Abstract
Group II introns are the putative progenitors of nuclear spliceosomal introns and use the same two-step splicing pathway. In the cell, the intron RNA forms a ribonucleoprotein (RNP) complex with the intron-encoded protein (IEP), which is essential for splicing. Although structures of spliced group II intron RNAs and RNP complexes have been characterized, structural insights into the splicing process remain enigmatic due to lack of pre-catalytic structural models. Here, we report two cryo-EM structures of endogenously produced group II intron RNPs trapped in their pre-catalytic state. Comparison of the catalytically activated precursor RNP to its previously reported spliced counterpart allowed identification of key structural rearrangements accompanying splicing, including a remodeled active site and engagement of the exons. Importantly, altered RNA–protein interactions were observed upon splicing among the RNP complexes. Furthermore, analysis of the catalytically inert precursor RNP demonstrated the structural impact of the formation of the active site on RNP architecture. Taken together, our results not only fill a gap in understanding the structural basis of IEP-assisted group II intron splicing, but also provide parallels to evolutionarily related spliceosomal splicing.
INTRODUCTION
Group II introns are ribozymes present in all domains of life as well as being putative ancestors of eukaryotic spliceosomal introns (1–4). The splicing process involves two consecutive transesterification steps giving rise to an intron lariat and the ligated exons (Figure 1A). This reaction can be carried out by the RNA itself under high salt concentrations in vitro (1,5,6). In the cell and under physiological conditions, splicing requires an intron-encoded protein (IEP), which is translated from the intron RNA (1,7). IEP binding allows stabilization of tertiary interactions in the absence of high salt, and splicing with a much higher efficiency (1,8–11). In addition to its maturase activity, which helps the intron fold properly and splice, the IEP possesses reverse transcriptase (RT) and, often, DNA endonuclease (EN) activities, which help the intron lariat invade target DNA sites and copy the intron into the host genome by reverse transcription (1,2,4,12).
Figure 1.
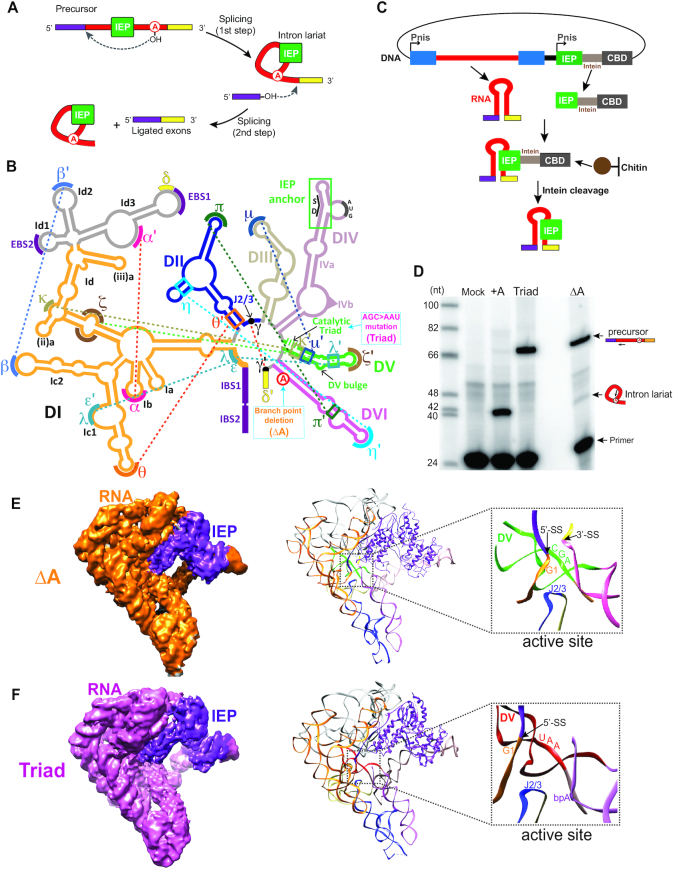
Overview of group II intron precursor RNPs. (A) The lariat-forming splicing pathway of group II introns. With the help of the intron-encoded protein (IEP, green), the hydroxyl group of the branch-point adenosine from DVI of the intron initiates a nucleophilic attack on the 5′-SS. This leads to a 2′-5′ phosphodiester bond between the first nucleotide of the intron and the branch point. The hydroxyl group of the 5′-exon then attacks the 3′-SS, yielding the intron in the lariat form as well as the ligated exons. The 5′-exon is shown in purple and the 3′ exon is shown in yellow. (B) Secondary structure of the group IIA intron Ll.LtrB. Mutations used in this work are illustrated in blue dotted boxes and key tertiary interactions are indicated by dashed lines. The triangle in DIV represents 169 nt in the ΔORF DIVb that are not shown. Exons are shown in the same color code as in panel A, with the IBSs indicated. SD in DIV marks the Shine-Dalgarno sequence. (C) Purification scheme of the precursor RNPs. The intron RNA and the IEP were co-expressed in the native host L. lactis. After cell lysis, the lysate was subjected to chitin resin, which binds the chitin-binding domain (CBD) to the C-terminal side of the IEP. The bound RNP was then released by DTT cleavage of an intein bridging the C-terminus of the IEP and the CBD, before two rounds of ultracentrifugation to yield the pure RNPs. (D) The two precursor RNPs are pre-catalytic. Primer extension analysis shows the unspliced nature of the two RNPs. The +A RNP was included in the analysis to indicate the band representing the spliced product (intron lariat). The intron-specific primer is 14-nt away from the 5′-SS and yields a 41-nt product for the spliced RNA and a 68-nt product for the unspliced. The primer extension products were analyzed in an 8% urea PAGE gel. (E-F) Cryo-EM structures of two pre-catalytic group II intron RNPs solved at ∼5.0 Å. Density maps (left) for the ΔA RNP (E) and the Triad RNP (F) were used to build their structural models (right). The close-up views show the active sites of the two precursor RNPs, with the first nucleotide of the intron (G1), the 5′-SS, the catalytic triad and J2/3 marked.
Group II introns are divided into three subgroups, IIA, IIB and IIC, based on structural variations. Although not conserved in their primary sequences, group II introns are highly conserved in secondary structures (13) (Figure 1B and Supplementary Figure S1). The intron RNA is organized into six domains stemming from a central wheel (13). Domain I (DI) is the largest and the first to be transcribed. DI not only provides a scaffold for proper folding of all subsequent domains, but also interacts with key motifs in other domains to promote catalysis. Among these motifs are the exon-binding sites (EBSs) that align the exons by Watson-Crick base pairing with the intron-binding sites (IBSs). DII and DIII serve as essential coeffectors for catalysis, whereas DIV contains the open reading frame (ORF) encoding the IEP, as well as a high-affinity binding site for the IEP (14,15). DV forms the core of the active site by providing the catalytic triad nucleotides (A2398, G2399 and C2400 of the Lactococcus lactis group II intron Ll.LtrB) that bind two magnesium ions (13,16), through contacting the dinucleotide bulge in DV (A2420 and C2421) and the junction of DII and DIII (J2/3: C469, G470, G471 and A472) (17). DVI harbors the branch-point adenosine which carries out the nucleophilic attack that initiates the first step of the transesterification. For the group IIA intron Ll.LtrB from L. lactis, the IEP is termed LtrA, which contains multiple domains including the N-terminal extension (NTE), the fingers-palm domain (also referred to as the RT domain), the thumb domain (also referred to as the X domain), the DNA-binding domain (DBD) and the EN domain (Figure 2A-B). The protein is anchored at DIVa via the NTE and makes additional contacts with DI via the fingers-palm and the thumb domains (10), which are both essential for splicing (1,18).
Figure 2.
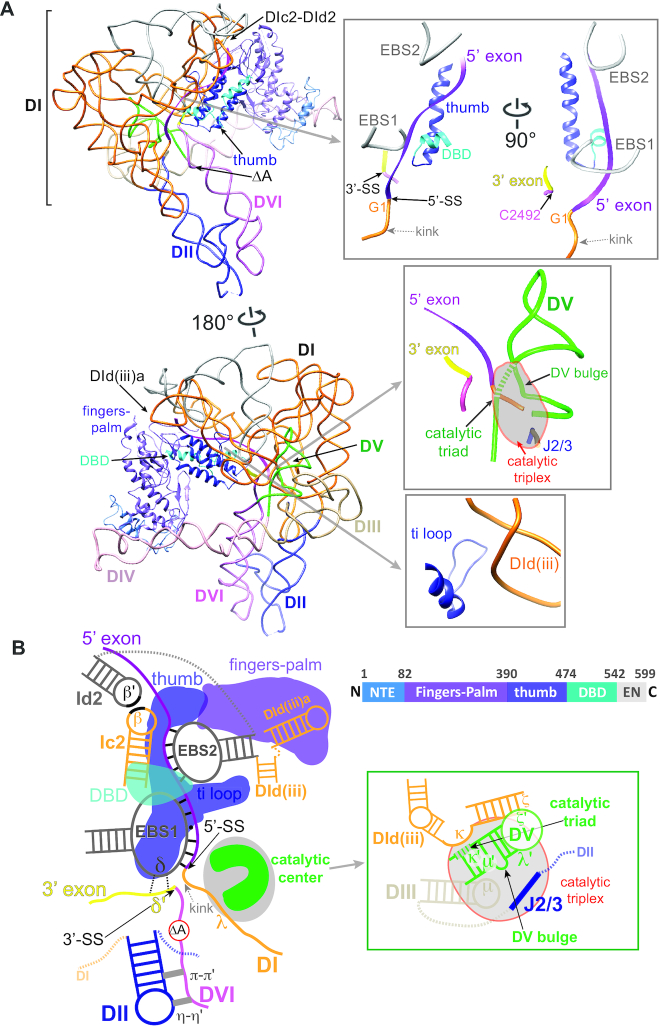
Distinctive features of the of the pre-catalytic RNP structure. (A) Two views of the ΔA RNP structure with key domains. Close-in views of distinctive regions are shown in boxes on the right. The fingers-palm, the thumb and the DBD domains of the IEP are indicated as ribbons in purple, blue and cyan, respectively, and shown linearly in panel (B). The color code of the intron RNA is the same as in Figure 1B. Boxes show the docked exon sequences buttressed by the protein (top), the active site (middle) and the ti loop-DId(iii) interaction (bottom). The catalytic triplex region is indicated by the grey-shaded area in the middle box. The catalytic triad nucleotides are shown as dashed tubes. (B) Schematic of 3D structure of the pre-catalytic ΔA RNP shown in 2D. The color code of the RNP is as above. Dotted grey lines denote omitted domain junctions. The π-π′ and η-η′ tetraloop-receptor interactions between DII and DVI are shown as grey bars. The dotted black lines between δ and δ′ nucleotides denote weak base-pairings. Domain arrangement of the IEP is shown on the right. NTE: N-terminal extension. DBD: DNA-binding domain. EN: DNA endonuclease domain.
Structures for various group II intron RNAs generated in vitro have so far been reported using X-ray crystallography (19–23). However, in cells, group II introns function as ribonucleoproteins (RNPs). Thus, it is of interest how functionalities of the IEP influence the activities of group II introns. Also, it is important to understand the molecular details of the holoenzyme structure. Recently, the post-catalytic structure of a group IIA intron Ll.LtrB holoenzyme was revealed by cryo-electron microscopy (cryo-EM) at 3.8 Å resolution (10). For this RNP, not only was the structure of the IEP solved for the first time, but the way that it interacts with the RNA was demonstrated, consistent with previous biochemical results (15,24).
In comparison to the spliceosome complex, which undergoes significant compositional remodeling during the same two transesterifications (3,25), the group II intron remains as a streamlined splicing machine with just the RNA and the IEP. It is thus likely that the intron RNA and the protein collectively exhibit versatility during catalysis in order to carry out the same series of events. Indeed, it was previously demonstrated biochemically using endogenously expressed RNPs that important changes in key RNA–RNA and RNA–protein interactions occur upon splicing, while the majority of the RNP architecture remains unchanged (9). Nevertheless, the structure of a pre-catalytic group II intron RNP has remained elusive, preventing the elucidation of spatial arrangements, particularly of the exons, in the holoenzyme before splicing.
In this study, we present the cryo-EM structures of the group II intron holoenzyme in two pre-catalytic forms. Through comparisons with the same RNP in the post-catalytic state, numerous structural rearrangements within the RNA and alterations in RNA–protein interactions were observed, providing a glimpse into conformational changes in the holoenzyme that dictate the catalytic process in intron RNA during splicing. Furthermore, the adjustments at the conserved active site as well as the highly coordinated RNA–protein interactions parallel interactions at the catalytic center of the eukaryotic spliceosome during the equivalent steps of splicing.
MATERIALS AND METHODS
RNP purification
The purification was carried out as described previously (9,10,26,27) using L. lactis strain IL1403 containing the plasmid pLNRK-nisLtrB (ΔORF-ΔA)+nisLtrA-intein-CBD (for ΔA RNP) or the plasmid pLNRK-nisLtrB (ΔORF-TriadAAU)+nisLtrA-intein-CBD (for Triad RNP). Briefly, the inoculated culture was induced with nisin for 3 h and the cells were collected. After the lysis, the lysate was loaded onto a chitin column and stringently washed. The RNP was released via intein cleavage by dithiothreitol (DTT) and the eluate was loaded onto a 10% sucrose cushion and subjected to ultracentrifugation. The pellet formed was then resuspended and loaded onto a 5–20% sucrose gradient for ultracentrifugation. The fractions of interest were buffer exchanged into RNP buffer (20 mM Tris–HCl pH 7.5, 200 mM KCl, 5 mM MgCl2) and snap-frozen for storage at –80°C.
Primer extension
The RNPs were subjected to phenol/chloroform extraction and ethanol precipitation for IEP removal. The primer (intron-specific primer IDT1073, GTACCTTAAACTACTTGACTTAACACC) was 5′-end labeled using [γ-32P] ATP (PerkinElmer) and T4 polynucleotide kinase (New England Biolabs). 20 ng of RNA from each RNP was applied in the primer extensions in which SuperScript III reverse transcriptase (Invitrogen) was used according to manufacturer's protocol in the presence of 0.4 pmol of the primer. The products were run in an 8% urea PAGE gel, exposed to a phosphor screen and analyzed by a GE Healthcare Typhoon Trio.
Electron microscopy
For cryo-EM specimen preparation, 4 μl RNP sample was pipetted onto freshly glow-discharged grids coated by home-made continuous thin carbon film, then the grids were blotted with filter paper (Ted Pella, Inc.) in FEI Vitrobot with a humidity of 100% and temperature at 12°C, followed by immediate plunge-freezing into liquid ethane. The frozen grids were stored in liquid nitrogen. For EM imaging, the specimens were transferred into a Titan Krios (300 kV) microscope equipped with a Gatan K2 Summit direct electron detector. Cryo-EM datasets were collected at a defocus range of –1.0 to –2.0 μm with a magnified physical pixel size of 1.307 Å, using the Etas automatic data collection software (developed by Bo Shen and Xueming Li, Tsinghua University). Every micrograph was dose-dependently fractionated into 32 frames and the total dose was ∼50 e/Å2.
Image processing of electron micrographs
We collected 8934, 9578, 9358 and 8741 micrographs for the ΔA dimer, the Triad dimer, the ΔA monomer and the Triad monomer, respectively (Supplementary Table S1). The beam-induced motion of these micrographs was corrected by MotionCor2 (28), after which CTFFIND4 package (29) was used to estimate the CTF parameters. Particles from the motion-corrected micrographs were auto-picked and two-dimensionally classified in Relion3.0 (30). The 2D classification results were manually evaluated to remove the junk particles grouped in the irregular classes. The remaining particles were further used to perform 3D classification and refinement. For the monomer datasets, we used the previously published +A structure (EMD: 3333) (10) low-pass filtered to 60 Å as the initial model and applied a monomeric mask to perform 3D classification and refinement. The cryo-EM structure of the ΔA monomer was determined at a resolution of 5.1 Å using 231 724 particles, and Triad monomer at 8.4 Å using 351 541 particles.
For the dimeric datasets, although the dimerization interface exhibited high flexibility, each monomer within the dimeric particle was relatively rigid. To separately reconstruct each monomer for final refinement, we used the published +A structure (EMD: 3333) (10) low-pass filtered to 60 Å as the initial model and applied a monomeric mask to perform the 3D classification and refinement to generate the angular distribution of each half of the dimeric particles, based on which the particle subtraction was performed in Relion3.0 (31,32). Subsequently, the subtracted residual particle signals were used for 3D classification and refinement to determine the angular distribution of the other half in the dimeric particles. Finally, these two datasets each containing a monomeric density signal were combined and centered to re-extract particles and to reconstruct 3D structures of the monomers within dimeric states. As a result, we solved ΔA structure in its dimeric state to a resolution of 5.0 Å using 203,373 particles, and Triad at 5.0 Å using 391 788 particles. The FSC profiles on three different directions were determined by Remote 3DFSC Processing Server (https://3dfsc.salk.edu/) (33). The density correlation was calculated by using the ‘fit-in-map’ command in UCSF Chimera with default settings, to demonstrate the overall similarity between EM densities of the pre- and post-catalytic RNPs.
Model building and comparison
Previously published +A RNP model (PDB: 5G2X) was firstly adjusted manually in COOT (34) and refined in PHENIX (35) based on its cryo-EM structure at 3.8 Å resolution (10). The revised +A model was docked into ΔA and Triad RNP EM maps as a rigid body using UCSF Chimera (36). The docked models were then rebuilt and adjusted manually in COOT, and finally refined in PHENIX in real space with secondary structure and geometry restraints. For model comparison, G3-A120, C161-A178 and C354-G391 in DI, where the structures are more rigid as judged by the 3D variability analysis of the EM densities, were used as references to align the precursor and +A models. The alignment was made by ‘MatchMaker’ in UCSF Chimera, restricting matching to the selected reference regions.
3D variability analysis
3D variability analysis (3DVA) was performed using cryoSPARC v2.9 (37), where individual particles were used to determine continuous as well as discrete heterogeneity to allow capturing of multiple conformers and structural flexibility within a single dataset. The ΔA and Triad particle images used for structure reconstruction in Relion3.0 previously were first used for homogeneous refinement in cryoSPARC, then the outputs were connected for a 3D variability job with a 8-Å filter resolution. Finally, the volume series were generated by a 3D variability display job with 20 frames and visualized in Chimera.
RESULTS
Overall architecture of two pre-catalytic group II intron RNPs
We expressed two ΔORF intron variants with mutations that would yield unspliced precursors of the Ll.LtrB intron RNP. Both were generated from the pLNRK plasmid with its IEP expressed in trans from a separate nisin promoter (Figure 1C) (10,38). The first mutant, referred to as ΔA (Figure 1B and Supplementary Figure S1), has the branch point adenosine in DVI deleted, rendering the active intron RNA largely unable to splice because of an inability to perform the initial nucleophilic attack (26). The second mutant, referred to as Triad (Figure 1B and Supplementary Figure S1), harbors a 2-nt mutation in the catalytic triad nucleotides in DV (G2399 and C2400, AGC>AAU), rendering the intron catalytically inert (9). We utilized an affinity-based purification approach coupled with sucrose gradient ultracentrifugation to obtain native precursor RNPs (Figure 1C, D and Supplementary Figure S2A), which were used for structural characterization by cryo-EM. As discussed previously, the ΔA RNP, with an intact DV, is catalytically competent, whereas the Triad mutant, lacking a properly formed active site, is catalytically incompetent (Figure 1D) (9,39).
In the purification of native RNPs from each of the two constructs, two peak fractions containing RNPs were recovered from the final sucrose gradient (Supplementary Figure S2A). We performed cryo-EM analysis on all of the four RNP populations. RNPs from the two relatively heavier fractions, with a large peak comprising the majority of the RNP population, were determined to be dimeric (Supplementary Figures S2–S4). In contrast, RNPs from the two lighter fractions were determined to be monomeric, and could be reconstructed at resolutions of 5.1 and 8.4 Å by cryo-EM for the ΔA and the Triad RNPs, respectively. The RNP dimers appeared intrinsically more rigid compared to their monomeric counterparts and the structures of the Triad and ΔA RNPs were both solved at a resolution of 5 Å by cryo-EM after applying a monomeric mask (31,32) (Supplementary Figure S4 and S5). The molar ratio of intron RNA to IEP in all four RNP populations was determined to be 1:1 (Supplementary Figures S3–S6). Importantly, a careful docking analysis comparing the densities of both dimeric RNPs to those of their monomeric counterparts showed no major structural differences in the RNP monomer relative to the dimer (Supplementary Figure S6). The correlation between the densities of monomeric and dimeric ΔA RNPs was ∼0.99, indicating that dimerization does not alter the intrinsic architecture of the RNP monomers.
In the dimer-derived density maps, the main chains of RNA as well as most IEP regions could be unambiguously traced, enabling us to manually build the models of the ΔA and the Triad RNPs (Figure 1E, F), based on the model of their previously published post-catalytic counterpart (10), referred to here as +A (Figure 1D). From the ΔA RNP models, we clearly see the catalytic center, comprising the catalytic triad in DV (AGC), juxtaposed to the 5′-splice site (5′-SS, next to G1, the first nucleotide of the intron RNA) and J2/3 (Figures 1E and 2A and Supplementary Figure S7A). In the Triad RNP, the active site is distorted, and there is a much larger distance between the 5′-SS and the position of the branch point, as clearly illustrated in both the models and in the EM densities (Figure 1F and Supplementary Figure S7B).
Distinctive features of the pre-catalytic RNP: exons, splice sites and IEP placement
The novelty of the ΔA RNP structure lies in the ability to view the 5′ and 3′ exons relative to the intron and the IEP. First, the exons are shown in close juxtaposition to the exon binding sites in the intron and the thumb domain of the IEP. The 5′ exon (IBS1 and IBS2) is paired with EBS1 and EBS2 (9), whereas the 3′ exon (δ′) is apposed to the δ sequence next to EBS1, with one of the α-helices of the thumb domain nestled alongside the EBS-IBS interactions (Figure 2A, top right, and Figure 2B). Second, the 5′ splice site is positioned in close proximity to the catalytic triplex formed between J2/3 and the DV active site (DV bulge and the catalytic triad) (Figure 2A, middle right). As seen in other pre-catalytic group II introns (17,20), the 5′ end of the intron (near G1) forms a notable kink, which possibly facilitates the initiation of branching.
The active site region including the 5′ splice site (5′-exon–intron boundary) is relatively free of interacting protein (Figure 2A, middle right, and Figure 2B), reinforcing the notion that the initiation of catalysis occurs by the RNA alone. Nevertheless, there are close interactions between protein and RNA in the immediate neighborhood of the active site, as exemplified by the ti loop-DId(iii) interaction, which is a highly variable motif across group II introns (Figure 2A, bottom right, and Figure 2B). These RNA–protein relationships likely mediate structural rearrangements during splicing, as delineated below.
Global similarities between the precursor RNPs and the post-catalytic RNP
To allow unbiased comparison of the two precursor models to the +A model, we updated the previously published +A model using the same modeling method as for the precursors in this work. The update resulted in little change compared to the previous model (Supplementary Figure S8). The comparison between spliced and unspliced RNPs reveals a similar overall tertiary structure across the catalytic stages, with density correlations of 0.95 and 0.88 between the ΔA and the +A RNPs and between the Triad and the +A RNPs, respectively (Supplementary Figure S9). This similarity suggests that the overall folding of the group II intron remains stable during the period that spans RNP maturation through splicing. In comparison to the +A RNP, the two precursors showed discrete differences upon splicing at the active site, the splice sites (SSs) as well as at the RNA–IEP interaction interface. In addition, local variations throughout the structure were observed between the two precursor RNPs, suggesting that the ΔA and the Triad RNPs represent two distinct pre-catalytic stages of maturation (Supplementary Figure S10). In the following sections, we will first focus on delineating the differences between the ΔA and the +A RNPs to infer the structural rearrangements in the RNP upon splicing (Figure 3). We will also analyze the common differences of the two catalytically competent ΔA and +A RNPs from the Triad RNP, focusing on the pivotal influence of the formation of the active site on the global architecture of the intron RNP.
Figure 3.
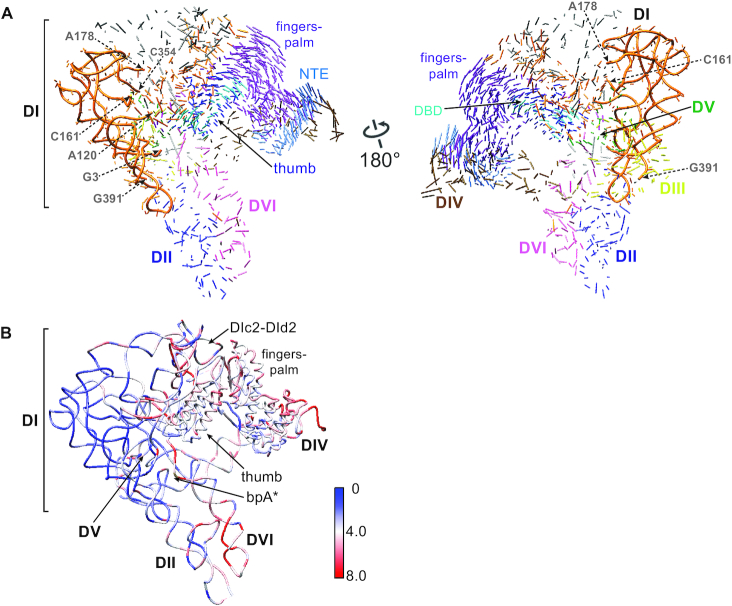
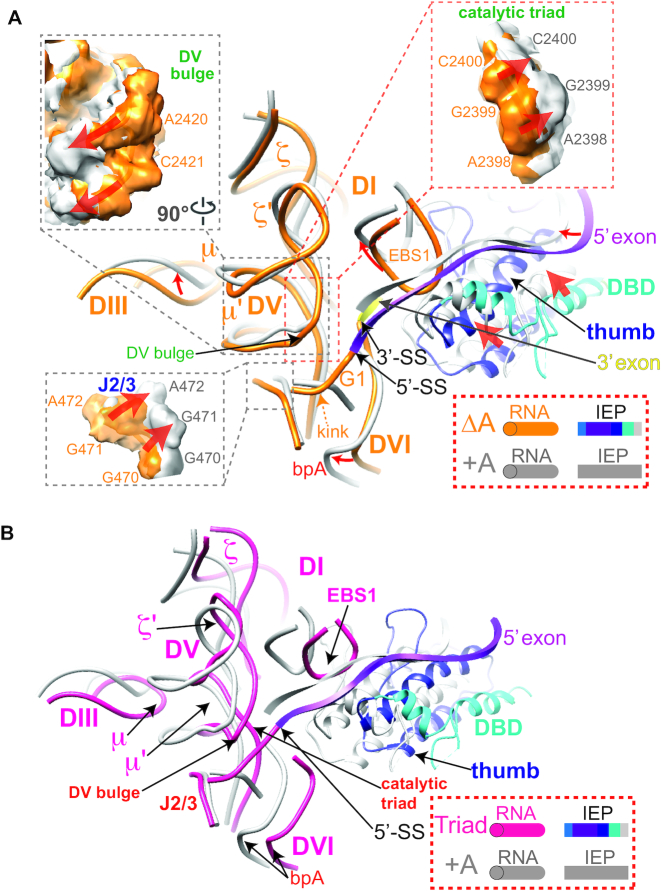
RNP conformational changes accompanying splicing. (A) 3D vector map showing structural rearrangements from ΔA to +A (PDB: 5G2X) RNPs. The docking is based on reference nucleotides G3-A120, C161-A178 and C354-G391 in DI of the RNA (tubes), where the RNA backbones are relatively invariable across the three RNPs. Vector lengths correlate to the scale of the movements across the two catalytic stages. Color code is the same as in Figure 2. (B) Distribution of the root-mean-square deviation (RSMD, Å) between the ΔA and +A RNPs mapped onto the ΔA RNP structure.
The IEP moves toward the active site upon splicing
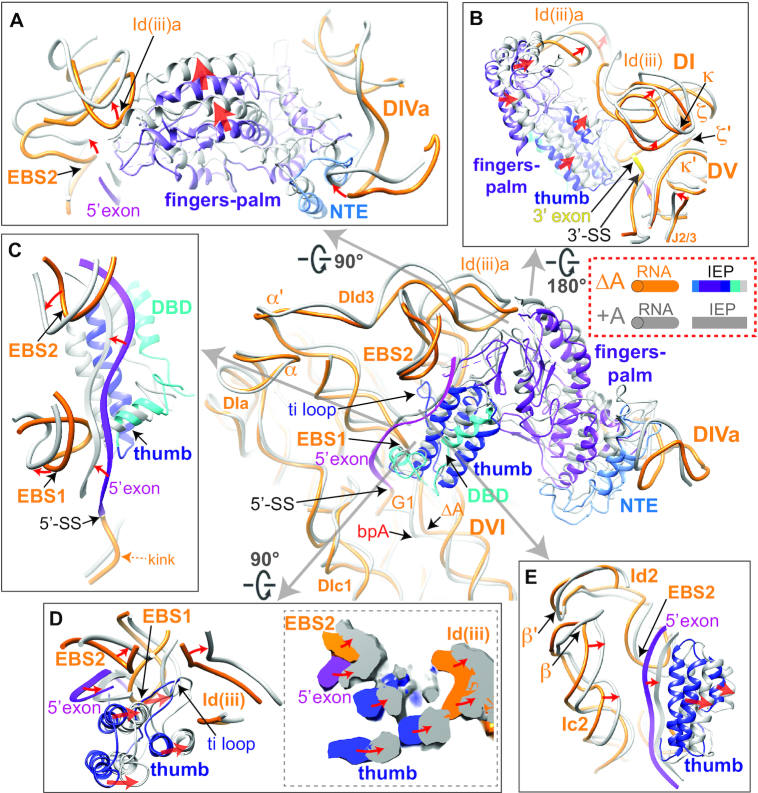
The overall IEP structure is similar when comparing the ΔA RNP with the +A RNP (Figure 3 and Supplementary Figure S11). However, multiple differences in RNA–protein interactions were observed between the two RNPs (Figures 3 and 4, center). First, the fingers-palm domain of the IEP, which interacts with the Id(iii)a stem-loop in DI of the RNA, is placed further towards the loop region of Id(iii)a in the post-catalytic RNP, resulting in displacement of the entire DId(iii)a (Figures 2B and 4A and Supplementary Movie 1). Additionally, the distortion in the basal stem region of DId(iii)a propagates to the κ and ξ motifs, communicating with κ′ and ξ′ in DV at the active site (Figure 4B and Supplementary Figure S12). Accordingly, EBS2, adjacent to the basal region of DId(iii)a, moves by ∼6 Å, pulling the 5′ exon in the direction of the thumb and the EBSs in the post-catalytic RNP (Figure 4C). Although a causal relationship cannot be inferred, it appears that the movements of the fingers-palm upon splicing correlate with adjustments at the active site and 5′ exon realignment.
Figure 4.
Rearrangements in RNA–protein interactions comparing the ΔA and +A RNPs The post-catalytic RNA and IEP are shown uniformly in gray, whereas the pre-catalytic RNA is shown in orange and its IEP shown in various colors as illustrated. Red arrows denote the directions of movement upon splicing. (A) RNA–protein interaction at the fingers-palm domain. DId(iii)a is distorted by the movement in the fingers-palm domain upon splicing. NTE: N-terminal extension. (B) RNA–protein interaction distorts DId(iii)a to remodel DV. (C) The RNA–protein interactions at the 5′ exon region showing 5′ exon being sandwiched by the EBSs, the thumb and DBD domains. (D) Enhanced RNA–protein interaction at the thumb domain upon splicing and the influence on exon placement. The cross-section of density maps of the ΔA and +A RNPs is shown in the dotted box on the right. (E) Coordinated movements of DIc2 accompanying the adjustments in thumb domain upon splicing. DIc2 moves towards the same direction as the thumb domain, contributing to stabilization of the exon in the new location.
The thumb domain, also required for splicing, likewise has its three essential α-helices advanced towards the intron ends in the post-catalytic RNP (Figure 4D and Supplementary Figure S13 and Movie 1). Simultaneously, the ti loop of the thumb domain interacts with DI’s Id(iii) to provide an additional anchor for the RNA–protein interaction (10,40). In the spliced RNP, the ti loop adopts different conformations, while maintaining the interactions with the neighboring RNA motifs DId(iii) and the EBSs (Figure 4D). The two α-helices of the thumb domain flanking the ti loop are placed closer to EBS1 and DId(iii) upon splicing (Figure 4D), while EBS1 in the +A RNP is translocated as much as ∼9 Å away from its position in the ΔA RNP (Figure 4C). Meanwhile, upon splicing, subdomains DIc2 and DId2, which are linked by the β–β′ interaction, together clamp the 5′ exon with the IEP (Figures 2B and 4E). Thereby, the thumb domain of the IEP exhibits enhanced interactions with cognate RNA motifs accompanying splicing, while the extensive network of altered RNA–protein interactions and movements of EBS1 potentially facilitate exon release after splicing.
IEP movement and rearrangement of the exons
Pre-catalytically, the IEP and EBS1 both stabilize the 5′-SS proximal to the active site (Figure 4C). Importantly, the 5′-SS exhibits a notable kink in the ΔA RNP (Figures 2B, 4C and 5A), similar to that of a pre-catalytic IIC intron previously reported (20). After splicing, the protein is further lodged into the core, while EBS1 retracts, pulling the 5′ exon away from the site of catalysis (Figure 4C and Supplementary Figure S14 and Movie 1). At the same time, the EBS2–IBS2 pair induces a torsion within the 5′ exon (Figure 4C). Meanwhile, the distances between some EBS nucleotides and their counterparts from the 5′ exon increase by as much as 2 Å (Supplementary Figure S14), indicative of weakened EBS1–IBS1 and EBS2–IBS2 interactions, consistent with previous biochemical data (9). Also, the δ nucleotides move as much as 9 Å upon splicing, leading to loosening of the δ-δ′ interaction and displacement of the 3′ exon (Figure 4C and Supplementary Figure S14). Such structural rearrangements at the exon-binding sites may facilitate forward splicing as well as the release of the mRNA after exon ligation.
Figure 5.
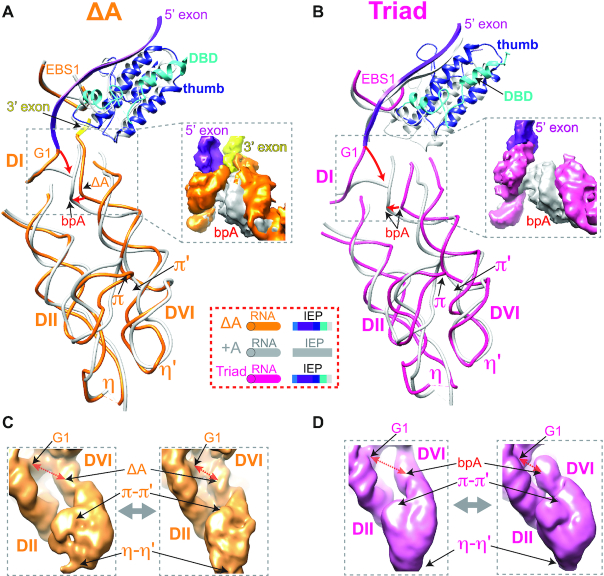
Rearrangements of regions key to catalysis. (A) The comparison of the ΔA RNP with the +A RNP (PDB: 5G2X) at DV and its interaction network. Arrows indicate movements in or adjacent to DV upon splicing. The post-catalytic RNA and IEP are shown uniformly in grey, whereas the pre-catalytic RNA is shown in orange and its IEP shown in various colors as illustrated. Density map comparisons for key motifs are shown in dashed boxes. The 5′ and 3′ exons are in magenta and yellow, respectively. (B) RNA structure comparison of the DV region in the Triad and the +A RNPs (PDB: 5G2X). Triad RNA is shown in dark pink.
Organization of the catalytic center and strengthening of tertiary interactions upon splicing
The network of tertiary interactions at the catalytic center encompassing DV becomes generally more compact upon splicing, with most DV-interacting motifs moving closer to the catalytic core (Figure 5A, B and Supplementary Figure S15 and Movie 2). DIII, which buttresses DV from the outside of the RNA scaffold via the tertiary interaction μ-μ′, pushes in the direction of ξ–ξ′ upon splicing, causing a torsion in DV which leads to rearrangements of the nearby catalytic triad nucleotides (Figures 2B and 5A, B and Supplementary Figure S15). Strengthening of tertiary interactions between DI and the 5′-SS pushes the DV bulge nucleotides (A2420 and C2421) upward, closer to the splice sites (Figure 5A). Meanwhile, rearrangements in two other tertiary interactions, ξ–ξ′ and κ–κ′ between DI and DV (Figure 4B), lead to a further compaction of the active site. As a result, the metal ion-binding pocket, comprised of the catalytic triad in DV, the DV bulge and J2/3, is remodeled upon splicing (Figures 4B and 5A).
Strong global effects of active site formation
The Triad RNP was made functionally inert by mutation of two of the catalytic triad nucleotides, abolishing its ability to bind metal ions essential for splicing (9,41). The Triad RNP exhibits a disrupted active site despite proper tertiary folding overall (Figure 1E, Supplementary Figure S7B and Movies 2–3). Importantly, this catalytically inactive Triad RNP has a globally less compact architecture than that of the ΔA and +A RNPs (Supplementary Figures S9–S10 and Movies 2–3).
The displacements of the IEP and the RNA–protein interactions in the Triad RNP are markedly more pronounced than those in the ΔA RNP when compared to the +A RNP (Supplementary Figure S16, center). The fingers-palm domain in the Triad RNP moved ∼17 Å away from the RNA compared with its location in +A, accompanied by a similar displacement of the interacting DId(iii)a (Supplementary Figure S16A). In addition, the EBSs and the 5′ exon are located further away from the active site relative to their locations in the +A RNP (Supplementary Figure S16B), while the density of the 3′ exon is not visible at all (Supplementary Figure S7B). The thumb domain is also dramatically farther away from the RNA compared to +A (Supplementary Figure S16C), indicating a looser structure that must become more organized prior to catalysis.
Further, comparison of the Triad RNP with the +A RNP indicates that all the DV-interacting elements, including ξ–ξ′, κ–κ′, μ–μ′ and λ–λ′ interactions, undergo compaction upon splicing, indicative of a multi-step adjustment in the active site during catalysis (Figure 5B and Supplementary Figure S17A–C). These movements are accompanied by a ∼6-Å increase in the distance between the mutated catalytic triad and the J2/3 compared to that in the +A RNP (Supplementary Figure S18), indicative of a disrupted metal-binding pocket in the Triad RNP. Moreover, movement of the μ motif in DIII by as much as 7 Å from its location in the +A RNP likely contributes to dislodging DV from the splice sites (Figure 5B and Supplementary Figure S17C). Taken together, the Triad RNP’s presumed inability to bind metal ions not only abolishes splicing (9,13), but also prevents proper formation of key tertiary interactions. Additionally, the greater structural differences between the Triad RNP and the +A RNP than those between the ΔA RNP and the +A RNP (Supplementary Figure S10) suggest an initial overall compaction of the RNP architecture with formation of the active site, followed by subtle and specific changes in RNP configuration upon splicing.
Flexibility of DII-DVI and structural rearrangements at the branch point
The tertiary pairings between the DII and DVI helices, the π–π′ and η–η′ interactions, exhibit considerable flexibility in the precursor RNPs (Supplementary Movie 4). While the π–π′ interaction stays engaged in both states (9), the π tetraloop is toggled upward towards the base of DII in the post-catalytic RNP (Figure 6A and Supplementary Movie 1). This toggling of the π nucleotides is likely a result of global twisting of DII and the interacting basal region of DVI before branching, which is less evident in the catalytically inert Triad RNP (Figure 6B). It is also interesting to note that the movement of DVI towards the 5′-SS is branch point-independent, and occurs with or without the branch point adenosine, albeit to a different extent (Figure 6A, B).
Figure 6.
Pre-catalytic configurations of the DII-DVI complex. (A, B) Structures of DII–DVI region in the ΔA (A) and the Triad (B) RNPs are compared to the +A RNP (PDB: 5G2X). RNAs of ΔA, +A and Triad are in orange, grey and dark pink, respectively. +A IEP is in grey and the precursors’ IEPs are shown in their corresponding colors. ‘bpA’ denotes the branch point adenosine and the orange ‘ΔA’ with arrow pointing to the branch point denotes adenosine deletion in the ΔA RNP. π-π′ and η-η′ motifs are indicated. The 5′ exons of the ΔA and Triad RNPs are in magenta. The 3′ exon, which is not visible in the Triad RNP, is in yellow of the ΔA RNP. (C, D) Intrinsic RNA flexibility of the DVI-DII pair as shown in Supplementary Movie 4. 3D variability analysis of ΔA (C) and Triad (D) RNPs identified conformers of DII-DVI region for each of the precursor RNPs, with representative conformations having the branch site (ΔA or bpA) away from or closer to the 5′ end of the intron (G1).
We further analyzed the 3D variability (37) of the DII-DVI region of the two precursors as well as the +A RNP (10). The results indicated that both precursor RNPs exhibit larger-scale movements compared to those of the +A RNP (Figure 6C, D and Supplementary Movie 4). In addition, two classes of conformers from each of the precursors were isolated, representing movement of the branch point relative to the 5′-SS (Figure 6C, D). Thus, in the group IIA Ll.LtrB intron, it is likely that, while the RNA–RNA interactions between DII and DVI remain engaged, the intrinsic flexibility of DVI contributes to the initiation of branching.
Active sites in the group II intron RNP and spliceosome are similar before splicing
Finally, we compared the catalytic centers of both precursor RNPs to those of two eukaryotic B* spliceosome conformers B*a1 and B*a2 (42), which exist immediately before the first transesterification step. There are significant parallels between the two strikingly different splicing machineries before initiation of catalysis, despite the spliceosome containing over 100 protein components and the group II intron RNP having only one (25) (Figure 7A). EBS1 in the group II intron and the snRNA U5 in the spliceosome both interact with the 5′ exon, which is right next to the kinked 5′-SS. Additionally, DVI and snRNA U2 help to position the branch point proximal to the active site, whereas DV and U6 provide the metal-binding pockets. Strikingly, the 3D conformations and protein interactions of these functionally related RNAs in group II intron precursors and eukaryotic spliceosome B* (Triad/B*a1 and ΔA/B*a2) are also almost identical. In addition, branch point rearrangements between pre- and post-catalytic states of group II intron and the spliceosome revealed a similar trend of movement for the bulged adenosine in DVI and U2 (42,43) (Figure 7B). It is thus interesting to postulate that the structural basis of the catalytic core and conformational rearrangements accompanying splicing remain largely unchanged during evolution from the relatively simple autocatalytic group II intron to the complex snRNP-assisted spliceosome.
Figure 7.
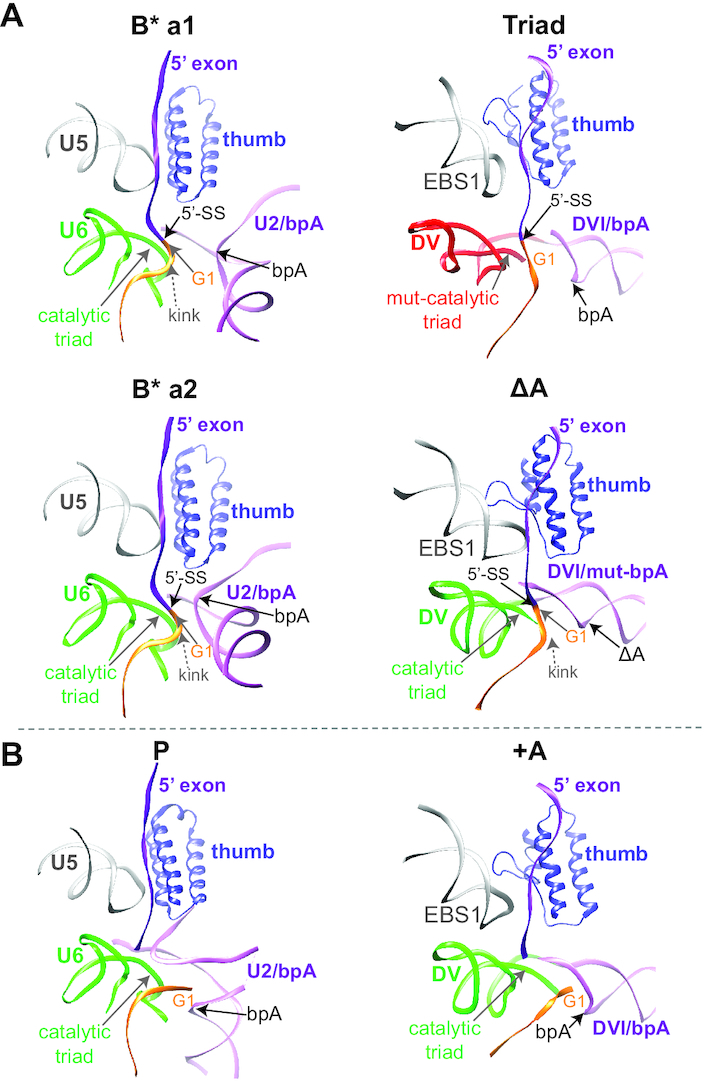
Comparison of group II intron and spliceosome at the active site. (A) Pre-catalytic structural arrangements at the active site of the group II intron RNP compared to the human B* spliceosome. Analogous arrangements at the active site are compared to the a1 and a2 conformations of the human B* spliceosome (PDB: 6J6H and 6J6G, respectively) in the presence of the thumb domain of the Prp8 protein. In the ΔA RNP and B*-a2 spliceosome, the branch point site is placed closer to the 5′-SS than that in the Triad RNP and B*-a1 spliceosome. The 5′-SS kink is indicated for the ΔA RNP and the B* spliceosome. (B) Comparison of the post-catalytic state between the group II intron RNP and human spliceosome. After splicing, the tertiary interactions at the active site remains stabilized in the +A RNP and the P complex (PDB: 5YLZ), while the kink at the 5′ end of the intron is relieved. ‘bpA’ denotes the branch site.
DISCUSSION
Despite being well known as autocatalytic ribozymes, group II introns rarely act alone in a living cell and must be chaperoned by an intron-encoded protein in order to execute splicing and retromobility. It is thus important to understand the structural basis for protein-facilitated splicing, which is the naturally occurring event. In this study, we present two pre-catalytic holoenzyme structures of a group II intron, in which we show important differences from their post-catalytic counterpart with respect to RNA organization as well as RNA–protein interactions. Our results imply a central role of the IEP in group II intron splicing, also shedding light on eukaryotic spliceosomal splicing, which is more highly dependent on protein factors.
Pre-catalytic stoichiometry of the group II intron RNP
The stoichiometry of a group II intron complexed with its IEP has been enigmatic (12,27,44). Retroviral reverse transcriptases (RTs), which are homologous to the thumb and fingers-palm domains of the IEPs, have been demonstrated to exist and function both as dimers and monomers, with examples from HIV-1 RT (45) and moloney murine leukemia virus (Mo-MLV) RT (46), respectively. Numerous group II intron IEPs have been biochemically and structurally reported as dimeric, and likely to bind to the intron RNA with a 2:1 ratio (11,40,47,48). However, the protein appeared monomeric in all three reported structures where the cognate nucleic acids were present, with a 1:1 ratio (10,49,50). It has been suggested that the RNP may adopt various stoichiometries at different catalytic stages (44).
In this study, we present a 1:1 or 2:2 stoichiometry for the pre-catalytic RNP. Although dimerization was shown to be weak and easily disrupted (9), the dimeric RNPs constitute the predominant portion of the population for both precursor RNPs. Consistent with these observations, the IEP has been reported to be able to dimerize by itself (11,40,47,48). Accordingly, our results suggest that the pre-catalytic dimerization is likely mediated by the IEP (Supplementary Figures S3 and S4). The possibility that dimerization occurs via the protein is also supported by the facts that the intron RNA alone has never been reported as dimeric, and that there was no difference between RNA structure profiles of the dimeric and monomeric RNPs (9). It remains uncertain whether pre-catalytic dimerization occurs in native cells or whether dimerization is a property of the overexpression and purification process. If dimerization does indeed occur in vivo at the precursor stage, this may increase local molecular crowding, which would facilitate the maturation and splicing of the intron RNA (51). Alternatively, IEP dimerization might facilitate branching by providing additional RNA–protein interactions at the branch point (52). During the process of reverse transcription, dimerization could circumvent dramatic conformational changes that may be required of the RT domain (7).
Loosely packed RNP precursors
Both pre-catalytic structures appear less compact than their post-catalytic counterpart, as shown by their larger spatial occupancy and higher flexibility (Figure 6C-D and Supplementary Figure S9). As a result of this pliability, we were unable to attain as high resolution in our reconstructions of the two precursors using similar numbers of particles collected as for the spliced +A particle (10) and with the same experimental conditions and parameters. The causes of the flexible nature of the two pre-catalytic RNPs are likely to be different. In the case of the Triad RNP, the failure to form the catalytic center results in a globally loose architecture (Figure 8A), whereas in the case of the ΔA RNP, the intron is active and in a dynamic state poised for branching (Figure 8B).
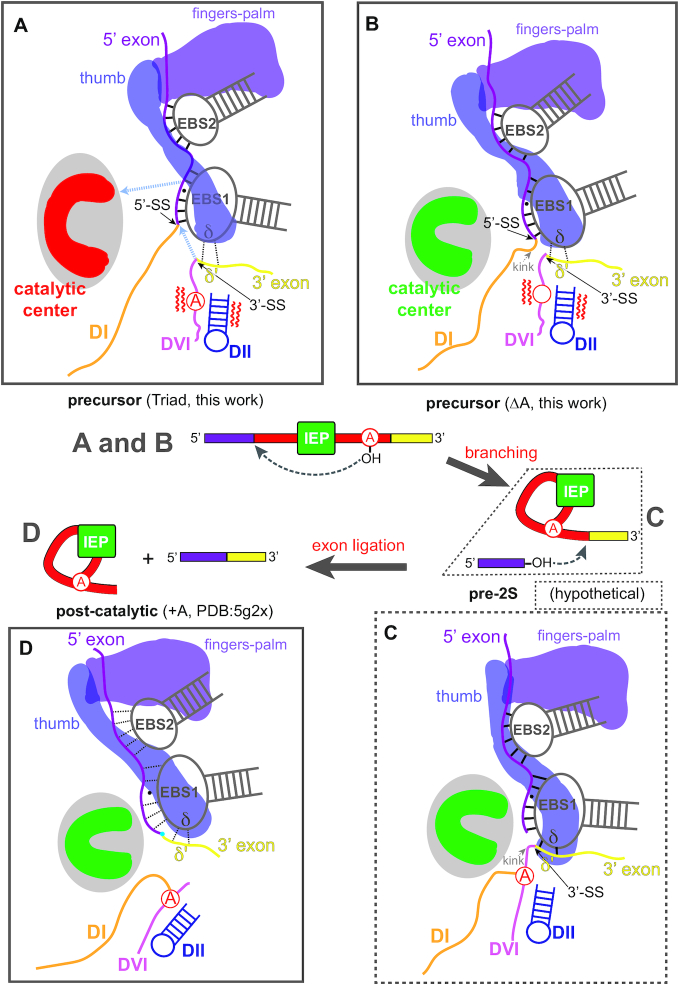
Figure 8.
Proposed model for IEP-promoted group II intron splicing Boxes A (Triad) and B (ΔA) correspond to structures solved in this work. Box C is a hypothetical structure that links B with Box D, the post-catalytic structure (+A, PDB: 5G2X). A, B, C and D in the middle splicing pathway diagram represent the boxed structures, respectively. (A) Pre-catalytic RNP before active site formation. Fingers-palm and thumb domains of the IEP loosely appose IBS-EBS and δ-δ′ pairings. Red denotes the inactive catalytic center. Grey dotted arrows denote direction of movement. (B) Pre-catalytic RNP after compaction and active site formation (red→green) proximal to the splice sites. The IEP moves closer to the active site. The thumb domain stabilizes the weak δ–δ′ interaction. Red wavy lines beside DII–DVI denote movements that present the branch point adenosine to the 5’-SS for the first step of splicing. (C) Hypothetical model after branching. It is postulated that the 3′ exon is further stabilized through enhanced δ–δ′ interaction. (D) RNP after second step of splicing (exon ligation) shows further movements of the IEP towards the catalytic center, and loosening of the EBS-IBS and δ-δ′ interactions, making the ligated exons prone to dissociation.
With the mutated catalytic triad, nucleotides at the very heart of the metal ion binding site, the RNP has significant localized differences from the ΔA and +A RNPs. The Triad RNP, which reflects a stage where the intron RNA has completed the tertiary collapse but is not yet fully committed to branching (53), exhibits a less structured active site as well as neighboring RNA–RNA interactions (Figures 5B and 8A and Supplementary Figure S17). In addition, the splice sites are not properly aligned in the Triad RNP (Figures 5B and 8A). Furthermore, the ϵ-ϵ′ and λ-λ′ interactions essential for placing the 5′-end of the intron (54) are not well-engaged in the Triad RNP, in agreement with a previous report on the interplay of bound metal ions with the ϵ–ϵ′ pairing (21). Likewise, the δ′ motif in the 3′ exon is disengaged from δ in DI (Figure 6B), These localized disruptions are in concert with a less compact structure throughout the entire RNP architecture, implicating a nucleating effect of proper formation of the active site on the entire RNP.
The IEP’s role in splicing
The IEP docks at DIVa of the intron RNA with high affinity and autoregulates its own translation (15). The fingers-palm (RT) and thumb domains contact DI and these interactions are required for catalysis (Figure 2B) (55). Although the IEP does not exhibit dramatic structural changes from RNP assembly through splicing (Figure 8), its functional interactions with key regions in the intron RNA undergo shifts, with a migration of key domains of the IEP, primarily the thumb domain, towards the active site (Figure 8). Given the fact that the thumb domain is indispensable for splicing (18,40), such proximity to the site of catalysis may assist the splicing process in a few ways. First, a more intimate RNA–protein interplay may stabilize the scaffold of the ribozyme's active site. Second, during splicing, the protein may help organize nucleotides at or near the 5′- and 3′-SSs as well as the exons (Figures 2, 4B, C and 5A). Particularly, the 3’ exon, lacking a strong RNA–RNA interaction, is likely guided by the protein during splicing (Figure 8C). Third, movements of the IEP may directly dictate the conformations of key RNA motifs such as EBS1, and/or indirectly influence RNA–protein interactions, as for the contact with DId(iii)a (Figure 4C). Finally, the non-conserved DNA binding domain (DBD) (56) also shifts toward the active site (Figure 5A), and thereby could buttress the active site. Additionally, such adjustments in the DBD might facilitate DNA binding in the retromobility process after the completion of forward splicing.
Active site formation and splicing
The Triad RNP, which is unable to properly form the active site, is not yet fully committed to splicing (Figure 8A). In contrast, the ΔA RNP represents a configuration further in the process, in which the catalytic center has been activated, immediately before the first nucleophilic attack that results in branching (Figure 8B). Commitment of the ΔA RNP is evidenced by a low level of splicing activity (Figure 1D) (9). Changes in the DV catalytic center upon splicing are attributable to strengthening of tertiary interactions (Figure 5B) (9), as well as by enhanced support from the IEP, which also has strengthened interactions with the 5′- and 3′-splice sites (Figure 4).
For a catalytically primed group II intron, the 5′ and 3′ exons are both engaged and their densities are obvious in the ΔA RNP (Figure 5A and 6A). Given that this is not the case for the Triad RNP, as described above, it would appear that the metal-binding pocket not only serves as the catalytic center, but also influences adjacent structures, including the placement of the exons. The ΔA RNP exhibits an arrangement of exon placement and EBS-IBS interactions similar to a IIC intron's pre-catalytic state, including a kink at the 5’-SS (Figures 2B, 4C, 5A and 7) (19,20,23,57). Additionally, the remodeling of the active site during splicing is in accord with reports on the IIC intron (17,19–22), suggesting that changes inside the catalytic core and surrounding the splice sites are uniform for all group II introns.
In such a catalytically active RNP, provided a branch-point, the splicing process would be initiated by the adenosine nucleophile at the 5′-SS in a way that is facilitated by the flexibility of DVI (Figures 6C, D and 8A, B) (58). As a result of the interplay between DII and DVI during splicing, DVI is rearranged to place the branch point adenosine at the 5′-SS to allow the first nucleophilic attack (Figures 1A and 6C, D). Curiously, DVI and branch point rearrangements in group IIB introns showed the π-π′ interaction as engaged (21,22) or disengaged (21,50) at different catalytic stages, although mutation in either π–π′ or η–η′ did not seem to affect the efficiency of branching (21). In contrast, our data with the group IIA intron suggest that both π–π′ and η–η′ interactions stay engaged before and after splicing. Such a discrepancy in stability of the π-π′ interaction may be explained by one or both of the following. First, whereas in the IIB intron the branch point is immediately adjacent to π′, in the IIA intron Ll.LtrB it is 12-nt from π′ and further away from the 3′-splice site. Second, IIA introns may use other factors, possibly including the IEP via its interaction with the intron's 3′ end, to govern activity of the branch point, somewhat analogous to spliceosomal introns (25). It is also possible that the previously-reported open structures near DVI were representations of the stage at which the RNA is not yet fully committed to catalysis.
Immediately after branching, it is postulated that the 3′ exon is further stabilized through IEP-mediated, enhanced δ–δ′ interaction, brought into close proximity to the active site as well as the reactive 5′ exon (Figure 8C). After the second transesterification forming ligated exons, the δ–δ′ interaction is again weakened, positioning the 3′ end of the intron away from the catalytic center (Figure 8D). Meanwhile, the 5′ exon adopts weaker interactions with the EBSs and relies more on interactions with the IEP for retention in the RNP complex, making the ligated exons prone to dissociation (Figures 4C and 8D, Supplementary Figure S14). Such a reconfigured active site may also favor the ensuing DNA targeting activities by the lariat RNP (1,59), as the EBSs are readjusted to dissociate from the exons to facilitate DNA binding (Figure 4C) (23).
The active site, from maturation to commitment, resembles the spliceosome
The spliceosome adopts multiple configurations during the two transesterification steps (25,60). Unlike the group II intron, deletion of the catalytic triad in the snRNA U6 of the spliceosome still allows formation of the pre-catalytic B* structure, with the help of ancillary RNA–RNA and RNA–protein interactions (Figure 7A) (61). This may be explained by spliceosomal active site formation being assisted by various proteins and snRNAs, whereas group II introns rely heavily on structures intrinsic to the RNA. Interestingly, while the eukaryotic splicing machinery is highly dynamic and undergoes extensive remodeling during splicing, the catalytic core composed of the snRNPs and more than 20 protein components remains relatively rigid (25). Such stability at the catalytic core is paralleled by the group II intron, as demonstrated by the comparison between the ΔA and +A RNPs (Figures 3 and 5).
Arrangement of key RNA components of the splice sites and branchpoint in DVI and the U2 snRNA of the spliceosome are highly similar (Figure 7). This is paralleled by the similarity between α helices of the thumb domain of the IEP and those of the Prp8 protein in the spliceosome. These findings support the hypothesis that the structure of the catalytic core and conformational rearrangements accompanying splicing remained largely unchanged during evolution from the group II intron-like ancestor to the spliceosome (25). In the more highly evolved spliceosome, the early RNA scaffold has been increasingly replaced by protein and/or snRNP components, such that splicing is regulated in a more complex and precise manner. Remarkably, for group II introns, extensive partnership between the RNA and the single protein provides both sufficient stability and versatility required for accurate and efficient splicing.
DATA AVAILABILITY
Cryo-EM maps have been deposited in the Electron Microscopy Data Bank with accession codes EMD-30532 and EMD-30541 for the dimeric and monomeric forms of the ΔA RNP, respectively, and with EMD-30533 and EMD-30540 for the dimeric and monomeric forms of the Triad RNP, respectively. The models based on the dimeric ΔA RNP and the dimeric Triad RNP, and the updated +A RNP model have been deposited in the Protein Data Bank with accession codes 7D0F, 7D0G and 7D1A, respectively. All other supporting data are included in the manuscript and the Supplementary Information.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Guosheng Qu and Carol Lyn Piazza for useful discussions, and Jianlin Lei, Xiaomin Li, Xiaomei Li, Danyang Li, Jie Wen, Xiaofeng Hu and Anbao Jia (Tsinghua University) for technical support in cryo-electron microscopy operation and data processing. We are thankful to Rui Bai, Ningning Li and Jie Xu for comments on the structural analysis.
Author contributions: X.D. and M.B. designed the RNP purification scheme and the biochemical experiments. N.L., X.D. and C.H. purified the RNPs. N.L., J.W. and H-W.W. collected and processed the cryo-EM data. J.Z., JW.W., N.L. and H-W.W. built the models. N.L., X.D., H-W.W. and M.B. analyzed the data and wrote the manuscript with input from all authors.
Contributor Information
Nan Liu, Ministry of Education Key Laboratory of Protein Sciences, Tsinghua-Peking Joint Center for Life Sciences, Beijing Advanced Innovation Center for Structural Biology, School of Life Sciences, Tsinghua University, Beijing 100084, China.
Xiaolong Dong, Department of Biological Sciences and RNA Institute, University at Albany, Albany, NY 12222, USA.
Cuixia Hu, Ministry of Education Key Laboratory of Protein Sciences, Tsinghua-Peking Joint Center for Life Sciences, Beijing Advanced Innovation Center for Structural Biology, School of Life Sciences, Tsinghua University, Beijing 100084, China.
Jianwei Zeng, Ministry of Education Key Laboratory of Protein Sciences, Tsinghua-Peking Joint Center for Life Sciences, Beijing Advanced Innovation Center for Structural Biology, School of Life Sciences, Tsinghua University, Beijing 100084, China.
Jiawei Wang, Ministry of Education Key Laboratory of Protein Sciences, Tsinghua-Peking Joint Center for Life Sciences, Beijing Advanced Innovation Center for Structural Biology, School of Life Sciences, Tsinghua University, Beijing 100084, China.
Jia Wang, Ministry of Education Key Laboratory of Protein Sciences, Tsinghua-Peking Joint Center for Life Sciences, Beijing Advanced Innovation Center for Structural Biology, School of Life Sciences, Tsinghua University, Beijing 100084, China.
Hong-Wei Wang, Ministry of Education Key Laboratory of Protein Sciences, Tsinghua-Peking Joint Center for Life Sciences, Beijing Advanced Innovation Center for Structural Biology, School of Life Sciences, Tsinghua University, Beijing 100084, China.
Marlene Belfort, Department of Biological Sciences and RNA Institute, University at Albany, Albany, NY 12222, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Natural Science Foundation of China [31825009 to H.-W.W.]; National Institutes of Health [GM39422, GM44844 to M.B.]. Funding for open access charge: NIH; Natural Science Foundation of China.
Conflict of interest statement. None declared.
REFERENCES
- 1. Lambowitz A.M., Zimmerly S.. Group II introns: mobile ribozymes that invade DNA. Cold Spring Harb. Perspect. Biol. 2011; 3:a003616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lambowitz A.M., Belfort M.. Mobile bacterial group II introns at the crux of eukaryotic evolution. Microbiol Spectr. 2015; 3:MDNA3–0050–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Will C.L., Luhrmann R.. Spliceosome structure and function. Cold Spring Harb. Perspect. Biol. 2011; 3:a003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Novikova O., Belfort M.. Mobile group II introns as ancestral eukaryotic elements. Trends Genet. 2017; 33:773–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jarrell K.A., Peebles C.L., Dietrich R.C., Romiti S.L., Perlman P.S.. Group II intron self-splicing. Alternative reaction conditions yield novel products. J. Biol. Chem. 1988; 263:3432–3439. [PubMed] [Google Scholar]
- 6. Daniels D.L., Michels W.J. Jr., Pyle A.M.. Two competing pathways for self-splicing by group II introns: a quantitative analysis of in vitro reaction rates and products. J. Mol. Biol. 1996; 256:31–49. [DOI] [PubMed] [Google Scholar]
- 7. Zhao C., Pyle A.M.. The group II intron maturase: a reverse transcriptase and splicing factor go hand in hand. Curr. Opin. Struct. Biol. 2017; 47:30–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Noah J.W., Lambowitz A.M.. Effects of maturase binding and Mg2+ concentration on group II intron RNA folding investigated by UV cross-linking. Biochemistry. 2003; 42:12466–12480. [DOI] [PubMed] [Google Scholar]
- 9. Dong X., Ranganathan S., Qu G., Piazza C.L., Belfort M.. Structural accommodations accompanying splicing of a group II intron RNP. Nucleic Acids Res. 2018; 46:8542–8556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Qu G., Kaushal P.S., Wang J., Shigematsu H., Piazza C.L., Agrawal R.K., Belfort M., Wang H.W.. Structure of a group II intron in complex with its reverse transcriptase. Nat. Struct. Mol. Biol. 2016; 23:549–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Saldanha R., Chen B., Wank H., Matsuura M., Edwards J., Lambowitz A.M.. RNA and protein catalysis in group II intron splicing and mobility reactions using purified components. Biochemistry. 1999; 38:9069–9083. [DOI] [PubMed] [Google Scholar]
- 12. Belfort M., Lambowitz A.M.. Group II intron RNPs and reverse transcriptases: from retroelements to research tools. Cold Spring Harb. Perspect. Biol. 2019; 11:a003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pyle A.M. The tertiary structure of group II introns: implications for biological function and evolution. Crit. Rev. Biochem. Mol. Biol. 2010; 45:215–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wank H., SanFilippo J., Singh R.N., Matsuura M., Lambowitz A.M.. A reverse transcriptase/maturase promotes splicing by binding at its own coding segment in a group II intron RNA. Mol. Cell. 1999; 4:239–250. [DOI] [PubMed] [Google Scholar]
- 15. Singh R.N., Saldanha R.J., D'Souza L.M., Lambowitz A.M.. Binding of a group II intron-encoded reverse transcriptase/maturase to its high affinity intron RNA binding site involves sequence-specific recognition and autoregulates translation. J. Mol. Biol. 2002; 318:287–303. [DOI] [PubMed] [Google Scholar]
- 16. Steitz T.A., Steitz J.A.. A general two-metal-ion mechanism for catalytic RNA. Proc. Natl. Acad. Sci. U.S.A. 1993; 90:6498–6502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Marcia M., Pyle A.M.. Visualizing group II intron catalysis through the stages of splicing. Cell. 2012; 151:497–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cui X., Matsuura M., Wang Q., Ma H., Lambowitz A.M.. A group II intron-encoded maturase functions preferentially in cis and requires both the reverse transcriptase and X domains to promote RNA splicing. J. Mol. Biol. 2004; 340:211–231. [DOI] [PubMed] [Google Scholar]
- 19. Toor N., Keating K.S., Taylor S.D., Pyle A.M.. Crystal structure of a self-spliced group II intron. Science. 2008; 320:77–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chan R.T., Robart A.R., Rajashankar K.R., Pyle A.M., Toor N.. Crystal structure of a group II intron in the pre-catalytic state. Nat. Struct. Mol. Biol. 2012; 19:555–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Robart A.R., Chan R.T., Peters J.K., Rajashankar K.R., Toor N.. Crystal structure of a eukaryotic group II intron lariat. Nature. 2014; 514:193–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chan R.T., Peters J.K., Robart A.R., Wiryaman T., Rajashankar K.R., Toor N.. Structural basis for the second step of group II intron splicing. Nat. Commun. 2018; 9:4676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Costa M., Walbott H., Monachello D., Westhof E., Michel F.. Crystal structures of a group II intron lariat primed for reverse splicing. Science. 2016; 354:6316. [DOI] [PubMed] [Google Scholar]
- 24. Matsuura M., Noah J.W., Lambowitz A.M.. Mechanism of maturase-promoted group II intron splicing. EMBO J. 2001; 20:7259–7270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yan C., Wan R., Shi Y.. Molecular mechanisms of pre-mRNA splicing through structural biology of the spliceosome. Cold Spring Harb. Perspect. Biol. 2019; 11:a032409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Huang T., Shaikh T.R., Gupta K., Contreras-Martin L.M., Grassucci R.A., Van Duyne G.D., Frank J., Belfort M.. The group II intron ribonucleoprotein precursor is a large, loosely packed structure. Nucleic Acids Res. 2011; 39:2845–2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gupta K., Contreras L.M., Smith D., Qu G., Huang T., Spruce L.A., Seeholzer S.H., Belfort M., Van Duyne G.D.. Quaternary arrangement of an active, native group II intron ribonucleoprotein complex revealed by small-angle X-ray scattering. Nucleic Acids Res. 2014; 42:5347–5360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zheng S.Q., Palovcak E., Armache J.P., Verba K.A., Cheng Y., Agard D.A.. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods. 2017; 14:331–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rohou A., Grigorieff N.. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 2015; 192:216–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Scheres S.H. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 2012; 180:519–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bai X.C., Rajendra E., Yang G., Shi Y., Scheres S.H.. Sampling the conformational space of the catalytic subunit of human gamma-secretase. Elife. 2015; 4:e11182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Scheres S.H. Processing of structurally heterogeneous cryo-EM data in RELION. Methods Enzymol. 2016; 579:125–157. [DOI] [PubMed] [Google Scholar]
- 33. Tan Y.Z., Baldwin P.R., Davis J.H., Williamson J.R., Potter C.S., Carragher B., Lyumkis D.. Addressing preferred specimen orientation in single-particle cryo-EM through tilting. Nat. Methods. 2017; 14:793–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Emsley P., Lohkamp B., Scott W.G., Cowtan K.. Features and development of Coot. Acta Crystallogr. D. Biol. Crystallogr. 2010; 66:486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Afonine P.V., Poon B.K., Read R.J., Sobolev O.V., Terwilliger T.C., Urzhumtsev A., Adams P.D.. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr. D Struct. Biol. 2018; 74:531–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E.. UCSF chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 2004; 25:1605–1612. [DOI] [PubMed] [Google Scholar]
- 37. Punjani A., Rubinstein J.L., Fleet D.J., Brubaker M.A.. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods. 2017; 14:290–296. [DOI] [PubMed] [Google Scholar]
- 38. Contreras L.M., Huang T., Piazza C.L., Smith D., Qu G., Gelderman G., Potratz J.P., Russell R., Belfort M.. Group II intron-ribosome association protects intron RNA from degradation. RNA. 2013; 19:1497–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Slagter-Jager J.G., Allen G.S., Smith D., Hahn I.A., Frank J., Belfort M.. Visualization of a group II intron in the 23S rRNA of a stable ribosome. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:9838–9843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Blocker F.J., Mohr G., Conlan L.H., Qi L., Belfort M., Lambowitz A.M.. Domain structure and three-dimensional model of a group II intron-encoded reverse transcriptase. RNA. 2005; 11:14–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Keating K.S., Toor N., Perlman P.S., Pyle A.M.. A structural analysis of the group II intron active site and implications for the spliceosome. RNA. 2010; 16:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wan R., Bai R., Yan C., Lei J., Shi Y.. Structures of the catalytically activated yeast spliceosome reveal the mechanism of branching. Cell. 2019; 177:339–351. [DOI] [PubMed] [Google Scholar]
- 43. Wilkinson M.E., Fica S.M., Galej W.P., Norman C.M., Newman A.J., Nagai K.. Postcatalytic spliceosome structure reveals mechanism of 3'-splice site selection. Science. 2017; 358:1283–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Agrawal R.K., Wang H.W., Belfort M.. Forks in the tracks: group II introns, spliceosomes, telomeres and beyond. RNA Biol. 2016; 13:1218–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sarafianos S.G., Marchand B., Das K., Himmel D.M., Parniak M.A., Hughes S.H., Arnold E.. Structure and function of HIV-1 reverse transcriptase: molecular mechanisms of polymerization and inhibition. J. Mol. Biol. 2009; 385:693–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Roth M.J., Tanese N., Goff S.P.. Purification and characterization of murine retroviral reverse transcriptase expressed in Escherichia coli. J. Biol. Chem. 1985; 260:9326–9335. [PubMed] [Google Scholar]
- 47. Zhao C., Pyle A.M.. Crystal structures of a group II intron maturase reveal a missing link in spliceosome evolution. Nat. Struct. Mol. Biol. 2016; 23:558–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Rambo R.P., Doudna J.A.. Assembly of an active group II intron-maturase complex by protein dimerization. Biochemistry. 2004; 43:6486–6497. [DOI] [PubMed] [Google Scholar]
- 49. Stamos J.L., Lentzsch A.M., Lambowitz A.M.. Structure of a thermostable group II intron reverse transcriptase with template-primer and its functional and evolutionary implications. Mol. Cell. 2017; 68:926–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Haack D.B., Yan X., Zhang C., Hingey J., Lyumkis D., Baker T.S., Toor N.. Cryo-EM structures of a group II intron reverse splicing into DNA. Cell. 2019; 178:612–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Paudel B.P., Fiorini E., Borner R., Sigel R.K.O., Rueda D.S.. Optimal molecular crowding accelerates group II intron folding and maximizes catalysis. Proc. Natl. Acad. Sci. U.S.A. 2018; 115:11917–11922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhao C., Pyle A.M.. Structural insights into the mechanism of group II intron splicing. Trends Biochem. Sci. 2017; 42:470–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Pyle A.M., Fedorova O., Waldsich C.. Folding of group II introns: a model system for large, multidomain RNAs. Trends Biochem. Sci. 2007; 32:138–145. [DOI] [PubMed] [Google Scholar]
- 54. Jacquier A., Michel F.. Base-pairing interactions involving the 5' and 3'-terminal nucleotides of group II self-splicing introns. J. Mol. Biol. 1990; 213:437–447. [DOI] [PubMed] [Google Scholar]
- 55. Gu S.Q., Cui X., Mou S., Mohr S., Yao J., Lambowitz A.M.. Genetic identification of potential RNA-binding regions in a group II intron-encoded reverse transcriptase. RNA. 2010; 16:732–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. San Filippo J., Lambowitz A.M.. Characterization of the C-terminal DNA-binding/DNA endonuclease region of a group II intron-encoded protein. J. Mol. Biol. 2002; 324:933–951. [DOI] [PubMed] [Google Scholar]
- 57. Toor N., Rajashankar K., Keating K.S., Pyle A.M.. Structural basis for exon recognition by a group II intron. Nat. Struct. Mol. Biol. 2008; 15:1221–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Plangger R., Juen M.A., Hoernes T.P., Nussbaumer F., Kremser J., Strebitzer E., Klingler D., Erharter K., Tollinger M., Erlacher M.D. et al.. Branch site bulge conformations in domain 6 determine functional sugar puckers in group II intron splicing. Nucleic. Acids. Res. 2019; 47:11430–11440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Cousineau B., Smith D., Lawrence-Cavanagh S., Mueller J.E., Yang J., Mills D., Manias D., Dunny G., Lambowitz A.M., Belfort M.. Retrohoming of a bacterial group II intron: mobility via complete reverse splicing, independent of homologous DNA recombination. Cell. 1998; 94:451–462. [DOI] [PubMed] [Google Scholar]
- 60. Hang J., Wan R., Yan C., Shi Y.. Structural basis of pre-mRNA splicing. Science. 2015; 349:1191–1198. [DOI] [PubMed] [Google Scholar]
- 61. Bao P., Boon K.L., Will C.L., Hartmuth K., Luhrmann R.. Multiple RNA-RNA tertiary interactions are dispensable for formation of a functional U2/U6 RNA catalytic core in the spliceosome. Nucleic Acids Res. 2018; 46:12126–12138. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Cryo-EM maps have been deposited in the Electron Microscopy Data Bank with accession codes EMD-30532 and EMD-30541 for the dimeric and monomeric forms of the ΔA RNP, respectively, and with EMD-30533 and EMD-30540 for the dimeric and monomeric forms of the Triad RNP, respectively. The models based on the dimeric ΔA RNP and the dimeric Triad RNP, and the updated +A RNP model have been deposited in the Protein Data Bank with accession codes 7D0F, 7D0G and 7D1A, respectively. All other supporting data are included in the manuscript and the Supplementary Information.