Abstract
Purpose:
SMARCA4 mutations are among the most common recurrent alterations in NSCLC, but the relationship to other genomic abnormalities and clinical impact has not been established.
Experimental Design:
To characterize SMARCA4 alterations in NSCLC, we analyzed the genomic, protein expression, and clinical outcome data of patients with SMARCA4 alterations treated at Memorial Sloan Kettering.
Results:
In 4813 tumors from patients with NSCLC, we identified 8% (n= 407) patients with SMARCA4-mutant lung cancer. We describe two categories of SMARCA4 mutations: Class 1 mutations (truncating mutations, fusions and homozygous deletion) and Class 2 mutations (missense mutations). Protein expression loss was associated with Class 1 mutation (81% vs 0%, (P < 0.001)). Both classes of mutation co-occured more frequently with KRAS, STK11, and KEAP1 mutations compared to SMARCA4 wildtype tumors (P < 0.001). In patients with metastatic NSCLC, SMARCA4 alterations were associated with shorter overall survival, with Class 1 alterations associated with shortest survival times (P < 0.001). Conversely, we found that treatment with immune checkpoint inhibitors was associated with improved outcomes in patients with SMARCA4-mutant tumors (P = 0.01), with Class 1 mutations having the best response to ICIs (p = 0.027).
Conclusions:
SMARCA4 alterations can be divided into two clinically relevant genomic classes associated with differential protein expression as well as distinct prognostic and treatment implications. Both classes co-occur with KEAP1, STK11, and KRAS mutations, but individually represent independent predictors of poor prognosis. Despite association with poor outcomes, SMARCA4-mutant lung cancers may be more sensitive to immunotherapy.
Introduction
Genomic abnormalities in the subunits of the SWI/SNF chromatin remodeling complex occur in approximately 20% of solid tumors and emerging data suggests that specific alterations within this complex might affect outcomes in certain solid tumors (1-3). For example, alterations in the SWI/SNF complex gene PBRM1 have been associated with improved outcomes in patients with renal cell carcinoma treated with immune checkpoint inhibitors (ICIs); refs. (3,4). In lung cancer, inactivation of the catalytic subunit SMARCA4 (BRG1), is the most common alteration within the SWI/SNF complex and has been associated with poor patient outcomes (1,5-10). SMARCA4 is one of two mutually exclusive DNA-dependent ATPases, along with SMARCA2, involved in transcriptional regulation of gene expression (11,12). Yet, the relationship between SMARCA4 and other alterations within the complex genomic landscape of lung cancer remains unclear.
Multiple studies have recently highlighted the importance of considering genes of interest within the context of commonly co-occurring mutations (13-18). For example, the identification of STK11, KEAP1, and TP53 mutant subgroups has changed the paradigm of classifying KRAS-mutant lung cancers and non-small cell lung cancers (NSCLCs) in general (13-15,18). These distinct subgroups correlate with differential responses to immunotherapy and long-term outcomes (13,14,17,18). Further, in EGFR-mutant lung cancer, mutations in TP53 and RB1 are associated with shorter response to TKIs and transformation to small cell carcinoma (15,16). Previous studies have shown that SMARCA4 alterations can co-occur in KRAS mutant tumors, yet they also occur independently and less commonly with other driver oncogenes such as epidermal growth factor receptor (EGFR); refs. (5,6). However, there are only limited data on SMARCA4’s relationship to these other co-occurring mutations (8,10) and the significance of SMARCA4 alterations among oncogene driven subsets of lung cancer is unknown.
Increased understanding of the relationship of SMARCA4 in lung cancer may enable new therapeutic opportunities in the future. Recently, SMARCA4 alterations have been shown to be oncogenic drivers in a highly aggressive subset of ovarian cancer, small cell carcinoma of the ovary, hypercalcemic type (SCCOHT) that shows increased susceptibility to ICIs (19). Further, there have been case reports of durable responses to ICIs in thoracic SMARCA4-deficient undifferentiated tumors and SMARCA4-deficient lung carcinoma (20,21), but no studies have comprehensively evaluated treatment outcomes in a large cohort of lung cancer patients. In this study, we characterize the clinical, molecular, and histologic relationships of SMARCA4 genomic and protein alterations in lung cancer.
Methods
We identified all patients with NSCLC of any stage with SMARCA4 alterations detected by MSK-IMPACT NGS (22) until April of 2019 who were treated at Memorial Sloan Kettering Cancer Center (MSK) for genomic analysis (Sup. Fig. 1).
SMARCA4 alterations were classified into two groups: SMARCA4 truncating mutations, fusions and homozygous deletions were deemed “Class 1 alteration” and 2) SMARCA4 missense mutations or variants of unknown significance, or “Class 2 alteration” based upon categorization in OncoKB (23). Tumors with concurrent Class 1 and Class 2 alterations were classified within the Class 1 category. A retrospective pathologic analysis of expression of SMARCA4 in all cases of with SMARCA4 molecular alterations was performed by immunohistochemistry using the previously described methods (10).
Somatic alterations were identified using the MSK-IMPACT assay as previously described (22). Individual genes were queried for distribution and enrichment among the patients with and without SMARCA4 alterations. Frequencies of gene alterations by SMARCA4 alteration were considered significant with a p-value < 0.05 and, to reduce false discovery in multiple testing, FDR q-value < 0.10. Tumor mutation burden (TMB) was normalized across each version of the MSK-IMPACT panel (341, 410, or 468 genes) and defined as the total number of mutations divided by the coding region captured reported as mutations/megabase in each panel (0.897 megabases (Mb) for 341-, 1.017 Mb for 410-, and 1.139 Mb for 468-gene panel). PD-L1 expression was scored as the percentage of tumor cells with membranous staining using predominantly E1L3N antibody, as previously described (24).
Medical, pharmacy, and pathology records for all patients with metastatic NSCLC and SMARCA4 alterations were reviewed to collect demographic, pathologic, and treatment data. A random sample of patients with metastatic NSCLC who had MSK-IMPACT without SMARCA4 alterations and were tested during the same time period was used as a comparator group. The response to anti-PD-(L)1 therapy was determined (database lock of April 1, 2019) using Response Evaluation Criteria in Solid Tumors (RECIST) version 1.1. by thoracic radiologists. This study was approved by the Institutional Review Board/Privacy Board at MSK and was in accordance with the Belmont report for retrospective review of records and waiver of consent.
Statistical Methods
Patient and tumor characteristics were compared across SMARCA4 mutation classes (Class 1, Class 2, wild type) using Chi-square tests and Kruskal-Wallis tests. Overall survival (OS) defined from the date of metastatic diagnosis to death and accounted for the left truncation time from metastatic diagnosis to IMPACT biopsy. Patients without events were censored at their last known visit date. Survival curves and estimates of the median survival time were generated using Kaplan-Meier methods and compared across the three mutation classes using log-rank tests. A Cox proportional hazards model was adjusted for age, sex, smoking status (never smoker, former light smoker, former heavy smoker and current smoker), histology (adenocarcinoma, squamous, other), as well as co-occurring STK11 and KEAP1 mutations, and TMB. Hazard ratios (HR) and 95% confidence intervals (CI) are reported. Sub-analyses of OS were performed among patients with KRAS mutations. Patients without follow-up after their IMPACT pathology date were excluded from analyses (n = 5).
The response to immunotherapy as characterized by progression-free survival (PFS), OS, and overall response rate (ORR) was examined among the subset of patients that received immunotherapy. PFS was defined as the time from start of PD-(L)1 inhibitor to clinical or radiographic progression, death, or the end of follow-up, and OS was defined as the time from the start of PD-(L)1 inhibitor to death or the end of follow-up. PFS and OS were analyzed using Kaplan Meier methods and Cox proportional hazards model accounting for left truncation, again adjusted for age, sex, smoking status, histology, TMB, and co-occurring STK11 and KEAP1 mutations. Best overall response was defined as complete or partial response. Multivariable logistic regression was applied to compare the likelihood of ORRR across SMARCA4 mutation classes adjusted for age, TMB, PD-L1, STK11, and KEAP1.
To assess whether immunotherapy is associated with improved survival among patients with Class 1 or 2 SMARCA4 mutations, we first calculated the propensity score, probability of receipt of ICIs based on available variables (mutation class, age, sex, race, smoking status, histology, TMB, co-occurring STK11 and KEAP1 mutations). We then adjusted for the propensity score when comparing OS for patients that received ICIs versus patients that did not via a Cox proportional hazards model accounting for left truncation. A p-value <0.05 was considered statistically significant for all analyses. Statistical analyses were performed with GraphPad Prism software version 7 (La Jolla, CA, www.graphpad.com) and R version 3.6.1 software (www.r-project.org).(25)
Results
Spectrum of SMARCA4 genomic alterations
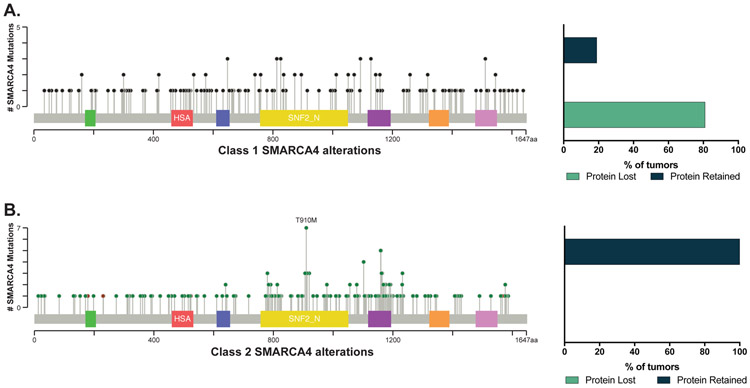
In patients with NSCLC tested by comprehensive next-generation sequencing (NGS), 8% (n = 407 of 4813) had a SMARCA4 alteration, with an array of SMARCA4 alterations identified (Fig. 1). SMARCA4 alterations were categorized into two groups based upon the type of genomic abnormality: 1) “Class 1 alterations” included truncating mutations deemed oncogenic, gene fusions, and homozygous deletions and 2) “Class 2 alterations” included all missense mutations and other variants of unknown significance based upon categorization in OncoKB (23). Tumors with concurrent Class 1 and Class 2 SMARCA4 alterations were categorized as Class 1 tumors. In total, 212 patients (4% of total, 52% of SMARCA4 variants) had tumors with Class 1 SMARCA4 alterations and 195 (4% of total, 48% of SMARCA4 variants) had tumors with Class 2 SMARCA4 alterations (Fig. 1).
Figure 1.
Spectrum of SMARCA4 alterations by class and association with SMARCA4 protein expression. (A) The distribution of Class 1 SMARCA4 alterations (n=212) and protein expression (n =62). (B) The distribution of Class 2 SMARCA4 alterations (n=95) and protein expression (n =24). Green QLQ, Gln, Leu, Gln motif; Red HSA, helicase/SANT-associated domain; Blue BRK, Brahma and Kismet domain; Yellow DEXDc, DEAD-like helicase superfamily domain; Purple SNF2_N, SNF2 family N-terminal domain; Orange HELICc, helicase superfamily C-terminal domain; Pink Bromo, bromodomain.
Relationship between class of SMARCA4 genomic alteration and protein expression
We next explored the relationship between the genomic class of SMARCA4 alteration and protein expression. Sufficient tissue for SMARCA4 immunohistochemical analysis was available for 86 cases, including 62 tumors with Class 1 (truncating) alterations and 24 tumors with Class 2 (missense) alterations. SMARCA4 expression loss was identified in 50 cases, all of which were tumors with Class 1 alterations (81% of Class 1 alterations). Overall, loss of SMARCA4 expression was significantly associated with Class 1 alterations (P < 0.001; Fig. 1).
Molecular landscape associated with SMARCA4 alterations
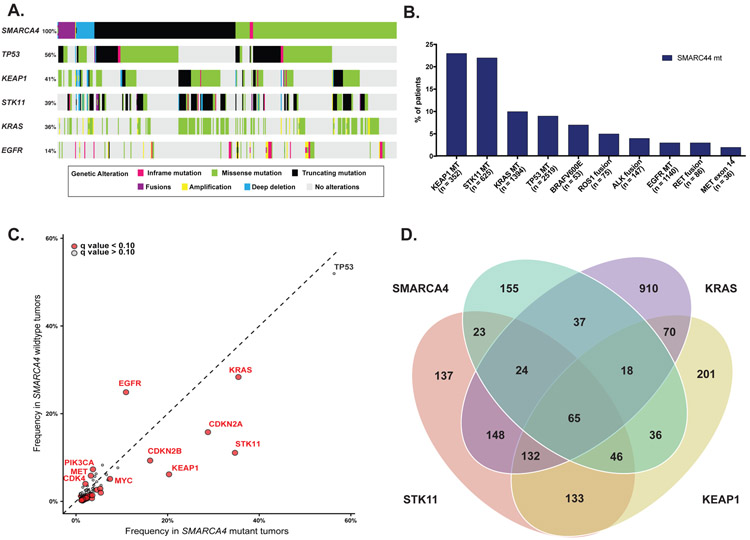
To evaluate the genomic context of SMARCA4 alterations, we evaluated genomic profiles of tumors harboring SMARCA4 alterations (n=407) and those without SMARCA4 alterations (n=4406). Among commonly altered genes in lung cancer, the most frequent co-occurring mutations with SMARCA4 alterations were TP53 (56%), KEAP1 (41%), STK11 (39%) and KRAS (36%) (Fig. 2A and 2B).
Figure 2.
Genomic context of SMARCA4 alterations. (A) Most frequent co-occurring alterations by SMARCA4 alteration. (B) Distribution of SMARCA4 alteration by commonly altered gene subgroups in NSCLC. (C) Frequency of altered individual genes within SMARCA4 mutant vs SMARCA4 wildtype subgroups. Genes labeled red were associated with significantly differential PD-L1 expression (q value <0.10). (D) Distribution of SMARCA4, STK11, KRAS, and KEAP1 alterations within NSCLC cohort.
We identified multiple genes that were associated with SMARCA4 alterations (Fig. 2C). Mutations in STK11 and KEAP1 had the strongest association with SMARCA4 mutant tumors compared to SMARCA4 wildtype tumors (P < 0.001, q < 0.001; P < 0.001, q < 0.001; Fig. 2B and 2C). Conversely EGFR alterations were strongly associated within SMARCA4 wildtype tumors compared to SMARCA4 mutants (P < 0.001, q < 0.001). SMARCA4 alterations occurred in the absence of KRAS, STK11, and KEAP1 alterations in 38% of cases (Fig. 2D). STK11 alterations occurred significantly more frequently with Class 1 than Class 2 alterations (P < 0.001, q = 0.08, Sup. Table 1). NKX2–1 and KEAP1 alterations also occurred more frequently with Class 1 alterations (P = 0.002, q = 0.19; P = 0.01, q = 0.34 respectively) and EGFR alterations were common with Class 2 alterations (P = 0.004, q = 0.19, Sup. Table 1).
Patient characteristics in advanced NSCLC by SMARCA4 alteration class
We then investigated how the findings from our molecular and expression analyses related to clinical outcomes in patients with advanced NSCLC. Patient characteristics among stage IV tumors with Class 1 (n=149) versus Class 2 (n=143) SMARCA4 alterations were generally similar (Table 1). The presence of a Class 1 or 2 SMARCA4 alteration was associated with history of smoking (P < 0.001) and non-adenocarcinoma histology (P < 0.001) compared to patients with SMARCA4 wildtype NSCLC (n=996) (Table 1). Among patients harboring either class of SMARCA4 mutation, 85% were smokers and 84% had adenocarcinoma; the rest had predominantly NSCLC, not otherwise classified.
Table 1:
Clinical characteristics of patients with advanced NSCLC by SMARCA4 alteration class.
| Characteristic |
SMARCA4 Class 1 (N = 149) |
SMARCA4 Class 2 (N = 143) |
SMARCA4 Wild type (N = 996) |
P-value |
|---|---|---|---|---|
| Median Age (Q1, Q3) | 65 (58, 72) | 65 (59, 72) | 65 (58, 73) | 0.7 |
| Sex | 0.052 | |||
| Female | 74 (50%) | 78 (55%) | 593 (60%) | |
| Male | 75 (50%) | 65 (45%) | 403 (40%) | |
| Race | 0.13 | |||
| White | 124 (83%) | 125 (87%) | 775 (78%) | |
| Black | 7 (5%) | 7 (5%) | 57 (6%) | |
| Asian | 10 (7%) | 6 (4%) | 101 (10%) | |
| Other | 8 (5%) | 5 (4%) | 63 (6%) | |
| Smoking | <0.001 | |||
| Never smoker | 17 (11%) | 26 (18%) | 315 (32%) | |
| Former light (<15 py) | 20 (13%) | 19 (13%) | 165 (17%) | |
| Former heavy (>15 py) | 80 (54%) | 71 (50%) | 373 (37%) | |
| Current smoker | 32 (21%) | 27 (19%) | 131 (13%) | |
| Histology | <0.001 | |||
| Adenocarcinoma | 121 (81%) | 125 (87%) | 914 (92%) | |
| Other | 28 (19%) | 18 (13%) | 82 (8%) |
py: pack years
Prognostic impact of Class 1 and Class 2 SMARCA4 alterations in advanced NSCLC
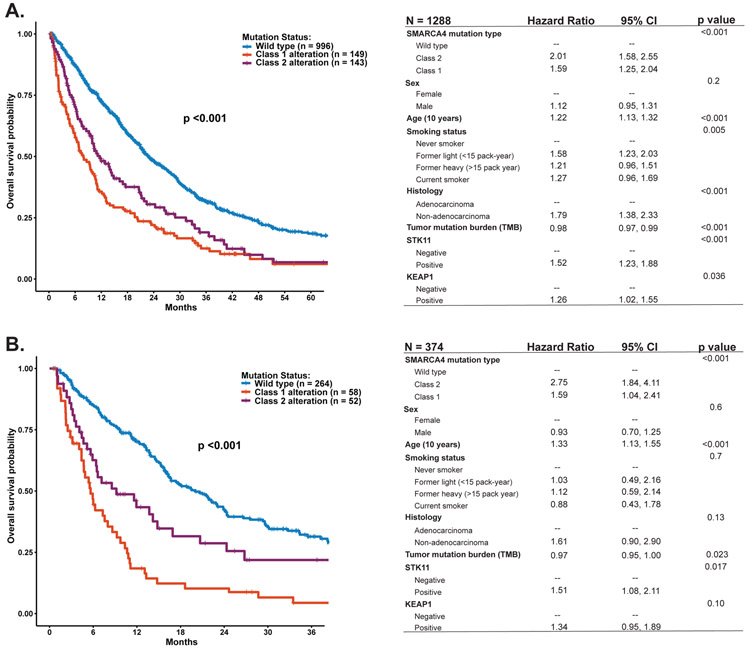
Overall, we found that patients with metastatic NSCLC harboring either Class 1 or Class 2 SMARCA4 alterations had shorter overall survival compared to patients with SMARCA4 wildtype NSCLC (p<0.001; Fig. 3A). Class 1 alterations were associated with the poorest outcomes (Fig. 3A). The differences in outcomes held in the multivariable survival analysis adjusted for age, sex, smoking status, histology, TMB, and the presence of STK11 and/or KEAP1 mutations (Fig. 3A).
Figure 3.
Survival by SMARCA4 alteration class. (A) Overall survival among all patients, with multivariate model (right). (B) Overall survival among patients with KRAS mutations, with multivariate model (right).
Given the heterogeneity of co-occurring mutations, we sought to further isolate the specific impact of SMARCA4 alterations by examining within the context of a single driver oncogene. We focused initially on 374 patients with tumors harboring KRAS mutations. In these patients, the presence of Class 1 or Class 2 SMARCA4 alterations was a poor prognostic factor and remained prognostic when accounting for age, sex, smoking status, histology, TMB, and the presence of STK11 or KEAP1 mutations (Fig. 3B). Further, the addition of STK11 and/or KEAP1 was associated with decreased survival, with patients with all three STK11, KEAP1, and SMARCA4 having the shortest survival (P <0.001, Sup. Fig. 2).
Association with benefit of immunotherapy
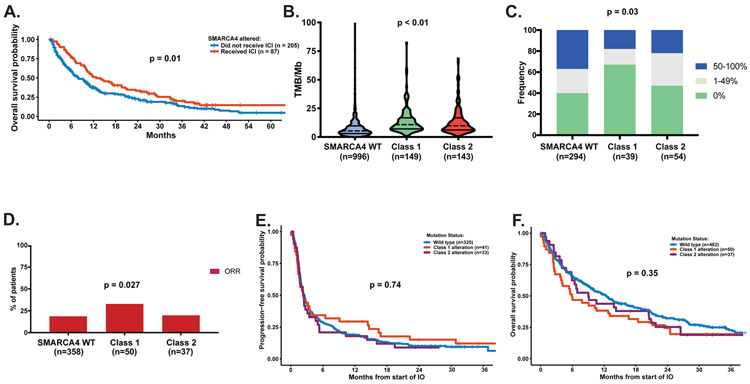
Next, we analyzed the impact of ICIs on patient outcomes. Among patients with SMARCA4 alterations, ICI use was associated with significantly improved survival from the start of ICIs (HR 0.67, 95% CI 0.48, 0.92, P = 0.01) (Fig. 4A). When evaluating known factors that predict outcomes to ICI, SMARCA4-mutant tumors had higher TMB (P < 0.001, Fig. 4B) but were more likely to be PD-L1 low or negative (P = 0.03, Fig. 4C). Class 1 alterations had lower expression of PD-L1 and higher median TMB compared to Class 2 alterations (Fig. 4B-C).
Figure 4.
PD-L1 expression, tumor mutational burden, and immune checkpoint inhibitor (ICI) outcomes. (A) Overall survival among patients with SMARCA4 alterations who did and did not receive ICIs. (B) Tumor mutational burden (TMB) by SMARCA4 alteration class. (C) PD-L1 expression frequency by SMARCA4 alteration class. (D) Overall response rate by SMARCA4 alteration class. (E) Progression-free survival by SMARCA4 alteration class. (F) Overall survival by SMARCA4 alteration class.
Finally, we sought to compare outcome among the two SMARCA4 mutant classes and SMARCA4 wildtype NSCLC in patients who had received ICI. Overall response was assessed in 445 out of 570 patients that received ICI. In unadjusted analyses, patients who harbored Class 1 alterations had a higher ORR in comparison to Class 2 alterations or SMARCA4 wild-type tumors (P = 0.027, Fig. 4D). There was no difference in progression-free survival (P = 0.74) or overall survival (P = 0.35) on ICIs by SMARCA4 alteration status (Fig. 4E-F).
Discussion
Here, we identify two specific classes of SMARCA4 alterations associated with distinct protein expression and differential negative clinical outcomes in patients with metastatic NSCLC. While both classes of SMARCA4 alterations are associated with poor clinical outcomes, Class 1 alterations, which are associated with protein loss, are the strongest independent negative prognostic factor for patients, but respond best to ICIs. Despite the negative prognostic impact compared to patients with SMARCA4 wildtype tumors, patients with SMARCA4 alterations who received ICIs had better outcomes than those who did not.
This study builds upon recent data that co-occurring STK11 and KEAP1 mutations in lung cancer can significantly impact prognosis and responsiveness to therapy. STK11 and KEAP1 alterations are linked with poor prognosis and lack of response to immunotherapy in KRAS-mutant tumors and more recently in all patients with NSCLC. We find that SMARCA4 alterations are associated with STK11 and KEAP1 mutations but are independent predictors of poor prognosis. SMARCA4 abnormalities in combination with STK11 and/or KEAP1 mutations have an additive impact on shortening survival. However, unlike STK11, SMARCA4 appears to be associated with increased sensitivity to immunotherapy. Future studies of STK11 and KEAP1 should incorporate exploration of SMARCA4 to further delineate the role of each co-occurring mutation in influencing patient outcomes and SMARCA4 should be identified and tested as a potential prognostic or predictive variable in prospective trials moving forward.
We observed that the spectrum of SMARCA4 alterations differentially impact protein expression. Our findings are consistent with other recent analyses that assessed the incidence of SMARCA4-mutant lung cancer and frequency of protein expression loss with truncating mutations, supporting our classification schema (8,10). Interestingly, while the effect of Class 1 (truncating) alterations was most profound, we also find that, unexpectedly, patients with Class 2 (mis-sense, non-truncating) SMARCA4 alterations had worse overall prognosis relative to patients with SMARCA4 wild-type tumors, suggesting that function may be compromised in the setting of intact expression. Recent preclinical work provides additional mechanistic support and reveals that missense mutations of SMARCA4 modify the open chromatin landscape and induce pro-oncogenic expression changes in MYC and its target genes, among others (26,27).
Our study is the first to evaluate how SMARCA4 alterations in NSCLC influence sensitivity to ICIs. Recent analyses have shown that SMARCA4 and PBRM1 could be associated with improved response to immunotherapy in subtypes of ovarian cancer and renal cell cancer (4,19) and case reports have described durable responses to ICIs in a patient with a thoracic SMARCA4-deficient undifferentiated tumor (also referred to as a SMARCA4-deficient thoracic sarcoma) and a patient with NSCLC (20,21). Despite high rates of PD-L1 negativity, patients with SMARCA4-mutant NSCLC appear to derive significant benefit from PD-(L)1 blockade. Therefore, SMARCA4 mutation status should be explored as a potentially novel biomarker of responsiveness to ICIs as a complement to PD-L1 expression and TMB in NSCLC.
While there are no known currently effective targeted treatments for SMARCA4-mutant NSCLCs, our study and others suggest SMARCA4 is a potential target in lung cancer with distinct therapeutic vulnerabilities. For example, CDK4/6, AURKA, ATR, and EZH2 inhibition have recently shown antitumor activity in preclinical models of SMARCA4 deficient tumors (1,16,25,28-33). SMARCA2 could be a synthetic lethal vulnerability in SMARCA4-mutant cancers. Prior reports have shown that SMARCA2 retains expression in SMARCA4-mutant NSCLC and several SMARCA2 inhibitors are currently in development to target this potential vulnerability (10,16). Future trials should explore use of these agents alone or in combination with ICIs given the efficacy of anti-PD-(L)1 antibodies in our analysis.
This study is a single-institution retrospective analysis and therefore has some inherent limitations. Unidentified factors associated with exposure and response to immunotherapy and overall survival could bias our results. Nevertheless, we accounted for all known potential variables that may influence outcomes. For example, we developed and incorporated a risk score to account for a patient’s likelihood of receiving anti-PD(L)1 therapy and used a Cox proportional hazards model for multivariate analysis using the variables available. Analyses adjusting for PD-L1 expression are limited by the modest number of patients with sufficient available tissue for retrospective staining for PD-L1 and SMARCA4. Future studies that incorporate zygosity are also needed to understand its impact on expression and clinical outcomes.
In sum, our report highlights that SMARCA4 alterations in lung cancer are uniquely linked to response to immunotherapy and patient outcomes. We found that the presence of SMARCA4 abnormalities is enriched in patients with KRAS, STK11, and KEAP1 mutations, but independently contributes to shortened overall survival with these co-occurring alterations. Despite these poor outcomes, patients with SMARCA4-mutant lung cancers may also be more sensitive to immunotherapy, which may enable new therapeutic options in the future.
Supplementary Material
Statement of translational relevance:
In this study, we characterize the clinical, molecular and histologic relationships of SMARCA4 genomic and protein alterations in lung cancer. SMARCA4 is the most commonly mutated member of the SWI/SNF complex, with mutations occurring in 8% of patients with NSCLC. Genomic, protein expression, and clinical outcome data identify two distinct classes of SMARCA4 alterations. SMARCA4 alterations often co-occur with STK11, KEAP1, and KRAS alterations, but they are a prognostic factor, independent of these alterations. Although patients whose tumors have Class 1 SMARCA4 alterations (associated with protein expression loss) have a very poor prognosis, they may have higher response rates to PD-(L)1 blockade despite low PD-L1 expression.
Acknowledgement of research support:
Supported by Memorial Sloan Kettering Cancer Center Support Grant/Core Grant (P30 CA008748) and the Druckenmiller Center for Lung Cancer Research at MSK. ACS Postdoctoral Fellowship 130361-PF-17–009-01-CDD. Koch Institute Quinquennial Postdoctoral Fellowship. MDH is a Damon Runyon Clinical Investigator supported (in part) by the Damon Runyon Cancer Research Foundation (CI-98–18) and is a member of the Parker Institute for Cancer Immunotherapy. This work was supported, in part, by a grant from John and Georgia DallePezze to Memorial Sloan Kettering Cancer Center.
Footnotes
Disclosures: AN was a consultant for Bayer MEA is a consultant for AstraZeneca and received support from Astrazeneca, Invivoscribe, and Raindance Technologies. JLS reports stock ownership in the following companies: Pfizer, Thermo Fischer Scientific, Inc., Merck & Co Inc and Chemed Corp. ML has received advisory board compensation from Boehringer Ingelheim, AstraZeneca, Bristol-Myers Squibb, Takeda, and Bayer, and research support from LOXO Oncology and Helsinn Healthcare. TJ is a member of the Board of Directors of Amgen and Thermo Fisher Scientific. He is also a co-Founder of Dragonfly Therapeutics and T2 Biosystems. T.J. serves on the Scientific Advisory Board of Dragonfly Therapeutics, SQZ Biotech, and Skyhawk Therapeutics. None of these affiliations represent a conflict of interest with respect to the design or execution of this study or interpretation of data presented in this manuscript. T.J. laboratory currently also receives funding from the Johnson & Johnson Lung Cancer Initiative and The Lustgarten Foundation for Pancreatic Cancer Research, but this funding did not support the research described in this manuscript. CMR has consulted regarding oncology drug development with AbbVie, Amgen, Ascentage, AstraZeneca, Bicycle, Celgene, Daiichi Sankyo, Genentech/Roche, Ipsen, Jansen, Jazz, Lilly/Loxo, Pfizer, PharmaMar, Syros, and Vavotek; he serves on the SAB for Bridge Medicines and Harpoon Therapeutics. MDH receives research support from Bristol-Myers Squibb; has been a compensated consultant for Merck, Bristol-Myers Squibb, AstraZeneca, Genentech/Roche, Nektar, Syndax, Mirati, Shattuck Labs, Immunai, Blueprint Medicines, Achilles, and Arcus; received travel support/honoraria from AstraZeneca, Eli Lilly, and Bristol-Myers Squibb; has options from Shattuck Labs, Immunai, and Arcus; has a patent filed by his institution related to the use of tumor mutation burden to predict response to immunotherapy (PCT/US2015/062208), which has received licensing fees from PGDx. GJR Memorial Sloan Kettering Cancer Center receives funding for research led by Dr. Riely from: Mirati, Merck, Takeda, Roche, Pfizer, and Novartis.
References
- 1.Helming KC, Wang X, Roberts CWM. Vulnerabilities of mutant SWI/SNF complexes in cancer. Cancer cell 2014;26(3):309–17 doi 10.1016/j.ccr.2014.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kadoch C, Hargreaves DC, Hodges C, Elias L, Ho L, Ranish J, et al. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nature genetics 2013;45(6):592–601 doi 10.1038/ng.2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abou Alaiwi S, Nassar AH, Xie W, Bakouny Z, Berchuck JE, Braun DA, et al. Mammalian SWI/SNF complex genomic alterations and immune checkpoint blockade in solid tumors. Cancer Immunology Research 2020:canimm.0866.2019 doi 10.1158/2326-6066.Cir-19-0866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miao D, Margolis CA, Gao W, Voss MH, Li W, Martini DJ, et al. Genomic correlates of response to immune checkpoint therapies in clear cell renal cell carcinoma. Science (New York, NY) 2018;359(6377):801–6 doi 10.1126/science.aan5951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lovly CM, Gupta A, Lipson D, Otto G, Brennan T, Chung CT, et al. Inflammatory myofibroblastic tumors harbor multiple potentially actionable kinase fusions. Cancer Discov 2014;4(8):889–95 doi 10.1158/2159-8290.CD-14-0377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Imielinski M, Berger AH, Hammerman PS, Hernandez B, Pugh TJ, Hodis E, et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell 2012;150(6):1107–20 doi 10.1016/j.cell.2012.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reisman DN, Sciarrotta J, Wang W, Funkhouser WK, Weissman BE. Loss of BRG1/BRM in human lung cancer cell lines and primary lung cancers: correlation with poor prognosis. Cancer research 2003;63(3):560–6. [PubMed] [Google Scholar]
- 8.Dagogo-Jack I, Schrock AB, Kem M, Jessop N, Lee J, Ali SM, et al. Clinicopathologic Characteristics of BRG1-Deficient NSCLC. J Thorac Oncol 2020. doi 10.1016/j.jtho.2020.01.002. [DOI] [PubMed] [Google Scholar]
- 9.Bell EH, Chakraborty AR, Mo X, Liu Z, Shilo K, Kirste S, et al. SMARCA4/BRG1 Is a Novel Prognostic Biomarker Predictive of Cisplatin-Based Chemotherapy Outcomes in Resected Non-Small Cell Lung Cancer. Clinical cancer research : an official journal of the American Association for Cancer Research 2016;22(10):2396–404 doi 10.1158/1078-0432.Ccr-15-1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rekhtman N, Montecalvo J, Chang JC, Alex D, Ptashkin RN, Ai N, et al. SMARCA4-Deficient Thoracic Sarcomatoid Tumors Represent Primarily Smoking-Related Undifferentiated Carcinomas Rather Than Primary Thoracic Sarcomas. J Thorac Oncol 2020;15(2):231–47 doi 10.1016/j.jtho.2019.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marquez-Vilendrer SB, Rai SK, Gramling SJ, Lu L, Reisman DN. Loss of the SWI/SNF ATPase subunits BRM and BRG1 drives lung cancer development. Oncoscience 2016;3(11–12):322–36 doi 10.18632/oncoscience.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Orvis T, Hepperla A, Walter V, Song S, Simon J, Parker J, et al. BRG1/SMARCA4 inactivation promotes non-small cell lung cancer aggressiveness by altering chromatin organization. Cancer research 2014;74(22):6486–98 doi 10.1158/0008-5472.Can-14-0061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arbour KC, Jordan EJ, Kim HR, Dienstag J, Yu H, Sanchez-Vega F, et al. Effects of co-occurring genomic alterations on outcomes in patients with KRAS-mutant non-small cell lung cancer. 2017:clincanres.1841.2017 doi 10.1158/1078-0432.CCR-17-1841 %J Clinical Cancer Research. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Skoulidis F, Byers LA, Diao L, Papadimitrakopoulou VA, Tong P, Izzo J, et al. Co-occurring Genomic Alterations Define Major Subsets of KRAS-Mutant Lung Adenocarcinoma with Distinct Biology, Immune Profiles, and Therapeutic Vulnerabilities. 2015;5(8):860–77 doi 10.1158/2159-8290.CD-14-1236 %J Cancer Discovery. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aggarwal C, Davis CW, Mick R, Thompson JC, Ahmed S, Jeffries S, et al. Influence of TP53 Mutation on Survival in Patients With Advanced EGFR-Mutant Non–Small-Cell Lung Cancer. 2018(2):1–29 doi 10.1200/po.18.00107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Papillon JPN, Nakajima K, Adair CD, Hempel J, Jouk AO, Karki RG, et al. Discovery of Orally Active Inhibitors of Brahma Homolog (BRM)/SMARCA2 ATPase Activity for the Treatment of Brahma Related Gene 1 (BRG1)/SMARCA4-Mutant Cancers. Journal of medicinal chemistry 2018;61(22):10155–72 doi 10.1021/acs.jmedchem.8b01318. [DOI] [PubMed] [Google Scholar]
- 17.Rizvi H, Sanchez-Vega F, La K, Chatila W, Jonsson P, Halpenny D, et al. Molecular Determinants of Response to Anti-Programmed Cell Death (PD)-1 and Anti-Programmed Death-Ligand 1 (PD-L1) Blockade in Patients With Non-Small-Cell Lung Cancer Profiled With Targeted Next-Generation Sequencing. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 2018;36(7):633–41 doi 10.1200/jco.2017.75.3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Skoulidis F, Arbour KC, Hellmann MD, Patil PD, Marmarelis ME, Awad MM, et al. Association of STK11/LKB1 genomic alterations with lack of benefit from the addition of pembrolizumab to platinum doublet chemotherapy in non-squamous non-small cell lung cancer. Journal of Clinical Oncology 2019;37(15_suppl):102– doi 10.1200/JCO.2019.37.15_suppl.102. [DOI] [Google Scholar]
- 19.Jelinic P, Ricca J, Van Oudenhove E, Olvera N, Merghoub T, Levine DA, et al. Immune-Active Microenvironment in Small Cell Carcinoma of the Ovary, Hypercalcemic Type: Rationale for Immune Checkpoint Blockade. Journal of the National Cancer Institute 2018;110(7):787–90 doi 10.1093/jnci/djx277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Naito T, Umemura S, Nakamura H, Zenke Y, Udagawa H, Kirita K, et al. Successful treatment with nivolumab for SMARCA4-deficient non-small cell lung carcinoma with a high tumor mutation burden: A case report. 2019;10(5):1285–8 doi 10.1111/1759-7714.13070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Henon C, Blay J-Y, Massard C, Mir O, Bahleda R, Dumont S, et al. Long lasting major response to pembrolizumab in a thoracic malignant rhabdoid-like SMARCA4-deficient tumor. Annals of Oncology 2019;30(8):1401–3 doi 10.1093/annonc/mdz160 %J Annals of Oncology. [DOI] [PubMed] [Google Scholar]
- 22.Cheng DT, Mitchell TN, Zehir A, Shah RH, Benayed R, Syed A, et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): A Hybridization Capture-Based Next-Generation Sequencing Clinical Assay for Solid Tumor Molecular Oncology. The Journal of molecular diagnostics : JMD 2015;17(3):251–64 doi 10.1016/j.jmoldx.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chakravarty D, Gao J, Phillips SM, Kundra R, Zhang H, Wang J, et al. OncoKB: A Precision Oncology Knowledge Base. JCO precision oncology 2017;2017 doi 10.1200/po.17.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schoenfeld AJ, Rizvi H, Bandlamudi C, Sauter JL, Travis WD, Rekhtman N, et al. Clinical and molecular correlates of PD-L1 expression in patients with lung adenocarcinomas✰. Annals of Oncology 2020;31(5):599–608 doi 10.1016/j.annonc.2020.01.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xue Y, Meehan B, Macdonald E, Venneti S, Wang XQD, Witkowski L, et al. CDK4/6 inhibitors target SMARCA4-determined cyclin D1 deficiency in hypercalcemic small cell carcinoma of the ovary. Nature communications 2019;10(1):558 doi 10.1038/s41467-018-06958-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hodges HC, Stanton BZ, Cermakova K, Chang CY, Miller EL, Kirkland JG, et al. Dominant-negative SMARCA4 mutants alter the accessibility landscape of tissue-unrestricted enhancers. Nat Struct Mol Biol 2018;25(1):61–72 doi 10.1038/s41594-017-0007-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stanton BZ, Hodges C, Calarco JP, Braun SM, Ku WL, Kadoch C, et al. Smarca4 ATPase mutations disrupt direct eviction of PRC1 from chromatin. Nature genetics 2017;49(2):282–8 doi 10.1038/ng.3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zernickel E, Sak A, Riaz A, Klein D, Groneberg M, Stuschke M. Targeting of BRM sensitizes BRG1 mutant lung cancer cell lines to radiotherapy. Molecular cancer therapeutics 2018. doi 10.1158/1535-7163.Mct-18-0067. [DOI] [PubMed] [Google Scholar]
- 29.Vangamudi B, Paul TA, Shah PK, Kost-Alimova M, Nottebaum L, Shi X, et al. The SMARCA2/4 ATPase Domain Surpasses the Bromodomain as a Drug Target in SWI/SNF-Mutant Cancers: Insights from cDNA Rescue and PFI-3 Inhibitor Studies. Cancer research 2015;75(18):3865–78 doi 10.1158/0008-5472.Can-14-3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fillmore CM, Xu C, Desai PT, Berry JM, Rowbotham SP, Lin YJ, et al. EZH2 inhibition sensitizes BRG1 and EGFR mutant lung tumours to TopoII inhibitors. Nature 2015;520(7546):239–42 doi 10.1038/nature14122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kurashima K, Kashiwagi H, Shimomura I, Suzuki A, Takeshita F, Mazevet M, et al. SMARCA4 deficiency-associated heterochromatin induces intrinsic DNA replication stress and susceptibility to ATR inhibition in lung adenocarcinoma. NAR Cancer 2020;2(2) doi 10.1093/narcan/zcaa005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lissanu Deribe Y, Sun Y, Terranova C, Khan F, Martinez-Ledesma J, Gay J, et al. Mutations in the SWI/SNF complex induce a targetable dependence on oxidative phosphorylation in lung cancer. Nature medicine 2018;24(7):1047–57 doi 10.1038/s41591-018-0019-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tagal V, Wei S, Zhang W, Brekken RA, Posner BA, Peyton M, et al. SMARCA4-inactivating mutations increase sensitivity to Aurora kinase A inhibitor VX-680 in non-small cell lung cancers. Nature communications 2017;8(1):14098 doi 10.1038/ncomms14098. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.