Abstract
Pseudomonas aeruginosa is a highly adaptable Gram-negative opportunistic pathogen, notably due to its large number of transcription regulators. The extracytoplasmic sigma factor (ECFσ) AlgU, responsible for alginate biosynthesis, is also involved in responses to cell wall stress and heat shock via the RpoH alternative σ factor. The SigX ECFσ emerged as a major regulator involved in the envelope stress response via membrane remodeling, virulence and biofilm formation. However, their functional interactions to coordinate the envelope homeostasis in response to environmental variations remain to be determined. The regulation of the putative cmaX-cfrX-cmpX operon located directly upstream sigX was investigated by applying sudden temperature shifts from 37°C. We identified a SigX- and an AlgU- dependent promoter region upstream of cfrX and cmaX, respectively. We show that cmaX expression is increased upon heat shock through an AlgU-dependent but RpoH independent mechanism. In addition, the ECFσ SigX is activated in response to valinomycin, an agent altering the membrane structure, and up-regulates cfrX-cmpX transcription in response to cold shock. Altogether, these data provide new insights into the regulation exerted by SigX and networks that are involved in maintaining envelope homeostasis.
Keywords: Pseudomonas aeruginosa, temperature, cell wall stress ECF sigma factor, regulation of transcription, membrane fluidity
Importance
Because temperature is one of the first physical signal that allows bacteria to distinguish a potential host from its environment, the response of P. aeruginosa has been more investigated in case of elevated rather than low temperature. Here, we show that the ECFσ SigX is an effector in the latter condition, which controls the transcription of the two members of an intriguing operon encoding the mechanosensitive channel CmpX and the hypothetical protein CfrX, while cmaX is under the control of AlgU. In addition, we show that SigX is activated following membrane structure alterations. More generally, we have identified temperature as a key environmental factor regulating antagonistically the expression and the activity of the two major cell wall stress response ECFσ in P. aeruginosa, suggesting that they are involved in specific but interconnected networks, in which the AlgU-dependent global transcriptional regulator AmrZ (alginate and motility regulator Z) could play a role by a mechanism that remains to be explored.
Introduction
Temperature shift is a ubiquitous signal perceived by living organisms to adapt their behaviors. The heat shock (HS) and cold shock (CS) responses (R), involve heat shock proteins (Hsps), and cold inducible proteins (Cips), respectively. Importantly, HS and CS pathways are also activated in response to antibiotics in bacteria (VanBogelen and Neidhardt, 1990; Kindrachuk et al., 2011), highlighting their importance in antibiotic resistance, noticeably of ESKAPE pathogens (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumanii, Pseudomonas aeruginosa, and Enterobacter species) (Esposito and De Simone, 2017). P. aeruginosa is a highly adaptable Gram-negative bacterium that can colonize a variety of ecological niches (Spiers et al., 2000) and infect various hosts (Rahme et al., 2000; Baldini et al., 2014). Being intrinsically resistant to a wide range of antibiotics and disinfectants, P. aeruginosa is responsible of both acute and chronic community- and hospital-acquired infections (Lyczak et al., 2002). Its remarkable versatility is associated with the over-representation of genes encoding sensors, signal transduction systems and regulators such as sigma factors (Stover et al., 2000; Chevalier et al., 2018).
A sudden temperature rise above 45°C activates the RpoH (σ32)-dependent HS response in P. aeruginosa (Allan et al., 1988) that, together with the extracytoplasmic sigma factor (ECFσ) AlgU, co-regulates the production of the alginate exopolysaccharide via over-expression of the algD operon leading to a mucoid phenotype (Schurr and Deretic, 1997). AlgU is a homolog of the Escherichia coli RpoE cell wall stress response ECFσ, whose expression is auto-regulated at the transcriptional level, and its activity modulated at the post-translational level by a conserved mechanism of regulated-intramembrane proteolysis (Chevalier et al., 2018). The AlgU regulon includes rpoH encoding the heat shock RpoH sigma factor (Schurr et al., 1995), amrZ (AlgU-dependent alginate and motility regulator Z) encoding a transcriptional regulator that activates the expression of genes involved in motility and in alginate production, including in particular algD (Tart et al., 2006), oprF encoding the major and multi-functional outer membrane porin (Chevalier et al., 2017), together with numerous genes involved in bacterial adaptation and cell wall synthesis (Wood and Ohman, 2009; Schulz et al., 2015). In agreement with its role in envelope homeostasis, AlgU is more expressed in response to hyperosmolarity (Aspedon et al., 2006), inhibition of cell wall synthesis (Wood and Ohman, 2009), exposure to microgravity stress (Crabbe et al., 2010), the absence of the Hfq RNA chaperone (Sonnleitner et al., 2006) and OprF (Bouffartigues et al., 2015). Except for hyperosmolarity, the same conditions also increase the expression of sigX, another envelope stress ECFσ, which responds to stimuli via unknown regulatory mechanisms. SigX is up-regulated in LB containing high sucrose concentration (Bouffartigues et al., 2014) and by hypo-osmolarity (Brinkman et al., 1999; Bouffartigues et al., 2012), two conditions affecting membrane homeostasis. Initially, SigX was shown to be involved in oprF transcription together with AlgU. However, the characterization of the transcriptome and proteome of sigX mutant and sigX overexpressing strains suggested its involvement in the expression of fatty acids biosynthetic genes (Gicquel et al., 2013; Blanka et al., 2014; Fléchard et al., 2018). Although a SigX-overexpressing strain is more resistant to a 2 h CS at 15°C (Boechat et al., 2013), highlighting its role in membrane fluidity, its expression in response to temperature variations has not been investigated.
In addition to OprF (Jaouen et al., 2004), the products of at least two genes located immediately upstream sigX, cmaX and cmpX, could be involved in temperature adaptation. Indeed, CmaX is homologous to the CorA Mg2+-dependent channel. Interestingly, the increase of these transport proteins enhanced the Salmonella enterica survival at high temperature (O’Connor et al., 2009), a condition that increased transcription of both Escherichia coli corA (Richmond et al., 1999) and P. aeruginosa cmaX (Chan et al., 2016). CmpX is predicted to be a small mechanosensitive ion channel (MscS). These membrane tensions-gated pores are involved in osmotic downshift adaptation (Martinac et al., 2014). Inversely to corA, the expression of mscS in E. coli, increased in response to low temperature (White-Ziegler et al., 2008). In addition, the MscS mechanosensitive properties are affected by temperature (Koprowski et al., 2015), suggesting that these proteins could be membrane fluidity sensors. cmaX and cmpX are predicted to form an operon together with cfrX (Database for prokaryotic OpeRons DOOR) (Mao et al., 2009). The latter gene is predicted to encode a small hypothetical protein of 84 amino acids, the cellular location and function of which have not yet been demonstrated, however, cfrX, like cmpX, have been shown to belong to the SigX regulon (Gicquel et al., 2013; Schulz et al., 2015). Altogether, these observations suggest that this putative operon might be differentially regulated by temperature.
Here, we studied the impact of temperature shifts on the cmaX-cfrX-cmpX operon expression and investigated the involvement of the sigma factors AlgU and SigX in its regulation. We showed that this operon expression is induced under HS from a cmaX-proximal promoter region regulated by AlgU and under CS from a SigX-dependent promoter upstream cfrX. In connection with this observation we also demonstrated that the level of sigX mRNA was increased following an exposure to valinomycin, a membrane modification promoting agent, suggesting that SigX responds to membrane alteration. Interestingly, we observed that SigX is involved in the regulation of amrZ in response to CS. Finally, our data suggest a fine-tuned interconnection between the two cell wall stress response SigX and AlgU ECFσ that could be relevant in others stress conditions affecting the membrane fluidity and structure.
Results
Members of the cmaX-cfrX-cmpX Operon Are Differentially Regulated in Response to HS or CS
To study the effects of temperature on the expression of the cmaX-cfrX-cmpX operon, we chose to expose the strain H103 to thermal down- and up-shifts from 37°C without causing lethal cell damages. To this aim, H103 cells were grown at 37°C until the mid-log phase, before being exposed to sudden shifts at 4°C or at 60°C, during 15 or 30 min, respectively. Then, the bacterial cell viability was evaluated by flow cytometry after a double labeling with SYTO 9 (alive cells) and propidium iodide (dead cells) (Supplementary Figure S1). Compared to the control condition at 37°C, a downshift at 4°C during 15 or 30 min had no significant effect on H103 wild-type [(WT); Hancock and Carey, 1979] viability while an upshift at 60°C increased the percentage of dead cells from 0.6 to 1.9% after 15 min and from 0.3 to 2.4% after 30 min of exposure. Noticeably, the bacterial population submitted to a 30 min HS was more heterogeneous compared to 15 min exposure, with 18.1 and 11.8% of injured cells, respectively. Therefore, the following sublethal conditions (HS: 15 min at 60°C; CS: 30 min at 4°C) were applied in the subsequent experiments. The impact of these temperature shifts on the expression of the predicted cmaX-cfrX-cmpX operon was evaluated using RT-qPCR. Following HS, cmaX, cfrX, and cmpX mRNA levels were increased (Figure 1A and Supplementary Table S1) with cmaX transcripts being the most elevated. Conversely, only cfrX and cmpX were significantly up-regulated under CS (Figure 1B and Supplementary Figure S1). These results suggest that cfrX-cmpX could be thermo-regulated at the transcriptional level from a promoter lying upstream of cfrX.
FIGURE 1.
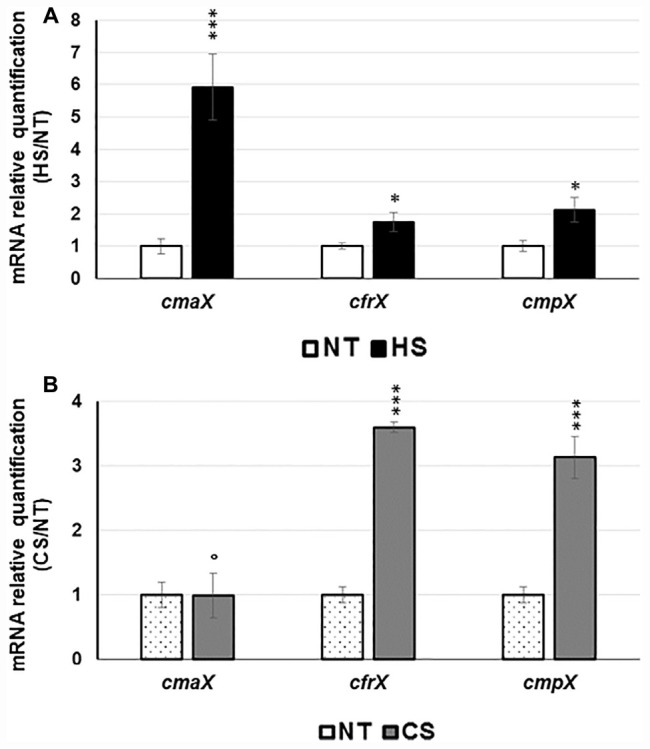
Effect of HS and CS on cmaX-cfrX-cmpX expression P. aeruginosa H103. Histograms show the means of normalized expression for each gene in HS (A) or CS (B) relatively to the control condition at 37°C as calibrator. Error bars indicate SEM of mean, n = 4. Significant differences between the sets of normalized expression obtained in each stress condition were calculated against those obtained at 37°C using a two-tailed t-test, and P-value are indicated by * or *** if < 0.05 or 0.0001, respectively. The symbol ° is indicated for non-significant results.
Identification of a Promoter Region Upstream of cfrX-cmpX
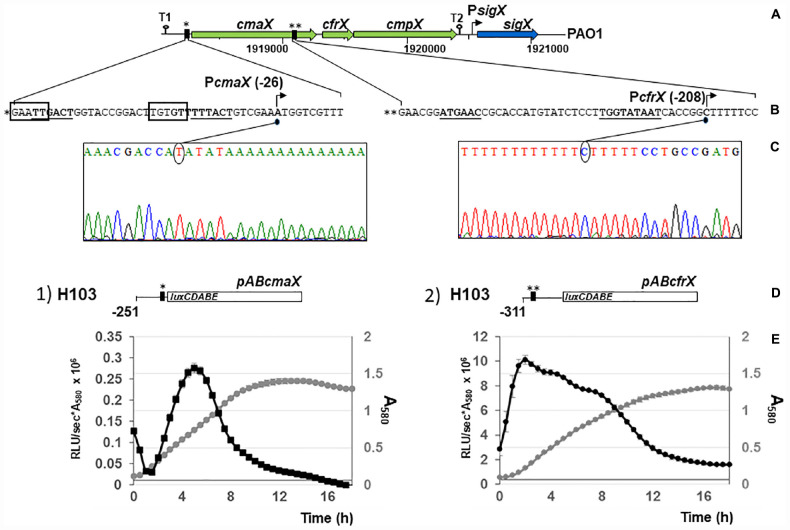
The 500 nucleotides DNA sequences upstream of each gene were analyzed using Bprom (Softberry). Two s70-like consensus promoters were predicted (Figures 2A,B), TTGACT(-35)-N17-TTTACT(-10) located in the intergenic region between rraA (PA1772) and cmaX, and ATGAAC(-35)-N17-TGGTATAAT(extended-10), within the cmaX encoding sequence, upstream of cfrX. No s70-like promoter was predicted within the 500 nucleotides upstream of cmpX. The consensus sequence of SigX-dependent promoters remains not precisely defined (Schulz et al., 2015) and we therefore could not search putative promoters by bioinformatic analysis. Using a 5′ RACE-PCR assay, the transcriptional start sites (TSS) of cmaX and cfrX were then identified at positions −26 for cmaX and −208 for cfrX relatively to the translational initiation site, downstream of the corresponding predictive −35 and −10 boxes (Figures 2B,C). The activity of each promoter region was measured during P. aeruginosa H103 growth in LB medium at 37°C using two bioluminescent reporters, pABcmaX and pABcfrX (Figures 2D,E). The relative luminescence (RL) signal from pABcmaX decreased at the beginning of the mid-log phase before increasing and finally reaching its maximum level of 0.27 RLU/sec∗A580 × 106 before decreasing at the end of the log phase. The pABcfrX promoter region was much more active, reaching a maximal level of 9.8 RLU/sec∗A580 × 106. The RL increased rapidly at the beginning, decreased slowly during the log phase and more steeply during the stationary phase. Confirming our sequence analysis, no transcriptional activity was detected from a transcriptional fusion harboring a 224 bp DNA fragment upstream of cmpX (data not shown). Our data confirmed the existence of a promoter region, which could differentially regulate the expression of cfrX and cmpX compared to that of the full-length cmaX-cfrX-cmpX operon in different environmental conditions.
FIGURE 2.
At least two promoter regions control the transcription of cmaX, cfrX, and cmpX in P. aeruginosa H103. (A) Schematic representation of the genomic environment of the predicted operon cmaX-cfrX-cmpX of P. aeruginosa. Large arrows indicate the direction of transcription of the ORFs, loops show the position of the putative rho-independent transcription terminators (T1 and T2), and the vertical black rectangles represent the two BPROM predicted promoters. (B) Sequences of the two putative promoters PcmaX and PcfrX. Predicted –35 and –10 motifs using BPROM are in bold case and underlined, the DNA-binding motif for AlgU is surrounded in black and TSSs are indicated by arrows. (C) Electrophoregrams obtained from 5′RACE-PCR experiments confirming the promoters upstream cmaX and cfrX genes. (D) Schematic representation of the construct harboring the pABcmaX or pABcfrX vectors. (A–C) * and ** correspond to the two distinct promoter regions. (E) Relative activity of the promoter regions upstream cmaX or cfrX in P. aeruginosa H103 during growth in LB medium at 37°C. Reporter constructs contain the PcmaX (construct 1, pABcmaX) or the PcfrX (construct 2, pABcfrX) promoter fused to the luxCDABE operon. The large white rectangle indicates the luxCDABE cassette, the vertical black rectangles represent promoters and a line shows the DNA sequence directly upstream cmaX or cfrX ORF. The position of the last nucleotide (relatively to the translational initiation site) included within the cloned DNA fragment is highlighted. The mean profile for growth and reporter assay from H103 transformed with construct 1 or 2 are shown and correspond to 3 independent experiments, ± SD. Transformants growth is presented in gray and the transcriptional activity pattern in black.
AlgU Is Involved in the Regulation of cmaX-cfrX-cmpX
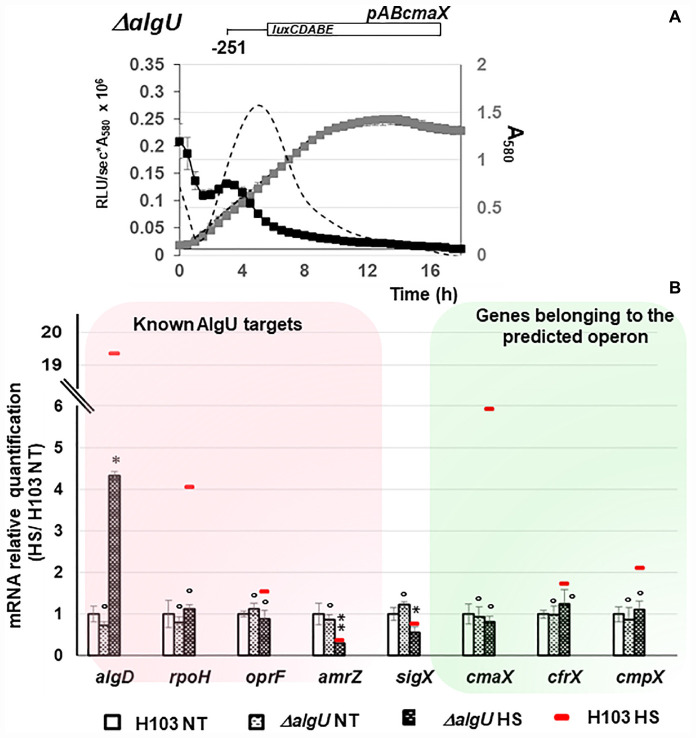
The expression of algU and its known targets algD, rpoH, oprF, and amrZ was quantified using RT-qPCR. The algU mRNA level was increased when bacteria were under HS (Supplementary Figure S2 and Supplementary Table S1). Accordingly, the mRNA levels of algD, rpoH, and oprF were significantly increased, while the expression of amrZ was reduced. The expression of the same genes was not significantly changed in response to a CS (Supplementary Figure S2 and Supplementary Table S1), confirming the involvement of P. aeruginosa H103 AlgU in the HS, but not in the CS response. By visual inspection, a putative AlgU recognition motif was detected [GAATT(-35)-N15-TGTGT(-10)] upstream of cmaX (Figure 2B), which is highly similar to the known AlgU consensus binding sequence (GAACTT-N16/17-TCTNN) (Firoved et al., 2002). To confirm the involvement of AlgU in this promoter region activity, the pABcmaX plasmid was introduced in an isogenic algU mutant and the bioluminescence signal was measured during growth in LB medium at 37°C (Figure 3A). The RL signal reached a maximum level of 0.21 RLU/sec∗A580 × 106 at the beginning of the mid-log phase, corresponding to an increase of about 50% compared to the WT, followed by a 50% decrease during exponential growth, suggesting an AlgU involvement in the control of this promoter region at this growth phase. The involvement of AlgU in the HS-inducible expression of the previously selected AlgU-regulated genes and of the cmaX-cfrX-cmpX operon was examined using RT-qPCR. Relatively to the expression measured in a WT strain similar expression levels were observed in the algU mutant grown at 37°C for all the genes tested, suggesting that AlgU only slightly contributes to their transcription in our control condition (Figure 3B and Supplementary Table S1). Although a residual heat-inducible expression of algD of about 30% was observed in the algU mutant, the rpoH and oprF expressions were unchanged when the algU mutant strain was exposed to HS, confirming that AlgU was required for their increased transcription in response to HS (Figures 3B, 6 and Supplementary Table S1). Interestingly, as observed for H103, the amrZ expression was down-regulated in the HS exposed algU mutant, suggesting an AlgU-independent mechanism of amrZ repression under HS. Similar to AlgU-targets rpoH and oprF, the heat-inducible expression of cmaX-cfrX-cmpX measured in H103 under HS was not observed in an algU mutant, indicating an AlgU involvement in the HS-induced increased expression of the operon (Figure 3B and Supplementary Table S1). To evaluate the involvement of RpoH in the heat-inducible expression of the operon a rpoH mutant in PAO1 was used. As previously observed in H103, the expression of cmaX was increased and in a lesser extent, those of cfrX and cmpX in PAO1 exposed to our HS condition (Supplementary Figure S3 and Table 1). A similar effect was observed in a rpoH mutant, suggesting that RpoH is not involved in the regulation of the operon in PAO1. Proteomic analysis of H103 under CS Total proteins from H103 cells harvested during mid-log phase and exposed or not to CS during 30 min were extracted and separated by two-dimensional gel electrophoresis (2D-E). Nine protein spots showing reproducible different intensities were identified by mass spectroscopy, among which some correspond to well-known markers of the CS response in other bacterial species (Beran and Simons, 2001; Gao et al., 2006; Table 1, Supplementary Figure S4, and Supplementary Table S2): two translation elongation factors (EF-Tu and Ef-Ts) and a well-described cold-inducible RNase (PNP)were up-regulated, and could together contribute to facilitate gene expression at the post-transcriptional level. Five of the identified proteins are involved in general metabolism and energy production. The arginine deiminase (ArcA), of the arginine fermentation pathway was down-regulated, while the 3-phosphoglycerate kinase (Pgk), the 4-hydroxyphenyl-pyruvate dioxygenase (Hpd), the electron transfer flavoproteins EtfA and EtfD were more abundant under CS, respectively (Table 1). Pgk, Hpd, and the Etf pathway contribute to oxidative energy production through their involvement in glycolysis, the phenylalanine/tyrosine catabolism and the utilization of branched-chain amino acids and fatty acids, respectively. In agreement with membrane lipid composition remodeling in response to CS, the abundance of the acetyl-coenzyme carboxylase A (AccA) was higher under CS (Figure 4 and Table 1). This protein is a subunit of the ACC complex and acts in concert with AccB, AccC, and AccD to provide the metabolic intermediate malonyl-CoA for de novo biosynthesis of fatty acids, which could contribute to restore the membrane fluidity upon a temperature downshift. Interestingly, the long chain fatty acid sensory regulator PsrA which negatively regulates the genes encoding the Etf pathway (Kojic et al., 2005; Kazakov et al., 2009) and acc genes, were previously shown to belong to the SigX regulon (Boechat et al., 2013; Gicquel et al., 2013; Blanka et al., 2014), suggesting that this ECFσ is able to respond to a sudden temperature downshift. The mRNA levels of accA and accB under CS condition were compared to those at 37°C in the WT strain (Figure 4A). In line with the increased abundance of AccA, accA and accB expression was increased. Although a decrease of the accB expression was previously shown to increase the c-di-GMP intracellular level via the Wsp system (Blanka et al., 2014), our CS conditions did not affect the c-di-GMP intracellular level (Supplementary Figure S5). Interestingly, the self-regulated psrA gene was down-regulated, indicating possibly a transcriptional de-repression of the Etf pathway (Figure 4). The observed positive effect of the temperature downshift on accA and accB mRNA abundances could result from the accumulation of SigX since its expression was up-regulated as well (Figure 4A). Consistently, CS caused an increase of the mRNA level of fabY (PA5174), a presumed direct SigX-target encoding the predominant FabY enzyme catalyzing the condensation of acetyl coenzyme A with malonyl-ACP (Yuan et al., 2012).
FIGURE 3.
Involvement of AlgU in cmaX, cfrX, and cmpX regulation under HS. (A) Activity pattern of the promoter region upstream cmaX in an algU mutant (black squares) or in H103 (dashed line) during growth in LB at 37°C of the H103 wild-type strain (black dashed line) or ΔalgU mutant (gray squares). n = 3, error bars indicate SD. (B) Histograms show the means of normalized expression relatively to H103 at 37°C as calibrator (white bars) for some known algU targets (red frame) and the studied operon (green frame) in H103 (red line) and in ΔalgU mutant exposed (dashed back bars) or not (dashed white bars) to HS. NT, not treated. Errors bars indicate SEM, n = 4. Significant differences between the sets of normalized expression obtained in HS condition were calculated against those obtained at 37°C for H103 using a two-tailed t-test and P-value, which are indicated by *, **, if < 0.05, 0.001, respectively. The symbol ° is indicated for non-significant results.
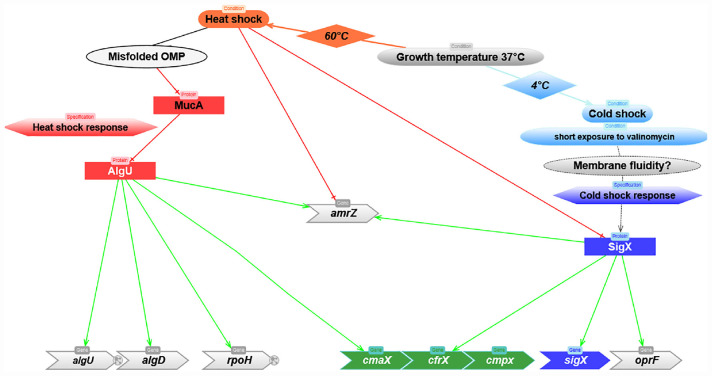
FIGURE 6.
Predicted model depicting the molecular mechanisms involved in the regulation of cmaX-cfrX-cmpX in response to HS and CS. Red and green lines correspond to negative or positive regulation, respectively.
TABLE 1.
List of identified proteins differentially expressed under CS in P. aeruginosa H103.
| Pseudocap | Spot number | PA number | Protein name | Putative function | Fold change CS/Control | Standard deviation |
| Energy metabolism | 1 | PA2951 | EtfA | Electron transfer flavoprotein, alpha subunit | 2.33 | 0.3 |
| 2 | PA0552 | Pgk | 3-phosphoglycerate kinase | 3.93 | 0.16 | |
| 3 | PA2953 | EtfD | Electron transfer flavoprotein-ubiquinone oxidoreductase | 2.85 | 0.5 | |
| Amino acid biosynthesis and metabolism | 4 | PA5171 | ArcA | Arginine deiminase | 0.29 | 0.14 |
| 5 | PA0865 | Hpd | 4-hydroxyphenylpyruvate dioxygenase | 3.25 | 1.43 | |
| Fatty acid and phospholipid metabolism | 6 | PA3639 | AccA | Acetyl-coenzyme A carboxylase carboxyl transferase (alpha subunit) | 3.21 | 0.44 |
| Translation, post-translational modification, degradation | 7 | PA3655 | Tsf | Elongation factor Ts | 1.69 | 0.03 |
| 8 | PA4265 | TufA | Elongation factor Tu | 2.43 | 0.18 | |
| 9 | PA4740 | Pnp | Polyribonucleotide nucleotidyltransferase | 1.61 | 0.09 |
Spots with an average fold change above 1.5 were considered. Standard deviations were calculated from three biological replicates.
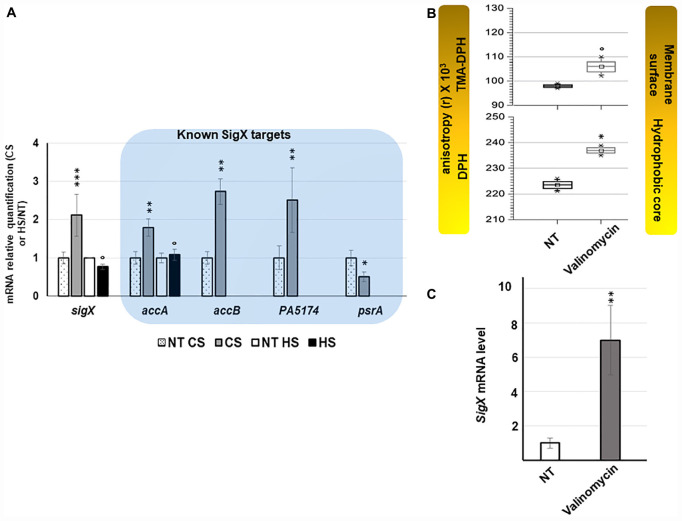
FIGURE 4.
sigX is up-regulated in response to CS and valinomycin promoting membrane stiffening. (A) Effect of thermal shocks on sigX expression and activity. Gray and black bars represent the mean normalized expression for sigX and its targets after CS or HS relatively to those obtained for H103 at 37°C (dotted and white bars). NT, not treated. Errors bars indicate SEM, n = 4. Significant differences between the sets of normalized expression obtained in each stress condition were calculated against those obtained at 37°C using a two-tailed t-test and P-value, which are indicated by *, **, or *** if < 0.05, 0.001, or 0.0001, respectively. The symbol ° is indicated for non-significant results. (B) TMA-DPH and DPH anisotropies was used to evaluate the effect of 45 μM valinomycin on membrane fluidity 2 h after the addition of the membrane acting-agent to mid-log phase cultures. Values obtained from unexposed cultures are indicated NT: not treated. Box plots correspond to each sets of data from three independent experiments. Means, medians and min/max anisotropies values are indicated by squares, lines and crosses, respectively. Significant differences between each condition were calculated using a two-tailed t-test and P-value, which are indicated by * if < 0.05. The symbol ° is indicated for non-significant results. (C) Relative expression of sigX after 2 h of exposure to 45 μM valinomycin. NT, not treated. Error bars indicate SEM, n = 3, P-value is calculated by two-tailed t-test and is indicated by ** if < 0.001.
Valinomycin Increases sigX Expression
To evaluate whether induced stiffening of the membrane could represent a stimulus increasing sigX expression, its mRNA level was measured after treatments of cells with sublethal concentrations of valinomycin, an antibacterial agent affecting the membrane. Alterations of membrane fluidity were evaluated by fluorescence anisotropy at the aqueous interface (1-[4-(trimethylamino)phenyl]-6-phenyl-1,3,5-hexatriene (TMA-DPH)) and at the hydrophobic core (1,6-diphenyl-1,3,5-hexatriene (DPH)) of the membrane. As previously reported (El Khoury et al., 2017) a significant increase of the anisotropy of both TMA-DPH and DPH probes was observed for valinomycin-exposed cells, confirming that valinomycin promotes membrane stiffening (Figure 4B and Supplementary Figure S6). Shorter exposition times were used to follow the early response of sigX to membrane perturbation (Figure 4C). After 2 h promoting the incorporation of valinomycin into the lipid bilayer, an increase of fluorescence anisotropy was observed with both membrane probes, showing that the membranes were more ordered and compact in this culture condition (Figure 4B) while a strong increase of sigX mRNA level was measured (Figure 4C). Taken together, this set of data shows that sigX responds to membrane stressors causing membrane stiffening (Figure 6).
SigX Is Involved in the Expression of cfrX-cmpX and amrZ Under CS
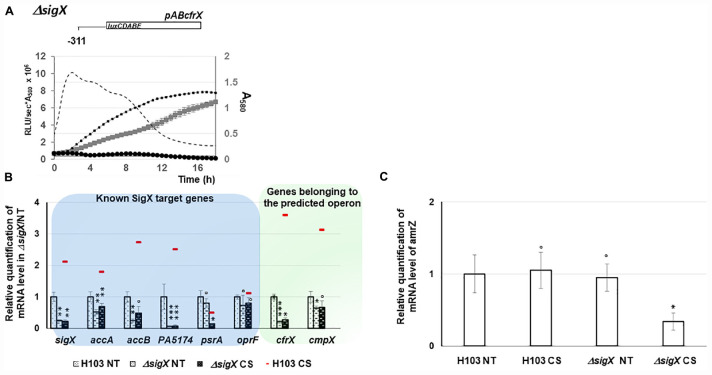
When comparing the relative bioluminescence signals measured from H103 or ΔsigX (PAOSX) harboring pABcfrX, only a residual background activity of about 8% was observed for ΔsigX during the log phase, revealing a major contribution of SigX to cfrX promoter activity (Figure 5A). The expression of the set of previously identified SigX targets was further evaluated in PAOSX at 37°C and under CS by RT-qPCR. Except for psrA, the expression of sigX as well as of the other SigX-regulated genes, accA, accB and PA5174 (fabY), was significantly down-regulated in ΔsigX compared to H103 (Figure 5B and Supplementary Table S1). Confirming that cfrX and cmpX are under the control of SigX in our control condition at 37°C, their expression was likewise down-regulated. Under CS, the mRNA levels of these genes, including cfrX and cmpX, were not significantly increased in ΔsigX (Figure 5B and Supplementary Table S1), confirming that their cold-inducible expression was SigX-dependent. The psrA mRNA level was lower in ΔsigX than in H103 when cells were exposed to a sudden temperature downshift (Figure 5B and Supplementary Table S1). This result might indicate that the repression of psrA in response to CS is likely related to a SigX-independent mechanism. Taken together, these data support that the cfrX-cmpX transcriptional unit is under the direct control of SigX, which is responsible for the increase of its transcription in response to CS.
FIGURE 5.
Involvement of SigX in cmaX, cfrX, and cmpX and amrZ regulation under CS. (A) Growth of H103 (black squares, dashed line) and PAOSX sigX mutant (gray squares) in LB at 37°C and activity pattern of the promoter region upstream cfrX in H103 (dashed line) and PAOSX (black circles) background. N = 3, error bars indicate SD. (B) Means of normalized expression relatively to H103 at 37°C (dotted white bars) of sigX and its targets (blue frame) and genes belonging to the operon (green frame), in H103 (red line) and in PAOSX exposed (dashed back bars) or not (dashed white bars) to CS. NT, not treated. (C) White bars show mean normalized amrZ expression (relative to H103 at 37°C) in H103 or PAOSX exposed or not to CS. (B,C) Error bars indicate SEM for n = 4, P-value were calculated by two-tailed t-test and are indicated by *, **, or *** if < 0.05, 0.001, or 0.0001, respectively. The symbol ° is indicated for non-significant results.
We next focused on AmrZ, whose expression was co-regulated with sigX during HS (Figure 3B), and which was shown by RNA-seq analysis to positively control cfrX and cmpX, as well as the CS-inducible SigX-target genes accA and PA5174 (fabY) (Jones et al., 2014). Consistently, AmrZ binding was identified upstream of sigX by ChIP-Seq in P. fluorescens F113 genome (Martínez-Granero et al., 2014). Altogether this set of data suggests that SigX and AmrZ could be involved in a feedback loop depending on the environmental condition. To evaluate the contribution of SigX in amrZ transcription in response to CS, the amrZ mRNA level was measured in the WT and ΔsigX strains exposed or not to CS. In H103, the amrZ expression was not upregulated in response to CS (Figure 5C). However, it was about three times down-regulated in PAOSX exposed to CS compared to untreated and CS treated H103, indicating that SigX could be involved in amrZ transcription in response to CS (Figure 6).
Discussion
Sudden temperature variations can trigger a cell wall stress response (CWSR) by altering the membrane composition, structure and functionality. Although the molecular mechanisms involved in the HS response are relatively well characterized in P. aeruginosa, involving the AlgU ECFσ via its regulon member RpoH, those involved in the CS response have only been scarcely described. Therefore, characterizing these mechanisms should highlight new effectors playing a key role in envelope homeostasis and in the adaptive capacity of P. aeruginosa under other chemical and physical stresses. In the present study, we focused on the description of the uncharacterized cmaX-cfrX-cmpX operon regulatory mechanisms by a combination of approaches, including bioinformatics, transcriptomics, proteomics and fluorescence polarization assays. Based on the identification of two promoter regions located upstream of cmaX and cfrX-cmpX, we demonstrated that AlgU and SigX differentially regulate the members of this cluster in function of temperature variations, and we therefore postulate that these two CWSR ECFσ respond to specific stimuli that can be caused by opposite temperature variations from 37°C (Figure 6). In agreement with previous results obtained by genome-wide transcription start site (TSS) mapped by RNA-seq (Wurtzel et al., 2012; Schulz et al., 2015), we confirmed using RACE-PCR that the cmaX-cfrX-cmpX transcription can be initiated from at least one promoter localized 26 nucleotides upstream of the cmaX translation initiation codon. Altogether, our results support cmaX as a member of AlgU regulon, but also highlight the existence of additional molecular mechanisms that could be involved in the cluster regulation, since a weaker increase of cfrX and cmpX mRNA levels was also measured under HS. This effect is in line with the previous transcriptomic studies aiming at characterizing P. aeruginosa AlgU regulon after a treatment with the D-cycloserine peptidoglycan synthesis inhibitor (Wood and Ohman, 2015), or the response to elevated temperature at 45°C (Chan et al., 2016). If a HS induction of cmaX was measured, neither cfrX nor cmpX were up-regulated, although these three genes have been previously shown to be co-transcribed by RT-PCR (Brinkman et al., 1999). We also showed that under HS the expression profile of the operon is not altered in a rpoH genetic background compared to the one obtained in the WT PAO1 strain, suggesting that RpoH is not involved in the HS-inducible expression of cmaX in our conditions and comforting the putative AlgU promoter sequence identified upstream the gene. In addition, RpoH does not seem to be involved in the regulation of cfrX and cmpX as well as sigX and oprF in our condition in PAO1. However, RNA-seq analyses showed a decrease in the expression of cfrX-cmpX, oprF, and sigX, which could be auto-regulated (Gicquel et al., 2013), as well as an increase of cmaX in a rpoH overexpressing strain (Schulz et al., 2015), suggesting a potential role of RpoH in the regulation of the operon that would be function of the P. aeruginosa strain and/or the condition used. By regulating the expression of many proteases and RNA chaperones such as Hfq that promotes RNA-RNA interactions (Schulz et al., 2015), RpoH could play a key role in the regulation of RNA metabolism by modulating the abundance of many proteins under stress conditions. Since an instability of SigX in a sigX overexpressing strain has been reported by Boechat et al. (2013), it could be of interest to better understand RpoH role. Interestingly, the recent development of the GRIL-seq tool to identify the targets of small RNAs has shown that the mRNAs of cmpX, but also of oprF, mucA encoding the AlgU anti-sigma factor, and amrZ are all targets of the sRNA PrrF1 in vivo (Han et al., 2016). This putative but intriguing regulatory mechanism could be involved in the low but reproducible decrease observed in our study of sigX and amrZ mRNA levels in P. aeruginosa H103 strain when exposed to HS. We propose thus that the interplay between the two major ECFσ controlling the envelope stress response could involve small regulatory RNAs and/or proteases. In line with this hypothesis, the expression of sigX was shown to be repressed in P. putida KT2440 exposed to elevated pressure, another condition that positively regulates HS but negatively the expression of CS proteins (Follonier et al., 2013).
Another major finding of this study is the increased sigX expression following an alteration of the membrane properties or CS. In addition, these results are in agreement with a recent publication, which shows that CspA1 contributes to SigX expression in Pseudomonas plecoglossicida (Huang et al., 2020). This result also reinforces the hypothesis of an atypical dependent regulatory mechanism requiring further characterization. Using bioinformatics, one of the main candidates suspected to be involved in SigX activity regulation is CfrX (Staroń et al., 2009). In agreement with previous transcriptomic data (Gicquel et al., 2013; Blanka et al., 2014), we here confirmed that cfrX and cmpX are two SigX targets. More precisely, we show that their expression is controlled by a large SigX-dependent promoter region, involving a promoter located at −208 nucleotides from the cfrX translation start site was identified. Remarkably, this promoter region has a more complex activity pattern than that of cmaX in the WT, suggesting the existence of several promoters, possibly under the control of different regulators. Interestingly, a 5′UTR of 13 nucleotides has also been identified by TSS mapping RNA-seq (Schulz et al., 2015), suggesting the existence of at least one additional proximal promoter, whose activity remains, however, to be experimentally validated. No specific TSS for cmpX was identified by this technique (Schulz et al., 2015), in agreement with our own unsuccessful attempts, confirming that cfrX and cmpX are co-transcribed (Brinkman et al., 1999). In view of the mechanisms known to regulate the activity of ECFσ, the role of SigX in the regulation of a large promoter region upstream of the cfrX-cmpX transcriptional unit supports the hypothesis of the involvement of CfrX, but also of the mechanosensitive ion channel CmpX in the regulation of SigX activity. In connection with CmpX putative activity as a membrane tension sensor, and the influence of temperature on membrane structure, we artificially altered the membrane fluidity using valinomycin to follow sigX response. Exposure to this compound led to an increased membrane stiffness after a relatively short time treatment, validating the chosen experimental strategy. Valinomycin was shown to directly induce alterations in the turnover of phospholipid fatty acids and in the membrane potential in human erythrocytes with respect to its ion channel function (Dise and Goodman, 1985). Following the valinomycin treatment, the sigX mRNA relative abundance increased. Similar results were previously observed when P. aeruginosa was grown in presence of high sucrose concentration (Bouffartigues et al., 2014), a treatment stabilizing the membrane composition during dehydration (Leslie et al., 1995). Although the molecular mechanism remains to be determined, our data strongly suggest that the membrane stiffness would trigger a bacterial response via SigX. Paradoxically, expression of sigX was also increased in response to hypo-osmolarity (Brinkman et al., 1999; Bouffartigues et al., 2012), a condition known for its tendency to increase membrane fluidity (Los and Murata, 2004). However, these effects have not yet been confirmed in P. aeruginosa and it would be interesting to evaluate them in hypo-osmotic shock conditions. In view of currently available data on sigX expression regulation (Chevalier et al., 2018) and the known effects of CS on bacteria (Barria et al., 2013), other mechanisms could be connected to our observations. Finally, our results confirm the involvement of SigX in controlling membrane fluidity since a sigX mutant displays a more rigid membrane, leading to impaired carbon metabolism (Fléchard et al., 2018). As depicted on Figure 6, the highly thermo-regulated processes described in this study highlight an interconnection between the two ECFσ AlgU and SigX to coordinate the CWSR in P. aeruginosa, in which the function of the members of the cmaX-cfrX-cmpX operon will now have to be clarified with regards to SigX regulation as well as envelope homeostasis. Moreover, we postulate that the expression and/or the activity of these two ECFσ would be coordinated by mechanisms that remain to be identified.
Materials and Methods
Bacterial Strains, Media and Growth Conditions
The bacterial strains used in this study are listed in the Supplementary Table S3. Liquid cultures were inoculated in LB medium (171 mM NaCl) at an initial absorbance at 580 nm (A580) of 0.08. Bacteria were grown at 37°C with orbital shaking at 180 rpm. Two solid media were used, LB containing 1.5% (w/v) technical agar (AES CHEMUNEX) and Pseudomonas Isolation Agar (PIA, Gibco-BRL, Grand Island, NY) for the selection of Pseudomonas strains. For CS or HS experiments, cells were grown as described above until the absorbance at 580 nm was between 0.5 and 0.6 and incubated at 4°C into iced water or at 60°C in stove for the indicated time. Control experiments were done by incubating cultures at 37°C in the same conditions. When necessary, the following antibiotics were added at the indicated concentration (μg.mL–1): gentamicin (Gm), 50; tetracycline (Tc), 250; carbenicillin (Cb), 300 for Pseudomonas strains, and Gm, 15; Tc, 15; ampicillin (Ap), 100 for E. coli strains.
Bacterial Cell Viability Assays by Flow Cytometry
To determine the viability of cells exposed at 37, 4, or 60°C, the LIVE/DEADTM BacLightTM Bacterial Viability and Counting Kit, for flow cytometry (Invitrogen, Molecular Probes, Carlsbad, CA) was used. Briefly, 105 cells exposed to CS or HS were diluted in 1 mL of PBS and stained following the manufacturer’s instructions. For analysis of the bacterial population using CytoFlex S flow cytometer (Beckman coulter Life science, Indianapolis, United States), red (propidium iodide, PI) and green (SYTO 9) fluorescence was recorded. An aliquot of cells was killed with 100% ethanol to act as a death control. The SYTO9 stained cells were detected by an excitation with 22 mW blue laser at 488 and with emission wavelength at 525 nm (green, with band pass filter of 40 nm), while the PI stained cells were detected by 690 nm (red, with band pass filter of 50 nm).
Genes Expression Analysis by Real Time Quantitative RT-PCR
Total RNA was extracted from bacterial cultures by the hot acid-phenol method as previously described (Bouffartigues et al., 2012) and tested by PCR for the absence of contaminating DNA using the primers pair FcfrX/RcfrX (Supplementary Table S4). The RNA concentration, the protein and solvent contaminations were determined by measuring the absorbance at 260, 280, and 230 nm, respectively. Total RNA quality was checked on a 2% agarose gel/1X-TAE electrophoresis buffer prior to use for cDNA synthesis. The mRNAs of interest were quantified by real-time PCR amplification of their cDNAs. Each primers pair was validated by verifying that the PCR efficiency was above 0.90, and that a single PCR product with the expected Tm was obtained. PCR were performed on cDNAs at least in triplicate and carried out in SYBR Green PCR Master MixTM (Applied Biosystems), with 300 nM of each primer (ABI 7500 Fast Q-PCR system, Applied Biosystems). The cDNA-generated signals were internally corrected with the 16S rRNA cDNA signal. The mRNA expression level was calculated by comparing threshold cycle (Ct) of target genes with control sample group and the relative quantification (RQ) data was determined with the 2–ΔΔCt method using the RQ manager v1.2.1 and the Microsoft-excel based softwares (Muller et al., 2002).
Bioinformatics Tools
DNA Sequences were extracted from the Pseudomonas genome database (Winsor et al., 2016) and analyzed using BPROM bacterial promoter prediction software to predict putative promoters1.
5′ RACE Assay
Predicted transcription start sites (TSSs) were confirmed using the 5′ RACE (rapid amplification of cDNA ends) procedure (3′/5′ RACE 2nd Generation kit; Roche Molecular Biochemicals) according to the manufacturer’s instructions. Briefly, cDNA was produced from 1 μg of total RNA using gene-specific primer AS1 (Supplementary Table S4) and further purified using the High Pure PCR product purification kit (Roche). Nested PCR was further achieved using a gene-specific AS2 primer (Supplementary Table S4) and oligo(dT) primer. Finally, the PCR product was cloned into pGEM-T Easy vector according to the manufacturer’s instructions (Promega) and 5 clones were sequenced to identify the 5′ end of the specific mRNA (Beckman Coulter Genomics, Villepinte, France).
Construction of Transcriptional Fusions
Promoter regions of interest were fused to the promoter-less luxCDABE cassette in the replicative pAB133 vector (Supplementary Table S3). DNA fragments containing the cmaX, cfrX, or cmpX promoter regions were PCR-amplified with the primers pairs FCmaX-RCmaX, FCFRX- RCFRX, or FCmpX-RCmpX (Supplementary Table S4). The SacI-SpeI-digested PCR product was inserted at the same restriction sites into pAB133 yielding pABcmaX, pABcfrX, and pABcmpX, respectively. The insert was verified by DNA sequencing.
Bioluminescence Assay
P. aeruginosa strains containing pAB-reporter vectors were grown in covered, white 96-well optiplates with a flat transparent bottom (BD Falcon, San Jose, CA). Bioluminescence and absorbance were simultaneously measured throughout bacterial growth using a thermo-controlled multi-mode plate reader (Xenius, SAFAS, Monaco). The bioluminescence values [in relative light units (RLU) × s–1] were divided by the absorbance values at 580 nm of the cultures, yielding relative bioluminescence values (in RLU/sec∗A580). The relative luminescence values of the negative P. aeruginosa control strain harboring the promoter-less vector pAB133 were subtracted from those containing the studied promoter, as described by Bazire et al. (2005). Each set of experiment was performed at least three times.
Construction of a P. aeruginosa H103 algU Deletion Mutant
To obtain the ΔalgU strain isogenic to H103, the gene was replaced by a truncated:Gm disrupted allele carried by the PEXUGL (derivated-PEX100Tlink vector (Quénée et al., 2005, Supplementary Table S3) using the previously described strategy by Bazire et al. (2010). Briefly, this construct was introduced into E. coli S17.1 (Simon et al., 1983) and mobilized into P. aeruginosa H103 by conjugational mating. Gm-trans conjugates were isolated, the Gm-resistance gene excised and the algU deletion confirmed by PCR using the primers pair algU1-algU4 (Bazire et al., 2010).
Membrane Fluidity Assays by Fluorescence Anisotropy
Bacterial cells grown in LB at 37°C to mid-log phase (A580 = 0.5) were exposed to 45 μM of valinomycin during the indicated time, and harvested by centrifugation, washed twice in sterile Tris-Cl buffer (15 mM, pH 7), and suspended in the same buffer to OD580 = 0.2. Membrane fluidity was measured using the probe 1,6-diphenyl-1,3,5-hexatriene (DPH, Sigma Aldrich) or its derivative N,N,N-Trimethyl-4-(6-phenyl-1,3,5-hexatrien-1-yl)phenylammonium p-toluenesulfonate (TMA-DPH) as previously described (Baysse et al., 2005).
Proteomic Analysis
Impact of a CS on the proteome of P. aeruginosa H103 was performed by 2D-electrophoresis as previously described by Massier et al. (2012) with minor modifications. Briefly, total proteins were extracted from cells, exposed or not to a CS during 30 min, as follow: bacteria were harvested from 10 mL of three independent control or CS-treated cultures by centrifugation (7,500 g for 20 min at 4°C) and suspended in 5 mL of lysis buffer (30 mM Tris-Cl pH8 and 0.1 mM phenylmethanesulfonylfluoride) prior to sonication. 350 μg of proteins were solubilized in 325 μL of re-hydratation buffer (7 M urea, 2 M thiourea, 4% CHAPS 3-((3-cholamidopropyl) dimethylammonio)-1-propanesulfonate, Bio-Rad Laboratories], 65 mM DTT, 0.5% carrier ampholytes mixture pH3-10 (Bio-Rad Laboratories) prior to loading on ReadyStripTM IPG strip (17 cm, pH 4–7, Bio-Rad Laboratories). After a passive rehydration step during 16 h, isoelectric focusing parameters were set as 50 μA/strip at 20°C and carried out using the following conditions: 0–100 V linear ramp for 45 min, 100–250 V rapid ramp for 15 min, 250–10,000 V gradual ramp for 2 h 45 and hold at 10,000 V until a total of 52,000 V × h was reached. The proteins were separated by SDS-PAGE 10% and coomassie blue stained. Two gels per protein sample were performed and scanned using a GS-800 densitometer (Bio-Rad Laboratories) to allow normalization and quantification of reproducible spots detected with PDQuest 2-DE analysis 7.4.0 software (Bio-Rad Laboratories). Protein spots were excised from gels and characterized through matrix-assisted laser desorption ionization time of flight mass spectrometry (MALDI-TOF/MS) analysis. Search parameters and statistical analyses of the sequences for proteins identification were determined by the probability-based Mowse score offered by MASCOT software as described previously (Barbey et al., 2012) using SwissProt 2020. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (Perez-Riverol et al., 2019) partner repository with the dataset identifier PXD021299.
Statistics and Reproducibility
All experiments were carried out at least three times independently. For RT-qPCR experiment P-values were calculated with the two-tailed t-test using qstats a complementary function of the Microsoft-excel based softwares previously mentioned (Muller et al., 2002; Figures 1, 3B,C, 4A,C, 5B,C and Supplementary Figure S2). Live/dead experiments, 2D-E and reporter-based assays were repeated three times with similar results as specify by the indicated standard deviation (Figures 2D, 3A, 5A and Supplementary Figures S1–S3). Box plot and statistical analyses for fluorescence anisotropy data were performed with Qtiplot software (Figure 4B and Supplementary Figure S4).
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (63) partner repository with the dataset identifier PXD021299.
Author Contributions
EB and SC designed the research and wrote the manuscript with input from all authors. EB, IS, OM, DT, JO, AD, AT, RD, CA, and MN performed the experiments.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding. DT and RD were recipients of doctoral fellowships from the Région Normandy, CA from Campus France and FUNAI (Nigeria), AT of a post-doctoral fellowship from European Union (FEDER) and Région Normandy. This work was supported by grants from the Evreux Portes de Normandie (France), the Conseil Général de l’Eure, the Région Normandy and European FEDER funds. MN and GB-W acknowledge support from the Helmholtz program BioInterfaces in Technology and Medicine (BIFTM) of KIT.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.579495/full#supplementary-material
Flow cytometry analysis of P. aeruginosa H103 viability exposed or not to CS or HS. Bacteria were grown in LB until the mid-log growth phase and subjected to CS or HS as indicated. 105 cells were stained using Syto9 and PI and then analyzed by flow cytometry. (A) Cytograms are representative of three independent experiments. Live and dead cells emitting a green or a red fluorescence are indicated within a red or purple frame, respectively. The green frame corresponds to cells with membrane damages. The mean percentage of live, dead and injured cells is indicated in each frame. (B) Histograms representing percent of cell viability from the experiments of live/dead assays (n = 3).
Effect of HS and CS on algU expression and activity. Black and gray bars show means normalized expression of algU and its targets after HS or CS relatively to the control condition at 37°C as calibrator (white and dotted bars). NT: not treated. Error bars indicate SEM, n = 4. Significant differences between the sets of normalized expression obtained in each stress condition were calculated against the those obtained at 37°C using a two-tailed t-test, and P-value are indicated by ∗, ∗∗, or ∗∗∗ if < 0.05, 0.001, or 0.0001, respectively. The symbol ° is indicated for non-significant results.
RpoH is not involved in the cmaX, cfrX, and cmpX regulation in response to HS in PAO1. Means of normalized expression for algU targets (red frame), the predicted operon (cmaX-cfrX-cmpX) and sigX in PAO1 (red line) or in an isogenic rpoH mutant exposed (dashed back bars) or not (dashed white bars) to HS.
2-DE total protein profiles of P. aeruginosa exposed to CS from 37 to 4°C or not (control). Gels are Coomassie blue stained and are representative of three independent proteins extractions. The protein spots satisfying both statistical biological variation analysis and mass spectroscopy identification scores were numbered according to Table 1 and Supplementary Table 2.
c-di-GMP level quantification by LC-MS/MS in H103 strain exposed or not to a CS. Intracellular c-di-GMP was extracted in H103 strain exposed (square) or not (point) to CS and quantified LC-MS/MS. Briefly, the absorbance at 580 nm of cultures were recorded and cells from 2 mL of culture were collected. Then cells were harvested by centrifugation, washed in 1 mL of sterile physiological water and c-di-GMP was extracted twice with extraction buffer without internal standard and quantified as previously described (Bouffartigues et al., 2015; Strehmel et al., 2015).
Effect of a sublethal dose of valinomycin on membrane fluidity measured by fluorescence polarization in stationary growth phase. TMA-DPH and DPH anisotropies were used to evaluate the effect of 45 μM valinomycin on membrane fluidity 22 h after the addition of the membrane acting-agent to mid-log phase cultures. Values obtained from unexposed cultures are indicated as NT: not treated. Box plots correspond to each sets of data from three independent experiments. Means, medians and min/max anisotropies values are indicated by squares, lines and crosses, respectively. Significant differences between each condition were calculated using a two-tailed t-test, and P-value are indicated by ∗ if < 0.05.
Normalized expression of the studied genes under HS or CS in the indicated genetic backgrounds relative to P. aeruginosa H103 at 37°C. Values are means ± SEM for at least three independent experiments.
Characteristics of proteins identified by Maldi-TOF analysis. MS and MS/MS correspond to the mass spectrometry method used for the protein identification.
Bacterial strains and plasmids used in this study. Cbr, carbenicillin resistance; Gmr, gentamycin resistance; Tcr, tetracycline resistance.
Primer sequences of the indicated genes used for quantitative RT-qPCR reactions, transcription start sites identification and transcriptional fusions construction.
References
- Allan B., Linseman M., MacDonald L. A., Lam J. S., Kropinski A. M. (1988). Heat shock response of Pseudomonas aeruginosa. J. Bacteriol. 170 3668–3674. 10.1128/jb.170.8.3668-3674.1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aspedon A., Palmer K., Whiteley M. (2006). Microarray analysis of the osmotic stress response in Pseudomonas aeruginosa. J. Bacteriol. 188 2721–2725. 10.1128/jb.188.7.2721-2725.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldini R. L., Starkey M., Rahme L. G. (2014). Assessing Pseudomonas virulence with the nonmammalian host model: Arabidopsis thaliana. Methods Mol. Biol. Clifton. N.J. 1149 689–697. 10.1007/978-1-4939-0473-0_53 [DOI] [PubMed] [Google Scholar]
- Barbey C., Crépin A., Cirou A., Budin-Verneuil A., Orange N., Feuilloley M., et al. (2012). Catabolic pathway of gamma-caprolactone in the biocontrol agent Rhodococcus erythropolis. J. Proteome Res. 11, 206–216. 10.1021/pr200936q [DOI] [PubMed] [Google Scholar]
- Barria C., Malecki M., Arraiano C. M. (2013). Bacterial adaptation to cold. Microbiol. Engl. 159 2437–2443. 10.1099/mic.0.052209-0 [DOI] [PubMed] [Google Scholar]
- Baysse C., Cullinane M., Dénervaud V., Burrowes E., Dow J. M., Morrissey J. P., et al. (2005). Modulation of quorum sensing in Pseudomonas aeruginosa through alteration of membrane properties. Microbiology (Reading, Engl.) 151, 2529–2542. 10.1099/mic.0.28185-0 [DOI] [PubMed] [Google Scholar]
- Bazire A., Dheilly A., Diab F., Morin D., Jebbar M., Haras D., et al. (2005). Osmotic stress and phosphate limitation alter production of cell-to-cell signal molecules and rhamnolipid biosurfactant by Pseudomonas aeruginosa. FEMS Microbiol. Lett. 253 125–131. 10.1016/j.femsle.2005.09.029 [DOI] [PubMed] [Google Scholar]
- Bazire A., Shioya K., Soum-Soutera E., Bouffartigues E., Ryder C., Guentas-Dombrowsky L., et al. (2010). The sigma factor AlgU plays a key role in formation of robust biofilms by nonmucoid Pseudomonas aeruginosa. J. Bacteriol. 192 3001–3010. 10.1128/jb.01633-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beran R. K., Simons R. W. (2001). Cold-temperature induction of Escherichia coli polynucleotide phosphorylase occurs by reversal of its autoregulation. Mol. Microbiol. 39 112–125. 10.1046/j.1365-2958.2001.02216.x [DOI] [PubMed] [Google Scholar]
- Blanka A., Schulz S., Eckweiler D., Franke R., Bielecka A., Nicolai T., et al. (2014). Identification of the alternative sigma factor SigX regulon and its implications for Pseudomonas aeruginosa pathogenicity. J. Bacteriol. 196 345–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boechat A. L., Kaihami G. H., Politi M. J., Lepine F., Baldini R. L. (2013). A novel role for an ECF sigma factor in fatty acid biosynthesis and membrane fluidity in Pseudomonas aeruginosa. PLoS One 8:e0084775. 10.1371/journal.pone.0084775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouffartigues E., Duchesne R., Bazire A., Simon M., Maillot O., Dufour A., et al. (2014). Sucrose favors Pseudomonas aeruginosa pellicle production through the extracytoplasmic function sigma factor SigX. FEMS Microbiol. Lett. 356 193–200. [DOI] [PubMed] [Google Scholar]
- Bouffartigues E., Gicquel G., Bazire A., Bains M., Maillot O., Vieillard J., et al. (2012). Transcription of the oprF gene of Pseudomonas aeruginosa is dependent mainly on the SigX sigma factor and is sucrose induced. J. Bacteriol. 194 4301–4311. 10.1128/jb.00509-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouffartigues E., Moscoso J. A., Duchesne R., Rosay T., Fito-Boncompte L., Gicquel G., et al. (2015). The absence of the Pseudomonas aeruginosa OprF protein leads to increased biofilm formation through variation in c-di-GMP level. Front. Microbiol. 6:630. 10.3389/fmicb.2015.00630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkman F. S., Schoofs G., Hancock R. E., De Mot R. (1999). Influence of a putative ECF sigma factor on expression of the major outer membrane protein, OprF, in Pseudomonas aeruginosa and Pseudomonas fluorescens. J. Bacteriol. 181 4746–4754. 10.1128/jb.181.16.4746-4754.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K.-G., Priya K., Chang C.-Y., Abdul Rahman A. Y., Tee K. K., Yin W.-F. (2016). Transcriptome analysis of Pseudomonas aeruginosa PAO1 grown at both body and elevated temperatures. PeerJ 4:e2223. 10.7717/peerj.2223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevalier S., Bouffartigues E., Bazire A., Tahrioui A., Duchesne R., Tortuel D., et al. (2018). Extracytoplasmic function sigma factors in Pseudomonas aeruginosa. 2019. Biochim. Biophys. Acta Gene. Regul. 7 706–721. 10.1016/j.bbagrm.2018.04.008 [DOI] [PubMed] [Google Scholar]
- Chevalier S., Bouffartigues E., Bodilis J., Maillot O., Lesouhaitier O., Feuilloley M. G. J., et al. (2017). Structure, function and regulation of Pseudomonas aeruginosa porins. FEMS Microbiol. Rev. 41 698–722. [DOI] [PubMed] [Google Scholar]
- Crabbe A., Pycke B., Van Houdt R., Monsieurs P., Nickerson C., Leys N., et al. (2010). Response of Pseudomonas aeruginosa PAO1 to low shear modelled microgravity involves AlgU regulation. Environ. Microbiol. 12 1545–1564. [DOI] [PubMed] [Google Scholar]
- Dise C. A., Goodman D. B. (1985). The relationship between valinomycin-induced alterations in membrane phospholipid fatty acid turnover, membrane potential, and cell volume in the human erythrocyte. J. Biol. Chem. 260 2869–2874. [PubMed] [Google Scholar]
- El Khoury M., Swain J., Sautrey G., Zimmermann L., Van Der Smissen P., Décout J.-L., et al. (2017). Targeting bacterial cardiolipin enriched microdomains: an antimicrobial strategy used by amphiphilic aminoglycoside antibiotics. Sci. Rep. 7:10697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito S., De Simone G. (2017). Update on the main MDR pathogens: prevalence and treatment options. Infez. Med. 25 301–310. [PubMed] [Google Scholar]
- Firoved A. M., Boucher J. C., Deretic V. (2002). Global genomic analysis of AlgU (sigma(E))-dependent promoters (sigmulon) in Pseudomonas aeruginosa and implications for inflammatory processes in cystic fibrosis. J. Bacteriol. 184 1057–1064. 10.1128/jb.184.4.1057-1064.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fléchard M., Duchesne R., Tahrioui A., Bouffartigues E., Depayras S., Hardouin J., et al. (2018). The absence of SigX results in impaired carbon metabolism and membrane fluidity in Pseudomonas aeruginosa. Sci. Rep. 8:17212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Follonier S., Escapa I. F., Fonseca P. M., Henes B., Panke S., Zinn M., et al. (2013). New insights on the reorganization of gene transcription in Pseudomonas putida KT2440 at elevated pressure. Microb. Cell Factor. 12:30. 10.1186/1475-2859-12-30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H., Yang Z. K., Wu L., Thompson D. K., Zhou J. (2006). Global transcriptome analysis of the cold shock response of Shewanella oneidensis MR-1 and mutational analysis of its classical cold shock proteins. J. Bacteriol. 188 4560–4569. 10.1128/jb.01908-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gicquel G., Bouffartigues E., Bains M., Oxaran V., Rosay T., Lesouhaitier O., et al. (2013). The extra-cytoplasmic function sigma factor sigX modulates biofilm and virulence-related properties in Pseudomonas aeruginosa. PLoS One 8:e080407. 10.1371/journal.pone.0080407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han K., Tjaden B., Lory S. (2016). GRIL-seq provides a method for identifying direct targets of bacterial small regulatory RNA by in vivo proximity ligation. Nat. Microbiol. 2:16239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock R. E., Carey A. M. (1979). Outer membrane of Pseudomonas aeruginosa: heat- 2-mercaptoethanol-modifiable proteins. J. Bacteriol. 140 902–910. 10.1128/jb.140.3.902-910.1979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L., Zhao L., Qi W., Xu X., Zhang J., Zhang J., et al. (2020). Temperature-specific expression of cspA1 contributes to activation of sigX during pathogenesis and intracellular survival in Pseudomonas plecoglossicida. Aquaculture 518:734861 10.1016/j.aquaculture.2019.734861 [DOI] [Google Scholar]
- Jaouen T., Dé E., Chevalier S., Orange N. (2004). Pore size dependence on growth temperature is a common characteristic of the major outer membrane protein OprF in psychrotrophic and mesophilic Pseudomonas species. Appl. Environ. Microbiol. 70 6665–6669. 10.1128/aem.70.11.6665-6669.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones C. J., Newsom D., Kelly B., Irie Y., Jennings L. K., Xu B., et al. (2014). ChIP-Seq and RNA-Seq reveal an AmrZ-mediated mechanism for cyclic di-GMP synthesis and biofilm development by Pseudomonas aeruginosa. PLoS Pathog. 10:e1003984 10.1371/journal.pone.1003984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazakov A. E., Rodionov D. A., Alm E., Arkin A. P., Dubchak I., Gelfand M. S. (2009). Comparative genomics of regulation of fatty acid and branched-chain amino acid utilization in Proteobacteria. J. Bacteriol. 191 52–64. 10.1128/jb.01175-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindrachuk K. N., Fernández L., Bains M., Hancock R. E. W. (2011). Involvement of an ATP-dependent protease, PA0779/AsrA, in inducing heat shock in response to tobramycin in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 55 1874–1882. 10.1128/aac.00935-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojic M., Jovcic B., Vindigni A., Odreman F., Venturi V. (2005). Novel target genes of PsrA transcriptional regulator of Pseudomonas aeruginosa. FEMS Microbiol. Lett. 246 175–181. 10.1016/j.femsle.2005.04.003 [DOI] [PubMed] [Google Scholar]
- Koprowski P., Sliwinska M. A., Kubalski A. (2015). Negative and positive temperature dependence of potassium leak in MscS mutants: implications for understanding thermosensitive channels. Biochim. Biophys. Acta 18 1678–1686. 10.1016/j.bbamem.2015.04.019 [DOI] [PubMed] [Google Scholar]
- Leslie S. B., Israeli E., Lighthart B., Crowe J. H., Crowe L. M. (1995). Trehalose and sucrose protect both membranes and proteins in intact bacteria during drying. Appl. Environ. Microbiol. 61 3592–3597. 10.1128/aem.61.10.3592-3597.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Los D. A., Murata N. (2004). Membrane fluidity and its roles in the perception of environmental signals. Biochim. Biophys. Acta 1666 142–157. 10.1016/j.bbamem.2004.08.002 [DOI] [PubMed] [Google Scholar]
- Lyczak J. B., Cannon C. L., Pier G. B. (2002). Lung infections associated with cystic fibrosis. Clin. Microbiol. Rev 15 194–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao F., Dam P., Chou J., Olman V., Xu Y. (2009). DOOR: a database for prokaryotic operons. Nucleic Acids Res. 37 D459–D463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinac B., Nomura T., Chi G., Petrov E., Rohde P. R., Battle A. R., et al. (2014). Bacterial mechanosensitive channels: models for studying mechanosensory transduction. Antioxid. Redox Signal. 20 952–969. 10.1089/ars.2013.5471 [DOI] [PubMed] [Google Scholar]
- Martínez-Granero F., Redondo-Nieto M., Vesga P., Martín M., Rivilla R. (2014). AmrZ is a global transcriptional regulator implicated in iron uptake and environmental adaption in P. fluorescens F113. BMC Genom. 15:237. 10.1186/1471-2164-15-237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massier S., Rincé A., Maillot O., Feuilloley M. G. J., Orange N., Chevalier S. (2012). Adaptation of Pseudomonas aeruginosa to a pulsed light-induced stress. J. Appl. Microbiol. 112 502–511. 10.1111/j.1365-2672.2011.05224.x [DOI] [PubMed] [Google Scholar]
- Muller P. Y., Janovjak H., Miserez A. R., Dobbie Z. (2002). Processing of gene expression data generated by quantitative real-time RT-PCR. Biotechniques 32 1372–1374. [PubMed] [Google Scholar]
- O’Connor K., Fletcher S. A., Csonka L. N. (2009). Increased expression of Mg2+ transport proteins enhances the survival of Salmonella enterica at high temperature. Proc. Natl. Acad. Sci. U.S.A. 106 17522–17527. 10.1073/pnas.0906160106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Riverol Y., Csordas A., Bai J., Bernal-Llinares M., Hewapathirana S., Kundu D. J., et al. (2019). The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 47 D442–D450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quénée L., Lamotte D., Polack B. (2005). Combined sacB-based negative selection and cre-lox antibiotic marker recycling for efficient gene deletion in Pseudomonas aeruginosa. Biotechniques 38 63–67. 10.2144/05381st01 [DOI] [PubMed] [Google Scholar]
- Rahme L. G., Ausubel F. M., Cao H., Drenkard E., Goumnerov B. C., Lau G. W., et al. (2000). Plants and animals share functionally common bacterial virulence factors. Proc. Natl. Acad. Sci. U.S.A. 97 8815–8821. 10.1073/pnas.97.16.8815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond C. S., Glasner J. D., Mau R., Jin H., Blattner F. R. (1999). Genome-wide expression profiling in Escherichia coli K-12. Nucleic Acids Res. 27 3821–3835. 10.1093/nar/27.19.3821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz S., Eckweiler D., Bielecka A., Nicolai T., Franke R., Dötsch A., et al. (2015). Elucidation of sigma factor-associated networks in Pseudomonas aeruginosa reveals a modular architecture with limited and function-specific crosstalk. PLoS Pathog. 11:e1004744 10.1371/journal.pone.1004744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schurr M. J., Deretic V. (1997). Microbial pathogenesis in cystic fibrosis: co-ordinate regulation of heat-shock response and conversion to mucoidy in Pseudomonas aeruginosa. Mol. Microbiol. 24 411–420. 10.1046/j.1365-2958.1997.3411711.x [DOI] [PubMed] [Google Scholar]
- Schurr M. J., Yu H., Boucher J. C., Hibler N. S., Deretic V. (1995). Multiple promoters and induction by heat shock of the gene encoding the alternative sigma factor AlgU (sigma E) which controls mucoidy in cystic fibrosis isolates of Pseudomonas aeruginosa. J. Bacteriol. 177 5670–5679. 10.1.1128/jb.177.19.5670-5679.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon R., Priefer U., Pühler A. (1983). A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Nat. Biotechnol. 1 784–791. 10.1038/nbt1183-784 [DOI] [Google Scholar]
- Sonnleitner E., Schuster M., Sorger-Domenigg T., Greenberg E. P., Bläsi U. (2006). Hfq-dependent alterations of the transcriptome profile and effects on quorum sensing in Pseudomonas aeruginosa. Mol. Microbiol. 59 1542–1558. 10.1111/j.1365-2958.2006.05032.x [DOI] [PubMed] [Google Scholar]
- Spiers A. J., Buckling A., Rainey P. B. (2000). The causes of Pseudomonas diversity. Microbiol. Engl. 146(Pt 10), 2345–2350. 10.1099/00221287-146-10-2345 [DOI] [PubMed] [Google Scholar]
- Staroń A., Sofia H. J., Dietrich S., Ulrich L. E., Liesegang H., Mascher T. (2009). The third pillar of bacterial signal transduction: classification of the extracytoplasmic function (ECF) sigma factor protein family. Mol. Microbiol. 74 557–581. 10.1111/j.1365-2958.2009.06870.x [DOI] [PubMed] [Google Scholar]
- Stover C. K., Pham X. Q., Erwin A. L., Mizoguchi S. D., Warrener P., Hickey M. J., et al. (2000). Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406 959–964. [DOI] [PubMed] [Google Scholar]
- Strehmel J., Neidig A., Nusser M., Geffers R., Brenner-Weiss G., Overhage J. (2015). Sensor kinase PA4398 modulates swarming motility and biofilm formation in Pseudomonas aeruginosa PA14. Appl. Environ. Microbiol. 81, 1274–1285. 10.1128/AEM.02832-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tart A. H., Blanks M. J., Wozniak D. J. (2006). The AlgT-dependent transcriptional regulator AmrZ (AlgZ) inhibits flagellum biosynthesis in mucoid, nonmotile Pseudomonas aeruginosa cystic fibrosis isolates. J. Bacteriol. 188 6483–6489. 10.1128/jb.00636-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanBogelen R. A., Neidhardt F. C. (1990). Ribosomes as sensors of heat and cold shock in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 87 5589–5593. 10.1073/pnas.87.15.5589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White-Ziegler C. A., Um S., Perez N. M., Berns A. L., Malhowski A. J., Young S. (2008). Low temperature (23 C) increases expression of biofilm-, cold-shock- and RpoS-dependent genes in Escherichia coli K-12. Microbiology 154 148–166. 10.1099/mic.0.2007/012021-0 [DOI] [PubMed] [Google Scholar]
- Winsor G. L., Griffiths E. J., Lo R., Dhillon B. K., Shay J. A., Brinkman F. S. L. (2016). Enhanced annotations and features for comparing thousands of Pseudomonas genomes in the Pseudomonas genome database. Nucleic Acids Res. 44 D646–D653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood L. F., Ohman D. E. (2009). Use of cell wall stress to characterize sigma 22 (AlgT/U) activation by regulated proteolysis and its regulon in Pseudomonas aeruginosa. Mol. Microbiol. 72 183–201. 10.1111/j.1365-2958.2009.9.06635.x [DOI] [PubMed] [Google Scholar]
- Wood L. F., Ohman D. E. (2015). Cell wall stress activates expression of a novel stress response facilitator (SrfA) under σ22 (AlgT/U) control in Pseudomonas aeruginosa. Microbiol. Engl. 161 30–40. 10.1099/mic.0.081182-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtzel O., Yoder-Himes D. R., Han K., Dandekar A. A., Edelheit S., Greenberg E. P., et al. (2012). The single-nucleotide resolution transcriptome of Pseudomonas aeruginosa grown in body temperature. PLoS Pathog. 8:e1002945 10.1371/journal.pone.1002945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y., Sachdeva M., Leeds J. A., Meredith T. C. (2012). Fatty acid biosynthesis in Pseudomonas aeruginosa is initiated by the FabY class of β-ketoacyl acyl carrier protein synthases. J. Bacteriol. 194 5171–5184. 10.1128/jb.00792-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Flow cytometry analysis of P. aeruginosa H103 viability exposed or not to CS or HS. Bacteria were grown in LB until the mid-log growth phase and subjected to CS or HS as indicated. 105 cells were stained using Syto9 and PI and then analyzed by flow cytometry. (A) Cytograms are representative of three independent experiments. Live and dead cells emitting a green or a red fluorescence are indicated within a red or purple frame, respectively. The green frame corresponds to cells with membrane damages. The mean percentage of live, dead and injured cells is indicated in each frame. (B) Histograms representing percent of cell viability from the experiments of live/dead assays (n = 3).
Effect of HS and CS on algU expression and activity. Black and gray bars show means normalized expression of algU and its targets after HS or CS relatively to the control condition at 37°C as calibrator (white and dotted bars). NT: not treated. Error bars indicate SEM, n = 4. Significant differences between the sets of normalized expression obtained in each stress condition were calculated against the those obtained at 37°C using a two-tailed t-test, and P-value are indicated by ∗, ∗∗, or ∗∗∗ if < 0.05, 0.001, or 0.0001, respectively. The symbol ° is indicated for non-significant results.
RpoH is not involved in the cmaX, cfrX, and cmpX regulation in response to HS in PAO1. Means of normalized expression for algU targets (red frame), the predicted operon (cmaX-cfrX-cmpX) and sigX in PAO1 (red line) or in an isogenic rpoH mutant exposed (dashed back bars) or not (dashed white bars) to HS.
2-DE total protein profiles of P. aeruginosa exposed to CS from 37 to 4°C or not (control). Gels are Coomassie blue stained and are representative of three independent proteins extractions. The protein spots satisfying both statistical biological variation analysis and mass spectroscopy identification scores were numbered according to Table 1 and Supplementary Table 2.
c-di-GMP level quantification by LC-MS/MS in H103 strain exposed or not to a CS. Intracellular c-di-GMP was extracted in H103 strain exposed (square) or not (point) to CS and quantified LC-MS/MS. Briefly, the absorbance at 580 nm of cultures were recorded and cells from 2 mL of culture were collected. Then cells were harvested by centrifugation, washed in 1 mL of sterile physiological water and c-di-GMP was extracted twice with extraction buffer without internal standard and quantified as previously described (Bouffartigues et al., 2015; Strehmel et al., 2015).
Effect of a sublethal dose of valinomycin on membrane fluidity measured by fluorescence polarization in stationary growth phase. TMA-DPH and DPH anisotropies were used to evaluate the effect of 45 μM valinomycin on membrane fluidity 22 h after the addition of the membrane acting-agent to mid-log phase cultures. Values obtained from unexposed cultures are indicated as NT: not treated. Box plots correspond to each sets of data from three independent experiments. Means, medians and min/max anisotropies values are indicated by squares, lines and crosses, respectively. Significant differences between each condition were calculated using a two-tailed t-test, and P-value are indicated by ∗ if < 0.05.
Normalized expression of the studied genes under HS or CS in the indicated genetic backgrounds relative to P. aeruginosa H103 at 37°C. Values are means ± SEM for at least three independent experiments.
Characteristics of proteins identified by Maldi-TOF analysis. MS and MS/MS correspond to the mass spectrometry method used for the protein identification.
Bacterial strains and plasmids used in this study. Cbr, carbenicillin resistance; Gmr, gentamycin resistance; Tcr, tetracycline resistance.
Primer sequences of the indicated genes used for quantitative RT-qPCR reactions, transcription start sites identification and transcriptional fusions construction.
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (63) partner repository with the dataset identifier PXD021299.