataset, while class 1, class 2, and class 5 predicted better prognosis (Figure 3D, Supplementary table S9). Since the Lund dataset had only a few stage I cases (n=58) the prognostic value of LMC signature could not be replicated in stage I disease.
LMC signature had independent prognostic value when compared with SNB
In the dataset derived from individuals who had a SNB, the prognostic value of combined LMC class signature and pre-SNB AJCC stage was similar to that of AJCC stage with SNB (i.e. stage post-SNB) (AUC 0.82 vs 0.80, P= 0.7, Figure 3E). Combining the LMC signature with AJCC stage post-SNB, patient’s sex, age at diagnosis and site of tumor increased the AUC to 0.88. Similarly, in the subset of patients at stage I pre-SNB, the LMC signature alone had comparable prognostic value to AJCC stage post-SNB (AUC=0.88 vs 0.83, P= 0.7, Figure 3F). In this stage I subset, addition of stage post-SNB, patient’s sex, age at diagnosis and site of tumor to the LMC signature further increased the AUC to 0.98. However, the limited sample size of stage I dataset and including so many variables clearly overfitted the model, giving near perfect classification and illustrating that independent datasets are needed to better assess performance.
Biological overlap between the LMC and existing signatures
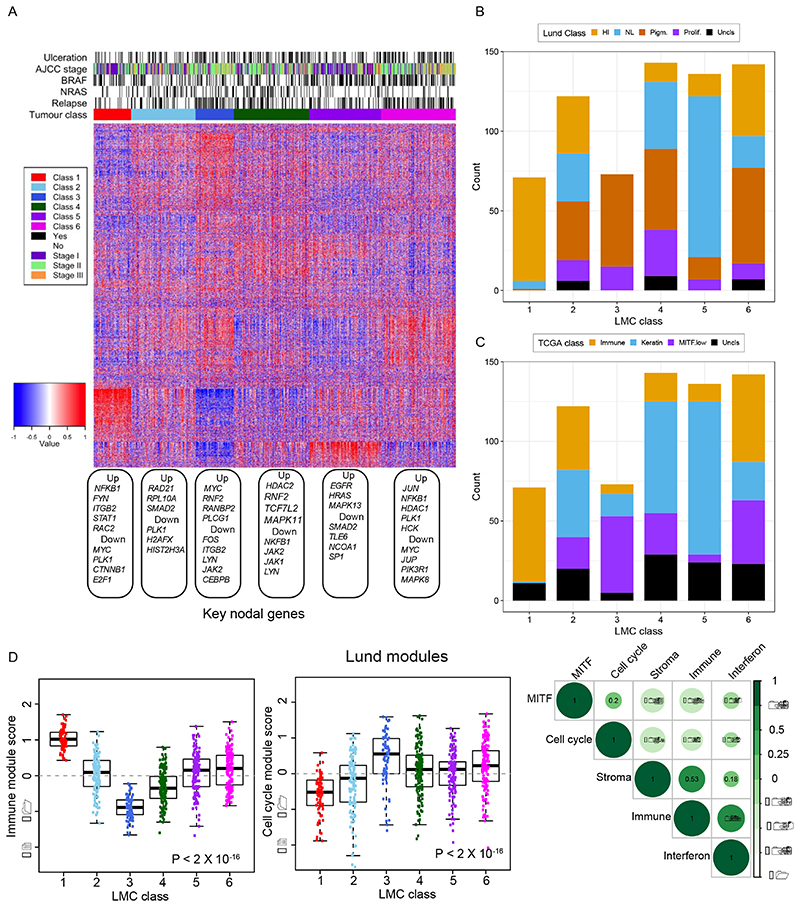
The six classes of LMC signature showed distinct gene expression profiles (Figure 4A) and showed partial overlap with the existing Lund and TCGA signatures. LMC classes 1, 3, and 5 overlapped substantially with the high-immune, pigmentation, and normal-like classes of the Lund 4-classes (Figure 4B), and with the immune, MITF low, and keratin classes of the TCGA 3-classes (Figure 4C). In contrast, LMC classes 2, 4, and 6 represented a mixture of the Lund 4-classes and TCGA 3-classes. Gene expression pathway enrichment analysis revealed distinctive biological features of the 6 LMC classes: notably class 2 was characterised by increased WNT signalling genes and metabolic pathways; class 4 by decreased expression of immune genes and class 6 by increased expression of cell cycle and consistent down-regulation of cell metabolism pathway genes (Supplementary table S14).
Figure 4. Biological characterization of the six LMC classes.
(A) The heatmap shows gene expression across the classes with tumor samples placed in columns and genes in rows. Blue depicts low expression and red depicts high expression. Each gene expression was standardized to mean 0 and standard deviation 1. The up- and down-regulated nodal genes identified in network analyses are shown under the heatmap. The barplot shows the overlap between the LMC classes and (B) Lund 4-classes (HI- high-immune, NL- normal-like, Pigm- pigmentation, Prolif- proliferative), and (C) TCGA 3-classes. The samples that could not be classified into the Lund 4-classes and TCGA 3-classes were labelled here as Uncls. (D) The modules (defined by a list of differentially upregulated genes) associated with melanoma-specific biological pathways as identified by the Lund group (29). Boxplots of immune and cell cycle module scores (standardized expressions) within the 6 LMC classes and correlation matrix of immune, cell cycle, MITF, stroma and interferon module scores. The module score variation across the classes was tested using the Kruskal-Wallis test.
When applied to the LMC 6 classes, the Lund modules (29) revealed discrimination consistent with enriched gene pathways: LMC class 1 tumors showed higher immune module activity, and class 3 tumors showed higher cell cycle module activity (Figure 4D). Interestingly, class 6 tumors had relatively higher cell cycle but also immune module activity and, as expected, the immune, stroma and interferon modules were positively correlated but they negatively correlated with cell cycle and MITF modules (Figure 4D). The tumor infiltrating immune cell populations imputed for each of the LMC classes (30) were consistent with the Lund immune module, as class 1 had the highest immune cell populations and class 3 the lowest, whilst class 6 appeared to maintain an intermediate level of immune cell populations, having the second highest scores on average (Supplementary figure S3).
A comparison with the Consensus Immunome Clusters (CICs), previously generated in the same LMC dataset based on 380 immune genes (13), showed that the 2 most prognostically contrasted LMC classes (class 1 and class 3) had a near perfect match with CIC 2 (high Immune) and CIC 3 (low immune/β-catenin high) respectively (Supplementary figure S4) while the rest of LMC classes were a mixture of CICs. Cluster 1 had correspondingly a higher proportion of tumors with histological evidence of brisk tumor infiltrating lymphocytes (36% compared with 8% in class 3). Analysing the correlation between the Gerami genes and LMC signature genes showed that the Gerami genes positively correlated with the genes upregulated in LMC class 5 tumors and negatively correlated with genes upregulated in LMC class 3 tumors (Supplementary figure S5). Consistent with this, Gerami clusters 1 and 2 highly overlapped with LMC classes 3 and 5 respectively (Supplementary figure S6).
JUN as marker of poor prognosis in class 6 tumors
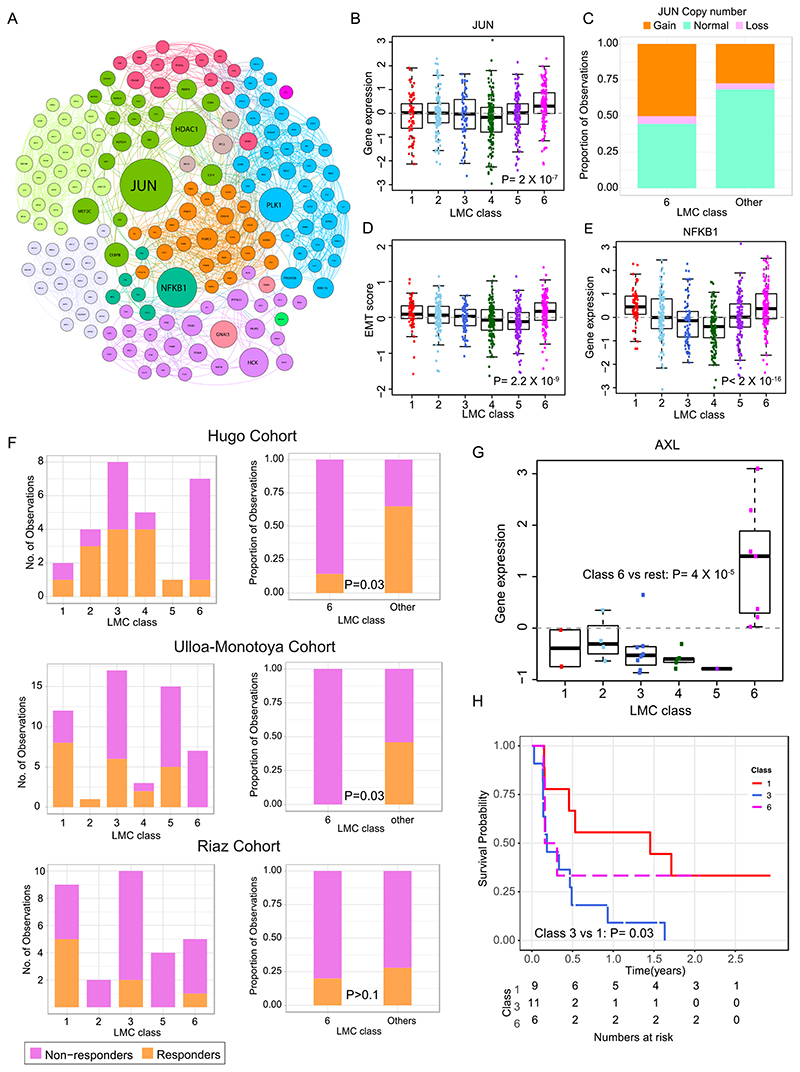
LMC class 6 predicted worse prognosis within AJCC stage I tumors. Further biological network analysis identified JUN as a key upregulated nodal gene in this class (Figure 5A-B). The NGS-based CNA data from a subset of LMC tumors (n=272) indicated that class 6 tumors were more likely to have DNA amplifications of JUN than other classes (P=0.003, Figure 5C, Supplementary figure S7). In melanoma, JUN has been reported to activate epithelial-to-mesenchymal transition (EMT), and accordingly a 6-gene based (31) and 200-gene based EMT signature (32) consistently scored higher in LMC class 6 than in all other LMC classes (Figure 5D, Supplementary figure S7). A secondary key nodal gene NFKB1 identified to be upregulated in class 6 had no copy number changes. Further examination of immunohistochemically stained sections, showed that all 4 tumors stained from class 6 were positive for NFKB1 protein expression, and this was similar to other LMC classes (P=0.4, Supplementary figure S7).
Figure 5. Biological characterization of LMC class 6 and association with response to immunotherapy.
(A) Network of upregulated genes in the LMC class 6 with key genes (highest betweenness centrality) shown as large circles. Sub-networks are shown in different colors. (B) Expression of JUN across the six LMC classes (Pvalue from Kruskal-Wallis test). (C) JUN copy number alterations in LMC class 6 vs other classes. (D) The 6-gene based EMT score in tumors across the six LMC classes (Pvalue from Kruskal-Wallis test). (E) The gene expression of NFKB1 across the 6 LMC classes (Pvalue from Kruskal-Wallis test). (F) The LMC classes association with response to immunotherapy in three cohorts (Pvalue from Fisher’s exact test). Patients in these cohorts were classified into the 6 LMC classes by the NCC method. (G) Expression of AXL across the six LMC classes in the Hugo Cohort dataset (Pvalue from Mann–Whitney U test). (H) Kaplan-Meier plot showing survival curves of LMC class 1, class 3, and class 6 in the Riaz Cohort. Other LMC classes had <5 samples and were excluded.
LMC signature as a potential predictor of response to immunotherapy
The value of the LMC signature in predicting outcome in patients treated with immunotherapy was assessed in three disparate clinical trial cohorts of metastatic melanoma (Figure 5F) (10–12). In the Hugo et al. cohort, tumors classified as class 6 were mainly non-responders to PD-1 blockade in comparison to the other LMC classes (P=0.03). Hugo et al. reported that expression of AXL predicts poor response to PD-1 blockade; the gene expression data revealed significantly higher AXL expression in class 6 tumors when compared to other classes within their cohort (Figure 5G). Similarly, for the cohort reported by Ulloa-Montoya et al., class 6 tumors showed a significantly higher proportion of non-responders to MAGE-A3 immunotherapy in comparison to other classes. The cohort reported by Riaz et al. was predominantly composed of non-responders to anti-CTLA-4 further treated with PD-1 blockade but LMC classes were not convincingly predictive but class 3 predicted poor prognosis, which was consistent with the LMC dataset when compared to good prognosis class 1 (Figure 5H).
Discussion
In this study, transcriptome classification was performed utilising a large population-ascertained cohort of primary melanomas, revealing classes having prognostic value in stage I disease. In stage I tumors, the LMC signature predicted outcome comparably to AJCC staging including SNB. Furthermore, evidence suggests that the signature predicted outcome in patients treated with immunotherapies.
Given the rising incidence of early stage tumors and the cost of adjuvant therapies to health services and to patients in terms of toxicity, there is an urgent need to identify better prognostic and predictive biomarkers for early stage disease. When previous gene signatures were applied to the LMC (3, 8), the signatures robustly predicted outcome when the dataset was analysed as a whole, but failed to do so in stage I tumors alone. Although the full Gerami signature was not available, analysing the prognostic value of genes reported in that study (6) showed that the genes were predictive of prognosis in the whole LMC dataset but not in stage I tumors. In this work, a six-class signature (Supplementary data file) was identified which was not only prognostic in the whole LMC dataset but also in patients diagnosed at AJCC stage I. The prognostic value of the LMC signature was validated in an independent cohort of primary melanoma built in Lund (4) although the number of stage I cases in this cohort was insufficient to allow replication of the signature’s prognostic value in stage I disease.
The LMC signature showed limited overlap with the Lund and TCGA signatures. When comparing it with previously identified immunome clusters by our group (13), two LMC classes strongly overlapped with immune subgroups. The non-overlapping classes could not be clearly discriminated using the immunome clusters suggesting that these LMC classes are driven by different genomic mechanisms. Comparison of LMC signature genes with Gerami genes indicated a biological pathway overlap as Gerami genes were found to be strongly correlated with LMC classes 3 and 5.
Although SNB is an important melanoma staging tool, the surgery is associated with morbidity (33, 34). In the LMC, SNB was observed to be of prognostic value in the whole dataset and in stage I tumors. However, the LMC signature performed just as well. Given the morbidity of SNB, it may be argued that the LMC signature should be tested in an independent study as a possible alternative to this procedure especially in stage I disease where the likelihood of a positive result is overall low and must be weighed against morbidity.
In melanoma, increased immune gene expression has been consistently shown to predict good prognosis (5, 9, 13, 35). However, a subset of tumors (LMC class 6) was observed which, despite showing immune gene expression, resulted in the patient’s early death. Further biological characterisation of this class identified copy number amplifications and increased expression of JUN. Ramsdale et al. have shown that JUN promotes an invasive cell phenotype through activation of the EMT pathway (36), and a higher scoring EMT signature in LMC class 6 confirmed increased activity of the EMT pathway in this class. Riesenberg et al. have reported that increased JUN expression leads to pro-inflammatory and stress signals that promote cytokine expression in coordination with NF-κB (37). Again, these findings are consistent with the presented transcriptomic observations of JUN and NFKB1 in defining LMC class 6 (Figure 5B, 5E). There was insufficient tissue to carry out immunohistochemistry for JUN, therefore JUN protein expression in the TCGA dataset was examined and confirmed a positive correlation between JUN gene transcription and protein expression (Supplementary figure S7). Collectively, these data are indicative of copy number gains resulting in both increased gene expression and transcriptional activity of JUN in LMC class 6 tumors, although further proteomic studies would be required to confirm this.
The LMC signature was associated with response to immunotherapies; specifically, class 6 associated with poor outcome in two of the three tested datasets. None of these data sets are sufficiently large to make clear inferences. It is of note that the expression of AXL, a known marker for immune evasion, was significantly upregulated in LMC class 6 in metastatic melanoma samples in the Hugo data set.
The inherent strength of this study is the relatively large size of the population ascertained cohort. A corresponding limitation is the lack of a well powered AJCC stage I dataset to allow independent replication of the signature in stage I melanoma. Another limitation of this study is that only one-third of LMC patients had a SNB, limiting the power to compare staging tests. The LMC recruitment period preceded the advent of both immunotherapy and targeted therapy, and only a very small number of the study participants have been treated with these drugs. Excluding the samples from these participants showed no modifying effect of such treatments on MSS in the LMC dataset (data not shown).
In conclusion, this study presents a novel signature with demonstrated prognostic value similar in magnitude to that of AJCC staging of melanoma, but having added value in stage I melanoma. The data further confirm that AJCC stage largely captures biological variation associated with survival. The LMC class signature prognostic value was similar to that of SNB in the whole dataset (where their effects were additive) and in stage I disease. The signature predicted poor outcome in patients receiving immunotherapies and in particular identified high-JUN/high-AXL as a tumor phenotype with poor prognosis in early and advanced stage melanoma albeit in very small datasets. This signature has the potential to be trialled as a biomarker in clinical monitoring programs and may help in early identification of patients who may or may not benefit from adjuvant therapies.
Supplementary Material
Translational relevance.
The introduction of adjuvant but toxic therapies for primary melanoma has highlighted the need to stratify patients based on improved prognostic and predictive biomarkers. We report a six-class transcriptomic signature generated from primary melanomas which predicted prognosis, notably in stage I disease. The signature demonstrated comparable prognostic value to that of sentinel node biopsy. When the six classes were applied to published transcriptomic datasets from patients treated with immunotherapy, one class consistently predicted poor outcome. This class was characterised by expression of JUN and AXL, both known determinants of poor therapeutic response in advanced melanoma. These findings suggest that the six-class signature should be applied to larger datasets as they become available, in order to further validate its clinical relevance as a prognostic/predictive biomarker in the adjuvant setting.
Acknowledgements
We thank all the participants of the LMC study and the research nurses who conducted the recruitment.
This work was funded by Cancer Research UK C588/A19167, C8216/A6129, and C588/A10721 and NIH CA83115. RT, JMSD and JP are supported by Horizon 2020 Research and Innovation Programme no. 641458 (MELGEN). Copy number data were generated using AICR grant 12-0023.
Footnotes
The authors have declared no conflicts of interest.
Accession number
The accession number for the microarray data reported in this paper is European Genome Archive accession number: EGAS00001002922.
References
- 1.Lo SN, Scolyer RA, Thompson JF. Long-term survival of patients with thin (T1) cutaneous melanomas: a Breslow thickness cut point of 0.8 mm separates higher-risk and lower-risk tumors. Annals of surgical oncology. 2018;25(4):894–902. doi: 10.1245/s10434-017-6325-1. [DOI] [PubMed] [Google Scholar]
- 2.Cancer research UK 21st September 2018. https://www.cancerresearchuk.org/health-professional/cancer-statistics/statistics-by-cancer-type/melanoma-skin-cancer/diagnosis-and-treatment#ref-1 .
- 3.Jonsson G, et al. Gene expression profiling-based identification of molecular subtypes in stage IV melanomas with different clinical outcome. Clin Cancer Res. 2010;16(13):3356–3367. doi: 10.1158/1078-0432.CCR-09-2509. [DOI] [PubMed] [Google Scholar]
- 4.Harbst K, et al. Molecular profiling reveals low- and high-grade forms of primary melanoma. Clin Cancer Res. 2012;18(15):4026–4036. doi: 10.1158/1078-0432.CCR-12-0343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nsengimana J, et al. Independent replication of a melanoma subtype gene signature and evaluation of its prognostic value and biological correlates in a population cohort. Oncotarget. 2015;6(13):11683–11693. doi: 10.18632/oncotarget.3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gerami P, et al. Development of a prognostic genetic signature to predict the metastatic risk associated with cutaneous melanoma. Clin Cancer Res. 2015;21(1):175–183. doi: 10.1158/1078-0432.CCR-13-3316. [DOI] [PubMed] [Google Scholar]
- 7.Ferris LK, et al. Identification of high-risk cutaneous melanoma tumors is improved when combining the online American Joint Committee on Cancer Individualized Melanoma Patient Outcome Prediction Tool with a 31-gene expression profile–based classification. Journal of the American Academy of Dermatology. 2017;76(5):818–825.:e813. doi: 10.1016/j.jaad.2016.11.051. [DOI] [PubMed] [Google Scholar]
- 8.The Cancer Genome Atlas Network. Genomic classification of cutaneous melanoma. Cell. 2015;161(7):1681–1696. doi: 10.1016/j.cell.2015.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lauss M, Nsengimana J, Staaf J, Newton-Bishop J, Jonsson G. Consensus of Melanoma Gene Expression Subtypes Converges on Biological Entities. J Invest Dermatol. 2016;136(12):2502–2505. doi: 10.1016/j.jid.2016.05.119. [DOI] [PubMed] [Google Scholar]
- 10.Hugo W, et al. Genomic and Transcriptomic Features of Response to Anti-PD-1 Therapy in Metastatic Melanoma. Cell. 2016;165(1):35–44. doi: 10.1016/j.cell.2016.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ulloa-Montoya F, et al. Predictive gene signature in MAGE-A3 antigen-specific cancer immunotherapy. J Clin Oncol. 2013;31(19):2388–2395. doi: 10.1200/JCO.2012.44.3762. [DOI] [PubMed] [Google Scholar]
- 12.Riaz N, et al. Tumor and Microenvironment Evolution during Immunotherapy with Nivolumab. Cell. 2017;171(4):934–949.:e916. doi: 10.1016/j.cell.2017.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nsengimana J, et al. beta-Catenin-mediated immune evasion pathway frequently operates in primary cutaneous melanomas. J Clin Invest. 2018;128(5):2048–2063. doi: 10.1172/JCI95351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clark WH, Jr, et al. Model predicting survival in stage I melanoma based on tumor progression. JNCI: Journal of the National Cancer Institute. 1989;81(24):1893–1904. doi: 10.1093/jnci/81.24.1893. [DOI] [PubMed] [Google Scholar]
- 15.Gershenwald JE, Scolyer RA. Melanoma Staging: American Joint Committee on Cancer (AJCC) 8th Edition and Beyond. Annals of Surgical Oncology. 2018;25(8):2105–2110. doi: 10.1245/s10434-018-6513-7. [DOI] [PubMed] [Google Scholar]
- 16.Du P, Kibbe WA, Lin SM. lumi: a pipeline for processing Illumina microarray. Bioinformatics. 2008;24(13):1547–1548. doi: 10.1093/bioinformatics/btn224. [DOI] [PubMed] [Google Scholar]
- 17.Lauss M, Visne I, Kriegner A, Ringner M, Jonsson G, Hoglund M. Monitoring of technical variation in quantitative high-throughput datasets. Cancer Inform. 2013;12:193–201. doi: 10.4137/CIN.S12862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wilkerson MD, Hayes DN. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics. 2010;26(12):1572–1573. doi: 10.1093/bioinformatics/btq170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Monti S, Tamayo P, Mesirov J, Golub T. Consensus clustering: a resampling-based method for class discovery and visualization of gene expression microarray data. Machine learning. 2003;52(1–2):91–118. [Google Scholar]
- 20.Therneau TM, Verze Lumley T. 2017 [Google Scholar]
- 21.Robin X, et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics. 2011;12(1):77. doi: 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sachs M. plotROC: Generate Useful ROC Curve Charts for Print and Interactive Use, 2016. R package version. 2(1):220. URL http://sachsmc.github.io/plotROC. [Google Scholar]
- 23.Wickham H. ggplot2: elegant graphics for data analysis. Springer; 2016. [Google Scholar]
- 24.Tibshirani R, Chu G, Narasimhan B, Li J. 2011 [Google Scholar]
- 25.Wu G, Dawson E, Duong A, Haw R, Stein L. ReactomeFIViz: a Cytoscape app for pathway and network-based data analysis. F1000Res. 2014;3:146. doi: 10.12688/f1000research.4431.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bastian M, Heymann S, Jacomy M. Gephi: an open source software for exploring and manipulating networks. Icwsm. 2009;8:361–362. [Google Scholar]
- 27.Durinck S, et al. BioMart and Bioconductor: a powerful link between biological databases and microarray data analysis. Bioinformatics. 2005;21(16):3439–3440. doi: 10.1093/bioinformatics/bti525. [DOI] [PubMed] [Google Scholar]
- 28.Durinck S, Spellman PT, Birney E, Huber W. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat Protoc. 2009;4(8):1184–1191. doi: 10.1038/nprot.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cirenajwis H, et al. Molecular stratification of metastatic melanoma using gene expression profiling: Prediction of survival outcome and benefit from molecular targeted therapy. Oncotarget. 2015;6(14):12297. doi: 10.18632/oncotarget.3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Angelova M, et al. Characterization of the immunophenotypes and antigenomes of colorectal cancers reveals distinct tumor escape mechanisms and novel targets for immunotherapy. Genome Biol. 2015;16(1):64. doi: 10.1186/s13059-015-0620-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang RY, et al. Functional relevance of a six mesenchymal gene signature in epithelial-mesenchymal transition (EMT) reversal by the triple angiokinase inhibitor, nintedanib (BIBF1120) Oncotarget. 2015;6(26):22098–22113. doi: 10.18632/oncotarget.4300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liberzon A, Birger C, Thorvaldsdottir H, Ghandi M, Mesirov JP, Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1(6):417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Balch CM, et al. Final version of 2009 AJCC melanoma staging and classification. J Clin Oncol. 2009;27(36):6199–6206. doi: 10.1200/JCO.2009.23.4799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morton DL, et al. Sentinel node biopsy for early-stage melanoma: accuracy and morbidity in MSLT-I, an international multicenter trial. Ann Surg. 2005;242(3):302–311. doi: 10.1097/01.sla.0000181092.50141.fa. discussion 311-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lauss M, et al. Mutational and putative neoantigen load predict clinical benefit of adoptive T cell therapy in melanoma. Nat Commun. 2017;8(1):1738. doi: 10.1038/s41467-017-01460-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ramsdale R, et al. The transcription cofactor c-JUN mediates phenotype switching and BRAF inhibitor resistance in melanoma. Sci Signal. 2015;8(390):ra82. doi: 10.1126/scisignal.aab1111. [DOI] [PubMed] [Google Scholar]
- 37.Riesenberg S, et al. MITF and c-Jun antagonism interconnects melanoma dedifferentiation with pro-inflammatory cytokine responsiveness and myeloid cell recruitment. Nature communications. 2015;6:8755. doi: 10.1038/ncomms9755. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.