Summary
CD19-directed immunotherapies are clinically effective for treating B-cell malignancies but also cause a high incidence of neurotoxicity. A subset of patients treated with chimeric antigen receptor (CAR) T cells or bispecific T-cell engager (BiTE) antibodies display severe neurotoxicity, including fatal cerebral edema associated with T cell infiltration into the brain. Here we report that mural cells, which surround the endothelium and are critical for blood-brain-barrier integrity, express CD19. We identify CD19 expression in brain mural cells using single-cell RNA-seq data and confirm perivascular staining at the protein level. CD19 expression in the brain begins early in development alongside the emergence of mural cell lineages and persists throughout adulthood across brain regions. Mouse mural cells demonstrate lower levels of Cd19 expression, suggesting limitations in preclinical animal models of neurotoxicity. These data suggest an on-target mechanism for neurotoxicity in CD19-directed therapies and highlight the utility of human single-cell atlases for designing immunotherapies.
Graphical Abstract
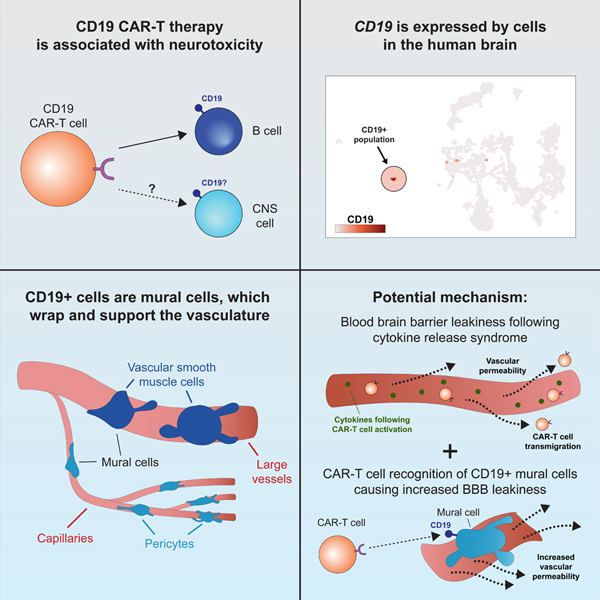
In brief
Single-cell RNA sequencing analysis shows that CD19, primarily considered as a B cell-specific surface antigen, is expressed in human brain mural cells that are critical for blood-bran-barrier integrity, suggesting that this cell population may contribute to the neurotoxicity of CD19-directed immunotherapy including CAR-T.
Introduction
CD19-targeting CAR-T cells have shown tremendous clinical efficacy in patients with B-cell leukemia and lymphoma, including those who have relapsed after receiving traditional chemotherapy regimens (Brentjens et al., 2003; Kochenderfer et al., 2010; Porter et al., 2011; Brentjens et al., 2013; Grupp et al., 2013). For example, in a recent phase II study of 111 patients with refractory B-cell lymphoma, of whom 101 were administered CD19 CAR-T cell therapy, 40% of patients showed a complete remission of disease 15 months after treatment (Neelapu et al., 2017). Similar results were observed in a separate study, with complete remission in 43% and 71% of patients with diffuse large B-cell and follicular lymphoma, respectively (Schuster et al., 2017). However, in addition to adverse effects related to cytokine release syndrome (CRS), a systemic inflammatory response caused by the massive activation of CAR-T cells after infusion, both studies reported a high incidence of neurotoxicity (64% and 39%, respectively). This is in agreement with previously reported rates of neurotoxicity in patients receiving CD19 CAR-T cell therapy as well as CD19/CD3 BiTE therapy, which uses bi-specific antibodies to recruit T cells to CD19-positive cells (Goebeler et al., 2016; Gust et al., 2017; Nagorsen et al., 2012).
Analyses of patients who received CD19 CAR-T therapy and displayed neurologic adverse reactions revealed edema, multifocal hemorrhage, and vascular disruption of the brain (Gust et al., 2017). Neuropathologic evaluation of brain tissue from a patient who developed fatal neurotoxicity revealed CAR-T cell infiltration into the brain, indicating that CAR-T cells are able to traffic to and ultimately infiltrate the brain (Gust et al., 2017). This is in agreement with a recent study which showed that in human patients, blinatumomab, a CD19/CD3 BiTE, leads to T cell adhesion and perivascular transmigration across brain endothelium (Klinger et al., 2020). However, the underlying mechanisms behind why neurotoxicity is observed with CD19-targeting immunotherapies have been harder to resolve (Ruella and June, 2018). One possible mechanism could be the unanticipated expression of CD19 on non-B cells in the brain.
This mechanism could explain the higher incidence of neurotoxicity in CD19-directed immunotherapies, compared to treatments targeting other B cell proteins, such as CD20. Notably, CD19 CAR-T cells are sensitive to even low levels of CD19 antigen density, emphasizing the importance of identifying any potential reservoir of CD19 other than B cells (Majzner et al., 2020). We therefore sought to investigate whether a population of CD19-expressing cells is present in the human brain.
Using single-cell RNA-sequencing data generated from human brain, we show that CD19 is expressed in mural cells. Mural cells are an integral part of the neurovascular unit (NVU), which surround endothelial cells and are critical for regulating the integrity of the BBB (Sweeney et al., 2019). Mural cells include pericytes and vascular smooth muscle cells (vSMCs), which are closely related cell types that differ anatomically: pericytes localize along capillaries, while vSMCs are found along larger vessels. Genetic mouse models have shown that pericytes are crucial components of the BBB, which does not properly develop in their absence (Armulik et al., 2010; Daneman et al., 2010). We found that CD19 expression in human mural cells was observed across multiple datasets, brain regions, and developmental timepoints. Importantly, the expressed isoform of CD19 in the brain contains the epitope targeted by clinical CAR-T cells and BiTEs (Sommermeyer et al., 2017; Sotillo et al., 2015). More broadly, this study highlights the utility of efforts like the Human Cell Atlas (Regev et al., 2017), whose large-scale generation of tissue-wide scRNA-seq data can be used to inform target selection in cancer immunotherapy.
Results
CD19-positive mural cells identified by scRNA-sequencing in human brain
We hypothesized that if a population of CD19-expressing cells is present in the human brain, it might be identifiable in data generated from recent efforts to map the human brain transcriptome with single-cell resolution. We first analyzed scRNA-seq data from 2,364 human prefrontal cortex cells (Zhong et al., 2018) Figure 1A). We clustered cells and identified broad populations, focusing subsequent analyses on non-neuronal, non-erythroid cells. These further segregated into astrocyte, lymphocyte, microglial, oligodendrocyte precursor, endothelial, and pericyte populations (Figure 1B–C, S1A). These populations were identified based on the expression canonical marker genes; mural cells specifically expressed expected marker genes, such as PDGFRB, FOXF2, RGS5, and CD248, while endothelial cells expressed a distinct set of markers, including CDH5 and PECAM1 (CD31; Figure 1D). Surprisingly, this analysis revealed a small population of cells (~1.5% of non-neuronal cells; ~0.2% including neuronal cells) that expressed CD19 and co-expressed the mural cell marker CD248 (Figure 1D, S1B). These cells were negative for the vSMC marker ACTA2, indicating that these cells were pericytes (Figure S1C). Importantly, this population was negative for the B cell marker CD79A, arguing against the possibility of artifactual CD19 expression due to B cell-pericyte doublets (Figure 1E). Additionally, these cells were positive for the tetraspanin CD81, which chaperones CD19 through secretory pathways to the plasma membrane and is required for surface expression of CD19 in B cells (Figure S1D; (Maecker and Levy, 1997; Miyazaki et al., 1997; Shoham et al., 2003; Tsitsikov et al., 1997; van Zelm et al., 2010)(Maecker and Levy, 1997; Miyazaki et al., 1997; Tsitsikov et al., 1997; van Zelm et al., 2010; Shoham et al., 2003; Braig et al., 2017). We also observed a separate population of lymphocytes, that expressed the marker genes CD45 (PTPRC), CD3, CD7, and IL2RG (CD132), representing a population of T cells (Figure 1E–F). Notably, the expression of lymphocyte markers and pericyte markers was mutually exclusive in the observed clusters, and the expression of CD19 was specific to the pericyte cluster (Figure 1F).
Figure 1. Identification of CD19 expressing pericytes in single-cell RNA-sequencing data.
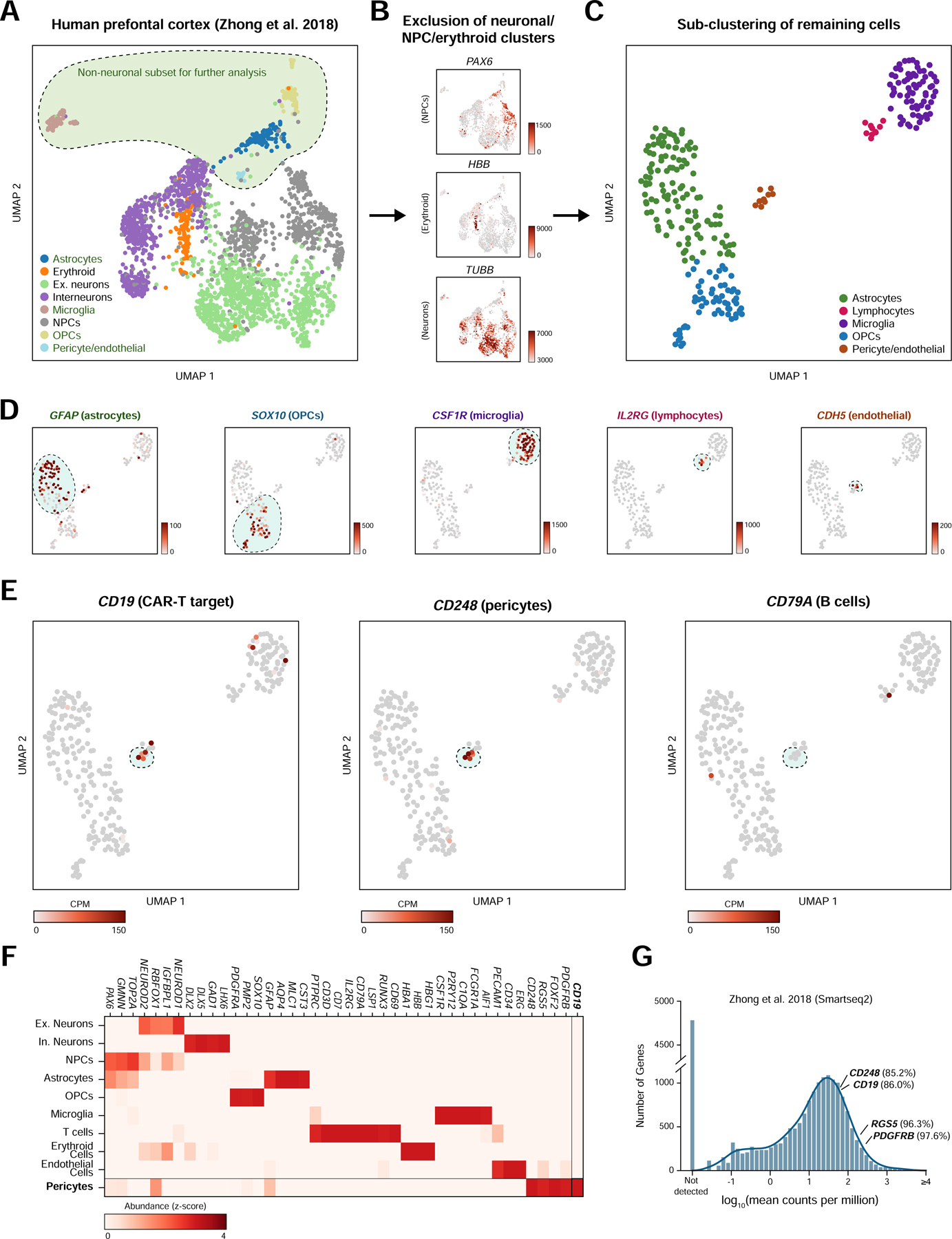
(A) UMAP projection of single-cell prefrontal cortex RNA-seq data. (B) The neuronal, neural progenitor cell, and erythroid clusters were identified as shown and subsequently excluded from further analysis. Unit, counts per million (CPM). (C) Non-neuronal cells are re-clustered and embedded using UMAP to more clearly distinguish populations. (D) Cell populations are identified by shown marker genes. Note that pericytes and endothelial cells are called as a single cluster but identifiable as separate populations by marker gene expression. Scale bar indicates CPM. (E) CD248-positive pericytes are also positive for the CAR-T target CD19 but negative for the B cell marker CD79A. (F) Heatmap showing relative gene abundances for specific marker genes across identified cell clusters. Pericytes are identified by PDGFRB, CD248, RGS5, and FOXF2 expression, and specifically express CD19. (G) Histogram of mean abundance of all genes in the human prefrontal cortex. Percentiles indicate the expression rank of a particular relative to all detected genes.
Due to the inherent sparsity of single-cell data, even highly expressed genes will not be detected in every cell, causing single-cell data to be zero-inflated (Butler et al., 2018). To ask at what level CD19 is expressed in pericytes, we compared the expression of CD19 to known pericyte marker genes, relative to the overall distribution of gene expression in pericytes. Mean-normalized gene expression values can provide a rough estimate of the relative expression of a given gene in a cell population of interest, despite the inherent sparsity of single-cell data. As expected, pericyte markers such as CD248, RGS5, and PDGFRB ranked in the top percentiles of gene expression in pericytes from the human prefrontal cortex (85th, 96th, and 98th, respectively) (Figure 1G; Methods). CD19 was similarly expressed highly, in the 86th percentile for gene expression.
We next sought to reproduce this observation in additional independent scRNA-seq datasets. In a dataset from human forebrain (La Manno et al., 2018), a mural cell population (48/7906 cells) positive for the markers CD248 and RGS5 also displayed CD19 expression (12/48 cells had measurable CD19) and the absence of the B-cell marker CD79A (Figure 2A). In a third dataset derived from human ventral forebrain (La Manno et al., 2016), we again observed expression of CD19 in a population of CD248-positive mural cells (195 mural cells/1977 cells, of which 25/195 had measurable CD19 (Figure 2B). As with other datasets, no RNA-sequencing counts for CD79A were identified in any mural cells. Although spurious non-mural CD19-positive cells were observable, these were likely the result of cell-cell doublets, such as mural-endothelial cell doublets observed due to the adhesion of mural to endothelial cells in the brain. In these datasets, CD19 ranked in the 86th and 71st percentiles for gene expression, suggesting relatively robust expression (Figure 2C).
Figure 2. Confirmation of mural cell CD19 expression in two independent datasets.
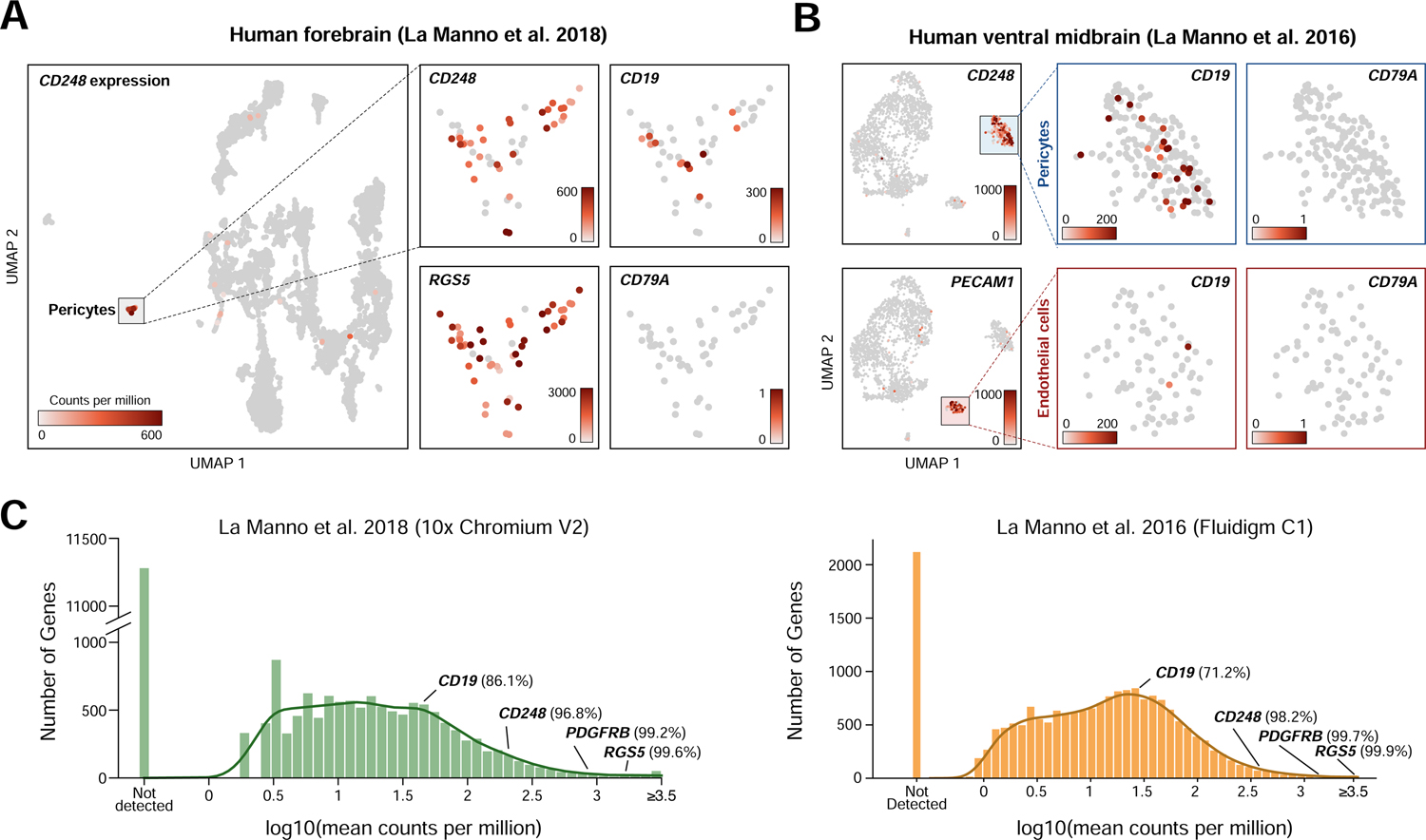
(A–B) UMAP plots showing single cell RNA-seq data from (a) human forebrain (La Manno et al., 2018) and (b) human ventral midbrain (La Manno et al., 2016), colored by gene expression value, showing CD19 expression in pericytes. (C) Histogram of mean gene expression values (log10 counts per million) in identified pericyte cells in La Manno et al. 2018 and La Manno et al. 2016. Relative gene expression percentiles are shown for indicated genes.
CD19 expression in adult human brain by IHC
To assess the expression of CD19 protein in human mural cells, we performed immunohistochemistry on several regions of the human brain using a clinically validated anti-human CD19 antibody (clone BT51E) on samples from healthy deceased subjects. This antibody recognizes the C-terminus of the CD19 protein, which is cytoplasmically localized. We found CD19 expression on cells present adjacent to the vessel basement membrane walls in perivascular areas (Figure 3). Abluminal CD19 expression was observed across multiple brain regions, with particular regions, such as the hippocampus, insula, temporal lobe, frontal lobe, and parietal lobe displaying a comparatively higher, albeit still rare, incidence of CD19-positive cells. In contrast, regions such as the pons and occipital lobe displayed lower rates of CD19-positive cells (Figure 3). Notably, CD19-positive cells were found along smaller capillaries (<8 µm) as well as larger vessels (>8 µm; majority of cells depicted), suggesting that in addition to pericytes, CD19 may also be expressed in vSMCs. It is possible that the CD19 staining observed by IHC may also be due to other cell types, such as passing B cells or glia. However, the abluminal localization along the vasculature of CD19+ cells is most consistent with staining of mural cells. While we found perivascular CD19 staining across multiple brain regions, abluminal CD19 staining was not found along all vessels, suggesting that there may be heterogeneity in the frequency of CD19+ mural cells. This may contribute to the inter-patient differences in severity and outcome of neurotoxicity observed clinically.
Figure 3. Perivascular staining of CD19 in human brain.
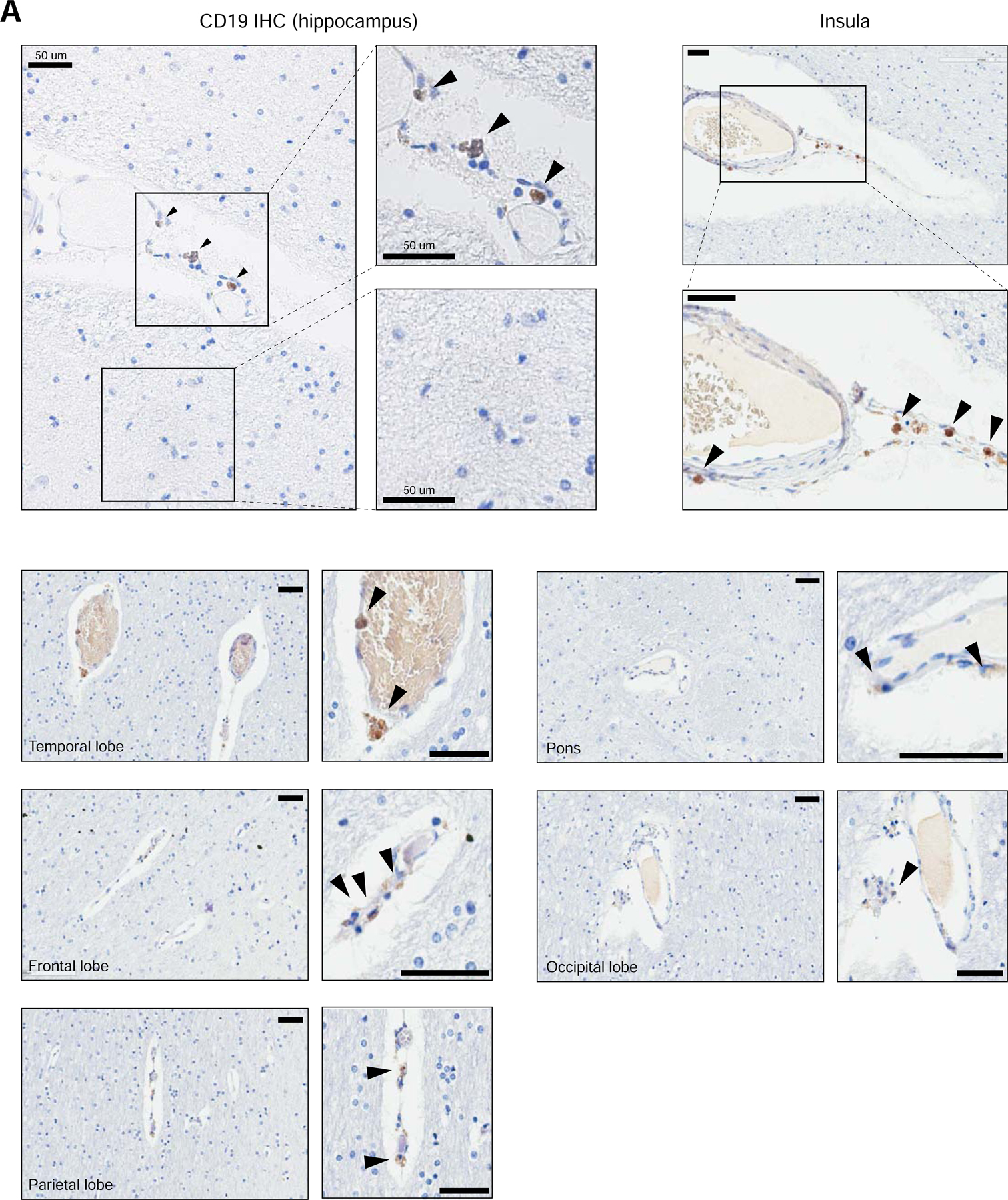
Representative immunohistochemistry staining for CD19 in human brain tissue. FFPE samples were stained for CD19 with a clinical protocol. Representative staining is shown for the hippocampus, insula, temporal lobe, frontal lobe, parietal lobe, pons, and occipital lobe. Scale bar = 50 µm. 5 slides were stained for the hippocampus, and 10 slides were stained for other brain regions.
Analysis of pericytes and vSMCs shows CD19 expression across mural cells.
Based on the perivascular staining of CD19 along both larger and smaller vessels, which suggested that CD19 expression might be a more general feature of mural cells, we sought to analyze the transcriptome of pericytes and vSMCs. We analyzed a large scRNA-seq dataset generated by the BRAIN Initiative Cell Census Network (BICCN, Kriegstein/UCSF), comprising scRNA-seq data from diverse human brain regions over many developmental timepoints spanning Carnegie Stage 12 (CS12), corresponding to gestational week (GW) 4, through GW25. This dataset included roughly 857,000 cells across 101 individual samples, affording us the power to perform a detailed analysis of the cell types within the NVU.
We first processed all 101 samples individually, using the same pipeline, removing any cells identified as putative doublets (Wolock et al., 2019) and then identifying distinct clusters of transcriptionally similar cells (Figure 4A, S2A). These clusters were aggregated into “meta-cell” transcriptomes (Methods), representing the average gene expression across the cells in that cluster, allowing us to efficiently identify populations of interest within the larger dataset. Together, this analysis identified 855 meta-cells, which were then analyzed together. We identified highly variable genes across the 855 meta-cells and then used these genes to perform dimensionality reduction and clustering (Figure 4B, S2B). Importantly, the clusters of meta-cells were largely similar on quality measures, such as the number of single cells per meta-cell, and mean number of counts per meta-cell (Figure S2C).
Figure 4. Meta-cell clustering identifies CD19 expression in human neurovascular meta-cells.
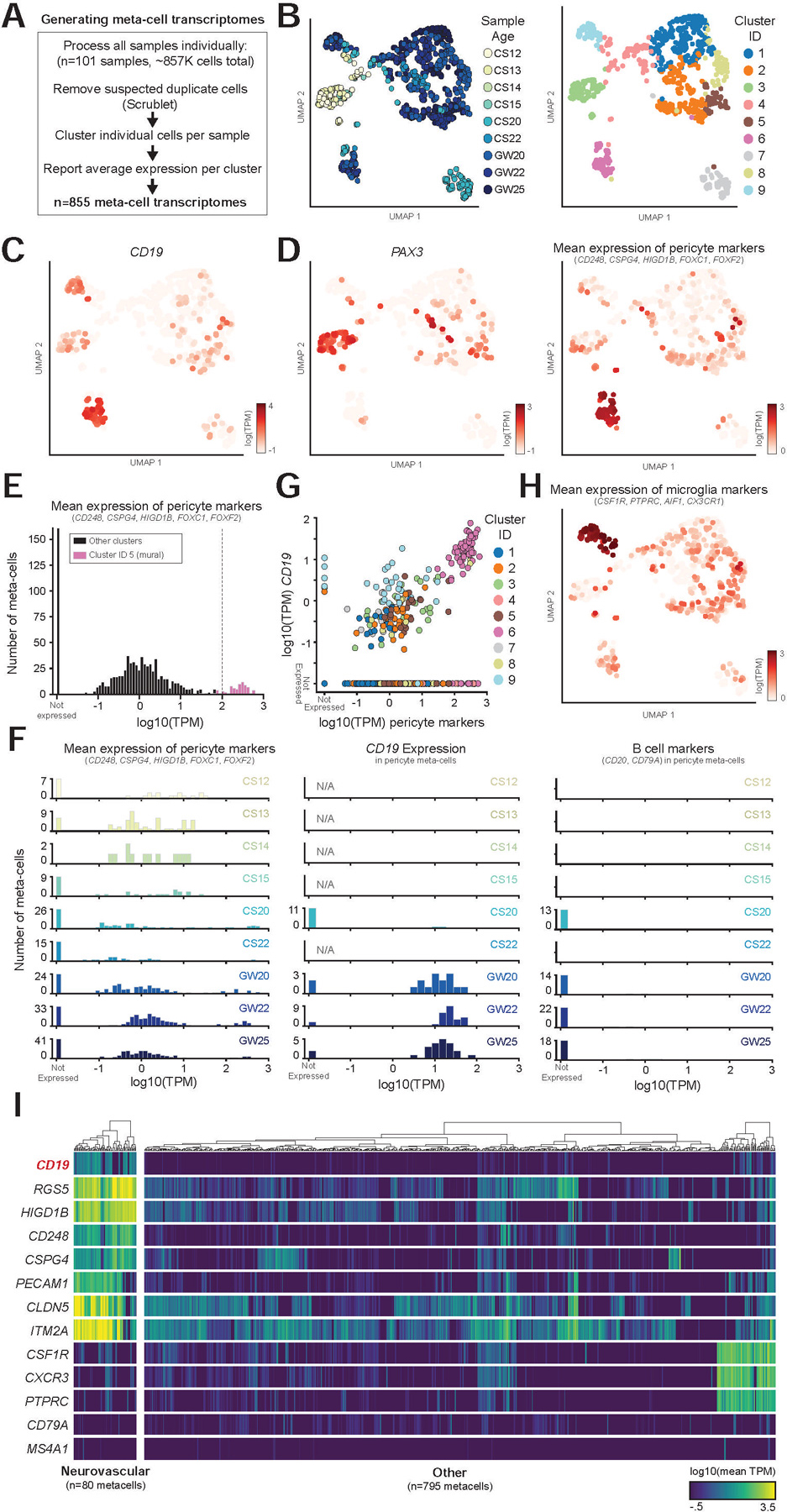
(A) Schematic showing how samples were processed (see Figure S2 for an example of a single dataset). (B) UMAP projection of meta-cells, colored by either sample age or cluster. (C–D) Expression of (c) CD19, (d) PAX3, marking undifferentiated progenitors, and mean expression of indicated pericyte marker genes. (E) Histogram of mean mural cell marker expression across all samples, showing separation of the identified neurovascular cell cluster. (F) Histogram as in (e), but separated by sample age, showing (left) pericyte marker gene expression, (middle) CD19 expression, (right) B cell marker gene expression. Note that CD19, but not B cell markers, are expressed in neurovascular meta-cells. (G) Scatter plot showing the correlation of pericyte marker genes with CD19 expression. Note the separation of the neurovascular cluster. (H) UMAP projection of meta-cells, colored by mean expression of microglia markers. (I) Heatmap showing log10 average TPM values for selected genes across meta-cells. Dendrograms indicate Ward’s hierarchical clustering of each of the two populations shown.
This analysis revealed that CD19 is highly expressed in neurovascular meta-cell clusters, consistent with our prior findings (Figure 4C–D). The meta-cell analysis does not show a clear separation between mural and endothelial cells (or other NVU cell types), since meta-cells may include closely clustering cell types in sparse individual samples. However, this clustering shows clear separation of meta-cells expressing mural markers, and the expression of CD19 and absence of B cell markers in these meta-cells (Figure 4E–F). Additionally, we observed a strong correlation between expression of mural cell marker genes and CD19 across the data (Figure 4G). Low-level CD19 expression was also observed in microglia clusters (Figure 4G–H). However, as mural cells and other cells of the NVU, which include perivascular macrophages, are tightly connected, this expression is likely the result of contamination from mural cells in microglia meta-cells.
Notably, mural meta-cells were not present in early timepoints (CS12-CS15), which had high expression of early developmental markers such as PAX3 and LIN28A (Figure 4D, S2E). In contrast, mural meta-cells at GW25 displayed high and specific expression of mural cell marker genes, such as CD248, RGS5, and FOXF2, in addition to specific expression of CD19 (Figure S2F). In this data, we are able to clearly separate undifferentiated progenitors and differentiated mural cells, which demonstrates that once mural cells emerge, they express CD19 (Figure 4E–F, S2E–F). Collectively, there was clear separation of meta-cells expressing neurovascular markers from all other meta-cells (Figure 4I), which allowed us to identify cells of interest for further analysis.
Pericytes are closely related to vSMCs, which together are the mural cells that line the brain vasculature (Uemura et al., 2020). These cells differ based on anatomical position; pericytes localize around capillaries and vSMCs localize around larger vessels, including arteries, arterioles, and venules. However, these cells are transcriptionally similar, sharing the identity of many marker genes and appearing to exist on a transcriptional lineage continuum (Vanlandewijck et al., 2018). ACTA2, encoding alpha-smooth muscle actin, is a canonical marker used to distinguish these two populations, which is significantly upregulated in vSMCs. Many pericyte markers, however, such as CSPG4 and RGS5, are also highly expressed in vSMCs, causing brain vSMCs to often be annotated as pericytes (Uemura et al., 2020).
We first asked whether the cells we had originally identified as CD19+ mural cells represented pericytes or vSMCs. We aggregated the three datasets from Zhong, 2018, La Manno, 2016, and La Manno, 2018, and analyzed them as a single integrated dataset (Figure S3A). We identified a subset of non-neuronal clusters enriched for mural marker genes (CD248, CSPG4) and confirmed that this population expressed both CD19 and CD81 (Figure S3B–C). We also identified subsets expressing the endothelial and microglial markers CD248 and CSF1R, respectively (Figure S3B). The non-neuronal subset (mural cells, endothelial cells, and microglia) was then re-analyzed and clustered to distinguish between transcriptional differences in cell types of the NVU (Figure 5A–B). We observed that these cells showed strong enrichment of reported pericyte markers, such as ABCC9 and KCNJ8, without enrichment of ACTA2 or other vSMC marker genes, suggesting that they represented bona-fide pericytes (Figure 5C; (Chasseigneaux et al., 2018; Vanlandewijck et al., 2018).
Figure 5. CD19 is expressed in both pericytes and vSMCs.
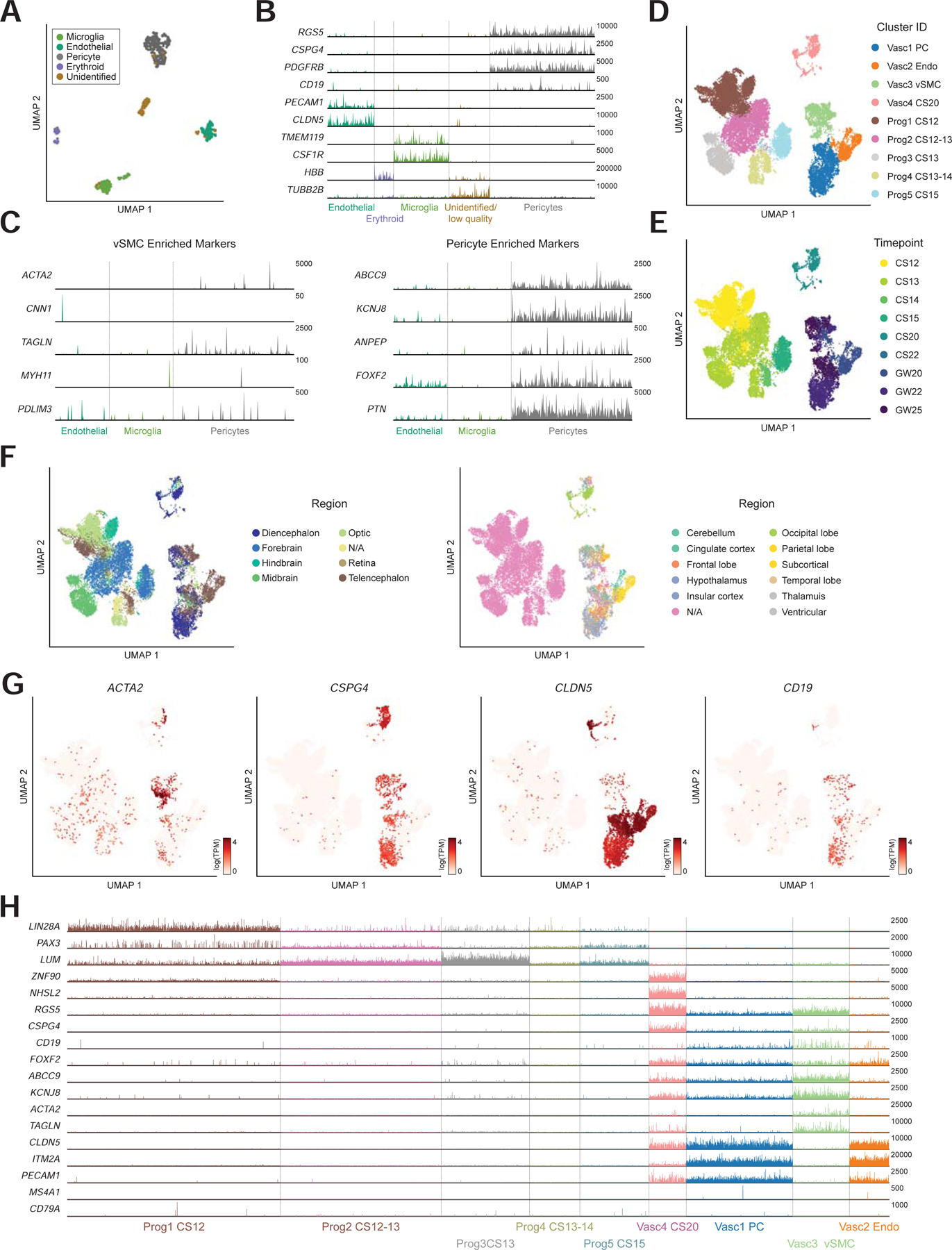
(A) Subset of non-neuronal cells from Zhong 2018, La Manno 2016, La Manno 2018. (B) expression of marker genes used for clustering. Max TPM per gene (y-axis) is indicated. (C) (left) low expression of vSMC marker genes and (right) high expression of pericyte marker genes. Y-axis labels indicate maximum TPM value shown. (D–F) Neurovascular and progenitor subset of BICCN data annotated by (d) cluster ID, (e) timepoint, or (f) region. Note that samples were annotated different levels of regional granularity, so resulting annotations are sometimes overlapping. (G) Expression of vSMC marker (ACTA2) as well as CSPG4 (pericyte) and CLDN5 (endothelial) markers. CD19 is expressed primarily in the vSMC and pericyte clusters. (H) Track plot showing lack of early developmental marker expression and distinguishing markers between vSMC and pericyte clusters. Y-axis labels indicate maximum TPM value shown.
We repeated this analysis using the cells from the BICCN dataset, performing a detailed analysis of neurovascular cells and related progenitors. Using the meta-cells as a guide, we identified ~92K single cells representing the NVU as well as early progenitors (Figure 4B, S3D). We noticed a bifurcation of progenitors from the early timepoints that appeared to be neural lineage-biased, and others that seemed biased toward non-neuronal lineages. The non-neuronal-biased progenitors, as well as the pericyte and endothelial cell clusters were then subset for further analysis (Figure S3E). These ~26K cells showed a clear separation between progenitors from CS12-CS15) and differentiated NVU cells from GW20-GW25 (Figure 5D–E). They were also representative of cells from many brain regions (Figure 5F). Note that samples were annotated at different levels of granularity, such as forebrain versus diencephalon or telencephalon; and only later samples had finer-grained regional resolution. Notably, we identified a smaller vSMC population in this dataset, likely due to the high number of cells in the BICCN data, as pericytes were more abundant than vSMCs (Figure 5G). These cells showed high expression of ACTA2 and TAGLN, as well as other pericyte markers such as RGS5 (Figure 5H). Importantly, CD19 was expressed in the vSMC population as well, consistent with the high transcriptional similarity between these two populations. This suggests that CD19 expression is not unique to pericytes, but a common feature in human brain mural cells, consistent with the staining pattern observed by IHC (Figure 3). We note that further assigning the identity of this vSMC population to venule, arterial, and arteriole sub-populations was challenging since canonical markers identified in mice did not perfectly align with the sub-populations identified in this human dataset (Vanlandewijck et al., 2018), suggesting transcriptional differences between mouse and human mural cells (Figure S3F).
Within the BICCN dataset, we observed a higher degree of endothelial marker gene expression in mural cell clusters, and vice-versa. For example, while endothelial and mural cell clusters are distinguishable, the expression of CLDN5, and endothelial cell marker, and CSPG4, a mural cell marker, is more overlapping than would be expected (Figure 5G). We postulate that is likely due to incomplete separation of these two cell types during tissue dissociation, as cells within the tightly-interacting NVU are challenging to fully dissociate, and cross-contamination is common (Vanlandewijck et al., 2018). However, the analysis of previous datasets, as well as analyses of individual BICCN datasets showed greater separation of neurovascular cells and clearly demonstrated that CD19 expression is primarily in mural cells and not endothelial cells (Figure 1F, 5B, S2A).
The CD19 isoform recognized by CAR-T cells is expressed in the adult human brain
We next sought to analyze the expression of CD19 in the adult human brain to complement the results observed previously by immunohistochemistry. scRNA-seq analysis of mural cells in adult human brain samples is challenging, due to difficulties in tissue acquisition and low numbers of neurovascular cells in the brain, relative to neuronal or glial populations (Figure S3A). Additionally, the majority of existing datasets are prospectively enriched for neuronal populations of interest. Therefore, to ask whether CD19 expression in pericytes is also present in adult samples, we utilized bulk RNA-sequencing data across human age and brain region generated by the Allen Institute Brainspan Project (Miller et al., 2014). This data contains more than 500 prenatal and postnatal samples from diverse brain regions (n=237 prenatal; n=287 postnatal). Since bulk tissue analyzed in different samples will have varying proportions of mural/vascular cells, the relative expression of mural genes across samples is a proxy for the underlying proportion of mural cells in the bulk tissue. For example, CD248 and ANPEP, two pericyte markers, are highly correlated in this data (Figure S4A).
We first confirmed that CD19 is expressed in both prenatal and postnatal samples at similar levels, and also expressed in samples from different brain regions (Figure 6A, Figure S6B). We next performed a genome-wide correlation analysis across only the postnatal samples to identify genes that are co-expressed with CD19. Strikingly, this analysis showed that CD248, CSPG4, ANPEP, FOXS1, and FN1 were in the top 1% of genes correlated with CD19 in this data (Figure 6B–C). Additionally, the top 200 genes most correlated with CD19 were enriched for GO terms associated with the NVU, such as angiogenesis, vasculogenesis, response to fluid shear stress, and multiple extracellular matrix related terms (Figure 6D). This suggests that the expression of CD19 in the adult brain is primarily the result of mural cell abundance, rather than B cell abundance. We then integrated the CD19-correlated gene module in adult brain with scRNA-seq data from the human brain and PBMCs (Methods). This allowed for the comparison of the gene module score (i.e., the enrichment of genes correlated with CD19 or another target) in brain pericytes, endothelial cells, B cells, in addition to other brain cells and PBMCs (Figure S4C). As a control, the gene modules associated with CD22 and CD74, two highly expressed B cell-enriched genes, were enriched in B cells, and the CSPG4 gene module was highly enriched in pericytes (Figure 6E, S4D–E). In contrast, the expression of the CD19-correlated gene module is highest in pericytes (Figure 6E). We noted that CD19 expression seemed to decrease with age (Figure 6A). This could be explained by two potential mechanisms: (1) either the proportion of CD19+ cells changes over time, or (2) the level of CD19 expression in mural cells changes over time. However, the concomitant decrease in CD248 expression with age, as well as prior studies (Erdő et al., 2017; Yang et al., 2020) support the former mechanism.
Figure 6. The CAR-T recognized CD19 isoform is expressed in the adult human brain.
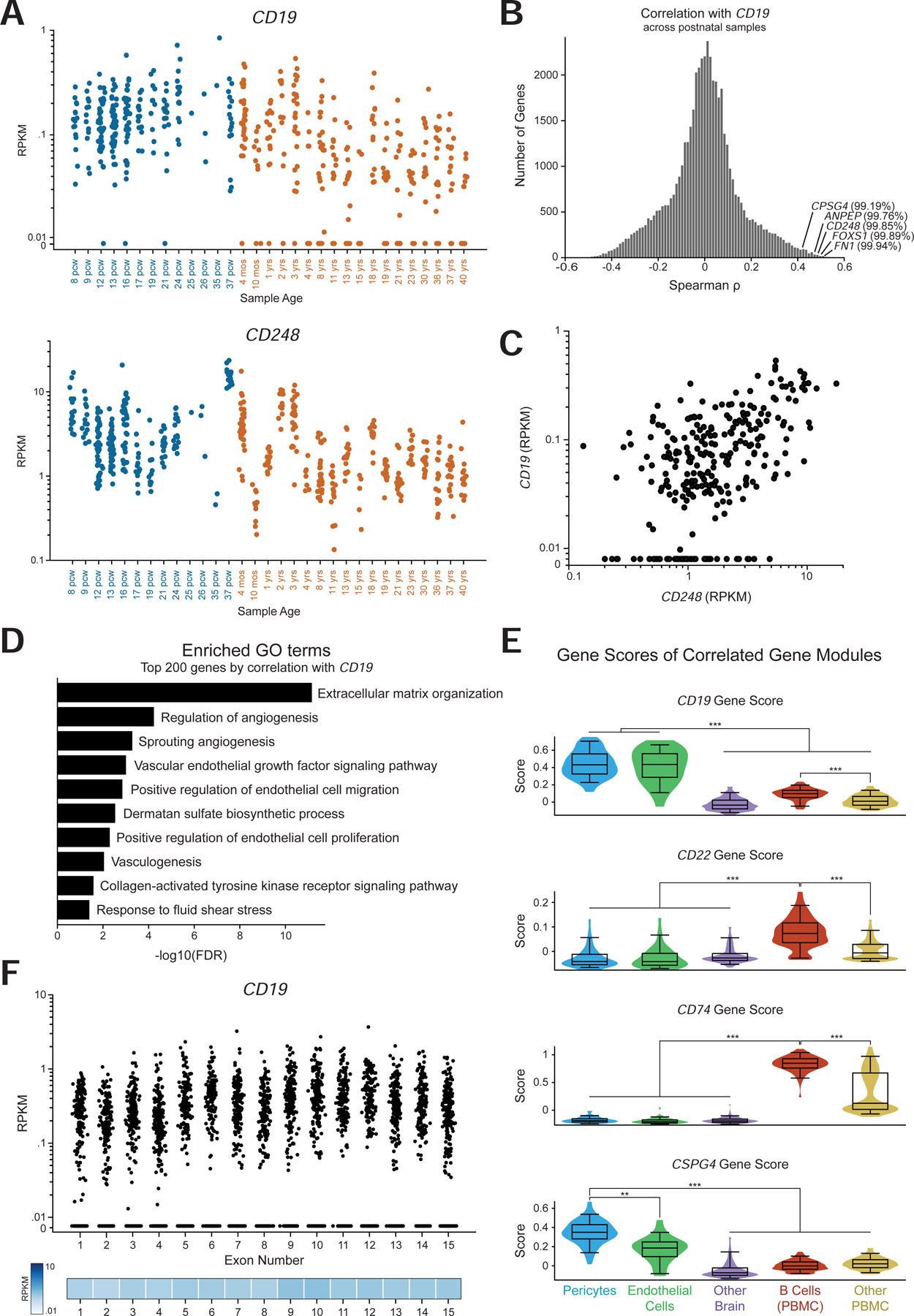
(A) Expression of CD19 (top) and CD248 (bottom) in data. Colors indicate prenatal and postnatal samples, and the different samples from distinct regions but the same age are plotted on the same x coordinate. (B) Histogram of the distribution of spearman correlation values for all genes with CD19 expression in only postnatal samples. The indicated percentiles indicate the percentile of that gene’s correlation. (C) Scatter plot of CD248 against CD19 RPKM values in only postnatal samples. (D) Enriched GO terms in the top 200 genes by spearman correlation with CD19. (E) Gene score distribution in single cells belonging to pericyte or endothelial clusters, as well as other brain cells; along with B cells and other PBMCs. Gene score was calculated with the top 30 genes by spearman correlation. (F) RPKM values per exon of CD19 in the Brainspan data, showing expression of the key exons 2 and 4 for CAR-T cell recognition.
Lastly, we asked whether the CD19 isoform that is expressed in the adult brain contains the specific CD19 epitope that is recognized by clinical CAR-T cells and BiTEs. The FMC63 scFv in clinical use recognizes an epitope encoded by exon 4 of CD19 (Sommermeyer et al., 2017). Additionally, variants skipping exon 2 may also result in a lack of CD19 trafficking to the cell surface, also allowing evasion of FMC63 detection (Sotillo et al., 2015). An analysis of the mean expression of each CD19 exon in the adult human brain showed an even distribution of expression across exons, which clear expression of the key exons 2 and 4 (Figure 6F).
Analysis of neurotoxicity in mouse models of CD19 CAR-T cell therapy
We next asked whether the expression of CD19 in human mural cells is conserved in mice, which would allow for the use of mouse models to study mechanisms of human neurotoxicity. First, we extracted and dissociated whole brains from healthy C57Bl/6J mice following a previously described protocol and analyzed the presence of CD19 by flow cytometry (Boroujerdi et al., 2014). This revealed the presence of a CD45-high CD19+ B cell population, as expected, and also revealed a rare population of CD45-CD19+ cells, which expressed CD19 at a similar level to B cells but did not express the B cell marker B220 (Figure S5A–C). Pericytes, identified as a CD13+CD45-population, were indeed positive for CD19, albeit rarely, in contrast with what was observed in human data (Figure S5D). To examine this at the transcriptional level, we performed scRNA-seq on cells isolated from dissociated whole mouse brain, followed by enrichment for CD19+, CD13+, and CD31+ cells. This analysis again confirmed low Cd19 expression in pericytes relative to CD19+ B cells, and in fewer cells compared to scRNA-seq of human mural cells (Figure S5E). Together, this data suggests that CD19+ mural cells are relatively less abundant in the mouse brain, compared to human brain.
Given the presence of non-B CD19+ cells in mice, we asked whether an infusion of CD19-directed CAR-T cells into immunodeficient, non-tumor bearing NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice would result in an observable neurologic phenotype. Since NSG mice do not develop B cells, any phenotype observed in this model is B cell independent and cannot be attributed to global effects related to CRS following B cell targeting. We generated either CD28 (28)-based or 4–1BB (BB)-based CAR-T cells, which differ in the signaling domain but are both used clinically, targeting either human (FMC63 scFv) or mouse CD19 (1D3 scFv, the same as used for flow cytometry analysis above). This comprised three conditions: hCD19BBz, representing a negative control that should not target murine CD19, mCD19BBz, and mCD1928z. We confirmed species-specific activation using human and murine CD19+ B-ALL cell lines (Figure S6A).
Seven days after infusion of CAR-T cells, NSG mice that had received murine-targeting, but not human-targeting, CD19 CAR-T cells displayed increased BBB permeability as measured by Evans Blue dye (EBD; Figure S6B–D). EBD binds to albumin, which normally remains in the bloodstream under physiologic conditions; when the BBB is disrupted, albumin is able to cross into the brain parenchyma. A fourth group of mice receiving no transfusion displayed no EBD staining, and a fifth group that received mannitol at the time of EBD infusion, representing a positive control, displayed high EBD staining, as expected (Figure S6C–D). The effect was higher in the mCD1928z compared to mCD19BBz CAR-T cell group, which is likely the result of stronger antigen receptor signaling provided by the CD28 domain (Salter et al., 2018). Indeed, prior studies have shown that the choice of costimulatory domain (CD28, 4–1BB, or others) strongly impacts sensitivity to antigen density with CD28-based CARs particularly sensitive to low levels of antigen density (Majzner et al., 2020). This experiment was repeated using a syngeneic, immunocompetent C57Bl/7J model with pretreatment with cyclophosphamide as a lymphodepleting preconditioning regimen. The syngeneic study recapitulated the pattern of BBB permeability observed in the NSG model, suggesting that the presence of murine CD19+ B cells in the syngeneic model did not affect the specific disruption of the BBB observed only in the murine-targeting conditions (Figure S6E–F). We also measured BBB integrity four days after CAR-T cell transfusion in NSG mice by measuring gadodiamide uptake using 9.4 tesla magnetic resonance imaging. This showed an increase in uptake in the mice that had received murine-targeting, but not human-targeting, CD19 CAR-T cells (Figure S6G–H). Finally, we dissociated brains from treated mice and performed flow cytometry to measure CD19+ pericyte depletion following CAR infusion. Mice receiving mCD1928z, but not hCD1928z, showed a decrease in both CD45+CD19+ B cells as well as CD45-CD13+CD19+ mural cells (Figure S6I). We did not specifically analyze any behavioral changes, but no overt behavioral phenotype was observed.
In summary, these experiments suggest that despite a lower frequency of CD19 expression in mouse mural cells, compared to human mural cells, administration of CD19-directed CAR-T cells can cause BBB leakiness and pericyte depletion in mice lacking B cells. It is possible that some of this effect may be mediated by targeting of microglia, or other CD19-expressing cell types not yet identified. Microglia were previously demonstrated to be depleted following CD19-directed CAR-T cell treatment in mice (Pennell et al., 2018), and also display low expression of Cd19 in our scRNA-seq data, as well as the Tabula Muris database (Tabula Muris Consortium et al., 2018). Therefore, these experiments demonstrate that while some aspects of CAR-T function and toxicity may be measured in preclinical mouse models, there are important limitations, since species-specific differences in cell type and transcriptional state may not perfectly match human-specific pathophysiology. Indeed, initial CAR-T cells studies in mouse models did not predict the degree of neurotoxicity that was later observed in human clinical trials (Giavridis et al., 2018; Ruella and June, 2018).
Human brain mural-specific expression of CD19
Since pericytes are present in multiple organs, we performed a comparative analysis of brain pericytes with pericytes and vSMCs from the lung, a highly vascularized tissue with high numbers of pericytes and endothelial cells (Travaglini et al., 2020). Although all mural cell populations showed shared expression of a core transcriptional identity, such as PDGFRB, RGS5, FOXS1, and KCNJ8, we identified numerous transcriptional differences between brain and lung pericytes (Figure 7A, S7A). Notably, CD19 is specifically expressed in brain, but not lung mural cells. Only 6/2724 lung mural cells had any detectable counts for CD19, showing no apparent enrichment over nonspecific counts found in all non-B cell clusters. This organ specificity may be explained by broader differences in lineage-specific transcription factors between the two cell types, which agrees with the distinct developmental origins of brain pericytes from neural crest cells (Etchevers et al., 2001). Namely, brain pericytes express BCL11A, and lowly express PAX5, two key developmental factors in brain development but also B cell development (Kozmik et al., 1992; Urbánek et al., 1997; Liu et al., 2003; Dias et al., 2016). Interestingly, EBF1, another transcription factor important for B cell development, was expressed by both populations. The expression of these B cell factors in brain mural cells may explain the specific expression of CD19 expression in these cells.
Figure 7. Brain pericyte-specific expression of CD19.
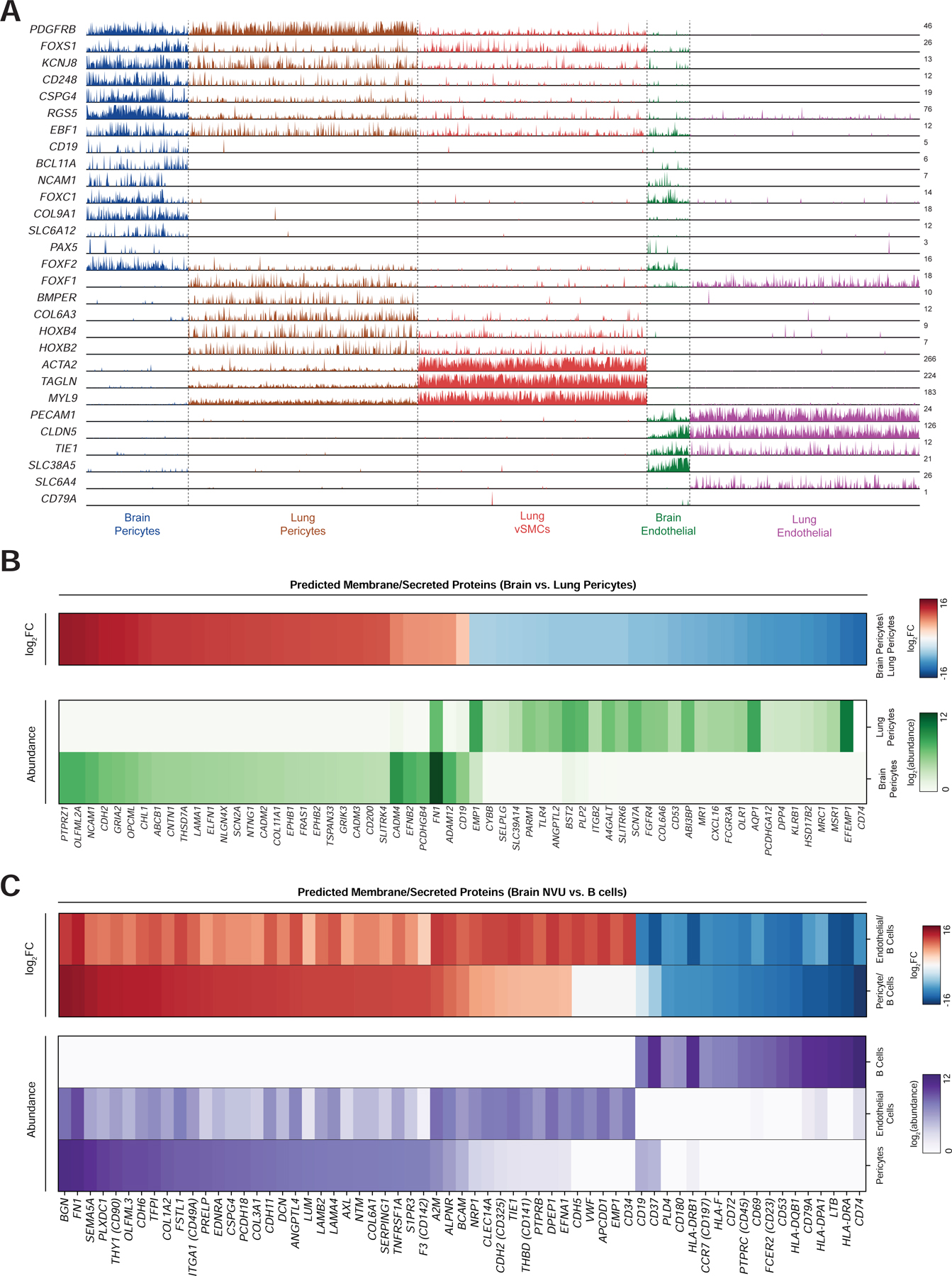
(A) Track plot showing expression of selected marker genes for each population. Note that CD19 expression is limited to brain pericytes, but not lung pericytes. Additionally, brain pericytes express certain transcription factors, such as BCL11A, that are enriched in B cells. Y-axis labels indicate maximum TPM value. (B) (top) heatmap of log2 fold-change in gene expression of surface/secreted genes between brain and lung pericytes. (bottom) heatmap of expression of the same genes. Abundance data has been quantile-normalized to improve comparison of relative expression between the two populations. (C) heatmaps showing (top) log2 fold-change and (bottom) quantile-normalized abundance, as in (b). Comparisons are made between brain pericytes, brain endothelial cells, and B cells (from PBMCs), and genes are ordered by log2 fold-change between pericytes and B cells.
Finally, we asked whether genes predicted to localize to the membrane or cell surface were differentially expressed in the NVU relative to lung pericytes as well as B cells and could be used to improve the safety of CD19-directed CAR-T (Figure 7B–C). For example, recent studies have shown the utility of “AND” logic gates to improve the specificity of CAR-T cells by requiring recognition of two distinct antigens (Roybal et al., 2016). We identified the most highly differentially expressed genes among those annotated as being secreted or located on the cell surface, in brain versus lung pericytes, as well as B cells versus brain pericytes and endothelial cells (Bausch-Fluck et al., 2015; Zheng et al., 2017). These could improve the specificity of B cell-directed CAR-T cells by requiring dual recognition of two antigens. This analysis revealed additional genes, such as CD74, HLA-DRA, and LTP, which are both highly expressed and highly enriched on B cells relative to brain pericytes (Figure 7C). An alternative approach to improving CAR-T cell specificity is to use “NOT” gates, where inhibitory CAR constructs recognize non-specific target proteins and prevent T cell activation (Fedorov et al., 2013). We identified numerous genes that are enriched in pericytes over B cells, such as BGN, FN1, and SEMA5A (Figure 7C).
Discussion
CD19-directed immunotherapies, such as CAR-T cells and BiTEs, have been highly effective in patients with B cell malignancies, achieving clinical remission in up to 90% of patients in some indications (June et al., 2018). However, neurotoxicity remains a major adverse effect experienced by these patients, which in some cases can lead to cerebral edema and death. Here, we describe a possible on-target mechanism for CD19 CAR-T cell mediated neurotoxicity, which may be caused by the previously unrecognized expression of CD19 in mural cells in the human brain. We show that CD19 expression is present across multiple independent datasets and present in pericytes as well as vSMCs. This finding was confirmed by IHC in adult tissue and by an analysis of a large database of RNA-sequencing data from adult samples from different ages and brain regions. Importantly, the CD19 isoform expressed in the adult brain contains the FMC63 epitope that is recognized by clinical-grade CD19 CAR-T cells and BiTEs. Surprisingly, we found that CD19 expression in mural cells is significantly higher in human than in mouse, highlighting the importance of studying these adverse effects in the context of human tissues. It is important to state that we are not advocating for the discontinuation of CD19-directed therapy use in cancer patients, particularly since acute neurotoxic adverse effects can be effectively managed in most cases, and CD19-directed immunotherapies are tremendously effective treatments for B cell cancers (Neelapu, 2019). However, these findings do provide a strong impetus for further investigation of mural cell targeting in the context of human clinical trials, as well as potential strategies to develop the next generation of therapies with improved safety profiles.
Previous studies in patients who received CD19 CAR-T therapy and had neurologic adverse reactions demonstrated edema, multifocal hemorrhage, and vascular disruption in the brain, and neuropathologic evaluation of the brain from patients who developed fatal CRS and/or neurotoxicity revealed CAR-T cell infiltration (Gust et al., 2017; Torre et al., 2018). These findings are consistent with a role of mural cells in mediating neurotoxicity; namely, CD19 CAR-T cell-mediated depletion of pericytes and vSMCs may lead to endothelial activation, disruption of tight junctions, increased BBB permeability, and cerebral edema. However, a caveat of the present study is that we have not determined the precise contributions or synergistic effects of mural cell death and CRS in this process. For example, vascular permeability changes related to CRS might increase permeability across the BBB and increase or enable CAR-T cell access to CD19-expressing mural cells. Along these lines, the CAR signaling motif also appears to play an important role in CAR-T cell-induced neurotoxicity; fatal cases of cerebral edemas have occurred predominantly in patients receiving CAR-T cells with the CD28 signaling domain (Table S1). This may be due to: 1) higher levels of cytokine release and CRS due to stronger antigen-receptor signaling (Ying et al., 2019), 2) increased sensitivity to low CD19 antigen density (Majzner et al., 2020), 3) higher cytolytic activity, or 4) a combination of these factors.
In addition to CAR-T cell-intrinsic effects, the clinical manifestation of severe neurotoxicity is likely a complex multifactorial process, where the lymphodepleting or chemotherapy regimen, scFv specificity (e.g., SJ25C1 of JCAR015 vs. FMC63 of tisagenlecleucel and axicabtagene ciloleucel), and co-localization of on- and off-target cells may all play a contributing role. For example, these findings may caution the use of intrathecal or intraventricular administration of anti-CD19 CAR-T cells for refractory primary CNS high-grade lymphoma where CAR-T cell activation could be triggered not only by the tumor itself but also by neighboring brain mural cells. Similarly, patient populations that are already at risk for encephalopathy after chemotherapy or whole brain radiotherapy may also be particularly vulnerable to neurotoxicity (Korfel and Schlegel, 2013). Future studies should investigate the connection between neurotoxicity associated with CD19 CAR-T cells and with chemotherapy agents such as fludarabine (Lowe et al., 2018), as well as any potential effects that system inflammation observed during CRS has on CD19 expression in mural cells.
There are several important limitations to our study. First, we have not demonstrated that mural cell CD19 expression is the cause of clinical neurotoxicity, and further studies are needed to determine whether CD19 CAR-T cells target and deplete human mural cells in patients, and whether this effect is independent or correlated with CRS. Second, it will be important to investigate whether there are inter-patient differences in mural cell frequency and/or CD19 expression in mural cells (for example, age-related) that could lead to differences in the incidence of neurotoxicity. Third, the IHC data suggests some degree of heterogeneity in CD19 expression, as not all vessels were stained positive, and as mentioned above, inter-patient differences in CD19 mural cell expression may contribute to differences in patient outcome. Neurovascular cell heterogeneity in humans remains poorly understood and will be important to study further (Faal et al., 2019, Uemura et al., 2020). Finally, while we have focused our discussion on the immediate clinical effects of mural cell depletion, there may also be long-term effects associated with targeting of these cells, which will require further studies in longitudinal patient cohorts.
The scRNA-seq data that we have presented here was obtained from prenatal human samples. We focused primarily on human data due to observed differences in CD19 expression between human and mouse (La Manno, Cell, 2016). Current data of the human brain is largely biased toward early or elderly timepoints, and there is a vast gulf of information missing on pediatric and middle-aged individuals. Thus, while our analysis clearly shows CD19+ mural cells in prenatal brain samples, and the analysis of bulk RNA-seq data shows that CD19+ expression persists throughout adulthood, future studies are needed to fully interrogate whether CD19 expression changes as a function of age or region. The majority of publicly-available adult human scRNA-seq data was sort-enriched for neuronal or other specific populations and therefore lack abundant mural cell populations to conclusively analyze. Two recently-published adult datasets do show a small mural cell population and include CD19+ mural cells (Hodge et al., 2019; Kim et al., 2020), but further datasets are needed due to the low numbers of mural cells and/or sparse expression of mural cell marker genes in existing adult data. Whether the comparatively low numbers of mural cells in adult datasets is due to technical challenges, such as sampling frequencies and/or loss of cells during dissociation, or biological differences associated with aging, and how this relates to the levels of CD19 expression observed in prenatal samples, is an important question for future study. Nonetheless, there is clear evidence that CD19 expression is present in brain mural cells in at least some timepoints and cases, which has thus far been overlooked. We hope that this finding will inform future studies in human samples, including in patients experiencing neurotoxicity.
More broadly, this research highlights one of many utilities for developing a comprehensive human single-cell atlas for clinical medicine (Regev et al., 2017). scRNA-seq is an unbiased, genome-wide measurement of gene expression that can capture even rare populations of cells. These rare cell types might otherwise be missed in measurements of bulk tissue due to their low frequency, but as we now demonstrate, could be critically important in the clinical effects of targeted therapy. While current CAR-T cells recognize only a single antigen, future generations of CAR-T cells may be able to discriminate between unique combinations of target antigens to improve their cell-type specificity. This may be particularly useful as CAR-T cells are increasingly deployed against solid tumors, which share most antigens with normal tissues. We envision that a comprehensive database of gene expression across all human cell types will enable the precise identification of cell type-specific target antigens which can be used to design safe and effective cellular immunotherapies. We hope that the analytical framework of comparing on-target and potential off-target cell types aids the future development of immunotherapies.
STAR Methods
Resource Availability
Lead Contact
Further information and requests for resources, data, and details of analysis should be directed to and will be fulfilled by the Lead Contact, Ansuman T. Satpathy (satpathy@stanford.edu).
Materials Availability
The anti-hCD19-BBz CAR was constructed as described previously (Milone et al., 2009). The mCD19-BBz and mCD19–28z CARs were constructed by ligating the mCD19 scFv (1D3) into the CAR backbone sequences of pTRPE –BBz and pTRPE – 28z. Third generation lentiviral vectors were produced as previously described (Milone et al., 2009).
Data and Code Availability
Raw data for mouse single-cell RNA-sequencing has been deposited in GEO and is available under accession number GSE153647. Details of analysis are provided below in the Methods, and any further questions should be directed to and will be fulfilled by the Lead Contact.
Processed sequencing data (gene counts per cell) were downloaded as follows: Zhong et al., 2018 (GEO GSE104276), La Manno et al., 2016 (GEO GSE76381), La Manno et al., 2018 (PanglaoDB database, Karolinska Institute, https://panglaodb.se/samples.html), Kriegstein/BICCN (NeMO archive, https://nemoarchive.org/), Brainspan (Allen Institute, https://www.brainspan.org/static/download.html). PBMC data was downloaded from the 10x Genomics reference datasets available on their website (https://support.10xgenomics.com/single-cell-gene-expression/datasets/3.0.0/pbmc_10k_v3). UMI counts for human lung (Travaglini et al., 2020) were downloaded from Synapse, accession syn21041850.
Experimental Model and Subject Details
Cell lines and cultures
The murine A-20 B cell leukemia cell line and human Nalm6 line were kindly provided by Dr. Jong Lee (University of Pennsylvania). Nalm6 and A20 tumor cell lines were cultured in RPMI-1640 media supplemented with 10% FBS, penicillin (100 U/mL; Gibco), streptomycin sulfate (100 mg/mL; Gibco), and 10 mmol/L HEPES (Gibco) (R10). 50 µM of 2-mercaptoethanol was added in the culture media of A20 cell line.
In vitro T cell transduction and cultures
Human CAR-T cells were produced from normal donor T cells provided by the University of Pennsylvania Human Immunology Core, as previously described (Milone et al., 2009). Cells were transduced with lentiviral vector encoding anti-human (h) or murine (m) CD19 scFv fused to CAR backbones containing either human 4–1BB or human CD28 and CD3zeta (CD247) signaling domains, as described and were expanded ex vivo for 11 days (Milone et al., 2009). Two cell expansions were produced from 2 different healthy human donors. The transduction efficiencies ranged from 20 to 40%.
Mouse models
All animal studies adhered to the NIH Guide for the Care and Use of Laboratory Animals and in accordance with protocols approved by The University of Pennsylvania Institutional Animal Care and Use Committee. NOD/scid/IL2rg (NSG) mice were obtained from the University of Pennsylvania Stem Cell and Xenograft Core and housed in the vivarium at the University of Pennsylvania under BSL2 and pathogen-free conditions. 1×107 T cells with 20% normalized CAR+ expression were administered to non-tumor bearing mice by one intravenous tail vein injection in 100µL of PBS. Adult mice of both sexes were used for experiments. For other experiments not using NSG mice, adult healthy C57Bl/6J mice of both sexes were obtained from Jackson Laboratory and housed in a BSL1 mouse facility in accordance with University protocols.
Method Details
Processing of human brain single-cell RNA-sequencing data and identification of CD19+ mural cells.
Sequence data were downloaded as described in Data and Code Availability. For data shown in Figures 1 and 2, samples were processed using Seurat (Butler et al., 2018) version 2.3.4. For data shown in Figures 4–7, samples were processed using Scanpy version 1.4.3 (Wolf et al., 2018) or Seurat version 3.1.4.
To process data from Zhong et al. 2018, La Manno et al. 2016, and La Manno et al. 2018, cells with fewer than 500 detected genes or UMI counts were excluded, and cell counts were normalized per cell using log(depth-normalized counts + 1). The ~1500–2500 most variable genes were used for clustering based on the variance to mean ratio. As the datasets include both post-mitotic and actively cycling cells, the cell cycle status was computed using the CellCycleScoring function and subsequently regressed out using the ScaleData function in Seurat. Principle component analysis was performed using the genes identified as highly variable for each dataset, and the top ~25–50 principle components were used for subsequent dimensionality reduction using the UMAP algorithm. Clusters were called using the FindClusters function 146 in Seurat, and marker genes for each cluster were identified using the FindMarkers function. Clusters were subsequently manually annotated by comparing highly enriched genes to known cell-type markers. For the analysis of Zhong et al. 2018 data, neuronal precursor cells, erythroid cells, and neuronal cells were identified and excluded, and the remaining cells were subsequently re-clustered. Gene expression data shown in Figure 1G and 2C represents mean expression across all cells with a given cluster label. Percentages and cell counts of a given label (e.g., pericytes) represent the total number of cells in a given cluster, not necessarily only those cells positive for an individual marker, unless otherwise indicated.
Generation of human brain meta-cells from Kriegstein/BICCN data
To identify meta-cells in the data from the Kriegstein lab/BICCN data, the processed UMI counts were first analyzed using Scrublet on a per-sample basis to identify potential doublets. The following parameters were used: expected_doublet_ratio=0.08, sim_doublet_ratio=2, min_counts=3, min_cells=3, min_gene_variability_pct1=75, n_prin_comps=30. A consistent threshold for doublet calling was set at 0.35 across all samples. The results of this were then saved and imported alongside the UMI counts into Seurat. Samples were all processed individually with the same pipeline, using the same parameters and clustering resolution. As a result, some samples may be under-clustered, as the clustering resolution was intentionally set to err on the side of merging similar cells rather than creating false positive clusters. Only cells with > 250 and < 4000 features were included, as well as less than 20% mitochondrial reads. A cell cycle difference score was calculated following the Seurat vignette. The SCTransform workflow was used, and percent mitochondrial reads, number of genes detected, and cell cycle difference were included as variables to regress out. 30 components were used for UMAP dimensionality reduction, with a random seed of 1, and n.neighbors=100. 30 components were used for FindNeighbors, and a resolution of .2 was used for FindClusters. Subsequently, predicted doublets were excluded, and the average depth-normalized expression of each cluster was calculated using the AverageExpression function. This information, representing the meta-cell transcriptome, alongside the individual cell metadata (e.g., cell barcode, cluster ID, UMAP coordinates, etc.), and the meta-cell metadata (e.g., number of cells per meta-cell, mean number of genes per cell for each meta-cell, etc.) was saved for each sample. A representative example of this output is shown in Figure S2A.
This information was then imported into Python and merged, to create a metacell x genes matrix, with each value representing mean(depth-normalized UMI counts). Variable genes were identified by fitting an exponential function to the log(mean) versus log(coefficient of variation) for each gene across meta-cells. The top 6000 genes, representing ~20%, based on distance from the fit line, were used for PCA. Data was log transformed, then z-normalized, prior to PCA. The scikit-learn decomposition.PCA() function was used with the svd_solver=‘arpack’. Then, the top 30 PCs were used as input to the UMAP reducer in the umap-learn package with the following parameters: n_neighbors=80, n_components=2, min_dist=.25, transform_seed=50, n_epochs=500. Last, the top 30 PCs were used as input to KMeans clustering in the scikit-learn package, with k=9.
Analysis of vSMC vs pericyte identity
To generate the integrated dataset shown in Figures 5A–C and S3A–C, the data from Zhong 2018, La Manno 2016, and La Manno 2018 was first processed using Scrublet to identify doublets with the following paramters: expected_doublet_rate=0.06, min_counts=2, min_cells=3, min_gene_variability_pct1=85, n_prin_comps=30. A threshold of 0.25 was used for doublet calling. The doublet information along with expression information was then imported into Seurat. Individual Seurat objects were created, using only the union of genes annotated in all three datasets (to account for small differences in upstream sample processing), and only including cells with at least 500 features detected. Cell cycle scoring was performed as described above, and each sample was processed using SCTransform with vars.to.regress = nFeature_RNA and percent.mt. 3000 features for integration were then chosen using the SelectIntegrationFeatures function, followed by PrepSCTIntegration, FindIntegrationAnchors, and IntegrateData using default parameters. For RunUMAP, the following parameters were used: dims=1:30, seed.use=20, min.dist=.3, spread=1. 50 PCs were used as input for FindNeighbors and resolution=1 was used for FindClusters. This output is shown in Figure S3A.
The subset of cells that were in clusters expressing pericyte/vSMC genes (RGS5, CSPG4, FOXF2, ACTA2), endothelial genes (PECAM1) or microglial genes (CSF1R) were included for downstream analysis. Predicted doublets were excluded from the cells falling into these clusters. These samples were re-processed using the same paramters for RunUMAP, FindNeighbors, and FindClusters as above, with the exception of dims=1:30 for FindNeighbors and resolution=1 for FindClusters.
To generate the dataset shown in Figures 5D–H and S3D–F, meta-cells from cluster ID=3, 4, 6, or 9 were included. The ~92K single cells that comprised these meta-cells were identified and imported into Scanpy. These were first processed, excluding cells with fewer than 500 genes, genes detected in fewer than 3 cells, and cells with more than 10% mitochondrial reads. Doublets had already been excluded in the generation of meta-cells, and so all cells analyzed here were not predicted doublets. Variable genes were identified with min_mean=0.0125, max_mean=3, min-dist=0.5. n_counts was regressed out, and max_value=10 was used for pp.scale(). 50 PCs were calculated and used for the pp.neighbors() calculation; default paramters were used for tl.umap() and resolution=0.65 was used for tl.leiden(). This output is shown in Figure S3D.
Then, clusters identified as pericytes/vSMCs/endothelial cells, as well as the subset of progenitor (early timepoints) cells that were SOX2 low and showed scattered expression of pericyte markers were analyzed. This comprised ~26K cells. These samples were re-processed using the same workflow, unless noted here: maximum percent_mito=8%, resolution=.25 for tl.leiden().
Analysis of Brainspan bulk RNA-sequencing data and integration of correlated gene sets with single-cell data
RPKM data processed at the gene- and exon-level was downloaded from the Allen Institute portal. For correlation analysis, spearman correlation was used. The top 200 genes by spearman correlation were analyzed using the PANTHER web tool.
PBMC data was integrated with the data from the Zhong 2018, La Manno 2016, and La Manno 2018 datasets. The BICCN data was not included due to the less clear separation between pericytes and endothelial cells observed in this data, presumably due to incomplete dissociation of the tightly interacting cell types leading to a high rate of false-positive endothelial transcripts being present in pericyte cells and vice-versa. These samples were merged and processed using Scanpy using join=outer. Cells with greater than 20% mitochondrial genes were excluded. To calculate a gene score, the top 30 genes identified based on spearman correlation in the Brainspan data were used as inputs to the tl.score_genes() function in Scanpy, with the following parameters: ctrl_size=50, random_state=1, n_bins=10. The ttest_ind() function in scipy.stats was used to calculate p-values for gene scores between the different clusters.
Processing of human lung single-cell RNA-sequencing data and comparison of gene expression with human brain mural cells
Lung data was first processed using Seurat to identify clusters with high expression of pericyte and endothelial marker genes. These clusters, which included ~19K cells, were then merged with the brain data from Zhong 2018, La Manno 2016, and La Manno 2018. The BICCN data was excluded for the same reason as above. These cells were then processed together to allow identification of the large vSMC, pericyte, and endothelial cell clusters in the lung data. The annotations of pericyte/endothelial cell identity were carried over from the above merged analysis of the three datasets described in the analysis of vSMC vs. pericyte identity of these datasets (Figures 5A–C). Diffxpy was used to perform differential gene expression testing between the different pairwise comparisons of interest (e.g., brain vs. lung pericytes). Due to the vast difference in the number of lung pericytes and endothelial cells versus brain cells, the lung data was sub-sampled to match the same number of cells in the respective brain cluster being analyzed in order to avoid confounding effects due to differences in cell number (i.e., the lung data had greater power to detect lowly-expressed genes).
Comparison of gene expression profiles of human brain mural cells with peripheral blood B cells
Single-cell gene expression data from B cells was generated from the PBMC data described in Data and Code Availability. A database of extracellular proteins based on mass spectrometry data was used for the analysis of putative cell-surface proteins (Bausch-Fluck et al., 2015). This database includes proteins that may be secreted and thus not strongly enriched at the cell surface, but in the interest of not excluding proteins that might be at the cell surface, this more comprehensive database was used. Gene expression was computed as the mean across all single B cells. To account for differences in the expected distribution of mean gene expression due to the inherent sparsity of single-cell RNA expression data, which is biased based on cell capture and library preparation technique, as well as sequencing depth, the distribution of gene expression values were quantile normalized using the normalize.quantiles function in R to facilitate comparisons across cell types. As such, the absolute gene expression values are pseudo-arbitrary, in that they have been transformed to be relatively comparable across different cell types from distinct single-cell sequencing techniques.
Generation of mouse single-cell RNA-sequencing data
Four mouse brains were dissected and dissociated following an established protocol (Lin, 2018). Briefly, whole brain was minced using a razor blade, and all tissue (not following the note to remove vessels as in the protocol) was pooled and transferred into a tube filled with an enzymatic dissociation solution containing 250 ug/mL STEMxyme I. This was incubated at 37 degrees for 30 minutes, then triturated, and filtered using a sucrose gradient to remove myelin debris. Red blood cell lysis was performed with ACK lysis buffer. The resulting cells were then resuspended in FACS buffer (HBSS without Ca/Mg, supplemented with 1% BSA), blocked at RT for 10 minutes using BioLegend TruStaing FcX (#101320), then stained for 20 minutes on ice with the following antibodies: CD19 (BioLegend 115512, diluted 1:100), CD13 (BD 558745, diluted 1:20), CD31 (BioLegend 102421, diluted 1:50), and CD45 (BioLegend 103113, diluted 1:100). CD19+, CD31+, and CD13+ cells were enriched by flow sorting prior to capture using the 10x Genomics 5’ scRNA kit following the manufacturer protocol. Libraries were sequenced using a HiSeq 2500.
Analysis of mouse single-cell RNA-sequencing data
Data was processed using CellRanger v3.0.2 using the prebuilt mm10 index v3.0.0. The resulting files were then processed using Scrublet to remove suspected doublets with the following settings: miin_counts=2, min_cells=3, min_gene_variability_pct1=85, n_prin_comps=30, threshold=0.25. Doublets were then excluded, and the remaining data was then processed using Scanpy, excluding cells with less than 250 genes or more than 10% mitochondrial reads. Variable genes were selected with the following parameters: min_mean=0.0125, max_mean=3, min_disp=0.5. 30 PCs were used for pp.neighbors(), and data was clustered using tl.leiden() with resolution=0.15.
Flow cytometry of mouse brain
For the mouse CNS neurovascular fraction characterization experiments, the following antibodies against mouse molecules were used: anti-mouse CD19 (clone 6D5 0.1mg/ml, BV650, Biolegend, dilution 1to 200), CD45 (clone 30-F11, APC-Fire750 Biolegend, dilution 1 to 100), CD31 (clone 390, PerCP-Cy5.5, Biolegend, dilution 1 to 400), B220 (clone RA3–6B2, PE/Dazzle 594 Biolegend, dilution 1 to 200), Zombie Aqua Fixable Viaiblity kit (dilution 1 to 400). Isolation and flow cytometric analysis of CNS perivascular cells was performed according to Crouch et. al, with modifications to digestion volumes to accommodate whole murine brain rather than microregions. Antibody staining was done using CD13-PE (BD Biosciences, cat. No. 558745), CD31-PECy7 (BD Biosciences, clone 390, cat no. 102418), CD41-APC (Biolegend, cat. no. 133914), CD45-APC (Biolegend, clone 30-F11, cat no.103112), CD19-BV421 (BD Biosciences, clone 1D3, cat. no. 562701), and BV421 Rat IgG2a κ isotype control (BD Biosciences, cat. no. 400549). Dead cells were excluded with 7-AAD (Biolegend, 420404). Fluorescence minus one (FMO) controls were used for each maker to define gates for each population. Samples were acquired on a BD 5-laser LSRFortessa or 4-laser BD Aria II. Note: epitope cleavage of CD13 and CD19 and loss of cell viability were observed with harsher digestion protocols using papain or higher concentrations of collagenase/dispase. Species-matched isotype controls were performed for each primary antibody to ensure the absence of non-specific binding. We also used a fluorophore minus one (FMO) control for each individual marker staining, to define gates for each population. An important challenge for these perivascular cell suspension preparations is the cell stress during dissociation due to trituration and digestion by enzymes, both known to impact cell viability and cause the low cell numbers we observed in our experiments. The dissociation itself is also known to cause significant cell surface marker epitope cleavage. We observed that CD13 and CD19 expression was present but variable according to the enzyme used (papain, collagenase, or hyaluronidase) for digestion.
Human T cell viability was determined by staining with LIVE/DEAD Fixable Violet Dead Cell Stain Kit (Invitrogen), followed by surface antibody staining. CAR expression was detected using biotinylated protein L (Genscript) followed by R-phycoerythrin-conjugated streptavidin (BD cat. #554061). All experiments were acquired on a BD LSR Fortessa flow cytometer (Becton Dickinson) and data was analyzed with FlowJo v10. s
Cytotoxicity assays
All cytotoxicity assays were flow cytometry-based and adapted from the Quah et al. protocol(Quah et al., 2007). Target cells were stained with CFSE (Thermo Fisher) and T cell effectors were stained with Cell Trace Far Red (Thermo Fisher) prior to co-culture. 24 hours after co-culture, cells were harvested, stained for viability with LIVE/DEAD Fixable Violet Dead Cell Stain Kit and analyzed by flow cytometry.
BBB permeability assays
Evans Blue dye (EBD) infusion was performed as previously described (Radu and Chernoff, 2013). Seven days after CAR-T cell infusion, mice were injected intravenously with EBD +/− mannitol. Thirty minutes after EBD infusion, mice were euthanized; brain tissues were excised and fixed in 10% formalin solution for at least 24 h. After washing and dehydrating, brains were embedded in paraffin and cross-sectioned at a thickness of 5µm. Sections were deparaffinized, mounted with DAPI-containing Vectashield, and analyzed by fluorescence microscopy.
C57BL/6J mice were obtained from Jackson Laboratory and housed in a BSL1 mouse facility at the University of Pennsylvania. Murine splenocytes from donor mice were harvested, activated with murine-specific anti-CD3/anti-CD28 magnetic beads (Thermo Fisher), and transduced with retroviral vector encoding mCD19mBBz CAR, mCD19mCD28z CAR, or hCD19mBBz CAR transgenes. Four days after transduction, 5×106 murine CAR-T cells were injected intravenously in recipient mice +/− (50mg/kg cyclophosphamide for lymphodepletion.
Immunohistochemistry of adult human brain
The human anti-CD19 antibody (clone BT51E) was used to stain normal human brain. Images were visualized and acquired using the Leica #NCL-L-CD19–163 microscope. Automated immunohistochemistry was performed with Ventana Benchmark XT following a clinically validated protocol for CD19. 4-micron FFPE tissue sections were deparaffinized and rehydrated. Antigen retrieval was performed using Standard Cell Condition 1 (pH 8.5) for 60 minutes (Ventana Medical Systems). Slides were incubated with anti-human CD19 (1:50, monoclonal, Abnova BT51E) at 37° for 30 minutes. The ultraView Universal DAB Detection system (Ventana) was used with 3,3’diaminobenzidine chromogen.
High resolution mouse brain MRI
3×107 T cells with 32% normalized CAR+ expression were counted using the ADAM-CellT (NanoEnTek, Seoul, Korea) and the Multisizer 4e (Beckman Coulter). T cells were administered to non-tumor bearing NSG mice (n=4 per group) by one intravenous tail vein injection in 100µL of PBS. All the MR imaging experiments were performed 4 days post CAR-T cell infusions. Mice were anesthetized by using isoflurane maintained at 1–1.5% in 1 liter/min air. Lateral tail vein was cannulated for administration of gadodiamide (287 mg/ml) or mannitol (25% w/v). Mice were placed in a 20 mm diameter commercial quadrature proton coil (m2m Imaging Corp., Cleveland, OH) and the probe was transferred into a 9.4T horizontal bore small animal MR scanner (Bruker, Billerica, MA). During the MRI scans, animal body temperature was maintained at 37 °C with the air generated and blowing through a heater (SA Instruments, Inc., Stony Brook, NY). Respiration and body temperature were continuously monitored using an MRI compatible small animal monitoring system (SA Instruments, Inc., Stony Brook, NY).
T1-mapping was performed pre- and post-gadodiamide injection on 0.7 mm thick coronal slice in the mid-brain using saturation recovery protocol with following parameters, number of averages=2, field of view=20×20 mm2, matrix size=192×192, echo time=8 ms, repetition times=200, 500, 800, 1200, 1500, 4000 and 9000 ms, scan time 30 minutes. Following the baseline T1 acquisition, gadodiamide was administered via tail vein as described previously (Ku et al., 2018; van Vliet et al., 2014). T1 acquisition scans were run consecutively after one another. Pre- and Post-gadodiamide administration T1 maps were generated using the image sequence analysis tool in Paravision 6.0.1 by exponential fitting of the signal recovery. An in-house MATLAB script was used to quantity the ΔT1 (T1Post – T1Pre) caused by Gd leakage in the brain.
Quantification and Statistical Analysis
Information on specific statistical tests used is provided in the figure legends and/or Method Details. Quantification of MRI ΔT1 was performed using a MATLAB script. For single-cell RNA-sequencing analysis, specific details regarding pre-processing steps and parameters are provided in the relevant section of the Method Details.
Supplementary Material
Supplementary Figure 1: Identification of non-neuronal clusters of interest. Related to Figure 1.
(A) UMAP plots showing expression of selected marker genes for mural cells, endothelial cells, and microglia/immune cells. The color represents counts per million of each gene for a given cell in all panels (a–d). (B) Separate clusters of endothelial cells (CD31/PECAM1+) and mural cells (RGS5+) within the central cluster of neurovascular cells. (C) Lack of ACTA2 expression indicating pericyte identity. (D) CD81 is expressed in mural cells.
Supplementary Figure 2: Meta-cell clustering of BICCN data. Related to Figure 4.
(A) Example processing of one sample (GW25 Thalamus). Clustering, doublet identification, as well as expression of selected marker genes is shown. Note that vSMCs, pericytes, and endothelial cells are clustered closely together, but are clearly distinguished by marker gene expression. (B) Identification of variable genes across meta-cells. Genes used for downstream analysis are shown in red. (C) Number of cells per metacell, grouped by cluster. (D) Mean number of genes (left) or counts (right) per cell in each metacell, grouped by cluster. (E) Mean expression of PAX3, PAX7, and LIN28A per meta-cell, showing the high expression at early timepoints and progressive loss throughout development. (F) Heatmap of selected marker genes for each meta-cell in the indicated ranges of sample ages, showing the absence of CD19 and mural cell gene expression at CS12–15 but the presence at GW25.
Supplementary Figure 4: Analysis of Allen Institute Brainspan data. Related to Figure 6.
(A) (top) Scatter plots showing correlation of CD248 and ANPEP (CD13), two pericyte markers and (bottom) CD248 and CLDN5, an endothelial cell marker gene, showing overall correlation of neurovascular-associated genes. (B) Expression of CD19 and CD248 by brain region. (C) Integrated clustering of brain and PBMC data, with B cell, pericyte, and endothelial cell clusters identified. (D) Comparison of CD19 expression and CD19-correlated gene score, showing high score expression in pericytes and endothelial cells, and smaller enrichment in B cell cluster, indicating that pericytes/vasculature is the primary driver of CD19 expression in the brain. Expression for genes is in counts per 10,000; unit for gene score is arbitrary unit. (E) Comparisons of additional genes/correlated gene scores, as controls, showing ability of the approach to selectively enrich for expected populations of cells. Units are as in (d).
Supplementary Figure 5: Analysis of CD19 expression in mouse brain. Related to Figure 6.
(A) Flow cytometric analysis of CNS neurovascular cells isolated from healthy C57BL/6J mice. The live cell fraction is shown with gating for the CD45+ and CD45− populations. CD19+ cells are present in the CD45− population as shown, along with CD31+ endothelial cells. CD19+ B220+ B-cells are found in the CD45+ fraction. (B) Fluorescence minus one (FMO) control staining for CD19, with the same gating as in (a). (C) (left) CD19 is expressed at roughly similar levels in the CD45− fraction as in the CD45+ fraction (B-cells); (right) B220 is expressed only by the CD45+ CD19+ B-cell fraction, but not in the CD45− CD19+ fraction. (D) Flow cytometry analysis of live, single cells, showing gating for CD45− CD41− CD13+ CD31− pericytes, and expression of CD19 against CD13. An isotype control for CD19 is shown. Data in (d) represents a separate experiment from (a-c). (E) scRNA-seq of dissociated mouse brain after enrichment for CD19+, CD31+, and CD19+ cells by FACS. (left) UMAP plot showing identified clusters, (right) expression of marker genes showing low CD19 expression in pericytes and moderate CD19 expression in microglia.
Supplementary Figure 3: Identification of sub-clusters of interest across brain datasets. Related to Figure 5.
(A) Integrated clustering of Zhong 2018, La Manno 2016, and La Manno 2018. (B) Expression of CD248 (pericyte), CSF1R (microglia), PECAM1 (endothelial), and CD19 in these datasets. (C) CD81 is expressed by CD19+ mural cells. (D) Analysis of cells comprising progenitors, vascular cells, and microglia from BICCN data. The cells that were included for subsequent analysis are indicated in the third panel. (E) Expression of selected marker genes that were used to identify clusters of cells. Cells are colored by TPM of indicated gene. (F) Expression of vascular SMC marker genes distinguishing venous, arterial, and arteriole SMCs. Cells are colored by TPM of indicated gene.
Supplementary Figure 7: Comparison of mural cell transcriptome from brain and lung. Related to Figure 7.
(A) Scatter plot comparing expression in brain pericytes and lung pericytes. Genes with at least an absolute log2 fold change of 2 are colored in blue, and selected genes are annotated, showing similar expression of markers such as PDGFRB, but distinct expression of other genes such as BCL11A and HOXB4. (B) as in (a) but comparing lung vSMCs with lung pericytes. (C) as in (a) but comparing brain endothelial cells and lung endothelial cells.
Supplementary Figure 6: Infusion of CD19-directed CAR-T cells into mice. Related to Figure 6.
(A) Flow cytometry based killing assays in human B-ALL cell line Nalm6 and murine CD19+ A20 B-ALL cell line in presence of human 4-1BB or CD28 based anti-human or murine CD19 CAR-T cells. (B) Representation of CAR constructs and animal study design using NSG mice. (C) Representative immunofluorescence imaging from the cortex showing nuclear staining (DAPI, blue) and Evans Blue dye (EBD) extravasation (red) for BBB disruption in NSG mice following murine-targeting CD19 CAR-T treatment (n=5 mice per treatment). Scale bar, 75 µm. (D) Quantification of EBD staining in (c) in each group of mice as determined by the product of the mean fluorescence intensity per field area. Representative sections (n=3 per mouse) were analyzed and graphed. *p<0.05, **p<0.005, 2 tailed unpaired t-test. (E) Syngeneic C57BL/6J mouse model recapitulates the murine CD19-specific neurotoxicity pattern observed in the immunodeficient model, showing staining for DAPI and EBD in cortical sections (n=5 mice per group). Scale bar, 75 µm. (F) Quantification of EBD staining for each group shown in (e). *p<0.05, **p<0.005, 2 tailed unpaired t-test. (G) In vivo imaging using high-resolution MRI with gadodiamide contrast shows BBB leakage after murine-targeting CD19 CAR-T cell therapy. All experiments were performed 4 days after CAR-T cell infusion. Anatomical reference image with selected region of interest (ROI) contour used to calculate delta T1 and T2 between each group is outlined. n=4 mice per group, mean and standard deviation across mice provided. (H). Quantification of mean delta T1 of MRI within ROI shown in (g), n=4 mice per group. *p<.05, two-tailed t-test. (I) (top) Decrease in CD45+CD19+ cells (n=5 mice per group) in mCD19-specific CAR-T treated mice as opposed to the hCD19 CAR-T group. (bottom) CD19 vs CD13 (n=5 mice per group) showing decrease in live brain CD45−CD41−CD31−CD13+CD19+ cells in the mCD19 specific CAR-T as opposed to the hCD19 CAR-T treated group. Error bars show the SEM, p-value using two-tailed t-test.
Single-cell RNA-seq reveals CD19 expression in human brain mural cells
Mural cells line blood vessels and maintain blood-brain barrier integrity
Brain mural cell CD19 expression is present across brain regions and human age
Targeting CD19+ mural cells may contribute to neurotoxicity of CAR-T therapy
Acknowledgements:
We thank members of the Chang, June, Posey, and Satpathy labs and Robbie Majzner for helpful discussion. This work was supported by the National Institutes of Health (NIH) P50-HG007735 (H.Y.C.), P50-NS062684 (T.J.M.), K08CA230188 (A.T.S.), U01MH105989 (A.R.K.), P41-EB015893 (R.R.), the Department of Veterans Affairs IK2BX004183 (A.D.P.), the Parker Institute for Cancer Immunotherapy (C.H.J., H.Y.C., A.D.P., and A.T.S.), and the Scleroderma Research Foundation (H.Y.C.). D.M. was supported by the Private Foundation of the Geneva University Hospital, Switzerland. K.R.P. was supported by a Stanford Graduate Fellowship. A.T.S. was supported by a Parker Bridge Scholar Award from the Parker Institute for Cancer Immunotherapy, a Technology Impact Award from the Cancer Research Institute, and a Career Award for Medical Scientists from the Burroughs Wellcome Fund. H.Y.C. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests:
C.H.J. and A.D.P. report intellectual property licensed to Novartis and Tmunity Therapeutics related to CAR-T cells. K.R.P., A.T.S., H.Y.C., D.M., A.D.P., and C.H.J. are listed as inventors on patent applications filed by Stanford University and the University of Pennsylvania. K.R.P. is a consultant for Maze Therapeutics. A.T.S. is a scientific founder of Immunai and receives research funding from Arsenal Biosciences. H.Y.C. is a co-founder of Accent Therapeutics and Boundless Bio, and an advisor to 10x Genomics, Arsenal Biosciences, and Spring Discovery.
References
- Armulik A, Genové G, Mäe M, Nisancioglu MH, Wallgard E, Niaudet C, He L, Norlin J, Lindblom P, Strittmatter K, et al. (2010). Pericytes regulate the blood–brain barrier. Nature 468, 557–561. [DOI] [PubMed] [Google Scholar]
- Bausch-Fluck D, Hofmann A, Bock T, Frei AP, Cerciello F, Jacobs A, Moest H, Omasits U, Gundry RL, Yoon C, et al. (2015). A mass spectrometric-derived cell surface protein atlas. PloS One 10, e0121314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boroujerdi A, Tigges U, Welser-Alves JV, and Milner R (2014). Isolation and culture of primary pericytes from mouse brain. Methods Mol. Biol. Clifton NJ 1135, 383–392. [DOI] [PubMed] [Google Scholar]
- Braig F, Brandt A, Goebeler M, Tony H-P, Kurze A-K, Nollau P, Bumm T, Böttcher S, Bargou RC, and Binder M (2017). Resistance to anti-CD19/CD3 BiTE in acute lymphoblastic leukemia may be mediated by disrupted CD19 membrane trafficking. Blood 129, 100–104. [DOI] [PubMed] [Google Scholar]
- Brentjens RJ, Latouche J-B, Santos E, Marti F, Gong MC, Lyddane C, King PD, Larson S, Weiss M, Rivière I, et al. (2003). Eradication of systemic B-cell tumors by genetically targeted human T lymphocytes co-stimulated by CD80 and interleukin-15. Nat. Med 9, 279–286. [DOI] [PubMed] [Google Scholar]
- Brentjens RJ, Davila ML, Riviere I, Park J, Wang X, Cowell LG, Bartido S, Stefanski J, Taylor C, Olszewska M, et al. (2013). CD19-targeted T cells rapidly induce molecular remissions in adults with chemotherapy-refractory acute lymphoblastic leukemia. Sci. Transl. Med 5, 177ra38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler A, Hoffman P, Smibert P, Papalexi E, and Satija R (2018). Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol 36, 411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chasseigneaux S, Moraca Y, Cochois-Guégan V, Boulay A-C, Gilbert A, Le Crom S, Blugeon C, Firmo C, Cisternino S, Laplanche J-L, et al. (2018). Isolation and differential transcriptome of vascular smooth muscle cells and mid-capillary pericytes from the rat brain. Sci. Rep 8, 12272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daneman R, Zhou L, Kebede AA, and Barres BA (2010). Pericytes are required for blood–brain barrier integrity during embryogenesis. Nature 468, 562–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias C, Estruch SB, Graham SA, McRae J, Sawiak SJ, Hurst JA, Joss SK, Holder SE, Morton JEV, Turner C, et al. (2016). BCL11A Haploinsufficiency Causes an Intellectual Disability Syndrome and Dysregulates Transcription. Am. J. Hum. Genet 99, 253–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdő F, Denes L, and de Lange E (2017). Age-associated physiological and pathological changes at the blood-brain barrier: A review. J. Cereb. Blood Flow Metab. Off. J. Int. Soc. Cereb. Blood Flow Metab 37, 4–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etchevers HC, Vincent C, Le Douarin NM, and Couly GF (2001). The cephalic neural crest provides pericytes and smooth muscle cells to all blood vessels of the face and forebrain. Dev. Camb. Engl 128, 1059–1068. [DOI] [PubMed] [Google Scholar]
- Faal T, Phan DTT, Davtyan H, Scarfone VM, Varady E, Blurton-Jones M, Hughes CCW, and Inlay MA (2019). Induction of Mesoderm and Neural Crest-Derived Pericytes from Human Pluripotent Stem Cells to Study Blood-Brain Barrier Interactions. Stem Cell Rep 12, 451–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorov VD, Themeli M, and Sadelain M (2013). PD-1- and CTLA-4-based inhibitory chimeric antigen receptors (iCARs) divert off-target immunotherapy responses. Sci. Transl. Med 5, 215ra172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner RA, Finney O, Annesley C, Brakke H, Summers C, Leger K, Bleakley M, Brown C, Mgebroff S, Kelly-Spratt KS, et al. (2017). Intent-to-treat leukemia remission by CD19 CAR T cells of defined formulation and dose in children and young adults. Blood 129, 3322–3331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giavridis T, van der Stegen SJC, Eyquem J, Hamieh M, Piersigilli A, and Sadelain M (2018). CAR T cell-induced cytokine release syndrome is mediated by macrophages and abated by IL-1 blockade. Nat. Med 24, 731–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goebeler M-E, Knop S, Viardot A, Kufer P, Topp MS, Einsele H, Noppeney R, Hess G, Kallert S, Mackensen A, et al. (2016). Bispecific T-Cell Engager (BiTE) Antibody Construct Blinatumomab for the Treatment of Patients With Relapsed/Refractory Non-Hodgkin Lymphoma: Final Results From a Phase I Study. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol 34, 1104–1111. [DOI] [PubMed] [Google Scholar]
- Gofshteyn JS, Shaw PA, Teachey DT, Grupp SA, Maude S, Banwell B, Chen F, Lacey SF, Melenhorst JJ, Edmonson MJ, et al. (2018). Neurotoxicity after CTL019 in a pediatric and young adult cohort. Ann. Neurol 84, 537–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grupp SA, Kalos M, Barrett D, Aplenc R, Porter DL, Rheingold SR, Teachey DT, Chew A, Hauck B, Wright JF, et al. (2013). Chimeric antigen receptor-modified T cells for acute lymphoid leukemia. N. Engl. J. Med 368, 1509–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gust J, Hay KA, Hanafi L-A, Li D, Myerson D, Gonzalez-Cuyar LF, Yeung C, Liles WC, Wurfel M, Lopez JA, et al. (2017). Endothelial Activation and Blood–Brain Barrier Disruption in Neurotoxicity after Adoptive Immunotherapy with CD19 CAR-T Cells. Cancer Discov 7, 1404–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodge RD, Bakken TE, Miller JA, Smith KA, Barkan ER, Graybuck LT, Close JL, Long B, Johansen N, Penn O, et al. (2019). Conserved cell types with divergent features in human versus mouse cortex. Nature 573, 61–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- June CH, O’Connor RS, Kawalekar OU, Ghassemi S, and Milone MC (2018). CAR T cell immunotherapy for human cancer. Science 359, 1361–1365. [DOI] [PubMed] [Google Scholar]
- Juno Therapeutics, a Subsidiary of Celgene (2020). A Phase 2, Single-arm, Multicenter Trial to Determine the Efficacy and Safety of JCAR015 in Adult Subjects With Relapsed or Refractory B-Cell Acute Lymphoblastic Leukemia (clinicaltrials.gov).
- Kim N, Kim HK, Lee K, Hong Y, Cho JH, Choi JW, Lee J-I, Suh Y-L, Ku BM, Eum HH, et al. (2020). Single-cell RNA sequencing demonstrates the molecular and cellular reprogramming of metastatic lung adenocarcinoma. Nat. Commun 11, 2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinger M, Zugmaier G, Nägele V, Goebeler M-E, Brandl C, Stelljes M, Lassmann H, von Stackelberg A, Bargou RC, and Kufer P (2020). Adhesion of T Cells to Endothelial Cells Facilitates Blinatumomab-Associated Neurologic Adverse Events. Cancer Res 80, 91–101. [DOI] [PubMed] [Google Scholar]
- Kochenderfer JN, Wilson WH, Janik JE, Dudley ME, Stetler-Stevenson M, Feldman SA, Maric I, Raffeld M, Nathan D-AN, Lanier BJ, et al. (2010). Eradication of B-lineage cells and regression of lymphoma in a patient treated with autologous T cells genetically engineered to recognize CD19. Blood 116, 4099–4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korfel A, and Schlegel U (2013). Diagnosis and treatment of primary CNS lymphoma. Nat. Rev. Neurol 9, 317–327. [DOI] [PubMed] [Google Scholar]
- Kozmik Z, Wang S, Dörfler P, Adams B, and Busslinger M (1992). The promoter of the CD19 gene is a target for the B-cell-specific transcription factor BSAP. Mol. Cell. Biol 12, 2662–2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku M-C, Waiczies S, Niendorf T, and Pohlmann A (2018). Assessment of Blood Brain Barrier Leakage with Gadolinium-Enhanced MRI. Methods Mol. Biol. Clifton NJ 1718, 395–408. [DOI] [PubMed] [Google Scholar]
- La Manno G, Gyllborg D, Codeluppi S, Nishimura K, Salto C, Zeisel A, Borm LE, Stott SRW, Toledo EM, Villaescusa JC, et al. (2016). Molecular Diversity of Midbrain Development in Mouse, Human, and Stem Cells. Cell 167, 566–580.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Manno G, Soldatov R, Zeisel A, Braun E, Hochgerner H, Petukhov V, Lidschreiber K, Kastriti ME, Lönnerberg P, Furlan A, et al. (2018). RNA velocity of single cells. Nature 560, 494–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DW, Kochenderfer JN, Stetler-Stevenson M, Cui YK, Delbrook C, Feldman SA, Fry TJ, Orentas R, Sabatino M, Shah NN, et al. (2015). T cells expressing CD19 chimeric antigen receptors for acute lymphoblastic leukaemia in children and young adults: a phase 1 dose-escalation trial. Lancet Lond. Engl 385, 517–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin G (2018). Brain Tissue Dissociation for Cell Sorting.
- Liu P, Keller JR, Ortiz M, Tessarollo L, Rachel RA, Nakamura T, Jenkins NA, and Copeland NG (2003). Bcl11a is essential for normal lymphoid development. Nat. Immunol 4, 525–532. [DOI] [PubMed] [Google Scholar]
- Locke FL, Neelapu SS, Bartlett NL, Siddiqi T, Chavez JC, Hosing CM, Ghobadi A, Budde LE, Bot A, Rossi JM, et al. (2017). Phase 1 Results of ZUMA-1: A Multicenter Study of KTE-C19 Anti-CD19 CAR T Cell Therapy in Refractory Aggressive Lymphoma. Mol. Ther 25, 285–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe KL, Mackall CL, Norry E, Amado R, Jakobsen BK, and Binder G (2018). Fludarabine and neurotoxicity in engineered T-cell therapy. Gene Ther 25, 176–191. [DOI] [PubMed] [Google Scholar]
- Maecker HT, and Levy S (1997). Normal lymphocyte development but delayed humoral immune response in CD81-null mice. J. Exp. Med 185, 1505–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majzner RG, Rietberg SP, Sotillo E, Dong R, Vachharajani VT, Labanieh L, Myklebust JH, Kadapakkam M, Weber EW, Tousley AM, et al. (2020). Tuning the Antigen Density Requirement for CAR T-cell Activity. Cancer Discov 10, 702–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maude SL, Laetsch TW, Buechner J, Rives S, Boyer M, Bittencourt H, Bader P, Verneris MR, Stefanski HE, Myers GD, et al. (2018). Tisagenlecleucel in Children and Young Adults with B-Cell Lymphoblastic Leukemia. N. Engl. J. Med 378, 439–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JA, Ding S-L, Sunkin SM, Smith KA, Ng L, Szafer A, Ebbert A, Riley ZL, Royall JJ, Aiona K, et al. (2014). Transcriptional landscape of the prenatal human brain. Nature 508, 199–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milone MC, Fish JD, Carpenito C, Carroll RG, Binder GK, Teachey D, Samanta M, Lakhal M, Gloss B, Danet-Desnoyers G, et al. (2009). Chimeric receptors containing CD137 signal transduction domains mediate enhanced survival of T cells and increased antileukemic efficacy in vivo. Mol. Ther. J. Am. Soc. Gene Ther 17, 1453–1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki T, Müller U, and Campbell KS (1997). Normal development but differentially altered proliferative responses of lymphocytes in mice lacking CD81. EMBO J 16, 4217–4225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagorsen D, Kufer P, Baeuerle PA, and Bargou R (2012). Blinatumomab: a historical perspective. Pharmacol. Ther 136, 334–342. [DOI] [PubMed] [Google Scholar]
- Neelapu SS (2019). Managing the toxicities of CAR T-cell therapy. Hematol. Oncol 37 Suppl 1, 48–52. [DOI] [PubMed] [Google Scholar]
- Neelapu SS, Locke FL, Bartlett NL, Lekakis LJ, Miklos DB, Jacobson CA, Braunschweig I, Oluwole OO, Siddiqi T, Lin Y, et al. (2017). Axicabtagene Ciloleucel CAR T-Cell Therapy in Refractory Large B-Cell Lymphoma. N. Engl. J. Med 377, 2531–2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JH, Rivière I, Gonen M, Wang X, Sénéchal B, Curran KJ, Sauter C, Wang Y, Santomasso B, Mead E, et al. (2018). Long-Term Follow-up of CD19 CAR Therapy in Acute Lymphoblastic Leukemia. N. Engl. J. Med [DOI] [PMC free article] [PubMed]
- Pennell CA, Barnum JL, McDonald-Hyman CS, Panoskaltsis-Mortari A, Riddle MJ, Xiong Z, Loschi M, Thangavelu G, Campbell HM, Storlie MD, et al. (2018). Human CD19-Targeted Mouse T Cells Induce B Cell Aplasia and Toxicity in Human CD19 Transgenic Mice. Mol. Ther. J. Am. Soc. Gene Ther 26, 1423–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter DL, Levine BL, Kalos M, Bagg A, and June CH (2011). Chimeric antigen receptor-modified T cells in chronic lymphoid leukemia. N. Engl. J. Med 365, 725–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quah BJC, Warren HS, and Parish CR (2007). Monitoring lymphocyte proliferation in vitro and in vivo with the intracellular fluorescent dye carboxyfluorescein diacetate succinimidyl ester. Nat. Protoc 2, 2049–2056. [DOI] [PubMed] [Google Scholar]
- Regev A, Teichmann SA, Lander ES, Amit I, Benoist C, Birney E, Bodenmiller B, Campbell PJ, Carninci P, Clatworthy M, et al. (2017). Science Forum: The Human Cell Atlas. ELife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roybal KT, Rupp LJ, Morsut L, Walker WJ, McNally KA, Park JS, and Lim WA (2016). Precision Tumor Recognition by T Cells With Combinatorial Antigen-Sensing Circuits. Cell 164, 770–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruella M, and June CH (2018). Predicting Dangerous Rides in CAR T Cells: Bridging the Gap between Mice and Humans. Mol. Ther. J. Am. Soc. Gene Ther 26, 1401–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salter AI, Ivey RG, Kennedy JJ, Voillet V, Rajan A, Alderman EJ, Voytovich UJ, Lin C, Sommermeyer D, Liu L, et al. (2018). Phosphoproteomic analysis of chimeric antigen receptor signaling reveals kinetic and quantitative differences that affect cell function. Sci. Signal 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster SJ, Svoboda J, Chong EA, Nasta SD, Mato AR, Anak Ö, Brogdon JL, Pruteanu-Malinici I, Bhoj V, Landsburg D, et al. (2017). Chimeric Antigen Receptor T Cells in Refractory B-Cell Lymphomas. N. Engl. J. Med 377, 2545–2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoham T, Rajapaksa R, Boucheix C, Rubinstein E, Poe JC, Tedder TF, and Levy S (2003). The Tetraspanin CD81 Regulates the Expression of CD19 During B Cell Development in a Postendoplasmic Reticulum Compartment. J. Immunol 171, 4062–4072. [DOI] [PubMed] [Google Scholar]
- Sommermeyer D, Hill T, Shamah SM, Salter AI, Chen Y, Mohler KM, and Riddell SR (2017). Fully human CD19-specific chimeric antigen receptors for T-cell therapy. Leukemia 31, 2191–2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotillo E, Barrett DM, Black KL, Bagashev A, Oldridge D, Wu G, Sussman R, Lanauze C, Ruella M, Gazzara MR, et al. (2015). Convergence of Acquired Mutations and Alternative Splicing of CD19 Enables Resistance to CART-19 Immunotherapy. Cancer Discov 5, 1282–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney MD, Zhao Z, Montagne A, Nelson AR, and Zlokovic BV (2019). Blood-Brain Barrier: From Physiology to Disease and Back. Physiol. Rev 99, 21–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabula Muris Consortium, Overall coordination, Logistical coordination, Organ collection and processing, Library preparation and sequencing, Computational data analysis, Cell type annotation, Writing group, Supplemental text writing group, and Principal investigators (2018). Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature 562, 367–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torre M, Solomon IH, Sutherland CL, Nikiforow S, DeAngelo DJ, Stone RM, Vaitkevicius H, Galinsky IA, Padera RF, Trede N, et al. (2018). Neuropathology of a Case With Fatal CAR T-Cell-Associated Cerebral Edema. J. Neuropathol. Exp. Neurol 77, 877–882. [DOI] [PubMed] [Google Scholar]
- Travaglini KJ, Nabhan AN, Penland L, Sinha R, Gillich A, Sit RV, Chang S, Conley SD, Mori Y, Seita J, et al. (2020). A molecular cell atlas of the human lung from single cell RNA sequencing. BioRxiv 742320. [DOI] [PMC free article] [PubMed]
- Tsitsikov EN, Gutierrez-Ramos JC, and Geha RS (1997). Impaired CD19 expression and signaling, enhanced antibody response to type II T independent antigen and reduction of B-1 cells in CD81-deficient mice. Proc. Natl. Acad. Sci. U. S. A 94, 10844–10849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turtle CJ, Hanafi L-A, Berger C, Hudecek M, Pender B, Robinson E, Hawkins R, Chaney C, Cherian S, Chen X, et al. (2016a). Immunotherapy of non-Hodgkin’s lymphoma with a defined ratio of CD8+ and CD4+ CD19-specific chimeric antigen receptor–modified T cells. Sci. Transl. Med 8, 355ra116–355ra116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turtle CJ, Hanafi L-A, Berger C, Gooley TA, Cherian S, Hudecek M, Sommermeyer D, Melville K, Pender B, Budiarto TM, et al. (2016b). CD19 CAR–T cells of defined CD4+:CD8+ composition in adult B cell ALL patients. J. Clin. Invest 126, 2123–2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turtle CJ, Hay KA, Hanafi L-A, Li D, Cherian S, Chen X, Wood B, Lozanski A, Byrd JC, Heimfeld S, et al. (2017). Durable Molecular Remissions in Chronic Lymphocytic Leukemia Treated With CD19-Specific Chimeric Antigen Receptor–Modified T Cells After Failure of Ibrutinib. J. Clin. Oncol 35, 3010–3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura MT, Maki T, Ihara M, Lee VMY, and Trojanowski JQ (2020). Brain Microvascular Pericytes in Vascular Cognitive Impairment and Dementia. Front. Aging Neurosci 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbánek P, Fetka I, Meisler MH, and Busslinger M (1997). Cooperation of Pax2 and Pax5 in midbrain and cerebellum development. Proc. Natl. Acad. Sci. U. S. A 94, 5703–5708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanlandewijck M, He L, Mäe MA, Andrae J, Ando K, Del Gaudio F, Nahar K, Lebouvier T, Laviña B, Gouveia L, et al. (2018). A molecular atlas of cell types and zonation in the brain vasculature. Nature 554, 475–480. [DOI] [PubMed] [Google Scholar]
- van Vliet EA, Otte WM, Gorter JA, Dijkhuizen RM, and Wadman WJ (2014). Longitudinal assessment of blood-brain barrier leakage during epileptogenesis in rats. A quantitative MRI study. Neurobiol. Dis 63, 74–84. [DOI] [PubMed] [Google Scholar]
- Wolf FA, Angerer P, and Theis FJ (2018). SCANPY: large-scale single-cell gene expression data analysis. Genome Biol 19, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolock SL, Lopez R, and Klein AM (2019). Scrublet: Computational Identification of Cell Doublets in Single-Cell Transcriptomic Data. Cell Syst 8, 281–291.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang AC, Stevens MY, Chen MB, Lee DP, Stähli D, Gate D, Contrepois K, Chen W, Iram T, Zhang L, et al. (2020). Physiological blood-brain transport is impaired with age by a shift in transcytosis. Nature. [DOI] [PMC free article] [PubMed]
- Ying Z, Huang XF, Xiang X, Liu Y, Kang X, Song Y, Guo X, Liu H, Ding N, Zhang T, et al. (2019). A safe and potent anti-CD19 CAR T cell therapy. Nat. Med 25, 947–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Zelm MC, Smet J, Adams B, Mascart F, Schandené L, Janssen F, Ferster A, Kuo C-C, Levy S, van Dongen JJM, et al. (2010). CD81 gene defect in humans disrupts CD19 complex formation and leads to antibody deficiency. J. Clin. Invest 120, 1265–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng GXY, Terry JM, Belgrader P, Ryvkin P, Bent ZW, Wilson R, Ziraldo SB, Wheeler TD, McDermott GP, Zhu J, et al. (2017). Massively parallel digital transcriptional profiling of single cells. Nat. Commun 8, 14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S, Zhang S, Fan X, Wu Q, Yan L, Dong J, Zhang H, Li L, Sun L, Pan N, et al. (2018). A single-cell RNA-seq survey of the developmental landscape of the human prefrontal cortex. Nature 555, 524–528. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1: Identification of non-neuronal clusters of interest. Related to Figure 1.
(A) UMAP plots showing expression of selected marker genes for mural cells, endothelial cells, and microglia/immune cells. The color represents counts per million of each gene for a given cell in all panels (a–d). (B) Separate clusters of endothelial cells (CD31/PECAM1+) and mural cells (RGS5+) within the central cluster of neurovascular cells. (C) Lack of ACTA2 expression indicating pericyte identity. (D) CD81 is expressed in mural cells.
Supplementary Figure 2: Meta-cell clustering of BICCN data. Related to Figure 4.
(A) Example processing of one sample (GW25 Thalamus). Clustering, doublet identification, as well as expression of selected marker genes is shown. Note that vSMCs, pericytes, and endothelial cells are clustered closely together, but are clearly distinguished by marker gene expression. (B) Identification of variable genes across meta-cells. Genes used for downstream analysis are shown in red. (C) Number of cells per metacell, grouped by cluster. (D) Mean number of genes (left) or counts (right) per cell in each metacell, grouped by cluster. (E) Mean expression of PAX3, PAX7, and LIN28A per meta-cell, showing the high expression at early timepoints and progressive loss throughout development. (F) Heatmap of selected marker genes for each meta-cell in the indicated ranges of sample ages, showing the absence of CD19 and mural cell gene expression at CS12–15 but the presence at GW25.
Supplementary Figure 4: Analysis of Allen Institute Brainspan data. Related to Figure 6.
(A) (top) Scatter plots showing correlation of CD248 and ANPEP (CD13), two pericyte markers and (bottom) CD248 and CLDN5, an endothelial cell marker gene, showing overall correlation of neurovascular-associated genes. (B) Expression of CD19 and CD248 by brain region. (C) Integrated clustering of brain and PBMC data, with B cell, pericyte, and endothelial cell clusters identified. (D) Comparison of CD19 expression and CD19-correlated gene score, showing high score expression in pericytes and endothelial cells, and smaller enrichment in B cell cluster, indicating that pericytes/vasculature is the primary driver of CD19 expression in the brain. Expression for genes is in counts per 10,000; unit for gene score is arbitrary unit. (E) Comparisons of additional genes/correlated gene scores, as controls, showing ability of the approach to selectively enrich for expected populations of cells. Units are as in (d).
Supplementary Figure 5: Analysis of CD19 expression in mouse brain. Related to Figure 6.
(A) Flow cytometric analysis of CNS neurovascular cells isolated from healthy C57BL/6J mice. The live cell fraction is shown with gating for the CD45+ and CD45− populations. CD19+ cells are present in the CD45− population as shown, along with CD31+ endothelial cells. CD19+ B220+ B-cells are found in the CD45+ fraction. (B) Fluorescence minus one (FMO) control staining for CD19, with the same gating as in (a). (C) (left) CD19 is expressed at roughly similar levels in the CD45− fraction as in the CD45+ fraction (B-cells); (right) B220 is expressed only by the CD45+ CD19+ B-cell fraction, but not in the CD45− CD19+ fraction. (D) Flow cytometry analysis of live, single cells, showing gating for CD45− CD41− CD13+ CD31− pericytes, and expression of CD19 against CD13. An isotype control for CD19 is shown. Data in (d) represents a separate experiment from (a-c). (E) scRNA-seq of dissociated mouse brain after enrichment for CD19+, CD31+, and CD19+ cells by FACS. (left) UMAP plot showing identified clusters, (right) expression of marker genes showing low CD19 expression in pericytes and moderate CD19 expression in microglia.
Supplementary Figure 3: Identification of sub-clusters of interest across brain datasets. Related to Figure 5.
(A) Integrated clustering of Zhong 2018, La Manno 2016, and La Manno 2018. (B) Expression of CD248 (pericyte), CSF1R (microglia), PECAM1 (endothelial), and CD19 in these datasets. (C) CD81 is expressed by CD19+ mural cells. (D) Analysis of cells comprising progenitors, vascular cells, and microglia from BICCN data. The cells that were included for subsequent analysis are indicated in the third panel. (E) Expression of selected marker genes that were used to identify clusters of cells. Cells are colored by TPM of indicated gene. (F) Expression of vascular SMC marker genes distinguishing venous, arterial, and arteriole SMCs. Cells are colored by TPM of indicated gene.
Supplementary Figure 7: Comparison of mural cell transcriptome from brain and lung. Related to Figure 7.
(A) Scatter plot comparing expression in brain pericytes and lung pericytes. Genes with at least an absolute log2 fold change of 2 are colored in blue, and selected genes are annotated, showing similar expression of markers such as PDGFRB, but distinct expression of other genes such as BCL11A and HOXB4. (B) as in (a) but comparing lung vSMCs with lung pericytes. (C) as in (a) but comparing brain endothelial cells and lung endothelial cells.
Supplementary Figure 6: Infusion of CD19-directed CAR-T cells into mice. Related to Figure 6.
(A) Flow cytometry based killing assays in human B-ALL cell line Nalm6 and murine CD19+ A20 B-ALL cell line in presence of human 4-1BB or CD28 based anti-human or murine CD19 CAR-T cells. (B) Representation of CAR constructs and animal study design using NSG mice. (C) Representative immunofluorescence imaging from the cortex showing nuclear staining (DAPI, blue) and Evans Blue dye (EBD) extravasation (red) for BBB disruption in NSG mice following murine-targeting CD19 CAR-T treatment (n=5 mice per treatment). Scale bar, 75 µm. (D) Quantification of EBD staining in (c) in each group of mice as determined by the product of the mean fluorescence intensity per field area. Representative sections (n=3 per mouse) were analyzed and graphed. *p<0.05, **p<0.005, 2 tailed unpaired t-test. (E) Syngeneic C57BL/6J mouse model recapitulates the murine CD19-specific neurotoxicity pattern observed in the immunodeficient model, showing staining for DAPI and EBD in cortical sections (n=5 mice per group). Scale bar, 75 µm. (F) Quantification of EBD staining for each group shown in (e). *p<0.05, **p<0.005, 2 tailed unpaired t-test. (G) In vivo imaging using high-resolution MRI with gadodiamide contrast shows BBB leakage after murine-targeting CD19 CAR-T cell therapy. All experiments were performed 4 days after CAR-T cell infusion. Anatomical reference image with selected region of interest (ROI) contour used to calculate delta T1 and T2 between each group is outlined. n=4 mice per group, mean and standard deviation across mice provided. (H). Quantification of mean delta T1 of MRI within ROI shown in (g), n=4 mice per group. *p<.05, two-tailed t-test. (I) (top) Decrease in CD45+CD19+ cells (n=5 mice per group) in mCD19-specific CAR-T treated mice as opposed to the hCD19 CAR-T group. (bottom) CD19 vs CD13 (n=5 mice per group) showing decrease in live brain CD45−CD41−CD31−CD13+CD19+ cells in the mCD19 specific CAR-T as opposed to the hCD19 CAR-T treated group. Error bars show the SEM, p-value using two-tailed t-test.
Data Availability Statement
Raw data for mouse single-cell RNA-sequencing has been deposited in GEO and is available under accession number GSE153647. Details of analysis are provided below in the Methods, and any further questions should be directed to and will be fulfilled by the Lead Contact.
Processed sequencing data (gene counts per cell) were downloaded as follows: Zhong et al., 2018 (GEO GSE104276), La Manno et al., 2016 (GEO GSE76381), La Manno et al., 2018 (PanglaoDB database, Karolinska Institute, https://panglaodb.se/samples.html), Kriegstein/BICCN (NeMO archive, https://nemoarchive.org/), Brainspan (Allen Institute, https://www.brainspan.org/static/download.html). PBMC data was downloaded from the 10x Genomics reference datasets available on their website (https://support.10xgenomics.com/single-cell-gene-expression/datasets/3.0.0/pbmc_10k_v3). UMI counts for human lung (Travaglini et al., 2020) were downloaded from Synapse, accession syn21041850.


