Summary
Inflammatory bowel disease (IBD) is a complex autoimmune disorder recently shown to be associated with SUMOylation, a post-translational modification mechanism. Here, we have identified a link between epithelial deSUMOylases and inflammation in IBD. DeSUMOylase SENP7 was seen to be upregulated specifically in intestinal epithelial cells in both human IBD and a mouse model. In steady state, but not IBD, SENP7 expression was negatively regulated by a direct interaction and ubiquitination by SIAH2. Upregulated SENP7 in inflamed tissue displayed a distinct interactome. These changes led to an expansion of localized proinflammatory γδ T cells. Furthermore, in vivo knockdown of SENP7 or depletion of γδ T cells abrogated dextran sulfate sodium (DSS)-induced gut inflammation. Strong statistical correlations between upregulated SENP7 and high clinical disease indices were observed in IBD patients. Overall, our data reveal that epithelial SENP7 is necessary and sufficient for controlling gut inflammation, thus highlighting its importance as a potential drug target.
Introduction
Inflammatory bowel disease (IBD) is a general term given for a group of gastrointestinal (GI) autoimmune diseases of unknown etiology. Among the two major forms of IBD, ulcerative colitis (UC) is characterized by inflammation restricted to the colon, while Crohn’s disease (CD) may manifest in any part of the GI tract. Epidemiologically IBD is on the rise, with more than 3.6 million people affected worldwide. IBD symptoms include diarrhea, abdominal cramps, rectal bleeding, and weight loss, together leading to a severely compromised lifestyle. Genetic background, an aberrant immune system, environmental factors, and composition of the microbiota have been implicated in IBD. Genome-wide association studies (GWASs) have led to the identification of >200 loci associated with IBD, the vast majority of which are shared between UC and CD (Jostins et al., 2012; Liu et al., 2015). The mucosa of IBD patients is densely infiltrated by immune cells of a proinflammatory nature that operate through complex crosstalk with stromal cells, epithelial cells, endothelial cells, and contents of the gastrointestinal lumen, including the commensal flora. Intestinal epithelial cells (IECs) perform both a barrier and signal transduction function in addition to their role in nutrient absorption. IECs have the capacity to sense luminal environment through their surface receptors and transduce the information to the immune system by secretion of a precise concoction of cytokines (Belkaid and Hand, 2014). Such conditioning of the immune system by IECs is critical for the maintenance of intestinal homeostasis. Immune cells, once recruited, carry out their function, which involves secretion of an array of interleukins (ILs). For instance, IL-17A is produced by T helper cells, natural killer (NK) T cells, and T cell receptor γδ (TCRγδ)+ cells and innate lymphoid cells (ILCs), and these cytokines have been strongly implicated in IBD (Lo Re et al., 2010). Prevailing evidence suggests that IECs can provide signals, thereby conditioning expansion of the TCRγδ leucocyte (intraepithelial lymphocyte [IEL]) compartment. TCRγδ IELs exhibit cytotoxic properties and are capable of producing cytokines such as tumor necrosis factor alpha (TNF-α), interferon γ (IFN-γ), transforming growth factor β (TGF-β), IL-10, IL-13, prothymosin β4, keratinocyte growth factor (KGF), and antimicrobial peptides. The function of TCRγδ+ cells has been rather controversial, with some studies demonstrating a protective role (Chen et al., 2002; Catalan-Serra et al., 2017) and others highlighting a negative role in the pathophysiology of IBD (Nanno et al., 2008; Do et al., 2011, 2017). TCRγδ IEL function and phenotype are likely to be determined by the specific milieu of cytokines and the cellular context. The precise signaling mechanisms operational in IECs during IBD that lead to the recruitment of proinflammatory immunocytes triggering disease pathophysiology are currently not well understood. Post-translational modification (PTM) mechanisms have been particularly implicated as crucial molecular systems that may shape the progression of inflammation (Ehrentraut and Colgan, 2012; Liu et al., 2016). SUMOylation, a reversible PTM, in particular has been linked to several inflammatory diseases (Seeler and Dejean, 2017; Yang et al., 2017). This may be attributed to the versatility, dynamicity, and reversibility of SUMOylation. Several reports from our group and others have highlighted the importance of SUMOylation in gut inflammation (Verma et al., 2015; Fritah et al., 2014; Mustfa et al., 2017). The reversibility of SUMOylation is achieved by deconjugation of SUMOylated protein by the action of deSUMOylases, a set of cysteine proteases that are also referred as SUMO-specific proteases or SUMO isopeptidases (Yeh et al., 2000). The human genome encodes for six deSUMOylases (SENP1, SENP2, SENP3, SENP5, SENP6, and SENP7). SENP6 knockdown animals display increased susceptibility to endotoxin shock (Liu et al., 2013), thus showing their role in the inflammatory cascade. The recent demonstration of alleviation of intestinal inflammation by inhibition of SUMO E3 ligase PIAS1 further establishes the intricate connection of SUMOylaiton to IBD progression (Yavvari et al., 2019). However, for possible clinical interventions for IBD, a much more detailed understanding of the tissue-specific functioning of the SUMOylation/deSUMOylation machinery is required. In the current work, we have investigated the possible involvement of epithelial deSUMOylases in governing the conditioning of immune cells. We demonstrate here a critical role for the epithelial deSUMOylase enzyme SENP7 in controlling inflammation via intestinal expansion of γδ T cells.
Results
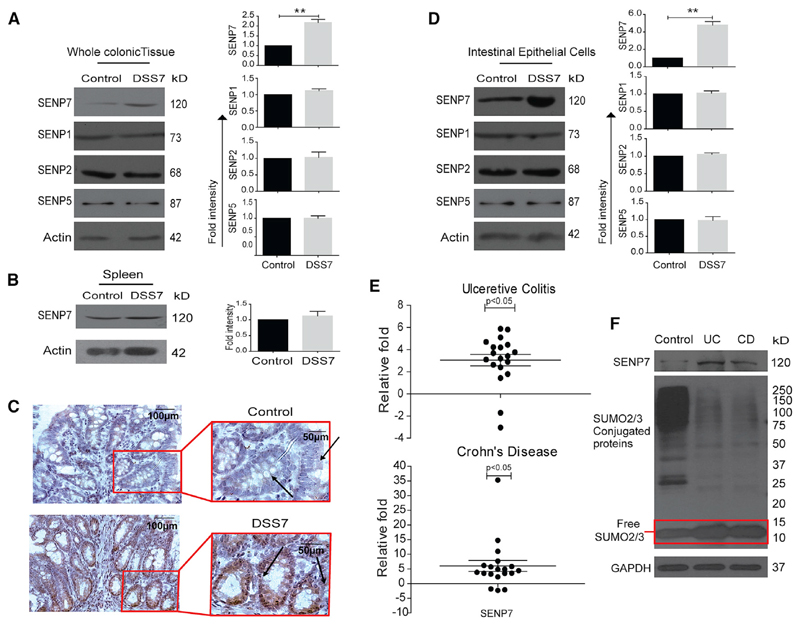
Upregulated Expression of DeSUMOylase SENP7 in Colonic Epithelial Cells in Murine and Human Colitis
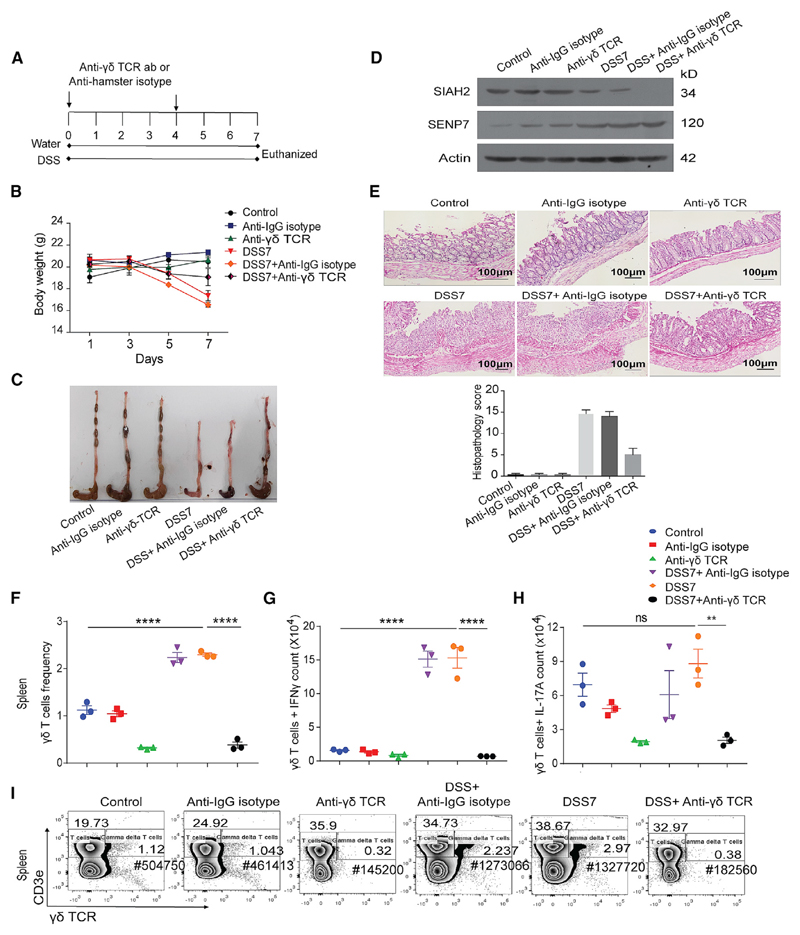
To investigate the possible role of deSUMOylases in IBD, a dextran sulfate sodium (DSS)-treated mouse model was used (Chassaing et al., 2014). DSS treatment of 7 days resulted in weight loss and the development of massive inflammation in the ceca and colon of these animals (hereafter DSS7 mice). This was evidenced by morphological and histopathological markers such as shortening of colon length, neutrophil infiltration, epithelial erosion, and crypt abscesses (Figures S1A and S1B). Expression analysis of deSUMOylases of colonic tissue of these mice revealed no change in the expression of SENP1, SENP2, and SENP5. Immunoblotting for SENP3 and SENP6 was also done, but these antibodies did not detect the desired proteins. However, the expression of SENP7 was significantly elevated in DSS7 mice compared to untreated control animals (Figure 1A). Notably, even a 3-day treatment of DSS resulted in altered expression of SENP7 (Figure 2H). Immunohistochemistry (IHC) of colonic tissue sections revealed a higher expression specifically in epithelial cells (see arrowheads in Figure 1C), crypts, and villi areas of DSS7 mice. Immunohistochemistry of fixed HCT8 cells with SENP7 either overexpressed or knocked down confirmed the specificity of the SENP7 antibody (Figure S1D). Furthermore, immunoblotting from lysates of isolated intestinal intraepithelial cells of DSS7 mice revealed a significant increase in expression of SENP7 (Figure 1D). The increase was specific to intestinal epithelium, while no change in other tissues was observed (Figure 1B). Together, these data suggested dysregulated expression of SENP7 in intestinal epithelial cells of a murine model of colitis. The relevance of these findings was investigated in humans. SENP7 expression of CD and UC endoscopic tissue samples was analyzed. These were part of an earlier study (Mustfa et al., 2017) in which a total of 19 samples each from CD, UC, and control individuals were included here. qRT-PCR using specific oligos for exons within the SENP7 catalytic domain revealed (Figure S1E) 3- and 5-fold higher SENP7 expression in UC and CD patients, respectively, compared to control patients (Figure 1E). SENP7 upregulation was also evidenced in immunoblots of these samples (Figure 1F). An overall decrease in SUMO2/3 modification, a key function of SENP7, was seen in UC and CD samples with a notable increase in the free SUMO2/3 protein (area with a red rectangle in Figure 1F). These outcomes were also seen in DSS7 mice (Figure S1C). Together, these data reveal that the expression of deSUMOylase enzyme SENP7 is altered in human and murine intestinal inflammation.
Figure 1. Upregulated Expression of deSUMOylase SENP7 in Colonic Epithelial Cells in Murine and Human Colitis.
(A) Expression analysis of SENP7, SENP1, SENP2, and SENP5 by immunoblotting from whole colonic tissue lysates of control mice and mice treated with 2.5% DSS for 7 days (DSS7) (n = 3). Densitometric analysis of expression by means + SEM from three independent experiments for the indicated protein (right panel). The quantitation panel was normalized to loading control (β actin) to calculate the fold intensity.
(B) Immunoblots from lysates of spleen showing no change in SENP7 expression. Densitometric analysis of values (means + SEM) from three independent experiments were plotted (right panel).
(C) Immunohistochemistry of SENP7 (brown color) from colonic sections (5 μm) showing expression in intestinal columnar epithelium and crypts (marked with arrows) and cell nuclei (blue color). Scale bar, 100 μm. Inset shows zoomed areas of the image.
(D) Immunoblot of indicated proteins from isolated intestinal epithelial cells lysates of control and DSS-treated mice (DSS7). Densitometric analysis of values (means + SEM) from three independent experiments were plotted (right panel).
(E) qRT-PCR-based fold-change expression of SENP7 from human CD (n = 19) and UC (n = 19) patient colonic epithelium relative to average control values (baseline zero) (n = 19) are plotted. Each dot represents data from one sample. GAPDH was used for normalization. Statistical testing was performed between the control and IBD (UC and CD) group using a Mann-Whitney U test (p value as indicated).
(F) SUMO2/3 immunoblot of lysates of human patient samples. GAPDH was used as a loading control. **p < 0.01.
See also Figure S1.
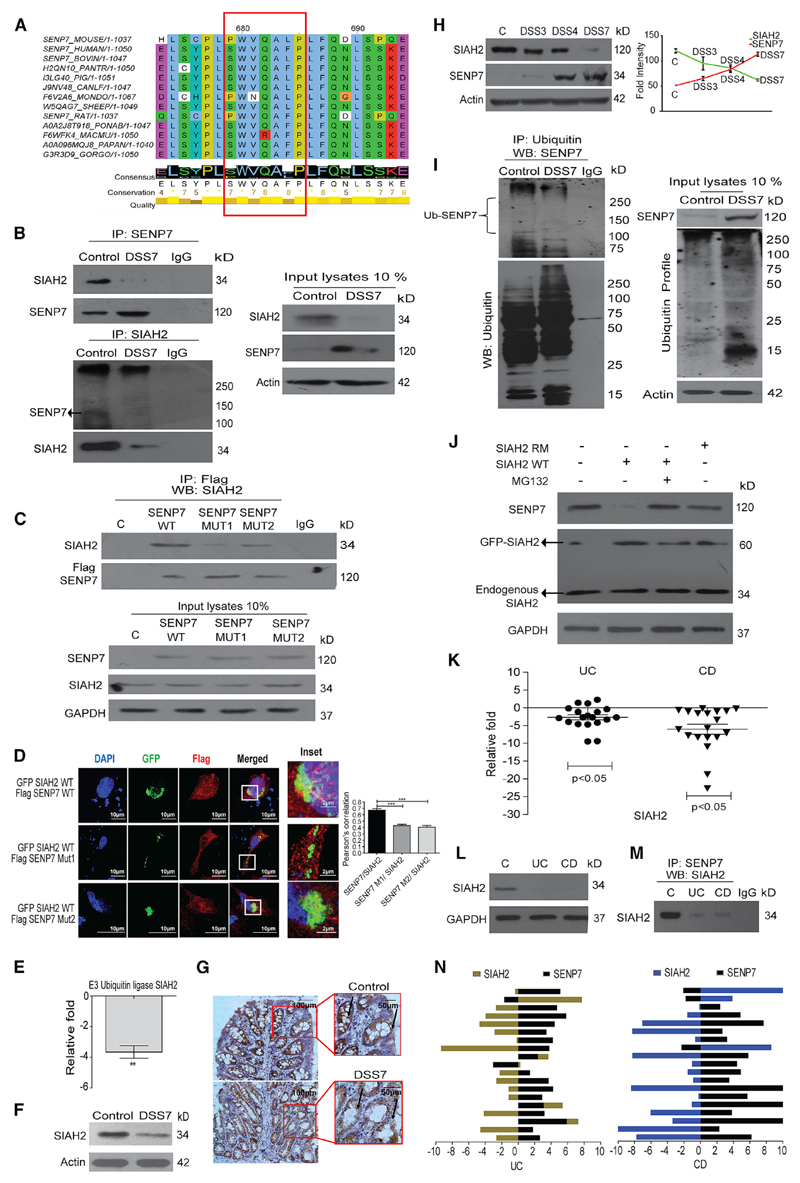
Figure 2. Altered Expression of SENP7 in the Inflamed Colon Is Negatively Regulated by SIAH2.
(A) Multiple sequence alignment of SENP7 primary sequence (from position 679 to 685 highlighting PxAxVxP, red box). Bottom of alignment displays the consensus annotation along with conservation and quality score (conservation depicted in quantitative value and quality by color-coded heatmap).
(B) Top: colonic lysates from untreated and DSS mice (DSS7) immunoprecipitated with anti-SENP7 antibody and immunoblotted. Bottom: reverse coIP using anti-SIAH2 antibody from the same lysates, with 10% of input loaded and immunoblotted. β-Actin was used as a loading control (n = 3).
(C) HCT8 cells transfected with either control FLAG plasmids or FLAG-SENP7 encoding wild-type (WT) or mutant (MUT1 or MUT2) plasmids lysed and immunoprecipitated with anti-FLAG antibody and immunoblotted. Bottom: 10% of input from lysates was loaded and immunoblotted. Isotype antibodies were used as a control (IgG) and GAPDH as a loading control (n = 3).
(D) Representative confocal images from cells transfected with GFP-SIAH2 encoding plasmids along with plasmids, either wild-type FLAG-SENP7 or mutant versions (SENP7 MUT 1 or SENP7 MUT 2). SIAH2 was visualized by GFP and SENP7 by anti-FLAG antibodies. Right: graph representing quantitation of fluorescence from images using Pearson’s correlation for SIAH2-SENP7 colocalization. Results are represented as mean + SEM (n = 3).
(E) Expression analysis of SIAH2 by qRT-PCR of colonic tissues of control and DSS mice (n = 3). The data were calculated as average fold change compared to a baseline obtained from values of the control group. β-Actin was used for normalization.
(F) Immunoblot showing SIAH2 expression in untreated control and DSS7 from lysates of colonic tissue (n = 3).
(G) Immunohistochemistry of colonic section (5 μm) showing SIAH2 expression (indicated by arrows) of DSS treated and control mice (scale bar, 100 μm). A magnified portion of relevance is shown in the inset.
(H) Expression dynamics of SIAH2 and SENP7 by immunoblotting (left panel) of colonic lysates of mice treated with DSS for different durations (DSS3, 3 days; DSS4, 4 days; and DSS7, 7 days). Correlation of SIAH2 and SENP7 levels as revealed by densitometric analysis using fold change from immunoblots (n = 3).
(I) Left: mouse colonic lysates were co-immunoprecipitated with anti-ubiquitin antibody and immunoblotted. Right: immunoblot showing 10% input. β-Actin was used as a loading control.
(J) Immunoblot showing SIAH2 (WT, wild-type; RM, Ring mutant) overexpression in HCT8 cells was sufficient for SENP7 degradation. Degradation dynamics of SENP7 from cells treated with MG132. GAPDH was used as a loading control.
(K) qRT-PCR analysis of SIAH2 from human colonic biopsy tissue samples of UC (n = 19) or CD (n = 19) patients or control individuals calculated as average fold change relative to control values (baseline zero) (n = 19) (each dot represents data from one individual). GAPDH was used for normalization. Statistical testing was performed between the control and IBD (UC and CD) groups using a Mann-Whitney U test (p value as indicated).
(L) Immunoblotting using SIAH2 antibodies of UC and CD mucosal epithelial lysates.
(M) Lysates of pooled biopsy samples from five patients were immunoprecipitated with anti-SENP7 antibody and immunoblotted.
(N) Histogram showing correlation analysis of SIAH2 and SENP7 expression from UC and CD (right panel) patient samples. Histogram prepared using relative fold change gene expression normalized to control. ***p < 0.001; **p < 0.01.
Altered Expression of SENP7 in the Inflamed Colon Is Mediated by the Ubiquitin Ligase SIAH2
To identify the underlying cause of SENP7 expression alteration, its primary sequence was analyzed. Interestingly, presence of a signature motif (PxAxVxP) was observed. This motif is known to be present in substrates of an E3 ubiquitin ligase, SIAH2 (House et al., 2003). However, its orientation in SENP7 was in the reverse direction (PxVxAxP) and was evolutionarily conserved in SENP7 (Figure 2A). Co-immunoprecipitation (coIP) of murine colonic lysates using SENP7-specific antibodies followed by immunoblotting revealed a strong interaction between SENP7 and SIAH2 (Figure 2B). The interaction between SENP7 and SIAH2 was seen in lysates from control, but not DSS-treated, animals (Figure 2B). Immunoprecipitation (IP) of SENP7 was confirmed by anti-SENP7 antibodies, and no interaction was seen with isotype immunoglobulin G (IgG) antibodies. The input blot revealed a decrease in SIAH2 expression in DSS7 animals. (Figure 2B). Reverse coIP also showed specific SENP7-SIAH2 interaction in epithelial lysates of control mice (Figure 2B).
To examine the importance of the PxVxAxP motif, two mutants SENP7-MUT1 (bearing NxVxNxP) and SENP7-MUT2 (bearing PxNxAxN) were created. coIP of lysates of epithelial cells (HCT-8) transfected with plasmids encoding these SENP7 variants showed the interaction between wild-type (WT) SENP7 with SIAH2. However, both SENP7-MUT1 and SENP7-MUT2 variants displayed very weak interaction (Figure 2C). These data revealed that although the PxVxAxP motif was critical for the interaction, it was not the only determinant. Tagged versions of SIAH2 (GFP-SIAH2) and SENP7 (FLAG-SENP7) or mutant SENP7 (SENP7-MUT1 or SENP7 MUT2) were overexpressed in HCT-8 cells followed by confocal imaging. The images revealed colocalization of SIAH2 with wild-type SENP7, but not with SENP7-MUT1 or SENP7-MUT2 (Figure 2D). Together, these data indicate a direct interaction of SENP7 with SIAH2.
Next, SIAH2 expression dynamics were probed. Compared to control animals, SIAH2 expression in DSS7 mice was significantly downregulated (Figures 2E and 2F). IHC revealed downregulation particularly in the intestinal epithelium and crypts of DSS7 mice (Figure 2G). The expression profiles of SIAH2 and SENP7 were investigated over the course of DSS treatment. Notably, SIAH2 protein levels showed a uniform decrease in DSS7 mice, while that of SENP7 was elevated (Figure 2H). Statistical analysis revealed an inverse correlation between these proteins (Figure 2H).
Next, possible ubiquitination of SENP7 by SIAH2 was investigated. coIP using anti-ubiquitin antibody showed bands higher than the regular size for SENP7, indicating possible polyubiquitination (Figure 2I). The higher sized bands were more intense in control lysates (lane 1). The lower molecular weight bands may represent truncated forms of SENP7. To test if SENP7 copy number was being controlled in cells via the ubiquitin-proteasome machinery, we overexpressed wild-type SIAH2 (SIAH2-WT) or its RING mutant, an ubiquitination-defective form (SIAH2-RM), in HCT8 cells followed by treatment with proteasome inhibitor MG132. Although overexpression of SIAH2-RM had no discernable effect on the expression of SENP7, SIAH2-WT showed a deleterious effect on SENP7 levels. Interestingly, treatment with MG132 abrogated SIAH2-mediated decrease of SENP7, suggesting a SIAH2-dependent proteasomal degradation of SENP7 (Figure 2J). Furthermore, expression of SIAH2 in UC and CD patient samples was significantly lowered in UC (−5- to −10-fold) and CD samples (−5- to −20-fold) (Figure 2K). Downregulation of SIAH2 was also evidenced by immunoblotting (Figure 2L). SIAH2-SENP7 physical association was also validated in human patient biopsy specimens (Figure 2M). Interestingly, correlation analysis in UC and CD samples showed a negative correlation between SIAH2 and SENP7 (Figure 2N). Together, these data revealed that in the healthy, but not inflamed, murine and human intestine, SENP7 interacts with SIAH2 and undergoes ubiquitination-dependent degradation.
SENP7 Displays an Altered Repertoire of Interactors in Inflamed Tissue
To understand the precise role of SENP7 in inflammation, we set forth to determine the SENP7 interactome through coIP of murine colonic lysates with SENP7 antibodies. Interactors of SENP7 would allow identification of colitis-specific regulators. These antibodies (Invitrogen PA5-36089) were very specific for SENP7, as revealed by a single band of the correct size obtained in the immunoblot (Figure S2A). Immunoprecipitated proteins using anti-SENP7 or isotype antibodies from colonic lysates of control and DSS7 animals (n = 5) (Figure 3A) were subjected to high-resolution tandem mass spectrometry qTOF analysis (MS/MS). The combined workflow enabled the identification of ~30,000 high-confidence total spectra using three search engines (Comet, Tandom, and SpectraST) and rescue spectras representing 1,758 proteins with a ProteinProphet probability of 0.99 and an error rate <0.00001. Further, to determine the quantitative values for the proteins identified, we used the Maxquant platform. The peptides identified in the negative control IgG isotype were subtracted manually in the later part of the analysis. The intensity-based absolute quantification (iBAQ)-derived differential proteome identified in control and DSS7 mice is represented in a Venn diagram (Figure 3B) showing the sum total of nonredundant proteins of 451 DSS7 mice compared to 385 mice in the control group, out of which 114 proteins were unique to the control group and 180 proteins were unique to DSS7 mice. We were able to identify the presence of SENP7 and SIAH2 in the lysates. Notably, the list includes several proteins that are known to be SUMOylated (Table 1).
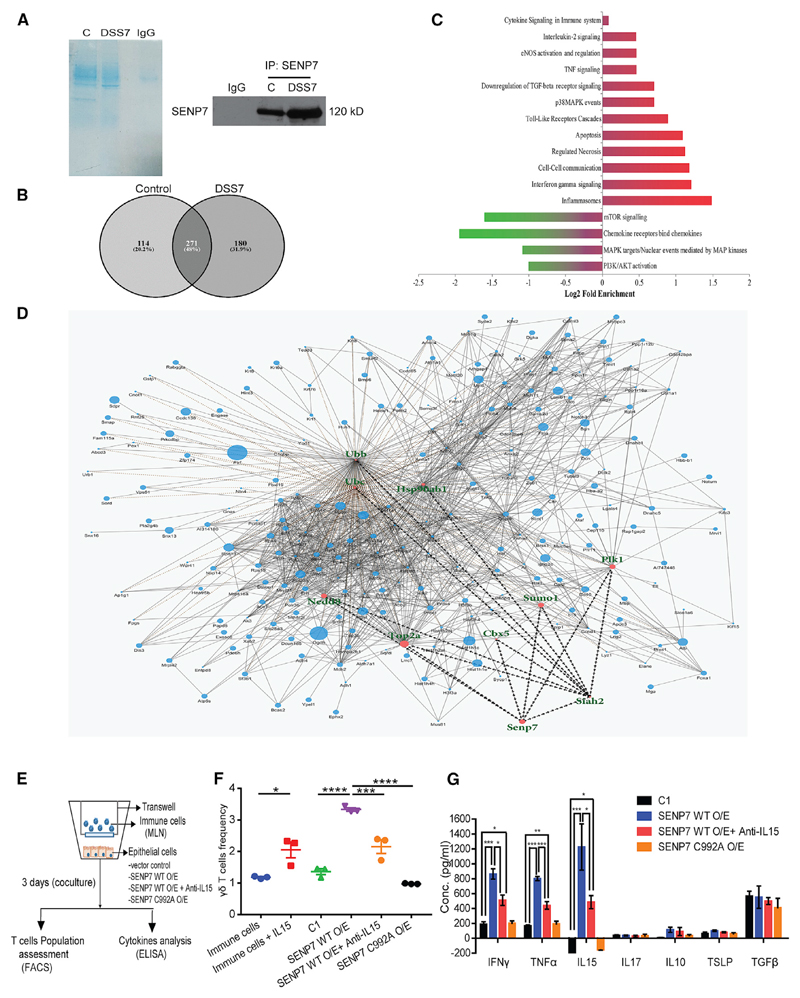
Figure 3. Altered Dynamics of the SENP7 Interactome in DSS Mice.
(A) Coomassie staining (left panel) and immunoblotting (right panel) of samples co-immunoprecipitated with anti-SENP7 or IgG from colonic lysates of control and DSS7 mice. Samples from (A) were in-gel digested and analyzed by ESI-tandem mass spectrometry (MS/MS) (n = 5 mice).
(B) Venn diagram displaying the number of individual proteins identified by MS/MS.
(C) The relative abundance of various signaling pathways based on MS/MS prepared using the Reactome database (shown as that of DSS-7 mice versus control). Gene Ontology is represented as fold enrichment. Probability was determined using a binomial statistic for the false discovery rate (FDR) and a p value cutoff of ≤ 0.05 significance level.
(D) Protein-protein interaction network for SENP7 and its interacting hub proteins based on the Boolean network model. The size of the node (circles) in the network represents the fold change identified by the LFQ algorithm, and the border of the nodes represents iBAQ-based fold change.
(E) Schematic of the steps involved in co-culture experiments using CT26 cells and total immune cells from MLNs.
(F) Population frequency γδ T cells was calculated and plotted (labeling: immune cells, cells from MLNs; immune cells + IL15, IL-15 treatment of cultured immune cells for 3 days; C1, immune cells co-cultured with CT26 cells transfected with empty FLAG vector; SENP7 WT O/E, SENP7 wild-type overexpressed CT26 in co-culture setup; SENP7 C992A O/E, catalytic mutant of SENP7; SENP7 WT O/E + anti-IL15, SENP7 overexpressed epithelial cells with IL-15 neutralization).
(G) ELISA for indicated cytokines from the supernatant of C1, SENP7 WT O/E, SENP7 WT O/E + Anti-IL15, and SENP C992A O/E cells described above. Each plot is representative of three independent experiments. Results are represented as mean+ SEM.
p values were determined by Student’s t test and one-way ANOVA with Tukey’s posttest (*p < 0.05; **p < 0.01; ***p < 0.001; ns, non-significant). See also Figure S2 and Tables S1, S2, and S3.
Table 1. SENP7-Interacting Proteins along with SUMOylation Site and Their Function in Disease.
| Protein Name | Condition of Identification | SUMOylation Site | SUMO Interaction Motif (SIM) | Function | Disease Reported |
|---|---|---|---|---|---|
| NCoR1a | DSS | 145/DAKK |
– | repress NF-κB and AP-1 genes (Tiefenbach et al., 2006; Perissi et al., 2004) | breast cancer |
| 1068/IKQE |
(Zhang et al., 2006) | ||||
| 1333IKFD | |||||
|
| |||||
| Notch1a | DSS | 1607/FKRD |
1575-1579/VLVVL | stem cell development, γδ T cell activation (Stier et al., 2002; Washburn et al., 1997) | IBD, cancer |
| 1712/VKSE |
(Dahan et al., 2011) | ||||
| 2242/AKPE | |||||
|
| |||||
| CBX5a | control/DSS | 84/EKSE | – | transcriptional activator | breast cancer (Bawa-Khalfe et al., 2012) |
|
| |||||
| PLK1a | control/DSS | 358/EKEE | – | cell cycle (Wen et al., 2017) | colorectal cancer (Takahashi et al., 2003) |
|
| |||||
| Vimentina | control/ DSS | 373/MKEE | – | epithelial-mesenchymal transition (Bawa-Khalfe et al., 2012) | IBD (Andoh et al., 2002) |
|
| |||||
| Birc2 | control/ DSS | 38/EKMK | 424-428/IVSVL | anti-apoptotic | IBD (Grabinger et al., 2017) |
|
| |||||
| 40/MKFD | 575-579/IVFIP | ||||
|
| |||||
| CARD9 | DSS | 165/LKEE | 22-26/ISVID | regulate gut microbiota | IBD (Lamas et al., 2016) |
|
| |||||
| 219/MAKE |
343-347/IEAIL | ||||
| 391/EKAD | |||||
|
| |||||
| S100A9 | DSS | 35/SKKE |
– | calcium-binding protein | IBD (Lee et al., 2012) |
| 51/MKKE | |||||
|
| |||||
| E3 ubiquitin ligase SIAH2 | control/DSS | – | 250-254/VLLIG | ubiquitination | prostate cancer (Qi et al., 2013) |
SUMOylation of these proteins is known.
Tables S1 and S2 list all of the proteins identified by all of the search engines. One of the representative proteins identified with a high score and peptide-to-spectra match (PSM) was NCoR1. We validated its interaction with SENP7 (Figure S2B). The results were further used to identify the gene-term enrichment analysis (Table S3). The highly significant biologically over-represented processes identified are associated either with immunological processes or related pathways (Figure 3C).
To infer the regulatory function of the data, we used the mathematical graph theory to understand the protein-protein interaction (PPI) network for SENP7 and its interacting hub proteins using a Boolean network model (details in STAR Methods), we identified that SENP7 is the direct interacting partner of CBX5, SIAH2, and SUMO-1 (Figure 3D). The presence of SIAH2 highlighted the reliability of the data. Based on the interactome, Gene Ontology (GO) enrichment, and previous results, we speculated that dysregulation of the SIAH2-SENP7 interaction in the epithelium was required for immune conditioning.
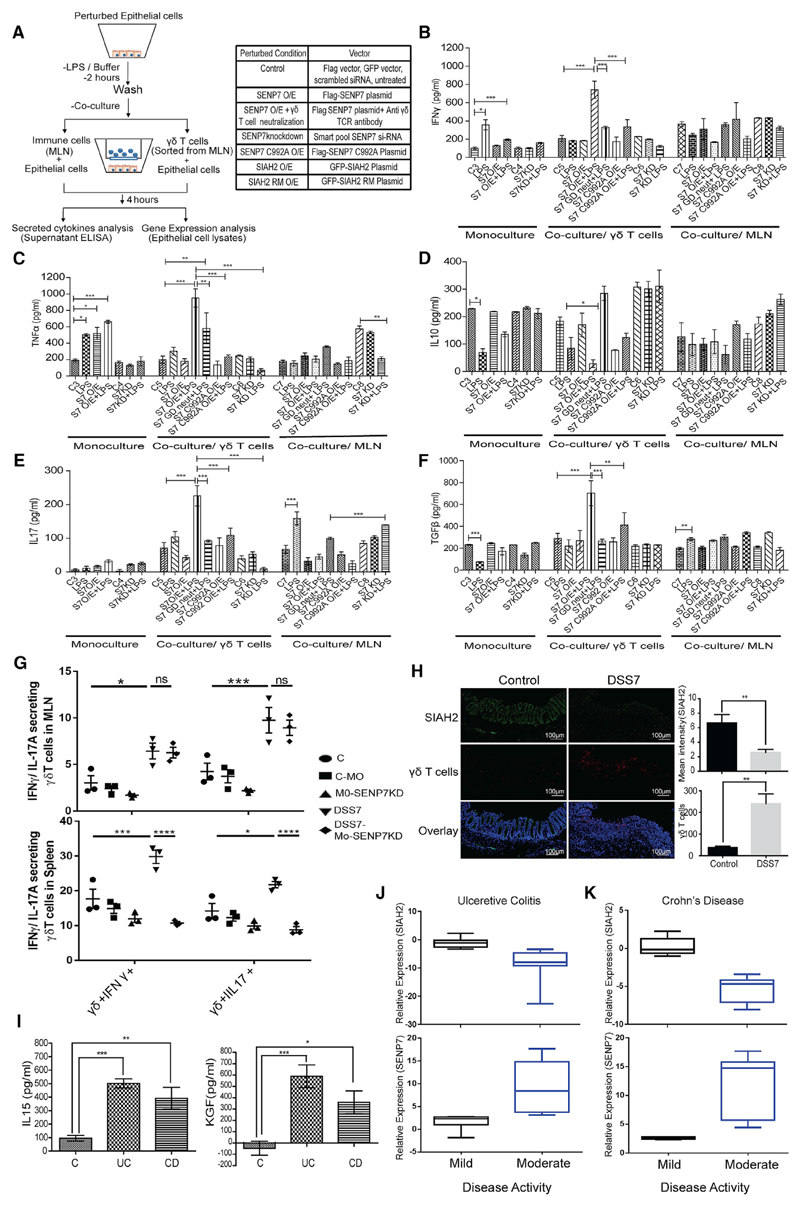
To test this possibility, we carried out co-culture experiments with murine colonic epithelial cells (CT26 line) grown in the bottom well and immunocytes (from mice mesenteric lymph nodes [MLNs]) grown in the inserts (upper well). The idea was to understand the effect of epithelial SENP7 perturbation on immune cells. SENP7 expression of CT26 cells (bottom well) was upregulated using a plasmid encoding SENP7 wild-type (WT) or catalytic mutant of SENP7 (C992A). Monocultures (epithelial cells only) were also included as a control group. In co-culture, the various perturbations were done as shown in Figure 3E. The cells were co-cultured for 3 days, followed by flow cytometry for immune phenotyping. We analyzed T cells (αβ T and γδ T cells), since they are important for IBD. A subtle expansion of the γδ T cell population in unperturbed epithelial cells (control 1) was observed. Remarkably γδ T cell frequency was increased by 3-fold when SENP7 was overexpressed (WT O/E) experimentally (Figures 3F and S2F), whereas an overexpressed catalytic mutant of SENP7 (C992A) was devoid of γδ T cell expansion (Figures 3F and S2F). Apart from this, no change in αβ CD4+ T cells and αβ CD8+ T cells was observed (Figures S2D, S2E, and S2G). Cytokine analysis of the supernatants from SENP7 WT O/E revealed a dramatic increase in IFN-γ, TNF-α, and IL-15, whereas no change on IL-17A was observed (Figure 3G). Remarkably overexpression of the SENP7 C992A mutant did not show any significant change in the expression of these cytokines, indicating a direct effect of SENP7 catalytic activity (Figure 3G). Monoculture of epithelial cells with SENP7 WT O/E, but not SENP7 C992A mutant, resulted in a significant increase in cytokine secretion levels of IFN-γ, TNF-α, and IL-15 (Figure S2I). It was previously reported that IL-15 is important for γδ T cell activation and proliferation and is highly expressed in IBD (Van Acker et al., 2016; Liu et al., 2000). Thus, we treated immune cells with recombinant IL-15, which led to an increase in the proliferation of γδ T cells (Figures 3F and S2F). Cytokine analysis from the collected supernatant of these samples showed a significant increase in the proinflammatory cytokines IFN-γ and TNF-α from IL-15-treated immune cells (Figure S2H). To test if the effect of IL-15 on γδ T cells was via SENP7, we neutralized IL-15 using anti-IL15 antibodies in co-culture. Flow cytometry analysis of immune cells revealed that IL-15 inhibition resulted in only a 2-fold increase in the γδ T cell population (Figures 3F and S2F). Moreover, IL-15 neutralization did not affect the cytokine secretion of IFN-γ, TNF-α, and IL-15 from SENP7-overexpressed epithelial cells but partially reduced them in comparison to the SENP7 O/E condition (Figure 3G) The levels of anti-inflammatory cytokines such as TGF-β, IL-10, and thymic stromal lymphopoietin (TSLP) appeared to be more or less similar in all conditions (Figure 3G). Together, these data revealed that epithelial SENP7 activation was sufficient for triggering inflammatory signaling leading to the accumulation of γδ T cells.
SENP7 Suppression In Vivo Relieves Mice from DSS-Induced Colitis
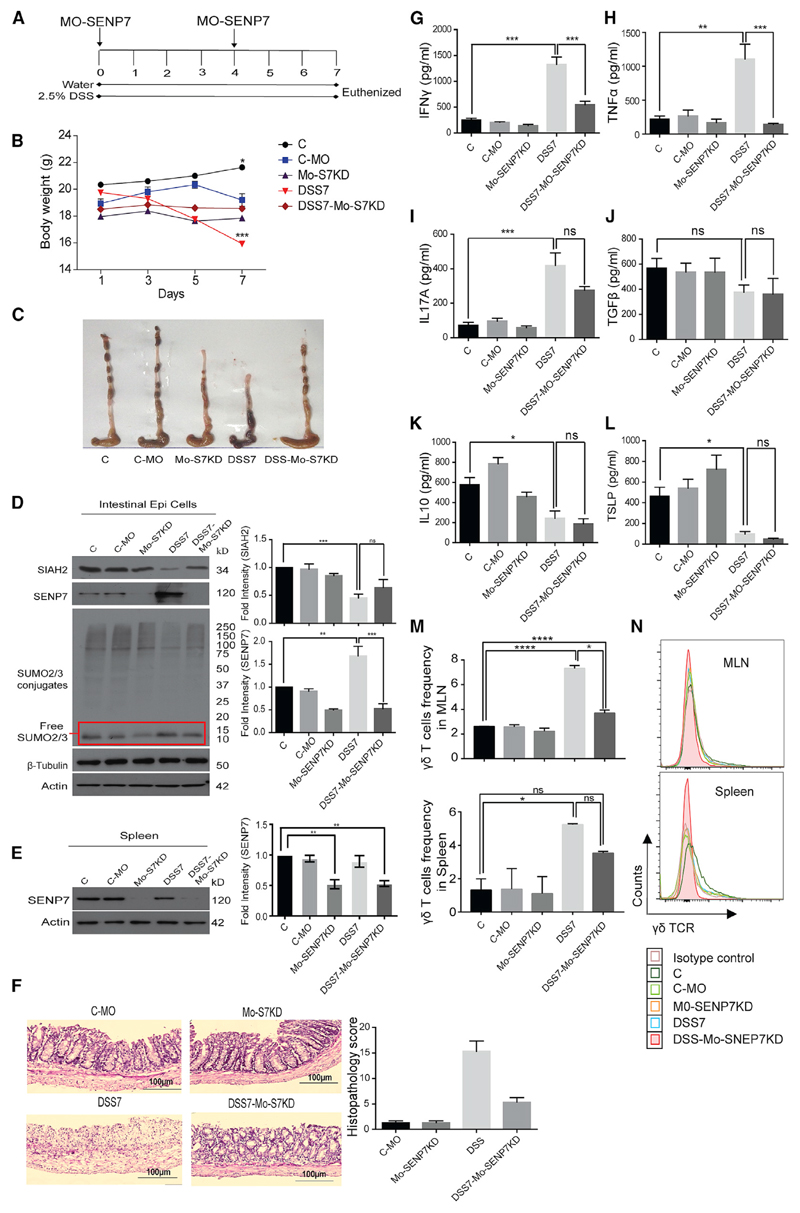
Since SENP7 knockout mice are nonviable (Huang et al., 2017), to gain a deeper understanding of the specific role of SENP7 in intestinal inflammation, an in vivo knockdown model of SENP7 was established. A customized specific morpholino was utilized (Gene Tools). The morpholino comprised two parts, a gene-specific oligomer and octa-guanidine dendrimer delivery moiety (mouse SENP7 Vivo morpholino). The strategy of SENP7 knockdown (hereafter Mo-SENP7KD) is depicted in Figure 4A. A standard morpholino group, targeting human beta-globin that is absent in mice, was also included as a negative control (hereafter C-MO). Treatment of the morpholino (C-MO or Mo-SENP7KD) did not result in weight loss or any other discernible change in animals (Figure 4B). The animals were euthanized after day 7 of DSS treatment, and various tissues of interest were isolated. In the colonic epithelium of Mo-SENP7KD mice, the levels of SENP7 were only half of those of C-MO mice, thus confirming knockdown (Figure 4D). Notably, even after DSS treatment in Mo-SENP7KD (hereafter DSS7-Mo-SENP7KD) mice, we did not observe any upregulation of SENP7, while we observed an increase in the DSS7-alone group. In line with our previous results, in these animals, SIAH2 expression was downregulated in the DSS7 and DSS7-Mo-SENP7KD groups, suggesting that SIAH2 expression was not dependent on SENP7 (Figure 4D). SUMO2/3 profiles from the colon revealed an increase in free SUMO2/3 in DSS-treated, but not DSS7-Mo-SENP7KD, mice (Figure 4D). Expression of SENP7 was downregulated in splenocytes in the DSS7-Mo-SENP7KD group, but we did not find any significant change upon DSS treatment (Figure 4E). Thus, based on these results, we confirmed the successful establishment of this in vivo SENP7 knockdown model. The morphology of intestines revealed that DSS7 treatment led to shortening of the ceca and colon, thereby confirming severe inflammation. Surprisingly, colon and ceca of the knockdown animals treated with DSS (DSS7-Mo-SENP7KD animals) showed no signs of inflammation (Figure 4C). Histopathology revealed the presence of several markers of inflammation in DSS7-treated animals, but not DSS7-Mo-SENP7KD animals, such as epithelial disruption, crypt abscess, and PMN infiltration (Figure 4F). The decrease in immune cell recruitment in Mo-SENP7KD animals was drastic, indicating that DSS administration did not lead to colitis. Such changes are often determined by the nature of cytokines that are secreted by the immune cells and the epithelium. Mucus secretions from intestines were collected for cytokine profiling by ELISA for a series of pro and anti-inflammatory cytokines. We observed elevated levels of several proinflammatory cytokines such as IFN-γ (5.22- ± 0.47-fold), TNF-α (5.01- ± 0.82-fold), and IL-17A (6- ± 0.86-fold) in DSS7 mice compared to either control or DSS7-Mo-SENP7KD animals (Figures 4G–4I). On the other hand, levels of anti-inflammatory cytokines were decreased in DSS7 mice, including TSLP (−5.46- ± 1.32-fold) and IL-10 (−2.7- ± 0.56-fold). The levels of TSLP and IL-10 were only marginally different in DSS7-Mo-SENP7KD mice group compared to DSS7 mice (Figures 4K and 4L). The mRNA expression analysis revealed the same results (Figures S3A and S3B), suggesting that knockdown of SENP7 abrogate inflammation.
Figure 4. SENP7 Suppression Ameliorates DSS Induced Colitis.
(A) Schematic representation of knockdown of SENP7 in mice. SENP7 morpholino was injected intraperitoneally (day 0) and on day 4, followed by euthanasia and tissue isolation (n = 3).
(B) Mouse body weight (C, untreated control; C-MO, negative control; Mo-SENP7KD, SENP7 knockdown; DSS7, 7 days of DSS treatment; DSS7-Mo-SENP7KD, SENP7 knockdown with 7 days of DSS treatment).
(C) Gross morphology of the colon.
(D) Immunoblot showing protein expression of SIAH2, SENP7, and SUMO2/3 from isolated intestinal epithelial cells. Densitometry analysis by means + SEM from three independent experiments (right panel).
(E) Immunoblot showing SENP7 expression in splenocytes of the indicated group. Actin was used as a loading control in (D) and (E). Densitometry analysis by means + SEM from three independent experiments (right panel).
(F) Histopathology along with scores (lower panel) of colonic sections.
(G–L) ELISAs of mucosal isolates from the intestines of mice: IFN-γ (G), TNF-α (H), IL-17 (I), TGF-β (J), IL-10 (K), and TSLP (L) (n = 3 mice). Data are presented as mean + SEM.
(M) Bar graphs showing frequency of γδ T cells in presence of morpholino-mediated SENP7 knockdown in DSS and recovery groups in MLNs and spleen.
(N) Histogram overlay for γδ T frequency for the above groups. Statistical significance was calculated using a one-way ANOVA with Tukey’s posttest.
*p < 0.05; **p < 0.01; ***p < 0.001; ns, non-significant. See also Figures S3–S5.
Next, to assess the immunological consequences of epithelial upregulation of SENP7, immune phenotyping was carried out. MLNs and spleens were isolated from various groups of mice described above followed by profiling of immune cells by flow cytometry using fluorochrome-conjugated antibodies. In both organs, DSS7 treatment did not result in any significant change in the total CD4+ T population (Figure S3G), but a minor subset of T cells (i.e., γδ T cells) showed a significant increase compared to control untreated mice or C-MO mice (Figures 4M and 4N). Strikingly, mice treated with DSS7-Mo-SENP7KD displayed a reduced number of γδ T cells from MLNs and spleens (Figures 4M and 4N). This was also confirmed by immunohistochemistry of the intraepithelial region, which revealed profound γδ T cell infiltration in DSS7, but not DSS-Mo-SENP7KD, mice (Figure S3C). Expression of SIAH2-SENP7 was checked directly on fluorescence-activated cell sorting (FACS)-sorted γδ T cells, total immune cells (from MLNs), and immune cells devoid of γδ T cells (from MLNs). No significant change in SIAH2 or SENP7 expression in any of these cell types was observed (Figure S3D). Together, these data indicated alteration of epithelial SENP7-SIAH2 expression during intestinal inflammation trigger an expansion of γδ T cells. We reasoned that γδ T cells might be key responders of SENP7-dependent epithelial signaling. γδ T cells are known to be regulated by a set of molecules, such as IL-15, aryl hydrocarbon receptor (AhR), and KGF (Khairallah et al., 2018). Therefore, the expression of these regulatory molecules linked to γδ T cells was examined. Significant IL-15 upregulation (~3.88- ± 0.11-fold) was observed in the colonic epithelium of DSS7 mice, but not in DSS7-treated Mo-SENP7KD animals (only ~1.09- ± 0.06-fold) (Figure S3E). We did not observe any significant change in the expression of AhR in DSS7 mice or DSS7-Mo-SENP7KD mice (Figure S3E). Notably, levels of KGF increased (~2-fold) in DSS7 mice but drastically downregulated in DSS7-Mo-SENP7KD mice. Further, direct examination of IL-15 secretion in the mucus layer by ELISA revealed a ~19.31- ± 3.35-fold increase in DSS7 mice compared to control animals. Notably, levels of this cytokine were significantly reduced in DSS7-Mo-SENP7KD mice (Figure S3F), suggesting an epithelial SENP7-dependent regulation of these T-cell-specific cytokine levels. These data led us to conclude that epithelial SENP7 expression modulates the secretion of IL-15, which in turn favors expansion of intestinal γδ T cells.
We tested the role of SIAH2 in the modulation of intestinal γδ T cells during inflammation. We used 17β-estradiol, which is known to induce SIAH2 (Frasor et al., 2005). Only male animals were included in the study to prevent steroidal interference in female mice. The overall methodology that was followed is described in Figure S4A. Post-treatment, the animals were euthanized, and expression analysis was carried out. The relative expression of SIAH2 went up (~3-fold), along with a concomitant decrease in SENP7 expression (2.5-fold) in the colonic tissue (Figures S4D and S4E), while the housekeeping genes β-actin and β-tubulin remained unaffected, thus negating the nonspecific effect of inducer for SIAH2 (Figures S4E). The impact of SIAH2-mediated SENP7 downregulation was further highlighted by a conspicuous loss in free SUMO2/3 and an increase in overall SUMO2/3-conjugation (Figure S4E). Notably, inducer-17βE-treated animals appeared to be resistant to DSS-induced colitis. This can be discerned by several parameters, including an increase in body weight of these animals and other relevant markers (Figure S4B). Specifically, while the colon and ceca of DSS7 mice were inflamed, the inducer-17βE-treated animals appeared to be normal and thus were similar to control animals (Figure S4B). Furthermore, histopathology of colonic sections revealed massive neutrophil infiltration (marked by an arrow in Figure S4C) in DSS7 mice that was absent in animals treated with inducer-17β E (Figure S4C). Together, our data revealed that experimental induction of SIAH2 or downregulation of SENP7 is sufficient for the abrogation of colitis.
To further understand the pathogenesis, immune phenotyping was performed using IELs, lamina propria (LP), MLNs, and spleens from these animals. Specifically, CD4+ T cells, CD8+ T cells, and γδ T cells were analyzed. Among the various cells examined CD4+ T cells or CD8+ T cells displayed no significant change during DSS treatment. Furthermore, subcellular staining of CD4+ and CD+ T cells with IFN-γ and IL-17A showed no significant change in DSS colitis in IELs (Figures S5A–S5G), LP, spleen, and MLNs (Suhail et al., 2019, Mendeley data), while γδ T cells upon DSS treatment showed a significant increase in IELs (Figure S4G), spleen (Figures S5L and S5O), and MLNs (Figures S5P and S5S) in comparison to control animals. No significant change was observed in the case of LP of these animals (Figures S5H and S5K). DSS treatment in animals pretreated with the inducer-17βE showed a much-reduced expansion of γδ T cell frequency in IELs (14-fold), spleen (3-fold), and MLNs (1.5-fold). Subcellular staining of γδ T cells with proinflammatory markers revealed that IFN-γ and IL-17A γδ T cells were significantly enhanced in DSS7 mice, whereas inducer-17βE-DSS-treated animals appeared to have reduced population of IFN-γ- and IL-17A-secreting γδ T cells in IELs (Figures S4H and S4I), spleen (Figures S5M and S5N), and MLNs (Figure S5Q and S5R). These changes at the cellular level were accompanied by a significant increase in the proinflammatory cytokines IFN-γ, IL-17A, and IL-9 and a concomitant decrease in the anti-inflammatory cytokine IL-10 (Figure S4J). Interestingly, expression levels of proinflammatory cytokines remained compromised in the inducer-17βE-DSS7 group (Figure S4J). In the intraepithelial region of colonic tissues of these mice, we observed a reduction in infiltration of γδ T cells compared to the DSS7 group (Figure S4F). These data led us to hypothesize that epithelial SIAH2-SENP7 signaling leads to an increase in γδ T cells during colitis.
γδ T Cell Depletion Prevents DSS-Induced Colitis
Since our data thus far pointed toward the involvement of γδ T cells in the progression of colitis, we investigated the effect of γδ T cell depletion on inflammation. An antibody against γδ TCR for neutralization was given intraperitoneally to mice. The overall methodology that was followed is described in Figure 5A. Post-treatment, the animals were euthanized, and various tissues were isolated. The relative γδ T cell population was found to be reduced by a factor of 5-fold in spleen (Figures 5F and 5I), IELs (Figures S6A and S6D), LPs (Figures S6E and S6H), and MLNs (Figures S6I and S6L) in comparison to control animals. Moreover, akin to our previous data in Figures S4H and S4I, IFN-γ - and IL-17A-secreting γδ T cells increased in spleen and IELs of DSS7 colitis mice, but not in γδ-T-cell-depleted animals (Figures 5G, 5H, S6B, and S6C). Similarly- IFN-γ- and IL-17A-secreting γδ T cells were depleted in LP and MLNs (Figures S6F, S6G, S6J, and S6K). CD4+ and CD8+ T cells are unaffected in γδ-T-cell-depleted mice in all tissues studied (Suhail et al., 2019, Mendeley Data). Expression of SIAH2 was reduced in DSS7-γδ-T-cell-depleted mice, similarly as in DSS7 mice. Also, SENP7 expression was upregulated in DSS7 mice as well as in γδ-T-cell-depleted mice (Figure 5D). Notably, γδ-T-cell-depleted mice appeared to be resistant to DSS-induced colitis (Figure 5B). Notably, the colon and ceca of DSS7 mice were shrunken, indicating inflammation, whereas the γδ-T-cell-depleted animals treated with DSS appeared to be normal, except for traces of rectal bleeding (Figure 5C). Furthermore, histopathology of colonic sections revealed massive neutrophil infiltration in DSS7 mice that was absent in animals treated with γδ T cell neutralizing antibody (Figure 5E). Together, our data revealed that in vivo depletion of γδ T cells, experimental induction of SIAH2, or downregulation of SENP7 is sufficient for preventing the development of colitis.
Figure 5. γδ T Cell Depletion Prevents DSS-Induced Colitis.
(A) Schematic of steps involved in γδ T cell depletion in mice. Anti-γδ TCR was injected intraperitoneally (two doses each of 0.5 mg/mouse) at the start of experiment (day 0) and at day 4, followed by euthanasia and tissue isolation (n = 3).
(B) Body weight of animals (Control, untreated control; Anti-IgG hamster isotype, mice given IgG isotype antibodies; anti-γδ TCR, γδ-T-cell-depleted mice; DSS7, 7 days of DSS treatment; DSS7 + anti-IgG isotype, non-targeting IgG isotype antibody with 7 days of DSS treatment; DSS7 + anti-γδ TCR, γδ-T-cell-depleted mice with 7 days of DSS treatment (n = 3).
(C) Gross morphology of mice colon of various categories.
(D) Immunoblot of SIAH2 and SENP7. β-Actin was used as a loading control.
(E) Histopathology of colonic cross-sections Scale bar, 100μm. Right: histopathology score calculated on the basis of inflammatory markers.
(F–I) Representative graph and contour plot showing immunophenotyping and intracellular cytokine staining of the indicated cell types from spleen (n = 3). For flow cytometry, cells were acquired on a BD FACS Canto, analyzed using FlowJo, and represented in the form of a graph (mean + SEM value). Statistical significance was calculated using a one-way ANOVA with Tukey’s posttest.
**p < 0.01; ****p < 0.001; ns, non-significant. See also Figure S6.
SENP7 Overexpression Triggers Activation of γδ T Cells, Leading to Proinflammatory Signaling
To explore the role of epithelial SENP7 during IBD in modulation of γδ T cell expansion, we carried out co-culture experiments with several different perturbations in epithelial cells. The overview of the strategy used is depicted in Figure 6A. Overexpression by SENP7 and SIAH2 encoding plasmids or knockdown by SENP7-specific small interfering RNA (siRNA) was done on epithelial cells by transfection (Figures S7G and S7H). These cells were either left untreated or stimulated by LPS for 2 h followed by co-culturing with immune cells from MLNs or FACS-sorted γδ T cells (right panel) of mice. As a negative control, these epithelial cells were also cultured separately (monoculture). As discerned by ELISA of culture supernatant, LPS stimulus in monoculture epithelial cells showed only a modest increase (~2-fold) in IFN-γ and TNF-α (Figures 6B and 6C, left panel) and a reduction in IL-10 and TGF-β (Figures 6D and 6F, left panel), while in the co-culture condition with γδ T cells in unperturbed epithelial cells with LPS stimulation, no change was observed. Notably, SENP7-overexpressed (SENP7 WT) epithelial cells and LPS stimulation showed exacerbated inflammatory response. We observed a significant increase in the secretion of IFN-γ (3- ± 0.2-fold), TNF-α (6.2- ± 0.8-fold), and IL-17A (2.5- ± 0.2-fold) (Figures 6B, 6C, and 6E, middle panel) in the culture supernatant of SENP7 WT and LPS-stimulated cells. Notably, in a catalytic mutant of SENP7 (SENP7 C992A), these changes were not observed, thus indicating a role in the induction of proinflammatory cytokine secretion. The anti-inflammatory cytokine IL-10 was decreased (−2- ± 0.1-fold) in the SENP7-overexpressed and stimulated condition (Figure 6D, middle panel), and TGF-β secretion was increased. Neutralization of γδ T cells using anti-γδ TCR antibodies resulted in a significant reduction of IFN-γ and IL-17A compared to control (Figures 6B and 6E, middle panel). The decrease in levels of TNF-α was only subtle (Figure 6C, middle panel). However, siRNA-mediated knockdown of SENP7 led to decreased secretion of IFN-γ, TNF-α, and IL-17A compared to those treated with control scrambled siRNA. Furthermore, SIAH2 overexpression mirrored these results (Figures S7B–S7F). Analysis by qRT-PCR in epithelial cells monocultured and co-cultured with γδ T cells showed significant upregulation of IFN-γ, TNF-α, and IL-15 (Figures S7I and S7J), and these cytokines are known to activate γδ T cells (Lahn et al., 1998). Epithelial cells with SENP7 WT overexpressed cultured with total immune cells did not show any change in IFN-γ, TNF-α, IL-10, and IL-17A secretion (Figures 6B–6E, right panel, and Figure S7K) compared to control. This could be attributed to the presence of other immune cells. Interestingly, subcellular staining of γδ T cells isolated from MLN and spleen of DSS7-treated animals revealed an increase in proinflammatory subtype IFN-γ- and IL-17A-secreting γδ T cells, while these were significantly reduced in DSS7-Mo-SENP7KD mice (Figures 6G). These results allowed us to conclude that SENP7 upregulation in epithelial cells was necessary and sufficient to trigger a proinflammatory environment via expansion of γδ T cells.
Figure 6. Epithelial SENP7 Overexpression Triggers γδ T Cell Activation and Inflammation.
(A) Schematic representation of co-culture experiments involving CT26 cells and total immune cells.
(B–F) Cytokines analysis from culture supernatant by ELISA for (B) IFN-γ, (C) TNF-α, (D) IL-10, (E) IL-17, and (F) TGF-β (C3, monoculture epithelial cells transfected with empty FLAG vector; C4, scrambled siRNA transfected monoculture; C5, empty-FLAG-vector-transfected epithelial cells with a γδ T cell; C6, scrambled siRNA transfected co-cultured epithelial cells with a γδ T cell; C7 and C8, epithelial cells with empty FLAG vector and scrambled siRNA co-cultured with total immune cells from MLNs. Each bar is representative of three independent experiments with three technical replicates. Data are presented as mean + SEM.
(G) Percentage of IFN-γ - and IL-17A-secreting γδ-TCR+ cells. Cultured immune cells from MLN or spleen of indicated group were surface stained for anti-γδTCR antibody followed by intracellular staining for anti-IL-17A and anti-IFN-γ (n = 3 mice).
(H) Fluorescence immunostaining of SIAH2 (green) and γδ T cells (red) in colonic sections of untreated and DSS7-treated mice. Scale bar, 100 um. Right panel represents mean intensity values of SIAH2 and number of γδ T cells.
(I) ELISA of IL-15 and KGF from mucosal extracts of UC (n = 8), CD (n = 8), and control (n = 5).
(J and K) Correlation analysis of SIAH2 and SENP7 expression with disease severity. Mild and moderate groups are divided on the basis of disease activity index (20 mild cases and 18 moderate cases). For flow cytometry, cells were acquired on a BD FACS Canto, analyzed using FlowJo, and represented in the form of a bar graph (mean + SEM values). Statistical differences were calculated using Prism by two-way ANOVA with Tukey’s test.
*p < 0.05; **p < 0.01; ****p < 0.001. See also Figure S7.
The in vivo relevance of these findings was next investigated by fluorescence co-immunostaining of SIAH2 and γδ T cells followed by confocal microscopy of colonic sections. SIAH2 expression was almost absent in DSS7 mice, while the proportion of γδ T cells increased significantly. Fluorescence intensity quantification revealed that SIAH2 levels were substantially higher in untreated animals than DSS-treated animals (Figure 6H).
We examined levels of γδ-T-cell-specific cytokines (IL-15 and KGF) in the biopsy samples of human UC, CD, and controls. Significantly elevated levels of both these cytokines were seen in UC and CD samples compared to the control group (Figure 6I). To determine whether these changes were connected to SENP7-SIAH2 activity, we looked at profiles of SIAH2 and SENP7 expression in relation to CD and UC disease severity. For this, the UDAI and CDI scores were taken from our earlier data (Mustfa et al., 2017) wherein the disease indices were categorized as mild (20 cases with scores 0.82 ± 0.33) or moderate (18 cases with scores above 5.48 ± 1.43). In the current work, we utilized the UDAI and CDI scores and compared them with profiles of SENP7-SIAH2 expression. Interestingly, both CD and UC samples with moderate disease indices formed a separate group with high expression of SENP7 and low SIAH2. In patients with mild disease, indices displayed low SENP7 and high SIAH2 levels (Figures 6J and 6K). These striking differences were also statistically significant, as revealed by strata analysis. Together, these data point toward crucial involvement of SENP7 and SIAH2 in epithelial signaling and immune cell recruitment leading to disease pathophysiology in IBD.
Discussion
In recent years, our overall understanding of the molecular mechanisms involved in IBD pathophysiology has greatly increased. Several of these events are due to overactivation of a wide range of immune cells that are guided by gut epithelial cells. Thus, signaling at the mucosal surface of the gut largely relies on epithelial cells that not only identify and sense the presence of foreign antigens but also efficiently transduce the information to the underlying immune system. In epithelial cells, a range of different signaling pathways operates in an orchestrated manner, and upstream initiation signal transduction often relies on PTMs such as SUMOylation (Mustfa et al., 2017; Hu and Shu, 2017; Treuter and Venteclef, 2011).
DeSUMOylases were identified nearly 20 years ago, but their role in cell signaling is not well explored. The current work reveals that increased expression of SENP7 mediates epithelial-immune crosstalk during inflammation. High expression of SENP7 has also been observed in pathophysiological conditions of other tissues such as prostate tumors (Zhu et al., 2015). SENP7 has been linked to very diverse functions (Barry et al., 2018; Romeo et al., 2015; Garvin et al., 2013; Bawa-Khalfe et al., 2012).
The fact that we observed a direct physical interaction of SENP7 with ubiquitin ligase SIAH2 was rather surprising, because the SIAH2 interaction motif is in reverse orientation. Nevertheless, the interaction was direct and very specific, as even a point mutation weakened the interaction. This highlights that protein-protein interaction depends on the context of interactors rather than the primary sequence alone. Ubiquitination-dependent degradation of SENP7 was previously reported by another group (Zhu et al., 2015). Thus, the choice of ligase and the adaptor may depend on the tissue microenvironment and context. The regulation of SIAH2 itself may depend on additional factors, and its copy number in the cell has been shown to be governed by the deubiquitinase enzyme Usp13 and miR335 (Scortegagna et al., 2011; Kim et al.2015). Ubc9 was earlier reported by us to be involved in AKT SUMOylation during IBD (Mustfa et al., 2017). In the present work, the AKT pathway was an important segment of the SENP7-interacting network (Figure 3C). Interactome analysis of colonic tissue revealed a very large and distinct set of interactors in control and DSS-treated mice.
Potentially for many of these interactors, SENP7 may be acting as a deSUMOylase and have a regulatory function. Interactome analysis revealed a strong connection between SENP7 and cellular immune pathways (Figure S2C). Interestingly, some of these pathways are known to be directly involved in intestinal inflammation and IBD. Our gene enrichment analysis of the immunological database for the SENP7-associated proteome ties the processes of adaptive immune responses like inflammation, T cell activation, T cell differentiation, and neutrophil migration (Figure S2C). This finding indicates that any disturbance in the regulation of SENP7 results in immune modulation in the microenvironment of the colon. The SENP7 is known to deconjugate poly-SUMO2/3 chains of the modified targets (Shen et al., 2009). However, in our proteomics data, we identified free SUMO-1 as an SENP7 interactor, because every SUM2/3 modified protein ends with a SUMO-1 at the C-terminus. Since SUMO-1 is part of the chain terminus, it is easily detected by MS/MS. Proteins such as vimentin, which was identified in our proteome studies, do have a role in many normal cellular processes, including development, differentiation, and inflammation. Its presence in our analysis is exciting and may be utilized as a platform for further studies to determine its relevance.
Several attempts have been made by various research groups to identify SENP-specific inhibitors, and this has led to the identification of compounds like the vinyl sulfone inhibitor, N-ethylmaleimide (NEM), and derivatives of various chemical derivative compounds (Kumar and Zhang, 2015). These compounds are good for biological-mechanism-related studies but have limitations in clinical settings because of off-target effects. In silico approaches have been successful in identifying chemical scaffolds that could be used to identify specific drugs for these targets (Kumar and Zhang, 2015; Yang et al., 2018). In the current work, we also generated a morpholino-based gut-specific SENP7 knockdown mouse model system. These animals were resistant to DSS-led inflammation. This model is very interesting from several different angles. First, it is important from point of our basic understanding of SENP7, as it tells the role of this protein in the context of the gut mucosa. Second, it highlights the possibility of using SENP7 and delivery vehicles similar to those used in the current study as a tool for combating intestinal inflammatory conditions not restricted to IBD. A dysregulated SUMOylome has been seen in several different forms of intestinal inflammation, including those resulting from infectious diseases (Ribet et al., 2010; Fritah et al., 2014; Verma et al., 2015; Sidik et al., 2015; Yadav et al., 2019). Lastly, gut-specific SENP7 knockdown or knockout may also have an effect on the composition of intestinal microbiota, an aspect that cannot be ignored if this will be evolved as a tool for intervention against inflammation.
This study also highlights the proinflammatory function of γδ T cells. The percentage of γδ T cells in the gut mucosa is very high, reaching almost 40% of the intestinal epithelial leukocytes. Their role in IBD has been rather controversial, with some studies highlighting their tissue-damaging functions (Simpson et al., 1997; Kawaguchi-Miyashita et al., 2001; Nanno et al., 2008; Do et al., 2011, 2017) and others highlighting their tissue repair and anti-inflammatory functions (Szczepanik et al., 2000; Chen et al., 2002; Kühl et al., 2007; Inagaki-Ohara et al., 2004; Hoffmann et al., 2001). In our study, by using both a murine model of colitis and human IBD patient samples, we have demonstrated an association between intestinal γδ T cells and inflammation occurring via epithelial-SENP7 singling.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-SUMO2/3 polyclonal antibody | Sigma-Aldrich | Cat#PA5-11319; RRID: AB_2286890 |
| Rabbit anti-SIAH2 polyclonal antibody | OriGENE | Cat# AP20493PU-N; RRID: AB_10757521 |
| Rabbit anti-SENP1 monoclonal antibody | Abcam | Cat#108981; RRID: AB_10862449 |
| Rabbit anti-SENP2 polyclonal antibody | Abcam | Cat#58418; RRID: AB_882485 |
| Rabbit anti-SENP5 polyclonal antibody | Sigma-Aldrich | Cat#SAB4500779; RRID: AB_10745479 |
| Rabbit anti-SENP7 polyclonal antibody | Invitrogen | Cat# PA5-36089; RRID: AB_2553364 |
| Rabbit anti-NCor1 polyclonal antibody | Invitrogen | Cat#PA1-844A; RRID: AB_2149004 |
| Rabbit anti-β-Actin monoclonal antibody | Abgenex | Cat#11-13012; RRID: NA |
| Mouse anti-β-Tubulin monoclonal antibody | Sigma-Aldrich | Cat#T7816; RRID: AB_261770 |
| Mouse anti-GAPDH monoclonal antibody | Invitrogen | Cat#398600; RRID: AB_2533438 |
| Rabbit monoclonal anti-Flag (DYKDDDDK Tag Antibody) | Cell Signaling Technology | Cat#2368S; RRID: AB_2217020 |
| Armenian Hamster anti-γδ TCR monoclonal antibody | BioLegend | Cat#118101; RRID: AB_313826 |
| Armenian Hamster FITC labeled secondary antibody | BioLegend | Cat#400906; RRID: NA |
| Goat anti-Rabbit HRP conjugated antibody | Invitrogen | Cat#65-6120; RRID: AB_2533967 |
| Mouse Anti-IL15 polyclonal antibody | R&D systems | Cat#AF447-SP; RRID: AB_2124437 |
| InVivoMAb anti-mouse TCR γ/δ monoclonal antibody (Clone UC7-13D5) | Bio X Cell, USA | Cat#BE0070; RRID: AB_1107751 |
| Anti-TCR γ/δ monoclonal antibody PE-Cyanine7 | eBioscience Invitrogen | Cat#25-5711-82; RRID: AB_2573464 |
| APC-Cy7 Hamster Anti-Mouse CD3e | BD Biosciences | Cat#557596; RRID: AB_396759 |
| PerCP/Cyanine5.5 anti-mouse CD4 monoclonal antibody | BioLegend | Cat#100434; RRID: AB_893324 |
| FITC anti-mouse CD8a monoclonal antibody | BioLegend | Cat#100804; RRID: AB_312765 |
| PE anti-mouse IL-17A monoclonal antibody | BioLegend | Cat#506904; RRID: AB_315464 |
| Alexa Fluor 647 anti-mouse IFN-γ monoclonal antibody | BioLegend | Cat#505814; RRID: AB_493314 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| 17β-Estradiol | Sigma-Aldrich | Cat#E8875-250MG |
| Recombinant mouse IL15 | R&D systems | Cat#447-ML-010 |
| Dextran Sulfate Sodium | Sigma-Aldrich | Cat#42867-100G |
| MG-132 | Sigma-Aldrich | Cat#M8699-1MG |
| Lipopolysaccharide | Sigma-Aldrich | Cat#LPS25 |
| Critical Commercial Assays | ||
| Co immunoprecipitation kit | Thermo | Cat#26149 |
| i-Taq Syber green master mix | Bio-Rad | Cat#1725124 |
| Nucleo Spin RNA-II Kit | MN | Cat#740955.50 |
| CBX™ protein assay kit | G-Bioscience | Cat#786-12X |
| Mouse IL-15 DuoSet ELISA | R&D systems | Cat#DY447-05 |
| Mouse IFN-gamma DuoSet ELISA | R&D systems | Cat#DY485-05 |
| Mouse TNF-alpha DuoSet ELISA | R&D systems | Cat#DY410-05 |
| Mouse IL-17 DuoSet ELISA | R&D systems | Cat#DY421-05 |
| Mouse TGF-beta DuoSet ELISA | R&D systems | Cat#DY1679-05 |
| Mouse IL-10 DuoSet ELISA | R&D systems | Cat#DY417-05 |
| Mouse TSLP DuoSet ELISA | R&D systems | Cat#DY555-05 |
| Human IL-15 DuoSet ELISA | R&D systems | Cat#DY247-05 |
| Human KGF/FGF-7 DuoSet ELISA | R&D systems | Cat#DY251 |
| Deposited Data | ||
| Raw files for proteomics data (SENP7 Interactome) | PRIDE (ProteomeXchange Consortium) | ProteomeXchange: PXD015994 and PXD016018 |
| Raw data files for CD4+ T and CD8+ T cells population frequency | Mendeley Data | https://doi.org/10.17632/z4xj6w9zxn.1 |
| Experimental Models: Cell Lines | ||
| HCT8 cells | ATCC | Cat#ATCC® CCL-244 |
| CT26 cells | ATCC | Cat#ATCC® CRL-2638 |
| Experimental Models: Organisms/Strains | ||
| Mouse: C57BL/6 | Institute small animal facility, Regional center for Biotechnology | NA |
| SMARTpool: ON-TARGETplus SENP7 siRNA | Dharmacon | Cat#L-006035-00-0005 5 nmol |
| Vivo-Morpholino: MO-SENP7 TGGCCCTGTCCATCTTCTCCTCC |
Gene Tools, LLC | NCBI Reference Sequence: NM_025483.4 |
| Vivo morpholino control non target TCCTAGAGGTCATCCTTGAAGCGCA |
Gene Tools, LLC | NA |
| Primers for real-time PCR, see Table S4 | This paper | NA |
| Primer SDM: SENP7 PAVP Mutant 1 Forward: CTTTCTTACCCGTTGAATTGGAATCAGGCATTTCCTTTG |
This paper | NA |
| Primer SDM: SENP7 PAVP Mutant 1 Reverse: CAAAGGAAATGCCTGATTCCAATTCAACGGGTAAGAAAG |
This paper | NA |
| Primer SDM: SENP7 PAVP Mutant 2 Forward: CCCGTTGTCTTGGAATCAGGCATTTAATTTGTTTCAGAAC |
This paper | NA |
| Primer SDM: SENP7 PAVP Mutant 2 Reverse: GTTCTGAAACAAATTAAATGCCTGATTCCAAGACAACGGG |
This paper | NA |
| Recombinant DNA | ||
| pCDNA5/FRT/TO SENP7 Flag | A Gift from Professor Jo Morris | Garvin et al., 2013 |
| pCDNA5/FRT/TO SENP7 C992A Flag | A Gift from Professor Jo Morris | Garvin et al., 2013 |
| pEGFP-N1 SIAH2 | A Gift from Professor Ze’ev A. Ronai | Kim et al., 2011 |
| pEGFP-N1 SIAH2 RM | A Gift from Professor Ze’ev A. Ronai | Kim et al., 2011 |
| Software and Algorithms | ||
| LAS X | Leica microsystems | https://www.leica-microsystems.com/products/microscope-software/p/leica-las-x-ls/ |
| Jalview Java alignment editor version 2.10. | Waterhouse et al., 2009 | http://www.jalview.org/ |
| Tcoffee | Notredame et al., 2000 | http://tcoffee.crg.cat/ |
| TransProteomics Pipeline (TPP). | Deutsch et al., 2010 | http://tools.proteomecenter.org/software.php |
| X! Tandem | MacLean et al., 2006 | https://www.thegpm.org |
| MaxQuant 1.5.2.8 | Cox and Mann, 2008 | https://www.maxquant.org |
| Cytoscape 3.2.1 | Shannon et al., 2003 | https://cytoscape.org |
| STRING v 10.0 | Szklarczyk et al., 2015 | https://string-db.org |
| GeneMANIA | Warde-Farley et al., 2010 | https://genemania.org/ |
| Reactome | Fabregat et al., 2018 | https://reactome.org/ |
| KEGG | Kanehisa and Goto, 2010 | https://www.genome.jp/kegg/kegg1.html |
| GPS-SUMO 1.0 | Zhao et al., 2014. | http://sumosp.biocuckoo.org/ |
| Gene Ontology Enrichment Analysis Software Toolkit (GOEAST) released 2016-07-15 Version 11 | Zheng and Wang, 2008 | http://omicslab.genetics.ac.cn/GOEAST/ |
| FlowJo Vx0.7 | Treestat | http://docs.flowjo.com/vx/ |
Lead Contact and Materials Availability
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Dr. C.V. Srikanth (cvsrikanth@rcb.res.in). There are restrictions on the availability of variants of SENP7 and SIAH2 plasmids as they are a gift from another source and require permission before sharing. All unique reagents generated in this study are available from the Lead contact.
Experimental Model and Subject Details
Animal strains
C57BL/6 mice aged 6-8 weeks used for the study were kept under pathogen-free conditions in properly ventilated cages with optimal temperature and humidity. Female mice were used in the experiments involving SENP7 Vivo-morpholino knockdown and γδ T cells depletion. Male mice were used in the experiments involving in-vivo SIAH2 induction using 17 β-estradiol in order to prevent steroidal interference on female mice. The influence of gender was not considered for this study. All the animal experiments were carried out in the Small Animal Facility of Regional Centre for Biotechnology (RCB). Animals ethics proposal was approved by the RCB Institutional Animal Ethics Committee (approval no. IAEC/2018/045).
For colitis study individual group of female mice were housed in proper ventilated cages. Experimental colitis was induced in an animal by orally feeding them with Dextran Sulfate Sodium (Cat No.42867-100G, Sigma, USA) dissolved in autoclaved drinking water at 2.5% concentration (w/v) for indicated days. Mice were monitored daily for their stress and rectal bleeding. Post DSS treatment animals were sacrificed by CO2 asphyxiation and organs were harvested for further analysis. Number of mice used in all experiments is three unless specified. In all mice experiments, untreated mice controls were given autoclaved drinking water. To knockdown SENP7, Vivo-morpholino was used, female mice were given SENP7 specific (TGGCCCTGTCCATCTTCTCCTCC) Vivo-morpholino (Gene Tools, LLC) and Control nontarget (TCCTAGAGGTCATCCTTGAAGCGCA) Morpholino (Gene Tools, LLC). Two doses of intraperitoneal injections were given on the first day and fourth day during the course of DSS treatment at the concentration of 10 mg/kg (20nM/mice). Different conditions included in the study were: (i) Control- untreated mice. (ii) Control morpholino- mice were given non-target morpholino, (iii) SENP7 knockdown- mice treated with SENP7 Vivo morpholino, (iv) DSS- 7 days DSS treated mice and (v) SENP7KD-DSS mice- SENP7 knockdown mice along with DSS treatment for 7 days. Mice were sacrificed after seven days and colon, MLN and spleen were collected for further analysis. For cytokines estimation, mucous was collected from the colon of each mice group and resuspended in 1XPBS. Cytokine analysis was done using ELISA. For in-vivo SIAH2 induction, male mice were used. 17β-Estradiol (Cat No. E8875-250MG, Sigma, USA) was given at a concentration of 5μg/ml in drinking water for continuous 14 days. During the course of treatment, post 7 days DSS was given in combination with 17β-Estradiol for next 7 days. Organs were later harvested for further study. For in vivo γδ TCR depletion, female mice were used. InVivoMAb anti-mouse TCR γ/δ monoclonal antibody (UC7-13D5) (Catalogue no.BE0070, Bioxcell, USA) were given two dosages intraperitoneally at the concentration of 0.5 mg per mice. Dosage was given on the first day and 4th day of DSS administration in mice. Mice were euthanized after 7 days and spleen, MLN and colon were harvested for further studies.
Human colonic biopsy samples
A total of 57 patients in the UC, CD and control groups were included in this study, from the All India Institute of Medical Sciences (AIIMS). These were part of an earlier study (Mustfa et al., 2017). Samples were collected from 19 patients in the UC, 19 patients CD and 19 control (IBD suspected) groups with age over 18 and below 60 years. The influence of gender was not considered for the study. Of the collected samples in UC, 12 were male and 7 were female and in CD, 13 were male and 6 were female. Inclusion criteria for UC were: (a) patients with suggestive history and characteristic endoscopic and histological findings of UC (ECCO consensus statement) as enrolled from the Inflammatory Bowel Disease.
Clinic at the Department of Gastroenterology, AIIMS; (b) patients with an ulcerative colitis disease activity index (UCDAI) of mild to moderate; and (c) patients with pancolitis or left-sided colitis. Exclusion criteria for UC were: (a) patients with proctitis (b) patients already initiated on steroid therapy and (c) patients with a coexistent disease such as HIV infection, tuberculosis or chronic renal failure. Inclusion criteria for CD were: (a) the diagnosis of CD established on the basis of the presence of characteristic clinical manifestations (chronic diarrhea, hematochezia, abdominal pain and intestinal obstructive manifestations), endoscopic features (skip lesions, asymmetrical involvement, longitudinal ulcers, aphthous ulcers) and histological evidence (acute or chronic colitis, the presence of inflammation extending beyond muscularis mucosae, lymphoid follicles and granuloma) (ECCO guidelines for diagnosis); and (b) adult patients with ileocolonic disease and Crohn’s disease activity index (CDAI) greater than 150 and less than 350. Exclusion criteria for CD were: (a) patients already initiated on steroid therapy and (b) patients with a coexistent disease such as HIV infection, tuberculosis or chronic renal failure. Adult patients suspected of having IBD but showing normal mucosa on colonoscopy were treated as controls. Four to six biopsies were collected from each patient and each biopsy from every individual were used for qPCR, western blotting and ELISA. Informed consent form was obtained from all the patients and the study protocol was submitted to the ethics committee of the institutes: RCB and AIIMS (approval no. IEC/NP/56/2014, OP-16/01.08.2015). Our fundamental step was to design a large value of computational power (at least 76%) with an effective sample size within the available resources and ethical considerations. The variance or standard deviation for sample size calculation was obtained from our pilot study. For every differential expression analysis, the p value was calculated. To this effect we required (n = 19, power = 76% and a = 5% per group), using SYSTAT software for two mean calculations. Our ultimate goal was to establish the role of the SUMO pathway by examining a large sample size to gain an understanding of the deSUMOylases in IBD.
Cells
Human HCT-8 and murine CT26 cell line were cultured in RPMI medium supplemented with 14 mM NaHCO3, 15 mM HEPES buffer (pH 7.4), 2 mM glutamine, 1 mM sodium pyruvate, 40 mg/liter penicillin, 90 mg/liter streptomycin, and 10% fetal bovine serum. One day before transfection, 2.5x105 cells were seeded in 24 or 12 well plates to obtain 60%–80% confluence and transfected with Lipofectamine 2000 (Invitrogen) or DharmaFECT (Dharmacon, USA) as per the manufacturer’s instructions. Briefly, 1 μg of SENP7 plasmids (wild-type and mutant form), SIAH2 plasmids (wild-type and ring mutant) or 20 pmol of SENP7 smart pool small interfering RNA (siRNA) (Dharmacon) were diluted in Opti-MEM (Invitrogen). Separately, Lipofectamine 2000 for plasmid and DharmaFECT for siRNA transfection were also diluted in Opti-MEM and incubated at room temperature for 5 min. Both samples were mixed and incubated at room temperature for 20 min. After 20 min, a complex mixture was added to cells with Opti-MEM and incubated without selection for 24 h. Cells were treated with different pharmacological inhibitors: LPS, 200ng/ml (Sigma) and MG-132, 20 mM for two hours (Sigma).
Method Details
Quantitative real-time PCR
Total RNA was isolated from mice tissue and indicated cell lines using Nucleo Spin RNA-II Kit (MN, Germany) according to the manufacturer’s protocol. One microgram of total RNA from each sample was used to synthesize cDNA using i-Script cDNA synthesis Kit (Bio-Rad, USA). Bio-Rad CFX 96™Real-Time Detection System was used for real-time PCR (qRT-PCR) using i-Taq Syber green (Bio-Rad, USA) according to manufacturer’s instruction CFX 96. For human samples, GAPDH gene was used as an endogenous control for normalization. Mouse samples were normalized with β-Actin. The list of primers is included in Table S4.
Western blot
Intestinal epithelial cells were isolated from the colon as previously described (Mustfa et al., 2017). Tissues and cells were lysed in RIPA lysis buffer (Sigma, USA) supplemented with Halt Protease and Phosphatase Inhibitor Cocktail (1X) (Thermo, USA) and 20mM NEM (Sigma, USA). Protein quantification was estimated using CBX™ protein assay kit (G-Bioscience, USA). Protein lysates in equal amounts were resolved on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to nitrocellulose membrane. Blots were probed with antibodies against SUMO2/3 (Invitrogen, USA), SIAH2 (Origene, USA), SENP1 (Abcam, USA), SENP2 (Abcam, USA), SENP5 (Sigma, USA), SENP7 (Invitrogen, USA), Actin (Abgenex, India), GAPDH (Invitrogen, USA), β-tubulin (Sigma, USA). Densitometry analysis of immunoblot band was done using a Bio-Rad gel documentation system (Bio-Rad, USA).
Multiple sequence alignment (MSA)
The protein sequence analysis of the SENP7 was performed through offline Jalview Java alignment editor version 2.10.5 (Waterhouse et al., 2009). Initially, the non-redundant and annotated SENP7 FASTA protein sequences were fetched using the Uniprot database of 13 organisms. The MSA was created using the Tcoffee alignment tool (Notredame et al., 2000) with default parameters and the generated alignments were colored according to the ClustalX coloring scheme. Below the alignment, the annotation was displayed describing the consensus, conservation, and the quality score. The consensus is described using the weblog information; conservation is depicted in quantitative value, * denotes identical amino acid residues, - denotes non-align amino acid residues, + denotes similar alignment, quality of the alignment is shown by the heatmap.
Immunoprecipitation
Mouse tissue and cell line cells were lysed in non-denaturing conditions using IP lysis buffer (Cat. No. 87788, Thermo, USA). Halt proteases complete inhibitor mix (1X) and 20mM NEM was added to the mixture. Cell debris was removed by centrifugation at 13,000 rpm for 10 min at 4°C. Standard Protocol of Co-immunoprecipitation kit (Thermo, USA) was followed. Briefly, respective antibodies were bound to Protein G-Sepharose beads and eluted in elution buffer followed by heating at 95°C for 10 minutes in Lane Marker buffer supplied with the kit. Six hundred microgram protein was used for immunoprecipitation. Isotype IgG (negative control) was prepared for control samples.
Protein Digestion
LC-MS/MS was done using intestinal epithelial lysates. Control and DSS mice lysates were immunoprecipitated with anti-SENP7 antibody (Invitrogen, USA) using six hundred micrograms of protein. Samples were later resolved on 10% SDS-PAGE and in-gel trypsin digestion was carried out using Trypsin Gold (Promega, USA) at 1:20 dilution. The peptides were extracted from the gel and purified using C18 SepPak columns (Thermo, USA). The samples were then dissolved in 1% acetonitrile in 0.1% formic acid. Tandem MS/MS analysis was performed using the 5600 triple TOF analyzer (ABsciex).
Electrospray ionization tandem mass spectrometry LC-MS/MS analysis
The peptides were analyzed using Triple TOF, ABsiex connected via trap column (Magic C18 AQ, 0.1 × 20 mm, 3 μm, 200 Å) and a nano-analytical column (Magic C18 AQ, 0.1 × 150 mm, 3 μm, 200 Å) coupled with electro spray ion source (ESI-tip) spray in triple-TOF (ABsiex) mass spectrometer (MS). The elution was performed with the flow rate of 300 nL/min in a continuous gradient of 5%–75% acetonitrile over 135 min. In the solvent system; Solvent A was 100% water in 0.1% formic acid, and solvent B was 100% acetonitrile in 0.1% formic acid. For acquisition, the data-dependent mode was used in mass spectrometer operated into automatically switch between MS and MS/MS acquisition. The precursor ion MS spectra scan range of 300-2000 (m/z) was used in the triple-TOF with resolution R = 75, 000. The six most abundant precursor ions were searched for detection of different masses in acquisition method and selected for fragmentation using collision-induced dissociation (CID) with a fixed cycle time of 3 s along with 2 min of release for exclusion filter.
Data Processing
The obtained raw (.wiff) files from the MS system were analyzed by TransProteomics Pipeline (TPP). Initially, the raw files were converted into the mzML format using the TPP msconvert tool using the default parameters. The converted mzML files were searched using the TPP version 5.1.0 released on 2017-11-03 to in-house combined UniProt (Mus musculus) database together with common contaminant sequences were provided for MS/MS spectra search. The database was also appended with an equal number of decoy sequences (reversed proteins sequences from the original database). Initially, for the analysis, the peptide assignments were performed using multiple search engines using X! Tandem (with the k-score plug-in; MacLean et al., 2006), SpectraST and Comet. For all the search engines, the parameters included semi-digested LysC and trypsin with two allowed miss cleavage, static modifications carbamidomethyl (Cys) and dynamic modification oxidation (Met), Gln-pyro Glu and Glu-pyro Glu remaining parameter were kept as default. The minimum peptide length parameter was set to 7 amino acid residue. Further for peptides probability assignments and validations, the PeptideProphet used for statistical validation of peptide-spectra-matches (PSM) and calculation of FDR at PSM level. However, the ProteinProphet algorithms were used in the pipeline to compute the probabilities score and fit a Gaussian distribution for the correct identifications for both individually searched peptides and the respective proteins (Deutsch et al., 2010). Another protein validation step was executed using both Peptide Prophet and Protein Prophet scores where the protein was authenticated if it contains minimum two top-ranked peptides with each peptide probability score above 95% (Tables S1 and S2). All the search engine results were merged and validated using another computational method, termed iProphet. This method takes as the input of PeptideProphet spectrum-level results from multiple LC-MS/MS runs and then computes a new probability at the level of a unique peptide sequence (or protein sequence; Nesvizhskii et al., 2003). This framework allows for the combination of results from multiple search tools and takes into account other supporting factors, including the number of sibling experiments identifying the same peptide ions, the number of replicate ion identifications, sibling ions, and sibling modification states. A model of iProphet performance with respect to the number of correct identifications versus error is shown in Tables S1 and S2. An iProphet probability of more than 0.99 was used as the cut-off for final identification of the protein. For protein quantitation, ≥ 2 unique peptides per protein were considered to ensure high-quality quantitation.
The same obtained raw (.wiff) files from the MS system from above-mentioned parameters were analyzed by MaxQuant software version 1.5.2.8 (Cox and Mann, 2008) for iBAQ (Intensity Based Absolute Quantification) algorithm analysis using the normalized XIC. The integrated Andromeda act as a search engine for protein identification and quantitation. The common contaminant sequences obtained from ftp://ftp.thegpm.org/fasta/cRAP and same in-house combined UniProt (Mus musculus) was used for the MS/MS spectra search. The false discovery rate (FDR) was set to < 1% to ensure only high-confidence protein identifications for PSM (peptide to spectra match), protein, and site decoy fraction. The enzyme specificity was set to trypsin with the maximum number of 2 missed cleavages. Additionally, in MaxQuant to validate and transfer identifications across different replicates, the ‘match between runs’ option was enabled with an extended retention time window of 0.7 minute and an alignment time window of 20 min. The subsequent downstream statistical analysis was performed in Perseus environment and two-sided t test was calculated at the statistical significance of p ≤ 0.05. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD015994 and PXD016018.
Bioinformatics analysis and PPI network construction
To examine the regulome and prediction of pathways for the identified proteins, the high confident proteins (DEPs) were employed to conceive protein-protein interaction (PPI) networks for SENP7 and its interacting hub proteins. The comprehensive attributes were collected, initially, the whole protein sequences were retrieved using an in-house made python script and other information from multiple PPI network databases including STRING v 10.0 (Szklarczyk et al., 2015), GeneMANIA (Warde-Farley et al., 2010), Reactome (Fabregat et al., 2018), and KEGG (Kanehisa and Goto, 2010). Based on the Boolean network model using degree, closeness, and betweenness centrality measures, SENP7 direct interacting partners were identified. These attributes were used at the common platform of Cytoscape 3.2.1 (Shannon et al., 2003) to visualize the network. The identified proteins in the PPI network was manually annotated using the Reactome database for biologically relevant processes. Also, the whole protein sequences were searched against the GPS-SUMO 1.0 engine for the prediction of potential SUMOylation sites and SUMO interaction motifs in the proteins (Zhao et al., 2014).
To further decipher the biological implication of IP-MS data the set of SENP7 associated proteome was annotated using Reactome and WikiPath for the target genes annotation. The gene-term enrichment analysis was performed with Gene Ontology Enrichment Analysis Software Toolkit (GOEAST) released 2016-07-15 Version 11 (Zheng and Wang, 2008). The fold enrichment was calculated using the expected value for a particular Gene Ontology (GO) category, based on the reference index within the database (Table S3). Consequently, the probability was circumscribed using a binomial statistic for p values with the cutoff of 0.05. Further, the statistical analysis was performed using the two-sided hypergeometric distribution tests along with Benjamini and Hochberg false discovery rate (FDR)-correction at < 0.05 significance level.
Immunohistochemistry and Immunocytochemistry
Colonic sections were fixed in 10% formalin buffer overnight and snap freeze in cryometrix (Cat. No.6769006, Thermo). 5 μm thick sections were cut onto glass slides and processed using Cryotome (Thermo). HCT8 cells transfected with SENP7 plasmids and siRNA were fixed with 10% formalin buffer for 20 minutes. Tissue sections or cells were washed using 1X PBS for three times (5 minutes each). Further, the endogenous peroxidase activity was quenched by treating the tissue with 3% H2O2 for 20 minutes. The tissue sample or cells were again washed using 1X PBS for two times (5 minutes each), then blocked with 5% goat serum at room temperature for one hour. The sections were incubated with anti-γδ TCR (1:100, Biolegend), anti SENP7 (1:50, Invitrogen) or anti-SIAH2 (1:100, Origene) antibodies prepared in 5% goat serum overnight in a moist chamber. Sections were washed with 1X PBS for five times at 5 minutes each and incubated with either secondary anti-Armenian Hamster FITC labeled secondary antibody (1:400) (Biolegend, USA) or anti-Rabbit HRP conjugate antibody (1:200, Invitrogen) for 2 hours prepared in 5% goat serum at room temperature in a moist chamber. The tissues were washed three times using 1X PBS. For immunohistochemistry, the slides were stained with DAB substrate (Sigma, USA) and counterstained with Hematoxylin. Images were taken on a Nikon Fluorescence Microscope using color camera. For immunocytochemistry, the slides were counterstained using DAPI for 5 minutes and then washed with 1X PBS and miliQ water and mounted using anti-fade reagent. The slides were visualized on a Leica SP8 confocal microscope.
Intraepithelial lymphocytes (IEL) and Lamina Propria (LP) Isolation
IEL was isolated by the method described in Little et al. (2005). Briefly, the colon was isolated from mice and fat and connective tissues were separated. Colon was cleaned by washing with ice-cold calcium and magnesium-free HBSS containing 2% FBS. It was cut open longitudinally and again washed twice to remove remaining feces. The opened intestine was then cut into 1 cm pieces. Tissues collected in 50ml falcon and washed thrice with HBSS (containing 2% FBS at 4 C). After washing, tissue was incubated for 20 minutes at 37 C in HBSS (10% FBS, 0.2 mmol/l EDTA, 1 mmol/l DTT, 100 U/ml penicillin/strep). Flask was shaken vigorously and filtered using 70μm nylon sieve filter. This step was repeated and cells were collected. Filtered cells were then pelleted and processed for flow cytometry. Tissue pieces were collected and further cut into 1 mm small pieces. After washing with HBSS with 2% FBS, they were incubated for 20 minutes with digestion buffer (Collagenase and DNase I in HBSS containing 10% HBSS). After every 5 minutes vigorous vortex was done. Digested minced tissue was then passed through nylon filter. Filtered cells were then centrifuge at 4000 rpm for 5 minutes and filtered again. Filtered cells then pelleted and resuspended in 1 mL 44% Percoll and transferred to another tube with 2 mL 67% Percoll. Centrifuged at 1600 rcf. break free. Buffy coat was collected and transferred to another tube with DMEM media with 10% FBS. IEL and LP collected were then proceed for immunophenotyping analysis by flow cytometry.
Immunophenotyping and intracellular cytokine staining
0.2 million cells isolated from single-cell suspension prepared from spleen, MLN, intraepithelial lymphocytes, lamina propria or cultured cells were surface stained with flourescenated anti-mouse-CD3 (BD Biosciences, USA), anti-mouse CD4 (BioLegend, USA), anti-mouse CD8a (Biolegend, USA) and anti-mouse γδ TCR antibodies (eBioscience Invitrogen, USA) for 20 minutes at room temperature and washed thereafter with PBS+1% FBS. For intracellular cytokine staining cells were stimulated by PMA (50ng/ml; Sigma, USA) and ionomycin (1 μg/ml: Sigma, USA) in presence of monensin containing protein transport inhibitor (Golgi stop, BD Biosceinces) for 4 h in 37°C CO2 incubator. FACS staining was then carried out by first doing surface marker staining as described above, thereafter, cells were fixed in Cyto fix and permeabilized with Perm Wash buffer (Fixation and Permeabilization Solution, BioLegend). Intracellular cytokine staining was then carried out by using flourescenated anti-mouse IL-17A and anti-mouse IFN-γ antibodies (BioLegend). Stained samples were then washed twice with PBS and acquired on BD FACS Canto (BD Biosciences) and data were analyzed on FlowJo Vx0.7 (Treestar).
Co-culture of intestinal epithelial cells with γδ T cells and immune cells
Mesenteric lymph nodes were collected from untreated mice and the single-cell suspension was prepared. γδ T cells were sorted using anti-γδ TCR antibody (Figure S7A). After sorting, γδ T cells were seeded on 24 well plate insert wells (Corning, USA). Separately CT26 mice intestinal epithelial cells were grown at the bottom of 24 well plates and transfected with wild-type SENP7 plasmid, catalytic mutant SENP7 C992A plasmid, SENP7 siRNA, SIAH2 wild-type plasmid and SIAH2 ring mutant plasmid. LPS treatment at the concentration of 200ng/ml was given for two hours and then cells were washed with PBS, followed by the addition of fresh media. Finally, epithelial cells and sorted γδ T cells or total immune cells were co-cultured for the next four hours. The effect of SENP7 over-expressed epithelial cells on total immune cells were analyzed by culturing SENP7 transfected CT26 cells for 3 days with and without immune cells. Anti-IL15 neutralizing antibody (R&D systems, USA) was given for consecutive 3 days at the concentration of 1μg/ml. Recombinant mouse IL-15 (R&D systems, USA) treatment at the concentration of 15 ng/ml was given on total immune cells grown as monoculture. The supernatant was collected for cytokines analysis using ELISA. Epithelial cells lysates were prepared for total RNA isolation.
Enzyme-linked immunosorbent assay
Colonic mucus and supernatant collected from cell culture experiment were transferred to a new tube. The supernatant was assayed by enzyme-linked immunosorbent assay (ELISA) for IL-15, IFNγ, TNF-α, IL-17A TGF-β, IL-10 and thymic stromal lymphopoietin (TSLP) using the manufacturer’s protocol (R&D systems, USA).
For human IBD patients, biopsies from each category were homogenized and incubated for 5 min at room temperature to permit the complete dissociation of nucleoprotein complexes. Thereafter, the lysates were centrifuged to remove cell debris and the supernatant was transferred to a new tube. The supernatant was assayed by enzyme-linked immunosorbent assay (ELISA) for IL-15 and KGF. (R&D systems, USA)
Quantification and Statistical Analysis
All results are expressed as the mean standard error from an individual experiment done in triplicate. Data were analyzed with one way ANOVA followed by Tukey’s post-test, standard two-tailed Student’s t test and the Mann–Whitney U-test where applicable, with p values of 0.05–0.001 considered statistically significant. We evaluated the statistics with SYSTAT and GraphPad PRISM. Correlation studies were analyzed with Spearman’s correlation
Data and Code Availability
The mass spectrometry proteomics data generated during this study are available at the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier: PXD015994 and PXD016018. The CD4+ and CD8+ T cells population data generated during this study are available at Mendeley Data. (https://data.mendeley.com/datasets/z4xj6w9zxn/1)
Supplementary Material
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2019.11.028.
Highlights.
The deSUMOylase SENP7 contributes to IBD pathophysiology
SENP7 function and interactome modulate epithelial-immune crosstalk
SIAH2 negatively regulates SENP7 by ubiquitination in healthy, but not inflamed, cells
Epithelial SENP7 upregulation triggers proinflammatory mechanisms via γδ T cells
Acknowledgments
We thank the RCB Mass-Spec facility and CIF. We are thankful to the THSTI FACS facility for flow cytometry experiments. We are thankful to Prof. Jo Morris for SENP7 plasmids. We also thank Prof. Ze’ev Ronai for the SIAH2 plasmids. This work was supported by the Wellcome Trust/DBT India Alliance Fellowship/Grant (grant number IA/I/11/2500284) awarded to C.V.S. and core funding from Regional Centre for Biotechnology, Faridabad, India.
Footnotes
Author Contributions
A.S., A.A., and C.V.S. wrote the manuscript; A.S., P.M., and P.G. conceived and performed experiments; C.V.S. designed the questions and experiments; Z.A.R., S.A.A., and M.S. contributed to analysis; and A.A., V.A., and C.V.S. provided feedback and expertise.
Declaration of Interests
The authors declare no competing interests
References
- Andoh A, Fujino S, Okuno T, Fujiyama Y, Bamba T. Intestinal subepithelial myofibroblasts in inflammatory bowel diseases. J Gastroenterol. 2002;37(Suppl 14):33–37. doi: 10.1007/BF03326410. [DOI] [PubMed] [Google Scholar]
- Barry R, John SW, Liccardi G, Tenev T, Jaco I, Chen CH, Choi J, Kasperkiewicz P, Fernandes-Alnemri T, Alnemri E, et al. SUMO-mediated regulation of NLRP3 modulates inflammasome activity. Nat Commun. 2018;9 doi: 10.1038/s41467-018-05321-2. 3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bawa-Khalfe T, Lu LS, Zuo Y, Huang C, Dere R, Lin FM, Yeh ET. Differential expression of SUMO-specific protease 7 variants regulates epithelial-mesenchymal transition. Proc Natl Acad Sci USA. 2012;109:17466–17471. doi: 10.1073/pnas.1209378109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkaid Y, Hand TW. Role of the microbiota in immunity and inflammation. Cell. 2014;157:121–141. doi: 10.1016/j.cell.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catalan-Serra I, Sandvik AK, Bruland T, Andreu-Ballester JC. Gammadelta T cells in Crohn’s disease: A new player in the disease pathogenesis? J Crohn’s Colitis. 2017;11:1135–1145. doi: 10.1093/ecco-jcc/jjx039. [DOI] [PubMed] [Google Scholar]
- Chassaing B, Aitken JD, Malleshappa M, Vijay-Kumar M. Dextran sulfate sodium (DSS)-induced colitis in mice. Curr Protoc Immunol. 2014;104:25. doi: 10.1002/0471142735.im1525s104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Chou K, Fuchs E, Havran WL, Boismenu R. Protection of the intestinal mucosa by intraepithelial gamma delta T cells. Proc Natl Acad Sci USA. 2002;99:14338–14343. doi: 10.1073/pnas.212290499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- Dahan S, Rabinowitz KM, Martin AP, Berin MC, Unkeless JC, Mayer L. Notch-1 signaling regulates intestinal epithelial barrier function, through interaction with CD4+ T cells, in mice and humans. Gastroenterology. 2011;140:550–559. doi: 10.1053/j.gastro.2010.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutsch EW, Mendoza L, Shteynberg D, Farrah T, Lam H, Tasman N, Sun Z, Nilsson E, Pratt B, Prazen B, et al. A guided tour of the Trans-Proteomic Pipeline. Proteomics. 2010;10:1150–1159. doi: 10.1002/pmic.200900375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do JS, Visperas A, Dong C, Baldwin WM, 3rd, Min B. Cutting edge: generation of colitogenic Th17 CD4 T cells is enhanced by IL-17+ γδ T cells. J Immunol. 2011;186:4546–4550. doi: 10.4049/jimmunol.1004021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do JS, Kim S, Keslar K, Jang E, Huang E, Fairchild RL, Pizarro TT, Min B. γδ T cells coexpressing gut homing α4β7 and αE integrins define a novel subset promoting intestinal inflammation. J Immunol. 2017;198:908–915. doi: 10.4049/jimmunol.1601060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrentraut SF, Colgan SP. Implications of protein post-translational modifications in IBD. Inflamm Bowel Dis. 2012;18:1378–1388. doi: 10.1002/ibd.22859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabregat A, Jupe S, Matthews L, Sidiropoulos K, Gillespie M, Garapati P, Haw R, Jassal B, Korninger F, May B, et al. The reactome pathway knowledgebase. Nucleic Acids Res. 2018;46(D1):D649–D655. doi: 10.1093/nar/gkx1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frasor J, Danes JM, Funk CC, Katzenellenbogen BS. Estrogen down-regulation of the corepressor N-CoR: mechanism and implications for estrogen derepression of N-CoR-regulated genes. Proc Natl Acad Sci USA. 2005;102:13153–13157. doi: 10.1073/pnas.0502782102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritah S, Lhocine N, Golebiowski F, Mounier J, Andrieux A, Jouvion G, Hay RT, Sansonetti P, Dejean A. Sumoylation controls host anti-bacterial response to the gut invasive pathogen Shigella flexneri. EMBO Rep. 2014;15:965–972. doi: 10.15252/embr.201338386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garvin AJ, Densham RM, Blair-Reid SA, Pratt KM, Stone HR, Weekes D, Lawrence KJ, Morris JR. The deSUMOylase SENP7 promotes chromatin relaxation for homologous recombination DNA repair. EMBO Rep. 2013;14:975–983. doi: 10.1038/embor.2013.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabinger T, Bode KJ, Demgenski J, Seitz C, Delgado ME, Kostadinova F, Reinhold C, Etemadi N, Wilhelm S, Schweinlin M, et al. Inhibitor of apoptosis protein-1 regulates tumor necrosis factor-mediated destruction of intestinal epithelial cells. Gastroenterology. 2017;152:867–879. doi: 10.1053/j.gastro.2016.11.019. [DOI] [PubMed] [Google Scholar]
- Hoffmann JC, Peters K, Henschke S, Herrmann B, Pfister K, Westermann J, Zeitz M. Role of T lymphocytes in rat 2,4,6-trinitrobenzene sulphonic acid (TNBS) induced colitis: increased mortality after gammadelta T cell depletion and no effect of alphabeta T cell depletion. Gut. 2001;48:489–495. doi: 10.1136/gut.48.4.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- House CM, Frew IJ, Huang H-L, Wiche G, Traficante N, Nice E, Catimel B, Bowtell DDL. A binding motif for Siah ubiquitin ligase. Proc Natl Acad Sci USA. 2003;100:3101–3106. doi: 10.1073/pnas.0534783100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu MM, Shu HB. Multifaceted roles of TRIM38 in innate immune and inflammatory responses. Cell Mol Immunol. 2017;14:331–338. doi: 10.1038/cmi.2016.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CJ, Wu D, Jiao XF, Khan FA, Xiong CL, Liu XM, Yang J, Yin TL, Huo LJ. Maternal SENP7 programs meiosis architecture and embryo survival in mouse. Biochim Biophys Acta Mol Cell Res. 2017;1864:1195–1206. doi: 10.1016/j.bbamcr.2017.03.005. [DOI] [PubMed] [Google Scholar]
- Inagaki-Ohara K, Chinen T, Matsuzaki G, Sasaki A, Sakamoto Y, Hiromatsu K, Nakamura-Uchiyama F, Nawa Y, Yoshimura A. Mucosal T cells bearing TCRgammadelta play a protective role in intestinal inflammation. J Immunol. 2004;173:1390–1398. doi: 10.4049/jimmunol.173.2.1390. [DOI] [PubMed] [Google Scholar]
- Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, Lee JC, Schumm LP, Sharma Y, Anderson CA, et al. International IBD Genetics Consortium (IIBDGC) Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2010;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi-Miyashita M, Shimada S, Kurosu H, Kato-Nagaoka N, Matsuoka Y, Ohwaki M, Ishikawa H, Nanno M. An accessory role of TCRgammadelta (+) cells in the exacerbation of inflammatory bowel disease in TCRalpha mutant mice. Eur J Immunol. 2001;31:980–988. doi: 10.1002/1521-4141(200104)31:4<980::aid-immu980>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- Khairallah C, Chu TH, Sheridan BS. Tissue adaptations of memory and tissue-resident gamma delta T cells. Front Immunol. 2018;9:2636. doi: 10.3389/fimmu.2018.02636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Scimia MC, Wilkinson D, Trelles RD, Wood MR, Bowtell D, Dillin A, Mercola M, Ronai ZA. Fine-tuning of Drp1/Fis1 availability by AKAP121/Siah2 regulates mitochondrial adaptation to hypoxia. Mol Cell. 2011;44:532–544. doi: 10.1016/j.molcel.2011.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Kim H, Park D, Jeoung D. miR-335 targets SIAH2 and confers sensitivity to anti-cancer drugs by increasing the expression of HDAC3. Mol Cells. 2015;38:562–572. doi: 10.14348/molcells.2015.0051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühl AA, Pawlowski NN, Grollich K, Loddenkemper C, Zeitz M, Hoffmann JC. Aggravation of intestinal inflammation by depletion/deficiency of gammadelta T cells in different types of IBD animal models. J Leukoc Biol. 2007;81:168–175. doi: 10.1189/jlb.1105696. [DOI] [PubMed] [Google Scholar]
- Kumar A, Zhang KY. Advances in the development of SUMO specific protease (SENP) inhibitors. Comput Struct Biotechnol J. 2015;13:204–211. doi: 10.1016/j.csbj.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahn M, Kalataradi H, Mittelstadt P, Pflum E, Vollmer M, Cady C, Mukasa A, Vella AT, Ikle D, Harbeck R, et al. Early preferential stimulation of gamma delta T cells by TNF-alpha. J Immunol. 1998;160:5221–5230. [PubMed] [Google Scholar]
- Lamas B, Richard ML, Leducq V, Pham HP, Michel ML, Da Costa G, Bridonneau C, Jegou S, Hoffmann TW, Natividad JM, et al. CARD9 impacts colitis by altering gut microbiota metabolism of tryptophan into aryl hydrocarbon receptor ligands. Nat Med. 2016;22:598–605. doi: 10.1038/nm.4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MJ, Lee JK, Choi JW, Lee CS, Sim JH, Cho CH, Lee KH, Cho IH, Chung MH, Kim HR, Ye SK. Interleukin-6 induces S100A9 expression in colonic epithelial cells through STAT3 activation in experimental ulcerative colitis. PLoS ONE. 2012;7:e38801. doi: 10.1371/journal.pone.0038801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little MC, Bell LV, Cliffe LJ, Else KJ. The characterization of intraepithelial lymphocytes, lamina propria leukocytes, and isolated lymphoid follicles in the large intestine of mice infected with the intestinal nematode parasite Trichuris muris. J Immunol. 2005;175:6713–6722. doi: 10.4049/jimmunol.175.10.6713. [DOI] [PubMed] [Google Scholar]
- Liu Z, Geboes K, Colpaert S, D’Haens GR, Rutgeerts P, Ceuppens JL. IL-15 is highly expressed in inflammatory bowel disease and regulates local T cell-dependent cytokine production. J Immunol. 2000;164:3608–3615. doi: 10.4049/jimmunol.164.7.3608. [DOI] [PubMed] [Google Scholar]
- Liu X, Chen W, Wang Q, Li L, Wang C. Negative regulation of TLR inflammatory signaling by the SUMO-deconjugating enzyme SENP6. PLoS Pathog. 2013;9:e1003480. doi: 10.1371/journal.ppat.1003480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JZ, van Sommeren S, Huang H, Ng SC, Alberts R, Takahashi A, Ripke S, Lee JC, Jostins L, Shah T, et al. International Multiple Sclerosis Genetics Consortium Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat Genet. 2015;47:979–986. doi: 10.1038/ng.3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Qian C, Cao X. Post-translational modification control of innate immunity. Immunity. 2016;45:15–30. doi: 10.1016/j.immuni.2016.06.020. [DOI] [PubMed] [Google Scholar]
- Lo Re S, Dumoutier L, Couillin I, Van Vyve C, Yakoub Y, Uwambayinema F, Marien B, van den Brûle S, Van Snick J, Uyttenhove C, et al. IL-17A-producing gammadelta T and Th17 lymphocytes mediate lung inflammation but not fibrosis in experimental silicosis. J Immunol. 2010;184:6367–6377. doi: 10.4049/jimmunol.0900459. [DOI] [PubMed] [Google Scholar]
- MacLean B, Eng JK, Beavis RC, McIntosh M. General framework for developing and evaluating database scoring algorithms using the TANDEM search engine. Bioinformatics. 2006;22:2830–2832. doi: 10.1093/bioinformatics/btl379. [DOI] [PubMed] [Google Scholar]
- Mustfa SA, Singh M, Suhail A, Mohapatra G, Verma S, Chakravorty D, Rana S, Rampal R, Dhar A, Saha S, et al. SUMOylation pathway alteration coupled with downregulation of SUMO E2 enzyme at mucosal epithelium modulates inflammation in inflammatory bowel disease. Open Biol. 2017;7 doi: 10.1098/rsob.170024. 170024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanno M, Kanari Y, Naito T, Inoue N, Hisamatsu T, Chinen H, Sugimoto K, Shimomura Y, Yamagishi H, Shiohara T, et al. Exacerbating role of gammadelta T cells in chronic colitis of T-cell receptor α mutant mice. Gastroenterology. 2008;134:481–490. doi: 10.1053/j.gastro.2007.11.056. [DOI] [PubMed] [Google Scholar]
- Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- Notredame C, Higgins DG, Heringa J. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J Mol Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- Perissi V, Aggarwal A, Glass CK, Rose DW, Rosenfeld MG. A corepressor/coactivator exchange complex required for transcriptional activation by nuclear receptors and other regulated transcription factors. Cell. 2004;116:511–526. doi: 10.1016/s0092-8674(04)00133-3. [DOI] [PubMed] [Google Scholar]
- Qi J, Tripathi M, Mishra R, Sahgal N, Fazli L, Ettinger S, Placzek WJ, Claps G, Chung LW, Bowtell D, et al. The E3 ubiquitin ligase Siah2 contributes to castration-resistant prostate cancer by regulation of androgen receptor transcriptional activity. Cancer Cell. 2013;23:332–346. doi: 10.1016/j.ccr.2013.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribet D, Hamon M, Gouin E, Nahori MA, Impens F, Neyret-Kahn H, Gevaert K, Vandekerckhove J, Dejean A, Cossart P. Listeria monocytogenes impairs SUMOylation for efficient infection. Nature. 2010;464:1192–1195. doi: 10.1038/nature08963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romeo K, Louault Y, Cantaloube S, Loiodice I, Almouzni G, Quivy JP. The SENP7 SUMO-protease presents a module of two HP1 interaction motifs that locks HP1 protein at pericentric heterochromatin. Cell Rep. 2015;10:771–782. doi: 10.1016/j.celrep.2015.01.004. [DOI] [PubMed] [Google Scholar]
- Scortegagna M, Subtil T, Qi J, Kim H, Zhao W, Gu W, Kluger H, Ronai ZA. USP13 enzyme regulates Siah2 ligase stability and activity via noncatalytic ubiquitin-binding domains. J Biol Chem. 2011;286:27333–27341. doi: 10.1074/jbc.M111.218214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeler JS, Dejean A. SUMO and the robustness of cancer. Nat Rev Cancer. 2017;17:184–197. doi: 10.1038/nrc.2016.143. [DOI] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen LN, Geoffroy M-C, Jaffray EG, Hay RT. Characterization of SENP7, a SUMO-2/3-specific isopeptidase. Biochem J. 2009;421:223–230. doi: 10.1042/BJ20090246. [DOI] [PubMed] [Google Scholar]
- Sidik SM, Salsman J, Dellaire G, Rohde JR. Shigella infection interferes with SUMOylation and increases PML-NB number. PLoS ONE. 2015;10:e0122585. doi: 10.1371/journal.pone.0122585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson SJ, Holländer GA, Mizoguchi E, Allen D, Bhan AK, Wang B, Terhorst C. Expression of pro-inflammatory cytokines by TCR alpha beta+ and TCR gamma delta+ T cells in an experimental model of colitis. Eur J Immunol. 1997;27:17–25. doi: 10.1002/eji.1830270104. [DOI] [PubMed] [Google Scholar]
- Stier S, Cheng T, Dombkowski D, Carlesso N, Scadden DT. Notch1 activation increases hematopoietic stem cell self-renewal in vivo and favors lymphoid over myeloid lineage outcome. Blood. 2002;99:2369–2378. doi: 10.1182/blood.v99.7.2369. [DOI] [PubMed] [Google Scholar]
- Suhail A, Rizvi ZA, Awasthi A, Srikanth CV. An SENP7 dependent epithelial signaling in IBD, Mendeley Data, v1. 2019 doi: 10.17632/z4xj6w9zxn.1. Published online November 4, 2019. [DOI] [Google Scholar]
- Szczepanik M, Gryglewski A, Bryniarski K, Stachura J, Ptak W. Experimental inflammatory bowel disease—role of T cells. J Physiol Pharmacol. 2000;51:333–346. [PubMed] [Google Scholar]
- Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi T, Sano B, Nagata T, Kato H, Sugiyama Y, Kunieda K, Kimura M, Okano Y, Saji S. Polo-like kinase 1 (PLK1) is overexpressed in primary colorectal cancers. Cancer Sci. 2003;94:148–152. doi: 10.1111/j.1349-7006.2003.tb01411.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiefenbach J, Novac N, Ducasse M, Eck M, Melchior F, Heinzel T. SUMOylation of the corepressor N-CoR modulates its capacity to repress transcription. Mol Biol Cell. 2006;17:1643–1651. doi: 10.1091/mbc.E05-07-0610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treuter E, Venteclef N. Transcriptional control of metabolic and inflammatory pathways by nuclear receptor SUMOylation. Biochim Biophys Acta. 2011;1812:909–918. doi: 10.1016/j.bbadis.2010.12.008. [DOI] [PubMed] [Google Scholar]
- Van Acker HH, Anguille S, Willemen Y, Van den Bergh JM, Berneman ZN, Lion E, Smits EL, Van Tendeloo VF. Interleukin-15 enhances the proliferation, stimulatory phenotype, and antitumor effector functions of human gamma delta T cells. J Hematol Oncol. 2016;9:101. doi: 10.1186/s13045-016-0329-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma S, Mohapatra G, Ahmad SM, Rana S, Jain S, Khalsa JK, Srikanth CV. Salmonella engages host microRNAs to modulate SUMOylation: a new arsenal for intracellular survival. Mol Cell Biol. 2015;35:2932–2946. doi: 10.1128/MCB.00397-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warde-Farley D, Donaldson SL, Comes O, Zuberi K, Badrawi R, Chao P, Franz M, Grouios C, Kazi F, Lopes CT, et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010;38(Web Server issue, Suppl):W214–20. doi: 10.1093/nar/gkq537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washburn T, Schweighoffer E, Gridley T, Chang D, Fowlkes BJ, Cado D, Robey E. Notch activity influences the alphabeta versus gammadelta T cell lineage decision. Cell. 1997;88:833–843. doi: 10.1016/s0092-8674(00)81929-7. [DOI] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview Version 2: a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen D, Wu J, Wang L, Fu Z. SUMOylation promotes nuclear import and stabilization of Polo-like kinase 1 to support its mitotic function. Cell Rep. 2017;21:2147–2159. doi: 10.1016/j.celrep.2017.10.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav K, Yavvari PS, Pal S, Kumar S, Mishra DK, Gupta S, Mitra M, Soni V, Khare N, Sharma P, et al. Oral delivery of cholic acid-derived amphiphile helps in combating Salmonella-mediated gut infection and inflammation. Bioconjug Chem. 2019;30:721–732. doi: 10.1021/acs.bioconjchem.8b00880. [DOI] [PubMed] [Google Scholar]
- Yang Y, He Y, Wang X, Liang Z, He G, Zhang P, Zhu H, Xu N, Liang S. Protein SUMOylation modification and its associations with disease. Open Biol. 2017;7 doi: 10.1098/rsob.170167. 170167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Xia Z, Wang X, Zhao X, Sheng Z, Ye Y, He G, Zhou L, Zhu H, Xu N, Liang S. Small-molecule inhibitors targeting protein SUMOylation as novel anticancer compounds. Mol Pharmacol. 2018;94:885–894. doi: 10.1124/mol.118.112300. [DOI] [PubMed] [Google Scholar]
- Yavvari PS, Verma P, Mustfa SA, Pal S, Kumar S, Awasthi AK, Ahuja V, Srikanth CV, Srivastava A, Bajaj A. A nanogel based oral gene delivery system targeting SUMOylation machinery to combat gut inflammation. Nanoscale. 2019;11:4970–4986. doi: 10.1039/c8nr09599j. [DOI] [PubMed] [Google Scholar]
- Yeh ETH, Gong L, Kamitani T. Ubiquitin-like proteins: new wines in new bottles. Gene. 2000;248:1–14. doi: 10.1016/s0378-1119(00)00139-6. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Yamashita H, Toyama T, Sugiura H, Ando Y, Mita K, Hamaguchi M, Hara Y, Kobayashi S, Iwase H. NCOR1 mRNA is an independent prognostic factor for breast cancer. Cancer Lett. 2006;237:123–129. doi: 10.1016/j.canlet.2005.05.046. [DOI] [PubMed] [Google Scholar]
- Zhao Q, Xie Y, Zheng Y, Jiang S, Liu W, Mu W, Liu Z, Zhao Y, Xue Y, Ren J. GPS-SUMO: a tool for the prediction of sumoylation sites and SUMO-interaction motifs. Nucleic Acids Res. 2014;42(Web Server issue, W1):W325–30. doi: 10.1093/nar/gku383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Q, Wang XJ. GOEAST: a web-based software toolkit for Gene Ontology enrichment analysis. Nucleic Acids Res. 2008;36:W358–63. doi: 10.1093/nar/gkn276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Ren S, Bitler BG, Aird KM, Tu Z, Skordalakes E, Zhu Y, Yan J, Sun Y, Zhang R. SPOP E3 ubiquitin ligase adaptor promotes cellular senescence by degrading the SENP7 deSUMOylase. Cell Rep. 2015;13:1183–1193. doi: 10.1016/j.celrep.2015.09.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data generated during this study are available at the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier: PXD015994 and PXD016018. The CD4+ and CD8+ T cells population data generated during this study are available at Mendeley Data. (https://data.mendeley.com/datasets/z4xj6w9zxn/1)