Summary
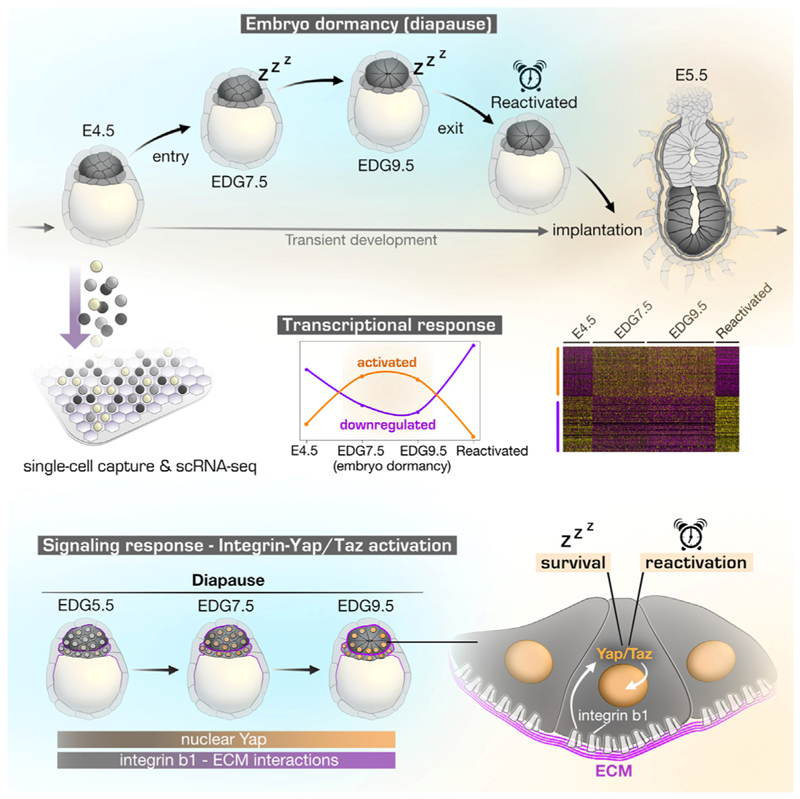
Embryonic diapause is a reproductive adaptation that enables some mammalian species to halt the otherwise continuous pace of embryonic development. In this dormant state, the embryo exploits poorly understood regulatory mechanisms to preserve its developmental potential for prolonged periods of time. Here, using mouse embryos and single-cell RNA sequencing, we molecularly defined embryonic diapause at single-cell resolution, revealing transcriptional dynamics while the embryo seemingly resides in a state of suspended animation. Additionally, we found that the dormant pluripotent cells rely on integrin receptors to sense their microenvironment and preserve their viability via Yap/Taz-mediated prosurvival signaling.
Graphical abstract.
Introduction
The implantation of the blastocyst into the uterine wall is a key step of the reproductive cycle that mediates the connection of the mammalian embryo to the maternal tissues during the early stages of pregnancy. Interestingly, in some species, this process can be put on hold, diverting embryonic development into a reversible state of dormancy, known as diapause. The dormant embryo can reside in this state over extended periods of time, without compromising its developmental potential. Although the process of transient embryogenesis has been intensively studied, the cellular and molecular mechanisms of embryonic diapause are still obscure.
Embryonic diapause is typically initiated at the late blastocyst stage, indicating that at this particular phase of development, the embryo is in a competent state for commencing implantation or dormancy. The process of implantation takes place in a restricted period of time known as the implantation window, which is determined primarily by the effects of two hormones, ovarian estrogen and progesterone. These hormones prepare the uterus to accommodate the mature blastocyst, synchronizing the timing of uterine receptivity with the developmental progression of the embryo.1
In mice, in the case of post-delivery fertilization during lactation, the development of the newly formed embryo proceeds normally until the blastocyst stage. Yet, the low level of estrogen in the lactating mother hinders the establishment of a receptive uterine environment; thus, the blastocyst is unable to initiate implantation. Remarkably, instead of fading away, the embryo “pauses” its transient developmental program and enters into a dormant state (facultative diapause). In this state, the mouse embryo remains viable and conserves its developmental capacity, whereas at any time, a surge of estrogen can swiftly establish a receptive uterine environment, enabling developmental reactivation and implantation.2
Diapause can also be experimentally induced in mice via surgical removal of the ovaries (ovariectomy) or injection of anti-estrogenic compounds.3 Conversely, the exit of diapause and initiation of implantation can be triggered by a single pulse of externally supplied estrogen.4 This provides a simple, temporally controlled experimental system to study embryo dormancy.
To decipher the cellular mechanisms of embryo dormancy, here, we characterized the transcriptome of murine embryos during entry, maintenance, and exit of diapause at single-cell resolution, in parallel with the “normal” pre-to post-implantation transition. Focusing on the pluripotent lineage, we uncovered dynamic modulations of signaling pathways and activation of a cryptic, diapause-specific transcriptional response. In particular, we found that integrin/Yap signaling is required for preserving the developmental capacity of the dormant embryo.
Results
Single-cell atlas of peri-implantation embryogenesis and embryo dormancy
At embryonic day 4.5 (E4.5), the mouse blastocyst consists of three fully specified lineages, namely, the epiblast (Epi), primitive endoderm (PrE), and trophectoderm (TE). In a receptive uterine environment, the E4.5 embryo initiates implantation and by E5.5 transforms into an early post-implantation conceptus (egg cylinder). Alternatively, in a non-receptive uterine environment, the embryo enters into a dormant state.
Our previous analysis using Wnt reporter mice revealed a peak of Wnt activity at day 3 of diapause, corresponding to an estimated day of gestation (EDG) 7.5. This was followed by a decrease in reporter expression by EDG9.5. Within this time frame, the Epi transformed from a simple ball of cells into an epithelial rosette-like structure.5 To decipher the underlying transcriptional dynamics, we sought to examine EDG7.5 and EDG9.5 embryos using single-cell RNA sequencing (scRNA-seq) alongside the transient E4.5 and E5.5 developmental stages (Figure 1A).
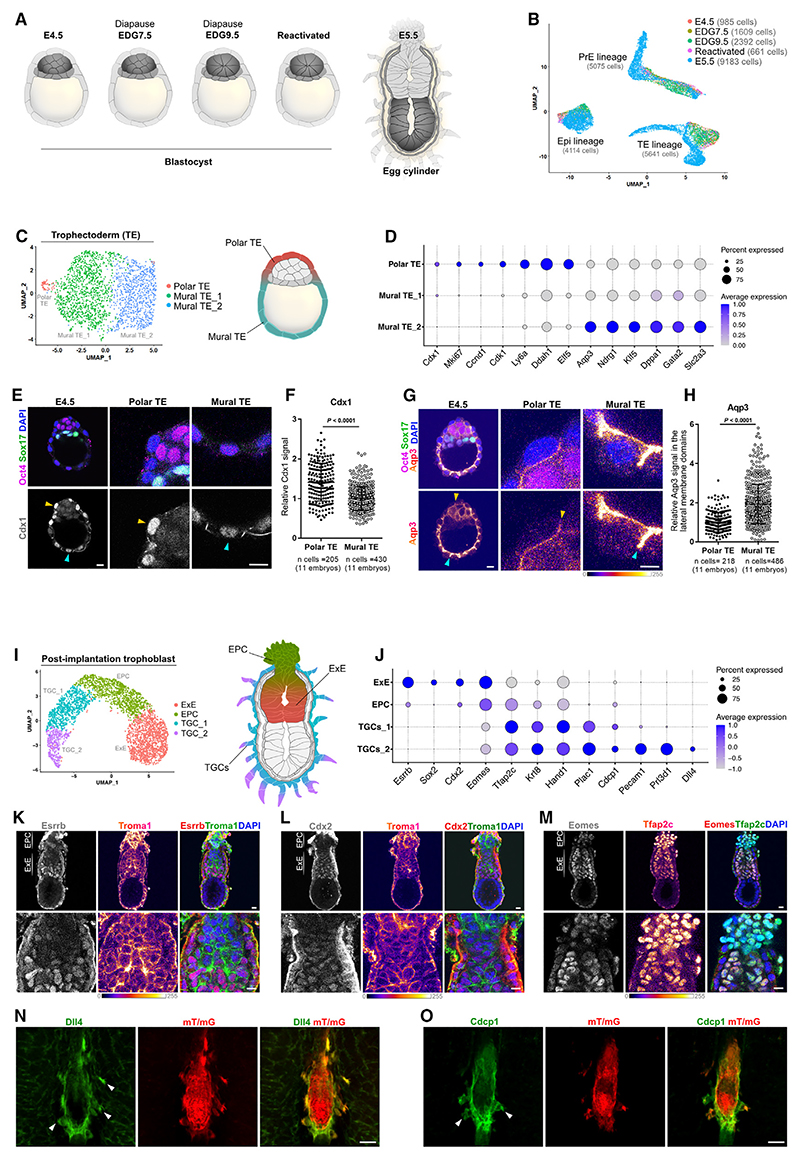
Figure 1. Cellular heterogeneity in the trophoblast lineage.
(A) Overview of the sampled developmental time points.
(B) UMAP plot of the examined transcriptomes.
(C) UMAP plot of the TE cells.
(D) Marker genes of the TE subpopulations.
(E) Cdx1 expression at E4.5. Yellow arrowheads, polar TE; cyan arrowheads, mural TE.
(F) Quantification of relative fluorescence intensity for Cdx1 in the TE nuclei. Data were normalized to the average Cdx1 signal in the mural TE and presented as mean ± SD, n = number of cells, two-tailed unpaired Student’s t test. Three independent experiments.
(G) Aqp3 expression at E4.5. Yellow arrowheads, polar TE; cyan arrowheads, the mural TE.
(H) Quantification of relative fluorescence intensity for Aqp3 in the lateral membranes of TE at E4.5. Data were normalized to the average Aqp3 signal in the polar TE and presented as mean ± SD, n = number of cells, two-tailed unpaired Student’s t test. Two independent experiments.
(I) UMAP plot of the E5.5 trophoblast subpopulations.
(J) Marker gene expression in the E5.5 trophoblast.
(K) E5.5 embryo stained for Esrrb, Troma1, and DAPI.
(L) E5.5 embryo stained for Cdx2, Troma1, and DAPI.
(M) E5.5 embryo stained for Eomes, Tfap2c, and DAPI.
(N) Staining for tdTomato and Dll4 in E5.5 mT/mG embryo within the maternal tissues. White arrowheads, TGCs.
(O) Staining for tdTomato and Cdcp1 in E5.5 mT/mG embryo within the maternal tissues. White arrowheads, TGCs. Scale bars: 10 μm in (E), (G), (K), (L), and (M) and 50 μm in (N) and (O). See also Figure S1.
To obtain a holistic view, we also included in the analysis embryos that exit diapause. To define the time of developmental reactivation, we induced diapause via ovariectomy and administered a single dose of estrogen (17β-estradiol, E2). We collected embryos after 18, 24, or 30 h and monitored the nascent RNA synthesis via the incorporation of 5-ethynyl uridine (EU). We found that at 30 h, both the TE and the inner cell mass (ICM) exhibited an EU signal, which is indicative for the exit of the dormant state (Figures S1A and S1B). Hereafter, we refer to these embryos as reactivated.
We collected blastocysts at E4.5 (206 embryos), EDG7.5 (247 embryos), EDG9.5 (223 embryos), reactivated (191 embryos) stages, and the egg cylinder stage (E5.5, 42 embryos) and performed scRNA-seq analysis (Figures 1A and 1B). The E4.5 dataset has also been included in another study from our lab.6 Uniform manifold approximation and projection (UMAP) clustering revealed three distinct groups of cells encompassing the Epi, PrE, and TE lineages and their derivatives (Figures 1B and S1C). Taking advantage of the relatively large number of cells in the scRNA-seq datasets (Figure 1B), we first examined and validated the emerging cellular heterogeneity during the blastocyst to egg cylinder transition, and then we focused on the transcriptional dynamics during embryo dormancy.
Cellular heterogeneity in the trophoblast lineage
Despite the different developmental timing, the E4.5, EDG7.5, EDG9.5, and reactivated embryos are all essentially blastocysts that consist of three cell lineages (Epi, PrE, and TE), and the derivatives of these tissues emerge after implantation (E5.5). Accordingly, the TE cells of the E4.5, EDG7.5, EDG9.5, and reactivated blastocysts clustered together apart from the post-implantation trophoblast (Figure S1D).
Focusing first on the TE, we delineated three subpopulations of cells corresponding to the polar TE and mural TE (mural TE_1 and mural TE_2) (Figure 1C). These subpopulations contained cells of the E4.5, EDG7.5, EDG9.5, and reactivated blastocysts (Figure S1E). The polar TE exhibited elevated expression of markers, such as Cdx1, Ccnd1, Mki67, Ly6a, Ddah1, and Elf5, whereas the mural TE (mural TE_1 and mural TE_2) gradually upregulated genes such as Aqp3, Ndrg1, Klf5, Dppa1, Gata2, and Slc2a3 (Figure 1D). The latter finding suggests that in comparison with mural TE_1, the mural TE_2 cells are more advanced in their differentiation (Figure 1C). On the protein level, we analyzed the expression pattern of Cdx1 and Aqp3 by immunohistochemistry, confirming their enrichment in the polar and mural TE, respectively (Figures 1E–1H).
The TE has two major functions: to mediate the implantation process and to give rise to the tissues of the placenta. This bifurcation of the TE functions becomes apparent after E4.5 during transient embryogenesis or following reactivation and implantation of the diapause embryo. The mural TE cells give rise to trophoblast giant cells (TGCs) that invade the uterine stroma, whereas the polar TE forms the extraembryonic ectoderm (ExE), which consists of multipotent stem cell progenitors of the placental tissues. Accordingly, the scRNA-seq analysis of the post-implantation trophoblast delineated the establishment of TGCs (TGCs_1 and TGCs_2), ExE, and the ectoplacental cone (EPC), which originates from the ExE (Figure 1I).
Marker gene analysis revealed the expression of trophoblast stem cell (TSC) factors, such as Esrrb, Sox2, Cdx2, and Eomes in the ExE, in line with previous reports.7,8 The TSC transcription factors were downregulated in the EPC, which exhibited an elevated expression of Tfap2c and Krt8 (Troma-1) (Figure 1J). Accordingly, we observed a similar expression pattern of these factors on the protein level in the ExE and EPC of E5.5 embryos (Figures 1K–1M and S1F).
The TGCs, which mediate implantation, also expressed Tfap2c and Krt8 and upregulated typical TGC markers, such as Prl3d1, Hand1, and Plac1 (Figure 1J). In addition, we detected the expression of endothelial genes, such as Pecam-1 and Dll4 (Figure 1J), as previously reported.9 Using whole-mount tissue clearing and staining of red fluorescent E5.5 mT/mG embryos within the maternal tissues, we confirmed the expression of Dll4 and Cdcp1 in the invasive TGCs (Figures 1N and 1O, arrow-heads). The enrichment of TGC-specific gene expression in the TGCs_2 cluster, in comparison with TGCs_1, suggests that these cells are developmentally more advanced and potentially located at the invasive front (Figures 1I and 1J).
Cellular heterogeneity in the PrE lineage
Next, we examined the cellular heterogeneity in the second extraembryonic lineage, the PrE. Similar to the pre-implantation trophoblast, the PrE cells of the E4.5, EDG7.5, EDG9.5, and reactivated blastocysts clustered together apart from the E5.5 cells (Figures 2A and S1G). During the implantation stages, the PrE gives rise to two main major cell types, namely, the parietal endoderm (PaE), which migrates over the mural TE/TGC compartment, and the visceral endoderm (VE), which surrounds the emerging egg cylinder. Accordingly, in addition to the PrE cluster of the blastocysts, we identified the PaE and VE cell populations of the post-implantation conceptus. Moreover, we could also clearly delineate the subcompartments of the VE, namely, the cells overlying the ExE (extraembryonic VE [exVE]), overlying the Epi (embryonic VE [emVE]) and the cells positioned at the distal tip (distal VE [DVE]) (Figures 2A, 2E, and S1I).
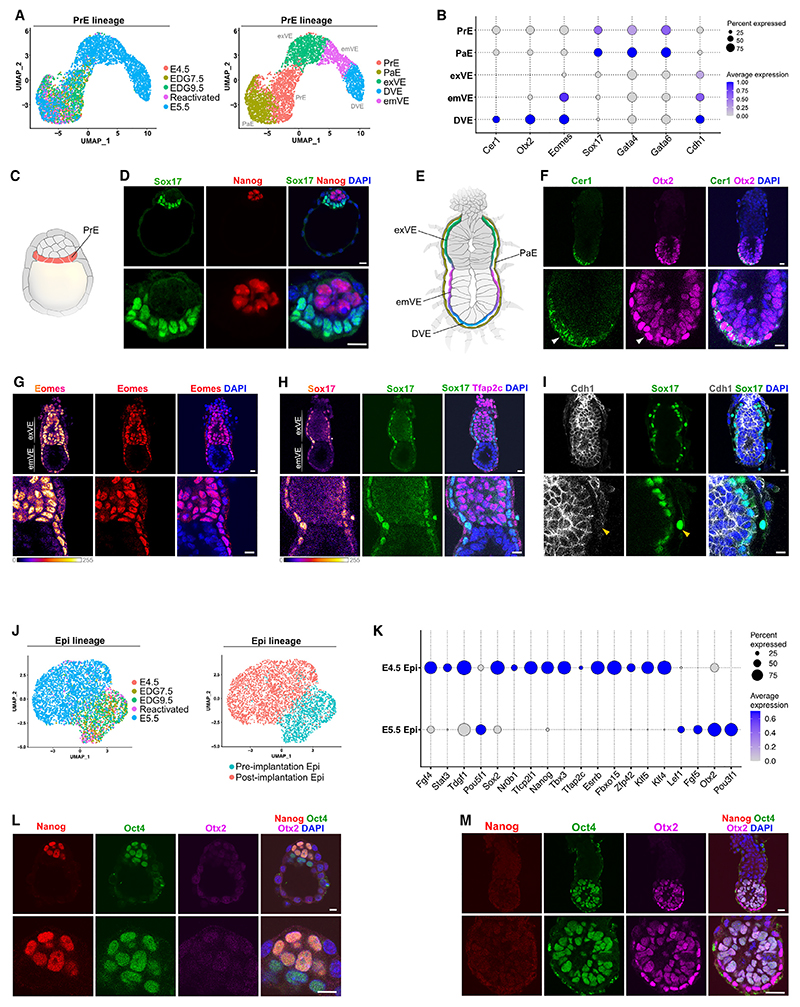
Figure 2. Cellular heterogeneity in the PrE lineage and pluripotent states transition in the Epi.
(A) UMAP plot of the PrE lineage cells (left) and subclustering (right).
(B) Marker gene expression in the PrE lineage.
(C) Schematic representation of blastocyst with PrE in orange.
(D) E4.5 embryo stained for Sox17, Nanog, and DAPI.
(E) Schematic representation of E5.5 mouse embryo with annotated DVE, emVE, exVE, and PaE.
(F) E5.5 embryo stained for Cer1, Otx2, and DAPI.
(G) E5.5 embryo stained for Eomes and DAPI.
(H) E5.5 embryo stained for Sox17, Tfap2c, and DAPI.
(I) E5.5 embryo stained for Cdh1, Sox17, and DAPI.
(J) UMAP plot of the Epi cells.
(K) Marker gene expression in the Epi.
(L) E4.5 embryo stained for Nanog, Oct4, Otx2, and DAPI.
(M) E5.5 embryo stained for Nanog, Oct4, Otx2, and DAPI.
Scale bars, 10 μm. See also Figure S1.
Marker gene analysis showed the expression of the master transcription factors Sox17, Gata4, and Gata6 in the PrE (Figures 2B–2D). In the VE at the egg cylinder stage, Sox17 expression was enriched in the exVE as previously shown,10 whereas Otx2 and Eomes were expressed predominantly in the emVE, and Gata4 was expressed in both the exVE and emVE. We validated this compartmentalized expression pattern also on the protein level in E5.5 embryos (Figures 2B–2H and S1H). In addition, the DVE cells, which are positioned within the emVE, expressed the Nodal antagonist Cer1 (Figures 2B and 2F), whereas the PaE cells downregulated the expression of the cell adhesion molecule Cdh1 (E-cadherin) (Figure 2I), in line with previous reports.6,11
Altogether, the analyses show that the PrE and TE lineages maintain their cell identity during diapause, exhibiting no major signs of differentiation. The latter is promptly initiated during the blastocyst to egg cylinder transition, leading to the emerging heterogeneity in the extraembryonic tissues.
Pre- to post-implantation transition in the Epi lineage
During the E4.5 to E5.5 transition, the naive pluripotent state is dismantled and transforms into more developmentally advanced (formative) pluripotency, which has been suggested to capacitate the Epi for further somatic and germ cell lineage specification.12 Accordingly, the scRNA-seq analysis showed the down-regulation of an array of naive transcription factors, such as Tbx3, Tfap2c, Esrrb, and Nanog, and the upregulation of post-implantation markers, such as Lef1, Fgf5, Pou3f1, and Otx2, in the Epi (Figures 2J and 2K).
To illustrate this transcriptional switch, we stained E4.5 and E5.5 embryos for Nanog, Otx2, and Oct4. Although the core pluripotency transcription factor Oct4 was expressed in the pre- and post-implantation Epi, Nanog, and Otx2 exhibited mutually exclusive expression patterns at the E4.5 and E5.5, respectively (Figures 2L and 2M), as previously shown.13,14
The UMAP analysis also showed that the Epi cells of the E4.5, EDG7.5, EDG9.5, and reactivated embryos clustered apart from the post-implantation Epi (Figures 2J and 2K). Although this indicates that the exit of naive pluripotency is halted in the dormant embryo, in the following experiments, we aimed to uncover potential “hidden” transcriptional changes during diapause.
Transcriptional dynamics during entry, maintenance, and exit of embryo dormancy
Based on the examined developmental stages, we delineated three main phases of diapause progression, namely, entry (E4.5–EDG7.5), maintenance (EDG7.5–EDG9.5), and exit (EDG9.5–reactivated) of diapause (Figures 3A, S2A, and S2C). We performed weighted gene co-expression network analysis (WGCNA)15 to construct networks of genes that show similar expression patterns across the three phases and to identify modules of co-expressed genes. Notably, we found two major co-expression modules showing up- or downregulation in the Epi (Figures 3A and 3B; Table S1).
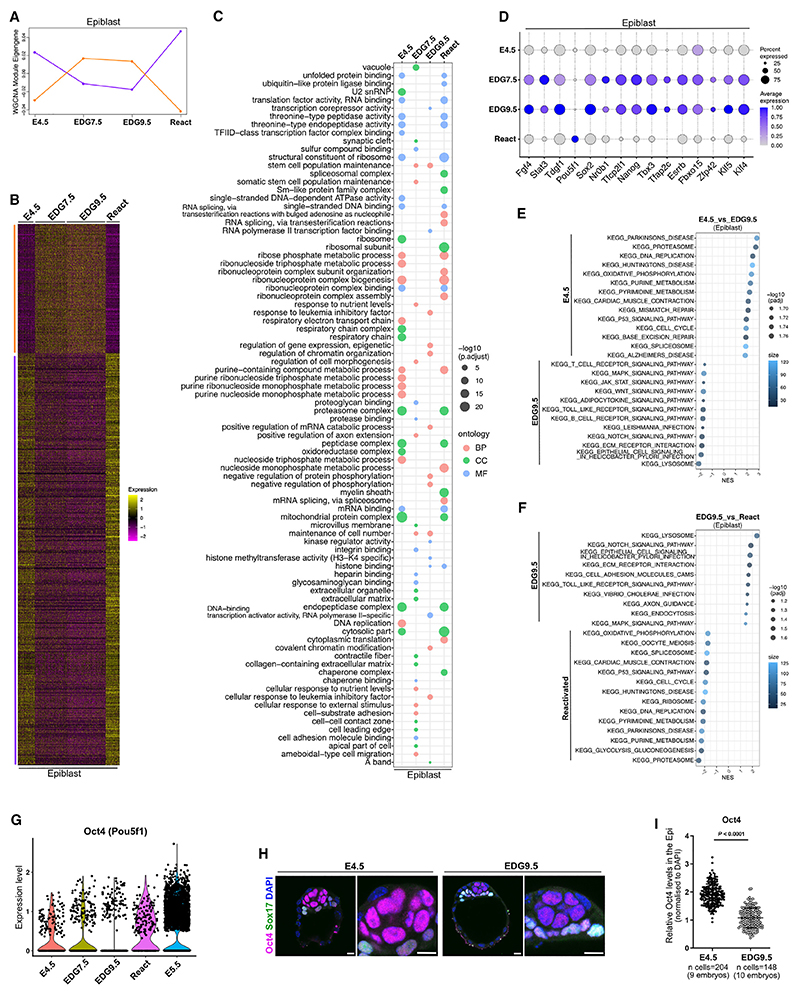
Figure 3. Transcriptional dynamics in the Epi.
(A) WGCNA identifies two modules of co-expressed genes in the Epi; x axis, developmental stages; y axis, log2-transformed, normalized intensity ratios in each stage.
(B) Heatmap plot of 1,127 genes in the two co-expression modules in the Epi.
(C) GO enrichment analysis in the Epi. MF, molecular function; BP, biological process; CC, cellular component.
(D) Pluripotency marker gene expression.
(E) KEGG pathways, E4.5 vs. EDG9.5 Epi.
(F) KEGG pathways, EDG9.5 vs. reactivated Epi.
(G) Oct4 expression in the Epi.
(H) Oct4 expression at E4.5 and EDG9.5.
(I) Quantification of relative fluorescence intensity for Oct4 in the Epi nuclei at E4.5 (two independent experiments) and EDG9.5 (three independent experiments) stages. The Epi cells are positive for Oct4 and negative for Sox17. Data were normalized to the average Oct4 signal in the EDG9.5 group and presented as mean ± SD, n = number of cells, two-tailed unpaired Student’s t test.
Scale bars, 10 μm. See also Figure S2.
The extraembryonic lineages also exhibited overall similar transcriptional dynamics. The majority of the active genes at E4.5 were downregulated during diapause, followed by their upregulation in the TE or PrE of the reactivated embryos. In addition, smaller groups of genes were transcriptionally activated at EDG7.5 and EDG9.5 or at EDG9.5 and reactivated embryos (Figures S2A–S2D; Table S1). This shows that both the embryonic and the extraembryonic lineages exhibit specific transcriptional signatures associated with the entry, progression, and exit of diapause.
Gene Onotology (GO) enrichment analysis revealed that the genes downregulated in the Epi during diapause were mainly related to replication (“single-stranded DNA binding” and “DNA replication”), transcription, and splicing (“TFIID-class transcription factor complex binding” and “spliceosomal complex”), as well as translation and protein folding (“translation factor activity, RNA binding,” “structural constituent of ribosome,” and “chaperone complex”) (Figure 3C; Table S2). Interestingly, in contrast to E4.5 and reactivated Epi cells, GO terms associated with protein degradation, such as “proteasome complex,” “peptidase complex,” and “endopeptidase complex,” were also downregulated at EDG7.5 and EDG9.5 (Figure 3C). Thus, although protein synthesis is reduced during diapause, this suggests that the opposite process of degradation might also be diminished, which may prolong the time of protein turnover.
Similarly, the TE and PrE also showed GO terms associated with reduced protein degradation at EDG7.5 and EDG9.5 (Figures S2E and S2F; Table S2). Interestingly, in contrast to the Epi, where gene sets associated with ribosomal biogenesis were downregulated during diapause, GO terms such as structural constituent of ribosome, “rRNA processing,” “rRNA binding,” “ribosomal small subunit biogenesis,” “ribosomal small subunit assembly,” “polysome,” and “cytosolic ribosome” were enriched at EDG9.5 in the TE and PrE (Figures S2E and S2F). This appears somewhat surprising because the blastocyst becomes biosynthetically quiescent during embryo dormancy. However, this response might not be required for the dormant state per se, but it could be a prerequisite for an adequate reactivation of the translational machinery during the exit of diapause.
Embryo dormancy is also associated with reduced energy metabolism.16 It is worth mentioning that the total amount of mitochondria in the embryo remains constant during the zygote-to-blastocyst transition. Thus, the cleavage divisions that generate smaller blastomeres also halve the number of mitochondria per cell with each division.17 Moreover, in comparison with the post-implantation embryo, the mitochondria in the pre-implantation embryo are relatively immature, exhibiting low oxygen consumption and reduced ATP production.18–20 Therefore, the pre-implantation embryos are referred to as metabolically quiet.21
Our scRNA-seq analysis revealed that GO terms associated with mitochondrial energy production were underrepresented at EDG7.5 and EDG9.5 compared with E4.5 and reactivated embryos. These gene sets were associated with the “respiratory chain complex” and “respiratory chain” (Epi, TE, and PrE), “cytochrome-c oxidase activity” and “cytochrome complex” (TE and PrE), “mitochondrial protein complex” (Epi and TE), “mitochondrial inner membrane” (TE), and “inner mitochondrial membrane protein complex” (PrE) (Figures 3C, S2E, and S2F). Moreover, GO terms related to purine nucleoside metabolic processes and energy production were associated only with E4.5 and/or reactivated embryos (Figures 3C, S2E, and S2F).
Altogether, this indicates that following the relatively low oxygen consumption and ATP production during the transient preimplantation embryogenesis, the metabolic rate further slows down during diapause until the embryo exits this state of metabolic quiescence upon reactivation.
Expression dynamics of the pluripotency transcription factors
During embryo dormancy, the pluripotent cells remain undifferentiated as the transition from naive to formative pluripotency is halted. Historically, this was used experimentally to increase the efficiency of embryonic stem cell (ESC) derivation.22,23 Accordingly, GO term analysis revealed the enrichment of gene sets associated with “maintenance of cell number” and “stem cell population maintenance” at EDG7.5 and EDG9.5 (Figure 3C). Gene expression analysis also showed that overall, the naive pluripotency transcription factors were not only maintained during diapause but also generally exhibited higher transcript levels compared with E4.5 and reactivated embryos (Figure 3D).
It has been well established that leukemia inhibitory factor (Lif)/Gp130-mediated activation of the Jak/Stat3 pathway is critical for ESC self-renewal.24,25 Although Gp130 function is dispensable for the pre- and early post-implantation embryogenesis,26,27 deletion of this receptor is detrimental to the maintenance of the pluripotent lineage during late diapause.28 Accordingly, we found GO terms “response to Lif” and “cellular response to Lif” enriched at EDG9.5 (Figure 3C). Moreover, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis of E4.5 vs. EDG9.5 Epi cells also showed the enrichment of Jak/Stat, as well as Wnt, Notch, and mitogen-activated protein kinase (MAPK) signaling during embryo dormancy (Figure 3E; Table S2). The MAPK cascade can be activated downstream of the Gp130 receptor and also as a response to Fgf stimulation.29 Interestingly, we found Fgf4 upregulation in the EDG7.5 and EDG9.5 Epi cells, which may supply this ligand to the extraembryonic tissues in a paracrine manner (Figure 3D).
Although, in general, the pluripotency transcription factors exhibited elevated transcript levels during diapause compared with E4.5 and reactivated embryos, Oct4 (Pou5f1) expression in the Epi was toned down (Figures 3D and 3G). We confirmed this also on the protein level, comparing Oct4 expression in E4.5 vs. EDG9.5 embryos (Figures 3H and 3I). At first glance, this is unexpected because Oct4 is a core pluripotency transcription factor. However, it has been shown that ESCs with reduced expression of Oct4 (heterozygous ESCs) exhibit a stabilized pluripotent state. These cells show increased Oct4 binding at pluripotency-associated enhancers, elevated Wnt expression, enhanced sensitivity to Lif, reduced Fgf responsiveness, and more homogeneous expression of pluripotency transcription factors. Similar to the diapause embryo (Figure 3D), the Fgf4 and Stat3 transcripts were also elevated in Oct4 heterozygous ESCs.30 Thus, the Oct4 reduction during diapause could be the physiological manifestation of the more stable pluripotent state of the Oct4 heterozygous ESCs in vitro.
Integrin signaling during embryo dormancy
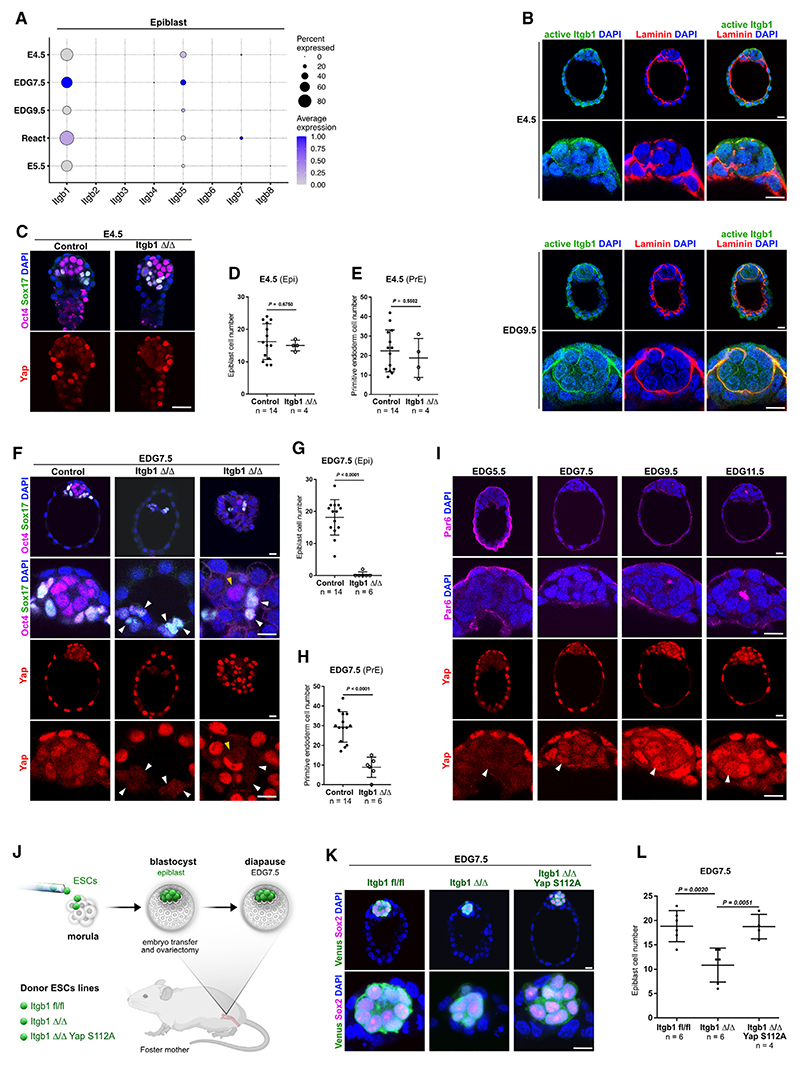
We have previously shown that during the EDG7.5 to EDG9.5 period of embryo dormancy, the Epi undergoes epithelialization forming a rosette-like structure. In accord with this morphogenetic transformation, the GO term analysis showed an enrichment of gene sets associated with epithelial polarity (“apical part of cell” and “microvillus membrane”), as well as integrin signaling and extracellular matrix (ECM) interactions (“integrin binding,” “collagen-containing ECM,” and “cell-substrate adhesion”) (Figure 3C). Similarly, the KEGG pathway analysis also indicated an enrichment of “ECM receptor interactions” in the Epi of the diapause embryos (Figures 3E and 3F). Thus, we sought to investigate the functional significance of integrin-ECM signaling during embryo dormancy.
First, we examined the expression of integrins and found that integrin beta-1 (Itgb1) was the predominantly expressed beta subunit in the Epi and PrE lineages (Figures 4A and S3A–S3C). We also confirmed the expression of Itgb1 in the ICM of E4.5 and EDG9.5 embryos on the protein level (Figure S3D). Because integrin-ECM binding depends on the protein conformation of the integrin receptors,31 we stained E4.5 and EDG9.5 embryos using an antibody that recognizes the open conformation of the Itgb1 extracellular domain (active Itgb1). To detect the ECM, we co-stained for laminin, which is a key component of the basement membrane (BM) in the embryo.32 At EDG9.5, we found an increased signal of the active form of Itgb1 co-localizing with laminin, indicating an ongoing engagement of this receptor with the ECM (Figure 4B).
Figure 4. Integrin/Yap signaling during embryo dormancy.
(A) Integrin beta subunits expression.
(B) E4.5 and EDG9.5 embryos stained for active Itgb1, laminin, and DAPI.
(C) E4.5 Itgb1 control and KO embryos stained for Oct4, Sox17, Yap, and DAPI.
(D) Epi cell number at E4.5. Three independent experiments.
(E) PrE cell number at E4.5. Three independent experiments.
(F) EDG7.5 Itgb1 control and KO embryos stained for Oct4, Sox17, Yap, and DAPI. Yellow arrowheads, Epi; white arrowheads, PrE.
(G) Epi cell number at EDG7.5. Five independent experiments.
(H) PrE cell number at EDG7.5. Five independent experiments.
(I) EDG5.5, EDG7.5, EDG9.5, and EDG11.5 embryos stained for Yap, Par6, and DAPI. Yap accumulation in the nuclei of the ICM is marked by arrowheads.
(J) Schematic representation of the morula aggregation experiments.
(K) EDG7.5 embryos harboring donor cells stained for Venus, Sox2, and DAPI. Separate channels are presented in Figure S4A.
(L) Epi cell number in EDG7.5 embryos harboring donor cells. Three independent experiments for Itgb1 fl/fl or Itgb1 Δ/Δ and two independent experiments for Itgb1 Δ/Δ Yap S112A group.
(D, E, G, and H) Data represent mean ± SD, n = number of embryos, two-tailed unpaired Student’s t test, p value is presented on the figures; (L) data represent mean ± SD, n = number of embryos, one-way ANOVA with Tukey’s post hoc test, p value is presented on the figures.
Scale bars, 10 μm. See also Figures S3 and S4.
To examine the functional significance of Itgb1, we generated Itgb1 knockout (KO) embryos via heterozygous intercrosses. We found no obvious alterations in the morphology of the Itgb1 KO blastocysts or changes in the cell number of the Epi and PrE at E4.5 compared with control littermates (Figures 4C–4E and S3E). However, after inducing diapause, we found that the loss of Itgb1 was detrimental for the Epi and PrE lineages (Figures 4F–4H). Accordingly, already at the onset of diapause, Itgb1 KO embryos showed increased levels of cleaved caspase-3-positive apoptotic cells in the ICM and scattered throughout the cavity (Figure S3F). Altogether, this suggests that Itgb1 plays a critical role in maintaining the ICM during embryo dormancy (Figures 4F–4H).
Integrin receptors bind to the ECM, establishing focal adhesions that transmit signaling cues, including mechanical force, to the cells.33 It has been shown that Yap and Taz, which are the central components of the Hippo pathway, can sense such mechanical inputs, which induce cytoplasmic to nuclear translocation of these factors.34 Therefore, we analyzed Yap localization during the progression of embryo dormancy (EDG5.5 to EDG11.5) and found that it accumulated in the nuclei of the Epi and PrE cells (Figures 4I and S3G). Moreover, the few remaining ICM cells in the dormant Itgb1 KO blastocysts failed to accumulate Yap in the nucleus (Figures 4F and S3H), indicating that this is an Itgb1-dependent process.
We next asked whether active Yap can compensate for the loss of Itgb1 in the Epi during diapause. To test this, we used our previously characterized ESCs harboring Itgb1 floxed allele (Itgb1 fl/fl) and Cre-ERT2-IRES-Venus transgene mediating Itgb1 excision (Itgb1 Δ/Δ) in the presence of 4-hydroxytamoxifen (4-OHT).35 In addition, we integrated a constitutively active form of Yap in which serine 112 was converted to alanine (Yap S112A).36 We aggregated morulae with these three cell lines (Itgb1 fl/fl, Itgb1 Δ/Δ, or Itgb1 Δ/Δ Yap S112A) and generated chimeric blastocysts comprising wild-type extraembryonic host cells and Venus-positive Epi cells derived from the donor ESCs (Figures S3I and S3J). The embryos were then transferred into the uteri of foster mothers, where diapause was induced by ovariectomy and subsequently isolated at EDG7.5 for analysis (Figure 4J). We found that chimeric embryos containing Itgb1 Δ/Δ cells exhibited a substantially reduced number of Epi cells, which was rescued by the expression of Yap-S112A (Figures 4K, 4L, and S4A). Thus, in the following experiments, we aimed to further investigate the potential role of Yap and Taz during embryo dormancy.
Yap/Taz function in the ICM during embryonic diapause
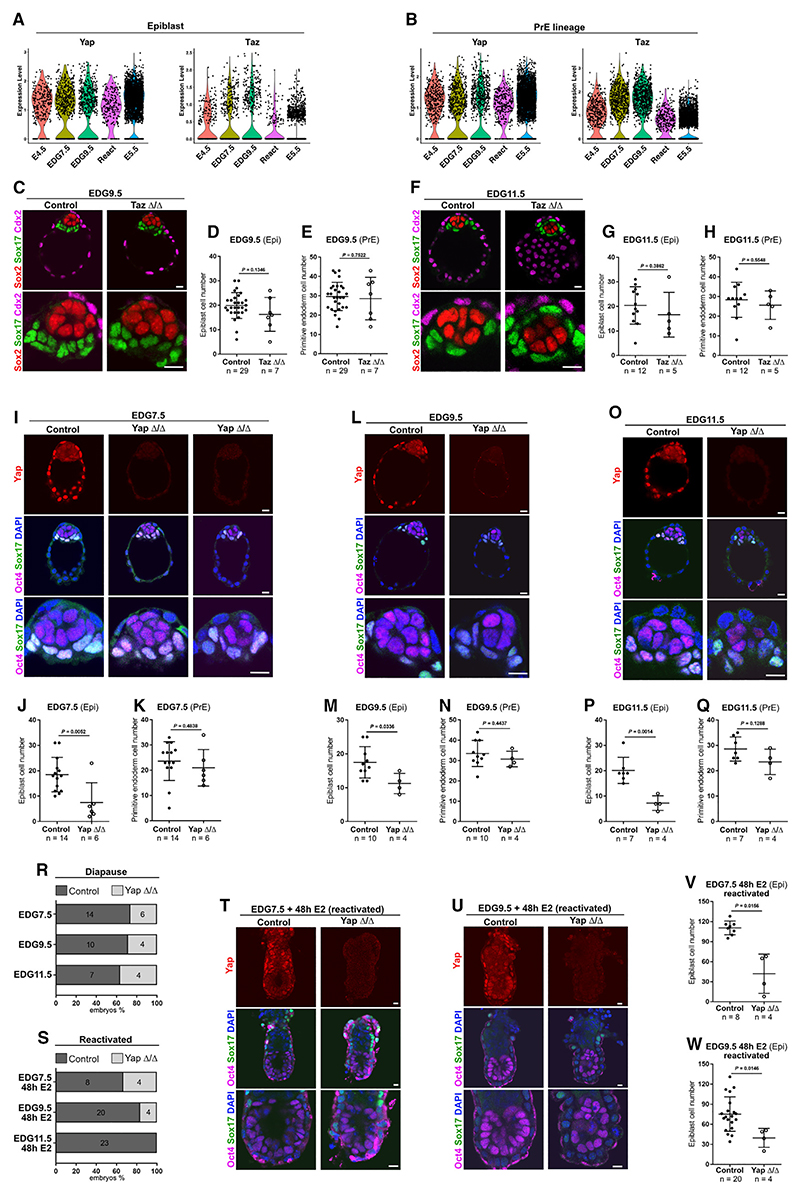
First, we examined Yap and Taz transcript levels in the Epi and PrE and found that Yap was the more predominantly expressed paralog in the Epi cells, whereas in the PrE lineage, both factors were expressed in a similar manner (Figures 5A and 5B). Because a combined genetic ablation of Yap and Taz results in a failure of TE specification and embryonic lethality at the morula stage,36 we analyzed the effects of individual Yap or Taz loss of function during embryo dormancy.
Figure 5. Yap/Taz function during embryo dormancy.
(A) Yap and Taz expression in the Epi.
(B) Yap and Taz expression in the PrE.
(C) Taz control and KO EDG9.5 embryos stained for Sox2, Sox17, and Cdx2.
(D) Epi cell number in Taz control and KO at EDG9.5. Six independent experiments.
(E) PrE cell number in Taz control and KO at EDG9.5. Six independent experiments.
(F) Taz control and KO EDG11.5 embryos stained for Sox2, Sox17, and Cdx2.
(G) Epi cell number in Taz control and KO at EDG11.5. Four independent experiments.
(H) PrE cell number in Taz control and KO at EDG11.5. Four independent experiments.
(I) Yap control and KO EDG7.5 embryos stained for Yap, Oct4, Sox17, and DAPI.
(J) Epi cell number in Yap control and KO at EDG7.5. Four independent experiments.
(K) PrE cell number in Yap control and KO at EDG7.5. Four independent experiments.
(L) Yap control (+/+) and KO (Δ/Δ) EDG9.5 embryos stained for Yap, Oct4, Sox17, and DAPI.
(M) Epi cell number in Yap control and KO at EDG9.5. Four independent experiments.
(N) PrE cell number in Yap control and KO at EDG9.5. Four independent experiments.
(O) Yap control and KO EDG11.5 embryos stained for Yap, Oct4, Sox17, and DAPI.
(P) Epi cell number in Yap control and KO embryos at EDG11.5. Three independent experiments.
(Q) PrE cell number in Yap control and KO at EDG11.5. Three independent experiments.
(R) Percentage of Yap control and KO at EDG7.5 (four independent experiments), EDG9.5 (four independent experiments), and EDG11.5 (three independent experiments) The number of embryos is indicated on the graph.
(S) Percentage of Yap control and KO after 48 h of E2 administration at EDG7.5 (two independent experiments), EDG9.5 (four independent experiments), or EDG11.5 (four independent experiments). The number of embryos is indicated on the graph.
(T) Yap control and KO embryos isolated after 48 h of E2 administration at EDG7.5 and stained for Yap, Oct4, Sox17, and DAPI.
(U) Yap control and KO embryos isolated after 48 h of E2 administration at EDG9.5 and stained for Yap, Oct4, Sox17, and DAPI.
(V) Epi cell number in Yap control and KO after 48 h of E2 administration at EDG7.5.
(W) Epi cell number in Yap control and KO after 48 h E2 administration at EDG9.5 based on Oct4 expression.
(D, E, G, H, J, K, M, N, P, Q, V, and W) Data represent mean ± SD, n = number of embryos, two-tailed unpaired Student’s t test, and p value is presented on the figures.
Scale bars, 10 μm. See also Figure S4.
It has been reported that Taz KO mice exhibit polycystic kidney disease and pulmonary emphysema.37,38 We examined Taz KO diapause embryos (EDG9.5 and EDG11.5) and found no substantial changes in the organization and the cell number of Epi and PrE lineages (Figures 5C–5H and S4B), suggesting that Yap expression might compensate for the Taz loss of function.
Yap deletion has been associated with defects in yolk sac vasculogenesis, embryonic axis elongation, and chorioallantoic attachment, resulting in developmental arrest at E8.5.39 We analyzed the effects of Yap ablation during the transient blastocyst to egg cylinder transition (E4.5–E5.5) and found that the Yap KO embryos were indistinguishable from the wild-type and heterozygous littermates (Figures S4C and S4D).
Next, we examined the effects of Yap deletion in the ICM during embryo dormancy. Marker gene expression analysis revealed that the PrE lineage in the Yap KO embryos was properly organized as a monolayer covering the pluripotent cells. Moreover, the number of PrE cells was similar to that of the control embryos at EDG7.5, EDG9.5, and EDG11.5 (Figures 5I, 5K, 5L, 5N, 5O, and 5Q). Thus, the loss of Yap did not affect the organization and maintenance of the PrE cells during embryo dormancy. Because both Yap and Taz exhibited similar expression levels (Figure 5B), this could govern the potential functional redundancy of these factors in the PrE.
Similar to in the PrE, the deletion of Yap also did not affect the organization of the Epi, which formed a polarized rosette-like structure, indicating that the loss of Yap is not associated with defects in the tissue-scale organization of the pluripotent lineage (Figures S4E and S4F). However, in comparison with control embryos, Yap mutants exhibited a substantial decrease in the number of Epi cells (Figures 5I, 5J, 5L, 5M, 5O, and 5P). At the same time, Yap KO embryos showed increased levels of cleaved caspase-3-positive apoptotic cells (Figure S4G). Thus, although dispensable for the transient (E4.5–E5.5) embryogenesis, Yap is required for the maintenance of the pluripotent lineage during embryo dormancy. Because Taz is also expressed in the Epi (Figure 5A), albeit at a lower level, one can speculate that Taz may partially compensate for the loss of Yap. Therefore, a putative deletion of both factors in the pluripotent cells may lead to more pronounced effects in the dormant embryo.
The analysis of Yap KO embryos showed that homozygous mutants were still present during diapause (Figure 5R). Interestingly, it has been reported that nuclear Yap is required for the re-entry of quiescent cells into the cell cycle.40 Thus, we examined the localization of Yap in implanted reactivated embryos and found an enrichment of nuclear Yap (Figures 5T and 5U, controls).
To understand whether Yap is involved in the re-entry into a proliferative phase, we administered E2 at EDG7.5, EDG9.5, and EDG11.5 to induce the exit of embryo dormancy, and after 48 h, we analyzed the number and morphology of the implanted embryos. We found that the number of implanted Yap KO embryos gradually decreased as reactivation was initiated from progressively deeper stages of embryo dormancy (Figure 5S). In addition, the number of Epi cells in the implanted Yap KO embryos was reduced compared with the controls (Figures 5T–5W), potentially reflecting the lower starting cell number at EDG7.5 and EDG9.5 (Figures 5J and 5M). Although Yap KO embryos were still present at EDG11.5, we found no mutant embryos following E2 administration (Figures 5R and 5S), indicating that Yap KO embryos reached a putative “point of no return” for efficient re-entry into transient embryogenesis.
Altogether, these analyses show that Yap is required to preserve the developmental capacity of the embryo during diapause. The failure of developmental re-entry from a deep state of dormancy could be a systemic defect because Yap is ubiquitously expressed in all tissues. Results from the Epi lineage show that Yap is required for the maintenance of the pluripotent cells, and this function may be shared to some extent with Taz, which may partially compensate for the Yap deficiency. Because the combined Yap and Taz deletion results in an earlier developmental defect, precluding analysis at the blastocyst stage and beyond,36 in the final set of experiments, we examined Yap/Taz loss of function in vitro in ESCs.
Yap/Taz function in ESCs
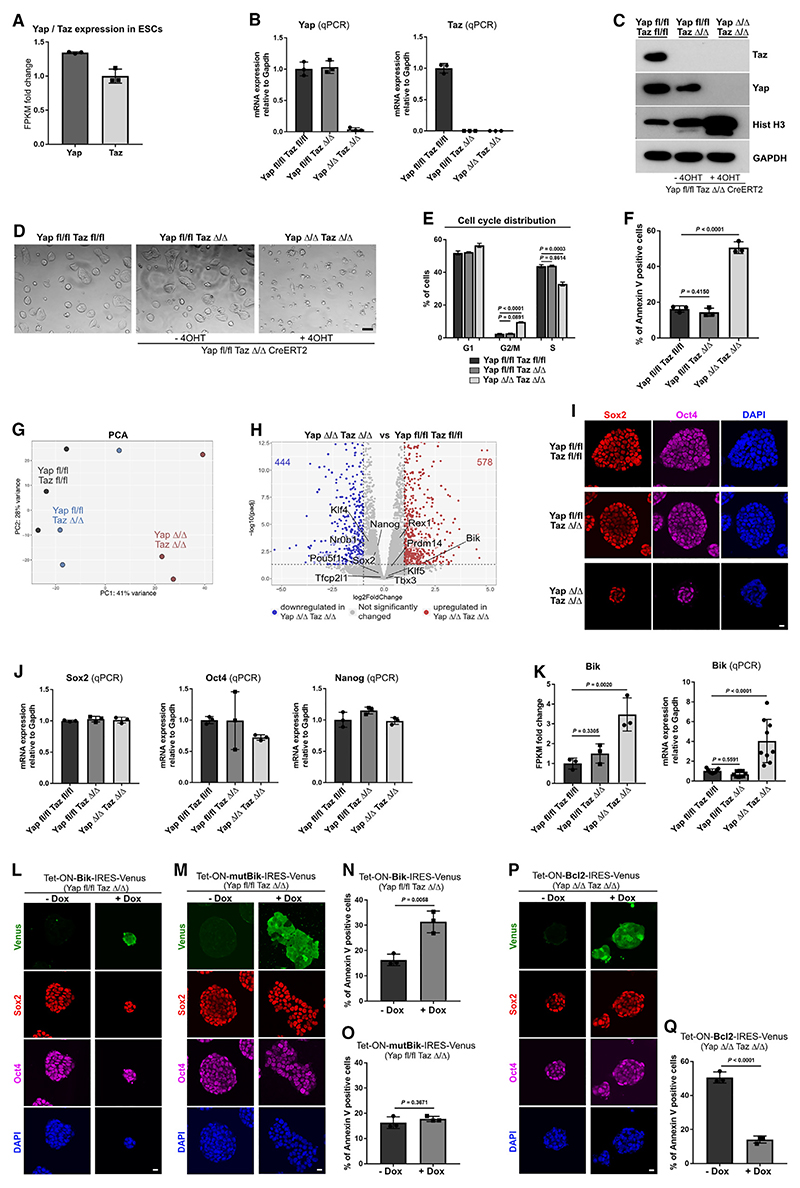
First, we analyzed the expression of Yap and Taz and found transcripts of both paralogs in ESCs (Figure 6A). Moreover, staining for Yap revealed nuclear accumulation in the pluripotent cells (Figure S5A). To decipher the functional significance of Yap and Taz, we established ESCs harboring conditional (floxed) alleles of both factors (Yap fl/fl Taz fl/fl) and ESCs harboring Yap floxed and Taz KO alleles (Yap fl/fl Taz Δ/Δ). The latter were stably transfected with a Cre-ERT2 transgene mediating Yap excision in the presence of 4-OHT. We confirmed the deletion of Yap on mRNA and protein levels after 48 h of treatment with 4-OHT, resulting in the efficient conversion of the Yap fl/fl Taz Δ/Δ into Yap Δ/Δ Taz Δ/Δ cells (Figures 6B and 6C).
Figure 6. Yap/Taz function in ESCs.
(A) Yap and Taz expression in ESCs, based on Fan et al.5 Data represent mean ± SD, n = three technical replicates.
(B) qPCR of Yap and Taz expression relative to GAPDH.
(C) Western blot analysis of Yap and Taz levels at day 2 of culture.
(D) Brightfield images on day 2 of culture.
(E) Cell cycle distribution determined by EdU assay. Data represent mean ± SEM, one-way ANOVA with Tukey’s post hoc test, p value is presented on the figures, n = three independent experiments.
(F) Annexin V assay at day 2 of culture.
(G) Principal component analysis (PCA) of the transcriptomes; n = three independent RNA-seq datasets for each condition.
(H) Volcano plot of gene expression.
(I) Sox2 and Oct4 expression analysis.
(J) qPCR of Sox2, Oct4, and Nanog expression relative to GAPDH.
(K) Gene expression of Bik based on the RNA-seq analysis (left) and qPCR relative to GAPDH (right).
(L) Control and ectopically expressing Bik Yap fl/fl Taz Δ/Δ ESCs at day 2, stained for Venus, Sox2, Oct4, and DAPI.
(M) Control and ectopically expressing mutBik Yap fl/fl Taz Δ/Δ ESCs at day 2, stained for Venus, Sox2, Oct4, and DAPI.
(N) Annexin V assay of control and ectopically expressing Bik Yap fl/fl Taz Δ/Δ ESCs at day 2.
(O) Annexin V assay of control and ectopically expressing mutBik Yap fl/fl Taz Δ/Δ ESCs at day 2.
(P) Control and ectopically expressing Bcl2 Yap Δ/Δ Taz Δ/Δ ESCs at day 2, stained for Venus, Sox2, Oct4, and DAPI.
(Q) Annexin V assay of control and ectopically expressing Bcl2 Yap Δ/Δ Taz Δ/Δ ESCs at day 2.
(B and J) Data represent mean ± SD, n = three independent experiments for each figure; (N, O, and Q) data represent mean ± SD, two-tailed unpaired Student’s t test, n = three independent experiments for each figure, p value is presented on the figures; (F and K) data represent mean ± SD, one-way ANOVA with Tukey’s post hoc test, n = three independent experiments for each figure, p value is presented on the figures.
Scale bars, 10 μm. See also Figures S5 and S6.
Next, we compared the morphology, cell cycle distribution, apoptosis, and expression of pluripotency markers in Yap fl/fl Taz fl/fl, Yap fl/fl Taz Δ/Δ, and the Yap Δ/Δ Taz Δ/Δ cells, which were cultured in the presence of 4-OHT for 48 h to induce Yap depletion. The double floxed and the Taz KO ESCs appeared morphologically indistinguishable, forming typical dome-shaped colonies. Similarly, the double KO ESCs also formed dome-shaped colonies, showing no obvious signs of differentiation. However, the colonies were smaller, and the cells appeared to exhibit an ongoing process of cell death (Figure 6D).
To understand whether proliferation and/or apoptosis were affected by the combined depletion of Yap and Taz, we analyzed the cell cycle distribution and the level of apoptotic cells. The cell cycle analysis revealed that the double KO ESCs did not exhibit a block in a particular phase of the cell cycle progression. However, compared with the double floxed and the Taz KO ESCs, the Yap Δ/Δ Taz Δ/Δ cells showed a reduction in the S phase and enrichment in the G2/M and slight enrichment in the G1 phases (Figures 6E and S5B). Thus, Yap/Taz may contribute to the cell cycle progression to the S phase, thereby promoting ESC proliferation.
To examine the level of apoptosis, we performed an annexin V assay and analyzed the signal via flow cytometry. We found no significant difference in the proportion of apoptotic cells comparing the double floxed vs. Taz KO ESCs. By contrast, there was a substantial increase in the annexin V-positive cells in the double KO (Figures 6F and S5C), indicating that Yap/Taz play a critical role in the survival and maintenance of the pluripotent cells.
Next, we performed RNA-seq analysis to get insight into the transcriptional response to Yap/Taz depletion. Principal component analysis (PCA) showed that the double floxed and Taz KO cells clustered closely together in relation to the double KO cells (Figure 6G). Accordingly, the number of deregulated genes in Taz KO vs. double-floxed cells was substantially lower in comparison with the Yap Δ/Δ Taz Δ/Δ vs. double-floxed ESCs (Figures 6H and S5D; Table S3).
Focusing first on the pluripotency markers, we found no significant changes in the expression of the core (Oct4 and Sox2) and the ancillary pluripotency transcription factors, such as Nanog, Rex1, Klf4, Nr0b1, Tfcp2l1, Tbx3, and Prdm14, in Taz KO or double KO cells in comparison with the double-floxed ESCs (Figures 6H and S5D). We confirmed the expression of Oct4 and Sox2 on the protein level and verified the overall similar transcript levels of Oct4, Sox2, and Nanog using quantitative PCR (qPCR) (Figures 6I and 6J).
Although the combined Yap/Taz deletion did not affect the pluripotency network, among the significantly upregulated genes, we found the Bcl-2-interacting killer (Bik) (Figures 6H and 6K; Table S3). Bik contains a Bcl-2 homology 3 (BH3) domain and belongs to the BH3-only group of pro-apoptotic factors. This factor triggers apoptosis through the mitochondrial pathway via the mobilization of Ca2+ from the endoplasmic reticulum and through the remodeling of the mitochondrial cristae.41
Although the loss of Yap and Taz most likely leads to a systemic failure of ESC survival, Bik could be one of the effectors contributing to the apoptotic response. To get insight into the effects of Bik, we stably transfected Yap fl/fl Taz Δ/Δ and Yap fl/fl Taz fl/fl ESCs with a Bik transgene followed by IRES-Venus sequence altogether under the control of doxycycline (Dox)-inducible Tet-ON promoter (Tet-ON-Bik-IRES-Venus). After 48 h of culture, the control ESCs (–Dox) and cells ectopically expressing Bik (+Dox) were analyzed for their annexin V signal. In accord with the pro-apoptotic functions of Bik, we observed elevated apoptosis in the Dox-treated Yap fl/fl Taz Δ/Δ and double-floxed cells. Importantly, this did not affect the expression of pluripotency factors (Figures 6L, 6N, and S6A–S6D), overall resembling the phenotype of the double KO ESCs (Figure 6I).
A critical role in Bik’s pro-apoptotic function is played by the BH3 domain, which enables interactions with other Bcl-2 family proteins.41,42 Thus, as an additional control, we generated a Yap fl/fl Taz Δ/Δ and Yap fl/fl Taz fl/fl ESC line that ectopically expresses a Dox-inducible Bik transgene harboring a mutated BH3 domain41 that disrupts protein binding (Tet-ON-mutBik-IRES-Venus). Accordingly, we found no substantial difference in the level of apoptosis in the control (–Dox) and Dox-treated Yap fl/fl Taz Δ/Δ and Yap fl/fl Taz fl/fl ESCs after 48 h of culture (Figures 6M, 6O, and S6E–S6H).
Finally, to understand whether countering the activation of the mitochondrial apoptotic pathway can improve the viability of the double KO cells, we ectopically expressed the anti-apoptotic factor Bcl-2. We generated Yap fl/fl Taz fl/fl and Yap Δ/Δ Taz Δ/Δ cells harboring a Dox-inducible Blc-2 transgene (Tet-ON-Bcl2-IRES-Venus) and examined the effects of Bcl-2 expression after 48 h of culture in the presence of Dox. In contrast to the control (–Dox) Yap Δ/Δ Taz Δ/Δ ESCs, which exhibited a high level of apoptosis, the percentage of annexin V-positive cells was substantially decreased only in the Dox-treated double KO ESCs, indicating partial rescue of the phenotype (Figures 6P, 6Q, S6I, and S6J).
Altogether, these analyses show that the loss of Yap/Taz function does not affect the undifferentiated status of the ESCs, but these factors play a pivotal role in the maintenance of the pluripotent cells by keeping the process of apoptosis at bay. During diapause, our results indicate that the cell signaling and transcriptional landscape define a dynamic state, sustaining the pluripotent cells of the Epi. In these physiological conditions, Itgb1-mediated activation of Yap safeguards cell viability and governs the developmental potential of the dormant embryo to re-enter transient embryogenesis.
Discussion
The E4.5 blastocyst is competent for commencing implantation or diapause, and the resolution to follow one of these routes is determined as a physiological response to the maternal environment. In a receptive uterus, the blastocyst initiates implantation to continue its transient embryogenesis, whereas in a non-receptive environment, the embryo enters a reversible state of dormancy. Because development arrests at the blastocyst stage, the embryo morphologically appears in a state of suspended animation. Concurrently, our scRNA-seq analysis revealed underlying transcriptional dynamics associated with the entry, progression, and exit of diapause.
The transcriptional analysis also showed that during diapause, both the embryonic and the extraembryonic tissues halted their differentiation programs, preserving their cell identity and stem cell properties. This has been implemented as an experimental approach to increase the efficiency of ESC derivation.22,23 Homogeneous ESC lines uniformly expressing pluripotency makers, such as Nanog and Esrrb, can be established and maintained in a medium supplemented with Mek and Gsk3 inhibitors and Lif (2i/Lif). Interestingly, ESCs with reduced expression of Oct4 (Oct4 +/Δ) exhibit a homogeneous expression of naive pluripotency transcription factors in the absence of pharmacological inhibitors. These cells upregulate the expression of Wnt ligands in an autocrine manner and show enhanced sensitivity to Lif/Stat3 stimulation.30 Accordingly, it has been previously reported that the canonical Wnt pathway activity upregulates Stat3 expression in ESCs.43
Interestingly, Oct4 expression was also reduced during diapause, while at the same time, the pluripotency transcription factor network was stabilized and generally upregulated. In line with the increased Wnt expression in Oct4 +/Δ ESCs, we have previously shown that the canonical Wnt signaling is activated during diapause in an autocrine manner.5 This pathway may promote Lif responsiveness, acting synergistically with the Gp130/Stat3 signaling. Accordingly, on a transcriptional level, our data showed an upregulation of Stat3 expression, whereas on the protein level, a substantial increase of Stat3 phosphorylation during diapause has been recently reported.44 Thus, the pathways modulated in vitro in 2i/Lif culture conditions may reflect, to some extent, the signaling activity in the dormant Epi.
Essentially, although both Wnt/β-catenin and Gp130 are dispensable for transient blastocyst development, their function is critical for maintaining the pluripotent lineage during embryo dormancy.5,28 In addition, we also observed increased expression of Fgf4 in the Epi similar to the Oct4 +/Δ ESCs in vitro. Because Fgf4 signaling has been shown to contribute to trophoblast and PrE survival,45,46 it is possible that the Fgf4 ligands produced by the pluripotent lineage may act in a paracrine manner in the extraembryonic tissues. Altogether, this indicates that the Oct4 reduction during diapause could be the physiological counterpart of the more stable pluripotent state observed in the Oct4 heterozygous ESCs.
The extraembryonic tissues may also provide cues supporting Epi homeostasis during diapause, thereby acting as a niche for the pluripotent cells. Our analyses indicate that a key component of this microenvironment is the ECM, which surrounds the Epi. We found that Itgb1 is the major receptor that mediates the interactions between the ECM and the Epi or PrE lineages, and deletion of Itgb1 resulted in a near-complete loss of the pluripotent cells and substantial reduction of the PrE. At the same time, Itgb1 deletion in the TE might be compensated by other members of the integrin family, as we have previously shown that the trophoblast co-expresses Itgb1, Itgb3, and Itgb5.8
The integrin receptors can read the rigidity and topology of the ECM and transmit the mechanical cues affecting actomyosin contractility and intracellular tension. The actin cytoskeleton integrates and converts these physical signals into a dynamic modulation of the Yap/Taz activity.34 We have also previously shown that Itgb1-ECM interactions are required for the establishment of epithelial polarity in the pluripotent lineage.35 In the context of pre-implantation embryogenesis, the formation of an apical domain in the TE leads to the inactivation of the Hippo pathway and nuclear translocation of Yap/Taz.36 Similarly, the establishment of a polarized Epi rosette may also contribute to Yap/Taz activation, promoting cell survival during embryo dormancy. Thus, multiple positive feedback mechanisms may reinforce Yap/Taz activation in the ICM, contributing to the gradual nuclear accumulation of these factors during embryo dormancy.
Deletion of Yap did not affect the normal E4.5 to E5.5 transition, whereas the dormant Yap KO embryos exhibited a substantial decrease in the number of Epi cells. This phenotype was less severe than upon the deletion of Itgb1, potentially due to the compensatory role of Taz. Accordingly, the combined ablation of both paralogs in ESCs resulted in a strong apoptotic response, indicating an essential role of Yap/Taz in the survival and self-renewal of the pluripotent cells. It has been shown that in the context of neoplasia, Yap/Taz activity also suppresses apoptosis and promotes tumor survival, which contributes to the chemoresistance of cancer stem cells.47 Moreover, Yap suppresses apoptosis in cancer cells that enter a senescence-like dormant state.48 Thus, similar to the epithelial-to-mesenchymal transition (EMT), which is a process observed in both development and cancer, one can speculate that cell dormancy may have physiological (embryonic diapause) and pathological (dormant cancer cells) manifestations with some shared basic principles.
Altogether, our study elucidates the dynamic transcriptional landscape and reveals the critical role of integrin/Yap signaling during embryo dormancy. This work can serve as a foundation to uncover other concealed regulatory mechanisms in the embryo and delineate parallels between embryonic diapause and the dormant state of particular cell types in the body.
Limitations of the study
The scRNA-seq analysis revealed transcriptional dynamics associated with the entry, progression, and exit of diapause. The majority of the active genes were downregulated during embryo dormancy, whereas other sets of genes were upregulated. The mechanisms governing this response require further investigation using complementary methods with single-cell resolution. The main limitation in such analyses is the collection of sufficient numbers of embryos under physiological conditions of dormancy and reactivation, particularly mutant embryos that arise following the Mendelian ratio. In addition, how Oct4 levels are fine-tuned during diapause, the role of other signaling cascades and factors in parallel to integrin-Yap/Taz, and the overarching impact of the uterine environment remain to be elucidated.
STAR ⋆ METHODS
Detailed methods are provided in the online version of this paper and include the following:
-
-
○
Lead contact
-
○
Materials availability
-
○
Data and code availability
-
○
-
EXPERIMENTAL MODEL AND STUDY PARTICIPANT DETAILS
-
○
Mice
-
○
Cell lines
-
○
-
-
○
Morula aggregation and embryo transfer
-
○
Induction of entry to and exit of diapause B Plasmids
-
○
Immunofluorescent staining
-
○
Whole-mount immunofluorescent staining
-
○
Tissue clearing
-
○
DNA isolation and PCR genotyping
-
○
RNA extraction and quantitative PCR analysis
-
○
Analysis of EU incorporation
-
○
Flow cytometry
-
○
Western blot
-
○
scRNA-seq and bioinformatics analysis
-
○
Bulk RNA-seq and bioinformatics analysis
-
○
Confocal microscopy and image analysis
-
○
Star ⋆ Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-Cdx1 | Novus Biologicals | Cat# NBP1-49538, RRID: AB_10011593 |
| Rabbit polyclonal anti-Aqp3 | Novus Biologicals | Cat# NBP2-33872, RRID: AB_2940888 |
| Mouse monoclonal anti-Esrrb | R&D systems | Cat# PPH6705-00, RRID: AB_2100412 |
| Mouse monoclonal anti-AP-2 gamma (6E4/4) | Santa Cruz | Cat# sc-12762, RRID: AB_667770 |
| Rabbit polyclonal anti-Eomes | Abcam | Cat# ab23345, RRID: AB_778267 |
| Mouse monoclonal anti-Cdx2 | Biogenex | Cat# MU392A-5UC, RRID: AB_2923402 |
| Rabbit monoclonal anti-Sox2 (D9B8N) | Cell signalling | Cat# 23064, RRID: AB_2714146 |
| Rat monoclonal anti-Cer1 | R&D systems | Cat# MAB1986, RRID: AB_2275974 |
| Goat polyclonal anti-Otx2 | R&D systems | Cat#AF1979, RRID: AB_2157172 |
| Goat polyclonal anti-Sox17 | R&D systems | Cat# AF1924, RRID: AB_355060 |
| Mouse monoclonal anti-Ecadherin, Alexa 647 conjugated | BD Biosciences | Cat# 560062, RRID: AB_1645407 |
| Rabbit polyclonal anti-Gata4 | Santa Cruz | Cat# sc-9053, RRID: AB_2247396 |
| Rabbit polyclonal anti-Nanog | Abcam | Cat# ab80892, RRID: AB_2150114 |
| Mouse monoclonal anti-Oct4A (D6C8T) | Cell signalling | Cat# 83932, RRID: AB_2721046 |
| Rabbit monoclonal anti-Yap (D8H1X) | Cell signalling | Cat# 14074, RRID: AB_2650491 |
| Rabbit monoclonal anti-Taz (E8E9G) | Cell signalling | Cat# 83669, RRID: AB_2800026 |
| Goat polyclonal anti-GFP | R&D systems | Cat# AF4240, RRID: AB_884445 |
| Rabbit polyclonal anti-RFP | Biomol | Cat# 600-401-379, RRID: AB_2209751 |
| Mouse monoclonal anti- PARD6B (B-10) | Santa Cruz | Cat# sc-166405, RRID: AB_2267890 |
| Rabbit monoclonal anti-Histone H3 (D1H2) | Cell Signaling | Cat# 4499, RRID: AB_10544537 |
| Rabbit monoclonal anti-GAPDH (D16H11) | Cell Signaling | Cat# 5174, RRID: AB_10622025 |
| Mouse polyclonal anti-Cdcp1 | R&D systems | Cat# AF4515-SP, RRID: AB_2078800 |
| Mouse polyclonal anti-Dll4 | R&D systems | Cat# AF1389, RRID: AB_354770 |
| Hamster monoclonal anti-CD29-FITC | BD Biosciences | Cat# 561796, RRID: AB_10894590 |
| Rat monoclonal anti-9EG7 (active beta1 integrin) | Home-made (kind gift from Prof.Dietmar Vestweber) | N/A |
| Rat anti-Troma1 | Home-made (kind gift from Prof. Rolf Kemler) |
N/A |
| Rabbit polyclonal anti-Laminin | Atlas Antibodies | Cat# L9393, RRID: AB_477163 |
| Rabbit monoclonal anti Cleaved Caspase-3 (5A1E) | Cell Signaling | Cat# 9664S, RRID: AB_2070042 |
| Alexa Flour 594 Donkey Anti-Rabbit IgG (H+L) | Thermo Fisher Scientific | Cat# A-21207, RRID: AB_141637 |
| Alexa Flour 647 Donkey Anti-Mouse IgG (H+L) | Thermo Fisher Scientific | Cat# A-31571, RRID: AB_162542 |
| Alexa Flour 488 Goat Anti-Rat IgG (H+L) | Thermo Fisher Scientific | Cat# A-21208, RRID: AB_2535794 |
| Alexa Flour 647 Donkey Anti-Goat IgG (H+L) | Thermo Fisher Scientific | Cat# A-21447, RRID: AB_2535864 |
| Alexa Flour 488 Donkey Anti-Goat IgG (H+L) | Thermo Fisher Scientific | Cat# A-11055, RRID: AB_2534102 |
| Alexa Flour 488 Donkey Anti-Mouse IgG (H+L) | Thermo Fisher Scientific | Cat# A-21202, RRID: AB_141607 |
| Chemicals, peptides, and recombinant proteins | ||
| DAPI | Carl Roth | Cat# 6335.1 |
| Fetal Bovine Serum | Biochrome | Cat# S0615 |
| DMEM high glucose | Sigma-Aldrich | Cat# D5671 |
| DMEM/F12 | Thermo Fisher Scientific | Cat# 21331046 |
| Neurobasal medium | Thermo Fisher Scientific | Cat# 21103049 |
| L-Glutamine | Sigma-Aldrich | Cat# G7513 |
| Sodium pyruvate | Sigma-Aldrich | Cat# S8636 |
| Non-essential amino acids | Sigma-Aldrich | Cat# M7145 |
| Penicillin-streptomycin | Sigma-Aldrich | Cat# P4333 |
| N2 | Thermo Fisher Scientific | Cat# 17502048 |
| B27 without vitamin A | Thermo Fisher Scientific | Cat# 12587001 |
| Trypsin-EDTA (0.25%) | Sigma-Aldrich | Cat# 25200056 |
| 2-mercaptoethanol | Sigma-Aldrich | Cat# M7522 |
| Formaldehyde (w/v), methanol-free 16% | Thermo Fisher | Cat# 28908 |
| Triton X-100 | Sigma-Aldrich | Cat# T9284 |
| PBS without Ca/Mg | Sigma-Aldrich | Cat# D8537 |
| iTag SYBR green supermix | Bio-Rad | Cat# 1725120 |
| GoTaq green master mix | Promega | Cat# M7123 |
| Precision Plus Protein Kaleidoscope protein standard | Bio-Rad | Cat# 161-0375 |
| PD0325901 | Cayman Chemical | Cat# 13034 |
| CHIR99021 | Tocris | Cat# 4423 |
| human Lif | homemade | A kind gift from Prof.Hans R. Schöler, Max Planck Institute for Molecular Biomedicine, Münster, Germany |
| Lipophectamine 2000 | Thermo Fisher Scientific | Cat# 11668027 |
| Doxycyclin | Sigma-Aldrich | Cat# D9891 |
| 4-hydroxytamoxifen (4-OHT) | Sigma-Aldrich | Cat# H7904 |
| Neomycin | Sigma-Aldrich | Cat# G8168 |
| Gelatin solution | Sigma-Aldrich | Cat# 1393 |
| ECL Western Blotting Detection Reagent | GE Healthcare | Cat# RPN2109 |
| ECL Prime Western Blotting Detection Reagent | GE Healthcare | Cat# RPN2232 |
| PVDF membrane | Millipore | Cat# IPVH00010 |
| Mineral oil, embryo-tested | Sigma-Aldrich | Cat# M5310 |
| PMSF | Sigma-Aldrich | Cat# P7626 |
| Donkey serum | VMR | Cat# S2170 |
| PMSG | Ceva | N/A |
| HCG | Ceva | N/A |
| Depo Clinovir | Pfizer | N/A |
| β-estradiol | Sigma-Aldrich | E8875 |
| M2 medium | Sigma-Aldrich | Cat# M7167 |
| KSOM | Millipore | Cat# MR-020P-5F |
| Tamoxifen | Sigma-Aldrich | Cat# T5648 |
| DMSO | Carl Roth | Cat# A994.1 |
| Tween 20 | Sigma-Aldrich | Cat# P7949 |
| Corn oil | Sigma-Aldrich | Cat# C8267 |
| Tyrode’s solution, Acidic | Sigma-Aldrich | Cat# T1788 |
| TrypLE Express Enzyme | Thermo Fisher Scientific | Cat# 12604013 |
| Methanol anhydrous 99.8% | Sigma-Aldrich | Cat# 322415-100ML |
| Benzyl Alcohol | Sigma-Aldrich | Cat# 24122-2.5L-M |
| Benzyl Benzoate 99+% | Thermo Fisher Scientific | Cat# 105860010 |
| Heparin sodium salt | Sigma-Aldrich | Cat# 3149 |
| Glycine | Sigma-Aldrich | Cat#G7126 |
| Sodium deoxycholate | Sigma-Aldrich | Cat# 30970 |
| Critical commercial assays | ||
| In-Fusion HD cloning kit | Clontech | Cat# 638909 |
| μ-Slide 8 Well | Ibidi | Cat# 80826 |
| RNeasy Mini Kit | Qiagen | Cat# 74106 |
| BCA Protein Assay Kit | Thermo Fisher Scientific | Cat# 23227 |
| Click-iT RNA Alexa Fluor 488 HCS Assay kit | Thermo Fisher Scientific | Cat# C10327 |
| Annexin V, Alexa Fluor 568 conjugate | Thermo Fisher Scientific | Cat# A13202 |
| Click-iT Plus EdU Alexa Fluor 647 Flow Cytometry Assay Kit | Thermo Fisher Scientific | Cat# C10634 |
| NEBNext Poly(A) mRNA Magnetic Isolation Module | New England Biolabs | Cat# E7490 |
| NEBNext Ultr II Directional RNA Library Prep with Sample Purification Beads | New England Biolabs | Cat# E7765 |
| NEBNext Multiplex Oligos for Illumina (96 Unique dual index primer pairs) | New England Biolabs | Cat# E6440 |
| BD Rhapsody WTA Reagent Kit | BD Biosciences | Cat# 633801 |
| BD Rhapsody Cartridge Kit | BD Biosciences | Cat# 633733 |
| BD Rhapsody cDNA Kit | BD Biosciences | Cat# 633773 |
| BD Rhapsody Cartridge Reagent Kit | BD Biosciences | Cat# 633731 |
| NextSeq 500/550 High Output Kit v2.5 (75 Cycles) | Illumina | Cat# 20024906 |
| NextSeq 500/550 High Output Kit v2.5 (150 cycles) | Illumina | Cat# 20024907 |
| NextSeq 500/550 Mid Output Kit (150cycles) | Illumina | Cat# 20024904 |
| Deposited data | ||
| bulk RNA-sequencing data | This paper | GEO: GSE241461 |
| scRNA-seq of EDG7.5, EDG9.5, Reactivated and E5.5 embryos | This paper | GEO: GSE241462 |
| E4.5 scRNA-seq dataset | Sathyanarayanan et al.6 | GEO: GSE159883 |
| Experimental models: Cell lines | ||
| Mouse: ESC_ Yap fl/fl Taz fl/fl | This paper | N/A |
| Mouse: ESC_ Yap fl/fl Taz fl/fl Tet-ON-Bik-IRES-Venus | This paper | N/A |
| Mouse: ESC_ Yap fl/fl Taz fl/fl Tet-ON-mutBik-IRES-Venus | This paper | N/A |
| Mouse: ESC_ Yap fl/fl Taz fl/fl Tet-ON-Bcl2-IRES-Venus | This paper | N/A |
| Mouse: ESC_ Yap fl/fl Taz Δ/Δ CreERT2 | This paper | N/A |
| Mouse: ESC_ Yap fl/fl Taz Δ/Δ CreERT2 Tet-ON-Bik-IRES-Venus | This paper | N/A |
| Mouse: ESC_ Yap fl/fl Taz Δ/Δ CreERT2 Tet-ON-mutBik-IRES-Venus | This paper | N/A |
| Mouse: ESC_ Yap fl/fl Taz Δ/Δ CreERT2 Tet-ON-Bcl2-IRES-Venus |
This paper | N/A |
| Mouse: ESC_ Itgb1 fl/fl CreERT2 | Kim et al.35 | N/A |
| Mouse: ESC_ Itgb1 fl/fl CreERT2 CAG-HA-IRES-Venus | This paper | N/A |
| Mouse: ESC_ Itgb1 fl/fl CreERT2 CAG-Yap-S112A-IRES-Venus |
This paper | N/A |
| Experimental models: Organisms/strains | ||
| Mouse: WT C57BL/6 | Bred in house | N/A |
| Mouse: WT CD1 | Bred in house | N/A |
| Mouse: WT B6C3F1 | Bred in house | N/A |
| Mouse: Zp3-Cre | de Vries et al.49 | JAX:003651, C57BL/ 6-Tg(Zp3-Cre)93Knw/J |
| Mouse: mT/mG | Muzumdar et al.50 | JAX:007576, Gt(ROSA)26Sortm4(ACTB-tdTomato,-EGFP)Luo/J |
| Mouse: Yapfl/fl/Tazfl/fl | Reginensi et al.51 | JAX:030532, C57BL/6-Wwtr1tm1Hmc Yap1tm1Hmc/WranJ |
| Mouse: Itgb1fl/fl | Raghavan et al.52 | JAX:004605, B6;129-Itgb1tm1Efu/J |
| Oligonucleotides | ||
| see Table S4 | N/A | |
| Recombinant DNA | ||
| PyCAG-PBase | Wang et al.53 | N/A |
| pPB-CAG-rtTAM2-IN | Adachi et al.54 | N/A |
| pPB-CAG-CreERT2-IRES-Neo | Adachi et al.54 | N/A |
| pPB-hCMV1-Bik-IRES-Venus | This paper | N/A |
| pPB-hCMV1-mutBik-IRES-Venus | This paper | N/A |
| pPB-hCMV1-Bcl2-IRES-Venus | This paper | N/A |
| pPB-CAG-HA-IRES-Venus | This paper | N/A |
| pPB-CAG-mYap mut aa112 S to A-IRES-Venus | This paper | N/A |
| pRA1473-CTV-YAP1mut (aa112 S to A) | Sivaraj et al.55 | N/A |
| pEGFP-Bik | Kagawa et al.56 | Addgene # 10952 |
| Venus-Bik-4E-pEGFP-C1 | Osterlund et al.42 | Addgene # 166744 |
| tetO-Bcl2-IRES-tdtomato (TET005) | Wu et al.57 | Addgene # 117857 |
| Software and algorithms | ||
| Snap Gene | GSL Biotech | RRID: SCR_015052 |
| Graphpad Prism | GraphPad | RRID: SCR_002798 |
| Fiji | Schindelin et al.58 | RRID: SCR_002285 |
| FlowJo | FlowJo | RRID: SCR_008520 |
| Seurat | Stuart et al.59 | RRID:SCR_016341 |
| DESeq2 | Liao et al.60 | RRID:SCR_015687 |
| UMI-tools | Smith et al.61 | RRID:SCR_017048 |
| STAR | Dobin et al.62 | RRID:SCR_004463 |
| Subread | Liao et al.63 | RRID:SCR_009803 |
| WGCNA | Langfelder and Horvath15 | RRID:SCR_003302 |
| TopHat2 | Kim et al.64 | RRID:SCR_013035 |
| HTSeq-count | Putri et al.65 | RRID:SCR_011867 |
Resource Availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Ivan Bedzhov (ivan.bedzhov@mpi-muenster.mpg.de).
Materials availability
This study did not generate new unique reagents.
Experimental Model and Study Participant Details
Mice
Animal experiments and husbandry were performed according to the German Animal Welfare guidelines and approved by the Landesamt für Natur, Umwelt und Verbraucherschutz Nordrhein-Westfalen (State Agency for Nature, Environment and Consumer Protection of North Rhine-Westphalia). The mice used in this study were at age from 6 weeks to 5 months. The animals were maintained under a 14-hour light/10-hour dark cycle with free access to food and water. Male mice were kept individually, whereas the female mice were housed in groups of up to four per cage. Embryos for experiments were obtained from wild-type and transgenic strains from matings using females with natural ovulation cycles or after superovulation. Red fluorescent mT/mG embryos were used for whole-mount tissue clearing and staining experiments at E5.5.50 Heterozygous mouse lines were generated from Itgb1fl/fl 52 and Yapfl/fl/Tazfl/fl 51 conditional knockout strains by crossings with Zp3-Cre49 mice. F1 generation of female mice with genotypes: Itgb1fl/+ Zp3-Cre or Yapfl/+/Tazfl/+ Zp3-Cre were crossed with C57Bl/6 males to generate Itgb1+/−, Yap+/-/Taz+/+ or Yap+/+/Taz+/- strains. Knockout embryos for experiments were derived from heterozygous intercrosses and compared to wild-type and heterozygous littermates.
Cell lines
The ESC lines were maintained in DMEM medium supplemented with 15% FBS, 2 mM l-glutamine, 1 mM sodium pyruvate, 0.1 mM non-essential amino acids, 50 U/ml penicillin-streptomycin, 0.1 mM 2-mercaptoethanol, 0.4 μM PD0325901 (Cayman Chemical, 13034), 3 μM CHIR99021 (Tocris, 4423) and 4 ng/ml Lif (prepared in house), on plastic dishes coated with 0.2% gelatin. The cells were grown at 37°C, 5% CO2 atmosphere in air, and passaged using 0.05% trypsin-EDTA. The ESCs were cultured in serum-free N2B27 medium (DMEM/F12 and Neurobasal medium mixed at a ratio of 1:1, 0.5% (v/v) N2, 0.5% (v/v) B27 without vitamin A, 2 μM L-glutamine, 50 U/ml penicillin-streptomycin, and 0.05 μM 2-mercaptoethanol) supplemented with 2i / Lif and then used for bulk RNA -seq. Yap/Taz double floxed ES cells were derived from Yap/Taz double floxed mouse strain.51 Briefly, E3.5 embryos were plated into individual wells of a 96-well plate containing inactivated mouse embryonic fibroblasts (MEFs) and cultured in ESC medium. After 4 days, the blastocyst outgrowths were trypsinised, transferred into fresh wells and the emerging colonies were further expanded. Cell lines stably expressing transgenes were generated using the PiggyBac (PB) transposon system.53 The transposase encoding plasmid (pIB1-PyCAG-PBase53) and PB transposon vector containing Cre-ERT2 (pIB58-PB-CAG-CreERT2-IRES-Neo54) were co-transfected in Yap/Taz double floxed ESCs. After 24 h, G418 (300 μg/ml) was added to the culture medium for 5 days to select stably transfected clones, which were then analysed by PCR and Western blot. Cre/loxP recombination was induced using 500 nM 4-OHT. The stable cell lines for inducible (Tet-ON) transgene expression were also generated using the PB transposon system. The transgenes were cloned into pIB12-PB-hCMV1-cHA-IRES-Venus plasmid that contains tetracycline response elements and co-transfected with transposase encoding plasmid (pIB1-PyCAG-PBase) and tetracycline-controlled transactivator (pIB11-PB-CAG-rtTAM2-IRES-Neo) plasmids using Lipophectamine 2000 (Invitrogen, 11668027) based on the manufacturer’s instructions, followed by selection with G418 (300 μg/ml). Tet-ON transgene expression was induced using 1 μg/ml of Dox. Stable cell lines for constitutive transgene expression were also generated using the PB transposon system. The transgenes were cloned into PB transposon vector (pIB235-PB-CAG-IRES-Venus) and co-transfected with a transposase-encoding plasmid (pIB1-PyCAG-PBase) in Itgb1 floxed CreERT2 ESCs using Lipophectamine 2000. After 24 h of transfection, G418 (300 mg/ml) was included in the culture medium for 7 days, and then the cells were sorted for Venus expression by flow cytometry.
Method Details
Morula aggregation and embryo transfer
E2.5 embryos were collected from the oviducts of superovulated B6C3F1 female mice by flushing with M2 medium. The Zona pellucida was removed by treatment with acidic Tyrode’s solution (Sigma-Aldrich). Each zona-free embryo was aggregated with ESCs in a depression well and cultured overnight in a droplet of KSOM medium under mineral oil at 37°C in 5% CO2. On the next morning, the embryos were washed several times and collected in M2 medium for embryo transfer.
Embryos were transferred into the uteri of pseudopregnant CD1 females that had been previously mated with vasectomised males. At the same time, the ovaries of the recipient females were removed to induce diapause. Starting the next morning, 3 mg of medroxyprogesterone 17-acetate was injected daily to maintain pregnancy. Embryos were collected at EDG7.5 for analysis.
Induction of entry to and exit of diapause
Surgical diapause was induced via ovariectomy of female mice on the morning of day 3.5 days post coitum (d.p.c). The pregnancy during diapause was maintained by a daily injection of 3 mg medroxyprogesterone 17-acetate (Depo-Clinovir). Non-surgical diapause was induced via injection of 10 μg tamoxifen (intraperitoneally) and 3 mg medroxyprogesterone 17-acetate (subcutaneously) at 1.5 and 2.5 d.p.c., followed by a daily injection of 3 mg medroxyprogesterone 17-acetate to maintain pregnancy. Dormant blastocysts were collected by uterine flushing with M2 medium at EDG5.5, EDG7.5, EDG9.5 or EDG11.5, respectively. The embryos were immediately processed for sequencing (EDG7.5, EDG9.5) or fixed and used for fluorescent staining. To induce the exit of diapause, ovariectomised pregnant mice were injected with 25 ng E2 and sacrificed at indicated time points.
Plasmids
All plasmids were generated using the In-Fusion HD Cloning Plus kit (Takara) based on the manufacturer’s instructions. pPB-hCMV1-Bik-IRES-Venus, pPB-hCMV1-mutBik-IRES-Venus and pPB-hCMV1-Bcl2-IRES-Venus were generated by inserting the Bik, mutBik or Bcl2, respectively, from the pEGFP-Bik56 (Addgene plasmid # 10952), Venus-Bik-4E-pEGFP-C142 (Addgene plasmid # 166744) or tetO-Bcl2-IRES-tdtomato (TET005)57 (Addgene plasmid # 117857) into PB transposon vector (pIB12 PB-hCMV1-cHA-IRES-Venus). pPB-CAG-Yap-S112A-IRES-Venus was generated by amplifying the Yap S112A coding sequence from pRA1473-CTV-YAP1mut (aa112 S to A) and the resulting amplicon was cloned into pIB235-PB-CAG-IRES-Venus.
Immunofluorescent staining
Preimplantation E4.5, diapause and Reactivated embryos were isolated by flushing uteri using M2 medium. Immediately after isolation, the embryos were fixed using 4% PFA for 10 min and then washed twice with a wash solution of 1% FCS in PBS. The fixed samples were permeabilised in buffer containing 0.1 M glycine, and 0.3% Triton X-100 in PBS for 5-10 min and then washed twice. Primary antibodies were applied in a blocking buffer of 2% FCS in PBS and incubated overnight at 4°C. The next day, the samples were washed twice with the washing solution and incubated overnight at 4°C with secondary antibodies and DAPI diluted in the blocking buffer. After three washes, the embryos were mounted on a glass bottom plate, in drops containing 1% FCS/PBS under mineral oil for imaging.
Post-implantation E5.5 embryos were dissected from the maternal tissues in M2 medium and the Reichert’s membrane was removed. The immunostaining protocol was applied as described above with modified incubation times for fixation (15 min) and permeabilization (10 -15 min).
Cells were fixed using 4% PFA for 20 min and after that washed twice in PBS. The fixed cells were permeabilised using 0.3% Triton X-100 in PBS for 5 min and then washed twice. The primary antibodies were applied in PBS and incubated overnight at 4°C. On the next day, the samples were washed twice in PBS and incubated overnight at 4°C with secondary antibodies and DAPI diluted in PBS. After three washes in PBS, the ESCs were directly imaged in the ibidi μ-plates used for cell culture. The antibodies used in this study are listed in the key resources table.
Whole-mount immunofluorescent staining
E5.5 deciduae were isolated from wild-type B6C3F1 females mated with mT/mG stud males. The samples were fixed with 4% PFA overnight at 4°C. The next day, the samples were washed twice with PBS for 1 h at room temperature followed by a wash with 0.2% Triton X-100 in PBS for 1 h at room temperature. The samples were incubated in Buffer-1 containing 0.2% Triton X-100 and 20% DMSO in PBS at 37°C overnight, followed by incubation in Buffer-2 containing 0.1% Tween-20, 0.1% Triton X-100, 0.1% deoxycholate, 0.1% NP40 and 20% DMSO in PBS at 37°C overnight. The samples were then permeabilised using a permeabilization buffer containing 0.2% Triton X-100, 0.3 M glycine and 20% DMSO in PBS overnight at 37°C. After that, the samples were blocked using a blocking solution containing 0.2% Triton X-100, 10% DMSO and 6% donkey serum in PBS overnight at 37°C. On the next day, the samples were transferred into primary antibodies solution, in buffer containing 5% DMSO and 3% donkey serum in PTwH buffer (0.2% Tween-20 and 10 mg/ml heparin in PBS) and incubated for 72 h at 37°C. The samples were washed with 0.1% Tween-20 in PBS 5-8 times over the course of 24 h at room temperature. After that the samples were incubated with the secondary antibodies (diluted in 3% donkey serum in PTwH) for 72 h at 37°C. The samples were washed with 0.1% Tween-20 in PBS 5-8 times over the course of 24 h and finally twice for 1 h in PBS at room temperature.
Tissue clearing
Whole-mount stained tissues were dehydrated by sequential incubation in increasing concentrations of methanol – 50% v/v methanol, 70% v/v methanol, 95% v/v methanol and 100% v/v methanol – at 1 h each. After that, the samples were incubated in 100% v/v methanol overnight. On the next day, the samples were transferred to a solution containing 1:1 ratio of 100% anhydrous methanol and BABB solution of 1:2 v/v benzyl alcohol (Sigma-Aldrich, 24122-2.5L-M) and benzyl benzoate (Thermo Fisher Scientific, 105860010) and incubated for a minimum of 4 h at room temperature. After that, the samples were transferred to 100% BABB solution and incubated overnight at room temperature. On the next day, the samples were transferred into fresh BABB solution before imaging.
DNA isolation and PCR genotyping
Samples were incubated with DNA lysis buffer containing 50 mM KCl, 10 mM Tris-HCl pH 8.0, 2 mM MgCl2, 0.45% NP-40, 0.45% Tween-20 and 0.4 mg/ml proteinase K at 55°C for at least 3 h, followed by 95°C incubation for 15 min. The volume of the lysis buffer was adjusted based on the amount of material: 100 μl lysis buffer was used for ear clips and ESCs, while 10 μl lysis buffer was used for pre-implantation embryos. PCR was performed using a GoTaq green master mix and the primers listed in the key resources table.
RNA extraction and quantitative PCR analysis
Total RNA was extracted using the NucleoSpin RNA (MACHEREY-NAGEL) Mini kit for RNA purification. The cDNA synthesis was performed using the M-MLV reverse transcriptase (MACHEREY-NAGEL). Transcript levels were detected using iTaq SYBR Green Supermix (Bio-Rad) with Quantstudio 3 (Applied Biosystems). Gene expression was normalised to the housekeeping genes GAPDH and calculated using the delta Ct algorithm. The primers used in this study are listed in the key resources table.
Analysis of EU incorporation
For the analysis of EU incorporation, isolated embryos were incubated in KSOM medium (Millipore, MR-020P-5F) supplemented with EU (1 mM final concentration) for 60 min. EU labelling of embryos was performed using the Click-iT RNA Alexa Fluor 488 HCS Assay kit (Thermo Fisher Scientific, C10327) according to the manufacturer’s instructions. Images were acquired using Leica SP8 confocal microscope.
Flow cytometry
ESCs were dissociated using 0.25% trypsin-EDTA and pelleted by centrifugation. After that the pellet was resuspended in 3% (v/v) FBS in PBS. FACSAria IIIu sorter (BD biosciences) was used for cell sorting and analysis. Single viable cells were first selected based on FSC and SSC gating and then Venus-positive cells were collected. Cell cycle analysis using EdU assay (Thermo Fisher Scientific, C10634) and cell death analysis using annexin V assay (Thermo Fisher Scientific, A13202) and DAPI (10 μg/ml) were performed according to the manufacturer’s instructions. Flowjo software (BD Biosciences) was used for data analysis.
Western blot
The ESCs were scraped, pelleted by centrifugation and lysed in ice-cold lysis buffer containing 10 mM Tris-HCl pH 7.6, 150 mM NaCl, 2 mM MgCl2, 2 mM EDTA, 0.1% Triton-X-100, 10% glycerol and 1x protease inhibitor cocktail (cOmplete ULTRA). The lysate was kept on ice for 20 min and the protein concentration was determined using the BCA protein assay kit (Thermo Fisher, 23227) based on the manufacturer’s instructions. The proteins were loaded in PAA gel and transferred to a PVDF membrane followed by blocking in 5% dry milk in PBST for 30 min. The membrane was incubated with primary antibodies at 4°C, overnight. On the next day, the membrane was washed with PBST and incubated with secondary antibodies conjugated to HRP for 2 h. The proteins were detected using ECL or ECL Plus and exposed to autoradiography films.
scRNA-seq and bioinformatics analysis
Blastocysts were collected at E4.5 (206 embryos), EDG7.5 (247 embryos), EDG9.5 (223 embryos), Reactivated (191 embryos) stages by flushing the uterine horns with M2 medium. E5.5 concepti (42 embryos) were isolated by dissecting deciduae in M2 medium. All samples were immediately processed as follows. The embryos were dissociated into single cells by incubating with TrypLE for 15 min at 37°C. All collected single cells were loaded directly onto a microwell cartridge of the BD Rhapsody Express system (BD) without undergoing cell counting. However, an estimated cell count between 1,000 ~ 2,000 cells was inferred. Single-cell whole transcriptome analysis libraries were prepared using BD Rhapsody WTA Reagent kit (BD,633802) with some modifications for the low number of input cells. First, we performed two rounds of random primer extension step with 100ul of total enzyme mix solution. Second, we repeated the purification step twice to ensure the maximum yield. And then anticipating a low amount of cDNA, we conducted a final round of PCR amplification with a total of 15 cycles. The final index PCR libraries were sequenced on the Illumina NextSeq 500 using Mid Output Kit v2.5 (150 cycles, Illumina) for 2 x 75bp paired-end reads with 8bp single index.
The FASTQ-format sequencing raw data was initially processed using UMI-tools (version 1.0.1). The data was then aligned to the mouse reference genome (mm10) using STAR (version 2.7.1a), and quantified with Subread featureCounts (version 1.6.4) to generate an expression matrix. Data normalization, dimensionality reduction and visualization were performed using the Seurat package (version 3.1.3), unless otherwise specified.
To ensure data quality, cells were filtered based on the criteria: having a number of genes per cell (nFeature_RNA) between 500 and 6000 and a percentage of mitochondrial genes (percent.mito) less than 25. Additionally, genes were filtered to include only those present in a minimum of 3 cells. After filtering, the matrices were normalized using the NormalizeData function with the LogNormalize method and a scale factor of 10,000. Variable genes were identified using the FindVariableFeatures function, selecting the top 2000 genes with the variance stabilizing transformation (VST) method, while also excluding genes related to the cell cycle (GO:0007049). Data integration was achieved through FindIntegrationAnchors and IntegrateData functions with default options. The integrated data was further processed using the ScaleData and RunPCA functions. Statistically significant principal components were identified using the JackStraw method, which were then used for UMAP non-linear dimensional reduction.
For unsupervised hierarchical clustering analysis, the FindClusters function in Seurat package was applied. Various resolutions between 0.1 and 0.9 were tested, and the final resolution was selected based on the most stable and relevant outcome, utilizing the clustree R package and considering prior knowledge. Cellular identity for each cluster was determined by finding cluster-specific marker genes using the FindAllMarkers function. Marker genes were considered with a minimum fraction of cells expressing the gene over 25% (min.pct=0.25), and comparisons were made against known cell type-specific genes from previous studies.
WGCNA R package was used to identify co-expressed gene modules for each cell type. First, we subset 10,000 the most variable genes from each dataset. A soft-threshold power, a tradeoff between scale free topology and mean connectivity was selected using pickSoftThreshold function with options of dataIsExpr = TRUE,powerVector = powers,corFnc = cor,corOptions = list(use = ‘p’),networkType = “signed”. Using the selected power, we generated adjacency and TOM similarity matrices, constructed the gene network and then identified co-expressed gene modules using blockwiseModules function. Pearson correlation coefficient was used with a relatively large minimum module size of 30 and a medium sensitivity (deepSplit=2) to cluster splitting. The threshold for merging of modules (mergeCutHeight) was 0.25.
EnrichGO function in R package clusterProfiler was used for GO over-representation test with qvalueCutoff = 0.1. The results were filter by removing redundancy using simplify function with similarity cutoff 0.7, Wang method for measure similarity and adjusted p value for representative term. Top 10 enriched terms for each ontology were demonstrated. Alternatively, PANTHER classification system (www.pantherdb.org) was used for GO biological process terms statistical overrepresentation test using selected gene lists.
Bulk RNA-seq and bioinformatics analysis
RNA quality was verified on an Agilent Bioanalyzer Nano Eukaryote chip. In all, 1 μg of the total RNA, with RIN (RNA integrity number) numbers above 7 (Agilent 2100 Bioanalyzer) were used. mRNA was enriched using NEBNext® Poly(A) mRNA Magnetic Isolation Module and cDNA library was prepared with NEBNext® Ultra™ II Directional RNA Library Prep Kit. Sequencing was performed on the NextSeq 500 system (75 cycles, high output, v2.5). The sequencing was performed in the Sequencing Core Facility of the Max Planck Institute of Molecular Biomedicine. The RNA sequencing reads were aligned to the mouse (mm10) reference genome with TopHat2 (version 2.1.1), and the aligned reads were used for the transcript quantification by using HTSeq-count (version 0.6.1). DESeq2 was used to identify differentially expressed genes across the samples.
Confocal microscopy and image analysis
The tissue-cleared samples were imaged using the Andor Dragonfly Spinning Disc confocal microscope with a Z-stack size of 5 μm through the whole decidua. Fluorescence images of the fixed embryos or cells were collected on Leica SP8 or Zeiss LSM 880 confocal microscopes. Z-sections were taken every 2 μm, using 20x or 40x oil objectives.
Image analysis was performed using Fiji software. To determine nuclear fluorescence intensities, nuclei were manually segmented based on the DAPI signal. We defined TE cells adjacent to the ICM as polar TE and the rest of the TE cells as mural TE. The mean fluorescence intensity of Cdx1 in the TE lineage was measured at a single optical plane containing the largest area of each nucleus. Cdx1 is expressed higher in the polar than the mural TE and its levels were presented as polar to mural TE ratio. The relative Cdx1 levels were calculated by dividing the mean fluorescence intensity of individual TE cells by the average Cdx1 intensity in the mural TE cells in each embryo.
Similarly, the mean fluorescence intensity of Aqp3 in the TE lineage was measured at a single optical plane containing the largest area of each lateral domain. Aqp3 is expressed higher in the mural than the polar TE and its levels were presented as mural to polar TE ratio. The relative Aqp3 levels were calculated by dividing the mean fluorescence intensity in individual TE cells by the average Aqp3 intensity of the lateral domains in the polar TE cells in each embryo.
For measuring Oct4 or nuclear Yap, nuclei were manually segmented based on the DAPI signal. The mean fluorescence intensity of Oct4 or nuclear Yap and DAPI was measured at a single optical plane containing the largest area of each nucleus. Normalised Oct4 or nuclear Yap levels were calculated by dividing the mean fluorescence intensity of Oct4 or nuclear Yap by the mean fluorescence intensity of DAPI in each nucleus.
Quantification and Statistical Analysis
Statistical analysis was performed using GraphPad Prism. Values are presented as means ± SEM or means ± SD. The exact p values are displayed in the figures. Statistical significance was calculated using a two-tailed unpaired Student’s t-test (two groups) or analysis of variance (ANOVA) (multiple groups) with a Tukey’s post hoc test.
Supplementary Material
In brief.
The underlying mechanisms maintaining the developmental potential of the dormant embryo are still obscure. Here, Chen and colleagues analyzed the single-cell transcriptomes of mouse embryos during diapause and transient development, revealing dynamic transcriptional landscapes and activation of integrin-Yap/Taz signaling, which preserves the developmental capacity of the dormant embryo.
Highlights.
Single-cell atlas of the pre-to post-implantation transition and embryo dormancy
Dynamic transcriptional landscapes during entry, progression, and exit of diapause
Diapause-specific transcriptional responses of the embryonic and extraembryonic tissues
Integrin-Yap/Taz signaling governs the survival and reactivation of the dormant embryo
Acknowledgments
We thank Prof. Dr. Hans R. Schöler, Prof. Dr. Sara Wickström, and Prof. Dr. Dietmar Vestweber (MPI-MB) for providing access to key infrastructure, equipment, and reagents; all members of the Embryonic Self-Organization research group for the constructive discussions and suggestions; and Dr. Celeste Brennecka for proofreading the manuscript. This work was supported by the European Research Council (ERC) Consolidator grant (MORPHEUS, 101043753) and the Collaborative Research Center 1348 “Dynamic Cellular Interfaces” grant (1348/2, B09) to I.B. Furthermore, R.C. received a scholarship from the China Scholarship Council and is a member of the International Max Planck Research School (IMPRS) Molecular Biomedicine, Münster, Germany. This work was funded/co-funded in part by the European Union (ERC, MORPHEUS, 101043753). Views and opinions expressed are, however, those of the author(s) only and do not necessarily reflect those of the European Union or the European Research Council. Neither the European Union nor the granting authority can be held responsible for them.
Footnotes
Author Contributions
I.B. and R.C. conceived the study, designed experiments, and interpreted the results. I.B. and R.C. wrote the manuscript. R.C. performed most of the experiments. I.B. and R.F. helped to collect embryos for sequencing. H.-W.J. performed the bioinformatic analysis. F.C. performed western blot experiments. N.G. performed tissue-clearing experiments. H.B. provided general technical support. M.S. performed FACS analysis. R.H.A. provided access to key infrastructure, reagents, and discussion of the results.
Declaration of Interests
The authors declare no competing interests.
Data and code availability
Single-cell and bulk RNA-seq data have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Ma WG, Song H, Das SK, Paria BC, Dey SK. Estrogen is a critical determinant that specifies the duration of the window of uterine receptivity for implantation. Proc Natl Acad Sci USA. 2003;100:2963–2968. doi: 10.1073/pnas.0530162100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Renfree MB, Shaw G. Diapause. Annu Rev Physiol. 2000;62:353–375. doi: 10.1146/annurev.physiol.62.1.353. [DOI] [PubMed] [Google Scholar]
- 3.MacLean Hunter SM, Evans M. Non-surgical method for the induction of delayed implantation and recovery of viable blastocysts in rats and mice by the use of tamoxifen and Depo-Provera. Mol Reprod Dev. 1999;52:29–32. doi: 10.1002/(SICI)1098-2795(199901)52:1<29∷AID-MRD4>3.0.CO;2-2. [DOI] [Google Scholar]
- 4.Yoshinaga K, Adams CE. Delayed implantation in the spayed, progesterone treated adult mouse. J Reprod Fertil. 1966;12:593–595. doi: 10.1530/jrf.0.0120593. [DOI] [PubMed] [Google Scholar]
- 5.Fan R, Kim YS, Wu J, Chen R, Zeuschner D, Mildner K, Adachi K, Wu G, Galatidou S, Li J, et al. Wnt/beta-catenin/Esrrb signalling controls the tissue-scale reorganization and maintenance of the pluripotent lineage during murine embryonic diapause. Nat Commun. 2020;11:5499. doi: 10.1038/s41467-020-19353-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sathyanarayanan A, Ing-Simmons E, Chen R, Jeong HW, Ozguldez HO, Fan R, Duethorn B, Kim KP, Kim YS, Stehling M, et al. Early developmental plasticity enables the induction of an intermediate extraembryonic cell state. Sci Adv. 2022;8:eabl9583. doi: 10.1126/sciadv.abl9583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Latos PA, Hemberger M. From the stem of the placental tree: trophoblast stem cells and their progeny. Development. 2016;143:3650–3660. doi: 10.1242/dev.133462. [DOI] [PubMed] [Google Scholar]
- 8.Ozguldez HO, Govindasamy N, Fan R, Long H, Mildner K, Zeuschner D, Trappmann B, Ranga A, Bedzhov I. Polarity inversion reorganizes the stem cell compartment of the trophoblast lineage. Cell Rep. 2023;42:112313. doi: 10.1016/j.celrep.2023.112313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Govindasamy N, Long H, Jeong HW, Raman R, Özcifci B, Probst S, Arnold SJ, Riehemann K, Ranga A, Adams RH, et al. 3D biomimetic platform reveals the first interactions of the embryo and the maternal blood vessels. Dev Cell. 2021;56:3276–3287.:e8. doi: 10.1016/j.devcel.2021.10.014. [DOI] [PubMed] [Google Scholar]
- 10.Kanai-Azuma M, Kanai Y, Gad JM, Tajima Y, Taya C, Kurohmaru M, Sanai Y, Yonekawa H, Yazaki K, Tam PPL, et al. Depletion of definitive gut endoderm in Sox17-null mutant mice. Development. 2002;129:2367–2379. doi: 10.1242/dev.129.10.2367. [DOI] [PubMed] [Google Scholar]
- 11.Filimonow K, Saiz N, Suwińska A, Wyszomirski T, Grabarek JB, Ferretti E, Piliszek A, Plusa B, Maleszewski M. No evidence of involvement of E-cadherin in cell fate specification or the segregation of Epi and PrE in mouse blastocysts. PLoS One. 2019;14:e0212109. doi: 10.1371/journal.pone.0212109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kinoshita M, Smith A. Pluripotency deconstructed. Dev Growth Differ. 2018;60:44–52. doi: 10.1111/dgd.12419. [DOI] [PubMed] [Google Scholar]
- 13.Acampora D, Di Giovannantonio LG, Garofalo A, Nigro V, Omodei D, Lombardi A, Zhang J, Chambers I, Simeone A. Functional antagonism between OTX2 and NANOG specifies a spectrum of heterogeneous identities in embryonic stem cells. Stem Cell Rep. 2017;9:1642–1659. doi: 10.1016/j.stemcr.2017.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Acampora D, Di Giovannantonio LG, Simeone A. Otx2 is an intrinsic determinant of the embryonic stem cell state and is required for transition to a stable epiblast stem cell condition. Development. 2013;140:43–55. doi: 10.1242/dev.085290. [DOI] [PubMed] [Google Scholar]
- 15.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fenelon JC, Renfree MB. The history of the discovery of embryonic diapause in mammals. Biol Reprod. 2018;99:242–251. doi: 10.1093/biolre/ioy112. [DOI] [PubMed] [Google Scholar]
- 17.Cree LM, Samuels DC, de Sousa Lopes SC, Rajasimha HK, Wonnapinij P, Mann JR, Dahl HHM, Chinnery PF. A reduction of mitochondrial DNA molecules during embryogenesis explains the rapid segregation of genotypes. Nat Genet. 2008;40:249–254. doi: 10.1038/ng.2007.63. [DOI] [PubMed] [Google Scholar]
- 18.Salewskij K, Gross-Thebing T, Ing-Simmons E, Duethorn B, Rieger B, Fan R, Chen R, Govindasamy N, Brinkmann H, Kremer L, et al. Ronin governs the metabolic capacity of the embryonic lineage for post-implantation development. EMBO Rep. 2021;22:e53048. doi: 10.15252/embr.202153048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lima A, Lubatti G, Burgstaller J, Hu D, Green A, Gregorio AD, Zawadzki T, Pernaute B, Mahammadov E, Dore M, et al. Differences in mitochondrial activity trigger cell competition during early mouse development. bioRxiv. 2020 doi: 10.1101/2020.01.15.900613. [DOI] [Google Scholar]
- 20.Fu Z, Wang B, Wang S, Wu W, Wang Q, Chen Y, Kong S, Lu J, Tang Z, Ran H, et al. Integral proteomic analysis of blastocysts reveals key molecular machinery governing embryonic diapause and re-activation for implantation in mice. Biol Reprod. 2014;90:52. doi: 10.1095/biolreprod.113.115337. [DOI] [PubMed] [Google Scholar]
- 21.Leese HJ. Metabolism of the preimplantation embryo: 40 years on. Reproduction. 2012;143:417–427. doi: 10.1530/REP-11-0484. [DOI] [PubMed] [Google Scholar]
- 22.Kawase E, Suemori H, Takahashi N, Okazaki K, Hashimoto K, Nakatsuji N. Strain difference in establishment of mouse embryonic stem (ES) cell lines. Int J Dev Biol. 1994;38:385–390. [PubMed] [Google Scholar]
- 23.Brook FA, Gardner RL. The origin and efficient derivation of embryonic stem cells in the mouse. Proc Natl Acad Sci USA. 1997;94:5709–5712. doi: 10.1073/pnas.94.11.5709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith AG, Heath JK, Donaldson DD, Wong GG, Moreau J, Stahl M, Rogers D. Inhibition of pluripotential embryonic stem cell differentiation by purified polypeptides. Nature. 1988;336:688–690. doi: 10.1038/336688a0. [DOI] [PubMed] [Google Scholar]
- 25.Williams RL, Hilton DJ, Pease S, Willson TA, Stewart CL, Gearing DP, Wagner EF, Metcalf D, Nicola NA, Gough NM. Myeloid leukaemia inhibitory factor maintains the developmental potential of embryonic stem cells. Nature. 1988;336:684–687. doi: 10.1038/336684a0. [DOI] [PubMed] [Google Scholar]
- 26.Yoshida K, Taga T, Saito M, Suematsu S, Kumanogoh A, Tanaka T, Fujiwara H, Hirata M, Yamagami T, Nakahata T, et al. Targeted disruption of gp130, a common signal transducer for the interleukin 6 family of cytokines, leads to myocardial and hematological disorders. Proc Natl Acad Sci USA. 1996;93:407–411. doi: 10.1073/pnas.93.1.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakashima K, Wiese S, Yanagisawa M, Arakawa H, Kimura N, Hisatsune T, Yoshida K, Kishimoto T, Sendtner M, Taga T. Developmental requirement of gp130 signaling in neuronal survival and astrocyte differentiation. J Neurosci. 1999;19:5429–5434. doi: 10.1523/JNEUROSCI.19-13-05429.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nichols J, Chambers I, Taga T, Smith A. Physiological rationale for responsiveness of mouse embryonic stem cells to gp130 cytokines. Development. 2001;128:2333–2339. doi: 10.1242/dev.128.12.2333. [DOI] [PubMed] [Google Scholar]
- 29.Burdon T, Smith A, Savatier P. Signalling, cell cycle and pluripotency in embryonic stem cells. Trends Cell Biol. 2002;12:432–438. doi: 10.1016/s0962-8924(02)02352-8. [DOI] [PubMed] [Google Scholar]
- 30.Karwacki-Neisius V, Göke J, Osorno R, Halbritter F, Ng JH, Weiße AY, Wong FCK, Gagliardi A, Mullin NP, Festuccia N, et al. Reduced Oct4 expression directs a robust pluripotent state with distinct signaling activity and increased enhancer occupancy by Oct4 and Nanog. Cell Stem Cell. 2013;12:531–545. doi: 10.1016/j.stem.2013.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lenter M, Uhlig H, Hamann A, Jenö P, Imhof B, Vestweber D. A monoclonal antibody against an activation epitope on mouse integrin chain beta 1 blocks adhesion of lymphocytes to the endothelial integrin alpha 6 beta 1. Proc Natl Acad Sci USA. 1993;90:9051–9055. doi: 10.1073/pnas.90.19.9051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bedzhov I, Zernicka-Goetz M. Self-organizing properties of mouse pluripotent cells initiate morphogenesis upon implantation. Cell. 2014;156:1032–1044. doi: 10.1016/j.cell.2014.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Humphrey JD, Dufresne ER, Schwartz MA. Mechanotransduction and extracellular matrix homeostasis. Nat Rev Mol Cell Biol. 2014;15:802–812. doi: 10.1038/nrm3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Totaro A, Panciera T, Piccolo S. YAP/TAZ upstream signals and downstream responses. Nat Cell Biol. 2018;20:888–899. doi: 10.1038/s41556-018-0142-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim YS, Fan R, Lith SC, Dicke AK, Drexler HCA, Kremer L, Kuempel-Rink N, Hekking L, Stehling M, Bedzhov I. Rap1 controls epiblast morphogenesis in sync with the pluripotency states transition. Dev Cell. 2022;57:1937–1956.:e8. doi: 10.1016/j.devcel.2022.07.011. [DOI] [PubMed] [Google Scholar]
- 36.Nishioka N, Inoue K, Adachi K, Kiyonari H, Ota M, Ralston A, Yabuta N, Hirahara S, Stephenson RO, Ogonuki N, et al. The Hippo signaling pathway components Lats and Yap pattern Tead4 activity to distinguish mouse trophectoderm from inner cell mass. Dev Cell. 2009;16:398–410. doi: 10.1016/j.devcel.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 37.Hossain Z, Ali SM, Ko HL, Xu J, Ng CP, Guo K, Qi Z, Ponniah S, Hong W, Hunziker W. Glomerulocystic kidney disease in mice with a targeted inactivation of Wwtr1. Proc Natl Acad Sci USA. 2007;104:1631–1636. doi: 10.1073/pnas.0605266104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Makita R, Uchijima Y, Nishiyama K, Amano T, Chen Q, Takeuchi T, Mitani A, Nagase T, Yatomi Y, Aburatani H, et al. Multiple renal cysts, urinary concentration defects, and pulmonary emphysematous changes in mice lacking TAZ. Am J Physiol Ren Physiol. 2008;294:F542–F553. doi: 10.1152/ajprenal.00201.2007. [DOI] [PubMed] [Google Scholar]
- 39.Morin-Kensicki EM, Boone BN, Howell M, Stonebraker JR, Teed J, Alb JG, Magnuson TR, O’Neal W, Milgram SL. Defects in yolk sac vasculogenesis, chorioallantoic fusion, and embryonic axis elongation in mice with targeted disruption of Yap65. Mol Cell Biol. 2006;26:77–87. doi: 10.1128/MCB.26.1.77-87.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Benham-Pyle BW, Pruitt BL, Nelson WJ. Cell adhesion. Mechanical strain induces E-cadherin-dependent Yap1 and beta-catenin activation to drive cell cycle entry. Science. 2015;348:1024–1027. doi: 10.1126/science.aaa4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chinnadurai G, Vijayalingam S, Rashmi R. BIK, the founding member of the BH3-only family proteins: mechanisms of cell death and role in cancer and pathogenic processes. Oncogene. 2008;27:S20–S29. doi: 10.1038/onc.2009.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Osterlund EJ, Hirmiz N, Pemberton JM, Nougaréde A, Liu Q, Leber B, Fang Q, Andrews DW. Efficacy and specificity of inhibitors of BCL-2 family protein interactions assessed by affinity measurements in live cells. Sci Adv. 2022;8:eabm7375. doi: 10.1126/sciadv.abm7375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hao J, Li TG, Qi X, Zhao DF, Zhao GQ. WNT/beta-catenin pathway up-regulates Stat3 and converges on LIF to prevent differentiation of mouse embryonic stem cells. Dev Biol. 2006;290:81–91. doi: 10.1016/j.ydbio.2005.11.011. [DOI] [PubMed] [Google Scholar]
- 44.Kraunsoe S, Azami T, Pei Y, Martello G, Jones K, Boroviak T, Nichols J. Requirement for STAT3 and its target, TFCP2L1, in self-renewal of naive pluripotent stem cells in vivo and in vitro. Biol Open. 2023;12:bio059650. doi: 10.1242/bio.059650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Molotkov A, Soriano P. Distinct mechanisms for PDGF and FGF signaling in primitive endoderm development. Dev Biol. 2018;442:155–161. doi: 10.1016/j.ydbio.2018.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang W, Klaman LD, Chen B, Araki T, Harada H, Thomas SM, George EL, Neel BG. An Shp2/SFK/Ras/Erk signaling pathway controls trophoblast stem cell survival. Dev Cell. 2006;10:317–327. doi: 10.1016/j.devcel.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 47.Johnson R, Halder G. The two faces of Hippo: targeting the Hippo pathway for regenerative medicine and cancer treatment. Nat Rev Drug Discov. 2014;13:63–79. doi: 10.1038/nrd4161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kurppa KJ, Liu Y, To C, Zhang T, Fan M, Vajdi A, Knelson EH, Xie Y, Lim K, Cejas P, et al. Treatment-induced tumor dormancy through YAP-mediated transcriptional reprogramming of the apoptotic pathway. Cancer Cell. 2020;37:104–122.:e12. doi: 10.1016/j.ccell.2019.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.de Vries WN, Binns LT, Fancher KS, Dean J, Moore R, Kemler R, Knowles BB. Expression of Cre recombinase in mouse oocytes: a means to study maternal effect genes. Genesis. 2000;26:110–112. [PubMed] [Google Scholar]
- 50.Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45:593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- 51.Reginensi A, Scott RP, Gregorieff A, Bagherie-Lachidan M, Chung C, Lim DS, Pawson T, Wrana J, McNeill H. Yap- and Cdc42-dependent nephrogenesis and morphogenesis during mouse kidney development. PLoS Genet. 2013;9:e1003380. doi: 10.1371/journal.pgen.1003380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Raghavan S, Bauer C, Mundschau G, Li Q, Fuchs E. Conditional ablation of beta1 integrin in skin. Severe defects in epidermal proliferation, basement membrane formation, and hair follicle invagination. J Cell Biol. 2000;150:1149–1160. doi: 10.1083/jcb.150.5.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang W, Lin C, Lu D, Ning Z, Cox T, Melvin D, Wang X, Bradley A, Liu P. Chromosomal transposition of PiggyBac in mouse embryonic stem cells. Proc Natl Acad Sci USA. 2008;105:9290–9295. doi: 10.1073/pnas.0801017105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Adachi K, Kopp W, Wu G, Heising S, Greber B, Stehling M, Araúzo-Bravo MJ, Boerno ST, Timmermann B, Vingron M, et al. Esrrb unlocks silenced enhancers for reprogramming to naive pluripotency. Cell Stem Cell. 2018;23:266–275.:e6. doi: 10.1016/j.stem.2018.05.020. [DOI] [PubMed] [Google Scholar]
- 55.Sivaraj KK, Dharmalingam B, Mohanakrishnan V, Jeong HW, Kato K, Schröder S, Adams S, Koh GY, Adams RH. YAP1 and TAZ negatively control bone angiogenesis by limiting hypoxia-inducible factor signaling in endothelial cells. eLife. 2020;9:e50770. doi: 10.7554/eLife.50770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kagawa S, He C, Gu J, Koch P, Rha SJ, Roth JA, Curley SA, Stephens LC, Fang B. Antitumor activity and bystander effects of the tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) gene. Cancer Res. 2001;61:3330–3338. [PubMed] [Google Scholar]
- 57.Wu Y, Ma L, Duyck K, Long CC, Moran A, Scheerer H, Blanck J, Peak A, Box A, Perera A, et al. A population of navigator neurons is essential for olfactory map formation during the critical period. Neuron. 2018;100:1066–1082.:e6. doi: 10.1016/j.neuron.2018.09.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM, Hao Y, 3rd, Stoeckius M, Smibert P, Satija R. Comprehensive integration of single-cell data. Cell. 2019;177:1888–1902.:e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 61.Smith T, Heger A, Sudbery I. UMI-tools: modeling sequencing errors in Unique Molecular Identifiers to improve quantification accuracy. Genome Res. 2017;27:491–499. doi: 10.1101/gr.209601.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liao Y, Smyth GK, Shi W. The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 2013;41:e108. doi: 10.1093/nar/gkt214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Putri GH, Anders S, Pyl PT, Pimanda JE, Zanini F. Analysing high-throughput sequencing data in Python with HTSeq 2.0. Bioinformatics. 2022;38:2943–2945. doi: 10.1093/bioinformatics/btac166. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Single-cell and bulk RNA-seq data have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.