Abstract
The three-dimensional configuration of the chromatin architecture is known to be crucial for alterations in the transcriptional network; however, the underlying mechanisms of epigenetic control of senescence-related gene expression by modulating the chromatin architecture remain unknown. Here, we demonstrate frequent chromosomal compartment switching during mouse embryonic fibroblasts (MEFs) replicative senescence as characterized by senescence-inactivated (SIAEs) and -activated enhancers (SAEs) in topologically associated domains (TADs). Mechanistically, SAEs are closely correlated with senescence-associated secretory phenotype (SASP) genes, which are a key transcriptional feature of an aging microenvironment that contributes to tumor progression, aging acceleration, and immunoinflammatory responses. Moreover, SAEs can positively regulate robust changes in SASP expression. The transcription factor CCAAT/enhancer binding protein α (C/EBPα) is capable of enhancing SAE activity, which accelerates the emergence of SAEs flanking SASPs and the secretion of downstream factors, contributing to the progression of senescence. Our results provide novel insight into the TAD-related control of SASP gene expression by revealing hierarchical roles of the chromatin architecture, transcription factors, and enhancer activity in the regulation of cellular senescence.
INTRODUCTION
Cellular senescence, a process that results in permanent proliferation defects, can be triggered by exogenous or endogenous stimuli (1,2). Initially defined as a phenotype in cultured fibroblasts, senescent cells can be identified by elevated lysosomal senescence-associated β-galactosidase (SA-β-gal) activity and expression of the tumor suppressor protein p16 (3,4). In addition to the exhaustive growth capacity attributed to telomere attrition, senescence can also be induced by DNA-damaging agents or aberrant oncogene activation governed by the retinoblastoma (Rb) and p53 signaling cascades (5,6).
Cellular senescence is recognized as a safeguard against cancer and therefore a potential basis for therapeutic regimens. Emerging evidence reinforces the concept that cellular senescence can extend beyond tumor suppression into many other biological processes, such as embryonic development, wound healing, tissue repair and aging-related diseases (7–9). Most importantly, cellular senescence is characterized by an array of secreted inflammatory factors known as the senescence-associated secretory phenotype (SASP) (10–12), including the well-described SASP factors, interleukins (ILs), chemokines (CXCL, CCL), and insulin-like growth factors (IGF), which are involved in the regulation of immune surveillance both pathologically and physiologically (13). The induced SASP factors can activate cell-surface receptors and signal transduction pathways to influence both extra- and intracellular microenvironments (14). Temporary secretion of SASP warns nearby cells of the potential danger and promotes immune clearance of damaged cells (9). However, persistent SASP secretion can cause systemic inflammation or the growth of malignant cells, displaying negative effects in numerous pathological states (15). Therefore, elucidation of the underlying mechanisms that control SASP gene expression during cellular senescence has become important for intervening in the aging process.
Indeed, certain morphological changes in chromatin occur in senescent cells, which affect chromosome condensation and distribution and are likely to affect the nuclear architecture (16–19), leading to alterations in nucleosome positioning, epigenetic landscapes, and transcriptional profiling. The use of Hi-C technology has revealed two major levels of topological organization across the mammalian genome. At the megabase (Mb) scale of chromatin organization, two spatially segregated compartments, A and B, have been shown to correlate with the euchromatic and heterochromatic regions, respectively. The submegabase scale consists of topologically associated domains (TADs) and chromatin loops (20–22) that restrict or facilitate interactions among cis-regulatory elements (23,24). Numerous studies have illustrated that dynamic alterations in higher-order chromatin organization are associated with distinct transcriptional profiles (25–28). For example, enhancers are cis-regulatory elements that reside far away from promoters yet are able to boost gene transcription effectively when bound to various transcription factors that regulate the expression of distal genes through specific 3D chromatin conformations (29,30). Cellular senescence is a multilayered process that displays hierarchical gene repression and activation mechanisms influenced by structural reorganization across the genome (31–33). Considering that the complex senescence regulatory network is involved in a variety of physiological processes (34), identification of senescence-driven enhancer repertoires across the genome is imperative.
In the present study, we report that the process of senescence can widely affect chromatin topological architecture as well as the epigenome and transcriptome. TAD-related spatial rearrangement of chromatin is accompanied by remodeling of the epigenetic enhancer repertoires in which H3K27ac-enriched senescence-activated enhancers (SAEs) modulate the kinetics of adjacent SASP genes. We further demonstrate that the transcription factor, CCAAT/enhancer binding protein α (C/EBPα), is recruited to the SAE region and acts as a key regulator during senescence, whereas suppression of C/EBPα reduces the activity of SAEs as well as the expression and secretion of SASP factors. In summary, our study reveals the spatiotemporal chromatin reorganization and enhancer redistribution that directs critical SASP gene regulatory networks during senescence.
MATERIALS AND METHODS
Generation of primary MEFs and REFs
MEFs were isolated from pregnant C57BL/6 mice and REFs were isolated from pregnant Sprague−Dawley rats. MEFs and REFs were cultured in DMEM supplemented with 10% fetal bovine serum (FBS, Gibco, 12800) under 5% CO2 at 37°C. Cells were serially passaged after reaching 80−90% confluence. The number for population doubling was calculated using the formula: PD = log2(Ne/N0), where Ne represents the number of counts and N0 represents the number of seeds. Cumulative PD numbers represent the sum of PDs from all previous passages, with cells derived from the primary culture considered PD = 0.
In situ Hi-C
Briefly, 2 × 106 cells fixed in 1% formaldehyde were suspended in Hi-C lysis buffer, to which 100 U MobI restriction enzyme (NEB, R0147) was added for overnight chromatin digestion. Biotin-14-dATP (Life Technologies, 19524016) was used to fill DNA restriction ends prior to DNA proximity ligation and crosslink reversal. To make the biotin-labeled DNA suitable for high-throughput sequencing using the Illumina platform, the DNA was sheared to 300−500 bp in size using a Covaris LE220 sonicator (Covaris, Woburn, MA) for 135 s. Sheared DNA was end-repaired and size-selected using AMPure XP beads (Beckman Coulter, A63882) prior to the dATP-tailing step. Biotin-labeled DNA was pulled down using Streptavidin T1 Dynabeads® (Life technologies, 65602). Bead-bound Hi-C DNA was amplified by 13 cycles of PCR. Hi-C libraries were constructed according to the NEBnext library preparation protocol (NEB, E7335) and sequenced on the Illumina Hiseq-PE150 platform (X, Corp). Further essential details regarding the in situ Hi-C experiment have been previously published (35).
Cellular senescence assays
In brief, cells for immunofluorescence were seeded on cover slides in 100-mm culture dishes. At a confluence of 70−80%, cells were washed twice with ice-cold 1 × PBS and fixed with 4% paraformaldehyde at room temperature for 10 min. Subsequently, the cells were blocked with 1% BSA at room temperature for 30 min and incubated overnight at 4°C with an anti-Ki67 primary antibody (Abcam, ab15580; 1:200). The following day, the cells were washed and incubated with 150 μl secondary antibody (Alexa Fluor 488 anti-rabbit; Alexa Fluor 594 anti-mouse, Life Technologies) at room temperature for 2 h. The DAPI (final concentration of 2 ng/μL) staining time was controlled at 3 min to avoid excessive background. Finally, the cover slides were washed, sealed with 5 μl Fluoromount-G® mounting medium, and observed under a fluorescence microscope 5 h later.
SA-β-gal assays were performed using senescence histochemical staining kits (Sigma-Aldrich, CAS0030) according to the manufacturer's instructions. Briefly, cells were incubated in fixation solution for 8 min, followed by staining solution containing the substrate X-gal at 37°C overnight. The following day, cells were washed prior to observation under a microscope. For long-term conservation, the plates were stored in 75% glycerin.
Expression analysis
For western blotting, cells were harvested using TRIzol reagent (Invitrogen, 10296028), and total protein was isolated according to the manufacturer's protocol. Total protein was dissolved in 1% SDS, subjected to SDS-PAGE, and transferred to nitrocellulose membrane. The membranes were incubated overnight at 4°C with the primary antibodies of interest, followed by washing three times with PBST (1 × PBS + 0.1% Tween-20). Subsequently, the membranes were incubated with a secondary antibody [IRDye 800CW goat anti-mouse (LI-COR, 926-32210)] for 2 h at room temperature and washed three times with PBST prior to visualization using the Odyssey infrared imaging system (Odyssey, LI-COR).
For total RNA analysis, cells were harvested using TRIzol reagent (Invitrogen, 10296028), and total RNA was isolated according to the manufacturer's protocol. For RT-qPCR analysis, 1 μg total RNA was reverse-transcribed using random hexamer DN6 and M-MLV reverse transcriptase (Invitrogen). A 10 μL reaction system including 2 μl reverse-transcribed product (1:50) and 8 μl SYBR Green I Master Mix (Takara) was evaluated using a LightCycler 450 instrument. The primers used are listed in the Supplementary Data. For RNA-seq analysis, strand-specific RNA libraries, from which ribosomes had been eliminated prior to sequencing, were implemented to acquire mRNA and lncRNA data on the Illumina Hiseq-PE150 platform (X, Corp).
For detection of soluble protein, the supernatants from serially passaged cells were collected and analyzed using a mouse cytokine array kit (R&D Systems, ARY028) according to the manufacturer's instructions.
ChIP-qPCR and ChIP-seq analyses
Cells were crosslinked with 1% formaldehyde solution (Sigma, F8775) for 10 min at room temperature, following which the fixation reaction was quenched for 5 min using glycine at a final concentration of 0.125 M. Subsequently, chromatin was sheared using a Bioruptor® until the DNA was fragmented to 300−500 bp. The supernatant was precleared with 20 μl salmon sperm DNA/Protein A sepharose beads (Sigma, GE17-5280). Next, 3 μg histone modification antibody (H3K27ac, Abcam, ab4729; H3K4me1, Abcam, ab8895; C/EBPα, Santa Cruz, sc365318X; C/EBPβ, Santa Cruz, sc7962X) was added to a 250-μg fraction of supernatant and agitated overnight at 4°C. Antibody–protein–DNA complexes were pulled down using 30 μl Protein A sepharose bead slurry. After washing, the eluted complexes were subsequently decrosslinked at 65°C for 5 h. Following the phenol chloroform extraction step, ChIP DNA was purified for further study. The primers used in ChIP-qPCR are listed in the Supplementary Data. For ChIP-seq analysis, sequencing libraries were established from ChIP DNA according to the Illumina protocol, and the libraries were sequenced on the Illumina Hiseq-PE150 platform (X, Corp).
Hi-C data processing
All paired-end reads were processed by HiC-Pro (version 2.7.8) to generate the interaction matrix. Subsequently, the matrix was normalized using Iterative Correction and Eigenvector decomposition (ICE) and library size. HiTC (R package) was used to visualize the interaction matrix and calculate the A/B compartments (at 100-kb resolution). When defining compartment B to A changes by comparing PD2 and PD14, we restricted the changes in PC1 values from ←0.2 to >0.2; and for A to B changes, we restricted the changes in PC1 values from >0.2 to ←0.2. The TADs were detected using the method described by Crane et al. (https://github.com/dekkerlab/crane-nature-2015).
RNA-seq data processing
Strand-specific RNA-seq reads were mapped to the mouse reference genome (version mm10) by hisat2 (version 2.0.5) with the parameter ‘–rna-strandness RF,’ and the gene expression level was measured by TPM using StringTie (version 1.3.1c). All mapped bam files were converted to bigwig using bedtools genomecov (version 2.24.0) for visualization in IGV.
ChIP-seq data processing
All ChIP-seq data were mapped to mm10 by Bowtie2 (version 2.3.2). Following removal of unmapped and nonunique mapped reads using SAMtools (version 1.3.1), duplicate PCR reads were filtered out by Picard Tools (version: 1.118). All peaks were called by MACS2 (version 2.1.1.20160309); however, histone modifications (H3K4me1, H3K27ac) and transcription factors (C/EBPα) were analyzed using different parameters. The former used ‘–broad -g mm -B –broad-cutoff 0.1,’ and the latter used ‘-f BAM -g mm -B -q 0.01.’ For comparison of the ChIP-seq and ENCODE data, H3K4me1, H3K27ac and CTCF ChIP-seq data generated in MEFs were downloaded from https://www.encodeproject.org/. Subsequently, reads were mapped to mm10 using the same process used for our ChIP-seq data. The correlations among all mapped bam files from our ChIP-seq and downloaded ENCODE data were calculated using Wigcorrelate, a tool downloaded from the UCSC website.
Identification of senescence-associated enhancers
A peak with both H3K4me1 and H3K27ac modifications that was at least 2-kb away from any gene TSS was defined as an enhancer. All enhancers from PD2, PD8, and PD14 were merged using BEDTools. To measure the activity of the enhancers, the H3K27ac read coverage for each merged enhancer was calculated and subsequently compared between PD2 and PD14 using edgeR (R package) to identify the senescence-associated enhancers. The enhancers were annotated using annotatePeaks.pl in homer2.
Statistical analysis
Data were analyzed by either a two-tailed unpaired Student's t-test or one-way ANOVA with Dunnett's multiple comparison test (GraphPad Prism software, version 5.01). The results are expressed as the mean ± S.D. Probability values (P) < 0.05 were considered statistically significant.
RESULTS
Chromatin structure reorganizes during cellular senescence
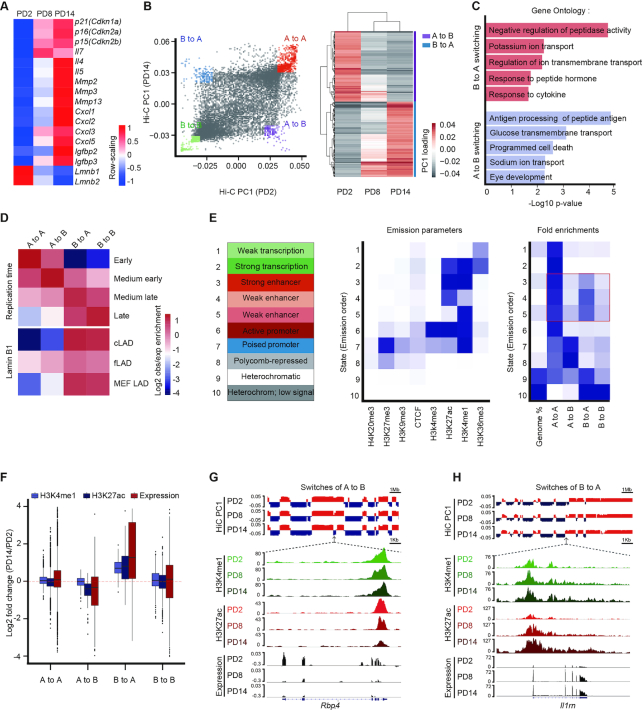
We established a cellular senescence model by the serial passage of primary mouse embryonic fibroblasts (MEFs). Late passage MEFs exhibited well-characterized senescence phenotypes, including limited cell growth capacity, elevated senescence-associated β-galactosidase (SA-β-gal) staining, and attenuated Mki67 signaling (Supplementary Figure S1A−C). To map the 3D organization of the genome, we performed in situ Hi-C in three population doublings (PDs) of MEFs: PD2, PD8 and PD14, representing the different stages from proliferation to senescence. Each population was sequenced using two replicate libraries, and the data showed high reproducibility (Supplementary Figure S1D). RNA-seq was performed in these three passages to detect genome-wide transcriptome profiling (Supplementary Figure S1E). The replicative senescent genes, including p16 (Cdkn2a), p21(Cdkn1a) and the other well-recognized senescent genes showed elevated expression during senescence (Figure 1A). Gene set enrichment analysis (GSEA) showed cell cycle checkpoint genes were down-regulated upon senescence entry, while the SASP-related interferon factor genes and top 100 up-regulated genes from public MEFs and WI38 data were positively correlated with our MEFs senescence data (36,37) (Supplementary Figure S1F and Supplementary Data).
Figure 1.
Remodeling of the enhancer landscape occurs during A and B compartment switching. (A) The heatmap displaying normalized genes expression among PD2, PD8 and PD14. Senescence-relevant genes are listed. The −1, +1 values are the t-statistic used to normalize the expression TPM values. (B) The first principal components values (PC1) from the Hi-C correlation matrix at 100-kb resolution represented the A (positive value) and B (negative value) compartment. Classification of four categories for A and B compartment switching as defined by PC1 values in a paired comparison of PD2 and PD14 (left) or comparison among PD2, PD8, and PD14 (right). (C) GO analysis of compartment switching from B to A (red, top) and A to B (blue, bottom) by GREAT. The top five most significant GO terms and their enrichment scores are displayed. (D) Overlapping enrichment for the genomic features of replication time and LAD distribution at the loci of the four categories of compartment switching. (E) Eight chromatin markers were used to train the chromatin states in MEFs by chromHMM. Heatmaps showing the distribution of the four compartment switching categories in epigenomic-marked chromatin states; enhancers are highlighted in a red box. (F) Violin plots displaying the H3K27ac, H3K4me1 and gene expression levels of PD2 versus PD14 in the four compartment switching categories. (G, H) Integrative Genomics Viewer (IGV) showing A to B compartment switching (G) and B to A compartment switching (H) dynamics accompanied by epigenetic enhancer profiling and gene expression analysis at the Rbp4 and Il1rn gene loci.
The Hi-C matrices displayed a typical checkerboard pattern of self-interacting topologically associated domains (TADs) in PD2, PD8, and PD14 cells. The interaction heatmaps displayed similar spatial chromatin interactions in the three PDs (Supplementary Figure S2A). In addition, the insulation scores (38) in the three PDs showed similar patterns, and the correlations between the conditions were significantly higher than those in other cell types (Supplementary Figure S2B−D), indicating stability of the TADs during senescence. Despite the conserved TADs and TAD boundaries during senescence, we performed pairwise subtraction of interaction frequency matrices between conditions to gain greater insight into the dynamics of 3D chromatin structures. The differences between PD2 and PD14 cells demonstrated prominent changes in the interaction frequency, with increased long-range interactions and decreased short-range interactions, while the other two pairwise comparisons exhibited similar but less dramatic trends (Supplementary Figure S3A). To investigate the changes in intrachromosomal interactions in more detail, we examined the mean contact probability and found that the contact probability ratio showed a >2.5-fold decrease in interactions within a 1-kb distance when comparing PD14 and PD2 (Supplementary Figure S3B). Moreover, the ratio of long-range interactions (≥ 2 Mb) versus short-range interactions (<2 Mb) in PD14 was significantly higher than that in PD2 (Supplementary Figure S3C). Further, in comparison with that of cis-interaction, a progressively increased level of trans-interaction was detected during the progression of senescence (Supplementary Figure S3D). Taken together, these results show the gain of long-range interactions and loss of short-range interactions at both the intra- and inter-chromosomal levels, which is in accordance with previous reports (39,40), suggesting profound nuclear chromatin remodeling during senescence.
A and B compartment switching occurs upon entry into senescence
Chromosomal compartments were defined by the first principal components scores (PC1) after matrix decomposition of the Hi-C correlation matrix. A positive PC1 represented an A compartment, which was correlated with euchromatin markers, active gene expression, and high gene density, while a negative PC1 represented a B compartment, which was correlated with heterochromatin markers, silent gene expression, and low gene density (Supplementary Figure S4A−C), consistent with a previous study (20). Subsequently, we examined the dynamics of chromosomal compartments and found that 11.2% of the genome changes compartment during senescence. We then classified compartment changes into four categories as defined by PC1 signals in PD2 and PD14: unchanged states in A (A to A) or B (B to B) and switched states between A and B (A to B and B to A) (Figure 1B). Using the GREAT webserver (41), several significant gene ontology terms were enriched in compartment-switching regions, such as the response to cytokines for B to A compartment switching and programmed cell death for A to B compartment switching (Figure 1C). Lamina-associated domains (LADs) are nuclear lamina-anchored large heterochromatic regions that display frequent changes during differentiation and senescence (42). LADs can be divided into two classes: constitutive LADs (cLADs), which are strongly conserved, and facultative LADs (fLADs), which exhibit cell type-dependent characteristics (43). Investigation into the relationship between LADs and compartment switching revealed modest enrichment of fLADs in compartment-switching regions as compared with cLADs (Figure 1D and Supplementary Figure S4D−F), suggesting the redistribution of LADs during senescence (18,39,40,44).
To understand the relationship between compartment switching and regulatory elements, we subsequently employed chromHMM to train 10 chromatin states of MEFs using eight epigenetic markers (45). The chromatin states were trained using ENCODE ChIP-seq data generated from proliferating MEFs. As expected, stable A compartments were strongly enriched with enhancer, promoter, and active transcription states. Surprisingly, we found a dominant rearrangement of enhancer signals within the A/B compartment switching loci (Figure 1E). Specifically, the chromatin states of the strong and weak enhancers more often resided in the B to A compartment-switching regions than in the stable B compartments. In contrast, A to B compartment switching was accompanied by a lower enrichment of enhancer signals as compared with that in stable A compartments (Figure 1E). These results indicate that 3D chromatin reorganization, in particular compartment switching, is correlated with the redistribution of enhancer elements during senescence.
Enhancers undergo global remodeling during senescence
Given that distinct enhancer landscape changes occurred during compartment switching, we subsequently asked the question whether reorganization of the enhancer landscape coordinates with chromatin architecture to affect the transcriptional profiles of downstream genes. We performed ChIP-seq of H3K4me1 and H3K27ac in PD2, PD8 and PD14 MEFs to profile global enhancer activities. Our data showed high correlations with ENCODE data, and the H3K4me1 and H3K27ac signals were enriched around gene transcription start sites (TSSs) (Supplementary Figure S5A and B), which is in accordance with previous studies (46,47). Using the MACS2 peak-calling software (48), we identified a total of ∼160 000 H3K4me1 and ∼100 000 H3K27ac peaks in the different PDs, with most peaks being distributed in distal intergenic and intronic regions, as previously reported (49) (Supplementary Figure S5C).
Integration of our genome topology architecture, epigenome, and transcriptome data provided us with a better understanding of the senescence process at different regulatory levels. Indeed, switching from compartment A to B was accompanied by prominently weakened H3K27ac and H3K4me1 signals and decreased gene expression (Figure 1F). In contrast, switching from compartment B to A was accompanied by gradual enhancement of H3K27ac and H3K4me1 signals and increased gene expression (Figure 1F). Furthermore, the Il1rn locus, a SASP gene involved in immune signaling (50), exhibited compartment switching from B to A accompanied by elevated enhancer signaling and gene expression during senescence. On the other hand, the Rbp4 locus, a member of the lipocalin family that induces mitochondrial damage and oxidative stress, exhibited compartment switching from A to B accompanied by attenuated enhancer signaling and gene expression during senescence (51) (Figure 1G and H). Collectively, these data demonstrate that chromatin compartment switching is accompanied by consistent changes in gene expression and histone modifications during senescence.
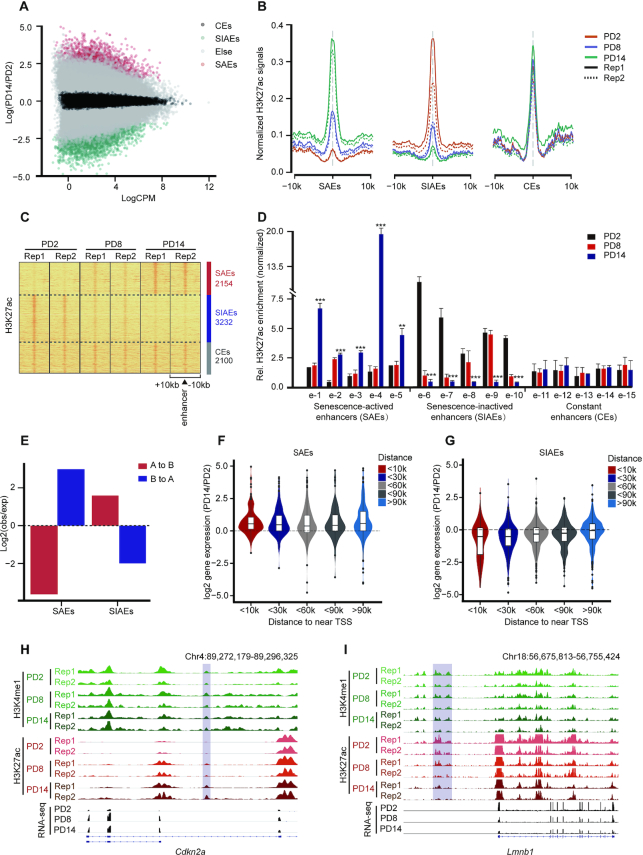
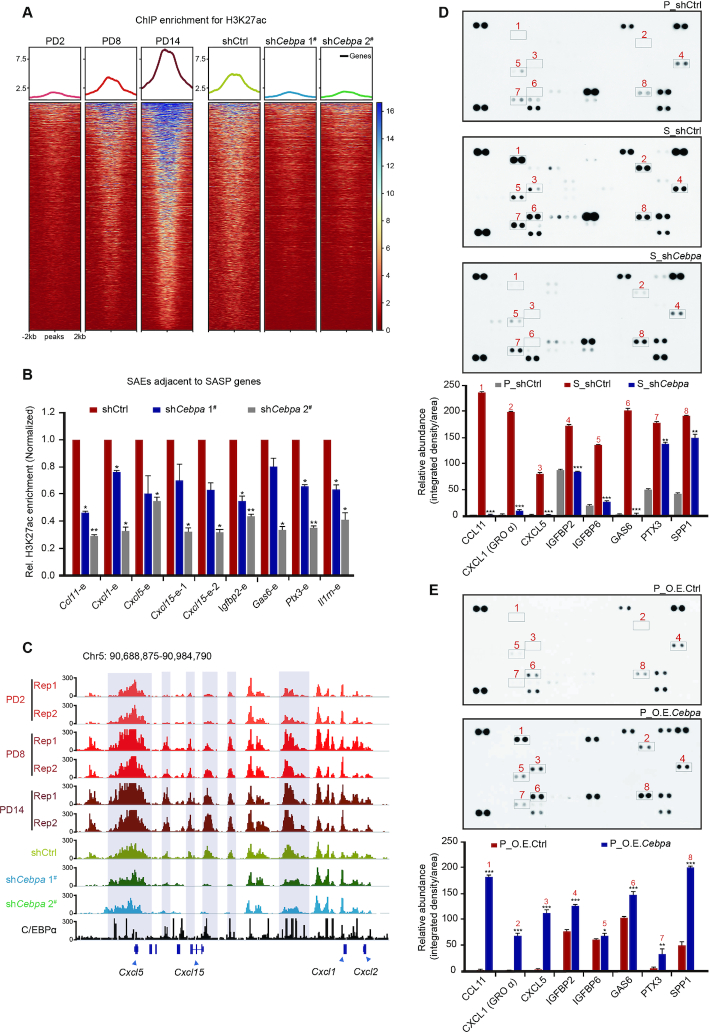
The H3K27ac marker can distinguish between active enhancers and inactive/poised enhancer elements containing H3K4me1 alone (52,53). Half the H3K4me1 peaks in the three PDs overlapped with the H3K27ac peaks (Supplementary Figure S5D). In addition, the H3K27ac levels changed to a much greater extent than the H3K4me1 levels between PD2 and PD14, especially at distal enhancers (Supplementary Figure S5E). Alterations in the levels of H3K27ac but not H3K4me1 suggest redistribution of active enhancers during senescence. Considering that the complex senescence regulatory network is involved in a variety of physiological processes, we were interested in identifying senescence-driven active enhancer repertoires across the genome. Accordingly, based on quantitative genome-wide changes in H3K27ac levels during senescence, we divided enhancers into three categories: senescence-activated enhancers (SAEs), senescence-inactivated enhancers (SIAEs), and constant enhancers (CEs) (Figure 2A−C). Randomly selected enhancers from different categories were further verified according to changes in their H3K27ac signal by ChIP-qPCR (Figure 2D). In addition, SAEs were preferentially located in the regions switching from B to A compartments, while SIAEs tended to be located in the regions switching from A to B compartments (Figure 2E). These findings reinforce the notion that reorganization of chromatin compartments is coupled to changes in enhancer activity.
Figure 2.
Genome-wide identification and characterization of senescence-associated enhancers. (A) Scatter plot showing the normalized H3K27ac signal dynamics in PD2 and PD14. SAEs, senescence-activated enhancers; SIAEs, senescence-inactivated enhancers; and CEs, constant enhancers. (B) Aggregate plots of H3K27ac showing the replicated SAEs, SIAEs and CEs in MEFs, at the enhancer peak center. (C) Heatmaps of H3K27ac signals surrounding the SAEs, SIAEs, and CEs. The number of enhancers in each of the three categories is listed on the right. Rows correspond to ±10-kb regions across the midpoint of each H3K27ac-enriched union enhancer. (D) The relative levels of H3K27ac for randomly selected SAEs and SIAEs in MEFs of different passage numbers were measured by ChIP-qPCR. The enrichment of H3K27ac was normalized to 10% input, and IgG was used as a negative control. Error bars represent the S.D. obtained from three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001. One-way ANOVA with Dunnett's multiple comparison test was performed. (E) The proportion of compartment switching clusters located in the different enhancer categories. (F, G) Violin plots showing the correlation between the expression of genes adjacent to the enhancers (SAEs: F, SIAEs: G) and the distances to the TSSs of the nearest genes during senescence. (H, I) H3K27ac ChIP-Seq occupancy profiles at representative SAE and SIAE loci for Cdkn2a and Lmnb1.
Senescence-activated enhancers (SAEs) regulate the expression of adjacent SASP genes
Since enhancers can activate the transcription of nearby and distal genes (54), we subsequently addressed the impact of enhancer remodeling during senescence on global gene expression. A total of 1452 and 2055 genes were adjacent to SAEs and SIAEs, respectively. Increased enhancer activity was associated with the upregulation of gene expression during senescence but was independent of the distance between enhancers and their target genes (Figure 2F). Furthermore, the genes near SIAEs were downregulated but CEs had no influence on gene expression (Figure 2G and Supplementary Figure S6A and B). p16 (encoded by Cdkn2a), a protein that regulates the cell cycle, was associated with SAEs and showed increased expression upon entry into senescence (Figure 2H). Lamin B1 (encoded by Lmnb1), a protein involved in maintaining nuclear architecture and lamina-anchored regions, was associated with SIAEs and showed decreased expression upon entry into senescence (Figure 2I).
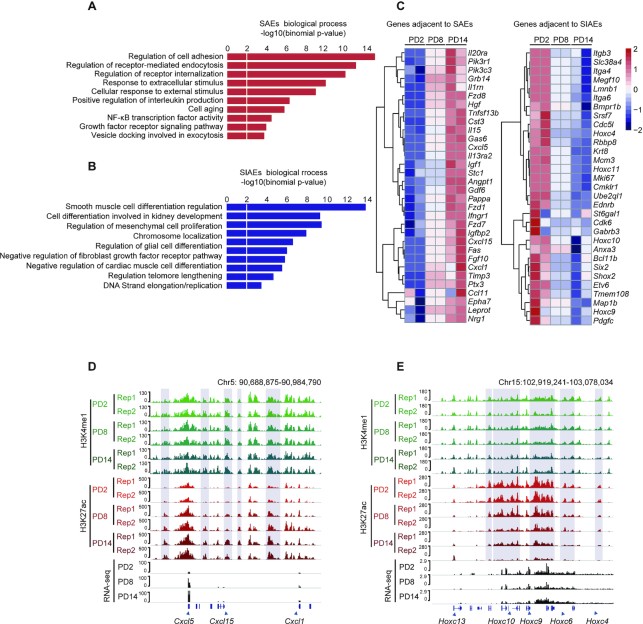
To substantiate the biological functions of genes adjacent to the senescence-associated enhancers, we performed gene ontology (GO) enrichment analysis using the GREAT algorithm. The pathways associated with SAEs included the positive regulation of cytokine secretion and NF-κB and growth factor signaling, which are crucial to the regulation of the SASP (Figure 3A). The SIAE-enriched pathways contributed to the senescence-associated exhaustion of growth capacity and dysregulation of the cell cycle (Figure 3B). In agreement with the GO enrichment analysis, many genes adjacent to SAEs were upregulated during senescence, including the well-described SASP gene family, Cxcl (Figure 3C and D). In contrast, Hox genes, key regulators of embryonic development (55,56), were flanked by SIAEs and showed decreased expression during senescence (Figure 3E). These results are consistent with the knowledge that senescence is a normal developmental mechanism found throughout the embryo, suggesting that enhancer dynamics contribute to, or are a prerequisite for, the stable repression of development-related genes. Taken together, these results demonstrate that numerous characterized SASP genes are adjacent to SAEs and the activation of SAEs may underlie the regulation of SASP expression upon entry into senescence.
Figure 3.
Senescence-activated enhancers (SAEs) affect adjacent SASP gene expression profiles. (A, B) Gene Ontology enrichments of SAEs (A) and SIAEs (B) by GREAT. The length of each bar represents the enriched P value for each biological process. (C) Heatmaps displaying normalized gene expression among PD2, PD8, and PD14. SAE- (left panel) and SIAE- (right panel) adjacent genes are listed. (D, E) H3K27ac ChIP-Seq occupancy profiles at representative SAE and SIAE loci for the Cxcl and Hoxc families. The genes in these two families that are adjacent to SAEs and SIAEs are marked by blue boxes.
C/EBPα is recruited to SAEs to accelerate senescence
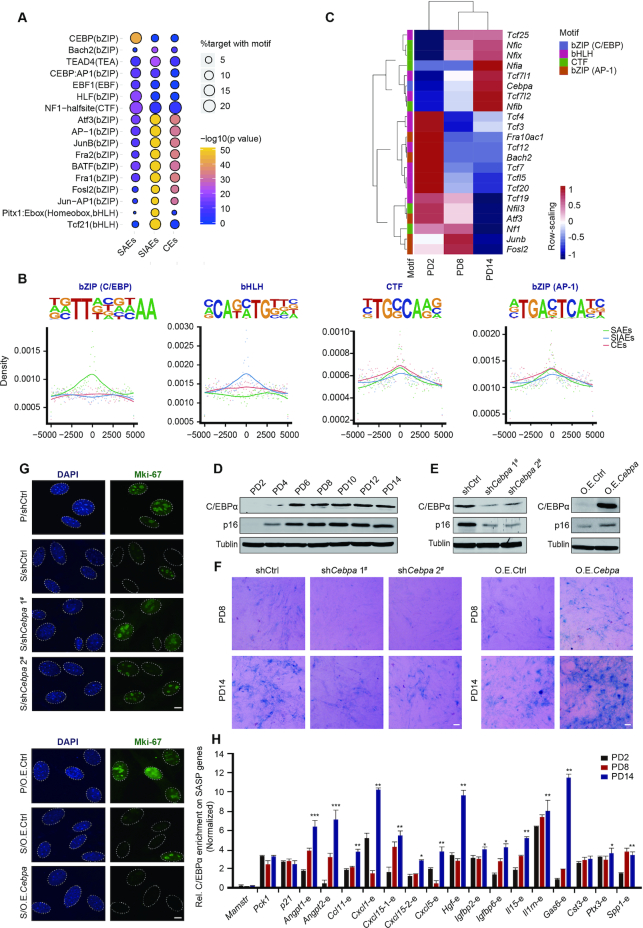
To investigate the transcriptional factors that regulate SAEs and influence the expression of downstream SASP genes, we performed motif enrichment analysis with a view to identifying putative transcription factors (TFs) that bind to SAEs. Several distinct motifs were significantly enriched in the three categories of enhancers (Figure 4A). These top-ranked motifs closely resembled consensus binding sites for basic leucine zipper domain (bZIP) factors, basic helix-loop-helix (bHLH) factors, and CCAAT box-binding transcription factors (CTF). Considering the bZIP family motif as two subclasses, bZIP in C/EBP (CCAAT/enhancer binding protein) and bZIP in AP-1 (Activator protein 1), the C/EBP motif was strongly enriched in SAEs, whereas the bHLH family motif was enriched in SIAEs (Figure 4B). The CTF and bZIP (AP-1) family motifs displayed no enrichment preference for SAEs or SIAEs.
Figure 4.
Recruitment of C/EBPα leads to premature cellular senescence. (A) Top-ranked enriched motif families among SAEs, SIAEs, and CEs (regions defined by Figure 2A−C) determined via Homer2 algorithms are listed. (B) Position-weight matrix showing the observed enhancer densities in four motif families containing C/EBP, AP-1, bHLH, and CTF sites. (C) Heatmaps showing the expression of filter genes from four motif families. The colorful modules are consistent with the classification in (B). (D) Western blotting analysis of total C/EBPα and p16 protein expression in serially passaged MEFs. Tubulin served as a loading control. (E) Western blotting showing the expression of C/EBPα and p16 after down- or upregulation of C/EBPα expression via lentiviral particles. Tubulin served as a loading control. (F) SA-β-gal staining in PD8 and PD14 after down- or upregulation of C/EBPα expression via lentiviral particles. Scale bar, 50 μm. (G) Immunofluorescence staining of Mki67 after down- or upregulation of C/EBPα expression. DAPI staining shows the position of the nucleolus. P represents proliferating cells, and S represents senescent cells. Scale bar, 10 μm. (H) The relative levels of C/EBPα and SASP genes flanking SAEs in MEFs of different passage numbers were measured by ChIP-qPCR. Specifically, C/EBPα enrichment for Mamstr served as a negative control, and C/EBPα enrichment for Pck1 and p21 served as positive controls, as previously described (68,69). The enrichment of C/EBPα was normalized to 10% input. Error bars represent the S.D. obtained from three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001. One-way ANOVA with Dunnett's multiple comparison test was performed.
Given the enrichment of the C/EBP motif in SAEs, we subsequently filtered TFs showing trending changes in expression in serially passaged MEFs to identify putative TFs that regulate these enhancer regions. Notably, only Cebpa, which encodes the C/EBPα protein, exhibited prominent upregulation in senescent cells at both the mRNA and protein levels (Figure 4C and D and Supplementary Figure S7A). C/EBPα is a tumor suppressor that constrains proliferation by inhibiting the transcriptional activity of E2F complexes (57) and its aberrant activity is involved in various cancers (58). Considering the role of C/EBPα in maintaining the nuclear architecture by ‘assisted regions’ that exhibit epigenetic features in breast cancer cells (59), we asked the question whether C/EBPα fulfils previously unknown functions as a modulator of senescence regulation. The expression of p16 is increased in replicative senescent cells and is considered a biomarker of senescence. Notably, shRNA-mediated knockdown of C/EBPα expression led to a reduced abundance of the p16 protein, while overexpression of C/EBPα restored p16 expression (Figure 4E). In addition, attenuated SA-β-gal staining was found in senescent C/EBPα-knockdown cells, while SA-β-gal staining in C/EBPα-overexpressing cells was increased (Figure 4F and Supplementary Figure S7B). Immunofluorescence staining of the proliferation marker, Mki67, was measured to assess the role of C/EBPα during the cell cycle. A decrease in Mki67 fluorescence intensity was seen following inhibition of C/EBPα expression in senescent cells, indicating reduced proliferation of late-stage MEFs (Figure 4G and Supplementary Figure S7C). Together, our results demonstrate that C/EBPα positively regulates senescence.
Considering the potential regulatory role of C/EBPα in senescence, we speculated that C/EBPα may be recruited to SAEs to regulate downstream SASP genes. Accordingly, we performed ChIP-qPCR analysis of C/EBPα on selected activated SASP genes adjacent to SAEs in cells of different passage numbers. Indeed, we revealed that C/EBPα could bind to SAEs, displaying a progressive increase in signal strength from PD2 to PD14 (Figure 4H). These results suggest that C/EBPα is recruited to a subset of SAEs flanking SASP genes during senescence, promoting the emergence of a senescence phenotype.
C/EBPα promotes the expression of SASP genes adjacent to SAEs
After confirming C/EBPα enrichment at SAEs during senescence, we next sought to determine the impact of this factor on the transcriptional output of senescent cells. Importantly, a myriad of SASP genes showed upregulated expression upon entry into senescence (Supplementary Figure S8A). Moreover, knockdown of C/EBPα in senescent cells caused a robust depression of SASP genes (Supplementary Figure S8B). Given the potential regulatory effect of C/EBPα on SASP genes, we examined whether C/EBPα affects the expression of SASP genes by regulating enhancer activity. Focusing on the elevated SAE peaks described above, we found overall reduced SAE activity following knockdown of C/EBPα expression (Figure 5A). In addition, using the ChIP-qPCR approach, we confirmed that knockdown of C/EBPα led to reduced activity of SAEs flanking SASP genes such as Cxcl1, Cxcl5, Cxcl15, Igfbp2 and Igfbp6 (Figure 5B). The H3K27ac signals for representative Cxcl genes are shown in Figure 5C.
Figure 5.
Activation of C/EBPα accelerates the secretion of SASP factors by modulating the flanking SAEs. (A) The enrichment of H3K27ac ChIP-Seq signals at SAEs during senescence (left) and after downregulation of C/EBPα expression (right). C/EBPα knockdown was performed in proliferating MEFs that were harvested at PD8. (B) ChIP-qPCR analysis of H3K27ac measuring the SAE activity adjacent to SASP genes after downregulation of C/EBPα expression. The enrichment of H3K27ac was normalized to 10% input, and IgG was used as a negative control. (C) IGV showing the H3K27ac ChIP-seq signals after inactivation of the C/EBPα and C/EBPα ChIP-seq signals at the Cxcl loci. The Cxcl genes adjacent to SAEs are marked by blue boxes. (D, E) Cytokine array analysis of secreted proteins and relative quantitation of SASP factors in proliferating (P), senescent (S), and C/EBPα-knockdown cells (D) or in control and C/EBPα-overexpressing cells (E). Error bars represent the S.D. obtained from three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001. Statistical analysis was performed using one-way ANOVA (D) and a Student's t-test (E).
Since the SASP comprises several soluble and insoluble factors that alter the cellular microenvironment, we investigated the influence of C/EBPα on the extracellular secretion of SASP factors. We examined a subset of the senescent cell secretome using commercially available cytokine arrays and observed dominant accumulation of CXCL1 (GROα), CXCL5, IGFBP2, IGFP6, GAS6 and CCL11 in the media of senescent cells as compared with those in the media of proliferating cells (Figure 5D). Importantly, SASP genes encoding these factors were modulated by C/EBPα-induced SAE activity. Cells cultured in senescent media following C/EBPα knockdown showed a prominent reduction in cytokine expression, while those cultured in proliferating media following C/EBPα overexpression showed elevated cytokine expression, demonstrating that C/EBPα controls the SASP secretome in senescent cells (Figure 5D, E). In summary, these results support the notion that C/EBPα promotes enhancer activity by being recruited to SAEs, which facilitates SASP gene expression and factor secretion.
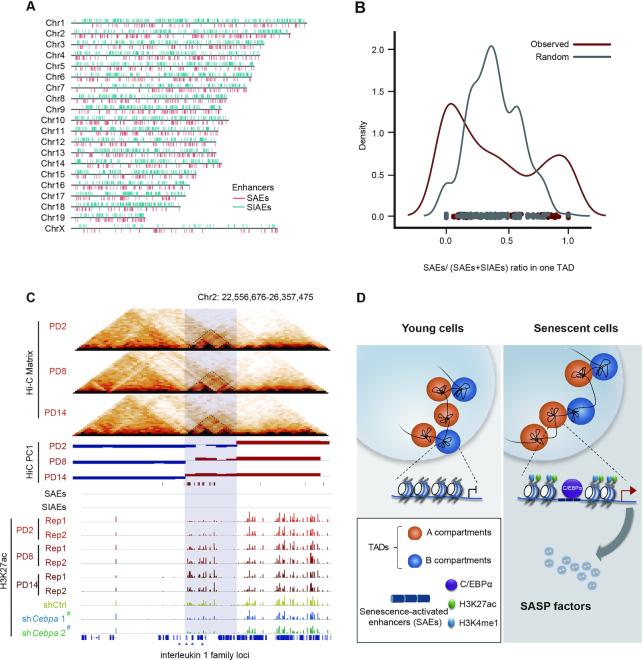
The enhancer landscape is reprogrammed at the TAD scale during senescence
Previous studies have shown that TAD is the basic unit of chromatin structure and contains boundaries that insulate communication between TADs (20–22). We questioned whether TAD is also a regulatory unit that mediates enhancer activity during senescence, resulting in the preferred location of certain SAEs/SIAEs in one TAD. We examined the genome-wide locations of SAEs and SIAEs and found that the same classes of enhancers were likely to group together (Figure 6A). We speculated that these enhancers grouped together at the TAD scale; therefore, we calculated the ratio of SAEs in each TAD. The bimodal distribution of SAE proportions in comparison with random genomic regions implies the preferred clustering of SAEs/SIAEs in one TAD (Figure 6B). The dynamic change in enhancers is restricted within a TAD unit, demonstrating broad enhancer landscape reprogramming during senescence. This feature indicates that C/EBPα regulates groups of enhancers at the TAD scale. As an example, the IL1 family of SASP proteins was located in one TAD that switched from the B to A compartment and was associated with elevated H3K27ac-decorated enhancer levels during senescence. Accordingly, the H3K27ac signals related to this TAD were greatly reduced following knockdown of C/EBPα, but the enhancer signals in nearby regions were not affected (Figure 6C). Other examples are provided in Supplementary Figure S9. Such SAE clustering in TAD units may help to ensure that C/EBPα affects specific downstream SASP genes. In summary, these results highlight that the enhancer landscape is reorganized at the TAD scale during senescence, in which C/EBPα binds to enhancer elements to promote SASP.
Figure 6.
Enhancer repertoires orchestrate gene expression in TAD units. (A) The distribution of SAE and SIAE clusters along each chromosome; each red bar represents an SAE location in the reference genome, and the blue bars represent SIAE locations. (B) The SAE density calculating the ratio of SAE to total enhancer (SAE + SIAE) in one TAD unit as compared with random sequences during senescence. (C) Visualization of the TAD structure, A/B compartments, and H3K27ac signals at interleukin 1 family loci. TADs are marked by dashed triangles. A compartments are colored red, and B compartments are colored blue. H3K27ac signals are shown as red bars during senescence and as blue bars following downregulation of C/EBPα expression. (D) Summary schematic depicting the chromatin architecture, epigenetic enhancer repertoires, and SASP gene kinetics mediated by C/EBPα during cellular senescence. The imaginary line under the compartments denotes the detailed enlargement at the transcriptional level.
DISCUSSION
Senescent cells dramatically increase the secretion of proinflammatory cytokines, chemokines, growth factors and proteases. These widespread changes can be subject to the epigenetic control of the 3D architecture of the genome. The Hi-C approach has been applied to explore changes in the 3D architecture of the genome in several senescence models (39,60), and a potential link between the 3D genome architecture and transcriptional senescent network control has been indicated. Considering the limited knowledge of the SASP to date, it is imperative to comprehensively elucidate the chromatin structure-dependent mechanism of SASP gene regulation. In the present study, we identified changes in the 3D genome structure at both the inter- and intrachromosomal levels during cellular senescence, in addition to the frequent transition of compartments. We also observed TAD-related redistribution of active enhancers associated with chromosomal compartment switching and defined SAE dynamics. Moreover, the transcription factor C/EBPα can be recruited to activate SAEs, which upregulate the expression of flanking SASP genes and promote the secretion of SASP factors (Figure 6D). Our study expands the role of C/EBPα in senescence and links chromatin reorganization to the SASP gene expression network.
Even though certain components of the SASP are conserved across various senescent cells, many SASP factors are expressed in a cell-context and tissue-specific manner in different senescence scenarios (61). For example, several well-described SASP factors, such as IL6, IL7 and GM-CSF, presumably help to establish persistent growth arrest in the context of oncogene-induced senescence (OIS) (10); however, IL6, IL1A and IL1B are not expressed in MEFs during senescence (61). Indeed, we reveal the role of C/EBPα in the control of SASP expression during MEFs senescence, while C/EBPα has been shown to either not be expressed or its expression fluctuates in IMR90 and WI38 cells of different passage number (58,62). Given that SASP factors vary in distinct cell types and in the presence of different senescence-inducing stimuli, the seemingly paradoxical alterations in C/EBPα abundance among models of senescence may be due to cell type specificity and context dependence. Consistent with this notion, the expression of C/EBPα was found to be increased in other rodent senescent cells, rat embryonic fibroblasts (REFs), which exhibit senescent phenotypic alterations in C/EBPα and SASP profiles similar to those of MEFs (Supplementary Figure S10A−C). In addition, knockdown of C/EBPα expression inhibited the expression of SASP genes flanking SAEs, while overexpression of C/EBPα led to the opposite phenotype (Supplementary Figure S10D and E). Thus, the activation of SAEs by C/EBPα stimulates SASP expression in MEFs and REFs, but not in IMR90, WI38, or oncogene-induced senescent cells, indicating that cell type and tissue context determine the specific response to C/EBPα. Hence, we propose that C/EBPα-mediated SASP activity at the TAD-scale contributes to senescence in a cellular-context and tissue-specific manner.
It is noteworthy that in addition to C/EBPα, another member of the C/EBP family, C/EBPβ, has also been demonstrated to be critical for the induction of certain SASP genes by binding to their promoters and activating gene expression during oncogene-induced senescence (11,12,63). However, unlike C/EBPα, we found that C/EBPβ exhibited limited regulation of the C/EBPα-targeted SAEs at both the transcriptional and secretory levels during MEFs senescence, and knockdown of C/EBPα or C/EBPβ did not affect the protein expression level of the other (Supplementary Figure S11A−E). Consistently, we observed profound changes in the SASP transcriptional repertoire including the Il1 and Cxcl clusters, but the well-described C/EBPβ-targeted SASP such as IL6, IL1A and IL1B were not expressed in the MEF senescence model. These results suggest that the divergent SASP programs are modulated in a C/EBP member-dependent manner in different senescence scenarios, and various transcription factors orchestrate the enhancer dynamics to regulate SASP expression profiles (64).
Moreover, several studies have revealed the cooperation between NF-κB signaling and C/EBPβ in the regulation of inflammatory factors; C/EBPβ can reduce NF-κB expression, thus increasing the levels of downstream secretory components (65). Here, during MEFs senescence, the NF-κb pathway displayed a higher P value as compared with that of the top-ranked transcription factor families with respect to SAE motif enrichment (Supplementary Figure S12A−C), indicating no binding of the NF-κb motif to SAE sequences. Furthermore, neither knockdown nor overexpression of C/EBPα had an impact on NF-κB levels (Supplementary Figure S12D), implying that NF-κB is not a target of C/EBPα. However, although we failed to find cooperation between NF-κB and C/EBPα, we cannot exclude the possibility that C/EBPα-activated SAEs may induce the binding of NF-κB to promoters of some SASP genes, resulting in their expression (66,67). Therefore, considering the well-known effects of NF-κB and C/EBPβ on SASP expression, the possibility that the NF-κB-C/EBPβ axis interactively or alternatively regulates the C/EBPα-mediated SASP mechanism needs to be further explored.
Cis-regulatory elements display cell type-specific patterns that are critical to transcriptional regulation. The epigenetic dynamics of chromatin remodeling often anticipate changes in chromosome compartmentalization and mediate switching between the A and B compartments. Our study, performed using multiple high-throughput sequencing technologies, illuminates the dynamic relationship among the chromatin architecture, enhancer repertoires, and gene expression. The 3D chromatin reorganization in senescent cells is associated with the widespread redistribution of active enhancers. Characterizing enhancer dynamics enables us to understand the chromatin-remodeling function of the tumor suppressor C/EBPα, which contributes to the SASP during cellular senescence. Elucidation of the regulatory mechanisms of SASP factors is essential for understanding how the physiology and behavior of senescence is regulated, and in the longer term, for developing therapeutic interventions.
DATA AVAILABILITY
All related sequencing data have been uploaded to NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE117210. Other data supporting the findings of the present study are available from the corresponding author upon reasonable request.
Supplementary Material
ACKNOWLEDGEMENTS
We are thankful to Dr Wei Wu for providing the lentivirus particle TRC 2.0 and shRNA constructs. We thank members of Dr Cheng Li's lab for expert technical assistance with the Hi-C experiments and suggestions regarding data analysis. We would also like to thank Dr Hendrik G. Stunnenberg for his helpful comments.
Contributor Information
Yiting Guan, Key Laboratory of Cell Proliferation and Differentiation, School of Life Sciences, Peking University, Beijing 100871, China; Institute of Clinical Medicine, Zhanjiang Central Hospital, Guangdong Medical University, Zhanjiang 524000, China.
Chao Zhang, Key Laboratory of Cell Proliferation and Differentiation, School of Life Sciences, Peking University, Beijing 100871, China; Center for Bioinformatics, School of Life Sciences and Center for Statistical Science, Peking University, Beijing 100871, China.
Guoliang Lyu, Key Laboratory of Cell Proliferation and Differentiation, School of Life Sciences, Peking University, Beijing 100871, China.
Xiaoke Huang, Key Laboratory of Cell Proliferation and Differentiation, School of Life Sciences, Peking University, Beijing 100871, China.
Xuebin Zhang, Key Laboratory of Cell Proliferation and Differentiation, School of Life Sciences, Peking University, Beijing 100871, China.
Tenghan Zhuang, Key Laboratory of Cell Proliferation and Differentiation, School of Life Sciences, Peking University, Beijing 100871, China.
Lumeng Jia, Center for Bioinformatics, School of Life Sciences and Center for Statistical Science, Peking University, Beijing 100871, China.
Lijun Zhang, Key Laboratory of Cell Proliferation and Differentiation, School of Life Sciences, Peking University, Beijing 100871, China.
Chen Zhang, China Department of Neurobiology, School of Basic Medical Sciences, Beijing Key Laboratory of Neural Regeneration and Repair, Advanced Innovation Center for Human Brain Protection, Capital Medical University, Beijing 100069, China.
Cheng Li, Center for Bioinformatics, School of Life Sciences and Center for Statistical Science, Peking University, Beijing 100871, China.
Wei Tao, Key Laboratory of Cell Proliferation and Differentiation, School of Life Sciences, Peking University, Beijing 100871, China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Key Research and Development Project [2020YFC2002900, 2017YFA0105201, 2016YFA0100103]; National Natural Science Foundation of China (NSFC) [31871312, 31671426, 31471205, 31670842, 71532001, 31871266]. Funding for open access charge: National Key Research and Development Project [2020YFC2002900, 2017YFA0105201, 2016YFA0100103]; National Natural Science Foundation of China (NSFC) [31871312, 31671426, 31471205, 31670842, 71532001, 31871266].
Conflict of interest statement. None declared.
REFERENCES
- 1. Collado M., Blasco M.A., Serrano M.. Cellular senescence in cancer and aging. Cell. 2007; 130:223–233. [DOI] [PubMed] [Google Scholar]
- 2. Bolden J.E., Lowe S.W.. Cellular senescence. Mol. Basis Cancer. 2015; 13:229–238. [Google Scholar]
- 3. Dimri G.P., Lee X., Basile G., Acosta M., Scott G., Roskelley C., Medrano E.E., Linskens M.H.K., Rubelj I., Pereirasmith O.. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. PNAS. 1995; 92:9363–9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Alcorta D.A., Xiong Y., Phelps D., Hannon G., Beach D., Barrett J.C.. Involvement of the cyclin-dependent kinase inhibitor p16 (INK4a) in replicative senescence of normal human fibroblasts. PNAS. 1996; 93:13742–13747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Halazonetis T.D., Gorgoulis V.G., Bartek J.. An oncogene-induced DNA damage model for cancer development. Science. 2008; 319:1352–1355. [DOI] [PubMed] [Google Scholar]
- 6. Serrano M., Lin A.W., Mccurrach M.E., Beach D., Lowe S.W.. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 1997; 88:593–602. [DOI] [PubMed] [Google Scholar]
- 7. Muñozespín D., Cañamero M., Maraver A., Gómezlópez G., Contreras J., Murillocuesta S., Rodríguezbaeza A., Varelanieto I., Ruberte J., Collado M.. Programmed cell senescence during mammalian embryonic development. Cell. 2013; 155:1104–1118. [DOI] [PubMed] [Google Scholar]
- 8. Demaria M., Ohtani N., Youssef S., Rodier F., Toussaint W., Mitchell J.R., Laberge R.-M., Vijg J., Van Steeg H., Dollé M.E.T. et al.. An essential role for senescent cells in optimal wound healing through secretion of PDGF-AA. Dev. Cell. 2014; 31:722–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Krizhanovsky V., Yon M., Dickins R.A., Hearn S., Simon J., Miething C., Yee H., Zender L., Lowe S.W.. Senescence of activated stellate cells limits liver fibrosis. Cell. 2008; 134:657–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Coppe J., Patil C.K., Rodier F., Sun Y., Munoz D.P., Goldstein J., Nelson P.S., Desprez P., Campisi J.. Senescence-Associated secretory phenotypes reveal Cell-Nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. PLoS Biol. 2008; 6:2853–2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Acosta J.C., O’Loghlen A., Banito A., Guijarro M.V., Augert A., Raguz S., Fumagalli M., Costa M.D., Brown C., Popov N.. Chemokine signaling via the CXCR2 receptor reinforces senescence. Cell. 2008; 133:1006–1018. [DOI] [PubMed] [Google Scholar]
- 12. Kuilman T., Michaloglou C., Vredeveld L.C.W., Douma S., Doorn R.V., Desmet C.J., Aarden L.A., Mooi W.J., Peeper D.S.. Oncogene-Induced senescence relayed by an Interleukin-Dependent inflammatory network. Cell. 2008; 133:1019–1031. [DOI] [PubMed] [Google Scholar]
- 13. Chien Y., Scuoppo C., Wang X., Fang X., Balgley B., Bolden J.E., Premsrirut P., Luo W., Chicas A., Cheng S.L.. Control of the senescence-associated secretory phenotype by NF-κB promotes senescence and enhances chemosensitivity. Genes Dev. 2011; 25:2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tchkonia T., Zhu Y., Van Deursen J.M., Campisi J., Kirkland J.L.. Cellular senescence and the senescent secretory phenotype: therapeutic opportunities. J. Clin. Invest. 2013; 123:966–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rodier F., Campisi J.. Four faces of cellular senescence. J. Cell Biol. 2011; 192:547–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Narita M., Nunez S., Heard E., Narita M., Lin A.W., Hearn S., Spector D.L., Hannon G.J., Lowe S.W.. Rb-Mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell. 2003; 113:703–716. [DOI] [PubMed] [Google Scholar]
- 17. Narita M., Narita M., Krizhanovsky V., Nunez S., Chicas A., Hearn S., Myers M.P., Lowe S.W.. A novel role for High-Mobility group A proteins in cellular senescence and heterochromatin formation. Cell. 2006; 126:503–514. [DOI] [PubMed] [Google Scholar]
- 18. Sadaie M., Salama R., Carroll T., Tomimatsu K., Chandra T., Young A.R.J., Narita M., Perezmancera P.A., Bennett D.C., Chong H.. Redistribution of the Lamin B1 genomic binding profile affects rearrangement of heterochromatic domains and SAHF formation during senescence. Genes Dev. 2013; 27:1800–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Shah P.P., Donahue G., Otte G.L., Capell B.C., Nelson D.M., Cao K., Aggarwala V., Cruickshanks H.A., Rai T.S., Mcbryan T.. Lamin B1 depletion in senescent cells triggers large-scale changes in gene expression and the chromatin landscape. Genes Dev. 2013; 27:1787–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dixon J.R., Selvaraj S., Yue F., Kim A., Li Y., Shen Y., Hu M., Liu J.S., Ren B.. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012; 485:376–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nora E.P., Lajoie B.R., Schulz E.G., Giorgetti L., Okamoto I., Servant N., Piolot T., van Berkum N.L., Meisig J., Sedat J.. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012; 485:381–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sexton T., Yaffe E., Kenigsberg E., Bantignies F., Leblanc B., Hoichman M., Parrinello H., Tanay A., Cavalli G.. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell. 2012; 148:458–472. [DOI] [PubMed] [Google Scholar]
- 23. Lupianez D.G., Kraft K., Heinrich V., Krawitz P., Brancati F., Klopocki E., Horn D., Kayserili H., Opitz J.M., Laxova R.. Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell. 2015; 161:1012–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Symmons O., Uslu V.V., Tsujimura T., Ruf S., Nassari S., Schwarzer W., Ettwiller L., Spitz F.. Functional and topological characteristics of mammalian regulatory domains. Genome Res. 2014; 24:390–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Andrey G., Montavon T., Mascrez B., Gonzalez F., Noordermeer D., Leleu M., Trono D., Spitz F., Duboule D.. A switch between topological domains underlies HoxD genes collinearity in mouse limbs. Science. 2013; 340:1234167–1234167. [DOI] [PubMed] [Google Scholar]
- 26. Deng W., Lee J., Wang H., Miller J.C., Reik A., Gregory P.D., Dean A., Blobel G.A.. Controlling Long-Range genomic interactions at a native locus by targeted tethering of a looping factor. Cell. 2012; 149:1233–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. De Laat W., Duboule D.. Topology of mammalian developmental enhancers and their regulatory landscapes. Nature. 2013; 502:499–506. [DOI] [PubMed] [Google Scholar]
- 28. Dixon J.R. A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature. 2013; 503:290–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Soucie E., Weng Z., Geirsdottir L., Molawi K., Maurizio J., Fenouil R., Mossadeghkeller N., Gimenez G., Vanhille L., Beniazza M.. Lineage-specific enhancers activate self-renewal genes in macrophages and embryonic stem cells. Science. 2016; 351:280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ingsimmons E., Seitan V.C., Faure A.J., Flicek P., Carroll T.J., Dekker J., Fisher A.G., Lenhard B., Merkenschlager M.. Spatial enhancer clustering and regulation of enhancer-proximal genes by cohesin. Genome Res. 2015; 25:504–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Criscione S.W., Teo Y.V., Neretti N.. The Chromatin landscape of cellular senescence. Trends Genet. 2016; 32:751–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Shin D., Kucia M., Ratajczak M.Z.. Nuclear and chromatin reorganization during cell senescence and aging – a mini-review. Gerontology. 2011; 57:76–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Iwasaki O., Tanizawa H., Kim K., Kossenkov A.V., Nacarelli T., Tashiro S., Majumdar S., Showe L.C., Zhang R., Noma K.. Involvement of condensin in cellular senescence through gene regulation and compartmental reorganization. Nat. Commun. 2019; 10:5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Munozespin D., Serrano M.. Cellular senescence: from physiology to pathology. Nat. Rev. Mol. Cell Biol. 2014; 15:482–496. [DOI] [PubMed] [Google Scholar]
- 35. Rao S.S.P., Huntley M.H., Durand N.C., Stamenova E., Bochkov I.D., Robinson J.T., Sanborn A.L., Machol I., Omer A.D., Lander E.S.. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014; 159:1665–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lee J., Mo Y., Gan H., Burgess R.J., Baker D.J., Van Deursen J.M., Zhang Z.. Pak2 kinase promotes cellular senescence and organismal aging. PNAS. 2019; 116:13311–13319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Casella G., Munk R., Kim K.M., Piao Y., De S., Abdelmohsen K., Gorospe M.. Transcriptome signature of cellular senescence. Nucleic Acids Res. 2019; 47:11476–11476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Crane E., Bian Q., Mccord R.P., Lajoie B.R., Wheeler B.S., Ralston E.J., Uzawa S., Dekker J., Meyer B.J.. Condensin-driven remodelling of X chromosome topology during dosage compensation. Nature. 2015; 523:240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chandra T., Ewels P.A., Schoenfelder S., Furlan-Magaril M., Wingett S.W., Kirschner K., Thuret J.Y., Andrews S., Fraser P., Reik W.. Global reorganization of the nuclear landscape in senescent cells. Cell Rep. 2015; 10:471–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sati S., Bonev B., Szabo Q., Jost D., Bensadoun P., Serra F., Loubiere V., Papadopoulos G.L., Rivera-Mulia J.C., Fritsch L. et al.. 4D genome rewiring during Oncogene-Induced and replicative senescence. Mol. Cell. 2020; 78:522–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. McLean C.Y., Bristor D., Hiller M., Clarke S.L., Schaar B.T., Lowe C.B., Wenger A.M., Bejerano G.. GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 2010; 28:495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lenain C., De C.G., Pagie L., Visser N.L., De M.H., De S.V., Perichupkes D., Van B.S., Peeper D.S.. Massive reshaping of genome-nuclear lamina interactions during oncogene-induced senescence. Genome Res. 2017; 27:1634–1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Meuleman W., Perichupkes D., Kind J., Beaudry J., Pagie L., Kellis M., Reinders M.J.T., Wessels L.F.A., Van Steensel B.. Constitutive nuclear lamina–genome interactions are highly conserved and associated with A/T-rich sequence. Genome Res. 2013; 23:270–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lenain C., De Graaf C.A., Pagie L., Visser N.L., De Haas M., De Vries S.S., Perichupkes D., Van Steensel B., Peeper D.S.. Massive reshaping of genome–nuclear lamina interactions during oncogene-induced senescence. Genome Res. 2017; 27:1634–1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ernst J., Kellis M.. ChromHMM: automating chromatin-state discovery and characterization. Nat. Methods. 2012; 9:215–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chepelev I., Wei G., Wangsa D., Tang Q., Zhao K.. Characterization of genome-wide enhancer-promoter interactions reveals co-expression of interacting genes and modes of higher order chromatin organization. Cell Res. 2012; 22:490–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sunkel B., Wu D., Chen Z., Wang C.M., Liu X., Ye Z., Horning A.M., Liu J., Mahalingam D., Lopeznicora H.. Integrative analysis identifies targetable CREB1/FoxA1 transcriptional co-regulation as a predictor of prostate cancer recurrence. Nucleic Acids Res. 2016; 44:4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W.. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008; 9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Adam R.C., Yang H., Rockowitz S., Larsen S.B., Nikolova M., Oristian D., Polak L., Kadaja M., Asare A., Zheng D.. Pioneer factors govern super-enhancer dynamics in stem cell plasticity and lineage choice. Nature. 2015; 521:366–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mosteiro L., Pantoja C., de Martino A., Serrano M.. Senescence promotes in vivo reprogramming through p16INK4a and IL-6. Aging Cell. 2018; 17:e12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wang J., Chen H., Liu Y., Zhou W., Sun R., Xia M.. Retinol binding protein 4 induces mitochondrial dysfunction and vascular oxidative damage. Atherosclerosis. 2015; 240:335–344. [DOI] [PubMed] [Google Scholar]
- 52. Creyghton M.P., Cheng A.W., Welstead G.G., Kooistra T.G., Carey B.W., Steine E.J., Hanna J., Lodato M.A., Frampton G.M., Sharp P.A.. Histone H3K27ac separates active from poised enhancers and predicts developmental state. PNAS. 2010; 107:21931–21936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Malik A.N., Vierbuchen T., Hemberg M., Rubin A.A., Ling E., Couch C.H., Stroud H., Spiegel I., Farh K.K., Harmin D.A.. Genome-wide identification and characterization of functional neuronal activity-dependent enhancers. Nat. Neurosci. 2014; 17:1330–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kim T.K., Shiekhattar R.. Architectural and functional commonalities between enhancers and promoters. Cell. 2015; 162:948–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Schworer S., Becker F., Feller C., Baig A.H., Kober U., Henze H., Kraus J.M., Xin B., Lechel A., Lipka D.B.. Epigenetic stress responses induce muscle stem-cell ageing by Hoxa9 developmental signals. Nature. 2016; 540:428–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lawrence H.J., Sauvageau G., Humphries R.K., Largman C.. The role of HOX homeobox genes in normal and leukemic hematopoiesis. Stem Cells. 1996; 14:281–291. [DOI] [PubMed] [Google Scholar]
- 57. Bo T.P., Pedersen T.Å., Xu X., Bo L., Wewer U.M., Friishansen L., Nerlov C.. E2F repression by C/EBPα is required for adipogenesis and granulopoiesis in vivo. Cell. 2001; 107:247–258. [DOI] [PubMed] [Google Scholar]
- 58. Lourenço A.R., Coffer P.J.. A tumor suppressor role for C/EBPα in solid tumors: more than fat and blood. Oncogene. 2017; 36:5221–5230. [DOI] [PubMed] [Google Scholar]
- 59. Nacht A.S., Ferrari R., Zaurin R., Scabia V., Carbonell-Caballero J., Le Dily F., Quilez J., Leopoldi A., Brisken C., Beato M. et al.. C/EBPα mediates the growth inhibitory effect of progestins on breast cancer cells. EMBO J. 2019; 38:e101426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zirkel A., Nikolic M., Sofiadis K., Mallm J.P., Brackley C.A., Gothe H., Drechsel O., Becker C., Altmüller J., Josipovic N.. HMGB2 loss upon senescence entry disrupts genomic organization and induces CTCF clustering across cell types. Mol. Cell. 2018; 70:730. [DOI] [PubMed] [Google Scholar]
- 61. Coppé J.-P., Patil C.K., Rodier F., Krtolica A., Beauséjour C.M., Parrinello S., Hodgson J.G., Chin K., Desprez P.-Y., Campisi J.. A human-like senescence-associated secretory phenotype is conserved in mouse cells dependent on physiological oxygen. PLoS One. 2010; 5:e9188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Marthandan S., Priebe S., Baumgart M., Groth M., Cellerino A., Guthke R., Hemmerich P., Diekmann S.. Similarities in gene expression profiles during in vitro aging of primary human embryonic lung and foreskin fibroblasts. Biomed. Res. Int. 2015; 2015:731938–731938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Huggins C.J., Malik R., Lee S., Salotti J., Thomas S., Martin N., Quiñones O.A., Alvord W.G., Olanich M.E., Keller J.R.. C/EBPγ suppresses senescence and inflammatory gene expression by heterodimerizing with C/EBPβ. Mol. Cell. Biol. 2013; 33:3242–3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Tasdemir N., Banito A., Roe J.S., Alonsocurbelo D., Camiolo M., Tschaharganeh D.F., Huang C.H., Aksoy O., Bolden J.E., Chen C.C.. BRD4 connects enhancer remodeling to senescence immune surveillance. Cancer Discov. 2016; 6:612–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Cappello C., Zwergal A., Kanclerski S., Haas S.C., Kandemir J.D., Huber R., Page S., Brand K.. C/EBPβ enhances NF–κB-associated signalling by reducing the level of IκB-α. Cell. Signal. 2009; 21:1918–1924. [DOI] [PubMed] [Google Scholar]
- 66. Orjalo A.V., Bhaumik D., Gengler B.K., Scott G.K., Campisi J.. Cell surface-bound IL-1α is an upstream regulator of the senescence-associated IL-6/IL-8 cytokine network. PNAS. 2009; 106:17031–17036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Stein B., Yang M.X.. Repression of the interleukin-6 promoter by estrogen receptor is mediated by NF-kappa B and C/EBP beta. Mol. Cell. Biol. 1995; 15:4971–4979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Jakobsen J.S., Waage J., Rapin N., Bisgaard H.C., Larsen F.S., Porse B.T.. Temporal mapping of CEBPA and CEBPB binding during liver regeneration reveals dynamic occupancy and specific regulatory codes for homeostatic and cell cycle gene batteries. Genome Res. 2013; 23:592–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. White P.S., Brestelli J., Kaestner K.H., Greenbaum L.E.. Identification of transcriptional networks during liver regeneration. J. Biol. Chem. 2005; 280:3715–3722. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All related sequencing data have been uploaded to NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE117210. Other data supporting the findings of the present study are available from the corresponding author upon reasonable request.